Abstract
Neurodegeneration in Huntington’s disease (HD) is accompanied by the aggregation of fragments of the mutant huntingtin protein, a biomarker of disease progression. A particular pathogenic role has been attributed to the aggregation-prone huntingtin exon 1 (HttEx1) fragment, whose polyglutamine (polyQ) segment is expanded. Unlike amyloid fibrils from Parkinson’s and Alzheimer’s diseases, the atomic-level structure of HttEx1 fibrils has remained unknown, limiting diagnostic and treatment efforts. We present and analyze the structure of fibrils formed by polyQ peptides and polyQ-expanded HttEx1. Atomic-resolution perspectives are enabled by an integrative analysis and unrestrained all-atom molecular dynamics (MD) simulations incorporating experimental data from electron microscopy (EM), solid-state NMR, and other techniques. Visualizing the HttEx1 subdomains in atomic detail helps explaining the biological properties of these protein aggregates, as well as paves the way for targeting them for detection and degradation.
1. Introduction
Huntington’s disease (HD) is one of a family of incurable neurological genetic disorders resulting from an aberrant expansion of a CAG trinucleotide repeat in a disease-specific gene.1,2 In HD, the mutation impacts the huntingtin (Htt) protein, which ends up with an expanded polyglutamine (polyQ) tract in its first exon (HttEx1, Fig. 1A).3 In HD patients and model animals, mutant Htt fragments matching HttEx1 form inclusions in brain areas affected by neurodegeneration. In cells, μm-sized inclusions are seen to contain fibrillar protein aggregates (Fig. 1B-C). The fibrillar HttEx1 adopts a morphology with all the common features of amyloid fibrils found in diseases such as Parkinson’s and Alzheimer’s disorders: highly stable protein deposits built around extended intermolecular β-sheets forming a characteristic cross-β architecture.4–6 The protein fibrils are of interest due to their potential involvement in pathogenic and neurotoxic processes—but also as biomarkers of disease onset and progression. Consequently, there is a need to resolve their atomic structures to facilitate the design of inhibitors, modulators, and diagnostic tools; e.g., a better understanding of HttEx1 fibril structure is essential to the development of ligands used to detect fibril formation in vivo.7 Recent years have seen important progress in amyloid structure determination through application of cryogenic electron microscopy (cryo-EM) and solid-state NMR spectroscopy (ssNMR):4,8,9 Breakthrough studies have produced structures of amyloid fibrils formed by tau, amyloid-β, and α-synuclein, associated with Alzheimer’s, Parkinson’s, and other amyloid disorders.9
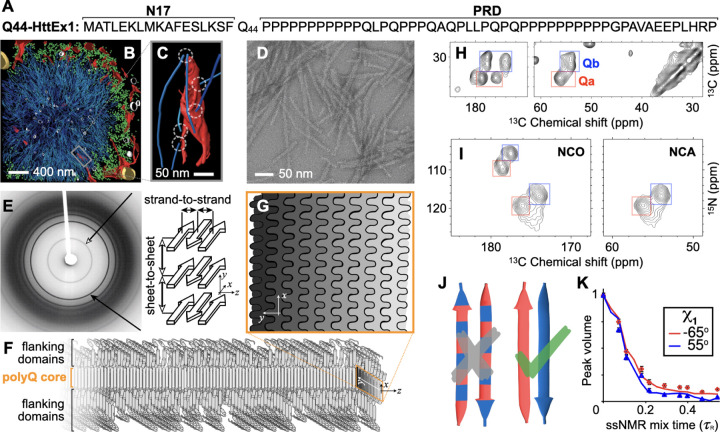
Figure 1. Huntingtin exon 1 sequence and aggregates.
(A) Sequence of Q44-HttEx1. N- and C-terminal flanking domains marked as N17 and PRD, respectively. (B) Colorized EM tomogram of a HttEx1 inclusion in mouse primary neuronal cells, showing protein fibrils (cyan) and mitochondria (gold). (C) Magnified region showing individual filaments interacting with ER membranes (red). (D) Negatively-stained TEM of Q44-HttEx1 aggregates formed in vitro; average fiber width ~6.5 nm. (E) X-ray fiber diffraction of K2Q31K2 fibrils, detecting the cross-β reflections of the amyloid core that represent the inter-strand and inter-sheet distances shown on the right. (F) Schematic model of a Q44-HttEx1 fibril, showing the N17 and PRD flanking domains outside the polyQ core. (G) Cross-section of the (6–7-nm wide) core showing the layering and interdigitation of the (differently shaded) β-sheets. (H) 2D 13C–13C ssNMR spectrum of 13C,15N-labeled Q44-HttEx1 fibrils showing backbone and sidechain cross-peaks. Peaks for Cα–CX correlations of two distinct “a” and “b” Gln conformers are marked with red and blue boxes, respectively. (I) 2D NCO and NCA ssNMR spectra of Q44-HttEx1 fibrils. (J) Pairs of antiparallel β-strands, schematically showing two previously proposed arrangements of the “a” (red) and “b” (blue) types of Gln. Given the ssNMR data (panels H–I), only the right one fits the experimental data. (K) ssNMR measurements sensitive of the side-chain dihedral angle χ1, with simulated curves for “a” (red) and “b” (blue) types of Gln. Panels B–C adapted with permission from Ref. 15, D–E and K from 5, F–G from 34, and I from Ref. 36.
The polyQ-based amyloid fibrils from HD and other CAG repeat disorders, however, have proved more challenging, and still lack truly atomic-level structures or even structural models.2 The aggregation propensity of polyQ proteins correlates strongly with their polyQ tract length, which inversely correlates with the age of onset.2,10 In their native state, polyQ segments are typically considered to lack a stable secondary structure, forming an intrinsically disordered region. This is also true for the polyQ-containing segment of Htt (HttEx1), which is invisible in cryo-EM of the full-length protein.11,12 However, in HD, N-terminal fragments of Htt can result from protease activity and missplicing, enabling subsequent aggregation into cellular inclusions.13,14 Cryogenic electron tomography in cellular context has revealed that μm-sized expanded inclusions contain nm-wide HttEx1 fibrils (Fig. 1B, C), which resemble those formed by purified HttEx1 in vitro (Fig. 1D).15,16
Prior structural studies have shown that HttEx1 fibrils comprise a highly ordered polyQ amyloid core surrounded by flexible non-amyloid flanking regions.17–20 Fiber X-ray diffraction studies showed the core itself to comprise anti-parallel β-sheets in a cross-β pattern (Fig. 1E);21–24 these experiments spurred proposals of various possible fibril models,21–24 which however lacked atomic-level detail. Moreover, all early X-ray-based models proved inconsistent with later work5,25–27 that revealed important new structural data via combinations of cryo-EM, ssNMR, and other techniques.5,17–20,27–32 A few recent studies are of particular interest. Firstly, an integration of ssNMR, X-ray, and EM measurements permitted the manual assembly of a schematic fibril architecture (Fig. 1F-G).5,18,25,26 Subsequently, a cryo-EM study of HttEx1 fibrils33 produced a medium-resolution density map, which—although it lacked the detail for a de-novo atomistic structure—was interpretable in terms of the said34 polyQ amyloid core architecture (Fig. 1F, G). The apparent success of such a unified (but as-yet qualitative) model underlines the need for a modern integrative-structural-biology approach35 to fully describe these fibrils that have much in common with other amyloids, but also a number of truly unique features.
Here we employ rigorous physics-based molecular modeling to integrate the rich set of collective experimental knowledge into the first atomic-resolution polyQ and HttEx1 fibril structures that are fully consistent with all the key experimental data, including sheet-to-sheet distances from fiber X-ray diffraction, fiber dimensions from EM, and structural constraints from ssNMR. The obtained structures are analyzed in the context of known structural features of HttEx1 amyloid fibrils, allowing a detailed explanation of their conformational and spectroscopic characteristics. Moreover, based on the disposition and accessibility of key residues and segments, the molecular architecture of these HD-related fibrils explains biological properties, such as the (in)ability for post-aggregation polyubiquitination and degradation.
2. Results
2.1. Architecture of the internal polyQ fibril core
We started the integrative structure determination of HttEx1 fibril from its polyQ amyloid core. Most experimental studies, in particular recent ssNMR studies, argue for a molecular architecture common to polyQ-containing peptide and protein fibrils.37 Various techniques show that polyQ amyloid features antiparallel β-sheets (Fig. 1E),23,24,27,30,38 in contrast to the parallel in-register fiber architecture of many other amyloid fibers.8,39 Another distinct feature of polyQ amyloid is that it forms long β-strands with few turns (Fig. 1F),5,24,26,33,34 unlike the majority of other amyloid structures that have short β-strands connected by turns or loops.8 The polyQ fiber core is multiple nanometers wide, devoid of water molecules, and forms a block-like structure featuring a stacking of less than ten β-sheets.24–26,33 Its cross-section must contain polyQ segments from multiple protein monomers (schematically illustrated in Fig. 1F-G).34 A large proportion of the Gln residues is buried, far away from the solvent, in a repetitive and pseudo-symmetric context. This ordered and repetitive nature is demonstrated by high-quality ssNMR spectra, with few and relatively narrow peaks (Fig. 1H).5,18,20,25,26,29,34 These ssNMR studies consistently show a characteristic spectroscopic fingerprint that identifies two dominant Gln residue conformations—see the two sets of peaks from so-called “a” and “b” conformers (red and blue boxes) present at roughly equal intensities in the 2D ssNMR spectra of Q44-HttEx1 fibrils (Fig. 1H-I). The two Gln conformers reflect two types of β-strand structures that co-assemble into a single antiparallel β-sheet (Fig. 1J).5,20,26
Integrating these experimental data, we concluded that the internal polyQ core structure can be captured by an infinitely-repeating eight-Gln unit that features antiparallel β-sheets (Fig. 2A). This construction has translational symmetry and lacks the twisting seen in many amyloid fibrils, since EM analyses of our and others’ HttEx1 fibrils lack strong signs of twisting.33 The repeating ‘unit cell’ (Fig. 2A) fulfills several a priori requirements: First, its peptide chain segments are two residues long, the minimum needed to capture the ‘odd/even’ side-chain alternation in a β-strand. Given that ssNMR shows only two ssNMR-detected conformers, in long strands, two residues should be sufficient to capture the structural variation within this amyloid core. Second, the unit cell contains two β-strands (that can differ in conformation), the minimum needed to describe an antiparallel β-sheet. This contrasts with parallel in-register β-sheets that could be represented with a single repeating β-strand.40 Third, the unit cell contains a stacking of two β-sheets, to permit the probing of distinct sheet–sheet dispositions. Fourth, certain ranges of side-chain dihedral angles (χ1, χ2, χ3) are required in order to fulfill a crucial feature of the interdigitating polyQ amyloid:5,23 the side-chain–side-chain hydrogen bonding that allows the polar Gln residues to be stowed in the waterless core environment (see Fig. 2A and Supplementary Fig. S1). Thus, e.g., the χ2 dihedral must be close to 180°, as also known from ssNMR and Raman experiments.5,41 This minimal model (Fig. 2A) fulfills the above-noted experimental constraints, and still yields a library of roughly 1 000 distinct architectures (see Methods for details).
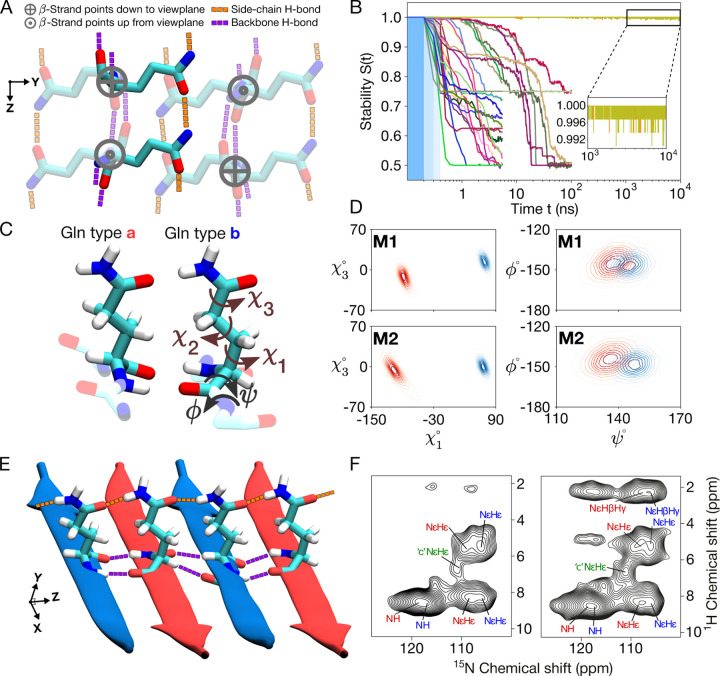
Figure 2. Structure of the internal polyQ amyloid core.
(A) The eight-Gln building block used to generate polyQ core candidates. Six residues are shown faded out to bring the two non-faded in focus: These two are on adjacent β-strands of an antiparallel β-sheet that is stabilized not just by the backbone hydrogen bonds (purple), but also by the continuous chains of side-chain hydrogen bonds (orange). The latter are crucial for packing the polar glutamines into the waterless amyloid core. When generating the core candidates, all the χ1 and χ3 dihedral angles were independently rotated to explore all potential hydrogen bond networks. (B) Stabilities of the 30 experimentally-feasible polyQ core candidates (represented by color-coded lines) as a function of MD simulation time. The three blue-shaded sections indicate position restraints of 1 000, 500, and 100 kJ/mol over consecutive 100-ps periods. Notably, only two candidates maintain stability throughout the 10-μs MD simulations; we denote these M1 and M2. (C) Atomic-level structures of the type “a” and “b” Glns in M2. Gln dihedral angle names shown on “b”. (D) The side-chain χ1–χ3 (left panels) and backbone ψ–ϕ (right) dihedral angle distributions of conformers “a” (red) and “b” (blue) for the final models M1 (top) and M2 (bottom). (E) Illustration of the inter-side-chain hydrogen-bond chain (orange) formed within β-sheet in M2. (F) 1H–15N 2D ssNMR spectra of K2Q11pGQ11K2 peptide fibrils with a contact time of 100 μs (left) and of 350 μs (right) showing core and surface signals from the polyQ. Peaks from the core conformers “a” (red) and “b” (blue) are indicated. The side chain peaks that participate in hydrogen bonding (NϵHϵ and NϵHβ/γ) are marked. Note that the Hϵ 1H shifts are indistinguishable for the “a” and “b” conformers. Conformer “c” (green) refers to glutamines outside the core, on the surface of the fibril.
A key additional consideration is that ssNMR has unambiguously shown sequential residues within each β-strand to have the same backbone conformations (based on identical chemical shifts). Thus the “a” and “b” conformers strictly occupy distinct strands, within which uniform backbone and χ1 dihedral angles are found (Fig. 1J). Moreover, these two distinct β-strand types differ in their ψ and χ1 dihedral angles (Fig. 1K). After applying this additional filtering, 30 distinct unit cell architectures still remained possible. To investigate the structural stability of these 30 models, we carried out all-atom MD simulations in a fully periodic setting on them; see Methods for details. Only two of the candidate models proved stable throughout the simulations (Fig. 2B and Supplementary Fig. S2).
Notably, these two stable 3D lattices (called M1 and M2) nicely capture known features of polyQ amyloid structure. For instance, the experimental constraints offered by ssNMR dihedral angle measurements are open to more than one interpretation, as is typical for such experiments.5,42 Previously, this ambiguity and angular uncertainty yielded qualitative models useful for illustrative purposes, but not robust upon close inspection. Here, we have narrowed down the viable and physically plausible models to only two, with these models identifying specific narrow regions within the equivocal dihedral angle space (Fig. 2D). In both models, the antiparallel β-sheet harbors strand-specific Gln structures occupying the side chain rotamers known as pt20° and mt-30°, according to Ref. 43. These conformations, as required, enable hydrogen bonding interactions between the side chains of the Gln conformers, in addition to typical strand-to-strand backbone hydrogen bonds (Fig. 2A,E). We used ssNMR to detect the protons involved in this hydrogen bonding (NϵH protons), in the form of 2D 1H-15N HETCOR spectra (Fig. 2F). The 1H chemical shifts are indistinguishable between the “a” and “b” Gln conformers. Structurally, this implies that the H-bonding distances should be very similar.44,45 This consideration was not imposed in the modelling, but is indeed observed in the stable structures (Supplementary Fig. S3), which demonstrate comparable side-chain–side-chain and side-chain–backbone distances in both the “a” and “b” conformers. In sum, we obtained valid atomic-resolution structures for the polyQ amyloid core, in two models that have many features in common. They formed the basis for further analysis and modeling, as will be discussed next.
2.2. Structure of polyQ15 peptide fibrils
The above 3D models capture the internal core of polyQ protein fibrils, but lack the defined width and water-facing surface features of the real polypeptide fibrils. To model a proper experimental system, we modified the ‘infinite cores’ M1 and M2 into models of fibrils formed by the polyQ15 peptide (Fig. 3A). These widely studied5,21,24,46 peptides contain a 15-residue polyQ segment, flanked by charged residues to enhance solubility. Experimental analysis by ssNMR spectroscopy and X-ray diffraction has shown that atomic conformations in the polyQ15 fibril core match those of the disease-relevant HttEx1 fibrils; e.g., the ssNMR signals from Gln within the D2Q15K2 fibrils (Fig. 3A) and the Q44-HttEx1 fibrils (Fig. 1H) are indistinguishable. Using both the M1 and M2 models, we constructed D2Q15K2 fibrils comprising seven anti-parallel β-sheets, consistent with the 5.5- to 6.5-nm fibril width seen experimentally by TEM (Fig. 3A); Fig. 3B shows the M2 D2Q15K2 fibril in cartoon representation. We then subjected both polyQ15 models to 1-μs atomistic MD simulations in explicit water; for details, see Methods. The structure of the buried Gln residues within the core remained stable throughout the simulations (Supplementary Fig. S4).
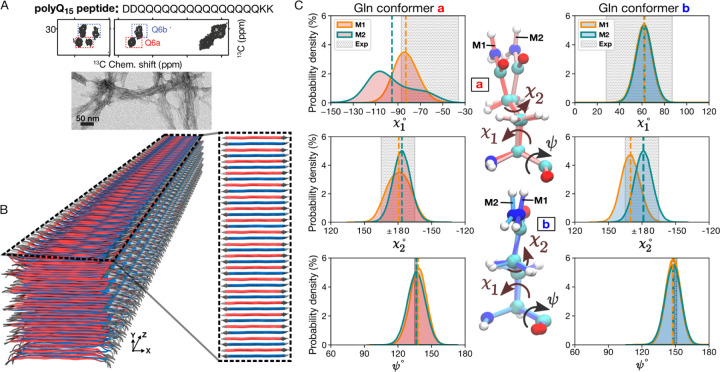
Figure 3. PolyQ peptide fibril structure.
(A) Amino acid sequence of D2Q15K2 peptide (top), 2D 13C–13C ssNMR spectrum (middle) of D2Q15K2 fibrils with single 13C-labeled Gln (Q6), and negatively-stained TEM of the peptide aggregates (bottom). The ssNMR data adapted from Ref. 47. Signals from type ”a” and ”b” Gln conformers are shown in red and blue boxes, respectively. (B) 3D cartoon representation of the structural model of the D2Q15K2 fibril. The alternation of β-strands of ”a” (red) and ”b” (blue) Gln conformers within a single sheet is shown on the right. (C) The χ1, χ2, and ψ dihedral angle distributions of “a” (left) and “b” (right) conformers in the context of the D2Q15K2 fibril for the M1 (orange) and M2 (green) models; dashed vertical lines represent mean values. The data were obtained from 1-μs MD simulations. Gray shading depicts the ssNMR constraints for the dihedral angles. The structures at the center show representative “a” (top) and “b” (bottom) conformers of the M1 and M2 models.
There is a high degree of agreement between the χ1, χ2, and ψ dihedral angle distributions observed in MD simulations of our polyQ15 models and the ssNMR angle constraints obtained via HCCH (χ1, χ2) and NCCN (ψ) experiments (Fig. 3C and Supplementary Fig. S5). As already noted, the “a” and “b” ssNMR signals reflect different χ1 and ψ values in the two Gln conformers, but a similar χ2 value. This is beautifully reproduced by the simulations of both polyQ15 models (Fig. 3C). The largest deviation affects the M2 model, in terms of its broader distribution of the conformer “a” χ1 angle, which in part lies outside the shaded region representing the (ambiguous) ssNMR constraint. Interestingly, experimental studies have shown5 that the conformer “a” is more dynamic than conformer “b”, reminiscent of the increased heterogeneity of this conformer in the MD simulations of both models (see also Supplementary Fig. S4). Also, the sheet-to-sheet and strand-to-strand distances within the models match well with repeat distances from X-ray diffraction (Supplementary Fig. S6). In sum, the MD simulations support the stability of these polyQ amyloid core structures and recapitulate key data from multiple experimental techniques.
Our modeling reveals an important new feature of polyQ15 fibrils, which, so far, has not been seen or studied: the solvent-facing external structure of the polyQ amyloid core. Fiber surface mediates interactions with cellular surroundings but also with any purposefully administered compounds, such as thioflavin-T or amyloid-specific tracers used in positron emission tomography (PET).7 The two outermost β-sheets exposed to water (top and bottom sheets in Fig. 3B) allow us to compare the surface-exposed Gln residues versus the residues buried in the fibril core. Figure 4A highlights the water-exposed Gln side-chains (green) in the polyQ15 model. These surface glutamines appeared as an averaged signal in ssNMR due to their increased dynamics (Fig. 2F green) and thus could not be resolved. Figure 4 shows the χ1–χ2 and χ3–χ2 dihedral angle distributions of the core Gln residues (panel B, gray) and compares them with the surface residues (panel C, green) for model M1 (see Supplementary Fig. S7 for M2). For the core residues, the rotamer states are similar to those discussed above (pt20° and mt-30°). For the surface residues, additional rotameric regions emerge, including Gln rotamers where χ2 deviates from 180°, close to rotamer pm0°.43 Interestingly, surface residues do not show fully free side-chain motion and retain defined structural features that determine the molecular profile of the corrugated polyQ amyloid core surface.
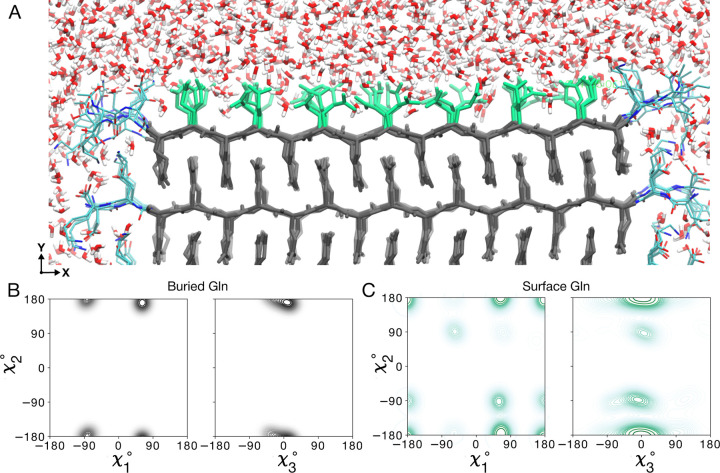
Figure 4. Water-facing surface of polyQ amyloid.
(A) Atomistic MD snapshot of the D2Q15K2 peptide fibril’s polyQ surface in contact with water. Exposed and buried Gln residues are colored green and gray, respectively. Note how the Gln side-chains internal to the (model M1) amyloid core are well-ordered, while the water-facing side-chains display more mobility. (B) Side-chain dihedral angle distributions for the buried Gln residues, and (C) for the Gln residues on the fibril surface. The surface-facing residues show more disorder, but are nonetheless constrained to eight varyingly prominent specific rotamer states.
2.3. Htt exon 1 fibril model
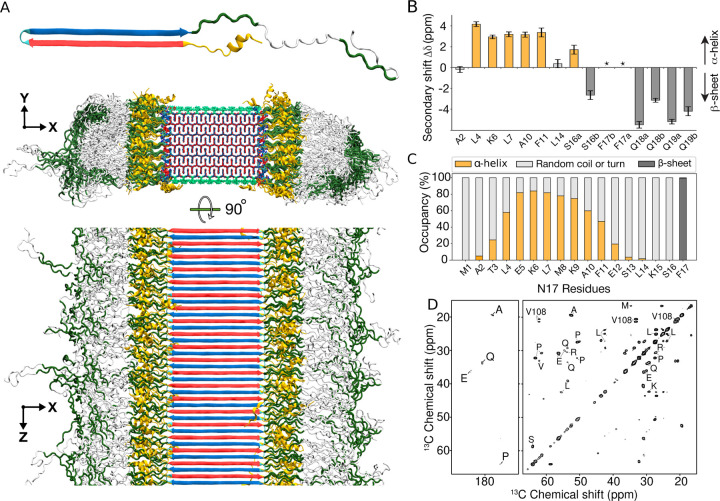
As already noted, the protein inclusions seen in HD patients and various HD model animals prominently feature the mutant HttEx1 protein fragment. Until now, no atomistic model of HttEx1 fibrils has been reported, although a diversity of schematic or cartoon-style models has been published over the years. Here, we build on our above-presented polyQ amyloid core model structures to construct an experiment-based molecular structure of Q44-HttEx1 fibrils. The Q44-HttEx1 construct is used to model the disease-relevant HD protein in a variety of experimental studies,18,19,34,36,48 as HD patients commonly have CAG repeat expansions that yield Htt proteins with approximately forty residues in their polyQ domain.2 Experimentally, similar to our analysis of the polyQ15 peptide fibrils above, the large majority of the polyQ residues are observed to be buried in the HttEx1 fibril core, in a multi-nm-size block-like core architecture.34 Different from fibrils made from short polyQ segments, the polyQ-expanded protein fibrils have protofilament architectures19,34 where the polyQ segment adopts a β-hairpin structure;5 the Q44-HttEx1 fibrils feature a β-hairpin with a single turn and two ~20 residue-long β-strands. Solid-state NMR studies with site-specific amino acid labels have indicated that the ordered β-sheet core extends from the final N17-domain residue (F17) to the penultimate glutamine in the polyQ segment.18,25 Unlike the polyQ segment, the two polyQ-flanking segments of HttEx1 (see Fig. 1A) are solvent exposed, lack β-structure, and display increased motion and disorder.17,19,20,33 The short N-terminal N17 segment has been reported to adopt a random coil or α-helical structure, depending on context;25,49 in fibrils, Electron Paramagnetic Resonance (EPR) and ssNMR have shown the N17 segment to display partial order, and partly α-helical structure.17,19,25 The longer C-terminal proline-rich domain (PRD) is disordered, except for the polyproline-II (PPII) helices at the locations of the two oligoproline segments.
Using these experimental data as input, we constructed the Q44-HttEx1 core base-architecture after the schematic model described earlier.5,18,19,34 The 44-residue polyQ segment has a β-hairpin conformation with a tight β-turn, containing an even number of residues, and two long β-strands extending from F17 to the penultimate glutamine. A notable feature of polyQ protein aggregates is that the polyQ domains can come together in many different orientations and alignments, yielding a large propensity for heterogeneity and stochatistic modes of assembly.18,33,34 Here, a configuration was selected to construct a model that mimics our previous schematic model,34 with flanking domains evenly distributed on both sides of the fibril core. To this end, we attached the appropriate HttEx1 flanking domains to the terminal regions of the polyQ section, thereby achieving the formation of monomer building blocks as illustrated in the top panel of Fig. 5A. The thus constructed HttEx1 fibril was subjected to unrestrained all-atom MD simulations to assess its stability and monitor its structural dynamics. The fibril conformation obtained after a 5-μs simulation is shown in Fig. 5A.
Figure 5. Structure of Q44-HttEx1 amyloid fibril.
(A) Atomic-level structural model for mutant HttEx1 fibril. The top image shows a representative monomer within the fibril, with its β-hairpin polyQ segment to the left and the largely disordered flanking segments to the right. The middle image shows a cross-section, and the bottom image a side view of the fibril. The polyQ core is shown with the conformer-identifying red (for “a”) and blue (for “b”) β-strands, the N-terminal flanking segments yellow, and the C-terminal polyproline II helices dark green. Surface residues of the polyQ amyloid core are light green. (B) Secondary chemical shift values for residues in the N-terminal end, indicating local α-helix (positive values) or β-sheet (negative) conformations. Doubled peaks indicating multiple co-existing conformations are marked with letters a and b. The asterisks mark F17, for which a peak (13Cβ) was not detected, but other resonances indicate β-sheet structure. (C) Secondary structure distribution of the 17 residues in the N-terminal flanking domain during the last 500 ns of 5-μs MD simulation. (D) A 2D TOBSY ssNMR spectrum of Q44-HttEx1 fibrils, in which observed cross-peaks correspond to highly flexible residues outside the fibril core. Most, but not all, peaks originate from the C-terminal tail of the PRD. Spectrum was acquired at 600 MHz at 8.33 kHz MAS.
2.4. Structural analysis of HttEx1 fibril
The resulting structure (Fig. 5A) reveals interesting features that permit comparison to experimental studies. Firstly, consistent with the polyQ15 fibril model, the HttEx1 polyQ core structure is found to be highly stable, with the bulk of the glutamine residues residing within the rigid and dehydrated core. A noteworthy observation lies in the β-turn conformation embedded within the β-hairpin structure: The simulation data underscores predominance of the type II turn over a type I’ conformation (Supplementary Fig. S8), a finding that concurs with the experimental evidence gathered from ssNMR study of such compact turns.5 As for polyQ15 fibrils, a minority population of glutamines (the light-green side-chains of the middle image in Fig. 5A) is exposed to the solvent; these surface side-chains, as analyzed in Fig. 4 above, show a semi-rigid behaviour, retaining a relatively high degree of structural order. The constrained dynamics of these solvent-exposed glutamines stand in large contrast to the dynamic disorder of both non-polyQ flanking domains—whose disposition and structure are of substantial interest, given that the flanking domains are known to govern key structural and pathogenic properties of the protein and its aggregates.17,19,34,47,50–52 Figure 5A illustrates the high level of disorder that appears in the MD ensemble of the flanking domains, manifested in both the N17 and the PRD. Although such pronounced dynamics interfere with the detailed experimental study of the flanking domains, their appearance in simulations fits with the dynamic disorder of the flanking domains in HttEx1 fibrils observed in many experimental studies, whether based on EPR, EM, or ssNMR.15,17,19,20,33,47
The secondary-structure preferences of N17, Q44, and PRD are summarized in Supplementary Fig. S9. There has been significant interest especially in the N17 segment, as it drives HttEx1 aggregation, but also harbors post-translational modifications that regulate HttEx1 (dis)aggregation and degradation.53–55 The fate of N17 in the fibrils has remained somewhat opaque, with seemingly conflicting reports of the presence and absence of (partial) α-helicity. The obtained HttEx1 fibril model may provide some insights, as its N17 segment displays a mixed secondary structure content, with much disorder (Fig. 5C and Supplementary Figs. S10–13). Virtually all N17 residues are seen to show some propensity for disorder, such that a subset of the proteins in the modelled fibril has an N17 devoid of α-helical structure (Fig. 6D). Yet, in close to half of the protein monomers, an α-helix is observed within N17. These findings match well to ssNMR analysis of the structure of N17 in fibrillar samples:19,25 On the one hand, signals from an α-helical N17 were detected, with helicity constrained to only part of the segment (Fig. 5B). The helical residues seen experimentally coincide remarkably well with the residues found to favour helicity in the MD fibril (Fig. 5C), providing further support for the validity of this structural ensemble. The observation that a significant part of N17 is not α-helical finds experimental support as well. Indeed, in some types of HttEx1 fibrils, no evidence of a structured or helical N17 has been found, indicating a propensity for substantial disorder.20,56 In other studies, N17 residues are observed to display both ordered and dynamic (ssNMR) signals, showing that different protein monomers have different degrees of (dis)order in the N17 segment.34 This same finding is clearly recapitulated in the obtained MD fibril model: It displays an innate heterogeneity in structure and dynamics of the N17 segment, even among monomers in the same fibril.
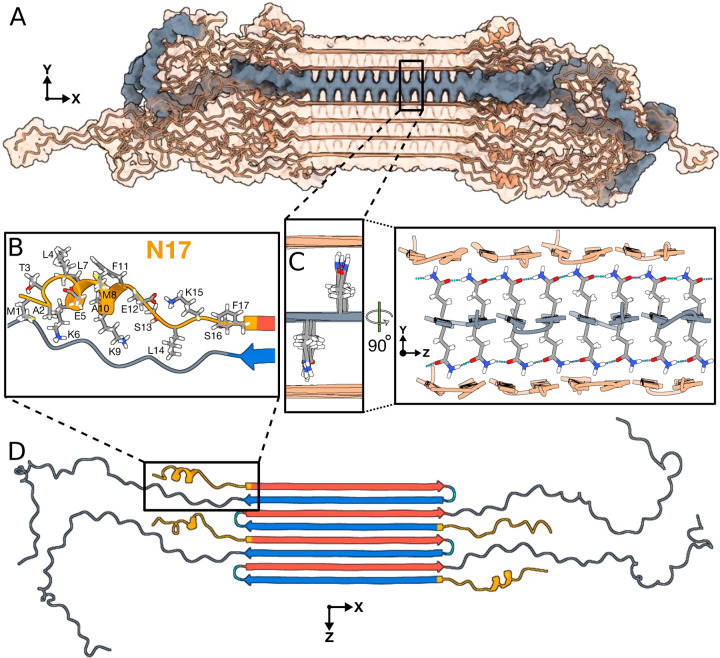
Figure 6. Structural analysis of the Q44-HttEx1 amyloid fibril.
For an illustrative 3D exploration of Q44-HttEx1 fibril structure, see the associated video content available in Extended Data 1. (A) A graphical depiction of the HttEx1 fibril. The region shaded in gray denotes a single sheet within the fibril’s architecture. (B) An atomic view of the N17 domain within the fibril, naming the specific amino acids. (C) An atomic depiction of the glutamine side-chains within the fibril. The high stability of the fibril structure is primarily attributed to the extensive hydrogen bonding interactions among the glutamines, as depicted in the right panel. (D) Side view representation of the β-sheet highlighted in panel A. A quartet of HttEx1 monomers is visible. The polyQ is color-coded for the type “a” (red) and “b” (blue) strands; the tight β-turn is cyan. The N17 and PRD domains are orange and gray, respectively. Note the structural variation between different monomers in the same fibril sheet, including in particular the range of helical content in the N17.
C-terminal to the polyQ segment, the PRD is known to display a gradient of dynamics.17–20,34,56 Experimentally, the PRD is relatively rigid proximal to the polyQ core, whilst its very tail end is highly flexible. The latter is evidenced by ssNMR measurements of NMR relaxation parameters and dynamics-selective spectra (Fig. 5D).19,20,28 The MD simulations indicate that the mobility of the PRD is not only constrained by its attachment to the rigid polyQ core, but also by pronounced PRD–PRD intermolecular interactions. Prior work has inferred a propensity for such interactions, especially in context of filament–filament interactions.19,29,34 The current model suggest that such interactions are also prominent in structuring the flanking domains of isolated protofilaments.
A notable feature of the HttEx1 fibril structural ensemble that was not a priori expected is that the flanking domains are not showing much interaction with the polyQ amyloid core surface. Throughout the simulation, the flanking domains display substantial dynamics, but preferentially cluster together. This behaviour seems reminiscent of the type of liquid phase separation seen for, e.g., the nuclear pore complex, mediated by the proline-rich segments. This leaves the outer polyQ core surface easily accessible not only to solvent, but also amyloid-binding molecules such as thioflavin-T (ThT) as well as PET ligands.7 An in-depth and comprehensive visualisation of the described HttEx1 fibril structure can be further enhanced by viewing the accompanying movie in Extended Data 1.
2.5. Implications for huntingtin biology
Cryogenic electron tomography analysis of cellular httEx1 aggregates have indicated that the fibrils may be structurally similar to those formed by purified proteins,15 which would suggest that our model also applies to cellular inclusions. Notably, structural features of our model appear to match, and thus rationalize, reported biological properties of HttEx1 fibrils. In particular, the surface-accessible features of amyloid fibrils are crucial for their biological, possibly pathological, properties. In our fibril architecture, the N17 segment is found to reside outside the fibril core, where it displays conformational and dynamic disorder. Despite these dynamics it is, however, tightly enclosed by the C-terminal PRD domains. Consequently, residues in N17 may be solvent accessible, but are nonetheless largely inaccessible to larger macromolecules such as chaperones, kinases, ubiquitinases, and other potential N17 interaction partners. Although in Fig. 6B the known phosphorylation sites T3, S13, and S16 may appear accessible, Fig. 6A shows this monomer in context, underlining that the N17 segment is fully surrounded by the longer PRD segments. Similarly, the only HttEx1 ubiquitination sites involve lysines K6 and K9 in N17, rendering them inaccessible. The buried nature of N17 also rationalizes the low level of engagement by the TRiC chaperone:31,57 Although known to bind this part of HttEx1, prior EM studies have shown TRiC to be unable engage HttEx1 fibrils, except near the fibril ends. N17 is also implicated in membrane interactions, such that its preferential exposure at fibril ends may explain the latter to be engaged with the ER membranes in Fig. 1C.52,58,59
Naturally—although we expect the obtained fibril structure to be a good representation of the fibrils present in our samples and also to have strong predictive and descriptive qualities for cellular HttEx1 aggregates—it cannot be excluded that cellular factors (ranging from chaperones to membrane interactions) may modulate the aggregation mechanism to the extent that the mature fibril architecture differs from the one obtained in vitro. Yet, unlike for other amyloid proteins, a remarkable feature of the HttEx1 ssNMR studies is that multiple groups have studied a variety of HttEx1 fibrils and always found the same signature spectra that are connected to the structural parameters used to construct our model. Indeed, there is little evidence for a qualitatively different fibril architecture. Thus, we are inclined to expect that while cellular conditions may change certain details in the fibril structure (for example, the fibril polymorph formed in presence of membranes may feature more exposed N17 segments52), they would not fundamentally change the protofilament architecture.
Yet, we do need to note that polyQ-based protein aggregates display a persistent and characteristic structural heterogeneity that is impossible to fully capture in practical MD simulations. Prior studies have discussed the predictable propensity for polyQ segments to self-assemble in a somewhat disordered or stochastic fashion, due to the lack of sequence variation.5,18,60 For instance, a polyQ chain extending an existing fibril can be added in different orientations. This variability is to some extent already displayed in our model: The alternating monomers in the single sheet in Fig. 6D have differently organized β-hairpins in their polyQ segment and their N17 segments on opposite sides of the filament. What is explicitly missing from our model is that incoming polyQ segments can add to the fibril with register mis-alignments, resulting in shorter or longer β-strands in slightly different sequence positions, without a major energetic cost. This would be expected to yield fibril cores with varying fibril widths, local defects, and protein-to-protein structural variations18,33,60 not accessible to more canonical amyloid fibrils formed by other proteins. This means that polyQ protein fibrils are expected (and observed33,34) to display inherent structural variations between fibrils in a single sample, and even within single fibrils. Thus it appears that, like snowflakes, each HttEx1 fibril is unique (which prohibits the canonization of a single definitive protofilament structure) but still characterizable by well-defined structural features. This indistinguishability in local (atomic) structure of differing fibril architectures manifests for instance in the near-identity of ssNMR peak positions reported for polymorphic HttEx1 fibrils.
Indeed, like other amyloids, polyQ proteins form different fibril polymorphs depending on experimental conditions.19,29,61 This structural variation imbues the fibrils with different degrees of cytotoxicity.29,61,62 Notably, the HttEx1 polymorphs often reflect a type of ‘supramolecular’ polymorphism, with different supramolecular assemblies formed from similarly structured protofilaments.34 The current model is expected to illuminate the atomic level conformation of one such protofilament, as it is based on experimental constraints that define its structure. While we do not explicitly probe the conformations of supramolecular polymorphs, the clustering of the dynamic flanking domains, seen for our single filaments, likely has some relevance to the inter-filament interactions—thought to be mediated by the PRD segments as well.
Finally, let us briefly discuss the implications of our models for HttEx1 fibrils formed by proteins with longer (and shorter) polyQ segments. Our HttEx1 fibril modelling focused on the HD-relevant Q44 that falls into the regime of common expansion lengths seen among patients. Yet, famously, mutant proteins can differ widely in their polyQ lengths. As illustrated in the polyQ15 peptide fibril, and discussed elsewhere,37,56,62 proteins with shorter polyQ lengths can still form fibrils (at least in vitro). However, in such fibrils the polyQ segment may not form a β-hairpin, but instead occupy a single extended β-strand. Naturally, this would modulate the disposition of flanking segments on the fibril surface. Nonetheless, the qualitative architecture would remain unchanged. Conversely, HttEx1 with longer polyQ lengths, such as those associated with juvenile HD, would be expected to form amyloid cores featuring multiple turns, unlike the single-turn structures analyzed here for Q44-HttEx1.
3. Conclusions
In this report we have described our construction of experimentally informed and validated structural models of polyQ15 and Q44-HttEx1 amyloid fibrils. These models provide the best atomistic views of these disease-associated protein inclusions to date, derived from a multi-technique structural analysis through integrative modelling. The obtained HttEx1 structure rationalizes a variety of experimental findings with notable biological and biomedical implications. The polyQ segment is mostly buried within the fibril, but the model reveals the structural and dynamical features of the minority of solvent-facing residues. These surface residues have proved challenging for experimental study, necessitating tailored and targeted approaches in, e.g., ssNMR.36 A better structural understanding of this special polyQ surface will be useful in efforts to design polyQ-amyloid-specific binders, e.g., for PET imaging.7 The visualization of the dynamically and structurally heterogeneous flanking domains enhance our understanding of their accessibility in the fibrils and pave the way for more in-depth analyses of their role in intracellular interactions with proteins and organelles.
4. Methods
Protein production and fibrillation
Mutant huntingtin exon 1 with a 44-residue polyQ core was expressed as part of a maltose binding protein (MBP) fusion protein as previously reported.18,19 The fusion protein MBP-Q44-HttEx1 was expressed in Escherichia coli BL21 (DE3) pLysS cells (Invitrogen, Grand Island, NY). Uniformly 13C and 15N labeled MBP-Q44-HttEx1 protein was expressed with 13C D-glucose and 15N ammonium chloride for MAS ssNMR studies. Then, cells were pelleted at 7 000 g, resuspended in phosphate buffered saline (PBS), pH 7.4 and lysed in presence of 1 mM phenylmethanesulfonyl fluoride (PMSF) by a HPL 6 maximator (Benelux BV, The Netherlands). After that, cells were centrifuged at 125 000 g for 1 h using an Optima LE-80K ultra-centrifuge (Beckmann Coulter). The supernatant was filtered over Millex-GP syringe-driven 0.22 μm PES membranes (Millipore Sigma, Burlington, MD). The MBP-Q44-HttEx1 protein was purified by fast protein liquid chromatography (FPLC) using a 5 ml HisTrap HP nickel column (GE Healthcare, Uppsala, Sweden) with 0.5 M imidazole gradient (SKU I5513–100G, Sigma, St. Louis, MO) on an AKTA system (GE Healthcare, Chicago, IL). The imidazole was removed from the purified protein using an Amicon Ultra centrifugal filter with a regenerated cellulose membrane (Millipore Sigma, Burlington, MA). At least 3 washes with imidazole-free PBS buffer were done. Protein concentration was calculated from the absorbance at 280 nm. According to ProtParam tool by ExPasy63 the extinction coefficient of the fusion protein is 66 350 M−1cm−1. Protein aggregation was initiated by addition of Factor Xa protease (SKU PR-V5581, Promega, Madison, WI) at 22°C, in order to cleave off the MBP fusion tag19,34 and release Q44-HttEx1. After 3 days, the obtained mature fibrils were washed with PBS to remove the cleaved MBP.
Transmission Electron Microscopy (TEM)
Transmission electron microscopy (TEM) was performed on mature D2Q15K2 peptide fibrils. The fibrils were re-suspended in MiliQ and then 5 μl of the sample was deposited on the plain carbon support film on 200 mesh copper grid (SKU FCF200-Cu-50, Electron Microscopy Sciences, Hatfield, PA). The grid was glow discharged for 0.5–1 min before adding the sample. After 30 s of the sample deposition, the excess MiliQ was removed by blotting, and immediately the negative staining agent 1% (w/v) uranyl acetate was applied. After 0.5–1 min, the excess stain was removed and the grid was air dried. The images were recorded on a Tecnai T12 electron microscope.
NMR experiments
The hydrated U-13C,15N-labeled Q44-HttEx1 fibrils were packed into a 3.2 mm ssNMR rotor (Bruker Biospin) using a previously described packing tool.64 The fibrils were packed in the rotor by centrifugation at ≈ 130 000 g in a Beckman Coulter Optima LE-80K ultracentrifuge equipped with an SW-32 Ti rotor for 1 hour. Experiments were performed on a wide-bore Bruker Avance-I 600 MHz (14.1 T) spectrometer or Bruker Avance Neo 600 MHz (14.1 T) spectrometer, using triple-channel (HCN) 3.2 mm MAS EFree probes. All experiments were acquired using two-pulse phase modulated (TPPM) proton decoupling of 83 kHz during acquisition65. The 2D 13C–13C DARR experiments66 were performed using a 3-μs 90° pulse on 1H, 4-μs 90° pulses on 13C, a 1H–13C CP contact time of 1 ms at 275 K, a DARR mixing time of 25 ms, and a recycle delay of 2.8 s. 2D 13C–15N NCO experiments were done at 277 K using a 3-μs 90° pulse on 1H, 8-μs 180° pulse on 13C, 1H-15N contact time of 1.5 ms, 15N–13C contact time of 4 ms and recycle delay of 2.8 s. 2D 13C–15N NCA experiments were done at 277 K using a recycle delay of 2.8 s, a 3-μs 90° pulse on 1H, 8-μs 180° pulse on 13C, 1.5 ms and 4 ms 1H–15N and 15N–13C contact times, respectively. In NCA and NCO experiments, the power levels for 15N and 13C during N–C transfer steps were 50 kHz and 62.5 kHz, respectively. The 2D refocused-INEPT 13C–13C 2D spectrum was obtained with total through-bond correlation spectroscopy (TOBSY; ) recoupling, measured at MAS rate of 8.3 kHz, using a 6 ms of TOBSY mixing time, a 3-μs 90° pulse on 1H, 4-μs 90° on 13C, at a temperature of 275 K.67 The 1H–15N heteronuclear correlation (HETCOR) experiments were done using a MAS rate of 13 kHz, 100 and 350 μs CP contact times, 4 s recycle delay, a 3-μs 90° 1H pulse, and at 275 K temperature. 100 kHz homonuclear FSLG 1H decoupling was applied during the t1 evolution time.
MD simulations
Calculations were carried out on the fast, free, and flexible Gromacs engine.68 All systems were first energy-minimized using steepest descent with one conjugate gradient step every 100 steps, then equilibrated through a 200-ps MD simulation in the NVT ensemble with positional restraints (1000 kJ/mol) on heavy atoms, followed by three 100-ps NPT runs with positional restraints (1000, 500, and 100 kJ/mol) on heavy atoms. The production MD simulations were done in the NPT ensemble, obtained through the Bussi–Donadio–Parrinello69 thermostat (T = 300 K, τT = 0.2 ps) and the isotropic (for the polyQ lattice systems) or semi-isotropic (Q15 and HttEx1 systems, xy and z coupled separately) Parrinello–Rahman70 (P = 1 bar, τP = 2 ps, κP = 4.5 × 10−5 bar−1 ) barostat. The van der Waals interactions were switched off between 1.0 and 1.2 nm; long-range electrostatics were treated via Particle Mesh Ewald71,72 with fourth-order interpolation, a real-space cut-off at 1.2 nm, and size-optimized fast Fourier transform parameters (grid spacing of roughly 0.16 nm). Covalent bonds involving hydrogens were constrained to their equilibrium lengths by (fourth-order double-iteration) parallel linear constraint solver (P-LINCS)73. Timestep was 2 fs, Verlet neighbour lists updated every 20 fs with the neighbour list radius automatically determined.
PolyQ amyloid core lattices
To build atomistic-resolution models of the internal structure of the polyQ amyloid core, we assumed it to comprise a lattice of antiparallel β-sheets stacked such that the Gln side-chains interdigitate. The pairs of side-chain dihedral angles (χ1, χ3) were set to satisfy the known characteristic of the interdigitating polyQ amyloid core:5,23 the existence of side-chain–side-chain hydrogen bond interactions. We took the fibril-axis direction to align with the Cartesian coordinate z, and to be perpendicular to the β-strands (aligned with x). To construct the 3D lattice, we considered a minimal “unit cell” consisting of eight Gln residues arranged in a 2 × 2 × 2 pattern (Fig. 2A): Along x, the unit cell contains a minimal peptide chain segment of two amino acids (2 × 2 × 2), representing the alternating (‘odd/even’, i.e., pointing ‘above’ and ‘below’ the β-sheet plane) residues of the β-strand. Along z, to describe an antiparallel β-sheet, minimum two β-strands are needed (2 × 2 × 2); in contrast to parallel in-register sheet structures that could be represented with a single repeating β-strand. Along y, the unit cell contains two distinct neighbouring β-sheets to permit and probe distinct sheet–sheet interfaces (2 × 2 × 2). Figure 2A illustrates how each eight-Gln unit cell contains four Gln–Gln pairs, which establish backbone hydrogen bonds (shown in purple) as well as side-chain hydrogen bonds (in orange). All the hydrogen bonds are aligned roughly along the fibril axis, z. For each Gln–Gln pair, there are 8 distinct classes of (χ1, χ3) orientations that permit side-chain–side-chain hydrogen bond chains along z (see Supplementary Fig. S1). As there are 4 Gln–Gln pairs in the unit cell, there are 84 = 4 096 possible plausible atomistic structures of the 8-Gln unit cell. Accounting for rotational and translational symmetry of the 3D lattice reveals, however, that at most 1 280 of these structures are unique. An important further consideration is that ssNMR experiments conclusively demonstrate that consecutive residues within each β-strand must adopt the same backbone conformations, as indicated by identical chemical shifts. As a result, each distinct strand exclusively contains either type “a” or type “b” Gln residues (Fig. 1I,J). After applying this filter, the number of possible distinct unit cells is reduced to 30. For each of these 30 unit cells candidates, we performed all-atom MD simulations. The fully periodic MD simulation box contained 40 identical unit cells, that is, a total of 8 × 40 = 320 Gln residues, organized as a stack (along y) of 4 antiparallel β-sheets, with each sheet comprising 8 β-strands of periodic (along x) Q10 peptides.
Acknowledging the limitations of a classical mechanics approximation (force field) of an inherently quantum system, we employed three state-of-the-art MD force fields in parallel: AMBER14SB,74 OPLSAA/M,75 and CHARMM36m.76 Gromacs version 2018.3 was used. During the 10-μs production runs, the stability of the 30 structural candidates was evaluated at 5 ns, 100 ns, 200 ns, and 1 μs. At each of these time points, only the candidates that maintained stability above 0.9 (see Eq. (1)) were further continued. The stability S(t) of the given structural candidate at a simulation time t was defined based on the χ1(t) and χ3(t) dihedral angles compared to the initial energy-minimized MD structure:
| (1) |
where N = 320 is the total number of residues, and Ni(t), i = {1, 3} is the number of residues whose χi(t) is within 90° of its initial value at time t.
Solvated polyQ15 fibrils
Q15 peptide fibrils were constructed from each of the two stable ssNMR-verified core models, M1 and M2. Two successive aspartic acid (DD) residues were added to the N-terminus and two lysines (KK) to the C-terminus of the Q15 peptide to mimic the D2Q15K2 peptide widely studied by experiments.5,21,24,46 The N-terminus was further capped with an acetyl (Ace) group, and the C-terminus was set uncharged (with –COOH capping) to match the peptides used in our experiments. A 7-sheet fibril structure was constructed, with each sheet comprising eight Ace-D2Q15K2 peptides. The simulation box was set up to form a quasi-infinite fibril (along z) under periodic boundary conditions and solvated with ∼ 9 800 water molecules in a cuboid box of ∼ 11 × 11 × 3.8 nm3.
State-of-the-art MD force fields AMBER14SB,74 OPLSAA/M,75 and the TIP3P77 water model were used for calculations carried out on the Gromacs version 2021.3. Production run length was 1 μs.
Solvated Q44-HttEx1 fibrils
The ssNMR data suggest that the HttEx1 aggregates display the same spectral patterns observed in polypeptide polyQ fibrils.5,37 This indicates the presence of a common atomic polyQ core structure among them. Consequently, we constructed an atomistic-resolution structure of the mutant HttEx1 fibril utilizing the polyQ core model described in the preceding section. In contrast to fibrils composed of short polyQ segments, the Q44-HttEx1 fibrils exhibited a distinct structural characteristic, consisting of a β-hairpin structure with a single turn. Hence, the constructed core domain of our Q44-HttEx1 encompasses a stack of seven antiparallel β-sheets composed of 44-residue polyQ hairpins (Q44). The hairpin arms consisted of “a” and “b” β-strands connected by a two-residue β-turn modeled based on a type I’ tight turn, known for its strong preference towards adopting a β-hairpin structure.78 The N-terminus of HttEx1, comprising 17 residues, was modeled utilizing the crystal structure of a single Htt(1–17) peptide in complex with the C4 single-chain Fv antibody (PDB ID: 4RAV).49 The residue F17 was included within the β-sheet core, pairing with the penultimate glutamine residue, as supported by the findings of ssNMR investigations.25 The PRD domain of the HttEx1, including 50 residues (P62–P111), was modelled as an end-to-end-distance-maximized random coil, with the two oligoproline (P62–P72 and P90–P99) segments in a polyproline-II-helix conformation. The termini were set to and COO− for the N- and C-terminal, respectively. The turns of neighboring Q44 hairpins were positioned on opposite sides of the fibril (Fig. 5A). The system was solvated with ∼840 000 water molecules, resulting in a total of ∼2 800 000 atoms, in a cuboid box of ∼ 37 × 37 × 19 nm3.
The production MD simulations were conducted for a duration of 5 μs on Gromacs version 2021.4 with AMBER14SB74 protein force field and TIP3P77 water.
Supplementary Material
6. Acknowledgments
This study was supported by funds from the CHDI foundation (contract A-17778), the CampagneTeam Huntington and prior funding via NIGMS R01 GM112678. Part of the work has been performed under the Project HPC-EUROPA3 (INFRAIA-2016-1-730897), with the support of the EC Research Innovation Action under the H2020 Programme, in particular, MBH gratefully acknowledges the support of Zernike Institute for Advanced Materials of the University of Groningen and the computer resources and technical support provided by SURF HPC. Majority of the calculations presented here were carried out on the MPG supercomputers RAVEN and COBRA hosted at MPCDF. MSM acknowledges financial support by the Volkswagen Foundation (86110) and by the Trond Mohn Foundation (BFS2017TMT01). We thank Dr. Alessia Lasorsa for recording some of the ssNMR experiments.
Footnotes
Data Availability
The data that support the findings of this study are openly available at Zenodo (DOI: 10.5281/zenodo.8143752).
Competing Interests
The authors declare no competing interests.
References
- [1].Ross C. A. Polyglutamine pathogenesis: emergence of unifying mechanisms for huntington’s disease and related disorders. Neuron 35, 819–822 (2002). [DOI] [PubMed] [Google Scholar]
- [2].Bates G. P. et al. Huntington disease. Nature reviews Disease primers 1, 1–21 (2015). [DOI] [PubMed] [Google Scholar]
- [3].DiFiglia M. et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277, 1990–1993 (1997). [DOI] [PubMed] [Google Scholar]
- [4].Sawaya M. R., Hughes M. P., Rodriguez J. A., Riek R. & Eisenberg D. S. The expanding amyloid family: Structure, stability, function, and pathogenesis. Cell 184, 4857–4873 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Hoop C. L. et al. Huntingtin exon 1 fibrils feature an interdigitated β-hairpin–based polyglutamine core. Proceedings of the National Academy of Sciences 113, 1546–1551 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Scherzinger E. et al. Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90, 549–558 (1997). [DOI] [PubMed] [Google Scholar]
- [7].Liu L. et al. Imaging mutant huntingtin aggregates: development of a potential pet ligand. Journal of medicinal chemistry 63, 8608–8633 (2020). [DOI] [PubMed] [Google Scholar]
- [8].van der Wel P. C. A. Insights into protein misfolding and aggregation enabled by solid-state NMR spectroscopy. Solid State Nucl Magn Reson 88, 1–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Fitzpatrick A. W. & Saibil H. R. Cryo-em of amyloid fibrils and cellular aggregates. Current opinion in structural biology 58, 34–42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Chen S., Ferrone F. A. & Wetzel R. Huntington’s disease age-of-onset linked to polyglutamine aggregation nucleation. Proceedings of the National Academy of sciences 99, 11884–11889 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Guo Q. et al. The cryo-electron microscopy structure of huntingtin. Nature 555, 117–120 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Harding R. J. et al. Huntingtin structure is orchestrated by hap40 and shows a polyglutamine expansion-specific interaction with exon 1. Communications Biology 4, 1374 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Lunkes A. et al. Proteases acting on mutant huntingtin generate cleaved products that differentially build up cytoplasmic and nuclear inclusions. Molecular cell 10, 259–269 (2002). [DOI] [PubMed] [Google Scholar]
- [14].Sathasivam K. et al. Aberrant splicing of htt generates the pathogenic exon 1 protein in huntington disease. Proceedings of the National Academy of Sciences 110, 2366–2370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Bäuerlein F. J. et al. In situ architecture and cellular interactions of polyq inclusions. Cell 171, 179–187 (2017). [DOI] [PubMed] [Google Scholar]
- [16].Peskett T. R. et al. A Liquid to Solid Phase Transition Underlying Pathological Huntingtin Exon1 Aggregation. Mol Cell (2018). [DOI] [PMC free article] [PubMed]
- [17].Bugg C. W., Isas J. M., Fischer T., Patterson P. H. & Langen R. Structural features and domain organization of huntingtin fibrils. Journal of Biological Chemistry 287, 31739–31746 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Hoop C. L. et al. Polyglutamine amyloid core boundaries and flanking domain dynamics in huntingtin fragment fibrils determined by solid-state nuclear magnetic resonance. Biochemistry 53, 6653–6666 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lin H.-K. et al. Fibril polymorphism affects immobilized non-amyloid flanking domains of huntingtin exon1 rather than its polyglutamine core. Nature communications 8, 1–12 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Isas J. M., Langen R. & Siemer A. B. Solid-state nuclear magnetic resonance on the static and dynamic domains of huntingtin exon-1 fibrils. Biochemistry 54, 3942–3949 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Perutz M., Staden R., Moens L. & De Baere I. Polar zippers. Current biology 3, 249–253 (1993). [DOI] [PubMed] [Google Scholar]
- [22].Perutz M. F., Finch J. T., Berriman J. & Lesk A. Amyloid fibers are water-filled nanotubes. Proceedings of the National Academy of Sciences 99, 5591–5595 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Sikorski P. & Atkins E. New model for crystalline polyglutamine assemblies and their connection with amyloid fibrils. Biomacromolecules 6, 425–432 (2005). [DOI] [PubMed] [Google Scholar]
- [24].Sharma D., Shinchuk L. M., Inouye H., Wetzel R. & Kirschner D. A. Polyglutamine homopolymers having 8–45 residues form slablike β-crystallite assemblies. Proteins: Structure, Function, and Bioinformatics 61, 398–411 (2005). [DOI] [PubMed] [Google Scholar]
- [25].Sivanandam V. et al. The aggregation-enhancing huntingtin n-terminus is helical in amyloid fibrils. Journal of the American Chemical Society 133, 4558–4566 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Schneider R. et al. Structural characterization of polyglutamine fibrils by solid-state nmr spectroscopy. Journal of molecular biology 412, 121–136 (2011). [DOI] [PubMed] [Google Scholar]
- [27].Buchanan L. E. et al. Structural motif of polyglutamine amyloid fibrils discerned with mixed-isotope infrared spectroscopy. Proceedings of the National Academy of Sciences 111, 5796–5801 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Caulkins B. G., Cervantes S. A., Isas J. M. & Siemer A. B. Dynamics of the Proline-Rich C-Terminus of Huntingtin Exon-1 Fibrils. J Phys Chem B 122, 9507–9515 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mario Isas J. et al. Huntingtin fibrils with different toxicity, structure, and seeding potential can be interconverted. Nat. Commun. 12, 4272 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Xiong K., Punihaole D. & Asher S. A. Uv resonance raman spectroscopy monitors polyglutamine backbone and side chain hydrogen bonding and fibrillization. Biochemistry 51, 5822–5830 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Shahmoradian S. H. et al. Tric’s tricks inhibit huntingtin aggregation. Elife 2, e00710 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wagner A. S. et al. Self-assembly of mutant huntingtin exon-1 fragments into large complex fibrillar structures involves nucleated branching. Journal of molecular biology 430, 1725–1744 (2018). [DOI] [PubMed] [Google Scholar]
- [33].Nazarov S., Chiki A., Boudeffa D. & Lashuel H. A. Structural basis of huntingtin fibril polymorphism revealed by cryogenic electron microscopy of exon 1 htt fibrils. Journal of the American Chemical Society (2022). [DOI] [PubMed]
- [34].Boatz J. C. et al. Protofilament structure and supramolecular polymorphism of aggregated mutant huntingtin exon 1. Journal of molecular biology 432, 4722–4744 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rout M. P. & Sali A. Principles for integrative structural biology studies. Cell 177, 1384–1403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Matlahov I., Boatz J. C. & van der Wel P. C. Selective observation of semi-rigid non-core residues in dynamically complex mutant huntingtin protein fibrils. Journal of structural biology: X 6, 100077 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Matlahov I. & van der Wel P. C. Conformational studies of pathogenic expanded polyglutamine protein deposits from huntington’s disease. Experimental Biology and Medicine 244, 1584–1595 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Sharma D., Sharma S., Pasha S. & Brahmachari S. K. Peptide models for inherited neurodegenerative disorders: conformation and aggregation properties of long polyglutamine peptides with and without interruptions. FEBS Lett 456, 181–185 (1999). [DOI] [PubMed] [Google Scholar]
- [39].Margittai M. & Langen R. Fibrils with parallel in-register structure constitute a major class of amyloid fibrils: molecular insights from electron paramagnetic resonance spectroscopy. Quarterly reviews of biophysics 41, 265–297 (2008). [DOI] [PubMed] [Google Scholar]
- [40].Sawaya M. R. et al. Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 447, 453–457 (2007). [DOI] [PubMed] [Google Scholar]
- [41].Punihaole D., Workman R. J., Hong Z., Madura J. D. & Asher S. A. Polyglutamine Fibrils: New Insights into Antiparallel β-Sheet Conformational Preference and Side Chain Structure. J Phys Chem B 120, 3012–3026 (2016). [DOI] [PubMed] [Google Scholar]
- [42].van der Wel P. C. Dihedral angle measurements for structure determination by biomolecular solid-state nmr spectroscopy. Frontiers in Molecular Biosciences 8, 791090 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lovell S. C., Word J. M., Richardson J. S. & Richardson D. C. The penultimate rotamer library. Proteins 40, 389–408 (2000). [PubMed] [Google Scholar]
- [44].Wagner G., Pardi A. & Wuethrich K. Hydrogen bond length and proton nmr chemical shifts in proteins. Journal of the American Chemical Society 105, 5948–5949 (1983). [Google Scholar]
- [45].Hori S., Yamauchi K., Kuroki S. & Ando I. Proton nmr chemical shift behavior of hydrogen-bonded amide proton of glycine-containing peptides and polypeptides as studied by ab initio mo calculation. International Journal of Molecular Sciences 3, 907–913 (2002). [Google Scholar]
- [46].Perutz M. F., Johnson T., Suzuki M. & Finch J. T. Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proceedings of the National Academy of Sciences 91, 5355–5358 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hoop C. L. et al. Polyglutamine amyloid core boundaries and flanking domain dynamics in huntingtin fragment fibrils determined by solid-state nuclear magnetic resonance. Biochemistry 53, 6653–6666 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Poirier M. A. et al. Huntingtin Spheroids and Protofibrils as Precursors in Polyglutamine Fibrilization. J Biol Chem 277, 41032–41037 (2002). [DOI] [PubMed] [Google Scholar]
- [49].De Genst E. et al. Structure of a single-chain fv bound to the 17 n-terminal residues of huntingtin provides insights into pathogenic amyloid formation and suppression. Journal of molecular biology 427, 2166–2178 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Duennwald M. L., Jagadish S., Muchowski P. J. & Lindquist S. L. Flanking sequences profoundly alter polyglutamine toxicity in yeast. Proc. Natl. Acad. Sci. USA 103, 11045–11050 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Caron N. S., Desmond C. R., Xia J. & Truant R. Polyglutamine domain flexibility mediates the proximity between flanking sequences in huntingtin. Proceedings of the National Academy of Sciences 110, 14610–14615 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Burke K. A., Kauffman K. J., Umbaugh C. S., Frey S. L. & Legleiter J. The interaction of polyglutamine peptides with lipid membranes is regulated by flanking sequences associated with huntingtin. J Biol Chem 288, 14993–15005 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Thakur A. K. et al. Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat Struct Mol Biol 16, 380–389 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Gu X. et al. Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron 64, 828–840 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].DeGuire S. M. et al. N-terminal Huntingtin (Htt) phosphorylation is a molecular switch regulating Htt aggregation, helical conformation, internalization, and nuclear targeting. J Biol Chem 293, 18540–18558 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Isas J. M., Langen A., Isas M. C., Pandey N. K. & Siemer A. B. Formation and Structure of Wild Type Huntingtin Exon-1 Fibrils. Biochemistry 56, 3579–3586 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Tam S. et al. The chaperonin TRiC blocks a huntingtin sequence element that promotes the conformational switch to aggregation. Nat Struct Mol Biol 16, 1279–1285 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Michalek M., Salnikov E. S., Werten S. & Bechinger B. Membrane interactions of the amphipathic amino terminus of huntingtin. Biochemistry 52, 847–858 (2013). [DOI] [PubMed] [Google Scholar]
- [59].Ceccon A. et al. Interaction of Huntingtin Exon-1 Peptides with Lipid-Based Micellar Nanoparticles Probed by Solution NMR and Q-Band Pulsed EPR. J Am Chem Soc 140, 6199–6202 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Phan T. T. & Schmit J. D. Thermodynamics of huntingtin aggregation. Biophysical Journal 118, 2989–2996 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Nekooki-Machida Y. et al. Distinct conformations of in vitro and in vivo amyloids of huntingtin-exon1 show different cytotoxicity. Proceedings of the National Academy of Sciences 106, 9679–9684 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Jain G. et al. Inhibitor-based modulation of huntingtin aggregation reduces fibril toxicity. bioRxiv 2023–04 (2023).
- [63].Gasteiger E. et al. Expasy: the proteomics server for in-depth protein knowledge and analysis. Nucleic acids research 31, 3784–3788 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Mandal A., Boatz J. C., Wheeler T. B. & van der Wel P. C. On the use of ultracentrifugal devices for routine sample preparation in biomolecular magic-angle-spinning nmr. Journal of biomolecular NMR 67, 165–178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Bennett A. E., Rienstra C. M., Auger M., Lakshmi K. & Griffin R. G. Heteronuclear decoupling in rotating solids. The Journal of chemical physics 103, 6951–6958 (1995). [Google Scholar]
- [66].Takegoshi K., Nakamura S. & Terao T. 13c–1h dipolar-assisted rotational resonance in magic-angle spinning nmr. Chemical physics letters 344, 631–637 (2001). [Google Scholar]
- [67].Baldus M. & Meier B. H. Total correlation spectroscopy in the solid state. The use of scalar couplings to determine the through-bond connectivity. J Magn Reson Ser A 121, 65–69 (1996). [Google Scholar]
- [68].Abraham M. J. et al. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25 (2015). [Google Scholar]
- [69].Bussi G., Donadio D. & Parrinello M. Canonical sampling through velocity rescaling. The Journal of chemical physics 126, 014101 (2007). [DOI] [PubMed] [Google Scholar]
- [70].Parrinello M. & Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. Journal of Applied physics 52, 7182–7190 (1981). [Google Scholar]
- [71].Darden T., York D. & Pedersen L. Particle mesh ewald: An n · log(n) method for ewald sums in large systems. The Journal of chemical physics 98, 10089–10092 (1993). [Google Scholar]
- [72].Essmann U. et al. A smooth particle mesh ewald method. The Journal of chemical physics 103, 8577–8593 (1995). [Google Scholar]
- [73].Hess B. P-lincs: A parallel linear constraint solver for molecular simulation. Journal of chemical theory and computation 4, 116–122 (2008). [DOI] [PubMed] [Google Scholar]
- [74].Maier J. A. et al. ff14sb: improving the accuracy of protein side chain and backbone parameters from ff99sb. Journal of chemical theory and computation 11, 3696–3713 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Robertson M. J., Tirado-Rives J. & Jorgensen W. L. Improved peptide and protein torsional energetics with the opls-aa force field. Journal of chemical theory and computation 11, 3499–3509 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Huang J. et al. Charmm36m: an improved force field for folded and intrinsically disordered proteins. Nature methods 14, 71–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W. & Klein M. L. Comparison of simple potential functions for simulating liquid water. The Journal of chemical physics 79, 926–935 (1983). [Google Scholar]
- [78].Kuhlman B., O’Neill J. W., Kim D. E., Zhang K. Y. & Baker D. Accurate computer-based design of a new backbone conformation in the second turn of protein l. Journal of molecular biology 315, 471–477 (2002). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.