Abstract
Following transcription, tRNAs undergo a series of processing and modification events to become functional adaptors in protein synthesis. Eukaryotes have also evolved intracellular transport systems whereby nucleus-encoded tRNAs may travel out and into the nucleus. In trypanosomes, nearly all tRNAs are also imported from the cytoplasm into the mitochondrion, which lacks tRNA genes. Differential subcellular localization of the cytoplasmic splicing machinery and a nuclear enzyme responsible for queuosine modification at the anticodon “wobble” position appear to be important quality control mechanisms for tRNATyr, the only intron-containing tRNA in T. brucei. Since tRNA-guanine transglycosylase (TGT), the enzyme responsible for Q formation, cannot act on an intron-containing tRNA, retrograde nuclear transport is an essential step in maturation. Unlike maturation/processing pathways, the general mechanisms of tRNA stabilization and degradation in T. brucei are poorly understood. Using a combination of cellular and molecular approaches, we show that tRNATyr has an unusually short half-life. tRNATyr, and in addition tRNAAsp, also show the presence of slow-migrating bands during electrophoresis; we term these conformers: alt-tRNATyr and alt-tRNAAsp, respectively. Although we do not know the chemical or structural nature of these conformers, alt-tRNATyr has a short half-life resembling that of tRNATyr; the same is not true for alt-tRNAAsp. We also show that RRP44, which is usually an exosome subunit in other organisms, is involved in tRNA degradation of the only intron-containing tRNA in T. brucei and is partly responsible for its unusually short half-life.
Keywords: actinomycin, half-life, tRNA, trypanosoma
INTRODUCTION
Transcription of tRNA genes in eukaryotes is catalyzed by RNA polymerase III (RNA Pol III), which generates molecules with 5′ and 3′ extensions. Depending on the organism, a variable number of tRNAs also contain introns, found between positions 37 and 38 of the anticodon loop. Maturation of tRNA precursors by termini definition, posttranscriptional modification, and splicing, shapes tRNA structure, function, and metabolic fate. For example, some modifications, particularly those occurring in the anticodon loop, are important for translational efficiency and/or accuracy; others may affect folding and/or the stability of the molecule. In trypanosomatids, which include members of the genus Trypanosoma and Leishmania, all tRNAs are encoded in the nucleus, while the mitochondrial genome is devoid of tRNA genes. Thus, mitochondria-destined tRNAs are exported to the cytoplasm and then imported into the mitochondrion, with initiator tRNAMet being the only exception. During intracellular transport, tRNAs can be modified in any of these compartments (Tan et al. 2002).
In T. brucei, only tRNATyr (Tb927.4.1219) contains an intron, which, similarly to S. cerevisiae, is spliced in the cytoplasm (Yoshihisa 2003; Shaheen and Hopper 2005; Lopes et al. 2016). Apparently, trypanosomes maintain the eukaryotic tRNA-splicing machinery to service a single tRNA, whose unconventional features include one of the shortest known introns (11 nt) that undergoes noncanonical base editing, a prerequisite for cleavage by the tRNA-splicing endonuclease (Rubio et al. 2013). The peculiarities extend to the mature molecule, which contains the modified nucleotide queuosine (Q) at the anticodon “wobble” position 34: the enzyme responsible for this modification is nuclear (Kessler et al. 2018). Hence, after splicing, this tRNA undergoes retrograde transport from the cytoplasm to the nucleus to receive Q, before being exported back into the cytoplasm (Kessler et al. 2018). Finally, both Q-modified and unmodified tRNATyr coexist in the cytoplasm and engage in translation, where the unmodified tRNA can decode the G-ending codons for tyrosine, while the Q-modified tRNA is necessary for the efficient translation of the U-ending codons (Dixit et al. 2021). Three other tRNAs (Asp, Asn, and His) also contain Q, but since these do not contain introns, they are exported from the nucleus after Q formation, with presumably no need for retrograde nuclear transport (Kessler et al. 2018). Lastly, the Q-containing tRNAs are preferentially imported into the mitochondrion, but how the mitochondrial import machinery discriminates their unmodified counterparts is unclear (Kulkarni et al. 2021).
Concomitantly to maturation, tRNAs are also subjected to quality surveillance. In S. cerevisiae, 5′–3′ exoribonucleases XRN1 (cytoplasmic), and XRN2 (RAT1, nuclear) protagonize the rapid tRNA decay (RTD) pathway which, eliminates hypomodified tRNAs to safeguard translation (Chernyakov et al. 2008). T. brucei encodes six orthologous XRN enzymes (A to F), but their functions in tRNA decay are unclear.
In many systems, the exosome, a multiprotein complex containing several 3′–5′ exoribonucleases, orchestrates general and controlled RNA degradation, with the latter contributing to maturation of rRNA and other noncoding RNAs (Allmang et al. 1999; LaCava et al. 2005). The S. cerevisiae exosome is formed by 10 core protein components and associated ribonucleases RRP6 and RRP44 (Mitchell and Tollervey 2000). The T. brucei exosome contains many homologs of the exosome components from other systems, with the exception of RRP44 (Estévez et al. 2001). In T. brucei, RRP44 is an autonomous exoribonuclease found in the nucleus and cytoplasm (Estévez et al. 2001; Cesaro et al. 2023). In the former, it participates in rRNA maturation, but it may also play a role in polycistronic mRNA processing (Estévez et al. 2001; Cesaro et al. 2023). However, little is known about tRNA stability determinants and decay mechanisms in trypanosomes. Because a stable structure and extensive modifications protect tRNAs from degradation, in other systems, tRNAs are relatively long-lived, with half-lives exceeding 60 h (Hanoune and Agarwal 1970; Abelson et al. 1974; Smith and Weinberg 1981; Sroga 1984; Karnahl and Wasternack 1992; Gudipati et al. 2012).
The unusual intracellular transport dynamics leading to persistence of “hypomodified” variants in the cytoplasm and nucleus raised the question of how tRNATyr escapes degradation mechanisms that, in other systems, mediate the removal of hypomodified tRNAs, such as the RTD pathway of S. cerevisiae mentioned above. We assessed the relative stability of several tRNAs in T. brucei including tRNATyr. Using northern hybridization analysis, we show that tRNATyr and tRNAAsp have unusual isoforms, which we term alt-tRNATyr and alt-tRNAAsp, respectively, as opposed to “canonical” tRNATyr and “canonical tRNAASP, which we referred to as simply tRNATyr and tRNAAsp. We also show that tRNATyr has a strikingly short half-life. Thus, we explored the contribution of the 5′–3′ RNA degradation pathway exemplified by XRN family members, and the 3′–5′ pathway, exemplified by RRP6, Dis3L2, and RRP44, to this phenomenon. We find that RRP44 plays a role in the steady-state levels of alt-tRNATyr and alt-tRNAAsp and tRNAAsp, but it has little effect on tRNATyr. However, following actinomycin D treatment, only the half-life of alt-tRNATyr and tRNATyr is increased, while the half-life of the equivalent tRNAAsp species remained unchanged. In addition, out of the XRNs tested, only XRNE had a modest impact on the levels of alt-tRNATyr but no effect on the other species. Taken together the decay kinetics of the different tRNA species suggest that the short half-life of tRNATyr may be due to its unusual intracellular transport dynamics, while alt-tRNATyr may serve as a repository for tRNATyr.
RESULTS
tRNATyr has a relatively short half-life in T. brucei
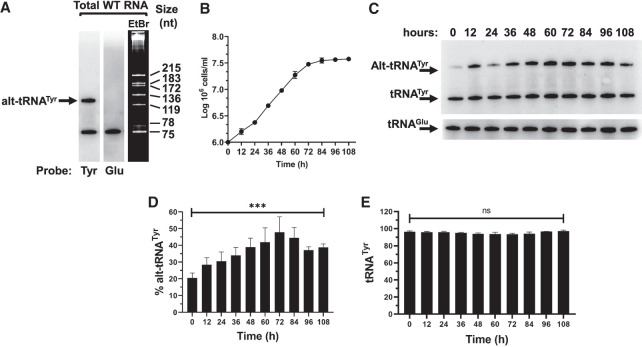
Recent studies have highlighted targeting of hypomodified tRNAs by the RTD pathway, orchestrated in S. cerevisiae by different nuclear and cytoplasmic enzymes (Frederick and Heinemann 2021). Given the elaborate intracellular transport dynamics of tRNATyr in T. brucei, we analyzed total RNA samples by northern blot hybridization with oligonucleotide probes for tRNATyr. We observed two distinct hybridization signals: a 75-nt band corresponding to the expected size of the mature molecule, and a higher molecular mass band, migrating with a size consistent with approximately 125-nt (Fig. 1A). Significantly, the size difference between the slower migrating band and the mature (∼45 nt) is larger than what can be accounted for by 3′ poly(U) extensions previously described (Schneider et al. 1993), and does not contain an intron (Lopes et al. 2016) and Figure 2, this work. We refer to it as an alternative isoform of the tRNA (alt-tRNATyr) (Fig. 1A). An alternative isoform was also detected in tRNAAsp (Tb927.7.6824), but not in other tRNAs tested (Fig. 2). To investigate the behavior of alt-tRNATyr, we grew T. brucei Lister 427 procyclic cells for 108 h, allowing the parasites to reach stationary phase (Fig. 1B). Samples were collected at 12 h time intervals and analyzed by northern blot hybridization, revealing that alt-tRNATyr relative amounts vary along with cell density, reaching a maximum of 45%–55% of the total tRNATyr at ∼72 h of growth, before stabilizing during the stationary phase, at ∼40% of the late logarithmic phase (Fig. 1C,D). However, the steady-state levels of the tRNATyr remained constant.
FIGURE 1.
T. brucei tRNATyr is present in two conformers. (A) Northern blot hybridization performed on total RNA sample extracted from T. brucei reveals two forms of tRNATyr, including the mature (75 nt) band and a higher (∼120 nt), alternate conformer. (B) Triplicate growth curve of WT T. brucei cells where total RNA was collected at every time point. (C) Northern blot hybridization performed on the extracted total RNA samples revealing the mature and alternate conformers of tRNATyr. The tRNAGlu was used as loading control. (D) Plotted amounts of alternate conformer (alt-tRNATyr) in relation to tRNATyr. (E) Plotted amounts of mature tRNATyr in D and E are the result of at least three biological replicas. Asterisks denote significance as determined by a one-way ANOVA yielding a P-value of 0.0003 for the data in D. “ns” stands for nonsignificant by the same test.
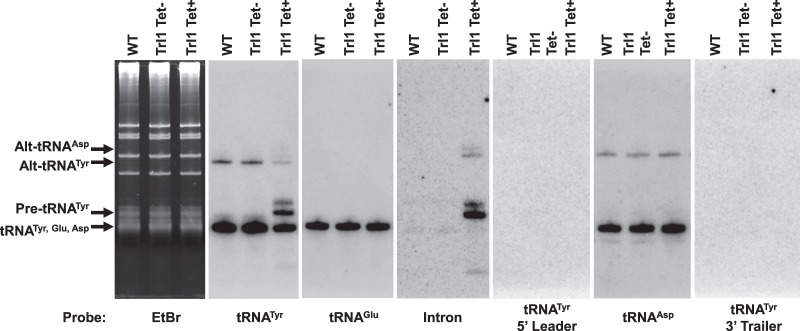
FIGURE 2.
Alt-tRNATyr does not contain an intron and neither alternative conformer contains 5′ or 3′ extensions corresponding to the genomic flanking regions. Northern blot hybridization performed on total RNA sample extracted from WT and Trl1 RNAi strains of T. brucei utilizing probes specific for tRNATyr (mature, intron-specific, 5′ leader, and 3′ trailer), tRNAGlu, and tRNAAsp.
It is unlikely that the slower migrating alt-tRNA band is due to the presence of the intron given the existence of alt-tRNAAsp, where the latter is not encoded with an intron. Nonetheless to rule this possibility, we performed northern blots using an intron probe specific for the edited intron of tRNATyr (Rubio et al. 2013). We also probed for the predicted 5′ and 3′ flaking regions. As a control, we used RNA isolated from down-regulation of TRL1, the putative splicing ligase of T. brucei, which we had shown to lead to accumulation of intro-containing tRNA (Lopes et al. 2016). Neither probe detected an intron signal in RNA from wild-type cells or the uninduced TRL1 control (Fig. 2). To further assess the nature of the 5′ and 3′ ends of tRNATyr and tRNAAsp, we also demethylated total tRNA from wild-type cells with the demethylase AlkB followed by Ordered Two-Template Relay (OTTR sequencing) as described in Materials and Methods. We plotted the sequences of CCA-containing mature tRNA reads from the +AlkB treated T. brucei OTTR-seq library. We found that all sequence reads start at the 5′ most end of the tRNA transcript and end with the -CCA tail; no extra nucleotides were observed at either end, ruling out the possibility that the alt-tRNAs are the result of nucleotide additions at either end (Supplemental Fig. S1). We also plotted the pre-tRNA reads from the +AlkB treated OTTR-seq library, where only extensions ±2 nt beyond the mature tRNA transcript were deemed a pre-tRNA read. In this case, some of the reads extend a few nucleotides into the leader sequence, and also contained extensions of ∼5–7 Us at the 3′ most end of the reads. The 3′-U extensions are consistent with what was previously described in the intron-containing tRNA. In the case of tRNATyr, one can see the presence of the intron in further support of these species as pre-tRNA reads (Supplemental Fig. S1A–C). Notably, in both cases manual searches for longer reads still containing the core tRNATyr and tRNAAsp sequence were performed, but no anomalous long sequences were detected. Furthermore, we also inspected tRNAAsn, which gets Q but does not have either an intron or an alternative form. This tRNA also does not contain any 5′ or 3′ extensions suggesting that the nature of the alt-tRNAs is not dependent on Q. Taken together, the finding that neither the intron-specific probe nor the flanking regions probes hybridized with either the mature or the alt-tRNA band in the wild-type or TRL1 uninduced control (Fig. 2) in agreement with the sequencing data in the Supplemental Figures.
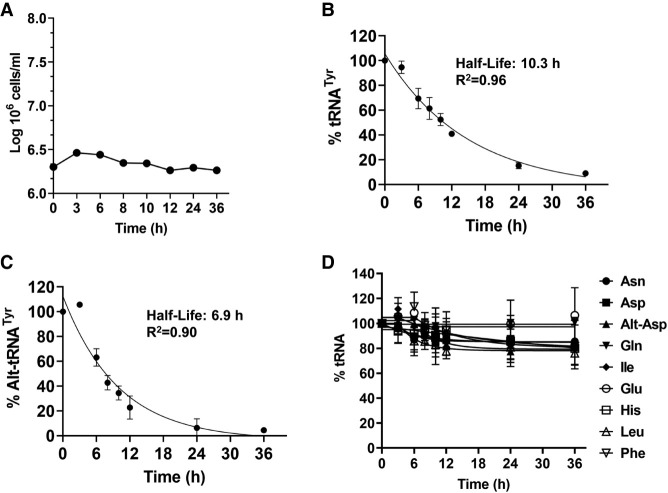
To investigate the relationship between the alt-tRNA and the canonical species, we enquired whether the alt-tRNATyr is an intermediate feeding into the overall pool after additional processing, or a “dead-end” product of aberrant tRNATyr maturation. Thus, we measured its general stability, we treated T. brucei cultures with the transcriptional elongation inhibitor actinomycin D. Although this inhibitor has been associated with inducing nucleolar stress in other systems, it has been the inhibitor of choice for these types of studies in the trypanosome system (Li et al. 2006; Kramer et al. 2008; Manful et al. 2011). Upon actinomycin treatment, we continued cell cultivation for 36 h. Total RNA was prepared from samples collected at the indicated time points (Figs. 3, 4). As shown, there is an observed increase in levels of alt-tRNATyr in the first 3 h of treatment. Similar behavior has been described for mRNA studies in T. brucei; this phenomenon was ascribed to the existence of a pool of precursors that feed into the system (Li et al. 2006). These experiments also revealed that tRNATyr and alt-tRNATyr have unusually short half-lives of 10.3 and 6.9 h, respectively (Figs. 3, 4). In contrast, the levels of other tested tRNAs remained stable for 36 h following actinomycin D treatment (Figs. 3, 4D). We performed similar experiments for tRNAAsp, which is the other tRNA displaying an alternative isoform. In contrast to tRNATyr and alt-tRNATyr, both tRNAAsp and alt-tRNAAsp remained stable for 36 h (Fig. 3).
FIGURE 3.
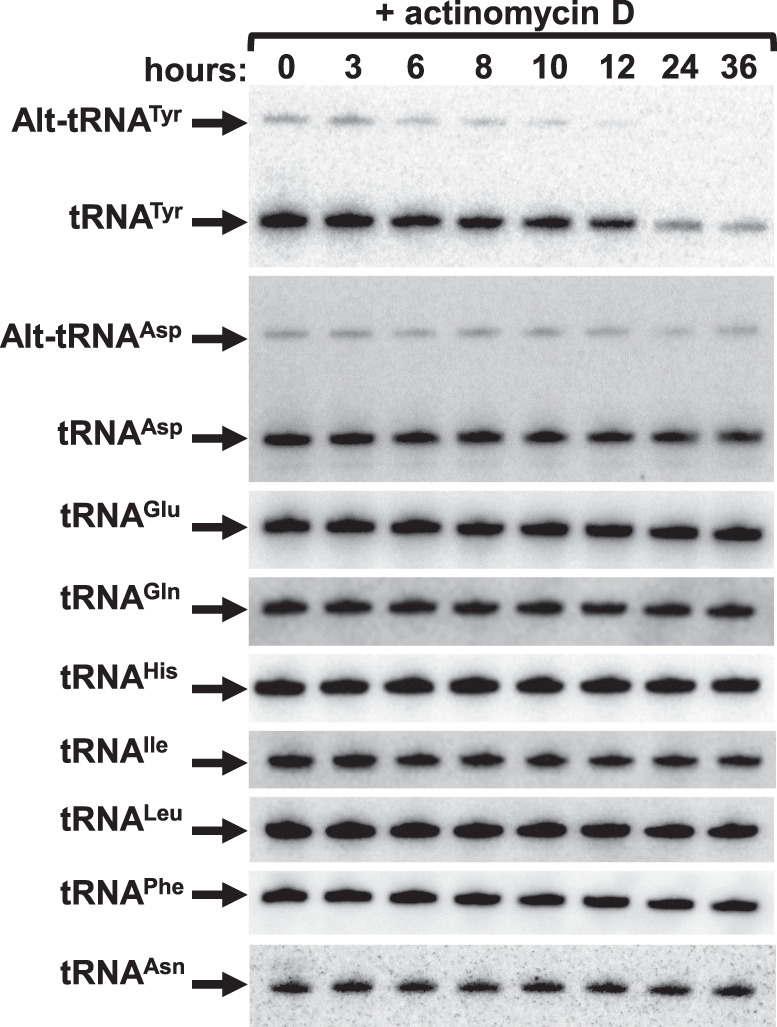
T. brucei tRNATyr is unusually unstable. Northern blot hybridization performed on the extracted total RNA samples from T. brucei cells subjected to Actinomycin D treatment revealing the mature and alternate conformers of tRNATyr. The same membrane was then stripped and probed for the tRNAs: Asp, Glu, Gln, His, Ile, Leu, Phe, and Asn. The tRNAGlu was used as loading control.
FIGURE 4.
T. brucei tRNATyr has an unusually short half-life, which is not shared by other tRNAs in the system. (A) Triplicate growth curve of WT T. brucei cells subjected to Actinomycin D treatment where total RNA was collected at the time points indicated. (B,C) Linear regression calculations utilizing the amounts of mature and alternate conformer tRNATyr. (D) Linear regression calculations utilizing the amounts of other tRNAs. Data were fit to a one-phase decay. All data plotted were the result of three biological replicas.
XRNE moderately impacts alt-tRNATyr stability
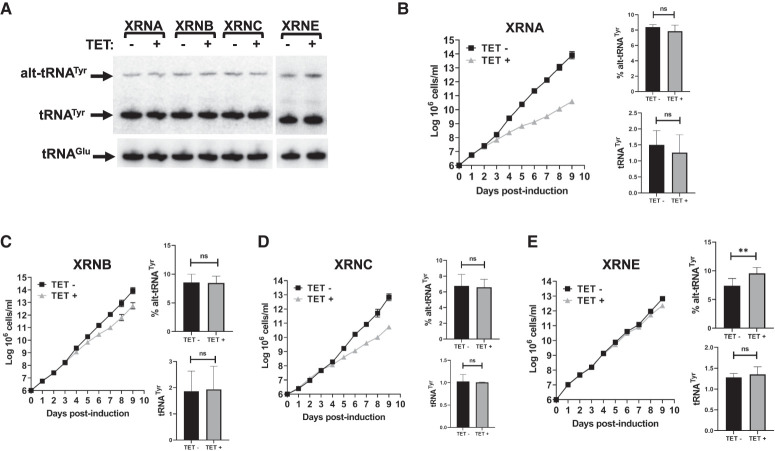
Despite their remarkable stability, tRNAs eventually decay via pathways that have been extensively studied in eukaryotes (Alexandrov et al. 2006; Chernyakov et al. 2008; Houseley and Tollervey 2009; Frederick and Heinemann 2021). RTD targets hypomodified tRNAs in S. cerevisiae, and involves 5′–3′ cytoplasmic XRN1 and the nuclear XRN2 (RAT1) exoribonucleases (Alexandrov et al. 2006; Chernyakov et al. 2008; Houseley and Tollervey 2009). The T. brucei genome encodes six XRN homologs: XRNA (Tb927.7.4900), XRNB (Tb927.5.2450), XRNC (Tb927.8.2810), XRND (Tb927.10.6220), XRNE (Tb927.5.3850), and XRNF (Tb927.11.1550) (Li et al. 2006). XRNA has been localized to the nucleus and the cytoplasm, and it is functionally linked to degradation of unstable (half-lives <30 min) developmentally regulated mRNAs (Li et al. 2006; Manful et al. 2011). XRNB and C localize to the cytoplasm, and apparently play negligible roles in RNA degradation (Li et al. 2006). The function of nuclear XRND remains unclear, as it is the case for XRNF (Li et al. 2006). XRNE has been localized to the nucleolus and has been implicated in rRNA processing and assembly (Sakyiama et al. 2013). To investigate whether the XRN homologs participate in tRNATyr decay, we systematically knocked down their expression by inducible RNAi. Induced and mock-treated transgenic RNAi cell lines were grown for 10 d (LaCount et al. 2000). Total RNA was collected after 6 d of RNAi induction for northern blot hybridization (Fig. 5; Supplemental Fig. S2). RNAi down-regulation of XRNA, B, and C caused varying growth defects, while XRNE RNAi displayed none. RNAi failed to appreciably down-regulate XRND and XRNF expression, and these proteins were not explored further. A minor, but statistically significant alt-tRNATyr accumulation was observed in XRNE RNAi (Fig. 5E), while XRNA, B, and C knockdowns did not affect the steady-state levels of either tRNATyr or alt-tRNATyr (Fig. 4B–D). We conclude that XRNE has a small effect in tRNATyr/alt-tRNATyr turnover, while XRNA, B, and C are not involved. XRND and F could not be evaluated, so their participation in tRNA turnover remains undisclosed.
FIGURE 5.
RNAi knockdown of XRN enzymes. (A) Northern blot hybridization performed on the extracted total RNA samples revealing the mature and alternate conformers of tRNATyr. The tRNAGlu was used as loading control. (B–E) Triplicate growth curve of T. brucei cells subjected to tetracycline-induced protein knockdown where total RNA was collected on day 6. Relative amounts of alternate conformer tRNATyr and mature tRNATyr. The graphs are the result of three biological replicas. The asterisks in “E” denote significance based on a two-tailed paired t-test with a P-value of 0.006. “ns” stands for nonsignificant by the same test.
RRP44 down-regulation stabilizes alt-tRNATyr, alt-tRNAAsp/mature tRNAAsp, but has little effect on mature tRNATyr
We next tested the possible involvement of exosome subunits in tRNATyr/alt-tRNATyr decay. The exosome of T. brucei has similar properties to that of S. cerevisiae, with two exceptions: (1) the 3′–5′ exoribonuclease RRP6 is present in stoichiometric amounts in both nuclear and cytoplasmic fractions, while in S. cerevisiae, it is nuclear only; and (2), the 3′–5′ exoribonuclease RRP44 is an autonomous enzyme and does not associate with the exosome (Estévez et al. 2001). Conversely, homologs for other subunits, do associate with the exosome (Estévez et al. 2001). The established roles for RRP44 and RRP6 in T. brucei are limited to rRNA processing (Estévez et al. 2001). Furthermore, we also assessed a potential role for Dis3L2 in tRNA degradation. Dis3L2 is a 3′–5′ endonuclease that is structurally similar to RRP44, but lacks the PIN domain, which is necessary for interaction with the exosome (Malecki et al. 2013). Dis3L2 is a cytoplasmic enzyme present in many eukaryotes, including T. brucei (Malecki et al. 2013; Clayton 2014; Towler et al. 2020). In S. pombe, Dis3L2 has been implicated in an RNA degradation pathway independent from RTD and the exosome, while in Drosophila, a role in cell proliferation and tissue growth has been suggested (Malecki et al. 2013; Towler et al. 2020).
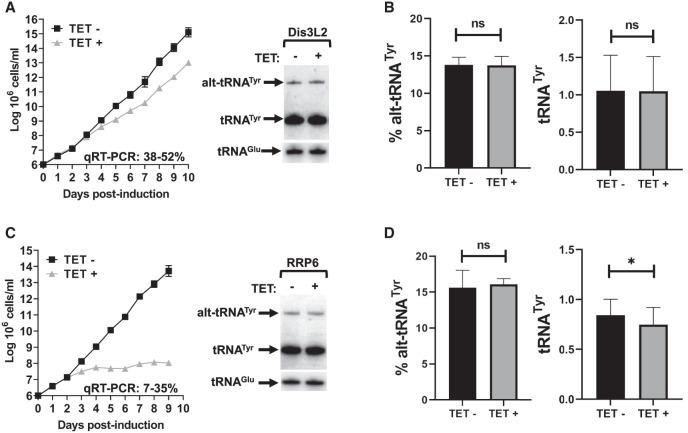
Down-regulation of Dis3L2 and RRP6 led to growth defects as compared to the RNAi uninduced control (TET-) but had little effect on the steady-state level of tRNA (Fig. 6). RRP44 down-regulation caused a growth defect and led to a statistically significant accumulation of alt-tRNATyr (Fig. 7). Remarkably, the steady-state levels of alt-tRNAAsp, and tRNAAsp were also increased (Fig. 7). But the levels of mature tRNATyr remained unchanged (Fig. 7). We note that the levels of alt-tRNATyr (∼20%) versus the mature form were lower than in Figure 1D. This can be explained by the experimental set up: time-resolved RNAi is performed by periodic dilution to keep the culture in exponential growth. In other words, cell density is kept at ∼3–5 × 106 cells/mL, comparable to 12–24 h in the growth curve from Figure 1B, and the bar graph in Figure 1D.
FIGURE 6.
RNAi knockdown of Dis3L2 and RRP6. (A,C) Triplicate growth curve of T. brucei cells subjected to tetracycline-induced protein knockdown where total RNA was collected on day 6, and northern blot hybridization performed on the extracted total RNA samples revealing the mature and alternate conformers of tRNATyr. (B,D) Relative amounts of alternate conformer tRNATyr and mature tRNATyr. The graphs are the result of three biological replicas. The asterisks in D denote significance based on a two-tailed paired t-test with a P-value of 0.03. “ns” stands for nonsignificant by the same test.
FIGURE 7.
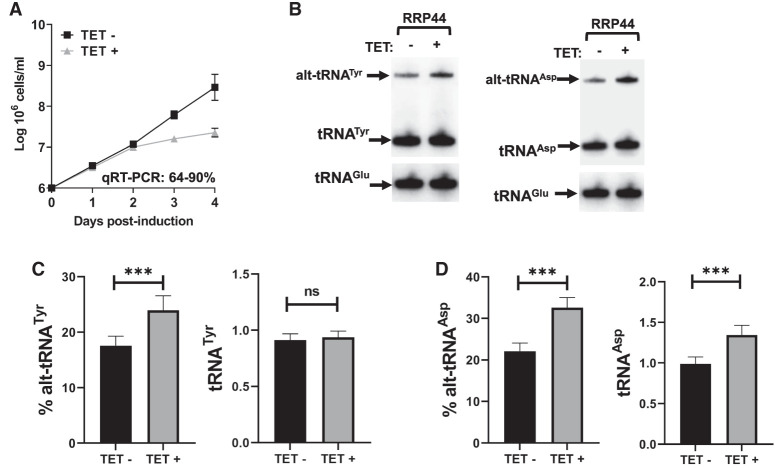
RNAi knockdown of RRP44. (A) Triplicate growth curve of T. brucei cells subjected to tetracycline-induced protein knockdown where total RNA was collected on day 4. (B) Northern blot hybridization performed on the extracted total RNA samples revealing the mature and alternate conformers of tRNATyr and tRNAAsp. (C,D) Relative amounts of alternate conformer tRNATyr and tRNAAsp, and mature tRNATyr and tRNAAsp. The graphs are the result of three biological replicas. The asterisks in C denote significance based on a two-tailed paired t-test with a P-value of 0.0009. The asterisks in D denote P-values of 0.0008 for both alt-tRNAAsp and tRNAAsp. “ns” stands for nonsignificant by the same test.
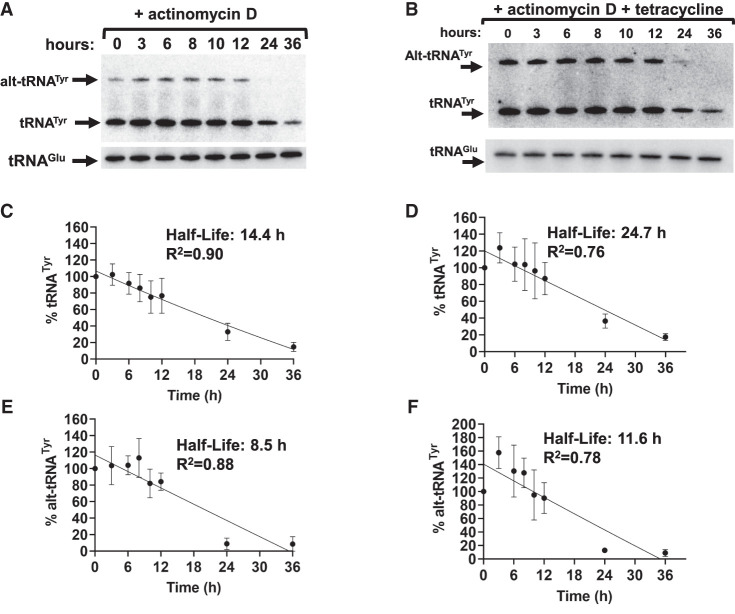
Considering alt-tRNATyr stabilization by RRP44 RNAi, we inquired whether RRP44 depletion would affect the half-life of this tRNA. We performed the transcription arrest assay as in Figure 2, but under RRP44 knockdown conditions, whereby actinomycin D was added on day 4 post RNAi induction. Cells were then harvested at various time points, and total RNA was analyzed by northern blot hybridization (Fig. 8). Down-regulation of RRP44 led to an increase in the half-life of both tRNATyr and alt-tRNATyr, from 14.4 and 8.5 h in the uninduced control (Fig. 8A,C,E) to 24.7 and 11.6 h, respectively, in RNAi-induced samples (Fig. 8B,D,F). Taken together, these experiments lead to the conclusion, that RRP44 is partially responsible for the unusually short half-life of tRNATyr and its alternate isoform.
FIGURE 8.
RNAi Knockdown of RRP44 stabilizes both tRNATyr conformers. (A,B) Northern blot hybridization performed on the extracted total RNA samples from RRP44 RNAi strain T. brucei cells subjected to Actinomycin D treatment with and without RRP44 depletion. The tRNAGlu was used as loading control. (C–F) Linear regression calculations utilizing the amounts of mature and alternate conformer tRNATyr with and without RRP44 depletion. Data were fit to a one-phase decay with R2 values as indicated. All data plotted were the result of three biological replicas.
Given the observed effect of XRNE down-regulation on the levels of alt-tRNATyr (Fig. 5E), we also performed similar actinomycin D experiments as above. As in the case of XRNE, actinomycin D had little effect on cell viability beyond growth arrest when comparing the uninduced to the RNAi-induced cells (Supplemental Fig. S3). Furthermore, down-regulation of XRNE had a negligible effect on the half-life of alt-tRNATyr (Supplemental Fig. S4). Taken together beyond redundancy in function among the nuclease tested here, only RRP44 has a significant contribution to the unusually short half-life of tRNATyr.
DISCUSSION
The rate of transcription, maturation and decay dictates the steady-state levels of tRNAs in cells, which are, in turn, crucial for protein synthesis. Conversely, the long half-lives of mature tRNAs are seemingly at odds with rapid responses to environmental changes that require translational reprograming. This places emphasis on tRNA decay pathways such as the 5′–3′ RTD and 3′–5′ exosome pathways, which actively scrutinize tRNAs (Estévez et al. 2001; Kadaba et al. 2006; Chernyakov et al. 2008). The frontline of tRNA scrutiny is nuclear surveillance, which eliminates incorrectly transcribed and hypomodified tRNAs by targeting aberrant molecules to the TRAMP complex, leading to A-tailing by Trf4 and 5 noncanonical poly(A) polymerases, followed by degradation by the nuclear exosome-associated 3′–5′ exoribonuclease RRP6 (Kadaba et al. 2006). Correctly processed tRNAs are further inspected by the nuclear and cytoplasmic RTD pathways (Chernyakov et al. 2008). In RTD, the major surveillance system for tRNA quality in eukaryotes, defective, damaged and hypomodified molecules are degraded by enzymes such as 5′–3′ exoribonucleases XRN1 and RAT1 (Chernyakov et al. 2008). These enzymes can either recognize the molecule directly, or be recruited by other proteins, such as the CCA-adding enzyme (CAE) (Wilusz et al. 2011). This enzyme, which has the canonical function of adding CCA to the 3′-end of newly synthesized tRNAs, can also recognize aberrant tRNAs and mark them for degradation by the RTD components by the addition of a CCACCA “degradation tag” (Wilusz et al. 2011). These essential functions have also been described in T. brucei, where CAE localizes to the nucleus, cytoplasm, and mitochondrion (Shikha and Schneider 2020). These pathways likely recognize uniform tRNA features, which makes the relative instability of T. brucei tRNATyr intriguing.
Considering the rapid turnover, tRNATyr may appear as a major target for the tRNA degradation pathways, including RTD and nuclear surveillance; however, our results provide no indication of their functioning in affecting tRNA steady-state levels. The XRN1 homolog, XRNE, was the only tested 5′–3′ exonuclease to exert a small, but statistically significant effect on tRNATyr stability. We cannot rule out, however, that some among the six XRNs have redundant roles, and that a more severe effect on stability may have been masked by the other homologs. Along these lines, we emphasize that since neither XRND or XRNF could be efficiently down-regulated by our RNAi approach, their involvement in tRNA stabilty will remain an open question. Moreover, RRP44 depletion affects both tRNATyr conformers, which leads to a statistically significant increase in tRNATyr stability. Unlike in other eukaryotes, T. brucei RRP44 does not associate with the exosome in any compartment (Estévez et al. 2001). T. brucei RRP44 reportedly performs canonical rRNA maturation function in the nucleus but has no assigned function in the cytoplasm (Estévez et al. 2001; Cesaro et al. 2019). Curiously, an effect on steady-state levels was also observed for tRNAAsp and alt-tRNAAsp under RRP44 knockdown conditions, but neither showed the shorter half-life observed with alt-tRNATyr/tRNATyr. Finally, T. brucei RRP44 is capable of degrading structured RNAs, such as stem–loops, in vitro (Cesaro et al. 2023). This activity is dependent on the RRP44 exonuclease domain, and can be observed even in structured substrates that lack of 3′ overhangs (Cesaro et al. 2023). Thus, it is imaginable that a biological function of RRP44 is in the turnover of tRNATyr, and potentially tRNAAsp.
In recent years, there have been many reports of tRNA fragments playing crucial roles in cell function. It is possible that the short half-life of tRNATyr may be due to fragment generation; however, we did not observe any tRNA fragments in our experiments, even after gross overexposure of the northern membranes, thus we think this possibility unlikely. T. brucei also encodes a homolog of the 3′–5′ exoribonuclease Dis3L2, which has been described in other eukaryotes as a stand-alone enzyme that is structurally similar to RRP44, and implicated on its own RNA degradation pathway (Malecki et al. 2013; Clayton 2014; Towler et al. 2020). We explored the possible involvement of the T. brucei homolog in tRNATyr degradation; however, our knockdown assays and northern blot analysis showed no effect on the tRNA stability, albeit we confirmed that the enzyme is essential for fitness. However, we cannot currently rule out the possibility of functional redundancy between these enzymes.
As stated above, the tRNATyr of T. brucei presents several unusual features that are extended in this study. We find that tRNATyr is present as two separate isoforms: the mature-sized 75-nt tRNATyr, and the ∼120-nt alt-tRNATyr, both of which are remarkably unstable for a tRNA, presenting short half-lives of only 10.3 and 6.9 h, respectively. This rapid turnover seemingly comes into conflict with the “high cost” of synthesizing a mature tRNATyr in T. brucei, as that involves use of the cytoplasmic splicing machinery, editing and modification events, and multidirectional transport. It also seems to be unrelated to the modification Q, as the other three Q-modified tRNAs in T. brucei did not show unusual stability by northern analysis. Furthermore, we had reported that the Q modification undergoes dynamic changes according to nutrient availability, meaning, when the concentration of certain nutrients present in the media are altered, the amount of Q-modified tRNATyr changes in a predictable way, affecting codon choice for tRNATyr, but seemingly not for other Q-tRNAs (Dixit et al. 2021). Taken together, these discoveries imply that, tRNATyr may function not only as a translation-competent molecule, but also as a nutrient sensor, in a network that regulates dynamic synthesis and removal of Q modification. Mature tRNAs act as nutrient sensing molecules in eukaryotes, with uncharged tRNAs signaling nutritional stress, leading to reduction in global protein biosynthesis during starvation of specific amino acids (Wek et al. 1995; Raina and Ibba 2014). Moreover, as mentioned above, tRNA fragments are also implicated in signaling pathways in bacteria and eukaryotes (Raina and Ibba 2014; Ren et al. 2019). Considering the rapid changes in nutrient availability that T. brucei may face when changing from vector to host, and vice-versa, we propose that the rapid turnover of tRNATyr ensures equally rapid changes in protein biosynthesis.
Currently, the identity of alt-tRNATyr remains unclear. Circularization would explain the observed differences in electrophoretic mobility between the alt-tRNA and mature tRNA bands. Circular RNAs (circRNAs) were first described over 40 years ago in eukaryotes, initially as either the products of self-splicing introns in single-cellular organisms, or somewhat rare, aberrant and poorly processed transcripts, that had no discernible function (Jeck et al. 2013b; Wilusz 2018). Recently, however, circRNAs have been brought to prominence, revealed as innate products of transcription that are generated through a mechanism dubbed “back splicing,” and shown to differ in stability and abundance from their linear counterparts (Jeck et al. 2013a). Some circRNAs accumulate in a tissue-specific manner in metazoans, sometimes possessing regulatory functions in specific cell types (Capel et al. 1993). Finally, tRNAs can generate their own abundant class of circular RNAs, called tRNA intronic circular RNAs (tricRNAs), which are produced during tRNA splicing (Lu et al. 2015; Noto et al. 2017; Schmidt et al. 2019). Considering this information, we wondered whether alt-tRNATyr was a circularized version of mature tRNATyr in T. brucei, and performed a two-dimensional gel northern blot hybridization assay as previously described (Blanc et al. 1999). This assay can reveal circular RNA bands based on aberrant migration patterns during the second phase of electrophoresis separation. In all our assays, however, alt-tRNATyr migrated in a straight line during this step, consistent with the migration expected for a linear molecule (Supplemental Fig. S5).
A second possibility is that the slower migration of alt-tRNAs is due to extensions at the 5′ or 3′ end; however, the OTTR sequencing provided partially rules out this scenario, although we cannot formally refute the argument of the existence of a yet to be identified modification that prevents sequencing for the true alt-tRNA and therefore skews what can be seen in our sequence analysis. Lastly, it is possible the alt-tRNA is a repository for the mature form of the molecule. Indeed, we notice that in the northern blots performed for the transcription arrest assay under RRP44 depletion, the bands corresponding to mature tRNATyr remains stable until the 12 h time point, when alt-tRNATyr is still present (Fig. 8B). After that threshold, alt-tRNATyr is no longer detectable in the membrane, and the mature tRNATyr finally starts to diminish (Fig. 8B). This would indicate that alt-tRNATyr may be converted into mature form, perhaps to help maintain appropriate levels of tRNATyr. However, currently it is not clear if the short half-lives of both tRNA species are just coincidental as we have failed to establish a strict precursor-product relationship with the assays provided.
Taken together, our results show that tRNATyr is indeed a unique molecule in T. brucei: a strikingly unstable tRNA present as two isoforms that is subjected to multiple transactions including intron editing, backbone modification and intracellular transport, and yet escaping most common RNA degradation pathways, to be regulated by RRP44 exonuclease.
MATERIALS AND METHODS
Cell culturing and RNAi knockdowns
Trypanosoma brucei 29-13 procyclic cells (Wirtz et al. 1999) were cultured at 27°C in SDM-79 media supplemented with 10% FBS, and growth was assessed by counting with Neubauer chambers. Gene knockdowns were performed with p2T7-177 plasmids as previously described (LaCount et al. 2000). RNAi was induced by addition of 1 µg/mL tetracycline to the media, generating a dsRNA from two head-to-head T7 promoters (LaCount et al. 2000). Target regions for knockdowns were selected with the RNAit online tool (https://dag.compbio.dundee.ac.uk/RNAit/) (Redmond et al. 2003), and oligos generated were as follows: XRNA: For TGCGATCTCATCTTCGGTCG; Rev TAGGAAGGAGAACCGAGCGA; XRNB: For GCGAACGCAAAAGTTGGACT; Rev CCGTGCATAGCCAATCAACG; XRNC: For CCAAGTTGACAACGGTGCAG; Rev GAACAACCCCCGTTTCGTTG; XRND: For ATTCGGATGGGACCCTGAGA; Rev GGGAAACTTCGCAGCTACCT; XRNE: For GCCAGCGAAAGATACTTGCG; Rev TGTCGGTTTCTCTCAGCACC; XRNF: For CCGACTTGGTGCTAGTTGGT; Rev CGATGGATCTGAGAGCCCAC; Dis3L2: For GTGTGATAGCCGTCCTCGAG; Rev CGTGGCAGGGTCGATACTAC; RRP6: For TCCCTTCAATTGAGCAGCGT; Rev ATGGCACCTTCCTTCAGCTC; RRP44: For AGAGTGAGGCTGCTGTTTCC; Rev GCCGATGCACAATTACGTCC.
Northern blot analysis
Cells at mid to late log (6 × 106 to 1 × 107 cells/mL) were harvested by centrifugation at 1400g for 10 min and washed twice with PBS. Total RNA was isolated from pellets using a guanidinium thiocyanate–phenol–chloroform protocol as previously described (Chomczynski and Sacchi 1987). Samples containing 5 µg of total RNA were resolved in denaturing 8M urea 8% polyacrylamide gels, electroblotted into Zeta-probe nylon membranes according to manufacturer protocol (Bio-Rad), then UV-cross-linked for 1 min. Northern blot hybridization was performed according to manufacturer specifications using 32P-labeled oligonucleotides. After hybridization, membranes were exposed overnight to a phosphorimager screen. Blots were analyzed using a Typhoon FLA 9000 scanner and the ImageQuant TL software (GE Healthcare). Probes used for Northern hybridization were as follows (5′–3′ orientation): tRNAAsn CTCCTCCCGTTGGATTCG; tRNAAsp CGGGTCACCCGCGTGACAGG; tRNAGln CAGGATTCGAACCTGGGTTTTCG; tRNAGlu TTCCGGTACCGGGAATCGAAC; tRNAHis GGGAAGACCGGGAATCGAAC; tRNAIle GGGGTTCGAACCCGCGATATTCGGT; tRNALeu AACCCACGCCTCCGGAGAG; tRNAPhe GCGACCCGGGATCGAACCAGGGACC; tRNATyr CCTTCCGGCCGGAATCGAACCAGCGAC; tRNATyr (Intron) GATACCTGCAAACTCTAC; tRNATyr (Leader) CTGACTGCCGCAGTAGTCGGG; tRNATyr (Trailer) CGTTTGTGGGCGTGAAAAAGT.
Transcription arrest assays
Cultures containing cells at early log (2 × 106 cells/mL) received actinomycin D (in DMSO) to a final concentration of 10 µg/mL, and were incubated at 27°C under transcription arrest while cells were harvested by centrifugation at scheduled time points, following protocol (Li et al. 2006). Total RNA was isolated from pellets using a guanidinium thiocyanate–phenol–chloroform protocol as previously described (Chomczynski and Sacchi 1987). All calculations and graphs were performed using GraphPad Prism version 8 for Windows (GraphPad Software, www.graphpad.com). Statistical analyses are as described in the figure legends.
Circular RNA analysis
Cells at mid to late log were harvested, and total RNA was isolated from pellets as described above. Samples containing 5 µg of total RNA were resolved first on a 5% polyacrylamide gel for the first dimension, then moved to a 10% polyacrylamide to be resolved in the second dimension. Samples were then electroblotted into Zeta-probe nylon membranes according to manufacturer protocol (Bio-Rad), then UV-cross-linked for 1 min. Northern blot hybridization was performed according to manufacturer specifications using 32P-labeled oligonucleotides. After hybridization, membranes were exposed overnight to a phosphoimager screen. Blots were analyzed using a Typhoon FLA 9000 scanner and the ImageQuant TL software (GE Healthcare).
RNA sequencing
Prior to library preparation, AlkB treatment of the RNA was carried out as previously described (Cozen et al. 2015) followed by RNA 3′ de-phosphorylation (Huppertz et al. 2014). Briefly, samples were treated with T4 Polynucleotide Kinase (T4PNK; New England Biolabs) in a modified 5× reaction buffer (350 mM Tris-HCl, pH 6.5, 50 mM MgCl2, 5 mM dithiothreitol) under low pH conditions in the absence of ATP for 30 min. This RNA was used to generate OTTR-seq as previously described (Upton et al. 2021). Briefly, total AlkB and PNK-treated RNA was 3′ tailed using mutant BoMoC RT in buffer containing only ddATP for 90 min at 30°C, with the addition of ddGTP for another 30 min at 30°C. This was then heat-inactivated at 65°C for 5 min, and unincorporated ddATP/ddGTP were hydrolyzed by incubation in 5 mM MgCl2 and 0.5 units of shrimp alkaline phosphatase (rSAP) at 37°C for 15 min. An amount of 5 mM EGTA was added and incubated at 65°C for 5 min to stop this reaction. Reverse transcription was then performed at 37°C for 30 min, followed by heat inactivation at 70°C for 5 min. The remaining RNA and RNA/DNA hybrids were then degraded using 1 unit of RNase A at 50°C for 10 min. cDNA was then cleaned up using a MinElute Reaction CleanUp Kit (Qiagen). To reduce adaptor dimers, cDNA was run on a 9% UREA page gel, and the size range of interest was cut out and eluted into gel extraction buffer (300 mM NaCl, 10 mM Tris; pH 8.0, 1 mM EDTA, 0.25% SDS) and concentrated using EtOH precipitation. Size-selected cDNA was then PCR amplified for 12 cycles using Q5 High-fidelity polymerase (NEB #M0491S). Amplified libraries were then run on a 6% TBE gel, and the size range of interest was extracted to reduce adaptor dimers further. Gel slices were eluted into gel extraction buffer (300 mM NaCl, 10 mM Tris; pH 8.0, 1 mM EDTA) followed by concentration using EtOH precipitation. Final libraries were pooled and sequenced on an Illumina NextSeq500 using a 150-cycle high-output kit.
Data processing
Sequencing adaptors were trimmed from raw reads using cutadapt, v1.18, and read counts were generated for tRNAs using tRAX (Holmes et al. 2022). Briefly, trimmed reads were mapped to the mature tRNAs of the Trypanosoma brucei Lister 427 genome assembly obtained from GtRNAdb and the reference genome sequence using Bowtie2 in very-sensitive mode with the following parameters to allow for a maximum of 100 alignments per read: –very-sensitive –ignore-quals –np 5 -k 100. Mapped reads were filtered to retain only the “best mapping” alignments. Raw read counts of tRNAs and pre-tRNAs were computed using tRNA annotations from GtRNAdb.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank all members of the Alfonzo laboratory for comments and discussions. The present work was funded in part by National Institutes of Health (NIH) grants GM132254 and GM084065-11 to J.D.A., RO1AI152408 to R.A., R01AI113157 to I.A., and by the National Human Genome Research Institute, National Institutes of Health (NHGRI, NIH) HG006753 grant to T.L.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079674.123.
REFERENCES
- Abelson HT, Johnson LF, Penman S, Green H. 1974. Changes in RNA in relation to growth of fibroblast 2. Lifetime of mRNA, rRNA, and tRNA in resting and growing cells. Cell 1: 161–165. 10.1016/0092-8674(74)90107-X [DOI] [PubMed] [Google Scholar]
- Alexandrov A, Chernyakov I, Gu W, Hiley SL, Hughes TR, Grayhack EJ, Phizicky EM. 2006. Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21: 87–96. 10.1016/j.molcel.2005.10.036 [DOI] [PubMed] [Google Scholar]
- Allmang C, Kufel J, Chanfreau G, Mitchell P, Petfalski E, Tollervey D. 1999. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J 18: 5399–5410. 10.1093/emboj/18.19.5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc V, Alfonzo JD, Aphasizhev R, Simpson L. 1999. The mitochondrial RNA ligase from Leishmania tarentolae can join RNA molecules bridged by a complementary RNA. J Biol Chem 274: 24289–24296. 10.1074/jbc.274.34.24289 [DOI] [PubMed] [Google Scholar]
- Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. 1993. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73: 1019–1030. 10.1016/0092-8674(93)90279-Y [DOI] [PubMed] [Google Scholar]
- Cesaro G, Carneiro FRG, Ávila AR, Zanchin NIT, Guimarães BG. 2019. Trypanosoma brucei RRP44 is involved in an early stage of large ribosomal subunit RNA maturation. RNA Biol 16: 133–143. 10.1080/15476286.2018.1564463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesaro G, da Soler HT, Guerra-Slompo EP, Haouz A, Legrand P, Zanchin NIT, Guimaraes BG. 2023. Trypanosoma brucei RRP44: a versatile enzyme for processing structured and non-structured RNA substrates. Nucleic Acids Res 51: 380–395. 10.1093/nar/gkac1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernyakov I, Whipple JM, Kotelawala L, Grayhack EJ, Phizicky EM. 2008. Degradation of several hypomodified mature tRNA species in Saccharomyces cerevisiae is mediated by Met22 and the 5′–3′ exonucleases Rat1 and Xrn1. Genes Dev 22: 1369–1380. 10.1101/gad.1654308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N. 1987. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159. 10.1016/0003-2697(87)90021-2 [DOI] [PubMed] [Google Scholar]
- Clayton CE. 2014. Networks of gene expression regulation in Trypanosoma brucei. Mol Biochem Parasitol 195: 96–106. 10.1016/j.molbiopara.2014.06.005 [DOI] [PubMed] [Google Scholar]
- Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. 2015. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods 12: 879–884. 10.1038/nmeth.3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit S, Kessler AC, Henderson J, Pan X, Zhao R, D'Almeida GS, Kulkarni S, Rubio MAT, Hegedusová E, Ross RL, et al. 2021. Dynamic queuosine changes in tRNA couple nutrient levels to codon choice in Trypanosoma brucei. Nucleic Acids Res 49: 12986–12999. 10.1093/nar/gkab1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estévez AM, Kempf T, Clayton C. 2001. The exosome of Trypanosoma brucei. EMBO J 20: 3831–3839. 10.1093/emboj/20.14.3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick MI, Heinemann IU. 2021. Regulation of RNA stability at the 3′ end. Biol Chem 402: 425–431. 10.1515/hsz-2020-0325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudipati RK, Xu Z, Lebreton A, Séraphin B, Steinmetz LM, Jacquier A, Libri D. 2012. Extensive degradation of RNA precursors by the exosome in wild-type cells. Mol Cell 48: 409–421. 10.1016/j.molcel.2012.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanoune J, Agarwal MK. 1970. Studies on the half life time of rat liver transfer RNA species. FEBS Lett 11: 78–80. 10.1016/0014-5793(70)80496-3 [DOI] [PubMed] [Google Scholar]
- Holmes AD, Howard JM, Chan PP, Lowe TM. 2022. tRNA Analysis of eXpression (tRAX): a tool for integrating analysis of tRNAs, tRNA-derived small RNAs, and tRNA modifications. bioRxiv 10.1101/2022.07.02.498565 [DOI] [Google Scholar]
- Houseley J, Tollervey D. 2009. The many pathways of RNA degradation. Cell 136: 763–776. 10.1016/j.cell.2009.01.019 [DOI] [PubMed] [Google Scholar]
- Huppertz I, Attig J, D'Ambrogio A, Easton LE, Sibley CR, Sugimoto Y, Tajnik M, König J, Ule J. 2014. iCLIP: protein-RNA interactions at nucleotide resolution. Methods 65: 274–287. 10.1016/j.ymeth.2013.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. 2013a. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19: 141–157. 10.1261/rna.035667.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. 2013b. Erratum: circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19: 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadaba S, Wang X, Anderson JT. 2006. Nuclear RNA surveillance in Saccharomyces cerevisiae: Trf4p-dependent polyadenylation of nascent hypomethylated tRNA and an aberrant form of 5S rRNA. RNA 12: 508–521. 10.1261/rna.2305406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnahl U, Wasternack C. 1992. Half-life of cytoplasmic rRNA and tRNA, of plastid rRNA and of uridine nucleotides in heterotrophically and photoorganotrophically grown cells of Euglena gracilis and its apoplastic mutant W3BUL. Int J Biochem 24: 493–497. 10.1016/0020-711X(92)90044-2 [DOI] [PubMed] [Google Scholar]
- Kessler AC, Kulkarni SS, Paulines MJ, Rubio MAT, Limbach PA, Paris Z, Alfonzo JD. 2018. Retrograde nuclear transport from the cytoplasm is required for tRNATyr maturation in T. brucei. RNA Biol 15: 528–536. 10.1080/15476286.2017.1377878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer S, Queiroz R, Ellis L, Webb H, Hoheisel JD, Clayton C, Carrington M. 2008. Heat shock causes a decrease in polysomes and the appearance of stress granules in trypanosomes independently of eIF2α phosphorylation at Thr169. J Cell Sci 121: 3002–3014. 10.1242/jcs.031823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni S, Rubio MAT, Hegedusová E, Ross RL, Limbach PA, Alfonzo JD, Paris Z. 2021. Preferential import of queuosine-modified tRNAs into Trypanosoma brucei mitochondrion is critical for organellar protein synthesis. Nucleic Acids Res 49: 8247–8260. 10.1093/nar/gkab567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. 2005. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 121: 713–724. 10.1016/j.cell.2005.04.029 [DOI] [PubMed] [Google Scholar]
- LaCount DJ, Bruse S, Hill KL, Donelson JE. 2000. Double-stranded RNA interference in Trypanosoma brucei using head-to-head promoters. Mol Biochem Parasitol 111: 67–76. 10.1016/S0166-6851(00)00300-5 [DOI] [PubMed] [Google Scholar]
- Li CH, Irmer H, Gudjonsdottir-Planck D, Freese S, Salm H, Haile S, Estévez AM, Clayton C. 2006. Roles of a Trypanosoma brucei 5′/3′ exoribonuclease homolog in mRNA degradation. RNA 12: 2171–2186. 10.1261/rna.291506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes RRS, de O Silveira G, Eitler R, Vidal RS, Kessler A, Hinger S, Paris Z, Alfonzo JD, Polycarpo C. 2016. The essential function of the Trypanosoma brucei Trl1 homolog in procyclic cells is maturation of the intron-containing tRNATyr. RNA 22: 1190–1199. 10.1261/rna.056242.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Filonov GS, Noto JJ, Schmidt CA, Hatkevich TL, Wen Y, Jaffrey SR, Gregory Matera A. 2015. Metazoan tRNA introns generate stable circular RNAs in vivo. RNA 21: 1554–1565. 10.1261/rna.052944.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malecki M, Viegas SC, Carneiro T, Golik P, Dressaire C, Ferreira MG, Arraiano CM. 2013. The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO J 32: 1842–1854. 10.1038/emboj.2013.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manful T, Fadda A, Clayton C. 2011. The role of the 5′–3′ exoribonuclease XRNA in transcriptome-wide mRNA degradation. RNA 17: 2039–2047. 10.1261/rna.2837311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P, Tollervey D. 2000. Musing on the structural organization of the exosome complex. Nat Struct Biol 7: 843–846. 10.1038/82817 [DOI] [PubMed] [Google Scholar]
- Noto JJ, Schmidt CA, Matera AG. 2017. Engineering and expressing circular RNAs via tRNA splicing. RNA Biol 14: 978–984. 10.1080/15476286.2017.1317911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina M, Ibba M. 2014. TRNAs as regulators of biological processes. Front Genet 5: 171. 10.3389/fgene.2014.00171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond S, Vadivelu J, Field MC. 2003. RNAit: an automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol Biochem Parasitol 128: 115–118. 10.1016/S0166-6851(03)00045-8 [DOI] [PubMed] [Google Scholar]
- Ren B, Wang X, Duan J, Ma J. 2019. Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science 365: 919–922. 10.1126/science.aav8907 [DOI] [PubMed] [Google Scholar]
- Rubio MAT, Paris Z, Gaston KW, Fleming IMC, Sample P, Trotta CR, Alfonzo JD. 2013. Unusual noncanonical intron editing is important for tRNA splicing in Trypanosoma brucei. Mol Cell 52: 184–192. 10.1016/j.molcel.2013.08.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakyiama J, Zimmer SL, Ciganda M, Williams N, Read LK. 2013. Ribosome biogenesis requires a highly diverged XRN family 5′→3′ exoribonuclease for rRNA processing in Trypanosoma brucei. RNA 19: 1419–1431. 10.1261/rna.038547.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt CA, Giusto JD, Bao A, Hopper AK, Gregory Matera A. 2019. Molecular determinants of metazoan tricRNA biogenesis. Nucleic Acids Res 47: 6452–6465. 10.1093/nar/gkz311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, McNally KP, Agabian N. 1993. Splicing and 3′-processing of the tyrosine tRNA of Trypanosoma brucei. J Biol Chem 268: 21868–21874. 10.1016/S0021-9258(20)80621-8 [DOI] [PubMed] [Google Scholar]
- Shaheen HH, Hopper AK. 2005. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci 102: 11290–11295. 10.1073/pnas.0503836102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikha S, Schneider A. 2020. The single CCA-adding enzyme of T. brucei has distinct functions in the cytosol and in mitochondria. J Biol Chem 295: 6138–6150. 10.1074/jbc.RA119.011877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DWE, Weinberg WC. 1981. Transfer RNA in reticulocyte maturation. Biochim Biophys Acta 655: 195–198. 10.1016/0005-2787(81)90009-5 [DOI] [PubMed] [Google Scholar]
- Sroga GE. 1984. Half-life of tRNA and the initial rate of tRNA accumulation in plant cells from cell suspension cultures of Lupinus angustifolius cv. Turkus and Petroselinum hortense cv. Berlińska growing at different rates. . J Plant Physiol 116: 81–89. 10.1016/S0176-1617(84)80086-3 [DOI] [PubMed] [Google Scholar]
- Tan THP, Bochud-Allemann N, Horn EK, Schneider A. 2002. Eukaryotic-type elongator tRNAMet of Trypanosoma brucei becomes formylated after import into mitochondria. Proc Natl Acad Sci 99: 1152–1157. 10.1073/pnas.022522299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towler BP, Pashler AL, Haime HJ, Przybyl KM, Viegas SC, Matos RG, Morley SJ, Arraiano CM, Newbury SF. 2020. Dis3L2 regulates cell proliferation and tissue growth though a conserved mechanism. PLoS Genet 16: 1–29. 10.1371/journal.pgen.1009297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton HE, Ferguson L, Temoche-Diaz MM, Liu XM, Pimentel SC, Ingolia NT, Schekman R, Collins K. 2021. Low-bias ncRNA libraries using ordered two-template relay: serial template jumping by a modified retroelement reverse transcriptase. Proc Natl Acad Sci 118: e2107900118. 10.1073/pnas.2107900118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wek SA, Zhu S, Wek RC. 1995. The histidyl-tRNA synthetase-related sequence in the eIF-2α protein kinase GCN2 interacts with tRNA and is required for activation in response to starvation for different amino acids. Mol Cell Biol 15: 4497–4506. 10.1128/MCB.15.8.4497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE. 2018. A 360° view of circular RNAs: from biogenesis to functions. Wiley Interdiscip Rev RNA 9: e1478. 10.1002/wrna.1478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Whipple JM, Phizicky EM, Sharp PA. 2011. tRNAs marked with CCACCA are targeted for degradation. Science 334: 817–821. 10.1126/science.1213671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirtz E, Leal S, Ochatt C, Cross GM. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol Biochem Parasitol 99: 89–101. 10.1016/S0166-6851(99)00002-X [DOI] [PubMed] [Google Scholar]
- Yoshihisa T. 2003. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell 14: 3266–3279. 10.1091/mbc.e02-11-0757 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.