Abstract
Three Candida albicans genes, designated FCR (for fluconazole resistance), have been isolated by their ability to complement the fluconazole (FCZ) hypersensitivity of a Saccharomyces cerevisiae mutant lacking the transcription factors Pdr1p and Pdr3p. Overexpression of any of the three FCR genes in the pdr1 pdr3 mutant resulted in increased resistance of the cells to FCZ and cycloheximide and in increased expression of PDR5, a gene coding for a drug efflux transporter of the ATP-binding cassette superfamily and whose transcription is under the control of Pdr1p and Pdr3p. Deletion of PDR5 in the pdr1 pdr3 strain completely abrogated the ability of the three FCR genes to confer FCZ resistance, demonstrating that PDR5 is required for FCR-mediated FCZ resistance in S. cerevisiae. The FCR1 gene encodes a putative 517-amino-acid protein with an N-terminal Zn2C6-type zinc finger motif homologous to that found in fungal zinc cluster proteins, including S. cerevisiae Pdr1p and Pdr3p. We have constructed a C. albicans CAI4-derived mutant strain carrying a homozygous deletion of the FCR1 gene and analyzed its ability to grow in the presence of FCZ. We found that the fcr1Δ/fcr1Δ mutant displays hyperresistance to FCZ and other antifungal drugs compared to the parental CAI4 strain. This hyperresistance could be reversed to wild-type levels by reintroduction of a plasmid-borne copy of FCR1 into the fcr1Δ/fcr1Δ mutant. Taken together, our results indicate that the FCR1 gene behaves as a negative regulator of drug resistance in C. albicans and constitute the first evidence that FCZ resistance can result from the inactivation of a regulatory factor such as Fcr1p.
Pleiotropic drug resistance (PDR) is characterized by the cross-resistance of cells to a large number of structurally and functionally unrelated cytotoxic compounds. PDR has been extensively studied in the yeast Saccharomyces cerevisiae and involves a network of membrane-associated transporters functioning as energy-dependent drug efflux pumps and of transcription factors regulating the expression of these pumps (reviewed in reference 6). For example, the overexpression of the PDR5, SNQ2, and YOR1 genes, encoding transporters of the ATP-binding cassette (ABC) superfamily, has been shown to result in PDR (16, 36, 38, 57). Although related in sequence, these transporters display distinct drug specificities: Pdr5p has been shown to confer resistance to cycloheximide (CYH), mycotoxins, and azole derivatives (9, 33, 38, 54); Snq2p has been shown to confer resistance to 4-nitroquinoline-N-oxide (4-NQO) and other chemicals (57); and Yor1p has been shown to confer resistance to oligomycin, reveromycin, and aureobasidin (16, 36, 46).
Transcription of PDR5, SNQ2, and YOR1 is controlled by Pdr1p and Pdr3p, two homologous transcription factors belonging to the Zn2C6 binuclear zinc cluster family (5, 7, 17–19, 21, 30, 34–36, 41, 43). Dominant hyperactive mutations at the PDR1 or PDR3 locus lead to the PDR phenotype, which correlates with the overexpression of PDR5, SNQ2, and YOR1 (7, 12, 19, 21, 36, 41, 44). Loss-of-function pdr1 and pdr3 mutants are hypersensitive to various drugs including CYH, 4-NQO, and oligomycin and display decreased levels of PDR5, SNQ2, and YOR1 expression (7, 12, 21, 36, 41). Finally, Pdr1p and Pdr3p have been shown to regulate the expression of other transporter-encoding genes such as HXT9 and HXT11, which code for hexose transporters of the major facilitator (MF) superfamily involved in PDR (45), as well as PDR10 and PDR15, which encode ABC transporters homologous to Pdr5p but whose role in PDR remains to be demonstrated (67).
Transcription factors of the bZip family such as Yap1p are also involved in PDR. Overexpression of Yap1p has been shown to confer resistance to toxic compounds such as CYH, 4-NQO, sulfometuron methyl, 1,10-phenanthroline, and various prooxidants (11, 32, 38, 56, 59, 63, 68). Yap1p has been shown to regulate the expression of the YCF1 gene, which encodes an ABC transporter that functions as a vacuolar glutathione-cadmium conjugate pump to confer cadmium resistance, and of the FLR1 and ATR1 genes, which code for two transporters of the MF superfamily involved in resistance to azole derivatives and to other unrelated drugs (1, 14, 63).
Candida albicans is an opportunistic yeast that causes severe infections in immunocompromised individuals (22). Among the different agents employed in antifungal therapy, the azole derivative fluconazole (FCZ) is the most widely used because of its low toxicity and its high efficacy (49). However, the successful treatment of candidosis by FCZ has been impaired by the emergence of drug-resistant strains in patients undergoing long-term or prophylactic treatment, mostly AIDS patients (24, 39, 49). Studies investigating the mechanisms of FCZ resistance in C. albicans have shown that a large number of resistant strains fail to accumulate FCZ due to an increased drug efflux. This correlates with the overexpression of the CDR1 and CDR2 genes, which encode two ABC transporters highly homologous to S. cerevisiae Pdr5p, and of the MDR1 gene, which codes for an MF transporter with high homology to S. cerevisiae Flr1p (25, 47, 53, 54). We have recently cloned and characterized the C. albicans CAP1 gene, which codes for a bZip transcription factor structurally and functionally similar to the S. cerevisiae Yap1p protein and which activates transcription of the FLR1 gene when overexpressed in S. cerevisiae (1). So far, the regulatory factors controlling the expression of CDR1, CDR2, and MDR1 in C. albicans have not been identified.
The isolation and characterization of a number of C. albicans genes involved in PDR have revealed that their protein products possess structural and functional homologues in S. cerevisiae, suggesting some similarity between the S. cerevisiae and the C. albicans PDR systems. By analogy to the well-studied PDR network of S. cerevisiae, we hypothesized that, in C. albicans, transcriptional regulators functionally homologous to S. cerevisiae Pdr1p and Pdr3p might control the expression of the PDR5 homologues CDR1 and CDR2, causing azole resistance. The aim of the present study was to identify such regulatory factors. This was performed by screening a C. albicans genomic DNA library for functional complementation of an S. cerevisiae pdr1 pdr3 mutant host. This strategy has enabled us to isolate three C. albicans genes able to restore FCZ tolerance in the pdr1 pdr3 strain. This report describes the structural and functional characterization of the FCR1 (for fluconazole resistance 1) gene, which codes for a transcription factor of the C6 zinc cluster family homologous to S. cerevisiae Pdr1p.
MATERIALS AND METHODS
Yeast strains and media.
The S. cerevisiae haploid strain KY320 (MATα ura3-52 ade2-101 trp1-81 lys2-801 his3-Δ200 leu2::PET56) and its isogenic derivative JY312 (MATα pdr1 pdr3::URA3 ade2-101 trp1-81 lys2-801 his3-Δ200 leu2::PET56) were obtained from Joseph Martens, University of Western Ontario, London, Ontario, Canada. The C. albicans strain CAI4 (Δura3::imm434/Δura3::imm434) was used in this study (26). Yeast cells were grown in yeast peptone dextrose (YPD) medium or in synthetic dextrose (SD) medium lacking uracil (SD−ura), leucine (SD−leu), or tryptophan (SD−trp) (58). Yeast transformations were performed according to the method described by Gietz and coauthors (27). Cultures were routinely grown at 30°C.
Isolation of the FCR1, FCR2, and FCR3 genes.
The YEp13-based C. albicans B792 genomic DNA library (a gift from Yigal Koltin, ChemGenics Pharmaceuticals Inc., Cambridge, Mass.) (51) was used to transform S. cerevisiae JY312 to leucine prototrophy. The transformants were grown on SD−leu plates, pooled, and plated onto solid SD−leu medium containing 4 μg of FCZ per ml, a concentration at which the growth of JY312 cells carrying only the empty vector YEp13 is completely inhibited. The viable colonies were scored as FCZ resistant. The plasmids were isolated from 12 resistant colonies and subjected to restriction mapping analysis. Secondary transformants were found to be resistant to FCZ, confirming the plasmid dependency of the resistance phenotype.
Drug resistance assays.
FCZ (a gift from Pfizer Canada Inc.) and ketoconazole (Medisca Inc.) were dissolved in water at concentrations of 10 and 20 mg/ml, respectively. Brefeldin A (Sigma) was dissolved in ethanol at 1 mg/ml. All stock solutions were kept at −20°C. For the MIC determination by plate assays, strains KY320 and JY312 transformed with plasmid YEp13 were streaked for single colonies on SD−leu plates containing increasing concentrations of CYH or FCZ. Cell growth was evaluated after 3 days of incubation at 30°C. The MIC was determined as the lowest concentration of the drug at which cell growth was completely inhibited in this assay. For microtiter plate assays, cells grown for 48 h on selective SD−leu medium were resuspended in a saline solution (0.85%) to an optical density at 600 nm (OD600) of 0.1. These cells were then diluted 100-fold in SD−leu medium. The diluted cell suspensions were added to round-bottom 96-well microtiter plates (50 μl/well, in duplicate) in wells containing equal volumes (50 μl) of medium with different concentrations of FCZ or drug-free medium. The plates were incubated at 30°C for 48 h. Cell growth was evaluated by reading the OD650 in a microplate reader (Vmax; Molecular Devices). The relative growth was calculated as the percent growth in drug-containing medium relative to the control growth in drug-free medium. For the spot assays with the S. cerevisiae transformants, aliquots of serially diluted cultures grown overnight in selective SD−leu medium were spotted onto SD−leu plates containing FCZ at 4 μg/ml or CYH at 0.04 mg/ml and incubated for 3 days at 30°C. For the C. albicans transformants, aliquots of serially diluted cultures grown overnight in selective SD−ura medium were spotted onto YPD plates containing different concentrations of FCZ, ketoconazole, or brefeldin A. The plates were photographed after 3 days of incubation at 30°C.
DNA sequencing and analysis.
Complete sequencing of the FCR1 gene on both DNA strands was performed with custom synthesized oligonucleotides, using the automated sequencing facilities of the Sheldon Biotechnology Centre (McGill University, Montreal, Canada). Sequence analyses were performed with the University of Wisconsin Genetics Computer Group programs (20) and the National Center for Biotechnology Information (NCBI) software.
RNA preparation and Northern blot analysis.
The S. cerevisiae and C. albicans strains were grown in the appropriate medium to an OD600 of 1.0. Total RNA was isolated by the glass-bead extraction method. RNA samples (20 μg) were electrophoresed on a 7.5% formaldehyde–1% agarose gel and transferred by capillarity onto a Zeta-Probe nylon membrane (Bio-Rad Laboratories, Mississauga, Ontario, Canada). Detection of specific RNAs was performed by hybridization at 65°C in 0.5 M NaPO4, pH 7.2–1 mM EDTA–7% sodium dodecyl sulfate–1% bovine serum albumin–100 μg of salmon sperm DNA per ml with 32P-labelled DNA probes, as previously described (1). The PDR5 probe was generated by PCR with primers 5′-CATACAGAAGCTCGAATC and 5′-CCACAGTTGACTGATAGG and overlaps a region from +111 to +447 of the PDR5 gene (positions are relative to the translation initiation codon) (7). The FCR1 probe was a 2.3-kb VspI DNA fragment isolated from clone pDTF5. A PDA1 probe, consisting of a 1.1-kb HindIII-SacII fragment isolated from plasmid pUC4E1α10 (65), and an ACT1 probe (provided by Beatrice Magee, University of Minnesota, St. Paul, Minn.) were used as internal controls to monitor S. cerevisiae and C. albicans RNA loading and transfer.
Construction of a triple pdr1 pdr3 pdr5 mutant strain.
The triple pdr1 pdr3 pdr5 mutant strain TY310 was obtained by deleting the chromosomal PDR5 gene in the JY312 strain by allele replacement, using the one-step PCR amplification method (8). A 900-bp fragment containing the TRP1 gene was generated by amplification with the following primers: 5′-GAAATTAAAGACCCTTTTAAGTTTTCGTATCCGCTCGTTCGAAAGACGGAGAGGGCCAAGAGGG and 5′-GAGCTGG TAAAT TCAAGAAAAT TGAAATGTAGAAAGC TCGC TGAATTCCTGCAGGCAAGTGCA. Each primer contains a sequence derived from the PDR5 open reading frame (ORF) (underlined) followed by a stretch of 17 nucleotides homologous to the TRP1 selectable marker. The PCR product was purified by using the QIAEX II gel extraction kit (QIAGEN Inc., Mississauga, Ontario, Canada), and 0.5 μg of DNA was used to transform the JY312 strain to tryptophan prototrophy. Southern blot analysis with PDR5 or TRP1 as probes indicated that three of five Trp+ transformants analyzed carried the expected pdr5Δ::TRP1 allele. One of these three mutants (TY310) was chosen for further experiments.
Deletion of FCR1.
The plasmid pGEM-7Zf(+) (Promega, Madison, Wis.) was digested with ClaI and XhoI and ligated to a 4-kb ClaI-SalI fragment containing the FCR1 gene isolated from plasmid pDTF1, producing plasmid pGEM-7Z/FCR1. This plasmid was digested with PacI and HincII to remove the entire FCR1 ORF, which was replaced with a 4-kb SalI-BglII fragment containing the hisG-CaURA3-hisG cassette from plasmid pMB-7 (26), to generate pGEM-7Z/fcr1Δ. A linear 6-kb fcr1Δ::hisG-CaURA3-hisG fragment was released from pGEM-7Z/fcr1Δ with SphI and SacI and used to transform C. albicans CAI4 to Ura+ prototrophy. Counter-selection of the URA3 gene was carried out on plates containing 5-fluoroorotic acid (5-FOA; 1 mg/ml) (10), with the exception that uracil was replaced by uridine (25 μg/ml). 5-FOA-resistant colonies were submitted to a second round of transformation with the fcr1Δ::hisG-CaURA3-hisG fragment, followed by counter-selection on 5-FOA. All the strains were analyzed by Southern blotting at each step of the process to confirm their genotype at the FCR1 locus.
Construction of C. albicans FCR1 expression plasmids.
The pVEC/FCR1 plasmid, which contains the complete FCR1 gene under the control of its own promoter, was constructed by cloning a 6.5-kb ClaI fragment, after it was isolated from pDTF5 and blunt-ended with T4 DNA polymerase, into the SmaI-cleaved pVEC vector, which carries a C. albicans autonomously replicating sequence and the CaURA3 gene as a selectable marker (a gift from Beatrice Magee, University of Minnesota) (40). The YPB-ADH/FCR1 plasmid was constructed by inserting a 2.3-kb VspI fragment, after it was isolated from pDTF1 and blunt-ended with T4 DNA polymerase, into the BglII-cleaved YPB-ADHpt vector which carries the C. albicans ADH1 promoter and terminator regions, a C. albicans autonomously replicating sequence, and the CaURA3 marker (a gift from Alistair Brown, University of Aberdeen, Aberdeen, United Kingdom) (3). This construct places the FCR1 structural gene, flanked by 137 bp of 5′ noncoding and 600 bp of 3′ of noncoding sequences, under the control of the ADH1 promoter.
Genomic DNA isolation and Southern blot analyses.
C. albicans genomic DNA was prepared essentially as described for S. cerevisiae (50), except that zymolase was added at a final concentration of 0.8 mg/ml. Genomic DNAs (2 μg) were digested with HindIII, electrophoresed on a 1% agarose gel, and transferred to a nylon membrane. Hybridization was performed as previously described (4), using a 2.3-kb VspI fragment from FCR1 or a 0.9-kb BamHI-BglII hisG fragment as probe.
Nucleotide sequence accession number.
The nucleotide sequence of the FCR1 gene has been deposited in the GenBank database under accession no. AF057038.
RESULTS
PDR1 and PDR3 are required for normal FCZ tolerance in S. cerevisiae.
Previous studies have demonstrated a prominent role for the S. cerevisiae regulatory factors Pdr1p and Pdr3p in the PDR phenotype (reviewed in reference 6). These transcription factors mediate their action by controlling the expression of different drug efflux pumps, including Pdr5p (7, 35). Cells carrying a pdr5 deletion are hypersensitive to the antifungal agent FCZ (54), suggesting that cells bearing a mutation in the PDR1 and PDR3 genes should display a similar phenotype. We have investigated the involvement of Pdr1p and Pdr3p in FCZ resistance by comparing the ability of the wild-type strain KY320 and its isogenic pdr1 pdr3 mutant derivative JY312 to grow in the presence of FCZ. To this end, both strains were transformed with plasmid YEp13 and assayed for resistance to FCZ and to CYH (a toxic compound known to belong to the PDR1, PDR3, and PDR5 spectrum of drugs) (6) by plating these transformants on SD−leu plates containing increasing concentrations of these drugs. We observed that KY320 cells were unable to grow at 0.1 μg of CYH per ml on synthetic medium, defined here as the MIC. As expected, this value was decreased to 0.025 μg/ml for strain JY312, which carries a pdr1 pdr3 double mutation. Interestingly, we found that the growth of KY320 cells was inhibited at the FCZ concentration of 50 μg/ml, whereas a concentration of FCZ of only 2 μg/ml was sufficient to prevent the growth of JY312 cells. These data clearly demonstrate a functional role for the PDR1 and PDR3 genes in maintaining normal levels of FCZ tolerance in S. cerevisiae, presumably by supporting normal levels of PDR5 expression.
Cloning of C. albicans genes complementing the pdr1 pdr3 mutation.
The marked hypersensitivity of strain JY312 to FCZ was used as a phenotype for functional complementation with a C. albicans genomic DNA library. JY312 cells were transformed with a C. albicans genomic library cloned in YEp13, a LEU2-based multicopy vector (51). Leu+ transformants were then plated on media containing 4 μg of FCZ per ml, a concentration at which the growth of JY312[YEp13] transformants is completely inhibited. The plasmids from 12 FCZ-resistant colonies were purified and retransformed to confirm the plasmid dependency of the resistant phenotype. Restriction mapping analysis showed that four groups of plasmids were obtained, and one representative plasmid from each group was chosen for further analysis. These plasmids contain 3.4-, 2.8-, 9-, and 11.5-kb C. albicans DNA fragments and were designated pDTF1, pDTF2, pDTF3, and pDTF5, respectively (data not shown). Further restriction mapping indicated that the genomic DNA insert of pDTF1 was identical to an internal segment of the large insert of pDTF5.
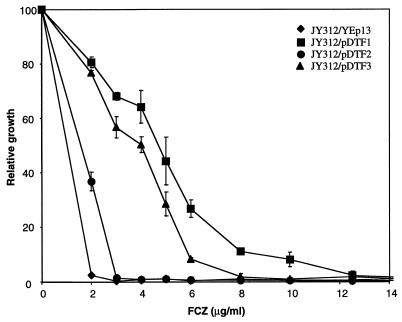
To quantify the level of resistance conferred by each plasmid, we used a microtiter plate assay to determine the MIC of FCZ for the JY312 secondary transformants carrying plasmids YEp13, pDTF1, pDTF2, and pDTF3. The growth of the pDTF1 transformant was inhibited at 12 μg of FCZ/ml, while cells transformed with pDTF2 and pDTF3 could grow in the presence of up to 3 and 8 μg of FCZ/ml, respectively (Fig. 1). The inability of cells transformed with pDTF2 to grow at concentrations of FCZ higher than 3 μg/ml in this assay even though pDTF2 was isolated in a screen employing 4 μg of FCZ/ml is probably due to differences between liquid assays and plate assays for drug tolerance. Nevertheless, these results confirmed that the FCZ resistance phenotype was indeed plasmid dependent. Southern blot analysis demonstrated that the genes carried by these plasmids were different from ERG16, CDR1, CDR2, MDR1, or CAP1 (1, 25, 37, 47, 53), indicating that they contain new C. albicans FCZ resistance genes (data not shown).
FIG. 1.
FCZ resistance phenotype of JY312 transformed with the control vector (YEp13) or with YEp13 carrying the different C. albicans genomic DNA fragments (pDTF1, pDTF2, and pDTF3). The degree of FCZ resistance was determined by a microtiter plate assay as described in Materials and Methods. The percentage of growth in different concentrations of FCZ is expressed relative to growth in drug-free medium (100%). The values are the averages of three independent experiments each performed in duplicate.
Transcriptional control of the S. cerevisiae PDR5 gene by the three different C. albicans clones.
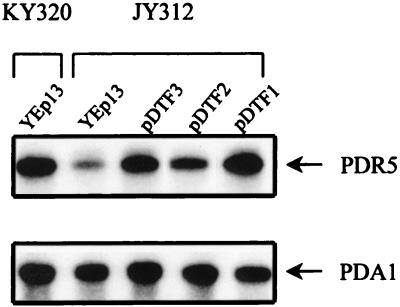
Previous studies have demonstrated the involvement of the transporter-encoding genes PDR5 and FLR1 in FCZ resistance in S. cerevisiae (1, 54). PDR5 expression is under the control of the two homologous zinc finger transcription factors Pdr1p and Pdr3p (35), whereas FLR1 expression is under the control of the bZIP transcription factor Yap1p (1). In order to determine whether the FCZ resistance conferred by pDTF1, pDTF2, or pDTF3 in strain JY312 was associated with increased PDR5 and/or FLR1 expression, we performed a Northern blot analysis. Total RNA extracted from strains KY320 and JY312 transformed with YEp13 or from JY312 transformed with plasmids pDTF1, pDTF2, or pDTF3 was subjected to Northern blot analysis with a probe for PDR5 (Fig. 2, top panel) or for PDA1 (Fig. 2, bottom panel), a gene which is constitutively expressed in S. cerevisiae under different growth conditions and which is used as a standard for mRNA quantitation (65). The amount of PDR5 transcripts was severely reduced in JY312[YEp13] cells lacking PDR1 and PDR3 as compared to the wild-type KY320[YEp13] cells, as expected from previous studies showing that PDR1 and PDR3 are required for maintaining normal levels of PDR5 expression (41, 43). When the JY312 strain was transformed with pDTF1, pDTF2, or pDTF3, a reproducible increase in PDR5 mRNA levels was observed compared to the JY312[YEp13] control (Fig. 2). Both pDTF1 and pDTF3 stimulate PDR5 transcription to levels similar to those detected in the wild-type KY320 cells transformed with the control plasmid, whereas only a slight increase in PDR5 mRNA is induced by pDTF2. This membrane was rehybridized with an FLR1 probe, showing that no FLR1 RNA was detected in KY320[YEp13] or in the JY312 strain carrying the pDTF1, pDTF2, or pDTF3 plasmid (data not shown). These data (i) support the hypothesis that the pDTF plasmids contain functional homologues of PDR1 and PDR3 which are able to activate PDR5 expression, either directly or indirectly, and (ii) suggest that the FCZ-resistant phenotype conferred by pDTF1, pDTF2, or pDTF3 is PDR5-mediated.
FIG. 2.
Northern blot analysis of total RNA extracted from the wild-type strain, KY320, transformed with YEp13 and from the isogenic pdr1 pdr3 mutant strain, JY312, transformed with YEp13, pDTF1, pDTF2, or pDTF3. The filter was hybridized sequentially with PDR5 and PDA1 32P-labelled probes. Autoradiography was carried out for 18 h for PDR5 and for 5 h for PDA1, with two intensifying screens at −80°C.
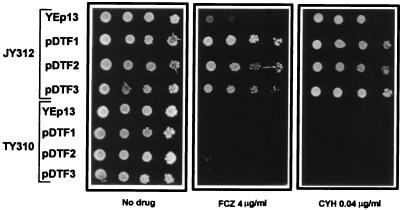
To test this hypothesis, we deleted PDR5 from JY312. The resulting strain, TY310, was transformed with the above plasmids, and the transformants were tested for FCZ resistance by spot assay. As shown in Fig. 3, JY312 cells transformed with plasmids pDTF1, pDTF2, or pDTF3 can grow on medium containing FCZ at 4 μg/ml (with the highest level of resistance conferred by pDTF1), unlike JY312[YEp13] control cells, which barely grow under these conditions (Fig. 3, middle panel). These results confirm our previous finding, obtained using a microtiter plate assay, that plasmids pDTF1, pDTF2, and pDTF3 can confer FCZ resistance to JY312 cells in liquid medium (Fig. 1). However, deletion of PDR5 in the pdr1 pdr3 strain completely abrogated the ability of the three plasmids to confer FCZ resistance, demonstrating that PDR5 is required for the FCZ resistance phenotype conferred by pDTF1, pDTF2, and pDTF3 in JY312 cells. Similar results were obtained when the different transformants were tested for their level of resistance to CYH, another substrate of the Pdr5p transporter (Fig. 3, right panel). These results are consistent with the hypothesis that pDTF1, pDTF2, and pDTF3 confer FCZ and CYH resistance through activation of PDR5 expression. The corresponding genes within these plasmids responsible for FCZ resistance have been named FCR1, FCR2, and FCR3 (for fluconazole resistance), respectively.
FIG. 3.
The PDR5 gene is required for the FCZ and CYH resistance phenotypes conferred by pDTF1, pDTF2, and pDTF3. Yeast strains JY312 (pdr1 pdr3) and TY310 (pdr1 pdr3 pdr5) were transformed with the plasmid YEp13, pDTF1, pDTF2, or pDTF3. The transformants were analyzed by spot assay for FCZ or CYH resistance on SD−leu plates containing the indicated concentrations of FCZ or CYH. The plates were photographed after 3 days of incubation at 30°C.
FCR1 encodes a putative regulatory factor, which is a member of the family of zinc cluster proteins.
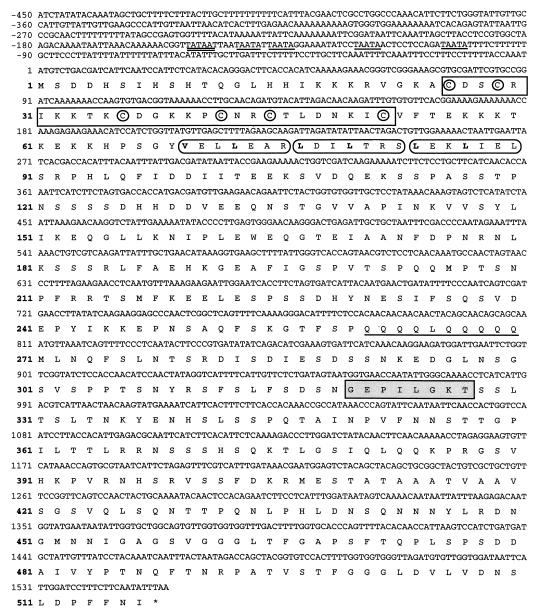
DNA sequence determination of the 3.4-kb insert in pDTF1 identified one ORF of 1,553 bp, whose sequence is presented in Fig. 4. The most upstream in-frame ATG codon of the FCR1 ORF has a conserved adenosine at position −3, in agreement with the consensus sequence for translation initiation in yeast (23). The 5′ noncoding region contains a putative TATA box at position −151 (5′-TATAAT [29]) followed by four pentanucleotide repeats (5′-TAATA) at positions −146, −137, −120, and −104 (positions are relative to the A of the ATG initiation codon set at +1). DNA sequence determination of a 1-kb region downstream of the stop codon identified the presence of consensus sequences for mRNA 3′-end formation in yeast (data not shown) (28).
FIG. 4.
Nucleotide and deduced amino acid sequences of FCR1. Nucleotide and amino acid (boldface type) numbers are indicated on the left. In the 5′ noncoding region, the putative TATA box is double underlined and the four TAATA repeats are underlined. The C6 zinc cluster motif is boxed, and the conserved cysteine residues are circled. A potential coiled-coil structure consisting of three heptad repeats is shown (oval boxes), with the aliphatic residues in the first and fourth positions of each repeat shown in bold. The stretch of glutamine residues is underlined. The consensus ATP and/or GTP binding motif is boxed and shaded in grey.
FCR1 codes for a Ser/Thr-rich protein (21%) with a predicted molecular weight of 57,000 and an estimated isoelectric point of 6.98. The N-terminal domain of the Fcr1p protein contains six cysteines with a spacing conforming to the pattern CX2CX6CX5–9CX2CX6C and extending from amino acid positions 26 to 52 (Fig. 4). This motif, referred to as the C6 zinc cluster or the Zn2Cys6 binuclear cluster motif, is present in the DNA-binding domain of several fungal transcriptional regulators, including the well characterized proteins Gal4p, Ppr1p, Put4p, Pdr1p, and Pdr3p (reviewed in reference 55). Upstream of the zinc cluster domain, a cluster of basic residues is found which could be involved in nuclear localization. Downstream of the zinc cluster, in the region spanning amino acid positions 69 to 92, there is a potential coiled-coil structure formed by three heptad repeats, each repeat containing an aliphatic residue at the first and fourth positions (Fig. 4). Based on the crystal structures of Gal4p and Ppr1p, this coiled-coil structure is predicted to form an amphipathic α-helix and to mediate homodimerization (55). The internal region of Fcr1p, overlapping residues 189 to 351, is highly charged and acidic (net charge, −8). It contains a stretch of nine glutamines interrupted by one leucine residue (Fig. 4). Glutamine-rich sequences are found in several transcription factors that function as activators as well as repressors (60, 61). The internal domain of Fcr1p also contains a potential ATP and/or GTP binding motif (known as Walker type A or P-loop motif) at amino acids 320 to 327 (GEPILGKT) (Fig. 4) that conforms to the consensus sequence GX4GKS/T/G (62). This motif is usually found in proteins that synthesize, bind, and/or hydrolyze ATP (62). However, functional ATP and/or GTP binding motifs have been recently identified in a small number of transcriptional regulators (13, 15), suggesting that this motif may be important for Fcr1p activity. Finally, the Fcr1p protein displays several putative phosphorylation sites for casein kinase II, protein kinase C, and cyclic-AMP-dependent protein kinase (not shown). The functionality of the different sequence motifs identified in Fcr1p remains to be determined.
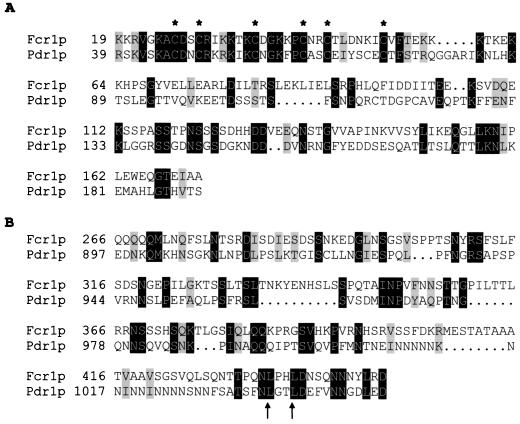
Searching the S. cerevisiae genome database with the Fcr1p protein sequence, using the FASTA program, revealed similarity with the zinc cluster domains of several yeast regulatory factors. A high level of similarity was detected with two putative transcriptional regulatory factors encoded by the ORFs YMR019w (32% identity over 108 amino acids) and YIL130w (26% identity over 233 amino acids) and with Pdr1p (24% identity over 176 amino acids) (data not shown). A sequence alignment of the N-terminal regions of Fcr1p and Pdr1p shows that the two proteins are highly homologous in their DNA-binding domains (overlapping the conserved cysteine residues) but that the sequence homology extends further downstream into the predicted linker and dimerization domains (55) (Fig. 5A). Our sequence analysis also revealed that the region spanning residues 266 to 449 in Fcr1p displays significant sequence homology with the C-terminal region of Pdr1p (Fig. 5B). This domain of Pdr1p has been shown to interact in vivo with the coactivator/repressor ADA complex, an association which inhibits the transactivation activity of Pdr1p (42). Furthermore, dominant gain-of-function mutations resulting in PDR due to the hyperactive transcription of the PDR5, SNQ2, and YOR1 genes in S. cerevisiae have been identified in the C-terminal domain of Pdr1p in the pdr1-8 and PDR1-12 mutants (L1036W and L1039Q; amino acid positions according to reference 5) (12, 64). Our sequence alignment indicates that these two leucine residues are conserved in Fcr1p (Fig. 5B). It will be interesting to test the effects of similar mutations in Fcr1p.
FIG. 5.
Sequence homologies between the N-terminal (A) and C-terminal (B) domains of Fcr1p and Pdr1p. The amino acid sequences of Fcr1p and Pdr1p were aligned with the BESTFIT program (20). Identical and conserved residues are shaded in black and grey, respectively, using the Boxshade program. The conserved cysteines in the C6 zinc cluster motif are indicated by asterisks, and the leucine residues mutated in the gain-of-function pdr1-8 and PDR1-12 mutants (L1036W and L1039Q [12, 66]) are indicated by arrows.
Construction of an FCR1 null mutant.
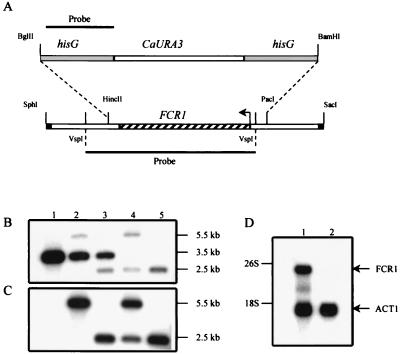
To investigate the role of the FCR1 gene in drug resistance, we deleted both FCR1 alleles from the Ura− auxotrophic CAI4 strain, using the “ura-blaster” strategy (26) (Fig. 6A). The hisG-CaURA3-hisG cassette was used to replace the entire FCR1 gene in pGEM-7Z/FCR1. The resulting construct was digested with SphI and SacI to produce a linear fragment consisting of the hisG-CaURA3-hisG selectable marker flanked on both sides by FCR1 noncoding sequences (Fig. 6A). This fcr1Δ::hisG-CaURA3-hisG deletion fragment was then used to transform the CAI4 strain. The replacement of one chromosomal copy of the FCR1 gene with the fcr1Δ::hisG-CaURA3-hisG fragment in Ura+ prototrophs was verified by Southern analysis, using an FCR1 (Fig. 6B) or a hisG (Fig. 6C) fragment as probe. The size of the 3.5-kb HindIII fragment corresponding to the intact allele of FCR1 (Fig. 6B, lane 1) was increased to 5.5 kb upon integration of the fcr1Δ::hisG-CaURA3-hisG cassette at this locus (Fig. 6B and C, lanes 2). Removal of the CaURA3 gene was achieved by counter-selection on 5-FOA. The size of the 5.5-kb fragment was decreased to 2.5 kb upon looping out of the CaURA3 gene via homologous recombination between the two hisG direct repeats (Fig. 6B and C, lanes 3). Deletion of the second FCR1 allele was performed as described above to generate a Ura+ fcr1Δ::hisG/fcr1Δ::hisG-CaURA3-hisG strain (Fig. 6B and C, lanes 4). Finally, excision of CaURA3 on 5-FOA yielded strain FM7 (fcr1Δ::hisG/fcr1Δ::hisG). As FM7 cells generated from the final step of the disruption are auxotrophic for uridine, they are thus suitable for transformation with a URA3-based vector for the reintroduction of the FCR1 gene in a null mutant background. The FM7 strain is viable, and we found that there was no substantial difference between FM7 and CAI4 cells with respect to colony size or growth rates on rich and minimal medium supplemented with different carbon sources (data not shown). These results indicate that the product of the FCR1 gene is not essential for growth of the CAI4 strain, at least under the growth conditions tested.
FIG. 6.
Chromosomal deletion of the FCR1 locus. (A) Schematic representation of the strategy used to generate the FCR1 deletion in the strain CAI4. The hisG-CaURA3-hisG cassette was used to replace the HincII-PacI fragment containing the FCR1 ORF. Only the relevant restriction sites are shown (the SphI and SacI sites are derived from the pGEM7Z-f(+) vector). The 0.9-kb hisG fragment (top) and the 2.3-kb VspI fragment (bottom) were used as probes to monitor the recombination events. (B and C) Southern blot analyses of genomic DNA from the parental CAI4 strain (lanes 1), the FCR1/fcr1Δ::hisG-CaURA3-hisG heterozygous strain (lanes 2), the FCR1/fcr1Δ::hisG strain after 5-FOA counter-selection (lanes 3), the fcr1Δ::hisG/fcr1Δ::hisG-CaURA3-hisG strain (lanes 4), and the fcr1Δ::hisG/fcr1Δ::hisG homozygous FM7 strain after the second 5-FOA counter-selection (lanes 5). The genomic DNA was digested with HindIII, electrophoresed in duplicate on an agarose gel, and transferred to nylon membranes. The blots were probed with the 2.3-kb VspI FCR1 fragment (B) or the 0.9-kb hisG fragment (C). Autoradiographies were for 10 and 15 h, respectively, with two intensifying screens at −80°C. The sizes of the fragments are indicated in kilobases. (D) Northern blot analysis of FCR1 in strains CAI4 and FM7. Total RNA (20 μg) prepared from the CAI4 (lane 1) and FM7 (lane 2) strains was electrophoresed on a 1% agarose gel, transferred to a nylon membrane, and probed simultaneously with an FCR1 probe and an ACT1 probe. Autoradiography was for 48 h, with two intensifying screens at −80°C.
A Northern blot analysis was performed to analyze the expression of FCR1 in C. albicans and to verify the absence of FCR1 mRNA in the FM7 strain (Fig. 6D). Total RNA was prepared from CAI4 and FM7 cells and analyzed with an FCR1 probe. This analysis revealed two FCR1-specific transcripts, a major transcript of 3 kb and a minor transcript of 2.1 kb (Fig. 6D, lane 1). No FCR1 transcript could be detected with the FCR1 probe in total RNA extracted from FM7 (Fig. 6D, lane 2), even after prolonged exposure of the blot (data not shown). These results demonstrate that the 3-kb and 2.1-kb transcripts are both FCR1 specific and confirm that the FCR1 gene has been successfully deleted in FM7 cells. Given that the size of the predicted FCR1 ORF is 1.5 kb, the results from the Northern blot analysis suggest that the FCR1 transcripts probably contain long 5′- and/or 3′-untranslated regions.
The loss of FCR1 results in hyperresistance of the cells to FCZ and other antifungal drugs.
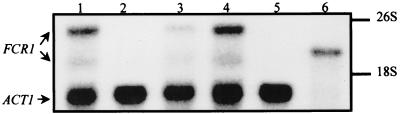
Since FCR1 is able to restore FCZ tolerance in a pdr1 pdr3 mutant strain through the activation of PDR5 expression in S. cerevisiae, we anticipated that the deletion of FCR1 in C. albicans would result in an increased susceptibility of the cells to FCZ. Unexpectedly, a comparison of the levels of FCZ susceptibility of the CAI4 (FCR1/FCR1) and FM7 (fcr1Δ/fcr1Δ) strains by spot assay demonstrated that the FM7 strain was more resistant to FCZ than the CAI4 strain (data not shown). The same phenotype was also observed with two additional homozygous fcr1Δ/fcr1Δ strains independently derived from CAI4, corroborating the results obtained with FM7. To confirm that the hyperresistant phenotype of FM7 was a consequence of the FCR1 deletion, we set out to determine if reintroduction of the FCR1 gene in the FM7 mutant would restore wild-type levels of FCZ susceptibility to the cells. To this end, two vectors, each carrying a functional copy of the FCR1 gene, were constructed. First, a 6.5-kb ClaI fragment isolated from pDTF5 and containing the entire FCR1 gene, including the promoter region, was cloned into the pVEC vector (40) to produce plasmid pVEC/FCR1. Second, a plasmid consisting of the FCR1 coding region under the control of the strong C. albicans ADH1 promoter was constructed by inserting a 2.3-kb VspI fragment isolated from pDTF1 into the vector YPB-ADHpt (3), generating plasmid YPB-ADH/FCR1. The pVEC/FCR1 and YPB-ADH/FCR1 plasmids were introduced into FM7, and the resulting transformants were analyzed by Northern blotting for their levels of FCR1 expression, together with CAI4 and FM7 transformed with the pVEC and YPB-ADH empty vectors as controls (Fig. 7). Total RNA was extracted from the different transformants grown in SD−ura selective media and analyzed simultaneously with the FCR1 and ACT1 probes. The results of this experiment confirmed the presence of the 3-kb and 2.1-kb FCR1 transcripts in the CAI4 cells transformed with the control vectors, transcripts which are absent in the FM7 transformants (Fig. 7; compare lanes 1 and 4 with lanes 2 and 5, respectively). FM7 cells transformed with pVEC/FCR1 were found to express both the 3-kb and 2.1-kb FCR1 transcripts, although at much lower levels than the CAI4[pVEC] transformants (Fig. 7, lane 3). In FM7 cells transformed with YPB-ADH/FCR1, a single FCR1 transcript of approximately 2.4 kb, which probably originates from the utilization of different transcription initiation and/or termination sites for FCR1 in the YPB-ADH/FCR1 construct, was detected (Fig. 7, lane 6). This transcript is expressed at very high levels, given that the amount of RNA loaded was 50 times less for this sample than for the other samples (thus resulting in the absence of ACT1 signal in lane 6).
FIG. 7.
Analysis of FCR1 expression in the C. albicans transformants CAI4[pVEC] (lane 1), FM7[pVEC] (lane 2), FM7[pVEC/FCR1] (lane 3), CAI4[YPB-ADH] (lane 4), FM7[YPB-ADH] (lane 5), and FM7[YPB-ADH/FCR1] (lane 6). Total RNA samples (lanes 1 to 5 contained 20 μg, and lane 6 contained 0.4 μg) were electrophoresed on an agarose gel and transferred to a nylon membrane. The membrane was hybridized simultaneously with an FCR1 probe and an ACT1 probe. Autoradiography was for 36 h, with two intensifying screens at −80°C.
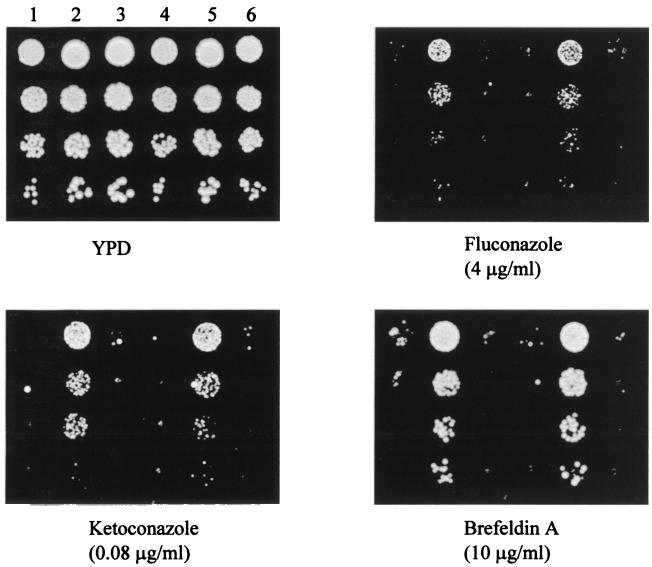
These transformants were analyzed by spot assay to determine their levels of FCZ resistance (Fig. 8). The results of this experiment confirmed that the FM7 strain, transformed with either pVEC or YPB-ADH, is more resistant to FCZ than the CAI4 strain transformed with the same plasmids (Fig. 8; compare lanes 2 and 5 with lanes 1 and 4, respectively). Moreover, introduction of pVEC/FCR1 in FM7 was found to revert the hyperresistant phenotype of the cells to a level of FCZ susceptibility similar to that observed in the CAI4[pVEC] transformants (Fig. 8; compare lanes 3 and 1), confirming that the hyperresistant phenotype of FM7 is indeed a consequence of the FCR1 deletion. Apparently, the low level of FCR1 expression detected in the FM7[pVEC/FCR1] transformants is sufficient to revert the FM7 hyperresistant phenotype (Fig. 7, lane 3). Similar results were also obtained with the YPB-ADH/FCR1 plasmid, confirming that the FCR1 ORF is sufficient for the reversion (Fig. 8). These transformants were also tested by spot assay for their levels of susceptibility to the antifungal drugs ketoconazole and brefeldin A and yielded essentially identical results to those obtained with FCZ (Fig. 8). Taken together, our results indicate that the FCR1 gene behaves as a negative regulator of drug resistance in C. albicans and demonstrate that azole resistance can result from the inactivation of a negative regulatory factor, such as Fcr1p.
FIG. 8.
Drug resistance phenotypes of the C. albicans transformants CAI4[pVEC] (lane 1), FM7[pVEC] (lane 2), FM7[pVEC/FCR1] (lane 3), CAI4[YPB-ADH] (lane 4), FM7[YPB-ADH] (lane 5), and FM7[YPB-ADH/FCR1] (lane 6). The different transformants were grown in SD−ura liquid medium and analyzed by spot assay on YPD plates in the absence (YPD) or in the presence of the indicated compounds (fluconazole, ketoconazole, and brefeldin A). Growth was recorded following incubation of the plates for two days at 30°C.
DISCUSSION
The heterologous expression of C. albicans genes in S. cerevisiae constitutes a powerful approach to identify of C. albicans genes involved in different biological processes, including PDR. Based on the apparent conservation of the PDR networks between these two yeasts, we hypothesized that, in C. albicans, transcriptional regulators functionally homologous to S. cerevisiae Pdr1p and Pdr3p might control the expression of transporter-encoding genes to cause drug resistance. In this work, we have used functional complementation of an S. cerevisiae pdr1 pdr3 strain to isolate three C. albicans genes, named FCR1, FCR2, and FCR3, capable of complementing the FCZ hypersusceptibility of the mutant strain. As judged from the results of the drug resistance assays, complementation of the FCZ hypersusceptibility of the pdr1 pdr3 mutant strain by the three genes was only partial. This could be explained by the inability of a single factor to substitute for the simultaneous absence of Pdr1p and Pdr3p in that strain. Indeed, it has been shown that Pdr3p itself can only partially complement the CYH hypersusceptibility of a double pdr1 pdr3 strain (18). Alternatively, this partial complementation could be a consequence of the heterologous expression of C. albicans genes in S. cerevisiae cells, as previously reported for other C. albicans genes (1). Northern blot analysis showed that each of these C. albicans genes is able to increase the expression of the PDR5 gene in the mutant strain and therefore is likely to encode a transcriptional regulatory factor. Overexpression of these genes in the wild-type PDR1 PDR3 parental strain did not result in increased FCZ resistance, consistent with the idea that they probably encode functional homologues of the Pdr1p and Pdr3p proteins (data not shown). For FCR1, this hypothesis was supported by nucleotide sequencing of the gene which was found to code for a regulatory factor belonging to the yeast zinc cluster family and which is homologous to S. cerevisiae Pdr1p (Fig. 5).
Sequencing of the S. cerevisiae genome has revealed the existence of a large family of regulatory factors characterized by the presence of a zinc cluster DNA-binding domain (55). This domain contains six highly conserved cysteine residues that bind to two zinc atoms, forming a structure (Zn2Cys6) which is required for the recognition of specific DNA sequences. Zinc cluster proteins have been shown to bind as homodimers to a pair of CGG triplets oriented either as direct, inverted, or everted repeats (reference 31 and references therein). The presence of a perfectly conserved zinc cluster motif clearly identifies Fcr1p as being a member of this family (Fig. 4). In addition to the zinc cluster DNA-binding domain, the general structure of these proteins includes a dimerization element, a linker region that connects the dimerization element to the zinc cluster, and various transactivation domains (48, 55). All of these domains were also found within Fcr1p (Fig. 4). In addition, many members of this family, including Gal4p, Pdr1p, and Pdr3p, also contain a weakly conserved internal region with potential regulatory functions, called the middle homology region, which is apparently absent in Fcr1p (55).
Sequence comparison analyses indicate that the zinc cluster domains of Fcr1p and Pdr1p are 53% identical and that this homology extends further downstream into the linker and dimerization domains. Also, the C-terminal domain of Fcr1p displays 29% identity with the C-terminal domain of Pdr1p, which has been shown to interact with the coactivator/repressor ADA complex and is believed to function as a transcriptional activating domain (42). Moreover, we find that two leucine residues, which are mutated in gain-of-function mutants of Pdr1p and which are associated with increased levels of PDR5 mRNA in these mutants, are conserved in Fcr1p (Fig. 5B) (5, 12, 64). Therefore, Fcr1p shares with Pdr1p two functional modules which could be involved in the transcriptional activation of PDR5 in S. cerevisiae: a DNA-binding domain to recognize upstream activating sequences in PDR5 and an activation domain to interact with the basal transcriptional machinery. Sequence comparison analysis did not identify other regions of homology between Fcr1p and Pdr1p outside these two domains. This is not surprising, given that the two proteins have quite different lengths (1,068 amino acids for Pdr1p versus 517 amino acids for Fcr1p). Consequently, it is difficult to conclude whether Fcr1p is the orthologue of Pdr1p in C. albicans. Nevertheless, our data clearly show that the two proteins are structurally and functionally related.
Different lines of evidence indicate that the ability of FCR1 to restore FCZ tolerance in the JY312 (pdr1 pdr3) strain is PDR5 mediated. First, expression of FCR1 in JY312 results in increased levels of PDR5 expression (Fig. 2). Second, deletion of PDR5 in JY312 completely abrogates the ability of FCR1 to restore FCZ resistance in the TY310 (pdr1 pdr3 pdr5) strain, indicating a functional interaction between FCR1 and PDR5 (Fig. 3). Third, the FCR1 gene product is homologous to Pdr1p and Pdr3p, which are involved in the transcriptional control of PDR5 (Fig. 5). Biochemical analyses have identified three sites in the PDR5 promoter, designated PDREs (for Pdr1p and/or Pdr3p response elements), with the consensus sequence 5′-TCCGCGGA, which are bound in vitro by both Pdr1p and Pdr3p (34, 35). Mutational analyses of these sequences have shown that each PDRE site is required for PDR5 promoter function and for drug resistance (35). It is thus possible that Fcr1p directly activates PDR5 transcription in S. cerevisiae by binding to the PDREs present in the PDR5 promoter. However, it is also possible that Fcr1p activates the expression of PDR5 through binding to DNA sequence elements distinct from the PDREs or that Fcr1p controls the expression of PDR5 in an indirect manner, by activating other factors involved in the regulation of PDR5.
We have investigated the role of Fcr1p in C. albicans drug resistance by deleting the FCR1 gene in strain CAI4. Surprisingly, we found that the resulting fcr1Δ/fcr1Δ deletion strain was more resistant to a number of structurally unrelated drugs, including FCZ, ketoconazole, and brefeldin A (Fig. 8) as well as fluphenazine and itraconazole (data not shown), than the wild-type CAI4 strain. This phenotype was confirmed by the facts that (i) three independently derived fcr1Δ/fcr1Δ deletion strains were found to display the same hyperresistant phenotype (data not shown) and that (ii) introduction of a plasmid-borne copy of the FCR1 gene in the fcr1Δ/fcr1Δ mutant was able to revert the hyperresistance of the cells to a level of susceptibility similar to that of the CAI4 parental strain (Fig. 8). Moreover, we found that the fcr1Δ/fcr1Δ mutant strain was not more resistant to other drugs, such as 4-nitroquinoline-N-oxide and 1,10-phenanthroline, demonstrating that the drug-resistant phenotype resulting from the fcr1Δ/fcr1Δ deletion is not generalized, but is indeed specific for certain types of drugs (data not shown). Taken together, these results clearly indicate that Fcr1p functions as a negative determinant of PDR in C. albicans. This finding was unexpected, in light of our demonstration that Fcr1p behaves as a positive regulator of both PDR5 and drug resistance in S. cerevisiae. A potential reason for this difference is that, as already shown for other C. albicans genes, FCR1 could behave as a mutant when expressed in a heterologous host such as S. cerevisiae (66). An explanation for the hyperresistance of the fcr1Δ/fcr1Δ mutant strain is that Fcr1p functions as an inhibitor of PDR in C. albicans by negatively regulating the expression of one or more target gene(s) mediating this resistance. Interestingly, we find that the set of toxic compounds to which the fcr1Δ/fcr1Δ mutant strain is hyperresistant overlaps with those to which CDR1- and CDR2-deleted strains are hypersensitive (52, 53). Consequently, one may postulate that Fcr1p negatively regulates the expression of CDR1, CDR2, or other genes conferring similar phenotypes in C. albicans. Alternatively, it is possible that Fcr1p positively regulates a gene whose expression confers drug sensitivity. Such a situation has been previously identified with the HXT9/HXT11 genes, which are under the control of PDR1 and PDR3 and whose deletion results in increased resistance to different drugs (45). The identification of the Fcr1p gene targets and the determination of whether Fcr1p acts as a positive or negative regulator should lead to a better understanding of the molecular mechanisms of drug resistance in this pathogenic yeast.
ACKNOWLEDGMENTS
We are very grateful to Joseph Martens for providing yeast strains, to Yigal Koltin for the C. albicans library, and to Beatrice Magee and Alistair Brown for the C. albicans vectors.
This work was supported by a joint research grant to M.R. from the Medical Research Council (MRC) of Canada and Pfizer Canada Inc. M.R. is supported by a scholarship from MRC.
REFERENCES
- 1.Alarco A-M, Balan I, Talibi D, Mainville N, Raymond M. AP1-mediated multidrug resistance in Saccharomyces cerevisiae requires FLR1 encoding a transporter of the major facilitator superfamily. J Biol Chem. 1997;272:19304–19313. doi: 10.1074/jbc.272.31.19304. [DOI] [PubMed] [Google Scholar]
- 2.Albertson G D, Niimi M, Cannon R D, Jenkinson H F. Multiple efflux mechanisms are involved in Candida albicans fluconazole resistance. Antimicrob Agents Chemother. 1996;40:2835–2841. doi: 10.1128/aac.40.12.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey D A, Feldmann P J, Bovey M, Gow N A, Brown A J. The Candida albicans HYR1 gene, which is activated in response to hyphal development, belongs to a gene family encoding yeast cell wall proteins. J Bacteriol. 1996;178:5353–5360. doi: 10.1128/jb.178.18.5353-5360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Balan I, Alarco A-M, Raymond M. The Candida albicans CDR3 gene codes for an opaque-phase ABC transporter. J Bacteriol. 1997;179:7210–7218. doi: 10.1128/jb.179.23.7210-7218.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Balzi E, Chen W, Ulaszewski S, Capieaux E, Goffeau A. The multidrug resistance gene PDR1 from Saccharomyces cerevisiae. J Biol Chem. 1987;262:16871–16879. [PubMed] [Google Scholar]
- 6.Balzi E, Goffeau A. Yeast multidrug resistance—the PDR network. J Bioenerg Biomembr. 1995;27:71–76. doi: 10.1007/BF02110333. [DOI] [PubMed] [Google Scholar]
- 7.Balzi E, Wang M, Leterme S, Van Dyck L, Goffeau A. PDR5, a novel yeast multidrug resistance conferring transporter controlled by the transcription regulator PDR1. J Biol Chem. 1994;269:2206–2214. [PubMed] [Google Scholar]
- 8.Baudin A, Ozier-Kalogeropoulos O, Denouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bissinger P H, Kuchler K. Molecular cloning and expression of the Saccharomyces cerevisiae STS1 gene product. A yeast ABC transporter conferring mycotoxin resistance. J Biol Chem. 1994;269:4180–4186. [PubMed] [Google Scholar]
- 10.Boeke J D, LaCroute F, Fink G R. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- 11.Bossier P, Fernandes L, Rocha D, Rodrigues-Pousada C. Overexpression of YAP2, coding for a new yAP protein, and YAP1 in Saccharomyces cerevisiae alleviates growth inhibition caused by 1,10-phenanthroline. J Biol Chem. 1993;268:23640–23645. [PubMed] [Google Scholar]
- 12.Carvajal E, van den Hazel H B, Cybularz-Kolaczkowska A, Balzi E, Goffeau A. Molecular and phenotypic characterization of yeast PDR1 mutants that show hyperactive transcription of various ABC multidrug transporter genes. Mol Gen Genet. 1997;256:406–415. doi: 10.1007/s004380050584. [DOI] [PubMed] [Google Scholar]
- 13.Chin K C, Li G G, Ting J P. Importance of acidic, proline/serine/threonine-rich, and GTP-binding regions in the major histocompatibility complex class II transactivator: generation of transdominant-negative mutants. Proc Natl Acad Sci USA. 1997;94:2501–2506. doi: 10.1073/pnas.94.6.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman S T, Tseng E, Moye-Rowley W S. Saccharomyces cerevisiae basic region-leucine zipper protein regulatory networks converge at the ATR1 structural gene. J Biol Chem. 1997;272:23224–23230. doi: 10.1074/jbc.272.37.23224. [DOI] [PubMed] [Google Scholar]
- 15.Crute B E, Lewis A F, Wu Z, Bushweller J H, Speck N A. Biochemical and biophysical properties of the core-binding factor α2 (AML1) DNA-binding domain. J Biol Chem. 1996;271:26251–26260. doi: 10.1074/jbc.271.42.26251. [DOI] [PubMed] [Google Scholar]
- 16.Cui Z, Hirata D, Tsuchiya E, Osada H, Miyakawa T. The multidrug resistance-associated protein (MRP) subfamily (Yrs1/Yor1) of Saccharomyces cerevisiae is important for the tolerance to a broad range of organic anions. J Biol Chem. 1996;271:14712–14716. doi: 10.1074/jbc.271.25.14712. [DOI] [PubMed] [Google Scholar]
- 17.Decottignies A, Lambert L, Catty P, Degand H, Epping E A, Moye-Rowley W S, Balzi E, Goffeau A. Identification and characterization of SNQ2, a new multidrug ATP binding cassette transporter of the yeast plasma membrane. J Biol Chem. 1995;270:18150–18157. doi: 10.1074/jbc.270.30.18150. [DOI] [PubMed] [Google Scholar]
- 18.Delahodde A, Delaveau T, Jacq C. Positive autoregulation of the yeast transcription factor Pdr3p, which is involved in control of drug resistance. Mol Cell Biol. 1995;15:4043–4051. doi: 10.1128/mcb.15.8.4043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delaveau T, Delahodde A, Carvajal E, Subik J, Jacq C. PDR3, a new yeast regulatory gene, is homologous to PDR1 and controls the multidrug resistance phenomenon. Mol Gen Genet. 1994;244:501–511. doi: 10.1007/BF00583901. [DOI] [PubMed] [Google Scholar]
- 20.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dexter D, Moye-Rowley W S, Wu A L, Golin J. Mutations in the yeast PDR3, PDR4, PDR7 and PDR9 pleiotropic (multiple) drug resistance loci affect the transcript level of an ATP binding cassette transporter encoding gene, PDR5. Genetics. 1994;136:505–515. doi: 10.1093/genetics/136.2.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dixon D M, McNeil M M, Cohen M L, Gellin B G, La Montagne J R. Fungal infections: a growing threat. Public Health Rep. 1996;111:226–235. [PMC free article] [PubMed] [Google Scholar]
- 23.Donahue T F, Cigan A M. Sequence and structural requirements for efficient translation in yeast. Methods Enzymol. 1990;185:366–372. doi: 10.1016/0076-6879(90)85032-j. [DOI] [PubMed] [Google Scholar]
- 24.Dupont B F, Dromer F, Improvisi L. The problem of azole resistance in Candida. J Mycol Med. 1996;6:12–19. [Google Scholar]
- 25.Fling M E, Kopf J, Tamarkin A, Gorman J A, Smith H A, Koltin Y. Analysis of a Candida albicans gene that encodes a novel mechanism for resistance to benomyl and methotrexate. Mol Gen Genet. 1991;227:318–329. doi: 10.1007/BF00259685. [DOI] [PubMed] [Google Scholar]
- 26.Fonzi W A, Irwin M Y. Isogenic strain construction and gene mapping in Candida albicans. Genetics. 1993;134:717–728. doi: 10.1093/genetics/134.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gietz R D, Schiestl R H, Willems A R, Woods R A. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- 28.Guo Z, Sherman F. 3′-end-forming signals of yeast mRNA. Trends Biochem Sci. 1996;21:477–481. doi: 10.1016/s0968-0004(96)10057-8. [DOI] [PubMed] [Google Scholar]
- 29.Hahn S, Buratowski S, Sharp P A, Guarente L. Yeast TATA-binding protein TFIID binds to TATA elements with both consensus and nonconsensus DNA sequences. Proc Natl Acad Sci USA. 1989;86:5718–5722. doi: 10.1073/pnas.86.15.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hallstrom T C, Moye-Rowley W S. Divergent transcriptional control of multidrug resistance genes in Saccharomyces cerevisiae. J Biol Chem. 1998;273:2098–2104. doi: 10.1074/jbc.273.4.2098. [DOI] [PubMed] [Google Scholar]
- 31.Hellauer K, Rochon M H, Turcotte B. A novel DNA binding motif for yeast zinc cluster proteins: the Leu3p and Pdr3p transcriptional activators recognize everted repeats. Mol Cell Biol. 1996;16:6096–6102. doi: 10.1128/mcb.16.11.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hertle K, Haase E, Brendel M. The SNQ3 gene of Saccharomyces cerevisiae confers hyper-resistance to several functionally unrelated chemicals. Curr Genet. 1991;19:429–433. doi: 10.1007/BF00312733. [DOI] [PubMed] [Google Scholar]
- 33.Hirata D, Yano K, Miyahara K, Miyakawa T. Saccharomyces cerevisiae YDR1, which encodes a member of the ATP-binding cassette (ABC) superfamily, is required for multidrug resistance. Curr Genet. 1994;26:285–294. doi: 10.1007/BF00310491. [DOI] [PubMed] [Google Scholar]
- 34.Katzmann D J, Burnett P E, Golin J, Mahe Y, Moye-Rowley W S. Transcriptional control of the yeast PDR5 gene by the PDR3 gene product. Mol Cell Biol. 1994;14:4653–4661. doi: 10.1128/mcb.14.7.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katzmann D J, Hallstrom T C, Mahe Y, Moye-Rowley W S. Multiple Pdr1p/Pdr3p binding sites are essential for normal expression of the ATP binding cassette transporter protein-encoding gene PDR5. J Biol Chem. 1996;271:23049–23054. doi: 10.1074/jbc.271.38.23049. [DOI] [PubMed] [Google Scholar]
- 36.Katzmann D J, Hallstrom T C, Voet M, Wysock W, Golin J, Volckaert G, Moye-Rowley W S. Expression of an ATP-binding cassette transporter-encoding gene (YOR1) is required for oligomycin resistance in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6875–6883. doi: 10.1128/mcb.15.12.6875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lai M H, Kirsch D R. Nucleotide sequence of cytochrome P450 L1A1 (lanosterol 14 α-demethylase) from Candida albicans. Nucleic Acids Res. 1989;17:804. doi: 10.1093/nar/17.2.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leppert G, McDevitt R, Falco S C, Van Dyk T K, Ficke M B, Golin J. Cloning by gene amplification of two loci conferring multiple drug resistance in Saccharomyces. Genetics. 1990;125:13–20. doi: 10.1093/genetics/125.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maenza J R, Merz W G, Romagnoli M J, Keruly J C, Moore R D, Gallant J E. Infection due to fluconazole-resistant Candida in patients with AIDS—prevalence and microbiology. Clin Infect Dis. 1997;24:28–34. doi: 10.1093/clinids/24.1.28. [DOI] [PubMed] [Google Scholar]
- 40.Magee B B, Magee P T. WO-2, a stable aneuploid derivative of Candida albicans strain WO-1, can switch from white to opaque and form hyphae. Microbiology (Reading) 1997;143:289–295. doi: 10.1099/00221287-143-2-289. [DOI] [PubMed] [Google Scholar]
- 41.Mahé Y, Parle-McDermott A, Nourani A, Delahodde A, Lamprecht A, Kuchler K. The ATP-binding cassette multidrug transporter Snq2 of Saccharomyces cerevisiae: a novel target for the transcription factors Pdr1 and Pdr3. Mol Microbiol. 1996;20:109–117. doi: 10.1111/j.1365-2958.1996.tb02493.x. [DOI] [PubMed] [Google Scholar]
- 42.Martens J A, Genereaux J, Saleh A, Brandl C J. Transcriptional activation by yeast PDR1p is inhibited by its association with NGG1p/ADA3p. J Biol Chem. 1996;271:15884–15890. doi: 10.1074/jbc.271.27.15884. [DOI] [PubMed] [Google Scholar]
- 43.Meyers S, Schauer W, Balzi E, Wagner M, Goffeau A, Golin J. Interaction of the yeast pleiotropic drug resistance genes PDR1 and PDR5. Curr Genet. 1992;21:431–436. doi: 10.1007/BF00351651. [DOI] [PubMed] [Google Scholar]
- 44.Nourani A, Papajova D, Delahodde A, Jacq C, Subik J. Clustered amino acid substitutions in the yeast transcription regulator Pdr3p increase pleiotropic drug resistance and identify a new central regulatory domain. Mol Gen Genet. 1997;256:397–405. doi: 10.1007/s004380050583. [DOI] [PubMed] [Google Scholar]
- 45.Nourani A, Wesolowski-Louvel M, Delaveau T, Jacq C, Delahodde A. Multiple-drug-resistance phenomenon in the yeast Saccharomyces cerevisiae: involvement of two hexose transporters. Mol Cell Biol. 1997;17:5453–5460. doi: 10.1128/mcb.17.9.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ogawa A, Hashida-Okado T, Endo M, Yoshioka H, Tsuruo T, Takesako K, Kato I. Role of ABC transporters in aureobasidin A resistance. Antimicrob Agents Chemother. 1998;42:755–761. doi: 10.1128/aac.42.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prasad R, Dewergifosse P, Goffeau A, Balzi E. Molecular cloning and characterization of a novel gene of Candida albicans, CDR1, conferring multiple resistance to drugs and antifungals. Curr Genet. 1995;27:320–329. doi: 10.1007/BF00352101. [DOI] [PubMed] [Google Scholar]
- 48.Reece R J, Ptashne M. Determinants of binding-site specificity among yeast C6 zinc cluster proteins. Science. 1993;261:909–911. doi: 10.1126/science.8346441. [DOI] [PubMed] [Google Scholar]
- 49.Rex J H, Rinaldi M G, Pfaller M A. Resistance of Candida species to fluconazole. Antimicrob Agents Chemother. 1995;39:1–8. doi: 10.1128/aac.39.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rose M D, Winston F, Hieter P. Methods in yeast genetics: a laboratory course manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 51.Rosenbluh A, Mevarech M, Koltin Y, Gorman J A. Isolation of genes from Candida albicans by complementation in Saccharomyces cerevisiae. Mol Gen Genet. 1985;200:500–502. doi: 10.1007/BF00425739. [DOI] [PubMed] [Google Scholar]
- 52.Sanglard D, Ischer F, Monod M, Bille J. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob Agents Chemother. 1996;40:2300–2305. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanglard D, Ischer F, Monod M, Bille J. Cloning of Candida albicans genes conferring resistance to azole antifungal agents: characterization of CDR2, a new multidrug ABC transporter gene. Microbiology. 1997;143:405–416. doi: 10.1099/00221287-143-2-405. [DOI] [PubMed] [Google Scholar]
- 54.Sanglard D, Kuchler K, Ischer F, Pagani J L, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob Agents Chemother. 1995;39:2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schjerling P, Holmberg S. Comparative amino acid sequence analysis of the C6 zinc cluster family of transcriptional regulators. Nucleic Acids Res. 1996;24:4599–4607. doi: 10.1093/nar/24.23.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schnell N, Krems B, Entian K D. The PAR1 (YAP1/SNQ3) gene of Saccharomyces cerevisiae, a c-jun homologue, is involved in oxygen metabolism. Curr Genet. 1992;21:269–273. doi: 10.1007/BF00351681. [DOI] [PubMed] [Google Scholar]
- 57.Servos J, Haase E, Brendel M. Gene SNQ2 of Saccharomyces cerevisiae, which confers resistance to 4-nitroquinoline-N-oxide and other chemicals, encodes a 169 kDa protein homologous to ATP-dependent permeases. Mol Gen Genet. 1993;236:214–218. doi: 10.1007/BF00277115. [DOI] [PubMed] [Google Scholar]
- 58.Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- 59.Stephen D W, Rivers S L, Jamieson D J. The role of the YAP1 and YAP2 genes in the regulation of the adaptive oxidative stress responses of Saccharomyces cerevisiae. Mol Microbiol. 1995;16:415–423. doi: 10.1111/j.1365-2958.1995.tb02407.x. [DOI] [PubMed] [Google Scholar]
- 60.Strich R, Surosky R T, Steber C, Dubois E, Messenguy F, Esposito R E. UME6 is a key regulator of nitrogen repression and meiotic development. Genes Dev. 1994;8:796–810. doi: 10.1101/gad.8.7.796. [DOI] [PubMed] [Google Scholar]
- 61.Treitel M A, Carlson M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci USA. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases, and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;8:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wemmie J A, Szczypka M S, Thiele D J, Moye-Rowley W S. Cadmium tolerance mediated by the yeast AP-1 protein requires the presence of an ATP-binding cassette transporter-encoding gene, YCF1. J Biol Chem. 1994;269:32592–32597. [PubMed] [Google Scholar]
- 64.Wendler F, Bergler H, Prutej K, Jungwirth H, Zisser G, Kuchler K, Hogenauer G. Diazaborine resistance in the yeast Saccharomyces cerevisiae reveals a link between YAP1 and the pleiotropic drug resistance genes PDR1 and PDR3. J Biol Chem. 1997;272:27091–27098. doi: 10.1074/jbc.272.43.27091. [DOI] [PubMed] [Google Scholar]
- 65.Wenzel T J, Teunissen A W, de Steensma H Y. PDA1 mRNA: a standard for quantitation of mRNA in Saccharomyces cerevisiae superior to ACT1 mRNA. Nucleic Acids Res. 1995;23:883–884. doi: 10.1093/nar/23.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Whiteway M, Dignard D, Thomas D Y. Dominant negative selection of heterologous genes: isolation of Candida albicans genes that interfere with Saccharomyces cerevisiae mating factor-induced cell cycle arrest. Proc Natl Acad Sci USA. 1992;89:9410–9414. doi: 10.1073/pnas.89.20.9410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wolfger H, Mahé Y, Parle-McDermott A, Delahodde A, Kuchler K. The yeast ATP binding cassette (ABC) protein genes PDR10 and PDR15 are novel targets for the Pdr1 and Pdr3 transcriptional regulators. FEBS Lett. 1997;418:269–274. doi: 10.1016/s0014-5793(97)01382-3. [DOI] [PubMed] [Google Scholar]
- 68.Wu A, Wemmie J A, Edgington N P, Goebl M, Guevara J L, Moye-Rowley W S. Yeast bZip proteins mediate pleiotropic drug and metal resistance. J Biol Chem. 1993;268:18850–18858. [PubMed] [Google Scholar]