Abstract
Purpose
Poor ovarian response (POR) affects approximately 9% to 24% of women undergoing in vitro fertilization (IVF) cycles, resulting in fewer eggs obtained and increasing clinical cycle cancellation rates. The pathogenesis of POR is related to gene variations. Our study included a Chinese family comprising two siblings with infertility born to consanguineous parents. Poor ovarian response (POR) was identified in the female patient who had multiple embryo implantation failures occurring in subsequent assisted reproductive technology cycles. Meanwhile, the male patient was diagnosed with non-obstructive azoospermia (NOA).
Methods
Whole-exome sequencing and rigorous bioinformatics analyses were conducted to identify the underlying genetic causes. Moreover, the pathogenicity of the identified splicing variant was assessed using a minigene assay in vitro. The remaining poor-quality blastocyst and abortion tissues from the female patient were detected for copy number variations.
Results
We identified a novel homozygous splicing variant in HFM1 (NM_001017975.6: c.1730-1G > T) in two siblings. Apart from NOA and POI, biallelic variants in HFM1 were also associated with recurrent implantation failure (RIF). Additionally, we demonstrated that splicing variants caused abnormal alternative splicing of HFM1. Using copy number variation sequencing, we found that the embryos of the female patients had either euploidy or aneuploidy; however, both harbored chromosomal microduplications of maternal origin.
Conclusion
Our results reveal the different effects of HFM1 on reproductive injury in males and females, extend the phenotypic and mutational spectrum of HFM1, and show the potential risk of chromosomal abnormalities under the RIF phenotype. Moreover, our study provides new diagnostic markers for the genetic counseling of POR patients.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10815-023-02761-8.
Keywords: Infertility, Recurrent implantation failure, Chromosomal abnormality, HFM1, CNV-seq
Introduction
Infertility is the absence of pregnancy after 12 months of regular, unprotected sexual intercourse [1]. Approximately 8%–12% of reproductive-aged couples are affected worldwide [1]. Recently, with assisted reproductive technology (ART), viable pregnancies have been achieved in many families with infertility. However, recurrent implantation failure (RIF) remains an intractable problem during the process. Currently, there is no accepted formal definition of RIF in the medical community. However, it is widely accepted that RIF occurs in women with infertility aged < 40 years who have failed clinical pregnancy after four good-quality embryo transfers, with at least three fresh or frozen in vitro fertilization (IVF) cycles [2]. Current research suggests prethrombotic state, embryo, uterine, and immune factors as possible etiologic factors in RIF [2]. Among them, the cause of abnormal embryo development is primarily attributed to the abnormal gametes produced by meiosis [2]. The etiology of gametogenesis defects is heterogeneous, including genetic and autoimmune diseases and iatrogenic factors [3]. Over the years, candidate gene studies based on knockout mouse models and high-throughput sequencing studies, especially whole exome sequencing (WES), have identified many genetic factors causing gametogenesis defects [3–5].
Meiosis is a special type of cell division in which the DNA replicates once, and the chromosome segregates twice, resulting in haploid gametes that offset the doubling of the number of chromosomes during fertilization. Both crossover and recombination occur during the first meiosis to ensure the diversity of genetic material [6]. However, meiotic errors often result in gametogenesis defects and the formation of abnormal embryos after fertilization, leading to aneuploidy, spontaneous abortion, and human infertility [7]. Using WES, researchers recently identified several meiosis genes linked to gametogenesis, such as C14orf39 (MIM:617307), MSH4 (MIM:602105), STAG3 (MIM:608489), SPATA22 (MIM:617673), DMC1 (MIM:602721), CCDC155 (MIM:618125), and HFM1 (MIM:615684) [8–16]. HFM1, a meiosis-specific gene located on chromosome 1q22 in humans, has 39 exons encoding a 1435-aa protein and is highly expressed in germline tissues. HFM1, which encodes an ATP-dependent DNA helicase, is required for crossover formation and the complete synapsis of homologous chromosomes during meiosis [17]. To date, pathogenic variants of HFM1 have been reported to cause POI, NOA, and oligozoospermia in humans. Phenotype heterogeneity can be observed in the clinical analyses of female patients with reported genetic defects, including severely impaired but present ova reserves. However, whether these patients have a successful pregnancy through ART needs to be further explored.
In this study, we performed WES and Sanger sequencing in a consanguineous Chinese family comprising two siblings with infertility, and we identified one novel homozygous splicing variant in HFM1 (NM_001017975.6: c.1730-1G > T). Based on rigorous bioinformatics analysis and minigene assay, we speculated that HFM1 was the pathogenic gene responsible for two infertility cases in this family. Both male and female patients had gametogenesis disorders due to meiotic abnormalities. In contrast to other female patients with HFM1 variations who are unable to achieve pregnancy, this female patient presented with incomplete oogenesis disorders and could obtain a small number of embryos by IVF. However, she presented with a phenotype of RIF after embryo transfer. Through embryo copy number variation sequencing (CNV-seq), we found that the embryos of female patient displayed either euploidy or aneuploidy, but both carried chromosomal microduplications of maternal origin. We believe this study is the first to report the variant in the HFM1 gene (NM_001017975.6: c.1730-1G > T) as a potential cause of RIF and further highlights the crucial role of meiotic gene HFM1 in human gametogenesis in both females and males.
Materials and methods
Study participants
A consanguineous Chinese family comprising two siblings with infertility was enrolled in this study. The female patient’s age was < 35, with antral follicle count (AFC) < 5, anti-mullerian hormone (AMH) < 1.2 ng/mL, and recurrent failure of IVF/ICSI cycles including three cycles of ART to retrieve eggs and a total of three cycles of transplants before being included in the study. Well-known factors leading to ovarian dysfunction, including ovarian surgery, radiation, chemotherapy, and chromosomal abnormalities, were excluded. Meanwhile, we also excluded possible genetic factors that could have contributed to recurrent failure of IVF/ICSI cycles, especially recurrent implantation failure, such as hereditary thrombophilia and abnormal uterine anatomy. Moreover, for the male patient (F II-2), no sperm was found in the semen analysis at the Department of Andrology, Reproductive Medicine Center, The First Affiliated Hospital of Anhui Medical University. We performed a serum reproductive hormone assay subsequently. Other factors associated with azoospermia, including chromosomal abnormalities, genomic azoospermia factor (AZF) deletions, testicular diseases, radiation, and chemotherapy, were excluded. Finally, a testicular biopsy was performed.
Ethical approval
This study was approved by the ethics committee of The First Affiliated Hospital of Anhui Medical University (PJ2020-13–10). Written informed consent was obtained from all participants before their inclusion in this study.
Whole-exome sequencing, bioinformatics analysis, and Sanger sequencing
Enriched with DNeasy Blood & Tissue Kit (Qiagen), the genomic DNA was extracted from the whole peripheral blood of all the family members for whole-exome sequencing (WES), bioinformatics analysis, and Sanger sequencing. According to the manufacturer’s instructions, we used the SureSelectXT Human All Exon Kit (Agilent Technologies, Santa Clara, CA, USA) to capture the whole exome, and sequencing was performed on the HiSeq X-Ten platform (Illumina, San Diego, CA, USA). The Burrows-Wheeler Aligner was used to map the original data to the human genome (hg19). Briefly, we annotated variants using allele frequency databases (1000G, ESP6500, ExAC, GnomAD, and GnomAD-EAS) and deleterious prediction tools (SIFT, Mutation Taster, Condel, SpliceAI, dbscSNV_RF, and dbscSNV_ADA). Variants with allele frequencies > 0.01 were excluded. We focused on missense and loss-of-function variants, including splicing (≤ 2 bp), stop-gain, stop-loss, and frameshift variants. Pathogenic variant genes specifically expressed in the reproductive system were used for further analysis. We paid more attention to the patient with homozygous and X-linked chromosomal variants. Lastly, we assessed the potential function and phenotype of the selected genes using the GeneCards database, the Online Mendelian Inheritance in Man database, the Mouse Genome Informatics database, and a PubMed-based literature review. Sanger sequencing was performed to identify the parental origins of the pathogenic variants (Supplementary Table S1).
Minigene assay for putative splicing variants
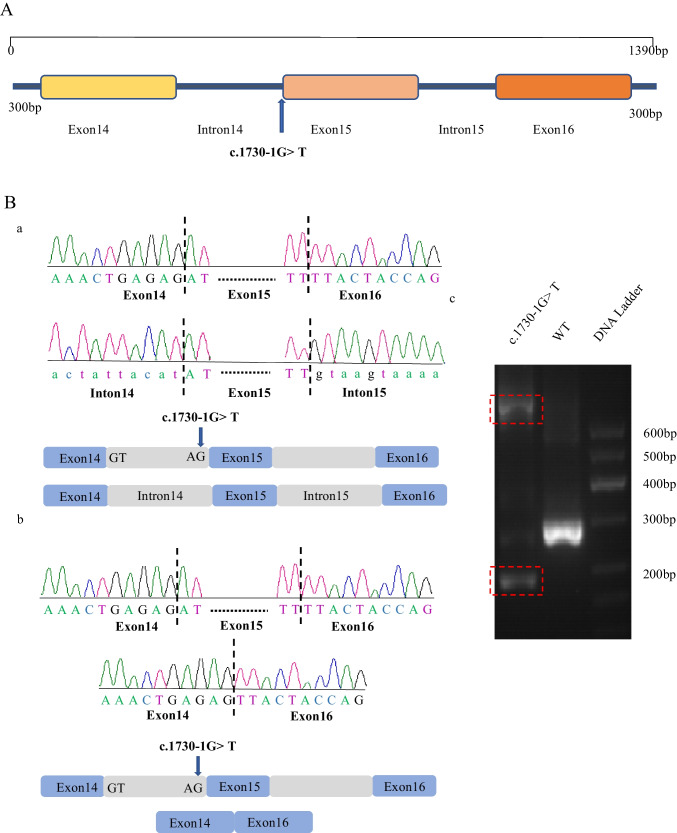
We performed a minigene splicing assay to verify whether the variant affected the splicing products. Thus, we designed a sequence containing exon 14, intron 14, exon 15, intron 15, exon 16, and 300 bp before exon 14 and after exon 16 of HFM1 (NM_001017975.6) using polymerase chain reaction (PCR) amplification and cloned the intact fragment into a modified PEGFP-C1 vector (Fig. 2A, Supplementary Table S2). Similarly, mutant plasmids were constructed via overlap-extension PCR with site-specific mutant primers. Lastly, we verified the wild type and variant constructs using Sanger sequencing.
Fig. 2.
Effects of the HFM1 variant in vitro. A Detailed map of plasmid construction, including exon 14, intron 14, exon 15, intron 15, exon 16, and the 300 bp base sequence before exon 14 and after exon 16. B The effect of the HFM1 splicing variant c.1730-1G > T. Compared to wild type, agarose gel electrophoresis (red dashed frame) showed a smaller and bigger cDNA fragment for the c.1730-1G > T variant (Fig. 2C-c). A possible scheme (Fig. 2C-a) shows that c.1730-1G > T provided no normal donor site, leading to keeping intron 14 and intron 15. Another kind of possible scheme (Fig. 2C-b) shows that donor c.1730-1G > T provided no normal donor site, leading to the skipping of exon 15 and resulting in premature termination
The HEK293 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) containing 10% fetal bovine serum and 1% penicillin/streptomycin stock solution at 37 °C, 5% CO2, and 95% humidity. Approximately 0.4 × 106 HEK293 cells per well were grown to 70% confluency in 6-well plates. Approximately 2.5 µg of plasmids (wild type or variants) were transfected into HEK293 cells using Lipofectamine 3000 reagent (Thermo Fisher Scientific, Waltham, MA, USA) according to standard protocols. After 48 h of culturing, TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) was used to extract cellular RNA according to the manufacturer’s protocols. Complementary DNA synthesis was carried out using 1 mg of RNA and the RevertAid First Strand cDNA Synthesis Kit following the standard protocol. Lastly, to evaluate splicing caused by mutations, we used High-Fidelity DNA Polymerase to perform PCR with the specialized primers shown in Supplementary Table S1. The PCR products were separated on a 3% agarose gel adding the nucleic acid developer, and the cDNA in the agarose gel from the two regions after separation was purified and Sanger sequenced.
Testis biopsy
After preoperative skin preparation in the supine position, with routine use of a disinfection towel, one testicle was placed in the left palm. Following 1% lidocaine infiltration anesthesia, testicular tissue was absorbed through a syringe and then sent to the reproductive laboratory to look for sperm.
IVF or intracytoplasmic sperm injection (ICSI) process
Since only one egg can be harvested in a natural cycle, women with infertility can obtain multiple eggs in a single cycle through controlled ovarian hyperstimulation to improve the rate of blastocyst acquisition. Ovulation-stimulating drugs were first used to promote the development of multiple follicles. When the follicles grew to an appropriate size, human chorionic gonadotropin (HCG) of 10000 IU was subcutaneously injected, and ultrasound-guided transvaginal oocyte retrieval was performed 24–36 h later. Sperm was obtained by masturbation or minimally invasive surgery on the day of oocyte retrieval. We first observed the appearance of eggs using an electron microscope before performing IVF or ICSI. For couples with infertility whose indication is IVF, eggs and sperm are fertilized naturally. For infertile couples with an ICSI indication, we injected a single sperm into the egg to help fertilization.
CNV-seq
DNA was extracted from the cell samples after lysis. Preamplification was first performed, followed by amplification. Because the number of cells in this experiment was less than three, the original 12 cycles were increased to 15 cycles during preamplification, and the original 14 cycles were increased to 18 cycles during amplification to increase the output of amplified products. After the whole genome assembly product was purified using the 1 × Agencourt AMPure XP kit (Beckman Coulter, CA, USA), the operation instructions were followed for every procedure, except for the transposition fragmentation time, which was extended from the original 5 to 10 min.
The original data generated by the Illumina Nextseq 550 sequencer were subjected to data-quality control, such as removing junctions and low-quality sequences. Using BWA software, high-quality reads were compared to the NCBI human reference genome (hg19) [18]. Picard software (2019 Broad Institute, https://broadinstitute.github.io/picard/) was then used to remove redundant sequences, and only unique reads were used for subsequent CNV calculations. Reads were divided into successive 600 Kb windows according to their genomic alignment locations, and the read count was used as the coverage of each window. The window coverage was then corrected: GC correction, mappability correction, and baseline correction [19]. The corrected window coverage was recognized by the hidden Markov model algorithm for the CNV boundary [20]. Different segments were segmented by the CNV boundary, and the median of the corrected coverage of all windows in the segment was counted as the copy ratio of the segment. Lastly, the status of each fragment was evaluated by the copy ratio; if the copy ratio was > 23, it was considered chromosome/fragment duplication, and if the copy ratio was between 23 and 27, it was considered repeated chimerism. When the copy ratio was < 17, chromosome/fragment deletion was considered, and deletion chimerism was considered when the copy ratio was between 13 and 17.
Analysis of the parental origin of abnormal chromosomes
DNA was extracted from the embryo samples and amplified by multiple displacement amplification to obtain a large amount of DNA. Genomic DNAs extracted from parental blood samples and embryos were used to construct libraries using the VeriSeq kit. Subsequently, the library data were sequenced on the NextSeq 550 sequencing platform to obtain sequencing data.
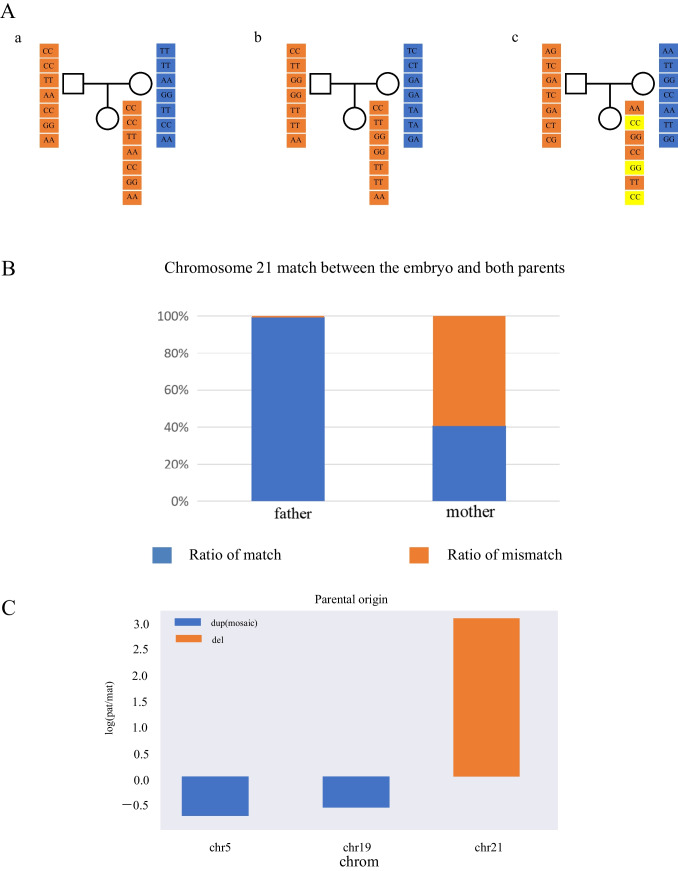
Genealogical sequencing data were filtered by reference genome alignment, detected genotypes, and site filtering (0.01 ≤ MAF ≤ 0.5). Based on the results of embryonic preimplantation genetic testing for aneuploidies, we speculated that there is only one chromosome 21; hence, we kept the embryo genotype homozygous for the site, selecting the effective sites (effective site refers to the homozygous site for either parent) and calculating the number of alleles shared between embryo and parents (Fig. 4A). The parental origin of chromosome 21 was inferred by analyzing the matching degree between the sequencing data of the embryo and the sequencing data of the parents. For parental origin analysis of copy number variation, the parental chromosome composition of the embryo was inferred by calculating the relative ratios of alleles shared by the parents based on Mendelian inheritance (Fig. 4C).
Fig. 4.
Verification of the parental origin of the copy number variation fragments. A-a Assuming that parents are homozygous, embryo genotype matching model. When both parents are homozygous, if the male genotype is AA and the female genotype is BB, the normal embryo will inherit the male allele A and the female allele B theoretically. Because the embryo chromosome is monosomy 21, if the embryo has genotype AA at this locus, it will be counted as the matched paternal chromosome; otherwise, it will be counted as the matched maternal chromosome. b Assuming that the male is homozygous and the female is heterozygous, embryo genotype matching model. When the male genotype is AA and the female genotype is AB, the normal embryo must inherit the male genotype A theoretically. Since the embryo chromosome is monosomy 21, if the genotype of this site in the embryo is AA, it will be considered as matching the paternal source. c Assuming that the male is heterozygous and the female is homozygous, embryo genotype matching model. When the male genotype is AB and the female genotype is AA, if the embryo genotype is AA, it is considered to match the maternal source. Orange square denotes maternal genotype. Blue square denotes paternal genotype. Yellow square denotes embryo genotype did not matching either parent. B The matching ratio was calculated according to the above method. Blue square denotes the matching ratio for the remaining chromosome 21 of the embryo for parents. Orange square denotes the mismatching ratio for the remaining chromosome 21 of the embryo for parents. C Above the 0 scale line, the chromosomal fragment is paternal, and below it, maternal. If the parental genotype is AA and BB, and the embryo is AA, so the maternal allele is 2 and the paternal allele is 0. Then, the parental ratio of the whole chromosome of the embryo was calculated. The parental ratio = the number of paternal alleles/the number of maternal alleles (for convenience of calculation, the parental ratio was taken as log). Finally, the parental chromosome composition of the embryo was inferred according to the parental ratio combined with the chromosome copy number. Theoretically, each pair of chromosomes of a normal individual was derived from the parents, and the parental ratio was 0. If the chromosome duplication exists, when the parental ratio is greater than 0, it means that there are superfluous paternal alleles in the embryo chromosome, and it is inferred that one chromosome duplication in the embryo is inherited from the father; otherwise, it is inherited from the mother. If a chromosome deletion is present, one of the remaining chromosomes in the embryo is presumed to have been inherited from the father when the parental ratio is greater than 0
Results
Novel homozygous splicing variant in HFM1 causes a POR sister and a NOA brother
Female patient (F II-1) and male patient (F II-2) were born in a consanguineous Chinese family. Based on age < 35, AFC < 5, and AMH < 1.2 ng/mL, female patient (F II-1) was diagnosed with poor ovarian response (POR) at the age of 31 according to the patient-oriented strategies encompassing individualized oocyte number (POSEIDON) criteria in the reproductive center of The First Affiliated Hospital of Anhui Medical University [21]. She experienced menarche at the age of 14 and had a 28–30-day menstrual cycle. In 2018, she underwent a biochemical pregnancy and had not been pregnant since then. Her basic hormone levels were as follows: follicle-stimulating hormone (FSH), 8.05 mIU/mL; luteinizing hormone (LH), 4.2 mIU/mL; estradiol (E2), 322 pmol/L; prolactin (PRL), 16.87 ng/mL; testosterone (T), 0.74 nmol/L; and anti-mullerian hormone (AMH), 0.5 ng/mL (Table 1). The sizes of the uterus and ovary (left ovary: 26 × 20 × 14 mm; right ovary: 27 × 17 × 17 mm), endometrial thickness, and chromosome karyotype were normal.
Table 1.
Clinical features and genetic information of HFM1 variations in F1 II-1 and F1 II-2
| Individuals | F1 II-1 | F1 II-2 |
|---|---|---|
| Clinical diagnosis | POR | NOA |
| Age | 31 | 25 |
| Secondary sexual characteristics | Normal | Normal |
| Testicular volume (left/right, ml) | - | 12/12 |
| Ovarian volume (left/right, mm) | 26 × 20 × 14/27 × 17 × 17 | - |
| Somatic karyotype | 46,XX | 46,XY |
| Sex hormone levels | ||
| FSH (mIU/ml) | 8.05 | 4.73 |
| LH (mIU/ml) | 4.2 | 5.06 |
| T (nmol/l) | 0.74 | 13.08 |
| E2 (pmol/l) | 322 | 141 |
| PRL (ng/ml) | 16.87 | 6.29 |
| AFC | 2–3 | - |
| AMH (ng/ml) | 0.5 | - |
| Y chromosome microdeletions | - | No |
| Information of HFM1 mutations | ||
| cDNA mutation | c.1730-1G > T | c.1730-1G > T |
| Mutation type | Splicing | Splicing |
| Allele frequency in human population | ||
| 1KGP | - | - |
| ESP6500 | - | - |
| ExAC | - | - |
| GnomAD | - | - |
| GnomAD-EAS | - | - |
| Functional prediction | ||
| SIFT | - | - |
| Mutation Taster | Damaging | Damaging |
| Condel | - | - |
| SpliceAI | Damaging | Damaging |
| dbscSNV_RF | Damaging | Damaging |
| dbscSNV_ADA | Damaging | Damaging |
RefSeq accession number of HFM1 is NM_001017975.6
Abbreviations: 1KGP, 1000 Genomes Project; ESP6500, NHLBI GO Exome Sequencing Project; ExAC, the Exome Aggregation Consortium; GnomAD, the Genome Aggregation Database; GnomAD-EAS, the Genome Aggregation Database-East Asian
A male patient (F II-2) was diagnosed with non-obstructive azoospermia (NOA) according to his semen analysis. His basic sex hormone levels were as follows: FSH, 4.73 mIU/mL; LH, 5.06 mIU/mL; E2, 141 pmol/L; PRL, 6.29 ng/mL; T, 13.08 nmol/L. The testicular volume (left testis: 12 mL; right testis: 12 mL) and chromosome karyotype were normal, and no Y chromosome microdeletion was detected (Table 1).
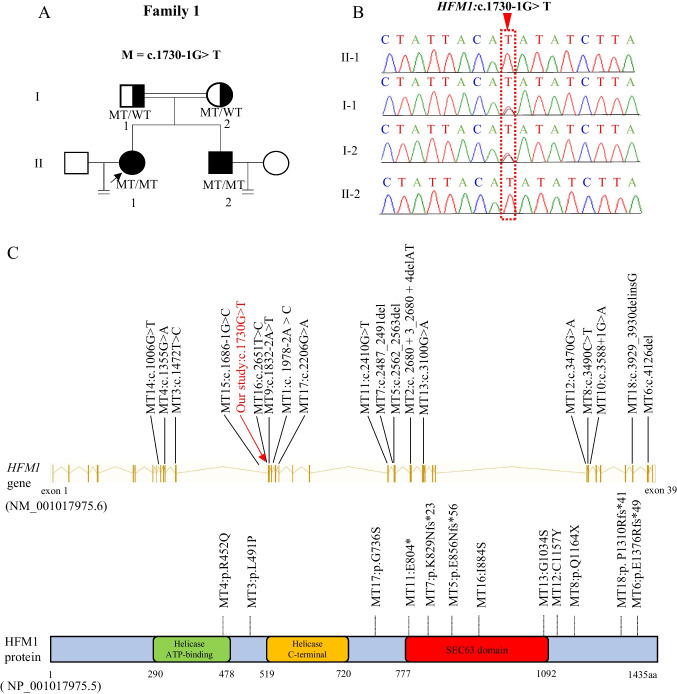
To determine the unknown genetic factors responsible for the female patient’s POR, WES was performed on her. After data screening, one homozygous splicing variant in HFM1 (NM_001017975.6: c.1730-1G > T) was identified, and this variant was not reported in public population databases, such as 1KGP, ESP6500, ExAC, GnomAD, and GnomAD-EAS, in terms of their frequencies (Table 1). The variant was predicted to be deleterious by four of the six tools, namely, SIFT, MutationTaster, Condel, SpliceAI, dbscSNV_RF, and dbscSNV_ADA (Table 1). HFM1 plays a key role in meiosis. A previous study showed that the homozygous HFM1 variant caused premature ovarian insufficiency (POI) and NOA in humans (Table 3, Fig. 1C), whereas the HFM1 knockout mouse model showed depletion of ovarian follicular reserve and subfertility in mice [14–17, 22]. Before this study, HFM1 variants had not been reported in infertile women with POR. Sanger sequencing confirmed that this variant was transmitted from the parental carriers of heterozygous mutations following a recessive inheritance pattern. The proband’s brother, diagnosed with NOA, also harbored this homozygous pathogenic variant (Fig. 1A, B).
Table 3.
Overview of the HFM1 variants observed in the reported individual
| PMID | Variant (NM_001017975.6) | Type of variant | Amino acid change | Female phenotype | Follicle | Male phenotype | Pregnancy outcomes | |
|---|---|---|---|---|---|---|---|---|
| Our study | - | c. [1730-1G > T] | Homozygous splice site | p.? | POR |
2–3 antral follicle was observed in each ovary |
NOA | Male infertility and female subfertility |
| Yu L et al. (2022) | 35881270 |
MT1c. [1978-2A > C] MT2 c. [2680 + 3_2680 + 4delAT] |
Compound heterozygous splice site + frameshift by deletion | p.? | DOR and RPL | Only one antral follicle was observed in each ovary | - |
Three pregnancy losses in natural conception; two of which were biochemical, while embryo stopped developing at 7–8 weeks in the remaining one pregnancy |
| Xie X et al. (2022) | 35526155 | MT3: c. [1472 T > C] | Homozygous missense | p. [L491P] | - | - | NOA | NA |
| MT4: c. [1355G > A] | Homozygous missense | p. [R452Q] | - | - | NOA | NA | ||
|
MT5: c. [2562_2563delAA]; MT6 c. [4126del] |
Compound heterozygous frameshift by deletion + frameshift by deletion |
p. [E856Nfs*56]; [E1376Rfs*49] |
- | - | NOA | NA | ||
|
MT7: c. [2487_2491del]; MT8: c. [3490C > T] |
Compound heterozygous frameshift by deletion + stop-gain |
p. [ K829Nfs*23]; [Q1164*] |
- | - | NOA | NA | ||
| Saebnia N et al. (2022) | 35486194 | MT9: c. [1832-2A > T] | Homozygous splice site | p.? | - | - | NOA | NA |
| Kherraf ZE et al. (2022) | 35172124 | MT10: c. [3588 + 1G > A] | Homozygous splice site | p.? | - | - | NOA | NA |
| Tucker EJ et al. (2022) | 34707299 | MT11: c. [2410G > T] | Homozygous stop-gain | p.[E804*] | POI | Absent | - | NA |
| Tang D et al. (2021) | 34429122 | MT8: c. [3490C > T] | Homozygous stop-gain | p.[Q1164*] | - | - | NOA | NA |
| MT12: c. [3470G > A] | Homozygous missense | p.[C1157Y] | - | - | NOA | NA | ||
| Liu H et al. (2020) | 32962729 |
MT13: c. [3100G > A]; MT14: c. [1006 + 1G > T] |
Compound heterozygous missense + splice site |
p. [G1034S]; ? | POI | Absent | - | NA |
| Zhe J et al. (2019) | 31279343 | MT12: c. [3470G > A] | Heterozygous missense | p. [C1157Y] | POI | Only one antral follicle was observed in the left ovary | - | Natural pregnancy, but the embryo stopped to develop at 8 weeks |
| Wang J et al. (2014) | 24597873 |
MT15: c. [1686-1G > C]; MT16: c. [2651 T > G] |
Compound heterozygous splice site + missense |
p.?; [I884S] | POI | NA | - | NA |
|
MT17: c. [2206G > A]; MT18: c. [3929_3930delinsG] |
Compound heterozygous missense + frameshift by deletion and insertion |
p.[G736S]; [P1310Rfs*41] | POI | NA | - | NA |
POR, poor ovarian response; DOR, diminished ovarian reserve; RPL, recurrent pregnancy loss; POI, premature ovarian insufficiency; NOA, non-obstructive azoospermia; NA, not applicable
Fig. 1.
Identification of novel homozygous splicing variant in HFM1. A, B Helicase for meiosis 1 (HFM1) variant was identified in a Chinese consanguineous family with a female patient (F1 II-1) diagnosed with poor ovarian response (POR) and a male patient (F1 II-2) diagnosed with non-obstructive azoospermia (NOA). The equal signs denote infertility, and the double lines between couples represent consanguinity. Squares denote male members, and circles denote female members. Solid symbols indicate the patient with infertility, and open symbols denote unaffected members. Member indicated by black arrow was selected for whole-exome sequencing. The nucleotides affected by the variant in the Sanger sequencing results are indicated by red dashed frame. MT, mutation type; WT, wild type. C Schematic representation of the variants has been reported positions in HFM1 at the genomic and protein levels. The red arrow shows the variant in our family. The schematic gene structure is based on the Ensembl database (GRCh37, transcript ID: NM_001017975.6). The yellow solid rectangles represent exons (exon 1 to 39), and the lines in yellow represent introns. The schematic protein structure is based on the UniProt database (A2PYH4)
Minigene assay evaluating the pathogenicity of this variation
We performed a minigene assay to evaluate the effect of the c.1730-1G > T variant. The c.1730-1G > T variant produced two kinds of cDNA fragments compared with the wild type, as seen by agarose gel electrophoresis (Fig. 2B-c), indicating abnormal splicing. A previous study showed that splicing variants could lead to exon jumps or intron retention. By designing primers for exons 14 and 16 (Fig. 2A, Supplementary Table S1), we obtained two possible splicing schemes using Sanger sequencing (Fig. 2B-a, b). A possible scheme shows that donor c.1730-1G > T provided no normal donor site, leading to intron 14 and intron 15 retention (Fig. 2B-a). Another possible scheme showed that the c.1730-1G > T variant provided an abnormal donor splice site, leading to the skipping of exon 15, which subsequently caused a frameshift and premature termination (Fig. 2B-b). Our results showed that the c.1730-1G > T variant caused aberrant alternative splicing, leading to abnormal transcripts and truncated HFM1 proteins.
Outcomes of siblings during ART cycles
The female patient (F II-1) underwent five cycles of ART to retrieve eggs, and a total of four cycles of transplants were performed from 2018 to 2021 (Table 2). The first two cycles were performed in other hospitals, and each cycle transplanted one cleaved embryo without implantation. However, other details are unknown. Three other cycles were performed at the reproductive center of The First Affiliated Hospital of Anhui Medical University. In the first two of the three cycles, two poor-quality blastocysts were obtained: one blastocyst transfer resulted in a non-pregnancy, and the other was frozen in a liquid nitrogen tank. In the third cycle, the proband obtained a fifth-day high-quality blastocyst after ICSI (Supplementary Fig. 1). Though pregnancy was confirmed by transvaginal ultrasound 22 days after transplantation with self-division into two gestational sacs, the female patient experienced embryo arrest 9 weeks after pregnancy (Table 2).
Table 2.
Clinical characteristics of IVF or ICSI attempts in F1 II-1
| IVF/ICSI attempts | IVF | IVF | IVF | ICSI | ICSI |
|---|---|---|---|---|---|
| No. of oocytes retrieved | 4 | 2 | 4 | 6 | 6 |
| No. of MII oocytes | N/A | N/A | 3 | 3 | 4 |
| Total fertilization rate (%) | N/A | N/A | 3/4(75) | 3/3(100) | 2/4(50) |
| Cleavage rate (%) | N/A | N/A | 3/3(100) | 3/3(100) | 2/2(100) |
| Good quality embryo number, day 5 | N/A | N/A | 0 | 0 | 1 |
| Blastocyst formation rate (%) | N/A | N/A | 1/3(33) | 1/3(33) | 1/2(50) |
| No. of embryos transferred | 1 | 1 | 1 | 0 | 1 |
| Clinical pregnancy | No | No | No | No | Yes |
| Live birth | No | No | No | No | No |
The first two assisted reproductive technology processes were carried out in other hospitals, and the specific data are unknown
N/A, not applicable; MII, metaphase II
IVF fertility rate = (number of fertilized egg/total number of eggs) × 100%
ICSI fertility rate = (number of fertilized egg/number of MII eggs) × 100%
Cleavage rate = (number of fertilized and cleaved embryos/(number of fertilized egg) × 100%
Blastocyst formation rate = (number of blastocysts formed/number of blastocysts cultured) × 100%
For the male patient (F II-2), sperm was still not detectable in his semen after a long period of drug treatment. Moreover, we failed to find any sperm in the testicular tissue by testis biopsy (Supplementary Fig. 2). Finally, he fathered a daughter through sperm donation.
The embryos obtained during ART in female patient carry chromosomal abnormalities leading to RIF
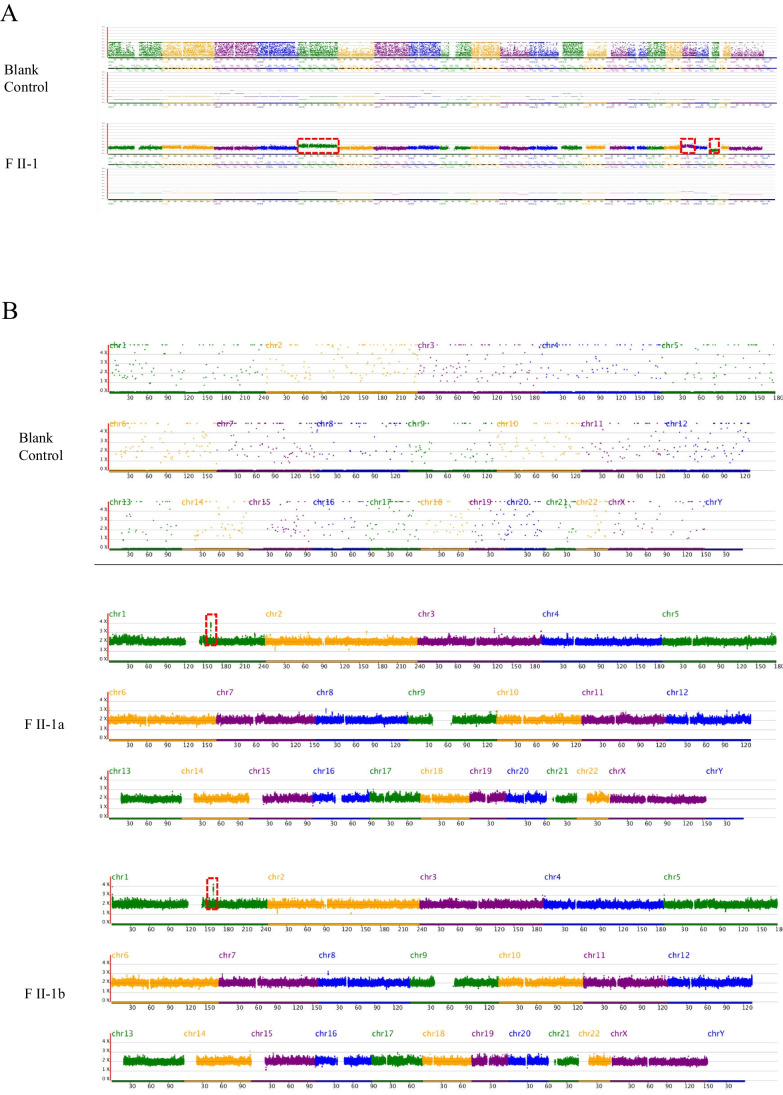
The female patient who obtained good-quality and poor-quality blastocyst in ART cycles and underwent multiple transfers did not become pregnant, diagnosing RIF. Considering that the outcome of previous ART sessions was not good and successful pregnancy has never been reported in female patients with biallelic variants of HFM1, we suggested that the patient perform CNV-seq on the frozen blastocyst before good-quality blastocyst transplantation. The poor-quality blastocyst was found to have one missing chromosome 21 and partial fragment duplication on chromosomes 5 and 19 (Fig. 3A, Supplementary Table S3). However, based on our findings regarding chromosome abnormalities, the proband insists on the direct transplantation of high-quality blastocysts after adequate clinical consultation. To explore the cause of embryo arrest after high-quality embryo transfer, we tested the abortion tissues and found that they were euploid but carrying 300 kb chromosomal microduplications (Fig. 3B, Supplementary Table S3). Moreover, a genotype alignment method was designed to explore the origin of abnormal chromosomes. According to the statistical results, the matching degree of chromosome 21 with the father and mother was 99.28% and 40.69%, respectively (Fig. 4B, Supplementary Table S4). Therefore, the remaining chromosome 21 came from the father. Meanwhile, microduplications on chromosomes 5 and 19 were confirmed to be inherited from the mother, which reinforces the maternal origin of the abnormal chromosomes (Fig. 4C).
Fig. 3.
Copy number variation sequencing (CNV-seq) results of the poor-quality blastocyst and abortion tissues. A The poor-quality blastocyst was monosomy 21 and partial fragment duplication on chromosomes 5 and 19 (180.68 Mb and 58.86 Mb, respectively. Red dashed frame). B The CNV-seq results-100 kb of spontaneous aborted products from the two embryos that divided from good-quality blastocyst were normal karyotypes and had partial fragment duplication on chromosomes 1 (red dashed frame)
Discussion
In the present study, we found a novel homozygous splicing variant in HFM1 (NM_001017975.6: c.1730-1G > T) in two siblings with infertility from a consanguineous family. The pathogenicity of this variant was confirmed to cause abnormal alternative splicing in vitro, including exon skipping and intron retention. The chromosomal abnormalities underlying RIF in the female patient harboring biallelic deleterious HFM1 variants have sparked concern over the potential opportunities and risks of pregnancy for the first time (Table 3). In all, our findings extend the phenotypic and mutational spectrum of HFM1.
In previous reports, Hfm1-/- adult mice are viable and show no abnormalities in systems other than the reproductive system [17]. Hfm1-/- male mice showed reduced testicular volume, hyperplasia of interstitial cells, and blocked spermatogenesis during meiosis I division [17]. Hfm1-/- female mice showed reduced ovarian volume, number of follicles, and corpora lutea [17]. Both male and female mice with biallelic Hfm1 deficiency are sterile because of gametogenic disorders [17]. Intriguingly, Wang et al. reported that Hfm1-/- female mice with knockout are subfertile [22]. We speculated that this phenomenon might be related to the strategy of constructing knockout mice, resulting in different functions of the residual HFM1 protein. These two different knockout methods lead to sterility and subfertility, corresponding to different variant sites in humans that lead to different degrees of egg reserves. Similarly, ovulation can occur in Hfm1-/- female mice in both reports, which remained the possibility for women to conceive naturally or with ART.
In the present study, despite showing decreased ovarian function, the female patient did not meet the criteria for POI but POR according to the POSEIDON criterion at the age of 31, inconsistent with previous reports [21]. Moreover, a few eggs (between 2 and 6) and corresponding blastocysts of various grades can be obtained. Intriguingly, during the most recent ART cycle, a clinically high-quality embryo was obtained. However, the high-quality embryo turned out to carry small fragments of copy number variations of unknown significance, which may be the reason of RIF in the female patient with HFM1 variation in ART cycles. Besides, the male patient sought artificial insemination with donor semen to achieve pregnancy due to the complete failure of sperm acquisition. The gender difference may be related to the different generation processes of male and female germ cells. Meiosis of female germ cells is initiated at the fetal stage, with maturation through meiotic divisions occurring at timed intervals in adults [7]. In contrast, meiosis in male germ cells is a continuous process initiated throughout most of adult life [7]. Here, the female patient was able to complete meiosis; the gametes produced by meiosis are abnormal according to the results of embryo CNV-seq and verification of the origin of abnormal chromosomes. Therefore, we believe that females with biallelic variants in HFM1 may have a dual risk of subfertility and RIF.
Previous reports have shown meiosis chromosome separation errors often occur in human females, especially during their first meiosis [23]. Another study showed that meiosis abnormalities in germ cells of Hfm1-/- mice mainly occurred during the first meiosis and that female Hfm1-/- mice had abnormal oocyte spindle morphology [22]. Accordingly, we demonstrated the paternal origin of the remaining chromosomes in the aneuploid embryo of the female patient. Therefore, we speculated that the patient’s biallelic variant in HFM1 resulted in abnormal spindle morphology, increasing the risk of erroneous division in the first meiotic division of the oocyte, resulting in an egg missing chromosome 21 (Fig. 5). Meanwhile, the microduplications of maternal origin on chromosomes 5 and 19 are likely resulted from a strand invasion error during the first meiotic division, leading to the incomplete exchange of alleles on homologous chromosomes (Fig. 5). For the abortion tissues, high-resolution CNV-seq revealed a euploid with 300 kb genome microduplications of undetermined clinical significance. However, due to technical limitations, the source of 300 kb microduplications cannot be determined, and other maternal factors leading to embryo arrest may have been excluded before embryo implantation. Therefore, 300 kb microduplication is recognized as the cause of embryo arrest.
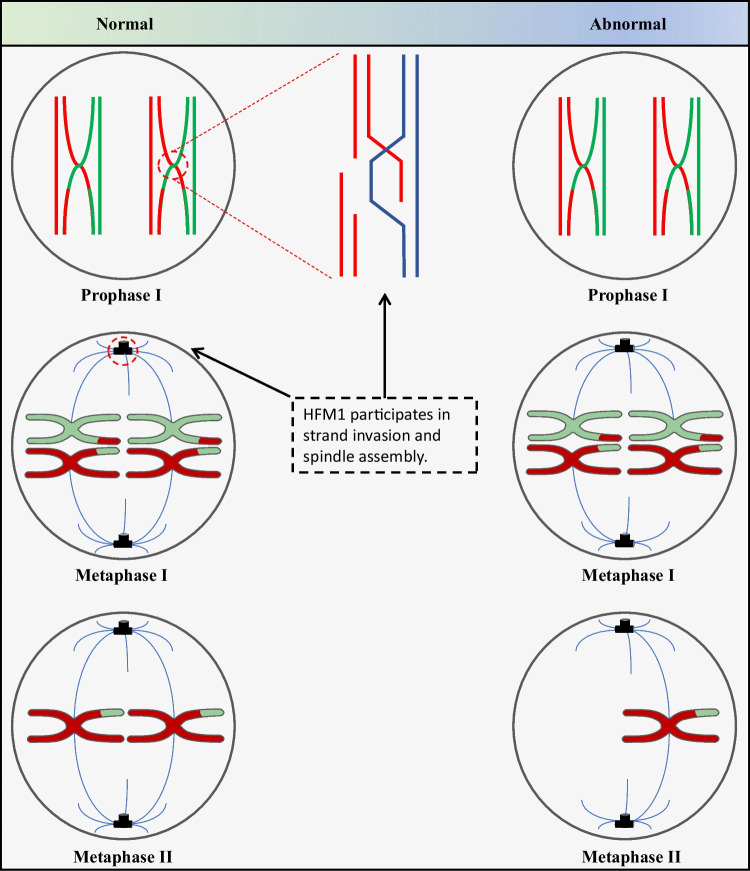
Fig. 5.
Meiosis process analysis. Homologous chromosomes separate during the first meiosis and sister chromatids separate during the second meiosis (left). HFM1 participates crossover formation and complete synapsis of homologous chromosomes of the first meiosis and participates spindle assemble. The formation of poor-quality blastocyst missing one chromosome 21 may be due to the ovum missing one chromosome 21 from patient carrying HFM1 variation. This is due to abnormality in the spindle, thus causing a pair of homologous chromosomes 21 to shift to the same pole during the first division of meiosis. Primary oocytes lacking chromosome 21 undergo second meiosis to form an ovum missing chromosome 21 (right)
In conclusion, comparison between two siblings with POR and NOA from a consanguineous family highlighted the difference in HFM1 variants on reproductive injury in males and females. Moreover, this is the first study to analyze the possible causes of RIF in female patients with novel homozygous pathogenic variants of HFM1 from an embryonic perspective, focusing on chromosomal abnormalities. Hence, our findings illustrate the possibility of pregnancy in female patients with HFM1 variants; however, preimplantation genetic diagnosis is needed to reduce the risk of RIF.
Our study has some limitations, such as the inability to perform immunofluorescence to verify oocyte spindle abnormalities in the female patient due to the limited number of eggs, and further studies in a larger population are needed.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank the patients and their families for participating in this study.
Author contribution
Y.C., X.H., and Z.Z. designed the experiments. F.T. and Y.G. performed the experiment and wrote the manuscript. K.L., D.T., Z.J., and J.F. analyzed the statistical data. Y.H. assisted embryo biopsy and vitrified thawing. Y.X. and C.W. collated the data of patients. H.C., M.L., and H.W. reviewed the data statistic. Z.W. and P.Z. revised the manuscript.
Funding
This work was supported by the National Key R&D Program of China (2021YFC2700901), the National Natural Science Foundation of China (grant numbers 82171607, 81901541), the University Synergy Innovation Program of Anhui Province (grant number GXXT-2021–071), the Research Fund of Anhui Institute of Major Science and Technology Projects (grant number 202003a07020012), the Institute Fund of Chinese Academy of Medical Sciences (grant number 2019PT310002), and the Research Fund of Anhui Institute of translational medicine (ZHYX2020A001).
Data availability
Data underlying this article will be shared with the corresponding authors upon reasonable request.
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Fei Tang, Yang Gao and KuoKuo Li contributed equally to this work.
Contributor Information
Zhiguo Zhang, Email: zhangzhiguo@ahmu.edu.cn.
Xiaojin He, Email: hxj0117@126.com.
Yunxia Cao, Email: caoyunxia5972@ahmu.edu.cn.
References
- 1.Carson SA, Kallen AN. Diagnosis and management of infertility: a review. JAMA. 2021;326(1):65–76. doi: 10.1001/jama.2021.4788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coughlan C, Ledger W, Wang Q, Liu F, Demirol A, et al. Recurrent implantation failure: definition and management. Reprod Biomed Online. 2014;28(1):14–38. doi: 10.1016/j.rbmo.2013.08.011. [DOI] [PubMed] [Google Scholar]
- 3.Yatsenko SA, Rajkovic A. Genetics of human female infertilitydagger. Biol Reprod. 2019;101(3):549–566. doi: 10.1093/biolre/ioz084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiao X, Ke H, Qin Y, Chen ZJ. Molecular genetics of premature ovarian insufficiency. Trends Endocrinol Metab. 2018;29(11):795–807. doi: 10.1016/j.tem.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Oud MS, Volozonoka L, Smits RM, Vissers L, Ramos L, et al. A systematic review and standardized clinical validity assessment of male infertility genes. Hum Reprod. 2019;34(5):932–941. doi: 10.1093/humrep/dez022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zickler D, Kleckner N. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol. 2015;7(6):a016626. doi: 10.1101/cshperspect.a016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bolcun-Filas E, Handel MA. Meiosis: the chromosomal foundation of reproduction. Biol Reprod. 2018;99(1):112–126. doi: 10.1093/biolre/ioy021. [DOI] [PubMed] [Google Scholar]
- 8.Fan S, Jiao Y, Khan R, Jiang X, Javed AR, et al. Homozygous mutations in C14orf39/SIX6OS1 cause non-obstructive azoospermia and premature ovarian insufficiency in humans. Am J Hum Genet. 2021;108(2):324–336. doi: 10.1016/j.ajhg.2021.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akbari A, Padidar K, Salehi N, Mashayekhi M, Almadani N, et al. Rare missense variant in MSH4 associated with primary gonadal failure in both 46, XX and 46. XY individuals Hum Reprod. 2021;36(4):1134–1145. doi: 10.1093/humrep/deaa362. [DOI] [PubMed] [Google Scholar]
- 10.Jaillard S, McElreavy K, Robevska G, Akloul L, Ghieh F, et al. STAG3 homozygous missense variant causes primary ovarian insufficiency and male non-obstructive azoospermia. Mol Hum Reprod. 2020;26(9):665–677. doi: 10.1093/molehr/gaaa050. [DOI] [PubMed] [Google Scholar]
- 11.Yao C, Hou D, Ji Z, Pang D, Li P, et al. Bi-allelic SPATA22 variants cause premature ovarian insufficiency and nonobstructive azoospermia due to meiotic arrest. Clin Genet. 2022;101(5–6):507–516. doi: 10.1111/cge.14129. [DOI] [PubMed] [Google Scholar]
- 12.He WB, Tu CF, Liu Q, Meng LL, Yuan SM, et al. DMC1 mutation that causes human non-obstructive azoospermia and premature ovarian insufficiency identified by whole-exome sequencing. J Med Genet. 2018;55(3):198–204. doi: 10.1136/jmedgenet-2017-104992. [DOI] [PubMed] [Google Scholar]
- 13.Wu H, Zhang X, Hua R, Li Y, Cheng L, et al. Homozygous missense mutation in CCDC155 disrupts the transmembrane distribution of CCDC155 and SUN1, resulting in non-obstructive azoospermia and premature ovarian insufficiency in humans. Hum Genet. 2022;141:1795–1809. doi: 10.1007/s00439-022-02459-4. [DOI] [PubMed] [Google Scholar]
- 14.Tang D, Lv M, Gao Y, Cheng H, Li K, et al. Novel variants in helicase for meiosis 1 lead to male infertility due to non-obstructive azoospermia. Reprod Biol Endocrinol. 2021;19(1):129. doi: 10.1186/s12958-021-00815-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang J, Zhang W, Jiang H, Wu B-L. Mutations in HFM1 in recessive. N Engl J Med. 2014;370(10):972–974. doi: 10.1056/NEJMc1310150. [DOI] [PubMed] [Google Scholar]
- 16.Xie X, Murtaza G, Li Y, Zhou J, Ye J, et al. Biallelic HFM1 variants cause non-obstructive azoospermia with meiotic arrest in humans by impairing crossover formation to varying degrees. Hum Reprod. 2022;37:1664–1677. doi: 10.1093/humrep/deac092. [DOI] [PubMed] [Google Scholar]
- 17.Guiraldelli MF, Eyster C, Wilkerson JL, Dresser ME, Pezza RJ. Mouse HFM1/Mer3 is required for crossover formation and complete synapsis of homologous chromosomes during meiosis. PLoS Genet. 2013;9(3):e1003383. doi: 10.1371/journal.pgen.1003383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Magi A, Tattini L, Pippucci T, Torricelli F, Benelli M. Read count approach for DNA copy number variants detection. Bioinformatics. 2012;28(4):470–478. doi: 10.1093/bioinformatics/btr707. [DOI] [PubMed] [Google Scholar]
- 20.Szatkiewicz JP, Wang W, Sullivan PF, Wang W, Sun W. Improving detection of copy-number variation by simultaneous bias correction and read-depth segmentation. Nucleic Acids Res. 2013;41(3):1519–1532. doi: 10.1093/nar/gks1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alviggi C, Andersen CY, Buehler K, Conforti A, De Placido G, et al. A new more detailed stratification of low responders to ovarian stimulation: from a poor ovarian response to a low prognosis concept. Fertil Steril. 2016;105(6):1452–1453. doi: 10.1016/j.fertnstert.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 22.Wang H, Zhong C, Yang R, Yin Y, Tan R, et al. Hfm1 participates in Golgi-associated spindle assembly and division in mouse oocyte meiosis. Cell Death Dis. 2020;11(6):490. doi: 10.1038/s41419-020-2697-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hassold T, Hunt P. To err (meiotically) is human: the genesis of human aneuploidy. Nat Rev Genet. 2001;2(4):280–291. doi: 10.1038/35066065. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data underlying this article will be shared with the corresponding authors upon reasonable request.