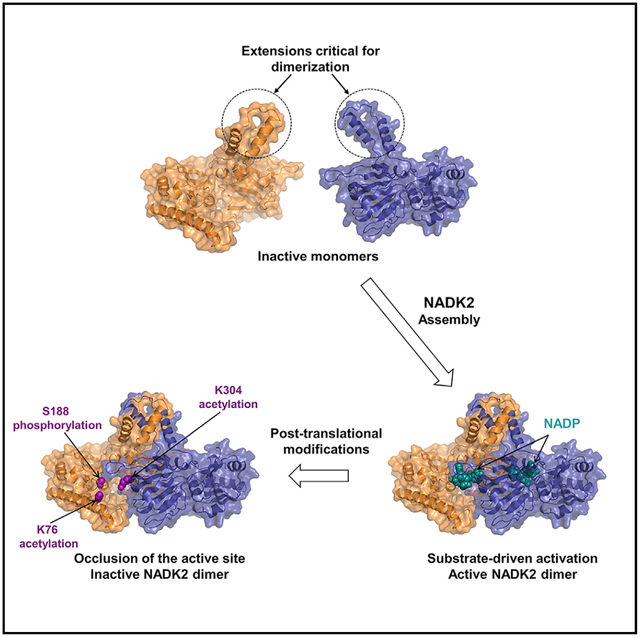
SUMMARY
NAD+ kinases (NADKs) are metabolite kinases that phosphorylate NAD+ molecules to make NADP+, a limiting substrate for the generation of reducing power NADPH. NADK2 sustains mitochondrial NADPH production that enables proline biosynthesis and antioxidant defense. However, its molecular architecture and mechanistic regulation remain undescribed. Here, we report the crystal structure of human NADK2, revealing a substrate-driven mode of activation. We find that NADK2 presents an unexpected dimeric organization instead of the typical tetrameric assemblage observed for other NADKs. A specific extended segment (aa 325–365) is crucial for NADK2 dimerization and activity. Moreover, we characterize numerous acetylation events, including those on Lys76 and Lys304, which reside near the active site and inhibit NADK2 activity without disrupting dimerization, thereby reducing mitochondrial NADP(H) production, proline synthesis, and cell growth. These findings reveal important molecular insight into the structure and regulation of a vital enzyme in mitochondrial NADPH and proline metabolism.
Graphical abstract
In brief
Mary and Soflaee et al. report a high-resolution structure of human mitochondrial NADK2 in complex with NADP+, revealing a substrate-assisted mode of catalysis. NADK2 displays a dimeric organization, with its substrate located at the dimer interface. The authors identify key acetylation and phosphorylation events critical for NADK2 function.
INTRODUCTION
Nicotinamide adenine dinucleotide phosphate (NADPH) is an essential reducing equivalent that provides a reservoir of electrons for anabolic reactions and defense against oxidative stress (DeBerardinis and Chandel, 2016; Fan et al., 2014; Lewis et al., 2014; Pollak et al., 2007). NADPH abundance is determined by the activity of NAD+ kinases (NADKs), enzymes that catalyze the phosphorylation of NAD+ to generate NADP+, the rate-limiting substrate for NADPH production (Pollak et al., 2007). Human cells encode two NADKs, cytosolic NADK and mitochondrial NADK2, that produce compartment-specific reducing power.
While cytosolic NADK is subject to phosphorylation and activation by Akt and PKC kinases to boost the NADP(H) production and support growth (Hoxhaj et al., 2019; Rabani et al., 2020; Schild et al., 2021), the regulation of mitochondrial NADK2 remains obscure.
NADK2 activity sustains the synthesis of mitochondrial NADPH required for counteracting reactive oxygen species (ROS) (Ohashi et al., 2012; Zhang et al., 2018). Furthermore, two recent studies revealed an essential role for NADK2 in enabling proline biosynthesis in proliferating cells (Tran et al., 2021; Zhu et al., 2021). Indeed, NADK2-deficient cells become proline auxotroph, therefore requiring proline supplementation to maintain cell growth and proliferation (Tran et al., 2021; Zhu et al., 2021). Mutations in NADK2 were found in patients exhibiting neurological and development symptoms, including encephalopathy, microcephaly, epilepsy, and in some cases, unremitting disease (Houten et al., 2014; Pomerantz et al., 2018), demonstrating the importance of this enzyme in human health.
NADKs belong to a small and peculiar superfamily of kinases that comprises the distantly related sphingosine kinases (SKs), soluble diacylglycerol kinases (DGKs), as well as the even more divergent 6-phosphofructokinases (PFKs), all of which share a “ggDgt” catalytic motif in their N-terminal α/β domain (Labesse et al., 2002). The C-terminal regions of NADKs, SKs, and DGKs contain a particular all-β domain—composed almost entirely of β sheets—that is involved in multimerization and ligand binding (Labesse et al., 2002). Conversely, the C terminus of PFKs possesses an α/β domain that is also involved in multimerization and ligand binding. While the catalytic mechanism of PFKs has been precisely described (Shirakihara and Evans, 1988), those of the other members of this superfamily remain still unknown.
Crystal structures of NADKs from various organisms, including bacteria (Mori et al., 2005b; Poncet-Montange et al., 2007), yeast (Ando et al., 2011), and human (deposited into protein data bank [PDB]: 3PFN) indicate an overall structure comprised of an N-terminal and a C-terminal domain. NADKs are homo-oligomers (tetrameric in most species) whose subunits interactions participate in substrate binding (Lerner et al., 2001; Pollak et al., 2007). However, there is currently no structural information on human NADK2 nor any molecular insight into the mechanisms that regulate its catalytic activity.
To gain better insights into the regulation and function of NADK2, we sought to define its structural and biochemical properties. Here, we report a high-resolution structure of human NADK2 bound to NADP+ and a catalytic dicationic metal (Mg2+, Ca2+, or Fe3+) that shows a new substrate-assisted catalysis mode. NADK2 appears to exist as a dimer that possesses specific and evolutionary conserved extensions critical for its dimerization and cellular functions. Finally, we map post-translational modifications (PTMs) of NADK2 in human cells and report key phosphorylation and acetylation events that are critical for NADK2 catalytic efficiency and necessary for mitochondrial NADP(H) production, proline synthesis, and cell proliferation.
RESULTS
Crystal structure of NADK2
Here, we reveal the crystal structure of the human mitochondrial NADK2 at 2.3 Å. We used a bacterial expression system to produce human NADK2 (aa 61–442 with an N terminus His-tag), only lacking the mitochondrial localization signal. We obtained crystal structures of NADK2 in a ligand-free form (apo-form) (Figure S1A) and in a complex with NADP+ (Figures 1A and S1B; Table 1). Both forms were highly similar, indicating no major structural changes upon ligand binding.
Figure 1. Crystal structure of NADK2.
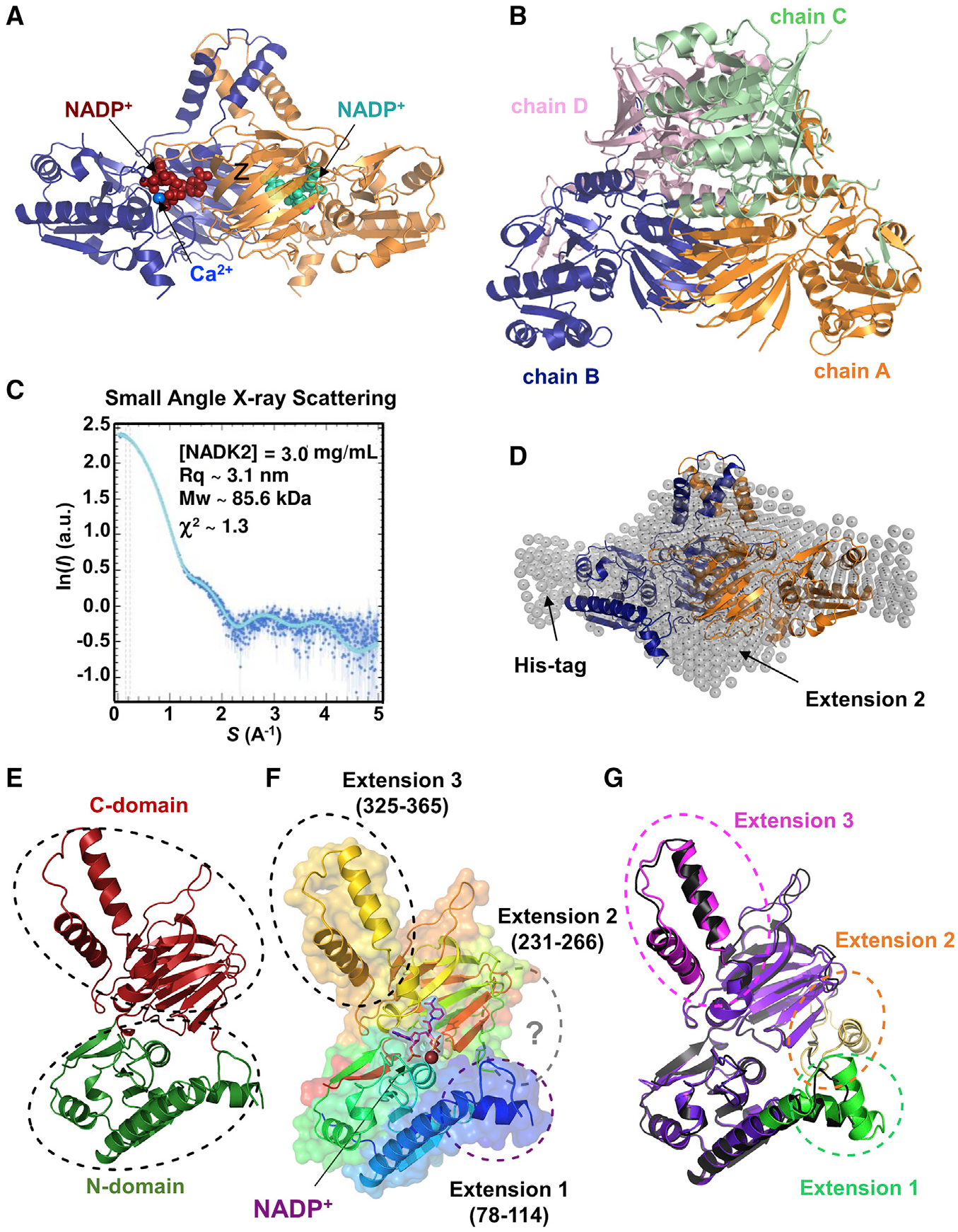
(A) Crystal structure of the NADK2 in complex with NADP+ at 2.3 Å. The dimeric structure of NADK2 as deduced from crystal symmetry is shown with chain A in blue ribbon and its crystal symmetric unit, chain B, in orange. NADP+ (red or cyan in different monomers) and bound calcium (blue sphere) are shown.
(B) Crystal structure of the tetrameric cytosolic NADK. The crystal structure of cytosolic human NADK deposited in PDB (PDB: 3PFN) in the same orientation as the crystal structure of NADK2 (shown in A). The four subunits (chains A–D) of cytosolic NADK are shown as ribbons and colored by chains (A, orange; B, blue; C, light green; D, light pink).
(C) SAXS profile of purified NADK2. SAXS experimental data were fitted to a theoretical SAXS curve that was computed from the NADK2 crystal structure (dimeric form) completed with the Histag and the 2nd extension. χ2 ~ 1.3 was calculated with Crysol. Guinier plot of the SAXS curve indicated a radius of gyration (Rq) ~ 3.1 nm and a molecular mass (MW) of ~85.6 kDa.
(D) Crystal structure of the NADK2 fitted in the SAXS envelope. The dimeric crystal structure (same coloring schemes as in A) was superimposed onto the SAXS envelope. Arrows indicate the positions of the polyhistidine (His) tag and the second NADK2 extension (aa 231–266), which was not observed in the crystal structure but deduced from the AlphaFold structure.
(E) Crystal structure of the NADK2 monomer indicating the N-terminal domain (green) and the C-terminal domain (red).
(F) Monomeric NADK2 shown in ribbon was colored in rainbow colors depicting specific NADK2 extensions (1–3). Extension 2 (aa 231–266) is not visible in the electron density, but the deduced model from AlphaFold is shown in gray. NADP+ is shown in violet stick, with its electron density in violet surface, and calcium ion is shown as a blue sphere.
(G) Comparison of the crystal structure and the AlphaFold model of NADK2. The monomeric NADK2 crystal structure is shown in black ribbon. The AlphaFold2 model is colored in violet ribbon, and the three extensions are indicated in green, brown, and pink.
Table 1.
Data collection and refinement statistics
| NADK2-NADP+-Ca2+ | NADK2-NADP+-Mg2+ | NADK2-NADP+-Fe3+ | NADK2+-Apo | |
|---|---|---|---|---|
| (PDB accession code) | (7R4M) | (7R4K) | (7R4L) | (7R4J) |
| Date collection | ||||
| Space group | P 43 21 2 | P 43 21 2 | P 43 21 2 | P 43 21 2 |
| Cell dimensions | ||||
| a, b, c (Å) | 68.47, 68.47, 223.42 | 67.58, 67.58, 221.81 | 67.58, 67.58, 221.81 | 68.47, 68.47, 221.62 |
| α, β, γ (°) | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 | 90.0, 90.0, 90.0 |
| Resolution range (Å) | 37.42–2.29 | 37.09–3.32 | 47.79–2.62 | 44.37–3.32 |
| R-mergea | 9.1 (104.3) | 27.6 (120.2) | 11.7 (92.7) | 15.6 (101.2) |
| R-pima | 5.5 (65.5) | 15.3 (67.1) | 6.5 (52.8) | 11.4 (73.5) |
| CC1/2a | 99.3 (38.6) | 97.0 (35.6) | 99.5 (53.0) | 98.9 (40.4) |
| Mean IσIa | 8.5 (0.9) | 3.9 (0.9) | 7.1 (0.9) | 5.9 (1.0) |
| Completeness (%)a | 98.0 (97.6) | 89.6 (90.2) | 97.5 (95.8) | 94.9 (96.1) |
| Redundancya | 3.2 (3.1) | 3.5 (3.2) | 3.4 (3.4) | 2.5 (2.5) |
| Refinement | ||||
| Resolution (Å) | 2.29 | 3.32 | 2.62 | 3.32 |
| No. reflections | 44,612 | 13,006 | 28,795 | 14,104 |
| Rwork/Rfree | 19.6/23.8 | 21.4/28.9 | 20.5/25.8 | 23.2/29.1 |
| No. atoms | ||||
| Protein | 2,668 | 2,578 | 2,710 | 2,638 |
| Ligand/ion | 49 | 49 | 49 | 2 |
| Water | 152 | 78 | 127 | 21 |
| B-factors | ||||
| Protein | 62.0 | 74.4 | 74.2 | 112.9 |
| Ligand/ion | 44.5 | 54.0 | 53.7 | 123.9 |
| Water | 56.1 | 58.3 | 63.9 | 88.4 |
| RMS deviations | ||||
| Bond lengths (Å) | 0.008 | 0.010 | 0.007 | 0.011 |
| Bond angles (°) | 0.99 | 1.34 | 0.97 | 1.42 |
Values in parentheses are for highest-resolution shell.
Notably, NADK2 formed a dimer with a perfect 2-fold symmetry, in contrast to the tetrameric structures observed in cytosolic NADK crystals (Figure 1B). The existence of NADK2 dimers was confirmed by both small-angle X-ray scattering (SAXS) (Figures 1C and 1D) and size-exclusion chromatography coupled to multi-angle light scattering (SEC-MALS) (Figures S1C and S1D), indicating an overall molecular weight ~ 86 kDa for the NADK2 dimer (monomeric NADK2 lacking aa 1–60 is ~44.6 kDa).
Importantly, the SAXS-derived shape envelope fit well with the crystal structure data, with a low discrepancy score χ2 of 1.3 (Figure 1C). Moreover, SAXS measurements show a stable NADK2 dimer (and no tetramerization) on a wide range of protein concentrations (χ2 ~ 2.1 when data merging was performed with 3, 6, and 12 mg/mL NADK2) (Figure S1E), in agreement with the crystal structure indicating a large inter-subunit interface of 7,520 Å2 assessed with PISA program (Figure 1A; Krissinel and Henrick, 2007).
The crystal structure of the core of the protein revealed a two-domain architecture, comprising an N-terminal (residues 66–219 and 436–442) and a C-terminal domain (residues 220–435) that are connected by a hinge comprising of residues 219–220 and 434–435 (Figure 1E). The overall NADK2 domain organization resembles that of NADKs, despite a significant sequence divergence (~20% sequence identity) (Figure S2; Ohashi et al., 2012).
We found that NADK2 exhibits two prominent helical extensions at aa 78–114 and aa 325–365 that are not present in the cytosolic NADK. The latter extension forms a long hairpin composed of two helical segments (327–340 and 350–364) that protrude away from a monomer to stack against its equivalent counterpart from the second monomer (Figures 1F, 1G, S1F, and S1G). As such, this segment appears to prevent a side-by-side dimerization of NADK2 dimers into a tetrameric assemblage, a characteristic of other NADKs (Figures 1B and S1G). The biological purpose, if any indeed exists, of such a tetramer-preventing structure remains to be determined.
Our structure is in perfect agreement with the AlphaFold prediction (Jumper et al., 2021), which also indicates the presence of another extended segment (residues 231–266) that was not fully resolved in our crystals (Figures 1G and S1F). Of note, NADK2 lacks the very C-terminal helix pointing into the active site that is conserved in archaeal, bacterial, and human cytosolic NADKs (Poncet-Montange et al., 2007). Hence, besides a rather conserved core structure, NADK2 possesses unique structural features that could be related to its specific regulations.
Substrate-driven catalysis mode of human NADK2
Mitochondrial NADK2 catalyzes the phosphorylation of NAD+ in the presence of Mg2+-ATP to generate NADP+, the precursor of NADPH (Ohashi et al., 2012). The NADP+-binding site sits at the interface of the NADK2 dimer, specifically in a groove formed between the N-terminal and C-terminal domains of one monomer and the C-terminal domain of the second monomer (Figures 1A and S1B). The adenine moiety of NADP+ is sandwiched between the catalytic aspartate D161, alanine A318, and asparagine N276, residues that are well conserved in NADKs (Figures 2A, 2B, and S2). A318 also belongs to the well-conserved NADK motif: TGSxAw motif (residues 314–319 on NADK2) (Figure S2). This motif provides an aromatic side chain (tryptophan W319 on NADK2 or a tyrosine on other NADKs) for stacking the nicotinamide moiety (Figure 2A). The latter forms hydrogen bonds with the side chains of asparagine N322 and serine S306′ (from a second monomer) and with the backbone carbonyl of glutamate E379′. This second monomer also contributes two hydrophobic side chains from the alanine A288′ and the isoleucine I381′, that help to tightly hold the nicotinamide moiety of NADP+. The ribose attached to the nicotinamide is hydrogenbonded to glutamate N277 (Figure 2A). Hence, the binding mode of the ligand in NADK2 strongly resembles the particular conformation it adopts in other NADKs (Figure 2B) (e.g., yeast mitochondrial NADK, PDB: 3AFO (Ando et al., 2011).
Figure 2. Substrate-driven catalysis mode of human NADK2.
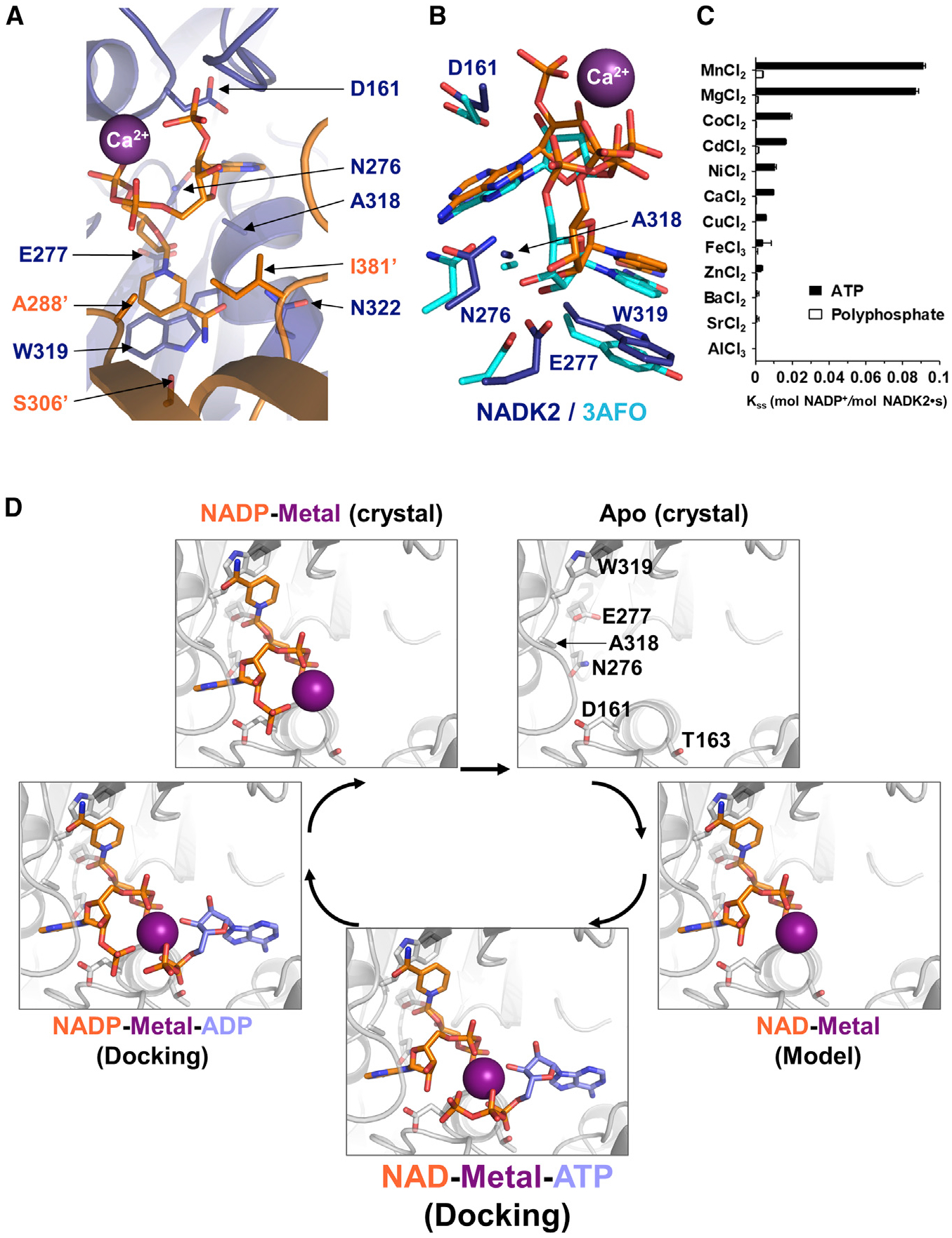
(A) Detailed view of the active site of crystalized NADK2 in complex with NADP+ (sticks) and calcium (Ca2+, violet sphere). The polypeptide chain is shown in ribbon, with one monomer in blue and the other one in orange. Side-chain interactions with NADP+ are shown in stick and colored with their corresponding monomeric color. Residues are numbered following the full-length sequence. The prime (′) symbol indicates residues belonging to the symmetry mate.
(B) Comparison of the active site of NADK2 and its yeast orthologue POS5 (crystal structure PDB: 3AFO), both in complex with NADP+. Side chains are shown in stick (dark blue, NADK2; cyan, 3AFO). For clarity, only the NADK2 residues are numbered.
(C) Enzymatic activity of human recombinant His-NADK2 in the presence of 6 mM of the indicated dicationic metals, 1 mM NAD+ and 5 mM ATP or polyphosphate as a phosphodonor.
(D) Enzymatic cycle of NADK2 as deduced from the crystal structures of the apo-form or the NADP+ bound form. The NAD+ complex was deduced from the NADP+-bound form by removing the 2′-phosphate group. The ADP or ATP molecules (colored in blue) were docked into the active site of NADP+ and NAD+-bound forms (colored in orange) using protein-ligand ant system (PLANTS) (Korb et al., 2009). Multiple metals (Mg2+, Ca2+, and Mn2+) were used during docking (see STAR Methods), and one representative metal is shown as a violet sphere. Side chains of catalytic residues (D161 and T163) within the conserved motif “ggDgt” and four other amino acids important for ligand binding (N276, E277, A318, and W319) are shown as gray sticks. For clarity, the amino acid labeling is shown for the apo-form.
Notably, our NADK2-NADP+ co-crystal structures allowed us to obtain the first reported visualization of the catalytic metal’s position in the active site of an NADK. The various crystal structures of NADK2 bound to NADP+ showed the presence of Mg2+, Ca2+, or Fe3+ metal ions (Figures 2A, 2B, and S1H–S1J). Because Mg2+ and the water molecule have the same number of electrons, to identify this metal in the X-ray diffraction (peak in the density at 4.0 σ), we compared it with the larger density of calcium (8.0 σ) or iron (8.3 σ) (Figures S1H–S1J). Specifically, the diphosphate group of NADP+ appeared to chelate each metal in the catalytic domain in the same manner (distances O–Ca2+ of 2.33 and 2.38 Å). This agrees with the rather poor specificity of all NADKs toward the nature of the catalytic metal. Profiling a variety of metal ions showed higher NADK2 activity in the presence of MnCl2 and MgCl2 and relatively lower activity in the presence of CoCl2, CdCl2, NiCl2, and CaCl2 when ATP was used as a phosphodonor rather than polyphosphates (Figure 2C).
Interestingly, the metals show no direct contact with NADK2 (Figures 2A and 2B). Instead, the ternary complexes, NADK2-NADP+-metals, show that the reaction product, NADP+, and certainly also the substrate itself, NAD+, chelate the catalytic metal instead of the catalytic aspartate (D161). The latter is buried (as in other NADKs) and only interacts with the 2′-hydroxyl group of NAD+ (Figure 2D). These data shed light on a previously proposed model of substrate-driven catalysis in which the catalytic aspartate does not directly accompany the transferred phosphate by chelating the metal, but instead activates the phospho-acceptor, here the 2′-hydroxyl group of NAD+ (Poncet-Montange et al., 2007). This activation mode is in stark contrast to other ATP-dependent kinases, including the distantly related PFKs, which harbors a similar “ggDgt” catalytic motif. Indeed, in most kinases the catalytic aspartate is involved in the chelation of the metal cation to activate the ATP phosphodonor (e.g., E. coli PFK) (Shirakihara and Evans, 1988). This substrate-driven catalysis mechanism allows for specific phosphorylation of NAD+ over other nucleotide phosphates (e.g., ADP) (Figure 2D). Thus, our high-resolution structure of NADK2 in complex with NADP+ and the metal dications enabled us to determine the structural basis for the NAD+-specificities of NADKs.
Extension 3 of NADK2 (aa 325–365) is required for its dimerization and function
To test whether NADK2 can exist as a dimer in cells, we expressed NADK2 in human embryonic kidney 293E (HEK293E) cells and subjected cells to chemical crosslinking with glutaraldehyde or disuccinimidyl suberate (DSS). Indeed, NADK2 was observed as a dimer in the presence of both crosslinkers (Figures 3A and 3B). These experiments confirmed that the NADK2 dimer observed in the crystal structure is not a crystallographic artifact and that NADK2 indeed exists as a dimer in cultured cells.
Figure 3. Extension 3 (aa 325–365) of NADK2 mediates its dimerization and is required for its catalytic activity.
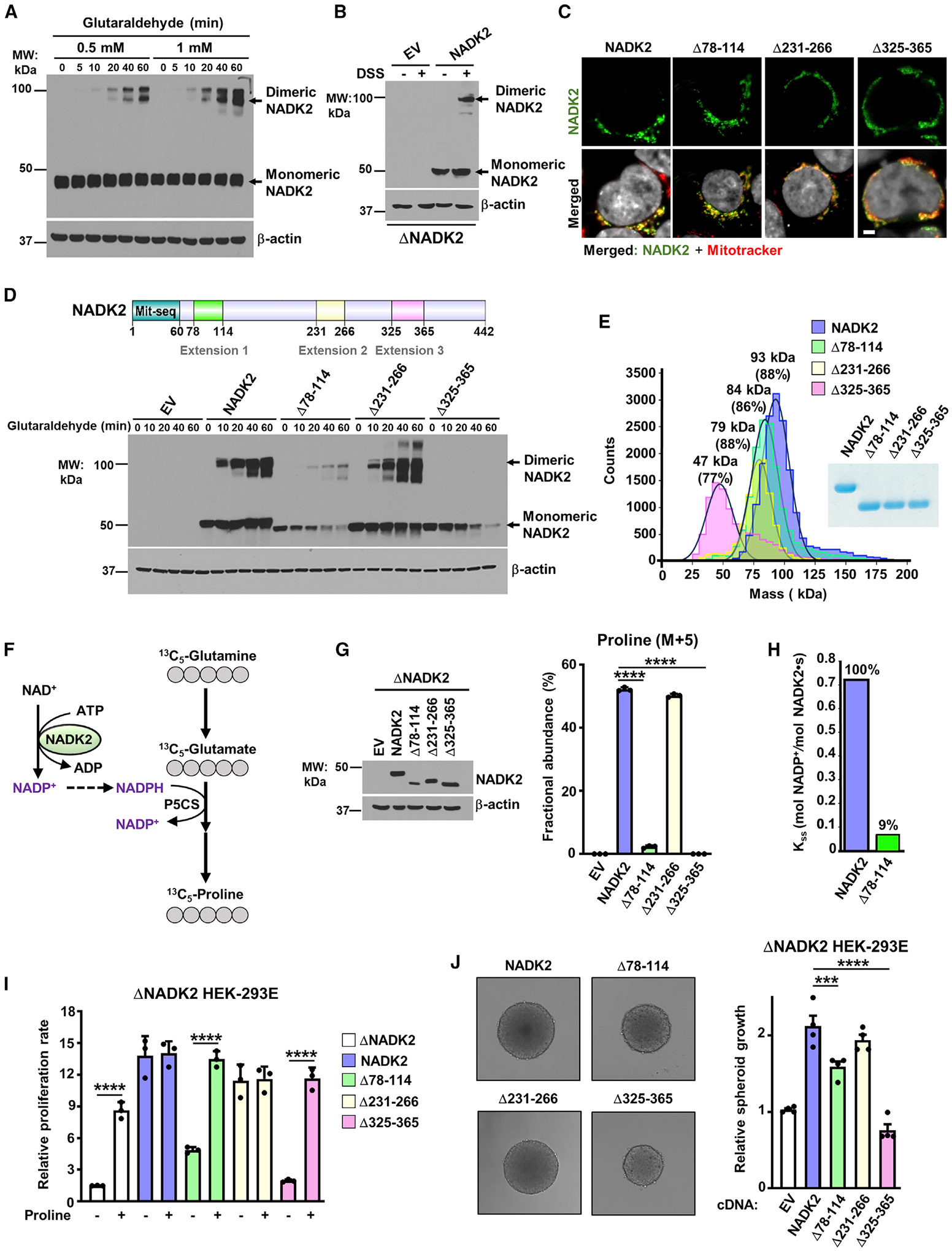
(A) Immunoblots from HEK293E cell extracts incubated with 0.5- or 1-mM glutaraldehyde for the indicated times. Monomeric and dimeric NADK2 and the β-actin control are shown.
(B) Immunoblots as in (A), but from ΔNADK2 HEK293E cells transiently transfected with either empty vector (EV) or NADK2 cDNA and treated with 1-mM DSS for 1 h.
(C) Mitochondrial localization of NADK2 extension-deficient variants. Representative images of localization of the indicated NADK2 variants (green) to mitochondria (red) detected by immunofluorescence with a NADK2 antibody. ΔNADK2 HEK293E cells were transiently transfected either empty vector (control), NADK2 wild type, or NADK2 variants with the indicated deleted extensions (Δ78–114, Δ231–266, and Δ325–365). Mitochondria were stained with MitoTracker (red) and nuclei with Hoechst (gray). Scale bars, 2 μm.
(D) Top: schematic of NADK2 depicting its mitochondrial localization sequence and the unique NADK2 extensions (aa 78–114, aa 231–266, and aa 325–365). Bottom: immunoblots as in (A), but from ΔNADK2 HEK293E cells that were transiently transfected with either empty vector (EV), NADK2 WT, or NADK2 variants with the indicated deleted extensions. Monomeric and dimeric NADK2 and the β-actin control are shown.
(E) Mass photometry of recombinant His-NADK2 (aa 61–442) or the indicated variants purified from E. coli, indicating that His-NADK2Δ325–365 exist in a monomeric state (calculated MW = 47 kDa for 77% of the sample), while NADK2, His-NADK2Δ78–114, and His-NADK2Δ231–266 are dimeric with the indicated molecular weight.
(F) Schematic of 13C5-Glutamine (M+5) tracing to assess the newly synthesized 13C5-proline. 13C-labeled carbons are shown in gray. The rate-limiting pyrroline-5-carboxylate synthase (P5CS) that requires NADK2 activity and mitochondrial NADPH is shown.
(G) Immunoblots and fractional abundance of proline (M+5) from ΔNADK2 HEK293E cells stably expressing either empty vector, NADK2 WT, or NADK2 variants with the indicated deleted extensions (Δ78–114, Δ231–266, Δ325–365) labeled for 3 h with 13C5 glutamine.
(H) Enzymatic activity of human recombinant His-NADK2 and NADK2Δ78–114 variant in the presence of 1 mM NAD+ and 5 mM MgATP.
(I) Cell proliferation of ΔNADK2 HEK293E cells stably expressing either empty vector, NADK2 WT, or the indicated NADK2 variants. Cells were grown for 72 h in 10% dialyzed serum in the presence or absence of 0.2 mM proline.
(J) Representative images and quantification of spheroid size from ΔNADK2 HEK293E cells stably expressing either NADK2 WT or the indicated NADK2 variants that were cultured in 10% dialyzed serum without proline. Quantification of the spheroid size was performed from 5 independent images.
Data are presented as the mean ± SD from n = 3 (G and I) or n = 4 (J) of biologically independent samples, and data are representative of at least two independent experiments. (G, I, and J) ***p < 0.001, ****p < 0.001 for multiple comparisons calculated using one-way ANOVA and Tukey’s post hoc test.
We then asked whether the helical extensions present solely in NADK2, but not other NADKs (aa 78–114, 231–266, or 325–365) could affect NADK2 dimerization, localization, and function. Specific deletion (Δ) of these segments did not affect mitochondrial localization of NADK2 (Figure 3C). However, NADK2Δ78–114 showed reduced protein stability when expressed in HEK293E cells (Figure 3D). Interestingly, deletion of most of the third extension of NADK2 (Δ325–365) drastically impaired its dimerization, while Δ78–114 and Δ231–266 showed no effect (Figure 3D). Mass measurements of single molecules in solution with mass photometry also revealed that NADK2Δ325–365 exists in its monomeric form while confirming a uniformly dimeric form for wild-type NADK2, as well as NADK2Δ78–114, and NADK2Δ231–266 variants (Figure 3E). These results are in accord with the structural architecture of the dimer, as the deletion Δ325–365 removes a large region of monomer-monomer interactions points (~2,160 Å2), whereas the 78–114 extension mediates much fewer contacts between the monomers (~870 Å2).
To assess the significance of these unique segments on NADK2 activity, we stably expressed the NADK2 deletion mutants in NADK2-deficient HEK293E cells and performed [13C5]-glutamine tracing to measure proline synthesis using liquid chromatography-mass spectrometry (LC-MS) (Figure 3F; Tran et al., 2021). Notably, similar to NADK2-deficient cells, NADK2Δ325–365 and NADK2Δ78–114 failed to synthesize proline from glutamine (Figures 3G and S3A). We did not observe noticeable changes in proline synthesis between expression of wild type versus NADK2Δ231–266. Since NADK2Δ78–114 expression yielded lower proline synthesis levels, possibly due to low protein abundance upon expression of this mutant in cells (Figure 3G), we compared its in vitro catalytic activity with the full-length NADK2. Interestingly, the NADK2Δ78–114 showed a significant decrease in catalytic activity, with only a 9% of residual activity when compared with the full-length NADK2 (Figure 3H), possibly due to its vicinity to the catalytic core (Figure 1F). Consistently, NADK2Δ325–365 expressing NADK-deficient cells were unable to proliferate in 2D (Figure 3I) or grow as 3D spheroids (Figure 3J) in the absence of proline, confirming the inability of these cells to produce proline de novo. Expression of NADK2Δ78–114 also showed growth defects both in 2D and 3D cell growth (Figures 3I and 3J), consistent with its reduced protein stability and catalytic activity. Together, our data suggest a crucial role for the 325–365 extended segment both for the dimerization and NADK2 function in the mitochondrial proline synthesis pathway. Moreover, the inability of a dimerization-deficient NADK2Δ325–365 mutant to support proline synthesis suggests that NADK2 dimerization may play a significant role in the activation and/or activity of NADK2.
The critical role of the dimerization interface for NADK2 function
To evaluate the role of the NADK2 dimer interface more precisely, we intended to disturb this interface by generating point mutants within extension 3, as well as within the NADK2 catalytic core (Figures 4A–4C). We choose buried and conserved positions pointing toward themselves through the dimerization to create a charge repulsion that would perturb this dimerization (Figures 4A and 4C). We substituted the small and hydrophobic valine V334 buried within the hydrophobic core at the interface of the two extensions 3 with a large and polar arginine residue (V334R) (Figure 4B). In parallel, we changed the arginine R378 that forms a buried salt bridge with the glutamate E379′ (from a second monomer) to glutamine (R378Q) (Figure 4C). Neither of these single-point mutations was sufficient to completely disrupt the dimeric structure, possibly because of the large and stable dimerization interface of NADK2 (Figure 4D). Therefore, these mutations were combined to create a double-mutant (V334R/R378Q), which resulted in monomeric NADK2 (Figure 4D) that was inactive and unable to synthesize proline in cells (Figures 4E and 4F). It is interesting to note that V334R mutation alone could partially destabilize the dimer and generate a small monomeric fraction. This mutation also caused the inactivation of NADK2 as evidenced by the substantial reduction of proline synthesis activity (Figure 4F). As expected, cells expressing V334R or the double-mutant V334R/R378Q could not proliferate without supplementation of exogenous proline (Figure 4G).
Figure 4. Characterization of the dimerization interface of NADK2.
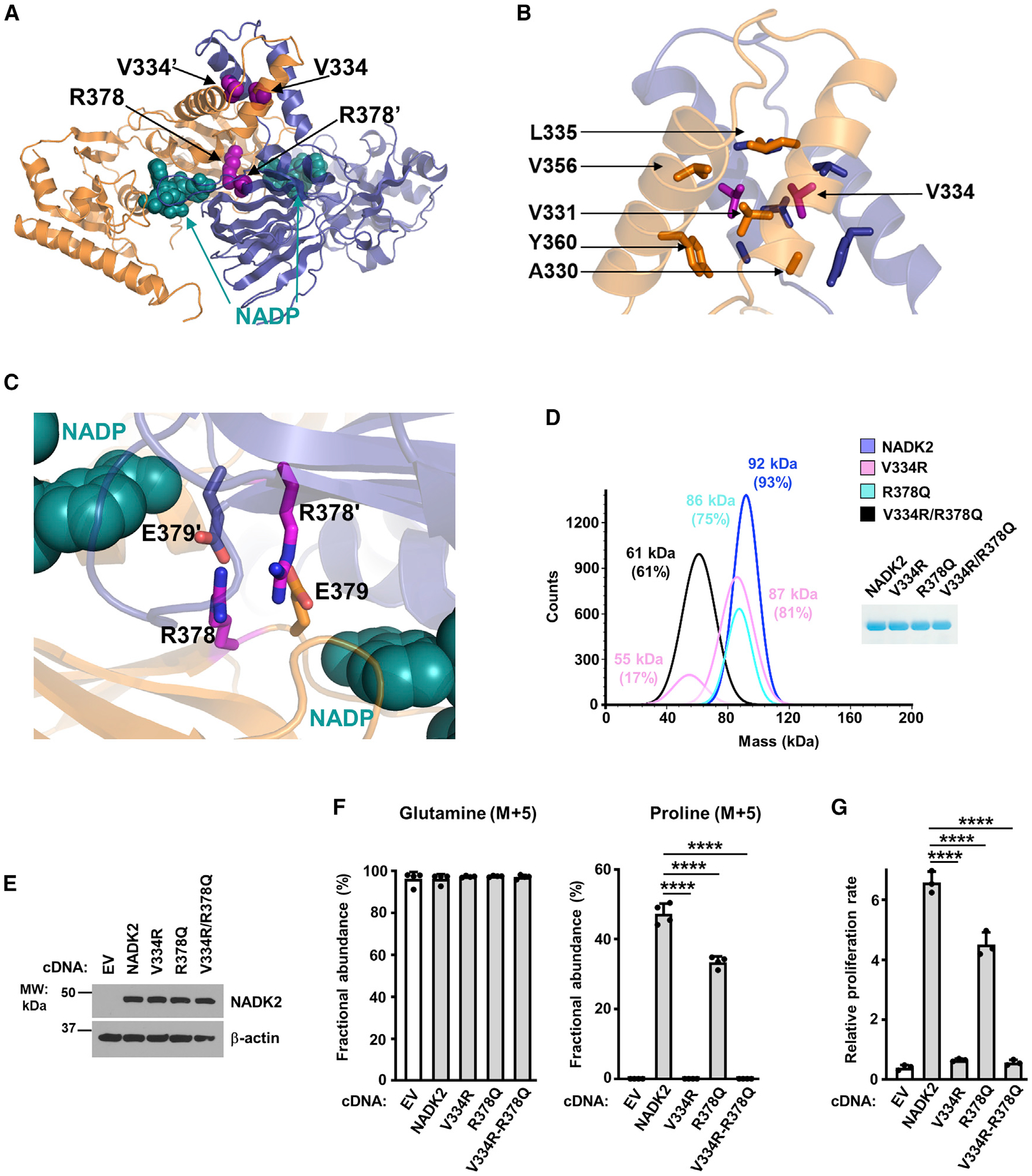
(A) Crystal structure of the dimeric NADK2 highlighting the position of two V334 and R378 residues at the dimerization interface. Monomers are shown in ribbon as in Figure 1A. The mutated residues are in pink spheres, and the NADP+ is in deep cyan spheres.
(B) Zoom in the crystal structure of the NADK2 highlighting the structural role of V334. The surrounding hydrophobic residues (A330, V331, L335, V356, and Y360) that directly interact with V334 are shown. Only one monomer is labeled for clarity.
(C) Zoom in the crystal structure of the NADK2 highlighting the structural role of R378. The two monomers are shown as orange and blue ribbons. The R378 and its interacting neighbor E379′ (from a second monomer) are shown in stick (colored as the monomer they belong to). The nicotinamide group of NADP+ is shown in a deep cyan sphere and labeled as in B.
(D) Mass photometry of recombinant His-NADK2 or the indicated variants purified from E. coli, showing that His-NADK2V334R/R378Q exists in a monomeric state (calculated MW = 61 kDa for 61% of the sample), while His-NADK2 or His-NADK2 R378Q are dimeric. His-NADK2V334R showed two pools corresponding to molecular weights of its monomeric and dimeric forms. Coomassie blue of purified NADK2 variants is shown on the right.
(E) Immunoblots from ΔNADK2 HEK293E cells stably expressing either empty vector, NADK2 WT, or the indicated NADK2 variants (V334R, R378Q, and V334R/R378Q).
(F) Fractional abundance of glutamine (M+5) and proline (M+5) from ΔNADK2 HEK293E cells stably expressing either empty vector (EV), NADK2 WT, or NADK2 variants (V334R, R378Q, and V334R/R378Q) labeled for 3 h with 13C5 glutamine.
(G) Relative cell proliferation of ΔNADK2 HEK293E cells stably expressing either empty vector, NADK2 WT, or the indicated NADK2 variants. Cells were grown for 72 h in 10% dialyzed serum in the absence of proline.
Data are presented as the mean ± SD from n = 3 (G) or n = 4 (F) of biologically independent samples, and data are representative of at least two independent experiments. (F and G) ****p < 0.001 for multiple comparisons calculated using one-way ANOVA and Tukey’s post hoc test.
NADK2 activity and functions are regulated by post-translational modifications
Mitochondrial enzymes are frequently subjected to regulation by PTMs, including acetylation, succinylation, and phosphorylation which can alter their activity and function (Carrico et al., 2018; Stram and Payne, 2016). Global high-throughput proteomics studies have identified multiple PTM events on NADK2 across tissues and species (Figures 5A and 5B; Table S1; Hornbeck et al., 2015). A study examined several of these phosphorylation sites for their in vitro NADK2 catalytic activity using recombinant protein produced in E. coli and found that phospho-mimetic or phospho-deficient mutant of Ser188 resulted in an inactive enzyme (Kawabata et al., 2015). We performed LC-MS/MS analysis of NADK2 immunopurified from HEK293E cells and found numerous new lysine acetylation, as well as multiple phosphorylation and succinylation events (Figures 5A and 5B; Table S1); however, we did not detect Ser 188 phosphorylation.
Figure 5. Regulation of NADK2 functions by post-translational modifications.
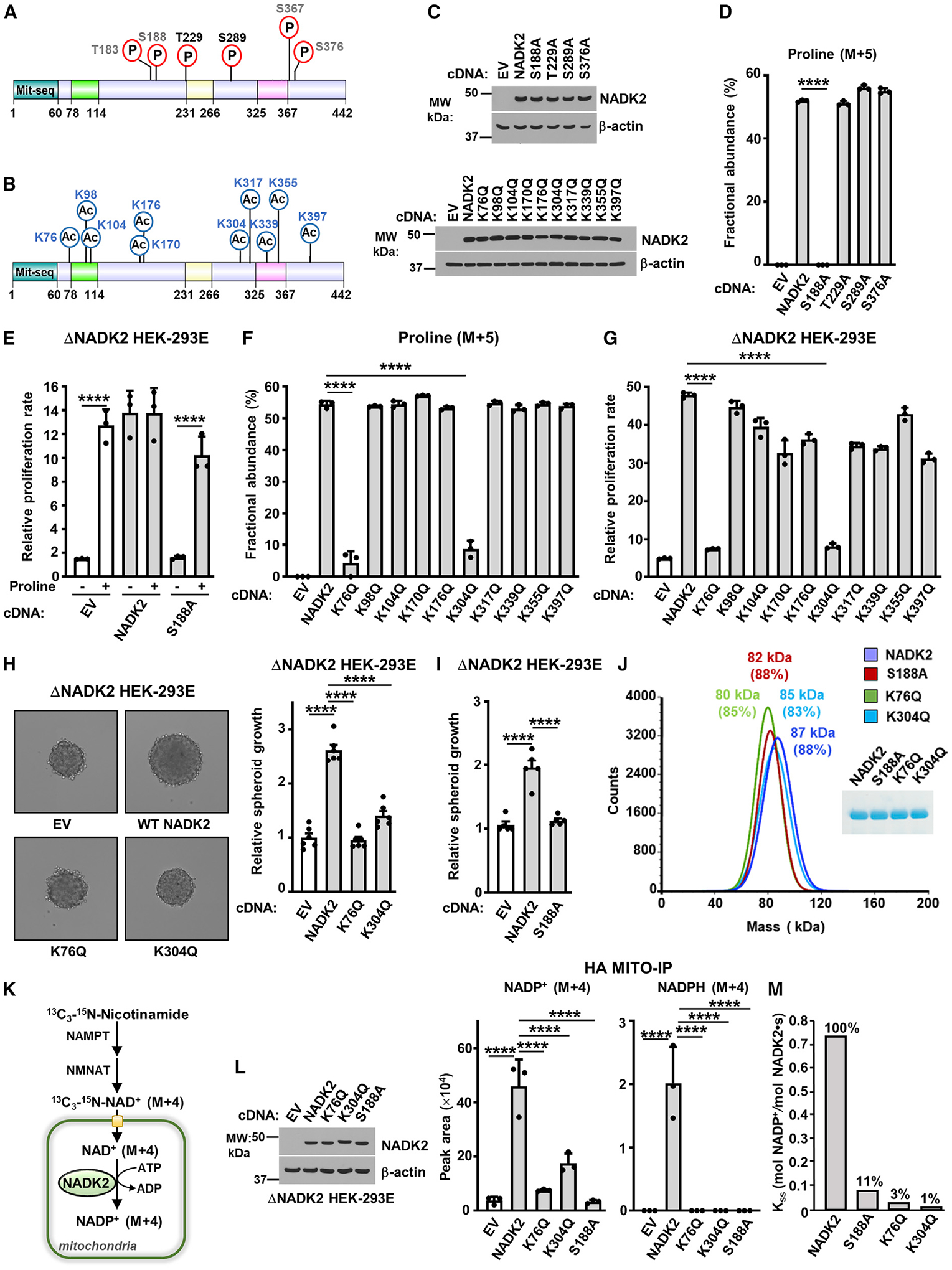
(A) Schematic of NADK2 as in Figure 3D, but showing phosphorylated residues identified in this study (black) or those curated from PhosphoSitePlus (gray). See Table 1.
(B) As in (A), but showing the acetylated residues found on NADK2 from HEK293E cells transiently expressing NADK2-FLAG (this study). See Table 1.
(C) Immunoblots of NADK2 variants from ΔNADK2 HEK293E cells stably expressing the indicated NADK2 phospho-mutants (S or T to A) or acetylation-mutants (K to Q).
(D) Fractional abundance of proline (M+5) as in Figure 3G, but from ΔNADK2 HEK293E cells stably expressing either empty vector, WT NADK2, or the indicated NADK2 variants (S or T to A) labeled for 3 h with 13C5-glutamine.
(E) Relative cell proliferation of ΔNADK2 HEK293E cells stably expressing either empty vector (EV), WT NADK2, or NADK2 S188A. Cells were grown for 72 h in 10% dialyzed serum in the presence or absence of 0.2 mM proline.
(F) Fractional abundance of proline (M+5) as in Figure 3G, but from ΔNADK2 HEK293E cells stably expressing either empty vector (EV), WT NADK2, or the indicated NADK2 acetylation-deficient mutants (K to Q) labeled for 3 h with 13C5-glutamine.
(G) Relative cell proliferation of ΔNADK2 HEK293E cells stably expressing either empty vector (EV), WT NADK2, or the indicated NADK2 acetylation-deficient mutants (K to Q). Cells were grown for 72 h in 10% dialyzed serum in the absence of 0.2 mM proline.
(H) Representative images and quantification of spheroid size from ΔNADK2 HEK293E cells stably expressing either empty vector, NADK2 WT, NADK2 K76Q, or K304Q mutants cultured without proline. Quantification of the spheroid size from 6 independent images for each condition is shown.
(I) Quantification of spheroid size from ΔNADK2 HEK293E cells stably expressing either WT NADK2 or NADK2 S188A cultured without proline. Quantification of the spheroid size from 5 independent images for each condition is shown.
(J) Mass photometry of recombinant His-NADK2 or the indicated variants purified from E. coli, showing that NADK2, His-NADK2 S188A, His-NADK2 K76Q, or His-NADK2 K304Q are dimers. Coomassie blue of purified NADK2 variants is shown on the right.
(K) Schematic of labeling with 13C3-15N-Nicotinamide (M+4), depicting the synthesis of mitochondrial NADP+ (M+4) from NADK2. NAD+ salvage enzymes nicotinamide phosphoribosyltransferase (NAMPT) and nicotinamide mononucleotide adenylyl transferase (NMANT) are shown.
(L) Immunoblots and normalized peak area of mitochondrial, NADP+ (M+4), and NADPH (M+4) from labeling with 13C3-15N-Nicotinamide (M+4), as illustrated in (K). ΔNADK2 HEK293E cells stably expressing HA-Mito were transiently transfected with EV, WT NADK2, or NADK2 K76Q, K304Q, or S188A mutants. HA-tagged mitochondria were immunoprecipitated (HA-Mito IP) from equal amount of protein for each condition.
(M) Enzymatic activity of human recombinant His-NADK2 and NADK2Δ78–114 variant in the presence of 1 mM NAD+ and 5 mM MgATP.
Data are presented as the mean ± SD from n = 3 (D–G and L) or n = 5 (H), n = 6 (I) of biologically independent samples. Data are representative of at least two independent experiments. (D–G, H, I, and L) ****p < 0.001 for multiple comparisons were calculated using one-way ANOVA and Tukey’s post hoc test.
To better understand the impact of NADK2 PTMs on its activity, we surveyed most of our identified lysine and serine/threonine modifications for their effect on NADK2 function. We reconstituted NADK2-deficient cells with either wild-type (WT) NADK2 or acetylation/succinylation-deficient mutants (lysine to glutamine, K/Q) or phospho-mutants (serine and threonine to alanine, S/A and T/A). We first confirmed that these mutations did not alter protein expression (Figure 5C) or mitochondrial localization of NADK2 (Figures S3B and S3C). Next, using 13C5-glutamine tracing, we showed that most of the S/A substitutions did not produce any noticeable effects on proline synthesis (Figures 5D and S3D), with the exception of S188A mutation, which completely blocked proline production. NADK2S188A-expressing cells, similar to NADK2-deficient cells, were unable to grow in the absence of proline (Figure 5E), further confirming the lack of activity of NADK2S188A and in accordance with a lack of in vitro enzymatic activity, as previously reported (Kawabata et al., 2015).
Profiling of the ten putative K/Q acetylation-deficient mutants demonstrated that K76Q and K304Q mutants profoundly reduced proline production (Figures 5F and S3E). Moreover, NADK2-deficient cells expressing these mutants were unable to grow in the absence of proline in 2D (Figure 5G) or 3D culture condition, similar to NADK2-deficient cells or NADK2S188A-expressing cells (Figures 5H, 5I, and S3F).
The K76Q, K304Q, and S188A mutations did not affect NADK2 dimerization (Figures 5J and S3G). We measured mitochondrial NADP+ synthesis using a rapid mitochondrial purification (MITO-Tag) method (Chen et al., 2016, 2017) that was coupled to a 13C3-15N-Nicotinamide (M+4) tracer (Figure 5K; Tran et al., 2021) and confirmed that NADK2-deficient cells expressing the K76Q, K304Q, and S188A mutants of NADK2 show a marked decrease in mitochondrial NADP+ and NADPH (Figure 5L), but not NAD+ levels (Figure S3H). In vitro kinase activity of these recombinant mutants only showed a residual catalytic activity (Figure 5M), further confirming the reduced NADK2 activity in these mutants. These findings demonstrate a significant role of K76, K304, and S188 residues for the catalytic activity of NADK2.
We then turned to the crystal structure for mechanistic insight on how these modifications exert such drastic effects on NADK2 function and activity. The active site groove harbors a number of positively charged residues, including the K76 from one subunit and K304 from the second subunit, as well as the S188, which appears to be readily available to stabilize the three phosphate groups of the phosphodonor ATP (Figure S4A). The hydroxyl group, Oγ, of the side chain of S188 is located in close proximity with the 2′-phosphate group of NADP+ (distance S188-Oγ—2′-PO4: 3.6 Å). Similarly, the two lysines—K76 from one monomer and K304′ from the second monomer—also point into the same NADP+-binding site upon NADK2 dimerization (Figure S4B). K76 is located in the vicinity of the metal ion (7.4 Å) and is predicted to lie next to the triphosphate moiety of ATP, while the K304’s amino-terminal group (Nζ) lies in the vicinity of the diphosphate group of NAD(P) (distance K304′-Nζ—PO4: 5.1 Å) and may participate to the stabilization of the reaction intermediate upon phosphodonor binding.
Therefore, from a structural standpoint, PTMs on these three residues (S188, K76, and K304) lying at the active site bring negative charges (i.e., phosphorylation, succinylation, and acetylation) that could serve either to neutralize the positively charged lysine side chains and/or to affect the local conformation of the active site, where the negatively charged phosphodonor (ATP), substrate (NAD+), and product (NADP+) bind (Figures S4A and S4B). Hence, phosphorylation, succinylation, or acetylation could generate a charge repulsion in the active site and interfere with the accommodation of the NAD+ substrate (K304 acetylation), the phosphodonor ATP (K76 acetylation), or with the phosphotransfer (S188 phosphorylation), thus impairing NADK2 enzymatic activity.
Together, our findings suggest that an intricate network of PTMs are at play near the active site of NADK2 that serves to regulate substrate binding and phosphorylation, and thereby NADK2 activity.
DISCUSSION
Here, we present the first high-resolution crystal structure of human mitochondrial NADK2 (2.3 Å), a crucial enzyme that drives the production of NADP(H) reducing cofactors in mitochondria for proline synthesis and redox homeostasis (Chandel, 2021; Fan et al., 2014; Ohashi et al., 2012; Tran et al., 2021; Zhu et al., 2021). The structure revealed a dimeric organization, in contrast to the tetrameric NADK structures solved thus far (Ando et al., 2011; Garavaglia et al., 2004; Mori et al., 2005b; Poncet-Montange et al., 2007). Biophysical characterization by SAXS and SEC-MALS confirmed the dimeric form of NADK2 in solution over a large range of concentrations, indicating the presence of a stable dimer.
The sequence of mitochondrial NADK2 is highly divergent from the cytosolic NADK and prokaryotic NADKs. NADK2 harbors three specific extensions: residues 77–114, 231–266, and 325–365, which are conserved in mammals, zebrafish, worms, and fruit flies, but not in the yeast homolog POS5 (Figure S2). Deletion of the third extension (aa 325–365) resulted in monomeric and inactive NADK2, which, when expressed in NADK2-deficient cells, resulted in proline auxotrophy due to its inability to enable proline production. Superimposition of the structures of NADK2 with that of human cytosolic NADK (unpublished, but deposited into PDB: 3PFN) suggests that the third extension would prevent the stacking of two NADK2 dimers to form a tetramer, as in the case of NADKs. Consistent with this notion, the yeast mitochondrial NADK2 homolog POS5, which lacks this extension, appears to have a tetrameric assemblage (Ando et al., 2011).
It still remains to be determined whether NADK2 dimerization is regulated in cells. Our attempts at disturbing the dimerization interface of NADK2 by employing single-point mutants within extension 3 (V334R) or the catalytic core (R378Q) failed, possibly because of the strong and large dimeric interface (7,520 Å2). However, the combination of these mutations resulted in monomeric and inactive NADK2, indicating that dimerization of NADK2 is critical for its activity.
We also captured crystals of NADK2 bound to NADP+ and to metal cations, including Ca2+, Mg2+, and Fe3+. To date, the position of the metals in any NADK structure has not been described. Our findings that the metal cations do not have direct contact with the catalytic aspartate (D161 in NADK2), but instead, the substrate itself (NAD+) is directly involved in chelating the metal ion, provides empirical evidence for the previously hypothesized substrate-driven catalysis mode for NADK kinases (Poncet-Montange et al., 2007). This mode of catalysis sheds light on why NADKs specifically phosphorylate NAD+ rather than other nucleotide phosphates.
Furthermore, our work provides the structural basis for the stringent regulation of human NADK2 by PTMs. We identified and characterized numerous acetylation and phosphorylation events under normal growth conditions (Table S1) and found that mutations that mimic acetylation (K/Q) of K76 and K304, and phospho-mutation S188A drastically decreased the NADK2-mediated cellular functions, including mitochondrial NADP(H) synthesis and proline production, rendering NADK2-deficient cells proline auxotroph. It is worth mentioning that disruption of these acetylations or phosphorylation events on NADK2 did not alter NADK2 dimerization, further indicating that the dimeric NADK2 is stable.
Our modeling analysis of the PTM events located in the vicinity of the active site of NADK2 suggested that acetylation of K304 may interfere with NAD+ binding, while acetylation of K76 would interfere with ATP binding. Moreover, S188 phosphorylation is expected to prevent the ATP to NAD+ phosphotransfer. These modeling observations are in agreement with the drastic effects of these mutations on NADK2 function (Figure S4). In contrast, the phosphorylation-mediated regulatory events in the cytosolic NADK are located in the N-terminal domain and serve as a rheostat that fine-tunes the activity of this enzyme (Hoxhaj et al., 2019; Love et al., 2015; Rabani et al., 2020; Schild et al., 2021).
Because mitochondrial acetyltransferases are yet to be discovered, protein acetylation in the mitochondria is thought to be largely non-enzymatic (Lombard et al., 2015). However, removal of these acetylation modifications is attributed to the activity of mitochondrial NAD-dependent SIRT3/SIRT5 deacetylases (Carrico et al., 2018). Further work is needed to define the upstream signals and enzymes (acetyltransferases, deacetylases, kinases, and phosphatases) that regulate acetylation of K76 and K304, and phosphorylation of S188 under physiological or pathological settings, such as cancer. This will illuminate the regulation of mitochondrial NADP(H) and its downstream processes, such as biosynthesis and redox.
NADK2 amplifications are found in ~10% of the non-small cell lung cancer, ~6%–8% of other cancers, including bladder, esophageal, stomach, ovarian, and melanoma (www.cbioportal.org) (Cerami et al., 2012; Gao et al., 2013). Increased NADK2 expression also correlates with decreased overall survival in patients with lung squamous cell carcinomas, uterine carcinoma, breast cancer, and sarcoma (Lánczky and Győrffy, 2021). Previous work has indicated that loss of NADK2 results in proline auxotrophy and reduces tumor growth (Tran et al., 2021; Zhu et al., 2021), suggesting that this proline auxotrophy can be harnessed in cancer by disrupting the NADK2-proline axis with a combination of NADK2 inhibition and proline-free diet. Thus, our crystal structure will allow for structure-based design of selective inhibitors of NADK2 that can be used for cancer therapies.
In summary, this study provides the first high-resolution structure of NADK2—a critical mitochondrial enzyme—and reveals its substrate-driven mode of activation. Our biochemical, structural, and functional analysis of NADK2 features also provides new mechanistic insight into a stringent regulation of NADK2 activity by PTMs.
Limitations of the study
In this study, we report the crystal structure of human NADK2, which revealed a dimeric organization. The substrate-binding site is located at the interface of the dimer, indicating a crucial role of this dimeric assembly for NADK2 activity. Indeed, NADK2 possesses three unique extension loops, where extension 3 (residues 325–367) mediates NADK2 dimerization. Deletion of extension 3 or point mutations in this region destabilizes the dimer and results in loss of NADK2 activity. While extension 1 (aa 78–114) of NADK2 is critical for NADK2 stability and its activity, extension 2 of NADK2 (aa 231–266) was not fully resolved in our crystals and did not affect NADK2 catalytic activity. It remains possible that extension 2 is involved in protein-protein interactions, however, further work is required to delineate its role.
NADK2 is regulated by PTMs, including acetylation events on K76 and K304 residues or phosphorylation of S188. While mutations of these residues abrogate the catalytic activity of NADK2 most likely because of the interference with NAD+ or ATP binding near the catalytic core, the dynamic regulation of these modifications in response to physiological or pathological cues, and identification of the upstream enzymes that directly regulate these sites remain to be elucidated. Future studies employing site-specific antibodies to detect these acetylation and phosphorylation events will enable the assessment of the regulation of mitochondrial NADP(H) metabolism under physiological or pathological contexts.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Requests of information and requests for reagents should be directed and will be fulfilled by the lead contact, Gerta Hoxhaj (gerta.hoxhaj@utsouthwestern.edu)
Materials availability
All unique reagents generated in this study will be made available by the lead contact.
Data and code availability
Original western blot and microscopy images have been deposited at Mendeley and are publicly available as of the date of publication. Validation reports of X-ray crystallography structures have been deposited at the PDB, and the refined models have been deposited in the Research Collaboratory for Structural Biology (http://www.rcsb.org) under the following accession numbers: 7R4J; 7R4K, 7R4L, and 7R4M, which are publicly available as of the date of publication. The DOIs and accession numbers are listed in the key resources table. Mass spectrometry data are presented in the Table S1.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| NADK2 Antibody | Abcam | Cat# ab181028 Lot# GR164717-1 |
| Anti-rabbit-secondary antibody | Cell Signaling Technology | Cat#7074 Lot# 29 |
| HRP-conjugated anti-mouse-secondary antibody | Cell Signaling Technology | Cat#7076 Lot# 34 |
| β-actin Antibody | Sigma-Aldrich | A5316 Lot# 059M4770V |
| Bacterial and virus strains | ||
| NEB Stable Competent E. coli | NEB | C3040 |
| BL21(DE3) Competent E. coli | NEB | C2527 |
| Chemicals, peptides, and recombinant proteins | ||
| DMSO | Sigma-Aldrich | D2650-100ML |
| Proline | Sigma-Aldrich | P5607-25G |
| 13C5 glutamine | Sigma-Aldrich | 605166-500MG |
| NEB-10-beta/Stable Outgrowth Media | NEB | B9035S |
| Crystal violet | Sigma-Aldrich | C6158-50G |
| Trypab Blue Soloution | Sigma-Aldrich | T8154-100ML |
| L-glutamine | Corning | MT25005Cl |
| Puromycin dichloride | Santa Cruz | sc-108071A |
| Polyethylenimine (PEI) | Polysciences | 23966-2 |
| Opti-MEM media | Thermo Fisher | 31985062 |
| Glutamine-free DMEM | Gibco | 11960-044 |
| SuperSignal™ West Femto Chemiluminescent | Thermo Fisher scientific | PI34096 |
| SuperSignal™ West Pico PLUS | Thermo Fisher scientific | PI34580 |
| DMEM | Corning CellGro | 10-017-CV |
| Fetal bovine serum (FBS) | R&D Systems a biotech brand | S11150 |
| Fetal Bovine Serum dialyzed | R&D Systems a biotech brand | S12850 |
| Microcystin-LR | Enzo Life Sciences | NC9580520 |
| Protease inhibitor cocktail | Sigma-Aldrich | P8340-5ML |
| Acrylamide Protogel 30% | Fisher Scientific | EC-890 |
| Bio-Rad Braford | Bio-Rad | 5000006 |
| Low melting Agarose | Lonza | 50101 |
| Tryptone | United States Biological Life sciences | 91079-40-2 |
| Yeast extract | Euromedex | UY2010 |
| Sodium Chloride | Euromedex | 1112-A |
| Tris Ultra Pure | Euromedex | 26-128-3094-B |
| Imidazole | Millipore | 288-32-4 |
| DTT | Euromedex | EU0006 |
| IPTG | Euromedex | EU0018 |
| InstantBlue® Coomassie Protein Stain | abcam | ab119211 |
| Complete™, EDTA-free Protease Inhibitor cocktail | Roche® Life Science Products | 05056489001 |
| Ampicillin | Euromedex | EU0400 |
| Chloramphenicol | Euromedex | 3886 |
| Tris Ultra Pure | Euromedex | 26-128-3094-B |
| NaCl | Euromedex | 1112-A |
| Sodium citrate tribasic dihydrate | Sigma-Aldrich | S4641 |
| Manganese(II) chloride tetrahydrate | Sigma-Aldrich | M8054 |
| Magnesium chloride hexahydrate | Sigma-Aldrich | M2670 |
| Calcium chloride | Sigma-Aldrich | C1016 |
| Cobalt(II) chloride | Sigma-Aldrich | 60818 |
| Cadmium chloride | Sigma-Aldrich | 202908 |
| Nickel(II) chloride hexahydrate | Sigma-Aldrich | 72247 |
| Copper(II) chloride dihydrate | Sigma-Aldrich | C6641 |
| Zinc chloride | Sigma-Aldrich | 31650-M |
| Barium chloride | Sigma-Aldrich | 202738 |
| Cadmium chloride | Sigma-Aldrich | 202908 |
| Iron(III) chloride | Sigma-Aldrich | F7134 |
| Aluminum chloride hexahydrate | Sigma-Aldrich | A0718 |
| NAD+ | Roche | 10127973001 |
| NADP+ disodium salt | Roche | 10128058001 |
| Adenosine 5′-triphosphate disodium salt hydrate | Sigma-Aldrich | A7699 |
| Sodium polyphosphate | Supelco | 1.06529 |
| D-Glucose 6-phosphate sodium salt | Sigma-Aldrich | G7879 |
| Glucose-6-phosphate Dehydrogenase from baker’s yeast (S. cerevisiae) | Sigma-Aldrich | G7877 |
| Critical commercial assays | ||
| HiSpeed Plasmid Maxi Kit (25) | Qiagen | 12663 |
| His TALON™ Buffer Set | TAKARA | 635651 |
| His TALON™ Gravity Column | TAKARA | 635655 |
| ZymoPURETM I Plasmid Midi Prep Kit | Zymo Research | D4201-B |
| Deposited data | ||
| NADK2 crystal structures | PDB IDs/accession numbers | 7R4J, 7R4K, 7R4L, 7R4M |
| A study by Mary and Soflaee et al., unprocessed files | Mendeley Data | https://doi.org/10.17632/8ybbnh7g3p.1 |
| Experimental models: Cell lines | ||
| HEK293E | Carol Mackintosh (University of Dundee) | N/A |
| ΔNADK2 HEK293E | Tran et al., 2021 | N/A |
| ΔNADK2 HA-Mito | Tran et al., 2021 | N/A |
| ΔNADK2 HEK293E +EV | This study | N/A |
| ΔNADK2 HEK293E +NADK2 | This study | N/A |
| ΔNADK2 HEK293E +Δ78-114-Flag | This study | N/A |
| ΔNADK2 HEK293E +Δ231-266 | This study | N/A |
| ΔNADK2 HEK293E +Δ325-365 | This study | N/A |
| ΔNADK2 HEK293E +S188A | This study | N/A |
| ΔNADK2 HEK293E +T229A | This study | N/A |
| ΔNADK2 HEK293E +S289A | This study | N/A |
| ΔNADK2 HEK293E +S376A | This study | N/A |
| ΔNADK2 HEK293E +K76Q | This study | N/A |
| ΔNADK2 HEK293E +K98Q | This study | N/A |
| ΔNADK2 HEK293E +K104Q | This study | N/A |
| ΔNADK2 HEK293E +K170Q | This study | N/A |
| ΔNADK2 HEK293E +K176Q | This study | N/A |
| ΔNADK2 HEK293E +K304Q | This study | N/A |
| ΔNADK2 HEK293E +K317Q | This study | N/A |
| ΔNADK2 HEK293E +K339Q | This study | N/A |
| ΔNADK2 HEK293E +K355Q | This study | N/A |
| ΔNADK2 HEK293E +K397Q | This study | N/A |
| ΔNADK2 HEK293E +V334R | This study | N/A |
| ΔNADK2 HEK293E +R378Q | This study | N/A |
| ΔNADK2 HEK293E +V334R-R378Q | This study | N/A |
| Recombinant DNA | ||
| pLenti III NADK2-Flag | Tran et al., 2021 | N/A |
| pLenti III NADK2-NADK2-Flag | This study | N/A |
| pLenti III NADK2-Δ78-114-Flag | This study | N/A |
| pLenti III NADK2-Δ231-266 | This study | N/A |
| pLenti III NADK2-Δ325-365 | This study | N/A |
| pLenti III NADK2-S188A-Flag | This study | N/A |
| pLenti III NADK2-T229A-Flag | This study | N/A |
| pLenti III NADK2-S289A-Flag | This study | N/A |
| pLenti III NADK2-S376A-Flag | This study | N/A |
| pLenti III NADK2-K76Q-Flag | This study | N/A |
| pLenti III NADK2-K98Q-Flag | This study | N/A |
| pLenti III NADK2-K104Q-Flag | This study | N/A |
| pLenti III NADK2-K170Q-Flag | This study | N/A |
| pLenti III NADK2-K176Q-Flag | This study | N/A |
| pLenti III NADK2-K304Q-Flag | This study | N/A |
| pLenti III NADK2-K317Q-Flag | This study | N/A |
| pLenti III NADK2-K339Q-Flag | This study | N/A |
| pLenti III NADK2-K355Q-Flag | This study | N/A |
| pLenti III NADK2-K397Q-Flag | This study | N/A |
| pLenti III NADK2-V334R-Flag | This study | N/A |
| pLenti III NADK2-R378Q-Flag | This study | N/A |
| pLenti III NADK2-V334R-R378Q-Flag | This study | N/A |
| pET15b Δ1-60 NADK2 | This study | N/A |
| pET15b Δ1-60 NADK2-Δ78-114 | This study | N/A |
| pET15b Δ1-60 NADK2-Δ231-266 | This study | N/A |
| pET15b Δ1-60 NADK2-Δ325-365 | This study | N/A |
| pET15b Δ1-60 NADK2-S188A | This study | N/A |
| pET15b Δ1-60 NADK2-K76Q | This study | N/A |
| pET15b Δ1-60 NADK2-K304Q | This study | N/A |
| pET15b Δ1-60 NADK2-V334R | This study | N/A |
| pET15b Δ1-60 NADK2-R378Q | This study | N/A |
| pET15b Δ1-60 NADK2-V334R/R378Q | This study | N/A |
| Software and algorithms | ||
| Discover MP 2.4.2 | Refeyn | https://www.refeyn.com/software-release-updates |
| Prism | Prism | https://www.graphpad.com/scientific-software/prism/ |
| TraceFinder 5.1 | ThermoScientific | https://www.thermofisher.com/us/en/home/industrial/mass-spectrometry/liquid-chromatography-mass-spectrometry-lc-ms/lc-ms-software/lc-ms-data-acquisition-software/tracefinder-software.html |
| ASTRA 6.1 | Waytt Technology | https://www.wyatt.com/products/software/astra.html |
| GraFit | Erithacus Software Ltd | v. 7.0.3 |
| Other | ||
| Mass Photometry TwoMP | Refeyn | N/A |
| SpectraMax iD3- Microplate reader | Molecular devices | N/A |
| Nitrocellulose membrane 0.2 μm | GE Healthcare | 10600001 |
| Blotting paper, 703 | VWR North America | 732-0591 |
| LUNA-II™ Automated Cell Counter | LUNA-II™ | N/A |
| Celigo image cytometer-4 Channel | Nexcelom Bioscience,UK | N/A |
| QExactive HF-X Orbitrap Mass Spectrometer | ThermoScientific | N/A |
| Orbitrap Exploris 480 Mass Spectromete | ThermoScientific | N/A |
| Amicon Ultra-15, PLTK, membrane Ultracel-PL, 30 kDa | Millipore | UFC903024 |
| Complete™ His-Tag Purification Column | Roche® Life Science Products | 06781535001 |
| Bolt™ 4 à 12 %, Bis-Tris | Invitrogen™ | NW04125BOX |
| CLARIOstar plate reader BMG LABTECH | BMG LABTECH | UV-Star® Half area |
| 96-well plates | Greiner | 655087 |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
HEK-293E cells (gift of Carol Mackintosh) were maintained in DMEM (Corning/Cellgro, 10–017-CV) containing 10% fetal bovine serum (FBS). NADK2-deficient HEK-293E cells were grown in 10% FBS supplemented with 0.2 mM proline. Metabolomics, cell proliferation, and 3D spheroid assays were performed in 10% dialyzed FBS in the presence or absence of 0.2 mM proline or as indicated in the figure legends. For transient transfections, HEK-293E cells in 6-well plate format were transfected with 2 μg of plasmid DNA using polyethylenimine (PEI) transfection method as previously described (Hoxhaj et al., 2019). Isogenic stable cell lines were generated via lentivirus delivery of either empty vector (EV) or NADK2 constructs in NADK2 knockout HEK-293E cells followed by 2 μg/ml of puromycin selection (Tran et al., 2021).
Expression and purification of human NADK2 from Escherichia coli
The bacterial strain described here is used for crystallography or in vitro experiments, rather than being experimental models. For protein production, the N-terminal His-tagged NADK2 pET15b plasmid coding the open reading frame for aa 61–442 was transformed in Escherichia coli (E. coli) strain BL21(DE3) (C2527 NEB). The bacteria carrying this expression plasmids were grown under aerobic condition in lysogeny broth (LB) at 37 °C with the appropriate antibiotics (ampicillin 100 μg/mL, chloramphenicol 34 μg/mL). The expression of the His-tagged protein was induced during the exponential phase (OD600 0.6–0.8) by adding 0.4 mM of isopropyl-β-D-thiogalactopyranoside (IPTG), 20 hours at 25 °C. Afterwards, bacterial cells were harvested by centrifugation at 6,000 g for 15 minutes at 4 °C, suspended in lysis buffer (50 mM Tris pH 7.8, 150 mM NaCl, 2 mM DTT) with 10 μg/mL lysozyme and a cocktail of protease inhibitors (cOmplete™, EDTA-free Protease Inhibitor cocktail Roche) and the pellet was frozen at −80 °C for 16 hours. Once thawed, the lysate was sonicated (6 min; 2 s pulses; amplitude 45; (Sonicator BIOBLOCK Scientific Vibra Cell 72405) and clarified by centrifugation (20,000 g, 30 min, 4 °C). The supernatant was collected, passed through a 0.45 μm filter and placed on the cOmplete™ His-Tag Purification Column (Roche® Life Science Products, 06781535001). The His-tagged protein was eluted with a linear gradient of imidazole (12 mM - 300 mM imidazole in 50 mM Tris pH 7.8, 150 mM NaCl, 2 mM DTT). The protein was subjected to size exclusion chromatography with a Superdex 200 column (GE Healthcare) equilibrated with 50 mM Tris pH 7.8, 150 mM NaCl, 2 mM DTT and the purity of NADK2 was confirmed by SDS-PAGE (Bolt™ 4–12 %, Bis-Tris, Invitrogen™, NW04125BOX).
METHOD DETAILS
Crystallization
Initial crystallization conditions of NADK2 with NADP+ were found by using a sitting-drop format and sparse-matrix screening strategy. Three screening kits were used to find promising conditions for crystal growth at 18 °C in 96-well plates. Seven hits were detected and refinement was performed on the two conditions providing with the best crystals. Finally, refined conditions that lead to suitable crystals were conducted with 100 mM to 250 mM Calcium chloride dihydrate, 0.1 M Sodium HEPES (pH 7.5), 28 % v/v PEG 400, 2–5 % Glycerol. The crystals were further improved using the Hampton Research 96-wells additive screen (Cat #; HR2–138). In a 96-wells plate, 5 μL of this additive screen were added to 40 μL of buffer containing 250 mM Calcium chloride dihydrate, 0.1 mM Sodium HEPES pH 7.5, 28 % v/v PEG 400, 2 % glycerol. Sitting-drops were made by mixing 100 nL of the NADK2 protein solution (9 mg/mL of NADK2 protein mixed with 10 mM NADP+) with an equal volume of the corresponding crystallization solution with additive present in each well of the plate. Several additives gave promising crystals, and one of them (NaBr 0.1 M) led to a single well-diffracting crystal (2.29 Å) of 220 μm × 90 mm of dimension.
Crystallographic studies
X-ray diffraction data sets were collected from frozen single crystals at the European Synchrotron Radiation Facility (Grenoble, France, beamline ID30B). Data were processed and scaled with the program suites installed at the ESRF (Monaco et al., 2013). The structure was solved by molecular replacement using the AutoMR procedure of the software PHENIX (Adams et al., 2010) using an ensemble-based model built using @TOME-2 (http://atome.cbs.cnrs.fr). A better MR solution was obtained using a monomer extracted from a dimeric model built using Alphafold-2 (https://colab.research.google.com/) with excellent statistics. Indeed, this model appeared afterward, very close to the final crystal structure with a RMSD of 0.9 Å over 172 Ca. Subsequently, iterative model rebuilding and refinement was performed first by using the program COOT (Emsley et al., 2010) and the program PHENIX (Adams et al., 2010). Final refinements were performed on the data set at 2.29 Å resolution. Some side-chains and several short segments of the protein (mainly small loops such as 344–346) were not clearly visible in the electron density. Figures of the protein, its ligands and the corresponding electron density were generated using PyMOL (http://pymol.sourceforge.net). The refined models and structure factors have been deposited in the Research Collaboratory for Structural Biology (http://www.rcsb.org) under the following accession numbers: 7R4J; 7R4K, 7R4L and 7R4M.
Small Angle X-ray Scattering data collection and analysis
The purified protein was concentrated using 15 mL Amicon® ultra 30 kDa (Millipore, UFC903024) and three different concentrations were prepared (3, 6 and 12 mg/mL). Data were recorded on BM29 beamline at the European Synchrotron Radiation Facility and analyzed using the ATSAS suite (Manalastas-Cantos et al., 2021). Kratky analysis revealed little disorder, and Guinier plot suggested a Rg of 3.1 nm. Within ATSAS, DAMMIN was used to derive an enveloped of the dimer and Crysol was used to evaluate the theoretical model built by Alphafold and then the crystal structure.
Size Exclusion Chromatography - Multi-Angle Light Scattering (SEC-MALS)
The measurements were done using Superdex 200 Increase 10/300 GL column (GE Healthcare) with a flow-rate of 0.5 mL/min, connected to Agilent™ Infinity II HPLC system coupled to miniDAWN TREOS (Waytt Technology) multi-angle light scattering detector and Optilab T-rEX (Waytt Technology) refractometer. A 20 μL sample of NADK2 at 3.5 mg/mL was injected and eluted in 50 mM Tris pH 7.8, 150 mM NaCl. The data were analyzed using the ASTRA software v. 6.1 (Wyatt Technology). The calculation of protein molecular weight was obtained from light scattering. The refractive index increment used was dn/dc 0.185 mL/g.
Ligand docking
Using the high-resolution crystal structure of NADK2 in complex with NADP+ and calcium we built several crude models of NADK2 with NAD+ (by replacing the 2’-phosphate group by a hydroxyl) or NADP+ in complex with magnesium, calcium or manganese. These templates were used to dock either ATP (in NAD+ complexes) or ADP (in NADP+ complexes) using the software PLANTS (Korb et al., 2009). 10 poses were visualized and a representative one was selected. Point mutations K76Q, S188A and K304Q were modeled using the software Coot. Similarly, modified side-chains for acetylated lysine or phosphorylated were modeled using the same tool. Then, the corresponding coordinates were used to resume the docking of ATP or ADP in the presence of metal-chelating NAD+ or NADP+. Figures of the resulting complexes were drawn using Pymol.
NADK2 enzymatic activity assays
In vitro enzymatic activity of NADK2 was assayed using the glucose 6-phosphate dehydrogenase coupling assay, as previously described for other NADKs (Mori et al., 2005a, 2005b). Experiments were carried out at 37 °C in a CLARIOstar plate reader (BMG LABTECH), using 96-well plates (UV-Star® Half area, Greiner bio-one) with a final volume of 100 μL. Buffer consisted of 50 mM Tris pH 7.5, 100 mM NaCl, 10 mM NaCitrate. Reaction mixture contained 6 mM metal ion, 1 mM NAD+, 5 mM ATP or polyphosphate, 5 mM glucose 6-phosphate, 1 U/mL glucose 6-phosphate dehydrogenase and 1 μM NADK2. Initial variation of the OD at 340 nm was converted into NADH concentration using an extinction coefficient of 6.22 M−1 cm−1 and fitted with a linear regression using GraFit software (Erithacus Software Ltd, v. 7.0.3). Results are expressed as steady-state rate constants, i.e. mol of NADP+ produced per mol of NADK per second. For the kinetic activity of the NADK2 variants, the concentration of the NADKs varied from 0.2 to 10 μM.
cDNA constructs for mammalian or bacterial expression
Full-length human NADK2 cDNA originally from Genscript (OHu24582) was subcloned into a lentivirus expressing construct (Lenti-III-PGK, Abmgood, G305) with a C-terminal FLAG-tag. For crystallography studies as well as in vitro enzymatic activities, a synthetic gene coding for an optimized sequence of human NADK2 lacking the mitochondrial localization signal (Δ1–60 NADK2) (EC 2.7.1.23; Uniprot ID: Q4G0N4) was subcloned in the plasmid pET15b that has an N-terminal His-tag. NADK variants (S/A, T/A, or K/Q) or deletions of specific amino acids regions (i.e., Δ78–114, Δ 231–266, Δ325–365) were generated by site-detected mutagenesis using KOD Xtreme™ Hot Start DNA Polymerase followed by DPN1 digestion of the parental DNA. All plasmids were verified by sequencing at the McDermott Center Sequencing Core at UT Southwestern Medical Center.
Cell proliferation assays
Isogenic ΔNADK2 HEK-293E cells stably expressing either NADK2 or Ser/Ala or Lys/Gln variants were plated in biological triplicates at a density of 50,000 cells/well in 24-well plates. Plates were coated with 0.01 % poly-L-lysine (Sigma, P4707) prior to cell seeding. Cell proliferation was assessed with Crystal violet assay. Cells were washed with 1 mL PBS prior to staining with 500 μL of Crystal violet staining solution (0.1 % crystal violet (Sigma, C6158) in 10 % ethanol) for 20 minutes at room temperature. Then, cells were washed three times with 1 mL PBS, air-dried for 1 hour, prior to dye extraction with 500 μL of 10 % acetic acid. Absorbance (590 nm) was measured directly from the 24-well plates using SpectraMax iD3 Multimode Microplate Detection Platform (Molecular Devices, LLC). Relative proliferation rate was measured 72 hours post-plating and normalized to Day 0 (12 hours post-plating).
Three-Dimensional (3D) Spheroid assay
3D spheroid growth was used to assess anchorage-independent cell growth (Vinci et al., 2015). Isogenic ΔNADK2 HEK-293E cells stably expressing either NADK2 or its variants (Δ78–114, Δ 231–266, Δ325–365, K76Q, K304Q, and S188A) were seeded on 96-well plates that were coated with 50 μL of 1.5 % low melting agarose (Lonza, 50101). Plates containing 1000 cells/well in a final volume of 150 μL of growth media (DMEM with 10 % dialyzed FBS) were centrifuged at 2000 rpm for 10 minutes using a Thermo Scientific™, Sorvall X1 Pro Centrifuge. Spheroid images were taken at Day 0 and at Day 6 with a Celigo image cytometer-4 Channel and software 5.1.0 from Nexcelom Bioscience). The spheroid area was analyzed using AxioVision Rel. 4.8 software (Carl Zeiss, Version AxioVs40 v. 4.8.2.0, Germany).
Immunofluorescence microscopy
Isogenic ΔNADK2 HEK-293E cells stably expressing either NADK2 or its variants 20,000 cells/well were seeded onto 8-Well Ibidi μ-Slide chamber (VWR, MSPP-8082690) in DMEM supplemented with 10 % FBS and 0.2 mM proline. 36 hours post-plating, mitochondrial staining was performed with 200 nM of MitoTracker® Red (CST, #9082) for 15 min. Cells were washed with PBS three times, fixed with 4 % paraformaldehyde (Santa Cruz Biotechnology, sc-281692) for 15 minutes at room temperature, permeabilized and blocked with 0.3 % Triton X-100 in 5 % Donkey Serum in PBS for 1 hour at room temperature. Cells were then incubated with primary antibodies (NADK2 (Abcam, ab181028; 1:1000), FLAG antibody (CST, 14793; 1:800)) overnight at 4 °C in antibody dilution buffer (PBS containing 0.1 % BSA and 0.1 % Triton × 100). Following three PBS washes (10 min each), cells were then incubated for 1 hour with goat anti-rabbit Alexa Fluor 488 (Invitrogen, A11008; 1:200) in the dark. Cells were subsequently washed three times with PBS, prior to staining with mounting media containing 10 μM Hoechst (Sigma-Merck, Germany, 62249) and 50 % Glycerol-PBS (Sigma-Aldrich, G6279). Images were taken with Zeiss LSM 780 Laser Scanning Microscope (Carl Zeiss, Germany).
Cell lysis, immunoblotting, and antibodies
For protein extracts, cells were washed with ice-cold PBS, prior to lysis with Triton lysis buffer (1 % Triton X-100, 40 mM HEPES, pH 7.4, 120 mM NaCl, 10 mM sodium pyrophosphate, 10 mM glycerol 2-phosphate, 50 mM NaF, 1 μM EDTA, 1 mM Microcystin-LR (Enzo life sciences, ALX-350–012-C500) and protease inhibitor cocktail (Sigma, P8340)). Lysates were clarified by centrifugation at 20,000 g for 15 min at 4°C, and protein concentrations were determined with a Bradford assay (Biorad, 500–0006). 20–30 μg protein lysates were subjected to SDS-PAGE followed by immunoblotting using the following primary antibodies: NADK2 (Abcam, ab181028; 1:1000), FLAG (CS, 14793; 11:1000), β-actin (Sigma, A5316; 1:5000), and HRP-conjugated anti-mouse (CST, 7076; 1:5000) and anti-rabbit (CST, 7074; 1:5000) secondary antibodies were used.
Crosslinking experiments
For glutaraldehyde crosslinking experiments (Sigma, G5882), HEK-293E ΔNADK2 cells (2.8 × 106) were seeded in 10 cm dishes. The next day, 40–50 % confluent cells were subjected to transient transfection with 10 μg of NADK2 wild-type or NADK2 variant plasmids. 48 hours post-transfection, cells were washed twice with cold PBS and lysed with 1 mL of CHAPS lysis buffer (0.3 % CHAPS, 40 mM HEPES, pH 7.4, 120 mM NaCl, 10 mM sodium pyrophosphate, 10 mM glycerol 2-phosphate, 50 mM NaF, 1 mM EDTA, 1 μM Microcystin-LR (Enzo life sciences, ALX-350–012-C500) and protease inhibitor cocktail (Sigma, P8340)). Cell lysates were divided into 50 μL aliquots and treated with either 0.5 mM or 1 mM of glutaraldehyde at room temperature for the indicated times. At the end-points, glutaraldehyde was quenched with Tris buffer pH 8.0 (final concentration 100 mM) for 15 minutes at room temperature.
For disuccinimidyl suberate (DSS) (Sigma, S1885) crosslinking, HEK-293E ΔNADK2 cells (2 × 105) plated in 6-well plates in DMEM containing 10 % FBS and 0.2 mM proline were subjected to transient transfection with 2 μg of NADK2 wild-type or NADK2 variants as indicated. After 48 hours post-transfection, cells were collected in 1.5 mL Eppendorf tubes, washed twice with PBS, and incubated for 1 hour on ice with 50 μL of PBS containing 1 mM DSS. Cells were gently vortexed every 10 minutes. DSS was quenched with Tris buffer pH 8.0 (final concentration 100 mM) for 15 minutes at room temperature.
Mass Photometry
His-tagged NADK2 (aa 61–442) or its variants in pET15b plasmid were transformed in BL21 (DE3) competent E. coli cells according to the manufacturer’s instructions. Protein expression was induced with 0.5 mM IPTG, and 500 mL of bacterial cultures were grown for 48 h at 18 °C. Protein purification was performed with a HisTALON™ Gravity Column Purification Kit (TakaraBio, 635654) according to the manufacturer’s instructions. NADK2 or its variants were eluted with 8 mL of the kit’s elution buffer that contains 150 mM imidazole. Different fractions of purified protein were concentrated using Amicon Ultra (EMD Millipore, UFC501024) - 0.5 mL filters. The protein purity was determined by western blotting. NADK2 protein diluted in water at final concentration of 0.1 μM was used for analysis for Mass Photometry (Refeyn TwoMP).
For each experiment microscope coverslips were washed three times with Milli-Q water (followed by two isopropanol washes. The coverslip was then dried with nitrogen stream to remove any liquid. Using a razor blade, gasket wells were cut and put it on the clean coverslip. The protein of interest was pre-prepared as 1 μM stock in PBS and diluted 1:10 in PBS (final 0.1 μM), just before starting the measurements. 2 μL of protein was used for each measurement and movies of 60 seconds were recorded. Data were analyzed using DiscoverMP (Refeyn).
Metabolite profiling and isotopic tracing by high resolution mass spectrometry (HRMS)
Metabolites were extracted with 80 % acetonitrile (prechilled at 4 °C) from nearly ~ 90 % confluent cells. Cells seeded in 24-well plates in biological triplicate or quadruplicates were scraped in 200 μL of 80 % acetonitrile, and subjected to centrifugation (20,000 g, 4 °C, 15 min). Supernatants containing soluble metabolites were run on an Orbitrap Exploris 480 Mass Spectrometer. Peak areas and fractional abundance from each metabolite SRM were integrated using Tracefinder 5.1 (Thermo Scientific). For 13C5-Glutamine isotopic tracing experiments, cells were incubated for 3 hours with glutamine-free DMEM (Thermo, 11-960-044), containing 10 % dialyzed FBS and 2 mM 13C5-Glutamine (Sigma, 605166) prior to metabolite extraction.
For isotopically labeled specimens, the mass spectra of metabolites were extracted and acquired using the Orbitrap Exploris 480 coupled to a Vanquish Duo UHPLC using methods and workflows that were described previously (Su et al., 2017; Tasdogan et al., 2020). Chromatographic separation was achieved with a Millipore ZIC-pHILIC column (5 μm, 2.1 × 150 mm) and a binary solvent system of 10 mM ammonium acetate in water, pH 9.8 (solvent A) and acetonitrile (solvent B) at a constant flow rate of 0.25 mL/min. For gradient separation, the column was equilibrated with 90 % solvent B. After injection, the gradient proceeded as follows: 0–15 min linear ramp from 90 % B to 30 % B; 15–18 min isocratic flow of 30 % B; 18–19 min linear ramp from 30 % B to 90 % B; 19–27 column regeneration with the isocratic flow of 90 % B. HRMS data were acquired with two separate acquisition methods. Individual samples were acquired with an HRMS full scan (precursor ion only) method switching between positive and negative polarities. The AGC target was set to 1 × 106 and a maximum injection time of 50 ms and resolving power of 240,000 full width at half-maximum (FWHM) and a mass range of 80–1,200 Da. For data-dependent tandem mass spectrometry (ddHRMS/MS) methods, precursor ion scans were acquired at a resolving power of 120,000 FWHM with a mass range of 80–1,200 Da. The AGC target value was set to 1 × 106 with a maximum injection time of 100 ms. Pooled samples were generated from an equal mixture of all individual samples and analyzed using individual positive- and negative-polarity spectrometry ddHRMS/MS acquisition methods for high-confidence metabolite ID. Product ion spectra were acquired at a resolving power of 15,000 FWHM without a fixed mass range. The AGC target value was set to 1 × 106 with a maximum injection time of 150 ms. Data-dependent parameters were set to acquire the top 10 ions with a dynamic exclusion of 25 s and a mass tolerance of 5 ppm. Isotope exclusion was turned on and a normalized collision energy of 30 was applied. Settings remained the same in both polarities.
Fractional enrichment was calculated using the theoretical mass of all 13C isotopologues of a metabolite and integrating the resulting peak in the extracted ion chromatogram. We used the following criteria to ensure the correct peaks were integrated for analysis: (1) the precursor ion m/z of the M+0 peak was matched within 5 ppm of the theoretical mass predicted by its chemical formula; (2) the retention time of the M+0 peak was within 5 % of the retention time of a purified chemical standard run with the same chromatographic method; (3) all isotopes of a potentially labeled metabolite were within 5 ppm of their predicted m/z by chemical formula; and (4) all isotopologues eluted simultaneously with the M+0 peak. An unlabeled sample was run alongside all isotopically labeled samples to acquire product ion spectra for additional verification.
Measurement of mitochondrial NAD(P)H by LC-MS/MS
3XHA-EGFP-OMP25 (HA-Mito) (Chen et al., 2016) was stably expressed in the NADK2-deficient cells (ΔNADK2 HEK293E) to generate ΔNADK2-HA-Mito HEK-293E cells (Tran et al., 2021). The latter were plated in 15-cm dishes and subjected to transient transfection with empty vector or NADK2 variants in Lenti-III-PGK plasmids. 48 hours post-transfection, cells were incubated for 4 hours in nicotinamide-free DMEM (US biologicals, D9800–17) containing 10 % dialyzed serum and 0.2 mg/mL 13C3-15N-nicotinamide (M+4) (Cambridge Isotope Laboratories, Inc, CNLM-9757–0.001). Rapid isolation of HA-tagged mitochondria was performed as previously described (Chen et al., 2016). Mitochondria isolated from equal amount of protein for each condition was resuspended in 50 μL of 80 % methanol. Mitochondrial metabolite extracts (20 μL) were directly injected to a QExactive HF-X hybrid quadrupole orbitrap high-resolution mass spectrometer (HRMS; Thermo Fisher Scientific) coupled to a Vanquish ultra-high-pressure liquid chromatography (UHPLC) system. We used a targeted selected ion monitoring (tSIM) scan and a resolving power of 60,000 FWHM and an AGC target of 1 × 105 with a maximum injection time of 100 ms for these scan events. An inclusion list for each M+0 isotopologue’s m/z of interest was created with an isolation window of 5 Da and an isolation offset of 1 Da for detection of the M+4 isotopologues (tracing from 3,13C −1,15N nicotinamide). Data analysis was performed with Tracefinder 5.1 (ThermoScientific).
PTM analysis by Mass Spectrometry
HEK-293E cells (8 × 15 cm dishes) were transiently transfected with NADK2-FLAG (20 μg/15 cm dish) and grown in normal growth media (DMEM with 10 % FBS). 48 hours post transfection, cells were subjected to lysis with 1 % Triton and NADK2-FLAG was immunoprecipitated with anti-FLAG M2 Affinity Gel (Sigma, A2220) for 4 hours (Hoxhaj et al., 2019). Beads were washed 4 times with lysis buffer and NADK2-FLAG was eluted with elution buffer (0.2 mg/mL 3xFLAG peptide, 10 mM HEPES, 50 mM NaCl, protease inhibitors and 1 μM Microcystin) for 1 hour at 4 °C. NADK2-FLAG eluate was subjected to Commassie Blue staining (Abcam, ab119211) and the NADK2 band was excised for PTM analysis.
Samples were digested overnight with trypsin (Pierce) following reduction and alkylation with DTT and iodoacetamide (Sigma–Aldrich). The samples then underwent solid-phase extraction cleanup with an Oasis MCX plate (Waters) and the resulting samples were injected onto an Orbitrap Fusion Lumos mass spectrometer coupled to an Ultimate 3000 RSLC-Nano liquid chromatography system. Samples were injected onto a 75 um i.d., 75-cm long EasySpray column (Thermo) and eluted with a gradient from 0–28 % buffer B over 90 min. Buffer A contained 2 % (v/v) ACN and 0.1 % formic acid in water, and buffer B contained 80 % (v/v) ACN, 10 % (v/v) trifluoroethanol, and 0.1 % formic acid in water. The mass spectrometer operated in positive ion mode with a source voltage of 2.2 kV and an ion transfer tube temperature of 275 °C. MS scans were acquired at 120,000 resolution in the Orbitrap and up to 10 MS/MS spectra were obtained in the ion trap for each full spectrum acquired using higher-energy collisional dissociation (HCD) for ions with charges 2–7. Dynamic exclusion was set for 25 s after an ion was selected for fragmentation.
Raw MS data files were analyzed using Proteome Discoverer v2.4 SP1 (Thermo), with peptide identification performed using Sequest HT searching against the mouse protein database from UniProt. Fragment and precursor tolerances of 10 ppm and 0.6 Da were specified, and three missed cleavages were allowed. Carbamidomethylation of Cys was set as a fixed modification, with oxidation of Met, phosphorylation of Ser, Thr, and Tyr, and acetylation and succinylation of Lys set as a variable modification. The false-discovery rate (FDR) cutoff was 1 % for all peptides.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis was performed using GraphPad Prism 8.4.3 software. For pairwise comparisons, two-tailed Student’s t-tests were used, and for multiple comparisons, one-way ANOVA test with Tukey’s post-hoc test. All error bars represent standard deviation (SD). All experimental data are representative of at least two independent experiments.
Supplementary Material
Highlights.
Structure of NADK2-NADP+-metal complex reveals a substrate-driven mode of catalysis
NADK2 displays a dimeric organization, with NAD+ substrate at the dimer interface
The extended loop (aa 325–365) is critical for dimerization and activity of NADK2
K76 and K304 acetylation impair NADK2 activity, proline synthesis, and cell growth
ACKNOWLEDGMENTS
We thank Amin Sagar for recording the SAXS data and the staff at the BioSAXS BM29 ESRF (Grenoble, France) for their help. We also acknowledge the staff of the ID30B beamline at the ESRF for the X-ray diffraction experiments. We also thank Chad Brautigam for his assistance with mass photometry, Feng Cai and Hieu Vu for assistance with MS analysis, and the UT Southwestern Proteomics Core for the help with the PTM analysis. The authors also acknowledge the UT Southwestern Quantitative Light Microscopy Core, a Shared Resource of the Harold C. Simmons Cancer Center, which is supported in part by an NCI Cancer Center Support grant (1P30 CA142543-01). Some data in this study were acquired with a mass photometer that was supported by award S10OD030312-01 from the National Institutes of Health. This research was supported by grants from the NIH: R01GM143236 (G.H.), a Welch foundation award (I-2067-20210327 [G.H.]), and a TS Alliance Research Grants Program award (885252 [G.H.]). C.M., G.L., and C.L. were supported from grants from the Agence Nationale de la Recherche NADKiller contract (ANR-17-CE18-0011-01) and Cov-H2L contract (ANR-20-CE18-25). G.H. is a recipient of a CPRIT Scholar (CPRIT; RR190087) and a V Scholar (V2021-019) award.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.molcel.2022.06.026.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). Phenix: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando T, Ohashi K, Ochiai A, Mikami B, Kawai S, and Murata K (2011). Structural determinants of discrimination of NAD+ from NADH in yeast mitochondrial NADH kinase Pos5. J. Biol. Chem 286, 29984–29992. 10.1074/jbc.M111.249011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrico C, Meyer JG, He W, Gibson BW, and Verdin E (2018). The mitochondrial acylome emerges: proteomics, regulation by sirtuins, and metabolic and disease implications. Cell Metab 27, 497–512. 10.1016/j.cmet.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404. 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandel NS (2021). NADPH-the forgotten reducing equivalent. Cold Spring Harb. Perspect. Biol 13, a040550. 10.1101/cshperspect.a040550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WW, Freinkman E, and Sabatini DM (2017). Rapid immunopurification of mitochondria for metabolite profiling and absolute quantification of matrix metabolites. Nat. Protoc 12, 2215–2231. 10.1038/nprot.2017.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WW, Freinkman E, Wang T, Birsoy K, and Sabatini DM (2016). Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism. Cell 166, 1324–1337.e11. 10.1016/j.cell.2016.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeBerardinis RJ, and Chandel NS (2016). Fundamentals of cancer metabolism. Sci. Adv 2, e1600200. 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K (2010). Features and development of coot. Acta Crystallogr. D Biol. Crystallogr 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Ye J, Kamphorst JJ, Shlomi T, Thompson CB, and Rabinowitz JD (2014). Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510, 298–302. 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. (2013). Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6, pl1. 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garavaglia S, Raffaelli N, Finaurini L, Magni G, and Rizzi M (2004). A novel fold revealed by Mycobacterium tuberculosis NAD kinase, a key allosteric enzyme in NADP biosynthesis. J. Biol. Chem 279, 40980–40986. 10.1074/jbc.M406586200. [DOI] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, and Skrzypek E (2015). PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res 43, D512–D520. 10.1093/nar/gku1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houten SM, Denis S, Te Brinke H, Jongejan A, van Kampen AH, Bradley EJ, Baas F, Hennekam RC, Millington DS, Young SP, et al. (2014). Mitochondrial NADP(H) deficiency due to a mutation in NADK2 causes dienoyl-CoA reductase deficiency with hyperlysinemia. Hum. Mol. Genet 23, 5009–5016. 10.1093/hmg/ddu218. [DOI] [PubMed] [Google Scholar]
- Hoxhaj G, Ben-Sahra I, Lockwood SE, Timson RC, Byles V, Henning GT, Gao P, Selfors LM, Asara JM, and Manning BD (2019). Direct stimulation of NADP(+) synthesis through Akt-mediated phosphorylation of NAD kinase. Science 363, 1088–1092. 10.1126/science.aau3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawabata Y, Murata K, and Kawai S (2015). Significance of Ser-188 in human mitochondrial NAD kinase as determined by phosphomimetic and phosphoresistant amino-acid substitutions. Biochem. Biophys. Res. Commun 468, 691–695. 10.1016/j.bbrc.2015.11.017. [DOI] [PubMed] [Google Scholar]
- Korb O, Stützle T, and Exner TE (2009). Empirical scoring functions for advanced protein-ligand docking with PLANTS. J. Chem. Inf. Model 49, 84–96. 10.1021/ci800298z. [DOI] [PubMed] [Google Scholar]
- Krissinel E, and Henrick K (2007). Inference of macromolecular assemblies from crystalline state. J. Mol. Biol 372, 774–797. 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Labesse G, Douguet D, Assairi L, and Gilles AM (2002). Diacylglyceride kinases, sphingosine kinases and NAD kinases: distant relatives of 6-phosphofructokinases. Trends Biochem. Sci 27, 273–275. 10.1016/s0968-0004(02)02093-5. [DOI] [PubMed] [Google Scholar]
- Lánczky A, and Győrffy B (2021). Web-based survival analysis tool tailored for medical research (KmPlot): development and implementation. J. Med. Internet Res 23, e27633. 10.2196/27633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner F, Niere M, Ludwig A, and Ziegler M (2001). Structural and functional characterization of human NAD kinase. Biochem. Biophys. Res. Commun 288, 69–74. 10.1006/bbrc.2001.5735. [DOI] [PubMed] [Google Scholar]
- Lewis CA, Parker SJ, Fiske BP, McCloskey D, Gui DY, Green CR, Vokes NI, Feist AM, Vander Heiden MG, and Metallo CM (2014). Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol. Cell 55, 253–263. 10.1016/j.molcel.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Dash BP, and Kumar S (2015). Acetyl-ed question in mitochondrial biology? EMBO J 34, 2597–2600. 10.15252/embj.201592927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love NR, Pollak N, Dölle C, Niere M, Chen Y, Oliveri P, Amaya E, Patel S, and Ziegler M (2015). NAD kinase controls animal NADP biosynthesis and is modulated via evolutionarily divergent calmodulin-dependent mechanisms. Proc. Natl. Acad. Sci. USA 112, 1386–1391. 10.1073/pnas.1417290112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manalastas-Cantos K, Konarev PV, Hajizadeh NR, Kikhney AG, Petoukhov MV, Molodenskiy DS, Panjkovich A, Mertens HDT, Gruzinov A, Borges C, et al. (2021). ATSAS 3.0: expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Crystallogr 54, 343–355. 10.1107/S1600576720013412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaco S, Gordon E, Bowler MW, Delagenière S, Guijarro M, Spruce D, Svensson O, McSweeney SM, McCarthy AA, Leonard G, et al. (2013). Automatic processing of macromolecular crystallography X-ray diffraction data at the ESRF. J. Appl. Crystallogr 46, 804–810. 10.1107/S0021889813006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Kawai S, Shi F, Mikami B, and Murata K (2005a). Molecular conversion of NAD kinase to NADH kinase through single amino acid residue substitution. J. Biol. Chem 280, 24104–24112. 10.1074/jbc. [DOI] [PubMed] [Google Scholar]
- Mori S, Yamasaki M, Maruyama Y, Momma K, Kawai S, Hashimoto W, Mikami B, and Murata K (2005b). NAD-binding mode and the significance of intersubunit contact revealed by the crystal structure of Mycobacterium tuberculosis NAD kinase-NAD complex. Biochem. Biophys. Res. Commun 327, 500–508. 10.1016/j.bbrc.2004.11.163. [DOI] [PubMed] [Google Scholar]
- Ohashi K, Kawai S, and Murata K (2012). Identification and characterization of a human mitochondrial NAD kinase. Nat. Commun 3, 1248. 10.1038/ncomms2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak N, Dölle C, and Ziegler M (2007). The power to reduce: pyridine nucleotides–small molecules with a multitude of functions. Biochem. J 402, 205–218. 10.1042/BJ20061638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz DJ, Ferdinandusse S, Cogan J, Cooper DN, Reimschisel T, Robertson A, Bican A, McGregor T, Gauthier J, Millington DS, et al. (2018). Clinical heterogeneity of mitochondrial NAD kinase deficiency caused by a NADK2 start loss variant. Am. J. Med. Genet. A 176, 692–698. 10.1002/ajmg.a.38602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poncet-Montange G, Assairi L, Arold S, Pochet S, and Labesse G (2007). NAD kinases use substrate-assisted catalysis for specific recognition of NAD. J. Biol. Chem 282, 33925–33934. 10.1074/jbc.M701394200. [DOI] [PubMed] [Google Scholar]
- Rabani R, Cossette C, Graham F, and Powell WS (2020). Protein kinase C activates NAD kinase in human neutrophils. Free Radic. Biol. Med 161, 50–59. 10.1016/j.freeradbiomed.2020.09.022. [DOI] [PubMed] [Google Scholar]
- Schild T, McReynolds MR, Shea C, Low V, Schaffer BE, Asara JM, Piskounova E, Dephoure N, Rabinowitz JD, Gomes AP, and Blenis J (2021). NADK is activated by oncogenic signaling to sustain pancreatic ductal adenocarcinoma. Cell Rep 35, 109238. 10.1016/j.celrep.2021.109238. [DOI] [PubMed] [Google Scholar]
- Shirakihara Y, and Evans PR (1988). Crystal structure of the complex of phosphofructokinase from Escherichia coli with its reaction products. J. Mol. Biol 204, 973–994. 10.1016/0022-2836(88)90056-3. [DOI] [PubMed] [Google Scholar]
- Stram AR, and Payne RM (2016). Post-translational modifications in mitochondria: protein signaling in the powerhouse. Cell. Mol. Life Sci 73, 4063–4073. 10.1007/s00018-016-2280-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Lu W, and Rabinowitz JD (2017). Metabolite spectral accuracy on orbitraps. Anal. Chem 89, 5940–5948. 10.1021/acs.anal-chem.7b00396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasdogan A, Faubert B, Ramesh V, Ubellacker JM, Shen B, Solmonson A, Murphy MM, Gu Z, Gu W, Martin M, et al. (2020). Metabolic heterogeneity confers differences in melanoma metastatic potential. Nature 577, 115–120. 10.1038/s41586-019-1847-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran DH, Kesavan R, Rion H, Soflaee MH, Solmonson A, Bezwada D, Vu HS, Cai F, Phillips JA 3rd, DeBerardinis RJ, and Hoxhaj G (2021). Mitochondrial NADP(+) is essential for proline biosynthesis during cell growth. Nat. Metab 3, 571–585. 10.1038/s42255-021-00374-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinci M, Box C, and Eccles SA (2015). Three-dimensional (3D) tumor spheroid invasion assay. J. Vis. Exp e52686. 10.3791/52686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Kim H, Fu Z, Qiu Y, Yang Z, Wang J, Zhang D, Tong X, Yin L, Li J, et al. (2018). Deficiency of the mitochondrial NAD kinase causes stress-induced hepatic steatosis in mice. Gastroenterology 154, 224–237. 10.1053/j.gastro.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Schwörer S, Berisa M, Kyung YJ, Ryu KW, Yi J, Jiang X, Cross JR, and Thompson CB (2021). Mitochondrial NADP(H) generation is essential for proline biosynthesis. Science 372, 968–972. 10.1126/science.abd5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Original western blot and microscopy images have been deposited at Mendeley and are publicly available as of the date of publication. Validation reports of X-ray crystallography structures have been deposited at the PDB, and the refined models have been deposited in the Research Collaboratory for Structural Biology (http://www.rcsb.org) under the following accession numbers: 7R4J; 7R4K, 7R4L, and 7R4M, which are publicly available as of the date of publication. The DOIs and accession numbers are listed in the key resources table. Mass spectrometry data are presented in the Table S1.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| NADK2 Antibody | Abcam | Cat# ab181028 Lot# GR164717-1 |
| Anti-rabbit-secondary antibody | Cell Signaling Technology | Cat#7074 Lot# 29 |
| HRP-conjugated anti-mouse-secondary antibody | Cell Signaling Technology | Cat#7076 Lot# 34 |
| β-actin Antibody | Sigma-Aldrich | A5316 Lot# 059M4770V |
| Bacterial and virus strains | ||
| NEB Stable Competent E. coli | NEB | C3040 |
| BL21(DE3) Competent E. coli | NEB | C2527 |
| Chemicals, peptides, and recombinant proteins | ||
| DMSO | Sigma-Aldrich | D2650-100ML |
| Proline | Sigma-Aldrich | P5607-25G |
| 13C5 glutamine | Sigma-Aldrich | 605166-500MG |
| NEB-10-beta/Stable Outgrowth Media | NEB | B9035S |
| Crystal violet | Sigma-Aldrich | C6158-50G |
| Trypab Blue Soloution | Sigma-Aldrich | T8154-100ML |
| L-glutamine | Corning | MT25005Cl |
| Puromycin dichloride | Santa Cruz | sc-108071A |
| Polyethylenimine (PEI) | Polysciences | 23966-2 |
| Opti-MEM media | Thermo Fisher | 31985062 |
| Glutamine-free DMEM | Gibco | 11960-044 |
| SuperSignal™ West Femto Chemiluminescent | Thermo Fisher scientific | PI34096 |
| SuperSignal™ West Pico PLUS | Thermo Fisher scientific | PI34580 |
| DMEM | Corning CellGro | 10-017-CV |
| Fetal bovine serum (FBS) | R&D Systems a biotech brand | S11150 |
| Fetal Bovine Serum dialyzed | R&D Systems a biotech brand | S12850 |
| Microcystin-LR | Enzo Life Sciences | NC9580520 |
| Protease inhibitor cocktail | Sigma-Aldrich | P8340-5ML |
| Acrylamide Protogel 30% | Fisher Scientific | EC-890 |
| Bio-Rad Braford | Bio-Rad | 5000006 |
| Low melting Agarose | Lonza | 50101 |
| Tryptone | United States Biological Life sciences | 91079-40-2 |
| Yeast extract | Euromedex | UY2010 |
| Sodium Chloride | Euromedex | 1112-A |
| Tris Ultra Pure | Euromedex | 26-128-3094-B |
| Imidazole | Millipore | 288-32-4 |
| DTT | Euromedex | EU0006 |
| IPTG | Euromedex | EU0018 |
| InstantBlue® Coomassie Protein Stain | abcam | ab119211 |
| Complete™, EDTA-free Protease Inhibitor cocktail | Roche® Life Science Products | 05056489001 |
| Ampicillin | Euromedex | EU0400 |
| Chloramphenicol | Euromedex | 3886 |
| Tris Ultra Pure | Euromedex | 26-128-3094-B |
| NaCl | Euromedex | 1112-A |
| Sodium citrate tribasic dihydrate | Sigma-Aldrich | S4641 |
| Manganese(II) chloride tetrahydrate | Sigma-Aldrich | M8054 |
| Magnesium chloride hexahydrate | Sigma-Aldrich | M2670 |
| Calcium chloride | Sigma-Aldrich | C1016 |
| Cobalt(II) chloride | Sigma-Aldrich | 60818 |
| Cadmium chloride | Sigma-Aldrich | 202908 |
| Nickel(II) chloride hexahydrate | Sigma-Aldrich | 72247 |
| Copper(II) chloride dihydrate | Sigma-Aldrich | C6641 |
| Zinc chloride | Sigma-Aldrich | 31650-M |
| Barium chloride | Sigma-Aldrich | 202738 |
| Cadmium chloride | Sigma-Aldrich | 202908 |
| Iron(III) chloride | Sigma-Aldrich | F7134 |
| Aluminum chloride hexahydrate | Sigma-Aldrich | A0718 |
| NAD+ | Roche | 10127973001 |
| NADP+ disodium salt | Roche | 10128058001 |
| Adenosine 5′-triphosphate disodium salt hydrate | Sigma-Aldrich | A7699 |
| Sodium polyphosphate | Supelco | 1.06529 |
| D-Glucose 6-phosphate sodium salt | Sigma-Aldrich | G7879 |
| Glucose-6-phosphate Dehydrogenase from baker’s yeast (S. cerevisiae) | Sigma-Aldrich | G7877 |
| Critical commercial assays | ||
| HiSpeed Plasmid Maxi Kit (25) | Qiagen | 12663 |
| His TALON™ Buffer Set | TAKARA | 635651 |
| His TALON™ Gravity Column | TAKARA | 635655 |
| ZymoPURETM I Plasmid Midi Prep Kit | Zymo Research | D4201-B |
| Deposited data | ||
| NADK2 crystal structures | PDB IDs/accession numbers | 7R4J, 7R4K, 7R4L, 7R4M |
| A study by Mary and Soflaee et al., unprocessed files | Mendeley Data | https://doi.org/10.17632/8ybbnh7g3p.1 |
| Experimental models: Cell lines | ||
| HEK293E | Carol Mackintosh (University of Dundee) | N/A |
| ΔNADK2 HEK293E | Tran et al., 2021 | N/A |
| ΔNADK2 HA-Mito | Tran et al., 2021 | N/A |
| ΔNADK2 HEK293E +EV | This study | N/A |
| ΔNADK2 HEK293E +NADK2 | This study | N/A |
| ΔNADK2 HEK293E +Δ78-114-Flag | This study | N/A |
| ΔNADK2 HEK293E +Δ231-266 | This study | N/A |
| ΔNADK2 HEK293E +Δ325-365 | This study | N/A |
| ΔNADK2 HEK293E +S188A | This study | N/A |
| ΔNADK2 HEK293E +T229A | This study | N/A |
| ΔNADK2 HEK293E +S289A | This study | N/A |
| ΔNADK2 HEK293E +S376A | This study | N/A |
| ΔNADK2 HEK293E +K76Q | This study | N/A |
| ΔNADK2 HEK293E +K98Q | This study | N/A |
| ΔNADK2 HEK293E +K104Q | This study | N/A |
| ΔNADK2 HEK293E +K170Q | This study | N/A |
| ΔNADK2 HEK293E +K176Q | This study | N/A |
| ΔNADK2 HEK293E +K304Q | This study | N/A |
| ΔNADK2 HEK293E +K317Q | This study | N/A |
| ΔNADK2 HEK293E +K339Q | This study | N/A |
| ΔNADK2 HEK293E +K355Q | This study | N/A |
| ΔNADK2 HEK293E +K397Q | This study | N/A |
| ΔNADK2 HEK293E +V334R | This study | N/A |
| ΔNADK2 HEK293E +R378Q | This study | N/A |
| ΔNADK2 HEK293E +V334R-R378Q | This study | N/A |
| Recombinant DNA | ||
| pLenti III NADK2-Flag | Tran et al., 2021 | N/A |
| pLenti III NADK2-NADK2-Flag | This study | N/A |
| pLenti III NADK2-Δ78-114-Flag | This study | N/A |
| pLenti III NADK2-Δ231-266 | This study | N/A |
| pLenti III NADK2-Δ325-365 | This study | N/A |
| pLenti III NADK2-S188A-Flag | This study | N/A |
| pLenti III NADK2-T229A-Flag | This study | N/A |
| pLenti III NADK2-S289A-Flag | This study | N/A |
| pLenti III NADK2-S376A-Flag | This study | N/A |
| pLenti III NADK2-K76Q-Flag | This study | N/A |
| pLenti III NADK2-K98Q-Flag | This study | N/A |
| pLenti III NADK2-K104Q-Flag | This study | N/A |
| pLenti III NADK2-K170Q-Flag | This study | N/A |
| pLenti III NADK2-K176Q-Flag | This study | N/A |
| pLenti III NADK2-K304Q-Flag | This study | N/A |
| pLenti III NADK2-K317Q-Flag | This study | N/A |
| pLenti III NADK2-K339Q-Flag | This study | N/A |
| pLenti III NADK2-K355Q-Flag | This study | N/A |
| pLenti III NADK2-K397Q-Flag | This study | N/A |
| pLenti III NADK2-V334R-Flag | This study | N/A |
| pLenti III NADK2-R378Q-Flag | This study | N/A |
| pLenti III NADK2-V334R-R378Q-Flag | This study | N/A |
| pET15b Δ1-60 NADK2 | This study | N/A |
| pET15b Δ1-60 NADK2-Δ78-114 | This study | N/A |
| pET15b Δ1-60 NADK2-Δ231-266 | This study | N/A |
| pET15b Δ1-60 NADK2-Δ325-365 | This study | N/A |
| pET15b Δ1-60 NADK2-S188A | This study | N/A |
| pET15b Δ1-60 NADK2-K76Q | This study | N/A |
| pET15b Δ1-60 NADK2-K304Q | This study | N/A |
| pET15b Δ1-60 NADK2-V334R | This study | N/A |
| pET15b Δ1-60 NADK2-R378Q | This study | N/A |
| pET15b Δ1-60 NADK2-V334R/R378Q | This study | N/A |
| Software and algorithms | ||
| Discover MP 2.4.2 | Refeyn | https://www.refeyn.com/software-release-updates |
| Prism | Prism | https://www.graphpad.com/scientific-software/prism/ |
| TraceFinder 5.1 | ThermoScientific | https://www.thermofisher.com/us/en/home/industrial/mass-spectrometry/liquid-chromatography-mass-spectrometry-lc-ms/lc-ms-software/lc-ms-data-acquisition-software/tracefinder-software.html |
| ASTRA 6.1 | Waytt Technology | https://www.wyatt.com/products/software/astra.html |
| GraFit | Erithacus Software Ltd | v. 7.0.3 |
| Other | ||
| Mass Photometry TwoMP | Refeyn | N/A |
| SpectraMax iD3- Microplate reader | Molecular devices | N/A |
| Nitrocellulose membrane 0.2 μm | GE Healthcare | 10600001 |
| Blotting paper, 703 | VWR North America | 732-0591 |
| LUNA-II™ Automated Cell Counter | LUNA-II™ | N/A |
| Celigo image cytometer-4 Channel | Nexcelom Bioscience,UK | N/A |
| QExactive HF-X Orbitrap Mass Spectrometer | ThermoScientific | N/A |
| Orbitrap Exploris 480 Mass Spectromete | ThermoScientific | N/A |
| Amicon Ultra-15, PLTK, membrane Ultracel-PL, 30 kDa | Millipore | UFC903024 |
| Complete™ His-Tag Purification Column | Roche® Life Science Products | 06781535001 |
| Bolt™ 4 à 12 %, Bis-Tris | Invitrogen™ | NW04125BOX |
| CLARIOstar plate reader BMG LABTECH | BMG LABTECH | UV-Star® Half area |
| 96-well plates | Greiner | 655087 |


