Abstract
Background
Patients with Epstein–Barr virus-positive gastric cancers or those with microsatellite instability appear to have a favourable prognosis. However, the prognostic value of the chromosomal status (chromosome-stable (CS) versus chromosomal instable (CIN)) remains unclear in gastric cancer.
Methods
Gene copy number aberrations (CNAs) were determined in 16 CIN-associated genes in a retrospective study including test and validation cohorts of patients with gastric cancer. Patients were stratified into CS (no CNA), CINlow (1–2 CNAs) or CINhigh (3 or more CNAs). The relationship between chromosomal status, clinicopathological variables, and overall survival (OS) was analysed. The relationship between chromosomal status, p53 expression, and tumour infiltrating immune cells was also assessed and validated externally.
Results
The test and validation cohorts included 206 and 748 patients, respectively. CINlow and CINhigh were seen in 35.0 and 15.0 per cent of patients, respectively, in the test cohort, and 48.5 and 20.7 per cent in the validation cohort. Patients with CINhigh gastric cancer had the poorest OS in the test and validation cohorts. In multivariable analysis, CINlow, CINhigh and pTNM stage III–IV (P < 0.001) were independently associated with poor OS. CIN was associated with high p53 expression and low immune cell infiltration.
Conclusion
CIN may be a potential new prognostic biomarker independent of pTNM stage in gastric cancer. Patients with gastric cancer demonstrating CIN appear to be immunosuppressed, which might represent one of the underlying mechanisms explaining the poor survival and may help guide future therapeutic decisions.
Introduction
The Cancer Genome Atlas (TCGA) identified four molecular gastric cancer subtypes: Epstein–Barr virus (EBV)-positive, microsatellite instable (MSI), genomically stable (GS), and chromosomal instable (CIN)1. Several studies2–7 have tried to establish the potential clinical value of the TCGA molecular classification in patients with gastric cancer. Patients with EBV-positive or MSI gastric cancer appear to have a favourable prognosis3,6–8. However, the relationship between CIN or GS subtype and patient prognosis is less clear. Some studies4,6,9 have suggested that patients with CIN gastric cancer have the poorest prognosis. Others have concluded that the GS subtype is related to poor prognosis3,7, or that there is no relationship between survival and GS or CIN subtype2,10,11. The CIN subtype is the most frequent molecular gastric cancer subtype1 and has been associated with intestinal-type histology, p53 mutation, and amplification of receptor tyrosine kinase (RTK) genes EGFR, ERBB2, FGFR1, FGFR2, cMET, and KRAS1,12. Furthermore, a link between the presence of CIN and tumour immune response has been suggested13–15. Given that RTK amplification16,17 and p53 mutation18,19 have been associated with poor prognosis in gastric cancer, the authors hypothesized that an increasing frequency of copy number aberrations (CNAs) of genes previously linked to the presence of CIN is related to poor prognosis in gastric cancer, and that the poor prognosis in patients with CIN gastric cancers is related to low levels of tumour-infiltrating immune cells as well as presence of high p53 expression. The aim of the present study was to investigate CNA frequency using multiplex ligation probe-dependent amplification (MLPA), and p53 expression and presence of tumour infiltrating immune cells by immunohistochemistry, initially in patients with resectable gastric cancer, and to validate the findings in an independent gastric cancer cohort with early and late stage disease as well as in data from TCGA—stomach adenocarcinoma (STAD).
Methods
Patients
This retrospective study comprised patients with gastric cancer from a test cohort (Kanagawa Cancer Center Hospital, Yokohama, Japan) and a validation cohort (Leeds Teaching Hospital NHS Trust, Leeds, UK). The study had ethical approval from the Leeds Research Ethics committee (CA01/122) and the Local Kanagawa Cancer Center Hospital Ethics Committee. Data from TCGA-STAD patients were extracted from public databases (p53 mutation status, gene copy number from https://www.cbioportal.org/ (440 samples), and CIBERSORT data from Thorsson et al.20) and used to externally validate findings of the present study.
Samples and data collection
Formalin-fixed paraffin-embedded tissue samples were retrieved from resected specimens or endoscopic biopsies from the histopathology archives. Clinicopathological data were extracted from histopathology reports or patients’ electronic records.
DNA extraction
All haematoxylin and eosin-stained slides from all specimens were reviewed. A primary tumour slide with the highest density of tumour cells per area was selected and outlined for microdissection as appropriate. DNA from resection specimens and endoscopic biopsies was extracted using a Qiagen genomic DNA extraction kit (Qiagen, Hilden, Germany)17,21 and TruXTRAC® (Covaris, Massachusetts, USA), respectively. DNA from 12 normal formalin-fixed paraffin embedded tonsils was extracted using the same method, pooled, and used as reference DNA.
Multiplex ligation-dependent probe amplification assay
Full experimental details, including sensitivity compared with single-nucleotide polymorphism arrays and fluorescence in situ hybridization using gastric cancer cell lines and formalin-fixed paraffin-embedded gastric cancer tissue, have been described previously17.
For analysis of the test cohort samples, MLPA probemix P458-A1 (MRC-Holland, Amsterdam, the Netherlands) including probes for EGFR (chromosomal locus 7p11.2), ERBB2 (17q12), FGFR2 (10q26.13), MET (7q31.2), TOP2A (17q21.2), KRAS (12p12.1), MYC (8q24.21), CSDM1 (8p23.2), PIK3CA (3q26.32), KLF5 (13q22.1), CCNE1 (19q12), and GATA6 (18q11.2) was used. Samples from the validation cohort were analysed with MLPA probemix P458-B1 (MRC-Holland, Amsterdam, the Netherlands), which also included probes for FGFR1 (8p12), GATA4 (8p23.1), CDK6 (7q21.2), and CCDN1 (11q13.2).
The MLPA data analysis used to determine chromosomal status has been described in detail previously22,23 (see also Supplementary material). Gene copy number thresholds were set according to published literature17,24: a gene copy number ratio of less than 0.80 was categorized as ‘deletion’, between 0.80 and 1.30 as ‘normal’, and above 1.30 as ‘amplification’. A gene classified as deleted or amplified was given a CNA score of 1, whereas a gene classified as normal was assigned a score of 0. The mean CNA score for each patient was calculated by adding the scores of all genes for that patient and dividing the result by the total number of genes investigated for that patient. Based on the mean (standard deviation) of the frequency of CNA scores for each cohort (1.0 (1.3) test cohort, 1.4 (1.4) validation cohort, 1.7 (1.7) for TCGA-STAD data set) (Fig. S1), cancers were classified as chromosome-stable (CS; CNA score 0), low CIN (CINlow; CNA score 1 or 2) or high CIN (CINhigh; CNA score 3 or higher).
Immunohistochemistry
Assessment of p53 expression and tumour immune cell infiltration by immunohistochemistry is described in the Supplementary material. Cancers were classified as immune cell high or low, based on median dichotomization.
Microsatellite instability and Epstein–Barr virus status
The MSI and EBV status of the test and validation cohorts were established previously25. MSI and EBV status were not determined in patients from the validation cohort with metastatic disease because of limited availability of material.
The Cancer Genome Atlas—stomach adenocarcinoma data extraction
The somatic copy number of the genes investigated by MLPA in this study, p53 mutation status, and the original TCGA gastric cancer classifier were extracted from the https://www.cbioportal.org/ website. The tumour-infiltrating lymphocyte score has been estimated computationally from RNA sequencing data using CIBERSORT26 by Thorsson and colleagues20, who made the raw data publicly available. TCGA-STAD data were used to validate the frequency of gene CNA and the relationship between chromosomal status, p53 mutation status, and tumour immune cell infiltration.
Statistical analysis
Statistical analyses were performed using PASW® Statistics version 26 (IBM, Armonk, New York, USA). Continuous variables are reported as median (range) and categorical variables as numbers with percentages. Univariable survival analyses for continuous variables were performed using Cox regression, and those for categorical variables using the Kaplan–Meier method and log rank test. Five-year overall survival (OS) was calculated from the date of surgery, or date of diagnosis for patients with non-resectable disease. The relationship between CNA score and OS was initially investigated in the test cohort and subsequently in the validation cohort. Univariable treatment interaction analysis was performed for CNA score and OS. As there was no significant treatment interaction (P > 0.050), treatment was not included in the multivariable analysis. The multivariable Cox regression survival analysis included age, sex, pTNM stage (7th edition), p53 expression, and chromosomal status in the model to identify independent prognostic factors for OS. Furthermore, among patients with gastric cancer and data on CNA score, EBV and MSI status, the relationship between OS and a TCGA-like classification was explored in an analysis comparing five groups of patients: EBV (all EBV-positive patients), MSI (EBV-negative, MSI), CS (EBV-negative, microsatellite-stable (MSS), CNA score 0), CINlow (EBV-negative, MSS, CNA score 1 or 2), and CINhigh (EBV-negative, MSS, CNA score 3 or more). The association between chromosomal status and clinicopathological data, p53 expression, and tumour immune cell infiltration status was analysed in all patients jointly using Pearson’s χ2 or Fisher’s exact tests, as appropriate. For more than two independent categorical variables, the Kruskal–Wallis test was used. For all analyses, P < 0.050 was considered significant. REMARK guidelines27 were followed.
Results
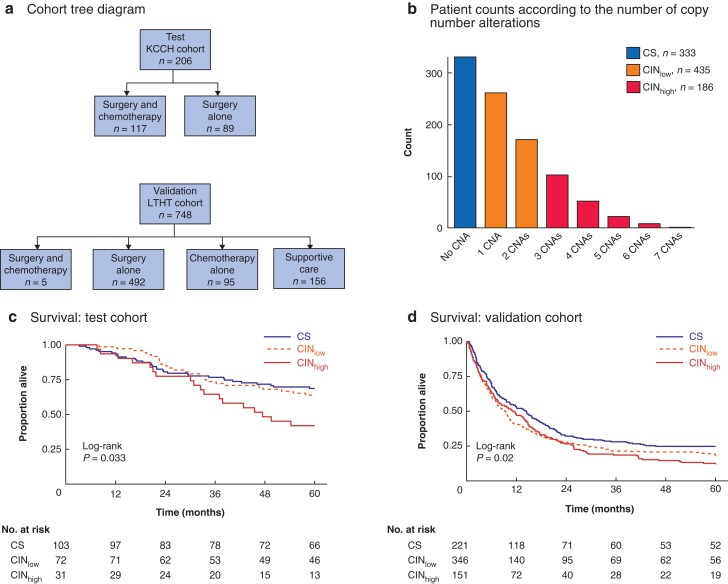
Tissue samples from 954 patients with gastric cancer were available for MLPA-based gene copy number analysis. There were 206 patients in the test cohort and 748 in the validation cohort (Fig. 1a). Clinical, demographic, and pathological data of the cohorts are shown in Table 1. Overall, the frequency of CS (no CNA), CINlow (1 or 2 genes with CNAs), and CINhigh (3 or more genes with CNAs) was 333 (34.9 per cent), 435 (45.5 per cent), and 186 (19.5 per cent), respectively (Fig. 1b). The frequency of CS, CINlow, and CINhigh was 103 (50.0 per cent), 72 (35.0 per cent), and 31 (15.0 per cent), respectively, in the test cohort, and 230 (30.7 per cent), 363 (48.5 per cent), and 155 (20.7 per cent), respectively, in the validation cohort.
Fig. 1.
Cohort distribution, chromosomal status classification, and its relationship to survival
a Cohort tree diagram. Two cohorts were formed for survival analyses: a test cohort comprising 206 patients with stage II–IV gastric cancer from Kanagawa Cancer Center Hospital, Yokohama, Japan, and a validation cohort comprising 748 patients with stage I–IV gastric cancer from Leeds Teaching Hospitals NHS Trust, Leeds, UK. Patients in the test cohort had locally advanced resectable gastric cancer treated by surgery alone (89) or surgery followed by adjuvant chemotherapy (117). In the validation cohort, patients with locally advanced gastric cancer received surgery and adjuvant chemotherapy (5) or surgery alone (492), and those with metastatic disease had chemotherapy alone (95) or best supportive care (156). b Bar chart showing frequency of patients with gastric cancer according to number of copy number aberrations (CNAs) determined in a preselected set of 16 genes. Chromosomal status was classified as chromosome-stable (CS; CNA score 0), chromosomal instability low (CINlow; CNA score 1 or 2) or chromosomal instability high (CINhigh). Kaplan–Meier analysis of overall survival in c test cohort and d validation cohort stratified by chromosomal status; 30 patients in the validation cohort were lost to follow-up. Survival was measured from the time of surgery, or time after diagnosis in patients who did not have surgery. c P = 0.033, d P = 0.020 (log rank test).
Table 1.
Demographics of test and validation gastric cancer cohorts
| Test cohort (n = 206) | Validation cohort (n = 748) | |||||||
|---|---|---|---|---|---|---|---|---|
| CS | CINlow | CINhigh | P* | CS | CINlow | CINhigh | P* | |
| Age (years) | ||||||||
| < 65 | 58 (28) | 34 (17) | 9 (4) | 0.027 | 66 (9) | 74 (10) | 41 (6) | 0.056 |
| ≥ 65 | 45 (22) | 38 (18) | 22 (11) | 163 (22) | 287 (38) | 114 (15) | ||
| Sex | ||||||||
| Males | 66 (32) | 46 (22) | 16 (8) | 0.424 | 105 (21) | 161 (32) | 59 (12) | 0.260 |
| Females | 37 (18) | 26 (13) | 15 (7) | 63 (13) | 72 (15) | 37 (7) | ||
| Treatment | ||||||||
| Surgery only | 37 (18) | 37 (18) | 15 (7) | 0.104 | 167 (22) | 231 (31) | 94 (13) | 0.758 |
| Surgery and chemotherapy | 66 (32) | 35 (17) | 16 (8) | 1 (0) | 2 (0) | 2 (0) | ||
| Chemotherapy only | – | – | – | 24 (3) | 45 (6) | 26 (4) | ||
| Supportive care only | – | – | – | 38 (5) | 85 (11) | 33 (4) | ||
| TNM stage | ||||||||
| I | – | – | – | 0.585 | 14 (2) | 28 (4) | 9 (1) | 0.144 |
| II | 37 (18) | 33 (16) | 12 (6) | 45 (6) | 56 (8) | 30 (4) | ||
| II | 63 (31) | 38 (18) | 19 (9) | 82 (11) | 106 (14) | 39 (5) | ||
| IV | 3 (2) | 1 (1) | 0 (0) | 89 (12) | 173 (23) | 77 (10) | ||
| Histological phenotype | ||||||||
| Intestinal | 29 (14) | 22 (11) | 16 (8) | 0.185 | 88 (18) | 142 (29) | 72 (15) | 0.006 |
| Diffuse | 68 (33) | 46 (22) | 14 (7) | 55 (11) | 58 (12) | 13 (2) | ||
| Mucinous/mixed | 6 (3) | 4 (2) | 1 (0) | 24 (5) | 31 (6) | 11 (2) | ||
| p53 immunohistochemistry | ||||||||
| High expression | 68 (33) | 51 (25) | 12 (6) | 0.005 | 104 (22) | 108 (23) | 37 (8) | <0.001 |
| Low expression | 35 (17) | 20 (10) | 19 (9) | 55 (12) | 111 (23) | 56 (12) | ||
Values in parentheses are percentages. *Fisher's Exact test.
The frequency of gene CNA across all study cohorts was similar to those in the TCGA-STAD database (Table S1). Tumours in 130 (88.4 per cent) of 147 TCGA-STAD patients originally classified as CIN by TCGA criteria were also classified as CIN using the authors’ 16 gene-based CNA thresholds. Tumours in 40 (70.1 per cent) of 57 TCGA-STAD patients originally classified as GS were classified as CS using the current study thresholds (Table S2).
Prognostic value of chromosomal status in test and validation cohorts
OS analysis was undertaken for 206 patients with stage II–IV gastric cancers in the test cohort. Stratification of patients by chromosomal status (CS versus CINlow versus CINhigh) demonstrated the poorest OS for patients with CINhigh gastric cancers (P = 0.033) (Fig. 1c). In the validation cohort of 718 patients (30 patients in this cohort were lost to follow-up), the HR for chromosomal status was 1.16 (1.04 to 1.30; P = 0.008). Kaplan–Meier analysis confirmed that patients with CINhigh gastric cancers had the poorest OS (P = 0.020) (Fig. 1d).
Prognostic value of Cancer Genome Atlas-like classification
In addition to chromosomal status, a TCGA-like classifier was created for all 924 patients with data on chromosomal status, EBV status, MSI status, and survival. Five groups of patients were created in a stepwise approach: EBV (18, 1.9 per cent), MSI (61, 6.6 per cent), CS (283, 30.6 per cent), CINlow (387, 41.9 per cent), and CINhigh (175, 18.9 per cent) (Fig. 2a).
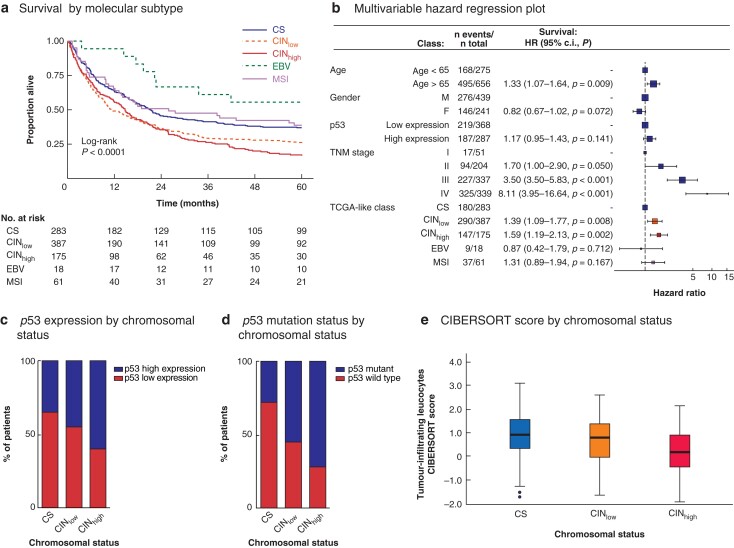
Fig. 2.
Survival by Cancer Genome Atlas-like classification in univariable and multivariable analyses, and association of chromosomal status with p53 expression, p53 mutation, and tumour immune cell infiltration
a Kaplan–Meier analysis of 5-year overall survival of patients with resectable gastric cancer by molecular subtype. CS, chromosome-stable; CINlow, chromosomal instability low; CINhigh, chromosomal instability high; EBV, Epstein–Bar virus; MSI, microsatellite instability. b Multivariable hazard regression plot; hazard ratios are shown with 95 per cent confidence intervals. c Stacked bar chart showing p53 expression level (high versus low) in relation to chromosomal status in test and validation cohorts combined (P < 0.001, Pearsons' Chi Squared test). d Stacked bar chart showing p53 mutation status in relation to chromosomal status in TGCA-STAD patients (P < 0.001, Pearsons' Chi Squared test). e Box plot showing CIBERSORT-derived lymphocyte infiltration signature score in TCGA-STAD patients according to chromosomal status. Median values (bold line), interquartile range (box), and range (error bars) excluding outliers (symbols) are shown (P < 0.001, Kruskal-Wallis test).
Kaplan–Meier analysis demonstrated the best OS for patients with EBV-positive gastric cancers (mean OS 43.1 months), followed by MSI (32.3 months), CS (30.4 months), CINlow (24.1 months), and CINhigh (22.6 months) gastric cancers (P < 0.001) (Fig. 2a).
In univariable analysis with CS as reference, the HR was 0.53 (95 per cent c.i. 0.26 to 1.08; P = 0.079) for patients with EBV-positive gastric cancers, 0.94 (0.66 to 1.35; P = 0.751) for patients with MSI gastric cancers, 1.41 (1.17 to 1.70; P <0.001) for patients with CINlow gastric cancers, and 1.58 (1.27 to 1.96; P < 0.001) for patients with CINhigh gastric cancers. In multivariable analysis, age over 65 years, TNM stage III–IV, CINlow and CINhigh were independent prognostic factors for poor OS (Fig. 2b).
Relationship between chromosomal status and clinicopathological variables
Intestinal-type gastric cancers more frequently showed CIN than diffuse-type lesions: 252 (68.3 per cent) versus 131 (51.6 per cent) (P < 0.001). Stage IV gastric cancers more frequently demonstrated CIN than stage I–III gastric cancers: 250 (73.7 per cent) versus 371 (60.3 per cent) (P < 0.001).
Relationship between chromosomal status and tumour infiltrating immune cells and p53 expression
Information on p53 expression was available for 676 patients. Some 296 gastric cancers (43.8 per cent) were classified as having high p53 expression based on p53 positivity in at least 50 per cent of tumour cells. Gastric cancers with high p53 expression were more frequently CINhigh (P < 0.001) (Fig. 2c). A similar relationship was seen using the MLPA–CNA-defined chromosomal status criteria (CS versus CINlow versus CINhigh) and p53 mutation status in TCGA-STAD patients (P < 0.001) (Fig. 2d).
Data on expression of immune cell markers CD45, CD3, CD8, CD68, and FOXP3 were available for 395 patients. Low levels of CD45-positive immune cells were seen in 45 diffuse-type gastric cancers with CIN (61.6 per cent) (P = 0.022). Among intestinal-type gastric cancers, CD3-positive lymphocyte levels were decreased in 60 gastric cancers (72.2 per cent) with CIN (P = 0.038). There was no relationship between chromosomal status and levels of CD8-, CD68- or FOXP3-positive immune cells (Table S3).
To further explore the relationship between chromosomal status and tumour immune cell infiltration, RNA sequencing data from TCGA-STAD previously analysed by CIBERSORT were used26. Using the 16-gene CNA-based chromosomal status classification, the CIBERSORT lymphocyte infiltration signature score decreased with increasing level of CIN (P < 0.001) (Fig. 2e).
Discussion
TCGA established a molecular classification of gastric cancers integrating results from multiple platforms1,28 and identified CIN as the commonest molecular gastric cancer subtype. Here, MLPA was used to investigate the gene copy number variation of 16 CIN-related genes and determine the chromosomal status in two independent gastric cancer cohorts, with a test and validation set approach. This study is the first to identify and subsequently validate an increasing frequency of CNAs in genes previously linked to the presence of CIN as an independent poor prognostic marker in patients with gastric cancer, irrespective of disease stage. It is also the first to associate the level of CIN with the anti-tumour immune response in gastric cancer. These results are potentially clinically relevant given that CIN is a common molecular subtype in patients with stage IV gastric cancer; the observed reduction in immune cell infiltration in CIN cancers might explain why immune checkpoint targeting therapy seems to be successful in only a subset of patients29.
The CIN frequency was 65.0 per cent in this study and 49.8 per cent in the TCGA-STAD data set1. The higher CIN frequency in this study may be due to differences in sample size, disease stage (more stage IV gastric cancers in the present study), and use of different methodology to identify chromosomal status. Despite this difference, the key findings were reproduced in the TCGA-STAD data set using the present CIN classification. This supports the value of assaying a preselected limited number of genes for CNA to determine chromosomal status. Furthermore, the present study showed an association between CIN and high p53 expression, which can be considered similar to the association between CIN and p53 mutation reported in TCGA-STAD1,28.
CIN frequency was higher in patients with stage IV gastric cancer, which is similar to findings in metastatic breast and head/neck cancers30. Similar to previous reports in gastric cancer1,28, the CIN frequency in the present study was higher in intestinal-type compared with diffuse-type gastric cancers.
Davoli and colleagues15 undertook a pan-cancer analysis of TCGA data, including the gastric cancer data set, and demonstrated that a large number of somatic CNAs were associated with lower expression of markers for cytotoxic immune cell infiltration. Furthermore, Kumagai et al.14 suggested that anti-tumour immune responses might be related to epidermal growth factor receptor signalling, and that aberrant RTK signalling often seems to be associated with CIN1,28. However, the present study is the first to suggest that even low frequency of CNA is associated with decreasing levels of infiltrating intratumour immune cells, which might be one of the underlying biological mechanisms contributing to the poor prognosis of patients with CIN gastric cancer.
Several studies2–7 have investigated the prognostic role of the proposed TCGA gastric cancer subtypes, with contradictory results. Here, minimal to no overlap was observed when gastric cancers were classified into EBV-positive, MSI, CIN, and CS subtypes, and survival by gastric cancer subtype showed that OS was best for patients with EBV-positive disease and poorest for those with CIN gastric cancer. These results are consistent with the literature suggesting a more favourable prognosis for EBV-positive or MSI gastric cancer3,6–8. In agreement with the present data, some previous studies4,6,9 suggested that patients with CIN cancer have the poorest prognosis. However, there are also studies suggesting that CS cancers have the poorest prognosis3,7 and others2,10 showing no difference in survival between TCGA subtypes. These differences could be related to differences in case mix and methodology used to identify patients with CIN or CS cancers. The strength of the present study results lies in the fact that similar findings were demonstrated in two independent cohorts, which also suggests that the association between chromosomal status and survival is similar in patients with gastric cancer from the East (Japan) and West (UK).
A recent study9 identified a subset of patients with non-CINhigh gastric cancer who seemed to benefit from neoadjuvant chemotherapy using different methodology to identify CIN status, but similarly concluded that CIN classification identified gastric cancer with different characteristics, with potential clinical implications. In summary, the present results suggest that CIN subtypes of gastric cancer are associated with a poorer prognosis and low tumour immune cell infiltration, and that this subset of patients may benefit from therapeutic agents that recruit and activate immune cells into the tumour microenvironment.
The present study has some limitations. It was a retrospective analysis that used material from patients with gastric cancer from two centres, which may have introduced bias. Although it was possible to compare frequency, and prognostic value between Asian and Caucasian patients with stage I–III gastric cancer, there was no access to Asian patients with stage IV gastric cancer. Furthermore, owing to an insufficient amount of material, it was not possible to investigate immune cell infiltration, p53 status, MSI, and EBV status in patients with stage IV disease.
Future work is needed to identify the underlying mechanisms by which CNAs of CIN-related genes cause immunosuppression in patients with gastric cancer. Chromosomal status measured by MLPA with a bespoke set of CIN-related genes may aid in personalization of treatment decisions, improving outcomes for patients with gastric cancer in the near future.
Supplementary Material
Contributor Information
Arnaldo N. S. Silva, Division of Pathology and Data Analytics, Leeds Institute of Medical Research at St James’s, University of Leeds, Leeds, UK Department of Surgery, University of Cambridge, Cambridge University Hospitals, Addenbrookes, Cambridge, UK; Cancer Research UK, Cambridge Institute, Cambridge, UK.
Yuichi Saito, Department of Surgery, Teikyo University School of Medicine, Tokyo, Japan; Department of Pathology, GROW School for Oncology and Reproduction, Maastricht University Medical Center+, Maastricht, the Netherlands.
Takaki Yoshikawa, Department of Gastric Surgery, National Cancer Center Hospital, Tokyo, Japan.
Takashi Oshima, Department of Gastrointestinal Surgery, Kanagawa Cancer Center Hospital, Yokohama, Japan.
Jeremy D. Hayden, Department of Upper Gastrointestinal Surgery, Institute of Oncology, Leeds Teaching Hospitals NHS Trust, Leeds, UK
Jan Oosting, Department of Pathology, Leiden University Medical Center, Leiden, the Netherlands.
Sophie Earle, Division of Pathology and Data Analytics, Leeds Institute of Medical Research at St James’s, University of Leeds, Leeds, UK.
Lindsay C. Hewitt, Division of Pathology and Data Analytics, Leeds Institute of Medical Research at St James’s, University of Leeds, Leeds, UK Department of Pathology, GROW School for Oncology and Reproduction, Maastricht University Medical Center+, Maastricht, the Netherlands.
Hayley L. Slaney, Division of Pathology and Data Analytics, Leeds Institute of Medical Research at St James’s, University of Leeds, Leeds, UK
Alex Wright, Division of Pathology and Data Analytics, Leeds Institute of Medical Research at St James’s, University of Leeds, Leeds, UK.
Imran Inam, Division of Pathology and Data Analytics, Leeds Institute of Medical Research at St James’s, University of Leeds, Leeds, UK.
Ruth E. Langley, MRC Clinical Trials Unit, University College London, London, UK
William Allum, Department of Surgery, Royal Marsden Hospital, London, UK.
Matthew G. Nankivell, MRC Clinical Trials Unit, University College London, London, UK
Gordon Hutchins, Division of Pathology and Data Analytics, Leeds Institute of Medical Research at St James’s, University of Leeds, Leeds, UK.
David Cunningham, Department of Medicine, Royal Marsden NHS Trust, London and Sutton, UK.
Heike I. Grabsch, Division of Pathology and Data Analytics, Leeds Institute of Medical Research at St James’s, University of Leeds, Leeds, UK Department of Pathology, GROW School for Oncology and Reproduction, Maastricht University Medical Center+, Maastricht, the Netherlands.
Funding
H.I.G. received funding from Cancer Research UK and Yorkshire Cancer Research. G.H. received funding from the Pathological Society of Great Britain and Ireland and the Academy of Medical Sciences. A.N.S.S. received funding from the Sasakawa Cancer Foundation and Association of Clinical Pathologists Student Bursary. T.Y. and T.O. recieved funding from non-profit organizations such as the Kanagawa Standard Anti-cancer Therapy Support System (Yokohama, Japan).
Disclosure. HG has received honoraria from Astra Zeneca and BMS for scientific advisory board activities not related to the current study. DC has received grant funding from MedImmune, Clovis, Eli Lilly, 4SC, Bayer, Celgene, Leap and Roche not related to the current study. The remaining authors declare no conflict of interest.
Supplementary material
Supplementary material is available at BJS online.
References
- 1. Cancer Genome Atlas Research Network . Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014;513:202–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Setia N, Agoston AT, Han HS, Mullen JT, Duda DG, Clark JW et al. A protein and mRNA expression-based classification of gastric cancer. Mod Pathol 2016;29:772–784 [DOI] [PubMed] [Google Scholar]
- 3. Ahn S, Lee SJ, Kim Y, Kim A, Shin N, Choi KU et al. High-throughput protein and mRNA expression-based classification of gastric cancers can identify clinically distinct subtypes, concordant with recent molecular classifications. Am J Surg Pathol 2017;41:106–115 [DOI] [PubMed] [Google Scholar]
- 4. Diaz Del Arco C, Estrada Munoz L, Molina Roldan E, Ceron Nieto MA, Ortega Medina L, Garcia Gomez de Las Heras S et al. Immunohistochemical classification of gastric cancer based on new molecular biomarkers: a potential predictor of survival. Virchows Arch 2018;473:687–695 [DOI] [PubMed] [Google Scholar]
- 5. Yoon JY, Sy K, Brezden-Masley C, Streutker CJ. Histo- and immunohistochemistry-based estimation of the TCGA and ACRG molecular subtypes for gastric carcinoma and their prognostic significance: a single-institution study. PLoS One 2019;14:e0224812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tsai JH, Jeng YM, Chen KH, Lee CH, Yuan CT, Liau JY. An integrative morphomolecular classification system of gastric carcinoma with distinct clinical outcomes. Am J Surg Pathol 2020;44:1017–1030 [DOI] [PubMed] [Google Scholar]
- 7. Wang Q, Xie Q, Liu Y, Guo H, Ren Y, Li J et al. Clinical characteristics and prognostic significance of TCGA and ACRG classification in gastric cancer among the Chinese population. Mol Med Rep 2020;22:828–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martinez-Ciarpaglini C, Fleitas-Kanonnikoff T, Gambardella V, Llorca M, Mongort C, Mengual R et al. Assessing molecular subtypes of gastric cancer: microsatellite unstable and Epstein–Barr virus subtypes. Methods for detection and clinical and pathological implications. ESMO Open 2019;4:e000470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kohlruss M, Krenauer M, Grosser B, Pfarr N, Jesinghaus M, Slotta-Huspenina J et al. Diverse ‘just-right’ levels of chromosomal instability and their clinical implications in neoadjuvant treated gastric cancer. Br J Cancer 2021;125:1621–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zheng X, Song X, Shao Y, Xu B, Hu W, Zhou Q et al. Prognostic role of tumor-infiltrating lymphocytes in esophagus cancer: a meta-analysis. Cell Physiol Biochem 2018;45:720–732 [DOI] [PubMed] [Google Scholar]
- 11. Cristescu R, Lee J, Nebozhyn M, Kim KM, Ting JC, Wong SS et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med 2015;21:449–456 [DOI] [PubMed] [Google Scholar]
- 12. Maleki SS, Rocken C. Chromosomal instability in gastric cancer biology. Neoplasia 2017;19:412–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bakhoum SF, Cantley LC. The multifaceted role of chromosomal instability in cancer and its microenvironment. Cell 2018;174:1347–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kumagai S, Koyama S, Nishikawa H. Antitumour immunity regulated by aberrant ERBB family signalling. Nat Rev Cancer 2021;21:181–197 [DOI] [PubMed] [Google Scholar]
- 15. Davoli T, Uno H, Wooten EC, Elledge SJ. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017;355:eaaf8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Deng N, Goh LK, Wang H, Das K, Tao J, Tan IB et al. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut 2012;61:673–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Silva ANS, Coffa J, Menon V, Hewitt LC, Das K, Miyagi Y et al. Frequent coamplification of receptor tyrosine kinase and downstream signaling genes in Japanese primary gastric cancer and conversion in matched lymph node metastasis. Ann Surg 2018;267:114–121 [DOI] [PubMed] [Google Scholar]
- 18. Wei K, Jiang L, Wei Y, Wang Y, Qian X, Dai Q et al. The prognostic significance of p53 expression in gastric cancer: a meta-analysis. J Cancer Res Clin Oncol 2015;141:735–748 [DOI] [PubMed] [Google Scholar]
- 19. Fisher OM, Lord SJ, Falkenback D, Clemons NJ, Eslick GD, Lord RV. The prognostic value of TP53 mutations in oesophageal adenocarcinoma: a systematic review and meta-analysis. Gut 2017;66:399–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Thorsson V, Gibbs DL, Brown SD, Wolf D, Bortone DS, Ou Yang TH et al. The immune landscape of cancer. Immunity 2018;48:812–830.e14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Grieken NC, Aoyama T, Chambers PA, Bottomley D, Ward LC, Inam I et al. KRAS and BRAF mutations are rare and related to DNA mismatch repair deficiency in gastric cancer from the East and the West: results from a large international multicentre study. Br J Cancer 2013;108:1495–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hewitt LC, Saito Y, Wang T, Matsuda Y, Oosting J, Silva ANS et al. KRAS status is related to histological phenotype in gastric cancer: results from a large multicentre study. Gastric Cancer 2019;22:1193–1203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Eijk R, Eilers PH, Natte R, Cleton-Jansen AM, Morreau H, van Wezel T et al. MLPAinter for MLPA interpretation: an integrated approach for the analysis, visualisation and data management of multiplex ligation-dependent probe amplification. BMC Bioinformatics 2010;11:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moelans CB, Holst F, Hellwinkel O, Simon R, van Diest PJ. ESR1 amplification in breast cancer by optimized RNase FISH: frequent but low-level and heterogeneous. PLoS One 2013;8:e84189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hewitt LC, Inam IZ, Saito Y, Yoshikawa T, Quaas A, Hoelscher A et al. Epstein–Barr virus and mismatch repair deficiency status differ between oesophageal and gastric cancer: a large multi-centre study. Eur J Cancer 2018;94:104–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol 2018;1711:243–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Altman DG, McShane LM, Sauerbrei W, Taube SE. Reporting recommendations for tumor marker prognostic studies (REMARK): explanation and elaboration. BMC Med 2012;10:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cancer Genome Atlas Research Network, Analysis Working Group: Asan University, BC Cancer Agency, Brigham, Women’s Hospital, Broad Institute et al. Integrated genomic characterization of oesophageal carcinoma. Nature 2017;541:169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Janjigian YY, Shitara K, Moehler M, Garrido M, Salman P, Shen L et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): a randomised, open-label, phase 3 trial. Lancet 2021;398:27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bakhoum SF, Ngo B, Laughney AM, Cavallo JA, Murphy CJ, Ly P et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018;553:467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.