Abstract
Eight different bacteriophages were isolated from leaves of Pisum sativum, Phaseolus vulgaris, Lycopersicon esculentum, Daucus carota sativum, Raphanus sativum, and Ocimum basilicum. All contain three segments of double-stranded RNA and have genomic-segment sizes that are similar but not identical to those of previously described bacteriophage φ6. All appear to have lipid-containing membranes. The base sequences of some of the viruses are very similar but not identical to those of φ6. Three of the viruses have little or no base sequence identity to φ6. Two of the viruses, φ8 and φ12, contain proteins with a size distribution very different from that of φ6 and do not package genomic segments of φ6. Whereas φ6 attaches to host cells by means of a pilus, several of the new isolates attach directly to the outer membrane. Although the normal hosts of these viruses seem to be pseudomonads, those viruses that attach directly to the outer membrane can establish carrier states in Escherichia coli or Salmonella typhimurium. One of the isolates, φ8, can form plaques on heptoseless strains of S. typhimurium.
Bacteriophage φ6 was isolated from bean straw infested with Pseudomonas syringae pv. phaseolicola (25). It contains a genome of three segments of double-stranded RNA (22) packaged inside a procapsid which is covered by a shell of protein P8 and a lipid-containing membrane containing additional viral proteins (25). The genome of φ6 has been cloned and sequenced, and the life cycle and structure of the phage have been subjects of considerable investigation (1, 8).
We have developed a model for the mechanism of genomic packaging in φ6 and accumulated evidence to support it (7, 15, 17). The model involves the binding of segment S to sites on the outside of the empty procapsid so as to position the 5′ end at an entry portal. Upon packaging of segment S, the binding sites are lost and new sites for segment M appear on the outside of the particle. The process repeats, and the sites for segment M disappear and those for segment L appear. We had shown previously that each of the segments contains a packaging sequence of about 200 nucleotides near the 5′ ends of the plus strands (5). We embarked upon the search for relatives of φ6 in the hope that a comparison of the sequences in the packaging regions of the segments would suggest important structural or sequence motifs. Up until now, φ6 has been alone in the family Cystoviridae and alone in the genus Cystovirus (11). It is now clear that this group is composed of many more phages, some very similar to φ6 and some rather distantly related.
MATERIALS AND METHODS
Bacterial strains, phage, and plasmids.
P. syringae pv. phaseolicola HB10Y (HB) (25) was used as the primary host for phage plating. LM2333 was selected as a mutant of HB resistant to several DNA phages. LM2489 is a derivative of LM2333. MPO.16 is a derivative of HB that lacks type IV pili and is resistant to φ6 (19, 20). MR is a derivative of P. aeruginosa that has the type IV pilus of P. syringae (19). LM2509 is a derivative of LM2489 that lacks pili and is resistant to φ6. Strain ERA is an isolate of P. pseudoalcaligenes. φ6 infects ERA when it has a mutation in gene 3 called h1 (9). S4 is a derivative of ERA that contains a nonsense suppressor mutation. S4M is a derivative of S4 that has mutated so as to be infected by φ6 without the h1 mutation. P. syringae pv. phaseolicola Ro49dRa1 is a rough derivative (21). SL1102 and SL3789 are strains of Salmonella typhimurium LT2 from the Salmonella Genetic Stock Centre, and they have lipopolysaccharide (LPS) defects due to mutations rfaE543 and rfaF511, respectively (18).
Plasmid pLM1454 is a derivative of the cloning vector pT7T319U (Pharmacia). It was used for the cloning of cDNA copies of phage DNA produced by reverse transcription (RT)-PCR.
Media.
The media used were LC and MB (23). Ampicillin plates contained 200 μg of ampicillin per ml in LC agar.
Enzymes and chemicals.
All restriction enzymes, T4 DNA ligase, T4 DNA polymerase, T4 polynucleotide kinase, Klenow enzyme, and exonuclease BAL 31 were purchased from Promega Corp. (Madison, Wis.), New England Biolabs (Boston, Mass.), and Boehringer GmbH (Mannheim, Germany).
Reverse transcriptase reaction.
About 3 μg of RNA in 20 μl of water was mixed with 200 ng of oligonucleotide primer OLM297, which is essentially identical to the first 16 nucleotides of the three genomic segments of φ6 and also carries a PstI site. Its sequence is GGGGGGCTGCAGAAAAAAACTTTATATA. The mixture was heated at 100°C for 4 min and quickly cooled to 42°C, and 12 μl was mixed with 4 μl of RT buffer (5×; Promega), 2 μl of deoxynucleoside triphosphates (2 mM each), and 2 μl of avian myeloblastosis virus reverse transcriptase (5 U). The mixture was incubated at 42°C for 1 h.
PCR.
A 10-μl volume of the RT mixture was supplemented with 0.2 mM deoxynucleoside triphosphates; 1 μg of OLM297; 1 μg of OLM15, OLM298, or OLM296; 2.5 U of Pwo DNA polymerase; and PCR buffer components (Boehringer GmbH) in 50 μl. The mixture was taken through 30 cycles of 94°C for 1 min, 45°C for 30 s, and 72°C for 2 min. The final product was extracted with phenol, ethanol precipitated, and dissolved in 20 μl of DNA buffer. Oligonucleotides OLM15, OLM298, and OLM296 are complementary to the plus strands of φ6 segments S (nucleotides 1158 to 1141), M (nucleotides 464 to 445), and L (nucleotides 656 to 634).
Cloning and sequencing.
The PCR products were cut with PstI and either SacI, MunI, or XhoI and ligated to vector pLM1454 that had been cut with PstI and SacI, EcoRI, or SalI, respectively. The ligates were then introduced into Escherichia coli JM109 by transformation, and white colonies were picked from Luria-Bertani plates containing ampicillin and 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Small DNA preparations were made, and the plasmids were analyzed to determine their restriction site patterns. Sequencing was done at the New York University Medical Center Sequencing Facility. In this way, we determined the sequences of the 5′ regions of the RNAs of φ7, φ9, φ10, and φ11.
LPS analysis.
A qualitative test for LPS structure is the gel electrophoresis procedure of de Kievit and Lam (5). Samples were prepared as previously described and stained with silver after periodate treatment.
Nucleotide sequence accession numbers.
Sequences have been deposited in GenBank with the following accession numbers: φ7 S, AF125682; φ7 M, AF125681; φ7 L, AF125680; φ9 S, AF125679; φ9 M, AF125678; φ9 L, AF125677; φ10 S, AF125676; φ10 M, AF125675; φ10 L, AF125674.
RESULTS AND DISCUSSION
Leaves of the various plants were incubated for several hours in Luria-Bertani broth at room temperature. The liquid was passed through a 0.2 μm filter, and aliquots of about 100 μl were plated on lawns of P. syringae pv. phaseolicola HB. The numbers of plaques appearing ranged from zero to several thousand. Most plaques contained virus that appeared to have DNA genomes. In order to screen out these phages, we isolated a mutant of HB that was resistant to several of the dominant plaque types. This strain was designated LM2333. φ6 infected LM2333 with the same efficiency as HB. The original strategy for the isolation of new phages was to plate the extracts on LM2333, test infection of HB, and then test for plaque formation on MPO.16, which is a derivative of HB that lacks type IV pili and cannot be infected by φ6. Phages that infected the first two strains and not MPO.16 would be tested for chloroform sensitivity and, if sensitive, would be then analyzed for the presence of a double-stranded RNA (dsRNA) genome.
Close relatives of φ6.
In this way, we isolated φ7, φ9, φ10, and φ11, all of which infected LM2333 and HB and not MPO.16. These phages contained genomes of three pieces of dsRNA. The RNA segments were of approximately the same sizes as those of φ6 (Table 1). RT-PCR analysis of this group, with primers derived from φ6 sequences, proved difficult but possible. When the resulting cDNA clones were sequenced, it was found that the nucleotide sequences differed from that of φ6 by about 15 to 20%. Within open reading frames (ORFs), the base sequence changes were concentrated in the third base of codon triplets so that the amino acid sequence remained highly conserved. The 5′ ends of the genomic plus strands contain the pac sequences, which are about 200 nucleotides in length and unique and specific for the packaging of each segment (6). The sequences of the pac regions in this group of phages were about 90% identical; however, the sequence changes were often complementary in regions where presumed stem-loop structures are present (27). Changes in one strand of the stems were often compensated by complementary changes in the other strand (7).
TABLE 1.
Properties of the new bacteriophages relative to φ6
| Phage | Ability to infect straind:
|
Genome segment sizes (kbp) | Source (reference) | ||||
|---|---|---|---|---|---|---|---|
| HB | MR | LM2333 | S4M | ERA | |||
| φ6 | + | + | + | + | −c | 2.9, 4.1, 6.4 | P. vulgaris (25) |
| φ7 | + | + | + | −c | 2.9, 4.1, 6.4 | P. sativum | |
| φ8 | −a | − | + | + | + | 3.2, 4.7, 7.1 | P. sativum |
| φ9 | + | + | + | −a | −a | 3.0, 4.4, 6.6 | P. vulgaris |
| φ10 | + | + | + | + | 3.0, 4.2, 6.8 | L. esculentum | |
| φ11 | + | + | + | 3.0, 4.2, 6.8 | L. esculentum | ||
| φ12 | −a | − | + | + | 3.0, 4.2, 6.8 | O. basilicum | |
| φ13 | −a | − | −b | + | 3.0, 4.2, 6.5 | Raphanus sativum | |
| φ14 | −a | −b | + | 3.0, 4.4, 6.6 | Daucus carota sativum | ||
No mutants for plating.
Mutant isolated that infected LM2333.
Mutant isolated that infected ERA.
Host strains are described in Materials and Methods.
The only observed exception was in the sequences preceding that of gene 14 (Fig. 1). Gene 14 is a small ORF preceding gene 7 in segment L of φ6 (2). It is not essential, is not a component of the virion, and probably plays a role in the regulation of expression of gene 7 (3). In φ6, gene 14 starts at nucleotide 270 (10). In φ7, gene 14 starts at nucleotide 473 with 75% base identity and 90% amino acid identity to the sequences of φ6. The sequence of gene 14 in φ7 is preceded by an ORF (ORF E) that starts at nucleotide 268, and that ORF has no similarity to the φ6 sequence. φ9 shows a similar arrangement, with the new ORF starting at nucleotide 268 and gene 14 starting at nucleotide 473. The base sequence of the new ORF in φ9 is similar (86%) but not identical to that in φ7. The pac region of φ6 segment L has been shown to end at about nucleotide 205 (6). It is interesting that the sequence divergence observed in φ7 and φ9 begins at about that point.
FIG. 1.
Comparison of the nucleotide sequences of the 5′ regions of segment L RNA plus strands in viruses closely related to φ6. Note that the pac sequence ends at about nucleotide 205. Gene 14 begins at nucleotide 270 in φ6 and φ10 and at nucleotide 473 in φ7 and φ9. ORF E is not present in φ6 or φ10 and starts at nucleotide 268 in φ7 and φ9.
The phages were tested for the ability to accept genomic segments from φ6. This was done by crossing the phages with a derivative of φ6 that carried a deletion of genes 7, 2, and 4 in segment L (14) and a Lacα reporter group in segment M (13). This phage forms plaques on a host containing plasmids that express genes 7, 2, and 4. The products of the cross were plated on strain LM1034, which carries the lacω gene so that blue plaques appear on X-Gal plates if the phage carries the α portion of the β-galactosidase gene. Since the φ6 deletion construct cannot infect this strain, any blue plaques indicate that segment M has been incorporated into the phage in question. In this way, it was found that φ7, φ9, φ10, and φ11 can accept the M segment of φ6. Similar experiments showed that they can also accept segment S of φ6.
Distant relatives of φ6.
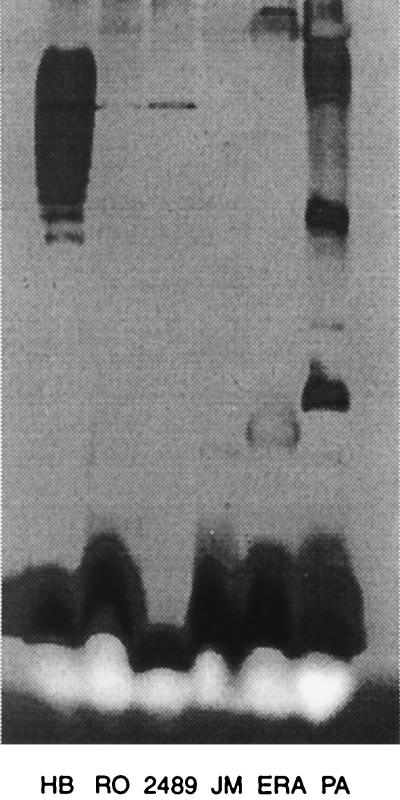
It was subsequently found that additional phages (φ8, φ12, and φ13) could be isolated that infect P. pseudoalcaligenes ERA, which is an alternative host for φ6 (9). These phages infected LM2333 but not HB, the normal host of φ6. The plating characteristics of the various isolates are shown in Table 1. These phages were able to infect a derivative of LM2333, designated LM2509, that is resistant to φ6 due to the loss of the specific type IV pilus. Various strains of P. syringae were tested for the ability to be infected by φ8, φ12, and φ13. The only positive candidate was a rough strain, Ro49dRa1. These results suggested that phages, φ8, φ12, and φ13 were attaching directly to rough LPS or some element exposed in this type of outer membrane. It also seemed reasonable that strain LM2333 and its derivatives were rough mutants of HB and that this was the reason for their general resistance to many DNA phages. E. coli JM109 is known to be somewhat rough in its LPS composition (24). Plating of concentrated suspensions of these phages on lawns of JM109 showed killing but not plaque formation. The phages did not show this killing of strain HB. A gel analysis of LPS of strains that plated this group of phages indicated that the strains on which they grew had truncated LPS (Fig. 2). A collection of S. typhimurium strains with various mutations causing changes in LPS was obtained from the Salmonella Genetic Stock Centre. The phages were plated on these strains. Phages φ12 and φ13 were able to kill strains that had lost O antigen, but they did not form plaques. Phage φ8 was able to kill the strains that had lost O antigen but was also able to form plaques on strains that harbored an rfaC, rfaE, or rfaF mutation (18). These strains have either no heptose or only one of the two normal heptose residues in their LPS. The efficiency of plating of φ8 on the heptoseless strains was similar to that on the pseudomonads, although the plaques were small and turbid.
FIG. 2.
Silver-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel of LPSs from various cultures. The samples are as follows: HB, smooth; RO, P. syringae rough; 2489, rough derivative of HB; JM, E. coli JM109; ERA, P. pseudoalcaligenes; PA, P. aeruginosa smooth.
Whereas RT-PCR on the RNAs of phages φ7, φ9, φ10, and φ11 was successful with primers derived from the φ6 sequence, similar attempts with the RNAs of φ8, φ12, and φ13 failed. The cloning and sequencing of the genomes of the latter three phages will be the subjects of separate reports. The three phages in this group were tested for the ability to acquire the M segment of φ6 in crosses. φ13 was able to pick up the M segment at low frequency; however, the other two phages could not acquire the φ6 M segment at a measurable frequency. Another way to test for the ability to acquire a new genomic segment is to plate the phage on a lawn of bacteria carrying a plasmid whose transcript contains a complete plus-strand copy of a genomic segment. If there is a strong enough selection, it is possible to screen for phages that have acquired the new genomic segment (14). Phages φ8, φ12, and φ13 cannot form plaques on HB and do not produce mutants that can infect this strain. If these phages could acquire segment M from φ6, they might be able to infect HB because the attachment proteins are coded by genes in the M segment. Phage was grown on lawns of strain LM2489 carrying plasmid pLM1084. Strain LM2489 can be infected by φ6 and all of its relatives. Plasmid pLM1084 produces a transcript that contains the entire M segment of φ6. We found that φ13 was able to acquire the M segment, while φ8 and φ12 were unable to do so. φ13 with a φ6 M segment was able to infect HB but not strain LM2509, which lacks the pilus receptor for φ6. Once we had a derivative of φ13 that could no longer form plaques on strain LM2509, we prepared strain LM2489 with a plasmid, pLM2440, containing a copy of segment M of φ13, with the φ6 pac region at the 5′ end and with kan inserted in the 3′ noncoding region. In this way, we were able to isolate a phage that had regained the φ13 host range but had kanamycin resistance as well. The sequencing and cloning of the cDNA copy of the φ13 genome are the subjects of another report (10a). It appears that φ8 and φ12 are quite distant genetically from φ6 and the closely related phages, while φ13 is between the two groups.
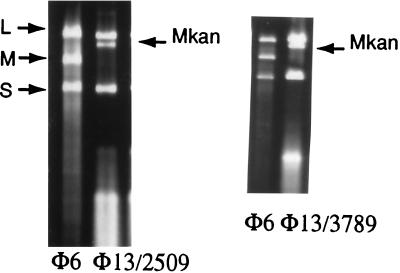
φ6 is able to form a stable carrier state in host cells (4, 12). In these cases, the phage reproduces in the cell without killing it. If the virus carries a gene such as kan, which confers resistance to kanamycin, the cells become resistant and will form colonies on agar containing kanamycin. The φ13 derivative with kan was able to form carrier state cells of strain LM2489. It was also able to form carrier state cultures of E. coli JM109 and S. typhimurium SL3789, although at a lower frequency than with Pseudomonas cells. If RNA is isolated from carrier state cells and subjected to electrophoresis, it is possible to see the dsRNA in ethidium-stained gels. The carrier state cells of JM109 and SL3789 contained viral RNA to about the same extent as the Pseudomonas carrier state cells (Fig. 3), indicating that the virus was able to propagate efficiently in the enteric bacteria once it had entered without killing the cells.
FIG. 3.
dsRNA extracted from cells resistant to kanamycin due to carrier state infections. Lanes: φ6, dsRNA isolated from φ6 virions; φ13/2509, RNA isolated from kanamycin-resistant cells of LM2509 (P. syringae without type IV pili); φ13/3789, RNA isolated from kanamycin-resistant S. typhimurium SL3789. φ13 contains a segment M with kan inserted in the 3′ noncoding region.
The host ranges of φ6 and its relatives are of interest in that they seem to constitute a niche that is left over after infection by more abundant phages. The rough-LPS mutants are generally resistant to many of the DNA phages (16), and the phages that use the HB type IV pilus are also able to infect cells that are resistant to the more common viruses that the host encounters. This type of strategy is seen in many bacteriophages; in members of the family Enterobacteriaceae, there are many phages that are specific to rough LPS (26), but the ability to infect rough-LPS mutants might also be a mechanism by which to enlarge the host range of the virus. The observation that φ8 can form plaques on Salmonella cells suggests that these phages can, in principle, propagate on many different gram-negative bacteria. It remains to be seen whether more phages of this family will be found among other gram-negative genera.
ACKNOWLEDGMENTS
This work was supported by grant GM31709 from the National Institutes of Health.
REFERENCES
- 1.Butcher S J, Dokland T, Ojala P M, Bamford D H, Fuller S D. Intermediates in the assembly pathway of the double-stranded RNA virus φ6. EMBO J. 1997;16:4477–4487. doi: 10.1093/emboj/16.14.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casini G, Revel H R. A new small low-abundant nonstructural protein encoded by the L segment of the dsRNA bacteriophage φ6. Virology. 1994;203:221–228. doi: 10.1006/viro.1994.1479. [DOI] [PubMed] [Google Scholar]
- 3.Casini G, Revel H R. Construction and analysis of a bacteriophage φ6 gene 14 nonsense mutant. Virology. 1996;216:455–458. doi: 10.1006/viro.1996.0084. [DOI] [PubMed] [Google Scholar]
- 4.Cuppels D A, Vidaver A K, Van Etten J L. Resistance to bacteriophage φ6 by Pseudomonas phaseolicola. J Gen Virol. 1979;44:493–504. [Google Scholar]
- 5.de Kievit T R, Lam J S. Isolation and characterization of two genes, waaC (rfaC) and waaF (rfaF), involved in Pseudomonas aeruginosa serotype O5 inner-core biosynthesis. J Bacteriol. 1997;179:3451–3457. doi: 10.1128/jb.179.11.3451-3457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gottlieb P, Qiao X, Strassman J, Frilander M, Mindich L. Identification of the packaging regions within the genomic RNA segments of bacteriophage φ6. Virology. 1994;200:42–47. doi: 10.1006/viro.1994.1160. [DOI] [PubMed] [Google Scholar]
- 7.Mindich L. Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage φ6. Microbiol Mol Biol Rev. 1999;63:149–160. doi: 10.1128/mmbr.63.1.149-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mindich L. Bacteriophage φ6: a unique virus having a lipid-containing membrane and a genome composed of three dsRNA segments. Adv Virus Res. 1988;35:137–176. doi: 10.1016/s0065-3527(08)60710-1. [DOI] [PubMed] [Google Scholar]
- 9.Mindich L, Cohen J, Weisburd M. Isolation of nonsense suppressor mutants in Pseudomonas. J Bacteriol. 1976;126:177–182. doi: 10.1128/jb.126.1.177-182.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mindich L, Nemhauser I, Gottlieb P, Romantschuk M, Carton J, Frucht S, Strassman J, Bamford D H, Kalkkinen N. Nucleotide sequence of the large double-stranded RNA segment of bacteriophage φ6: genes specifying the viral replicase and transcriptase. J Virol. 1988;62:1180–1185. doi: 10.1128/jvi.62.4.1180-1185.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10a.Mindich, L., et al. Unpublished data.
- 11.Murphy F A. Virus taxonomy: sixth report of the International Committee on Taxonomy of Viruses. New York, N.Y: Springer-Verlag; 1995. [Google Scholar]
- 12.Onodera S, Olkkonen V M, Gottlieb P, Strassman J, Qiao X, Bamford D H, Mindich L. Construction of a transducing virus from double-stranded RNA bacteriophage φ6: establishment of carrier states in host cells. J Virol. 1992;66:190–196. doi: 10.1128/jvi.66.1.190-196.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Onodera S, Qiao X, Gottlieb P, Strassman J, Frilander M, Mindich L. RNA structure and heterologous recombination in the double-stranded RNA bacteriophage φ6. J Virol. 1993;67:4914–4922. doi: 10.1128/jvi.67.8.4914-4922.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onodera S, Qiao X, Qiao J, Mindich L. Acquisition of a fourth genomic segment in bacteriophage φ6: a bacteriophage with a genome of three segments of dsRNA. Virology. 1995;212:204–212. doi: 10.1006/viro.1995.1469. [DOI] [PubMed] [Google Scholar]
- 15.Onodera S, Qiao X, Qiao J, Mindich L. Directed changes in the number of dsRNA genomic segments in bacteriophage φ6. Proc Natl Acad Sci USA. 1998;95:3920–3924. doi: 10.1073/pnas.95.7.3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Picken R N, Beacham I R. Bacteriophage-resistant mutants of Escherichia coli K12 with altered lipopolysaccharide. Studies with concanavalin A. J Gen Microbiol. 1977;102:319–326. doi: 10.1099/00221287-102-2-319. [DOI] [PubMed] [Google Scholar]
- 17.Qiao X, Qiao J, Mindich L. Stoichiometric packaging of the three genomic segments of dsRNA bacteriophage φ6. Proc Natl Acad Sci USA. 1997;94:4074–4079. doi: 10.1073/pnas.94.8.4074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roantree R J, Kuo T-T, MacPhee D G. The effect of defined lipopolysaccharide core defects upon antibiotic resistances of Salmonella typhimurium. J Gen Microbiol. 1977;103:223–234. doi: 10.1099/00221287-103-2-223. [DOI] [PubMed] [Google Scholar]
- 19.Roine E, Raineri D M, Romantschuk M, Wilson M, Nunn D N. Characterization of type IV pilus genes in Pseudomonas syringae pv. tomato DC3000. Mol Plant-Microbe Interact. 1998;11:1048–1056. doi: 10.1094/MPMI.1998.11.11.1048. [DOI] [PubMed] [Google Scholar]
- 20.Romantschuk M, Bamford D H. Function of pili in bacteriophage φ6 penetration. J Gen Virol. 1985;66:2461–2469. doi: 10.1099/0022-1317-66-11-2461. [DOI] [PubMed] [Google Scholar]
- 21.Romantschuk M, Olkkonen V M, Bamford D H. The nucleocapsid of bacteriophage φ6 penetrates the host cytoplasmic membrane. EMBO J. 1988;7:1821–1829. doi: 10.1002/j.1460-2075.1988.tb03014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Semancik J S, Vidaver A K, Van Etten J L. Characterization of a segmented double-helical RNA from bacteriophage φ6. J Mol Biol. 1973;78:617–625. doi: 10.1016/0022-2836(73)90283-0. [DOI] [PubMed] [Google Scholar]
- 23.Sinclair J F, Cohen J, Mindich L. The isolation of suppressible nonsense mutants of bacteriophage φ6. Virology. 1976;75:198–208. [PubMed] [Google Scholar]
- 24.Stevenson G, Neal B, Liu D, Hobbs M, Packer N H, Batley M, Redmond J W, Lindquist L, Reeves P. Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb gene cluster. J Bacteriol. 1994;176:4144–4156. doi: 10.1128/jb.176.13.4144-4156.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vidaver A K, Koski R K, Van Etten J L. Bacteriophage φ6: a lipid-containing virus of Pseudomonas phaseolicola. J Virol. 1973;11:799–805. doi: 10.1128/jvi.11.5.799-805.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilkinson R G, Gemski P, Jr, Stocker B A D. Non-smooth mutants of Salmonella typhimurium: differentiation by phage sensitivity and genetic mapping. Can J Microbiol. 1972;70:527–554. doi: 10.1099/00221287-70-3-527. [DOI] [PubMed] [Google Scholar]
- 27.Zuker M. On finding all suboptimal foldings of an RNA molecule. Science. 1989;244:48–52. doi: 10.1126/science.2468181. [DOI] [PubMed] [Google Scholar]