Summary
Plants mount coordinated immune responses to defend against pathogens. However, the cellular components required for plant immunity are not fully understood. The jasmonate-mimicking coronatine (COR) toxin produced by Pseudomonas syringae pv. tomato (Pst) DC3000 functions to overcome plant immunity. We previously isolated eight Arabidopsis (scord) mutants that exhibit increased susceptibility to a COR-deficient mutant of Pst DC3000. Among them, the scord6 mutant exhibits defects both in stomatal closure response and in restricting bacterial multiplication inside the apoplast. However, the identity of SCORD6 remained elusive.
In this study, we aim to identify the SCORD6 gene.
We identified SCORD6 via next-generation sequencing and found it to be MURUS1 (MUR1), which is involved in the biosynthesis of GDP-L-fucose.
Discovery of SCORD6 as MUR1 led to a series of experiments that revealed a multi-faceted role of L-fucose biosynthesis in stomatal and apoplastic defenses as well as in pattern-triggered immunity and effector-triggered immunity, including glycosylation of pattern-recognition receptors. Furthermore, compromised stomatal and/or apoplastic defenses were observed in mutants of several fucosyltransferases with specific substrates (e.g., O-glycan, N-glycan or the DELLA transcriptional repressors). Collectively, these results uncover a novel and broad role of L-fucose and protein fucosylation in plant immuinity.
Keywords: coronatine, fucosylation, jasmonate, L-fucose, plant immunity, stomata
Introduction
In nature, plants are exposed to a wide variety of microbes, including pathogens. To protect against pathogen attacks, plants have developed various defense strategies, including preformed physical barriers (e.g., cuticles and cell walls) and antimicrobial compounds, as well as an inducible innate immune system (Jones & Dangl 2006; Bigeard et al. 2015). Research on Arabidopsis immune responses suggests that the inducible plant innate immune system consists of two signaling branches. The first branch involves recognition of conserved pathogen/microbe-associated molecular patterns (PAMPs/MAMPs) by pattern-recognition receptors (PRRs), resulting in pattern-triggered immunity (PTI) (Macho & Zipfel 2014). For example, perception of bacterial flagellin or a highly conserved 22 amino acid epitope, flg22, is mediated by the FLAGELLIN-SENSITIVE2 (FLS2) receptor (Gomez-Gomez & Boller 2000; Chinchilla et al. 2006). Similarly, elongation factor-Tu (EF-Tu) or a 26-amino acid epitope of EF-Tu, elf26, is recognized by the EF-Tu receptor (EFR) (Kunze et al. 2004; Zipfel et al. 2006). The second branch of inducible innate immunity, designated effector-triggered immunity (ETI), is triggered by direct or indirect recognition of pathogen virulence proteins (“effectors”) by cognate disease resistance (R) proteins (Cui et al. 2015). For example, the effector protein AvrRpt2 from the bacterial pathogen Pseudomonas syringae pv. tomato (Pst) is indirectly perceived by the RESISTANCE TO P. syringae 2 (RPS2) protein and activates downstream ETI signaling and defense outputs, including the hypersensitive cell death response (Axtell & Staskawicz 2003; Mackey et al. 2003).
As foliar pathogens, strains of P. syringae often go through an initial epiphytic phase on the surfaces of the above-ground parts (collectively called the phyllosphere) of plants and a subsequent endophytic phase inside the apoplast (intercellular spaces between mesophyll cells), where aggressive multiplication takes place (Melotto et al. 2008; Xin et al. 2018). The transition from the epiphytic phase to endophytic phase involves the entry of P. syringae into the plant apoplast via wounds or natural openings, such as stomata, in the epidermis (Hirano & Upper 2000). In response, plants have evolved elaborate defense responses to counter bacteria during invasion through stomata and inside the apoplast. As the first line of defense, plants respond to live bacteria or MAMPs (such as flg22 and elf26) by closing stomatal aperture (Melotto et al. 2006; Arnaud & Hwang 2015; Ye & Murata 2016). Specific PRRs are necessary for plants to sense MAMPs and induce stomatal closure (Melotto et al. 2006; Zhang et al. 2008; Mersmann et al. 2010). In addition, salicylic acid (SA) biosynthesis and signaling are also required for pathogen-induced stomatal closure (Arnaud & Hwang 2015; Ye & Murata 2016; Melotto et al. 2017). Moreover, both MAMP perception/ signaling and SA biosynthesis/signaling are also important for plant defense against bacterial multiplication inside the apoplast (Heese et al. 2007; Genger et al. 2008; Nekrasov et al. 2009; Boutrot et al. 2010; Zeng & He 2010; Zeng et al. 2011; Uddin et al. 2017).
The phytotoxin coronatine (COR) plays multiple roles in Pst DC3000 infection of Arabidopsis and is implicated in overcoming both stomatal and apoplastic defenses to facilitate bacterial invasion and multiplication, respectively (Zeng et al. 2011). COR is a remarkable structural mimic of the active form of plant hormone jasmonoyl-isoleucine (JA-Ile), and its virulence roles appear to be mediated through activating jasmonate (JA) signaling (Zhang et al. 2017) and other mechanisms (Kim et al. 2017). COR interferes with MAMP-induced stomatal closure and triggers stomatal reopening (Melotto et al. 2006; Zheng et al. 2012; Montillet et al. 2013; Melotto et al. 2017). COR also suppresses ABA-, oxylipin- and dark-induced stomatal closure (Melotto et al. 2006; Montillet et al. 2013; Panchal et al. 2016). In addition, COR is required for Pst DC3000 multiplication in the apoplast, as demonstrated by infiltration-based inoculation that delivers bacteria directly into the apoplast, bypassing stomatal defense (Brooks et al. 2004; Zeng & He 2010). Importantly, research indicates that COR suppresses stomatal closure and apoplastic defense through antagonizing SA signaling (Brooks et al. 2005; Melotto et al. 2006; Zeng & He 2010; Zheng et al. 2012).
The multiple roles of COR in Pst DC3000 infection was also demonstrated in an unbiased genetic screen by Zeng and colleagues for Arabidopsis mutants with increased susceptibility to COR-deficient Pst DC3000 (scord) (Zeng et al. 2011). Of the eight scord mutants, two were affected only in stomatal response, two were affected only in apoplastic defense, and the other four, including scord6, were affected in both. In this study, we identified the SCORD6 gene. SCORD6 encodes an isoform of GDP-D-mannose-4,6-dehydratase (GMD), GMD2 (also named MURUS1 [MUR1]; Reiter et al. 1993), which has been shown to be involved in the de novo synthesis of GDP-L-fucose, but has not been reported to play a role in plant-bacterial interactions. We found that a defect in L-fucose synthesis has a broad effect on multiple aspects of plant defense, including stomatal defense, apoplastic defense, PTI and ETI. Further analysis of known alleles of SCORD6 as well as mutants of specific fucosyltransferases showed a requirement of intact N-glycans, O-glycans and mono-O-fucosylated proteins for proper plant immune responses. Overall, the identification of the SCORD6 gene highlights an important and previously underappreciated role of L-fucose biosynthesis and fucosylation of polysaccharides and proteins in multiple immune responses in Arabidopsis.
Materials and methods
Plant materials and growth condition
Arabidopsis mutants were obtained from the Arabidopsis Biological Resource Center (Alonso et al. 2003), including mur1-1 (CS6243) (Reiter et al. 1993; Bonin et al. 1997), mur2-1 (CS8565) (Reiter et al. 1997; Vanzin et al. 2002), fut4 (SAIL_284_B05) (Liang et al. 2013), fut6 (SALK_099500) (Liang et al. 2013), cgl1-3 (SALK_073650) (Frank et al. 2008), fucTa (SALK_087481) (Strasser et al. 2004), fucTb (SALK_063355) (Strasser et al. 2004), fut13-2 (SALK_067444) (Anderson et al. 2012), stt3a-2 (SALK_058814) (Koiwa et al. 2003), stt3b-1 (SALK_033391) (Koiwa et al. 2003), spy-3 (CS6268) (Jacobsen & Olszewski 1993) and spy-5 (CS8094) (Wilson & Somerville 1995). Arabidopsis plants used for stomatal closure assays and bacterial infection assays were grown under 12-h light/12-h dark photoperiod at 22-23 °C, 100 μmol m−2 s−1 and ~50% humidity for 4 to 5 weeks.
Identification of the SCORD6 gene
Genomic DNA of the scord6 mutant and the parental Col-7 plant was extracted using PowerPlant Pro DNA Isolation Kit (Mo-Bio) and sent to the Michigan State University (MSU) Research Technology Support Facility (RTSF) Genomic Core for paired-end sequencing using Illumina’s HiSeq 2500 next-generation sequencer. A total of ~3 Gbp genome sequences for each sample were obtained and the coverage was 21- and 26-fold for the scord6 mutant and Col-7 wild-type, respectively. The read quality control was assessed using FastQC (Andrew 2010). The reads of Col-7 and the scord6 mutant were assembled based on Col-0 TAIR10 genome sequence and the sequence of the activation-tagging vector pSKI015 (Weigel et al. 2000) using Bowtie2 (Langmead & Salzberg 2012). Sequence variations including SNPs and INDELs were identified using SAMtools and VCFtools (H. Li et al. 2009; Danecek et al. 2011). Homozygous variations (QUAL value >= 30) detected only in the scord6 genome, but not in the parental Col-7 genome, were selected and mapped back to the previously identified candidate regions by physical mapping (Zeng et al. 2011). Only nonsynonymous SNPs or INDELs located within the open-reading frame of a gene in the candidate regions of the scord6 genome were selected and confirmed by Integrative Genomics Viewer (IGV) (Robinson et al. 2011) and PCR. Primers used in targeted PCR for confirming the presence of deletion in scord6 mutant were described in Supplemental Table 1.
Generation of transgenic Arabidopsis
The coding DNA sequence (CDS) of the MUR1 (AT3G51160) gene was amplified from a total mRNA extract of Col-0 leaf tissue and cloned into the donor vector pDONR207 via BP Clonase II (Invitrogen), generating the entry clone. The entry clone was then recombined with the destination vector pGWB517 (Nakagawa et al. 2007) using LR Clonase II (Invitrogen) to create 35Spro:MUR1-4xMyc. The confirmed construct was introduced into Agrobacterium tumefaciens (GV3101) by electroporation. 35Spro:MUR1-4xMyc construct was then transformed into the scord6 mutant via A. tumefaciens-mediated Arabidopsis transformation (Clough & Bent 1998). Half-strength Murashige and Skoog (MS) medium with 50 μg ml−1 hygromycin was used to select transgenic seedlings expressing the 35Spro:MUR1-4xMyc transgene.
Bacterial infection assays
P. syringae infection assays in Arabidopsis were performed as described previously (Yao et al. 2013). Briefly, four- to five-week-old Arabidopsis plants were dip-inoculated with bacterial suspension (1 × 108 cfu ml−1 Pst DC3118 [OD600 = 0.1; Spectronic 20D+, Thermo Scientific] in 0.25 mM MgCl2 containing 0.025% Silwet-77 solution) or syringe-infiltrated with a bacterial suspension (Pst DC3118 [1 × 105 ~ 1 × 106 cfu ml−1] or Pst DC3000 (avrRpt2) [1 × 106 cfu ml−1] in 0.25 mM MgCl2 solution). Bacterial populations were determined by serial dilutions of plant homogenates after two or three days post-inoculation. A total of 12 leaf discs (diameter of 4 mm) from three fully expanded leaves of one plant were collected as one biological replicate. For MAMP-induced protection assays, plant leaves were syringe-infiltrated with a 0.1 μM flg22 (EZBiolab), 0.5 μM elf26 (EZBiolab), or 0.1% DMSO (mock) for 22-24 hours followed by infiltration of bacterial suspension (1 × 106 cfu ml−1 Pst DC3000 in 0.25 mM MgCl2 solution).
To assess bacterial numbers at day 0 in various mutant plants, bacterial populations were recorded 1 hour after infiltration-inoculation with 5 × 105 cfu ml−1 Pst DC3118 (Fig. S1)
Stomatal closure assays
Leaf discs (~3 mm × 3 mm) were collected one hour after lights turned on in the growth chamber and were submerged in MES buffer (25mM MES-KOH pH6.15, 10mM KCl) with 100 μM SA (Sigma-Aldrich), 10 μM ABA (Sigma-Aldrich) or 0.1% DMSO (mock) for one hour. For measuring pathogen-induced stomatal closure, leaf discs were submerged in water or bacterial suspension (1x108 cfu ml−1 Pst DC3118 in water) for two hours. Stomatal apertures were captured using Olympus FluoView 1000 Spectral-based Laser Scanning Confocal Microscope with excitation/emission at 405/460 nm. The length and width of the pore aperture were measured using ImageJ (https://imagej.nih.gov/ij/). Stomata from four different plants per genotype (1-2 leaf discs per plant, ~8 stomata per leaf disc) were imaged. At least 30 stomata per genotype were measured for each treatment and the ratio of width/length was graphed to represent the stomatal aperture status.
Phytohormone extraction and quantification
Phytohormones were extracted and quantified as described previously (Zeng et al. 2011) with minor modifications. In brief, approximately 100 mg leaf tissues from four- to five-week old plants, were flash-frozen in liquid nitrogen, grounded and extracted at 4 °C overnight using 1 ml of ice-cold extraction buffer containing methanol:water (80:20 v/v), 0.1% v/v formic acid, and 0.1 g l−1 butylated hydroxytoluene supplemented with 100 nM deuterated-ABA (ABA-2H6) as an internal standard. After overnight incubation at 4 °C with gentle agitation, samples were cleared by centrifugation (12000 × g) and filtered through a 0.2 μm PTFE membrane (Millipore) before being transferred to autosampler vials. ABA (Sigma-Aldrich) was used to prepare calibration standards and generate standard curves (3.9 nM-1000 nM). 10 uL injections of plant extracts were separated using an Acquity UPLC module (Waters) equipped with an Ascentis Express fused-core C18 Column (2.1×50 mm, 2.7 μm; Supelco) heated to 50 °C. The aqueous phase A consisted of 0.15% formic acid in water and the organic phase B was methanol. The separation consisted of a linear increase from A:B (49:1, v/v) to 100% B over 2.5 minutes at a flow rate of 0.4 ml minute−1. Transitions from deprotonated molecules to characteristic product ions were monitored for ABA (m/z 263.1>153.1) and ABA-2H6 (m/z 269.1>159.1) on a Quattro Premier tandem mass spectrometer (Waters) in negative ion mode. The capillary voltage was 3500 V, cone voltage 25 V, and extractor voltage 5 V. The source temperature was maintained at 120 °C and desolvation temperature was 350 °C. Nitrogen was used as desolvation and cone gas at a flow rate of 600 l h−1 and 50 l h−1, respectively. The collision energies and source cone potentials were optimized for each transition using QuanOptimize software. MassLynx 4.1 equipped with application manager QuanLynx was used for data acquisition and processing.
Scanning electron microscopy (SEM)
SEM samples were prepared and scanned at the Center for Advanced Microscopy, MSU. Leaf samples were fixed at 4 °C for one and a half hours in 4% glutaraldehyde buffered with 0.1 M sodium phosphate at pH 7.4. Following a brief rinse in the buffer, samples were dehydrated in an ethanol series (25%, 50%, 75%, 95%) for 50-60 min each, and then in 100% ethanol for one hour three times. Samples were dried in a Leica Microsystems model EM CPD300 critical point dryer (Leica Microsystems) using liquid carbon dioxide as the transitional fluid. Samples were then mounted on aluminum stubs using high vacuum carbon tabs (SPI Supplies) and coated with osmium (~ 10 nm thickness) in an NEOC-AT osmium coater (Meiwafosis Co., Ltd.). Finally, samples were examined with a JEOL JSM-6610LV scanning electron microscope with SEI mode at 12 kV accelerating voltage, 30 spot size and ~15 mm working distance (JEOL Ltd.).
Transmission electron microscopy (TEM)
TEM samples were prepared and scanned at the Center for Advanced Microscopy, MSU. After primary fixation (formaldehyde/glutaraldehyde, 2.5% each in 0.1 M sodium cacodylate buffer, pH 7.4, Electron Microscopy Sciences), samples were washed with 0.1 M cacodylate buffer and postfixed with 1% osmium tetroxide in 0.1 M cacodylate buffer, dehydrated in a gradient series of acetone and infiltrated and embedded in Spurr. 70-nm thin sections were obtained using a Power Tome Ultramicrotome (RMC, Boeckeler Instruments) and post-stained with uranyl acetate and lead citrate. Images were taken with JEOL 100CXII Transmission Electron Microscope (JEOL Ltd.) at an accelerating voltage of 100kV.
Protein immunoblot analysis
Proteins were extracted from four- to five-week-old plants using protein extraction buffer (50 mM Tris-HCl, pH7.5, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS with 100 μM MG132 proteasome inhibitor (Cayman Chemical), 1% protease inhibitor (Sigma-Aldrich) and 1 mM DTT). Protein concentrations were measured using the RC/DC protein assay kit (BioRad) with bovine serum albumin (BSA; BioRad) as standards. Protein samples were adjusted to the same concentrations of total proteins and immunoblotted with the corresponding antibodies. For evaluating the expression of MUR1-4xMyc from 35Spro:MUR1-4xMyc plants, rabbit polyclonal anti-c-Myc primary antibodies (1:5,000; Clontech) and goat anti-rabbit secondary antibodies (1:20,000; Agrisera) were used. For detecting α1,3-fucosylated protein N-glycans, rabbit anti-fucose primary antibodies (1:4,000; Agrisera) and goat anti-rabbit secondary antibodies (1:20,000) were used. For detecting loading controls, the membranes were stained with staining solution (40% methanol, 10 % acetic acid, 0.1% (w/v) Naphthol Blue Black (Sigma-Aldrich)) for 20 min, and destained twice with destaining solution (20% methanol, 7.5% acetic acid) for 10 min each.
Immunoblot of FLS2 and BRI1-ASSOCIATED KINASE1 (BAK1) proteins, was performed as previously described (Tateda et al. 2014) with minor modifications. Briefly, 3-week old plants were harvested in liquid nitrogen and ground to fine powders before being taken up in a 3 x volume (3 ml g−1 tissue) of protein extraction buffer (50 mM Tris-HCl, pH 7.6, 150 mM NaCl, 10% glycerol, 1% Igepal CA-630 (Sigma-Aldrich), 0.5% sodium deoxycholate, and 1 x cOmplete mini protease inhibitor (1 tablet per 10 ml; Roche)). Crude extracts were subsequently centrifuged at 10,000 x g to pellet insoluble materials. Solubilized protein was normalized via Bradford assay (BioRad). For relative abundance comparisons, approximately 60 ug of solubilized protein was separated by SDS-PAGE on 4-12% NuPAGE bis-tris gels (ThermoFisher). For deglycosylation experiments, 20 ug of solubilized protein was treated with N-glycosidase F (PNGase F) (New England Biolabs) according to the manufacturer’s instructions and separated by SDS-PAGE on 3-8% NuPAGE tris-acetate gels (ThermoFisher). SDS-PAGE gels were transferred to PVDF membranes using an iBlot 2 dry blotting system (ThermoFisher) and proteins were analyzed by immunoblotting with anti-FLS2 (1:5000; Agrisera) and anti-BAK1 (1:8000; Agrisera) antibodies. Horseradish peroxidase-conjugated anti-rabbit IgG antibody (1:60000; Agrisera) was used as a secondary antibody and visualized with either SuperSignal West Dura or SuperSignal West Pico PLUS chemiluminescent substrates.
RNA isolation and qPCR assays
Col-7 and scord6 leaves were collected after elf26, flg22, or 0.1% DMSO (mock) treatment. Total RNA was extracted using RNeasy Plant Mini kit (Qiagen). M-MLV Reverse transcriptase (Life Technologies) and SYBR Green master mix (Life Technologies) were used for reverse transcription and real-time PCR. Primers were described in Supporting Information Table S1.
ROS assays
Leaf discs (diameter = 4 mm) from five-week old plants were harvested and floated (the adaxial side facing up) in white 96-well microplate filled with 200 μl water per well. Water was removed 16-18 hours later. 100 μl solution (34 μg ml−1 luminol (Sigma-Aldrich), 20 μg ml−1 horseradish peroxidase (Sigma-Aldrich) in water) with 0.1 μM flg22 or 0.3% DMSO (mock) were added into each well and luminescence was measured immediately at 470 nm using SpectraMax L Luminescence Microplate Reader (Molecular Devices).
Qualification of bacterial type III effector translocation
Bacterial effector translocation assays were performed and quantified as described previously (Huot et al. 2017). Briefly, leaves of four- to five-week old Col-7 plants were infiltration-inoculated with 5-10 × 107 cfu ml−1 of Pst DC3000, which carries the P nptII ::avrPto-CyaA plasmid (Schechter et al. 2004). In order to achieve similar bacterial numbers between Col-7 and the scord6 mutant after 7 hours of inoculation, leaves of scord6 plants were infiltration-inoculated with 1-5 × 107 cfu ml−1 of Pst DC3000 (P nptII ::avrPto-CyaA). Leaf discs were collected seven hours post inoculation for both bacterial populations and cAMP quantification. Bacterial populations were determined by serial dilutions of plant homogenates. cAMP was extracted and quantified using the Direct cAMP ELISA kit (ENZO) and normalized by total plant protein, which was quantified using a Quickstart Bradford assay (BioRad). In order to eliminate the influence of different bacterial populations, cAMP amounts (pmol cAMP μg−1 protein) were further divided by the corresponding bacterial populations (cfu cm−2) in each plant.
Results
Identification of the SCORD6 gene
Although the scord6 mutant was previously isolated from the activation-tagging T-DNA (pSKI015) insertion lines (Weigel et al. 2000), the T-DNA insertion site could not be recovered using plasmid rescue or inverse genomic polymerase chain reaction (iPCR) cloning (Zeng et al. 2011). Furthermore, no T-DNA (pSKI015)-associated glufosinate/Basta resistance was detected in the scord6 mutant (Fig. S2), indicating loss or mutation of the T-DNA sequence in this mutant. Consistent with these observations, we could not locate the intact T-DNA sequence during the analysis of scord6 genomic sequences. We therefore hypothesized that the scord6 mutant phenotypes may be caused by other types of sequence variants, including single-nucleotide polymorphisms (SNPs) or insertions/deletions (INDELs), in the scord6 genome. Using SAMtools and VCFtools, homozygous variations detected only in the scord6 genome, but not in the parental Col-7 genome, were selected and mapped back to the previously identified candidate regions by physical mapping (Zeng et al. 2011). Among the SNPs and INDELs detected, only a 25-bp deletion was located to the candidate region (18.84 Mb-19.03 Mb/III) (Zeng et al. 2011). Specifically, the deletion was found near the 3′ end of the AT3G51160 gene, previously named MUR1 (Reiter et al. 1993) and GMD2 (Bonin et al. 1997) (hereinafter called MUR1 gene) (Fig. 1a). The deletion was further confirmed using IGV (Robinson et al. 2011) and genomic PCR.
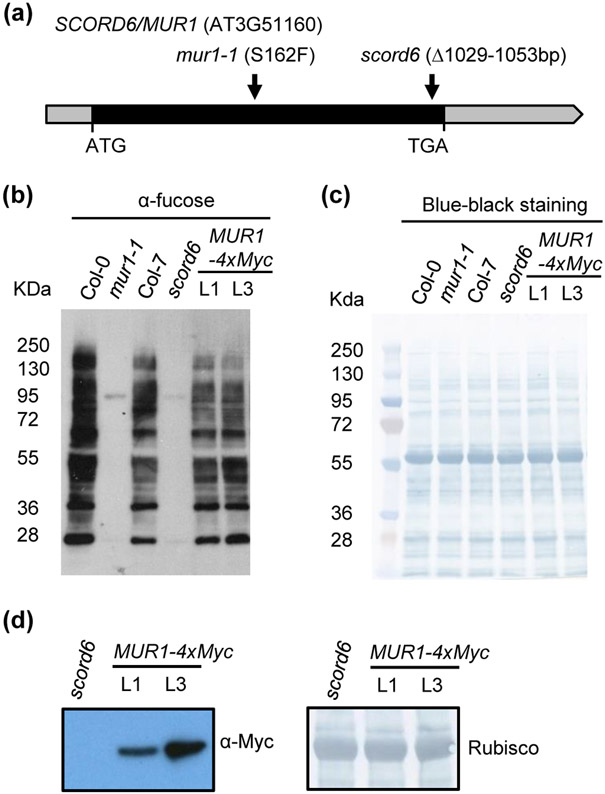
Fig. 1: Identification of the SCORD6 gene.
(a) Schematic depiction of the Arabidopsis SCORD6/MUR1 (AT3G51160) gene. Exon is depicted as black box, and untranslated regions (5’ UTR and 3’ UTR) are shown as gray boxes. Arrows indicate the positions of a SNP or deletion for different allelic mutant lines.
(b) Western blot analysis of total leaf proteins of Col-0, mur1-1, Col-7, scord6 and 35Spro:MUR1-4xMyc transgenic plants using an anti-fucose antibody.
(c) Naphthol Blue Black staining of the same total leaf protein samples to illustrate equal loading.
(d) Western blot to detect the expression of MUR1-4xMyc protein in transgenic 35Spro:MUR1-4xMyc lines in the scord6 background.
Similar L-fucose deficiency and stomatal phenotypes in scord6 and mur1-1 mutants
MUR1 encodes an isoform of GDP-D-mannose-4,6-dehydratase and catalyzes the first step of the de novo synthesis of GDP-L-fucose from GDP-D-mannose (Bonin et al. 1997). L-Fucose is involved in the biosynthesis of several cell wall polymers (e.g., pectin, xyloglucan), and in the sugar-mediated modification of plant proteins (e.g., glycosylation and mono-O-fucosylation of DELLAs) (Reiter et al. 1993; Zentella et al. 2017). Arabidopsis mur1 mutants (Reiter et al. 1993) exhibit an almost complete loss of L-fucose in shoot derived cell wall materials and glycoproteins (Reiter et al. 1993; Zablackis et al. 1996; Rayon et al. 1999; O'Neill et al. 2001; van Hengel & Roberts 2002). We used anti-fucose antibodies to detect α1,3-fucosylated protein N-glycans in scord6 and mur1-1 (with a point mutation, S162F, in the MUR1 gene; Fig. 1a) mutants and their corresponding wild-type Col-7 and Col-0 plants. Compared to Col-7 plants, significantly less α1,3-fucosylated N-glycans were detected in the scord6 leaves (Fig. 1b,c). The level of α1,3-fucosylated N-glycan in the scord6 mutant was comparable to the level in the mur1-1 mutant. We also produced stable T2 lines of the scord6 mutant expressing MUR1-4xMyc under the control of the constitutive cauliflower mosaic virus 35S promoter 35Spro. These lines produced the MUR1-4xMyc protein (Fig. 1d) and restored the production of α1,3-fucosylated N-glycans in the scord6 mutant (Fig. 1b,c).
The scord6 mutant was previously observed to have an abnormal stomatal morphology (Zeng et al. 2011). In this study, we found that, like the scord6 mutant, the mur1-1 mutant also showed markedly reduced central ridges surrounding the stomatal aperture (Fig. S3). TEM examination revealed that the collapsed central ridges of stomatal aperture are associated with an altered pattern of the outer cuticular ledges, which exhibited smaller upward angles in the scord6 and mur1-1 mutants compared to Col-0 plants (Fig. S3). This indicates a tight association between production of L-fucose and formation of stomatal cuticular ledges.
Mutations in the MUR1 gene affect Arabidopsis immunity
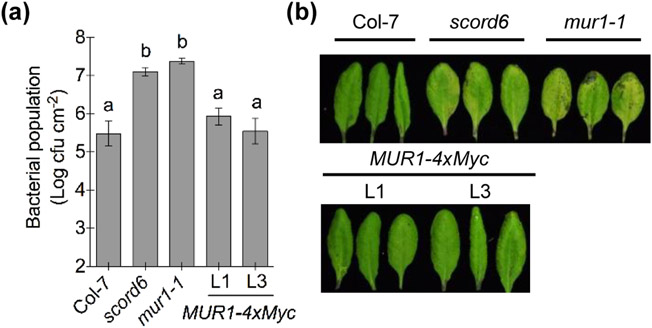
The identification of SCORD6 as MUR1 prompted us to examine the disease susceptibility of the mur1-1 mutant, by monitoring its stomatal response and apoplastic immune capacity. For overall disease susceptibility assays, we infected Col-7, the scord6 mutant and the mur1-1 mutant by dip-inoculation with the COR-deficient mutant Pst DC3118. Like scord6 plants (Zeng et al. 2011), mur1-1 plants showed increased susceptibility to Pst DC3118, illustrated by more severe disease symptoms and higher bacterial populations, compared to wild-type plants (Fig. 2). Importantly, the transgenic lines of 35Spro:MUR1-4xMyc in the scord6 background showed recovery of wild-type resistance against Pst DC3118 (Fig. 2). This genetic complementation experiment demonstrates that the 25-bp deletion mutation in the MUR1 gene is indeed responsible for the enhanced disease susceptibility to Pst DC3118 in the scord6 mutant.
Fig. 2: Mutations in the MUR1 gene affect Arabidopsis defenses.
(a, b) Bacterial populations (a) and disease symptoms (b) three days after dip-inoculation with 1 × 108 cfu ml−1 Pst DC3118. Different letters above the columns indicate significant differences (P < 0.05) of bacterial populations between genotypes by one-way ANOVA with Tukey’s test (n = 4, error bars, ± standard error of the mean [SEM]).
Next, we examined the mur1-1 mutant for bacteria-, SA- or ABA-induced stomatal closure. As shown in Fig. S4, wild-type Col-7 and 35Spro:MUR1-4xMyc plants exhibited significantly reduced stomatal apertures in response to Pst DC3118 inoculation (Fig. S4a,b). In contrast, stomatal apertures of the scord6 and mur1-1 mutants were less closed after inoculation with Pst DC3118 (Fig. S4a), suggesting compromised stomatal defense. Similarly, in response to SA treatment, wild-type Col-7 showed significant stomatal closure, whereas the scord6 and mur1-1 mutants exhibited almost no reduction in stomatal aperture (Fig. S4c). Interestingly, wild-type Col-7, the scord6 mutant and the mur1-1 mutant showed normal stomatal closure in response to ABA treatment (Fig. S4d). Furthermore, the basal levels of ABA were also comparable between the scord6 mutant and wild-type Col-7 plants (Fig. S5), in contrast to significantly lower basal levels of SA in the scord6 mutant (Zeng et al. 2011). Thus, mur1 mutations do not pleiotropically affect ABA-induced stomatal closure, but they specifically affect pathogen- and SA-induced stomatal closure.
To determine whether the mur1-1 mutant also exhibited increased susceptibility in the apoplast, leaves of these mutants were syringe-infiltrated with Pst DC3118 to bypass stomatal defense. Both the scord6 and mur1-1 mutant plants showed increased bacterial growth and disease symptoms (Fig. S6). In contrast, 35Spro:MUR1-4xMyc plants restricted the multiplication of Pst DC3118 inside the leaf apoplast to a level that was comparable to wild-type plants (Fig. S6b,c).
Taken together, the results from our genomic, molecular genetics, and phenotypic analyses are consistent with the conclusion that SCORD6 is MUR1, implicating a novel role of MUR1 in plant immunity against bacterial pathogens.
Mutations in the MUR1 gene affect PTI and ETI in the Arabidopsis apoplast
The increased bacterial multiplication inside the apoplast of the scord6 mutant could be caused by a defect in a canonical defense pathway(s) or some other physiological processes, which was a significant uncertainty in our previous study of the scord6 mutant (Zeng et al. 2011). In this study, we conducted a series of experiments to gain insight into the apoplast hyper-susceptibility of scord6/mur1 mutants. First, we performed experiments to determine whether a lack of L-fucose in the cell walls of scord6 and mur1-1 plants affects bacterial type III secretion of effectors in the apoplast, we performed translocation assays using the AvrPto-CyaA reporter (Schechter et al. 2004). Slightly higher levels of cAMP were detected in the scord6 mutant compared to Col-7 plants. However, this slight difference was not significant in two of the three experimental repeats (Fig. S7). Furthermore, analysis of pooled results from all three experiments did not show a significant difference between the scord6 mutant and wild-type Col-7 plants (Fig. S7), suggesting that increased bacterial multiplication in the apoplast in the scord6 and mur1-1 plants is not likely caused by increased translocation of bacterial effectors.
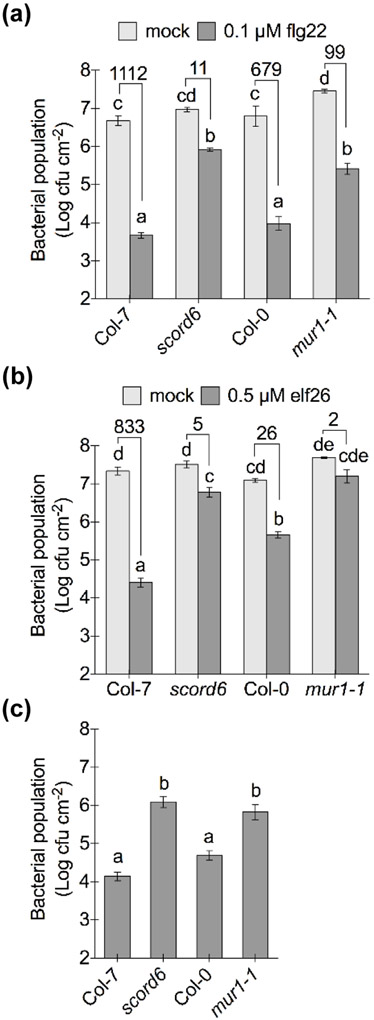
Next, we examined the competency of the scord6 and mur1-1 mutants for PTI and ETI inside the apoplast. The MAMPs flg22 and elf26 were used to induce FLS2- and EFR-mediated PTI, respectively, followed by Pst DC3000 infection via infiltration-inoculation directly into the apoplast. Bacterial populations in the scord6 mutant were found to be significantly higher than those in wild-type plants with 0.1 μM flg22 or 0.5 μM elf26 pre-treatment (Fig. 3a,b), indicating compromised flg22/elf26-induced PTI in the apoplast of the scord6 mutant. Similarly, with 0.1 μM flg22 or 0.5 μM elf26 treatment, the mur1-1 mutant harbored significantly higher levels of bacteria compared to wild-type plants (Fig. 3a,b). In particular, bacterial populations in the mur1-1 mutant pre-treated with 0.5 μM elf26 reached a level similar to those with mock pre-treatment, indicating an almost complete loss of elf26-induced PTI (Fig. 3b). Unlike Col-7 and the scord6 mutant, differences in bacterial populations were noted between Col-0 and the mur1-1 mutant with mock treatment; however, the degree of bacterial growth inhibition with flg22/elf26 treatment was much larger in wild-type Col-0 plants than that in the mur1-1 mutant (Fig. 3a,b).
Fig. 3: Mutations in the MUR1 gene affect flg22-, elf26- and AvrRpt2-induced immunity in Arabidopsis.
(a) Bacterial populations two days after infiltration-inoculation with 1 × 106 cfu ml−1 Pst DC3000. Plants were pre-treated with 0.1 μM flg22 or 0.1% DMSO (mock) for 22 hours. Different letters above columns indicate significant differences (P < 0.05) between bacterial populations (n = 4, error bars, ± SEM); analyzed by two-way ANOVA with Tukey’s test. Numbers above the columns are fold changes of bacterial populations with flg22 treatment compared to mock treatment, indicating the inhibition of bacterial growth with flg22 treatment.
(b) Bacterial populations two days after infiltration-inoculation with 1 × 106 cfu ml−1 Pst DC3000. Plants were pretreated with 0.5 μM elf26 or 0.1% DMSO (mock) for 22 hours. Different letters above columns indicate significant differences (P < 0.05) between bacterial populations (n = 4, error bars, ± SEM), analyzed by two-way ANOVA with Tukey’s test. Numbers above the columns are fold changes of bacterial populations with elf26 treatment compared to mock treatment, indicating the inhibition of bacterial growth with elf26 treatment.
(c) Bacterial populations two days after infiltration-inoculation with 1 x 106 cfu ml−1 Pst DC3000 (avrRpt2). Different letters above columns indicate significant differences (P < 0.05) of bacterial population between plant genotypes (n = 4, error bars, ± SEM); analyzed by one-way ANOVA with Tukey’s test.
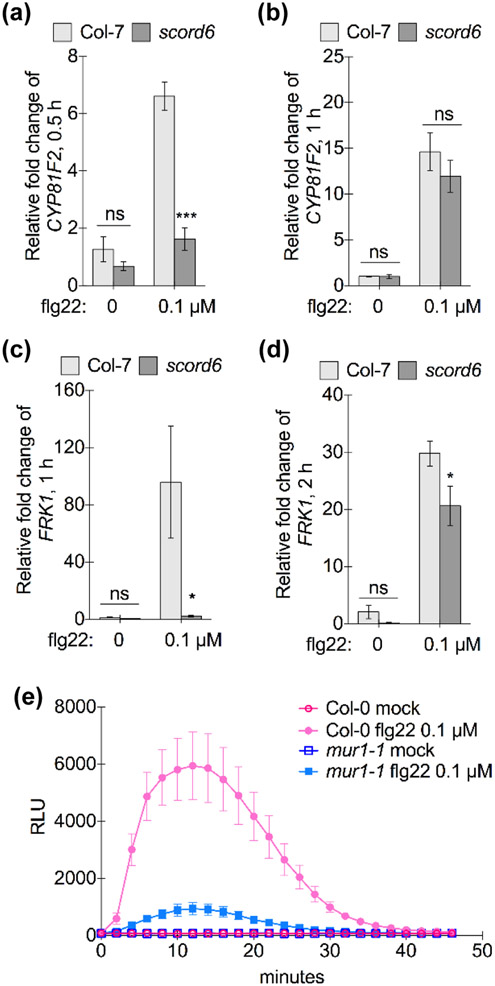
To further clarify the involvement of PTI signaling components in the compromised flg22/elf26-induced protection of the scord6 and mur1-1 mutants, we characterized MAMP-induced expression of PTI early response genes, CYP81F2 (AT5G57220), and FLG22-INDUCED RECEPTOR-LIKE KINASE1 (FRK1, AT2G19190), in the scord6 mutant (Asai et al. 2002; J. Li et al. 2009). 0.1 μM flg22 induced expression of the CYP81F2 gene at 30 min in wild-type plants, while the scord6 mutant had a significantly lower level of CYP81F2 gene expression (Fig. 4a). One hour after flg22 treatment, however, the scord6 mutant exhibited a similar level of CYP81F2 expression compared to wild-type plants (Fig. 4b), indicating delayed PTI response gene expression in the scord6 mutant. Similarly, compromised expression of the FRK1 gene was detected in the scord6 mutant (Fig. 4c,d). We also compared induction of ROS burst in the mur1-1 mutant and wild-type Col-0 plants. With 0.1 μM flg22 treatment, the mur1-1 mutant showed lower levels of ROS production compared to Col-0 (Fig. 4e). Taken together, these results indicate that the scord6 and mur1-1 mutants are compromised in PTI signaling.
Fig. 4: Mutations in the MUR1 gene affect flg22-induced PTI signaling in Arabidopsis.
(a, b) Expression of PTI early response gene CYP81F2 30 min (a) or one hour (b) after 0.1 μM flg22 or 0.1% DMSO (mock) treatment. *** P < 0.001 indicates significant differences of relative fold changes between wild-type Col-7 and the scord6 mutant (n = 4, error bars, ± SEM), analyzed by Student's t-test (ns: not significant).
(c, d) Expression of the PTI response gene, FRK1, one (c) or two hours (d) after 0.1 μM flg22 or 0.1% DMSO (mock) treatment. * 0.01 < P < 0.05 indicates significant differences of relative fold changes between wild-type Col-7 and the scord6 mutant (n = 4, error bars, ± SEM), analyzed by Student's t-test (ns: not significant).
(e) ROS production in Col-0 and the mur1-1 mutant after 0.1 μM flg22 or 0.3% DMSO (mock) treatment. Relative light units (RLU) indicates the level of ROS production (n = 4, error bars, ± SEM).
We further investigated the abundance and the glycosylation forms of FLS2 and BAK1 in Col-7, Col-0, scord6 and mur1-1 plants. Compared to wild-type plants, there were no significant changes in FLS2 and BAK1 protein levels in the two mutant plants (Fig. S8). Interestingly, however, consistent with a defect in fucose biosynthesis in the scord6 and mur1-1 mutants, these proteins showed sensitivity to N-glycosidase F (PNGase F), which cleaves plant N-glycans without α1,3-fucose in the core N-acetylglucosamine (GlcNAc; Tretter et al. 1991), as indicated by a larger decrease in the molecular weights (MWs) of FLS2 and BAK1 after PNGase F. In contrast, FLS2 and BAK1 in wild-type plants were less sensitive to PNGase F treatment, as indicated by a smaller decrease in the MWs of FLS2 and BAK1. This result is consistent with the presence of α1,3-fucose in the N-glycans attached to FLS2 and BAK1 in wild-type plants.
To investigate whether the apoplast of the scord6 and mur1-1 mutants could mount an effective ETI, Pst DC3000 (avrRpt2) was infiltrated into the apoplast of the scord6 and mur1-1 mutants and their respective wild-type plants. Bacterial populations of Pst DC3000 (avrRpt2) were significantly higher in the scord6 and mur1-1 mutants, compared to their respective wild-type plants (Fig. 3c). Taken together, these results show that the apoplast of the scord6 and mur1-1 mutants is defective in mounting full-scale PTI or ETI against bacterial infection.
Fucosyltransferases specific for N-glycans affect Arabidopsis apoplastic and stomatal defense
In plants, L-fucose is involved in fucosylation of a diverse set of substrates, including pectin, xyloglucan, glycoproteins, and DELLA proteins (Reiter et al. 1993; Zentella et al. 2017). Therefore, we hypothesized that the observed defect in immune responses in the scord6 and mur1-1 mutants could result from a defect in fucosylation of one or more of these classes of substrates. To examine this possibility, we analyzed Arabidopsis mutants that are specifically affected in each of these known fucosylation processes.
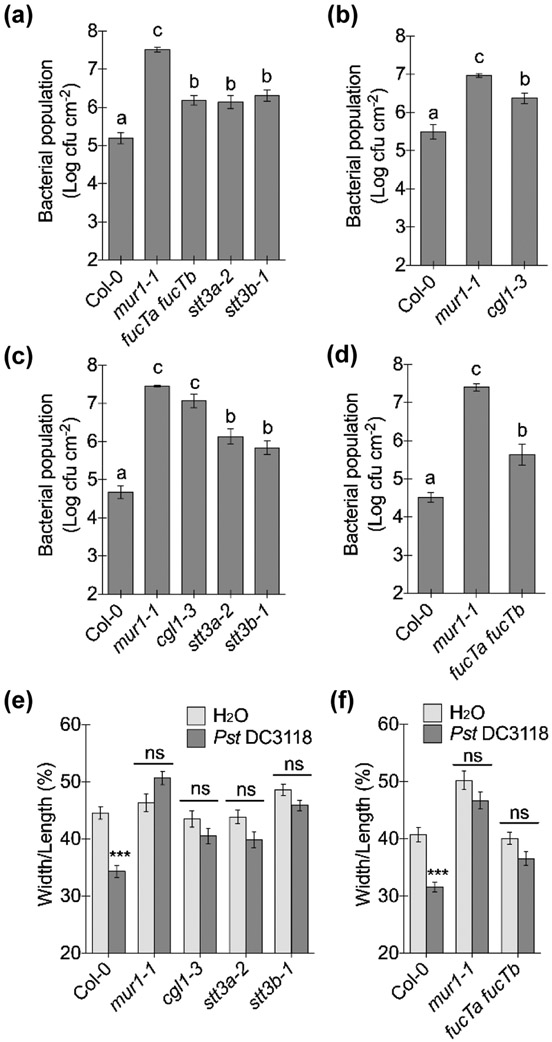
We first focused on fucosylation of N-glycan, as L-fucose is an important component of the carbohydrate chains of glycoproteins (Strasser 2016). Similar to the mur1-1 mutant (Rayon et al. 1999), the scord6 mutant is affected in the profiles of N-glycosylated proteins (Fig. 1b). Arabidopsis mutants defective in two core α1,3-fucosyltransferases, FucTA/FUT11 and FucTB/FUT12 (hereinafter called FucTA and FucTB), which modify N-glycans redundantly (Fig. S9a; Strasser et al. 2004; Strasser 2016), were examined for apoplastic and/or stomatal defenses. The fucTa fucTb double mutant plants showed compromised stomatal closure in response to Pst DC3118 inoculation (Fig. 5) and allowed increased bacterial multiplication after dip- or infiltration-inoculation, compared to wild-type plants. The increased susceptibility of the fucTa fucTb double mutant suggests that the core fucosyltransferases, FucTA and FucTB, are required for Arabidopsis to mount normal apoplastic and stomatal defenses. Consistent with the redundant function of FucTA and FucTB (Bakker et al. 2001), fucTa and fucTb single mutants did not exhibit enhanced susceptibility to Pst DC3118 infection (Fig. S10a).
Fig. 5: N-glycosylation is required for Arabidopsis stomatal closure and apoplastic defense.
(a, b) Bacterial populations three days after dip-inoculation (onto the leaf surface) with 1 × 108 cfu ml−1 Pst DC3118. Different letters above the columns indicate significant differences (P < 0.05) of bacterial populations between plant genotypes by one-way ANOVA with Tukey’s test (n = 4, error bars, ± SEM).
(c, d) Bacterial populations three days after infiltration-inoculation (into the leaf apoplast) with 5 × 105 cfu ml−1 Pst DC3118. Different letters above the columns indicate significant differences (P < 0.05) of bacterial populations between plant genotypes by one-way ANOVA with Tukey’s test (n = 4, error bars, ± SEM).
(e, f) Stomatal apertures two hours after leaves were inoculated with 1 × 108 cfu ml−1 Pst DC3118 or water (mock). Different letters above columns indicate significant differences (P < 0.05) between stomatal apertures (n > 50, error bars, ± SEM), analyzed by two-way ANOVA with Tukey’s test.
Next, we examined FucTC/FUT13, which is an α1,4-fucosyltransferase involved in the synthesis of the Lewis A-glycoepitopes of N-glycans (Fig. S9a; Leonard et al. 2002; Strasser 2016). We found that, unlike the fucTa fucTb double mutant, the fut13-2 mutant (Anderson et al. 2012) did not show increased susceptibility to Pst DC3118, compared to wild-type plants after dip-inoculation with Pst DC3118 (Fig. S10a), indicating that, unlike the core α1,3-linked fucose, the integrity of the Lewis A-type structure of N-glycans is not required for Arabidopsis defenses.
The requirement of FucTA and FucTB for Arabidopsis defenses prompted us to further examine the involvement of other steps of N-glycan processing (Fig. S9a) in plant immunity. Arabidopsis stt3a-2 and stt3b-1 mutants are defective in the putative subunits of the oligosaccharyltransferase (OST) complex, which catalyzes the transfer of the pre-assembled oligosaccharide to an asparagine residue (Koiwa et al. 2003). These two mutants showed increased susceptibility to Pst DC3118 both in dip- and infiltration-inoculation experiments. Moreover, compared to wild-type plants, stomatal responses to Pst DC3118 were also compromised in the stt3a-2 and stt3b-1 mutants (Fig. 5). Additionally, β1,2-N-acetylglucosaminyltransferase I (GnTI) initiates the formation of complex and hybrid N-glycans in the Golgi (von Schaewen et al. 1993). An Arabidopsis GnTI mutant, complex glycan1 (cgl1-3) (Frank et al. 2008), also exhibited compromised apoplastic and stomatal defenses (Fig. 5). These results clearly showed that multiple steps of N-glycan processing affect apoplastic and stomatal defenses in Arabidopsis.
Fucosyltransferases specific for O-glycans and xyloglucan exhibit differential effects on Arabidopsis immunity
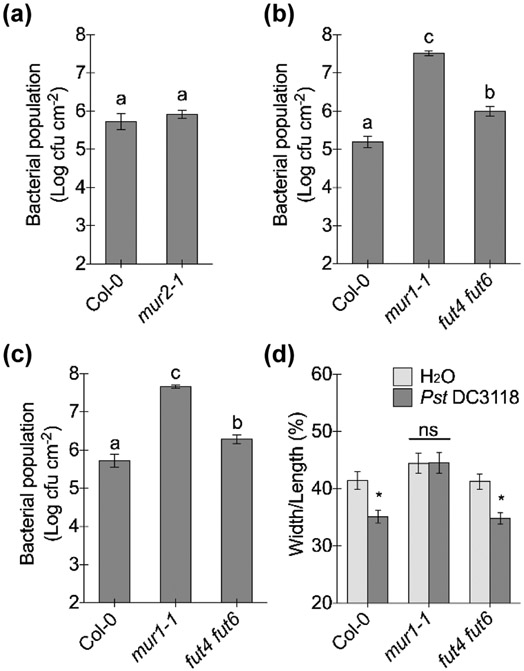
We next carried out experiments to determine whether defects in fucosylation of cell wall polymers or O-glycan play a role in Arabidopsis immunity. As no pectin-specific fucosyltransferase has been found so far, disease assays and stomatal assays were conducted with Arabidopsis mutants mur2-1 (defective in fucosylation of xyloglucan; Perrin et al. 1999; Faik et al. 2000; Vanzin et al. 2002) and fut4 fut6 (defective in fucosylation of O-glycan chains of arabinogalactan proteins; Wu et al. 2010; Liang et al. 2013) (Fig. S9b,c). mur2-1, fut4 and fut6 single mutants allowed similar levels of bacterial growth as wild-type plants after Pst DC3118 dip-inoculation (Fig. 6a; S10b). However, the fut4 fut6 double mutant exhibited increased bacterial growth compared to wild-type plants after both dip- and infiltration-inoculations while maintaining a normal stomatal closure response upon Pst DC3118 inoculation (Fig. 6b-d). These results indicate that a defect in fucosylation of xyloglucan (in the mur2-1 mutant) does not affect Arabidopsis defenses, whereas a defect in fucosylation of O-glycan (in the fut4 fut6 mutant) affects apoplastic but not stomatal defense.
Fig. 6: Arabidopsis fucosyltransferase mutants differentially affect apoplastic and/or stomatal defenses.
(a, b) Bacterial populations three days after dip-inoculation with 1 × 108 cfu ml−1 Pst DC3118. Different letters above the columns indicate significant differences (P < 0.05) of bacterial populations between plant genotypes by Student's t-test (a) or one-way ANOVA with Tukey’s test (b) (n = 4, error bars, ± SEM). Note: data from Fig. 5a and 6b were collected in the same experiment.
(c) Bacterial populations three days after infiltration-inoculation with 5 × 105 cfu ml−1 Pst DC3118. Different letters above the columns indicate significant differences (P < 0.05) of bacterial populations between plant genotypes by one-way ANOVA with Tukey’s test (n = 8, error bars, ± SEM).
(d) Stomatal apertures two hours after leaves were inoculated with 1 × 108 cfu ml−1 Pst DC3118 or water (mock). Different letters above columns indicate significant differences (P < 0.05) between stomatal apertures (n > 50, error bars, ± SEM), analyzed by two-way ANOVA with Tukey’s test.
The O-fucosyltransferase SPY affects Arabidopsis immunity
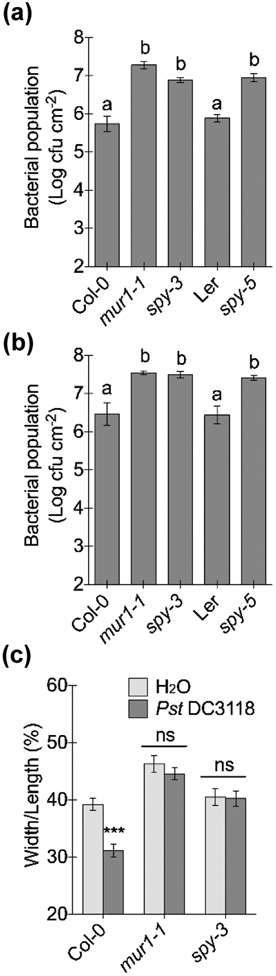
Recently, a protein O-fucosyltransferase, SPINDLY (SPY), was shown to catalyze mono-O-fucosylation (attaching mono fucose to specific Ser/Thr residues) of DELLA transcriptional repressors (Zentella et al. 2017). This fucosylation activates DELLA repressors, which negatively regulate gibberellin (GA) hormone signaling (Zentella et al. 2017). Interestingly, DELLA proteins have been shown to be involved in plant immunity. For example, a quadruple loss-of-function mutant of DELLAs showed higher resistance to Pst DC3000 compared to wild-type plants (Navarro et al. 2008). Hence, we examined the immune competence of Arabidopsis SPY mutants, spy-3 and spy-5 (Jacobsen & Olszewski 1993; Wilson & Somerville 1995; Jacobsen et al. 1996). Based on pathogen inoculation (both dip and apoplastic infiltration) and stomatal closure assays, spy mutants exhibited compromised apoplastic and stomatal defenses (Fig. 7), compared to the corresponding wild-type plants, Col-0 and Landsberg erecta (Ler), respectively. Strikingly, Pst DC3118 populations in spy-3 and spy-5 mutants after dip- or infiltration-inoculation reached levels comparable to those in the mur1-1 mutants. This phenotype contrasts with those of other fucosyltransferase mutants (e.g., fucTa fucTb and fut4 fut6), which exhibited only moderately enhanced disease susceptibility. This finding suggests that lack of SPY-mediated mono-O-fucosylation of proteins might be a significant contributor to compromised Arabidopsis immunity in the scord6 and mur1-1 mutants.
Fig. 7: The Arabidopsis mono-O-glucosyltransferase spy mutant is affected in apoplastic and stomatal defenses.
(a) Bacterial populations three days after dip-inoculation with 1 × 108 cfu ml−1 Pst DC3118. Different letters above the columns indicate significant differences (P < 0.05) of bacterial populations between plant genotypes by one-way ANOVA with Tukey’s test (n = 4, error bars, ± SEM).
(b) Bacterial populations three days after infiltration-inoculation with 1 × 106 cfu ml−1 Pst DC3118. Different letters above the columns indicate significant differences (P < 0.05) of bacterial populations between plant genotypes by one-way ANOVA with Tukey’s test (n = 4, error bars, ± SEM).
(c) Stomatal apertures two hours after leaves were inoculated with 1 × 108 cfu ml−1 Pst DC3118 or water (mock). Different letters above columns indicate significant differences (P < 0.05) between stomatal apertures (n > 50, error bars, ± SEM), analyzed by two-way ANOVA with Tukey’s test.
Discussion
In this study, we have identified the SCORD6 gene and provided multiple lines of evidence that SCORD6 is in fact MUR1. Prior to this study, MUR1 has been known as an isoform of GDP-D-mannose-4,6-dehydratase catalyzing the de novo biosynthesis of L-fucose and being involved in fucosylation of xyloglucan, pectin, glycoproteins and DELLA proteins (Reiter et al. 1993; Bonin et al. 1997; Zentella et al. 2017). In this study we found that the scord6/mur1 mutants are affected in multiple aspects/pathways of plant defense, including stomatal defense, apoplastic defense, PTI and ETI, suggesting a multifaceted role of L-fucose in plant immunity (Table S2).
Because Arabidopsis mur1 mutants were originally isolated in a study of plant cell walls (Reiter et al. 1993), most studies of MUR1 have since focused on its function in cell wall composition, integrity, mechanical performance and cell-wall related plant growth and development (Reiter et al. 1993; Zablackis et al. 1996; Reiter et al. 1997; Rayon et al. 1999; O'Neill et al. 2001; van Hengel & Roberts 2002; Freshour et al. 2003; Ryden et al. 2003; Abasolo et al. 2009; Dumont et al. 2015; Goncalves et al. 2017; Voxeur et al. 2017). However, two studies have implicated a role of MUR1 in plant responses to biotic or abiotic stresses. The mur1-1 mutant was found to exhibit significantly decreased resistance to the penetration of the non-host barley powdery mildew fungus Blumeria graminis f. sp. hordei (Bgh), which was attributed to a defect in the physical barrier of the primary cell wall (Assaad et al. 2004). Similarly, Feng and colleagues (Feng et al. 2018) found that mur1 mutants showed hypersensitivity to salinity stress, which was again proposed to result from a defect in pectin cross-linking.
It is plausible that the requirement of MUR1 for bacterium-triggered stomatal movements resides in part through its role in building a proper plant cell wall. Indeed, it is generally believed that guard cell walls need to be both strong and elastic in order to sustain the high internal pressure and reversible movements (Jones et al. 2005). Cell wall components, like cellulose, hemicellulose and pectin, have all been shown to be involved in basic guard cell movement in response to abiotic stress or chemical treatments (Jones et al. 2003; Jones et al. 2005; Choi et al. 2011; Merced & Renzaglia 2014; Amsbury et al. 2016; Rui & Anderson 2016). In our previous study, we reported that the scord6 mutant has greatly reduced outer cuticular ledges of guard cells (Zeng et al. 2011). This striking stomatal phenotype is also present in the mur1-1 mutant, further implicating an important role of MUR1 in the formation of normal outer cuticular ledges of guard cells (Fig. S3). The outer cuticular ledges of guard cells have long been reported to function by preventing water loss (Schonherr & Ziegler 1975; Lu et al. 2012). Moreover, Pautov and colleagues recently proposed that the outer ledges prevent wide opening of the stomatal pore in woody plants (Pautov et al. 2017). Further studies involving new cuticular ledge mutants that do not affect other cellular processes could be used to determine whether an intact cuticular ledge is necessary for stomatal defense against pathogen invasion. It is important to note that mur1 and scord6 mutations do not pleiotropically affect stomatal movements, as ABA-triggered stomatal closure remains normal (Fig. S4). Therefore, a nonspecific effect of cell wall defects on stomatal movements would not explain the specific effect of mur1 and scord6 mutations on bacterium-triggered stomatal closure.
Indeed, our results suggested that MUR1 likely also functions in mediating plant immunity via other mechanisms that are indicative of immune and hormone signaling pathways (i.e., besides the requirement of MUR1 for plant immunity through the function of physical barriers or guard cell responses). We observed delayed/reduced expression of PTI early response genes and compromised ROS burst in the scord6 or mur1-1 mutant, respectively (Fig. 4). We found this result intriguing as previous studies showed that the EFR receptor is highly glycosylated and this glycosylation is necessary for the stability and function of EFR (J. Li et al. 2009; Nekrasov et al. 2009; Saijo et al. 2009; Haweker et al. 2010; Sun et al. 2012). An N-glycan processing mutant, stt3a-2, harbors reduced levels of EFR protein and exhibited compromised defense against Pst DC3000 with spray-inoculation (Saijo et al. 2009; Haweker et al. 2010). In this study, we found that stt3a-2 and other N-glycan processing mutants (stt3b-1, cgl1-3 and fucTa fucTb) showed compromised defense against Pst DC3118 in both dip- and infiltration-inoculations (Fig. 5) and that both FLS2 and BAK1 in the scord6 and mur1-1 mutants were sensitive to PNGase F, which cleaves plant N-glycans without α1,3-fucose in the core GlcNAc (Fig. S8). However, the involvement of fucosylation in PTI likely goes beyond affecting receptor glycosylation. This is because the triple mutant fucTa fucTb xylT, defective in α1,3-fucosyltransferases and β1,2-xylosyltransferase, was reported to have a wild-type level of EFR protein (Haweker et al. 2010), but we observed compromised resistance against Pst DC3118 in the fucTa fucTb double mutant (Fig. 5a,d).
It is worth noting that possible structural changes in cell wall, due to lack of fucose, might have an impact on plant immune signaling. For example, FERONIA (FER), the malectin-like receptor kinase, maintains cell-wall integrity during salt stress and appears to act as a scaffold to modulate FLS2-BAK1 receptor kinase complex assembly and plant immunity (Stegmann et al. 2017; Feng et al. 2018). Moreover, internalization of flg22-activated FLS2 depends on cytoskeleton (Robatzek et al. 2006) and is required for long distance transport of flg22 (Jelenska et al. 2017). Chemicals that perturb cell wall deposition or cell-wall related mutants have been reported to affect cytoskeleton organization (Baluska et al. 2003; Tolmie et al. 2017). Hence, it is possible that cell wall integrity may play a role in plant immunity through regulation of cell wall-associated receptors and the cytoskeleton.
Another interesting observation made in this study is the significantly compromised plant immunity in spy mutants. In particular, the immune defect in the spy mutant against Pst DC3118 infection was severe and comparable to that observed for the mur1-1 mutant (Fig. 7). As other fucosyltransferase mutants (e.g., fucTa fucTb and fut4 fut6) tested in our study only exhibited moderately enhanced bacterial growth, it is likely that SPY-mediated mono-O-fucosylation of proteins plays a critical role in mediating plant immunity. Although SPY-mediated mono-O-fucosylation activates DELLAs and in turn suppresses GA signaling (Zentella et al. 2017), the increased susceptibility to Pst DC3118 of spy mutants probably result from a defect in a pathway other than GA signaling, because a quintuple mutant of DELLA proteins, della (Feng et al. 2008), showed resistance to Pst DC3118, similar to wild-type Ler plants (Fig. S10c). Moreover, higher resistance to Pst DC3000 was detected in the quadruple DELLA (Navarro et al. 2008), further suggesting that loss of DELLAs at least does not affect Arabidopsis immunity against P. syringae. These findings therefore raise the possibility that the SPY protein mediates plant immunity through pathways other than GA signaling, and besides DELLAs, there are likely other substrates of the SPY protein.
In summary, identification of SCORD6 as MUR1 led us to uncover a novel and multifaceted role of L-fucose biosynthesis and protein fucosylation genes in plant immunity against bacterial pathogen P. syringae and connect L-fucose to central plant immunity pathways, including PTI and ETI. These results open new avenues for future studies to identify immune-related targets of fucosylation and has potential to reveal new insights into immune-related polysaccharides or proteins in plants.
Supplementary Material
Fig. S1: Bacterial populations 1 hour after infiltration-inoculation with Pst DC3118.
Fig. S2: Loss of Basta resistance in the scord6 mutant.
Fig. S3: SEM and TEM images of stomatal apertures.
Fig. S4: Mutations in the MUR1 gene affect pathogen- and SA-induced stomatal closure in Arabidopsis.
Fig. S5: ABA levels in Col-7 and the scord6 mutant.
Fig. S6: Mutations in the MUR1 gene affect Arabidopsis apoplastic defense.
Fig. S7: Bacterial effector translocation in Col-7 and the scord6 mutant plants.
Fig. S8: FLS2 and BAK1 abundance and sensitivity to PNGase F.
Fig. S9: Simplified diagrams of N-glycan processing and modification of O-glycan and xyloglucan.
Fig. S10: Disease assays of Arabidopsis mutants of fucosyltransferases and quintuple della mutant.
Table S1: List of primers.
Table S2: Summary of key assay results of the Arabidopsis mutants analyzed in this study.
Acknowledgements
TEM and SEM images were generated with the assistance of Dr. Alicia Withrow and Carol Flegler (Center for Advanced Microscopy, MSU), respectively. The protocol of stomatal closure assay was provided by Dr. Yi-Ju Lu (MSU). qPCR primers for FRK1 were obtained from Dr. Yuti Cheng (MSU). Phytohormone quantification was accomplished with the technical support and the ABA-d6 internal standard from Dr. Tony Schilmiller and Dr. Dan Jones (RTSF Mass Spectrometry and Metabolomics Core, MSU). This research was funded by US National Science Foundation (IOS-1557437 to B.D. and S.Y.H.), US Department of Agriculture – NIFA (2015-67017-23360 and 2017-67017-26180 to S.Y.H), and US Department of Energy (the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science; DE–FG02–91ER20021 for infrastructural support to S.Y.H.).
References
- Abasolo W, Eder M, Yamauchi K, Obel N, Reinecke A, Neumetzler L, Dunlop JW, Mouille G, Pauly M, Hofte H, et al. 2009. Pectin may hinder the unfolding of xyloglucan chains during cell deformation: implications of the mechanical performance of Arabidopsis hypocotyls with pectin alterations. Molecular Plant 2: 990–999 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Amsbury S, Hunt L, Elhaddad N, Baillie A, Lundgren M, Verhertbruggen Y, Scheller HV, Knox JP, Fleming AJ, Gray JE. 2016. Stomatal function requires pectin de-methyl-esterification of the guard cell wall. Current Biology 26: 2899–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson CT, Wallace IS, Somerville CR. 2012. Metabolic click-labeling with a fucose analog reveals pectin delivery, architecture, and dynamics in Arabidopsis cell walls. Proceedings of the National Academy of Sciences of the United States of America 109: 1329–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrew S. 2010. FastQC: a quality control tool for high throughput sequence data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc [Google Scholar]
- Arnaud D, Hwang I. 2015. A sophisticated network of signaling pathways regulates stomatal defenses to bacterial pathogens. Molecular Plant 8: 566–581 [DOI] [PubMed] [Google Scholar]
- Asai T, Tena G, Plotnikova J, Willmann MR, Chiu WL, Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. 2002. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature 415: 977–983 [DOI] [PubMed] [Google Scholar]
- Assaad FF, Qiu JL, Youngs H, Ehrhardt D, Zimmerli L, Kalde M, Wanner G, Peck SC, Edwards H, Ramonell K, et al. 2004. The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Molecular Biology of the Cell 15: 5118–5129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Staskawicz BJ. 2003. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell 112: 369–377 [DOI] [PubMed] [Google Scholar]
- Bakker H, Schijlen E, de Vries T, Schiphorst WE, Jordi W, Lommen A, Bosch D, van Die I. 2001. Plant members of the α1→3/4-fucosyltransferase gene family encode an α1→4-fucosyltransferase, potentially involved in Lewisa biosynthesis, and two core α1→3-fucosyltransferases. FEBS Letters 507: 307–312 [DOI] [PubMed] [Google Scholar]
- Baluska F, Samaj J, Wojtaszek P, Volkmann D, Menzel D. 2003. Cytoskeleton-plasma membrane-cell wall continuum in plants. Emerging links revisited. Plant Physiology 133: 482–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigeard J, Colcombet J, Hirt H. 2015. Signaling mechanisms in pattern-triggered immunity (PTI). Molecular Plant 8: 521–539 [DOI] [PubMed] [Google Scholar]
- Bonin CP, Potter I, Vanzin GF, Reiter WD. 1997. The MUR1 gene of Arabidopsis thaliana encodes an isoform of GDP-D-mannose-4,6-dehydratase, catalyzing the first step in the de novo synthesis of GDP-L-fucose. Proceedings of the National Academy of Sciences of the United States of America 94: 2085–2090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutrot F, Segonzac C, Chang KN, Qiao H, Ecker JR, Zipfel C, Rathjen JP. 2010. Direct transcriptional control of the Arabidopsis immune receptor FLS2 by the ethylene-dependent transcription factors EIN3 and EIL1. Proceedings of the National Academy of Sciences of the United States of America 107: 14502–14507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks DM, Bender CL, Kunkel BN. 2005. The Pseudomonas syringae phytotoxin coronatine promotes virulence by overcoming salicylic acid-dependent defences in Arabidopsis thaliana. Molecular Plant Pathology 6: 629–639 [DOI] [PubMed] [Google Scholar]
- Brooks DM, Hernandez-Guzman G, Kloek AP, Alarcon-Chaidez F, Sreedharan A, Rangaswamy V, Penaloza-Vazquez A, Bender CL, Kunkel BN. 2004. Identification and characterization of a well-defined series of coronatine biosynthetic mutants of Pseudomonas syringae pv. tomato DC3000. Molecular Plant-Microbe Interactions 17: 162–174 [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Bauer Z, Regenass M, Boller T, Felix G. 2006. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. The Plant Cell 18: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi JY, Seo YS, Kim SJ, Kim WT, Shin JS. 2011. Constitutive expression of CaXTH3, a hot pepper xyloglucan endotransglucosylase/hydrolase, enhanced tolerance to salt and drought stresses without phenotypic defects in tomato plants (Solanum lycopersicum cv. Dotaerang). Plant Cell Reports 30: 867–877 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. 1998. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cui H, Tsuda K, Parker JE. 2015. Effector-triggered immunity: from pathogen perception to robust defense. Annual Review of Plant Biology 66: 487–511 [DOI] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27: 2156–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont M, Lehner A, Bardor M, Burel C, Vauzeilles B, Lerouxel O, Anderson CT, Mollet JC, Lerouge P. 2015. Inhibition of fucosylation of cell wall components by 2-fluoro 2-deoxy-L-fucose induces defects in root cell elongation. The Plant Journal 84: 1137–1151 [DOI] [PubMed] [Google Scholar]
- Faik A, Bar-Peled M, DeRocher AE, Zeng W, Perrin RM, Wilkerson C, Raikhel NV, Keegstra K. 2000. Biochemical characterization and molecular cloning of an α-1,2-fucosyltransferase that catalyzes the last step of cell wall xyloglucan biosynthesis in pea. The Journal of Biological Chemistry 275: 15082–15089 [DOI] [PubMed] [Google Scholar]
- Feng S, Martinez C, Gusmaroli G, Wang Y, Zhou J, Wang F, Chen L, Yu L, Iglesias-Pedraz JM, Kircher S, et al. 2008. Coordinated regulation of Arabidopsis thaliana development by light and gibberellins. Nature 451: 475–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W, Kita D, Peaucelle A, Cartwright HN, Doan V, Duan Q, Liu MC, Maman J, Steinhorst L, Schmitz-Thom I, et al. 2018. The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Current Biology 28: 666–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Kaulfurst-Soboll H, Rips S, Koiwa H, von Schaewen A. 2008. Comparative analyses of Arabidopsis complex glycan1 mutants and genetic interaction with staurosporin and temperature sensitive3a. Plant Physiology 148: 1354–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freshour G, Bonin CP, Reiter WD, Albersheim P, Darvill AG, Hahn MG. 2003. Distribution of fucose-containing xyloglucans in cell walls of the mur1 mutant of Arabidopsis. Plant Physiology 131: 1602–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genger RK, Jurkowski GI, McDowell JM, Lu H, Jung HW, Greenberg JT, Bent AF. 2008. Signaling pathways that regulate the enhanced disease resistance of Arabidopsis "defense, no death" mutants. Molecular Plant-Microbe Interactions 21: 1285–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Gomez L, Boller T. 2000. FLS2: an LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Molecular Cell 5: 1003–1011 [DOI] [PubMed] [Google Scholar]
- Goncalves B, Maugarny-Cales A, Adroher B, Cortizo M, Borrega N, Blein T, Hasson A, Gineau E, Mouille G, Laufs P, et al. 2017. GDP-L-fucose is required for boundary definition in plants. Journal of Experimental Botany 68: 5801–5811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haweker H, Rips S, Koiwa H, Salomon S, Saijo Y, Chinchilla D, Robatzek S, von Schaewen A. 2010. Pattern recognition receptors require N-glycosylation to mediate plant immunity. The Journal of Biological Chemistry 285: 4629–4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heese A, Hann DR, Gimenez-Ibanez S, Jones AM, He K, Li J, Schroeder JI, Peck SC, Rathjen JP. 2007. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proceedings of the National Academy of Sciences of the United States of America 104: 12217–12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano SS, Upper CD. 2000. Bacteria in the leaf ecosystem with emphasis on Pseudomonas syringae-a pathogen, ice nucleus, and epiphyte. Microbiology and Molecular Biology Reviews 64: 624–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot B, Castroverde CDM, Velasquez AC, Hubbard E, Pulman JA, Yao J, Childs KL, Tsuda K, Montgomery BL, He SY. 2017. Dual impact of elevated temperature on plant defence and bacterial virulence in Arabidopsis. Nature Communications 8: 1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Binkowski KA, Olszewski NE. 1996. SPINDLY, a tetratricopeptide repeat protein involved in gibberellin signal transduction in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America 93: 9292–9296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen SE, Olszewski NE. 1993. Mutations at the SPINDLY locus of Arabidopsis alter gibberellin signal transduction. The Plant Cell 5: 887–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenska J, Davern SM, Standaert RF, Mirzadeh S, Greenberg JT. 2017. Flagellin peptide flg22 gains access to long-distance trafficking in Arabidopsis via its receptor, FLS2. Journal of Experimental Botany 68: 1769–1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. 2006. The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Jones L, Milne JL, Ashford D, McCann MC, McQueen-Mason SJ. 2005. A conserved functional role of pectic polymers in stomatal guard cells from a range of plant species. Planta 221: 255–264 [DOI] [PubMed] [Google Scholar]
- Jones L, Milne JL, Ashford D, McQueen-Mason SJ. 2003. Cell wall arabinan is essential for guard cell function. Proceedings of the National Academy of Sciences of the United States of America 100: 11783–11788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, Kim HY, Jeon JY, Kim DM, Zhou Y, Lee JS, Lee H, Choi HK. 2017. Effects of coronatine elicitation on growth and metabolic profiles of Lemna paucicostata culture. PLoS ONE 12: e0187622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koiwa H, Li F, McCully MG, Mendoza I, Koizumi N, Manabe Y, Nakagawa Y, Zhu J, Rus A, Pardo JM, et al. 2003. The STT3a subunit isoform of the Arabidopsis oligosaccharyltransferase controls adaptive responses to salt/osmotic stress. The Plant Cell 15: 2273–2284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. 2004. The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. The Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nature Methods 9: 357–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard R, Costa G, Darrambide E, Lhernould S, Fleurat-Lessard P, Carlue M, Gomord V, Faye L, Maftah A. 2002. The presence of Lewis a epitopes in Arabidopsis thaliana glycoconjugates depends on an active α4-fucosyltransferase gene. Glycobiology 12: 299–306 [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, Genome Project Data Processing S. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Zhao-Hui C, Batoux M, Nekrasov V, Roux M, Chinchilla D, Zipfel C, Jones JD. 2009. Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proceedings of the National Academy of Sciences of the United States of America 106: 15973–15978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Basu D, Pattathil S, Xu WL, Venetos A, Martin SL, Faik A, Hahn MG, Showalter AM. 2013. Biochemical and physiological characterization of fut4 and fut6 mutants defective in arabinogalactan-protein fucosylation in Arabidopsis. Journal of Experimental Botany 64: 5537–5551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S, Zhao H, Des Marais DL, Parsons EP, Wen X, Xu X, Bangarusamy DK, Wang G, Rowland O, Juenger T, et al. 2012. Arabidopsis ECERIFERUM9 involvement in cuticle formation and maintenance of plant water status. Plant Physiology 159: 930–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macho AP, Zipfel C. 2014. Plant PRRs and the activation of innate immune signaling. Molecular Cell 54: 263–272 [DOI] [PubMed] [Google Scholar]
- Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. 2003. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell 112: 379–389 [DOI] [PubMed] [Google Scholar]
- Melotto M, Underwood W, He SY. 2008. Role of stomata in plant innate immunity and foliar bacterial diseases. Annual Review of Phytopathology 46: 101–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY. 2006. Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Melotto M, Zhang L, Oblessuc PR, He SY. 2017. Stomatal defense a decade later. Plant Physiology 174: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merced A, Renzaglia K. 2014. Developmental changes in guard cell wall structure and pectin composition in the moss Funaria: implications for function and evolution of stomata. Annals of Botany 114: 1001–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mersmann S, Bourdais G, Rietz S, Robatzek S. 2010. Ethylene signaling regulates accumulation of the FLS2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiology 154: 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montillet JL, Leonhardt N, Mondy S, Tranchimand S, Rumeau D, Boudsocq M, Garcia AV, Douki T, Bigeard J, Lauriere C, et al. 2013. An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biology 11: e1001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Suzuki T, Murata S, Nakamura S, Hino T, Maeo K, Tabata R, Kawai T, Tanaka K, Niwa Y, et al. 2007. Improved Gateway binary vectors: high-performance vectors for creation of fusion constructs in transgenic analysis of plants. Bioscience, Biotechnology, and Biochemistry 71: 2095–2100 [DOI] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lison P, Nemri A, Harberd NP, Jones JD. 2008. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Current Biology 18: 650–655 [DOI] [PubMed] [Google Scholar]
- Nekrasov V, Li J, Batoux M, Roux M, Chu ZH, Lacombe S, Rougon A, Bittel P, Kiss-Papp M, Chinchilla D, et al. 2009. Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. The EMBO Journal 28: 3428–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill MA, Eberhard S, Albersheim P, Darvill AG. 2001. Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294: 846–849 [DOI] [PubMed] [Google Scholar]
- Panchal S, Roy D, Chitrakar R, Price L, Breitbach ZS, Armstrong DW, Melotto M. 2016. Coronatine facilitates Pseudomonas syringae infection of Arabidopsis leaves at night. Frontiers in Plant Science 7: 880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pautov A, Bauer S, Ivanova O, Krylova E, Sapach Y, Gussarova G. 2017. Role of the outer stomatal ledges in the mechanics of guard cell movements. Trees 31: 125–135 [Google Scholar]
- Perrin RM, DeRocher AE, Bar-Peled M, Zeng W, Norambuena L, Orellana A, Raikhel NV, Keegstra K. 1999. Xyloglucan fucosyltransferase, an enzyme involved in plant cell wall biosynthesis. Science 284: 1976–1979 [DOI] [PubMed] [Google Scholar]
- Rayon C, Cabanes-Macheteau M, Loutelier-Bourhis C, Salliot-Maire I, Lemoine J, Reiter WD, Lerouge P, Faye L. 1999. Characterization of N-glycans from Arabidopsis. Application to a fucose-deficient mutant. Plant Physiology 119: 725–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter WD, Chapple C, Somerville CR. 1997. Mutants of Arabidopsis thaliana with altered cell wall polysaccharide composition. The Plant Journal 12: 335–345 [DOI] [PubMed] [Google Scholar]
- Reiter WD, Chapple CC, Somerville CR. 1993. Altered growth and cell walls in a fucose-deficient mutant of Arabidopsis. Science 261: 1032–1035 [DOI] [PubMed] [Google Scholar]
- Robatzek S, Chinchilla D, Boller T. 2006. Ligand-induced endocytosis of the pattern recognition receptor FLS2 in Arabidopsis. Genes Dev 20: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nature Biotechnology 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui Y, Anderson CT. 2016. Functional analysis of cellulose and xyloglucan in the walls of stomatal guard cells of Arabidopsis. Plant Physiology 170: 1398–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryden P, Sugimoto-Shirasu K, Smith AC, Findlay K, Reiter WD, McCann MC. 2003. Tensile properties of Arabidopsis cell walls depend on both a xyloglucan cross-linked microfibrillar network and rhamnogalacturonan II-borate complexes. Plant Physiology 132: 1033–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saijo Y, Tintor N, Lu X, Rauf P, Pajerowska-Mukhtar K, Haweker H, Dong X, Robatzek S, Schulze-Lefert P. 2009. Receptor quality control in the endoplasmic reticulum for plant innate immunity. The EMBO Journal 28: 3439–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schechter LM, Roberts KA, Jamir Y, Alfano JR, Collmer A. 2004. Pseudomonas syringae type III secretion system targeting signals and novel effectors studied with a Cya translocation reporter. Journal of Bacteriology 186: 543–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonherr J, Ziegler H. 1975. Hydrophobic cuticular ledges prevent water entering the air pores of liverwort Thalli. Planta 124: 51–60 [DOI] [PubMed] [Google Scholar]
- Stegmann M, Monaghan J, Smakowska-Luzan E, Rovenich H, Lehner A, Holton N, Belkhadir Y, Zipfel C. 2017. The receptor kinase FER is a RALF-regulated scaffold controlling plant immune signaling. Science 355: 287–289 [DOI] [PubMed] [Google Scholar]
- Strasser R 2016. Plant protein glycosylation. Glycobiology 26: 926–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser R, Altmann F, Mach L, Glossl J, Steinkellner H. 2004. Generation of Arabidopsis thaliana plants with complex N-glycans lacking β1,2-linked xylose and core α1,3-linked fucose. FEBS Letters 561: 132–136 [DOI] [PubMed] [Google Scholar]
- Sun W, Cao Y, Jansen Labby K, Bittel P, Boller T, Bent AF. 2012. Probing the Arabidopsis flagellin receptor: FLS2-FLS2 association and the contributions of specific domains to signaling function. The Plant Cell 24: 1096–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateda C, Zhang Z, Shrestha J, Jelenska J, Chinchilla D, Greenberg JT. 2014. Salicylic acid regulates Arabidopsis microbial pattern receptor kinase levels and signaling. The Plant Cell 26: 4171–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolmie F, Poulet A, McKenna J, Sassmann S, Graumann K, Deeks M, Runions J. 2017. The cell wall of Arabidopsis thaliana influences actin network dynamics. Journal of Experimental Botany 68: 4517–4527 [DOI] [PubMed] [Google Scholar]
- Tretter V, Altmann F, Marz L. 1991. Peptide-N4-(N-acetyl-β-glucosaminyl)asparagine amidase F cannot release glycans with fucose attached α1→3 to the asparagine-linked N-acetylglucosamine residue. European Journal of Biochemistry 199: 647–652 [DOI] [PubMed] [Google Scholar]
- Uddin MN, Akhter S, Chakraborty R, Baek JH, Cha JY, Park SJ, Kang H, Kim WY, Lee SY, Mackey D, et al. 2017. SDE5, a putative RNA export protein, participates in plant innate immunity through a flagellin-dependent signaling pathway in Arabidopsis. Scientific Reports 7: 9859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hengel AJ, Roberts K. 2002. Fucosylated arabinogalactan-proteins are required for full root cell elongation in Arabidopsis. The Plant Journal 32: 105–113 [DOI] [PubMed] [Google Scholar]
- Vanzin GF, Madson M, Carpita NC, Raikhel NV, Keegstra K, Reiter WD. 2002. The mur2 mutant of Arabidopsis thaliana lacks fucosylated xyloglucan because of a lesion in fucosyltransferase AtFUT1. Proceedings of the National Academy of Sciences of the United States of America 99: 3340–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Schaewen A, Sturm A, O'Neill J, Chrispeels MJ. 1993. Isolation of a mutant Arabidopsis plant that lacks N-acetyl glucosaminyl transferase I and is unable to synthesize Golgi-modified complex N-linked glycans. Plant Physiology 102: 1109–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voxeur A, Soubigou-Taconnat L, Legee F, Sakai K, Antelme S, Durand-Tardif M, Lapierre C, Sibout R. 2017. Altered lignification in mur1-1 a mutant deficient in GDP-L-fucose synthesis with reduced RG-II cross linking. PLoS ONE 12: e0184820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Ahn JH, Blazquez MA, Borevitz JO, Christensen SK, Fankhauser C, Ferrandiz C, Kardailsky I, Malancharuvil EJ, Neff MM, et al. 2000. Activation tagging in Arabidopsis. Plant Physiology 122: 1003–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RN, Somerville CR. 1995. Phenotypic suppression of the gibberellin-insensitive mutant (gai) of Arabidopsis. Plant Physiology 108: 495–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Williams M, Bernard S, Driouich A, Showalter AM, Faik A. 2010. Functional identification of two nonredundant Arabidopsis α(1,2)fucosyltransferases specific to arabinogalactan proteins. The Journal of Biological Chemistry 285: 13638–13645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin XF, Kvitko B, He SY. 2018. Pseudomonas syringae: what it takes to be a pathogen. Nature Reviews. Microbiology 16: 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J, Withers J, He SY. 2013. Pseudomonas syringae infection assays in Arabidopsis. Methods in Molecular Biology 1011: 63–81 [DOI] [PubMed] [Google Scholar]
- Ye W, Murata Y. 2016. Microbe associated molecular pattern signaling in guard cells. Frontiers in Plant Science 7: 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zablackis E, York WS, Pauly M, Hantus S, Reiter WD, Chapple CC, Albersheim P, Darvill A. 1996. Substitution of L-fucose by L-galactose in cell walls of Arabidopsis mur1. Science 272: 1808–1810 [DOI] [PubMed] [Google Scholar]
- Zeng W, Brutus A, Kremer JM, Withers JC, Gao X, Jones AD, He SY. 2011. A genetic screen reveals Arabidopsis stomatal and/or apoplastic defenses against Pseudomonas syringae pv. tomato DC3000. PLoS Pathogens 7: e1002291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng W, He SY. 2010. A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv. tomato DC3000 in Arabidopsis. Plant Physiology 153: 1188–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Sui N, Barnhill B, Hsieh WP, Hu J, Shabanowitz J, Boyce M, Olszewski NE, Zhou P, Hunt DF, et al. 2017. The Arabidopsis O-fucosyltransferase SPINDLY activates nuclear growth repressor DELLA. Nature Chemical Biology 13: 479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang F, Melotto M, Yao J, He SY. 2017. Jasmonate signaling and manipulation by pathogens and insects. Journal of Experimental Botany 68: 1371–1385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, He SY, Assmann SM. 2008. The plant innate immunity response in stomatal guard cells invokes G-protein-dependent ion channel regulation. The Plant Journal 56: 984–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XY, Spivey NW, Zeng W, Liu PP, Fu ZQ, Klessig DF, He SY, Dong X. 2012. Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host and Microbe 11: 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. 2006. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell 125: 749–760 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1: Bacterial populations 1 hour after infiltration-inoculation with Pst DC3118.
Fig. S2: Loss of Basta resistance in the scord6 mutant.
Fig. S3: SEM and TEM images of stomatal apertures.
Fig. S4: Mutations in the MUR1 gene affect pathogen- and SA-induced stomatal closure in Arabidopsis.
Fig. S5: ABA levels in Col-7 and the scord6 mutant.
Fig. S6: Mutations in the MUR1 gene affect Arabidopsis apoplastic defense.
Fig. S7: Bacterial effector translocation in Col-7 and the scord6 mutant plants.
Fig. S8: FLS2 and BAK1 abundance and sensitivity to PNGase F.
Fig. S9: Simplified diagrams of N-glycan processing and modification of O-glycan and xyloglucan.
Fig. S10: Disease assays of Arabidopsis mutants of fucosyltransferases and quintuple della mutant.
Table S1: List of primers.
Table S2: Summary of key assay results of the Arabidopsis mutants analyzed in this study.