Abstract
In Pseudomonas aeruginosa (P. aeruginosa), transcription factors (TFs) are important mediators in the genetic regulation of adaptability and pathogenicity to respond to multiple environmental stresses and host defences. The P. aeruginosa genome harbours 371 putative TFs; of these, about 70 have been shown to regulate virulence-associated phenotypes by binding to the promoters of their target genes. Over the past three decades, several techniques have been applied to identify TF binding sites on the P. aeruginosa genome, and an atlas of TF binding patterns has been mapped. The virulence-associated regulons of TFs show complex crosstalk in P. aeruginosa's regulatory network. In this review, we summarise the recent literature on TF regulatory networks involved in the quorum-sensing system, biofilm formation, pyocyanin synthesis, motility, the type III secretion system, the type VI secretion system, and oxidative stress responses. We discuss future perspectives that could provide insights and targets for preventing clinical infections caused by P. aeruginosa based on the global regulatory network of transcriptional regulators.
Keywords: Crosstalk, Pseudomonas aeruginosa, Regulatory network, Transcriptional regulators, Virulence
Introduction
Pseudomonas aeruginosa (P. aeruginosa) is a gram-negative, opportunistic, human-infecting pathogen that can cause chronic lung infections in susceptible hosts, lung inflammation, infections of soft tissues and burn wounds and other types of infection.1 P. aeruginosa accounts for 10% of nosocomial infections.2 Chronic infections and lung inflammation caused by P. aeruginosa are key causes of death in patients with cystic fibrosis.1 The respiratory tract is a frequent site of infection with a wide range of viruses, bacteria, or co-infection. The microbiota promotes resistance to respiratory infection. Viral or bacterial infections can induce alterations of the microbiome and secondary bacterial pneumonia.3 The invasion of pathogenic bacteria or viruses also affects the ability of each other to infect. Respiratory viruses (such as influenza, respiratory syncytial virus, and parainfluenza) co-infected with P. aeruginosa have a higher prevalence (16.6%) and death rate (10.5%) among the patients compared with the infection without P. aeruginosa.4 P. aeruginosa isolates with extensive biofilm production also enhance their antibiotic resistance and in vivo colonization in COVID-19 patients.5 P. aeruginosa employs diverse metabolic and virulence systems, such as the quorum-sensing system (QS), type III secretion system (T3SS), type VI secretion system (T6SS), flagella and type IV pili, to adapt to internal and external environmental challenges.6,7,8 Other virulence factors (e.g., biofilm, exopolysaccharide, lipopolysaccharide, elastase, protease, esterase and rhamnolipids) form a highly complex but ordered virulence system.9, 10, 11, 12 Although investigations of the pathogenic mechanisms of P. aeruginosa have led to the development of therapeutic approaches, only a few drugs have been approved to treat P. aeruginosa infections.13, 14, 15, 16, 17 We previously proposed that targeting key virulence regulators has great potential for developing novel anti-P. aeruginosa drugs.18 Here, we review the literature on P. aeruginosa's integrative virulence-associated regulatory networks, which are mediated by a group of transcription factors (TFs).
DNA-binding TFs play a crucial role in mediating the adaptability and pathogenicity of pathogenic bacteria. The P. aeruginosa genome harbours 371 putative TFs that belong to 29 families, such as the LysR family (113 TFs), Arc family (56 TFs), LuxR family (30 TFs), OmpR family (24 TFs), TetR family (23 TFs) and GntR family (23 TFs).19 Several experimental techniques, such as chromatin immunoprecipitation in conjunction with protein-binding microarray (PBM),20 DNA affinity purification sequencing (DAP-seq),21, 22, 23 chromatin immunoprecipitation coupled with high-throughput sequencing (ChIP-seq),24, 25, 26, 27, 28, 29 high-throughput analysis of in vitro protein–DNA interactions using massively parallel sequencing (Bind-n-seq)30 and high-throughput systematic evolution of ligands by exponential enrichment (HT-SELEX) assay, have been applied to study the bacterial TFs of P. aeruginosa.31 We and our collaborators have characterized eight regulators (VqsR, VqsM, AlgR, CdpR, CysB, RsaL, AnvM and RpoN) and elucidated their molecular pathogenic mechanisms.25,27,28,32, 33, 34, 35, 36 A genomic virulence-associated network containing 20 key TFs and their targets is mapped, providing an online analysis platform for peer researchers.26 An online regulatory-interaction database of P. aeruginosa, named RegulomePA, is also developed.37 RegulomePA contains 4,827 interactions stemming from the interactions of 27.27% of TFs and 54.16% of TFs with their target genes (50.8% of the total genes). An additional transcriptional regulatory network containing 364 transcriptomes of P. aeruginosa, identifying 104 independently modulated sets of genes involved in biosynthetic gene clusters and associated secretion systems, was reconstructed.38 We recently mapped an atlas of the binding specificities of 182 TFs in P. aeruginosa using HT-SELEX.31 In addition, we construct nine virulence-associated regulatory networks and found 32 newly identified TFs involved in multiple virulence pathways and physiological processes. More recently, the regulatory atlas of two-component systems (TCSs) containing 55 TCS response regulators is mapped using DAP-seq, revealing numerous new regulatory interactions in several virulence-associated and metabolic pathways.22
In this review, we integrate the virulence-associated transcriptional regulators, especially TFs, and map seven regulatory networks, including the QS, pyocyanin synthesis, motility, biofilm formation, T3SS, T6SS and oxidative stress response networks. We also discuss the challenges and trends in studying the pathogenesis, drug targets and infection prevention of P. aeruginosa, which will provide insights for preventing clinical infections caused by P. aeruginosa based on the global regulatory network of transcription regulators.
QS-related regulatory networks control P. aeruginosa communication and virulence
The QS of P. aeruginosa, which controls most virulence factors, has been continuously studied for three decades. Although virulence phenotypes are mainly determined by four QSs (Las, Rhl, Pqs and Iqs),12,39 a group of regulators mediates the integrated regulatory network-associated QS. For instance, LasR, acting as the master regulator of the QS, directly controls the gene expression of Las, Rhl, Pqs, and Iqs.3,40, 41, 42, 43, 44 LasR binds to the lasI promoter to activate the production of 3-oxo-dodecanoyl-HSL (C12). C12 can bind to the N-terminal of LasR to enhance the DNA-binding affinity of LasR to the lasI promoter as well as the promoters of Rhl and Pqs via a positive-feedback mechanism. LasR also directly binds the ambB-E promoter to control IQS.20 Vfr and AmpR specifically regulate the expression of lasR.45,46 PtxR inhibits the transcription levels of the pqsA-E operon and rhlI and activates lasI.47 QscR inhibits the expression of lasI and thus delays the transcription of QS genes.48 QteE can lock the QS by destabilizing LasR and independently reducing RhlR levels when the cell density (OD600) is higher than 2.3.49 In addition, CspC can upregulate Las-QS by binding to the 5′ UTR (untranslated regions) of rsaL mRNA.50
We and our collaborators have determined the direct binding sites of eight QS-related regulators, VqsR, VqsM, AlgR, CdpR, RsaL, CysB, AnvM and RpoN.25,27,28,32, 33, 34, 35, 36 The functions of these regulators overlap with other virulence-related pathways. For instance, VqsM simultaneously mediates Las-QS, the T3SS and antibiotic resistance by binding to the promoters of lasI, exsA, and nfxB, respectively.28 CdpR, an important QS regulator, interacts with the ClpAS-P system to regulate virulence factors and pathogenicity.34 RpoN has a direct regulatory effect on all three QS systems as well as T6SS.27 AnvM, encoding a hypothetical protein, mediates pathogenicity under anaerobic conditions and host defences.51
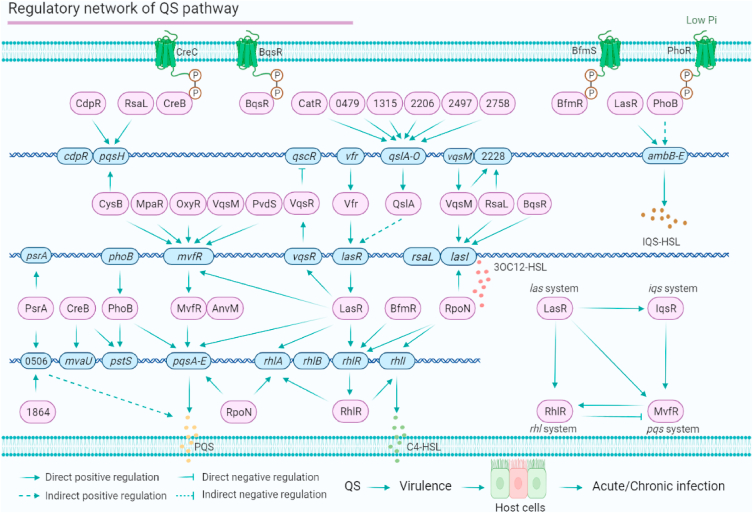
We used a genome-wide network-based approach to uncover the crosstalk of 20 key virulence-related TFs and constructed a genomic regulatory network (PAGnet) that includes 347 direct target genes and their regulatory relationships.26 Our HT-SELEX data show that PA2497 directly regulates the expression of qslA, which encodes a QS anti-activator. QslA is a negative regulator of multiple virulence phenotypes, including QS (lasR, rhlR and rhlI), H2-T6SS (hsiA2), T2SS (xcpP), elastase and pyocyanin in PAO1.52 PA2206, PA2758, PA1315, PA0479, and CatR are identified as upstream regulators of qslA,31 revealing a new regulatory network upstream of the QS. PA1864 and PsrA directly co-regulate PA0506, which encodes a probable acyl-CoA dehydrogenase involved in PQS,31,53,54 CreB can bind to the promoter of pstS. Thus, we conclude that the QS architecture is controlled by a regulatory network containing 28 regulators (Fig. 1). Among these QS-related regulators, 12 co-target genes (pqsH, qslA, PA2228, mvfR, lasR, lasI, PA0506, pstS, pqsA, rhlA, rhlR and rhlI) are regulated by at least two regulators, indicating multiple co-regulatory networks in the QS (Fig. 1). For example, qslA is co-regulated by six TFs, and mvfR is co-regulated by five TFs.
Figure 1.
The QS pathway regulatory network of P. aeruginosa. The QS consists of four systems, the Las system, Rhl system, Pqs system and Iqs system, which are regulated by more than 28 transcriptional regulators with daedal networks. This regulatory network indicates the binding target genes and the crosstalk among these regulators.
Pyocyanin biosynthesis is controlled by 19 TFs
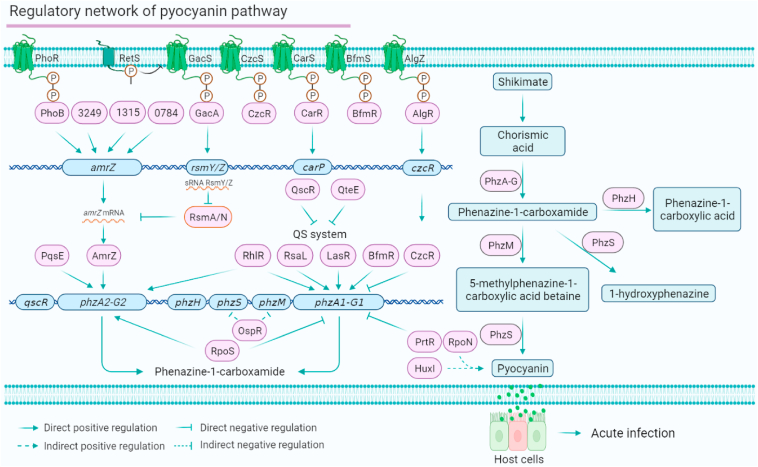
Phenazines are a class of antibiotics secreted by multiple Pseudomonas spp., displaying a broad spectrum of antimicrobial activity to various organisms.12 The phenazine blue pyocyanin is only secreted by P. aeruginosa. Pyocyanin is one of the key virulence factors in P. aeruginosa and is involved in both acute and chronic infections.55,56 Pyocyanin biosynthesis is encoded by two gene clusters, phzA1-phzG1 and phzA2-phzG2 (Fig. 2). It has been reported that phzA1 is directly regulated by LasR, RhlR, RsaL, BfmR and CzcR,22,57,58 and phzA2 is directly bound by RhlR and AmrZ.29 Some pyocyanin repressors, such as QscR, PtxR, QteE and sigma factor RpoS, have also been reported. QscR, a quorum-sensing control repressor, negatively regulates pyocyanin production.48 PtxR only inhibits phzA1-G1 expression but not phzA2-G2 expression.47 Our SELEX assay showed that PA1314 directly binds to the promoter of phzH (encoding a potential phenazine-modifying enzyme).31 CreB binds to the promoter of mvaU, which is involved in pyocyanin synthesis.59 HuxI activates the production of pyocyanin.60 QteE inhibits pyocyanin production and QS-related phenotypes in P. aeruginosa.61 RpoS inhibits the expression of phzA1 but activates the expression of phzA2 at the post-transcriptional level, which might be due to the diverse host environments.36 Because the production of pyocyanin is regulated by QS, the crosstalk among multiple pyocyanin-related regulators is common in P. aeruginosa's regulatory network. For instance, LasR, RhlR, RsaL, BfmR, CzcR, PtxR and QteE share their targets in the QS, T3SS and T6SS. In conclusion, the two gene clusters (phzA1-G1 and phzA2-G2) involved in pyocyanin biosynthesis are stringently controlled by these global regulators (Fig. 2). Similar to biocontrol bacteria, such as Bacillus spp., Lysobacter spp. and Pseudomonas spp., pyocyanin produced by P. aeruginosa can also be called a ‘long-range weapon’ as it is indirectly toxic to host cells, independent of cell–cell contact.62
Figure 2.
The pyocyanin synthesis pathway regulatory network of P. aeruginosa. (Left panel) The pyocyanin synthesis regulatory network is mediated by 19 regulators (PhoB, PA3249, PA1315, PA0784, GacA, CarR, PqsE, AmrZ, QscR, QteE, RhlR, RsaL, LasR, BfmR, CzcR, RpoS, PrtR, AlgR and RsmA/N) and three co-target genes (amrZ, phzA1-G1 and phzA2-G2). (Right panel) The biosynthesis pathway of phenazines (including pyocyanin, 1-hydroxyphenazine and phenazine-1-carboxylic acid).
P. aeruginosa employs a regulatory network to regulate motility
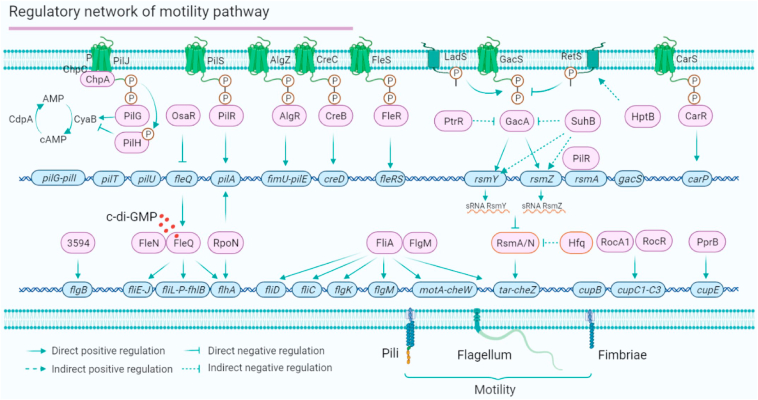
P. aeruginosa has a full set of motility-related genes that control flagella and type IV pili, which are required for swarming, swimming, and twitching motility. Six TCSs and a chemosensory system are involved in the motility-related regulatory network, including FleS/R, GacS/A, CreC/B, CarS/R, PilS/R, AlgZ/AlgR and the Chp system (Fig. 3). In the motility regulatory network, the Gac/Rsm signalling pathway functions as a global regulatory system that mediates P. aeruginosa motility. PtrR, SuhB and HptB inhibit the GacA/S system. Sigma factors and anti-sigma factors provide another level of motility regulation. Sigma factor FliA directly binds to the promoters of fliD, fliC, flgK, motA-cheW operon and tar-cheZ operon.63,64 The function of FliA is also affected by the anti-sigma factor FlgM.65 The sigma factor and global regulator RpoN directly regulate the expression of pilA and flhA to control flagellar motility. Vfr directly binds to the promotor of fleQ (encoding the major flagellar regulator).66 FleQ binds to the promoters of flhA, fliE-J operon and fliL-flhB operon. The binding affinity of FleQ is mediated by the anti-sigma factor FleN and the intracellular c-di-GMP level.67 PA3594 can directly regulate the transcription of flgB.31 Thus, 16 regulators directly regulate the motility behaviours of P. aeruginosa (Fig. 3). Three target genes (such as pilA, flhA and tar-cheZ) are co-regulated by at least two regulators, such as FleQ, RpoN, FliA and RsmA/N, which have been proposed to be master regulators in the P. aeruginosa motility regulatory network.
Figure 3.
The motility regulatory network of P. aeruginosa. P. aeruginosa motility includes swarming, swimming and twitching. Swarming motility is driven by flagella, which are controlled by FleQ, two sigma factors (FliA and RpoN), two anti-sigma factors (FleN and FlgM), PA3594, and the Gac/Rsm signaling system. Swimming and twitching motility are driven by the type VI pili, which is regulated by PilRS, AlgRZ, RpoN and the Chp system. RocA1/RocR and PprB regulate the fimbriae, which are required for adherence. The motility network is mediated by 16 direct transcription regulators (such as FleQ, RpoN and PilR) and shares three co-target genes (rsmY/Z, flhA and tar-cheZ).
Twenty-eight TFs form a complex biofilm-related regulatory network
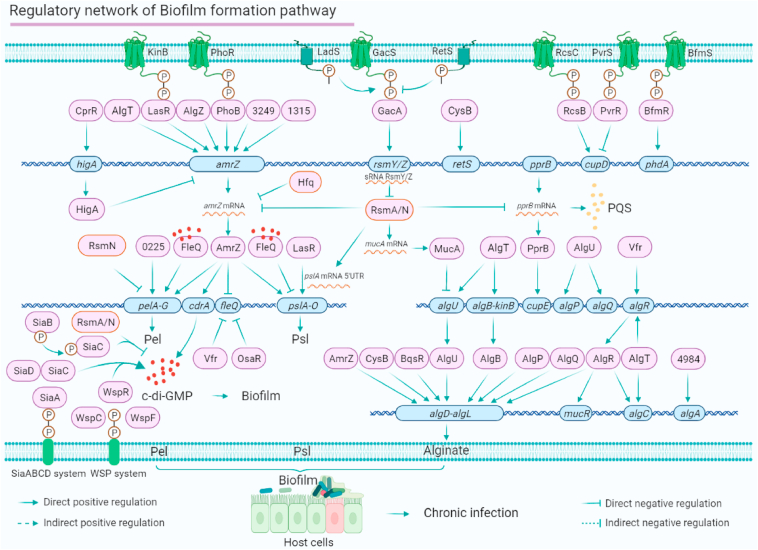
P. aeruginosa biofilm formation is one of the chief culprits of chronic infection in the respiratory tract of cystic fibrosis patients and has adaptive resistance to antibiotics in hospitals. P. aeruginosa biofilms act as a helper to improve the living environment and assist the virulence system to adapt to the complex microbial gaseous-fluid environment of the respiratory tract. P. aeruginosa biofilm formation is coordinated by multiple mechanisms involving more than 20 regulators, such as PhoB, GacA, AmrZ, FleQ, RsmA/N, BfmR, MucA, AlgU, AlgR, QsaR, PA4547 and PprB. These regulators have been individually studied, and their crosstalk has also been analysed.18,26 BfmR positively regulates biofilm formation and cell death by activating the expression of phdA, which encodes prevent-host-death protein A.68 All of these regulators are involved in TCS, QS, and c-di-GMP networks, which respond to the regulation and synthesis of biofilm matrix components alginate, Psl and Pel (Fig. 4).
Figure 4.
The biofilm formation pathway regulatory network of P. aeruginosa. The P. aeruginosa biofilm matrix mainly consists of three kinds of extracellular polysaccharides (Pel, Psl and alginate extracellular polysaccharide), which are synthesised by the pelA-G operon, pslA-O operon and algD-L operon, and is regulated by 29 direct transcription regulators. We have summarised the direct regulators involved in biofilm formation and re-mapped the regulatory network underlying these regulators. The network shows 10 co-target genes: amrZ, cupD, pelA-G, fleQ, pslA-O, algU, algR, algA, algD-L and algC.
The production of Psl and Pel is dynamically regulated by multiple virulence-related TFs. In Figure 4, we map the P. aeruginosa biofilm regulatory network, visualising the whole atlas constructed by several virulence-related TFs. The Gac/Rsm signalling pathway is the master mediator of P. aeruginosa biofilm formation. GacS senses the environmental signals to rapidly activate its kinase activity and then transfers its phosphate group to GacA. The phosphorylated GacA directly controls the expression of small RNAs (RsmY and RsmZ) that directly bind to RsmA to form a RsmYZ–RsmA complex. RsmYZ–RsmA subsequently binds to the mRNA of amrZ or the 5′ UTR of pslA mRNA to inhibit Psl production. The pslA promoter is also bound by LasR and the sigma factor RpoS. AmrZ binds to the pelA promoter to directly control Pel production. The expression of the pelA-G operon is positively regulated by PA0225.69 The PA0225-deletion mutant produces less biofilm than the wild-type strain PAO1. The intracellular c-di-GMP concentration dynamically regulates P. aeruginosa biofilm formation. The chemotactic system Wsp controls c-di-GMP production by sensing the extracellular environmental signals. WspR, a diguanylate cyclase, can receive phosphate groups from the Wsp system to enhance the intracellular concentration of c-di-GMP. Meanwhile, AmrZ directly regulates cdrA, which encodes an intracellular c-di-GMP concentration reporter. Interestingly, OsaR directly binds to the promoter of fleQ and inhibits its expression.70 FleQ is a master regulator in the c-di-GMP-controlled biofilm pathway.71,72 FleQ binds to the promoters of pelA, pslA and cdrA. FleQ and c-di-GMP can form a complex that mediates the bacterial behaviour between motility and biofilm formation. At low c-di-GMP concentrations, FleQ which is not bound by c-di-GMP interacts with the promoter of flagellar genes (such as fleS and fliF) to activate the flagella to promote bacterial motility.71 At high c-di-GMP concentrations, more c-di-GMP/FleQ complex is formed, which shows stronger affinity to the promoters of biofilm synthesis genes, such as pslA and pelA.71 As a result, c-di-GMP/FleQ positively regulates the expression of the pel operon, and negatively regulates the psl operon, showing that c-di-GMP/FleQ is a double-edged sword for biofilm formation.71,73
Alginate biosynthesis is another major biofilm matrix component that is directly regulated by eight TFs. RsmA binds to the mucA mRNA to control the expression of MucA, which can directly bind to the promoter of algU.74,75 AlgU directly controls alginate biosynthesis by binding to the algD-L operon promoter. More than 10 TFs directly regulate the transcription activity of algD. Among them, the global regulator AmrZ binds to the algD promoter. TCS response regulators AlgB, AlgR and BqsR also directly control algD expression via the various signals transmitted by their corresponding sensor kinases. The TCS AlgB-KinB is required for biofilm formation in different regulatory pathways.76,77 Meanwhile, sigma factors AlgU, AlgT and RhlR co-regulate algB-kinB expression. Other alginate-related genes can be directly regulated by several TFs. For instance, algA is regulated by PA4984, and AlgU co-regulates the expression of algP and algQ.
Adherence is an early step of biofilm formation. Adhesive factors required for biofilm formation are globally regulated by RsmA/N, which binds to pprB mRNA to positively regulate its expression.78 PprB regulates bacterial adherence and biofilm maturation by directly binding to the cupE promoter. TCSs RcsBC and PvrRS show opposite effects on cupD expression,79 which may be caused by their opposite sensing signals. Here, we show that 30 regulators can directly regulate the expression of biofilm-related genes (Fig. 4). Additionally, 10 co-target genes, amrZ, cupD, pelA-G, fleQ, pslA-O, algU, algR, algD-L, algC and algA, are included in this network (Fig. 4).
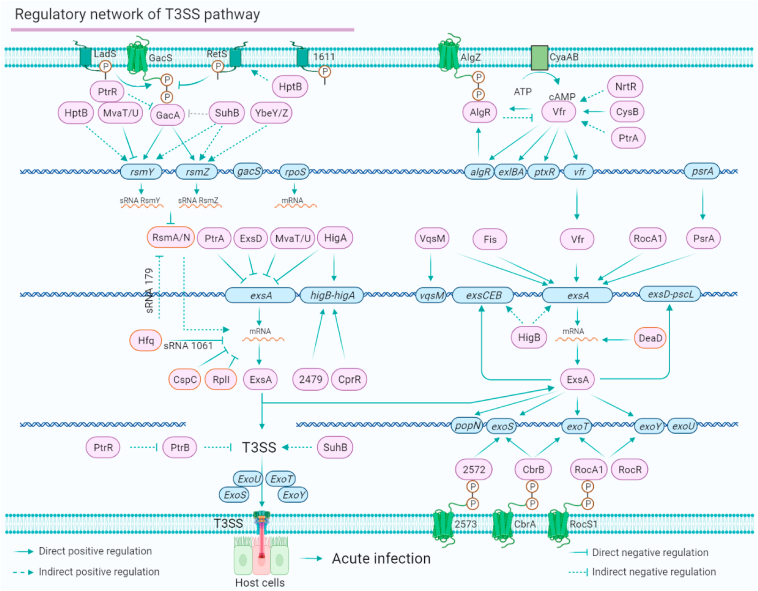
The regulatory network of the T3SS mediates P. aeruginosa virulence
P. aeruginosa injects T3SS exoenzyme effectors into host cells, causing ventilator-associated pneumonia.80, 81, 82 Although previous studies have investigated the regulation of the Vfr–ExsA–T3SS pathway and the functions of effectors, the complete regulatory network of the T3SS has yet to be uncovered. Here, we map an up-to-date regulatory network of the T3SS in P. aeruginosa based on the direct regulation between TFs and their targets involved in the T3SS. In the complex T3SS regulatory network, a group of TFs play important roles in mediating T3SS gene transcription, post-transcriptional modification and effector secretion. As shown in Figure 5, the T3SS is regulated by two main pathways, namely the Gac/Rsm–T3SS pathway and the Vfr–ExsA–T3SS pathway. In the Gac/Rsm–T3SS pathway, the global regulator GacA is strictly regulated by its sensor kinase GacS, which can be inhibited by RetS, but HptB (histidine phosphotransfer protein) is an activator of RetS.83 PtrR and SuhB are two GacA repressors. MvaT/U represses the expression of rsmY,84, 85, 86 and rsmZ is controlled by GacA, SuhB, and YbeY/Z.87 In the Vfr–ExsA–T3SS pathway, the DNA-binding affinity of Vfr is affected by the level of cAMP, which is controlled by CyaAB. Vfr is also positively regulated by NrtR,88 CysB and AlgR.
Figure 5.
The T3SS pathway regulatory network of P. aeruginosa. The T3SS in P. aeruginosa is regulated by two main regulatory signalling systems, Gac/Rsm-ExsA-T3SS and cAMP/Vfr-ExsA-T3SS. ExsA is the master regulator of the T3SS. The expression of ExsA is inhibited by PtrA, ExsD, MvaT/U and HigA at the transcriptional level and by Hfq, CspC and RplI at the post-transcriptional level. ExsA acts as a co-target that can be directly activated by five regulators, VqsM, Fis, Vfr, RocA1 and PsrA. The downstream T3SS effectors (such as exoS, exoT and exoY) are also directly regulated by ExsA, PA2572/PA2573, CbrAB and RocS1/A1. The T3SS is directly mediated by 21 direct regulators and six co-target genes (rsmY/Z, exsA, higB-A, exoS, exoT and exoY).
As the master regulator of the T3SS in P. aeruginosa, the self-feedback TF ExsA is mediated by six positive regulators (Vfr, VqsM, Fis, RcoA1, PsrA, and HigB) and four negative regulators (PtrA, ExsD, MvaT/U, and HigA) at the transcriptional level. ExsA is positively regulated by DeaD89 but negatively regulated by Hfq,90 CspC,91 and RplI69 at the translational level. In addition, the transcription of T3SS effectors can be directly regulated by a group of TFs. TCSs PA2573/PA2572 and CbrAB directly regulate the transcription of exoS.22 RocS1-RocA1/RocR and CbrAB directly regulate the transcription of exoT, and PA4080 directly regulates the expression of exoU.22 Therefore, the T3SS is mediated by a complex TF-dependent network that controls the T3SS device assembly, gene regulation, and effector secretion by responding to signals from host cells (Fig. 5). Twenty-one regulators directly regulate the expression of T3SS genes. Six target genes (rsmY/Z, exsA, higB-A, exoS, exoT, and exoY) are co-regulated by more than two regulators.
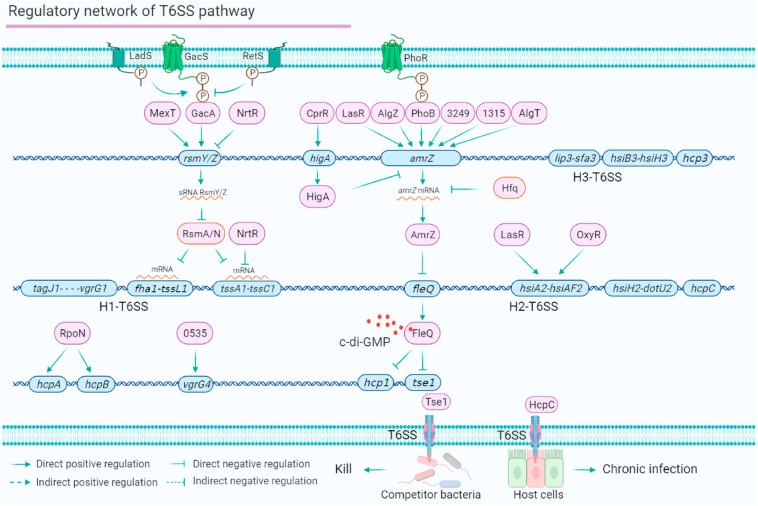
The T6SS regulatory network enhances P. aeruginosa virulence and competitiveness
The bacterial T6SS is a powerful weapon that injects toxic effectors into eukaryotic host cells and prokaryotic competitor bacteria, causing host damage or providing a competitive advantage in diverse microbial environments. The P. aeruginosa genome harbours three T6SS gene islands,92 H1-T6SS, H2-T6SS, and H3-T6SS. Here, we review the TF-dependent networks involved in T6SS regulation (Fig. 6). First, the T6SS is mainly regulated by the Gac/Rsm-T6SS signalling system. The RNA-binding proteins RsmA/N directly bind to tssA1 and fha1 mRNAs.78,93 Meanwhile, NrtR inhibits H1-T6SS expression by binding to the promoters of rsmY, rsmZ and tssA1.94 Second, the T6SS is regulated by the PhoB-AmrZ-T6SS pathway. AmrZ is regulated by a group of TFs, including PhoB, PA3249, PA1315, AlgZ and AlgT. AmrZ directly binds to the promoter of fleQ, and FleQ directly inhibits the expression of two H1-T6SS effector genes (hcp1 and tse1) via a c-di-GMP-dependent mechanism.95 In addition, our recent study reveals that AnvM positively regulates the expression of hsiA2,51 suggesting the regulatory role of AnvM in H2-T6SS. RpoN directly binds to the promotors of T6SS effectors hcpA and hcpB to positively regulate their expression.27 Additionally, PA0535 directly regulates the expression of vgrG4. Eight regulators (RsmA/N, NrtR, AmrZ, LasR, OxyR, RpoN, PA0535 and FleQ) directly regulate T6SS genes (Fig. 6). The T6SS is directly mediated by relatively few regulators compared with the regulatory networks described in previous sections. These TFs are regulators of H1-T6SS and H2-T6SS; however, the regulators involved in H3-T6SS remain largely elusive.
Figure 6.
The T6SS pathway regulatory network of P. aeruginosa. The T6SS signalling system is mainly controlled by Gac/Rsm-H1-T6SS and AmrZ-FleQ-H1-T6SS, and is responsible for providing competitive advantages in environments with diverse bacteria. RpoN directly binds to the promoter of two effector genes (hcpA and hcpB). H2-T6SS is directly regulated by LasR and OxyR. Effector HcpC can be secreted in host cells to cause chronic infection. The T6SS network is mediated by 18 regulators and four co-target genes (rsmY/Z, tssA1-C1, amrZ and hisA2-F2).
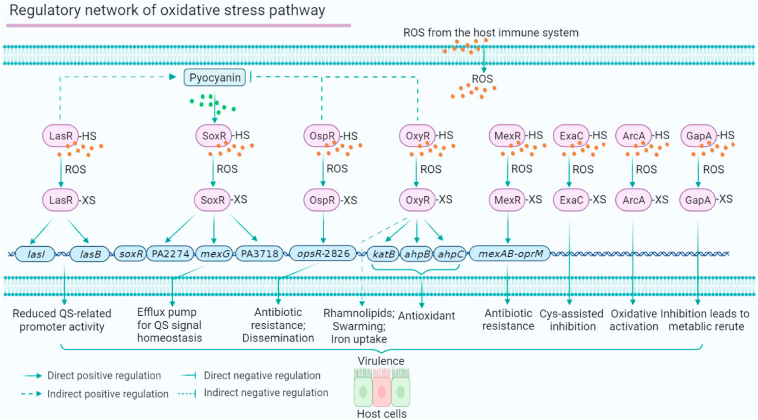
The regulatory network of P. aeruginosa oxidative stress response
To maintain sustained virulence, bacterial pathogens must overcome the reactive oxygen species (ROS) barriers produced by the host immune system, such as H2O2 and –OH. Many ROS-sensing components have been reported, including various ROS-deactivating enzymes, such as catalase, superoxide dismutase, thioredoxin, and glutaredoxin. Pathogenic bacteria use multiple TFs that harbour ROS-sensing cysteines, such as OxyR, MgrA, OhrR, and SarA, to respond to and overcome host-derived ROS.96,97,98 In P. aeruginosa, five regulators and three metabolic proteins have been reported to sense oxidation stress, including LasR, SoxR, MexR, OxyR, OspR, ExaC, ArcA and GapA (Fig. 7). SoxR directly regulates the expression of three genes, including mexGHI-ompD operon (involved in QS signal homeostasis), PA3718, and PA2274.99 SoxR can be activated by the redox-active antibiotic pyocyanin.100 OxyR directly upregulates antioxidant genes to defend against H2O2 stress as well as iron uptake.101,102 OxyR also positively regulates swarming motility and rhamnolipid production and negatively regulates pyocyanin production, which may have a protective effect on the oxyR mutant.103 OspR is also found to regulate phenazine biosynthesis, beta-lactam resistance, and dissemination in a mouse acute infection model.104 The other oxidation-sensing regulator, MexR, regulates antibiotic resistance by binding to the promoter of the mexAB-oprM operon.105 In addition, we have discovered ∼200 ROS-sensing proteins in P. aeruginosa and identified four roles of oxidation-sensitive cysteines: i) Cys79 is required for the oxidation response of LasR; ii) H2O2 inhibits the activity of ExaC; iii) H2O2 activates the activity of ArcA; iv) GapA functions as a redox-sensing metabolic switch. Pathogenic bacteria use multi-layered responses to ROS to rapidly adapt to oxidative stress.106 Cys201 and Cys203 are also required for LasR to sense oxidative stress.107,108 Taken together, the ROS regulatory network is essential for maintaining P. aeruginosa virulence and overlaps with metabolic pathways and antibiotic resistance.
Figure 7.
The oxidative stress response regulatory network of P. aeruginosa. Five TFs (LasR, SoxR, MexR, OxyR and OspR) involved in the QS, antibiotic resistance and motility sense ROS and translocate the signals to the downstream virulence-related phenotypes. Three metabolic proteins (ExaC, ArcA and GapA) also sense and respond to ROS from the host immune system.
SELEX and DAP-seq predict interactions between novel TFs and virulence
Although the regulatory atlas of virulence-related transcription regulators in P. aeruginosa has been studied for more than three decades, the direct and specific binding targets and binding patterns have still not been fully uncovered. Recently, we have used an HT-SELEX assay to predict 32 uncharacterized TFs with new regulatory roles in multiple virulence pathways.31 Moreover, the binding motifs of 55 TCS response regulators and their crosstalk were revealed using DAP-seq.22 Here, we summarise the potential TF-related regulatory network involved in these virulence pathways. We mainly focus on the predicted regulatory networks involved in the QS, biofilm formation, motility, T3SS, and T6SS.
We recently predict the new functions of five TFs involved in the QS.31 For example, PA1241 is identified as a potential QS regulator that binds to the promoters of anr, anvM, pqsH, and cdpR. SouR can bind to the promoters of lecB, cdpR, pqsH, phzA2 and lasR, suggesting potential roles in the QS. DAP-seq also reveals that 14 response regulators are involved in QS regulation.22 For example, the mvfR promoter harbours the binding motifs of BqsR, KdpE, and AmgR.22 CzcR is predicted to bind to the phzA1 promoter, and seven TFs are predicted to mediate biofilm formation.31 rsmY/Z was predicted to be regulated by a group of TFs identified by DAP-seq, including CopR, PirR, AgtR and CzcR.22 The new functions of the 10 TFs involved in T6SS are predicted by SELEX.31 For example, SouR is predicted to bind to the promoters of tssA1, fha1, and hsiH2. PA1241 is predicted to bind to the promoters of anvM, hcpB, hsiA2, and lipA. Eight TFs are predicted to regulate motility.31 DAP-seq results indicate that PirR is a direct regulator of fleQ.22 DAP-seq also suggests new functions of 7 TFs related to T3SS. RocA1, ErbR, BfmR, PilR, PA4080, AgtR and PirR have conserved potential binding sites in the exsA promoter of P. aeruginosa PA14.22 In summary, SELEX and DAP-seq predict the involvement of 19, 13, 10, 15, and 7 potential TFs in the pathways of the QS, biofilm formation, T6SS, motility, and T3SS, respectively.
Conclusions and future perspectives
In this review, we summarise the seven main virulence-associated regulatory networks of P. aeruginosa, the QS, pyocyanin synthesis, motility, biofilm formation, T3SS, T6SS and oxidative stress response networks, revealing the global network and crosstalk between multiple pathways. We conclude that the virulence regulatory network is dynamically regulated according to environmental signals and the host immune system. The binding sites for half of the TFs have been identified using technologies such as PBM, ChIP-seq, HT-SELEX, Bind-n-seq, and DAP-seq,22,26,31 but the binding sites of the remaining TFs remain elusive.
Nevertheless, the abovementioned technologies each have shortcomings: i) ChIP-seq data are limited by antibody specificity and indirect protein-DNA binding; ii) HT-SELEX and Bind-n-seq data do not generate native genomic motif-generating sequences and lack secondary DNA modifications in vitro; and iii) although DAP-seq uses the genomic DNA library, the binding reaction is performed in vitro. Therefore, it is worth exploring how to accurately identify binding sites by establishing in vitro bacterial cell models that better mimic in vivo environments. For instance, TFs can be expressed in the native bacterial cells, and cell lysates can be used as reaction conditions for binding between TFs and the genomic DNA library. This strategy not only mimics the in vivo environment but identifies transcriptional enhancers, which are short DNA sequences with TF-binding sites.109 TF binding may be controlled by specific conditions, such as carbon or nitrogen resources, microelements, ROS, and host defences. A recent review noted that when host cells perceive pathogen invasion, immune cells affect a change in the metabolic pathway.110 Immune cells promote anti-inflammatory and antibacterial reactions, which can act as host immune signals to activate virulence gene expression in pathogens. This interaction between immune cells and pathogens suggests that the functional study of P. aeruginosa TFs and pathogenic mechanisms should not be limited to the culture environment or in vitro biochemical reactions but also explore co-interactions with host immune cells. A comprehensive analysis of binding sites and transcription regulator functions would provide new insights into P. aeruginosa virulence-associated and metabolic networks.
Based on these virulence-associated TFs, we propose the following directions to develop effective strategies against P. aeruginosa infection. i) Metabolic networks are related to virulence-associated pathways in P. aeruginosa,111,112 thus, targeting metabolic networks is a promising antimicrobial strategy.111,113, 114, 115 ii) Targeting virulence-associated regulators is another approach to prevent infections caused by P. aeruginosa.18 High-throughput screening assays based on microtiter plates or transcriptional reporters are well used for identifying new drugs to target specific virulence-associated TFs.116, 117, 118, 119, 120, 121 In addition, in silico computational screening method is also a good approach to identify new inhibitors for virulence factors, such as LasB, LasR, PqsA, PqsR, and SagS.122, 123, 124, 125, 126, 127, 128 A master regulator analysis based on the virulence networks (such as PAGnet) can be used to prioritize these transcription regulators.26 The obtained master regulators can be preferentially used to screen antimicrobial drugs via the high-throughput screening assays abovementioned. The antimicrobial activities of these candidate drugs can be evaluated by phenotypic analysis. Besides, the combined therapy of candidate drugs and existing antibiotics may have a more synergistic effect on P. aeruginosa infections. iii) The regulation of post-transcriptional modification is a promising technique for preventing infections caused by prokaryotic pathogens. Inducing RNA methylation, such as by N6-methyladenosine, is another possible approach for overcoming the virulence of P. aeruginosa.129 iv) Secondary structure modifications (such as G-quadruplex) provide new perspectives for studying the infections caused by P. aeruginosa. The specific secondary structures of RNA affect the transcription and translation of virulence genes. One hundred and sixty-one RNA G-quadruplex (rG4) sites at the genome-wide level have been identified using rG4-seq. Among them, 6.9% of rG4 sites are located in virulence-related genes, such as those involved in the T3SS, T6SS, QS, motility and biofilm formation.130 For instance, rG4 mediates biofilm production and motility by enhancing bswR expression.130 Thus, RNA secondary structure in virulence-related TFs warrant further study. Nevertheless, the above proposed potential strategies are still in laboratory trials and are far from clinical application, especially in the regulation of post-transcriptional and secondary structure modifications. Many QS-deficient variant strains, such as lasR, rpoN, mucA, rhlRI and mexT mutants, have been isolated clinically, and they still remain virulent.3,131, 132, 133, 134, 135, 136, 137, 138, 139 Although QS inhibitors may not be suitable antimicrobial targets for P. aeruginosa clinical isolates, specific anti- P. aeruginosa therapies are needed for these clinical isolates.18 For example, combination therapies of antimicrobials and antivirulents also show promising synergistic anti-biofilm effects.140, 141, 142
Overall, this review summarises the seven main virulence-associated regulatory networks in P. aeruginosa and the direct regulatory network of transcription regulators. Many TFs are predicted to be involved in virulence pathways, which will provide potential insight into new interactions among TFs and target genes. We propose that the whole metabolic network, in combination with post-transcriptional RNA secondary structure, contributes to the regulation of P. aeruginosa virulence.
Conflict of interests
The authors declare no conflict of interests.
Funding
This work was supported by Project of the Dominant Discipline in Jiangsu Province, China (No. 80900246 to X.S.), General Research Fund of Hong Kong, China (No. 11102720, 21103018, 11101619, 11103221 and 11103221 to X.D.), National Natural Science Foundation of China (No. 32272619 to X.S., No. 31870116 and 32172358 to X.D.) and Tung Biomedical Sciences Centre, China (No. 9609313 to X.D.).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Contributor Information
Xiaolong Shao, Email: xlshao@njau.edu.cn.
Xin Deng, Email: xindeng@cityu.edu.hk.
References
- 1.Deretic V., Schurr M.J., Yu H. Pseudomonas aeruginosa, mucoidy and the chronic infection phenotype in cystic fibrosis. Trends Microbiol. 1995;3(9):351–356. doi: 10.1016/s0966-842x(00)88974-x. [DOI] [PubMed] [Google Scholar]
- 2.Impey R.E., Panjikar S., Hall C.J., et al. Identification of two dihydrodipicolinate synthase isoforms from Pseudomonas aeruginosa that differ in allosteric regulation. FEBS J. 2020;287(2):386–400. doi: 10.1111/febs.15014. [DOI] [PubMed] [Google Scholar]
- 3.Kostylev M., Kim D.Y., Smalley N.E., Salukhe I., Greenberg E.P., Dandekar A.A. Evolution of the Pseudomonas aeruginosa quorum-sensing hierarchy. Proc Natl Acad Sci U S A. 2019;116(14):7027–7032. doi: 10.1073/pnas.1819796116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu Y., Ling L., Wong S.H., et al. Outcomes of respiratory viral-bacterial co-infection in adult hospitalized patients. EClinicalMedicine. 2021;37 doi: 10.1016/j.eclinm.2021.100955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qu J., Cai Z., Liu Y., et al. Persistent bacterial coinfection of a COVID-19 patient caused by a genetically adapted Pseudomonas aeruginosa chronic colonizer. Front Cell Infect Microbiol. 2021;11 doi: 10.3389/fcimb.2021.641920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goodman A.L., Kulasekara B., Rietsch A., Boyd D., Smith R.S., Lory S. A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev Cell. 2004;7(5):745–754. doi: 10.1016/j.devcel.2004.08.020. [DOI] [PubMed] [Google Scholar]
- 7.Hauser A.R. The type III secretion system of Pseudomonas aeruginosa: infection by injection. Nat Rev Microbiol. 2009;7(9):654–665. doi: 10.1038/nrmicro2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hogardt M., Heesemann J. Microevolution of Pseudomonas aeruginosa to a chronic pathogen of the cystic fibrosis lung. Curr Top Microbiol Immunol. 2013;358:91–118. doi: 10.1007/82_2011_199. [DOI] [PubMed] [Google Scholar]
- 9.Olejnickova K., Hola V., Ruzicka F. Catheter-related infections caused by Pseudomonas aeruginosa: virulence factors involved and their relationships. Pathog Dis. 2014;72(2):87–94. doi: 10.1111/2049-632X.12188. [DOI] [PubMed] [Google Scholar]
- 10.Schuster M., Greenberg E.P. A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int J Med Microbiol. 2006;296(2–3):73–81. doi: 10.1016/j.ijmm.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 11.Smith R.S., Iglewski B.H.P. Aeruginosa quorum-sensing systems and virulence. Curr Opin Microbiol. 2003;6(1):56–60. doi: 10.1016/s1369-5274(03)00008-0. [DOI] [PubMed] [Google Scholar]
- 12.Jimenez P.N., Koch G., Thompson J.A., Xavier K.B., Cool R.H., Quax W.J. The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev. 2012;76(1):46–65. doi: 10.1128/MMBR.05007-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chastre J., Wunderink R., Prokocimer P., Lee M., Kaniga K., Friedland I. Efficacy and safety of intravenous infusion of doripenem versus imipenem in ventilator-associated pneumonia: a multicenter, randomized study. Crit Care Med. 2008;36(4):1089–1096. doi: 10.1097/CCM.0b013e3181691b99. [DOI] [PubMed] [Google Scholar]
- 14.Luyt C.E., Aubry A., Lu Q., et al. Imipenem, meropenem, or doripenem to treat patients with Pseudomonas aeruginosa ventilator-associated pneumonia. Antimicrob Agents Chemother. 2014;58(3):1372–1380. doi: 10.1128/AAC.02109-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walkty A., Adam H., Baxter M., et al. In vitro activity of plazomicin against 5,015 gram-negative and gram-positive clinical isolates obtained from patients in canadian hospitals as part of the CANWARD study, 2011-2012. Antimicrob Agents Chemother. 2014;58(5):2554–2563. doi: 10.1128/AAC.02744-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pankuch G.A., Lin G., Kubo A., Armstrong E.S., Appelbaum P.C., Kosowska-Shick K. Activity of ACHN-490 tested alone and in combination with other agents against Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2011;55(5):2463–2465. doi: 10.1128/AAC.01390-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cigana C., Bernardini F., Facchini M., et al. Efficacy of the novel antibiotic POL7001 in preclinical models of Pseudomonas aeruginosa pneumonia. Antimicrob Agents Chemother. 2016;60(8):4991–5000. doi: 10.1128/AAC.00390-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao X., Xie Y., Zhang Y., et al. Novel therapeutic strategies for treating Pseudomonas aeruginosa infection. Expet Opin Drug Discov. 2020;15(12):1403–1423. doi: 10.1080/17460441.2020.1803274. [DOI] [PubMed] [Google Scholar]
- 19.Winsor G.L., Griffiths E.J., Lo R., Dhillon B.K., Shay J.A., Brinkman F.S. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44(D1):D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilbert K.B., Kim T.H., Gupta R., Greenberg E.P., Schuster M. Global position analysis of the Pseudomonas aeruginosa quorum-sensing transcription factor LasR. Mol Microbiol. 2009;73(6):1072–1085. doi: 10.1111/j.1365-2958.2009.06832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartlett A., O'Malley R.C., Huang S.C., et al. Mapping genome-wide transcription-factor binding sites using DAP-seq. Nat Protoc. 2017;12(8):1659–1672. doi: 10.1038/nprot.2017.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Trouillon J., Imbert L., Villard A.M., Vernet T., Attrée I., Elsen S. Determination of the two-component systems regulatory network reveals core and accessory regulations across Pseudomonas aeruginosa lineages. Nucleic Acids Res. 2021;49(20):11476–11490. doi: 10.1093/nar/gkab928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trouillon J., Sentausa E., Ragno M., et al. Species-specific recruitment of transcription factors dictates toxin expression. Nucleic Acids Res. 2020;48(5):2388–2400. doi: 10.1093/nar/gkz1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaini S., Lyubetskaya A., Gomes A., et al. Transcription factor binding site mapping using ChIP-seq. Microbiol Spectr. 2014;2(2):1–21. doi: 10.1128/microbiolspec.MGM2-0035-2013. [DOI] [PubMed] [Google Scholar]
- 25.Kang H., Gan J., Zhao J., et al. Crystal structure of Pseudomonas aeruginosa RsaL bound to promoter DNA reaffirms its role as a global regulator involved in quorum-sensing. Nucleic Acids Res. 2017;45(2):699–710. doi: 10.1093/nar/gkw954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang H., Shao X., Xie Y., et al. An integrated genomic regulatory network of virulence-related transcriptional factors in Pseudomonas aeruginosa. Nat Commun. 2019;10(1):2931. doi: 10.1038/s41467-019-10778-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shao X., Zhang X., Zhang Y., et al. RpoN-dependent direct regulation of quorum sensing and the type VI secretion system in Pseudomonas aeruginosa PAO1. J Bacteriol. 2018;200(16):e00205–e00218. doi: 10.1128/JB.00205-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang H., Deng X., Li X., Ye Y., Wu M. Molecular mechanisms of master regulator VqsM mediating quorum-sensing and antibiotic resistance in Pseudomonas aeruginosa. Nucleic Acids Res. 2014;42(16):10307–10320. doi: 10.1093/nar/gku586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones C.J., Newsom D., Kelly B., et al. ChIP-Seq and RNA-Seq reveal an AmrZ-mediated mechanism for cyclic di-GMP synthesis and biofilm development by Pseudomonas aeruginosa. PLoS Pathog. 2014;10(3) doi: 10.1371/journal.ppat.1003984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.An S.Q., Valvano M.A., Yu Y.H., Webb J.S., Lopez Campos G. An improved bind-n-seq strategy to determine protein-DNA interactions validated using the bacterial transcriptional regulator YipR. BMC Microbiol. 2020;20(1):1. doi: 10.1186/s12866-019-1672-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang T., Sun W., Fan L., et al. An atlas of the binding specificities of transcription factors in Pseudomonas aeruginosa directs prediction of novel regulators in virulence. Elife. 2021;10 doi: 10.7554/eLife.61885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liang H., Deng X., Ji Q., Sun F., Shen T., He C. The Pseudomonas aeruginosa global regulator VqsR directly inhibits QscR to control quorum-sensing and virulence gene expression. J Bacteriol. 2012;194(12):3098–3108. doi: 10.1128/JB.06679-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kong W., Zhao J., Kang H., et al. ChIP-seq reveals the global regulator AlgR mediating cyclic di-GMP synthesis in Pseudomonas aeruginosa. Nucleic Acids Res. 2015;43(17):8268–8282. doi: 10.1093/nar/gkv747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao J., Yu X., Zhu M., et al. Structural and molecular mechanism of CdpR involved in quorum-sensing and bacterial virulence in Pseudomonas aeruginosa. PLoS Biol. 2016;14(4) doi: 10.1371/journal.pbio.1002449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Song Y., Yang C., Chen G., et al. Molecular insights into the master regulator CysB-mediated bacterial virulence in Pseudomonas aeruginosa. Mol Microbiol. 2019;111(5):1195–1210. doi: 10.1111/mmi.14200. [DOI] [PubMed] [Google Scholar]
- 36.Sun L., Chi X., Feng Z., et al. phz1 contributes much more to phenazine-1-carboxylic acid biosynthesis than phz2 in Pseudomonas aeruginosa rpoS mutant. J Basic Microbiol. 2019;59(9):914–923. doi: 10.1002/jobm.201900165. [DOI] [PubMed] [Google Scholar]
- 37.Galán-Vásquez E., Luna-Olivera B.C., Ramírez-Ibáñez M., Martínez-Antonio A. RegulomePA: a database of transcriptional regulatory interactions in Pseudomonas aeruginosa PAO1. Database (Oxford) 2020;2020:baaa106. doi: 10.1093/database/baaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajput A., Tsunemoto H., Sastry A.V., et al. Machine learning from Pseudomonas aeruginosa transcriptomes identifies independently modulated sets of genes associated with known transcriptional regulators. Nucleic Acids Res. 2022;50(7):3658–3672. doi: 10.1093/nar/gkac187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee J., Wu J., Deng Y., et al. A cell-cell communication signal integrates quorum sensing and stress response. Nat Chem Biol. 2013;9(5):339–343. doi: 10.1038/nchembio.1225. [DOI] [PubMed] [Google Scholar]
- 40.Schuster M., Lostroh C.P., Ogi T., Greenberg E.P. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol. 2003;185(7):2066–2079. doi: 10.1128/JB.185.7.2066-2079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner V.E., Bushnell D., Passador L., Brooks A.I., Iglewski B.H. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J Bacteriol. 2003;185(7):2080–2095. doi: 10.1128/JB.185.7.2080-2095.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Waters C.M., Bassler B.L. Quorum sensing: cell-to-cell communication in bacteria. Annu Rev Cell Dev Biol. 2005;21:319–346. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 43.Williams P., Camara M. Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr Opin Microbiol. 2009;12(2):182–191. doi: 10.1016/j.mib.2009.01.005. [DOI] [PubMed] [Google Scholar]
- 44.Schuster M., Sexton D.J., Diggle S.P., Greenberg E.P. Acyl-homoserine lactone quorum sensing: from evolution to application. Annu Rev Microbiol. 2013;67:43–63. doi: 10.1146/annurev-micro-092412-155635. [DOI] [PubMed] [Google Scholar]
- 45.Albus A.M., Pesci E.C., Runyen-Janecky L.J., West S.E., Iglewski B.H.H. Vfr controls quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1997;179(12):3928–3935. doi: 10.1128/jb.179.12.3928-3935.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Balasubramanian D., Kumari H., Jaric M., et al. Deep sequencing analyses expands the Pseudomonas aeruginosa AmpR regulon to include small RNA-mediated regulation of iron acquisition, heat shock and oxidative stress response. Nucleic Acids Res. 2014;42(2):979–998. doi: 10.1093/nar/gkt942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carty N.L., Layland N., Colmer-Hamood J.A., Calfee M.W., Pesci E.C., Hamood A.N. PtxR modulates the expression of QS-controlled virulence factors in the Pseudomonas aeruginosa strain PAO1. Mol Microbiol. 2006;61(3):782–794. doi: 10.1111/j.1365-2958.2006.05269.x. [DOI] [PubMed] [Google Scholar]
- 48.Chugani S.A., Whiteley M., Lee K.M., D'Argenio D., Manoil C., Greenberg E.P. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2001;98(5):2752–2757. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Siehnel R., Traxler B., An D.D., Parsek M.R., Schaefer A.L., Singh P.K. A unique regulator controls the activation threshold of quorum-regulated genes in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2010;107(17):7916–7921. doi: 10.1073/pnas.0908511107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li S., Gong X., Yin L., et al. Acetylation of CspC controls the las quorum-sensing system through translational regulation of rsaL in Pseudomonas aeruginosa. mBio. 2022;13(3) doi: 10.1128/mbio.00547-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Y., Zhou C.M., Pu Q., et al. Pseudomonas aeruginosa regulatory protein AnvM controls pathogenicity in anaerobic environments and impacts host defense. mBio. 2019;10(4) doi: 10.1128/mBio.01362-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sana T.G., Lomas R., Gimenez M.R., et al. Differential modulation of quorum sensing signaling through QslA in Pseudomonas aeruginosa strains PAO1 and PA14. J Bacteriol. 2019;201(21) doi: 10.1128/JB.00362-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kojic M., Jovcic B., Vindigni A., Odreman F., Venturi V. Novel target genes of PsrA transcriptional regulator of Pseudomonas aeruginosa. FEMS Microbiol Lett. 2005;246(2):175–181. doi: 10.1016/j.femsle.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 54.Wells G., Palethorpe S., Pesci E.C. PsrA controls the synthesis of the Pseudomonas aeruginosa quinolone signal via repression of the FadE homolog, PA0506. PLoS One. 2017;12(12) doi: 10.1371/journal.pone.0189331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mahajan-Miklos S., Tan M.W., Rahme L.G., Ausubel F.M. Molecular mechanisms of bacterial virulence elucidated using a Pseudomonas aeruginosa-Caenorhabditis elegans pathogenesis model. Cell. 1999;96(1):47–56. doi: 10.1016/s0092-8674(00)80958-7. [DOI] [PubMed] [Google Scholar]
- 56.Wilson R., Sykes D.A., Watson D., Rutman A., Taylor G.W., Cole P.J. Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infect Immun. 1988;56(9):2515–2517. doi: 10.1128/iai.56.9.2515-2517.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rampioni G., Schuster M., Greenberg E.P., et al. RsaL provides quorum sensing homeostasis and functions as a global regulator of gene expression in Pseudomonas aeruginosa. Mol Microbiol. 2007;66(6):1557–1565. doi: 10.1111/j.1365-2958.2007.06029.x. [DOI] [PubMed] [Google Scholar]
- 58.Whiteley M., Greenberg E.P. Promoter specificity elements in Pseudomonas aeruginosa quorum-sensing-controlled genes. J Bacteriol. 2001;183(19):5529–5534. doi: 10.1128/JB.183.19.5529-5534.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li C., Wally H., Miller S.J., Lu C.D. The multifaceted proteins MvaT and MvaU, members of the H-NS family, control arginine metabolism, pyocyanin synthesis, and prophage activation in Pseudomonas aeruginosa PAO1. J Bacteriol. 2009;191(20):6211–6218. doi: 10.1128/JB.00888-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cai Z., Yang F., Shao X., et al. ECF sigma factor HxuI is critical for in vivo fitness of Pseudomonas aeruginosa during infection. Microbiol Spectr. 2022;10(1) doi: 10.1128/spectrum.01620-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang H., Duan J., Sibley C.D., Surette M.G., Duan K. Identification of mutants with altered phenazine production in Pseudomonas aeruginosa. J Med Microbiol. 2011;60(Pt 1):22–34. doi: 10.1099/jmm.0.022350-0. [DOI] [PubMed] [Google Scholar]
- 62.Lin L., Xu K., Shen D., Chou S.H., Gomelsky M., Qian G. Antifungal weapons of Lysobacter, a mighty biocontrol agent. Environ Microbiol. 2021;23(10):5704–5715. doi: 10.1111/1462-2920.15674. [DOI] [PubMed] [Google Scholar]
- 63.Starnbach M.N., Lory S. The fliA (rpoF) gene of Pseudomonas aeruginosa encodes an alternative sigma factor required for flagellin synthesis. Mol Microbiol. 1992;6(4):459–469. doi: 10.1111/j.1365-2958.1992.tb01490.x. [DOI] [PubMed] [Google Scholar]
- 64.Dasgupta N., Wolfgang M.C., Goodman A.L., et al. A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol Microbiol. 2003;50(3):809–824. doi: 10.1046/j.1365-2958.2003.03740.x. [DOI] [PubMed] [Google Scholar]
- 65.Frisk A., Jyot J., Arora S.K., Ramphal R. Identification and functional characterization of flgM, a gene encoding the anti-sigma 28 factor in Pseudomonas aeruginosa. J Bacteriol. 2002;184(6):1514–1521. doi: 10.1128/JB.184.6.1514-1521.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dasgupta N., Ferrell E.P., Kanack K.J., West S.E., Ramphal R. fleQ, the gene encoding the major flagellar regulator of Pseudomonas aeruginosa, is sigma70 dependent and is downregulated by Vfr, a homolog of Escherichia coli cyclic AMP receptor protein. J Bacteriol. 2002;184(19):5240–5250. doi: 10.1128/JB.184.19.5240-5250.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chanchal, Banerjee P., Raghav S., Goswami H.N., Jain D. The antiactivator FleN uses an allosteric mechanism to regulate sigma(54)-dependent expression of flagellar genes in Pseudomonas aeruginosa. Sci Adv. 2021;7(43) doi: 10.1126/sciadv.abj1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Petrova O.E., Schurr J.R., Schurr M.J., Sauer K. The novel Pseudomonas aeruginosa two-component regulator BfmR controls bacteriophage-mediated lysis and DNA release during biofilm development through PhdA. Mol Microbiol. 2011;81(3):767–783. doi: 10.1111/j.1365-2958.2011.07733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wang D., Zhang X., Yin L., et al. RplI interacts with 5' UTR of exsA to repress its translation and type III secretion system in Pseudomonas aeruginosa. PLoS Pathog. 2022;18(1) doi: 10.1371/journal.ppat.1010170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ma Y., Liu Y., Bi Y., et al. OsaR (PA0056) functions as a repressor of the gene fleQ encoding an important motility regulator in Pseudomonas aeruginosa. J Bacteriol. 2021;203(20) doi: 10.1128/JB.00145-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baraquet C., Harwood C.S. FleQ DNA binding consensus sequence revealed by studies of FleQ-dependent regulation of biofilm gene expression in Pseudomonas aeruginosa. J Bacteriol. 2015;198(1):178–186. doi: 10.1128/JB.00539-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hickman J.W., Harwood C.S. Identification of FleQ from Pseudomonas aeruginosa as a c-di-GMP-responsive transcription factor. Mol Microbiol. 2008;69(2):376–389. doi: 10.1111/j.1365-2958.2008.06281.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Baraquet C., Murakami K., Parsek M.R., Harwood C.S. The FleQ protein from Pseudomonas aeruginosa functions as both a repressor and an activator to control gene expression from the pel operon promoter in response to c-di-GMP. Nucleic Acids Res. 2012;40(15):7207–7218. doi: 10.1093/nar/gks384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wu W., Badrane H., Arora S., Baker H.V., Jin S. MucA-mediated coordination of type III secretion and alginate synthesis in Pseudomonas aeruginosa. J Bacteriol. 2004;186(22):7575–7585. doi: 10.1128/JB.186.22.7575-7585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jones A.K., Fulcher N.B., Balzer G.J., et al. Activation of the Pseudomonas aeruginosa AlgU regulon through mucA mutation inhibits cyclic AMP/Vfr signaling. J Bacteriol. 2010;192(21):5709–5717. doi: 10.1128/JB.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chand N.S., Lee J.S., Clatworthy A.E., Golas A.J., Smith R.S., Hung D.T. The sensor kinase KinB regulates virulence in acute Pseudomonas aeruginosa infection. J Bacteriol. 2011;193(12):2989–2999. doi: 10.1128/JB.01546-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mukherjee S., Jemielita M., Stergioula V., Tikhonov M., Bassler B.L. Photosensing and quorum sensing are integrated to control Pseudomonas aeruginosa collective behaviors. PLoS Biol. 2019;17(12) doi: 10.1371/journal.pbio.3000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Romero M., Silistre H., Lovelock L., et al. Genome-wide mapping of the RNA targets of the Pseudomonas aeruginosa riboregulatory protein RsmN. Nucleic Acids Res. 2018;46(13):6823–6840. doi: 10.1093/nar/gky324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mikkelsen H., Ball G., Giraud C., Filloux A. Expression of Pseudomonas aeruginosa CupD fimbrial genes is antagonistically controlled by RcsB and the EAL-containing PvrR response regulators. PLoS One. 2009;4(6) doi: 10.1371/journal.pone.0006018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hauser A.R., Cobb E., Bodi M., et al. Type III protein secretion is associated with poor clinical outcomes in patients with ventilator-associated pneumonia caused by Pseudomonas aeruginosa. Crit Care Med. 2002;30(3):521–528. doi: 10.1097/00003246-200203000-00005. [DOI] [PubMed] [Google Scholar]
- 81.Roy-Burman A., Savel R.H., Racine S., et al. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J Infect Dis. 2001;183(12):1767–1774. doi: 10.1086/320737. [DOI] [PubMed] [Google Scholar]
- 82.Crouch Brewer S., Wunderink R.G., Jones C.B., Leeper K.V., Jr. Ventilator-associated pneumonia due to Pseudomonas aeruginosa. Chest. 1996;109(4):1019–1029. doi: 10.1378/chest.109.4.1019. [DOI] [PubMed] [Google Scholar]
- 83.Bordi C., Lamy M.C., Ventre I., et al. Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol Microbiol. 2010;76(6):1427–1443. doi: 10.1111/j.1365-2958.2010.07146.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Williams McMackin E.A., Marsden A.E., Yahr T.L., H-Ns Family members MvaT and MvaU regulate the Pseudomonas aeruginosa type III secretion system. J Bacteriol. 2019;201(14):e00054. doi: 10.1128/JB.00054-19. 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li K., Xu C., Jin Y., et al. SuhB is a regulator of multiple virulence genes and essential for pathogenesis of Pseudomonas aeruginosa. mBio. 2013;4(6):e00419-13. doi: 10.1128/mBio.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li K., Yang G., Debru A.B., et al. SuhB regulates the motile-sessile switch in Pseudomonas aeruginosa through the Gac/Rsm pathway and c-di-GMP signaling. Front Microbiol. 2017;8:1045. doi: 10.3389/fmicb.2017.01045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xia Y., Xu C., Wang D., et al. YbeY controls the type III and type VI secretion systems and biofilm formation through RetS in Pseudomonas aeruginosa. Appl Environ Microbiol. 2020;87(5):e02171-20. doi: 10.1128/AEM.02171-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jin Y., Zhang M., Zhu F., et al. NrtR regulates the type III secretion system through cAMP/Vfr pathway in Pseudomonas aeruginosa. Front Microbiol. 2019;10:85. doi: 10.3389/fmicb.2019.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Intile P.J., Balzer G.J., Wolfgang M.C., Yahr T.L. The RNA helicase DeaD stimulates ExsA translation to promote expression of the Pseudomonas aeruginosa type III secretion system. J Bacteriol. 2015;197(16):2664–2674. doi: 10.1128/JB.00231-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Janssen K.H., Corley J.M., Djapgne L., et al. Hfq and sRNA 179 inhibit expression of the Pseudomonas aeruginosa cAMP-Vfr and type III secretion regulons. mBio. 2020;11(3) doi: 10.1128/mBio.00363-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li S., Weng Y., Li X., et al. Acetylation of the CspA family protein CspC controls the type III secretion system through translational regulation of exsA in Pseudomonas aeruginosa. Nucleic Acids Res. 2021;49(12):6756–6770. doi: 10.1093/nar/gkab506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mougous J.D., Cuff M.E., Raunser S., et al. A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science. 2006;312(5779):1526–1530. doi: 10.1126/science.1128393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Marden J.N., Diaz M.R., Walton W.G., et al. An unusual CsrA family member operates in series with RsmA to amplify posttranscriptional responses in Pseudomonas aeruginosa. Proc Natl Acad Sci U S A. 2013;110(37):15055–15060. doi: 10.1073/pnas.1307217110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang X., Yin L., Liu Q., et al. NrtR mediated regulation of H1-T6SS in Pseudomonas aeruginosa. Microbiol Spectr. 2022;10(1) doi: 10.1128/spectrum.01858-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhou T., Huang J., Liu Z., Lin Q., Xu Z., Zhang L.H. The two-component system FleS/FleR represses H1-T6SS via cyclic di-GMP signaling in Pseudomonas aeruginosa. Appl Environ Microbiol. 2022;88(2) doi: 10.1128/AEM.01655-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen P.R., Bae T., Williams W.A., et al. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat Chem Biol. 2006;2(11):591–595. doi: 10.1038/nchembio820. [DOI] [PubMed] [Google Scholar]
- 97.Fuangthong M., Helmann J.D. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine-sulfenic acid derivative. Proc Natl Acad Sci U S A. 2002;99(10):6690–6695. doi: 10.1073/pnas.102483199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Fujimoto D.F., Higginbotham R.H., Sterba K.M., et al. Staphylococcus aureus SarA is a regulatory protein responsive to redox and pH that can support bacteriophage lambda integrase-mediated excision/recombination. Mol Microbiol. 2009;74(6):1445–1458. doi: 10.1111/j.1365-2958.2009.06942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Palma M., Zurita J., Ferreras J.A., et al. Pseudomonas aeruginosa SoxR does not conform to the archetypal paradigm for SoxR-dependent regulation of the bacterial oxidative stress adaptive response. Infect Immun. 2005;73(5):2958–2966. doi: 10.1128/IAI.73.5.2958-2966.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dietrich L.E., Teal T.K., Price-Whelan A., Newman D.K. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science. 2008;321(5893):1203–1206. doi: 10.1126/science.1160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Vinckx T., Matthijs S., Cornelis P. Loss of the oxidative stress regulator OxyR in Pseudomonas aeruginosa PAO1 impairs growth under iron-limited conditions. FEMS Microbiol Lett. 2008;288(2):258–265. doi: 10.1111/j.1574-6968.2008.01360.x. [DOI] [PubMed] [Google Scholar]
- 102.Ochsner U.A., Vasil M.L., Alsabbagh E., Parvatiyar K., Hassett D.J. Role of the Pseudomonas aeruginosa oxyR-recG operon in oxidative stress defense and DNA repair: OxyR-dependent regulation of katB-ankB, ahpB, and ahpC-ahpF. J Bacteriol. 2000;182(16):4533–4544. doi: 10.1128/jb.182.16.4533-4544.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Vinckx T., Wei Q., Matthijs S., Cornelis P. The Pseudomonas aeruginosa oxidative stress regulator OxyR influences production of pyocyanin and rhamnolipids: protective role of pyocyanin. Microbiology (Read) 2010;156(Pt 3):678–686. doi: 10.1099/mic.0.031971-0. [DOI] [PubMed] [Google Scholar]
- 104.Lan L., Murray T.S., Kazmierczak B.I., He C. Pseudomonas aeruginosa OspR is an oxidative stress sensing regulator that affects pigment production, antibiotic resistance and dissemination during infection. Mol Microbiol. 2010;75(1):76–91. doi: 10.1111/j.1365-2958.2009.06955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen H., Hu J., Chen P.R., et al. The Pseudomonas aeruginosa multidrug efflux regulator MexR uses an oxidation-sensing mechanism. Proc Natl Acad Sci U S A. 2008;105(36):13586–13591. doi: 10.1073/pnas.0803391105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Deng X., Weerapana E., Ulanovskaya O., et al. Proteome-wide quantification and characterization of oxidation-sensitive cysteines in pathogenic bacteria. Cell Host Microbe. 2013;13(3):358–370. doi: 10.1016/j.chom.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Xuan G., Lv C., Xu H., et al. Sulfane sulfur regulates LasR-mediated quorum sensing and virulence in Pseudomonas aeruginosa PAO1. Antioxidants. 2021;10(9):1498. doi: 10.3390/antiox10091498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kafle P., Amoh A.N., Reaves J.M., et al. Molecular insights into the impact of oxidative stress on the quorum-sensing regulator protein LasR. J Biol Chem. 2016;291(22):11776–11786. doi: 10.1074/jbc.M116.719351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shlyueva D., Stampfel G., Stark A. Transcriptional enhancers: from properties to genome-wide predictions. Nat Rev Genet. 2014;15(4):272–286. doi: 10.1038/nrg3682. [DOI] [PubMed] [Google Scholar]
- 110.Rosenberg G., Riquelme S., Prince A., Avraham R. Immunometabolic crosstalk during bacterial infection. Nat Microbiol. 2022;7(4):497–507. doi: 10.1038/s41564-022-01080-5. [DOI] [PubMed] [Google Scholar]
- 111.Bartell J.A., Blazier A.S., Yen P., et al. Reconstruction of the metabolic network of Pseudomonas aeruginosa to interrogate virulence factor synthesis. Nat Commun. 2017;8 doi: 10.1038/ncomms14631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Panayidou S., Georgiades K., Christofi T., Tamana S., Promponas V.J., Apidianakis Y. Pseudomonas aeruginosa core metabolism exerts a widespread growth-independent control on virulence. Sci Rep. 2020;10(1):9505. doi: 10.1038/s41598-020-66194-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chavali A.K., D'Auria K.M., Hewlett E.L., Pearson R.D., Papin J.A. A metabolic network approach for the identification and prioritization of antimicrobial drug targets. Trends Microbiol. 2012;20(3):113–123. doi: 10.1016/j.tim.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mienda B.S., Salihu R., Adamu A., Idris S. Genome-scale metabolic models as platforms for identification of novel genes as antimicrobial drug targets. Future Microbiol. 2018;13:455–467. doi: 10.2217/fmb-2017-0195. [DOI] [PubMed] [Google Scholar]
- 115.Oberhardt M.A., Puchałka J., Fryer K.E., Martins dos Santos V.A., Papin J.A. Genome-scale metabolic network analysis of the opportunistic pathogen Pseudomonas aeruginosa PAO1. J Bacteriol. 2008;190(8):2790–2803. doi: 10.1128/JB.01583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Müh U., Schuster M., Heim R., Singh A., Olson E.R., Greenberg E.P. Novel Pseudomonas aeruginosa quorum-sensing inhibitors identified in an ultra-high-throughput screen. Antimicrob Agents Chemother. 2006;50(11):3674–3679. doi: 10.1128/AAC.00665-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Starkey M., Lepine F., Maura D., et al. Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity. PLoS Pathog. 2014;10(8) doi: 10.1371/journal.ppat.1004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Imperi F., Massai F., Facchini M., et al. Repurposing the antimycotic drug flucytosine for suppression of Pseudomonas aeruginosa pathogenicity. Proc Natl Acad Sci U S A. 2013;110(18):7458–7463. doi: 10.1073/pnas.1222706110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Borlee B.R., Geske G.D., Blackwell H.E., Handelsman J. Identification of synthetic inducers and inhibitors of the quorum-sensing regulator LasR in Pseudomonas aeruginosa by high-throughput screening. Appl Environ Microbiol. 2010;76(24):8255–8258. doi: 10.1128/AEM.00499-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.van Tilburg Bernardes E., Charron-Mazenod L., Reading D.J., Reckseidler-Zenteno S.L., Lewenza S. Exopolysaccharide-repressing small molecules with antibiofilm and antivirulence activity against Pseudomonas aeruginosa. Antimicrob Agents Chemother. 2017;61(5):e01997-16. doi: 10.1128/AAC.01997-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Aiello D., Williams J.D., Majgier-Baranowska H., et al. Discovery and characterization of inhibitors of Pseudomonas aeruginosa type III secretion. Antimicrob Agents Chemother. 2010;54(5):1988–1999. doi: 10.1128/AAC.01598-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zhu J., Cai X., Harris T.L., et al. Disarming Pseudomonas aeruginosa virulence factor LasB by leveraging a Caenorhabditis elegans infection model. Chem Biol. 2015;22(4):483–491. doi: 10.1016/j.chembiol.2015.03.012. [DOI] [PubMed] [Google Scholar]
- 123.Abelyan N., Grabski H., Tiratsuyan S. In silico screening of flavones and its derivatives as potential inhibitors of quorum-sensing regulator LasR of Pseudomonas aeruginosa. Mol Biol. 2020;54(1):153–163. doi: 10.31857/S0026898420010024. [DOI] [PubMed] [Google Scholar]
- 124.Shaker B., Ahmad S., Thai T.D., Eyun S.I., Na D. Rational drug design for Pseudomonas aeruginosa PqsA enzyme: an in silico guided study to block biofilm formation. Front Mol Biosci. 2020;7 doi: 10.3389/fmolb.2020.577316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vetrivel A., Natchimuthu S., Subramanian V., Murugesan R. High-throughput virtual screening for a new class of antagonist targeting LasR of Pseudomonas aeruginosa. ACS Omega. 2021;6(28):18314–18324. doi: 10.1021/acsomega.1c02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Baloyi I.T., Adeosun I.J., Yusuf A.A., Cosa S. In silico and in vitro screening of antipathogenic properties of melianthus comosus (Vahl) against Pseudomonas aeruginosa. Antibiotics. 2021;10(6):679. doi: 10.3390/antibiotics10060679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tajani A.S., Jangi E., Davodi M., et al. Anti-quorum sensing potential of ketoprofen and its derivatives against Pseudomonas aeruginosa: insights to in silico and in vitro studies. Arch Microbiol. 2021;203(8):5123–5132. doi: 10.1007/s00203-021-02499-w. [DOI] [PubMed] [Google Scholar]
- 128.Behera S.K., Panda A.K., Mishra R., Mahanty A., Bisht S.S. Structure based virtual screening and molecular dynamics of natural anti-biofilm compounds against SagS response regulator/sensor kinase in Pseudomonas aeruginosa. J Biomol Struct Dyn. 2022:1–16. doi: 10.1080/07391102.2022.2100482. [DOI] [PubMed] [Google Scholar]
- 129.Deng X., Chen K., Luo G.Z., et al. Widespread occurrence of N6-methyladenosine in bacterial mRNA. Nucleic Acids Res. 2015;43(13):6557–6567. doi: 10.1093/nar/gkv596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shao X., Zhang W., Umar M.I., et al. RNA G-quadruplex structures mediate gene regulation in bacteria. mBio. 2020;11(1) doi: 10.1128/mBio.02926-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Smith E.E., Buckley D.G., Wu Z., et al. Genetic adaptation by Pseudomonas aeruginosa to the airways of cystic fibrosis patients. Proc Natl Acad Sci U S A. 2006;103(22):8487–8492. doi: 10.1073/pnas.0602138103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yang L., Jelsbak L., Marvig R.L., et al. Evolutionary dynamics of bacteria in a human host environment. Proc Natl Acad Sci U S A. 2011;108(18):7481–7486. doi: 10.1073/pnas.1018249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hoffman L.R., Kulasekara H.D., Emerson J., et al. Pseudomonas aeruginosa lasR mutants are associated with cystic fibrosis lung disease progression. J Cyst Fibros. 2009;8(1):66–70. doi: 10.1016/j.jcf.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Folkesson A., Jelsbak L., Yang L., et al. Adaptation of Pseudomonas aeruginosa to the cystic fibrosis airway: an evolutionary perspective. Nat Rev Microbiol. 2012;10(12):841–851. doi: 10.1038/nrmicro2907. [DOI] [PubMed] [Google Scholar]
- 135.Valentini M., Gonzalez D., Mavridou D.A., Filloux A. Lifestyle transitions and adaptive pathogenesis of Pseudomonas aeruginosa. Curr Opin Microbiol. 2018;41:15–20. doi: 10.1016/j.mib.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 136.Ahmed S.A.K.S., Rudden M., Elias S.M., et al. Pseudomonas aeruginosa PA80 is a cystic fibrosis isolate deficient in RhlRI quorum sensing. Sci Rep. 2021;11(1):5729. doi: 10.1038/s41598-021-85100-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Boşgelmez-Tinaz G., Ulusoy S. Characterization of N-butanoyl-L-homoserine lactone (C4-HSL) deficient clinical isolates of Pseudomonas aeruginosa. Microb Pathog. 2008;44(1):13–19. doi: 10.1016/j.micpath.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 138.Martínez-Carranza E., García-Reyes S., González-Valdez A., Soberón-Chávez G. Tracking the genome of four Pseudomonas aeruginosa isolates that have a defective Las quorum-sensing system, but are still virulent. Access Microbiol. 2020;2(7) doi: 10.1099/acmi.0.000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Shang Z., Wang H., Zhou S., Chu W. Characterization of N-Acyl-homoserine lactones (AHLs)-deficient clinical isolates of Pseudomonas aeruginosa. Indian J Microbiol. 2014;54(2):158–162. doi: 10.1007/s12088-014-0449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Baker P., Hill P.J., Snarr B.D., et al. Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci Adv. 2016;2(5) doi: 10.1126/sciadv.1501632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Das T., Simone M., Ibugo A.I., Witting P.K., Manefield M., Manos J. Glutathione enhances antibiotic efficiency and effectiveness of DNase I in disrupting Pseudomonas aeruginosa biofilms while also inhibiting pyocyanin activity, thus facilitating restoration of cell enzymatic activity, confluence and viability. Front Microbiol. 2017;8:2429. doi: 10.3389/fmicb.2017.02429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Singh N., Romero M., Travanut A., et al. Dual bioresponsive antibiotic and quorum sensing inhibitor combination nanoparticles for treatment of Pseudomonas aeruginosa biofilms in vitro and ex vivo. Biomater Sci. 2019;7(10):4099–4111. doi: 10.1039/c9bm00773c. [DOI] [PubMed] [Google Scholar]