Abstract
Mycobacteria are nonflagellated gram-positive microorganisms. Previously thought to be nonmotile, we show here that Mycobacterium smegmatis can spread on the surface of growth medium by a sliding mechanism. M. smegmatis spreads as a monolayer of cells which are arranged in pseudofilaments by close cell-to-cell contacts, predominantly along their longitudinal axis. The monolayer moves away from the inoculation point as a unit with only minor rearrangements. No extracellular structures such as pili or fimbriae appear to be involved in this process. The ability to translocate over the surface correlates with the presence of glycopeptidolipids, a mycobacterium-specific class of amphiphilic molecules located in the outermost layer of the cell envelope. We present evidence that surface motility is not restricted to M. smegmatis but is also a property of the slow-growing opportunistic pathogen M. avium. This form of motility could play an important role in surface colonization by mycobacteria in the environment as well as in the host.
Although most mycobacteria are free-living saprophytic organisms, much of the research on this genus has focused on those species that are pathogenic to humans. These include obligate pathogens such as the leprosy bacillus, M. leprae, and the tubercule bacillus, M. tuberculosis, which kills more than 3 million people per year and infects one-third of the world population (8, 23). Others are opportunistic pathogens which occur naturally in the environment but can occasionally cause disease, especially in immunocompromised individuals. The most important of the opportunistic pathogens are the members of the M. avium-M. intracellulare complex, which are a leading cause of bacteremia in AIDS patients (21).
One of the most striking characteristics of mycobacteria is the enormous complexity of their cell envelope (reviewed in references 9 and 14). Extensive chemical analyses have shown that the cell wall of mycobacteria consists of three components. The outside layer is composed of mycolic acids, a complex mixture of long-chain α-branched β-hydroxy fatty acids which are arranged as a densely packed monolayer. The mycolic acids are covalently linked to arabinogalactan, which is in turn attached to the peptidoglycan layer. This complex cell wall is surrounded by a capsule of noncovalently bound polysaccharides, proteins, and a small amount of lipids, which include the species- and type-specific glycopeptidolipids (GPLs) and phenolic glycolipids. This unusual envelope provides mycobacteria with remarkable impermeability to external substances, a critical virulence determinant for these organisms.
While much effort has been placed on studying the functions of cell wall components in pathogenesis, little attention has been focused on the biological significance of the cell wall architecture for free-living mycobacteria. In nature most bacteria are associated with surfaces (12). The type of interaction between a bacterium and a surface, whether it attaches to it or moves on it, is largely determined by the nature of the bacterial cell surface. Bacteria have evolved a wide array of surface translocation modes (20), all of which require special surface structures or components, including flagella, pili and fimbriae, surfactants, slime, and capsules. Here we report for the first time that the fast-growing saprophytic species M. smegmatis and the slow-growing opportunistic pathogen M. avium have the ability to translocate on solid surfaces by a flagellum-independent spreading mechanism known as sliding (20). Spreading appears to require the presence of GPLs on the cell surface since rough strains of both species, which lack GPLs, do not exhibit this form of translocation. This form of motility is likely to play a significant role in the ability of mycobacteria to colonize surfaces in the environments as well as in the host.
MATERIALS AND METHODS
Strains and growth media.
M. smegmatis mc2155 (35) and its morphological variants were routinely grown in M63 salts medium (31) supplemented with 1 mM MgCl2, glucose (0.2 or 2%), Casamino Acids (0.5%), FeCl2 (10 μM), and a micronutrient solution (28), as indicated. Middlebrook 7H9 and 7H10 media (Difco) supplemented with ADC (22) were used to grow M. avium. M. avium 2151-SmD, SmT, Rg-0, and Rg-4 (7) were provided by J. Belisle.
Surface spreading assays.
M63 or 7H9 medium supplemented as indicated were solidified with 0.3% agar (Difco) or 0.1 to 0.8% ultrapure SeaKem LE agarose (FMC Bioproducts). Twenty-five milliliters of sterile medium that had been cooled to 65°C was dispensed per plate (9-cm diameter). Plates were allowed to sit at room temperature overnight prior to inoculation and were inoculated from single colonies by poking with a sterile toothpick or from liquid cultures after cells had been washed in M63 salts. Spreading was evaluated visually after incubation of parafilm-sealed plates at 37°C in a humidified incubator (50% relative humidity) for the indicated period of time.
Phase-contrast microscopy.
Cells on the surface of the growth medium were visualized with a Nikon Diaphot 200 inverted microscope. The images were captured with a black and white CCD72 camera integrated with a power Macintosh 8600-300 computer with video capability (Cupertino). Images were processed using Scion Image (Scion Corporation) and Photoshop 4.0.1 (Adobe) software.
Electron microscopy.
Formvar carbon-coated copper grids were gently placed on the surface of the solid growth medium directly over the spreading cells. After 1 min, the grids were carefully removed, rinsed twice in distilled water, and stained with 1% uranyl acetate or 2% phosphotungstic acid, as indicated, for 1 min. Negatively stained cells were visualized by using a JEOL 1200 EX, 80-kV transmission electron microscope.
Mixing experiments with GFP-labeled cells.
M. smegmatis mc2155 was transformed by standard procedures (22) with pGFP, a vector carrying a promoterless gfp gene (38) cloned into the shuttle vector pMVI203 (11a) or pGFP/O, a plasmid carrying a transcriptional fusion to gfp which results in detectable levels of green fluorescent protein (GFP) expression (25a). Four-day-old cultures of cells grown in M63–0.2% glucose–kanamycin (25 μg/ml) medium were mixed 1:100 (1 GFP-labeled cell per every 100 unlabeled cells), centrifuged, and washed twice in M63 salts. Twenty-five microliters of a 10−4 dilution was inoculated onto the surface of 0.3% agarose–M63 salts and –7H9 basal medium (without glycerol) plates. Phase-contrast and fluorescence microscopy analyses (200× magnification) were performed using a Nikon microscope equipped with episcopic-fluorescence attachment EFD-3 and a fluorescein isothiocyante filter. Images were captured with an Optronics DEI-750 color camera and processed with Scion Image and Photoshop software.
Isolation of GPLs and TLC.
GPLs were isolated from cells grown on the surface of 7H9–ADC–0.3% agarose plates as previously described (10). GPL profiles were analyzed by thin-layer chromatography (TLC) on silica plates (Alltech), using as developing solvent chloroform-methanol-water (90:10:1 by volume). After chromatography, lipids were visualized by spraying with 10% H2SO4 in ethanol and heating at 120°C. M. smegmatis GPLs were identified by comparison with published patterns of GPLs analyzed under the same conditions (17).
RESULTS
M. smegmatis spreads on the surface of semisolid agar plates.
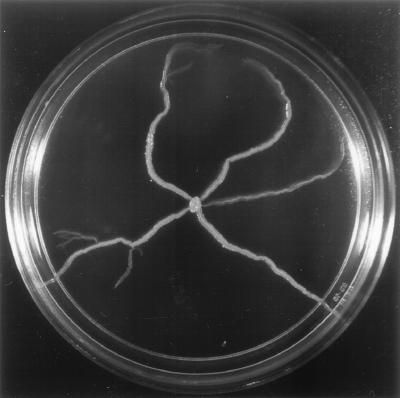
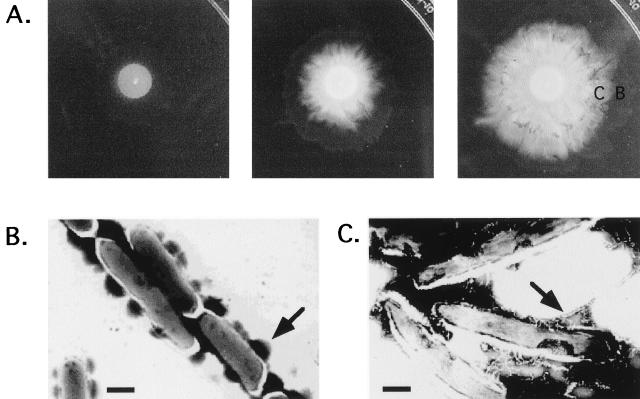
When M. smegmatis mc2155 was inoculated on a semisolid motility agar plate (0.3% agar) containing low levels of nutrients (such as M63 salts or 7H9 basal medium with no added carbon source), two distinct phases of growth were observed. Initially, bacterial growth occurred on the surface at the point of inoculation, as expected from nonswimming bacteria. After 3 to 4 days, however, a striking change occurred. Finger-like extensions appeared in the periphery of the colony and spread outwards from the initial inoculation point (Fig. 1). Phase-contrast microscopy revealed that the tips of the spreading fingers consist of a monolayer of cells translocating on the surface as a compact group. No discrete movement of individual cells was observed, in contrast to the jerky movements of twitching Pseudomonas aeruginosa or the forward/backwards movement of gliding Myxococcus xanthus (20). Concentrations of agar above 0.6% completely inhibited the spreading of mycobacteria, while replacing the agar by ultrapure agarose allowed reproducible spreading under a variety of conditions. Therefore, we used agarose as a solidifying agent in the medium for all the subsequent experiments. The ability of mycobacteria to translocate over surfaces had not been previously reported.
FIG. 1.
Macroscopic morphology of M. smegmatis mc2155 strain spreading on the surface of a motility agar plate. mc2155 was grown in 7H10, and a single colony was transferred with a toothpick to the center of a 0.3% agar plate containing 7H9 basal medium without any added carbon source. The plate was sealed with parafilm and incubated at 37°C for 2 weeks.
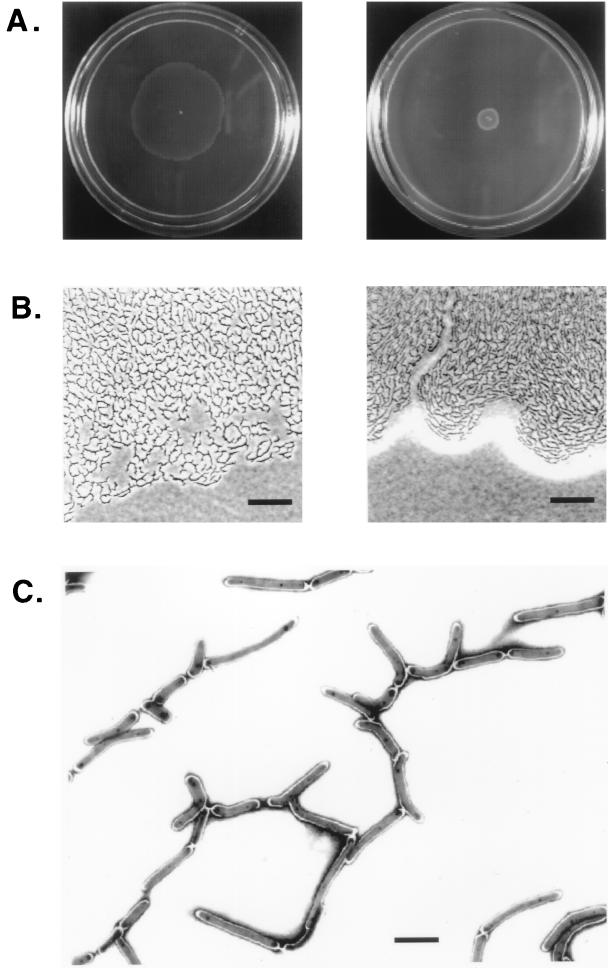
The extent of spreading of M. smegmatis on the surface of agarose plates depends on the degree of wetness.
M. smegmatis was able to spread on the surface of M63 salts plates (with no added carbon source) over a wide range of agarose concentrations (0.1 to 0.8%). In 0.1% agarose, the spreading cells appeared to sink into grooves on the soft surface and expanded as fingers reminiscent of those observed in motility agar plates (data not shown). Plates prepared with concentrations of agarose equal or above 0.2% were rigid enough to allow the spreading fronts to extend over a large surface, eventually surrounding the inoculation site with a circular halo (Fig. 2A). The diameter of the halo was inversely related to the agarose concentration, indicating that the wetness of the medium is a critical parameter affecting surface spreading of M. smegmatis. Indeed, the use of freshly prepared plates and a humidified incubator are critical for optimum spreading. The diameter of the halo correlates with the density at which cells are packed within the monolayer. Phase microscopy revealed that the halos produced in 0.3 and 0.8% agarose plates consist entirely of a monolayer of cells arranged as pseudofilaments which are more tightly packed at 0.8% agarose (Fig. 2B). A phase-bright slime covering the spreading halo was particularly noticeable in the high-percentage agarose plate. Perhaps the slime extracts moisture from the medium to create an appropriate surface for the cells to slide on, as has been proposed for other surface-translocating bacteria (11, 36).
FIG. 2.
Macroscopic and microscopic analysis of mc2155 spreading on the surface of agarose plates. (A) Halo formation on 0.3% (left) and 0.8% (right) agarose plates containing M63 salts with no added source of carbon. Plates were inoculated by poking a single colony from a 0.2% glucose M63 agar plate and transferring it to the center of the plate. The photograph was taken after 5 days of incubation at 37°C. (B) Phase-contrast images of the edges of the spreading halos shown in panel A. Bar, 25 μm. (C) Electron micrograph of cells spreading on a 0.3% agarose–M63 salts plate. A Formvar carbon-coated grid was placed directly over the spreading halo and cells were stained with 1% uranyl acetate. Bar, 2 μm.
A more detailed view of the arrangement of the spreading cells was obtained by electron microscopical analysis of grids that had been placed directly over the halo. Spreading cells are arranged in pseudofilaments by end-to-end connections along their longitudinal axis (Fig. 2C). However, the contact points between cells do not always coincide with the cell poles as would be expected if septum separation after cell division had not been complete. Rods are frequently curved, and no surface structures such as pili or fimbriae are observed. Rather, the whole mass of cells seems to be encased in an electron-light layer within which an amorphous material connecting groups of cells can be occasionally observed.
Spreading of M. smegmatis on a solid surface is accompanied by growth.
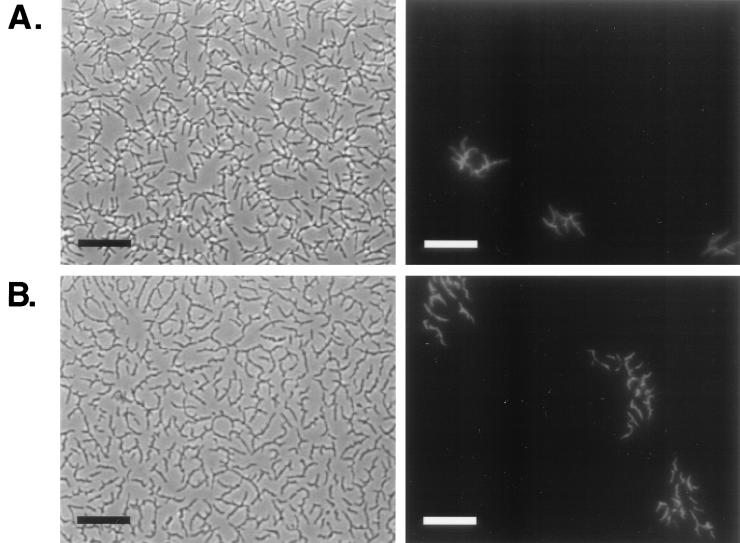
In order to address questions regarding growth rates and movement of individual cells within an expanding halo, we performed a series of mixing experiments in which a minority of the cells used as inoculum were labeled with GFP, allowing their identification by fluorescence microscopy. M. smegmatis cells were transformed with either pGFP, a plasmid containing a promotorless gfp, or pGFP/O, a plasmid with a transcriptional fusion of an M. smegmatis gene to gfp that results in detectable GFP expression. Cells containing pGFP/O exhibited uniform detectable GFP levels in all the cells of the population when growing as a halo in 0.3% agarose–M63 salts plates (data not shown). GFP-labeled cells were mixed 1:100 with unlabeled cells and plated in triplicate on 0.3% agarose–M63 and –7H9 (without glycerol or ADC) plates. Immediately after inoculation, GFP-labeled cells were present mostly as single cells within the inoculum, although small clumps (two to four cells) were also visible (data not shown). After 2 days of incubation the diameters of the halos were 2.4 ± 0.1 cm in M63 and 3.2 ± 0.3 cm in 7H9. At that time (Fig. 3), no single fluorescent cells were observed, but instead green cells were arranged in discrete small groups within the monolayers, indicating that growth had occurred in both media. On average the groups of green cells contained approximately 16 to 20 cells in M63 and 50 to 70 cells in 7H9, which corresponds to four and six doublings, respectively, after 2 days of incubation. A small number of larger groups of fluorescent cells, probably resulting from the growth of the clumps in the inoculum, were also observed. These results show that the formation of halos is accompanied by growth and that the faster growth observed in 7H9 correlates with a higher spreading rate. The carbon and energy sources supporting this growth are unknown but are unlikely to consist of carried-over liquid medium components, since the cells used in these experiments were washed repeatedly in M63 buffer prior to inoculation. These results also show that cells within the monolayer remain in the vicinity of their siblings. This very limited rearrangement of the spreading cells markedly contrasts with the high fluidity of cell-cell interactions in swarming Serratia liquefaciens or gliding M. xanthus, where similar mixing experiments showed isolated GFP-labeled cells within the moving population (16, 37).
FIG. 3.
Growth accompanies mycobacterial spreading. A 1:100 mix of GFP-labeled (light) and unlabeled (dark) mc2155 cells grown as described in Materials and Methods were plated on the surface of 0.3% M63 salts– (A) and 7H9 (with no added carbon source)– (B) agarose plates. Photographs were taken after 2 days of incubation at 37°C. Phase-contrast images showing the continuous spreading halo are on the left, and fluorescent micrographs of the same fields showing the locations of GFP-labeled cells are on the right. Bars, 25 μm.
M. smegmatis colony morphology variants exhibit altered spreading phenotypes.
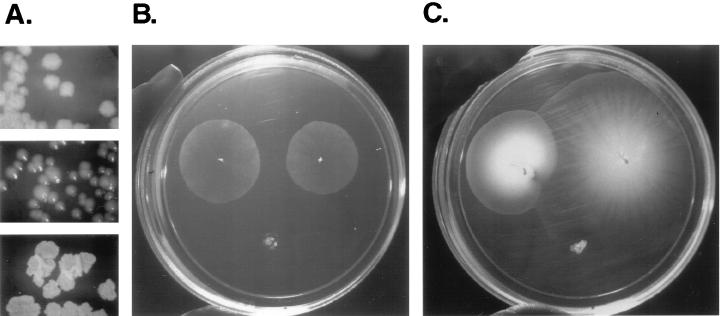
The capacity of the cells to spread over the growth surface is likely to be determined in part by the surface properties of the cells. Since differences in colony morphology in many bacterial species are associated with changes in cell surface components, we analyzed the spreading phenotypes of a collection of uncharacterized spontaneous M. smegmatis mutants previously isolated in our laboratory on the basis of their altered colony appearance. We chose two clones, Sm-1 and Rg-1, as representatives of the most severe morphological changes (Fig. 4A). The original M. smegmatis strain, mc2155, appears rugose but moist in 7H10 agar plates. In contrast, under the same conditions Sm-1 is moist and smooth while Rg-1 is rough and extremely dry. A comparison of the spreading phenotypes of these strains in M63 salts–0.3% agarose plates with no added carbon source is shown in Fig. 4B. Sm-1 was able to spread on the surface, producing halos that were very similar to those of mc2155. In contrast, Rg-1 was completely unable to spread and grew at the inoculation point as a densely packed mass of clumped cells.
FIG. 4.
Spreading phenotype of M. smegmatis colony morphology variants. (A) Colony morphology of mc2155, Sm-1, and Rg-1 on 7H10 agar plates. (B and C) Spreading phenotype of the morphology variants in 0.3% agarose plates containing M63 salts (B) or M63 with 2% glucose and 0.5% Casamino Acids (C). For both plates: mc2155, top left; Sm-1, top right; Rg-1, bottom.
Addition of nutrients (2% glucose and 0.5% Casamino Acids) had profound effects on the spreading behavior (Fig. 4C). While Rg-1 grew but did not spread, mc2155 and Sm-1 maintained their spreading capacity but the spreading zone was multilayered. While mc2155 appeared smooth and uniform, a star-like pattern irradiating from the inoculation point appeared in the Sm-1 halos, which were consistently larger than those of mc2155. By the time nutrients had been exhausted, the plate was covered by very dense masses of the spreading strains, while the growth of the nonspreading Rg-1 had been severely limited (results not shown). These results indicate first that the surface properties of the cells can severely affect their ability to spread on solid surfaces. In addition, they demonstrate that the ability to spread confers competitive advantage for surface colonization and access to nutrients since all three strains grow at similar rates in standard 2% agar plates of the same composition (which do not allow spreading).
Time-lapse movies of spreading halos under phase microscopy.
Addition of high levels of all nutrients to M63 plates (glucose, Casamino Acids, and iron and other micronutrients) led to a faster halo expansion. Under these conditions, it was possible to record time-lapse movies of phase-contrast images of the edge of a spreading halo. These movies (one of which is available at http://gasp.med.harvard.edu/smegmatis/sliding.html) clearly show a compact mass of cells sliding over the agar surface away from the inoculation point, with only minor rearrangements. The approximate speeds of spreading were 1.6 μm/min for mc2155 and 2.5 μm/min for Sm-1.
Pattern formation in the spreading halos growing in nutrient-rich medium.
In a nutrient-rich environment, first the halo spreads as a monolayer and then further growth converts it into a multilayer of densely packed cells. These sequential rounds of expansion result in the appearance of ring-like patterns in the halos, which are particularly conspicuous for Sm-1 spreading in 7H9–ADC–0.3% agarose plates (Fig. 5A). This pattern is reminiscent of that observed for swarming Proteus mirabilis colonies, which are known to result from developmental cycles of spreading and consolidation (39). Significant morphological differentiation exists between the cells in the monolayer surrounding the spreading colony and those in the densely packed areas of the interior. Under the electron microscope, the arrangement of cells within the periphery of the monolayer in rich medium appears similar to that observed in minimal M63 or 7H9 salts except for the abundance of negatively stained material irregularly associated with the cell surface (Fig. 5B). Cells in the interior of the colony, however, are completely surrounded by negatively stained material of fibrous appearance which can be seen connecting groups of cells (Fig. 5C).
FIG. 5.
Pattern formation in a spreading Sm-1 colony. A 25-μl aliquot of a saturated Sm-1 culture was plated onto a 7H9–ADC–0.3% agarose plate. (A) Pictures of the spreading colony taken 1, 2, and 3 days after inoculation. (B and C) Electron micrographs of cells taken at day 3 from the transparent periphery (B) and opaque interior (C) of a spreading colony. Cells were negatively stained with 2% phosphotungstic acid. Bar, 1 μm. Arrows mark the structures discussed in the text.
M. smegmatis strains unable to spread lack GPLs.
Previous studies with a variety of surface-translocating microorganisms have revealed that amphiphilic surface-active molecules (such as polysaccharides, peptidolipids, and sulfonolipids) secreted into the medium or present on the cell surface are often required for movement (1, 18, 26). We hypothesized that some amphiphilic substance produced by mc2155 and Sm-1 but missing in Rg-1 could be required for the spreading behavior. GPLs were possible candidates for this function (for reviews see references 9 and 14). The general structure of M. smegmatis GPLs is as follows:
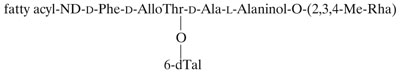 |
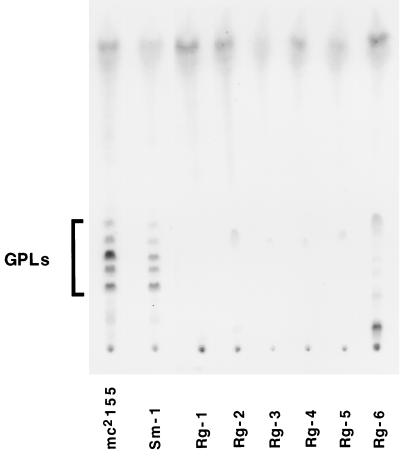
GPLs consist of a mixture of 3-hydroxy and 3-methoxy long-chain fatty acids amidated by a tripeptide (d-Phe-d-alloThr-d-Ala) terminated by an l-alaninol. The alaninol is glycosylated by an O-methylated rhamnosyl residue, and a 6-deoxytalose is attached to the d-alloThr residue. GPLs are known to be surface exposed (30) and have been extensively correlated with colony morphology variations in members of the M. avium complex (2, 7). The results of TLC analysis of the GPL profiles of cells grown on the surface of 0.3% agarose–7H9–ADC plates are shown in Fig. 6. Equal weights of lipid extract were loaded for all strains. mc2155 and Sm-1 show the characteristic profile of GPLs for M. smegmatis (17). Only minor diferences in relative amounts can be discerned between the two strains. In contrast, Rg-1 shows complete absence of GPLs under the same conditions, even when the TLC was overloaded (results not shown). To further strengthen the correlation between the lack of GPLs and the spread-deficient phenotype, we analyzed five other independently isolated rough strains. All five were unable to spread in all the media tested (data not shown). TLC analysis of GPL preparations (Fig. 6) revealed that four of them showed complete lack of GPLs, while a fifth one (Rg-6) gave an anomalous GPL profile, with one major brightly yellow staining band of significantly slower mobility under these conditions. The identity of this compound has not been determined. These results indicate that the presence of GPLs is required for M. smegmatis to spread over the growth surface.
FIG. 6.
GPL profiles of M smegmatis colony morphology variants. From left to right mc2155, Sm-1 (smooth), and six rough strains, Rg-1, Rg-2, Rg-3, Rg-4, Rg-5, and Rg-6. Approximately equal dry weights of lipid extract were loaded in each lane. The TLC was developed in chloroform-methanol-water (90:10:1), dried, sprayed with 10% H2SO4 in ethanol, and heated to 120°C.
M. avium also exhibits surface spreading motility.
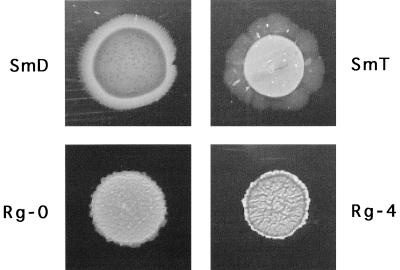
Since GPLs are also produced by a large number of mycobacteria other than M. smegmatis, we decided to test whether other mycobacterial species are also capable of surface translocation. We chose to test the opportunistic pathogen M. avium as a representative of the slow-growing mycobacteria. M. avium A4, A5, MAC101, and 920A6 were all able to spread on the surface of 7H9 salts–0.3% agarose and 7H9–ADC–0.3% agarose plates, producing halos of similar morphology to those of M. smegmatis (results not shown). In order to correlate GPL synthesis, colony morphology, and spreading motility of M. avium, we also tested four partially characterized morphological variants derived from the 2151 strain (6, 7). 2151-SmD and -SmT, are smooth opaque and transparent variants, respectively, which produce GPLs. Rg-0 and Rg-4 are spontaneous rough variants deficient in the synthesis of GPL to different degrees. Rg-4 is completely devoid of any GPL structure, while Rg-0 can synthesize the lipopeptide core of the GPL. After 2 weeks of incubation at 37°C on 7H9–ADC–0.3% agarose plates, all four strains showed different patterns of surface spreading (Fig. 7). Strain 2151-SmT produced the largest halos, very similar to those of mc2155. 2151-SmD also spread, but it formed halos of radial appearance. Finally, Rg4 did not spread, while RgO produced very reduced and compact spreading areas. These results show that surface translocation is not restricted to the rapid-growing M. smegmatis but is also present in at least one slow-growing species and, furthermore, that in both species the ability to synthesize GPLs correlates with efficient surface spreading.
FIG. 7.
Spreading phenotype of M. avium colony morphology variants 2151-SmD, 2151-SmT, Rg-O, and Rg-4 on 7H9–ADC–0.3% agarose plates. Photographs were taken 3 weeks after inoculation.
DISCUSSION
The results presented in this paper constitute the first report of motility in mycobacteria, which are traditionally defined as nonmotile organisms (19). Although the term motility has been often used to describe only the capacity of bacteria to swim in liquid media, it is now recognized that the ability to move on solid surfaces is widespread among bacteria. Swarming of P. mirabilis, gliding of myxobacteria and cyanobacteria, and twitching of pseudomonads are well-known examples of bacterial surface translocation. The mode of mycobacterial surface translocation reported here should be classified as sliding as defined by Henrichsen (20): “a kind of surface translocation produced by the expansive forces in a growing culture in combination with special surface properties of the cells resulting in reduced friction between cell and substrate. The micromorphological pattern is that of a uniform sheet of closely packed cells in a single layer. The sheet moves slowly as a unit.” Examples of sliding bacteria include members of the genera Alcaligenes, Flavobacterium, Acinetobacter, Streptococcus, and Corynebacterium (20). The mechanism underlying sliding in these organisms has not been characterized in detail. In addition, the flagellum-independent surface spreading of Serratia (26) also satisfies the definition of sliding given by Henrichsen (20).
The arrangement of the translocating mycobacteria as a sliding sheet is most clearly shown in the time-lapse movies, where it is evident that cohesive groups of cells are pushed away from the inoculation site. Importantly, movement of individual cells relative to others, which is one of the main differences between sliding and twitching or gliding, is not observed. The restricted fluidity within the monolayer is also confirmed by the close grouping of siblings in the GFP labeling experiments. The nature of the cell-to-cell contacts within the monolayer is not known. Groups of cells appear to be arranged as pseudofilaments, mostly along their longitudinal axis, but contacts are not restricted to the cell poles. Extracellular structures such as pili or fimbriae, which have been implicated in a variety of cell-to-cell contacts (34, 38), were not observed in preparations of spreading cells negatively stained with uranyl acetate or phosphotugstic acid.
Bacterial translocation over surfaces requires reduced friction between the cells and the substratum. In particular, movement of cells over the surface of an agar or agarose plate should be facilitated by a reduction of the hydrophilic interactions between cells and the surface. Bacterial surface-active compounds are known to have a profound effect in the interaction of bacteria with interfaces, and a variety of these compounds, such as lipopeptides, sulfonolipids, and polysaccharides, have been implicated in surface translocation in several systems (29). By analogy, we hypothesized that sliding motility of mycobacteria was therefore likely to involve some kind of surface-active compound. The capsular GPLs (reviewed in references 9 and 14) could play such a role since they are surface-exposed amphiphilic molecules whose absence has been extensively correlated with rough colony morphology, the phenotype of strains deficient for surface spreading. Our data show that six independently isolated rough mutants of M. smegmatis tested were unable to spread on agarose plates and were defective in GPLs (five completely lack GPLs, and one showed an obviously altered profile). Furthermore, the two rough strains of M. avium we have tested, which are also GPL−, exhibited spreading-deficient phenotypes. Therefore, there is a strong correlation between the lack of GPL and the inability to move on a surface.
The involvement of GPLs in surface motility in mycobacteria is reminiscent of the role of serrawettings in Serratia spreading (reviewed in reference 16). Serrawettings are a family of cyclic lipopeptides with surfactant activity required for flagellum-dependent and -independent surface translocation (25, 26). They are secreted into the medium, where they form a hydrophobic conditioning film over the hydrophilic agar surface, thereby reducing the interactions at the interface and promoting spreading. Mycobacterial GPLs, however, are present on the cell surface of intact M. smegmatis and M. avium (30) and have been found to be the major components of the superficial layer of smooth variants of M. avium and M. intracellulare (2, 3). Freeze fracture analysis of intramacrophagic M. avium has shown that the bacilli are surrounded by a discontinuous multilamellar capsule-like structure where each lamella is made up of paralell fibers of GPL (33). We have observed discontinuous capsular structures in negatively stained preparations of M. smegmatis strains spreading in rich medium (Fig. 5B). These structures are present in the GPL-producing strains mc2155 and Sm-1 but are absent in Rg-1, the GPL− strain, and might therefore represent accumulations of GPL in the surface of translocating bacteria. GPLs could render the bacterial surface more hydrophobic and therefore decrease interactions with the agarose surface, facilitating spreading growth. GPLs might also be released in some proportion, creating a conditioning film on the agarose surface for the cells to slide on, as is the case for Serratia.
GPLs are likely not to be the only components affecting mycobacterial spreading motility. For example, spreading bacteria appear to be surrounded by a mucoid clear material or slime layer of unknown composition. In addition, two of our M. smegmatis strains, mc2155 and Sm-1, which in our analysis appear similar in their GPL components, show differences in their spreading phenotypes in rich media, where Sm-1 spreads faster and forms halos with a complex radial pattern absent in mc2155. There are also obvious differences between the spreading phenotypes of M. avium 2151-SmD and 2151-SmT, both of which produce GPLs (7). These strains differ in the amount of capsular polysaccharide, which is decreased in the SmD strain (32).
On rich medium plates, the morphology of the spreading colony becomes complex. What in poor medium is a fairly uniform spreading of cells as a monolayer, in rich medium appears to turn into cycles of spreading followed by conversion of the monolayer into a dense cell mass. This switch is accompanied by changes in the appearance of the cell surface: fibers connecting groups of cells are present in the densely packed areas but missing in the spreading front. The result is a series on concentric zones of growth surrounded in the periphery by a monolayer of cells. The cause of the switch between forms of growth is unlikely to be starvation since we observed it in small isolated microcolonies growing in small numbers on very rich moist plates, but could be due to cell density. Similar successive rounds of expansion have been reported in swarming colonies of Bacillus subtilis (27) and P. mirabilis (reviewed in reference 4). In the case of P. mirabilis it is well documented that the terraces are the result of rounds of swarming followed by consolidation, where cells “dedifferentiate” into the nonmotile vegetative cells. The mechanism that synchronizes these changes is not completely understood. A membrane sensor histidine kinase has been recently found to be involved in the process (5), and differences in fimbria and pilus expression levels have been observed among areas of a colony (24). Similarly, differential gene expression within an expanding colony is likely to cause the cycles observed in the expansion of a mycobacterial colony in rich medium.
The most obvious advantage of surface translocation is that it results in fast colonization of the available surface by the motile bacteria. We have shown that under conditions that allow spreading, motile strains of M. smegmatis quickly colonize the growth surface and outcompete the nonmotile strains for access to the available nutrients. Thus, surface translocation is likely to play an important role in the evolutionary success of free-living mycobacteria in the environment as most bacterial growth is likely to occur on a surface (12). In addition, surface translocation could play another role for M. avium. Infections by this opportunistic pathogen are acquired through the gastrointestinal and respiratory tracts (21). The capacity of M. avium strains to spread over surfaces might play an important role in mucosal colonization and thus could be a virulence determinant. Interestingly, fresh isolates of M. avium strains from patients are SmT (13, 15), and under our conditions, strain 2151-SmT showed the most pronounced spreading phenotype.
We have shown that mycobacterial spreading motility is not restricted to M. smegmatis but also occurs with M. avium. Interestingly, GPLs are synthesized by a large number of mycobacterial species, and other classes of amphiphilic lipids that could play a similar role are present in the outermost layer of other mycobacteria (14). The ability to translocate over surfaces might thus be a general characteristic of mycobacteria.
ACKNOWLEDGMENTS
We thank John Belisle and Michael Starnbach for providing M. avium strains, Maria Ericsson for assistance with the electron microscope, and members of the Kolter lab for valuable discussions and comments on the manuscript.
This work was supported by a postdoctoral fellowship to A.M. from the Heiser Program for Research in Leprosy and Tuberculosis and NIH grant GM58213 to R.K.
REFERENCES
- 1.Abbanat D R, Leadbetter E R, Godchaux III W, Escher A. Sulphonolipids are molecular determinants of gliding motility. Nature. 1986;324:367–369. [Google Scholar]
- 2.Barrow W W, Brennan P J. Isolation in high frequency of rough variants of Mycobacterium intracellulare lacking C-mycoside glycopeptidolipid antigens. J Bacteriol. 1982;150:381–384. doi: 10.1128/jb.150.1.381-384.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrow W W, Ullom B P, Brennan P J. Peptidoglycolipid nature of the superficial cell-wall sheath of smooth-colony-forming mycobacteria. J Bacteriol. 1980;144:814–822. doi: 10.1128/jb.144.2.814-822.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belas R. Proteus mirabilis and other swarming bacteria. 183–219. In: Shapiro M J A and D., editor. Bacteria as multicellular organisms. Oxford, England: Oxford University Press; 1996. [Google Scholar]
- 5.Belas R, Schneider R, Melch M. Characterization of Proteus mirabilis precocious swarming mutants: identification of rsbA, encoding a regulator of swarming behavior. J Bacteriol. 1998;180:6126–6139. doi: 10.1128/jb.180.23.6126-6139.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belisle J T, Klaczkiewicz K, Brennan P J, Jacobs W R, Jr, Inamine J. Rough morphological variants of Mycobacterium avium. Characterization of genomic deletions resulting in the loss of glycopetidolipid expression. J Biol Chem. 1993;268:10517–10523. [PubMed] [Google Scholar]
- 7.Belisle J T, McNeil M R, Chatterjee D, Inamine J, Brennan P J. Expression of the core lipopetide of the glycopeptidolipid surface antigens in rough mutants of Mycobacterium avium. J Biol Chem. 1993;268:10510–10516. [PubMed] [Google Scholar]
- 8.Bloom B R, Murray C J L. Tuberculosis: commentary on a reemergent killer. Science. 1992;257:1055–1064. doi: 10.1126/science.257.5073.1055. [DOI] [PubMed] [Google Scholar]
- 9.Brennan P J, Nikaido H. The envelope of Mycobacteria. Annu Rev Biochem. 1995;64:29–63. doi: 10.1146/annurev.bi.64.070195.000333. [DOI] [PubMed] [Google Scholar]
- 10.Brennan P J, Souhrada M, Ullom B, McClatchy J K, Goren M B. Identification of atypical mycobacteria by thin-layer chromatography of their surface antigens. J Clin Microbiol. 1978;8:374–379. doi: 10.1128/jcm.8.4.374-379.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burchard R P. Gliding motility of prokaryotes: ultrastructure, physiology and genetics. Annu Rev Microbiol. 1981;35:497–529. doi: 10.1146/annurev.mi.35.100181.002433. [DOI] [PubMed] [Google Scholar]
- 11a.Connel, N., and B. Jacobs. Unpublished results.
- 12.Costerton J W, Lewandowski D E, Cladweil D E, Korber D R, Lappin-Scott H M. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 13.Crowle A J, Tsang A Y, Vatter A E, May M H. Comparison of 15 laboratory and patient-derived strains of Mycobacterium avium for ability to infect and multiply in cultured human macrophages. J Clin Microbiol. 1986;24:812–821. doi: 10.1128/jcm.24.5.812-821.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daffe M, Draper P. The envelope layers of Mycobacteria with reference to their pathogenicity. Adv Microb Physiol. 1998;39:131–203. doi: 10.1016/s0065-2911(08)60016-8. [DOI] [PubMed] [Google Scholar]
- 15.Dunbar F P, Pejovic I, Cacciatore R, Peric-Golia L, Runyon E H. Mycobacterium intracellulare maintenance of pathogenicity in relationship to lyophilization and colony form. Scand J Respir Dis. 1968;49:153–162. [PubMed] [Google Scholar]
- 16.Eberl L, Molin S, Givskov M. Surface motility of Serratia liquefaciens MG1. J Bacteriol. 1999;181:1703–1712. doi: 10.1128/jb.181.6.1703-1712.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eckstein T M, Silbaq F S, Chaterjee D, Kelly N J, Brennan P J, Belisle J T. Identification and recombinant expression of a Mycobacterium avium rhamnosyltransferase gene (rtfA) involved in glycopeptidolipid biosynthesis. J Bacteriol. 1998;180:5567–5573. doi: 10.1128/jb.180.21.5567-5573.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Godchaux W, III, Lynes M A, Leadbetter E R. Defects in gliding motility in mutants of Cytophaga johnsonae lacking a high-molecular-weight cell surface polysaccharide. J Bacteriol. 1991;173:7607–7614. doi: 10.1128/jb.173.23.7607-7614.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goodfellow M, Cross T. Classification. In: Goodfellow M, Mordarski M, Williams S T, editors. The biology of actinomycetes. London, England: Academic Press; 1983. pp. 8–99. [Google Scholar]
- 20.Henrichsen J. Bacterial surface translocation: a survey and classification. Bacteriol Rev. 1972;36:478–503. doi: 10.1128/br.36.4.478-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Inderlied C B, Kemper C A, Bermudez L E. The Mycobacterium avium complex. Clin Microbiol Rev. 1993;6:266–310. doi: 10.1128/cmr.6.3.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs W R, Jr, Kalpana G V, Cirillo J D, Pascopella L, Snapper S B, Udani R A, Jones W, Barletta R G, Bloom B R. Genetic systems for mycobacteria. Methods Enzymol. 1991;204:537–555. doi: 10.1016/0076-6879(91)04027-l. [DOI] [PubMed] [Google Scholar]
- 23.Kochi A. Government intervention programs in HIV/tuberculous infection. Outline of guidelines for national tuberculosis control programs in view of the HIV epidemic. Bull Int Union Tuberc Lung Dis. 1991;66:33–66. [PubMed] [Google Scholar]
- 24.Latta R K, Grondin A, Jarrel H C, Nichols G R, Berube L R. Differential expression of nonagglutinating fimbriae and MR/P pili in swarming colonies of Proteus mirabilis. J Bacteriol. 1999;181:3220–3225. doi: 10.1128/jb.181.10.3220-3225.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindum P W, Anthoni C, Christoffersen C, Eberl L, Molin S, Givskov M. N-Acyl-l-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J Bacteriol. 1998;180:6384–6388. doi: 10.1128/jb.180.23.6384-6388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25a.Martinez, A., S. Torello, and R. Kolter. Unpublished results.
- 26.Matsuyama T, Kaneda K, Nakagawa Y, Isa K, Hara-Hotta H, Yano I. A novel extracellular cyclic lipopeptide which promotes flagellum-dependent and -independent spreading growth of Serratia marcesens. J Bacteriol. 1992;174:1769–1776. doi: 10.1128/jb.174.6.1769-1776.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendelson N H, Salhi B. Patterns of reporter gene expression in the phase diagram of Bacillus subtilis colony forms. J Bacteriol. 1996;178:1980–1989. doi: 10.1128/jb.178.7.1980-1989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Neidhardt F C, Bloch P L, Smith D F. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neu T R. Significance of bacterial surface-active compounds in interaction of bacteria with interfaces. Microbiol Rev. 1996;60:151–166. doi: 10.1128/mr.60.1.151-166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ortalo-Magne A, Lemassu A, Laneelle M, Bardou F, Silve G, Gounon P, Marchal G, Daffe M. Identification of surface-exposed lipids on the cell envelopes of Mycobacterium tuberculosis and other mycobacterial species. J Bacteriol. 1996;178:456–461. doi: 10.1128/jb.178.2.456-461.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pardee A B, Jacob F, Monod J. The genetic control and cytoplasmic expression of “inducibility” in the synthesis of β-galactosidase in E. coli. J Mol Biol. 1959;1:165–178. [Google Scholar]
- 32.Rastogi N, Frehel C, Ryter A, Ohanyon H, Lesourd M, David H L. Multiple drug resistance in Mycobacterium avium: is the wall architecture responsible for the exclusion of antimicrobial agents? Antimicrob Agents Chemother. 1981;20:666–667. doi: 10.1128/aac.20.5.666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rulong S, Aguas A P, Pinto Da Silva P, Silva M T. Intramacrophagic Mycobacterium avium bacilli are coated by a multiple lamellar structure: freeze fracture analysis of infected mouse liver. Infect Immun. 1991;59:3895–3902. doi: 10.1128/iai.59.11.3895-3902.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shimkets L J. Social and developmental biology of the myxobacteria. Microbiol Rev. 1990;54:473–501. doi: 10.1128/mr.54.4.473-501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snapper S, Lugosi L, Jekkel A, Melton R, Kieser T, Bloom B R, Jacobs W R., Jr Lysogeny and transformation in Mycobacteria: stable expression of foreign genes. Proc Natl Acad Sci USA. 1988;85:6987–6991. doi: 10.1073/pnas.85.18.6987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stahl S J, Stewart K R, Williams F D. Extracellular slime associated with Proteus mirabilis during swarming. J Bacteriol. 1983;154:930–937. doi: 10.1128/jb.154.2.930-937.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wall D, Kaiser D. Alignment enhances the cell-to-cell transfer of pilus phenotype. Proc Natl Acad Sci USA. 1998;95:3054–3058. doi: 10.1073/pnas.95.6.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whittaker C J, Klier C M, Kolenbrander P E. Mechanisms of adhesion by oral bacteria. Annu Rev Microbiol. 1996;50:513–552. doi: 10.1146/annurev.micro.50.1.513. [DOI] [PubMed] [Google Scholar]
- 39.Williams F D, Schwarzhoff R H. Nature of the swarming phenomenon in Proteus. Annu Rev Microbiol. 1978;32:101–122. doi: 10.1146/annurev.mi.32.100178.000533. [DOI] [PubMed] [Google Scholar]