Abstract
Glucose, the most abundant monosaccharide in nature, is the principal carbon and energy source for nearly all cells. The first, and rate-limiting, step of glucose metabolism is its transport across the plasma membrane. In cells of many organisms glucose ensures its own efficient metabolism by serving as an environmental stimulus that regulates the quantity, types, and activity of glucose transporters, both at the transcriptional and posttranslational levels. This is most apparent in the baker’s yeast Saccharomyces cerevisiae, which has 20 genes encoding known or likely glucose transporters, each of which is known or likely to have a different affinity for glucose. The expression and function of most of these HXT genes is regulated by different levels of glucose. This review focuses on the mechanisms S. cerevisiae and a few other fungal species utilize for sensing the level of glucose and transmitting this information to the nucleus to alter HXT gene expression. One mechanism represses transcription of some HXT genes when glucose levels are high and works through the Mig1 transcriptional repressor, whose function is regulated by the Snf1-Snf4 protein kinase and Reg1-Glc7 protein phosphatase. Another pathway induces HXT expression in response to glucose and employs the Rgt1 transcriptional repressor, a ubiquitin ligase protein complex (SCFGrr1) that regulates Rgt1 function, and two glucose sensors in the membrane (Snf3 and Rgt2) that bind glucose and generate the intracellular signal to which Rgt1 responds. These two regulatory pathways collaborate with other, less well-understood, pathways to ensure that yeast cells express the glucose transporters best suited for the amount of glucose available.
Glucose is the preferred carbon and energy source for most cells. In addition to being a major nutrient, glucose can act as a “growth hormone” to regulate several aspects of cell growth, metabolism, and development. How a eukaryotic cell senses glucose and signals its presence, how this signal affects cellular processes, and how optimal utilization of the sugar is achieved are fundamental, unanswered questions. Defects in glucose sensing, signaling, and metabolism cause the severe and prevalent metabolic disorders in mammals known as diabetes. Thus, it is of major interest to understand these processes.
The first and limiting step of glucose metabolism is its transport across the plasma membrane. Thus, it is not surprising that in many different kinds of cells glucose ensures its own efficient metabolism by serving as an environmental stimulus that regulates the quantity, types, and activity of glucose transporters, both at the transcriptional and post-translational levels. This review focuses on regulation of expression and function of the several hexose transporter (HXT) genes of the baker’s yeast Saccharomyces cerevisiae, which has proven useful for studying the mechanisms of glucose sensing, signaling, and utilization in a eukaryotic cell. It is now apparent that multiple pathways regulate the transcription of several members of the HXT gene family in response to different levels of extracellular glucose. Two members of the glucose transporter gene family (RGT2 and SNF3) seem to encode proteins that function not as transporters but as sensors of extracellular glucose that generate an intracellular signal for glucose-induced transcription of the HXT genes. These mechanisms of transcriptional regulation of the HXT genes by glucose are the major focus of this review. Several recent reviews focus on other aspects of yeast glucose transporters (12, 18, 28, 53, 75, 77).
HEXOSE TRANSPORTER PROTEINS
Function and Transport Kinetics
Saccharomyces cerevisiae has 20 genes that encode proteins similar to glucose (hexose) transporters (HXT1 to HXT17, GAL2, SNF3, and RGT2) (12, 18, 28, 75). These Hxt proteins belong to the major facilitator superfamily (MFS) of transporters (89, 117). S. cerevisiae has the largest number of MFS transporters of any organism. MFS proteins transport their substrates by passive, energy-independent facilitated diffusion, with glucose moving down a concentration gradient (12). Because many prokaryotic and mammalian sugar transporters are MFS members, studies performed with the yeast hexose transporters will be valuable in understanding the structure, function, and regulation of glucose transporters from a wide variety of other organisms.
Two uptake systems were described in S. cerevisiae: a constitutive, low-affinity system (high Km, 15 to 20 mM) and a glucose-repressed, high-affinity system (low Km, 1 to 2 mM) (8, 9, 11, 12, 125). It now seems clear that low-affinity and high-affinity glucose transport represent the sum of several transporters rather than being the result of individual transporters. None of these transporters are essential for growth on glucose, indicating their functional redundancy. The presence of multiple hexose transporters with different affinities for glucose in baker’s yeast is not surprising, given the fact that it grows well on a broad range of glucose concentrations (from a few μM to 2 M). Indeed, the amount of glucose available dictates the expression of the appropriate glucose transporters by closely regulating HXT gene expression.
Because of the large number of functionally redundant HXT genes the organism possesses, it was difficult to isolate mutants of yeast defective in glucose uptake. The isolation of the first yeast mutants defective in glucose transport (13), and the recognition that they are defective in a gene (SNF3) encoding a protein similar to glucose transporters of mammalian cells (25), stimulated further efforts to identify yeast glucose transporters. Molecular insight into glucose transporter function and regulation was forthcoming once the HXT1 to HXT4 hexose transporter genes were isolated (72, 76, 79, 140), but most of these genes were not recognized until the sequence of the yeast genome was completed (2, 75). Of the 20 members of the HXT gene family, only 7 are known to encode functional glucose transporters. A strain lacking these seven HXT genes (HXT1 through HXT7, called the hxt null mutant [hxt1Δ-hxt7Δ]) is unable to grow on glucose, fructose, or mannose and has no glycolytic flux (18, 82, 128). Introduction of any one of the seven HXT genes into the hxt null mutant is sufficient to allow it to grow on glucose. HXT2, HXT6, or HXT7 are sufficient for growth on 0.1% glucose, suggesting that they encode high-affinity transporters. HXT1, HXT3, or HXT4 enable growth only on higher glucose concentrations (more than 1%), suggesting that these genes encode low-affinity glucose transporters (128). GAL2, which encodes a galactose transporter similar to the Hxt proteins (100, 145), is also able to complement the glucose growth defect of the hxt null mutant (82), consistent with the finding that galactose-grown cultures transport glucose with the same affinity as glucose-grown cells (see references cited in reference 28). The fact that the hxt null mutant does not grow on glucose indicates that the remaining 10 HXT genes (HXT8 through HXT17) encode proteins that either are unable to transport glucose or are not expressed under the conditions tested. Indeed, we observed very low level expression of genes HXT8 to HXT17 (see Table 3).
TABLE 3.
Transcriptional regulation of HXT-like genes by glucose
| Gene | Mean β-galactosidase activity (Miller units)
|
|||
|---|---|---|---|---|
| 5% glycerol | 5% glycerol + 0.1% glucose | 4% glucose | Fold regulationa | |
| HXT11 | 0.7 | 0.9 | 0.6 | Not regulated |
| HXT12 | 0.3 | 0.5 | 0.3 | Not regulated |
| HXT10 | 13 | 11 | 0.8 | 16× GR |
| HXT16 | 2.5 | 3 | 0.22 | 10× GR |
| HXT17 | 3.7 | 5.6 | 0.8 | 4× GR |
| HXT5 | 217 | 746 | 68 | 3× GR, 3× LGI |
| HXT8 | 1.1 | 6.4 | 0.3 | 3× GR, 6× LGI |
| HXT13 | 41 | 163 | 4.2 | 10× GR, 4× LGI |
| HXT14 | 1.5 | 6 | 0.6 | 2× GR, 4× LGI |
| HXT15 | 3.9 | 11 | 1 | 4× GR, 3× LGI |
| RAG1 | 1.9 | 11 | 508 | 254× HGI |
| KHT2 | 113 | 440 | 469 | 4× GI |
GI, glucose induced; LGI, low glucose induced; HGI, high glucose induced; GR, glucose repressed.
S. cerevisiae cells express only the glucose transporters appropriate for the amount of extracellular glucose available. This is due to the combined action of different regulatory mechanisms, including transcriptional regulation of various HXT genes in response to extracellular glucose (109, 110, 157), and to inactivation of Hxt proteins under certain conditions (18, 54, 74, 124, 130). Also, modulation of the affinity of certain glucose transporters for glucose and interaction between different transporters may contribute to the ability of yeast cells to adapt to different extracellular glucose concentrations. Based on measurements of the kinetics of glucose transport, it has been proposed that the rate of glucose transport (Vmax) is constant and that only the affinity of the transporter for glucose changes when cells are shifted from a high to low levels of glucose (154). From this it was suggested that expression of the transporters is constitutive and that regulation of glucose transport is mediated by a factor that modifies their affinity for glucose (154). However, as described below, it is clear that transcription of the genes encoding the metabolically relevant glucose transporters (HXT1 to HXT7) is regulated by glucose (12, 18, 82, 109, 136). While the affinity of the different transporters for glucose may be modified posttranslationally, transcriptional regulation of the HXT genes is undoubtedly a major reason why the kinetics of glucose transport varies in response to the amount of glucose available.
Correlation between Transporter Regulation and Function
The way in which the HXT genes are transcriptionally regulated in response to glucose is consistent with their function as low- or high-affinity transporters. HXT1 transcription is induced only by high concentrations of glucose, suggesting that it encodes a low-affinity (high Km) transporter. Indeed, expression of HXT1 in the hxt null mutant restores growth only on high concentrations of glucose (more than 1%) and provides low-affinity glucose transport (Km = 100 mM) (128). HXT3 expression is induced by both low and high levels of glucose, and it confers relatively low-affinity glucose transport (Km = 60 mM) on the hxt null mutant, suggesting that it is a low-affinity glucose transporter. Thus, Hxt1 and Hxt3 are probably responsible for transporting glucose in cells growing on high concentrations of glucose (18, 128). HXT2, HXT6, and HXT7 expression, on the other hand, is induced only by low concentrations of glucose, suggesting that they encode high-affinity transporters. Indeed, HXT2, HXT6, and HXT7 enable the hxt null mutant to grow on low glucose concentrations (around 0.1%). HXT6 and HXT7 behave as high-affinity glucose transporters (Km = 1 to 2 mM) when expressed in the hxt null mutant, while HXT2 confers on this mutant biphasic uptake kinetics (Km = 1.5 mM and Km = 60 mM) (128). Perhaps the affinity of the Hxt2 transporter is modulated in response to different glucose concentrations, or perhaps Hxt2 has a regulatory role in the hxt null mutant and activates the expression of other transporters with low and high affinities for glucose. In any case, it is likely that Hxt2, Hxt6, and Hxt7 are responsible for transporting glucose when it is scarce. It is unclear why yeast cells need several transporters with the same affinity.
The transcriptional regulation of HXT4, however, is inconsistent with its predicted function. Its induction only by low levels of glucose suggests that it encodes a high-affinity transporter, but HXT4 does not restore growth of the hxt null mutant on low concentrations of glucose (5 mM = 0.09%), and encodes a protein with intermediate affinity for glucose (Km = 9 mM) (128). This inconsistency could be explained if transcriptional regulation of the HXT genes was altered in the hxt null mutant. It is known that the hxt null mutant, which is completely impaired for glucose uptake, is defective in glucose repression of SUC2 (128). It is possible that glucose repression of HXT4 is also abolished in the hxt null mutant. Alternatively, HXT4 could have a role in regulating expression of the HXT genes remaining in this mutant, so the glucose transport properties of the hxt null mutant expressing HXT4 could be due to other Hxt proteins.
Since most of the data on the kinetics of glucose transport mediated by the individual Hxt proteins was obtained by their expression in the hxt null mutant, the results do not necessarily reflect the in vivo function of these Hxt transporters. If the affinity of some of the Hxt proteins is modulated by their interaction with other transporters, a single Hxt protein might behave differently in this mutant than in a wild-type strain. In addition, some of the missing HXT genes could be necessary for regulation of other HXT genes. Thus, vigilance is required in interpreting the results obtained from expression of individual hexose transporters in this strain.
TRANSCRIPTIONAL REGULATION OF HXT GENE EXPRESSION BY GLUCOSE
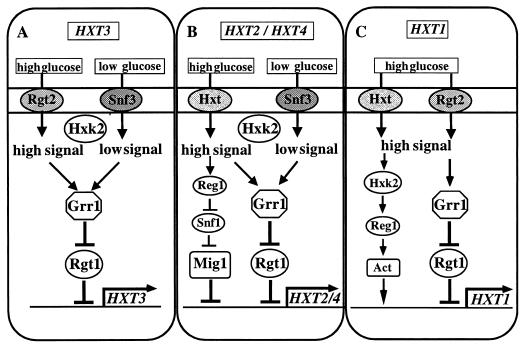
Initial studies suggested that expression of some HXT genes is regulated by glucose (12, 113, 115, 156, 157). Indeed, transcription of HXT1 through HXT4 is between 10- and 300-fold induced by glucose, depending on the gene (109) (Table 1). These four HXT genes exhibit three different types of regulation by glucose: (i) induction by glucose independent of sugar concentration (HXT3), (ii) induction only by low levels of glucose (HXT2 and HXT4), and (iii) induction only by high concentrations of glucose (HXT1).
TABLE 1.
Regulation of HXT1 to HXT4 gene expression by different levels of glucosea
| Relevant genotype | Mean β-galactosidase activity (Miller units)
|
||
|---|---|---|---|
| 2% galactose | 2% galactose + 0.1% glucose | 4% glucose | |
| HXT1::lacZ | 0.6 | 1.6 | 254 |
| HXT2::lacZ | 21 | 145 | 32 |
| HXT3::lacZ | 18 | 116 | 210 |
| HXT4::lacZ | 19 | 163 | 8 |
Data are from reference 109.
These three responses to glucose are due to the action of three overlapping regulatory pathways (Fig. 1). First, glucose induction of all four genes is due to a repression mechanism mediated by the Rgt1 repressor, which inhibits expression of the HXT genes in the absence of glucose. Both low and high concentrations of glucose induce HXT transcription by inhibiting Rgt1 repressor function. The Grr1 protein is required for glucose inhibition of Rgt1 function. The intracellular glucose signal responsible for Grr1-mediated inhibition of Rgt1 repressor function seems to be generated by Snf3 and Rgt2, two glucose transporter-like proteins that serve as glucose sensors for low and high concentrations of glucose, respectively. Second, HXT2 and HXT4 are subject to glucose repression, mediated by the Mig1 repressor, which acts upon many other glucose-repressed genes, and the Snf1 protein kinase, which regulates Mig1 function. Superimposition of this regulatory pathway upon the Rgt1-mediated pathway at the HXT2 and HXT4 promoters results in these genes being induced only by low concentrations of glucose. Third, maximal HXT1 expression requires a high glucose-induced mechanism whose components have not yet been identified. Coupling of this regulatory pathway with the Rgt1-mediated mechanism causes HXT1 to be expressed only in cells growing on high levels of glucose. In the next sections we describe in detail these regulatory mechanisms and how they lead to the three different responses to glucose.
FIG. 1.
Three different modes of induction of HXT gene transcription by different levels of glucose. An arrow implies positive regulation; a line with a bar denotes negative regulation.
Expression of genes encoding glycolytic enzymes and ribosomal proteins are also induced by glucose. However, this involves pathways that are different from the pathway responsible for glucose induction of the HXT genes: different regulatory proteins act upon these two types of genes, and glycolytic metabolites seem to be responsible for glucose-induced transcription of the glycolytic genes (16, 47, 98), while glucose metabolism is not required for the induction of the HXT genes by glucose (114).
HXT3: Induction of Transcription by Glucose Independent of Sugar Concentration
Mutations in HXT3 were identified as suppressors of the potassium uptake defect caused by mutation of the genes encoding potassium transporters (TRK1 and TRK2) (presumably due to symport of potassium ions and glucose) (72). HXT3 also suppresses the growth defect of snf3 mutants on raffinose when overexpressed (12, 72). Expression of HXT3 is induced about 10-fold by both low and high concentrations of glucose (Fig. 1A) (109). This is because only the Rgt1-mediated regulatory pathway seems to act upon HXT3: deletion of RGT1 causes constitutive (glucose-independent) expression of HXT3. Grr1 is required for glucose induction of HXT3, and thus plays a positive role in HXT3 induction, by inactivating the Rgt1 repressor. The modest (about threefold) increase in HXT3 expression caused by inactivation of various genes involved in glucose repression (such as mig1, ssn6, and tup1) suggests that glucose repression plays a minor role in regulating HXT3 expression (109). The HXT3 promoter contains several potential binding sites for Mig1, suggesting that this modest repression may be mediated directly by Mig1.
It has been reported that HXT3 expression is maximal when cells enter the stationary-growth phase (72). While it is an interesting possibility that a stationary-phase-specific regulatory mechanism may act upon HXT3, our current knowledge about glucose induction of HXT3 expression leads us to believe that this is due to repression of HXT3 expression by the initial high levels of glucose (4%) in the medium into which the cells were inoculated. As the cells consume the glucose, the extracellular concentration decreases and reaches an optimal concentration where HXT3 transcription is maximally induced.
HXT2 and HXT4: Induction of Transcription by Low Levels of Glucose
HXT2 and HXT4 were cloned as multicopy suppressors of the high-affinity glucose uptake defect of a snf3Δ mutant (12, 76, 140). HXT4 was also obtained in a screen for S. cerevisiae genes that can complement the low-affinity glucose uptake defect of the Kluyveromyces lactis rag1 mutant (121). HXT4 is the only HXT gene that can complement the galactose growth defect of a gal2 mutant (121), suggesting that Hxt4 has relaxed substrate specificity.
HXT2 and HXT4 are expressed at a low level in cells growing either in the absence of glucose or on high glucose concentrations. Their expression is induced approximately 10-fold by low levels of glucose or fructose (0.1% or 5.6 mM), and by raffinose (a trisaccharide, consisting of fructose-glucose-galactose, that is equivalent to low glucose, because most laboratory strains of S. cerevisiae can only inefficiently cleave the fructose-glucose bond [catalyzed by invertase] and thus obtain only low levels of fructose from it) (Fig. 1B). The induction of HXT2 transcription by low levels of glucose correlates with a 20-fold increase in protein levels, indicating that the transcriptional regulation is indeed reflected in the amount of protein produced (156). This regulation seems physiologically relevant, because it causes these high-affinity glucose transporters to be made only when they are most useful to the cell (i.e., when glucose is scarce).
This pattern of regulation is due to the action of two independent repression mechanisms (Fig. 1B). In the absence of glucose Rgt1 binds to the HXT2 and HXT4 promoters and represses their expression. At high concentrations of glucose, their expression is repressed by the Mig1 repressor (99, 101, 109, 110), which is responsible for repression of many glucose-repressed genes, such as SUC2 and GAL (45, 63, 133, 144). Only at low concentrations of glucose (∼0.05 to ∼0.4%) are both repressor proteins inactive, resulting in expression of HXT2 and HXT4. Deletion of RGT1 causes HXT2 and HXT4 to be expressed in the absence of glucose but has no effect on Mig1-mediated repression of these genes at high glucose concentrations. Conversely, deletion of MIG1 causes expression of HXT2 and HXT4 to become inducible by high levels of glucose but has no effect on Rgt1-mediated repression in the absence of glucose. Thus, the combination of the two different repression mechanisms results in an 8- to 10-fold induction of HXT2 and HXT4 expression only by low levels of glucose (0.1%). The regulation of HXT2 and HXT4 provides a simple example of how multiple regulatory proteins that respond differently to environmental signals can combine to provide a specific and unique pattern of gene expression.
Repression by Rgt1 and Mig1 requires Ssn6 and Tup1 (109), which form a complex that functions as a general repressor of gene expression (69, 144, 159). As shown in Table 2, expression of HXT2 and HXT4 is constitutive (carbon source independent) in an ssn6 mutant, because this mutation relieves repression by both Rgt1 and Mig1. Ssn6 and Tup1 are not DNA binding proteins but are recruited to diverse promoters by several different DNA binding proteins, including Rgt1 and Mig1 (69, 142, 148, 149, 159). Ssn6 and Tup1 may repress transcription by organizing repressive regions of chromatin and/or by directly interacting with the transcriptional machinery (32, 55, 69, 126).
TABLE 2.
Expression of HXT1 and HXT2 in rgt1, mig1, snf1, and ssn6 mutantsa
| Relevant genotype | Mean β-galactosidase activity (Miller units)
|
|||||
|---|---|---|---|---|---|---|
|
HXT1
|
HXT2
|
|||||
| Gal | Gal + Glu | Glu | Gal | Gal + Glu | Glu | |
| Wild type | 0.6 | 1.6 | 254 | 21 | 195 | 32 |
| rgt1 | 14 | 14 | 42 | 191 | 334 | 20 |
| mig1 | 0.3 | 2.3 | 221 | 23 | 235 | 82 |
| snf1 | ND | ND | 195 | ND | 18 | ND |
| ssn6 | 221 | 214 | 461 | 281 | 385 | 488 |
Data are from reference 109. Gal, galactose; Glu, glucose; ND, not determined.
Glucose regulates the nuclear localization of the Mig1 repressor: in the absence of glucose it is in the cytoplasm; high concentrations of glucose cause it to move into the nucleus (36). Mig1 function is regulated by Snf1 (also known as Cat1 or Ccr1), a serine-threonine protein kinase that is required for derepression of glucose-regulated genes (22–24, 27, 48, 135). In cells growing on low levels of glucose, Snf1 is active and inhibits Mig1 repressor function by causing it to reside in the cytoplasm. High levels of glucose inhibit Snf1 activity (48), causing Mig1 to move into the nucleus and repress transcription. It is likely that Snf1 directly phosphorylates Mig1 (35, 143), though this has not been rigorously tested. Consistent with the role of Mig1 in glucose repression, induction of HXT2 and HXT4 by low levels of glucose is completely abolished in a snf1 mutant, due to constant repression by Mig1p (109) (Table 2). This explains why Snf1 is required for depression of high-affinity glucose transport (8). Snf1 does not function in the glucose induction pathway, because expression of the non-glucose-repressible HXT1 gene is not decreased in snf1 mutants (109).
HXT6 and HXT7: Repression by High Levels of Glucose
HXT6 and HXT7 are regulated similarly and encode nearly identical proteins (differing in only two amino acids). These two genes are arranged in tandem, immediately downstream of HXT3. In some strains HXT6 and HXT7 are fused into one gene in which the promoter is derived from HXT7 and the coding sequence from the HXT6 gene (82). Interestingly, a yeast strain selected for growth under glucose-limited conditions for 450 generations appears to contain more than three copies of this chimeric gene as a result of multiple tandem duplication events (20). Like HXT2 and HXT4, HXT6 expression is repressed by high concentrations of glucose (82). However, unlike HXT2 and HXT4, HXT6 expression is only modestly induced by glucose: it is high in cells growing on nonfermentable carbon sources such as glycerol and ethanol and is only about two- to threefold higher in cells growing on low concentrations of glucose or raffinose. That is, HXT6 (and HXT7) have a higher basal level of expression than HXT1 to HXT4 (18, 82, 136).
Regulation of HXT6 differs from that of HXT2 and HXT4 in another significant way: SNF3 has a negative effect on expression of HXT6, being required for glucose repression of its expression (82), in contrast to its positive regulatory role in HXT2 and HXT4 expression (109). This is curious, because HXT6 is the only gene whose glucose repression is known to require SNF3. Since SNF3 is not required for glucose repression of other genes (e.g., ADH2, SUC2, and GAL1), it is unlikely to encode a component of the glucose repression pathway. Snf3p also has a positive role in HXT6 expression, because it is required for the modest induction of HXT6 expression by raffinose or low glucose (82).
HXT1: Induction of Transcription by High Glucose Concentrations
HXT1 was identified as a multicopy suppressor of the growth defect of snf3 mutants (79). Mutations in HXT1 also suppress the potassium transport defect of the trk1 trk2 double mutant, which lacks potassium transporters (72). HXT1 expression is induced about 300-fold at high concentrations (4% or 222 mM) of glucose (Table 1). Rgt1 represses the expression of HXT1 in the absence of glucose, as it does for HXT2, HXT3, and HXT4. In a strain lacking Rgt1, HXT1 expression is about 20-fold derepressed in the absence of glucose. In addition to inhibiting HXT1 expression in the absence of glucose, Rgt1 activates transcription at high glucose concentrations (112), and this accounts for about fivefold activation of HXT1 transcription by high levels of glucose. Since high levels of glucose still induce HXT1 expression in rgt1 mutants (about 5- to 10-fold, see Table 2), another regulatory mechanism (whose components have not yet been identified but which requires HXK2 and REG1) mediates induction of HXT1 expression by high levels of glucose. Thus, three different pathways, repression by Rgt1 in the absence of glucose, activation by Rgt1 at high levels of glucose, and activation by an unidentified mechanism at high levels of glucose, are responsible for high glucose induction of HXT1 expression.
Recently, HXT1 expression was found to be inducible by hyperosmotic stress (1 M NaCl or 1.5 M sorbitol) (49). This requires HOG1, a MAP kinase required for adaptation to hyperosmotic stress (19). In addition, Sln1, a histidine kinase that inhibits Hog1 kinase activity (88), negatively regulates HXT1 (49). One possibility to explain this phenomenon is that increased glucose uptake is necessary for the synthesis of glycerol, a major osmoprotectant in yeast. Consistent with this idea, expression of two other genes encoding key enzymes of glycerol synthesis (GLK1, encoding glucokinase, and GPD1, encoding NAD+-dependent glycerol-3-phosphate dehydrogenase) is also induced by hyperosmotic stress (49).
Transcriptional Regulation of Other HXT Genes
Only limited information is available on the expression of the remaining HXT genes (HXT5 and HXT8 to HXT17). To test whether expression of these genes is also regulated by glucose, their upstream regulatory regions (up to the ATG) were fused to lacZ and their expression and regulation was monitored by different levels of glucose (Table 3) (108). Since these promoter-lacZ fusions are in multicopy plasmids, it is difficult to extrapolate their true expression levels, but their levels of expression can be compared to those of HXT1-HXT4 promoter-lacZ fusions in the same reporter plasmid. Except for HXT5 and HXT13, these HXT genes are expressed at very low levels, being expressed 30- to 300-fold less than HXT1 and HXT2. These genes are subjected to several different modes of regulation by glucose. Expression of HXT12 is very low and not regulated by glucose, as is expression of HXT11, which is 97% identical to HXT9 (both within their coding and promoter regions). HXT10, HXT16, and HXT17 are repressed by glucose to various degrees (4- to 16-fold). HXT5, HXT8, HXT13, HXT14, and HXT15 are induced three- to sixfold by low levels of glucose and repressed to various degrees by high levels of glucose. Regulation of HXT5 and HXT13 transcription resembles that of HXT2 and HXT4, except that high glucose repression of HXT5 is not as strong (about threefold) and low glucose induction of HXT13 is more modest (about fourfold). Thus, none of these HXT genes exhibit the same regulation as HXT1 to HXT4.
Although these genes were named HXT because of their homology to the members of the HXT gene family, it remains to be determined whether they indeed encode glucose transporters. Some of these genes might be involved in the transport of sugars other than glucose (though there are not many candidates since S. cerevisiae grows on few sugars), and some could have only a regulatory function. One strategy that has been used to test whether these HXT-like genes encode glucose transporters is to express them in a strain that is missing all known genes encoding glucose transporters (hxt1Δ through hxt7Δ) and that is therefore unable to grow on glucose (128). Introduction of any one of the seven missing HXT genes supports growth of this hxt null mutant on glucose. HXT8 appears to function as a glucose transporter since it is able to partially complement the glucose growth defect of the hxt null mutant (but only when overexpressed). Surprisingly, HXT10, which encodes a protein approximately 80% identical to Hxt2, does not complement the hxt null mutant, so it appears unable to transport a significant amount of glucose (18, 128).
A clue to the function of some of these HXT-like genes comes from the different ways some of them were obtained. HXT11 was cloned as a multicopy suppressor of a mutation in RAG1, which encodes a low-affinity glucose transporter of the yeast K. lactis that is required for growth of this yeast at high glucose concentrations (107). This suggests that Hxt11 functions as a glucose transporter.
Hxt9 and Hxt11 may play an important role in pleiotropic drug resistance (107). Pleiotropic drug resistance in yeast is caused by overexpression of ATP-binding cassette (ABC) transporters such as Pdr5, Snq2, and Yor1 and resembles the multidrug-resistance (MDR) phenotype of mammalian cells (5, 33). The MDR phenotype of tumor cells, which is due to increased expression of an ABC transporter, is resistance to a variety of chemotherapeutic agents. Expression of the yeast ABC transporter genes is controlled by two homologous zinc-finger-containing transcription factors, Pdr1 and Pdr3. An in vivo screen designed to identify target proteins of Pdr1 and Pdr3 yielded HXT9 and HXT11. Expression of both genes appears to be regulated by Pdr1 and Pdr3, and Pdr3 was shown to bind directly to the HXT11 promoter (107). Interestingly, deletion of HXT9 and/or HXT11 causes resistance to several drugs such as cycloheximide and sulfometuron methyl, and overexpression of HXT11 (but not of HXT1) increases sensitivity to these drugs. These results suggest that Hxt11 is required to transport drugs into the cell, a function opposite to that of the ABC transporters. This is curious, since expression of HXT11 and PDR5 (which transports drugs out of cells) is activated by the same transcription factors. It has been speculated that these hexose transporters could negatively regulate ABC transporter function, providing a negative feedback regulation. Alternatively, Hxt11 and Hxt9 might be directly involved in the uptake of drugs (107).
The promoter region of HXT13 was obtained in a screen for targets of the transcription factor Hap2, a transcriptional regulator of genes such as that encoding cytochrome c involved in respiration (28). Interestingly, HXT13 appears to be expressed at higher levels than HXT8 to HXT17 and, like all Hap2-regulated genes, is also about 10-fold repressed by high concentrations of glucose. However, no difference in HXT13 mRNA levels was observed in cells grown on ethanol compared to cells grown on 2% glucose (18).
Transcriptional Regulation of GAL2
The galactose permease (Gal2) is more than 60% identical to the Hxt proteins and thus belongs to the Hxt protein family. A strain lacking GAL2 grows poorly on media containing galactose (145). GAL2 is expressed only when galactose is available, because its transcription requires the galactose-activated transcription factor Gal4 (61, 63), and is repressed at high concentrations of glucose (mediated by Mig1) (63, 145). Gal2 is also subject to glucose-induced inactivation (also known as catabolite inactivation). This inactivation process involves the glucose-induced internalization of Gal2 by endocytosis and its subsequent degradation in the vacuole (54). Gal2 (expressed from a constitutive promoter) can also transport glucose with almost the same affinity as galactose (Km = 2 mM) (82, 106, 128). This explains why glucose transport in galactose-grown cells is strongly inhibited by galactose (104) and why 6-deoxy-glucose is transported with much higher affinity by galactose-grown cells than by glucose-grown cells (73).
In contrast to glucose induction of HXT gene expression, which is regulated by a receptor-mediated process (where glucose serves as a ligand for the Snf3 and Rgt2 glucose receptors), stimulation of transcription by galactose requires the presence of intracellular galactose (61, 84). Galactose induction of GAL2 is mediated by the Gal3 protein, which functions as the sensor and transducer of the galactose signal. Gal3 is similar to galactokinase (Gal1) but lacks any detectable galactokinase activity (84). Gal3 seems to bind galactose (in an ATP-dependent fashion) and inhibit Gal80 (a repressor of GAL genes), which is thought to enable Gal4 to activate transcription of the GAL genes (15, 120, 138, 162, 163).
Transcriptional Regulation of SNF3 and RGT2 Genes
Snf3 and Rgt2 are about 70% similar to each other but are less than 30% similar to the other members of the Hxt family (18, 75, 114, 160). As discussed in the next section, Snf3 and Rgt2 appear not to transport glucose but to serve as sensors of extracellular glucose that generate the intracellular signal for induction of HXT1 to HXT4 expression. Both genes are expressed at very low levels: about 100- to 300-fold lower than the HXT1 to HXT4 genes (114). Consistent with its proposed role as a high-affinity glucose sensor, SNF3 transcription is repressed at high concentrations of glucose (91, 103, 109). Rgt2 is proposed to function as a low-affinity glucose sensor, and consistent with this role, its expression is independent of the glucose concentration (114).
COMPONENTS OF THE GLUCOSE INDUCTION SIGNALING PATHWAY
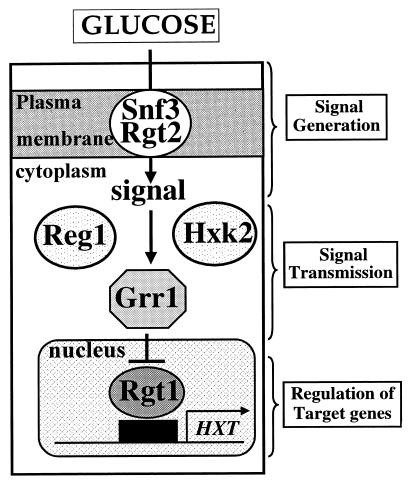
Three key components of the pathway responsible for glucose induction of HXT gene expression have been identified (Fig. 2): (i) the glucose sensors Snf3 and Rgt2, which are plasma membrane proteins that sense the presence of extracellular glucose and generate an intracellular signal for induction of HXT gene expression; (ii) the Rgt1 repressor, a C6 zinc finger DNA binding protein that binds to the promoters of the HXT genes and represses their transcription in the absence of glucose and activates HXT1 transcription when high levels of glucose are present; and (iii) Grr1, which is required for glucose regulation of Rgt1 function and is a component of the SCFGrr1 complex of proteins that have been implicated in protein modification by ubiquitin. In addition, Hxk2 and Reg1, two other proteins known to be involved in glucose signaling, may play a role in glucose induction of the HXT genes.
FIG. 2.
The glucose induction pathway of the HXT genes and its components. The arrow implies positive regulation; a line with a bar denotes negative regulation.
Snf3 and Rgt2: Glucose Sensors
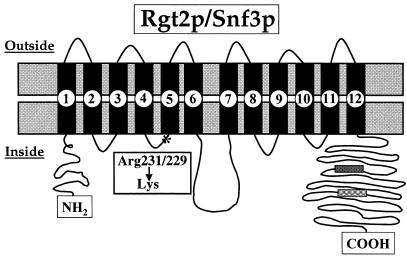
SNF3 and RGT2 encode proteins with 12 predicted transmembrane-spanning domains that are about 60% identical to each other (Fig. 3). They are the most divergent members of the glucose transporter protein family, being only 26 to 30% identical to the other Hxt proteins (25, 91, 114, 160). SNF3 was identified as a mutant that does not grow on sucrose or raffinose (102); RGT2 mutations were obtained as dominant suppressors of the raffinose growth defect of snf3 mutants (92).
FIG. 3.
The predicted transmembrane topology of the Rgt2 and Snf3 glucose transporters in the plasma membrane based on the model for Glut1 (97). The predicted transmembrane domains are numbered 1 to 12. The asterisk shows the position of the Arg-231 (in Rgt2) and Arg-229 (in Snf3) that is mutated to a lysine in the dominant mutants RGT2-1 and SNF3-1, respectively. The boxes indicate the 25-amino-acid repeat in the Snf3 and Rgt2 carboxyl-terminal tail. Snf3 has two copies and Rgt2 has only one copy of this repeat.
Mutants of snf3 were found to be defective in high-affinity glucose transport, which initially led to the conclusion that Snf3 is a high-affinity glucose transporter (13, 25). This was supported by the observation that multiple copies of HXT1 and HXT2 restore the raffinose growth defect of snf3 mutants (76, 79). However, several subsequent pieces of evidence led to the view that Snf3 has a regulatory rather than a metabolic role in glucose transport. First, SNF3 is expressed at a very low level relative to other glucose transporter genes (about 300-fold less than HXT1) (12, 103, 114). Second, SNF3 has a negative effect on the growth of an hxt1-hxt4Δ strain on intermediate levels (0.5%) of glucose, rather than the positive effect that would be expected for a glucose transporter (72). Third, analysis of transport kinetics in a snf3Δ mutant suggested that the decrease in high-affinity glucose uptake is not due to loss of a single transporter (30). Finally, Snf3 was found to be required for induction of transcription of the HXT2, HXT3, and HXT4 genes by low levels of glucose (109) (Table 4). This last observation strongly suggests that snf3 mutants are defective in high-affinity glucose transport because they are unable to express genes encoding high-affinity glucose transporters.
TABLE 4.
Expression of HXT1 and HXT2 in grr1, rgt1, snf3, rgt2, RGT2-1 and snf3 rgt2 mutantsa
| Relevant genotype | Mean β-galactosidase activity (Miller units)
|
|||||
|---|---|---|---|---|---|---|
|
HXT1
|
HXT2
|
|||||
| Gal | Gal + Glu | Glu | Gal | Gal + Glu | Glu | |
| Wild type | 0.6 | 1.6 | 254 | 21 | 195 | 32 |
| grr1 | 0.2 | 0.2 | 0.3 | 20 | 18 | 18 |
| rgt1 | 14 | 14 | 42 | 191 | 334 | 20 |
| snf3 | 0.7 | 1.2 | 227 | 16 | 23 | 7 |
| rgt2 | 0.8 | 4.2 | 62 | 19 | 397 | 34 |
| RGT2-1 | 30 | 223 | 393 | 213 | 415 | 48 |
| snf3 rgt2 | 0.6 | ND | 0.9 | 6 | 14 | ND |
Three key pieces of evidence suggest that Snf3 and Rgt2 are not glucose transporters but glucose receptors that bind glucose outside the cell and generate a signal inside the cell for induction of HXT gene expression (114). First, they are required for glucose induction of expression of several HXT genes. Snf3 is required for induction of HXT2 and HXT4 expression by low levels of glucose but not for induction of HXT1 expression by high levels of glucose (Table 4). This suggests that it functions as a sensor of low levels of glucose. This conclusion is consistent with the observation that transcription of SNF3 is maximal when glucose levels are low (SNF3 expression is repressed about fivefold by high levels of glucose) (114). Rgt2, on the other hand, appears to be a sensor of high levels of glucose, because it is required for maximal induction of HXT1 expression by high concentrations of glucose but not for induction of HXT2 and HXT4 expression by low levels of glucose (Table 4). It is appropriate, then, that RGT2 is expressed in cells growing on high levels of glucose (it is expressed constitutively, being neither repressed nor induced by glucose) (114). Second, Snf3 and Rgt2 are apparently unable to transport glucose. Expression of SNF3 or RGT2 in the hxt null mutant (hxt1-hxt7Δ) does not provide growth of this mutant on glucose, even when they are overexpressed from high-copy-number plasmids (82, 111). Thus, in contrast to the Hxt proteins, Snf3 and Rgt2 appear to have little or no ability to transport glucose.
Perhaps the most compelling observation that supports the view that Snf3 and Rgt2 are glucose sensors comes from the identification of a dominant mutation in these genes (RGT2-1 and SNF3-1) that causes constitutive expression of HXT1 to HXT4 (i.e., in the absence of the inducer glucose) (114) (Table 4). We imagine that this mutation converts Snf3 and Rgt2 into their glucose-bound forms, thereby causing them always to generate the glucose signal that activates HXT expression. This and several other observations (30, 31, 72, 82, 150) led to the proposal that Snf3 and Rgt2 are membrane receptors that bind glucose outside the cell and generate a signal inside the cell for activation of gene expression (111, 114). In this view, glucose signaling by Snf3 and Rgt2 is a receptor-mediated process similar to hormone signaling in mammalian cells.
Snf3 and Rgt2 are composed of two functional domains: the 12 predicted transmembrane domains and a long C-terminal tail that is predicted to reside in the cytoplasm (91) (Fig. 3). The 12 transmembrane domains are similar to those of bona fide glucose transporters and almost certainly form the glucose-binding pocket. The considerable divergence of the transmembrane segments of Snf3 and Rgt2 from the hexose transporters probably accounts for their inability to transport glucose. The amino acid altered in the dominant, constitutive-signaling mutants lies in this region of the protein (SNF3-1, R229K; RGT2-1, R231K) (Fig. 3). This arginine, which is in the cytoplasmic loop between predicted transmembrane segments 4 and 5, is conserved in all other glucose transporters that have been identified, suggesting that it is crucial for binding or translocation of glucose. We believe that changing this residue to lysine converts the receptors into their glucose-bound and therefore glucose-signaling forms.
The C-terminal segments of Snf3 and Rgt2 that are predicted to be in the cytoplasm are unusually long (341 amino acids in Snf3 and 218 in Rgt2). They differ in this respect from all other members of the hexose transporter family (in any organism), none of which have a predicted cytoplasmic C-terminal tail longer than 60 amino acids (except for two potential glucose sensors from Neurospora crassa and K. lactis, described below). The sequences of the Snf3 and Rgt2 cytoplasmic tails are only similar to one another in a stretch of 25 amino acids. Snf3 contains two of these 25-amino-acid sequences; Rgt2 contains only one (Fig. 3). These three sequences are identical at 16 of 25 positions (111).
Two observations indicate that the cytoplasmic tails of Snf3 and Rgt2 are involved in generating the signal for induction of HXT expression upon glucose binding. First, the tails are required for glucose induction of HXT expression: the Snf3 tail is required for low glucose induction of HXT2 and HXT4 expression (31, 111, 150), and the Rgt2 tail is required for high glucose induction of HXT1 expression (111). Second, the cytoplasmic tail of Snf3 is sufficient for glucose-inducible signaling: attaching it to the Hxt1 or Hxt2 glucose transporters creates a chimeric protein that is able to complement the defect in glucose induction of HXT gene expression of snf3 and rgt2 mutants. That is, these modified glucose transporters sense glucose and generate an intracellular signal for induction of HXT expression. Another observation that implicates the C-terminal tail of Snf3 in glucose signaling is that overexpression of just the Snf3 tail suppresses the growth defect of snf3 mutants on low levels of glucose (31) and seems to restore the low glucose induction defect of the snf3 mutant (150). However, it is curious that this suppression does not appear to be accompanied by restoration of glucose transport (31). The 25-amino-acid sequences seem to be the functional regions of the tails, because deletions that remove them but leave the rest of the tail intact abolish signaling (12, 31, 91, 111). Furthermore, the magnitude of signaling by Snf3 correlates with the number of 25-amino-acid elements it possesses in its cytoplasmic tail: two elements (the full complement) lead to normal glucose induction of HXT2 expression, and one element provides for partial induction (111).
The nature of the intracellular signal generated by Rgt2 and Snf3 in response to glucose is not known. Nevertheless, we can state with confidence that the primary signal is neither glucose nor one of its metabolites. The strongest justification for this statement is the observation that the dominant SNF3-1 and RGT2-1 mutants generate the glucose induction signal in the absence of glucose (114). In other words, these mutants do not need to transport glucose into the cell to generate the signal. Further support for this conclusion comes from analysis of a snf3 rgt2 double mutant, which is unable to grow on glucose because of its inability to express any HXT genes. While expression of HXT1 (from the ADH1 promoter) in this mutant corrects its glucose growth defect, it does not restore glucose induction of expression of the other HXT genes, indicating that glucose transport and metabolism are not sufficient for generation of the glucose induction signal (111). Conversely, the hxt1-hxt7Δ null mutant is defective in glucose uptake but exhibits normal glucose induction of HXT1 and HXT2 expression (measured by using fusions of these promoters to lacZ) (108), further supporting the idea that glucose transport and metabolism are not necessary for generation of the glucose induction signal.
How Snf3 and Rgt2 generate the signal for induction of HXT gene expression in response to extracellular glucose remains to be elucidated. We imagine that the binding of glucose to the transmembrane-spanning domain induces a conformational change that is transmitted to the C-terminal signaling domain and affects its interaction with the next component(s) of the signal transduction pathway. Since the domains that are likely responsible for signaling (the 25-amino-acid repeats) are the same in both proteins, we believe both proteins interact with the same or a similar component of the signal transduction pathway. Identifying this component is of utmost importance. A potential candidate is the protein encoded by SKS1 (suppressor kinase of snf3), which was isolated as a multicopy suppressor of the snf3 growth defect on raffinose (161). It is a serine/threonine protein kinase that is homologous to Ran1 of Schizosaccharomyces pombe, which regulates the switch between meiosis and vegetative growth (93). Deletion of SKS1 has no phenotype, but perhaps this is not surprising because it has a homologue (YDR247) in the yeast genome. Since the SKS1 promoter appears to contain Rgt1 binding sites (multiple copies of SKS1 are able to suppress snf3 mutants, presumably by titrating Rgt1 [161]), it is possible that SKS1 itself is regulated by the glucose induction pathway.
We strongly suspect that Snf3 and Rgt2 generate the same signal, because the SNF3-1 and RGT2-1 dominant mutants both exhibit constitutive expression of both the low-glucose-induced genes (HXT2 et al.) and high-glucose-induced genes (HXT1). We believe the different roles of Snf3 and Rgt2 in low- and high-glucose-induced gene expression is simply due to their different affinities for glucose. We imagine that Rgt2 is only involved in induction of HXT1 expression in response to high levels of glucose because it is a low-affinity glucose receptor and that Snf3 is only involved in induction of HXT2 expression by low levels of glucose because it is a high-affinity glucose receptor. Snf3, being a high-affinity receptor, would also be expected to bind high levels of glucose and generate a signal, but this ability is attenuated by the low levels of Snf3 that are present under this condition (due to the approximately fivefold repression of SNF3 expression by high levels of glucose) (12, 91, 109). In fact, Snf3 does seem to contribute to high glucose signaling, because induction of HXT1 expression is only reduced about fivefold in an rgt2 mutant but is completely defective in the snf3 rgt2 double mutant (Table 4). The basal level of Snf3 that is produced when glucose is abundant must be sufficient to provide for some glucose sensing and signaling in the rgt2 mutant under these conditions. The lack of any significant reduction of HXT1 induction by high levels of glucose in a snf3 mutant suggests that Rgt2 is sufficient for full induction of HXT1 expression.
Two other transporters in yeast may play a similar role in nutrient sensing and signaling. Expression of several different amino acid transporters requires Ssy1, which is similar to amino acid permeases but is apparently unable to transport amino acids into the cell (37, 64). Like the glucose sensors, Ssy1 possesses an unusually long cytoplasmic tail but in this case at the N terminus of the protein. Recently it has been shown that Ssy1 is required for transcriptional induction of the AGP1 gene (which encodes a low-affinity, broad-specificity amino acid permease) and of five other genes, in response to multiple amino acids (57). Interestingly, induction of the AGP1 gene by amino acids is abolished in a grr1 mutant, indicating that Grr1 also plays an important role in this amino acid signal transduction pathway (57). From these data it seems likely that Ssy1 is an amino acid receptor that functions as a sensor of external amino acids analogous to the glucose receptors Snf3 and Rgt2. Another example is the high-affinity ammonium ion transporter Mep2, which is required for pseudohyphal differentiation at limiting concentrations of ammonium (85). It has been suggested that Mep2 is a sensor of ammonia that is involved in generating the signal for filamentous growth in response to nitrogen limitation. Mep2 does not appear to possess a long cytoplasmic tail, but a small intracellular loop of Mep2 required for mediating filamentous growth has been identified (85). What is lacking from these two stories is conclusive evidence that these transporter-like proteins act as sensors, like that provided for the glucose sensors by their dominant, constitutive signaling mutations.
Grr1: Inhibitor of the Repressor
Mutants of GRR1 (Glucose Repression Resistant) were originally isolated as resistant to 2-deoxyglucose (4). These mutants were found to be defective in glucose repression of several enzymes, including invertase, maltase, galactokinase, and the mitochondrial enzyme cytochrome c oxidase. GRR1, which is expressed constitutively at very low levels (43, 80), encodes a 132-kDa protein that is present in a large cytoplasmic protein complex. It appears to play several roles in yeast cells, because grr1 mutations have pleiotropic effects, including elongated cell morphology, increased resistance to heavy metals and sulfite, increased sensitivity to osmotic stress and nitrogen starvation, loss of aromatic amino acid transport, decreased glucose uptake, and insensitivity to glucose inactivation of the maltose permease (4, 29, 43, 57, 59, 152).
Mutants of grr1 are unable to induce HXT expression in response to glucose and thus are impaired in glucose uptake (109, 115, 152) (Table 4). This is due to their inability to inactivate Rgt1 in response to glucose, because mutations in RGT1 restore expression of the HXT genes to grr1 mutants (42, 92, 109). Thus, Grr1 is only indirectly involved in glucose repression: the glucose repression defect of grr1 mutants is simply a consequence of their inability to transport significant amounts of glucose (109, 152). Other phenotypes of grr1 mutants, such as their elongated cell morphology, are not suppressed by mutations in RGT1 (6, 42), indicating that Grr1 acts upon other proteins in yeast.
Mutations in grr1 convert the normally glucose-repressed gene SUC2 into a glucose-induced gene (43, 151). Low levels of glucose induce expression of SUC2, and GRR1 is required for this (116). In a grr1 mutant, the signal for induction of SUC2 expression appears to be shifted from low to high concentrations of glucose. Thus, grr1 mutants are defective in growth on raffinose for two reasons: they have reduced levels of invertase due to reduced derepression of SUC2 and reduced ability to transport the fructose that is liberated from raffinose by the action of invertase (152).
Grr1 has two protein interaction domains that are essential for its function: 12 leucine-rich repeats, preceded by an F-box motif. The F-box interacts with Skp1 (71, 80), which is part of several different but related enzyme complexes that direct protein ubiquitination (3, 50, 118, 137). These protein complexes are named SCF for their constituents: Skp1, the Cullin Cdc53 and the ubiquitin-conjugating enzyme Cdc34, and one of several F-box proteins (137). At least three different SCF complexes have been identified in yeast: SCFCdc4, SCFGrr1, and SCFMet30 (118). They seem to differ in the F-box protein they contain, which is thought to be responsible for recruiting substrates to the complex for Cdc34-catalyzed ubiquitination. The F-box-containing protein Cdc4 recruits the cyclin-dependent kinase inhibitor Sic1 to SCFCdc4 (3, 137); Grr1 recruits the G1 cyclins Cln1 and Cln2, and possibly Rgt1, to SCFGrr1 (6, 137). It is thought that the leucine-rich repeats of Grr1 are its substrate-recruiting domain (80).
Because it is part of an SCF complex, it seems likely that Grr1 regulates glucose-induced gene expression via a ubiquitin-directed or related process. The fact that SKP1 and CDC53 are required for glucose-induced HXT1 expression (80) is consistent with this idea. It is possible that SCFGrr1 does not direct modification of proteins with ubiquitin but rather with one of the two ubiquitin-related proteins (Smt3 and Rub1), whose attachment to proteins is catalyzed by conjugating enzymes specific for these proteins (60, 78). One simple view is that SCFGrr1 directs the modification of Rgt1 in response to glucose. However, it is also possible that SCFGrr1 modifies a yet unidentified regulator of Rgt1. How modification of Rgt1 (or its regulator) with ubiquitin (or with Smt3 or Rub1) alters Rgt1 function is not known. Whatever the mechanism, it is unlikely to be by degradation: Rgt1 must be present in cells grown both in the presence and absence of glucose, because it functions as a transcriptional activator in the former and a transcriptional repressor in the latter (112). Moreover, Rgt1 levels are similar in cells grown in glucose or glycerol (81).
The pleiotropic defects associated with grr1 mutants are consistent with the idea that Grr1 recruits diverse targets to the SCFGrr1 complex. Among these are the G1 cyclins Cln1 and Cln2 (137), whose degradation requires Grr1 (6). The abnormal cell morphology of grr1 mutants is probably due to overaccumulation of G1 cyclins (6). Another target of Grr1 is Gic2, an effector of the GTP-binding protein Cdc42, which is involved in actin polarization and bud emergence (58). Thus, Grr1 acts at the intersection of gene expression, cell cycle progression, and nutrient availability and could play a key role in integrating these important processes. Interestingly, the Grr1-Skp1 interaction is enhanced by high concentrations of extracellular glucose (80), which suggests a simple model for how glucose could affect these diverse cellular processes.
Rgt1: Transcriptional Repressor
RGT1 was identified as a gene whose inactivation suppresses the high-affinity glucose transport defect of snf3 mutants (42, 92, 152). Mutants of SNF3 are unable to grow on low levels of glucose because they cannot increase expression of the high-affinity glucose transporters encoded by HXT2 and HXT4 under these conditions. This must be due to failure of snf3 mutants to inactivate Rgt1 in response to glucose, because RGT1 mutations suppress the low-glucose growth defect of snf3 mutants. Multicopy plasmids carrying Rgt1 binding sites also suppress the low-glucose growth defect of snf3 mutants, probably by titrating Rgt1 (113, 140). Mutations in RGT1 also suppress the glucose repression defect of grr1 mutants by restoring HXT gene expression, thereby reinstating transport of glucose and, consequently, generation of the glucose repression signal (42, 151, 152) (Table 4).
Rgt1 is a DNA binding protein with an amino-terminal C6 zinc cluster motif (Cys6 Zn2) like that found in the several members of the Gal4 family of transcription factors (1, 112, 123). The C6 zinc finger of Rgt1 contains the same residues that in Gal4 make base-specific contacts to DNA (Lys17 and Lys18) (90). Interestingly, Rgt1 differs from most other members of the Gal4 family in having a glycine in place of the proline residue of Gal4 (Pro26) that is critical for zinc binding (62). Another significant difference between Rgt1 and the other Gal4 family members is that Rgt1 appears to lack the dimerization domain that follows the zinc finger of nearly all of these proteins (90), suggesting that it binds to DNA as a monomer. This is unlike most of the members of this protein family, which bind as dimers to a CGG palindrome (CGGnCCG, with a different number of bases [n] separating the CGG repeats in the binding site of each protein), each subunit of the dimer recognizing one CGG sequence (1, 127). Hap1 recognizes two CGG sequences as direct repeats (119). By contrast, all four binding sites of Rgt1 that have been identified contain only one CGG sequence, supporting the idea that Rgt1 binds to DNA as a monomer. ArgRII is the only other member of the C6 zinc cluster family that appears to bind to only one CGG sequence (34, 39). The apparent simplicity of the DNA binding sites of Rgt1 and ArgRII makes one wonder how they can achieve site specificity of DNA binding.
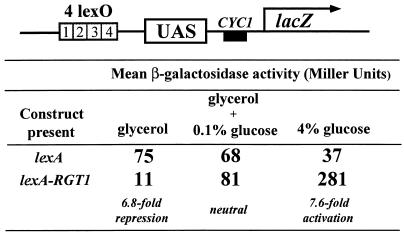
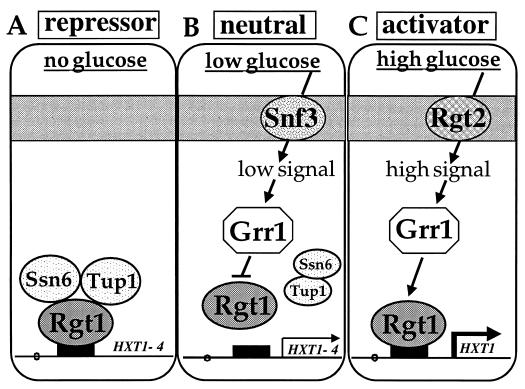
Rgt1 is a bifunctional transcription factor that displays three different transcriptional modes in response to glucose (Fig. 4). It functions as a transcriptional repressor in the absence of glucose, it is a transcriptional activator at high concentrations of glucose, and it is neutral (neither represses nor activates transcription) in cells growing on low levels of glucose (112). It must act directly to effect repression and activation of gene expression, because a lexA-Rgt1 fusion protein has these functions. In the absence of glucose Rgt1 binds to the HXT1-HXT4 promoters and represses their expression (Fig. 4). Rgt1 probably represses transcription by recruiting the general repressors Ssn6 and Tup1, because repression of the HXT genes in the absence of glucose requires SSN6 and TUP1 (109) (Fig. 5). Low levels of glucose inactivate Rgt1 repressor function and thus derepress HXT gene expression. At high concentrations of glucose, Rgt1 is converted into a transcriptional activator that is required for full induction of HXT1 expression (112). As expected, the activator function of Rgt1 is independent of SSN6 and TUP1 (Fig. 5).
FIG. 4.
Rgt1 is a bifunctional transcription factor that represses or activates transcription in response to glucose.
FIG. 5.
Three different modes of transcriptional activity of Rgt1 in response to glucose. In the absence of glucose, Rgt1 works as a transcriptional repressor (A); at low levels of glucose, Rgt1 has no transcriptional activity (B); and at high concentrations of glucose, Rgt1 activates transcription (C).
Glucose alters Rgt1 function through a pathway that requires the Snf3 and Rgt2 glucose sensors and Grr1. Grr1 is required both for conversion of Rgt1 into an activator and for inactivation of Rgt1 repressor function (112) (Fig. 5). Hxk2, which plays a major role in glucose repression of gene expression (45, 63), is also required for maximal induction of HXT1 expression by high levels of glucose, but the ability of Rgt1 to activate transcription at high concentrations of glucose is not significantly reduced in hxk2 mutants (112). Thus, the glucose repression pathway does not seem to be required for conversion of Rgt1 into an activator.
It is not known how glucose regulates Rgt1 function. Since transcription of RGT1 is not regulated in response to glucose (112), the activity of Rgt1 is likely regulated posttranslationally. A lexA-Rgt1 chimera lacking the DNA binding domain of Rgt1 is regulated like intact Rgt1, so it is unlikely that glucose regulates its DNA binding activity. It is more likely that Rgt1 is modified in response to glucose and that this affects either its nuclear localization or its transcriptional repression and activation abilities.
Htr1/Mth1 and Msn3
MTH1 (also known as HTR1) may encode a component of the glucose induction mechanism that regulates HXT gene expression. A dominant mutation in MTH1 (HTR1-23) causes defective transcription of several (probably all) HXT genes (113). Because of this, HTR1-23 mutants have impaired glucose transport and consequently grow poorly on glucose and have defective glucose repression of gene expression (18, 113, 136). These phenotypes closely resemble those of grr1 mutants (43, 115). The dominant DGT1-1 mutation results in phenotypes similar to that of HTR1-23 (44), suggesting that these mutations may be allelic. Because mutations in RGT1, SSN6, and TUP1 suppress the growth defect of HTR1-23 on glucose (18, 136), Mth1/Htr1 may be involved in regulating Rgt1 activity.
MTH1 encodes a protein similar to Msn3 (56) (also known as Std1). It has been suggested that Msn3/Std1 is involved in the glucose repression pathway, because multiple copies of MSN3/STD1 suppress the sucrose growth defect of snf1 mutants (56, 141). However, we suspect that Mth1 and Msn3 are involved only in glucose induction of SUC2 expression, since a strain lacking both MTH1 and MSN3/STD1 is defective in induction of SUC2 by low levels of glucose (56).
The function(s) of Msn3/Std1 and Mth1 are mysterious, though several tantalizing clues have recently been uncovered. Msn3/Std1 is found in two locations in yeast cells: in (or close to) the nucleus and in the plasma membrane (17, 18, 134). The nuclear location of Msn3/Std1 may be a reflection of the fact that it physically interacts with the TATA-box binding protein (TBP) (141), which raises the possibility that it is directly involved in transcription. Interestingly, Msn3 has been found to interact with the C-terminal tails of Rgt2 and Snf3, and Mth1 interacts with the C-terminal tail of Snf3 (134), suggesting that it (and possibly also Mth1/Htr1) could be involved in transducing the glucose signal from the plasma membrane to the nucleus. However, the plasma membrane location of Msn3/Std1 is not dependent on SNF3 and RGT2 (17, 18, 134). Mutations in MTH1 suppress the growth defect of snf3 and snf3 rgt2 mutants, probably by increasing the expression of hexose transporter genes. We imagine that the dominant HTR1-23 mutation in MTH1 interferes with glucose signaling by Snf3 and Rgt2. These findings are consistent with the idea that Msn3/Std1 and Mth1 might play a direct role in glucose signaling, but the precise role they play remains to be determined.
Other Components
Glucose kinases appear to play at least two roles in glucose transport. Yeast possesses three such enzymes: hexokinase 1 (Hxk1), hexokinase 2 (Hxk2), and glucokinase (Glk1) (41, 63). Hxk2 is the main glucose-phosphorylating enzyme in cells growing with abundant glucose; Hxk1 serves cells when glucose is scarce. A triple kinase mutant (hxk1 hxk2 glk1) lacks high-affinity glucose uptake (11), which is restored by introduction of either HXK1 or HXK2 or GLK1 (9). This suggests that glucose phosphorylation is necessary for high-affinity uptake. However, high-affinity uptake of the nonphosphorylable glucose analog 6-deoxyglucose was shown to be also kinase-dependent, suggesting that the hexokinases may have a role in high-affinity glucose uptake that is different from their catalytic function (10). Hxk2 is partially required for full induction of HXT expression by both low and high levels of glucose (109), suggesting that it might be involved in generating or transducing an intracellular glucose signal. It is interesting in this regard that the sole hexokinase of K. lactis (Rag5) is essential for glucose-induced transcription of the RAG1 gene, which encodes a low-affinity glucose transporter (122). The different possible roles for the glucose kinases in glucose transporter function remain to be sorted out.
REG1 is a gene that appears to be involved in both glucose repression and glucose induction of gene expression. It encodes a regulatory subunit of protein phosphatase type 1 (Glc7) (146) that seems to target Glc7 to the Snf1 protein kinase for subsequent dephosphorylation (86). Mutants of REG1 are defective in glucose repression of gene expression (45) and also in RNA processing (147). In addition, REG1 is required for full induction of HXT1 expression by high concentrations of glucose (109). It remains to be established whether Reg1 is a component of the glucose induction pathway or affects HXT1 expression indirectly.
POSTTRANSLATIONAL REGULATION
The function of several sugar transporters is regulated posttranslationally. The best-understood example of this is the glucose-induced degradation of the galactose (Gal2) and maltose (Mal62) transporters (a phenomenon also called catabolite inactivation), which helps to ensure that yeast cells utilize these two sugars only if glucose is unavailable (52, 95, 124, 131, 132). Glucose-induced inactivation of Gal2 appears to be mediated by its ubiquitination, which targets it to the vacuole where it is degraded (54). Degradation of the maltose permease in response to glucose involves two signaling pathways, one dependent on glucose transport, the other one independent. The glucose-transport-independent inactivation pathway works through Rgt2p and Snf3p and requires Grr1p and Rgt1p. The glucose-transport-dependent pathway requires the function of the Hxt proteins and thus the transport and metabolism of glucose (59). Interestingly, it has recently been shown that the glucose-induced degradation of the maltose permease is ubiquitin dependent (94). In contrast to these well-studied examples, little is known about posttranslational regulation of glucose transport. Glucose inactivation of glucose transport (both low- and high-affinity components) was observed in nitrogen-starved cells (21), but the hexose transporters that are subject to glucose-induced inactivation have yet to be identified. Recent data on glucose inactivation of Hxt6 and Hxt7 indicate that these transporters are degraded in the vacuole after internalization by endocytosis. Moreover, the components of the ubiquitin machinery are required for high-glucose-induced degradation of Hxt6 and Hxt7 (74). Finally, Hxt2 transport activity might be regulated by glucose. HXT2 expression (caused by mutation of SSN6) does not lead to increased glucose transport if glucose is not present in the growth medium, suggesting that low levels of glucose are required to activate Hxt2 transporter function (157).
Most of the Hxt proteins contain consensus sites for N-linked glycosylation and phosphorylation by protein kinase A and casein kinase II (12, 18, 53, 75), but it is not known if any of them are so modified. Hxt2 does not appear to be glycosylated, and likely is not phosphorylated (12, 157). Many glucose transporters, including the Hxt proteins, contain a leucine zipper motif, which is located in or near the second putative transmembrane domain. Perhaps this sequence motif, which is known to mediate protein-protein interactions, is involved in oligomerization of the hexose transporters. The mammalian glucose transporter Glut1 exists as dimers and tetramers, but oligomerization is not required for Glut1 transporter function (97).
TRANSPORTER STRUCTURE AND GENE ORGANIZATION
The mammalian glucose transporter Glut1 has 12 transmembrane domains, with its N and C termini located in the cytoplasm (97). Because of their clear similarity to Glut1, it is likely that the yeast hexose transporters have a similar topology. Mutational analysis of glucose transporters has been of limited utility for defining their functional domains, but one promising strategy for this is the construction of hybrids of different transporters. Analysis of chimeras of Hxt2 and the galactose transporter shed light on how Gal2 recognizes galactose (106). Even though glucose and galactose are nearly identical molecules (they are epimers of the C4 carbon), Hxt2 transports only glucose, while Gal2 transports galactose and (with slightly lower efficiency) glucose. Replacement of just transmembrane domains 10 to 12 (101 amino acids) of Hxt2 with those of Gal2 is sufficient to convert it into a galactose transporter (106). Further analysis revealed that the galactose recognition domain of Gal2 resides in transmembrane domain 10, which differs from that of Hxt2 in only 12 of 35 amino acids (67). Two of these (Hxt2 residues Phe431 and Tyr440) appear to be key for substrate binding, because replacing them with the corresponding Gal2 residues (Tyr446 and Trp455) enables Hxt2 to transport galactose (albeit with a lower Km than that of Gal2) (68).
Replacement of Tyr446 in Gal2 with any of the other 19 amino acids completely abolishes galactose transport activity, suggesting that this residue is essential for the transport or recognition of galactose. By contrast, Trp455 is important but not essential for galactose recognition by Gal2 (68). More detailed mutational analysis of the two critical aromatic amino acids of Gal2 (Tyr446 and Trp455) and of Hxt2 (Phe431 and Tyr440) suggests that the presence of an aromatic amino acid in the middle of transmembrane domain 10 is essential for substrate recognition by Gal2 and Hxt2 transporters (66).
Trp455 of Gal2 corresponds to Trp388 of Glut1, which has been shown to be important for the levels, targeting, and forskolin binding of Glut1 (46, 153). A recent analysis of the two aromatic amino acids of Glut1 (Phe379 and Trp388) in transmembrane domain 10 that correspond to Tyr446 and Trp455 of Gal2 revealed a critical role for Trp388 in glucose transport (65). Thus, in contrast to Gal2, the residue at the cytoplasmic end of transmembrane domain 10 of Glut1 (Trp388, which corresponds to Trp455 of Gal2) is essential for transport activity. Another amino acid in transmembrane segment 10 of Glut1 that appears to be particularly important for Glut1 function is Pro385 (139).
The genomic organization of the HXT genes suggests that some of them arose by relatively recent gene duplication. Two clusters of three HXT genes are organized in tandem: HXT4-HXT1-HXT5 are together on chromosome VIII, and HXT3-HXT6-HXT7 are together on chromosome IV (129). All the others are scattered. Some are near telomeres, reminiscent of the organization of genes for sugar utilization (e.g., SUC and MAL).
REGULATION OF GLUCOSE TRANSPORTERS FROM OTHER YEASTS AND FUNGI
Genes encoding sugar transporters have also been identified in other yeasts, such as K. lactis (milk yeast), Schizosaccharomyces pombe (fission yeast), Pichia stipitis (a xylose-fermenting yeast) and the fungus N. crassa. None of these fungal species appear to have as many glucose transporters as S. cerevisiae.
Kluyveromyces lactis
K. lactis, like S. cerevisiae, has two glucose uptake systems: a constitutive one with high affinity for glucose (Km of about 1 mM) and a glucose-inducible one with low affinity for glucose (Km of about 20 to 50 mM) (158). Only three glucose transporter genes have been identified in K. lactis. The RAG1 and KHT2 genes encode low-affinity glucose transporters that are about 75% identical to each other and up to 70% identical to the Hxt proteins of S. cerevisiae. RAG1 is required for growth on high concentrations of glucose and fructose (158). HGT1 encodes a high-affinity glucose transporter that is only about 30% identical to the Hxt proteins (7). HGT1 is required for growth on low concentrations of glucose, and overexpression of HGT1 does not allow rag1 mutants to grow on high concentrations of glucose (7).
Transcription of the RAG1 gene is induced by high concentrations of glucose, fructose, mannose, and raffinose (note that raffinose is equivalent to a high level of glucose for K. lactis, because this organism can cleave it into its constituent monosaccharides, unlike S. cerevisiae, for which this trisaccharide is equivalent to a low level of glucose, because it can only obtain low levels of fructose from it) (26). Curiously, galactose and lactose also induce RAG1 transcription, even though these sugars are not substrates of the Rag1 protein. RAG4, RAG5, and RAG8 are involved in regulating RAG1 transcription. RAG5 encodes the only hexokinase of K. lactis, which is essential for glucose-induced transcription of RAG1 (122). RAG8 encodes a casein kinase I very similar to the two casein kinases I of S. cerevisiae, Yck1 and Yck2. Indeed, overexpressed RAG8 complements the temperature-sensitive growth defect of a yck1 yck2 double mutant (14). RAG4 encodes a low-affinity glucose sensor strikingly similar to Snf3 and Rgt2. It has a C-terminal extension containing the 25-amino-acid element found in the C-terminal tails of Rgt2 and Snf3 that is thought to be involved in glucose signaling. It is likely that Rag4 functions as a low-affinity glucose sensor that generates the high glucose signal required for glucose-induced transcription of the RAG1 gene. Transcription of HGT1 does not seem to be regulated by carbon source, but Rag5 (the hexokinase) appears to have a positive role and Rag4 (the glucose sensor) a negative role in HGT1 expression (7).
Since the transcriptional regulation of RAG1 by high levels of glucose resembles that of HXT1 in S. cerevisiae, we tested whether high levels of glucose in S. cerevisiae could induce RAG1 expression. High levels of glucose induce RAG1 expression more than 200-fold in S. cerevisiae (Table 3). This is due to the Grr1-Rgt1 pathway, because deletion of RGT1 causes a 10- to 20-fold depression when glucose is absent, and induction of RAG1 is abolished in a grr1 mutant (108). These results strongly suggest that the glucose induction pathway is conserved in K. lactis and S. cerevisiae. Consistent with this view, K. lactis appears to have a glucose sensor similar to Snf3 and Rgt2 (Rag4) that likely generates the high glucose signal for induction of RAG1 expression. In addition, KHT2 expression is more than fourfold induced by glucose, independent of the sugar concentration (Table 3), which resembles regulation of HXT3 of S. cerevisiae (though it is not known if this is also mediated by the Grr1-Rgt1 pathway).
Schizosaccharomyces pombe
Glucose uptake mutants of S. pombe were isolated in a screen for mutants that are able to utilize gluconate in the presence of high concentrations of 2-deoxyglucose (51, 96). This screen was based on the fact that 2-deoxyglucose, whose uptake is mediated by a glucose transporter(s), prevents growth on gluconate-containing media, presumably because it represses expression of the gluconate transporter. The mutants obtained grew poorly on glucose and lacked any measurable glucose uptake. The gene affected in these mutants is GHT1, which encodes a protein with over 30% identity to the Hxt proteins. Multiple copies of GHT1 restores the growth defect of the hxt null mutant of S. cerevisiae on 2% glucose, suggesting that it indeed encodes a glucose transporter (83).
Systematic sequencing of the S. pombe genome has revealed three additional genes (GHT2, GHT3, and GHT4), which encode proteins that are 55 to 75% identical to Ght1 and 40 to 45% identical to the Hxt proteins (18). The existence of four different GHT genes suggests that the original mutant of S. pombe defective in glucose transport either carries a mutation in a regulatory gene required for expression of the other GHT genes or that GHT2, GHT3, and GHT4 are not expressed or may not function as glucose transporters.
Pichia stipitis
The yeast P. stipitis is able to ferment xylose (derived from the hydrolysis of hemicelluloses of plants). It has low-affinity and high-affinity proton symport systems for this sugar which appear to be expressed constitutively (38, 70). The low-affinity xylose transport system has been shown to facilitate the uptake of glucose with a Km of 0.3 to 1 mM (18, 70).
Three different genes, SUT1 through SUT3, have been recently identified in the yeast P. stipitis and shown to encode glucose transporters (155). All three transporters are able to restore the growth defect of the hxt null mutant, indicating that Sut1 through Sut3 function as glucose transporters in S. cerevisiae. These transporters are also able to transport xylose and other monosaccharides when expressed in S. cerevisiae but with much lower affinities than for glucose (Km for glucose is in the millimolar range). The Sut2 protein differs from Sut3 only in one amino acid (155). A strain lacking SUT1 is not defective in growth on glucose but appears to have lost the low-affinity glucose uptake system. While transcription of the SUT1 gene is strongly induced by glucose, SUT2 and SUT3 are expressed only under aerobic conditions and independent of carbon source (155).
Neurospora crassa
The glucose transporter Rco3 of the filamentous fungus N. crassa displays a significant homology to the Snf3 and Rgt2 sensors. It is required for glucose repression of gene expression, for conidiation, and for expression of glucose transporter activity (87). Based on these observations, it has been proposed that Rco3, like Snf3 and Rgt2, functions as a nutrient sensor. Interestingly, Rco3 also has a long C-terminal extension of 119 amino acids, but it lacks the conserved 25-amino-acid motif involved in glucose signaling that is present in the Snf3, Rgt2, and Rag4 C-terminal tails.
CONCLUSIONS AND PERSPECTIVES
Glucose induction of its own transport in S. cerevisiae is reminiscent of glucose-induction of glucose transport in mammalian cells. In both cases, glucose increases the number of glucose transporters in the plasma membrane. In yeast, glucose does this directly (by increasing expression of the HXT genes), but in mammalian cells it is accomplished indirectly through the action of the hormone insulin, which stimulates insertion of the Glut4 glucose transporter into the plasma membrane of fat and muscle cells (97). The cells that measure the concentration of glucose in the blood are mainly the insulin-producing β-cells of the pancreas. Glucose increases the amount of insulin these cells secrete in at least two ways: it rapidly stimulates secretion of insulin that is stored in vesicles and, more slowly, increases insulin gene transcription. These events require the glucose transporter Glut2 and glucokinase (105). The signal for stimulation of insulin secretion is the ATP that is generated from glucose metabolism. Because glucokinase catalyzes the rate-limiting step of glucose metabolism in β-cells, it serves as the glucose sensor (40). Thus, it is not clear whether functional analogs of Snf3 and Rgt2 exist in β-cells. It is significant that neither Caenorhabditis elegans nor mammals contain proteins that have the 25-amino-acid repeat found in the C-terminal cytoplasmic tails of Snf3 and Rgt2 proteins. Furthermore, none of the candidate glucose transporters of C. elegans seem to have unusually long C-terminal tails. Thus, metazoans may not possess glucose sensors like those found in yeast.
A major challenge is to determine the function of the Hxt proteins that are not required for growth of yeast on glucose (Hxt8 to Hxt17). Another important goal is to determine the affinities of all the Hxt proteins for glucose and to determine whether they are modified posttranslationally. Several important questions remain regarding the issue of the transcriptional regulation of HXT gene expression by glucose. Paramount among these is how Snf3 and Rgt2 generate the glucose signal and how it is transmitted to the nucleus. How does Rgt1, the apparent ultimate target of the glucose-signaling pathway, display different activities in response to different levels of glucose? How does Grr1 inhibit Rgt1 function? Does Grr1 respond to the glucose signal or is some other component of the regulatory pathway sensitive to the glucose signal? It seems likely that analysis of this pathway will inform the mechanisms by which glucose regulates gene expression in other organisms.
ACKNOWLEDGMENTS
We thank Martin Schmidt and Eckhardt Boles for sharing information prior to publication. We are grateful to all our colleagues who sent reprints and preprints important for this review and who have shared information, ideas, and reagents with us in the past. We also thank the two reviewers of this manuscript for their valuable comments.
Work in our laboratory was supported by NIH grant GM32540 and by funds provided by the James S. McDonnell Foundation.
REFERENCES
- 1.Andre B. The UGA3 gene regulating the GABA catabolic pathway in Saccharomyces cerevisiae codes for a putative zinc-finger protein acting on RNA amount. Mol Gen Genet. 1990;220:269–276. doi: 10.1007/BF00260493. [DOI] [PubMed] [Google Scholar]
- 2.Andre B. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast. 1995;11:1575–1611. doi: 10.1002/yea.320111605. [DOI] [PubMed] [Google Scholar]
- 3.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J W, Elledge S J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86:263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 4.Bailey R B, Woodward A. Isolation and characterization of a pleiotropic glucose repression resistant mutant of Saccharomyces cerevisiae. Mol Gen Genet. 1984;193:507–512. doi: 10.1007/BF00382091. [DOI] [PubMed] [Google Scholar]
- 5.Balzi E, Goffeau A. Yeast multidrug resistance: the PDR network. J Bioenerg Biomembr. 1995;27:71–76. doi: 10.1007/BF02110333. [DOI] [PubMed] [Google Scholar]
- 6.Barral Y, Jentsch S, Mann C. G1 cyclin turnover and nutrient uptake are controlled by a common pathway in yeast. Genes Dev. 1995;9:399–409. doi: 10.1101/gad.9.4.399. [DOI] [PubMed] [Google Scholar]
- 7.Billard P, Menart S, Blaisonneau J, Bolotin-Fukuhara M, Fukuhara H, Wesolowski-Louvel M. Glucose uptake in Kluyveromyces lactis: role of the HGT1 gene in glucose transport. J Bacteriol. 1996;178:5860–5866. doi: 10.1128/jb.178.20.5860-5866.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bisson L F. High-affinity glucose transport in Saccharomyces cerevisiae is under general glucose repression control. J Bacteriol. 1988;170:4838–4845. doi: 10.1128/jb.170.10.4838-4845.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bisson L F, Fraenkel D G. Involvement of kinases in glucose and fructose uptake by Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1983;80:1730–1734. doi: 10.1073/pnas.80.6.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bisson L F, Fraenkel D G. Transport of 6-deoxyglucose in Saccharomyces cerevisiae. J Bacteriol. 1983;155:995–1000. doi: 10.1128/jb.155.3.995-1000.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bisson L F, Fraenkel D G. Expression of kinase-dependent glucose uptake in Saccharomyces cerevisiae. J Bacteriol. 1984;159:1013–1017. doi: 10.1128/jb.159.3.1013-1017.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bisson L F, Coons D M, Kruckeberg A L, Lewis D A. Yeast sugar transporters. Crit Rev Biochem Mol Biol. 1993;28:259–308. doi: 10.3109/10409239309078437. [DOI] [PubMed] [Google Scholar]
- 13.Bisson L F, Neigeborn L, Carlson M, Fraenkel D G. The SNF3 gene is required for high-affinity glucose transport in Saccharomyces cerevisiae. J Bacteriol. 1987;169:1656–1662. doi: 10.1128/jb.169.4.1656-1662.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blaisonneau J, Fukuhara H, Wesolowski-Louvel M. The Kluyveromyces lactis equivalent of casein kinase I is required for transcription of the gene encoding the low-affinity glucose permease. Mol Gen Genet. 1997;253:469–477. doi: 10.1007/s004380050345. [DOI] [PubMed] [Google Scholar]
- 15.Blank T E, Woods M P, Lebo C M, Xin P, Hopper J E. Novel Gal3 proteins showing altered Gal80p binding cause constitutive transcription of Gal4p-activated genes in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:2566–2575. doi: 10.1128/mcb.17.5.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boles E, Zimmermann F K. Induction of pyruvate decarboxylase in glycolysis mutants of Saccharomyces cerevisiae correlates with the concentrations of three-carbon glycolytic metabolites. Arch Microbiol. 1993;160:324–328. doi: 10.1007/BF00292085. [DOI] [PubMed] [Google Scholar]
- 17.Boles, E. Personal communication.
- 18.Boles E, Hollenberg C P. The molecular genetics of hexose transport in yeasts. FEMS Microbiol Rev. 1997;21:85–111. doi: 10.1111/j.1574-6976.1997.tb00346.x. [DOI] [PubMed] [Google Scholar]
- 19.Brewster J L, Valoir T D, Dwyer N D, Winter E, Gustin M C. An osmosensing signal transduction pathway in yeast. Science. 1993;259:1760–1763. doi: 10.1126/science.7681220. [DOI] [PubMed] [Google Scholar]
- 20.Brown C J, Todd K M, Rosenzweig R F. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol Biol Evol. 1998;15:931–942. doi: 10.1093/oxfordjournals.molbev.a026009. [DOI] [PubMed] [Google Scholar]
- 21.Busturia A, Lagunas R. Catabolite inactivation of the glucose transport system in Saccharomyces cerevisiae. J Gen Microbiol. 1986;132:379–385. doi: 10.1099/00221287-132-2-379. [DOI] [PubMed] [Google Scholar]
- 22.Carlson M. Regulation of glucose utilization in yeast. Curr Opin Genet Dev. 1998;8:560–564. doi: 10.1016/s0959-437x(98)80011-7. [DOI] [PubMed] [Google Scholar]
- 23.Celenza J L, Carlson M. Cloning and genetic mapping of SNF1, a gene required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol Cell Biol. 1984;4:49–53. doi: 10.1128/mcb.4.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Celenza J L, Carlson M. A yeast gene that is essential for release from glucose repression encodes a protein kinase. Science. 1986;233:1175–1180. doi: 10.1126/science.3526554. [DOI] [PubMed] [Google Scholar]
- 25.Celenza J L, Marshall-Carlson L, Carlson M. The yeast SNF3 gene encodes a glucose transporter homologous to the mammalian protein. Proc Natl Acad Sci USA. 1988;85:2130–2134. doi: 10.1073/pnas.85.7.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X J, Wesolowski-Louvel M, Fukuhara H. Glucose transport in the yeast Kluyveromyces lactis. II. Transcriptional regulation of the glucose transporter gene RAG1. Mol Gen Genet. 1992;233:97–105. doi: 10.1007/BF00587566. [DOI] [PubMed] [Google Scholar]
- 27.Ciriacy M. Isolation and characterization of yeast mutants defective in intermediary carbon metabolism and in carbon catabolite derepression. Mol Gen Genet. 1977;154:213–220. doi: 10.1007/BF00330840. [DOI] [PubMed] [Google Scholar]
- 28.Ciriacy M, Reifenberger E. Hexose transport. In: Zimmermann F K, Entian K-D, editors. Yeast sugar metabolism: biochemistry, genetics, biotechnology, and applications. Lancaster, Pa: Technomic; 1997. pp. 45–65. [Google Scholar]
- 29.Conklin D S, Kung C, Culbertson M R. The COT2 gene is required for glucose-dependent divalent cation transport in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:2041–2049. doi: 10.1128/mcb.13.4.2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coons D M, Boulton R B, Bisson L F. Computer-assisted nonlinear regression analysis of the multicomponent glucose uptake kinetics of Saccharomyces cerevisiae. J Bacteriol. 1995;177:3251–3258. doi: 10.1128/jb.177.11.3251-3258.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Coons D M, Vagnoli P, Bisson L F. The c-terminal domain of Snf3p is sufficient to complement the growth defect of snf3 null mutations in Saccharomyces cerevisiae: SNF3 functions in glucose recognition. Yeast. 1997;13:9–20. doi: 10.1002/(SICI)1097-0061(199701)13:1<9::AID-YEA51>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 32.Cooper J P, Roth S Y, Simpson R T. The global transcriptional regulators, SSN6 and TUP1, play distinct roles in the establishment of a repressive chromatin structure. Genes Dev. 1994;8:1400–1410. doi: 10.1101/gad.8.12.1400. [DOI] [PubMed] [Google Scholar]
- 33.Decottignies A, Goffeau A. Complete inventory of the yeast ABC proteins. Nat Genet. 1997;15:137–145. doi: 10.1038/ng0297-137. [DOI] [PubMed] [Google Scholar]
- 34.De Rijcke M, Seneca S, Punyammalee B, Glansdorff N, Crabeel M. Characterization of the DNA target site for the yeast ARGR regulatory complex, a sequence able to mediate repression or induction by arginine. Mol Cell Biol. 1992;12:68–81. doi: 10.1128/mcb.12.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DeVit, M. J., and M. Johnston. 1998. Unpublished data.
- 36.DeVit M J, Waddle J A, Johnston M. Regulated nuclear translocation of the Mig1 glucose repressor. Mol Biol Cell. 1997;8:1603–1618. doi: 10.1091/mbc.8.8.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Didion T, Regenberg B, Jorgensen M U, Kielland-Brandt M C, Andersen H A. The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol Microbiol. 1998;27:643–650. doi: 10.1046/j.1365-2958.1998.00714.x. [DOI] [PubMed] [Google Scholar]
- 38.Does A L, Bisson L F. Comparison of glucose uptake kinetics in different yeasts. J Bacteriol. 1989;171:1303–1308. doi: 10.1128/jb.171.3.1303-1308.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dubois E, Messenguy F. In vitro studies of the binding of the ARGR proteins to the ARG5,6 promoter. Mol Cell Biol. 1991;11:2162–2168. doi: 10.1128/mcb.11.4.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Efrat S, Tal M, Lodish H F. The pancreatic beta-cell glucose sensor. Trends Biochem Sci. 1994;19:535–538. doi: 10.1016/0968-0004(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 41.Entian K-D, Kopetzki E, Fröhlich K-U, Mecke D. Cloning of hexokinase isoenzyme PI from Saccharomyces cerevisiae: PI transformants confirm the unique role of hexokinase isoenzyme PII for glucose repression in yeasts. Mol Gen Genet. 1984;198:50–54. doi: 10.1007/BF00328699. [DOI] [PubMed] [Google Scholar]
- 42.Erickson J R, Johnston M. Suppressors reveal two classes of glucose repression genes in the yeast Saccharomyces cerevisiae. Genetics. 1994;136:1271–1278. doi: 10.1093/genetics/136.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Flick J S, Johnston M. GRR1 of Saccharomyces cerevisiae is required for glucose repression and encodes a protein with leucine-rich repeats. Mol Cell Biol. 1991;11:5101–5112. doi: 10.1128/mcb.11.10.5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gamo F-J, Lafuente M J, Gancedo C. The mutation in DGT1-1 decreases glucose transport and alleviates carbon catabolite repression in Saccharomyces cerevisiae. J Bacteriol. 1994;176:7423–7429. doi: 10.1128/jb.176.24.7423-7429.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gancedo J M. Yeast carbon catabolite repression. Microbiol Mol Biol Rev. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Garcia J C, Strube M, Leingang K, Keller K, Mueckler M M. Amino acid substitutions at tryptophan 388 and tryptophan 412 of the HepG2 (Glut1) glucose transporter inhibit transport activity and targeting to the plasma membrane in Xenopus oocytes. J Biol Chem. 1992;267:7770–7776. [PubMed] [Google Scholar]
- 47.Goncalves P M, Griffioen G, Bebelman J P, Planta R J. Signaling pathways leading to transcriptional regulation of genes involved in the activation of glycolysis in yeast. Mol Microbiol. 1997;25:483–493. doi: 10.1046/j.1365-2958.1997.4811847.x. [DOI] [PubMed] [Google Scholar]
- 48.Hardie D G, Carling D, Carlson M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu Rev Biochem. 1998;67:821–855. doi: 10.1146/annurev.biochem.67.1.821. [DOI] [PubMed] [Google Scholar]
- 49.Hirayama T, Maeda T, Saito H, Shinozaki K. Cloning and characterization of seven cDNAs for hyperosmolarity-responsive (HOR) genes of Saccharomyces cerevisiae. Mol Gen Genet. 1995;249:127–138. doi: 10.1007/BF00290358. [DOI] [PubMed] [Google Scholar]
- 50.Hochstrasser M. Protein degradation or regulation: Ub the judge. Cell. 1996;84:813–815. doi: 10.1016/s0092-8674(00)81058-2. [DOI] [PubMed] [Google Scholar]
- 51.Hoever M, Milbradt B, Höfer M. d-Gluconate is an alternative growth substrate for cultivation of Schizosaccharomyces pombe mutants. Arch Microbiol. 1992;157:191–193. [Google Scholar]
- 52.Holzer H. Catabolite inactivation in yeast. Trends Biochem Sci. 1976;1:178–181. [Google Scholar]
- 53.Horak J. Yeast nutrient transporters. Biochim Biophys Acta. 1997;1331:41–79. doi: 10.1016/s0304-4157(96)00015-9. [DOI] [PubMed] [Google Scholar]
- 54.Horak J, Wolf D H. Catabolite inactivation of the galactose transporter in the yeast Saccharomyces cerevisiae: ubiquitination, endocytosis, and degradation in the vacuole. J Bacteriol. 1997;179:1541–1549. doi: 10.1128/jb.179.5.1541-1549.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang L, Zhang W, Roth S Y. Amino termini of histones H3 and H4 are required for a1-α2 repression in yeast. Mol Cell Biol. 1997;17:6555–6562. doi: 10.1128/mcb.17.11.6555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hubbard E J A, Jiang R, Carlson M. Dosage-dependent modulation of glucose repression by MSN3 (STD1) in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:1972–1978. doi: 10.1128/mcb.14.3.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iraqui I, Vissers S, Bernard F, de Craene J-O, Boles E, Urrestarazu A, Andre B. Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1 are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol Cell Biol. 1999;19:989–1001. doi: 10.1128/mcb.19.2.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jaquenoud M, Gulli M P, Peter K, Peter M. The Cdc42p effector Gic2p is targeted for ubiquitin-dependent degradation by the SCFGrr1 complex. EMBO J. 1998;17:5360–5373. doi: 10.1093/emboj/17.18.5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jiang H, Medintz I, Michels C A. Two glucose sensing/signaling pathways stimulate glucose-induced inactivation of maltose permease in Saccharomyces. Mol Biol Cell. 1997;8:1293–1304. doi: 10.1091/mbc.8.7.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson E S, Blobel G. Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J Biol Chem. 1997;272:26799–26802. doi: 10.1074/jbc.272.43.26799. [DOI] [PubMed] [Google Scholar]
- 61.Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987;51:458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Johnston M. Genetic evidence that zinc is an essential co-factor in the DNA binding domain of GAL4 protein. Nature. 1987;328:353–355. doi: 10.1038/328353a0. [DOI] [PubMed] [Google Scholar]
- 63.Johnston M, Carlson M. Regulation of carbon and phosphate utilization. In: Jones E W, Pringle J, Broach J R, editors. The molecular and cellular biology of the yeast Saccharomyces. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1992. pp. 193–281. [Google Scholar]
- 64.Jorgensen M U, Bruun M B, Didion T, Kielland-Brandt M C. Mutations in five loci affecting GAP1-independent uptake of neutral amino acids in yeast. Yeast. 1998;14:103–114. doi: 10.1002/(SICI)1097-0061(19980130)14:2<103::AID-YEA203>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 65.Kasahara T, Kasahara M. Tryptophan 388 in putative transmembrane segment 10 of the rat glucose transporter Glut1 is essential for glucose transport. J Biol Chem. 1998;273:29113–29117. doi: 10.1074/jbc.273.44.29113. [DOI] [PubMed] [Google Scholar]
- 66.Kasahara M, Maeda M. Contribution to substrate recognition of two aromatic amino acid residues in putative transmembrane segment 10 of the yeast sugar transporters Gal2 and Hxt2. J Biol Chem. 1998;273:29106–29112. doi: 10.1074/jbc.273.44.29106. [DOI] [PubMed] [Google Scholar]
- 67.Kasahara M, Shimoda E, Maeda M. Transmembrane segment 10 is important for substrate recognition in Gal2 and Hxt2 sugar transporters in the yeast Saccharomyces cerevisiae. FEBS Lett. 1996;389:174–178. doi: 10.1016/0014-5793(96)00567-4. [DOI] [PubMed] [Google Scholar]
- 68.Kasahara M, Shimoda E, Maeda M. Amino acid residues responsible for galactose recognition in yeast Gal2 transporter. J Biol Chem. 1997;272:16721–16724. doi: 10.1074/jbc.272.27.16721. [DOI] [PubMed] [Google Scholar]
- 69.Keleher C A, Reed M J, Schultz J, Carlson M, Johnson A D. Ssn6-Tup1 is a general repressor of transcription in yeast. Cell. 1992;68:709–719. doi: 10.1016/0092-8674(92)90146-4. [DOI] [PubMed] [Google Scholar]
- 70.Kilian S G, van Uden N. Transport of xylose and glucose in the xylose-fermenting yeast Pichia stipitis. Appl Microbiol Biotechnol. 1988;27:545–548. [Google Scholar]
- 71.Kishi T, Seno T, Yamao F. Grr1 functions in the ubiquitin pathway in Saccharomyces cerevisiae through association with Skp1. Mol Gen Genet. 1998;257:143–148. doi: 10.1007/s004380050633. [DOI] [PubMed] [Google Scholar]
- 72.Ko C H, Liang H, Gaber R F. Roles of multiple glucose transporters in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:638–648. doi: 10.1128/mcb.13.1.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kotyk A, Michaljanicova D, Veres K, Soukupova V. Transport of 4-deoxy- and 6-deoxy-glucose in baker’s yeast. Folia Microbiol. 1975;20:496–503. doi: 10.1007/BF02891709. [DOI] [PubMed] [Google Scholar]
- 74.Krampe S, Stamm O, Hollenberg C P, Boles E. Catabolite inactivation of the high-affinity hexose transporters Hxt6 and Hxt7 of Saccharomyces cerevisiae occurs in the vacuole after internalization by endocytosis. FEBS Lett. 1998;441:343–347. doi: 10.1016/s0014-5793(98)01583-x. [DOI] [PubMed] [Google Scholar]
- 75.Kruckeberg A L. The hexose transporter family of Saccharomyces cerevisiae. Arch Microbiol. 1996;166:283–292. doi: 10.1007/s002030050385. [DOI] [PubMed] [Google Scholar]
- 76.Kruckeberg A L, Bisson L F. The HXT2 gene of Saccharomyces cerevisiae is required for high-affinity transport. Mol Cell Biol. 1990;10:5903–5913. doi: 10.1128/mcb.10.11.5903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lagunas R. Sugar transport in Saccharomyces cerevisiae. FEMS Microbiol Rev. 1993;104:229–242. doi: 10.1016/0378-1097(93)90598-v. [DOI] [PubMed] [Google Scholar]
- 78.Lammer D, Mathias N, Laplaza J M, Jiang W, Liu Y, Callis J, Goebl M, Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lewis D A, Bisson L F. The HXT1 gene product of Saccharomyces cerevisiae is a new member of the family of hexose transporters. Mol Cell Biol. 1991;11:3804–3813. doi: 10.1128/mcb.11.7.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li F N, Johnston M. Grr1 of Saccharomyces cerevisiae is connected to the ubiquitin proteolysis machinery through Skp1: coupling glucose sensing to gene expression and the cell cycle. EMBO J. 1997;16:5629–5638. doi: 10.1093/emboj/16.18.5629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Li, F., J. Polish, and M. Johnston. 1998. Unpublished data.
- 82.Liang H, Gaber R F. A novel signal transduction pathway in Saccharomyces cerevisiae defined by Snf3-regulated expression of HXT6. Mol Biol Cell. 1996;7:1953–1966. doi: 10.1091/mbc.7.12.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lichtenberg-Frate H, Naschen T, Heiland S, Hofer M. Properties and heterologous expression of the glucose transporter GHT1 from Schizosaccharomyces pombe. Yeast. 1997;13:215–224. doi: 10.1002/(SICI)1097-0061(19970315)13:3<215::AID-YEA80>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 84.Lohr D, Venkov P, Zlantanova J. Transcriptional regulation in the yeast GAL gene family: a complex genetic network. FASEB J. 1995;9:777–787. doi: 10.1096/fasebj.9.9.7601342. [DOI] [PubMed] [Google Scholar]
- 85.Lorenz M C, Heitman J. The MEP2 ammonium permease regulates pseudohyphal differentiation in Saccharomyces cerevisiae. EMBO J. 1998;17:1236–1247. doi: 10.1093/emboj/17.5.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ludin K, Jiang R, Carlson M. Glucose-regulated interaction of a regulatory subunit of protein phosphatase 1 with the Snf1 protein kinase in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1998;95:6245–6250. doi: 10.1073/pnas.95.11.6245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Madi L, McBride A, Bailey L A, Ebbole D J. rco-3, a gene involved in glucose transport and conidiation in Neurospora crassa. Genetics. 1997;146:499–508. doi: 10.1093/genetics/146.2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maeda T, Wurgler-Murphy S M, Saito H. A two-component system that regulates an osmosensing MAP kinase cascade in yeast. Nature. 1994;369:242–245. doi: 10.1038/369242a0. [DOI] [PubMed] [Google Scholar]
- 89.Marger M D, Saier M H J. A major superfamily of transmembrane facilitators that catalyze uniport, symport and antiport. Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- 90.Marmorstein R, Carey M, Ptashne M, Harrison S C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- 91.Marshall-Carlson L, Celenza J L, Laurent B C, Carlson M. Mutational analysis of the SNF3 glucose transporter of Saccharomyces cerevisiae. Mol Cell Biol. 1990;10:1105–1115. doi: 10.1128/mcb.10.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Marshall-Carlson L, Neigeborn L, Coons D, Bisson L, Carlson M. Dominant and recessive suppressors that restore glucose transport in a yeast snf3 mutant. Genetics. 1991;128:505–512. doi: 10.1093/genetics/128.3.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.McLeod M, Beach D. Homology between ran1+ gene of fission and protein kinases. EMBO J. 1986;5:3665–3671. doi: 10.1002/j.1460-2075.1986.tb04697.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Medintz I, Jiang H, Michels C A. The role of ubiquitin conjugation in glucose-induced proteolysis of Saccharomyces maltose permease. J Biol Chem. 1998;273:34454–34462. doi: 10.1074/jbc.273.51.34454. [DOI] [PubMed] [Google Scholar]
- 95.Medintz I, Jiang H, Han E K, Cui W, Michels C A. Characterization of the glucose-induced inactivation of maltose permease in Saccharomyces cerevisiae. J Bacteriol. 1996;178:2245–2254. doi: 10.1128/jb.178.8.2245-2254.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Milbradt B, Höfer M. Glucose-transport-deficient mutants of Schizosaccharomyces pombe: phenotype, genetics and use for genetic complementation. Microbiology. 1994;140:2617–2623. doi: 10.1099/00221287-140-10-2617. [DOI] [PubMed] [Google Scholar]
- 97.Mueckler M. Facilitative glucose transporters. Eur J Biochem. 1994;219:713–725. doi: 10.1111/j.1432-1033.1994.tb18550.x. [DOI] [PubMed] [Google Scholar]
- 98.Müller S, Boles E, May M, Zimmermann F K. Different internal metabolites trigger the induction of glycolytic gene expression in Saccharomyces cerevisiae. J Bacteriol. 1995;177:4517–4519. doi: 10.1128/jb.177.15.4517-4519.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nehlin J O, Ronne H. Yeast MIG1 repressor is related to the mammalian early growth response and Wilms’ tumour finger proteins. EMBO J. 1990;9:2891–2898. doi: 10.1002/j.1460-2075.1990.tb07479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nehlin J O, Carlberg M, Ronne H. Yeast galactose permease is related to yeast and mammalian glucose transporters. Gene. 1989;85:313–319. doi: 10.1016/0378-1119(89)90423-x. [DOI] [PubMed] [Google Scholar]
- 101.Nehlin J O, Carlberg M, Ronne H. Control of yeast GAL genes by MIG1 repressor: a transcriptional cascade in the glucose response. EMBO J. 1991;10:3373–3377. doi: 10.1002/j.1460-2075.1991.tb04901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Neigeborn L, Schwartzberg P, Reid R, Carlson M. Null mutations in the SNF3 gene of Saccharomyces cerevisiae cause a different phenotype than do previously isolated missense mutations. Mol Cell Biol. 1986;6:3569–3574. doi: 10.1128/mcb.6.11.3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nevado J, Navarro M A, Heredia C F. Galactose inhibition of the constitutive transport of hexoses in Saccharomyces cerevisiae. Yeast. 1993;9:111–119. doi: 10.1002/yea.320090202. [DOI] [PubMed] [Google Scholar]
- 105.Newgard C B, McGarry J D. Metabolic coupling factors in pancreatic beta-cell signal transduction. Annu Rev Biochem. 1995;64:689–719. doi: 10.1146/annurev.bi.64.070195.003353. [DOI] [PubMed] [Google Scholar]
- 106.Nishizawa K, Shimoda E, Kasahara M. Substrate recognition domain of the Gal2 galactose transporter in yeast Saccharomyces cerevisiae as revealed by chimeric galactose-glucose transporters. J Biol Chem. 1995;270:2423–2326. doi: 10.1074/jbc.270.6.2423. [DOI] [PubMed] [Google Scholar]
- 107.Nourani A, Wesolowski-Louvel M, Delaveau T, Jacq C, Delahodde A. Multi-drug-resistance phenomenon in the yeast Saccharomyces cerevisiae: involvement of two hexose transporters. Mol Cell Biol. 1997;17:5453–5460. doi: 10.1128/mcb.17.9.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Özcan, S., and M. Johnston. 1998. Unpublished data.
- 109.Özcan S, Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Özcan S, Johnston M. Two different repressors collaborate to restrict expression of the yeast glucose transporter genes HXT2 and HXT4 to low levels of glucose. Mol Cell Biol. 1996;16:5536–5545. doi: 10.1128/mcb.16.10.5536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Özcan S, Dover J, Johnston M. Glucose sensing and signaling by two glucose receptors in the yeast S. cerevisiae. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Özcan S, Leong T, Johnston M. Rgt1p of Saccharomyces cerevisiae, a key regulator of glucose-induced genes, is both an activator and a repressor of transcription. Mol Cell Biol. 1996;16:6419–6426. doi: 10.1128/mcb.16.11.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Özcan S, Freidel K, Leuker A, Ciriacy M. Glucose uptake and catabolite repression in dominant HTR1 mutants of Saccharomyces cerevisiae. J Bacteriol. 1993;175:5520–5528. doi: 10.1128/jb.175.17.5520-5528.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Özcan S, Dover J, Rosenwald A G, Wölfl S, Johnston M. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc Natl Acad Sci USA. 1996;93:12428–12432. doi: 10.1073/pnas.93.22.12428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Özcan S, Schulte F, Freidel K, Weber A, Ciriacy M. Glucose uptake and metabolism in grr1/cat80 mutants of Saccharomyces cerevisiae. Eur J Biochem. 1994;224:605–611. doi: 10.1111/j.1432-1033.1994.00605.x. [DOI] [PubMed] [Google Scholar]
- 116.Özcan S, Vallier L G, Flick J S, Carlson M, Johnston M. Expression of the SUC2 gene of Saccharomyces cerevisiae is induced by low levels of glucose. Yeast. 1997;13:127–137. doi: 10.1002/(SICI)1097-0061(199702)13:2<127::AID-YEA68>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 117.Pao S S, Paulsen I T, Saier M H., Jr Major facilitator superfamily. Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Patton E E, Willems A R, Tyers M. Combinatorial control in ubiquitin-dependent proteolysis: don’t Skp the F-box hypothesis. Trends Genet. 1998;14:236–243. doi: 10.1016/s0168-9525(98)01473-5. [DOI] [PubMed] [Google Scholar]
- 119.Pfeifer K, Kim K S, Kogan S, Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989;56:291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- 120.Platt A, Reece R J. The yeast galactose genetic switch is mediated by the formation of a Gal4p-Gal80p-Gal3p complex. EMBO J. 1998;14:4086–4091. doi: 10.1093/emboj/17.14.4086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Prior C, Fukuhara H, Blasisonneau J, Wesolowski-Louvel M. Low-affinity glucose carrier gene LGT1 of Saccharomyces cerevisiae, a homolog of the Kluyveromyces lactis RAG1 gene. Yeast. 1993;9:1373–1377. doi: 10.1002/yea.320091211. [DOI] [PubMed] [Google Scholar]
- 122.Prior C, Mamessier P, Fukuhara H, Chen X J, Wesolowski-Louvel M. The hexokinase gene is required for transcriptional regulation of the glucose transporter gene RAG1 in Kluyveromyces lactis. Mol Cell Biol. 1993;13:3882–3889. doi: 10.1128/mcb.13.7.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Purnelle B, Skala J, Dyck L V, Goffeau A. The sequence of a 12 kb fragment on the left arm of yeast chromosome XI reveals five new open reading frames, including a zinc finger protein and a homolog of the UDP-glucose pyrophosphorylase from potato. Yeast. 1992;8:977–986. doi: 10.1002/yea.320081108. [DOI] [PubMed] [Google Scholar]
- 124.Ramos J, Cirillo V P. Role of cyclic-AMP-dependent protein kinase in catabolite inactivation of the glucose and galactose transporters in Saccharomyces cerevisiae. J Bacteriol. 1989;171:3545–3548. doi: 10.1128/jb.171.6.3545-3548.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ramos J, Szkutnicka K, Cirillo V P. Relationship between the low- and high-affinity glucose transport systems in Saccharomyces cerevisiae. Bacteriol. 1988;170:3575–3577. doi: 10.1128/jb.170.11.5375-5377.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Redd M J, Arnaud M B, Johnson A D. A complex composed of tup1 and ssn6 represses transcription in vitro. J Biol Chem. 1997;272:11193–11197. doi: 10.1074/jbc.272.17.11193. [DOI] [PubMed] [Google Scholar]
- 127.Reece R J, Ptashne M. Determinants of binding-site specificity among yeast C6 zinc cluster proteins. Science. 1993;261:909–911. doi: 10.1126/science.8346441. [DOI] [PubMed] [Google Scholar]
- 128.Reifenberger E, Boles E, Ciriacy M. Kinetic characterization of individual hexose transporters of Saccharomyces cerevisiae and their relation to the triggering mechanisms of glucose repression. Eur J Biochem. 1997;245:324–333. doi: 10.1111/j.1432-1033.1997.00324.x. [DOI] [PubMed] [Google Scholar]
- 129.Reifenberger E, Freidel K, Ciriacy M. Identification of novel HXT genes in Saccharomyces cerevisiae reveals the impact of individual hexose transporters on glycolytic flux. Mol Microbiol. 1995;16:157–167. doi: 10.1111/j.1365-2958.1995.tb02400.x. [DOI] [PubMed] [Google Scholar]
- 130.Riballo E, Lagunas R. Involvement of endocytosis in catabolite inactivation of the K+ and glucose transport systems in Saccharomyces cerevisiae. FEMS Microbiol Lett. 1994;121:77–80. doi: 10.1111/j.1574-6968.1994.tb07078.x. [DOI] [PubMed] [Google Scholar]
- 131.Riballo E, Mazon M J, Lagunas R. cAMP-dependent protein kinase is not involved in catabolite inactivation of the transport of sugars in Saccharomyces cerevisiae. Biochim Biophys Acta. 1994;1192:143–146. doi: 10.1016/0005-2736(94)90154-6. [DOI] [PubMed] [Google Scholar]
- 132.Robertson J L, Halvorson H O. The components of maltozymase in yeast and their behavior during deadaptation. J Bacteriol. 1957;73:186–198. doi: 10.1128/jb.73.2.186-198.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ronne H. Glucose repression in fungi. Trends Genet. 1995;11:12–17. doi: 10.1016/s0168-9525(00)88980-5. [DOI] [PubMed] [Google Scholar]
- 134.Schmidt M C, McCartney R R, Zhang X, Tillman T S, Solimeo H, Wölfl S, Almonte C, Watkins S C. Std1 and Mth1 proteins interact with the glucose sensors to control glucose-regulated gene expression in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4561–4571. doi: 10.1128/mcb.19.7.4561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Schüller H-J, Entian K-D. Isolation and expression analysis of two yeast regulatory genes involved in the derepression of glucose-repressible enzymes. Mol Gen Genet. 1987;209:366–373. doi: 10.1007/BF00329667. [DOI] [PubMed] [Google Scholar]
- 136.Schulte F, Ciriacy M. HTR1/MTH1 encodes a repressor for HXT genes. Yeast. 1995;11:S239. [Google Scholar]
- 137.Skowyra D, Craig K L, Tyers M, Elledge S J, Harper J W. F-box proteins are receptors that recruit phosphorylated substrates to the SCF ubiquitin-ligase complex. Cell. 1997;91:209–219. doi: 10.1016/s0092-8674(00)80403-1. [DOI] [PubMed] [Google Scholar]
- 138.Suzuki-Fujimoto T, Fukuma M, Yano K I, Sakurai H, Vonika A, Johnston S A, Fukasawa T. Analysis of the galactose signal transduction pathway in Saccharomyces cerevisiae: interaction between Gal3p and Gal80p. Mol Cell Biol. 1996;16:2504–2508. doi: 10.1128/mcb.16.5.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tamori Y, et al. Substitution at Pro385 of GLUT1 perturbs the glucose transport function by reducing conformational flexibility. J Biol Chem. 1994;269:2982–2986. [PubMed] [Google Scholar]
- 140.Theodoris G, Fong N M, Coons D M, Bisson L F. High-copy suppression of glucose transport defects by HXT4 and regulatory elements in the promoters of the HXT genes in Saccharomyces cerevisiae. Genetics. 1994;137:957–966. doi: 10.1093/genetics/137.4.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Tillman T S, Ganster R W, Jiang R, Carlson M, Schmidt M C. STD1 (MSN3) interacts directly with the TATA-binding protein and modulates transcription of the SUC2 gene of Saccharomyces cerevisiae. Nucleic Acids Res. 1995;23:3174–3180. doi: 10.1093/nar/23.16.3174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Treitel M A, Carlson M. Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci USA. 1995;92:3132–3136. doi: 10.1073/pnas.92.8.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Treitel M A, Kuchin S, Carlson M. Snf1 protein kinase regulates phosphorylation of the Mig1 repressor in Saccharomyces cerevisiae. Mol Cell Biol. 1998;18:6273–6280. doi: 10.1128/mcb.18.11.6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Trumbly R J. Glucose repression in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1992;6:15–21. doi: 10.1111/j.1365-2958.1992.tb00832.x. [DOI] [PubMed] [Google Scholar]
- 145.Tschopp J, Emr S, Field C, Schekman R. GAL2 codes for a membrane-bound subunit of the galactose permease in Saccharomyces cerevisiae. J Bacteriol. 1986;166:313–318. doi: 10.1128/jb.166.1.313-318.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tu J, Carlson M. The GLC7 type 1 protein phosphatase is required for glucose repression in Saccharomyces cerevisiae. EMBO J. 1995;14:5939–5946. doi: 10.1002/j.1460-2075.1995.tb00282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tung K-S, Norbeck L L, Nolan S L, Atkinson N S, Hopper A K. SRN1, a yeast gene involved in RNA processing, is identical to HEX2/REG1, a negative regulator in glucose repression. Mol Cell Biol. 1992;12:2673–2680. doi: 10.1128/mcb.12.6.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Tzamarias D, Struhl K. Functional dissection of the yeast Cyc8-Tup1 transcriptional co-repressor complex. Nature. 1994;369:758–761. doi: 10.1038/369758a0. [DOI] [PubMed] [Google Scholar]
- 149.Tzamarias D, Struhl K. Distinct TPR motifs of Cyc8 are involved in recruiting the Cyc8-Tup1 corepressor complex to differentially regulated promoters. Genes Dev. 1995;9:821–831. doi: 10.1101/gad.9.7.821. [DOI] [PubMed] [Google Scholar]
- 150.Vagnoli P, Coons D M, Bisson L F. The c-terminal domain of Snf3p mediates glucose-responsive signal transduction in Saccharomyces cerevisiae. FEMS Microbiol Lett. 1998;160:31–36. doi: 10.1111/j.1574-6968.1998.tb12886.x. [DOI] [PubMed] [Google Scholar]
- 151.Vallier L G, Carlson M. New SNF genes, GAL11 and GRR1, affect SUC2 expression in Saccharomyces cerevisiae. Genetics. 1991;129:675–684. doi: 10.1093/genetics/129.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Vallier L G, Coons D, Bisson L F, Carlson M. Altered regulatory responses to glucose are associated with a glucose transport defect in grr1 mutants of Saccharomyces cerevisiae. Genetics. 1994;136:1279–1285. doi: 10.1093/genetics/136.4.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wadzinski B E, Shanahan M F, Seamon K B, Ruoho A E. Localization of the forskolin photolabelling site within the monosaccharide transporter of human erythrocytes. Biochem J. 1990;272:151–158. doi: 10.1042/bj2720151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Walsh M C, Smits H P, Scholte M, van Dam K. Affinity of glucose transport in Saccharomyces cerevisiae is modulated during growth on glucose. J Bacteriol. 1994;176:953–958. doi: 10.1128/jb.176.4.953-958.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Weierstall T, Hollenberg C P, Boles E. Cloning and characterization of three genes (SUT1-3) encoding glucose transporters of the yeast Pichia stipitis. Mol Microbiol. 1999;31:871–883. doi: 10.1046/j.1365-2958.1999.01224.x. [DOI] [PubMed] [Google Scholar]
- 156.Wendell D L, Bisson L F. Physiological characterization of putative high-affinity glucose transport protein Hxt2 of Saccharomyces cerevisiae by use of antisynthetic peptide antibodies. J Bacteriol. 1993;175:7689–7696. doi: 10.1128/jb.175.23.7689-7696.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wendell D L, Bisson L F. Expression of high-affinity glucose transport protein Hxt2p of Saccharomyces cerevisiae is both repressed and induced by glucose and appears to be regulated posttranslationally. J Bacteriol. 1994;176:3730–3737. doi: 10.1128/jb.176.12.3730-3737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Wesolowski-Louvel M, Goffrini P, Ferrero I, Fukuhara H. Glucose transport in the yeast Kluyveromyces lactis. I. Properties of an inducible low-affinity glucose transporter gene. Mol Gen Genet. 1992;233:97–105. doi: 10.1007/BF00587565. [DOI] [PubMed] [Google Scholar]
- 159.Williams F E, Varanasi U, Trumbly R J. The CYC8 and TUP1 proteins involved in glucose repression in Saccharomyces cerevisiae are associated in a protein complex. Mol Cell Biol. 1991;11:3307–3316. doi: 10.1128/mcb.11.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wölfl S, Hanemann V, Saluz H P. Analysis of a 26,756bp segment from the left arm of yeast chromosome IV. Yeast. 1996;12:1549–1554. doi: 10.1002/(sici)1097-0061(199612)12:15<1549::aid-yea42>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 161.Yang Z, Bisson L F. The SKS1 protein kinase is a multicopy suppressor of the snf3 mutation of Saccharomyces cerevisiae. Yeast. 1996;12:1407–1419. doi: 10.1002/(SICI)1097-0061(199611)12:14%3C1407::AID-YEA36%3E3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 162.Yano K-I, Fukasawa T. Galactose-dependent reversible interaction of Gal3p with Gal80p in the induction pathway of Gal4p-activated genes of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1997;94:1721–1726. doi: 10.1073/pnas.94.5.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zenke F T, Engles R, Vollenbroich V, Meyer J, Hollenberg C P, Breunig K D. Activation of Gal4p by galactose-dependent interaction of galactokinase and Gal80p. Science. 1996;272:1662–1665. doi: 10.1126/science.272.5268.1662. [DOI] [PubMed] [Google Scholar]