Abstract
Recent advances in structural DNA nanotechnology have been facilitated by design tools that continue to push the limits of structural complexity while simplifying an often-tedious design process. We recently introduced the software MagicDNA, which enables design of complex 3D DNA assemblies with many components; however, the design of structures with free-form features like vertices or curvature still required iterative design guided by simulation feedback and user intuition. Here, we present an updated design tool, MagicDNA 2.0, that automates the design of free-form 3D geometries, leveraging design models informed by coarse-grained molecular dynamics simulations. Our GUI-based, stepwise design approach integrates a high level of automation with versatile control over assembly and subcomponent design parameters. We experimentally validated this approach by fabricating a range of DNA origami assemblies with complex free-form geometries, including a 3D Nozzle, G-clef, and Hilbert and Trifolium curves, confirming excellent agreement between design input, simulation, and structure formation.
Design algorithms informed by simulations allow users to arrange and bend DNA into complex 3D structures and assemblies.
INTRODUCTION
Since its inception in the 1980s (1), structural DNA nanotechnology has found applications across a vast array of fields, including biosensing, nanoelectronics, gene and drug delivery, computing, optics, and plasmonics (2–6). The unique and exact molecular programmability inherent in the antiparallel and complementary base pairing of double-stranded DNA (dsDNA) enables the realization of nanoscale devices of high precision and geometric complexity. In addition, the ability to integrate single-stranded DNA (ssDNA) and dsDNA with rigid bundles of dsDNA helices enables tailoring of both the dynamic and mechanical properties of these devices (7–9). This ability to precisely design structures with tunable stiffness and dynamics makes DNA nanotechnology highly suited for translating macroscopic mechanisms and machine and material design concepts to the nanoscale. However, realizing advanced design concepts, such as compliant mechanisms and architected materials that contain intricate features like bends and vertices in three-dimensional (3D) (“free-form”) geometries, remains challenging, often requiring substantial design iterations even for experts. Here, we introduce a design algorithm and tool to automate the design of free-form structures, and we validate them both with current computational tools and experimental fabrication. Our results establish a powerful computer-aided design (CAD) approach that will allow lay users to create complex 3D structures and assemblies without needing to learn the underlying molecular design concepts.
Scaffolded DNA origami is one approach particularly well suited for the design of complex 3D DNA nanostructures (10, 11). In this approach, many short oligonucleotides [~20 to 60 nucleotides (nt) long] called “staples” bind to a long (typically ~7000 to 8000 nt long) ssDNA termed “scaffold” to drive folding of the scaffold into a compact defined shape. The staples drive folding by binding to and bridging multiple distant contiguous sites of the scaffold to form dsDNA helices connected by migrationally immobile Holliday junctions that, if appropriately positioned, yield bundles of parallel dsDNA helices that can be arranged into a huge variety of 2D and 3D geometries.
As the DNA origami technique matured, CAD tools have become integral to facilitating a rational design process. A recent review article by Dey et al. (12) categorized available software tools for designing DNA origami structures into three generations. The first-generation design tools like caDNAno (13) and Tiamat (14) implemented graphical user interfaces (GUIs) to allow users to manually specify the routing and base pairing relationship among DNA strands to generate staple strand sequence lists for folding. The second-generation design tools leveraged commercial CAD software to specify input geometries and developed algorithms to automate the underlying strand routings. These tools largely circumvent user inputs other than the original geometry, making structure design simpler for nonexperts but limiting the ability to tune local mechanical and dynamic properties (15–21). Last, the third-generation software tools combine features from the first- and second-generation tools to improve versatility for both expert and nonexperts (22, 23). Within this realm, we recently introduced the tool MagicDNA (24), which combines the advantage of GUI and inherited routing algorithm to design complex DNA nanostructures.
However, one limitation of all these CAD tools is that they inherently build up structures from straight segments of dsDNA helices or their bundles. Features like vertices and bends are achieved by coupling helices of different lengths together to form either bundles with angled edges (i.e., gradients) that can be connected to form a vertex or bundles that accumulate continuous bending stresses across their length to form curved features (25, 26). The integration of CAD tools with simulation tools (27–32) has been critical to enabling accurate design of these complex geometric features. In particular, the recently developed CAD tool MagicDNA facilitates integration with oxDNA coarse-grained simulations (30, 31, 33, 34) to acquire 3D conformation feedback for design iteration. MagicDNA’s combination of graphical interfaces for 3D design manipulation and design parameter input, automated routing algorithms, and coupling to simulation to facilitate iterative design enables realization of complex multicomponent assemblies with user control over local mechanical and dynamic properties. In addition, the recently developed tool DNAxiS leverages simulation to design shapes with curvature but is currently limited to structures with revolved symmetry, either axisymmetric or periodic circular symmetry (35). However, even with these advanced design tools, achieving true free-form geometric designs is still challenging and often requires many iterations to tune the desired geometry.
To achieve true free-form design capability, we introduce here a simulation-guided algorithm for automated vertex and curvature design that we experimentally validate and implement in a GUI in the MagicDNA package. Our algorithm takes sketched free-form spline curves as user input and converts these mathematical splines into physical DNA bundles. We introduce an analytical algorithm called "extrude" to automate the design of a vertex of defined vertex angle formed by the connection between two neighboring wedge-shaped bundles. This algorithm is informed by a series of oxDNA simulations of vertex joint designs that model the relationship between bending angle and vertex design parameters (i.e., edge gradients and bundle cross-sectional geometry). We also introduce an approach called "sweep" to design continuous, curved shapes in 3D from a series of subtly bending segments following similar simulation-based analytical models. These two algorithms are coupled to other useful features of MagicDNA such as 3D multicomponent assembly, scaffold and staple routing algorithms, multiscaffold design, and coarse-grained simulation feedback, providing a powerful platform for rapid design of free-form DNA architectures. To illustrate the versatility of our approach, we fabricated a range of 3D curved DNA origami structures designed using our platform and found an excellent agreement between experiments and design predictions. Our results demonstrate outstanding control over the 3D geometry of DNA origamis through CAD and leverages the design versatility of integrated CAD and engineering through MagicDNA (24), allowing for rapid realization of complex DNA nanostructures, even by researchers from other fields.
RESULTS
Overall approach: From free-form splines to DNA bundles
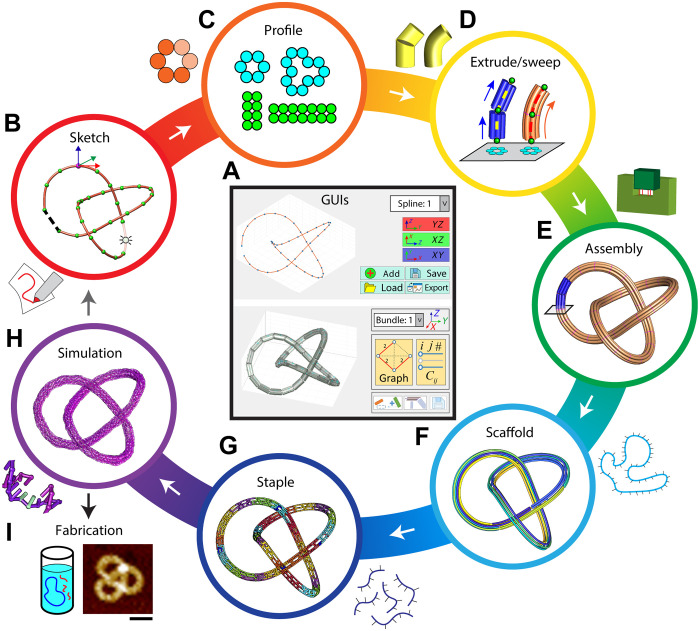
To realize free-form DNA origami structures, we developed a GUI where users can manually define their design using a series of points (Fig. 1, A and B). These “control” points are connected either by straight discrete segments with the input points defining vertices (extrude) or by smooth splines (of fourth-order polynomials), which are then broken into smaller segments with small relative angles to closely approximate the continuous curvature (sweep). Next, the mathematical straight-segment or smooth spline is converted into a DNA nanostructure of physical dimensions taking the dsDNA length and bundle cross section into account (Fig. 1C). The cross section can consist of any even number of duplexes in a square or honeycomb lattice. This conversion between conceptual lines to tangible DNA bundles involves the calculation of edge gradients for bending angles, local orientations for bundles, or nonlinear duplex lengths within a bundle for continuous geometries (Fig. 1D, details discussed in following sections). Once DNA bundles are created with approximate positions and orientations in the assembly model (Fig. 1E), the connectivity matrix based on distances between connection sites serves as a high-level bridge between the user-defined bundle layout and the scaffold routing algorithm that connects all the DNA bundles via scaffold routing (Fig. 1F). The staple strand routing with frequent, periodic crossovers is mostly adapted from caDNAno (13), as in the original implementation of MagicDNA (24), to locally define the shape of the individual DNA bundles (Fig. 1G), with a few adaptions for continuous geometries and higher-order assembly through overhangs. Last, oxDNA simulations (Fig. 1H) provide a rapid feedback on the 3D conformation of the structure for fine-tuning the design before fabrication (Fig. 1I).
Fig. 1. Flow chart of the key steps involved in the design of free-form DNA origami structures.
(A) GUI of the software, showing the spline and bundle panel. (B to H) Steps involved in free-form design. (I) Experimental fabrication of the design for validation. The trefoil knot used here is purely for illustrative purposes: to show 3D spline curves and continuous geometries. In reality, this structure exhibited low experimental yields, likely due to kinetic traps arising from its unique topology. Scale bar, 50 nm.
Extrude method for generating piecewise curved structures with vertices
The extrude tool allows users to connect individual bundle components with desired angles by automating the design of vertices according to the defined input parameters (vertex angle and bundle cross section). This involves the calculation of (i) bundle orientations (for projecting cross-sectional profiles along the helical axis direction) and (ii) edge gradients for the two bending directions. The 3D orientation of each bundle is specified using two orthogonal unit vectors: One vector is normal to the cross-sectional profile (i.e., the helical axis), and the other points along the cross section describing its rotation about the normal vector. Using a straight-line representation (connecting the control points of splines in a chain; Fig. 2A, left), the algorithm takes the orientation of the first bundle as the reference frame and keeps propagating the orientations of the subsequent bundle relative to the prior bundle. Once all bundle orientations are identified, the user can define the cross section for individual bundle components.
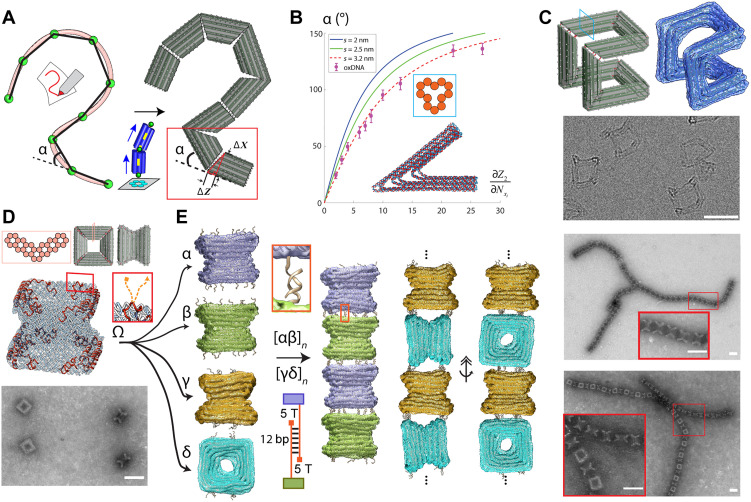
Fig. 2. Extrude tool for creating piecewise curved structures from straight bundles that can be connected using overhangs to form larger arrays.
(A) Conversion of a spline into a series of connected DNA origami bundle components with edge gradients to approximate the target shape. (B) Single-joint oxDNA simulation to determine bending angles based on the input edge gradient (i.e., dZ/dX and dZ/dY) and cross section. (C) Bundle model (left), oxDNA simulation average rendered with the outer surface (right), and cryo–electron microscopy (cryo-EM) image of the Hilbert structure (bottom). (D) From top to bottom: Cross section, bundle model, helical routing model, and transmission electron microscopy (TEM) image of the Nozzle structure. Inset shows overhang positions (orange). (E) oxDNA simulation averages with surface rendering of versions α, β, γ, and δ, showing the different overhang positions (left). The inset shows the formed, stable duplex of two complementary overhangs of the multimer [αβ]n. The sketch depicts our used overhang design, i.e., using a 5-T spacer between the DNA origami interface and the 12 base pair (bp) sequence used to form the higher ordered structure. Results of the simulation of multimeric Nozzle filaments [αβ]n and [γδ]n and their respective, experimental validation. Inset show zoomed-in structures. Scale bars, 50 nm.
The bending angle α at a vertex is related to the edge gradients of the two adjoining bundles on either side of the vertex, where the edge gradient is defined as the ratio of the difference in duplex lengths between neighboring layers of dsDNA helices to the layer, i.e., the center-to-center distance between duplexes in successive layers (Fig. 2B). Although one could use the geometry of DNA duplexes to derive an analytical relationship between the symmetric edge gradients and α, previous work has shown that the layer width is larger than the nominal 2-nm diameter of the DNA helix. Thus, to evaluate the relationship between α and the edge gradient parameters, we performed oxDNA simulations of a single DNA origami joint with four commonly used cross sections (Fig. 2B and figs. S1 and S2). Our results show that larger cross sections allow for more precise control of the bending angle. We also compared the simulation results to a geometric model of the vertex angle that depends on the difference in length between successive layers of dsDNA helices and the interhelical spacing (figs. S1 and S2). We found that an effective helical spacing of 3.2 nm near vertices best captures the vertex geometry (Fig. 2B). While prior work has found an effective interhelical spacing of 2.1 to 2.4 nm (36) in DNA origami bundles, we attribute our slightly larger spacing to base pair fraying at the bundle edges commonly observed in regions with lower crossover densities (fig. S1) (27, 37, 38). Therefore, we implemented this geometric vertex model with the 3.2-nm spacing to automate the vertex design process in our algorithm.
We implemented this vertex design model into the MagicDNA software tool and integrated it with the bundle location, orientation, and scaling calculations to automate the design of “extruded” 3D geometries consisting of straight segments with user-defined length and cross section connected by vertices. To illustrate the robustness of our automated extrude design approach, we designed and fabricated two structures: a Hilbert curve structure with a 12-helix cross section, which contains eight well-defined vertices forming a first-order Hilbert curve in 3D (Fig. 2C, top), and a Nozzle structure, which contains four well-defined vertices adjoining four bundles with a V-shaped cross section to form a 3D nozzle geometry (Fig. 2D, top). Implementing common DNA origami protocols (12, 39), we realized high yields of properly folded structures (Fig. 2, C and D). Transmission electron microscopy (TEM) imaging revealed a homogeneous set of structures for both the Hilbert curve and Nozzle designs. As the Hilbert structure has a more open geometry that could collapse upon surface deposition, we used cryo–electron microscopy (cryo-EM) to reveal multiple orientations of this structure and confirm its 3D geometry. For the Nozzle structure, we observed various orientations in negative-stain TEM clearly illustrating different design features and validating successful folding.
We next used the Nozzle structure to demonstrate and validate an overhang design tool in MagicDNA, where ssDNA overhangs can be positioned at precise locations on the surface of the structure for connecting them into higher-order assemblies. We folded four versions (α, β, γ, and δ) of the Nozzle structure with unique overhangs. Versions α and β were designed with corresponding patterns of mutually complementary overhangs on the ends of the structure, while γ was designed with an overhang pattern on its ends that matches an overhang pattern on the side of δ. We then performed an ABAB-type multimerization by mixing these structures. We realized two varieties of 1D filaments: one where both subunits are oriented similarly (Fig. 2E, [αβ]n) with the Nozzle axis aligned along the length of the filament, and another where the Nozzles alternate between aligned and perpendicular orientations with respect to the filament axis (Fig. 2E, [γδ]n). TEM imaging revealed that proper multimerization of both filaments (Fig. 2E, right) and agarose gel electrophoresis shows that multimerization works over a broad range of MgCl2 concentrations (fig. S14). Gel electrophoresis analysis, folding yields, and additional TEM images are provided in table S1 and figs. S4, S5, S7 to S13, and S15 to S18 for all extrude designs explored in this work.
Sweep method for design of continuously curved structures
Although one could adopt the extrude method and keep adding control points in splines to form increasingly shorter bundles with more subtle bending angles for close to continuous curved geometries. A less tedious alternative is to exploit the smoothness of the splines to automatically subdivide splines into DNA bundles that closely track the curved geometry. We refer to this approach as the sweep method (Fig. 3A). In this method, the algorithm treats the spline as a parametric curve, in terms of variable s, and takes the first derivative of the positions p(s) ≡ [x(s), y(s), z(s)] on the spline to obtain the unit tangent vector êk(s) describing the normal vector of the cross-sectional profile in a continuous manner. Then, the algorithm calculates the second derivative of p(s) to obtain the unit normal vector êi(s), a reference vector lying on the cross-sectional plane, and the binormal vector êj(s) from the vector product êk × êi (Fig. 3A). Because the spline describes the centroid of the cross section, the 3D locations of each dsDNA duplex along the spline curve can be calculated by vector addition of the spline position p and linear combinations of êi and êj. In this manner, the conformation of all duplexes in the target structure can be described using a single variable s, allowing the creation of a continuous 3D model of the structure satisfying the inputs of the spline path and the desired cross-sectional profile. Through user-defined slicing (magenta slices in Fig. 3A, right), the entire 3D model can be discretized into multiple DNA bundles of desired duplex lengths.
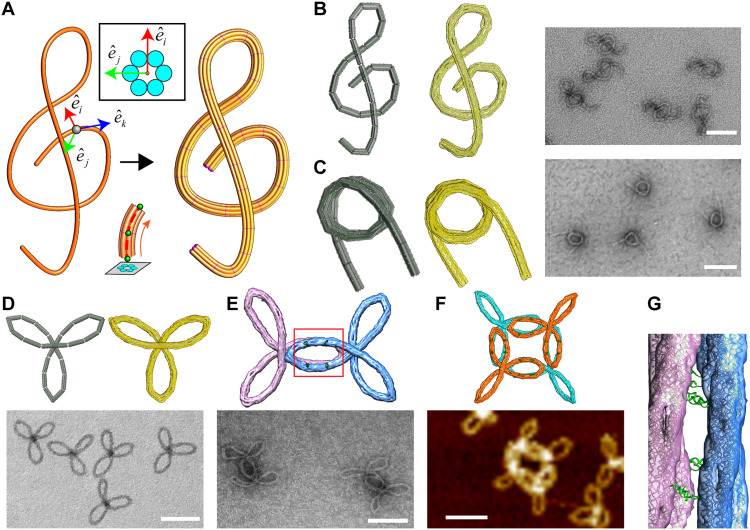
Fig. 3. Sweep tool for designs with continuous free-form curvature.
(A) Conversion of a mathematical spline into a bundle model (B) G-clef structure (from left to right): Bundle model, oxDNA simulation, and TEM image. (C) Nucleosome-like spring structure (from left to right): Bundle model, oxDNA simulation, and TEM image. (D) Monomeric version of the Trifolium structure (bundle model and oxDNA simulation on top; TEM image on bottom). (E) Dimeric version of the Trifolium structure (oxDNA simulation on top; TEM image on bottom). (F) Closed-ring tetrameric version of the Trifolium structure (oxDNA simulation on top; AFM image on bottom). (G) Zoom-in on the red square from the overlapping arms in (E) showing the hybridization of overhangs on different bundles. Scale bars, 100 nm.
To validate the sweep algorithm, we designed and fabricated three distinct free-form structures, namely, a G-clef, a Nucleosome-like spring, and a Trifolium structure, each with a six-helix bundle cross section. Beginning with the G-clef structure, Fig. 3A shows the conversion of the initial sketched spline into a continuous 3D bundle model, which was then discretized into 25 shorter components. The lengths of dsDNA duplexes in each segment were calculated by integrating the parametric curves for duplex axes between consecutive slice points (indicated by red lines in Fig. 3). The interfaces between consecutive bundles were held tight and parallel via multiple scaffold connections (typically zero base long). We also implemented specific scaffold double connections between bundles 1 and 19, 6 and 21, and 11 and 18 (fig. S19) to constrain the bundles into the G-clef shape. OxDNA simulations revealed that the structure closely approximates the free-form G-clef design (Fig. 3B, middle), and TEM images of the fabricated structures confirmed a successful folding into the predicted structure (Fig. 3B, right). The Nucleosome-like spring was designed to form a 3D curve with 1.5 turns, forming a ~60-nm–nominal diameter core with ~60- to 80-nm straight extensions on both sides (Fig. 3C). Simulations and images (Fig. 3C, middle) again revealed that the design accurately captures the desired 3D free-form geometry. Last, we designed the Trifolium design and showed successful folding of the structures according to design predictions (Fig. 3D). As in the case of the Nozzle structure, we used the Trifolium structure to demonstrate integration of sweep tool with the overhang tool for higher-order assembly. To this end, we designed dimeric and tetrameric assemblies of the Trifolium structures using the overhang tool and confirmed successful fabrication of both assemblies using simulations and imaging (Fig. 3, E to G). Gel electrophoresis analysis, folding yields, and additional TEM images are provided in table S1 and figs. S20, S21, S23, S24, and S32 to S37 for all sweep designs.
Integrating with MagicDNA features to expand design space
The extrude and sweep approaches allow for the automated design of complex 3D geometries. Implementing this free-form design automation in MagicDNA allows users to leverage other features of this software to expand on free-form design. Here, we expand on free-form designs by leveraging two specific features of MagicDNA: (i) control over component-level cross section and (ii) versatile scaffold routing algorithm including multiscaffold design. We used the extrude approach to design a crown structure resembling the symbol for the queen chess piece (Fig. 4A). We carried the line model design (Fig. 4A, left) through the MagicDNA design workflow to assign distinct cross sections to individual components, including a six-helix bundle honeycomb cross section to the spikes of the crown, an eight-helix bundle square lattice cross section to the base, and a two-helix bundle cross section to the struts that connect the base to the spikes (Fig. 4A, middle). The versatile scaffold routing algorithm and assembly operations of the MagicDNA framework allowed us to place scaffold connections to the base and the outer frame on either side of the struts to stabilize the overall geometry. OxDNA simulations (Fig. 4A, right) and TEM images (Fig. 4B) revealed that the design accurately captured the target geometry and that the structure folded efficiently.
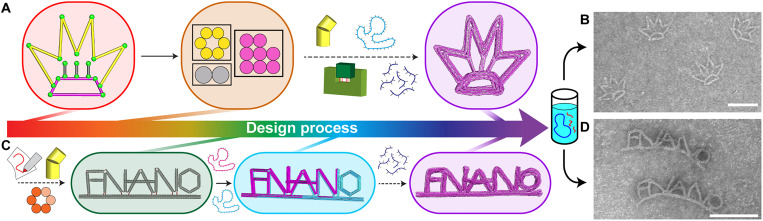
Fig. 4. Combining free-form design with variable cross section and multiscaffold features of MagicDNA.
(A) The crown design workflow starts with the line model comprising multiple splines totaling 15 bundle components. Three different cross sections were assigned to the yellow (crown spikes, six-helix bundle), pink (base, eight-helix bundle), and gray (struts, two-helix bundle) components. Components were assembled followed by automated scaffold and staple routing, and the final design was simulated in oxDNA. (B) TEM images illustrate well-folded crown structures. (C) FNANO design workflow follows similar steps, with the bundle model and multiscaffold routing highlighted, culminating in the final design simulated in oxDNA. (D) TEM images reveal well-folded FNANO script structures. Scale bars, 100 nm.
These advances in design capability are synergistic with recent advances in fabrication methods, namely, the ability to incorporate multiple orthogonal sequence scaffolds to fold larger structures in a single-pot folding reaction (40). We previously implemented multiscaffold design into MagicDNA. To demonstrate integration of multiscaffold design with free-form automation, we designed an FNANO script structure composed of five letters that are assembled and connected to a stiff support bundle (Fig. 4C). All letters have an eight-helix bundle cross section, and the support bundle has a six-helix bundle cross section. The multiscaffold routing algorithm in MagicDNA automatically divided the design into two scaffolds with the structure formed from a total of 14,778 base pairs. Successful realization of the FNANO structure is shown in Fig. 4D. Gel electrophoresis analysis, folding yields, and additional TEM images are provided in table S1 and figs. S26, S27, S29, and S30 for structures with different cross sections and multiple scaffolds.
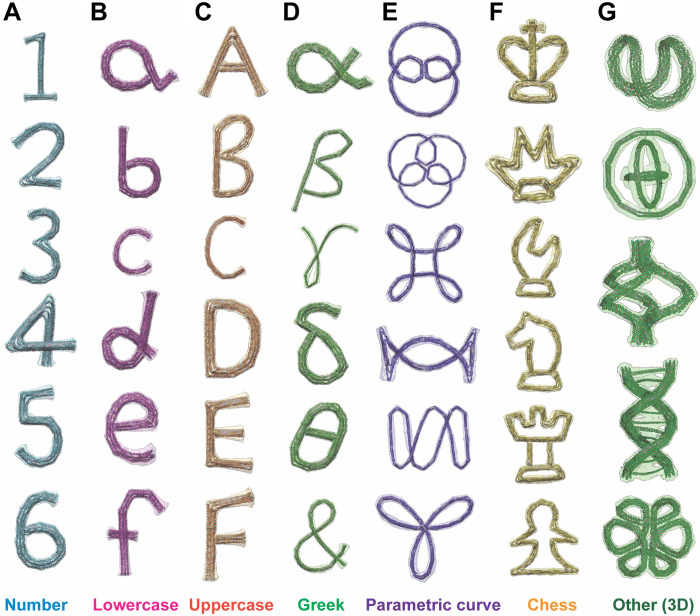
Lastly, we highlight the capability of the automated free-form design implemented in MagicDNA with an assorted gallery of sophisticated DNA origami nanostructures presented in Fig. 5 including numbers, lowercase, uppercase, and Greek letters, parametric curves, chess pieces, and other 3D free-form designs, demonstrating the versatility of our design approach and software tool. All designs were simulated in oxDNA, and the molecular structure of the average configuration is overlaid with a semitransparent surface view that envelops the maximum conformational fluctuations, giving an indication of the local flexibility and the overall structural stability. As expected, structures with open topology, small cross sections (e.g., six-helix bundles), and lengthy components display larger fluctuations around the mean configuration (e.g., the letter c) (fig. S44).
Fig. 5. Design gallery of free-form structures created using our software.
Conformations of additional free-form designs obtained using oxDNA simulations. The designs include (A) numbers, (B) lowercase letters, (C) uppercase letters, (D) Greek letters and the ampersand symbol, (E) parametric curves, (F) six chess pieces, and (G) other 3D structures. Each structure is shown by its mean conformation computed from the simulation with a bounding surface representing the SD of fluctuations that the structure exhibits in the simulation.
DISCUSSION
Design philosophy
We present a versatile design framework that interweaves GUIs (Fig. 1A) with algorithmic automations to enable the design of arbitrary free-form DNA origami structures and assemblies based on user-defined design parameters. The design process starts with the user sketching the geometry of the envisioned structure as free-form dimensionless splines by taking advantage of a GUI using which one can define and manipulate the shape of the splines and visualize them in real time. This real-time sketching allows users to rely on gradual modifications to achieve the target design. Realizing the 3D form of the structural design requires additional inputs such as cross-sectional profiles and edge gradients. Typically, the structures are composed of many dsDNA bundles, which would make manual input of bundle design parameters tedious and error prone. To address this problem, we implemented two new algorithms: the extrude and sweep methods. Both algorithms rely on a series of oxDNA simulations that allowed us to derive a geometric model for relating bundle design parameters to bending angle, which was implemented as an algorithm to automatically convert the spline model to the assembly model (i.e., geometry composed of connected bundles) in MagicDNA. Having the full sketch and assembly design process implemented in one software (unlike the second-generation design tools that import geometry models from exterior CAD software) allows for flexible iteration between the sketch and the assembly step to rapidly define and tune the geometry while still leveraging the algorithms implemented in MagicDNA for scaffold and staple strand routing to iterate and complete designs. These designs can be evaluated computationally by coarse-grained molecular dynamics simulations such as oxDNA with automatically generated simulation input files to achieve an iterative robust design framework before moving forward to experimental fabrication (see Supplementary Text for note on design iteration). Although the underlying algorithms for scaffold and staple routing and for edge gradients (leading to the extrude and sweep methods) have proven to work robustly, designs with complex topology might require additional considerations, including control over the folding pathway (41, 42) to achieve high yields. For example, such control over folding could improve the yield of the trefoil knot topology for the design presented in Fig. 1I (additional images in fig. S67). One additional restriction in the scaffold routing algorithm of our software prevents the use of an odd number of helices because of a requisite step of pairing helices, as discussed previously (24).
Versatile design framework and hierarchical assembly
We sacrificed full automation of the design process to allow users to customize structures for intended applications. For example, the same geometry can be realized with different bundle cross sections to modulate the structure stiffness, as demonstrated with the DNA crown structure (Fig. 4A). The choice of bundle cross section depends on various factors, including the desired thickness and flexibility of the structure and the amount of available scaffold. On the basis of our results, vertex fluctuations also depend on the cross section of the vertex arms, with larger cross sections leading to more precise vertex angles. In general, larger cross sections lead to stiffer structures that better maintain the desired shape, but this comes at the expense of using more scaffold for individual components, which could mean reducing the size or number of components in the overall structure or using multiple scaffolds or larger scaffolds to fold the structure. Furthermore, a semiautomatic software allows the user to examine the design status at several stages, modify details from default settings, and seamlessly move forward and backward through the design steps, all of which is facilitated by rapid feedback in GUIs to visualize results of design choices. Furthermore, the design paradigm introduced here can fully leverage the growing library of scaffold sources (40, 43–45), removing restrictions in spatial dimensions and allowing users to realize even more diverse structures. For example, we used the multiscaffold feature of MagicDNA to achieve large structures, such as the “FNANO” script (Fig. 4C), which was designed from two scaffolds with orthogonal sequences (44) to honor this annual conference that has played a seminal role in fostering the field of DNA nanotechnology.
Expanded design scope with free-form features
The design algorithms and associated software and GUIs developed here substantially expand the capability of the MagicDNA design framework for designing free-form, curved features. Our results demonstrate a vast array of free-form designs, fabricated with high yields (>80%; table S1) and with an excellent agreement between oxDNA simulations and experiments. Apart from validating our design strategy, these results also illustrate the importance of coarse-grained simulations to computationally support the immense design space available for free-form structures with customizable features, allowing users to control both geometry and properties. In particular, stiffness can be increased by using larger cross sections (which might require a multiscaffold design) or, for cases with overlapping components, introducing interbundle connections. For example, the bottom structure in the “other 3D” series (Fig. 5) where a continuous free-form six-helix bundle routes through the six faces of a cube (46) includes six interbundle connections along its edges. One could also exploit the versatility of the semiautomatic framework to design dynamic devices with multiple components, connected by single-stranded scaffold segments, to engineer predefined motions, as shown in the gyroscope structure (Fig. 5G, second from top) where two rotational degrees of freedoms exist between the inner and outer rings. While certain design concepts might be already covered by existing software, such as TALOS for 3D polyhedral wireframe structures (15), our approach is complementary, allowing users to diversify those designs, add irregular structural motifs, or combine wireframe components with non-wireframe components into a larger assembly.
The wide range of DNA origami designs presented illustrates the realization of advanced free-form features within the general categories of static, compliant, and dynamic DNA nanostructures. The free-form design algorithms integrate seamlessly with other features in MagicDNA such as hierarchical assembly and multiscaffold designs to further broaden the design space for DNA self-assembly and provide a foundation for the realization of advanced materials, including assemblies of compliant mechanisms (47), curved polymeric DNA origami designs (48), and meta-materials (49). Moving forward, we envision enhancing the design capabilities of our software by directly including structure-property relationships into the design tools, where emerging tools like machine learning are expected to play a useful role (50).
MATERIALS AND METHODS
Materials
Oligonucleotides for folding of the DNA origami structures were purchased at 10 nmol synthesis scale from Eurofins Genomics (https://eurofinsgenomics.com) or at 25 nmol synthesis scale from Integrated DNA Technologies Inc. (www.IDTDNA.com). DNA oligonucleotides were purchased with salt-free purification at 100-μM concentration in ribonuclease-free water and were used without further purification. The single-stranded scaffold (p8064) was prepared as described previously (51), and the single-stranded scaffold (CS03) was purchased from tilibit (www.tilibit.com/).
DNA origami folding and purification
Folding of free-form DNA origami structures was performed by mixing 10-nM scaffold DNA with 100-nM corresponding staple strands in Tris-EDTA buffer, supplemented with MgCl2 [tris, 5 mM; EDTA, 1 mM; MgCl2, 10 mM (pH 8)]. Samples were folded in a Bio-Rad PCR cycler by first heating them up to 65°C for 15 min and then cooling them down to 20°C over the course of 14 hours. The Nozzle structure was folded over 2.5 days. The detailed protocols for thermal annealing and all staple strand sequences are provided in the Supplementary Materials. Purification of DNA origami structures was performed by gel extraction (Freeze ‘N Squeeze, Bio-Rad) or polyethylene glycol (PEG) precipitation (52). Hierarchical assembly of the Trifolium structure was performed by first folding monomers with different sets of overhangs, purifying them individually, mixing them in an equimolar ratio, and incubating them at temperatures between 40° and 55°C for 20 hours. Unless stated otherwise, the magnesium concentration for these hierarchical assemblies was adjusted to 20 mM. Hierarchical assembly of the Nozzle multimers was performed by folding structures with different sets of overhangs, purifying them individually, mixing them in an equimolar ratio, and incubating them at 40°C for 20 hours.
AFM imaging
Gel-purified structures (around 1 nM) were used for atomic force microscopy (AFM) imaging by adsorbing 6 μl of sample onto freshly cleaved mica (V1, Ted Pella). After 3 min of incubation, the mica was rinsed carefully with Milli-Q H2O and dried with a gentle flow of air. Samples were subsequently imaged in ScanAsyst Mode using a Bruker BioScope Resolve microscope equipped with a Nanoscope V controller. ScanAsyst Air probes (Bruker) with a nominal spring constant of 0.4 N/m were used for scanning. Height information was recorded in the retrace channel.
Negative-stain TEM
Purified DNA origami structures (1 to 10 nM) were adsorbed onto freshly glow-discharged copper grids (Electron Microscopy Sciences, Hatfield, PA) and incubated for 4 min. Excess sample solution was subsequently wicked off with filter paper (Whatman #4), and the grid was stained with two 6-μl drops of 1% aqueous Uranyl acetate (SPI Supplies) solution. Samples were dried for at least 30 min before imaging. Imaging was performed on a FEI Tecnai G2 Spirit operated at 80-kV acceleration.
Cryo–electron microscopy
The Hilbert structure used for cryo-EM analysis was subjected to two rounds of PEG precipitation to ensure sufficient purity. Furthermore, the second round was used to increase the DNA origami concentration to 500 nM. Glow-discharged grids (Ted Pella) were used with a Thermo Scientific Vitrobot at 22°C, 0-s drain time, 3-s blot time, 0 blot force at 100% humidity. Imaging was done on a Thermo Scientific Glacios cryo-TEM, equipped with a Falcon III direct electron detector and 200-kV field emission gun.
Coarse-grained molecular dynamics simulations
For performing oxDNA2 simulations, the relevant topology and initial configuration files were generated using the refined version of MagicDNA developed here. The initial configuration was relaxed in a manner similar to our previous study. Briefly, this involved substituting the DNA backbone potential with linear springs, gradually increasing the force constants of these springs, and then applying mutual traps between paired scaffold and staple bases over a period of 100,000 time steps. Next, the backbone potential was restored, and a further 1 million time steps of simulations were carried out, still retaining the mutual traps to finalize the relaxation process. During both these relaxation steps, we used a small time step of 3.03 fs. Last, we removed the mutual traps and continued the simulation for an additional period of 20 million time steps of size 15.15 fs for calibrating the edge gradients at bundle vertex and 10 million time steps of size 3.03 fs for the other simulations providing feedback on structure conformation for each design. We used a John thermostat (with diffusion coefficient and Newtonian step settings of 2.5 and 103) to maintain a constant temperature of 30°C. A monovalent salt concentration at 0.5 M was chosen to mimic standard Mg-induced folding conditions. GPU acceleration was used whenever available. The trajectory files were analyzed using functions built into MagicDNA, including visualization of configurations and calculation of root mean square deviation and root mean square fluctuations. The average configurations were exported to the UCSF Chimera software and rendered to obtain high-quality images. We typically report the mean configuration of each structure and a surface map enveloping its conformational fluctuations.
Acknowledgments
We acknowledge resources from the Campus Microscopy and Imaging Facility (CMIF) at The Ohio State University for negative-stain TEM imaging. Cryo-EM was performed at the Center of Electron Microscopy and Analysis (CEMAS) at The Ohio State University. We are thankful to G. Agarwal (The Ohio State University) for providing access to the Bruker BioScope Resolve AFM.
Funding: This work was funded by the National Science Foundation, award numbers: 1921881 and 1933344 to C.E.C. and M.G.P. and 1921955 to G.A. AFM imaging was supported by the National Institutes of Health grant 1S10OD025096-01A1 to G. Agarwal. Computational resources were provided by the Duke Computing Cluster (DCC) and the XSEDE Program supported by the National Science Foundation (grant no. ACI-1053575).
Author contributions: W.G.P. performed all experiments. C.-M.H. wrote the MATLAB code and performed oxDNA simulations. W.G.P. and C-.M.H. wrote the initial draft of the manuscript and prepared the figures. All authors discussed results. All authors reviewed and edited the final manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data and code used in the study are available through Open Science Framework (DOI 10.17605/OSF.IO/N284U) and at GitHub (https://github.com/cmhuang2011/MagicDNA).
Supplementary Materials
This PDF file includes:
Supplementary Text: Folding protocols
Supplementary Text: Design iterations
Table S1
Figs. S1 to S67
Legend for data S1
Other Supplementary Material for this manuscript includes the following:
Data S1
REFERENCES AND NOTES
- 1.N. C. Seeman, Nucleic acid junctions and lattices. J. Theor. Biol. 99, 237–247 (1982). [DOI] [PubMed] [Google Scholar]
- 2.D. Daems, W. Pfeifer, I. Rutten, B. Sacca, D. Spasic, J. Lammertyn, Three-dimensional DNA origami as programmable anchoring points for bioreceptors in fiber optic surface plasmon resonance biosensing. ACS Appl. Mater. Interfaces 10, 23539–23547 (2018). [DOI] [PubMed] [Google Scholar]
- 3.E. Lin-Shiao, W. G. Pfeifer, B. R. Shy, M. Saffari Doost, E. Chen, V. S. Vykunta, J. R. Hamilton, E. C. Stahl, D. M. Lopez, C. R. Sandoval Espinoza, A. E. Deyanov, R. J. Lew, M. G. Poirer, A. Marson, C. E. Castro, J. A. Doudna, CRISPR-Cas9-mediated nuclear transport and genomic integration of nanostructured genes in human primary cells. Nucleic Acids Res. 50, 1256–1268 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.C. R. Lucas, P. D. Halley, A. A. Chowdury, B. K. Harrington, L. Beaver, R. Lapalombella, A. J. Johnson, E. K. Hertlein, M. A. Phelps, J. C. Byrd, C. E. Castro, DNA origami nanostructures elicit dose-dependent immunogenicity and are nontoxic up to high doses in vivo. Small 18, e2108063 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.M. Dass, F. N. Gür, K. Kołątaj, M. J. Urban, T. Liedl, DNA origami-enabled plasmonic sensing. J. Phys. Chem. C 125, 5969–5981 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.B. R. Aryal, D. R. Ranasinghe, C. Pang, A. E. F. Ehlert, T. R. Westover, J. N. Harb, R. C. Davis, A. T. Woolley, Annealing of polymer-encased nanorods on DNA origami forming metal–semiconductor nanowires: Implications for nanoelectronics. ACS Appl. Nano Mater. 4, 9094–9103 (2021). [Google Scholar]
- 7.M. Darcy, K. Crocker, Y. Wang, J. V. Le, G. Mohammadiroozbahani, M. A. S. Abdelhamid, T. D. Craggs, C. E. Castro, R. Bundschuh, M. G. Poirier, High-force application by a nanoscale DNA force spectrometer. ACS Nano 16, 5682–5695 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.W. Pfeifer, P. Lill, C. Gatsogiannis, B. Sacca, Hierarchical assembly of DNA filaments with designer elastic properties. ACS Nano 12, 44–55 (2018). [DOI] [PubMed] [Google Scholar]
- 9.F. N. Gür, S. Kempter, F. Schueder, C. Sikeler, M. J. Urban, R. Jungmann, P. C. Nickels, T. Liedl, Double- to single-strand transition induces forces and motion in DNA origami nanostructures. Adv. Mater. 33, 2101986 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.X. Yan, Y. Wang, N. Ma, Y. Yu, L. Dai, Y. Tian, Dynamically reconfigurable DNA origami crystals driven by a designated path diagram. J. Am. Chem. Soc. 145, 3978–3986 (2023). [DOI] [PubMed] [Google Scholar]
- 11.A.-K. Pumm, W. Engelen, E. Kopperger, J. Isensee, M. Vogt, V. Kozina, M. Kube, M. N. Honemann, E. Bertosin, M. Langecker, R. Golestanian, F. C. Simmel, H. Dietz, A DNA origami rotary ratchet motor. Nature 607, 492–498 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.S. Dey, C. Fan, K. V. Gothelf, J. Li, C. Lin, L. Liu, N. Liu, M. A. D. Nijenhuis, B. Saccà, F. C. Simmel, H. Yan, P. Zhan, DNA origami. Nat. Rev. Methods Primers 1, 13 (2021). [Google Scholar]
- 13.S. M. Douglas, A. H. Marblestone, S. Teerapittayanon, A. Vazquez, G. M. Church, W. M. Shih, Rapid prototyping of 3D DNA-origami shapes with caDNAno. Nucleic Acids Res. 37, 5001–5006 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.S. Williams, K. Lund, C. Lin, P. Wonka, S. Lindsay, H. Yan, in DNA Computing, A. Goel, F. C. Simmel, P. Sosík, Eds. (Springer Berlin Heidelberg, 2009), pp. 90–101. [Google Scholar]
- 15.H. Jun, T. R. Shepherd, K. Zhang, W. P. Bricker, S. Li, W. Chiu, M. Bathe, Automated sequence design of 3D polyhedral wireframe DNA origami with honeycomb edges. ACS Nano 13, 2083–2093 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.H. Jun, X. Wang, W. P. Bricker, M. Bathe, Automated sequence design of 2D wireframe DNA origami with honeycomb edges. Nat. Commun. 10, 5419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.H. Jun, F. Zhang, T. Shepherd, S. Ratanalert, X. Qi, H. Yan, M. Bathe, Autonomously designed free-form 2D DNA origami. Sci. Adv. 5, eaav0655 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R. Veneziano, S. Ratanalert, K. Zhang, F. Zhang, H. Yan, W. Chiu, M. Bathe, Designer nanoscale DNA assemblies programmed from the top down. Science 352, 1534–1534 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.E. Benson, A. Mohammed, A. Bosco, A. I. Teixeira, P. Orponen, B. Högberg, Computer-aided production of scaffolded DNA nanostructures from flat sheet meshes. Angew. Chem. Int. Ed. Engl. 55, 8869–8872 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.E. Benson, A. Mohammed, J. Gardell, S. Masich, E. Czeizler, P. Orponen, B. Högberg, DNA rendering of polyhedral meshes at the nanoscale. Nature 523, 441–444 (2015). [DOI] [PubMed] [Google Scholar]
- 21.M. Lolaico, S. Blokhuizen, B. Shen, Y. Wang, B. Högberg, Computer-aided design of A-trail routed wireframe DNA nanostructures with square lattice edges. ACS Nano 17, 6565–6574 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.H. Jun, X. Wang, M. F. Parsons, W. P. Bricker, T. John, S. Li, S. Jackson, W. Chiu, M. Bathe, Rapid prototyping of arbitrary 2D and 3D wireframe DNA origami. Nucleic Acids Res. 49, 10265–10274 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.E. de Llano, H. Miao, Y. Ahmadi, A. J. Wilson, M. Beeby, I. Viola, I. Barisic, Adenita: Interactive 3D modelling and visualization of DNA nanostructures. Nucleic Acids Res. 48, 8269–8275 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.C.-M. Huang, A. Kucinic, J. A. Johnson, H.-J. Su, C. E. Castro, Integrated computer-aided engineering and design for DNA assemblies. Nat. Mater. 20, 1264–1271 (2021). [DOI] [PubMed] [Google Scholar]
- 25.H. Dietz, S. M. Douglas, W. M. Shih, Folding DNA into twisted and curved nanoscale shapes. Science 325, 725–730 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Y. Suzuki, I. Kawamata, K. Mizuno, S. Murata, Large deformation of a DNA-origami nanoarm induced by the cumulative actuation of tension-adjustable modules. Angew. Chem. Int. Ed. 59, 6230–6234 (2020). [DOI] [PubMed] [Google Scholar]
- 27.D. N. Kim, F. Kilchherr, H. Dietz, M. Bathe, Quantitative prediction of 3D solution shape and flexibility of nucleic acid nanostructures. Nucleic Acids Res. 40, 2862–2868 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.C. Maffeo, A. Aksimentiev, MrDNA: A multi-resolution model for predicting the structure and dynamics of DNA systems. Nucleic Acids Res. 48, 5135–5146 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. Y. Lee, J. G. Lee, G. Yun, C. Lee, Y.-J. Kim, K. S. Kim, T. H. Kim, D.-N. Kim, Rapid computational analysis of DNA origami assemblies at near-atomic resolution. ACS Nano 15, 1002–1015 (2021). [DOI] [PubMed] [Google Scholar]
- 30.T. E. Ouldridge, A. A. Louis, J. P. K. Doye, Structural, mechanical, and thermodynamic properties of a coarse-grained DNA model. J. Chem. Phys. 134, 085101 (2011). [DOI] [PubMed] [Google Scholar]
- 31.B. E. K. Snodin, F. Randisi, M. Mosayebi, P. Šulc, J. S. Schreck, F. Romano, T. E. Ouldridge, R. Tsukanov, E. Nir, A. A. Louis, J. P. K. Doye, Introducing improved structural properties and salt dependence into a coarse-grained model of DNA. J. Chem. Phys. 142, 234901 (2015). [DOI] [PubMed] [Google Scholar]
- 32.W. T. Kaufhold, W. Pfeifer, C. E. Castro, L. Di Michele, Probing the mechanical properties of DNA nanostructures with metadynamics. ACS Nano 16, 8784–8797 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.L. Rovigatti, P. Šulc, I. Z. Reguly, F. Romano, A comparison between parallelization approaches in molecular dynamics simulations on GPUs. J. Comput. Chem. 36, 1–8 (2015). [DOI] [PubMed] [Google Scholar]
- 34.P. Šulc, F. Romano, T. E. Ouldridge, L. Rovigatti, J. P. Doye, A. A. Louis, Sequence-dependent thermodynamics of a coarse-grained DNA model. J. Chem. Phys. 137, 135101 (2012). [DOI] [PubMed] [Google Scholar]
- 35.D. Fu, R. Pradeep Narayanan, A. Prasad, F. Zhang, D. Williams, J. S. Schreck, H. Yan, J. Reif, Automated design of 3D DNA origami with non-rasterized 2D curvature. Sci. Adv. 8, eade4455 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.S. M. Douglas, H. Dietz, T. Liedl, B. Högberg, F. Graf, W. M. Shih, Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–418 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Z. Shi, C. E. Castro, G. Arya, Conformational dynamics of mechanically compliant DNA nanostructures from coarse-grained molecular dynamics simulations. ACS Nano 11, 4617–4630 (2017). [DOI] [PubMed] [Google Scholar]
- 38.B. E. K. Snodin, J. S. Schreck, F. Romano, A. A. Louis, J. P. K. Doye, Coarse-grained modelling of the structural properties of DNA origami. Nucleic Acids Res. 47, 1585–1597 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.K. F. Wagenbauer, F. A. S. Engelhardt, E. Stahl, V. K. Hechtl, P. Stommer, F. Seebacher, L. Meregalli, P. Ketterer, T. Gerling, H. Dietz, How we make DNA origami. Chembiochem 18, 1873–1885 (2017). [DOI] [PubMed] [Google Scholar]
- 40.X. Liu, F. Zhang, X. Jing, M. Pan, P. Liu, W. Li, B. Zhu, J. Li, H. Chen, L. Wang, J. Lin, Y. Liu, D. Zhao, H. Yan, C. Fan, Complex silica composite nanomaterials templated with DNA origami. Nature 559, 593–598 (2018). [DOI] [PubMed] [Google Scholar]
- 41.F. Schneider, N. Möritz, H. Dietz, The sequence of events during folding of a DNA origami. Sci. Adv. 5, eaaw1412 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.A. E. Marras, L. Zhou, H.-J. Su, C. E. Castro, Programmable motion of DNA origami mechanisms. Proc. Natl. Acad. Sci. 112, 713–718 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.T. Aksel, Z. Yu, Y. Cheng, S. M. Douglas, Molecular goniometers for single-particle cryo-electron microscopy of DNA-binding proteins. Nat. Biotechnol. 39, 378–386 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.F. A. S. Engelhardt, F. Praetorius, C. H. Wachauf, G. Brüggenthies, F. Kohler, B. Kick, K. L. Kadletz, P. N. Pham, K. L. Behler, T. Gerling, H. Dietz, Custom-size, functional, and durable DNA origami with design-specific scaffolds. ACS Nano 13, 5015–5027 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.X. Chen, B. Jia, Z. Lu, L. Liao, H. Yu, Z. Li, Aptamer-integrated scaffolds for biologically functional DNA origami structures. ACS Appl. Mater. Interfaces 13, 39711–39718 (2021). [DOI] [PubMed] [Google Scholar]
- 46.C. Kimberling, P. J. C. Moses, Closed space curves made from circles on polyhedra. J. Geom. Graph. 15, 29–43 (2011). [Google Scholar]
- 47.H.-J. Su, C. Castro, A. Marras, L. Zhou, The Kinematic Principle for Designing DNA Origami Mechanisms: Challenges and Opportunities. J. Mech. Des. 139(6): 06230 (2017).
- 48.M. W. Grome, Z. Zhang, F. Pincet, C. Lin, Vesicle tubulation with self-assembling DNA nanosprings. Angew. Chem. Int. Ed. Engl. 57, 5330–5334 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.R. Li, H. Chen, J. H. Choi, Auxetic two-dimensional nanostructures from DNA**. Angew. Chem. Int. Ed. 60, 7165–7173 (2021). [DOI] [PubMed] [Google Scholar]
- 50.M. DeLuca, S. Sensale, P.-A. Lin, G. Arya, Prediction and control in DNA nanotechnology. ACS Appl. Bio Mater., (2023). [DOI] [PubMed] [Google Scholar]
- 51.C. E. Castro, F. Kilchherr, D. N. Kim, E. L. Shiao, T. Wauer, P. Wortmann, M. Bathe, H. Dietz, A primer to scaffolded DNA origami. Nat. Methods 8, 221–229 (2011). [DOI] [PubMed] [Google Scholar]
- 52.E. Stahl, T. G. Martin, F. Praetorius, H. Dietz, Facile and scalable preparation of pure and dense DNA origami solutions. Angew. Chem. Int. Ed. Engl. 53, 12735–12740 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Text: Folding protocols
Supplementary Text: Design iterations
Table S1
Figs. S1 to S67
Legend for data S1
Data S1