Abstract
The transformer-based models, such as GPT-31 and DALL-E2, have achieved unprecedented breakthroughs in the field of natural language processing and computer vision. The inherent similarities between natural language and biological sequences have prompted a new wave of inferring the grammatical rules underneath the biological sequences. In genomic study, it is worth noting that DNA sequences alone cannot explain all the gene activities due to epigenetic mechanism. To investigate this problem, we propose EpiGePT, a new transformer-based language pretrained model in epigenomics, for predicting genome-wide epigenomic signals by considering the mechanistic modeling of transcriptional regulation. Specifically, EpiGePT takes the context-specific activities of transcription factors (TFs) into consideration, which could offer deeper biological insights comparing to models trained on DNA sequence only. In a series of experiments, EpiGePT demonstrates state-of-the-art performance in a diverse epigenomic signals prediction tasks as well as new prediction tasks by fine-tuning. Furthermore, EpiGePT is capable of learning the cell-type-specific long-range interactions through the self-attention mechanism and interpreting the genetic variants that associated with human diseases. We expect that the advances of EpiGePT can shed light on understanding the complex regulatory mechanisms in gene regulation. We provide free online prediction service of EpiGePT through https://health.tsinghua.edu.cn/epigept/.
Introduction
One of the fundamental problems in genomic study is how to decode and interpret the human genome sequences in a complex manner. Progress toward this goal is largely hindered by the vast majority of non-coding regions3. For example, it remains unclear how the genomic variants in the noncoding regions lead to malfunctions of regulatory elements by disrupting the underlying regulatory syntax of DNA4. Inspired from the field of natural language processing, there exists a natural analogy between human language and DNA sequence where texts are made of words and DNA sequence can be characterized by nucleotides or k-mers. The inherent similarities between natural language and biological sequences provide new perspectives towards better understanding the complex DNA language.
Recently, generative pre-trained transformer (GPT) models have achieved unprecedented success in various domains, including computer vision and natural language processing (NLP)1, 5. Such pre-trained models can be readily tailored or adapted to various downstream tasks. To date, the application of generative pre-trained models in genomic study remains largely unexplored. It is noticeable that a number of machine learning-based approaches have been proposed for predicting various genomic and epigenomic signals, such as chromatin accessibility6, 7, histone modification8 or chromatin interactions9, 10. However, these methods are rather scattered, with specific models designed for specific prediction tasks. It is in an urgent need to develop a foundation model to facilitate multiple genomic and epigenomic prediction tasks and unveil the universal gene regulation rules.
To design a genomic foundation model, it is worth noting that the existing large language models (LLMs) purely rely on the language context consisting of words and sentences while the DNA sequences cannot explain all the heritable and stable changes in gene activity due to epigenetic mechanisms. In other words, the genomic foundation model based on pure DNA sequence may largely ignore the context-specific information, thus lacking mechanistic interpretation of context-specific gene regulation. For example, using transformer-based language model to decode genome sequence has been attempted by a recent work Enformer11. However, Enformer is not capable of predicting the function of sequences in new cellular contexts, which largely limits its generalization power.
To overcome the above limitation, we proposed EpiGePT, a new transformer-based deep learning framework, to predict genome-wide epigenomic signals by taking the mechanistic modeling of transcriptional regulation into consideration. With EpiGePT, we are able to investigate how to utilize the power of transformer-based language model to help researchers uncover how trans-regulatory factors (e.g., TFs) regulate target genes by interacting with cis-regulatory elements and further lead to changes in different chromatin states. After pretraining on a diverse panel of cell line and tissue level data from the Encode database12, EpiGePT is able to directly predict the genome-wide chromatin states in any new cellular context given the expression profile of a few hundreds of TFs or facilitate new prediction tasks (e.g., 3D genome interaction) with finetuning.
To the best of our knowledge, EpiGePT is the first pretrained Transformer model for epigenomics with mechanistic modeling of transcriptional regulation. EpiGePT differs from existing methods in the following three aspects. First, unlike the methods that take pure DNA sequence as input, EpiGePT additionally takes the context-specific information (e.g., TF activities) as input, thus enabling genome-wide prediction power in any new cellular context. Second, instead of using task-specific model to predict a single genomic and epigenomic signal, EpiGePT is designed for simultaneously predicting multiple epigenomic signals of the same genomic region through multi-task learning, thus improving learning efficiency and prediction accuracy compared to the task-specific models. Third, many methods typically take short DNA sequence (e.g., a few hundred or a thousand base pair) as input, which may not be adequate to capture the complex syntax of DNA due to truncation. The long input DNA sequence (e.g., 128kb) for EpiGePT greatly enhances the ability for the model to capture the long-range interaction in the genome, which are crucial for understanding the gene regulation mechanism.
In a series of experiments, we illustrate that our method is superior to existing methods in a various tasks of chromatin states prediction, as well as the variant effect prediction. We also show that the self-attention mechanism greatly helps unveil the complex code in the conformation of long-range chromatin interactions, such as promoter-enhancer interactions and promoter-silencer interactions. EpiGePT is an example of how transformer-based language model and large-scale pretrain can be used in genomics research to provide biological insights. With the help of EpiGePT model, it is expected that researchers can dissect the comprehensive genomic regulatory code given the cellular context information and accelerate research findings in genomic study.
Results
Overview of EpiGePT model
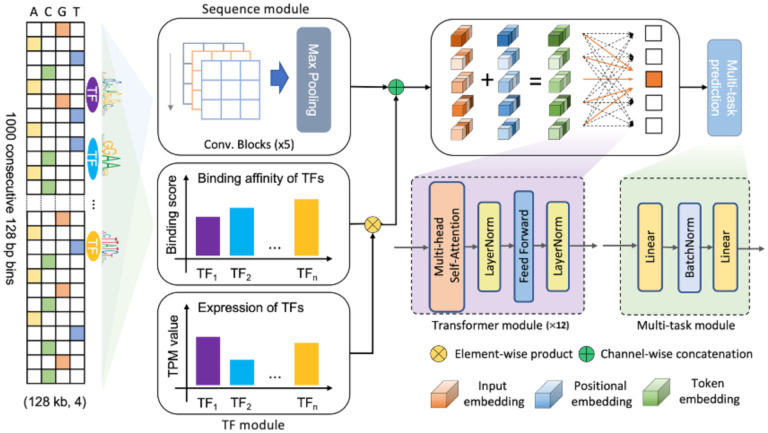
We developed a novel Transformer13-based language model named EpiGePT to predict multiple chromatin states across different cell types. EpiGePT is a language model for cross-cell-type prediction of chromatin states by multi-task learning based on genome-wide pre-training on epigenomic data (Fig. 1 and Fig. S6). EpiGePT is composed of four modules, including a sequence module, a TF module, a transformer module, and a prediction module. The sequence module is responsible for processing the long DNA sequence of interest (e.g., 128 kb) by employing a series of convolutional and pooling blocks (e.g., 5) to extract a comprehensive set of sequence features. By reducing the input length by 25=128 times through pooling operations, this module effectively compresses the input information while retaining essential features. The TF module is specifically designed to extract cell-type-specific features by taking the expression of transcription factors in the given context, as well as their corresponding motif score into account. This module helps capture the unique characteristics of each cell type by considering the binding status of TFs involved in gene regulation. In the transformer module, each token corresponds to a genomic bin in the original DNA sequence and has hybrid features derived from both sequence and TFs. The module leverages self-attention mechanisms to learn the comprehensive relationships among the input bins, enabling the model to make predictions of multiple chromatin states under the given context cellular. By taking advantage of this approach, EpiGePT provides a powerful tool for predicting multiple chromatin states and enables researchers to gain insights into the underlying regulatory mechanisms of the genome.
Fig. 1. Overview of the EpiGePT model for multiple epigenomic signals prediction.
The model consists of four modules, namely the Sequence module, the TF module, the Transformer module, and the Multi-task module. The sequence module comprises multiple layers of convolution applied to the one-hot encoded DNA sequence input. The input sequence length consists of 1000 genomic bins of 128bp for the prediction of multiple signals and 50 bins of 200bp for the prediction of DNase signal alone. The TF module encompasses the binding status and expression of 711 transcription factors. The Transformer module consists of a series of consecutive transformer encoders, while the multi-task module is composed of a fully connected layer.
EpiGePT enables genome-wide prediction of chromatin states
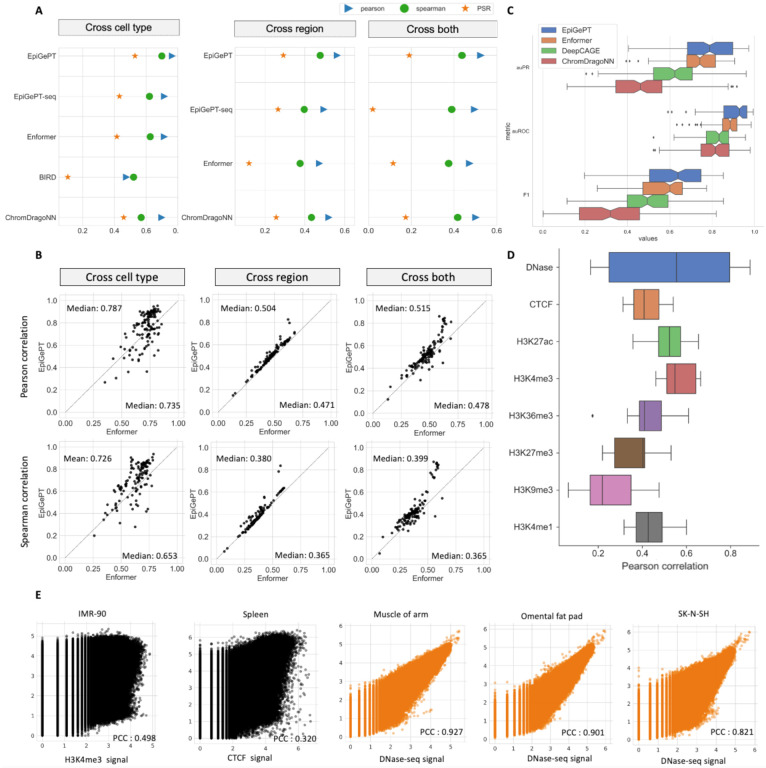
To assess the predictive performance for epigenomic signals of EpiGePT, specifically in predicting chromatin accessibility, a comprehensive evaluation was conducted. We first applied EpiGePT to predict the chromatin accessibility based on the widely available public DNase-seq14 data across diverse cell types or tissues. In brief, DNase-seq data across 129 cell types were collected from the ENCODE12 project. After data preprocessing and normalization (see Methods), 1,175,374 genomic regions were extracted where each pair of cell type and genomic regions constitutes a training instance. We meticulously devised comprehensive experimental settings by partitioning the training and test sets based on either genomic regions or cell types. In detail, we employed the following three data partitioning settings for a comprehensive evaluation (Fig. S1, Text S1). For “cross-cell-type” prediction, we partitioned the data into training and testing sets based on cell types. For “cross-region” prediction, we partitioned the data into training and testing sets based on the genomic regions. For “cross-both” prediction, we conducted rigorous data split to ensure that both the cell types and genomic regions in the test stage are unseen during the training process. We employed three evaluation metrics, namely Pearson correlation coefficient, Spearman correlation coefficient and prediction square error, to assess the similarity between the predicted and true values of DNase signals (See Methods). It is shown that EpiGePT consistently outperforms other competing methods, including Enformer11, BIRD15, and ChromDragoNN16 by a relatively large margin under the above experimental settings (Fig. 2A and Fig. S2). EpiGePT achieves 5.0%, 8.9%, and 5.2 % higher performance than Enformer, the best baseline method, in terms of the mean Pearson correlation coefficient under three data-partition settings, respectively (Fig. 2B). Besides the chromatin accessibility regression task, we also designed binary chromatin accessibility status prediction task by assessing whether a peak exists within the corresponding genomic bin (>50% overlap). We made slight adjustments to the regression model by modifying the activation function and loss function to accommodate the binary classification task (See methods). The results show that EpiGePT achieves an average auPRC (area under the precision-recall curve) of 0.767 compared to 0.727 of Enformer11, 0.623 of DeepCAGE17 and 0.476 of ChromDragoNN16 (Fig. 2C).
Fig. 2. Performance of EpiGePT and baseline methods on the benchmark experiment.
(A) EpiGePT and baseline methods were compared in terms of their regression performance for DNase signal regression across cell types, genomic regions, and combined cell type and genomic regions. (B) Comparison of EpiGePT and Enformer performance. Each point in the scatter plot represents the performance of Enformer on the data of a specific cell type (x-axis) compared to the performance of EpiGePT (y-axis). (C) EpiGePT and baseline methods’ performance on binary prediction of DNase-seq signals. (D) EpiGePT demonstrates excellent performance in predicting diverse epigenetic signals across various cell types, including DNase-seq, CTCF, and histone modifications. (E) EpiGePT predictions compared to experimental signals visualized for a representative example. Genome-wide multi-signal predictions (black) and DNase-specific predictions (orange).
Next, we extended the chromatin state prediction task from a single target to multiple targets by predicting multiple chromatin states, including chromatin accessibility, CTCF18, ChIP-seq, and six types of different histone modifications19 (See Methods). When considering eights different chromatin states, only 28 cell types have the corresponding available data simultaneously. After preprocessing, 13,300 genomic regions each with a length of 128 kbp were extracted, which cover 56.7% of the whole genome. However, compared to the data in the DNase-only prediction experiment, the correlation coefficient was reduced due to the prevalence of a substantial number of zero signals in the genomic regions being predicted. Using a similar data split strategy as the single target for cross-cell-type prediction, EpiGePT demonstrated a mean Pearson correlation coefficient between 0.259 to 0.566 of different chromatin states in the test cell types (Fig. 2D). Specifically, EpiGePT achieves remarkably high performance in predicting chromatin state signals for certain cell types, such as the colon tissue, with a Pearson correlation coefficient of 0.888. Furthermore, as it shown in Fig. S3C, it significantly outperformed Enformer in terms of performance across these tested cell types and different signals (one-side p-value < 2.79e-10 under binomial hypothesis test). To make the chromatin states prediction task more illustrative, several tracks of predicted chromatin states and the corresponding ground truth chromatin states were displayed. For instance, at the position of from 61,056,000 to 61,184,000 on chromosome 20, we used the UCSC genome browser20 to show the predicted values and true values of CTCF (Pearson correlation coefficient of 0.518) and DNase signals (Pearson correlation coefficient of 0.869), as well as the regulatory relationships within this region. (Fig. S2A). In addition, we also compared EpiGePT with ChromDragoNN16 on binary and quaternary classification tasks based on ChromHMM21 annotations (Fig. S3). EpiGePT achieved an average auROC (area under the receiver operating characteristic curve) of 0.855 in binary classification, significantly higher than that of ChromDragoNN16 (0.774). In quaternary classification, EpiGePT achieved a macro-auROC of 0.879, also significantly higher than ChromDragoNN16 (0.856, one-side p-value < 0.001). These results demonstrate the effectiveness and accuracy of EpiGePT in predicting multiple chromatin states, leveraging the four modules. The effectiveness and prediction power achieved, in conjunction with the self-attention mechanism, lays the foundation for deciphering regulatory relationships.
To further verify the roles of the main modules in the model, we conducted the ablation experiments on the model architecture. For TF module ablation, the above experimental results compared to EpiGePT without TF module (EpiGePT-seq) and EpiGePT have demonstrated that EpiGePT outperforms EpiGePT-seq in cross-cell-type prediction of DNase signals, with an average Pearson correlation coefficient of 0.714 and a median of 0.74 for EpiGePT-seq, while EpiGePT achieves 0.756 (average) and 0.787 (median). In addition, the inclusion of the TF module enables EpiGePT to predict chromatin states at the locus level for different cell types. However, like Enformer, EpiGePT-seq predicts the same values for different cell types at the same locus, resulting in a zero correlation for cross-cell-type prediction. We examined the impact of the TF module on multi-task prediction by employing three methods, namely replacing TF scores with zero, adding random noise to TF, and removing motif binding scores. The results indicated that when the expression of TFs was set to zero, the prediction of H3K27ac yielded a Pearson correlation coefficient of 0.190. However, incorporating the TF module significantly improved the coefficient to 0.543, demonstrating a beneficial impact of the TF module.
For sequence module ablation, we randomly subsampled 10,000 genomic bins and 20 cell types to train a TF-only model. The results indicated that removing the sequence module resulted in an average decrease of 0.084 in the Pearson correlation coefficients of the eight signals on a cell-type wise basis, and with a particularly significant decrease of 0.13 in predicting H3K4me3 signals (Fig. S4A).
For multi-task module, we predicted the eight chromatin states involved in training using eight individual models for single-task prediction. The results were evaluated on a cross-cell type prediction manner. In the case of predicting the signal of H3K4me1, the average Pearson correlation decreased from 0.408 to 0.329. When predicting the H3K4me1 signal, the average Pearson correlation coefficient decreased from 0.408 to 0.329. Similarly, the overall prediction performance for the eight signals declined by 0.074 (Fig. 4B). This decrease may be attributed to the intricate nature of gene regulation. The distinct chromatin states can complement and synergize with each other through multi-task learning, allowing the model to gain deeper biological insights compared to a single-target prediction model.
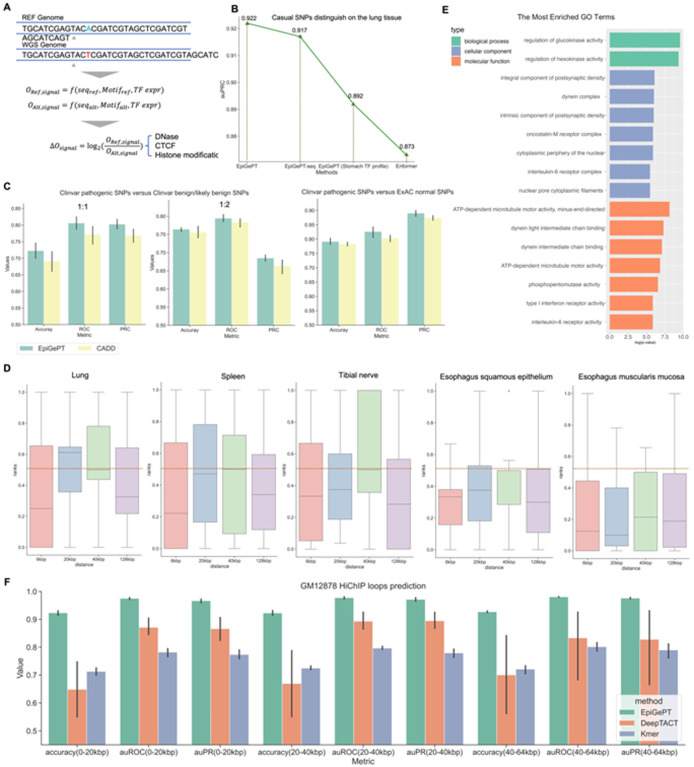
Fig. 4. Variant effect prediction of EpiGePT.
(A) The LOS score for each epigenomic signal is calculated by the log change fold of the predicted epigenomic signal for reference genome and WGS genome. (B) The performance of EpiGePT and Enformer in discriminating causal SNPs on the Lung tissue. (C) The three subplots from left to right respectively depict the classification results for disease-related SNPs and benign SNPs down-sampled sourced from the ClinVar database, with balanced positive and negative samples (1:1 and 1:2 ratio), as well as normal SNPs sourced from the ExAC database with a MLP classifier. (D) The ranked position of COVID-19 related GWAS data among surrounding benign SNPs based on their LOS scores, as determined using different tissue or cell-type expression data. The results were stratified based on the distance range of the risk region. The resulting mean and median ranks were both below 0.5. (E) Enrichment result (Biological process, Cellular component and Molecular function) of the nearest genes of the COVID-19 associated SNPs with the max LOS scores. (F) The performance (auROC and auPRC) of the fine-tuned EpiGePT model and baseline methods (DeepTACT and Kmer) in distinguishing enhancer-gene pairs at various distance ranges (0–20 kbp, 20–40 kbp and 40–64 kbp) on K562 cell line.
Furthermore, we performed additional experiments to investigate the effect of the number of cell lines on the prediction performance. Specifically, we focused on DNase predictions and randomly downsampled the training cell types from 103 to 75, 50, 25 for each of the five folds in the cross-validation experiment. The results demonstrated a strong positive correlation between the number of cell lines and the prediction performance. Under five-fold cross-validation, the median Pearson correlation coefficient on the test set across 129 cell types decreased from 0.793 to 0.790, 0.761, and 0.732, respectively. These findings suggest that our current model has potential room for improvement and additional training with more cell lines will lead to even better predictive performance, thereby offering more comprehensive insights into the regulatory mechanisms for researchers (Fig. S4C). In summary, EpiGePT demonstrated superior performance in predicting both single and multiple epigenomic signals over existing methods, providing a robust foundation for decoding the complex landscape of gene regulation.
EpiGePT facilitates long-range chromatin interaction identification
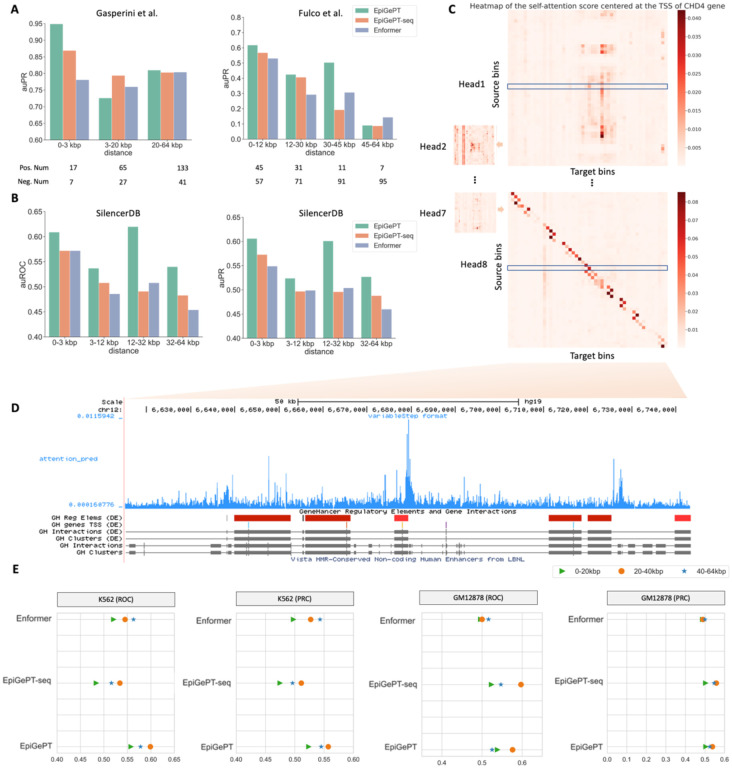
We examined the capacity of EpiGePT for predicting long-range chromatin interactions, which play a critical role in preserving chromatin architecture and elucidating 3D contacts between distal regulatory elements and target genes. Traditional methods typically take short DNA sequence (e.g., 1kbp) as input, thus cannot take the long-range chromatin interactions into consideration. During the training of EpiGePT model, the self-attention mechanism in the transformer module plays an important role in capturing the potential interactions between different DNA bins. We utilized the cell-type specific self-attention scores to predict chromatin interactions, including enhancer-promoter and silencer-promoter interactions (see Methods). We initially investigated whether EpiGePT can differentiate experimentally validated enhancer-promoter interactions from other interactions. Two datasets containing 664 and 5,091 candidate enhancer-promoter interactions or element-TSS interactions obtained by CRISPRi22 experiments were used and further filtered and stratified by the distance. In the Gasperini et al23. dataset, EpiGePT consistently outperform EpiGePT-seq and Enformer by achieving the highest auPRC in all groups. For instance, EpiGePT achieved auPRC of 0.949, 0.726, and 0.810 for identifying enhancer-promoter pairs in the 0–3 kbp, 3–20 kbp, and 20–64 kbp ranges, respectively (Fig. 3A and Fig. S8). In the Fulco et al.24 dataset, EpiGePT obtains better performance than EpiGePT-seq in most groups, which illustrates the positive benefit of the cell-type-specificity brought in the TF module. EpiGePT consistently outperforms Enformer across different groups by a relatively large margin. As shown in Fig. 3A, EpiGePT achieves an auPRC of 0.618, compared to 0.531 of Enformer, and 0.568 of EpiGePT-seq in 0–12kbp group.
Fig. 3. Application of self-attention mechanism in EpiGePT for long-range chromatin interaction identification.
(A) The performance (auPRC) of attention score of EpiGePT in distinguishing enhancer-gene pairs at different distance ranges on two different datasets. (B) The performance (auROC and auPRC) of attention score of EpiGePT in distinguishing silencer-gene pairs at different distance ranges based on the data from SilencerDB25. (C) Heatmap of the self-attention matrix of each attention head centered at the TSS of the CHD4 gene, the (i, j) element in the matrix denotes the average attention score between the ith genomic bin and the jth genomic bin across all layers. (D) Attention scores centered at the TSS of the CHD4 gene, and putative enhancer regions in its vicinity. (E) The performance (auROC and auPRC) of attention score of EpiGePT in distinguishing HiChIP loops of H3K27ac at different distance ranges on K562 cell line and GM12878 cell line.
Next, we explored whether EpiGePT is also capable of predicting the promoter-silencer interactions. Since there is very limited experiment-validated silencer-promoter interactions, we downloaded putative silencers from the SilencerDB25 and used the promoter of annotated nearest gene as the potential target. For negative silencer-promoter pairs, we selected the same promoter and equidistant genomic regions in the opposite direction to ensure the consistency of distance distributions between positive and negative sample pairs at different distance levels. As a result, EpiGePT achieves a better performance in discerning positive silencer-promoter pairs from negative pairs than Enformer by a relatively large margin. For instance, EpiGePT displays an auROC of 0.575 in long-range interactions (32–64kbp) with positive-to-negative ratio 1:1 setting, compared to 0.547 of EpiGePT-seq, and 0.483 of Enformer (Fig. 3B). According to these results, the self-attention mechanism significantly enhances the ability to identify potential chromatin interactions and increases the interpretability of the model.
The HiChIP26 sequencing technology provides unprecedented opportunities to uncover 3D genomic interactions. We aim to investigate the predictive performance of EpiGePT on 3D genome interaction based on HiChIP data. Here, we employ the same strategy as described above to calculate attention scores for the regulatory element-promoter pairs and collected HiChIP loops on K562 and GM12878 cell lines from the HiChIPdb27. The results demonstrate that incorporating TF expression data into EpiGePT leads to enhanced predictive performance for HiChIP loops compared to the pure sequence models Enformer and EpiGePT-seq, across diverse distance ranges and in two distinct cell lines. Specifically, within the 20–40 kbp distance range on K562 cell line, EpiGePT achieves an auROC of 0.599 for the 1:1 positive-to-negative ratio, surpassing Enformer’s performance of 0.545 (Fig. 3E). These findings suggest that, even without any fine-tuning, EpiGePT’s attention scores encompass more accurate and comprehensive biological information, underscoring its potential for capturing intricate genomic interactions.
To better understand the self-attention mechanism of EpiGePT and bridge the gap between the model and its interpretability, we visualized the attention matrices after normalization (Fig. 3C). The visualization shows prominent scores between certain genomic bins, indicating the potential presence of interactions. We centered on the transcription start site (TSS) of the CHD4 gene and calculated the self-attention scores between the genomic bins within its upstream and downstream 128kbp. The attention scores exhibited peaks near the regulatory elements in the vicinity of the TSS, which further validates the feasibility and accuracy of our prediction of enhancer-promoter interactions (Fig. 3D).
EpiGePT improves variant effect prediction
One of the most essential tasks for EpiGePT is to dissect the effect of genetic variants that occur in different genomic regions. As most of the variants identified by the GWAS studies lie in the non-coding regions of the genome, which makes it difficult to interpret the variant effect, most sequence-based computational models directly take the alleles sequence as input and compare the difference in the predicted regulatory activity. The advantage of EpiGePT model comes from the TF module where variant effect can be estimated under any given cellular context. This is extremely helpful when predicting the effect of the disease- or phenotype-associated SNPs. To test the ability of EpiGePT in variant effect prediction, we first collected an eQTLs dataset28 that contains 20,913 causal and non-causal variant-gene pairs in total across 49 different tissues from the supplementary data of Wang et al28. EpiGePT and EpiGePT-seq are then applied to estimate the log-odds scores (LOS) given both the reference and alterative DNA sequence and the corresponding relevant TF profile (see Methods, Fig. 4A). Finally, a random forest classifier is trained based on the LOS scores across different chromatin states. The experimental results show that in the lung tissue, EpiGePT demonstrates and auPRC of 0.922, compared to 0.873 of Enformer in distinguishing casual SNPs. To verify the effectiveness of TF module, we replace the TF profile of lung with stomach, which is much less relevant to the lung tissue. The auPRC decreases from 0.922 to 0.892 (Fig. 4B). Similarly, EpiGePT-seq can achieve an average auPRC of 0.910, compared to 0.898 of Enformer using 5-fold cross-validation for predicting causal variants on 48 extracted tissues (Fig. S3D). In the adrenal gland tissue, EpiGePT-seq demonstrates an average auPRC of 0.883, compared to only 0.842 of Enformer. The above experiments show the predictive power of EpiGePT in estimating the variant effect.
To further evaluate the performance of EpiGePT in predicting disease-associated variants, we extracted 52, 876 pathogenic SNPs from the ClinVar29 database and 418, 863 benign SNPs from the ClinVar database, also with 84, 095 benign SNPs from the ExAC database30 as positive and negative sets, respectively. We defined a 64kbp region surrounding each pathogenic SNP as the risk region. We screened all benign and likely benign SNPs that fall within the risk region from the negative sets for classification. As the relevant tissue or cell type information is not available, we concatenated the LOS of the eight epigenomic signals and self-attention scores across 28 cell types into a single 252-dimensional vector and then train a classifier for predicting whether the given SNP is pathogenic (see Methods). To assess the whether the 252-dimensional features are beneficial in predicting pathogenic SNPs, we concatenated it with 52 annotations from CADD31, resulting in a comprehensive feature vector. Subsequently, we compared the performance of this combined feature vector with that of the individual features derived exclusively from CADD. We then utilized these two sets of features to train multi-layer perceptron (MLP) classifiers separately. The results demonstrate that incorporating EpiGePT’s variant effect features from multiple cell types significantly enhances the performance of the classifier in predicting pathogenic SNPs. Specifically, when the positive-to-negative sample ratio was set to 1:1, the average auROC increased from 0.772 to 0.806, and the average accuracy increased from 0.690 to 0.723 (Fig. 4C). This observation indicates that features extracted by EpiGePT provide a valuable complement to CADD annotation, enabling a more comprehensive depiction of variant characteristics, and thereby facilitating the discovery of disease-associated variants.
EpiGePT prioritizes potential SNPs associated with comorbidities of COVID-19
We investigated whether the ability of EpiGePT to predict variant effects could help in the discovery of key SNPs related to COVID-19. COVID-19 is an infectious disease caused by the SARS-CoV-2 virus, which emerged in late 2019 and quickly spread around the world, causing a global pandemic32. In order to validate the ability of EpiGePT in identifying key SNPs, we collected GWAS data from the COVID-19 host genetics33, including 9,484 variants. These variants were derived from 4,933 patients with confirmed severe respiratory symptoms and 1,398,672 control individuals without COVID-19 symptoms. To validate the ability of the model to identify COVID-19-related SNPs, we firstly defined a risk region around the selected COVID-19-related SNPs and computed the rank of the variant score of pathogenic SNPs within the surrounding benign SNPs from the ClinVar database. The expected rank for random guessing (uniform distribution) is 0.5. Interestingly, we found that the average rank of COVID-19-related SNPs was significantly lower than 0.5 across several tissues or cell types (Fig. 4D). For instance, when lung expression data was employed and a 6-kbp risk region was examined, the median rank was 0.250, and when expression data of esophagus squamous epithelium was used the median rank was 0.333, significantly lower than 0.5 (one-side p-value of 0.013 under one-sided Binomial Test). However, when we employed the expression data from smooth muscle cells, which are a more widespread cell type with lower relevance to COVID-19, the median rank exhibited a notable decrease to 0.381. Notably, when focusing on the 40-kbp risk region, the median rank further declined to 0.850, higher than 0.5. These findings suggest that EpiGePT model is able to prioritize the COVID-19-related SNPs thus shedding lights on finding the potential disease-associated variants with our pretrained large language model.
Next, based on the aforementioned findings, we aimed to use EpiGePT to identify genes that are highly related to COVID-19. Since the genetic pathology of COVID-19 is not yet clear and the earliest lesion is in the lungs, we ranked all 9,484 possible SNPs using lung expression data. We then identified the SNPs with the highest ranks and performed gene ontology enrichment analysis on nearest genes of the 100 top ranked SNPs (Fig. 4E). The enrichment results revealed potential biological processes that are relevant to COVID-19, such as the regulation of glucokinase activity which is associated with the homeostasis of human blood glucose34. Notably, diabetes mellitus, a condition closely associated with hyperglycemia, is a typical comorbidity of COVID-1935. Besides, among the top 10 potential genes that scored the highest, we identified that the TBC1D4 gene, which regulates glucose homeostasis, is potentially associated with COVID-19 comorbidities. Our findings are consistent with previous research by Pellegrina et al.36 and highlights the potential of our EpiGePT approach in discovering new genetic markers that may be implicated in the pathogenesis of COVID-19. Overall, our EpiGePT model provides new perspectives for understanding how the genetic variants could contribute to the COVID-19 susceptibility and severity.
Fine-tuning on EpiGePT enables accurate prediction of regulatory interactions
Fine-tuning is an strategy that transfers the knowledge of a pretrained model to new tasks, which is particularly prevalent in language models such as GPT37 and BERT38. Here, we finetuned a pretrained EpiGePT model on a new task for predicting the 3D genome interaction. Given the HiChIP H3K27ac data from K562 and GM12878 cell lines, the features of two anchors were extracted from a pretrained EpiGePT model and then fed to a finetune network to predict whether it is a HiChIP loop. We compared EpiGePT with pretrain and finetuning strategy to two baselines, DeepTACT39 and a k-mer frequency40 based method (see Methods). The results illustrate that EpiGePT exhibits a superior classification performance across diverse distance ranges compared to baselines. For example, in the GM12878 cell line within the 20–40kbp distance range, EpiGePT demonstrates a significantly improved predictive performance with an auROC of 0.949, surpassing 0.866 of DeepTACT39 and 0.771 of Kmer (Fig. 4F and Fig. S7). This significant improvement achieved through fine-tuning EpiGePT on a limited dataset aligns well with the concept of few-shot learners1, highlighting the power of the pretrained EpiGePT model.
EpiGePT encompasses the regulatory relationships between TFs and target genes.
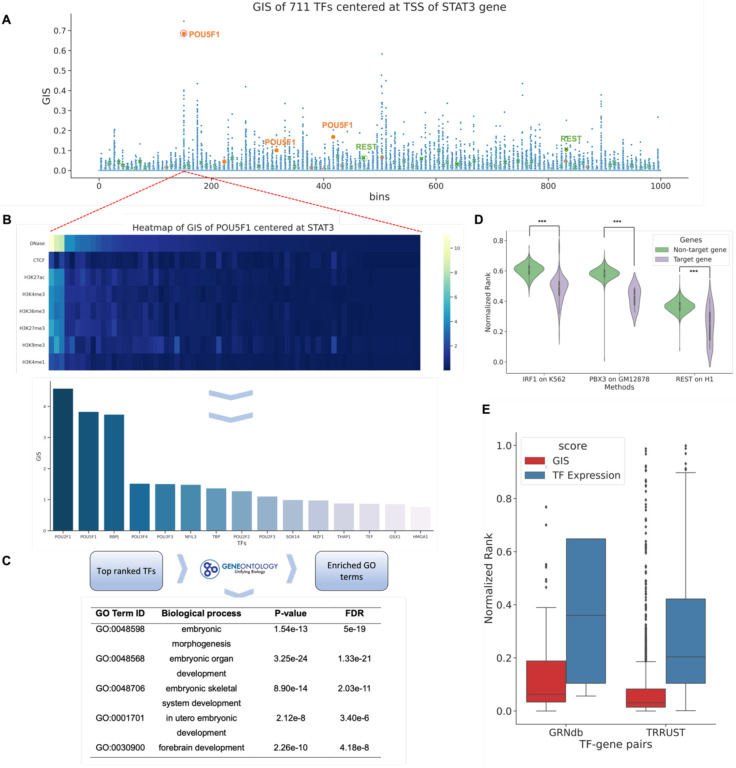
One of the key differentiating factors of EpiGePT compared to other sequence models lies in its integration of TF binding status and TF expression. This unique feature empowers EpiGePT to capture potential regulatory relationships embedded within the genomic sequence. In this study, we specifically aimed to validate whether EpiGePT learns the regulatory relationships between TFs and target genes (TGs). We defined gradient importance scores (GIS) based on the absolute gradient values of predicted epigenomic signals w.r.t. TF profile to rank TFs give a TG (see Methods). Particularly, we collected 15 TFs that play critical regulatory roles in embryonic stem cells (ESC)41, 42 and validated their interaction relationships using computed GIS. As an example, for a target gene STAT3 that plays essential role for ESC pluripotency43, we computed GIS for each core TF across 1000 genomic bins and find other key TFs in ESC ranked 1st (REST) and 2nd (POU5F1) at specific bins (Fig. 5A–B). Interestingly, the GO terms enriched by the top 10% prioritized TF coding genes also included biological processes of embryonic cell differentiation and development when we focus on the genomic bin that POU5F1 ranked 2nd (Fig. 5C). We selected a TF as the target gene and calculated the integrated GIS (IGIS) score for another key TF across eight epigenomic signals. Multiple TF-gene pairs identified by IGIS that showed significant associations with POU5F1, such as ESRRB-POU5F144 (rank 2nd), and ETV5-POU5F145 (rank 5th). Furthermore, we use TF-TG relationships from either ChIP-seq data or external databases as ground truth to validate whether IGIS is effective in prioritizing the TFs given a TG. First, we defined potential TF-target gene pairs based on TF ChIP-seq data specific to certain cell types among all human genes (see Methods). The results demonstrated a significant difference in rank between TF-target gene pairs and TF-non-target gene pairs based on the IGIS score (Fig. 5D), with the former exhibiting considerably higher ranks (one-side p-value < 0.001). Second, we collected TF-target regulatory network data from two publicly available databases. We obtained a total of 1,066 TF-gene pairs from the GRNdb46 database based on liver-specific GTEx data, and 2,705 TF-gene pairs from the TRRUST47 database after filtering. Then we calculated the rank of each TF based on GIS of the TF expression and a genomic bin-level mask for each pair. Interestingly, when using liver expression data, we found that the average rank of TFs from TRRUST was 7.9%, significantly lower than the rank based on expression values (one-side p-value < 1e-5). Similarly, based on the GRNdb data, the majority of TF-gene pairs obtained had TF ranks within the range of 20%, and the mean of this distribution was significantly lower than the rank based on expression values with one-side p-value < 1e-5 (Fig. 5E). For instance, TMEM55B plays a significant role in regulating lysosome movement, and is regulated by sterol response element binding factor 2 (SREBF2)48, while GIS enable the identification of SREBF2 as the top-ranked TF associated with TMEM55B, further validating the role of GIS in prioritizing functional TFs. The comprehensive validation from both ChIP-seq datasets and external databases further support the effectiveness of GIS in identifying context-specific TF-TG relationships.
Fig. 5. Gradient importance scores (GIS) uncover regulatory transcription factors.
(A) Genomic regions around TSS of the STAT3 gene and expression data on ESC were fed into EpiGePT. The scatter plot represents the GIS scores of core TFs on each genomic bin. Each dot represents the GIS score of a core TF on a specific genomic bin. In specific bins, key TFs in ESC, such as NR5A2 and POU1F5 highlighted in the figure, exhibit high ranks in the GIS scores. (B) Heatmap of GIS for 10% important TFs surrounding STAT3 gene, specifically focusing on bins with POU1F5 ranked 3rd. Each row represents a predicted epigenomic signal, and the TFs are sorted based on their GIS on DNase signals. Bar plot of the top 15 TFs with the highest GIS scores. (C) Based on the top 10% ranked TFs mentioned above, gene ontology enrichment analysis revealed significant enrichment in biological processes related to embryonic development and cellular differentiation. (D) Based on TF ChIP-seq data, all 23,635 human genes were classified into target genes and non-target genes. The results revealed that TFs exhibited significantly higher ranks on potential target genes compared to non-target genes. (E) The distribution of the rank of TFs in the GIS and expression value among the 2,705 TF-gene pairs from the TRRUST database and 1,066 TF-gene pairs derived from genotype-tissue expression (GTEx) data of the liver sourced from the GRNdb database. The analysis reveals that the median rank of TFs from the TRRUST database is significantly lower than 0.06 (one-side p-value < 3.12 e-18) and the median rank of TFs obtained from the GRNdb database is significantly lower than 0.5 (one-side p-value < 2.50 e-140).
Online prediction tool for EpiGePT
In order to facilitate the utilization of EpiGePT for the prediction of multiple chromatin states of any cellular context and any genomic regions, especially for research personnel who lack coding expertise, we have developed a user-friendly online web server, named EpiGePT-online (https://health.tsinghua.edu.cn/epigept/) (Text S2). The online web server was developed using PHP, JavaScript and HTML, which provides an interactive web interface for online prediction of 8 chromatin states of specific genomic regions (Fig. 6). Users can obtain the predicted signals of multiple genomic regions by submitting a region file and a TF expression file of 711 selected TFs (Supplementary Table S3), or obtain predicted signals of specific regions directly by selecting genomic locus. As to the two types of input files, we provide example files to demonstrate their formats, and accept expression files stored in either numpy or csv formats, to increase the universality of the web server (Fig. S5). The web server outputs the results in a web summary html, which saves significant amount of time for installation and implementation. Furthermore, we provide a detailed tutorial to enable users to quickly learn how to use our website. We anticipate that this web server will assist researchers in predicting chromatin states of specific cell types and further deepening their understanding of gene regulatory mechanisms.
Fig. 6. Overview of the online prediction web server of EpiGePT.
We collected eight types of epigenetic genome modification signals and corresponding expression data of transcription factors in different cell types or tissues from the ENCODE project. Based on these data, we trained the EpiGePT model and deployed it as a built-in kernel on an Apache server. Users without much coding experience can also access the web server in two ways to obtain the eight types of epigenetic genome modification signals for specified cell types and genomic regions without programming or installation.
Discussion
In this paper, we introduced EpiGePT, a transformer-based large language model, for predicting the chromatin states given any cellular context. Compared with the existing machine learning based computational methods, EpiGePT takes transcription factor profile and DNA sequence information as inputs by multitask learning and self-attention mechanism within a unified model. With these two types of input information and four modules of network architecture, EpiGePT overcomes the limitation of the existing models and demonstrates state-of-art performance in prediction of multiple chromatin signals in diverse experimental settings. With the superior predictive performance of EpiGePT, we are able to investigate one of the fundamental questions in functional genomics: how transcription factors and cis-regulatory elements regulate gene activity. In this work, we investigated this question from two aspects: 1) identifying the interactions of cis-regulatory elements and their target genes with the help of self-attention mechanism in EpiGePT; 2) estimating the variant effect based on the LOS scores computed by the outputs of EpiGePT to assist in discovering human disease-associated SNPs. First, the self-attention scores between tokens can provides us with an intuitive and quantitative measure of the interaction level between different genomic regions, which offers new opportunity to discover the target gene of cis-regulatory elements and find the interpretability of EpiGePT. Second, the LOS scores of the multiple chromatin signals from different tissue or cell lines are complementing each other, which provides us with a more comprehensive characterization of the variant and enables accurate prediction of the variant impact. Such variant effect prediction by EpiGePT establishes a foundation for understanding the underlying relationship between genetic variations and disease mechanisms.
There exist several extensions and refinements that can be applied to further improve the EpiGePT model. Firstly, the incorporation of chromatin regulators (CRs) as trans-acting factors into the TF module could enhance the modeling of regulated transcription processes, thereby increasing the accuracy of the predictions. Secondly, the inclusion of high-order interactions between TFs in the framework could provide a more comprehensive representation of the regulatory relationships, and potentially enhance the predictive performance. Third, the application of EpiGePT to single-cell genomics could enable the profiling of chromatin signals at single-cell resolution, facilitating a holistic understanding of regulatory heterogeneity in different cell subpopulations for researchers.
Based on EpiGePT, users are able to predict multiple chromatin profiles in different cell lines or tissues, which could provide a foundation for biological discovery, decoding transcriptional regulation mechanisms, and investigating disease mechanisms. We anticipate that EpiGePT can provide valuable insights to researchers in understanding regulatory mechanisms.
Methods
Data processing
Chromatin accessibility data and Expression data
We used three different datasets in the experiments. For chromatin accessible data, we downloaded DNase bam files and narrow peaks across 129 human biosamples from ENCODE12 project (Supplementary table S1 and S2). We divided the human hg19 genome into 200bp non-overlapping locus (use bin instead), and we assigned the label for each locus in each cell type. For the regression design, we pooled the bam files of multiple replicates for a cell type (Supplementary table S1 and S2), and obtain the raw read count nlk for locus l in cell type k. We normalized the raw read count in order to eliminate the effect of sequencing depths, in the form of ñlk = Nnlk/Nk, where Nk denotes the total number of pooled reads for cell type k and denotes the minimal number of pooled reads across all cell types. The normalized read counts are further log transformed with pseudo count 1, which represent the continuous level of chromatin accessibility. For binary classification design, we assigned a binary label ylk to 1 if the number of raw read counts of the locus l in the cell type k greater than 30, which represent the locus is an accessible region in this cell type, resulting in the identification of regions as accessible in 13% on average and 8% at median in the screened genomic regions across 129 cell types. The proportion of open regions varies among different cell types, and the average openness level mentioned above is generally consistent with that maintained in ChromDragoNN16.
RNA-seq data of the 711 human transcription factors were downloaded and extracted from the ENCODE project (Supplementary table S5 and S6). We perform log transformation with pseudo count 1 and quantile normalization based on TPM values. The normalized TPM values were averaged across replicates and mean expression profile of each cell type was finally used to calculated of the transcription feature.
Multiple chromatin signals data
DNase-seq, RNA-seq and ChIP-seq data were also downloaded from ENCODE project (Supplementary table S3, S4 and S6). We applied the same process to these data as above, and finally we obtained the 8 chromatin signals of 13,300,000 bins of 128bp in 28 cell types. The continuous level of chromatin signals we extracted were ‘DNase’, ‘CTCF’, ‘H3K27ac’, ‘H3K4me3’, ‘H3K36me3’, ‘H3K27me3’, ‘H3K9me3’ and ‘H3K4me1’, which includes crucial epigenetic modifications and markers for gene regulation and transcription.
Chromatin states data
We downloaded the 15-state ChromHMM21 annotations across 127 epigenomes from the ROADMAP project. The state of chromatin is annotated for each 200bp bin in a specific cell type. RNA-seq data across 56 cell types of TFs was download and extracted from the ROADMAP49 project (Supplementary table S7 and S8). Subsequently, we mapped these 711 transcription factors to the downloaded RNA-seq data, resulting in the identification of RNA-seq data for 642 transcription factors. In the subsequent experiments, we utilized the expression data of these 642 transcription factors. We finally calculated the normalized TPM values of the 642 TFs on 56 cell types we extracted for the using in the classification model. For coarse grain chromatin state prediction, we took the state ‘Quies’ as low signal regions and other states as signal regions. For fine grain chromatin state prediction, we extracted the state ‘TssA’, ‘TssAFInk’, ‘TssBiv’ and ‘BivFInk’ as TSS regions, state ‘EnhG’, ‘Enh’ and ‘EnhBiv’ as enhancer regions, ‘Quies’ as low signal regions and other state as other regions. To balance the number of different chromatin states, we downsampled the low signal regions and obtained 921,074 locus each cell line finally.
Model architecture
Sequence module and Transformer module
As shown in Figure 1 and Fig. S6A, the sequence module receives a one-hot matrix (A = [0,0,0,1], C = [0,1,0,0], G = [0,0,1,0], T = [0,0,0,1]) of size (128000,4) as input, representing a sequence of 128 kilobase pairs (kbps) and contains five 1-dimentional convolutional blocks to extract DNA sequence features. Each block includes a convolutional layer and a maxpooling layer (Fig. S6B). The first convolutional layer considers the input channels as 4 and performs convolution along the sequence direction. The input sequence features are one-hot embeddings of size L × 4, where L denotes the length of the input long range DNA sequence. After 5 maxpooling layers, the output size of sequence feature is L/N × C, where C denotes the hyperparameter for sequence embedding and N denotes the length of locus to predict. We set C to 256 in the pre-training stage of chromatin accessibility prediction experiments. Rectified linear units (ReLU) are used after each convolution operation for keeping positive activations and setting negative activation values to zeros. Sequence features were then concatenated with transcriptomics features, and we finally obtained a vector of size L/N × (C + nTF), where nTF denotes the dimension of the transcription factors features after padding. In our model, after adding padding to the 711 TFs, the nTF is set to 712. Therefore, the input token number for the transformer module is 1000, and each token embedding has a dimensionality of 968.
We utilize the transformer module to integrate information from both the sequence and transcription factors (TFs), enabling the capturing of long-range interactions between genomic bins. We applied Nt layers of Transformer encoder with nhead different attention heads to the token embedding sequence. The input X of the transformer encoder is a genomic bin sequence with dimensions (Sequence length, embedding dim). Specifically, this dimension is (1000, 968) in EpiGePT, indicating that input genomic bin sequence has a length of 1000, and each genomic bin has an embedded representation that combines the sequence information with cell-type-specific features with dimension of 968. Each Transformer encoder includes a multi-head self-attention mechanism and a feed-forward neural network. For self-attention in each head, the calculation is based on the matrix operation.
For multi-head attention, Transformer encoder learns parameter matrices, and for the ith head and concatenate the multiple heads to do the projection.
Where dmodel denotes the dimension of token in the input sequence X, which is 968 in EpiGePT and dQ = dK = dV = 512. The matrices Q, K, and V are obtained by the application of mapping functions represented by, and, followed by concatenating of Qi to Q, Ki to K, and V to V. These mapping functions serve to transform the concatenated embeddings into the resulting matrices. We set Nt to 16 for the chromatin accessible prediction experiments, Nt to 12 for the chromatin state classification and multiple chromatin signals prediction experiments, and set nhead to 8 for all experiments.
The regression model, the output layer uses a linear transformation and use mean square error (MSE) as the loss function. For classification model, the output layer uses a linear transformation combined with a sigmoid function, and use the cross-entropy loss for classification experiments.
TF module
For binding status, we scanned the input bins for potential binding sites for a set of 711 human transcription factors from HOCOMOCO database50 with the tool Homer51 (Table S5). We then selected the maximum score of reported binding status for each transcription factor to obtain a vector of 711 dimensions as the motif feature for each DNA bin. For gene expression, we focused on log-transformed TPM values of the 711 transcription factors and obtained a vector of 711 dimensions after quantile normalization as the expression feature. With these data, we combined the two vectors of motif and expression features by taking the element-wise product, and we concatenated the result to the output of sequence module.
Model evaluation
To evaluate our model, we applied five-fold cross-validation in the different experiments on cell-type level. For chromatin accessible experiments, the 129 cell lines are partitioned into a training set and a testing set randomly.
Cell-type-wise metrics are defined to evaluate our method in different experiments, which were calculate with the data within a test cell type across all genomic locus. For binary classification design, we used cell-type-wise auPRC and auROC to evaluate our EpiGePT. Let YL×K and be the true and predicted matrix, where L denotes the number of locus and K denotes the number of test cell types. We calculated the auPRC and auROC for each (y1i, y2i, ⋯ , yLi) and (, , ⋯ ,). For multiple classification, we use macro average of the auPRC and auROC to evaluate the classification performance, which compute the metric independently for each class and then take the average hence treating all classes equally. For regression design, we used two metrics for model evaluation, which are cell-type-wise Pearson correlation coefficient and prediction squared error. Prediction square error (PSR) is calculated as, where denotes the mean of the true level of the response in the cell type k.
To compare the performance of our method with other baseline methods, we conducted hypothesis testing on the metrics based on cell types. Since the metrics on a given cell type across different methods are paired data and the statistical distribution is unknown, we employed both Binomial and Wilcoxon tests, with the alternative hypothesis being that EpiGePT outperforms the other methods. If we reject the null hypothesis, it provides compelling evidence to support the claim that EpiGePT performs better than the other methods.
To evaluate the computational efficiency, we recorded the running time of a single epoch of EpiGePT and baseline methods (Supplementary Text S3). Compared to traditional CNN models such as DeepCAGE17 and ChromDragoNN16, as well as larger sequence models like Enformer, EpiGePT demonstrates a balance between high computational efficiency and performance.
Model fine-tunning
For the fine-tuning process, we kept the parameters of the pre-trained model fixed without making any updates. For the specific fine-tuning task of chromatin interaction prediction based on HiChIP data, the multi-task module was replaced with a two-layer MLP network, containing 256 hidden nodes for each layer. During the training process, only the weights in the MLP network were updated. Notably, when utilizing HiChIP data at a resolution of 5k, both the enhancer and promoter anchors spanned 5kbp. Then we input a region extending 128kbp from the center of the anchor of the neighboring gene into the EpiGePT. Consequently, a 968-dimensional feature vector for each genomic bin was derived from the output of the last transformer encoder layer. These feature vectors from all bins within the two anchors were concatenated, resulting in a high-dimensional vector of size 76,472.
Baseline methods
Four baselines were introduced for epigenetic signals prediction. BIRD15 is a multiple linear regression model that only takes gene expression data as input and makes predictions on a fixed locus. ChromDragoNN16 is a deep neural network that takes gene expression of 1630 TFs and DNA sequence as input. Specifically, ChromDragoNN16 uses a ResNet52 to extract sequence features and use linear transformation to combine the TF gene expression feature and sequence feature to make the final prediction. DeepCAGE17 Integrating regulatory DNA sequence is a deep densely connected convolutional network for predicting chromatin accessibility. The dense-connected neural network architecture used by DeepCAGE17 may struggle to capture the complex interactions between genomic regions. Enformer11 is a deep neural network that integrates convolutional neural network and transformer, and only takes DNA sequence as input. Enformer takes DNA sequence of length 196kbp as input to predict 5,313 genomic tracks of human and 1,643 tracks of mouse genome simultaneously. However, one of the limitations of Enformer is that it can only model and predict cell types in the training data and cannot be applied to new cell types. In order to ensure the fairness of the benchmark experiment, we retrained the Enformer model with the same input and output data as EpiGePT when reproduce the Enformer model (Text S4).
Two baseline methods were introduced for predicting HiChIP interaction. DeepTACT39 is a deep learning method for predicting 3D chromatin contacts using both DNA sequence and chromatin accessibility. We adopted the structure of DeepTACT39 and kept the anchor length at 5k. The input to the model consists of two anchor sequences represented as one-hot matrices and the two openness scores of the two anchors on the corresponding cell type extracted from OpenAnnotate53. Regarding the Kmer features40, K is chosen as 5 to extract sequence features. For each anchor, a vector of dimension 45 = 1024 was obtained. Further training was performed using an MLP with a hidden layer dimension of 256.
Enhancer, Silencer and HiChIP loop prioritization
We collected cis-regulatory elements-gene pairs in K562 cells from other studies and public database to demonstrate the interpretability of self-attention mechanisms in the EpiGePT. Enhancers and silencers are typical cis-regulatory elements known play important roles in transcriptional control during normal development and disease. For enhancers, we downloaded enhancer-gene pairs from two studies: Gasperini et al.23 and Fulco et al.24, both of which were tested using a CRISPRi22 assay perturbation. Two datasets contain 664 and 5,091 enhancer-promoter interactions or element-TSS interactions. For silencers, we obtained and random sampled 831 validated silencers-gene pairs with distance within 64kbp in K562 cells curated from high-throughput experiments from SilencerDB. As there are no experimentally validated interaction relationships between these silencers and genes, we generated silencer-gene pairs by associating the nearest neighbor genes for classification purposes. Similarly, negative samples were generated by constructing DNase-seq, ATAC-seq and nearest genes using the same approach. Ultimately, we obtained a dataset comprising 1,662 silencer-gene pairs, encompassing both positive and negative instances.
To obtain scores for regulatory element-gene pairs, we first used the region extending 128kbp from the center of the enhancer as input and extracted the token where the interacting genes reside, so that we could filter out regulatory element-gene pairs that were located further than 64kbp apart. Subsequently, we stratified the remaining pairs based on their distance. Since the positive and negative sample ratios varied across datasets, we adopted different stratification strategies for different distance ranges (Fig. 3). Next, we averaged the attention matrices of the Transformer encoder across all layers and heads. The summed attention scores from other tokens to the key token containing the gene TSS were used as the attention score of this element-gene pair. This score represents the attention value that the enhancer-centered region receives for the transcription start site (TSS) of the gene. We also calculated the attention score from the bin containing the center of the regulatory element to the bin containing the TSS, which only slightly affects the experimental results of regulatory element prioritization.
We collected 5k resolution data from the HiChIPdb (http://health.tsinghua.edu.cn/hichipdb/) database, specifically from K562 and GM12878 cell lines. We filtered the data to include only loops where at least one anchor falls within a gene region. We stratified the loops based on distance into three categories: 0–20kbp, 20–40kbp, and 40–64kbp. For each distance category, we selected 2000 positive pairs with most significant q-value. To ensure consistency in the distance distribution, we selected negative pairs by fixing a gene and choosing anchors at equidistant locations in the opposite direction.
Gradient importance scores
EpiGePT possesses the capability to assign priority rankings to transcription factors by utilizing gradient importance scores (GIS), taking into account specific cell types and chromatin regions. The GIS were employed to identify potential functional relationships between specific transcription factors (TFs) and target genes. Specifically, for a given TF-target gene pair, the transcription start sites (TSS) of genes were used as central loci, and the regions spanning 128 kbp upstream and downstream of the TSS were selected as input. Next, we filtered out bins with motif binding scores indicating potential binding for the given TF. For these selected bins, we calculated the GIS for the predictions of eight epigenomic signals across the 711 core TFs.
Where, i denotes the ith TF in the set of core TFs, j denotes the jth cell type, k denotes the kth predicted epigenomic signal, and ζ denotes the set of genomic bins that have binding for the given TF. In the calculation of the gradient, denotes the predicted value of the kth epigenomic signal by the model using the expression in the jth cell type at the lth bin. On the other hand, tfij denotes the product of the expression of ith TF in the jth cell type and the corresponding TF binding score.
If we consider the GIS for the prediction of all 8 epigenomic signals simultaneously, we can prioritize the TFs by calculating their ranks based on each signal separately. Then, we can calculate an integrated gradient importance score (IGIS) for each TF by aggregating the ranks from all 8 signals.
Both the GIS and the IGIS are capable of capturing the significance of a transcription factor (TF) in regulating a specific gene within the context of a specific cell type. Consequently, these scores hold potential value in the discovery of TFs that play crucial roles in the regulation of specific genes, thereby contributing to our understanding of essential regulatory mechanisms.
In the context of validating TF-TG pairs in the GRNdb and TRRUST databases, we opted to utilize liver expression data as a representative example due to the unavailability of cell type information for TRRUST. Furthermore, in this experimental setup, the tfij denotes the expression of ith TF in the jth cell type and ζ denotes the set of genomic bins that have binding for the TF of the given TF-target gene pair.
Potential TF-target gene pairs from ChIP-seq data
In this study, we utilized three distinct cell types to conduct a comprehensive screening of TF-target gene pairs and non-target gene pairs across the human genome. Initially, we obtained the narrow peak files (ENCFF388AJH, ENCFF717IXP, and ENCFF885KLR) from ChIP-seq experiments across three cell types from the ENCODE project. Subsequently, we meticulously examined the number of peaks within a 128kbp region both upstream and downstream of the transcription start site (TSS) for each gene. Different thresholds were applied to the ChIP-seq data of various TFs. Genes lacking any peaks within the defined region were classified as non-target genes, while genes surpassing the threshold in terms of peak counts were designated as target genes. Specifically, for the aforementioned three cell types, threshold values of 10, 15, and 6 were respectively employed. Finally, the IGIS approach was employed to determine the corresponding ranks of TFs in the TF-target gene pairs.
Pathogenic SNPs prioritization
We collected single nucleotide polymorphisms (SNPs) data from the ClinVar and ExAC databases, which include both potentially pathogenic and benign SNPs. To evaluate the ability of EpiGePT to predict variant effects, we computed the log-odds scores (LOS) for multiple chromatin signals using EpiGePT on these SNPs. Subsequently, we utilized these scores to distinguish between pathogenic and benign SNPs. The LOS score for each chromatin signal was defined by computing a forward pass through the model using the reference and alternative alleles.
Each chromatin epigenomic profile in each cell line or tissue predicted by EpiGePT can be used to compute a specific variant score. We did not take the absolute value in this calculation, so the resulting LOS score indicates the direction of change in the model output after the appearance of the variant. In addition to the predicted chromatin signals output by the eight models, attention score changes based on self-attention are also noteworthy. We computed the log-odds scores for attention by summing the attention scores of the 10 bins upstream and downstream of the locus of the SNP, to evaluate the effect of the variant.
Where i represents the index of the neighboring bins relative to the locus of the SNP. To avoid the variant effects of different bins from cancelling each other out during the summation process, we computed the absolute value of the change in attention scores for each bin and then summed the scores of the 10 adjacent bins centered at the SNP position. For the classification of pathogenic SNPs, we calculated these nine LOS for attention separately for each of the 28 tissues or cell lines in training data. As a result, we obtained a feature vector of 252 dimensions for each SNP. Then a classifier with 252 features computed by EpiGePT and 52 annotations from CADD score as inputs are used to predict pathogenic SNPs against benign or likely benign SNPs. Here, we employed MLP as classifier to validate the effectiveness of the features we obtained. A five-fold cross-validation experiment is employed for validation, and we utilize two different positive-to-negative sample ratios, namely 1:1 and 1:2. For each sample ratio, we randomly sample 32,000 positive samples. The effectiveness of the variant score in identifying pathogenic SNPs is evaluated using the area under the auROC and the auPRC. Additionally, we also utilized the logistic regression (LR) as the classifier, consistent with the LR classifier used in CADD, and found a similar improvement when predicting pathogenic SNPs.
We applied the same method to calculate the LOS scores of the 8 predicted chromatin signals for the COVID-19 GWAS data. The absolute values of the scores were summed as the overall score for each SNP. For each significant SNP associated with COVID-19 severity obtained from the GWAS data, we selected normal SNPs within a 64kb region around the SNP as background to calculate the rank of the LOS score for the COVID-19 associated SNP in this region. Furthermore, we calculated the LOS scores for all 9, 484 COVID-19 associated SNPs and ranked them accordingly. The top 10 SNPs with the highest LOS scores were selected, which are considered to have potential genetic associations with COVID-19 severity and complications.
GTEx classification
We collected eQTL data from the supplementary materials of Wang et al28. In their study, the authors identified causal eQTLs through statistical fine-mapping, using a posterior inclusion probability (PIP) threshold of >0.9 for putative causal variants based on expression modifier score (EMS), and a PIP threshold of <0.9 for putative non-causal variants. To validate the ability of EpiGePT to distinguish potential causal variants, we perform a classification task on these variants. For each variation, 128kbp sequence regions near it were selected as the input of the model, and a score of variation was given by EpiGePT model. For each variant under each tissue, we can obtain an 8-dimensional vector of genomic features including DNase, CTCF and other ChIP-seq signals. Based on the LOS score, separate random forest classifiers consisting of 10 decision trees are trained for each tissue in order to distinguish between causal and non-causal variants. The models are evaluated using 5-fold validation on each tissue, with area under the auPRC and auROC as metrics for assessing their ability to distinguish between causal and non-causal variants.
Supplementary Material
Fig. S1. Three data partitioning strategies for model training and testing.
Fig. S2. EpiGePT's performance in predicting DNase-seq and other epigenetic signals.
Fig. S3. Performance of EpiGePT and baseline methods on chromatin states classification,multiple epigenomic profiles prediction and causal variants classification.
Fig. S4. Ablation analysis of the EpiGePT model.
Fig. S5. Case application of the EpiGePT-online.
Fig S6. Model architecture of EpiGePT for multiple epigenomic signals prediction.
Fig. S7. The fine-tuning performance of the EpiGePT model on predicting potential enhancer-promoter regulatory networks.
Fig. S8. The performance (auROC) of attention score of EpiGePT in distinguishing regulatory element-gene pairs at different distance ranges.
Text S1. Data splitting strategy for model training.
Text S2. System design and implementation of the web server.
Text S3. Running time of the EpiGePT and baseline methods.
Text S4. Implementation of Enformer model and Enformer+.
Table S1. The information of DNase-seq bam file across 129 biosamples from the ENCODE project.
Table S2. The information of RNA-seq tab-separated values (tsv) file across 129 biosamples from the ENCODE project.
Table S3. The information of DNase-seq, CTCF and other six Histone markers bam file across 28 cell lines or tissues from the ENCODE project.
Table S4. The information of RNA-seq tab-separated values (tsv) file across 28 cell lines or tissues from the ENCODE project.
Table S5. The preprocessed expression data of 711 human transcription factors from the ENCODE project across 129 biosamples.
Table S6. The preprocessed expression data of 711 human transcription factors from the ENCODE project across 28 cell lines or tissues.
Table S7. The order and names of epigenomes of the expression matrices across 56 epigenomes from the ROADMAP project.
Table S8. The preprocessed expression data of 642 human transcription factors across 56 epigenomes from the ROADMAP project.
Acknowledgments
Z.G and R.J. was supported by the National Key Research and Development Program of China [2021YFF1200902] and the National Natural Science Foundation of China [62203236 and 62273194]. Q.L., W.Z and W.H.W were supported by NIH grants R01 HG010359, P50 HG007735 and NSF DMS 1952386.
Footnotes
Code availability
All components of EpiGePT are freely available at https://github.com/ZjGaothu/EpiGePT. Here, users can access the code for reproducing EpiGePT, as well as the data collection and preprocessing pipelines used for model training in benchmark experiments.
Competing interests
The authors have declared no competing interests.
Data availability
Information and processed data of multiple chromatin signals of whole genome, motif binding status and expression data of TFs in the corresponding cell lines/tissues, which are used in EpiGePT are available at Supplementary Tables. The information about the cell lines/tissues used and the 711 filtered transcription factors is available in the supplementary table. The High throughput validated silencers of K562 cell line are download from SilencerDB (http://health.tsinghua.edu.cn/silencerdb) database. The HiChIP data of K562 cell line and GM12878 cell line are downloaded from HiChIPdb (http://health.tsinghua.edu.cn/hichipdb/) database. The DNase-seq peak and ATAC-seq peak data are obtained from the ENCODE project. Enhancer-gene pairs of CRISPRi24 experiments are obtained from the supplementary information of Gasperini et al. and Fulco et al. The regulatory network data for transcription factors and target genes were obtained from the TRRUST47 database (https://www.grnpedia.org/trrust/) and the GRNdb46 database (http://www.grndb.com). The annotated chromatin states for whole genome are downloaded from the ROADMAP epigenomics project (https://egg2.wustl.edu/roadmap/web_portal/chr_state_learning.html). The RNA-seq read counts matrix for protein coding genes used for the prediction of the chromatin 15-states annotated by ChromHMM are downloaded from the ROADMAP project (https://egg2.wustl.edu/roadmap/data/byDataType/rna/expression/57epigenomes.N.pc.gz). The GWAS data of COVID-19 are download from the COVID-19 Host Genetics Initiative (https://www.covid19hg.org/).
Reference
- 1.Brown T. et al. Language models are few-shot learners. Advances in neural information processing systems 33, 1877–1901 (2020). [Google Scholar]
- 2.Ramesh A. et al. in International Conference on Machine Learning 8821–8831 (PMLR, 2021). [Google Scholar]
- 3.Alexander R.P., Fang G., Rozowsky J., Snyder M. & Gerstein M.B. Annotating non-coding regions of the genome. Nature Reviews Genetics 11, 559–571 (2010). [DOI] [PubMed] [Google Scholar]
- 4.O’Malley R.C. et al. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165, 1280–1292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yenduri G. et al. Generative Pre-trained Transformer: A Comprehensive Review on Enabling Technologies, Potential Applications, Emerging Challenges, and Future Directions. arXiv preprint arXiv:2305.10435 (2023). [Google Scholar]
- 6.Liu Q., Xia F., Yin Q. & Jiang R. Chromatin accessibility prediction via a hybrid deep convolutional neural network. Bioinformatics 34, 732–738 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Min X., Zeng W., Chen N., Chen T. & Jiang R. Chromatin accessibility prediction via convolutional long short-term memory networks with k-mer embedding. Bioinformatics 33, i92–i101 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin Q., Wu M., Liu Q., Lv H. & Jiang R. DeepHistone: a deep learning approach to predicting histone modifications. BMC genomics 20, 11–23 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Q., Lv H. & Jiang R. hicGAN infers super resolution Hi-C data with generative adversarial networks. Bioinformatics 35, i99–i107 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng W., Wu M. & Jiang R. Prediction of enhancer-promoter interactions via natural language processing. BMC Genomics 19, 84(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Avsec Ž. et al. Effective gene expression prediction from sequence by integrating long-range interactions. Nature methods 18, 1196–1203 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Consortium E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57(2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vaswani A. et al. in Advances in neural information processing systems 5998–6008 (2017). [Google Scholar]
- 14.Song L. & Crawford G.E. DNase-seq: a high-resolution technique for mapping active gene regulatory elements across the genome from mammalian cells. Cold Spring Harbor Protocols 2010, pdb. prot5384 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou W. et al. Genome-wide prediction of DNase I hypersensitivity using gene expression. Nature communications 8, 1–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nair S., Kim D.S., Perricone J. & Kundaje A. Integrating regulatory DNA sequence and gene expression to predict genome-wide chromatin accessibility across cellular contexts. Bioinformatics 35, i108–i116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q., Hua K., Zhang X., Wong W.H. & Jiang R. DeepCAGE: incorporating transcription factors in genome-wide prediction of chromatin accessibility. Genomics, Proteomics & Bioinformatics 20, 496–507 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holwerda S.J.B. & de Laat W. CTCF: the protein, the binding partners, the binding sites and their chromatin loops. Philosophical Transactions of the Royal Society B: Biological Sciences 368, 20120369(2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bannister A.J. & Kouzarides T. Regulation of chromatin by histone modifications. Cell research 21, 381–395 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karolchik D. et al. The UCSC genome browser database. Nucleic acids research 31, 51–54 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ernst J. & Kellis M. Chromatin-state discovery and genome annotation with ChromHMM. Nature protocols 12, 2478–2492 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Larson M.H. et al. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nature protocols 8, 2180–2196 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gasperini M. et al. A genome-wide framework for mapping gene regulation via cellular genetic screens. Cell 176, 377–390. e319 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fulco C.P. et al. Activity-by-contact model of enhancer–promoter regulation from thousands of CRISPR perturbations. Nature genetics 51, 1664–1669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zeng W. et al. SilencerDB: a comprehensive database of silencers. Nucleic acids research 49, D221–D228 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mumbach M.R. et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nature methods 13, 919–922 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng W., Liu Q., Yin Q., Jiang R. & Wong W.H. HiChIPdb: a comprehensive database of HiChIP regulatory interactions. Nucleic Acids Research 51, D159–D166 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Q.S. et al. Leveraging supervised learning for functionally informed fine-mapping of cis-eQTLs identifies an additional 20,913 putative causal eQTLs. Nature Communications 12, 3394(2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Landrum M.J. et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic acids research 44, D862–D868 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karczewski K.J. et al. The ExAC browser: displaying reference data information from over 60 000 exomes. Nucleic acids research 45, D840–D845 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rentzsch P., Witten D., Cooper G.M., Shendure J. & Kircher M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic acids research 47, D886–D894 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li J., Lai S., Gao G.F. & Shi W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature 600, 408–418 (2021). [DOI] [PubMed] [Google Scholar]
- 33.org C.-H.G.I. a.b. The COVID-19 host genetics initiative, a global initiative to elucidate the role of host genetic factors in susceptibility and severity of the SARS-CoV-2 virus pandemic. European Journal of Human Genetics 28, 715–718 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agius L. Targeting hepatic glucokinase in type 2 diabetes: weighing the benefits and risks. Diabetes 58, 18–20 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Singh A.K., Gupta R., Ghosh A. & Misra A. Diabetes in COVID-19: Prevalence, pathophysiology, prognosis and practical considerations. Diabetes & Metabolic Syndrome: Clinical Research & Reviews 14, 303–310 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pellegrina D., Bahcheli A.T., Krassowski M. & Reimand J. Human phosphor-signaling networks of SARS-CoV-2 infection are rewired by population genetic variants. Molecular Systems Biology 18, e10823(2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Radford A., Narasimhan K., Salimans T. & Sutskever I. Improving language understanding by generative pre-training. (2018).
- 38.Devlin J., Chang M.-W., Lee K. & Toutanova K. Bert: Pre-training of deep bidirectional transformers for language understanding. arXiv preprint arXiv:1810.04805 (2018). [Google Scholar]
- 39.Li W., Wong W.H. & Jiang R. DeepTACT: predicting 3D chromatin contacts via bootstrapping deep learning. Nucleic acids research 47, e60–e60 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chor B., Horn D., Goldman N., Levy Y. & Massingham T. Genomic DNA k-mer spectra: models and modalities. Genome biology 10, 1–10 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Q., Chipperfield H., Melton D.A. & Wong W.H. A gene regulatory network in mouse embryonic stem cells. Proceedings of the National Academy of Sciences 104, 16438–16443 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharov A.A. et al. Identification of Pou5f1, Sox2, and Nanog downstream target genes with statistical confidence by applying a novel algorithm to time course microarray and genome-wide chromatin immunoprecipitation data. BMC genomics 9, 1–19 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raz R., Lee C.-K., Cannizzaro L.A., d’Eustachio P. & Levy D.E. Essential role of STAT3 for embryonic stem cell pluripotency. Proceedings of the National Academy of Sciences 96, 2846–2851 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.van den Berg D.L. et al. An Oct4-centered protein interaction network in embryonic stem cells. Cell stem cell 6, 369–381 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J. et al. The oncogene Etv5 promotes MET in somatic reprogramming and orchestrates epiblast/primitive endoderm specification during mESCs differentiation. Cell death & disease 9, 224(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang L. et al. GRNdb: decoding the gene regulatory networks in diverse human and mouse conditions. Nucleic acids research 49, D97–D103 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Han H. et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic acids research 46, D380–D386 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willett R. et al. TFEB regulates lysosomal positioning by modulating TMEM55B expression and JIP4 recruitment to lysosomes. Nature communications 8, 1580(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bernstein B.E. et al. The NIH roadmap epigenomics mapping consortium. Nature biotechnology 28, 1045–1048 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kulakovskiy I.V. et al. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic acids research 46, D252–D259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heinz S. et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Molecular cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He K., Zhang X., Ren S. & Sun J. in Proceedings of the IEEE conference on computer vision and pattern recognition 770–778 (2016). [Google Scholar]
- 53.Chen S. et al. OpenAnnotate: a web server to annotate the chromatin accessibility of genomic regions. Nucleic Acids Research 49, W483–W490 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Three data partitioning strategies for model training and testing.
Fig. S2. EpiGePT's performance in predicting DNase-seq and other epigenetic signals.
Fig. S3. Performance of EpiGePT and baseline methods on chromatin states classification,multiple epigenomic profiles prediction and causal variants classification.
Fig. S4. Ablation analysis of the EpiGePT model.
Fig. S5. Case application of the EpiGePT-online.
Fig S6. Model architecture of EpiGePT for multiple epigenomic signals prediction.
Fig. S7. The fine-tuning performance of the EpiGePT model on predicting potential enhancer-promoter regulatory networks.
Fig. S8. The performance (auROC) of attention score of EpiGePT in distinguishing regulatory element-gene pairs at different distance ranges.
Text S1. Data splitting strategy for model training.
Text S2. System design and implementation of the web server.
Text S3. Running time of the EpiGePT and baseline methods.
Text S4. Implementation of Enformer model and Enformer+.
Table S1. The information of DNase-seq bam file across 129 biosamples from the ENCODE project.
Table S2. The information of RNA-seq tab-separated values (tsv) file across 129 biosamples from the ENCODE project.
Table S3. The information of DNase-seq, CTCF and other six Histone markers bam file across 28 cell lines or tissues from the ENCODE project.
Table S4. The information of RNA-seq tab-separated values (tsv) file across 28 cell lines or tissues from the ENCODE project.
Table S5. The preprocessed expression data of 711 human transcription factors from the ENCODE project across 129 biosamples.
Table S6. The preprocessed expression data of 711 human transcription factors from the ENCODE project across 28 cell lines or tissues.
Table S7. The order and names of epigenomes of the expression matrices across 56 epigenomes from the ROADMAP project.
Table S8. The preprocessed expression data of 642 human transcription factors across 56 epigenomes from the ROADMAP project.
Data Availability Statement
Information and processed data of multiple chromatin signals of whole genome, motif binding status and expression data of TFs in the corresponding cell lines/tissues, which are used in EpiGePT are available at Supplementary Tables. The information about the cell lines/tissues used and the 711 filtered transcription factors is available in the supplementary table. The High throughput validated silencers of K562 cell line are download from SilencerDB (http://health.tsinghua.edu.cn/silencerdb) database. The HiChIP data of K562 cell line and GM12878 cell line are downloaded from HiChIPdb (http://health.tsinghua.edu.cn/hichipdb/) database. The DNase-seq peak and ATAC-seq peak data are obtained from the ENCODE project. Enhancer-gene pairs of CRISPRi24 experiments are obtained from the supplementary information of Gasperini et al. and Fulco et al. The regulatory network data for transcription factors and target genes were obtained from the TRRUST47 database (https://www.grnpedia.org/trrust/) and the GRNdb46 database (http://www.grndb.com). The annotated chromatin states for whole genome are downloaded from the ROADMAP epigenomics project (https://egg2.wustl.edu/roadmap/web_portal/chr_state_learning.html). The RNA-seq read counts matrix for protein coding genes used for the prediction of the chromatin 15-states annotated by ChromHMM are downloaded from the ROADMAP project (https://egg2.wustl.edu/roadmap/data/byDataType/rna/expression/57epigenomes.N.pc.gz). The GWAS data of COVID-19 are download from the COVID-19 Host Genetics Initiative (https://www.covid19hg.org/).