Abstract
The cytotoxic effects of reactive oxygen species are largely mediated by iron. Hydrogen peroxide reacts with iron to form the extremely reactive and damaging hydroxyl radical via the Fenton reaction. Superoxide anion accelerates this reaction because the dismutation of superoxide leads to increased levels of hydrogen peroxide and because superoxide elevates the intracellular concentration of iron by attacking iron-sulfur proteins. We found that regulators of the Escherichia coli responses to oxidative stress, OxyR and SoxRS, activate the expression of Fur, the global repressor of ferric ion uptake. A transcript encoding Fur was induced by hydrogen peroxide in a wild-type strain but not in a ΔoxyR strain, and DNase I footprinting assays showed that OxyR binds to the fur promoter. In cells treated with the superoxide-generating compound paraquat, we observed the induction of a longer transcript encompassing both fur and its immediate upstream gene fldA, which encodes a flavodoxin. This polycistronic mRNA is induced by paraquat in a wild-type strain but not in a ΔsoxRS strain, and SoxS was shown to bind to the fldA promoter. These results demonstrate that iron metabolism is coordinately regulated with the oxidative stress defenses.
Reactive oxygen species can damage DNA, lipid membranes, and proteins and have been implicated in numerous diseases. As a defense, both prokaryotic and eukaryotic cells have inducible responses that protect against oxidative damage. These antioxidant defense systems have been best characterized in Escherichia coli (reviewed in reference 29). Hydrogen peroxide activates the transcription factor OxyR through the oxidation of two cysteines and formation of an intramolecular disulfide bond (34). Activated OxyR then induces transcription of a set of antioxidant genes, including katG (hydroperoxidase I), ahpCF (alkylhydroperoxidase), dps (a nonspecific DNA binding protein), gorA (glutathione reductase), grxA (glutaredoxin I), and oxyS (a regulatory RNA). Superoxide-generating compounds, such as paraquat, activate the transcription factor SoxR by oxidizing the 2Fe-2S cluster in the protein through an unknown mechanism (11, 16). Oxidized SoxR then induces the expression of the second transcription factor SoxS, which directly activates the transcription of sodA (manganese superoxide dismutase), fpr (ferredoxin/flavodoxin-NADP+ reductase), zwf (glucose 6-phosphate dehydrogenase), fumC (fumarase C), nfo (endonuclease IV), acnA (aconitase A), and micF (a regulatory RNA).
There is an intimate relationship between iron metabolism and oxidative stress. Iron is an indispensable element for living cells, since many metabolic enzymes have iron as a cofactor in their active sites. On the other hand, through the Fenton reaction, iron also promotes the formation of hydroxyl radicals, which indiscriminately damage all cellular components. Thus, cells have evolved regulatory systems to ensure the sufficient uptake of iron to meet their physiological requirements yet at the same time minimize iron toxicity. The regulation of iron homeostasis in both eukaryotic and prokaryotic cells is the subject of intense study, and much is known. In prokaryotic cells, a transcription factor denoted Fur (ferric uptake regulation) negatively regulates many genes involved in ferric iron uptake from the environment (reviewed in references 6 and 7). Most Fur-regulated genes are derepressed in growth at low iron and are repressed under conditions of high iron, and in vitro DNA binding assays suggest that high levels of iron favor Fur association with DNA (2, 3, 9, 15). Thus, Fur is considered to be an iron-dependent repressor. The findings that Δfur mutants are sensitive to hydrogen peroxide and show increased oxidative DNA damage and mutations under aerobic conditions have implicated Fur in the defenses against oxidative stress (32). However, Fur expression in response to oxidative stress has not been examined.
As part of our continuous effort to better define the physiological role of OxyR, we initiated a computational approach to identify additional OxyR-regulated genes. We used an algorithm based on information theory (26) that uses previously identified OxyR binding sites as a model to search through the entire E. coli genome for new OxyR binding sites. This approach predicted an OxyR binding site in the promoter region of the fur gene. Subsequent experiments confirmed OxyR binding to this region and showed that fur expression is induced by OxyR after hydrogen peroxide treatment. We also found that SoxRS regulates fur by activating the expression of a transcript encoding both flavodoxin and Fur. These results show that the control of iron metabolism in E. coli is an integral part of the antioxidant defense response and that the regulation of Fur by OxyR and SoxRS directly reflects the chemistry between iron and reactive oxygen species.
MATERIALS AND METHODS
Computer search program.
The seven OxyR target sequences analyzed previously (25) and two new sites (grxA at position 207 in GenBank entry M13449 and a second Mu phage mom site at position 59 in GenBank entry V01463) were used to generate an individual information weight matrix (26). The matrix was scanned across the entire E. coli genomic sequence (5). The sites identified were sorted by information content so that the strongest sites could be investigated further. Local regions of the genome surrounding the strongest sites were displayed using the Lister program (version 9.02) (Fig. 1) to show coding regions along with sequence walkers representing potential binding sites (27). Detailed Lister maps of the fldA-fur region including ribosome binding sites and cyclic AMP receptor protein sites are available online (28).
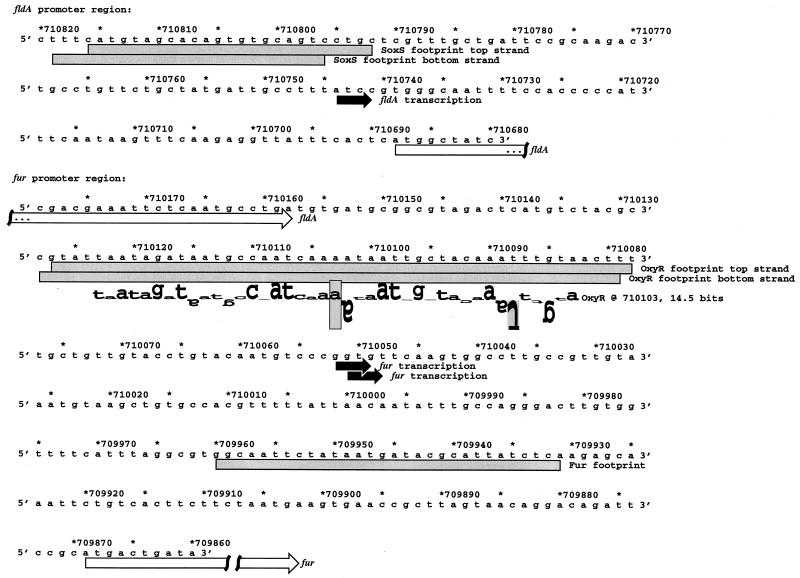
FIG. 1.
Sequences of fur and fldA promoter regions. The DNA sequences and coordinates are for E. coli from GenBank accession no. U00096 (5). The transcriptional starts are marked by black arrows, the fldA and fur open reading frames are indicated by white arrows, and the DNase I footprints are denoted by gray boxes. The location of the predicted OxyR site is shown by a sequence walker (27), in which the rectangle surrounding the adenine at position 710103 indicates the center (zero base) of the binding site. A sequence walker consists of a string of letters in which the height of each letter shows the contribution that the corresponding base would make to the average sequence conservation shown by the sequence logo of all binding sites (25). The sequence walker is given on a scale of bits of information. The rectangle surrounding the adenine gives the scale, from −3 bits up to the maximum conservation at +2 bits. Positively contributing bases are above the zero line, while negatively contributing ones are below the line. By using bits, the heights of all letters can be added together to obtain the total sequence conservation of a site. The fur site is 14.5 bits, and the average OxyR site is 16.7 ± 1.9 bits. The rectangle surrounding the inverted thymine at position 710088 indicates that no T residues were found at this position in the OxyR sites used to construct the model.
Strains and plasmids.
The plasmids used in this study were constructed as follows. An 840-bp fragment carrying the fldA gene was amplified by PCR from genomic DNA (using the primers 5′-GCC ACT TGA ATT CCG GGA CAT TG and 5′-GAA CGG ATC CAA GAG ATG TTA ATG C [underlined sequences are restriction sites]) and cloned into the EcoRI and BamHI sites of pUC18 to generate pGSO96. A 250-bp fragment containing the fur promoter region was amplified by PCR from genomic DNA (using primers 5′-AAT GAA TTC CAC AAG TCC CTG GC and 5′-CCG CGG ATC CTG TAG AAA AAT GGG) and cloned into the EcoRI and BamHI sites of pUC18 to generate pGSO97. A 200-bp fragment containing the fldA promoter was PCR amplified from genomic DNA (using primers 5′-GCC GTA GCG AAC GGA TCC AAG AG and 5′-GCC AGT GAA AGC TTT GAG TG) and subcloned into the HindIII and BamHI sites of pUC18 to generate pGSO98. The integrity of all clones was verified by DNA sequencing. Standard cloning techniques were used. The ΔoxyR::kan (GSO9 [32]), ΔsoxRS-zjc2205 zjc2204::Tn10kan (DJ901 [14]), and Δfur::kan (QC1732 [32]) mutant alleles were moved into MC4100 by P1 transduction to generate GSO47, GSO71, and GSO72, respectively.
RNA isolation.
Cultures were grown under aeration at 37°C in LB (Luria-Bertani) rich medium or M63 minimal medium supplemented with 2 mg of glucose and 20 μg of vitamin B1 per ml. The cell pellet from 25 ml of culture grown to an optical density at 600 nm of 0.2 to 0.3 was resuspended in 1 ml of TRIZOL (Gibco BRL). All subsequent purification steps were carried out according to the TRIZOL reagent manual (based on reference 8). RNA yields were typically 100 to 200 μg.
Primer extension assay.
RNA samples were subjected to primer extension assays as described elsewhere (30), using primers specific to fur (5′-CAA GTC CCT GGC AAA TAT TG and 5′-TAT TGT TAT CAG TCA TGC GG) and fldA (5′-CCA AGC TGT TTT TGA ATC AT).
Northern blotting.
The RNA samples (10 μg) were denatured at 65°C in 0.5× Tris-borate-EDTA buffer–70% formamide, separated on 6% urea-polyacrylamide gels, and transferred to nylon membranes by electroblotting. The membranes were probed with the EcoRI-BamHI fragment of pGSO97 (32P labeled at the EcoRI site) and the EcoRI-BamHI fragment of pGSO96 (random primer labeled with 32P).
Protein purification.
OxyR protein was purified as described elsewhere (30). Strain XA90 harboring the SoxS overexpression plasmid (pKOXS) was obtained from B. Demple. Crude extracts of cells overexpressing the SoxS protein were prepared as described by Li and Demple (21).
DNase I footprinting.
The DNase I footprinting assay of OxyR binding to the fur promoter, contained on the BamHI-EcoRI fragment of pGSO97, was carried out as described previously (31). The DNase I footprint assay of SoxS binding to the fldA promoter, contained on the BamHI-HindIII fragment of pGSO98, was carried out as described previously (21).
Immunoblot assays.
Proteins were separated on a sodium dodecyl sulfate (SDS)–12% polyacrylamide gels and transferred to a nitrocellulose filter by electroblotting. The filter was then probed with a 1:500 dilution of Fur antiserum, provided by M. Vasil. Bound antibody was visualized with rabbit antiserum by using an enhanced chemiluminescence Western blotting kit from Amersham. The α-Fur antiserum was precleaned as follows. The cell pellet of 20 ml of GSO72 (Δfur::kan strain) grown to optical density at 600 nm of 0.2 was resuspended in 100 μl of phosphate-buffered saline, mixed with 6 μl of lysozyme (10 mg/ml), and then frozen and thawed three times. The lysed cells were spread onto a nitrocellulose filter, and the filter was allowed to dry for 10 min. The filter was then incubated with the diluted Fur antiserum for 2 h. After the GSO72 filter was removed, the diluted serum was used to probe the experimental filters. The purified Fur protein was provided by C. Outten and T. O’Halloran.
RESULTS
OxyR activation of fur.
This work was initiated by a search for additional OxyR binding sites. Rather than carry out traditional genetic or biochemical screens, we used a computational approach based on information theory (26). With this approach, previously identified OxyR binding sites (31) were used as a model to search the E. coli genome sequence (5). The computer search indicated an OxyR binding site at coordinate 710103, centered at 234 bp upstream of the start codon of the fur gene (Fig. 1). This site has an information content of 14.5 bits, which is higher than those of the oxyR (13.9 bits), dps (12.3 bits), and gorA (11.2 bits) sites but lower than those of the katG (19.1 bits), ahpC (23.1 bits), and grxA (26.0 bits) sites.
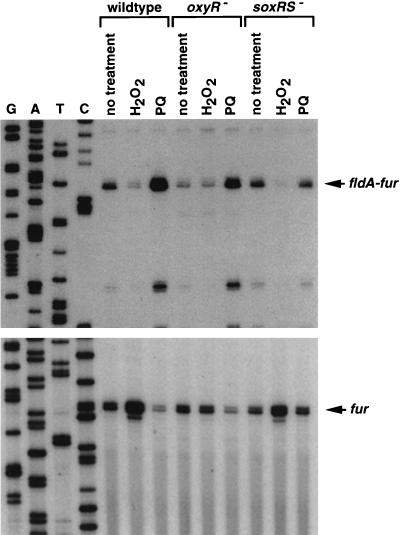
To determine whether fur is regulated by OxyR, we examined fur expression in wild-type and ΔoxyR::kan mutant strains treated with hydrogen peroxide. Primer extension assays carried out with two different primers showed that fur mRNA levels are induced by 1 mM hydrogen peroxide and that this induction is oxyR dependent (Fig. 2, bottom, and data not shown). Similar 10-fold induction was observed in rich (LB) and minimal (M63) medium. The start of the fur message was mapped to two adjacent G residues located more than 100 bases upstream of the two transcription starts reported by de Lorenzo et al. (10). The RNA sample analyzed in this previous study was derived from a fur-lacZ fusion plasmid carrying only 80 bp of the fur promoter. The previously mapped transcription start might correspond to a weak promoter or is an artifact of the reverse transcription reaction.
FIG. 2.
Primer extension assays of fur and fldA expression in wild-type, ΔoxyR, and ΔsoxRS strains grown in LB. Exponential-phase cultures were split into three aliquots; one aliquot was left untreated, one was treated with 1 mM hydrogen peroxide, and the third was treated with 0.1 mM paraquat (PQ). The cells were then harvested after 10 min. Total RNA was isolated, and primer extension assays were carried out with primers specific to fldA (5′-CCA AGC TGT TTT TGA ATC AT; top) and fur (5′-CAA GTC CCT GGC AAA TAT TG; bottom). The neighboring sequencing reactions were carried out with the same primers.
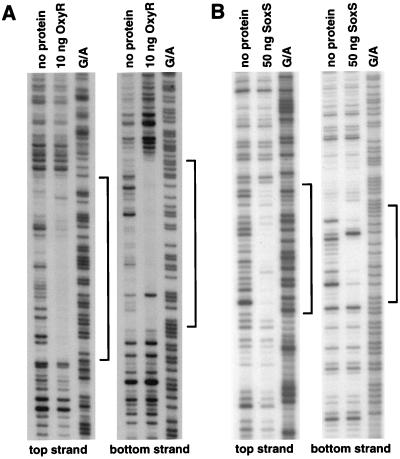
To test for OxyR binding to the promoter region of fur, we carried out DNase I footprinting experiments. As shown in Fig. 3A, the region protected from DNase I digestion directly corresponds to the binding site predicted by information theory (Fig. 1). The OxyR binding site is immediately upstream of the −35 region of the promoter, an arrangement that has been observed at other OxyR-activated promoters (31).
FIG. 3.
DNase I footprinting assays of OxyR binding to the fur promoter and SoxS binding to the fldA promoter. Protected regions on both strands of each promoter are indicated by the brackets. For the OxyR binding to the fur promoter, the BamHI-EcoRI fragment of pGSO97 was 32P labeled at either the BamHI site (top strand) or the EcoRI site (bottom strand), and the DNase I footprinting was carried out as described elsewhere (31). For the SoxS binding to the fldA promoter, the BamHI-HindIII fragment of pGSO98 was 32P labeled at either the BamHI site (top strand) or the HindIII site (bottom strand), and the DNase I footprinting was carried out as described elsewhere (21). The samples were run in parallel with Maxam-Gilbert G/A sequencing ladders.
SoxRS activation of fldA-fur.
The results described above and the intimate relationship between oxidative stress and iron metabolism prompted us to investigate the effect of paraquat treatment on expression of the fur gene. We observed a decrease rather than an increase of the main primer extension product detected with the fur-specific primer (Fig. 2, bottom). However, we noticed the presence of a longer extension product induced by paraquat in a soxRS-dependent fashion. We mapped the start of this extension product to the promoter region of the fldA gene, which is directly upstream of and in the same orientation as the fur gene (Fig. 2, top). Treatment with 0.1 mM paraquat led to a similar 10-fold induction of the fldA transcript in both rich and minimal media (data not shown).
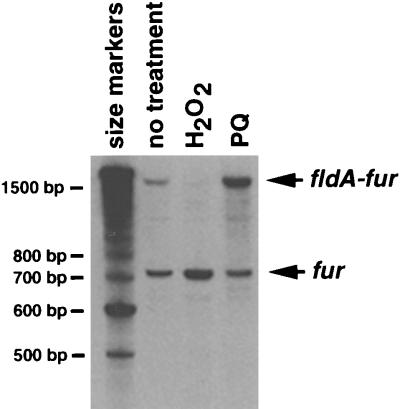
To determine whether the long extension product is from an mRNA that encompasses both fldA and fur, we carried out Northern blot analysis on the RNA samples used for the primer extension assays shown in Fig. 2. A transcript that is detected by both fldA (data not shown) and fur probes (Fig. 4) is induced by paraquat. Interestingly, hydrogen peroxide treatment leads to a significant decrease in the levels of the fldA-fur transcript while paraquat treatment leads to a slight decrease in the levels of the fur transcript. We do not know the reasons for this regulation.
FIG. 4.
Northern blot of total cellular RNA isolated from wild-type cells grown in LB which were left untreated or were exposed to 1 mM hydrogen peroxide or 0.1 mM paraquat (PQ). The blot was probed with a 32P-labeled fragment carrying the fur promoter (pGSO97). A 32P-labeled 100-bp DNA ladder was electrophoresed alongside the RNA to determine the approximate sizes of the RNA species. Correspondingly, the lengths of the upper and lower transcripts are estimated to be 1.5 and 0.7 kb, respectively.
To test for SoxS binding to the promoter region of fldA, we again carried out DNase I footprinting assays (Fig. 3B). The region protected by SoxS spans 25 bases on the coding strand (Fig. 1 and 3B) and contains a GCAC sequence which matches the previously determined SoxS binding motif (22).
High levels of Fur protein.
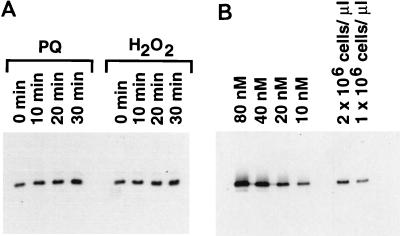
To assess whether the OxyR and SoxRS induction of fur transcription leads to changes in Fur protein levels, we carried out immunoblot analysis. As shown in Fig. 5A, both hydrogen peroxide treatment and paraquat treatment led to a twofold increase in the intracellular Fur protein levels within 30 min. By comparing the cellular Fur levels with defined amounts of purified Fur protein, we estimate that the intracellular Fur concentration in nontreated cells during exponential growth to be approximately 5,000 molecules/cell (Fig. 5B). Thus, oxidative stress leads to a concentration of approximately 10,000 Fur molecules/cell. These levels are similar to the 2,500 Fur molecules/cell determined for Vibrio cholerae (33).
FIG. 5.
Immunoblot analysis of Fur protein levels. (A) Wild-type cells (MC4100) grown to exponential phase in LB were treated with 0.2 mM hydrogen peroxide or 0.1 mM paraquat (PQ), and samples were taken at the indicated times. The cell pellets were then resuspended in protein loading buffer to give a final concentration of 2 × 106 cells/μl. An aliquot (5 μl) of each sample was loaded on an SDS-polyacrylamide gel and probed with Fur antiserum. (B). Purified Fur protein was suspended in protein loading buffer to give final concentrations of 80, 40, 20, and 10 nM. Aliquots (5 μl) of each purified Fur sample together with aliquots (5 and 2.5 μl) of the sample from the untreated cells were loaded in the indicated lanes of an SDS-polyacrylamide gel and probed with Fur antiserum.
DISCUSSION
We have shown that the primary regulators of the E. coli response to oxidative stress, OxyR and SoxRS, modulate the levels of the ferric uptake regulator Fur. OxyR, which senses elevated levels of hydrogen peroxide, binds to the fur promoter and induces the expression of a transcript encoding Fur. SoxR and SoxS, which modulate the response to superoxide-generating compounds, activate the expression of a transcript encoding both flavodoxin and Fur. This is achieved by SoxS binding to the promoter region of fldA. The induction of fur transcription by OxyR and SoxRS leads to an increase in Fur protein concentration. We propose that this regulatory network may also exist in other bacterial organisms, especially since Fur is ubiquitous among prokaryotes. We have observed that the fldA-fur gene arrangement is conserved in Klebsiella pneumoniae (1), Haemophilus influenzae, Yersinia pestis, and Actinobacillus actinomycetemcomitans.
It is well documented that a major portion of the hydrogen peroxide toxicity in E. coli can be attributed to DNA damage which is caused by hydroxyl radicals generated by hydrogen peroxide and intracellular iron via the Fenton reaction (18). In addition, recent work has demonstrated that superoxide toxicity is mainly due to its role in accelerating the Fenton reaction (20, 24). Superoxide attacks iron-sulfur proteins, leading to the release of iron that can participate in the Fenton reaction. Since the toxicities of both hydrogen peroxide and superoxide are exacerbated by iron, Fur induction by both OxyR and SoxRS reflects the chemistry of oxidative stress. The finding that Fur is induced by oxidative stress is also consistent with the observation that Fur provides protection against oxidative damage and mutagenesis (32). However, the exact consequences of the twofold increase in Fur levels are unclear. Possibly, new Fur synthesis is required to replace Fur that is damaged by oxidants reacting with metals bound by the repressor. The increase in Fur levels may also be required for increased repression of iron uptake.
In general, the very high levels of the Fur protein raise questions as to the role of this transcription factor. We have found that there are approximately 5,000 Fur molecules/cell in exponentially growing E. coli cultures and approximately 10,000 Fur molecules/cell after oxidative stress. The levels that we have determined for E. coli are comparable with the 2,500 Fur molecules/cell reported for V. cholerae (33) but much higher than the levels of other transcription factors. For example, there are estimated to be 10 to 20 copies of the LacI repressor and 50 to 300 copies of the Trp repressor per cell (12, 19). One possible reason for the high concentration of Fur protein is that there are many predicted Fur sites in the E. coli genome (20a). The regulation of iron uptake genes, however, may not be the sole function of Fur. Possibly, Fur sequesters iron in order to reduce the levels of DNA-bound iron. This function would be similar to a function ascribed to the ferritin-like, nonspecific DNA binding protein Dps (13), whose expression is also regulated by OxyR. Another role of Fur may be to catalyze the breakdown of hydrogen peroxide. The dismutation of hydrogen peroxide is a thermodynamically favorable reaction (ΔE = +1.06 V) and could be catalyzed by the Fur-bound iron cycling between the Fe2+ and Fe3+ oxidation states. Roles in addition to transcriptional regulation should be considered in further studies of Fur function.
We have also shown that the expression of flavodoxin A is coinduced with Fur when cells are treated with paraquat. Flavodoxin together with ferredoxin/flavodoxin-NADP+ reductase (encoded by fpr) functions as a versatile reduction system for many metalloproteins. It is required for the conversion of dethiobiotin to biotin and the activation of anaerobic ribonucleotide reductase, anaerobic pyruvate formate-lyase, and vitamin B12-dependent methionine synthase (reviewed in reference 17). Interestingly, the expression of the ferredoxin/flavodoxin-NADP+ reductase is also regulated by SoxRS (23). In addition, fpr deletion strains are hypersensitive to paraquat killing and ferredoxin/flavodoxin-NADP+ reductase-overexpressing strains are resistant (4). However, the roles of flavodoxin and the ferredoxin/flavodoxin-NADP+ reductase in protecting against oxidative stress are not clear. We speculate that one role of the flavodoxin reduction system might be to keep Fe-S clusters reduced and thus resistant to superoxide attack. Another possible role of flavodoxin and the ferredoxin/flavodoxin-NADP+ reductase may be to modulate the redox state of the SoxR protein. In fact, compared to a wild-type control strain, we have observed stronger initial soxS induction followed by a faster decay of soxS levels after paraquat treatment of an flavodoxin overexpression strain (data not shown).
Our findings, together with the report that Fur binds to its own promoter (10), suggest that the regulation of fur expression is complex. This conclusion is reinforced by additional computation searches which indicate that the cyclic AMP receptor protein and MarA regulator may also bind in the fldA-fur region (28). Our results also illustrate the coordination between different regulatory pathways in the response to oxidative stress. Given the wide range of oxidative damage to the cell, it is conceivable that DNA repair, protein degradation, metabolic energy generation, cell division, and other cellular activities are all coordinately regulated. Together, these activities form an oxidative stress response network. Mapping out the connectivity of this network and quantitatively modelling the regulation is one important direction for future studies of cellular defenses against oxidative stress.
ACKNOWLEDGMENTS
We thank B. Demple and L. Rosner for strains, T. O’Halloran and C. Outten for purified Fur, and M. Vasil for Fur antiserum. We also appreciate the editorial comments of T. O’Halloran and D. Touati.
M.Z. acknowledges a postdoctoral fellowship from American Cancer Society.
REFERENCES
- 1.Achenbach L A, Genova E G. Transcriptional regulation of a second flavodoxin gene from Klebsiella pneumoniae. Gene. 1997;194:235–240. doi: 10.1016/s0378-1119(97)00168-6. [DOI] [PubMed] [Google Scholar]
- 2.Bagg A, Neilands J B. Mapping of a mutation affecting regulation of iron uptake systems in Escherichia coli K-12. J Bacteriol. 1985;161:450–453. doi: 10.1128/jb.161.1.450-453.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagg A, Neilands J B. Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry. 1987;26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 4.Bianchi V, Haggård-Ljungquist E, Pontis E, Reichard P. Interruption of the ferredoxin (flavodoxin) NADP+ oxidoreductase gene of Escherichia coli does not affect anaerobic growth but increases sensitivity to paraquat. J Bacteriol. 1995;177:4528–4531. doi: 10.1128/jb.177.15.4528-4531.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blattner F R, Plunkett G, Bloch C A, Perna N T, Burland V, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Braun V. Avoidance of iron toxicity through regulation of bacterial iron transport. Biol Chem. 1997;378:779–786. [PubMed] [Google Scholar]
- 7.Braun V, Hantke K, Köster W. Bacterial iron transport: mechanisms, genetics, and regulation. In: Sigel A, Sigel H, editors. Metal ions in biological systems. New York, N.Y: Marcel Dekker; 1998. pp. 67–145. [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.De Lorenzo V, Wee S, Herrero M, Neilands J B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol. 1987;169:2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Lorenzo V, Herrero M, Giovannini F, Neilands J B. Fur (ferric uptake regulation) protein and CAP (catabolite-activator protein) modulate transcription of fur gene in Escherichia coli. Eur J Biochem. 1988;173:537–546. doi: 10.1111/j.1432-1033.1988.tb14032.x. [DOI] [PubMed] [Google Scholar]
- 11.Gaudu P, Moon N, Weiss B. Regulation of the soxRS oxidative stress regulon. Reversible oxidation of the Fe-S centers of SoxR in vivo. J Biol Chem. 1997;272:5082–5086. doi: 10.1074/jbc.272.8.5082. [DOI] [PubMed] [Google Scholar]
- 12.Gilbert W, Müller-Hill B. Isolation of the lac repressor. Proc Natl Acad Sci USA. 1966;56:1891–1898. doi: 10.1073/pnas.56.6.1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grant R A, Filman D J, Finkel S E, Kolter R, Hogle J M. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Biol. 1998;5:294–303. doi: 10.1038/nsb0498-294. [DOI] [PubMed] [Google Scholar]
- 14.Greenberg J T, Monach P, Chou J H, Josephy P D, Demple B. Positive control of a global antioxidant defense regulon activated by superoxide-generating agents in Escherichia coli. Proc Natl Acad Sci USA. 1990;87:6181–6185. doi: 10.1073/pnas.87.16.6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hantke K. Regulation of ferric iron transport in Escherichia coli K12: isolation of a constitutive mutant. Mol Gen Genet. 1981;182:288–292. doi: 10.1007/BF00269672. [DOI] [PubMed] [Google Scholar]
- 16.Hidalgo E, Ding H, Demple B. Redox signal transduction: mutations shifting [2Fe-2S] centers of the SoxR sensor-regulator to the oxidized form. Cell. 1997;88:121–129. doi: 10.1016/s0092-8674(00)81864-4. [DOI] [PubMed] [Google Scholar]
- 17.Hoover D M, Ludwig M L. A flavodoxin that is required for enzyme activation: the structure of oxidized flavodoxin from Escherichia coli at 1.8 Å resolution. Protein Sci. 1997;6:2525–2537. doi: 10.1002/pro.5560061205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imlay J A, Chin S M, Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988;240:640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- 19.Kelley R L, Yanofsky C. trp aporepressor production is controlled by autogenous regulation and inefficient translation. Proc Natl Acad Sci USA. 1982;79:3120–3124. doi: 10.1073/pnas.79.10.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Keyer K, Imlay J A. Superoxide accelerates DNA damage by elevating free-iron levels. Proc Natl Acad Sci USA. 1996;93:13635–13640. doi: 10.1073/pnas.93.24.13635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20a.Lewis, K., M. Zheng, G. Storz, and T. D. Schneider. Unpublished observation.
- 21.Li Z, Demple B. SoxS, an activator of superoxide stress genes in Escherichia coli. Purification and interaction with DNA. J Biol Chem. 1994;269:18371–18377. [PubMed] [Google Scholar]
- 22.Li Z, Demple B. Sequence specificity for DNA binding by Escherichia coli SoxS and Rob proteins. Mol Microbiol. 1996;20:937–945. doi: 10.1111/j.1365-2958.1996.tb02535.x. [DOI] [PubMed] [Google Scholar]
- 23.Liochev S I, Hausladen A, Beyer W F, Fridovich I. NADPH:ferredoxin oxidoreductase acts as a paraquat diaphorase and is a member of the soxRS regulon. Proc Natl Acad Sci USA. 1994;91:1328–1331. doi: 10.1073/pnas.91.4.1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCormick M L, Buettner G R, Britigan B E. Endogenous superoxide dismutase levels regulate iron-dependent hydroxyl radical formation in Escherichia coli exposed to hydrogen peroxide. J Bacteriol. 1998;180:622–625. doi: 10.1128/jb.180.3.622-625.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schneider T D. Reading of DNA sequence logos: prediction of major groove binding by information theory. Methods Enzymol. 1996;274:445–455. doi: 10.1016/s0076-6879(96)74036-3. [DOI] [PubMed] [Google Scholar]
- 26.Schneider T D. Information content of individual genetic sequences. J Theor Biol. 1997;189:427–441. doi: 10.1006/jtbi.1997.0540. [DOI] [PubMed] [Google Scholar]
- 27.Schneider T D. Sequence walkers: a graphical method to display how binding proteins interact with DNA or RNA sequences. Nucleic Acids Res. 1997;25:4408–4415. doi: 10.1093/nar/25.21.4408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider, T. D. 30 March 1999, posting date. Fur region lister maps. [Online.] http://www.lecb.ncifcrf.gov/∼toms/paper/oxyrfur/. [16 June 1999, last date accessed.]
- 29.Storz G, Imlay J A. Oxidative stress. Curr Opin Microbiol. 1999;2:188–194. doi: 10.1016/s1369-5274(99)80033-2. [DOI] [PubMed] [Google Scholar]
- 30.Storz G, Altuvia S. OxyR regulon. Methods Enzymol. 1994;234:217–223. doi: 10.1016/0076-6879(94)34088-9. [DOI] [PubMed] [Google Scholar]
- 31.Toledano M B, Kullik I, Trinh F, Baird P T, Schneider T D, Storz G. Redox-dependent shift of OxyR-DNA contacts along an extended DNA-binding site: a mechanism for differential promoter selection. Cell. 1994;78:897–909. doi: 10.1016/s0092-8674(94)90702-1. [DOI] [PubMed] [Google Scholar]
- 32.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Watnick P I, Eto T, Takahashi H, Calderwood S B. Purification of Vibrio cholerae Fur and estimation of its intracellular abundance by antibody sandwich enzyme-linked immunosorbent assay. J Bacteriol. 1997;179:243–247. doi: 10.1128/jb.179.1.243-247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng M, Åslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]