Abstract
Objective
Inflammatory bowel disease (IBD) is a multifactorial immune-mediated inflammatory disease of the intestine, comprising Crohn’s disease and ulcerative colitis. By characterising metabolites in faeces, combined with faecal metagenomics, host genetics and clinical characteristics, we aimed to unravel metabolic alterations in IBD.
Design
We measured 1684 different faecal metabolites and 8 short-chain and branched-chain fatty acids in stool samples of 424 patients with IBD and 255 non-IBD controls. Regression analyses were used to compare concentrations of metabolites between cases and controls and determine the relationship between metabolites and each participant’s lifestyle, clinical characteristics and gut microbiota composition. Moreover, genome-wide association analysis was conducted on faecal metabolite levels.
Results
We identified over 300 molecules that were differentially abundant in the faeces of patients with IBD. The ratio between a sphingolipid and L-urobilin could discriminate between IBD and non-IBD samples (AUC=0.85). We found changes in the bile acid pool in patients with dysbiotic microbial communities and a strong association between faecal metabolome and gut microbiota. For example, the abundance of Ruminococcus gnavus was positively associated with tryptamine levels. In addition, we found 158 associations between metabolites and dietary patterns, and polymorphisms near NAT2 strongly associated with coffee metabolism.
Conclusion
In this large-scale analysis, we identified alterations in the metabolome of patients with IBD that are independent of commonly overlooked confounders such as diet and surgical history. Considering the influence of the microbiome on faecal metabolites, our results pave the way for future interventions targeting intestinal inflammation.
Keywords: IBD, CROHN'S DISEASE, STOOL MARKERS, INTESTINAL MICROBIOLOGY
WHAT IS ALREADY KNOWN ON THIS TOPIC
The gut microbiome is increasingly recognised as a metabolic organ that affects host health.
Accumulating evidence suggests that gut microbiota-derived metabolites are critical mediators between microbiota and immune response.
While dysbiosis in gut communities has been extensively explored in inflammatory bowel disease (IBD), unravelling the mechanisms behind host–microbiota interactions remains complex and requires insights into the metabolic activity in the gut.
WHAT THIS STUDY ADDS
Gut metabolism in patients with IBD is characterised by lower levels of molecules derived from saccharolytic fermentation and increased metabolites from proteolytic fermentation.
Gut microbiota composition is the main determinant of faecal metabolite content, as compared with host lifestyle, genetic and clinical phenotypes.
In the gut of patients with IBD, the expansion of pathobionts co-occurs with an increased concentration of sphingolipids, ethanolamine and primary bile acids.
Intestinal resections have a long-lasting effect on intestinal bile acid and lipid metabolism.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
Future studies investigating host metabolism should account for microbial composition and history of intestinal surgeries.
The faecal metabolome offers new avenues for identifying biomarkers for IBD and other immune-mediated inflammatory diseases.
Understanding the gut microbiota’s contribution to human metabolism in health and diseases is essential for designing future dietary interventions.
Introduction
Characterisation of the host–microbiota symbiosis is crucial in the context of intestinal disorders such as inflammatory bowel disease (IBD) in which the gut environment is severely perturbed, yet the disease-causing mechanisms are still largely unknown. IBD is a chronic inflammatory disorder of the gastrointestinal tract that consists of two main subtypes: ulcerative colitis (UC) and Crohn’s disease (CD).1 2 In IBD, periods of active disease are characterised by loss of strictly anaerobic bacteria, blooming of facultative anaerobes and alterations in the chemical environment in the gut.3 For example, reductions of gut barrier-protecting short-chain fatty acids (SCFA) and alterations in bile acids, sphingolipids and tryptophan-derived metabolites have been consistently reported in faeces of patients with IBD.4 5 However, a large number of molecules in the human body remain uncharacterised, and thus their implications for human health remain unknown. Considering that a subset of small molecules, including microbiome-derived metabolites, have been shown to regulate the immune response, it is crucial to characterise these metabolites and understand which factors determine their concentrations in the gut.
Recent technological advances in mass spectrometry techniques have enabled high-throughput characterisation and quantification of a wide range of known and chemically unannotated molecules.6 In this context, the characterisation of faecal metabolites holds great potential for discovering non-invasive biomarkers and therapeutic targets. To date, however, studies performing untargeted metabolomics on the faeces of patients with IBD have been scarce, limited in sample size and lacking in-depth information on host genetics, lifestyle, diet and clinical characteristics.4 7
In this study, we aimed to determine alterations in the gut metabolism of patients with IBD to pinpoint factors influencing faecal metabolite levels. Our findings highlight the potential of faecal metabolites as biomarkers for IBD and show that, despite the influence of lifestyle, genetics and disease, faecal microbes are a strong predictor of the levels and composition of metabolites in the gut.
Methods
Cohort and metadata description
Samples were obtained from two established cohorts: LifeLines,8 a population biobank from the north of the Netherlands, and 1000IBD,9 a cohort of patients with IBD from the University Medical Centre of Groningen. In this study, we included 255 non-IBD controls, 238 patients with CD and 174 patients with UC. Sample collection and storage are described in online supplemental methods and cohort characteristics are summarised in online supplemental table 1 and table 1.10 11
Table 1.
Cohort description
| Phenotypes | Control | IBD | P value |
| 255 | 424 | ||
| Age (mean (SD)) | 46.83 (12.08) | 42.61 (14.69) | <0.001 |
| Body mass index (mean (SD)) | 25.17 (3.84) | 25.44 (5.02) | 0.462 |
| Sex=male (%) | 114 (44.7) | 170 (40.1) | 0.272 |
| Diagnosis (%) | |||
| CD | NA | 238 (56.1) | |
| IBDU | NA | 12 (2.8) | |
| UC | NA | 174 (41.0) | |
| Disease location (%) | |||
| Both | NA | 97 (22.8) | |
| Colon | NA | 228 (53.7) | |
| Ileum | NA | 69 (16.2) | |
| Disease acitvity (%) | |||
| Active | NA | 107 (25.2) | |
| Not active | NA | 311 (73.3) | |
| Estimated bowel movement a day (mean (SD)) | 1.33 (0.66) | 2.73 (2.37) | <0.001 |
| Smoker=yes (%) | 38 (14.9) | 87 (20.5) | 0.084 |
| Faecal calprotectin >200 = yes (%) | 8 (3.2) | 168 (45.0) | <0.001 |
| Montreal B (%) | |||
| B1 | NA | 129 (54.2) | |
| B2 | NA | 76 (31.9) | |
| B3 | NA | 28 (11.7) | |
| Montreal B perianal (%) | |||
| No | NA | 326 (76.8) | |
| Yes | NA | 68 (16.0) | |
| Montreal S (%) | |||
| S0 | NA | 7 (4.0) | |
| S1 | NA | 56 (32.1) | |
| S2 | NA | 69 (39.6) | |
| S3 | NA | 27 (15.5) | |
| Montreal A (%) | |||
| A1 | NA | 54 (12.7) | |
| A2 | NA | 245 (57.7) | |
| A3 | NA | 85 (20.0) | |
| Montreal L (%) | |||
| L1 | NA | 63 (14.8) | |
| L2 | NA | 203 (47.8) | |
| L3 | NA | 94 (22.1) | |
| L4 | NA | 21 (4.9) | |
| Montreal E (%) | |||
| E1 | NA | 21 (12.0) | |
| E2 | NA | 57 (32.7) | |
| E3 | NA | 80 (45.9) | |
| Microbiome Shannon Index (mean (SD)) | 3.05 (0.33) | 2.89 (0.41) | <0.001 |
CD, Crohn’s disease; IBD, inflammatory bowel disease; IBDU, inflammatory bowel disease unclassified; NA, not available; UC, ulcerative colitis.
gutjnl-2022-328048supp001.pdf (126.2KB, pdf)
gutjnl-2022-328048supp002.xlsx (226.3MB, xlsx)
Metabolite quantification
Metabolomics measurements performed by Metabolon (North Carolina, USA) detected 1684 faecal metabolites (online supplemental table 2). The concentrations of eight short-chain and branched-chain fatty acids were measured using liquid chromatography with tandem mass spectrometry (online supplemental table 3).
Metabolic data processing
Metabolomic data were handled as a compositional dataset and transformed using centered log-ratios. Metabolites were split into three categories based on their prevalence. The first group consisted of metabolites present in more than 70% of the samples in both the cases and controls (x=854). Missing values were imputed using k-Nearest Neighbour Imputation.12 We set the number of nearest neighbours to 10 (k=10) for the imputation and Euclidean distance as a metric. The second group of metabolites (prevalence <70% and >20%, x=514) were transformed into binary traits (metabolite presence/absence). Rare metabolites (prevalence <20%, x=316) were excluded from analyses.
Identification of metabolites associated with IBD
We performed linear regression analysis using the lm function in R. The abundance of each metabolite was compared between disease groups (IBD/CD/UC) and controls. Technical factors (storage time, input grams of faeces and sample batch), host characteristics (age, sex, body mass index (BMI) and bowel movements per day), intestinal integrity (any resection: yes/no) and 24 dietary patterns that were significantly different between cases and controls were included as covariates in the regression models (online supplemental table 4). Less prevalent metabolites (prevalence <70% and >20% of samples) were evaluated using logistic regressions. All p values were adjusted for multiple testing using Benjamini-Hochberg. A false discovery rate (FDR) <0.05 was used as the threshold for statistical significance.
Prediction of IBD based on metabolomics profiles
We used CoDaCoRe13 (V.0.0.1) to identify ratios of metabolites and bacterial abundances that could predict IBD and its subphenotypes (see online supplemental methods).
Association between metabolites and phenotypes
An association analysis between phenotypes and metabolites was performed within each cohort (controls, CD and UC). Each phenotype–metabolite combination was tested using linear regression, including age, sex, BMI, bowel movements per day and technical factors as covariates.
The results of the metabolite–phenotype analyses were combined in a meta-analysis using random-effects models implemented in R package meta (V.4.8). Results were considered statistically significant when the meta-FDR<0.05 (online supplemental methods).
Genome-wide association analysis on faecal metabolites
Exome sequencing and genomic array data were available for both cohorts (see online supplemental methods). Linear regression was used for metabolites present in >70% of the samples and logistic regression for those present in between 70% and 20% of the samples. Analyses were performed per cohort, and results were combined in a meta-analysis, as previously described.14 In addition to accounting for the confounders described above (see the Identification of metabolites associated with IBD section), we included population genetic structure as a covariate in the analysis. To determine the statistical significance of our findings, we adopted two thresholds: a genome-wide significance (p<5e–08), and a more conservative cut-off, a study-wide significance (p<2.97e–11). The study-wide significance threshold was determined by dividing the genome-wide threshold by the total number of metabolites (5.0e–08/1684).
Co-occurrence patterns between bacteria and metabolites
The QIIME 15 implementation of mmvec V.1.0.616 was used to estimate the co-occurrence probabilities between highly prevalent metabolites and bacteria. Furthermore, we assessed the associations between individual microbiome features (taxa, gene clusters and metabolic pathways) and metabolites using regression models considering the interaction between bacteria and dysbiosis (online supplemental methods).
Differential abundance analyses of faecal microbiome features
Linear regression analysis was used to identify microbiome features that differed between controls and IBD. Age, sex, BMI, average bowel movements per day, history of intestinal resections (yes/no) and raw sequencing read depth were included as covariates in the regression models (online supplemental methods).
Association between dysbiosis and faecal metabolites
Phenotypic differences between patients with dysbiotic and eubiotic microbiota were established using χ2 or Wilcoxon-rank test for categorical and continuous variables, respectively. Differences in the abundance of faecal metabolites between the two groups of patients were tested using linear regression. Age, sex, BMI, intestinal resection (yes/no), ileocecal valve in situ (yes/no), average bowel movements per day and differences in 12 dietary patterns (online supplemental table 5) were added as covariates in the regression models.
Metabolite-level prediction
To predict the levels of metabolites in faeces, we performed regression models with L1 regularisation (lasso) using the glmnet R package17 (see online supplemental methods).
Results
Patients with IBD have a distinct faecal metabolite profile
Metabolites were assessed in the faecal samples from 238 patients with CD, 174 patients with UC and 255 non-IBD controls (table 1). On average, 1011 metabolites were detected per sample, ranging from 784 to 1241 molecules.
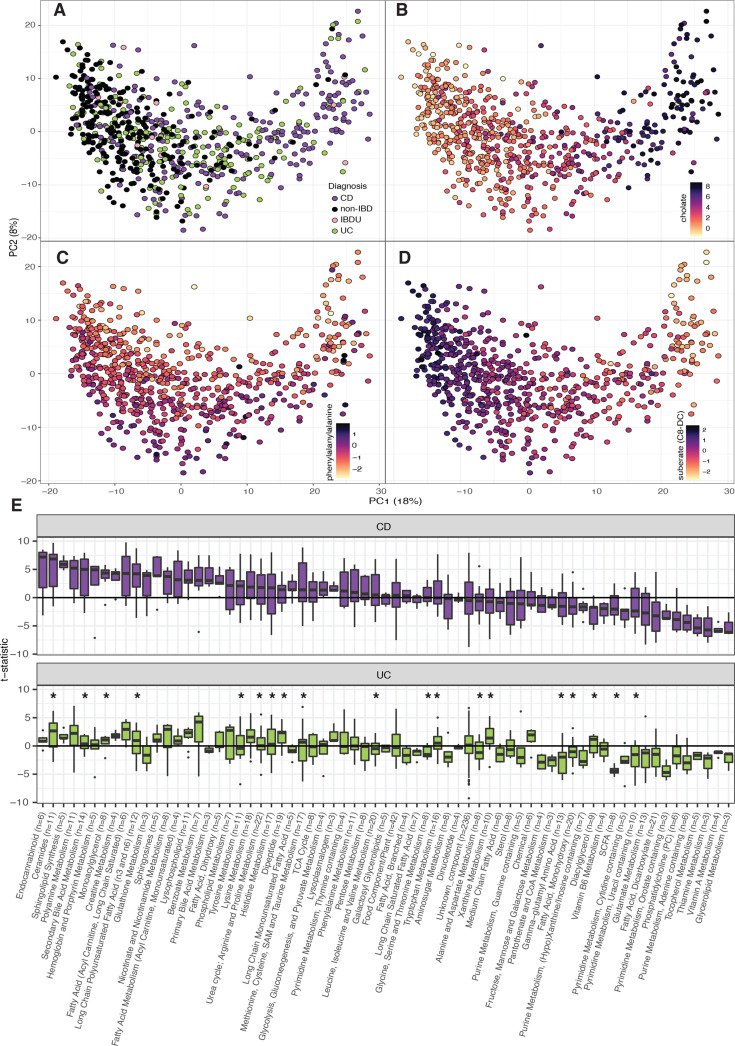
Principal coordinate analysis (PCoA) based on metabolites levels showed that IBD samples are dispersed across a cluster that partially overlaps with controls (figure 1A). The first component of the PCoA captured 18% of the variation and was driven by the levels of carnitine and bile and fatty acids, while the second component, representing 8% of cohort variation, was driven by the abundance of dipeptides and several unclassified metabolites (online supplemental table 6, figure 1B–D).
Figure 1.
Faecal metabolite alterations in patients with Crohn’s disease and ulcerative colitis. (A–D). Principal coordinate analyses depicting the clustering of 255 non-IBD (black), 238 CD (purple), 174 UC (green) and 12 IBDU (pink) samples according to their metabolomic composition. The first principal component is mainly driven by the levels of cholic acid and suberate (B, D) and the second component by the concentrations of phenylalanylalanine (panel C). Light–dark colour gradient represents low–high metabolite values. Metabolite concentrations are expressed as centred log-ratio (clr) of the AUC raw values. (E). Metabolite differences between cases and controls grouped into metabolomic pathways. For clarity, only categories with three or more metabolites are shown (number of metabolites per categories are indicated on the x-axis). The y-axis represents the t-statistic value from the linear regression model (see online supplemental methods). Asterisks indicate significant differences between CD and UC (FDR<0.05, online supplemental tables 7–9). AUC, area under the curve; CD, Crohn’s disease; FDR, false discovery rate; IBD, inflammatory bowel disease; IBDU, inflammatory bowel disease unclassified; UC, ulcerative colitis.
Differential abundance analysis revealed 324 associations when comparing patients with CD to controls and 308 associations when comparing patients with UC to controls (FDR<0.05) (online supplemental table 7, online supplemental figure 1A–E). Moreover, when looking into lower prevalence metabolites, we found that products of the metabolism of bile acids, ceramides and steroids were more prevalent in faeces of patients with IBD than in controls (182 and 119 molecules associated with CD and UC, respectively, online supplemental table 7).
gutjnl-2022-328048supp004.pdf (1.1MB, pdf)
A prominent signal in both disease groups was the depletion of vitamins and fatty acid-related molecules compared with controls (figure 1E). Patients with IBD presented higher levels of the phenolic compound p-cresol sulphate. The level of indole-propionic acid was decreased in UC, while tryptamine and kynurenine were increased in both CD and UC (FDR<0.05, online supplemental figure 1D). Patients with IBD also showed higher levels of arachidonic acid (20:4n6) and a lower ratio of omega-6/omega-3 fatty acids (online supplemental table 8, online supplemental figure 1E).
We also found that 106 metabolites were differentially abundant between CD and UC. For example, patients with UC presented higher levels of diaminopimelate (DAPA), an alpha-amino acid present in the cell membrane of gram-negative bacteria. Interestingly, DAPA-containing peptidoglycans can trigger the immune response mediated by NOD1 18 (online supplemental figure 1C, online supplemental table 9).
Patients with UC show the lowest concentrations of SCFAs in faeces
The concentrations of SCFAs are essential for immune modulation, and their synthesis is dependent on colonic bacterial fermentation of polysaccharides.19 After correcting for potential confounding effects, including anthropometric measurements, batch and sample storage time (see online supplemental methods, online supplemental table 4), acetate, propionate and butyrate were found in lower concentrations in patients with UC when compared to controls (FDRUC<0.05). No significant differences in these metabolites were observed between CD and controls. In contrast, levels of hexanoic and valeric acids were significantly lower in both groups of patients (online supplemental figure 2, online supplemental table 7).
gutjnl-2022-328048supp005.pdf (1.2MB, pdf)
Faecal metabolomic profiles correctly classify IBD samples
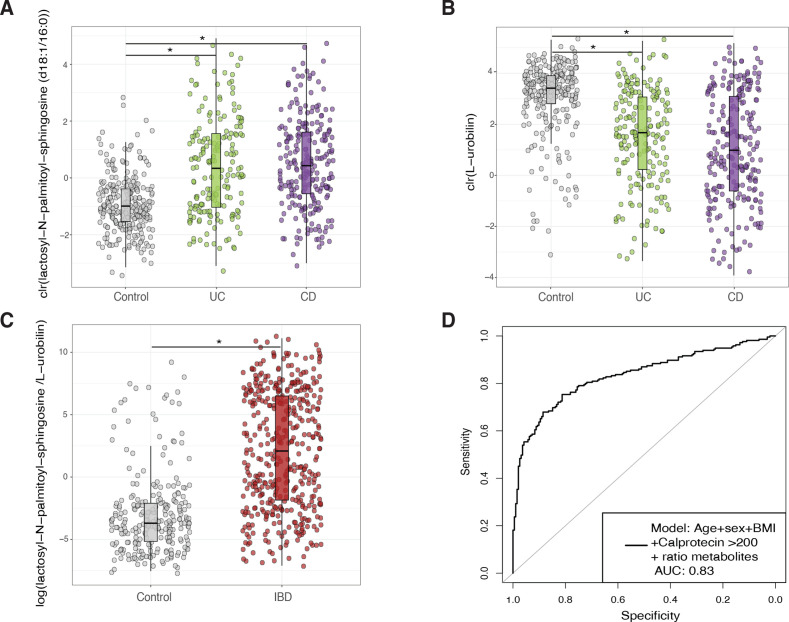
Given the substantial variations observed in the metabolite levels between patients with IBD and non-IBD controls, we investigated the possibility of enhancing the accuracy of the faecal calprotectin test by combining multiple metabolites. To identify potential biomarkers, we employed a machine learning approach to predict disease phenotypes (see online supplemental methods). Including the ratio between the sphingolipid lactosyl-N-palmitoyl-sphingosine (d18:1/16:0) and L-urobilin improved the accuracy of age, sex, BMI and faecal calprotectin levels as disease predictors (AUCcv=0.85, AUCtest=0.83, p=9.89e–13, figure 2, online supplemental table 10). In addition, the ratio between these two metabolites was higher in patients with a long-term remission compared with controls (no flare-ups registered 1 year before and after sample collection, n=61, Wilcoxon test, p=0.0036) although significantly lower when compared with samples from other individuals with IBD in our cohort (Wilcoxon test, p=5.05e–5, online supplemental figure 3A). A similar performance was achieved with bacteria abundances (AUCcv=0.86, AUCtest=0.84, p=6.04e–14, online supplemental figure 3B, C). Combining metabolite and microbiome ratios led to a modest but significant increase in model performance (AUCtest=0.85, p=4.34 e–09). Within patients with IBD, metabolites showed a limited power to correctly classify CD or UC samples (AUCcv=0.78, AUCtest=0.67) and active disease versus remission (AUCcv=0.72, AUCtest=0.60) (online supplemental table 10).
Figure 2.
Biomarker discovery for the diagnosis of IBD. (A, B) Show the abundance of the metabolites with the highest potential to discriminate between samples from non-IBD (grey) and IBD (UC in green and CD in purple). (C). Boxplots depict the value of a potential biomarker for IBD. The y-axis is the log-transformed value of the ratio constructed from the levels of lactosyl-N-palmitoyl-sphingosine (d18:1/16:0) and L-urobilin. Boxplot in grey depicts values in non-IBD controls. Boxplot in red depicts values in patients with IBD. (D). Receiver operating characteristic curve (ROC curve) of the prediction model based on patient characteristics (age, sex and BMI), the levels of faecal calprotectin (expressed as a binary trait (yes/no) if levels of this marker were >200 µg/g of faeces) and the ratio between metabolites. The prediction value, expressed as the area under the curve (AUC), reached a value of 0.83 in the test dataset. Metabolite values are clr-transformed. Boxplot shows the median and interquartile range (25th and 75th). Whiskers show the 1.5*IQR range. Asterisks indicate significant differences between groups (FDR<0.05). BMI, body mass index; CD, Crohn’s disease; FDR, false discovery rate; UC, ulcerative colitis.
gutjnl-2022-328048supp006.pdf (543.7KB, pdf)
Intestinal resections are associated with long-term metabolic alterations
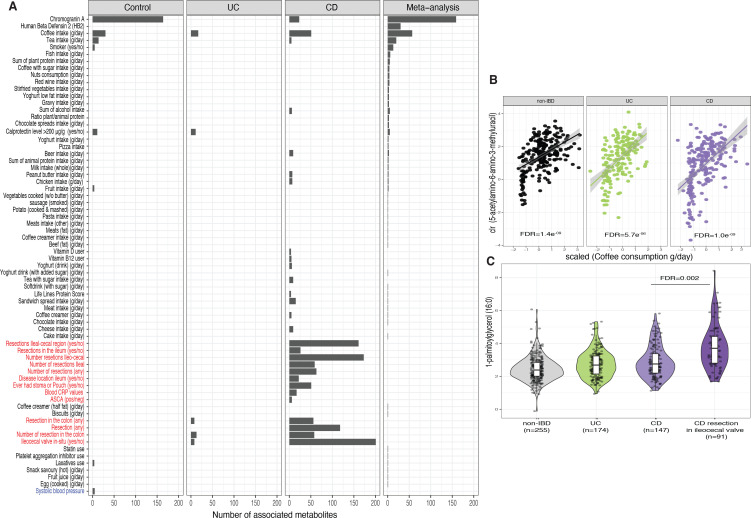
After identifying alterations in the faecal metabolome in individuals with IBD, we aimed to describe which lifestyle, dietary and clinical factors contributed to the levels of faecal metabolites. We assessed the association between faecal metabolites and 229 host characteristics, including dietary habits, medication use and clinical features. We carried out association analysis per condition (i.e., CD only, UC only and controls only) and combined the results of overlapping metadata in a meta-analysis.
In patients with CD, resection of the ileocecal valve was associated with changes in the abundance of 212 metabolites, including cholic acid and several monoacylglycerols. Colonic resection was associated with modifications in the levels 56 molecules in CD and 8 molecules in UC (online supplemental table 11, figure 3 A-C). For example, colonic resection negatively correlated with the faecal levels of pyridoxamine (vitamin B6).
Figure 3.
Potential determinants of faecal metabolite levels. (A) Bar plot showing the number of significant associations between phenotypes and metabolites in each of the cohorts and in the meta-analysis (online supplemental table 11). Only phenotypes with more than three associations are shown. Red labels indicate phenotypes exclusively available for cases and blue labels for controls. (B). Correlation plot showing the relation between AAMU (expressed as clr-transformed AUC values) and coffee consumption (x-axis) per cohort. Coffee consumption is represented as the estimated consumption per day (grams/day) adjusted by overall individual calorie intake (see online supplemental methods). (C). Boxplots showing the levels of 1-palmitoylglycerol (16:0). Boxplot shows the median and IQR (25th and 75th). Whiskers show the 1.5*IQR range. Data distribution is represented by background violin-plot. Lines in the correlation plot show linear regression and shadows indicate the 95% CI. AAMU, 5-acetylamino-6-amino-3-methyluracil; AUC, area under the curve.
There were no significant differences in metabolites between different groups of disease behaviour or disease severity after statistically adjusting for gut surgery (resected vs non-resected). However, we did observe several interesting trends (p<0.05, FDR>0.05, online supplemental table 12). Patients with CD and penetrating diseases had lower butyrate levels (B1 vs B3). Disease severity (Montreal S score) positively correlated with tyramine faecal abundance. In patients with UC, participants with proctitis (E1 classification) had lower levels of 2R-3R-hydroxybutyrate and higher levels of cytidine compared with patients with extensive inflammation in the colon (E3 classification) (online supplemental figure 4A, B).
gutjnl-2022-328048supp007.pdf (392.8KB, pdf)
The levels of chromogranin A showed the largest number of associations with faecal metabolites in non-IBD controls, including positive associations with N-formylmethionine, cholesterol and secondary bile acids. Chromogranin A has been reported as a potential biomarker of gut health, showing a strong correlation with the microbiota composition in the gut.10 Furthermore, participants with calprotectin >200 µg/g showed lower levels of cytidine in faeces and increased sphingosines and ceramides in UC but not CD (figure 3A, online supplemental table 11).
Furthermore, the detection of several metabolites reflected aspects of the lifestyle of participants in our cohort. We found 158 associations between metabolites and dietary patterns (FDRmeta<0.05, online supplemental table 11), however, approximately one-third of these were related to the consumption of coffee (n=57), including positive correlations between coffee intake and the levels of picolinate and 5-acetylamino-6-amino-3-methyluracil (AAMU), a major caffeine metabolite (figure 3B) (FDRMeta<0.05, online supplemental table 11). Cotinine, an alkaloid found in tobacco plant, was found in faeces of self-reported smokers (FDRmeta=1.31e–06, online supplemental table 13) and O-desmethyltramadol, the primary metabolite of the opioid tramadol, was detected in several patients with CD using opioids (FDR=0.009, online supplemental table 13).
To investigate if the observed associations between lifestyle, clinical factors and faecal metabolites were driven by alterations in the gut microbiota we conducted a mediation analysis. We observed evidence for 119, 38 and 695 mediated effects in controls, UC and CD, respectively. Specifically, we found that Lawsonibacter asaccharolyticus mediated the relationship between coffee intake and several caffeine derivatives, such as 1,3-dimethylruic acid. In patients with CD, we observed that the resection of the ileocecal valve resulted in a decline in the abundance of Faecalibacterium prausnitzii, which negatively impacted the levels of anti-inflammatory metabolites, including butyrate (online supplemental table 14, online supplemental figure 5).
gutjnl-2022-328048supp008.pdf (1.2MB, pdf)
NAT2 genotype strongly associated with coffee metabolism
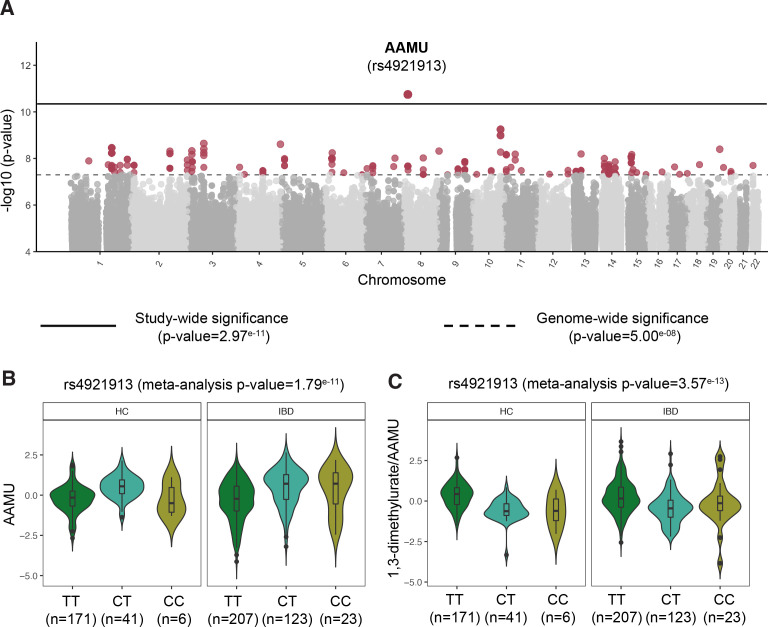
Next, we carried out a faecal metabolome genome-wide association analysis to examine the correlation between host genetics and levels of faecal metabolites. Overall, genetics showed a relatively small impact on the faecal metabolite levels compared to the impact of genetics on blood metabolite levels reported in other studies.20–22 At a study-wide significance level, we found an association between a genetic polymorphism located closely to NAT2 (rs4921913) and AAMU (p meta=1.79e–11). This genetic variant is in linkage disequilibrium with a SNP reported to be associated with the ratio between 1,3-dimethylurate and AAMU (rs35246381, r2>0.8).23 As expected, we could also replicate this finding in our cohort (p IBD=8.46e–09, p controls=4.17e–09, p meta=3.57e–13, figure 4). AAMU is a metabolite derived from coffee, and its levels in faeces correlate with coffee consumption. Nonetheless, this gene–metabolite association remained significant even after adjusting for coffee intake (p IBD=2.2e–16, p controls=2.0e–09, online supplemental table 15).
Figure 4.
Genome-wide association between genetic polymorphisms and faecal metabolites. (A) Manhattan plot shows the strong association between a single nucleotide polymorphism located near the NAT2 gene and AAMU, a metabolite derived from caffeine. Solid horizontal line signifies the significance threshold corrected by multiple hypothesis testing. Dashed line indicates the classic genome-wide significance threshold. Metabolites passing this threshold (in red) are considered suggestive associations (online supplemental table 15). (B) Boxplot depicting the levels of AAMU in non-IBD controls and IBD, stratified by SNP rs4921913 genotype. (C) Boxplot showing the relation between SNP rs4921913 and the ratio of 1,3-dimethylurate to AAMU. This association was previously described in the TwinsUK cohort.23 Metabolite values are presented as the residuals of the model regressing the covariates age, sex, BMI and technical confounders. Boxplot shows the median and IQR (25th and 75th). Whiskers show the 1.5*IQR range. Data distribution is represented by background violin-plot. AAMU, 5-acetylamino-6-amino-3-methyluracil; BMI, body mass index; IBD, inflammatory bowel disease.
Gut microbiota composition is linked to metabolomic profiles
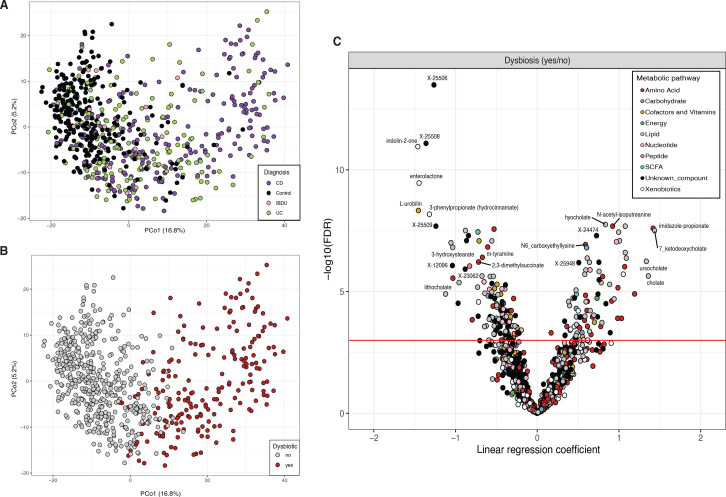
The gut microbiota of individuals with IBD often undergoes transitions from a healthy state (eubiosis) to an unhealthy state (dysbiosis).3 24 Understanding the metabolic changes that accompany dysbiosis may provide crucial insights into the pathomechanisms of IBD.
In our cohort, participants with dysbiosis were more likely to have CD (n=130, χ2 test FDR=2.45e–04) and ileocecal valve resections (n=76, χ2 test, FDR=2.93e–10), but no significant differences were found in faecal calprotectin levels (proportion of individuals with calprotectin >200 µg/g, χ2 test FDR=0.65). A significant increase in the abundance of pathobionts such as Clostridium boltae, Erysipelatoclostridium ramosum and Ruminococcus gnavus was observed, as well as a decreased abundance of 52 bacterial species in dysbiotic communities (online supplemental table 5, figure 5A, B).
Figure 5.
Metabolic signature of patients with intestinal dysbiosis. (A) Principal coordinate analysis on microbiome composition per sample (dots). Colours indicate disease phenotypes: CD (purple), UC (green), IBD-undetermined (pink), non-IBD (black). (B) Red dots depict samples considered to be dysbiotic based on the median distance to non-IBD samples. (C) Volcano plot showing the p value (y-axis) and regression coefficients (x-axis, positive values indicate enrichment in dysbiosis) of the association analyses between dysbiotic and non-dysbiotic IBD samples (online supplemental table 5). Dot colour indicates pathway annotations provided by Metabolon (online supplemental table 2). CD, Crohn’s disease; IBD, inflammatory bowel disease; UC, ulcerative colitis.
Comparing the metabolite composition of IBD samples from patients with dysbiosis to those with eubiotic microbial communities revealed the enrichment of 202 metabolites and the depletion of 258 metabolites. In dysbiotic samples, we observed reduced levels of indolin-2-one and 3-phenylpropionate and increased levels of imidazole propionate, long-chain polyunsaturated fatty acids (PUFAs) and primary bile acids (FDR<0.05, online supplemental table 5, figure 5C). Alterations in the bile acids pools were also reflected in a higher prevalence of taurine-conjugated and sulphated bile acids in dysbiotic samples (FDR<0.05, online supplemental table 5).
Next, we investigated the correlation between gut microbiota and metabolites while correcting for disease phenotypes (non-IBD, CD or UC) and dysbiotic status (yes/no) (see online supplemental methods). We found a total of 13 761 significant associations between bacteria presence/absence and metabolites levels, and 5942 significant associations between bacterial abundances and metabolites (online supplemental tables 16 and 17, figure 6A, online supplemental figure 6, FDR<0.05).
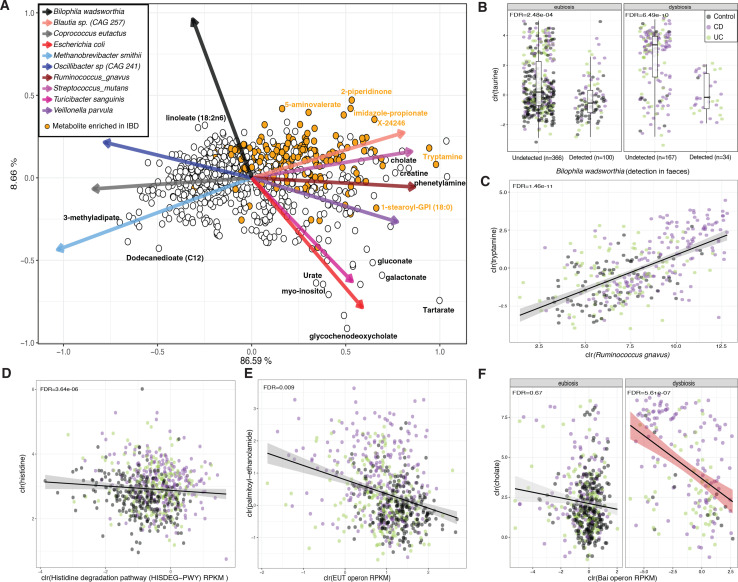
Figure 6.
Metabolite co-occurrence with faecal microbes. (A) Biplot representing conditional probabilities of co-occurrence between metabolites (dots) and microbes (arrows). Distances between dots and arrow tips represent the probability of co-occurrence of each metabolite and microbe (online supplemental table 21). Orange dots highlight metabolites enriched in samples from patients with IBD in the linear regression analysis (online supplemental table 7). Arrow direction indicates the probability of microbes co-occurring with the levels of metabolites To enhance interpretability, names of only a few metabolites are shown and only the top-10 species explaining the largest amount of variation are visualised. (B) Taurine levels stratified by the presence or absence of Bilophila wadsworthia in faecal metagenomes. (C) Correlation between levels of tryptamine and abundance of Ruminococcus gnavus. Only samples in which the bacterium had a non-zero relative abundance are shown (n=339). (D–F) The relation between histidine and MetaCyc Histidine degradation pathway (D), between oleoyl-ethanolamide and the eut operon (E) and between cholic acid and the bai operon (F) are shown as examples of the correlation between microbiota metabolic potential and metabolite levels. Metabolite, bacteria and pathway values are clr-transformed. Boxplot shows the median and IQR (25th and 75th). Whiskers show the 1.5*IQR range.Correlation plot lines show linear regression. Shadows indicate the 95% CI. IBD, inflammatory bowel disease.
gutjnl-2022-328048supp009.pdf (536.5KB, pdf)
Of these associations, 1137 showed a significant interaction effect with dysbiosis status, with only 56 associations exhibiting different directionality between dysbiotic and eubiotic samples. For instance, the detection of Ruthenibacterium lactatiformans in eubiotic samples showed a negative association with butyrate levels, while in dysbiotic samples, this correlation was positive. Regardless of dysbiosis, the presence of Akkermansia municiphila and Oscillibacter spp (CAG 241) in faeces were associated with higher levels of dicarboxylic acids, sebacate (C10-DC) and dodecanedioate (C12)), and the presence of Bilophila wadsworthia was associated with lower levels of taurine and N, N, N-trimethyl-alanyl-proline betaine (figure 6B, FDR<0.05). Furthermore, our results revealed strong positive correlations between F. prausnitzii and hypoxanthine abundances, (FDR=1.46e–11), R. gnavus and tryptamine (FDR=1.46e–11) (figure 6C), as well as imidazole propionate and Streptococcus parasanguinis (FDR=0.007).
Additionally, the abundance of specific microbial metabolic pathways and gene clusters were found to be linked with the metabolic profiles (figure 6D–F). Positive correlations were observed between the abundance of bile acid-inducible operons (bai operon) and levels of lithocholic acid (FDR=9.76e–19), as well as negative correlations with cholic acid (FDR=8.90e–06, online supplemental table 18, figure 6F). However, these effects were more pronounced in dysbiotic samples (FDRinteraction dysbiosis<0.05). On the other hand, reductions in the levels of palmitoyl-ethanolamide and oleoyl-ethanolamide were associated with an increase in the abundance of ethanolamine utilisation operons (eut operon, FDR<0.05, figure 6E). The eut operon is known to be carried by several gut pathobionts, allowing the use of ethanolamine as a source of carbon and nitrogen.25 Moreover, a higher abundance of genes involved in the L-histidine degradation pathway I (MetaCyc ID: HISDEG-PWY) was associated with lower levels of histidine, a metabolite found to be increased in samples from patients with IBD (FDR=3.64e–06, online supplemental table S19, figure 6D).
Microbiome composition predicts metabolite levels in faeces
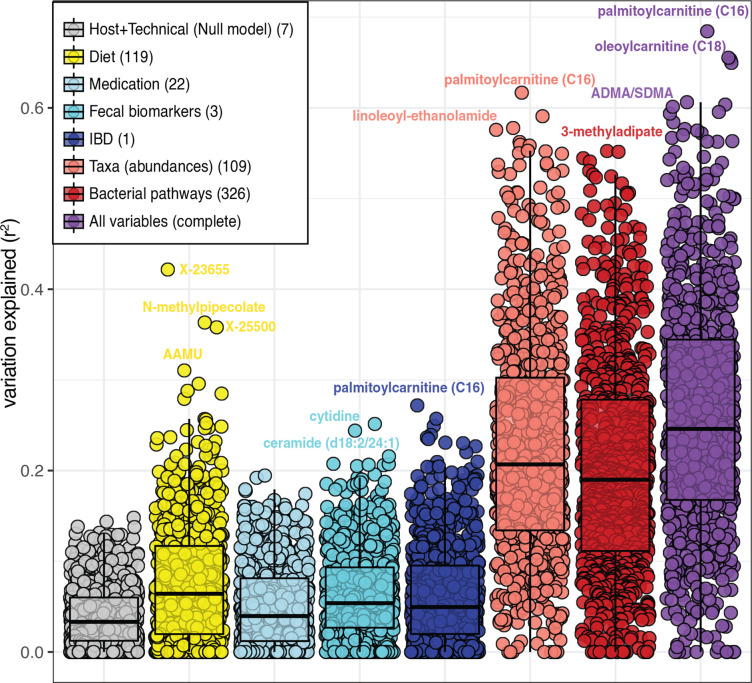
Finally, the predictability of each metabolite was estimated using a combination of host information, dietary habits and the faecal microbiome. Dietary intake predicted the levels of 37 metabolites (>20% of explained variation), with the top 10 dietary-predicted metabolites being 7 unclassified molecules and 3 coffee-related metabolites. Meanwhile, bacterial abundances were a strong predictor of 82 metabolites (>40% of the variation), including the levels of molecules such as lithocholate (41%, s.d. 18%) and dimethylarginine (ADMA/SDMA, 53%, s.d. 4%). Adding diet and participants’ characteristics slightly improved microbiome-based models (paired Wilcoxon-test, p <2.2×10–6) (figure 7, online supplemental table 20).
Figure 7.
Metabolite prediction. Microbial abundances (light red) and bacterial pathways (dark red) show the largest potential to predict the levels of metabolites. Boxplots show the ability to predict metabolites levels of eight different models using seven types of data. Dots represent metabolites, and values in the y-axis represent the percentage of variation explained from cross-validated penalised regression methods using different sets of predictors (see online supplemental methods). The number of features in each model are indicated in parentheses in the legend (online supplemental table 20).
Discussion
We comprehensively characterised faecal metabolites in samples from patients with IBD and representatives of the Dutch population. Our results revealed alterations in the levels of more than 300 highly prevalent faecal metabolites in patients with IBD. Additionally, we described potential determinants of faecal metabolome composition by integrating untargeted metabolomics with extensive information on dietary habits, host genetics, clinical characteristics and gut microbiota composition.
The drastic alteration in faecal metabolite composition in patients with IBD suggests a shift from saccharolytic to proteolytic fermentation metabolism,26 as evidenced by increased levels of metabolites derived from the metabolism of aromatic amino acids, such as p-cresol sulphate (FDRIBD=8.29e–06) and 3-indoxyl sulphate27–29 (FDRIBD=0.04). (online supplemental table 7). The accumulation of these compounds has been linked to various health conditions, such as chronic kidney disease30 and colorectal cancer31 32; suggesting that higher presence of these molecules and lower levels of saccharolytic products, like SCFAs, may indicate an unhealthy gut milieu.
The overlap in the faecal metabolite signatures between patients with CD and UC suggests a common underlying alteration in gut metabolism. In total, 58% of the metabolites significantly associated with UC were also found to be associated with CD. When comparing the faecal metabolite profiles of patients with CD and UC, we observed significant differences in the levels of 106 metabolites. For instance, alterations in the bile acid pool were a distinctive feature of CD, while a reduction in the concentrations of SCFAs was a common characteristic of UC.
In patients with CD, we observed a marked increase in the faecal levels of sphingolipids, including several sphingomyelins and ceramides. Sphingolipids are components of the intestinal cell membrane and are produced either by the de novo condensation of serine to palmitoyl-CoA or the uptake of endogenous and dietary sphingolipids. In addition to their structural role, sphingolipids can act as signalling molecules, mediating cell differentiation, apoptosis, and inflammation.33 Previous studies have shown an accumulation of sphingolipids in colitis mouse models and in faeces of patients with IBD34; however, the mechanisms underlying this dysregulation and whether it precedes the development of inflammation are still unclear.
Experimental evidence suggests that an increase in ceramides levels, either due to the activation of ceramide synthetase or the increased breakdown of sphingomyelins into ceramides, can activate the proinflammatory transcription factor NF-κB, leading to the production of prostaglandin E2 via the induction of COX-2 gene expression.35 Ceramides can also be converted into ceramide 1-phosphate or further degraded into sphingosine, which can also be phosphorylated to form sphingosine 1-phosphate (S1P). These molecules play a key role in regulating inflammatory processes, making S1P a promising target for IBD due to its role in modulating lymphocyte migration from lymph nodes.36
Contrary to the proinflammatory effects of host-produced sphingolipids, it has been shown that sphingolipids produced by bacteria like Bacteroides can exert anti-inflammatory effects,31 emphasising the importance of microbial-produced molecules in maintaining intestinal health and the delicate balance between pro-inflammatory and anti-inflammatory molecules.
In addition to the increased levels of sphingolipids, we also report higher levels of N-acylethanolamines (palmitoyl-ethanolamide, linoleoyl-ethanolamide, oleoyl-ethanolamide and stearoyl-ethanolamide) in the faeces of patients with CD compared with non-IBD controls. Although the mechanism behind the accumulation of these atypical endocannabinoids still needs to be elucidated, current evidence suggests that ethanolamides might shape the gut microbiota during inflammation.37
Our study found that patients with IBD also have elevated levels of long-chain fatty acids (LCFAs) and PUFAs, such as acylcarnitines and arachidonic acid, in their faeces. Previous research by Smith et al has identified palmitoylcarnitine (C16) as a faecal biomarker for IBD, linking acylcarnitine accumulation in the intestinal lumen to a reduced LCFA uptake in colonocytes during inflammation.38 Furthermore, increasing evidence suggests that diets high in PUFAs can contribute to intestinal inflammation. Exposure to omega-3 and omega-6 PUFA can trigger an inflammatory response in intestinal organoids from patients with CD and in mice models with an impaired glutathione peroxidase 4 (GPX4) gene expression.
We also observed a significant increase in the levels of amino acids and their derivatives in patients with IBD. These findings align with previous research conducted on a cohort of newly diagnosed patients with IBD (n=78), where the levels of several amino acids could differentiate IBD samples from controls with high accuracy.39 In particular, we found a strong increase in taurine levels in IBD samples.
It has been shown that bacteria in the colon can use taurine as a substrate, releasing sulfite, which can be further converted to hydrogen sulfide.40 This accumulation of hydrogen sulfide has been linked to epithelial damage and colitis. B. wadsworthia is a sulfate-reducing bacterium that has been shown to have the capability to metabolise taurine, which provides a potential explanation for the inverse relationship observed between the detection of B. wadsworthia and taurine levels in our cohort.
Furthermore, faeces from patients with IBD exhibited depletion of nucleotides, enterolactone (a bacterial product produced from the breakdown of dietary lignans) and biotin (vitamin B7). These findings suggest that the loss of bacterial diversity and biomass in the gut of patients with IBD41 could drive the reduction in essential functions such as fibre digestion and vitamin production. The restoration of microbial production of these metabolites through dietary administration of their precursors could serve as a potential strategy to prevent flare-ups and address the dysregulation of the gut microbiome in IBD.
The impact of genetics and lifestyle on the faecal metabolite levels
Along with the influence of IBD, diet and lifestyle are determinants of the abundance of small molecules in the human body. By correlating metabolites to dietary data, medication use and lifestyle factors, we found that daily habits such as smoking or coffee and tea consumption strongly correlated with their derivative molecules (online supplemental tables 11–13). Despite these associations, long-term dietary habits were moderately associated with faecal metabolome composition. Our prediction model revealed that only a few faecal metabolites could be predicted using dietary information (15 metabolites, explained variance >25%, online supplemental table 20), including several unclassified metabolites, coffee and derivatives (AAMU, N-methyl pipecolate, theophylline) and enterolactone (a lignan derivative). Conversely, recent studies have reported more substantial impact of dietary intake on the levels of circulating metabolites.21 We hypothesise that our dietary data underestimates the contribution of food intake to levels of faecal metabolites since it is based on food frequency questionnaires. Future studies should consider the use of 24-hour dietary recalls to capture daily dietary variations when aiming to explore relations between food intake and faecal metabolites and food–microbiome interactions. Furthermore, the impact of the host’s absorption rates, metabolism and biotransformations in the gastrointestinal tract should be considered when studying the relationship between dietary habits and faecal metabolites.
Our mediation analysis provided statistical evidence for the role of the gut microbiota as a mediator between faecal metabolites and clinical and lifestyle factors associations. For example, the levels of several coffee-related metabolites in faeces partially depended on the abundance of L. asaccharolyticus. Although this specie has been associated with coffee intake before,42 its capacity to metabolise molecules found in coffee, such as AAMU and 1,3-dimethylruic acid, remains unknown. However, relations between exposures, microbiota and metabolites are complex, for example, metabolites can shape the gut microbiota composition and bacteria can establish cross-feeding metabolic networks; therefore, functional validations are needed to better estimate the directionality of these interactions.
Host genetics showed a small impact on metabolite levels in faeces. The only association that passed our significance threshold was between a single nucleotide polymorphism near the NAT2 gene and AAMU, a caffeine metabolism product (online supplemental table 15). NAT2 encodes an N-acetyltransferase enzyme that detoxifies several xenobiotics, including coffee and certain types of medication. A study in the TwinsUK biobank also reported this association and estimated that host genetics has a moderate effect on faecal metabolites, with an average heritability of ~18%.23 This relatively low heritability contrasts with the impact of host genetics on the levels of circulating metabolites43 44 and might be explained by the fact that faecal metabolites are primarily influenced by microbial transformations occurring in the colon, which can potentially mask genetic effects. Moreover, sample sizes are still a limiting factor for discovering metabolite–genome associations. In fact, when using a looser significance cut-off (p <5×10−8), we found >200 suggestive associations pointing to the metabolism of cholesterol and serotonin. For example, LRP5L was associated with serotonin and PNLIPRP2 with 1-palmitoyl-2-linoleoyl-digalactosyl glycerol (16:0/18:2) (online supplemental table 15). LRP5L belongs to the low-density lipoprotein (LDL) receptor family found to be involved in controlling serotonin levels in the duodenum.45 Both PNLIPRP2 and 1-palmitoyl-2-linoleoyl-digalactosyl glycerol (a choline derivative) are linked to cholesterol metabolism, supporting that choline supplements maintain blood cholesterol homoeostasis,46 and PNLIPRP2 has been associated with LDL levels in blood.47
The relation between gut microbiota composition and gut metabolism
The strong relationship between the microbiome and metabolites enabled us to estimate the levels of faecal metabolites using metagenomic sequencing data (online supplemental table 20). In line with previous studies23 43 48 49, well-predicted molecules included putrescine, urobilin, bile salts and fatty acids. However, further functional evidence is necessary to verify that all these well-predicted molecules can indeed be products of microbial metabolism. Notably, models trained on controls and tested on IBD samples showed lower prediction accuracy compared with models trained on both IBD and non-IBD datasets. This low cross-predictability between cases and controls has also been reported by Muller et al 49 and implies that some microbiota–metabolite associations may be context-specific or become more evident when microbial communities are perturbed. For instance, patients with IBD often exhibit alterations in their gut microbiota composition, leading to dysbiosis. In line with this, our analysis revealed 1137 metabolite–microbiota associations significantly influenced by dysbiosis.
Patients with IBD and dysbiosis displayed an enrichment of 202 highly prevalent metabolites, including a significant increase in primary and conjugated bile acid levels compared with eubiotic IBD samples (online supplemental table 5). The accumulation of cholic acid in the colon has been shown to exert selective pressure on the gut ecosystem due to its antimicrobial properties,50 which could explain the expansion of bile-resistant bacteria, such as C. boltae and R. gnavus in dysbiosis. Additionally, the loss of certain bacteria may also contribute to the accumulation of primary bile acids, as evidenced by the decreased abundance of bai operons and the increased ratio of primary to secondary bile acids in dysbiotic samples compared with eubiotic samples. Furthermore, bile acids play a pivotal role in regulating metabolism, exerting signalling effects in preserving the intestinal barrier and regulating the host’s immune system,51 thereby making them a highly attractive target for therapeutic intervention in IBD.
In our cohort, dysbiosis was associated with ileum disease involvement and an ileocecal valve resection (online supplemental table 5). Accumulating literature demonstrates that disruptions in the small intestine due to inflammation or surgery can significantly impact faeces’ metabolite and microbial composition.52 53 For example, Halfvarson et al 24 showed that patients with intestinal surgery in the ileum had a less stable microbiota and more frequently transitioned between eubiotic and dysbiotic states. Given the critical role of the small intestine in nutrient absorption, it is plausible that disruptions in this section of the gut lead to persistent alterations in the concentrations of bile acids, amino acids and lipids in the colon, which might reshape the microbial composition towards a dysbiotic state. The stratification of patients based on their disease location and microbiome composition should be considered in future metabolomic studies and clinical interventions, as it can potentially uncover more targeted and effective treatments for the disease.
Furthermore, the coabundance analysis performed in this study provides insight into the relationship between bacteria and their associated metabolic products. This information can serve as a basis for identifying potential therapeutic targets for treating IBD. For instance, F. prausnitzii, a specie which is depleted in the faeces of patients with IBD, was positively correlated with SCFAs and hypoxanthine levels (online supplemental table 17). Hypoxanthine can be produced by F. prausnitzii through the metabolisation of adenine54 and plays a role in maintaining the intestinal epithelium.55 Similarly, R. gnavus, which is highly abundant in dysbiotic samples from patients with IBD,56 was positively correlated with the levels of tryptamine (online supplemental table 17). R. gnavus is capable of producing tryptamine via the decarboxylation of tryptophan.57 Accumulation of tryptamine may increase gut motility via activation of serotonin receptor-4, which could explain why some patients experience decreased intestinal transit times during flares.58 In line with our findings, higher levels of tryptamine have been reported in individuals with irritable bowel syndrome and diarrhoea and have been associated with the metabolic activity of R. gnavus.59 Additionally, the positive correlation we observed between S. parasanguinis and imidazole propionate could be explained by the capacity of this bacterium to degrade histidine.60 Imidazole propionate has been linked to the risk of developing type 2 diabetes and regulates activation of the mTORC1 signalling pathway,61 62 which is implicated in IBD.63
Overall, the substantial variations in metabolite composition between eubiotic and dysbiotic microbial communities, and the strong co-occurrence between metabolites and bacterial species, support the notion that faecal metabolomics partially reflects the metabolic activity of the gut microbiota. Further functional validation and longitudinal monitoring of microbial–metabolite associations are necessary to determine the direction of these relationships and assess their impact on disease progression.
Faecal metabolites as novel biomarkers for IBDs
As opposed to colonoscopies, the current invasive gold standard for diagnosing IBD, we demonstrated the potential of faecal metabolites as a non-invasive method for disease diagnosis. The ratio between the levels of two metabolites, lactosyl-N-palmitoyl-sphingosine (d18:1/16:0) and L-urobilin was identified as a biomarker for IBD in our cohort (online supplemental table 10). Other studies have also observed reduced levels of L-urobilin and increased sphingolipids in patients with IBD faeces.3 5 In a North American longitudinal cohort,3 we observed that the ratio between a sphingolipid (ceramide (18:1/16:0)), and L-urobilin were consistently higher in patients with IBD compared with non-IBD controls underscoring our findings(online supplemental figure 7) . Faecal measurements targeting these two molecules could be relatively easy to implement in combination with the faecal calprotectin test.
gutjnl-2022-328048supp010.pdf (62.1KB, pdf)
It is important to note that our study cohort primarily consisted of subjects with a prolonged history of IBD. Therefore, the validity of the identified biomarker must be confirmed in newly diagnosed patients with IBD, as well as in individuals with other gastrointestinal disorders.
Finally, it is also important to acknowledge the limitations of untargeted metabolomics approaches. This study focused on annotated molecules with relatively high prevalence, but a substantial number of metabolites remained unidentified, and their physiological significance is unknown. Approximately one-third of the metabolites detected in our dataset (492 out of 1684 metabolites) could not be linked to a previously characterised compound, emphasising the need for further efforts to fully characterise the molecular diversity in the human body. Additionally, the semiquantitative nature of untargeted metabolomics limits the ability to establish the normal concentration range of each metabolite in faeces.
In conclusion, this study provides a detailed characterisation of the faecal metabolites in the context of health and intestinal inflammation, replicating known disease-relevant molecules and expanding our knowledge of disease heterogeneity. In addition, we pinpoint multiple associations between microbiota, diet and faecal metabolite levels, which we believe provide valuable resources for further investigation of metabolite-based or microbiota-based interventions and treatment in IBD.
gutjnl-2022-328048supp003.zip (5.2MB, zip)
gutjnl-2022-328048supp011.pdf (822.2KB, pdf)
Acknowledgments
The authors thank Kate Mc Intyre for substantive English editing.
Footnotes
Twitter: @arnauvich, @CollijValerie, @HannahAugustijn, @SashaZhernakova, @RGacesa, @jingyuan_fu
SH, SA-S and VC contributed equally.
Correction notice: This article has been corrected since it published Online First. The supplementary figures have been replaced.
Contributors: AVV and RKW designed the study. AVV, SH, SA-S, RG, HEA contributed to the data analysis. AVV and SH wrote the manuscript. BHDJ handled the samples in the laboratory. LAB, SH, SA-S, VC, HEA, JF, AZ, AAG, JP, JS, CG, GA-A, RAAAR and RKW critically reviewed the manuscript. AVV and RKW are responsible for the overall content as guarantor.
Funding: This study was funded by Takeda Development Center Americas. JF is supported by the Dutch Heart Foundation IN-CONTROL (CVON2018-27), the ERC Consolidator grant (grant agreement No. 101001678), NWO-VICI grant VI.C.202.022, and the Netherlands Organ-on-Chip Initiative, an NWO Gravitation project (024.003.001) funded by the Ministry of Education, Culture and Science of the government of The Netherlands. AZ is supported by the Dutch Heart Foundation IN-CONTROL (CVON2018-27), the ERC Starting Grant 715772, NWO-VIDI grant 016.178.056, ZONMW MEMORABEL grant 733050814 and the NWO Gravitation grant Exposome-NL (024.004.017). RKW is supported by HORIZON-HLTH-2022-STAYHLTH-02-0 and the Seerave Foundation.
Competing interests: This study was funded by Takeda Development Center Americas. RKW acted as a consultant for Takeda and received unrestricted research grants from Takeda and Johnson and Johnson pharmaceuticals and speaker fees from AbbVie, MSD, Olympus and AstraZeneca. GA-A, CG, JS, JP and AAG are or were employees of Takeda Pharmaceuticals at the time this study was conducted.No disclosures: All other authors have nothing to disclose.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request. Tables containing the levels of faecal metabolites and bacterial taxa abundances are provided with the manuscript. The raw metagenomics, host genomics and phenotypic data used in this study are available from the European Genome–Phenome Archive data repository: 1000 Inflammatory bowel disease (IBD) cohort (https://www.ebi.ac.uk/ega/datasets/EGAD00001004194), Lifelines DEEP cohort (https://www.ebi.ac.uk/ega/datasets/EGAD00001001991). This includes submitting a letter of intent to the corresponding data access committees. Codes are publicly available at: https://github.com/GRONINGEN-MICROBIOME-CENTRE/Fecal_Metabolites_IBD
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study involves human participants and was approved by M12.1139652008.338. Participants gave informed consent to participate in the study before taking part.
References
- 1. Kaplan GG, Ng SC. Understanding and preventing the global increase of inflammatory bowel disease. Gastroenterology 2017;152:313–21. 10.1053/j.gastro.2016.10.020 [DOI] [PubMed] [Google Scholar]
- 2. Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 2014;146:1489–99. 10.1053/j.gastro.2014.02.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lloyd-Price J, Arze C, Ananthakrishnan AN, et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 2019;569:655–62. 10.1038/s41586-019-1237-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lavelle A, Sokol H. Gut microbiota-derived metabolites as key actors in inflammatory bowel disease. Nat Rev Gastroenterol Hepatol 2020;17:223–37. 10.1038/s41575-019-0258-z [DOI] [PubMed] [Google Scholar]
- 5. Franzosa EA, Sirota-Madi A, Avila-Pacheco J, et al. Author correction: gut microbiome structure and metabolic activity in inflammatory bowel disease. Nat Microbiol 2019;4:898. 10.1038/s41564-019-0442-5 [DOI] [PubMed] [Google Scholar]
- 6. Johnson CH, Ivanisevic J, Siuzdak G. Metabolomics: beyond biomarkers and towards mechanisms. Nat Rev Mol Cell Biol 2016;17:451–9. 10.1038/nrm.2016.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bauset C, Gisbert-Ferrándiz L, Cosín-Roger J. Metabolomics as a promising resource identifying potential biomarkers for inflammatory bowel disease. J Clin Med 2021;10:622. 10.3390/jcm10040622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tigchelaar EF, Zhernakova A, Dekens JAM, et al. Cohort profile: lifelines deep, a prospective, general population cohort study in the Northern Netherlands: study design and baseline characteristics. BMJ Open 2015;5:e006772. 10.1136/bmjopen-2014-006772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Imhann F, Van der Velde KJ, Barbieri R, et al. The 1000IBD project: multi-omics data of 1000 inflammatory bowel disease patients; data release 1. BMC Gastroenterol 2019;19:5. 10.1186/s12876-018-0917-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhernakova A, Kurilshikov A, Bonder MJ, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science 2016;352:565–9. 10.1126/science.aad3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vich Vila A, Imhann F, Collij V, et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med 2018;10:eaap8914. 10.1126/scitranslmed.aap8914 [DOI] [PubMed] [Google Scholar]
- 12. Kowarik A, Templ M. Imputation with the R package VIM. J Stat Softw 2016;74:1–16. 10.18637/jss.v074.i07 [DOI] [Google Scholar]
- 13. Gordon-Rodriguez E, Quinn TP, Cunningham JP. Learning sparse log-ratios for high-throughput sequencing data. Bioinformatics 2021;38:157–63. 10.1093/bioinformatics/btab645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kurilshikov A, Medina-Gomez C, Bacigalupe R, et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat Genet 2021;53:156–65. 10.1038/s41588-020-00763-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 2019;37:852–7. 10.1038/s41587-019-0209-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morton JT, Aksenov AA, Nothias LF, et al. Learning representations of microbe-metabolite interactions. Nat Methods 2019;16:1306–14. 10.1038/s41592-019-0616-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Friedman JH, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw 2010;33:1–22. 10.18637/jss.v033.i01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chamaillard M, Hashimoto M, Horie Y, et al. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol 2003;4:702–7. 10.1038/ni945 [DOI] [PubMed] [Google Scholar]
- 19. Parada Venegas D, De la Fuente MK, Landskron G, et al. Corrigendum: short chain fatty acids (SCFAs) -mediated gut epithelial and immune regulation and its relevance for inflammatory bowel diseases. Front Immunol 2019;10:1486. 10.3389/fimmu.2019.01486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Diener C, Dai CL, Wilmanski T, et al. Genome-microbiome interplay provides insight into the determinants of the human blood metabolome. Nat Metab 2022;4:1560–72. 10.1038/s42255-022-00670-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen L, Zhernakova DV, Kurilshikov A, et al. Influence of the microbiome, diet and genetics on inter-individual variation in the human plasma metabolome. Nat Med 2022;28:2333–43. 10.1038/s41591-022-02014-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yin X, Chan LS, Bose D, et al. Genome-wide association studies of metabolites in finnish men identify disease-relevant loci. Nat Commun 2022;13:1644. 10.1038/s41467-022-29143-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zierer J, Jackson MA, Kastenmüller G, et al. The fecal metabolome as a functional readout of the gut microbiome. Nat Genet 2018;50:790–5. 10.1038/s41588-018-0135-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Halfvarson J, Brislawn CJ, Lamendella R, et al. Dynamics of the human gut microbiome in inflammatory bowel disease. Nat Microbiol 2017;2:17004. 10.1038/nmicrobiol.2017.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Garsin DA. Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat Rev Microbiol 2010;8:290–5. 10.1038/nrmicro2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Korpela KD. Diet, microbiota, and metabolic health: trade-off between saccharolytic and proteolytic fermentation. Annu Rev Food Sci Technol 2018;9:65–84. 10.1146/annurev-food-030117-012830 [DOI] [PubMed] [Google Scholar]
- 27. Gryp T, Vanholder R, Vaneechoutte M, et al. P-cresyl sulfate. Toxins (Basel) 2017;9:52. 10.3390/toxins9020052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kikuchi K, Saigusa D, Kanemitsu Y, et al. Gut microbiome-derived phenyl sulfate contributes to albuminuria in diabetic kidney disease. Nat Commun 2019;10:1835. 10.1038/s41467-019-09735-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Edamatsu T, Fujieda A, Itoh Y. Phenyl sulfate, indoxyl sulfate and p-cresyl sulfate decrease glutathione level to render cells vulnerable to oxidative stress in renal tubular cells. PLoS One 2018;13:e0193342. 10.1371/journal.pone.0193342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu W-C, Tomino Y, Lu K-C. Impacts of indoxyl sulfate and p-cresol sulfate on chronic kidney disease and mitigating effects of AST-120. Toxins (Basel) 2018;10:367. 10.3390/toxins10090367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brown DG, Rao S, Weir TL, et al. Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab 2016;4:11. 10.1186/s40170-016-0151-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Al Hinai EA, Kullamethee P, Rowland IR, et al. Modelling the role of microbial p-cresol in colorectal genotoxicity. Gut Microbes 2019;10:398–411. 10.1080/19490976.2018.1534514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abdel Hadi L, Di Vito C, Riboni L. Fostering inflammatory bowel disease: sphingolipid strategies to join forces. Mediators Inflamm 2016;2016:3827684. 10.1155/2016/3827684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Maceyka M, Spiegel S. Sphingolipid metabolites in inflammatory disease. Nature 2014;510:58–67. 10.1038/nature13475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nixon GF. Sphingolipids in inflammation: pathological implications and potential therapeutic targets. Br J Pharmacol 2009;158:982–93. 10.1111/j.1476-5381.2009.00281.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Danese S, Furfaro F, Vetrano S. Targeting S1P in inflammatory bowel disease: new avenues for modulating intestinal leukocyte migration. J Crohns Colitis 2018;12:S678–86. 10.1093/ecco-jcc/jjx107 [DOI] [PubMed] [Google Scholar]
- 37. Fornelos N, Franzosa EA, Bishai J, et al. Growth effects of N-acylethanolamines on gut bacteria reflect altered bacterial abundances in inflammatory bowel disease. Nat Microbiol 2020;5:486–97. 10.1038/s41564-019-0655-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Smith SA, Ogawa SA, Chau L, et al. Mitochondrial dysfunction in inflammatory bowel disease alters intestinal epithelial metabolism of hepatic acylcarnitines. J Clin Invest 2021;131:e133371. 10.1172/JCI133371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Jagt JZ, Struys EA, Ayada I, et al. Fecal amino acid analysis in newly diagnosed pediatric inflammatory bowel disease: a multicenter case-control study. Inflamm Bowel Dis 2022;28:755–63. 10.1093/ibd/izab256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Walker A, Schmitt-Kopplin P. The role of fecal sulfur metabolome in inflammatory bowel diseases. Int J Med Microbiol 2021;311:151513. 10.1016/j.ijmm.2021.151513 [DOI] [PubMed] [Google Scholar]
- 41. Vieira-Silva S, Sabino J, Valles-Colomer M, et al. Quantitative microbiome profiling disentangles inflammation- and bile duct obstruction-associated microbiota alterations across PSC/IBD diagnoses. Nat Microbiol 2019;4:1826–31. 10.1038/s41564-019-0483-9 [DOI] [PubMed] [Google Scholar]
- 42. Asnicar F, Berry SE, Valdes AM, et al. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med 2021;27:321–32. 10.1038/s41591-020-01183-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bar N, Korem T, Weissbrod O, et al. A reference map of potential determinants for the human serum metabolome. Nature 2020;588:135–40. 10.1038/s41586-020-2896-2 [DOI] [PubMed] [Google Scholar]
- 44. Shin S-Y, Fauman EB, Petersen A-K, et al. An atlas of genetic influences on human blood metabolites. Nat Genet 2014;46:543–50. 10.1038/ng.2982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yadav VK, Ryu J-H, Suda N, et al. LRP5 controls bone formation by inhibiting serotonin synthesis in the duodenum. Cell 2008;135:825–37. 10.1016/j.cell.2008.09.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Al Rajabi A, Castro GSF, da Silva RP, et al. Choline supplementation protects against liver damage by normalizing cholesterol metabolism in Pemt/Ldlr knockout mice fed a high-fat diet. J Nutr 2014;144:252–7. 10.3945/jn.113.185389 [DOI] [PubMed] [Google Scholar]
- 47. Liu DJ, Peloso GM, Yu H, et al. Exome-wide association study of plasma lipids in > 300,000 individuals. Nat Genet 2017;49:1758–66. 10.1038/ng.3977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mallick H, Franzosa EA, Mclver LJ, et al. Predictive metabolomic profiling of microbial communities using amplicon or metagenomic sequences. Nat Commun 2019;10:3136. 10.1038/s41467-019-10927-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Muller E, Algavi YM, Borenstein E. A meta-analysis study of the robustness and universality of gut microbiome-metabolome associations. Microbiome 2021;9:203. 10.1186/s40168-021-01149-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wahlström A, Sayin SI, Marschall H-U, et al. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab 2016;24:41–50. 10.1016/j.cmet.2016.05.005 [DOI] [PubMed] [Google Scholar]
- 51. Thomas JP, Modos D, Rushbrook SM, et al. The emerging role of bile acids in the pathogenesis of inflammatory bowel disease. Front Immunol 2022;13:829525. 10.3389/fimmu.2022.829525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Fang X, Vázquez-Baeza Y, Elijah E, et al. Gastrointestinal surgery for inflammatory bowel disease persistently lowers microbiome and metabolome diversity. Inflamm Bowel Dis 2021;27:603–16. 10.1093/ibd/izaa262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Jansson J, Willing B, Lucio M, et al. Metabolomics reveals metabolic biomarkers of Crohn’s disease. PLoS ONE 2009;4:e6386. 10.1371/journal.pone.0006386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Heinken A, Khan MT, Paglia G, et al. Functional metabolic map of Faecalibacterium prausnitzii, a beneficial human gut microbe. J Bacteriol 2014;196:3289–302. 10.1128/JB.01780-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lee JS, Wang RX, Alexeev EE, et al. Hypoxanthine is a checkpoint stress metabolite in colonic epithelial energy modulation and barrier function. J Biol Chem 2018;293:6039–51. 10.1074/jbc.RA117.000269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hall AB, Yassour M, Sauk J, et al. A novel Ruminococcus gnavus clade enriched in inflammatory bowel disease patients. Genome Med 2017;9:103. 10.1186/s13073-017-0490-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Williams BB, Van Benschoten AH, Cimermancic P, et al. Discovery and characterization of gut microbiota decarboxylases that can produce the neurotransmitter tryptamine. Cell Host Microbe 2014;16:495–503. 10.1016/j.chom.2014.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bhattarai Y, Williams BB, Battaglioli EJ, et al. Gut microbiota-produced tryptamine activates an epithelial G-protein-coupled receptor to increase colonic secretion. Cell Host Microbe 2018;23:775–85. 10.1016/j.chom.2018.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhai L, Huang C, Ning Z, et al. Ruminococcus gnavus plays a pathogenic role in diarrhea-predominant irritable bowel syndrome by increasing serotonin biosynthesis. Cell Host Microbe 2023;31:33–44. 10.1016/j.chom.2022.11.006 [DOI] [PubMed] [Google Scholar]
- 60. Molinaro A, Bel Lassen P, Henricsson M, et al. Imidazole propionate is increased in diabetes and associated with dietary patterns and altered microbial ecology. Nat Commun 2020;11:5881. 10.1038/s41467-020-19589-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Koh A, Molinaro A, Ståhlman M, et al. Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell 2018;175:947–61. 10.1016/j.cell.2018.09.055 [DOI] [PubMed] [Google Scholar]
- 62. Koh A, Mannerås-Holm L, Yunn N-O, et al. Microbial imidazole propionate affects responses to metformin through p38γ-dependent inhibitory AMPK phosphorylation. Cell Metab 2020;32:643–53. 10.1016/j.cmet.2020.07.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lin X, Sun Q, Zhou L, et al. Colonic epithelial mTORC1 promotes ulcerative colitis through COX-2-mediated Th17 responses. Mucosal Immunol 2018;11:1663–73. 10.1038/s41385-018-0018-3 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2022-328048supp001.pdf (126.2KB, pdf)
gutjnl-2022-328048supp002.xlsx (226.3MB, xlsx)
gutjnl-2022-328048supp004.pdf (1.1MB, pdf)
gutjnl-2022-328048supp005.pdf (1.2MB, pdf)
gutjnl-2022-328048supp006.pdf (543.7KB, pdf)
gutjnl-2022-328048supp007.pdf (392.8KB, pdf)
gutjnl-2022-328048supp008.pdf (1.2MB, pdf)
gutjnl-2022-328048supp009.pdf (536.5KB, pdf)
gutjnl-2022-328048supp010.pdf (62.1KB, pdf)
gutjnl-2022-328048supp003.zip (5.2MB, zip)
gutjnl-2022-328048supp011.pdf (822.2KB, pdf)
Data Availability Statement
Data are available on reasonable request. Tables containing the levels of faecal metabolites and bacterial taxa abundances are provided with the manuscript. The raw metagenomics, host genomics and phenotypic data used in this study are available from the European Genome–Phenome Archive data repository: 1000 Inflammatory bowel disease (IBD) cohort (https://www.ebi.ac.uk/ega/datasets/EGAD00001004194), Lifelines DEEP cohort (https://www.ebi.ac.uk/ega/datasets/EGAD00001001991). This includes submitting a letter of intent to the corresponding data access committees. Codes are publicly available at: https://github.com/GRONINGEN-MICROBIOME-CENTRE/Fecal_Metabolites_IBD