Abstract
Objectives
Somatic mutations may predict prognosis, therapeutic response, or cancer progression. We evaluated targeted sequencing of oral rinse samples (ORS) for non-invasive mutational profiling of oral squamous cell carcinomas (OSCC).
Materials and Methods
A custom hybrid capture panel targeting 42 frequently mutated genes in OSCC was used to identify DNA sequence variants in matched ORS and fresh-frozen tumors from 120 newly-diagnosed patients. Receiver operating characteristic (ROC) curves determined the optimal variant allele fraction (VAF) cutoff for variant discrimination in ORS. Behavioral, clinical, and analytical factors were evaluated for impacts on assay performance.
Results
Half of tumors involved oral tongue (50%), and a majority were T1-T2 tumor stage (55%). Median depth of sequencing coverage was 260X for OSCC and 1,563X for ORS. Frequencies of single nucleotide variants (SNVs) at highly mutated genes (including TP53, FAT1, HRAS, NOTCH1, CDKN2A, CASP8, NFE2L2, and PIK3CA) in OSCC were highly correlated with TCGA data (R=0.96, p=2.5E-22). An ROC curve with area-under-the-curve (AUC) of 0.80 showed that, at an optimal VAF cutoff of 0.10%, ORS provided 76% sensitivity, 96% specificity, but precision of only 2.6E-4. At this VAF cutoff, 206 of 270 SNVs in OSCC were detected in matched ORS. Sensitivity varied by patient, T stage and target gene. Neither downsampled ORS as matched control nor a naïve Bayesian classifier adjusting for sequencing bias appreciably improved assay performance.
Conclusion
Targeted sequencing of ORS provides moderate assay performance for noninvasive detection of SNVs in OSCC. Our findings strongly rationalize further clinical and laboratory optimization of this assay, including strategies to improve precision.
Keywords: Oral cavity cancer, molecular genetic profiling, noninvasive assay, squamous cell carcinoma
INTRODUCTION
Approximately 355,000 new cases and 177,000 deaths from oral cavity squamous cell carcinoma (OSCC) occurred worldwide in 2018 [1]. For a majority of patients, the main course of treatment involves primary surgical resection followed by risk-based adjuvant therapy, i.e., radiation with or without chemotherapy [2–4]. Unfortunately, 5-year overall survival for early and local-regionally advanced OSCC is only 80% and 55%, respectively [5, 6]. A majority of deaths is due to cancer progression at the primary site or at regional lymph nodes.
Genetic mutation profiling of cancers including head and neck squamous cell carcinomas (HNSCC) has improved understanding of the molecular basis for cancer formation and has revealed novel candidate therapeutic targets. Comparison of next-generation sequencing of human papillomavirus (HPV)-positive vs. HPV-negative HNSCC has confirmed that they are genetically distinct diseases with regard to mutational spectra, copy number alterations and gene expression profiles [7, 8]. Profiling of OSCC, a subset of HNSCC which is mostly HPV-negative, has identified marked enrichment of mutations in tumor suppressor genes (e.g., TP53, CDKN2A, and FAT1), whereas potentially targetable activating mutations are infrequent. However, preliminary data indicate tumor TP53 mutation status may confer differential sensitivity of OSCC to adjuvant radiation therapy combined with platinum-based drugs versus taxanes [9]. Although infrequent, particular mutations in genes such as HRAS, NOTCH1, and TP53 may confer sensitivity to specific small molecule inhibitors, a subject of clinical trials in patients with recurrent disease (e.g. NCT03719690, NCT02383927) [10]. Moreover, total mutational burden may be associated with response to immunotherapy [11, 12]. Thus, genetic profiling of OSCC offers the potential to improve clinical decision making in the near future.
While genetic profiling of cancers is increasingly available, it remains expensive and requires repeated biopsies to identify mutations associated with resistance to therapies. Moreover, the tumor specimens that are studied may underrepresent cancer heterogeneity. To overcome these limitations, recent efforts have focused on targeted sequencing of tumor DNA extracted from body fluids such as saliva and blood (i.e., cell-free, fragmented circulating tumor DNA [ctDNA]). Although the presence of germline DNA in specimens may reduce assay sensitivity and specificity [13–15], differential DNA methylation and distinct fragmentation patterns in tumor versus normal tissue may improve analysis of ctDNA and facilitate increased clinical value [14, 16].
We recently reported that HPV DNA detection in oral rinse samples (ORS) from patients with oral cavity or oropharyngeal cancers has moderate sensitivity (80–84%) and high specificity (88–100%) for the diagnosis of HPV-positive HNSCC [17]. Persistence of oral HPV DNA after completion of primary therapy was associated with high rates of local-regional recurrence and poor overall survival [17]. Similarly, in additional studies, detection of persistent cell-free HPV DNA (cfDNA) in plasma or serum identified patients at high risk for recurrence, and demonstrated considerable promise for post-treatment surveillance of recurrent disease [18–20]. Thus, HPV DNA persistence may identify patients with minimal residual disease (MRD) who may benefit from additional, adjuvant therapy.
This ability to detect cancer cells shed from the oropharynx in ORS as a surrogate for biopsies suggests even greater potential for noninvasive molecular profiling of OSCC, since the oral cavity is anatomically more accessible. However, OSCCs are almost always HPV-negative, so tumor-associated somatic variant detection is required, rather than monitoring of virus sequences. Wang and colleagues provided preliminary data for genetic profiling of HNSCC by analyzing tumor DNA extracted from saliva or plasma [21]. Here we describe results of a genomics study designed to investigate the use of targeted sequencing of ORS DNA for mutational profiling of OSCC. As part of this investigation, we probed clinical and analytical factors that affect assay performance.
MATERIALS AND METHODS
Study population
Patients with newly diagnosed OSCC at the Ohio State University Medical Center from July 2011 to December 2014 provided written informed consent to participate in our prospective cohort study to evaluate molecular profiling of ORS DNA [8]. ORS were collected at time of diagnosis by means of a 30 second oral rinse and gargle with normal saline. Fresh-frozen tumor samples were collected at the time of primary surgical resection. Demographic and behavioral risk-factor profiles were collected by use of audio computer-assisted self-interview [22]. TNM pathologic stage was determined from the surgical pathology report according to the AJCC 7th edition. Analysis was restricted to enrolled patients with available matched ORS and fresh-frozen OSCC sample pairs (n = 120) that had adequate depth of sequencing coverage (n= 118). The protocol was approved by the institutional review boards of Ohio State University and the University of Texas MD Anderson Cancer Center.
Specimen Processing
Tumor samples were snap-frozen in liquid nitrogen within 30 minutes of resection and then stored at −80°C. Frozen tissues were submerged in O.C.T. compound (TissueTek) and sectioned using a cryostat at 10-micron thickness. One of every 10 sections was assessed for tumor content using hematoxylin and eosin stained slides with light microscopy. Macro-dissection was used to ensure >70% cancer cell content. Frozen curls were digested in proteinase K overnight, followed by DNA extraction with phenol-chloroform-isoamyl alcohol (25:24:1) precipitation or RNA extraction using Trizol (Invitrogen). Purified DNA and RNA were stored at −80°C.
ORS samples were centrifuged, and resulting cell pellets were washed twice in phosphate-buffered saline and resuspended into two equal aliquots for storage at −80°C. ORS DNA was purified by use of the DSP virus/pathogen Midi kit with a QIAsymphony SP (Qiagen) running the Pathogen Complex 800 protocol [23].
Tumor HPV status
To determine virus status, tumors were evaluated for both type-specific, high-risk HPV DNA and E6/E7 mRNA expression [24]. The Roche Linear Array was used to screen for 15 high-risk HPV types in purified tumor DNA [25]. In HPV-positive samples, virus copy numbers per cell were quantified using HPV type-specific real-time TaqMan PCR assays upon normalization to ERV3, a single copy human gene. Purified tumor RNA was reverse transcribed, and resulting cDNA was assayed for viral E6/E7 expression by type-specific PCR. Tumors were called HPV-positive when HPV DNA copy number ≥1 copy per cell and >3 E6/E7 transcripts per cell were detected [24].
Targeted sequencing panel design
We set out to design a custom gene panel capable of detecting at least one somatic mutation in >90% of HPV-negative OSCC samples. To identify recurrently mutated genes for inclusion in our custom Agilent SureSelect XT hybrid capture panel, we analyzed whole exome sequencing (WES) data from 335 HPV-negative OSCC cases, comprising 26 Ohio cohort samples and 309 TCGA samples. Data were downloaded from TCGA (https://www.cbioportal.org) and analyzed as previously described [8]. SNVs were called using MuTect v. 1.1.7 [26]. To focus on genes that are highly mutated, we used MutSigCV (v1.4) [27], OncoDrive-fm, OncoClust v1.0.0 [28], and DrGaP (v0.1.0) [29]. We selected genes having at least seven non-synonymous variants (>2% samples), median gene expression level > 1 FpKM, and significant adjusted p-values (i.e., q-values) as determined using at least one of these tools. A total of 42 significantly mutated genes were chosen for inclusion in our panel, including 24 genes identified from MutSig, q < 0.2; 33 genes from OncoDrive-fm, q < 0.2; 13 genes from OncoClust, q < 0.05; and 26 genes from DrGaP, q < 1E-5 (i.e., some genes were chosen based on these various criteria and on more than one software package; Supplemental Table S1). Our hybrid capture bait design included all exons from the 32 genes that were called by MutSig (q-value < 0.1) and/or OncoDrive-fm (q-value < 0.1). To minimize the baits’ footprint, only exons with coding-change mutations were included for the other 10 genes. Agilent SureDesign software (https://earray.chem.agilent.com) was used to design the target probe regions, to which we added 10-bp flanking segments. The resulting custom 42-gene panel was predicted to detect at least one mutation in 99% of OSCC [7, 8].
Sequencing of OSCC and matched ORS
Sequencing libraries were prepared from fresh-frozen OSCC and matched ORS DNA using custom Agilent SureSelect XT baits for hybrid capture using automation on an Agilent Bravo liquid handling instrument, following the manufacturer’s protocol. Sample and library concentrations were measured with a Qubit fluorimeter (Thermo-Fisher). Size distributions of DNA samples were determined with a TapeStation or Bioanalyzer (Agilent). Samples with compatible barcodes were pooled, and pairedend sequencing (2 × 150 bp) was performed on an Illumina HiSeq 4000.
Data processing and somatic variant calling
Sequence reads were aligned against the hg19 (GRCh37) human reference genome using BWA (version 0.7.9a) [30]. Duplicate reads with the same start and stop alignment coordinates were removed using Picard tools for deduping (http://broadinstitute.github.io/picard). A matched ORS and OSCC pair was considered evaluable if the depth of sequencing coverage was >300x (ORS) and >150x (OSCC). All potential sequence variants were identified by comparison with the reference genome using the VARSCAN “mpileup2cns” function with parameter “--min-reads2=1 –p-value=1 –min-var-freq=0” and were annotated with ANNOVAR version 521 [31]. To maximize sensitivity of variant detection, we assessed various minimum thresholds of read counts, thereby allowing detection of variants each supported by at least one, two or five alternative reads.
Matched normal tissue controls (e.g., blood leukocytes), in which only germline sequence variants would be expected, were not included in this study. Therefore, from the set of all sequence variants detected in OSCC samples, we first identified germline variants by screening population variation databases, including dbSNP138 [32, 33], 1000 Genomes Phase3 v5 [34], and gnomAD v2.1.1 [35]; variants tabulated at any frequency > 0 in these databases were called germline variants. In addition, SNVs in OSCC samples with variant allele fraction (VAF) greater than 80% were considered germline variants.
After filtering these germline variants, remaining variants supported by one or more independent sequencing read(s) (Fig. 2) were considered as somatic variant candidates, which in turn were filtered further by removing OSCC somatic candidates with VAF < 10%. Synonymous somatic variants were also removed. The remaining somatic variants identified in fresh-frozen OSCC were taken as a “gold standard” for comparison with ORS variants.
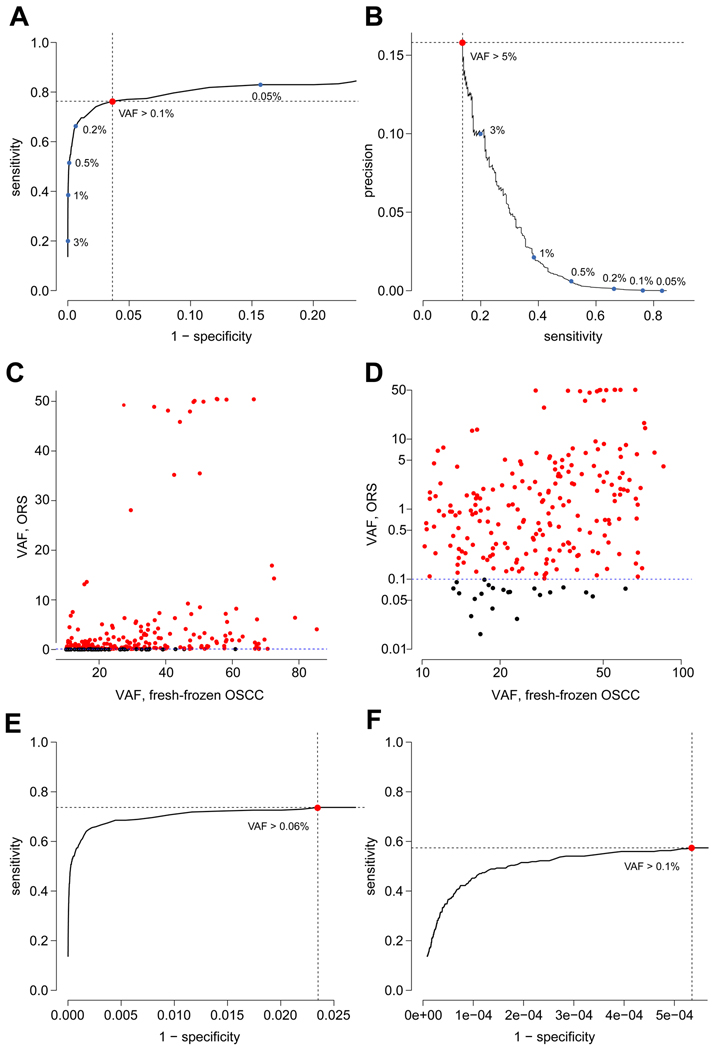
Figure 2. Optimizing the detection of variants in ORS as a surrogate for OSCC.
(A) ROC curve for somatic mutation detection in ORS, based on a cutoff of a minimum of one sequencing read to call each single nucleotide variant (SNV), with AUC of 0.795. Red dot, optimal sensitivity and specificity were determined using the Youden Index, revealing a minimal VAF cutoff of 0.1% with sensitivity, 76.3%; specificity, 96.4%; and precision, 2.6E-4. Blue dots, other points along the ROC curve with suboptimal VAF cutoffs as indicated. (B) Precision-recall curve for variant detection in ORS shows that (red dot) the optimal precision of 0.158 is reached at sensitivity of 0.137 when VAF cutoff is 5.0%. Blue dots, other points along the PR curve with corresponding VAF cutoffs as indicated. (C, D) Scatter plots show association between VAFs of non-synonymous somatic mutations in ORS (based on a minimum of one sequencing read to call each SNV; panel A) vs. in matched, fresh-frozen OSCC samples, plotted as percentages on (C) linear and (D) log scales. Of the 270 non-synonymous variants (dots) in OSCC, 206 were identified in ORS as “true positives” (red dots); others were “false negatives” (black dots). The Spearman correlation coefficient between SNV VAFs in ORS vs. in OSCC was 0.41, p=1.2E-12. (E) An additional ROC curve, based on ≥2 sequencing reads required per SNV, with AUC of 0.703, reveals an optimized VAF cutoff of 0.06%; sensitivity, 73.7%; specificity, 97.6%; and precision, 3.9E-4. (F) An additional ROC curve, based on ≥5 sequencing reads required per SNV, with AUC of 0.499, reveals an optimized VAF cutoff of 0.11%; sensitivity, 57.4%; specificity, 99.9%; and precision, 0.0133.
TCGA somatic variants, called previously from WES data with VAF > approximately 5% (https://www.cbioportal.org) [8], were restricted to the same genomic sequences represented in our custom hybrid capture panel (Supp. Table S1).
Clinical and genomic sensitivity analysis
We conducted genetic profiling of ORS to optimize utility as a surrogate to detect somatic variants in paired OSCC. To calculate sensitivity, specificity, and precision, ORS variants that passed filtering were considered candidate somatic variants and were compared to true-positive somatic variants identified in matched fresh-frozen OSCC. True-negative somatic variants were called at each reference nucleotide in the targeted genomic region (n = 182,135 nucleotides captured) and in each sample (n = 118) for which no alternative alleles were identified at or above the VAF cutoff. The optimal VAF cutoff for ORS somatic variant detection was determined by maximizing the sensitivity and specificity of the receiver operating characteristics (ROC) curve based on the Youden index, calculated as specificity+sensitivity-1 [36]. The area under the curve (AUC) also was calculated.
We used two alternative analytical approaches to call somatic variants. In the first approach, we identified germline variants upon downsampling of ORS variants. In the second, we developed a naïve Bayesian classifier. Both approaches are described in detail in Supplemental Information. Clinical characteristics were grouped into categorical variables to test their associations with sensitivities of somatic variant detection in the ORS, using one-way ANOVA. Statistical significance was defined at p < 0.05.
RESULTS
Study population
The study population consisted of 120 OSCC patients from whom matched ORS and fresh-frozen OSCC tumor were collected. A summary of patient characteristics is shown in Table 1 (N = 118, after filtering for adequate sample pair sequencing depth of coverage). Median age was 61 years (range, 19–89), and 56% were male. Oral tongue was the most common primary site. Tumor stage as per the AJCC 7th edition was distributed evenly between T1-T2 and T3-T4. A majority (60%) of patients had cervical nodal metastases. Approximately one third of patients were never or light smokers, or never regular users of alcohol, respectively. Five (4.2%) of 118 tumors were HPV-positive.
Table 1.
Clinical features of 118 patients analyzed after filtering for adequate read counts in sample pairs. NA, not available. Anatomic subsites are annotated per sample in Supp. Fig. S6.
| N=118 | |
|---|---|
| age, median (range), years | 61 (19–89) |
| gender, N (%) | |
| male | 66 (56) |
| female | 52 (44) |
| primary site, N (%) | |
| alveolar process | 2 (1.7) |
| buccal mucosa | 5 (4.2) |
| floor of mouth | 16 (13.6) |
| gingiva | 1 (0.8) |
| lip | 3 (2.5) |
| mandible | 11 (9.3) |
| oral tongue | 59 (50.0) |
| palate | 4 (3.4) |
| retromolar trigone | 9 (7.6) |
| synchronous primaries | 8 (6.8) |
| tumor grade – N (%) | |
| well-differentiated | 27 (23) |
| moderately differentiated | 82 (70) |
| poorly differentiated | 9 (8) |
| T stage, N (%) | |
| T1 | 23 (19) |
| T2 | 42 (36) |
| T3 | 25 (21) |
| T4 | 28 (24) |
| N stage, N (%) | |
| N0 | 42 (36) |
| N1 | 27 (23) |
| N2 | 42 (36) |
| N3 | 1 (1) |
| NA | 6 (5) |
| smoking, N (%) | |
| never / light smoker | 41 (35) |
| former smoker | 41 (35) |
| current smoker | 26 (22) |
| NA | 10 (8) |
| alcohol consumption, N (%) | |
| never / never regular | 40 (34) |
| former regular | 35 (30) |
| current regular | 32 (27) |
| NA | 11 (9) |
Detection of somatic variants in fresh-frozen OSCC
We designed our custom Agilent SureSelect hybrid capture panel to target 42 of the most frequently mutated cancer genes in HPV-negative OSCC (i.e., capturing ~182,000 nucleotides in the target region). The panel was used to identify somatic mutations in fresh-frozen OSCC, which served as “gold standard” calls. The median number of targeted sequence reads was 1.3 million (range: 0.7–2.5 million) per tumor, with a median depth of coverage of 260x (range: 155–476x; Supp. Fig. S1A). We excluded results for HLA-B and HLA-C from further analysis because intrinsic sequence similarities across orthologous gene templates resulted in a high rate of misalignments and false variant calls. We also excluded results for KMT2C and KMT2D because of high false variant calls caused by pseudogene sequences in the reference human genome.
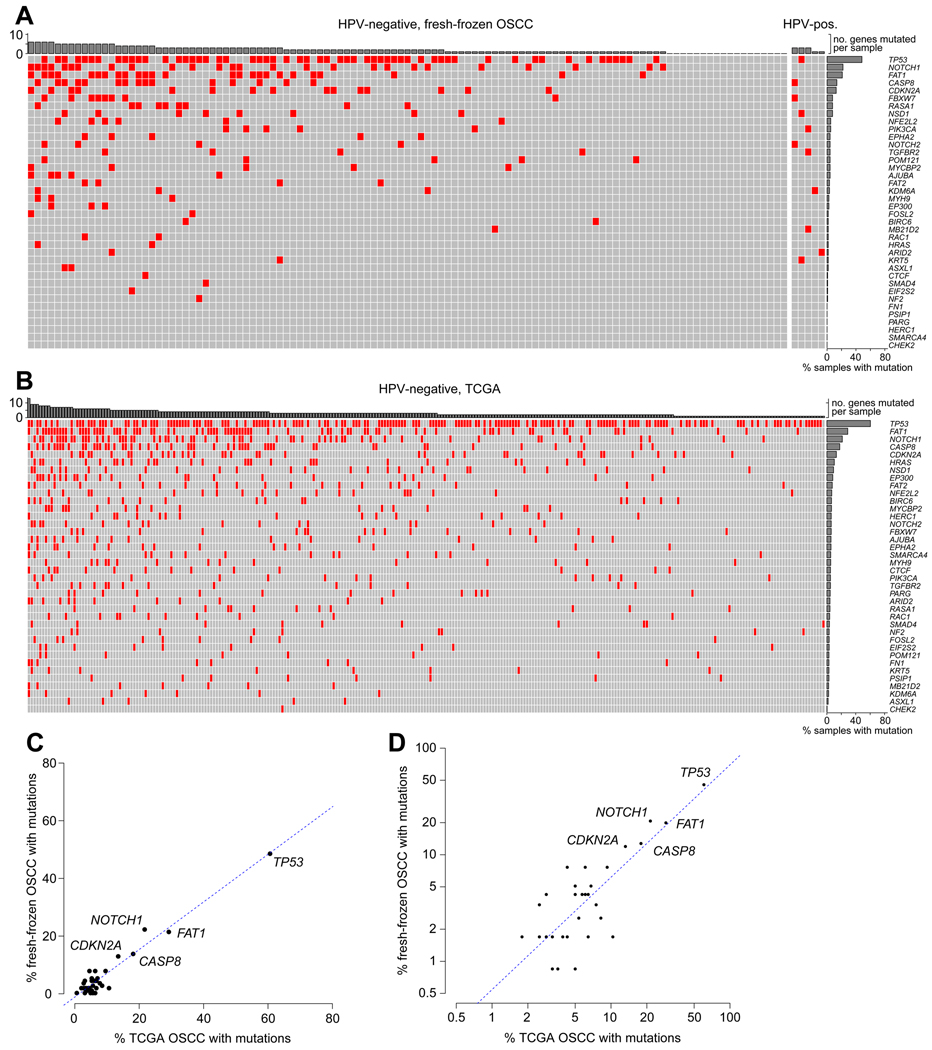
Upon alignment of sequence data against the reference human genome, a total of 1,308,131 variants were identified across 118 fresh-frozen OSCC samples. A variant was categorized as germline and thus filtered if present in population variation databases at frequency > 0% or if a SNV with a VAF > 80% (Supp. Fig. S2). Remaining variants were considered to be somatic variant candidates and were filtered further by applying a cutoff of VAF < 10% to remove poorly supported variants and minor subclones. Synonymous somatic variants also were excluded from further analysis. The remaining 270 nonsynonymous somatic variants in fresh-frozen OSCCs were taken as “gold standard” SNVs against which ORS variants could be compared. These OSCC variants included 128 nonsynonymous SNVs, 50 stop gain/loss variants, 24 splice variants, 57 frameshift insertion/deletions and 11 in-frame insertion/deletions. At least one OSCC variant was identified in 100 of 118 patients (84.7%; median 2 per patient; range 0–8) and in 32 of 38 genes (84%) analyzed (Fig. 1A, Supp. Table S2).
Figure 1. Somatic mutations detected in fresh-frozen OSCC.
(A) Pattern of somatic mutations in OSCC samples in the study population. An OncoPrint plot displays 270 non-synonymous somatic mutations (red) disrupting (y-axis) genes as detected by targeted sequencing in (x-axis) fresh-frozen OSCC assayed. At least one OSCC variant was identified in 100 of 118 patients (84.7%; median 2 per patient; range 0–8) and in 32 of 38 genes (84%) analyzed. Bar graphs, top: count of genes disrupted by at least one mutation in each sample (x-axis); right: fraction of samples each with at least one mutation in the indicated genes (y-axis). (B) Somatic mutations in WES data from 309 TCGA OSCC samples, mapped onto the same target gene panel. (C, D) Scatter plots show fraction of fresh-frozen OSCC samples harboring at least one mutation in each assayed gene (labeled or unlabeled, individual dots, n = 38) in the study population (y-axis) vs. TCGA (x-axis) cohorts, plotted on (C) linear or (D) log scales. Dotted line, linear fit model; correlation R=0.96, p=2.5E-22.
Gene mutation frequencies in the OSCC studied here were compared to those reported in TCGA samples [7]. Mutations in tumor suppressor genes were the most frequent in both cohorts (Fig. 1A, 1B). The most commonly mutated genes in our study population included TP53 (60 mutations), FAT1 (34 mutations), NOTCH1 (31 mutations), CASP8 (16 mutations), and CDKN2A (17 mutations). These rankings closely matched those of the most highly mutated genes in the TCGA cohort (Fig. 1B). A significant correlation was observed between frequencies of samples harboring mutations in targeted genes in the study population and in TCGA (R = 0.96, p=2.5E-22, n = 38 genes), plotted either on linear or log scales (Figs. 1C, 1D). We conclude that the OSCC cases in this study are representative of OSCC overall, based on TCGA data as a reference.
Detection of candidate somatic variants in ORS
To identify somatic variants in ORS, targeted sequencing libraries were prepared by hybridization of genomic DNA using the same custom bait panel used for matched OSCC samples. Normal buccal mucosal cells are expected at high abundance in ORS [37], so germline alleles would be expected to occur predominantly in heterozygosity, i.e., at relatively high VAFs of approximately 50%. ORS would be expected to include a sampling of tumor cells in addition to these normal cells shed into saliva and the oral rinses. For this reason, true positive somatic variants were anticipated at relatively low VAFs. To detect such somatic variants sensitively, we sequenced ORS more deeply than the tumor tissue. The median number of reads per ORS was 27.7 million (range, 9.3–39 million). The median depth of coverage at targeted genomic regions was 1,563x (range, 201–3,546x) (Supp. Fig. S1B). Two patients whose ORS had inadequate sequencing depth of coverage were removed from further analysis, resulting in 118 matched pairs of fresh-frozen OSCC and ORS. Non-reference variants in ORS were identified by aligning sequence reads against the hg19 reference assembly. To maximize sensitivity, even single reads alone were counted in documenting alternative alleles.
We used deep sequencing data to assay all 182,135 nucleotides in the targeted genomic regions in each of the 118 ORS samples. After removing germline and synonymous variants from the set of all variants identified in the dataset, 5,046,099 candidate somatic variants remained, each supported by at least one sequencing read from an ORS sample. A large majority had relatively low VAFs < 1% (Supp. Fig. S3). We inferred that many of these may be due to sequencing errors, given the high depth of sequencing coverage achieved per ORS (Supp. Fig. S1B). Therefore, we plotted an ROC curve to optimize a VAF cutoff so that true positive somatic variants could be distinguished from others. The ROC curve had an AUC of 0.80. This analysis found that a VAF cutoff of 0.10% provided optimal assay performance by maximizing the sum of sensitivity and specificity (Fig. 2A). Of the ORS somatic variant candidates, 4,264,269 (84.5%) had VAFs below this cutoff, so they were called negative. The remaining 781,830 variants identified from at least one read support each in ORS were counted as either true positives or false positives. Of the 270 SNVs in fresh-frozen OSCC, 206 also were detected in the matched ORS as true positives. These results yielded a sensitivity of 76% and a specificity of 96% (Fig. 2A). As expected, assay precision was low (2.6E-4), due to large numbers of false positive variant calls at many nucleotide positions across the samples compared with small numbers of true positives. A precision-recall curve showed that optimal assay precision was 0.158, but sensitivity was only 0.137 when VAF cutoff was 5.0% (Fig. 2B). In comparing VAFs of ORS and matched OSCC SNVs, the Spearman correlation coefficient was 0.41 (p-value = 1.2E-12; Fig. 2C and 2D).
We evaluated the impact on assay sensitivity and specificity by requiring additional, independent reads supporting each somatic SNV. We generated additional ROC curves based on a required minimum of two reads or of five reads supporting each SNV (Fig. 2E and 2F). The results confirmed that increasing the number of independent reads supporting each variant call improved assay specificity and marginally the precision, but at the detriment of AUC and of sensitivity. Applying a minimal requirement of 5 reads per variant was too stringent to be useful at the read depths achieved here, as shown by the corresponding ROC curve’s AUC =0.499 (Fig. 2F).
We hypothesized that the detection of true-positive variants in ORS, each with VAF > 0.10%, is associated with higher sequencing depths of coverage per variant sample when compared with the false-negative variant calls. Comparison of these distributions confirmed a significant association between sequencing depth of coverage at each variant and the distinction betwen true-positive and false-negative variants in ORS samples (p = 0.040; Supp. Fig. S4).
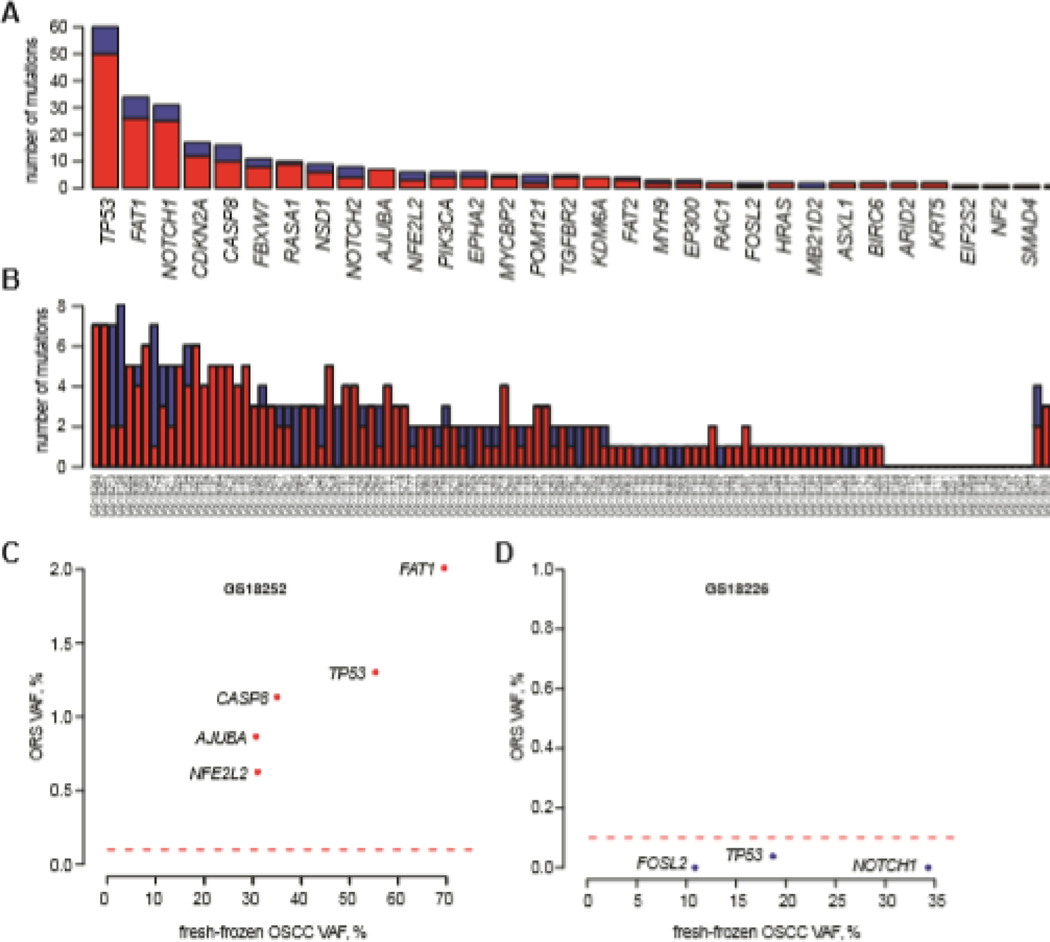
Sensitivity of mutation detection in ORS varies by gene and by patient
We sought to identify other factors that potentially could affect the sensitivity of SNV detection in ORS when compared with variants in matched OSCC. Based on an optimized VAF cutoff of 0.10%, we observed differences in ORS variant detection across the individual genes included in the custom bait panel. TP53, FAT1, NOTCH1, NOTCH2, CASP8, and CDKN2A harbored the highest numbers of detected variants. By contrast, the highest sensitivity of detection was observed for variants in HRAS, BIRC6, KDM6A, KRT5, SMAD4, EIF2S2, ARID2, and NF2 (Fig. 3A). The median sensitivity of variant detection per gene was 80% (range, 0–100%) in the 32 genes in which variants were detected in fresh-frozen OSCCs (Supp. Fig. S5A). By contrast, the specificity of variant detection per gene was uniformly high across the targeted genes (Supp. Fig. S5B). As expected, assay precision was low across the targeted genes, with the highest level of precision observed in detecting variants at TP53 (Supp. Fig. S5C).
Figure 3. Comparisons of ORS mutation detection and sensitivity across assayed genes and across samples.
(A) Comparisons of sensitivity of mutation detection in ORS samples across assayed genes. Barplot of numbers of mutations that (red) were and (blue) were not detected in the ORS samples for each gene (x-axis) found to harbor mutations in OSCC samples. Of the 32 genes mutated across the OSCC samples, mutations in 31 (97%) were identified in at least one corresponding ORS sample. (B) Comparisons of mutation detection and sensitivity across ORS samples. Barplot of numbers of mutations that (red) were and (blue) were not detected for each ORS sample (x-axis). Of the 100 OSCC samples harboring at least one mutation in the assayed genes, 89 (89%) of the paired ORS also had the same mutations. (C) Scatterplot showing VAFs of mutations in six indicated genes as detected in (y-axis) ORS vs. (x-axis) matched fresh-frozen OSCC from one patient (i.e. GS18252). Red dotted line, minimal VAF cutoff = 0.10%. These ORS variants (red dots) were considered true positives. (D) Scatterplot showing VAFs of mutations in three indicated genes as detected in ORS (y-axis) vs. matched fresh-frozen OSCC (x-axis) from another patient (i.e. GS18226). Red dotted line, minimal VAF cutoff = 0.10%. These ORS variants were considered false negatives (red dots).
Substantial variability in sensitivity per individual ORS also was observed. The median count of variants detected was 1 per sample, with variant counts ranging from 0 to 9. The mean and median sensitivities for variant detection across all patient-derived ORS samples were 64.9% and 100% (range, 0–100%), respectively (Fig. 3B). This interpatient variability is illustrated by distinct patterns of VAFs detected in two patients when comparing ORS vs. primary OSCC samples ( Figs. 3C and D). In one patient’s ORS, sensitivity was optimal because each variant exceeded the established VAF cutoff (Fig. 3C). In contrast, sensitivity was very poor in a second case because the VAFs for each somatic variant detected in the ORS sample each fell below this threshold (Fig. 3D). This inter-patient variability in sensitivity was attributable in part to differences in depths of sequencing coverage across the ORS (Supp. Fig. S1B, Supp. Fig. S4). Median depths of coverage were significantly higher for true-positives SNVs above the VAF cutoff, compared with false-negatives below that cutoff (p=0.0398; Supp. Fig. S4). As before, the specificity of variant detection was ~1 across all patients (Supp. Fig. S6). As expected, assay precision was low across almost all patients, with higher levels of precision (>0.1%) observed in only a few samples (Supp. Fig. S6).
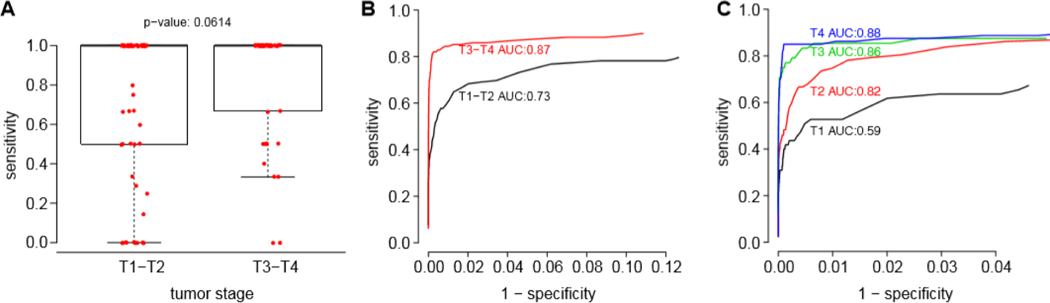
We sought to determine if various clinical characteristics also affected the sensitivity of mutation detection in ORS. Regarding tumor stage, we observed a trend for improved assay sensitivity with more advanced T stage tumors (T3, T4; p = 0.06). For T3 and T4 tumors, mean sensitivity was 84.2%, compared with 70.3% for T1-T2 stage tumors. ROC curves were generated to compare tumor stages; the AUC for T3 or T4 tumors was 0.87, versus 0.73 for T1 or T2 tumors (Fig. 4A, 4B). Moreover, the AUCs of ROC curves for each increasing tumor stage displayed progressively higher values, ranging from only 0.59 for T1 tumors to 0.88 for T4 tumors (Fig. 4C). In contrast, primary tumor site (p=0.88), nodal stage (p=0.89), smoking status (p=0.71), alcohol consumption (p=0.16), and status of recurrence (p=0.69) did not affect assay sensitivity (Supp. Fig. S7).
Figure 4. Increased sensitivity of ORS mutation detection in higher tumor stage (T3 and T4) patients.
(A) Box and whiskers plots display distribution of mutation detection sensitivity for each ORS (red dots) according to T-stage (x-axis). Heavy black line, median of distribution. (B, C) ROC curves calculated for samples with (B) T-stages T1 and T2 (black), vs. T3 and T4 (red); and for (C) individual T-stages as indicated, respectively; y-axis, sensitivity; x-axis, 1 - specificity.
Alternative analytical approaches to variant calling did not improve assay performance
We evaluated two alternative analytical approaches to determine their effects on assay performance, as detailed in Supplemental Information. In the first approach, we noted that ORS would be likely to include large numbers of normal buccal mucosal cells, which are routinely sampled for germline variant detection (particularly in the context of other cancer types). Therefore, we downsampled ORS sequence reads to identify germline variants. These were then used as an alternative reference for comparisons to call somatic SNVs in fresh-frozen OSCC. In so doing, we detected 364 nonsynonymous SNVs in the OSCC, a count higher than results from our primary analytic approach. We then recalculated the ROC curve for variant calls across ORS, showing an AUC of 0.76 (Supp. Fig. S8). At an optimized VAF cutoff of 0.10%, somatic mutation profiling of ORS had 73% sensitivity, 94% specificity and precision of 2.9E-4 (Supp. Fig. S8). When optimized separately, precision reached 9.9E-3 at a VAF cutoff of 1.4% and a sensitivity of only 19%. We concluded that overall assay performance with the downsampling approach was similar to our primary approach.
In another alternate approach, we developed naive Bayesian models to evaluate various clinical and genomic factors that could introduce biases or sequencing errors into ORS data. These included behavioral characteristics, e.g., never, former, or current tobacco smoking and alcohol consumption. Also investigated were anatomical site of the cancer, nodal status, and disease recurrence. We also evaluated genomic features such as site-specific nucleotide changes, changes in codons, and gene-specific sequence characteristics in the Bayesian models (Supp. Fig. S9A). Models involving various possible combinations of these factors did not substantially enhance assay performance in detecting somatic variants in ORS. Across all models, the sensitivity (median, 60%; average, 46.5%; range, 9–62%), specificity (median, 99.6%; average, 99.7%; range, 99.6–100%), and precision (median, 0.07%; average, 0.12%; range, 0.06–3.8%) all varied, but were not improved. Even optimized Bayesian models that each included nucleotide changes and functional amino acid changes (Supp. Fig. S9A) had ROC curves with a low AUC of only 0.58 and optimized VAF cutoff of 0.03% (Supp. Fig. S9B–C). The sensitivity, specificity, and precision of these optimized Bayesian models were only 62% 99.6%, and 6.8E-4, respectively. The relatively poor sensitivity observed with the naïve Bayesian model was attributable to false negative variant calls in ORS, improperly designated as sequencing biases by the model (Supp. Fig. S9B–C). The very poor precision from this model again was due to high numbers of false positive variants in ORS.
In sum, when comparing the sensitivity, specificity, and precision resulting from these two alternative approaches, no substantial improvements in assay performance were observed over our primary approach as described above.
DISCUSSION
In this study, we investigated the feasibility and optimization of targeted sequencing of ORS to profile somatic mutations in newly diagnosed OSCC. We considered the somatic variants identified from targeted sequencing of the fresh-frozen OSCC as a “gold standard” and used them as a basis for comparison with variants detected by targeted sequencing of ORS. Because ORS collections are noninvasive, they comprise an easily accessible, practical, and promising source of OSCC cells to detect or monitor somatic mutations in key genes.
In the absence of matched normal samples, we evaluated multiple analytical methods to define germline variants in the ORS and OSCC, which then allowed us to identify somatic mutations. Our principal approach involved comparisons of sequencing data against the reference human genome, followed by screening against large population-level variant databases. We also defined variants with VAF > 80% as germline variants. Based on these criteria, we then focused on identification of somatic mutations.
We acknowledge that inclusion of matched normal tissue would improve calls of germline and thus somatic variants in ORS. Nevertheless, we omitted inclusion of matched normal control specimens such as blood as a basis to define germline variants for several reasons. These included cost savings; evaluation of this assay under “real world” conditions comparable to other cancer genomics assays, which also omit such controls; and the fact that buccal mucosal cells are used commonly as a source of normal germline DNA. For this latter reason, we explored downsampling of the ORS variants as an alternative approach to detect germline SNVs (Supp. Fig. S8).
Based on ROC curves, we optimized the VAF cutoffs of ORS variants in order to maximize both sensitivity and specificity of their detection. The ROC curve from our main approach had an AUC of 0.80. At an optimized VAF cutoff of 0.10%, somatic mutation profiling of ORS by targeted sequencing had 76% sensitivity, 96% specificity and precision of only 2.6E-4 (Fig. 2). Under conditions requiring at least one sequencing read and VAF>0.1%, limited ability to discriminate between false-positive SNVs having low VAFs (arising from sequencing errors) vs. true-positive somatic mutations in tumors severely limited assay precision. Alternative approaches including downsampling of ORS sequence reads, to provide a surrogate of matched normal controls, and use of naïve Bayesian models, which were applied to identify and filter false-positive calls, each marginally improved precision but compromised sensitivity (Supp. Fig. S8 and S9). In addition to sequencing errors, we cannot exclude additional sources of low frequency variants in the ORS, including mutations attributable to tobacco-associated field carcinogenesis or from rare subclonal variants present in the tumor.
We observed a trend for higher assay sensitivity for patients with advanced T stage primary tumors (i.e., T3 and T4; Fig. 4). This trend is plausible, since larger, more invasive primary tumors would be expected to have more exposed surface and to shed more cancer cells into saliva and ORS, thereby making them more readily detectable by increasing somatic mutations’ VAFs. Sensitivity of mutation profiling of ORS also was significantly influenced by the depth of sequencing coverage (Supp. Fig. S1 and S3), suggesting potential effects of specimen collection quality on assay performance. We speculate that some specimens may not adequately represent tumor cells shed into saliva, perhaps due to anatomical considerations or due to poor individual subject compliance with the ORS collection protocol.
These results prompted us to consider potential methodological adjustments that may improve ORS assay performance in the future. For example, incorporation of unique molecular identifier (UMI) barcodes into sequencing libraries could facilitate improved discrimination of true somatic variants from sequencing errors [38]. The higher assay sensitivity observed at higher depth of sequencing coverage for ORS suggested that implementation of quality control metrics to require relatively high minimum numbers of sequencing reads would improve assay performance [39]. This would need to be balanced against costs of deep sequencing, although those costs continue to drop over time. Adding direct cytobrush sampling of smaller, T1 or T2 tumors would likely result in higher assay sensitivity and increase ROC AUCs toward those observed with advanced T stage tumors. Addition of methods to enrich tumor cells from saliva specimens also would improve assay performace. Modifications in bait design and/or replacement of highly variable genes (e.g., HLA, KMT2C) also may yield improvements. New sequencing methods that add DNA methylation data could enhance assay sensitivity and specificity [14, 16, 40], as performed for HPV-positive HNSCC [41]. In the absence of matched normal tissue controls, comparison of candidate variants with databases tabulating clinically relevant variants could improve identification of both germline and somatic variants in ORS. Assay precision could be increased by markedly restricting the numbers of nucleotides assessed, to focus on established nucleotides that undergo somatic mutagenesis, as tabulated in such databases, which would reduce false positive calls. Previously undetected variants could be added to a growing, comprehensive database of somatic variants, only in cases when supporting data met stringent criteria including validation. Refinements to our approach are necessary before initiating a study with training and testing sets, but together with other recent reports (Table 2), our study provides strong proof-of-principle data demonstrating that molecular profiling of OSCC by targeted sequencing of ORS is technically feasible and promising.
Table 2.
Comparison of recent studies of somatic mutation detection in head and neck cancer surrogate biospecimens.
| Author | N | Surrogate sample | Targets | Test Method |
Gold standard method | Baseline Assay Performance |
|---|---|---|---|---|---|---|
| This study | 118 ND OSCC |
ORS | 42 genes | HCS | T, 42 gene HCS | Variant-specific sensitivity 76%, specificity 0.80, precision 2.6E-4 |
| Wang, 2015 [21] | 71 ND HNSCC |
ORS or S or P | HPV16/18 or 6 genes or 1 patient-specific SNV |
ddPCR Safe-Seq |
T/N, WGS or WES in a subset | Sensitivity only: 18/21 HPV-positive positive in saliva 21/26 positive for ≥ 1 variant in saliva or comparison to gold standard. |
| Lee, 2021 [44] | 7 ND HNSCC |
S | 12–14 patient-specific SNVs |
HCS | T, WES or 277 gene HCS |
Sensitivity only: 6/7 positive for ≥ SNV |
| Galot, 2020 [45] | 39 RM HNSCC |
P | 604 genes | HCS | T/N, 604 gene HCS in a subset | Sensitivity only: 20/39 positive for ≥ 1 variant 40/209 variants detected in 18 patients tumor. |
| Shanmugam, 2021 [43] | 121 OSCC | ORS | 7 genes | HCS | T, 7 gene HCS | Sensitivity: 116/121 positive for ≥ 1 variant Lower limit of assay detection: 0.25% VAF Intra-assay agreement on SNV calls: 99% subset, 88% in ORS subset |
| Cui, 2021 [46] | 11 ND OSCC |
ORS, P | 308 genes | HCS | T/N, WES | Not reported. |
| Wu, 2021 [42] | 27 HNSCC | S, P | 1021 genes | HCS | T/N 1021 gene HCS | Sensitivity: 19/27 positive for ≥ 1 variant in saliva. |
N, number of patients; ND, newly diagnosed; RM, recurrent or metastatic; OSCC oral squamous cell carcinoma; HNSCC, head and neck squamous cell carcinoma; ORS, saliva; P, plasma; HPV, human papillomavirus; SNV, single nucleotide variant; PCR, polymerase chain reaction; ddPCR, digital droplet PCR; HCS, hybrid capture sequencing; matched normal blood; WES, whole exome sequencing; WGS, whole genome sequencing, AUC, area under the receiver-operating curve. Reference number indicated
Other studies investigating use of ORS or other body fluids as surrogates for OSCC have been published recently [21, 42–46] (Table 2). Here we report one of the largest collections of ORS and OSCC sample pairs studied to date. We used a relatively large and unbiased target gene panel focused on OSCC mutations for hybrid capture sequencing, as did others [42, 45, 46]. Unique to our analysis was the use of ROC curves, Bayesian analysis and downsampling to optimize sensitivity and specificity of variant-specific detection in ORS versus OSCC. Shanmugam and colleagues confirmed 88% of somatic variants detected in saliva samples by resequencing a subset of them [43]; we did not perform this step here. By contrast, our target gene panel was comparatively larger [43] (Table 2).
Analysis of biomarkers in saliva (e.g., ORS, whole or stimulated saliva), including protein, DNA, RNA, and metabolites, has been used to discriminate benign from malignant oral lesions [47]. These assays aim to facilitate oral cancer screening, but have not been recommended by the US Preventive Services Task Force due to insufficient evidence of efficacy. Given that saliva is derived in part from serum, salivary proteomics for noninvasive detection of non-oral cancers also has been explored [48]. Molecular profiling of oral pre-malignant lesions has identified mutations (e.g., in TP53) that increase risk of progression to malignancy, but the utility of ORS profiling may be limited by small precancerous lesion size [49]. Here, our intent was not to develop a screening test, but to conduct rapid, noninvasive mutational profiling of newly diagnosed OSCC to facilitate research, therapeutic decision making, detection of MRD, and molecular surveillance. As a majority of OSCC recurrences comprise local-regional failures, and as OSCC biopsies are not necessarily collected at all centers, ORS profiling at diagnosis could facilitate prospective monitoring of patient-specific somatic mutations in readily accessible body fluids including saliva and plasma. Such profiling also could facilitate design of neoantigen vaccines for adjuvant administration.
Noninvasive molecular profiling of solid tumors has demonstrated clinical utility in several settings in addition to HNSCC. Our ORS assay performance is comparable to PCR-based, noninvasive assays for detection of EGFR mutations in non-small cell lung cancers [50]. In those cancers, ctDNA profiling is used for targeted treatment selection and to determine clinical trial eligibility and mechanisms of resistance to treatment. For example, profiling has identified the EGFR T790M mutation as the principal cause of non-small cell lung cancer resistance to first and second generation EGFR tyrosine kinase inhibitors [51–53]. In colon cancer, persistence of variants in ctDNA after surgery and adjuvant chemotherapy has been associated with higher recurrence rates, shorter relapse-free survival, and shorter overall survival [54]. Higher ctDNA VAFs prior to colorectal cancer surgery have been correlated with shorter progression free-survival and overall survival [15]. Similar results have been observed for lung and breast cancers, where post-treatment blood samples positive for ctDNA were associated with shorter survival [55, 56]. These findings highlight the potential role of MRD detection, indicated by persistence of tumor DNA either in ORS or plasma, as an important early predictor of recurrence. Given this potential, further development of noninvasive assays with improved performance is ongoing, with a goal to facilitate translation to clinical decision making.
In summary, targeted sequencing of ORS for molecular profiling of OSCC has assay performance comparable to that of early PCR-based ctDNA profiling of lung cancer, and offers several important potential clinical applications. This study will inform adjustments to our methodology with potential for further improvements in assay performance.
Supplementary Material
Highlights.
Evaluation of genetic profiling of oral rinse samples as surrogates for oral cancers Assay comprised targeted sequencing of 42 genes frequently mutated in oral cancers We used ROC curves, Bayesian analysis, and downsampling to optimize assay performance Assay yielded good sensitivity and excellent specificity but poor precision Methods to improve assay performance -- especially assay precision -- were identified
ACKNOWLEDGMENTS
We thank the patients with oral cavity squamous cell cancers at Ohio State University (OSU) who enrolled in our research study; members of the Gillison and Symer laboratories and Dr. Xiayu Rao for insightful comments; TCGA for access to whole exome sequencing; and the Genomics Shared Resource at OSU Comprehensive Cancer Center (OSUCCC), supported by NCI CCSG grant P30CA016058. This study was funded by the Oral Cancer Foundation (M.L.G.); OSUCCC (M.L.G., D.E.S.); University of Texas MDACC (M.L.G., D.E.S.); Ohio Supercomputer Center (PAS0425, D.E.S.); Ohio Cancer Research Associate grant (GRT00024299, K.A.); Cancer Prevention Research Institute of Texas (CPRIT, RR170005, M.L.G.); National Institute of Dental and Craniofacial Research Institute grant DE023175 (M.L.G.); and National Cancer Institute grant R50CA211533 (K.A.). Dr. Gillison is a CPRIT Scholar in Cancer Research. The funding sources had no role in study design; in the collection, analysis or interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Dr. Negrao reported consulting for Mirati, Merck/MSD, and Genentech and research grants from Mirati, Novartis, Checkmate, Alaunos, AstraZeneca, Pfizer, and Genentech.
Dr. Gillison reported consulting for Roche; Bayer Healthcare Pharmaceuticals; BioNTech AG; EMD Sereno, Inc.; Shattuck Labs, Inc.; Kura Oncology; Gilead Sciences, Inc.; Ipsen Biopharmaceuticals, Inc.; Debiopharm; Bicara Therapeutics; LLX Solutions, LLC.; Eisai Medical Research; Nektar Therapeutics; Mirati Therapeutics; Sensei Biotherapeutics, Inc.; Seagen; OncLive; Istari Oncology, Inc.; iTeos Therapeutics; Coherus Biosciences; Caladrius Biosciences; and Exelixis, Inc.
Footnotes
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- [1].Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- [2].Bernier J, Cooper JS, Pajak TF, van Glabbeke M, Bourhis J, Forastiere A, et al. Defining risk levels in locally advanced head and neck cancers: a comparative analysis of concurrent postoperative radiation plus chemotherapy trials of the EORTC (#22931) and RTOG (# 9501). Head Neck. 2005;27:843–50. doi: 10.1002/hed.20279 [DOI] [PubMed] [Google Scholar]
- [3].Bernier J, Domenge C, Ozsahin M, Matuszewska K, Lefebvre JL, Greiner RH, et al. Postoperative irradiation with or without concomitant chemotherapy for locally advanced head and neck cancer. N Engl J Med. 2004;350:1945–52. doi: 10.1056/NEJMoa032641 [DOI] [PubMed] [Google Scholar]
- [4].Cooper JS, Pajak TF, Forastiere AA, Jacobs J, Campbell BH, Saxman SB, et al. Postoperative concurrent radiotherapy and chemotherapy for high-risk squamous-cell carcinoma of the head and neck. N Engl J Med. 2004;350:1937–44. doi: 10.1056/NEJMoa032646 [DOI] [PubMed] [Google Scholar]
- [5].Bossi P, Lo Vullo S, Guzzo M, Mariani L, Granata R, Orlandi E, et al. Preoperative chemotherapy in advanced resectable OCSCC: long-term results of a randomized phase III trial. Ann Oncol. 2014;25:462–6. doi: 10.1093/annonc/mdt555 [DOI] [PubMed] [Google Scholar]
- [6].Ganly I, Goldstein D, Carlson DL, Patel SG, O’Sullivan B, Lee N, et al. Long-term regional control and survival in patients with “low-risk,” early stage oral tongue cancer managed by partial glossectomy and neck dissection without postoperative radiation: the importance of tumor thickness. Cancer. 2013;119:1168–76. doi: 10.1002/cncr.27872 [DOI] [PubMed] [Google Scholar]
- [7].Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82. doi: 10.1038/nature14129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gillison ML, Akagi K, Xiao W, Jiang B, Pickard RKL, Li J, et al. Human papillomavirus and the landscape of secondary genetic alterations in oral cancers. Genome Res. 2019;29:1–17. doi: 10.1101/gr.241141.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Michikawa C, Torres-Saavedra PA, Silver NL, Harari PM, Kies MS, Rosenthal DI, et al. Evolutionary action score of TP53 analysis in pathologically high-risk HPV-negative head and neck cancer from a phase II clinical trial: NRG Oncology RTOG 0234. Journal of Clinical Oncology. 2019;37:6010-. doi: 10.1101/gr.241141.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ho AL, Brana I, Haddad R, Bauman J, Bible K, Oosting S, et al. Tipifarnib in Head and Neck Squamous Cell Carcinoma With HRAS Mutations. J Clin Oncol. 2021;39:1856–64. doi: 10.1200/JCO.20.02903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Cristescu R, Mogg R, Ayers M, Albright A, Murphy E, Yearley J, et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science. 2018;362. doi: 10.1126/science.aar3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ott PA, Bang YJ, Piha-Paul SA, Razak ARA, Bennouna J, Soria JC, et al. T-Cell-Inflamed Gene-Expression Profile, Programmed Death Ligand 1 Expression, and Tumor Mutational Burden Predict Efficacy in Patients Treated With Pembrolizumab Across 20 Cancers: KEYNOTE-028. J Clin Oncol. 2019;37:318–27. doi: 10.1200/JCO.2018.78.2276 [DOI] [PubMed] [Google Scholar]
- [13].Forshew T, Murtaza M, Parkinson C, Gale D, Tsui DW, Kaper F, et al. Noninvasive identification and monitoring of cancer mutations by targeted deep sequencing of plasma DNA. Sci Transl Med. 2012;4:136ra68. doi: 10.1126/scitranslmed.3003726 [DOI] [PubMed] [Google Scholar]
- [14].Cristiano S, Leal A, Phallen J, Fiksel J, Adleff V, Bruhm DC, et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature. 2019;570:385–9. doi: 10.1038/s41586-019-1272-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Phallen J, Sausen M, Adleff V, Leal A, Hruban C, White J, et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci Transl Med. 2017;9. doi: 10.1126/scitranslmed.aan2415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Kim S-T, Raymond VM, Park JO, Zotenko E, Park YS, Schultz M, et al. Abstract 916: Combined genomic and epigenomic assessment of cell-free circulating tumor DNA (ctDNA) improves assay sensitivity in early-stage colorectal cancer (CRC). Cancer Research. 2019;79:916-. doi: 10.1158/1538-7445.Am2019-916 [DOI] [Google Scholar]
- [17].Fakhry C, Blackford AL, Neuner G, Xiao W, Jiang B, Agrawal A, et al. Association of Oral Human Papillomavirus DNA Persistence With Cancer Progression After Primary Treatment for Oral Cavity and Oropharyngeal Squamous Cell Carcinoma. JAMA Oncol. 2019;5:985–92. doi: 10.1001/jamaoncol.2019.0439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chera BS, Kumar S, Shen C, Amdur R, Dagan R, Green R, et al. Plasma Circulating Tumor HPV DNA for the Surveillance of Cancer Recurrence in HPV-Associated Oropharyngeal Cancer. J Clin Oncol. 2020;38:1050–8. doi: 10.1200/JCO.19.02444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hanna GJ, Supplee JG, Kuang Y, Mahmood U, Lau CJ, Haddad RI, et al. Plasma HPV cell-free DNA monitoring in advanced HPV-associated oropharyngeal cancer. Ann Oncol. 2018;29:1980–6. doi: 10.1093/annonc/mdy251 [DOI] [PubMed] [Google Scholar]
- [20].Siravegna G, O’Boyle CJ, Varmeh S, Queenan N, Michel A, Stein J, et al. Cell-Free HPV DNA Provides an Accurate and Rapid Diagnosis of HPV-Associated Head and Neck Cancer. Clin Cancer Res. 2022;28:719–27. doi: 10.1158/1078-0432.CCR-21-3151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Wang Y, Springer S, Mulvey CL, Silliman N, Schaefer J, Sausen M, et al. Detection of somatic mutations and HPV in the saliva and plasma of patients with head and neck squamous cell carcinomas. Sci Transl Med. 2015;7:293ra104. doi: 10.1126/scitranslmed.aaa8507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].D’Souza G, Kreimer AR, Viscidi R, Pawlita M, Fakhry C, Koch WM, et al. Case-control study of human papillomavirus and oropharyngeal cancer. N Engl J Med. 2007;356:1944–56. doi: 10.1056/NEJMoa065497 [DOI] [PubMed] [Google Scholar]
- [23].Broutian TR, He X, Gillison ML. Automated high throughput DNA isolation for detection of human papillomavirus in oral rinse samples. J Clin Virol. 2011;50:270–5. doi: 10.1016/j.jcv.2010.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Jordan RC, Lingen MW, Perez-Ordonez B, He X, Pickard R, Koluder M, et al. Validation of methods for oropharyngeal cancer HPV status determination in US cooperative group trials. Am J Surg Pathol. 2012;36:945–54. doi: 10.1097/PAS.0b013e318253a2d1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Coutlee F, Rouleau D, Petignat P, Ghattas G, Kornegay JR, Schlag P, et al. Enhanced detection and typing of human papillomavirus (HPV) DNA in anogenital samples with PGMY primers and the Linear array HPV genotyping test. J Clin Microbiol. 2006;44:1998–2006. doi: 10.1128/JCM.00104-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–8. doi: 10.1038/nature12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gonzalez-Perez A, Lopez-Bigas N. Functional impact bias reveals cancer drivers. Nucleic Acids Res. 2012;40:e169. doi: 10.1093/nar/gks743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hua X, Xu H, Yang Y, Zhu J, Liu P, Lu Y. DrGaP: a powerful tool for identifying driver genes and pathways in cancer sequencing studies. Am J Hum Genet. 2013;93:439–51. doi: 10.1016/j.ajhg.2013.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Sherry ST, Ward M, Sirotkin K. dbSNP-database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999;9:677–9. [PubMed] [Google Scholar]
- [33].Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–11. doi: 10.1093/nar/29.1.308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Genomes Project C, Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alfoldi J, Wang Q, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–43. doi: 10.1038/s41586-020-2308-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Youden WJ. Index for rating diagnostic tests. Cancer. 1950;3:32–5. doi: [DOI] [PubMed] [Google Scholar]
- [37].Harty LC, Shields PG, Winn DM, Caporaso NE, Hayes RB. Self-collection of oral epithelial cell DNA under instruction from epidemiologic interviewers. Am J Epidemiol. 2000;151:199–205. doi: 10.1093/oxfordjournals.aje.a010188 [DOI] [PubMed] [Google Scholar]
- [38].Odegaard JI, Vincent JJ, Mortimer S, Vowles JV, Ulrich BC, Banks KC, et al. Validation of a Plasma-Based Comprehensive Cancer Genotyping Assay Utilizing Orthogonal Tissue- and Plasma-Based Methodologies. Clin Cancer Res. 2018;24:3539–49. doi: 10.1158/1078-0432.CCR-17-3831 [DOI] [PubMed] [Google Scholar]
- [39].Cheng SJ, Chang CF, Ko HH, Lee JJ, Chen HM, Wang HJ, et al. Hypermethylated ZNF582 and PAX1 genes in mouth rinse samples as biomarkers for oral dysplasia and oral cancer detection. Head Neck. 2018;40:355–68. doi: 10.1002/hed.24958 [DOI] [PubMed] [Google Scholar]
- [40].Lanman RB, Mortimer SA, Zill OA, Sebisanovic D, Lopez R, Blau S, et al. Analytical and Clinical Validation of a Digital Sequencing Panel for Quantitative, Highly Accurate Evaluation of Cell-Free Circulating Tumor DNA. PLoS One. 2015;10:e0140712. doi: 10.1371/journal.pone.0140712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Giuliano AR, Nedjai B, Lorincz AT, Schell MJ, Rahman S, Banwait R, et al. Methylation of HPV 16 and EPB41L3 in oral gargles: Associations with oropharyngeal cancer detection and tumor characteristics. Int J Cancer. 2020;146:1018–30. doi: 10.1002/ijc.32570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wu P, Xie C, Yang L, Liu Y, Zeng J, Li X, et al. The genomic architectures of tumour-adjacent tissues, plasma and saliva reveal evolutionary underpinnings of relapse in head and neck squamous cell carcinoma. Br J Cancer. 2021;125:854–64. doi: 10.1038/s41416-021-01464-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Shanmugam A, Hariharan AK, Hasina R, Nair JR, Katragadda S, Irusappan S, et al. Ultrasensitive detection of tumor-specific mutations in saliva of patients with oral cavity squamous cell carcinoma. Cancer. 2021;127:1576–89. doi: 10.1002/cncr.33393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Lee YC, Kim SI, Kwon H, Bae JS, Lee S. Circulating tumor DNA in the saliva of patients with head and neck cancer: A pilot report. Oral Dis. 2021;27:1421–5. doi: 10.1111/odi.13683 [DOI] [PubMed] [Google Scholar]
- [45].Galot R, van Marcke C, Helaers R, Mendola A, Goebbels RM, Caignet X, et al. Liquid biopsy for mutational profiling of locoregional recurrent and/or metastatic head and neck squamous cell carcinoma. Oral Oncol. 2020;104:104631. doi: 10.1016/j.oraloncology.2020.104631 [DOI] [PubMed] [Google Scholar]
- [46].Cui Y, Kim HS, Cho ES, Han D, Park JA, Park JY, et al. Longitudinal detection of somatic mutations in saliva and plasma for the surveillance of oral squamous cell carcinomas. PLoS One. 2021;16:e0256979. doi: 10.1371/journal.pone.0256979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Khurshid Z, Zafar MS, Khan RS, Najeeb S, Slowey PD, Rehman IU. Role of Salivary Biomarkers in Oral Cancer Detection. Adv Clin Chem. 2018;86:23–70. doi: 10.1016/bs.acc.2018.05.002 [DOI] [PubMed] [Google Scholar]
- [48].Kaczor-Urbanowicz KE, Wei F, Rao SL, Kim J, Shin H, Cheng J, et al. Clinical validity of saliva and novel technology for cancer detection. Biochim Biophys Acta Rev Cancer. 2019;1872:49–59. doi: 10.1016/j.bbcan.2019.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].William WN, Lee W-C, Lee JJ, Lin HY, Eterovic AK, El-Naggar AK, et al. Genomic and transcriptomic landscape of oral pre-cancers (OPCs) and risk of oral cancer (OC). Journal of Clinical Oncology. 2019;37:6009-. doi: 10.1200/JCO.2019.37.15_suppl.6009 [DOI] [Google Scholar]
- [50].Li C, He Q, Liang H, Cheng B, Li J, Xiong S, et al. Diagnostic Accuracy of Droplet Digital PCR and Amplification Refractory Mutation System PCR for Detecting EGFR Mutation in Cell-Free DNA of Lung Cancer: A Meta-Analysis. Front Oncol. 2020;10:290. doi: 10.3389/fonc.2020.00290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. N Engl J Med. 2005;352:786–92. doi: 10.1056/NEJMoa044238 [DOI] [PubMed] [Google Scholar]
- [52].Goss G, Tsai CM, Shepherd FA, Bazhenova L, Lee JS, Chang GC, et al. Osimertinib for pretreated EGFR Thr790Met-positive advanced non-small-cell lung cancer (AURA2): a multicentre, open-label, single-arm, phase 2 study. Lancet Oncol. 2016;17:1643–52. doi: 10.1016/S1470-2045(16)30508-3 [DOI] [PubMed] [Google Scholar]
- [53].Mok TS, Wu YL, Ahn MJ, Garassino MC, Kim HR, Ramalingam SS, et al. Osimertinib or Platinum-Pemetrexed in EGFR T790M-Positive Lung Cancer. N Engl J Med. 2017;376:629–40. doi: 10.1056/NEJMoa1612674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346ra92. doi: 10.1126/scitranslmed.aaf6219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Chaudhuri AA, Chabon JJ, Lovejoy AF, Newman AM, Stehr H, Azad TD, et al. Early Detection of Molecular Residual Disease in Localized Lung Cancer by Circulating Tumor DNA Profiling. Cancer Discov. 2017;7:1394–403. doi: 10.1158/2159-8290.CD-17-0716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015;7:302ra133. doi: 10.1126/scitranslmed.aab0021 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.