Abstract
Angelman syndrome (AS) is a rare neurodevelopmental disorder caused by loss of function mutations in maternally expressed UBE3A. No gene-specific treatment is available for patients so far. Although intact and transcriptionally active, paternally inherited UBE3A is silenced by elongation of antisense long noncoding RNA UBE3A-ATS in neurons. Here, we demonstrated that RNA targeting of paternal Ube3a-ATS with a high-fidelity CRISPR-Cas13 (hfCas13x.1) system could restore Ube3a expression to similar levels as that of maternal Ube3a in the cultured mouse neurons. Furthermore, injection into lateral ventricles with neuron-specific hSyn1 promoter-driven hfCas13x.1 packaged in adeno-associated virus (AAV-PHP.eb) could restore paternal Ube3a expression in cortex and hippocampus of neonatal AS mice for up to 4 months after treatment. Behavioral tests showed that expression of paternal Ube3a significantly alleviated AS-related symptoms, including obesity and motor function. Our results suggested that hfCas13x.1-mediated suppression of the Ube3a-ATS lncRNA potentially serves as a promising targeted intervention for AS.
Keywords: Angelman syndrome, RNA editing, high-fidelity Cas13, paternal Ube3a, motor function
Graphical abstract
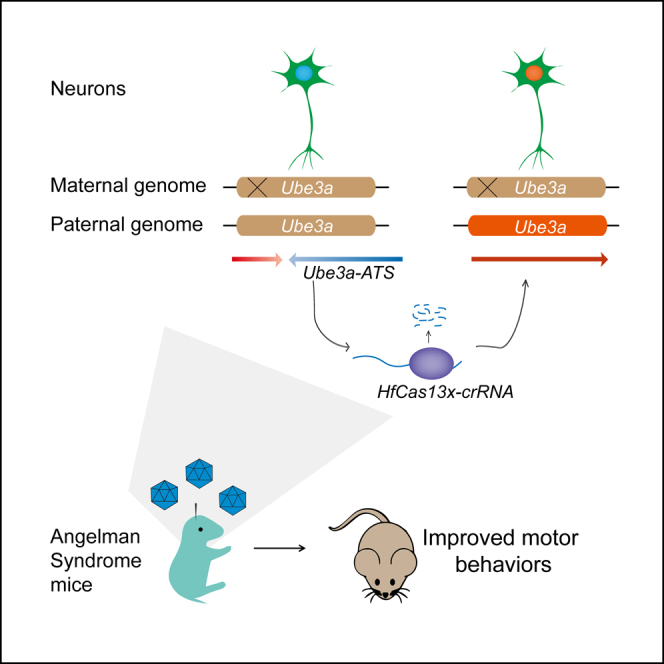
Yang et al. demonstrated that RNA targeting of paternal Ube3a-ATS with a high-fidelity CRISPR-Cas13 (hfCas13x.1) system could restore Ube3a expression in the cultured mouse neurons and in vivo, and also improve several behavioral deficits in AS mice.
Introduction
Angelman syndrome (AS) is a severe neurodevelopmental disorder, with an incidence of approximately 1 in 12,000–1/20,000 worldwide, which is characterized by seizures and severely impaired motor, cognitive, and language skills.1 AS is mainly attributable to loss of maternally inherited ubiquitin protein ligase E3A (UBE3A), which mediates the targeted degradation of several protein substrates.2,3,4,5,6 Only the maternally inherited UBE3A allele in neurons is active due to selective silencing of paternal UBE3A by its antisense transcript, UBE3A-ATS. UBE3A-ATS is a polyadenylated transcript processed from SNHG14 small nucleolar RNA host gene 14 (SNHG14) transcription and it overlaps with the UBE3A gene locus, resulting in early termination and degradation of UBE3A transcripts.7,8,9,10 Approximately 75% of AS cases are reportedly caused by deletions of the maternal chromosomal region 15q11.2–q13. The remaining cases are attributable to other mutations in maternal UBE3A, paternal uniparental disomy, or imprinting defects.1
There are currently no specific therapies available for the treatment of AS. However, activation of the intact, functional, paternal copy of UBE3A by eliminating the UBE3A-ATS long non-coding RNA (lncRNA) represents a promising potential therapeutic strategy. Genetic restoration of Ube3a by interferences with Ube3a-ATS has been reported to dramatically correct the aberrant neurobehavioral phenotypes in AS model mice.10 Antisense oligonucleotides have been developed to reactivate Ube3a expression by inhibiting Ube3a-ATS in AS mice and are now in phase I and II clinical trials.11,12 However, this treatment strategy requires frequent administration through the lumbar. Directed targeting of Ube3a-ATS with SaCas9 can also effectively decrease Ube3a-ATS transcription in mice by introducing double-stranded breaks (DSBs), insertions and deletions (indels), or adeno-associated virus (AAV) vector integration,13,14 which may induce unexpected genomic change.15
CRISPR-Cas13, a class 2 type VI RNA endonuclease, can bind and cleave single-stranded RNA guided by an engineered CRISPR RNA (crRNA).16,17,18,19,20 Gene therapies based on RNA-targeting nucleases are currently under exploration as a safer alternative to DNA editing Cas endonucleases, since they are accompanied by a lower risk of introducing permanent genomic alterations. To date, Cas13x.1 is the smallest known crRNA-guided RNA endonuclease, with only 775 amino acids, and a high-fidelity variant of this Cas, hfCas13x.1, has been shown to exhibit high on-target activity with markedly lower collateral activity. Here, we showed that treatment with hfCas13x.1 guided by crRNA targeting the intergenic region between Ube3a and Snord115, expressed under the neuron-specific hSyn1 promoter and packaged in an AAV-PHP.eb vector effectively induced the expression of paternal Ube3a, resulting in significant improvements to behavioral impairments in AS model mice.
Results
A high-fidelity Cas13x.1 reinstates paternal Ube3a expression in primary cultured mouse neurons
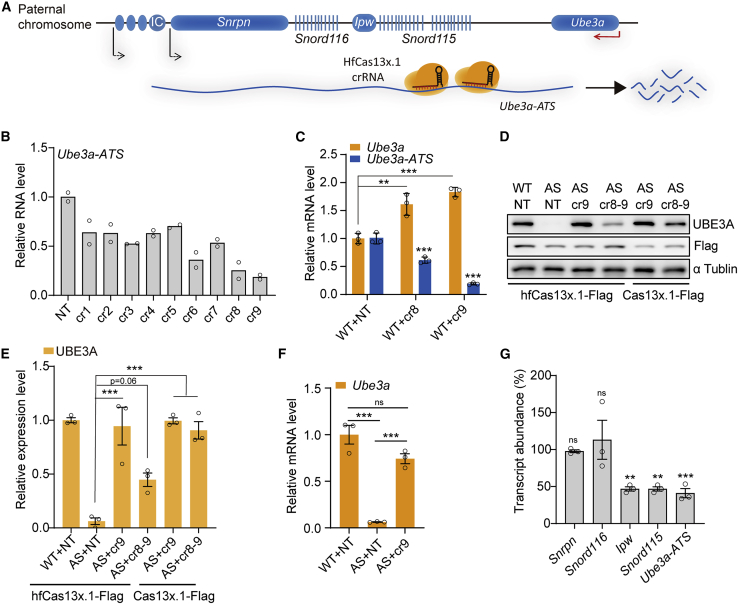
To detect whether Cas13x.1 could inhibit the transcription of Ube3a-ATS lncRNA, we designed none crRNAs targeting premature Ube3a-ATS transcripts in mice, with target sites between the Snord115 cluster and the Ube3a locus (Figure 1A and Table S1), and then constructed elongation factor 1 alpha short promoter (EFS)-Nls-Cas13x.1-Nls-Flag-T2A-EGFP and each crRNA individually into all-in-one plasmid (Figure S1A). We transduced N2a neuroblastoma cells with the constructs above, and then sorted the EGFP+ cells by flow cytometry for quantification of RNA level of Ube3a-ATS regions downstream of Snord115 genes by reverse transcriptase quantitative PCR (RT-qPCR). The results indicated that cr8 and cr9 provided the highest knockdown efficiency (i.e., >70%), compared with a non-targeting (NT) control (Figure 1B and Table S1). To evaluate the knockdown efficiency of Ube3a expression in primary cultured neurons, we isolated primary cortical neurons from C57 mouse embryos and infected these cells with lentivirus expressing Cas13x.1 and crRNA (Figure S1A). Quantification of Ube3a-ATS RNA and Ube3a mRNA in infected primary neurons showed that Cas13x.1/cr8 and Cas13x.1/cr9 induced significant up-regulation of Ube3a transcription concurrent with Ube3a-ATS knockdown (Figure 1C). Previous study showed that Cas13x.1 could well tolerate a single-nucleotide mismatch.20 We predicted that cr9 could target multiple target sites with one or two base pair mismatches in the intergenic Snord115-flanking sequence (Table S1).
Figure 1.
Screen of crRNAs targeting the lncRNA Ube3a-ATS to unsilence paternal Ube3a
(A) Schematic diagram of the Ube3a locus in the mouse genome. IC, imprinting center. (B) Ube3a-ATS mRNA expression levels under targeting by different crRNAs in N2a cells (n = 2 for all groups). (C) RT-qPCR analysis of mRNA expression in wild-type (WT) primary cultured neurons of C57 mice. The WT neurons were infected with lentivirus containing EFS-hfCas13x.1/U6-cr8 (WT + cr8), EFS-hfCas13x.1/U6-cr9 (WT + cr9), or the non-targeted control crRNA EFS-hfCas13x.1/U6-NT (WT + NT) (n = 3 for all groups). (D and E) Western blot analysis (D) and quantification of band density (E) of UBE3A protein expression in cultured primary neurons of WT or AS mice infected with lentivirus-containing hSyn1-hfCas13x.1/U6-NT or hSyn1-hfCas13x.1/U6-cr9 (n = 3 for all groups). (F) RT-qPCR analysis of Ube3a mRNA levels in cultured primary neurons of WT or AS mice infected with hSyn1-hfCas13x.1/U6-NT or hSyn1-hfCas13x.1/U6-cr9 (n = 3 for all groups). (G) Expression of the indicated genes in cultured primary neurons of AS mice infected with hSyn1-hfCas13x.1/U6-cr9 to silence the paternal lncRNA Ube3a-ATS (n = 3 for all groups). Levels were relative to that in primary neurons of AS mice infected with the hSyn1-hfCas13x.1/U6-NT non-targeted control. Statistical significance was assessed by one-way ANOVA followed by Tukey’s multiple comparison test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. ns, not significant.
We also isolated and infected AS (Ube3am–/p+) primary cortical neurons with lentivirus expressing Cas13x.1-Flag to investigate whether neuron-specific promoter, human Syn1 (hSyn1) (Figures S1D and S1E), induced high efficiency of Ube3a-ATS silencing compared with the EFS (Figures S1B and S1C) promoter under the same conditions. Western blot results showed that the hSyn1-driven Cas13x.1 with cr8 reinstated UBE3A protein to 52.5% compared with the expression level of maternal UBE3A in WT cells, while EFS-driven Cas13x.1 reinstated UBE3A protein to 34.8% (Figures S1B and S1C). This result indicated that hSyn1-Cas13x.1 with crRNA could restore UBE3A expression more effectively in AS primary neurons. Since high-fidelity Cas13x.1 (hfCas13x.1) has markedly lower collateral cleavage activity compared with wild-type (WT) Cas13x.1,21 we examined UBE3A protein (Figures 1D, quantified in 1E) under Ube3a-ATS silencing by WT Cas13x.1 and hfCas13x.1 in primary AS mouse neurons. The results showed that hfCas13x.1 with cr9 could restore UBE3A protein expression through the suppression of Ube3a-ATS as effectively as WT Cas13x.1 with cr9 (Figures 1D and 1E). The Ube3a mRNA is also significantly reinstated by hfCas13x.1 (Figure 1F). Additionally, we found that the cr8–9 tandem crRNA with hfCas13x.1 could not restore UBE3A as efficiently as cr9 alone, indicating that the addition of cr8 did not positively affect hfCas13x.1 function (Figures 1D and 1E). To evaluate the localization of unsilenced paternal UBE3A, we also isolated and infected AS (Ube3am–/p+) primary cortical neurons with lentivirus expressing Cas13x.1-Flag. We co-stained UBE3A and Flag (hfCas13x.1-Flag) and found that the UBE3A expression restored by hfCas13x.1/cr9 was enriched in the nucleus of AS mouse primary neurons (Figure S1F).
Snrpn, Snord116, Ipw, and Snord115 are all processed from the same Snhg14 transcript. To detect the effects on these genes by hfCas13x.1/cr9, we evaluated the expression of Snrpn, Snord116, Ipw, and Snord115 in AS neurons infected with lentivirus expressing hfCas13x.1 and cr9 by RT-qPCR, which indicated that the expression of Ipw and Snord115, relatively close to the crRNA target sites, were affected by the cr9, while Snrpn and Snord116 were not (Figure 1G). Whole transcriptome RNA sequencing (RNA-seq) of infected AS mouse primary neurons indicated that relatively few genes were differentially expressed between the hfCas13x.1/U6-cr9 treatment group and neurons infected with the NT crRNA (Figure S1G and Table S2). We also checked the expression of predicted potential off-target sites and found no significantly decreased expression of these transcripts in RNA-seq data (Table S3), which suggested high targeting specificity for hfCas13x.1/cr9. Collectively, these data indicated that hSyn1-driven hfCas13x.1 coordinated by U6-driven cr9 crRNA effectively and specifically unsilences Ube3a in neurons.
AAV-packaged hfCas13x.1 guided by cr9 unsilenced paternal Ube3a expression in mouse cortex and hippocampus in vivo
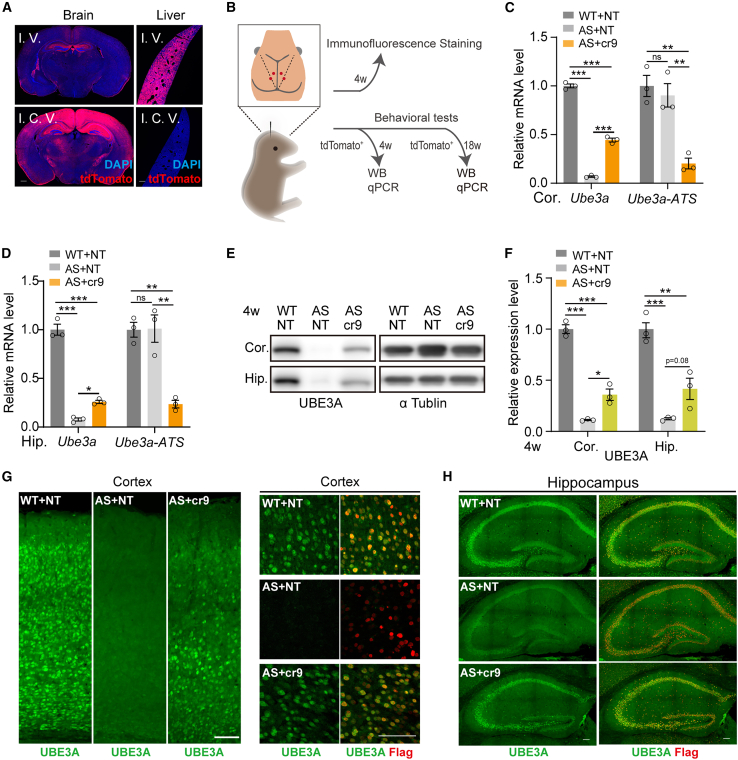
Previous studies have shown that restoring Ube3a expression during early developmental stages could rescue most AS-associated phenotypes in AS model mice,22 while the AAV-PHP.eb capsid has been shown to efficiently cross the blood-brain barrier to infect cells in the central nervous system.23 Based on these findings, we individually performed intracerebroventricular (I.C.V.) and intravenous (I.V.) injections with AAV-PHP.eb containing a CAG promoter-driven tdTomato reporter in neonatal mice. One facial vein was administered 1.9 × 1011 virus particles, or 0.3 × 1011 virus particles were injected into the bilateral ventricle of the neonatal mouse. When examined histologically at 3 weeks, animals injected by I.V. infusion showed remarkably high tdTomato signal in liver tissue, while I.C.V. injection with fewer virus particles led to obvious tdTomato expression in the central nervous system (Figure 2A).
Figure 2.
AAV delivery of the CRISPR-hfCas13x system restores paternal Ube3a expression in AS mouse brain
(A) TdTomato signal in brain and liver of WT mouse at 3 weeks after I.C.V. injection of 1.9 × 1011 or I.V. injection of 0.3 × 1011 AAV-PHP.eb particles containing CAG-tdTomato at P0. Scale bar, 500 μm. (B) Experimental timeline for evaluation in WT and AS (Ube3am−/p+) mice after I.C.V. injection of AAV-PHP.eb containing hSyn1-hfCas13x.1/U6-NT or hSyn1-hfCas13x.1/U6-cr9 at P0. (Cand D) RT-qPCR analysis of mRNA expression of Ube3a and Ube3a-ATS in the cerebral cortex (Cor.) (C) and hippocampus (Hip.) (D) of WT and AS mice with indicated treatment at 4 weeks (n = 3 for all groups). (E and F) Western blot (E) and quantification (F) of protein expression in the cerebral cortex (cor.) and hippocampus (hip.) of WT and AS mice with indicated treatment at 4 weeks (n = 3 for all groups). (G and H) Immunofluorescence staining for UBE3A with FLAG in cortex (G) or hippocampus (H) of WT or AS mice with indicated treatment at 4 weeks. Scale bar, 100 μm. Statistical significance was assessed by one-way ANOVA followed by Tukey’s multiple comparison test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. ns, not significant.
In light of these results, we then packaged AAV-PHP.eb with hSyn1-hfCas13x.1 and U6 promoter-NT or cr9 (Figure S1A). To further determine whether hfCas13x.1/cr9 could unsilence paternal Ube3a, we also made Ube3am+/pYFP knock-in mice, which harbors a paternal Ube3a-YFP knock-in allele, and primary neurons from Ube3am+/pYFP embryos were isolated and infected with the packaged AAV with hfCas13x.1/NT or hfCas13x.1/cr9. RT-qPCR-based detection of YFP showed that AAV-packaged hSyn1-hfCas13x.1/U6-cr9 significantly induced the expression of paternal Ube3a-YFP in neurons (Figure S2A). To evaluate the efficiency of Ube3a-ATS silencing in brain, we co-injected 2.5 × 109 AAV-PHP.eb-CAG-tdTomato with a total of 5 × 1010 AAV-PHP.eb-hSyn1-hfCas13x.1/U6-cr9 or AAV-PHP.eb-hSyn1-hfCas13x.1/U6-NT particles I.C.V. into postnatal day 1 (P1) AS or WT mice (Figure 2B). We dissected the tdTomato+ cortex, hippocampus, spinal cord, or cerebellum tissues for quantification of Ube3a-ATS and Ube3a expression at 4 weeks. RT-qPCR results showed that treatment with hSyn1-hfCas13x.1/U6-cr9 led to a 77.5% knockdown of Ube3a-ATS transcripts in the cortex tissues (Figure 2C) and 76.6% knockdown in hippocampus tissues (Figure 2D) of AS mice compared with that in AS control mice administered with hSyn1-hfCas13x.1/U6-NT. Furthermore, paternal Ube3a mRNA levels increased to 44.3% in cortex tissues and 17.6% in hippocampus tissues of AS mice compared with expression levels of maternal Ube3a in C57 WT control mice (Figures 2C and 2D).
Western blots showed that paternal UBE3A protein expression reached 35.9% in cortex tissues and 41.6% in hippocampus tissues of AS mice at 4 weeks after infection with AAV-PHP.eb-hSyn1-hfCas13x.1/U6-cr9 (Figures 2E and 2F), compared with that in WT mice. By contrast, no paternal UBE3A protein expression could be detected in spinal cord or cerebellum samples of any mice at 4 weeks (Figures S2B and S2C). Paternal UBE3A protein expression maintained 21.1% and 39.1% expression in cortex and hippocampus, respectively, at 18 weeks; however, the difference in statistical analysis was not significant (Figures S2D and S2E). RT-qPCR-based detection of Snord115 target gene transcripts, including Crhr1, Ralgps, Taf1, Pbrm1, and Dpm2, indicated no significant changes in the expression of any of these genes in cortical tissues of AS mice, despite partial inhibition of Snord115 expression by the AAV-PHP.eb-hSyn1-hfCas13x.1/U6-cr9 system (Figure S2F). Moreover, the detection of hfCas13x.1 transcription driven by the hSyn1 promoter in the cortex and liver of AS mice indicated that it was exclusively expressed in the brain (Figure S2G). When examined histologically at 4 weeks, mice showed uneven expression of hfCas13x.1-Flag in cortex and hippocampus cells (Figure S3A). AS mice treated with AAV-PHP.eb-hSyn1-hfCas13x.1/U6-cr9 mice had a significant expression of unsilenced UBE3A in the nucleus of cortex and hippocampus (Figures 2G, 2H, and S3B), and we also found the cells with strong Flag staining showed higher expression of UBE3A (Figure S3B). Co-staining of UBE3A and NeuN in treated mice indicated that UBE3A was specifically recovered in neurons (Figure S4). Moreover, we observed no obvious structural change in brain slices and no abnormal NeuN signal in immunostaining images (Figures 2G, S3, and S4). Collectively, these results indicated that hfCas13x.1/cr9 was delivered by the AAV-PHP.eb vector and could successfully suppress Ube3a-ATS transcripts to unsilence paternal Ube3a in AS mice in vivo.
CRISPR-hfCas13x.1 in the AAV-PHP.eb vector improves impaired motor function in AS mice
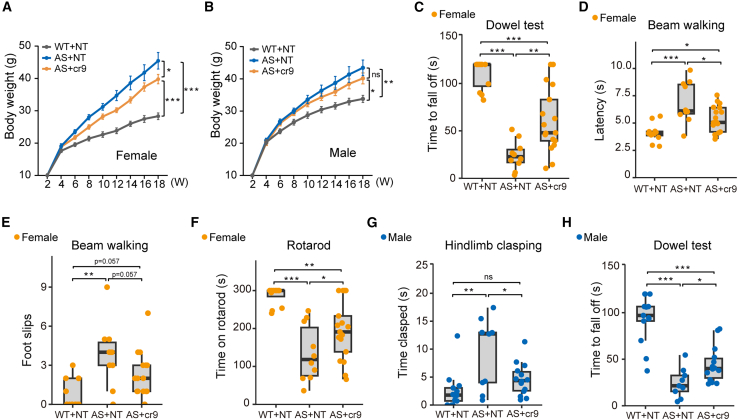
Previous reports have shown that AS mouse models exhibit deficiencies in motor coordination, obesity, hypoactivity, and memory deficit.24,25,26,27 To evaluate the phenotypic improvement in treated AS mice, we then conducted several behavioral tests for phenotypic analysis of WT or AS model mice I.C.V. with AAV-PHP.eb-hSyn1-hfCas13x.1/U6-cr9 or AAV-PHP.eb-hSyn1-hfCas13x.1/U6-NT control (Figures S5A, S5C–S5J). We found that WT mice injected with hfCas13x.1/U6-NT control exhibited a sex effect in body weight (Figure S5B), which was also observed in a previous investigation.13 In hindlimb clasping and beam walking tests, female AS mice with hfCas13x.1/NT showed more pronounced deficits when compared with sex-matched WT mice with hfCas13x.1/NT (Figures S5D, S5H, and S5I). In an open field test, the distance traveled showed no significant difference between WT and AS mice injected with hfCas13x.1/NT (Figure S5E). Moreover, a significant difference was observed between female and male AS mice with hfCas13x.1/NT in a rotarod test (Figure S5J). Hence, body weight and behavior results for each gender were analyzed separately. We found that obesity in female AS mice administrated with hfCas13x.1/cr9 was significantly improved (Figure 3A), but not in male AS mice (Figure 3B). Furthermore, female AS mice treated with hfCas13x.1/cr9 also showed improvement in the dowel test (Figure 3C), beam walking test (Figures 3D and 3E), and rotarod test (Figure 3F), while male mice showed improvement in the hindlimb clasping test (Figure 3G) and dowel test (Figure 3H), indicating that motor coordination and balance were improved in AS mice treated with hfCas13x.1/cr9 compared with NT-treated AS control mice.
Figure 3.
HfCas13x.1/cr9 improves motor functions in an AS mouse model
(A) Body weight of female mice was measured biweekly over 18 weeks. (B) Body weight of male mice was measured biweekly over 18 weeks. (C) Dowel test data in 13-week-old female mice. (D and E) Beam walking assays in 14-week-old female mice. Time to traverse the beam (D) and the number of foot slips (E) are shown. (F) Accelerating rotarod test data in 15-week-old female mice. (G) Hindlimb clasping assays in 7-week-old male mice. (H) Dowel test data in 13-week-old male mice. N = 12 for WT + NT; n = 10 for AS + NT; n = 17 for AS + cr9 in female mice. N = 13 for WT + NT; n = 9 for AS + NT; n = 14 for AS + cr9 in male mice. Statistical significance was assessed by one-way ANOVA followed by the Holm-Sidak comparison test. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. ns, not significant.
However, female AS mice injected with hfCas13x.1/cr9 showed no improvement in the hindlimb clasping test (Figure S6A), and male AS mice injected with hfCas13x.1/cr9 showed slight improvement in the rotarod test (Figure S6B). Female AS mice injected with hfCas13x.1/cr9 showed the trend of entering the center more times than AS + NT control mie (Figure S6C), but not the male AS mice (Figure S6D). Also, AS mice treated with the hfCas13x.1/cr9 vector showed no improvement in the marble burying test (Figures S6E and S6F), and the microcephaly phenotype was also not rescued (Figures S6G and S6H). In our hands, AS mice showed no deficits in contextual-based and cue-based learning during fear conditioning assays (Figures S6I–S6K), consistent with some reported results13,28; therefore, this assay was not performed in hfCas13x.1/cr9-treated mice. We also compared the body weight between non-injected WT and WT with hfCas13x.1/NT group, and no difference in body weight was observed between the two groups (Figures S6L and S6M), indicating that AAV-packaged hfCas13x.1 was well tolerated in mice. In summary, these results demonstrate that the unsilencing of paternal Ube3a at the P1 stage by AAV-PHP.eb-hSyn1-hfCas13x.1/U6-cr9 significantly improved several AS-related behavioral impairments in mice.
Discussion
In this study, we examined different combinations of tissue-specific promoters, modes of administration, Cas endonucleases, and crRNAs to explore the specificity, delivery efficiency, and therapeutic effects of silencing paternal Ube3a-ATS in vivo. We proved that an RNA-guided CRISPR-hfCas13x.1 system can effectively recover the expression of UBE3A via knockdown of the lncRNA Ube3a-ATS in vitro and in vivo. An increasing number of AAV-based gene therapies have been approved by the US Food and Drug Administration to treat inherited diseases.29,30 RNA-guided endonucleases have facilitated the development of precisely targeted gene therapies, and numerous clinical trials exploring the safety and efficacy of CRISPR/Cas-based strategies are currently in progress.31 CRISPR-hfCas13x.1 system will be a promising and safe treatment strategy for AS.
Other studies have reported a CRISPR-SaCas9 system that could unsilence paternal Ube3a by introducing indels, through AAV vector integration, or by inducing the formation of secondary or tertiary genomic structures at the Ube3a-ATS locus.13,14 However, the end fragments generated by DSBs can induce chromosomal translocations, rearrangements, or large fragment deletions.15,32,33 CRISPR-Cas13 enzymes do not contain the RuvC and HNH domains responsible for DNA cleavage, which avoid genomic alterations and off-targets.17 Therefore, using a high-specificity Cas13x system packaged in AAV may represent a safe and potential long-lasting therapeutic option for targeted treatment of AS.
However, we found that the expression of hfCas13x.1-Flag varies in each neuron cell and the UBE3A was significantly reinstated in cells with high expression of hfCas13x.1-Flag (Figure S3), indicating the uneven spreading of the AAV vector in the brain. This could explain that the recovered protein levels were not high enough in cortex and hippocampus, and AAV-PHP.eb-hSyn1-hfCas13x.1/U6-cr9 treatment partially improved motor functions of AS mouse model. By comparing male and female mice injected with NT control, we found that WT or AS mice showed sex effects in several behavioral tests (Figures S5B–S5K), which might explain gender-dependent improvements in the obesity phenotype and motor functions in AS mice treated with hfCas13x.1-Flag/cr9 (Figures 3A–3H and S6A–S6H). It is reasonable to believe that the further optimization of AAV vector and drug component will improve the therapeutic effect in disease model and promote translational research of this approach.
Snhg14, as a single long RNA, is co-transcriptionally spliced into several host genes that were further processed into Snrpn mRNA, Snord116, Ipw, Snord115, and Ube3a-ATS. In this study, we found that hfCas13x.1/cr9 also decreased the expression of Snor115 and Ipw in primary cells. As mentioned above, cr9 has multiple target sites with one or two mismatches (Table S1) between Snord115 clusters, and these target sites are also close to the Ipw location in mice. Therefore, Snord115 and Ipw might have no enough time to be spliced from the nascent RNA when hfCas13x.1/crRNA bound and degraded the nascent RNAs. This crRNA-specific effect could be solved by switching crRNA.
Adolescent reactivation in Ube3aStop/p+;CreERT+ mice only partially rescued motor coordination, while no improvement was seen in the adult reactivation group.22 This indicated earlier reinstatement of Ube3a is critical for AS treatment. However, humans have a very long childhood compared with mice, and until now the precise treatment window in AS patients remains uncertain.22 Therefore, whether patients of different ages can benefit from gene therapy needs to be thoroughly studied. Although the bilateral I.C.V. route is commonly used in preclinical and clinical studies for CNS diseases and the patient ages ranged from 1 day to 84 years,34,35 the same AAV serotype usually displayed variable delivery efficacy and safety among mice and primates; hence, preclinical studies in non-human primates with the AAV vector need to be conducted in the future.
Overall, we demonstrated that the hfCas13x.1-crRNA system effectively and specifically relieves Ube3a silencing by degrading Ube3a-ATS in primary neurons and in vivo. Moreover our data showed that one-time injection of AAV with hfCas13x.1-crRNA significantly improved several motor functions in AS mice, indicating its potential for treatment of AS.
Materials and methods
Plasmid construction
The coding sequences of Cas13x.1 and crRNAs were constructed into different backbones with the NEBuilder HiFi DNA assembly Master Mix (New England BioLabs), as previously described.20 All crRNA sequences are provided in Table S1. The plasmid diagram is shown in Figure S1A.
N2a culture, transient transfection, and cell sorting
The N2a cell line was purchased from the cell bank of the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, and cultured in an incubator at 37°C with 5% CO2. The medium of N2a is DMEM (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco), 1% penicillin/streptomycin (Gibco), and 1× non-essential amino acids (Gibco). Cells were transfected with plasmids and PEI (Polyscience) in a 1:2 ratio 12 h later after being plated. Two days after transfection, cells were digested with 0.05% trypsin (Thermo Fisher Scientific) and sorted by MoFlo XDP. GFP-positive cells are kept in DMEM and then in Trizol for RNA isolation.
Animals
All animal experiments were approved by the Animal Care and Use Committee of the Institute of Neuroscience, Chinese Academy of Sciences, Shanghai, China. Ube3am−/p+ mice and Ube3am+/pYFP mice were maintained on the C57Bl/6 background. All mice used in this study were housed in 12 h light/dark cycle. Mice were randomly assigned to the corresponding treatments. All experimental protocols were approved by the Animal Care and Use Committee of the Institute of Neuroscience, Chinese Academy of Sciences, Shanghai, China.
AS mouse model and Ube3aYFP mice
Ube3a KO mice were generated by Jiang et al.36 Ube3aYFP mice were generated as previously described.37 AS mouse models were generated by crossing the Ube3am+/p- heterogeneous females among Ube3a KO mice with C57BL/6 WT males from Shanghai SLAC Laboratory Animal Co., Ltd. Ube3am+/pYFP were generated by crossing the Ube3aYFP heterogeneous male mice with C57BL/6 WT females. Ube3amatYFP/P+ were generated by crossing the Ube3aYFP heterogeneous female mice with C57BL/6 WT males.
Recombinant lentivirus particles preparation
Lentiviral transgene plasmids were co-transfected with package plasmids and envelope plasmids into HEK293 cells to produce recombinant lentiviral particles delivering the hfCas13x.1 system. The HEK293 cell supernatant was collected for ultracentrifugation (27,000 rpm, CP90NX, Hitachi). The DNA genome titer of lentiviral vector genomes was determined by detecting lentivirus vectors' copy numbers in cells with RT-qPCR.
Neuronal culture and lentivirus infection
The cortex and hippocampus were dissected from E14–E16 embryos, dissociated in digestion buffer with papain (Worthington Bio Corp), as described previously.38 The digestion buffer contains (in mM): 161 NaCl, 5.0 KCl, 2.9 CaCl2, 5.0 HEPES, 5.5 glucose, 0.53 MgSO4, and 0.0056 phenol red, pH 7.4, with additional (in mM) 1.7 cysteine, 1 CaCl2, and 0.5 EDTA. The tissues were digested for 30 min at 37°C, then plated on PDL (Sigma-Aldrich) in neuronal medium (Thermo Fisher Scientific) containing 2 mM Glutamax-I (Thermo Fisher Scientific), B27 (Thermo Fisher Scientific), and 5% FBS. Four hours later, the neurons were maintained in the same medium without FBS. On days in vitro 2, the virus was added to well for 24 h, and then half-changing the medium was carried out for three consecutive days. The subsequent experiments were performed 7–9 days later, after virus infection.
Quantitative PCR and RNA-seq analysis
RNA was extracted from cells or dissected tissues using TRIZOL (Invitrogen) and converted to cDNA using HiScript Q RT SuperMix for qPCR (Vazyme) according to the manufacturer’s instructions. We performed qPCRon a Roche 480 II-A using AceQ Universal SYBR qPCR Master Mix (Vazyme). The primers used in qPCR are shown in Table S5. For transcriptome-wide RNA-seq, RNA was extracted and then reverse transcribed to cDNA, which was used for transcriptome-wide RNA-seq. An average of the three repeats is presented. Raw data from the sequencer were first undergone quality control using FastQC. Next, reads are cleaned using trim galore, where the Illumina adapter sequence and low quality (Phred score <20) at the 3′ end were trimmed, and the paired reads were removed if any of the two reads did not meet the minimum length (20 bp). Salmon (v1.5) was then used to quantify the expression of transcripts. Differential analysis between the groups was done by a count-based method limma, which is implemented in R and voom is involved for normalization.34 Significantly expressed genes were screened by BH-adjusted p value (<0.05) and the fold-change (>2).
Western blotting
Cells or dissected brain regions were homogenized in SDS lysis buffer (Beyotime) containing a 1× Protease inhibitor cocktail (Beyotime). We loaded 40–60 μg total proteins and separated by SDS-PAGE (Epizyme) and transferred to polyvinylidene fluoride membrane (Merk Millipore). The membrane was blocked in 5% skim milk powder in Tris-buffered saline with Tween buffer (Epizyme). The following antibodies were diluted and incubated overnight: anti-UBE3A (1:1,000; A300-352A, Bethyl), anti-Flag (1:3,000; F1804, Sigma), anti-α-Tubulin (1:5,000; AF0001, Beyotime). After primary antibody incubation, membranes were probed with goat anti-mouse HPR (1:5,000; A9044, Sigma-Aldrich) or goat anti-rabbit HRP (1:5,000; A0545, Sigma), and images were collected using AmershamImager600 (GE) and analyzed by ImageJ software.
AAV virus production
AAV-PHP.eb serotype was used in this study.23,35 The CAG-tdTomato, hSyn1-hfCas13x.1/U6-cr9, or hfCas13x.1/U6-NT plasmids were constructed and sequenced before packaging into AAV-PHP.eb vehicle; and the AAV vectors were packaged by PackGene Biotech. The vector titer was determined by qPCR specific for the inverted terminal repeat.
I.C.V. injection and I.V. injection
For I.C.V. administration, neonatal mice were placed on ice for hypothermia anesthesia and injected with AAV virus into bilateral ventricles at four sites.39,40 A total of 5 × 1010 AAV (containing hSyn1-hfCas13x.1/U6-cr9 or hSyn1-hfCas13x.1/U6-NT) particles plus 2.5 × 109 AAV (CAG-tdTomato) in PBS was injected into each mouse within 24 h of birth using Nanoject III (Warner Instruments). For immunofluorescence experiments, no AAV-containing CAG-tdTomato was injected. Tdtomato+ cortex or hippocampus tissues were dissected for RT-qPCR and western blotting. For I.V. injection, AAV solution was infused into the facial vein of neonatal mice as previously described.39
Immunofluorescence
Primary neurons were fixed with 4% paraformaldehyde (PFA) for 1 h. Mice were perfused with PBS and then 4% PFA; brain tissues were removed for further fixation overnight and then dehydrated in 30% sucrose for 3 consecutive days. Tissues were sectioned to a thickness of 40 μm with Thermo HM525-NX and were performed antigen retrieval at 70°C in sodium citrate solution for an hour. Antibodies were diluted in primary antibody dilution buffer (Epizyme) and incubated as following ratio: anti-Ube3a (1:500; A300-352A, Bethyl), anti-Flag (1:500; F1084, Sigma-Aldrich), anti-NeuN (1:2,000,MAB377,Chemicon) overnight at 4°C, and then incubated with Alexa Fluor 488 or Alexa Fluor 647 goat secondary antibody to rabbit IgG and Alexa Fluor 568 goat antibody to mouse IgG (1:500, Invitrogen) for 2 h at 4°C. Slides were sealed with fluoromount-G mounting medium. All images were visualized under a Nikon C2.
Behavioral tests
All behavioral experiments were performed blind to genotypes and injection treatment of animals. Mice were placed in the room for 30 min to acclimate to the environment before the test.
Hindlimb clasping
Each mouse was suspended 10 cm above the table for 20 s by being held by the tail. The video was recorded and time was scored offline. Hindlimb clasping time is the total time spent on clasping. Clasping was defined by the behavior of incomplete splay with one or both hind limbs.
Open field
Each mouse was placed on the edge of a 40 cm × 40 cm open-field box and allowed to explore for 15 min. The center area is 20 cm × 20 cm square in the center of the arena. The distance traveled is the total travel distance by each mouse in the arena, which was recorded and analyzed by camera and EthoVision software (Noldus Wageningen). Center frequency means the number of center entries of each mouse for 15 min.
Dowel test
The 1-m-long dowel (diameter of 9 mm) was placed parallel to the ground at a height of 30 cm. Mice were individually placed on dowel and recorded the time on the dowel. The longest experimental record is 120 s.10
Beam walking test
The 1-m-long dowel (diameter of 16 mm) was placed parallel to the ground at a height of 30 cm. There was a safe platform at one end of the dowel. After 2 days of training, latency was quantified by measuring the time it took for the mouse to walk over the beam, as well as the number of footsteps was counted. The longest experimental record is 60 s.
Accelerating rotarod test
The test was performed on a rotarod system with the rod rotating from 5 rpm to 30 rpm over 5 min. Mice were trained for 2 days. Two trials were performed for each day with more than 1 h inter-trial interval. On the test day, mice were given two trials and recorded the time of remaining on the rod until falling off or making two consecutive turns. The average time of the two trials was calculated.
Marble burying test
Each mouse is placed in a mouse cage containing 20 small (1.5 cm) clean black marbles on top of 4 inches of corn cob bedding, forming 4 rows of 5 columns. After a period of 30 min of exploration, the mouse is removed from the cage and the number of marbles that are buried at least 50% is recorded.
Fear conditioning
On the training day, after 2 min in the conditioning chamber, mice received two pairings of a tone (2,800 Hz, 85 db, 30 s) with a co-terminating foot-shock (0.7 mA, 2 s), after which they remained in the chamber for 1 additional min and then were returned to their home cages. At 24 h after training, mice were returned to the conditioning chamber for 5 min and tested for freezing in response to the training context. The percent of time spent freezing was taken as an index of contextual learning and memory. At 48 h after training, mice were returned to the conditioning chamber with new walls, mice received a tone (2,800 Hz, 85 db, 30 s) without a foot shock; the percent of time spent freezing was taken as an index of cue learning and memory.
Statistical analyses
All results from analyses are presented as the mean ± standard error of the mean, except for behavioral tests. Two-tailed, unpaired Student’s t-test was used for comparisons between two normally distributed groups, and differences were considered significant when the p values was less than 0.05. For data consisting of more than two groups except for behavioral tests, varying in a single factor, one-way ANOVA and Tukey’s multiple comparison tests were used and differences were considered significant when the p value was less than 0.05. Behavioral tests are presented as boxplot whiskers with the middle line showing the median. For behavioral data consisting of more than two groups, varying in a single factor, one-way ANOVA and Holm-Sidak comparison test were used and differences were considered significant when the p values was less than 0.05.
Acknowledgments
We thank the FACS facility, H. Wu, and L. Quan at ION.
This work was supported by the following funding: Lingang Lab (LG202106-01-02); Lingang Lab (LG202106-01-04); National Natural Science Foundation of China (82101552); STI2030-Major Projects (2021ZD0200900); Strategic Priority Research Program of Chinese Academy of Sciences (XDB32060000); National Natural Science Foundation of China (31925016,82021001); Basic Frontier Scientific Research Program of Chinese Academy of Sciences From 0 to 1 original innovation project (ZDBS-LY-SM001); Shanghai Municipal Science and Technology Major Project (2018SHZDZX05); Project of Shanghai Municipal Science and Technology Commission (20MC1920400); Technology Major Project (Grant No. 2018SHZDZX05); and Shanghai Science and Technology Development Funds (21QA1409900).
Author contributions
Conceiving the project: C.Z. and H.Y.; designing experiments: J.L., Z.S., and Y.L.; conducting experiments: J.L., Z.S., Y.L., Z.Y., X.L., J.T., R.L., G.G., and F.Y.; bioinformatic analysis: Y.L.; funding acquisition: H.Y. and C.Z.; writing – original draft: J.L. and Z.S.; writing – review and editing: J.L., Z.S., Y.L., H.Y., C.Z., and Z.X.
Declaration of interests
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2023.02.015.
Contributor Information
Zhi-Qi Xiong, Email: xiongzhiqi@ion.ac.cn.
Changyang Zhou, Email: zhouchangyang@ion.ac.cn.
Hui Yang, Email: huiyang@ion.ac.cn.
Supplemental information
Data and code availability
All RNA-seq data have been deposited in the NCBI SRA under project accession number PRJNA842160. Source data are provided in this paper.
References
- 1.Buiting K., Williams C., Horsthemke B. Angelman syndrome - insights into a rare neurogenetic disorder. Nat. Rev. Neurol. 2016;12:584–593. doi: 10.1038/nrneurol.2016.133. [DOI] [PubMed] [Google Scholar]
- 2.Reiter L.T., Seagroves T.N., Bowers M., Bier E. Expression of the Rho-GEF Pbl/ECT2 is regulated by the UBE3A E3 ubiquitin ligase. Hum. Mol. Genet. 2006;15:2825–2835. doi: 10.1093/hmg/ddl225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huibregtse J.M., Scheffner M., Howley P.M. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mishra A., Godavarthi S.K., Jana N.R. UBE3A/E6-AP regulates cell proliferation by promoting proteasomal degradation of p27. Neurobiol. Dis. 2009;36:26–34. doi: 10.1016/j.nbd.2009.06.010. [DOI] [PubMed] [Google Scholar]
- 5.Greer P.L., Hanayama R., Bloodgood B.L., Mardinly A.R., Lipton D.M., Flavell S.W., Kim T.K., Griffith E.C., Waldon Z., Maehr R., et al. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140:704–716. doi: 10.1016/j.cell.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Margolis S.S., Salogiannis J., Lipton D.M., Mandel-Brehm C., Wills Z.P., Mardinly A.R., Hu L., Greer P.L., Bikoff J.B., Ho H.Y.H., et al. EphB-mediated degradation of the RhoA GEF Ephexin5 relieves a developmental brake on excitatory synapse formation. Cell. 2010;143:442–455. doi: 10.1016/j.cell.2010.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rougeulle C., Cardoso C., Fontés M., Colleaux L., Lalande M. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat. Genet. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- 8.Runte M., Hüttenhofer A., Gross S., Kiefmann M., Horsthemke B., Buiting K. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum. Mol. Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- 9.Meng L., Person R.E., Beaudet A.L. Ube3a-ATS is an atypical RNA polymerase II transcript that represses the paternal expression of Ube3a. Hum. Mol. Genet. 2012;21:3001–3012. doi: 10.1093/hmg/dds130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meng L., Person R.E., Huang W., Zhu P.J., Costa-Mattioli M., Beaudet A.L. Truncation of Ube3a-ATS unsilences paternal Ube3a and ameliorates behavioral defects in the Angelman syndrome mouse model. PLoS Genet. 2013;9:e1004039. doi: 10.1371/journal.pgen.1004039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meng L., Ward A.J., Chun S., Bennett C.F., Beaudet A.L., Rigo F. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–412. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dindot S.V., Christian S., Murphy W.J., Berent A., Panagoulias J., Schlafer A., Ballard J., Radeva K., Robinson R., Myers L., et al. Development of an ASO therapy for Angelman syndrome by targeting an evolutionarily conserved region at the start of the UBE3A-AS transcript. BioRvix. 2021 doi: 10.1101/2021.07.27.453820. Preprint at. [DOI] [PubMed] [Google Scholar]
- 13.Wolter J.M., Mao H., Fragola G., Simon J.M., Krantz J.L., Bazick H.O., Oztemiz B., Stein J.L., Zylka M.J. Cas9 gene therapy for Angelman syndrome traps Ube3a-ATS long non-coding RNA. Nature. 2020;587:281–284. doi: 10.1038/s41586-020-2835-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmid R.S., Deng X., Panikker P., Msackyi M., Breton C., Wilson J.M. CRISPR/Cas9 directed to the Ube3a antisense transcript improves Angelman syndrome phenotype in mice. J. Clin. Invest. 2021;131:e142574. doi: 10.1172/JCI142574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosicki M., Tomberg K., Bradley A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 2018;36:765–771. doi: 10.1038/nbt.4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abil Z., Zhao H. Engineering reprogrammable RNA-binding proteins for study and manipulation of the transcriptome. Mol. Biosyst. 2015;11:2658–2665. doi: 10.1039/c5mb00289c. [DOI] [PubMed] [Google Scholar]
- 17.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B.T., Shmakov S., Makarova K.S., Semenova E., Minakhin L., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.East-Seletsky A., O'Connell M.R., Knight S.C., Burstein D., Cate J.H.D., Tjian R., Doudna J.A. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538:270–273. doi: 10.1038/nature19802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yan W.X., Chong S., Zhang H., Makarova K.S., Koonin E.V., Cheng D.R., Scott D.A. Cas13d is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol. Cell. 2018;70:327–339.e5. doi: 10.1016/j.molcel.2018.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu C., Zhou Y., Xiao Q., He B., Geng G., Wang Z., Cao B., Dong X., Bai W., Wang Y., et al. Programmable RNA editing with compact CRISPR-Cas13 systems from uncultivated microbes. Nat. Methods. 2021;18:499–506. doi: 10.1038/s41592-021-01124-4. [DOI] [PubMed] [Google Scholar]
- 21.Tong H., Huang J., Xiao Q., He B., Dong X., Liu Y., Yang X., Han D., Wang Z., Wang X., et al. High-fidelity Cas13 variants for targeted RNA degradation with minimal collateral effects. Nat. Biotechnol. 2023;41:108–119. doi: 10.1038/s41587-022-01419-7. [DOI] [PubMed] [Google Scholar]
- 22.Silva-Santos S., van Woerden G.M., Bruinsma C.F., Mientjes E., Jolfaei M.A., Distel B., Kushner S.A., Elgersma Y. Ube3a reinstatement identifies distinct developmental windows in a murine Angelman syndrome model. J. Clin. Invest. 2015;125:2069–2076. doi: 10.1172/JCI80554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan K.Y., Jang M.J., Yoo B.B., Greenbaum A., Ravi N., Wu W.L., Sánchez-Guardado L., Lois C., Mazmanian S.K., Deverman B.E., Gradinaru V. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 2017;20:1172–1179. doi: 10.1038/nn.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnstone K.A., DuBose A.J., Futtner C.R., Elmore M.D., Brannan C.I., Resnick J.L. A human imprinting centre demonstrates conserved acquisition but diverged maintenance of imprinting in a mouse model for Angelman syndrome imprinting defects. Hum. Mol. Genet. 2006;15:393–404. doi: 10.1093/hmg/ddi456. [DOI] [PubMed] [Google Scholar]
- 25.Cattanach B.M., Barr J.A., Beechey C.V., Martin J., Noebels J., Jones J. A candidate model for Angelman syndrome in the mouse. Mamm. Genome. 1997;8:472–478. doi: 10.1007/s003359900479. [DOI] [PubMed] [Google Scholar]
- 26.Huang H.S., Burns A.J., Nonneman R.J., Baker L.K., Riddick N.V., Nikolova V.D., Riday T.T., Yashiro K., Philpot B.D., Moy S.S. Behavioral deficits in an Angelman syndrome model: effects of genetic background and age. Behav. Brain Res. 2013;243:79–90. doi: 10.1016/j.bbr.2012.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allensworth M., Saha A., Reiter L.T., Heck D.H. Normal social seeking behavior, hypoactivity and reduced exploratory range in a mouse model of Angelman syndrome. BMC Genet. 2011;12:7. doi: 10.1186/1471-2156-12-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sonzogni M., Wallaard I., Santos S.S., Kingma J., du Mee D., van Woerden G.M., Elgersma Y. A behavioral test battery for mouse models of Angelman syndrome: a powerful tool for testing drugs and novel Ube3a mutants. Mol. Autism. 2018;9:47. doi: 10.1186/s13229-018-0231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao Z., Anselmo A.C., Mitragotri S. Viral vector-based gene therapies in the clinic. Bioeng. Transl. Med. 2022;7:e10258. doi: 10.1002/btm2.10258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodrigues G.A., Shalaev E., Karami T.K., Cunningham J., Slater N.K.H., Rivers H.M. Pharmaceutical development of AAV-based gene therapy products for the eye. Pharm. Res. 2018;36:29. doi: 10.1007/s11095-018-2554-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Serajian S., Ahmadpour E., Oliveira S.M.R., Pereira M.d.L., Heidarzadeh S. CRISPR-cas Technology: emerging applications in clinical microbiology and infectious diseases. Pharmaceuticals. 2021;14:1171. doi: 10.3390/ph14111171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yin J., Lu R., Xin C., Wang Y., Ling X., Li D., Zhang W., Liu M., Xie W., Kong L., et al. Cas9 exo-endonuclease eliminates chromosomal translocations during genome editing. Nat. Commun. 2022;13:1204. doi: 10.1038/s41467-022-28900-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adikusuma F., Piltz S., Corbett M.A., Turvey M., McColl S.R., Helbig K.J., Beard M.R., Hughes J., Pomerantz R.T., Thomas P.Q. Large deletions induced by Cas9 cleavage. Nature. 2018;560:E8–E9. doi: 10.1038/s41586-018-0380-z. [DOI] [PubMed] [Google Scholar]
- 34.Law C.W., Chen Y., Shi W., Smyth G.K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu X., Wang J., Yao X., Xiao Q., Xue Y., Wang S., Shi L., Shu Y., Li H., Yang H. Screened AAV variants permit efficient transduction access to supporting cells and hair cells. Cell Discov. 2019;5:49. doi: 10.1038/s41421-019-0115-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiang Y.H., Armstrong D., Albrecht U., Atkins C.M., Noebels J.L., Eichele G., Sweatt J.D., Beaudet A.L. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998;21:799–811. doi: 10.1016/s0896-6273(00)80596-6. [DOI] [PubMed] [Google Scholar]
- 37.Huang H.S., Allen J.A., Mabb A.M., King I.F., Miriyala J., Taylor-Blake B., Sciaky N., Dutton J.W., Jr., Lee H.M., Chen X., et al. Topoisomerase inhibitors unsilence the dormant allele of Ube3a in neurons. Nature. 2011;481:185–189. doi: 10.1038/nature10726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu X., Malenka R.C. Beta-catenin is critical for dendritic morphogenesis. Nat. Neurosci. 2003;6:1169–1177. doi: 10.1038/nn1132. [DOI] [PubMed] [Google Scholar]
- 39.Glascock J.J., Osman E.Y., Coady T.H., Rose F.F., Shababi M., Lorson C.L. Delivery of therapeutic agents through intracerebroventricular (ICV) and intravenous (IV) injection in mice. J. Vis. Exp. 2011:2968. doi: 10.3791/2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim J.Y., Grunke S.D., Levites Y., Golde T.E., Jankowsky J.L. Intracerebroventricular viral injection of the neonatal mouse brain for persistent and widespread neuronal transduction. J. Vis. Exp. 2014:51863. doi: 10.3791/51863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq data have been deposited in the NCBI SRA under project accession number PRJNA842160. Source data are provided in this paper.