Abstract
Background
Gut microbiota, particularly Oxalobacter formigenes, has been previously reported to be associated with kidney stones. However, the conflicting results from both observational and intervention studies have created substantial uncertainty regarding the contribution of Oxalobacter formigenes to the formation of kidney stone.
Methods
We employed a two-sample MR analysis to investigate the causal relationship between gut microbiota and kidney stones using GWASs summary statistics obtained from the MiBioGen and FinnGen consortia. Moreover, we conducted a reserve MR analysis to assess the direction of the causal associations between gut microbiota and kidney stones. The inverse variance weighted (IVW) approach represents the primary method of Mendelian Randomization (MR) analysis.
Results
Our analyses do not yield supportive evidence for a causal link between the genus Oxalobacter (OR = 0.99, 95% CI: 0.90–1.09, p = 0.811) and the formation of kidney stones. The order Actinomycetales (OR = 0.79, 95% CI: 0.65–0.96, p = 0.020), family Actinomycetaceae (OR = 0.79, 95% CI: 0.65–0.96, p = 0.019), family Clostridiaceae 1 (OR = 0.80, 95% CI: 0.67–0.96, p = 0.015), genus Clostridiumsensustricto 1 (OR = 0.81, 95% CI: 0.67–0.98, p = 0.030) and genus Hungatella (OR = 0.86, 95% CI: 0.74–0.99, p = 0.040) had protective effects on kidney stones, and the genus Haemophilus (OR = 1.16, 95% CI: 1.01–1.33, p = 0.032), genus Ruminococcaceae (UCG010) (OR = 1.38, 95% CI: 1.04–1.84, p = 0.028), genus Subdoligranulum (OR = 1.27, 95% CI: 1.06–1.52, p = 0.009) were risk factors for kidney stones. Differential abundance analysis provide no evidence of a association between Oxalobacter formigenes and kidney stones, and showed genus Subdoligranulum were risk factors for kidney stones. Reverse MR analysis did not indicate any causal association of kidney stones on gut microbiota. No considerable heterogeneity of instrumental variables or horizontal pleiotropy was observed.
Conclusion
Our two-sample MR study did not find any causal relationship between genus Oxalobacter and kidney stones. The association between gut microbiota and kidney stones does not solely depend on the presence of genus Oxalobacter/Oxalobacter formigenes. A more integrated approach using multiple omics platforms is needed to better understand the pathogenesis of kidney stones in the context of complex gene–environment interactions over time.
Keywords: kidney stones, gut microbiota, Oxalobacter formigenes, Mendelian randomization, genus Subdoligranulum
Introduction
Kidney stones are a common chronic disease with a prevalence of 5–10%, and the rates appear to be rising in almost every country (Singh et al., 2022). Furthermore, the 10-year recurrence rate of kidney stones is 50% (Pearle et al., 2014), causing a significant burden on healthcare systems and having a substantial impact on patients’ quality of life. The etiology of kidney stones is highly complex and multifactorial, encompassing a range of factors including genetics, ethnicity, environment, dietary habits and metabolism. Despite significant research efforts, the precise mechanisms underlying the formation of kidney stone remain poorly understood. Thus, there is an urgent need to gain deeper insights into the pathogenesis of kidney stones and identify novel therapeutic targets that can be leveraged to develop more efficacious treatment strategies for this condition.
Gut microbiota, the complex community of microorganisms residing in the gastrointestinal tract, plays a crucial role in shaping the development and maintenance of the host’s health status through various interactions with the host, such as nutrient metabolism and immune regulation. Kidney stone is significantly influenced by various metabolic diseases, including hypertension, diabetes, and obesity, which have been found to be associated with alterations in the gut microbiota. In hypertensive patients, there is a significant reduction in microbial richness, diversity, and evenness, accompanied by a notable increase in the Firmicutes/Bacteroidetes ratio (Yang et al., 2015; Li et al., 2017). A study conducted on Chinese adolescents revealed a significant decrease in the abundance of Bacteroides thetaiotaomicron in obese individuals (Liu R. et al., 2017). A cohort study conducted in Sweden discovered a substantial reduction in gut microbial diversity among patients with type 2 diabetes mellitus (T2DM) (Wu et al., 2020). Recent researches have provided compelling evidence supporting a link between kidney stones and the gut microbiota (Mehta et al., 2016; Ticinesi et al., 2018; Stanford et al., 2020; Bostanghadiri et al., 2021). Studies have uncovered significant differences in the abundance of gut microbiota between kidney stone patients and healthy individuals (Ticinesi et al., 2018; Yuan et al., 2022). In fact, current researches on the relationship between kidney stones and gut microbiota have primarily focused on Oxalobacter formigenes, a bacterium that metabolizes oxalate as its principal energy source. This bacterium has been isolated from the fecal samples of individuals who have never experienced kidney stones, and its presence has been correlated with a reduction in urinary oxalate excretion (Siener et al., 2013). However, clinical studies have yielded inconsistent findings. Oxalobacter formigenes have been detected also in the fecal samples of individuals with a history of recurrent kidney stone formation. Interventions involving administration of Oxalobacter formigenes or other probiotic strains with oxalate-degrading capabilities have not consistently demonstrated a significant reduction in urinary oxalate excretion (Siva et al., 2009).
Therefore, further investigation is necessary to determine the specific contribution of Oxalobacter formigenes and other gut microbiota taxa to kidney stones. Mendelian randomization (MR) studies, akin to randomized controlled trials (RCTs), is an epidemiological approach for exploring the causal association between environmental exposures and diseases (Smith and Ebrahim, 2003; Swanson et al., 2017). Two-sample MR analysis allows for the integration of single-nucleotide polymorphisms (SNPs) data on exposure and outcome variables from independent genome-wide association studies (GWASs) to generate a unified causal estimate. SNPs adhere to the fundamental principle of random allocation of genetic variants during meiosis, which circumvents the influence of confounding factors and mitigates the risk of reverse causation, given that genetic variants precede the onset of the disease (Lawlor et al., 2008). This study employed a two-sample MR analysis to investigate the causal relationship between gut microbiota and kidney stones using GWASs summary statistics obtained from the MiBioGen and FinnGen consortia.
Methods
Exposure data
The instrumental variables utilized in our study were SNPs that were strongly associated with the composition of the human gut microbiome. These SNPs were selected from the largest GWASs dataset from the international consortium MiBioGen, involving 18,340 samples of 16S rRNA gene sequencing data from 24 population-based cohorts of various ancestries (Kurilshikov et al., 2021). A total of 211 gut microbiome taxa were included in the analysis, including 131 genera, 35 families, 20 orders, 16 classes, and 9 phyla.
Outcome data
GWAS summary statistics for kidney stones were available from the 7th release of the FinnGen consortium, which consisted of 7,433 cases and 301,094 controls with the adjustment for age, sex, 10 principal components and genotyping batch. The cases were identified by the diagnosis codes of N20 in the International Classification of Diseases, 10th Revision (ICD-10) and 592 in ICD-8 and ICD-9.
Instrumental variable selection
The three assumptions of MR are as follows: (1) Genetic variants have a strong and reliable association with the risk factors being studied; (2) Genetic variants are not associated with any confounding factors that could influence both the risk factors and the outcome; (3) Genetic variants only affect the outcome through their impact on the risk factors being investigated. SNPs associated with each gut microbiome were screened using the genome-wide significance threshold (p < 1 × 10−5) by referring to current MR studies on gut microbiome (Sanna et al., 2019). To ensure statistical independence, a linkage disequilibrium (LD) analysis (R2 < 0.001, clumping distance = 10,000 kb) was executed based on the European-based 1,000 Genome Projects. Furthermore, to ensure that the effects of the SNPs on each gut microbiota taxon correspond to the same allele as the effects on kidney stones, palindromic SNPs were removed. The F-statistic of instrumental variables was calculated to evaluate the extent of weak instrumental bias (F = beta2/se2). A F-statistic >10 was deemed as indicative of no bias caused by weak instrumental variables. To mitigate the correlation between SNPs and confounding factors, we utilized PhenoScanner1 to examine all incorporated instrumental variables, and subsequently removed SNPs that were associated with confounding factors.
Statistical analysis
We employed five commonly used MR methods to investigate whether there was a causal relationship between gut microbiota and kidney stones, namely, the inverse-variance weighted (IVW), MR-Egger regression, weighted mode, weighted median and simple mode (Burgess et al., 2013; Bowden et al., 2015, 2016; Hartwig et al., 2017). IVW method was employed as the primary MR method to infer causality. IVW method assumes that all instrumental variables have a common causal effect on the outcome through the exposure. It combines the effect estimates of each instrumental variable in a meta-analysis-like framework, using the inverse of the variance of each effect estimate as a weight, to obtain a summary causal estimate. MR-Egger regression is a method used when there is horizontal pleiotropy, which occurs when the instrumental variables affect the outcome through pathways that are not mediated by the exposure. The MR-PRESSO analysis was also used to detect the presence of horizontal pleiotropy. After removing these outliers, the MR-PRESSO method re-estimates the causal effect using the remaining instrumental variables. By identifying and adjusting for horizontal pleiotropy in this way, the MR-PRESSO analysis can provide a more accurate and robust estimate of the causal effect in Mendelian randomization studies. Moreover, we conducted a reserve MR analysis to assess the direction of the causal associations between gut microbiota and kidney stones. Cochrane’s Q test was computed to evaluate the degree of heterogeneity observed among the effect estimates of SNPs. The scatter plots and funnel plots were created for visualizing the results of MR analyses and identifying potential outliers. “Leave-one-out” analysis was performed to identify potential heterogeneous SNPs. This approach involves sequentially removing one SNP at a time from the instrumental variable set and re-estimating the causal effect estimate. All statistical analyses in this study were conducted using the R software, version 4.1.3. The “TwoSampleMR” R package and the “MRPRESSO” R package were utilized in our MR study.
Differential abundance analysis
We downloaded the raw data using the sequence read archive (SRA) accession numbers SRP140641, SRP140933, SRP066940, SRP103884, SRP125171 at https://github.com/amill017/USD_metaanalysis_2020 (Kachroo et al., 2021). The raw sequencing files of each sample were subjected to quality control using the fastp software. The quality-controlled sequencing data were processed using the Vsearch software to perform deduplication, denoising, and removal of chimeric sequences, resulting in a non-redundant Operational Taxonomic Units (OTU) table. The generated OTU table was imported into Qiime2, and species annotation was performed using available tools in Qiime2. Differential species analysis was conducted using the DESeq2 and Qiime2 packages in R software.
Results
SNPs selection
We obtained 102, 178, 215, 375, and 1,381 SNPs (p < 1 × 10−5) at the phylum, class, order, family, and genus levels, respectively. Namely, 2,251 SNPs were selected as instrumental variables. All F-statistics of instrumental variables were greater than 10.
MR analyses
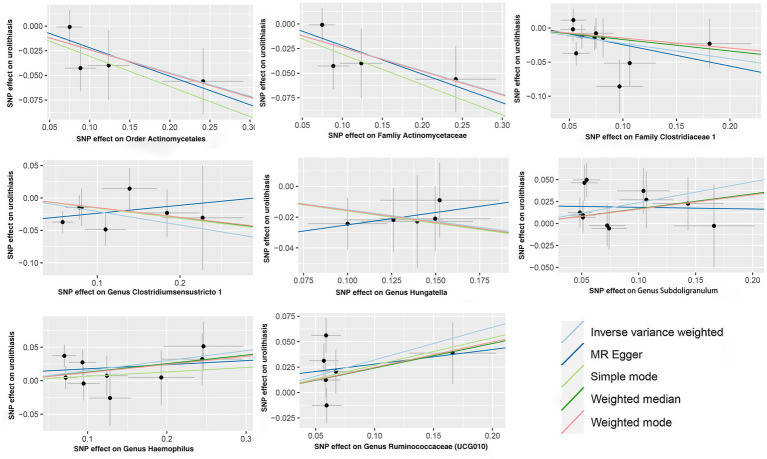
IVW analysis showed that the order Actinomycetales, family Actinomycetaceae, family Clostridiaceae 1, genus Clostridiumsensustricto 1, genus Haemophilus, genus Hungatella, genus Ruminococcaceae (UCG010), genus Subdoligranulum were associated with kidney stones, while there is no causal relationship between genus Oxalobacter (OR = 0.99, 95% CI: 0.90–1.09, p = 0.811) and kidney stones. The order Actinomycetales (OR = 0.79, 95% CI: 0.65–0.96, p = 0.020), family Actinomycetaceae (OR = 0.79, 95% CI: 0.65–0.96, p = 0.019), family Clostridiaceae 1 (OR = 0.80, 95% CI: 0.67–0.96, p = 0.015), genus Clostridiumsensustricto 1 (OR = 0.81, 95% CI: 0.67–0.98, p = 0.030) and genus Hungatella (OR = 0.86, 95% CI: 0.74–0.99, p = 0.040) had protective effects on kidney stones, and the genus Haemophilus (OR = 1.16, 95% CI: 1.01–1.33, p = 0.032), genus Ruminococcaceae (UCG010) (OR = 1.38, 95% CI: 1.04–1.84, p = 0.028), genus Subdoligranulum (OR = 1.27, 95% CI: 1.06–1.52, p = 0.009) were risk factors for kidney stones (Table 1). The scatter plots for the causal relationship between gut microbiota and kidney stones was presented in Figure 1. The specific instrumental variables used in MR analysis were listed in Supplementary Table S1. Supplementary Table S2 compiled the comprehensive results of all MR analyses conducted. Reverse MR analysis did not indicate any causal association of kidney stones on gut microbiota (Supplementary Table S3).
Table 1.
MR results of causal links between gut microbiome and kidney stones risk.
| Group | Gut microbiota | MR method | No.SNP | OR (95% CI) | p-value | F-statistic | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genus | Oxalobacter | Inverse variance weighted | 11 | 0.99 (0.90–1.09) | 0.811 | 21.35 | MR Egger | 11 | 0.78 (0.50–1.24) | 0.319 | Weighted median | 11 | 0.97 (0.85–1.11) | 0.667 | Simple mode | 11 | 0.96 (0.76–1.21) | 0.729 | Weighted mode | 11 | 0.96 (0.77–1.20) | 0.730 | ||||
| Order | Actinomycetales | Inverse variance weighted | 4 | 0.79 (0.65–0.96) | 0.020 | 21.51 | MR Egger | 4 | 0.75 (0.48–1.16) | 0.322 | Weighted median | 4 | 0.79 (0.61–1.02) | 0.066 | Simple mode | 4 | 0.74 (0.52–1.04) | 0.178 | Weighted mode | 4 | 0.79 (0.60–1.03) | 0.183 | ||||
| Family | Actinomycetaceae | Inverse variance weighted | 4 | 0.79 (0.65–0.96) | 0.019 | 21.57 | MR Egger | 4 | 0.75 (0.48–1.15) | 0.320 | Weighted median | 4 | 0.79 (0.61–1.01) | 0.058 | Simple mode | 4 | 0.74 (0.52–1.05) | 0.188 | Weighted mode | 4 | 0.79 (0.60–1.03) | 0.180 | ||||
| Family | Clostridiaceae1 | Inverse variance weighted | 10 | 0.80 (0.67–0.96) | 0.015 | 20.28 | MR Egger | 10 | 0.73 (0.43–1.24) | 0.276 | Weighted median | 10 | 0.84 (0.66–1.08) | 0.184 | Simple mode | 10 | 0.86 (0.61–1.22) | 0.430 | Weighted mode | 10 | 0.86 (0.63–1.18) | 0.386 | ||||
| Genus | Clostridiumsensustricto 1 | Inverse variance weighted | 7 | 0.81 (0.67–0.98) | 0.030 | 20.60 | MR Egger | 7 | 1.13 (0.70–1.83) | 0.637 | Weighted median | 7 | 0.86 (0.66–1.12) | 0.256 | Simple mode | 7 | 0.85 (0.58–1.26) | 0.455 | Weighted mode | 7 | 0.86 (0.62–1.20) | 0.417 | ||||
| Genus | Haemophilus | Inverse variance weighted | 9 | 1.16 (1.01–1.33) | 0.032 | 23.42 | MR Egger | 9 | 1.06 (0.78–1.44) | 0.705 | Weighted median | 9 | 1.14 (0.95–1.36) | 0.172 | Simple mode | 9 | 1.07 (0.83–1.37) | 0.626 | Weighted mode | 9 | 1.13 (0.89–1.42) | 0.338 | ||||
| Genus | Hungatella | Inverse variance weighted | 5 | 0.86 (0.74–0.99) | 0.040 | 20.43 | MR Egger | 5 | 1.17 (0.49–2.83) | 0.744 | Weighted median | 5 | 0.85 (0.71–1.03) | 0.098 | Simple mode | 5 | 0.85 (0.68–1.07) | 0.243 | Weighted mode | 5 | 0.86 (0.68–1.07) | 0.242 | ||||
| Genus | Ruminococcaceae (UCG010) | Inverse variance weighted | 6 | 1.38 (1.04–1.84) | 0.028 | 21.43 | MR Egger | 6 | 1.15 (0.49–2.72) | 0.759 | Weighted median | 6 | 1.27 (0.94–1.72) | 0.115 | Simple mode | 6 | 1.31 (0.85–2.02) | 0.280 | Weighted mode | 6 | 1.28 (0.92–1.79) | 0.203 | ||||
| Genus | Subdoligranulum | Inverse variance weighted | 11 | 1.27 (1.06–1.52) | 0.009 | 21.12 | MR Egger | 11 | 0.98 (0.62–1.55) | 0.936 | Weighted median | 11 | 1.18 (0.93–1.51) | 0.179 | Simple mode | 11 | 1.18 (0.85–1.63) | 0.348 | Weighted mode | 11 | 1.18 (0.87–1.60) | 0.317 |
Figure 1.
Scatter plots for the causal links between gut microbiota and kidney stones.
Sensitivity analyses
According to the results of Cochran’s Q test, there was no statistically significant heterogeneity observed among these instrumental variables (Supplementary Table S4). MR-Egger regression intercept analysis showed that there was no directional horizontal pleiotropy for gut microbiota in kidney stones (Supplementary Table S5). Visual examination of scatter plots (Supplementary Figure S1) and leave-one-out plots (Supplementary Figure S2) revealed the possible existence of outliers among the instrumental variables of genus Hungatella, genus Haemophilus and genus Ruminococcaceae (UCG010). However, MR-PRESSO analysis indicated no outliers in the results (Supplementary Table S6). As a result, the available evidence did not support the presence of horizontal pleiotropy in the relationship between these bacteria and kidney stones.
Differential abundance analysis
Differential abundance analysis of the gut microbiota revealed that, compared to the control group, the stone former group showed no significant difference in the abundance of Oxalobacter formigenes. The abundance of the genus Anaerostipes, genus Bifidobacterium, genus Dialister, genus Erysipelotrichaceae_UCG-003, genus Muribaculaceae and genus Klebsiella were decreased, while the abundance of genus Acidaminococcus, genus Alloprevotella, genus Catenibacterium, genus Faecalitalea, genus Megasphaera, genus Parabacteroides, genus Prevotellaceae_NK3B31_group, genus Pseudomonas and genus Subdoligranulum were increased in the stone former group. Additionally, there were some variations in the abundance of certain bacterial species within the genera Bacteroides, Faecalibacterium, and Prevotella, with some showing an increase and others showing a decrease (Supplementary Figure S3; Supplementary Tables S8, S9). The Pearson correlation coefficient for the correlation analysis is 0.694, indicating the presence of differences in gut microbiota between the stone former group and the control group.
Discussion
As far as we are aware, this is the first MR analysis conducted to explore the potential causal relationship between gut microbiota and kidney stones. Neither MR analysis nor differential abundance analysis provide evidence of a causal association between genus Oxalobacter/Oxalobacter formigenes and kidney stones. Both MR analysis and differential abundance analysis showed genus Subdoligranulum were risk factors for kidney stones.
Calcium oxalate stones, comprising approximately 80% of all kidney stone varieties, represent the most commom classification of kidney stones (Khan et al., 2016). The gastrointestinal tract assumes a pivotal function in oxalate metabolism, as it exerts significant impact on the urinary oxalate excretion and, by extension, the proclivity for the development of kidney stones (Robijn et al., 2011). The existence of gut microbiota capable of degrading oxalate has the potential to impede oxalate absorption and reduce oxalate excretion. In recent decades, there has been a focused investigation into the contribution of gut microbiota to the physiopathologic of the gut–kidney axis. In fact, many of these studies have been predominantly centered on Oxalobacter formigenes, a type of oxalate-degrading bacteria via the expression of oxalyl-CoA decarboxylase and formyl-CoA transferase (Ellis et al., 2016). Several studies have provided evidence that patients with kidney stones have a lower incidence of Oxalobacter formigenes in their stool than healthy control subjects (Kumar et al., 2002, 2004; Kaufman et al., 2008). There also exists evidence indicating that patients with kidney stones who have Oxalobacter formigenes in their fecal matter experience a reduced level of urinary oxalate excretion in comparison to those normal individuals (Neuhaus et al., 2000; Kwak et al., 2003; Troxel et al., 2003). However, the conflicting results from both observational and intervention studies have created substantial uncertainty regarding the contribution of Oxalobacter formigenes to the formation of kidney stone (Ticinesi et al., 2019).
Numerous fact ors have been identified as being associated with the Oxalobacter colonization in the gut microbiota, including ethnicity, country of residence, age, education level, recent antibiotic usage, body weight, and nutritional imbalances (Liu M. et al., 2017; Ticinesi et al., 2020). By utilizing genomic shotgun data and shotgun community profiling analysis, a study showed a total of 31% of healthy young adults residing in the United States demonstrated a positive presence of Oxalobacter formigenes, which is much lower than that detected in tribal populations from Venezuela and Tanzania (Barnett et al., 2016; PeBenito et al., 2019). Some studies suggested lifetime exposure to antibiotics, especially long-duration treatments initiated at a younger age, is significantly associated with an increased risk of developing kidney stones (Tasian et al., 2018; Ferraro et al., 2019). A study indicated that prior exposure to antibiotics, even those that are not typically effective against Oxalobacter formigenes, can impact the colonization rates of this bacterium (Kaufman et al., 2008). Dietary habits and nutrition may constitute partial etiologies of the observed differences in gut microbiota composition between individuals with nephrolithiasis and the control group. Kaufman et al. have noted a positive correlation between oxalate consumption and the prevalence of Oxalobacter formigenes in the control group, due to the fact that dietary oxalate serves as a significant energy substrate for this bacterium, in addition to endogenously-produced oxalate (Kaufman et al., 2008). Intestinal oxalate absorption is also influenced by the ratio of dietary calcium to oxalate, and an optimal ratio promotes the formation of oxalate-calcium complexes in the gut lumen, thereby inhibiting intestinal absorption and promoting renal excretion, ultimately limiting the formation of kidney stones (Siener et al., 2003; Knight et al., 2006). In fact, most studies investigating the relationship between kidney stones and gut microbiota have not effectively integrated these factors. Furthermore, in a large case–control study on the relationship between Oxalobacter formigenes and kidney stone formation, no difference was observed in urinary oxalate excretion and the presence of Oxalobacter formigenes colonization (Kaufman et al., 2008).
In addition, two distinct studies have found significant correlations between the average relative abundance of certain specific taxa, including Sutterella, Veillonella and Peptococcus, and urinary oxalate excretion levels (Suryavanshi et al., 2016; Ticinesi et al., 2018), indicating that the gut-kidney axis may not solely depend on the presence of Oxalobacter formigenes. Subsequent investigations have indicated that, in healthy individuals, the presence of Oxalobacter formigenes is linked to an intricate network of bacteria that may possess oxalate-degrading capabilities themselves or promote the metabolic activity of Oxalobacter formigenes (Suryavanshi et al., 2018; Miller et al., 2019). This notion has been validated in mice that were transplanted with human fecal samples colonized by Oxalobacter formigenes, as the transplantation process led to a specific expansion of the bacteria network associated with Oxalobacter formigenes (Pebenito et al., 2019).
Subdoligranulum is a genus of anaerobic, spore-free Gram-negative bacteria. In studies investigating various diseases, genus Subdoligranulum has been consistently linked to chronic inflammation-related immune markers (Shi et al., 2021). Currently, it is widely believed that there is a close association between gut microbiota and obesity, as well as related metabolic diseases. A study has revealed that the abundance of genus Subdoligranulum is positively correlated with body weight and BMI (Kim et al., 2014). Li et al. conducted a study on the alterations in gut microbiota in individuals with hypertension and observed a positive correlation between the abundance of genus Subdoligranulum and both systolic and diastolic blood pressure (Li et al., 2019). Genus Subdoligranulum is also enriched in the diabetes mellitus individuals (Zhao et al., 2020). Therefore, the potential mechanisms may involve the involvement of gut microbiota in chronic inflammatory responses, as well as the mediation of obesity and metabolic dysregulation, which contribute to the formation of kidney stones.
Previous studies have indicated that certain bacterial genera, including Prevotella, Acinetobacter, Pseudomonas, Staphylococcus, Megamonas, and Cetobacterium, have been detected in individuals with kidney stones (Jandhyala et al., 2015; Liu et al., 2020). These genera are known to be associated with inflammatory diseases, implying that inflammation might play a role in the development of kidney stones. It is noteworthy that Faecalibacterium possesses the capability to synthesize short-chain fatty acids (SCFAs), particularly butyrate, which plays a crucial role in modulating inflammation. Hence, the depletion of this bacterium could contribute to the formation of kidney stones (Gambaro et al., 2017).
This study has several notable strengths. Firstly, previous case–control studies were unable to establish the temporal sequence between gut microbiota colonization and kidney stone formation, as stool samples were collected after episode occurred. While the MR analysis is an appropriate method for investigating causal relationships, as it can eliminate potential confounding factors and reverse causation, and differential abundance analysis helps improve the stability of our results. Secondly, the genetic variants of gut microbiota used in the study were obtained from the largest available GWAS meta-analysis, which ensured the strength and reliability of the instruments used in the MR analysis. Thirdly, horizontal pleiotropy was detected and addressed through the use of MR-PRESSO and MR-Egger regression intercept term tests.
The present study also has some limitations. Firstly, since the exposure dataset only provided information at the genus taxonomic level, we were unable to investigate the causal association between gut microbiota and kidney stones at the species level. Secondly, kidney stone composition was not available in the FinnGen consortium, thereby impeding our ability to conduct subgroup analyses. Further studies with subgroup analysis stratified by the composition of kidney stones may yield more reliable results. Thirdly, although the majority of participants in the GWAS meta-analysis for gut microbiota data were of European ancestry, there is still potential for population stratification bias, and the findings of this study may not be fully generalizable to individuals of non-European ancestry. Finally, We were unable to conduct correlation analyses of gut microbiota abundance between kidney stone patients and healthy individuals.
Conclusion
Our two-sample MR study did not find any causal relationship between genus Oxalobacter and kidney stones. The association between gut microbiota and kidney stones does not solely depend on the presence of Oxalobacter formigenes. A more integrated approach using multiple omics platforms is needed to better understand the pathogenesis of kidney stones in the context of complex gene–environment interactions over time.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
The patients/participants provided their written informed consent to participate in this study.
Author contributions
ML: project development, data analysis, and manuscript writing. YZ, JW, and MG: data collection and analysis. YZ: data collection. JW and MG: manuscript editing. ZZ and HC: project development and manuscript editing. All authors contributed to the article and approved the submitted version.
Funding
Funding was provided by the Fundamental Research Funds for the Central Universities of Central South University (2022ZZTS0294 to ML; 2021zzts0348 to ZZ), the National Natural Science Foundation of China (82170781 to HC) and Natural Science Foundation of Hunan Province (2021JJ31050 to HC).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We express our gratitude to the participants and investigators of the MiBioGen consortium and FinnGen study.
Glossary
Abbreviations
- MR
Mendelian randomization
- RCTs
randomized controlled trials
- SNPs
single-nucleotide polymorphisms
- GWASs
genome-wide association studies
- IVW
inverse-variance weighted
Footnotes
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1204311/full#supplementary-material
References
- Barnett C., Nazzal L., Goldfarb D. S., Blaser M. J. (2016). The Presence of Oxalobacter formigenes in the Microbiome of Healthy Young Adults. J. Urol. 195, 499–506. doi: 10.1016/j.juro.2015.08.070, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostanghadiri N., Ziaeefar P., Sameni F., Mahmoudi M., Hashemi A., Darban-Sarokhalil D. (2021). The controversial association of gut and urinary microbiota with kidney stone formation. Microb. Pathog. 161:105257. doi: 10.1016/j.micpath.2021.105257, PMID: [DOI] [PubMed] [Google Scholar]
- Bowden J., Davey Smith G., Burgess S. (2015). Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 44, 512–525. doi: 10.1093/ije/dyv080, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowden J., Davey Smith G., Haycock P. C., Burgess S. (2016). Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 40, 304–314. doi: 10.1002/gepi.21965, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess S., Butterworth A., Thompson S. G. (2013). Mendelian randomization analysis with multiple genetic variants using summarized data. Genet. Epidemiol. 37, 658–665. doi: 10.1002/gepi.21758, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis M. E., Mobley J. A., Holmes R. P., Knight J. (2016). Proteome Dynamics of the Specialist Oxalate Degrader Oxalobacter formigenes. J. Proteomics Bioinform. 9, 19–24. doi: 10.4172/jpb.1000384, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro P. M., Curhan G. C., Gambaro G., Taylor E. N. (2019). Antibiotic Use and Risk of Incident Kidney Stones in Female Nurses. Am. J. Kidney Dis. 74, 736–741. doi: 10.1053/j.ajkd.2019.06.005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambaro G., Croppi E., Bushinsky D., Jaeger P., Cupisti A., Ticinesi A., et al. (2017). The Risk of Chronic Kidney Disease Associated with Urolithiasis and its Urological Treatments: A Review. J. Urol. 198, 268–273. doi: 10.1016/j.juro.2016.12.135, PMID: [DOI] [PubMed] [Google Scholar]
- Hartwig F. P., Davey Smith G., Bowden J. (2017). Robust inference in summary data Mendelian randomization via the zero modal pleiotropy assumption. Int. J. Epidemiol. 46, 1985–1998. doi: 10.1093/ije/dyx102, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jandhyala S. M., Talukdar R., Subramanyam C., Vuyyuru H., Sasikala M., Nageshwar R. D. (2015). Role of the normal gut microbiota. World J. Gastroenterol. 21, 8787–8803. doi: 10.3748/wjg.v21.i29.8787, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kachroo N., Lange D., Penniston K. L., Stern J., Tasian G., Bajic P., et al. (2021). Meta-analysis of Clinical Microbiome Studies in Urolithiasis Reveal Age, Stone Composition, and Study Location as the Predominant Factors in Urolithiasis-Associated Microbiome Composition. MBio 12:e0200721. doi: 10.1128/mBio.02007-21, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman D. W., Kelly J. P., Curhan G. C., Anderson T. E., Dretler S. P., Preminger G. M., et al. (2008). Oxalobacter formigenes may reduce the risk of calcium oxalate kidney stones. J. Am. Soc. Nephrol. 19, 1197–1203. doi: 10.1681/ASN.2007101058, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan S. R., Pearle M. S., Robertson W. G., Gambaro G., Canales B. K., Doizi S., et al. (2016). Kidney stones. Nat. Rev. Dis. Prim. 2:16008. doi: 10.1038/nrdp.2016.8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.-S., Song M.-Y., Kim H. (2014). The anti-obesity effect of Ephedra sinica through modulation of gut microbiota in obese Korean women. J. Ethnopharmacol. 152, 532–539. doi: 10.1016/j.jep.2014.01.038, PMID: [DOI] [PubMed] [Google Scholar]
- Knight J., Jiang J., Assimos D. G., Holmes R. P. (2006). Hydroxyproline ingestion and urinary oxalate and glycolate excretion. Kidney Int. 70, 1929–1934. doi: 10.1038/sj.ki.5001906, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Ghoshal U. C., Singh G., Mittal R. D. (2004). Infrequency of colonization with Oxalobacter formigenes in inflammatory bowel disease: possible role in renal stone formation. J. Gastroenterol. Hepatol. 19, 1403–1409. doi: 10.1111/j.1440-1746.2004.03510.x, PMID: [DOI] [PubMed] [Google Scholar]
- Kumar R., Mukherjee M., Bhandari M., Kumar A., Sidhu H., Mittal R. D. (2002). Role of Oxalobacter formigenes in calcium oxalate stone disease: a study from North India. Eur. Urol. 41, 318–322. doi: 10.1016/s0302-2838(02)00040-4, PMID: [DOI] [PubMed] [Google Scholar]
- Kurilshikov A., Medina-Gomez C., Bacigalupe R., Radjabzadeh D., Wang J., Demirkan A., et al. (2021). Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 53, 156–165. doi: 10.1038/s41588-020-00763-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak C., Kim H. K., Kim E. C., Choi M. S., Kim H. H. (2003). Urinary oxalate levels and the enteric bacterium Oxalobacter formigenes in patients with calcium oxalate urolithiasis. Eur. Urol. 44, 475–481. doi: 10.1016/s0302-2838(03)00318-x, PMID: [DOI] [PubMed] [Google Scholar]
- Lawlor D. A., Harbord R. M., Sterne J. A. C., Timpson N., Davey S. G. (2008). Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat. Med. 27, 1133–1163. doi: 10.1002/sim.3034, PMID: [DOI] [PubMed] [Google Scholar]
- Li H., Liu B., Song J., An Z., Zeng X., Li J., et al. (2019). Characteristics of Gut Microbiota in Patients with Hypertension and/or Hyperlipidemia: A Cross-Sectional Study on Rural Residents in Xinxiang County, Henan Province. Microorganisms 7:399. doi: 10.3390/microorganisms7100399, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhao F., Wang Y., Chen J., Tao J., Tian G., et al. (2017). Gut microbiota dysbiosis contributes to the development of hypertension. Microbiome 5:14. doi: 10.1186/s40168-016-0222-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Hong J., Xu X., Feng Q., Zhang D., Gu Y., et al. (2017). Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med. 23, 859–868. doi: 10.1038/nm.4358, PMID: [DOI] [PubMed] [Google Scholar]
- Liu Y., Jin X., Hong H. G., Xiang L., Jiang Q., Ma Y., et al. (2020). The relationship between gut microbiota and short chain fatty acids in the renal calcium oxalate stones disease. FASEB J. 34, 11200–11214. doi: 10.1096/fj.202000786R, PMID: [DOI] [PubMed] [Google Scholar]
- Liu M., Koh H., Kurtz Z. D., Battaglia T., PeBenito A., Li H., et al. (2017). Oxalobacter formigenes-associated host features and microbial community structures examined using the American Gut Project. Microbiome 5:108. doi: 10.1186/s40168-017-0316-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta M., Goldfarb D. S., Nazzal L. (2016). The role of the microbiome in kidney stone formation. Int. J. Surg. 36, 607–612. doi: 10.1016/j.ijsu.2016.11.024, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A. W., Choy D., Penniston K. L., Lange D. (2019). Inhibition of urinary stone disease by a multi-species bacterial network ensures healthy oxalate homeostasis. Kidney Int. 96, 180–188. doi: 10.1016/j.kint.2019.02.012, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus T. J., Belzer T., Blau N., Hoppe B., Sidhu H., Leumann E. (2000). Urinary oxalate excretion in urolithiasis and nephrocalcinosis. Arch. Dis. Child. 82, 322–326. doi: 10.1136/adc.82.4.322, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearle M. S., Goldfarb D. S., Assimos D. G., Curhan G., Denu-Ciocca C. J., Matlaga B. R., et al. (2014). Medical management of kidney stones: AUA guideline. J. Urol. 192, 316–324. doi: 10.1016/j.juro.2014.05.006, PMID: [DOI] [PubMed] [Google Scholar]
- Pebenito A. M., Liu M., Nazzal L., Blaser M. J. (2019). Development of a Humanized Murine Model for the Study of Oxalobacter formigenes Intestinal Colonization. J. Infect. Dis. 220, 1848–1858. doi: 10.1093/infdis/jiz370, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- PeBenito A., Nazzal L., Wang C., Li H., Jay M., Noya-Alarcon O., et al. (2019). Comparative prevalence of Oxalobacter formigenes in three human populations. Sci. Rep. 9:574. doi: 10.1038/s41598-018-36670-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robijn S., Hoppe B., Vervaet B. A., D’Haese P. C., Verhulst A. (2011). Hyperoxaluria: a gut-kidney axis? Kidney Int. 80, 1146–1158. doi: 10.1038/ki.2011.287, PMID: [DOI] [PubMed] [Google Scholar]
- Sanna S., van Zuydam N. R., Mahajan A., Kurilshikov A., Vich Vila A., Võsa U., et al. (2019). Causal relationships among the gut microbiome, short-chain fatty acids and metabolic diseases. Nat. Genet. 51, 600–605. doi: 10.1038/s41588-019-0350-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T.-T., Xin Z., Hua L., Wang H., Zhao R.-X., Yang Y.-L., et al. (2021). Comparative assessment of gut microbial composition and function in patients with Graves’ disease and Graves’ orbitopathy. J. Endocrinol. Investig. 44, 297–310. doi: 10.1007/s40618-020-01298-2, PMID: [DOI] [PubMed] [Google Scholar]
- Siener R., Bangen U., Sidhu H., Hönow R., von Unruh G., Hesse A. (2013). The role of Oxalobacter formigenes colonization in calcium oxalate stone disease. Kidney Int. 83, 1144–1149. doi: 10.1038/ki.2013.104 [DOI] [PubMed] [Google Scholar]
- Siener R., Ebert D., Nicolay C., Hesse A. (2003). Dietary risk factors for hyperoxaluria in calcium oxalate stone formers. Kidney Int. 63, 1037–1043. doi: 10.1046/j.1523-1755.2003.00807.x, PMID: [DOI] [PubMed] [Google Scholar]
- Singh P., Harris P. C., Sas D. J., Lieske J. C. (2022). The genetics of kidney stone disease and nephrocalcinosis. Nat. Rev. Nephrol. 18, 224–240. doi: 10.1038/s41581-021-00513-4 [DOI] [PubMed] [Google Scholar]
- Siva S., Barrack E. R., Reddy G. P. V., Thamilselvan V., Thamilselvan S., Menon M., et al. (2009). A critical analysis of the role of gut Oxalobacter formigenes in oxalate stone disease. BJU Int. 103, 18–21. doi: 10.1111/j.1464-410X.2008.08122.x, PMID: [DOI] [PubMed] [Google Scholar]
- Smith G. D., Ebrahim S. (2003). “Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease? Int. J. Epidemiol. 32, 1–22. doi: 10.1093/ije/dyg070, PMID: [DOI] [PubMed] [Google Scholar]
- Stanford J., Charlton K., Stefoska-Needham A., Ibrahim R., Lambert K. (2020). The gut microbiota profile of adults with kidney disease and kidney stones: a systematic review of the literature. BMC Nephrol. 21:215. doi: 10.1186/s12882-020-01805-w, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryavanshi M. V., Bhute S. S., Gune R. P., Shouche Y. S. (2018). Functional eubacteria species along with trans-domain gut inhabitants favour dysgenic diversity in oxalate stone disease. Sci. Rep. 8:16598. doi: 10.1038/s41598-018-33773-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryavanshi M. V., Bhute S. S., Jadhav S. D., Bhatia M. S., Gune R. P., Shouche Y. S. (2016). Hyperoxaluria leads to dysbiosis and drives selective enrichment of oxalate metabolizing bacterial species in recurrent kidney stone endures. Sci. Rep. 6:34712. doi: 10.1038/srep34712, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson S. A., Tiemeier H., Ikram M. A., Hernán M. A. (2017). Nature as a Trialist?: Deconstructing the Analogy Between Mendelian Randomization and Randomized Trials. Epidemiology 28, 653–659. doi: 10.1097/EDE.0000000000000699, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasian G. E., Jemielita T., Goldfarb D. S., Copelovitch L., Gerber J. S., Wu Q., et al. (2018). Oral Antibiotic Exposure and Kidney Stone Disease. J. Am. Soc. Nephrol. 29, 1731–1740. doi: 10.1681/ASN.2017111213, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticinesi A., Milani C., Guerra A., Allegri F., Lauretani F., Nouvenne A., et al. (2018). Understanding the gut-kidney axis in nephrolithiasis: an analysis of the gut microbiota composition and functionality of stone formers. Gut 67, 2097–2106. doi: 10.1136/gutjnl-2017-315734, PMID: [DOI] [PubMed] [Google Scholar]
- Ticinesi A., Nouvenne A., Chiussi G., Castaldo G., Guerra A., Meschi T. (2020). Calcium Oxalate Nephrolithiasis and Gut Microbiota: Not just a Gut-Kidney Axis. A Nutritional Perspective. Nutrients 12:548. doi: 10.3390/nu12020548, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ticinesi A., Nouvenne A., Meschi T. (2019). Gut microbiome and kidney stone disease: not just an Oxalobacter story. Kidney Int. 96, 25–27. doi: 10.1016/j.kint.2019.03.020 [DOI] [PubMed] [Google Scholar]
- Troxel S. A., Sidhu H., Kaul P., Low R. K. (2003). Intestinal Oxalobacter formigenes colonization in calcium oxalate stone formers and its relation to urinary oxalate. J. Endourol. 17, 173–176. doi: 10.1089/089277903321618743 [DOI] [PubMed] [Google Scholar]
- Wu H., Tremaroli V., Schmidt C., Lundqvist A., Olsson L. M., Krämer M., et al. (2020). The Gut Microbiota in Prediabetes and Diabetes: A Population-Based Cross-Sectional Study. Cell Metab. 32, 379–390.e3. doi: 10.1016/j.cmet.2020.06.011, PMID: [DOI] [PubMed] [Google Scholar]
- Yang T., Santisteban M. M., Rodriguez V., Li E., Ahmari N., Carvajal J. M., et al. (2015). Gut dysbiosis is linked to hypertension. Hypertension 65, 1331–1340. doi: 10.1161/HYPERTENSIONAHA.115.05315, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C., Jin X., He Y., Liu Y., Xiang L., Wang K. (2022). Association of dietary patterns with gut microbiota in kidney stone and non-kidney stone individuals. Urolithiasis 50, 389–399. doi: 10.1007/s00240-022-01325-2, PMID: [DOI] [PubMed] [Google Scholar]
- Zhao X., Zhang Y., Guo R., Yu W., Zhang F., Wu F., et al. (2020). The Alteration in Composition and Function of Gut Microbiome in Patients with Type 2 Diabetes. J. Diabetes Res. 2020, 8842651–8842613. doi: 10.1155/2020/8842651, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.