Abstract
Viruses are critical for regulating microbial communities and biogeochemical processes affecting carbon/nutrient cycling. However, the role of soil phages in controlling microbial physiological traits and intrinsic dissolved organic matter (DOM) properties remains largely unknown. Herein, microcosm experiments with different soil phage concentrates (including no-added phages, inactive phages, and three dilutions of active phages) at two temperatures (15 °C and 25 °C) were conducted to disclose the nutrient and DOM dynamics associated with viral lysing. Results demonstrated three different phases of viral impacts on CO2 emission at both temperatures, and phages played a role in maintaining Q10 within bounds. At both temperatures, microbial nutrient limitations (especially P limitation) were alleviated by viral lysing as determined by extracellular enzyme activity (decreased Vangle with active phages). Additionally, the re-utilization of lysate-derived DOM by surviving microbes stimulated an increase of microbial metabolic efficiency and recalcitrant DOM components (e.g., SUV254, SUV260 and HIX). This research provides direct experimental evidence that the “viral shuttle” exists in soils, whereby soil phages increase recalcitrant DOM components. Our findings advance the understanding of viral controls on soil biogeochemical processes, and provide a new perspective for assessing whether soil phages provide a net “carbon sink” vs. “carbon source” in soils.
Subject terms: Biogeochemistry, Biogeochemistry, Microbial ecology
Introduction
Soil microorganisms are major agents of soil organic carbon (SOC) cycling, both as decomposers and contributors to the long-term persistence of SOC [1–3]. Annually, about 60 Pg SOC are mineralized to CO2 by soil microorganisms and released into the atmosphere, thereby being a major regulator of global warming [4, 5]. To date, the role of abiotic factors in regulating microbes and thereby the SOC decomposition rate has been well established [6, 7]. However, the impact of microbial predators on microbial communities and their functions, through top-down control, remains elusive.
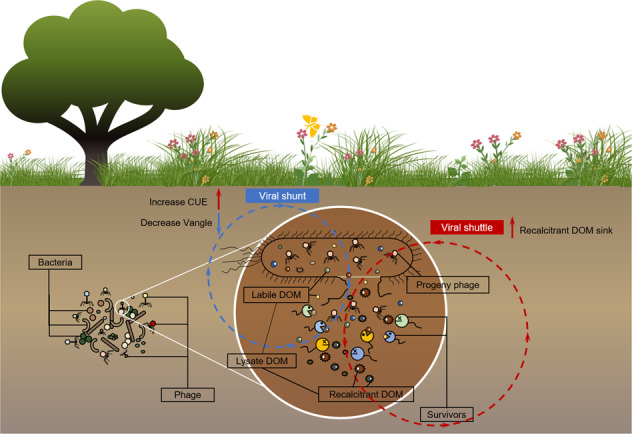
Viruses as the most abundant biological entity can dramatically influence microbial abundance and community structure through viral lysing [8–10]. In oceans, phages (viruses infecting prokaryotes) lyse 20–40% of prokaryotes each day, which significantly affects the marine carbon cycle where they account for an estimated 6–26% of marine carbon exchange [11, 12]. Although there is no direct evidence to what extent soil bacteriophages affect soil carbon cycling, the abundance of soil phages (roughly 107–1010 g-1 soil) and the frequency of phage infection of bacterial cells are both higher than those found in aquatic environments [13–15]. The “viral shunt” mechanism refers to the cellular matter liberated by viral lysing that can be reutilized by surviving heterotrophic microbes. This further stimulates the recycling and mineralization of dissolved organic matter (DOM) into CO2, and hinders nutrient and energy flows to higher trophic levels [16, 17]. This process in the marine system can release 145 Gt carbon and boost the microbial utilization of DOM by up to 27% [18–20]. The viral shunt has been documented in the soil environment using a 13C labeling approach, whereas the quantity of carbon involved remains unexplored [21]. Another pathway by which phages influence carbon fluxes is termed the “viral shuttle”. This mechanism describes the cell debris and transparent exopolymeric polysaccharides, as well as other lysis products, which are relatively recalcitrant to microbial decomposition, and therefore tend to aggregate and be retained in the soil environment for long time periods [22]. This concept is only discussed in a few review articles and is believed to act as a carbon sink in oceans [23]. To date, the viral shuttle has never been documented in soils, not to mention the possible mechanisms involved therein.
According to their life cycle, phages can be divided into virulent and temperate categories, whose infective properties are markedly different [24]. Virulent phages only act as predators controlling bacterial communities via lysing cells, based on the Kill-the-Winner model (KtW) [8]. In contrast, temperate phages can integrate their DNA or RNA into the host’s chromosomes and replicate with their host, known as the lysogenic cycle [25]. When host cells cannot feed temperate phages for the replication of progeny, they switch to the lytic cycle [26]. Different from KtW, lysogenic phages can help their host survive in harsh environments by integrating auxiliary metabolic genes (AMGs) into the host’s genome, referred to as the Piggyback-the-Winner model (PtW) [10, 27, 28]. Additionally, temperature can alter interactions between phages and hosts, by altering the viral latent period, viral and host production rates, burst size, and so on [29, 30]. Moreover, different microbial species have different affinities for various DOM chemical components [31–34]. Based on the impact of diverse viral life models on bacterial communities and different microbial affinities for DOM components, we posit that the presence of viruses will alter the fractionation of DOM chemical components, thereby affecting the biogeochemical cycling of carbon.
Recent studies demonstrated that soil nutrient limitations are a crucial factor controlling microbial metabolism and carbon cycling in terrestrial ecosystems, especially phosphorus limitation [35]. This leads to the question: Can soil nutrient limitation be alleviated by release of intracellular materials by viral lysing? Moreover, the role of temperature in regulating the viral lysis rate and infection process maybe a critical factor in a rapidly warming world [29, 30, 36]. To address these knowledge gaps, we hypothesize that microbial nutrient limitation will be alleviated by active phages and the microbial metabolic efficiency will be increased due to lysate DOM, for which microbes have a strong affinity. Further, we posit that these processes will vary as a function of temperature.
There are few studies examining the impact of soil viruses on soil carbon and nitrogen cycling [37–39], let alone on soil carbon dynamics from the perspective of microbial trait-based strategies and DOM components. To test our hypotheses and disentangle the impacts of soil phages on SOM dynamics, soil bacteria and phages were reinoculated in sterilized soil and incubated at two different temperatures (15 °C and 25 °C). The objectives of this study were to (1) explore how soil phages influence microbial respiration; (2) disclose the impact of phages on the bacterial community and trait-based strategies; and (3) reveal if soil phages promote the accumulation of recalcitrant DOM components, consistent with the viral shuttle mechanism.
Materials and methods
Soil sampling
Soil was collected from a natural forest in Jiangxi Ecological Science and Technology Park of Soil and Water Conservation (115°42′08″-115°43′06″E, 29°16′37″-29°17′40″N). This is the center of the red soil region of China, located in De’an County, Jiangxi Province; additional details of the study area can be found in elsewhere [40]. Five 2 × 2 m subplots were established at each corner and the center of three randomly established large plots (20 × 20 m), and the 15 subplot composite samples (0–20 cm) were combined to form a single equal-weight composite representative of the study area. The sample was transported to the laboratory on ice. Plant residues and coarse fragments were removed from the soil sample by passing through a 2-mm sieve, and the <2-mm soil fraction divided into two parts: (1) air-dried for physicochemical analysis and (2) stored fresh at 4 °C for extraction of soil bacteria and phages.
Preparation of phage and bacterial suspensions from soil
Soil phage and bacterial suspensions were extracted from 1.2 kg of soil suspended in 1.2 L saline magnesium buffer (SM; per L containing 5.8 g NaCl, 2 g MgSO4·7H2O, 50 mL 1 M Tris-HCl (pH 7.5)). The suspension was vortexed for 3 min at maximum intensity, shaken for 2 h at 250 rpm and then centrifuged at 4 °C for 20 min (2500 × g) [41, 42]. The supernatant was passed through a 2-μm cellulose acetate filter, before being filtered and concentrated using tangential flow filtration (TFF) with two different pore-size filters (0.22 μm and 100 kDa) [17, 37]. The retentates obtained (>0.22 μm) were used as bacterial suspensions, and the concentrated filtrates were used as phage suspensions (100 kDa–0.22 μm) and no-phage suspensions (<100 kDa) [38]. The final volumes of retentates and filtrates were 500 mL, 80 mL and 620 mL, respectively. In order to simulate the in-situ environment, we added 1.2 ml of phage suspensions and 3.6 ml bacterial suspensions to each serum bottle containing 35 sterilized air-dried soil, which approximated real-world phage and bacteria concentrations. We then adjusted the soil moisture to 40% of field capacity with 4.9 ml of no-phage water (filtered through 100 kDa).
Experimental design
Experiments were conducted in 250-mL serum bottles with 35 g sterilized air-dried soil (50 kGy γ-radiation over 24 h). Microcosm treatments consisted of (1) bacterial suspensions with no added phage (SM); (2) bacterial suspensions with autoclaved dead phage (DV) [38]; and (3) bacterial suspensions with different phage dilutions (no dilution (V), twofold dilution (2-1 V) and tenfold dilution (10-1 V)). All treatments were incubated at two different temperatures (15 °C and 25 °C) with four replicates. The total number of microcosms were 5 soil phage states (no phage, inactive phage, and active phage with 0-, 2- and tenfold dilution) × 2 temperatures × 3 sampling times (days 7, 28 and 98) × 4 replicates = 120 total microcosms.
Soil properties and gas emission analysis
A pH meter was applied to measure soil pH in a 1:2.5 soil: distilled water ratio after a 30 min equilibration. Soil organic carbon (SOC) and soil total nitrogen (TN) were quantified by an elemental analyzer. Available P (Bray-P) was extracted with 0.03 M NH4F/0.025 M HCl and quantified using the molybdenum blue spectroscopy method [43, 44]. Microbial biomass carbon (MBC), nitrogen (MBN) and phosphorus (MBP) were measured by CHCl3 fumigation-extraction [45, 46]. Soil total P (TP) was measured using a digestion method [47, 48]. Dissolved organic matter (DOM) as characterized by organic carbon (DOC) and nitrogen (DON) was extracted with Milli-Q water at a ratio of 1:10 (w/v) and determined using a Multi C/N TOC analyzer. Extracted DOM samples were kept in the dark at 4 °C for further characterization. Soil NH4+ and NO3- were extracted by 1 M KCl from fresh soil, and determined using a flow injection analyzer [49]. The measurement of extracellular enzyme activity, putative microbial physiological traits and calculation of microbial metabolic efficiency are described in Supplementary Information.
Soil CO2 fluxes from the incubation studies were determined at 2, 5, 7, 14, 17, 21, 28, 35, 42, 49, 56, 63, 70, 77, 84, 91 and 98 days using gas chromatography (GC-200 Plus Shimadzu, Japan) as described by [50–52]. The respiration rate (R) of CO2 (mg CO2 kg soil-1 d-1) was calculated as [52]:
| 1 |
where ρ, V, M, dc/dt and T denote the density of CO2 at standard temperature and pressure (1.98 g/L), volume of incubation jar (L), mass of soil (g), slope of the linear regression for gas concentration gradient through time and incubation temperature (K), respectively. Cumulative CO2 emission was calculated as the sum of the area bounded by the rate using the trapezoidal rule.
DOM spectral analysis
DOM concentration of incubated soil was expressed as the DOC concentration determined by a total organic carbon (TOC) analyzer [53]. The UV-Vis spectra of DOM were assessed for a series of indices as proxies for chemical characteristics of the DOM (e.g., SUV254, SUV260 and spectral slope ratio (SR) representing the aromaticity, hydrophobicity and molecular weight, respectively), via an ultraviolet-visible spectrophotometer [54–56]. SUV254 and SUV260 were calculated as the ratio of UV absorption at 254 and 260 nm to the DOC concentration [57, 58].
After diluting DOC concentrations to 8 mg C L–1, excitation-emission matrix (EEM) spectra of DOM were measured by fluorescence spectrometry, with a scan rate of 2000 nm min–1 [59]. The excitation wavelength range was 200–500 nm (5-nm sampling interval), whereas the emission wavelength range was 250–600 nm (2-nm sampling interval). EEM fluorescence spectra were visualized in MATLAB 2012b with DOM Fluor toolbox [56, 58]. The humification index (HIX) was calculated for an excitation wavelength of 254 nm, by dividing the peak area of the emission wavelengths between 435 and 480 nm by the sum of the areas between 435–480 nm and 300–345 nm [60, 61].
High-throughput sequencing and analysis of bacterial community
Soil DNA was extracted using the MoBio PowerSoil DNA Isolation Kit (MP Biomedicals, USA) following manufacturer’s instructions. The primers 515 F/907 R were used to amplify the V4-V5 region of the 16 S rRNA gene and sequenced on an HiSeq PE250 sequencing platform (Illumina, USA) [31, 62]. Bacteria reads were in the range of 48,477–89,042 per sample, and the screened sequences were clustered into operational taxonomic units (OTUs) using UPARSE, with a similarity cutoff of 97% [48, 49]. For each representative sequence, annotated taxonomic information was determined using the SILVA (version 138) database [63]. The obtained sequences were used to assess bacterial α-diversity (i.e., Shannon index) and principal coordinates analysis (PCoA) was applied to represent the shift in the bacterial community based on a Bray–Curtis dissimilarity matrix, with the vegan package (version 2.5–7) [32].
Statistical analyses
Redundancy analysis (RDA) was applied to reveal relationships between bacterial communities and various soil/microbial parameters using the vegan package in R 3.6.3. Spearman heatmap plots and other plots were visualized by Origin. One-way ANOVAs with subsequent least-significant-difference (LSD) tests were used to determine differences among measured parameters (e.g., Vangle, Vlength, CO2 emission, CUE, SUV254, HIX SUV260). The main statistically significant factors for predicting CO2, qCO2 and CUE were identified by random forest analysis (R package: rfPermute), whereby a higher percentage of mean squared error (%IncMSE) indicates a more important factor [64, 65]. A correlation network between bacterial taxa and measured parameters was used to identify soil taxa strongly interacting with Vangle and recalcitrant DOM (e.g., SUV254, SUV260 and HIX). Spearman correlations were considered as strong when r > 0.5 and significant when p < 0.05; correlation matrices were visualized with Gephi [66, 67]. SPSS Version 24 (SPSS, Chicago, IL, USA) and R version 3.6.3 were used for statistical analysis with a significant level of p < 0.05, unless otherwise stated.
Results
Microbial respiration dynamics at different temperatures
All five soil virus treatments (i.e., SM, DV, V, 2–1 V and 10–1 V) displayed three phases of CO2 released during soil incubation (Fig. 1). In the first phase (first two weeks after microbe/virus inoculation), the CO2 efflux rate at both 15 °C and 25 °C was lower in active viral groups, compared to the SM and DV groups. During the second and third phases, the trend for CO2 efflux rates varied between temperatures. At 25 °C, the second phase (days 14 to 42) CO2 efflux rate with active viruses was higher than that in SM and DV groups, although the CO2 emission rate was different among active viral dilutions. During the third phase (days 42–98), the CO2 efflux rate in the SM and DV groups again exceeded those of the active groups (Fig. 1A). In contrast to the 25 °C treatments, the phase 2 (days 15 to 35) CO2 efflux rate at 15 °C for the 10–1 V group with active viruses was higher than the SM and DV groups. For phase 3 at 15 °C, the CO2 efflux rate in all active viral groups was significantly higher than in the SM and DV groups, which persisted for the duration of the experiment (Fig. 1B).
Fig. 1. Dynamics of CO2 emission rate (mean ± std dev) with added viruses incubated over 98 days.
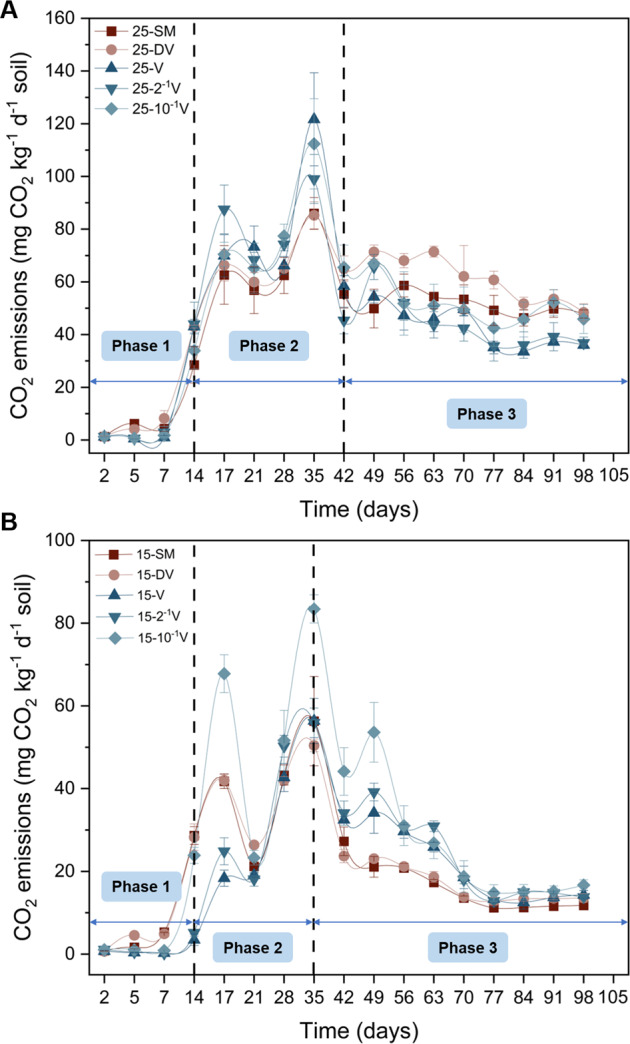
25 °C (A) and 15 °C (B). SM, no added viruses; DV, inactive viruses; V, active viruses with no dilution; 2–1 V, active viruses with twofold dilution; 10–1 V, active viruses with tenfold dilution; 25 and 15 signify different temperatures (25 °C and 15 °C) during the incubation.
Total cumulative CO2 fluxes during the incubation are shown in Fig. S1. To assess the mechanisms associated with the three phases of CO2 effluxes, three reference time points were selected to characterize each phase: Days 7, 28 and 98. Cumulative CO2 emissions with active viruses at 25 °C were significantly lower than the SM and DV groups (p < 0.05) in the first phase (Fig. S2a). During the second phase, cumulative CO2 efflux in active viral groups increased and exceeded those of the SM and DV groups. Cumulative CO2 production in active viral groups followed the order: 25-2–1 V > 25-V > 25–10–1 V (Fig. S2c). By the end of the incubation, cumulative CO2 production changed to the order: 25-DV > 25–10–1 V > 25-SM ≈ 25-V ≈ 25-2–1 V (Fig. S2e). In contrast, cumulative CO2 production with active viruses at 15 °C showed the order of 15–10–1 V > 15-2–1 V > 15-V throughout the entire incubation period (Fig. S2b, d, f). Compared to the 15-SM and 15-DV groups, cumulative CO2 production in active viral groups was lower at the early stage. For the entire incubation period, cumulative CO2 production followed: 15–10–1 V > 15-2–1 V > 15-V > 15-DV > 15-SM (Fig. S2f).
The temperature sensitivity of CO2 emissions (Q10) was calculated, to investigate how soil phages influence Q10. The Q10 values of active phage groups were significantly lower than those in SM and DV groups, and were maintained within the bounds of 1–3 after the cultivation was stable (Fig. S3).
Soil microbial metabolic efficiency and resource acquisition traits
To illustrate the influence of soil viruses on microbial metabolic efficiency and resource acquisition traits, qCO2 and CUE were used to quantify the metabolic efficiency, and resource acquisition traits were assessed by Vlength and Vangle. For microbial metabolic efficiency, qCO2 showed no observable differences among different groups or temperatures (Fig. S4), whereas CUE significantly changed after the addition of active viruses (Fig. 2). At 25 °C, the CUE of groups inoculated with active viruses was higher than the SM and DV groups and decreased with increasing dilution (Fig. 2A). At 15 °C, only the CUE of the 15-V group was higher than the 15-SM group, even though the trend of CUE with active viruses declined with dilution. The addition of inactive viruses seemed to increase the CUE relative to the no virus addition group (SM) at 15 °C (Fig. 2B).
Fig. 2. Carbon use efficiency (CUE) (mean ± std dev).
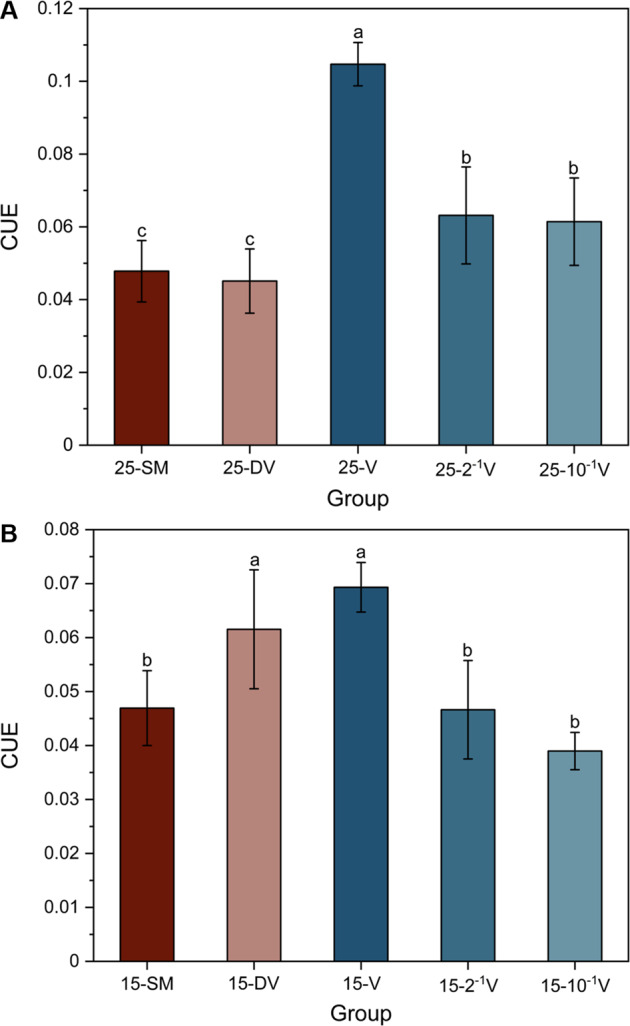
25 °C (A) and 15 °C (B). SM, no added viruses; DV, inactive viruses; V, active viruses with no dilution; 2–1 V, active viruses with twofold dilution; 10–1 V, active viruses with tenfold dilution; 25 and 15 signify different temperatures (25 °C and 15 °C) during the incubation. Different lowercase letters represent significant differences at p < 0.05.
Microbial acquisition strategies can be elucidated from Vlength and Vangle, where the former is used to represent the relative C vs. nutrient (N and P)-acquiring enzyme activities and the latter expresses the relative P vs. N-acquiring enzyme activities. All groups were deemed plentiful with respect to organic carbon (Vlength was less than 1), albeit a higher Vlength occurred for active viral groups (Fig. S5). Adding active viruses into the soil seemed to alleviate nutrient constraints, especially for phosphorus, based on the decrease in Vangle (Fig. 3). The effect of alleviated nutrient constraints depended on temperature, with the lowest Vangle occurring for the tenfold dilution group at 25 °C, whereas the opposite was found at 15 °C.
Fig. 3. Microbial resource acquisition strategies.
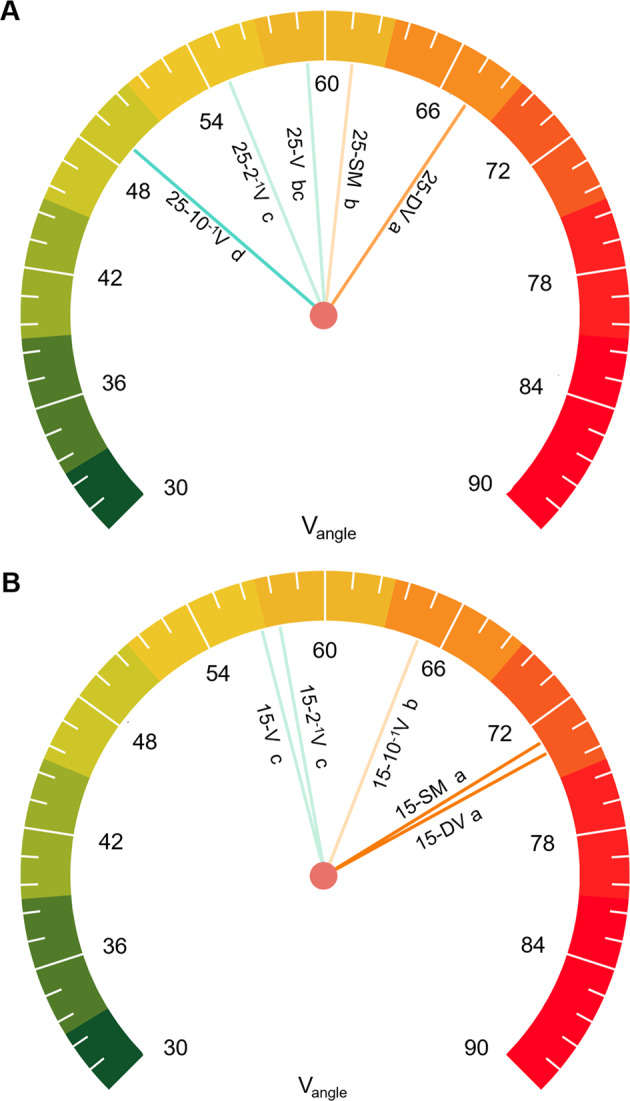
25 °C (A) and 15 °C (B). Vangle quantifies the relative P vs. N-acquiring enzyme activities; higher Vangle represents relatively higher P vs. N acquisition strategies. SM, no added viruses; DV, inactive viruses; V, active viruses with no dilution; 2–1 V, active viruses with twofold dilution; 10–1 V, active viruses with tenfold dilution; 25 and 15 signify different temperatures (25 °C and 15 °C) during the incubation. Different lowercase letters represent significant differences at p < 0.05.
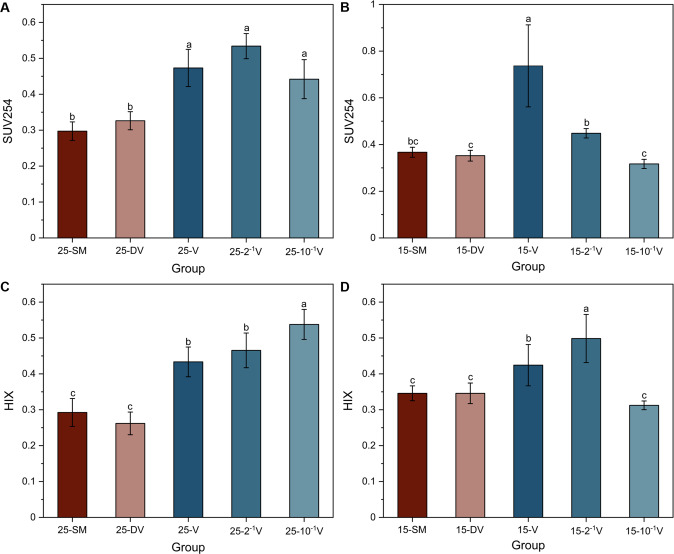
Accumulation of recalcitrant DOM components
The DOC concentration across the different groups showed no significant differences. In contrast, the optical properties of DOM were diverse among treatments, as measured by 3D-EEM fluorescence spectroscopy and UV−vis spectroscopy (Figs S6, S7). After addition of soil viruses at 25°C, SUV254 and HIX showed a considerable increase of 37.9–98.1% and 48.3–105.3%, respectively (Fig. 4A, C). This indicates a higher degree of humification and aromaticity, characteristics that make DOM more resistant to microbial decomposition. At 15 °C, only the 15-V group with the highest viral inoculation produced higher SUV254 (p < 0.05), as compared to the 15-SM and 15-DV groups (Fig. 4B). Inoculating soil viruses increased HIX at 15 °C, except for the 15–10–1 V group, which indicates that the DOM aromaticity increased by 22.5% in the 15-V group and 44.2% in the 15–2–1 V group (Fig. 4D). SUV260, as a proxy for DOM hydrophobicity, showed changes in treatment groups similar to SUV254 (Fig. S7).
Fig. 4. Changes of recalcitrant DOM components (mean ± std dev) after incubation.
SUV254 (A, B) represents the aromaticity degree of DOM, and HIX (C, D) indicates the humification degree of DOM. SM, no added viruses; DV, inactive viruses; V, active viruses with no dilution; 2–1 V, active viruses with twofold dilution; 10–1 V, active viruses with twnfold dilution; 25 and 15 signify different temperatures (25 °C and 15 °C) during the incubation. Different lowercase letters represent significant differences at p < 0.05.
Linking microbial properties to microbial metabolic activity and recalcitrant DOM components
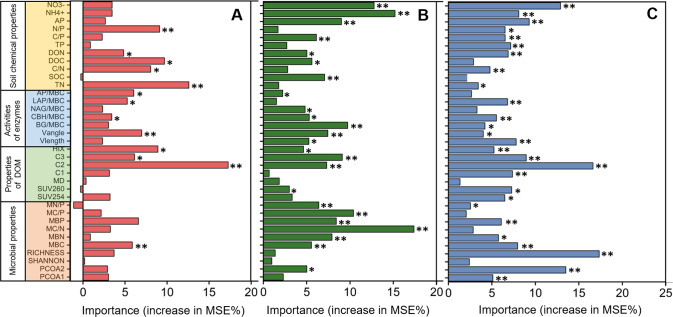
A correlation analysis was conducted to assess whether the decreased Vangle (index of alleviated nutrient constraints by viral lysis) was related to microbial metabolic activity and DOM optical properties. All of the measured indexes (e.g., CO2 emission, SUV254, SUV260, HIX) showed significant negative correlations with Vangle (Fig. S8). These associations could indicate that the nutrients released by viral lysis increased metabolic activity (shown as CO2 emission) and enriched recalcitrant DOM components (represented by SUV254, SUV260 and HIX). Further, random forest analysis indicated that both Vangle and the DOM indexes (e.g., HIX, C1, C2, C3) could explain the differences in CO2 emission, qCO2 and CUE (Fig. 5).
Fig. 5. Mean predictor importance (percent increased mean square error, MSE) for microbial metabolic indexes, besed on Random Forest analysis.
The metabolic quotient (qCO2), (A), carbon use efficiency (CUE), (B) and CO2 emission (C). NO3-, soil nitrate-N; NH4 + , soil ammonium-N; AP available phosphorus, TP total phosphorus, SOC soil organic carbon, TN total nitrogen, DOC dissolved organic carbon, DON dissolved organic nitrogen, N/P the ratio of total nitrogen and total phosphorus, C/P the ratio of soil organic carbon and total phosphorus, C/N the ratio of soil organic carbon and total nitrogen, BG β-1,4-glucosidase, CBH cellobiohydrolase, NAG N-acetyl-β-D-glucosaminidase, LAP leucine aminopeptidase, AP/MBC means normalizing acid phosphatase activities to per MBC. Enzyme/MBC represents specific enzyme activities calculated by normalizing activities to units/mg MBC; Ecoenzymatic vector Vlength (Vlength, relative C:nutrient acquiring ratio) and Vangle (Vangle, relative P:N-acquiring ratio); HIX, humification index of DOM; C1 (humic-like), C2 (quinone-like), C3 (protein-like), 3D-fluorescence components of DOM; MD, represents the relative molecular mass of DOM; SUV254, represents the aromatic degree of DOM; SUV260, represents the hydrophobicity of DOM; MBC microbial biomass carbon, MBN microbial biomass nitrogen, MBP microbial biomass phosphorus, RICHNESS richness of bacterial community, SHANNON alpha diversity of bacterial community, PCoA1 PCoA2 are the two-dimensional PCoA ordinations of taxonomic composition for the bacterial community. *p < 0.05; **p < 0.01.
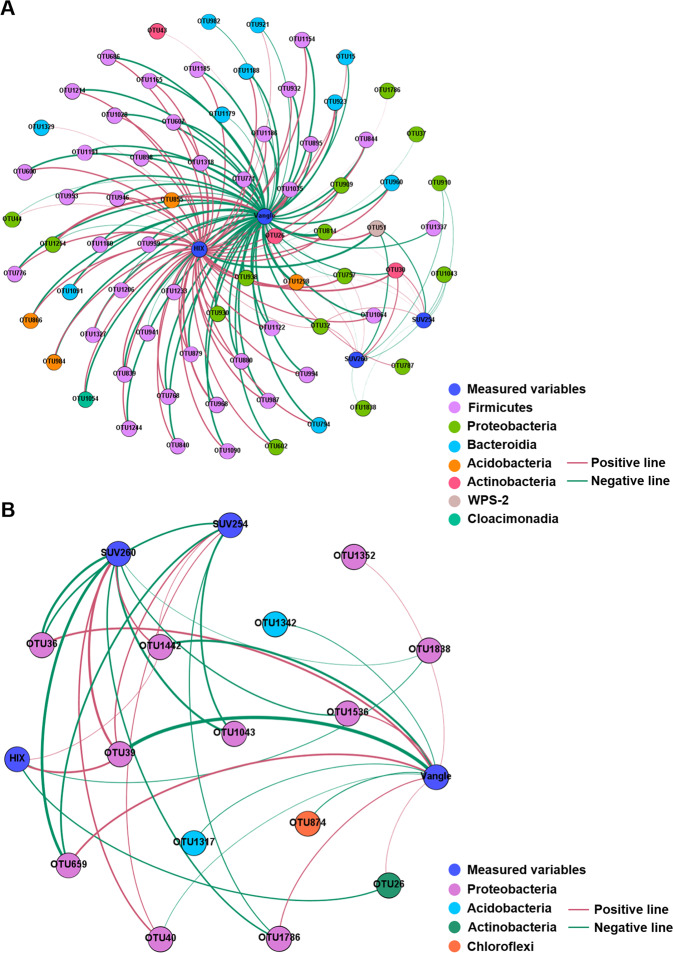
Bacterial communities were significantly altered by soil phages regardless of temperature (Adonis, p < 0.05; Fig. S9). Redundancy analysis was applied to explore the potential effects of edaphic properties (filtered by a variance inflation factor (VIF < 10)) on bacterial community composition (Fig. S10). Results showed that the selected factors explained 55.0% of the variance in the bacterial community. Most factors were related to DOM properties and indexes related to bacterial physiological traits (e.g., HIX, SUV260, C3, DOC, Vangle and Vlength). Bacterial taxa associated with Vangle and the optical properties of DOM (including SUV254, SUV260 and HIX) were identified based on Spearman correlations (r > 0.5, p < 0.05; Fig. 6). The correlation network between bacterial taxa and various factors suggested that nearly all bacterial species were negatively associated with Vangle, whereas they were positively correlated with HIX, SUV254 and SUV260 (Table S1).
Fig. 6. Correlation networks between measured variables (e.g., Vangle, SUV254, SUV260 and HIX) and associated bacterial taxa.
25 °C (A) and 15 °C (B). Different color of nodes represents different species (colored at the phylum level), and measured variables (labeled with characters). Red lines indicate positive interactions and green lines indicate negative interactions, at p < 0.05 and r > 0.5. The thickness of the lines indicates the strength of the correlations.
Discussion
The three phases of microbial respiration affected by phages
Three phases of microbial respiration were observed in both temperature regimes (Fig. 1). We attribute these phase dynamics to a tradeoff between the rate of bacterial mortality caused by viruses and the utilization of nutrients acquired by survivors, which is consistent with previous studies [17]. However, the phases differed with temperature. Compared with control groups (SM and DV) at 25 °C, CO2 released from the soil with active phage additions was initially lower, and then increased before finally decreasing again (Fig. 1A). This CO2-release pathway can be explained as follow: (1) Active phages lysed bacterial cells resulting in a decreased bacterial population and a subsequent reduction in CO2 emission. (2) Released intracellular organic matter from viral lysing was reutilized by the surviving microbes, which stimulated bacterial activity and increased microbial respiration. (3) Finally, continued viral lysing resulted in a lower microbial abundance and decreased CO2 fluxes. In the last phase, the number of survivors were too few to support higher CO2 fluxes relative to the control groups [39]. Furthermore, at 15 °C, phases 2 and 3 contrasted with the phenomena at 25 °C (Fig. 1B). In phase 2 at 15 °C, only microbial respiration from the lowest concentration of active phages (15–10–1 V) was higher than that in control groups. During phase 3, CO2 emissions from all active phage groups were higher throughout the remainder of the incubation. These results from both temperature regimes are in full agreement with the viral shunt mechanism as interpreted by previous studies [17, 22].
The phase dynamics of microbial respiration are differentially affected by the temperature of the soil environment. The highest CO2 emission for all active phage groups occurred in phase 2 at 25 °C, whereas the highest CO2 emissions were somewhat delayed at 15 °C. This might results from the warmer temperature advancing the onset of the viral shunt and/or shortening the viral latent period [17]. Additionally, the difference in microbial respiration rates at different temperatures during phase 3 may be partially due to increased bacterial activity at 25 °C, which increased CO2 emission in control groups and was not evident at 15 °C. This demonstrates that microbial activity increased at the higher temperature and without the influence of predators, thereby accelerating respiration rates [68–70]. Moreover, temperature could change interactions between phages and their hosts, such as altering the viral latent period, viral and host production rates, and burst size [29, 30].
Apart from temperature, cumulative CO2 fluxes were significantly associated with the concentration of phages. Inoculating higher concentrations of active phages resulted in a slight increase or no obvious change in CO2 fluxes, whereas adding a lower concentration of phages stimulated CO2 emission (Fig. S2e, S2f). This infers that high inoculation rates caused increased microbial mortality, which subsequently decreased SOC mineralization, even though a more nutritionally-enriched environment is presumed relative to the low inoculation rate [39]. In addition, the Q10 values for active phage treatments appeared to play a role in maintaining Q10 values within the bounds of 1–3, which is in accordance with the global average Q10 value of 2.56 ± 0.86 [71]. However, the Q10 of SM and DV groups were significantly higher than those in active phages groups after the cultivation was stable, with Q10 values exceeding 3 at Day 98 (Fig. S3). These changes of Q10 among treatment groups suggest that soil phages could decrease the temperature sensitivity of CO2 to warming, which could attenuate SOC mineralization affects due to global warming. However, the specific mechanisms warrant future investigation.
Lysate DOM alleviates microbial nutrient limitations
Previous studies revealed that viral lysate-derived DOM is relatively more bioavailable for microbial utilization, and can serve as an important substrate for microbial metabolism, particularly in harsh environments experiencing nutrient deprivation [72–75]. These studies found approximately 75% of lysate DOM was reused by surviving microbes, with 50–80% identified as bioavailable compounds, such as amino acids and carbohydrates [73, 74, 76]. Hence, the components of bioavailable DOM would have a higher affinity by microbes [34, 77]. This is consistent with our findings that microbial CUE increased in active phage groups (Fig. 2).
Recent studies report that soil microorganisms are often subject to P and N limitations at the global scale [78–80]. In pure culture systems, the release of phosphorus from host cells by viral lysing has been demonstrated, which suggests that viral lysates could be an important source of inorganic nutrients and markedly promote P regeneration [72]. Thus, to test our hypothesis that microbial nutrient limitation will be alleviated with active phages in the soil environment, the microbial physiological traits were assessed based on extracellular enzyme activity. The results supported our hypothesis by indicating that nutrient constraints (especially phosphorus limitation) were alleviated by lysate DOM, as demonstrated by a decrease in Vangle (Fig. 3).
Several previous studies found that microbial P limitation can have prominent impacts on soil carbon cycling [35, 81, 82]. For example, P limitation can stimulate more CO2 released from soil owing to more enzymes being secreted by microbes in an attempt to obtain additional P from SOM [83–86]. Conversely, P limitation may constrain microbial activity, thereby increasing soil C stocks due to reduced SOC mineralization [87, 88]. The results of our study support the former premise, that alleviating phosphorus limitation by increasing lysate DOM increases microbial CUE. To our knowledge, this is the first report assessing nutrient restriction of microorganisms from the perspective of soil virus interactions with the microbial community.
Viruses contribute to accumulation of recalcitrant DOM in the soil environment
In our study, the alleviation of microbial P limitation appeared to promote SOC sequestration through accumulation of recalcitrant DOM (as indicated by higher SUV254, SUV260 and HIX; Fig. 4 and S7). We found that Vangle showed a significant, negative correlation with several indexes of recalcitrant DOM. Moreover, the coupling of correlation and random forest analyses highlighted these DOM parameters as effective predictors of microbial metabolic efficiency and activity (Fig. 5 and S8). Transformation of microbial carbon into recalcitrant DOM components could represent an important soil carbon sink via a biological pump mechanism, with the concomitant re-utilization of the bioavailable intracellular organic matter increasing microbial activity, which is known as the viral shuttle mechanism [22, 23, 89, 90].
CDOM and FDOM are important fractions of environmental DOM (accounting for 20–70% of the overall DOM) that have been utilized to assess the quality and transformations of DOM [55, 57, 58]. Various studies investigated the relationships between viruses and DOM in aquatic environments and found that high viral lytic activity resulted in the production and accumulation of FDOM, especially humic-like FDOM [73, 91–94]. These results are in agreement with our study that found a higher degree of humification and aromaticity in the accumulated DOM generated upon addition of active phages (Fig. 4, S6 and S7). Additionally, protists, phytoplankton and heterotrophic microbes are crucial factors regulating DOM dynamics [95–97].
Based on bacterial community composition and the correlation network between bacterial taxa and soil indexes, our results showed that phages have an appreciable impact on the bacterial community, and interactions between lysate DOM and bacteria play a strong role in determining the accumulation of recalcitrant DOM in soils (Fig. 6). Previous studies in oceans indicated a large accumulation of viral lysate DOM that is not readily utilized by microbes and therefore buildups in the recalcitrant DOM pool [23, 55, 60, 77, 98]. A possible explanation is that viral lysis suppresses the abundant microbial taxa and gives rare taxa time and space to propagate, which increases the utilization of bioavailable DOM and alters the microbial community [75, 99]. Additionally, viruses could change the microbial pathway for uptake of DOM compounds by introducing their genetic information into the host and reprogramming their metabolic pathways [16, 23, 100]. Our findings highlight that viral lysate-derived DOM is not only metabolized and respired as CO2, but other fractions are transformed into recalcitrant DOM that accumulates in the soil environment [22, 89]. This is the first study to provide direct evidence for the existence of the viral shuttle mechanism in soils.
Conclusion
This study contributes a more comprehensive understanding of viral effects on microbial respiration and DOM alteration in the soil environment. Microbial respiration displayed three distinct phases over the 98-day incubation period that were highly dependent on temperature and phage concentration. We ascribe these findings to the tradeoff between the rate of bacterial mortality caused by viruses and utilization of the nutrients released by the surviving microbial community. The viral lysate-derived DOM can alleviate soil nutrient limitations, especially P limitation, as reflected by a lower Vangle. Furthermore, our results highlight that the bioavailable fraction of the lysate-derived DOM can be re-utilized by the surviving microbes, whereas the recalcitrant DOM fraction (e.g., SUV254, SUV260 and HIX) can contribute to carbon sequestration in soils. This is the first study to demonstrate that both the viral shunt and viral shuttle mechanisms are active in the soil environment. This research provides new insights to guide future investigations that incorporate the role of viruses and the viral shuttle mechanism into nutrient cycling in soil environments.
Supplementary information
Acknowledgements
This study was supported by the National Natural Science Foundation of China (41721001, 41991334), the Science and Technology Program of Zhejiang Province (2022C02046), the 111 Project (B17039), and China Agriculture Research System (CARS-01).
Author contributions
Design of experiment: JX; soil sampling and microcosm incubation: DT, YW and HY; measurement of soil properties: DT and HS; statistical analysis: DT; visualization: DT; writing—original draft: DT and JX; writing—review and editing: JX, RA. Dahlgren and DT.
Data availability
The original sequence data were deposited into the National Center for Biotechnology Information Sequence Read Archive database under project ID PRJNA974558. Experimental data have been included as a supplement to this publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41396-023-01438-5.
References
- 1.Wang C, Qu L, Yang L, Liu D, Morrissey E, Miao R, et al. Large-scale importance of microbial carbon use efficiency and necromass to soil organic carbon. Glob Change Biol. 2021;27:2039–48. doi: 10.1111/gcb.15550. [DOI] [PubMed] [Google Scholar]
- 2.Liang C, Schimel JP, Jastrow JD. The importance of anabolism in microbial control over soil carbon storage. Nat Microbiol. 2017;2:1–6. doi: 10.1038/nmicrobiol.2017.105. [DOI] [PubMed] [Google Scholar]
- 3.Ni H, Jing X, Xiao X, Zhang N, Wang X, Sui Y, et al. Microbial metabolism and necromass mediated fertilization effect on soil organic carbon after long-term community incubation in different climates. ISME J. 2021;15:2561–73. doi: 10.1038/s41396-021-00950-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hursh A, Ballantyne A, Cooper L, Maneta M, Kimball J, Watts J. The sensitivity of soil respiration to soil temperature, moisture, and carbon supply at the global scale. Glob Change Biol. 2017;23:2090–103. doi: 10.1111/gcb.13489. [DOI] [PubMed] [Google Scholar]
- 5.Lei J, Guo X, Zeng Y, Zhou J, Gao Q, Yang Y. Temporal changes in global soil respiration since 1987. Nat Commun. 2021;12:403. doi: 10.1038/s41467-020-20616-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, Xu H, Wu Y, Ai Z, Zhang J, Liu G, et al. Effects of natural vegetation restoration on dissolved organic matter (DOM) biodegradability and its temperature sensitivity. Water Res. 2021;191:116792. doi: 10.1016/j.watres.2020.116792. [DOI] [PubMed] [Google Scholar]
- 7.Geisen S, Hu S, dela Cruz TEE, Veen GF. Protists as catalyzers of microbial litter breakdown and carbon cycling at different temperature regimes. ISME J. 2021;15:618–21. doi: 10.1038/s41396-020-00792-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuzyakov Y, Mason-Jones K. Viruses in soil: nano-scale undead drivers of microbial life, biogeochemical turnover and ecosystem functions. Soil Biol Biochem. 2018;127:305–17. [Google Scholar]
- 9.Roossinck MJ. Plants, viruses and the environment: ecology and mutualism. Virology. 2015;479:271–77. doi: 10.1016/j.virol.2015.03.041. [DOI] [PubMed] [Google Scholar]
- 10.Chevallereau A, Pons BJ, van Houte S, Westra ER. Interactions between bacterial and phage communities in natural environments. Nat Rev Microbiol. 2022;20:49–62. doi: 10.1038/s41579-021-00602-y. [DOI] [PubMed] [Google Scholar]
- 11.Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiol Rev. 2004;28:127–81. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 12.Suttle CA. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–12. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi R, Bowatte S, Taki K, Ohashi Y, Asakawa S, Kimura M. High frequency of phage-infected bacterial cells in a rice field soil in Japan. Soil Sci Plant Nutr. 2011;57:35–39. [Google Scholar]
- 14.Guemes AGC, Youle M, Cantu VA, Felts B, Nulton J, Rohwer F. Viruses as Winners in the Game of Life. In: Enquist LW, editor. Annual Review of Virology, Vol 3. Annual Review of Virology. 32016. p. 197–214. [DOI] [PubMed]
- 15.Williamson KE, Fuhrmann JJ, Wommack KE, Radosevich M. Viruses in Soil Ecosystems: An Unknown Quantity Within an Unexplored Territory. In: Enquist L, editor. Annual Review of Virology, Vol 4. Annual Review of Virology. 42017. p. 201–19. [DOI] [PubMed]
- 16.Suttle CA. Viruses in the sea. Nature. 2005;437:356–61. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 17.Wang S, Yu S, Zhao X, Zhao X, Mason-Jones K, Zhu Z, et al. Experimental evidence for the impact of phages on mineralization of soil-derived dissolved organic matter under different temperature regimes. Sci Total Environ. 2022;846:157517. doi: 10.1016/j.scitotenv.2022.157517. [DOI] [PubMed] [Google Scholar]
- 18.Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–48. doi: 10.1038/21119. [DOI] [PubMed] [Google Scholar]
- 19.Danovaro R, Dell’Anno A, Corinaldesi C, Magagnini M, Noble R, Tamburini C, et al. Major viral impact on the functioning of benthic deep-sea ecosystems. Nature. 2008;454:1084–27. doi: 10.1038/nature07268. [DOI] [PubMed] [Google Scholar]
- 20.Lara E, Vaque D, Sa EL, Boras JA, Gomes A, Borrull E, et al. Unveiling the role and life strategies of viruses from the surface to the dark ocean. Sci Adv. 2017;3:e1602565. doi: 10.1126/sciadv.1602565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li Y, Watanabe T, Murase J, Asakawa S, Kimura M. Identification of the major capsid gene (g23) of T4-type bacteriophages that assimilate substrates from root cap cells under aerobic and anaerobic soil conditions using a DNA-SIP approach. Soil Biol Biochem. 2013;63:97–105. [Google Scholar]
- 22.Zimmerman AE, Howard-Varona C, Needham DM, John SG, Worden AZ, Sullivan MB, et al. Metabolic and biogeochemical consequences of viral infection in aquatic ecosystems. Nat Rev Microbiol. 2020;18:21–34. doi: 10.1038/s41579-019-0270-x. [DOI] [PubMed] [Google Scholar]
- 23.Chen X, Weinbauer MG, Jiao N, Zhang R. Revisiting marine lytic and lysogenic virus-host interactions: Kill-the-Winner and Piggyback-the-Winner. Sci Bull. 2021;66:871–74. doi: 10.1016/j.scib.2020.12.014. [DOI] [PubMed] [Google Scholar]
- 24.Dou C, Xiong J, Gu Y, Yin K, Wang J, Hu Y, et al. Structural and functional insights into the regulation of the lysis-lysogeny decision in viral communities. Nat Microbiol. 2018;3:1285–94. doi: 10.1038/s41564-018-0259-7. [DOI] [PubMed] [Google Scholar]
- 25.Howard-Varona C, Hargreaves KR, Abedon ST, Sullivan MB. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 2017;11:1511–20. doi: 10.1038/ismej.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kronheim S, Daniel-Ivad M, Duan Z, Hwang S, Wong AI, Mantel I, et al. A chemical defence against phage infection. Nature. 2018;564:283–86. doi: 10.1038/s41586-018-0767-x. [DOI] [PubMed] [Google Scholar]
- 27.Silveira CB, Rohwer FL. Piggyback-the-Winner in host-associated microbial communities. Npj Biofilms Microbiomes. 2016;2:16010. doi: 10.1038/npjbiofilms.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang D, Yu P, Ye M, Schwarz C, Jiang X, Alvarez PJJ. Enhanced mutualistic symbiosis between soil phages and bacteria with elevated chromium-induced environmental stress. Microbiome. 2021;9:150. doi: 10.1186/s40168-021-01074-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demory D, Arsenieff L, Simon N, Six C, Rigaut-Jalabert F, Marie D, et al. Temperature is a key factor in Micromonas-virus interactions. ISME J. 2017;11:601–12. doi: 10.1038/ismej.2016.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Padfield D, Castledine M, Buckling A. Temperature-dependent changes to host-parasite interactions alter the thermal performance of a bacterial host. ISME J. 2020;14:389–98. doi: 10.1038/s41396-019-0526-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Yang S, Semenov MV, Yao F, Ye J, Bu R, et al. Temperature sensitivity of SOM decomposition is linked with a K-selected microbial community. Glob Change Biol. 2021;27:2763–79. doi: 10.1111/gcb.15593. [DOI] [PubMed] [Google Scholar]
- 32.Tong D, Li Z, Xiao H, Nie X, Liu C, Zhou M. How do soil microbes exert impact on soil respiration and its temperature sensitivity? Environ Microbiol. 2021;23:3048–58. doi: 10.1111/1462-2920.15520. [DOI] [PubMed] [Google Scholar]
- 33.Xiao HB, Li ZW, Chang XF, Huang JQ, Nie XD, Liu C, et al. Soil erosion-related dynamics of soil bacterial communities and microbial respiration. Appl Soil Ecol. 2017;119:205–13. [Google Scholar]
- 34.Ling L, Luo Y, Jiang B, Lv J, Meng C, Liao Y, et al. Biochar induces mineralization of soil recalcitrant components by activation of biochar responsive bacteria groups. Soil Biol Biochem. 2022;172:108778. [Google Scholar]
- 35.Cui Y, Moorhead DL, Wang X, Xu M, Wang X, Wei X, et al. Decreasing microbial phosphorus limitation increases soil carbon release. Geoderma. 2022;419:104463. [Google Scholar]
- 36.Mojica KDA, Huisman J, Wilhelm SW, Brussaard CPD. Latitudinal variation in virus-induced mortality of phytoplankton across the North Atlantic Ocean. ISME J. 2016;10:500–13. doi: 10.1038/ismej.2015.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Braga LPP, Spor A, Kot W, Breuil M-C, Hansen LH, Setubal JC, et al. Impact of phages on soil bacterial communities and nitrogen availability under different assembly scenarios. Microbiome. 2020;8:52. doi: 10.1186/s40168-020-00822-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albright MBN, Gallegos-Graves LV, Feeser KL, Montoya K, Emerson JB. Dunbar MSaJ. Experimental evidence for the impact of soil viruses on carbon cycling during surface plant litter decomposition. ISME Commun. 2022;2:24. doi: 10.1038/s43705-022-00109-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei X, Ge T, Wu C, Wang S, Mason-Jones K, Li Y, et al. T4-like phages reveal the potential role of viruses in soil organic matter mineralization. Environ Sci Technol. 2021;55:6440–48. doi: 10.1021/acs.est.0c06014. [DOI] [PubMed] [Google Scholar]
- 40.Ma Y, Li Z, Deng C, Yang J, Tang C, Duan J, et al. Effects of tillage-induced soil surface roughness on the generation of surface-subsurface flow and soil loss in the red soil sloping farmland of southern China. Catena. 2022;213:106230. [Google Scholar]
- 41.Liang X, Wagner RE, Zhuang J, DeBruyn JM, Wilhelm SW, Liu F, et al. Viral abundance and diversity vary with depth in a southeastern United States agricultural Ultisol. Soil Biol Biochem. 2019;137:107546. [Google Scholar]
- 42.Narr A, Nawaz A, Wick LY, Harms H, Chatzinotas A. Soil viral communities vary temporally and along a land use transect as revealed by virus-like particle counting and a modified community fingerprinting approach (fRAPD) Front Microbiol. 2017;8:1975. doi: 10.3389/fmicb.2017.01975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murphy J, Riley JP. A modified single solution mthoed for determination of phosphate in natural waters. Anal Chim Acta. 1962;26:31–36. [Google Scholar]
- 44.Huang Y, Dai Z, Lin J, Li D, Ye H, Dahlgren RA, et al. Labile carbon facilitated phosphorus solubilization as regulated by bacterial and fungal communities in Zea mays. Soil Biol Biochem. 2021;163:108465. [Google Scholar]
- 45.Spohn M, Klaus K, Wanek W, Richter A. Microbial carbon use efficiency and biomass turnover times depending on soil depth - implications for carbon cycling. Soil Biol Biochem. 2016;96:74–81. [Google Scholar]
- 46.Brookes PC, Powlson DS, Jenkinson DS. Measurement of microbial biomass phosphorus in soil. Soil Biol Biochem. 1982;14:319–29. [Google Scholar]
- 47.Bray RH, Kurtz LT. Determination of total organic and available forms of phosphorus in soils. Soil Sci. 1945;59:39–45. [Google Scholar]
- 48.Feng J, Zeng X-M, Zhang Q, Zhou X-Q, Liu Y-R, Huang Q. Soil microbial trait-based strategies drive metabolic efficiency along an altitude gradient. ISME Commun. 2021;1:71. doi: 10.1038/s43705-021-00076-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X, Huang X, Cheng J, Cheng Z, Yang Q, Hu L, et al. Diversity-triggered bottom-up trophic interactions impair key soil functions under lindane pollution stress. Environ Pollut. 2022;314:120293–93. doi: 10.1016/j.envpol.2022.120293. [DOI] [PubMed] [Google Scholar]
- 50.Zhao HC, Lin JH, Wang XH, Shi JC, Dahlgren RA, Xu JM. Dynamics of soil microbial N-cycling strategies in response to cadmium stress. Environ Sci Technol. 2021;55:14305–15. doi: 10.1021/acs.est.1c04409. [DOI] [PubMed] [Google Scholar]
- 51.Jiao PP, Li ZW, Yang L, He JJ, Chang XF, Xiao HB, et al. Bacteria are more sensitive than fungi to moisture in eroded soil by natural grass vegetation restoration on the Loess Plateau. Sci Total Environ. 2021;756:143899. doi: 10.1016/j.scitotenv.2020.143899. [DOI] [PubMed] [Google Scholar]
- 52.Zhang L, Song L, Wang B, Shao H, Zhang L, Qin X. Co-effects of salinity and moisture on CO2 and N2O emissions of laboratory-incubated salt-affected soils from different vegetation types. Geoderma. 2018;332:109–20. [Google Scholar]
- 53.Huang M, Li Z, Luo N, Yang R, Wen J, Huang B, et al. Application potential of biochar in environment: insight from degradation of biochar-derived DOM and complexation of DOM with heavy metals. Sci Total Environ. 2019;646:220–28. doi: 10.1016/j.scitotenv.2018.07.282. [DOI] [PubMed] [Google Scholar]
- 54.Shi W, Zhuang W-E, Hur J, Yang L. Monitoring dissolved organic matter in wastewater and drinking water treatments using spectroscopic analysis and ultra-high resolution mass spectrometry. Water Res. 2021;188:116406. doi: 10.1016/j.watres.2020.116406. [DOI] [PubMed] [Google Scholar]
- 55.Chen X, Wei W, Xiao X, Wallace D, Hu C, Zhang L, et al. Heterogeneous viral contribution to dissolved organic matter processing in a long-term macrocosm experiment. Environ Int. 2022;158:106950. doi: 10.1016/j.envint.2021.106950. [DOI] [PubMed] [Google Scholar]
- 56.Ding X, Xu W, Li Z, Huang M, Wen J, Jin C, et al. Phosphate hinders the complexation of dissolved organic matter with copper in lake waters. Environ Pollut. 2021;276:116739. doi: 10.1016/j.envpol.2021.116739. [DOI] [PubMed] [Google Scholar]
- 57.Helms JR, Stubbins A, Ritchie JD, Minor EC, Kieber DJ, Mopper K. Absorption spectral slopes and slope ratios as indicators of molecular weight, source, and photobleaching of chromophoric dissolved organic matter. Limnol Oceanogr. 2008;53:955–69. [Google Scholar]
- 58.Huang M, Zhou M, Li Z, Ding X, Wen J, Jin C, et al. How do drying-wetting cycles influence availability of heavy metals in sediment? A perspective from DOM molecular composition. Water Res. 2022;220:118671. doi: 10.1016/j.watres.2022.118671. [DOI] [PubMed] [Google Scholar]
- 59.Wu J, Zhang H, Yao Q-S, Shao L-M, He P-J. Toward understanding the role of individual fluorescent components in DOM-metal binding. J Hazard Mater. 2012;215:294–301. doi: 10.1016/j.jhazmat.2012.02.075. [DOI] [PubMed] [Google Scholar]
- 60.Wang L, Lin Y, Ye L, Qian Y, Shi Y, Xu K, et al. Microbial roles in dissolved organic matter transformation in full-scale wastewater treatment processes revealed by reactomics and comparative genomics. Environ Sci Technol. 2021;55:11294–307. doi: 10.1021/acs.est.1c02584. [DOI] [PubMed] [Google Scholar]
- 61.Ohno T. Fluorescence inner-filtering correction for determining the humification index of dissolved organic matter. Environ Sci Technol. 2002;36:742–46. doi: 10.1021/es0155276. [DOI] [PubMed] [Google Scholar]
- 62.Zhao K, Yu H, Xue R, Stirling E, Wang Y, Xu J, et al. The only constant is change: endogenous circadian rhythms of soil microbial activities. Soil Biol Biochem. 2022;173:108805. [Google Scholar]
- 63.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:590–96. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jiao S, Chen W, Wang J, Du N, Li Q, Wei G. Soil microbiomes with distinct assemblies through vertical soil profiles drive the cycling of multiple nutrients in reforested ecosystems. Microbiome. 2018;6:146. doi: 10.1186/s40168-018-0526-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Z, Tong D, Nie X, Xiao H, Jiao P, Jiang J, et al. New insight into soil carbon fixation rate: the intensive co-occurrence network of autotrophic bacteria increases the carbon fixation rate in depositional sites. Agric Ecosyst Environ. 2021;320:107579. [Google Scholar]
- 66.Zhou H, Gao Y, Jia XH, Wang MM, Ding JJ, Cheng L, et al. Network analysis reveals the strengthening of microbial interaction in biological soil crust development in the Mu Us Sandy Land, northwestern China. Soil Biol Biochem. 2020;144:107782. [Google Scholar]
- 67.Liang X, Zhuang J, Loffler FE, Zhang Y, DeBruyn JM, Wilhelm SW, et al. Viral and bacterial community responses to stimulated Fe(III)-bioreduction during simulated subsurface bioremediation. Environ Microbiol. 2019;21:2043–55. doi: 10.1111/1462-2920.14566. [DOI] [PubMed] [Google Scholar]
- 68.Wang C, Morrissey EM, Mau RL, Hayer M, Pineiro J, Mack MC, et al. The temperature sensitivity of soil: microbial biodiversity, growth, and carbon mineralization. ISME J. 2021;15:2738–47. doi: 10.1038/s41396-021-00959-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nottingham AT, Scott JJ, Saltonstall K, Broders K, Montero-Sanchez M, Puspok J, et al. Microbial diversity declines in warmed tropical soil and respiration rise exceed predictions as communities adapt. Nat Microbiol. 2022;7:1650–60. doi: 10.1038/s41564-022-01200-1. [DOI] [PubMed] [Google Scholar]
- 70.Yang J, Jia X, Ma H, Chen X, Liu J, Shangguan Z, et al. Effects of warming and precipitation changes on soil GHG fluxes: a meta-analysis. Sci Total Environ. 2022;827:154351. doi: 10.1016/j.scitotenv.2022.154351. [DOI] [PubMed] [Google Scholar]
- 71.Li JQ, Pei JM, Pendall E, Fang CM, Nie M. Spatial heterogeneity of temperature sensitivity of soil respiration: a global analysis of field observations. Soil Biol Biochem. 2020;141:107675. [Google Scholar]
- 72.Haaber J, Middelboe M. Viral lysis of Phaeocystis pouchetii: implications for algal population dynamics and heterotrophic C, N and P cycling. ISME J. 2009;3:430–41. doi: 10.1038/ismej.2008.125. [DOI] [PubMed] [Google Scholar]
- 73.Heinrichs ME, Tebbe DA, Wemheuer B, Niggemann J, Engelen B. Impact of viral lysis on the composition of bacterial communities and dissolved organic matter in deep-sea sediments. Viruses-Basel. 2020;12:922. doi: 10.3390/v12090922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sebastian M, Auguet J-C, Restrepo-Ortiz CX, Sala MM, Marrase C, Gasol JM. Deep ocean prokaryotic communities are remarkably malleable when facing long-term starvation. Environ Microbiol. 2018;20:713–23. doi: 10.1111/1462-2920.14002. [DOI] [PubMed] [Google Scholar]
- 75.Zhang L, Chen M, Chen X, Wang J, Zhang Y, Xiao X, et al. Nitrifiers drive successions of particulate organic matter and microbial community composition in a starved macrocosm. Environ Int. 2021;157:106776. doi: 10.1016/j.envint.2021.106776. [DOI] [PubMed] [Google Scholar]
- 76.Aristegui J, Gasol JM, Duarte CM, Herndl GJ. Microbial oceanography of the dark ocean’s pelagic realm. Limnol Oceanogr. 2009;54:1501–29. [Google Scholar]
- 77.Zhao Z, Gonsior M, Schmitt-Kopplin P, Zhan Y, Zhang R, Jiao N, et al. Microbial transformation of virus-induced dissolved organic matter from picocyanobacteria: coupling of bacterial diversity and DOM chemodiversity. ISME J. 2019;13:2551–65. doi: 10.1038/s41396-019-0449-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Du E, Terrer C, Pellegrini AFA, Ahlstrom A, van Lissa CJ, Zhao X, et al. Global patterns of terrestrial nitrogen and phosphorus limitation. Nat Geosci. 2020;13:221–26. [Google Scholar]
- 79.Hou E, Wen D, Jiang L, Luo X, Kuang Y, Lu X, et al. Latitudinal patterns of terrestrial phosphorus limitation over the globe. Ecol Lett. 2021;24:1420–31. doi: 10.1111/ele.13761. [DOI] [PubMed] [Google Scholar]
- 80.Li Y, Niu S, Yu G. Aggravated phosphorus limitation on biomass production under increasing nitrogen loading: a meta-analysis. Glob Change Biol. 2016;22:934–43. doi: 10.1111/gcb.13125. [DOI] [PubMed] [Google Scholar]
- 81.Exbrayat JF, Pitman AJ, Zhang Q, Abramowitz G, Wang YP. Examining soil carbon uncertainty in a global model: response of microbial decomposition to temperature, moisture and nutrient limitation. Biogeosciences. 2013;10:7095–108. [Google Scholar]
- 82.Ma B, Stirling E, Liu Y, Zhao K, Zhou J, Singh BK, et al. Soil biogeochemical cycle couplings inferred from a function-taxon network. Research. 2021;2021:7102769. doi: 10.34133/2021/7102769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Feng J, Tang M, Zhu B. Soil priming effect and its responses to nutrient addition along a tropical forest elevation gradient. Glob Change Biol. 2021;27:2793–806. doi: 10.1111/gcb.15587. [DOI] [PubMed] [Google Scholar]
- 84.Fang Y, Nazaries L, Singh BK, Singh BP. Microbial mechanisms of carbon priming effects revealed during the interaction of crop residue and nutrient inputs in contrasting soils. Glob Change Biol. 2018;24:2775–90. doi: 10.1111/gcb.14154. [DOI] [PubMed] [Google Scholar]
- 85.Cui Y, Zhang Y, Duan C, Wang X, Zhang X, Ju W, et al. Ecoenzymatic stoichiometry reveals microbial phosphorus limitation decreases the nitrogen cycling potential of soils in semi-arid agricultural ecosystems. Soil Tillage Res. 2020;197:104463. [Google Scholar]
- 86.Cui Y, Wang X, Zhang X, Ju W, Duan C, Guo X, et al. Soil moisture mediates microbial carbon and phosphorus metabolism during vegetation succession in a semiarid region. Soil Biol Biochem. 2020;147:107814. [Google Scholar]
- 87.Zhu Z, Ge T, Luo Y, Liu S, Xu X, Tong C, et al. Microbial stoichiometric flexibility regulates rice straw mineralization and its priming effect in paddy soil. Soil Biol Biochem. 2018;121:67–76. [Google Scholar]
- 88.Ochoa-Hueso R, Borer ET, Seabloom EW, Hobbie SE, Risch AC, Collins SL, et al. Microbial processing of plant remains is co-limited by multiple nutrients in global grasslands. Glob Change Biol. 2020;26:4572–82. doi: 10.1111/gcb.15146. [DOI] [PubMed] [Google Scholar]
- 89.Jiao N, Herndl GJ, Hansell DA, Benner R, Kattner G, Wilhelm SW, et al. Microbial production of recalcitrant dissolved organic matter: long-term carbon storage in the global ocean. Nat Rev Microbiol. 2010;8:593–99. doi: 10.1038/nrmicro2386. [DOI] [PubMed] [Google Scholar]
- 90.Breitbart M, Bonnain C, Malki K, Sawaya NA. Phage puppet masters of the marine microbial realm. Nat Microbiol. 2018;3:754–66. doi: 10.1038/s41564-018-0166-y. [DOI] [PubMed] [Google Scholar]
- 91.Lonborg C, Middelboe M, Brussaard CPD. Viral lysis of Micromonas pusilla: impacts on dissolved organic matter production and composition. Biogeochemistry. 2013;116:231–40. [Google Scholar]
- 92.Xiao X, Guo W, Li X, Wang C, Chen X, Lin X, et al. Viral lysis alters the optical properties and biological availability of dissolved organic matter derived from Prochlorococcus Picocyanobacteria. Appl Environ Microbiol. 2021;87:e02271–20. doi: 10.1128/AEM.02271-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhao Z, Gonsior M, Luek J, Timko S, Ianiri H, Hertkorn N, et al. Picocyanobacteria and deep-ocean fluorescent dissolved organic matter share similar optical properties. Nat Commun. 2017;8:15284. doi: 10.1038/ncomms15284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kinsey JD, Corradino G, Ziervogel K, Schnetzer A, Osburn CL. Formation of chromophoric dissolved organic matter by bacterial degradation of phytoplankton-derived aggregates. Front Mar Sci. 2018;4:430. [Google Scholar]
- 95.Hu SK, Herrera EL, Smith AR, Pachiadaki MG, Edgcomb VP, Sylva SP, et al. Protistan grazing impacts microbial communities and carbon cycling at deep-sea hydrothermal vents. Proc Natl Acad Sci USA. 2021;118:e2102674118. doi: 10.1073/pnas.2102674118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dittmar T, Lennartz ST, Buck-Wiese H, Hansel DA, Santinelli C, Vanni C, et al. Enigmatic persistence of dissolved organic matter in the ocean. Nat Rev Earth Env. 2021;2:570–83. [Google Scholar]
- 97.Rastelli E, Corinaldesi C, Dell’Anno A, Tangherlini M, Martorelli E, Ingrassia M, et al. High potential for temperate viruses to drive carbon cycling in chemoautotrophy-dominated shallow-water hydrothermal vents. Environ Microbiol. 2017;19:4432–46. doi: 10.1111/1462-2920.13890. [DOI] [PubMed] [Google Scholar]
- 98.Ma X, Coleman ML, Waldbauer JR. Distinct molecular signatures in dissolved organic matter produced by viral lysis of marine cyanobacteria. Environ Microbiol. 2018;20:3001–11. doi: 10.1111/1462-2920.14338. [DOI] [PubMed] [Google Scholar]
- 99.Chen X, Ma R, Yang Y, Jiao N, Zhang R. Viral regulation on bacterial community impacted by lysis-lysogeny switch: a microcosm experiment in eutrophic coastal waters. Front Microbiol. 2019;10:1763. doi: 10.3389/fmicb.2019.01763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Howard-Varona C, Lindback MM, Bastien GE, Solonenko N, Zayed AA, Jang H, et al. Phage-specific metabolic reprogramming of virocells. ISME J. 2020;14:881–95. doi: 10.1038/s41396-019-0580-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original sequence data were deposited into the National Center for Biotechnology Information Sequence Read Archive database under project ID PRJNA974558. Experimental data have been included as a supplement to this publication.