Abstract
Purpose
Proliferative vitreoretinopathy (PVR) is the most common cause of failure of retinal reattachment surgery, and the molecular changes leading to this aberrant wound healing process are currently unknown. Our ultimate goal is to study PVR pathogenesis by employing single-cell transcriptomics to dissect cellular heterogeneity.
Design
Here we aimed to compare single-cell RNA sequencing (scRNA-seq) and single-nucleus RNA-sequencing (snRNA-seq) of retinal PVR samples in the rabbit model.
Participants
Unilateral induction of PVR lesions in rabbit eyes with contralateral eyes serving as controls.
Methods
Proliferative vitreoretinopathy was induced unilaterally in Dutch Belted rabbits. At different timepoints after PVR induction, retinas were dissociated into either cells or nuclei suspension and processed for scRNA-seq or snRNA-seq.
Main Outcome Measures
Single cell and nuclei transcriptomic profiles of retinas after PVR induction.
Results
Single-cell RNA sequencing and snRNA-seq were conducted on retinas at 4 hours and 14 days after disease induction. Although the capture rate of unique molecular identifiers and genes were greater in scRNA-seq samples, overall gene expression profiles of individual cell types were highly correlated between scRNA-seq and snRNA-seq. A major disparity between the 2 sequencing modalities was the cell type capture rate, however, with glial cell types overrepresented in scRNA-seq, and inner retinal neurons were enriched by snRNA-seq. Furthermore, fibrotic Müller glia were overrepresented in snRNA-seq samples, whereas reactive Müller glia were overrepresented in scRNA-seq samples. Trajectory analyses were similar between the 2 methods, allowing for the combined analysis of the scRNA-seq and snRNA-seq data sets.
Conclusions
These findings highlight limitations of both scRNA-seq and snRNA-seq analysis and imply that use of both techniques together can more accurately identify transcriptional networks critical for aberrant fibrogenesis in PVR than using either in isolation.
Financial Disclosure(s)
Proprietary or commercial disclosure may be found after the references.
Keywords: Single cell RNA-sequencing, Single nucleus RNA-sequencing, Proliferative vitreoretinopathy, Fibrosis
Proliferative vitreoretinopathy (PVR) is among the most important causes of visual morbidity in patients with retinal diseases, and its treatment is a major unmet need. Proliferative vitreoretinopathy is analogous to an aberrant wound healing process that stiffens and distorts the normally pliant retina and disrupts its natural conformance to the curved posterior eye wall.1 Early-stage PVR is characterized by retinal stiffening and contraction; retinal pigment epithelium (RPE) cells and activated Müller glia are thought to play important roles at this stage.2,3 Disease progress involves retinal traction and detachment and, ultimately, vision loss. Triggers of PVR include retinal detachment, infection or inflammation, penetrating injury, hemorrhage, and, most frustratingly, the incisional retinal surgery that is often used to treat these conditions.4 Multiple episodes of reparative surgery are often required to reposition the retina and mitigate against further vision loss.
Aside from RPE cells and Müller glia, multiple other retinal cell types may be involved in early and/or late PVR development, yet the temporal cascade of molecular changes in various cell types remain poorly understood. This knowledge gap is further compounded by processes that cause cells to substantially alter their gene expression profiles, such as epithelial-to-mesenchymal transition in which RPE cells assume contractile properties.5 In general, molecular data regarding PVR have come from bulk analysis.6,7 The lack of information on temporally resolved, cell-specific gene expression changes has impeded research on developing rational pharmacologic treatment to target the specific pathways that initiate or drive progression of PVR.
To address this knowledge gap, we aimed to study PVR pathogenesis using single-cell transcriptomic analysis. Over the last few years, this approach has been used in multiple species to identify molecular markers of virtually all retinal cell subtypes,8, 9, 10 identify gene regulatory networks controlling retinal development and regeneration,11, 12, 13, 14 and identify molecular changes associated with onset and progression of disease.15, 16, 17 Single-cell transcriptomics can be conducted using single-cell RNA sequencing (scRNA-seq), in which entire dissociated cells are profiled, or using single-nucleus RNA sequencing (snRNA-seq) on isolated cell nuclei.18,19 Both approaches have been reported to have specific advantages and disadvantages.20, 21, 22 Single-cell RNA sequencing captures larger numbers of fully spliced mRNA species, capturing both cytoplasmic and nuclear transcripts, but is not usable in analysis of frozen or fixed tissue. SnRNA-seq, on the other hand, is suitable for analysis of archival material and efficiently captures nuclear transcripts but is enriched for un- or partially spliced transcripts.23,24 Selective loss of individual cell types has also been reported for the 2 approaches, with loss of large, mature neuronal subtypes in brain and retina, and fibrotic cells in the kidney with scRNA-seq.12,25, 26, 27 In contrast, depletion of active microglia has been reported in brain snRNA-seq libraries.28
Although both scRNA-seq and snRNA-seq have been extensively used in the retina, a systematic comparison of the 2 approaches using the same input material has not been conducted. As a result, it is not clear to what extent the limitations of the 2 approaches apply, particularly in the study of complex retinal disease. Likewise, little is known about the relative accuracy and efficiency of these methods in the retina of adult mammalian species other than mice. In this study, we use both scRNA-seq and snRNA-seq to globally profile cellular transcriptomic changes in a rabbit surgical model of PVR. In addition to large differences between the 2 approaches in the relative abundance of major cell types, we also observe differences in the efficiency of capture of fibrotic and reactive Müller glia, differences in the overall levels of cell–cell contamination, and differential expression of many individual genes. These findings highlight the limitations of each approach and imply their combined use may prove more effective at profiling global transcriptional changes in disease.
Methods
PVR Induction
All animal experiments were performed in accordance with the guidelines for the Use of Animals in Ophthalmic and Vision Research of the Association for Research in Vision and Ophthalmology, approved by the Johns Hopkins University’s Institutional Animal Care and Use Committee and in adherence with the ARRIVE 2.0 Guidelines. Dutch Belted rabbits (Robinson Services Inc) weighing 1.0 to 4.0 kg were maintained in a temperature-controlled, 12-hour light cycle environment with ad libitum food and water. Briefly, animals were anesthetized with ketamine (35 mg/kg IM) and xylazine (5 mg/kg IM), intubated, and maintained on isoflurane. Unilateral induction of PVR lesions was achieved via lensectomy, pars plana vitrectomy retinotomy, and extensive retinal detachment induction followed by retinal cryotherapy and intravitreal autologous platelet-rich plasma injection; contralateral eyes served as controls.
Cell Dissociation
Rabbits were euthanized, and eyes were enucleated and placed in Hank's Balanced Salt Solution. Control tissue or PVR lesions were dissected en bloc and transferred to ice-cold media containing Hibernate A, 1× B27 supplement, and 1× GlutaMAX (ThermoFisher). The tissue was then dissociated using the Papain Dissociation System (Worthington). Briefly, samples were incubated in the papain/DNAse mixture for 30 minutes at 37°C, with gentle inversion every 5 minutes until most of the tissue broke apart. The cell suspension was triturated 10 times using a wide-bore pipette before being mixed in a 1:1 ratio with 0.1× albumin-ovomucoid inhibitor and DNAse in EBSS. The cell:inhibitor mixture was carefully layered on top of 1× albumin-ovomucoid inhibitor in EBSS and centrifuged at 70× g for 6 minutes at room temperature. The cell pellet was resuspended in ice-cold, PBS-containing, 0.04% bovine serum albumin (BSA) and 0.5 U/μL RNasin Plus RNase inhibitor (Promega) to achieve a concentration of ∼750–1200 cells/μL. Cell concentration and viability were assessed using a combination of trypan blue and DAPI staining.
Nuclei Lysis
Nuclei were isolated following a modified version of 10× Genomics’ isolation of nuclei from embryonic mouse brain tissue. Harvested PVR lesions were first homogenized in 1 ml of chilled lysis buffer (10 mMTris-HCl, 10 mM NaCl, 3 mM MgCl2, 0.01% Nonidet P40) using an RNase-free pestle. This suspension was incubated on ice for 15 minutes and then triturated 10 times using a wide-bore pipette. The sample was passed through a 70 μm Flowmi filter and centrifuged at 600× g for 5 minutes at 4°C. The supernatant was discarded and the nuclei pellet was resuspended in 1 ml nuclei wash and resuspension buffer (1× PBS with 1% BSA and 0.2 U/μL RNase inhibitor) and filtered through a 40 μm Flowmi filter to remove cell debris and large clumps. The centrifugation, resuspension, and filtering was repeated once more before the nuclei were resuspended in the proper volume of nuclei wash and resuspension buffer to achieve a concentration of ∼750–1200 nuclei/μL. Nuclei concentration was assessed using a combination of trypan blue and DAPI staining.
scRNA-seq Library Construction and Sequencing
Single-cell RNA-sequencing and snRNA-seq were performed on dissociated retinal cells or nuclei using the Chromium Next GEM Single-Cell 3ʹ Reagent Kits v3.1 (10× Genomics). Briefly, retinal cells or nuclei (∼16 000 cells per sample) were loaded into the 10× Chromium controller and downstream scRNA-seq or snRNA-seq libraries were generated and indexed by following the manufacturer’s instructions. Libraries were pooled and sequenced on Illumina NovaSeq 6000 targeting 50 000 reads per cell.
scRNA-seq Analysis
Sequencing reads were demultiplexed and aligned to the OryCun2.0 rabbit reference genome using the Cell Ranger 6.1.2 mkfastq and count pipeline (10× Genomics, Pleasanton, CA), using default parameters for scRNA-seq or using the --include-introns flag for snRNA-seq and for scRNA-seq with intron mapping analysis. The generated cell-by-gene count matrix was then used as the input for downstream analysis. The count matrix was analyzed using the Seurat 4.0 R package.29 Metadata corresponding to sample, injury time point, and modality were added and merged into cell-only, nuclei-only, or combined Seurat objects. Cells and nuclei that had <500 unique molecular identifiers (UMIs) or > 50 000 UMIs along with cells that had > 25% mitochondrial content or nuclei that had > 2% mitochondrial content were filtered out. Doublets were identified and removed using the scDblFinder R package.30
The cell, nuclei, and combined data sets underwent normalization, variable feature selection, and scaling using the default parameters in Seurat’s NormalizeData, FindVariableFeatures, and ScaleData functions. Principal components were then calculated based on the top 2000 variable features and batch corrected using the Harmony R package.31 Uniform Manifold Approximation and Projection (UMAP) dimension reduction was performed on the top 10 corrected principal components, and clusters were computed using Seurat’s FindNeighbors and FindClusters functions. Cell types were then identified in the cell and nuclei data sets using a list of known marker genes that were used previously and then transferred into the combined data sets.13,32,33 Cell type similarities between the cell and nuclei data sets were then calculated on all variable features using the MetaNeighbor R package.34
Before identifying differentially expressed genes between the cells and nuclei, the combined data set was randomly downsampled so that each cell type had equal numbers from either sequencing modality to reduce bias due to over representation of certain cell types in either data set. Differentially expressed genes between cells and nuclei were determined using the Wilcoxon rank sum test. The clusterProfiler R package was used to perform gene ontology enrichment analysis of biological processes found in the differentially expressed gene lists.35
To determine the different cell states of Müller glia, the Müller glia from the combined data set was subsetted and randomly downsampled so that each sequencing modality had equal contribution. The data were renormalized and scaled with the gene number, and UMI variables were regressed out. Principal components were calculated based on the top 2000 variable features and Harmony batchcorrected using the sample identification and modality variables for removal. The cell state scores of Müller glia were calculated by the UCell R package using known genes of resting and reactive glia and fibrotic cells identified in previous studies.32,36,37 Using the control cells as the root, the Slingshot R package was used to order the cells along a pseudotime lineage and a negative binomial generalized additive model was fitted using the fitGAM function of the tradeSeq R package.38,39 An association test was used to determine gene expression changing along the pseudotime lineage. The pseudotime analysis was also replicated and compared using the monocle3 R package, and differentially expressed genes along pseudotime were grouped into modules using the find_gene_modules function.40 RNA velocity was used to infer transcriptional dynamics and predict the future state and transition of individual cells and nuclei in the data set using the spliced and unspliced counts data generated by velocyto python package and velocity calculated and visualized using the scVelo python package.41,42
Data and Code Availability
All rabbit scRNA-seq and snRNA-seq data can be accessed at GEO accession numbers GSE217333. Code used to analyze the data sets in this study can be found at https://github.com/csanti88/pvr_singlecell_vs_singlenucleiRNAseq_2022.
Results
To globally profile gene expression changes that occur during the progression of PVR, we conducted scRNA-seq and snRNA-seq on retina dissected from rabbits in which PVR-like lesions had been induced by vitrectomy, retinotomy, and retinal detachment induced by saline injection, platelet-enriched plasma injection, and cryotherapy to induce scar formation.43,44 Retina tissue was extracted from the lesion site, dissociated, split into 2 equal portions, and then processed and analyzed using scRNA-seq or snRNA-seq, respectively (Fig 1A and Table S1, available at www.ophthalmologyscience.org). Three different treatment conditions were profiled (uninjured control and both 4 hours and 14 days after PVR induction), thereby ensuring a broad representation of disease progression.
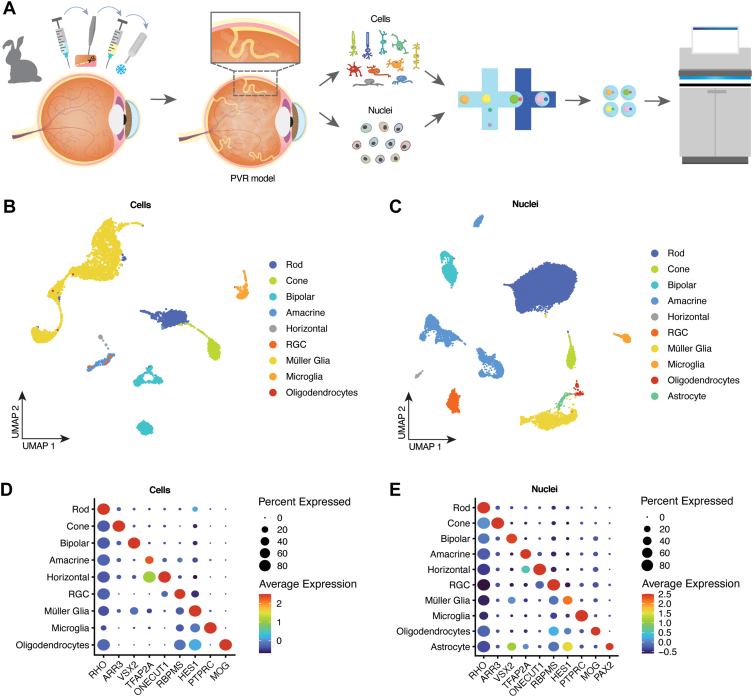
Figure 1.
All major retinal cell types are captured by single-cell RNA-sequencing (scRNA-seq) and single-nucleus RNA-sequencing (snRNA-seq). A, Schematic summary of the study. Proliferative vitreoretinopathy (PVR) lesions were induced via lensectomy, pars plana vitrectomy, retinotomy, and extensive retinal detachment induction followed by retinal cryotherapy and intravitreal autologous platelet-rich plasma injection. At different time points, the lesions were dissected out, and cells were dissociated, or nuclei were isolated before profiling by scRNA-seq or snRNA-seq. B, C, Combined Uniform Manifold Approximation and Projection (UMAP) projection of all cells (left) or nuclei (right) profiled in this study. D, E, Examples of gene expression levels for selected cell type-specific genes for cells (left) or nuclei (right). RGC = retinal ganglion cell.
We then combined data sets from all 3 timepoints for scRNA-seq (Fig 1B) and snRNA-seq (Fig 1C) separately, visualizing different cell types using UMAP analysis. Using well-characterized cell type-specific marker genes, we were able to readily identify every retina major cell type in each data set, with the exception of astrocytes, which could be clearly resolved in the scRNA-seq data set (Fig 1D, E; Fig S1A, B, available at www.ophthalmologyscience.org). Oligodendrocytes, which are not present in mouse or human retina, are abundant in the rabbit retina and myelinate the axons of retinal ganglion cells in structures known as the medullary rays.45
We next combined these 2 data sets and quantified the abundance of each major cell type. Overall, gene expression profiles were very similar for individual cell types, and combined UMAP analysis revealed that common cell type-specific clusters detected by scRNA-seq and snRNA-seq were essentially superimposable in the combined UMAP plot (Fig 2A). Direct comparison of cell type-specific gene expression profiles likewise showed very high correlation between scRNA-seq and snRNA-seq profiles of individual cell types, with these invariably being more closely correlated than profiles of other cell types (Fig 2B).
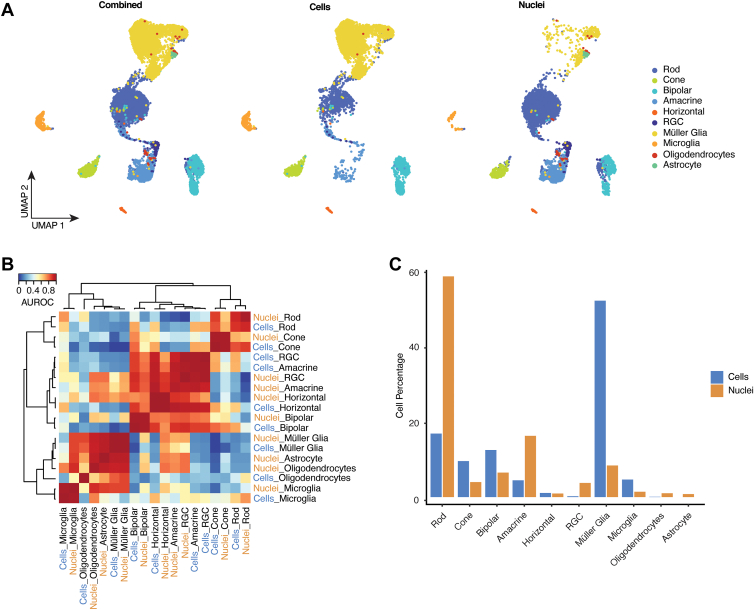
Figure 2.
Single-nucleus RNA-sequencing (snRNA-seq) has improved capture efficiencies for neuronal cell types. A, Uniform Manifold Approximation and Projection (UMAP) embedding of single-cell RNA-sequencing (scRNA-seq) and snRNA-seq profiles showing profiles of the combined data set (left) scRNA-seq (middle), and snRNA-seq (right), colored by the assigned cell type signatures. B, Heatmap of area under the receiver operating characteristics (AUROC) scores of the combined data set between all retinal cell types based on highly variable genes. All replicating cell types cluster together after applying hierarchical clustering. C, Bar plots of cell type proportions from each sequencing modality. RGC = retinal ganglion cell.
However, we observed major differences in the relative abundance of each cell type. Müller glia, cone photoreceptors, bipolar cells, and microglia were all overrepresented in the scRNA-seq data set, whereas rod photoreceptors, amacrine cells, retinal ganglion cells, and oligodendrocytes were enriched in the snRNA-seq data set. Horizontal cells were roughly equally abundant in both samples (Fig 2C; Fig S2A, available at www.ophthalmologyscience.org). Although a comprehensive histological analysis of cell type ratios has yet to be conducted in rabbit retina, it is clear that the snRNA-seq data set provides, overall, a more accurate reflection of the true abundance of these various cell types. For instance, the rod:cone ratio as observed histologically is 20:146 versus 13:1 as measured by snRNA-seq and 1.7:1 as measured by scRNA-seq (Fig S2C, available at www.ophthalmologyscience.org). Likewise, the histologically defined relative ratios of inner retinal cells are 27 (bipolar):21 (amacrine):17 (Müller glia):1 (horizontal cells).47 We observed that these were 12:4:48:1 for scRNA-seq and 7:17:9:1 for snRNA-seq (Fig 2C; Fig S2B, available at www.ophthalmologyscience.org). Because roughly half of all amacrine cells exist as displaced amacrine cells in the ganglion cell layer,48 the ratios of cells profiled using snRNA-seq closely match those defined using histological approaches.
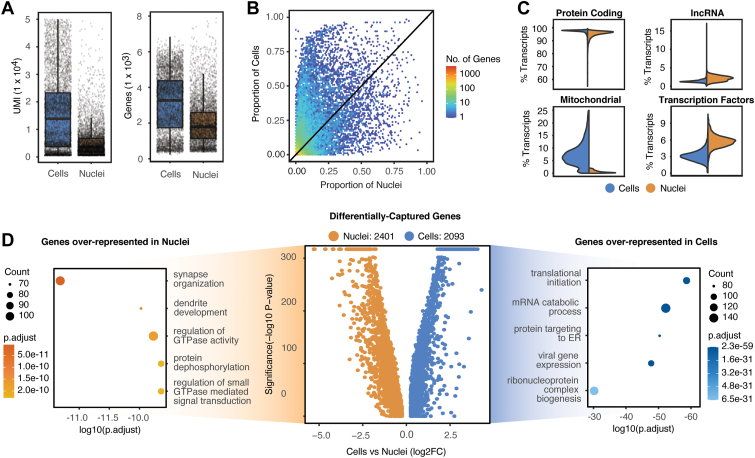
Next, we more closely examined differences in the gene expression levels between the scRNA-seq and snRNA-seq data sets. As expected, when data from all individual cellular or nuclear profiles were aggregated, we observed significantly greater numbers of UMI and individual genes detected in scRNA-seq relative to snRNA-seq profiles (Fig 3A). We likewise observed that expression of individual genes was detected in a larger fraction of individual scRNA-seq than snRNA-seq profiles (Fig 3B). As expected, we also observed a vastly larger number of spliced reads in the scRNA-seq data set when compared with the snRNA-seq data set (Fig S3A, available at www.ophthalmologyscience.org). Correspondingly, we observed both common and cell type-specific differences in the expression levels of mRNAs corresponding to specific functional classes of genes in the scRNA-seq relative to the snRNA-seq data set (Fig 3C). Transcripts encoded by genes controlling protein synthesis, oxidative phosphorylation, viral gene expression, synapse formation, and RNA metabolism and splicing were enriched in scRNA-seq samples from all cell types, whereas snRNA-seq samples were enriched for transcripts encoding genes regulating histone modification, GTPase activity, cellular morphogenesis, and dendrite development. Notable cell type-specific differences in measured gene expression levels included enrichment of transcripts regulating protein dephosphorylation and ciliogenesis in cone snRNA-seq, polyubiquitination in microglial snRNA-seq, and neutrophil degranulation in microglial scRNA-seq data sets. Overall, we observed 2401 genes showing significantly higher expression in the snRNA-seq data set, whereas in the scRNA-seq data set we observed 2093 that showed higher expression in ≥ 1 cell types (Fig 3D; Fig S3C–I, available at www.ophthalmologyscience.org). We also noted substantial differences in the level of contamination of individual cell types by genes specifically expressed in other cell types in the 2 data sets, with snRNA-seq overall showing lower levels of contamination relative to scRNA-seq data, particularly for rod photoreceptor-enriched genes (Fig S3B, available at www.ophthalmologyscience.org). A recent study showed that inclusion of intronic reads improved the sensitivity of scRNA-seq data sets.49 Including intron mapping in our scRNA-seq data set allowed for increased UMI and gene capture rates and lower mapping percentage of reads to mitochondrial genes (Fig S3J–L, available at www.ophthalmologyscience.org). However, 97% of the differentially captured genes between cells with intron mapping and nuclei overlapped with differentially captured genes when using cells without intron mapping (Fig S3M, available at www.ophthalmologyscience.org).
Figure 3.
Differences between single-cell and single-nucleus RNA-sequencing modalities. A, Boxplots showing that the unique molecular identifier (UMI) capture rate (left) and the gene capture rate (right) is higher in cells versus nuclei. B, Binned scatterplot of the proportion of genes detected in cells vs. nuclei. C, Split violin plots displaying the distribution of percentage transcripts that map to protein coding (top left), long noncoding RNA (top right), mitochondrial (bottom left), and transcription factor genes (bottom right). D, Four thousand four hundred ninety-four genes are differentially captured in the combined data set with 2401 genes expressed higher in nuclei and 2093 expressed higher in cells. The top 5 overrepresented gene ontology terms (GO) are displayed on the left (nuclei) and right (cells) of the volcano plot.
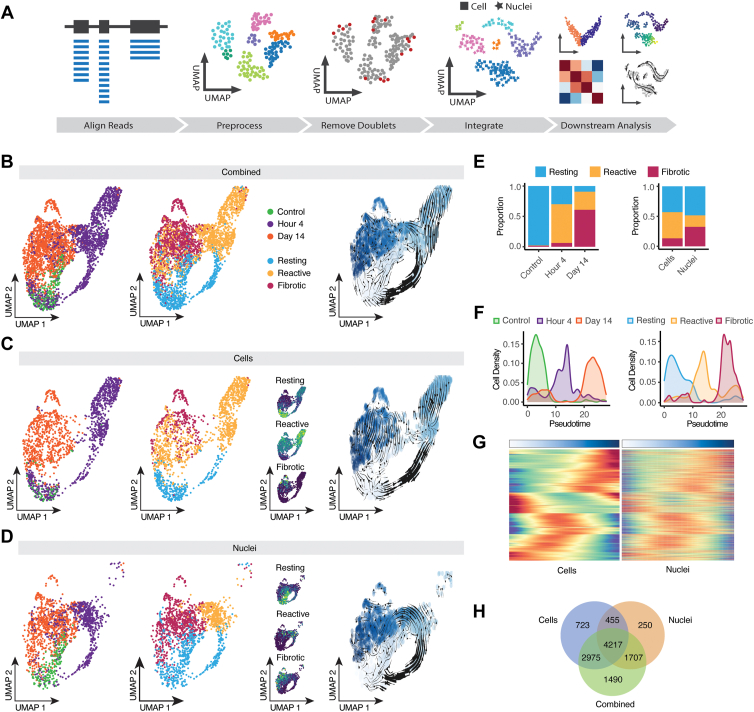
By analyzing multiple timepoints of PVR progression, we were also able to investigate the efficiency with which scRNA-seq and snRNA-seq captured differences between resting, activated, and fibrotic Müller glial cells. We investigated this by conducting combined UMAP analysis of Müller glia from both scRNA-seq and snRNA-seq data sets, incorporating equal number of cells from each data set generated and timepoint profiled (Fig 4A). Using well-characterized molecular markers,32,50, 51, 52 we then grouped glia into resting, reactive, and fibrotic subgroups (Fig 4B). Müller glia profiled by scRNA-seq showed much higher overall enrichment for reactive glia (Fig 4D, E), whereas glia profiled by snRNA-seq were much more likely to be fibrotic (Fig 4D, E) and somewhat more likely to be in a resting state, even after controlling for injury state. Significant differences in the expression in the relative levels of marker genes of specific glial states are also observed between scRNA-seq and snRNA-seq data sets. Pseudotime lineage analysis and RNA velocity analysis of the individual data sets and the combined data set show overlap of cellular transitions between glial cell states, starting with control cells, followed by reactive and terminating with fibrotic cells (Fig 4B–D,F; Fig S4A–C), available at www.ophthalmologyscience.org. Differential gene expression along pseudotime of the combined data set also captures the majority of gene changes seen in the individually analyzed data sets (Fig 4G, H). The analysis of differential expression between postinjured and fibrotic Müller glia, compared with control glia, using pseudobulk methods, revealed that only approximately 25% to 35% of the significant genes were shared between scRNA-seq and snRNA-seq data sets. Notably, scRNA-seq identified genes related to NADH metabolism, protein translation, and cell adhesion, whiereas snRNA-seq identified genes associated with kinase and GTPase activity (Fig S4D–G, available at www.ophthalmologyscience.org). However, despite this discrepancy, around 60% of the unique genes that were differentially expressed in the nuclei data set exhibited a similar trend in the cell data set, and roughly 65% of the unique differentially expressed genes identified in the cell data set displayed a comparable trend in the nuclei data set.
Figure 4.
Integration of single-cell RNA sequencing (scRNA) and single-nucleus RNA sequencing (snRNA-seq) captures Müller glia transitional states after proliferative vitrreoretinopathy induction. A, Schematic summary of integrating and combined analysis of scRNA and snRNA-seq data. B–D, Uniform Manifold Approximation and Projection (UMAP) embeddings of the combined integrated (B), cells (C), and nuclei (D). Dots are colored by injury timepoints (left), cell state with the respective combined expression of cell state specific genes (middle) and pseudotime with RNA velocity trajectory predictions overlayed. E, Comparison of Müller glia transitional states proportions between injury timepoints of the combined data set (left) and sequencing modalities (right). F, Density plots of injury timepoints (left) or cell states (right) plotted along pseudotime in the combined data set. G, Heatmap comparing gene expression dynamics of common highly variable differentially expressed genes along pseudotime between scRNA-seq (left) and snRNA-seq (right). H, Comparison of combined or modality-specific analysis of differentially expressed genes along pseudotime.
Discussion
In this study, we performed a systematic comparison of scRNA-seq and snRNA-seq data sets obtained from a rabbit model of PVR across multiple stages of disease progression. Although both scRNA-seq and snRNA-seq captured and accurately profiled gene expression in abundant retinal cell types, we observed major differences between these data sets. Overall, cell type proportions captured by snRNA-seq much more accurately reflected cell composition data obtained from histological analysis. Single-cell RNA sequencing overrepresented Müller glia and microglia, as well as cone photoreceptor and bipolar interneurons, but captured few amacrine cells and hardly any retinal ganglion cells, oligodendrocytes, and astrocytes. This may have reflected relative differences in viability during the dissociation and microfluidic steps of library construction and has been observed in other tissue sample preparations.26,53,54 Optimized dissociation protocols such as gentle papain treatment or fixative approaches such as rapid methanol fixation may be used to partially overcome this sampling bias and improve the capture efficiency of retinal neurons and rare cell types.55,56 Furthermore, despite the greater overall efficiency of Müller glia capture with scRNA-seq, snRNA-seq more efficiently captures fibrotic glia. Improved recovery of fibrotic cells with snRNA-seq has previously been reported in kidneys26 and may reflect greater difficulty in removing extracellular matrix associated with these cells and thereby obtaining clean dissociation. The greater level of rod and cone photoreceptor contamination seen in scRNA-seq data sets likely reflects inclusion of mRNA-containing inner segment fragments, which may be less likely to contaminate nuclear preparations, whereas the greater levels of inner retinal cell contamination seen in snRNA-seq may simply reflect the overall more efficient recovery of these cell types.
Although these findings generally weigh in favor of routine use of snRNA-seq for these studies, scRNA-seq offers several potential overall advantages. The number of transcripts and genes detected in each cellular profile are, as expected, considerably higher than observed with snRNA-seq. We likewise observe many differences in the expression of individual genes between cell-specific scRNA-seq and snRNA-seq profiles. Transcripts encoding certain functional categories of genes are consistently enriched in either scRNA-seq or snRNA-seq data sets both across all cell types and in individual cell types. Likewise, several well-characterized marker genes for different Müller glia activation states show dramatic differences in expression levels between scRNA-seq and snRNA-seq data obtained from identical biological samples. If resources allow, combined use of both approaches may thus provide a more comprehensive and useful data set for analyzing progression of PVR than using either technique in isolation.
Acknowledgments
The authors would like to thank the Johns Hopkins Transcriptomics and Deep Sequencing Core for use of the 10× Genomics Chromium controller and library sequencing and all the members of the Singh and Blackshaw labs for comments on the manuscript.
Manuscript no. XOPS-D-22-00260R2.
Footnotes
Supplemental material available atwww.ophthalmologyscience.org.
Disclosure(s}: All authors have completed and submitted the ICMJE disclosures form.
The authors have made the following disclosures: S.B.: Financial support – Genentech; Consultant – Third Rock Ventures, Tenpoint Therapeutics; Other – CDI Labs, LLC.
M.S.S.: Consultant – Revision Therapeutics, Johnson & Johnson, Novartis Pharmaceuticals, W. L. Gore & Associates, Kala Pharmaceuticals, Acucela; Consultant and financial support – Bayer AG; Other – Agnos Therapeutics.
This project was funded by Bayer AG as part of a sponsored research collaboration with the Wilmer Eye Institute, Johns Hopkins Hospital (M.S.S.), and a Visual Sciences Training grant 2T32EY007143 (C.P.S.).
HUMAN SUBJECTS: No human subjects were included in this study. Data from the IRIS® Registry are deidentified and do not require patient-level consent. Therefore, institutional review board approval was not required. Nonhuman animals were used in this study. All animal experiments were performed in accordance with the guidelines for the Use of Animals in Ophthalmic and Vision Research of the Association for Research in Vision and Ophthalmology. Protocol was approved by the Johns Hopkins University’s Institutional Animal Care and Use Committee and in adherence with the ARRIVE 2.0 Guidelines. All research adhered to the tenets of the Declaration of Helsinki.
Author Contributions:
Conception and design: Santiago, Blackshaw, Singh
Data collection: Santiago, Gimmen, Lu, McNally, Creamer, Orzolek, Blackshaw, Singh
Analysis and interpretation: Santiago, Gimmen, Blackshaw, Singh
Obtained funding: Singh
Overall responsibility: Santiago, Gimmen, Lu, McNally, Duncan, Creamer, Orzolek, Blackshaw, Singh
Contributor Information
Seth Blackshaw, Email: sblack@jhmi.edu.
Mandeep S. Singh, Email: singhcorrespauth@gmail.com.
Supplementary Data
Feature plots of marker gene expression in (A) scRNA-seq and (B) snRNA-seq data sets.
Cell type proportions of the current data set. A, Counts and cell type proportion of in the scRNA-seq and snRNA-seq data. B, C Comparison of the observed cell and nuclei cell type percentage to previous histological studies for cell types found in the rabbit (B) inner nuclear layer and (C) outer nuclear layer.
Differences between single-cell and single nuclei RNA-sequencing modalities for each cell type. A, Boxplots showing spliced (left) and unspliced RNA counts between scRNA and snRNA-seq. B, Expression levels of enriched bipolar, amacrine, rod, and Müller glia genes in each cell type. C–I, Differentially captured genes and top 5 overrepresented GO terms between scRNA and snRNA-seq for (C) rod, (D) cone, (E) bipolar, (F) amacrine, (G) horizontal, (H) Müller glia, and (I) microglial cell types. J–L, Boxplots depicting the comparison of UMI capture rate (J) and gene capture rate (K) is higher in cell data set with included intronic reads versus cell data sets where the intronic reads have been discarded while also having lower percent of reads mapped to mitochondrial genes (L). The data for the cells and nuclei boxplots (J–K) are obtained from Figure 3A. M, Venn diagrams comparing the overlap of genes differentially captured in cells with or without intron mappings versus nuclei. GO = gene ontology; UMI = unique molecular identifier.
Combined analysis of injured Müller glia from the scRNA and snRNA-seq data sets. A, RNA velocity trajectory predictions overlayed on Müller glia from the combined (top), cells (middle), or bottom (nuclei) data sets, with each cell colored by cell state. B, C Density plots of injury timepoint (B) or cell state (C) plotted along pseudotime separated by sequencing modality. D, Comparison of combined or modality-specific analysis of differentially expressed genes between control and injury by Wilcoxon rank sum test on pseudobulk gene profiles separated by genes that are upregulated (top) or downregulated (bottom) after injury. E, Scatterplots of differentially expressed genes between control and fibrotic Müller glia comparing log2 fold change of significant genes captured by both cells and nuclei (left), captured only by nuclei (middle), and captured only by cells (right). Numbers specify the number of genes in each quadrant. F, Enrichment map of overrepresented GO terms comparing enrichment results of differentially expressed genes between control and fibrotic Müller glia in cells and nuclei data sets. Similar terms are connected and are clustered together, and the proportion of each term is determined by the number of genes from each modality contributing to that term. G, Feature plots of known genes expressed in resting (left) reactive (middle) and fibrotic (right) Müller glia split by modality. GO = gene ontology.
References
- 1.Idrees S., Sridhar J., Kuriyan A.E. Proliferative vitreoretinopathy: a review. Int Ophthalmol Clin. 2019;59:221–240. doi: 10.1097/IIO.0000000000000258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eastlake K., Banerjee P.J., Angbohang A., et al. Müller glia as an important source of cytokines and inflammatory factors present in the gliotic retina during proliferative vitreoretinopathy. Glia. 2016;64:495–506. doi: 10.1002/glia.22942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pennock S., Haddock L.J., Eliott D., et al. Is neutralizing vitreal growth factors a viable strategy to prevent proliferative vitreoretinopathy? Prog Retin Eye Res. 2014;40:16–34. doi: 10.1016/j.preteyeres.2013.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Nagasaki H., Shinagawa K., Mochizuki M. Risk factors for proliferative vitreoretinopathy. Prog Retin Eye Res. 1998;17:77–98. doi: 10.1016/s1350-9462(97)00007-4. [DOI] [PubMed] [Google Scholar]
- 5.Casaroli-Marano R.P., Pagan R., Vilaró S. Epithelial-mesenchymal transition in proliferative vitreoretinopathy: intermediate filament protein expression in retinal pigment epithelial cells. Invest Ophthalmol Vis Sci. 1999;40:2062–2072. [PubMed] [Google Scholar]
- 6.Asato R., Yoshida S., Ogura A., et al. Comparison of gene expression profile of epiretinal membranes obtained from eyes with proliferative vitreoretinopathy to that of secondary epiretinal membranes. PLoS One. 2013;8 doi: 10.1371/journal.pone.0054191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yao J., Hu L.-L., Li X.-M., et al. Comprehensive circular RNA profiling of proliferative vitreoretinopathy and its clinical significance. Biomed Pharmacother. 2019;111:548–554. doi: 10.1016/j.biopha.2018.12.044. [DOI] [PubMed] [Google Scholar]
- 8.Shekhar K., Lapan S.W., Whitney I.E., et al. Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell. 2016;166:1308–1323.e30. doi: 10.1016/j.cell.2016.07.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macosko E.Z., Basu A., Satija R., et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peng Y.-R., Shekhar K., Yan W., et al. Molecular classification and comparative taxonomics of foveal and peripheral cells in primate retina. Cell. 2019;176:1222–1237.e22. doi: 10.1016/j.cell.2019.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lyu P., Hoang T., Santiago C.P., et al. Gene regulatory networks controlling temporal patterning, neurogenesis, and cell-fate specification in mammalian retina. Cell Rep. 2021;37 doi: 10.1016/j.celrep.2021.109994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu Y., Shiau F., Yi W., et al. Single-cell analysis of human retina identifies evolutionarily conserved and species-specific mechanisms controlling development. Dev Cell. 2020;53:473–491.e9. doi: 10.1016/j.devcel.2020.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Clark B.S., Stein-O’Brien G.L., Shiau F., et al. Single-cell RNA-seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron. 2019;102:1111–1126.e5. doi: 10.1016/j.neuron.2019.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Finkbeiner C., Ortuño-Lizarán I., Sridhar A., et al. Single-cell ATAC-seq of fetal human retina and stem-cell-derived retinal organoids shows changing chromatin landscapes during cell fate acquisition. Cell Rep. 2022;38 doi: 10.1016/j.celrep.2021.110294. [DOI] [PubMed] [Google Scholar]
- 15.Kruczek K., Qu Z., Gentry J., et al. Gene therapy of dominant CRX-Leber congenital amaurosis using patient stem cell-derived retinal organoids. Stem Cell Rep. 2021;16:252–263. doi: 10.1016/j.stemcr.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas E.D., Timms A.E., Giles S., et al. Cell-specific cis-regulatory elements and mechanisms of non-coding genetic disease in human retina and retinal organoids. Dev Cell. 2022;57:820–836.e6. doi: 10.1016/j.devcel.2022.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Koren E.G., Yu C., Klingeborn M., et al. Microglial function is distinct in different anatomical locations during retinal homeostasis and degeneration. Immunity. 2019;50:723–737.e7. doi: 10.1016/j.immuni.2019.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grindberg R.V., Yee-Greenbaum J.L., McConnell M.J., et al. RNA-sequencing from single nuclei. Proc Natl Acad Sci U S A. 2013;110:19802–19807. doi: 10.1073/pnas.1319700110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang F., Barbacioru C., Wang Y., et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- 20.Ding J., Adiconis X., Simmons S.K., et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat Biotechnol. 2020;38:737–746. doi: 10.1038/s41587-020-0465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Selewa A., Dohn R., Eckart H., et al. Systematic comparison of high-throughput single-cell and single-nucleus transcriptomes during cardiomyocyte differentiation. Sci Rep. 2020;10:1535. doi: 10.1038/s41598-020-58327-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Denisenko E., Guo B.B., Jones M., et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 2020;21:130. doi: 10.1186/s13059-020-02048-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta A., Shamsi F., Altemose N., et al. Characterization of transcript enrichment and detection bias in single-nucleus RNA-seq for mapping of distinct human adipocyte lineages. Genome Res. 2022;32:242–257. doi: 10.1101/gr.275509.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer J., Ayers T. Single nucleus RNA-sequencing: how it’s done, applications and limitations. Emerg Top Life Sci. 2021;5:687–690. doi: 10.1042/ETLS20210074. [DOI] [PubMed] [Google Scholar]
- 25.Bakken T.E., Hodge R.D., Miller J.A., et al. Single-nucleus and single-cell transcriptomes compared in matched cortical cell types. PLoS One. 2018;13 doi: 10.1371/journal.pone.0209648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu H., Kirita Y., Donnelly E.L., Humphreys B.D. Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol. 2019;30:23–32. doi: 10.1681/ASN.2018090912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tran N.M., Shekhar K., Whitney I.E., et al. Single-cell profiles of retinal ganglion cells differing in resilience to injury reveal neuroprotective genes. Neuron. 2019;104:1039–1055.e12. doi: 10.1016/j.neuron.2019.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thrupp N., Frigerio C.S., Wolfs L., et al. Single-nucleus RNA-seq is not suitable for detection of microglial activation genes in humans. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.108189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hao Y., Hao S., Andersen-Nissen E., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Germain P.-L., Lun A., Meixide C.G., et al. Doublet identification in single-cell sequencing data using scDblFinder. F1000Research. 2021;10:979. doi: 10.12688/f1000research.73600.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korsunsky I., Millard N., Fan J., et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoang T., Wang J., Boyd P., et al. Gene regulatory networks controlling vertebrate retinal regeneration. Science. 2020:370. doi: 10.1126/science.abb8598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ling J.P., Wilks C., Charles R., et al. ASCOT identifies key regulators of neuronal subtype-specific splicing. Nat Commun. 2020;11:137. doi: 10.1038/s41467-019-14020-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crow M., Paul A., Ballouz S., et al. Characterizing the replicability of cell types defined by single cell RNA-sequencing data using MetaNeighbor. Nat Commun. 2018;9:884. doi: 10.1038/s41467-018-03282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu G., Wang L.-G., Han Y., He Q.-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A J Integr Bio. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andreatta M., Carmona S.J. UCell: robust and scalable single-cell gene signature scoring. Comput Struct Biotechnol J. 2021;19:3796–3798. doi: 10.1016/j.csbj.2021.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dorrier C.E., Aran D., Haenelt E.A., et al. CNS fibroblasts form a fibrotic scar in response to immune cell infiltration. Nat Neurosci. 2021;24:234–244. doi: 10.1038/s41593-020-00770-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Street K., Risso D., Fletcher R.B., et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics. 2018;19:477. doi: 10.1186/s12864-018-4772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van den Berge K., Roux de Bézieux H., Street K., et al. Trajectory-based differential expression analysis for single-cell sequencing data. Nat Commun. 2020;11:1201. doi: 10.1038/s41467-020-14766-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cao J., Spielmann M., Qiu X., et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manno G.L., La Manno G., Soldatov R., et al. RNA velocity of single cells. Nature. 2018;560:494–498. doi: 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergen V., Lange M., Peidli S., et al. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol. 2020;38:1408–1414. doi: 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 43.Goldaracena M.B., Begoña Goldaracena M., Garcia-Layana A., et al. The role of retinotomy in an experimental rabbit model of proliferative vitreoretinopathy. Curr Eye Res. 1997;16:422–427. doi: 10.1076/ceyr.16.5.422.7052. [DOI] [PubMed] [Google Scholar]
- 44.Wong C.W., Cheung N., Ho C., et al. Characterisation of the inflammatory cytokine and growth factor profile in a rabbit model of proliferative vitreoretinopathy. Sci Rep. 2019;9 doi: 10.1038/s41598-019-51633-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schnitzer J. Distribution and immunoreactivity of glia in the retina of the rabbit. J Comp Neurol. 1985;240:128–142. doi: 10.1002/cne.902400203. [DOI] [PubMed] [Google Scholar]
- 46.Famiglietti E.V., Sharpe S.J. Regional topography of rod and immunocytochemically characterized “blue” and “green” cone photoreceptors in rabbit retina. Vis Neurosci. 1995;12:1151–1175. doi: 10.1017/s0952523800006799. [DOI] [PubMed] [Google Scholar]
- 47.Strettoi E., Masland R.H. The organization of the inner nuclear layer of the rabbit retina. J Neurosci. 1995;15:875–888. doi: 10.1523/JNEUROSCI.15-01-00875.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strettoi E., Masland R.H. The number of unidentified amacrine cells in the mammalian retina. Proc Natl Acad Sci U S A. 1996;93:14906–14911. doi: 10.1073/pnas.93.25.14906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pool A-H, Poldsam H, Chen S, et al. Enhanced recovery of single-cell RNA-sequencing reads for missing gene expression data. bioRxiv. 10.1101/2022.04.26.489449. In press. [DOI]
- 50.Kanda A., Noda K., Hirose I., Ishida S. TGF-β-SNAIL axis induces Müller glial-mesenchymal transition in the pathogenesis of idiopathic epiretinal membrane. Sci Rep. 2019;9:673. doi: 10.1038/s41598-018-36917-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Friedlander M. Fibrosis and diseases of the eye. J Clin Invest. 2007;117:576–586. doi: 10.1172/JCI31030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mallone F., Costi R., Marenco M., et al. Understanding drivers of ocular fibrosis: current and future therapeutic perspectives. Int J Mol Sci. 2021:22. doi: 10.3390/ijms222111748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wen F., Tang X., Xu L., Qu H. Comparison of single-nucleus and single-cell transcriptomes in hepatocellular carcinoma tissue. Mol Med Rep. 2022;26 doi: 10.3892/mmr.2022.12855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Andrews T.S., Atif J., Liu J.C., et al. Single-cell, single-nucleus, and spatial RNA sequencing of the human liver identifies cholangiocyte and mesenchymal heterogeneity. Hepatol Commun. 2022;6:821–840. doi: 10.1002/hep4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alles J., Karaiskos N., Praktiknjo S.D., et al. Cell fixation and preservation for droplet-based single-cell transcriptomics. BMC Biol. 2017;15:44. doi: 10.1186/s12915-017-0383-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Fadl B.R., Brodie S.A., Malasky M., et al. An optimized protocol for retina single-cell RNA sequencing. Mol Vis. 2020;26:705–717. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Feature plots of marker gene expression in (A) scRNA-seq and (B) snRNA-seq data sets.
Cell type proportions of the current data set. A, Counts and cell type proportion of in the scRNA-seq and snRNA-seq data. B, C Comparison of the observed cell and nuclei cell type percentage to previous histological studies for cell types found in the rabbit (B) inner nuclear layer and (C) outer nuclear layer.
Differences between single-cell and single nuclei RNA-sequencing modalities for each cell type. A, Boxplots showing spliced (left) and unspliced RNA counts between scRNA and snRNA-seq. B, Expression levels of enriched bipolar, amacrine, rod, and Müller glia genes in each cell type. C–I, Differentially captured genes and top 5 overrepresented GO terms between scRNA and snRNA-seq for (C) rod, (D) cone, (E) bipolar, (F) amacrine, (G) horizontal, (H) Müller glia, and (I) microglial cell types. J–L, Boxplots depicting the comparison of UMI capture rate (J) and gene capture rate (K) is higher in cell data set with included intronic reads versus cell data sets where the intronic reads have been discarded while also having lower percent of reads mapped to mitochondrial genes (L). The data for the cells and nuclei boxplots (J–K) are obtained from Figure 3A. M, Venn diagrams comparing the overlap of genes differentially captured in cells with or without intron mappings versus nuclei. GO = gene ontology; UMI = unique molecular identifier.
Combined analysis of injured Müller glia from the scRNA and snRNA-seq data sets. A, RNA velocity trajectory predictions overlayed on Müller glia from the combined (top), cells (middle), or bottom (nuclei) data sets, with each cell colored by cell state. B, C Density plots of injury timepoint (B) or cell state (C) plotted along pseudotime separated by sequencing modality. D, Comparison of combined or modality-specific analysis of differentially expressed genes between control and injury by Wilcoxon rank sum test on pseudobulk gene profiles separated by genes that are upregulated (top) or downregulated (bottom) after injury. E, Scatterplots of differentially expressed genes between control and fibrotic Müller glia comparing log2 fold change of significant genes captured by both cells and nuclei (left), captured only by nuclei (middle), and captured only by cells (right). Numbers specify the number of genes in each quadrant. F, Enrichment map of overrepresented GO terms comparing enrichment results of differentially expressed genes between control and fibrotic Müller glia in cells and nuclei data sets. Similar terms are connected and are clustered together, and the proportion of each term is determined by the number of genes from each modality contributing to that term. G, Feature plots of known genes expressed in resting (left) reactive (middle) and fibrotic (right) Müller glia split by modality. GO = gene ontology.
Data Availability Statement
All rabbit scRNA-seq and snRNA-seq data can be accessed at GEO accession numbers GSE217333. Code used to analyze the data sets in this study can be found at https://github.com/csanti88/pvr_singlecell_vs_singlenucleiRNAseq_2022.