SUMMARY
The ability of stem cells to build and replenish tissues depends on support from their niche. While niche architecture varies across organs, its functional importance is unclear. During hair follicle growth, multipotent epithelial progenitors build hair via crosstalk with their remodeling fibroblast niche, the dermal papilla, providing a powerful model to functionally interrogate niche architecture. Through mouse intravital imaging, we show that dermal papilla fibroblasts remodel individually and collectively to form a morphologically polarized, structurally robust niche. Asymmetric TGFβ signaling precedes morphological niche polarity, and loss of TGFβ signaling in dermal papilla fibroblasts leads them to progressively lose their stereotypic architecture, instead surrounding the epithelium. The reorganized niche induces redistribution of multipotent progenitors, but nevertheless supports their proliferation and differentiation. However, the differentiated lineages and hairs produced by progenitors are shorter. Overall, our results reveal that niche architecture optimizes organ efficiency, but is not absolutely essential for organ function.
Graphical Abstract
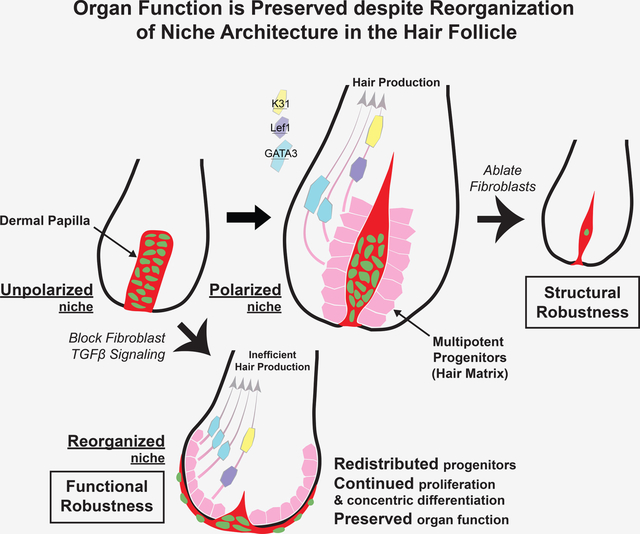
eTOC Blurb:
Wei et al. combine 3D analyses, a signaling reporter, genetic manipulations, and intravital imaging to resolve the organization and function of the hair follicle niche architecture. They show that the hair follicle niche acquires structural and functional robustness to continue supporting progenitors even when its architecture is reorganized.
INTRODUCTION
Interaction between stem cells and their niche lays the foundation for organ architecture and function.1,2 The niche creates a microenvironment with the correct signaling and physical characteristics to support stem cell survival and production of differentiated daughter cells.3,4 Niches across mammalian tissues take a variety of architecture types, including ensheathing stem cells (intestine, lung, mammary gland, etc) and enclosed by stem cells (hair follicle, tooth, tongue, etc).5–9 This implies that niche architecture is important for stem cell function, but few tools exist to test the function of niche architecture in vivo.
The hair follicle is a useful model for studying the impact of organ architecture on function because hair follicles recurrently grow and regress in each hair cycle throughout adulthood.10 This cycling requires concurrent rearrangement of hair follicle stem cells and associated niche fibroblasts in the dermal papilla and dermal sheath (Fig.S1A).11–14 Dermal papilla fibroblasts form a single cluster at the bottom of the hair follicle and remodel dramatically during hair follicle growth.15 First, the dermal papilla is enclosed by the proliferating progeny of hair stem cells, a pool of multipotent progenitors (also called hair matrix), then significantly elongates with negligible fibroblast proliferation while secreting growth factors to instruct the proliferation and differentiation of multipotent progenitors.12,16 Consequently, seven concentrically organized epithelial lineages are generated, fueling hair production (Fig.1A). Thus, dermal papilla remodeling during hair follicle regeneration provides an outstanding model to study how different niche architectures impact their crosstalk with stem cells and overall organ function.
Figure 1. Niche cells undergo collective polarization to build a morphologically polarized architecture.
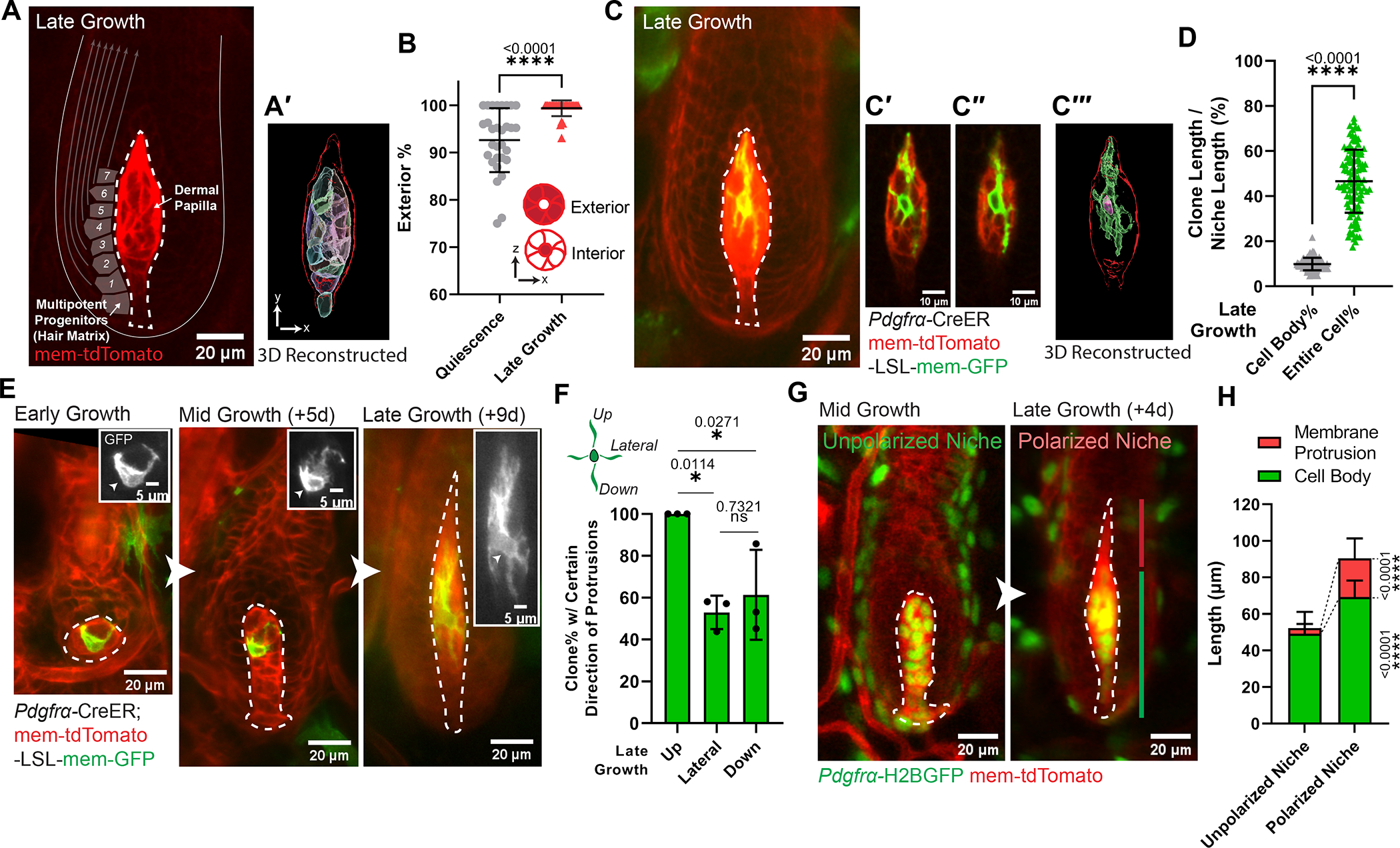
A. Intravitally imaged dermal papilla (DP, outlined) at the hair follicle late-growth stage (n=3 mice). DP fibroblasts are labeled with membrane-tdTomato. Gray squares illustrate adjacent multipotent progenitors, with arrows indicating their differentiation routes. (A′) 3D reconstructed DP (red outlined) based on its membrane-tdTomato signal. Fibroblast cell bodies are rendered into surfaces with different colors.
B. Percentage of fibroblasts whose cell bodies are located at DP exterior. Exterior-localized: any fibroblasts whose cell body directly contacts DP edge; interior-localized: a fibroblast cell body surrounded by other fibroblasts’ cell bodies. n=30 quiescent and 30 late-growth HFs from 3 mice.
C. Intravitally imaged single fibroblast at late-growth stage (n=3 mice). (C′-C″) Long membrane protrusions extend through other fibroblasts. (C‴) 3D reconstructed single fibroblast based on its membrane-GFP signal. Within the red-outlined DP, this fibroblast’s cell body is rendered into a pink surface and its entire membrane in a green surface.
D. Coverage percentage of DP fibroblast clones at late growth, as the length of fibroblast cell body or entire membrane relative to the entire DP length. n=103 fibroblast clones from 3 mice.
E. Longitudinal imaging of a representative DP fibroblast from early to late-growth (n=3 mice). Insets highlight membrane-GFP (gray) and fibroblast cell body (arrowheads).
F. Percentage of DP fibroblasts harboring certain direction of membrane protrusions at late growth. n=77 fibroblast clones from 3 mice.
G. Longitudinally imaging of the same DP from mid- to late-growth. The membrane protrusion compartment (red line) on top of the cell body compartment (green line) organizes a morphologically polarized niche at late-growth (Anagen IIIc-VI), in contrast to an unpolarized niche at mid-growth (late Anagen II-IIIb).
H. Length of DP compartments at mid-growth (unpolarized niche) and late-growth stage (polarized niche). Lengths of membrane protrusion and cell body compartments are significantly different between the polarized and unpolarized niche stages. n=117 unpolarized DPs and 102 polarized DPs from 3 mice.
DPs are dash-lined. In Fig.1C, E, a single fibroblast is labeled by mosaic recombined membrane-GFP (mTmG) under the fibroblast driven CreER (Pdgfrα-CreER). In Fig.1G, fibroblast nuclei are labeled in green (Pdgfrα-H2BGFP) and cell membranes are in red (membrane-tdTomato). All data are presented as mean ± S.D. Unpaired two-tailed t-test is used in Fig.1B, D, H. Tukey’s multiple comparisons test is used in Fig.1F. See also Figure S1, Data S1 (Movie).
As multipotent progenitors reposition along the dermal papilla, they flexibly adopt different fates depending on the timing and location of their disassociation from the niche.17–20 Signaling crosstalk between the dermal papilla and multipotent progenitors has been relatively well characterized,21–34 but the direct impacts of the dermal papilla’s three-dimensional (3D) organization on hair follicle growth is not well understood. To address this question, we leveraged our ability to manipulate and longitudinally track the same hair follicle niche fibroblasts in live mice (Fig.S1A′)35,36, and uncovered how the structural features of a remodeling niche are formed and optimize stem cell activities and organ function.
RESULTS
Niche cells undergo collective polarization to confer a structurally robust architecture
The dermal papilla is a small cluster when it supports epithelial proliferation at early growth stages (Anagen I to early Anagen II), and elongates as it begins supporting epithelial differentiation at mid-growth (late Anagen II to IIIb) and late growth stages (Anagen IIIc to VI) (Fig.S1A–A′).10 To understand whether fibroblasts also change their organization within the dermal papilla during remodeling, we labeled them with membrane-tdTomato and 3D reconstructed their cell bodies (Fig.1A–A′). At the quiescent stage, we found that the dermal papilla contains a population of interior-localized fibroblasts surrounded by exterior-localized fibroblasts (Fig.1B). In contrast, at the late growth stage, there is a significantly increased percentage of fibroblasts positioning their cell bodies along the exterior of the dermal papilla, directly interfacing with adjacent multipotent progenitors (Fig.1A–B; Data S1–Movie). This suggests that the dermal papilla at late growth is organized to optimally communicate with multipotent progenitors.
To understand how fibroblast morphology contributes to these organizational changes within the dermal papilla, we resolved individual fibroblasts by mosaic labeling with membrane-GFP (Pdgfrα-CreER; mTmG). Surprisingly, we found that at the late growth stage, individual dermal papilla fibroblasts extend long membrane protrusions across the entire niche (Fig.1C–D; Data S1–Movie), allowing them to contact distant fibroblasts and multipotent progenitors. This contrasts with the much shorter membrane protrusions on dermal papilla fibroblasts at mid-growth (Fig.1E; Fig.S1B). Interestingly, at the late growth stage, only a subset have lateral or downward-oriented membrane protrusions, but all fibroblasts have upward-oriented protrusions (Fig.1F; Fig.S1B′). By differentially labeling all fibroblast nuclei and cell membranes (Pdgfrα-H2BGFP; mem-tdTomato), we found that dermal papilla fibroblast membrane protrusions are close to their cell bodies at mid-growth, but at the late growth stage, the dermal papilla develops a distinct membrane protrusion compartment over its cell body compartment (Fig.1G–H). Altogether, this indicates that dermal papilla fibroblasts collectively form a morphologically polarized niche architecture at the late growth stage by extending membrane protrusions and spatially compartmentalizing them from their cell bodies. We hereafter call dermal papilla at the mid-growth stages (late Anagen II to IIIb) “unpolarized niche,” and at the late growth stages (Anagen IIIc to VI) “polarized niche” to refer to this tissue-level architectural change.
To better understand the structural properties of dermal papilla architecture throughout its polarization process, we asked how different niche architectures respond to fibroblast removal. We genetically induced fibroblast apoptosis by expressing diphtheria toxin (Pdgfrα-H2BGFP; Pdgfrα-CreER; mTmG; LSL-DTA) at the unpolarized (mid-growth) and polarized (late-growth) niche stages, and revisited the same dermal papillae over time. The loss of Pdgfrα-H2BGFP positive nuclei confirmed fibroblast death across different regions and showed that niche fibroblasts died at similar kinetics for both stages (Fig.2A–B; Fig.S1C–G). Interestingly, the polarized and unpolarized dermal papillae that remained enclosed but lost more than half of their constituent fibroblasts still extended membranes to occupy a similar overall dermal papilla length (Fig.2A, 2C), indicating a structural robustness that maintains an enclosed niche architecture even under loss of many constituent cells. However, unpolarized niches lost their enclosed architecture much earlier and more frequently, in contrast to the majority of polarized niches remaining enclosed by the epithelium even 6–7 days after fibroblast ablation (Fig.2D). This data suggests that a morphologically polarized niche becomes more structurally robust to maintain its enclosed architecture (Fig.2E).
Figure 2. Polarized niche confers a structurally robust architecture despite the loss of its constituent cells.
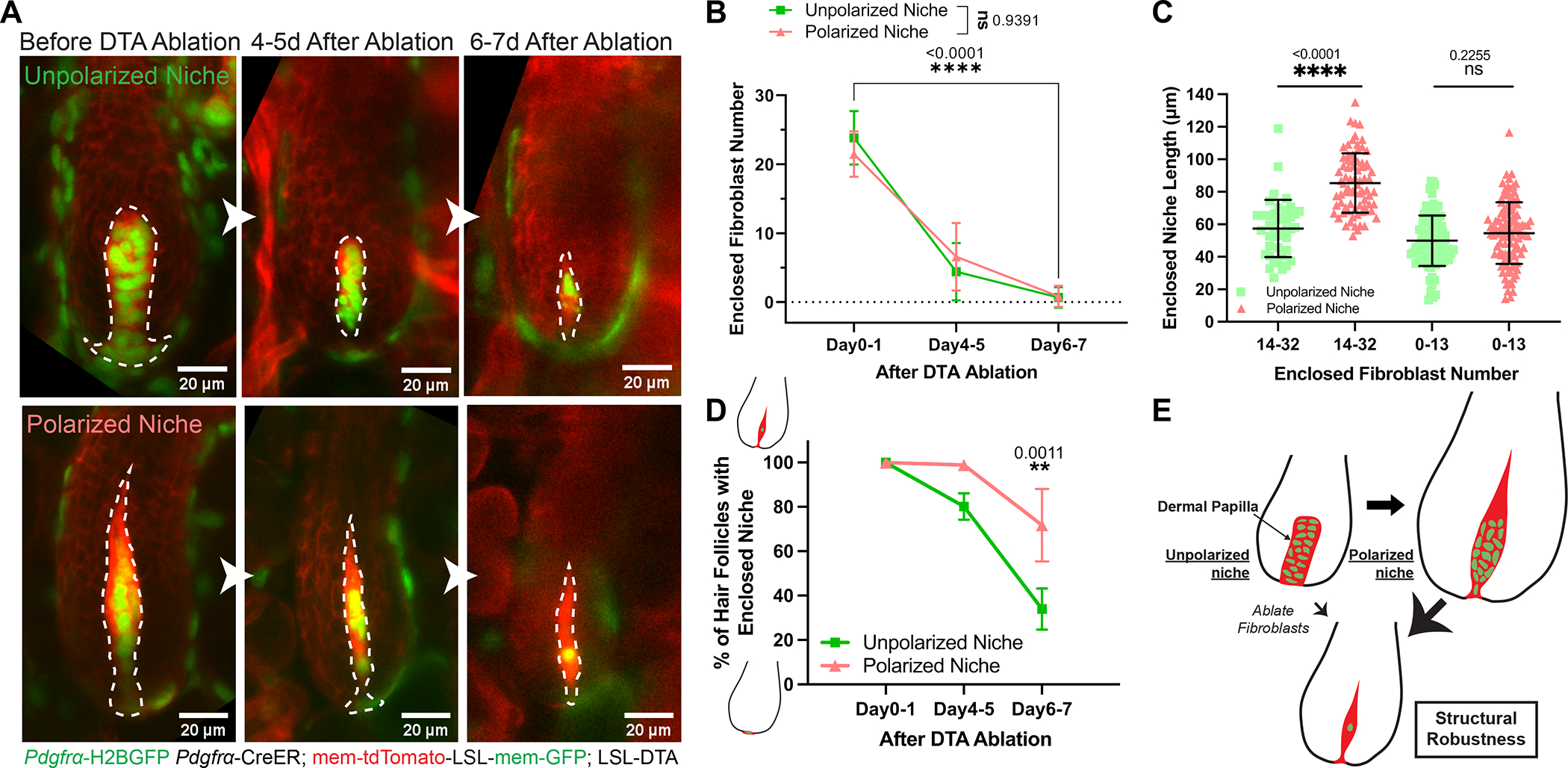
A. The same unpolarized and polarized dermal papillae (DPs, dash-lined) longitudinally imaged before and after diphtheria toxin (DTA) induced fibroblast ablation. Ablation is achieved by tamoxifen activated Cre in fibroblasts (Pdgfrα-CreER) and LSL-DTA. Fibroblast nuclei are labeled in green (Pdgfrα-H2BGFP) and cell membranes are in red (membrane-tdTomato).
B. The number of fibroblasts that remain enclosed within the hair follicle (HF) epithelium after DTA-induced fibroblast ablation. Fibroblast number is counted based on green fibroblast nuclear signal labeled by Pdgfrα-H2BGFP. For unpolarized DPs at mid-growth, n=41 (Day0–1), 53 (Day4–5), and 60 (Day6–7) from 3 mice; for polarized DPs at late-growth, n=72 (Day0–1), 78 (Day4–5), and 71 (Day6–7) from 3 mice.
C. Length of entire DPs that remain enclosed after ablation. Based on the reported critical threshold of DP fibroblast number,11 we classified DP as either with abundant (14 to 32) or few (0 to 13) enclosed fibroblasts. For unpolarized DPs, n=42 HFs (14–32 fibroblasts) and 99 HFs (0–13 fibroblasts) from 3 mice; For polarized DPs, n=77 HFs (14–32 fibroblasts) and 110 HFs (0–13 fibroblasts) from 3 mice.
D. Percentage of HFs that remain enclosed DPs after fibroblast ablation. For HFs with unpolarized DPs, n=3 mice (68 HFs at Day0–1, 68 HFs at Day4–5, and 66 HFs at Day6–7); for HFs with polarized DPs, n=3 mice (102 HFs at Day0–1, 108 HFs at Day4–5, and 89 HFs at Day6–7).
E. Schematic of forming a polarized DP niche architecture that is structurally more robust even after fibroblast ablation. Green indicates DP fibroblast nuclei and red indicates their membranes.
All data are presented as mean ± S.D. Mixed-effects analysis is used in Fig.2B. Tukey’s multiple comparisons test is used in Fig.2C, D. See also Figure S1.
Niche architecture and location are actively maintained through fibroblast TGFβ signaling
The differential effects of fibroblast ablation on unpolarized and polarized niches suggest a link between niche architectural polarity and maintenance. The expression of TGFβ receptor 1 (TGFβRI) begins in dermal papilla fibroblasts right before architectural polarization,37 making it a promising candidate mediator of dermal papilla polarity and/or structural robustness. To investigate the role of TGFβ signaling in the dermal papilla, we first examined when and where TGFβ signaling is activated in dermal papilla fibroblasts. We built a TGFβ reporter mouse line by fluorescently tagging endogenous Smad4 (mNeonGreen-Smad4, Fig.S2A–B), which accumulates in the nucleus upon TGFβ signaling.38 Mouse embryonic fibroblasts from our reporter mouse showed nuclear localization of mNeonGreen-Smad4 in response to TGFβ addition (Fig.S2C), confirming that this mouse line reflects TGFβ signaling. Excitingly, tracking signaling activity in vivo revealed that nuclear Smad4 is first detectable in upper dermal papilla fibroblasts in the unpolarized niche, and this asymmetric signaling activation is sustained as the dermal papilla becomes morphologically polarized (Fig.3A).
Figure 3. Niche architecture and location are actively maintained through fibroblast TGFβ signaling.
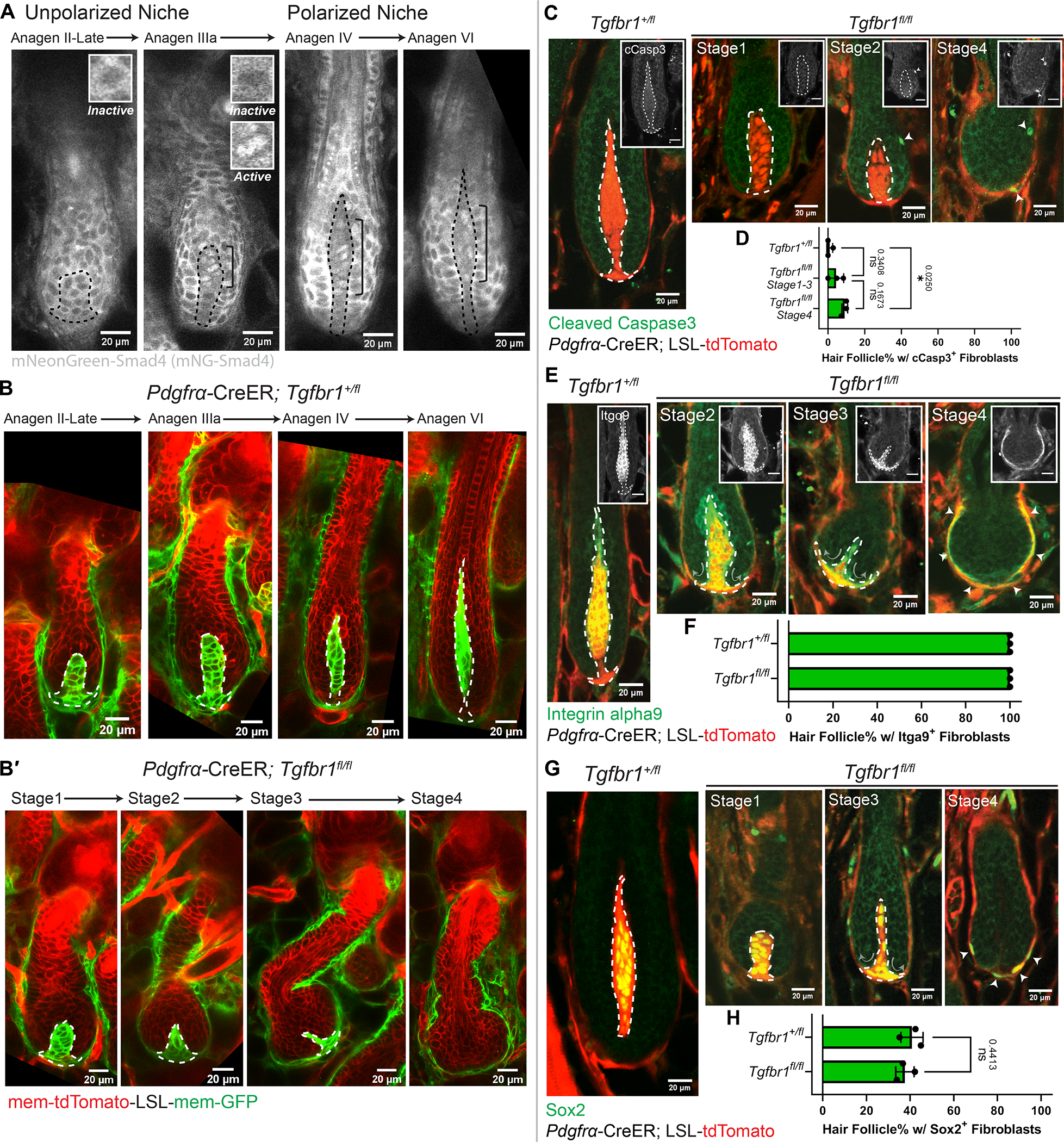
A. Intravital imaging of TGFβ signaling reporter, mNeonGreen-Smad4 (gray), in hair follicles (HFs) at different stages (n=3 mice). Nuclear-localized Smad4 is present in dermal papilla (DP) fibroblasts within the bracket. Insets show inactive (cytoplasmic) or active (nuclear) Smad4 signals.
B-B′. Intravitally imaged Tgfbr1+/fl and Tgfbr1fl/fl HFs at mid and late-growth stages (B, Tgfbr1+/fl late Anagen II-VI; B′, Tgfbr1fl/fl Stage1–4) after CreER induction at the quiescent stage (n=3 mice). Fibroblast membranes are in green membrane-GFP and other membranes are in red membrane-tdTomato.
C. Immunostaining for cleaved Caspase3 (cCasp3, green) in thick back skin sections to detect apoptosis at different stages. Arrowheads indicate cCasp3 signal. Insets show cCasp3 in gray (Scale bar=20μm).
D. Percentage of HFs containing cCasp3+ fibroblasts. N=3 Tgfbr1+/fl mice (280 late-growth HFs), n=3 Tgfbr1fl/fl mice (49 HFs at Stage1–3, 81 HFs at Stage4). Tukey’s multiple comparisons test.
E. Immunostaining for Integrin alpha9 (Itgα9, green) in thick back skin sections to detect DP identity at different stages. Arrowheads indicate Itgα9+ regions in Tgfbr1fl/fl Stage4. Arrows indicate the reorganizing direction of DP fibroblasts. Insets show Itgα9 in gray (Scale bar=20μm).
F. Percentage of HFs containing Itgα9+ fibroblasts. N=3 Tgfbr1+/fl mice (234 late-growth HFs), n=3 Tgfbr1fl/fl mice (144 HFs from stages shown).
G. Immunostaining for Sox2 (green) in thick back skin sections to detect DP identity at different stages. Arrowheads indicate Sox2+ regions in Tgfbr1fl/fl Stage4. Arrows indicate the reorganizing direction of DP fibroblasts.
H. Percentage of HFs containing Sox2+ fibroblasts. N=3 Tgfbr1+/fl mice (594 late-growth HFs), n=3 Tgfbr1fl/fl mice (226 HFs from stages shown). Unpaired two-tailed t-test.
DPs are dash-lined. In Fig.3C, E, G, fibroblasts are labeled in red by Pdgfrα-CreER; LSL-tdTomato. All data are presented as mean ± S.D. See also Figure S2–3, Data S2 (Movie).
The spatiotemporal coupling of signaling asymmetry and niche polarization prompted us to functionally examine the consequences of TGFβ signaling loss on dermal papilla architecture. To this end, we knocked out TGFβRI in fibroblasts from the quiescent stage via Cre induction at postnatal day 18–20 and used fluorescent reporters to identify fibroblasts that expressed Cre (Pdgfrα-CreER; Tgfbr1fl/fl combined with either membrane-GFP or cytoplasmic tdTomato reporter). Revisits of the same mice showed that Tgfbr1fl/fl mutant niches proceeded to the unpolarized stage like Tgfbr1+/fl controls (Fig.3B–B′, Tgfbr1fl/fl Stage1 and Tgfbr1+/fl late Anagen II-IIIa; Fig.S2D). However, they failed both to elongate and to polarize, and gradually lost their enclosed architecture (Fig.3B–B′, Tgfbr1fl/fl Stage2–4). Negligible fibroblast apoptosis was detected prior to loss of tdTomato+ (Tgfbr1fl/fl) fibroblast enclosure (Fig.3C–D, Tgfbr1fl/fl Stage1–3), indicating that fibroblast apoptosis is not the cause of niche architecture loss.
To better understand the effects of TGFβRI deletion prior to loss of dermal papilla architecture, we asked whether dermal papilla fibroblasts lost their identity markers. We stained for two dermal papilla identity markers: integrin alpha9 and Sox2.39,40 Surprisingly, we found that dermal papilla fibroblasts largely retained identity markers in Tgfbr1fl/fl mutants (Fig.3E–H; Fig.S3A–D). Despite slightly reduced Sox2 positive fibroblast numbers (Fig.S3B), the position of integrin alpha9 and Sox2 positive fibroblasts progressively transitioned from enclosed within the epithelium to ensheathing around the epithelium (Fig.3E–H; Fig.S3A, S3C–D; Data S2–Movie). Further molecular analyses of TGFβ signaling downstream transcription factors confirmed the effectiveness of our genetic ablation (reduced nuclear phospho-Smad2-positive fibroblasts in Tgfbr1fl/fl mutants, Fig.S2E). We also observed a loss of the normal organization of fibronectin and alpha-smooth muscle actin (α-SMA), another two known downstream targets of TGFβ signaling,41,42 in fibroblasts that ensheath the Tgfbr1fl/fl mutant hair follicle (Fig.S3E–F′). Excitingly, despite these molecular changes, this reorganized sheath-like niche was still capable of re-clustering during the regression phase, much like counterpart wild-type follicles, to sustain the subsequent hair follicle cycle, and even recapitulated its architectural reorganization (i.e. ensheathing the epithelium) in the new cycle (Fig.S2F–G). Overall, this repeated niche reorganization indicates that fibroblast TGFβ signaling is not only necessary to induce a morphologically polarized niche, but is also required to actively maintain niche’s enclosed architecture and location.
Reorganized niche induces redistribution of multipotent progenitors but largely supports their function
The reorganized niche architecture in the Tgfbr1fl/fl mutant provided us with an excellent opportunity to investigate how epithelial multipotent progenitors are impacted by niche architecture, which has been understudied compared to their signaling crosstalk. We first interrogated the proliferation status of multipotent progenitors in the Tgfbr1fl/fl mutant by staining for the proliferation marker Ki67. Despite the changed niche architecture, mutant hair follicles had an overall bulb-like structure containing a pool of proliferative epithelial cells, similar to control hair follicles (Fig.4A–B). However, only epithelial cells in the outer bulb layers were proliferative in mutant hair follicles, indicating a spatial reorganization of the bulb region (Fig.4C). Furthermore, the pattern of cells in the proliferation zone expressing Lef1, a multipotent progenitor marker,19 also redistributed to the outer epithelial layers upon niche reorganization (Fig.4D–E), further demonstrating that upon niche reorganization, multipotent progenitors remained proliferative but changed location.
Figure 4. Reorganized niche induces redistribution of multipotent progenitors but largely supports their function.
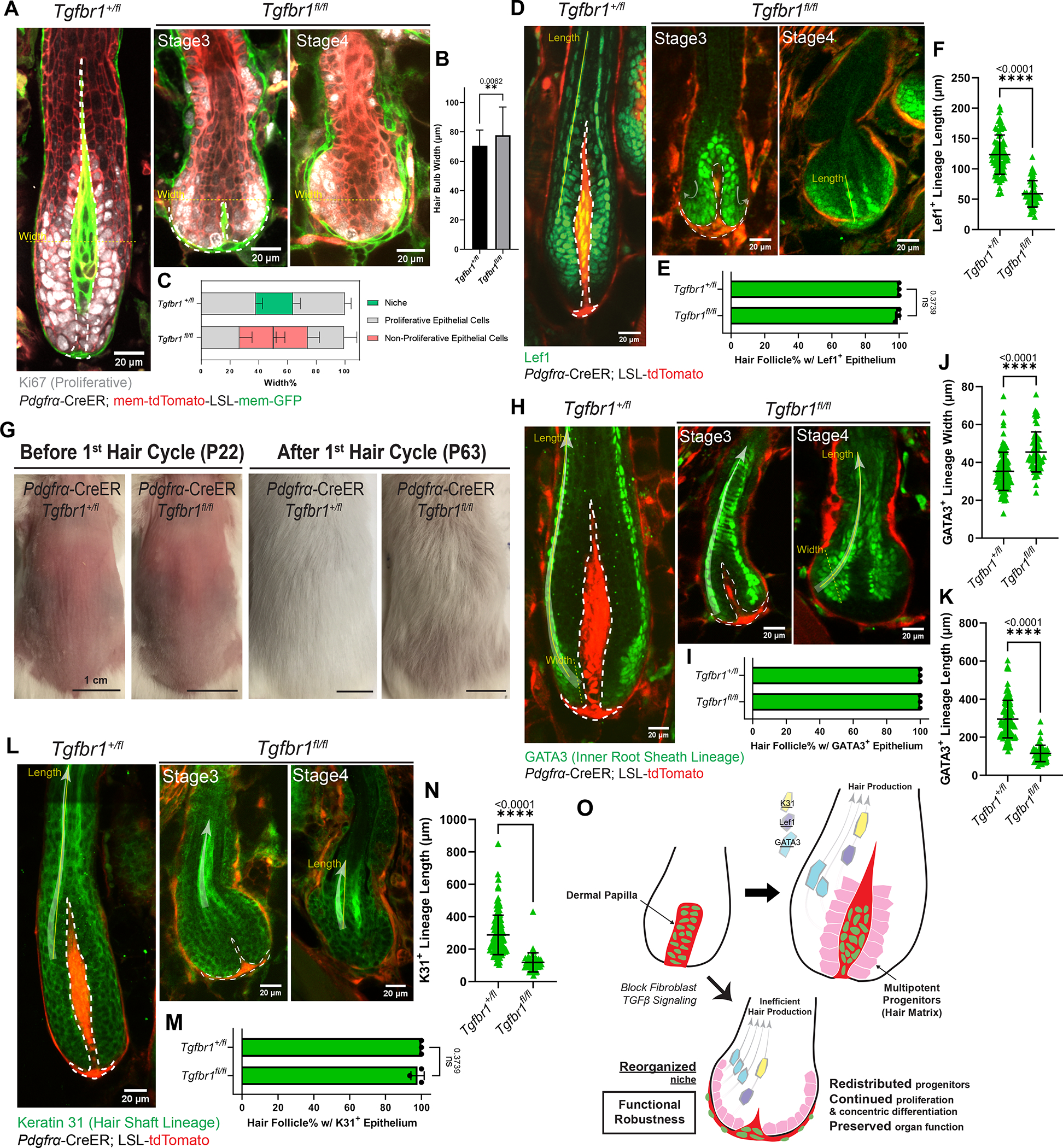
A. Immunostaining for Ki67 (gray) in ear skin whole mounts to detect proliferative epithelial cells in hair follicles (HFs) at different stages. Fibroblast membranes are in green (Pdgfrα-CreER; membrane-GFP) and other membranes are in red (membrane-tdTomato).
B. Hair bulb maximal width in Tgfbr1+/fl and Tgfbr1fl/fl at yellow dash-line locations in Fig.4A. n=75 HFs from three Tgfbr1+/fl mice, n=62 HFs from three Tgfbr1fl/fl mice.
C. Spatial distribution of cell populations (niche, proliferative epithelial cells, non-proliferative epithelial cells) in the bulb region, measured as the width percentage of each population at maximal hair bulb width (yellow dash-lines in Fig.4A). n=75 HFs from three Tgfbr1+/fl mice, n=62 HFs from three Tgfbr1fl/fl mice.
D. Immunostaining for Lef1 (green) in thick back skin sections to detect multipotent progenitors and Wnt/β-catenin signaling at different stages. The redistribution directions are marked by arrows.
E. Percentage of HFs containing Lef1+ epithelial populations in the bulb region. n=3 Tgfbr1+/fl mice (215 late-growth HFs), n=3 Tgfbr1fl/fl mice (93 HFs from stages shown).
F. Length of Lef1+ differentiating lineage along the differentiation routes (yellow solid line in Fig.4D). n=81 late-growth HFs from three Tgfbr1+/fl mice, n=49 Stage4 HFs from three Tgfbr1fl/fl mice.
G. Representative photos of mice with back skin shaved at the quiescent stage, and revisited after an entire first hair cycle. n=3 Tgfbr1+/fl and 3 Tgfbr1fl/fl mice.
H. Immunostaining for GATA3 (green) in thick back skin sections to detect the differentiation of inner root sheath lineages (Cuticle and Huxley’s layer) at different stages. The differentiation routes are marked with arrows.
I. Percentage of HFs containing GATA3+ epithelial populations in concentric organizations. n=3 Tgfbr1+/fl mice (328 late-growth HFs), n=3 Tgfbr1fl/fl mice (103 HFs from stages shown).
J. Width of GATA3+ differentiating lineage at the beginning of differentiation routes (yellow dash line in Fig.4H). n=109 late-growth HFs from three Tgfbr1+/fl mice, n=59 Stage4 HFs from four Tgfbr1fl/fl mice.
K. Length of GATA3+ differentiating lineage along the differentiation routes (yellow solid line in Fig.4H). n=112 late-growth HFs from three Tgfbr1+/fl mice, n=51 Stage4 HFs from four Tgfbr1fl/fl mice.
L. Immunostaining for Keratin 31 (K31, green) in thick back skin sections to detect the differentiation of hair shaft lineage (Cortex) at different stages. The differentiation routes are marked with arrows.
M. Percentage of HFs containing K31+ epithelial populations in concentric organizations. n=3 Tgfbr1+/fl mice (243 late-growth HFs), n=3 Tgfbr1fl/fl mice (109 HFs from stages shown).
N. Length of K31+ differentiating lineage along the differentiation routes (yellow solid line in Fig.4L). n=132 late-growth HFs from three Tgfbr1+/fl mice, n=44 Stage4 HFs from three Tgfbr1fl/fl mice.
O. Schematic illustrating that reorganized niche architecture continues supporting redistributed proliferative multipotent progenitors. However, progenitor differentiation is rerouted and generates shorter lineages for less efficient hair production.
Dermal papillae are dash-lined. In Fig.4D, H, L, fibroblasts are labeled in red by Pdgfrα-CreER; LSL-tdTomato. All data are presented as mean ± S.D. and analyzed with unpaired two-tailed t-test. See also Figure S2, S4.
We next asked whether the reorganized niche and epithelial progenitors can still support hair production. We shaved the mouse back skin at the quiescent stage (postnatal day 18–20) and tracked hair regrowth after an entire hair cycle. Interestingly, we found that hair shafts were still produced, though shorter in length, in Tgfbr1fl/fl mutants (Fig.4G; Fig.S4A–D). To understand what caused this hair shortening, we stained for multiple differentiation lineage markers, including inner root sheath (GATA3) and hair shaft (Keratin 31).30 Notably, both lineages were still present and concentrically organized, like in Tgfbr1+/fl controls (Fig.4H–I; Fig.4L–M). However, GATA3+ epithelial cells in the Tgfbr1fl/fl mutants expanded their differentiation pool (increased lineage width, Fig.4J) and adopted a concave rather than a convex path (Fig.4H, arrow). This indicates that multipotent progenitors spatially reroute their differentiation path upon niche reorganization. Furthermore, both GATA3+ and Keratin 31+ lineages showed precocious loss of their differentiation markers along the differentiation path (indicated by decreased lineage length, Fig.4H, 4L, Tgfbr1fl/fl Stage4; Fig.4K, 4N). Staining for other lineage markers (Trichohyalin, Keratin 32, Fig.S4I–J), differential interference contrast (Fig.S4B) and transmission electron microscopy (Fig.S4E–H″) further supported this observation that Tgfbr1fl/fl mutant hair follicles maintained their concentric differentiation organization but their progenitors produced shorter differentiated lineages relative to controls. Similarly, Lef1 and phospho-Smad2 activities in the Tgfbr1fl/fl mutant differentiating progenitors were also maintained in concentric organization and got precociously lost (Fig.4D, 4F; Fig.S2E, Tgfbr1fl/fl Stage4). Given that these markers reflect Wnt and TGFβ signaling activation above the proliferation zone, this further suggests that while defective, retained signaling may maintain a basic level of hair progenitor functions. These results together indicate that multipotent progenitors under the instruction of a reorganized niche can still largely proliferate and differentiate, but generate shorter differentiated lineages and produce hairs less efficiently.
DISCUSSION
Overall, we elucidated that that dermal papilla fibroblasts remodel collectively to form a morphologically polarized and structurally robust niche architecture, and that this architecture dictates multipotent progenitor location and differentiation for optimal hair production, which is possible but inefficient in a sheath-like niche architecture (Fig.4O).
Our 3D reconstruction and longitudinal analyses reveal that dermal papilla fibroblasts acquire long membrane protrusions by the late growth (polarized) stage (Fig.1), allowing them to contact several adjacent and distant hair progenitors. This raises the question of whether and how dermal papilla fibroblasts could instruct fate specifications of positionally distant hair progenitors. One possibility is that these extended fibroblast membrane protrusions could generate directional transport of signaling factors to facilitate different progenitor fate specifications, as in fly wing imaginal disc and germline stem cells.43,44 Alternatively, the potential of fate specifications could differ between large (cell body) and smaller (membrane protrusion) fibroblast-progenitor contact areas, similar to fate specification in ascidian embryogenesis.45 The latter model could explain why cell bodies and cell membranes generally occupy different compartments within the polarized dermal papilla (Fig.1G). Future investigation into the formation and function of niche polarity will illuminate how niche cells remodel different compartments to perform diverse functions.
We also found that a polarized niche is structurally robust to maintain its enclosed architecture in the face of extreme damage, such as loss of many constituent cells (Fig.2). This structural robustness may be achieved by elongated membrane protrusions, which could generate deeper and stronger adhesions within the epithelium. Alternatively, a polarized niche may build a more rigid cytoskeleton and extracellular matrix to resist remodeling.46–48 The dermal sheath may also contribute to the structural robustness; indeed, hair follicles with unpolarized niches (which are less structurally robust by our measurement) lost more dermal sheath cells upon fibroblast ablation compared to those with polarized niches (Fig.S1D–G). Investigating how fibroblast ablation affects niche composition and the function of different fibroblast subpopulations would better pinpoint the mechanism of structural robustness.
Here, we discovered a role for TGFβ signaling in hair follicle fibroblasts to actively maintain niche enclosure. Previous manipulations of dermal papilla signaling status and identity signatures altered multipotent progenitors’ activities, but did not noticeably affect the niche architecture.32,49–51 In contrast, we show that genetically deleting fibroblast Tgfbr1 affected multipotent progenitor activity by reorganizing niche fibroblasts which continue to express most niche markers (Fig.3). However, we noted that the number of Sox2 and Lef1 positive cells is decreased, but they are still present in the reorganized dermal papilla (Fig.S3A–B; Fig.4D), similar to gene changes in 2D cultured human dermal papillae,52 implying that niche identity is at least partially separable from its architecture. We also detected cytoskeleton and extracellular matrix changes in the dermal sheath after niche reorganization (Fig.S3E–F′), suggesting that dermal sheath TGFβ signaling may also contribute to niche remodeling. Future studies across fibroblast subpopulations will disentangle the functional relevance of multiple aspects of niche architecture and progenitor organization.
Finally, organ architecture and function are thought of as intimately connected, so it is surprising that dermal papillae which are reorganized into a sheath are nevertheless able to support hair progenitor proliferation and differentiation (Fig.4). Most strikingly, reorganized dermal papillae support progenitor differentiation in a concentric organization resembling wild-type hair follicles and mirroring the concentric organization of prospective epithelial stem cells during hair follicle morphogenesis.53 Since many progenitor lineages are established before niche polarization,10 our results suggest that at least at the late-growth stage, niche architecture mainly reinforces and optimizes pre-established progenitor differentiation programs. Therefore, the purpose of dermal papilla enclosure and polarized elongation may be to maintain proximity to differentiating multipotent progenitors, and a sheath-like niche would only be proximal to progenitors at the beginning of their differentiation journey. This explains why sheath-like dermal papillae only produce short hairs, similar to the compromised hair produced from transplanted 3D spheroid cultured human dermal papillae,52 together implying that a polarized and elongated niche architecture is still necessary for optimal organ function. Overall, these findings prompt a framework in which to scrutinize the functional importance of niche architectures in different organs (e.g., sheath-like niches in sweat glands, lung, and intestines54–59) and across species (e.g., sheath-like niches in the Drosophila germline, and enclosed niches in feather follicles and human epidermal rete ridges60–62). Learning the functional principles of niche architecture from these diverse biological systems will not only deepen our understanding of when and how organ architecture affects its function, but also provide insights about designing cell therapies and engineered tissues with efficiently coupled architecture and function.
Limitations of study:
The fibroblast cell body exterior/interior analyses focused on smaller hair follicles in the ear skin that have narrower dermal papillae than the larger dermal papillae in guard hair follicles, which constitute ~1% of total hair follicles in the back skin. The percentage of exterior-localized dermal papilla fibroblasts is lower (~70%) in guard hair dermal papillae than small ear hair follicles. Pdgfrα-CreER is a pan-fibroblast driver, which limits our ability to understand specific fibroblast populations’ contributions to dermal papilla architecture. Future studies using fibroblast-lineage restricted drivers would clarify the roles of the dermal papilla and the dermal sheath in regulating niche architecture. In addition, it remains unclear how the polarized niche becomes more structurally robust than the unpolarized niche; profiling niche cellular and acellular compositions (e.g., extracellular matrix) after ablation will provide more mechanistic insights. Furthermore, some fibroblasts retained nuclear phospho-Smad2 after fibroblast Tgfbr1fl/fl deletion, indicating that our CreER induction may not have excised both Tgfbr1 alleles in every fibroblast. Novel genetic reporters that reliably report cells with full gene deletions would allow behavioral tracing and molecular profiling of both mutant and wild-type fibroblasts to understand their interactions during niche reorganization. Lastly, further comprehensive spatial molecular profiling and intravital imaging-based behavior tracing on hair progenitors and their differentiated progenies would better link functional hair production with progenitors’ dynamic proliferation, differentiation, and molecular changes to deepen our understanding of functional robustness after Tgfbr1fl/fl deletion.
STAR★Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Valentina Greco (valentina.greco@yale.edu).
Materials availability
The mouse line (mNeonGreen-Smad4) generated in this study will be shared by the lead contact upon request.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Mice
Pdgfrα-H2BGFP (JAX Strain #007669),63 Pdgfrα-CreER (JAX Strain #018280),64 Rosa26-loxP-membraneTomato-(stop)-membraneGFP (mTmG) (JAX Strain #007676),65 ROSA-LSL-DTA (JAX Strain #009669),66 and R26-LSL-tdTomato mice (JAX Strain #007909)67 were obtained from The Jackson Laboratory. Tgfbr1fl/fl mice were obtained from V. Kaartinen.68 All mice were bred into a CD1 background.
mNeonGreen-Smad4 (mNG-Smad4) mice were generated as follows. CRISPR-Cas9 guide RNA was targeted to a site 43bp downstream from the endogenous Smad4 translation start site in exon 2. The linear DNA gBlock repair template (Integrated DNA Technologies) consisted of mNeonGreen (708bp), a linker (12bp), a CRISPR target site recoded with silent mutations to prevent recombination of the edited sequence, flanked by 36bp homology arms. Following PCR amplification and purification of the gBlocks, DNA, CRISPR-Cas9 nuclease, and guide RNAs were introduced by pronuclear injection into B6SJL embryos then transferred to pseudo-pregnant recipient CD1 females (JHUSOM Transgenic Core). Pups carrying the integrated mNG-Smad4 sequence were identified by genomic PCR amplification of the insertion site. As homozygote progeny could not be produced, founder lines were bred into a CD1 background and maintained as heterozygotes. Sequence information for mNeonGreen-Smad4 mice is summarized below.
gRNA for CRISPR:
ACTATGTACAATGCTCAGAC (20bp, REV)
HDR template (837bp linear DNA gBlock, from Integrated DNA Technologies):
Capitalized sequences at either end denote the 36bp 5′ & 3′ homology arms.
Lowercase denotes the inserted mNeonGreen and linker sequences.
Capitalized bases preceding the 3′ homology arm denote silent mutations in the CRISPR-Cas9 target area, specifically residues downstream of the original SMAD4 start codon up to the gRNA PAM site.
TTGGAGAGTTGGATTTAAAAGAAAAAACTTGAACAAatggtgagcaagggcgaggaggataacatggcctctctcccagcgacacatgagttacacatctttggctccatcaacggtgtggactttgacatggtgggtcagggcaccggcaatccaaatgatggttatgaggagttaaacctgaagtccaccaagggtgacctccagttctccccctggattctggtccctcatatcgggtatggcttccatcagtacctgccctaccctgacgggatgtcgcctttccaggccgccatggtagatggctccggctaccaagtccatcgcacaatgcagtttgaagatggtgcctcccttactgttaactaccgctacacctacgagggaagccacatcaaaggagaggcccaggtgaaggggactggtttccctgctgacggtcctgtgatgaccaactcgctgaccgctgcggactggtgcaggtcgaagaagacttaccccaacgacaaaaccatcatcagtacctttaagtggagttacaccactggaaatggcaagcgctaccggagcactgcgcggaccacctacacctttgccaagccaatggcggctaactatctgaagaaccagccgatgtacgtgttccgtaagacggagctcaagcactccaagaccgagctcaacttcaaggagtggcaaaaggcctttaccgatgtgatgggcatggacgagctgtacaagtccggatccagaatggaTaaCatgtcGatTacCaaCacCccGacCagCaaTgaCgcATGTCTGAGCATTGTACATAGTTTGATGTGTCATAGA
PCR primers
DNA oligos (ordered from Integrated DNA Technologies, stored as 200μM stock)
FWD: GCC TTT GCA ATA GTG AGG AGA CCG G
REV: GCA TCT GGG AAT GCT CTC TTC TCG CC
Both male and female mice between two weeks and two months of age (for the specific age used in each experiment, see Method details) were used in this study. Mice of either sex were randomly selected for experiments. No blinding was done. All studies and procedures involving animal subjects were approved by the Institutional Animal Care and Use Committee at Yale School of Medicine and were conducted in accordance with the approved animal handling protocol no. 11303. This study is compliant with all relevant ethical regulations regarding animal research.
Primary cultures
Primary mouse embryonic fibroblasts (MEFs) were isolated from heterozygous mNG-Smad4 mice embryos and wild-type littermates using previously described protocols.69 In general, E13.5 embryos were dissected in cold PBS to remove the brain and viscera; tails were collected for genotyping. Remaining tissues were finely minced and incubated in 0.25% trypsin-EDTA at 4°C overnight. Dissociated cells were suspended in warmed MEF culture medium (DMEM with 15% fetal bovine serum (FBS) (SH30071.02HI, Hyclone), 100U/ml penicillin-streptomycin (P0781, Sigma)) by vigorous pipetting. Larger clumps were removed by allowing sedimentation for 1 minute then collecting supernatant, twice. Cells from embryos of either sex were pooled by genotype before plating. Prior to use, MEFs were cultured in 37°C in 5% CO2, 2% O2 in MEF medium and passaged maximum twice.
METHOD DETAILS
CreER induction
To label fibroblasts in a mosaic pattern, we benefited from the endogenous leakage of Pdgfrα-CreER; mTmG mice within the fibroblast population. Therefore, even without tamoxifen induction, a small population of fibroblasts was labeled and we could define their morphology. Mosaic labeling analyses span across fibroblasts at different dermal papilla positions, but the very bottom stalk cells are underrepresented in the quantification. To achieve diphtheria toxin-induced fibroblast ablation, five doses of 100mg/kg tamoxifen (T5648-5G, Sigma) in corn oil (C8267-2.5L, Sigma) were delivered via intraperitoneal injection into Pdgfrα-H2BGFP; Pdgfrα-CreER; mTmG; ROSA-LSL-DTA mice. Doses were given twice per day for three days starting at postnatal day 24–25 (including both mid- and late-growth hair follicles). To achieve conditional knockout of the Tgfbr1 allele in fibroblasts, three doses of 100mg/kg tamoxifen in corn oil were delivered via intraperitoneal injection into Pdgfrα-CreER; Tgfbr1+/fl and Tgfbr1fl/fl mice. Doses were given once per day for three days, starting at postnatal day 18–20 (quiescent telogen hair follicles). Phenotype penetrance of Tgfbr1fl/fl mutant hair follicles is on average 83% across all hair follicles.
Intravital imaging and tracking hair growth
Procedures of intravital imaging were similar to those previously described35 and are summarized below. During the experiment, we maintained mice anaesthetized via vaporized 1.5% isoflurane and 1.5% oxygen in air and placed them on a warming pad to maintain body temperature. The mouse ear was mounted on a speculum with a glass coverslip on top. We used a LaVision TriM Scope II (LaVision Biotec) microscope equipped with a Chameleon Vision II (Coherent) two-photon laser (940nm) and a Chameleon Discovery (Coherent) two-photon laser (1120nm) for intravital imaging. Either a Zeiss ×20/1.0, Nikon ×25/1.10, or a Nikon ×40/1.15 water immersion objective was used to scan a field of view of 0.25mm2 (20X), 0.24mm2 (25X), or 0.09mm2 (40X). Via a motorized stage, we acquired multiple tiles of z-stacks that reach up to 200μm deep and vary each z step from 1 to 10μm depending on the required spatial resolution. Patterns of hair follicles were used as landmarks to revisit the same skin area longitudinally.
Timing of hair cycle initiation varies slightly between different mice. When necessary, we kept imaging until the mice reached the stage of interest. Wild-type and Tgfbr1+/fl control hair follicle stages were identified based on the morphological staging system previous literature reported.10 To summarize, early-growth stages include Anagen I to early Anagen II; mid-growth stages include late Anagen II to Anagen IIIb; late-growth stages include Anagen IIIc to Anagen VI. The “early-growth, mid-growth, and late-growth stages” terminologies are used to describe the growth of hair follicles rather than the actual hairs. Tgfbr1fl/fl mutant hair follicle stages were identified based on dermal papilla morphology. Mutant dermal papillae at Stage1 look similar to Anagen IIIa-b in control littermates. By contrast, mutant dermal papillae at Stage2 are triangle-shaped, at Stage3 are very thin, and disappear from within the epithelium at Stage4. The frequency and length of longitudinal revisits depended on the purpose of each experiment and are indicated in the figures. In particular, for DTA ablation experiments, after tamoxifen induced DTA ablation at postnatal day 24–25, mice were intravitally imaged 0–1 days, 4–5 days, 6–7 days later to revisit the same hair follicles for analyses. After DTA ablation, dermal papillae could be identified based on their retained stronger membrane-tdTomato fluorescent signals and different membrane organizations compared to the hair follicle epithelium. For mNG-Smad4 and Tgfbr1 deletion experiments (Fig.3A–B′), intravital longitudinal imaging were performed every three to five days, and representative examples of hair follicles were selected to represent different stages. The transition time between each Tgfbr1fl/fl mutant stage takes on average one to two days. Representative images in Fig.3A–B′ are not necessarily the same revisited hair follicles, but the sequence and timing of Tgfbr1fl/fl mutant stage progression was identified based on the information provided by both revisiting the same hair follicle over time and tracking the hair follicle population along the wave of ear hair cycle from multiple mice. To track the hair follicle throughout the regression phase and the next hair cycle, hair follicles from the same ear skin region in tamoxifen-induced Pdgfrα-CreER; Tgfbr1fl/fl mice were intravitally imaged starting at postnatal day 33–36 and revisited every five to seven days until they reached the next growth cycle at mid- to late-growth stage.
To track hair growth, we shaved the back skin of tamoxifen-induced Pdgfrα-CreER; Tgfbr1+/fl and Tgfbr1fl/fl mice and removed their hairs using Nair Hair Removal Cream once during postnatal day 18–21. Then, these mice were photographed on the next day and every week until the end of first hair cycle at postnatal day 60–63. During all procedures, mice were anaesthetized and kept on a warming pad, similar to intravital imaging.
Hair plucking and quantification
We shaved the back skin and removed hairs from tamoxifen-induced Pdgfrα-CreER; Tgfbr1+/fl and Tgfbr1fl/fl mice using Nair Hair Removal Cream once during postnatal day 18–21. Then back skins of Pdgfrα-CreER; Tgfbr1+/fl and Tgfbr1fl/fl mice were collected at the end of first hair cycle (postnatal day 63–65) and stored in 20mM EDTA (in PBS, AB00502-01000, AmericanBio) at 4°C. To quantify the frequency and length of different hair types, clumps of phenotypic representative hairs from the middle back skin region were plucked with forceps under the dissection microscope. Individual hairs were mounted in anhydrous glycerol (AB00751-00500, AmericanBio) and imaged under ZEISS SteREO Discovery.V12 equipped with a digital camera (AxioCam MR R3). Images were taken via differential interference contrast (DIC) channel in ZEN 2.3 pro software. Hair lengths were measured in Fiji software. Hair types were classified based on different morphologies of hair types, as previously reported.11 In general, guard hairs are straight and the longest and contain one to two medulla columns; awl hairs are straight but shorter than guard hair and contain two to three medulla columns; auchene hairs have a single bend in the middle and contain two to three medulla columns; zigzag hairs have multiple alternating bends and contain only one medulla column.
Cell culture and imaging
For mNG-Smad4 validation experiments, wild-type and mNG-Smad4 heterozygous MEFs were seeded 7×103/well onto fibronectin (FC010, EMD-Millipore)-coated 96-well glass-bottom plates (CellVis). Cells were imaged at 5-minute intervals; vehicle control or TGF-β1 (7754-BH, R&D, to an end concentration of 10ng/μl) was added to media after 20 minutes of imaging onset. Live specimens were maintained at 37°C in 5% CO2 in a humidified environmental control chamber (H301-K-FRAME, OKOlab) during imaging. Images were acquired on a Metamorph-controlled Nikon Eclipse Ti epifluorescence microscope fitted with a 40X oil objective and a Photometric Prime 95B CMOS camera.
Immunofluorescence
Based on hair follicle stages of interests, Tgfbr1+/fl control back skin samples were collected at postnatal day 27–39, and Tgfbr1fl/fl mutant back skin samples were collected at postnatal day 29–40 to cover different hair follicle stages. For thick tissue sections, back skins were dissected, then either freshly embedded in optimal cutting temperature compound (OCT) and frozen on dry ice, or first fixed in 4% paraformaldehyde (PFA) in PBS at room temperature for one hour and then 30% sucrose at 4°C overnight. The OCT blocks were later sectioned at 30μm thickness to preserve 3D architecture. These sections were further fixed in 4% PFA for 20 minutes and washed in 2% Triton X-100 in PBS three times (each 10 minutes) at room temperature or 37°C. Then the sections were incubated in the blocking solution (5% normal donkey serum, 1% BSA and 2% Triton X-100 in PBS) at room temperature or 37°C overnight. Later, the sections were incubated with primary antibodies diluted in the blocking solution at room temperature or 37°C for one day, and then washed in 2% Triton X-100 in PBS three times (each 10 minutes) at room temperature or 37°C. The sections were further incubated with secondary antibody diluted in the blocking solution at room temperature or 37°C overnight and then washed in 2% Triton X-100 in PBS three times (each 10 minutes). If needed, the sections were further stained in 3μM/ml SiR-DNA (SKU CY-SC007, Cytoskeleton, Inc.) in HBSS for one day at 37°C. The stained sections were mounted in SlowFade Diamond Antifade Mountant (S36972, Invitrogen) and imaged under the two-photon microscope.
The following primary antibodies were used: human/mouse active Caspase3 antibody (AF835, R&D Systems, 1:200), mouse integrin alpha9 antibody (AF3827, R&D Systems, 1:200), Sox2 monoclonal antibody, eBioscience (14-9811-82, Invitrogen, 1:400), Lef1 monoclonal antibody (E.281.8) (MA5-14966, Invitrogen, 1:400), recombinant anti-GATA3 antibody [EPR16651] (ab199428, Abcam, 1:200), anti-acidic hair keratin K31 guinea pig polyclonal antibody (GP-hHa1, Progen, 1:200), Phospho-SMAD2 (Ser465/Ser467) (E8F3R) Rabbit mAb (#18338, Cell Signaling, 1:50), anti-fibronectin antibody (AB2033, Sigma-Aldrich, 1:200), anti-acidic hair keratin K32 guinea pig polyclonal antibody (GP-HHA2, Progen, 1:100), and TCHH polyclonal antibody (PA5-120392, Invitrogen, 1:200).
The following secondary antibodies were used: goat anti-rabbit Alexa 488 (A32731, Invitrogen, 1:200), donkey anti-goat Alexa 488 (A-11055, Invitrogen, 1:200), donkey anti-rat Alexa 488 (A-21208, Invitrogen, 1:200), goat anti-guinea pig Alexa 488 (A-11073, Invitrogen, 1:200). Phospho-SMAD2 and TCHH immunostaining required Alexa Fluor™ 488 Tyramide SuperBoost™ Kit (goat anti-rabbit IgG, B40922, Invitrogen).
For whole-mount staining, Tgfbr1+/fl and Tgfbr1fl/fl ear skin was collected at postnatal day 26–39 to cover different hair follicle stages. Ear skin was dissected and fixed in 4% PFA in PBS at 4°C overnight. Fixed tissues were cut into small pieces and incubated in the blocking solution (5% normal donkey serum, 1% BSA and 2% Triton X-100 in PBS) at 37°C for 3 days. Blocked tissues were incubated with anti-Ki67 antibody (ab15580, Abcam, 1:200), or mouse integrin alpha9 (AF3827, R&D Systems, 1:100), or Sox2 monoclonal antibody, eBioscience (14-9811-82, Invitrogen, 1:150), or anti-alpha smooth muscle actin antibody (ab5694, Abcam, 1:200) at 37°C for 6 days. Tissues were then washed in PBS with 2% Triton X-100 at 37°C for 4 days and incubated with goat anti-rabbit Alexa 633 (A-21071, Invitrogen, 1:100), or goat anti-rabbit Alexa 488 (A32731, Invitrogen, 1:200), or donkey anti-goat Alexa 488 (A-11055, Invitrogen, 1:200) at 37°C for 5 days. After staining, tissues were washed in PBS with 2% Triton X-100 at 37°C for 5 days and mounted in SlowFade Diamond Antifade Mountant (S36972, Invitrogen). The stained whole mounts were imaged under the two-photon microscope.
Transmission electron microscopy
For transmission electron microscopy (TEM), back skin samples from tamoxifen-induced Pdgfrα-CreER; Tgfbr1+/fl and Tgfbr1fl/fl mice were collected at postnatal day 26–42 to cover different hair follicle stages. Samples were fixed in 2% glutaraldehyde (16120, Electron Microscopy Sciences), 4% PFA (15710, Electron Microscopy Sciences), and 2 mM CaCl2 in 0.1 M sodium cacodylate buffer, pH 7.2 (11652, Electron Microscopy Sciences), at room temperature for more than an hour, dehydrated in acetone, postfixed in 1% osmium tetroxide (19150, Electron Microscopy Sciences), and processed for Eponate 12 (18012, Ted Pella, Inc) embedding. Resin blocks were sectioned with a Leica EM UC6 ultramicrotome. Semithin sections (500nm) were stained with toluidine blue (22050, Electron Microscopy Sciences) and examined under a bright field using a Zeiss Axio Scope A1 microscope. Ultrathin sections (60–65nm) of selected areas were counterstained with uranyl acetate (22400, Electron Microscopy Sciences) and lead citrate (15326, Fluka). Images were taken with a transmission electron microscope (Tecnai G2-12; FEI, Hillsboro, OR) equipped with a digital camera (AMT BioSprint29).
QUANTIFICATION AND STATISTICAL ANALYSIS
Image analysis
Z-stacks of images were imported into Fiji (ImageJ, NIH) or Imaris (BitPlane) for analysis. Tiles of different z-stacks were stitched in Fiji. Either selected single optical z slice or summed z slices were used. In main figures, images are single optical z slice in Fig.1A, C′–C″, Fig.3, and Fig.4, and summed optical z slices in Fig.1C, E, G, Fig.2A. Optical section properties of images in supplemental figures are indicated in legends. Quantification of percentages, measurement of length and width were performed in Fiji based on z-stack’s 3D information. 3D dermal papilla reconstructions were performed in Imaris via its surface rendering function.
Statistics and reproducibility
Statistical calculations were performed with Prism 9 (GraphPad). All data are presented as mean ± S.D. P value <0.05 is considered significant; the precise P values can be found in figures. For specific statistical tests in main figures, unpaired two-tailed t-test is used in Fig.1B, D, H, Fig.3H, and Fig.4. Tukey’s multiple comparisons test is used in Fig.1F, Fig.2C, D, Fig.3D. Specific statistical tests in supplemental figures can be found in their legends. Sample sizes are indicated in figure legends. No statistical method was used to predetermine sample size. All experiments were repeated at least three times.
Supplementary Material
The entire dermal papilla at late-growth is 3D reconstructed based on its membrane-tdTomato signal. Within the red outlined dermal papilla, cell bodies of individual fibroblasts are rendered into surfaces with different colors. Further, a single fibroblast is 3D reconstructed based on its membrane-GFP signal. The cell body of this fibroblast is rendered into a pink surface and its entire membrane in a green surface.
Representative z-stack movies of immunostained Sox2 and Integrin alpha9 (Itgα9) (green) in ear skin whole mounts to detect fibroblasts with dermal papilla identity at different stages. Fibroblasts are labeled in red by Pdgfrα-CreER; LSL-tdTomato. Scale bar sizes are indicated. See also Figure S3A–D.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Human/Mouse Active Caspase-3 Antibody | R&D Systems | Cat#AF835 |
| Mouse Integrin alpha 9 Antibody | R&D Systems | Cat#AF3827 |
| SOX2 Monoclonal Antibody (Btjce), eBioscience™ | Invitrogen | Cat#14-9811-82; RRID: AB_11219471 |
| LEF1 Monoclonal Antibody (E.281.8) | Invitrogen | Cat#MA5-14966; RRID: AB_10987030 |
| Recombinant Anti-GATA3 antibody [EPR16651] - ChIP Grade | Abcam | Cat#ab199428 |
| anti-acidic Hair Keratin K31 guinea pig polyclonal, serum | Progen | Cat#GP-hHa1 |
| Phospho-SMAD2 (Ser465/Ser467) (E8F3R) Rabbit mAb | Cell Signaling | Cat#18338 |
| Anti-Fibronectin Antibody | Sigma-Aldrich | Cat#AB2033 |
| anti-acidic Hair Keratin K32 guinea pig polyclonal, serum | Progen | Cat#GP-HHA2 |
| TCHH Polyclonal Antibody | Invitrogen | Cat#PA5-120392; RRID: AB_2913964 |
| Anti-Ki67 antibody | Abcam | Cat#ab15580 |
| Anti-alpha smooth muscle actin antibody | Abcam | Cat#ab5694 |
| Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ Plus 488 | Invitrogen | Cat#A32731; RRID: AB_2633280 |
| Donkey anti-Goat IgG (H+L) Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 488 | Invitrogen | Cat#A-11055; RRID: AB_2534102 |
| Donkey anti-Rat IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 488 | Invitrogen | Cat#A-21208; RRID: AB_2535794 |
| Goat anti-Guinea Pig IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 488 | Invitrogen | Cat#A-11073; RRID: AB_2534117 |
| Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor™ 633 | Invitrogen | Cat#A-21071; RRID: AB_2535732 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| CRISPR-spCas9 nuclease | JHUSOM Transgenic Mouse Core Facility | RRID:SCR_017809 |
| Fetal Bovine Serum (FBS) | Hyclone | Cat#SH30071.02HI |
| Penicillin-Streptomycin | Sigma | Cat#P0781 |
| Tamoxifen | Sigma | Cat#T5648-5G |
| Corn oil | Sigma | Cat#C8267-2.5L |
| GLYCEROL, ANHYDROUS “SUPER GLYCEROL” | AmericanBio | Cat#AB00751-00500 |
| EDTA, 0.5M SOLUTION, PH 8.0 | AmericanBio | Cat#AB00502-01000 |
| Human Plasma Fibronectin | EMD Millipore | Cat#FC010 |
| TGF-β1 | R&D | Cat#7754-BH |
| SlowFade Diamond Antifade Mountant | Invitrogen | Cat#S36972 |
| Paraformaldehyde (PFA) 16% Aqueous SOL. EM GRADE | Electron Microscopy Sciences | Cat#15710 |
| Aqueous Glutaraldehyde EM Grade, 10% 10x10ml | Electron Microscopy Sciences | Cat#16120 |
| Sodium Cacodylate Buffer, 0.2M | Electron Microscopy Sciences | Cat#11652 (500ML) |
| Osmium Tetroxide 4% solution | Electron Microscopy Sciences | Cat#19150 |
| Toluidine Blue O | Electron Microscopy Sciences | Cat#22050 |
| Uranyl Acetate | Electron Microscopy Sciences | Cat#22400 |
| Lead Citrate | Fluka | Cat#15326 |
| Critical Commercial Assays | ||
| goat anti-rabbit IgG Alexa Fluor™ 488 Tyramide SuperBoost™ Kit | Invitrogen | Cat#B40922 |
| SiR-DNA Kit | Cytoskeleton, Inc | Cat#SKU CY-SC007 |
| PELCO® Eponate 12™ Kit with BDMA | Ted Pella, Inc. | Cat#18012 |
| Experimental Models: Organisms/Strains | ||
| Mouse: Pdgfrα-H2BGFP: B6.129S4-Pdgfratm11(EGFP)Sor/J | The Jackson Laboratory | RRID:IMSR_JAX:007669 |
| Mouse: Pdgfrα-CreER: B6N.Cg-Tg(Pdgfra-cre/ERT)467Dbe/J | The Jackson Laboratory | RRID:IMSR_JAX:018280 |
| Mouse: Rosa26-loxP-membraneTomato-(stop)-membraneGFP(mTmG): B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J | The Jackson Laboratory | RRID:IMSR_JAX:007676 |
| Mouse: ROSA-LSL-DTA: B6.129P2-Gt(ROSA)26Sortm1(DTA)Lky/J | The Jackson Laboratory | RRID:IMSR_JAX:009669 |
| Mouse: R26-LSL-tdTomato: B6.Cg-Gt(ROSA)26Sortm9(CAG-tdTomato)Hze/J | The Jackson Laboratory | RRID:IMSR_JAX:007909 |
| Mouse: Tgfbr1fl/fl: STOCK Tgfbr1tm1.1Kar/KulJ | V. Kaartinen (Larsson et al.68) | RRID:IMSR_JAX:028701 |
| Mouse: B6SJL/F2 | JHUSOM Transgenic Mouse Core Facility | RRID:SCR_017809 |
| Mouse: mNeonGreen-Smad4: B6SJL-Smad4tm1(mNG)+/− | This Paper | N/A |
| Mouse: CD1: Crl:CD1(ICR) | Charles River | Strain #022 |
| Oligonucleotides | ||
| CRISPR gRNA: 5’ ACTATGTACAATGCTCAGAC 3’ | This Paper | N/A |
| PCR primer for gBlock amplification, Forward: 5’ GCC TTT GCA ATA GTG AGG AGA CCG G | This Paper | N/A |
| PCR primer for gBlock amplification, Reverse: 5’ GCA TCT GGG AAT GCT CTC TTC TCG CC | This Paper | N/A |
| Recombinant DNA | ||
| Linear dsDNA gBlock (837bp) HDR template: See Methods: Mice for sequence information |
This Paper | N/A |
| Software and Algorithms | ||
| ZEN 2.3 pro | ZEISS | micro-shop.zeiss.com/en/de/softwarefinder/software-categories/zen-blue/zen-blue-pro/ |
| Fiji (ImageJ) | NIH | fiji.sc |
| GraphPad Prism 9 | GraphPad Software, inc | graphpad.com |
| Imaris | Bitplane/Oxford Instruments | imaris.oxinst.com |
| Adobe Illustrator | Adobe | adobe.com |
| Adobe Premiere Pro | Adobe | adobe.com |
Highlights:
Hair follicle niche cells collectively build a morphologically polarized architecture
The polarized niche architecture is structurally robust even upon cell loss
Niche architecture and location are maintained through fibroblast TGFβ signaling
Niche reorganization induces hair progenitor redistribution but supports function
ACKNOWLEDGMENTS
We thank Greco lab members, thesis committee members of H.W. (Stefania Nicoli, Haifan Lin, and Scott Holley), as well as Maria Jose Ortuno Romero, Emily Forrest, Andrew Lee Cox, and Caroline Hendry for thoughtful feedbacks on the manuscript. H.W. is supported by China Scholarship Council Grant. V.G. is supported by an HHMI Scholar award (55108527) and NIH grants (R01AR063663, R01AR067755). Research reported in this publication is supported by the National Institute on Aging of the National Institutes of Health under Award Number DP1AG066590. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
INCLUSION AND DIVERSITY
We support inclusive, diverse, and equitable conduct of research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Xin T, Greco V, and Myung P (2016). Hardwiring Stem Cell Communication through Tissue Structure. Cell 164, 1212–1225. 10.1016/j.cell.2016.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuchs E, and Blau HM (2020). Tissue Stem Cells: Architects of Their Niches. Cell Stem Cell 27, 532–556. 10.1016/j.stem.2020.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Brien LE, and Bilder D (2013). Beyond the niche: tissue-level coordination of stem cell dynamics. Annu Rev Cell Dev Biol 29, 107–136. 10.1146/annurev-cellbio-101512-122319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lane SW, Williams DA, and Watt FM (2014). Modulating the stem cell niche for tissue regeneration. Nat Biotechnol 32, 795–803. 10.1038/nbt.2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Plikus MV, Wang X, Sinha S, Forte E, Thompson SM, Herzog EL, Driskell RR, Rosenthal N, Biernaskie J, and Horsley V (2021). Fibroblasts: Origins, definitions, and functions in health and disease. Cell 184, 3852–3872. 10.1016/j.cell.2021.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCarthy N, Kraiczy J, and Shivdasani RA (2020). Cellular and molecular architecture of the intestinal stem cell niche. Nat Cell Biol 22, 1033–1041. 10.1038/s41556-020-0567-z. [DOI] [PubMed] [Google Scholar]
- 7.Goodwin K, and Nelson CM (2020). Branching morphogenesis. Development 147. 10.1242/dev.184499. [DOI] [PubMed] [Google Scholar]
- 8.Mao JJ, and Prockop DJ (2012). Stem cells in the face: tooth regeneration and beyond. Cell Stem Cell 11, 291–301. 10.1016/j.stem.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mistretta CM, and Bradley RM (2021). The Fungiform Papilla Is a Complex, Multimodal, Oral Sensory Organ. Curr Opin Physiol 20, 165–173. 10.1016/j.cophys.2021.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MuÈller-RoÈver S, Foitzik K, Paus R, Handjiski B, van der Veen C, Eichmüller S, McKay IA, and Stenn KS (2001). A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. Journal of Investigative Dermatology 117, 3–15. 10.1046/j.0022-202x.2001.01377.x. [DOI] [PubMed] [Google Scholar]
- 11.Chi W, Wu E, and Morgan BA (2013). Dermal papilla cell number specifies hair size, shape and cycling and its reduction causes follicular decline. Development 140, 1676–1683. 10.1242/dev.090662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan BA (2014). The Dermal Papilla: An Instructive Niche for Epithelial Stem and Progenitor Cells in Development and Regeneration of the Hair Follicle. Cold Spring Harbor Perspectives in Medicine 4, a015180. 10.1101/cshperspect.a015180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahmani W, Abbasi S, Hagner A, Raharjo E, Kumar R, Hotta A, Magness S, Metzger D, and Biernaskie J (2014). Hair Follicle Dermal Stem Cells Regenerate the Dermal Sheath, Repopulate the Dermal Papilla, and Modulate Hair Type. Developmental Cell 31, 543–558. 10.1016/j.devcel.2014.10.022. [DOI] [PubMed] [Google Scholar]
- 14.Heitman N, Sennett R, Mok KW, Saxena N, Srivastava D, Martino P, Grisanti L, Wang Z, Ma’ayan A, Rompolas P, and Rendl M (2020). Dermal sheath contraction powers stem cell niche relocation during hair cycle regression. Science 367, 161–166. 10.1126/science.aax9131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Driskell RR, Clavel C, Rendl M, and Watt FM (2011). Hair follicle dermal papilla cells at a glance. Journal of Cell Science 124, 1179–1182. 10.1242/jcs.082446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tobin DJ, Gunin A, Magerl M, and Paus R (2003). Plasticity and cytokinetic dynamics of the hair follicle mesenchyme during the hair growth cycle: Implications for growth control and hair follicle transformations. Journal of Investigative Dermatology Symposium Proceedings 8, 80–86. 10.1046/j.1523-1747.2003.12177.x. [DOI] [PubMed] [Google Scholar]
- 17.Legue E, and Nicolas JF (2005). Hair follicle renewal: organization of stem cells in the matrix and the role of stereotyped lineages and behaviors. Development 132, 4143–4154. 10.1242/dev.01975. [DOI] [PubMed] [Google Scholar]
- 18.Legue E, Sequeira I, and Nicolas JF (2010). Hair follicle renewal: authentic morphogenesis that depends on a complex progression of stem cell lineages. Development 137, 569–577. 10.1242/dev.044123. [DOI] [PubMed] [Google Scholar]
- 19.Mesler AL, Veniaminova NA, Lull MV, and Wong SY (2017). Hair Follicle Terminal Differentiation Is Orchestrated by Distinct Early and Late Matrix Progenitors. Cell Rep 19, 809–821. 10.1016/j.celrep.2017.03.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xin T, Gonzalez D, Rompolas P, and Greco V (2018). Flexible fate determination ensures robust differentiation in the hair follicle. Nat Cell Biol 20, 1361–1369. 10.1038/s41556-018-0232-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jahoda CA, Horne KA, and Oliver RF (1984). Induction of hair growth by implantation of cultured dermal papilla cells. Nature 311, 560–562. 10.1038/311560a0. [DOI] [PubMed] [Google Scholar]
- 22.Botchkarev VA, Botchkareva NV, Roth W, Nakamura M, Chen LH, Herzog W, Lindner G, McMahon JA, Peters C, Lauster R, et al. (1999). Noggin is a mesenchymally derived stimulator of hair-follicle induction. Nat Cell Biol 1, 158–164. 10.1038/11078. [DOI] [PubMed] [Google Scholar]
- 23.Panteleyev AA, Jahoda CA, and Christiano AM (2001). Hair follicle predetermination. J Cell Sci 114, 3419–3431. 10.1242/jcs.114.19.3419. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt-Ullrich R, and Paus R (2005). Molecular principles of hair follicle induction and morphogenesis. Bioessays 27, 247–261. 10.1002/bies.20184. [DOI] [PubMed] [Google Scholar]
- 25.Rendl M, Lewis L, and Fuchs E (2005). Molecular dissection of mesenchymal-epithelial interactions in the hair follicle. PLoS Biol 3, e331. 10.1371/journal.pbio.0030331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greco V, Chen T, Rendl M, Schober M, Pasolli HA, Stokes N, dela Cruz-Racelis J, and Fuchs E (2009). A two-step mechanism for stem cell activation during hair regeneration. Cell stem cell 4, 155–169. 10.1016/j.stem.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sennett R, and Rendl M (2012). Mesenchymal-epithelial interactions during hair follicle morphogenesis and cycling. Seminars in Cell & Developmental Biology 23, 917–927. 10.1016/j.semcdb.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hsu YC, Li LS, and Fuchs E (2014). Transit-Amplifying Cells Orchestrate Stem Cell Activity and Tissue Regeneration. Cell 157, 935–949. 10.1016/j.cell.2014.02.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rezza A, Wang ZC, Sennett R, Qiao WL, Wang DM, Heitman N, Mok KW, Clavel C, Yi R, Zandstra P, et al. (2016). Signaling Networks among Stem Cell Precursors, Transit-Amplifying Progenitors, and their Niche in Developing Hair Follicles. Cell Reports 14, 3001–3018. 10.1016/j.celrep.2016.02.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H, Adam RC, Ge YJ, Hua ZL, and Fuchs E (2017). Epithelial-Mesenchymal Micro-niches Govern Stem Cell Lineage Choices. Cell 169, 483–+. 10.1016/j.cell.2017.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adam RC, Yang H, Ge YJ, Lien WH, Wang P, Zhao YL, Polak L, Levorse J, Baksh SC, Zheng DY, and Fuchs E (2018). Temporal Layering of Signaling Effectors Drives Chromatin Remodeling during Hair Follicle Stem Cell Lineage Progression. Cell Stem Cell 22, 398–+. 10.1016/j.stem.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harshuk-Shabso S, Dressler H, Niehrs C, Aamar E, and Enshell-Seijffers D (2020). Fgf and Wnt signaling interaction in the mesenchymal niche regulates the murine hair cycle clock. Nat Commun 11, 5114. 10.1038/s41467-020-18643-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Joost S, Annusver K, Jacob T, Sun X, Dalessandri T, Sivan U, Sequeira I, Sandberg R, and Kasper M (2020). The Molecular Anatomy of Mouse Skin during Hair Growth and Rest. Cell Stem Cell 26, 441–457 e447. 10.1016/j.stem.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 34.Shin W, Rosin NL, Sparks H, Sinha S, Rahmani W, Sharma N, Workentine M, Abbasi S, Labit E, Stratton JA, and Biernaskie J (2020). Dysfunction of Hair Follicle Mesenchymal Progenitors Contributes to Age-Associated Hair Loss. Dev Cell 53, 185–198 e187. 10.1016/j.devcel.2020.03.019. [DOI] [PubMed] [Google Scholar]
- 35.Pineda CM, Park S, Mesa KR, Wolfel M, Gonzalez DG, Haberman AM, Rompolas P, and Greco V (2015). Intravital imaging of hair follicle regeneration in the mouse. Nature protocols 10, 1116. 10.1038/nprot.2015.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marsh E, Gonzalez DG, Lathrop EA, Boucher J, and Greco V (2018). Positional Stability and Membrane Occupancy Define Skin Fibroblast Homeostasis In Vivo. Cell 175, 1620–1633 e1613. 10.1016/j.cell.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paus R, Foitzik K, Welker P, BulfonePaus S, and Eichmuller S (1997). Transforming growth factor-beta receptor type I and type II expression during murine hair follicle development and cycling. Journal of Investigative Dermatology 109, 518–526. 10.1111/1523-1747.ep12336635. [DOI] [PubMed] [Google Scholar]
- 38.Warmflash A, Zhang Q, Sorre B, Vonica A, Siggia ED, and Brivanlou AH (2012). Dynamics of TGF-beta signaling reveal adaptive and pulsatile behaviors reflected in the nuclear localization of transcription factor Smad4. Proc Natl Acad Sci U S A 109, E1947–1956. 10.1073/pnas.1207607109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Driskell RR, Giangreco A, Jensen KB, Mulder KW, and Watt FM (2009). Sox2-positive dermal papilla cells specify hair follicle type in mammalian epidermis. Development 136, 2815–2823. 10.1242/dev.038620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai SY, Clavel C, Kim S, Ang YS, Grisanti L, Lee DF, Kelley K, and Rendl M (2010). Oct4 and klf4 reprogram dermal papilla cells into induced pluripotent stem cells. Stem Cells 28, 221–228. 10.1002/stem.281. [DOI] [PubMed] [Google Scholar]
- 41.Hinz B (2015). The extracellular matrix and transforming growth factor-beta1: Tale of a strained relationship. Matrix Biol 47, 54–65. 10.1016/j.matbio.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 42.Meng XM, Nikolic-Paterson DJ, and Lan HY (2016). TGF-beta: the master regulator of fibrosis. Nat Rev Nephrol 12, 325–338. 10.1038/nrneph.2016.48. [DOI] [PubMed] [Google Scholar]
- 43.Ramirez-Weber FA, and Kornberg TB (1999). Cytonemes: cellular processes that project to the principal signaling center in Drosophila imaginal discs. Cell 97, 599–607. 10.1016/s0092-8674(00)80771-0. [DOI] [PubMed] [Google Scholar]
- 44.Inaba M, Buszczak M, and Yamashita YM (2015). Nanotubes mediate niche-stem-cell signalling in the Drosophila testis. Nature 523, 329–332. 10.1038/nature14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guignard L, Fiuza UM, Leggio B, Laussu J, Faure E, Michelin G, Biasuz K, Hufnagel L, Malandain G, Godin C, and Lemaire P (2020). Contact area-dependent cell communication and the morphological invariance of ascidian embryogenesis. Science 369. 10.1126/science.aar5663. [DOI] [PubMed] [Google Scholar]
- 46.Couchman JR, and Gibson WT (1985). Expression of basement membrane components through morphological changes in the hair growth cycle. Dev Biol 108, 290–298. 10.1016/0012-1606(85)90033-8. [DOI] [PubMed] [Google Scholar]
- 47.Jahoda CA, Mauger A, Bard S, and Sengel P (1992). Changes in fibronectin, laminin and type IV collagen distribution relate to basement membrane restructuring during the rat vibrissa follicle hair growth cycle. J Anat 181 (Pt 1), 47–60. [PMC free article] [PubMed] [Google Scholar]
- 48.Tsutsui K, Machida H, Nakagawa A, Ahn K, Morita R, Sekiguchi K, Miner JH, and Fujiwara H (2021). Mapping the molecular and structural specialization of the skin basement membrane for inter-tissue interactions. Nat Commun 12, 2577. 10.1038/s41467-021-22881-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Enshell-Seijffers D, Lindon C, Kashiwagi M, and Morgan BA (2010). beta-catenin activity in the dermal papilla regulates morphogenesis and regeneration of hair. Dev Cell 18, 633–642. 10.1016/j.devcel.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clavel C, Grisanti L, Zemla R, Rezza A, Barros R, Sennett R, Mazloom AR, Chung CY, Cai XQ, Cai CL, et al. (2012). Sox2 in the Dermal Papilla Niche Controls Hair Growth by Fine-Tuning BMP Signaling in Differentiating Hair Shaft Progenitors. Developmental Cell 23, 981–994. 10.1016/j.devcel.2012.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chi W, Morgan O, Wu E, and Morgan BA (2017). Stabilization of beta-Catenin Does Not Increase Dermal Papilla Cell Number in the Hair Follicle. J Invest Dermatol 137, 245–246. 10.1016/j.jid.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Higgins CA, Chen JC, Cerise JE, Jahoda CA, and Christiano AM (2013). Microenvironmental reprogramming by three-dimensional culture enables dermal papilla cells to induce de novo human hair-follicle growth. Proc Natl Acad Sci U S A 110, 19679–19688. 10.1073/pnas.1309970110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morita R, Sanzen N, Sasaki H, Hayashi T, Umeda M, Yoshimura M, Yamamoto T, Shibata T, Abe T, Kiyonari H, et al. (2021). Tracing the origin of hair follicle stem cells. Nature 594, 547–552. 10.1038/s41586-021-03638-5. [DOI] [PubMed] [Google Scholar]
- 54.Shyer AE, Huycke TR, Lee C, Mahadevan L, and Tabin CJ (2015). Bending gradients: how the intestinal stem cell gets its home. Cell 161, 569–580. 10.1016/j.cell.2015.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu CP, Polak L, Keyes BE, and Fuchs E (2016). Spatiotemporal antagonism in mesenchymal-epithelial signaling in sweat versus hair fate decision. Science 354. 10.1126/science.aah6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shoshkes-Carmel M, Wang YJ, Wangensteen KJ, Toth B, Kondo A, Massasa EE, Itzkovitz S, and Kaestner KH (2018). Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature 557, 242–246. 10.1038/s41586-018-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rao-Bhatia A, Zhu M, Yin WC, Coquenlorge S, Zhang X, Woo J, Sun Y, Dean CH, Liu A, Hui CC, et al. (2020). Hedgehog-Activated Fat4 and PCP Pathways Mediate Mesenchymal Cell Clustering and Villus Formation in Gut Development. Dev Cell 52, 647–658 e646. 10.1016/j.devcel.2020.02.003. [DOI] [PubMed] [Google Scholar]
- 58.Cordero-Espinoza L, Dowbaj AM, Kohler TN, Strauss B, Sarlidou O, Belenguer G, Pacini C, Martins NP, Dobie R, Wilson-Kanamori JR, et al. (2021). Dynamic cell contacts between periportal mesenchyme and ductal epithelium act as a rheostat for liver cell proliferation. Cell Stem Cell 28, 1907–1921 e1908. 10.1016/j.stem.2021.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Goodwin K, Jaslove JM, Tao H, Zhu M, Hopyan S, and Nelson CM (2022). Patterning the embryonic pulmonary mesenchyme. iScience 25, 103838. 10.1016/j.isci.2022.103838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Losick VP, Morris LX, Fox DT, and Spradling A (2011). Drosophila stem cell niches: a decade of discovery suggests a unified view of stem cell regulation. Dev Cell 21, 159–171. 10.1016/j.devcel.2011.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Solanas G, and Benitah SA (2013). Regenerating the skin: a task for the heterogeneous stem cell pool and surrounding niche. Nat Rev Mol Cell Biol 14, 737–748. 10.1038/nrm3675. [DOI] [PubMed] [Google Scholar]
- 62.Wu P, Jiang TX, Lei M, Chen CK, Hsieh Li SM, Widelitz RB, and Chuong CM (2021). Cyclic growth of dermal papilla and regeneration of follicular mesenchymal components during feather cycling. Development 148. 10.1242/dev.198671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hamilton TG, Klinghoffer RA, Corrin PD, and Soriano P (2003). Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol Cell Biol 23, 4013–4025. 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kang SH, Fukaya M, Yang JK, Rothstein JD, and Bergles DE (2010). NG2+ CNS glial progenitors remain committed to the oligodendrocyte lineage in postnatal life and following neurodegeneration. Neuron 68, 668–681. 10.1016/j.neuron.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Muzumdar MD, Tasic B, Miyamichi K, Li L, and Luo L (2007). A global double-fluorescent Cre reporter mouse. Genesis 45, 593–605. 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 66.Voehringer D, Liang HE, and Locksley RM (2008). Homeostasis and effector function of lymphopenia-induced “memory-like” T cells in constitutively T cell-depleted mice. J Immunol 180, 4742–4753. 10.4049/jimmunol.180.7.4742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140. 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Larsson J, Goumans MJ, Sjostrand LJ, van Rooijen MA, Ward D, Leveen P, Xu X, ten Dijke P, Mummery CL, and Karlsson S (2001). Abnormal angiogenesis but intact hematopoietic potential in TGF-beta type I receptor-deficient mice. EMBO J 20, 1663–1673. 10.1093/emboj/20.7.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pokrass MJ, Ryan KA, Xin T, Pielstick B, Timp W, Greco V, and Regot S (2020). Cell-Cycle-Dependent ERK Signaling Dynamics Direct Fate Specification in the Mammalian Preimplantation Embryo. Dev Cell 55, 328–340 e325. 10.1016/j.devcel.2020.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The entire dermal papilla at late-growth is 3D reconstructed based on its membrane-tdTomato signal. Within the red outlined dermal papilla, cell bodies of individual fibroblasts are rendered into surfaces with different colors. Further, a single fibroblast is 3D reconstructed based on its membrane-GFP signal. The cell body of this fibroblast is rendered into a pink surface and its entire membrane in a green surface.
Representative z-stack movies of immunostained Sox2 and Integrin alpha9 (Itgα9) (green) in ear skin whole mounts to detect fibroblasts with dermal papilla identity at different stages. Fibroblasts are labeled in red by Pdgfrα-CreER; LSL-tdTomato. Scale bar sizes are indicated. See also Figure S3A–D.
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.


