Significance
Metastasis is responsible for most of cancer-related deaths, which requires dramatic remodeling of the cytoskeleton. Circular RNA ASH2 (circASH2) functions as a powerful tumor suppressor via directly modulating stability of microfilament. The matchmaker function of circASH2 in delivering Tropomyosin 4 (TPM4) transcripts to the Y-box binding protein 1 (YBX1)/heterogeneous nuclear ribonucleoproteins (hnRNP) complex and then accelerating nonsense-mediated messenger RNA decay (NMD) of TPM4 is unexpected but should have a significant impact on our understanding of posttranscriptional regulation mediated by noncoding RNA (ncRNA). This study has provided convincing data on how a circular RNA (circRNA) regulates RNA-binding protein (RBP)’s function in a liquid–liquid phase separation (LLPS)-dependent manner, which represents a significant conceptual advancement in circRNA and RBP-related research.
Keywords: liquid–liquid phase separation, mRNA decay, cytoskeleton, circASH2, metastasis
Abstract
Metastasis, especially intrahepatic, is a major challenge for hepatocellular carcinoma (HCC) treatment. Cytoskeleton remodeling has been identified as a vital process mediating intrahepatic spreading. Previously, we reported that HCC tumor adhesion and invasion were modulated by circular RNA (circRNA), which has emerged as an important regulator of various cellular processes and has been implicated in cancer progression. Here, we uncovered a nuclear circRNA, circASH2, which is preferentially lost in HCC tissues and inhibits HCC metastasis by altering tumor cytoskeleton structure. Tropomyosin 4 (TPM4), a critical binding protein of actin, turned out to be the major target of circASH2 and was posttranscriptionally suppressed. Such regulation is based on messenger RNA (mRNA)/precursormRNA splicing and degradation process. Furthermore, liquid–liquid phase separation of nuclear Y-box binding protein 1 (YBX1) enhanced by circASH2 augments TPM4 transcripts decay. Together, our data have revealed a tumor-suppressive circRNA and, more importantly, uncovered a fine regulation mechanism for HCC progression.
Liver cancer is the fourth leading cause for cancer-related mortality globally, while hepatocellular carcinoma (HCC) accounts for the majority of primary liver cancers (1). Despite a comprehensive regimen available, the prognosis of patients with advanced HCC remains poor (2). Early intrahepatic metastasis, especially micrometastases, is a huge challenge for HCC treatment and prognosis improvement (2). An ascensive learning of the HCC mechanism of progression could be the key to pharmaceutical development.
Cell migration is an essential step for tumor metastasis, especially in tumors from the epithelium. During this process, cytoskeletal reorganization and remodeling are indispensable (3, 4). The mammalian cytoskeleton is one of the most complicated and functionally versatile structures, involved in processes such as motility, adhesion, reaction to external forces, and adaptation of cell shape (5). Previous studies, including ours, demonstrated that adhesion with extracellular matrix (ECM) is closely related to HCC metastasis (6, 7). Meanwhile, adhesion of tumor cells has been proven to be the start of cytoskeleton formation and downstream signals activation (8, 9). Furthermore, the cytoskeleton state [such as actin filaments (F-actin), paxillin, etc.] could influence the interaction between cells and ECM (8, 9). Among cytoskeletal proteins, tropomyosin 4 (TPM4) is a member of actin-binding proteins involved in the cytoskeleton of nonmuscle cells, which could provide stability to the F-actin and regulate other actin-binding proteins (10). Although TPM4 has been reported as an oncoprotein in several malignancies, its upstream mechanism remains unclear (11, 12).
Precursor messenger RNA (pre-mRNA) splicing is a fundamental process in eukaryotes’ gene expression (13). In this process, alternative splicing is a major mechanism to shape the expression pattern, and splicing regulators, like heterogeneous nuclear ribonucleoproteins (hnRNPs) and serine/arginine-rich splicing factors, modulate the former (13). In addition, lots of coregulators also participate in this process. Y-box binding protein 1 (YBX1), a well-known RNA-binding protein (RBP), could work with hnRNPs to form a functional complex (14, 15) and regulate pre-mRNA splicing and mRNA decay (15–17). YBX1 is a highly disordered protein with a fixed structure in the middle, which is suitable for interactions with RNAs (18, 19). Intriguingly, intrinsically disordered was identified to be the key characteristic for liquid–liquid phase separation (LLPS) of protein (20).
Active LLPS mediates membraneless compartment formation, thereby spatiotemporally regulating many critical cellular processes, including pre-mRNA splicing (20). LLPS is usually driven by weak and dynamic multivalent interactions, correlated with intrinsically disordered regions (IDR) and scaffolding nuclear acid molecules (21). Meanwhile, noncoding RNAs (ncRNAs), such as NEAT1 and SNHG9, could coordinate with RBPs and induce LLPS in a concentration- or structure-dependent manner (22, 23). A recent study also indicated that circular RNA (circRNA) VAMP3 drives LLPS of CAPRIN1 and promotes stress granule formation in the cytoplasm (24). How ncRNAs influence LLPS has raised extensive attention in researchers. But more functional RNAs need to be identified in order to draw the whole picture.
circRNAs are covalently closed single-stranded RNAs and proved to be a widespread and functional RNA species (25). Depending on their localization and interaction with other molecules, circRNAs modulate transcription and splicing, regulate stability and translation of mRNA, influence function and metabolism of proteins, and even directly serve as translational templates in different biological and pathophysiological conditions (25). In cancer research, accumulated evidence showed a considerable role of circRNA during tumor initiation and progression (26, 27). However, there is still a long way to go before circRNA could be applied in clinical settings.
Here, we performed in-depth analyses of circRNomics from human HCC samples and identified circular RNA ASH2 (circRNA) as a metastasis-suppressive circRNA. Phenomenologically, circASH2 sabotaged tumor cytoskeleton and attenuated cell adhesion. Clinically, down-regulated circASH2 expression indicated a worse prognosis of HCC patients. Mechanistically, circASH2 induced LLPS of YBX1 and then regulated the hnRNPs/YBX1 complex. In this way, circASH2 targeted TPM4 transcripts and specifically promoted its decay. Finally, HCC progression was inhibited by a TPM4-related cytoskeleton/tumor adhesion pathway.
Results
Identification of circASH2 as a circRNA Involved in HCC Metastasis.
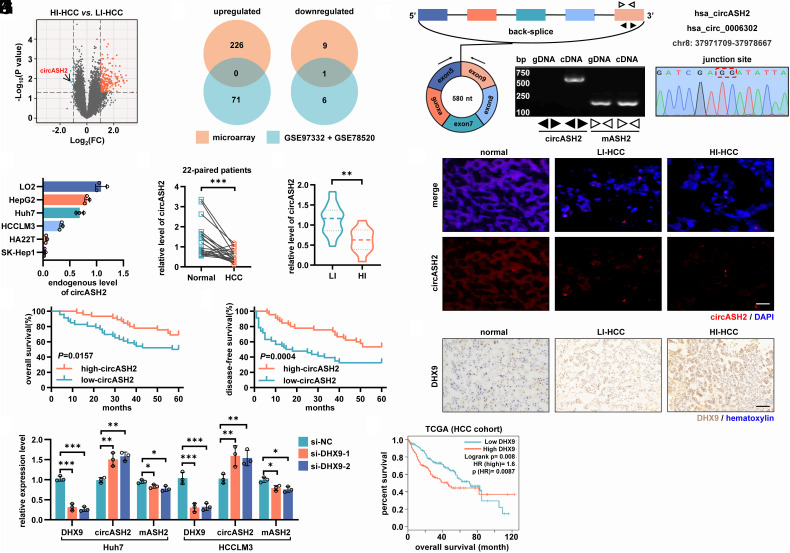
Emerging pieces of evidence show that circRNAs are key signaling regulators in HCC (27). To identify functional circRNAs involved in HCC metastasis, we performed circRNA microarray analysis of 10 primary HCC tissues, including 5 highly invasive (HI: a. tumor diameters were smaller than 3 cm; b. diagnosed as postsurgical metastasis or recurrence) HCC tissues and 5 low-invasive (LI: a. tumor diameters were larger than 3 cm; b. postsurgical metastasis or recurrence negative) HCC tissues (SI Appendix, Table S1). Here, we detected a total of 236 differentially expressed circRNAs (|log2(fold-change)|≥ 1, P < 0.05) between the two groups. Among the identified circRNAs, 226/10 circRNAs were up-regulated/down-regulated significantly in HI HCC (Fig. 1A).
Fig. 1.
The microarray analysis identified circASH2 as a metastasis-suppressive role and regulated by DHX9 in HCC. (A) circRNA microarray analysis of 5 HI and 5 LI HCC tissues. Differentially expressed circRNAs (|log2(fold-change)|≥ 1, P < 0.05) were highlighted (up-regulated in HI: orange points; down-regulated in HI: green points). circASH2 is denoted by a black arrow. (B) A Venn diagram showing differentially expressed circRNAs within our microarray data and GSE78520 + GSE97332 dataset. (C) Genomic loci and validation of circASH2. The unique backspliced junction fragment of circASH2 (divergent primers) was detected in Huh7 cells by RT-qPCR and validated by Sanger sequencing. (D) Endogenous level of circASH2 in several human HCC cell lines (HepG2, Huh7, HCCLM3, HA22T, and SK-Hep1) and a human liver cell line (LO-2). (E and F) The expression of circASH2 was measured by RT-qPCR in 22-paired matched HCC and nontumor tissues (E, cohort 1, ***P < 0.001, paired Student’s t test), and 32 HI/LI HCC tissues (F, cohort 2, **P < 0.01, unpaired Student’s t test). (G) Representative RNA FISH images of circASH2 (red, 40 nM, 55 °C, 2 h) in nontumor tissues, LI HCC tissues, and HI HCC tissues. The nuclei were labeled with DAPI (blue, 1:100, R.T., 5 min). (Scale bars, 100 μm.) (H and I) Kaplan–Meier curves showing overall survival (H) and disease-free survival (I) of 91 HCC patients (cohort 3) followed up to 60 mo. Patients were separated by the median expression level of circASH2, using the Gehan–Breslow test. (J) The expression levels of DHX9, circASH2, and mASH2 were detected by RT-qPCR after being treated with DXH9 siRNAs in Huh7 and HCCLM3 cells (mean ± SD, *P < 0.05, **P < 0.01, ***P < 0.001, one-way ANOVA test). (K) Representative IHC images of DHX9 (brown, 1:200, 4 °C, 2 h) in nontumor tissues, LI HCC tissues, and HI HCC tissues. The nuclei were labeled with hematoxylin (blue, R.T., 5 min). (Scale bars, 200 μm.) (L) Kaplan–Meier curves showing overall survival of HCC patients from the TCGA cohort. Patients were separated by the median expression level of DHX9, using the Gehan–Breslow test.
Also, we analyzed published HCC circRNA microarray datasets [Gene Expression Omnibus (GEO), GSE78520 and GSE97332, SI Appendix, Fig. S1A]. We chose the top 250 circRNAs with the most significant differences in expression in each dataset for subsequent combined analysis (|log2(fold-change)|≥ 1, P < 0.05, SI Appendix, Fig. S1B). By combining this result with our HI/LI microarray analysis, we identified one circRNA, circASH2 (hsa_circ_0006302), was declining while tumor cell becoming more aggressive, implying a possible tumor suppressive role of circASH2. (Fig. 1 B and C).
We then validated circASH2 and its junction site in HCC cells with designed specific divergent primers and Sanger sequencing (Fig. 1C). Like most circRNAs, circASH2 also exhibits higher stability than ASH2 mRNA (mASH2) (SI Appendix, Fig. S1 C and D). These results demonstrated that circASH2 is a genuine circRNA with the expected molecular structure and biochemical features.
Next, we examined the endogenous level of circASH2 in several human HCC cell lines and human liver cell line (LO2). Interestingly, circASH2 was expressed at a relatively higher level in the nonmetastatic lines (HepG2, Huh7) than in the metastatic cell lines [HCCLM3 (high-circASH2HCC cell), HA22T, and SK-Hep1, Fig. 1D]. In addition, ∼49, 34, 13, and 8 circASH2 copies per cell were estimated in Huh7, HCCLM3, HA22T, and SK-Hep1 cells, respectively (SI Appendix, Fig. S1E).
Thus, it is reasonable to speculate that circASH2, as a circRNA, might play a significant role in HCC metastasis.
Decreased circASH2 Expression Correlates with Poor Prognosis in HCC.
To verify the function of circASH2 in HCC, we analyzed its expression in a clinical sample from a small group of 22 patients (cohort 1). Consistent with the GEO dataset results above, circASH2 was hardly detectable in most HCC tumor tissue, compared to matched nontumor tissues (Fig. 1E). Then, we reached a similar conclusion with cohort 2 (11 HI and 21 LI HCC tissues), in which HI tissues presented significantly lower levels of circASH2 than LI tissues (Fig. 1F). Consistent results were observed by FISH (Fig. 1G). In a third cohort of 91 HCC patients (cohort 3) with follow-up records (SI Appendix, Table S2), lower circASH2 expression in tumors strongly correlated with shortened overall survival and disease-free survival (DFS) (Fig. 1 H and I).
Collectively, these findings indicated a tumor-suppressive role of circASH2 in HCC.
circASH2 Expression Is Negatively Regulated by DHX9 in HCC.
mASH2 was generally considered a tumor promoter and overexpressed in HCC tissues (SI Appendix, Fig. S1 F and G) (28, 29). Therefore, we presumed that a posttranscriptional regulation led to an adverse expression pattern of mASH2 and circASH2 in HCC. Our previous research identified that ADAR1 could influence certain circRNAs generation (30), and enzymes sharing similar function have been reported (31). Next, we detected the expression of circASH2 and mASH2 after individually silencing these enzymes, including DHX9, ADAR1, and QKI (SI Appendix, Fig. S1H). Only DHX9 silence, instead of ADAR1 or QKI, enhanced circASH2 expression and consequently lowered mASH2 (Fig. 1J and SI Appendix, Fig. S1I). Notably, DHX9 was obviously up-regulated in HCC evidenced by the data from cohort 1 and The Cancer Genome Atlas Program (TCGA) cohort (SI Appendix, Fig. S1 J and K) and further overexpressed in HI HCC tissues (SI Appendix, Fig. S1L). Additionally, a high expression level of DHX9 correlated with poor prognosis of HCC patients (Fig. 1L).
Taken together, the downregulation of circASH2 was, at least partially, caused by the overexpressed DHX9 in HCC.
circASH2 Suppresses HCC Metastasis In Vivo.
To verify the impact of circASH2 in liver cancer, we conducted animal experiments using three canonical HCC metastasis mouse models. First, we constructed an orthotopic xenograft mouse model according to our previous report (6). By detecting circASH2 expression levels in injected cells and in situ tumors, we demonstrated that the stably transfected cell lines could consistently overexpress circASH2 (SI Appendix, Fig. S2A). In the 9th week, most mice (6/7, 85.7%) from the control group developed multifocal lesions in the liver, which were significantly inhibited (2/7, 28.6%) in the circASH2 group (Fig. 2 A and B). When the mice were killed in the 12th week, mice from the control group received more lung/gastrointestinal tract (GI) metastasis, compared to the circASH2 group (Fig. 2C and SI Appendix, Fig. S2 B and C).
Fig. 2.
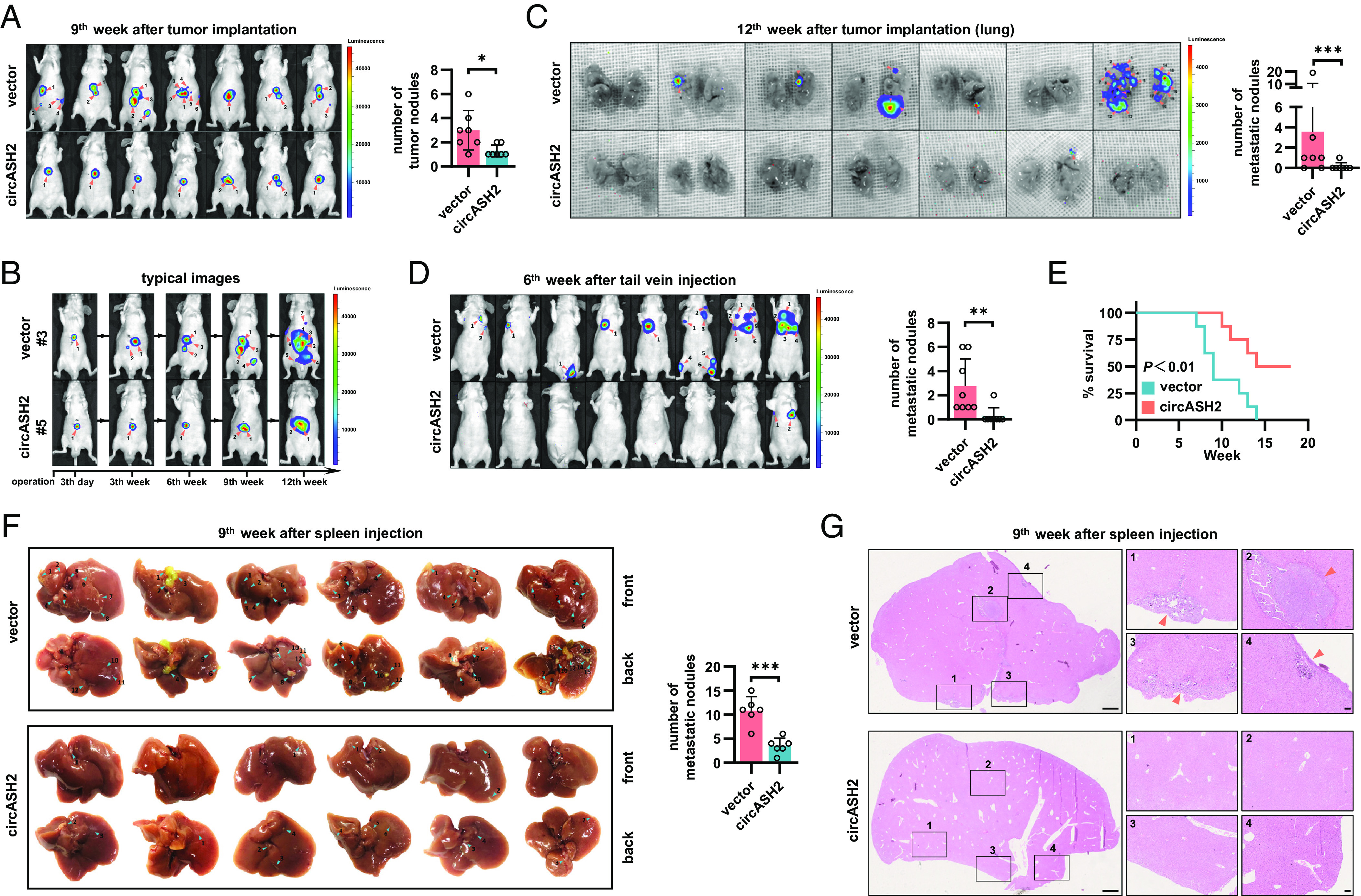
circASH2 suppresses HCC metastasis in vivo. (A, Left) In vivo image system (IVIS) images of HA22T tumors in the 9th week after transplantation (both local growth and metastases are marked with orange arrows, n = 7). (Right) Tumor foci are quantified (mean ± SD, **P < 0.01, unpaired Student’s t test). (B) Typical images of HA22T tumors (#3 mouse in the control group and #5 the mouse in the circASH2 group) at the indicated time (both local growth and metastases are marked with orange arrows). (C, Left) IVIS images showing pulmonary metastasis in mice scarified in the 12th week (metastases are marked with orange arrows, n = 7). (Right) Pulmonary metastases are quantified (mean ± SD, ***P < 0.001, unpaired Student’s t test). (D, Left) IVIS images of HA22T tumors in the 6th week after rapid tail vein injection (metastases are marked with orange arrows, n = 8). (Right) Tumor foci are quantified (mean ± SD, **P < 0.01, unpaired Student’s t test). (E) Kaplan–Meier curves showing overall survival of 16 mice with indicated HA22T tumors followed up to 18 wk (**P < 0.01, Gehan–Breslow test). (F, Left) Liver images of HA22T tumors in the 9th week after spleen injection (intrahepatic metastases are marked with blue arrows, n = 6). (Right) Intrahepatic metastases are quantified (mean ± SD, ***P < 0.001, unpaired Student’s t test). (G) Typical HE staining images of HA22T tumors in the 9th week after spleen injection. Left: (Scale bar, 1 mm.) Right: (Scale bar, 100 μm.)
Second, we established a rapid tail vein injection model, by injecting HA22T cells (1 × 106 cells per mouse) into the lateral tail vein of the mouse in the control or circASH2 group (n = 8). In the 6th week after injection, only 1/8 mouse (12.5%) in the circASH2 group developed a lung metastatic nodule, whereas all eight mice (100.0%) in the control group had evidence of lung or bone metastasis (Fig. 2D). At 12th weeks, the circASH2 group showed an obvious better prognosis, including an overall survival and metastasis ratio (Fig. 2E and SI Appendix, Fig. S2D).
Third, consistent results were found in the intrasplenic injection model. By injecting HCC tumor cells into the mouse spleen by a surgical approach, liver metastasis was significantly prohibited in the circASH2 group, while the spleen orthotopic tumor showed no difference in size (Fig. 2 F and G and SI Appendix, Fig. S2E).
Together, animal experiments strongly suggested that circASH2 could suppress HCC metastasis, including intrahepatic metastasis, hematogenous metastasis, and implantation.
circASH2 Impairs the Cytoskeleton Assembly in HCC.
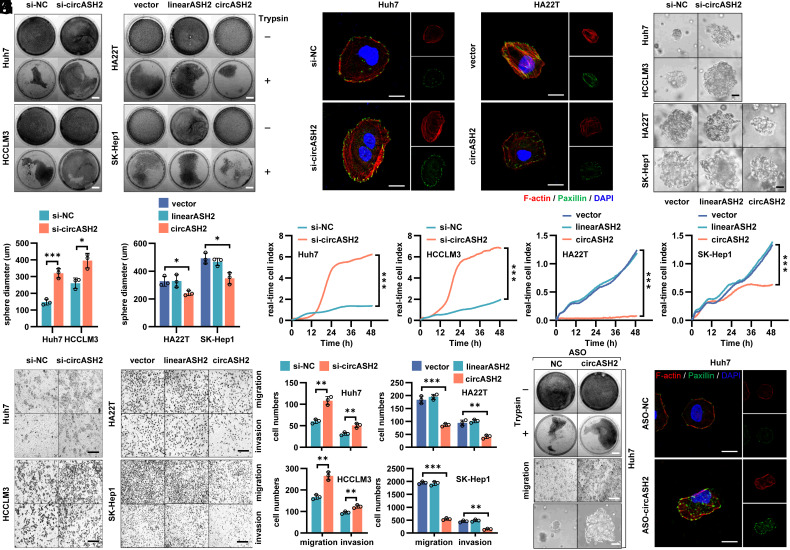
Next, according to endogenous circASH2 levels, we manipulated circASH2 expression in four HCC cell lines by lentiviral overexpression, small-interfering RNA (siRNA) knockdown, or antisense oligonucleotide (ASO) silence (SI Appendix, Fig. S3 A–C). Intriguingly, during routine cell culture, we noticed that circASH2 overexpression markedly promoted cell detachment after trypsinization, while circASH2 silence rendered cells more resistant to trypsin digestion (Fig. 3A and SI Appendix, Fig. S3D). According to our previous work, these data suggested modified adhesion function of tumor cells (6). Considering the close connection between cell adhesion and cytoskeleton formation (5), we analyzed cytoskeleton status in circASH2-munipulated HCC cells. The results showed that circASH2 could strongly suppress F-actin and paxillin in tumor cells, which implied an impaired cytoskeleton (Fig. 3B and SI Appendix, Fig. S3E).
Fig. 3.
circASH2 impairs the cytoskeleton assembly in HCC. (A) Trypsin digestion assay on 4 HCC cell lines with circASH2 overexpression [linearASH2 (580 bp) as a negative control] or knockdown. Cells without (−) or with (+) trypsinization were fix-stained with crystal violet (0.1%, R.T., 5 min) and photographed. (Scale bars, 1 cm.) (B) Cytoskeleton formation in Huh7 cells with circASH2 knockdown and HA22T cells with circASH2 overexpression was shown by phalloidin staining of F-actin (red, 1:200, R.T., 10 min) and immunofluorescence of paxillin (green, 1:200, R.T., 1 h), while DAPI was used to stain the nucleus. (Scale bar, 10 μm.) (C and D) In vitro 3D invasion assays were performed using 4 HCC cell lines with circASH2 overexpression or knockdown. The representative pictures are shown (C), and the diameter of the sphere was measured and analyzed (D, *P < 0.05, ***P < 0.001, unpaired Student’s t test or one-way ANOVA test). (Scale bar, 100 μm.) (E) Real-time capture of 4 HCC cell lines (circASH2 overexpression or knockdown) migration was performed on the xCELLigence System and Real-time Cell Analysis (RTCA) Dual Purpose (DP) Instrument over 48 h. Data were processed by RTCA Software 2.0 (***P < 0.001, two-way ANOVA test). (F and G) Transwell assays with 4 HCC cell lines (circASH2 overexpression or knockdown). Representative fields of the porous membranes are shown (F, scale bar, 100 μm), and cell numbers per field are quantified (G, means ± SD, **P < 0.01, and ***P < 0.001, unpaired Student’s t test or one-way ANOVA test). (H) Trypsin digestion assay on Huh7 cells with circASH2 knockdown (ASO-circASH2). Cells without (−) or with (+) trypsinization were fix-stained with crystal violet and photographed. (Scale bars, 1 cm.) (I) Transwell migration assays with circASH2 knockdown (ASO-circASH2) Huh7 cells. Representative fields of the porous membranes are shown. (Scale bar, 100 μm.) (J) 3D invasive ability of circASH2 knockdown (ASO-circASH2) Huh7 cells was measured, and the representative pictures are shown. (Scale bar, 100 μm.) (K) Cytoskeleton formation in Huh7 cells with circASH2 knockdown (ASO-circASH2) was shown by phalloidin staining of F-actin (red) and immunofluorescence of paxillin (green), while DAPI was used to stain the nucleus. (Scale bar, 10 μm.)
To further verify our finding, we constructed a 3-dimensional (3D) invasion system with Matrigel and found that circASH2 could cripple the tumor cell by down-regulating its invasion ability (Fig. 3 C and D). Meanwhile, the dynamic metastasis curve measured by the xCELLigence real-time cell analysis (RTCA) system indicated that circASH2 reduced HCC cell mobility (Fig. 3E). An analogous result was observed in transwell assays (Fig. 3 F and G). Alternatively, circASH2 silence triggered by specific ASOs also impaired the cytoskeleton assembly and exhibited promotive effects on HCC cell mobility (Fig. 3 H–K and SI Appendix, Fig. S3 F–I).
These in vitro results supported our findings about HCC metastasis in animal and patient’s sample study.
TPM4 Is Repressed by circASH2 and Functionally Relevant.
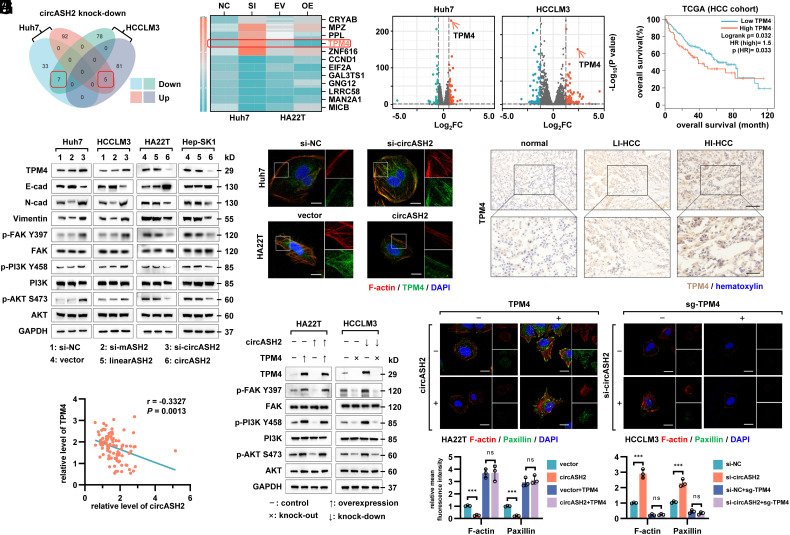
To address the mechanism of circASH2-regulated cytoskeleton assembly, we generated a series of cell lines and performed transcriptome analyses subsequently (SI Appendix, Fig. S4A). In total, seven genes were up-regulated, and five genes were down-regulated in two si-circASH2 HCC cell lines (Fig. 4A). We then validated these candidates in different circASH2-overexpression and circASH2-knockdown HCC cells by RT-qPCR (Fig. 4B). Among them, only TPM4 [the excluded candidates either did not have the desired expression trend (e.g., CRYAB) or were not associated with the phenotype (e.g., CCND1)], a member of the tropomyosin family of actin-binding proteins involved in stability of F-actin (10), presented an accordant change in the mRNA level with the transcriptome sequencing (up-regulated in the knockdown group but down-regulated in the overexpression group, Fig. 4 B and C). Moreover, TPM4 was also negatively correlated with the HCC prognosis (Fig. 4D).
Fig. 4.
TPM4 is repressed by circASH2 and functionally relevant. (A) A Venn diagram showing differentially expressed genes in circASH2 knock-down HCC cells (Huh7 and HCCLM3) compared with the control group. The overlapped gene numbers are highlighted with red boxes. (B) A heatmap showing the expression levels of 12 candidates detected by RT-qPCR after overexpressing or silencing circASH2. TPM4 is highlighted with a red box. (C) A volcano map showing overall changes in gene expression (up-regulated in the si-circASH2 group: orange points; down-regulated in the si-circASH2 group: green points), and TPM4 was one of the most up-regulated genes both in circASH2 knock-down Huh7 and HCCLM3 cells (denoted by orange arrows). (D) Kaplan–Meier curves showing overall survival of HCC patients from the TCGA cohort. Patients were separated by the expression level of TPM4, using the Gehan–Breslow test. (E) HCC cells with circASH2 overexpression or knockdown were analyzed for TPM4 expression and other related signaling by immunoblotting with the indicated antibodies (1:1,000, 4 °C, overnight). GAPDH was used as an internal reference. (F) Representative fluorescence images showing the expression levels and locations of TPM4 (green, 1:200, R.T., 1 h) and F-actin (red) after circASH2 overexpression or knockdown, while DAPI (blue) was used to stain the nucleus. (Scale bar, 10 μm.) (G) Representative IHC images of TPM4 (brown, 1:250, 4 °C, 2 h) in nontumor tissues, LI HCC tissues, and HI HCC tissues. The nuclei were labeled with hematoxylin (blue). (Scale bars, 200 μm.) (H) Correlations between expression levels of circASH2 and TPM4 in HCC tissues (cohort 3). RNA levels were determined using RT-qPCR and normalized to GAPDH. The r values and P values were calculated using Pearson correlation analysis. (I and J) Reexpression of TPM4 markedly abolished circASH2-mediated inhibition on the cytoskeleton, while knockout of TPM4 largely revoked the function of circASH2 (I) Immunoblots with indicated antibodies; (J, Up) Representative fluorescence images of the cytoskeleton, (scale bar, 10 μm.) (Down) Quantitative analysis on the fluorescence photos by mean fluorescence intensity (means ± SD, ***P < 0.001, one-way ANOVA test).
Next, circASH2-suppressed TPM4 expression was verified with immunoblots in multiple cell lines (Fig. 4E). Meanwhile, TPM4 combined with F-actin as previously reported (10), and circASH2 inhibited intensity of both in fluorescence staining (Fig. 4F). We also confirmed the expression level of circASH2, and TPM4 was negatively correlated in patient samples analysis (cohort 2, Figs. 1G and 4G; cohort 3, Fig. 4H). These results suggested a negative regulation of TPM4 from circASH2.
Cytoskeleton status was closely related to cell adhesion, focal adhesion formation, and mobility (5–8). We analyzed the relevant pathway by gene set enrichment analysis, and the result showed that the FAK pathway, PI3K/AKT pathway, and the ECM–receptor interaction pathway were significantly activated after circASH2 silence (SI Appendix, Fig. S4 B–D). Activation of these signals could trigger a series of cellular biological behaviors which results in tumor progression and invasion (4, 7). Similar results were obtained by immunoblots (Fig. 4E).
To prove that TPM4 was indispensable for the circASH2-induced function, we designed relevant assays by ectopic expression/deletion of TPM4. The results showed that ectopic TPM4 restored the FAK/PI3K/AKT pathway and strengthened the cytoskeleton and completely discarded the effect of circASH2 (Fig. 4 I and J). A similar conclusion was reached when we deleted TPM4 in circASH2 knockdown cells, using the CRISPR/Cas9 system (Fig. 4 I and J). Furthermore, circASH2 inhibited cell adhesion, migration and invasion, which could be reversed by TPM4 manipulation both in vitro and in vivo (SI Appendix, Fig. S4 E–G).
Together, these epistasis analyses strongly suggested that circASH2 inhibits HCC cell adhesion and mobility by repressing its downstream effector TPM4.
circASH2 Promotes mRNA Decay by Physically Connecting TPM4 Transcripts.
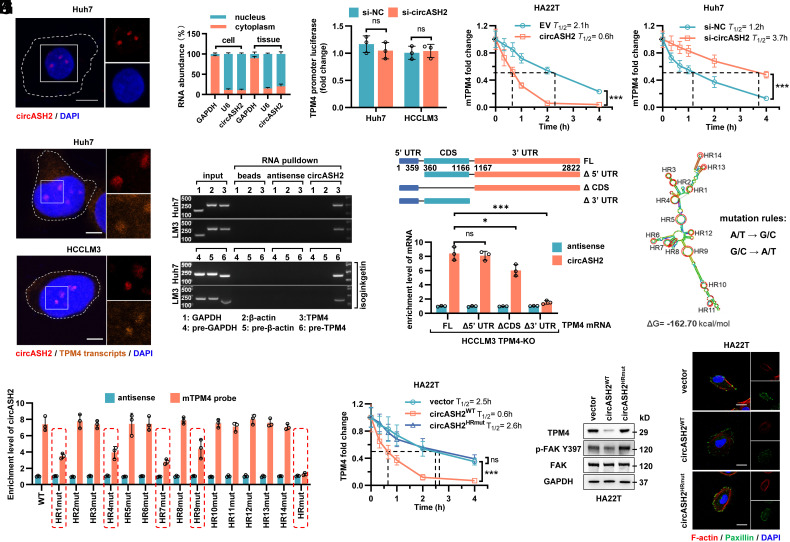
To dissect the mechanism by which circASH2 regulates TPM4 expression, we first determined the subcellular localization of circASH2 by FISH and nuclear–cytoplasm fractionation. Although circASH2 is an exonic circRNA, a predominant nuclear distribution of circASH2 was found in HCC (Fig. 5 A and B). Meanwhile, Argonaute 2 (AGO2) RIP further verified that circASH2 might not function as a miRNA sponge (SI Appendix, Fig. S5A). Besides, circASH2 did not possess the ability to encode a polypeptide (SI Appendix, Fig. S5B). Moreover, considering that both TPM4 mRNA and protein were prohibited by circASH2 (Fig. 4 B and E), we believed that a transcriptional or posttranscriptional regulation on TPM4 is induced by circASH2. However, comparable TPM4 promoter luciferase activity was observed when circASH2 was modulated, implying that a posttranscriptional regulation is more likely (Fig. 5C).
Fig. 5.
circASH2 promotes mRNA decay by physically connecting TPM4 transcripts. (A) Subcellular localization of circASH2 (red) in Huh7 cells revealed by FISH. Nuclei were labeled with DAPI (blue). (Scale bars, 10 μm.) (B) RT-qPCR detection of circASH2 from the indicated compartments of Huh7 cells and HCC tissues (n = 3). GAPDH mRNA and U6 RNA were used as reference RNAs from the cytoplasm and nucleus, respectively. (C) Dual-luciferase reporter assay of the TPM4 promoter (−2,000~0 bp) in si-NC and si-circASH2 HCC cells (n.s., not significant, unpaired Student’s t test). The firefly luciferase activity is normalized to the Renilla luciferase activity (firefly luciferase/Renilla luciferase) and presented as relative luciferase activity. (D) RT-qPCR detection of TPM4 mRNA in circASH2 overexpression or knockdown HCC cells treated with 2 μg/mL actinomycin D for the indicated times. The dashed line shows the half-life of TPM4 mRNA (mean ± SD, ***P < 0.001, two-way ANOVA test). (E) Representative FISH images of circASH2 (red) and TPM4 transcripts (orange, 40 nM, 55 °C, 2 h) showing a marked colocalization of them in Huh7 and HCCLM3 cells. DAPI (blue) was used to stain the nucleus. (Scale bars, 10 μm.) (F) The circRNA pulldown assay confirmed that circASH2 could bind with TPM4 mRNA and TPM4 pre-mRNA (treated with 10 μM isoginkgetin for 48 h). mRNA/pre-mRNA of GAPDH and β-actin were used as a negative control. (G, Top) Cartoon depicting the structure and sequence of TPM4 mRNA (including full-length transcript and 3 different truncations). (Bottom) TPM4-knockout HCCLM3 cells were reconstituted with TPM4FL, TPM4Δ5′UTR, TPM4ΔCDS, and TPM4Δ3′ UTR, respectively. RT-qPCR was used to detect the circASH2-enriched TPM4 mRNA levels in indicated cells (means ± SD, *P < 0.05, ***P < 0.001 and n.s., not significant, unpaired Student’s t test). (H) The predicted secondary structure and HR (1 to 14, denoted by red circles) of circASH2 was made by Mfold 2.3 software. Meanwhile, a schematic illustration of circASH2 mutations used in this study is shown. I. HA22T carrying indicated circASH2 plasmids (WT or HRmut) were used for detection of circASH2-TPM4 mRNA interaction. The enrichment level of circASH2 coisolated by TPM4 mRNA pulldown was detected by RT-qPCR. (J) RT-qPCR detection of TPM4 mRNA in indicated HA22T cells treated with 2 μg/mL actinomycin D for the indicated times. The dashed line shows the half-life of TPM4 mRNA (mean ± SD, ***P < 0.001 and n.s., not significant, two-way ANOVA test). (K) Endogenous TPM4 level and FAK activation in HA22T cells expressing WT or HRmut circASH2 were analyzed by immunoblots. GAPDH was used as an internal reference. (L) The state of cytoskeleton in indicated HA22T cells was shown by phalloidin staining of F-actin (red) and immunofluorescence of paxillin (green), while DAPI was used to stain the nucleus. (Scale bar, 10 μm.)
Recent studies suggest that N6-methyladenosine (m6A), a most abundant mRNA modification, could cause mRNA instability, while circRNA might influence this process (32, 33). Intriguingly, we found that circASH2 indeed promoted TPM4 mRNA degradation; however, in an m6A-independent manner (Fig. 5D and SI Appendix, Fig. S5C). According to our previous study, certain circRNA might function as a molecular scaffold, thereby influencing the substrate’s stability (6, 27). Inspired by nucleus localization of circASH2 (Fig. 5 A and B), we performed FISH and identified a marked colocalization of circASH2 and TPM4 transcripts (Fig. 5E). Next, we tested possible physical interaction between circASH2 and TPM4 transcripts. Biotin-labeled enrichment assays indicated that circASH2 could combine with not only TPM4 mRNA but also TPM4 pre-mRNA (Fig. 5F). These data strongly delivered an idea that circASH2 could specifically target TPM4 transcripts and disturb its stability.
Considering the consensus sequence of TPM4 mRNA and pre-mRNA, we designed a truncation map and identified that the deletion of the 3′ UTR nucleotide fragment of TPM4 mRNA (TPM4Δ3′UTR) markedly impaired the interaction of TPM4 mRNA with circASH2, while the absence of CDS domain (TPM4ΔCDS) also weakened the binding (Fig. 5G). RNA loops are known to be the key structure to mediate the RNA interactome (34). Hence, we constructed a structural model of circASH2 with the lowest free energy according to a previous described method (6) (Fig. 5H and SI Appendix, Fig. S5D). Hairpins (HR) mutation based on the predicted model revealed that HR 1, HR4, HR7, and HR9 are related to circASH2 combination (Fig. 5I). Mutation of all these four HRs (circASH2HRmut) completely suspended the interaction between circASH2 and TPM4 mRNA (Fig. 5I) and consequently attenuated the prodegradative effect induced by circASH2 (Fig. 5 J and K). Foreseeably, circASH2HRmut also lost the capability of weakening cytoskeleton formation and adhesion of tumor (Fig. 5L and SI Appendix, Fig. S5E). Meanwhile, according to sequence analysis, four fragments of TPM4 mRNA (AATCATTGT, ATTTAC, TTCTTTC, and TAATAT) were required for interaction with HR1, HR4, HR7, and HR9, which are majorly localized in 3′ UTR regions (SI Appendix, Fig. S5F).
In general, these epistasis analyses markedly indicated that circASH2 physically binds to TPM4 transcripts and facilitates its degradation, which represents a circRNA-induced posttranscriptional regulation.
The circASH2/hnRNP Complex Accelerates mRNA Degradation via Nonsense-Mediated Decay (NMD).
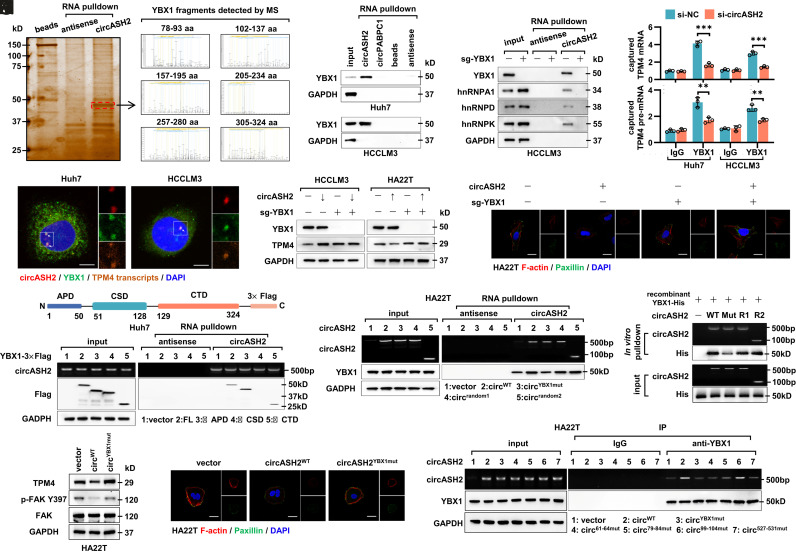
Since circASH2 directly bound to TPM4 mRNA/pre-mRNA and possibly functioned as a complex scaffold (25, 35–37), we supposed there should be other functional cofactors to mediate TPM4 mRNA/pre-mRNA decay. To prove our hypothesis, we successfully enriched circASH2 and coisolated a number of its potential binding proteins for mass spectrometry (MS) analysis (Fig. 6A and SI Appendix, Table S3). Gene ontology analysis implied circASH2 combined proteins notably related to mRNA splicing and stabilization (Fig. 6B). Interestingly, a series of hnRNPs were identified in MS and further verified by immunoblot (Fig. 6 A and C and SI Appendix, Fig. S6 A and B).
Fig. 6.
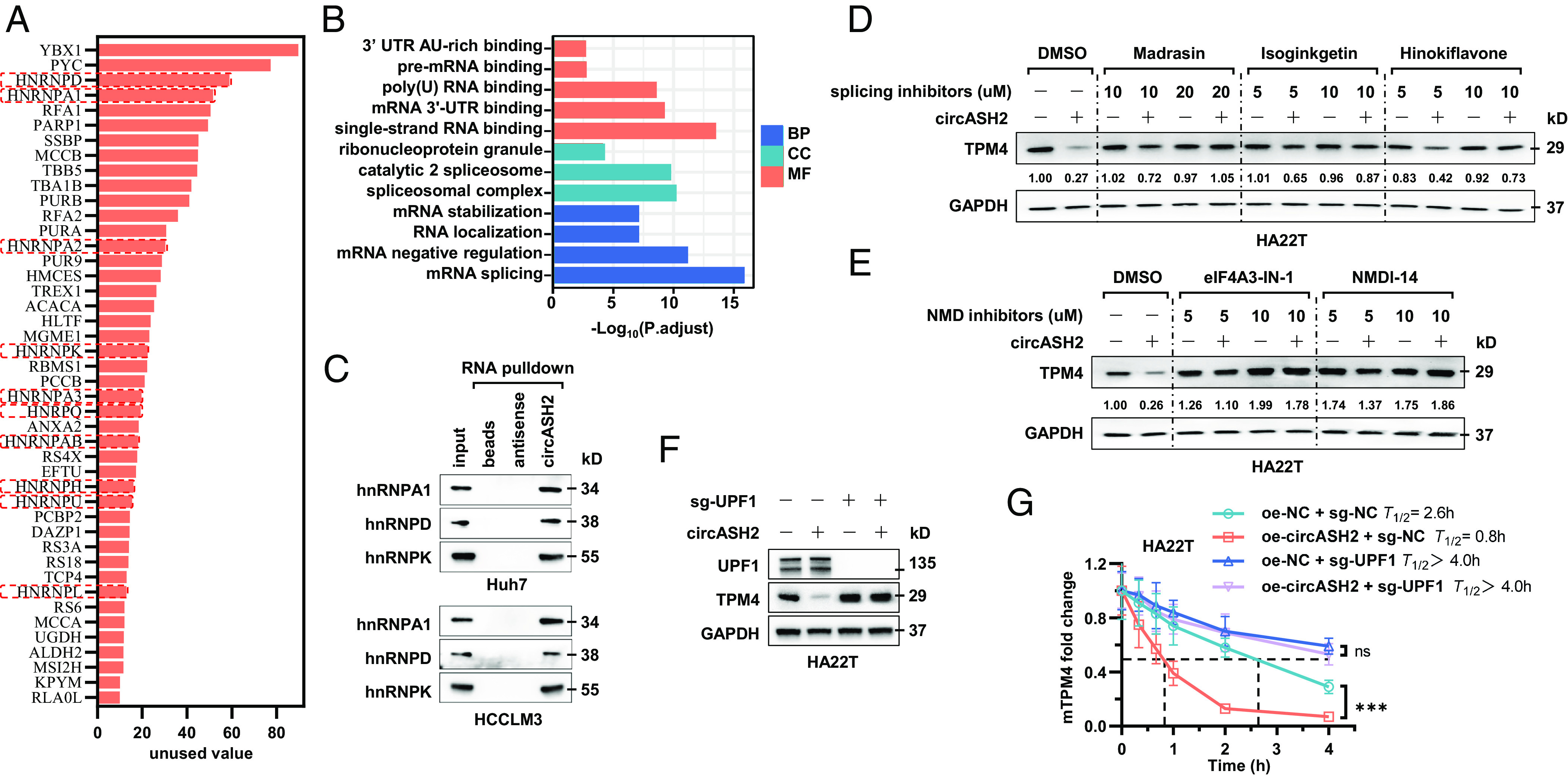
circASH2/hnRNP complex accelerates mRNA degradation via nonsense-mediated decay. (A) A diagram showing potential circASH2-binding proteins identified by MS (listed in the order of unused value). hnRNPs are highlighted with red boxes. (B) Gene ontology (GO) analysis of circASH2-interacting proteins, including molecular function (MF), biological process (BP), and cellular component (CC). (C) Three representative hnRNPs (hnRNPA1, hnRNPD, and hnRNPK) were confirmed as circASH2-binding proteins by immunoblot analysis in Huh7 and HCCLM3 cells. (D) HA22T cells overexpressing circASH2 or control vector were treated with Dimethylsulfoxide (DMSO), Madrasin (10 to 20 μM), isoginkgetin (5 to 10 μM), or hinokiflavone (5 to 10 μM) for 48 h. The endogenous TPM4 level was probed, while GAPDH was used as an internal reference. (E) HA22T cells overexpressing circASH2 or control vector were treated with DMSO, eIF4A3-IN-1 (5 to 10 μM), NMDl-14 (5 to 10 μM) for 48 h. The endogenous TPM4 level was probed, while GAPDH was used as an internal reference. (F) UPF1 was depleted in HA22T cells, and circASH2-induced downregulation of endogenous TPM4 was assessed by immunoblot analysis. GAPDH was used as an internal reference. (G) RT-qPCR detection of TPM4 mRNA at the indicated times in indicated HA22T cells treated with 2 μg/mL actinomycin D. The dashed line shows the half-life of TPM4 mRNA (mean ± SD, ***P < 0.001 and n.s., not significant, two-way ANOVA test).
hnRNPs, including more than 20 major proteins (hnRNPA0-hnRNPU), are the key parts of the spliceosome and responsible for regulating splicing of freshly transcribed RNAs (38). A remarkable combination between circASH2 and hnRNPs implied that circASH2 might interfere TPM4 transcripts’ stability via splicing regulation. To prove this, we applied three mRNA splicing inhibitors [Madrasin, isoginkgetin, and hinokiflavone] and found that TPM4 suppression induced by circASH2 is significantly attenuated, in a dose-dependent manner (Fig. 6D and SI Appendix, Fig. S6C). In most scenarios, mRNA that has undergone aberrant splicing tends to enter the process of NMD instead of translation (39, 40) and results in a rapid degradation of mRNA and suppression of the corresponding gene. To verify this, we blocked the NMD pathway by specific inhibitors [eIF4A3-IN-1 and NMDl14] and found that circASH2-induced TPM4 downregulation is impeded (Fig. 6E and SI Appendix, Fig. S6D). Furthermore, deletion of UPF1, the key component of the NMD pathway (40), could lead to a similar conclusion (Fig. 6 F and G).
These data demonstrated that, in HCC cells, circASH2-induced TPM4 downregulation relied on the pre-mRNA splicing and NMD process, in which the hnRNPs complex functions as a key regulator.
circASH2/hnRNP-Induced TPM4 mRNA Degradation Is YBX1 Dependent.
MS analysis of the circASH2 interactome revealed YBX1, a well-known RBP implicated in pre-mRNA splicing (16, 17) and mRNA decay (17, 41, 42), ranked as the top circASH2-binding protein (Figs. 6A and 7A). YBX1 is one of the first recognized prespliceosome associated proteins (43) which usually functions coordinately with hnRNPs inside the nucleus (14, 15). During splicing, YBX1 could function in recognition of a specific sequence of pre-mRNA and recruit splicing factors (e.g., hnRNPs) (19). To further illustrate the role of YBX1 in our model, we conducted a series of experiments.
Fig. 7.
circASH2/hnRNP-induced TPM4 mRNA degradation is YBX1-dependent. (A, Left) circASH2-binding proteins purified by circASH2 pulldown were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by silver staining. The indicated protein bands (denoted by the red box) were cut from gel and used for mass spectrometry (MS) analysis. (Right) Several YBX1 peptide fragments were identified by MS. (B) Detection of endogenous YBX1 following circASH2 pulldown from Huh7 and HCCLM3 cells, while circPABPC1 was used as a negative control. (C) Immunoblot analysis showed that YBX1 KO impeded the interaction between circASH2 and hnRNPs (represented by hnRNPA1, hnRNPD, and hnRNPK) in HCCLM3 cells. (D) RT-qPCR was applied for detection of endogenous TPM4 transcripts immunoprecipitated with YBX1 (1:100, 4 °C, overnight), while circASH2 knockdown can weaken the efficiency (**P < 0.01, and ***P < 0.001, unpaired Student’s t test). (E) circASH2 FISH (red), TPM4 transcripts FISH (orange), and anti-YBX1 immunofluorescence (green, 1:200, R.T., 1 h) staining showing colocalization of these three molecules (arrowheads) in Huh7 and HCCLM3 cells. Nuclei were labeled with DAPI (blue). (Scale bars, 10 μm.) (F) HCC cells with or without YBX1 KO were transduced with the circASH2 vector (HA22T) or si-circASH (HCCLM3). TPM4 expression was determined by immunoblot analysis, while GAPDH was used as an internal reference. (G) Cytoskeleton formation of indicated HCC cells was shown by phalloidin staining of F-actin (red) and immunofluorescence of paxillin (green), while DAPI was used to stain the nucleus. (Scale bar, 10 μm.) (H, Top) Cartoon depicting the structure and sequence of YBX1-3×Flag. (Bottom) Huh7 cells were transduced with YBX1FL-3×Flag, YBX1ΔAPD-3×Flag, YBX1ΔCSD-3×Flag, and YBX1ΔCTD-3×Flag, respectively. Immunoblot analysis was used to detect the YBX1-3×Flag enriched by circASH2 pulldown. (I) HA22T cells were transduced with circASH2WT, circASH2YBX1mut (mutation in the YBX1-binding site), or 2 random-mutant circASH2 (the mutation sites were distant from the YBX1 binding site), respectively. Immunoblot analysis was used to detect endogenous YBX1 enriched by circASH2 pulldown. (J) For the in vitro binding assay, 2 ng recombinant YBX1-His was incubated with circASH2WT, circASH2YBX1mut, or 2 random-mutant circASH2 (purified from HA22T cells overexpressing different types of circASH2 and immobilized on 20 µL beads), respectively. After coincubation, the beads were magnetically separated and washed 5 times with wash buffer. circASH2-bound YBX1-His was detected by immunoblots. (K) The endogenous TPM4 level and FAK activation in HA22T cells expressing circASH2WT or circASH2YBX1mut were determined by immunoblot analysis. (L) Cytoskeleton formation in indicated HA22T cells was shown by phalloidin staining of F-actin (red) and immunofluorescence of paxillin (green), while DAPI was used to stain the nucleus. (Scale bar, 10 μm.) (M) HA22T cells were transduced with circASH2WT, circASH2YBX1mut, circASH261-64mut, circASH279-84mut, circASH299-104mut, and circASH2527-531mut, respectively. RT-qPCR was used to detect the circASH2 enriched by anti-YBX1 immunoprecipitation.
First, we confirmed the interaction of circASH2 and YBX1 by circRNA-pulldown immunoblot (Fig. 7B), RNA immunoprecipitation (SI Appendix, Fig. S7A) and colocalization staining (SI Appendix, Fig. S7B). Second, YBX1 knockout completely suspended the connection between circAHS2 and several major hnRNPs, suggesting that YBX1 is necessary for circASH2-hnRNPs complex assembly (Fig. 7C). Third, YBX1 immunoprecipitation successfully enriched TPM4 mRNA/pre-mRNA, and such enrichment was impaired after circASH2 knockdown (Fig. 7D). Fourth, YBX1, circASH2, and TPM4 transcripts strictly overlapped in the staining image of the HCC nucleus (Fig. 7E). Finally, deletion of YBX1 completely abrogated the biological effects of the circASH2/TPM4 axis on HCC cells, including the cytoskeleton and cell adhesion (Fig. 7 F and G and SI Appendix, Fig. S7 C and D). It should be noted that deletion of YBX1 up-regulated the TPM4 protein level in HCCLM3 but failed to do so in low-circASH2 ones (HA22T, Figs. 1D and 7F). Meanwhile, the protein level and subcellular localization of YBX1 remained intact after modulating circASH2 (SI Appendix, Fig. S7 E and F).
To further detail the interaction mechanism, we truncated YBX1 as YBX1Δalanine/proline-rich domain (APD), YBX1Δcold shock domain (CSD), and YBX1ΔC-terminal domain (CTD). We identified that the CSD fragment was necessary for circASH2 binding (Fig. 7H and SI Appendix, Fig. S7G). Meanwhile, interaction simulation implied that YBX1 and TPM4 bound to the allopatric region of circASH2, respectively (SI Appendix, Figs. S5F, S7H, and S7I). Moreover, mutation on predicted binding sites of circASH2 (circASH2YBX1mut), not random mutation (circASH2random1 and circASH2random2, the same number of bases distant from the predicted binding site are mutated), impaired the connection with YBX1, in vivo and in vitro (Fig. 7 I and J). Similarly, circASH2YBX1mut also led to circASH2 function loss (Fig. 7 K and L and SI Appendix, Fig. S7J). Further mapping of the circASH2 mutant fragment indicated that 61 to 64 nt, 80 to 84 nt, and 526 to 530 nt regions of circASH2 were indispensable for YBX1 binding (Fig. 7M and SI Appendix, Fig. S7K).
The above-mentioned data demonstrated YBX1 featured in the circASH2/hnRNPs/TPM4 axis and, according to the YBX1 knockout assay (Fig. 7C) and in vitro binding assay between recombinant YBX1 and circASH2 (Fig. 7K), we believed that circASH2 directly interacted with YBX1, followed by recruiting hnRNPs to form a functional complex to regulate TPM4 transcript splicing.
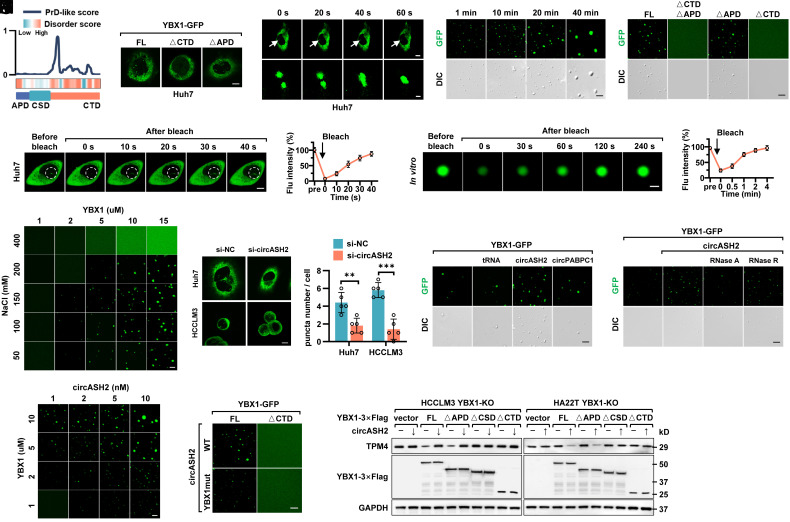
Intranuclear YBX1 Undergoes LLPS.
A very recent publication reported that certain protein would concentrate on specific mRNA via LLPS and regulate its translation (44). On the other hand, hnRNPs are closely related to LLPS (45). In our study, we noticed that YBX1 formed obvious puncta (dia. >0.5 um) and overlapped with circASH2 and TPM4 transcripts in the nucleus (Fig. 7E and SI Appendix, Fig. S7B). Therefore, we wondered whether such concentrating puncta were induced by LLPS.
According to previous reports, IDRs could be one of the decisive factors to mediate LLPS (21). By analyzing the YBX1 protein sequence, we found that CTD of YBX1 is highly disordered, both for a disorder score and a prion-like domain (PrLD) score (PrD-like, one of the most typical disorder sequences, Fig. 8A and SI Appendix, Table S4). Next, we expressed YBX1full-length (FL)-GFP, YBX1ΔAPD-GFP, and YBX1ΔCTD -GFP ectopically based on the disorder score in HCC cells (SI Appendix, Fig. S8A), and the results showed YBX1 condensate in the nucleus observed in the YBX1FL-GFP and YBX1ΔAPD-GFP groups but disappeared in the YBX1ΔCTD -GFP group (Fig. 8B and SI Appendix, Fig. S8B).
Fig. 8.
Intranuclear YBX1 undergoes LLPS enhanced by circASH2 and is indispensable for circASH2 signals. (A) Intrinsically disordered region (IDR) prediction of YBX1. (Top) Predictions of prion-like domains (PrLDs) and disordered regions. (Bottom) Schematic illustration of YBX1 structural domains. (B) Huh7 cells transfected with YBX1FL-GFP, YBX1△CTD-GFP, or YBX1△APD-GFP were analyzed by confocal microscopy. Representative pictures are shown. (Scale bar, 10 μm.) (C) Time-series fluorescence microscopy analysis of YBX1-GFP puncta in Huh7 cells. The Bottom row shows a zoom-in view of two fusing puncta. [Scale bar, 10 μm (Top) and 2 μm (Bottom).] (D, Left) Representative micrographs of YBX1-GFP puncta before and after photobleaching in Huh7 cells. (Scale bar, 10 μm.) (Right) Quantification of fluorescence intensity recovery in the bleached region of YBX1 puncta. (E) Small droplets fused into larger ones over time in vitro. (Scale bar, 5 μm.) (F) Phase separation assay of truncation mutants of YBX1 in vitro. The YBX1FL-GFP and YBX1△APD-GFP phase separated into liquid-like droplets, whereas YBX1△CTD+APD-GFP and YBX1△CTD-GFP failed. (Scale bar, 5 μm.) (G, Left) Representative micrographs of YBX1-GFP puncta before and after photobleaching in vitro. (Scale bar, 2 μm.) (Right) Quantification of fluorescence intensity recovery in the bleached region of YBX1 puncta. (H) Representative fluorescent images of phase separation behaviors of different concentrations of purified YBX1-GFP in different concentrations of NaCl (with 5% PEG8000). (Scale bar, 5 μm.) (I, Left) Representative fluorescent images of YBX1-GFP in control and circASH2-silenced HCC cells. (Scale bar, 10 μm.) (Right) Quantification of YBX1-GFP puncta number in control and circASH2-silenced HCC cells (mean ± SD, **P < 0.01 and ***P < 0.001, unpaired Student’s t test). (J) In vitro phase separation of purified YBX1-GFP with or without addition of 50 ng/μL different kinds of RNAs. circPABPC1 and tRNA were used as a negative control. (Scale bar, 5 μm.) (K) In vitro phase separation of purified YBX1-GFP with 50 ng/μL circASH2 with or without addition of 20U RNaseA or 20U RNaseR. (Scale bar, 5 μm.) (L) In vitro phase separation assay showing that circASH2 promotes YBX1 LLPS in a dose-dependent manner. (Scale bar, 5 μm.) (M) In vitro phase separation assay showing that circASH2WT facilitates YBX1WT-GFP, instead of YBX1△CTD-GFP, phase separation, whereas circASH2YBX1mut loses the function. (Scale bar, 5 μm.) (N) YBX1-KO HCC cells (HCCLM3 and HA22T) were reconstituted with YBX1FL, YBX1△APD, YBX1△CSD, and YBX1△CTD, respectively. Immunoblots were used to detect the endogenous TPM4 level in the indicated cells.
Also, we dynamically observed that two YBX1 puncta coalesced into a larger one (Fig. 8C and SI Appendix, Fig. S8C), indicating that YBX1 puncta possesses the liquid-like property. A similar LLPS feature was obtained by fluorescence recovery after photobleaching (FRAP) (Fig. 8D and SI Appendix, Fig. S8D). Additionally, 1,6-hexanediol, a LLPS inhibitor, led to disassembly of YBX1 puncta (SI Appendix, Fig. S8E). These findings suggested that YBX1 undergoes LLPS in vivo.
Next, we purified YBX1-GFP from stable transfected cells and performed phase separation assays with polyethylene glycol 8000 (PEG8000, a molecular crowding agent). As expected, YBX1-GFP formed droplets over time in vitro (Fig. 8E), which was also disrupted by 1,6-hexanediol (SI Appendix, Fig. S8F). Consistently, the CTD fragment was also indispensable for LLPS of YBX1 in vitro (Fig. 8F). To further verify the liquid-like properties of these condensates, we performed a FRAP assay, confirming that photobleaching the YBX1-GFP droplets resulted in a rapid recovery of fluorescence within 4 min (Fig. 8G). In addition, the orthogonal experiment indicated that an increased YBX1 concentration and proper NaCl concentration promoted droplet formation, in which YBX1 started to form liquid-like droplets at 2 µM (Fig. 8H and SI Appendix, Fig. S8G).
Overall, these results suggested that YBX1 could undergo LLPS both in vivo and in vitro. Importantly, the CTD fragment of YBX1, as a highly disordered region, is indispensable for such a biological process.
YBX1-LLPS Is Enhanced by circASH2 and Is a Prerequisite of circASH2 Signals.
It has been reported that RNAs could participate in LLPS-related events (22, 23). Our data indicated that circASH2 specifically colocalized with YBX1 condensates (Fig. 7E and SI Appendix, Fig. S7B), and we logically investigated whether circASH2 regulated YBX1-LLPS. In vivo, we found that YBX1 puncta formation was attenuated in circASH2 knockdown cells (Fig. 8I), while overexpressed circASH2 increased YBX1 puncta and circASHYBX1mut failed to do so (SI Appendix, Fig. S8H). In vitro, circASH2YBX1-GFP was coincubated with circASH2, circPABPC1, and tRNA, respectively, and only circASH2 successfully promoted YBX1 condensation formation (Fig. 8J). Moreover, such enhancement could be prohibited by RNase A, not RNase R, suggesting that YBX1 droplet formation was facilitated by circRNA (Fig. 8K). Meanwhile, the orthogonal experiment demonstrated a positive correlation between the circASH2 concentration and droplet formation of YBX1 (Fig. 8L and SI Appendix, Fig. S8I). Finally, such enhancement could be eliminated by either circASH2YBX1mut or YBX1ΔCSD-GFP (Fig. 8M and SI Appendix, Fig. S8I).
Then, we investigated whether YBX1-LLPS is the requirement for circASH2-induced signals and function. We constructed and reexpressed YBX1FL-Flag, YBX1ΔAPD-Flag (retains the ability to occur LLPS as well as to combine with circASH2), YBX1ΔCSD-Flag (cannot bind to circASH2), and YBX1ΔCTD-Flag (loses the ability to occur LLPS) in YBX1-KO HCC cells, respectively. The result indicated that only when YBX1retains the ability to occur LLPS and to combine with circRNA could circASH2 suppress TPM4 expression (Fig. 8N). Consistently, HCC cytoskeleton staining and digestion assay verified that YBX1-LLPS is a prerequisite of circASH2-induced signals (SI Appendix, Fig. S8 K and L).
Collectively, circASH2 could accelerate the occurrence of YBX1 LLPS both in vivo and in vitro, which is also essential for the transmission of the circASH2-TPM4 signals.
Discussion
Metastasis is responsible for most of cancer-related deaths, which is a multistep process and requires dramatic remodeling of the cytoskeleton (4). Cells could communicate with the external environment through the cytoskeleton, thereby receiving external signals and guiding behaviors such as pseudopodia formation, migration, and invasion (5). Once the state of the cytoskeleton is changed, the conduction of various signaling pathways would be affected, such as the FAK-PI3K-AKT pathway, Hippo-YAP/TAZ pathway, Rho pathway, etc (7). For instance, the cytoskeleton directly regulates the interaction between integrins and ECM, followed by affecting the phosphorylation state of FAK (7). Then, phosphorylated FAK could drive downstream signaling cascades, such as the PI3K-AKT pathway observed in our model. As one of the major cytoskeletal networks, the actin cytoskeleton is the collection of F-actins with their accessory and regulatory proteins (e.g., TPM4) (10, 46), functioning as primary force-generating machinery in cells (47). In this study, we identified TPM4 as an oncoprotein that promoted HCC metastasis by stabilizing the actin cytoskeleton.
Subcellular localization of circRNAs is a cue for finding their function and mediated mechanism (25). Generally, circRNAs containing intron-derived sequences prefer to localize in the nucleus, whereas exonic circRNAs are largely localized to the cytoplasm (48, 49). Recent studies already demonstrated that exonic circRNAs could also function in the nucleus (50–52). In our study, circASH2 was mainly located in the HCC nucleus and interacted with several critical RBPs to modulate mRNA/pre-mRNA splicing. Considering YBX1 enriched more in the cytoplasm, circASH2-induced LLPS of nuclear YBX1 is more specific and informative for future research. Meanwhile, how exonic circRNAs enriched in the nucleus or shuttled to the cytoplasm remains unclear. According to Huang et al., deletion of Drosophila DExH/D box helicase (Hel25E) could lead to nuclear accumulation of long (>800 nt) circRNAs, suggesting that the length might be one of the determinants (53). Another case is mitochondria-encoded circRNAs (mecciRNAs), which were transcribed and largely accumulated in mitochondria (a portion could be exported to cytosol) (54). Although evidence suggests that PNPase might participate in mecciRNA-related transportation (54), further study is still urgently needed. Overall, distribution and transportation of circRNA are not well investigated yet and require more attention.
Intracellular mRNA homeostasis is essential for cell survival and function, among which NMD is one of the best characterized and most evolutionarily conserved mechanisms (40). Basically, mRNAs harboring a premature termination codon (PTC) are selectively degraded by NMD (39, 40). In our study, we found that specific inhibitors or UPF1 deletion could suspend circASH2-induced signal, indicating that NMD should be the way out of modulated TPM4 transcripts. Whether circASH2-YBX1-hnRNPs induced a PTC in TPM4 transcripts is worth further study. Besides, NMD prefers to influence mRNAs with an unusually long 3′ UTR (>1,000 bp) (40). Therefore, major TPM4 transcripts that naturally owned a long 3′ UTR (>1,600 bp) could be one of the reasons for its NMD-related degradation, and circASH2-induced splicing might be able to enhance such a scenario. Among the hnRNPs, hnRNP L was the one that mostly correlated with NMD, according to recent publications (55). Generally, hnRNP L could protect mRNAs from NMD degradation, especially in tumors (55). In our study, a bunch of hnRNPs was identified to interact with circASH2. Although there is a lack of direct evidence in other hnRNPs-NMD, hnRNP-induced splicing events, such as alternative splicing, are very likely followed by NMD or alternative splicing coupled to NMD.
In this study, circASH2 functions as a scaffold to assemble YBX1, hnRNPs, and TPM4 transcripts to form a complex and lead to TPM4 transcripts’ decay. An intriguing question is whether such a complex was TPM4 transcript specific. According to immunofluorescence experiments of YBX1, only nuclear YBX1 was distributed in several locations, while the cytoplasmic one was widely spread. With the almost perfect colocalization of circASH2, YBX1, and TPM4 transcripts, we believed that circASH2 induced a very specific regulation of TPM4 here. This also provided a possibility for other circRNAs to modulate such regulation in different genes, through a similar complex (YBX1/hnRNPs) or a diverse one.
Intracellular LLPS provides an explanation for how a cell efficiently organizes various molecules to perform a specific reaction or activity, in a temporal and spatial manner. LLPS participates in many cellular processes and in cancer research; accumulated evidence indicates that LLPS participated in various tumor biological processes (20, 56). RBPs, such as YBX1, are prone to undergo LLPS (57). We first reported that intranuclear YBX1 could undergo LLPS, and more importantly, this process was promoted by circASH2. Similarly, Long noncoding RNA (lncRNA) has also been reported to mediate LLPS in certain circumstances (23). In our model, phase separation strengthened the concentration of YBX1 in the TPM4 transcript region and logically enhanced YBX1/hnRNP complex–mediated editing. Moreover, we dissected the mechanism detail of YBX1-LLPS in our study, including the necessary fragment of YBX1 for LLPS and possible function for downstream cellular behaviors. These results highlighted our findings and provided insights for deep cancer mechanism dissection.
circRNA-based research has reached a very significant achievement over the past decade (25). In cancer biology, circRNAs serve as tumor suppressors or oncogenic drivers through various mechanisms (26). Here, we revealed that circASH2, a nuclear circRNA, was down-regulated in HCC, which conforms to the knowledge that cancer cells often exhibit a reduction in global circRNA abundance (25). More importantly, circASH2 remodeled tumor cytoskeleton via suppressing TPM4 and exerts a significant antimetastatic activity in vivo and in vitro. Mechanistically, circASH2 facilitates the LLPS of nuclear YBX1 and targets TPM4 transcripts by assembling a complex with hnRNPs. Such a circRNA/YBX1/hnRNP complex interfered TPM4 transcript splicing and led to its accelerated degradation (SI Appendix, Fig. S9). Our research provided a circRNA-induced tumor regulation mechanism, which linked cytoskeleton, mRNA splicing, and LLPS together. With more confidence for circRNA-related drug development (58), we believed that our study would benefit liver cancer treatment in the future.
Materials and Methods
Details of this section are included in SI Appendix, Materials.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Dr. Ruihua Li (Hengrui Pharmaceutical LTD, CN) for her suggestions on LLPS-related experiments and Dr. Jingxin Li (University of Science and Technology of China, CN) for her technical assistance on determination of circRNA copy numbers. We appreciate Aksomics Co., Ltd. (human circRNA microarray), Novegene Co., Ltd. (MS), and TSINGKE Biological Technology Co., Ltd. (next-generation sequencing) for their technology service. Boqiang Liu receives funding support from Postdoctoral Science Fund of China. Professor L.S. receives funding support from National Natural Science Foundation of China (No. 82203426), Zhejiang Natural Science Foundation (No. LY23H160021), and Clinical Top-Notch Personnel Support of Zhejiang University. Professor X.C. receives funding support from Zhejiang Clinical Research Center of Minimally Invasive Diagnosis and Treatment of Abdominal Diseases (No. 2018E50003) and Key Research and Development Project of Zhejiang Province (No. 2018C03083). Professor G.S. receives funding support from National Key R&D Program of China (2019YFA0802600) and National Natural Science Foundation of China (No. 31725016, No. 31930019, and No. 91940303). Professor H.Y. receives funding support from “Ten Thousand Plan” Innovation Leader of Zhejiang Province (No. 2020R52007), Major Science and Technology Project of Zhejiang Province of China (No. 2021C306), and Key Project of Province and Ministry (No. WKJ-ZJ-2124).
Author contributions
B.L., L.S., and X.C. designed research; B.L., H.S., J.H., B.J., Y.T., W.L., W.Z., J.N., and J.Z. performed research; J.H., J.S., H.Y., and Y.W. contributed new reagents/analytic tools; B.L. and H.S. analyzed data; J.S. H.Y., and Y.W. collected and provided clinical samples and data; G.S. administrative support and revision suggestion; and B.L., L.H., and L.S. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. Y.G.C. is a guest editor invited by the Editorial Board.
Contributor Information
Liang Shi, Email: Liang_Shi@zju.edu.cn.
Xiujun Cai, Email: srrsh_cxj@zju.edu.cn.
Data, Materials, and Software Availability
RNA-seq data and circRNA microarray data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE236039 (59) and GSE235991 (60), respectively. All other study data are included in the article and/or SI Appendix.
Supporting Information
References
- 1.Sung H., et al. , Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249 (2021). [DOI] [PubMed] [Google Scholar]
- 2.Llovet J. M., et al. , Hepatocellular carcinoma. Nat. Rev. Dis. Primers 7, 6 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Stuelten C. H., Parent C. A., Montell D. J., Cell motility in cancer invasion and metastasis: Insights from simple model organisms. Nat. Rev. Cancer 18, 296–312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seetharaman S., Etienne-Manneville S., Cytoskeletal crosstalk in cell migration. Trends Cell Biol. 30, 720–735 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Hohmann T., Dehghani F., The cytoskeleton-A complex interacting meshwork. Cells 8, 362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi L., et al. , A tumor-suppressive circular RNA mediates uncanonical integrin degradation by the proteasome in liver cancer. Sci. Adv. 7, eabe5043 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geiger B., Bershadsky A., Pankov R., Yamada K. M., Transmembrane crosstalk between the extracellular matrix–cytoskeleton crosstalk. Nat. Rev. Mol. Cell Biol. 2, 793–805 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Bachir A. I., Horwitz A. R., Nelson W. J., Bianchini J. M., Actin-based adhesion modules mediate cell interactions with the extracellular matrix and neighboring cells. Cold Spring Harb. Perspect. Biol. 9, a023234 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lopez-Colome A. M., Lee-Rivera I., Benavides-Hidalgo R., Lopez E., Paxillin: A crossroad in pathological cell migration. J. Hematol. Oncol. 10, 50 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gateva G., et al. , Tropomyosin isoforms specify functionally distinct actin filament populations in vitro. Curr. Biol. 27, 705–713 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao X., et al. , Cytoplasmic localization isoform of cyclin Y enhanced the metastatic ability of lung cancer via regulating tropomyosin 4. Front. Cell Dev. Biol. 9, 684819 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caporali S., et al. , The miR-133a, TPM4 and TAp63gamma role in myocyte differentiation microfilament remodelling and colon cancer progression. Int. J. Mol. Sci. 22, 9818 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wahl M. C., Will C. L., Luhrmann R., The spliceosome: Design principles of a dynamic RNP machine. Cell 136, 701–718 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Ghanawi H., et al. , Loss of full-length hnRNP R isoform impairs DNA damage response in motoneurons by inhibiting Yb1 recruitment to chromatin. Nucleic Acids Res. 49, 12284–12305 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nasrin F., et al. , HnRNP C, YB-1 and hnRNP L coordinately enhance skipping of human MUSK exon 10 to generate a Wnt-insensitive MuSK isoform. Sci. Rep. 4, 6841 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayavelu A. K., et al. , Splicing factor YBX1 mediates persistence of JAK2-mutated neoplasms. Nature 588, 157–163 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Deng M., et al. , YBX1 mediates alternative splicing and maternal mRNA decay during pre-implantation development. Cell Biosci. 12, 12 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Calabretta S., Richard S., Emerging roles of disordered sequences in RNA-binding proteins. Trends Biochem. Sci. 40, 662–672 (2015). [DOI] [PubMed] [Google Scholar]
- 19.Lyabin D. N., Eliseeva I. A., Ovchinnikov L. P., YB-1 protein: Functions and regulation. Wiley Interdiscip. Rev. RNA 5, 95–110 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Hahn S., Phase separation, protein disorder, and enhancer function. Cell 175, 1723–1725 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Alberti S., Gladfelter A., Mittag T., Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell 176, 419–434 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clemson C. M., et al. , An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 33, 717–726 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li R. H., et al. , A phosphatidic acid-binding lncRNA SNHG9 facilitates LATS1 liquid-liquid phase separation to promote oncogenic YAP signaling. Cell Res. 31, 1088–1105 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S., et al. , circVAMP3 drives CAPRIN1 phase separation and inhibits hepatocellular carcinoma by suppressing c-Myc translation. Adv. Sci. (Weinh) 9, e2103817 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu C. X., Chen L. L., Circular RNAs: Characterization, cellular roles, and applications. Cell 185, 2016–2034 (2022). [DOI] [PubMed] [Google Scholar]
- 26.Kristensen L. S., Jakobsen T., Hager H., Kjems J., The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 19, 188–206 (2022). [DOI] [PubMed] [Google Scholar]
- 27.Shen H., et al. , Circular RNAs: Characteristics, biogenesis, mechanisms and functions in liver cancer. J. Hematol. Oncol. 14, 134 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luscher-Firzlaff J., et al. , The human trithorax protein hASH2 functions as an oncoprotein. Cancer Res. 68, 749–758 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Cheng X., et al. , MKL1 potentiates lung cancer cell migration and invasion by epigenetically activating MMP9 transcription. Oncogene 34, 5570–5581 (2015). [DOI] [PubMed] [Google Scholar]
- 30.Shi L., et al. , Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis. 8, e3171 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu J., et al. , Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J. Hepatol. 68, 1214–1227 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Zaccara S., Ries R. J., Jaffrey S. R., Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 20, 608–624 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Huang R., et al. , N(6)-methyladenosine modification of fatty acid amide hydrolase messenger RNA in circular RNA STAG1-Regulated astrocyte dysfunction and depressive-like behaviors. Biol. Psychiatry 88, 392–404 (2020). [DOI] [PubMed] [Google Scholar]
- 34.Zhu P., et al. , IL-13 secreted by ILC2s promotes the self-renewal of intestinal stem cells through circular RNA circPan3. Nat. Immunol. 20, 183–194 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Zhao Q., et al. , Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell 183, 76–93.e22 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Du W. W., et al. , Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 44, 2846–2858 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gong X., et al. , Circular RNA circEsyt2 regulates vascular smooth muscle cell remodeling via splicing regulation. J. Clin. Invest. 131, e147031 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Geuens T., Bouhy D., Timmerman V., The hnRNP family: Insights into their role in health and disease. Hum. Genet. 135, 851–867 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Supek F., Lehner B., Lindeboom R. G. H., To NMD or not to NMD: Nonsense-mediated mRNA decay in cancer and other genetic diseases. Trends Genet. 37, 657–668 (2021). [DOI] [PubMed] [Google Scholar]
- 40.Kurosaki T., Popp M. W., Maquat L. E., Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 20, 406–420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y., et al. , RNA 5-Methylcytosine facilitates the maternal-to-zygotic transition by preventing maternal mRNA decay. Mol. Cell 75, 1188–1202.e11 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Chen X., et al. , 5-methylcytosine promotes pathogenesis of bladder cancer through stabilizing mRNAs. Nat. Cell Biol. 21, 978–990 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Hartmuth K., et al. , Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl. Acad. Sci. U.S.A. 99, 16719–16724 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kang J. Y., et al. , LLPS of FXR1 drives spermiogenesis by activating translation of stored mRNAs. Science 377, eabj6647 (2022). [DOI] [PubMed] [Google Scholar]
- 45.Molliex A., et al. , Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Janco M., et al. , The impact of tropomyosins on actin filament assembly is isoform specific. Bioarchitecture 6, 61–75 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Svitkina T., The actin cytoskeleton and actin-based motility. Cold Spring Harb. Perspect. Biol. 10, a018267 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Conn V. M., et al. , A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 3, 17053 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Guo J. U., Agarwal V., Guo H., Bartel D. P., Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 15, 409 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Guarnerio J., et al. , Intragenic antagonistic roles of protein and circRNA in tumorigenesis. Cell Res. 29, 628–640 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia P., et al. , A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity 48, 688–701.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Wang X., et al. , CircURI1 interacts with hnRNPM to inhibit metastasis by modulating alternative splicing in gastric cancer. Proc. Natl. Acad. Sci. U.S.A. 118, e2012881118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Huang C., Liang D., Tatomer D. C., Wilusz J. E., A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 32, 639–644 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu X., et al. , Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci. China Life Sci. 63, 1429–1449 (2020). [DOI] [PubMed] [Google Scholar]
- 55.Kishor A., Ge Z., Hogg J. R., hnRNP L-dependent protection of normal mRNAs from NMD subverts quality control in B cell lymphoma. EMBO J. 38, e99128 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mehta S., Zhang J., Liquid-liquid phase separation drives cellular function and dysfunction in cancer. Nat. Rev. Cancer 22, 239–252 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu X. M., Ma L., Schekman R., Selective sorting of microRNAs into exosomes by phase-separated YBX1 condensates. Elife 10, e71982 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.He A. T., Liu J., Li F., Yang B. B., Targeting circular RNAs as a therapeutic approach: Current strategies and challenges. Signal Transduct. Target Ther. 6, 185 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu B., Effect of knock-down of circASH2 on gene expression in HCC cells. Gene Expression Omnibus database. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE236039. Deposited 28 June 2023.
- 60.Liu B., Shi L., circRNA expression level in highly/low invasive HCC tissues. Gene Expression Omnibus database. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE235991. Deposited 27 June 2023.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
RNA-seq data and circRNA microarray data have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE236039 (59) and GSE235991 (60), respectively. All other study data are included in the article and/or SI Appendix.