Abstract
Uveitis, a vision-threatening inflammatory disease worldwide, is closely related to resident microglia. Retinal microglia are the main immune effector cells with strong plasticity, but their role in uveitis remains unclear. N6-methyladenosine (m6A) modification has been proven to be involved in the immune response. Therefore, we in this work aimed to identify the potentially crucial m6A regulators of microglia in uveitis. Through the single-cell sequencing (scRNA-seq) analysis and experimental verification, we found a significant decrease in the expression of fat mass and obesity-associated protein (FTO) in retinal microglia of uveitis mice and human microglia clone 3 (HMC3) cells with inflammation. Additionally, FTO knockdown was found to aggravate the secretion of inflammatory factors and the mobility/chemotaxis of microglia. Mechanistically, the RNA-seq data and rescue experiments showed that glypican 4 (GPC4) was the target of FTO, which regulated microglial inflammation mediated by the TLR4/NF-κB pathway. Moreover, RNA stability assays indicated that GPC4 upregulation was mainly regulated by the downregulation of the m6A “reader” YTH domain family protein 3 (YTHDF3). Finally, the FTO inhibitor FB23-2 further exacerbated experimental autoimmune uveitis (EAU) inflammation by promoting the GPC4/TLR4/NF-κB signaling axis, and this could be attenuated by the TLR4 inhibitor TAK-242. Collectively, a decreased FTO could facilitate microglial inflammation in EAU, suggesting that the restoration or activation of FTO function may be a potential therapeutic strategy for uveitis.
Keywords: Fat mass and obesity-associated protein, Glypican 4, Microglia, N6-methyladenosine, Uveitis, YTH domain Family protein 3
Introduction
Uveitis is a vision-threatening ocular inflammatory disease that affects multiple systems with complicated etiology. Autoimmune uveitis accounts for the majority of noninfectious uveitis and involves aberrant immune recognition of unique self-antigens in target tissues.1 Previous studies have focused on peripheral immune cells in the development of autoimmune uveitis. Vogt-Koyanagi-Harada disease and Behcet's disease, the most common uveitis entities in China, have been reported to be associated with the Th17 response.2,3 Experimental autoimmune uveitis (EAU) is regarded as a classical animal model of human autoimmune uveitis, and both Th1 and Th17 cells are considered as essential inducers of EAU.4 Furthermore, activated gamma delta T cells and dendritic cells promote inflammatory progression in EAU.5 However, little is known about retinal innate immune cells in autoimmune uveitis.
With the rapid development of molecular biology techniques, such as single-cell sequencing (scRNA-seq), the crucial role of resident microglia in the retina has been determined.6 Microglia are characterized as the main actors in the inflammation of uveitis on account of their multiple functions.7 Microglial activation can be mainly categorized into the M1 proinflammatory state termed “classical activation” and the M2 anti-inflammatory state termed “alternative activation”.8 Resident microglia could migrate to the subretinal space and adhere to the retinal pigment epithelium in a mouse model of photoreceptor degeneration.9,10 Besides, microglia are required to amplify retinal inflammation by mediating immune cell entry into the retina. After depletion of microglia, circulating immune cells could not enter the retina to initiate EAU.11 Hence, exploring the underlying functions and mechanisms of microglia is essential for understanding the pathological process of uveitis.
As the most abundant internal mRNA modification in eukaryotes, m6A is critical in multiple physiological and pathological biological processes.12, 13, 14, 15, 16 Specific ablation of ALKBH5 (AlkB homolog 5) in T cells could confer protection against experimental autoimmune encephalomyelitis (EAE), a classical animal model for studying multiple sclerosis.17 T cell-specific METTL14 deficiency prevented the differentiation of naïve T cells into Tregs, leading to an imbalance in spontaneous colitis.18 These dynamic processes are modulated by methyltransferases (writers), demethylases (erasers), and readers.19 For m6A modification, the “writer” complex can deposit methyl on adenosine. For m6A removal, “erasers”, such as FTO (the first m6A “eraser” identified), can remove methyl from adenosine.20 In recent years, strong evidence has indicated that FTO could regulate the genesis and development of autoimmune diseases.21 Associations between FTO mRNA expression and some indicators of rheumatoid arthritis were identified by high-throughput m6A sequencing.22 Latent autoimmune diabetes in adults has been reported to be linked to the high-risk genotype of FTO SNP rs9939609.23 Although the effects of FTO in other autoimmune diseases are well documented, the mechanism of m6A demethylase in uveitis are poorly understood.
IRBP (interphotoreceptor retinoid-binding protein)-induced EAU mice and LPS/IFN-γ-induced inflammatory HMC3 cells were utilized in this study. FTO expression was specifically decreased in microglia both in vivo and in vitro, as confirmed by scRNA-seq data and validation with RT-qPCR and Western blotting. However, this significant change was not observed for other m6A methylases and demethylases. Furthermore, compared with normal HMC3 cells, FTO-depleted ones were more likely to induce inflammation, manifested by elevated expression of inflammatory factors, enhanced cell mobility, and chemotaxis to CD4+ T cells. Consistently, FTO-deficient mice with EAU exhibited amplified expression of inflammatory factors and more obvious intraocular inflammation. Subsequent experiments revealed that FTO knockdown triggered the upregulation of GPC4, which was mediated by reduced YTHDF3 expression. Moreover, GPC4 was identified as the key enhancer of the TLR4/NF-κB pathway. Collectively, these data suggest that decreased FTO exacerbated microglial inflammation in EAU by targeting GPC4, and the retention of FTO function may be helpful in identifying a strategy to slow the progression of uveitis.
Methods and materials
Animals
Female C57BL/6J mice (6–8 weeks old) were provided by the Experimental Animal Center of Chongqing Medical University (Chongqing, China) and kept in a specific pathogen-free environment. The protocol was approved by the Ethics Committee of the First Affiliated Hospital of Chongqing Medical University, Chongqing, China.
Reagents
IRBP651-670 (LAQGAYRTAVDLESLASQLT) peptide was synthesized and provided by Sangon Biological Engineering Technology & Services Co., Ltd. (Shanghai, China). Both complete Freund's adjuvant and pertussis toxin were purchased from Sigma–Aldrich (St. Louis, MO, USA). Heat-killed Mycobacterium tuberculosis strain H37Ra was ordered from BD Biosciences (NJ, USA).
Induction of EAU
Female C57BL/6J mice were subcutaneously immunized with human IRBP651–670 (500 μg) dissolved in 0.1 mL of PBS containing 20% DMSO (V900090 MSDS, Sigma–Aldrich, USA) and emulsified with an equal volume of complete Freund's adjuvant containing M. tuberculosis strain H37Ra. Mice were injected intraperitoneally with 1 μg of Bordetella pertussis toxin.
Clinical and histopathological assessment
On the 14th day after modeling, the clinical symptoms of EAU were assessed by slit lamp examination. The clinical severity of ocular inflammation was evaluated in a blinded manner by three independent experienced observers, and half-point increments were scored on a scale of 0–5 according to five independent criteria and Caspi's criteria.24 Mice were sacrificed after clinical scoring, and their eyeballs were enucleated and fixed. Then, paraffin sections were prepared along the papillary-optic nerve axis. The sections were stained with hematoxylin and eosin for histopathological evaluation at grades 0–4 according to Caspi's criteria.24
Intravitreal injection of FB23-2
On the 9th day of EAU, the mice were injected intravitreally with FB23-2 (10 μmol/μL), which is a potent and selective m6A demethylase FTO inhibitor (Sigma–Aldrich, St. Louis, USA).25 FB23-2 was dissolved in DMSO and diluted in PBS. DMSO diluted proportionally with PBS was injected as a control.
Intraperitoneal injection of TAK-242 (resatorvid)
After EAU induction for 9 days, all immunized mice were examined with a slit lamp to guarantee successful modeling. The mice were randomly divided into three groups according to the different treatments, the EAU + Vehicle group, the EAU + FB23-2 group (receiving once FB23-2 intravitreal administration on the 9th day), and the EAU + FB23-2+TAK-242 group (receiving intraperitoneal administration of TAK-242 at a dose of 3 mg/kg per day for three times with a one-day interval). The TAK-242 was purchased from MedChemExpress, and the doses and administration times were selected based on several relevant articles.26,27
Flow cytometric analysis
The mouse retinas were removed, crushed, and filtered in a cell strainer (70 μm nylon, BD Falcon, San Jose, CA, USA) to obtain single-cell suspensions. The cells were stained with live/dead dye (Thermo Fisher Scientific, Waltham, MA, USA) at 4 °C for 10 min. After two washes with PBS, the cells were stained with antibodies specific to surface markers CD4 (1 μL/106 cells, BioLegend, USA) and CD11b (1 μL/106 cells, BioLegend, USA). Then, the cells were fixed with fixation buffer (BioLegend, USA) at room temperature for 20 min and permeabilized with intracellular staining (BioLegend, USA). The harvested cells were stained with intracellular cytokine iNOS (0.5 μL/106 cells, BioLegend, USA). The cells were subsequently analyzed with BD FACSAria™ fusion flow cytometer (USA), and the data analysis was performed with FlowJo version X software.
Immunofluorescence
For retinal flat mounts, the eyeballs were immersed in 4% paraformaldehyde for 2 h, cut into flat mounts, blocked with 3% goat serum and 0.3% Triton X-100 for 1 h, and then incubated with IBA1 (Wako, 019–19741, 1:1,000) at 4 °C overnight. Next, the retinas were washed three times with PBS and incubated with Cy3-labeled goat anti-rabbit IgG (H + L) (Beyotime, A0516, 1:5,000) for 1 h. For paraffin sections, 5 μm paraffin slides were incubated at 75 °C for 90 min and deparaffinized in xylene. After rehydration in a descending series of ethanol, the slides were washed in distilled water for 5 min. Antigens were repaired in citrate antigen repair buffer (pH 6.0) (Servicebio, Wuhan, China). After preparation, staining was performed, the method of which was the same as that for retinal spreading. In contrast, we used CD4 (Invitrogen, MA1-146, 1:400), IBA1 (WAKO, 019–19741, 1:1,000), and DAPI for co-staining. Images were taken by confocal microscopy (Zeiss, Germany).
Cell culture
The human microglial cell line HMC3 was purchased from the American Type Culture Collection (ATCC, USA) and maintained in our laboratory with Eagle's minimum essential medium (EMEM, ATCC, USA) supplemented with 10% fetal bovine serum (FBS, Gibco, USA) and 1% penicillin/streptomycin (Gibco, USA). All these cells were cultured in 5% CO2 at 37 °C.
Quantitative real-time PCR
TRIzol reagent (Invitrogen, San Diego, California, USA) was used to extract total RNA from EAU tissues and HMC3 cells as per the manufacturer's instructions. The mRNA levels were measured by RT Master Mix for qPCR (MedChemExpress, USA) and SYBR Green qPCR Master Mix (MedChemExpress, USA). All results were normalized to the β-actin expression. The primers for the targeted genes used are shown in Table S1.
Cell transfection
Short hairpin RNAs (shRNAs) targeting FTO and GPC4 and negative control were synthesized and cloned into the pGLVH1/GFP/Puro vector by GeneChem (Shanghai, China). Puromycin (1 μg/mL; Beyotime) was added after three days for selection. The transfection efficiency was confirmed via RT-qPCR and Western blotting. Small interfering RNA (siRNA) targeting YTHDF3 (Thermo Fisher Scientific) was used for YTHDF3 knockdown in HMC3 cells. The sequences of shRNAs and siRNAs are shown in Table S2.
Western blot
Cell and retinal tissues were extracted with lysis buffer (Beyotime, Shanghai, China) and centrifuged at 4 °C and 14,000 rpm for 10 min. Protein samples were separated via 7.5% (wt/vol) SDS-PAGE and transferred onto PVDF membranes (Millipore, Billerica, MA, USA). The membranes were blocked with 5% skim milk for 1.5 h at room temperature. Then, the primary antibodies were incubated overnight at 4 °C. The membranes were washed three times and incubated with secondary antibodies for 1 h at room temperature. An ECL kit (Thermo Scientific, CA, USA) was used to visualize the band signal, and ImageJ software was used to quantify the signal. The primary antibodies used in this study were as follows: iNOS (ab178945, Abcam, 1:800), TNFα (ab183218, Abcam, 1:500), IL6 (ab233706, Abcam, 1:500), FTO (ab126605, Abcam, 1:800), GPC4 (sc-517403, Santa Cruz, 1:800), NF-κB p65 (ab32536, Abcam, 1:1,000), NF-κB p-p65 S536 (3033, Cell Signaling Technology, 1:1,000), TLR4 (ab22048, Abcam, 1:1,000), MYD88 (ab133739, Abcam, 1:1,000), CD14 (221678, Abcam, 1:1,000), CXCL10 (14969, Cell Signaling Technology, 1:1,000), and β-actin (20536-1-AP, Proteintech, 1:3,000). The secondary antibodies used included HRP-conjugated AffiniPure goat anti-mouse IgG (SA00001-1, Proteintech, 1:6,000) and HRP-conjugated AffiniPure goat anti-rabbit IgG (SA00001-2, Proteintech, 1:6,000).
Cell proliferation assay
The cell proliferation was evaluated using a CCK-8 detection kit as per the manufacturer's instructions (HY–K0301, MedChemExpress, USA). In brief, the cells were seeded in 96-well plates at a density of 5000 cells/100 μL. The dye solution was added at the indicated times (0, 12, 24, 48, 72 h), and the plate was incubated at 37 °C for 2 h. Then, the absorbance was measured at 450 nm.
Enzyme-linked immunosorbent assay
The cell supernatant was harvested, centrifuged, and then stored at −80 °C. The concentration of tumor necrosis factor-α (TNFα) was measured using ELISA kits (Elabscience, Wuhan, China).
Culturing of primary microglia
Primary retinal microglia were cultured following the procedures modified based on the previously described.28,29 Briefly, the tissues from postnatal (8–10 days) C57BL/J mice were pulled together and were digested with the papain for 15 min (Sigma, USA). The pooled single-cell suspensions were washed in DMEM/F12 + 10%FBS (Gibco) and filtered through a 70 μm cell strainer (Jiete, China). After centrifugation (1000 rpm for 5 min), the supernatant was discarded and the isolated cells were then cultured in a 5% CO2 incubator at 37 °C with DMEM/F12 + 10%FBS + 10 μg/mL M-CSF (novaprotein, CB34). Microglial purification was conducted when mixed retinal cells were cultured for 13–14 days. After 14 days of culture, microglia were subjected to mild trypsinization (0.0625%) for 30 min at 37 °C to remove macroglia. 0.25% trypsin was then used to digest microglia for 10 min at 37 °C. The detached cells, composed of 95% microglia, were seeded into the Transwell system.
Transwell assay
The Corning™ Transwell systems with inserts (5-μm pores for primary microglia and 8-μm pores for HMC3 cells) were applied. HMC3 cells (100,000) from different groups were seeded in a medium consisting of 2% FBS in the upper compartment; the lower chamber consisted of 10% FBS. After 24-h incubation at 37 °C, LPS (1 μg/mL, Sigma) and IFN-γ (500 ng/mL, PeproTech) were added to the lower chamber. One day later, the HMC3 cells were washed with PBS twice and fixed with 4% paraformaldehyde for 20 min, and stained with crystal violet (Beyotime, China). The remaining cells in the upper compartment were removed, and the images of the stained cells were quantified by ImageJ (USA).
Wound healing assay
Cells were incubated until they formed a 90%–100% confluent monolayer. Next, the cells from different groups were scratched by a sterile pipette tip in an FBS-free medium. After 24-h incubation at 37 °C, the images of cell migration into the scratched area were obtained by a microscope, and the distances were analyzed by ImageJ (USA).
Quantification of m6A RNA levels
The colorimetric method was used to detect the overall level of m6A RNA modification according to the manufacturer's instructions for the EpiQuik m6A RNA Methylation Quantification Kit (EpiGentek, CA, P-9008-48).30 In brief, 100 ng of total RNA was bound to each well, and the antibody capture solution and antibody detection solution were added. Finally, the absorbance of the m6A modification was detected at 450 nm.
Single-cell RNA sequencing
RNA was extracted using TRIzol™ reagent. RNA quality was strictly controlled by Novogene Technology Co., Ltd. (Beijing, China). Library construction and transcriptome analysis were completed by Novogene Technology Co., Ltd. Differentially expressed genes (DEGs) were identified based on Padj < 0.05 and | log2 fold change (FC) | > 1.
Methylated RNA immunoprecipitation-quantitative PCR
MeRIP-qPCR was conducted according to a protocol slightly modified based on the previously described.31,32 In brief, the TRIzol method was used to extract total RNA. Fifty nanograms of total RNA was saved, and one-tenth was used as input. The remaining RNA was used for m6A immunoprecipitation with m6A antibody (Synaptic Systems, USA) in immunoprecipitation (IP) buffer (500 μL) to obtain m6A pulldown components (m6A IP portion). RNA from immunoprecipitated m6A was immunoprecipitated using Dynabeads Protein A (Thermo Fisher Scientific, USA), followed by elution. The RNA concentration of m6A IP RNAs recovered by ethanol precipitation was determined by spectrophotometry (Thermo Fisher Scientific, USA). Next, the input and m6A IP RNA (1 ng) were subjected to RT-qPCR, as described above.
RNA stability
The cells were pre-stimulated with LPS and IFN-γ in the experimental groups. Actinomycin D (Sigma, USA) at 4 μg/mL was added to the medium. The cells were harvested after incubation at specific time points (0, 2, 4, and 6 h), and the RNA was quantified via RT-qPCR.
Coculture of microglia and primary CD4+ T cells
Whole blood was provided by healthy donors from The First Affiliated Hospital of Chongqing Medical University. Peripheral blood mononuclear cells (PBMCs) were separated from whole blood using Ficoll density gradient centrifugation (TBD Science). CD4+ T cells from PBMCs were isolated following the manufacturer's instructions of the EasySep™ Cell Separation Kit (17952; STEMCELL Technologies, Canada). Isolated CD4+ T cells were plated in the top chamber at a density of 1 × 105 cells/well in 500 μL of RPMI 1640 medium consisting of 2% FBS, whereas HMC3 cells were added to the lower chamber in advance with 1 mL of EMEM consisting of 10% FBS. After the microglia were stimulated with LPS and IFN-γ for 24 h, we collected cell suspensions from the lower chamber for cell counting.
Statistical analysis
All the data were expressed as the mean ± SD and analyzed by IBM SPSS Statistics 20.0 (Chicago, USA). All the figures were constructed using GraphPad Prism version 8.0 software (San Diego, USA). For data conforming to normality and homogeneity of variance, unpaired Student's t-test was employed for comparisons of two sets of samples, and one-way ANOVA followed by Bonferroni's test was used in multiple groups. Otherwise, a nonparametric test was applied via the Mann–Whitney U test. The differences between groups at P < 0.05 were considered statistically significant.
Results
Microglial FTO was decreased during EAU progression
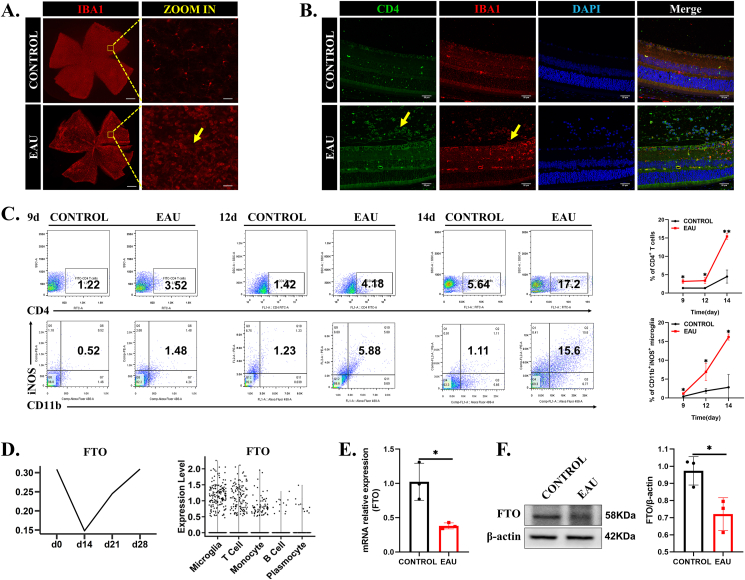
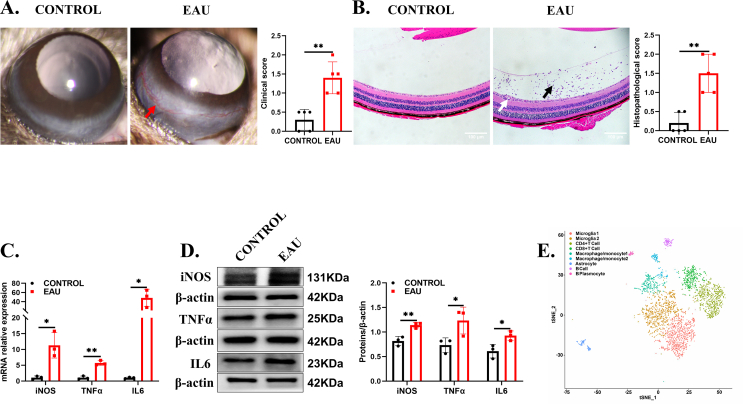
First, we stably generated EAU according to a previous study.33 The EAU mice exhibited conjunctival and ciliary hyperemia, infiltration of anterior chamber inflammatory cells, and more retinal folds than the control mice. Clinical scores and pathology scores were consistent with the representative characteristics (Fig. S1A, B). Furthermore, retinal inducible NOS (iNOS), tumor necrosis factor-α (TNFα), and interleukin 6 (IL6) levels were increased in the EAU mice (Fig. S1C, D). To further explore the functioning cells in the retina, we analyzed single-cell sequencing data of retinal cells on the 14th day (unpublished data) and found that among the CD45+ retinal immune cells, microglia played a key role in EAU development (Fig. S1E). To further test the changes of microglia in EAU, we performed immunofluorescence staining of whole retinas from the EAU mice and the control mice. Retinal flat mounts indicated that microglial numbers increased significantly on the 14th day, and the cell bodies became larger, showing an activated amoebic morphology (Fig. 1A). In addition, both CD4+ T cells and microglia infiltrated the entire retina at the peak of EAU inflammation (Fig. 1B). Given that microglia, but not macrophages, predominate in the retina, we performed flow cytometry to detect the number of CD4+ T cells and microglia (CD11b+iNOS+) during EAU progression. The results showed that CD4+ T cells and CD11b+iNOS+ microglia were increased from the 9th day to the 14th day (inflammation peak), and microglial activation and proliferation preceded CD4+ T cell infiltration, suggesting that activated microglia may play an important chemotactic role in EAU development (Fig. 1C). Based on the aforementioned scRNA-seq data, we compared the expression of m6A modification genes in retinal microglia at four different periods and found that the expression of m6A demethylase FTO was decreased on the 14th day, recovered on the 21st and 28th days, and the expression was negatively correlated with the severity of inflammation (Fig. 1D). To verify it, we also measured retinal FTO expression on the 14th day. Similarly, FTO expression was significantly reduced in the EAU mice (Fig. 1E, F). Taken together, these data indicated that microglial FTO was involved in the progression of EAU.
Figure 1.
Microglial FTO was decreased during EAU progression. (A) Retinal flat-mount staining with IBA1 (red)-labeled microglia of the control and EAU groups (n = 5). The arrows indicate activated microglia. Scale bar, 500 μm (left) and 25 μm (right). (B) CD4 (green) and IBA1 (red) staining of paraffin-embedded retinal cross-sections between the EAU mice and the control mice. The arrows indicate proliferating microglia and CD4+ T cells. Scale bar, 25 μm. (C) Retinal flow charts and quantification of CD4+ T cells (upper) and microglia (lower) at 9, 12, and 14 days of EAU. Microglia markers: CD11b and iNOS (n = 3). (D) scRNA-seq was used to detect FTO expression at 0, 14, 21, and 28 days of EAU. (E, F) The RNA and protein expression levels of FTO in the control and EAU groups (n = 3). Values are analyzed using the unpaired Student's t-test. ∗P < 0.05, ∗∗P < 0.01.
Downregulated FTO was involved in microglia-elicited inflammation and migration in vitro
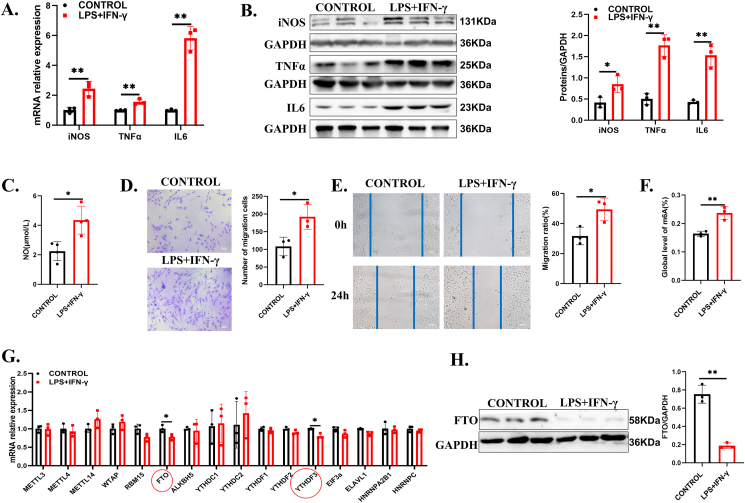
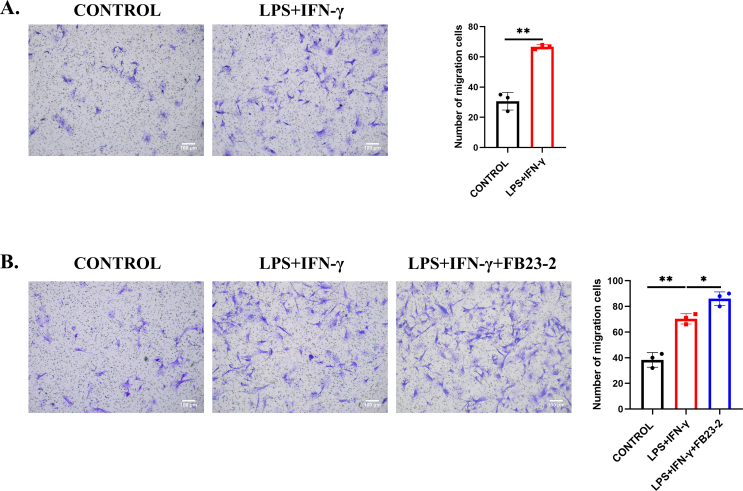
To further investigate the role of m6A modification in regulating microglial function, we established an inflammation model in HMC3 cells using LPS (1 μg/mL) and IFN-γ (500 ng/mL), the pro-inflammatory agonists were confirmed to activate microglia to inflammation state.34,35 RT-qPCR and Western blotting showed that the expression of M1 microglial markers, including iNOS, TNFα, and IL6, was upregulated significantly after 24-h stimulation, showing a tendency of microglial M1 polarization (Fig. 2A, B). In addition, the production of NO, which iNOS catalyzes from L-arginine, was obviously increased (Fig. 2C). Additionally, the Transwell assay was used to examine the phenotype of microglia. These results showed that the migration ability of HMC3 cells was significantly enhanced after inflammatory stimulation compared with those of the controls (Fig. 2D), and this result was similarly validated in primary microglia (Fig. S2A). Furthermore, the mobility of HMC3 cells was also enhanced in experimental groups by using the wound-healing model (Fig. 2E). To investigate the effect of m6A modification on inflammation, we examined the level of total m6A and found that m6A methylation was increased in the LPS- and IFN-γ-treated HMC3 cells compared with the control cells (Fig. 2F). We also tested the RNA expression of m6A-associated genes. The mRNA expression levels of the eraser FTO and the reader YTHDF3 were decreased significantly with LPS and IFN-γ, but we did not find any significant changes in other known m6A writers, erasers, or m6A-binding proteins in HMC3 cells (Fig. 2G). Consistently, FTO protein expression was also significantly decreased after LPS and IFN-γ stimulation (Fig. 2H). These data suggested that FTO downregulation aggravated inflammation in HMC3 cells.
Figure 2.
Downregulated FTO was involved in microglia-elicited inflammation and migration in vitro. (A, B) RT-qPCR and Western blot for detecting the expression of iNOS, TNFα, and IL6 with or without LPS + IFN-γ treatment for 24 h (n = 3). (C) Griess assay for detecting NO production in the supernatant with or without LPS + IFN-γ treatment for 24 h (n = 3). (D) Transwell images and corresponding quantifications in the control group and the LPS + IFN-γ group (n = 3). Scale bar, 100 μm. (E) Representative photomicrographs of the wound-healing assay performed on HMC3 cells before and after scratching. (F) The overall m6A% level of the control group and the LPS + IFN-γ group (n = 3). (G) RT-qPCR for testing the relative mRNA expression of m6A methylase, demethylase, and readers in the control group and the LPS + IFN-γ group (n = 3). (H) FTO protein expression in the control group and the LPS + IFN-γ group (n = 3). Values are analyzed using the unpaired Student's t-test. ∗P < 0.05, ∗∗P < 0.01.
FTO knockdown upregulated inflammation and promoted microglial migration
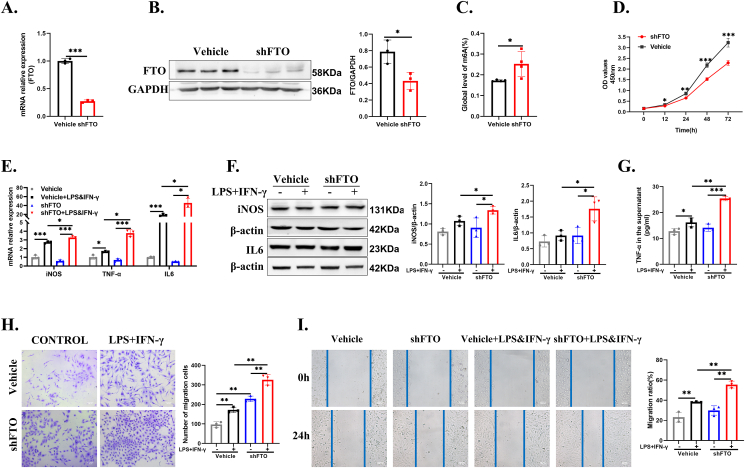
To verify the functional role of FTO in the development of inflammation in vitro, we transfected FTO shRNA into HMC3 cells to reduce its expression, and the knockdown efficiency was validated by RT-qPCR and Western blotting (Fig. 3A, B). Besides, the overall m6A modification rate was increased after inhibiting FTO expression (Fig. 3C). Then, we used CCK-8 analysis to evaluate cell proliferation, and the results showed that decreasing FTO expression reduced the proliferation of HMC3 cells (Fig. 3D). Next, the RNA and protein expression levels of iNOS, IL6, and TNFα were detected, and the data suggested that inflammatory factors in the shFTO group were significantly higher than those in the vehicle group (Fig. 3E–G). In addition, Transwell migration assays showed that HMC3 cell migration was significantly increased in response to LPS and IFN-γ and was obviously enhanced after FTO knockdown (Fig. 3H). To further explore the migration ability of microglia, a selective FTO inhibitor (FB23-2) was applied in primary microglia. The results revealed that FB23-2 aggravated the migration of primary microglia (Fig. S2B). Next, we assessed the cell motility of microglia by performing a scratch wound-healing assay. More invading cells were found in wells containing shFTO lentivirus than that in the vehicle (Fig. 3I). These results suggested that the downregulation of FTO was likely responsible for the inflammatory phenotypes in microglia cells.
Figure 3.
FTO knockdown upregulated inflammation and promoted microglial migration. (A,B) RT-qPCR and Western blot for determining FTO knockdown efficiency (n = 3, unpaired Student's t-test). (C) Percentage of m6A content of the total RNA in the vehicle group and the shFTO group (n = 3, unpaired Student's t-test). (D) CCK-8 assay for determining the proliferation of the vehicle and shFTO groups (n = 3; unpaired Student's t-test). (E, F) The mRNA and protein levels of iNOS, TNFα, and IL6 in the vehicle- and shFTO-treated cells with or without LPS + IFN-γ stimulation (n = 3; one-way ANOVA). (G) ELISAs for testing TNFα expression in the vehicle- or shFTO-treated cells with or without LPS + IFN-γ stimulation (n = 3; one-way ANOVA). (H) Transwell images and corresponding quantifications of the vehicle group, vehicle + LPS&IFN-γ group, shFTO group, and shFTO + LPS&IFN-γ group (n = 3; one-way ANOVA). (I) Representative images of the wound-healing assay performed in the vehicle group, vehicle + LPS&IFN-γ group, shFTO group, and shFTO + LPS&IFN-γ group (n = 3; one-way ANOVA). ∗P < 0.05, ∗∗P < 0.01.
GPC4 might be the downstream target of FTO
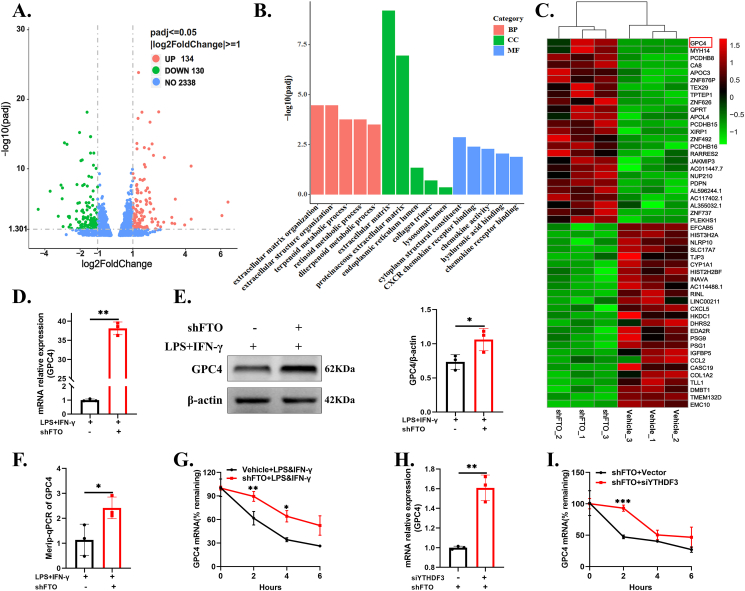
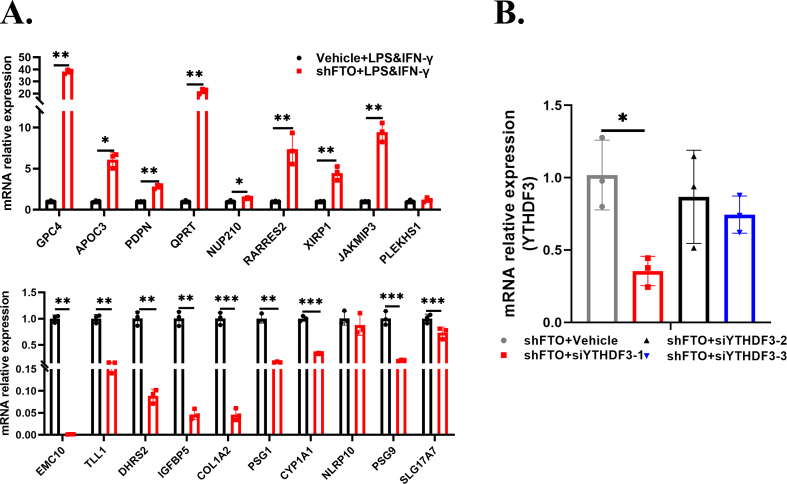
To investigate the molecular mechanism through which FTO knockdown aggravated microglia-mediated inflammation, we mapped RNA transcriptomics in the LPS- and IFN-γ-stimulated HMC3 cells with stable FTO reduction and the control cells. The volcano plot showed a total of 134 upregulated mRNAs and 130 downregulated mRNAs (| log2FC | > 1, Padj < 0.05) (Fig. 4A). Gene Ontology (GO) functional enrichment analysis was performed on the DEGs (Fig. 4B). The heatmap showed 25 upregulated genes and 25 downregulated genes (| log2FC | > 2, Padj < 0.05) (Fig. 4C). As inflammation was more severe after FTO knockdown, we hypothesized that anti-inflammatory genes were downregulated or proinflammatory genes were upregulated. Based on this assumption, 19 genes were selected as candidate genes (including 9 upregulated and 10 downregulated genes) from the heatmap by literature mining combined with GeneCards, BioGPS, and UCSC database analysis, and were verified using RT-qPCR (Fig. S3A). Among these candidates, through RT-qPCR, we found that GPC4 had the most obviously increased expression after FTO knockdown (Fig. 4D). Given these findings, we then evaluated the protein levels of GPC4 and found that GPC4 was obviously increased in the FTO knockdown cells (Fig. 4E). GPC4, a glycosylphosphatidylinositol (GPI)-anchored heparan sulfate proteoglycan (HSPG), is an evolutionarily conserved synaptic organizing protein.36 We further investigated whether FTO-mediated GPC4 upregulation was caused by transcriptional modification. MeRIP-qPCR and mRNA stability assays were conducted to determine the intrinsic mechanism by which FTO affects GPC4 mRNA. As expected, FTO knockdown markedly increased the m6A methylation of GPC4 total RNA (Fig. 4F) and enhanced the stability of this protein (Fig. 4G). Combined with the results of RNA-qPCR in Figure 2G, these findings showed that YTHDF3 may play an important role in the shFTO HMC3 cells because m6A readers can directly or indirectly recognize and bind to m6A-modified RNAs to play specific roles.37 Then, we transfected three different YTHDF3 siRNAs and the corresponding vector into the FTO-deficient cells, with siYTHDF3-1 having the highest knockdown efficiency (Fig. S3B). GPC4 mRNA expression was significantly increased in the shFTO + siYTHDF3 group compared with the control group (Fig. 4H). As a member of the YTH (YT521-B homology) domain protein family, YTHDF3 can affect mRNA stability. Therefore, we measured the half-life of GPC4 mRNA in the YTHDF3-depleted FTO-deficient cells. YTHDF3 silencing obviously increased the stability of GPC4 mRNA (Fig. 4I). Collectively, these findings suggested that FTO regulated GPC4 expression in a YTHDF3-dependent manner.
Figure 4.
GPC4 might be the downstream target of FTO. (A,B) The volcano plot of DEGs in the vehicle + LPS&IFN-γ and shFTO + LPS&IFN-γ cells (| log2FC | > 1, Padj < 0.05). (C) The heatmap of significant DEGs in the abovementioned groups (| log2FC | > 1, Padj < 0.05). (D,E) The RT-qPCR and Western blot for validating GPC4 expression in the abovementioned groups. (F) m6A enrichment of GPC4 mRNA was detected by MeRIP-qPCR in the abovementioned groups. (G) RT-qPCR for showing GPC4 mRNA expression after the administration of ActD (5 μg/mL) for 0, 2, 4, and 6 h in the abovementioned groups. (H) RT-qPCR for detecting GPC4 mRNA levels in the shFTO + siYTHDF3 group and the shFTO + vector group. (I) RNA stability analysis in the shFTO + siYTHDF3 group and shFTO + vector group (n = 3). Values are analyzed using the unpaired Student's t-test. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
FTO-mediated GPC4 regulated microglia through the TLR4/NF-κB signaling pathway
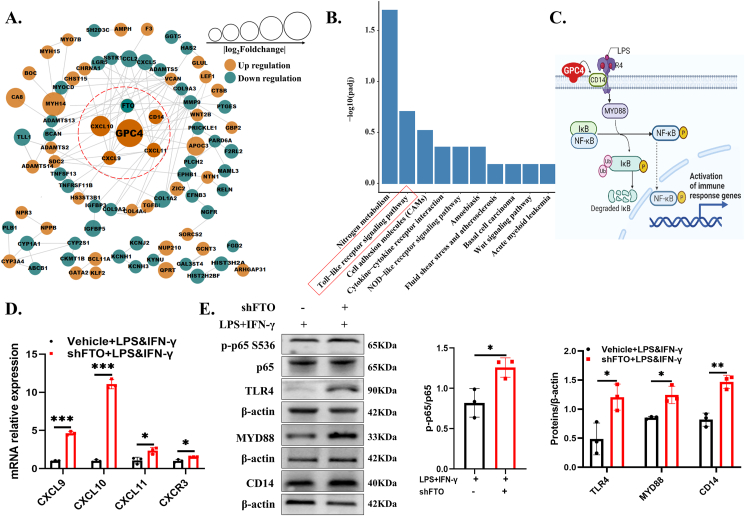
To determine the relationships among FTO, GPC4, and other genes, we used the STRING database and Cytoscape to map the protein–protein interaction (PPI) network and combined the results with Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment data to identify downstream signaling pathways. Construction of the PPI network revealed the potential connection between FTO, GPC4, CD14 and chemokines (including CXCL9, CXCL10 and CXCL11) (Fig. 5A). Since CD14, CXCL9, CXCL10, and CXCL11 were all enriched in the Toll-like receptor signaling pathway (KEGG hsa04510) and this pathway was ranked second among the 10 most significantly enriched pathways according to KEGG analysis (Padj < 0.05) (Fig. 5B), we proposed that the interaction between GPC4 and CD14 (an LPS receptor of the TLR4 pathway anchored to the cell membrane by GPI) might promote the TLR4/NF-kB pathway in the shFTO microglia (Fig. 5C). We verified the importance of this pathway, and our results showed that FTO knockdown increased the expressions of chemokines including CXCL9, CXCL10, CXCL11 and their co-receptor CXCR3 (Fig. 5D). We further found that FTO depletion facilitated the phosphorylation of NF-κB p65 (Ser536) and increased the expression of TLR4, MYD88 and CD14 in the LPS- and IFN-γ-treated cells (Fig. 5E), indicating that disruption of FTO may promote the expression of key proteins in the TLR4/NF-κB pathway mediated by GPC4.
Figure 5.
FTO-mediated GPC4 regulated microglia through the TLR4/NF-κB signaling pathway. (A) Identification of the most significant module from the PPI network using the STRING database. (B) KEGG pathway enrichment analysis of DEGs. (C) Schematic diagram of the conjectured pathway. (D) The mRNA levels of CXCL9, CXCL10, CXCL11, CXCR3, TLR4, and CD14 in the vehicle + LPS&IFN-γ group, and the shFTO + LPS&IFN-γ group were measured via RT-qPCR. (E) Western blotting for showing the protein expression of NF-κB p-p65 (S536), p65, TLR4, MYD88, and CD14 in the vehicle + LPS&IFN-γ group and the shFTO + LPS&IFN-γ group (n = 3). Values are analyzed using the unpaired Student's t-test. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001.
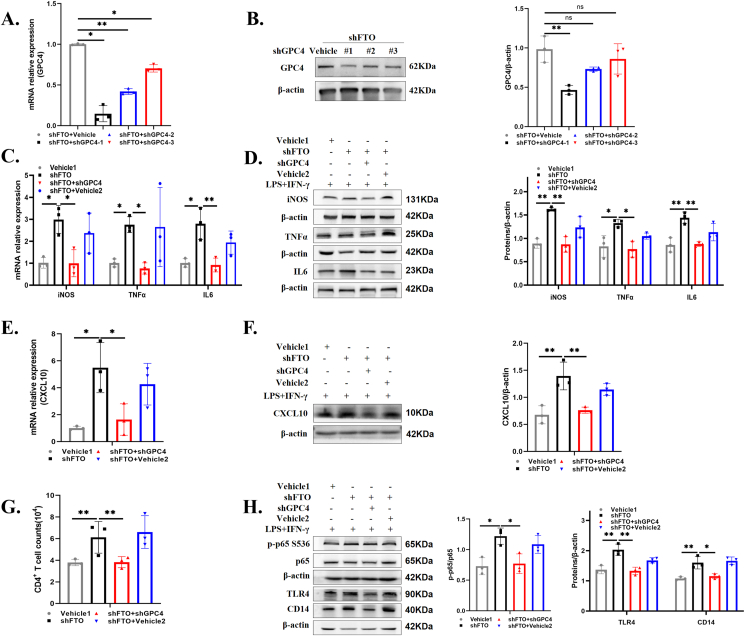
GPC4 knockdown reversed the effects of FTO in microglia
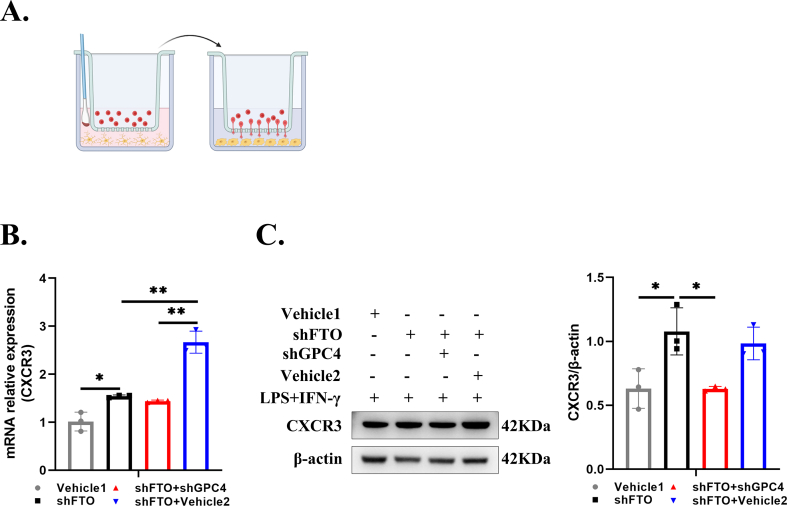
To further verify whether FTO-modulated GPC4 is a potential activator of the TLR4/NF-κB pathway that mediates inflammation and migration, we stably transfected FTO-deficient HMC3 cells with lentiviral vehicle or three lentiviral shGPC4 vectors. RT-qPCR and Western blotting were performed, and the results showed that shGPC4-1 had the strongest inhibitory efficiency, while shGPC4-2/3 had a weak inhibitory effect on GPC4 with FTO knockdown (Fig. 6A, B). Subsequently, GPC4 downregulation alleviated the increase in the mRNA expression of inflammatory factors caused by shFTO in HMC3 cells stimulated with LPS and IFN-γ (Fig. 6C), and protein expression was proven by Western blotting (Fig. 6D). Furthermore, GPC4 silencing effectively reduced the upregulation of CXCL10 in the shFTO cells (Fig. 6E, F). CXCL10 acts as a chemoattractant for T cells and regulates T cell functions such as adhesion.38 Hence, we isolated CD4+ T cells from freshly prepared PBMCs and co-cultured them with different groups of HMC3 cells primed with LPS and IFN-γ (Fig. S4A). We found that the number of migrated CD4+ T cells cocultured with shFTO HMC3 cells was prominently increased, which was observably reversed by GPC4 knockdown (Fig. 6G). In addition, GPC4 knockdown partially down-regulated the expression of CXCR3, which was increased by FTO downregulation (Fig. S4B, C). Here, we found that FTO silencing facilitated NF-κB p65 protein phosphorylation and increased the protein levels of CD14 and TLR4 with LPS and IFN-γ stimulation, while GPC4 knockdown partially reversed the activation of this pathway induced by FTO knockdown (Fig. 6H). Our data indicated that FTO knockdown exerted an injury-promoting effect and activated TLR4/NF-κB signaling in HMC3 cells, and this promotion could be reversed by the depletion of GPC4.
Figure 6.
GPC4 knockdown reversed the effects of FTO on microglia. (A,B) RT-qPCR and Western blot for detecting GPC4 expression in the FTO-deficient HMC3 cells transfected with vehicle lentivirus, shGPC4-1, shGPC4-2, or shGPC4-3 lentivirus. (C,D) RT-qPCR and Western blot for showing the expression of iNOS, TNFα, and IL6 in vehicle 1 (corresponding to shFTO) +LPS&IFN-γ, shFTO + LPS&IFN-γ, shFTO + shGPC4+LPS&IFN-γ, and shFTO + vehicle 2 (corresponding to shGPC4) +LPS&IFN-γ groups. (E, F) CXCL10 mRNA and protein expression in different groups with the abovementioned stimulation. (G) The number of human CD4+ T cells that migrated into lower HMC3 cells in the abovementioned groups. (H) Expression levels of key proteins involved in the TLR4 pathway (TLR4, CD14, and CXCL10) in the abovementioned groups (n = 3). Values are analyzed using the one-way ANOVA. ∗P < 0.05; ∗∗P < 0.01.
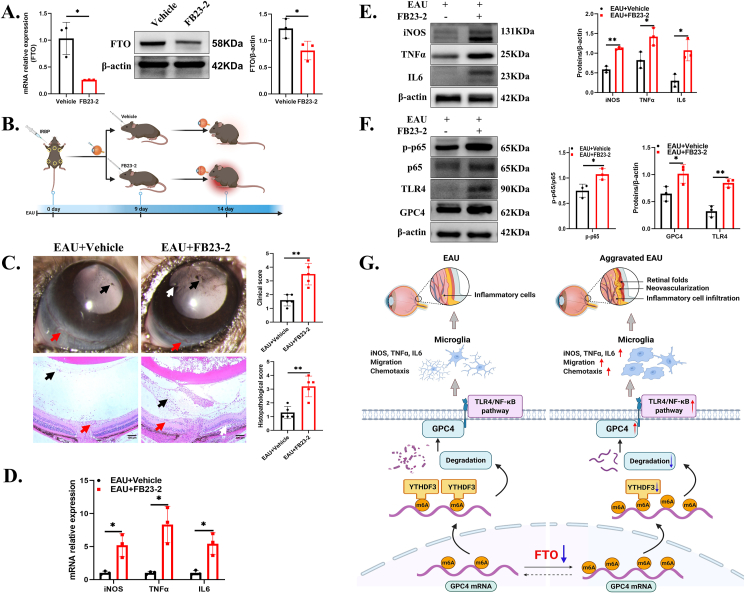
FTO inhibition aggravated EAU progression in vivo
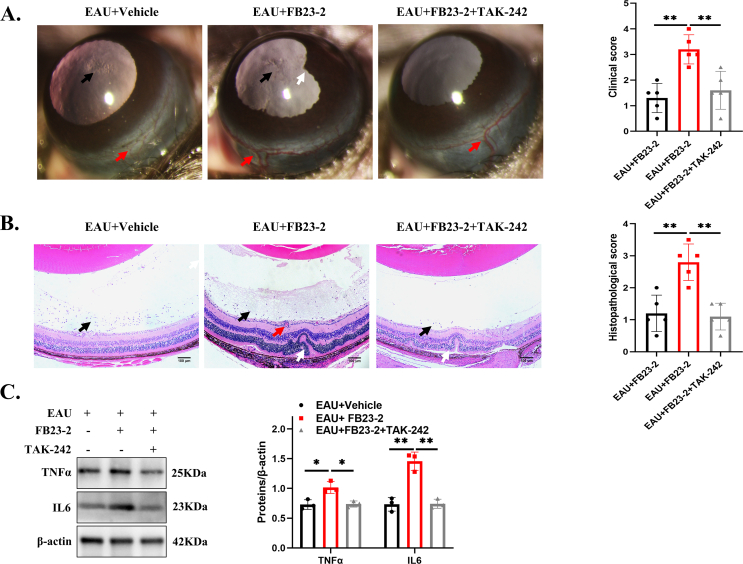
To validate the effect of FTO, we injected 10 μM FB23-2 (a potent and selective FTO inhibitor) into the vitreous cavity. Mice were sacrificed after 5 days, and retinal RNA and proteins were extracted. Our results showed that FB23-2 significantly decreased the mRNA and protein expression of retinal FTO (Fig. 7A). Furthermore, to determine the role of FTO in EAU progression, we treated the EAU mice with FB23-2 via an intravitreal injection after 9 days of IRBP immunization (Fig. 7B). The EAU + FB23-2 group showed higher clinical and histopathological scores on day 14 than the EAU + vehicle group (Fig. 7C). Furthermore, higher expression levels of retinal inflammatory factors were found in the FB23-2 group than in the vehicle group, including iNOS, TNFα, and IL6 (Fig. 7D, E). Next, the results showed that the phosphorylation of NF-κB p65 was increased significantly. Similarly, the GPC4 and TLR4 protein levels markedly increased (Fig. 7F). It has been reported that the NF-κB inhibitors could alleviate EAU progression.39,40 Additionally, we used the TLR4 inhibitor TAK-242 (resatorvid) after FB23-2 intravitreal injection to verify the FTO inhibitor's effect and found that EAU could be alleviated by inhibiting this pathway according to clinical and histopathological scores (Fig. S5A, B). The protein expression of TNFα and IL6 showed a significant decrease in the EAU + FB23-2+TAK-242 group (Fig. S5C). Based on the above evidence, FTO inhibition likely aggravated EAU progression through the activation of the GPC4/TLR4/NF-κB pathway.
Figure 7.
FTO inhibition aggravated EAU progression in vivo. (A) Retinal FTO mRNA and protein expression in the FB23-2-treated mice and the PBS + DMSO-treated mice (n = 3). (B) Experimental flowchart describing the modeling process and drug injection. (C) Representative images and quantification of the ocular anterior chamber (upper) and eyeball sections (lower) (n = 5). Black arrows indicate inflammatory cells. Red arrows indicate conjunctival or ciliary hyperemia, or vasculitis. White arrows indicate posterior synechiae, or retinal folds. Scale bar, 100 μm. (D, E) RT-qPCR and Western blot for showing the expression levels of iNOS, TNFα, and IL6 in the EAU + vehicle and EAU + FB23-2 groups (n = 3). (F) Protein expression levels of GPC4, TLR4, p-p65, and p65 in the EAU + vehicle and EAU + FB23-2 groups (n = 3). Values are analyzed using the unpaired Student's t-test. ∗P < 0.05; ∗∗P < 0.01. (G) Graphical summary of this study.
Discussion
Our work has provided strong evidence for the critical role of FTO in the development of EAU. First, microglial FTO expression was decreased significantly in EAU mice, which was parallel to the results in HMC3 cells. Second, FTO knockdown aggravated the microglia-elicited inflammatory response, including secretion of inflammatory factors as well as mobility and chemotaxis of CD4+ T cells in vitro. Third, RNA-seq and subsequent rescue validation showed that FTO targeted GPC4 to activate the TLR4/NF-κB axis, which further mediated microglial inflammation. For reader proteins, the data indicated that FTO-induced GPC4 increased mainly in an m6A-YTHDF3-dependent manner. Finally, the FTO inhibitor FB23-2 increased the production of inflammatory factors via activation of the GPC4/TLR4/NF-κB pathway and then exacerbated retinal inflammation in vivo. In summary, our study indicated that FTO-mediated m6A modification regulated microglial plasticity via the GPC4/TLR4/NF-κB signaling axis in uveitis (Fig. 7G).
Uveitis represents a group of immune-mediated diseases, but the exact pathogenesis is obscure.41 Previous studies have shown that epigenetic factors such as DNA methylation could play crucial roles in the progression of uveitis.42 Qiu et al. proved that the methylation of TBX21 and RORC (both are T cell transcription factors) obviously changed during the progression of EAU.43,44 Another study found that the promoter methylation levels of GATA3, TGF-β, and IL-4 were significantly upregulated in patients with Vogt-Koyanagi-Harada disease.45 We provided evidence that m6A RNA modification of microglia would be helpful to identify new pathogenic risk genes causing uveitis. Our previous study showed that YTHDC1, an m6A modification reader, played a critical role in microglial inflammatory response regulation and then participated in the development of autoimmune ocular diseases.46 In the study, we focused on exploring the relationship between FTO and autoimmune uveitis. Furthermore, our RNA-seq analysis revealed that FTO knockdown impacted multiple aspects of inflammation, including migration, retinoid metabolic processes, and cytokine–cytokine receptor interactions. The results described the roles of FTO in the autoimmune process and helped identify and discover key regulators of autoimmune uveitis.
GPC4, which belongs to the HSPG family,47 was identified as a target of FTO and may participate in the TLR4/NF-κB pathway by anchoring CD14. GPC4 was reported to be an essential downstream effector of CD36, and ectopic GPC4 could reverse the tumor suppressive function of CD36 by activating the β-catenin/c-Myc pathway in CRC cells.48 As a GPI-anchored protein, GPC4 can be released from producing cells through different mechanisms, including GPI anchor cleavage, proteolytic shedding, and vesicular release.49 Therefore, the specific mechanism by which GPC4 interacts with CD14 in the TLR4 pathway needs further study. In addition, our findings indicated that the half-life of GPC4 mRNA in the FTO-silenced HMC3 cells was longer than that in the controls, and this effect was mainly dependent on YTHDF3. YTHDF3, a member of the YTH family,50,51 promoted protein translation together with YTHDF1, and could promote mRNA degradation through YTHDF2.52 Another study has shown that YTHDF1 enhances RNA translation, DF2 promotes RNA degradation, and DF3 enhances translation and degradation.53,54 YTHDF3 knockdown augmented the GPC4 expression level and prolonged the GPC4 half-life in microglia, indicating that YTHDF3 could promote the degradation of GPC4. However, whether YTHDF3 affects the translation of GPC4 and how it functions with other readers require further study.
We used a similar approach to construct the EAU model using female mice referred to previous studies on EAU,33,55,56 but whether our conclusions could be extrapolated to microglia of male mice requires further study. Since microglia account for most of the CD45+ retinal immune cell populations and play important roles in EAU,57,58 we performed the flow cytometry using CD11b and iNOS to label microglia according to previous studies.59,60 In addition, LPS- and IFN-γ-treated HMC3 cells were used to simulate the EAU based on the previous studies.61,62 There were other cell models to simulate EAU, such as immune T-cells producing high IFN-γ and low IL-4 in vitro when stimulated with IRBP.63 More defined cell models for simulating uveitis should be explored. Furthermore, we interfere with GPC4 only using in vitro EAU model, mainly due to the absence of GPC4 inhibitors or activators, the low transfection efficiency of microglia,64,65 the high costs, and time commitment of transgenic mice. All these above issues will be further explored in our future studies.
Collectively, an important proinflammatory role for decreased FTO in autoimmune uveitis was identified, which was involved in the upregulation of the target GPC4 modification via the CD14/TLR4/NF-κB pathway. Our findings expand the field of RNA epigenetic regulation. More importantly, the possibility that FTO may have other roles in uveitis progression should be explored.
Conclusions
In summary, decreased FTO regulates microglial plasticity, including secretion of inflammatory factors, cell mobility, and chemotaxis, accompanied by increased ocular inflammation in EAU mice. These processes are driven by YTHDF3 downregulation and subsequent upregulation of the GPC4/TLR4/NF-κB pathway. This study indicates that m6A modification might be a potentially effective therapeutic target for autoimmune diseases such as uveitis.
Author contributions
S.Y.H. and W.Q.L. designed the project, performed the experiments, analyzed and interpreted the data, and wrote the whole manuscript. G.Q.W., X.T.W., W.F., and Z.Z. helped perform some of the experiments. S.P.H and N.L. supervised the research program and edited the manuscript.
Data availability
The original contributions presented in this study are included in the supplementary material, and any further data not included in this article are available from the corresponding author upon reasonable request.
Conflict of interests
The authors declare that they have no conflict of interests in this work.
Funding
This study was supported by the National Natural Science Foundation Project of China (No. 82070951, 82271078 and 81873678); the Innovative Research Group Project of Chongqing Education Commission (China) (No. CXQT19015); the Natural Science Foundation Project of Chongqing, China (No. cstc2019jcyjmsxmX0120); the Innovation Supporting Plan of Overseas Study of Chongqing, China (No. cx2018010); the Chongqing Education Commission (China) (No. KJQN202000406); the National Key Clinical Specialties Construction Program of China, Chongqing Branch of National Clinical Research Center for Ocular Diseases; the Chongqing Key Laboratory of Ophthalmology (China) (CSTC, No. 2008CA5003); and Natural Science Foundation Project of Chongqing Medical University (China) (No. W0047).
Footnotes
Peer review under responsibility of Chongqing Medical University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gendis.2022.09.008.
Contributor Information
Na Li, Email: 102600@cqmu.edu.cn.
Shengping Hou, Email: sphou828@163.com.
Abbreviations
- ALKBH5
AlkB homolog 5
- ANOVA
Analysis of variance
- CXCL9
Chemokine (C-X-C motif) ligand 9
- CXCL10
Chemokine (C-X-C motif) ligand 10
- CXCL11
Chemokine (C-X-C motif) ligand 11
- DCs
Dendritic cells
- DEGs
Differentially expressed genes
- DMEM
Dulbecco's modified Eagle's medium
- DMSO
Dimethyl sulfoxide
- EAU
Experimental autoimmune uveitis
- FBS
Fetal bovine serum
- FTO
Fat mass and obesity-associated protein
- GO
Gene Ontology
- GPC4
Glypican 4
- GPI
Glycosylphosphatidylinositol
- HMC3
Human microglia clone 3
- HSPG
Heparan sulfate proteoglycan
- IBA-1
Ionized calcium binding adapter molecule 1
- IFN-γ
Interferon γ
- IL6
Interleukin-6
- iNOS
Inducible nitric oxide synthase
- IRBP
Interphotoreceptor retinoid binding protein
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LPS
Lipopolysaccharide
- MeRIP-qPCR
m6A RNA immunoprecipitation quantitative polymerase chain reaction
- METTL3
Methyltransferase 3
- METTL14
Methyltransferase 14
- m6A
N6-methyladenosine
- NF-κB
Nuclear factor κB
- NO
Nitric oxide
- PBS
Phosphate-buffered saline
- PPI
Protein–protein interaction
- RBM15
RNA binding motif protein 15
- RNA-seq
RNA sequencing
- RPE
Retinal pigment epithelium
- RT-qPCR
Reverse transcription quantitative polymerase chain reaction
- scRNA-seq
Single cell RNA sequencing
- TBST
Tris-buffered saline plus Tween-20
- TLR4
Toll-like receptor 4
- TNFα
Tumor necrosis factor-α
- Th1
T helper 1 cells
- Th17
T helper 17 cells
- WTAP
Wilms' tumor 1-associating protein
- YTHDF3
YTH domain family protein 3
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Figure S1.
Construction of EAU mice and unbiased classification of retinal immune cells via scRNA-seq. (A, B) Retinal anterior chamber inflammation, retinal HE staining, and corresponding clinical scores and pathology scores. Red arrows indicate conjunctival and ciliary hyperemia. Black arrows indicate inflammatory cells of eyeball sections. White arrow indicates vasculitis. n = 5. Mann–Whitney U test. ∗∗P < 0.01. (C, D) RT-qPCR and Western blot for showing the expression levels of iNOS, TNFα, and IL6 in the control and EAU groups. n = 3. Unpaired Student's t-test. ∗P < 0.05; ∗∗P < 0.01. (E) t-SNE embedding of scRNA-seq for showing different cell-type clusters in EAU retinas.
Figure S2.
The migration ability of primary microglia in different groups. (A) Representative Transwell images and quantifications of primary microglia in the control group and the LPS + IFN-γ group (n = 3; unpaired Student's t-test). Scale bar, 100 μm. (B) Representative images and quantifications of primary microglia in the control group, LPS&IFN-γ group, and LPS&IFN-γ + FB23-2 group (n = 3; one-way ANOVA). Scale bar, 100 μm ∗P < 0.05; ∗∗P < 0.01.
Figure S3.
Validation of DEGs according to the RNA-seq data and knockdown efficiency of YTHDF3. (A) RNA expression of DEGs detected by RT-qPCR. Upper, upregulated DEGs. Lower, downregulated DEGs. n = 3. Unpaired Student's t-test. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. (B) Knockdown efficiency of YTHDF3 determined by RT-qPCR. n = 3. Unpaired Student's t-test. ∗P < 0.05.
Figure S4.
GPC4 knockdown reversed the expression of CXCR3 in HMC3 cells. (A) Experimental flowchart of coculture. (B, C) RT-qPCR and Western blot for showing the expression of CXCR3 in four different groups with LPS and IFN-γ stimulation. n = 3. One-way ANOVA. ∗P < 0.05; ∗∗P < 0.01.
Figure S5.
TLR4 inhibitor could partially rescue the severe inflammation of EAU caused by decreased FTO. (A) The anterior chamber inflammation and clinical scores of EAU + Vehicle, EAU + FB23-2, and EAU + FB23-2 + TAK-242 groups. Red arrows indicate conjunctival and/or ciliary congestion. Black arrows indicate infiltration of inflammatory cells. White arrow indicates posterior synechiae. (B) The retinal histopathological staining and corresponding scores of EAU + Vehicle, EAU + FB23-2, and EAU + FB23-2 + TAK-242 groups. Red arrows indicate vasculitis. Black arrows indicate infiltration of inflammatory cells. White arrow indicates retinal folds. Scale bar, 100 μm. (C) Western blot for showing the expression of TNFα and IL6 in the abovementioned three groups. n = 3. One-way ANOVA. ∗P < 0.05; ∗∗P < 0.01.
References
- 1.Chong W.P., Mattapallil M.J., Raychaudhuri K., et al. The cytokine IL-17A limits Th17 pathogenicity via a negative feedback loop driven by autocrine induction of IL-24. Immunity. 2020;53(2):384–397. doi: 10.1016/j.immuni.2020.06.022. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang C., Tian Y., Lei B., et al. Decreased IL-27 expression in association with an increased Th17 response in Vogt-Koyanagi-Harada disease. Invest Ophthalmol Vis Sci. 2012;53(8):4668–4675. doi: 10.1167/iovs.12-9863. [DOI] [PubMed] [Google Scholar]
- 3.Stadhouders R., Lubberts E., Hendriks R.W. A cellular and molecular view of T helper 17 cell plasticity in autoimmunity. J Autoimmun. 2018;87:1–15. doi: 10.1016/j.jaut.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 4.Lee R.W., Nicholson L.B., Sen H.N., et al. Autoimmune and autoinflammatory mechanisms in uveitis. Semin Immunopathol. 2014;36(5):581–594. doi: 10.1007/s00281-014-0433-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang B., Tian Q., Guo D., et al. Activated γδ T cells promote dendritic cell maturation and exacerbate the development of experimental autoimmune uveitis (EAU) in mice. Immunol Invest. 2021;50(2–3):164–183. doi: 10.1080/08820139.2020.1716786. [DOI] [PubMed] [Google Scholar]
- 6.Stratoulias V., Venero J.L., Tremblay M.È., et al. Microglial subtypes: diversity within the microglial community. EMBO J. 2019;38(17) doi: 10.15252/embj.2019101997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fan W., Huang W., Chen J., et al. Retinal microglia: functions and diseases. Immunology. 2022;166(3):268–286. doi: 10.1111/imm.13479. [DOI] [PubMed] [Google Scholar]
- 8.Lan X., Han X., Li Q., et al. Modulators of microglial activation and polarization after intracerebral haemorrhage. Nat Rev Neurol. 2017;13(7):420–433. doi: 10.1038/nrneurol.2017.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heng J.S., Hackett S.F., Stein-O'brien G.L., et al. Comprehensive analysis of a mouse model of spontaneous uveoretinitis using single-cell RNA sequencing. Proc Natl Acad Sci U S A. 2019;116(52):26734–26744. doi: 10.1073/pnas.1915571116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.O'Koren E.G., Yu C., Klingeborn M., et al. Microglial function is distinct in different anatomical locations during retinal homeostasis and degeneration. Immunity. 2019;50(3):723–737.e7. doi: 10.1016/j.immuni.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Okunuki Y., Mukai R., Nakao T., et al. Retinal microglia initiate neuroinflammation in ocular autoimmunity. Proc Natl Acad Sci U S A. 2019;116(20):9989–9998. doi: 10.1073/pnas.1820387116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y., Li L., Li J., et al. The emerging role of m6A modification in regulating the immune system and autoimmune diseases. Front Cell Dev Biol. 2021;9 doi: 10.3389/fcell.2021.755691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin Y., Li B., Arumugam S., et al. m6A mRNA methylation-directed myeloid cell activation controls progression of NAFLD and obesity. Cell Rep. 2021;37(6) doi: 10.1016/j.celrep.2021.109968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi H., Wei J., He C. Where, when, and how: context-dependent functions of RNA methylation writers, readers, and erasers. Mol Cell. 2019;74(4):640–650. doi: 10.1016/j.molcel.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meng J., Liu X., Tang S., et al. METTL3 inhibits inflammation of retinal pigment epithelium cells by regulating NR2F1 in an m6A-dependent manner. Front Immunol. 2022;13 doi: 10.3389/fimmu.2022.905211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang S., Meng J., Tan J., et al. N6-methyladenosine demethylase FTO regulates inflammatory cytokine secretion and tight junctions in retinal pigment epithelium cells. Clin Immunol. 2022;241 doi: 10.1016/j.clim.2022.109080. [DOI] [PubMed] [Google Scholar]
- 17.Zhou J., Zhang X., Hu J., et al. m6A demethylase ALKBH5 controls CD4+ T cell pathogenicity and promotes autoimmunity. Sci Adv. 2021;7(25) doi: 10.1126/sciadv.abg0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu T.X., Zheng Z., Zhang L., et al. A new model of spontaneous colitis in mice induced by deletion of an RNA m 6 A methyltransferase component METTL14 in T cells. Cell Mol Gastroenterol Hepatol. 2020;10(4):747–761. doi: 10.1016/j.jcmgh.2020.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bechara R., Amatya N., Bailey R.D., et al. The m6A reader IMP2 directs autoimmune inflammation through an IL-17- and TNFα-dependent C/EBP transcription factor axis. Sci Immunol. 2021;6(61) doi: 10.1126/sciimmunol.abd1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang J., Liu J., Zhao S., Tian F. N 6-methyladenosine METTL3 modulates the proliferation and apoptosis of lens epithelial cells in diabetic cataract. Mol Ther Nucleic Acids. 2020;20:111–116. doi: 10.1016/j.omtn.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paramasivam A., Priyadharsini J.V., Raghunandhakumar S. Implications of m6A modification in autoimmune disorders. Cell Mol Immunol. 2020;17(5):550–551. doi: 10.1038/s41423-019-0307-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Q., Gao Y., Zhang L., et al. Decreased ALKBH5, FTO, and YTHDF2 in peripheral blood are as risk factors for rheumatoid arthritis. Biomed Res Int. 2020;2020 doi: 10.1155/2020/5735279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hjort R., Ahlqvist E., Andersson T., et al. Physical activity, genetic susceptibility, and the risk of latent autoimmune diabetes in adults and type 2 diabetes. J Clin Endocrinol Metab. 2020;105(11):e4112–e4123. doi: 10.1210/clinem/dgaa549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang Y., He J., Liang H., et al. Aryl hydrocarbon receptor regulates apoptosis and inflammation in a murine model of experimental autoimmune uveitis. Front Immunol. 2018;9:1713. doi: 10.3389/fimmu.2018.01713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y., Su R., Sheng Y., et al. Small-molecule targeting of oncogenic FTO demethylase in acute myeloid leukemia. Cancer Cell. 2019;35(4):677–691. doi: 10.1016/j.ccell.2019.03.006. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng D.W., Deng W.W., Song W.F., et al. Biomaterial-mediated modulation of oral microbiota synergizes with PD-1 blockade in mice with oral squamous cell carcinoma. Nat Biomed Eng. 2022;6(1):32–43. doi: 10.1038/s41551-021-00807-9. [DOI] [PubMed] [Google Scholar]
- 27.Ono Y., Maejima Y., Saito M., et al. TAK-242, a specific inhibitor of Toll-like receptor 4 signalling, prevents endotoxemia-induced skeletal muscle wasting in mice. Sci Rep. 2020;10(1):694. doi: 10.1038/s41598-020-57714-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu M.X., Zhao G.L., Hu X., et al. P2X7/P2X4 receptors mediate proliferation and migration of retinal microglia in experimental Glaucoma in mice. Neurosci Bull. 2022;38(8):901–915. doi: 10.1007/s12264-022-00833-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu S.T., Zhong S.M., Li X.Y., et al. EphrinB/EphB forward signaling in Müller cells causes apoptosis of retinal ganglion cells by increasing tumor necrosis factor alpha production in rat experimental glaucomatous model. Acta Neuropathol Commun. 2018;6(1):111. doi: 10.1186/s40478-018-0618-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen X., Gong W., Shao X., et al. METTL3-mediated m 6 A modification of ATG7 regulates autophagy-GATA4 axis to promote cellular senescence and osteoarthritis progression. Ann Rheum Dis. 2022;81(1):87–99. doi: 10.1136/annrheumdis-2021-221091. [DOI] [PubMed] [Google Scholar]
- 31.Lin X., Chai G., Wu Y., et al. RNA m6A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat Commun. 2019;10(1):2065. doi: 10.1038/s41467-019-09865-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Li Z., Weng H., Su R., et al. FTO plays an oncogenic role in acute myeloid leukemia as a N 6-methyladenosine RNA demethylase. Cancer Cell. 2017;31(1):127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tan J., Liu H., Huang M., et al. Small molecules targeting RORγt inhibit autoimmune disease by suppressing Th17 cell differentiation. Cell Death Dis. 2020;11(8):697. doi: 10.1038/s41419-020-02891-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qin C., Liu Q., Hu Z.W., et al. Microglial TLR4-dependent autophagy induces ischemic white matter damage via STAT1/6 pathway. Theranostics. 2018;8(19):5434–5451. doi: 10.7150/thno.27882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y., Zhao C., Meng J., et al. Galectin-3 regulates microglial activation and promotes inflammation through TLR4/MyD88/NF-kB in experimental autoimmune uveitis. Clin Immunol. 2022;236 doi: 10.1016/j.clim.2022.108939. [DOI] [PubMed] [Google Scholar]
- 36.Anderson D.J., Le Moigne R., Djakovic S., et al. Targeting the AAA ATPase p97 as an approach to treat cancer through disruption of protein homeostasis. Cancer Cell. 2015;28(5):653–665. doi: 10.1016/j.ccell.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S., Chai P., Jia R., Jia R. Novel insights on m 6 A RNA methylation in tumorigenesis: a double-edged sword. Mol Cancer. 2018;17(1):101. doi: 10.1186/s12943-018-0847-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mikolajczyk T.P., Szczepaniak P., Vidler F., Maffia P., Graham G.J., Guzik T.J. Role of inflammatory chemokines in hypertension. Pharmacol Ther. 2021;223 doi: 10.1016/j.pharmthera.2020.107799. [DOI] [PubMed] [Google Scholar]
- 39.Kitamei H., Iwabuchi K., Namba K., et al. Amelioration of experimental autoimmune uveoretinitis (EAU) with an inhibitor of nuclear factor-kappaB (NF-kappaB), pyrrolidine dithiocarbamate. J Leukoc Biol. 2006;79(6):1193–1201. doi: 10.1189/jlb.0805453. [DOI] [PubMed] [Google Scholar]
- 40.Hsu S.M., Yang C.H., Shen F.H., Chen S.H., Lin C.J., Shieh C.C. Proteasome inhibitor bortezomib suppresses nuclear factor-kappa B activation and ameliorates eye inflammation in experimental autoimmune uveitis. Mediat Inflamm. 2015;2015 doi: 10.1155/2015/847373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Napier R.J., Lee E.J., Davey M.P., et al. T cell-intrinsic role for Nod2 in protection against Th17-mediated uveitis. Nat Commun. 2020;11(1):5406. doi: 10.1038/s41467-020-18961-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hughes T., Ture-Ozdemir F., Alibaz-Oner F., Coit P., Direskeneli H., Sawalha A.H. Epigenome-wide scan identifies a treatment-responsive pattern of altered DNA methylation among cytoskeletal remodeling genes in monocytes and CD4+ T cells from patients with Behçet's disease. Arthritis Rheumatol. 2014;66(6):1648–1658. doi: 10.1002/art.38409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu Y., Yu H., Zhu Y., et al. Hypermethylation of interferon regulatory factor 8 (IRF8) confers risk to vogt-koyanagi-harada disease. Sci Rep. 2017;7(1):1007. doi: 10.1038/s41598-017-01249-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qiu Y., Zhu Y., Yu H., Zhou C., Kijlstra A., Yang P. Dynamic DNA methylation changes of Tbx21 and Rorc during experimental autoimmune uveitis in mice. Mediat Inflamm. 2018;2018 doi: 10.1155/2018/9129163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu Y., Yu H., Qiu Y., et al. Promoter hypermethylation of GATA3, IL-4, and TGF-β confers susceptibility to vogt-koyanagi-harada disease in Han Chinese. Invest Ophthalmol Vis Sci. 2017;58(3):1529–1536. doi: 10.1167/iovs.16-21188. [DOI] [PubMed] [Google Scholar]
- 46.Zhou H., Xu Z., Liao X., Tang S., Li N., Hou S. Low expression of YTH domain-containing 1 promotes microglial M1 polarization by reducing the stability of sirtuin 1 mRNA. Front Cell Neurosci. 2021;15 doi: 10.3389/fncel.2021.774305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fico A., Maina F., Dono R. Fine-tuning of cell signaling by glypicans. Cell Mol Life Sci. 2011;68(6):923–929. doi: 10.1007/s00018-007-7471-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fang Y., Shen Z.Y., Zhan Y.Z., et al. CD36 inhibits β-catenin/c-myc-mediated glycolysis through ubiquitination of GPC4 to repress colorectal tumorigenesis. Nat Commun. 2019;10(1):3981. doi: 10.1038/s41467-019-11662-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Müller G.A. The release of glycosylphosphatidylinositol-anchored proteins from the cell surface. Arch Biochem Biophys. 2018;656:1–18. doi: 10.1016/j.abb.2018.08.009. [DOI] [PubMed] [Google Scholar]
- 50.Chen X., Xu M., Xu X., et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer. 2020;19(1):106. doi: 10.1186/s12943-020-01220-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tong J., Flavell R.A., Li H.B. RNA m6A modification and its function in diseases. Front Med. 2018;12(4):481–489. doi: 10.1007/s11684-018-0654-8. [DOI] [PubMed] [Google Scholar]
- 52.Shi H., Wang X., Lu Z., et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res. 2017;27(3):315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang Q., Zhang Q., Fei P., et al. Ranibizumab injection as primary treatment in patients with retinopathy of prematurity: anatomic outcomes and influencing factors. Ophthalmology. 2017;124(8):1156–1164. doi: 10.1016/j.ophtha.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 54.Zaccara S., Jaffrey S.R. A unified model for the function of YTHDF proteins in regulating m 6 A-modified mRNA. Cell. 2020;181(7):1582–1595. doi: 10.1016/j.cell.2020.05.012. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caspi R.R., Roberge F.G., Chan C.C., et al. A new model of autoimmune disease. Experimental autoimmune uveoretinitis induced in mice with two different retinal antigens. J Immunol. 1988;140(5):1490–1495. [PubMed] [Google Scholar]
- 56.Chan C.C., Caspi R.R., Ni M., et al. Pathology of experimental autoimmune uveoretinitis in mice. J Autoimmun. 1990;3(3):247–255. doi: 10.1016/0896-8411(90)90144-h. [DOI] [PubMed] [Google Scholar]
- 57.Chandler L.C., McClements M.E., Yusuf I.H., et al. Characterizing the cellular immune response to subretinal AAV gene therapy in the murine retina. Mol Ther Methods Clin Dev. 2021;22:52–65. doi: 10.1016/j.omtm.2021.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lipski D.A., Dewispelaere R., Foucart V., et al. MHC class II expression and potential antigen-presenting cells in the retina during experimental autoimmune uveitis. J Neuroinflammation. 2017;14(1):136. doi: 10.1186/s12974-017-0915-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lückoff A., Scholz R., Sennlaub F., et al. Comprehensive analysis of mouse retinal mononuclear phagocytes. Nat Protoc. 2017;12(6):1136–1150. doi: 10.1038/nprot.2017.032. [DOI] [PubMed] [Google Scholar]
- 60.Ma W., Silverman S.M., Zhao L., et al. Absence of TGFβ signaling in retinal microglia induces retinal degeneration and exacerbates choroidal neovascularization. Elife. 2019;8 doi: 10.7554/eLife.42049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang G., Li X., Li N., et al. Icariin alleviates uveitis by targeting peroxiredoxin 3 to modulate retinal microglia M1/M2 phenotypic polarization. Redox Biol. 2022;52 doi: 10.1016/j.redox.2022.102297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J., Wu L.Y., Wu G.S., et al. Differential expression of nitric oxide synthase in experimental uveoretinitis. Invest Ophthalmol Vis Sci. 1999;40(9):1899–1905. [PubMed] [Google Scholar]
- 63.Bansal S., Barathi V.A., Iwata D., et al. Experimental autoimmune uveitis and other animal models of uveitis: an update. Indian J Ophthalmol. 2015;63(3):211–218. doi: 10.4103/0301-4738.156914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maes M.E., Colombo G., Schulz R., et al. Targeting microglia with lentivirus and AAV: recent advances and remaining challenges. Neurosci Lett. 2019;707 doi: 10.1016/j.neulet.2019.134310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang X., Mosser D.M. Macrophage activation by endogenous danger signals. J Pathol. 2008;214(2):161–178. doi: 10.1002/path.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in this study are included in the supplementary material, and any further data not included in this article are available from the corresponding author upon reasonable request.