Abstract
Objective
Defining the regulators of cell metabolism and signaling is essential to design new therapeutic strategies in obesity and NAFLD/NASH. E3 ubiquitin ligases control diverse cellular functions by ubiquitination-mediated regulation of protein targets, and thus their functional aberration is associated with many diseases. The E3 ligase Ube4A has been implicated in human obesity, inflammation, and cancer. However, its in vivo function is unknown, and no animal models are available to study this novel protein.
Methods
A whole-body Ube4A knockout (UKO) mouse model was generated, and various metabolic parameters were compared in chow- and high fat diet (HFD)-fed WT and UKO mice, and in their liver, adipose tissue, and serum. Lipidomics and RNA-Seq studies were performed in the liver samples of HFD-fed WT and UKO mice. Proteomic studies were conducted to identify Ube4A's targets in metabolism. Furthermore, a mechanism by which Ube4A regulates metabolism was identified.
Results
Although the body weight and composition of young, chow-fed WT and UKO mice are similar, the knockouts exhibit mild hyperinsulinemia and insulin resistance. HFD feeding substantially augments obesity, hyperinsulinemia, and insulin resistance in both sexes of UKO mice. HFD-fed white and brown adipose tissue depots of UKO mice have increased insulin resistance and inflammation and reduced energy metabolism. Moreover, Ube4A deletion exacerbates hepatic steatosis, inflammation, and liver injury in HFD-fed mice with increased lipid uptake and lipogenesis in hepatocytes. Acute insulin treatment resulted in impaired activation of the insulin effector protein kinase Akt in liver and adipose tissue of chow-fed UKO mice. We identified the Akt activator protein APPL1 as a Ube4A interactor. The K63-linked ubiquitination (K63-Ub) of Akt and APPL1, known to facilitate insulin-induced Akt activation, is impaired in UKO mice. Furthermore, Ube4A K63-ubiquitinates Akt in vitro.
Conclusion
Ube4A is a novel regulator of obesity, insulin resistance, adipose tissue dysfunction and NAFLD, and preventing its downregulation may ameliorate these diseases.
Keywords: Ube4A, Ubiquitination, Obesity, NAFLD, Insulin/Akt signaling, APPL1
Highlights
-
•
The U-box containing E3 ubiquitin ligase Ube4A is a novel regulator of metabolism.
-
•
Deletion of Ube4A exacerbates high fat diet-induced obesity, hyperinsulinemia, and insulin resistance in both sexes of mice.
-
•
Ube4A deletion augments hepatic steatosis and enhances lipid uptake and lipogenesis in hepatocytes.
-
•
Ube4A facilitates insulin signaling by K63-ubiquitination of Akt and APPL1.
-
•
Maintaining Ube4A's activity may ameliorate obesity, T2DM and NAFLD.
1. Introduction
Obesity and type-2 diabetes mellitus (T2DM) are global health concerns as these diseases significantly increase the risk of other co-morbidities such as non-alcoholic fatty liver disease/steatohepatitis (NAFLD/NASH) and cardiovascular diseases [[1], [2], [3]]. Obesity-induced insulin resistance and metabolic dysfunction in the adipose tissue upregulates inflammatory adipokines and downregulates metabolism-promoting adipokines [4]. Moreover, the fat-storing ability of dysfunctional adipocytes is reduced, leading to an increase in serum levels of non-esterified fatty acids (NEFA) that are taken up by the liver [5,6]. In addition, obesity-induced hyperinsulinemia causes selective hepatic insulin resistance [[1], [2], [3]] wherein both gluconeogenesis and de novo lipogenesis processes are increased in hepatocytes, contributing to hyperglycemia, lipotoxic stress, and steatohepatitis [[1], [2], [3],[5], [6], [7]]. A complete knowledge of the mechanisms that regulate metabolism and insulin sensitivity in health and diseases is lacking.
E3 ubiquitin ligases modulate various cellular functions by ubiquitinating target proteins. Aberrant activation of E3 ligases is associated with many diseases such as metabolic diseases, cancer, and neurodegeneration [8]. In the process of protein ubiquitination, the ubiquitin-loaded E2 (E2-Ub) binds to the catalytic domain of an E3 ligase. The E3 ligase also interacts with the target protein via its substrate binding site and promotes transfer of Ub from E2 onto the targets. This process is repeated to transfer more Ub monomers to one of the seven lysine (K) residues (K6, K11, K27, K29, K33, K48 and K63) of the terminally attached Ub, forming distinct poly-Ub chains. Of these linkages, K48- and K63-linked-Ub (K48-Ub and K63-Ub) are well studied. K48-Ub primarily facilitates protein degradation via the ubiquitin proteasomal system (UPS), whereas K63-Ub mostly regulates activity, localization, and protein–protein interaction of the targets [9,10]. In mammals, about 600–700 E3 ligases are known that contain E2-binding RING- and HECT-domains, and many of these enzymes are well characterized [10,11]. A distinct family of E3 ligases contain the E2-binding U-box domain that resembles RING but lacks the Zn2+-coordinating residues [12]. U-box E3 ligases are prevalent in plants where they regulate stress responses [13]. Mammals only have seven U-box ligases, of which carboxy-terminus of HSC70 interacting protein (CHIP), pre-mRNA processing factor 19 (PRP19) and ubiquitin conjugation factor E4B (Ube4B) have been partially characterized. These ligases regulate protein quality control, DNA repair and apoptosis [[14], [15], [16], [17]]. Moreover, neuron-specific expression of Ube4B has been shown to augment obesity and hepatic steatosis in mice [18].
Ube4A, a member of the mammalian U-box domain E3 ligase family, has been implicated in metabolic and gastrointestinal diseases, and cancer [[19], [20], [21], [22], [23], [24], [25]]. Loss-of-stability mutations in Ube4A were identified in a population of obese and intellectually disabled subjects and the promoter region of Ube4A was found to be hypermethylated in severely obese children, indicting its downregulation in this disease [21,22]. Downregulation of Ube4A was also noted in the pancreatic islets of a population of diabetic patients [23]. Moreover, Ube4A has also been identified as an intestinal autoantigen in Crohn's disease [24].
Cell biological and biochemical studies showed that Ube4A regulates various protein targets via ubiquitination. It facilitates degradation of the antiviral protein viperin and the tumor suppressor protein P53, the transcription factor nuclear factor erythroid 2 like 1 (NRF1) [[26], [27], [28]]. Ube4A also mediates K63- and K48-Ub of proteins that facilitate DNA repair [29]. However, in vivo functions of Ube4A are unknown, primarily due to lack of appropriate mouse models to study this novel protein. Thus, by generating whole-body Ube4A knockout (UKO) mice, here we investigated the impact of Ube4A deletion on the adverse metabolic effects of high fat diet (HFD) and sought to identify the mechanisms by which this novel E3 ligase regulates metabolism and insulin signaling.
2. Methods
2.1. Animal studies
All animal studies were approved (approval numbers: 2732 and 2739) by the Saint Louis University Institutional Animal Care and Use Committee (IACUC). Mice were housed at 23 °C under 12-hour light/dark cycles and fed ad libitum with free access to water and were in good health condition throughout the duration of treatment. Eight-week-old, chow-fed mice were fed HFD (Kcals: fat 61.6% carbohydrates 20.3% and protein 18.1%, details in the materials section) for 10 or 16 weeks as indicated. After the feeding period, mice were fasted for 5 h followed by euthanasia by carbon dioxide asphyxiation. Serum and tissues were collected, weighed, and preserved following standard procedures for the assessment of various parameters [[30], [31], [32]].
2.2. Generation of UKO mice
The Ube4A gene is in the chromosome 9 of Mus musculus. Using CRISPR/Cas9 technology [33,34], two LoxP sites were inserted at the 5′- and 3′-ends of the last three exons, encoding the U-Box domain of Ube4A. The insertions were confirmed by next generation sequencing. Joining of the 5′- and 3′-ends of the targeting locus generated two groups of C57BL6J mice: i) Ube4A-Flox (LoxP sites encompassing the targeting locus) and ii) UKO (complete deletion of the targeting locus of 9859 base pairs). Multiple lines of chimeric UKO mice were cross-bred with WT C57BL6J mice to obtain heterozygous Ube4A-Flox or UKO mice, which were inbred to generate respective homozygous lines. For genotyping, Ub1, Ub3 and Ub4 primers were used. The WT and UKO mice showed band sizes of 345 (Ub3-Ub4) and 482 (Ub1-Ub4) base pairs, respectively, and the UHet mice showed both bands. Both lines were confirmed by sequencing the genotyped bands. UKO mice on C57BL6J background and their age-matched WT littermates were used in this study. Primer sequences used were:
Ub1 – GTAAAATGCTTGCCACACAGACATGAGTG
Ub3 - CCAGTCCAGGCATCTCATCAAGACATTTC
Ub4 - CATGTTATGATGACCCCAGCCATGAAACG.
2.3. Body weight and composition
Weekly body weight was measured and fat, lean, and fluid mass of mice were measured before and after the HFD feeding using the Minispec LF-NMR (Brucker Optics) analyzer [30].
2.4. Glucose, insulin, and glucagon tolerance tests (GTT, ITT and GgTT)
These tests were performed in male mice on the 4th, 6th, and 8th week of HFD-feeding, respectively to monitor the progress of insulin resistance and hyperglycemia. For GTT, glucose (2 g/kg BW, i.p.) was injected in 16 h-fasted animals. For ITT or GgTT, human insulin (0.75 U/kg BW, i.p.) or glucagon (15 μg/kg BW, i.p.) was injected in 5 h-fasted mice. In female mice, GTT and ITT were performed on the 12th and 14th week of HFD feeding. GTT was also performed in young (2-month-old), chow-fed, male and female mice. In all the experiments, blood glucose levels were measured by glucometer by puncturing tail veins of mice before and after the indicated time periods of injection [30].
2.5. Whole-body energy expenditure
Energy expenditure was measured in a cohorts of male mice on the 6th week of HFD-feeding and in young (2-month-old), chow-fed mice following a published method [32]. Mice were placed individually in metabolic cages at 23 °C with a precise thermostatic control in a Comprehensive Laboratory Monitoring System (CLAMS; Columbus Instruments) and were acclimatized for 2 days. Afterwards, oxygen consumption, (VO2), carbon dioxide release (VCO2), respiratory exchange ratio (RER), Energy expenditure (EE) and locomotor activity were measured for indicated time periods. VO2 and EE were calculated with and without normalizing with lean mass. Average food intake was assessed by measuring the food before and after the study. The average daily intake of kcals per mouse was also calculated [30].
2.6. Tissue collection and assessment of metabolic and liver injury parameters
Adipose tissue, liver and other tissues were isolated following standard procedures [[30], [31], [32]]. Blood was collected from 4 h-fasted mice by cardiac puncture, and serum was prepared following standard procedure. Serum TAG, NEFA, and total cholesterol were measured at the Mouse Metabolic Phenotyping Centers (MMPC), University of Cincinnati, College of Medicine Pathology & Laboratory Medicine. Serum concentrations of insulin, AST and ALT were determined using commercial kits [30]. TAG from liver was measured using a commercial assay kit after isolating liver lipids following a standard procedure [35].
2.7. Hematoxylin and Eosin (H&E) staining and NAFLD feature scoring
These studies were done following our previously published protocols [30,32]. NAFLD features were scored using a semiquantitative rodent scoring system [36]. Hepatic steatosis and hypertrophy were scored as: 0 (<5%), 1 (5%–33%), 2 (33%–66%), 3 (>66%). Number of inflammatory foci/200 X field was also scored. The numbers shown in the table denote an average of the individual scores in N = 5 mice/cohort.
2.8. Sirius red staining and analysis of total collagen content
Paraffin-embedded liver sections were stained with 0.1% Sirius Red in saturated picric acid for 2 h. Slides were then washed in water, dehydrated with ethanol and xylene, and mounted. The degree of collagen accumulation was assessed by morphometric analysis, as we described previously [30]. Briefly, about 15–20 nonoverlapping images randomly selected from each liver section were captured at 20X magnification. The same threshold was applied to all images. Sirius Red staining was quantified by digital image analysis using ImageJ software (NIH). The amount of collagen in the UKO livers was expressed relative to the amount of collagen in corresponding WT control group.
2.9. Lipidomics studies
Lipids, extracted from liver tissues [37], were spiked with an internal lipid standard mix that consists of phosphatidylcholine (20:0/20:0), phosphatidylethanolamine (14:0/14:0), sphingomyelin (d18:1/17:0), cholesteryl ester (17:0), ceramide (17:0), lysophosphatidylcholine (17:0), phosphatidylserine (14:0/14:0), triglyceride (17:0/17:0/17:0), phosphatidylglycerol (14:0/14:0) and diglyceride (20:0/20:0). Lipid extracts were dried under nitrogen and were dissolved in methanol: isopropanol (1:1) for lipidomic analysis. Lipids were separated on an Accucore C30 column (2.1 × 150 mm) at 35 °C. Mobile phase A had 60% acetonitrile, 40% water, 10 mM ammonium formate and 0.1% formic acid. Mobile phase B was comprised of 90% isopropanol, 10% acetonitrile with 2 mM ammonium formate, and 0.02% formic acid with a gradient. Untargeted lipidomics was performed using a Q-Exactive mass spectrometer (Thermo Scientific). Data-dependent mass spectrometry-mass spectrometry (ddMS2), the top 10 most abundant peaks from a full MS1 scan were acquired in both positive and negative ion modes. Full scan MS1 was performed with chromatogram peak width set at 7s, scan range 200–1200 m/z, AGC target 1 × 106, resolution 70000, and maximum injection time 246 ms. For ddMS2 negative ion analyses, parameters were resolution 17500, maximum injection time 54 ms; AGC target 2 × 105; isolation window 1.0 m/z; normalized collision energy (NCE) 20, 30, and 40; and dynamic exclusion at 10s. Similar parameters were used for positive ion mode ddMS2 analyses, except NCE was set to 20 and 40. Ions present in blank injections were excluded. MS data were analyzed using Xcalibur Qual Browser (Thermo Scientific). Untargeted LC-MS data were processed using LipidSearch 4.1 (Thermo Scientific) [38]. Both positive and negative MSdd.raw (data-dependent) files were used for lipid identification. Peak areas were normalized to internal lipid standards for each lipid class. Only the lipid classes that were normalized to internal standards were selected for further analysis. Each identified lipid was manually validated by investigating MS/MS fragmentation, the compatible retention time for lipid class, and acceptable peak shape [39]. Data scaling was set to autoscaling (mean-centered and divided by the standard deviation of each variable). The significant species were selected using the cut-off of 1.5-fold change and p-value <0.05.
2.10. RNA-Seq and quantitative RT-PCR (qRT-PCR) studies
For RNA-Seq, total RNA samples were prepared, indexed, pooled, and sequenced on an Illumina NovoSeq [30]. Gene expression was quantified using Kallisto [40]. Differential gene expression analysis was performed using an R edgeR package with a raw gene count matrix and were normalized using the TMM (The trimmed mean of M-values) method. Differential expressions were examined using a negative binomial generalized linear model. p-values were reported with likelihood test. For Gene Set Enrichment Analysis (GSEA), genes were ranked based on fold changes and p-values derived from the edgeR analysis. Enrichment analysis was performed using an R package “fgsea” along with the MSigDB C2 V7.5.1 gene sets [41]. qRT-PCR studies were done following a standard, SYBR green-based, ΔΔCt method, using our previously published protocol [30].
2.11. Primary hepatocyte isolation and hepatic glucose production assay
Glucose production was assessed following a standard protocol [42]. Briefly, primary hepatocytes from 8-week-old, chow fed WT and UKO male mice were isolated by perfusing the liver in situ with 1 mg/ml collagenase via the portal vein [30,42]. Hepatocytes, counted and plated onto collagen coated 24-well plates (100,000 cells/well), were washed with PBS, and incubated with DMEM without glucose, glutamine, pyruvate, and phenol red. After 2 h, the media was replaced with basal DMEM with glucagon (100 ng/ml) and sodium pyruvate (5 mM) for 3 h. Thereafter, media was collected, and glucose concentration was assessed with a kit. Glucose levels are expressed as μg glucose/mg cell protein/h.
2.12. Fatty acid oxidation and glycerolipid synthesis in hepatocytes
Fatty acid oxidation in WT and UKO primary hepatocytes was measured by the production of [3H]2O from [3H]-palmitate [43]. Briefly, PBS-washed primary hepatocytes (5 × 105 cells/well in a 12-well plate) were incubated with 200 μl of PBS containing [3H]-palmitate (10 μCi, bound to fatty acid free albumin - 125 μM, palmitate:albumin 2:1 with carnitine - 100 μM) for 2 h. Afterwards, the media was collected and 200 μl of cold 10% trichloroacetic acid was added. After centrifugation, the supernatant (350 μl) was mixed with freshly prepared NaOH (6 M−55 μl), loaded onto an ion-exchange resin column (DOWEX-400) and washed with 1.7 ml of water. The eluate was counted in a liquid scintillation counter. Counts were normalized by the protein content per well and expressed as fold change.
After removal of media for quantifying fatty acid oxidation, cell lysates were collected, resuspended in PBS (200 μl), and lipids were extracted as described [37]. Aliquots of the organic fraction dissolved in CHCl3/CH3OH (2/1: vol/vol) were applied to thin layer chromatography (TLC) with standards for TAG and DAG (TLC: solvent - Hexane: Diethyl ether: Acetic acid – 80:20:1 vol/vol). After 30 min, bands containing TAG and DAG were scraped from the TLC plate into scintillation vials and radioactivity was counted [44]. Counts were normalized to the cellular protein content and expressed as fold change.
2.13. Hepatic fatty acid uptake assay
Hepatocytes (2.5 × 105 cells/well) were serum starved for 2 h, washed with Krebs Ringer buffer (KRB with 0.1% BSA) and incubated in KRB containing a radioactive mix ([3H]-oleic acid - 2 μCi/ml, oleate - 100 μM and BSA - 200 μM) for indicated time periods. Afterwards, cells were washed with KRB (+0.5% BSA) and 0.1 M NaOH (1 ml) was added. After 30 min, radioactivity of the cell suspension (800 μl) was counted and normalized with total protein. Data was presented as fold change, considering the average change in WT-5 min as 1 [45].
2.14. Mitochondrial oxygen consumption rate (OCR) in brown adipocytes
Stromal vascular fractions (SVF), isolated from the brown adipose tissue (BAT) of 8-week-old mice, were cultured in DMEM/F12 containing FBS (10%) and penicillin/streptomycin and were differentiated to adipocytes using media containing insulin (5 μg/ml), indomethacin (125 μM), dexamethasone (2 μg/ml), 3-isobutyl-1-methylxanthine (0.5 mM), rosiglitazone (0.5 μM), and 3,3′,5-triiodo-l-thyronine (T3, 1 nM). After 2 days, the media was replaced with a media containing insulin and T3, which was replaced every 2 days until full differentiation [32]. OCR was measured in mature adipocytes in a Seahorse Extracellular Flux Analyzer in an XF assay medium supplemented with pyruvate (5 mM) and glucose (2.5 mM). After baseline measurements, 3 sequential injections were applied to the wells: oligomycin (complex V inhibitor, 1 μg/ml), FCCP (uncoupler, 1 μM), and antimycin A (complex III inhibitor, 0.8 μM) combined with rotenone (complex I inhibitor, 3 μM) [32].
2.15. Insulin signaling studies in mice, hepatocytes, and adipocytes
Overnight (16 h)fasted WT and UKO mice were injected once with human insulin (2U/kg BW, i.p.) or PBS. After 15 min, mice were euthanized, and tissues were collected for immunoblot and immunoprecipitation studies. To test insulin signaling in primary cells, primary hepatocytes and SVF from BAT were isolated as described previously [30,32]. SVF was differentiated to adipocytes in vitro following previously published methods [32]. Six hour-serum-starved hepatocytes and 16 h-serum-starved BAT-adipocytes were treated with insulin (10 nM) for indicated time periods, after which cells were lysed and proteins isolated and quantified. Proteins of interest were checked by immunoblotting.
2.16. Immunoprecipitation and immunoblotting studies
These studies were performed following our published protocols [[30], [31], [32]]. For immunoblotting studies, tissues or cells were lysed in a lysis buffer (20 mM Tris–HCl, pH 7.4; 150 mM NaCl; 1% Triton with protease and phosphatase inhibitors). Equal amounts (30 μg) of total protein from the lysates were loaded on a 10% SDS-PAGE and proteins of interest were detected by immunoblotting. Although antibodies, used for immunoprecipitation, are standard validated commercial antibodies, prior validation experiments were done to confirm their specificity by comparing them with IgG-control antibodies. For immunoprecipitation studies, 3 mg of total protein from mouse liver lysates were precleared with IgG-control antibodies and Protein A/G-agarose beads. The precleared lysates were incubated with primary antibodies for 16 h. After that, the lysates were incubated with Protein A/G-agarose beads for 2 h. Thereafter, beads were washed (3X) with the lysis buffer, and proteins were eluted by boiling the beads with LDS buffer. Proteins were resolved by 4–20% SDS-PAGE. Immunoprecipitated and coimmunoprecipitated proteins were determined by appropriate antibodies. To detect K63-Ub modification of Akt and APPL1, the membranes at and above the immunoprecipitated proteins were incubated with a K63-Ub antibody. Protein bands were detected in a Bio-Rad Chemidoc Imaging System. Densitometric analyses of protein bands were performed using the ImageJ software. For phosphorylation analysis, the band-intensity of the phospho-protein was normalized against total level of the same protein. For total protein analysis, the band intensity was normalized against a loading control.
2.17. Proteomic studies
The Ube4A-interactome from BAT was identified using our published methods [30]. Briefly, protein lysates were prepared from BAT tissue of young (2-month-old), chow-fed mice. Equal (2 mg) amounts of protein were used for immunoprecipitation (after preclearing) with Ig-control or Ig-Ube4A antibodies. Proteins co-precipitated with both antibodies were identified by LC-MS/MS and proteins that are specifically co-precipitated with Ube4A were considered as putative Ube4A interactors.
2.18. Immunofluorescence studies
WT and UKO fibroblasts were plated on poly-d-lysine coated coverslips placed inside a 6-well cell culture plate and grown to 80% confluency. Serum starved (16 h) cells were treated with PBS or insulin (10 nM) for 15 min. The cells were rinsed in 1xPBS immediately after the treatment, fixed in 4% PFA. Fixed cells were washed 3X in PBS and blocked (1 h) with a blocking buffer (1xPBS/5% normal goat serum/0.3% Triton X-100). Cells were then incubated (16 h) with p-AKT-S473 antibody (1:250) at 4 °C. After that, cells were washed 3X in PBS and incubated (1 h) with a secondary antibody (Alexa Fluor 488, 1:1000 + DAPI 1:1000) solution in the dark at room temperature. Thereafter, cells were washed 3X in PBS, and the coverslips were mounted on slides using Prolong Gold Antifade mounting media. Images were captured in a Confocal microscope (Leica SP8 TCS STED 3X) at 63X magnification.
2.19. Cloning of Ube4A in various expression vectors and production of adenovirus (AV)
All clones were used after sequence confirmation. The human Ube4A cDNA clone, purchased commercially, was PCR amplified and cloned into a GST-tagged mammalian and bacterial expression vectors pCMV-GST and pGEX-6P2 using SalI-NotI restriction sites [32]. Human HA-ubiquitin (Ub) [46] was also cloned into the pGEX-6P2 vector. Ube4A was cloned into an adenoviral vector using the AdEasy system [47]. Briefly, the Ube4A cDNA was cloned into the pShuttle-CMV plasmid [48] using the XbaI-HindIII restriction sites. BJ5183-Ad-1 bacterial cells, containing the necessary adenoviral genes, were transformed with the recombinant pShuttle-CMV + Ube4A plasmid. The recombinant adenoviral plasmid was purified and transfected to HEK293-Ad cells to produce the virus.
2.20. Adenovirus (AV)-Ube4A overexpression in UKO fibroblasts and hepatocytes
For AV transfection in primary hepatocytes, media was replaced with fresh complete DMEM 2 h after plating. At this time, control-AV and Ube4A-AV treatments were done (10 μl AV supernatant/well in a 6 well plate). Twenty-four hours later, cells were serum starved for 16 h and treated with insulin (10 nM) for the indicated time. Cells were then harvested, and proteins were isolated for immunoblotting studies. For primary fibroblasts, control-AV and Ube4A-AV were treated similarly for 48 h. After that, cells were serum starved for 16 h. Insulin treatment and downstream processing were done following the procedure described for hepatocytes.
2.21. Ube4A overexpression and depletion in HEK293 cells
HEK293 cells, at 85% confluency, were transfected with GST-control and GST-Ube4A plasmid constructs in 10 cm dishes. Cells were lysed 48 h post-transfection, APPL1 was immunoprecipitated and proteins of interest were detected by immunoblotting. For Ube4A depletion studies, 85% confluent HEK293 cells were transfected with control siRNA or Ube4A siRNA (100 nM each). After 16 h, cells were co-transfected with the same amount of siRNA and 10 μg of pcDNA3 flag HA human Akt1 [49] plasmid. After 24 h, cells were lysed, and HA-Akt was immunoprecipitated with a HA-antibody magnetic bead conjugate. The immunoprecipitated proteins were extracted from the beads by heating the samples at 95 °C with the LDS buffer and β-mercaptoethanol and resolved in a 4–20% SDS-PAGE.
2.22. Purification of recombinant full length (FL) and U-box deleted (ΔU-box) Ube4A and HA-ubiquitin (Ub)
GST-tagged FL-Ube4A, ΔU-box-Ube4A and HA-Ub containing pGEX-6P2 plasmids were transformed separately into the E. coli strain BL21 (DE3). Recombinant proteins were purified by slightly modifying a published method [29]. Recombinant proteins were either eluted from the beads and detached from the GST-tag using the elution buffer (50 mM Tris pH 7.4, 150 mM NaCl and PreScission protease) or were kept as GST-tagged proteins on the beads, depending on the requirements. Purified soluble proteins were stored at −80 °C. Proteins on beads were kept at 4 °C and were used fresh in the experiments.
2.23. In vitro ubiquitination assay
The reaction mixture contained recombinant proteins (Ub - 800 ng; E1/Uba1 - 350 ng; E2/UbcH5a [50] - 500 ng; GST-Ube4A – 70 ng, ΔU-box Ube4A – 140 ng, and Akt – 300 ng), Tris-Cl (25 mM, pH 7.4), NaCl (50 mM), Mg-ATP (5 mM), and dithiothreitol (DTT – 1 mM). GST-Ube4A or ΔUbox-Ube4A was immobilized on glutathione-agarose beads. The reaction mixture was incubated for 4 h at 37 °C. After that, the mixture was separated from Ube4A/ΔU-box-Ube4A-beads by low centrifugation. The supernatant was carefully removed, mixed with Laemmli buffer, and set to boil at 95 °C for 4 min. K63-Ub and AKT were detected by immunoblotting analysis.
2.24. Statistics
Exploratory experiments were used to determine sample size with ample statistical power. Animals were excluded if they showed any signs of sickness. Numbers of mice (n) used in experiments are indicated in the legends. Immunoblots were quantified using ImageJ software (NIH). Data are presented as mean ± s.e.m within dot plots. Each symbol represents an individual sample. For multiple comparisons, one-way or two-way ANOVA with Holm-Šidák multiple comparison test and for 2 independent data sets, Two-tailed unpaired Student's t-test were used. Area under curve analysis and statistical significance was calculated in GraphPad Prism, 8.2.1.
3. Results
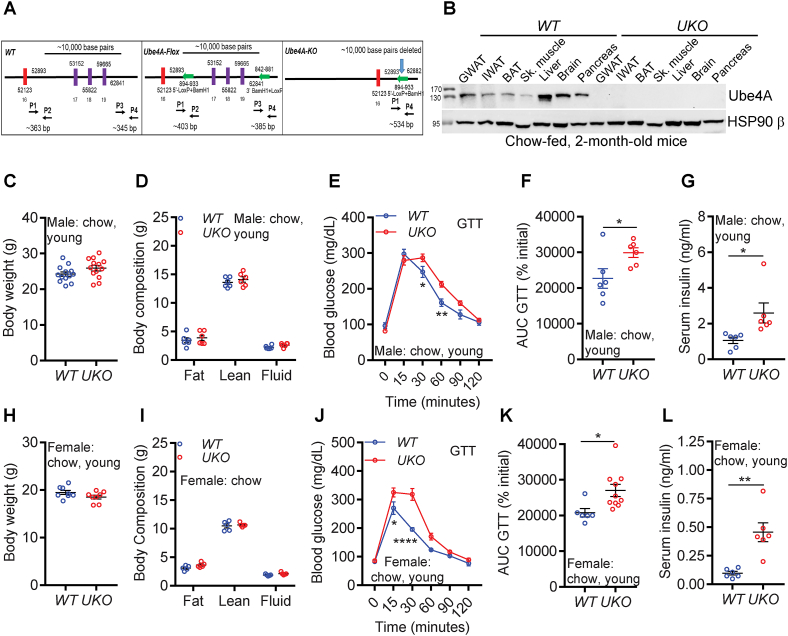
Ube4A deletion does not alter body weight but causes mild insulin resistance in young, chow-fed mice:UKO mice were generated on a C57BL6J background by CRISPR/Cas9-mediated targeting of U-box and 3′-UTR of the Ube4A locus in chromosome 9 (Figure 1A). Genotyping and DNA sequencing confirmed deletion of the targeted locus (Figs. S1A and B). Among metabolic tissues, Ube4A is highly expressed in the gonadal adipose tissue (GWAT) and liver in young (2-month-old), chow-fed mice (Figures 1B and S1C). Ube4A protein is absent in tissues from UKO mice (Figs. 1B and S1D). UKO mice are viable, fertile, active and do not exhibit any noticeable health concerns.
Figure 1.
Ube4A deletion does not alter body weight but causes mild insulin resistance in young, chow-fed mice.
A. CRISPR/Cas9-mediated targeting of the U-Box and part of the 3′-UTR of the Ube4A locus in the mouse chromosome 9.
B. Expression levels of the Ube4A protein in tissues of young, chow-fed, WT and UKO mice. Figure represents data from N = 3 mice.
C. Body weight of young, chow-fed male mice (n = 13/cohort).
D. Body composition of young, chow-fed male mice (n = 6/cohort).
E. Raw values of the GTT in young, chow-fed male mice (n = 6/cohort).
F. Area under curve (AUC) values of the GTT (% initial) (n = 6/cohort).
G. Serum insulin levels in 5 h-fasted, young, chow-fed male mice (N = 6/cohort).
H. Body weight of young, chow-fed female mice (n = 8/cohort).
I. Body composition (fat, lean and fluid mass) of young, chow-fed female mice (n = 5/cohort).
J. Raw values of the GTT in young, chow-fed female mice (n = 6–10/cohort).
K. AUC values of the GTT (% initial) in young, chow-fed female mice (n = 6–10/cohort).
L. Serum insulin levels in 5 h-fasted young, chow-fed female mice (N = 6/cohort).
Young, chow-fed male WT and UKO mice exhibit similar body weights and composition (Figure 1C,D). Fasting (16 h) blood glucose levels are also similar in both genotypes (Fig. S1E, time 0). However, the clearance of injected glucose (glucose tolerance test, GTT) is slightly slower in male UKO mice (Figures. 1E,F and S1E). Conversely, serum insulin levels are significantly higher in the UKO mice (Figure 1G).
Characterization of young, chow-fed female mice obtained similar results. In general, body weight of female mice are slightly lower than male mice although no genotype specific differences in body weight or composition are observed in females ((Figure 1,H,I). However, like males, female UKO mice also exhibit slightly diminished uptake of exogenous glucose (GTT) despite the presence of higher levels of insulin in their serum (Figure 1J-L and S1F). These results suggest that Ube4A deletion causes mild hyperinsulinemia and insulin resistance in both sexes of young, chow-fed mice.
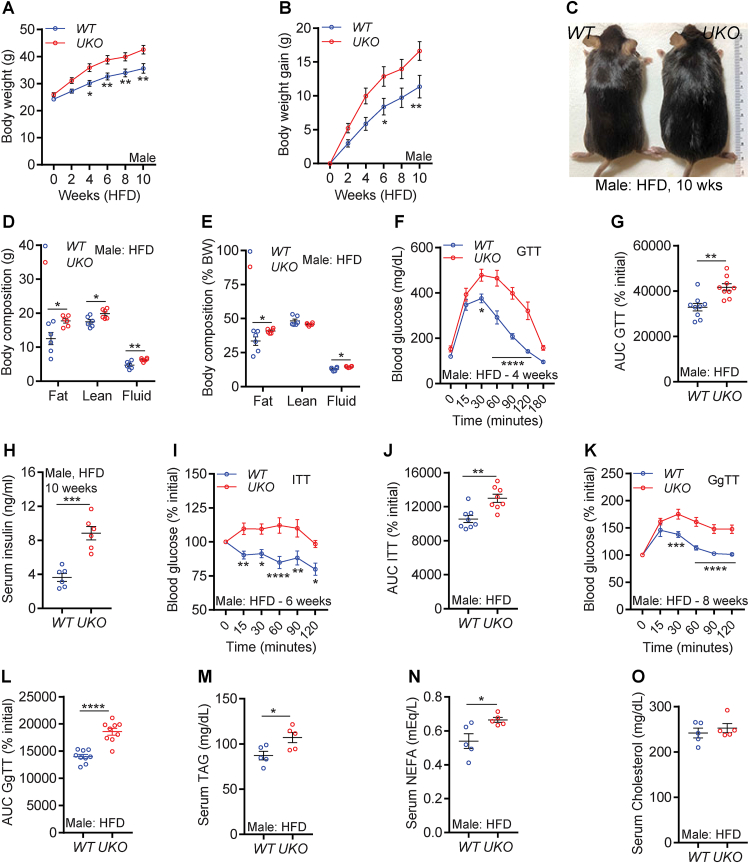
Ube4A deletion augments HFD-induced obesity and insulin resistance in male mice: Mild hyperinsulinemia and modest reduction in glucose uptake was observed in young, chow-fed UKO mice, which prompted us to test whether HFD feeding aggravates these traits in the knockouts. To examine this, male WT and UKO mice were fed a HFD (60% kcals from fat) for 10 weeks [31,32]. Interestingly, HFD-fed UKO mice gain body weight faster than the WT littermates (Figure 2A–C). HFD feeding augments increase in total and percent (of total body mass) fat mass in UKO mice compared to WT mice (Figures 2D and S2A). Total lean mass is increased in UKO mice, although the percentage lean mass is similar in both genotypes (Figure 2D,E). Fluid mass, a minor component of the total body mass, is slightly higher in the knockouts (Figure 2D,E). Thus, male UKO mice are prone to HFD-induced weight gain primarily due to increased fat mass.
Figure 2.
Ube4A deletion augments HFD-induced obesity and insulin resistance in male mice.
A-B. Weekly body weight and weight gain of HFD-fed male mice (n = 13/cohort).
C. Representative image of HFD-fed male mice after conclusion of the study.
D-E. Total and percent (over total body weight) body composition of HFD-fed male mice (N = 6/cohort).
F. Raw values of the GTT in HFD-fed male mice (n = 9/cohort). GTT was performed in 4-week-HFD-fed mice.
E. AUC values of the rate (percent of 0 time point) of decrease in blood glucose levels in a GTT in HFD-fed male mice (n = 9/cohort).
G. Serum insulin levels in 5 h-fasted male mice after 10-week of HFD feeding (n = 6/cohort).
H. Rate (percent of 0 time point) of decrease in blood glucose levels in an insulin tolerance test (ITT) in HFD-fed male mice (n = 8/cohort). ITT was performed in 6-week-HFD-fed mice.
I. AUC values of the ITT presented in Figure 2H (n = 9/cohort).
sJ. Rate (percent of 0 time point) of increase in blood glucose levels in a glucagon tolerance test (GgTT) in HFD-fed male mice (n = 9/cohort). GgTT was performed in 8-week-HFD-fed mice.
K. AUC values of the GgTT presented in Figure 2J (n = 9/cohort).
L-N. Serum levels of TAG, NEFA and total cholesterol in HFD-fed male mice.
The number of mice (n) used are presented as individual datapoints. Mean ± s.e.m. shown within dot plots. For multiple comparisons, two-way ANOVA with Holm-Šidák multiple comparison test and for two independent data sets, Two-tailed unpaired Student's t-test. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
Next, we tracked the progression of hyperglycemia in HFD-fed mice. HFD feeding exacerbates glucose intolerance in UKO mice (Figures 2F,G and S2A,B, assessed on the 4th week of HFD). Serum insulin levels (measured in 10-week-HFD-fed mice after euthanasia) were substantially higher in UKO mice (Figure 2H). Moreover, exogenous insulin treatment (insulin tolerance test, ITT, performed on the 6th week of HFD) reduces blood glucose concentrations much less efficiently in HFD-fed UKO mice (1.5%) compared to WT controls (20%) (Figures 2I,J and S2C). The injected concentrations of glucose and insulin in GTT and ITT were normalized to body weight of mice. Thus, it is conceivable that part of the observed changes in GTT could arise due to administration of more glucose in the HFD-fed knockout cohorts [51]. However, hyperinsulinemia or even higher concentration of injected insulin (normalized to body weight) in ITT failed to decrease blood glucose levels in HFD-fed UKO mice. Furthermore, chow-fed UKO mice, despite receiving the same concentration of glucose as WT mice, exhibit impaired glucose uptake (Figure 1E,J). These results together suggest that insulin sensitivity is impaired in UKO mice.
Conversely, HFD-fed UKO mice are hypersensitive to glucagon-induced hepatic glucose production, evidenced by augmented increase in blood glucose levels in the knockouts following glucagon injection (glucagon tolerance test, GgTT, evaluated on the 8th week of HFD) (Figure 2K,L and S2D). Accordingly, glucose production is increased in hepatocytes isolated from chow-fed, young, UKO mice, suggesting Ube4A regulates glucagon signaling in a cell autonomous manner (Fig. S2E). HFD-induced increases in serum levels of other metabolites such as TAG and NEFA are higher in male UKO mice whereas cholesterol levels are similar in both genotypes (Figure 2M−O). HFD feeding upregulates Ube4A expression in GWAT and liver (Figs. S2F and G), indicating that Ube4A may be having a protective role in the early stage of obesity and insulin resistance.
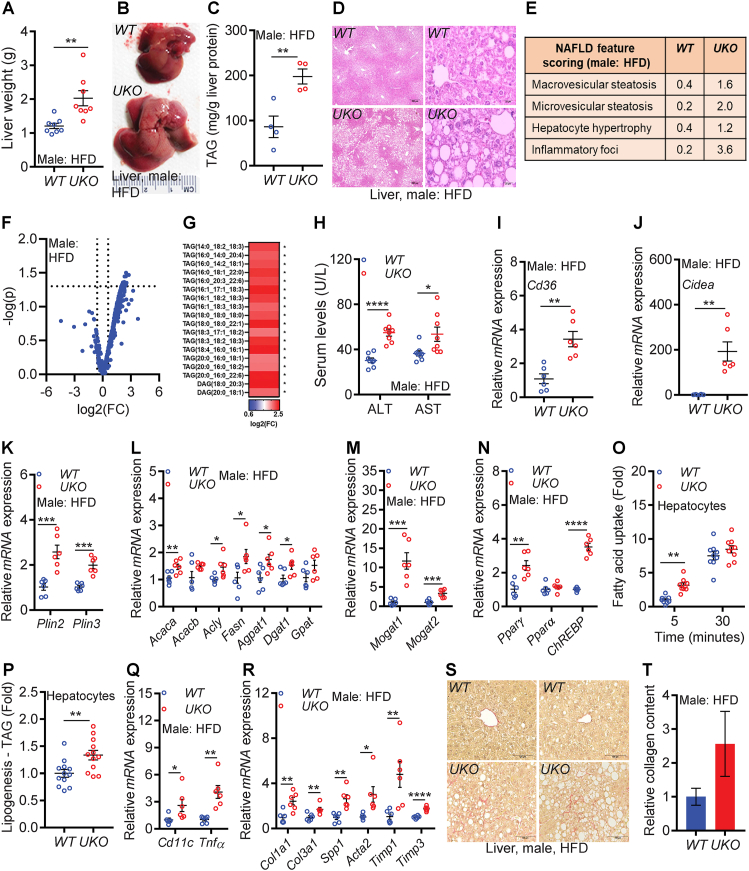
Ube4A deletion exacerbates HFD-induced hepatic steatosis and liver injury in male mice due to increased fatty acid uptake, lipogenesis, and lipid droplet stabilization: Due to high expression of Ube4A in metabolic tissues, especially in liver and adipose tissue, these tissues were thoroughly characterized in HFD-fed male UKO mice after euthanasia (Figure 3, Figure 4). Ten weeks of HFD feeding caused hepatomegaly to a greater extent in UKO mice compared to WT mice (Figure 3A,B). HFD feeding for 10–12 weeks has been shown to cause only mild hepatomegaly in male WT mice [52,53], which is in line with our observation (Figure 3A,B). Liver triglyceride levels are higher in UKO compared to WT mice (Figure 3C). Histology and NAFLD feature scoring detected substantially more macro- and micro-steatosis, hepatocyte hypertrophy, and inflammatory foci in UKO livers compared to WT controls (Figure 3D,E). Untargeted lipidomic analyses showed that multiple glycerolipid species of intrahepatic TAG and DAG (diacylglycerol) are increased in UKO livers (Figure 3F,G and Table S1). Serum levels of the liver injury markers, aspartate, and alanine aminotransferase (AST and ALT) are also higher in male UKO mice (Figure 3H).
Figure 3.
Ube4A deletion exacerbates HFD-induced hepatic steatosis and liver injury in male mice with increased fatty acid uptake, lipogenesis, and lipid droplet stabilization.
A. Weight of liver in HFD-fed male mice (N = 8/cohort).
B. Representative image of liver of HFD-fed male mice.
C. Liver triglyceride (TAG) levels in HFD-fed male mice (N = 4/cohort).
D. Representative images of liver histology in HFD-fed male mice. Images were taken at 4X (left panel, scale bar: 250 μm) and 40X (right panel, scale bar: 25 μm) magnifications.
E. Average values of NAFLD feature scores in HFD-fed male mouse livers (N = 5/cohort).
F. Volcano plot from the lipidomics study in HFD-fed male mouse livers (N = 5/cohort).
G. Species of TAG and DAG in HFD-fed male WT and UKO livers (N = 5/cohort).
H. Serum levels of AST and ALT in HFD-fed male mice (N = 8/cohort).
I–N. Relative mRNA expression of lipogenic genes and transcription factors in HFD-fed male mice (N = 6/cohort).
O. Uptake of [3H]-oleate in hepatocytes under basal conditions (N = 4, experimental replicates from N = 3 male mice/cohort).
P. Incorporation of [3H]-palmitate into TAG in hepatocytes (N = 12 experimental replicates from N = 4 male mice/cohort).
Q-R. Inflammatory and fibrogenic gene expression in HFD-fed male mice (N = 6/cohort).
S-T. Representative images (20X, scale bar: 100 μM) of Sirius-Red staining in HFD-fed mouse livers. For quantification, the mean value of WT was set as 1 (n = 5/cohort).
The number of mice (n) are presented as individual datapoints. Mean ± s.e.m. shown within dot plots. For multiple comparisons, two-way ANOVA with Holm-Šidák multiple comparison test and for two independent data sets, Two-tailed unpaired Student's t-test. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
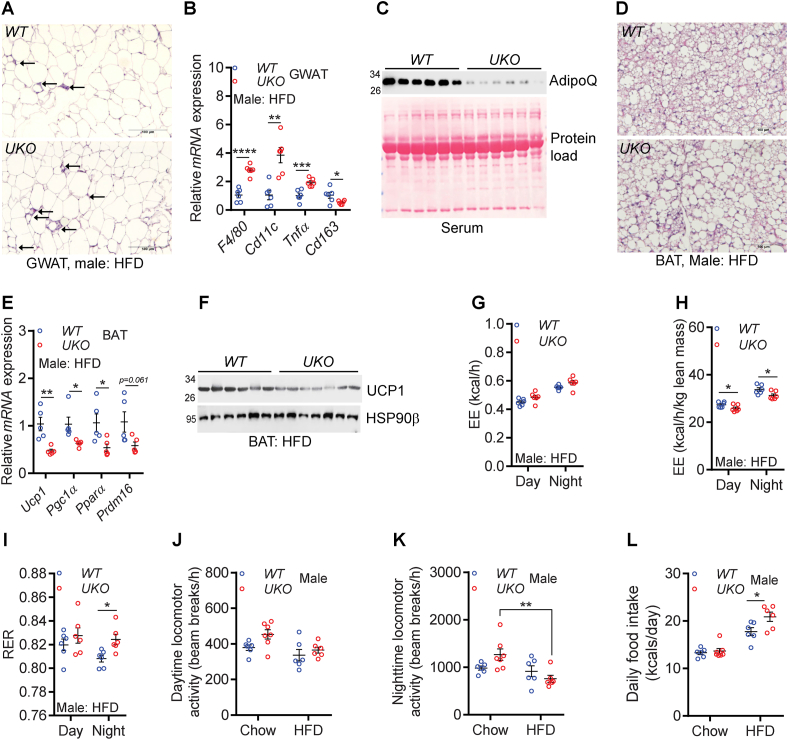
Figure 4.
HFD-fed male UKO mice exhibit increased adipose tissue metabolic dysfunction.
A. Representative histological images (20X, scale bar: 100 μm) of GWAT of HFD-fed mice. Arrows indicate crown-like structures.
B. Relative mRNA levels of M1 and M2 macrophage markers in the GWAT of HFD-fed male mice (N = 6/cohort).
C. Serum AdipoQ levels in HFD-fed male mice (N = 6/cohort). Serum samples were denatured and run on a denatured SDS-PAGE to detect monomeric AdipoQ.
D. Representative histological images (20X, scale bar: 100 μm) of HFD-fed mouse BAT.
E. Relative mRNA expression of the thermogenic machinery in the BAT of HFD-fed male mice (N = 5–6/cohort).
F. Levels of UCP1 protein in the BAT of HFD-fed male mice (N = 6–7/cohort).
G. EE (whole mouse) in HFD-fed mice (N = 6/cohort).
H. EE (normalized by lean mass) in HFD-fed mice (N = 6/cohort).
I. RER in HFD-fed mice (N = 6/cohort).
J-K. Average daytime and nighttime activity in chow- and HFD-fed mice (N = 6–7/cohort).
L. Average daily caloric intake in chow- and HFD-fed mice (N = 6–7/cohort).
The number of mice (n) used are presented as individual datapoints. Mean ± s.e.m. shown within dot plots. For multiple comparisons, two-way ANOVA with Holm-Šidák multiple comparison test and for two independent data sets, Two-tailed unpaired Student's t-test. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
Ube4A, through its E3 ubiquitin ligase activity, may alter stability and activity of transcription factors, which could change the expression of metabolic and other genes. Thus, to determine the pathways that are altered in the HFD-fed liver by Ube4A deletion, RNA-Seq and confirmatory qRT-PCR studies were performed. Genes involved in lipid metabolism and injury are altered in HFD-fed male UKO livers (Fig. S3A and Tables S2 and 3). qRT-PCR experiments confirmed that genes involved in lipid uptake (e.g., Cd36), lipid storage (e.g., Cidea, Plin2 and Plin3) and lipogenesis (e.g., Acaca, Acly, Fasn, Agpat1, Dgat1 and Mogat1 and 2) pathways and the lipogenic transcription factors (e.g., Pparγ and ChREBP) are upregulated in the knockout livers (Figure 3I-N). Consequently, UKO hepatocytes take up fatty acids and incorporate them into TAG more efficiently than WT hepatocytes in vitro (Figure 3O,P). Although genes encoding the mitochondrial energy oxidation pathway are generally downregulated (e.g., Cox7a2L and Atp5L) in the livers of HFD-fed knockouts, Ube4A does not appear to regulate fatty acid oxidation in hepatocytes in vitro (Figs. S3B and C and Tables S2 and 3).
Chronic intrahepatic fat accumulation is associated with inflammation in NAFLD [1,3]. The inflammatory genes (e.g., Cd11c and TNFα) and pathways are upregulated in HFD-fed UKO livers (Figures 3Q and S3A and Tables S2 and 3). TNFα is upregulated at the protein levels in the UKO livers, evidenced by increased expression of its transmembrane form (Figs. S3D and E). Besides generating the soluble, secretory form of TNFα, the transmembrane form can directly promote inflammatory responses by functioning as a ligand of the TNFα receptor of adjacent cells [54]. Expression of the macrophage marker F4/80 is largely similar in the liver of both genotypes (Fig. S3F). Perhaps immune cells other than macrophages such as neutrophils, T cells, B cells and dendritic cells, also known to accumulate in NAFLD livers [55], contribute to the inflammation observed in the UKO livers. Moreover, substantial upregulation of genes and pathways involved in hepatic stellate cell activation and fibrogenesis (e.g., Col1a1, Col3a1, Spp1, Acta2, Timp1 and Timp3) are observed in UKO livers (Figure 3R, Fig. S3A and Tables S2 and 3). Accordingly, ∼40% of UKO mice show moderate fibrosis (Figure 3S,T). Thus, Ube4A deletion exacerbates HFD-induced hepatic steatosis and liver injury in mice.
HFD-fed male UKO mice exhibit increased adipose tissue metabolic dysfunction: Whole-body Ube4A deletion aggravates metabolic dysfunction in adipose tissue. The GWAT of HFD-fed UKO mice accumulates more crown-like structures (indicative of inflammatory macrophage accumulation) and exhibit increased expression of total (F4/80) and pro-inflammatory macrophage (Cd11c, Tnfα) markers (Figure 4A,B). In contrast, expression of the anti-inflammatory and insulin-sensitizing macrophage marker Cd163 is reduced in the knockouts (Figure 4B). Levels of the insulin-sensitizing adipokine, adiponectin (AdipoQ), are also substantially diminished in serum and GWAT of UKO mice (Figures 4C and S4A-C). BAT depots of HFD-fed UKO mice accumulate more fat and express reduced levels of genes that promote energy expenditure (EE) and thermogenesis including the uncoupling protein 1 (UCP1) (Figure 4D–F and Fig. S4D). However, WT and UKO BAT-adipocytes (differentiated in vitro from stromal vascular fractions - SVF) exhibit similar OCR profiles, indicating Ube4A may not directly influence energy metabolism in healthy BAT-adipocytes in a cell autonomous manner under the tested conditions (Fig. S4E).
To test whether dysfunction of metabolic tissues alters energy expenditure (EE) in UKO mice, CLAMS studies were conducted in chow- and HFD-fed animals. Raw (whole mouse) values of oxygen consumption (VO2) and EE are similar in young, chow-fed WT and UKO male mice (Figs. S4F–H). The conclusion is same even when these values were normalized to lean mass, the largest contributor of EE (Figs. S4I–K). Respiratory exchange ratio (RER) is marginally lower in young, chow-fed UKO mice (Fig. S4L). HFD-fed WT and UKO mice also exhibit similar raw (whole mouse) values of VO2 and EE (Figures 4G and S4M,N). However, in this case, lean mass-normalized values of VO2 and EE are significantly lower in UKO mice (Figure 4H and Fig. S4O,P). HFD-fed UKO mice exhibit slightly higher nighttime RER (Figure 4I). Metabolic dysfunction in HFD-fed UKO mice may also arise from reduced locomotor activity as HFD feeding reduces activity in UKO but not in WT mice, especially during nighttime (Figure 4J,K). Total energy intake is similar in young, chow-fed WT and UKO mice (Figure 4L - chow). Raw values of total energy intake are slightly increased in HFD-fed knockouts, although it is slightly lower in the knockouts when normalized to total body weight (Figure 4L - HFD and Fig. S4Q). Thus, HFD-fed male UKO mice exhibit increased adipose tissue dysfunction, and reduced energy disposal and activity.
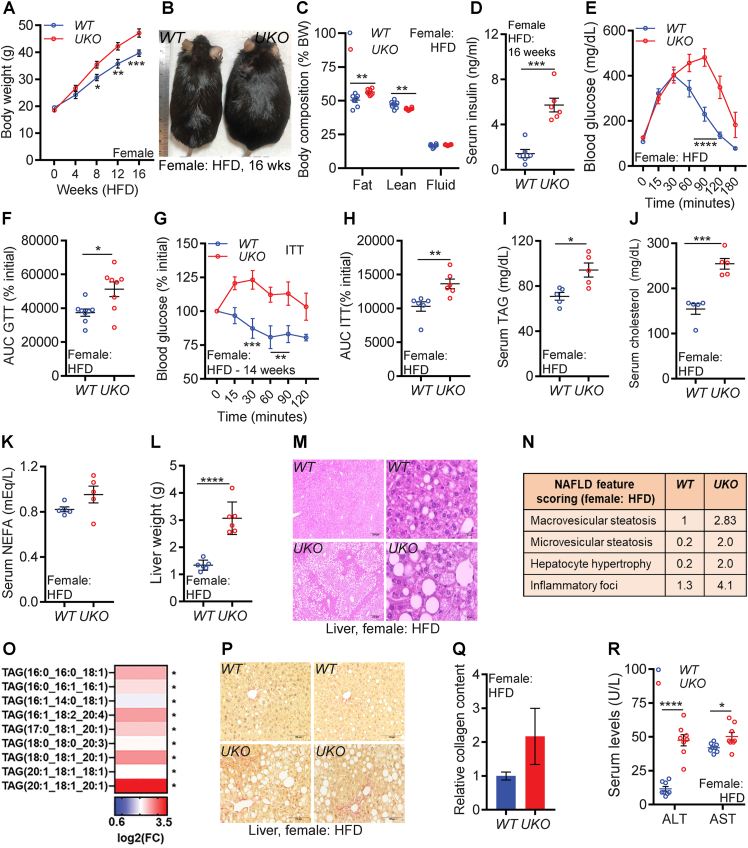
In female UKO mice, HFD feeding aggravates obesity, insulin resistance, hepatic steatosis, and liver injury: The current understanding of sex differences in obesity and NAFLD/NASH is insufficient and most mouse studies are conducted in male mice due to a markedly attenuated dysmetabolic phenotype in female mice fed a HFD. Therefore, we tested whether Ube4A deletion impacts HFD-induced metabolic parameters in female mice. Generally, HFD-fed WT C57BL6/J female mice exhibit milder insulin resistance and NAFLD compared to male mice. For example, the initial rate of HFD-induced weight gain (up to 4 weeks) in male mice was found to be faster than female mice [31]. Moreover, 16 weeks of high fat and high fructose containing diet induced hyperinsulinemia, insulin resistance, hepatic TAG accumulation and liver injury to a higher extent in male mice compared to female mice [56]. We observed that 8-week-HFD-fed female and 4-week-HFD-fed male WT mice exhibit similar glucose tolerance profiles, indicating that HFD-fed female mice take longer time to become insulin resistant compared to male mice (Fig. S5A, male-data is from Fig. S2B). Thus, to induce a full spectrum of metabolic dysfunction in female mice, we fed the HFD to female mice for 16 weeks.
HFD-induced gain in body weight is greater in the female knockouts due to increased (total and percent) fat mass (Figure 5A–C and Figs. S5B and C). Although total lean mass is increased, percent lean mass is decreased in HFD-fed female UKO mice. Total but not percent fluid mass is increased in female UKO mice (Figure 5A–C and Figs. S5B and C). Thus, increased fat but not lean or fluid mass corresponds to increased body weight in female knockouts. HFD caused hyperinsulinemia and insulin resistance to a greater extent in female UKO mice than WT mice. Serum insulin concentration is increased in HFD-fed female UKO mice (Figure 5D). Moreover, HFD-fed female knockouts exhibit reduced clearance of blood glucose in GTT and ITT (Figure 5E–H and Figs. S5D and E). Serum levels of TAG and cholesterol are significantly higher in female UKO mice, and NEFA is mildly increased in the knockouts in this condition (Figure 5I–K).
Figure 5.
In female UKO mice, HFD feeding aggravates obesity, insulin resistance, hepatic steatosis, and liver injury.
A. Weekly body weight in female mice (n = 8/cohort).
B. Representative image of HFD-fed mice.
C. Percent body composition of mice after 16 weeks of HFD-feeding (n = 6/cohort).
D. Serum insulin levels in 5 h-fasted mice after 16 weeks of HFD-feeding (n = 6/cohort).
E. Raw values of the GTT in HFD-fed mice (n = 8/cohort). GTT was performed in 12-week-HFD-fed mice.
F. AUC values of the GTT (% initial) in HFD-fed mice (n = 8/cohort).
G. Rate (percent of 0 time point) of decrease in blood glucose levels in an ITT in HFD-fed mice (n = 6/cohort). ITT was performed in 14-week-HFD-fed mice.
H. AUC values of the ITT presented in Figure 5G (n = 6/cohort).
I–K. Serum levels of TAG, NEFA and total cholesterol and in HFD-fed mice (n = 5/cohort).
L. Weight of liver in HFD-fed mice (n = 6/cohort).
M. Representative images of liver histology from HFD-fed mice. Images were taken at 4X (left panel, scale bar: 250 μm) and 40X (right panel, scale bar: 25 μm) magnifications.
N. Average values of NAFLD feature scores in HFD-fed mouse livers (N = 5/cohort).
O. Species of TAG in HFD-fed mouse livers (N = 5/cohort).
P-Q. Representative images (20X, scale bar: 100 μM) of Sirius-Red staining in HFD-fed mouse livers. For quantification, the mean value of WT was set as 1 (n = 5/cohort).
R. Serum levels of AST and ALT in HFD-fed mice (N = 8/cohort).
The number of mice (n) used are presented as individual datapoints. Mean ± s.e.m. shown within dot plots. For multiple comparisons, two-way ANOVA with Holm-Šidák multiple comparison test and for two independent data sets, Two-tailed unpaired Student's t-test. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, ∗∗∗∗P < 0.0001.
HFD feeding leads to robust hepatomegaly and steatosis in female UKO but not in WT mice (Figure 5L,M). NAFLD feature scoring showed that macro- and micro-steatosis, hepatocyte hypertrophy, and the inflammatory foci are substantially increased in female UKO livers (Figure 5N). Untargeted lipidomics revealed increased levels of TAG species in the female UKO livers (Figures 5O and S5F and Table S4). Surprisingly, some species of DAG and phosphatidyserine (PS) are decreased in female UKO livers (Fig. S5G). As was observed in males, hepatic collagen content is mildly increased in female UKO livers (Figure 5P,Q). Increased serum levels of AST and ALT in female UKO mice suggest that HFD feeding caused liver injury in the knockouts to a greater extent than WT mice (Figure 5R). Ube4A deletion also enhances lipid accumulation in various adipose tissue depots of female mice (Figs. S5H–J). Thus, HFD feeding aggravates obesity, insulin resistance, hepatic steatosis, and liver injury in female UKO mice.
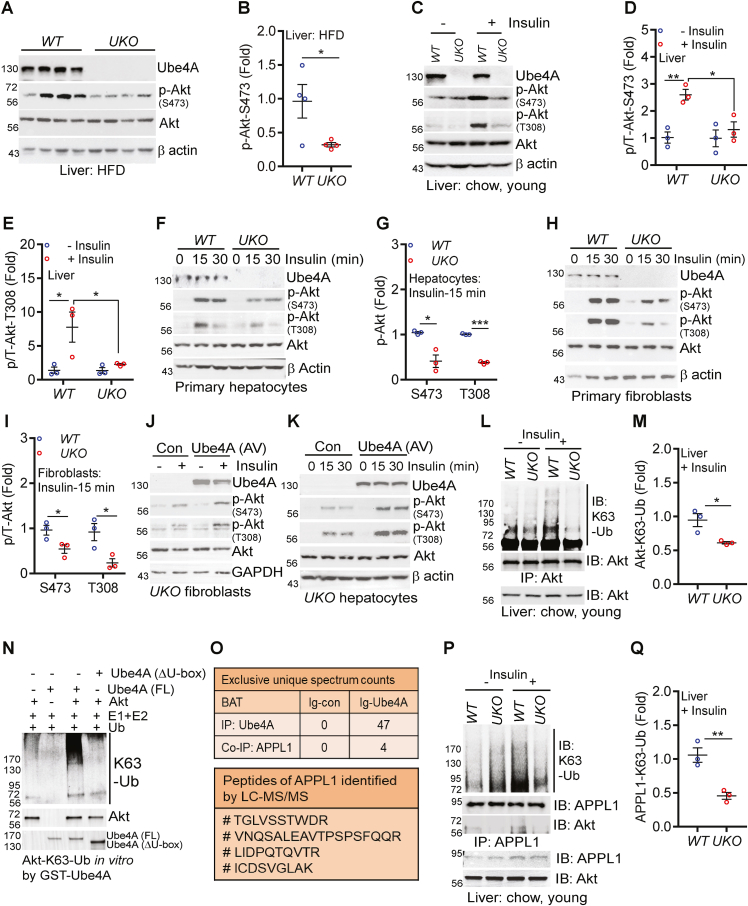
Ube4A mediates insulin signaling via K63-Ub of Akt and APPL1: The above results establish that Ube4A maintains metabolic homeostasis and insulin sensitivity presumably by ubiquitinating multiple proteins in key metabolic pathways. Here, we focused on the insulin signaling pathway as deletion of Ube4A impairs this process even in young, chow-fed mice. Insulin enhances glucose uptake and reduces gluconeogenesis and the protein kinase Akt is a major effector in this pathway [31]. Insulin activates Akt via its membrane translocation and subsequent phosphorylation at threonine 308 (T308) and serine 473 (S473) by phosphoinositide-dependent kinase (PDK1) and mammalian target of rapamycin complex 2 (mTORC2), although the S473 phosphorylation can be partially insulin-independent [57,58]. Stimulatory phosphorylation of Akt (S473) is diminished in liver and adipose tissue of HFD-fed knockouts (Figures 6A–B and S6A,B). Acute insulin treatment enhances Akt phosphorylation (S473 and T308) in the liver of young, chow-fed WT but to a much lower level in UKO mice (Figure 6C–E). Insulin-induced augmentation of phosphorylation of the Akt targets FOXO1 (Forkhead Box O1, regulates gluconeogenesis) and AS160 (Akt substrate of 160 kDa, regulates glucose uptake) are also diminished in the insulin-treated UKO liver (Fig. S6C). Furthermore, insulin-induced Akt phosphorylation is impaired in UKO primary hepatocytes and fibroblasts in vitro (Figure 6F–I). Immunofluorescence studies further indicated that insulin-induced S473 phosphorylation of membrane- and cytosol-Akt are decreased in UKO fibroblasts (Fig. S6D). Conversely, adenovirus (AV)-mediated overexpression of Ube4A restores insulin-induced Akt phosphorylation in fibroblasts and hepatocytes (Figure 6J,K).
Figure 6.
Ube4A mediates insulin signaling via K63-Ub of Akt and APPL1.
A-B. Stimulatory phosphorylation (S473) of Akt in the liver of HFD-fed mice (N = 4/cohort).
C-E. Acute insulin-induced stimulatory phosphorylation of Akt (S473 and T308) in the liver of young, chow-fed mice. Quantification of 3 independent experiments is shown.
F-G. Acute insulin-induced stimulatory phosphorylation of Akt (S473 and T308) in hepatocytes in vitro. Quantification of 3 independent experiments is shown.
H–I. Acute insulin-induced stimulatory phosphorylation of Akt (S473 and T308) in primary fibroblasts in vitro. Quantification of 3 independent experiments is shown.
J-K. Effects of adenovirus (AV)-mediated overexpression of Ube4A on acute insulin-induced stimulatory phosphorylation of Akt (S473 and T308) in primary fibroblasts and hepatocytes. In fibroblasts, the Akt-T308 antibody detected a non-specific band at a lower molecular weight, which did not respond to insulin.
L-M. K63-Ub of Akt in the liver of untreated and acute insulin-treated young, chow-fed mice. Data represents 3 independent experiments.
N. Ube4A (FL)-mediated K63-Ub of Akt in vitro. Ube4A-ΔU-box was used as a negative control. Data represents at least 3 independent experiments.
O. Top panel: LC-MS/MS-based identification of the exclusive unique spectrum counts of immunoprecipitated Ube4A and co-immunoprecipitated APPL1 from the BAT of young, chow-fed WT mice. Ig-control samples did not show any spectrum. Bottom panel: LC-MS/MS-based identification of the APPL1 peptides in the above experiment.
P-Q. Acute insulin-induced K63-Ub of APPL1 and APPL1-Akt binding in the liver of UKO mice. K63-Ub was detected in immunoprecipitated APPL1. Co-precipitated Akt was detected by immunoblotting in APPL1-immunoprecipitated samples. Data represents 3 independent experiments.
Each figure represents the conclusion from at least three independent experiments. Mean ± s.e.m. shown within dot plots. For multiple comparisons, two-way ANOVA with Holm-Šidák multiple comparison test and for 2 independent data sets, Two-tailed unpaired Student's t-test. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001.
We next sought to determine the mechanisms by which Ube4A facilitates Akt activation. Insulin and other growth factors stimulate various E3 ubiquitin ligases that modify Akt via K63-Ub, which facilitates its membrane translocation, phosphorylation, and activation [59,60]. Ube4A is known to regulate function of proteins via K63-Ub [29]. Therefore, we hypothesized that Ube4A activates Akt via K63-Ub modification of this kinase. Insulin stimulates K63-Ub modification of Akt in the liver of young, chow-fed, WT mice (Figure 6L). However, this modification is decreased in the liver of insulin-treated UKO littermates (Figure 6L,M). Moreover, Ube4A depletion (siRNA) reduces K63-Ub of overexpressed (HA-Akt) in HEK293 cells under basal conditions (Fig. S6E). Furthermore, bacterially purified full length, active (FL) Ube4A but not the catalytically inactive enzyme (ΔU-box) K63-ubiquitinates recombinant Akt in vitro (Figures 6N and S6F).
Signaling and proteomic studies in BAT from young, chow-fed mice provided additional mechanistic insights into this process. Like liver and hepatocytes, insulin-induced Akt activation is severely impaired in BAT tissues of UKO mice in vivo and UKO BAT-SVF-adipocytes in vitro (Figure, S6G-I). To detect the Ube4A-interacome in vivo, proteins immunoprecipitated from BAT lysates with the Ube4A antibody and a control antibody were detected by liquid chromatography-tandem mass spectrometry (LC-MS/MS). The Ube4A-interactome (proteins that were co-precipitated with the Ube4A antibody but not with the control antibody) contains several proteins that regulate cell metabolism and signaling. The APPL1 (adaptor protein phosphotyrosine interacting with PH-domain and leucine zipper 1) protein, which enhances insulin/Akt and adiponectin signaling [61,62], was among the top hits (Figure 6O). Insulin-induced Akt activation requires K63-Ub modification of APPL1, which increases its binding with Akt, leading to membrane translocation, phosphorylation, and activation of the kinase [61,63]. Therefore, we assessed K63-Ub modification of APPL1 in the UKO liver tissue. Insulin enhances K63-Ub of APPL1 and its binding with Akt in the liver of young, chow-fed WT but these processes are substantially hampered in UKO mice (Figure 6P,Q). Conversely, overexpression of GST-Ube4A increases these endogenous processes in HEK293 cells under basal conditions (Fig. S6J). These results strongly suggest that Ube4A is a bona fide E3 ubiquitin ligase of both Akt and APPL1, which K63-ubiquitinates these proteins upon insulin treatment, leading to the activation of Akt to promote insulin signaling.
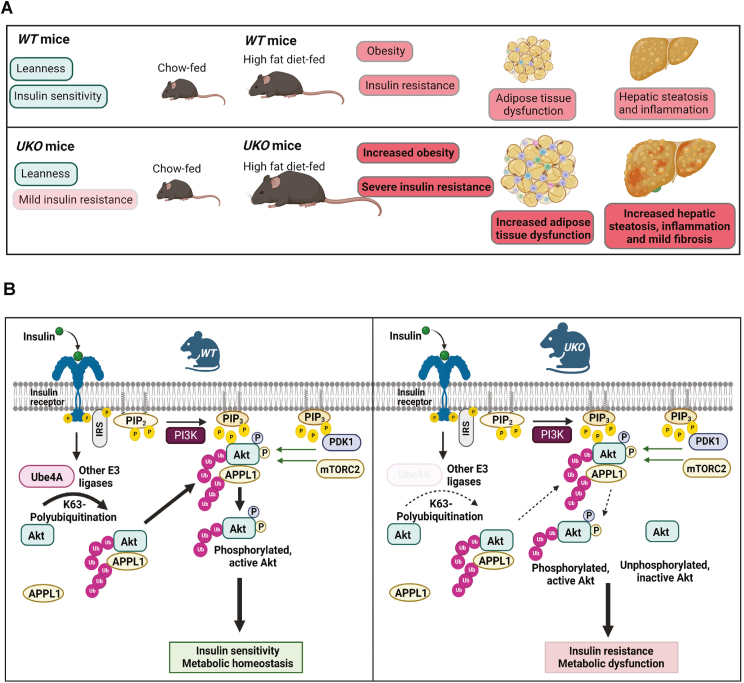
4. Discussion
We discovered that the U-box domain containing E3 ubiquitin ligase Ube4A is a novel regulator of metabolism and insulin signaling. Whole-body Ube4A deletion does not alter body weight but causes mild hyperinsulinemia and impairment in glucose uptake in young, chow-fed mice. HFD feeding aggravates insulin resistance in the knockouts and the HFD-fed UKO mice also exhibit increased obesity, adipocyte dysfunction, hepatic steatosis, and liver injury (Figure 7A). Thus, UKO mice could be used as a model to rapidly develop obesity, insulin resistance and NAFLD. Removal of Ube4A reduces insulin signaling in metabolic tissues and cells of young, chow-fed mice, indicating that metabolic dysfunction in UKO mice is caused at least partly by augmented insulin resistance. Mechanistically, Ube4A mediates K63-Ub modification of Akt and APPL1, promotes insulin-induced Akt activation whereas Ube4A deletion reduces K63-Ub modification of these proteins, leading to insulin resistance (Figure 7B).
Figure 7.
Ube4A maintains metabolic homeostasis by mediating insulin signaling.
A. Whole-body Ube4A deletion causes mild hyperinsulinemia and moderate impairment in glucose uptake in young, chow-fed mice. Moreover, HFD feeding develops severe insulin resistance in the knockouts. HFD-fed UKO mice also exhibit increased obesity, adipocyte dysfunction, hepatic steatosis, and liver injury.
B. In WT conditions, Ube4A mediates K63-Ub of Akt and APPL1, leading to Akt activation and insulin-induced metabolic effects. In UKO mice, K63-Ub of these proteins is disrupted, affecting insulin signaling and metabolism. Solid and open arrows indicate fully and partially active processes, respectively.
Ube4A is ubiquitously expressed in mice with its highest expression in the liver [19]. Both previous and current studies identified minimal expression of Ube4A in the mouse skeletal muscle, whereas in fetal humans, Ube4A is highly expressed in this tissue [20]. Ube4A is also expressed in the brain [19,20] (and this study). Thus, our study is in line with previous studies with additional information on Ube4A's expression in adipose tissue and pancreas. Based on the tissue expression profiles of Ube4A, the broad range of the observed phenotypes such as hyperinsulinemia, adipose tissue metabolic dysfunction, inflammation, activation of hepatic fibrogenic pathways, mild hyperphagia and reduced activity in HFD-fed UKO mice suggest that Ube4A may directly regulate functions of key metabolic tissues. Future studies will determine the effects of cell-specific Ube4A deletion on obesity, insulin resistance and NAFLD. For example, impact of hepatocyte-specific Ube4A deletion on NAFLD/NASH, adipocyte-specific Ube4A deletion on adipokine secretion and cold or β3 adrenergic receptor mediated thermogenesis with indirect effects on liver injury and pancreatic endocrine cell-specific deletion of Ube4A on insulin and/or glucagon secretion and signaling will be explored.
Both sexes of HFD-fed UKO mice exhibit substantially increased metabolic dysfunction and liver injury, although a few notable sex-specific differences were also observed. For example, decreased liver concentrations of some species of DAG and phosphatidyserine (PS) species and increased serum levels of cholesterol were observed specifically in female UKO mice. Lipid species, stereoisomers, or subcellular compartmentalization of DAGs exert distinct metabolic functions [64,65]. PS, an abundant membrane lipid, is essential for cell growth and survival while exofacial membrane-PS facilitates clearing of apoptotic cells [66]. PS, synthesized in the endoplasmic reticulum (ER), is transferred to mitochondria [66]. Interestingly, deficient transfer of PS between these organelles induces ER stress, NASH, and liver cancer in mice [67]. Thus, decreased PS may hamper clearing of apoptotic hepatocytes, induce ER stress, and aggravate liver injury in the female UKO mice [66]. Sex-specific differences in serum cholesterol may arise due to alterations in cholesterol synthesis, uptake, and/or lipoprotein metabolism in male and female UKO mice. A thorough comparison of all the metabolic parameters in both sexes of WT and UKO mice, fed chow, HFD or other forms of unhealthy diets (high fat, high fructose, high cholesterol etc.) for the same duration, is necessary to obtain direct comprehensive knowledge about sex-specific metabolic functions of Ube4A.
Akt regulates growth and metabolism via multiple mechanisms [68]. Akt-mediated hepatocyte survival reduces liver injury [69]. Moreover, it regulates brown fat organogenesis and thermogenesis via isoform- and cell specific-mechanisms [70,71]. APPL1 also has pleiotropic effects on metabolism [72]. APPL1's interaction with the adiponectin receptor mediates adiponectin signaling, which is crucial for metabolic health [62]. Adiponectin expression and secretion are severely diminished in HFD-fed UKO mice (this study). Therefore, impaired Akt and APPL1 activities may cause metabolic dysfunction via other mechanisms in addition to impacting insulin signaling. Whether K63-Ub regulates Akt's survival functions and APPL1-mediated adiponectin signaling in metabolic tissues will be assessed in future studies.
Further studies are required to obtain additional insights on the molecular mechanisms by which Ube4A regulates metabolism. The E3 ligases TRAF6 and SKP2-SCF mediate K63-Ub modification of Akt and/or APPL1 [59,60,63]. Ube4A can function as an E4 ligase to increase the length of ubiquitin chains on proteins, initiated by other E3 ligases [29]. Therefore, Ube4A may independently polyubiquitinate APPL1 and Akt in vivo and/or it may polyubiquitinate these proteins following their ubiquitination by TRAF6 and SKP2-SCF. Moreover, K63-Ub activates TRAF6 [73]. Thus, Ube4A may also activate Akt via this mechanism. Ube4A is localized in cytosol and nucleus [20,29], but it's in vivo targets in these compartments are unknown. We observed alterations in energy metabolic, inflammatory and fibrogenic genes in the UKO liver, suggesting that Ube4A may regulate relevant transcription factors via ubiquitination. Ube4A-interacting transcription factors and regulators, identified by the LC-MS/MS studies, will be validated to obtain further insights into its mechanism of actions.
Downregulation of Ube4A is associated with human obesity and diabetes [[21], [22], [23]], which may be consistent with our observation that deletion of Ube4A in mice promotes obesity and insulin resistance, especially when fed a HFD. We also observed that Ube4A is upregulated in 10-week-HFD-fed mice. Whether preventing Ube4A downregulation or increasing its expression might prevent or delay progression of metabolic disease still needs to be established. In pre-clinical mouse studies, levels of mRNA do not always correlate with protein expression, and protein levels often do not always correspond to functional consequences. Thus, future studies will be directed at determining activity, post-translational modifications, localization, and protein interactions of Ube4A in multiple tissues from patients with various stages of NAFLD and other diseases as well as healthy control subjects to obtain a more comprehensive understanding of the role of this novel protein in metabolism and metabolic diseases.
5. Conclusions
This study revealed that Ube4A protects mice from developing obesity, insulin resistance and NAFLD and determined the mechanism by which it contributes to normal insulin signaling. Thus, preventing the loss of Ube4A activity may prevent or reverse metabolic diseases.
Author contributions
A.C. conceived, outlined, and directed the project, and provided critical inputs throughout the study; M.C. generated UKO mice from chimeras and maintained mouse colonies; S.M., M.C. and E.N.M. performed majority of the experiments; J.H., M.J.J., H.L.C., K.P., B.U., A.J.L., D.C., and K.S.M. also performed experiments; K.S.M. shared reagents; A.C., S.M., M.C., E.N.M. and J.H. analyzed majority of the data; J.Z. analyzed RNA-Seq data; B.N-T., B.U. and D.C. analyzed and quantified liver histology data; D.A.F. directed the lipidomics study and analyzed the data; B.N.F., S.M. and A.J.L. analyzed lipid uptake, lipid oxidation and lipogenesis data; A.C. prepared final figures and wrote the manuscript. All the authors provided critical comments and approved the manuscript prior to submission. A.C. finalized the manuscript and submitted it to the journal.
Declaration of competing interest
B.T. has been an advisor or consultant for Alimentiv, Allergan, Allysta, Alnylam, Amgen, Arrowhead, Axcella, Boehringer Ingelheim, BMS, Coherus, Cymabay, Enanta, Fortress, Genfit, Gilead, High Tide, HistoIndex, Innovo, Intercept, Ionis, LG Chem, Lipocine, Madrigal, Medimmune, Merck, Mirum, NGM, NovoNordisk, Novus Therapeutics, pHPharma, Sagimet, Target RWE, 89Bio; he has stock options in HepGene; he has institutional research grants from Allergan, BMS, Cirius, Enanta, Genfit, Gilead, Intercept, Madrigal, NGM. B.N.F is a shareholder and a member of the Scientific Advisory Board for Cirius Therapeutics. The other authors do not have any conflict of interest.
Acknowledgements
This research was funded by the NIH grants R01DK103746 and R01DK132162; and startup fund, Liver Center Grant, and President Research Grant from Saint Louis University (SLU) to Anutosh Chakraborty. It was also supported by NIH shared instrumentation grant S10OD025246 to David Ford and Saint Louis University Liver Center support for Barbara Ulmasov. Work in the Finck lab was funded by NIH grants R01DK117657 and R01DK104995. The Core services of the Diabetes Research Center (P30 DK020579), Digestive Diseases Research Cores Center (P30 DK052574), and the Nutrition Obesity Research Center (P30 DK56341) at the Washington University School of Medicine also supported this work. We thank the Washington University Proteomics Shared Resource (WU-PSR) (R. Reid Townsend MD. Ph.D., director). The expert technical assistance of Petra Erdmann-Gilmore, Yiling Mi, and Rose Connors is gratefully acknowledged. WU-PSR is supported in part by the WU Institute of Clinical and Translational Sciences (NCATS UL1 TR000448), the Mass Spectrometry Research Resource (NIGMS P41 GM103422), and the Siteman Comprehensive Cancer Center Support Grant (NCI P30 CA091842). We thank Mike White, Monica Sentmanat and other members of the Genome engineering and iPSC Center and Transgenic, Knockout and Microinjection core facilities at the Washington University in Saint Louis for generating the Ube4A-Flox and UKO mouse lines. We thank the Genome Technology Access Center (GTAC) at the McDonnell Genome Institute at Washington University in Saint Louis for the RNA-Seq studies. The Center is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center. We also thank Dr. Nada Abumrad's lab for sharing the protocol for fatty acid uptake assay. We thank Dr. Grant Kolar and Caroline Murphy at the histology core facility for preparing the histology slides; Kathleen Donovan and colleagues from Comparative Medicine for helping with mouse maintenance; Dhruv Nagesh, Rashini Jayawardene and Ameya Padakanti for their assistance in protein purification and generating Ube4A constructs; and all members of the Department of Pharmacology and Physiology for their support. Serum levels of TAG, NEFA and cholesterol were measured at the Mouse Metabolic Phenotyping Centers (MMPC), University of Cincinnati, College of Medicine Pathology & Laboratory Medicine. pCDNA-HA-Ub, pShuttle-CMV and pcDNAflag-HA-Akt1 plasmids were purchased from Addgene (Yeh, Vogelstein and Sellers labs, respectively). The schematic models (Figure 7) were created with Biorender.com.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2023.101767.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Chakravarthy M.V., Neuschwander-Tetri B.A. The metabolic basis of nonalcoholic steatohepatitis. Endocrinol Diabetes Metab. 2020;3 doi: 10.1002/edm2.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loomba R., Friedman S.L., Shulman G.I. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell. 2021;184:2537–2564. doi: 10.1016/j.cell.2021.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman S.L., Neuschwander-Tetri B.A., Rinella M., Sanyal A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24:908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kusminski C.M., Bickel P.E., Scherer P.E. Targeting adipose tissue in the treatment of obesity-associated diabetes. Nat Rev Drug Discov. 2016;15:639–660. doi: 10.1038/nrd.2016.75. [DOI] [PubMed] [Google Scholar]
- 5.Brown M.S., Goldstein J.L. Selective versus total insulin resistance: a pathogenic paradox. Cell Metabol. 2008;7:95–96. doi: 10.1016/j.cmet.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 6.Vatner D.F., Majumdar S.K., Kumashiro N., Petersen M.C., Rahimi Y., Gattu A.K., et al. Insulin-independent regulation of hepatic triglyceride synthesis by fatty acids. Proc Natl Acad Sci U S A. 2015;112:1143–1148. doi: 10.1073/pnas.1423952112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuchs M., Sanyal A.J. Lipotoxicity in NASH. J Hepatol. 2012;56:291–293. doi: 10.1016/j.jhep.2011.05.019. [DOI] [PubMed] [Google Scholar]
- 8.Popovic D., Vucic D., Dikic I. Ubiquitination in disease pathogenesis and treatment. Nat Med. 2014;20:1242–1253. doi: 10.1038/nm.3739. [DOI] [PubMed] [Google Scholar]
- 9.Wang G., Gao Y., Li L., Jin G., Cai Z., Chao J.I., et al. K63-linked ubiquitination in kinase activation and cancer. Front Oncol. 2012;2:5. doi: 10.3389/fonc.2012.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Komander D., Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 11.Berndsen C.E., Wolberger C. New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol. 2014;21:301–307. doi: 10.1038/nsmb.2780. [DOI] [PubMed] [Google Scholar]
- 12.Patterson C. A new gun in town: the U box is a ubiquitin ligase domain. Sci STKE 2002. 2002:pe4. doi: 10.1126/stke.2002.116.pe4. [DOI] [PubMed] [Google Scholar]
- 13.Trenner J., Monaghan J., Saeed B., Quint M., Shabek N., Trujillo M. Evolution and functions of plant U-box proteins: from protein quality control to signaling. Annu Rev Plant Biol. 2022;73:93–121. doi: 10.1146/annurev-arplant-102720-012310. [DOI] [PubMed] [Google Scholar]
- 14.Paul I., Ghosh M.K. A CHIPotle in physiology and disease. Int J Biochem Cell Biol. 2015;58:37–52. doi: 10.1016/j.biocel.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 15.Idrissou M., Maréchal A. The PRP19 ubiquitin ligase, standing at the cross-roads of mRNA processing and genome stability. Cancers. 2022;14 doi: 10.3390/cancers14040878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Antoniou N., Lagopati N., Balourdas D.I., Nikolaou M., Papalampros A., Vasileiou P.V.S., et al. The role of E3, E4 ubiquitin ligase (UBE4B) in human pathologies. Cancers. 2019;12 doi: 10.3390/cancers12010062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaneko-Oshikawa C., Nakagawa T., Yamada M., Yoshikawa H., Matsumoto M., Yada M., et al. Mammalian E4 is required for cardiac development and maintenance of the nervous system. Mol Cell Biol. 2005;25:10953–10964. doi: 10.1128/mcb.25.24.10953-10964.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Susaki E., Kaneko-Oshikawa C., Miyata K., Tabata M., Yamada T., Oike Y., et al. Increased E4 activity in mice leads to ubiquitin-containing aggregates and degeneration of hypothalamic neurons resulting in obesity. J Biol Chem. 2010;285:15538–15547. doi: 10.1074/jbc.M110.105841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hatakeyama S., Yada M., Matsumoto M., Ishida N., Nakayama K.I. U box proteins as a new family of ubiquitin-protein ligases. J Biol Chem. 2001;276:33111–33120. doi: 10.1074/jbc.M102755200. [DOI] [PubMed] [Google Scholar]
- 20.Contino G., Amati F., Pucci S., Pontieri E., Pichiorri F., Novelli A., et al. Expression analysis of the gene encoding for the U-box-type ubiquitin ligase UBE4A in human tissues. Gene. 2004;328:69–74. doi: 10.1016/j.gene.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 21.Melo U.S., Bonner D., Kent Lloyd K.C., Moshiri A., Willis B., Lanoue L., et al. Biallelic UBE4A loss-of-function variants cause intellectual disability and global developmental delay. Genet Med. 2021;23:661–668. doi: 10.1038/s41436-020-01047-z. [DOI] [PubMed] [Google Scholar]
- 22.Huang R.C., Garratt E.S., Pan H., Wu Y., Davis E.A., Barton S.J., et al. Genome-wide methylation analysis identifies differentially methylated CpG loci associated with severe obesity in childhood. Epigenetics. 2015;10:995–1005. doi: 10.1080/15592294.2015.1080411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vinod M., Patankar J.V., Sachdev V., Frank S., Graier W.F., Kratky D., et al. MiR-206 is expressed in pancreatic islets and regulates glucokinase activity. Am J Physiol Endocrinol Metab. 2016;311:E175–E185. doi: 10.1152/ajpendo.00510.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakiyama T., Fujita H., Tsubouchi H. Autoantibodies against ubiquitination factor E4A (UBE4A) are associated with severity of Crohn's disease. Inflamm Bowel Dis. 2008;14:310–317. doi: 10.1002/ibd.20328. [DOI] [PubMed] [Google Scholar]
- 25.Caren H., Holmstrand A., Sjoberg R.M., Martinsson T. The two human homologues of yeast UFD2 ubiquitination factor, UBE4A and UBE4B, are located in common neuroblastoma deletion regions and are subject to mutations in tumours. Eur J Cancer. 2006;42:381–387. doi: 10.1016/j.ejca.2005.09.030. [DOI] [PubMed] [Google Scholar]
- 26.Yuan Y., Miao Y., Qian L., Zhang Y., Liu C., Liu J., et al. Targeting UBE4A revives viperin protein in epithelium to enhance host antiviral defense. Mol Cell. 2019 doi: 10.1016/j.molcel.2019.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Xie C., Long F., Li L., Li X., Ma M., Lu Z., et al. PTBP3 modulates P53 expression and promotes colorectal cancer cell proliferation by maintaining UBE4A mRNA stability. Cell Death Dis. 2022;13:128. doi: 10.1038/s41419-022-04564-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu X., Zou R., Zhang Z., Ji J., Li J., Huo X.Y., et al. UBE4A catalyzes NRF1 ubiquitination and facilitates DDI2-mediated NRF1 cleavage. Biochim Biophys Acta Gene Regul Mech. 2023;1866 doi: 10.1016/j.bbagrm.2023.194937. [DOI] [PubMed] [Google Scholar]
- 29.Baranes-Bachar K., Levy-Barda A., Oehler J., Reid D.A., Soria-Bretones I., Voss T.C., et al. The ubiquitin E3/E4 ligase UBE4A adjusts protein ubiquitylation and accumulation at sites of DNA damage, facilitating double-strand break repair. Mol Cell. 2018;69:866–878.e7. doi: 10.1016/j.molcel.2018.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukherjee S., Chakraborty M., Ulmasov B., McCommis K., Zhang J., Carpenter D., et al. Pleiotropic actions of IP6K1 mediate hepatic metabolic dysfunction to promote nonalcoholic fatty liver disease and steatohepatitis. Mol Metabol. 2021;54 doi: 10.1016/j.molmet.2021.101364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakraborty A., Koldobskiy M.A., Bello N.T., Maxwell M., Potter J.J., Juluri K.R., et al. Inositol pyrophosphates inhibit Akt signaling, thereby regulating insulin sensitivity and weight gain. Cell. 2010;143:897–910. doi: 10.1016/j.cell.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhu Q., Ghoshal S., Rodrigues A., Gao S., Asterian A., Kamenecka T.M., et al. Adipocyte-specific deletion of Ip6k1 reduces diet-induced obesity by enhancing AMPK-mediated thermogenesis. J Clin Invest. 2016;126:4273–4288. doi: 10.1172/jci85510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sentmanat M.F., Peters S.T., Florian C.P., Connelly J.P., Pruett-Miller S.M. A survey of validation strategies for CRISPR-Cas9 editing. Sci Rep. 2018;8:888. doi: 10.1038/s41598-018-19441-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Modzelewski A.J., Chen S., Willis B.J., Lloyd K.C.K., Wood J.A., He L. Efficient mouse genome engineering by CRISPR-EZ technology. Nat Protoc. 2018;13:1253–1274. doi: 10.1038/nprot.2018.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu H., Rimbert A., Palmer A.E., Toyohara T., Xia Y., Xia F., et al. GPR146 deficiency protects against hypercholesterolemia and atherosclerosis. Cell. 2019;179:1276–1288.e14. doi: 10.1016/j.cell.2019.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liang W., Menke A.L., Driessen A., Koek G.H., Lindeman J.H., Stoop R., et al. Establishment of a general NAFLD scoring system for rodent models and comparison to human liver pathology. PLoS One. 2014;9 doi: 10.1371/journal.pone.0115922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 38.Contrepois K., Mahmoudi S., Ubhi B.K., Papsdorf K., Hornburg D., Brunet A., et al. Cross-platform comparison of untargeted and targeted lipidomics approaches on aging mouse plasma. Sci Rep. 2018;8 doi: 10.1038/s41598-018-35807-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Breitkopf S.B., Ricoult S.J.H., Yuan M., Xu Y., Peake D.A., Manning B.D., et al. A relative quantitative positive/negative ion switching method for untargeted lipidomics via high resolution LC-MS/MS from any biological source. Metabolomics. 2017;13 doi: 10.1007/s11306-016-1157-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 41.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McCommis K.S., Chen Z., Fu X., McDonald W.G., Colca J.R., Kletzien R.F., et al. Loss of mitochondrial pyruvate Carrier 2 in the liver leads to defects in gluconeogenesis and Compensation via pyruvate-alanine cycling. Cell Metabol. 2015;22:682–694. doi: 10.1016/j.cmet.2015.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Djouadi F., Bonnefont J.P., Munnich A., Bastin J. Characterization of fatty acid oxidation in human muscle mitochondria and myoblasts. Mol Genet Metabol. 2003;78:112–118. doi: 10.1016/s1096-7192(03)00017-9. [DOI] [PubMed] [Google Scholar]
- 44.Kaibori M., Kwon A.H., Oda M., Kamiyama Y., Kitamura N., Okumura T. Hepatocyte growth factor stimulates synthesis of lipids and secretion of lipoproteins in rat hepatocytes. Hepatology. 1998;27:1354–1361. doi: 10.1002/hep.510270523. [DOI] [PubMed] [Google Scholar]
- 45.Smith J., Su X., El-Maghrabi R., Stahl P.D., Abumrad N.A. Opposite regulation of CD36 ubiquitination by fatty acids and insulin: effects on fatty acid uptake. J Biol Chem. 2008;283:13578–13585. doi: 10.1074/jbc.M800008200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kamitani T., Kito K., Nguyen H.P., Yeh E.T. Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem. 1997;272:28557–28562. doi: 10.1074/jbc.272.45.28557. [DOI] [PubMed] [Google Scholar]
- 47.McCommis K.S., Douglas D.L., Krenz M., Baines C.P. Cardiac-specific hexokinase 2 overexpression attenuates hypertrophy by increasing pentose phosphate pathway flux. J Am Heart Assoc. 2013;2 doi: 10.1161/jaha.113.000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He T.C., Zhou S., da Costa L.T., Yu J., Kinzler K.W., Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci U S A. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hsieh A.C., Bo R., Manola J., Vazquez F., Bare O., Khvorova A., et al. A library of siRNA duplexes targeting the phosphoinositide 3-kinase pathway: determinants of gene silencing for use in cell-based screens. Nucleic Acids Res. 2004;32:893–901. doi: 10.1093/nar/gkh238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhu Q., Yu T., Gan S., Wang Y., Pei Y., Zhao Q., et al. TRIM24 facilitates antiviral immunity through mediating K63-linked TRAF3 ubiquitination. J Exp Med. 2020;217 doi: 10.1084/jem.20192083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ayala J.E., Samuel V.T., Morton G.J., Obici S., Croniger C.M., Shulman G.I., et al. Standard operating procedures for describing and performing metabolic tests of glucose homeostasis in mice. Dis Model Mech. 2010;3:525–534. doi: 10.1242/dmm.006239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bar A., Kieronska-Rudek A., Proniewski B., Suraj-Prażmowska J., Czamara K., Marczyk B., et al. In vivo magnetic resonance imaging-based detection of heterogeneous endothelial response in thoracic and abdominal aorta to short-term high-fat diet ascribed to differences in perivascular adipose tissue in mice. J Am Heart Assoc. 2020;9 doi: 10.1161/jaha.120.016929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hussain A., Yadav M.K., Bose S., Wang J.H., Lim D., Song Y.K., et al. Daesiho-tang is an effective herbal formulation in attenuation of obesity in mice through alteration of gene expression and modulation of intestinal microbiota. PLoS One. 2016;11 doi: 10.1371/journal.pone.0165483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horiuchi T., Mitoma H., Harashima S., Tsukamoto H., Shimoda T. Transmembrane TNF-alpha: structure, function and interaction with anti-TNF agents. Rheumatology. 2010;49:1215–1228. doi: 10.1093/rheumatology/keq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huby T., Gautier E.L. Immune cell-mediated features of non-alcoholic steatohepatitis. Nat Rev Immunol. 2022;22:429–443. doi: 10.1038/s41577-021-00639-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ganz M., Csak T., Szabo G. High fat diet feeding results in gender specific steatohepatitis and inflammasome activation. World J Gastroenterol. 2014;20:8525–8534. doi: 10.3748/wjg.v20.i26.8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.White M.F., Kahn C.R. Insulin action at a molecular level - 100 years of progress. Mol Metabol. 2021;52 doi: 10.1016/j.molmet.2021.101304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petersen M.C., Shulman G.I. Mechanisms of insulin action and insulin resistance. Physiol Rev. 2018;98:2133–2223. doi: 10.1152/physrev.00063.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yang W.L., Wang J., Chan C.H., Lee S.W., Campos A.D., Lamothe B., et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science. 2009;325:1134–1138. doi: 10.1126/science.1175065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chan C.H., Li C.F., Yang W.L., Gao Y., Lee S.W., Feng Z., et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell. 2012;149:1098–1111. doi: 10.1016/j.cell.2012.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cheng K.K., Iglesias M.A., Lam K.S., Wang Y., Sweeney G., Zhu W., et al. APPL1 potentiates insulin-mediated inhibition of hepatic glucose production and alleviates diabetes via Akt activation in mice. Cell Metabol. 2009;9:417–427. doi: 10.1016/j.cmet.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 62.Mao X., Kikani C.K., Riojas R.A., Langlais P., Wang L., Ramos F.J., et al. APPL1 binds to adiponectin receptors and mediates adiponectin signalling and function. Nat Cell Biol. 2006;8:516–523. doi: 10.1038/ncb1404. [DOI] [PubMed] [Google Scholar]
- 63.Cheng K.K., Lam K.S., Wang Y., Wu D., Zhang M., Wang B., et al. TRAF6-mediated ubiquitination of APPL1 enhances hepatic actions of insulin by promoting the membrane translocation of Akt. Biochem J. 2013;455:207–216. doi: 10.1042/bj20130760. [DOI] [PubMed] [Google Scholar]
- 64.Eichmann T.O., Lass A. DAG tales: the multiple faces of diacylglycerol--stereochemistry, metabolism, and signaling. Cell Mol Life Sci. 2015;72:3931–3952. doi: 10.1007/s00018-015-1982-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Finck B.N., Hall A.M. Does diacylglycerol accumulation in fatty liver disease cause hepatic insulin resistance? BioMed Res Int. 2015;2015 doi: 10.1155/2015/104132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kay J.G., Fairn G.D. Distribution, dynamics and functional roles of phosphatidylserine within the cell. Cell Commun Signal. 2019;17:126. doi: 10.1186/s12964-019-0438-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hernández-Alvarez M.I., Sebastián D., Vives S., Ivanova S., Bartoccioni P., Kakimoto P., et al. Deficient endoplasmic reticulum-mitochondrial phosphatidylserine transfer causes liver disease. Cell. 2019;177:881–895.e17. doi: 10.1016/j.cell.2019.04.010. [DOI] [PubMed] [Google Scholar]
- 68.Schultze S.M., Jensen J., Hemmings B.A., Tschopp O., Niessen M. Promiscuous affairs of PKB/AKT isoforms in metabolism. Arch Physiol Biochem. 2011;117:70–77. doi: 10.3109/13813455.2010.539236. [DOI] [PubMed] [Google Scholar]
- 69.Liu W., Jing Z.T., Xue C.R., Wu S.X., Chen W.N., Lin X.J., et al. PI3K/AKT inhibitors aggravate death receptor-mediated hepatocyte apoptosis and liver injury. Toxicol Appl Pharmacol. 2019;381 doi: 10.1016/j.taap.2019.114729. [DOI] [PubMed] [Google Scholar]
- 70.Sanchez-Gurmaches J., Martinez Calejman C., Jung S.M., Li H., Guertin D.A. Brown fat organogenesis and maintenance requires AKT1 and AKT2. Mol Metabol. 2019;23:60–74. doi: 10.1016/j.molmet.2019.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sostre-Colón J., Uehara K., Garcia Whitlock A.E., Gavin M.J., Ishibashi J., Potthoff M.J., et al. Hepatic AKT orchestrates adipose tissue thermogenesis via FGF21-dependent and -independent mechanisms. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Deepa S.S., Dong L.Q. APPL1: role in adiponectin signaling and beyond. Am J Physiol Endocrinol Metab. 2009;296:E22–E36. doi: 10.1152/ajpendo.90731.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Talreja J., Samavati L. K63-Linked polyubiquitination on TRAF6 regulates LPS-mediated MAPK activation, Cytokine production, and bacterial clearance in toll-like receptor 7/8 primed murine macrophages. Front Immunol. 2018;9:279. doi: 10.3389/fimmu.2018.00279. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.