Key Words: axonal regeneration, conditional knockout, neurotransmitter, optic nerve injury, presynaptic neuron, retinal network, synaptic connection, synaptic vesicles, visual acuity, zinc transporter 3
Abstract
Vision depends on accurate signal conduction from the retina to the brain through the optic nerve, an important part of the central nervous system that consists of bundles of axons originating from retinal ganglion cells. The mammalian optic nerve, an important part of the central nervous system, cannot regenerate once it is injured, leading to permanent vision loss. To date, there is no clinical treatment that can regenerate the optic nerve and restore vision. Our previous study found that the mobile zinc (Zn2+) level increased rapidly after optic nerve injury in the retina, specifically in the vesicles of the inner plexiform layer. Furthermore, chelating Zn2+ significantly promoted axonal regeneration with a long-term effect. In this study, we conditionally knocked out zinc transporter 3 (ZnT3) in amacrine cells or retinal ganglion cells to construct two transgenic mouse lines (VGATCreZnT3fl/fl and VGLUT2CreZnT3fl/fl, respectively). We obtained direct evidence that the rapidly increased mobile Zn2+ in response to injury was from amacrine cells. We also found that selective deletion of ZnT3 in amacrine cells promoted retinal ganglion cell survival and axonal regeneration after optic nerve crush injury, improved retinal ganglion cell function, and promoted vision recovery. Sequencing analysis of reginal ganglion cells revealed that inhibiting the release of presynaptic Zn2+ affected the transcription of key genes related to the survival of retinal ganglion cells in postsynaptic neurons, regulated the synaptic connection between amacrine cells and retinal ganglion cells, and affected the fate of retinal ganglion cells. These results suggest that amacrine cells release Zn2+ to trigger transcriptomic changes related to neuronal growth and survival in reginal ganglion cells, thereby influencing the synaptic plasticity of retinal networks. These results make the theory of zinc-dependent retinal ganglion cell death more accurate and complete and provide new insights into the complex interactions between retinal cell networks.
Introduction
The optic nerve (ON), formed from the axons of retinal ganglion cells (RGCs), is a crucially important structure in the central nervous system that transmits visual information obtained by the retina to the brain. The ON is frequently researched in the study of the central nervous system because of its accessibility, anatomy, and functional importance. Because of their length, trajectory, and space limitations, RGC axons are highly vulnerable to damage, such as from head trauma and traumatic brain injury (Williams et al., 2020). And glaucoma, the world’s most common irreversible blinding eye disease with reduction in the visual field, is also due to the damage of the ON (Quigley, 2016). Similar to most mature neurons in the central nervous system, RGCs are unable to regenerate axons once they are injured (Benowitz et al., 2017; Tran et al., 2019; Tian et al., 2022), and their fate are influenced not only by cell intrinsic mechanisms, but also by the complex circuitry of the retina, which includes changes in inhibitory interneurons, such as amacrine cells (ACs) (Sergeeva et al., 2021). ACs modulate inhibitory inputs into RGCs by γ-aminobutyric acid or glycine (Vígh et al., 2000; Kim et al., 2015). Growing evidence suggests that this complex AC-RGC retinal circuitry plays an important role in RGC survival and axon regeneration.
Zinc is essential for the structures and functions of countless proteins, with roles in neurotransmission, gene regulation, and enzymatic activity (Frederickson et al., 2000). Zinc is also involved in synaptic plasticity, information processing, memory, learning, and the regulation of neuronal development (Kumar et al., 2021). As a neurosecretory product and cofactor in the central nervous system, zinc is highly concentrated in synaptic vesicles (Frederickson et al., 2000). To maintain Zn2+ concentrations within a narrow range for normal physiological functions and avoid mismetallation and Zn2+ toxicity, a precise homeostatic machinery consisting of metal-buffering proteins, metallothioneins, and zinc transporters (ZnTs and ZIPs) has evolved (Aras and Aizenman, 2011; Hara et al., 2022). Zinc transporter 3 (ZnT3) loads Zn2+ into synaptic vesicles (Palmiter et al., 1996), and thus ZnT3 levels reflect the level of intercellular Zn2+.
Our previous study reported an interesting phenomenon in which mobile Zn2+ suppressed RGC survival and regenerative ability. Evidence suggests that ACs produce this inhibitory signal immediately when the ON is injured, owing to the strong co-localization among zinc, zinc transporter, and ACs (Li et al., 2017). In response to optic nerve injury, both mobile zinc and ZnT3 levels increase rapidly in the inner plexiform layer (IPL) of the retina, where ACs and bipolar cells synapse on the dendrites of RGCs (Li et al., 2017). These results indicate that ZnT3 packages Zn2+ into the synaptic vesicles and is released from AC terminals and that presynaptic zinc negatively affects the fate of RGCs. Whether deleting ZnT3, chelating Zn2+, or locking the synaptic release from AC terminals (for example, with Clostridium tetani neurotoxin) can preserve RGC survival and ON regeneration remains uncertain.
In this study, we investigated whether synaptic zinc came from ACs and deletion them in ACs can promote RGCs survival and optic nerve regeneration. We generated and analyzed two strains of transgenic mice with conditional knockout of ZnT3 in ACs and RGCs, determine the cellular localization of synaptic zinc, confirm that only ZnT3 deletion in AC rather than RGC plays a protective role and precludes the postsynaptic effect of zinc in RGC’s survival.
Methods
Animals
The use of animals in this study was approved by the Animal Use and Care Committee of Zhongshan Ophthalmic Center at Sun Yat-sen University, Guangzhou, China (approval No. 2019-178) on December 10, 2019. All breeding and experimental procedures were conducted in accordance with the guidelines of the Association for Research in Vision and Ophthalmology (Association for Research in Vision and Ophthalmology, 2021).
ZnT3fl/wt (Slc30a3fl/wt) mice were generated by GemPharmatech (Nanjing, China). VGAT-Cre (Strain # 028862; RRID: IMSR_JAX:028862) mice were obtained from Dr. Larry I. Benowitz, Boston Children’s Hospital, Harvard Medical School. VGLUT2-Cre (Strain # 028863; RRID: IMSR_JAX:028863) mice were obtained from GemPharmatech. All mice were maintained on a C57BL/6J background.
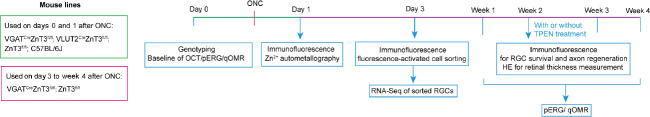
ZnT3fl/fl mice were generated from ZnT3fl/wt mice. ZnT3fl/wt mice were crossed with VGAT-Cremut/wt transgenic mice to generate ZnT3fl/wt VGAT-Cremut/wt mice and then crossed with ZnT3fl/fl mice to generate experimental animals, including ZnT3fl/flVGAT-Cremut/wt mice and the littermate control ZnT3fl/flVGAT-Crewt/wt mice. ZnT3fl/flVGLUT2-Cremut/wt mice were generated using the same strategy. The animals were randomly divided into different experimental groups (details of the sample size are described in the figure legends). At least four animals in each group were examined at each time point. Male and female mice were at ratios dependent on litters available and with equal distributions across experiments. The animals were housed in temperature (22°C) and humidity (60%) controlled cages, with a 12/12-hour light/dark cycle and access to water and normal diet food ad libitum. The experimental overview is shown in Figure 1.
Figure 1.
Flowchart of experimental procedures.
The animals (including VGATCreZnT3fl/fl; VLUT2CreZnT3fl/fl; ZnT3fl/fl; and C57BL/6J mice) were examined randomly. Timeline of experiments is presented on a horizontal line and the animal lines we used were list beside. HE: Hematoxylin and eosin staining; OCT: fundus color photography and optical coherence tomography; ONC: optic nerve crush; pERG: pattern electroretinogram; qOMR: optomotor response test; RGC: retinal ganglion cell; TPEN: N,N,N′,N′-tetrakis (2-pyridylmethyl) ethylenediamine; VGAT: vesicular γ-aminobutyric acid transporter; ZnT3: zinc transporter 3.
Genotyping
Fresh mouse tail clips were dissolved in tail lysis buffer at 55°C for 12 hours. Genomic DNA was extracted using absolute ethanol and 70% ethanol. Genotyping was performed by polymerase chain reaction using DNA polymerase (Vazyme, Nanjing, China, Cat# P222). To verify the deletion of Slc30a3 in the retina of ACs or RGCs, genomic DNA was extracted and amplified using the primers GPS00001161-Slc30a3-5wt-tF1 and GPS00001161-Slc30a3-5wt-tR1 (primer information is presented in Additional Table 1). The amplicon size for Slc30a3 wild-type (WT) was 202 bp and Slc30a3 was 304 bp (Additional Table 1). The presence of VGAT-Cre was verified using 12785 and oIMR8292 primers (amplicon size 200 bp), and primers 12786 and 12785 were used to identify the WT allele (323 bp) (Additional Table 1). We used C57BL/6J (wt/wt) mice, VGATCreZnT3fl/fl (mut/wt; Slc30a3fl/fl) mice, and blank control (water) for grouping. The presence of VGLUT2-Cre was determined by polymerase chain reaction using primers 13007 and 32667, with amplicon sizes of 124 (mutant) and 245 (WT) bp (Additional Table 1). We used C57BL/6J (wt/wt) mice, VGLUT2CreZnT3fl/fl (mut/wt; Slc30a3fl/fl) mice, and blank control (water) for grouping.
Additional Table 1.
Primer Information of VGATCreZnT3fl/fl and VGLUT2CreZnT3fl/fl mice
| No. | Primer name | Sequence (5’-3’) size | Expected band | Primer illustration |
|---|---|---|---|---|
| VGATCreZnT3fl/fl mice | ||||
| 1 | GPS00001161-Slc30a3-5wt-tF1 | ACT TGG ACC CAT CTC TGT CTC AG |
wt = 202 bp; fl = 304 bp | To detect Slc30a3 |
| GPS00001161-Slc30a3-5wt-tR1 | AGC GAG TGG ATG CTC TTC TTG G |
|||
| 2 | 12786 | CAG GGC GAT GTG GAA TAG AAA |
wt = 323 bp | To detect Vgat-cre |
| 12785 | CTT CGT CAT CGG CGG CAT CTG | |||
| 3 | 12785 | CTT CGT CAT CGG CGG CAT CTG | Mutant = ~200 bp | |
| oIMR8292 | CCA AAA GAC GGC AAT ATG GT | |||
| VGLUT2CreZnT3fl/fl mice | ||||
| 1 | 13007 | ACA CCG GCC TTA TTC CAA G | Mutant = 124 bp; | To detect |
| 32667 | AAG AAG GTG CGC AAG ACG | wt = 245 bp | Vglut2-cre | |
| 32668 | CTG CCA CAG ATT GCA CTT GA | |||
| 2 | GPS00001161-Slc30a3-5wt-tF1 | ACT TGG ACC CAT CTC TGT CTC AG |
wt = 202 bp; fl = 304 bp |
To detect Slc30a3 |
| GPS00001161-Slc30a3-5wt-tR1 | AGC GAG TGG ATG CTC TTC TTG G |
|||
fl: Slc30a3 flox; VGAT: vesicular γ-aminobutyric acid transporter; VGLUT2: vesicular-glutamate transporter 2; wt: wild type.
ONC
ON surgeries were performed on mice aged 8 weeks (average body weight, 20–25 g) under general anesthesia with a mixture of ketamine (100 mg/kg, Gutian Pharmaceutical, Gutian, China) and xylazine (10 mg/kg, Sigma-Aldrich, Shanghai, China) administered by intraperitoneal injection. The ON was exposed under an operating microscope after an incision was made in the right orbit. The ON was crushed 0.5 mm behind the nerve head for 5 seconds with angled forceps. In some experiments, animals were injected intravitreally with 3 µL of vehicle (phosphate-buffered solution) or N,N,N′,N′-tetrakis (2-pyridylmethyl) ethylenediamine (TPEN; 100 µM in phosphate-buffered solution; Merck Millipore, Shanghai, China, Cat# 616394) after nerve injury.
Pattern electroretinogram
Pattern electroretinogram (pERG) was performed using Roland-RETI scan (Electrophysiological Diagnostic Systems, Brandenburg, Germany, RETI-Scan21). On days 0 (baseline), 7, 14 and 21 post-ON injury, the mice were anesthetized with ketamine (100 mg/kg) and xylazine (10 mg/kg); the pupils were dilated with tropicamide (5 mg/mL, Shenyang Sinqi Pharmaceutical Co., Ltd., Shenyang, China) and eye gel was applied to protect the cornea. The mice were kept under dim red light throughout the procedure and held on a heated platform to maintain their body temperature at 37°C. The pERG electrode was placed on the cornea and positioned to encompass the pupil without obstructing views. A visual stimulus was generated using a black-and-white reversing checkerboard pattern. In total, 100 contrast reversals of the pERG were repeated twice in each eye, and 200 cycles were segmented, averaged, and recorded. The averaged pERGs were analyzed to evaluate the peak-to-trough P50 to N95 amplitude.
Fundus color photography and optical coherence tomography
Fundus color photography and optical coherence tomography were performed on intact mice (ZnT3fl/fl and VGATCreZnT3fl/fl mice), as previously described (Lin et al., 2022). Images were taken using a Phoenix Micron IV system (Phoenix Research Laboratories Inc., Pleasanton, CA, USA). The animals were anesthetized as described above; their eyes were treated with a drop of tropicamide solution to dilate their pupils and eye gel to keep the lens hydrated. Twenty images were averaged to eliminate projection artifacts and were taken near the ON head; the imaging location was marked on the ocular fundus with a green line.
Optomotor response test
On days 0 (baseline), 7, 14, and 28 post-ONC, the optomotor response test was performed as previously described (Lin et al., 2022). A spatial frequency threshold test (OptoDrum, Striatech, Germany) using optomotor reflex was used to measure visual acuity. Mice were placed on a pedestal located in the center of four monitors arranged in a quadrangle with moving vertical black-and-white stripes and allowed to freely move in the center of this area. The rotation speed (12°/s) and contrast (100%) on the monitors were kept constant. A movement of the head in the direction of the stripes or rotation of the body in a direction concordant with the stimulus was considered positive tracking. The right and left eyes were tested separately on the basis of the direction of rotation of the grating. The reflex was not triggered until the tests eventually reached the threshold for mouse vision. The stimulus pattern was adjusted continuously and automatically during the experiment to determine the visual threshold.
Immunofluorescence
ZnT3 and βIII-tubulin were detected at days 1 and 3 post-injury, and vesicular γ-aminobutyric acid transporter and Calbindin D-28k were detected at day 1 post-injury. Following perfusion, the eyes were enucleated with the anterior segments removed; the samples were fixed for 1 hour using 4% paraformaldehyde, transferred to phosphate-buffered solution containing 30% sucrose overnight, and then sectioned frozen at 14 µm. Frozen sections were blocked with appropriate serum and Triton at 4°C for 1 hour and then incubated with the following primary antibodies at 4°C overnight: ZnT3 (rabbit polyclonal, 1:500, Synaptic Systems, Goettingen, Germany, Cat# 197002, RRID: AB_2189664), vesicular γ-aminobutyric acid transporter (VGAT; guinea pig polyclonal, 1:500, Synaptic Systems, Cat# 131004, RRID: AB_887873), Calbindin D-28k (mouse monoclonal, 1:1000, Swant, Marly, Switzerland, Cat# 300, RRID: AB_10000347), and βIII-tubulin (TUJ-1; mouse polyclonal, 1:500, BioLegend, San Diego, CA, USA, Cat# 801202, RRID: AB_10063408). The sections were incubated with secondary antibodies at room temperature (25°C) for 2 hours: Goat anti-Rabbit IgG H&L Alexa Fluor 647 (1:500, Abcam, Shanghai, China, Cat# ab150079, RRID: AB_2722623), Goat anti-Guinea Pig IgG H&L Alexa Fluor 555 (1:500, Thermo Fisher, Shanghai, China, Cat# A-21435, RRID: AB_2535856), and Goat anti-Mouse IgG H&L Alexa Fluor 488 (1:500, Abcam, Cat# ab150113, RRID: AB_2576208). Nuclei were stained with 4′,6-diamidino-2-phenylindole (Bioss, Beijing, China, C02-04002). Images were captured using a Zeiss LSM980 confocal (Carl Zeiss, Jena, Thuringia, Germany) or Leica DMi8 microscope (Leica Microsystems, Wetzlar Deutschland, Germany) and analyzed using ImageJ (version 1.52) (National Institutes of Health, Bethesda, MD, USA) (Schneider et al., 2012).
For quantitation of RGC survival and axon regeneration, at 2 weeks after ONC, the eyes were fixed in 4% paraformaldehyde for 1 hour, and retinas were isolated, permeabilized, and incubated with primary antibodies against tubulin-βIII (TUJ-1, mouse polyclonal, 1:500, BioLegend, Cat# 801202, RRID: AB_10063408) and RNA binding protein, mRNA processing factor (RBPMS; rabbit polyclonal, 1:500, GeneTex, Irvine, CA, USA, Cat# GTX118619, RRID: AB_10720427), followed by staining with Alexa Fluor 555-conjugated secondary antibody (1:500, Abcam, Cat# ab150074, RRID: AB_2636997) to rabbit IgG and Alexa Fluor 488-conjugated secondary antibody (1:500, Abcam, Cat# ab150113, RRID: AB_2576208) to mouse IgG. ImageJ was used to count tubulin-βIII+ or RBPMS+ cells from eight images per retina (200×, DMi8; Leica) taken at prespecified areas, four at 1 mm and four at 2 mm from the ON head. The results were averaged to estimate overall RGC survival per square millimeter. For evaluation of axon regeneration, ONs were embedded in optical coherence tomography Tissue-Tek Medium (Sakura Finetek, Tokyo, Japan), frozen, and cryostat-sectioned longitudinally at 10 µm. Regenerating axons were stained with growth associated protein 43 (GAP-43) antibody (1:200, Cell Signaling Technology, Trask Lane, Danvers, MA, USA, Cat# 8945, RRID: AB_10860076) at 4°C overnight, followed by Goat anti-Rabbit IgG H&L Alexa Fluor 488 (1:500, Abcam, Cat# ab150077, RRID: AB_2630356) at room temperature (25°C) for 2 hours. To estimate the total number of regenerating axons per nerve, axons were manually counted in at least four longitudinal sections per case at predetermined distances from the crush site.
Hematoxylin and eosin staining
Eyes were collected 14 days post-injury, and paraffin-embedded slices were prepared through the optic disc. The slices were stained with hematoxylin and eosin and photographed under an optical microscope (200×, DMi8; Leica). The thickness of ganglion cell layer, IPL, inner nuclear layer, outer plexiform layer, and outer nuclear layer was calculated using ImageJ.
Zn2+ autometallography
Autometallography was performed on day 1 post-ONC as described previously (Li et al., 2017). Briefly, mice were injected intraperitoneally with sodium selenite (1.5 mg/mL in distilled water; Sigma, Shanghai, China, 15 mg/kg) 2 hours before transcardial perfusion with isotonic saline (Sigma) and 2.5% (v/v) glutaraldehyde (ECOTOP, Guangzhou, China, ED-8484) in 0.1 M phosphate buffer (pH, 7.4) under inhalation anesthesia with isoflurane (3% for 2 minutes, RWD Life Science Co., Ltd., Shenzhen, China). At least three images from different areas of each retinal section were captured under bright-field illumination (400×, DMi8; Leica). The intensity of the Zn2+-autometallography signal in the IPL was analyzed using ImageJ.
Fluorescence-activated cell sorting
Fluorescence-activated cell sorting (FACS) was performed as previously described (Chintalapudi et al., 2017). Briefly, retinas from mice (intact control, ZnT3fl/fl, and VGATCreZnT3fl/fl mice on day 3 post-ONC) were dissected in a dish containing phosphate-buffered solution/1% fetal bovine serum, and the cornea, sclera, and lens were discarded. Eyes were visually inspected for damage, blood, or inflammation, which were used as the exclusion criteria. Retinas were digested by enzymatic digestion with a combination of 15 IU/mL papain (Solarbio, Beijing, China, G8430), 1 mg/mL collagenase (Sigma, C0130), and 200 U/mL DNase I (Sigma, D5025); samples were dissociated into single-cell suspensions and incubated in phosphate-buffered solution/10% fetal bovine serum. Samples were incubated with an antibody cocktail using fluorescently-tagged CD90.2 AF-700 (BioLegend, Cat# 105320, RRID: AB_493725), CD48 PE-Cyanine7 (BioLegend, Cat# 103424, RRID: AB_2075049), CD15 PE (BioLegend, Cat# 125606, RRID: AB_1089192), and un-tagged CD57 (Sigma-Aldrich, Cat# C6680, RRID: AB_1078474) for 30 minutes. Cells were washed with media and labeled with the secondary antibody Brilliant Violet 421, Goat Anti-mouse (BioLegend, Cat# 405317, RRID: AB_10900419) for 30 minutes. Immediately prior to FACS, the live cell marker Fixable Viability Stain 780 (BD Horizon, San Diego, CA, USA, 565388) was added. Cellular debris and dead cells (780 negative cells) were excluded from the analysis. RGCs were collected on the basis of CD90.2 positive expression and CD48, CD15, and CD57 negative expressions by flow cytometry (BD, FACSAria III, BD Life Sciences, San Jose, CA, USA).
RNA-sequencing library preparation
RNA from FACS-sorted RGCs was isolated, and complementary DNA amplification products were prepared using the Discover-sc WTA Kit V2 (Vazyme, Nanjing, China, #N711), following the manufacturer’s protocols. Briefly, RNA from approximately 1000 FACS-sorted RGCs randomly selected from each group (eight retinas for FACS of each group) were isolated. Oligo dT Primer was used as the reverse transcription primer for cDNA synthesis, and the Discover-sc Reverse Transcriptase (Vazyme) was used to add an adapter sequence to the 3′ end of the cDNA; the full-length cDNA amplification product was obtained by polymerase chain reaction amplification of the adaptor. The TruePrep DNA Library Prep Kit V2 (Vazyme, #TD502) and TruePrep Index Kit V2 for Illumina (Vazyme, #TD202) were used for cDNA fragmentation, end repair, adaptor ligation, and library amplification following the manufacturer’s instructions. The library size distribution and molar concentration were determined using the Agilent 2100 Bioanalyzer High-Sensitivity DNA Assay (Agilent Technologies, Inc. Waldbronn, Germany). Three libraries were constructed for each treatment group.
Processing of RNA-sequencing data
Library products 250–450 bp in size were enriched, quantified, and sequenced on the Illumina HiSeqTM 2000 platform (Illumina, Inc. San Diego, CA, USA). Sequence reads were mapped to the mouse genome assembly (GRCm38/mm10) using STAR version 2.5.3a (Dobin et al., 2013) (http://code.google.com/p/rna-star), and differentially expressed genes were identified using the edgeR package. The false discovery rate cutoff value was set at 0.05, and the minimum fold change was 2. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes enrichment analyses were performed on the genes upregulated in VGATCreZnT3fl/fl mice and downregulated in ZnT3fl/fl mice after injury and genes downregulated in VGATCreZnT3fl/fl mice and upregulated in ZnT3fl/fl mice after injury using cluster Profiler R package (Yu et al., 2012). The above analysis and data visualization were conducted in R version 4.0.0 (https://www.r-project.org/). Gene Ontology enrichment analysis was also performed on synaptic genes upregulated in VGATCreZnT3fl/fl mice and downregulated in ZnT3fl/fl mice after injury using SynGO (https://syngoportal.org/) (Koopmans et al., 2019).
Statistical analysis
All tissue processing, quantification, and data analyses were performed in a blinded manner throughout the study. Biological replicates (n) followed the standards accepted in the literature (Xie et al., 2022) and prior experience from our laboratory. Data were analyzed by one-way analysis of variance with Tukey’s multiple comparisons or unpaired two-tailed Student’s t-test with GraphPad Prism for Windows (version 8.0.1, GraphPad Software, San Diego, CA, USA, www.graphpad.com). Data are presented as mean ± standard error of the mean (SEM). Statistical significance was set at P < 0.05.
Results
Generation of ZnT3 conditional knockout mice
Because we have preliminarily explored that ZnT3 was localized at inner plexiform layer (Li et al., 2017), and ZnT3 is responsible for loading zinc into synaptic vesicles, we generated ZnT3 conditional knockout mice to verify the origin of Zn2+. ZnT3 is encoded by Slc30a3, which has six transcripts (Ensembl, 2018). Analysis of the structure of the Slc30a3 gene indicated that exon 2 through exon 7 of the Slc30a3-201 (ENSMUST00000031037.13) transcript, a 923 bp region, was suitable as the knockout region. Deletion of this region leads to a frameshift mutation, resulting in disruption of protein function. We used clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) technology to modify the Slc30a3 gene (Figure 2A). VGLUT2-Cre mice selectively express Cre in excitatory neurons, such as RGCs (Gong et al., 2006; Wässle et al., 2006; Lu et al., 2020; Wang et al., 2020), whereas VGAT-Cre mice were reported to express Cre in ACs and horizontal cells (Cueva et al., 2002; Lu et al., 2020; Figure 2B). Because VGAT-Cre is often considered to be selectively expressed in AC when researchers focused on ON regeneration for horizontal cells do not make direct contact with RGCs (Zhang et al., 2019; Lu et al., 2020), and the results of our previous studies (Li et al., 2017, 2022), RNA-seq of our dataset and immunostaining of ZnT3 and Calbindin (a horizontal cell marker) in retina with or without injury confirmed ZnT3 was expressed in ACs rather than horizontal cells (Additional Figure 1 (996.5KB, tif) ). Therefore, the ZnT3fl/flVGATCre mice are considered to delete ZnT3 in ACs. We mated ZnT3fl/fl (Slc30a3fl/fl) mice with VGLUT2-Cre transgenic mice to generate RGC ZnT3-knockout mice (VGLUT2CreZnT3fl/fl) and VGAT-Cre mice to generate AC ZnT3-knockout mice (VGATCreZnT3fl/fl). Genotyping with tail DNA confirmed the presence of Slc30a3fl/fl, VGAT-Cre, and VGLUT2-Cre in the respective mice (see the Methods section for details) (Figure 2C and D).
Figure 2.
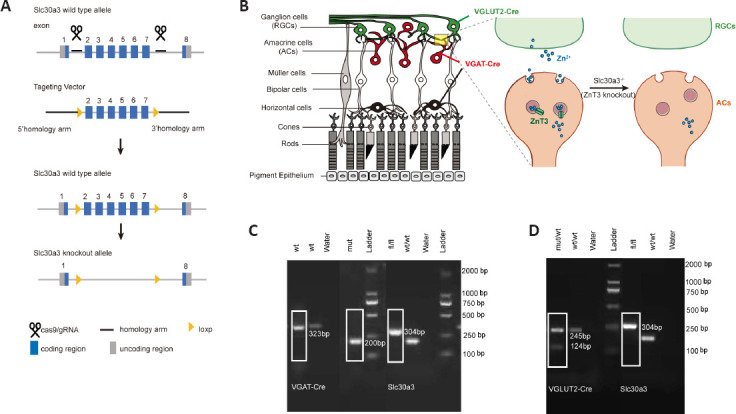
Specific knockout of ZnT3 in ACs and RGCs.
(A) Schematic illustration of the ZnT3 (Slc30a3) knockout strategy by CRISPR/Cas9 technology. (B) Schematic of the retinal structure (left); VGLUT2-Cre mice selectively express Cre in RGCs (green), whereas VGAT-Cre mice selectively express Cre in amacrine (red) and horizontal cells (black). Synaptic connections between RGCs and ACs are shown on the right; Zn2+ is encapsulated in the synaptic vesicles in ACs, ZnT3 transports the vesicles to the synaptic cleft, and Zn2+ released into RGCs. (C) Genotyping of VGAT-Cre (mut/wt), Slc30a3fl/fl, and Slc30a3fl/flVGAT-Cre mice (n = 4). The white boxes indicate the genotyping result of Slc30a3fl/fl VGAT-Cre mice. VGAT-Cre: Mutant = 200 bp, wt = 323 bp; Slc30a3: fl = 304 bp, wt = 202 bp. (D) Genotyping of VGLUT2-Cre (mut/wt), Slc30a3fl/fl and Slc30a3fl/flVGLUT2-Cre mice (n = 4). The white boxes indicate the genotyping result of Slc30a3fl/flVGLUT2-Cre mice. VGUT2-Cre: Mutant = 124 bp, wt = 245 bp; Slc30a3: fl = 304 bp, wt = 202 bp. AC: Amacrine cell; CRISPR/Cas9: clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9; RGCs: retinal ganglion cells; wt or wt/wt: C57BL/6J wild-type mice; VGAT: vesicular γ-aminobutyric acid transporter; VGLUT2: vesicular-glutamate transporter 2; ZnT3: zinc transporter 3.
AC-specific ZnT3 deletion inhibits Zn2+ increase in the retina after injury
We first assessed the expression levels of ZnT3 and Zn2+ in mouse strains after ONC. In intact mice, a faint ZnT3 signal in the IPL was observed, and it significantly increased (P = 0.003) by the first day after ONC; on the third day, ZnT3 level decreased but was still significantly higher (P = 0.0402) than that in controls (Figure 3A and B). This is consistent with our previous study (Li et al., 2017), which showed that Zn2+ levels were highest in the IPL on the first day after injury. We therefore next examined the expression of ZnT3 at 1 day post-ONC. On the first day after ONC, strong staining of ZnT3 was observed in littermate control (ZnT3fl/fl) and VGLUT2CreZnT3fl/fl mice; however, VGATCreZnT3fl/fl mice did not show ZnT3 expression with or without injury (Figure 3C). These results suggest that ZnT3 accumulates primarily in vesicles in ACs. This was supported by the strong overlap between ZnT3 and vesicular γ-aminobutyric acid transporter VGAT, an AC synapse marker, but not Tuj1, an RGC marker (Figure 3C).
Figure 3.
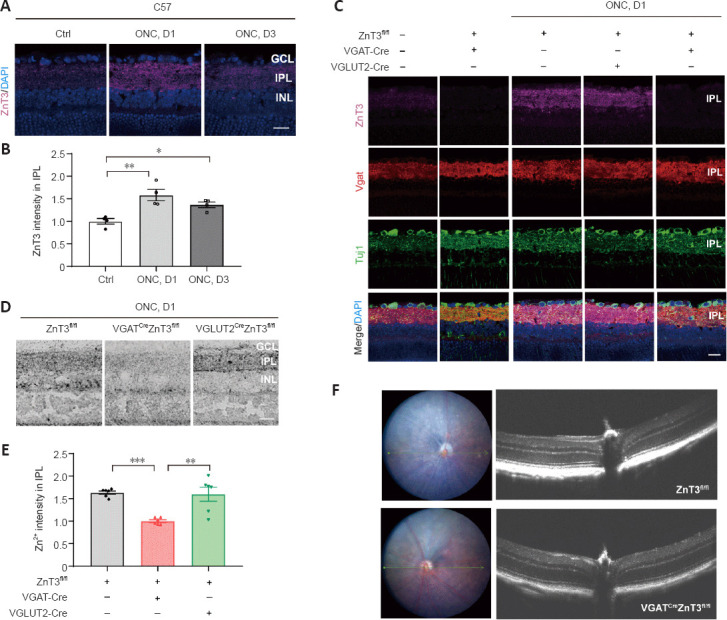
Conditional knockout of ZnT3 in ACs inhibits Zn2+ increase in the retina after injury.
(A) ZnT3 (purple) immunostaining in the retinas of C57BL/6J wild-type mice. Scale bar: 20 µm. (B) Quantitation of ZnT3 immunostaining in the IPL before and at days 1 and 3 after ONC in wild-type mice (normalized to the normal control, n = 4). (C) ZnT3 (purple), Vgat (red), Tuj1 (green), and DAPI (blue) immunostaining in the retinas of C57BL/6J wild-type, VGATCreZnT3fl/fl, VGLUT2CreZnT3fl/fl, and ZnT3fl/fl mice. C57 mice showed a weak ZnT3 signal when uninjured, strong staining of ZnT3 was observed in ZnT3fl/fl and VGLUT2CreZnT3fl/fl mice after ONC, and VGATCreZnT3fl/fl mice did not show any staining with or without injury. Scale bar: 20 µm. (D) AMG staining in the IPL of VGATCreZnT3fl/fl, VGLUT2CreZnT3fl/fl, and ZnT3fl/fl mice. The level of Zn2+ was reduced in VGATCreZnT3fl/fl mice after ONC, whereas the levels of zinc were similar in VGLUT2CreZnT3fl/fl and ZnT3fl/fl mice. Scale bar: 20 µm. (E) Quantification of AMG staining after ONC at day 1 (normalized to VGATCreZnT3fl/fl, n = 6). Data are presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 (one-way analysis of variance followed by Tukey’s multiple comparisons). (F) Retinal imaging and optical coherence tomography scanning (taken at the reference point (green arrow)) of VGATCreZnT3fl/fl mice and their littermates (ZnT3fl/fl mice). No pathological changes were observed. AC: Amacrine cell; AMG: autometallography; DAPI: 4′,6-diamidino-2-phenylindole; GCL: ganglion cell layer; INL: inner nuclear layer; IPL: inner plexiform layer; ONC: optic nerve crush; VGAT: vesicular γ-aminobutyric acid transporter; VGLUT2: vesicular-glutamate transporter 2; ZnT3: zinc transporter 3.
Autometallography was performed to visualize mobile zinc in the IPL because of its selectivity for Zn2+ (Danscher and Stoltenberg, 2005). While the control group (ZnT3fl/fl) and VGLUT2CreZnT3fl/fl mice (conditional knockout of ZnT3 in RGCs) showed similar levels of zinc after ONC, VGATCreZnT3fl/fl mice, with ZnT3 deletion in ACs, showed lower levels of Zn2+ (Figure 3D and E). Thus, these results reveal that selective deletion of ZnT3 in ACs resulted in reduced Zn2+ after injury. No architectural pathological changes were observed in the transgenic mice via fundus color photography and optical coherence tomography (Figure 3F), demonstrating that the gene knockout did not affect the normal structural features of the retina.
Deletion of ZnT3 in ACs attenuates RGC death and retinal thinning after ONC
We next assessed RGC count after injury by staining for RBPMS and Tuj1 (Figure 4A). We selected eight regions in each retina, including the central and peripheral parts, and counted the RGCs of the whole-mount, central, and peripheral retina (Figure 4B). ONC led to significant RGC death at 2 weeks. ZnT3 knockout in ACs nearly doubled RGC survival (Figure 4C), and the survival rate of RGC in both the peripheral and central retina was improved (Figure 4D and E). Combination treatment with TPEN, a high-affinity, membrane-permeable Zn2+ chelator (Lees et al., 1998), did not increase the survival rate of RGCs in VGATCreZnT3fl/fl mice (Additional Figure 2A (623.2KB, tif) –D (996.5KB, tif) ). RGC-specific deletion of ZnT3 did not prevent RGC death after injury (Additional Figure 2E (623.2KB, tif) –H (623.2KB, tif) ).
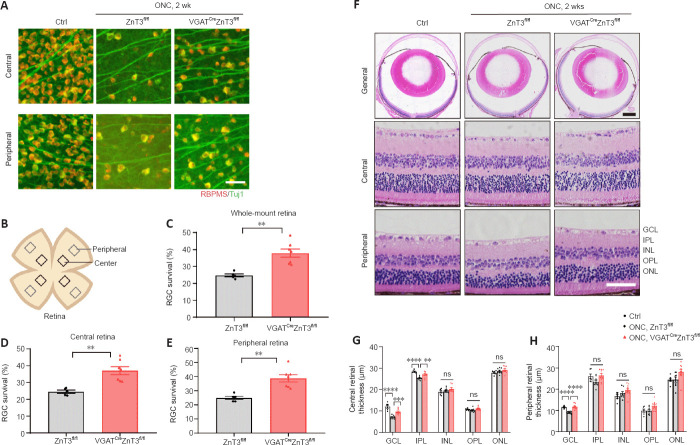
Figure 4.
Knockout of ZnT3 in ACs promotes RGC survival.
(A) Retinal whole-mounts immunostained for RNA binding protein with RBPMS (red) and Tuj1 (green) to visualize RGCs in the retinas of VGATCreZnT3fl/fl mice and the littermate control ZnT3fl/fl mice 2 weeks post-injury. RGC survival was increased in VGATCreZnT3fl/fl mice. Scale bar: 50 µm. (B) Schematic diagram of the whole-mount retina, showing the eight areas from the central and peripheral areas of the retina used for quantification. (C–E) Quantitative RGC counts in whole-mount, peripheral, and central retinal samples (n = 5, 7). Animals without ONC were used to be comparison group (n = 6). (F) Representative low- and high-power images of hematoxylin and eosin-stained cross sections. Compared with ZnT3fl/fl mice, VGATCreZnT3fl/fl mice showed attenuated retinal thinning. Scale bar: 50 µm for high-power images and 500 µm for low-power images. (G, H) Quantitative analyses of retinal thickness in the central and peripheral region of the retina (n = 6, 8, 10). Data are presented as mean ± SEM. **P < 0.01, ***P < 0.001, ****P < 0.0001 (unpaired two-tailed Student’s t-test (C–E) or one-way analysis of variance followed by Tukey’s multiple comparisons (G, H)). AC: Amacrine cell; Ctrl: control; GCL: ganglion cell layer; INL: inner nuclear layer; IPL: inner plexiform layer; ns: not significant; ONC: optic nerve crush; ONL: outer nuclear layer; OPL: outer plexiform layer; RBPMS: RNA binding protein, mRNA processing factor; RGC: retinal ganglion cell; VGAT: vesicular γ-aminobutyric acid transporter; ZnT3: zinc transporter 3.
We also evaluated retinal thickness by hematoxylin and eosin staining of retinal cross-sections, which can be affected by early glaucomatous damage (Hood et al., 2013). The retinal thickness, particularly the inner retinal layer in the central retina (including the retinal nerve fiber layer and local ganglion cell plus IPL), showed minimal ONC injury in VGATCreZnT3fl/fl mice compared with ZnT3fl/fl littermates (Figure 4F–H).
Zn2+ depletion in ACs promotes axon regeneration
We next investigated whether ZnT3 knockout in ACs influences the ability of RGCs to regenerate axons. We used GAP-43 to quantify the number of axons extending selected distances beyond the injury site 2 weeks after injury (Leon et al., 2000). Only a few axons regenerated beyond the injury site after ONC in control mice (ZnT3fl/fl), whereas a strong increase in regeneration was observed in VGATCreZnT3fl/fl mice (Figure 5A and B).
Figure 5.
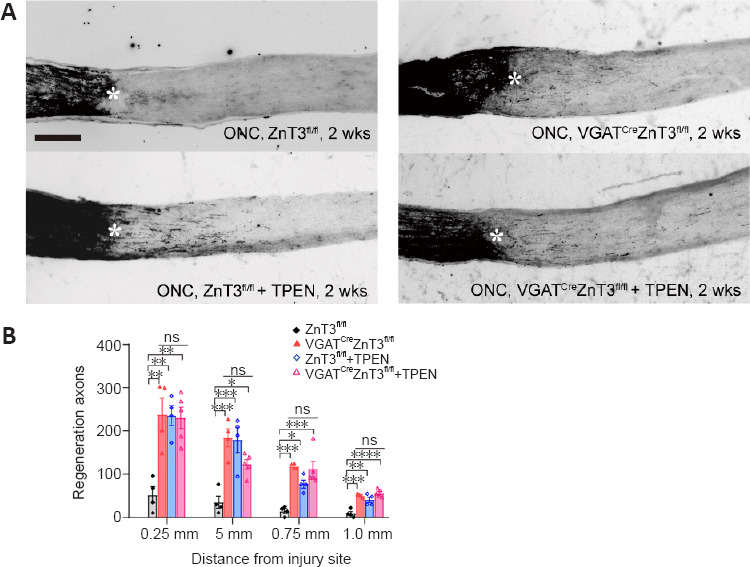
ZnT3 knockout in ACs promotes axon regeneration.
(A) Longitudinal sections through mouse optic nerve immunostained for GAP-43 at 2 weeks post-injury in VGATCreZnT3fl/fl and ZnT3fl/fl mice and the groups treated with TPEN. Few axons regenerated beyond the injury site after ONC in control mice (ZnT3fl/fl), while VGATCreZnT3fl/fl mice and mice (ZnT3fl/fl and VGATCreZnT3fl/fl) with TPEN treatment showed a strong increase in regeneration. Asterisks indicate the injury site. Scale bar: 200 µm. (B) Quantification of regenerated axons at different selected distances beyond the injury site 2 weeks post-injury. Data are presented as mean ± SEM (n = 4, 4, 4, 5). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (one-way analysis of variance followed by Tukey’s multiple comparisons). There was no significance among VGATCreZnT3fl/fl, ZnT3fl/fl + TPEN, and VGATCreZnT3fl/fl + TPEN groups. AC: Amacrine cell; GAP-43: growth associated protein 43; ns: not significant; ONC: optic nerve crush; TPEN: N,N,N’,N’-tetrakis (2-pyridylmethyl) ethylenediamine; VGAT: vesicular γ-aminobutyric acid transporter; ZnT3: zinc transporter 3.
To investigate whether knockout of ZnT3 in ACs could prevent all mobile Zn2+ caused by optic injury in the retina, we evaluated the therapeutic effect of TPEN in transgenic mice. Intraocular injection of TPEN immediately after ONC chelates all the free zinc (Li et al., 2017). Intense axonal regeneration was observed at 2 weeks after ONC, with no difference from that of VGATCreZnT3fl/fl mice (Figure 5A and B). The combination of ZnT3 deletion in ACs with TPEN-mediated Zn2+ chelation immediately after ONC treatment resulted in a regeneration level similar to that of either treatment alone (Figure 5A and B). These experimental results demonstrate that knockout of ZnT3 in AC can suppress the increase in zinc levels and promote axon regeneration after injury.
Zn2+ depletion recovers visual function and behavioral function
PERG is a well-established marker of RCG function (Bach et al., 2018). There was no significant difference in the PERG waveform between the VGATCreZnT3fl/fl mice and the control group at baseline (Figure 6A and B), and the P50-N95 amplitude of the pERG waveform, which was markedly reduced after injury, was also similar in both groups 7 days after ONC. The amplitude was well preserved in VGATCreZnT3fl/fl mice at 14 days and even further at 21 days (Figure 6A and B). Our results indicate that ZnT3-specific knockout attenuates the loss of RGC function in ONC injury.
Figure 6.
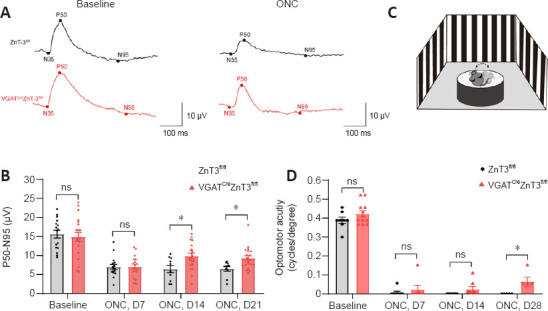
Retinal ganglion cell function and visual acuity were confirmed by pattern electroretinogram and optomotor response test.
(A) Representative images of pERG waveform with VGATCreZnT3fl/fl and ZnT3fl/fl mice. The P50-N95 amplitude of pERG was well preserved in VGATCreZnT3fl/fl mice at days 14 and 21 after ONC. (B) Quantification of the pERG P50–N95 amplitudes (n = 16, 20 for baseline; n = 16, 18 for day 7; n = 10, 20 for day 14; and n = 10, 20 for day 21). (C) Schematic diagram of the optomotor response test. The head movements of the mice were recorded and analyzed. (D) Quantification of optomotor response (n = 8, 12 for baseline; n = 8, 7 for day 7; n = 8, 8 for day 14; and n = 5, 5 for day 28). Data are presented as mean ± SEM. *P < 0.05 (unpaired two-tailed Student’s t-test). ns: Not significant; ONC: optic nerve crush; pERG: pattern electroretinogram; pERG: pattern electroretinogram; VGAT: vesicular γ-aminobutyric acid transporter; ZnT3: zinc transporter 3.
The optomotor response is the head-turning response to the rotation of a grating. The highest spatial frequency eliciting this response provides an estimate of visual acuity and this was measured using an optokinetic virtual reality system (Lu et al., 2020; Figure 6C). Four weeks after injury, VGATCreZnT3fl/fl mice showed a significantly higher visual acuity compared with the control mice after injury (Figure 6D). This suggests that the VGATCreZnT3fl/fl mice were able to remain a part of visual function after damage.
Deficiency of AC-released Zn2+ influences expression of genes related to neuronal growth and survival in RGC
Our results indicated that knocking out ZnT3 in ACs promoted the survival and functional improvement of RGCs. To further explore the potential mechanism, we evaluated the transcriptomic changes in the RGCs. RGCs from VGATCreZnT3fl/fl and ZnT3fl/fl mice with ONC on day 3 and intact WT mice were purified by FACS (Figure 7A and Additional Figure 3A (580.7KB, tif) ) and subjected to RNA-sequencing. The whole retina RNA sequencing results are shown in Additional Figure 4 (545.3KB, tif) .
Figure 7.
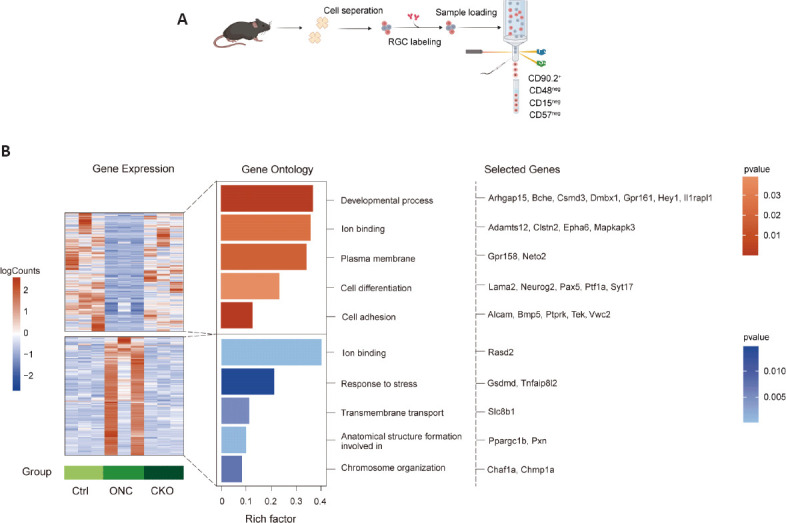
RNA-sequencing for RGCs sorted by FACS.
(A) Schematic diagram of the experimental flow. RGCs were sorted by FACS at 0 (no crush) and 3 days following optic nerve crush (n = 8 retinas in each group), and RNA sequencing was performed. (B) mRNA expression in the three groups. Colors in the heatmap indicate the mRNA expression levels. Bar plot colors represent P-values of the top Gene Ontology terms associated with genes in each cluster. Representative genes for each term are shown. CKO: Conditional knockout; Ctrl: control; FACS: fluorescence-activated cell sorting; ONC: optic nerve crush; RGC: retinal ganglion cell; VGAT: vesicular γ-aminobutyric acid transporter.
Comparative analysis of the differentially expressed genes among each group (Additional Figure 3B (580.7KB, tif) ) revealed two patterns of gene expression in response to ON injury (Figure 7B and Additional Tables 2 and 3 (212.3KB, pdf) ). The first cluster includes genes with upregulated expression in response to injury. Functionally, these genes are associated with developmental processes, ion binding, plasma membranes, cell differentiation, and cell adhesion (Figure 7B and Additional Table 2). The cluster included Alcam and Tek, which have been implicated in RGC axon elongation and maintaining normal aqueous humor circulation to avoid glaucoma (Thelen et al., 2012; Thomson et al., 2020). The second cluster includes genes with downregulated expression in response to injury. These genes are involved in ion binding, stress response, transmembrane transport, anatomical structure formation, and chromosome organization (Figure 7B and Additional Table 3 (212.3KB, pdf) ). Several genes, such as Gsdmd, Chaf1a, and Pxn, have previously been implicated in stress/injury response and neuronal death (Gotenstein et al., 2010; Tao et al., 2021; Yao et al., 2022). Kyoto Encyclopedia of Genes and Genomes analysis analysis was performed on the differentially expressed genes to explore potential pathways (Additional Figure 3C (580.7KB, tif) and D (580.7KB, tif) ). These results reveal that changes in the presynaptic neurons of RGCs affect the transcription levels of genes in RGCs that may play important roles in their survival and regeneration.
Additional Table 2.
Gene Ontology of the up-regulated cluster related to Figure 7
| GO ID | Description | generatio | BgRatio | Pvalue | Gene_name |
|---|---|---|---|---|---|
| Cellular component | |||||
| GO:0005886 | Plasma membrane | 84 | 5576 | 0.018367 | Alcam, Art3, Abcb11, Slc51a, Pcdhb18, Vwc2, Cd247, Acvr1, Unc5c, Olfr1437, Arhgap45, Pcdhb5, Pcdhb16, Kctd16, Evc, Nkain4, Chrna5, Npc1l1, Rgs13, C8g, Vmn1r4, Ackr4, Tmprss9, Ace, Slc22a27, Btla, Tshr, Utrn, Cd79a, Gpr161, Csmd3, Gabrq, Clec5a, Pcdhb9, Plcb1, Frk, Tmem65, Gab2, Fmnl3, Clec2g, Amhr2, Pcdhb19, Kcnmb4, Pcsk9, Adcy10, Pcdhb11, Nkain3, Pcdha8, Fhit, Pcdhb8, Gng10, Astn1, Tgfbr3, Pcdh20, Pcdhgc3, Trpm4, Ptprk, Clstn2, Pcdhgb1, Neto2, Aoc3, 1700019D03Rik, Olfr1254, Ptprcap, Ocln, Pcdhb4, Tek, Vmn1r200, Slc7a3, Ptgdr2, Dsg2, Stbd1, Gpr158, Car15, H2-M3, Pik3cg, Hspb1, Lama2, Lat, Scn9a, Il1rapl1, Tyrobp, Pcdha11, Pcdhga5 |
| GO:0016020 | Membrane | 84 | 5576 | 0.018367 | Alcam, Art3, Abcb11, Slc51a, Pcdhb18, Vwc2, Cd247, Acvr1, Unc5c, Olfr1437, Arhgap45, Pcdhb5, Pcdhb16, Kctd16, Evc, Nkain4, Chrna5, Npc1l1, Rgs13, C8g, Vmn1r4, Ackr4, Tmprss9, Ace, Slc22a27, Btla, Tshr, Utrn, Cd79a, Gpr161, Csmd3, Gabrq, Clec5a, Pcdhb9, Plcb1, Frk, Tmem65, Gab2, Fmnl3, Clec2g, Amhr2, Pcdhb19, Kcnmb4, Pcsk9, Adcy10, Pcdhb11, Nkain3, Pcdha8, Fhit, Pcdhb8, Gng10, Astn1, Tgfbr3, Pcdh20, Pcdhgc3, Trpm4, Ptprk, Clstn2, Pcdhgb1, Neto2, Aoc3, 1700019D03Rik, Olfr1254, Ptprcap, Ocln, Pcdhb4, Tek, Vmn1r200, Slc7a3, Ptgdr2, Dsg2, Stbd1, Gpr158, Car15, H2-M3, Pik3cg, Hspb1, Lama2, Lat, Scn9a, Il1rapl1, Tyrobp, Pcdha11, Pcdhga5 |
| GO:0071944 | Cell periphery | 84 | 5578 | 0.01853 | Alcam, Art3, Abcb11, Slc51a, Pcdhb18, Vwc2, Cd247, Acvr1, Unc5c, Olfr1437, Arhgap45, Pcdhb5, Pcdhb16, Kctd16, Evc, Nkain4, Chrna5, Npc1l1, Rgs13, C8g, Vmn1r4, Ackr4, Tmprss9, Ace, Slc22a27, Btla, Tshr, Utrn, Cd79a, Gpr161, Csmd3, Gabrq, Clec5a, Pcdhb9, Plcb1, Frk, Tmem65, Gab2, Fmnl3, Clec2g, Amhr2, Pcdhb19, Kcnmb4, Pcsk9, Adcy10, Pcdhb11, Nkain3, Pcdha8, Fhit, Pcdhb8, Gng10, Astn1, Tgfbr3, Pcdh20, Pcdhgc3, Trpm4, Ptprk, Clstn2, Pcdhgb1, Neto2, Aoc3, 1700019D03Rik, Olfr1254, Ptprcap, Ocln, Pcdhb4, Tek, Vmn1r200, Slc7a3, Ptgdr2, Dsg2, Stbd1, Gpr158, Car15, H2-M3, Pik3cg, Hspb1, Lama2, Lat, Scn9a, Il1rapl1, Tyrobp, Pcdha11, Pcdhga5 |
| Molecular function | |||||
| GO:0043167 | Ion binding | 90 | 5936 | 0.025628 | Gm2004, Ttll9, Abcb11, Zfp429, Pcdhb18, Acvr1, Peg10, Snx33, Arhgap45, Pcdhb5, Fblim1, Pcdhb16, Lrp1b, Mos, 1110008L16Rik, Zfp975, Crip3, Chrna5, Zfp979, Nlrp4f, Zbtb7c, Epha6, Atp13a5, Ddx43, Osbpl10, Fstl4, Qpct, Adamts12, Zfp92, Ace, Gm38394, Nr1h4, Rasef, Gm10324, Utrn, Glis3, Pcdhb9, Bche, Gm32687, Plcb1, Col5a2, Frk, Fthl17f, Efhc1, Slc22a21, Plekhf1, Gm14322, Mapkapk3, Amhr2, Pla2g4a, Pcdhb19, Zfp931, Mex3c, Galnt18, Adcy10, Pcdhb11, Gm19410, Gstm7, Pcdha8, Fhit, Pcdhb8, Klf11, Tgfbr3, Pcdh20, Pcdhgc3, Trpm4, Hmgcll1, Plcz1, Hapln4, Rnf19b, Npr3, Clstn2, Stk32b, Pcdhgb1, Alpk1, Aoc3, Zfp850, Reps2, Pcdhb4, Tek, Dsg2, Gm6871, Car15, Pik3cg, Scn9a, Zfp595, Zfp119b, Papolb, Pcdha11, Pcdhga5 |
| GO:0032182 | Ubiquitin-like protein binding | 4 | 108 | 0.048709 | Rnf19b, Ddi1, Hspb1, Simc1 |
| Biological process | |||||
| GO:0007155 | Cell adhesion | 32 | 1234 | 8.88E-05 | Alcam, Pcdhb18, Vwc2, Acvr1, Pcdhb5, Fblim1, Pcdhb16, Adamts12, Utrn, Bmp5, Pcdhb9, Plcb1, Pcdhb19, Pcdhb11, Pcdha8, Pcdhb8, Astn1, Pcdh20, Pcdhgc3, Hapln4, Ptprk, Clstn2, Pcdhgb1, Aoc3, Pcdhb4, Tek, Dsg2, H2-M3, Lama2, Lat, Pcdha11, Pcdhga5 |
| GO:0022610 | Biologicaladhesion | 32 | 1234 | 8.88E-05 | Alcam, Pcdhb18, Vwc2, Acvr1, Pcdhb5, Fblim1, Pcdhb16, Adamts12, Utrn, Bmp5, Pcdhb9, Plcb1, Pcdhb19, Pcdhb11, Pcdha8, Pcdhb8, Astn1, Pcdh20, Pcdhgc3, Hapln4, Ptprk, Clstn2, Pcdhgb1, Aoc3, Pcdhb4, Tek, Dsg2, H2-M3, Lama2, Lat, Pcdha11, Pcdhga5 |
| GO:0032502 | Developmental process | 93 | 5644 | 0.001767 | Alcam, Csrnp1, Cnmd, Shcbp1l, Hnmt, Dmbx1, Vwc2, Syt17, Acvr1, Unc5c, Peg10, Sct, Armc4, Fblim1, Rai14, Pcdhb16, Evc, Id4, Bend6, Dnaic1, C1galt1c1, Zbtb7c, Evi2, Pnpt1, Ttc9, Fstl4, Adamts12, Ehf, Cdkn2b, Ace, Gm38394, Nr1h4, Tshr, Utrn, Cd79a, Gpr161, Csmd3, Bmp5, Clec5a, Bche, Plcb1, Col5a2, Frk, Tmem65, Mycl, Efhc1, Gab2, Fmnl3, Clec2g, Amhr2, Pla2g4a, Pcsk9, Mex3c, Pax5, Klf1, Mospd1, Otor, Dusp6, Astn1, Tgfbr3, Etl4, Trpm4, Plcz1, Sema3a, Npr3, Maf, Ptprk, Clstn2, Tbx6, Kbtbd8, Ptf1a, Neurog2, Dazl, Pgf, Arhgap15, Tek, Nat1, Dsg2, Klhl10, H2-M3, Hey1, Pik3cg, Dnajb9, Hspb1, Lama2, Hesx1, Scn9a, Tal2, Il1rapl1, Tyrobp, Trappc2, Tdrd5, Dact1 |
| GO:0048856 | Anatomical structure development | 88 | 5284 | 0.001824 | Alcam, Csrnp1, Cnmd, Hnmt, Dmbx1, Vwc2, Syt17, Acvr1, Unc5c, Peg10, Sct, Armc4, Fblim1, Pcdhb16, Evc, Id4, Bend6, Dnaic1, C1galt1c1, Evi2, Pnpt1, Ttc9, Fstl4, Adamts12, Ehf, Cdkn2b, Ace, Gm38394, Nr1h4, Tshr, Utrn, Cd79a, Gpr161, Csmd3, Bmp5, Clec5a, Bche, Plcb1, Col5a2, Tmem65, Mycl, Efhc1, Gab2, Fmnl3, Clec2g, Amhr2, Pla2g4a, Pcsk9, Mex3c, Pax5, Klf1, Otor, Dusp6, Astn1, Tgfbr3, Etl4, Trpm4, Plcz1, Sema3a, Npr3, Maf, Ptprk, Clstn2, Tbx6, Kbtbd8, Ptf1a, Neurog2, Dazl, Pgf, Arhgap15, Tek, Nat1, Dsg2, Klhl10, H2-M3, Hey1, Pik3cg, Dnajb9, Hspb1, Lama2, Hesx1, Scn9a, Tal2, Il1rapl1, Tyrobp, Trappc2, Tdrd5, Dact1 |
| GO:0048869 | Cellular developmental process | 62 | 3861 | 0.022863 | Alcam, Cnmd, Shcbp1l, Vwc2, Syt17, Acvr1, Unc5c, Peg10, Fblim1, Rai14, Id4, Bend6, C1galt1c1, Zbtb7c, Evi2, Fstl4, Adamts12, Ehf, Cdkn2b, Ace, Tshr, Cd79a, Csmd3, Bmp5, Clec5a, Bche, Plcb1, Col5a2, Frk, Mycl, Gab2, Fmnl3, Clec2g, Pcsk9, Mex3c, Pax5, Klf1, Mospd1, Dusp6, Astn1, Tgfbr3, Trpm4, Sema3a, Maf, Ptprk, Tbx6, Kbtbd8, Ptf1a, Neurog2, Dazl, Pgf, Arhgap15, Tek, Klhl10, H2-M3, Hey1, Dnajb9, Hspb1, Lama2, Tyrobp, Tdrd5, Dact1 |
| GO:0044767 | Single-organism developmental process | 69 | 4385 | 0.024452 | Alcam, Cnmd, Shcbp1l, Vwc2, Syt17, Acvr1, Unc5c, Peg10, Sct, Fblim1, Rai14, Id4, Bend6, C1galt1c1, Zbtb7c, Evi2, Pnpt1, Fstl4, Adamts12, Ehf, Cdkn2b, Ace, Gm38394, Tshr, Cd79a, Gpr161, Csmd3, Bmp5, Clec5a, Bche, Plcb1, Col5a2, Frk, Mycl, Gab2, Fmnl3, Clec2g, Pcsk9, Mex3c, Pax5, Klf1, Mospd1, Dusp6, Astn1, Tgfbr3, Etl4, Trpm4, Sema3a, Maf, Ptprk, Tbx6, Kbtbd8, Ptf1a, Neurog2, Dazl, Pgf, Arhgap15, Tek, Nat1, Klhl10, H2-M3, Hey1, Dnajb9, Hspb1, Lama2, Hesx1, Tyrobp, Tdrd5, Dact1 |
| GO:0030154 | Cell differentiation | 59 | 3757 | 0.039332 | Alcam, Cnmd, Shcbp1l, Vwc2, Syt17, Acvr1, Unc5c, Peg10, Rai14, Id4, Bend6, C1galt1c1, Zbtb7c, Evi2, Fstl4, Adamts12, Ehf, Cdkn2b, Ace, Tshr, Cd79a, Csmd3, Bmp5, Clec5a, Bche, Plcb1, Col5a2, Frk, Mycl, Gab2, Clec2g, Pcsk9, Mex3c, Pax5, Klf1, Mospd1, Dusp6, Astn1, Tgfbr3, Trpm4, Sema3a, Maf, Ptprk, Tbx6, Kbtbd8, Ptf1a, Neurog2, Dazl, Pgf, Tek, Klhl10, H2-M3, Hey1, Dnajb9, Hspb1, Lama2, Tyrobp, Tdrd5, Dact1 |
AC-RGC synaptic connection regulated by Zn2+
Visual circuit development leads to precise connectivity and identification of synaptic loci (Hooks and Chen, 2020), and synaptic plasticity contributes to the alignment of the retinal networks. To better understand the synaptic communication between ACs and RGCs influenced by zinc and examine whether the ZnT3 conditional knockout affects the RGC synaptic biological process (Koopmans et al., 2019), we performed GO enrichment analysis with synapse Gene Ontology (SynGO) (https://syngoportal.org), a curated knowledge base of the synapse. Previous studies showed that ONC decreases the transport of proteins involved in synaptic plasticity (Sharma et al., 2015). We thus focused on genes upregulated in conditional knockout (CKO) mice (VGATCreZnT3fl/fl mice) (CKO/ONC; fold change > 2; P < 0.05). The genes were locationally enriched for postsynaptic specialization, including postsynaptic density (GO: 0014069), such as Clstn2, Grin3a, and Neto2, and postsynaptic specialization membranes (GO: 0099634), such as Chrne (Figure 8A). Additionally, functional enrichment was observed for genes involved in synapse organization (GO:0050808), such as Chrna5 and Plcb1, and trans-synaptic signaling (GO:0099537), such as Sema3a and Il1rapl1 (Figure 8B). These results suggest that conditional knockout of ZnT3 may restore AC-RGC synapse assembly and cell-cell signaling within or from synapses, without affecting RGC synaptic metabolism and transport. This upregulated gene expression signature suggests that AC-derived vesicular zinc may affect the synaptic link between ACs and RGCs, which further regulates the fate of RGCs.
Figure 8.
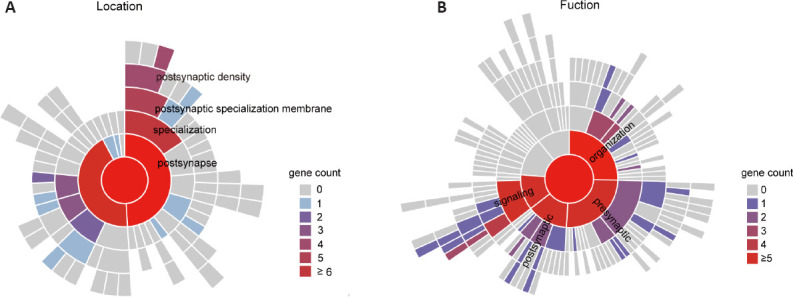
SynGO enrichment analysis of genes upregulated in ZnT3 conditional knockout RGCs.
(A) Sunburst plots of SynGO-enrichment analyses of locations (cellular components) for 560 CKO upregulated genes comparing the transcriptomes of ONC only group (CKO/ONC, fold change > 2, P < 0.05). (B) Sunburst plots of SynGO-enrichment analyses of functions (biological processes) for 560 CKO-upregulated genes when comparing the transcriptomes of ONC only group (CKO/ONC, fold change > 2, P < 0.05). Color code denotes the gene count, including child terms. CKO: Conditional knockout; ONC: optic nerve crush; RGC: retinal ganglion cell; SynGO: synapse Gene Ontology; ZnT3: zinc transporter 3.
Discussion
Studies in conditional knockout mice have identified genes such as Pannexin1 and Rac1 that play a positive role in protecting RGCs (Dvoriantchikova et al., 2012; Zhang et al., 2020). Our results showed for the first time that knocking out ZnT3 in ACs can play a protective role in RGCs. This result indicates that interneurons influence the activity of their target neurons through the release of appropriate neurochemicals, such as zinc, and that Zn2+ acts as a potent modulator of neuronal signaling, as reported previously (Paul et al., 2017). Notably, different neurotransmitter transporters colocalize on synaptic vesicles, and the co-release of zinc and other vesicular contents (such as glutamate) may have a mixed effect on target cells (Upmanyu et al., 2022). In our previous study, either global deletion of ZnT3 or chelation with TPEN led to regeneration of RGC axons after crush and promote RGC survival for up to 3 months after the injury, and strong colocalization of ZnT3 with ACs suggested ACs as the origin of zinc (Li et al., 2017). In this study, we established two different types of conditional knockout mice, and our results provide direct and strong evidence for ACs as the cellular location of zinc origin. Knowledge of the cellular localization of zinc supports our hypothesis that zinc plays a key role in RGC death after injury.
Manipulating the activity of ACs or their synaptic connections with RGCs provides a powerful strategy for regulating the regenerative ability of RGCs. The retina is a thin but complex structure that includes 10 layers of cells, which constitute a complex network of cells with a preliminary visual information-processing function. Neurons are located in the final segment of the retina, and the status of RGCs is closely related to their interaction with other cells, including interneurons, glial cells, and inflammatory cells (Zhang et al., 2022). Therefore, retinal circuits can affect RGC survival and axon regeneration. ACs are predominant inhibitory interneurons that diminish RGC activity state by inhibitory neurotransmitters, including γ-aminobutyric acid, glycine, and dopamine (Lee et al., 2014; Tien et al., 2016). Purified ACs can induce neonatal RGC to shift their growth pattern from axonogenesis to dendritogenesis in culture, as contact-mediated or membrane-associated signaling is involved in this inhibition and synaptic interactions play a role in both cell types (Goldberg et al., 2002; Goldberg, 2004). Moreover, ACs become hyperactive when RGC axons are injured, and AC silencing doubles the survival of axotomized RGCs (Zhang et al., 2019). Importantly, our study confirms that ACs respond rapidly after RGC injury by Zn2+ accumulation within hours, and selective deletion of ZnT3 in ACs reduces RGCs’ death. Notably, the knockout of ZnT3 in RGCs did not rescue the damage.
Our results showed that zinc chelator treatment of conditional knockout mice did not increase the survival and regeneration-promoting effect, suggesting that knocking out ZnT3 in the AC alone controls almost all of the mobile zinc between the synapses. This reflects the cell specificity of zinc localization and the importance of ZnT3 among zinc transporter families in the retina. Either Slc30a3 deletion or zinc chelator treatment doubled the survival rate of RGCs. In addition, zinc and zinc transporters (ZnT1, ZnT3, and ZnT7) are located in different layers of the retina; they are involved in metal metabolism and maintaining certain concentrations and are necessary for normal functions (Márquez García et al., 2022). As a neuromodulator of synaptic neurotransmission (Choi et al., 2020; Niu et al., 2020; Baltaci et al., 2022), Zn2+ may have mixed positive and negative effects on RGCs.
To verify how deletion ZnT3 in AC affected RGC survival status after injury, we sorted RGCs from retinas by FACS, whether injured or not. It is a reliable technology for isolating primary mouse RGCs because of its sensitivity, repeatability, and immediate recognition of viable cells, on the ground of protocols that contain both positive and negative selection criteria (Chintalapudi et al., 2017). RNA-sequencing of three groups (intact mice, VGATCreZnT3fl/fl mice, and littermate control ZnT3fl/fl mice with injury) identified upregulated and downregulated genes in response to injury. The results provide some mechanistic insights. As expected, deletion of ZnT3 in ACs upregulates several neuroprotective genes in RGCs; some genes regulate the development of the retina, such as Bche, Dmbx1, Gpr161, Neurog2, and Hey1 (Mack and Robitzki, 2000; Matteson et al., 2008; Wong et al., 2010; Kowalchuk et al., 2018; Kaufman et al., 2021; Sahu et al., 2021; Zhang et al., 2021). Alcam is related to cell adhesion; it is present in extending axons and crucial for the elongation and navigation of RGC axons (Thelen et al., 2012). Tek is relevant to glaucoma; insufficient TEK can develop a hypomorphic Schlemm’s canal insufficient for aqueous humor outflow (Thomson et al., 2020). Furthermore, Neto2, EphA6, Adamts12, Vwc2, Bmp5, Ptf1a, Ptprk, Gpr158, Lama2, and Pax5 encode proteins that promote neurite outgrowth and neurogenesis and regulate neural progenitor cells (Drosopoulos et al., 1999; Koike et al., 2007; Das et al., 2016; Vernon and Swanson, 2017; Wu et al., 2017; Jovanovic et al., 2018; Ahmed et al., 2019; Çetereisi et al., 2019; Jin and Xiang, 2019; Mohamedi et al., 2021; Vitic et al., 2021). Clstn2, Csmd3, Il1rapl1, Syt17, and Mapkapk3 encode modulators of synaptogenesis that support the demands of synaptic communication (Eales et al., 2014; Montani et al., 2019; Ruhl et al., 2019; Ranneva et al., 2020; Song et al., 2022). Considering the importance of synaptic connections in the retinal neural network, we performed GO enrichment analysis with SynGO between the CKO and ONC groups. The enrichment of upregulated genes in the VGATCreZnT3fl/fl group revealed an improvement in synaptic plasticity.
In contrast, RGCs in the VGATCreZnT3fl/fl mice downregulated several genes negatively associated with neuronal survival. Chmp1a is related to the proliferation and differentiation of retinal progenitor cells (Sun et al., 2020); Chaf1a, Pxn, and Rasd2 encode proteins that block neuronal differentiation and axonal regeneration and promote neurodegeneration (Gotenstein et al., 2010; Tao et al., 2021; Ramírez-Jarquín et al., 2022). Slc8b1 is related to impaired synaptic transmission (Stavsky et al., 2021), and Ppargc1b encodes a protein that reduces the mammalian target of rapamycin expression (Liu et al., 2017), which is a powerful signal for enhancing axonal regeneration in RGCs (Lim et al., 2016). Moreover, pyroptosis and apoptosis play important roles in RGC injury (Yao et al., 2022). Gsdmd and Tnfaip8l2 encode key molecules in these pathways. Remarkably, our analysis revealed that modifying genes in ACs causes the reprogramming of RGCs at a significantly early stage. Thus, our results highlight the potential importance of manipulating the targets of presynaptic neurons for effective neuroprotection in RGCs.
We validated the positive effect of the conditional knockout mice: an AC-specific deletion of ZnT3 can morphologically and functionally protect RGCs. The sequencing results provide several insights into the potential mechanisms underlying these phenomena, and we are currently explore the zinc-triggered neurodegeneration. However, we also noted differences in axon regeneration between ZnT3 global knockout (ZnT3–/–) mice and conditional knockout mice (VGATCreZnT3fl/fl). Global knockout appears to be more effective in axon regeneration (Li et al., 2017); the difference in results may be because of the different background of the mice (129S for ZnT3–/– and C57BL/6J for VGATCreZnT3fl/fl) or the effect of postsynaptic Zn2+ on RGCs. Multiple models, including chronic experimental glaucoma models, are required to confirm our findings in future studies. Moreover, further exploration of the specific molecular mechanism for zinc-triggered damage is required.
In conclusion, the current study clarifies the cellular localization pattern of zinc and provides convincing evidence that conditional deletion of ZnT3 in ACs promotes RGC survival and ON regeneration, improves intersynaptic plasticity, and results in vision recovery in response to ONC. RGC-specific sequencing revealed potential factors that may be involved in the underlying mechanisms in RGCs. This study presents new insights into ON injury and neuronal damage and enriches the theory of intercellular interaction in retinal network.
Additional files:
Additional Figure 1 (996.5KB, tif) : Expression of Slc30a3 in retinal cells and immunostaining of ZnT3 and Calbindin in retina.
Expression of Slc30a3 in retinal cells and immunostaining of ZnT3 and Calbindin in retina.
(A) tSNE plot showing clusters of retinal cell populations. (B) Feature plot showing global expression of Slc30a3 across normal retinal cells. (C) Dotplot showing the expression of Slc30a3 in different retinal cell types. (D) Immunostaining of ZnT3 (red: Alexa Fluor 555), Calbindin (green: Alexa Fluor 488) and DAPI (blue) in the intact retinas of and retinas after ONC at day 1. There was no overlap between ZnT3 and the maker of horizontal cells (Calbindin) Scale bar: 20 μm. DAPI: 4′,6-Diamidino-2-phenylindole; GCL: ganglion cell layer; IPL: inner plexiform layer; INL: inner nuclear layer; ONC: optic nerve crush; tSNE:t-distributed stochastic neighbor embedding; VGAT: vesicular γ-aminobutyric acid transporter; ZnT3: zinc transporter 3.
Additional Figure 2 (623.2KB, tif) : The combination treatment with Zn2+ chelator does not increase the survival rate of RGCs in VGATCreZnT3fl/fl mice and knockout ZnT3 in RGCs has no effect on RGC survival.
The combination treatment with Zn2+ chelator does not increase the survival rate of RGCs in VGATCreZnT3fl/fl mice and knockout ZnT3 in RGCs has no effect on RGC survival.
(A) Retinal whole mounts immunostained Tuj1 (green: Alexa Fluor 488) to visualize RGCs in the retinas of ZnT3fl/fl (VGAT-Crewt/wt; ZnT3fl/fl) mice, VGATCreZnT3fl/fl mice, and their combination treatment with TPEN 2 weeks post-injury. VGATCreZnT3fl/fl mice increased the RGC survival rate, but combination treatment with TPEN can not improve the survival rate of VGATCreZnT3fl/fl mice further. Scale bar: 50 μm. (B-D) Quantitative RGC counts in whole-mount, peripheral, and central retinal samples (n = 5, 7, 4). *P < 0.05, **P < 0.01, ***P < 0.001, compared with ZnT3fl/fl mice post-injury (one-way analysis of variance followed by Tukey’s multiple comparisons). ns: no significance compared with VGATCreZnT3fl/fl mice 2 post-injury. (E) Retinal whole mounts immunostained Tuj1 (green: Alexa Fluor 488) to visualize RGCs in the retinas of VGLUT2CreZnT3fl/fl mice and littermate control ZnT3fl/fl (VGLUT2-Crewt/wt; ZnT3fl/fl) mice 2 weeks post-injury. VGLUT2CreZnT3fl/fl mice cannot increase the RGC survival rate. Scale bar: 50 μm. (F-H) Quantitative RGC counts in whole-mount, peripheral, and central retinal samples (n = 4, 4). **P < 0.01 (unpaired two-tailed Student’s t-test). ns: no significance compared with ZnT3fl/fl mice 2 weeks post-injury. All data are presented as mean ± SEM. Ctrl: Control; ONC: optic nerve crush; RGC: retinal ganglion cell; TPEN: N,N,N’,N’-tetrakis (2-pyridylmethyl) ethylenediamine; VGAT: vesicular γ-aminobutyric acid transporter; VGLUT2: vesicular-glutamate transporter 2; ZnT3: zinc transporter 3.
Additional Figure 3 (580.7KB, tif) : Identification and sequencing of sorted RGCs.
Identification and sequencing of sorted RGCs
(A) Cells were immunostained for RBPMS (red: Alexa Fluor 555) and DAPI (blue) to identify sorted cells by fluorescence-activated cell sorting. Scale bar: 50 μm. (B) Statistical bar chart of differentially expressed genes between the groups (fold change > 2, P < 0.05). (C) Top 10 KEGG pathway enrichments of CKO-upregulated clusters. (D) Top 10 KEGG pathway enrichments of the CKO-downregulated cluster. Ctrl: Control; CKO: conditional knockout; DAPI: 4′,6-diamidino-2-phenylindole; KEGG: Kyoto Encyclopedia of Genes and Genomes; RBPMS: RNA binding protein, mRNA processing factor; VGAT: vesicular γ-aminobutyric acid transporter.
Additional Figure 4 (545.3KB, tif) : The expression of genes in the whole retina is based on whole retina RNA sequencing.
The expression of genes in the whole retina is based on whole retina RNA sequencing.
(A-E) The expression of genes upregulated after optic nerve injury in RGCs of CKO group. (F) The expression of genes downregulated after optic nerve injury in RGCs of CKO group. *P < 0.05. (G) The expression of genes associated with synaptic connections. All data are presented as mean ± SEM (n = 4) (one-way analysis of variance followed by Tukey’s multiple comparisons (A-F) or unpaired t-test (G)). Ctrl: Control; CKO: conditional knockout; ONC: optic nerve crush; ns: no significance; RGC: retinal ganglion cell.
Additional Table 1: Primer Information of VGATCreZnT3fl/fl and
VGLUT2CreZnT3fl/fl mice.
Additional Table 2: Gene Ontology of the up-regulated cluster related to Figure 7.
Additional Table 3 (212.3KB, pdf) : Gene Ontology of the down-regulated cluster related to Figure 7.
Gene Ontology of the down-regulated cluster related to Figure 7
Acknowledgments:
We thank Prof. Larry I. Benowitz, Prof. Paul A. Rosenberg, and Hui-Ya Gilbert (all from Boston Children’s Hospital, Harvard Medical School) for their advice and VGAT-Cre mice from their laboratory.
Footnotes
Funding: This study was supported by the National Key R&D Project of China, No. 2020YFA0112701 (to YZ); the National Natural Science Foundation of China, Nos. 82171057 (to YZ), 81870657 (to YL); Science and Technology Program of Guangzhou of China, No. 202206080005 (to YZ); and the Natural Science Foundation of Guangdong Province of China, No. 2022A1515012168 (to YL).
Conflicts of interest: The authors declare that they have no competing interests.
Data availability statement: All data relevant to the study are included in the article or uploaded as Additional files.
Editor’s evaluation: The study was well designed, including the use of VGATCreZnT3fl/fl conditional knockout mice to demonstrate the role of Zn2+ in amacrine cells/RGCs after optic nerve crush. The article is well-written and the data support the conclusion.
C-Editor: Zhao M; S-Editors: Yu J, Li CH; L-Editors: Yu J, Song LP; T-Editor: Jia Y
References
- 1.Ahmed M, Marziali LN, Arenas E, Feltri ML, Ffrench-Constant C. Laminin α2 controls mouse and human stem cell behaviour during midbrain dopaminergic neuron development. Development. 2019;146:dev172668. doi: 10.1242/dev.172668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aras MA, Aizenman E. Redox regulation of intracellular zinc:molecular signaling in the life and death of neurons. Antioxid Redox Signal. 2011;15:2249–2263. doi: 10.1089/ars.2010.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Association for Research in Vision and Ophthalmology (2021) ARVO statement for the use of animals in ophthalmic and vision research [Google Scholar]
- 4.Bach M, Cuno AK, Hoffmann MB. Retinal conduction speed analysis reveals different origins of the P50 and N95 components of the (multifocal) pattern electroretinogram. Exp Eye Res. 2018;169:48–53. doi: 10.1016/j.exer.2018.01.021. [DOI] [PubMed] [Google Scholar]
- 5.Baltaci SB, Unal O, Gulbahce-Mutlu E, Gumus H, Pehlivanoglu S, Yardimci A, Mogulkoc R, Baltaci AK. The role of zinc status on spatial memory, hippocampal synaptic plasticity, and insulin signaling in icv-stz-induced sporadic Alzheimer's-like disease in rats. Biol Trace Elem Res. 2022;200:4068–4078. doi: 10.1007/s12011-021-02999-2. [DOI] [PubMed] [Google Scholar]
- 6.Benowitz LI, He Z, Goldberg JL. Reaching the brain:advances in optic nerve regeneration. Exp Neurol. 2017;287:365–373. doi: 10.1016/j.expneurol.2015.12.015. [DOI] [PubMed] [Google Scholar]
- 7.Çetereisi D, Kramvis I, Gebuis T, van der Loo RJ, Gouwenberg Y, Mansvelder HD, Li KW, Smit AB, Spijker S. Gpr158 deficiency impacts hippocampal CA1 neuronal excitability, dendritic architecture, and affects spatial learning. Front Cell Neurosci. 2019;13:465. doi: 10.3389/fncel.2019.00465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chintalapudi SR, Patel NN, Goldsmith ZK, Djenderedjian L, Wang XD, Marion TN, Jablonski MM, Morales-Tirado VM. Isolation of primary murine retinal ganglion cells (RGCs) by flow cytometry. J Vis Exp. 2017:55785. doi: 10.3791/55785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choi BY, Hong DK, Jeong JH, Lee BE, Koh JY, Suh SW. Zinc transporter 3 modulates cell proliferation and neuronal differentiation in the adult hippocampus. Stem Cells. 2020;38:994–1006. doi: 10.1002/stem.3194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cueva JG, Haverkamp S, Reimer RJ, Edwards R, Wässle H, Brecha NC. Vesicular gamma-aminobutyric acid transporter expression in amacrine and horizontal cells. J Comp Neurol. 2002;445:227–237. doi: 10.1002/cne.10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Danscher G, Stoltenberg M. Zinc-specific autometallographic in vivo selenium methods:tracing of zinc-enriched (ZEN) terminals, ZEN pathways, and pools of zinc ions in a multitude of other ZEN cells. J Histochem Cytochem. 2005;53:141–153. doi: 10.1369/jhc.4R6460.2005. [DOI] [PubMed] [Google Scholar]
- 12.Das G, Yu Q, Hui R, Reuhl K, Gale NW, Zhou R. EphA5 and EphA6:regulation of neuronal and spine morphology. Cell Biosci. 2016;6:48. doi: 10.1186/s13578-016-0115-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR:ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drosopoulos NE, Walsh FS, Doherty P. A soluble version of the receptor-like protein tyrosine phosphatase kappa stimulates neurite outgrowth via a Grb2/MEK1-dependent signaling cascade. Mol Cell Neurosci. 1999;13:441–449. doi: 10.1006/mcne.1999.0758. [DOI] [PubMed] [Google Scholar]
- 15.Dvoriantchikova G, Ivanov D, Barakat D, Grinberg A, Wen R, Slepak VZ, Shestopalov VI. Genetic ablation of Pannexin1 protects retinal neurons from ischemic injury. PLoS One. 2012;7:e31991. doi: 10.1371/journal.pone.0031991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eales KL, Palygin O, O'Loughlin T, Rasooli-Nejad S, Gaestel M, Müller J, Collins DR, Pankratov Y, Corrêa SA. The MK2/3 cascade regulates AMPAR trafficking and cognitive flexibility. Nat Commun. 2014;4701;5 doi: 10.1038/ncomms5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ensembl (2018) Gene:Slc30a3 ENSMUSG00000029151. [accessed June 29, 2018]. http://asia.ensembl.org/Mus_musculus/Gene/Summary?g=ENSMUSG00000029151;r=5:31243450-31265581.
- 18.Frederickson CJ, Suh SW, Silva D, Frederickson CJ, Thompson RB. Importance of zinc in the central nervous system:the zinc-containing neuron. J Nutr. 2000;130:1471s–1483s. doi: 10.1093/jn/130.5.1471S. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg JL. Intrinsic neuronal regulation of axon and dendrite growth. Curr Opin Neurobiol. 2004;14:551–557. doi: 10.1016/j.conb.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Goldberg JL, Klassen MP, Hua Y, Barres BA. Amacrine-signaled loss of intrinsic axon growth ability by retinal ganglion cells. Science (New York, NY) 2002;296:1860–1864. doi: 10.1126/science.1068428. [DOI] [PubMed] [Google Scholar]
- 21.Gong J, Jellali A, Mutterer J, Sahel JA, Rendon A, Picaud S. Distribution of vesicular glutamate transporters in rat and human retina. Brain Res. 2006;1082:73–85. doi: 10.1016/j.brainres.2006.01.111. [DOI] [PubMed] [Google Scholar]
- 22.Gotenstein JR, Swale RE, Fukuda T, Wu Z, Giurumescu CA, Goncharov A, Jin Y, Chisholm AD. The C. elegans peroxidasin PXN-2 is essential for embryonic morphogenesis and inhibits adult axon regeneration. Development. 2010;137:3603–3613. doi: 10.1242/dev.049189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hara T, Yoshigai E, Ohashi T, Fukada T. Zinc transporters as potential therapeutic targets:An updated review. J Pharmacol Sci. 2022;148:221–228. doi: 10.1016/j.jphs.2021.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Hood DC, Raza AS, de Moraes CG, Liebmann JM, Ritch R. Glaucomatous damage of the macula. Prog Retin Eye Res. 2013;32:1–21. doi: 10.1016/j.preteyeres.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hooks BM, Chen C. Circuitry underlying experience-dependent plasticity in the mouse visual system. Neuron. 2020;106:21–36. doi: 10.1016/j.neuron.2020.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin K, Xiang M. Transcription factor Ptf1a in development, diseases and reprogramming. Cell Mol Life Sci. 2019;76:921–940. doi: 10.1007/s00018-018-2972-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jovanovic VM, Salti A, Tilleman H, Zega K, Jukic MM, Zou H, Friedel RH, Prakash N, Blaess S, Edenhofer F, Brodski C. BMP/SMAD pathway promotes neurogenesis of midbrain dopaminergic neurons in vivo and in human induced pluripotent and neural stem cells. J Neurosci. 2018;38:1662–1676. doi: 10.1523/JNEUROSCI.1540-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kalappa BI, Anderson CT, Goldberg JM, Lippard SJ, Tzounopoulos T. AMPA receptor inhibition by synaptically released zinc. Proc Natl Acad Sci U S A. 2015;112:15749–15754. doi: 10.1073/pnas.1512296112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaufman ML, Goodson NB, Park KU, Schwanke M, Office E, Schneider SR, Abraham J, Hensley A, Jones KL, Brzezinski JA. Initiation of Otx2 expression in the developing mouse retina requires a unique enhancer and either Ascl1 or Neurog2 activity. Development. 2021;148:dev199399. doi: 10.1242/dev.199399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim T, Soto F, Kerschensteiner D. An excitatory amacrine cell detects object motion and provides feature-selective input to ganglion cells in the mouse retina. Elife. 2015;4:e08025. doi: 10.7554/eLife.08025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koike N, Kassai Y, Kouta Y, Miwa H, Konishi M, Itoh N. Brorin, a novel secreted bone morphogenetic protein antagonist, promotes neurogenesis in mouse neural precursor cells. J Biol Chem. 2007;282:15843–15850. doi: 10.1074/jbc.M701570200. [DOI] [PubMed] [Google Scholar]
- 32.Koopmans F, van Nierop P, Andres-Alonso M, Byrnes A, Cijsouw T, Coba MP, Cornelisse LN, Farrell RJ, Goldschmidt HL, Howrigan DP, Hussain NK, Imig C, de Jong APH, Jung H, Kohansalnodehi M, Kramarz B, Lipstein N, Lovering RC, MacGillavry H, Mariano V, et al. SynGO:an evidence-based, expert-curated knowledge base for the synapse. Neuron. 2019;103:217–234.e4. doi: 10.1016/j.neuron.2019.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kowalchuk AM, Maurer KA, Shoja-Taheri F, Brown NL. Requirements for Neurogenin2 during mouse postnatal retinal neurogenesis. Dev Biol. 2018;442:220–235. doi: 10.1016/j.ydbio.2018.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kumar V, Kumar A, Singh K, Avasthi K, Kim JJ. Neurobiology of zinc and its role in neurogenesis. Eur J Nutr. 2021;60:55–64. doi: 10.1007/s00394-020-02454-3. [DOI] [PubMed] [Google Scholar]
- 35.Lee S, Chen L, Chen M, Ye M, Seal RP, Zhou ZJ. An unconventional glutamatergic circuit in the retina formed by vGluT3 amacrine cells. Neuron. 2014;84:708–715. doi: 10.1016/j.neuron.2014.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lees GJ, Cuajungco MP, Leong W. Effect of metal chelating agents on the direct and seizure-related neuronal death induced by zinc and kainic acid. Brain Res. 1998;799:108–117. doi: 10.1016/s0006-8993(98)00483-1. [DOI] [PubMed] [Google Scholar]
- 37.Leon S, Yin Y, Nguyen J, Irwin N, Benowitz LI. Lens injury stimulates axon regeneration in the mature rat optic nerve. J Neurosci. 2000;20:4615–4626. doi: 10.1523/JNEUROSCI.20-12-04615.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y, Wen Y, Liu X, Li Z, Lin B, Deng C, Yu Z, Zhu Y, Zhao L, Su W, Zhuo Y. Single-cell RNA sequencing reveals a landscape and targeted treatment of ferroptosis in retinal ischemia/reperfusion injury. J Neuroinflammation. 2022;19:261. doi: 10.1186/s12974-022-02621-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Andereggen L, Yuki K, Omura K, Yin Y, Gilbert HY, Erdogan B, Asdourian MS, Shrock C, de Lima S, Apfel UP, Zhuo Y, Hershfinkel M, Lippard SJ, Rosenberg PA, Benowitz L. Mobile zinc increases rapidly in the retina after optic nerve injury and regulates ganglion cell survival and optic nerve regeneration. Proc Natl Acad Sci U S A. 2017;114:E209–218. doi: 10.1073/pnas.1616811114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lim JH, Stafford BK, Nguyen PL, Lien BV, Wang C, Zukor K, He Z, Huberman AD. Neural activity promotes long-distance, target-specific regeneration of adult retinal axons. Nat Neurosci. 2016;19:1073–1084. doi: 10.1038/nn.4340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin J, Xue J, Xu Q, Liu Z, Zhao C, Tang J, Han J, A S, Wang W, Zhuo Y, Li Y. In situ-crosslinked hydrogel-induced experimental glaucoma model with persistent ocular hypertension and neurodegeneration. Biomater Sci. 2022;10:5006–5017. doi: 10.1039/d2bm00552b. [DOI] [PubMed] [Google Scholar]
- 42.Liu YC, Gao XX, Zhang ZG, Lin ZH, Zou QL. PPAR gamma coactivator 1 beta (PGC-1β) reduces mammalian target of rapamycin (mTOR) expression via a SIRT1-dependent mechanism in neurons. Cell Mol Neurobiol. 2017;37:879–887. doi: 10.1007/s10571-016-0425-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu Y, Brommer B, Tian X, Krishnan A, Meer M, Wang C, Vera DL, Zeng Q, Yu D, Bonkowski MS, Yang JH, Zhou S, Hoffmann EM, Karg MM, Schultz MB, Kane AE, Davidsohn N, Korobkina E, Chwalek K, Rajman LA, et al. Reprogramming to recover youthful epigenetic information and restore vision. Nature. 2020;588:124–129. doi: 10.1038/s41586-020-2975-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mack A, Robitzki A. The key role of butyrylcholinesterase during neurogenesis and neural disorders:an antisense-5'butyrylcholinesterase-DNA study. Prog Neurobiol. 2000;60:607–628. doi: 10.1016/s0301-0082(99)00047-7. [DOI] [PubMed] [Google Scholar]
- 45.Márquez García A, Salazar V, Lima Pérez L. Consequences of zinc deficiency on zinc localization, taurine transport, and zinc transporters in rat retina. Microsc Res Tech. 2022;85:3382–3390. doi: 10.1002/jemt.24193. [DOI] [PubMed] [Google Scholar]
- 46.Matteson PG, Desai J, Korstanje R, Lazar G, Borsuk TE, Rollins J, Kadambi S, Joseph J, Rahman T, Wink J, Benayed R, Paigen B, Millonig JH. The orphan G protein-coupled receptor, Gpr161, encodes the vacuolated lens locus and controls neurulation and lens development. Proc Natl Acad Sci U S A. 2008;105:2088–2093. doi: 10.1073/pnas.0705657105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mohamedi Y, Fontanil T, Cal S, Cobo T, Obaya ÁJ. ADAMTS-12:functions and challenges for a complex metalloprotease. Front Mol Biosci. 2021;8:686763. doi: 10.3389/fmolb.2021.686763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montani C, Gritti L, Beretta S, Verpelli C, Sala C. The synaptic and neuronal functions of the X-linked intellectual disability protein interleukin-1 receptor accessory protein like 1 (IL1RAPL1) Dev Neurobiol. 2019;79:85–95. doi: 10.1002/dneu.22657. [DOI] [PubMed] [Google Scholar]
- 49.Niu L, Li L, Yang S, Wang W, Ye C, Li H. Disruption of zinc transporter ZnT3 transcriptional activity and synaptic vesicular zinc in the brain of Huntington's disease transgenic mouse. Cell Biosci. 2020;10:106. doi: 10.1186/s13578-020-00459-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Palmiter RD, Cole TB, Quaife CJ, Findley SD. ZnT-3, a putative transporter of zinc into synaptic vesicles. Proc Natl Acad Sci U S A. 1996;93:14934–14939. doi: 10.1073/pnas.93.25.14934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paul A, Crow M, Raudales R, He M, Gillis J, Huang ZJ. Transcriptional architecture of synaptic communication delineates GABAergic neuron identity. Cell. 2017;171:522–539.e20. doi: 10.1016/j.cell.2017.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quigley HA. Understanding glaucomatous optic neuropathy:the synergy between clinical observation and investigation. Annu Rev Vis Sci. 2016;2:235–254. doi: 10.1146/annurev-vision-111815-114417. [DOI] [PubMed] [Google Scholar]
- 53.Ramírez-Jarquín UN, Sharma M, Shahani N, Li Y, Boregowda S, Subramaniam S. Rhes protein transits from neuron to neuron and facilitates mutant huntingtin spreading in the brain. Sci Adv. 2022;8:eabm3877. doi: 10.1126/sciadv.abm3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ranneva SV, Maksimov VF, Korostyshevskaja IM, Lipina TV. Lack of synaptic protein, calsyntenin-2, impairs morphology of synaptic complexes in mice. Synapse. 2020;74:e22132. doi: 10.1002/syn.22132. [DOI] [PubMed] [Google Scholar]
- 55.Ruhl DA, Bomba-Warczak E, Watson ET, Bradberry MM, Peterson TA, Basu T, Frelka A, Evans CS, Briguglio JS, Basta T, Stowell MHB, Savas JN, Roopra A, Pearce RA, Piper RC, Chapman ER. Synaptotagmin 17 controls neurite outgrowth and synaptic physiology via distinct cellular pathways. Nat Commun. 2019;10:3532. doi: 10.1038/s41467-019-11459-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sahu A, Devi S, Jui J, Goldman D. Notch signaling via Hey1 and Id2b regulates Müller glia's regenerative response to retinal injury. Glia. 2021;69:2882–2898. doi: 10.1002/glia.24075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ:25 years of image analysis. Nat Methods. 2012;9:671–675. doi: 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sergeeva EG, Rosenberg PA, Benowitz LI. Non-cell-autonomous regulation of optic nerve regeneration by amacrine cells. Front Cell Neurosci. 2021;15:666798. doi: 10.3389/fncel.2021.666798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharma TP, Liu Y, Wordinger RJ, Pang IH, Clark AF. Neuritin 1 promotes retinal ganglion cell survival and axonal regeneration following optic nerve crush. Cell Death Dis. 2015;6:e1661. doi: 10.1038/cddis.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song W, Li Q, Wang T, Li Y, Fan T, Zhang J, Wang Q, Pan J, Dong Q, Sun ZS, Wang Y. Putative complement control protein CSMD3 dysfunction impairs synaptogenesis and induces neurodevelopmental disorders. Brain Behav Immun. 2022;102:237–250. doi: 10.1016/j.bbi.2022.02.027. [DOI] [PubMed] [Google Scholar]
- 61.Stavsky A, Stoler O, Kostic M, Katoshevsky T, Assali EA, Savic I, Amitai Y, Prokisch H, Leiz S, Daumer-Haas C, Fleidervish I, Perocchi F, Gitler D, Sekler I. Aberrant activity of mitochondrial NCLX is linked to impaired synaptic transmission and is associated with mental retardation. Commun Biol. 2021;4:666. doi: 10.1038/s42003-021-02114-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun N, Zhang D, Ni N, Tang Z, Gao H, Ju Y, Dai X, Wang J, Gu P, Ji J. miR-17 regulates the proliferation and differentiation of retinal progenitor cells by targeting CHMP1A. Biochem Biophys Res Commun. 2020;523:493–499. doi: 10.1016/j.bbrc.2019.11.108. [DOI] [PubMed] [Google Scholar]
- 63.Tao L, Moreno-Smith M, Ibarra-García-Padilla R, Milazzo G, Drolet NA, Hernandez BE, Oh YS, Patel I, Kim JJ, Zorman B, Patel T, Kamal AHM, Zhao Y, Hicks J, Vasudevan SA, Putluri N, Coarfa C, Sumazin P, Perini G, Parchem RJ, et al. CHAF1A blocks neuronal differentiation and promotes neuroblastoma oncogenesis via metabolic reprogramming. Adv Sci (Weinh) 2021;8:e2005047. doi: 10.1002/advs.202005047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thelen K, Maier B, Faber M, Albrecht C, Fischer P, Pollerberg GE. Translation of the cell adhesion molecule ALCAM in axonal growth cones - regulation and functional importance. J Cell Sci. 2012;125:1003–1014. doi: 10.1242/jcs.096149. [DOI] [PubMed] [Google Scholar]
- 65.Thomson BR, Grannonico M, Liu F, Liu M, Mendapara P, Xu Y, Liu X, Quaggin SE. Angiopoietin-1 knockout mice as a genetic model of open-angle glaucoma. Transl Vis Sci Technol. 2020;9:16. doi: 10.1167/tvst.9.4.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tian F, Cheng Y, Zhou S, Wang Q, Monavarfeshani A, Gao K, Jiang W, Kawaguchi R, Wang Q, Tang M, Donahue R, Meng H, Zhang Y, Jacobi A, Yan W, Yin J, Cai X, Yang Z, Hegarty S, Stanicka J, et al. Core transcription programs controlling injury-induced neurodegeneration of retinal ganglion cells. Neuron. 2022;110:2607–2624.e8. doi: 10.1016/j.neuron.2022.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tien NW, Kim T, Kerschensteiner D. Target-specific glycinergic transmission from VGluT3-expressing amacrine cells shapes suppressive contrast responses in the retina. Cell Rep. 2016;15:1369–1375. doi: 10.1016/j.celrep.2016.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tran NM, Shekhar K, Whitney IE, Jacobi A, Benhar I, Hong G, Yan W, Adiconis X, Arnold ME, Lee JM, Levin JZ, Lin D, Wang C, Lieber CM, Regev A, He Z, Sanes JR. Single-cell profiles of retinal ganglion cells differing in resilience to injury reveal neuroprotective genes. Neuron. 2019;104:1039–1055.e12. doi: 10.1016/j.neuron.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Upmanyu N, Jin J, Emde HV, Ganzella M, Bösche L, Malviya VN, Zhuleku E, Politi AZ, Ninov M, Silbern I, Leutenegger M, Urlaub H, Riedel D, Preobraschenski J, Milosevic I, Hell SW, Jahn R, Sambandan S. Colocalization of different neurotransmitter transporters on synaptic vesicles is sparse except for VGLUT1 and ZnT3. Neuron. 2022;110:1483–1497.e7. doi: 10.1016/j.neuron.2022.02.008. [DOI] [PubMed] [Google Scholar]
- 70.Vernon CG, Swanson GT. Neto2 assembles with kainate receptors in DRG neurons during development and modulates neurite outgrowth in adult sensory neurons. J Neurosci. 2017;37:3352–3363. doi: 10.1523/JNEUROSCI.2978-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vígh J, Bánvölgyi T, Wilhelm M. Amacrine cells of the anuran retina:morphology, chemical neuroanatomy, and physiology. Microsc Res Tech. 2000;50:373–383. doi: 10.1002/1097-0029(20000901)50:5<373::AID-JEMT6>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 72.Vitic Z, Safory H, Jovanovic VM, Sarusi Y, Stavsky A, Kahn J, Kuzmina A, Toker L, Gitler D, Taube R, Friedel RH, Engelender S, Brodski C. BMP5/7 protect dopaminergic neurons in an α-synuclein mouse model of Parkinson's disease. Brain. 2021;144:e15. doi: 10.1093/brain/awaa368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang L, Klingeborn M, Travis AM, Hao Y, Arshavsky VY, Gospe SM., 3rd Progressive optic atrophy in a retinal ganglion cell-specific mouse model of complex I deficiency. Sci Rep. 2020;10:16326. doi: 10.1038/s41598-020-73353-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wässle H, Regus-Leidig H, Haverkamp S. Expression of the vesicular glutamate transporter vGluT2 in a subset of cones of the mouse retina. J Comp Neurol. 2006;496:544–555. doi: 10.1002/cne.20942. [DOI] [PubMed] [Google Scholar]
- 75.Williams PR, Benowitz LI, Goldberg JL, He Z. Axon regeneration in the mammalian optic nerve. Annu Rev Vis Sci. 2020;6:195–213. doi: 10.1146/annurev-vision-022720-094953. [DOI] [PubMed] [Google Scholar]
- 76.Wong L, Weadick CJ, Kuo C, Chang BS, Tropepe V. Duplicate dmbx1 genes regulate progenitor cell cycle and differentiation during zebrafish midbrain and retinal development. BMC Dev Biol. 2010;10:100. doi: 10.1186/1471-213X-10-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu Q, Tang W, Luo Z, Li Y, Shu Y, Yue Z, Xiao B, Feng L. DISC1 regulates the proliferation and migration of mouse neural stem/progenitor cells through Pax5, Sox2, Dll1 and Neurog2. Front Cell Neurosci. 2017;11:261. doi: 10.3389/fncel.2017.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xie L, Cen LP, Li Y, Gilbert HY, Strelko O, Berlinicke C, Stavarache MA, Ma M, Wang Y, Cui Q, Kaplitt MG, Zack DJ, Benowitz LI, Yin Y. Monocyte-derived SDF1 supports optic nerve regeneration and alters retinal ganglion cells'response to Pten deletion. Proc Natl Acad Sci U S A. 2022;119:e2113751119. doi: 10.1073/pnas.2113751119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yao Y, Xu Y, Liang JJ, Zhuang X, Ng TK. Longitudinal and simultaneous profiling of 11 modes of cell death in mouse retina post-optic nerve injury. Exp Eye Res. 2022;222:109159. doi: 10.1016/j.exer.2022.109159. [DOI] [PubMed] [Google Scholar]
- 80.Yu G, Wang LG, Han Y, He QY. clusterProfiler:an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhang ML, Zhao GL, Hou Y, Zhong SM, Xu LJ, Li F, Niu WR, Yuan F, Yang XL, Wang Z, Miao Y. Rac1 conditional deletion attenuates retinal ganglion cell apoptosis by accelerating autophagic flux in a mouse model of chronic ocular hypertension. Cell Death Dis. 2020;11:734. doi: 10.1038/s41419-020-02951-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang Q, Li Y, Zhuo Y. Synaptic or non-synaptic?Different intercellular interactions with retinal ganglion cells in optic nerve regeneration. Mol Neurobiol. 2022;59:3052–3072. doi: 10.1007/s12035-022-02781-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zhang X, Mandric I, Nguyen KH, Nguyen TTT, Pellegrini M, Grove JCR, Barnes S, Yang XJ. Single cell transcriptomic analyses reveal the impact of bHLH factors on human retinal organoid development. Front Cell Dev Biol. 2021;9:653305. doi: 10.3389/fcell.2021.653305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang Y, Williams PR, Jacobi A, Wang C, Goel A, Hirano AA, Brecha NC, Kerschensteiner D, He Z. Elevating growth factor responsiveness and axon regeneration by modulating presynaptic inputs. Neuron. 2019;103:39–51.e5. doi: 10.1016/j.neuron.2019.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Expression of Slc30a3 in retinal cells and immunostaining of ZnT3 and Calbindin in retina.
(A) tSNE plot showing clusters of retinal cell populations. (B) Feature plot showing global expression of Slc30a3 across normal retinal cells. (C) Dotplot showing the expression of Slc30a3 in different retinal cell types. (D) Immunostaining of ZnT3 (red: Alexa Fluor 555), Calbindin (green: Alexa Fluor 488) and DAPI (blue) in the intact retinas of and retinas after ONC at day 1. There was no overlap between ZnT3 and the maker of horizontal cells (Calbindin) Scale bar: 20 μm. DAPI: 4′,6-Diamidino-2-phenylindole; GCL: ganglion cell layer; IPL: inner plexiform layer; INL: inner nuclear layer; ONC: optic nerve crush; tSNE:t-distributed stochastic neighbor embedding; VGAT: vesicular γ-aminobutyric acid transporter; ZnT3: zinc transporter 3.
The combination treatment with Zn2+ chelator does not increase the survival rate of RGCs in VGATCreZnT3fl/fl mice and knockout ZnT3 in RGCs has no effect on RGC survival.
(A) Retinal whole mounts immunostained Tuj1 (green: Alexa Fluor 488) to visualize RGCs in the retinas of ZnT3fl/fl (VGAT-Crewt/wt; ZnT3fl/fl) mice, VGATCreZnT3fl/fl mice, and their combination treatment with TPEN 2 weeks post-injury. VGATCreZnT3fl/fl mice increased the RGC survival rate, but combination treatment with TPEN can not improve the survival rate of VGATCreZnT3fl/fl mice further. Scale bar: 50 μm. (B-D) Quantitative RGC counts in whole-mount, peripheral, and central retinal samples (n = 5, 7, 4). *P < 0.05, **P < 0.01, ***P < 0.001, compared with ZnT3fl/fl mice post-injury (one-way analysis of variance followed by Tukey’s multiple comparisons). ns: no significance compared with VGATCreZnT3fl/fl mice 2 post-injury. (E) Retinal whole mounts immunostained Tuj1 (green: Alexa Fluor 488) to visualize RGCs in the retinas of VGLUT2CreZnT3fl/fl mice and littermate control ZnT3fl/fl (VGLUT2-Crewt/wt; ZnT3fl/fl) mice 2 weeks post-injury. VGLUT2CreZnT3fl/fl mice cannot increase the RGC survival rate. Scale bar: 50 μm. (F-H) Quantitative RGC counts in whole-mount, peripheral, and central retinal samples (n = 4, 4). **P < 0.01 (unpaired two-tailed Student’s t-test). ns: no significance compared with ZnT3fl/fl mice 2 weeks post-injury. All data are presented as mean ± SEM. Ctrl: Control; ONC: optic nerve crush; RGC: retinal ganglion cell; TPEN: N,N,N’,N’-tetrakis (2-pyridylmethyl) ethylenediamine; VGAT: vesicular γ-aminobutyric acid transporter; VGLUT2: vesicular-glutamate transporter 2; ZnT3: zinc transporter 3.
Identification and sequencing of sorted RGCs
(A) Cells were immunostained for RBPMS (red: Alexa Fluor 555) and DAPI (blue) to identify sorted cells by fluorescence-activated cell sorting. Scale bar: 50 μm. (B) Statistical bar chart of differentially expressed genes between the groups (fold change > 2, P < 0.05). (C) Top 10 KEGG pathway enrichments of CKO-upregulated clusters. (D) Top 10 KEGG pathway enrichments of the CKO-downregulated cluster. Ctrl: Control; CKO: conditional knockout; DAPI: 4′,6-diamidino-2-phenylindole; KEGG: Kyoto Encyclopedia of Genes and Genomes; RBPMS: RNA binding protein, mRNA processing factor; VGAT: vesicular γ-aminobutyric acid transporter.
The expression of genes in the whole retina is based on whole retina RNA sequencing.
(A-E) The expression of genes upregulated after optic nerve injury in RGCs of CKO group. (F) The expression of genes downregulated after optic nerve injury in RGCs of CKO group. *P < 0.05. (G) The expression of genes associated with synaptic connections. All data are presented as mean ± SEM (n = 4) (one-way analysis of variance followed by Tukey’s multiple comparisons (A-F) or unpaired t-test (G)). Ctrl: Control; CKO: conditional knockout; ONC: optic nerve crush; ns: no significance; RGC: retinal ganglion cell.
Gene Ontology of the down-regulated cluster related to Figure 7