Abstract
Background
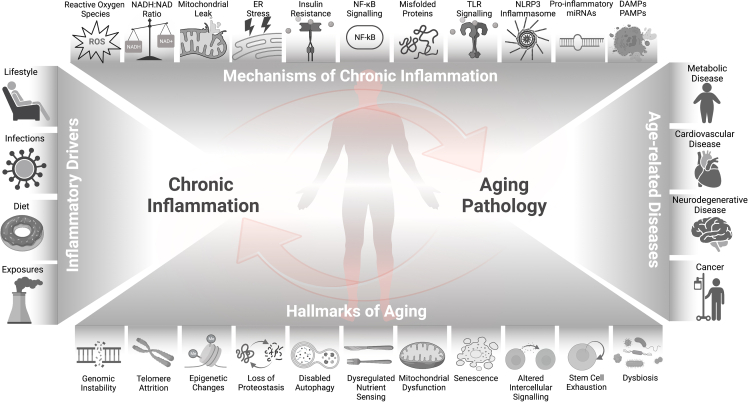
Recently, the hallmarks of aging were updated to include dysbiosis, disabled macroautophagy, and chronic inflammation. In particular, the low-grade chronic inflammation during aging, without overt infection, is defined as “inflammaging,” which is associated with increased morbidity and mortality in the aging population. Emerging evidence suggests a bidirectional and cyclical relationship between chronic inflammation and the development of age-related conditions, such as cardiovascular diseases, neurodegeneration, cancer, and frailty. How the crosstalk between chronic inflammation and other hallmarks of aging underlies biological mechanisms of aging and age-related disease is thus of particular interest to the current geroscience research.
Scope of review
This review integrates the cellular and molecular mechanisms of age-associated chronic inflammation with the other eleven hallmarks of aging. Extra discussion is dedicated to the hallmark of “altered nutrient sensing,” given the scope of Molecular Metabolism. The deregulation of hallmark processes during aging disrupts the delicate balance between pro-inflammatory and anti-inflammatory signaling, leading to a persistent inflammatory state. The resultant chronic inflammation, in turn, further aggravates the dysfunction of each hallmark, thereby driving the progression of aging and age-related diseases.
Main conclusions
The crosstalk between chronic inflammation and other hallmarks of aging results in a vicious cycle that exacerbates the decline in cellular functions and promotes aging. Understanding this complex interplay will provide new insights into the mechanisms of aging and the development of potential anti-aging interventions. Given their interconnectedness and ability to accentuate the primary elements of aging, drivers of chronic inflammation may be an ideal target with high translational potential to address the pathological conditions associated with aging.
Keywords: Aging, Ageing, Inflammation, Inflammaging, Nutrient sensing, Senescence
1. Introduction
Aging is a complex process that affects humans at the molecular, cellular, tissue, and systemic levels. It results, in part, from the compounding accumulation of damage and waning repair mechanisms at each hierarchical level, altogether contributing to the development of age-related diseases (ARDs) later in life. Therefore, to no surprise, aging is the single most important risk factor for many chronic diseases contributing to morbidity and mortality. While aging predisposes older populations to more severe infection resulting from communicable diseases, such as influenza and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1,2], this review will focus on the interplay of inflammaging and aging hallmarks associated with non-communicable diseases such as cardiovascular diseases (CVDs), cancer, osteoarthritis, type 2 diabetes (T2D), and neurodegenerative diseases such as Alzheimer's disease (AD) and Parkinson's disease (PD) [3,4].
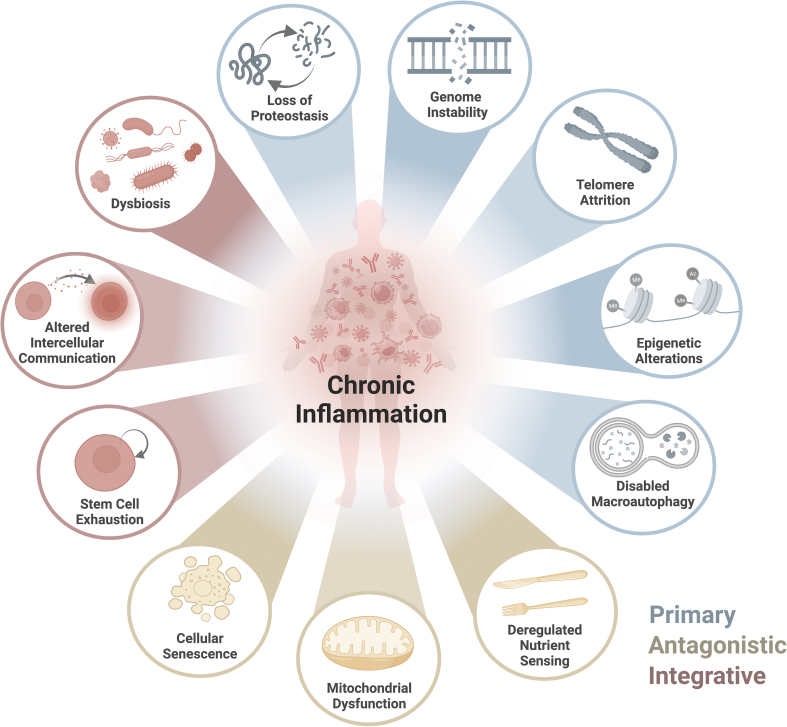
Despite many proposed theories, none alone comprehensively captures the aging process. Age-related changes were traditionally depicted by nine cellular and molecular hallmarks of aging that include 1) genomic instability, 2) telomere attrition, 3) epigenetic alterations, 4) loss of proteostasis, 5) deregulated nutrient sensing, 6) mitochondrial dysfunction, 7) cellular senescence, 8) stem cell exhaustion and 9) altered intercellular communication [5] (Figure 1). More recently, several concepts have emerged in light of advances in high throughput multi-omics analysis, such as transcriptomics, proteomics, and epigenomics. In a recent review, López-Otin and colleagues expanded on the original nine hallmarks of aging, including disabled macroautophagy, dysbiosis, and chronic inflammation [6]. Each hallmark not only manifests during normal aging but the corresponding aging pathology can be exacerbated or reduced through experimental aggravation or amelioration of the hallmark [5,6]. The large degree of mechanistic interplay between the hallmarks makes the hallmark relationships best characterized as a web of bidirectional loop interactions rather than stand-alone pillars.
Figure 1.
Chronic Inflammation and the Hallmarks of Aging: Relationships of chronic inflammation and other hallmarks of aging: genomic instability, telomere attrition, epigenetic changes, loss of proteostasis, and disabled autophagy grouped as the primary hallmarks; deregulated nutrient sensing, mitochondrial dysfunction, and cellular senescence grouped as the antagonistic hallmarks; altered intercellular communication, stem cell exhaustion, and dysbiosis grouped as the integrative hallmarks (López-Otín, 2023).
Under the geroscience hypothesis, failure in this “network of aging” and their homeostatic mechanisms accelerates the pace of aging and susceptibility to ARDs. An integrated and holistic approach to aging biology emphasizes the significance of influential factors across the network of hallmarks, including diet, exercise, and other lifestyle factors. The human immune system, specifically inflammation, is also largely considered one of these ubiquitous factors that, to some degree, impacts each of these processes [7]. Inflammation is a broad term referring to the defense mechanisms that evolved to protect an organism from infection and injury. While acute inflammation is a cascade of steps in response to infection or injury that ultimately clears invading pathogens and incites wound healing, chronic inflammation is a potentially pathologic process arising from the perpetuity of the initial trigger or the dysregulation of signaling pathways that is harmful to health. “Inflammaging” describes this non-resolving, low-grade, and chronic inflammation process that progresses with age [8,9].
This review emphasizes the broad overview of bidirectional relationships between chronic inflammation and the other 11 hallmarks of aging, which contribute to “inflammaging.” We will dedicate additional discussion on altered nutrient sensing.
2. Primary hallmarks
The primary hallmarks, either individually or through synergy, can lead to damage on a molecular and cellular level. The cumulative effects of this damage are universally negative and believed to be involved in the instigation and progression of ARDs.
2.1. Genomic instability
Genomic instability refers to DNA alterations, including changes in nucleic acid sequences and aneuploidies [10]. These events can irreversibly alter the content of the genome, contributing to detrimental changes in gene expression, cell division, senescence, and death. Altogether, these cell outcomes conspire towards functional impairment at the tissue level and ultimately manifest accelerated aging pathology and ARDs as cells accumulate nuclear and mitochondrial genomic damage with age. Maintenance of genomic stability is essential for cellular integrity to prevent errors from DNA replication by endogenous and exogenous challenges. Each day, endogenous insults - including replication errors and free radical damage during normal metabolism - result in as many as 104–105 DNA lesions per human cell [11,12]. While aberration from exogenous exposures (such as ultraviolet light (UV), ionizing radiation, mutagenic chemicals, etc.) are largely unique to each individual's biography of exposures, the relatively high baseline rate of DNA damage suggests that the stability of the genome largely hinges on effective DNA repair processes which diminish with age rather than avoiding damage altogether. This decline in repair capacity is thought to contribute to the accumulation of genomic insults and accelerated aging pathology later in life, as well as the stepwise increase in cancer with age [12,13].
Chronic inflammation and oxidative stress are interconnected pathological processes that lead to genomic damage and instability (Figure 2). This process is most keenly characterized in the inflammation-mediated initiation and progression of cancers, where chronic inflammation has been shown to account for 20–25% of cancers [14] and influence mitogenicity [15]. However, the inflammation-induced genomic instability that potentiates the fitness of cancer cells can also result in the cellular and tissue debility characteristic of aging decline [16]. Therefore, the relationship between chronic inflammation and genomic instability is multifaceted and bidirectional.
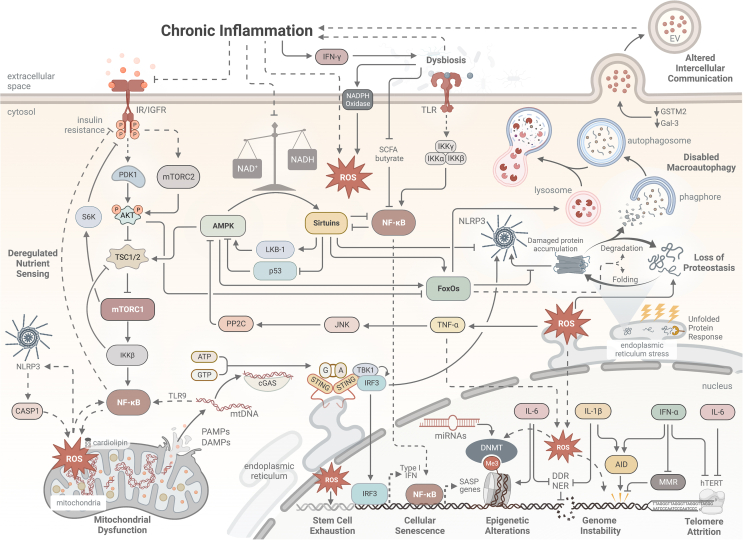
Figure 2.
Proposed Mechanisms of Relationships Underlying Chronic Inflammation and the Hallmarks of Aging: Chronic inflammation interacts with other hallmarks of aging via a complex network of intra- and extracellular signalings, including but not limited to nutrient-sensing, danger-sensing, and autophagy pathways, acting as both a cause and a result of aging. A = adenine, AID = activation-induced cytidine deaminase, AKT = Protein kinase B, AMPK = 5′-adenosine monophosphate-activated protein kinase, ATP = adenosine triphosphate, CASP1 = caspase-1, cGAS = cyclic GMP–AMP synthase (cGAS), DAMPs = deoxyribonucleic acid damage-associated molecular patterns, DDR = deoxyribonucleic acid damage response, DNMT = methyltransferase, EV = extracellular vesicle, FoxO = forkhead box protein O, G = guanine, GSTM2 = Glutathione S-transferase Mu 2, GTP = guanine triphosphate, hTERT = human telomerase reverse transcriptase, IFN = interferon, IGFR = insulin-like growth factor receptor, IKK = inhibitor of nuclear factor-κB (IκB) kinase, IL = interleukin, IR = insulin receptor, IRF = Interferon regulatory factors, JNK = jun n-terminal kinase, LKB-1 = Liver Kinase B1, Me3 = methyl, MMR = mismatch repair, mtDNA = mitochondrial eoxyribonucleic acid, mTORC = mechanistic target of rapamycin complex, NAD/NADH = nicotinamide adenine dinucleotide, NER = nucleotide excision repair, NF-κB = nuclear factor kappa B, NLRP3 = nod-like receptor family pyrin domain-containing 3, PAMPs = pathogen-associated molecular pattern molecules, PDK1 = 3-phosphoinositide-dependent protein kinase-1, PP2C = Protein phosphatase 2C, ROS = reactive oxygen species, S6K = Ribosomal protein S6 kinase, SASP = senescence-associated secretory phenotype, SCFA = short chain fatty acid, STING = stimulator of interferon genes, TBK = tank-binding kinase 1, TLR = toll-like receptor, TNF = tumor necrosis factor, TSC = tuberous sclerosis complex.
2.1.1. Genomic instability leads to inflammation
Chronic inflammation can also be caused by genomic damage and instability by several mechanisms generating a bidirectional feedback loop. Firstly, reactive oxygen species (ROS) induced single-stranded breaks, which can be detected by the DNA-break sensor, poly (adenosine diphosphate-ribose) polymerase 1 (PARP1) [17]. Persistent PARP1 activation, however, depletes NAD+, which dampens downstream sirtuin (Sirt) activities [18,19]. The resultant Sirt3 and Sirt5 depletion can mediate the accruement of mitochondrial DNA damages, leading to the pro-inflammatory cytokine profile of mitochondrial dysfunction-associated senescence [20]. Impaired functions of other Sirts, including Sirt2, also lead to higher nod-like receptor family pyrin domain-containing 3 (NLRP3) inflammasome activity, which is associated with accelerated aging [21] and altered Sirt-dependent nutrient-sensing pathways [22]. These intertwined impairments fuel accelerated aging pathology by promoting inflammation and senescence through mechanisms introduced in the following sections (Inflammation & Cellular Senescence).
DNA damage can also directly trigger inflammation through leakage of DNA into the cytoplasm, which can occur through ruptured micronuclei or DNA end resection [23]. Cytoplasmic DNA can prompt the cyclic GMP–AMP synthase (cGAS) stimulator of interferon gene (STING) pathway to activate tank-binding kinase 1 (TBK1) and interferon regulatory factor 3 (IRF3) to induce a type I interferon (IFN) response and nuclear factor kappa B (NF-κB) cascade. DNA damage response to single and double-stranded breaks also recruits ataxia telangiectasia and Rad3-related protein (ATR) and ataxia-telangiectasia mutated (ATM), which activate NF-κB through GATA4 stabilization [24,25] or directly activate tumor necrosis factor receptor-associated factor 6 (TRAF6) or STING [26,27]. Other events associated with DNA damage, including DNA repair deficiencies, activation of transposons, cellular senescence, R-loop formation, and changes in chromatin structure that link DNA damage to inflammation and aging, have been recently reviewed in detail [24,28].
Finally, if not corrected, genomic instability can lead to neoantigens in cells, which predispose cells to be targeted by the immune system and contribute to inflammation. This phenomenon is exemplified in the susceptibility of microsatellite-unstable tumors, which are mismatch repair (MMR)-deficient, to treatment with immunotherapies, such as immune checkpoint inhibitors [29].
2.1.2. Inflammation causes genomic instability
Chronic inflammation has been shown to induce genomic instability through multiple mechanisms. Among pro-inflammatory cytokines, tumor necrosis factor-alpha (TNF-α) is commonly elevated with age [30] and induces ROS through cell-specific and common mechanisms. Specifically, binding of TNF-α to its receptors induces downstream phosphorylation of p47phox (phox: phagocyte oxidase), which recruits TRAF4. This complex then translocates to the plasma membrane, facilitating nicotinamide adenine dinucleotide phosphate hydrogen (NADPH) oxidase activity [31,32]. Despite the effectiveness of ROS production in pathogen defense and cell signaling, ROS can directly cause damage to macromolecules within cells, and DNA is one of the most susceptible biological targets of oxidative stress due to its limited structural stability [33]. ROS-mediated deleterious effects are thought to contribute to human age-related degenerative conditions, including AD [34] and PD [35], CVD [36], pulmonary fibrosis [37], nonalcoholic fatty liver disease (NAFLD) [38], and reproductive decline [39,40]. A more comprehensive review of the impact of inflammation-induced ROS and reactive nitrogen species (RNS) on DNA base damage, as well as strand breaks, has been published recently [41].
Chronic inflammation also downregulates MMR proteins, such as MutL homolog 1 (MLH1), through various mechanisms attributable to TNF-α, interleukin (IL)-1β, and Prostaglandin E1 (PGE1) signaling (Figure 2) [42]. Mutations or epigenetic silencing of MMR members is associated with increased genetic instability, termed microsatellite instability (MSI) which accentuates the accumulation of DNA replication errors throughout the genome [43]. Additionally, IL-6, a marker of chronic inflammation, appears to negatively impact the nucleotide excision repair pathway, which is responsible for repairing a wide variety of DNA lesions caused by UV radiation, mutagenic chemicals, and chemotherapeutic agents [44]. The underlying mechanisms, however, remain unclear.
Furthermore, activation-induced cytidine deaminase (AID) is a DNA mutator enzyme induced by inflammation [45]. Evolutionarily, AID functions in activated B cells to generate immune diversity by inducing somatic hypermutation and class-switch recombination in immunoglobulin genes [46]. Under chronic inflammation, however, AID can be aberrantly expressed in epithelial cells and induce somatic mutations and chromosomal aberrations [46]. The expression of AID is induced by age-associated inflammatory cytokines, including NF-κB, IL-4, and TGF-β (transforming growth factor-beta) [47,48] and overexpressed in several inflammatory-mediated cancers and non-cancer-related inflammatory diseases such as chronic gastritis [49], ulcerative colitis [50], and pancreatitis [51].
Finally, inducible nitric oxide synthase (NOS2), which produces NO, and whose expression is associated with inflammation, was recently shown to reduce DNA methyltransferase 1 (DNMT1), resulting in long interspersed nuclear element 1 (LINE1) retrotransposon hypomethylation, expression, and DNA damage [52]. In addition, LINE1 induction can drive type I IFN secretion in senescent murine cells, thereby contributing to the formation of a cycle where inflammation further facilitates DNA damage, maintenance of senescence secretory products and inflammaging [53]. The interplay between epigenetics, inflammation and DNA damage is discussed further in subsequent sections (Inflammation & Epigenetic Changes).
2.2. Telomere attrition
Somatic cells, including B cells and T cells, have a finite capacity to divide due to the inability of DNA polymerase to completely synthesize the distal ends, or telomeres, of eukaryotic DNA [54]. Thus, after a certain number of cell cycles known as the “Hayflick Limit” [55], telomere length decreases to a critical length, after which the cell ceases to replicate and instead diverts to cellular senescence or apoptosis through DNA damage response (DDR) signaling [56]. In humans, the lack of somatic expression of telomerase, the enzyme necessary for the maintenance of telomere length, in most cells accelerates the progression of telomere erosion, which has been shown to contribute to a pro-inflammatory milieu, the development of chronic diseases, and shorter life expectancy [57]. Evidence suggests that age-associated inflammation also worsens telomeric loss [58,59]. Shortened telomeres or altered telomere structures can ultimately lead to apoptosis [60], cellular senescence [61], and a state of extensive genomic instability known as “telomere crisis,” linking telomere attrition to other hallmarks of aging [62,63]. This section will briefly discuss links between telomere attrition and chronic inflammation in aging.
2.2.1. Telomere attrition promotes inflammation
Inflammation can be triggered and maintained on several levels by telomere dysfunction. For example, the telomere-mitochondria axis has recently been identified as a novel contributor to inflammation [64]. In addition, during aging, DNA damage and telomere dysfunction activate p53, which suppresses peroxisome proliferator-activated receptor gamma coactivator 1 alpha (PGC-1α) and mitochondrial Sirts (Sirt3/4/5) [65,66], thus impairing mitochondrial function and leading to over-production of ROS [67]. The resultant oxidative stress promotes inflammation and further damages telomere structures [68]. Additionally, in macrophages from late-generation telomerase-deficient mice, downregulation of PGC-1α induces activation of the NLRP3 inflammasome, resulting in an amplified inflammatory response upon bacterial challenge [69]. Increased caspase-1 from NLRP3 exacerbates inflammation by facilitating the conversion of pro-IL-18 to mature IL-18, which is sourced from telomere attrition-induced ATM/cABL activation and yes-associated protein 1 (YAP1) nuclear translocation [62]. Furthermore, telomere dysfunction resulting from shelterin loss also triggers autophagic cell death by activating the cGAS-STING pathway [70], which can initiate an inflammatory cascade via IRF3-TBK1–mediated upregulation of Type I interferons, leading to enhanced cross-priming activity and recruitment of IFNγ-producing CD8+ T cells [71].
Aging is also associated with telomere shortening in T cells [72], which is associated with a loss of naive T cell pool, lower T cell receptor diversity, and accumulation of antigen-experienced and even senescent T cells exhibiting a pro-inflammatory profile, such as increased IL-6 and TNF-α [[73], [74], [75]]. Overall, these changes lead to impaired functional responsiveness of the aged immune systems to emerging antigens, such as SARS-CoV-2 [76,77]. Interestingly, some CD4+ (especially naive and central memory) T cells can elongate their telomeres by acquiring telomere-containing vesicles from antigen-presenting cells (APCs), thus gaining protection from immunosenescence and conferring long-lasting immune response [78]. However, it remains to be studied if the vesicle-mediated telomere acquisition can be harnessed for its therapeutic potential to ameliorate systemic inflammaging.
2.2.2. Inflammation drives telomere attrition
In multiple pathological conditions of aging, inflammation can also contribute to telomere attrition. On the tissue level, studies have demonstrated a direct link between local inflammation and telomere attrition, including gastritis [79], NAFLD [80], and chronic obstructive pulmonary disease (COPD) [81]. Chronic inflammation markers, such as TNF-α and IL-6, are associated with shortened telomeres in non-cancerous cells [82], though this association in malignant cells is less understood [83]. TNF-α, specifically, has been shown to exhibit a causal role in downregulating telomerase activity through downstream phosphorylation of ATF7 by p38, resulting in accelerated telomere shortening associated with aging in mice [84]. The oxidative stress resulting from age-associated mitochondrial dysfunction and ROS production can also accelerate telomere shortening by generating irreparable single-strand breaks in telomere regions [85,86], which in itself can incite more inflammation. Similarly, ROS produced by neutrophils when fighting pathogens facilitates the spreading of telomere attrition and senescence to neighboring non-immune cells through paracrine signaling [58]. In addition, age-associated inflammation depletes cellular NAD+ levels, thus limiting the availability of Sirts [[87], [88], [89]]. Reduced NAD + levels may downregulate Sirt6 activity, lowering telomere stability and increasing attrition [65,66,90]; however, further research is needed to further characterize this mechanism as direct or indirect.
Type I IFNs, pro-inflammatory cytokines secreted during viral infection, are drivers for T cell senescence [91] and have also been shown to inhibit telomerase activity and promote telomere erosion in stimulated CD8+ T cells [92] and CD4+ T cells [93]. Specifically, IFN-α is suggested to inhibit human telomerase reverse transcriptase (hTERT) activity by decreasing activation of NF-κB that can activate hTERT, despite its pro-inflammatory potential [92] (Figure 2). Persistent activation of NF-κB, however, contributes to telomere shortening in certain cell types, including muscle stem cells (MuSCs) and hepatocytes [94,95].
2.3. Epigenetic alterations
The interconnection between epigenetics and inflammaging is an important area in contemporary aging research. Age-associated epigenetic modulations are increasingly shown to favor the formation of a pro-inflammatory environment, which can, in turn, remodel epigenetic patterns [96]. Changes in DNA methylation and histone modifications during aging can lead to inflammatory consequences, while age-dependent alterations in non-coding RNA (ncRNA) and retrotransposons (RTPs) [97] can regulate genes involved in inflammaging, exhibiting potentials for novel therapeutic targets for ARDs [98].
2.3.1. DNA methylation
Aging has been associated with abnormal patterns of DNA hypo- and hypermethylation at promoter CpG islands [96,99]. The DNA hypomethylation during aging can be partially attributable to the decreased activity of DNMTs, such as DNMT-1 and DNMT-3B [100] (Figure 2). During aging, loss of DNA methylation usually activates genes involved in inflammatory responses, such as NF-κB signaling, macrophage activation, and IFN signaling [101]. Age-associated DNA hypomethylation, caused by tissue necrosis and incomplete clearance of apoptotic cells, also impacts cell-free DNA (cfDNA) [102]. The unmethylated cfDNA resembles microbial DNA, as sensed by DNA-sensing receptors, such as cGAS and toll-like receptor (TLR)-9, leading to inflammatory responses [103].
However, persistent inflammation can also contribute to aberrant DNA methylation. Recently, a large multi-ethnic epigenome-wide association study suggests that the altered CpG methylation patterns can result from elevated C-reactive protein (CRP) during chronic inflammation. This inflammatory methylation signature can significantly increase the risk for cardiometabolic diseases and COPD [104]. At the molecular level, oxidative stress triggered by chronic inflammation can relocalize DNMTs to guanine and cytosine-rich regions, causing DNA methylation in potentially undesirable genes [105]. For example, DNA hypermethylation at the promoter regions of autophagy-related 5 (ATG5) or microtubule-associated protein 1A/1B-light chain 3 (LC3-B) can downregulate autophagy in aging murine macrophages [106]. Inflammaging can also alter epigenetic patterns in several chronic diseases, including neurodegeneration [107], ulcerative colitis [108], Helicobacter pylori infection [109], and hepatitis [110].
2.3.2. Histone modification
The N terminal tails of the four core histones (H2A, H2B, H3, and H4) are subjected to post-translational modifications, including acetylation, methylation, and phosphorylation, and depending on histones residues and modification types, gene transcription can be activated or repressed [100]. The activated, accessible euchromatin is associated with increased acetylation and trimethylation of histone H3 lysine (H3K)-4, H3K-36, and H3K-79, whereas transcriptionally inactive heterochromatin has low acetylation and increased methylation of H3K-9, H3K-27, and H4K-20 [111]. Specifically, the global increase of H3K-27me3 in multiple tissues of various organisms has been newly identified as a common epigenetic signature of aging [112]. Generally, inflammatory genes are activated and repressed by histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively [113]. Correspondingly, cell senescence can be delayed via the inhibition of HATs and induced by inhibiting HDACs [[114], [115], [116]].
During aging and cell senescence, profound chromatin rearrangement can occur and cause global heterochromatin loss [96,117]. Compromised chromatin architecture at specific domains, including centromere, telomeres, and retrotransposons, results in chromatin relaxation, reactivation of deleterious genes, and genomic instability that fuel a pro-inflammatory environment [118,119]. For example, Sirt6 shows HDAC activity at H3K-9 and H3K-56, while Sirt6-deficient mice exhibited not only genomic instability and shortened lifespan but also accumulation of LINE1 cDNA, which triggers type I interferon response via cGAS-STING [90,120]. Aging is also associated with a global reduction in histone trimethylation at H3K-9me3, which is implicated in suppressing the reactivation of human endogenous retrovirus (HERV) [121,122]. Indeed, human endogenous retrovirus-K (HERV-K) is found to increase in patients with AD and cause subsequent neurodegeneration and microglial accumulation through TLR-8 activation [123,124]. Notably, HERV-K can also drive cell senescence through cGAS-STING activation [125], further emphasizing the causal role of DNA hypomethylation in inflammaging.
2.3.3. microRNAs (miRNAs)/non-coding RNA
Micro-ribonucleic acids (miRNAs) are small, non-coding RNAs that bind to the 3′ untranslated region of messenger RNA (mRNA) and regulate gene expression [126]. miRNAs can act as either repressors or activators of gene expression, depending on the target mRNA and the miRNA's corresponding seed sequence [126]. By doing so, miRNAs regulate aging by altering the expression of genes involved in the aging process, such as those involved in DNA damage repair, cell cycle control, and apoptosis. miRNAs can also regulate inflammation by targeting cytokines and other molecules involved in inflammatory pathways. This miRNA subset has been coined “inflammamiRs” owing to their ability to influence NF-κB/NLRP3 and IL-6 inflammatory pathways (particularly miR-21–5p and miR-146a-5p) [127].
In addition, miRNAs can interact with epigenetic machinery, such as DNA methylation and histone modification, to modulate gene expression and aging-related processes. For example, miR-17 mediates DNMT-1 downregulation in neurotoxin-induced PD and leads to an aberrant DNA methylation pattern in PD [128]. Therefore, miRNA may serve as a promising therapeutic target for ARDs, including neurodegenerative disease [129], CVD [130], degenerative musculoskeletal disease [131], metabolic disease [132], and ovarian aging [133].
2.4. Loss of proteostasis
Protein homeostasis, or “proteostasis,” is a network of intra- and extracellular interactions that ensure the proper translation and folding of newly synthesized proteins, as well as the refolding and degradation of misfolded proteins [134]. Derangement of proteostasis, such as by proteasomal deregulation [135], is a primary manifestation of aging, underlying several ARDs [134]. A growing body of evidence indicates that immune reactions are induced by proteostatic stress, and excessive inflammation may contribute to the age-related decline in proteostasis [136], indicating that proteostasis and inflammation are mutually influenced [137]. With aging, both aspects of proteostasis—folding and degradation—are reduced, resulting in an accumulation of damaged proteins and organelles, termed “garb-aging,” which can exacerbate inflammaging (Figure 2) [138].
2.4.1. Reduced proteostasis promotes inflammation
During aging, the robustness of the proteostasis network is challenged by continuous exposure to mutagens that cause misfolded proteins and damaged organelles, such as mitochondria [139]. The perturbed proteostasis can promote inflammation as misfolded proteins form proteotoxic aggregates, which initiate excessive unfolded protein response (UPR) through activation of the inositol-requiring transmembrane kinase/endoribonuclease 1α (IRE1α), the activating transcription factor 6 (ATF6), and the protein kinase R-like endoplasmic reticulum kinase (PERK), eventually leading to NF-κB activation [140,141]. The dysregulation of UPR in immune cells thus contributes to multiple pathologies, including autoimmunity and metabolic disorders [142]. Proteotoxic aggregates also can be recognized, as damage-associated molecular patterns (DAMPs), by pattern-recognition receptors (PRRs) like TLRs and nod-like receptors (NLRs), leading to the assembly of the NLRP3 inflammasome, increased IL-18 and IL-1β secretion by caspase-1 cleavage [143], and eventually pyroptosis-mediated cell death [144]. These deleterious effects can be amplified in postmitotic cells, such as neurons and cardiomyocytes, which are not actively dividing to dilute cellular debris into daughter cells, increasing inflammaging [145].
Proteostasis depends on the proper functioning of proteolytic systems, namely, the ubiquitin-proteolytic system (UPS) and the autophagy-lysosome system (ALS) [134]. However, as cells age, damaged cellular content accumulates and overwhelms the cellular proteolytic systems, contributing to diverse types of tissue inflammation, such as the neuroinflammation in PD [146] and senescence-associated secretory phenotype (SASP)-induced inflammation [147]. Reduced proteasome function also can potentially lead to proteasome-associated autoinflammatory syndrome (PRAAS), characterized by prominent type I IFN gene signatures [148]. In mice with T cell-specific knockout of Rpn13, a ubiquitin receptor critical for proteasome induction, there is a marked increase in programmed cell death protein 1 (PD-1)+CD44highCD4+ T cell frequency upon TCR stimulation, accompanied by profound production of pro-inflammatory cytokines and chemokines [149]. Besides UPS dysfunction, impaired ALS function, as seen in age-dependent downregulation of autophagy genes [106,150], can also contribute to inflammaging (discussed in depth in Disabled Macroautophagy).
2.4.2. Inflammation impairs proteostasis
Systemic and chronic inflammation during aging can deregulate proteostasis via multiple mechanisms, undermining both protein folding and degradation. The inflammation-induced ER stress can activate UPR, thus altering the protein synthesis/folding arm of proteostasis by 1) PERK-dependent phosphorylation of eukaryotic translation initiation factor 2 (eIF2α) to hinder translation of misfolded protein; and 2) decreasing influx of translated proteins into the endoplasmic reticulum (ER) by degrading ER membrane-associated mRNAs with regulated IRE1α-dependent decay (RIDD) [151]. Inflammation in itself can lead to increased levels of ROS and oxidative stress [137,152], which impair protein folding by exposing hydrophobic cores that facilitate the formation of misfolded, cytotoxic, and degradation-resistant protein aggregates [153,154]. In addition, “oxi-inflamm-aging,” a newly coined aging pathology [155], describes that oxidative stress elicited by inflammation promotes the accumulation of a variety of proteotoxic cellular products, including lipofuscins, advanced glycation end-products (AGEs), Tau protein aggregates, α-synuclein fibrils, and β-amyloid, all contributing to deregulated proteostasis [138]. Specifically, lipofuscin, a type of highly oxidized and cross-linked protein or lipid aggregate, contributes to the failure of lysosomal enzymes and inefficient clearance of autophagosomes [134], leading to enhanced inflammation, ROS production, and even senescence in postmitotic cells, such as cardiomyocytes [156].
In the presence of oxidative stress or chronic inflammation, TNF-α and IFN-γ can promote the conversion of normal proteasome into an inducible isoform, or “immunoproteasome” (IP) [157]. IP is constitutively expressed in hematopoietic cells and participates in antigen processing for the presentation by major histocompatibility complex I (MHC I) molecules. It can degrade oxidized proteins more efficiently, thus playing a critical role in cytokine signaling and proteostasis [148]. However, the IP can also promote pro-inflammatory T helper 1 (Th1) and T helper 17 (Th17) cells and suppress the Tregs cells [157], while malfunctioning of the IP is involved in the development of autoimmune diseases [158,159] and accelerated aging [160,161], indicating that inflammation and derailed proteostasis is interdependently contributing to the pathogenesis of aging.
2.5. Disabled macroautophagy
Due to its increasingly broad importance in aging, macroautophagy has emerged as its own hallmark of aging, despite its strong connection to proteostasis [6]. With autophagosome-lysosome fusion, macroautophagy processes not only protein cargos but also non-protein macromolecules (including lipid vesicles and glycogen) and organelles (such as “mitophagy” for mitochondria, “ribophagy” for ribosomes) [162], thus controlling cytoplasmic quality, cellular metabolism, as well as innate and adaptive immunity [163]. However, a general decline in autophagy function, which has different subtypes such as macroautophagy, microautophagy, and chaperone-mediated autophagy (CMA), is observed in older individuals to accelerate aging by promoting chronic inflammation [164]. Therefore, this section focuses on how diverse crosstalk, beyond the scope of macroautophagy, between chronic inflammation and autophagy can contribute to inflammaging (Figure 2).
2.5.1. Autophagy removes sources of chronic inflammation
The reduction of autophagic flux can contribute to poor cytosolic quality through the accumulation of protein aggregates, cytosolic DNA, dysfunctional organelles, and reduced elimination of pathogens – all of which contribute to inflammation signaling [165]. Thus, a failure of adequate autophagy often manifests as dysregulated inflammation in animal models and human diseases [166] through the accumulation of cytosolic protein aggregates and condensates [167], which act as DAMPs [168]. Given that microbes, damaged organelles, and organic or inorganic crystals are all sources of inflammatory signals (PAMPs and DAMPs), the cytoplasmic clean-up function of autophagy is typically anti-inflammatory in cells capable of activating cell-autonomous inflammatory responses [166]. For instance, CMA is recently found to assist in removing palmitoylated NLRP3, thus preventing sustained inflammation in human and mouse macrophage models [169]. The age-related decline in CMA function is associated with increased NLRP3 activation and IL-1β secretion, leading to vascular inflammation and accelerated atherosclerosis progression [170].
A declining capacity to remove damaged organelles also contributes to inflammaging. For instance, “MitophAging” refers to the age-related pathogenesis resulting from the loss of mitophagy [171]. Aging human CD4+ T cells are found to be burdened with declining mitophagy, shown as accumulating autophagosomes consisting of undigested and damaged mitochondria, which stimulate ROS generation and the NF-κB pathway, triggering chronic inflammation, immunosenescence, and dysfunctional adaptive immunity in older individuals [172]. As well, immunosenescence can be ameliorated by autophagy during aging. In old human B cells, spermidine-induced synthesis of the autophagosomal and lysosomal master regulator, transcription factor EB (TFEB), leads to enhanced autophagy and reversed B cell senescence [173]. However, although autophagy plays a crucial role in removing senescent cells [174], it is worth noting that autophagy may also provide building blocks for SASP during some forms of senescence, thus complicating the relationship between autophagy and age-related senescence [175].
Notably, caloric restriction-mimetic therapies, such as metformin [176], rilmenidine [177], rapamycin, and “rapalogs” [178], as well as senolytic drugs [179,180], are proposed to exert anti-inflammaging effect by activating autophagic processes, including mitophagy and CMA, thus promoting the proper disposal of damaged cellular components, which can otherwise trigger a cascade of inflammatory pathways [138]. The mechanistic target of rapamycin 1 (mTORC1), located on lysosomes, is of particular interest as a modulator linking metabolism, autophagy, inflammation, and aging biology (see the section on Deregulated nutrient sensing).
2.5.2. Chronic inflammation impairs autophagy
Chronic inflammation is also understood to impair autophagy. This effect is often seen in neurodegenerative disorders such as PD [181], AD [182], Huntington's Disease [183], and Amyotrophic Lateral Sclerosis [184]. The role of neuroinflammation in impairing efficient autophagy was demonstrated as lipopolysaccharide (LPS)-induced inflammatory stress substantially increased cytokine production (IL-1β, TNF-α, and IL-6) and decreased the levels of autophagy markers (Beclin-1, p62, and LC3 II) [185]. Similarly, TNF-α can inhibit microglial autophagy via the mTOR pathway, while enhanced autophagy can promote microglial M2 polarization, promoting the resolution of inflammation in PD [186]. Recent studies also suggest that microglia-induced neuroinflammation can promote the accumulation and transmission of α-syn and Tau proteins to aggravate PD and AD, respectively [187]. Specifically, constitutive microglial activation, for example, by LPS, can increase α-syn accumulation and transmission to the substantia nigra and striatum, leading to increased autophagy burden and worsened PD [[188], [189], [190]]. In a murine AD model, constitutive activation of microglial NF-κB exacerbates Tau seeding and spreading in young PS19 mice, while NF-κB inactivation rescues microglial autophagy [191]. Taken together, chronic inflammation impacts autophagy and may be particularly relevant in the proteinopathies conferred by aging.
3. Antagonistic hallmarks
The antagonistic hallmarks are a response to damages caused by the primary hallmarks. Initially, the response helps mitigate the damage and has benefits. However, incidentally or with prolonged pathway stimulation, the mechanisms become harmful, contributing to the aging process.
3.1. Deregulated nutrient sensing
Nutrient sensing is essential in regulating the aging process. Current evidence suggests prolonged overnutrition accelerates aging, while caloric restriction favors longevity [192]. In addition, nutrient sensing can be deregulated under pathogenic conditions, including multiple metabolic illnesses associated with chronic, low-grade inflammation, such as T2D and atherosclerosis, common chronic diseases of age. Classical pathways that regulate nutrient sensing include the insulin and the insulin-like growth factor 1 (IGF-1) signaling pathways (collectively known as the IIS pathway), the mTOR pathway, the AMPK pathway, and the Sirts. This section will focus on the bidirectional relationships between the deregulation of these pathways and the chronic inflammation that compounds the aging process and related pathology (Figure 2).
3.1.1. IIS deregulation induces inflammation
During aging, the IIS pathway can be deregulated and mediate chronic inflammation. Aging is usually associated with declined insulin clearance, leading to hyperinsulinemia and insulin resistance (IR) in peripheral metabolic tissues [193,194]. IR also induces dyslipidemia, represented as high plasma levels of triglycerides and free fatty acids (FFAs) [195]. Triglycerides and FFAs can trigger macrophage inflammatory responses by inducing NF-κB, culminating in a pro-inflammatory environment that worsens IR [196]. Chronic hyperinsulinemia and elevated FFA also support chronic mTOR activation [197], a major driver of glycolysis in immune cells [198]. Increased glycolysis in innate immune cells can facilitate M1-like macrophage polarization with cytokines linked to inflammaging, like TNF, IL-1, and IL-6 [199]. In addition, insulin inhibits forkhead box protein O1 (FoxO1) activity, while FoxO1 deficiency impairs proteostasis in aged T cells and causes pro-inflammatory, senescence-like phenotypes [200].
Insulin itself can also directly act on the immune system to fuel inflammaging. For instance, insulin receptor signaling has been shown to acutely promote murine immune cell functions, including T cell effector responses, through supporting nutrient uptake and glycolytic capacities, key metabolic necessities of inflammation [201,202]. In addition, mice deficient in insulin receptors in T regulatory (Tregs) cells are protected from age-induced glucose intolerance and adipose tissue inflammation, highlighting an inflammatory role for insulin in altering Treg responses [203]. Interestingly, age-associated B cells (ABCs), a T-bet + memory-like B cell subset linked with increased autoimmune diseases and aging in mice [204,205], can promote inflammatory T cells potentially through increased glycolysis [206,207], though it remains unknown if insulin/IGFs is a necessary input. Thus, the immune homeostasis supported by proper IIS signaling is likely essential, especially in the elderly, who have a higher susceptibility to infectious diseases and lower capacity to generate adaptive immune responses against pathogens, such as SARS-CoV-2 and vaccination [2,208]. Altogether, IIS deregulation may form a vicious loop that fuels both inflammatory processes and potential changes linked to immunosenescence, exacerbating ARDs.
3.1.2. mTOR pathway deregulation induces inflammation
The mTOR kinase is essential in maintaining metabolic homeostasis by sensing amino acid levels and modulating insulin signaling [209]. mTOR in mammals exists within the mTOR complex 1 (mTORC1) and 2 (mTORC2). mTORC1 responds to IIS signaling through Akt (protein kinase B), senses amino acid abundance, and controls cell growth and senescence [210]. At the same time, mTORC2 can be activated through insulin-like growth factor receptor (IGFR)- and insulin receptor substrate (IRS)-mediated signaling, controlling metabolism and actin organization [211]. When triggered by prolonged IIS signaling, mTORC1 can be hyperactivated, while several ARDs, including T2D, have mTORC1 hyperactivation as a risk factor [212,213], which fuels inflammation and accelerates aging progression [214]. The chronic, low-grade inflammation induced by mTORC1 hyperfunction and its resulting inhibitor of nuclear factor-κB (IκB) kinase β (IKKβ) overactivation (Figure 2) [215,216] is associated with decreased lifespan [217]. Reducing mTORC1 signaling and its associated tissue inflammation can also benefit tissue healthspan in aging mice [218], whereas mice with constitutive mTORC1 overactivation showed accelerated aging that can be rescued by rapamycin treatment [219].
The mTOR signaling also impacts aging progression by modulating inflammation through cellular senescence and immune cell responses [209], [220]. mTORC1 inhibition with rapamycin can blunt the production of pro-inflammatory and pro-oxidant products from senescent cells exhibiting SASP [221,222]. In murine macrophages, reduction of mTORC1 through Raptor (regulatory-associated protein of TOR) knockdown decreases inflammatory gene expression, and overexpression of mTORC1 signaling through knockdown of mTORC1 inhibitor, TSC1 (Tuberous Sclerosis 1), leads to elevated glycolysis and shift to pro-inflammatory M1-like macrophages [223,224]. Intriguingly, in healthy aging individuals, mTORC1 blockade exhibited enhanced antiviral immunity toward influenza infection [225]. This data is especially significant under the ongoing pandemic of coronavirus disease 2019 (COVID-19), which in itself increases mTORC1 activity [226]. Thus, manipulation of mTOR signaling may provide a novel intervention to enhance the adaptive immunity in aging individuals and reduce mortality from viral infections, thus providing a potential axis to improve human longevity [227].
3.1.3. AMPK-sirtuin pathway deregulation induces inflammation
By acting in the opposite direction to the IIS and the mTOR pathways, the AMPK-sirtuin pathway signals nutrient scarcity and catabolism. The AMPK pathway is activated by an increased AMP:ATP ratio during starvation or caloric restriction [228], while sirtuins are a family of (NAD+)-dependent deacetylases [229]. Mechanistically, AMPK activation can increase the cellular NAD + concentration, leading to the expression of Sirt1 that deacetylates the liver kinase B1 (LKB-1), an upstream activator of AMPK [230], forming a positive feedback loop (Figure 2). Functionally, AMPK activation can inhibit mTORC1 by activating TSC1/2 and stimulate Sirts to deacetylate FoxOs or autophagy-related proteins (ATG5 and ATG7) [231,232], leading to 1) enhanced expression of PGC-1α for better mitochondrial biogenesis [233] and 2) inhibition of cytoplasmic p53 for autophagy [234,235]. Recently, AMPK is also found to directly phosphorylate folliculin-interacting protein 1 (FNIP1), which forms a complex with folliculin to suppress the complex's function; through this interaction, AMPK is able to promote TFEB-mediated autophagy and induce lysosomal and mitochondrial biogenesis [236]. Of note, metformin, an AMPK activator, normalizes mitochondrial function and alleviates age- and senescence-associated inflammation by enhancing T cell autophagy [237]. Therefore, the AMPK-Sirt pathway plays an important role in regulating chronic inflammation.
As seen in the decreased NAD + level and the associated metabolic dysfunction during aging, the declining AMPK-Sirt responsiveness with age is gaining increasing attention for its proinflammatory potential [[238], [239], [240]]. Sustained AMPK activation is essential for the survival and functions of both human and murine in vitro induced Treg cells [241]. In line with Tregs’ function in inhibiting chronic inflammation induced by self-antigens in aged persons [242], mice with Treg-specific AMPK deletion developed autoimmune and inflammatory liver diseases [243]. In murine macrophages, AMPKα1 deletion induces M1 hyperpolarization upon LPS challenge, whereas metformin increases IL-10 and reduces IL-6 and IL-12 secretion in wild-type M1 macrophages [244]. Loss of AMPK in other tissues also causes inflammation and age-associated pathologies, such as NAFLD [245] and chronic kidney disease [246]. Similarly, loss of Sirt6 in the aging mouse brain leads to decreased mitochondrial function and increased ROS production that potentiates inflammaging [247]. In contrast, an intact AMPK-Sirt pathway can suppress NLRP3 (Figure 2), thus quenching chronic inflammation and contributing to a healthier aging process [[248], [249], [250]]. In murine hematopoietic stem cells (HSCs) with age-related defects in autophagy, Sirt3 can restore mitophagy, reduce oxidative stress, and slow aging progression [251]. This evidence indicates that the AMPK-Sirt pathway, if deregulated during aging, can cause profound disruption in energy homeostasis and aberrant inflammatory responses.
3.1.4. Inflammation deregulates the IIS pathway
Local and systemic inflammation accompanying aging and ARDs can also deregulate nutrient-sensing pathways [252]. Aging deregulates the IIS pathway by promoting a pro-inflammatory environment in metabolic tissues, such as adipose tissues [253,254], muscles [255], and digestive tracts [256,257]. This chronic, low-grade metabolic inflammation is characterized by increased FFAs, ROS, and UPR linked to ER stress, which can activate the JNK pathway, mediating the serine phosphorylation of IRS1/2 and thus blockade the IIS pathway [258]. Likewise, older mice express higher levels of inflammatory markers, such as TNF-α and IL-1β in skeletal muscles [259], leading to activated IKKβ/NF-κB pathway that phosphorylates IRS1 and contributes to IR [260] (Figure 2). Notably, the reduction of inflammation is implicated in the extension of life/health span observed under caloric restriction. For example, methionine restriction (MR) has been found to reduce basal levels of serum IGF1 and glucose, associated with downregulation of multiple pathways with pro-inflammatory potential, such as TNF signaling via NF-κB, IL-6, and JAK/STAT3 signaling in LmnaG609G/G609G mice, a mouse model of Hutchinson–Gilford progeria syndrome (HGPS) [261]. Similarly, inhibiting methionine catabolism by the pluripotency factor NANOG in LmnaG609G/G609G mice restores insulin sensitivity, glucose uptake and glycolysis, and significantly enhances the force-generating ability of aged mice [262].
In digestive tracts, especially the gut, recent studies indicate that microbial dysbiosis can increase with aging in the elderly, where pro-inflammatory commensal bacteria are enriched at the expense of beneficial microbes [263,264]. A pro-inflammatory gut environment due to dysbiosis, such as with aging, can worsen IR in mice by increasing gut permeability, facilitating bacterial product leakage into circulation, which contributes to chronic inflammation in tissues, including in the intestine [265,266]. Chronic inflammation, in turn, interferes with the IIS signaling, propelling age-related metabolic diseases [257,267]. A mucosal source of inflammation may also be related to altered IgA responses with age, as IgA is one link governing inflammation associated with the impairment of nutrient-sensing pathways [268,269].
Remarkably, the insulin receptor can also function through multiple non-canonical pathways, such as acting as a transcription factor by binding to gene promoters [270], controlling senescence through a ligand and tyrosine kinase-independent (LYK-I) pathway [271], and modulating hepatocyte and adipocyte functions by forming dynamic clusters on the cell surface [272]. Since all these pathways are interrupted in insulin-resistant cells or by ROS accumulation [272], future work is needed to tease out precise mechanistic connections between their deregulation and metabolic inflammation in aging populations.
3.1.5. Inflammation deregulates the mTOR pathway
Characterized by increased levels of pro-inflammatory cytokines, particularly IL-1β, IL-6, and TNF-α [273], inflammaging also feeds into the deregulation of the mTOR pathway. Pro-inflammatory cytokines resulting from age-induced ER stress can interact with mTORC1, perturbing its normal function and expediting aging. For example, IKKβ, a downstream effector of the TNF-α pathway activated during aging [274], has been shown to phosphorylate and inhibit TSC1 directly, constitutively activating mTORC1 [275]. Consequently, mTORC1/S6K hyperactivation promotes IRS1 degradation and generates a feedback inhibition of the phosphatidylinositol-3-kinase (PI3K)/Akt pathway, leading to peripheral IR (Figure 2) [276,277]. In addition, viral infection and chronic inflammation during aging can activate nucleic acid sensors, such as cGAS, retinoic acid-inducible gene I (RIG-I), and anti-melanoma differentiation-associated gene 5 (MDA5) [[278], [279], [280]], whose downstream target, TBK1, has been shown to activate mTORC1 [281] directly and activate mTORC2 signaling indirectly [282]. The inflammaging-induced deregulation of mTOR signaling is also observed in aged murine peritoneal macrophages (PM). With metabololipidomics and proteomic profiling, a recent study found a significant upregulation of pro-inflammatory pathways, such as integrin signaling, mTOR, NF-κB, NO and ROS production, p38 MAPK and TNFR1/2 signaling in old M2a-PM [283]. These changes are accompanied by a downregulation of pathways for inflammatory function of M1 macrophages, such as glycolysis, nuclear factor erythroid 2-related factor 2 (NRF2), and hypoxia-inducible factor 1-alpha (HIF-1α) signaling [283], indicating that the dichotomy of pro-inflammatory (M1-like) and pro-resolving (M2-like) macrophages during inflammaging can be metabolically obscure.
Interestingly, sestrins (SESNs), a family of stress-inducible proteins, are emerging as metabolic regulators that inhibit inflammaging mainly through activating AMPK and inhibiting mTORC1 [284,285]. For example, SESN1/2 inhibits liver mTORC1 signaling in a leucine-sensitive manner, thus ameliorating age-induced reduction in ketogenesis [286,287]. However, SESNs themselves also can be a target reduced during inflammation, leading to mTOR hyperactivation [288]. For example, the induction of SESN2 has been shown to require the PI3K/Akt pathway, which is frequently obstructed during inflammaging and obesity due to IRS1 serine phosphorylation [289]. In support of this, SESNs levels are lower in healthy aging men than younger men [290], while SESN1-SESN2-knockout mice showed hyperactivation of mTORC1 in aged knockout mice [291]. However, how inflammation directly alters SESNs expression warrants further research. These findings indicate that inflammation can exert a potent impact on aging physiology through cytokine-mTOR interaction and crosstalk with other components in age-associated stress responses.
3.1.6. Inflammation deregulates AMPK-sirtuin pathway
Although the AMPK pathway integrates the IIS, mTOR, and Sirt pathways, acting as a master regulator of metabolism and inflammation, inflammaging can also reversely impose a profound impact on the AMPK-Sirt pathway by preventing it from maintaining proper energy homeostasis. The activation capacity of this pathway declines under age-induced ER stress, oxidative stress, and chronic inflammation [238,292,293], while high glucose/lipids/amino acid levels, LPS, and pro-inflammatory cytokines, such as TNF-α and NF-κB, can also inhibit AMPK activity [[294], [295], [296]]. Particularly, LPS can mediate inflammation by dose-dependently facilitating AMPK dephosphorylation [297]. Intriguingly, AMPK exhibits viral-sensing capability by directly phosphorylating TBK1 and promoting type I interferon gene expression in response to viral nucleic acids detected by the cGAS-STING pathway [279,298]. The genetic ablation of AMPK reduced Sendai virus sensing and damage-induced DNA sensing, underlying a compromised innate immunosurveillance [298]. Thus, during aging, which may be linked to increased circulating bacterial and viral products, an inflammatory environment can distort AMPK signaling, reducing the beneficial effects of AMPK-dependent autophagy, mitochondrial biogenesis, and antiviral immune responses, the reduction of which are all linked to frailty and chronic disease in the elderly.
Moreover, age-associated metabolic inflammation can deplete NAD+, leading to age-dependent loss of Sirt activity [88], [89], [299]. Meanwhile, reduced Sirt1 reduces mitochondrial biogenesis, reduced mitochondrial Sirt3 reduces mitochondrial antioxidant and repair systems, and reduced NAD + may also impair Sirt2, enhancing NLRP3 inflammasome activity [22]. Thus, inflammaging can induce a positive feedback loop through altered Sirt function to drive ROS production, mitochondrial dysfunction, and more inflammation. In addition, as a natural by-product of amino acid metabolism and urea cycle, ammonia levels in the brain increase with age, contributing to neuroinflammation through p38 MAPK/NF-κB pathway and blood–brain-barrier breakdown [300]. In human and murine skeletal muscle/myotubes, hyperammonemia can inhibit the mitochondrial electron transport chain complex I that oxidizes NADH to NAD+, leading to a lower redox ratio, Sirt3 dysfunction, and postmitotic senescence in myotubes, accompanied by increased acetyl–NF–κB p65 expression, an activating post-translational modification [301]. Thus, multiple metabolic mechanisms contribute to Sirt decline with age.
Collectively, the major nutrient-sensing pathways, including the IIS, mTOR, Sirts, and AMPK pathways, orchestrate the physiology of longevity. Their deregulation, as a hallmark of aging, fuels the metabolic inflammation that impacts the healthspan and lifespan of aging populations. Early detection of immunometabolic dysregulation with multi-omics approaches, such as lipidomics, metabolomics, and glycomics [302], permits the early diagnosis of metabolic and ARDs and represents a future direction of personalized geroprotective treatments.
3.2. Mitochondrial dysfunction
Mitochondria developed through the merging of a prokaryotic organism into a eukaryotic cell [303]. From these origins, mitochondria maintain their own DNA (mtDNA) and DNA replication process. They act to supply the cell with ATP and synthesize key molecules in the processes of inflammation, oxidation, and metabolism. In recent years surmounting evidence suggests that mitochondria not only play an influential role in the progression of the inflammaging phenotype but are also heavily impaired by chronic inflammatory states [85]. Indeed, mitochondrial function declines with aging, and this change is associated with increased ROS, reduced ATP production, elevated mitochondrial damage, and age-related epiphenomena (Figure 2) [[304], [305], [306]]. While multiple mechanisms link mitochondrial dysfunction with aging, one such mechanism relates to age-related depletion of NAD+, which causes loss of efficient sirtuin activity, resulting in impaired mitochondrial SIRT3. Impaired Sirt3 leads to dysregulation in mitochondrial antioxidant systems, mtDNA repair, and mitochondrial quality control and biogenesis pathways [307]. Here, we examine the reciprocal links between inflammation and mitochondria with age.
3.2.1. mtDNA promotes chronic inflammation
Because of their endosymbiotic origins, mitochondria carry a milieu of potential DAMPs that may be uniquely immuno-stimulating [308]. Some of the most putative mitochondrial DAMPs include the release of mtDNA, N-formyl peptides, and unique mitochondrial lipid species such as cardiolipin [309]. During aging, there is increased mitochondrial dysfunction, reduced genome integrity, and damage [310], allowing for the increased potential of age-related interactions with mitochondrial DAMPs.
The phospholipid cardiolipin is essential for maintaining mitochondrial structure and function and normally siloed to the inner mitochondrial membrane. However, when exposed to the cytoplasm, it stimulates mitophagy, the process in which dysfunctional mitochondria are eliminated and new mitochondria are formed [311]. Apoptosis is precipitated in most severe cases when cardiolipin stimulates the release of cytochrome c [312]. Mitochondrial proteins, including N-Formyl peptides, primarily induce inflammation through binding to formyl peptide receptor-1 [313].
In the case of mtDNA, immune activation can be mediated by TLR signaling. Indeed, mtDNA binds TLR-9, a receptor capable of detecting unmethylated DNA with CpG motifs derived from bacteria and viruses and initiating the innate immune response [314]. Activation of TLR-9 leads to NF-κB signaling, which induces the expression of several pro-inflammatory cytokine genes [315] and is aberrant in many chronic diseases [316]. In this way, mitochondria not only contribute to the transmission of danger signals within the cell but are also a major source of molecules capable of activating the innate immune system. For reasons not entirely understood, circulating mtDNA appears to increase gradually with age after the fifth decade of life [317], and the abundance of unhoused mtDNA is associated with accelerated aging pathology [318]. Various inflammatory pathways are activated when ROS-damaged mtDNA and cardiolipin are released into the cytosol. Responses that have been best studied involve the NLRP3 inflammasome, the cGAS-STING pathway, and NF-κB [309,319]. Cytosolic mitochondrial RNA has similar effects, stimulating TBK1/IKK and NF-κB through RIG-1, MDA5, and mitochondrial antiviral-signaling protein (MAVS) [320]. Mitochondrial damage also activates other inflammasomes such as NLRP10, and AIM2 in keratinocytes and macrophages, respectively [321,322], thus implicating their potential role during mitochondrial damage-induced inflammation in aging. Finally, mitochondrial-derived phosphocreatine and ATP can also instigate inflammation through inflammasome activation or, in the case of ATP, by also binding extracellular P2X purinoceptor 7 on innate immune cells [322,323].
In addition to mtDNA and ROS, mitochondrial dysfunction releases TCA cycle intermediate metabolites into the cytosol with immunomodulatory effects [324]. Fumarate has been shown to impose immunomodulatory effect through controlling chromatin modifications and regulating protein succination [325]. Specifically, pro-inflammatory insults can lead to the accumulation of fumarate through glutamine anaplerosis which has been shown to be necessary for trained immunity and inflammation by inhibiting lysine-specific demethylase 5 A (KDM5) histone demethylase activity; inhibition of KDM5 increases the levels of H3K4me3, a marker of active gene transcription at the promoters of TNFα and IL6 cytokines [326]. Furthermore, elevated levels of succinate, resulting from mitochondrial dysfunction, can potentially contribute to chronic inflammation by activating the HIF-1α pathway through stabilization of HIF-1α. Succinate inhibits prolyl hydroxylases, enzymes responsible for the degradation of HIF-1α. Consequently, stabilized HIF-1α translocates into the nucleus and initiates the transcription of key inflammatory cytokines, particularly IL-1β [327]. It remains to be seen if such mechanisms occur during inflammaging.
Itaconate has been shown to inhibit the nuclear factor-kappa B (NF-κB) pathway, a key regulator of inflammation [328]. Itaconate acts by directly modifying specific cysteine residues on key proteins involved in NF-κB signaling, such as IκB kinase (IKK) and Kelch-like ECH-associated protein 1 (Keap1). This modification disrupts the activation of NF-κB and promotes its nuclear export, thereby suppressing the expression of pro-inflammatory genes. Additionally, itaconate can also promote anti-inflammatory responses by activating the anti-inflammatory transcription factor NRF2, which induces the expression of antioxidant and anti-inflammatory genes including GPX1 and SOD1 which decrease ROS [328,329]. Itaconate may be induced during aging inside immune cells like macrophages, where it may act as a compensatory factor to help dampen inflammation or maintain tissue homeostasis [330].
3.2.2. Inflammation impairs mitochondrial function
Conversely, chronic inflammation has been shown to impair mitochondrial function through inflammasome activation and other pro-inflammatory immune signaling engagements, such as the exaggerated generation of ROS, which produce damage in mitochondrial proteins, lipids, and mtDNA [331]. An array of ARDs, including CVDs, cancer, neurodegenerative disease, and metabolic diseases, has been linked to mitochondrial dysfunction [[332], [333], [334]]. Similar to DNA damage, mitochondrial machinery, and compartments accumulate damage over time and rely on repair and replacement mechanisms to maintain performance. However, instead of repairing damaged entities, dysfunctional mitochondrial proteins and even entire organelles are selectively removed through mitophagy [335,336]. This quality control process selects for healthy mitochondria to replicate and govern homeostasis. However, both excessive chronic inflammation and aging impair autophagic mechanisms. For instance, excessive inflammation due to the NLRP3 inflammasome can trigger CASP1-dependent mitochondrial damage, ROS production, dissipation of mitochondrial membrane potential, permeabilization, and inhibition of mitophagy through cleavage of Parkin [319]. The resultant accumulation of damaged mitochondria leads to uncontrolled ROS and mtDNA leakage and the ensuing IL-1β- and IL-6-dependent inflammation orchestrated by cGAS-STING signaling (Figure 2) [337].
3.3. Cellular senescence
Cellular senescence is a state of proliferative arrest in response to cell stressors [338]. Senescence can be induced by various stimuli, including DNA damage, oxidative stress, replicative stress, mitochondrial signaling, and altered expression of certain oncogenes [339]. Senescent cells remain mechanically intact, metabolically operational, and involved in intercellular signaling while still arresting growth and replication. Senescence growth and replicative arrest have been shown to be mediated by major tumor–suppressor pathways such as p16INK4a, pRB (retinoblastoma protein), and by p53 [340,341]. Senescent cells and their secretory products are now widely accepted as important contributors to aging and age-related disease. Cell-specific effects of cellular senescence and inflammaging have been recently reviewed elsewhere [342].
3.3.1. Cell senescence and its SASP promotes inflammation
The secretion profile of factors specific to senescent cells has been termed the “senescence-associated secretory phenotype” or SASP [343,344]. The SASP is a bioactive secretome thought to promote the recruitment and activation of immune cells that would clear senescent cells. When clearance fails, the result is the accumulation of senescent cells and SASP factors which leads to diminished tissue function and steadily elevated proinflammatory tone. SASP factors are also thought to be critical mediators of tissue repair [345]. However, they may also potentially facilitate senescence spreading into neighboring cells [346]. Some of the most robustly induced secreted pro-inflammatory SASP proteins include, but are not limited to, IL-6, IL-8, IL-1, granulocyte-macrophage colony-stimulating factor (GM-CSF), growth-regulated oncogene (GRO)α, monocyte chemotactic protein (MCP)-2, MCP-3, matrix metallopeptidase (MMP)-9, −1, −3 and several IGF-binding proteins. SASP factors that have been proposed to be biomarker candidates of aging include markers such as growth differentiation factor (GDF)-15, stanniocalcin 1, MMP-1, Inhibin Subunit Beta A (INHBA), and serpin family E member 1 (SERPINE1) [343,347].
In addition to pro-senescence mechanisms, such as persistent DDR signaling, GATA4 stabilization, and ensuing NF-κB and C/EBPβ cascades [25,348,349], the secretion of SASP factors can also be initiated or maintained by two microRNAs, mir-146a/b. These microRNAs orchestrate a negative feedback loop dampening the escalation of NF-κB activity and maintaining a chronic low-grade inflammatory phenotype [350]. Despite the induction of mir-146a/b, the SASPs persist in promoting the low-grade inflammation thought to drive chronic pathologies associated with aging [351]. Interestingly, the genetic determinants of senescence also leads to unique SASPs and inflammatory profiles. For instance, p21-induced senescence induces a distinct SASP characterized by C-X-C motif chemokine 14 (CXCL14) expression, which puts stressed cells under immunosurveillance [352]. In addition to recruiting immune cells, SASP factors can also induce markers of aging, such as CD38, on immune cells. Macrophages expressing CD38 in aged tissues deplete NAD+, which further drives aging phenotypes [87,89,299]. Thus, it will be interesting in the future to map out how underlying instigators of senescence contribute to specific inflammatory profiles across the chronic diseases of aging and to better understand distinct impacts on immune cell populations.
Of note, not all SASP immunological factors should be considered pathogenic with age. For instance, senescent cells can also secrete factors that stimulate tissue repair and epithelial regeneration [345]. As a result, the use of senolytics that target senescent cells should be used with caution, as indiscriminate senolysis can interfere with less appreciated benefits of senescent cells.
3.3.2. Inflammation promotes senescence
Senescent cells have been shown to accumulate over the life span of humans and predominantly in renewable tissues and tissues that experience prolonged inflammation [353,354]. Many inflammatory diseases, such as atherosclerosis, osteoarthritis, idiopathic pulmonary fibrosis, and insulin resistance, also show elevations in senescent cells associated with inflammation [22]. As previously mentioned, molecular damage, including DNA and structural damage induced by oxidative stress associated with chronic inflammation, can trigger senescence. The accumulation of oxidative stress, DNA damage, and even reactivation of HERV can exacerbate inflammation in aging individuals by activating the cGAS-STING pathway, resulting in increased interferons or SASP signaling (Figure 2) [125,355]. In addition, as discussed in section 3.1., mTORC1 signaling mediates the production of various pro-inflammatory cytokines including IL-1A, while rapamycin can suppress the translation of the membrane-bound cytokine IL-1A, leading to diminished NF-κB transcriptional activity and reduced SASP [356]. Moreover, viral infection, such as by SARS-CoV-2, can evoke “cytokine storms” characterized as pro-inflammatory cytokines, particularly in the elderly, and amplify SASP from pre-existing senescent cells. The resulted increase in SASP can further exacerbate tissue damage in addition to the direct cellular damage due to virus itself [357]. The innate immune sensing of viral RNA or cytosolic chromatin fragments associated with aging through cGAS-STING and its downstream mediators, predominantly IRF3, NF-κB and C/EBPβ, may also promote both induction and spreading of senescence [[357], [358], [359]].
4. Integrative hallmarks
The integrative hallmarks arise when the homeostatic machinery of aging tissues fails to resolve the accumulated damage from the primary and antagonistic hallmarks [6]. Because of the broad, tissue-specific etiology underlying the integrative hallmarks, their initiation, and progression can exacerbate the primary and antagonistic hallmarks, engendering a large set of age-related pathologies at the systemic level [360].
4.1. Altered intercellular communication
Intercellular signaling, as a hallmark of aging, encompasses many modes of cell-to-cell signaling, including endocrine, neuronal, and neuroendocrine pathways, as well as cell-to-cell contact and the exchange of vesicle-packaged and free soluble factors, including inflammatory signals [6]. Because of the vast role of aberrant intercellular signaling in aging pathology and the degree to which chronic inflammation contributes, these two hallmarks were differentiated in the 2023 Hallmarks of Aging update [6]. Here we briefly highlight links between chronic inflammation and other forms of intercellular communication, including hormones and extracellular vesicles.
4.1.1. Hormones and chronic inflammation
In a similar age-dependent manner, secretory patterns of the hormones produced by the hypothalamic–pituitary axis (HPA) [361] and gut–pancreas–liver axis (GPLA) change [362], as does the sensitivity of the axis response to feedback. The HPA is a master regulator of the body's systemic homeostasis, such as energy balance, body temperature, sleep, blood pressure, and circadian rhythms. Background-level inflammation in the hypothalamus, concisely termed “hypothalamic microinflammation,” is thought to contribute to the deviation from homeostasis, particularly metabolic dysregulation, with age and the tonal systemic inflammation that ensues [363].
In addition to decreased insulin sensitivity and clearance, aging is associated with reductions in fasting glucagon-like peptide-1 (GLP-1) and glucose-dependent insulinotropic polypeptide (GIP), which may predispose to the development of glucose intolerance and T2DM with age [362]. The consequence of dysregulated insulin is the inflammatory reprogramming of immunometabolism and immune cell function (discussed in the section IIS deregulation induces inflammation). Furthermore, the age-associated downregulation of sex hormones, estrogen, and testosterone, may also promote inflammation. Estrogen can induce Th2 responses [364], and testosterone receptors in T cells promote forkhead box protein P3 (Foxp3) expression and Treg fates [365].
4.1.2. Extracellular vesicles as mediators of inflammaging
EVs are lipid membrane vesicles derived from multivesicular bodies or plasma membrane containing proteins, lipids, and nucleotide species, sharing a snapshot of the donor cell state with the recipient [366]. Released by most cell types, they are a newly appreciated form of intercellular communication, linking hallmarks of aging through inflammatory mediators [367,368]. In cancer and autoimmunity, EVs promote inflammation [369], for instance, in the delivery of TLR ligands to plasmacytoid dendritic cells [370]. EVs also exhibit regenerative potential as stem cell-derived or “young” EVs can improve healthspan, though they can lose their regenerative potential over time. For example, mesenchymal stem cell (MSC)-derived small EVs reduce endothelial cell senescence via delivery of miR-146a [371], while reduced levels of EVs containing galectin-3 (Gal-3) in aging plasma donors correlate with a reduced ability to induce osteoblasts from MSCs [372] and may also potentiate PAMP-TLR responses [373]. Similarly, EVs from young human donors were found to have higher levels of glutathione-related protein (GSTM2) than old EVs, which promotes antioxidant activity [374] (Figure 2). EV research is a burgeoning field, and the modes by which EVs partake in inflammaging require further study, including their immunosuppressive role in expanding senescence [375] and their ability to serve as a biomarker for age-related conditions [376].
4.2. Stem cell exhaustion
Adult stem cells function as self-renewal pools that facilitate tissue repair by replacing damaged cells in various tissues and organs [377]. During aging, multiple factors, including telomere attrition, DNA damage, epigenetic dysregulation, and chronic inflammation, cause a decline in both the regenerative and proliferative capacities of stem cells, leading to stem cell exhaustion and accumulation of senescent cells [378,379]. Senescent cells can in turn facilitate additional stem cell dysfunction. For instance, in aged progeroid INK-ATTAC mice, senescent fat progenitors inhibit adipogenesis in non-senescent progenitors through activin A secretion [380]. Chronic inflammation, even early in life, can contribute to the depletion of HSCs, causing phenotypes of premature aging [381], whereas the inhibition of NF-κB activation demonstrates a functional rejuvenation of aged skeletal stem/progenitor cells (SSPC) [382]. Meanwhile, senescent cells can exacerbate stem cell exhaustion via SASP cytokines that favor the infiltration of immune cells, creating an inflamed niche that further impedes stem cell function [[383], [384], [385]]. For example, aged skeletal stem cells (SSCs) become pro-inflammatory and contribute to an inflamed bone marrow niche, which can promote the dysfunction of HSCs [386], while aged HSCs can also activate NF-κB, facilitating the myeloid-skewed differentiation and the eventual loss of self-renewal [387,388]. Additionally, the regenerative capacity of differentiated cells in response to injury is also weakened in aged animals, indicating that the injury-induced cellular plasticity also may be impaired during aging [389]. Although acute inflammation can provoke somatic stemness and promote tissue recovery by IL-6-mediated epigenetic modifications [390], chronic inflammation can lock cells in epigenetically plastic states disabled for reparative differentiation, leading to the accumulation of damaged cells and even tumorigenesis [391].
Recent advances have illustrated the importance of stem cell-related factors that rejuvenate tissues by transiently reprogramming somatic cells. Specifically, the Yamakana factors (Oct3/4, Sox2, Klf4, c-Myc, or OSKM) can induce aging somatic cells to transiently de-differentiate into a younger state, after which the somatic identity is regained with age-related epigenetic patterns erased and cellular functions rejuvenated [392]. Multiple in vivo and in vitro studies have proved the efficacy of OSKM- or OSK-mediated transient reprogramming in murine and human cells, leading to the marked reversal of epigenetic clocks, reduction in inflammatory profiles, and restoration of lost functionality in diseased or aged cells [[393], [394], [395]]. Notably, as transient reprogramming recapitulates features of natural tissue repair, which often declines with age due to accumulated senescent cells, the removal of senescent cells by senolysis extends Drosophila lifespan synergistically with OSKM expression [395]. Other means for stem cell renewal include fecal microbiota transplantation from young mice to aged mice, which can enhance hematopoietic repopulation capacity, thus exhibiting the potential to rejuvenate HSCs [396].
4.3. Dysbiosis
The gut microbiome plays a vital role in maintaining human health through nutrient digestion and absorption, protection against pathogens, and the production of important metabolites such as vitamins, as well as amino acid and fatty acid derivatives [397]. Dysbiosis refers to an imbalance in the gut microbiome, which can occur for various reasons and negatively impact health and serve as a catalyst for fueling inflammaging. However, the contribution of dysbiosis in the context of the human microbiome interaction, particularly regarding its impact on systemic immune function or deterioration of this function among the elderly as it relates to ARDs, is an ongoing area of research [398]. A recent study by Galkin and colleagues shows that microbiome profiles can predict the age of healthy individuals, and patients with T1D exhibit age acceleration according to the microbiome clock [399]. Furthermore, a growing body of literature implicates age-related dysbiosis of the gut microbiome as contributing to tissue-specific (such as in the liver, adipose, and intestine) as well as a global inflammatory state in the elderly [257]. Interestingly, besides the gut–brain axis, oral dysbiosis also plays a role in mediating age-related neurodegenerative diseases, expanding the term into the “oral–gut–brain axis”. Metabolites produced by gut and oral microbes (particularly Streptococcus mutans and Porphyromonas gingivalis) have been linked to the formation of β-amyloid and its accumulation in the brain, tau protein phosphorylation, and neuroinflammation in individuals with Alzheimer's disease [400].
Short-chain fatty acids (SCFA), such as butyrate, are considered a primary microbial anti-inflammatory metabolite that is known to inhibit the NF-κB pathway [401] and regulate Th17/Treg differentiation by inhibiting IL-6/signal transducer and the activator of transcription 3 (STAT3)/IL-17 pathway and promoting Foxp3 [402,403]. Faecalibacterium prausnitzii has been identified as a butyrate-producing bacterium with an inverse relationship with different proinflammatory markers [402].
Another feature of the centenarians' gut ecosystem is the increase in facultative anaerobes, such as bacteria belonging to the Micrococcaceae family, the Fusobacterium, Bacillus, Staphylococcus, and Corynebacterium genera, and many members of the phylum Proteobacteria. Such opportunistic microbes, especially the Enterobacteriaceae family, thrive in an inflamed environment and are known to increase in the elderly and are associated with a decrease in F. prausnitzii [404]. Overall, studies with centenarian populations typically show reduced core taxa, such as Bacteroides and Roseburia, associated with increases in Akkermansia and Bifidobacterium, which may have pro-longevity effects [405]. Akkermansia can be modulated by local levels of IFN-γ [406], a cytokine linked to inflammaging, and has immunomodulatory effects in the host, possibly through its pili structures [407]. Other mechanisms by which changes in the gut microbiota can intersect with aging or immunity include the production of amino acid metabolites, such as from tryptophan (e.g., indoles that bind the aryl hydrocarbon receptor [408] and phenylalanine/tyrosine [409], and through the production of secondary bile acids [410]. Despite these advances, more work, such as through fecal microbial transplantation studies [411], will undoubtedly improve our knowledge of mechanisms by which the gut microbiota intersects with the immune system to control healthy aging. A full scope of the recent advances in characterizing the role of the gut microbiome in aging and longevity has been systematically reviewed here [412].
5. Conclusions and future perspectives
This review highlights the bidirectional relationships between chronic inflammation and the hallmarks of aging by summarizing the underlying molecular mechanisms (Figure 3). Immune dysfunction and changes in inflammatory pathways are transversal contributors to the aging process, propagating a rippling effect through the other drivers of aging. Due to its interconnectedness and ability to accentuate aging, drivers of chronic inflammation may be the ideal target to address aging with high translational potential. While their clinical application and long-term safety remain to be proven in clinical trials, current evidence suggests that immune-targeting approaches have the potential to revolutionize longevity research and treatment, as they have done in cancer therapies. Boosting and targeting the immune system to clear senescent cells is just one example, as we have discussed. Progress in this field will require technological improvements in the measurement and analysis of multi-omics and rapid cycles of translation from model organisms to humans and vice versa to identify new promising therapeutic targets.
Figure 3.
Clinical and Molecular Drivers Underlying the Loop of Chronic Inflammation and the Hallmarks of Aging: Through various molecular mechanisms and environmental triggers, chronic inflammation can drive aging and age-related diseases, which in turn can exacerbate chronic inflammation along with other hallmarks of aging. DAMPs = deoxyribonucleic acid damage-associated molecular patterns, ER = endoplasmic reticulum, NAD/NADH = nicotinamide adenine dinucleotide, NF-κB = nuclear factor kappa B, NLRP3 = nod-like receptor family pyrin domain-containing 3, PAMPs = pathogen-associated molecular pattern molecules, TLR = toll-like receptor.
As our understanding of aging continues to expand, new hallmarks of aging will inevitably emerge. Foreseeable future hallmarks of aging include those linked to the extracellular matrix (ECM) and mechanoimmunology, both up-and-coming areas of basic aging science research [[413], [414], [415]]. Indeed, aging and ARDs are associated with changes in the ECM and mechanical cell surroundings, including immune cells. Mechanosensing by immune cells, such as through YAP/TAZ or PIEZO1 signaling [416,417], alters immune cell function, maturation, and metabolism to drive aberrant inflammation [418]. Thus, one future direction of inflammaging research is to link physical cues with chemical cues, thus generating a better understanding of immunological changes with age.
Author contributions
All authors contributed to the writing of this manuscript.
Declaration of competing interest
DF is co-founder and & Chairman of the Scientific Advisory Board of Edifice Health, a company that utilizes Inflammatory Age (iAge) to quantify chronic inflammation and immune health. The remaining authors declare no competing interests.
Acknowledgements
This work was funded in part by funds from NIH grants R01DK128435 (DAW) and 5T32AG000266-23 (JJB), P01AI153559 (DF), U54AG075932 (DF, DAW), as well as the Buck Bioinformatics Core (DF) and CIHR grant PJT186165 (DAW).
Contributor Information
David Furman, Email: DFurman@buckinstitute.org.
Daniel A. Winer, Email: DWiner@buckinstitute.org.
Abbreviations
Abbreviation Definition
- ABCs
age-associated B cells
- AD
Alzheimer's Disease
- AGEs
advanced glycation end-products
- AID
activation-induced cytidine deaminase
- ALS
autophagy-lysosome system
- AMPK
5′-adenosine monophosphate-activated protein kinase
- AP-1
activator protein 1
- APCs
antigen-presenting cells
- ARDs
age-related diseases
- ATF6
ATG5 activating transcription factor 6 Autophagy-related 5
- ATM
ataxia-telangiectasia mutated
- ATR
ataxia telangiectasia and Rad3-related protein
- BAX
Bcl-2 Associated X-protei
- C/EBP
CCAAT-enhancer-binding proteins
- CBP
cyclic adenosine monophosphate response element binding protein binding protein
- cfDNA
cell-free DNA
- cGAS
cyclic GMP–AMP synthase
- CHOP
CCAAT-enhancer-binding proteins homologous protein
- CMA
chaperone-mediated autophagy
- COPD
chronic obstructive pulmonary disease
- COVID-19
coronavirus disease 2019
- CRP
c-reactive protein
- CVD
cardiovascular disease
- DAMPs
damage-associated molecular patterns
- DDR
DNA damage response
- DNA
deoxyribonucleic acid
- DNMT
DNA methyltransferase
- eIF2α
eukaryotic translation initiation factor 2
- ER
endoplasmic reticulum
- FFAs
Free fatty acids
- FoxO
forkhead box protein O
- GDF
growth differentiation factor
- GM-CSF
granulocyte-macrophage colony-stimulating factor
- GRO
growth-regulated oncogene
- H3K
H3 lysine
- HATs
histone acetyltransferases
- HDACs
histone deacetylases
- HERV-K
human endogenous retrovirus-K
- HSC
hematopoietic stem cells
- hTERT
human telomerase reverse transcriptase
- IFN
interferon
- IGF-1
insulin-like growth factor 1
- IGFR
insulin-like growth factor receptor
- IKKβ
inhibitor of nuclear factor kappa-B kinase subunit beta
- IL
interleukin
- INHBA
inhibin Subunit Beta A
- IP
immunoproteasome
- IR
insulin resistance
- IRE1α
inositol-requiring transmembrane kinase/endoribonuclease 1α
- IRF3
interferon regulatory factor 3
- IRS
insulin receptor substrate
- IIS
insulin and insulin-like growth factor 1 signaling
- JNK
jun n-terminal kinase
- LC3B
microtubule-associated protein 1 A/1 B-light chain 3
- LINE1
long interspersed nuclear element 1
- LKB-1
liver kinase B1
- LPS
lipopolysaccharide
- MAVS
mitochondrial antiviral-signaling protein
- MCP
monocyte chemotactic protein
- MDA5
anti-melanoma differentiation-associated gene 5
- miRNAs
microribonucleic acid
- MLH1
MutL homolog 1
- MMP
matrix metallopeptidase
- MMR
mismatch repair
- mRNA
messenger ribonucleic acid
- mtDNA
mitochondrial DNA
- mTOR
mammalian target of rapamycin
- mTORC1
mammalian target of rapamycin complex 1
- mTORC2
mammalian target of rapamycin complex 2
- MuSCs
muscle stem cells
- NAD
nicotinamide adenine dinucleotide
- NADPH
nicotinamide adenine dinucleotide phosphate hydrogen
- NAFLD
nonalcoholic fatty liver disease
- ncRNA
non-coding RNA
- NF-κB
nuclear factor kappa B
- NLR
nod-like receptor
- NLRP3
nod-like receptor family pyrin domain-containing 3
- NOS2
nitric oxide synthase
- PAMPs
pathogen-associated molecular pattern molecules
- PARP1
poly(adenosine diphosphate-ribose) polymerase 1
- PD
Parkinson's Disease
- PERK
protein kinase R-like endoplasmic reticulum kinase
- PGC
peroxisome proliferator-activated receptor gamma coactivator
- Phox
phagocyte oxidase
- PI3K
phosphatidylinositol-3-kinase
- PRAAS
proteasome-associated autoinflammatory syndrome
- PRRs
pattern-recognition receptors
- RIDD
regulated IRE1α-dependent decay
- RIG-I
retinoic acid-inducible gene I
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- RTPs
retrotransposons
- SARS-CoV-2
severe acute respiratory syndrome coronavirus 2
- SASP
senescence-associated secretory phenotype
- SERPINE1
serpin family E member 1
- SESNs
sestrins
- Sirt
sirtuin
- SSPC
skeletal stem/progenitor cells
- STING
stimulator of interferon genes
- T2D
type 2 diabetes
- TBK1
tank-binding kinase 1
- TCR
T cell receptor
- TGFβ
transforming growth factor-beta
- TLR
toll-like receptor
- TNF
tumor necrosis factor
- TRAF6
tumor necrosis factor receptor-associated factor 6
- UPR
unfolded protein response
- UPS
ubiquitin-proteolytic system
- UV
ultraviolet light
- VAT
visceral adipose tissue
- YAP1
yes-associated protein 1
Data availability
No data was used for the research described in the article.
References
- 1.Keilich S.R., Bartley J.M., Haynes L. Diminished immune responses with aging predispose older adults to common and uncommon influenza complications. Cell Immunol. 2019;345(103992) doi: 10.1016/j.cellimm.2019.103992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartleson J.M., Radenkovic D., Covarrubias A.J., Furman D., Winer D.A., Verdin E. SARS-CoV-2, COVID-19 and the ageing immune system. Nat Aging. 2021;1(9):769–782. doi: 10.1038/s43587-021-00114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Niccoli T., Partridge L. Ageing as a risk factor for disease. Curr Biol. 2012;22(17):R741–R752. doi: 10.1016/j.cub.2012.07.024. [DOI] [PubMed] [Google Scholar]
- 4.Jiang X., Holmes C., McVean G. The impact of age on genetic risk for common diseases. PLoS Genet. 2021;17(8) doi: 10.1371/journal.pgen.1009723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153(6):1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G. Hallmarks of aging: an expanding universe. Cell. 2023;186(2):243–278. doi: 10.1016/j.cell.2022.11.001. [DOI] [PubMed] [Google Scholar]
- 7.Furman D., Campisi J., Verdin E., Carrera-Bastos P., Targ S., Franceschi C., et al. Chronic inflammation in the etiology of disease across the life span. Nat Med. 2019;25(12):1822–1832. doi: 10.1038/s41591-019-0675-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melov S. Mitochondrial oxidative stress. Physiologic consequences and potential for a role in aging. Ann N Y Acad Sci. 2000;908(1):219–225. doi: 10.1111/j.1749-6632.2000.tb06649.x. [DOI] [PubMed] [Google Scholar]
- 9.Fulop T., Larbi A., Pawelec G., Khalil A., Cohen A.A., Hirokawa K., et al. Immunology of aging: the birth of inflammaging. Clin Rev Allergy Immunol. 2023;64(2):109–122. doi: 10.1007/s12016-021-08899-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saxena S., Zou L. Hallmarks of DNA replication stress. Mol Cell. 2022;82(12):2298–2314. doi: 10.1016/j.molcel.2022.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362(6422):709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 12.Giglia-Mari G., Zotter A., Vermeulen W. DNA damage response. Cold Spring Harb Perspect Biol. 2011;3(1):a000745. doi: 10.1101/cshperspect.a000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Y., Geng A., Zhang W., Qian Z., Wan X., Jiang Y., et al. Fight to the bitter end: DNA repair and aging. Ageing Res Rev. 2020;64(101154) doi: 10.1016/j.arr.2020.101154. [DOI] [PubMed] [Google Scholar]
- 14.Crusz S.M., Balkwill F.R. Inflammation and cancer: advances and new agents. Nat Rev Clin Oncol. 2015;12(10):584–596. doi: 10.1038/nrclinonc.2015.105. [DOI] [PubMed] [Google Scholar]
- 15.Guo S., Zhu X., Huang Z., Wei C., Yu J., Zhang L., et al. Genomic instability drives tumorigenesis and metastasis and its implications for cancer therapy. Biomed Pharmacother. 2023;157(114036) doi: 10.1016/j.biopha.2022.114036. [DOI] [PubMed] [Google Scholar]
- 16.Hanahan D. Hallmarks of cancer: new dimensions. Cancer Discov. 2022;12(1):31–46. doi: 10.1158/2159-8290.CD-21-1059. [DOI] [PubMed] [Google Scholar]
- 17.Abbotts R., Wilson D.M., 3rd Coordination of DNA single strand break repair. Free Radic Biol Med. 2017;107:228–244. doi: 10.1016/j.freeradbiomed.2016.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verdin E. NAD+ in aging, metabolism, and neurodegeneration. Science. 2015;350(6265):1208–1213. doi: 10.1126/science.aac4854. [DOI] [PubMed] [Google Scholar]
- 19.Lee J.H., Hussain M., Kim E.W., Cheng S.J., Leung A.K.L., Fakouri N.B., et al. Mitochondrial PARP1 regulates NAD+-dependent poly ADP-ribosylation of mitochondrial nucleoids. Exp Mol Med. 2022;54(12):2135–2147. doi: 10.1038/s12276-022-00894-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wiley C.D., Velarde M.C., Lecot P., Liu S., Sarnoski E.A., Freund A., et al. Mitochondrial dysfunction induces senescence with a distinct secretory phenotype. Cell Metabol. 2016;23(2):303–314. doi: 10.1016/j.cmet.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andina N., de Meuron L., Schnegg-Kaufmann A.S., Sarangdhar M.A, Ansermet C., Bombaci G., et al. Increased inflammasome activation is associated with aging and chronic myelomonocytic leukemia disease severity. J Immunol. 2023;210(5):580–589. doi: 10.4049/jimmunol.2200412. [DOI] [PubMed] [Google Scholar]
- 22.Walker K.A., Basisty N., Wilson D.M., 3rd, Ferrucci L. Connecting aging biology and inflammation in the omics era. J Clin Invest. 2022;132(14) doi: 10.1172/JCI158448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erdal E., Haider S., Rehwinkel J., Harris A.L., McHugh P.J. A prosurvival DNA damage-induced cytoplasmic interferon response is mediated by end resection factors and is limited by Trex1. Genes Dev. 2017;31(4):353–369. doi: 10.1101/gad.289769.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao J., Zhang L., Lu A., Han Y., Colangelo D., Bukata C., et al. ATM is a key driver of NF-κB-dependent DNA-damage-induced senescence, stem cell dysfunction and aging. Aging (Albany NY) 2020;12(6):4688–4710. doi: 10.18632/aging.102863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kang C., Xu Q., Martin T.D., Li M.Z., Demara M., Aron L., et al. The DNA damage response induces inflammation and senescence by inhibiting autophagy of GATA4. Science. 2015;349(6255):aaa5612. doi: 10.1126/science.aaa5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dunphy G., Flannery S.M., Almine J.F., Connolly D.J., Paulus C., Jonsson K.L., et al. Non-canonical activation of the DNA sensing adaptor STING by ATM and IFI16 mediates NF-κB signaling after nuclear DNA damage. Mol Cell. 2018;71(5):745–760. doi: 10.1016/j.molcel.2018.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hinz M., Stilmann M., Arslan S.Ç., Khanna K.K., Dittmar G., Scheidereit C. A cytoplasmic ATM-TRAF6-cIAP1 module links nuclear DNA damage signaling to ubiquitin-mediated NF-κB activation. Mol Cell. 2010;40(1):63–74. doi: 10.1016/j.molcel.2010.09.008. [DOI] [PubMed] [Google Scholar]
- 28.Zhao Y., Simon M., Seluanov A., Gorbunova V. DNA damage and repair in age-related inflammation. Nat Rev Immunol. 2023;23(2):75–89. doi: 10.1038/s41577-022-00751-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cercek A., Lumish M., Sinopoli J., Weiss J., Shia J., Lamendola-Essel M., et al. PD-1 blockade in mismatch repair-deficient, locally advanced rectal cancer. N Engl J Med. 2022;386(25):2363–2376. doi: 10.1056/NEJMoa2201445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.St Sauver J., Rocca W., LeBrasseur N., Chamberlain A., Olson J., Jacobson D., et al. Inflammatory biomarkers, multi-morbidity, and biologic aging. J Int Med Res. 2022;50(7) doi: 10.1177/03000605221109393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X., Andresen1 B.T., Hill M., Zhang J., Booth F., Zhang C. Role of reactive oxygen species in tumor necrosis factor-alpha induced endothelial dysfunction. Curr Hypertens Rev. 2008;4(4):245–255. doi: 10.2174/157340208786241336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blaser H., Dostert C., Mak T.W., Brenner D. TNF and ROS crosstalk in inflammation. Trends Cell Biol. 2016;26(4):249–261. doi: 10.1016/j.tcb.2015.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Dizdaroglu M., Jaruga P. Mechanisms of free radical-induced damage to DNA. Free Radic Res. 2012;46(4):382–419. doi: 10.3109/10715762.2011.653969. [DOI] [PubMed] [Google Scholar]
- 34.Ionescu-Tucker A., Cotman C.W. Emerging roles of oxidative stress in brain aging and Alzheimer's disease. Neurobiol Aging. 2021;107:86–95. doi: 10.1016/j.neurobiolaging.2021.07.014. [DOI] [PubMed] [Google Scholar]
- 35.Kolodkin A., Sharma R.P., Colangelo A.M., Ignatenko A., Martorana F., Jennen D., et al. ROS networks: designs, aging, Parkinson’s disease and precision therapies. NPJ Syst Biol Appl. 2020;6(1):34. doi: 10.1038/s41540-020-00150-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pignatelli P., Menichelli D., Pastori D., Violi F. Oxidative stress and cardiovascular disease: new insights. Kardiol Pol. 2018;76(4):713–722. doi: 10.5603/KP.a2018.0071. [DOI] [PubMed] [Google Scholar]
- 37.Otoupalova E., Smith S., Cheng G., Thannickal V.J. Oxidative stress in pulmonary fibrosis. Compr Physiol. 2020;10(2):509–547. doi: 10.1002/cphy.c190017. [DOI] [PubMed] [Google Scholar]
- 38.Tang S.P., Mao X.L., Chen Y.H., Yan L.L., Ye L.P., Li S.W. Reactive oxygen species induce fatty liver and ischemia-reperfusion injury by promoting inflammation and cell death. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.870239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang L., Tang J., Wang L., Tan F., Song H., Zhou J., et al. Oxidative stress in oocyte aging and female reproduction. J Cell Physiol. 2021;236(12):7966–7983. doi: 10.1002/jcp.30468. [DOI] [PubMed] [Google Scholar]
- 40.Nago M., Arichi A., Omura N., Iwashita Y., Kawamura T., Yumura Y. Aging increases oxidative stress in semen. Investig Clin Urol. 2021;62(2):233–238. doi: 10.4111/icu.20200066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kay J., Thadhani E., Samson L., Engelward B. Inflammation-induced DNA damage, mutations and cancer. DNA Repair. 2019;83(102673) doi: 10.1016/j.dnarep.2019.102673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koi M., Tseng-Rogenski S.S., Carethers J.M. Inflammation-associated microsatellite alterations: mechanisms and significance in the prognosis of patients with colorectal cancer. World J Gastrointest Oncol. 2018;10(1):1–14. doi: 10.4251/wjgo.v10.i1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neri S., Gardini A., Facchini A., Oliveri F., Franceschi C., Ravaglia G., et al. Mismatch repair system and aging: microsatellite instability in peripheral blood cells from differently aged participants. J Gerontol A Biol Sci Med Sci. 2005;60(3):285–292. doi: 10.1093/gerona/60.3.285. [DOI] [PubMed] [Google Scholar]
- 44.Mayer-Hamblett N., Kronmal R.A., Gibson R.L., Rosenfeld M., Retsch-Bogart G., Treggiari M.M., et al. Initial Pseudomonas aeruginosa treatment failure is associated with exacerbations in cystic fibrosis. Pediatr Pulmonol. 2012;47(2):125–134. doi: 10.1002/ppul.21525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mechtcheriakova D., Svoboda M., Meshcheryakova A., Jensen-Jarolim E. Activation-induced cytidine deaminase (AID) linking immunity, chronic inflammation, and cancer. Cancer Immunol Immunother. 2012;61(9):1591–1598. doi: 10.1007/s00262-012-1255-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimizu T., Marusawa H., Endo Y., Chiba T. Inflammation-mediated genomic instability: roles of activation-induced cytidine deaminase in carcinogenesis. Cancer Sci. 2012;103(7):1201–1206. doi: 10.1111/j.1349-7006.2012.02293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Çakan E., Gunaydin G. Activation induced cytidine deaminase: an old friend with new faces. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.965312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Qiao Y., Han X., Guan G., Wu N., Sun J., Pak V., et al. TGF-β triggers HBV cccDNA degradation through AID-dependent deamination. FEBS Lett. 2016;590(3):419–427. doi: 10.1002/1873-3468.12058. [DOI] [PubMed] [Google Scholar]
- 49.Chiba T., Marusawa H., Ushijima T. Inflammation-associated cancer development in digestive organs: mechanisms and roles for genetic and epigenetic modulation. Gastroenterology. 2012;143(3):550–563. doi: 10.1053/j.gastro.2012.07.009. [DOI] [PubMed] [Google Scholar]
- 50.Gushima M., Hirahashi M., Matsumoto T., Fujita K., Ohuchida K., Oda Y., et al. Expression of activation-induced cytidine deaminase in ulcerative colitis-associated carcinogenesis. Histopathology. 2011;59(3):460–469. doi: 10.1111/j.1365-2559.2011.03965.x. [DOI] [PubMed] [Google Scholar]
- 51.Sawai Y., Kodama Y., Shimizu T., Ota Y., Maruna T., Eso Y., et al. Activation-induced cytidine deaminase contributes to pancreatic tumorigenesis by inducing tumor-related gene mutations. Cancer Res. 2015;75(16):3292–3301. doi: 10.1158/0008-5472.CAN-14-3028. [DOI] [PubMed] [Google Scholar]
- 52.Switzer C.H., Cho H.J., Eykyn T.R., Lavender P., Eaton P. NOS2 and S-nitrosothiol signaling induces DNA hypomethylation and LINE-1 retrotransposon expression. Proc Natl Acad Sci U S A. 2022;119(21) doi: 10.1073/pnas.2200022119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Cecco M., Ito T., Petrashen A.P., Elias A.E., Skvir N.J., Criscione S.W., et al. L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature. 2019;566(7742):73–78. doi: 10.1038/s41586-018-0784-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bellon M., Nicot C. Telomere dynamics in immune senescence and exhaustion triggered by chronic viral infection. Viruses. 2017;9(10) doi: 10.3390/v9100289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hayflick L. The limited in vitro lifetime of human diploid cell strains. Exp Cell Res. 1965;37(3):614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 56.Sławińska N., Krupa R. Molecular aspects of senescence and organismal ageing-DNA damage response, telomeres, inflammation and chromatin. Int J Mol Sci. 2021;22(2):590. doi: 10.3390/ijms22020590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rossiello F., Jurk D., Passos J.F., d'Adda di Fagagna F. Telomere dysfunction in ageing and age-related diseases. Nat Cell Biol. 2022;24(2):135–147. doi: 10.1038/s41556-022-00842-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lagnado A., Leslie J., Ruchaud-Sparagano M.H., Victorelli S., Hirsova P., Ogrodnik M., et al. Neutrophils induce paracrine telomere dysfunction and senescence in ROS-dependent manner. EMBO J. 2021;40(9) doi: 10.15252/embj.2020106048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bloom S.I., Tucker J.R., Lim J., Thomas T.J., Stoddard G.J., Lesniewski L.A., et al. Aging results in DNA damage and telomere dysfunction that is greater in endothelial versus vascular smooth muscle cells and is exacerbated in atheroprone regions. GeroScience. 2022;44(6):2741–2755. doi: 10.1007/s11357-022-00681-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang J., Thomas H.R., Li Z., Yeo N.C.F., Scott H.E., Dang N., et al. Puma, noxa, p53, and p63 differentially mediate stress pathway induced apoptosis. Cell Death Dis. 2021;12(7):659. doi: 10.1038/s41419-021-03902-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kumari R., Jat P. Mechanisms of cellular senescence: cell cycle arrest and senescence associated secretory phenotype. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.645593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chakravarti D., LaBella K.A., DePinho R.A. Telomeres: history, health, and hallmarks of aging. Cell. 2021;184(2):306–322. doi: 10.1016/j.cell.2020.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Maciejowski J., de Lange T. Telomeres in cancer: tumour suppression and genome instability. Nat Rev Mol Cell Biol. 2017;18(3):175–186. doi: 10.1038/nrm.2016.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nassour J., Aguiar L.G., Correia A., Schmidt T.T., Mainz L., Przetocka S., et al. Telomere-to-mitochondria signalling by ZBP1 mediates replicative crisis. Nature. 2023;614(7949):767–773. doi: 10.1038/s41586-023-05710-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Amano H., Chaudhury A., Rodriguez-Aguayo C., Lu L., Akhanov V., Catic A., et al. Telomere dysfunction induces sirtuin repression that drives telomere-dependent disease. Cell Metabol. 2019;29(6):1274–1290. doi: 10.1016/j.cmet.2019.03.001. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Amano H., Sahin E. Telomeres and sirtuins: at the end we meet again. Mol Cell Oncol. 2019;6(5) doi: 10.1080/23723556.2019.1632613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Heba A.C., Toupance S., Arnone D., Peyrin-Biroulet L., Benetos A., Ndiaye N.C. Telomeres: new players in immune-mediated inflammatory diseases? J Autoimmun. 2021;123(102699) doi: 10.1016/j.jaut.2021.102699. [DOI] [PubMed] [Google Scholar]
- 68.Van Der Stukken C., Nawrot T.S., Alfano R., Wang C., Langie S.A.S., Plusquin M., et al. The telomere-mitochondrial axis of aging in newborns. Aging (Albany NY) 2022;14(4):1627–1650. doi: 10.18632/aging.203897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kang Y., Zhang H., Zhao Y., Wang Y., Wang W., He Y., et al. Telomere dysfunction disturbs macrophage mitochondrial metabolism and the NLRP3 inflammasome through the PGC-1α/TNFAIP3 axis. Cell Rep. 2018;22(13):3493–3506. doi: 10.1016/j.celrep.2018.02.071. [DOI] [PubMed] [Google Scholar]
- 70.Nassour J., Radford R., Correia A., Fuste J.M., Schoelle B., Jauch A., et al. Autophagic cell death restricts chromosomal instability during replicative crisis. Nature. 2019;565(7741):659–663. doi: 10.1038/s41586-019-0885-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mender I., Zhang A., Ren Z., Han C., Deng Y., Siteni S., et al. Telomere stress potentiates STING-dependent anti-tumor immunity. Cancer Cell. 2020;38(3):400–411. doi: 10.1016/j.ccell.2020.05.020.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mittelbrunn M., Kroemer G. Hallmarks of T cell aging. Nat Immunol. 2021;22(6):687–698. doi: 10.1038/s41590-021-00927-z. [DOI] [PubMed] [Google Scholar]
- 73.Laphanuwat P., Gomes D.C.O., Akbar A.N. Senescent T cells: beneficial and detrimental roles. Immunol Rev. 2023 doi: 10.1111/imr.13206. Published online 25 April. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marrella V., Facoetti A., Cassani B. Cellular senescence in immunity against infections. Int J Mol Sci. 2022;23(19) doi: 10.3390/ijms231911845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Matthe D.M., Thoma O.M., Sperka T., Neurath M.F., Waldner M.J. Telomerase deficiency reflects age-associated changes in CD4+ T cells. Immun Ageing. 2022;19(1):16. doi: 10.1186/s12979-022-00273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ruiz A., Flores-Gonzalez J., Buendia-Roldan I., Chavez-Galan L. Telomere shortening and its association with cell dysfunction in lung diseases. Int J Mol Sci. 2021;23(1):425. doi: 10.3390/ijms23010425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Aviv A. The bullwhip effect, T-cell telomeres, and SARS-CoV-2. Lancet Healthy Longev. 2022;3(10):e715–e721. doi: 10.1016/S2666-7568(22)00190-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lanna A., Vaz B., D’Ambra C., Valvo S., Vuotto C., Chiurchiù V., et al. An intercellular transfer of telomeres rescues T cells from senescence and promotes long-term immunological memory. Nat Cell Biol. 2022;24(10):1461–1474. doi: 10.1038/s41556-022-00991-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tahara T., Shibata T., Kawamura T., Ishizuka T., Okubo M., Nagasaka M., et al. Telomere length in non-neoplastic gastric mucosa and its relationship to H. pylori infection, degree of gastritis, and NSAID use. Clin Exp Med. 2016;16(1):65–71. doi: 10.1007/s10238-014-0335-0. [DOI] [PubMed] [Google Scholar]
- 80.Laish I., Mannasse-Green B., Hadary R., Biron-Shental T., Konikoff F.M., Amiel A., et al. Telomere dysfunction in nonalcoholic fatty liver disease and cryptogenic cirrhosis. Cytogenet Genome Res. 2016;150(2):93–99. doi: 10.1159/000454654. [DOI] [PubMed] [Google Scholar]
- 81.Lim H.F., Tan N.S., Dehghan R., Shen M., Liew M.F., Bee S.W.L., et al. Shortened telomere length in sputum cells of bronchiectasis patients is associated with dysfunctional inflammatory pathways. Lung. 2022;200(3):401–407. doi: 10.1007/s00408-022-00535-0. [DOI] [PubMed] [Google Scholar]
- 82.Slusher A.L., Zúñiga T.M., Acevedo E.O. Inflamm-aging is associated with lower plasma PTX3 concentrations and an impaired capacity of PBMCs to express hTERT following LPS stimulation. Mediat Inflamm. 2019;2019 doi: 10.1155/2019/2324193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chung S.S., Wu Y., Okobi Q., Adekoya D., Atefi M., Clarke O., et al. Proinflammatory cytokines IL-6 and TNF-α increased telomerase activity through NF-κB/STAT1/STAT3 activation, and withaferin A inhibited the signaling in colorectal cancer cells. Mediat Inflamm. 2017;2017:5958429. doi: 10.1155/2017/5958429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maekawa T., Liu B., Nakai D., Yoshida K., Nakamura K.I., Yasukawa M., et al. ATF7 mediates TNF-α-induced telomere shortening. Nucleic Acids Res. 2018;46(9):4487–4504. doi: 10.1093/nar/gky155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Conte M., Martucci M., Chiariello A., Franceschi C., Salvioli S. Mitochondria, immunosenescence and inflammaging: a role for mitokines? Semin Immunopathol. 2020;42(5):607–617. doi: 10.1007/s00281-020-00813-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jacome Burbano M.S., Gilson E. The power of stress: the Telo-hormesis hypothesis. Cells. 2021;10(5):1156. doi: 10.3390/cells10051156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Covarrubias A.J., Perrone R., Grozio A., Verdin E. NAD+ metabolism and its roles in cellular processes during ageing. Nat Rev Mol Cell Biol. 2021;22(2):119–141. doi: 10.1038/s41580-020-00313-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zeidler J.D., Kashyap S., Hogan K.A., Chini E.N. Implications of the NADase CD38 in COVID pathophysiology. Physiol Rev. 2022;102(1):339–341. doi: 10.1152/physrev.00007.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zeidler J.D., Hogan K.A., Agorrody G., Peclat T.R., Kashyap S., Kanamori K.S., et al. The CD38 glycohydrolase and the NAD sink: implications for pathological conditions. Am J Physiol Cell Physiol. 2022;322(3):C521–C545. doi: 10.1152/ajpcell.00451.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Guo Z., Li P., Ge J., Li H. SIRT6 in aging, metabolism, inflammation and cardiovascular diseases. Aging Dis. 2022;13(6):1787–1822. doi: 10.14336/AD.2022.0413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Abbas A.A., Akbar A.N. Induction of T cell senescence by cytokine induced bystander activation. Front Aging. 2021;2 doi: 10.3389/fragi.2021.714239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lanna A., Coutavas E., Levati L., Seidel J., Rustin M.H., Henson S.M., et al. IFN-α inhibits telomerase in human CD8+ T cells by both hTERT downregulation and induction of p38 MAPK signaling. J Immunol. 2013;191(7):3744–3752. doi: 10.4049/jimmunol.1301409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Reed J.R., Vukmanovic-Stejic M., Fletcher J.M., Soares M.V., Cook J.E., Orteu C.H., et al. Telomere erosion in memory T cells induced by telomerase inhibition at the site of antigenic challenge in vivo. J Exp Med. 2004;199(10):1433–1443. doi: 10.1084/jem.20040178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tichy E.D., Ma N., Sidibe D., Loro E., Kocan J., Chen D.Z., et al. Persistent NF-κB activation in muscle stem cells induces proliferation-independent telomere shortening. Cell Rep. 2021;35(6) doi: 10.1016/j.celrep.2021.109098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jurk D., Wilson C., Passos J.F., Oakley F., Correia-Melo C., Greaves L., et al. Chronic inflammation induces telomere dysfunction and accelerates ageing in mice. Nat Commun. 2014;2(1):4172. doi: 10.1038/ncomms5172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nardini C., Moreau J.F., Gensous N., Ravaioli F., Garagnani P., Bacalini M.G. The epigenetics of inflammaging: the contribution of age-related heterochromatin loss and locus-specific remodelling and the modulation by environmental stimuli. Semin Immunol. 2018;40:49–60. doi: 10.1016/j.smim.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 97.Gorbunova V., Seluanov A., Mita P., McKerrow W., Fenyö D., Boeke J.D., et al. The role of retrotransposable elements in ageing and age-associated diseases. Nature. 2021;596(7870):43–53. doi: 10.1038/s41586-021-03542-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang L., Lu Q., Chang C. Epigenetics in health and disease. Adv Exp Med Biol. 2020;1253:3–55. doi: 10.1007/978-981-15-3449-2_1. [DOI] [PubMed] [Google Scholar]
- 99.Zhang J., Wang S., Liu B. New insights into the genetics and epigenetics of aging plasticity. Genes. 2023;14(2):329. doi: 10.3390/genes14020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Saul D., Kosinsky R.L. Epigenetics of aging and aging-associated diseases. Int J Mol Sci. 2021;22(1):401. doi: 10.3390/ijms22010401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sarkar A., Liu N.Q., Magallanes J., Tassey J., Lee S., Shkhyan R., et al. STAT3 promotes a youthful epigenetic state in articular chondrocytes. Aging Cell. 2023;22(2) doi: 10.1111/acel.13773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Teo Y.V., Capri M., Morsiani C., Pizza G., Faria A.M.C., Franceschi C., et al. Cell-free DNA as a biomarker of aging. Aging Cell. 2019;18(1) doi: 10.1111/acel.12890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Urban L.A., Trinh A., Pearlman E., Siryaporn A., Downing T.L. The impact of age-related hypomethylated DNA on immune signaling upon cellular demise. Trends Immunol. 2021;42(6):464–468. doi: 10.1016/j.it.2021.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wielscher M., Mandaviya P.R., Kuehnel B., Joehanes R., Mustafa R., Robinson O., et al. DNA methylation signature of chronic low-grade inflammation and its role in cardio-respiratory diseases. Nat Commun. 2022;13(1):2408. doi: 10.1038/s41467-022-29792-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhao J., Huai J. Role of primary aging hallmarks in Alzheimer's disease. Theranostics. 2023;13(1):197–230. doi: 10.7150/thno.79535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Khalil H., Tazi M., Caution K., Ahmed A., Kanneganti A., Assani K., et al. Aging is associated with hypermethylation of autophagy genes in macrophages. Epigenetics. 2016;11(5):381–388. doi: 10.1080/15592294.2016.1144007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Dyer M.L., Khouja J.N., Jackson A.R., Havill M.A., Dockrell M.J., Munafo M.R., et al. Effects of electronic cigarette e-liquid flavouring on cigarette craving. Tobac Control. 2023;32(e1):e3–e9. doi: 10.1136/tobaccocontrol-2021-056769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Issa J.P., Ahuja N., Toyota M., Bronner M.P., Brentnall T.A. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001;61(9):3573–3577. https://www.ncbi.nlm.nih.gov/pubmed/11325821 [PubMed] [Google Scholar]
- 109.Niwa T., Tsukamoto T., Toyoda T., Mori A., Tanaka H., Maekita T., et al. Inflammatory processes triggered by Helicobacter pylori infection cause aberrant DNA methylation in gastric epithelial cells. Cancer Res. 2010;70(4):1430–1440. doi: 10.1158/0008-5472.CAN-09-2755. [DOI] [PubMed] [Google Scholar]
- 110.Nishida N., Nagasaka T., Nishimura T., Ikai I., Boland C.R., Goel A. Aberrant methylation of multiple tumor suppressor genes in aging liver, chronic hepatitis, and hepatocellular carcinoma. Hepatology. 2008;47(3):908–918. doi: 10.1002/hep.22110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vakoc C.R., Sachdeva M.M., Wang H., Blobel G.A. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26(24):9185–9195. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yang N., Occean J.R., Melters D.P., Shi C., Wang L., Stransky S., et al. A hyper-quiescent chromatin state formed during aging is reversed by regeneration. bioRxivorg. 2023 doi: 10.1101/2023.02.14.528512. Published online 15 February. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hyun K., Jeon J., Park K., Kim J. Writing, erasing and reading histone lysine methylations. Exp Mol Med. 2017;49(4):e324. doi: 10.1038/emm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhu X., Chen Z., Shen W., Huang G., Sedivy J.M., Wang H., et al. Inflammation, epigenetics, and metabolism converge to cell senescence and ageing: the regulation and intervention. Signal Transduct Targeted Ther. 2021;6(1):245. doi: 10.1038/s41392-021-00646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Soriano-Cantón R., Perez-Villalba A., Morante-Redolat J.M., Marqués-Torrejón M.Á., Pallás M., Pérez-Sánchez F., et al. Regulation of the p19(Arf)/p53 pathway by histone acetylation underlies neural stem cell behavior in senescence-prone SAMP8 mice. Aging Cell. 2015;14(3):453–462. doi: 10.1111/acel.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sen P., Lan Y., Li C.Y., Sidoli S., Donahue G., Dou Z., et al. Histone acetyltransferase p300 induces DE Novo super-enhancers to drive cellular senescence. Mol Cell. 2019;73(4):684–698. doi: 10.1016/j.molcel.2019.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Andrenacci D., Cavaliere V., Lattanzi G. The role of transposable elements activity in aging and their possible involvement in laminopathic diseases. Ageing Res Rev. 2020;57(100995) doi: 10.1016/j.arr.2019.100995. [DOI] [PubMed] [Google Scholar]
- 118.Pappalardo X.G., Barra V. Losing DNA methylation at repetitive elements and breaking bad. Epigenet Chromatin. 2021;14(1):25. doi: 10.1186/s13072-021-00400-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yushkova E., Moskalev A. Transposable elements and their role in aging. Ageing Res Rev. 2023;86 doi: 10.1016/j.arr.2023.101881. [DOI] [PubMed] [Google Scholar]
- 120.Simon M., Van Meter M., Ablaeva J., Ke Z., Gonzalez R.S., Taguchi T., et al. LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metabol. 2019;29(4):871–885. doi: 10.1016/j.cmet.2019.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Geis F.K., Goff S.P. Silencing and transcriptional regulation of endogenous retroviruses: an overview. Viruses. 2020;12(8):884. doi: 10.3390/v12080884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang K., Liu H., Hu Q., Wang L., Liu J., Zheng Z., et al. Epigenetic regulation of aging: implications for interventions of aging and diseases. Signal Transduct Targeted Ther. 2022;7(1):374. doi: 10.1038/s41392-022-01211-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lee M.Y., Lee J., Hyeon S.J., Cho H., Hwang Y.J., Shin J.Y., et al. Epigenome signatures landscaped by histone H3K9me3 are associated with the synaptic dysfunction in Alzheimer’s disease. Aging Cell. 2020;19(6) doi: 10.1111/acel.13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dembny P., Newman A.G., Singh M., Hinz M., Szczepek M., Krüger C., et al. Human endogenous retrovirus HERV-K(HML-2) RNA causes neurodegeneration through Toll-like receptors. JCI Insight. 2020;5(7) doi: 10.1172/jci.insight.131093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liu X., Liu Z., Wu Z., Ren J., Fan Y., Sun L., et al. Resurrection of endogenous retroviruses during aging reinforces senescence. Cell. 2023;186(2):287–304. doi: 10.1016/j.cell.2022.12.017. e26. [DOI] [PubMed] [Google Scholar]
- 126.Kinser H.E., Pincus Z. MicroRNAs as modulators of longevity and the aging process. Hum Genet. 2020;139(3):291–308. doi: 10.1007/s00439-019-02046-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Olivieri F., Prattichizzo F., Giuliani A., Matacchione G., Rippo M.R., Sabbatinelli J., et al. miR-21 and miR-146a: the microRNAs of inflammaging and age-related diseases. Ageing Res Rev. 2021;70(101374):101374. doi: 10.1016/j.arr.2021.101374. [DOI] [PubMed] [Google Scholar]
- 128.Zhang H.Q., Wang J.Y., Li Z.F., Cui L., Huang S.S., Zhu L.B., et al. DNA methyltransferase 1 is dysregulated in Parkinson’s disease via mediation of miR-17. Mol Neurobiol. 2021;58(6):2620–2633. doi: 10.1007/s12035-021-02298-w. [DOI] [PubMed] [Google Scholar]
- 129.Wang X., Zhou Y., Gao Q., Ping D., Wang Y., Wu W., et al. The role of exosomal microRNAs and oxidative stress in neurodegenerative diseases. Oxid Med Cell Longev. 2020;2020:3232869. doi: 10.1155/2020/3232869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jusic A., Thomas P.B., Wettinger S.B., Dogan S., Farrugia R., Gaetano C., et al. Noncoding RNAs in age-related cardiovascular diseases. Ageing Res Rev. 2022;77(101610):101610. doi: 10.1016/j.arr.2022.101610. [DOI] [PubMed] [Google Scholar]
- 131.Zheng Y.L., Song G., Guo J.B., Su X., Chen Y.M., Yang Z., et al. Interactions among lncRNA/circRNA, miRNA, and mRNA in musculoskeletal degenerative diseases. Front Cell Dev Biol. 2021;9:753931. doi: 10.3389/fcell.2021.753931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li C., Ni Y.Q., Xu H., Xiang Q.Y., Zhao Y., Zhan J.K., et al. Roles and mechanisms of exosomal non-coding RNAs in human health and diseases. Signal Transduct Targeted Ther. 2021;6(1):383. doi: 10.1038/s41392-021-00779-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Colella M., Cuomo D., Peluso T., Falanga I., Mallardo M., De Felice M., et al. Ovarian aging: role of pituitary-ovarian axis hormones and ncRNAs in regulating ovarian mitochondrial activity. Front Endocrinol. 2021;12:791071. doi: 10.3389/fendo.2021.791071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Frankowska N., Lisowska K., Witkowski J.M. Proteolysis dysfunction in the process of aging and age-related diseases. Front Aging. 2022;3 doi: 10.3389/fragi.2022.927630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rai M., Curley M., Coleman Z., Demontis F. Contribution of proteases to the hallmarks of aging and to age-related neurodegeneration. Aging Cell. 2022;21(5) doi: 10.1111/acel.13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Miles J., Scherz-Shouval R., van Oosten-Hawle P. Expanding the organismal proteostasis network: linking systemic stress signaling with the innate immune response. Trends Biochem Sci. 2019;44(11):927–942. doi: 10.1016/j.tibs.2019.06.009. [DOI] [PubMed] [Google Scholar]
- 137.Ruano D. Proteostasis dysfunction in aged mammalian cells. The stressful role of inflammation. Front Mol Biosci. 2021;8 doi: 10.3389/fmolb.2021.658742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Franceschi C., Garagnani P., Vitale G., Capri M., Salvioli S. Inflammaging and ‘garb-aging’. Trends Endocrinol Metabol. 2017;28(3):199–212. doi: 10.1016/j.tem.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 139.Cuanalo-Contreras K., Schulz J., Mukherjee A., Park K.W., Armijo E., Soto C. Extensive accumulation of misfolded protein aggregates during natural aging and senescence. Front Aging Neurosci. 2022;14 doi: 10.3389/fnagi.2022.1090109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Chipurupalli S., Samavedam U., Robinson N. Crosstalk between ER stress, autophagy and inflammation. Front Med. 2021;8 doi: 10.3389/fmed.2021.758311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Salminen A., Kaarniranta K., Kauppinen A. ER stress activates immunosuppressive network: implications for aging and Alzheimer's disease. J Mol Med. 2020;98(5):633–650. doi: 10.1007/s00109-020-01904-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Di Conza G., Ho P.C., Cubillos-Ruiz J.R., Huang S.C.C. Control of immune cell function by the unfolded protein response. Nat Rev Immunol. 2023 doi: 10.1038/s41577-023-00838-0. Published online 8 February. [DOI] [PubMed] [Google Scholar]
- 143.Li D., Wu M. Pattern recognition receptors in health and diseases. Signal Transduct Targeted Ther. 2021;6(1):291. doi: 10.1038/s41392-021-00687-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Tan Y., Chen Q., Li X., Zeng Z., Xiong W., Li G., et al. Pyroptosis: a new paradigm of cell death for fighting against cancer. J Exp Clin Cancer Res. 2021;40(1):153. doi: 10.1186/s13046-021-01959-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tabibzadeh S. Cell-centric hypotheses of aging. Front Biosci. 2021;26(1):1–49. doi: 10.2741/4888. [DOI] [PubMed] [Google Scholar]
- 146.Kulkarni A., Preeti K., Tryphena K.P., Srivastava S., Singh S.B., Khatri D.K. Proteostasis in Parkinson's disease: recent development and possible implication in diagnosis and therapeutics. Ageing Res Rev. 2023;84(101816) doi: 10.1016/j.arr.2022.101816. [DOI] [PubMed] [Google Scholar]
- 147.Abbadie C., Pluquet O. Unfolded protein response (UPR) controls major senescence hallmarks. Trends Biochem Sci. 2020;45(5):371–374. doi: 10.1016/j.tibs.2020.02.005. [DOI] [PubMed] [Google Scholar]
- 148.Goetzke C.C., Ebstein F., Kallinich T. Role of proteasomes in inflammation. J Clin Med. 2021;10(8):1783. doi: 10.3390/jcm10081783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Arata Y., Watanabe A., Motosugi R., Murakami R., Goto T., Hori S., et al. Defective induction of the proteasome associated with T-cell receptor signaling underlies T-cell senescence. Gene Cell. 2019;24(12):801–813. doi: 10.1111/gtc.12728. [DOI] [PubMed] [Google Scholar]
- 150.Xu F., Zhang C., Zou Z., Fan E.K.Y., Chen L., Li Y., et al. Aging-related Atg5 defect impairs neutrophil extracellular traps formation. Immunology. 2017;151(4):417–432. doi: 10.1111/imm.12740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Colombini B., Dinu M., Murgo E., Lotti S., Tarquini R., Sofi F., et al. Ageing and low-level chronic inflammation: the role of the biological clock. Antioxidants. 2022;11(11) doi: 10.3390/antiox11112228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Zuo L., Prather E.R., Stetskiv M., Garrison D.E., Meade J.R., Peace T.I., et al. Inflammaging and oxidative stress in human diseases: from molecular mechanisms to novel treatments. Int J Mol Sci. 2019;20(18):4472. doi: 10.3390/ijms20184472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Lévy E., El Banna N., Baïlle D., Heneman-Masurel A., Truchet S., Rezaei H., et al. Causative links between protein aggregation and oxidative stress: a review. Int J Mol Sci. 2019;20(16):3896. doi: 10.3390/ijms20163896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Fani G., La Torre C.E., Cascella R., Cecchi C., Vendruscolo M., Chiti F. Misfolded protein oligomers induce an increase of intracellular Ca2+ causing an escalation of reactive oxidative species. Cell Mol Life Sci. 2022;79(9):500. doi: 10.1007/s00018-022-04513-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Martínez de Toda I., Ceprián N., Díaz-Del Cerro E., De la Fuente M. The role of immune cells in oxi-inflamm-aging. Cells. 2021;10(11):2974. doi: 10.3390/cells10112974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Li W.W., Wang H.J., Tan Y.Z., Wang Y.L., Yu S.N., Li Z.H. Reducing lipofuscin accumulation and cardiomyocytic senescence of aging heart by enhancing autophagy. Exp Cell Res. 2021;403(1) doi: 10.1016/j.yexcr.2021.112585. [DOI] [PubMed] [Google Scholar]
- 157.Tubío-Santamaría N., Ebstein F., Heidel F.H., Krüger E. Immunoproteasome function in normal and malignant hematopoiesis. Cells. 2021;10(7):1577. doi: 10.3390/cells10071577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Papendorf J.J., Krüger E., Ebstein F. Proteostasis perturbations and their roles in causing sterile inflammation and autoinflammatory diseases. Cells. 2022;11(9):1422. doi: 10.3390/cells11091422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Sarrabay G., Méchin D., Salhi A., Boursier G., Rittore C., Crow Y., et al. PSMB10, the last immunoproteasome gene missing for PRAAS. J Allergy Clin Immunol. 2020;145(3):1015–1017. doi: 10.1016/j.jaci.2019.11.024. e6. [DOI] [PubMed] [Google Scholar]
- 160.Pickering A.M., Lehr M., Miller R.A. Lifespan of mice and primates correlates with immunoproteasome expression. J Clin Invest. 2015;125(5):2059–2068. doi: 10.1172/JCI80514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Tripodi L., Molinaro D., Fortunato F., Mella C., Cassani B., Torrente Y., et al. Immunoproteasome inhibition ameliorates aged dystrophic mouse muscle environment. Int J Mol Sci. 2022;23(23):14657. doi: 10.3390/ijms232314657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Levine B., Kroemer G. Biological functions of autophagy genes: a disease perspective. Cell. 2019;176(1–2):11–42. doi: 10.1016/j.cell.2018.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Lőrincz P., Juhász G. Autophagosome-lysosome fusion. J Mol Biol. 2020;432(8):2462–2482. doi: 10.1016/j.jmb.2019.10.028. [DOI] [PubMed] [Google Scholar]
- 164.Tabibzadeh S. Role of autophagy in aging: the good, the bad, and the ugly. Aging Cell. 2023;22(1) doi: 10.1111/acel.13753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Aman Y., Schmauck-Medina T., Hansen M., Morimoto R.I., Simon A.K., Bjedov I., et al. Autophagy in healthy aging and disease. Nat Aging. 2021;1(8):634–650. doi: 10.1038/s43587-021-00098-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Deretic V., Levine B. Autophagy balances inflammation in innate immunity. Autophagy. 2018;14(2):243–251. doi: 10.1080/15548627.2017.1402992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Noda N.N., Wang Z., Zhang H. Liquid-liquid phase separation in autophagy. J Cell Biol. 2020;219(8) doi: 10.1083/jcb.202004062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Rubinsztein D.C., Bento C.F., Deretic V. Therapeutic targeting of autophagy in neurodegenerative and infectious diseases. J Exp Med. 2015;212(7):979–990. doi: 10.1084/jem.20150956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Wang L., Cai J., Zhao X., Ma L., Zeng P., Zhou L., et al. Palmitoylation prevents sustained inflammation by limiting NLRP3 inflammasome activation through chaperone-mediated autophagy. Mol Cell. 2023;83(2):281–297. doi: 10.1016/j.molcel.2022.12.002. e10. [DOI] [PubMed] [Google Scholar]
- 170.Qiao L., Ma J., Zhang Z., Sui W., Zhai C., Xu D., et al. Deficient chaperone-mediated autophagy promotes inflammation and atherosclerosis. Circ Res. 2021;129(12):1141–1157. doi: 10.1161/CIRCRESAHA.121.318908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bakula D., Scheibye-Knudsen M. MitophAging: mitophagy in aging and disease. Front Cell Dev Biol. 2020;8:239. doi: 10.3389/fcell.2020.00239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Bektas A., Schurman S.H., Gonzalez-Freire M., Dunn C.A., Singh A.K., Macian F., et al. Age-associated changes in human CD4+ T cells point to mitochondrial dysfunction consequent to impaired autophagy. Aging (Albany NY) 2019;11(21):9234–9263. doi: 10.18632/aging.102438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhang H., Alsaleh G., Feltham J., Sun Y., Napolitano G., Riffelmacher T., et al. Polyamines control eIF5A hypusination, TFEB translation, and autophagy to reverse B cell senescence. Mol Cell. 2019;76(1):110–125. doi: 10.1016/j.molcel.2019.08.005. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Abdullah Marzoog B. Autophagy as an anti-senescent in aging neurocytes. Curr Mol Med. 2023:23. doi: 10.2174/1566524023666230120102718. [DOI] [PubMed] [Google Scholar]
- 175.Carosi J.M., Fourrier C., Bensalem J., Sargeant T.J. The mTOR-lysosome axis at the centre of ageing. FEBS Open Bio. 2022;12(4):739–757. doi: 10.1002/2211-5463.13347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Liu J.K. Antiaging agents: safe interventions to slow aging and healthy life span extension. Nat Products Bioprospect. 2022;12(1):18. doi: 10.1007/s13659-022-00339-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bennett D.F., Goyala A., Statzer C., Beckett C.W., Tyshkovskiy A., Gladyshev V.N., et al. Rilmenidine extends lifespan and healthspan in Caenorhabditis elegans via a nischarin I1-imidazoline receptor. Aging Cell. 2023;22(2) doi: 10.1111/acel.13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hodzic Kuerec A., Maier A.B. Why is rapamycin not a rapalog? Gerontology. 2023 doi: 10.1159/000528985. Published online 7 January. [DOI] [PubMed] [Google Scholar]
- 179.Reed R., Miwa S. Cellular senescence and ageing. Subcell Biochem. 2023;102:139–173. doi: 10.1007/978-3-031-21410-3_7. [DOI] [PubMed] [Google Scholar]
- 180.Chaib S., Tchkonia T., Kirkland J.L. Cellular senescence and senolytics: the path to the clinic. Nat Med. 2022;28(8):1556–1568. doi: 10.1038/s41591-022-01923-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhuang X.X., Wang S.F., Tan Y., Song J.X., Zhu Z., Wang Z.Y., et al. Pharmacological enhancement of TFEB-mediated autophagy alleviated neuronal death in oxidative stress-induced Parkinson’s disease models. Cell Death Dis. 2020;11(2):128. doi: 10.1038/s41419-020-2322-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Zhang Z., Yang X., Song Y.Q., Tu J. Autophagy in Alzheimer's disease pathogenesis: therapeutic potential and future perspectives. Ageing Res Rev. 2021;72(101464) doi: 10.1016/j.arr.2021.101464. [DOI] [PubMed] [Google Scholar]
- 183.Croce K.R., Yamamoto A. A role for autophagy in Huntington's disease. Neurobiol Dis. 2019;122:16–22. doi: 10.1016/j.nbd.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Amin A., Perera N.D., Beart P.M., Turner B.J., Shabanpoor F. Amyotrophic lateral sclerosis and autophagy: dysfunction and therapeutic targeting. Cells. 2020;9(11):2413. doi: 10.3390/cells9112413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Zhang L., Dai L., Li D. Mitophagy in neurological disorders. J Neuroinflammation. 2021;18(1):297. doi: 10.1186/s12974-021-02334-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Zhu R., Luo Y., Li S., Wang Z. The role of microglial autophagy in Parkinson's disease. Front Aging Neurosci. 2022;14 doi: 10.3389/fnagi.2022.1039780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Wang Z., Wang Q., Li S., Li X.J., Yang W., He D. Microglial autophagy in Alzheimer's disease and Parkinson's disease. Front Aging Neurosci. 2022;14 doi: 10.3389/fnagi.2022.1065183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Niu H., Wang Q., Zhao W., Liu J., Wang D., Muhammad B., et al. IL-1β/IL-1R1 signaling induced by intranasal lipopolysaccharide infusion regulates alpha-Synuclein pathology in the olfactory bulb, substantia nigra and striatum. Brain Pathol. 2020;30(6):1102–1118. doi: 10.1111/bpa.12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Lv Q.K., Tao K.X., Wang X.B., Yao X.Y., Pang M.Z., Liu J.Y., et al. Role of α-synuclein in microglia: autophagy and phagocytosis balance neuroinflammation in Parkinson’s disease. Inflamm Res. 2023;72(3):443–462. doi: 10.1007/s00011-022-01676-x. [DOI] [PubMed] [Google Scholar]
- 190.Karpowicz R.J., Jr., Trojanowski J.Q., Lee V.M.Y. Transmission of α-synuclein seeds in neurodegenerative disease: recent developments. Lab Invest. 2019;99(7):971–981. doi: 10.1038/s41374-019-0195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Wang C., Fan L., Khawaja R.R., Liu B., Zhan L., Kodama L., et al. Microglial NF-κB drives tau spreading and toxicity in a mouse model of tauopathy. Nat Commun. 2022;13(1):1969. doi: 10.1038/s41467-022-29552-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.López-Otín C., Galluzzi L., Freije J.M.P., Madeo F., Kroemer G. Metabolic control of longevity. Cell. 2016;166(4):802–821. doi: 10.1016/j.cell.2016.07.031. [DOI] [PubMed] [Google Scholar]
- 193.Marmentini C., Soares G.M., Bronczek G.A., Piovan S., Mareze-Costa C.E., Carneiro E.M., et al. Aging reduces insulin clearance in mice. Front Endocrinol. 2021;12 doi: 10.3389/fendo.2021.679492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Evans J.L., Goldfine I.D. Aging and insulin resistance: just say iNOS. Diabetes. 2013;62(2):346–348. doi: 10.2337/db12-1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Ormazabal V., Nair S., Elfeky O., Aguayo C., Salomon C., Zuñiga F.A. Association between insulin resistance and the development of cardiovascular disease. Cardiovasc Diabetol. 2018;17(1):122. doi: 10.1186/s12933-018-0762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Ieronymaki E., Theodorakis E.M., Lyroni K., Vergadi E., Lagoudaki E., Al-Qahtani A., et al. Insulin resistance in macrophages alters their metabolism and promotes an M2-like phenotype. J Immunol. 2019;202(6):1786–1797. doi: 10.4049/jimmunol.1800065. [DOI] [PubMed] [Google Scholar]
- 197.Guillén C., Benito M. mTORC1 overactivation as a key aging factor in the progression to type 2 diabetes mellitus. Front Endocrinol. 2018;9:621. doi: 10.3389/fendo.2018.00621. Published 2018 Oct 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Cheng S.C., Quintin J., Cramer R.A., Shepardson K.M., Saeed S., Kumar V., et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345(6204):1250684. doi: 10.1126/science.1250684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Viola A., Munari F., Sánchez-Rodríguez R., Scolaro T., Castegna A. The metabolic signature of macrophage responses. Front Immunol. 2019;10:1462. doi: 10.3389/fimmu.2019.01462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Jin J., Li X., Hu B., Kim C., Cao W., Zhang H., et al. FOXO1 deficiency impairs proteostasis in aged T cells. Sci Adv. 2020;6(17) doi: 10.1126/sciadv.aba1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Tsai S., Clemente-Casares X., Zhou A.C., Lei H., Ahn J.J., Chan Y.T., et al. Insulin receptor-mediated stimulation boosts T cell immunity during inflammation and infection. Cell Metabol. 2018;28(6):922–934. doi: 10.1016/j.cmet.2018.08.003. [DOI] [PubMed] [Google Scholar]
- 202.Makhijani P., Basso P.J., Chan Y.T., Chen N., Baechle J., Khan S., et al. Regulation of the immune system by the insulin receptor in health and disease. Front Endocrinol. 2023;14:1128622. doi: 10.3389/fendo.2023.1128622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wu D., Wong C.K., Han J.M., Orban P.C., Huang Q., Gillies J., et al. T reg-specific insulin receptor deletion prevents diet-induced and age-associated metabolic syndrome. J Exp Med. 2020;217(8) doi: 10.1084/jem.20191542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Ma S., Wang C., Mao X., Hao Y. B cell dysfunction associated with aging and autoimmune diseases. Front Immunol. 2019;10:318. doi: 10.3389/fimmu.2019.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Phalke S., Rivera-Correa J., Jenkins D., Flores Castro D., Giannopoulou E., Pernis A.B. Molecular mechanisms controlling age-associated B cells in autoimmunity. Immunol Rev. 2022;307(1):79–100. doi: 10.1111/imr.13068. [DOI] [PubMed] [Google Scholar]
- 206.Li K., Romero M., Cañardo M., Garcia D., Diaz A., Blomberg B.B., et al. B cells from old mice induce the generation of inflammatory T cells through metabolic pathways. Mech Ageing Dev. 2023;209(111742) doi: 10.1016/j.mad.2022.111742. [DOI] [PubMed] [Google Scholar]
- 207.Frasca D., Romero M., Garcia D., Diaz A., Blomberg B.B. Hyper-metabolic B cells in the spleens of old mice make antibodies with autoimmune specificities. Immun Ageing. 2021;18(1):9. doi: 10.1186/s12979-021-00222-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Shirakawa K., Sano M. T cell immunosenescence in aging, obesity, and cardiovascular disease. Cells. 2021;10(9):2435. doi: 10.3390/cells10092435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Szwed A., Kim E., Jacinto E. Regulation and metabolic functions of mTORC1 and mTORC2. Physiol Rev. 2021;101(3):1371–1426. doi: 10.1152/physrev.00026.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Laplante M., Sabatini D.M. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Walters H.E., Cox L.S. MTORC inhibitors as broad-spectrum therapeutics for age-related diseases. Int J Mol Sci. 2018;19(8) doi: 10.3390/ijms19082325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Haythorne E., Lloyd M., Walsby-Tickle J., Tarasov A.I., Sandbrink J., Portillo I., et al. Altered glycolysis triggers impaired mitochondrial metabolism and mTORC1 activation in diabetic β-cells. Nat Commun. 2022;13(1):6754. doi: 10.1038/s41467-022-34095-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Chrienova Z., Nepovimova E., Kuca K. The role of mTOR in age-related diseases. J Enzym Inhib Med Chem. 2021;36(1):1679–1693. doi: 10.1080/14756366.2021.1955873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Mota-Martorell N., Jové M., Pamplona R. MTOR complex 1 content and regulation is adapted to animal longevity. Int J Mol Sci. 2022;23(15):8747. doi: 10.3390/ijms23158747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Xu T., Liu J., Li X.R., Yu Y., Luo X., Zheng X., et al. The mTOR/NF-κB pathway mediates neuroinflammation and synaptic plasticity in diabetic encephalopathy. Mol Neurobiol. 2021;58(8):3848–3862. doi: 10.1007/s12035-021-02390-1. [DOI] [PubMed] [Google Scholar]
- 216.Li Y., Yang L., Dong L., Yang Z.W., Zhang J., Zhang S.L., et al. Crosstalk between the Akt/mTORC1 and NF-κB signaling pathways promotes hypoxia-induced pulmonary hypertension by increasing DPP4 expression in PASMCs. Acta Pharmacol Sin. 2019;40(10):1322–1333. doi: 10.1038/s41401-019-0272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Liu G.Y., Sabatini D.M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol. 2020;21(4):183–203. doi: 10.1038/s41580-019-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ham D.J., Börsch A., Chojnowska K., Lin S., Leuchtmann A.B., Ham A.S., et al. Distinct and additive effects of calorie restriction and rapamycin in aging skeletal muscle. Nat Commun. 2022;13(1):2025. doi: 10.1038/s41467-022-29714-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Correia-Melo C., Birch J., Fielder E., Rahmatika D., Taylor J., Chapman J., et al. Rapamycin improves healthspan but not inflammaging in nfκb1-/- mice. Aging Cell. 2019;18(1) doi: 10.1111/acel.12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Collins S.L., Oh M.H., Sun I.H., Chan-Li Y., Zhao L., Powell J.D., et al. MTORC1 signaling regulates proinflammatory macrophage function and metabolism. J Immunol. 2021;207(3):913–922. doi: 10.4049/jimmunol.2100230. [DOI] [PubMed] [Google Scholar]
- 221.Lesniewski L.A., Seals D.R., Walker A.E., Henson G.D., Blimline M.W., Trott D.W., et al. Dietary rapamycin supplementation reverses age-related vascular dysfunction and oxidative stress, while modulating nutrient-sensing, cell cycle, and senescence pathways. Aging Cell. 2017;16(1):17–26. doi: 10.1111/acel.12524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Pan H., Finkel T. Key proteins and pathways that regulate lifespan. J Biol Chem. 2017;292(16):6452–6460. doi: 10.1074/jbc.R116.771915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Zhu L., Yang T., Li L., Sun L., Hou Y., Hu X., et al. TSC1 controls macrophage polarization to prevent inflammatory disease. Nat Commun. 2014;5(1):4696. doi: 10.1038/ncomms5696. [DOI] [PubMed] [Google Scholar]
- 224.Jiang H., Westerterp M., Wang C., Zhu Y., Ai D. Macrophage mTORC1 disruption reduces inflammation and insulin resistance in obese mice. Diabetologia. 2014;57(11):2393–2404. doi: 10.1007/s00125-014-3350-5. [DOI] [PubMed] [Google Scholar]
- 225.Mannick J.B., Morris M., Hockey H.U.P., Roma G., Beibel M., Kulmatycki K., et al. TORC1 inhibition enhances immune function and reduces infections in the elderly. Sci Transl Med. 2018;10(449):eaaq1564. doi: 10.1126/scitranslmed.aaq1564. [DOI] [PubMed] [Google Scholar]
- 226.Mullen P.J., Garcia G., Jr., Purkayastha A., Matulionis N., Schmid E.W., Momcilovic M., et al. SARS-CoV-2 infection rewires host cell metabolism and is potentially susceptible to mTORC1 inhibition. Nat Commun. 2021;12(1):1876. doi: 10.1038/s41467-021-22166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Geier C., Perl A. Therapeutic mTOR blockade in systemic autoimmunity: implications for antiviral immunity and extension of lifespan. Autoimmun Rev. 2021;20(12) doi: 10.1016/j.autrev.2021.102984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Moldakozhayev A., Gladyshev V.N. Metabolism, homeostasis, and aging. Trends Endocrinol Metabol. 2023;34(3):158–169. doi: 10.1016/j.tem.2023.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Sharma A., Chabloz S., Lapides R.A., Roider E., Ewald C.Y. Potential synergistic supplementation of NAD+ promoting compounds as a strategy for increasing healthspan. Nutrients. 2023;15(2):445. doi: 10.3390/nu15020445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Chen Y., Liu Y., Jiang K., Wen Z., Cao X., Wu S. Linear ubiquitination of LKB1 activates AMPK pathway to inhibit NLRP3 inflammasome response and reduce chondrocyte pyroptosis in osteoarthritis. J Orthop Translat. 2023;39:1–11. doi: 10.1016/j.jot.2022.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Sadria M., Layton A.T. Interactions among mTORC, AMPK and SIRT: a computational model for cell energy balance and metabolism. Cell Commun Signal. 2021;19(1):57. doi: 10.1186/s12964-021-00706-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Kim J.K., Silwal P., Jo E.K. Sirtuin 1 in host defense during infection. Cells. 2022;11(18):2921. doi: 10.3390/cells11182921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Desjardins E.M., Smith B.K., Day E.A., Ducommun S., Sanders M.J., Nederveen J.P., et al. The phosphorylation of AMPKβ1 is critical for increasing autophagy and maintaining mitochondrial homeostasis in response to fatty acids. Proc Natl Acad Sci U S A. 2022;119(48) doi: 10.1073/pnas.2119824119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Lee I.H. Mechanisms and disease implications of sirtuin-mediated autophagic regulation. Exp Mol Med. 2019;51(9):1–11. doi: 10.1038/s12276-019-0302-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Ziętara P., Dziewięcka M., Augustyniak M. Why is longevity still a scientific mystery? Sirtuins-past, present and future. Int J Mol Sci. 2022;24(1):728. doi: 10.3390/ijms24010728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Malik N., Ferreira B.I., Hollstein P.E., Curtis S.D., Trefts E., Weiser Novak S., et al. Induction of lysosomal and mitochondrial biogenesis by AMPK phosphorylation of FNIP1. Science. 2023;380(6642) doi: 10.1126/science.abj5559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Bharath L.P., Agrawal M., McCambridge G., Nicholas D.A., Hasturk H., Liu J., et al. Metformin enhances autophagy and normalizes mitochondrial function to alleviate aging-associated inflammation. Cell Metabol. 2020;32(1):44–55. doi: 10.1016/j.cmet.2020.04.015. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Salminen A., Kaarniranta K. AMP-activated protein kinase (AMPK) controls the aging process via an integrated signaling network. Ageing Res Rev. 2012;11(2):230–241. doi: 10.1016/j.arr.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 239.Oka S.I., Titus A.S., Zablocki D., Sadoshima J. Molecular properties and regulation of NAD+ kinase (NADK) Redox Biol. 2023;59(102561) doi: 10.1016/j.redox.2022.102561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zhou H., Liu S., Zhang N., Fang K., Zong J., An Y., et al. Downregulation of Sirt6 by CD38 promotes cell senescence and aging. Aging (Albany NY) 2022;14(23):9730–9757. doi: 10.18632/aging.204425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.de Candia P., Procaccini C., Russo C., Lepore M.T., Matarese G. Regulatory T cells as metabolic sensors. Immunity. 2022;55(11):1981–1992. doi: 10.1016/j.immuni.2022.10.006. [DOI] [PubMed] [Google Scholar]
- 242.Deng B., Zhang W., Zhu Y., Li Y., Li D., Li B. FOXP3+ regulatory T cells and age-related diseases. FEBS J. 2022;289(2):319–335. doi: 10.1111/febs.15743. [DOI] [PubMed] [Google Scholar]
- 243.Zhu H., Liu Z., An J., Zhang M., Qiu Y., Zou M.H. Activation of AMPKα1 is essential for regulatory T cell function and autoimmune liver disease prevention. Cell Mol Immunol. 2021;18(12):2609–2617. doi: 10.1038/s41423-021-00790-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Phair I.R., Nisr R.B., Howden A.J.M., Sovakova M., Alqurashi N., Foretz M., et al. AMPK integrates metabolite and kinase-based immunometabolic control in macrophages. Mol Metabol. 2023;68(101661) doi: 10.1016/j.molmet.2022.101661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Yan C., Tian X., Li J., Liu D., Ye D., Xie Z., et al. A high-fat diet attenuates AMPK α1 in adipocytes to induce exosome shedding and nonalcoholic fatty liver development in vivo. Diabetes. 2021;70(2):577–588. doi: 10.2337/db20-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Ma H., Guo X., Cui S., Wu Y., Zhang Y., Shen X., et al. Dephosphorylation of AMP-activated protein kinase exacerbates ischemia/reperfusion-induced acute kidney injury via mitochondrial dysfunction. Kidney Int. 2022;101(2):315–330. doi: 10.1016/j.kint.2021.10.028. [DOI] [PubMed] [Google Scholar]
- 247.Smirnov D., Eremenko E., Stein D., Kaluski S., Jasinska W., Cosentino C., et al. SIRT6 is a key regulator of mitochondrial function in the brain. Cell Death Dis. 2023;14(1):35. doi: 10.1038/s41419-022-05542-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.He H., Xu P., Zhang X., Liao M., Dong Q., Cong T., et al. Aging-induced IL27Ra signaling impairs hematopoietic stem cells. Blood. 2020;136(2):183–198. doi: 10.1182/blood.2019003910. [DOI] [PubMed] [Google Scholar]
- 249.Li Y.Q., Jiao Y., Liu Y.N., Fu J.Y., Sun L.K., Su J. PGC-1α protects from myocardial ischaemia-reperfusion injury by regulating mitonuclear communication. J Cell Mol Med. 2022;26(3):593–600. doi: 10.1111/jcmm.16236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Nam B.Y., Jhee J.H., Park J., Kim S., Kim G., Park J.T., et al. PGC-1α inhibits the NLRP3 inflammasome via preserving mitochondrial viability to protect kidney fibrosis. Cell Death Dis. 2022;13(1):31. doi: 10.1038/s41419-021-04480-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Fang Y., An N., Zhu L., Gu Y., Qian J., Jiang G., et al. Autophagy-Sirt3 axis decelerates hematopoietic aging. Aging Cell. 2020;19(10) doi: 10.1111/acel.13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Salvestrini V., Sell C., Lorenzini A. Obesity may accelerate the aging process. Front Endocrinol. 2019;10:266. doi: 10.3389/fendo.2019.00266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Khan S., Chan Y.T., Revelo X.S., Winer D.A. The immune landscape of visceral adipose tissue during obesity and aging. Front Endocrinol. 2020;11:267. doi: 10.3389/fendo.2020.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Song J., Farris D., Ariza P., Moorjani S., Varghese M., Blin M., et al. Age-associated adipose tissue inflammation promotes monocyte chemotaxis and enhances atherosclerosis. Aging Cell. 2023;22(2) doi: 10.1111/acel.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Shou J., Chen P.J., Xiao W.H. Mechanism of increased risk of insulin resistance in aging skeletal muscle. Diabetol Metab Syndrome. 2020;12(1):14. doi: 10.1186/s13098-020-0523-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Ragonnaud E., Biragyn A. Gut microbiota as the key controllers of ‘healthy’ aging of elderly people. Immun Ageing. 2021;18(1):2. doi: 10.1186/s12979-020-00213-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Shemtov S.J., Emani R., Bielska O., Covarrubias A.J., Verdin E., Andersen J.K., et al. The intestinal immune system and gut barrier function in obesity and ageing. FEBS J. Published online. June 2022;21 doi: 10.1111/febs.16558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Yung J.H.M., Giacca A. Role of c-jun N-terminal kinase (JNK) in obesity and type 2 diabetes. Cells. 2020;9(3):706. doi: 10.3390/cells9030706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Rivas D.A., McDonald D.J., Rice N.P., Haran P.H., Dolnikowski G.G., Fielding R.A. Diminished anabolic signaling response to insulin induced by intramuscular lipid accumulation is associated with inflammation in aging but not obesity. Am J Physiol Regul Integr Comp Physiol. 2016;310(7):R561–R569. doi: 10.1152/ajpregu.00198.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Tilg H., Moschen A.R. Inflammatory mechanisms in the regulation of insulin resistance. Mol Med. 2008;14(3–4):222–231. doi: 10.2119/2007-00119.Tilg. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Bárcena C., Quirós P.M., Durand S., Mayoral P., Rodríguez F., Caravia X.M., et al. Methionine restriction extends lifespan in progeroid mice and alters lipid and bile acid metabolism. Cell Rep. 2018;24(9):2392–2403. doi: 10.1016/j.celrep.2018.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Rajabian N., Ikhapoh I., Shahini S., Choudhury D., Thiyagarajan R., Shahini A., et al. Methionine adenosyltransferase2A inhibition restores metabolism to improve regenerative capacity and strength of aged skeletal muscle. Nat Commun. 2023;14(1):886. doi: 10.1038/s41467-023-36483-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Xu C., Zhu H., Qiu P. Aging progression of human gut microbiota. BMC Microbiol. 2019;19(1):236. doi: 10.1186/s12866-019-1616-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Martino C., Dilmore A.H., Burcham Z.M., Metcalf J.L., Jeste D., Knight R. Microbiota succession throughout life from the cradle to the grave. Nat Rev Microbiol. 2022;20(12):707–720. doi: 10.1038/s41579-022-00768-z. [DOI] [PubMed] [Google Scholar]
- 265.Luck H., Tsai S., Chung J., Clemente-Casares X., Ghazarian M., Revelo X.S., et al. Regulation of obesity-related insulin resistance with gut anti-inflammatory agents. Cell Metabol. 2015;21(4):527–542. doi: 10.1016/j.cmet.2015.03.001. [DOI] [PubMed] [Google Scholar]
- 266.Thevaranjan N., Puchta A., Schulz C., Naidoo A., Szamosi J.C., Verschoor C.P., et al. Age-associated microbial dysbiosis promotes intestinal permeability, systemic inflammation, and macrophage dysfunction. Cell Host Microbe. 2017;21(4):455–466. doi: 10.1016/j.chom.2017.03.002. e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Winer D.A., Winer S., Dranse H.J., Lam T.K.T. Immunologic impact of the intestine in metabolic disease. J Clin Invest. 2017;127(1):33–42. doi: 10.1172/JCI88879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Luck H., Khan S., Kim J.H., Copeland J.K., Revelo X.S., Tsai S., et al. Gut-associated IgA+ immune cells regulate obesity-related insulin resistance. Nat Commun. 2019;10(1):3650. doi: 10.1038/s41467-019-11370-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Ciocca M., Zaffina S., Fernandez Salinas A., Bocci C., Palomba P., Conti M.G., et al. Evolution of human memory B cells from childhood to old age. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.690534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Hancock M.L., Meyer R.C., Mistry M., Khetani R.S., Wagschal A., Shin T., et al. Insulin receptor associates with promoters genome-wide and regulates gene expression. Cell. 2019;177(3):722–736. doi: 10.1016/j.cell.2019.02.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Nagao H., Jayavelu A.K., Cai W., Pan H., Dreyfuss J.M., Batista T.M., et al. Unique ligand and kinase-independent roles of the insulin receptor in regulation of cell cycle, senescence and apoptosis. Nat Commun. 2023;14(1):57. doi: 10.1038/s41467-022-35693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Dall’Agnese A., Platt J.M., Zheng M.M., Friesen M., Dall’Agnese G., Blaise A.M., et al. The dynamic clustering of insulin receptor underlies its signaling and is disrupted in insulin resistance. Nat Commun. 2022;13(1):7522. doi: 10.1038/s41467-022-35176-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Bleve A., Motta F., Durante B., Pandolfo C., Selmi C., Sica A. Immunosenescence, inflammaging, and frailty: role of myeloid cells in age-related diseases. Clin Rev Allergy Immunol. 2023;64(2):123–144. doi: 10.1007/s12016-021-08909-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Davizon-Castillo P., McMahon B., Aguila S., Bark D., Ashworth K., Allawzi A., et al. TNF-α-driven inflammation and mitochondrial dysfunction define the platelet hyperreactivity of aging. Blood. 2019;134(9):727–740. doi: 10.1182/blood.2019000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Lee D.F., Kuo H.P., Chen C.T., Wei Y., Chou C.K., Hung J.Y., et al. IKKbeta suppression of TSC1 function links the mTOR pathway with insulin resistance. Int J Mol Med. 2008;22(5):633–638. doi: 10.3892/ijmm_00000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Tzatsos A., Kandror K.V. Nutrients suppress phosphatidylinositol 3-kinase/Akt signaling via raptor-dependent mTOR-mediated insulin receptor substrate 1 phosphorylation. Mol Cell Biol. 2006;26(1):63–76. doi: 10.1128/MCB.26.1.63-76.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Yoneyama Y., Inamitsu T., Chida K., Iemura S.I., Natsume T., Maeda T., et al. Serine phosphorylation by mTORC1 promotes IRS-1 degradation through SCFβ-TRCP E3 ubiquitin ligase. iScience. 2018;5:1–18. doi: 10.1016/j.isci.2018.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Brisse M., Ly H. Comparative structure and function analysis of the RIG-I-like receptors: RIG-I and MDA5. Front Immunol. 2019;10:1586. doi: 10.3389/fimmu.2019.01586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Decout A., Katz J.D., Venkatraman S., Ablasser A. The cGAS-STING pathway as a therapeutic target in inflammatory diseases. Nat Rev Immunol. 2021;21(9):548–569. doi: 10.1038/s41577-021-00524-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Di Giorgio E., Xodo L.E. Endogenous retroviruses (ERVs): does RLR (RIG-I-like receptors)-MAVS pathway directly control senescence and aging as a consequence of ERV DE-repression? Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.917998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Bodur C., Kazyken D., Huang K., Ekim Ustunel B., Siroky K.A., Tooley A.S., et al. The IKK-related kinase TBK1 activates mTORC1 directly in response to growth factors and innate immune agonists. EMBO J. 2018;37(1):19–38. doi: 10.15252/embj.201696164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Tooley A.S., Kazyken D., Bodur C., Gonzalez I.E., Fingar D.C. The innate immune kinase TBK1 directly increases mTORC2 activity and downstream signaling to Akt. J Biol Chem. 2021;297(2) doi: 10.1016/j.jbc.2021.100942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Schädel P., Czapka A., Gebert N., Jacobsen I.D., Ori A., Werz O. Metabololipidomic and proteomic profiling reveals aberrant macrophage activation and interrelated immunomodulatory mediator release during aging. Aging Cell. 2023 doi: 10.1111/acel.13856. Published online 26 April. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Fan X., Zeng Y., Song W., Li J., Ai S., Yang D., et al. The role of Sestrins in the regulation of the aging process. Mech Ageing Dev. 2020;188(111251) doi: 10.1016/j.mad.2020.111251. [DOI] [PubMed] [Google Scholar]
- 285.Chen Y., Huang T., Yu Z., Yu Q., Wang Y., Hu J., et al. The functions and roles of sestrins in regulating human diseases. Cell Mol Biol Lett. 2022;27(1):2. doi: 10.1186/s11658-021-00302-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Cangelosi A.L., Puszynska A.M., Roberts J.M., Armani A., Nguyen T.P., Spinelli J.B., et al. Zonated leucine sensing by Sestrin-mTORC1 in the liver controls the response to dietary leucine. Science. 2022;377(6601):47–56. doi: 10.1126/science.abi9547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Gu X., Jouandin P., Lalgudi P.V., Binari R., Valenstein M.L., Reid M.A., et al. Sestrin mediates detection of and adaptation to low-leucine diets in Drosophila. Nature. 2022;608(7921):209–216. doi: 10.1038/s41586-022-04960-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Wang L.X., Zhu X.M., Yao Y.M. Sestrin2: its potential role and regulatory mechanism in host immune response in diseases. Front Immunol. 2019;10:2797. doi: 10.3389/fimmu.2019.02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Ben-Sahra I., Dirat B., Laurent K., Puissant A., Auberger P., Budanov A., et al. Sestrin2 integrates Akt and mTOR signaling to protect cells against energetic stress-induced death. Cell Death Differ. 2013;20(4):611–619. doi: 10.1038/cdd.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Zeng N., D'Souza R.F., Mitchell C.J., Cameron-Smith D. Sestrins are differentially expressed with age in the skeletal muscle of men: a cross-sectional analysis. Exp Gerontol. 2018;110:23–34. doi: 10.1016/j.exger.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 291.Yang B.A., Castor-Macias J., Fraczek P., Cornett A., Brown L.A., Kim M., et al. Sestrins regulate muscle stem cell metabolic homeostasis. Stem Cell Rep. 2021;16(9):2078–2088. doi: 10.1016/j.stemcr.2021.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Muller F.L., Lustgarten M.S., Jang Y., Richardson A., Van Remmen H. Trends in oxidative aging theories. Free Radic Biol Med. 2007;43(4):477–503. doi: 10.1016/j.freeradbiomed.2007.03.034. [DOI] [PubMed] [Google Scholar]
- 293.Naidoo N. ER and aging-Protein folding and the ER stress response. Ageing Res Rev. 2009;8(3):150–159. doi: 10.1016/j.arr.2009.03.001. [DOI] [PubMed] [Google Scholar]
- 294.Sag D., Carling D., Stout R.D., Suttles J. Adenosine 5’-monophosphate-activated protein kinase promotes macrophage polarization to an anti-inflammatory functional phenotype. J Immunol. 2008;181(12):8633–8641. doi: 10.4049/jimmunol.181.12.8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Nath N., Khan M., Rattan R., Mangalam A., Makkar R.S., de Meester C., et al. Loss of AMPK exacerbates experimental autoimmune encephalomyelitis disease severity. Biochem Biophys Res Commun. 2009;386(1):16–20. doi: 10.1016/j.bbrc.2009.05.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Viollet B., Horman S., Leclerc J., Lantier L., Foretz M., Billaud M., et al. AMPK inhibition in health and disease. Crit Rev Biochem Mol Biol. 2010;45(4):276–295. doi: 10.3109/10409238.2010.488215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Fan K., Lin L., Ai Q., Wan J., Dai J., Liu G., et al. Lipopolysaccharide-induced dephosphorylation of AMPK-activated protein kinase potentiates inflammatory injury via repression of ULK1-dependent autophagy. Front Immunol. 2018;9:1464. doi: 10.3389/fimmu.2018.01464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Zhang Q., Liu S., Zhang C.S., Wu Q., Yu X., Zhou R., et al. AMPK directly phosphorylates TBK1 to integrate glucose sensing into innate immunity. Mol Cell. 2022;82(23):4519–4536. doi: 10.1016/j.molcel.2022.10.026. [DOI] [PubMed] [Google Scholar]
- 299.Covarrubias A.J., Kale A., Perrone R., Lopez-Dominguez J.A., Pisco A.O., Kasler H.G., et al. Senescent cells promote tissue NAD+ decline during ageing via the activation of CD38+ macrophages. Nat Metab. 2020;2(11):1265–1283. doi: 10.1038/s42255-020-00305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Bobermin L.D., Roppa R.H.A., Gonçalves C.A., Quincozes-Santos A. Ammonia-induced glial-inflammaging. Mol Neurobiol. 2020;57(8):3552–3567. doi: 10.1007/s12035-020-01985-4. [DOI] [PubMed] [Google Scholar]
- 301.Mishra S., Welch N., Karthikeyan M., Bellar A., Musich R., Singh S.S., et al. Dysregulated cellular redox status during hyperammonemia causes mitochondrial dysfunction and senescence by inhibiting sirtuin-mediated deacetylation. Aging Cell. 2023:e13852. doi: 10.1111/acel.13852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Franceschi C., Garagnani P., Parini P., Giuliani C., Santoro A. Inflammaging: a new immune-metabolic viewpoint for age-related diseases. Nat Rev Endocrinol. 2018;14(10):576–590. doi: 10.1038/s41574-018-0059-4. [DOI] [PubMed] [Google Scholar]
- 303.Dyall S.D., Brown M.T., Johnson P.J. Ancient invasions: from endosymbionts to organelles. Science. 2004;304(5668):253–257. doi: 10.1126/science.1094884. [DOI] [PubMed] [Google Scholar]
- 304.Zampino M., Brennan N.A., Kuo P.L., Spencer R.G., Fishbein K.W., Simonsick E.M., et al. Poor mitochondrial health and systemic inflammation? Test of a classic hypothesis in the Baltimore Longitudinal Study of Aging. GeroScience. 2020;42(4):1175–1182. doi: 10.1007/s11357-020-00208-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Tian Q., Mitchell B.A., Zampino M., Fishbein K.W., Spencer R.G., Ferrucci L. Muscle mitochondrial energetics predicts mobility decline in well-functioning older adults: the baltimore longitudinal study of aging. Aging Cell. 2022;21(2) doi: 10.1111/acel.13552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Bencivenga L., Strumia M., Rolland Y., Martinez L., Cestac P., Guyonnet S., et al. Biomarkers of mitochondrial dysfunction and inflammaging in older adults and blood pressure variability. GeroScience. 2023;45(2):797–809. doi: 10.1007/s11357-022-00697-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Bugga P., Alam M.J., Kumar R., Pal S., Chattopadyay N., Banerjee S.K. Sirt3 ameliorates mitochondrial dysfunction and oxidative stress through regulating mitochondrial biogenesis and dynamics in cardiomyoblast. Cell Signal. 2022;94(110309) doi: 10.1016/j.cellsig.2022.110309. [DOI] [PubMed] [Google Scholar]
- 308.Kroemer G., Galassi C., Zitvogel L., Galluzzi L. Immunogenic cell stress and death. Nat Immunol. 2022;23(4):487–500. doi: 10.1038/s41590-022-01132-2. [DOI] [PubMed] [Google Scholar]
- 309.Marchi S., Guilbaud E., Tait S.W.G., Yamazaki T., Galluzzi L. Mitochondrial control of inflammation. Nat Rev Immunol. 2023;23(3):159–173. doi: 10.1038/s41577-022-00760-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Picca A., Lezza A.M.S., Leeuwenburgh C., Pesce V., Calvani R., Landi F., et al. Fueling inflamm-aging through mitochondrial dysfunction: mechanisms and molecular targets. Int J Mol Sci. 2017;18(5) doi: 10.3390/ijms18050933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Chu C.T., Ji J., Dagda R.K., Jiang J.F., Tyurina Y.Y., Kapralov A.A., et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013;15(10):1197–1205. doi: 10.1038/ncb2837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Li M., Mandal A., Tyurin V.A., DeLucia M., Ahn J., Kagan V.E., et al. Surface-binding to cardiolipin nanodomains triggers cytochrome c pro-apoptotic peroxidase activity via localized dynamics. Structure. 2019;27(5):806–815. doi: 10.1016/j.str.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Rabiet M.J., Huet E., Boulay F. Human mitochondria-derived N-formylated peptides are novel agonists equally active on FPR and FPRL1, while Listeria monocytogenes-derived peptides preferentially activate FPR. Eur J Immunol. 2005;35(8):2486–2495. doi: 10.1002/eji.200526338. [DOI] [PubMed] [Google Scholar]
- 314.Mallavia B., Liu F., Lefrançais E., Cleary S.J., Kwaan N., Tian J.J., et al. Mitochondrial DNA stimulates TLR9-dependent neutrophil extracellular trap formation in primary graft dysfunction. Am J Respir Cell Mol Biol. 2020;62(3):364–372. doi: 10.1165/rcmb.2019-0140OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315.Shen J., Dai Z., Li Y., Zhu H., Zhao L. TLR9 regulates NLRP3 inflammasome activation via the NF-kB signaling pathway in diabetic nephropathy. Diabetol Metab Syndrome. 2022;14(1):26. doi: 10.1186/s13098-021-00780-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Revelo X.S., Ghazarian M., Chng M.H.Y., Luck H., Kim J.H., Zeng K., et al. Nucleic acid-targeting pathways promote inflammation in obesity-related insulin resistance. Cell Rep. 2016;16(3):717–730. doi: 10.1016/j.celrep.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Pinti M., Cevenini E., Nasi M., De Biasi S., Salvioli S., Monti D., et al. Circulating mitochondrial DNA increases with age and is a familiar trait: implications for ‘inflamm-aging. Eur J Immunol. 2014;44(5):1552–1562. doi: 10.1002/eji.201343921. [DOI] [PubMed] [Google Scholar]
- 318.Picca A., Guerra F., Calvani R., Romano R., Coelho-Junior H.J., Damiano F.P., et al. Circulating mitochondrial DNA and inter-organelle contact sites in aging and associated conditions. Cells. 2022;11(4):675. doi: 10.3390/cells11040675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Yu J., Nagasu H., Murakami T., Hoang H., Broderick L., Hoffman H.M., et al. Inflammasome activation leads to Caspase-1-dependent mitochondrial damage and block of mitophagy. Proc Natl Acad Sci U S A. 2014;111(43):15514–15519. doi: 10.1073/pnas.1414859111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Tigano M., Vargas D.C., Tremblay-Belzile S., Fu Y., Sfeir A. Nuclear sensing of breaks in mitochondrial DNA enhances immune surveillance. Nature. 2021;591(7850):477–481. doi: 10.1038/s41586-021-03269-w. [DOI] [PubMed] [Google Scholar]
- 321.Próchnicki T., Vasconcelos M.B., Robinson K.S., Mangan M.S.J., De Graaf D., Shkarina K., et al. Mitochondrial damage activates the NLRP10 inflammasome. Nat Immunol. 2023;24(4):595–603. doi: 10.1038/s41590-023-01451-y. [DOI] [PubMed] [Google Scholar]
- 322.Ghiringhelli F., Apetoh L., Tesniere A., Aymeric L., Ma Y., Ortiz C., et al. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1beta-dependent adaptive immunity against tumors. Nat Med. 2009;15(10):1170–1178. doi: 10.1038/nm.2028. [DOI] [PubMed] [Google Scholar]
- 323.Billingham L.K., Stoolman J.S., Vasan K., Rodriguez A.E., Poor T.A., Szibor M., et al. Mitochondrial electron transport chain is necessary for NLRP3 inflammasome activation. Nat Immunol. 2022;23(5):692–704. doi: 10.1038/s41590-022-01185-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Martínez-Reyes I., Chandel N.S. Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun. 2020;11(1):102. doi: 10.1038/s41467-019-13668-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Jové M., Pradas I., Mota-Martorell N., Cabré R., Ayala V., Ferrer I., et al. Succination of protein thiols in human brain aging. Front Aging Neurosci. 2020;12:52. doi: 10.3389/fnagi.2020.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.Arts R.J.W., Novakovic B., Ter Horst R., Carvalho A., Bekkering S., Lachmandas E., et al. Glutaminolysis and fumarate accumulation integrate immunometabolic and epigenetic programs in trained immunity. Cell Metabol. 2016;24(6):807–819. doi: 10.1016/j.cmet.2016.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Tannahill G.M., Curtis A.M., Adamik J., Palsson-McDermott E.M., McGettrick A.F., Goel G., et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496(7444):238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 328.Wang L, He C. Nrf2-mediated anti-inflammatory polarization of macrophages as therapeutic targets for osteoarthritis. Front Immunol. 2022;13:967193. doi: 10.3389/fimmu.2022.967193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Mills E.L., Ryan D.G., Prag H.A., Dikovskaya D., Menon D., Zaslona Z., et al. Nature. 2018 Apr;556(7699):113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Wang Y., Li S., Zhao L., Cheng P., Liu J., Guo F., et al. Aging relevant metabolite itaconate inhibits inflammatory bone loss. Front Endocrinol. 2022;13:885879. doi: 10.3389/fendo.2022.885879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331.Nakahira K., Haspel J.A., Rathinam V.A., Lee S.J., Dolinay T., Lam H.C., et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12(3):222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 332.Cloonan S.M., Choi A.M.K. Mitochondria in lung disease. J Clin Invest. 2016;126(3):809–820. doi: 10.1172/JCI81113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Dromparis P., Michelakis E.D. Mitochondria in vascular health and disease. Annu Rev Physiol. 2013;75(1):95–126. doi: 10.1146/annurev-physiol-030212-183804. [DOI] [PubMed] [Google Scholar]
- 334.Nunnari J., Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148(6):1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Fischer F., Hamann A., Osiewacz H.D. Mitochondrial quality control: an integrated network of pathways. Trends Biochem Sci. 2012;37(7):284–292. doi: 10.1016/j.tibs.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 336.Szklarczyk R., Nooteboom M., Osiewacz H.D. Control of mitochondrial integrity in ageing and disease. Philos Trans R Soc Lond B Biol Sci. 2014;369(1646) doi: 10.1098/rstb.2013.0439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Zhong W., Rao Z., Xu J., Sun Y., Hu H., Wang P., et al. Defective mitophagy in aged macrophages promotes mitochondrial DNA cytosolic leakage to activate STING signaling during liver sterile inflammation. Aging Cell. 2022;21(6) doi: 10.1111/acel.13622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338.Palmer A.K., Tchkonia T., Kirkland J.L. Targeting cellular senescence in metabolic disease. Mol Metabol. 2022;66(101601) doi: 10.1016/j.molmet.2022.101601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Gorgoulis V., Adams P.D., Alimonti A., Bennett D.C., Bischof O., Bishop C., et al. Cellular senescence: defining a path forward. Cell. 2019;179(4):813–827. doi: 10.1016/j.cell.2019.10.005. [DOI] [PubMed] [Google Scholar]
- 340.Rayess H., Wang M.B., Srivatsan E.S. Cellular senescence and tumor suppressor gene p16. Int J Cancer. 2012;130(8):1715–1725. doi: 10.1002/ijc.27316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Kastenhuber E.R., Lowe S.W. Putting p53 in context. Cell. 2017;170(6):1062–1078. doi: 10.1016/j.cell.2017.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Uyar B., Palmer D., Kowald A., Murua Escobar H., Barrantes I., et al. Single-cell analyses of aging, inflammation and senescence. Ageing Res Rev. 2020;64(101156):101156. doi: 10.1016/j.arr.2020.101156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Basisty N., Kale A., Jeon O.H., Kuehnemann C., Payne T., Rao C., et al. A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol. 2020;18(1) doi: 10.1371/journal.pbio.3000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Saul D., Kosinsky R.L., Atkinson E.J., Doolittle M.L., Zhang X., LeBrasseur N.K., et al. A new gene set identifies senescent cells and predicts senescence-associated pathways across tissues. Nat Commun. 2022;13(1):4827. doi: 10.1038/s41467-022-32552-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Reyes N.S., Krasilnikov M., Allen N.C., Lee J.Y., Hyams B., Zhou M., et al. Sentinel p16INK4a+ cells in the basement membrane form a reparative niche in the lung. Science. 2022;378(6616):192–201. doi: 10.1126/science.abf3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Jeon O.H., Mehdipour M., Gil T.H., Kang M., Aguirre N.W., Robinson Z.R., et al. Systemic induction of senescence in young mice after single heterochronic blood exchange. Nat Metab. 2022;4(8):995–1006. doi: 10.1038/s42255-022-00609-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Schafer M.J., Zhang X., Kumar A., Atkinson E.J., Zhu Y., Jachim S., et al. The senescence-associated secretome as an indicator of age and medical risk. JCI Insight. 2020;5(12) doi: 10.1172/jci.insight.133668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Pezone A., Olivieri F., Napoli M.V., Procopio A., Avvedimento E.V., Gabrielli A. Inflammation and DNA damage: cause, effect or both. Nat Rev Rheumatol. 2023;19(4):200–211. doi: 10.1038/s41584-022-00905-1. [DOI] [PubMed] [Google Scholar]
- 349.Rodier F., Coppé J.P., Patil C.K., Hoeijmakers W.A., Muñoz D.P., Raza S.R., et al. Persistent DNA damage signalling triggers senescence-associated inflammatory cytokine secretion. Nat Cell Biol. 2009;11(8):973–979. doi: 10.1038/ncb1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Olivieri F., Albertini M.C., Orciani M., Ceka A., Cricca M., Procopio A.D., et al. DNA damage response (DDR) and senescence: shuttled inflamma-miRNAs on the stage of inflamm-aging. Oncotarget. 2015;6(34):35509–35521. doi: 10.18632/oncotarget.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Suh N. MicroRNA controls of cellular senescence. BMB Rep. 2018;51(10):493–499. doi: 10.5483/bmbrep.2018.51.10.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Sturmlechner I., Zhang C., Sine C.C., van Deursen E.J., Jeganathan K.B., Hamada N., et al. P21 produces a bioactive secretome that places stressed cells under immunosurveillance. Science. 2021;374(6567):eabb3420. doi: 10.1126/science.abb3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Xu P., Wang M., Song W.M., Wang Q., Yuan G.C., Sudmant P.H., et al. The landscape of human tissue and cell type specific expression and co-regulation of senescence genes. Mol Neurodegener. 2022;17(1):5. doi: 10.1186/s13024-021-00507-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Sayed N., Huang Y., Nguyen K., Krejciova-Rajaniemi Z., Grawe A.P., et al. An inflammatory aging clock (iAge) based on deep learning tracks multimorbidity, immunosenescence, frailty and cardiovascular aging. Nat Aging. 2021;1(7):598–615. doi: 10.1038/s43587-021-00082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 355.Skopelja-Gardner S., An J., Elkon K.B. Role of the cGAS-STING pathway in systemic and organ-specific diseases. Nat Rev Nephrol. 2022;18(9):558–572. doi: 10.1038/s41581-022-00589-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 356.Laberge R.M., Sun Y., Orjalo A.V., Patil C.K., Freund A., Zhou L., et al. MTOR regulates the pro-tumorigenic senescence-associated secretory phenotype by promoting IL1A translation. Nat Cell Biol. 2015;17(8):1049–1061. doi: 10.1038/ncb3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 357.Schmitt C.A., Tchkonia T., Niedernhofer L.J., Robbins P.D., Kirkland J.L., Lee S. COVID-19 and cellular senescence. Nat Rev Immunol. 2023;23(4):251–263. doi: 10.1038/s41577-022-00785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 358.Li T., Chen Z.J. The cGAS-cGAMP-STING pathway connects DNA damage to inflammation, senescence, and cancer. J Exp Med. 2018;215(5):1287–1299. doi: 10.1084/jem.20180139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 359.Yang H., Wang H., Ren J., Chen Q., Chen Z.J. cGAS is essential for cellular senescence. Proc Natl Acad Sci U S A. 2017;114(23):E4612–E4620. doi: 10.1073/pnas.1705499114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 360.Alle Q., Le Borgne E., Milhavet O., Lemaitre J.M. Reprogramming: emerging strategies to rejuvenate aging cells and tissues. Int J Mol Sci. 2021;22(8):3990. doi: 10.3390/ijms22083990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 361.Cai D., Khor S. ‘hypothalamic microinflammation’ paradigm in aging and metabolic diseases. Cell Metabol. 2019;30(1):19–35. doi: 10.1016/j.cmet.2019.05.021. [DOI] [PubMed] [Google Scholar]
- 362.Pham H., Marathe C.S., Phillips L.K., Trahair L.G., Hatzinikolas S., Huynh L., et al. Longitudinal changes in fasting and glucose-stimulated GLP-1 and GIP in healthy older subjects. J Clin Endocrinol Metab. 2019;104(12):6201–6206. doi: 10.1210/jc.2019-01262. [DOI] [PubMed] [Google Scholar]
- 363.Tang Y., Purkayastha S., Cai D. Hypothalamic microinflammation: a common basis of metabolic syndrome and aging. Trends Neurosci. 2015;38(1):36–44. doi: 10.1016/j.tins.2014.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 364.Priyanka H.P., Nair R.S. Neuroimmunomodulation by estrogen in health and disease. AIMS Neurosci. 2020;7(4):401–417. doi: 10.3934/Neuroscience.2020025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Walecki M., Eisel F., Klug J., Baal N., Paradowska-Dogan A., Wahle E., et al. Androgen receptor modulates Foxp3 expression in CD4+CD25+Foxp3+ regulatory T-cells. Mol Biol Cell. 2015;26(15):2845–2857. doi: 10.1091/mbc.E14-08-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 366.Kalluri R., LeBleu V.S. The biology, function, and biomedical applications of exosomes. Science. 2020;367(6478) doi: 10.1126/science.aau6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 367.Romero-García N., Huete-Acevedo J., Mas-Bargues C., Sanz-Ros J., Dromant M., Borrás C. The double-edged role of extracellular vesicles in the hallmarks of aging. Biomolecules. 2023;13(1):165. doi: 10.3390/biom13010165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.van Niel G., D'Angelo G., Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213–228. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 369.Makhijani P., McGaha T.L. Myeloid responses to extracellular vesicles in health and disease. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.818538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Salvi V., Gianello V., Busatto S., Bergese P., Andreoli L., D’Oro U., et al. Exosome-delivered microRNAs promote IFN-α secretion by human plasmacytoid DCs via TLR7. JCI Insight. 2018;3(10) doi: 10.1172/jci.insight.98204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Xiao X., Xu M., Yu H., Wang L., Li X., Rak J., et al. Mesenchymal stem cell-derived small extracellular vesicles mitigate oxidative stress-induced senescence in endothelial cells via regulation of miR-146a/Src. Signal Transduct Targeted Ther. 2021;6(1):354. doi: 10.1038/s41392-021-00765-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 372.Weilner S., Keider V., Winter M., Harreither E., Salzer B., Weiss F., et al. Vesicular Galectin-3 levels decrease with donor age and contribute to the reduced osteo-inductive potential of human plasma derived extracellular vesicles. Aging (Albany NY) 2016;8(1):16–33. doi: 10.18632/aging.100865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Díaz-Alvarez L., Ortega E. The many roles of galectin-3, a multifaceted molecule, in innate immune responses against pathogens. Mediat Inflamm. 2017;2017 doi: 10.1155/2017/9247574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Fafián-Labora J.A., Rodríguez-Navarro J.A., O'Loghlen A. Small extracellular vesicles have GST activity and ameliorate senescence-related tissue damage. Cell Metabol. 2020;32(1):71–86. doi: 10.1016/j.cmet.2020.06.004. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 375.Salminen A., Kaarniranta K., Kauppinen A. Exosomal vesicles enhance immunosuppression in chronic inflammation: impact in cellular senescence and the aging process. Cell Signal. 2020;75(109771) doi: 10.1016/j.cellsig.2020.109771. [DOI] [PubMed] [Google Scholar]
- 376.Ludwig N., Whiteside T.L., Reichert T.E. Challenges in exosome isolation and analysis in health and disease. Int J Mol Sci. 2019;20(19):4684. doi: 10.3390/ijms20194684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Spehar K., Pan A., Beerman I. Restoring aged stem cell functionality: current progress and future directions. Stem Cell. 2020;38(9):1060–1077. doi: 10.1002/stem.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 378.Guo J., Huang X., Dou L., Yan M., Shen T., Tang W., et al. Aging and aging-related diseases: from molecular mechanisms to interventions and treatments. Signal Transduct Targeted Ther. 2022;7(1):391. doi: 10.1038/s41392-022-01251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 379.Mejia-Ramirez E., Florian M.C. Understanding intrinsic hematopoietic stem cell aging. Haematologica. 2020;105(1):22–37. doi: 10.3324/haematol.2018.211342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Xu M., Palmer A.K., Ding H., Weivoda M.M., Pirtskhalava T., White T.A., et al. Targeting senescent cells enhances adipogenesis and metabolic function in old age. Elife. 2015;4 doi: 10.7554/eLife.12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 381.Bogeska R., Mikecin A.M., Kaschutnig P., Fawaz M., Büchler-Schäff M., Le D., et al. Inflammatory exposure drives long-lived impairment of hematopoietic stem cell self-renewal activity and accelerated aging. Cell Stem Cell. 2022;29(8):1273–1284. doi: 10.1016/j.stem.2022.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Josephson A.M., Bradaschia-Correa V., Lee S., Leclerc K., Patel K.S., Muinos Lopez E., et al. Age-related inflammation triggers skeletal stem/progenitor cell dysfunction. Proc Natl Acad Sci U S A. 2019;116(14):6995–7004. doi: 10.1073/pnas.1810692116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 383.Brunet A., Goodell M.A., Rando T.A. Ageing and rejuvenation of tissue stem cells and their niches. Nat Rev Mol Cell Biol. 2023;24(1):45–62. doi: 10.1038/s41580-022-00510-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 384.Moiseeva V., Cisneros A., Sica V., Deryagin O., Lai Y., Jung S., et al. Senescence atlas reveals an aged-like inflamed niche that blunts muscle regeneration. Nature. 2023;613(7942):169–178. doi: 10.1038/s41586-022-05535-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Cai Y., Song W., Li J., Jing Y., Liang C., Zhang L., et al. The landscape of aging. Sci China Life Sci. 2022;65(12):2354–2454. doi: 10.1007/s11427-022-2161-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 386.Ambrosi T.H., Marecic O., McArdle A., Sinha R., Gulati G.S., Tong X., et al. Aged skeletal stem cells generate an inflammatory degenerative niche. Nature. 2021;597(7875):256–262. doi: 10.1038/s41586-021-03795-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Yang D., de Haan G. Inflammation and aging of hematopoietic stem cells in their niche. Cells. 2021;10(8):1849. doi: 10.3390/cells10081849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Chen Z., Amro E.M., Becker F., Hölzer M., Rasa S.M.M., Njeru S.N., et al. Cohesin-mediated NF-κB signaling limits hematopoietic stem cell self-renewal in aging and inflammation. J Exp Med. 2019;216(1):152–175. doi: 10.1084/jem.20181505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Zhang B., Gladyshev V.N. How can aging be reversed? Exploring rejuvenation from a damage-based perspective. Adv Genet. 2020;1(1) doi: 10.1002/ggn2.10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 390.Zhu H., Liu X., Ding Y., Tan K., Ni W., Ouyang W., et al. IL-6 coaxes cellular dedifferentiation as a pro-regenerative intermediate that contributes to pericardial ADSC-induced cardiac repair Stem. Cell Res Ther. 2022;13(1):44. doi: 10.1186/s13287-021-02675-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Menendez J.A., Alarcón T. Senescence-inflammatory regulation of reparative cellular reprogramming in aging and cancer. Front Cell Dev Biol. 2017;5:49. doi: 10.3389/fcell.2017.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 392.Simpson D.J., Olova N.N., Chandra T. Cellular reprogramming and epigenetic rejuvenation. Clin Epigenet. 2021;13(1):170. doi: 10.1186/s13148-021-01158-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 393.Yang J.H., Hayano M., Griffin P.T., Amorim J.A., Bonkowski M.S., Apostolides J.K., et al. Loss of epigenetic information as a cause of mammalian aging. Cell. 2023;186(2):305–326. doi: 10.1016/j.cell.2022.12.027. e27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Sarkar T.J., Quarta M., Mukherjee S., Colville A., Paine P., Doan L., et al. Transient non-integrative expression of nuclear reprogramming factors promotes multifaceted amelioration of aging in human cells. Nat Commun. 2020;11(1):1545. doi: 10.1038/s41467-020-15174-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Gill D., Parry A., Santos F., Okkenhaug H., Todd C.D., Hernando-Herraez I., et al. Multi-omic rejuvenation of human cells by maturation phase transient reprogramming. Elife. 2022;11 doi: 10.7554/eLife.71624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Zeng X., Li X., Li X., Wei C., Shi C., Hu K., et al. Fecal microbiota transplantation from young mice rejuvenates aged hematopoietic stem cells by suppressing inflammation. Blood. 2023;141(14):1691–1707. doi: 10.1182/blood.2022017514. [DOI] [PubMed] [Google Scholar]
- 397.Ghosh T.S., Shanahan F., O’Toole P.W. The gut microbiome as a modulator of healthy ageing. Nat Rev Gastroenterol Hepatol. 2022;19(9):565–584. doi: 10.1038/s41575-022-00605-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Amsterdam D., Ostrov B.E. The impact of the microbiome on immunosenescence. Immunol Invest. 2018;47(8):801–811. doi: 10.1080/08820139.2018.1537570. [DOI] [PubMed] [Google Scholar]
- 399.Galkin F., Mamoshina P., Aliper A., Putin E., Moskalev V., Gladyshev V.N., et al. Human gut microbiome aging clock based on taxonomic profiling and deep learning. iScience. 2020;23(6):101199. doi: 10.1016/j.isci.2020.101199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Molinero N., Antón-Fernández A., Hernández F., Ávila J., Bartolomé B., Moreno-Arribas M.V. Gut Microbiota, an additional hallmark of human aging and neurodegeneration. Neuroscience. 2023;518:141–161. doi: 10.1016/j.neuroscience.2023.02.014. [DOI] [PubMed] [Google Scholar]
- 401.Segain J.P., Raingeard de la Blétière D., Bourreille A., Leray V., Gervois N., Rosales C., et al. Butyrate inhibits inflammatory responses through NFkappaB inhibition: implications for Crohn’s disease. Gut. 2000;47(3):397–403. doi: 10.1136/gut.47.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Zhou L., Zhang M., Wang Y., Dorfman R.G., Liu H., Yu T., et al. Faecalibacterium prausnitzii produces butyrate to maintain Th17/Treg balance and to ameliorate colorectal colitis by inhibiting histone deacetylase 1. Inflamm Bowel Dis. 2018;24(9):1926–1940. doi: 10.1093/ibd/izy182. [DOI] [PubMed] [Google Scholar]
- 403.Ang Q.Y., Alexander M., Newman J.C., Tian Y., Cai J., Upadhyay V., et al. Ketogenic diets alter the gut microbiome resulting in decreased intestinal Th17 cells. Cell. 2020;181(6):1263–1275. doi: 10.1016/j.cell.2020.04.027.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Biagi E., Nylund L., Candela M., Ostan R., Bucci L., Pini E., et al. Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One. 2010;5(5) doi: 10.1371/journal.pone.0010667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Biagi E., Franceschi C., Rampelli S., Severgnini M., Ostan R., Turroni S., et al. Gut Microbiota and extreme longevity. Curr Biol. 2016;26(11):1480–1485. doi: 10.1016/j.cub.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 406.Greer R.L., Dong X., Moraes A.C., Zielke R.A., Fernandes G.R., Peremyslova E., et al. Akkermansia muciniphila mediates negative effects of IFNγ on glucose metabolism. Nat Commun. 2016;7:13329. doi: 10.1038/ncomms13329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Ottman N., Reunanen J., Meijerink M., Pietilä T.E., Kainulainen V., Klievink J., et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS One. 2017;12(3) doi: 10.1371/journal.pone.0173004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Lin Y.H., Luck H., Khan S., Schneeberger P.H.H., Tsai S., Clemente-Casares X., et al. Aryl hydrocarbon receptor agonist indigo protects against obesity-related insulin resistance through modulation of intestinal and metabolic tissue immunity. Int J Obes. 2019;43(12):2407–2421. doi: 10.1038/s41366-019-0340-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Ghosh T.S., Das M., Jeffery I.B., O’Toole P.W. Adjusting for age improves identification of gut microbiome alterations in multiple diseases. Elife. 2020;9 doi: 10.7554/eLife.50240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 410.Campbell C., McKenney P.T., Konstantinovsky D., Isaeva O.I., Schizas M., Verter J., et al. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature. 2020;581(7809):475–479. doi: 10.1038/s41586-020-2193-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 411.Fransen F., van Beek A.A., Borghuis T., Aidy S.E., Hugenholtz F., van der Gaast-de Jongh C., et al. Aged gut Microbiota contributes to systemical inflammaging after transfer to germ-free mice. Front Immunol. 2017;8:1385. doi: 10.3389/fimmu.2017.01385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 412.Badal V.D., Vaccariello E.D., Murray E.R., Yu K.E., Knight R., Jeste D.V., et al. The gut microbiome, aging, and longevity: a systematic review. Nutrients. 2020;12(12):3759. doi: 10.3390/nu12123759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Du H., Bartleson J.M., Butenko S., Alonso V., Liu W.F., Winer D.A., et al. Tuning immunity through tissue mechanotransduction. Nat Rev Immunol. 2023;23(3):174–188. doi: 10.1038/s41577-022-00761-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Park S., Kim B. Aging-related structural change in 3D extracellular matrix affects its mechanics. Med Eng Phys. 2022;106(103843) doi: 10.1016/j.medengphy.2022.103843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Schmauck-Medina T., Molière A., Lautrup S., Zhang J., Chlopicki S., Madsen H.B., et al. Vol. 14. Aging (Albany; NY: 2022. pp. 6829–6839. (New hallmarks of ageing: a 2022 Copenhagen ageing meeting summary). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Lee M., Du H., Winer D.A., Clemente-Casares X., Tsai S. Mechanosensing in macrophages and dendritic cells in steady-state and disease. Front Cell Dev Biol. 2022;10:1044729. doi: 10.3389/fcell.2022.1044729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Sladitschek-Martens H.L., Guarnieri A., Brumana G., Zanconato F., Battilana G., Xiccato R.L., et al. YAP/TAZ activity in stromal cells prevents ageing by controlling cGAS-STING. Nature. 2022;607(7920):790–798. doi: 10.1038/s41586-022-04924-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Chakraborty M., Chu K., Shrestha A., Revelo X.S., Zhang X., Gold M.J., et al. Mechanical stiffness controls dendritic cell metabolism and function. Cell Rep. 2021;34(2):108609. doi: 10.1016/j.celrep.2020.108609. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.