Abstract
Background
Vacuolar type H+-ATPases play a critical role in the maintenance of vacuolar homeostasis in plant cells. V-ATPases are also involved in plants' defense against environmental stress. This research examined the expression and regulation of the catalytic subunit of the vacuolar type H+-ATPase in Arabidopsis thaliana and the effect of environmental stress on multiple transcripts generated by this gene.
Results
Evidence suggests that subunit A of the vacuolar type H+-ATPase is encoded by a single gene in Arabidopsis thaliana. Genome blot analysis showed no indication of a second subunit A gene being present. The single gene identified was shown by whole RNA blot analysis to be transcribed in all organs of the plant. Subunit A was shown by sequencing the 3' end of multiple cDNA clones to exhibit multi site polyadenylation. Four different poly (A) tail attachment sites were revealed. Experiments were performed to determine the response of transcript levels for subunit A to environmental stress. A PCR based strategy was devised to amplify the four different transcripts from the subunit A gene.
Conclusions
Amplification of cDNA generated from seedlings exposed to cold, salt stress, and etiolation showed that transcript levels for subunit A of the vacuolar type H+-ATPase in Arabidopsis were responsive to stress conditions. Cold and salt stress resulted in a 2–4 fold increase in all four subunit A transcripts evaluated. Etiolation resulted in a slight increase in transcript levels. All four transcripts appeared to behave identically with respect to stress conditions tested with no significant differential regulation.
Background
Vacuolar-type H+-ATPases are enzymes responsible for the energization of membranes and the acidification of compartments within the eukaryotic cell via the establishment of proton and electrochemical gradients at the expense of ATP. The vacuolar type H+-ATPase in plants is a large multimeric enzyme complex whose function is to pump protons across a membrane via primary active transport. Vacuolar type ATPases are homologs of the F-type ATP synthases and probably convert the free energy of hydrolysis of the high-energy phosphate bond into rotational motion [1-9]. Vacuolar type H+-ATPases are critical for the maintenance of homeostasis in eukaryotic cells [10,11].
In plant cells V-ATPases are responsible for the deacidification of the cytosol and the energization of secondary transport processes across the tonoplast, as well as in the endocytotic and secretory pathways [12]. In addition, the vacuolar type ATPase is thought to be primarily responsible for the acidification and expansion of the large central vacuole [12-15]. In plants a critical event in growth and development is the maturation and expansion of the central vacuole. Upon maturation, the vacuolar contents can comprise more than eighty percent of the total cell volume [16]. Primarily filled with water, the vacuole also is a repository for a wide variety of solutes fulfilling numerous important metabolic functions. The influx of water and metabolites into the vacuole is dependent in part on the generation of a proton motive force across the tonoplast.
Vacuolar type H+-ATPases are large, multimeric enzyme complexes of 500–750 kDa consisting of 15 or more different proteins. Structurally, they can be divided into integral (V0) and peripheral (V1) membrane sectors [17]. The peripheral membrane sector is located on the cytoplasmic side of the membrane and is composed of at least five different subunits including the catalytic subunit, subunit A. The V1 peripheral sector is further divided into a head group and stalk region. The V0 integral sector is embedded in the membrane and consists of at least 4 different subunits.
Subunit A of the vacuolar-type H+-ATPase is the catalytic subunit. The subunit A protein is approximately 70 kDa in most organisms studied and is a hydrophilic peptide located in the head group of the V1 peripheral sector in three copies per holoenzyme [18]. This subunit contains a nucleotide binding motif and functions to bind and hydrolyze ATP [19]. In addition, subunit A contains a highly conserved cysteine residue located within the enzymes' catalytic center that may be involved in regulation of the holoenzyme [20-23]. Cloning, sequencing and characteristics of subunit A of the V-Type ATPase from Arabidopsis thaliana have been previously reported [24].
Small multigene families of V-ATPase subunit A are known to exist in flowering plants and algae. The catalytic subunit exists as two distinct genes with highly conserved exons and intron boundaries in twelve species of plants [25]. In the flowering plant Daucus carota (carrot) evidence exists for two distinct isoforms for the catalytic subunit, one of which is tonoplast specific while the other is localized to the Golgi [26]. In addition, two distinct mRNAs have been isolated from cotton (Gossypium hirsutum) [27]. Two subunit A mRNAs are expressed in the unicellular alga Acetabularia acetabulum[28,29]. Isoforms of other V-ATPase subunits are also known from plants including subunit B [27,30,32], the proteolipid subunit [33-37] and subunits D and E [38].
What is lacking in our knowledge of vacuolar type ATPases is a sense of what purpose these multigene family proteins may serve in the cellular milieu and how they participate in the growth, development, and metabolism of higher plants. It has been proposed that multigene families might be encoding functionally distinct isoforms of V-ATPase subunits. These isoforms may be required to provide unique functions or regulatory constraints within the varied micro-compartments requiring V-ATPases [26,27,34,39,43]. In addition, different isoforms may be responsible for targeting to varied subcellular compartments [10,26,28,36,41,44,47]. Furthermore, it is possible that gene family members may behave in a differential manner with respect to environmental stress [36].
Three enzymes are primarily responsible for the maintenance of proton flux across the tonoplast in plants, the vacuolar H+-ATPase, the H+ pumping pyrophosphatase [48] and the Na+/H+ antiporter [49,50]. All three enzymes have been implicated in allowing plants to cope with environmental stress conditions.
Several studies have been conducted in an attempt to evaluate the role vacuolar type ATPases play in allowing plants to cope with environmental stress. These endeavors have primarily concentrated on salt and cold stress. Cold tolerant Brassica napus, a close relative of Arabidopsis thaliana, demonstrated an increase in both subunit A mRNA and protein in response to chilling at 2°C [50]. This chilling also resulted in a concomitant increase in cell sap osmotic pressure and endogenous ABA accumulation. These researchers indicated that Arabidopsis also was a cold tolerant plant which exhibited a similar response with respect to V-ATPase subunit A mRNA, however they provided no data. In rice (Oryza sativa) chilling plants at 10°C resulted in a two fold increase in vacuolar type ATPase activity evaluated by in vitro assay of isolated membranes [51]. This study did not analyze changes in protein level in the isolated membranes in response to chilling. Cold sensitive mung bean seedlings (Vigna radiata) subjected to chilling at 4°C for 48 hours contrastingly showed no increase in vacuolar type ATPase activity in isolated membranes [52]. In addition, these researchers found no change in the amount of V-ATPase protein levels in response to chilling.
The facultative CAM plant Mesembryanthemum crystallinum displays a differential organ level response to salt stress while still engaged in C3 photosynthesis [53]. Subjecting plants to 400 mM NaCl resulted in an approximately two fold increase in the transcript levels of subunits A, B, and the proteolipid in roots prior to the shift to Crassulacean acid metabolism. Fully expanded leaves exhibited an increase only for the proteolipid mRNA, an increase of approximately three fold over non salt stressed plants [54]. Cell suspension cultures when subjected to salt stress exhibited increase in activity of both P-Type and V-Type ATPases without concomitant increase in protein levels [55]. Transcript levels of subunit E of the V-ATPase in Mesembryanthemum crystallinum were recently shown to increase in leaves but not roots of salt stressed plants with a concomitant increase in protein levels in leaves [56]. On the other hand, subunit E in Hordeum vulgare showed no effect on mRNA level in response to salt stress [57]. The effects of salt stress on V-ATPases has also been investigated in tobacco (Nicotiana tabacum) suspension cells. Subjecting NaCl adapted cells to salt stress resulted in an approximately four fold increase in both proton transport and ATP hydrolysis on isolated membranes compared to unadapted cells [58]. Remarkably this increase in overall V-ATPase activity coincided with an apparent reduction in the amount of enzyme on the tonoplast. This apparently contradictory finding may be the result of physical changes in the enzyme itself or changes in the lipid environment [58]. Especially confounding is the finding that mRNA levels for subunit A increase two to four fold in response to the same levels of salt stress in tobacco suspension cells [59]. Experiments with tomato (Lycopersicon esculentum) showed similar results with respect to induction of mRNA for subunit A with an approximately two to four fold increase seen with respect to salt stress [60]. Recent work in the resurrection plant Tortula ruralis indicated that transcript levels of the proteolipid (subunit c) increased in response to salt stress but that there was no corresponding increase in protein levels [61].
The limited work done to evaluate the role of V-ATPases in drought stress has provided conflicting results. Drought stress in winter Brassica napus resulted in a very similar response as cold stress, that is a dramatic increase in subunit A mRNA [50]. However, drought stress in Lycopersicon esculentum resulted in no apparent increase in mRNA levels for subunit A [60].
None of the previously mentioned efforts into evaluating the role of vacuolar type ATPases in environmental stress acclimation take into account the fact that subunits A, B, and the proteolipid are most certainly present as small gene families in the plants studied. Furthermore, it is possible that these gene family members may behave in a differential manner with respect to stress. The only work published previously on V-ATPase subunit isoforms and stress is concerned with the proteolipid gene family in Arabidopsis thaliana. Three proteolipid genes were evaluated for their response to salt stress as well as etiolation [36]. All three genes exhibited an approximately two fold increase in mRNA levels in response to salt stress (50 mM). However, with respect to etiolation two isoforms showed no response while the third demonstrated an increase in mRNA levels.
A number of genes in plants have been found to contain multiple poly (A) attachment sites yielding different transcripts from a single gene. Several examples have recently emerged where differentially polyadenylated genes have transcripts that are post transcriptionally regulated based on sugar sensing. In rice (Oryza sativa), two transcripts generated off an α amylase gene exhibit differential stability based on sucrose availability [65]. A cell wall invertase gene (Incw-1) in Maize (Zea mays) is expressed as two transcripts differing in 3' ends. The two transcripts are differentially expressed in response to sugars [66]. In this study we explore the possibility that differentially polyadenylated transcripts of the V-ATPase subunit A encoding gene show different responses with respect to etiolation, salt, or chilling stress.
Results and discussion
A vacuolar type H+-ATPase subunit A cDNA has been previously identified and sequenced (GenBank accession number U65638) [24]. The cDNA was identified as subunit A of the vacuolar type H+-ATPase via identity with other previously sequenced subunit A genes from other organisms. The primary nucleotide and amino acid sequence of the A. thaliana subunit A cDNA showed a high degree of identity with subunit A cDNA sequenced from other organisms. This cDNA encoded a putative open reading frame consistent with that of other subunit A genes and was shown to have all the motifs common to subunit A [67]. This high degree of conservation was evident across taxa as diverse as animals, plants, fungi, and protozoa [68].
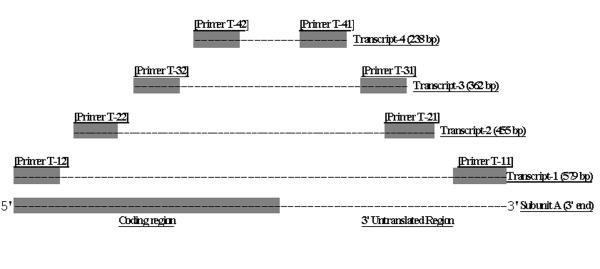
Genome blot analysis was utilized to determine the number of genes in the A. thaliana genome corresponding to subunit A of the vacuolar type ATPase. Genomic DNA was isolated as described, subjected to restriction enzyme digestion, and immobilized on a positively charged nylon membrane. Ten single restriction enzyme digestions were performed. Hybridization was conducted at moderate stringency with a 300 base pair homologous A. thaliana cDNA probe corresponding to subunit A of the vacuolar type ATPase. Results indicated hybridization to only a single band for nine out of the ten enzymes (Fig. 1). The HaeIII digest showed faint hybridization to a second band that was later shown to be the result of a HaeIII site located in the region of the probe (results not shown).
Figure 1.
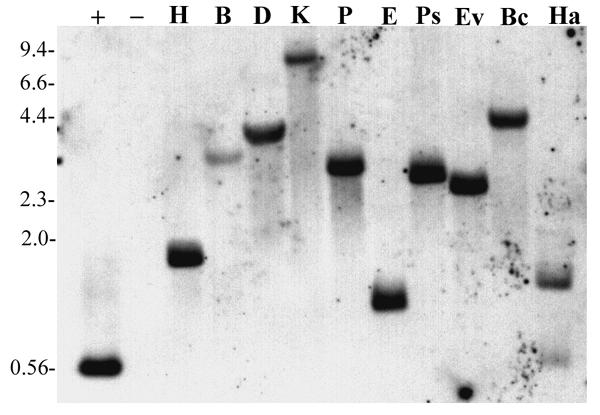
Whole genome blot analysis of vacuolar type H+-ATPase subunit A gene in A. thaliana. Each lane represents 5 μg genomic DNA digested with the indicated restriction enzyme. Restriction enzymes used were; H, HindIII, B, BglII, D, DraI, K, KpnI, P, PvuII, E, EcoRI, Ps, PstI, Ev, EcoRv, Bc, BclI, and Ha, HaeIII. Hybridization was to a single band in each lane except for HaeIII due to the presence of a recognition site in the region corresponding to the probe. Positive hybridization control was 20 pg of unlabeled probe DNA generated via PCR. HindIII digested lambda DNA (2 μg) was used as negative control. Numbers to the left of the blot indicate the approximate size of DNA fragments (in kilobases) as determined by HindIII digestion of Lambda DNA.
All evidences acquired to date failed to indicate the presence of a second V-ATPase subunit A gene in A. thaliana. Whole genome blot analysis detected a single gene in the A. thaliana genome. Extensive screening of a cDNA library derived from all organs and multiple developmental stages of the plant revealed only a single subunit A cDNA. A polymerase chain reaction based screen failed to detect an intron length polymorphism (results not shown) between two subunit A genes found to be present in other plants [25]. Lastly, extensive database searches of GenBank, the Arabidopsis EST database, and the Arabidopsis Genome Initiative (TAIR, http://www.arabidopsis.org) report only a single subunit A gene in the current version of the Arabidopsis genome (chromosome 1 BAC F9K20).
This situation of a single subunit A gene appears to be unusual in plants. Admittedly only a small subset of plants has been examined in detail. Of the species examined to date three have only a single subunit A gene detected, the alga Coleochaete scutata, sugar beet (Beta vulgarise[63,64], and A. thaliana[25]. Flowering plants shown to have at least two subunit A genes include oat (Avena sativa), carrot (Daucus carota), tobacco (Nicotiana tabacum), tomato (Lycopersicon esculentum), cotton (Gossypium hirsutum), magnolia (Magnolia virginana), Hydrastis canadensis, Chenopodium rubrum, and clematis (Clematis liguisticfolio). In addition, the unicellular alga Acetabularia acetabulum has two V-ATPase subunit A genes.
RNA blot analysis was performed to determine the organ level expression pattern of the subunit A gene. Vacuolar type ATPases are housekeeping enzymes critical for the maintenance of homeostasis. Therefore, it is expected that all living plant cells require V-ATPase function. Since A. thaliana seemed to possess only a single gene for subunit A of the V-ATPase it was expected that this gene should be expressed in all organs tested. Whole RNA was harvested from leaves, bolts, siliques, flowers, axenically grown seedlings, and axenically grown roots. RNA was denatured with glyoxal, fractionated via agarose gel electrophoresis, and immobilized on a positively charged nylon membrane. Hybridization was conducted at high stringency with a 1700 base pair homologous cDNA probe labeled with digoxigenin. In agreement with our expectations, results indicated high levels of expression in all organs tested (Fig. 2) with slightly higher levels of subunit A mRNA present in seedlings. This is most likely due to the fact that seedlings are the most rapidly growing and metabolizing stage of the plant tested. Seedlings are undergoing dramatic and rapid vacuole biogenesis to allow for rapid growth at this stage of development and therefore would be expected to require a higher level of subunit A expression than other more terminally differentiated organs. Roots, leaves, bolts, and flowers showed approximately equivalent amounts of signal with somewhat lower levels in siliques.
Figure 2.
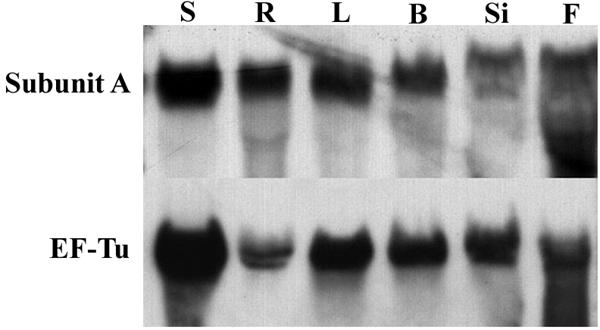
RNA blot analysis of subunit A in A. thaliana. The probe utilized was approximately 1700 base pairs in length corresponding to the vacuolar type H+-ATPase subunit A cDNA from Arabidopsis thaliana. Probe DNA was labeled with digoxigenin via the Polymerase Chain Reaction. Each lane contained 20 μg of whole RNA. Lanes correspond to the following organs: S-seedlings, R-roots, L-leaves, B-bolts, Si-siliques, and F-flowers. EF-Tu is a nuclear encoded chloroplast gene transcribed in all organs of the plant. Hybridization to a digoxigenin labeled probe corresponding to EF-Tu was performed to evaluate the RNA isolated from each organ to determine its suitability for RNA blot analysis.
Most eukaryotic mRNAs are characterized by the presence of a poly (A) tail, a series of adenine residues post transcriptionally attached to the 3' untranslated region. The primary function of the poly (A) tail is thought to be in message stability assisted by a series of poly (A) binding proteins [69]. In addition, the poly (A) tail in conjunction with the 5' cap may be required for efficient translation of plant mRNAs [70].
Subunit A of the vacuolar type H+-ATPase in A. thaliana was shown to exhibit multi site polyadenylation. Sequencing the 3' ends of seven independent clones isolated from the PRL-2 cDNA library and one EST obtained through the Arabidopsis EST database revealed four different length transcripts produced from the single subunit A gene based on different poly (A) tail attachment sites (Fig. 3). Multiple polyadenylation sites are fairly common in plant mRNAs but this phenomenon is rare in animal genes [71,72]. Since the A. thaliana V-ATPase subunit A gene produced at least four different transcripts (all producing the same protein) and is one of two flowering plants observed to date not to have a multigene family for this subunit, experiments were designed to determine if muiti site polyadenylation might be a mechanism for gene regulation in plants.
Figure 3.
Vacuolar type H+-ATPase subunit A 3' untranslated region nucleotide sequence. Nucleotide sequence is depicted in the 5' to 3' direction. The diamond (♦) indicates the coding region stop (TAA). Arrows (↓) indicate sites of Poly (A) tail attachment for the four different transcripts amplified. A total of eight cDNA clones were sequenced from the 3' end, in each case the poly (A) tail was added at an adenine residue in the DNA sequence. Bold underlined motifs correspond to putative polyadenylation signals.
V-ATPase proteolipid isoforms were shown to exhibit differential transcript expression levels in response to environmental stresses [36]. This fact implies a possible role for these isoforms in allowing the plant to deal with stress. Experiments were designed to determine if, in A. thaliana, differentially polyadenylated transcripts could be regulated in response to stress.
First, it was critical to confirm that subunit A transcript levels were at all responsive to environmental stress conditions. Three environmental stresses were chosen including 100 mM NaCl, etiolation for four days, and chilling at 6 degrees for four days. In all three situations axenic, seven-day-old seedlings grown on agar were evaluated. Seven day old seedlings were chosen for their short growth time, limited space requirements, and ease in culturing. In addition, the facts that they are rapidly growing and metabolizing, were shown to produce ample quantities of subunit A transcript, and possess several organs including roots, hypocotyls, and cotyledons made them an attractive choice for analysis.
Whole RNA blot analysis was used first to evaluate the overall response of subunit A to stress conditions. RNA was extracted from stress treated seedlings and non-treated control plants. Hybridization was performed as previously described. Results indicated a subunit A transcript response to stress. An approximate 2–4 fold increase in transcript level was detected in seedlings subjected to salt and chilling stress compared to control plants (Fig. 4). Etiolated seedlings showed essentially equivalent amounts of subunit A transcript compared to controls. Since etiolation results in rapid elongation of seedlings it was expected that there would be a concomitant increase in subunit A messenger RNA as seen with proteolipid subunit mRNA in A. thaliana[36] and subunit A message levels in rapidly expanding cotton (Gossypium hirsutum) fibers [73]. Equivalent loading of whole RNA was evaluated via spectrophotometry measuring absorption at 260 nm using a Perkin-Elmer Lambda-3 spectrophotometer and by visual confirmation using non denaturing ethidium bromide stained agarose gel electrophoresis (results not shown).
Figure 4.

Subunit A transcriptional response to environmental stress conditions in Arabidopsis seedlings. Each lane contained 20 μg whole RNA. The probe used was the same for organ level RNA blot analysis (1700 base pair subunit A cDNA labeled with digoxigenin). Lanes correspond to the following: N-normal, untreated control plants grown at 23°C with 16 hour photoperiod, S-sodium chloride treated plants subjected to 100 mM sodium chloride in the medium, E-etiolation, where plants were germinated in light for three days then transferred to total darkness for four days, C-cold, where plants were germinated under normal conditions for three days and then transferred to a cold room at 6°C for four days.
Having established that overall subunit A transcript levels were responsive to environmental stress conditions an assay was designed to examine the response of the four individual lengthed transcripts to the same stress conditions. The goal was to determine if any of the four were differentially expressed in response to stress. A PCR based approach was taken to evaluate transcript levels for the four different subunit A mRNAs. Primer pairs were designed (Table 1) that would only amplify one of the four transcripts. This was accomplished by anchoring the downstream primer in the poly (A) tail. The upstream primer was located in the coding region of the subunit A cDNA. Anchoring the downstream primer in the poly (A) tail prevented that oligonucleotide from priming at any other site since 10 out of 18 nucleotides were T's at the 5' end. The primer pairs were designed such that the four amplification products would differ in size by approximately 100 base pairs for ease of visualization on agarose gels and blots (Fig. 5 and Fig. 6). Primer pair T-11 and T-12 corresponds to transcript-1 and produce an amplification product of 579 base pairs. Primer pair T-21 and T-22 corresponds to transcript-2 and amplify a band of 455 base pairs. Primers T-31 and T-32 correspond to transcript-3 and amplify a 362 base pair product. Lastly, primer pair T-41 and T-42 corresponds to transcript-4 and produce a 238 base pair amplification product. The transcript-4 amplification product, which is the smallest and nested within the other three, acted as probe to detect all four transcripts. This nesting was accomplished to avoid differences in signal during hybridization based on different probe lengths and labeling efficiency.
Table 1.
Polymerase Chain Reaction primers. Primers SM-2, SM-3 and VA-1, VA-9 were utilized in the generation of digoxigenin labeled probes for hybridization. Primers T-11 through T-42 were utilized to amplify subunit A transcripts based on the position of the poly A tail.
| Primer | 5' to 3' sequence | Amplification product | 5' nucleotide position in cDNA |
| SM-2 | CCTGGTGCATTTGGTTGTGGAAA | Subunit A cDNA (300 bp probe) | 779 |
| SM-3 | CATCATACTAACATTGTAACCCAT | Subunit A cDNA (300 bp probe) | 1055 |
| VA-1 | TCTTCGAACACACAAGCCACTTT | Subunit A cDNA (1700 bp probe) | 274 |
| VA-9 | AGAGACCATGAATGAAACCCTCT | Subunit A cDNA (1700 bp probe) | 1959 |
| T-11 | TTTTTTTTTTACAAAATG | Subunit A, 3' end transcript-1 | 2264 |
| T-12 | TGAGAGAGCAGCTGGAAT | Subunit A, 3' end transcript-1 | 1702 |
| T-21 | TTTTTTTTTTATCCCCAA | Subunit A, 3' end transcript-2 | 2227 |
| T-22 | TCAGAAGTTCGAAGACCC | Subunit A, 3' end transcript-2 | 1789 |
| T-31 | TTTTTTTTTTGGAGATAA | Subunit A, 3' transcript-3 | 2210 |
| T-32 | CTGGATTCCGTGCTTTGG | Subunit A, 3' transcript-3 | 1866 |
| T-41 | TTTTTTTTTTCAAATCAA | Subunit A, 3' transcript-4 | 2173 |
| T-42 | GGAGAAAGAGGGTTTCAT | Subunit A, 3' transcript-4 | 1952 |
Figure 5.

Subunit A 3' end transcript control amplification. Whole RNA was extracted from seedlings and cDNA was generated as described in materials and methods. Polymerase Chain Reaction amplifications were performed with the appropriate primer pairs. Amplification products were fractionated by agarose gel electrophoresis, transferred to solid support and hybridized with a digoxigenin labeled probe corresponding to the transcript-4 amplification product. All four transcripts were successfully amplified. Bands produced from the PCR reaction were all of the appropriate, expected size. In addition, the transcript-4 probe successfully detected all four amplification products.
Figure 6.
Overlapping nature of the four transcript amplification products. Transcript-4 is the smallest amplification product nested within the other three to be used as a probe to equally detect all four transcripts. Downstream primers are anchored in the poly (A) tail of the message by a series of ten T residues. Thus only the first 8 nucleotides of each downstream primer correspond to nucleotides in the 3' end of the subunit A gene. Primer pair T-11 and T-12 amplifies a 579 base pair fragment corresponding to transcript-1. Primer pair T-21 and T-22 amplifies a 455 base pair fragment corresponding to transcript-2. Primer pair T-31 and T-32 amplifies a 362 base pair fragment corresponding to transcript-3. Primer pair T-41 and T-42 amplifies a 238 base pair fragment corresponding to transcript-4.
A control experiment was performed prior to the actual stress analysis to ensure that the primer pairs amplified the correct transcript only and that the transcript-4 probe would detect all four amplification products. Whole RNA was extracted from seedlings and cDNA was generated using oligo dT as described in materials and methods. Subunit A transcripts were amplified from the generated cDNA with the appropriate primer pairs and transferred to solid support. Hybridization was performed at high stringency with the transcript-4 amplification product as probe. The results of this experiment indicated that all four primer pairs amplified only a single band of the appropriate and expected size and that the transcript-4 probe adequately hybridized to all four amplification products (Fig. 5).
The differential expression of subunit A transcripts was evaluated in response to salt, cold stress, and etiolation. Complementary DNA's were generated from seven-day-old seedlings subjected to individual stress conditions. Transcripts 1–4 were evaluated via RT-PCR using transcript specific primers. Amplification products were immobilized on solid support and hybridized with the transcript-4 amplification product. The results of this experiment indicate that all four transcripts behaved identically with respect to individual stress conditions (Fig. 7). All four transcripts showed approximately 2–4 fold increases in response to 100 mM sodium chloride stress compared to untreated controls. Similarly, treatment of plants at six degrees for four days resulted in a similar increase in signal for all four transcripts. Etiolation resulted in slightly higher message levels for transcripts -1, -2, and -3 compared to controls. The lesser degree of increase for transcript-3 might be due to near saturation of the signal. The latter might be due to the higher abundance of the transcript, or due to more efficient amplification of the transcript-3 primer set. Transcript-4 seems to show slightly less signal than control plants but the autoradiograph is somewhat under developed. These results are in agreement with the whole RNA stress blot analysis. Apparent quantitative differences in upregulation between transcripts shown in (Fig. 7) might be due to differential amplification efficiency of the four different primer sets and do not necessarily represent true differential upregulation of the four transcripts. The data can only be evaluated quantitatively between samples amplified with the same primer set. In summation, no significant differential regulation of the four subunit A transcripts was detected in seedlings in response to the three stress conditions evaluated.
Figure 7.
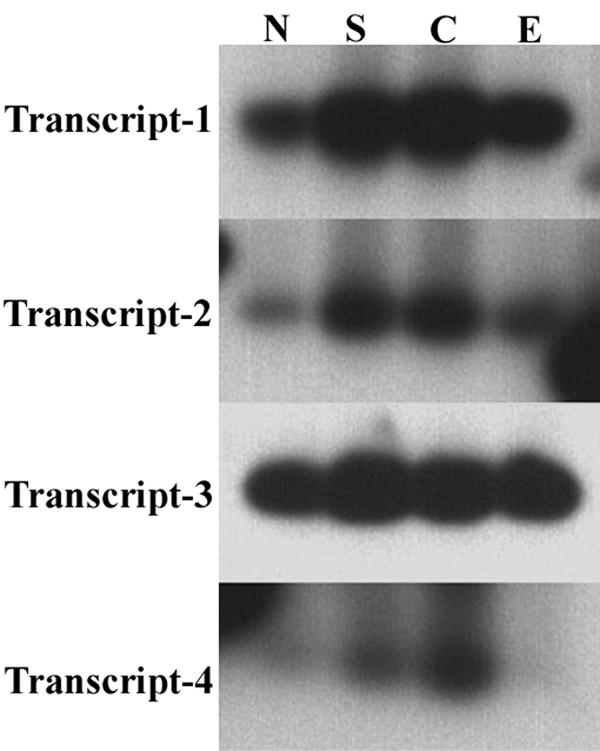
Differential expression of subunit A transcripts in response to environmental stress conditions. Whole RNA was extracted from stress treated seedlings and control plants. Complementary DNA's were generated and transcripts amplified by PCR as described in materials and methods. All sixteen amplification products were fractionated on a single agarose gel, transferred to a solid support and hybridized with a digoxigenin labeled probe corresponding to the transcript-4 amplification product. Multiple time exposures were taken and the best was used for each panel, thus this figure does not represent equivalent time exposures for all four transcripts. These results are of a single experiment which has not been replicated. Lanes correspond to the following: N-normal, untreated control plants, S-sodium chloride treated plants, C-cold, E-etiolation.
Conclusions
Evidence gathered to date indicates the presence of a single gene encoding subunit A of the vacuolar type H+-ATPase in A. thaliana. The search for additional subunit A genes was conducted using whole genome blot analysis, screening a lambda PRL-2 cDNA library, intron length polymorphism analysis, and extensive computer searches of the Arabidopsis Expressed Sequence Tag database, GenBank, and the Arabidopsis Genome Initiative. Most flowering plants evaluated to date have a small multigene family encoding subunit A of the vacuolar type H+-ATPase. The small size of the A. thaliana genome may be indicative of a trend toward streamlining of DNA in this plant, which may have eliminated a redundant subunit A gene over evolutionary time. Subunit A genes and subsequently proteins are highly conserved. Therefore, it is not likely that a subunit A gene would have escaped the screen by having a low degree of nucleotide identity.
Vacuolar type ATPases are housekeeping enzymes probably needed in all living plant cells. The single A. thaliana subunit A gene was shown by whole RNA blot analysis to be transcribed in all organs of the plant tested. The organs analyzed included roots, leaves, bolts, siliques, and flowers. In addition, relatively high levels of transcript were found in whole seedlings. These results support the notion of a single subunit A gene in A. thaliana that is transcribed in all cells of the plant.
Numerous examples are present in the literature [see [14] and references therein] of changes in transcript, protein, or activity levels of vacuolar type H+-ATPases in response to environmental stress. These results indicate a potential role for the vacuolar type H+-ATPase in maintenance of homeostasis during times of environmental stress. Whole RNA blot analysis performed on seedlings exposed to environmental stress conditions indicated that transcript levels of subunit A were responsive to salt and cold stress treatments. Transcript levels were shown to increase approximately 2–4 fold in seedlings treated with cold and salt stress. Etiolation did not produce as significant a change in transcript level as salt and cold stress.
The single vacuolar type H+-ATPase subunit A gene detected in A. thaliana can produce at least four different length transcripts by using different polyadenylation sites. These transcripts differ only in their 3' untranslated region and produce identical proteins. The presence of regulatory elements in the 3'end of the subunit A gene could be a potential mechanism by which this protein is post transcriptionally regulated.
However, the evidence gathered in this work indicates that the four different transcripts generated off the single subunit A gene are not significantly differentially regulated in response to salt, chilling, or etiolation in axenically grown seedlings. Multi site polyadenylation does not appear to be a mechanism for posttranscriptional control of gene expression for subunit A of the vacuolar type H+-ATPase in A. thaliana for the stress conditions and developmental stage tested.
Materials and methods
Plant materials
Arabidopsis thaliana ecotype Columbia seeds were obtained from the Arabidopsis Biological Resource Center, Ohio State University, Columbus OH. Seeds were germinated in 4-inch pots according to standard protocols. Axenic seedlings, and soil grown plants were maintained in a growth chamber at 23°C under a 16-hour photoperiod until appropriate organs were mature. Axenic seedling cultures were prepared by placing surface sterilized seeds on 90 mm germination plates containing 1% agar and 4.4 g/L MS salts with minimal organics (Sigma Chemical Co. St. Louis, MO), pH 6.0. Seedlings were harvested when 7 days old. Axenic roots were obtained by plating surface sterilized seeds on Nunc square Bio-Assay dishes (Fisher Scientific, Pittsburgh, PA). Plates were allowed to dry under a laminar flow hood, sealed with paraflim and incubated vertically as above. Roots were harvested 7–14 days later.
Growth effects of salt stress were evaluated by germinating surface sterilized seeds on agar containing various concentrations of sodium chloride. The optimal concentration chosen to evaluate the effects of salt stress on V-ATPase subunit A transcript levels was 100 mM. Higher concentrations resulted in failure to germinate or extremely poor growth. At 100 mM salt most seeds germinated but required more time to do so than controls. Plantlets did grow but were less robust than non-treated controls (results not shown). Seedlings were harvested after seven days of growth.
In the case of etiolation stress, surface sterilized seeds were germinated on agar for three days under light and temperature conditions described above. After three days the plates were placed in a light sealed box at the same temperature conditions. Seedlings were harvested four days later. Plantlets exhibited the typical characteristics of etiolation, the hypocotyls were lengthened and the cotyledons reduced compared to light grown controls.
Cold stress was evaluated by germinating surface sterilized seeds as described for three days under normal light and temperature. Agar plates containing seedlings were then transferred to a cold room at 6°C. Seedlings were maintained at this temperature for four days before being harvested.
Isolation of nucleic acids
Genomic DNA was extracted from mature leaves using a miniprep protocol [74] modified by the addition of a CTAB extraction [75]. A. thaliana leaves were harvested, hand washed and then ground to a fine powder in liquid nitrogen. Ground material was used immediately for DNA extraction or stored at -80°C. Post extraction samples were treated with RNAse A at 37°C for 15 minutes to remove contaminating RNA.
Organ specific whole RNA extraction was performed as described [75]. Specific organs were harvested and immediately ground to a fine powder in liquid nitrogen. Special care was taken in the case of siliques and flowers to harvest the organs directly into liquid nitrogen to prevent degradation of the RNA. Standard precautions were taken to eliminate ribonuclease contamination in solutions and glassware including baking all glassware at 200°C for six hours, and treating all solutions (except those containing Tris) with 0.1% diethylpyrocarbonate for 12 hours followed by autoclaving.
Nucleic acids were quantitated by spectrophotometry and via inspection of ethidium bromide stained agarose gels.
Genome blot analysis
Genomic DNA (5–10 μg) was digested with restriction enzymes according to the manufacturers recommendations. Digested DNA was size fractionated via gel electrophoresis in 12 cm gels containing 0.8% agarose run in IX TAE or TBE at 25 volts for 12 hours. Fractionated DNA in gels was depurinated, denatured, and neutralized according to standard protocols [76]. DNA was transferred to positively charged nylon membranes (Hybond N+) (Amersham Pharmacia Biotech, Inc., Piscataway, NJ) by the wick transfer method using 20X SSC overnight. Membranes were baked at 80°C for 1.5 hours.
Hybridization was performed according to a procedure from [77] modified as described below. Membranes were first washed for 5 minutes in 5X SSC at room temperature followed by 1 hour at 65°C in 1X SSC/0.1% SDS. Membranes were then pre-wet for 10 minutes in wash buffer (20 mM Na2HPO4 pH-7.2, 1 mM EDTA, 1% SDS). Prehybridization was performed at 67°C for two hours in a seal-a-meal bag (Dazey Corporation, Industrial Airport, KS) with hybridization solution (0.25 M Na2HPO4 pH-7.2, 1 mM EDTA, 20% SDS with 0.5% blocking reagent-Boehringer Mannheim, Biochemical Products, Indianapolis IN) at a volume of 125 μl/sq. cm. of membrane and with formamide added to a final concentration of 25% and 15 μg/ml boiled salmon sperm DNA. Hybridization was performed overnight at 67°C in a sealed bag with 125 μl/sq cm fresh hybridization solution containing formamide and salmon sperm DNA as above with the addition of 40 ng/ml digoxigenin labeled probe denatured by boiling for 5 minutes. The probe utilized was a 300 base pair digoxigenin labeled PCR product.
Membranes were washed after hybridization four times at 65°C for 30 minutes each with vigorous agitation in wash buffer. Membranes were then soaked for 10 minutes in maleate buffer (0.1 M maleic acid, 3 M NaCl, 0.3% Tween 20, pH 8) at room temperature followed by blocking for two hours at room temperature in maleate buffer with 0.5% blocking reagent (Boehringer Mannheim, Biochemical Products, Indianapolis IN). Blocked membranes were sealed in a bag with 125 μl/sq. cm. of maleate buffer with 0.5% blocking reagent to which was added anti digoxigenin Fab fragment antibody (diluted 20,000:1) conjugated with alkaline phosphatase (Boehringer Mannheim, Biochemical Products, Indianapolis IN). After incubation for 30 minutes at room temperature membranes were washed in maleate buffer four times for 15 minutes each at room temperature. Next, membranes were soaked for 10 minutes in substrate buffer (100 mM TRIS, 100 mM NaCl, 50 mM MgCl2 pH 9.5) at room temperature. Incubation was then performed for ten minutes at room temperature with CDP-Star chemiluminescent substrate (Boehringer Mannheim, Biochemical Products, Indianapolis IN) diluted 1:100 in substrate buffer. Membranes were then exposed to X-ray film.
Whole RNA blot analysis
RNA was fractionated via denaturing agarose gel electrophoresis using glyoxal according to standard protocols [76]. Immediately before use the glyoxal was deionized by vortexing in the presence of mixed bed resin beads (Sigma Chemical Co. St. Louis, MO) until the pH was greater than 5. Determination of pH was made using Hydrion pH sensitive paper (Fisher Scientific, Pittsburgh, PA).
Agarose gels (0.8%, 12 cm) cast in 10 mM sodium phosphate buffer were run for 2 hours at 100 volts with the buffer recirculated by a peristaltic pump. RNA gels were blotted using the same procedure as DNA gels omitting the depurination, denaturation, and neutralization steps. In addition, hybridization and chemiluminescent immunodetection were also performed using the same protocol for genome blot analysis except for the following. Just prior to prewetting in wash buffer for prehybridization RNA membranes were given two deglyoxylation washes in 20 mM Tris pH 8 for twenty minutes each at 65°C. The probe used for whole RNA blot analysis was a 1700 base pair PCR product.
Generation of complementary DNA
Synthesis of cDNA was accomplished using MMLV reverse transcriptase (Gibco BRL Life Technologies, Bethesda, MA) and oligo dT according to the manufacturers instructions. Whole RNA was diluted to a concentration of 0.67 μg/μl in a total volume of 10–15 μl. The sample was heated at 90°C for 5 minutes and chilled on ice. The reverse transcription reaction mixture included the following components in a total volume of 20 μl. Nucleotides (dNTP's) (Amersham Pharmacia Biotech, Piscataway, NJ) were added to a final concentration of 0.5 mM. Forty units of a ribonuclease inhibitor (RNAsin) (Promega, Madison, WI) were added. Denatured, whole RNA (2 μg) was added in a volume of 3 μl. Oligo dT primer (1.1 μl) was added to a final concentration of 1.5 μM. Dithiothreitol was added to a final concentration of 10 mM. Dilution buffer (5X concentration, provided by the manufacturer) was added to a final concentration of 1X. After all the components were assembled, 200 units of MMLV reverse transcriptase (Gibco BRL Life Technologies, Bethesda, MA) were added. The sample was incubated at 42°C for 1 hour followed by heat denaturation of the enzyme at 95°C for 10 minutes. Samples were stored at -20°C.
Polymerase Chain Reaction amplifications
In general, PCR reactions were performed in a volume of 50 μl with 50 picomoles of each primer, dNTP's (Amersham Pharmacia Biotech, Inc., Piscataway, NJ) at a final concentration of 0.5 mM, 1 μl of template, 1X PCR buffer (10X PCR buffer contained 700 mM Tris pH 8.8, 20 mM MgCl2, 1% Triton X-100, and 0.1% Tween 20) and Taq polymerase (Perkin-Elmer Inc., Wellesley MA) diluted to the manufacturers recommendations. The reaction components were assembled except Taq polymerase and heated at 95°C for 5 minutes. Polymerase was added and the PCR allowed to cycle for 1 minute at 94°C, 1 minute at 55°C and 1.5 minutes at 72°C for 30 cycles followed by 10 minutes at 72°C.
All digoxigenin labeled probes for non-radioactive detection were generated using the Polymerase Chain Reaction. Probes were labeled using 10X PCR labeling mix (Boehringer Mannheim, Biochemical Products, Indianapolis IN) at a concentration of 1X replacing the normal dNTP mix. In each case 1 μl of template was added to the reaction mix. Template was created by amplifying a band off an Expressed Sequence Tag (EST G5B3T7) from the Arabidopsis expressed sequence tag database [78], fractionating the product on a low melting point agarose gel, and excising the band. The excised, diluted band was re-amplified to ensure fidelity of the PCR. After successful re-amplification the template was utilized for probe generation.
A 300 base pair cDNA probe corresponding to nucleotide residues 779–1078 of the A. thaliana vacuolar type H+-ATPase subunit A gene was generated using primers SM-2 and SM-3 (Table 1). This probe was used for whole genome blot analysis. An approximately 1700 base pair cDNA probe corresponding to nucleotide residues 274–1981 of subunit A was generated using primers VA-1 and VA-9. This probe was used for multi organ RNA blot analysis and stress response northerns. In both cases template was generated off EST G5B3T7. Cycling parameters for both probes were identical to those described above.
Probes were precipitated by adding 1 μl of tRNA (10 mg/ml) or 1/50th volume of glycogen (10 mg/ml in sterile distilled water) to the PCR reaction followed by 1/10th volume of sterile 4 M lithium chloride. Next, 3 volumes of ice-cold absolute ethanol were added and the DNA precipitated for 30 minutes at -80°C. DNA was pelleted by centrifugation at 4°C for 15 minutes. Pellets were washed with ice cold 70% ethanol, air dried for 15 minutes and resuspended in 50 μl TE buffer/0.1 % SDS. Probes were quantitated by fractionation of 1/10th volume in a 0.8% agarose gel compared to a known quantity of HindIII digested lambda DNA. Probes were stored at -20°C until use.
Four different subunit A transcripts were amplified using RT-PCR. Primer pairs were designed with one upstream coding region primer and one primer anchored in the poly (A) tail of the transcript. In all four cases template was 1 μl of seedling cDNA. Amplification reaction mixtures were assembled as described, with PCR buffer modified to contain only 1 mM magnesium chloride. Transcript 1 was amplified using primers T-11 and T-12 (Table 1). Transcript 2 was amplified using primers T-21 and T-22. Transcript 3 was amplified using primers T-31 and T-32. Transcript 4 was amplified using primers T-41 and T-42. The PCR was allowed to proceed in each case for 30 cycles of 1 minute at 94°C, 1 minute at the annealing temperature, and 1 minute at 72°C. Primers amplifying transcripts 1 and 2 were annealed at 35°C, primers amplifying transcripts 3 and 4 were annealed at 40°C. Following 30 cycles, primer extensions were completed at 72°C for 10 minutes. PCR products were then fractionated on 2.5% agarose gels.
The transcript 4 amplification product was used as template for the generation of a digoxigenin labeled probe for use in the subunit A transcript stress response experiment. The transcript 4 amplification product was purified via low melting point agarose gel electrophoresis and used as template for probe generation. The digoxigenin labeled probe was generated as described using the transcript 4 cycling parameters and primers T-41 and T-42.
DNA sequence of all PCR primers used in this research can be found in Table 1. A diagrammatic representation of the PCR amplification products generated from transcript specific primers is shown in fig. 6.
Abbreviations
H+-ATPase-proton pumping adenosine triphosphatase, V-ATPase-vacuolar type proton pumping adenosine triphosphatase, EST-expressed sequence tag, PCR-Polymerase Chain Reaction, RT-PCR-reverse transcriptase-Polymerase Chain Reaction.
Acknowledgments
Acknowledgments
The authors wish to thank Shenell Antrobus and Alex Delcampo for their excellent technical assistance. This research was supported through NSF grant BSR 9020868 and by the University of Connecticut Research Foundation.
Contributor Information
Scot M Magnotta, Email: smagnotta@mail.hartford.edu.
Johann Peter Gogarten, Email: gogarten@uconnvm.uconn.edu.
References
- Junge W, Lill H, Engelbrecht S. ATP Synthase: An Electrochemical Transducer with Rotatory Mechanics. Trends Biochem Sci. 1997;22:420–423. doi: 10.1016/S0968-0004(97)01129-8. [DOI] [PubMed] [Google Scholar]
- Fillingame RH. Molecular Rotary Motors. Science. 1999;286:1687–1688. doi: 10.1126/science.286.5445.1687. [DOI] [PubMed] [Google Scholar]
- Fillingame RH. Coupling H+ Transport and ATP Synthesis in F1F0-ATP Synthases: Glimpses of Interacting Parts in a Dynamic Molecular Machine. J Exp Biol. 1997;200:217–224. doi: 10.1242/jeb.200.2.217. [DOI] [PubMed] [Google Scholar]
- Sabbert D, Engelbrecht S, Junge W. Intersubunit Rotation in Active F-ATPase. Nature. 1996;381:623–625. doi: 10.1038/381623a0. [DOI] [PubMed] [Google Scholar]
- Wang H, Oster G. Energy Transduction in the F1 Motor of ATP Synthase. Nature. 1998;396:279–282. doi: 10.1038/24409. [DOI] [PubMed] [Google Scholar]
- Elston T, Wang H, Oster G. Energy Transduction in ATP Synthase. Nature. 1998;391:510–513. doi: 10.1038/35185. [DOI] [PubMed] [Google Scholar]
- Boyer PD. The Binding Change Mechanism for ATP Synthase-Some Probabilities and Possibilities. Biochim Biophys Acta. 1993;1140:215–250. doi: 10.1016/0005-2728(93)90063-L. [DOI] [PubMed] [Google Scholar]
- Yasuda R, Noji H, Kinosita K, Yoshida M. F1-ATPase is a Highly Efficient Molecular Motor that Rotates with Discrete 120° Steps. Cell. 1998;93:1117–1124. doi: 10.1016/s0092-8674(00)81456-7. [DOI] [PubMed] [Google Scholar]
- Noji H. The Rotary Enzyme of the Cell: The Rotation of F1-ATPase. Science. 1998;282:1844–1845. doi: 10.1126/science.282.5395.1844. [DOI] [PubMed] [Google Scholar]
- Gluck SL. The Vacuolar H+-ATPases: Versatile Proton Pumps Participating in Constitutive and Specialized Functions in Eukaryotic Cells. Int Rev Cytol. 1993;137:105–137. [PubMed] [Google Scholar]
- Harvey WR, Wieczorek H. Animal Plasma Membrane Energization by Chemiosmotic H+-ATPases. J Exp Biol. 1997;200:203–216. doi: 10.1242/jeb.200.2.203. [DOI] [PubMed] [Google Scholar]
- Liittge U, Ratajczak R. The Physiology, Biochemistry, and Molecular Biology of the Plant Vacuolar ATPase. In: RA Leigh, D Sanders, editor. The Plant Vacuole, Academic Press, San Diego. 1997. pp. 253–285. [Google Scholar]
- Marty F. Plant Vacuoles. Plant Cell. 1999;11:587–599. doi: 10.1105/tpc.11.4.587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkla BJ, Pantoja O. Physiology of Ion Transport across the Tonoplast of Higher Plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:159–184. doi: 10.1146/annurev.arplant.47.1.159. [DOI] [PubMed] [Google Scholar]
- Taiz L. The Plant Vacuole. J Exp Biol. 1992;172:113–122. doi: 10.1242/jeb.172.1.113. [DOI] [PubMed] [Google Scholar]
- Martinoia E, Ratajczak R. Transport of Organic Molecules Across the Tonoplast. In: RA Leigh, D Sanders, editor. The Plant Vacuole, Academic Press, San Diego. 1997. pp. 366–390. [Google Scholar]
- Bowman BJ, Bowman EJ. Mitochondrial and Vacuolar ATPases. The Mycota III, Biochemistry and Molecular Biology. 1996. pp. 57–83.
- Forgac M. Structure and Function of Vacuolar Class of ATP-Driven Proton Pumps. Physiological Reviews. 1989;69:765–796. doi: 10.1152/physrev.1989.69.3.765. [DOI] [PubMed] [Google Scholar]
- Randall SK, Sze H. Probing the Catalytic Subunit of the Tonoplast H+-ATPase from Oat Roots. J Biol Chem. 1987;262:7135–7141. [PubMed] [Google Scholar]
- Feng Y, Forgac M. A Novel Mechanism for Regulation of Vacuolar Acidification. J Biol Chem. 1992;267:19769–19772. [PubMed] [Google Scholar]
- Feng Y, Forgac M. Inhibition of Vacuolar H+-ATPase by Disulfide Bond Formation between Cysteine 254 and Cysteine 532 in Subunit A. J Biol Chem. 1994;269:13224–13230. [PubMed] [Google Scholar]
- Hager A, Lanz C. Essential Sulfhydryl Groups in the Catalytic Center of the Tonoplast H+-ATPase from Coleoptiles of Zea mays L. as Demonstrated by the Biotin-Streptavidin-Peroxidase System. Planta. 1989;180:116–122. doi: 10.1007/BF02411417. [DOI] [PubMed] [Google Scholar]
- Taiz L, Nelson H, Maggert K, Morgan L, Yatabe B, Taiz SL, Rubinstein B, Nelson N. Functional Analysis of Conserved Cysteine Residues in the Catalytic Subunit of the Yeast Vacuolar H+-ATPase. Biochim Biophys Acta. 1994;1194:329–334. doi: 10.1016/0005-2736(94)90315-8. [DOI] [PubMed] [Google Scholar]
- Magnotta S, Gogarten JP. Characterization and Isolation of a Vacuolar Type H+-ATPase Subunit A cDNA from Arabidopsis thaliana. Plant Physiol. 1997;115:1730. [Google Scholar]
- Starke T, Gogarten JP. A Conserved Intron in the V-ATPase A Subunit Genes of Plants and Algae. FEBS Lett. 1993;315:252–258. doi: 10.1016/0014-5793(93)81174-X. [DOI] [PubMed] [Google Scholar]
- Gogarten JP, Fichmann J, Braun Y, Morgan L, Styles P, Taiz SL, DeLapp K, Taiz L. The Use of Antisense mRNA to Inhibit the Tonoplast H+-ATPase in Carrot. Plant Cell. 1992;4:851–864. doi: 10.1105/tpc.4.7.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins TA, Wan C-Y, Lu C-C. Ancient Origin of the Vacuolar H+-ATPase 69-kilodalton Catalytic Subunit Superfamily. Theor Appl Genet. 1994;89:514–524. doi: 10.1007/BF00225389. [DOI] [PubMed] [Google Scholar]
- Konishi K, Moritani C, Rahman MH, Kadowaki H, Ohmori S, de Groot EJ, Oesterhelt D, Ikeda M. Molecular Cloning of cDNAs encoding Acetabularia acetabulum V-type ATPase, A Subunit. Plant Physiol. 1995;109:337. doi: 10.1104/pp.109.1.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Konishi K, Kadowaki H, Moritani C, Watanabe Y. Molecular Cloning of cDNAs Encoding Acetabularia acetabulum V-type ATPase, A and B Subunits. Plant Physiol. 1996;111:651. [Google Scholar]
- DuPont FM, Tanaka CK, Hurkman WJ. Separation and Immunological Characterization of Membrane Fractions from Barley Roots. Plant Physiol. 1988;86:717–724. doi: 10.1104/pp.86.3.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkelman T, Houtchens KA, DuPont FM. Two cDNA Clones Encoding Isoforms of the B Subunit of the Vacuolar ATPase from Barley Roots. Plant Physiol. 1994;104:287–288. doi: 10.1104/pp.104.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hortensteiner S, Martinoia E, Amrhein N. Factors Affecting the Reformation of Vacuoles in Evacuolated Protoplasts and the Expression of the Two Vacuolar Proton Pumps. Planta. 1994;192:395–403. doi: 10.1007/BF00198576. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Umami K, Rahman MH, Moritani C, Watanabe Y. Primary Structures of Four Additional cDNA Clones Coding for Acetabularia acetabulum V-type ATPase, Proteolipid Subunit. Plant Physiol. 1997;115:864. [Google Scholar]
- Lai S, Watson JC, Hansen JN, Sze H. Molecular Cloning and Sequencing of cDNAs Encoding the Proteolipid Subunit of the Vacuolar H+-ATPase from a Higher Plant. J Biol Chem. 1991;266:16078–16084. [PubMed] [Google Scholar]
- Hasenfratz M-P, Tsou C-L, Wilkins T. Expression of Two Related Vacuolar H+-ATPase 16-Kilodalton Proteolipid Genes is Differentially Regulated in a Tissue-Specific Manner. Plant Physiol. 1995;108:1395–1404. doi: 10.1104/pp.108.4.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perera IY, Li X, Sze H. Several Distinct Genes Encode Nearly Identical 16 kDa Proteolipids of the Vacuolar H+-ATPase from Arabidopsis thaliana. Plant Mol Biol. 1995;29:227–244. doi: 10.1007/BF00043648. [DOI] [PubMed] [Google Scholar]
- Hirata R, Graham LA, Takatsuki A, Stevens TH, Anraku Y. VMA11 and VMA16 Encode Second and Third Proteolipid Subunits of the Saccharomyces cerevisiae Vacuolar Membrane H+-ATPase. J Biol Chem. 1997;272:4795–4803. doi: 10.1074/jbc.272.17.11344. [DOI] [PubMed] [Google Scholar]
- Kawamura Y, Arakawa K, Maeshima M, Yoshida S. Tissue Specificity of E Subunit Isoforms of Plant Vacuolar H+-ATPase and Existence of Isotype Enzymes. J Biol Chem. 2000;275:6515–6522. doi: 10.1074/jbc.275.9.6515. [DOI] [PubMed] [Google Scholar]
- Bartkiewicz M, Hernando H, Reddy SV, Roodman GD, Baron R. Characterization of the Osteoclast Vacuolar H+-ATPase B-subunit. Gene. 1995;160:157–164. doi: 10.1016/0378-1119(95)00228-X. [DOI] [PubMed] [Google Scholar]
- Herman EM, Li X, Su RT, Larsen P, Hsu H-T, Sze H. Vacuolar-Type H+-ATPases are Associated with the Endoplasmic Reticulum and Provacuoles of Root Tip Cells. Plant Physiol. 1994;106:1313–1324. doi: 10.1104/pp.106.4.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando N, Bartkiewicz M, Collin-Osdoby P, Osdoby P. Alternative Splicing Generates a Second Isoform of the Catalytic A Subunit of the Vacuolar H+-ATPase. Proc Natl Acad Sci USA. 1995;92:6087–6091. doi: 10.1073/pnas.92.13.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson N. Evolution of Organellar Proton-ATPases. Biochim Biophys Acta. 1992;1100:109–124. doi: 10.1016/0005-2728(92)90072-a. [DOI] [PubMed] [Google Scholar]
- Nelson RD, Guo X-L, Masood K, Brown D, Kalkbrenner M, Gluck S. Selectively Amplified Expression of an Isoform of the Vacuolar H+-ATPase 56 Kilodalton Subunit in Renal Intercalated Cells. Proc Natl Acad Sci USA. 1992;89:3541–3545. doi: 10.1073/pnas.89.8.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley D, Taiz L. Immunological Localization of the Vacuolar H+-ATPase in Maize Root Tip Cells. Plant Physiol. 1989;89:391–395. doi: 10.1104/pp.89.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolson MF, Wu B, Proteau D, Taillon BE, Roberts BT, Hoyt MA, Jones EW. STV1 Gene Encodes Functional Homologue of 95-kDa Yeast Vacuolar H+-ATPase Subunit Vph1p. J Biol Chem. 1994;269:14064–14074. [PubMed] [Google Scholar]
- Moriyama Y, Nelson N. H+-translocating ATPase in Golgi Apparatus. J Biol Chem. 1989;264:18445–18450. [PubMed] [Google Scholar]
- Puopolo K, Kumamoto C, Adachi I, Magner R, Forgac M. Differential Expression of the "B" Subunit of the Vacuolar H+-ATPase in Bovine Tissues. J Biol Chem. 1992;267:3696–3706. [PubMed] [Google Scholar]
- Zhen RG, Kim EJ, Rea PA. The Molecular and Biochemical Basis of Pyrophosphate-Energized Proton Translocation at the Vacuolar Membrane. In: RA Leigh, D Sanders, editor. The Plant Vacuole, Academic Press, San Diego, pp. 297–337. [Google Scholar]
- Gaxiola RA, Rao R, Sherman A, Grisafi P, Alper SL, Fink GR. The Arabidopsis thaliana Proton Transporters AtNhx1 and Avp1, can Function in Cation Detoxification in Yeast. Proc Natl Acad Sci USA. 1999;96:1480–1485. doi: 10.1073/pnas.96.4.1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt Tolerance Conferred by Overexpression of a Vacuolar Na+/H+ Antiport in Arabidopsis. Science. 1999;285:1256–1258. doi: 10.1126/science.285.5431.1256. [DOI] [PubMed] [Google Scholar]
- Orr W, White TC, Iu B, Robert L, Singh J. Characterization of a Low Temperature Induced cDNA from Winter Brassica napus Encoding the 70 kDa Subunit of Tonoplast ATPase. Plant Mol Biol. 1995;28:943–948. doi: 10.1007/BF00042078. [DOI] [PubMed] [Google Scholar]
- Darley CP, Davies JM, Sanders D. Chill-Induced Changes in the Activity and Abundance of the Vacuolar Proton-Pumping Pyrophosphatase from Mung Bean Hypocotyls. Plant Physiol. 1995;109:659–665. doi: 10.1104/pp.109.2.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carystinos GD, MacDonald HR, Monroy AF, Dhindsa RS, Poole RJ. Vacuolar H+-Translocating Pyrophosphatase is Induced by Anoxia or Chilling in Seedlings of Rice. Plant Physiol. 1995;108:641–649. doi: 10.1104/pp.108.2.641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löw R, Rockel B, Kirsch M, Ratajczak R, Hörtensteiner S, Martinoia E, Liittge U, Rausch T. Early Salt Stress Effects on the Differential Expression of Vacuolar H+-ATPase Genes in Roots and Leaves of Mesembryanthemum crystallinum. Plant Physiol. 1996;110:259–265. doi: 10.1104/pp.110.1.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiantis MS, Bartholomew DM, Smith JAC. Salt Regulation of Transcript Levels for the c Subunit of a Leaf Vacuolar H+-ATPase in the Halophyte Mesembryanthemum crystallinum. Plant J. 1996;9:729–736. doi: 10.1046/j.1365-313X.1996.9050729.x. [DOI] [PubMed] [Google Scholar]
- Golldack D, Dietz KJ. Salt-induced Expression of the Vacuolar H+-ATPase in the Common Ice Plant is Developmentally Controlled and Tissue Specific. Plant Physiol. 2001;125:1643–1654. doi: 10.1104/pp.125.4.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietz K-J, Rudloff S, Ageorges A, Eckerskorn C, Fischer K, Arbinger B. Subunit E of the Vacuolar H+-ATPase of Hordeum vulgare L.: cDNA Cloning, Expression and Immunological Analysis. Plant J. 1995;8:521–529. doi: 10.1046/j.1365-313X.1995.8040521.x. [DOI] [PubMed] [Google Scholar]
- Reuveni M, Bennett AB, Bressan RA, Hasegawa PM. Enhanced H+ Transport Capacity and ATP Hydrolysis Activity of the Tonoplast H+-ATPase after NaCl Adaptation. Plant Physiol. 1990;94:525–530. doi: 10.1104/pp.94.2.524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera-Estrella R, Barkla BJ, Bohnert HJ, Pantoja O. Salt Stress in Mesembryanthemum crystallinum L. Cell Suspensions Activates Adaptive Mechanisms Similar to those Observed in the Whole Plant. Planta. 1999;207:426–435. doi: 10.1007/s004250050501. [DOI] [PubMed] [Google Scholar]
- Narasimhan ML, Binzel ML, Perez-Prat E, Chen Z, Nelson DE, Singh NK, Bressan RA, Hasegawa PM. NaCl Regulation of Tonoplast ATPase 70 Kilodalton Subunit mRNA in Tobacco Cells. Plant Physiol. 1991;97:562–568. doi: 10.1104/pp.97.2.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Kanokporn T, Zeng Q, Wilkins TA, Wood AJ. Characterization of the V-type H+-ATPase in the Resurrection Plant Tortula ruralis: Accumulation and Polysomal Recruitment of the Proteolipid c Subunit in Response to Salt-stress. J Exp Bot. 2002;53:225–232. doi: 10.1093/jexbot/53.367.225. [DOI] [PubMed] [Google Scholar]
- Binzel ML, Dunlap JR. Abscisic Acid does not Mediate Nad-Induced Accumulation of 70-kDa Subunit Tonoplast H+-ATPase Message in Tomato. Planta. 1995;197:563–568. [Google Scholar]
- Lehr A, Kirsch M, Viereck R, Schiemann J, Rausch T. cDNA and Genomic Cloning of Sugar Beet V-type H+-ATPase Subunit A and c Isoforms: Evidence for Coordinate Expression during Plant Development and Coordinate Induction in Response to High Salinity. Plant Mol Biol. 1999;39:463–475. doi: 10.1023/A:1006158310891. [DOI] [PubMed] [Google Scholar]
- Kirsch M, Zhigang A, Viereck R, Löw R, Rausch T. Salt Stress Induces an Increased Expression of V-type H+-ATPase in Mature Sugar Beet Leaves. Plant Mol Biol. 1996;32:543–547. doi: 10.1007/BF00019107. [DOI] [PubMed] [Google Scholar]
- Chan M-T, Yu S-M. The 3' Untranslated Region of a Rice α-Amylase Gene Functions as a Sugar-Dependent mRNA Stability Determinant. Proc Natl Acad Sci USA. 1998;95:6543–6547. doi: 10.1073/pnas.95.11.6543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng W-H, Taliercio EW, Chourey PS. Sugars Modulate an Unusual Mode of Control of the Cell-Wall Invertase Gene (Incw1) through its 3' Untranslated Region in a Cell Suspension Culture of Maize. Proc Natl Acad Sci USA. 1999;96:10512–10517. doi: 10.1073/pnas.96.18.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnotta S. Identification and Molecular Characterization of Vacuolar Type H+-ATPase Subunit A and Subunit B isoforms in Arabidopsis thaliana. PhD Dissertation University of Connecticut, Storrs, CT, USA. 2000.
- Hilario E, Gogarten JP. The V-ATPase A Subunit Gene (vma-1) from Giardia lamblia. Biochim Biophys Acta. 1995;128:94–98. doi: 10.1016/0005-2736(95)00130-u. [DOI] [PubMed] [Google Scholar]
- Green PJ. Control of mRNA Stability in Higher Plants. Plant Physiol. 1993;102:1065–1070. doi: 10.1104/pp.102.4.1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. Posttranscriptional Regulation of Gene Expression in Plants. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:77–105. doi: 10.1146/annurev.pp.44.060193.000453. [DOI] [Google Scholar]
- Dean C, Tamaki S, Dunsmuir P, Faureau M, Katayama C, Dooner H, Bedbrook J. mRNA Transcripts of Several Plant Genes are Polyadenylated at Multiple Sites In Vivo. Nucl Acids Res. 1986;14:2229–2240. doi: 10.1093/nar/14.5.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Ueda T, Messing J. The Formation of mRNA 3'-Ends in Plants. Plant J. 1995;8:323–329. doi: 10.1046/j.1365-313X.1995.08030323.x. [DOI] [PubMed] [Google Scholar]
- Smart LB, Vojdani F, Maeshima M, Wilkins TA. Genes Involved in Osmoregulation during Turgor-Driven Cell Expansion of Developing Cotton Fibers are Differentially Regulated. Plant Physiol. 1998;116:1539–1549. doi: 10.1104/pp.116.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A Plant DNA Miniprep Protocol. Plant Mol Biol Rep. 1983;1:19–21. [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. Current Protocols in Molecular Biology. New York: Greene Publ Assoc/Wiley Inter science. 1987.
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor Press. 1989.
- Engler-Blum G, Meier M, Frank J, Müller GA. Reduction of Background Problems in Nonradioactive Northern and Southern Blot Analyses Enables Higher Sensitivity Than 32P-Based Hybridizations. Anal Biochem. 1993;210:235–244. doi: 10.1006/abio.1993.1189. [DOI] [PubMed] [Google Scholar]
- Newman T, de Bruijn FJ, Green J, Keegstra K, Kende H, McIntosh L, Ohlrogge J, Raikhel N, Sommerville S, Thomashow M, Retzel E, Sommerville C. Genes Galore: A Summary of Methods for Accessing Results from Large Scale Partial Sequencing of Anonymous Arabidopsis cDNA Clones. Plant Physiol. 1994;106:1241–1255. doi: 10.1104/pp.106.4.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]