Abstract
Flavonols are plant specialized metabolites with important functions in plant growth and development. Isolation and characterization of mutants with reduced flavonol levels, especially the transparent testa mutants in Arabidopsis thaliana, have contributed to our understanding of the flavonol biosynthetic pathway. These mutants have also uncovered the roles of flavonols in controlling development in above- and below-ground tissues, notably in the regulation of root architecture, guard cell signaling, and pollen development. In this review, we present recent progress made towards a mechanistic understanding of flavonol function in plant growth and development. Specifically, we highlight findings that flavonols act as reactive oxygen species (ROS) scavengers and inhibitors of auxin transport in diverse tissues and cell types to modulate plant growth and development and responses to abiotic stresses.
Keywords: Flavonols, Flavonoids, Reactive oxygen species, Root development, Stomatal closure, Pollen development
Biosynthesis of flavonols
Flavonoids are a group of plant specialized metabolites that have multiple important functions including conferring flower color to attract pollinators and acting as antioxidants that regulate levels of reactive oxygen species (ROS) to control plant growth, development, and fertility [1–3]. The general structure of flavonoids consists of two benzene rings linked by a heterocyclic ring. Flavonoids are divided into subgroups based on modifications of the heterocyclic ring, with the major groups being flavonones, flavanols, flavonols, anthocyanins, and proanthocyanins [1]. The biosynthetic pathway of flavonoids has become a topic of broad interest due to their importance in modulating plant form and function, as well as their pharmacological and nutritional benefits [4–6].
This review focuses on flavonols due to recent evidence suggesting they have unique functions in controlling plant signaling and development. Flavonols are distinguished from other groups of flavonoids by the hydroxylation of one of the benzene rings. Each flavonol has a distinct pattern of hydroxylation of the benzene ring [7]. Three core flavonols are the most prevalent across the plant kingdom, which are the monohydroxylated kaempferol, the dihydroxylated quercetin, and the trihydroxylated myricetin, shown in Figure 1. Flavonols are further modified by the addition of diverse moieties to the hydroxyl groups, with carbohydrate modifications being the most frequent [8]. These decorations contribute to the immense chemodiversity found in the flavonol subgroup. Another well-characterized modification is the methylation of quercetin that yields isorhamnetin [9].
Figure 1. The flavonoid biosynthetic pathway.
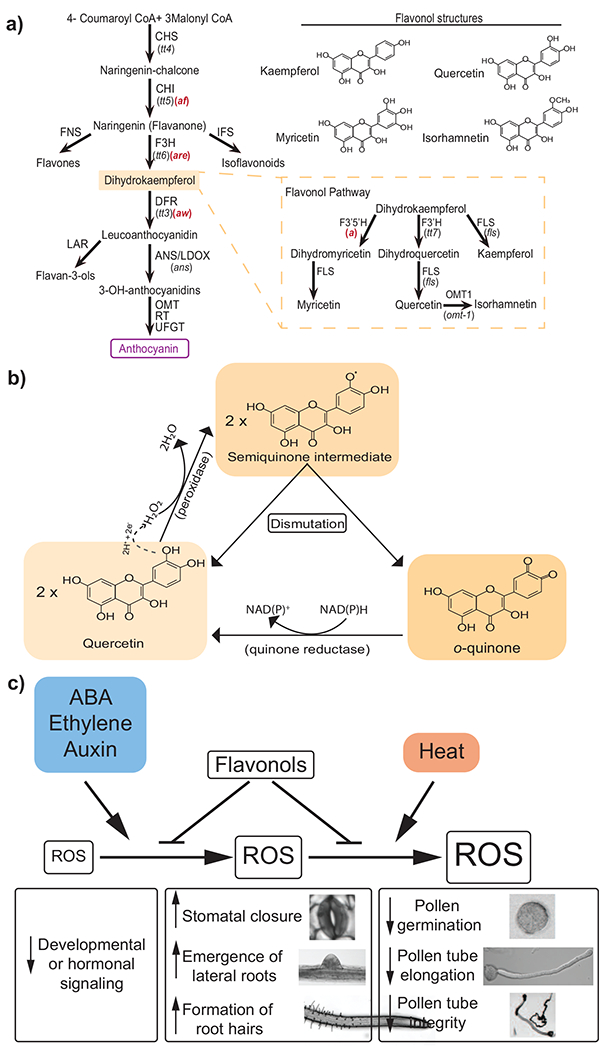
Enzymes are indicated next to arrows with mutant names in parentheses under the enzyme abbreviation. Arabidopsis mutants are in black and tomato mutants are in red. The flavonol biosynthesis branch is illustrated in the yellow dashed box. Chemical structure of specific flavonols are presented on the upper right. Abbreviations: CHS, chalcone synthase; CHI, chalcone isomerase; FNS, flavone synthase; F3H, flavanone 3-hydroxylase; IFS, isoflavone synthase; DFR, dihydroflavonol 4-reductase; LAR, leucoanthocyanidin reductase; ANS/LDOX, anthocyanidin synthase/leucoanthocyanidin dioxygenase; RT, rhamnosyl transferase; UFGT, UDP-glucose flavonoid 3-O-glucotransferase; F3’5’H, flavonoid 3’5’-hydroxylase; F3’H, flavonoid 3’-hydroxylase; FLS, flavonol synthase; OMT-1, O-methyltransferase-1. Figure adapted from [64 & 66], B. The predicted mechanism by which the flavonol quercetin donates electrons to convert hydrogen peroxide to water is shown, along with a potential mechanism by which reduced flavonols are regenerated. The transfer of electrons and protons to hydrogen peroxide are indicated above the dashed arrow. Types of enzymes that may catalyze these chemical reactions are presented in parentheses. It is also possible that the quinone reductase may convert the quinone back to a semiquinone. C. This diagram summarizes environmental or hormonal factors that increase levels of reactive oxygen species (ROS) and the role of flavonols as modulators of ROS to control developmental and physiological responses. Factors affecting ROS levels are indicated in colored boxes and the resulting changes in ROS concentration are depicted by increased size of boxes. Hormonal and environment increases in ROS levels are indicated by arrows and the ability of flavonols to reduce ROS accumulation is indicated by a blunt-end arrow. Observed developmental or physiological changes linked to the different ROS levels are indicated below the ROS changes. Abbreviation: ABA, abscisic acid.
The enzymes of the flavonoid biosynthetic pathway in Arabidopsis were identified through isolation and characterization of mutants that lacked brown proanthocyanin pigmentation in their seed coat, or testa [10]. The resulting transparent testa (tt) mutants were used to elucidate the biochemical pathway in Arabidopsis by mapping each mutation to genes encoding pathway enzymes. This genetic approach was feasible in Arabidopsis as most pathway enzymes are encoded by single genes [11]. Studies in other species have revealed similar enzymatic sequences, with multiple genes encoding isoenzymes that catalyze most steps of this biosynthetic pathway [12,13].
Flavonoids are synthesized from 4-coumaroyl-CoA, a phenylalanine derivative, and three molecules of malonyl-CoA, a fatty acid derivative, to produce naringenin chalcone through a reaction catalyzed by chalcone synthase (CHS), as shown in Figure 1. The cysteine residue in the active site of CHS has evolved to enhance its nucleophilicity and this increase in catalytic activity is suggested to have contributed to the diversification of the flavonoid biosynthetic pathway in terrestrial plants [14]. Naringenin chalcone is isomerized to naringenin by the enzyme chalcone isomerase. Naringenin is the precursor for flavonol biosynthesis and is converted to dihydrokaempferol by flavanone 3-hydroxylase (F3H). The flavonol precursor dihydrokaempferol can be converted to three different products by three different enzymes. Dihydrokaempferol can be converted to the other flavonol biosynthesis intermediates dihydroquercetin and dihydromyricetin by flavonoid 3-hydroxylase (F3’H) and flavonoid 3’5-hydroxylase (F3’5’H), respectively. FLS then oxidizes dihydrokaempferol, dihydroquercetin, and dihydromyricetin to the flavonols kaempferol, quercetin, and myricetin, respectively.
Pathway enzymes have multiple subcellular localizations to control where specific pathway products accumulate. For example, CHS and CHI enzymes were localized in the nucleus of cortical and epidermal cells of the root tip in Arabidopsis using immunofluorescence [15], and flavonol accumulation was detected in the nucleus [16]. In grape berry, CHS has also been found associated with the plastid and rough endoplasmic reticulum [17]. Given the many functional roles of flavonoids, their biosynthesis may occur in different tissues to allow distinct functions within these tissues. Most striking is the synthesis of anthocyanins, which is turned off in roots, pollen, and other locations, allowing higher accumulation of flavonols to modulate development, since dihydroflavonols are not converted to these downstream anthocyanin metabolites [16,18]. Genes encoding flavonoid biosynthetic pathway enzymes are highly regulated to allow both tissue specific expression and to allow pathway activity to be regulated by changes in environmental and hormonal signals.
Flavonoid biosynthesis is regulated by intrinsic and extrinsic factors
The flavonoid biosynthetic pathway is tightly regulated by intrinsic factors such as hormones and diverse external biotic and abiotic factors, which control the transcription of pathway enzymes. Expression of flavonoid biosynthetic genes depends on complexes of transcription factors. Members of the R2R3-MYB and bHLH families are required for all pathway enzymes to be expressed and a WD40 protein is a third component of complexes that regulate the downstream pathway enzymes that produce anthocyanins [19]. In Arabidopsis, three R2R3-MYB family transcription factors, MYB11, MYB12, and MYB111, activate the expression of the biosynthetic genes encoding CHS, CHI, F3H, and FLS in a tissue-specific manner [20]. Mutations mMYB12 block flavonol synthesis in roots, while MYB111 is required for pathway activity in cotyledon and primary leaves [21]. Other cell types show different transcription factor requirements. In Arabidopsis pollen, the triple mutant myb11 myb12 myb111 [22] accumulates flavonols, suggesting flavonol synthesis is independent of these three transcription factors in this cell type. However, leaves of this triplHe mutant or a single mutant in myb111 were incapable of synthesizing flavonols while maintaining anthocyanin production in leaves. This suggests that dihydroflavonols were still synthesized but were directly channeled into anthocyanins due to the absence of FLS activity [21].
Flavonol biosynthesis can be regulated by multiple plant hormones. Ethylene increases flavonol accumulation in guard cells to regulate stomatal aperture [23] and in roots to regulate gravitropism [16,24]. In roots, ethylene and auxin increase flavonol synthesis by increasing the abundance of transcripts encoding multiple flavonol biosynthetic enzymes [16]. These responses are dependent on MYB12 as neither hormone alters flavonol accumulation in a myb12 mutant [16]. Auxin and ethylene increase transcripts encoding CHS, CHI, and FLS, but F3H is only induced by auxin [16]. This results in different flavonol accumulation patterns with kaempferol increasing in response to both hormones, while auxin, but not ethylene, increases quercetin, the product of F3’H. The WRKY23 transcription factor is also required for flavonol induction by auxin, as a wrky23 mutant has reduced MYB12 transcripts and no induction of flavonol biosynthesis in roots [25].
One notable function of flavonols is protecting plants from environmental fluctuations that may be stressful. The flavonol biosynthetic pathway is thought to have evolved in plants during their colonization of land, when plants were first faced with new environmental stresses including ultraviolet (UV) light [14]. Many studies have reported increased flavonol levels in plants exposed to UV-B [26]. While the exact mechanism of flavonols in UV-B protection remains to be established, studies have demonstrated that excess flavonols provide protection against UV-B, such as the Arabidopsis transgenic line over-expressing flavonol synthase [27] and the UV-B tolerant Arabidopsis uvt1 mutant, which does not sustain leaf damage under UV-B stress [28]. Studies have also reported increased expression of the genes encoding flavonol biosynthetic enzymes CHS, F3H and FLS in different plants exposed to UV-B [28–30]. Flavonol biosynthesis under visible light in the absence of UV-B wavelengths is regulated through COP1, an E3 ubiquitin ligase. COP1 ubiquitinates HY5, a TF that activates expression of R2R3-MYB proteins [31]. Additionally, red and blue light induce flavonol synthesis through actions of HY5 and cryptochromes, respectively [32,33].
In addition to UV-B radiation, flavonols offer protection from a host of other abiotic stresses, which are increasing with climate change [34]. For example, drought conditions have been shown to induce accumulation of quercetin and kaempferol in Arabidopsis [35] and several tomato varieties [36]. External stimuli that result in increased flavonol production also include heat, high salinity [37]. and ozone; the latter of which increases flavonols by increasing nitrate reductase activity to elevate nitric oxide, which acts as a signaling molecule [38], These stresses are often tied to increased flavonol production to prevent stress-induced inhibition of plant growth and development [39].
Flavonols modulate reactive oxygen species (ROS) homeostasis and auxin transport
Flavonols have been shown to regulate plant growth, development, and physiology through two distinct mechanisms: maintenance of ROS homeostasis and inhibition of auxin transport. The tightly regulated production of ROS is necessary for molecular signaling involved in different plant processes [40]. However, excess ROS accumulation, e.g. during abiotic stresses, generates oxidative stress that damages cellular macromolecules [41]. As a result, plants need intricate mechanisms to maintain ROS homeostasis. ROS levels are a function of production by biosynthetic enzymes, transport, and scavenging [42]. Plants have evolved different enzymatic and non-enzymatic systems for ROS scavenging and flavonols are important players in mediating this homeostasis [43]. Indeed, their chemical structure, with its many hydroxyl groups, can act as a powerful electron donor, making flavonols potent antioxidants/reductants [44]. Compared to other plant antioxidants such as phenolic acids, flavonols have a more potent antioxidant capacity, based on their ability to neutralize singlet oxygen and hydrogen peroxide [45,46]. The chemistry by which flavonols are predicted to donate electrons to convert hydrogen peroxide to water is shown in Figure 1. as modified from [46]. The resulting semiquinone intermediate is predicted to dismutate back to quercetin or to a quinone that may be reduced back to quercetin via a NAD(P)H dependent quinone reductase. The following sections review the linkages between flavonol regulation of development through modulation of ROS homeostasis.
A second activity of flavonols is to modulate polar auxin transport, which moves auxin long distance in plant tissues [47]. Polar transport is mediated by auxin influx carriers, including AUX1 and LAX (like AUX1) proteins, and efflux carrier proteins, which include both PIN (Pin formed) and ABCB (ATP Binding Cassette B) proteins [48–50]. The polarity of auxin transport is largely driven by asymmetric location of the PIN proteins [51], the activity of which is highly regulated through transcriptional controls, covalent protein modification, and elaborate cell biological mechanisms that control the amount, activity, and localization of these proteins [52].
The ability of flavonols to reduce polar auxin transport has been demonstrated through genetic approaches using Arabidopsis tt mutants [53]. Auxin transport is elevated in inflorescences, hypocotyls, and roots of Arabidopsis plants with tt4 mutations [16,54–56]. In contrast. tt7 and tt3 mutants, which have elevated kaempferol or elevated kaempferol and quercetin, respectively, show reduced polar auxin transport [16,55,57]. The increased polar auxin transport in the tt4 mutant can be reversed by chemical complementation with naringenin, which is a downstream pathway intermediate [55]. Flavonols also regulate auxin distribution by redirecting PIN-mediated auxin efflux during root gravitropic response [58,59], to facilitate asymmetries in auxin at the root tip needed for root gravitropism. A recent report suggests that the flavonol quercetin competes with the key auxin efflux inhibitor N-1-naphthylphthalamic (NPA) acid by directly interacting with PIN protein [60].
Additional evidence suggests that flavonols modulate auxin transport mediated by ABCB transporters [61]. When ABCB proteins were expressed in a heterologous system, transport of auxin across the membrane was reduced by quercetin [62–64]. Additionally, the effect of tt4 on root gravitropism is lost in an abcb4 double mutant, consistent with flavonoids targeting ABCB proteins in vivo [65]. Quercetin, and to a lesser extent kaempferol, disrupts the protein complex between ABCB1 and TWISTED DWARF1 (TWD1), an immunophilin that regulates ABCB1 transport activity [66], suggesting that flavonols may inhibit auxin transport by altering protein complexes needed for maximal auxin transport. The resulting differences in auxin transport in these flavonol-deficient mutants have been tied to changes in growth and development, as described below.
Flavonols regulate root development
The role of flavonols in controlling root development has been demonstrated using genetic approaches. Arabidopsis has been utilized in a majority of these studies due to the genetic simplicity of its flavonol biosynthetic pathway, where single genes encoding pathway enzymes has allowed identification of mutant phenotypes in single mutant lines [67]. Developmental roles of flavonols are best characterized using tt4 mutants with defects in the gene encoding the first enzyme in the flavonol pathway, chalcone synthase (CHS) [68]. Multiple tt4 alleles have been examined with reported root phenotypes that include increased numbers of root hairs [69] and lateral roots [40,55,70], as well as impaired root gravitropism [16,24,56]. However, not all of these mutant alleles are equivalently well characterized, with the genetic and biochemical activities of the point mutant alleles in the Ler background being less well characterized than the Col-0 alleles [68], so this review will focus on insight from the Col-0 alleles. The root developmental phenotypes in tt4 are similar in fls mutants, which catalyze the final step in flavonol synthesis, consistent with reduced flavonol levels controlling mutant phenotypes [40,69]. Additional studies have examined the developmental function of specific flavonols by using mutants that are impaired in branch point enzymes, revealing distinct functions of different flavonols in aspects of root development [16,40,69]. In roots, flavonols modulate development via antioxidant activity [71] or as modulators of auxin transport [53].
In Arabidopsis, flavonols are negative regulators of root hair formation [69]. In both tt4 and fls mutants, there are significant increases in the number of root hairs that form from trichoblast (root hair forming) cells near the root tip [69]. The effect of the tt4 mutation can be reversed by genetic complementation with a CHS-GFP transgene and chemical complementation with naringenin, a metabolite downstream of CHS in the flavonol pathway, as shown in Figure 2. An examination of the root hair phenotype of mutants in genes encoding branchpoint enzymes revealed that quercetin is the active flavonol controlling this process, as the tt7 mutant, defective in F3’H, the enzyme that produces dihydroquercetin, has an identical phenotype to tt4 (Figure 2). LC-MS was used to demonstrate that this mutant has elevated kaempferol and no detectable quercetin [69].
Figure 2. The flavonol biosynthetic machinery is localized to the root epidermis to regulate root hair initiation.
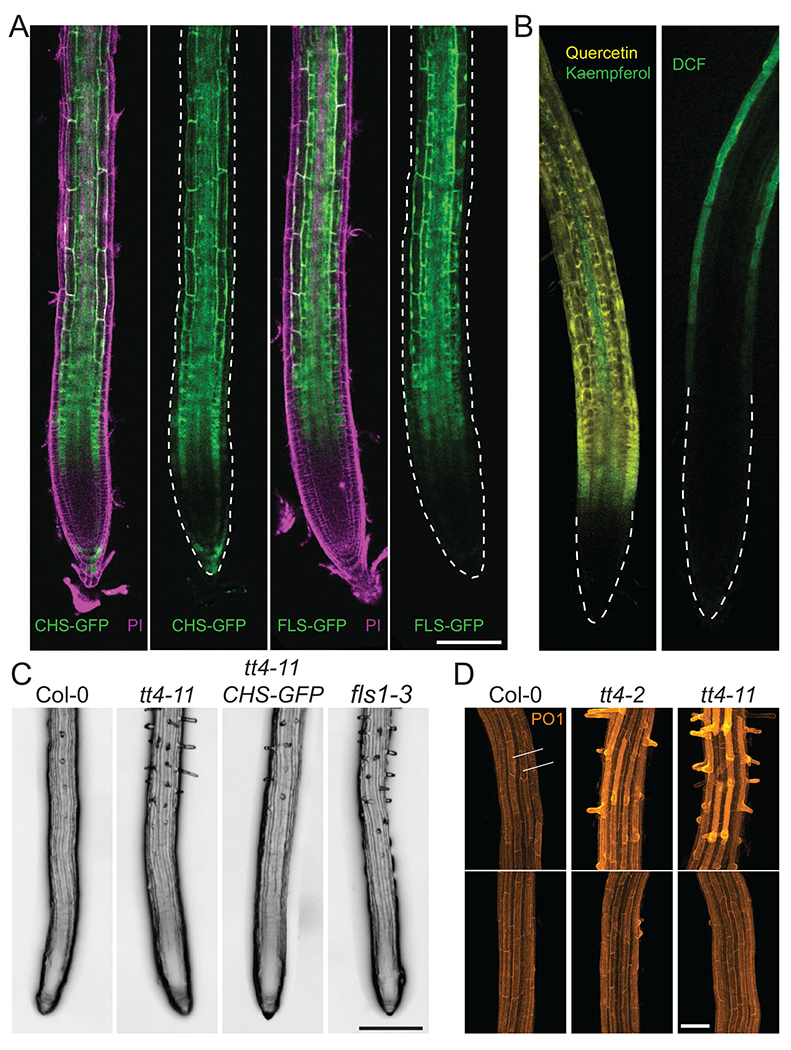
(a) Fluorescence of transgenic Arabidopsis lines containing CHS-GFP and FLS1-GFP reporters (green) stained with the cell wall probe propidium iodide (PI) (magenta) show that flavonol biosynthetic enzymes localize to the root epidermis. Images without PI channel have epidermal tissues outlined by dashed lines. Scale bar =100 μm (b) LSCM images of WT Arabidopsis roots treated with either the flavonol-selective probe, DPBA, with yellow fluorescence of quercetin and green fluorescence of kaempferol (left image), or the fluorescence of the general ROS sensor, dichlorofluorescein (DCF) (green, right image), reveals ROS accumulate in higher levels in root epidermal tissues where flavonols are less abundant. (c) Representative images of root hair number in WT Arabidopsis and mutants with impaired flavonol accumulation, tt4-ll and fls1-3, and tt4-11 complemented with a CHS-GFP transgene (ttd-11 CHS-GFP) show increased root hair numbers in mutants with defects in flavonol synthesis. Scale bar = 200 μm (d) Confocal images of WT, tt4-2, and tt4-11 Arabidopsis lines stained with the H2O2 probe, Peroxy orange 1, display elevated H2O2 accumulation in root hair forming cells (denoted as 1, 3, and 5) relative to nonhair cells (2 and 4). H2O2 accumulation is increased in tt4-2 and tt4-11, though this phenotype can be restored to WT levels through treatment with the flavonol precursor, naringenin. Scale bar = 50 μm. Adapted from Ref. [64].
In root hairs, flavonols function by controlling the levels of reactive oxygen species. Two tt4 alleles have elevated ROS in root hair forming cells, as revealed with a generic ROS sensor and with a hydrogen peroxide selective reporter [69]. ROS levels using these reporters are returned to wild-type levels upon chemical complementation with naringenin [69]. Flavonol synthesis and accumulation is highest in root cortical cells, which are adjacent to the epidermal tissues from which root hairs form, as demonstrated by staining with the flavonol-specific dye, diphenylboric acid 2-aminoethyl ester (DPBA), and localization of FLS-GFP and CHS-GFP reporters. Cortical cells have low levels of ROS, while root hair forming epidermal cells have high levels of ROS and lower levels of flavonols. Together these results suggest a model in which flavonols modulate root hair formation by controlling ROS in Arabidopsis root hairs [69]. Similarly, in the are mutant of tomato, which has a defect in the gene encoding F3H and reduced flavonol levels, there is an increased number of root hairs with elevated ROS in these root hairs, suggesting that this flavonol function may be conserved across species [72].
Lateral root formation is another process that is negatively regulated by flavonols, however studies have demonstrated that kaempferol controls this process, which differs from the active flavonol controlling root hair formation. Flavonols accumulate in lateral root primordia, as judged by DPBA staining and by visualization of GFP fusions to either CHS or FLS [40]. Mutants in either tt4 orfls have increased numbers of lateral roots [40,55,70] when plants are grown under sucrose conditions that enhance flux through the flavonoid pathway [70]. This phenotype is also reversible by genetic and chemical complementation [40]. In contrast to increased lateral root formation in tt4, the tt7 mutant has a reduced number of emerged lateral roots. Since the combined number of lateral root primordia and emerged lateral roots are similar in tt7, this phenotype is due to a defect in lateral root emergence [67]. Using LC-MS, the levels of kaempferol are elevated by ~2-fold in roots of tt7, with kaempferol accumulation increased within the lateral root primordia, as detected with DPBA [40]. This suggests that kaempferol, rather than quercetin, is the active flavonol regulating the emergence of lateral roots. Examination of ROS accumulation in these mutants, revealed reduced superoxide accumulation in the lateral root primordia of tt7 consistent with a role of flavonols in regulating ROS to modulate lateral root formation [40]. These findings, combined with those described above on root hair initiation, provide evidence for the functional specificity of flavonol intermediates controlling different developmental responses.
Earlier experiments examined whether these lateral root phenotypes in flavonol deficient mutants were tied to altered auxin transport. Polar auxin transport from the shoot toward the root tip (rootward transport, which was formerly called acropetal transport) is required for lateral root formation as mutations or auxin transport inhibitor treatments that block this polarity of auxin transport impaired lateral root formation [73]. Rootward auxin transport in tt4 is elevated consistent with the absence of a negative regulator of transport [56].
Several studies have implicated flavonols in controlling root gravitropism via modulation of auxin transport, which is required for gravitropism. Consistent with flavonols negatively regulating auxin transport, the tt4 mutant has increased auxin transport in roots using radioactive auxin transport assays, which measured shootward transport (formerly called basipetal transport) [16,56], which is the polarity of auxin transport that controls root gravitropism [74]. The tt4 mutant fails to establish an asymmetric auxin gradient at the root tip which is needed for root gravitropic curvature [16,56]. Like root hair formation, the tt4 and tt7 mutants have similarly impaired gravitropic responses, consistent with quercetin regulating both responses [16]. As this flavonol-regulated auxin movement occurs through epidermal cells [16], which also give rise to root hairs, these findings suggest two quercetin functions in this cell type. It is important to note the possibility that these functions are linked, as multiple studies have shown that auxin transport is regulated by reactive oxygen and reactive nitrogen species [75,76] suggesting that auxin transport regulation by flavonols may be tied to their antioxidant activity.
Flavonol are involved in above-ground vegetative development and in gas exchange
In addition to their role in root architecture, flavonols have profound effects on leaf development and leaf physiology, including gas exchange through stomata. The absence of flavonols in the tt4-1 (CHS) mutant contributes to a smaller leaf area and slower inflorescence growth rate [77]. On the leaf surface, several striking phenotypes have been tied to altered flavonol biosynthesis, including changes in the morphology of pavement cells and trichomes and the aperture of stomata. Rhamnosylated flavonols were shown to regulate the shape of pavement cells in Arabidopsis, with the flavonol rhamnosyl-deficient rol1 mutant having brick-shaped instead of jigsaw-shaped pavement cells [78]. This mutant also displayed deformed trichomes [78]. While the flavonol biosynthetic mutant af in tomato, which is deficient in CHI1, had normally formed trichomes, it showed a reduced density of glandular trichomes [79]. These trichomes failed to produce the high levels of flavonols found in the wildtype trichomes and had elevated levels of ROS [80]. Additionally, trichomes of the af mutant accumulated reduced levels of terpenoids, which are plant specialized metabolites that mediate the defense against herbivores in leaves. The reduction in terpenoid synthesis was attributed to elevated ROS levels [80]. Due to the lower terpenoid pools, the af mutant was more susceptible to diverse herbivores [79].
The stomatal pores, which are found on the leaf surface, regulate entry of CO2 for photosynthesis and exit of water during transpiration. Flavonols accumulate within the two guard cells, which flank the stomata, as demonstrated by localization with the flavonol dye DPBA in Arabidopsis, tomato, and apple [23,81,82]. Flavonols accumulate in both the cytosol and nuclei of guard cells [23,82], at substantially higher levels than the surrounding pavement cells, as shown in Figure 3. Flavonol accumulation appears to be the result of flavonol synthesis in guard cells as CHS-GUS and FLS-GFP reporters are expressed within this cell type [23,83].
Figure 3. Flavonols accumulate within guard cells to influence stomatal aperture.
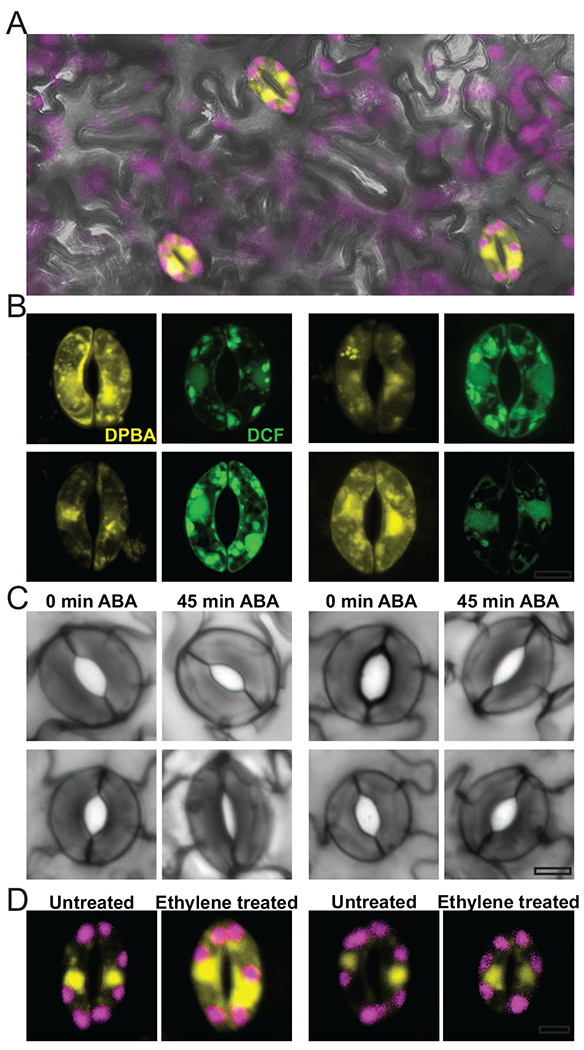
(a) Confocal micrograph of a VF36 tomato leaf stained with the flavonol-selective probe DPBA (yellow) reveals flavonol accumulation in guard cells. Chlorophyll autofluorescence is visualized in magenta, (b) Fluorescence of DPBA (yellow) or the general ROS sensor dichlorofluorescein (DCF) (green) in guard cells of the are and aw tomato mutants and their respective wild-type parental lines, VF36 and AC, reveals an inverse relationship between flavonol levels and general ROS accumulation, (c) Representative images of stomatal aperture of guard cells treated with ABA show that the are (anthocyanin reduced) mutant with decreased flavonols and elevated ROS levels has enhanced rates of stomatal closure, while the anthocyanin without (aw) mutant with increased flavonols and decreased ROS accumulation has reduced rates of stomatal closure, (d) DPBA fluorescence is increased in guard cells following treatment with ethylene gas, while they remain unchanged in the ethylene insensitive Neverripe mutant. Scale bars = 5 μm. (Adapted from Ref. [78]. Copyright 2017 American Society of Plant Biologists.
Stomatal closure is highly regulated by light levels and the hormone ABA, which signals reduced water availability [84]. In response to elevated ABA, localized ROS accumulation in guard cells drives stomatal closure [85,86]. Consistent with the role of flavonols as antioxidants, these molecules function in guard cells to regulate stomatal aperture through the scavenging of ROS [43]. The Arabidopsis CHS mutant allele, tt4-2, and the tomato F3H mutant are (anthocyanin reduced), both have reduced flavonol levels and accumulate higher levels of ROS in guard cells and display an enhanced rate of stomatal closure [23,82]. In contrast, a flavonol overproducing tomato mutant, aw (with a defect in the gene encoding DFR), has a dampened response to ABA-dependent stomatal closure. Elevated flavonol levels appear to confer drought tolerance in Arabidopsis, as shown by a transgenic line overexpressing the transcription factors MYB12 and PAP1 that drive flavonol synthesis [35]. This improved drought response was also seen in a maize mutant containing elevated levels of flavonols [87].
Flavonol synthesis is upregulated in guard cells in several species in response to hormones such as ethylene [23], the growth promoting molecule 5-Aminolevulinic acid (ALA) [88], and the drought hormone abscisic acid (ABA) [89]. Both ethylene and ALA were found to increase flavonol levels, as judged by DPBA staining, and led to more open stomata consistent with flavonols reducing ROS levels. In contrast, an untargeted metabolomic study in guard cell protoplasts from Brassica napus found that ABA increased flavonol metabolites under conditions where ABA induced stomatal closure [89]. Since ABA-induced stomatal closure requires an initial ROS burst [90,91], it would be interesting to evaluate the kinetics of flavonol upregulation in the same species in response to these hormones as ABA might trigger a delayed synthesis of flavonols following the elevated ROS burst after ABA treatment as a protective mechanism to prevent ROS from reaching damaging levels.
Flavonols enhance reproduction
Flavonols are known to play an important role in plant sexual reproduction in multiple species [92]. Successful sexual reproduction requires the development of viable pollen, the male gametophyte, within the locules of anthers. These locules are lined by the tapetum, a cell layer that contributes nutrients and cell wall materials to the developing pollen. During pollination, mature pollen is released from anthers and lands on a stigma where it germinates a pollen tube that navigates the stigma and style to fertilize ovules [93]. Evidence for the role of flavonols throughout the developmental sequence of plant sexual reproduction has come from studies with mutants in multiple species that have defects in pollen development or tube growth. These species include tomato, petunia, tobacco, maize, rice and apple [18,94]. Mutations in CHS block flavonol synthesis, which has enabled researchers to uncover the role of flavonols in reproduction. For example, the white pollen in a flavonol deficient maize mutant is unable to pollinate [95]. In petunia, the white anther (wa) CHS-mutant produces pollen that is unable to germinate. The sterility in this mutant is rescued by exogenous addition of kaempferol and genetic complementation with a CHS transgene [96]. In vitro germination assays showed that addition of kaempferol to flavonol deficient petunia pollen rescues tube growth and germination [97]. The essential role of flavonols in pollen tube formation is further highlighted in rice, as evidenced in the CHS mutant line that shows lower pollen germination rate and an inability to produce functional pollen tube, while maintaining normal pollen morphology [98]. RNAi silencing of the CHS gene in tomato results in the production of fruits with complete absence of seeds [99]. Similarly, RNAi silencing of FLS in tobacco results in reduced pollen germination and pollen tube elongation [100]. However, the role of flavonols in reproduction are not limited to pollen-specific accumulation, as flavonols localized to the pistil have been shown to aid in pollen tube growth and seed set in apples [94]. Intriguingly, there is one species in which flavonols have not been found to control reproduction; Arabidopsis tt mutants do not exhibit reproductive defects [101]. However, a putative flavonol transport Arabidopsis mutant showed reduced pollen viability and seed set [102].
Insight into how flavonols aid in pollen development was revealed through examination of the flavonol-deficient tomato mutant are, which produces fewer viable pollen and has reduced pollen tube germination, elongation, and higher rates of pollen tube rupture [18], as shown in Figure 4. The reduced pollen tube growth and the pollen tube integrity defects of this mutant were rescued with genetic complementation with an F3H transgene [18]. The levels of reactive oxygen species (ROS) in this mutant were elevated, consistent with flavonols acting as antioxidants in pollen [18]. Addition of ascorbic acid and the ROS synthesis inhibitor diphenylene iodinium both reduced ROS and increased pollen tube growth [18], consistent with a role of flavonols in promoting pollen viability by scavenging excess ROS.
Figure 4. Flavonols promote pollen development and tube growth in tomato.
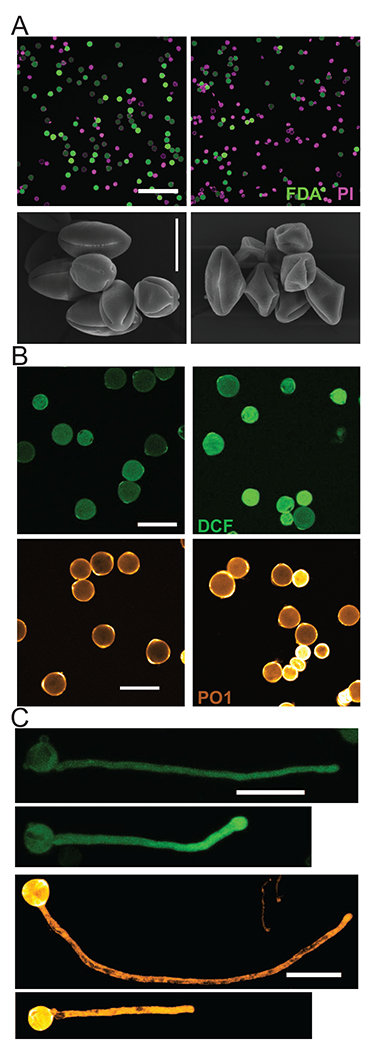
(a) Confocal images of VF36 and flavonol-deficient are mutant pollen grains stained with fluorescein diacetate (FDA, green; live grains) and propidium iodide (PI, magenta; dead grains) and scanning electron micrographs show that the are mutant produces less viable pollen. Scale bars = 200 μm for confocal images and 20 μm for electron micrographs, (b) The general ROS sensor, DCF, and the H2O2-selective chemical probe, Peroxy orange 1 (PO1) reveal increased ROS accumulation in pollen grains of the are mutant. Scale bar = 50 μm (c) Confocal micrographs display significantly impaired pollen tube growth as well as increased levels of ROS in the are mutant. Scale bar = 50 μm (top) and 100 μm (bottom). Adapted from Ref. [18]. Copyright 2018 Proceedings of the National Academy of Sciences.
ROS can increase to damaging levels during diverse abiotic stresses, including heat stress [41], and flavonols may function to prevent this damage [18]. The pronounced reduction in pollen viability and pollen tube growth was linked to overaccumulation of ROS during heat stress [18]. Metabolomic analysis of tomato pollen at different developmental stages revealed that microspores accumulate high levels of flavonols after acute heat stress of 38°C [103], suggesting that flavonol increases may reduce the negative effects of heat stress on pollen viability. Additionally, transcriptomic analysis of a thermotolerant and thermosensitive rice variety during meiosis showed higher expression of flavonol biosynthetic genes in the thermotolerant variety in response to heat stress [104].
Despite the importance of flavonols for pollen development and tube growth, it is not clear yet whether flavonols are produced within pollen or whether these molecules move into pollen from surrounding tissues. Indeed, during pollen development of Brassica, flavonols first accumulate in the tapetum and are then deposited on the pollen coat after tapetal programmed cell death [105,106]. In tomato, DPBA staining has shown that the flavonols accumulate both within pollen grains and on the surface of grains [18]. Transcriptomic studies of developing pollen have shown that the abundance of transcripts encoding some flavonol biosynthetic genes is relatively low [107] which aligns with the observation that at least part of the flavonols accumulating in mature pollen are likely derived from the tapetum [105,106].
Conclusions
Recent evidence has revealed critical functions for flavonols in the regulation of plant growth and development in a variety of tissues across many species. Genetic and chemical approaches have been combined to yield insight into the localization of the synthesis and accumulation of products of the flavonol biosynthetic pathway, providing insight into controls of their synthesis and their functions in specific tissues. Mechanistically, flavonols can influence growth and development through inhibition of auxin transport, regulating distribution of this hormone to modulate development. Additionally, flavonols can act as scavengers of reactive oxygen species to maintain homeostasis to ensure productive signaling while preventing excess levels from accumulating, resulting in cellular damage. The antioxidant capacity of flavonols provides a protective role in plant responses to environmental stresses, including elevated temperature and drought, which are increasing in prevalence as our climate changes. Altogether this evidence provides important insight into flavonol-dependent regulation of plant growth and development, as well as a potential avenue by which to enhance plant tolerance to detrimental environmental stress.
Acknowledgements and funding sources
We apologize to authors of literature we could not cite due to space constraints. We appreciate the insight from Troy Stich on flavonol redox chemistry and the confocal images shared by Sheena Gayomba and Emily Martin. This project was supported by the National Science Foundation (IOS-1558046 and MCB-1716279 to GKM), USDA-NIFA (2020-67013-30907 to GKM and JKM), KU Leuven (STG/21/019 to JKM) and a fellowship from the Wake Forest University Center for Molecular Signaling and an NIH T32 fellowship (GM127261 to A.E.P).
Abbreviations
- CHS
chalcone synthase
- CHI
chalcone isomerase
- FNS
flavone synthase
- F3H
flavanone 3-hydroxylase
- IFS
isoflavone synthase
- DFR
dihydroflavonol 4-reductase
- LAR
leucoanthocyanidin reductase
- ANS/LDOX
anthocyanidin synthase/leucoanthocyanidin dioxygenase
- OMT
O-methyltransferase
- RT
rhamnosyl transferase
- UFGT
UDP-glucose flavonoid 3-O-glucotransferase
- F3′5′H
flavonoid 3′5′-hydroxylase
- F3′H
flavonoid 3′-hydroxylase
- FLS
flavonol synthase
- OMT-1
O-methyltransferase-1
- tt
transparent testa
- ROS
reactive oxygen species
- LAX
like AUX1
- PIN
Pin formed
- ABCB
ATP Binding Cassette B
- NPA
N-1-naphthylphthalamic acid
- TWD1
TWISTED DWARF1
- DPBA
diphenylboric acid 2-aminoethyl ester
- are
anthocyanin reduced
- rol1
repressor of Irx1
- af
anthocyanin free
- aw
anthocyanin without
- ALA
5-Aminolevulinic acid
- ABA
abscisic acid
- wa
white anther
- DCF
dichlorofluorescein
- H2O2
hydrogen peroxide
- PO1
peroxy orange 1
- FDA
fluorescein diacetate
- PI
propidium iodide
- AC
Ailsa Craig
- nr
neverripe
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Annotated Bibliography
- 1.Kumar S, Pandey AK: Chemistry and biological activities of flavonoids: an overview. Scientific World Journal 2013, 2013:162750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gayomba SR, Watkins JM, Muday GK: Flavonols Regulate Plant Growth and Development through Regulation of Auxin Transport and Cellular Redox Status. In Recent Advances in Polyphenol Research. 2017:143–170. [Google Scholar]
- 3.Winkel-Shirley B: Flavonoid Biosynthesis. A Colorful Model for Genetics, Biochemistry, Cell Biology, and Biotechnology. Plant Physiol 2001, 126:485–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barreca D, Trombetta D, Smeriglio A, Mandalari G, Romeo O, Felice MR, Gattuso G, Nabavi SM: Food flavonols: Nutraceuticals with complex health benefits and functionalities. Trends Food Sci Technol 2021, 117:194–204. [Google Scholar]
- 5.Kothari D, Lee W-D, Kim S-K: Allium Flavonols: Health Benefits, Molecular Targets, and Bioavailability. Antioxidants 2020, 9 (9):888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butelli E, Titta L, Giorgio M, Mock H-P, Matros A, Peterek S, Schijlen EGWM, Hall RD, Bovy AG, Luo J, et al. : Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat Biotechnol 2008, 26:1301–1308. [DOI] [PubMed] [Google Scholar]
- 7.Falcone Ferreyra ML, Rius S, Casati P: Flavonoids: biosynthesis, biological functions, and biotechnological applications. Frontiers in Plant Science 2012, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Heim KE, Tagliaferro AR, Bobilya DJ: Flavonoid antioxidants: chemistry, metabolism and structure-activity relationships. J Nntr Biochem 2002, 13:572–584. [DOI] [PubMed] [Google Scholar]
- 9.Zhang H, Wang J, Goodman HM: An Arabidopsis gene encoding a putative 14-3-3-interacting protein, caffeic acid/5-hydroxyferulic acid O-methyltransferasel Biochimica et Biophysica Acta (BBA) - Gene Structure and Expression 1997, 1353:199–202. [DOI] [PubMed] [Google Scholar]
- 10.Koornneef M: Mutations affecting the testa colour in Arabidopsis. Arabidopsis Information Service 1990, 27:1–4. [Google Scholar]
- 11.Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR: The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiology and Biochemistry 2013, 72:21–34. [DOI] [PubMed] [Google Scholar]
- 12.Anguraj Vadivel AK, Krysiak K, Tian G, Dhaubhadel S: Genome-wide identification and localization of chalcone synthase family in soybean (Glycine max [L]Merr). BMC Plant Biol 2018, 18:325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu X, Zhang S, Liu X, Shang J, Zhang A, Zhu Z, Zha D: Chalcone synthase (CHS) family members analysis from eggplant (Solanum melongena L.) in the flavonoid biosynthetic pathway and expression patterns in response to heat stress. PLoS One 2020, 15:e0226537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.*Liou G, Chiang Y-C, Wang Y, Weng J-K: Mechanistic basis for the evolution of chalcone synthase catalytic cysteine reactivity in land plants. J Biol Chem 2018, 293:18601–18612. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article investigates the structure of CHS enzymes in the five major lineages of land plants. It unveils structural differences in CHS between these lineages and suggests that evolutionary changes in the CHS active site might have contributed to the diversification of the flavonoid pathway.
- 15.Saslowsky DE, Warek U, Winkel BSJ: Nuclear Localization of Flavonoid Enzymes in Arabidopsis. Journal of Biological Chemistry 2005, 280:23735–23740. [DOI] [PubMed] [Google Scholar]
- 16.Lewis DR, Ramirez MV, Miller ND, Vallabhaneni P, Ray WK, Helm RF, Winkel BSJ, Muday GK: Auxin and Ethylene Induce Flavonol Accumulation through Distinct Transcriptional Networks. Plant Physiol 2011, 156:144–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian L, Wan S-B, Pan Q-H, Zheng Y-J, Huang W-D: A novel plastid localization of chalcone synthase in developing grape berry. Plant Science 2008, 175:431–436. [Google Scholar]
- 18.*Muhlemann JK, Younts TLB, Muday GK: Flavonols control pollen tube growth and integrity by regulating ROS homeostasis during high-temperature stress. Proceedings of the National Academy of Sciences 2018, 115: E11188–E11197. [DOI] [PMC free article] [PubMed] [Google Scholar]; This article uncovers the mechanism by which flavonols promote pollen development and tube growth, especially during heat stress. It shows that heat stress leads to substantial increases in ROS and that flavonols act as ROS scavengers to protect pollen from the oxidative stress caused by elevated temperatures.
- 19.Li S: Transcriptional control of flavonoid biosynthesis: fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal Behav 2014, 9:e27522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu W, Dubos C, Lepiniec L: Transcriptional control of flavonoid biosynthesis by MYB–bHLH–WDR complexes. Trends Plant Sci 2015, 20:176–185. [DOI] [PubMed] [Google Scholar]
- 21.Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B: Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. The Plant Journal 2007, 50:660–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stracke R, Jahns O, Keck M, Tohge T, Niehaus K, Fernie AR, Weisshaar B: Analysis of PRODUCTION OF FLAVONOL GLYCOSIDES-dependent flavonol glycoside accumulation in Arabidopsis thaliana plants reveals MYB11-, MYB12- and MYB111-independent flavonol glycoside accumulation. New Phytol 2010, 188:985–1000. [DOI] [PubMed] [Google Scholar]
- 23.Watkins JM, Hechler PJ, Muday GK: Ethylene-Induced Flavonol Accumulation in Guard Cells Suppresses Reactive Oxygen Species and Moderates Stomatal Aperture. Plant Physiol 2014, 164:1707–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buer CS, Sukumar P, Muday GK: Ethylene modulates flavonoid accumulation and gravitropic responses in roots of Arabidopsis. Plant Physiol 2006, 140:1384–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grunewald W, de Smet I, Lewis DR, Löfke C, Jansen L, Goeminne G, vanden Bossche R, Karimi M, de Rybel B, Vanholme B, et al. : Transcription factor WRKY23 assists auxin distribution patterns during Arabidopsis root development through local control on flavonol biosynthesis. Proc Natl Acad Sci U S A 2012, 109:1554–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.*Ferreyra MLF, Serra P, Casati P: Recent advances on the roles of flavonoids as plant protective molecules after UV and high light exposure. Physiol Plant 2021, 173:736–749. [DOI] [PubMed] [Google Scholar]; A comprehensive review of the role flavonoids play in the protection of plants from UV exposure and high light irradiation. The authors provide detail on the diversity of flavonoids upregulated in different species, how they are distributed in plants, and mechanisms by which they protect plants during exposure to high light and UV.
- 27.Emiliani J, Grotewold E, Ferreyra MLF, Casati P: Flavonols protect arabidopsis plants against UV-B deleterious effects. Mol Plant 2013, 6:1376–1379. [DOI] [PubMed] [Google Scholar]
- 28.Bieza K, Lois R: An Arabidopsis Mutant Tolerant to Lethal Ultraviolet-B Levels Shows Constitutively Elevated Accumulation of Flavonoids and Other Phenolics. Plant Physiol 2001,126:1105–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu M, Li X, Liu Y, Cao B: Regulation of flavanone 3-hydroxylase gene involved in the flavonoid biosynthesis pathway in response to UV-B radiation and drought stress in the desert plant, Reaumuria soongorica. Plant Physiology and Biochemistry 2013, 73:161–167. [DOI] [PubMed] [Google Scholar]
- 30.Zhao B, Wang L, Pang S, Jia Z, Wang L, Li W, Jin B: UV-B promotes flavonoid synthesis in Ginkgo biloba leaves. Ind Crops Prod 2020, 151:112483. [Google Scholar]
- 31.Bhatia C, Gaddam SR, Pandey A, Trivedi PK: COP1 mediates light-dependent regulation of flavonol biosynthesis through HY5 in Arabidopsis. Plant Science 2021, 303:110760. [DOI] [PubMed] [Google Scholar]
- 32.Cominelli E, Gusmaroli G, Allegra D, Galbiati M, Wade HK, Jenkins GI, Tonelli C: Expression analysis of anthocyanin regulatory genes in response to different light qualities in Arabidopsis thaliana. J Plant Physiol 2008, 165:886–894. [DOI] [PubMed] [Google Scholar]
- 33.Das PK, Geul B, Choi SB, Yoo SD, Park Y il: Photosynthesis-dependent anthocyanin pigmentation in arabidopsis. Plant Signal Behav 2011, 6:23–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.*Laoué J, Fernandez C, Ormeño E: Plant Flavonoids in Mediterranean Species: A Focus on Flavonols as Protective Metabolites under Climate Stress. Plants (Basel) 2022, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]; A comprehensive review that describes the different protective roles flavonols play in response to external stresses associated with climate change such as warming, drought, UV radiation, and salinity. The review also highlights the ROS scavenging capacity of flavonols in relationship to their protective role during the described abiotic stresses.
- 35.Nakabayashi R, Mori T, Saito K: Alternation of flavonoid accumulation under drought stress in Arabidopsis thaliana. Plant Signal Behav 2014, 9:e29518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sánchez-Rodríguez E, Moreno DA, Ferreres F, Rubio-Wilhelmi M del M, Ruiz JM: Differential responses of five cherry tomato varieties to water stress: Changes on phenolic metabolites and related enzymes. Phytochemistry 2011, 72:723–729. [DOI] [PubMed] [Google Scholar]
- 37.Martinez V, Mestre TC, Rubio F, Girones-Vilaplana A, Moreno DA, Mittler R, Rivero RM: Accumulation of Flavonols over Hydroxycinnamic Acids Favors Oxidative Damage Protection under Abiotic Stress. Front Plant Sci 2016, 7:838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu M, Zhu Y, Dong J, Jin H, Sun L, Wang Z, Lu Z, Zhang M, Lu D: Ozone induces flavonol production of Ginkgo biloba cells dependently on nitrate reductase-mediated nitric oxide signaling. Environ Exp Bot 2012, 75:114–119. [Google Scholar]
- 39.Chapman JM, Muhlemann JK, Gayomba SR, Muday GK: RBOH-Dependent ROS Synthesis and ROS Scavenging by Plant Specialized Metabolites To Modulate Plant Development and Stress Responses. Chem Res Toxicol 2019, 32:370–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.**Chapman JM, Muday GK: Flavonols modulate lateral root emergence by scavenging reactive oxygen species in Arabidopsis thaliana. J Biol Chem 2021, 296:100222. [DOI] [PMC free article] [PubMed] [Google Scholar]; This work examined multiple flavonoid biosynthetic pathway mutants to show that mutants that make no flavonols, have increased lateral root emergence. A kaempferol overproducing mutant, tt7, had reduced numbers of lateral roots, as well as increased levels of kaempferol and decreased ROS accumulation in lateral root primordia. This implicates kaempferol as the active flavonol in this process.
- 41.Baxter A, Mittler R, Suzuki N: ROS as key players in plant stress signalling. J Exp Bot 2014, 65:1229–1240. [DOI] [PubMed] [Google Scholar]
- 42.Mittler R, Zandalinas SI, Fichman Y, van Breusegem F: Reactive oxygen species signaling in plant stress responses. Nat Rev Mol Cell Biol 2022, 23:663–679. [DOI] [PubMed] [Google Scholar]
- 43.Agati G, Azzarello E, Pollastri S, Tattini M: Flavonoids as antioxidants in plants: Location and functional significance. Plant Science 2012, 196:67–76. [DOI] [PubMed] [Google Scholar]
- 44.Ghiotto RCT, Lavarda FC, Ferreira FJB: Antioxidant activity of flavonols. Int J Quantum Chem 2004, 97:949–952. [Google Scholar]
- 45.Csepregi K, Hideg É: Phenolic Compound Diversity Explored in the Context of Photo-Oxidative Stress Protection. Phytochemical Analysis 2018, 29:129–136. [DOI] [PubMed] [Google Scholar]
- 46.Pourcel L, Routaboul J-M, Cheynier V, Lepiniec L, Debeaujon I: Flavonoid oxidation in plants: from biochemical properties to physiological functions. Trends Plant Sci 2007, 12:29–36. [DOI] [PubMed] [Google Scholar]
- 47.Naramoto S: Polar transport in plants mediated by membrane transporters: focus on mechanisms of polar auxin transport. Carr Opin Plant Biol 2017, 40:8–14. [DOI] [PubMed] [Google Scholar]
- 48.Geisler M, Aryal B, di Donato M, Hao P: A Critical View on ABC Transporters and Their Interacting Partners in Auxin Transport. Plant Cell Physiol 2017, 58:1601–1614. [DOI] [PubMed] [Google Scholar]
- 49.Swarup R, Bhosale R: Developmental Roles of AUX1/LAX Auxin Influx Carriers in Plants. Front Plant Sci 2019, 10:1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sauer M, Kleine-Vehn J: PIN-FORMED and PIN-LIKES auxin transport facilitators. Development 2019, 146:dev168088. [DOI] [PubMed] [Google Scholar]
- 51.Adamowski M, Friml J: PIN-Dependent Auxin Transport: Action, Regulation, and Evolution. Plant Cell 2015, 27:20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Semeradova H, Montesinos JC, Benkova E: All Roads Lead to Auxin: Post-translational Regulation of Auxin Transport by Multiple Hormonal Pathways. Plant Commun 2020, 1:100048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Peer WA, Murphy AS: Flavonoids and auxin transport: modulators or regulators? Trends Plant Sci 2007, 12:556–563. [DOI] [PubMed] [Google Scholar]
- 54.Murphy A, Peer WA, Taiz L: Regulation of auxin transport by aminopeptidases and endogenous flavonoids. Planta 2000, 211:315–324. [DOI] [PubMed] [Google Scholar]
- 55.Brown DE, Rashotte AM, Murphy AS, Normanly J, Tague BW, Peer WA, Taiz L, Muday GK: Flavonoids Act as Negative Regulators of Auxin Transport in Vivo in Arabidopsis. Plant Physiol 2001, 126:524–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Buer CS, Muday GK: The transparent testa4 mutation prevents flavonoid synthesis and alters auxin transport and the response of Arabidopsis roots to gravity and light. Plant Cell 2004, 16:1191–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peer WA, Bandyopadhyay A, Blakeslee JJ, Makam SN, Chen RJ, Masson PH, Murphy AS: Variation in Expression and Protein Localization of the PIN Family of Auxin Efflux Facilitator Proteins in Flavonoid Mutants with Altered Auxin Transport in Arabidopsis thaliana. Plant Cell 2004, 16:1898–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Santelia D, Henrichs S, Vincenzetti V, Sauer M, Bigler L, Klein M, Bailly A, Lee Y, Friml J, Geisler M, et al. : Flavonoids redirect PIN-mediated polar auxin fluxes during root gravitropic responses. J Biol Chem 2008, 283:31218–31226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kuhn BM, Nodzyński T, Errafi S, Bucher R, Gupta S, Aryal B, Dobrev P, Bigler L, Geisler M, Zažímalová E, et al. : Flavonol-induced changes in PIN2 polarity and auxin transport in the Arabidopsis thaliana roll-2 mutant require phosphatase activity. Sci Rep 2017, 7:41906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teale WD, Pasternak T, Dal Bosco C, Dovzhenko A, Kratzat K, Bildl W, Schwörer M, Falk T, Ruperti B, v Schaefer J, et al. : Flavonol-mediated stabilization of PIN efflux complexes regulates polar auxin transport. EMBO J 2021, 40:el04416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zaziímalová E, Murphy AS, Yang H, Hoyerová K, Hosek P: Auxin transporters--why so many? Cold Spring Harb Perspect Biol 2010, 2:a001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, Bandyopadhyay A, Titapiwatanakun B, Peer WA, Bailly A, Richards EL, et al. : Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGPl. The Plant Journal 2005, 44:179–194. [DOI] [PubMed] [Google Scholar]
- 63.Bouchard R, Bailly A, Blakeslee JJ, Oehring SC, Vincenzetti V, Lee OR, Paponov I, Palme K, Mancuso S, Murphy AS, et al. : Immunophilin-like TWISTED DWARF1 modulates auxin efflux activities of Arabidopsis P-glycoproteins. J Biol Chem 2006, 281:30603–30612. [DOI] [PubMed] [Google Scholar]
- 64.Blakeslee JJ, Bandyopadhyay A, Lee OR, Mravec J, Titapiwatanakun B, Sauer M, Makam SN, Cheng Y, Bouchard R, Adamec J, et al. : Interactions among PIN-FORMED and P-Glycoprotein Auxin Transporters in Arabidopsis. Plant Cell 2007, 19:131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lewis DR, Miller ND, Splitt BL, Wu G, Spalding EP: Separating the Roles of Acropetal and Basipetal Auxin Transport on Gravitropism with Mutations in Two Arabidopsis Multidrug Resistance-Like ABC Transporter Genes. Plant Cell 2007, 19:1838–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bailly A, Sovero V, Vincenzetti V, Santelia D, Bartnik D, Koenig BW, Mancuso S, Martinoia E, Geisler M: Modulation of P-glycoproteins by auxin transport inhibitors is mediated by interaction with immunophilins. J Biol Chem 2008, 283:21817–21826. [DOI] [PubMed] [Google Scholar]
- 67.Saslowsky D, Winkel-Shirley B: Localization of flavonoid enzymes in Arabidopsis roots. The Plant Journal 2001, 27:37–48. [DOI] [PubMed] [Google Scholar]
- 68.Bowerman PA, Ramirez M v, Price MB, Helm RF, Winkel BSJ: Analysis of T-DNA alleles of flavonoid biosynthesis genes in Arabidopsis ecotype Columbia. BMC Res Notes 2012, 5:485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.**Gayomba SR, Muday GK: Flavonols regulate root hair development by modulating accumulation of reactive oxygen species in the root epidermis. Development 2020, 147:dev185819. [DOI] [PubMed] [Google Scholar]; The authors use mutants in genes encoding flavonol biosynthetic enzymes to show that flavonols act as negative regulators of root hair formation. Mutants at multiple steps in the flavonol biosynthetic pathway implicate quercetin as the active flavonol regulating root hair initiation. Flavonols show opposite localization to ROS accumulation and flavonol deficient mutants were shown to have increased accumulation of the H2O2-selective dye peroxy orange 1 in root hair forming cells, consistent with ROS driving root hair initiation.
- 70.Buer CS, Djordjevic MA: Architectural phenotypes in the transparent testa mutants of Arabidopsis thaliana. J Exp Bot 2009, 60:751–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.*Martin RE, Postiglione AE, Muday GK: Reactive oxygen species function as signaling molecules in controlling plant development and hormonal responses. Carr Opin Plant Biol 2022, 69:102293. [DOI] [PMC free article] [PubMed] [Google Scholar]; A detailed review that focuses on the role that reactive oxygen species plays in the modulation of plant hormonal and developmental signaling pathways across numerous tissues. The authors highlight enzymes responsible for localized ROS synthesis, as well as antioxidant systems that keep these molecules balanced. A comprehensive list of ROS detection methods is provided.
- 72.Maloney GS, DiNapoli KT, Muday GK: The anthocyanin reduced tomato mutant demonstrates the role of flavonols in tomato lateral root and root hair development. Plant Physiol 2014, 166:614–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reed RC, Brady SR, Muday GK: Inhibition of Auxin Movement from the Shoot into the Root Inhibits Lateral Root Development in Arabidopsis. Plant Physiol 1998, 118:1369–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Rashotte AM, Brady SR, Reed RC, Ante SJ, Muday GK: Basipetal Auxin Transport Is Required for Gravitropism in Roots of Arabidopsis1. Plant Physiol 2000, 122:481–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parveen N, Kandhol N, Sharma S, Singh VP, Chauhan DK, Ludwig-Müller J, Corpas FJ, Tripathi DK: Auxin Crosstalk with Reactive Oxygen and Nitrogen Species in Plant Development and Abiotic Stress. Plant Cell Physiol 2022, doi: 10.1093/pcp/pcacl38. [DOI] [PubMed] [Google Scholar]
- 76.Fernández-Marcos M, Sanz L, Lewis DR, Muday GK, Lorenzo O: Control of Auxin Transport by Reactive Oxygen and Nitrogen Species BT - Polar Auxin Transport. In Edited by Chen R, Baluška F. Springer Berlin Heidelberg; 2013:103–117. [Google Scholar]
- 77.Buer CS, Kordbacheh F, Truong TT, Hocart CH, Djordjevic MA: Alteration of flavonoid accumulation patterns in transparent testa mutants disturbs auxin transport, gravity responses, and imparts long-term effects on root and shoot architecture. Planta 2013, 238:171–189. [DOI] [PubMed] [Google Scholar]
- 78.Kuhn BM, Errafi S, Bucher R, Dobrev P, Geisler M, Bigler L, Zažímalová E, Ringli C: 7-Rhamnosylated Flavonols Modulate Homeostasis of the Plant Hormone Auxin and Affect Plant Development. J Biol Chem 2016, 291:5385–5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kang J-H, McRoberts J, Shi F, Moreno JE, Jones AD, Howe GA: The Flavonoid Biosynthetic Enzyme Chalcone Isomerase Modulates Terpenoid Production in Glandular Trichomes of Tomato. Plant Physiol 2014, 164:1161–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sugimoto K, Zager JJ, Aubin BS, Lange BM, Howe GA: Flavonoid deficiency disrupts redox homeostasis and terpenoid biosynthesis in glandular trichomes of tomato. Plant Physiol 2022, 188:1450–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.LIU L, XIONG L, AN Y, ZHENG J, WANG L: Flavonols Induced by 5-Aminolevulinic Acid Are Involved in Regulation of Stomatal Opening in Apple Leaves. Hortic Plant J 2:323–330. [Google Scholar]
- 82.**Watkins JM, Chapman JM, Muday GK: Abscisic Acid-Induced Reactive Oxygen Species Are Modulated by Flavonols to Control Stomata Aperture. Plant Physiol 175:1807–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]; Tomato mutants with either impaired flavonol synthesis or that overproduce flavonols were used to show that flavonols modulate ABA-induced stomatal closure. Confocal microscopy was used to show that flavonols accumulate specifically within guard cells. Mutants with decreased flavonol levels display higher levels of ROS, while the overproduction of flavonols decreases the amount of ROS that accumulates in guard cells. Ethylene treatment increased guard cell flavonol levels and reduced rates of ABA-dependent stomatal closure.
- 83.Pandey S, Wang R-S, Wilson L, Li S, Zhao Z, Gookin TE, Assmann SM, Albert R: Boolean modeling of transcriptome data reveals novel modes of heterotrimeric G-protein action. Mol Syst Biol 2010, 6:372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hsu P-K, Dubeaux G, Takahashi Y, Schroeder JI: Signaling mechanisms in abscisic acid-mediated stomatal closure. Plant J 2021, 105:307–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Postiglione AE, Muday GK: The Role of ROS Homeostasis in ABA-Induced Guard Cell Signaling. Frontiers in Plant Science 2020,11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Postiglione AE, Muday GK: Abscisic Acid Increases Hydrogen Peroxide in Mitochondria to Facilitate Stomatal Closure. Plant Physiol 2022, doi: 10.1093/plphys/kiac601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.*Li B, Fan R, Sun G, Sun T, Fan Y, Bai S, Guo S, Huang S, Liu J, Zhang H, et al. : Flavonoids improve drought tolerance of maize seedlings by regulating the homeostasis of reactive oxygen species. Plant Soil 2021, 461:389–405. [Google Scholar]; This work characterized a maize mutant that displays enhanced drought tolerance. This mutant was identified through a leaf temperature screen after drought treatment and displayed upregulation of genes in the flavonol biosynthetic pathway. Guard cells of this mutant accumulated higher levels of flavonols and decreased ROS accumulation, allowing the mutant to maintain photosynthetic rate and stomatal conductance during decreased water availability.
- 88.An Y, Feng X, Liu L, Xiong L, Wang L: ALA-Induced Flavonols Accumulation in Guard Cells Is Involved in Scavenging H202 and Inhibiting Stomatal Closure in Arabidopsis Cotyledons. Frontiers in Plant Science 2016,7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.*Zhu M, Assmann SM: Metabolic Signatures in Response to Abscisic Acid (ABA) Treatment in Brassica napus Guard Cells Revealed by Metabolomics. Sci Rep 2017, 7:12875. [DOI] [PMC free article] [PubMed] [Google Scholar]; A metabolomics approach was used to elucidate changes in metabolic signatures following ABA treatment of Brassica napus Guard Cell protoplasts. Flavonoids were found to be ABA responsive, with quercetin found to be strongly upregulated following ABA treatment.
- 90.Kwak JM, Mori IC, Pei Z-M, Leonhardt N, Torres MA, Dangl JL, Bloom RE, Bodde S, Jones JDG, Schroeder JI: NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J 2003, 22:2623–2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pei ZM, Murata Y, Benning G, Thomine S, Klüsener B, Allen GJ, Grill E, Schroeder JI: Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 2000, 406:731–734. [DOI] [PubMed] [Google Scholar]
- 92.Taylor LP, Grotewold E: Flavonoids as developmental regulators. Curr Opin Plant Biol 2005, 8:317–323. [DOI] [PubMed] [Google Scholar]
- 93.Johnson MA, Harper JF, Palanivelu R: A Fruitful Journey: Pollen Tube Navigation from Germination to Fertilization. Annu Rev Plant Biol 2019, 70:809–837. [DOI] [PubMed] [Google Scholar]
- 94.Chen W, Xiao Z, Wang Y, Wang J, Zhai R, Lin-Wang K, Espley R, Ma F, Li P: Competition between anthocyanin and kaempferol glycosides biosynthesis affects pollen tube growth and seed set of Malus. Hortic Res 2021, 8:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Coe EH, McCormick SM, Modena SA: White pollen in maize. Journal of Heredity 1981, 72:318–320. [Google Scholar]
- 96.Napoli CA, Fahy D, Wang HY, Taylor LP: white anther: A petunia mutant that abolishes pollen flavonol accumulation, induces male sterility, and is complemented by a chalcone synthase transgene. Plant Physiol 1999, 120:615–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Guyon VN, Astwood JD, Gamer EC, Dunker AK, Taylor LP: Isolation and Characterization of cDNAs Expressed in the Early Stages of Flavonol-Induced Pollen Germination in Petunial. Plant Physiol 2000, 123:699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang L, Lam PY, Lui ACW, Zhu F-Y, Chen M-X, Liu H, Zhang J, Lo C: Flavonoids are indispensable for complete male fertility in rice. J Exp Bot 2020, 71:4715–4728. [DOI] [PubMed] [Google Scholar]
- 99.Schijlen EGWM, de Vos CHR, Martens S, Jonker HH, Rosin FM, Molthoff JW, Tikunov YM, Angenent GC, van Tunen AJ, Bovy AG: RNA Interference Silencing of Chalcone Synthase, the First Step in the Flavonoid Biosynthesis Pathway, Leads to Parthenocarpic Tomato Fruits. Plant Physiol 2007, 144:1520–1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Mahajan M, Ahuja PS, Yadav SK: Post-Transcriptional Silencing of Flavonol Synthase mRNA in Tobacco Leads to Fruits with Arrested Seed Set. PLoS One 2011, 6:e28315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Burbulis IE, Iacobucci M, Shirley BW: A null mutation in the first enzyme of flavonoid biosynthesis does not affect male fertility in Arabidopsis. Plant Cell 1996, 8:1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Thompson EP, Wilkins C, Demidchik V, Davies JM, Glover BJ: An Arabidopsis flavonoid transporter is required for anther dehiscence and pollen development. J Exp Bot 2010, 61:439–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Paupière MJ, Müller F, Li H, Rieu I, Tikunov YM, Visser RGF, Bovy AG: Untargeted metabolomic analysis of tomato pollen development and heat stress response. Plant Reprod2017, 30:81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cai Z, He F, Feng X, Liang T, Wang H, Ding S, Tian X: Transcriptomic Analysis Reveals Important Roles of Lignin and Flavonoid Biosynthetic Pathways in Rice Thermotolerance During Reproductive Stage. Front Genet 2020, 11:562937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hsieh K, Huang AHC: Tapetosomes in Brassica Tapetum Accumulate Endoplasmic Reticulum–Derived Flavonoids and Alkanes for Delivery to the Pollen Surface. Plant Cell 2007, 19:582–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li Y, Suen DF, Huang C-Y, Rung S-Y, Huang AHC: The Maize Tapetum Employs Diverse Mechanisms to Synthesize and Store Proteins and Flavonoids and Transfer Them to the Pollen Surface . Plant Physiol 2012, 158:1548–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Loraine AE, Blakley IC, Jagadeesan S, Harper J, Miller G, Firon N: Analysis and visualization of RNA-Seq expression data using RStudio, Bioconductor, and Integrated Genome Browser. Methods Mol Biol 2015, 1284:481–501. [DOI] [PMC free article] [PubMed] [Google Scholar]


