Abstract
In the late 1970s, on the basis of rRNA phylogeny, Archaea (archaebacteria) was identified as a distinct domain of life besides Bacteria (eubacteria) and Eucarya. Though forming a separate domain, archaea display an enormous diversity of lifestyles and metabolic capabilities. Many archaeal species are adapted to extreme environments with respect to salinity, temperatures around the boiling point of water, and/or extremely alkaline or acidic pH. This has posed the challenge of studying the molecular and mechanistic bases on which these organisms can cope with such adverse conditions. This review considers our cumulative knowledge on archaeal mechanisms of primary energy conservation, in relationship to those of bacteria and eucarya. Although the universal principle of chemiosmotic energy conservation also holds for Archaea, distinct features have been discovered with respect to novel ion-transducing, membrane-residing protein complexes and the use of novel cofactors in bioenergetics of methanogenesis. From aerobically respiring archaea, unusual electron-transporting supercomplexes could be isolated and functionally resolved, and a proposal on the organization of archaeal electron transport chains has been presented. The unique functions of archaeal rhodopsins as sensory systems and as proton or chloride pumps have been elucidated on the basis of recent structural information on the atomic scale. Whereas components of methanogenesis and of phototrophic energy transduction in halobacteria appear to be unique to archaea, respiratory complexes and the ATP synthase exhibit some chimeric features with respect to their evolutionary origin. Nevertheless, archaeal ATP synthases are to be considered distinct members of this family of secondary energy transducers. A major challenge to future investigations is the development of archaeal genetic transformation systems, in order to gain access to the regulation of bioenergetic systems and to overproducers of archaeal membrane proteins as a prerequisite for their crystallization.
Ever since archaea have been studied, their ability to thrive in unusual habitats under extremely harsh conditions has stimulated interest in the molecular mechanisms that confer heat stability on proteins at temperatures above 100°C, tolerance of extreme pH values and salt concentrations, and unique metabolic functions not found in bacteria, such as methanogenesis or rhodopsin-linked energy and signal transduction. Actually, ever since Archaea was identified as a third evolutionary kingdom (606–608) presumably located relatively near the hypothetical root of the evolutionary tree, it has been speculated that the structural organization and metabolic pathways of archaea might reflect more ancestral organisms whose essential properties differ from those of bacteria and eucarya. In this regard, one has to realize with respect to biological energy conservation that all existing forms of life rely on the universal principle of chemiosmotic energy transduction (366, 367), which in phylogenetic terms should have evolved very early. In fact, the origin of cellular life must have been connected with the permanent manifestation of mechanisms allowing the transduction of energy between exergonic and endergonic processes and with the development of transitory and long-term energy stores.
The definition of Archaea as a separate domain of organisms was based on the comparative analysis of 16S rRNA sequences (405), which led to a result different from classical taxonomy. The term Archaea reflects an earlier idea that these organisms descended from life forms that existed prior to the division into the bacterial and eukaryal domains. However, based on the sequences of universally present proteins (176), Archaea has been placed on the branch also leading to Eucarya. A feature that distinguishes Archaea from Bacteria is the structure of archaeal ribosomes (435), which in halophiles were first recognized to contain acidic rather than basic proteins (42). In addition, the transcriptional machinery is unique as to the structure of DNA-dependent RNA polymerases (133, 626). With respect to subunit structure, a closer relationship to eukaryotes than to bacteria was found (300). Another feature distinguishing Archaea from Bacteria is the specific composition of archaeal surface layers (266), which do not contain peptidoglycans. Their glycoprotein surface layers can form quasicrystalline structures (97, 360) that are firmly attached to the plasma membrane, thus leaving practically no periplasmic space (39).
Nevertheless, although archaea are located on a distinct evolutionary branch as depicted in Fig. 1, they represent, with regard to their primary energy-transducing mechanisms, a very heterogeneous domain comprising chemolithoautotrophic as well as organotrophic species. In addition to obligate anaerobes such as the methanogens, a second group that performs various types of aerobic or anaerobic respiration can be distinguished. Further, for some halobacteria we have to consider archaeal phototrophic energy transformation in addition to respiratory mechanisms.
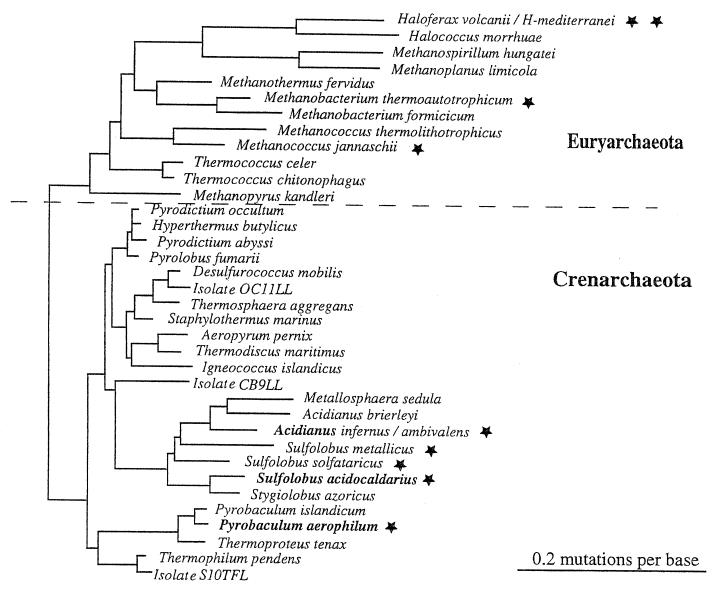
FIG. 1.
Phylogenetic tree. The scheme demonstrates the division into Crenarchaeota and Euryarchaeota and shows the position of the major archaeal genera. The tree was redrawn according to references 88, 175, and 608. Stars denote archaeal species for which specific bioenergetic information has been found.
In contrast to this diversity, archaeal membrane structures reveal a comparatively homogeneous phenotype, significantly different from that of other prokaryotes. Diether and tetraether lipids are uniformly used as building blocks for archaeal plasma membranes (266). Obviously, the low ion permeability of membranes formed from these bipolar monolayer-forming lipids (136, 571) contributes significantly to the stability of chemiosmotic charge separation in archaea, particularly at high temperatures and/or at extremely low pH values.
Interestingly, neither oxygenic nor anoxygenic “green” photosynthesis has been found in the archaeal kingdom. This latter observation served as an argument in favor of the respiration-first hypothesis, suggesting that the formation of the basic structure of terminal oxidase complexes preceded the occurrence of chlorophyll-based water-splitting and charge-separating systems (93, 94).
It is the aim of this review to introduce the reader to archaeal bioenergetics by a critical state-of-the-art report and to demonstrate similarities and distinguishing features by contrast to bacterial and eucaryal systems. It will not be possible in all cases to give an unambiguous answer to the question of what is typical or genuine for the archaeal domain, because, especially within respiratory electron transport, we will find a number of chimeric functional complexes. In fact, one has to assume that during early evolution, i.e., prior to the division into three urkingdoms, the barriers against lateral gene transfer were much lower than they are now (605).
Of the various bioenergetic mechanisms in archaeal organisms, the present review focuses specifically on the primary energy conservation that involves membrane-residing chemiosmotic processes. Therefore, purely fermentative energy transduction by substrate-level phosphorylation as well as secondary active-transport systems for solutes will not be discussed. As an exception among secondary energy transducers, the ATP synthase complexes will be dealt with because they apparently possess a unique and ubiquitously conserved mechanism, irrespective of the primary energy converter which provides the electrochemical ion gradient to be used as the driving force for high-energy bond formation during ADP phosphorylation.
Another limitation is the great diversity of the archaeal domain, much greater than that suggested by Fig. 1. What we presently know about diversity within the archaeal domain is probably only the tip of the iceberg. By means of molecular studies based on rRNA-directed probes, new archaea are constantly being discovered not only in deep-sea vents or solfataric fields (37, 89, 379) but also in mesophilic and even low-temperature environments. Unfortunately, only a few of these isolates or new species identified by DNA hybridization will prove amenable to cultivation. Thus, the diversity of bioenergetic systems may well exceed the number of classes reviewed in this comprehensive study.
CHEMIOSMOSIS IN ARCHAEA
Primary energy conservation by membrane-residing systems is characterized by the formation of an electrochemical potential of hydrogen ions or sodium ions. According to the work of Mitchell (367), the free energy stored in this gradient is described by equations 1 and 2 for the proton motive force:
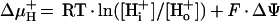 |
1 |
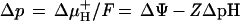 |
2 |
The primary pumps may be driven by redox systems, by methyl transfer reactions as in methanogenesis, or by light as in photophosphorylating halobacteria.
As illustrated by some representative examples below, whole-cell experiments with various genera of Archaea have proven that ATP synthesis is driven, according to the chemiosmotic theory, at the expense of such ion gradients. These experiments were of significance because minimal systems such as inverted plasma membrane vesicles or spheroplasts which are easily prepared from several bacterial organisms are essentially inaccessible in the case of Archaea. The rigid structure and extremely tight adhesion or interdigitation of the glycoprotein cell walls covering archaeal plasma membranes (39, 98, 181, 270, 514) represent an invincible obstacle. For the same reason, the preparation of intact complexes of energy-transducing membrane proteins is quite difficult. In addition, other factors are frequently responsible for the failure to purify catalytically active complexes, such as ATP synthase or terminal oxidases. Such factors can be the absence of the high pH gradient to which membrane proteins are exposed in vivo, extreme salt concentrations, hypersensitivity toward oxygen, and cold dissociation even at room temperature. Also, the determination of energetic parameters such as ΔpH or ΔΨ by direct monitoring or by distribution of diffusible molecular probes is very limited at ambient pH values below 3, the physiological environmental pH for many extreme acidophiles.
Extremely acidophilic organisms, including the archaeon Thermoplasma, were shown to create an inverted membrane potential (30, 364) in order to prevent acidification of the cytosol by influx of H+ at the prevailing ΔpH. This does not generally apply to all acidophiles, however. The membrane potential of thermoacidophilic archaea such as Sulfolobus may be rather low (approximately 30 mV), and most of the proton motive force is maintained by a large pH gradient of >3 (335, 370, 467). Chemiosmotic H+ cycling (370) with H+/O ratios of 3 and a strict correlation of proton motive force (Δp) with cellular ATP levels could be established for Sulfolobus. Dissipation of Δp by protonophores caused an immediate collapse of ATP synthesis; at Δp ≅ 0, a persisting residual ΔpH was counterbalanced by an inverted membrane potential, in which the inside was positive (335). In the same experiments, external proton pulses that lowered the pH from 6.1 to 3.4 produced an increase of Δp from −94 to −170 mV with a concomitant rise of intracellular ATP. For experimental reasons, the reported data was determined at 45°C at an ambient pH of 3.5 and thus may assume slightly different values at the optimal growth temperature of the cells. A review of strategies to cope with extremely low pH values is given in reference 465.
With Halobacterium halobium, the coupling of either a light- or a respiration-induced electrochemical proton gradient with intracellular ATP has been established (361–363). Interestingly, by cation counter transport considerable energy can be stored in the form of a potassium gradient also. Photophosphorylation is a backup system under oxygen limitation in extremely halophilic archaea. This is corroborated by recent studies of the haloalkaliphile Natronobacterium pharaonis (604) demonstrating full recovery of Δp under oxygen-limiting conditions during illumination. Actually, in these latter archaea the main contribution to the proton motive force is made by the membrane potential of ΔΨ = −225 to −280 mV, and ΔpH is influenced only marginally by oxygen limitation. Under these conditions, the high membrane potential is generated by an outwardly directed chloride gradient produced by the light-activated chloride pump halorhodopsin (HR); it is insensitive to protonophores and uncouplers and can even be increased by the Cl−/OH− exchanger triphenyltin (604).
The membrane potential can contribute approximately 90% to the proton motive force (56) in methanogenic archaea also, as shown with Methanosarcina barkeri. Evidence for H+- and Na+-mediated chemiosmotic energy transduction in methanogens has been compiled previously (124); thereby, the sodium and proton gradients may be linked by Na+-H+ antiporters (382). A methanogenic strain, Gö1 (now classified as Methanosarcina mazei), is the only known case in which the successful preparation of archaeal vesicular membrane systems has provided a useful experimental model for the study of energy transduction (58, 59).
The coexistence of proton- and sodium ion-coupled energy converters in anaerobes as well as the branching of electron transport pathways in aerobic archaea is difficult to resolve because these organisms either lack or are insensitive to the site-specific inhibitors known to function in bacteria or eucarya. In addition, genetic systems for directed mutagenesis or gene disruption in archaea have scarcely been developed or are unavailable.
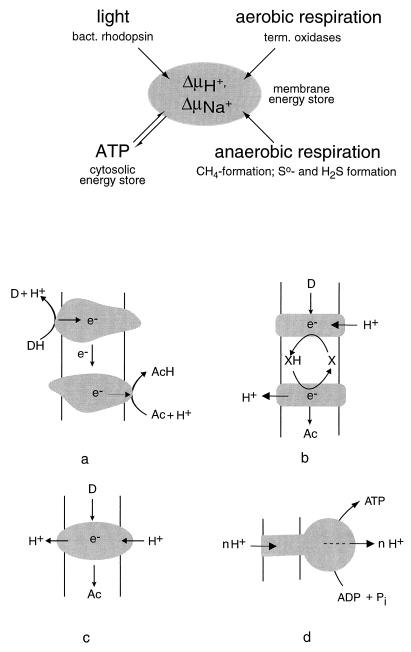
The scheme of Fig. 2 illustrates the generation of ion gradients by primary pumps and their utilization by secondary processes. In the following sections, molecular properties of the known functional complexes are discussed separately for methanogenic, respiring, or photophosphorylating archaea; it should be noted, however, that current complete genome projects have predicted the existence of additional functional complexes which have not yet been verified at the protein or mRNA level.
FIG. 2.
Primary energy-transducing processes and coupling principles in membrane bioenergetics. The top scheme illustrates the processes found in archaea that contribute to the formation of either proton or sodium ion potentials across the plasma membrane. Details are discussed throughout this review. The bottom schemes illustrate three mechanisms by which an ion gradient can be produced: (a) chemical charge separation (only electrons are transferred through the membrane); (b) a mobile membrane-integral cofactor like the quinones or methanophenazine functioning as proton transporter (examples are bc1 complexes); and (c) redox-driven pumps like cyt c oxidase. All schemes are drawn for an H+/e− ratio of 1. Scheme d illustrates the proton-driven ATP synthase of the FoF1 or A1Ao type as an example for a secondary energy transducer. D, electron donor; Ac, electron acceptor.
ENERGETICS OF METHANOGENESIS
Methanogens are a phylogenetically diverse but nutritionally rather uniform group of strictly anaerobic archaea. They are able to grow by the conversion of a small number of compounds to methane. This rather simple pathway is not coupled to substrate-level phosphorylation but, instead, to the generation of ion gradients across the membrane that are used to drive the synthesis of ATP. Interestingly, the pathway of methane formation is coupled to the simultaneous generation of primary gradients of both protons and sodium ions. Although methanogens are nutritionally rather similar and employ identical pathways, they differ significantly with respect to the components involved in the proton motive electron transport chain and, therefore, most likely employ different mechanisms to generate the proton gradient. For example, methylotrophic methanogens, such as M. mazei Gö1, contain cytochromes, whereas hydrogenotrophic methanogens, such as Methanobacterium thermoautotrophicum, do not. Since most of our current knowledge derives from studies using methylotrophic methanogens, in particular M. barkeri and M. mazei, this section focuses on these organisms. For a more thorough discussion of the pathways and the biochemistry of methanogenesis, the reader is referred to recent reviews (58, 124, 381, 561).
Proton Motive Electron Transport Chains in Methanogens
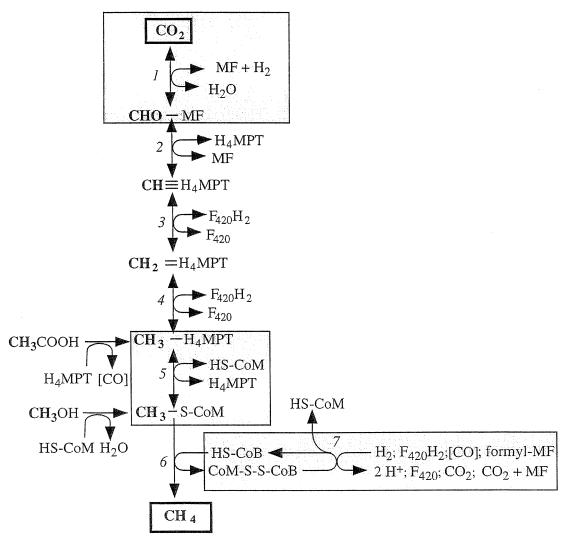
Central to all pathways of methane formation is the intermediate methyl coenzyme M (2-methylthioethanesulfonate; CoM), the ultimate precursor of methane (Fig. 3). It is reductively demethylated by the methyl-CoM reductase with electrons derived from reduced CoB (7-mercaptoheptanoylthreonine phosphate), to give rise to methane and a heterodisulfide of CoM and CoB (CoM-S-S-CoB; henceforth referred to as the heterodisulfide), in a reaction involving the cofactor F430 (reaction 6 in Fig. 3). To complete the cycle, the heterodisulfide is reduced by the heterodisulfide reductase complex (reaction 7 in Fig. 3); this reaction is most important in terms of energy conservation (561). The heterodisulfide reductase is membrane bound and operates as the final limb of several membrane-bound electron transport chains (124). Depending on the substrate, the electron donor used is different. Hydrogenase is employed during growth on H2 plus CO2, CO dehydrogenase (or reduced ferredoxin:heterodisulfide oxidoreductase) is used during growth on acetate, and F420 dehydrogenase (F420, a 5′-deazaflavin, is the universal electron carrier in methanogens) and formylmethanofuran (formyl-MF) dehydrogenase are used during growth on methyl group-containing C1 substrates.
FIG. 3.
Pathways of methanogenesis. Reactions involved in energy conservation are boxed. The reduction of methyl-CoM (reactions 6 and 7) is common to all methanogenic substrates. During methane formation from H2 plus CO2, reactions 1 to 5 proceed in the direction of CO2 reduction. The methyl groups of methanol and acetate enter the central pathway at the level of H4MPT. During methanogenesis from methanol, one-fourth of the methanol is oxidized to CO2 by the reversal of reactions 1 to 5; the six reducing equivalents gained are used to reduce 3 mol of methanol to methane. During methanogenesis from acetate, the carboxyl group is oxidized to CO2 and the electrons gained are used to reduce the methyl group to acetate. F420, oxidized form of coenzyme F420; F420H2, reduced form of F420; HS-CoM, CoM (2-mercaptoethanesulfonate); HS-CoB, CoB (7-mercaptoheptanoylthreonine phosphate); CoM-S-S-CoB, heterodisulfide of HS-CoM and HS-CoB. Enzymes: 1, formyl-MF dehydrogenase; 2, formyl-MF:H4MPT formyltransferase and methenyl-H4MPT cyclohydrolase; 3, F420-dependent methylene-H4MPT dehydrogenase; 4, F420-dependent methylene-H4MPT reductase; 5, methyl-H4MPT:CoM-methyltransferase; 6, methyl-CoM reductase; 7, heterodisulfide reductase system (different electron donor systems are indicated).
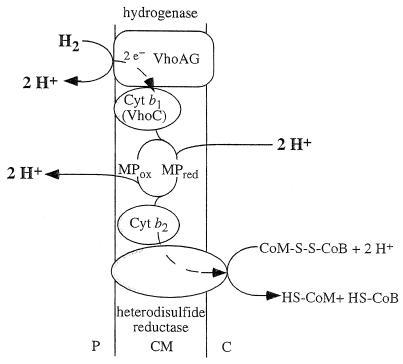
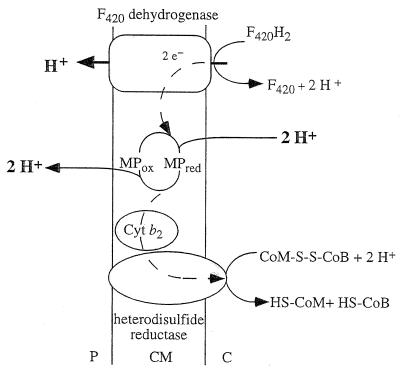
H2-dependent reduction of the heterodisulfide, as catalyzed by inverted vesicles of M. mazei Gö1, was accompanied by H+ translocation into the lumen of the vesicles (Fig. 4). Protonophores inhibited ATP formation but stimulated electron transport, i.e., heterodisulfide reduction. Electron transport and ATP synthesis were inhibited by the ATPase inhibitor N,N′-dicyclohexylcarbodiimide (DCCD), but inhibition was relieved by the addition of protonophores. These effects are clearly reminiscent of respiratory control as observed in mitochondria and can be taken as evidence that the ΔμH+ generated drives the synthesis of ATP from ADP and Pi. Washed everted vesicles exhibited stringent coupling between heterodisulfide reduction and ATP synthesis, with maximal stoichiometries of 1H+ translocated/e− and 1ATP synthesized/4e− (120).
FIG. 4.
Tentative scheme of electron flow and proton translocation during heterodisulfide reduction with H2 as electron donor. This reaction sequence is part of methanogenesis from H2-CO2. This scheme is valid for methylotrophic methanogens only, for hydrogenotrophic methanogens do not contain cytochromes and the presence of methanophenazine (MP) has not been verified. The heterodisulfide reductase is not indicated to be a proton pump, but this cannot be ruled out a priori. This scheme is based on the experimentally derived stoichiometry of 3 to 4 H+ translocated/methyl group reduced. P, periplasm; CM, cytoplasmic membrane; C, cytoplasm. For explanations, see the text.
The F420H2-dependent heterodisulfide reduction was also shown to drive proton translocation into the lumen of everted vesicles of M. mazei Gö1, resulting in the generation of a ΔμH+ (Fig. 5) Protonophores stimulated the heterodisulfide reduction but prevented ΔμH+ formation and ATP synthesis. The ATP synthase inhibitor DCCD decreased the rate of F420H2-dependent heterodisulfide reduction. The reversal of this DCCD-mediated inhibition by protonophores and the stimulation of the F420H2-dependent heterodisulfide reduction by ADP indicate stringent coupling between electron transport and ATP synthesis. The F420H2-dependent heterodisulfide reductase system displayed stoichiometries of 1 H+ translocated/e− and 0.8 ATP synthesized/4e− (122).
FIG. 5.
Tentative scheme of electron flow and proton translocation during heterodisulfide reduction with F420H2 as electron donor. This reaction sequence is part of methanogenesis from methanol, methylamines, and formate. This scheme is valid for methylotrophic methanogens only (see the legend to Fig. 4). F420, coenzyme F420; MP, methanophenazine; P, periplasm; CM, cytoplasmic membrane; C, cytoplasm. For explanations, see the text.
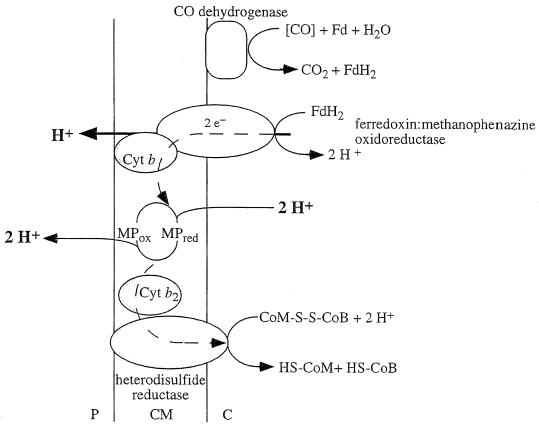
Evidence that the conversion of CO to CO2 and H2 (ΔG°′ = −20 kJ/mol) by resting cells of M. barkeri is coupled to the synthesis of ATP has been presented (70, 71). The cleavage of acetyl-CoA as catalyzed by carbon monoxide dehydrogenase yields enzyme-bound CO and an enzyme-bound methyl group (150, 308). The latter is transferred via a corrinoid protein to tetrahydromethanopterin (H4MPT). Enzyme-bound CO undergoes ferredoxin-dependent oxidation to carbon dioxide, catalyzed by carbon monoxide dehydrogenase (151, 560). In a reconstituted system consisting of purified CO dehydrogenase, heterodisulfide reductase, and ferredoxin, CO oxidation was coupled to heterodisulfide reduction. However, the rate of heterodisulfide reduction was increased 10-fold by addition of membranes, indicating a membrane-bound electron transport chain from ferredoxin to the heterodisulfide (Fig. 6) (420, 512).
FIG. 6.
Tentative scheme of electron flow and proton translocation coupled to heterodisulfide reduction with CO as electron donor. This reaction sequence is part of methanogenesis from acetate. The presence of methanophenazine (MP) in acetate-grown cells has not been verified. Fd, ferredoxin; P, periplasm; CM, cytoplasmic membrane; C, cytoplasm. For explanations, see the text.
Methyl group oxidation proceeds via the reversal of CO2 reduction (reactions 1 to 5 of Fig. 3 in the oxidative direction). There are indications that formyl-MF oxidation is accompanied by the generation of an electrochemical ion potential across the membrane, either protons or sodium ions (262, 603). The formyl-MF-dependent heterodisulfide reduction is associated with a large ΔG°′ of −58 kJ/mol. In contrast, ΔG°′ of the electron transfer from F420H2 to the heterodisulfide is considerably smaller (−29 kJ/mol). Since the physiological electron acceptor employed in the oxidation of formyl-MF to CO2 is unknown, the ΔG°′ of the formyl-MF oxidation cannot be calculated. However, the midpoint potential at pH 7 (Em,7) of the CO2–formyl-MF couple of −500 mV indicates that a low-potential electron carrier can be reduced (47).
Components of the Electron Transport Chain
Since little is known about the formyl-MF-dependent heterodisulfide reduction, only the F420, the H2-, and the CO-dependent systems will be considered here.
Heterodisulfide reductase.
The reaction catalyzed by the heterodisulfide reductase resembles a polysulfide reduction catalyzed by some bacteria and archaea. The S-S bonds of polysulfide can be reduced by H2 as external electron donor, and this reaction is coupled with energy conservation (480).
Heterodisulfide reductase was first purified from H2-CO2-grown M. thermoautotrophicum. It contained three subunits with apparent molecular masses of 80 (HdrA), 36 (HdrB), and 21 (HdrC) kDa and (per mol of heterotrimer) approximately 1 mol of flavin adenine dinucleotide (FAD), 20 mol of nonheme iron, and 20 mol of acid-labile sulfur (205, 508). The encoding genes have been cloned and sequenced (206). Sequence comparisons indicated that HdrA harbors four [4Fe-4S] clusters and binds FAD. HdrC is considered to be an electron carrier protein with two [4Fe-4S] clusters and a short stretch of hydrophobic amino acids that could anchor the complex to the membrane. Interestingly, HdrB is similar to subunit C of the succinate dehydrogenase (SDH) of Acidianus ambivalens and Sulfolobus acidocaldarius.
From membranes of acetate-grown M. barkeri, a heterodisulfide reductase complex which also contained the electron donor, the F420-nonreactive hydrogenase, was purified. This complex contained nine subunits of 46, 39, 28, 25, 23, 21, 20, 16, and 15 kDa, three of which are subunits of the F420-nonreactive hydrogenase. The monomeric heterodisulfide reductase contained 0.7 mol of cytochrome b (cyt b) and 18 mol of nonheme iron and acid-labile sulfur. The 23-kDa subunit carried cyt b (211).
Heterodisulfide reductase itself was purified from membranes of methanol-grown M. barkeri, and the encoding genes were cloned and sequenced (212, 296). The reductase was composed of only two subunits with apparent molecular masses of 46 (HdrD) and 23 (HdrE) kDa. The enzyme contained 0.6 mol of cyt b and 20 mol of nonheme iron and acid-labile sulfur per mol of heterodimer. Biochemical and molecular data revealed that HdrE is a b-type cytochrome with five potentially membrane-spanning helices. HdrD contains two [4Fe-4S] clusters, and its N and C termini are similar to HdrC and HdrB from M. thermoautotrophicum, respectively, indicating that HdrD of M. barkeri and HdrC and HdrB of M. thermoautotrophicum are functionally equivalent. Although small amounts of FAD were found in the heterodisulfide reductase from M. barkeri, it was shown later that heterodisulfide reduction did not depend on FAD. Moreover, no FAD binding site was found in the deduced amino acid sequence of the enzyme (296), which is in contrast to the enzyme from hydrogenotrophic methanogens.
From membranes of acetate-grown Methanosarcina thermophila, a two-subunit heterodisulfide reductase (53 and 27 kDa) was isolated. The small subunit contained 2 mol of cytochrome; the large subunit contained two distinct [Fe4S4]2+/1+ clusters. One heme is a high-spin heme with a midpoint potential of −23 mV, whereas the low-spin heme has a midpoint potential of −180 mV. The midpoint potentials for the two clusters are −100 and −400 mV (512).
Hydrogenases.
The hydrogenases are the entry point for electrons derived from molecular hydrogen. Of the four types of hydrogenases isolated from methanogens to date, one was clearly shown to be involved in energy conservation. The F420-reactive hydrogenase reacts with F420 and viologen dyes, whereas the F420-nonreactive hydrogenase reacts with viologen dyes only. The latter enzyme is therefore often referred to as methyl viologen-reactive hydrogenase. The function of the F420-reactive hydrogenase in energy metabolism is still a matter for debate (14, 78, 338, 385). On the other hand, there is clear evidence that the F420-nonreactive hydrogenase is the electron donor for a membrane-bound electron transport chain. In methylotrophic methanogens, the F420-nonreactive hydrogenase is found in the particulate fraction. The enzyme as purified from M. mazei Gö1 was composed of only two subunits containing redox-active Ni and iron-sulfur clusters (123). A molecular analysis revealed that M. mazei Gö1 contains two operons encoding isoenzymes designated vho for viologen-reactive hydrogenase 1 and vht for viologen-reactive hydrogenase 2. Both operons encode the structural subunits of the hydrogenase (VhoG, VhtG, VhoA, and VhtA) and a gene coding for cyt b (VhoC and VhtC); this indicates that these b cytochromes are the natural electron acceptors of the two F420-nonreactive isoenzymes. The small subunit contains a leader peptide, which suggests that the catalytic part of the enzyme faces the periplasm (121). Interestingly, the C termini of the two b cytochromes are not homologous, indicating that they interact with different proteins. Indeed, Northern blot analysis revealed that the expression of the isoenzymes is substrate dependent. vho was apparently constitutively expressed, whereas vht was expressed only during growth on H2-CO2 or methanol (119). Therefore, it was speculated that the vho gene products are part of the heterodisulfide reductase system whereas the vht gene products are involved in electron flow to and from CO2 in the course of the formyl-MF dehydrogenase reaction (124).
F420 dehydrogenase.
The F420H2 dehydrogenase is the entry point for the electrons derived from F420H2 oxidation. The enzyme was first isolated from Methanolobus tindarius after solubilization from membranes with detergents (182). The apparent molecular mass of the native enzyme was 120 kDa; it consisted of five different subunits of 45, 41, 22, 18, and 17 kDa. The enzyme contained 16 mol of nonheme iron and 16 mol of acid-labile sulfur per mol, but flavin was not detected. The gene encoding the 40-kDa subunit (ffdB) was cloned (600). Sequence analysis, primer extension, and reverse transcription-PCR indicated that ffdB is part of an operon harboring three additional genes (ffdA, ffdC, and ffdD). FfdA is similar to the F420-dependent methylene-H4MPT reductase. The first 90 amino acids of FfdB are similar to numerous ferredoxins, suggesting the likely presence of at least two iron-sulfur centers. FfdC and FfdD are similar to proteins of unknown function of Methanococcus jannaschii and Archaeoglobus fulgidus. FfdD appears to be very hydrophobic and is likely to be the membrane anchor. Recently, F420 dehydrogenases were purified from M. mazei Gö1 and the sulfate-reducing archaeon A. fulgidus (3, 297). Flavin was detected in both enzymes. Therefore, it is likely that flavin is also present in the enzyme from M. tindarius but lost during purification.
Membrane-Integral Electron Carriers
With respect to their membrane-integral electron carriers, and thus probably with respect to the mechanism of proton translocation, methanogens can be divided into two groups: the methylotrophic organisms, in which a variety of b- and c-type cytochromes were found, and the hydrogenotrophic methanogens, which are devoid of cytochromes (260, 295). In hydrogenotrophic methanogens, the situation is far from settled; polyferredoxins described above and a recently described flavoprotein encoded by the gene fpaA (394) are the only electron carriers identified so far.
In methylotrophic methanogens, there are several lines of evidence for the involvement of cytochromes in electron transport from the F420-nonreactive hydrogenase to the heterodisulfide in methylotrophic methanogens. First, membranes of acetate-grown cells catalyze an H2-dependent reduction of cytochromes (275, 560), and second, hdrE (designated cyt b2) is part of the heterodisulfide reductase operon and was expressed during growth on H2-CO2 (296). The vho operon encoding cyt b1 along with the structural subunits of the F420-nonreactive hydrogenase was also expressed during growth on H2-CO2 (119). Therefore, an electron flow from the F420-nonreactive hydrogenase via cyt b1 and b2 to the heterodisulfide can be envisaged (Fig. 4).
Experiments performed with M. mazei Gö1 strongly suggest that one or several cytochromes also participate in electron transport from F420H2 to the heterodisulfide (264). Membranes of M. mazei Gö1 contain two b- and two c-type cytochromes with midpoint potentials (Em,7) of −135 and −240 mV (b-type cytochromes) and −140 and −230 mV (c-type cytochromes). The cytochromes were reduced by F420H2 and oxidized by the heterodisulfide at high rates. Addition of the heterodisulfide to reduced cytochromes and subsequent low-temperature spectroscopy showed the oxidation of cyt b564. This indicates the involvement of cytochromes in electron transport from F420 via cyt b to the heterodisulfide (Fig. 5).
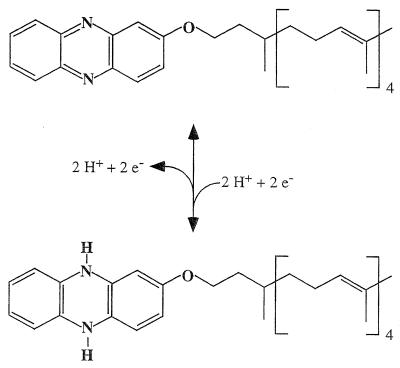
A different class of membrane-bound electron carriers was discovered recently (1). Membranes of methanogens do not contain typical quinones found in bacteria or aerobic archaea. However, extraction of membranes from methanol-grown M. mazei Gö1 with isooctane yielded a fraction containing a redox-active, low-molecular-weight compound identified as a phenazine derivative, methanophenazine. The structure and reactivity of methanophenazine are given in Fig. 7. Methanophenazine is reduced by F420 dehydrogenase or hydrogenase, and reduced methanophenazine then reduces the heterodisulfide (2, 40). In a reconstituted system consisting of purified F420 dehydrogenase and heterodisulfide reductase, methanophenazine mediated the electron transfer from F420 to the heterodisulfide (40). Methanophenazine was isolated from methanol-grown M. mazei Gö1, but it is probably also involved in electron transport to the heterodisulfide from other donors, i.e., formyl-MF and CO. The most interesting question, whether methanophenazine is also present in hydrogenotrophic methanogens, remains to be solved.
FIG. 7.
Structure and reactivity of methanophenazine, a membrane-integral electron and hydrogen carrier of methanogens.
Possible Mechanisms of ΔμH+ Formation Coupled to Electron Transport Reactions
In methylotrophic methanogens, the F420-nonreactive hydrogenase is localized in the periplasm, as inferred from its leader sequence and its homology to membrane-bound, cytochrome-containing bacterial hydrogenases. Formation of a proton potential can be easily envisaged, because the uptake of H2 and transfer of electrons to an electron acceptor would lead to the liberation of scalar protons on the outside of the cytoplasmic membrane. However, the H+/CH4 stoichiometry of 3 to 4 (measured in M. barkeri during methanogenesis from methanol-H2 [57]) cannot be accounted for by scalar protons only. That leaves us with the question of the nature of the vectorial proton pump. Electron flow from F420H2 to methanophenazine, as well as from the reduced methanophenazine to the heterodisulfide, is coupled to proton translocation, indicating the presence of two coupling sites (2). The F420H2 dehydrogenase and the H+-translocating bacterial NADH dehydrogenase have in common a complex structure and the presence of flavins and iron-sulfur centers. Therefore, it is tempting to speculate that the F420H2 dehydrogenase, like the NADH dehydrogenase, is a proton pump. It is not known whether the heterodisulfide reductase itself is a proton pump. The genes encoding the hydrogenase, the heterodisulfide reductase, and part of the F420 dehydrogenase are known, but the similarities of the deduced proteins to subunits of NADH dehydrogenases or cytochrome oxidases are too low to allow identification of polypeptides involved in proton transport.
With the discovery of methanophenazine, another possibility has emerged. By analogy with the ubiquinone cycle in the bc1 complex, it is likely that electron transfer from cyt b1 to methanophenazine is coupled to proton uptake from the cytoplasm. The reduced methanophenazine then donates its electrons to cyt b2, and the protons are liberated into the periplasm. The H+/e− stoichiometry of such a mechanism would be fixed at 1H+/e−.
Sodium Bioenergetics of Methanogenesis
Apart from the proton motive electron transport chain, methanogens have a primary sodium ion pump, the methyl-H4MPT:CoM methyltransferase (44, 149, 384). This enzyme is part of the central pathway, and therefore Na+ transport is obligatory for methane formation. This enzyme represents the first example of a methyltransferase catalyzing ion transport across a membrane. Because the central pathway is reversible, this enzyme functions as generator of a sodium ion potential during methanogenesis from CO2 or acetate but as an endergonic reaction driven by the sodium ion potential in the course of methyl group oxidation, which has to be carried out during methanogenesis from methyl group-containing C1 compounds (380). Unlike the cytochromes and the resulting differences in the electron transport chains, the methyltransferase is found in every methanogen, and there is no reason to assume different reaction mechanisms.
The energetics of the methyltransferase was first investigated by using cell suspensions of M. barkeri and the substrate combination H2-HCHO. Upon addition of the substrate, sodium ions were actively extruded from the cytoplasm, resulting in the generation of a transmembrane Na+ gradient of −60 mV. Na+ translocation was not inhibited by protonophores or inhibitors of the Na+-H+ antiporter, indicating a primary mechanism. This process resulted in the generation of a protonophore-resistant membrane potential of −60 mV; correspondingly, protonophores elicited formation of a reversed ΔpH (inside acidic) of the same magnitude as the ΔΨ (384). A Na+-formaldehyde stoichiometry of 3 to 4 was determined with cell suspensions (261). By the use of everted vesicles of M. mazei Gö1, the methyltransferase was identified as a Na+ pump (44); this was later corroborated with the purified enzyme reconstituted into liposomes. These proteoliposomes catalyzed an electrogenic Na+ transport with a stoichiometry of 1.7 mol of Na+ per mol of methyl-H4MPT demethylated (317).
The methyltransferase contains the cofactor Coα–[α-(5-hydroxybenzimidazolyl)]-cobamide (factor III), which is involved in methyl transfer (163, 164, 427). The cofactor in its superreduced Co(I) form accepts the methyl group from methyl-H4MPT, giving rise to a methyl-Co(III) intermediate. In the second partial reaction, this methyl-Co(III) is subjected to a nucleophilic attack, probably by the thiolate anion of CoM, to give rise to methyl-CoM and regenerated Co(I) (149, 164):
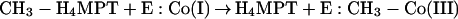 |
3 |
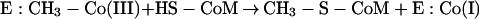 |
4 |
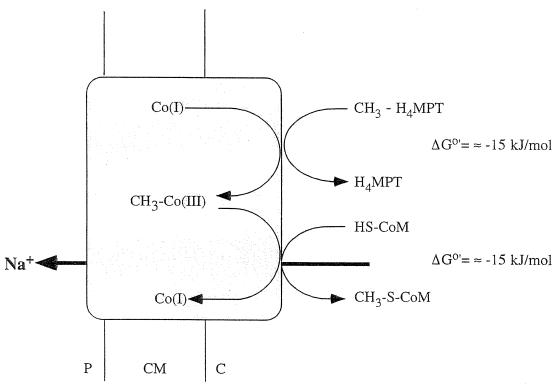
Reaction 3 has a free-energy change of −15 kJ/mol and was not stimulated by sodium ions. On the other hand, demethylation of the enzyme-bound corrinoid (reaction 4) is also accompanied by a free-energy change of −15 kJ/mol, and this reaction was sodium ion dependent, with half-maximal activity obtained at approximately 50 μM Na+. This finding indicates that the demethylation of the enzyme-bound corrinoid is coupled to sodium ion translocation (599) (Fig. 8).
FIG. 8.
Tentative scheme of the reaction mechanism of the Na+-translocating methyl-H4MPT:CoM-methyltransferase. The enzyme is a multisubunit enzyme consisting of eight nonidentical subunits in unknown stoichiometry. The reaction can be divided into two partial reactions, methylation and demethylation of an enzyme-bound corrinoid cofactor. The demethylation reaction is apparently coupled to Na+ transport. Co(I) and Co(III) denote different valence states of the enzyme-bound corrinoid cofactor. HS-CoM, CoM (2-mercaptoethanesulfonate); P, periplasm; CM, cytoplasmic membrane; C, cytoplasm. For explanations, see the text.
The methyltransferase was purified from M. thermoautotrophicum and M. mazei Gö1 (163, 317). In the latter, six subunits were found, with apparent molecular masses of 34, 28, 20, 13, 12, and 9 kDa; it contains a [4Fe-4S] cluster with an E0′ of −215 mV and a base-on cobamide with a standard reduction potential of −426 mV for the Co2+/1+ couple (324). In M. thermoautotrophicum, eight subunits were found, with apparent molecular masses of 34 (MtrH), 28 (MtrE), 24 (MtrC), 23 (MtrA), 21 (MtrD), 13 (MtrG), 12.5 (MtrB), and 12 (MtrF) kDa. The purified enzyme contains 2 mol of corrinoid, 8 mol of nonheme iron, and 8 mol of acid-labile sulfur (163, 192). The encoding genes have been sequenced from a number of methanogens; they are organized in an operon in the order mtrEDCBAFGH. Hydrophobicity plots indicate that all of the subunits except MtrA and MtrH are hydrophobic and potentially membrane bound. Very recently, the membrane localization of MtrD was confirmed experimentally for M. mazei Gö1, M. thermoautotrophicum, and M. jannaschii (456). This subunit may be directly involved in Na+ transport (318).
MtrA was overexpressed, purified from Escherichia coli, and successfully reconstituted with cobalamin. Electron paramagnetic resonance (EPR) spectroscopic studies indicate that the cobalamin is in the base-off form and that the axial ligand is a histidine residue of MtrA (191). From this observation, a hypothetical mechanism was formulated for coupling the methyl transfer reaction to ion transport via a long-range conformational change in the protein (191). It is known that cob(II)alamin and cob(III)alamin, but not cob(I)alamin, carry an axial ligand. Methylation of cob(I)alamin gives rise to a methylcob(III)alamin, which is then able to ligate the histidine residue; demethylation leads to a reversal of this reaction. It is easily conceivable that binding and dissociation of the histidine residue with the corrinoid lead to a conformational change in the hydrophilic part of the enzyme. This change is then transmitted to the membrane-bound subunits, giving rise to Na+ transport. Work on the structure and function of this interesting enzyme is just emerging but is apparently well on its way.
ATP Synthesis in Methanogens
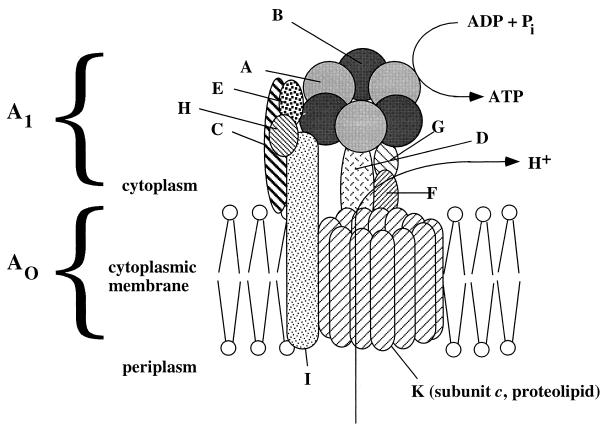
Methanogens are the only microorganisms known to produce two primary ion gradients, ΔμNa+ and ΔμH+, at the same time. They are, therefore, confronted with the problem of coupling both ion gradients to the synthesis of ATP (124). How this is achieved is still a matter for debate. There have been conflicting reports regarding ΔμNa+-driven ATP synthesis in the hydrogenotrophic archaeon M. thermoautotrophicum. Smigan and coworkers (515, 516) had indications of a Na+-ATPase along with a H+-ATPase, whereas Kaesler and Schönheit favored a mechanism in which the ΔμNa+ established by the methyltransferase reaction is converted to a secondary proton gradient that then drives synthesis of ATP via a H+-translocating A1Ao ATPase (262). The latter hypothesis is supported by the finding that the genomes of the hydrogenotrophic methanogens M. jannaschii and M. thermoautotrophicum contain genes that encode the A1Ao ATPase but lack those of F1Fo ATPase (87, 517). On the other hand, differential inhibitor studies indicated the simultaneous presence of both A1Ao and F1Fo ATP synthases in M. mazei Gö1 (43). The A1Ao enzyme may be coupled to H+ transport, whereas the F1Fo ATPase may be Na+ coupled. However, no F1Fo ATPase could be purified from M. mazei Gö1, nor have the encoding genes been detected. In another organism, M. barkeri MS, a gene cluster encoding an F1Fo ATPase has been identified in addition to the archaeal A1Ao ATPase genes (549); however, the deduced γ subunit is very unusual and presumably nonfunctional, and no gene encoding subunit δ was found. Since an mRNA transcript could not be detected in cells grown on methanol, it is doubtful that the F1Fo-like genes are expressed in M. barkeri (312). The presence of both F1Fo and A1Ao was also proposed for halobacteria (227, 230). Most likely, the F1Fo ATPase genes definitely present at least in M. barkeri MS have arisen from horizontal gene transfer. In line with this argument is the discovery of V1Vo ATPases in bacteria (433, 620). However, the presence of the F1Fo ATPases in Methanosarcina species still has to be proven biochemically, and, if present, their contribution to energy metabolism has to be clarified. Apparently, the mechanism for ΔμNa+-driven ATP synthesis differs among the methanogens. The structure and function of the A1Ao ATPases are discussed below (“Secondary Energy Converters”).
Bioenergetics of the Acetyl-CoA Pathway in Archaea and Bacteria: Differences and Similarities
One of the major differences between the anaerobic bacteria and the archaea that employ the acetyl-CoA pathway is the way that CO2 is activated. In bacteria, this requires the action of formate dehydrogenase and formyltetrahydrofolate synthase, at the expense of ATP hydrolysis. In the reverse reaction, oxidation of formyltetrahydrofolate is coupled to ATP synthesis by substrate-level phosphorylation (320). In methanogens, the low redox potential of the [CO2 + MF]/[formyl-MF] couple of approximately −500 mV (47) is overcome, not by ATP hydrolysis, but by a reversed electron flow driven by the transmembrane ion (H+ or Na+) potential (262).
Whereas all methanogens tested so far require Na+ for growth and methane formation (422, 423), homoacetogens can be divided into two groups with respect to their energy metabolism, the proton organisms and the sodium ion organisms. In the latter, an as yet unidentified primary sodium ion pump is operative. Since these organisms have membrane-bound corrinoids, it is speculated that the methyltransferase is the sodium ion pump (382). If this is the case, this would allow a study of the evolution of Na+-translocating methyltransferases.
The Na+ gradient established in methanogens is coupled to ATP synthesis, but the mechanisms involved are still controversial and may differ among the various methanogens. In homoacetogens, H+-translocating ATPases are found in proton organisms (112, 113), but a Na+-translocating F1Fo ATP synthase was found in the Na+-dependent homoacetogen Acetobacterium woodii (214, 439). The finding of Na+-ATPases in homoacetogens strengthens the assumption that Na+-ATPases are also present in methanogens.
ENERGETICS OF RESPIRATION
Aerobiosis and Other Respiration Forms in Archaea
Obligate aerobes are relatively uncommon among the archaea. Given the phylogenetic position of Archaea, this may reflect the prevalence of anaerobic energy-transducing reactions at early stages of evolution; likewise, all organisms branching off at the bottom of the phylogenetic tree are hyperthermophiles, in conformity with the assumption that life originated in hot environments (6, 540).
Table 1 gives an overview of the archaea that grow obligately or facultatively with oxygen or other high-potential terminal electron acceptors. Only those for which sufficient data is available are included. The genus Acidianus displays obligate chemolithoautotrophic growth with CO2 as the sole carbon source. Most other species are facultative or obligate heterotrophs. The autotrophic growth of S. acidocaldarius with sulfur as electron donor, as reported for the original isolates (82), has to be questioned since the deposited type strains (DSM 639 and ATCC 33909) are incapable of such growth.
TABLE 1.
Overview of growth conditions and energy sources for aerobic archaea
| Species | Aerobiosisa | Growthb | Tmax (°C)c | pH ranged | e− acceptor(s) | e− donor(s)e | End product(s) |
|---|---|---|---|---|---|---|---|
| Sulfolobus acidocaldarius | o | 85 | 1–5 | O2 | Organic [H], cell | ||
| Sulfolobus solfataricus | o | h | 87 | 3–5 | O2 | Sugars, amino acids | H2O; H2SO4; CO2 |
| Sulfolobus shibatae | o (f) | h | 86 | 3–5 | O2; (Fe3+, Mo4+) | H2S/[H2] | |
| Sulfolobus metallicus | o | f/(f) | 75 | ? | O2 | Sulfidic ores; S2−/S0 | H2SO4 |
| Metallosphaera sedula | o | a | 80 | 1–4.5 | O2 | Sulfidic ores/S2−/S0 | H2SO4 |
| Acidianus infernus | f | a | 95 | 1.5–4 | O2/S0 | Sulfidic ores/S2−/S0/H2 | H2SO4; H2S |
| Acidianus brierleyi | o (f) | a | 75 | 1.5–4 | O2/S0 | S2−/S0 | H2SO4; H2S |
| Acidianus ambivalens | f | a | 95 | 1–4 | O2/S0 | S2−/S0 | H2SO4; H2S |
| Stygiolobus azoricus | an | a | 89 | 1.5–5 | S0 | H2 | H2S |
| Pyrobaculum aerophilum | f | h | 103 | 5.8–9 | O2/NO3− | Peptone/yeast/H2 | H2O; CO2?; N2? |
| Pyrobaculum islandicum | an | f/(h) | 103 | 5–7 | S0/S2− | H2/cell/peptone | H2S |
| Pyrobaculum organotrophicum | an | h | 104 | 5–7 | S0/S2−/S2O3− | Yeast/peptone | H2S |
| Thermoplasma acidophilum | o | h | 65 | 0.5–4 | O2 | Yeast/sugars | H2O; CO2 |
| Thermoplasma volcanium | o | h | 65 | 0.5–4 | O2/S0 | Yeast/sugars | H2O; CO2 |
| Picrophilus oshimae | o | h | 60 | 0.5–2.2 | O2 | Yeast + sugar | H2O; CO2 |
| Picrophilus torridus | o | h | 60 | 0.5–2.2 | O2 | Yeast + sugar | H2O; CO2 |
| Halobacterium salinarum | o/hν | h | m | n | O2 | Cell/yeast | H2O; CO2 |
| Halobacterium saccharovorum | o/hν | h | m | n | O2 | Cell/yeast | H2O; CO2 |
| Haloferax mediterranei | o/hν | h | m | n | O2 | Cell/yeast | H2O; CO2 |
| Haloferax volcanii | o (f) | h | m | n | O2/NO3− | Cell/yeast | H2O; N2; CO2 |
| Haloferax denitrificans | f | h | m | n | NO3−/O2 | Cell/yeast | N2; H2O; CO2 |
| Natronobacterium pharaonis | o/hν | h | 45 | 7.5–9.5 | O2 | Amino acids/carbonic acids | H2O; CO2 |
o, obligate aerobe; f, facultative respiration with acceptors other than oxygen; an, obligate anaerobe; hν, auxiliary phototrophic energy conservation.
h, heterotrophic; a, autotrophic; f, facultatively heterotrophic.
Maximum temperature at which growth occurs. m, mesophilic.
n, neutrophilic.
cell, cell extract; yeast, yeast extract.
Members of the genus Sulfolobus are obligate aerobes. Interestingly, some Sulfolobus isolates were found to reduce ferric ions or molybdate as terminal acceptors under low oxygen tension (80, 83). In addition, Sulfolobus and Acidianus strains have been shown to grow aerobically by the oxidation of molecular hydrogen (Knallgas reaction) (236) at low oxygen concentrations (0.2 to 0.5%).
Alternatively, Acidianus species can derive energy from the reduction of sulfur by molecular hydrogen under anoxic conditions. This sulfur respiration appears to be the preferential energy source: Acidianus is thus classified as a facultative aerobe.
With the exception of Pyrobaculum, the extreme thermophiles in Table 1 are also extreme acidophiles (optimal growth at pH 1 to 3.5), which imposes a bioenergetic challenge regarding the maintenance of a nearly neutral cytosol. In contrast, the members of the aerobic order Halobacteriales are either neutrophilic or alkaliphilic. However, only halobacteria can synthesize purple membranes and use light as an additional energy source. It has been proposed that respiration was the primary energy-transducing mechanism of halobacteria and that the light-driven ion pumps might reflect later adaptations to low oxygen tension (513), an inevitable consequence of the extremely high salinity of their natural habitat.
Not only oxygen reduction but also various forms of anaerobic respiration have been reported as energy sources for archaea. Table 2 summarizes the free-energy changes of the respiratory redox systems used as primary energy sources in archaea. Haloferax mediterranei, Haloferax denitrificans, and Haloferax volcanii are capable of reducing nitrate (as terminal acceptor) to nitrogen (407, 566, 567); several halobacterial nitrate reductases have been described (16, 50, 229). The hyperthermophile Pyrobaculum aerophilum also reduces nitrate under anoxic conditions (586). Moreover, some members of the halobacterial family perform fumarate respiration (406) or grow fermentatively on arginine (193). Purely anaerobic respiration was reported for the genera Thermoproteus, Pyrodictium, Desulfurococcus, Archaeoglobus, and Thermodiscus (7, 483, 540).
TABLE 2.
Standard free energy changes of aerobic and anaerobic respiratory reactions identified as energy sources for growth of archaeaa
| Reaction | Redox system | −ΔG°′ (kJ/mol) |
|---|---|---|
| 1 | H2 + 1/2O2 → H2O | 236.6 |
| 2 | 2S0 + 2H2O + 3O2 → 2H2SO4 | 1,014.0 |
| 3 | 2FeS2 + 2H2O + 7O2 → 2FeSO4 + 2H2SO4 | 1,498.9 |
| 4 | [H]2-X + 1/2O2 → H2O + X (X = NADH2, QH2 succinate, etc.) | 219.0 |
| 5 | S0 + 2[H] → H2S | 33.5 |
| 6 | SO42− + 8[H] + 2H+ → H2S + 4H2O | 151.7 |
| 7 | NO3− + 8[H] + 2H+ → NH4+ + 3H2O | 598.7 |
| 8 | 2NO + 2H+ + 2e− → N2O + H2O | 305.9 |
| 9 | N2O + 2H+ + 2e− → N2 + H2O | 341.1 |
The free energy values were calculated from redox potentials or the energies of formation as described in reference 562.
In contrast to oxygen respiration, none of the alternate electron transport systems has yet been elucidated in detail at the level of genes or proteins.
Components of Aerobic Electron Transfer
The paradigm derived from mitochondrial respiratory chains and from studies of purple bacteria suggests the presence of four major complexes for optimal energy conservation. In this scheme, complex I acts as an energy-transducing NADH dehydrogenase on the low-potential side, and complex II serves as SDH; both are Q reductases. Reduced quinones are reoxidized by complex III, the so-called bc1 complex, which transfers electrons to complex IV, the terminal oxidase, via cyt c. In contrast to the classical concept, membrane-integral archaeal electron transfer complexes connected by mobile carriers can be fused to supercomplexes and, in some cases, have unusual compositions. Both membrane-integral redox complexes and mobile electron carriers are discussed in the following sections.
Membrane-residing quinone reductases.
NADH dehydrogenases (complex I) and SDHs are the major reductants of quinones in all respiratory chains. Two types of the former dehydrogenase are known, NDH-I and NDH-II; only NDH-I types are capable of H+ or Na+ pumping. Whereas membrane-associated NDH-II activities and NADH-dependent respiration have been found in archaea, nothing equivalent to an energy-transducing complex I has been detected.
(i) NADH dehydrogenases.
Older reports on the characterization and partial purification of NADH dehydrogenase activities from halobacteria are reviewed in reference 226. These activities were usually measured with redox dyes as electron acceptors; however, inhibition by the quinone analog 2-heptyl-4-hydroxyquinoline-N-oxide has been interpreted to indicate the in vivo transfer of electrons to the quinone pool (302). Sulfolobus membranes oxidize NADH with low activity in a cyanide-sensitive reaction (21, 592). None of these activities was sensitive to rotenone, amytal, piericidine, or antimycin. From the inhibition of cell respiration by acridone carbonic acid derivatives (402), it was concluded that S. acidocaldarius has an NDH-II type enzyme (463). This activity is only loosely associated with the membrane.
An NADH:acceptor oxidoreductase from the cytosol of Sulfolobus sp. strain 7 has been purified as a dimeric, water-soluble, 95-kDa protein with two molecules of FAD/molecule of protein (595); it may represent the membrane-peripheral fraction of a larger complex, because there was virtually no activity with caldariella quinone (Qcal) as acceptor. This has not yet been confirmed. Interestingly, a similar enzyme has been isolated from aerobically grown A. ambivalens and characterized (177). The monomeric, 76-kDa protein contains FAD and has NADH:acceptor oxidoreductase activity. However, from EPR spectroscopic studies on its interaction with ferredoxin, the authors conclude that the protein might function in vivo as an NADH:ferredoxin oxidoreductase.
Numerous inhibitors of respiratory electron transport in membranes of Halobacterium salinarum have been tested, confirming that this NADH-oxidizing activity is also due to an NDH-II type enzyme which is not involved in energy conservation (531). In contrast to Sulfolobus, the membrane-bound enzyme is capable of transferring reducing equivalents to horse heart cyt c as experimental acceptor.
Sequence data is not available for any of these NADH dehydrogenases from halobacteria or members of the Sulfolobales. The ongoing genome sequencing projects may help to clarify whether the genes for an energy-conserving NDH-I complex are completely absent from archaea. Surprisingly, open reading frames in the genome of the obligate anaerobe A. fulgidus could be attributed by similarity to genes of NADH dehydrogenase (subunit 1) and of cytochrome oxidase (281). Similar attributions have been made for genes from M. thermoautotrophicum (517), M. jannaschii (87), or P. aerophilum (153). However, these alignments are only fragments of the analogous bacterial or eucaryal genes, respectively; moreover, they usually concern only a single polypeptide. Furthermore, there is no indication of the presence of any of the remaining (up to 14) polypeptides, e.g., the iron-sulfur proteins, required for completion of a functional complex I.
In conclusion, attempts to isolate an archaeal integrated NADH:quinone reductase complex have failed so far. Cumulative evidence for the absence of a complex I analog in membranes from Sulfolobus and H. salinarum was mainly derived from three observations: (i) none of the characteristic iron-sulfur centers of complex I-type NADH dehydrogenases could be detected by EPR spectroscopy (309, 310), (ii) no sensitivity to complex I inhibitors could be demonstrated, and (iii) hybridization with NDH-I-directed DNA probes (against NDH-I-encoding genes of Paracoccus denitrificans) failed.
(ii) SDHs and a novel complex II.
Succinate-stimulated respiration and SDH activities with redox dyes as electron acceptors have been reported for several aerobic archaeal species (10, 20, 477, 592).
An unusually high concentration of SDH was found in membranes of Thermoplasma acidophilum (20); this allowed direct characterization of the three iron-sulfur clusters, S1 [2Fe-2S], S2 [4Fe-4S], and S3 [3Fe-4S], with redox potentials of +60 and +68 mV for clusters S3 and S1, respectively. The S2 cluster was indirectly detected by spin-spin interaction. The physiological significance of the high SDH activity in T. acidophilum remains unexplained. The enzyme complex has not been purified to homogeneity.
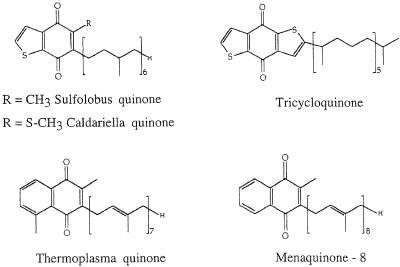
Well-characterized preparations were obtained from the thermoacidophiles S. acidocaldarius (371) and Sulfolobus sp. strain 7 (250) and from the haloalkaliphile N. pharaonis (477). Complete genetic analyses of archaeal SDH operons are presently available for only S. acidocaldarius and N. pharaonis (254, 477). Figure 9 shows a comparison of gene arrangements in archaeal SDH operons, with reference to E. coli as a bacterial representative. While the order of the genes varies, the basic polypeptide composition is in agreement with the known complex II (SDH-fumarate reductase) (184, 207). The catalytic core consists of a larger (approximately 65 kDa) FAD-containing subunit and a smaller polypeptide (26 to 37 kDa) that bears three FeS clusters. This is accompanied by two smaller subunits which usually function as a membrane anchor. One of these may host a heme B. Its participation in electron transport is not entirely certain; it may also have structural functions (184). Heme B was found in archaeal SDH complexes from T. acidophilum (29) and N. pharaonis (477) but not in those from members of the Sulfolobales.
FIG. 9.
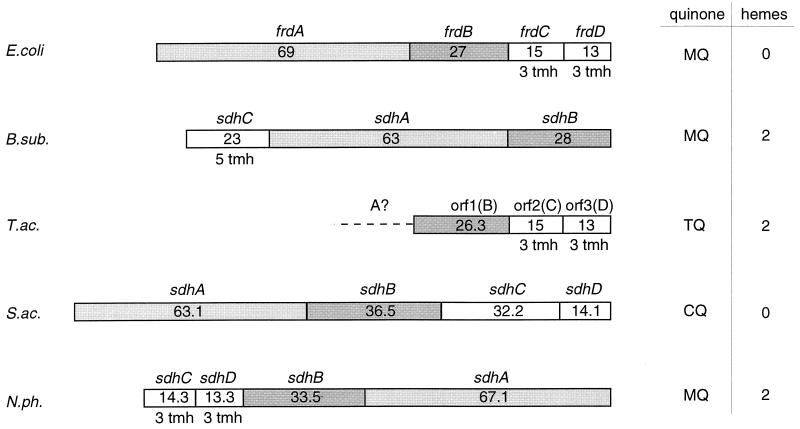
Gene organization within archaeal SDH operons. Numbers indicate the calculated molecular masses (in kilodaltons) of the gene products. In all cases, subunit A is the flavin-containing dehydrogenase polypeptide; subunit B refers to the iron-sulfur protein in complex II or fumarate reductase, respectively. The columns on the right indicate the type of quinone used as terminal electron acceptor in vivo and the number of heme B molecules present in the complex. MQ, menaquinone; TQ, Thermoplasma quinone, CQ, caldariella quinone. tmh, putative transmembrane helices. E.coli, E. coli (256); B.sub., B. subtilis (353); T.ac., T. acidophilum (29); S.ac., S. acidocaldarius (254) (accession no. Y09041); N.ph., N. pharaonis (accession no. Y07709). The assignment of the open reading frames was done by analyzing sequence similarities. From Thermoplasma, only a partially sequenced operon has been reported; ORF-1 has homology to sdhB and frdB; ORF-2 is a putative diheme cyt b.
Two complex II preparations from Sulfolobus (250, 294) deserve special attention. In contrast to other archaeal SDH complexes, that from Sulfolobus sp. strain 7 was reported to reduce Qcal readily; succinate respiration could be restored in liposomes by reconstitution of the preparation with a terminal Q-oxidizing complex (250). This is the only direct evidence that in Sulfolobus reducing equivalents are transferred from succinate to terminal oxidases via CoQ.
This observation is in obvious contrast to the unusual properties of a novel complex II from S. acidocaldarius (254, 371). The purified complex is inactive with Qcal. In isolated membranes, electron transfer from succinate to respiratory Rieske FeS proteins is evident from EPR spectroscopy (471), but the exact electron path is unclear. The involvement of a radical species was indicated by the strong inhibitory interaction with the free radicals of 2,3,5,6-tetrachlorobenzoquinone (371). The complex dissociates easily from the membrane. Genetic analysis of the SDH operon (254) suggests that the two small polypeptides are essentially hydrophilic; indeed, none of the polypeptides contained a clear membrane-spanning α helix. In addition, subunit B, which hosts the FeS clusters in all SDHs and fumarate reductases (184), is likely to bear in S. acidocaldarius a second [4Fe-4S] cluster instead of the regular [3Fe-4S] cluster S3. This might be indicative of a different electron pathway that eventually involves thiols, because subunit C also exhibits a novel and unusual sequence pattern containing a repetitive, cysteine-rich motif, CX31–37CCX26–35CX2C. This motif has also been detected in a hypothetical protein sequence of Synechocystis and in the heterodisulfide reductase of M. thermoautotrophicum (206). Its functional significance is not understood, but it may explain the failure of S. acidocaldarius SDH to directly reduce Qcal in vitro. The recent finding in methanogen membranes of a phenazine isoprenoid (1, 40) which functions as an intermediate electron carrier has encouraged the search for an equivalent in aerobic archaea. It may bridge the gap in electron transfer from SDH in S. acidocaldarius to Qcal. A dual function of the complex also appears possible: it may function as a fumarate reductase under autotrophic growth conditions, with a hitherto unknown source of reducing equivalents, analogous to a thiol-driven fumarate reductase of M. thermoautotrophicum (206). The properties of the two well-characterized SDH complexes are compiled in Table 3.
TABLE 3.
Succinate:acceptor oxidoreductasesa
| Parameter | S. acidocaldarius (DSM 639) | Sulfolobus sp. strain 7 |
|---|---|---|
| No. of subunits | 4 | 4 |
| Mr (103) | 138 | 148 |
| Mr subunits (103) | 66, 31, 28, 12.8 | 66, 37, 33, 12 |
| Genes | sdhABCD | NA |
| Flavin (nmol/mg) | 4.6 | 5.6 |
| λmax (nm) | 455 (ox-red) | 445 |
| Fe (nmol/mg) | 102 | 83 |
| S0 (nmol/mg) | 150 | — |
| Cyt b | None | None |
| Km (succinate) (mM) | 1.4 | 0.28 |
| Km (DCPIP) (μM) | 65.4 | 89 |
| Km (Qcal) (μM) | — | 60 |
| Vmax (μmol/min/mg)b | 7.8 (55°C) | 13 (50°C) |
| — | 14 (Q1) | |
| — | 1.8 (Qcal) | |
| Turnover (s−1) | 154 (81°C) | — |
| EPR | ||
| (S-3) oxidized | g = 2.02 | g = 2.02 |
| g = 2.08 (satell.) | — | |
| (S-1) succinate reduced | gz = 2.05 | gz = 2.03 |
| gy = 1.935 | gy = 1.94 | |
| gx = 1.904 | gx = 1.90 | |
| pH optimum | 6.5 | 6.5–6.8 |
| Ea (kJ/mol) | 59–64 | — |
| Ki | ||
| Malonate (mM) | 3.1 | — |
| Oxaloacetate (mM) | 0.28 | — |
| TCBQc (μM) | 1.5 | — |
Comparison of structural and kinetic data for the only two archaeal SDH preparations (complex II). Data were taken from references 250, 369, and 371. NA, not available; Qcal, caldariella quinone; Q1, ubiquinol-1; (satell.), satellite band; —, not tested or not reported.
Activity with artificial electron acceptors phenazine methosulfate (PMS) and 2,6-dichlorophenolindophenol (DCPIP).
TCBQ, tetrachlorobenzoquinone.
Membrane-integral quinol-oxidizing complexes.
Quinol oxidation in aerobic respiration is usually accomplished by integral membrane-protein complexes acting sequentially as redox-driven proton pumps. The identification and mechanisms of the respective archaeal proton pumps are still under debate. Detailed investigations have been reported for just a few extremely thermoacidophilic archaea and two halobacterial species. The latter appear to possess unbranched quinol-oxidizing respiratory chains, with complexes essentially similar to those from mitochondria or purple bacteria. In contrast, unusual and novel supercomplexes from members of the Sulfolobales have been isolated and characterized.
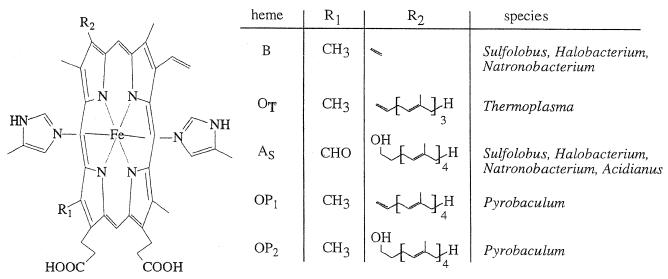
Membrane-bound quinol-oxidizing systems contain multiple hemoproteins detectable by difference spectroscopy in visible light. Indeed, archaebacterial membranes were shown to reveal composite (reduced-oxidized) difference spectra by superimposing various cytochrome spectra. Table 4 gives a comprehensive listing of cytochromes from aerobically growing archaea, together with their redox potentials, heme types, and tentative functions.
TABLE 4.
Properties of cytochromes from thermoacidophilic aerobic archaeaa
| Cytochrome type and source | Molecular mass (kDa) | Gene(s) | Cofactor(s) | E0 (mV) | Function(s) | Reference(s) |
|---|---|---|---|---|---|---|
| b558/566, S. acidocaldarius (DSM 639) | 66 (PAGE) | cbsA, cbsB | 1 heme B | >300 | Highly glycosylated; unknown function; exposed to periplasm | 45, 220 |
| 55 (deglyc.) | ||||||
| 50.5 (DNA) | ||||||
| b562, S. acidocaldarius (DSM 639) | 45 (PAGE) | soxM | 1 heme B | ND | Alternate terminal ba3 oxidase complex | 327, 328 |
| 87.1 (DNA) | 1 heme As | |||||
| 1 Cu | ||||||
| b563, Sulfolobus sp. strain 7 | 37 (PAGE) | NA | 1 heme B | 146 | Component of respiratory complex | 246, 247 |
| a587 I, S. acidocaldarius (DSM 639) | 39/64b (PAGE) | soxC | 2 heme AS | 210 | Component of terminal oxidase SoxABCD | 92, 173, 175 |
| 62.8 (DNA) | 270 | |||||
| a587 II, S. acidocaldarius (DSM 639) | 37 (PAGE) | soxG | 2 heme AS | 20 | Component of SoxM alternative heme-Cu oxidase | 92, 327 |
| 56.7 (DNA) | 100 | |||||
| a583, Sulfolobus sp. strain 7 | Unknown | NA | 1 heme AS | 270 | Terminal oxidase complex; cyt c function assumed | 246,247, 250 |
| aa3, S. acidocaldarius (DSM 639) | 38 (PAGE) | soxB | 2 heme AS | 220 | Subunit I of heme-Cu quinol oxidase SoxABCD | 22, 23, 329 |
| 57.9 (DNA) | 1 Cu | 365 | ||||
| aa3, Sulfolobus sp. strain 7 | 40 (PAGE) | NA | 2 heme AS | 117 | Putative subunit I of terminal oxidase complex (c oxidase activity) | 247 |
| 1 Cu | 325 | |||||
| aa3, A. ambivalens | 40 (PAGE) | doxB | 2 heme AS | 235 | Subunit I of terminal quinol oxidase | 24 |
| 65 (DNA) | 1 Cu | 330 | ||||
| b562/aa3, S. solfataricus (DSM 1616) | ND | NA | Heme B | ND | Unresolved components; CO-reactive respiratory complex | 594 |
| ND | Heme AS | |||||
| b558/562, T. acidophilum | 18 (PAGE) | Orf-2 | 2 heme B | 75 | cyt b of putative fumarate dehydrogenase | 162 |
| −150 |
Abbreviations: deglyc., deglycosylated protein; AS, A-type heme as described for Fig. 14; NA, no genetic data available; ND, not determined; PAGE, Mr determined by PAGE; DNA, molecular mass derived from gene sequence. Further details may be extracted from the references given.
Molecular mass determined by PAGE in presence of 1% i-butanol.
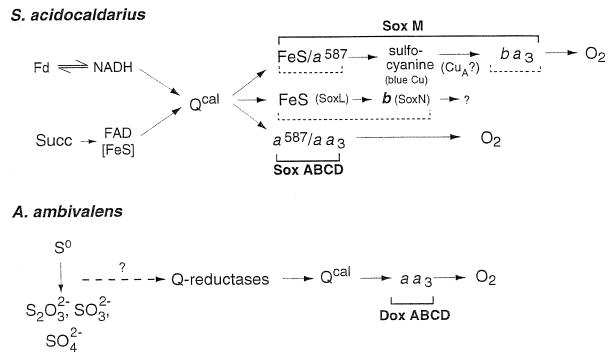
(i) The SoxABCD complex.
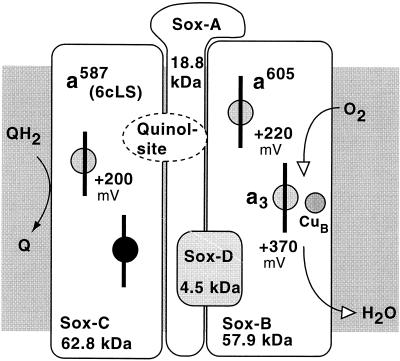
Figure 10 illustrates schematically the composition of a novel terminal oxidase complex, SoxABCD, discovered in S. acidocaldarius (92, 329). It was found to combine features of what are usually separate respiratory complexes III and IV, although its cyt aa3 subunit (SoxB) could be isolated as a functional quinol oxidase by harsh detergent treatment (23). The integrated complex is transcribed from an operon encoding four polypeptides, designated SoxA to -D. Of these, SoxB represents a subunit I equivalent of typical heme-Cu oxidases; its binuclear center contains two hemes A and one CuB, which have been well characterized by optical, EPR, and resonance-Raman spectroscopy (168, 173, 224). SoxA, like subunit II in the quinol oxidase of E. coli (339, 388), lacks the typical mixed-valence binuclear CuA ligands of cyt c oxidases. In total, the complex contains four hemes AS. The two additional hemes are the reaction centers of SoxC; judged by its primary and secondary structural elements, it represents a cyt b homolog and can be identified spectroscopically as cyt a587-I. When examined in intact membranes, the complex reveals CO binding kinetics similar to those of mitochondrial cyt c oxidase; the cyt a587 and cyt aa3 centers exhibit coordinated redox behavior on the millisecond time scale (172). Interestingly, with tetramethyl-p-phenylenediamine (TMPD) as electron donor, the turnover of the integrated complex (1,300 s−1) is about threefold higher than that of the purified subunit I equivalent, SoxB. The small hydrophobic subunit, SoxD, has no redox centers.
FIG. 10.
Composition of the SoxABCD terminal oxidase of S. acidocaldarius. The shaded areas represent the membrane. Each box represents a single constituent polypeptide with its type of redox center and the redox potential measured at pH 6.5. The dashed oval insert illustrates the proposed location of the binding site for caldariella quinone at an interface between subunit II (SoxA) and the cyt b analog SoxC. Further details are given in the text.
The presence of a diheme cyt b analog is reminiscent of a complex III equivalent, although there is no Rieske-type iron-sulfur protein in SoxABCD. On that basis, the possibility of a proton-pumping Q cycle analogous to quinol:cyt c reductases has been proposed: the function of cyt c1 is replaced by direct electron transfer to the low-spin cyt a in the terminal oxidase segment, SoxB (92).
The SoxABCD complex, the only example known from aerobic archaea, was reconstituted into liposomes and shown to act as an electrogenic proton pump by a yet unknown mechanism (175). By applying an artificial electron-donating system that does not produce scalar protons outside the vesicles, H+/e− ratios of >1 were measured. This would be in agreement with the proposed Q cycle. Details of this mechanism are discussed below (“Proton Pathways in Terminal Oxidases of Archaea”).
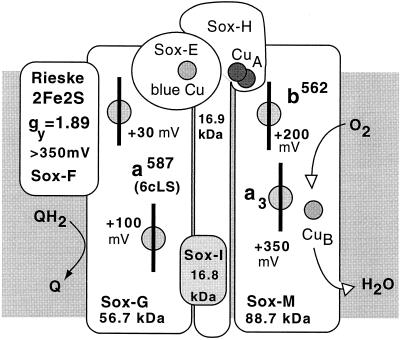
(ii) The SoxM complex.
Even larger than SoxABCD, the so-called SoxM complex of S. acidocaldarius comprises in its purified state seven functional redox centers involving a total of 10 metal binding sites (92, 327). Figure 11 shows a sketch of the complex, including the redox potentials of its components. All subunits are encoded by a gene cluster, which suggests coordinated transcription of a supercomplex composed of two functional substructures. One complex is made up of the gene products of soxH and soxM. SoxM is a ba3-type heme-Cu terminal oxidase with α bands at 562 and 605 nm; the polypeptide has homologies to a fusion protein of subunits I and III of typical cyt c oxidases. Subunit II (SoxH) is supplied by the second gene cluster and, in contrast to quinol oxidases, provides all the ligands for the formation of a CuA mixed-valence redox center. At this site, electrons may be provided by the polypeptide SoxE (sulfocyanine), a homolog to blue copper proteins similar to rusticyanine (91). This mobile electron carrier with low membrane affinity is thought to serve as electron acceptor for a bc1-analogous part of the complex composed of cyt a587-II, another cyt b analog and product of the soxG gene, and the Rieske FeS protein, SoxF.
FIG. 11.
Composition of the respiratory supercomplex SoxM. Each box of the scheme represents a constituent polypeptide together with its redox centers and the respective redox potentials (where known). The location of the blue copper protein sulfocyanine is hypothetical, as a possible link between the partial complexes of SoxG-SoxF and SoxM-SoxH. For details, see the text.
It is important to note that two Rieske FeS proteins have been identified in S. acidocaldarius (487). Both have been characterized, sequenced, and expressed heterologically in E. coli (26, 489, 490). Whereas Rieske II (SoxF) has been clearly identified as a component of the SoxM supercomplex, Rieske I (SoxL) (488) has not been attributed to any of the known respiratory pathways and is assumed to belong to a novel complex of the quinol-oxidizing respiratory system which has not yet been characterized at the protein level (491). Both Rieske proteins are expressed constitutively in Sulfolobus, as are the genes of both complexes, SoxABCD and SoxM.
Nevertheless, the electron transport activity of the isolated SoxM complex is extremely low (327). Neither Qcal nor artificial donors produced a significant turnover. This is most likely due to the lack of sulfocyanine as an intermediate carrier, which is obviously lost during membrane preparation and has so far been identified only on a genetic basis, not chemically. It is likely to be present in vivo because transcription into a single RNA has been demonstrated (286).
(iii) Other archaeal terminal oxidases.
A similar quinol-oxidizing supercomplex has been reported for Sulfolobus sp. strain 7 (246). However, primary sequence data and gene organization are unknown. This supercomplex contains at least four different heme centers and also a Rieske FeS protein (gxyz = 2.02, 1.89, and 1.79). The attribution of redox potentials to individual heme centers appears very preliminary and uncertain in terms of the proposed electron transport pathways and proton-pumping mechanisms. The supercomplex acts as a caldariella quinol oxidase (246) and was functionally reconstituted into detergent micelles together with complex II preparations from the same Sulfolobus strain to serve as quinone reductase (250). A Q-cycle-like mechanism for electron transport and proton translocation has been proposed. However, the typical Q-cycle inhibitors myxothiazole and stigmatellin did not inhibit electron transport activity. Moreover, the Rieske protein would not function in a Q cycle with a single-heme b-type cytochrome (b562; E0 = 146 mV), while cyt a583 may well assume a cyt c-like function with E0 = +270 mV. A c-type cytochrome or a blue copper protein was not detected in the preparation. These obvious differences from the SoxM complex of S. acidocaldarius necessitate the complete sequencing of the genes in order to disclose the real number of heme centers involved.
The quinol-oxidizing complex of A. ambivalens (24) has the simplest structure in terms of redox centers. It is composed of six subunits encoded by two separate operons (430), both of which are present in duplicate in the A. ambivalens genome. Only one polypeptide (the doxB gene product) has redox centers, which identify it as a heme-Cu oxidase. This 64.9-kDa polypeptide contains two hemes AS as cyt a and a3, respectively; the latter is associated with one Cu in the binuclear center. Functional assignments of the other subunits (20.4, 18.8, 38, 7.1, and 7.2 kDa) could not be made because they lack any detectable similarity to known membrane proteins. However, all polypeptides have several putative transmembrane helices; deviating from typical heme-Cu oxidases, the catalytic polypeptide has 14 instead of 12 membrane-spanning helices, with the additional pair at the C-terminal end.
The integrated complex oxidizes Qcal with high activity and is completely inhibited by cyanide; it is also highly sensitive to inhibition by quinolone derivatives such as 2-methyl-3-decyl-quinolone (356). Preparations which have lost the 45-kDa subunit display significant TMPD oxidase activity, which is negligible in the intact complex (430).
This single complex represents the entire respiratory system of A. ambivalens. Its redox centers have been characterized as low- and high-spin hemes by EPR spectroscopy (24), with redox potentials recently actualized as +215 and +415 mV, respectively, displaying a redox-Bohr effect with a pKa of 5.4 (171). The noncompetitive inhibition by quinolones suggests two quinone-quinol binding sites (356), one of which might serve as an intermediate electron storage site as revealed also by the persistent appearance of a stable radical in the enzyme. This has been supported by rapid kinetic studies suggesting that a tightly bound quinone might mimic the function of the CuA site of cyt c oxidases (171), thus forming a fourth redox center.
Most halobacterial terminal oxidases barely survive standard purification procedures: therefore, none of the isolated complexes have been functionally characterized. An aa3-type oxidase has been isolated from H. salinarum as an inactive complex (158), and its amino acid sequence is now available (117). In H. salinarum, a spectral feature was reported (531), indicating the likely presence of a cytochrome bd analog (408). A terminal oxidase complex of N. pharaonis (349, 477) was first isolated as a catalytically incompetent protein fraction. Though its function is not well characterized, it is assumed to act as a cyt c oxidase, and its structure has been deduced from the completely sequenced operon.
The oxidase complex of N. pharaonis is spectroscopically characterized as a ba3-type oxidase with two major polypeptides of 36 and 40 kDa. In subunit I, the six heme and CuB ligands were identified at the canonically conserved positions. In membranes, the potentials of the heme centers were determined as +268 mV (cyt b) and +358 mV (cyt a3). Subunit II provides all the ligands to form the mixed-valence CuA center (348) and thus may function physiologically either as a cyt c oxidase or as the oxidase for halocyanine (see above). Information on a complex III equivalent is rudimentary, although a cyt bc fraction could be isolated from membrane solubilizates (477). At least, supercomplexes of the kind found in thermoacidophilic archaea are absent in N. pharaonis.
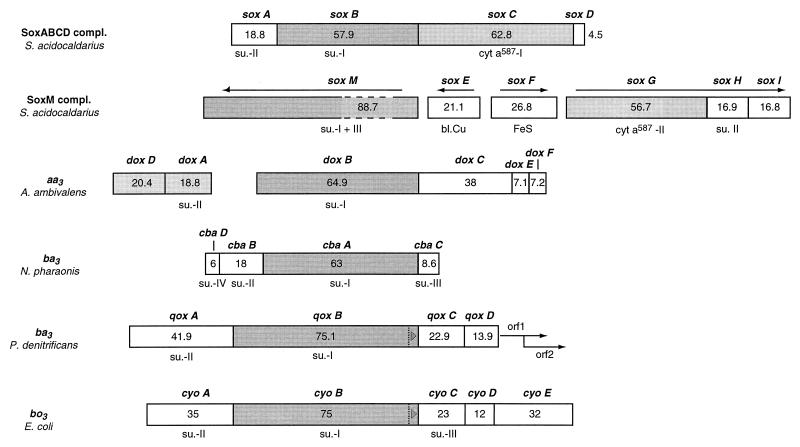
For a comparison of the compositions of archaeal quinol-oxidizing complexes, Fig. 12 illustrates the organization of the respective operons.
FIG. 12.
Gene organization of archaeal terminal oxidases. The operon structure is compared to those of E. coli and P. denitrificans. Numbers within the boxes refer to the molecular masses (in kilodaltons) of the gene products; arrows indicate the direction of transcription. The assignment of genes to subunits of the oxidase complexes is given below the boxes; the darkly shaded box in all cases signifies subunit I, identifying the complex as a member of the heme-Cu oxidase family. In the case of A. ambivalens, the two operons are located far apart on the genome, whereas the soxM locus of S. acidocaldarius forms a close gene cluster. Data were compiled from references 92, 99, 327, 329, 348, 430, 443, 470, and 471 and/or the EMBL data bank. su, subunit.
Mobile electron carriers, hemes, and small metal proteins.
Mobile electron carriers include membrane-integral as well as membrane-associated and soluble redox systems. For example, the Rieske iron-sulfur proteins are anchored in the membrane by a single membrane-spanning polypeptide chain, but their functional center is located within a mobile domain that shuttles between different electron-accepting sites (610, 625). Small blue copper proteins are associated with the membrane surface, as are c-type cytochromes, while ferredoxins are soluble in the aqueous phase. In contrast, the quinones are strictly membrane-embedded, lipid-soluble, redox mediators. All of these components, including the soluble compounds, are necessary to constitute functional respiratory electron transport systems. The typical archaeal examples are discussed below together with the specific hemes of archaeal cytochromes.
(i) Quinones.
Electron flow between dehydrogenases and cytochrome components of the respiratory system is mediated by quinones. The structures of archaeal quinones are depicted in Fig. 13. Halobacteria contain menaquinones with unsaturated isoprenoid side chains, predominantly MK-8 (104, 106, 107), whereas partially or fully reduced side chains are found in other archaea (Archaeoglobus, Thermoproteus, and Pyrobaculum). T. acidophilum contains thermoplasma quinone that is oxidized 10 times faster than is reduced Q10 (105, 234).
FIG. 13.
Structures of respiratory quinones isolated from membranes of thermoacidophilic and halophilic archaea.
Sulfolobus and Acidianus have unique quinones that are derivatives of benzo-[b]-thiophen-4,7-quinone. The novel caldariella quinone (126) was detected in all members of the Sulfolobales as a major constituent, making up approximately 60% of the total quinones (106), which corresponds to 0.28 to 0.38% of cell dry weight. Three variants were detected, of which the tricyclic quinone (2-polyprenyl-benzo[1,2-b;4,5-b′]dithiophene-4,8-quinone) is present only in traces (392, 573). In Sulfolobus solfataricus, the ratio between Sulfolobus quinone and caldariella quinone (Qcal) was reported to vary considerably with growth temperature (392), which shifts the relative content of Qcal between 60 and 99% of the total quinone. In A. ambivalens, the ratio between Qcal and Sulfolobus quinone is reversed by transition from aerobic to anaerobic growth. The function of quinone in anaerobically growing cells has not been studied so far.
Electrochemical properties were reported only for caldariella quinone (QcalH2), extracted from S. acidocaldarius (DSM 639) (23). In vitro, QcalH2 acts as a substrate of the isolated cyt aa3 of Sulfolobus; in vivo, it is the reductant of the SoxABCD complex and presumably also of the SoxM supercomplex (327) (see below). Stabilized in Triton micelles at an ambient pH of 6.5 and at 25°C, it has a half-reduction potential of +100 to +106 mV. The reduced-minus-oxidized difference spectrum shows a minimum at 351 nm, a maximum at 325 nm, and an isosbestic point at 341 nm (e351–341 = 1.77 × 103 M−1 cm−1). At temperatures above 60°C and at pH > 6.5, considerable autoxidation occurs.
(ii) Hemes.
Prenylated hemes, the typical cofactors of terminal heme-copper oxidases, have been analyzed for a variety of archaeal membranes (325, 332). Neither unmodified heme A nor heme O (protoheme IX) is present in terminal oxidases from archaea. Instead, several novel prenylated hemes have been identified as depicted in Fig. 14. The substituents at position 8 are identical to those in the usual heme A or O (formyl or methyl, respectively), but the oxidation state and the length of the isoprenoid side chain at position 2 are altered. In all cases, the farnesyl residue is extended by one isoprene unit. The increased hydrophobicity of the geranylgeranyl chain may be relevant to the stabilization of the heme cofactor in oxidases of extreme thermophiles. It has been proposed (332) that the large pool of the precursor, geranylgeranyl-pyrophosphate, might affect the preferential formation of these new hemes. As discovered by resonance-Raman spectroscopy, unusual profiles of vibrational modes result from the immediate environment of the chromophore in the enzyme-bound state but not from altered porphyrin substituents (see below and references 167, 168, and 210).
FIG. 14.
Structures of hemes found in archaeal terminal oxidases. The hemes are assigned according to references 332 and 336.
Heme AS was found in the aa3 and ba3 oxidases of Sulfolobus, Acidianus, Halobacterium, and Natronobacterium; Thermoplasma has heme OT, while Pyrobaculum has two similar heme OP species as well as heme AS. T. acidophilum may also contain heme D (332). Surprisingly, Thermus thermophilus is the only representative of the bacterial domain that also contains heme AS (typical for archaea) in both terminal oxidases (caa3 and ba3).
Based on spectroscopic analysis (187, 264, 301, 477), one concludes that heme B is ubiquitous in archaea, whereas heme C has been found in methanogens and halophiles but is apparently absent from thermoacidophiles. There have been contradictory reports on T. acidophilum. The presence of heme C (46, 233, 502) has been demonstrated, whereas recent heme composition studies of archaeal cytochromes detected no heme C in membranes of either T. acidophilum or P. aerophilum (332).
(iii) Ferredoxins.
In sulfur-metabolizing archaea, an abundance of ferredoxins has been observed (559). Their high thermostability (27) may not only indicate the use of iron-sulfur clusters as primordial redox systems in early evolution (589) but also reflect their advantage over NAD+ as a primary hydrogen-transducing coenzyme at high temperatures. In fact, archaeal ferredoxins have functionally replaced pyridine nucleotides in several key enzymes such as 2-oxoacid dehydrogenases (60, 276, 621), aldehyde dehydrogenase (166, 377), or glyceraldehyde dehydrogenase (486). Although ferredoxins are not directly involved in primary energy conservation, they assume a key role in channeling hydrogen to NADH as the actual respiratory substrate. Ferredoxins have the advantage of providing reducing equivalents at a potential negative enough to drive hydrogenase-catalyzed reactions (E0 = −450 mV), NAD(P)+ reduction (E0 = −320 mV), or the reduction of sulfur (E0 = −270 mV). In addition, a ferredoxin (E0 = −342 mV) has been purified from N. pharaonis (477). The primary structures of several archaeal ferredoxins are known (276, 365, 425); the three-dimensional structure of one of the Sulfolobus ferredoxins (156) reveals a Zn2+ binding site in addition to the [3Fe-4S] and the [4Fe-4S] clusters. Recent genetic and sequencing data (558) suggests that multiple genes in members of the Sulfolobales may code for highly similar ferredoxins. Despite the presence of an additional metal center, no differences in thermostability have been found. Thus, the hypothesis that the ferredoxins differ with respect to electron acceptor specificity may be considered. Both ferredoxins from A. ambivalens have been cloned and expressed in E. coli (253), which should allow this hypothesis to be tested in the near future. Physicochemical and EPR spectroscopic properties of archaeal ferredoxins from hyperthermophiles are discussed in references 248 and 559. A more detailed analysis of redox-linked protonation equilibria was presented for the Sulfolobus ferredoxin (79). A [3Fe-4S] cluster with a reduction potential of −275 mV shows uptake of one proton with a pKa of 5.8 upon a one-electron reduction; the similarity to the intracellular pH of S. acidocaldarius may indicate a physiological role of this reversible protonation, which may influence the electron transfer properties of this ferredoxin. The [4Fe-4S] cluster has a redox potential of −529 mV at pH 6.4.
The ferredoxin-reoxidizing iron flavoprotein (IFP) isolated from Sulfolobus sp. strain 7 may be important for hydrogen flow in oxidative bioenergetic reactions (249). The dimer of the three-subunit protein contains two flavin mononucleotide centers (E0 = −57 mV) and two [2Fe-2S] centers (E0 = −260 mV). A possible electron pathway, ferredoxin ⇒ IFP[2Fe-2S] ⇒ FMN ⇒ X, has been proposed. Whether X represents an entry point into a respiratory chain is speculative. The isolated IFP does not react with pyridine nucleotides (249) and may be a fragment of a yet-unresolved new electron pathway in archaeal bioenergetics. As mentioned previously, a 76-kDa flavoprotein from A. ambivalens may act as an NADH:ferredoxin oxidoreductase and therefore also play an important role in the bioenergetics of hydrogen flow (177).
(iv) Rieske iron-sulfur proteins.
Rieske FeS proteins are characteristic components of complex III in respiratory chains (so-called bc1 complexes) and also of the structurally related b6f complexes in photosynthetic electron transport. A common feature of respiratory Rieske proteins is the Cys-Cys-His-His ligation of the iron atoms in the [2Fe-2S] cluster, which shows a high redox potential and two typical pK values (for a review, see reference 574). Their function is to accept one electron from a reduced quinone (QH2) which is channeled to the next higher acceptor, while the other electron is transferred to a low-potential heme of a b-type cytochrome as a quinone reductant in the Q cycle (575).
Archaeal Rieske proteins have been detected by EPR spectroscopy only in Sulfolobus and H. salinarum (19, 531). Two Rieske proteins were isolated from S. acidocaldarius, and their sequences were determined (487).
One Rieske protein, the product of the soxL gene, has a pronounced ubiquinol:cyt c reductase activity (488) and only 29% similarity to the SoxF Rieske protein (component of the SoxM complex; see above). Both exhibit the typical bis-Cys/bis-His ligation (in contrast to ferredoxin-type FeS clusters) but reveal an insertion between the cluster binding sites much larger than that of any other Rieske-type FeS protein (487). Nevertheless, in membranes, their EPR signals (gxyz = 2.031, 1.89, and 1.725) and the redox potential (E0 = −325 mV) identify these archaeal iron-sulfur proteins as typical respiratory Rieske proteins; two redox-dependent pKa values (6.2 and 8.5) can be titrated (25). Both proteins have been expressed in E. coli in a truncated form without the membrane-anchoring N terminus (490). The recombinant SoxF Rieske protein has been crystallized (69).
A reading frame in the fully sequenced genome of P. aerophilum was identified as coding for a Rieske protein, which was then produced as a recombinant protein in E. coli (491). The product has all the functional properties of a respiratory Rieske protein.
Nevertheless, since representatives of Sulfolobales do not contain c-type cytochromes, classical bc1 complexes are absent, although the association of these [2Fe-2S] proteins with cyt b or cyt b homologs suggests an essentially similar function. Consequently, it seems appropriate to define cyt b-Rieske complexes as the actual core of complex III analogs, irrespective of the low-potential electron acceptor.
A novel type of iron cluster, which may act as a functional analog of Rieske iron-sulfur proteins, was found in Sulfolobus metallicus (601). The characteristic EPR signals, which could be detected in intact cells and in membrane preparations, suggest a spin-coupled cluster with at least two iron atoms; in the ground state, it has a spin of S = 1/2. The center has a high redox potential of +350 mV and is located in succinate-reducible membranes. Interestingly, the presence of a typical Rieske center could also be detected by EPR spectroscopy in this organism.
(v) Small copper proteins.
Blue copper proteins, which may functionally substitute for cyt c, have been detected in archaea. Halocyanine was isolated from membranes of N. pharaonis as a 15.5-kDa protein by treatment with mild detergents (473). The soluble protein contains one Cu2+ ion per mol and has a broad absorption band at 600 nm. EPR spectra revealed a close resemblance to plastocyanine, suggesting a type I copper ligation with two His, one Met, and one Cys. This is in line with primary sequence data, which also predicts an N-terminal membrane anchor (350). The Cu binding site, located at the C-terminal end, has a high degree of sequence identity with other small copper proteins (446a). The potentials for blue copper proteins range from +183 to +680 mV (551), whereas the redox potential of +183 mV at neutral pH is well suited to assume a cyt c-like function in respiratory electron transport. The redox potential changes from +333 mV at pH 4 to +119 mV at pH 10: there are three pK values within this range, which are presumably associated with three histidine residues that interact with the redox site. Fourier transform infrared (FTIR) spectra point to a special C ⩵ O peptide group that serves as a fifth ligand to copper. Upon oxidation at a low pH, the uptake of a proton by a carboxyl moiety of Glu or Asp was inferred from strong bands at 1,398 and 1,557 cm−1: this reaction can be written as H+ ⇒ Cured + R-COO− ⇒ CuOX + R-COOH (81). The pH dependence of the midpoint potential emerging from FTIR spectral changes was not seen in resonance-Raman spectroscopy. A pronounced similarity to the coordination geometry of azurin was determined. The protein bands strongly suggest a predominant β-sheet structure for this new copper protein.
Sulfocyanine was first predicted as a putative blue copper protein in S. acidocaldarius from the sequence of the soxE gene of the SoxM complex (92). Northern blotting revealed that the gene is transcribed, but the protein has never been isolated from the native organism. Recently, it could be expressed in E. coli in amounts which will allow its biophysical characterization (286). This is quite significant because sulfocyanine is thought to be an essential intermediate in electron transport through the SoxM complex of S. acidocaldarius and is thus expected to restore activity to the isolated supercomplex. Membrane binding of sulfocyanine is apparently very weak, resulting in its loss during purification of Sulfolobus plasma membranes.
Organization of Archaeal Respiratory Chains
Oxygen respiration.
Spectroscopic investigation of respiratory pigments in membranes suggested that archaeal respiratory chains are quite similar to those of bacteria or mitochondria; however, the discovery of supercomplexes and of unusual associations between cytochromes revealed a number of specific archaeal features which are described below. A full set of the energy-transducing respiratory complexes I to IV has never been found in archaea.
However, individual respiratory catalysts and fairly complete respiratory chains have been characterized so far only in Sulfolobus, Acidianus, and some Halobacterium spp. Their electron transport systems were mainly identified by absorption spectroscopy of intact cells, of membranes, or of detergent-solubilized complexes (21, 157, 186, 326, 409, 593). The sequence of electron transport was derived from redox titrations of absorption bands assigned to specific cytochromes or to mobile electron carriers with the aid of standard redox potentials (E0).
As mentioned above (“Components of Aerobic Electron Transfer”), no equivalent to complex I (the proton-pumping NADH:CoQ reductase) has been found. The strongest evidence for this was the absence of the characteristic iron-sulfur centers of complex I-type NADH dehydrogenases, usually detectable by EPR spectroscopy (309, 310, 585).
SDH activities, i.e., complex II equivalents, were found in all organisms from Table 1 (20, 29, 179, 477) that have been investigated, and several enzymes have been partially or completely purified (250, 369). The presence of this enzyme does not necessarily indicate a role within the respiratory system but rather its metabolic function in the citric acid cycle. Recall that much of this cycle operates anaerobically (504) and that it can even operate in reverse for CO2 fixation (267, 499).
The occurrence of a complex III, usually organized as a bc1 complex, is a crucial issue in aerobic archaea. These bc1 complexes are quinol:cyt c oxidoreductases, which typically contain Rieske iron-sulfur proteins (576). Such proteins have been unequivocally detected only in S. acidocaldarius (26, 487), P. aerophilum (491), and H. salinarum (531). In other organisms, a firmly bound c-type cytochrome serves as the usual electron acceptor. Since no c-type cytochromes are present in members of the Sulfolobales, however, the association of these iron-sulfur proteins and their electron acceptors has to be different from that of known bc1 complexes. In other cases, such as N. pharaonis, b- and c-type cytochromes were detected but an associated Rieske iron-sulfur protein was not. It seems appropriate to define cyt b-FeS protein complexes with the option of variable or alternate electron acceptors as possible equivalents to complex III in archaea. Hence, the absence of a classical complex III suggests that energy conservation by proton pumping in this part of the archaeal respiratory chain may also be accomplished by novel complexes, if it occurs at all.
Terminal oxidases (complex IV equivalents) are ubiquitously present in aerobic archaea, comprising quinol oxidases and cytochrome oxidases of the aa3 or ba3 type (as described above and in references 325, 430, 470, and 471). The actual terminal oxidase polypeptides are, in several cases, constituent parts of the novel archaeal supercomplexes. Although the terminal section of archaeal respiratory systems has to be considered an energy-conserving segment, it may function either by chemical charge separation or as an electrogenic vectorial proton pump; this is discussed in the following section.
Figure 15 presents tentative schemes for respiratory electron flux for S. acidocaldarius and A. ambivalens. The split electron transport chain of S. acidocaldarius was postulated earlier (21, 466) from spectra of CO-reduced membrane extracts; one branch consists of the SoxABCD complex. An electron pathway has been envisaged for SoxABCD (109, 325), Qcal ⇒ cyt a587 ⇒ cyt aa3 ⇒ O2, possibly involving a proton motive Q cycle (575). cyt a587 (SoxG) would, in this mechanism, function like the b cytochromes in bc1 complexes. Another possible function would be that of an electron store for the transition from two-electron to one-electron transport reactions in respiration, i.e., as a feeder to the terminal aa3-type oxidase. Notably, however, the isolated polypeptide SoxB (as the actual heme-Cu oxidase) also has quinol-oxidizing activity.
FIG. 15.
Schematic representation of the electron transport chains of S. acidocaldarius and A. ambivalens. The brackets encompass components coded for by a single gene cluster or operon. Brackets with dashed lines signify functional units which may operate as proton pumps. The position of sulfocyanine in the upper scheme is hypothetical at present. The presence of CuA is inferred from the binding motif in subunit II of SoxM. The question mark in the A. ambivalens chain underscores the lack of information concerning the path of electrons from sulfur oxidation to caldariella quinone.
The supercomplex SoxM may also function as a quinol oxidase but is apparently composed of two sequential segments bridged by the blue copper protein, sulfocyanine. Presumably, the latter has a redox potential similar to that of halocyanine (81). Thus, this supercomplex may represent a genetic as well as a functional fusion of an analog to complex III with a ba3-type terminal oxidase utilizing sulfocyanine instead of cyt c. In fact, the presence of a putative CuA binding site in its subunit II has been deduced from the DNA-derived sequence (92). The respective protein could be expressed in E. coli, and Cu2+ can be correctly inserted (286). The association of the Rieske iron-sulfur protein (SoxF) with cyt a587 (SoxG) and sulfocyanine (SoxE) would mimic a bc1 complex. Actually, a catalytically active form of the complex SoxM has not been obtained yet.
A third QH2-oxidizing pathway, involving the alternate Rieske FeS protein SoxL of S. acidocaldarius (487, 488), can be postulated on a genetic basis because the respective gene cluster also encodes a novel b-type cytochrome. This suggests the coordinated expression of an additional cyt b-Rieske complex hitherto unidentified on the protein level (491a). Actually, the predicted b-type cytochrome, a product of the newly discovered soxN gene, may again host heme AS and therefore not be easily detectable under the huge absorption peak (587 nm) that dominates the visible spectrum of the reduced membranes.
A similar but unbranched electron pathway was proposed for the supercomplex of Sulfolobus sp. strain 7 (an S. solfataricus strain) (247). The authors suggested the following electron pathway: Qcal ⇒ cyt b562 ⇒ Rieske FeS ⇒ cyt a583 ⇒ cyt aa3 ⇒ O2. In this proposal, cyt a583 assumes a cyt c-like function whereas cyt b562 is associated in a complex III-like mechanism with a Rieske iron-sulfur protein (246, 247). The available data is insufficient, however, to confirm this hypothesis; in particular, the lack of primary structural data on the components has prevented the proof that cyt a583 in S. solfataricus really is a single-heme cytochrome, as has been postulated.
A. ambivalens has the simplest respiratory system ever detected. Its terminal oxidase functions as a quinol oxidase and is the only cytochrome identified in the plasma membrane. However, the reduction of Qcal during aerobic growth of the obligate chemoautotrophic sulfur oxidizer is enigmatic. A sulfur oxygenase-reductase with a complicated reaction mechanism has been described (282, 283). The enzyme catalyzes a partial disproportionation of sulfur into oxidized and reduced species with a broken stoichiometry. It is certain that electrons flow to Qcal in the oxidative branch of the reaction, but the nature of the reductases and the possible intermediates involved are unknown. There is no indication of alternate electron pathways with oxygen as electron acceptor.
There is little detailed functional information on respiratory chains of halobacteria. Catalytically intact respiratory complexes have not been isolated. Spectroscopic investigations of H. salinarum (H. halobium) membranes have revealed the presence of b-type cytochromes, which may possibly be involved in a bc1-like complex (178, 187). A partial purification has been described (186) along with c-type cytochromes and even an aa3-type terminal oxidase (157, 159, 160). The gene for subunit I of the latter has been sequenced, clearly placing the enzyme in the heme-Cu oxidase family (117).
A full set of electron transport components, purified from N. pharaonis and characterized spectroscopically (477), may be arranged according to their redox potentials into a respiratory chain. It includes b- and c-type cytochromes, the blue copper protein halocyanine (E0 = +128 mV) (350, 473), and a ba3-type terminal oxidase, which has been sequenced (349). Electrons enter the respiratory chain via SDH and/or NADH oxidase. The reducing equivalents may be transported via a cyt bc complex to halocyanine, which is presumably reoxidized by the terminal ba3-type oxidase. An FeS protein that is absent from the preparation would be required to restore the proposed bc complex to a fully active complex III equivalent. In addition, on the basis of reported redox potentials, the transfer of electrons from succinate to this complex would require energy under standard conditions, a discrepancy which remains to be explained. Table 5 summarizes the properties of components constituting the respiratory chain of N. pharaonis (477).
TABLE 5.
Components of the electron transport system of N. pharaonisa
| Protein | Localization | Mr (103) | Cofactor(s) | E0 (mV) |
|---|---|---|---|---|
| Ferredoxin | Cytosol | 15.2 | 2Fe-2S | −342 |
| SDH complex | Membrane | 90 | FAD, FeS, heme B | +8 (FAD) |
| cyt c | Cytosol | 75 | Heme C | −142 |
| cyt bc | Membrane | 14, 18 | Heme B, heme C | −117, −44 |
| Halocyanine | Membrane | 15.4 | Cu | +128 |
| cyt ba3 | Membrane | 40, 36 | Heme B, heme AS, CuA, CuB | +268, +358 |
Alternate types of respiration.
Anaerobic respiratory enzymes such as nitrate reductases have been described for archaea elsewhere (50, 406, 566, 586) and referred to briefly above (“Aerobiosis and Other Respiration Forms in Archaea”). Complete electron transport pathways and energy conservation sites with electron acceptors other than oxygen have not yet been worked out. However, a purified sulfur-reducing membrane-protein complex from Pyrococcus abyssi (129), comprising hydrogenase and sulfur reductase activities, is a candidate for energy conservation by sulfur respiration. The organism gains energy by reduction of elemental sulfur with H2 to H2S. The Triton-solubilized complex of approximately 520 kDa contains nine polypeptides ranging in molecular mass from 24 to 82 kDa. The presence of acid-labile sulfur indicates the participation of iron-sulfur centers; in addition, nickel, copper, and a b-type cytochrome were identified as functional components. The orientation in the membrane of oxidative and reductive partial reactions, i.e., hydrogenase activity and sulfur reductase activity, is not known. However, a scheme (Fig. 16) illustrates the postulated transduction of energy from hydrogen oxidation into a proton motive force, on the assumption that sulfur reduction and hydrogen oxidation occur on opposite sides of the membrane. The critical step would be the electrogenic transport of negative charge from outside to inside. Although this is only a hypothesis, it encourages us to examine the isolated complex in reconstituted membranes with an eye to the orientation of its activities. If hydrogenase releases protons outside the plasma membrane and sulfur reduction consumes protons on the inside, a proton gradient could be formed. This would require the presence of a soluble form of elemental sulfur inside the cell. The volatility of H2S, as a driving force for the reaction, may play a significant role; at high temperatures, H2S would escape into the gas phase and thus shift the reaction equilibrium toward formation of a proton gradient.
FIG. 16.
Possible mechanisms for ΔμH+ generation by sulfur reduction. (Left) H2 oxidation and S0 reduction both occur on the outside. (Right) The redox reactions take place on opposite sides with respect to the membrane. For details, see the text. Hy, hydrogenase; SR, sulfur reductase.
Proton Pathways in Terminal Oxidases of Archaea
Generation of a proton motive force across a membrane by the quinol-oxidizing segment of respiratory chains can occur by three different mechanisms. The simplest one is chemical charge separation. A second, more sophisticated mechanism is a Q cycle. Finally, active pumping by heme-copper oxidases is the third mechanism that, in addition to the proton gradient produced by chemical charge separation, translocates one proton per electron transferred to oxygen (1H+/e−). The latter process makes optimal use of the large change in free energy of oxygen reduction. All three mechanisms presumably function also in archaea.
Generation of a proton motive force via chemical charge separation results from the uptake of protons for water formation from inside the cell, coupled with the release of protons from quinol oxidation outside the plasma membrane, without pumping of extra protons. For reasons described below, the latter mechanism was proposed to occur in A. ambivalens (234). Its energetic inefficiency may be the reason for the slow aerobic growth of this archaeon, despite rapid oxygen turnover and sulfur consumption.
Proton pumping by means of a Q cycle has been proposed for the SoxABCD complex (329) and was demonstrated experimentally when the complex was coreconstituted in liposomes together with the SoxL-Rieske protein, which allows the liposomes to be energized by cyt c (175). This artificial system made use of the fact that reduced cyt c can deliver electrons to the Rieske protein, and the latter, via coreconstituted endogenous caldariella quinone, reduces the cyt a587 of the SoxABCD complex. Thus, the quinol oxidase is energized from the outside of the liposomes by cyt c. The procedure circumvents two serious obstacles: first, reduced caldariella quinone (the specific substrate of the complex) cannot be added in aqueous solution, and second, the random orientation of reconstituted oxidase also excludes an energization by TMPD as reductant. The latter is an effective substrate for the solubilized complex (174) but, due to its significant membrane permeability, would reduce both orientations of the complex and prevent the determination of H+/e− ratios. The interpretation of the results of these sophisticated experiments was that the SoxABCD complex performs a Q cycle, assuming that the SoxB subunit of the complex is not itself a proton pump. The reasons are the same as those for the A. ambivalens oxidase, explained below on the basis of primary sequence data.
Do archaeal heme-copper oxidases provide the structural prerequisites for the third mechanism, i.e., proton pumping by the heme- and copper-containing polypeptide of terminal oxidases? To answer this question, the following argument must be considered. From extensive mutational studies of terminal oxidases from E. coli, Rhodobacter sphaeroides (146, 161, 563, 564), and P. denitrificans (215, 426) and especially from the three-dimensional structure of cyt c oxidases from P. denitrificans (251) and mitochondria (578, 579), it has been concluded that different pathways exist for protons consumed during oxygen reduction and those that are pumped. Two channels in subunit I, designated the K channel (involving an invariantly conserved Lys residue) and the D channel (involving a conserved Asp residue) are characterized by two distinct sequence motifs in transmembrane helix XII and in the helix II-III loop of heme-Cu oxidases. In other respects, these share a strictly conserved topology of their heme and CuB ligands (90, 165) in transmembrane helices II-X and VI-VII, respectively. In addition, a strictly conserved Glu residue (Glu-274 in P. denitrificans) at the end of the proton uptake pathway through the D channel is definitely required for pumping. Recent FTIR studies and molecular modelling underline the significance of the glutamate residue within a strongly apolar environment as a critical proton gate, presumably involving a chain of fixed water molecules (431, 445). Thus, besides the specific -N-(X)10-D-(X)6-N-motif of the helix II-III loop of the K channel, the glutamate in the -HPE- motif in helix VI was declared indispensable for proton pumping (165).
Both motifs are missing in several archaeal terminal oxidase complexes and in that of the eubacterium T. thermophilus, strongly suggesting that these complexes are incapable of proton pumping. They contrast with oxidases from P. aerophilum, S. acidocaldarius (ba3 [SoxM]), and H. salinarum. Accordingly, the terminal quinol oxidase of A. ambivalens has been designated as nonpumping but capable of generating a proton motive force by chemical charge separation. Similarly, the SoxB subunit of the SoxABCD complex from S. acidocaldarius and the aa3 oxidase from N. pharaonis would not be competent as proton pumps. Figure 17 depicts the relevant amino acids participating in the formation of the D and K channels in subunit I of the compared terminal oxidases. Earlier reports of the generation of a proton motive force by purified SoxB reconstituted into liposomes and capable of oxidizing TMPD-ascorbate (174) can easily be explained by chemical charge separation without electrogenic pumping. Therefore, the demonstration of proton pumping (with H+/e− >1) by the integrated SoxABCD complex (175) described above suggests that the operation of a Q cycle is most likely the mechanism of proton translocation.
FIG. 17.
Amino acid residues of subunit I involved in proton pumping by terminal oxidases. The boxed area encloses the archaeal terminal oxidases which are compared to the bacterial ones from T. thermophilus and P. denitrificans; the latter serves as a reference enzyme with known three-dimensional structure. Column 1 identifies the positions of the critical amino acids within individual transmembrane helices or the helix II-III connecting loop; + and − denote the presence and absence, respectively, of a specific residue in the enzymes; letters and numbers in parentheses describe positions and types of the replacement amino acids in the archaeal organisms. All oxidases which lack the motif in loop II-III are considered to be incapable of pumping protons by the mechanism employed by cyt c oxidase. Details and conclusions from this analysis are described in the text. “a,” numbering according to P. denitrificans; “b” refers to entire motif (-Nx10Dx6N-). P.den., P. denitrificans; H.sal., H. salinarum; S.ac., S. acidocaldarius; A.amb., A. ambivalens; N.pha., N. pharonis; T.th., T. thermophilus.
Whether the inability of an oxidase to pump protons can be deduced simply from structural comparisons remains to be demonstrated experimentally in each individual case. The reconstituted cyt c oxidase ba3 from T. thermophilus appears to pump protons with low stoichiometry (0.5H+/e−) when energized with a photoactivated ruthenium II complex (269). This result makes the above conclusions questionable and, in fact, strongly suggests the possibility of alternate proton pathways. Assuming that the SoxABCD complex can perform a Q cycle and, in addition, that the SoxB subunit would pump protons, this archaeal oxidase complex should be able to achieve H+/e− ratios of >2, which has not been observed. Irrespective of these arguments, the results with the T. thermophilus oxidase confirm the necessity of the strictly conserved K channel for the redox reaction, while possibly alternate pathways to the D channel may exist but have not been resolved structurally.
Archaea, like bacteria, can possess simultaneously both types of terminal oxidases (e.g., S. acidocaldarius and T. thermophilus), which may or may not contain the sequence motifs discussed above. Therefore, terminal oxidases may have evolved by an early gene duplication of subunit I, prior to the division into the archaeal and bacterial domains (608). The physiological meaning of the coexistence of two oxidases is not yet understood. For example, greatly differing oxygen affinities would permit adaptation to varying micro-oxygenic conditions, as in Bradyrhizobium japonicum (429). Another possibility would be the preferential operation of specific oxidases at certain pH values. Since no data is available, these possibilities are moot pending further investigation.
LIGHT-DRIVEN ENERGETICS
General Overview
In the early 1970s, the so-called purple membrane from H. salinarum (55) was found to contain only a single protein (401). It was later demonstrated that a retinal chromophore attached to this protein gives rise to the purple color. Because of the obvious resemblance to vertebrate rhodopsins, the authors named this retinylidene protein bacteriorhodopsin (BR). It was also immediately clear that, upon light activation, a proton is transferred across the membrane from the inside of the cell to the extracellular medium. The proton potential generated thereby is utilized by H. salinarum for the synthesis of ATP (432).
During the elucidation of the proton-pumping mechanism of BR and the physiology of H. salinarum, three other bacterial rhodopsins were discovered (an extensive progress report on the photophysics and photochemistry of retinal proteins is presented in reference 413). These pigments are functionally quite distinct. HR, like BR an ion pump, is an inwardly directed chloride pump. The two other bacterial rhodopsins turned out to be photoreceptors that trigger the photophobic and/or photoattractive response of bacteria. All four bacterial rhodopsins are strikingly similar with respect to structure, the retinal chromophore and its attachment site, and the light-activated reaction cycle.
The four pigments are membrane proteins with an approximate molecular mass of 26 kDa (see reference 503 for a comparison of their amino acid sequences). In each case, the polypeptide chain folds into seven membrane-spanning helices (indexed helices A to G) with the C terminus positioned on the cytoplasmic side. The retinal chromophore is attached in the all-trans configuration to a Lys residue via a protonated Schiff base in the C-terminal helix G. The structural model of BR (216) revealed the locations in the sequence that are involved in the chromophore binding site. This was confirmed by a comparison of the amino acid sequences of all archaeal rhodopsins known at the time that exhibited distinct homologies at those particular positions (503, 519).
In a direct comparison of amino acid sequences of rhodopsins from different archaeal species, clusters of conserved residues are found in helices C and D, containing three Trp residues, which enclose the retinal chromophore. The Lys residue of helix G, which marks the retinal attachment site, and an Asp residue situated one helical turn upstream are conserved throughout. This aspartic acid residue is part of the counterion complex of the protonated Schiff base. Another interesting stretch of homologies was detected on helix C, where an Arg residue also contributes to the Schiff base environment.
Upon light excitation, all four microbial rhodopsins undergo cyclic reaction sequences. During the photocycle, the physiological response, either vectorial ion transport or initiation of the signal transduction chain, is activated. The reversible molecular events associated with this include the isomerization of the retinal chromophore from all-trans to 13-cis, the deprotonation of the Schiff base (HR is an exception), and conformational changes of the protein backbone. Protonation-deprotonation reactions of internal aspartyl carboxyl groups are functionally also quite important (see below). All these light-triggered thermal reactions have to be reversed because, once a cycle is completed, the protein regains its original state.
The photocycle has been studied in great detail for BR and less stringently for the other three pigments. The general picture emerging from these investigations can be summarized as follows: the intermediates of the BR photocycle are named in alphabetical order, starting with J for the earliest reaction product. J is followed by five other intermediates, K, L, M, N, and O. These letters were chosen by analogy to the reaction sequence observed in visual pigments, with Lumi and Meta identified as physiologically important states (323, 542).
When the photocycles of the other pigments were compared to those from BR, it became apparent that they follow a common scheme with some variations, which depend on the initial state and the nature of the counterion complex (see below). The long-lived intermediate M, which is formed on the microsecond time scale, is physiologically quite relevant. In the case of the sensory rhodopsins (SRs), it represents the signaling state (615, 617), whereas in BR, M separates the proton release pathway from the proton uptake steps. The chloride pump HR is an exception because in the naturally occurring photocycle an intermediate with a deprotonated Schiff base (a typical property of the M state) is missing.
The ion pumps return to the original state on the millisecond time scale. The SRs are slower, requiring about 100 ms. The reason for the difference is likely to be related to the functions of these proteins. Whereas the efficiency of the ion pumps is dependent on a high turnover rate, the SRs have to remain in the signaling state long enough to relay the signal to cytoplasmic proteins.
BR
Structure of BR.
The observation that BR forms a hexagonal two-dimensional lattice within the purple membrane (55) facilitated further analysis of its structure. A low-resolution map of the electron-scattering density of the purple membrane revealed seven peaks, which can be attributed to seven helices of BR that are oriented almost perpendicularly to the membrane surface (217). From the primary structure of BR (277, 414), it became evident that the protein is a chain of 248 amino acids, which folds into a bundle of seven helices with short nonhelical interconnecting loops. Several potentially charged amino acids are found in the hydrophobic interior of the protein. Two Asp residues (Asp-85 and Asp-96) were located in helix C, Asp-115 was in helix D, and finally Asp-212 and Lys-216 were in helix G. The latter amino acid was determined to be the binding site for retinal (41, 311). In subsequent work, Engelman and coworkers assigned the helices to the structural map of BR (141, 572). However, an unequivocal designation became available only with a more refined structural model of BR, based on high-resolution electron cryomicroscopy of the purple membrane and on reconsideration of earlier biophysical and biochemical data (216). This report also offers a thorough review of the literature prior to 1990. A structural resolution of 3.5 Å in the membrane plane and of 10 Å in the perpendicular direction was obtained. Improved structural maps are now available (180, 279). During the past year, structures of still-higher resolution were deduced from analysis of three-dimensional BR crystals (142, 337, 416). The procedures which led to the successful crystallization differed considerably. Pebay-Peyroula et al. (416) were able to grow small hexagonal crystals from cubic lipid phases and to obtain a resolution of 2.5 Å. This method was also applied by Luecke et al. (337), who improved the structural resolution to 2.3 Å. An interesting new approach, perhaps applicable to other membrane proteins, uses successive fusions of BR-containing vesicles (556). The birefringent hexagonal crystals diffract X rays beyond 2.5 Å. Another crystallization method, which may also be generally applicable, is the heterogeneous nucleation of BR on benzamidine crystals (142). The epitaxially growing BR crystals are monoclinic and diffract at 2.9 Å. This latter work is of special interest because the interaction of the native lipids with the BR trimer was determined. The general structural features of the molecule published so far are quite similar; however, there still are discrepancies in the loop regions and in the proton conduction channels. These latter discrepancies make it difficult to establish the molecular mechanism of proton pumping. In the following paragraphs, progress in determining the BR structure is explained beginning with the original Henderson structure (216).
The helices are arranged in a kidney-shaped array of seven rods that cross the membrane almost vertically. Due to low resolution, the connecting loops between the helices were not resolved. However, based on this model calculation of the molecular dynamics provided more detailed insight into the structure of the ground state (134, 237, 252, 395) and of intermediate states (238, 321, 322, 611, 612). The contributions of molecular dynamics studies to the structure and function of BR are reviewed in reference 501. The structure allows for two half-channels that connect the cytoplasm and the extracellular side with the Schiff base environment (Fig. 18). A hydrophobic pore with a smaller diameter opens to the cytoplasm. This channel contains only a few cavities that might harbor water molecules (142, 337, 416). The other, broader channel connects the interior of the protein with the extracellular side. Retinal is located at the interface between the two channels, with the proton of the Schiff base facing the extracellular channel (EC channel). In their original report, Henderson et al. (216) proposed that these channels constitute the path of the protons across the membrane; this has now been corroborated by the higher-resolution structures. FTIR experiments demonstrated that Asp residues within the hydrophobic interior of the protein are proton donor and acceptor sites (135, 137, 511). These sites have been assigned to specific Asp residues by site-directed mutagenesis studies (76, 170) and by solid-state nuclear magnetic resonance (NMR) studies of [4-13C]Asp-labeled BR (138, 139, 354, 355). These experiments showed that Asp-85 and Asp-212, which are located in the EC channel, are deprotonated in the resting state. The protonated carboxyl group of Asp-96 (354), a component of the cytoplasmic channel (CP channel), donates its proton to the deprotonated Schiff base during the M ⇒ N transition (170). Asp-115, not directly located in the cytoplasmic pore, is also protonated (354). The pKs of these two residues were determined to be greater than 11 (354, 552). Further investigations of the structural properties of the CP channel using FTIR spectroscopic techniques in combination with site-directed mutagenesis of BR have provided evidence for a hydrogen-bonded chain that includes Asp-96, Thr-46, Val-49, Pro-50, Ala-53, and water molecules (183, 216, 613, 614). The existence of water in this part of the molecule has also been suggested by structural analysis (142, 337). During proton uptake, this hydrogen-bonded chain apparently bridges the carboxyl group of Asp-96 with the Schiff base.
FIG. 18.
Structural model of BR (A) and proton transfer steps (B). (A) Structural model of BR adapted from reference 279. The retinal chromophore is depicted as dark gray circles. The charged residues on the cytoplasmic side may function as proton collectors that funnel a proton to residue Asp-96. The EC channel comprises several charged amino acids, including Asp-212, Asp-85, and Arg-82. (B) Correlation of proton transfer steps with the spectroscopically determined transitions of intermediates. The two proton transfers occurring during the L ⇒ M transition (≈10 μs) depict the deprotonation of the Schiff base concomitant with the protonation of Asp-85 and the ejection of a proton to the extracellular side. In the subsequent steps, a proton from Asp-96 reprotonates the Schiff base (M ⇒ N). Asp-96 receives a proton from the cytoplasm during the N ⇒ O transition. Finally, the proton reservoir in the CP channel (H+ in the dotted circle) is replenished by the protonated Asp-85, which completes the cycle. The thermal reisomerization of 13-cis retinal to all-trans retinal occurs during the N ⇒ O transition. The protein switch that separates the accessibility between the CP and EC channels is thought to occur between two different M intermediates.
The EC channel constitutes the proton release pathway, broader and more hydrophilic than the CP channel. There are several charged amino acids (Asp-212, Asp-85, Arg-82, Glu-194, and Glu-204) and two Tyr residues (Tyr-57 and Tyr-185) that contribute to the interior of the channel and to the environment of the Schiff base. Asp-85 has a special role as the primary proton acceptor. The positive charge of the Schiff base is neutralized by the charged residues mentioned above, which form a diffuse counterion complex (115, 235). Structural models of the two channels, including the pattern of hydrogen bonds, were provided in recent papers on the structure of BR. Since various aspects of these proposals still differ, the molecular mechanism of proton pumping will remain the subject of further experiments.
Five helices (B, C, E, F, and G) form the environment of the retinal chromophore. Retinal is tilted relative to the membrane normal, with the ionone ring pointing away from the cytoplasm. The configuration and the position of retinal within the BR molecule have been determined by a variety of biophysical methods, including solid-state NMR spectroscopy, neutron diffraction, and resonance-Raman and FTIR spectroscopy (reviewed in reference 413; see also reference 372 for a more recent publication). The retinal chromophore is located near the center of the protein and assumes an all-trans configuration. The isomerization around the 6s bond that connects the ionone ring with the unsaturated hydrocarbon chain is in the trans conformation, which allows for the chromophore to be planar (108, 352). This observation has important implications for the wavelength regulation in BR and may also apply to the other rhodopsins. Protonated Schiff bases usually absorb at approximately 460 nm; BR has an absorption maximum at 568 nm. The difference calculated in wave numbers is called the opsin shift because the specific interaction of the protein with the chromophore leads to this bathochromic displacement of the absorption maximum. Solid-state NMR experiments (235) and the spectroscopic properties of BR reconstituted with retinal analogs (as well as similar experiments with model substances [319]) led to an explanation of the opsin shift. According to this model, two effects, i.e., the C6-C7 single-bond configuration and the weak counterion strength (115), influence each other synergistically and are sufficient to explain the large opsin shift of BR (235). The data from NMR spectroscopy and the analysis of chromophore models clearly indicate that the interaction of the protonated Schiff base with the counterion is weak. The NMR data also suggests that this counterion cannot be a point charge, e.g., a single, negative carboxylate group from an aspartic acid residue, but must be a complex consisting of charged groups in the neighborhood of the Schiff base. This picture has been substantiated by recent data on the structure of BR.
Mechanism of proton pumping.
The mechanism of vectorial proton transfer was elucidated by analysis of the photocycle. After absorption of light, the all-trans retinal Schiff base isomerizes to a 13-cis conformation. The first subpicosecond event (500 fs) in the primary photoreaction is the formation of the J intermediate (128, 245, 397, 509). It is quite striking that this photoisomerization is so rapid. The protein apparently provides a vacuum-like environment in which the ultrafast reaction can occur. With a quantum yield of approximately 0.6, this process is quite efficient (568). K, the next intermediate in the reaction sequence, is formed within 3 ps (130, 397, 424, 428, 509). As determined by FTIR spectroscopy, the protein responds only slightly to this altered situation, which results in a strained, distorted chromophore (75, 144). The chromophore relaxes during the transition from K to L, which is reached on the microsecond time scale. In L, the conditions for the release of the proton to the extracellular side are established. During the L ⇒ M transition, the proton of the Schiff base is transferred to Asp-85. Concomitantly, another proton is released to the bulk phase. Recent data, although still controversial (142, 337, 436), indicates that this proton originates from Glu-204 (440, 479), the pKa of which decreases upon protonation of Asp-85 (31, 440). During the lifetime of the M intermediate, which has an absorption maximum at 410 nm, the transition between the extracellular and the cytoplasmic accessibility of the Schiff base must take place (Fig. 18). The nature of this transition is not known, but conformational changes of the protein are probably responsible (198, 285, 546). The Schiff base is then reprotonated by Asp-96, forming the N intermediate (219, 520). The reisomerization of the retinal chromophore from the 13-cis to the all-trans configuration first occurs in the next step of the reaction sequence, in which the O intermediate is formed. At the same time, Asp-96 receives a proton from Asp-38, which has obtained a proton from the cytoplasm (444). The O intermediate absorbs at 620 nm. This redshift of the absorption maximum may be due to the still-protonated Asp-85. A similar redshift is observed for the so-called blue membrane (278), for which protonation of Asp-85 could also be demonstrated (355). In the last step of the photocycle, Asp-85 loses its proton. Since all steps of the photocycle need to be reversed in order to regain the original state (195), it is also necessary that access to the Schiff base returns to the extracellular side. That the first transition does occur and that it occurs between the M and N states has been demonstrated in voltage-clamp experiments (386). In this work, BR was expressed in the plasma membrane of oocytes (387), and the voltage-dependent photocurrent was measured. Double-flash experiments with blue light (which photoconverts the M intermediate directly back to the initial ground state) at pump-inhibiting potentials provide the first experimental data on M states with different accessibilities, thus supporting the idea of a switching event between two different forms of M.
Several other experiments have attempted to identify the switch at the molecular level. For example, amide I and amide II vibrational modes have been interpreted as indicative of conformational changes of the protein backbone (for example, see reference 218). Time-resolved EPR measurements directly showed differences in mobility at defined positions of the BR molecule (446, 539). A more direct approach has been chosen by a few groups, who used X-ray (263, 285, 389), electron (218, 545, 546, 587), and neutron diffraction (116) studies to characterize structural changes occurring at different stages of the photocycle. These experiments show that helices F and G are predominantly affected during the lifetimes of the M and N intermediates, interpreted as an increase in the structural order of helix G and an outward tilting of the cytoplasmic part of helix F. This latter finding is of significance because it bears on a possible mechanism of signal transfer from SR to its transducer (see below).
The photocycle has been the subject of numerous reports. The original model of Lozier et al. (323), with the sequence BR ⇒ K ⇒ L ⇒ M ⇒ N ⇒ O ⇒ BR, is now generally accepted, though the existence of the N intermediate has sometimes been neglected. The kinetic data of the photocycle can be satisfactorily approximated with seven exponentials (101), indicating that at least seven distinct intermediates must exist; of these, only the five spectrally distinguishable species K to O have been identified. Therefore, the simple kinetic scheme BR ⇒ K ⇒ L ⇒ M ⇒ N ⇒ O ⇒ BR has had to be modified; back reactions, side reactions, new intermediates, or even parallel photocycles have been introduced. The most controversial part of the photocycle involves the M ⇒ N ⇒ O transition, for which two (and as many as five) M intermediates have been proposed, as have also back reactions such as M ⇔ N and/or N ⇔ O (for a review, see reference 306). In an effort to reconcile the published data with their own measurements, Chizhov et al. (101) argued for a scheme of irreversible sequential transitions between quasiequilibria of intermediates (Fig. 19). The quasiequilibria are controlled by rate-limiting dynamics of the protein and/or proton transfer steps. P3 and P4 both represent L540 ⇔ M410 equilibria and are distinguished by the relative percentages of the two components L540 and M410, which shift from 20% M410 to 70% M410. The irreversible transition of (L540 ⇔ M410) ⇒ M410 may be considered the molecular switch proposed in references 195 and 528 and verified by the work of Nagel et al. (386), in which the access to the Schiff base changes from the extracellular side to the CP channel. In the second part of the photocycle, a proton from the CP channel reprotonates the Schiff base. These steps are also characterized by quasiequilibria (P6 and P7) that are quite sensitive to the pH of the buffer and the temperature. This model implies that the ion-translocating steps are not rate limiting. Apparently, the conformational changes of the protein and/or the isomerization of the retinal chromophore determines the time course of the vectorial proton transfer.
FIG. 19.
Scheme of the BR photocycle based on a sequential irreversible model according to reference 101. P0 to P7 denote the spectroscopically defined intermediates. The four states P3, P4, P7, and P6 are quasiequilibria between M and L, M and N, and N and O. The decay rates (at 20°C) of the intermediates are depicted between the wings.
It should be noted that Lanyi and coworkers have proposed a mechanism for the proton pump (84). According to this local-access model, the retinal Schiff base retains connectivity to both the EC and the CP channels throughout the photocycle. Vectoriality is obtained by a shift in the pKa values of the groups in the proton conduction chains. In wild-type BR, an initial increase of the pKa of Asp-85 facilitates proton transfer from the Schiff base. Conversely, the subsequent decrease of the pKa of Asp-96 allows the rapid reprotonation of the Schiff base.
HR
Structure of HR.
HR, the second light-driven ion pump of H. salinarum (for a recent comprehensive review, see reference 398), has now been found in other halophilic archaebacteria (411, 519). First described and named by Mukohata and colleagues (347), HR has an absorption maximum at 576 nm and was first sequenced by Blanck and Oesterhelt (53). Upon light excitation, chloride ions are transferred from the medium into the cell (35, 65, 496, 534). The ion selectivity of the transport is not restricted to chloride; bromide, iodide, and nitrate are also transported (203, 208, 496). Compared to that of BR, the absorption maximum of HR has a longer wavelength.
The amino acid sequences of several HRs reveal structures quite similar to those of BR, including the omnipresent main motif of seven helical transmembrane chains. Further structural information was gained through electron cryomicroscopy of membrane sheets. The projection map of HR at 7-Å resolution was very similar to that of BR at the same resolution (199, 200). These structural similarities match the close resemblance of the primary sequences of the two pigments. One interesting difference is found on the cytoplasmic side of the helices B, C, F, and G. As mentioned above, these helices form the CP channel. In HR, the cross-sectional area is larger than in BR, indicating a pore wider than would be required to transport halides. The EC channels, of similar dimensions in BR and HR, would easily accommodate chloride anions as indicated by functional studies (102, 152).
Most of the conserved residues are associated with the retinal binding site. Compared to BR, interesting differences are found in positions of functional importance for proton translocation. Asp-96 (numbered according to the BR sequence) is replaced by an Ala residue, and Asp-85 is replaced by a Thr residue. This latter substitution is of particular interest because a negative charge is removed from the counterion complex, thereby creating an anion binding site (410, 474). Indeed, chloride binds to a mutant of BR in which Asp-85 is replaced by Thr. This mutated BR pumps chloride actively upon light excitation (461).
The putative binding of anions has been studied extensively. Ogurusu et al. (403) concluded that HR has a single binding site, because the absorption spectrum of HR is dependent on the chloride concentration. Further spectroscopic analyses of the binding of different anions by HR in Na2SO4 (which itself does not bind to the protein) indicate that anions occupy two different sites (305, 498). These two sites may participate in the uptake and release of chloride during transport. However, new FTIR spectroscopic results indicate only a single binding site (597). The discrepancies between this view and the proposed two sites may be resolved by assuming a one-site, two-state model (597).
As already mentioned, the replacement of an Asp residue in BR by a Thr residue creates an anion binding site. Data from resonance-Raman spectroscopy of HR supports the idea that chloride binds near the Schiff base (127, 169, 341, 415); it was concluded that the positive Schiff base cation is weakly hydrogen bonded to a complex counterion made up of chloride, Asp-238, and Arg-108 (18). Braiman and coworkers demonstrated that the C⩵N stretching vibration is affected by the type of anion, indicating a binding site near the Schiff base (597). Furthermore, vibrational modes from the Arg residue were also perturbed (77). In elegant experiments, these modes were assigned to Arg-108; it was also shown that the guanidinium ion can mimic the Arg residue in the Arg108Gln mutant (450). The similarity between the HR and BR counterion complexes is quite evident from the observation of a so-called acid purple form of BR (125, 152), the creation of an anion binding site in the BR mutant Asp96Thr, and the observation that HR from N. pharaonis (132, 304) forms a blue pigment upon anion removal (474). Even the proton-pumping capability of BR could be reestablished in HR from N. pharaonis (582).
N. pharaonis, a haloalkaliphilic bacterium from the Wadi Natrun of Upper Egypt, expresses two rhodopsin-like pigments, HR (pHR) and the photophobic receptor-SR II (pSRII) (52). The blue form of pHR is spectroscopically quite similar to the cation-depleted so-called blue membrane of BR. Removal of a negative charge from the counterion complex of the parent state leads to this bathochromic shift. In BR, the protonation or neutralization of Asp-85 (355, 412, 547) removes the negative charge, whereas in pHR the anion can be extracted by size-exclusion chromatography (474). The purple coloration of pHR can be restored by adding anions, useful in examining the effect of specific anions on spectroscopic properties and photocycle kinetics (474). All monovalent anions such as halides, azide, or nitrate are able to bind to the protein. The anion binding kinetics are compatible with a single binding site (102), thus confirming the interpretation of Walter and Braiman (597) for HR. It should be noted that azide-reconstituted pHR displays a photocycle very similar to that of BR, including an M-like intermediate. Indeed, transport studies clearly demonstrated that this modified pHR acts as a proton pump (582).
The chromophore of HR, all-trans retinal, binds via a protonated Schiff base to Lys-242 on helix G. The pK of the Schiff base varies from 7.4 to 8.9, depending on the anion present (474, 498, 535). It is noteworthy that light adaptation does not lead to a 100% all-trans configuration of the retinal chromophore in HR, as observed for BR (265, 306, 584, 628). This somewhat complicates an analysis of the photocycle because of the contribution of the transport-inactive cis cycle.
Mechanism of chloride pumping.
The functional and physiological properties of HR have been investigated intensively (reviewed in reference 398). The chromophore adopts an all-trans configuration, including a C6-C7 trans single bond (38). After absorption of light, the electronically excited state relaxes within the picosecond time scale to a redshifted photoproduct with maximal absorption at 600 nm (HR600). The quantum yield of this reaction, about 0.3, is much lower than that determined for BR (268, 400). The primary photoreaction involves an all-trans→13-cis isomerization of retinal (127, 154, 202, 303, 399), and according to the FTIR difference spectrum between HR600 and HR, this is quite similar to the K-BR difference spectrum (448). The following sequence of events involves at least two additional intermediates, which absorb at 520 nm (HR520) and 640 nm (HR640). These intermediates absorb above 500 nm and must therefore possess a protonated Schiff base. Unlike the BR photocycle, the functionally active HR photocycle does not include the M intermediate, although this is observed in a side reaction which is catalyzed by azide (209). The photocycle has been analyzed with transient-absorption spectroscopy, resonance-Raman spectroscopy, and FTIR spectroscopy (reviewed in references 15 and 398).
A structural analysis of the retinal chromophore by resonance-Raman spectroscopy (127, 154) revealed that HR520 and HR640 are both characterized by a 13-cis, 15-anti-retinylidene Schiff base. However, recent data from FTIR experiments using HR from N. pharaonis indicated an all-trans configuration of the retinal chromophore (583).
Until now, there has been no comprehensive model of the photocycle which explains the effects of temperature, anion, and pH upon the reaction kinetics. In a recent proposal, Váró and coworkers (85) ascribe HR640 to the so-called cis cycle, which originates from the photoactivation of (13-cis retinal)-HR. On the other hand, the HR640 intermediate is clearly seen in HR from N. pharaonis (474, 583). By analogy to the model of the BR photocycle (101), the HR photocycle may also represent a sequence of irreversible steps between quasiequilibria of spectroscopically identified species (100). Closely related to the problem of the photocycle model is that of the steps of chloride uptake and release. Based on the dependence of the kinetics on the chloride concentration, it has been concluded that chloride is released during the lifetime of HR520 and taken up during the reformation of the ground state (307, 399, 400, 627). Electrical measurements by the black lipid membrane technique placed the ion movements within the same time ranges. However, it is possible that the actual ion-translocating steps are considerably faster than the apparent rate constants. If this is the case, the observed rate constants may reflect only rate-limiting changes in protein conformation or retinal isomerization. In new experiments, Chizhov et al. (100a) showed that this accessibility switch between the CP and the EC channels involves two different HR640 species, which rapidly equilibrate with HR520 species. The unsettled questions of the photocycle of HR and the mechanism of chloride transfer across the membrane require the combined efforts of time-resolution techniques (e.g., visual absorption and FTIR spectroscopy).
HR can be converted into a proton pump in a two-photon process (36). Green light alone causes the transfer of chloride ions into the cell. Additional blue light, which excites the HR410 formed by the side reaction mentioned above, converts HR into a proton pump in the direction opposite the one in BR, i.e., protons are pumped into the cell; this process can be enhanced by the addition of azide. Similar effects have been observed for BR mutants in which Asp-85 is replaced by a neutral amino acid such as Thr or Asn (569).
HR from N. pharaonis behaves quite differently. The photocycle of pHR in the presence of azide is quite similar to the BR photocycle with a pronounced transient concentration of an M-like intermediate (474). This observation prompted Váró et al. (582) to analyze the ion specificity of the one-photon-activated transport, revealing that this modified HR acts as a perfect proton pump in which protons are transferred into the extracellular medium. This result is in line with reports that the Asp85Thr BR mutant can function as a chloride pump (461) and that chloride transport activities are restored in the so-called acid purple membrane, which is obtained at very low pH values (125).
SRs
The phototactic activity of the archaeon H. salinarum is mediated by two photoreceptors that have a close structural relationship to the ion pumps BR and HR (for recent reviews on the photochemistry and function of microbial SRs, see references 232 and 530). The archaea are attracted to light of >520 nm and repelled by light of <500 nm. The response to a photoattractant is mediated by SRI (62, 204). In a two-photon reaction, this receptor can also trigger a negative response to near-UV light (527). Moreover, repellent light of <500 nm in wavelength is recognized by a second pigment, SRII, which absorbs at 490 nm. SRI and SRII are expressed under different conditions. The biosynthesis of SRI, like that of the two ion pumps, is induced by anoxic conditions. SRII is produced constitutively by the cell, suggesting that SRII appeared quite early in the evolution of retinal pigments (554), which thus protected the respiring and fermenting halobacteria from exposure to photooxidative conditions. By contrast, SRI enables the halobacteria to locate light conditions optimal for the operation of the two ion pumps, BR and HR, without being harmed by UV light.
Biochemical data (521, 523) and the sequence determination of SRI and SRII revealed sequences, upstream of both the sopI and sopII gene loci, that correspond to the halobacterial transducer of rhodopsin (Htr) (503, 619, 623). HtrI and HtrII show distinct similarities to the bacterial chemoreceptors, especially in the signal domain region and in the flanking methylation-demethylation sites. Apparently, archaea have developed information transfer chains that possess extended structural similarities to those of bacteria.
SRI.
SRI, first described in 1982 by Bogomolni and Spudich (62), was isolated (482) and subsequently sequenced (54). It contains an all-trans, 6s-trans protonated Schiff base (155) which is attached to a Lys residue on helix G (Fig. 18). The chromophore-binding site accepts only all-trans retinal and not 13-cis retinal, as was also observed for BR (616). In comparison to the other archaeal rhodopsins, the absorption maximum of SRI is shifted down to 587 nm. This bathochromic shift was explained by the protonation of the side chain carboxyl group of Asp-76 (437, 438), which essentially results in the removal of a negative charge from the Schiff base counterion complex. The deprotonated pigment absorbs at 550 nm; a pK of 7.2 has been determined for this transition (64). A similar effect is observed for BR, in which the protonation of Asp-85 leads to the so-called blue membrane (278), and also for HR from N. pharaonis, in which the removal of the chloride anion (474) leads to chromophores absorbing at 600 nm. It should be noted that the pK of Asp-76 rises to 8.5 upon binding of the halobacterial transducer of rhodopsin I (HtrI) (64), an observation that may be significant for the mechanism of signal transduction (see below).
The photocycle of SRI closely resembles that of BR (63). Upon light excitation, a redshifted intermediate (S610) is obtained and then decays to an L-like intermediate (S560) within approximately 90 μs. The next intermediate (S373), which is formed within 300 μs, is characterized by an unprotonated Schiff base and a 13-cis configuration of the retinal chromophore (196, 577, 616). It constitutes the physiologically active state that triggers positive phototaxis (343, 615). It slowly decays, returning to the original ground state within approximately 800 ms. These kinetic data imply a high transient concentration of S373, which makes an additional two-photon process possible. This photoactivated branch of the photocycle leads to reversal of the flagellar motor, thus enabling the bacteria to avoid harmful UV light (527). It is certainly a striking feature of SRI to allow a positive response in a one-photon process and a negative response in a two-photon reaction; the aspartate receptor (Tar), among others, also displays this dual capacity (9).
It has already been mentioned that Asp-85 and Asp-96 play a crucial role in the mechanism of the BR proton pump. In SRI, the corresponding sites are Asp-76 and Tyr-87. As outlined above, in SRI Asp-76 is protonated. The protonation of the corresponding amino acid in BR (Asp-85) leads to a nonfunctional protein. Mutations of Asp-96 to neutral amino acid residues impair the proton pump activity, which can be restored by the addition of azide (570). This data suggests that SRI can also function as a proton pump. Indeed, during light excitation, a vectorial proton transport directed to the outside of envelope vesicles was observed (64). The action spectrum of the proton pump activity has a maximum at 540 to 550 nm, not at 587 nm as in the parent SRI. This shift toward the blue region has been explained by the presence of a deprotonated Asp-76. Apparently, protonation of Asp-76 does not lead to functional (proton pump) activity, as found with the acid blue form of BR. The binding of HtrI to SRI apparently abolishes the ability to pump protons (summarized in references 524 and 525). In contrast to these results, Haupts et al. (195) provided evidence that the HtrI-SRI complex also pumps protons. The question whether the proton pump activity observed for SRI and its transducer is required for signal transduction has not yet been answered. In this context, it should be noted that the Asp76Asn mutant is fully functional in phototaxis (437).
SRII.
The fourth retinal protein of H. salinarum, SRII or phoborhodopsin, was first described by Takahashi et al. (553). This receptor had eluded a thorough analysis due to its very low concentration in the cell, coupled with its instability at low ionic strength and in the presence of certain detergents. Although a partial purification was accomplished (475), information about SRII mostly was restricted to spectroscopic data and physiological properties (346, 617). The absorption maximum at 490 nm with a shoulder at 460 nm suggests a planar structure of the retinal chromophore (554). The photocycle of SRII, like that of BR and SRI, includes an intermediate with a blue-shifted absorption maximum characteristic of a deprotonated Schiff base (241, 475).
The difficulties encountered in isolating SRII from H. salinarum stimulated a search for SRII-like pigments in other species. Such a blue-light receptor was detected in N. pharaonis (52) and characterized spectroscopically (225, 239, 240, 368). This SR from N. pharaonis (pSRII) is functionally identical to SRII but stable under low salt concentrations (476, 478), allowing its purification and amino acid sequencing (503). This latter work showed that pSRII and pHtrII are transcribed as a single transcript, indicating the existence of a pSRII-pHtrII protein complex similar to SRI-HtrI. Comparison of the amino acid sequences revealed strong homologies to the other archaeal proteins, particularly SRI. Calculations of the phylogenetic relationship of 22 amino acid sequences in archaeal rhodopsins and correlation with functional properties have revealed four members of this family. (i) Proton pumps are characterized by Asp-85 and Asp-96 (numbering according to the BR sequence); (ii) in chloride pumps, these sites are occupied by Thr and Ala residues, respectively; (iii) in SRI, the Asp residue in position 85 is conserved whereas Asp-96 is replaced by an aromatic residue; and (iv) in the distinct additional member, pSRII, the indicator positions 85 and 96 are homologous to those of SRI, but Met-118 is replaced by a Val residue. The amino acid sequence of SRII from H. salinarum, recently analyzed (623), is also quite similar to that of pSRII.
The photoreaction of pSRII from N. pharaonis has been studied by several groups (103, 225, 239, 240, 368, 476). Like that of SRII, the pSRII photocycle contains K-, L-, M-, and O-like species. The K- and M-like intermediates have been analyzed by FTIR difference spectroscopy (140), indicating that considerable conformational movements of pSRII already take place at the stage of the early K-like intermediate. These changes are more intense than those described for SRI (437). Concomitant with the deprotonation of the Schiff base, a carboxyl group which may arise from Asp-75 is protonated. The protonation reaction is pH independent over the pH range from 5 to 8, in contrast to the situation in SRI, in which a pK of 7.2 for the corresponding residue Asp-76 was observed (64). Unpublished data from measurements with black lipid membranes indicates that pSRII is an outwardly directed proton pump (492).
The extensive homologies of the archaeal transducers with those from the chemotactic signal transduction chain described for E. coli raise questions about the phylogenetic origin of the transducers and their receptors. Retinal protein structures are shared by archaea and eucarya with regard to the photochemistry. However, the 11-cis/trans isomerization found in several vertebrate rhodopsins differs from the trans/13-cis thermoreversible photoexcitation in the archaeal rhodopsins, as do the coupled proton reactions. The transducers in many representative eucarya are G proteins, whereas archaeal transducers resemble bacterial chemoreceptors-transducers. Recent studies analyzing transcription factors suggest that the archaea may have evolved after the bacteria branched from the main stem (449). If this is true, rhodopsins would have appeared after Bacteria branched from the evolutionary tree that leads to Eucarya. The organisms of the archaeal branch could have combined proteins of the bacterial signal transduction chain with the newly developed photoreceptors.
Proton Transport in Archaeal Rhodopsins: a Common Property of Ion Pumps and Photoreceptors
Present knowledge about archaeal rhodopsins warrants the development of general models combining ion transport and sensory transduction. The Oesterhelt group proposed the so-called isomerization-switch-transfer (IST) model (195, 197), which reconciles the different one- and two-photon ion translocations in BR, HR, and SRI. The authors proposed that the selectivity of BR for protons and of HR for chloride ions can be switched to chloride ions and protons, respectively. Since the overall reaction sequence of the archaeal rhodopsins leads back to the original ground state, it is cyclic in nature, and the three different functional elements must occur at least twice to reset the system. The photochemical or thermal isomerization of retinal has to be reversed in order to restore the original state. The same holds true for the switch that modulates the access to the CP or periplasmic channels and also the charge transfer. The model postulates that ion transfer (T) and change of accessibility (S) are kinetically independent; both are induced by the isomerization of retinal (I). It is noteworthy that all ion transfer steps observed so far in archaeal rhodopsins can be explained by the IST model. To illustrate the model, we describe proton transport in BR.
After photoisomerization of BR (I*) (the asterisk denotes photoisomerization), a proton is transferred from the Schiff base to Asp-85 (T); this step resembles the L ⇒ M transition. The accessibility of the Schiff base is then changed from the EC to the CP channel (S). A proton from the cytoplasm is now able to reprotonate the Schiff base via Asp-96 (T), and retinal isomerizes thermally from a 13-cis to an all-trans configuration (I). In the final step, the Schiff base regains access to the EC channel (S), resulting in the overall sequence I*-T-S-T-I-S. For a more detailed description of the IST model, the reader is referred to reference 197.
In the other (constrained-relaxed [CR]) model, proposed by Spudich and Lanyi (528), ion transport and signal transduction are explained by two conformations of the archaeal rhodopsins. In the relaxed state (R), the Schiff base has access to the CP channel, whereas in the constrained conformation (C), the accessibility is reversed and the Schiff base is connected to the EC channel. According to this proposal, BR retains a constrained conformation in its unphotolyzed state. Upon light excitation, a volume increase (500) indicates the transition to the relaxed state (263, 271) and the change of accessibility from the EC to the CP channel. The back reaction to the ground state returns the protein to its constrained form. In contrast to BR, HR in the unphotolyzed state adopts a relaxed conformation, which switches upon light excitation to the constrained form and therefore causes chloride transport into the cytoplasm. The authors note that this assumption is supported by the projection map of HR (200), which resembles that of the relaxed BR.
For the unphotolyzed state of SRI, a mixture of the two conformations is assumed, representing the attractant-repellent functions of the protein. The one- or two-photon process shifts the state to the relaxed or constrained conformation, respectively, thus causing the organism to be attracted or repelled. Recently, the CR model has been reviewed and explained in more detail by Spudich (526).
The IST and the CR models certainly do not exclude each other. They provide working hypotheses to inspire new experiments. However, for a more detailed understanding of the mechanisms of ion transport and signal transduction, high-resolution structural data of the four pigments and their photocycle intermediates will be critical. Unfortunately, this information is presently not available.
Phototaxis and Chemotaxis
The phototactic and chemotactic behavior of archaeal species has been studied chiefly for H. salinarum. Quite early, methylation-demethylation reactions were shown to be involved in the phototaxis of H. salinarum (396, 484). The substrate involved is Htr, a 94-kDa protein (523). Subsequently, the amino acid sequence of HtrI revealed close similarities to the chemotactic receptors (145, 619). Additional information about the coupling of the SRs with the chemotactic signal transduction chain in bacteria was gained from the sequence analysis of the genes for pSRII (503) and SRII (623). Biochemical data indicated that SR and Htr form tightly bound protein complexes (292). HtrI associates with SRI as a dimer (624), and the same stoichiometry is assumed for the SRII-HtrII complex. The photochemistries of SRI and SRII are altered upon binding to their respective transducers (462, 522). These transducer-SR complexes provide intriguing model systems for the elucidation of signal transduction across membranes. Indeed, functional and physiological analysis of the interaction of SRI with its transducer, HtrI, has already provided considerable insight into the signal transduction mechanism. A scheme of the signal transduction chain taken from reference 526 is shown in Fig. 20.
FIG. 20.
The four rhodopsins of H. salinarum. The two ion pumps, BR and HR, convert light energy into a proton and a chloride gradient, respectively, which are utilized by the cell for its energy-requiring steps, e.g., ATP synthesis, metabolism, and/or ion transport. The two other retinylidene pigments, SRI and SRII, function as photoreceptors, which direct the bacteria to optimal light conditions. The external stimuli, light and chemoeffectors, activate their corresponding receptors, i.e., the SRs, SRI and SRII, or the chemoreceptors. The SRs form complexes with their homodimeric transducers, HtrI and HtrII. After the absorption of light, the signal is transferred to the cytoplasmic domain of the transducers, which consists of three parts. Directly following the two membrane-spanning helices are small domains that are specific for the Htrs. The methylation sites downstream are involved in processes of adaptation to constant stimuli. Finally, the signaling domain interacts with a His kinase (CheA), which then transfers this information via the phosphoregulator (CheY) to the flagellar motor switch. (The figure is reproduced from reference 526 with the kind permission of J. L. Spudich.)
An analysis of the phototaxis of SR and Htr mutants showed that protonatable residues in the Htr-SR contact loop are essential for the interaction of HtrI and SRI and influence the photochemistry and function of SRI (258). The minimal fragment of HtrI able to carry out a wild-type, pH-insensitive SRI photocycle consists of residues 1 to 147, including the two transmembrane helices (TM1 and TM2) and a short cytoplasmic tail. The authors proposed that the HtrI cytoplasmic region extending from the C-terminal end of TM2 to residue 147 forms part of an electrostatic network with SRI, which also influences the Schiff base-counterion complex. The alteration of this network by the photochemical reaction cycle shifts HtrI back and forth between attractant and repellent conformations. The equilibrium between these conformations is sensitive to the extracellular pH and to mutations in HtrI.
The experimental removal of the cytoplasmic domain of HtrI, which directly follows the two transmembrane helices, interrupted signal transduction (291). The authors therefore proposed that this domain, which is absent from bacterial transducer homologs, serves to activate the downstream signaling chain. Removal of both the signaling domain and the methylation sites resulted in similar findings (618). However, a direct signaling contact of the SRs with the transmembrane helices of their corresponding transducers cannot be ruled out.
Signal transduction chain.
The discovery of a bacterial signal transduction component in H. salinarum prompted the search for the other members of the halobacterial signal relay chain. In bacteria, the receptors for the external stimuli are the methyl-accepting chemotaxis proteins (MCPs), e.g., the aspartate or serine receptor, Tar or Tsr, respectively (for a review of bacterial chemotaxis, see reference 201). MCPs have periplasmic (ligand) binding domains, two transmembrane helices, and a cytoplasmic loop that contains the signaling and the flanking methylation sites involved in the adaptive response of the bacteria. The signaling domain interacts with the first component, the autophosphorylating histidinyl kinase, CheA, of the so-called two-component system (reviewed by Stock et al. [541]). CheA phosphorylates the response regulator, CheY, which undergoes an increase in affinity for the switch of the flagellar motor, thus controlling the reversal frequency. The adaptive response is mediated by CheR and CheB, which function as methylation and demethylation enzymes, respectively.
The archaeal signal transduction chain has close similarities to that of bacterial chemotaxis (11, 12, 396, 484, 521). As outlined above, the transducers HtrI and HtrII resemble the cytoplasmic domains of the MCPs. Recently, the amino acid sequences of other soluble and membrane transducer proteins from H. salinarum have been genetically determined (451, 622). These proteins belong to three subfamilies (622); (i) a bacterial type of chemotaxis transducer with two transmembrane helices connecting a periplasmic and a cytoplasmic domain, (ii) a protein with a single cytoplasmic domain connected to two or more transmembrane helices, and (iii) a protein with a soluble cytoplasmic domain. Furthermore, a cytoplasmic arginine receptor (Car) has been identified (544). It is interesting to note that HtrII possesses, in contrast to pHtrII, an extra periplasmic loop (623). These receptor molecules must account for the ability of H. salinarum cells to respond to external stimuli. Halobacteria are attracted by several substances (e.g., glucose, sodium acetate, histidine, asparagine, and leucine) and repelled by chemicals such as sodium phenolate (373, 485). The cells are also attracted by oxygen (543). Furthermore, it was shown that BR mediates an attractant response (48, 49).
The similarities of the transducers of halobacterial phototaxis to the bacterial MCPs (313) prompted the search for cytosolic chemotactic proteins. Rudolph and Oesterhelt cloned and sequenced proteins with considerable sequence identities (30 to 35%) to the bacterial CheA (452) and CheB and CheY (454). They belong to an archaeal che operon, which includes cheA, cheB, cheY, and the novel cheJ (453). The function of CheJ is presently unknown. The extensive functional and structural similarities between the bacterial and archaeal systems suggest that the switching process will likewise be similar (453). Both cases can be kinetically described by a four-state model (298, 344).
Evidence for another component of the signal transduction system was found when fumarate “rescued” an H. salinarum mutant that was unable to reverse its direction of swimming (346). Furthermore, biochemical evidence for a membrane-bound fumarate binding protein was presented (345). However, the relationship among this factor, the CheA-CheY signal transduction chain, and the motor switch has yet to be determined.
Flagella and the flagellar motor.
The motility of halobacteria is derived from the rotation of the flagellar bundles driven by a biochemical motor. In the absence of external stimuli, halobacteria switch spontaneously from counterclockwise to clockwise rotation of the flagellar motor. This, together with Brownian movement, results in the random distribution of the organisms in their environment (342). A sudden increase of an attractant (e.g., orange light) blocks the switch for a few seconds, which ultimately results in movement oriented toward the light source. Conversely, an increased repellent concentration or a decreased attractant concentration produces shorter switching periods, allowing the bacteria to escape unfavorable conditions. The different signals sensed by the halobacterial cell are integrated by the signal relay and then transferred to the motor switch. Continuous input of a signal leads to adaptation to these conditions, the mechanism of which has been analyzed (343). On the other hand, deadaptation can occur by altering the strength of the signal input (222, 223, 529).
The right-handed flagellar bundle of H. salinarum is inserted into one pole or both of the rod-shaped cell. Interestingly, the flagella do not unbundle during a change in the rotational orientation (13); this contrasts with E. coli cells, whose flagellar bundles fly apart in the clockwise rotational mode (340). The flagella of H. salinarum have been isolated; they consist of several proteins that form three major bands by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (PAGE) (13). The assembly of halobacterial flagellar structures has recently been studied in greater detail (288, 289, 507, 557), and archaeal flagellar structures and assembly have been reviewed (143).
SECONDARY ENERGY CONVERTERS
Energy stored in electrochemical gradients of ions across plasma membranes is utilized for various secondary processes, such as the synthesis of ATP. Recent molecular and biochemical data revealed the unique structure and function of archaeal ATPases. Archaeal adenylate kinases and cytosolic pyrophosphatases (PPases) also are very different from their bacterial and eucaryal counterparts. The former enzyme catalyzes the equilibration of cellular energy charge at the nucleotide level, while the latter enzyme provides a thermodynamic sink to remove the pyrophosphate produced in several substrate activation equilibria.
The Family of ATPases
A single family of ATPases embraces the complex enzymes designated F1Fo, V1Vo (vacuolar), and A1Ao (archaeal). The P-type ATPases (417–419) belong to another family, distinguished in structure, mechanism, and evolutionary origin. F-type ATPases can reversibly link the equilibrium of ATP hydrolysis (ATP synthesis) to the flux of cations (H+ or Na+) through the membrane channel (Fo), whereas V-type ATPases, under physiological conditions, act only as ATP-driven ion pumps. Judged by their subunit composition and primary sequences, the archaeal enzymes are more closely related to the V-type ATPases, but they function as ATP synthases, i.e., like the F1Fo ATPases, at least in methanogens, halophiles, and S. acidocaldarius (464, 468, 472). Some hyperthermophilic archaea are strictly fermentative; an ATP hydrolase would be sufficient to satisfy the cellular requirements of such organisms, but the enzymes in question have not yet been investigated.
ATPases are integral membrane complexes with a peripheral protein moiety representing the catalytic headpiece (F1, A1, and V1). This is connected to an ion channel (Fo, Ao, and Vo) by a stalk that operates as a conformational energy transducer (for reviews, see references 73, 147, 417, 421, 505, 506, and 598). The recent elucidation of the high-resolution, three-dimensional structure of the mitochondrial F1 domain (4, 5, 581) and the demonstration that it functions as a molecular rotary engine (393, 459) have established a master image for the whole family of ATP synthases. The electrochemical ion gradient across the membrane drives the rotation of a membrane subunit assembly. The rotational energy is mechanically transferred to the catalytic sites in F1 to liberate preformed ATP. Concurrently, ADP and Pi are bound and occluded at another site (for details, the reader is referred to references 259 and 280). For every four protons translocated, the rotor moves by 120° inside the stator (the headpiece of ATP synthase) and one molecule of ATP is synthesized (194, 460). The stalk linking Fo to F1 is composed of two structural elements: one functions as part of the rotor (subunit γ in F-type ATPases) extending into the center of F1; the other functions as part of the stator and holds the nonmobile domains of Fo and F1 in place (61, 72, 601).
Archaeal ATPases from Sulfolobales members, methanogens, and halophiles have been isolated and studied. The number of polypeptides found in solubilized ATPases varied from two to seven (226, 228, 468, 533), but recent molecular studies indicate that 9 to 10 genes are required for ATPase function-assembly (457, 517, 602). The heterogeneity in the preparations is probably the result of differences in isolation conditions and in the physicochemical properties of the cytoplasmic membrane. In the case of extreme thermophiles, the protein complexes may be cold labile and/or certain polypeptides may escape solubilization. The isolation of an intact ATP synthase complex from archaeal plasma membranes has not yet been accomplished. Although many details of the molecular structure of the membrane domain of archaeal ATP synthases remain unknown, it is possible to propose a structural model based on the gene sequences.
The ATPases of Sulfolobus
Isolated plasma membranes of Sulfolobus (DSM 639) exhibit ATPase activities with two widely separated pH optima, 2.5 and 6.5 (333, 591, 592). The acid ATPase activity is actually due to a nonspecific PPase (an ectoenzyme), whereas the ATPase activity at pH 6.5 reflects that of the ATP synthase complex.
Both membrane-bound and solubilized ATPases of S. acidocaldarius (330, 334) are activated by sulfite and, in its presence, display a pH optimum of 6.3; the ATPase is insensitive to azide, oligomycin, bafilomycin, or vanadate but is inhibited by nitrate, p-HO-mercuribenzoate, mersalyl, and NBD-Cl. The ATPase can be released from the plasma membrane by pyrophosphate treatment as a 380-kDa particle containing four subunits of 65, 51, 20, and 12 kDa (331). The molecular properties of the solubilized ATPase from S. acidocaldarius (DSM 639) are summarized in Table 6. It has six nucleotide binding sites: three high-affinity sites and three very low affinity sites (468), reminiscent of typical F1 ATPases (for reviews, see references 73, 109, 110, 147, and 421). Photoaffinity labeling with 2-N3-ATP irreversibly inhibits the enzyme. The analog serves as a substrate (Km = 400 μM) and competes with ATP for binding. Both subunits, A and B, are labeled with a preference for the A polypeptide. Titration of the enzyme with 2-N3-ATP indicates (by extrapolation) that inhibition is complete when three sites have been blocked, but the reagent can bind covalently to all six (68, 468, 469).
TABLE 6.
Molecular and catalytic properties of A1 ATPase from S. acidocaldariusa
| Property | Value |
|---|---|
| Molecular mass (kDa) of A1 preparation | 380 |
| Subunits and stoichiometry | A3B3GD |
| Apparent molecular mass of polypeptides (kDa) | 65 (A), 51 (B), 20 (G), 12 (D) |
| Substrate specificity | ATP > GTP > ITP > CTP > UTP |
| Relative sp act (%) | 100b, 77, 72, 37, 31 |
| Apparent Km (Mg-ATP) (μM) | 180–200 |
| Activating cations | Mg2+, Mn2+ |
| Activating anion | Sulfite |
| pH optimum | 6.25 (sulfite present) |
| Temp optimum (°C) | 75–80 |
| Mg-ADP binding (μM) | Kd1 (70°C), 0.2; Kd1 (70°C), 1.8 |
| Activation energy (kJ/mol) | 67 |
| Inhibitors | Nitrate, NBD-Cl, erythrosin B, p-HO-mercuribenzoate |
| Insensitive toward: | N3−, o-vanadate, oligomycin, bafilomycin A1 |
The finding of six nucleotide binding sites is in agreement with the subunit stoichiometry (331) of A3B3 for the large polypeptides, which is supported also by the appearance of the Sulfolobus ATPase in high-resolution electron micrographs (472). The large subunits show the pseudohexagonal arrangement of six peripheral globular masses typical of all ATPases, reflecting the alternating arrangement of α/β-subunit pairs exemplified by the chloroplast enzyme (565).
Solubilized ATPases with essentially similar properties have also been reported for two other Sulfolobus strains, Sulfolobus sp. strain 7 (591) and S. solfataricus 98/2 (231). Besides, A. ambivalens was recently shown to possess a membrane-bound ATPase of the same kind (213).
The Halobacterial ATPases
ATP synthesis in cells (362, 533) or membrane vesicles (374, 375) of halobacteria can be driven by light, respiration, or artificially induced pH gradients. ATP synthesis and also ATP hydrolysis by isolated membranes are all sensitive to DCCD (293), suggesting that they are mediated by a proton-translocating ATPase.
Four species have been analyzed for ATPases: H. salinarum, Halobacterium saccharovorum, H. volcanii, and H. mediterranei. A complete ATP synthase complex could not be isolated from any of these organisms. The existence of a typical DCCD binding proteolipid has been demonstrated by [14C]DCCD labeling, but there have been contradictory reports of its apparent molecular mass: 9.7 (538), 14 (111), or 42 (287) kDa. Sequence data, available for only H. salinarum, clearly demonstrates an 8-kDa proteolipid with two transmembrane helices.
Kinetic studies of the ATPase from H. saccharovorum (493, 494) suggested nonlinear kinetics and inhibition by ADP (as also observed with F1 ATPase). Tight binding of ADP and Pi has been demonstrated, but the total number of nucleotide sites has not been determined for any of the halobacterial enzymes. A common feature of the enzymes from H. salinarum and H. volcanii is the preference for Mn2+ over Mg2+ as an activating ion (51, 111, 390). The catalytic properties and inhibitor responses of halobacterial ATPases are compared in Table 7; additional details are reviewed below.
TABLE 7.
Characteristics of soluble A1 ATPases from halophilic archaeaa
| Characteristic | Result or value for the following species:
|
|||
|---|---|---|---|---|
| H. saccharovorum | H. salinarum | H. volcanii | H. mediterranei | |
| Molecular mass (kDa) | 350 | 320 | 420b | ND |
| No. of subunit polypeptides | 4 | >2 | 4 | 4–7 |
| Sequenced genes | None | 2 (A, B) | A–E | None |
| Km [ATP] (mM) | 2.9 | 1.4 | 0.2 [Mn], 1.2 [Mg] | 5 |
| Ki [ADP] (μM) | 25–50 | 80 | 2.5 × 103 | ND |
| pH optimum | 8.5–10 | 5.8 | 9 (60°C) | 9 (60°C) |
| Azide inhibition | No/(yes)c | No | No | Yes |
| Nitrate inhibition | Yes | Yes | No (yes)e | Yes |
| Activating anion(s) | Cl−; SO3− | SO4− | NR | NR |
| NEM inhibition | Yes (A)d | Yes | Yes | Yes |
| NBD-Cl inhibition | Yes (B) | Yes | No | Yes |
| Inhibitory cation | ND | ND | Mn2+ | Mn2+ |
Data were compiled from references 111, 376, 493–495, 532, and 536; for comparison, detailed data on enzymatic properties of ATPases from Sulfolobus are contained in Table 6 and in reference 464. ND, not determined; NR, not reported.
Molecular mass calculated assuming an A3B3GDE stoichiometry.
Different in membrane-bound and solibilized states.
Indicates mainly labeled polypeptide.
Conflicting reports.
H. saccharovorum (ATCC 29253) ATPase, solubilized by octylglucoside as a 350-kDa complex, is composed of polypeptides with apparent molecular masses of 87, 60, 29, and 20 kDa (228, 533). As in many other halophilic proteins, an excess of acidic amino acids is present in polypeptides A and B, probably an adaptation to the high salt concentration of their environment (66). The enzyme is inhibited by N-ethylmaleimide (NEM) and NBD-Cl (a typical covalent inhibitor of F1 ATPases). Nucleotides protect the enzyme from inhibition by NEM and NBD-Cl. NBD-Cl binds preferentially to the 60-kDa polypeptide, whereas NEM labels the 87-kDa subunit (548).
A cryptic, detergent-inducible ATPase activity from membranes of H. saccharovorum (M6) exhibits strong inhibition by ADP (293). The preparation is activated by dithiothreitol and contains polypeptides with molecular masses of 98, 71, 31, and 22 kDa (67, 497). Direct photolabeling with [α-32P]ATP revealed the presence of a high-affinity site on subunit A (98 kDa) with a Kd of 0.6 μM, indicating that A is the catalytic subunit. A noncatalytic site was postulated for subunit B (71 kDa) (67). The stoichiometry of sites and subunits was not determined. A reaction mechanism analogous to that of F1 ATPase was inferred from the nonlinear kinetics of ATP hydrolysis and from the inhibition by tight binding of ADP and Mg2+ in the absence of Pi (493). Development of this inhibition is turnover dependent (transition into a slow state). Like F-type ATPases, this enzyme is inhibited by azide. In addition, a second binding site for Mg2+ separate from the catalytic site has been proposed (494). Occupation of the second site is thought to stimulate both the reaction rate and affinity at the catalytic site.
The H. salinarum ATPase in its membrane-bound state is inhibited by NEM, NBD-Cl, and NO3− but not by azide (376). Treatment with alkaline EDTA releases a 320-kDa complex from the membrane; the complex contains two polypeptides of 86 and 64 kDa and retains the inhibitor profile (390). In addition to ATP, the enzyme hydrolyzes ITP, GTP, and CTP; sulfite and sulfate strongly stimulate activity; and Mn2+ is three times more effective than Mg2+ in stimulating activity. For ATP, a Km of 1.4 mM was determined; the Ki for ADP (competitive) was 80 μM.
H. volcanii and H. mediterranei are closely related to each other but distantly related to the genus Halobacterium (536, 538). The membrane-bound enzymes from both species have maximal activities at pH 9 and 60°C; both ATPases require high salt concentrations (2 and 3.5 M, respectively) and are inhibited by free Mn2+ ions (111). Isolated ATPase from H. mediterranei is inhibited by azide, nitrate, NBD-Cl, and NEM (very weakly). DCCD did not inhibit the membrane-bound ATPase, but it labeled a 14-kDa polypeptide of the detergent-solubilized enzyme; apparent molecular masses of the putative subunits in this preparation were 54, 49, 22, 14, 12, and 7.5 kDa (111). The ATPase from H. volcanii is specific for ATP (Km = 0.24 mM for Mn-ATP or 1 mM for Mg-ATP; corrected for the dissociation equilibria at the applied concentrations of Mn2+ and Mg2+) (537). The soluble enzyme is sensitive to NEM but not to azide, bafilomycin, NBD-Cl, or nitrate. The insensitivity toward nitrate correlates with the ability of H. volcanii to grow with nitrate as terminal electron acceptor (16, 50).
The ATPases of Methanogens
Cellular function of the A1Ao ATPase from methanogens.
A1Ao ATPases have been purified from various methanogens, but there is no direct experimental proof that the archaeal ATPase is indeed an ATP synthase. However, there is overwhelming circumstantial evidence in favor of this assumption (383). Furthermore, recently published genome sequences of two methanogens, M. jannaschii (87) and M. thermoautotrophicum (517), revealed the presence of genes encoding the A1Ao ATPase but no genes encoding an F1Fo ATPase. Since ion gradient-driven phosphorylation is the only way for these organisms to synthesize ATP, this is the most conclusive evidence that the A1Ao ATPase does function as an ATP synthase in vivo.
Features of ATPases from methanogens.
The H+-translocating ATPase from M. mazei is the best-investigated methanogen ATPase, so it is described in detail. Information on other ATPases from methanogens is fragmentary, and the integrity of the preparations is variable, often deviating from the polypeptide compositions predicted by the DNA sequences. In addition, the reported inhibitor sensitivities have been determined under nonuniform conditions, with either the membrane-bound or the solubilized forms. Kinetic data (Km, Kd, and Ki values) and titrations of the number of nucleotide binding sites are not available. Apparently, the low efficiency of solubilization and the poor stability of the entirely intact ATPase complex have hampered detailed functional investigations.
Table 8 summarizes some fundamental properties of ATPases from methanogens, either determined on isolated proteins or derived from genetic information.
TABLE 8.
Comparison of basic properties of ATPases from methanogenic archaeaa
| Organism | No. of polypeptides (kDa) | No. of identified genes | Coupling ion | Inhibitors | Molecular mass of proteolipid (kDa) | Reference |
|---|---|---|---|---|---|---|
| M. barkeri | 2–6 (420) | 2 | H+ | DCCD, NO3− | 6 | 244 |
| M. thermophila | 5 (540) | ND | H+ | DCCD, NO3− | (?) | 242 |
| M. tindarius | 4 (445) | ND | H+ | DCCD, NO3− | 5.5 | 481 |
| M. mazei | 6 | 10 | H+ | DCCD, NO3− | 7 | 602 |
| M. thermoautotrophicum | 9–10b | H+ | NR | 15.6b | 290 | |
| M. jannaschii | 9–10b | ? | ? | 21.3 | 87 | |
| M. voltae | 3 | NA+ (?) | DCCD, NO3− | ND | 96 |
Compared to that for Sulfolobus and Halobacterium, knowledge of the catalytic properties is still rudimentary. Inhibitory function of DCCD refers to the membrane-bound ATPase activities. ND, not determined; NR, not reported.
Data extracted from genome sequences.
An active ATPase consisting of only the two subunits, A (63 kDa) and B (50 kDa), was purified from M. barkeri (243). Extraction of the membranes with octylglucoside allowed the isolation of a more complex, DCCD-sensitive form of this ATPase, with additional subunits of 40, 27, 23, and 6 kDa (244). The 6-kDa polypeptide was identified as the DCCD binding, membrane-integral subunit.
The marine methanogen M. tindarius has a membrane ATPase which is composed of four polypeptides (481) and probably resembles the hydrophilic headpiece of an ATP synthase complex; this ATPase is strongly inhibited by nitrate, a typical inhibitor of vacuolar ATPases. Neither this nor any of the other ATPases from methanogens was sensitive to azide, a typical inhibitor of F-type ATPases.
The acetoclastic methanogen Methanothrix thermophila has an A-type ATPase that was extracted by detergents; it was stimulated sixfold by sulfite and had a temperature optimum of 70°C (242). DCCD strongly inhibited the activity of both the membrane-bound and the soluble forms. N-terminal sequencing of the five constituent polypeptides of 67 (A), 52 (B), 37, 28, and 22 kDa revealed high homology to other members of the archaeal ATPase family. The same applies to the three polypeptides (80, 55, and 25 kDa) of a fragmentary ATP synthase isolated from Methanococcus voltae (96), which is assumed to perform ΔμNa+-driven ATP synthesis.
Genetic Organization of Known A1Ao ATPases
Genes encoding A1Ao ATPases have to date been cloned from three methanogens, M. jannaschii (87), M. thermoautotrophicum ΔH (517), and M. mazei (457, 602) and also from the archaea A. fulgidus (281) and Pyrococcus horikoshii (273, 274). Some of the genes encoding the A1Ao ATPase from the hyperthermophile S. acidocaldarius (118), from Desulfurococcus sp. (510), and the halophile H. volcanii (537, 538) are also known.
As can be seen from Fig. 21, the overall genetic organization of the A1Ao ATPase genes in the three methanogens, and also in A. fulgidus and P. horikoshii, is almost identical. A gene encoding a hydrophilic polypeptide is followed by two genes encoding hydrophobic subunits. The remaining genes encode hydrophilic subunits. The last gene in the aha operon of M. mazei, ahaG, has a homolog at the same position in the V1Vo ATPase operon from Enterococcus hirae (ntpH) (555) and in the A1Ao ATPase operon from S. acidocaldarius (atpE) (118), but there are no unequivocal homologs in other representatives of the Archaea. Therefore, it is still an open question whether ahaG is related to ATPase function or assembly.
FIG. 21.
Structure of A1Ao ATPase-encoding genes in archaea. Genes encoding similar polypeptides are indicated by the same pattern. Genes encoding hydrophobic polypeptides are marked by asterisks. It is suggested but not supported by experimental data that MT0952 and MJ0614 are related to ATPase function or assembly. For gene-polypeptide correspondence, see the text.
The entire aha gene cluster of M. mazei is transcribed as a single 9-kb message. Upstream of ahaH is an AT-rich region which contains two potential archaeal promoter sequences. Interestingly, there is an additional message of 0.64 kb that covers only ahaK, the gene encoding the proteolipid (457). This is of particular importance since the proteolipid is present in 9 to 12 copies per enzyme molecule, and therefore the organisms have had to develop mechanisms to ensure enhanced synthesis of the proteolipid relative to others. In E. coli, the ATPase-encoding genes form a polycistronic operon, and the high level of synthesis of the proteolipid is achieved by enhancement of translation (351). The proteolipid-encoding gene of M. mazei is also part of a polycistronic message, but no translational enhancer motif is present. In this case, the relatively high copy number of the proteolipid is apparently achieved entirely by the additional transcription of the proteolipid-encoding gene.
The ATPase genes of M. jannaschii are organized into two clusters which are separated by 334 kbp (Fig. 21). This should be kept in mind when deducing the minimal number of subunits required for ATPase function and assembly in archaea from gene sequences. Apparently, only a single cluster of ATPase genes has been found in S. acidocaldarius and H. volcanii. At present, we can assume for archaea that 9 to 10 genes are related to ATPase function and assembly.
Properties and Functions of the Polypeptides Involved in ATPase Function and Assembly
The information given below is based on the enzymes from methanogens (457, 602), unless otherwise stated.
Subunit H (Mr, 12,000) is hydrophilic and highly charged. Database searches have revealed no homologs in V1Vo and F1Fo ATPases.
Subunit I is very similar to subunit a of V1Vo ATPases but ranges in Mr from 72,000 to 76,000. It has a hydrophilic N-terminal domain with a structure rich in α helices, resembling subunit b of F1Fo ATPases. Interestingly, the similarity of the hydrophilic domain of subunit b of F1Fo ATPases to the hydrophilic domain of subunit I is 22 to 31%. The hydrophobic C terminus of subunit I is predicted to have seven transmembrane helices; similarity to subunit a of F1Fo ATPases is below 20%. The approximately 30% difference in apparent molecular mass between subunit I and subunit a of V1Vo ATPases is explained by an extreme shortage of hydrophilic loops connecting the transmembrane helices.
Subunit K is synonymous with proteolipid. Proteolipids from several archaeal organisms have been purified and characterized and shown to have an Mr of 8,000 and two transmembrane helices. This size, which is similar to that of the proteolipid from F1Fo ATPases, was always held to explain the F1Fo-like properties of the A1Ao ATPases, i.e., their function as ATP synthases. However, the recently published genome sequences of methanogens indicate, at least on the DNA level, the presence of proteolipids with four (M. thermoautotrophicum ΔH) and six (M. jannaschii) transmembrane helices, originating by gene multiplication with subsequent fusion of the genes (Fig. 22). For M. jannaschii, this was verified by matrix-assisted laser desorption ionization mass spectrometry with the purified protein (455). The active carboxylate is conserved in helices two and four of the M. thermoautotrophicum proteolipid, but in the M. jannaschii proteolipid it is conserved in only helices four and six, not helix two. The ATP synthases of M. thermoautotrophicum and M. jannaschii are the first ones known to have proteolipids with four and six transmembrane helices. Most important, the proteolipid from M. jannaschii has only two active carboxylates per six transmembrane helices, a matter of particular importance for the coupling of ion transport to ATP synthesis. The proteolipids from representatives of Archaea are similar to those of V1Vo ATPases from representatives of Bacteria or Eucarya. A leader sequence is present in H. salinarum and S. acidocaldarius.
FIG. 22.
Gene-polypeptide correspondence in proteolipids from methanoarchaea. The proteolipid-encoding gene of M. mazei (atpK) codes for an 8-kDa polypeptide with two transmembrane helices connected by a loop. It is suggested to fold in the membrane like a hairpin. Helix 2 contains the proton-translocating residue. The proteolipid-encoding genes (atpK) of M. thermoautotrophicum and M. jannaschii arose by duplication and triplication, respectively, of an ancestral gene with subsequent fusion of the genes. The proteolipids from M. thermoautotrophicum and M. jannaschii are predicted to have two and three predicted hairpins, respectively. The duplicated proteolipid from M. thermoautotrophicum contains the proton-translocating residue in both hairpins, but the triplicated form of M. jannaschii lacks it in the first of three hairpins.
Subunit E (Mr = 20,400 to 22,900) is similar to subunit E of V1Vo ATPases, and according to its size, it could well be the homolog of subunit δ of F1Fo ATPases.
Subunit C (Mr = 41,300 to 42,400) is similar to subunit d of V1Vo ATPases. Apparently, there is no homologous polypeptide in F1Fo ATPases. Subunit d of V1Vo ATPases is a hydrophilic polypeptide, but it copurifies with the Vo domain and is therefore regarded as a Vo polypeptide. Currently, there is no information on the localization of subunit C in A1Ao ATPases.
Subunit F is a hydrophilic polypeptide with an apparent molecular mass of 10.8 to 11.8 kDa, which suggests that it is the homolog of subunit F of V1Vo ATPases and subunit ɛ of F1Fo ATPases.
Subunit A (Mr = 63,800 to 66,400) is similar to subunit A of V1Vo ATPases. On the other hand, 25 to 30% of the residues are identical with those of subunit β of the F1Fo ATPase of E. coli, indicating that these two subunits have a common evolutionary precursor. Subunit A contains the Walker motifs A and B (596), which are part of the nucleotide binding domain, indicating that it is the catalytic subunit. All of the residues involved in nucleotide binding in the mitochondrial F1Fo ATPase are conserved in subunit A of A1Ao ATPase, indicating a common mechanism for nucleotide binding.
Subunit B (Mr = 50,600 to 51,900) is closely related to subunit B from V1Vo ATPases and similar to subunit α from F1Fo ATPase, but sequence analysis and experimental data suggest that subunit B is noncatalytic. The same is apparently true for subunit B from V1Vo ATPases.
Subunit D is similar to subunit D of V1Vo ATPase. Twenty-two percent of the residues of subunit D from M. mazei are identical to subunit γ of F1Fo ATPases; the N and C termini are well conserved. These regions are predicted to be α-helical, a striking homology to subunit γ of F1Fo ATPases. Thus, AhaD and the corresponding subunits in V1Vo and A1Ao ATPases might be homologs of subunit γ of F1Fo ATPases.
AhaG (Mr = 6,135) is a hydrophilic polypeptide. Database searches have not revealed homologs in V1Vo and F1Fo ATPases. It is unknown whether the gene product is related to ATPase function and assembly.
A Structural Model
It is evident from the genetic data that the A1Ao ATPase is composed of at least 9 to 10 nonidentical subunits. A structural model is presented in Fig. 23. A1Ao ATPases, like F1Fo and V1Vo ATPases, are composed of a head and a base connected by a stalk. The traditional stalk is assumed to be part of the rotor, whereas a second stalk, visualized in the V1Vo ATPase of Clostridium fervidus (61), is the stator of the rotatory enzyme. The dimensions of the domains and the complex in its entirety are comparable to those of F1Fo and V1Vo ATPases. The subunit composition of each domain is still speculative.
FIG. 23.
Hypothetical structure of the A1Ao ATPase from M. mazei. The model is based on experimental data and by analogy to F1Fo and V1Vo ATPases. It is not clear whether AhaG and AhaH are indeed part of the structure. The localization of AhaC, AhaD, AhaE, AhaF, AhaG, and AhaH is speculative. Note that this model does not apply to all methanoarchaeal ATPases, for the number of proteolipid monomers is likely to differ, depending on the size of the monomer. Furthermore, no homologs of AhaG can be identified with certainty in other archaea.
The headpiece A1 is composed of subunits A and B; electron microscopy suggests that three copies of each are arranged alternately around a central mass. In F1Fo ATPases, this central mass is formed by subunit γ, which plays an important role in transmitting energy from the membrane domain to the catalytic domain. Subunit D of A1Ao is assumed to be the homolog of γ (i.e., part of the rotor). Subunits E and F are likely to be the homologs of subunits δ and ɛ of F1Fo ATPases and, therefore, part of the stator. Nothing is known about the function of subunit C, except that its homolog from V1Vo ATPases copurifies with the membrane domain. Subunit C could be part of the stalk and associated with the membrane domain.
The membrane domain Ao is composed of two subunits, the proteolipid and subunit I, and it is likely that both subunits are involved in ion transport. In the membrane, the 8-kDa proteolipid from E. coli folds like a hairpin; it forms two transmembrane helices and contains one protonatable group (in helix two) that catalyzes H+ transport. Twelve copies of the 8-kDa proteolipid are present per F1Fo ATPase molecule (257), arranged in a ringlike structure packed front to back in the membrane (148). By analogy to F1Fo ATPases, it is assumed that the proteolipid of archaea also has a ringlike structure with a constant number of 24 helices per enzyme. This would require 12 copies of the two-helix proteolipid from M. mazei, 6 copies of the four-helix proteolipid from M. thermoautotrophicum, and 4 copies of the six-helix proteolipid from M. jannaschii.
It was always thought that the inability of the V1Vo ATPases to synthesize ATP was due to their 16-kDa proteolipid. However, recent findings of duplicated and triplicated versions of proteolipids in A1Ao ATP synthases have made it clear that the size of the proteolipid is not what determines whether a given enzyme can catalyze ATP synthesis in addition to hydrolysis. The important factor is the number of protonatable groups per enzyme, which (based on a constant number of 24 helices per enzyme) is 8 in M. jannaschii, 12 in all other A1Ao and F1Fo ATPases presently known, and 6 in V1Vo ATPases. Assuming three catalytic centers per enzyme, this would lead to a stoichiometry of 2 H+/ATP in V1Vo ATPase and 4 H+/ATP in F1Fo ATPase and almost all A1Ao ATPases, but 2.7 H+/ATP in the A1Ao ATPase of M. jannaschii. Since the enzyme from M. jannaschii is an ATP synthase, a stoichiometry of 2.7 H+/ATP is apparently sufficient for ATP synthesis. The stoichiometry of 2 H+/ATP observed in V1Vo ATPase is too low to sustain ATP synthesis, but it does allow the generation of a large proton gradient across the membrane.
Subunit I is a two-domain subunit; it is very tempting to speculate that the two domains have distinct functions which, in F1Fo ATPases, are associated with two separate polypeptides, subunits a and b. The hydrophobic domains of subunit a of V1Vo and of subunit a of F1Fo ATPases are involved in H+ translocation (114, 148, 314, 315). The relatively large and α-helical hydrophilic domain of subunit I could serve as the stator in A1Ao ATPases.
The novel features of archaeal ATP synthases.
The same structure, i.e., two domains connected by a stalk, is found in ATPases from all three branches of the evolutionary tree, and it is assumed that all ATPases arose from a common ancestor (176, 391, 550). The major polypeptides (A and B) originated by the duplication of a single ancestral gene. In the line leading to the A1Ao and V1Vo ATPases, a number of deletions and insertions occurred. These changes had two consequences: one subunit of the A3B3 core particle lost catalytic activity, and the catalytic subunit was enlarged relative to the noncatalytic one (the reverse is the case for F1Fo ATPases).
One hypothesis states that the divergence of the A1Ao ATPases and the V1Vo ATPases took place by a duplication and subsequent fusion of the genes encoding the proteolipid. This would explain the apparent inability of V1Vo ATPases to synthesize ATP. However, multiplied proteolipids have been found in ATP synthases from Archaea. Interestingly, the multiplied genes were fused in M. thermoautotrophicum and M. jannaschii but not in A. fulgidus, which apparently represents an intermediate in evolution (Fig. 22). Very recently, duplications and triplications of the two-helix proteolipid from E. coli were generated by molecular methods. Interestingly, transformants were able to grow on succinate, thus demonstrating that these F1Fo ATPases with multiplied proteolipids had retained the ability to synthesize ATP (257). Taken together, these experiments supply clear evidence that the ability of a given enzyme to synthesize ATP is not determined simply by the size of the proteolipid per se, but rather by structure conservation and by the capacity of each hairpin to participate in ion translocation. In the duplicated four-helix proteolipids of V1Vo ATPases, hairpin 1 is apparently degenerate, having lost its protonatable group and the capability to pump protons; perhaps for that reason, V1Vo ATPases are unable to synthesize ATP.
It is difficult to decide whether the discovery of multiplied proteolipids in archaeal organisms calls into question the hypothesis that the divergence of A1Ao from V1Vo took place by duplication of the proteolipid-encoding gene. However, it should be kept in mind that most archaeal proteolipids have a molecular mass of approximately 8 kDa, and only a few have undergone multiplication. Recently, it was proposed that duplication of the eucaryal and that of the archaeal proteolipid genes were independent events (221). Multiplication of a proteolipid-encoding gene (with and without subsequent fusion of the multiplied genes) was also found in the operon encoding the Na+-F1Fo ATPase of the gram-positive bacterium A. woodii (434). The evolution of ATPases is complicated by the possibility of a horizontal gene transfer, which could explain the occurrence of multiplied proteolipids found in some A1Ao ATPases. The occurrence of V1Vo ATPases in some bacterial species, as well as the coexistence of A1Ao and F1Fo ATPase genes in M. barkeri, has also been interpreted as the result of horizontal gene transfer (404). What could be the reason for multiplication of the proteolipid-encoding gene, which is apparently found in all domains of life? The copy number of proteolipids is 4 to 12 times higher than that of the other subunits. As long as the proteolipid-encoding gene is part of a polycistronic message (as is apparently the case in all prokaryotes), only a few mechanisms for enhanced synthesis of the proteolipid are conceivable: first, enhancement of translation as observed for E. coli (351); second, separate, additional transcription of the proteolipid-encoding gene only, as observed for M. mazei; and third, multiplication of the proteolipid-encoding gene and insertion of the copies into the same polycistronic message, as observed for A. woodii. In any case, further regulation via degradation of mRNA is conceivable.
Auxiliary Energy Transducers
Two soluble enzymes involved in bioenergetics are fundamental to the life of all organisms: adenylate kinases and PPases. Archaeal examples of these enzymes have recently been characterized and crystallized, revealing unusual features unique to the archaea.
Regardless of the source of ATP, whether it is generated by fermentation or by chemiosmotic processes, adenylate kinase operation functions as an energy buffer within the adenine nucleotide pool (28). This is facilitated by its low specificity for nucleotide triphosphates as phosphate donors. Deletion of the gene for inorganic PPase is lethal, as shown in E. coli (95), because the hydrolytic cleavage of pyrophosphate does not take place. This reaction serves as a thermodynamic sink for numerous activation and polymerization reactions that liberate inorganic pyrophosphate.
The unique structure of archaeal adenylate kinases.
Adenylate kinases from Sulfolobus (272, 299), Methanococcus (87, 458), and H. halobium (518) have been described elsewhere. According to genetically derived sequences, the latter adenylate kinase belongs to the long type found in many eukaryotes; the former two enzymes are members of the short type, apparently forming a novel family typical of archaea (188). This has been confirmed by the high-resolution, three-dimensional structure of the Sulfolobus enzyme (588), which turns out to be trimeric (in contrast to the monomeric adenylate kinases from bacterial and eucaryal sources). The four methanococcal enzymes have a high degree of homology (about 80%), suggesting that the oligomeric structure is characteristic of the whole family. Trimerization is mediated by hydrophobic interactions and salt bridges between α helices in the core of the oligomer and by hydrogen bonds between a projecting extra loop (not found in any other adenylate kinase) and a β strand of the neighboring monomer. The canonical P-loop motif of adenylate kinases in the pyrophosphate binding fold, GxPGxGKGT (131, 378), is conserved, with two significant deviations: the replacement of the underlined glycine residue by a hydroxylated residue of serine (Sulfolobus) or threonine (methanogens) and the absence of the adjacent lysine residue in sequences from methanogens. This lysine residue represents an essential residue in the phosphate binding region and may be compensated for by another basic residue from outside the P-loop domain in adenylate kinases from methanogens.
Trimerization is considered to contribute essentially to the extreme thermostability (86) of the adenylate kinase from S. acidocaldarius.
The enzyme from Sulfolobus is the only archaeal example that has been functionally characterized after purification to homogeneity (299). The catalytic trimer has a temperature optimum of 90°C; the pH optimum of 5.5 to 6.0 reflects the cytosolic pH of Sulfolobus. It is absolutely specific for AMP as phosphate acceptor, with nearly identical apparent Km values of 0.6 × 10−3 to 0.7 × 10−3 M for ATP, ADP, and AMP. In contrast to the mammalian enzymes, its sensitivity to the inhibitor diadenosine-5,5′-pentaphosphate is extremely low (50% inhibitory concentration >300 μM).
A phylogenetic dendrogram including all nucleotide monophosphate kinases known clearly reveals that archaeal adenylate kinases form a distinct branch, with the pyrimidine nucleotide monophosphate kinases as their closest relatives.
Archaeal PPases.
Inorganic pyrophosphate as a possible primordial energy source has been discussed in a number of reviews (32–34). It serves as an energy donor in various microbial reactions (185, 609); an archaeal example is the phosphofructokinase from Thermoproteus tenax (8). In addition to its role as an inorganic energy donor, pyrophosphate (when converted to higher polyphosphates) may serve also as a phosphate store in low-salinity habitats. On the other hand, hydrolytic removal of cytosolic pyrophosphate is fundamental for the shift of the reaction equilibria toward product formation, for example during DNA replication or transcription and many other pyrophosphate-liberating processes.
Mg2+-dependent cytosolic inorganic PPases from methanogens (255, 447, 580) and thermoacidophiles (17, 357, 358, 441, 442, 590), but not from halobacteria, have been characterized. The discovery of membrane-bound ecto-PPases in members of the Sulfolobales was surprising. The fact that bacitracin inhibits both the PPase of S. acidocaldarius and cell growth (358, 359) suggests that this enzyme acts as a dolichol PPase and is involved in the glycosylation of cell wall components. Dolichol PPases do not require divalent cations for activity and cleave pyrophosphate bonds with low specificity (including those of ATP and ADP); this explains the second pH optimum at pH 2.5 to 3.0 for membrane-bound ATPase activities in Sulfolobus (333, 592). By contrast, the PPase from M. thermoautotrophicum (DSM 3590) requires Mg2+ but is further activated when Fe2+, Ni2+, or Zn2+ ions are also present. Table 9 summarizes the properties of archaeal PPases.
TABLE 9.
Properties of soluble and membrane-bound archaeal PPasesa
| Organism | Location | Mass (kDa) | Oligomeric statec | pH optimum | Temp optimum (°C) | Gene sequenced | Km (μM) | Inhibitor(s) | Activator | Reference(s) |
|---|---|---|---|---|---|---|---|---|---|---|
| S. acidocaldarium | Cytosolb | 19 | Hex | 6.20 | 85 | Yes | ? | F−, methylglyoxal | Mg2+ | 357 |
| Sulfolobus sp. strain 7 | Cytosol | 21 | ? | 6.50 | ? | No | ND | ND | Mg2+ | 590 |
| T. acidophilum | Cytosol | 21 | Hex | ? | 85 | Yes | 7 | F− | Mg2+ | 441, 442 |
| M. thermoautotrophicum | Cytosol | 25 | Hex | 7.70 | 70 | No | 160 | F−, Mn2+ | Mg2+ | 580 |
| M. soehngenii | Cytosol | 35, 33 | 139 | 8.00 | ND | no | 100 | ND | Mg2+ | 255 |
| M. thermoautotrophicum | Membrane | 31 | ND | 8.00 | 60 | No | ND | ND | Fe, Ni, Zn2+ | 447 |
| S. acidocaldarius | Membrane | 17 | 70 | 3.00 | ND | No | ? | Bacitracin | Noneb | 358 |
| Sulfolobus sp. strain 7 | Membrane | 19 | 35 | 2.50 | ND | No | ND | ND | None | 17 |
The data are compiled from the references cited in the rightmost column. ND, not determined.
In solubilized form, this enzyme requires a partially purified anionic membrane lipid fraction for maximal activation (358).
If the oligomeric state is not known, the apparent molecular mass of the solubilized enzyme is given. Hex, hexameric.
A number of sequences have been derived from the genomes of M. jannaschii (87) and A. fulgidus (281), but surprisingly, homologous genes were not identified, even though enzymatic assays clearly revealed the presence of a cytosolic PPase with a temperature optimum of 85°C in cell extracts of the former (190). Very recent data suggests, however, that a novel species of inorganic PPases exists in Methanococcus and Archaeoglobus, a species which is homologous to the enzyme from Bacillus subtilis (190, 239) and unrelated to the classical cytosolic PPases discussed above.
The extent of knowledge on cytosolic archaeal PPases varies. The enzyme from Methanothrix soehngenii is composed of two different polypeptides, a clear exception to the rule, because all other cytosolic PPases are homo-oligomers. Several enzymes were previously described as tetrameric, e.g., those from Sulfolobus (357, 590). However, crystallization and determination of the high-resolution three-dimensional structure of PPase from S. acidocaldarius (316), together with sedimentation analysis, revealed a hexameric structure. Thus, it is likely that all hyperthermophilic archaeal PPases are hexameric. The tight packing of the oligomer may cause irregular migration in gel-exclusion chromatography. Interestingly, this phenomenon resembles the situation with adenylate kinase and superoxide dismutase from Sulfolobus (284, 588).
PPase from S. acidocaldarius is the example best investigated (189) with respect to thermostability. In the presence of Mg2+ the midpoint temperature of thermal unfolding is 98°C; with Mn2+ it shifts to above 100°C. Binding of a single bivalent cation per monomer is sufficient to enhance thermostability. Moreover, X-ray crystallography has shown that the number of intersubunit contacts and of intramolecular hydrogen bonds is also higher than those in mesophilic PPases. The overall folding is surprisingly similar to that of other prokaryotic PPases (316). Fourteen amino acid residues involved in the binding of substrates and metal ions and in catalysis are invariantly conserved in all known sequences of homo-oligomeric PPases, although the degree of identity varies from 13 to 83%. In contrast to adenylate kinases, there appears to be no class of PPases typical of Archaea. It may be the case that archaeal PPases retain the folding pattern of an ancestral enzyme and also the spatial position of catalytic residues, while tolerating numerous mutations that did not affect these parameters.
CONCLUSIONS AND PERSPECTIVES
The study of archaeal bioenergetics has made a number of unexpected discoveries. These include the detection of new coenzyme factors in methanogenesis, new energy-conserving pathways, the unusual composition of archaeal respiratory complexes (which may include novel mechanisms of proton pumping), and novel light-induced signaling mechanisms (not to mention rhodopsin-dependent energy conservation in members of the Halobacteriales). In addition, investigation of the archaea in general has contributed largely to the elucidation of microbial evolution, protein phylogeny, and bioenergetic mechanisms.
Nevertheless, a number of interesting phenomena still elude experimental access. For example, the mechanisms of proton pumping by heterodisulfide reductase and other archaeal membrane protein complexes remain to be understood at the molecular level. Similarly, the function of the cysteine-rich subunit of Sulfolobus SDH in electron transport is entirely enigmatic, as are the primary energy-transducing mechanisms in archaeal sulfur metabolism. Another example is the highly glycosylated cyt b which is exposed on the outer surface of the S. acidocaldarius plasma membrane. Its function is unknown but may afford new insight into the periplasmic redox systems of members of the Sulfolobales. The participation of multiple isoforms of ferredoxins in archaeal redox chemistry opens the prospect of metabolic compartmentation by donor-acceptor specificity. Many other unresolved details of bioenergetics and related metabolic reactions could be listed. Actually, the fact that less than 40% of the reading frames identified in various archaeal genomes can presently be attributed to known proteins illustrates that much further research work will be necessary, shifting the focus of biochemistry from pure genomics to protein biochemistry.
The answers to many of the open questions will grow from the development of procedures for the genetic transformation of archaea. Thus far, transformation has been successful only with Halobacterium and Haloferax, and genetic tools for methanogens are available to only a limited extent; no practical procedures are available for members of the Sulfolobales. However, only such procedures will allow us to genetically disrupt individual members of parallel electron transport pathways and work out the function of each limb. Likewise, it will be necessary to construct overproducers in order to overcome the problem of accessibility of certain archaeal membrane protein complexes, either for in vitro reconstitution or for crystallization. The major challenge in this respect is the development of stable shuttle vectors which replicate in both E. coli (for example) and specific archaeal hosts and which tolerate extreme temperatures or ionic strength within the latter. Besides their importance for the further elucidation of archaeal bioenergetics, such systems will open a wide field of applications including the biotechnological use of hyperthermophiles as cell factories for production of extremely thermostable, halotolerant, or specifically glycosylated archaeal proteins.
ACKNOWLEDGMENTS
We are most grateful to J. Kraus for his help with computer graphics and the management of the reference data library, to H. Lorenz and A. Scholz for secretarial assistance, and to many colleagues and friends who provided recent results prior to publication as well as stimulating discussions and criticism. We are grateful to L. Rogers-Blaut for language editing.
For continuous support of our research by generous grants, we acknowledge the support of the Deutsche Forschungsgemeinschaft.
ADDENDUM IN PROOF
After the manuscript had been accepted for publication, additional evidence for the mechanisms of energy conservation by sulfur respiration was reported (R. Hedderich, O. Klimmek, A. Kröger, R. Dirmeier, M. Keller, and K. O. Stetter, FEMS Microbiol. Rev. 22:353–381, 1999).
REFERENCES
- 1.Abken H-J, Tietze M, Brodersen J, Bäumer S, Beifuss U, Deppenmeier U. Isolation and characterization of methanophenazine and function of phenazines in membrane-bound electron transport of Methanosarcina mazei Gö1. J Bacteriol. 1998;180:2027–2032. doi: 10.1128/jb.180.8.2027-2032.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abken H J, Bäumer S, Brodersen J, Murakami E, Ragsdale S W, Gottschalk G, Deppenmeier U. Membrane-bound electron transport and H+ translocation in Methanosarcina barkeri Gö1. Biospectrum, special issue, March. 1998. p. 38. [Google Scholar]
- 3.Abken H J, Deppenmeier U. Purification and properties of an F420H2 dehydrogenase from Methanosarcina mazei Gö1. FEMS Microbiol Lett. 1997;154:231–237. [Google Scholar]
- 4.Abrahams J P, Buchanan S K, Van Raaij M J, Fearnley I M, Leslie A G W, Walker J E. The structure of bovine F1-ATPase complexed with the peptide antibiotic efrapeptin. Proc Natl Acad Sci USA. 1996;93:9420–9424. doi: 10.1073/pnas.93.18.9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abrahams J P, Leslie A G W, Lutter R, Walker J E. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 6.Achenbach-Richter L, Gupta R, Stetter K O, Woese C R. Were the original eubacteria thermophiles? Syst Appl Microbiol. 1987;9:34–39. doi: 10.1016/s0723-2020(87)80053-x. [DOI] [PubMed] [Google Scholar]
- 7.Adams M W W. Enzymes and proteins from organisms that grow near and above 100°C. Annu Rev Microbiol. 1993;47:627–658. doi: 10.1146/annurev.mi.47.100193.003211. [DOI] [PubMed] [Google Scholar]
- 8.Adelroth P, Gennis R B, Brzezinski P. Role of the pathway through K(I-362) in proton transfer in cytochrome c oxidase from Rhodobacter sphaeroides. Biochemistry. 1998;37:2470–2476. doi: 10.1021/bi971813b. [DOI] [PubMed] [Google Scholar]
- 9.Adler J. Chemotaxis in bacteria. Annu Rev Biochem. 1975;44:341–356. doi: 10.1146/annurev.bi.44.070175.002013. [DOI] [PubMed] [Google Scholar]
- 10.Aitken D M, Brown A D. Citrate and gloxylate cycles in the halophile, Halobacterium salinarum. Biochim Biophys Acta. 1969;177:351–354. doi: 10.1016/0304-4165(69)90148-2. [DOI] [PubMed] [Google Scholar]
- 11.Alam M, Hazelbauer G L. Structural features of methyl-accepting taxis proteins conserved between archaebacteria and eubacteria revealed by antigenic cross-reaction. J Bacteriol. 1991;173:5837–5842. doi: 10.1128/jb.173.18.5837-5842.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alam M, Lebert M, Oesterhelt D, Hazelbauer G L. Methyl-accepting taxis proteins in Halobacterium halobium. EMBO J. 1989;8:631–639. doi: 10.1002/j.1460-2075.1989.tb03418.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alam M, Oesterhelt D. Morphology function and isolation of halobacterial flagella. J Mol Biol. 1984;176:459–475. doi: 10.1016/0022-2836(84)90172-4. [DOI] [PubMed] [Google Scholar]
- 14.Alex L A, Reeve J N, Orme-Johnson W H, Walsh C T. Cloning, sequence determination, and expression of the genes encoding the subunits of the nickel-containing 8-hydroxy-5-deazaflavin reducing hydrogenase from Methanobacterium thermoautotrophicum strain DH. Biochemistry. 1990;24:7237–7244. doi: 10.1021/bi00483a011. [DOI] [PubMed] [Google Scholar]
- 15.Althaus T, Eisfeld W, Lohrmann R, Stockburger M. Application of raman spectroscopy to retinal proteins. Isr J Chem. 1995;35:227–251. [Google Scholar]
- 16.Alvarez-Ossorio M C, Muriana F J G, de la Rossa F F, Relimpio A M. Purification and characterization of a nitrate reductase from the halophilic archaebacterium Haloferax mediterranei. Z Naturforsch. 1992;47c:670–676. [Google Scholar]
- 17.Amano T, Wakagi T, Oshima T. An ecto-enzyme from Sulfolobus acidocaldarius strain 7 which catalyzes hydrolysis of inorganic pyrophosphate, ATP, and ADP: purification and characterization. J Biochem (Tokyo) 1993;114:329–333. doi: 10.1093/oxfordjournals.jbchem.a124176. [DOI] [PubMed] [Google Scholar]
- 18.Ames J B, Raap J, Lugtenburg J, Mathies R A. Resonance Raman study of halorhodopsin photocycle kinetics, chromophore structure, and chloride-pumping mechanism. Biochemistry. 1992;31:12546–12554. doi: 10.1021/bi00165a002. [DOI] [PubMed] [Google Scholar]
- 19.Anemüller, S. 1997. Personal communication.
- 20.Anemüller S, Hettmann T, Moll R, Teixeira M, Schäfer G. EPR characterization of an archaeal succinate dehydrogenase in the membrane bound state. Eur J Biochem. 1995;232:563–568. [PubMed] [Google Scholar]
- 21.Anemüller S, Lübben M, Schäfer G. The respiratory system of Sulfolobus acidocaldarius, a thermoacidophilic archaebacterium. FEBS Lett. 1985;193:83–87. [Google Scholar]
- 22.Anemüller S, Schäfer G. Cytochrome aa3 from the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. FEBS Lett. 1989;244:451–455. doi: 10.1016/0014-5793(93)81328-w. [DOI] [PubMed] [Google Scholar]
- 23.Anemüller S, Schäfer G. Cytochrome aa3 from Sulfolobus acidocaldarius. Eur J Biochem. 1990;191:297–305. doi: 10.1111/j.1432-1033.1990.tb19123.x. [DOI] [PubMed] [Google Scholar]
- 24.Anemüller S, Schmidt C L, Pacheco I, Schäfer G, Teixeira M. A cytochrome aa3-type quinol oxidase from Desulfurolobus ambivalens, the most acidophilic archaeon. FEMS Microbiol Lett. 1994;117:275–280. [Google Scholar]
- 25.Anemüller S, Schmidt C L, Schäfer G, Bill E, Trautwein A X, Teixeira M. Evidence for a two proton dependent redox equilibrium in an archaeal Reiske iron-sulfur cluster. Biochem Biophys Res Commun. 1994;202:252–257. doi: 10.1006/bbrc.1994.1920. [DOI] [PubMed] [Google Scholar]
- 26.Anemüller S, Schmidt C L, Schäfer G, Teixeira M. Evidence for a Rieske-type FeS center in the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. FEBS Lett. 1993;318:61–64. doi: 10.1016/0014-5793(93)81328-w. [DOI] [PubMed] [Google Scholar]
- 27.Aono S, Bryant F O, Adams M W W. A novel and remarkably thermostable ferredoxin from the hyperthermophilic archaebacterium Pyrococcus furiosus. J Bacteriol. 1989;171:3433–3439. doi: 10.1128/jb.171.6.3433-3439.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atkinson D E. The energy charge of the adenylate pool as a regulatory parameter. Interaction with feedback modifiers. Biochemistry. 1968;7:4030–4034. doi: 10.1021/bi00851a033. [DOI] [PubMed] [Google Scholar]
- 29.Bach M, Reiländer H, Gärtner P, Lottspeich F, Michel H. Nucleotide sequence of a putative succinate dehydrogenase operon in Thermoplasma acidophilum. Biochim Biophys Acta. 1993;1174:103–107. doi: 10.1016/0167-4781(93)90101-i. [DOI] [PubMed] [Google Scholar]
- 30.Bakker E P. The role of alkali-cation transport in energy coupling of neutrophilic and acidophilic Bacteria: an assessment of methods and concepts. FEMS Microbiol Rev. 1991;75:319–334. [Google Scholar]
- 31.Balashov S P, Govindjee R, Imasheva E S, Misra S, Ebrey T G, Feng Y, Crouch R K, Menick D R. The two pKa’s of aspartate-85 and control of thermal isomerization and proton release in the arginine-82 to lysine mutant of bacteriorhodopsin. Biochemistry. 1995;34:8820–8834. doi: 10.1021/bi00027a034. [DOI] [PubMed] [Google Scholar]
- 32.Baltscheffsky H, Baltscheffsky M. Energy-rich phosphate compounds and the origin of life. In: Poglazov P, editor. Evolutionary biochemistry. Moscow, Russia: ANKO; 1995. pp. 191–199. [Google Scholar]
- 33.Baltscheffsky H, Lundin M, Luxemburg C, Nyren P. Inorganic pyrophosphate and the molecular evolution of biological energy coupling. Chem Scr. 1986;26B:259–262. [Google Scholar]
- 34.Baltscheffsky M, Baltscheffsky H. Inorganic pyrophosphate and inorganic pyrophosphatases. Mol Mechan Bioenerg. 1992;14:331–348. [Google Scholar]
- 35.Bamberg E, Hegemann P, Oesterhelt D. The chromoprotein of halorhodopsin is the light-driven electrogenic chloride pump in Halobacterium halobium. Biochemistry. 1984;23:6216–6221. doi: 10.1021/bi00320a050. [DOI] [PubMed] [Google Scholar]
- 36.Bamberg E, Tittor J, Oesterhelt D. Light-driven proton or chloride pumping by halorhodopsin. Proc Natl Acad Sci USA. 1993;90:639–643. doi: 10.1073/pnas.90.2.639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barns S M, Delwiche C F, Palmer J D, Pace N R. Perspectives on archaeal diversity, thermophily and monophyly from environmental rRNA sequences. Proc Natl Acad Sci USA. 1996;93:9188–9193. doi: 10.1073/pnas.93.17.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baselt D R, Fodor S P, van der Steen R, Lugtenburg J, Bogomolni R A, Mathies R A. Halorhodopsin and sensory rhodopsin contain a C6-C7 s-trans retinal chromophore. Biophys J. 1989;55:193–196. doi: 10.1016/S0006-3495(89)82791-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baumeister W, Lembcke G. Structural features of archaebacterial cell envelopes. J Bioenerg Biomembr. 1992;24:567–575. doi: 10.1007/BF00762349. [DOI] [PubMed] [Google Scholar]
- 40.Bäumer S, Murakami E, Brodersen J, Gottschalk G, Ragsdale S W, Deppenmeier U. The F420H2:heterodisulfide oxidoreductase system from Methanosarcina species: 2-hydroxyphenazine mediates electron transfer from F420H2 dehydrogenase to heterodisulfide reductase. FEBS Lett. 1998;428:295–298. doi: 10.1016/s0014-5793(98)00555-9. [DOI] [PubMed] [Google Scholar]
- 41.Bayley H, Huang K S, Radhakrishnan R, Ross A H, Takagaki Y, Khorana H G. Site of attachment of retinal in bacteriorhodopsin. Proc Natl Acad Sci USA. 1981;78:2225–2229. doi: 10.1073/pnas.78.4.2225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bayley S T, Kushner D J. The ribosomes of the extremely halophilic bacterium, Halobacterium cutirubrum. J Mol Biol. 1974;9:654–669. doi: 10.1016/s0022-2836(64)80173-x. [DOI] [PubMed] [Google Scholar]
- 43.Becher B, Müller V. ΔμNa+ drives the synthesis of ATP via an ΔμNa+-translocating F1F0-ATP synthase in membrane vesicles of the archaeon Methanosarcina mazei Gö1. J Bacteriol. 1994;176:2543–2550. doi: 10.1128/jb.176.9.2543-2550.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Becher B, Müller V, Gottschalk G. N5-Methyl-tetrahydromethanopterin:coenzyme M methyltransferase of Methanosarcina strain Gö1 is an Na+-translocating membrane protein. J Bacteriol. 1992;174:7656–7660. doi: 10.1128/jb.174.23.7656-7660.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becker M, Schäfer G. Purification and spectral characterization of a b-type cytochrome from the plasma membrane of the archaebacterium Sulfolobus acidocaldarius. FEBS Lett. 1991;291:331–335. doi: 10.1016/0014-5793(91)81314-x. [DOI] [PubMed] [Google Scholar]
- 46.Belly R T, Bohlool B B, Brock T D. The genus Thermoplasma. Ann N Y Acad Sci. 1973;225:94–107. [Google Scholar]
- 47.Bertram P A, Thauer R K. Thermodynamics of the formylmethanofuran dehydrogenase reaction in Methanobacterium thermoautotrophicum. Eur J Biochem. 1994;226:811–818. doi: 10.1111/j.1432-1033.1994.t01-1-00811.x. [DOI] [PubMed] [Google Scholar]
- 48.Bibikov S I, Grishanin R N, Kaulen A D, Marwan W, Oesterhelt D, Skulachev V P. Bacteriorhodopsin is involved in halobacterial photoreception. Proc Natl Acad Sci USA. 1993;90:9446–9450. doi: 10.1073/pnas.90.20.9446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bibikov S I, Grishanin R N, Marwan W, Oesterhelt D, Skulachev V P. The proton pump bacteriorhodopsin is a photoreceptor for signal transduction in Halobacterium halobium. FEBS Lett. 1991;295:223–226. doi: 10.1016/0014-5793(91)81423-6. [DOI] [PubMed] [Google Scholar]
- 50.Bickel-Sandkötter S, Ufer M. Properties of a dissimilatory nitrate reductase from the halophilic archaeon Haloferax volcanii. Z Naturforsch. 1995;50c:365–372. [Google Scholar]
- 51.Bickel-Sandkötter S, Wagner V, Schumann D. ATP-synthesis in archaea: structure-function relations of the halobacterial A-ATPase. Photosynth Res. 1998;57:335–345. [Google Scholar]
- 52.Bivin D B, Stoeckenius W. Photoactive retinal pigments in haloalkaliphilic bacteria. J Gen Microbiol. 1986;132:2167–2177. doi: 10.1099/00221287-132-8-2167. [DOI] [PubMed] [Google Scholar]
- 53.Blanck A, Oesterhelt D. The halo-opsin gene. II: Sequence primary structure of halorhodopsin and comparison with bacteriorhodopsin. EMBO J. 1987;6:265–273. doi: 10.1002/j.1460-2075.1987.tb04749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blanck A, Oesterhelt D, Ferrando E, Schegk E S, Lottspeich F. Primary structure of sensory rhodopsin I, a prokaryotic photoreceptor. EMBO J. 1989;8:3963–3971. doi: 10.1002/j.1460-2075.1989.tb08579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Blaurock A E, Stoeckenius W. Structure of the purple membrane. Nat New Biol. 1971;233:152–155. doi: 10.1038/newbio233152a0. [DOI] [PubMed] [Google Scholar]
- 56.Blaut M, Gottschalk G. Coupling of ATP synthesis and methane formation from methanol and molecular hydrogen in Methanosarcina barkeri. Eur J Biochem. 1984;141:217–222. doi: 10.1111/j.1432-1033.1984.tb08178.x. [DOI] [PubMed] [Google Scholar]
- 57.Blaut M, Müller V, Gottschalk G. Proton translocation coupled to methanogenesis from methanol + hydrogen in Methanosarcina barkeri. FEBS Lett. 1987;215:53–57. [Google Scholar]
- 58.Blaut M, Müller V, Gottschalk G. Energetics of methanogenesis studied in vesicular systems. J Bioenerg Biomembr. 1992;24:529–546. doi: 10.1007/BF00762346. [DOI] [PubMed] [Google Scholar]
- 59.Blaut M, Peinemann S, Deppenmeier U, Gottschalk G. Energy transduction in vesicles of the methanogenic strain Gö1. FEMS Microbiol Rev. 1990;87:367–372. [Google Scholar]
- 60.Bock A-K, Kunow J, Glasemacher J, Schönheit P. Catalytic properties, molecular composition and sequence alignments of pyruvate:ferredoxin oxidoreductase from the methanogenic archaeon Methanosarcina barkeri (strain Fusaro) Eur J Biochem. 1996;237:35–44. doi: 10.1111/j.1432-1033.1996.0035n.x. [DOI] [PubMed] [Google Scholar]
- 61.Boekema E J, Ubbink-Kok T, Lolkema J S, Brisson A, Konings W N. Visualization of a peripheral stalk in V-type ATPase: evidence for the stator structure essential to rotational catalysis. Proc Natl Acad Sci USA. 1997;94:14291–14293. doi: 10.1073/pnas.94.26.14291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bogomolni R A, Spudich J L. Identification of a third rhodopsin-like pigment in phototactic Halobacterium halobium. Proc Natl Acad Sci USA. 1982;79:6250–6254. doi: 10.1073/pnas.79.20.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bogomolni R A, Spudich J L. The photochemical reactions of bacterial sensory rhodopsin-I. Biophys J. 1987;52:1071–1075. doi: 10.1016/S0006-3495(87)83301-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bogomolni R A, Stoeckenius W, Szundi I, Perozo E, Olson K D, Spudich J L. Removal of transducer HtrI allows electrogenic proton translocation by sensory rhodopsin I. Proc Natl Acad Sci USA. 1994;91:10188–10192. doi: 10.1073/pnas.91.21.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bogomolni R A, Taylor M E, Stoeckenius W. Reconstitution of purified halorhodopsin. Proc Natl Acad Sci USA. 1984;81:5408–5411. doi: 10.1073/pnas.81.17.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Böhm G, Jaenicke R. A structure-based model for the halophilic adaptation of dihydrofolate reductase from Halobacterium volcanii. Protein Eng. 1994;7:213–230. doi: 10.1093/protein/7.2.213. [DOI] [PubMed] [Google Scholar]
- 67.Bonet M L, Schobert B. The catalytic site is located on subunit I of the ATPase from Halobacterium saccharovorum—a direct photoaffinity labeling study. Eur J Biochem. 1992;207:369–376. doi: 10.1111/j.1432-1033.1992.tb17059.x. [DOI] [PubMed] [Google Scholar]
- 68.Bönisch H. Dipl. thesis. Lübeck, Germany: University of Lübeck; 1992. Bindungsstudien mit dem photoreaktiven Substratanalogen 2-azido-ATP an der ATPase aus Sulfolobus acidocaldarius; pp. 1–38. [Google Scholar]
- 69.Bönisch, H. E. 1998. Personal communication.
- 70.Bott M, Eikmanns B, Thauer R K. Coupling of carbon monoxide oxidation to CO2 and H2 with the phosphorylation of ADP in acetate-grown Methanosarcina barkeri. Eur J Biochem. 1986;159:393–398. doi: 10.1111/j.1432-1033.1986.tb09881.x. [DOI] [PubMed] [Google Scholar]
- 71.Bott M, Thauer R K. Proton translocation coupled to the oxidation of carbon monoxide to CO2 and H2 in Methanosarcina barkeri. Eur J Biochem. 1989;179:469–472. doi: 10.1111/j.1432-1033.1989.tb14576.x. [DOI] [PubMed] [Google Scholar]
- 72.Böttcher B, Schwarz L, Gräber P. Direct indication for the existence of a double stalk in CF0F1. J Mol Biol. 1998;281:757–762. doi: 10.1006/jmbi.1998.1957. [DOI] [PubMed] [Google Scholar]
- 73.Boyer P D. The binding change mechanism for ATP synthase—some probabilities and possibilities. Biochim Biophys Acta. 1993;1140:215–250. doi: 10.1016/0005-2728(93)90063-l. [DOI] [PubMed] [Google Scholar]
- 74.Boyer P D. Energy, life, and ATP. Biosci Rep. 1998;18:97–117. doi: 10.1023/a:1020188311092. [DOI] [PubMed] [Google Scholar]
- 75.Braiman M S, Mathies R A. Resonance Raman spectra of bacteriorhodopsins: primary photoproduct evidence for a distorted 13 cis retinal chromophore. Proc Natl Acad Sci USA. 1982;79:403–407. doi: 10.1073/pnas.79.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Braiman M S, Mogi T, Marti T, Stern L J, Khorana H G, Rothschild K J. Vibrational spectroscopy of bacteriorhodopsin mutants: light-driven proton transport involves protonation changes of aspartic acid residues 85, 96, and 212. Biochemistry. 1988;27:8516–8520. doi: 10.1021/bi00423a002. [DOI] [PubMed] [Google Scholar]
- 77.Braiman M S, Walter T J, Briercheck D M. Infrared spectroscopic detection of light-induced change in chloride-arginine interaction in halorhodopsin. Biochemistry. 1994;33:1629–1635. doi: 10.1021/bi00173a003. [DOI] [PubMed] [Google Scholar]
- 78.Braks I, Hoppert M, Roge S, Mayer F. Structural aspects and immunolocalization of the F420-reducing and non-F420-reducing hydrogenases from Methanobacterium thermoautotrophicum Marburg. J Bacteriol. 1994;176:7677–7687. doi: 10.1128/jb.176.24.7677-7687.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Breton J L, Duff J L C, Butt J N, Armstrong F A, George S J, Pétillot Y, Forest E, Schäfer G, Thomson A J. Identification of the iron-sulfur clusters in a ferredoxin from the archaeon Sulfolobus acidocaldarius—evidence for a reduced [3Fe-4S] cluster with pH-dependent electronic properties. Eur J Biochem. 1995;233:937–946. doi: 10.1111/j.1432-1033.1995.937_3.x. [DOI] [PubMed] [Google Scholar]
- 80.Brierley C L, Brierley J A. Anaerobic reduction of molybdenum by Sulfolobus species. Zentbl Bakteriol Hyg Abt 1 Orig C. 1982;3:289–294. [Google Scholar]
- 81.Brischwein M, Scharf B, Engelhard M, Mäntele W. Analysis of the redox reaction of an archaeal copper protein, halocyanin, by electrochemistry and FTIR spectroscopy. Biochemistry. 1993;32:13710–13717. doi: 10.1021/bi00212a041. [DOI] [PubMed] [Google Scholar]
- 82.Brock T D, Brock K M, Belly R T, Weiss R L. Sulfolobus, new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch Microbiol. 1972;84:54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- 83.Brock T D, Gustafson J. Ferric ion reduction by sulfur- and iron-oxidizing bacteria. Appl Environ Microbiol. 1976;32:567–571. doi: 10.1128/aem.32.4.567-571.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Brown L S, Dioumaev A K, Needleman R, Lanyi J K. Local-access model for proton transfer in bacteriorhodopsin. Biochemistry. 1998;37:3982–3993. doi: 10.1021/bi9728396. [DOI] [PubMed] [Google Scholar]
- 85.Brown L S, Váró G, Hatanaka M, Sasaki J, Kandori H, Maeda A, Friedman N, Sheves M, Needleman R, Lanyi J K. The complex extracellular domain regulates the deprotonation and reprotonation of the retinal schiff base during the bacteriorhodopsin photocycle. Biochemistry. 1995;34:12903–12911. doi: 10.1021/bi00039a053. [DOI] [PubMed] [Google Scholar]
- 86.Buchanan S K, Walker J E. Large-scale chromatographic purification of F1F0-ATPase and complex I from bovine heart mitochondria. Biochem J. 1996;318:343–349. doi: 10.1042/bj3180343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bult C J, White O, Olsen G J, Zhou L X, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow W P, Hanna M C, Cotton M C, Roberts K M, Hurst A M, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 88.Burggraf S, Huber H, Stetter K O. Reclassification of the crenarchaeal orders and families in accordance with 16S rRNA sequence data. Int J Syst Bacteriol. 1997;47:657–660. doi: 10.1099/00207713-47-3-657. [DOI] [PubMed] [Google Scholar]
- 89.Burggraf S, Mayer T, Amann R, Schadhauser S, Woese C R, Stetter K O. Identifying members of the domain Archaea with rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1994;60:3112–3119. doi: 10.1128/aem.60.9.3112-3119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Calhoun M W, Thomas J W, Gennis R B. The cytochrome oxidase superfamily of redox-driven proton pumps. Trends Biochem Sci. 1994;19:325–330. doi: 10.1016/0968-0004(94)90071-x. [DOI] [PubMed] [Google Scholar]
- 91.Casimiro D R, Toy-Palmer A, Blake II R C, Dyson H J. Gene synthesis, high-level expression, and mutagenesis of Thiobacillus ferrooxidans rusticyanin: his 85 is a ligand to the blue copper center. Biochemistry. 1995;34:6640–6648. doi: 10.1021/bi00020a009. [DOI] [PubMed] [Google Scholar]
- 92.Castresana J, Lübben M, Saraste M. New archaebacterial genes coding for redox proteins: implications for the evolution of aerobic metabolism. J Mol Biol. 1995;250:202–210. doi: 10.1006/jmbi.1995.0371. [DOI] [PubMed] [Google Scholar]
- 93.Castresana J, Lübben M, Saraste M, Higgins D G. Evolution of cytochrome oxidase, an enzyme older than atmospheric oxygen. EMBO J. 1994;13:2516–2525. doi: 10.1002/j.1460-2075.1994.tb06541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Castresana J, Saraste M. Evolution of energetic metabolism: the respiration-early hypothesis. Trends Biochem Sci. 1995;20:443–448. doi: 10.1016/s0968-0004(00)89098-2. [DOI] [PubMed] [Google Scholar]
- 95.Chen J, Brevet A, Froman M, Leveque F, Schmitter J-M, Blanquet S, Plateau P. Pyrophosphatase is essential to the growth of Escherichia coli. J Bacteriol. 1990;172:5686–5689. doi: 10.1128/jb.172.10.5686-5689.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen W, Konisky J. Characterization of a membrane-associated ATPase from Methanococcus voltae, a methanogenic member of the archaea. J Bacteriol. 1993;175:5677–5682. doi: 10.1128/jb.175.17.5677-5682.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cheong G-W, Guckenberger R, Fuchs K-H, Gross H, Baumeister W. The structure of the surface layer of Methanoplanus limicola obtained by a combined electron microscopy and scanning tunneling microscopy approach. J Struct Biol. 1993;111:125–134. [Google Scholar]
- 98.Cheong G W, Typke D, Baumeister W. Projection structure of the surface layer of Methanoplanus limicola at 10Å resolution obtained by electron cryomicroscopy. J Struct Biol. 1996;117:138–144. [Google Scholar]
- 99.Chepuri V, Lemieux L, Au D C-T, Gennis R B. The sequence of the cyo operon indicates substantial structural similarities between the cytochrome o ubiquinol oxidase of Escherichia coli and the aa3-type family of cytochrome c oxidases. J Biol Chem. 1990;265:11185–11192. [PubMed] [Google Scholar]
- 100.Chizhov, I. 1998. Personal communication.
- 100a.Chizhov, I., et al. Unpublished data.
- 101.Chizhov I, Chernavskii D S, Engelhard M, Müller K H, Zubov B V, Hess B. Spectrally silent transitions in the bacteriorhodopsin photocycle. Biophys J. 1996;71:2329–2345. doi: 10.1016/S0006-3495(96)79475-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chizhov I, Geeves M, Scharf B, Hess B, Engelhard M. Anion binding to halorhodopsin from Natronobacterium pharaonis. Biophys J. 1995;68:A441. [Google Scholar]
- 103.Chizhov I, Schmies G, Seidel R, Sydor J R, Lüttenberg B, Engelhard M. The photophobic receptor from Natronobacterium pharaonis—temperature and pH dependencies of the photocycle of sensory rhodopsin II. Biophys J. 1998;75:999–1009. doi: 10.1016/S0006-3495(98)77588-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Collins M D. Analysis of isoprenoid quinones. Methods Microbiol. 1985;18:329–366. [Google Scholar]
- 105.Collins M D. Structure of Thermoplasma quinone from Thermoplasma acidophilum. FEMS Microbiol Lett. 1985;28:21–23. [Google Scholar]
- 106.Collins M D, Langworthy T A. Respiratory quinone composition of some acidophilic bacteria. Syst Appl Microbiol. 1983;4:295–304. doi: 10.1016/S0723-2020(83)80016-2. [DOI] [PubMed] [Google Scholar]
- 107.Collins M D, Ross H N M, Tindall B J, Grant W D. Distribution of isoprenoid quinones in halophilic bacteria. J Appl Bacteriol. 1981;50:559–565. [Google Scholar]
- 108.Copié V, McDermott A E, Beshah K, Williams J C, Spijker-Assink M, Gebhard R, Lugtenburg J, Herzfeld J, Griffin R G. Deuterium solid-state nuclear magnetic resonance studies of methyl group dynamics in bacteriorhodopsin and retinal model compounds: evidence for a 6-s-trans chromophore in the protein. Biochemistry. 1994;33:3280–3286. doi: 10.1021/bi00177a019. [DOI] [PubMed] [Google Scholar]
- 109.Cross R L. The reaction mechanism of FoF1-ATP synthases. Mol Mech Bioenerg. 1992;13:317–330. [Google Scholar]
- 110.Cross R L. Our primary source of ATP. Nature. 1994;370:594–595. doi: 10.1038/370594a0. [DOI] [PubMed] [Google Scholar]
- 111.Dane M, Steinert K, Esser K, Bickel-Sandkötter S, Rodriguez-Valera F. Properties of the plasma membrane ATPase of the halophilic archaebacteria Haloferax mediterranei and Haloferax volcanii. Z Naturforsch. 1992;47c:835–844. [Google Scholar]
- 112.Das A, Ivey D M, Ljungdahl L G. Purification and reconstitution into proteoliposomes of the F1F0 ATP synthase from the obligately anaerobic gram-positive bacterium Clostridium thermoautotrophicum. J Bacteriol. 1997;179:1714–1720. doi: 10.1128/jb.179.5.1714-1720.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Das A, Ljungdahl L G. Composition and primary structure of the F1F0 ATP synthase from the obligately anaerobic bacterium Clostridium thermoaceticum. J Bacteriol. 1997;179:3746–3755. doi: 10.1128/jb.179.11.3746-3755.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Deckers-Hebestreit G, Altendorf K. The F0F1-type ATP synthases of bacteria: structure and function of the F0 complex. Annu Rev Microbiol. 1996;50:791–824. doi: 10.1146/annurev.micro.50.1.791. [DOI] [PubMed] [Google Scholar]
- 115.De Groot H J M, Harbison G S, Herzfeld J, Griffin R G. Nuclear magnetic resonance study of the schiff base in bacteriorhodopsin: counterion effects on the 15N shift anisotropy. Biochemistry. 1989;28:3346–3353. doi: 10.1021/bi00434a033. [DOI] [PubMed] [Google Scholar]
- 116.Dencher N A, Dresselhaus D, Zaccai G, Büldt G. Structural changes in bacteriorhodopsin during proton translocation revealed by neutron diffraction. Proc Natl Acad Sci USA. 1989;86:7876–7879. doi: 10.1073/pnas.86.20.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Denda K, Fujiwara T, Seki M, Yoshida M, Fukumori Y, Yamanaka T. Molecular cloning of the cytochrome aa3 gene from the archaeon (archaebacterium) Halobacterium halobium. Biochem Biophys Res Commun. 1991;181:316–322. doi: 10.1016/s0006-291x(05)81420-2. [DOI] [PubMed] [Google Scholar]
- 118.Denda K, Konishi J, Hajiro K, Oshima T, Date T, Yoshida M. Structure of an ATPase operon of an acidothermophilic archaebacterium, Sulfolobus acidocaldarius. J Biol Chem. 1990;265:21509–21513. [PubMed] [Google Scholar]
- 119.Deppenmeier U. Different structure and expression of the operons encoding the membrane-bound hydrogenases from Methanosarcina mazei Gö1. Arch Microbiol. 1995;164:370–376. doi: 10.1007/BF02529985. [DOI] [PubMed] [Google Scholar]
- 120.Deppenmeier U, Blaut M, Gottschalk G. H2:heterodisulfide oxidoreductase, a second energy-conserving system in the methanogenic strain Gö1. Arch Microbiol. 1991;155:272–277. [Google Scholar]
- 121.Deppenmeier U, Blaut M, Lentes S, Herzberg C, Gottschalk G. Analysis of the vhoGAC and vhtGAC operons from Methanosarcina mazei strain Gö1, both encoding a membrane-bound hydrogenase and a cytochrome b. Eur J Biochem. 1995;227:261–269. doi: 10.1111/j.1432-1033.1995.tb20383.x. [DOI] [PubMed] [Google Scholar]
- 122.Deppenmeier U, Blaut M, Mahlmann A, Gottschalk G. Reduced coenzyme F420:heterodisulfide oxidoreductase, a proton-translocating redox system in methanogenic bacteria. Proc Natl Acad Sci USA. 1990;87:9449–9453. doi: 10.1073/pnas.87.23.9449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Deppenmeier U, Blaut M, Schmidt B, Gottschalk G. Purification and properties of a F420-nonreactive, membrane-bound hydrogenase from Methanosarcina strain Gö1. Arch Microbiol. 1992;157:505–511. doi: 10.1007/BF00276770. [DOI] [PubMed] [Google Scholar]
- 124.Deppenmeier U, Müller V, Gottschalk G. Pathways of energy conservation in methanogenic archaea. Arch Microbiol. 1996;165:149–163. [Google Scholar]
- 125.Dér A, Száraz S, Tóth-Boconádi R, Tokaji Z, Keszthelyi L, Stoeckenius W. Alternative translocation of protons and halide ions by bacteriorhodopsin. Proc Natl Acad Sci USA. 1991;88:4751–4755. doi: 10.1073/pnas.88.11.4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.De Rosa M, De Rosa S, Gambacorta A, Minale L, Thomson R H, Worthington R D. Caldariella quinone, a unique benzo-b-thiophen-4,7-quinone from Caldariella acidophila, an extremely thermophilic and acidophilic bacterium. J Chem Soc Perkin Trans. 1977;1:653–657. doi: 10.1039/p19770000653. [DOI] [PubMed] [Google Scholar]
- 127.Diller R, Stockburger M, Oesterhelt D, Tittor J. Resonance Raman study of intermediates of the halorhodopsin photocycle. FEBS Lett. 1987;217:297–304. [Google Scholar]
- 128.Dinur U, Honig B, Ottolenghi M. Analysis of primary photochemical processes in bacteriorhodopsin. Photochem Photobiol. 1981;33:523–527. [Google Scholar]
- 129.Dirmeier R, Keller M, Frey G, Huber H, Stetter K O. Purification and properties of an extremely thermostable membrane-bound sulfur-reducing complex from the hyperthermophilic Pyrodictium abyssi. Eur J Biochem. 1998;252:486–491. doi: 10.1046/j.1432-1327.1998.2520486.x. [DOI] [PubMed] [Google Scholar]
- 130.Dobler J, Zinth W, Kaiser W, Oesterhelt D. Excited-state reaction dynamics of bacteriorhodopsin studied by femtosecond spectroscopy. Chem Phys Lett. 1988;144:215–220. [Google Scholar]
- 131.Dreusicke D, Schulz G E. The glycine-rich loop of adenylate kinase forms a giant anion hole. FEBS Lett. 1986;208:301–304. doi: 10.1016/0014-5793(86)81037-7. [DOI] [PubMed] [Google Scholar]
- 132.Duschl A, Lanyi J K, Zimányi L. Properties and photochemistry of a halorhodopsin from the haloalkalophile, Natronobacterium pharaonis. J Biol Chem. 1990;265:1261–1267. [PubMed] [Google Scholar]
- 133.Edgell D R, Klenk H-P, Doolittle W F. Gene duplications in evolution of archaeal family B DNA polymerases. J Bacteriol. 1997;179:2632–2640. doi: 10.1128/jb.179.8.2632-2640.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Edholm O, Berger O, Jähnig F. Structure and fluctuations of bacteriorhodopsin in the purple membrane: a molecular dynamics study. J Mol Biol. 1995;250:94–111. doi: 10.1006/jmbi.1995.0361. [DOI] [PubMed] [Google Scholar]
- 135.Eisenstein L, Lin S L, Dollinger G, Odashima K, Ding W D, Nakanishi K. FTIR difference studies on apoproteins; protonation states of aspartic and glutamic acid residues during the photocycle of bacteriorhodopsin. J Am Chem Soc. 1987;109:6860–6862. [Google Scholar]
- 136.Elferink M G L, De Wit J G, Driessen A J M, Konings W N. Stability and proton-permeability of liposomes composed of archaeal tetraether lipids. Biochim Biophys Acta. 1994;1193:247–254. doi: 10.1016/0005-2736(94)90160-0. [DOI] [PubMed] [Google Scholar]
- 137.Engelhard M, Gerwert K, Hess B, Kreutz W, Siebert F. Light-driven protonation changes of internal aspartic acids of bacteriorhodopsin: an investigation by static and time-resolved infrared difference spectroscopy using [4-13C] aspartic acid labeling purple membrane. Biochemistry. 1985;24:400–407. doi: 10.1021/bi00323a024. [DOI] [PubMed] [Google Scholar]
- 138.Engelhard M, Hess B, Emeis D, Metz G, Kreutz W, Siebert F. Magic angle sample spinning 13C nuclear magnetic resonance of isotopically labeled bacteriorhodopsin. Biochemistry. 1989;28:3967–3975. doi: 10.1021/bi00435a051. [DOI] [PubMed] [Google Scholar]
- 139.Engelhard M, Hess B, Metz G, Kreutz W, Siebert F, Soppa J, Oesterhelt D. High resolution 13C-solid state NMR of bacteriorhodopsin: assignment of specific aspartic acids and structural implications of single site mutations. Eur Biophys J. 1990;18:17–24. doi: 10.1007/BF00185416. [DOI] [PubMed] [Google Scholar]
- 140.Engelhard M, Scharf B, Siebert F. Protonation changes during the photocycle of sensory rhodopsin II from Natronobacterium pharaonis. FEBS Lett. 1996;395:195–198. doi: 10.1016/0014-5793(96)01041-1. [DOI] [PubMed] [Google Scholar]
- 141.Engelman D M, Henderson R, McLachlan A D, Wallace B A. Path of the polypeptide in bacteriorhodopsin. Proc Natl Acad Sci USA. 1980;77:2023–2027. doi: 10.1073/pnas.77.4.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Essen L O, Siegert R, Lehmann W D, Oesterhelt D. Lipid patches in membrane protein oligomers—crystal structure of the bacteriorhodopsin-lipid complex. Proc Natl Acad Sci USA. 1998;95:11673–11678. doi: 10.1073/pnas.95.20.11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Faguy D M, Jarrell K F, Kuzio J, Kalmokoff M L. Molecular analysis of archael flagellins: similarity to the type IV pilin-transport superfamily widespread in bacteria. Can J Microbiol. 1994;40:67–71. doi: 10.1139/m94-011. [DOI] [PubMed] [Google Scholar]
- 144.Fahmy K, Siebert F, Großjean M F, Tavan P. Photoisomerization in bacteriorhodopsin studied by FTIR, linear dichroism and photoselection experiments combined with quantum chemical theoretical analysis. J Mol Struct. 1989;214:257–288. [Google Scholar]
- 145.Ferrando-May E, Krah M, Marwan W, Oesterhelt D. The methyl-accepting transducer protein Htrl is functionally associated with the photoreceptor sensory rhodopsin I in the archaeon Halobacterium salinarum. EMBO J. 1993;12:2999–3005. doi: 10.1002/j.1460-2075.1993.tb05968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fetter J R, Qian J, Shapleigh J, Thomas J W, Garcia-Horsman A, Schmidt E, Hosler J P, Babcock G T, Gennis R B, Ferguson-Miller S. Possible proton relay pathways in cytochrome c oxidase. Proc Natl Acad Sci USA. 1995;92:1604–1608. doi: 10.1073/pnas.92.5.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fillingame R H, Girvin M E, Fraga D, Zhang Y. Correlations of structure and function in H+ translocating subunit c of F1F0 ATP synthase. Ann N Y Acad Sci. 1993;671:323–334. doi: 10.1111/j.1749-6632.1992.tb43806.x. [DOI] [PubMed] [Google Scholar]
- 148.Fillingame R H, Jones P C, Jiang W, Valiyaveetil F I, Dmitriev O Y. Subunit organization and structure in the F0 sector of Escherichia coli F1F0 ATP synthase. Biochim Biophys Acta. 1998;1365:135–142. doi: 10.1016/s0005-2728(98)00053-x. [DOI] [PubMed] [Google Scholar]
- 149.Fischer R, Gärtner P, Yeliseev A, Thauer R K. N5-Methyltetrahydromethanopterin:coenzyme M methyltransferase in methanogenic archaebacteria is a membrane protein. Arch Microbiol. 1992;158:208–217. doi: 10.1007/BF00290817. [DOI] [PubMed] [Google Scholar]
- 150.Fischer R, Thauer R K. Methyltetrahydromethanopterin as an intermediate in methanogenesis from acetate in Methanosarcina barkeri. Arch Microbiol. 1989;151:459–465. [Google Scholar]
- 151.Fischer R, Thauer R K. Ferredoxin-dependent methane formation from acetate in cell extracts of Methanosarcina barkeri (strain MS) FEBS Lett. 1990;269:368–372. doi: 10.1016/0014-5793(90)81195-t. [DOI] [PubMed] [Google Scholar]
- 152.Fischer U, Oesterhelt D. Chromophore equilibria in bacteriorhodopsin. Biophys J. 1979;26:211–230. doi: 10.1016/S0006-3495(79)85172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Fitz-Gibbon S, Choi A J, Miller J H, Stetter K O, Simon M I, Swanson R, Kim U. A fosmid-based genomic map and identification of 474 genes of the hyperthermophilic archaeon Pyrobaculum aerophilum. Extremophiles. 1997;1:36–51. doi: 10.1007/s007920050013. [DOI] [PubMed] [Google Scholar]
- 154.Fodor S P A, Bogomolni R A, Mathies R A. Structure of the retinal chromophore in the hRL intermediate of halorhodopsin from resonance Raman spectroscopy. Biochemistry. 1987;26:6775–6778. doi: 10.1021/bi00395a029. [DOI] [PubMed] [Google Scholar]
- 155.Fodor S P A, Gebhard R, Lugtenburg J, Bogomolni R A, Mathies R A. Structure of the retinal chromophore in sensory rhodopsin I from resonance Raman spectroscopy. J Biol Chem. 1989;264:18280–18283. [PubMed] [Google Scholar]
- 156.Fujii T, Hata Y, Wakagi T, Tanaka N, Oshima T. Novel zinc-binding centre in thermoacidophilic archaeal ferredoxins. Nat Struct Biol. 1996;3:834–837. doi: 10.1038/nsb1096-834. [DOI] [PubMed] [Google Scholar]
- 157.Fujiwara T, Fukumori Y, Yamanaka T. aa3-type cytochrome c oxidase occurs in Halobacterium halobium and its activity is inhibited by higher concentrations of salt. Plant Cell Physiol. 1987;28:29–36. [Google Scholar]
- 158.Fujiwara T, Fukumori Y, Yamanaka T. Purification and properties of Halobacterium halobium “cytochrome aa3” which lacks CuA and CuB. J Biochem. 1989;105:287–292. doi: 10.1093/oxfordjournals.jbchem.a122655. [DOI] [PubMed] [Google Scholar]
- 159.Fujiwara T, Fukumori Y, Yamanaka T. The separation between cytochrome a and cytochrome a3 in the absolute spectrum. In: Kim C H, Ozawa T, editors. Bioenergetics. New York, N.Y: Plenum Press; 1990. pp. 155–159. [Google Scholar]
- 160.Fujiwara T, Fukumori Y, Yamanaka T. Halobacterium halobium cytochrome b-558 and cytochrome b-562: purification and some properties. J Biochem (Tokyo) 1993;113:48–54. doi: 10.1093/oxfordjournals.jbchem.a124002. [DOI] [PubMed] [Google Scholar]
- 161.Garcia-Horsman J A, Puustinen A, Gennis R B, Wikström M. Proton transfer in cytochrome bo3 ubiquinol oxidase of Escherichia coli: second-site mutations in subunit I that restore proton pumping in the mutant Asp135→Asn. Biochemistry. 1995;34:4428–4433. doi: 10.1021/bi00013a035. [DOI] [PubMed] [Google Scholar]
- 162.Gärtner P. Characterization of a quinole-oxidase activity in crude extracts of Thermoplasma acidophilum and isolation of an 18-kDa cytochrome. Eur J Biochem. 1991;200:215–222. doi: 10.1111/j.1432-1033.1991.tb21070.x. [DOI] [PubMed] [Google Scholar]
- 163.Gärtner P, Ecker A, Fischer R, Linder D, Fuchs G, Thauer R K. Purification and properties of N5-methyltetrahydromethanopterin:coenzyme M methyltransferase from Methanobacterium thermoautotrophicum. Eur J Biochem. 1993;213:537–545. doi: 10.1111/j.1432-1033.1993.tb17792.x. [DOI] [PubMed] [Google Scholar]
- 164.Gärtner P, Weiss D S, Harms U, Thauer R K. N5-methyltetrahydromethanopterin:coenzyme M methyltransferase from Methanobacterium thermoautotrophicum. Catalytic mechanism and sodium ion dependence. Eur J Biochem. 1994;226:465–472. doi: 10.1111/j.1432-1033.1994.tb20071.x. [DOI] [PubMed] [Google Scholar]
- 165.Gennis R B, Ferguson-Miller S. Protein structure: proton-pumping oxidases. Curr Biol. 1996;6:36–38. doi: 10.1016/s0960-9822(02)00416-5. [DOI] [PubMed] [Google Scholar]
- 166.George G N, Prince R C, Mukund S, Adams M W W. Aldehyde ferredoxin oxidoreductase from the hyperthermophilic archaebacterium Pyrococcus furiosus contains a tungsten oxo-thiolate center. J Am Chem Soc. 1992;114:3521–3523. [Google Scholar]
- 167.Gerscher S, Döpner S, Hildebrandt P, Gleissner M, Schäfer G. Resonance Raman spectroscopic study of the terminal oxidase complex of Sulfolobus acidocaldarius. J Inorg Biochem. 1995;59:283. doi: 10.1021/bi960896d. [DOI] [PubMed] [Google Scholar]
- 168.Gerscher S, Döpner S, Hildebrandt P, Gleissner M, Schäfer G. Resonance Raman spectroscopy of the integral quinol oxidase complex of Sulfolobus acidocaldarius. Biochemistry. 1996;35:12796–12803. doi: 10.1021/bi960896d. [DOI] [PubMed] [Google Scholar]
- 169.Gerscher S, Mylrajan M, Hildebrandt P, Baron M H, Müller R, Engelhard M. Chromophore-anion interactions in halorhodopsin from Natronobacterium pharaonis probed by time-resolved resonance raman spectroscopy. Biochemistry. 1997;36:11012–11020. doi: 10.1021/bi970722b. [DOI] [PubMed] [Google Scholar]
- 170.Gerwert K, Hess B, Soppa J, Oesterhelt D. Role of aspartate-96 in proton translocation by bacteriorhodopsin. Proc Natl Acad Sci USA. 1989;86:4943–4947. doi: 10.1073/pnas.86.13.4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Giuffre A, Gomes C, Antonini G, D’Itri E, Teixeira M, Brunori M. Functional properties of the quinol oxidase from Acidianus ambivalens and the possible role of its electron donor: studies in the membrane-integrated and purified enzyme. Eur J Biochem. 1997;250:383–388. doi: 10.1111/j.1432-1033.1997.0383a.x. [DOI] [PubMed] [Google Scholar]
- 172.Giuffrè A, Antonini G, Brunori M, D’Itri E, Malatesta F, Nicoletti F, Anemüller S, Gleissner M, Schäfer G. Sulfolobus acidocaldarius terminal oxidase. A kinetic investigation and its structural interpretation. J Biol Chem. 1994;269:31006–31011. [PubMed] [Google Scholar]
- 173.Gleissner M. Ph.D. thesis. Lübeck, Germany: University of Lübeck; 1996. Der terminale oxidase Komplex aus Sulfolobus acidocaldarius; pp. 1–119. [Google Scholar]
- 174.Gleissner M, Elferink M G L, Driessen A J M, Konings W N, Anemüller S, Schäfer G. Generation of proton-motive force by an archaeal terminal quinol oxidase from Sulfolobus acidocaldarius. Eur J Biochem. 1994;224:983–990. doi: 10.1111/j.1432-1033.1994.00983.x. [DOI] [PubMed] [Google Scholar]
- 175.Gleissner M, Kaiser U, Antonopuolos E, Schäfer G. The archaeal SoxABCD-complex is a proton pump in Sulfolobus acidocaldarius. J Biol Chem. 1997;272:8417–8426. doi: 10.1074/jbc.272.13.8417. [DOI] [PubMed] [Google Scholar]
- 176.Gogarten J P, Taiz L. Evolution of proton pumping ATPases: rooting the tree of life. Photosynth Res. 1992;33:137–146. doi: 10.1007/BF00039176. [DOI] [PubMed] [Google Scholar]
- 177.Gomes C M, Teixeira M. The NADH oxidase from the thermoacidophilic archaea Acidianus ambivalens: isolation and physicochemical characterisation. Biochem Biophys Res Commun. 1998;243:412–415. doi: 10.1006/bbrc.1998.8111. [DOI] [PubMed] [Google Scholar]
- 178.Gradin C H, Colmsjö A. Oxidation-reduction potentials and absorption spectra of two b-type cytochromes from the halophilic archaebacterium, Halobacterium halobium. Arch Biochem Biophys. 1987;256:515–522. doi: 10.1016/0003-9861(87)90609-6. [DOI] [PubMed] [Google Scholar]
- 179.Gradin C H, Hederstedt L, Baltscheffsky H. Soluble succinate dehydrogenase from the halophilic archaebacterium, Halobacterium halobium. Arch Biochem Biophys. 1985;239:200–205. doi: 10.1016/0003-9861(85)90827-6. [DOI] [PubMed] [Google Scholar]
- 180.Grigorieff N, Ceska T A, Downing K H, Baldwin J M, Henderson R. Electron-crystallographic refinement of the structure of bacteriorhodopsin. J Mol Biol. 1996;259:393–421. doi: 10.1006/jmbi.1996.0328. [DOI] [PubMed] [Google Scholar]
- 181.Grogan D W. Organization and interactions of cell envelope proteins of the extreme thermoacidophile Sulfolobus acidocaldarius. Can J Microbiol. 1996;42:1163–1171. [Google Scholar]
- 182.Haase P, Deppenmeier U, Blaut M, Gottschalk G. Purification and characterization of F420H2-dehydrogenase from Methanolobus tindarius. Eur J Biochem. 1992;203:527–531. doi: 10.1111/j.1432-1033.1992.tb16579.x. [DOI] [PubMed] [Google Scholar]
- 183.Haberkorn R A, Herzfeld J, Griffin R G. High resolution 31P and 13C nuclear magnetic resonance spectra of unsonicated model membranes. J Am Chem Soc. 1978;100:1296–1298. [Google Scholar]
- 184.Hägerhäll C. Succinate: quinone oxidoreductases—variations on a conserved theme. Biochim Biophys Acta. 1997;1320:107–141. doi: 10.1016/s0005-2728(97)00019-4. [DOI] [PubMed] [Google Scholar]
- 185.Halestrap A P. The regulation of the matrix volume of mammalian mitochondria in vivo and in vitro and its role in the control of mitochondrial metabolism. Biochim Biophys Acta. 1989;973:355–382. doi: 10.1016/s0005-2728(89)80378-0. [DOI] [PubMed] [Google Scholar]
- 186.Hallberg Gradin C, Baltscheffsky H. Solubilization and separation of two b-type cytochromes from a carotenoid mutant of Halobacterium halobium. FEBS Lett. 1981;125:201–204. doi: 10.1016/0014-5793(81)80718-1. [DOI] [PubMed] [Google Scholar]
- 187.Hallberg Gradin C, Colmsjö A. Four different b-type cytochromes in the halophilic archaebacterium, Halobacterium halobium. Biochim Biophys Acta. 1989;272:130–136. doi: 10.1016/0003-9861(89)90203-8. [DOI] [PubMed] [Google Scholar]
- 188.Haney P J, Berk H, Lynn D, Konisky J. The adenylate kinase genes of M. voltae, M. thermolithotrophicus, M. jannaschii, and M. igneus define a new family of adenylate kinases. Gene. 1997;185:239–244. doi: 10.1016/s0378-1119(96)00651-8. [DOI] [PubMed] [Google Scholar]
- 189.Hansen T, Leppänen V-M, Goldman A, Brandenburg K, Schäfer G. The extreme thermostable pyrophosphatase from Sulfolobus acidocaldarius: enzymatic and comparative biophysical characterization. Arch Biochem Biophys. 1999;363:135–147. doi: 10.1006/abbi.1998.1072. [DOI] [PubMed] [Google Scholar]
- 190.Hansen, T., and G. Schäfer. 1997. Personal communication.
- 191.Harms U, Thauer R K. The corrinoid-containing 23-kDa subunit MtrA of the energy-conserving N5-methyltetrahydromethanopterin:coenzyme M methyltransferase complex from Methanobacterium thermoautotrophicum—EOR spectroscopic evidence for a histidine residue as a cobalt ligand of the cobamide. Eur J Biochem. 1996;241:149–154. doi: 10.1111/j.1432-1033.1996.0149t.x. [DOI] [PubMed] [Google Scholar]
- 192.Harms U, Weiss D S, Gärtner P, Linder D, Thauer R K. The energy conserving N5-methyltetrahydromethanopterin:coenzyme M methyltransferase complex from Methanobacterium thermoautotrophicum is composed of eight different subunits. Eur J Biochem. 1995;228:640–648. doi: 10.1111/j.1432-1033.1995.0640m.x. [DOI] [PubMed] [Google Scholar]
- 193.Hartmann R, Sickinger H D, Oesterhelt D. Anaerobic growth of halobacteria. Proc Natl Acad Sci USA. 1980;77:3821–3825. doi: 10.1073/pnas.77.7.3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Häsler K, Engelbrecht S, Junge W. Three-stepped rotation of subunits gamma and ɛ in single molecules of F-ATPase as revealed by polarized, confocal fluorometry. FEBS Lett. 1998;426:301–304. doi: 10.1016/s0014-5793(98)00358-5. [DOI] [PubMed] [Google Scholar]
- 195.Haupts U, Bamberg E, Oesterhelt D. Different modes of proton translocation by sensory rhodopsin I. EMBO J. 1996;15:1834–1841. [PMC free article] [PubMed] [Google Scholar]
- 196.Haupts U, Eisfeld W, Stockburger M, Oesterhelt D. Sensory rhodopsin I photocycle intermediate SRI380 contains 13-cis retinal bound via an unprotonated schiff base. FEBS Lett. 1994;356:25–29. doi: 10.1016/0014-5793(94)01226-1. [DOI] [PubMed] [Google Scholar]
- 197.Haupts U, Tittor J, Bamberg E, Oesterhelt D. General concept for ion translocation by halobacterial retinal proteins—the isomerization/switch/transfer (IST) model. Biochemistry. 1997;36:2–7. doi: 10.1021/bi962014g. [DOI] [PubMed] [Google Scholar]
- 198.Hauss T, Büldt G, Heyn M P, Dencher N A. Light-induced isomerization causes an increase in the chromophore tilt in the M intermediate of bacteriorhodopsin: a neutron diffraction study. Proc Natl Acad Sci USA. 1994;91:11854–11858. doi: 10.1073/pnas.91.25.11854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Havelka W A, Henderson R, Heymann J A W, Oesterhelt D. Projection structure of halorhodopsin from Halobacterium halobium at 6 A resolution obtained by electron cryo-microscopy. J Mol Biol. 1993;234:837–846. doi: 10.1006/jmbi.1993.1629. [DOI] [PubMed] [Google Scholar]
- 200.Havelka W A, Henderson R, Oesterhelt D. Three-dimensional structure of halorhodopsin at 7 Å resolution. J Mol Biol. 1995;247:726–738. doi: 10.1006/jmbi.1995.0176. [DOI] [PubMed] [Google Scholar]
- 201.Hazelbauer G L, Yaghmai R, Burrows G G, Baumgartner J W, Dutton D P, Morgan D G. Transducers: transmembrane receptor proteins involved in the bacterial chemotaxis signal transduction pathway. Symp Soc Gen Microbiol. 1990;46:107–134. [Google Scholar]
- 202.Hazemoto N, Kamo N, Kobatake Y. Suggestion of existence of two forms of halorhodospin in alkaline solution. Biochem Biophys Res Commun. 1984;118:502–507. doi: 10.1016/0006-291x(84)91331-7. [DOI] [PubMed] [Google Scholar]
- 203.Hazemoto N, Kamo N, Kobatake Y, Tsuda M, Terayama Y. Effect of salt on photocycle and ion-pumping of halorhodopsin and third rhodopsinlike pigment of Halobacterium halobium. Biophys J. 1984;45:1073–1077. doi: 10.1016/S0006-3495(84)84254-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Hazemoto N, Kamo N, Terayama Y, Kobatake Y, Tsuda M. Photochemistry of two rhodopsinlike pigments in bacteriorhodopsin-free mutant of Halobacterium halobium. Biophys J. 1983;44:59–64. doi: 10.1016/S0006-3495(83)84277-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hedderich R, Berkessel A, Thauer R K. Purification and properties of heterodisulfide reductase from Methanobacterium thermoautotrophicum (strain Marburg) Eur J Biochem. 1990;193:255–261. doi: 10.1111/j.1432-1033.1990.tb19331.x. [DOI] [PubMed] [Google Scholar]
- 206.Hedderich R, Koch J, Linder D, Thauer R K. The heterodisulfide reductase from Methanobacterium thermoautotrophicum contains sequence motifs characteristic of pyridine-nucleotide dependent thioredoxin reductases. Eur J Biochem. 1994;225:253–261. doi: 10.1111/j.1432-1033.1994.00253.x. [DOI] [PubMed] [Google Scholar]
- 207.Hederstedt L, Ohnishi T. Progress in succinate: quinone oxidoreductase research. Mol Mech Bioenerg. 1992;7:163–198. [Google Scholar]
- 208.Hegemann P, Oesterhelt D, Bamberg E. The transport activity of the light-driven chloride pump halorhodopsin is regulated by green and blue light. Biochim Biophys Acta. 1985;819:195–205. [Google Scholar]
- 209.Hegemann P, Oesterhelt D, Steiner M. The photocycle of the chloride pump halorhodopsin. I. Azide-catalyzed deprotonation of the chromophore is a side reaction of photocycle intermediates inactivating the pump. EMBO J. 1985;4:2347–2350. doi: 10.1002/j.1460-2075.1985.tb03937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Heibel G E, Anzenbacher P, Hildebrandt P, Schäfer G. Unusual heme structure in cytochrome aa3 from Sulfolobus acidocaldarius: a resonance Raman investigation. Biochemistry. 1993;32:10878–10884. doi: 10.1021/bi00091a043. [DOI] [PubMed] [Google Scholar]
- 211.Heiden S, Hedderich R, Setzke E, Thauer R K. Purification of a cytochrome b containing H2-heterodisulfide oxidoreductase complex from membranes of Methanosarcina barkeri. Eur J Biochem. 1993;213:529–535. doi: 10.1111/j.1432-1033.1993.tb17791.x. [DOI] [PubMed] [Google Scholar]
- 212.Heiden S, Hedderich R, Setzke E, Thauer R K. Purification of a two-subunit cytochrome b-containing heterodisulfide reductase from methanol-grown Methanosarcina barkeri. Eur J Biochem. 1994;221:855–861. doi: 10.1111/j.1432-1033.1994.tb18800.x. [DOI] [PubMed] [Google Scholar]
- 213.Heinrich, M. E. 1999. Personal communication.
- 214.Heise R, Müller V, Gottschalk G. Acetogenesis and ATP synthesis in Acetobacterium woodii are coupled via a transmembrane primary sodium gradient. FEMS Microbiol Lett. 1993;112:261–268. [Google Scholar]
- 215.Hellwig P, Behr J, Ostermeier C, Richter O M H, Pfitzner U, Odenwald A, Ludwig B, Michel H, Mäntele W. Involvement of glutamic acid 278 in the redox reaction of the cytochrome c oxidase from Paracoccus denitrificans investigated by FTIR spectroscopy. Biochemistry. 1998;37:7390–7399. doi: 10.1021/bi9725576. [DOI] [PubMed] [Google Scholar]
- 216.Henderson R, Baldwin J M, Ceska T A, Zemlin F, Beckmann E, Downing K H. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J Mol Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 217.Henderson R, Unwin P N T. Three-dimensional model of purple membrane obtained by electron microscopy. Nature. 1975;257:28–32. doi: 10.1038/257028a0. [DOI] [PubMed] [Google Scholar]
- 218.Hendrickson F M, Burkard F, Glaeser R M. Structural characterization of the L-to-M transition of the bacteriorhodopsin photocycle. Biophys J. 1998;75:1446–1454. doi: 10.1016/S0006-3495(98)74063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Hessling B, Souvignier G, Gerwert K. A model-independent approach to assigning bacteriorhodopsin’s intramolecular reactions to photocycle intermediates. Biophys J. 1993;65:1929–1941. doi: 10.1016/S0006-3495(93)81264-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Hettmann T, Schmidt C L, Anemüller S, Zähringer U, Moll H, Petersen A, Schäfer G. Cytochrome b-558/566 from the archaeon Sulfolobus acidocaldarius: a novel, highly glycosylated, membrane bound B-type hemoprotein. J Biol Chem. 1998;273:12032–12040. doi: 10.1074/jbc.273.20.12032. [DOI] [PubMed] [Google Scholar]
- 221.Hilario E, Gogarten J P. The prokaryote-to-eukaryote transition reflected in the evolution of the V/F/A-ATPase catalytic and proteolipid subunits. J Mol Evol. 1998;46:703–715. doi: 10.1007/pl00006351. [DOI] [PubMed] [Google Scholar]
- 222.Hildebrand E, Dencher N A. Two photosystems controlling behavioural responses in Halobacterium; phobic response; motility. Nature. 1975;257:46–49. doi: 10.1038/257046a0. [DOI] [PubMed] [Google Scholar]
- 223.Hildebrand E, Schimz A. Behavioral pattern and its sensory control in Halobacterium halobium. In: Eisenbach M, Balaban M, editors. Sensing and response in microorganisms. Amsterdam, The Netherlands: Elsevier; 1985. pp. 129–142. [Google Scholar]
- 224.Hildebrandt P, Heibel G, Anemüller S, Schäfer G. Resonance Raman study of cytochrome aa3 from Sulfolobus acidocaldarius. FEBS Lett. 1991;283:131–134. doi: 10.1016/0014-5793(91)80570-s. [DOI] [PubMed] [Google Scholar]
- 225.Hirayama J, Imamoto Y, Shichida Y, Kamo N, Tomioka H, Yoshizawa T. Photocycle of phoborhodopsin from haloalkaliphilic bacterium (Natronobacterium pharaonis) studied by low-temperature spectrophotometry. Biochemistry. 1992;31:2093–2098. doi: 10.1021/bi00122a029. [DOI] [PubMed] [Google Scholar]
- 226.Hochstein L I. The membrane bound enzymes in archaea. In: Kates M, Kushner D J, Matheson A T, editors. The biochemistry of archaea. Amsterdam, The Netherlands: Elsevier; 1993. pp. 297–323. [Google Scholar]
- 227.Hochstein L I. Is the Paracoccus halodenitrificans ATPase a chimeric enzyme. FEMS Microbiol Lett. 1996;140:55–60. doi: 10.1111/j.1574-6968.1996.tb08314.x. [DOI] [PubMed] [Google Scholar]
- 228.Hochstein L I, Kristjansson H, Altekar W. The purification and subunit structure of a membrane-bound ATPase from the archaebacterium Halobacterium saccharovorum. Biochem Biophys Res Commun. 1987;147:295–300. doi: 10.1016/s0006-291x(87)80120-1. [DOI] [PubMed] [Google Scholar]
- 229.Hochstein L I, Lang F. Purification and properties of a dissimilatory nitrate reductase from Haloferax denitrificans. Arch Biochem Biophys. 1991;288:380–385. doi: 10.1016/0003-9861(91)90210-a. [DOI] [PubMed] [Google Scholar]
- 230.Hochstein L I, Lawson D. Is ATP synthesized by a vacuolar-ATPase in the extremely halophilic Bacteria. Experientia. 1993;49:1059–1063. [Google Scholar]
- 231.Hochstein L I, Stan-Lotter H. Purification and properties of an ATPase from Sulfolobus solfataricus. Arch Biochem Biophys. 1992;295:153–160. doi: 10.1016/0003-9861(92)90501-m. [DOI] [PubMed] [Google Scholar]
- 232.Hoff W D, Jung K H, Spudich J L. Molecular mechanism of photosignaling by archaeal sensory rhodopsins. Annu Rev Biophys Biomol Struct. 1997;26:223–258. doi: 10.1146/annurev.biophys.26.1.223. [DOI] [PubMed] [Google Scholar]
- 233.Holländer R. The cytochromes of Thermoplasma acidophilum. J Gen Microbiol. 1978;108:165–167. [Google Scholar]
- 234.Holländer R, Wolf G, Mannheim W. Lipoquinones of some bacteria and mycoplasmas, with considerations on their functional significance. Antonie Leeuwenhoek. 1977;43:177–185. doi: 10.1007/BF00395672. [DOI] [PubMed] [Google Scholar]
- 235.Hu J, Griffin R G, Herzfeld J. Synergy in the spectral tuning of retinal pigments: complete accounting of the opsin shift in bacteriorhodopsin. Proc Natl Acad Sci USA. 1994;91:8880–8884. doi: 10.1073/pnas.91.19.8880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Huber G, Drobner E, Huber H, Stetter K O. Growth by aerobic oxidation of molecular hydrogen in archaea—a metabolic property so far unknown for this domain. Syst Appl Microbiol. 1992;15:502–504. [Google Scholar]
- 237.Humphrey W, Logunov I, Schulten K, Sheves M. Molecular dynamics study of bacteriorhodopsin and artificial pigments. Biochemistry. 1994;33:3668–3678. doi: 10.1021/bi00178a025. [DOI] [PubMed] [Google Scholar]
- 238.Humphrey W, Xu D, Sheves M, Schulten K. Molecular dynamics study of the early intermediates in the bacteriorhodopsin photocycle. J Phys Chem. 1995;99:14549–14560. [Google Scholar]
- 239.Imamoto Y, Shichida Y, Hirayama J, Tomioka H, Kamo N, Yoshizawa T. Nanosecond laser photolysis of phoborhodopsin: from Natronobacterium pharaonis appearance of KL and L intermediates in the photocycle at room temperature. Photochem Photobiol. 1992;56:1129–1134. [Google Scholar]
- 240.Imamoto Y, Shichida Y, Hirayama J, Tomioka H, Kamo N, Yoshizawa T. Chromophore configuration of pharaonis phoborhodopsin and its isomerization on photon absorption. Biochemistry. 1992;31:2523–2528. doi: 10.1021/bi00124a012. [DOI] [PubMed] [Google Scholar]
- 241.Imamoto Y, Shichida Y, Yoshizawa T, Tomioka H, Takahashi T, Fujikawa K, Kamo N, Kobatake Y. Photoreaction cycle of phoborhodopsin studied by low-temperature spectrophotometry. Biochemistry. 1991;30:7416–7424. doi: 10.1021/bi00244a008. [DOI] [PubMed] [Google Scholar]
- 242.Inatomi K-I, Kamagata Y, Nakamura K. Membrane ATPase from the aceticlastic methanogen Methanothrix thermophila. J Bacteriol. 1993;175:80–84. doi: 10.1128/jb.175.1.80-84.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Inatomi K I. Characterization and purification of the membrane-bound ATPase of the archaebacterium Methanosarcina barkeri. J Bacteriol. 1986;167:837–841. doi: 10.1128/jb.167.3.837-841.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Inatomi K I, Maeda M, Futai M. Dicyclohexylcarbodiimide-binding protein is a subunit of the Methanosarcina barkeri ATPase complex. Biochem Biophys Res Commun. 1989;162:1585–1590. doi: 10.1016/0006-291x(89)90856-5. [DOI] [PubMed] [Google Scholar]
- 245.Ippen E P, Shank C V. Subpicosecond spectroscopy of bacteriorhodopsin. Science. 1978;200:1279–1281. doi: 10.1126/science.663607. [DOI] [PubMed] [Google Scholar]
- 246.Iwasaki T, Matsuura K, Oshima T. Resolution of the aerobic respiratory system of the thermoacidophilic archaeon, Sulfolobus sp. strain 7. 1. The archaeal, terminal oxidase supercomplex is a functional fusion of respiratory complexes III and IV with no c-type cytochromes. J Biol Chem. 1995;270:30881–30892. doi: 10.1074/jbc.270.52.30881. [DOI] [PubMed] [Google Scholar]
- 247.Iwasaki T, Wakagi T, Isogai Y, Iizuka T, Oshima T. Resolution of the aerobic respiratory system of the thermoacidophilic archaeon, Sulfolobus sp. strain 7.2. Characterization of the archaeal terminal oxidase subcomplexes and implication for the intramolecular electron transfer. J Biol Chem. 1995;270:30893–30901. doi: 10.1074/jbc.270.52.30893. [DOI] [PubMed] [Google Scholar]
- 248.Iwasaki T, Wakagi T, Isogai Y, Tanaka K, Iizuka T, Oshima T. Functional and evolutionary implications of a [3Fe-4S] cluster of the dicluster-type ferredoxin from the thermoacidophilic archaeon, Sulfolobus sp. strain 7. J Biol Chem. 1994;269:29444–29450. [PubMed] [Google Scholar]
- 249.Iwasaki T, Wakagi T, Oshima T. Ferredoxin-dependent redox system of a thermoacidophilic archaeon, Sulfolobus sp. strain 7. Purification and characterization of a novel reduced ferredoxin-reoxidizing iron-sulfur flavoprotein. J Biol Chem. 1995;270:17878–17883. doi: 10.1074/jbc.270.30.17878. [DOI] [PubMed] [Google Scholar]
- 250.Iwasaki T, Wakagi T, Oshima T. Resolution of the aerobic respiratory system of the thermoacidophilic archaeon, Sulfolobus sp. strain 7. 3. The archaeal novel respiratory complex II (succinate:caldariellaquinone oxidoreductase complex) inherently lacks heme group. J Biol Chem. 1995;270:30902–30908. doi: 10.1074/jbc.270.52.30902. [DOI] [PubMed] [Google Scholar]
- 251.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 Å resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 252.Jähnig F, Edholm O. Modeling of the structure of bacteriorhodopsin. A molecular dynamics study. J Mol Biol. 1992;226:837–850. doi: 10.1016/0022-2836(92)90635-w. [DOI] [PubMed] [Google Scholar]
- 253.Jannsen, S., and G. Schäfer. 1999. Personal communication.
- 254.Jannsen S, Schäfer G, Moll R. A succinate dehydrogenase with novel structure and properties from the hyperthermophilic archaeon Sulfolobus acidocaldarius: genetic and biophysical characterization. J Bacteriol. 1997;179:5560–5569. doi: 10.1128/jb.179.17.5560-5569.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Jetten M S M, Fluit T J, Stams A J M, Zehnder A J B. A fluoride-insensitive inorganic pyrophosphatase isolated from Methanothrix soehngenii. Arch Microbiol. 1992;157:284–289. doi: 10.1007/BF00245163. [DOI] [PubMed] [Google Scholar]
- 256.Jones H M, Gunsalus R P. Transcription of the Escherichia coli fumarate reductase genes (frdABCD) and their coordinate regulation by oxygen, nitrate, and fumarate. J Bacteriol. 1985;164:1100–1109. doi: 10.1128/jb.164.3.1100-1109.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Jones P C, Fillingame R H. Genetic fusions of subunit-c in the Fo sector of H+ transporting ATP synthase. Functional dimers and trimers and determination of stoichiometry by cross-linking. J Biol Chem. 1998;273:29701–29705. doi: 10.1074/jbc.273.45.29701. [DOI] [PubMed] [Google Scholar]
- 258.Jung K H, Spudich J L. Protonatable residues at the cytoplasmic end of transmembrane helix-2 in the signal transducer HtrI control photochemistry and function of sensory rhodopsin I. Proc Natl Acad Sci USA. 1996;93:6557–6561. doi: 10.1073/pnas.93.13.6557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Junge W, Lill H, Engelbrecht S. ATP synthase: an electrochemical transducer with rotary mechanics. Trends Biochem Sci. 1997;22:420–423. doi: 10.1016/s0968-0004(97)01129-8. [DOI] [PubMed] [Google Scholar]
- 260.Jussofie A, Gottschalk G. Further studies on the distribution of cytochromes in methanogenic bacteria. FEMS Microbiol Lett. 1986;22:15–18. [Google Scholar]
- 261.Kaesler B, Schönheit P. The sodium cycle in methanogenesis. CO2 reduction to the formaldehyde level in methanogenic bacteria is driven by a primary electrochemical potential of Na+ generated by formaldehyde reduction to CH4. Eur J Biochem. 1989;186:309–316. doi: 10.1111/j.1432-1033.1989.tb15210.x. [DOI] [PubMed] [Google Scholar]
- 262.Kaesler B, Schönheit P. The role of sodium ions in methanogenesis. Formaldehyde oxidation to CO2 and 2 H2 in methanogenic bacteria is coupled with primary electrogenic Na+ translocation at a stoichiometry of 2—3 Na+/CO2. Eur J Biochem. 1989;184:223–232. doi: 10.1111/j.1432-1033.1989.tb15010.x. [DOI] [PubMed] [Google Scholar]
- 263.Kamikubo H, Kataoka M, Váró G, Oka T, Tokunaga F, Needleman R, Lanyi J K. Structure of the N intermediate of bacteriorhodopsin revealed by x-ray diffraction. Proc Natl Acad Sci USA. 1996;93:1386–1390. doi: 10.1073/pnas.93.4.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Kamlage B, Blaut M. Characterization of cytochromes from Methanosarcina strain Gö1 and their involvement in electron transport during growth on methanol. J Bacteriol. 1992;174:3921–3927. doi: 10.1128/jb.174.12.3921-3927.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Kamo N, Hazemoto N, Kobatake Y, Mukohata Y. Light and dark adaptation of halorhodopsin. Arch Biochem Biophys. 1985;238:90–96. doi: 10.1016/0003-9861(85)90144-4. [DOI] [PubMed] [Google Scholar]
- 266.Kandler O, König H. Cell envelopes of archaea: structure and chemistry. In: Kates M, Kushner D J, Matheson A T, editors. The biochemistry of archaea. 1993. pp. 223–259. Elsiever, Amsterdam, The Netherlands. [Google Scholar]
- 267.Kandler O, Stetter K O. Evidence for autotrophic CO2 assimilation in Sulfolobus brierleyi via a reductive carboxylic acid pathway. Zentbl Bakteriol Hyg Abt 1 Orig C. 1981;2:111–121. [Google Scholar]
- 268.Kandori H, Yoshihara K, Tomioka H, Sasabe H. Primary photochemical events in halorhodopsin studied by subpicosecond time-resolved spectroscopy. J Phys Chem. 1992;96:6066–6071. [Google Scholar]
- 269.Kannt A, Soulimane T, Buse G, Becker A, Bamberg E, Michel H. Electrical current generation and proton pumping catalyzed by the ba3-type cytochrome c oxidase from Thermus thermophilus. FEBS Lett. 1998;434:17–22. doi: 10.1016/s0014-5793(98)00942-9. [DOI] [PubMed] [Google Scholar]
- 270.Karrasch S, Hegerl R, Hoh J H, Baumeister W, Engel A. Atomic force microscopy produces faithful high-resolution images of protein surfaces in an aqueous environment. Proc Natl Acad Sci USA. 1994;91:836–838. doi: 10.1073/pnas.91.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Kataoka M, Kamikubo H, Tokunaga F, Brown L S, Yamazaki Y, Maeda A, Sheves M, Needleman R, Lanyi J K. Energy coupling in an ion pump. The reprotonation switch of bacteriorhodopsin. J Mol Biol. 1994;243:621–638. doi: 10.1016/0022-2836(94)90037-x. [DOI] [PubMed] [Google Scholar]
- 272.Kath T, Schmid R, Schäfer G. Identification, cloning, and expression of the gene for adenylate kinase from the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. Arch Biochem Biophys. 1993;307:405–410. doi: 10.1006/abbi.1993.1607. [DOI] [PubMed] [Google Scholar]
- 273.Kawarabayasi Y, Sawada M, Horikawa Y, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka H, Nakazawa M, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Kikuchi H. Complete sequence and gene organization of the genome of a hyperthermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 1998;5(Suppl.):147–155. doi: 10.1093/dnares/5.2.147. [DOI] [PubMed] [Google Scholar]
- 274.Kawarabayasi Y, Sawada M, Horikawa Y, Haikawa Y, Hino Y, Yamamoto S, Sekine M, Baba S, Kosugi H, Hosoyama A, Nagai Y, Sakai M, Ogura K, Otsuka H, Nakazawa M, Takamiya M, Ohfuku Y, Funahashi T, Tanaka T, Kudoh Y, Yamazaki J, Kushida N, Oguchi A, Aoki K, Kikuchi H. Complete sequence and organization of the genome of a hyperthermophilic archaebacterium, Pyrococcus horikoshii. DNA Res. 1998;5:55–76. doi: 10.1093/dnares/5.2.55. [DOI] [PubMed] [Google Scholar]
- 275.Kemner J M, Krzycki J A, Prince R C, Zeikus J G. Spectroscopic and enzymatic evidence for membrane-bound electron transport carriers and hydrogenase and their relation to cytochrome b function in Methanosarcina barkeri. FEMS Lett. 1987;48:267–272. [Google Scholar]
- 276.Kerscher L, Nowitzki S, Oesterhelt D. Thermoacidophilic archaebacteria contain bacterial-type ferredoxins acting as electron acceptors of 2-oxoacid:ferredoxin oxidoreductases. Eur J Biochem. 1982;128:223–230. doi: 10.1111/j.1432-1033.1982.tb06955.x. [DOI] [PubMed] [Google Scholar]
- 277.Khorana H G, Gerber G E, Herlihy W C, Gray C P, Anderegg R J, Nihei K, Biemann K. Amino acid sequence of bacteriorhodopsin. Proc Natl Acad Sci USA. 1979;76:5046–5050. doi: 10.1073/pnas.76.10.5046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Kimura Y, Ikegami A, Stoeckenius W. Salt and pH-dependent changes of the purple membrane absorption spectrum. Photochem Photobiol. 1984;40:641–646. doi: 10.1111/j.1751-1097.1984.tb05353.x. [DOI] [PubMed] [Google Scholar]
- 279.Kimura Y, Vassylyev D G, Miyazawa A, Kidera A, Matsushima M, Mitsuoka K, Murata K, Hirai T, Fujiyoshi Y. Surface of bacteriorhodopsin revealed by high-resolution electron crystallography. Nature. 1997;389:206–211. doi: 10.1038/38323. [DOI] [PubMed] [Google Scholar]
- 280.Kinosita K, Jr, Yasuda R, Noji H, Ishiwata S, Yoshida M. F1-ATPase: a rotary motor made of a single molecule. Cell. 1998;93:21–24. doi: 10.1016/s0092-8674(00)81142-3. [DOI] [PubMed] [Google Scholar]
- 281.Klenk H-P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, Richardson D L, Kerlavage A R, Graham D E, Kyrpides N C, Fleischmann R D, Quackenbush J, Lee N H, Sutton G G, Gill S, Kirkness E F, Dougherty B A, McKenney K, Adams M D, Loftus B. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 282.Kletzin A. Coupled enzymatic production of sulfite, thiosulfate, and hydrogen sulfide from sulfur: purification and properties of a sulfur oxygenase reductase from the facultatively anaerobic archaebacterium Desulfurolobus ambivalens. J Bacteriol. 1989;171:1638–1643. doi: 10.1128/jb.171.3.1638-1643.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Kletzin A. Sulfur oxidation and reduction in archaea: sulfur oxygenase/-reductase and hydrogenases from the extremely thermophilic and facultatively anaerobic archaeon Desulfurolobus ambivalens. Syst Appl Microbiol. 1994;16:534–543. [Google Scholar]
- 284.Knapp S, Kardinahl S, Hellgren N, Tibbelin G, Schäfer G, Ladenstein R. Refined crystal structure of superoxide dismutase from the hyperthermophilic archaeon Sulfolobus acidocaldarius at 2.2 A resolution. J Mol Biol. 1999;285:689–702. doi: 10.1006/jmbi.1998.2344. [DOI] [PubMed] [Google Scholar]
- 285.Koch M H J, Dencher N A, Oesterhelt D, Plöhn H-J, Rapp G, Büldt G. Time-resolved X-ray diffraction study of structural changes associated with the photocycle of bacteriorhodopsin. EMBO J. 1991;10:521–526. doi: 10.1002/j.1460-2075.1991.tb07978.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Komorowski, L. 1999. Personal communication.
- 287.Konishi T, Murakami N. Detection of two DCCD binding components in the envelope membrane of H. halobium. FEBS Lett. 1984;169:283–286. [Google Scholar]
- 288.Kostyukova A S, Polosina Y Y, Pyatibratov M G, Tiktopulo E I, Fedorov O V. Possible participation of chaperones in archaebacterial flagella assembly. Dokl Akad Nauk. 1994;339:544–546. . (In Russian.) [Google Scholar]
- 289.Kostyukova A S, Pyatibratov M G, Tarasov V Y, Gongadze G M, Fedorov O V. Structural and functional peculiarities of the archaeal apparatus of motility connected with extremal environment. Biofizika. 1996;41:164–168. . (In Russian.) [Google Scholar]
- 290.Kovbasnjuk O N, Antonenko Y N, Yaguzhinsky L S. Proton dissociation from nigericin at the membrane-water interface, the rate-limiting step of K+/H+ exchange on the bilayer lipid membrane. FEBS Lett. 1991;289:176–178. doi: 10.1016/0014-5793(91)81063-e. [DOI] [PubMed] [Google Scholar]
- 291.Krah M, Marwan W, Oesterhelt D. A cytoplasmic domain is required for the functional interaction of SRI and HtrI in archaeal signal transduction. FEBS Lett. 1994;353:301–304. doi: 10.1016/0014-5793(94)01068-4. [DOI] [PubMed] [Google Scholar]
- 292.Krah M, Marwan W, Verméglio A, Oesterhelt D. Phototaxis of Halobacterium salinarum requires a signalling complex of sensory rhodopsin I and its methyl-accepting transducer HtrI. EMBO J. 1994;13:2150–2155. doi: 10.1002/j.1460-2075.1994.tb06491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Kristjansson H, Hochstein L I. Dicyclohexylcarbodiimide-sensitive ATPase in Halobacterium saccharovorum. Arch Biochem Biophys. 1985;241:590–595. doi: 10.1016/0003-9861(85)90584-3. [DOI] [PubMed] [Google Scholar]
- 294.Krulwich T A, Guffanti A A. Alkalophilic bacteria. Annu Rev Microbiol. 1989;43:435–463. doi: 10.1146/annurev.mi.43.100189.002251. [DOI] [PubMed] [Google Scholar]
- 295.Kühn W, Gottschalk G. Characterization of the cytochromes occurring in Methanosarcina species. Eur J Biochem. 1983;135:89–94. doi: 10.1111/j.1432-1033.1983.tb07621.x. [DOI] [PubMed] [Google Scholar]
- 296.Künkel A, Vaupel M, Heim S, Thauer R K, Hedderich R. Heterodisulfide reductase from methanol-grown cells of Methanosarcina barkeri is not a flavoenzyme. Eur J Biochem. 1997;244:226–234. doi: 10.1111/j.1432-1033.1997.00226.x. [DOI] [PubMed] [Google Scholar]
- 297.Kunow J, Linder D, Stetter K O, Thauer R K. F420H2:quinone oxidoreductase from Archaeoglobus fulgidus—characterization of a membrane-bound multisubunit complex containing FAD and iron-sulfur clusters. Eur J Biochem. 1994;223:503–511. doi: 10.1111/j.1432-1033.1994.tb19019.x. [DOI] [PubMed] [Google Scholar]
- 298.Kuo S C, Koshland D E., Jr Multiple kinetic states for the flagellar motor switch. J Bacteriol. 1989;171:6279–6287. doi: 10.1128/jb.171.11.6279-6287.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Lacher K, Schäfer G. Archaebacterial adenylate kinase from the thermoacidophile Sulfolobus acidocaldarius: purification, characterization, and partial sequence. Arch Biochem Biophys. 1993;302:391–397. doi: 10.1006/abbi.1993.1229. [DOI] [PubMed] [Google Scholar]
- 300.Langer D, Hain J, Thuriaux P, Zillig W. Transcription in archaea: similarity to that in eucarya. Proc Natl Acad Sci USA. 1995;92:5768–5772. doi: 10.1073/pnas.92.13.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Lanyi J K. Studies on the electron transport chain of extremely halophilic Bacteria. I. Spectrophotometric identification of the cytochromes of Halobacterium cutirubrum. Arch Biochem Biophys. 1968;128:716–724. doi: 10.1016/0003-9861(68)90080-5. [DOI] [PubMed] [Google Scholar]
- 302.Lanyi J K. Studies of the electron transport chain of extremely halophilic bacteria. II. Salt dependence of reduced diphosphopyridine nucleotide oxidase. J Biol Chem. 1969;244:2864–2869. [PubMed] [Google Scholar]
- 303.Lanyi J K. Light-dependent trans to cis isomerization of the retinal in halorhodopsin. FEBS Lett. 1984;2:337–342. doi: 10.1016/0014-5793(84)80764-4. [DOI] [PubMed] [Google Scholar]
- 304.Lanyi J K, Duschl A, Hatfield G W, May K, Oesterhelt D. The primary structure of halorhodopsin from Natronobacterium pharaonis. J Biol Chem. 1990;265:1253–1260. [PubMed] [Google Scholar]
- 305.Lanyi J K, Duschl A, Váró G, Zimányi L. Anion binding to the chloride pump, halorhodopsin, and its implications for the transport mechanism. FEBS Lett. 1990;265:1–6. doi: 10.1016/0014-5793(90)80869-k. [DOI] [PubMed] [Google Scholar]
- 306.Lanyi J K, Varo G. The photocycles of bacteriorhodopsin. Isr J Chem. 1995;35:365–385. [Google Scholar]
- 307.Lanyi J K, Vodyanoy V. Flash spectroscopic studies of the kinetics of the halorhodopsin photocycle. Biochemistry. 1986;25:1465–1470. doi: 10.1021/bi00354a042. [DOI] [PubMed] [Google Scholar]
- 308.Laufer K, Eikmanns B, Frimmer U, Thauer R K. Methanogenesis from acetate by Methanosarcina barkeri: catalysis of acetate formation by methyl iodide, CO2 and H2 by the enzyme system involved. Z Naturforsch. 1987;42:360–372. [Google Scholar]
- 309.Leif H, Sled V D, Ohnishi T, Weiss H, Friedrich T. Isolation and characterization of the proton-translocating NADH:ubiquinone oxidoreductase from Escherichia coli. Eur J Biochem. 1995;230:538–548. doi: 10.1111/j.1432-1033.1995.tb20594.x. [DOI] [PubMed] [Google Scholar]
- 310.Leif H, Weidner U, Berger A, Spehr V, Braun M, Van Heek P, Friedrich T, Ohnishi T, Weiss H. Escherichia coli NADH dehydrogenase I, a minimal form of the mitochondrial complex I. Biochem Soc Trans. 1993;21:998–1001. doi: 10.1042/bst0210998. [DOI] [PubMed] [Google Scholar]
- 311.Lemke H D, Oesterhelt D. Lysine 216 is a binding site of the retinyl moiety in bacteriorhodopsin. FEBS Lett. 1981;128:255–260. doi: 10.1016/0014-5793(81)80093-2. [DOI] [PubMed] [Google Scholar]
- 312.Lemker, T., and V. Müller. 1998. Unpublished data.
- 313.Le Moual H, Koshland D E. Molecular evolution of the c-terminal cytoplasmic domain of a superfamily of bacterial receptors involved in taxis. J Mol Biol. 1996;265:568–585. doi: 10.1006/jmbi.1996.0483. [DOI] [PubMed] [Google Scholar]
- 314.Leng X H, Manolson M F, Forgac M. Site-directed mutagenesis of the 100 kDa subunit (Vph1p) of the yeast vacuolar H+ ATPase. J Biol Chem. 1996;271:22487–22493. doi: 10.1074/jbc.271.37.22487. [DOI] [PubMed] [Google Scholar]
- 315.Leng X H, Manolson M F, Forgac M. Function of the COOH terminal domain of Vph1p in activity and asesmbly of the yeast V-ATPase. J Biol Chem. 1998;273:6717–6723. doi: 10.1074/jbc.273.12.6717. [DOI] [PubMed] [Google Scholar]
- 316.Leppänen V-M, Nummelin H, Hansen T, Schäfer G, Goldman A. Refined crystal structure and basis of thermostability in Sulfolobus acidocaldarius inorganic pyrophosphatase. Protein Sci. 1999;8:1218–1231. doi: 10.1110/ps.8.6.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317.Lienard T, Becher B, Marschall M, Bowien S, Gottschalk G. Sodium ion translocation by N5-methyltetrahydromethanopterin:coenzyme M methyltransferase from Methanosarcina mazei GÜ1 reconstituted in ether lipid liposomes. Eur J Biochem. 1996;239:857–864. doi: 10.1111/j.1432-1033.1996.0857u.x. [DOI] [PubMed] [Google Scholar]
- 318.Lienard T, Gottschalk G. Cloning, sequencing and expression of the genes encoding the sodium translocating N5-methyltetrahydromethanopterin:coenzyme M methyltransferase of the methylotrophic archaeon Methanosarcina mazei Go1. FEBS Lett. 1998;425:204–208. doi: 10.1016/s0014-5793(98)00229-4. [DOI] [PubMed] [Google Scholar]
- 319.Livnah N, Sheves M. The schiff base bond configuration in bacteriorhodopsin and in model compounds. Biochemistry. 1993;32:7223–7228. doi: 10.1021/bi00079a019. [DOI] [PubMed] [Google Scholar]
- 320.Ljungdahl L G. The autotrophic pathway of acetate synthesis in acetogenic bacteria. Annu Rev Microbiol. 1986;40:415–450. doi: 10.1146/annurev.mi.40.100186.002215. [DOI] [PubMed] [Google Scholar]
- 321.Logunov I, Humphrey W, Schulten K, Sheves M. Molecular dynamics study of the 13-cis form (bR548) of bacteriorhodopsin and its photocycle. Biophys J. 1995;68:1270–1282. doi: 10.1016/S0006-3495(95)80301-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Logunov I, Schulten K. Quantum chemistry-molecular dynamics study of the dark-adaptation process in bacteriorhodopsin. J Am Chem Soc. 1996;118:9727–9735. [Google Scholar]
- 323.Lozier R H, Bogomolni R A, Stoeckenius W. Bacteriorhodopsin: a light-driven proton pump in Halobacterium halobium. Biophys J. 1975;15:955–962. doi: 10.1016/S0006-3495(75)85875-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Lu W-P, Becher B, Gottschalk G, Ragsdale S W. Electron paramagnetic resonance spectroscopic and electrochemical characterization of the partially purified N5-methyltetrahydromethanopterin:coenzyme M methyltransferase from Methanosarcina mazei Gö1. J Bacteriol. 1995;177:2245–2250. doi: 10.1128/jb.177.9.2245-2250.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Lübben M. Cytochromes of archaeal electron transfer chains. Biochim Biophys Acta. 1995;1229:1–22. doi: 10.1016/0005-2728(94)00174-4. [DOI] [PubMed] [Google Scholar]
- 326.Lübben M, Anemüller S, Schäfer G. Investigation of the bioenergetic system of Sulfolobus acidocaldarius DSM 639. Syst Appl Microbiol. 1986;7:425–426. [Google Scholar]
- 327.Lübben M, Arnaud S, Castresana J, Warne A, Albracht S P J, Saraste M. A second terminal oxidase in Sulfolobus acidocaldarius. Eur J Biochem. 1994;224:151–159. doi: 10.1111/j.1432-1033.1994.tb20006.x. [DOI] [PubMed] [Google Scholar]
- 328.Lübben M, Castresana J, Warne A. Terminal oxidases of Sulfolobus: genes and proteins. Syst Appl Microbiol. 1994;16:556–559. [Google Scholar]
- 329.Lübben M, Kolmerer B, Saraste M. An archaebacterial terminal oxidase combines core structures of two mitochondrial respiratory complexes. EMBO J. 1992;11:805–812. doi: 10.1002/j.1460-2075.1992.tb05117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Lübben M, Lünsdorf H, Schäfer G. The plasma membrane associated ATPase of the thermoacidophilic archaebacterium Sulfolobus acidocaldarius: purification and immunological relationships to F1-ATPases. Eur J Biochem. 1987;167:211–219. doi: 10.1111/j.1432-1033.1987.tb13325.x. [DOI] [PubMed] [Google Scholar]
- 331.Lübben M, Lünsdorf H, Schäfer G. Archaebacterial ATPase: studies on subunit composition and quaternary structure of the F1-analogous ATPase from Sulfolobus acidocaldarius. Biol Chem Hoppe-Seyler. 1988;369:1259–1266. doi: 10.1515/bchm3.1988.369.2.1259. [DOI] [PubMed] [Google Scholar]
- 332.Lübben M, Morand K. Novel prenylated hemes as cofactors of cytochrome oxidases. J Biol Chem. 1994;269:21473–21479. [PubMed] [Google Scholar]
- 333.Lübben M, Schäfer G. A plasma membrane associated adenosine triphosphatase of heterotrophically grown Sulfolobus acidocaldarius. EBEC Short Rep. 1985;3:336–337. [Google Scholar]
- 334.Lübben M, Schäfer G. A plasma membrane associated ATPase from the thermoacidophilic archaebacterium Sulfolobus acidocaldarius (DSM 639) Eur J Biochem. 1987;164:533–540. doi: 10.1111/j.1432-1033.1987.tb11159.x. [DOI] [PubMed] [Google Scholar]
- 335.Lübben M, Schäfer G. Chemiosmotic energy conversion of the archaebacterial thermoacidophile Sulfolobus acidocaldarius: oxidative phosphorylation and the presence of an F0-related N,N′-dicyclohexylcarbodiimide-binding proteolipid. J Bacteriol. 1989;171:6106–6116. doi: 10.1128/jb.171.11.6106-6116.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 336.Lübben M, Warne A, Albracht S P J, Saraste M. The purified SoxABCD quinol oxidase complex of Sulfolobus acidocaldarius contains a novel haem. Mol Microbiol. 1994;13:327–335. doi: 10.1111/j.1365-2958.1994.tb00426.x. [DOI] [PubMed] [Google Scholar]
- 337.Luecke H, Richter H T, Lanyi J K. Proton transfer pathways in bacteriorhodopsin at 2.3 Angstrom resolution. Science. 1998;280:1934–1937. doi: 10.1126/science.280.5371.1934. [DOI] [PubMed] [Google Scholar]
- 338.Lünsdorf H, Niedrig M, Fiebig K. Immunocytochemical localization of the coenzyme F420-reducing hydrogenase in Methanosarcina barkeri Fusaro. J Bacteriol. 1991;173:978–984. doi: 10.1128/jb.173.3.978-984.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339.Ma J X, Katsonouri A, Gennis R B. Subunit II of the cytochrome bo3 ubiquinol oxidase from Escherichia coli is a lipoprotein. Biochemistry. 1997;36:11298–11303. doi: 10.1021/bi9709710. [DOI] [PubMed] [Google Scholar]
- 340.MacNab R M, Ornston M K. Normal-to-curly flagellar transitions and their role in bacterial tumbling. Stabilization of an alternative quaternary structure by mechanical force. J Mol Biol. 1977;112:1–30. doi: 10.1016/s0022-2836(77)80153-8. [DOI] [PubMed] [Google Scholar]
- 341.Maeda A, Ogurusu T, Yoshizawa T. Resonance Raman study on binding of chloride to the chromophore of halorhodopsin. Biochemistry. 1985;24:2517–2521. [Google Scholar]
- 342.Marwan W, Alam M, Oesterhelt D. Rotation and switching of the flagellar motor assembly in Halobacterium halobium. J Bacteriol. 1991;173:1971–1977. doi: 10.1128/jb.173.6.1971-1977.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Marwan W, Bibikov S I, Montrone M, Oesterhelt D. Mechanism of photosensory adaptation in Halobacterium salinarum. J Mol Biol. 1995;246:493–499. doi: 10.1006/jmbi.1994.0101. [DOI] [PubMed] [Google Scholar]
- 344.Marwan W, Oesterhelt D. Signal formation in the halobacterial photophobic response mediated by a fourth retinal protein/P480. J Mol Biol. 1987;195:333–342. doi: 10.1016/0022-2836(87)90654-1. [DOI] [PubMed] [Google Scholar]
- 345.Marwan W, Oesterhelt D. Light-induced release of the switch factor during photophobic responses of Halobacterium halobium. Naturwissenschaften. 1991;78:127–129. [Google Scholar]
- 346.Marwan W, Schäfer W, Oesterhelt D. Signal transduction in Halobacterium depends on fumarate. EMBO J. 1990;9:355–362. doi: 10.1002/j.1460-2075.1990.tb08118.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 347.Matsubo-Yagi A, Mukohata Y. ATP synthesis linked to light-dependent proton uptake in a red mutant strain of Halobacterium lacking bacteriorhodopsin. Arch Biochem Biophys. 1980;199:297–303. doi: 10.1016/0003-9861(80)90284-2. [DOI] [PubMed] [Google Scholar]
- 348.Mattar S, Engelhard M. Primary structure of cytochrome ba3 from the haloalkaliphilic archaeon Natronobacterium pharaonis; phylogenetic and functional implications. EBEC Short Rep. 1996;9:85. [Google Scholar]
- 349.Mattar S, Engelhard M. Cytochrome ba3 from Natronobacterium pharaonis—an archaeal four-subunit cytochrome-c-type oxidase. Eur J Biochem. 1997;250:332–341. doi: 10.1111/j.1432-1033.1997.0332a.x. [DOI] [PubMed] [Google Scholar]
- 350.Mattar S, Scharf B, Kent S B H, Rodewald K, Oesterhelt D, Engelhard M. The primary structure of halocyanin, an archaeal blue copper protein, predicts a lipid anchor for membrane fixation. J Biol Chem. 1994;269:14939–14945. [PubMed] [Google Scholar]
- 351.McCarthy J E G, Schairer H U, Sebald W. Translational initiation frequency of atp genes from Escherichia coli: identification of an intercistronic sequence that enhances translation. EMBO J. 1985;4:519–526. doi: 10.1002/j.1460-2075.1985.tb03659.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.McDermott A E, Creuzet F, Gebhard R, van der Hoef K, Levitt M H, Herzfeld J, Lugtenburg J, Griffin R G. Determination of internuclear distances and the orientation of functional groups by solid-state NMR: rotational resonance study of the conformation of retinal in bacteriorhodopsin. Biochemistry. 1994;33:6129–6136. doi: 10.1021/bi00186a012. [DOI] [PubMed] [Google Scholar]
- 353.Melin L, Magnusson K, Rutberg L. Identification of the promoter of the Bacillus subtilis sdh operon. J Bacteriol. 1987;169:3232–3236. doi: 10.1128/jb.169.7.3232-3236.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Metz G, Siebert F, Engelhard M. High-resolution solid state 13C NMR of bacteriorhodopsin: characterization of [4-13C]Asp resonances. Biochemistry. 1992;31:455–462. doi: 10.1021/bi00117a022. [DOI] [PubMed] [Google Scholar]
- 355.Metz G, Siebert F, Engelhard M. Asp85 is the only internal aspartic acid that gets protonated in the M intermediate and the purple-to-blue transition of bacteriorhodopsin: a solid-state 13C CP-MAS NMR investigation. FEBS Lett. 1992;303:237–241. doi: 10.1016/0014-5793(92)80528-o. [DOI] [PubMed] [Google Scholar]
- 356.Meunier B, Madgwick S A, Reil E, Oettmeier W, Rich P R. New inhibitors of the quinol oxidation sites of bacterial cytochromes bo and bd. Biochemistry. 1995;34:1076–1083. doi: 10.1021/bi00003a044. [DOI] [PubMed] [Google Scholar]
- 357.Meyer W, Moll R, Kath T, Schäfer G. Purification, cloning, and sequencing of archaebacterial pyrophosphatase from the extreme thermoacidophile Sulfolobus acidocaldarius. Arch Biochem Biophys. 1995;319:149–156. doi: 10.1006/abbi.1995.1277. [DOI] [PubMed] [Google Scholar]
- 358.Meyer W, Schäfer G. Characterization and purification of a membrane-bound archaebacterial pyrophosphatase from Sulfolobus acidocaldarius. Eur J Biochem. 1992;207:741–746. doi: 10.1111/j.1432-1033.1992.tb17104.x. [DOI] [PubMed] [Google Scholar]
- 359.Meyer, W., and G. Schäfer. 1993. Personal communication.
- 360.Michel H, Neugebauer D C, Oesterhelt D. The 2D-crystalline cell wall of Sulfolobus acidocaldarius: structure, solubilization and reassembly. In: Baumeister W, Vogell W, editors. Electron microscopy at molecular dimensions. Berlin, Germany: Springer-Verlag; 1980. pp. 27–35. [Google Scholar]
- 361.Michel H, Oesterhelt D. Electrochemical proton gradient across the cell membrane of Halobacterium halobium: comparison of the light-induced increase with the increase of intracellular ATP under steady state illumination. Biochemistry. 1980;19:4615–4619. doi: 10.1021/bi00561a012. [DOI] [PubMed] [Google Scholar]
- 362.Michel H, Oesterhelt D. Electrochemical proton gradient across the cell membrane of Halobacterium halobium: effect of DCCD, relation to intracellular ATP, ADP and phosphate concentration, and influence of the potassium gradient. Biochemistry. 1980;19:4607–4614. doi: 10.1021/bi00561a011. [DOI] [PubMed] [Google Scholar]
- 363.Michel H, Oesterhelt D. Light-induced changes of the pH gradient and the membrane potential in Halobacterium halobium. FEBS Lett. 1976;2:175–178. doi: 10.1016/0014-5793(76)80473-5. [DOI] [PubMed] [Google Scholar]
- 364.Michels M, Bakker E P. Generation of a large, protonophore-sensitive proton motive force and pH difference in the acidophilic bacteria Thermoplasma acidophilum and Bacillus acidocaldarius. J Bacteriol. 1985;161:231–237. doi: 10.1128/jb.161.1.231-237.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 365.Minami Y, Wakabayashi S, Wada K, Matsubara H, Kerscher L, Oesterhelt D. Amino acid sequence of a ferredoxin from thermoacidophilic archaebacterium, Sulfolobus acidocaldarius. Presence of an N6-monomethyllysine and phyletic consideration of archaebacteria. J Biochem. 1985;97:745–753. doi: 10.1093/oxfordjournals.jbchem.a135114. [DOI] [PubMed] [Google Scholar]
- 366.Mitchell P. Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature. 1961;191:144–148. doi: 10.1038/191144a0. [DOI] [PubMed] [Google Scholar]
- 367.Mitchell P. Chemiosmotic coupling in oxidative and photosynthetic phosphorylation. Glynn; 1966. , Bodmin, Cornwall, United Kingdom. [DOI] [PubMed] [Google Scholar]
- 368.Miyazaki M, Hirayama J, Hayakawa M, Kamo N. Flash photolysis study on pharaonis phoborhodopsin from a haloalkaliphilic bacterium (Natronobacterium pharaonis) Biochim Biophys Acta. 1992;1140:22–29. [Google Scholar]
- 369.Moll R. Isolierung und Charakterisierung des Succinat Dehydrogenase Komplexes aus Sulfolobus acidocaldarius. Ph.D. thesis. Lübeck, Germany: University of Lübeck; 1991. [Google Scholar]
- 370.Moll R, Schäfer G. Chemiosmotic H+ cycling across the plasma membrane of the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. FEBS Lett. 1988;232:359–363. [Google Scholar]
- 371.Moll R, Schäfer G. Purification and characterisation of an archaebacterial succinate dehydrogenase complex from the plasma membrane of the thermoacidophile Sulfolobus acidocaldarius. Eur J Biochem. 1991;201:593–600. doi: 10.1111/j.1432-1033.1991.tb16319.x. [DOI] [PubMed] [Google Scholar]
- 372.Moltke S, Nevzorov A A, Sakai N, Wallat I, Job C, Nakanishi K, Heyn M P, Brown M F. Chromophore orientation in bacteriorhodopsin determined from the angular dependence of deuterium nuclear magnetic resonance spectra of oriented purple membranes. Biochemistry. 1998;34:11821–11835. doi: 10.1021/bi980676v. [DOI] [PubMed] [Google Scholar]
- 373.Moracci M, La Volpe A, Pulitzer J F, Rossi M, Ciaramella M. Expression of the thermostable β-galactosidase gene from the archaebacterium Sulfolobus solfataricus in Saccharomyces cerevisiae and characterization of a new inducible promoter for heterologous expression. J Bacteriol. 1992;174:873–882. doi: 10.1128/jb.174.3.873-882.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 374.Mukohata Y, Isoyama M, Fuke A. ATP synthesis in cell envelope vesicles of Halobacterium halobium driven by membrane potential and/or base-acid transition. J Biochem. 1986;99:1–8. doi: 10.1093/oxfordjournals.jbchem.a135448. [DOI] [PubMed] [Google Scholar]
- 375.Mukohata Y, Yoshida M. Activation and inhibition of ATPase synthesis in cell envelope vesicles of Halobacterium halobium. J Biochem. 1987;101:311–318. doi: 10.1093/oxfordjournals.jbchem.a121915. [DOI] [PubMed] [Google Scholar]
- 376.Mukohata Y, Yoshida M. The H+-translocating ATP synthase in Halobacterium halobium differs from F0F1-ATPase/synthase. J Biochem. 1987;102:797–802. doi: 10.1093/oxfordjournals.jbchem.a122118. [DOI] [PubMed] [Google Scholar]
- 377.Mukund S, Adams M W W. Characterization of a novel tungsten-containing formaldehyde ferredoxin oxidoreductase from the hyperthermophilic archaeon, Thermococcus litoralis. A role for tungsten in peptide catabolism. J Biol Chem. 1993;268:13592–13600. [PubMed] [Google Scholar]
- 378.Müller C W, Schulz G E. Structure of the complex of adenylate kinase from Escherichia coli with the inhibitor P1,P5-di(adenosine5′-)pentaphosphate. J Mol Biol. 1988;202:909–912. doi: 10.1016/0022-2836(88)90567-0. [DOI] [PubMed] [Google Scholar]
- 379.Müller E C, Wittmann-Liebold B. Phylogenetic relationship of organisms obtained by ribosomal protein comparison. Cell Mol Life Sci. 1997;53:34–50. doi: 10.1007/PL00000578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 380.Müller V, Blaut M, Gottschalk G. The transmembrane electrochemical gradient of Na+ as driving force for methanol oxidation in Methanosarcina barkeri. Eur J Biochem. 1988;172:601–606. doi: 10.1111/j.1432-1033.1988.tb13931.x. [DOI] [PubMed] [Google Scholar]
- 381.Müller V, Blaut M, Gottschalk G. Bioenergetics of methanogenesis. In: Ferry J G, editor. Methanogenesis. New York, N.Y: Chapman and Hall; 1993. pp. 360–406. [Google Scholar]
- 382.Müller V, Gottschalk G. The sodium ion cycle in acetogenic and methanogenic bacteria; generation and utilization of a primary electrochemical sodium ion gradient. In: Drake H L, editor. Acetogenesis. New York, N.Y: Chapman and Hall; 1994. pp. 127–156. [Google Scholar]
- 383.Müller V, Ruppert C, Lemker T. Structure and function of the A0A1 ATPases from methanogenic Archaea. J Bioenerg Biomembr. 1999;31:15–27. doi: 10.1023/a:1005451311009. [DOI] [PubMed] [Google Scholar]
- 384.Müller V, Winner C, Gottschalk G. Electron transport-driven sodium extrusion during methanogenesis from formaldehyde + H2 by Methanosarcina barkeri. Eur J Biochem. 1988;178:519–525. doi: 10.1111/j.1432-1033.1988.tb14478.x. [DOI] [PubMed] [Google Scholar]
- 385.Muth E. Localization of the F420-reducing hydrogenase in Methanococcus voltae cells by immunogold technique. Arch Microbiol. 1988;150:205–207. [Google Scholar]
- 386.Nagel G, Kelety B, Möckel B, Büldt G, Bamberg E. Voltage dependence of proton pumping by bacteriorhodopsin is regulated by the voltage-sensitive ratio of M-1 to M-2. Biophys J. 1998;74:403–412. doi: 10.1016/S0006-3495(98)77797-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 387.Nagel G, Möckel B, Büldt G, Bamberg E. Functional expression of bacteriorhodopsin in oocytes allows direct measurement of voltage dependence of light induced H+ pumping. FEBS Lett. 1995;377:263–266. doi: 10.1016/0014-5793(95)01356-3. [DOI] [PubMed] [Google Scholar]
- 388.Nakamura H, Saiki K, Mogi T, Anraku Y. Assignment and functional roles of the cyoABCDE gene products required for the Escherichia coli bo-type quinol oxidase. J Biochem (Tokyo) 1997;122:415–421. doi: 10.1093/oxfordjournals.jbchem.a021769. [DOI] [PubMed] [Google Scholar]
- 389.Nakasako M, Kataoka M, Amemiya Y, Tokunaga F. Crystallographic characterization by X-ray diffraction of the M-intermediate from the photo-cycle of bacteriorhodopsin at room temperature. FEBS Lett. 1991;292:73–75. doi: 10.1016/0014-5793(91)80837-s. [DOI] [PubMed] [Google Scholar]
- 390.Nanba T, Mukohata Y. A membrane-bound ATPase from Halobacterium halobium: purification and characterization. J Biochem. 1987;102:591–598. doi: 10.1093/oxfordjournals.jbchem.a122092. [DOI] [PubMed] [Google Scholar]
- 391.Nelson N. Evolution of organellar proton-ATPases. Biochim Biophys Acta. 1992;1100:109–124. doi: 10.1016/0005-2728(92)90072-a. [DOI] [PubMed] [Google Scholar]
- 392.Nicolaus B, Trincone A, Lama L, Palmieri G, Gambacorta A. Quinone composition in Sulfolobus acidocaldarius grown under different conditions. Syst Appl Microbiol. 1992;15:18–20. [Google Scholar]
- 393.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Direct observation of the rotation of F1-ATPase. Nature. 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 394.Nölling J, Ishii M, Koch J, Pihl T D, Reeve J N, Thauer R K, Hedderich R. Characterization of a 45-kDa flavoprotein and evidence for a rubredoxin, two proteins that could participate in electron transport from H2 to CO2 in methanogenesis in Methanobacterium thermoautotrophicum. Eur J Biochem. 1995;231:628–638. doi: 10.1111/j.1432-1033.1995.0628d.x. [DOI] [PubMed] [Google Scholar]
- 395.Nonella M, Windemuth A, Schulten K. Structure of bacteriorhodopsin and in situ isomerization of retinal: a molecular dynamics study. Photochem Photobiol. 1991;54:937–948. [Google Scholar]
- 396.Nordmann B, Lebert M R, Alam M, Nitz S, Kollmannsberger H, Oesterhelt D, Hazelbauer G L. Identification of volatile forms of methyl groups released by Halobacterium salinarum. J Biol Chem. 1994;269:16449–16454. [PubMed] [Google Scholar]
- 397.Nuss M C, Zinth W, Kaiser W, Koelling E, Oesterhelt D. Femtosecond spectroscopy of the first events of the photochemical cycle in bacteriorhodopsin. Chem Phys Lett. 1985;117:1–7. [Google Scholar]
- 398.Oesterhelt D. Structure and function of halorhodopsin. Isr J Chem. 1995;35:475–494. [Google Scholar]
- 399.Oesterhelt D, Hegemann P, Tavan P, Schulten K. Trans-cis isomerization of retinal and a mechanism for ion translocation in halorhodopsin. Eur Biophys J. 1986;14:123–129. [Google Scholar]
- 400.Oesterhelt D, Hegemann P, Tittor J. The photocycle of the chloride pump halorhodopsin II: quantum yields and a kinetic model. EMBO J. 1985;4:2351–2356. doi: 10.1002/j.1460-2075.1985.tb03938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Oesterhelt D, Stoeckenius W. Rhodopsin-like protein from the purple membrane of Halobacterium halobium. Nature. 1971;233:149–152. doi: 10.1038/newbio233149a0. [DOI] [PubMed] [Google Scholar]
- 402.Oettmeier W, Massin K, Soll M, Reil E. Acridones and quinolones as inhibitors of ubiquinone functions in the mitochondrial respiratory chain. Biochem Soc Trans. 1994;22:213–216. doi: 10.1042/bst0220213. [DOI] [PubMed] [Google Scholar]
- 403.Ogurusu T, Maeda A, Sasaki N, Yoshizawa T. Effects of chloride on the absorption spectrum and photoreactions of halorhodopsin. Biochim Biophys Acta. 1982;682:446–451. [Google Scholar]
- 404.Olendzenski L E, Hilario E, Gogarten J P. Horizontal gene transfer and fusing lines of descent: the archaebacteria—a chimera? In: Syvanen M, Kado C I, editors. Horizontal gene transfer. New York, N.Y: Chapman & Hall; 1998. pp. 63–74. [Google Scholar]
- 405.Olsen G J, Pace N R, Nuele M, Kaine B P, Gupta R, Woese C R. Sequence of the 16S rRNA gene from the thermoacidophilic archaebacterium Sulfolobus solfataricus and its evolutionary implications. J Mol Evol. 1985;22:301–307. doi: 10.1007/BF02115685. [DOI] [PubMed] [Google Scholar]
- 406.Oren A. Anaerobic growth of halophilic archaebacteria by reduction of fumarate. J Gen Microbiol. 1991;137:1387–1390. [Google Scholar]
- 407.Oren A. The ecology of the extremely halophilic archaea. FEMS Microbiol Rev. 1994;13:415–440. doi: 10.1111/j.1574-6941.2002.tb00900.x. [DOI] [PubMed] [Google Scholar]
- 408.Osborne J P, Gennis R B. Sequence analysis of cytochrome bd oxidase suggests a revised topology for subunit I. Biochim Biophys Acta. 1999;1410:32–50. doi: 10.1016/s0005-2728(98)00171-6. [DOI] [PubMed] [Google Scholar]
- 409.Oshima T, Ohba M, Wakagi T. Some biochemical properties of an acido-thermophilic archaebacterium. Origins Life. 1984;14:665–669. [Google Scholar]
- 410.Otomo J. Anion selectivity and pumping mechanism of halorhodopsin. Biophys Chem. 1995;56:137–141. doi: 10.1016/0301-4622(95)00025-s. [DOI] [PubMed] [Google Scholar]
- 411.Otomo J, Tomioka H, Sasabe H. Bacterial rhodopsins of newly isolated halobacteria. J Gen Microbiol. 1992;138:1027–1037. doi: 10.1099/00221287-138-11-2389. [DOI] [PubMed] [Google Scholar]
- 412.Otto H, Marti T, Holz M, Mogi T, Stern L J, Engel F, Khorana H G, Heyn M P. Substitution of amino acids Asp-85, Asp-212, and Arg-82 in bacteriorhodopsin affects the proton release phase of the pump and the pK of the schiff base. Proc Natl Acad Sci USA. 1990;87:1018–1022. doi: 10.1073/pnas.87.3.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Ottolenghi M, Sheves M. Photophysics and photochemistry of retinal proteins. In: Levanon H, editor. Laser Pages Publishing Ltd. 1995. pp. 3–4. Jerusalem, Israel. [Google Scholar]
- 414.Ovchinnikov Y A, Abdulaev N G, Feigina M Y, Kiselev A V, Lobanov N A. The structural basis of the functioning of bacteriorhodopsin: an overview. FEBS Lett. 1979;100:219–224. doi: 10.1016/0014-5793(79)80338-5. [DOI] [PubMed] [Google Scholar]
- 415.Pande C, Lanyi J K, Callender R H. Effects of various anions on the Raman spectrum of halorhodopsin. Biophys J. 1989;55:425–431. doi: 10.1016/S0006-3495(89)82836-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Pebay-Peyroula E, Rummel G, Rosenbusch J P, Landau E M. X-ray structure of bacteriorhodopsin at 2.5 Angstroms from microcrystals grown in lipidic cubic phases. Science. 1997;277:1676–1681. doi: 10.1126/science.277.5332.1676. [DOI] [PubMed] [Google Scholar]
- 417.Pedersen P L, Amzel L M. ATP synthases. Structure, reaction center, mechanism, and regulation of one of nature’s most unique machines. J Biol Chem. 1993;268:9937–9940. [PubMed] [Google Scholar]
- 418.Pedersen P L, Carafoli E. Ion motive ATPases. I. Ubiquity, properties, and significance to cell function. Trends Biochem Sci. 1987;12:146–150. [Google Scholar]
- 419.Pedersen P L, Carafoli E. Ion motive ATPases. II. Energy coupling and work output. Trends Biochem Sci. 1987;12:186–189. [Google Scholar]
- 420.Peer C W, Painter M H, Rasche M E, Ferry J G. Characterization of a CO:heterodisulfide oxidoreductase system from acetate-grown Methanosarcina thermophila. J Bacteriol. 1994;176:6974–6979. doi: 10.1128/jb.176.22.6974-6979.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 421.Penefsky H S, Cross R L. Structure and mechanism of F0F1 type ATP synthase and ATPases. Adv Enzymol. 1991;64:173–214. doi: 10.1002/9780470123102.ch4. [DOI] [PubMed] [Google Scholar]
- 422.Perski H J, Moll J, Thauer R K. Sodium dependence of growth and methane formation in Methanobacterium thermoautotrophicum. Arch Microbiol. 1981;130:319–321. [Google Scholar]
- 423.Perski H J, Schönheit P, Thauer R K. Sodium dependence of methane formation in methanogenic Bacteria. FEBS Lett. 1982;143:323–326. [Google Scholar]
- 424.Petrich J W, Breton J, Martin J L, Antonetti A. Femtosecond absorption spectroscopy of the light-adapted and dark-adapted bacteriorhodopsin. Chem Phys Lett. 1987;137:369–375. [Google Scholar]
- 425.Pfeifer F, Griffig J, Oesterhelt D. The fdx gene encoding the (2Fe-2S)ferredoxin of Halobacterium salinarum (H. halobium) Mol Gen Genet. 1993;239:66–71. doi: 10.1007/BF00281602. [DOI] [PubMed] [Google Scholar]
- 426.Pfitzner U, Odenwald A, Ostermann T, Weingard L, Ludwig B, Richter O M H. Cytochrome c oxidase (heme aa3) from Paracoccus denitrificans: analysis of mutations in putative proton channels of subunit I. J Bioenerg Biomembr. 1998;30:89–97. doi: 10.1023/a:1020515713103. [DOI] [PubMed] [Google Scholar]
- 427.Poirot C M, Kengen S W, Valk E, Keltjens J T, Van der Drift C, Vogels G D. Formation of methylcoenzyme M from formaldehyde by cell free extracts of Methanobacterium thermoautotrophicum. Evidence for the involvement of a corrinoid-containing methyltransferase. FEMS Microbiol Lett. 1987;40:7–13. [Google Scholar]
- 428.Polland H J, Franz M A, Zinth W, Kaiser W, Koelling E, Oesterhelt D. Early picosecond events in the photocycle of bacteriorhodopsin. Biophys J. 1986;49:651–662. doi: 10.1016/S0006-3495(86)83692-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 429.Preisig O, Zufferey R, Thony-Meyer L, Appleby C A, Hennecke H. A high-affinity cbb3-type cytochrome oxidase terminates the symbiosis-specific respiratory chain of Bradyrhizobium japonicum. J Bacteriol. 1996;178:1532–1538. doi: 10.1128/jb.178.6.1532-1538.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 430.Purschke W, Schmidt C L, Petersen A, Anemüller S, Schäfer G. The terminal quinol oxidase of the hyperthermophilic archaeon Desulfurolobus ambivalens exhibits unusual subunit structure and gene organization. J Bacteriol. 1997;179:1344–1353. doi: 10.1128/jb.179.4.1344-1353.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Puustinen A, Bailey J A, Dyer R B, Mecklenburg S L, Wikström M, Woodruff W H. Fourier transform infrared evidence for connectivity between CuB and glutamic acid 286 in cytochrome bo3 from Escherichia coli. Biochemistry. 1997;36:13195–13200. doi: 10.1021/bi971091o. [DOI] [PubMed] [Google Scholar]
- 432.Racker E, Stoeckenius W. Reconstitution of purple membrane vesicles catalyzing light driven proton uptake and adenosine triphosphate formation. J Biol Chem. 1974;249:662–663. [PubMed] [Google Scholar]
- 433.Radax C, Sigurdson O, Hreggvidson G O, Aichinger N, Gruber C, Kristjansson J K, Stan-Lotter H. F- and V-type ATPases in the genus Thermus and related species. Syst Appl Microbiol. 1998;21:12–22. doi: 10.1016/S0723-2020(98)80003-9. [DOI] [PubMed] [Google Scholar]
- 434.Rahlfs, S., and V. Müller. 1999. Personal communication.
- 435.Ramirez C, Köpke A K E, Yang D C, Boeckh T, Matheson A T. The structure, function and evolution of archaeal ribosomes. In: Kates M, Kushner D J, Matheson A T, editors. Biochemistry of Archaea. Amsterdam, The Netherlands: Elsevier; 1993. pp. 439–466. [Google Scholar]
- 436.Rammelsberg R, Huhn G, Lübben M, Gerwert K. Bacteriorhodopsin’s intramolecular proton-release pathway consists of a hydrogen-bonded network. Biochemistry. 1998;37:5001–5009. doi: 10.1021/bi971701k. [DOI] [PubMed] [Google Scholar]
- 437.Rath P, Olson K D, Spudich J L, Rothschild K J. The schiff base counterion of bacteriorhodopsin is protonated in sensory rhodopsin I: spectroscopic and functional characterization of the mutated proteins D76N and D76A. Biochemistry. 1994;33:5600–5606. doi: 10.1021/bi00184a032. [DOI] [PubMed] [Google Scholar]
- 438.Rath P, Spudich E, Neal D D, Spudich J L, Rothschild K J. Asp76 is the schiff base counterion and proton acceptor in the proton-translocating form of sensory rhodopsin I. Biochemistry. 1996;35:6690–6696. doi: 10.1021/bi9600355. [DOI] [PubMed] [Google Scholar]
- 439.Reidlinger J, Müller V. Purification of ATP-synthase from Acetobacterium woodii and identification as a Na+ translocating F1F0-type enzyme. Eur J Biochem. 1994;223:275–283. doi: 10.1111/j.1432-1033.1994.tb18992.x. [DOI] [PubMed] [Google Scholar]
- 440.Richter H T, Brown L S, Needleman R, Lanyi J K. A linkage of the pKa’s of asp-85 and glu-204 forms part of the reprotonation switch of bacteriorhodopsin. Biochemistry. 1996;35:4054–4062. doi: 10.1021/bi952883q. [DOI] [PubMed] [Google Scholar]
- 441.Richter O-M H, Schäfer G. Cloning and sequencing of the gene for the cytoplasmic inorganic pyrophosphatase from the thermoacidophilic archaebacterium Thermoplasma acidophilum. Eur J Biochem. 1992;209:351–355. doi: 10.1111/j.1432-1033.1992.tb17296.x. [DOI] [PubMed] [Google Scholar]
- 442.Richter O-M H, Schäfer G. Purification and enzymic characterization of the cytoplasmic pyrophosphatase from the thermoacidophilic archaebacterium Thermoplasma acidophilum. Eur J Biochem. 1992;209:343–349. doi: 10.1111/j.1432-1033.1992.tb17295.x. [DOI] [PubMed] [Google Scholar]
- 443.Richter O-M H, Tao J, Turba A, Ludwig B. A cytochrome ba3 functions as a quinol oxidase in Paracoccus denitrificans. Purification, cloning, and sequence comparison. J Biol Chem. 1994;269:23079–23086. [PubMed] [Google Scholar]
- 444.Riesle J, Oesterhelt D, Dencher N A, Heberle J. D38 is an essential part of the proton translocation pathway in bacteriorhodopsin. Biochemistry. 1996;35:6635–6643. doi: 10.1021/bi9600456. [DOI] [PubMed] [Google Scholar]
- 445.Riistama S, Hummer G, Puustinen A, Dyer R B, Woodruff W H, Wikström M. Bound water in the proton translocation mechanism of the haem-copper oxidases. FEBS Lett. 1997;414:275–280. doi: 10.1016/s0014-5793(97)01003-x. [DOI] [PubMed] [Google Scholar]
- 446.Rink T, Riesle J, Oesterhelt D, Gerwert K, Steinhoff H J. Spin-labeling studies of the conformational changes in the vicinity of D36, D38, T46, and E161 of bacteriorhodopsin during the photocycle. Biophys J. 1997;72:983–993. doi: 10.1016/S0006-3495(97)78131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446a.Ronk M, Shively J E, Shute E A, Blake R C. Amino acid sequence of the blue copper protein rusticyanin from Thiobacillus ferrooxidans. Biochemistry. 1991;30:9435–9442. doi: 10.1021/bi00103a007. [DOI] [PubMed] [Google Scholar]
- 447.Roth R, Bachofen R. Characterization and partial purification of an ATPase and inorganic pyrophosphatase of the archaebacterium Methanobacterium thermoautotrophicum. Biochim Biophys Acta. 1994;1201:271–283. doi: 10.1016/0304-4165(94)90051-5. [DOI] [PubMed] [Google Scholar]
- 448.Rothschild K J, Bousché O, Braiman M S, Hasselbacher C A, Spudich J L. Fourier transform infrared study of the halorhodopsin chloride pump. Biochemistry. 1988;27:2420–2424. doi: 10.1021/bi00407a026. [DOI] [PubMed] [Google Scholar]
- 449.Rowlands T, Baumann P, Jackson S P. The TATA-binding protein: a general transcription factor in eukaryotes and archaebacteria. Science. 1994;264:1326–1329. doi: 10.1126/science.8191287. [DOI] [PubMed] [Google Scholar]
- 450.Rüdiger M, Haupts U, Gerwert K, Oesterhelt D. Chemical reconstitution of a chloride pump inactivated by a single point mutation. EMBO J. 1995;14:1599–1606. doi: 10.1002/j.1460-2075.1995.tb07148.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 451.Rudolph J, Nordmann B, Storch K F, Gruenberg H, Rodewald K, Oesterhelt D. A family of halobacterial transducer proteins. FEMS Microbiol Lett. 1996;139:161–168. doi: 10.1111/j.1574-6968.1996.tb08197.x. [DOI] [PubMed] [Google Scholar]
- 452.Rudolph J, Oesterhelt D. Chemotaxis and phototaxis require a CheA histidine kinase in the archaeon Halobacterium salinarum. EMBO J. 1995;14:667–673. doi: 10.1002/j.1460-2075.1995.tb07045.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 453.Rudolph J, Oesterhelt D. Deletion analysis of the che operon in the archaeon Halobacterium salinarum. J Mol Biol. 1996;258:548–554. doi: 10.1006/jmbi.1996.0267. [DOI] [PubMed] [Google Scholar]
- 454.Rudolph J, Tolliday N, Schmitt C, Schuster S C, Oesterhelt D. Phosphorylation in halobacterial signal transduction. EMBO J. 1995;14:4249–4257. doi: 10.1002/j.1460-2075.1995.tb00099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 455.Ruppert, C., H. Kavermann, S. Wimmers, R. Schmid, J. Kellermann, F. Lottspeich, H. Huber, K. O. Stetter, and V. Müller. The proteolipid of the A1A0 ATP synthase from Methanococcus jannaschii has six transmembrane helices but only two proton-translocating carboxyl groups. J. Biol. Chem., in press. [DOI] [PubMed]
- 456.Ruppert, C., and V. Müller. 1998. Unpublished data.
- 457.Ruppert C, Wimmers S, Lemker T, Müller V. The A1A0 ATPase from Methanosarcina mazei: cloning of the 5′ end of the aha operon encoding the membrane domain and expression of the proteolipid in a membrane-bound form in Escherichia coli. J Bacteriol. 1998;180:3448–3452. doi: 10.1128/jb.180.13.3448-3452.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 458.Rusnak P, Haney P, Konisky J. The adenylate kinases from a mesophilic and three thermophilic methanogenic members of the Archaea. J Bacteriol. 1995;177:2977–2981. doi: 10.1128/jb.177.11.2977-2981.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 459.Sabbert D, Engelbrecht S, Junge W. Intersubunit rotation in active F-ATPase. Nature. 1996;381:623–625. doi: 10.1038/381623a0. [DOI] [PubMed] [Google Scholar]
- 460.Sabbert D, Engelbrecht S, Junge W. Functional and idling rotatory motion within F1-ATPase. Proc Natl Acad Sci USA. 1997;94:4401–4405. doi: 10.1073/pnas.94.9.4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Sasaki J, Brown L S, Chon Y-S, Kandori H, Maeda A, Needleman R, Lanyi J K. Conversion of bacteriorhodopsin into a chloride ion pump. Science. 1995;269:73–75. doi: 10.1126/science.7604281. [DOI] [PubMed] [Google Scholar]
- 462.Sasaki J, Spudich J L. The transducer protein HtII modulates the lifetimes of sensory rhodopsin II photointermediates. Biophys J. 1998;75:2435–2440. doi: 10.1016/S0006-3495(98)77687-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 463.Schäfer, G. 1996. Personal communication.
- 464.Schäfer G. Bioenergetics of the archaebacterium Sulfolobus. Biochim Biophys Acta. 1996;1277:163–200. doi: 10.1016/s0005-2728(96)00104-1. [DOI] [PubMed] [Google Scholar]
- 465.Schäfer G. How can Archaea cope with extreme acidity? In: Poole R, editor. Mechanisms by which bacteria respond to pH. London, United Kingdom: John Wiley and Sons; 1998. pp. 131–151. [Google Scholar]
- 466.Schäfer G, Anemüller S, Moll R, Gleissner M, Schmidt C L. Has Sulfolobus an archaic respiratory system? Structure, function, and genes of its components. Syst Appl Microbiol. 1994;16:544–555. [Google Scholar]
- 467.Schäfer G, Anemüller S, Moll R, Meyer W, Lübben M. Electron transport and energy conservation in the archaebacterium Sulfolobus acidocaldarius. FEMS Microbiol Rev. 1990;75:335–348. [Google Scholar]
- 468.Schäfer G, Meyering-Vos M. The plasma membrane ATPase of archaebacteria. A chimeric energy converter. Ann N Y Acad Sci. 1992;671:293–309. doi: 10.1111/j.1749-6632.1992.tb43804.x. [DOI] [PubMed] [Google Scholar]
- 469.Schäfer G, Meyering-Vos M. F-type or V-type? The chimeric nature of archaebacterial ATP synthase. Biochim Biophys Acta. 1992;1101:232–235. doi: 10.1016/0005-2728(92)90233-r. [DOI] [PubMed] [Google Scholar]
- 470.Schäfer G, Purschke W, Schmidt C L. On the origin of respiration: electron transport proteins from archaea to man. FEMS Microbiol Rev. 1996;18:173–188. doi: 10.1111/j.1574-6976.1996.tb00235.x. [DOI] [PubMed] [Google Scholar]
- 471.Schäfer G, Purschke W G, Gleissner M, Schmidt C L. Respiratory chains of archaea and extremophiles. Biochim Biophys Acta. 1996;1275:16–20. doi: 10.1016/0005-2728(96)00043-6. [DOI] [PubMed] [Google Scholar]
- 472.Schäfer G, Weber J, Tiedge H, Lübben M. Proton-ATPases: universal catalysts in biological energy transfer. In: Stein W, editor. The ion pumps: structure, function and regulation. New York, N.Y: Alan R. Liss, Inc.; 1988. pp. 57–66. [PubMed] [Google Scholar]
- 473.Scharf B, Engelhard M. Halocyanin, the first example of an archaebacterial blue copper protein from Natronobacterium pharaonis. Biochemistry. 1993;32:12894–12900. doi: 10.1021/bi00210a043. [DOI] [PubMed] [Google Scholar]
- 474.Scharf B, Engelhard M. Blue halorhodopsin from Natronobacterium pharaonis: wavelength regulation by anions. Biochemistry. 1994;33:6387–6393. doi: 10.1021/bi00187a002. [DOI] [PubMed] [Google Scholar]
- 475.Scharf B, Hess B, Engelhard M. Chromophore of sensory rhodopsin II from Halobacterium halobium. Biochemistry. 1992;31:12486–12492. doi: 10.1021/bi00164a027. [DOI] [PubMed] [Google Scholar]
- 476.Scharf B, Pevec B, Hess B, Engelhard M. Biochemical and photochemical properties of the photophobic receptors from Halobacterium halobium and Natronobacterium pharaonis. Eur J Biochem. 1992;206:359–366. doi: 10.1111/j.1432-1033.1992.tb16935.x. [DOI] [PubMed] [Google Scholar]
- 477.Scharf B, Wittenberg R, Engelhard M. Electron transfer proteins from a haloalkaliphilic archaebacterium: main components of the respiratory chain of Natronobacterium pharaonis are cytochrome bc and cytochrome ba3. Biochemistry. 1997;36:4471–4479. doi: 10.1021/bi962312d. [DOI] [PubMed] [Google Scholar]
- 478.Scharf B, Wolff E K. Phototactic behaviour of the archaebacterial Natronobacterium pharaonis. FEBS Lett. 1994;340:114–116. doi: 10.1016/0014-5793(94)80183-5. [DOI] [PubMed] [Google Scholar]
- 479.Scharnagl C, Hettenkofer J, Fischer S F. Proton release pathway in bacteriorhodopsin: molecular dynamics and electrostatic calculations. Int J Quant Chem. 1994;21:33–56. [Google Scholar]
- 480.Schauder R, Kröger A. Bacterial sulphur respiration. Arch Microbiol. 1993;159:491–497. [Google Scholar]
- 481.Scheel E, Schäfer G. Chemiosmotic energy conversion and the membrane ATPase of Methanolobus tindarius. Eur J Biochem. 1990;187:727–735. doi: 10.1111/j.1432-1033.1990.tb15360.x. [DOI] [PubMed] [Google Scholar]
- 482.Schegk E S, Oesterhelt D. Isolation of a prokaryotic photoreceptor: sensory rhodopsin from halobacteria. EMBO J. 1988;7:2925–2933. doi: 10.1002/j.1460-2075.1988.tb03151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 483.Schicho R N, Ma K, Adams M W W, Kelly R M. Bioenergetics of sulfur reduction in the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol. 1993;175:1823–1830. doi: 10.1128/jb.175.6.1823-1830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Schimz A. Methylation of membrane proteins is involved in chemosensory and photosensory behavior of Halobacterium halobium. FEBS Lett. 1981;125:205–207. doi: 10.1016/0014-5793(81)80719-3. [DOI] [PubMed] [Google Scholar]
- 485.Schimz A, Hildebrand E. Chemosensory responses of Halobacterium halobium. J Bacteriol. 1979;140:749–753. doi: 10.1128/jb.140.3.749-753.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 486.Schlegl J, Hardt W-D, Erdmann V A, Hartmann R K. Contribution of structural elements to Thermus thermophilus ribonuclease P RNA function. EMBO J. 1994;13:4863–4869. doi: 10.1002/j.1460-2075.1994.tb06813.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 487.Schmidt C L, Anemüller S, Schäfer G. Two different respiratory Rieske proteins are expressed in the extreme thermoacidophilic crenarchaeon Sulfolobus acidocaldarius: cloning and sequencing of their genes. FEBS Lett. 1996;388:43–46. doi: 10.1016/0014-5793(96)00511-x. [DOI] [PubMed] [Google Scholar]
- 488.Schmidt C L, Anemüller S, Teixeira M, Schäfer G. Purification and characterization of the Rieske iron-sulfur protein from the thermoacidophilic crenarchaeon Sulfolobus acidocaldarius. FEBS Lett. 1995;359:239–243. doi: 10.1016/0014-5793(94)00052-w. [DOI] [PubMed] [Google Scholar]
- 489.Schmidt C L, Hatzfeld O, Link T A, Schäfer G. Expression of the Rieske proteins I and II from the thermoacidophilic crenarchaeon Sulfolobus acidocaldarius in Escherichia coli. Biol Chem Hoppe-Seyler. 1997;378:124. [Google Scholar]
- 490.Schmidt C L, Hatzfeld O M, Petersen A, Link T A, Schäfer G. Expression of the Sulfolobus acidocaldarius Rieske iron sulfur protein II (SoxF) with the correctly inserted [2Fe2S] cluster in Escherichia coli. Biochem Biophys Res Commun. 1997;234:283–287. doi: 10.1006/bbrc.1997.6599. [DOI] [PubMed] [Google Scholar]
- 491.Schmidt, C. L., and G. Schäfer. 1999. Personal communication.
- 491a.Schmidt, C. L., et al. 1997. Personal communication.
- 492.Schmies, G., B. Lüttenberg, I. Chizhov, M. Engelhard, A. Becker, and E. Bamberg. Sensory rhodopsin II from the haloalkaphilic Natronobacterium pharaonis: light activated proton transfer reactions. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 493.Schobert B. F1-like properties of an ATPase from the archaebacterium Halobacterium saccharovorum. J Biol Chem. 1991;266:8008–8014. [PubMed] [Google Scholar]
- 494.Schobert B. The binding of a second divalent metal ion is necessary for the activation of ATP hydrolysis and its inhibition by tightly bound ADP in the ATPase from Halobacterium saccharovorum. J Biol Chem. 1992;267:10252–10257. [PubMed] [Google Scholar]
- 495.Schobert B. The effects of sulfite or nitrate on turnover-dependent inhibition in the ATPase from Halobacterium saccharovorum are related to the binding of the second metal ion. Biochemistry. 1993;32:13204–13211. doi: 10.1021/bi00211a032. [DOI] [PubMed] [Google Scholar]
- 496.Schobert B, Lanyi J K. Halorhodopsin is a light-driven chloride pump. J Biol Chem. 1982;257:10306–10313. [PubMed] [Google Scholar]
- 497.Schobert B, Lanyi J K. Hysteretic behavior of an ATPase from the archaebacterium, Halobacterium saccharovorum. J Biol Chem. 1989;264:12805–12812. [PubMed] [Google Scholar]
- 498.Schobert B, Lanyi J K, Oesterhelt D. Effects of the anion binding on the deprotonation reactions of halorhodopsin. J Biol Chem. 1986;261:2690–2696. [PubMed] [Google Scholar]
- 499.Schönheit P, Schäfer T. Metabolism of hyperthermophiles. World J Microbiol. 1995;11:26–57. doi: 10.1007/BF00339135. [DOI] [PubMed] [Google Scholar]
- 500.Schulenberg P J, Gärtner W, Braslavsky S E. Time-resolved volume changes during the bacteriorhodopsin photocycle: a photothermal beam deflection study. J Phys Chem. 1995;99:9617–9624. [Google Scholar]
- 501.Schulten K, Humphrey W, Logunov I, Sheves M, Xu D. Molecular dynamics studies of bacteriorhodopsin photocycles. Isr J Chem. 1995;35:447–464. [Google Scholar]
- 502.Searcy D G, Whatley F R. Thermoplasma acidophilum: glucose degradative pathways and respiratory activities. Syst Appl Microbiol. 1984;5:30–40. [Google Scholar]
- 503.Seidel R, Scharf B, Gautel M, Kleine K, Oesterhelt D, Engelhard M. The primary structure of sensory rhodopsin II: a member of an additional retinal protein subgroup is coexpressed with its transducer, the halobacterial transducer of rhodopsin II. Proc Natl Acad Sci USA. 1995;92:3036–3040. doi: 10.1073/pnas.92.7.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 504.Selig M, Schönheit P. Oxidation of organic compounds to CO2 with sulfur or thiosulfate as electron acceptor in the anaerobic hyperthermophilic archaea Thermoproteus tenax and Pyrobaculum islandicum proceeds via the citric acid cycle. Arch Microbiol. 1994;162:286–294. [Google Scholar]
- 505.Senior A E. The proton-translocating ATPase of Escherichia coli. Annu Rev Biophys Biophys Chem. 1990;19:7–41. doi: 10.1146/annurev.bb.19.060190.000255. [DOI] [PubMed] [Google Scholar]
- 506.Senior A E. Catalytic sites of Escherichia coli F1-ATPase. J Bioenerg Biomembr. 1992;24:479–483. doi: 10.1007/BF00762365. [DOI] [PubMed] [Google Scholar]
- 507.Serganova I S, Polosina Y Y, Kostyukova A S, Metlina A L, Pyatibratov M G, Fedorov O V. Flagella of halophilic archaea—biochemical and genetic analysis. Biochemistry-Russia. 1995;60:953–957. [PubMed] [Google Scholar]
- 508.Setzke E, Hedderich R, Heiden S, Thauer R K. H2:heterodisulfide oxidoreductase complex from Methanobacterium thermoautotrophicum—composition and properties. Eur J Biochem. 1994;220:139–148. doi: 10.1111/j.1432-1033.1994.tb18608.x. [DOI] [PubMed] [Google Scholar]
- 509.Sharkov A V, Pakulev A V, Chekalin S V, Matveetz Y A. Primary events in bacteriorhodopsin probed by subpicosecond spectroscopy. Biochim Biophys Acta. 1985;808:94–102. [Google Scholar]
- 510.Shibui H, Hamamoto T, Yohda M, Kagawa Y. The stabilizing residues and the functional domains in the hyperthermophilic V-ATPase of Desulfurococcus. Biochem Biophys Res Commun. 1997;234:341–345. doi: 10.1006/bbrc.1997.6644. [DOI] [PubMed] [Google Scholar]
- 511.Siebert F, Maentele W, Kreutz W. Evidence for the protonation of two internal carboxylic groups during the photocycle of bacteriorhodopsin. FEBS Lett. 1982;141:82–87. [Google Scholar]
- 512.Simianu M, Murakami E, Brewer J M, Ragsdale S W. Purification and properties of the heme- and iron-sulfur-containing heterodisulfide reductase from Methanosarcina thermophila. Biochemistry. 1998;37:10027–10039. doi: 10.1021/bi9726483. [DOI] [PubMed] [Google Scholar]
- 513.Skulachev V P. Bioenergetics of extreme halophiles. In: Kates M, Kushner D J, Matheson A T, editors. The biochemistry of Archaea. Amsterdam, The Netherlands: Elsevier; 1993. pp. 25–39. [Google Scholar]
- 514.Sleytr U B, Sára M. Bacterial and archaeal S-layer proteins: structure-function relationships and their biotechnological applications. Trends Biotechnol. 1997;15:20–26. doi: 10.1016/S0167-7799(96)10063-9. [DOI] [PubMed] [Google Scholar]
- 515.Smigan P, Majernik A, Greksak M. Na-driven ATP synthesis in Methanobacterium thermoautotrophicum and its differentiation from H+-driven ATP synthesis by rhodamine 6G. FEBS Lett. 1994;349:424–428. doi: 10.1016/0014-5793(94)00716-0. [DOI] [PubMed] [Google Scholar]
- 516.Smigan P, Majernik A, Polak P, Hapala I, Greksak M. The presence of H+ and Na+-translocating ATPases in Methanobacterium thermoautotrophicum and their possible function under alkaline conditions. FEBS Lett. 1995;371:119–122. doi: 10.1016/0014-5793(95)00866-8. [DOI] [PubMed] [Google Scholar]
- 517.Smith D R, Doucette-Stamm L A, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, Harrison D, Hoang L, Keagle P, Lumm W, Pothier B, Qiu D, Spadafora R, Vicaire R, Wang Y, Wierzbowski J, Gibson R, Jiwani N, Caruso A, Bush D, Safer H, Patwell D, Prabhakar S, McDougall S, Shimer G, Goyal A, Pietrokovski S, Church G M, Daniels C J, Mao J-I, Rice P, Nölling J, Reeve J N. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 518.Song S, Inouye S, Kawai M, Fukami-Kobayashi K, Go M, Nakazawa A. Cloning and characterization of the gene encoding Halobacterium halobium adenylate kinase. Gene. 1996;175:65–70. doi: 10.1016/0378-1119(96)00121-7. [DOI] [PubMed] [Google Scholar]
- 519.Soppa J, Duschl J, Oesterhelt D. Bacterioopsin, haloopsin, and sensory opsin I of the halobacterial isolate Halobacterium sp. strain SG1: three new members of a growing family. J Bacteriol. 1993;175:2720–2726. doi: 10.1128/jb.175.9.2720-2726.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Souvignier G, Gerwert K. Proton uptake mechanism of bacteriorhodopsin as determined by time-resolved stroboscopic-FTIR-spectroscopy. Biophys J. 1992;63:1393–1405. doi: 10.1016/S0006-3495(92)81722-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 521.Spudich E N, Hasselbacher C A, Spudich J L. Methyl-accepting protein associated with bacterial sensory rhodopsin I. J Bacteriol. 1988;170:4280–4285. doi: 10.1128/jb.170.9.4280-4285.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 522.Spudich E N, Spudich J L. The photochemical reactions of sensory rhodopsin I are altered by its transducer. J Biol Chem. 1993;268:16095–16097. [PubMed] [Google Scholar]
- 523.Spudich E N, Takahashi T, Spudich J L. Sensory rhodopsins I and II modulate a methylation/demethylation system in Halobacterium halobium phototaxis. Proc Natl Acad Sci USA. 1989;86:7746–7750. doi: 10.1073/pnas.86.20.7746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 524.Spudich J L. Protein-protein interaction converts a proton pump into a sensory receptor. Cell. 1994;79:747–750. doi: 10.1016/0092-8674(94)90064-7. [DOI] [PubMed] [Google Scholar]
- 525.Spudich J L. Transducer protein HtrI controls proton movements in sensory rhodopsin I. Biophys Chem. 1995;56:165–169. doi: 10.1016/0301-4622(95)00029-w. [DOI] [PubMed] [Google Scholar]
- 526.Spudich J L. Variations on a molecular switch—transport and sensory signalling by archaeal rhodopsins. Mol Microbiol. 1998;28:1051–1058. doi: 10.1046/j.1365-2958.1998.00859.x. [DOI] [PubMed] [Google Scholar]
- 527.Spudich J L, Bogomolni R A. Mechanism of colour discrimination by a bacterial sensory rhodopsin. Nature. 1984;312:509–513. doi: 10.1038/312509a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Spudich J L, Lanyi J K. Shuttling between two protein conformations—the common mechanism for sensory transduction and ion transport. Curr Opin Cell Biol. 1996;8:452–457. doi: 10.1016/s0955-0674(96)80020-2. [DOI] [PubMed] [Google Scholar]
- 529.Spudich J L, Stoeckenius W. Photosensory and chemosensory behaviour of Halobacterium halobium. Photobiochemistry. 1979;1:43–53. [Google Scholar]
- 530.Spudich J L, Zacks D N, Bogomolni R A. Microbial sensory rhodopsins—photochemistry and function. Isr J Chem. 1995;35:495–513. [Google Scholar]
- 531.Sreeramulu K, Schmidt C L, Schäfer G, Anemüller S. Studies on the electron transport chain of the euryarchaeon Halobacterium salinarum: indications for a type-II NADH dehydrogenase and a complex-III analog. J Bioenerg Biomembr. 1998;179:155–160. doi: 10.1023/a:1020538129400. [DOI] [PubMed] [Google Scholar]
- 532.Stan-Lotter H, Hochstein L I. A comparison of an ATPase from the archaebacterium Halobacterium saccharovorum with the F1 moiety from the Escherichia coli ATP synthase. Eur J Biochem. 1989;179:155–160. doi: 10.1111/j.1432-1033.1989.tb14533.x. [DOI] [PubMed] [Google Scholar]
- 533.Stan-Lotter H, Sulzner M, Egelseer E, Norton C F, Hochstein L I. Comparison of membrane ATPases from extreme halophiles isolated from ancient salt deposits. Origins Life Evol Biosph. 1993;23:53–64. doi: 10.1007/BF01581990. [DOI] [PubMed] [Google Scholar]
- 534.Steiner M, Oesterhelt D. Isolation and properties of the native chromoprotein halorhodopsin. EMBO J. 1983;2:1379–1385. doi: 10.1002/j.1460-2075.1983.tb01595.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 535.Steiner M, Oesterhelt D, Ariki M, Lanyi J K. Halide binding by the purified halorhodopsin chromoprotein I: effects on the chromophore. J Biol Chem. 1984;259:2179–2184. [PubMed] [Google Scholar]
- 536.Steinert K, Bickel-Sandkötter S. Isolation, characterization and substrate specificity of the plasma membrane ATPase of the halophilic archaeon Haloferax volcanii. Z Naturforsch. 1996;51c:29–39. doi: 10.1515/znc-1996-1-207. [DOI] [PubMed] [Google Scholar]
- 537.Steinert K, Kroth-Pancic P G, Bickel-Sandkötter S. Nucleotide sequence of the ATPase A- and B-subunits of the halophilic archaebacterium Haloferax volcanii and characterization of the enzyme. Biochim Biophys Acta. 1995;1249:137–144. doi: 10.1016/0167-4838(95)00033-q. [DOI] [PubMed] [Google Scholar]
- 538.Steinert K, Wagner V, Kroth-Pancic P G, Bickel-Sandkötter S. Characterization and subunit structure of the ATP synthase of the halophilic archaeon Haloferax volcanii and organization of the ATP synthase genes. J Biol Chem. 1997;272:6261–6269. doi: 10.1074/jbc.272.10.6261. [DOI] [PubMed] [Google Scholar]
- 539.Steinhoff H J, Mollaaghababa R, Altenbach C, Hideg K, Krebs M, Khorana H G, Hubbell W L. Time-resolved detection of structural changes during the photocycle of spin-labeled bacteriorhodopsin. Science. 1994;266:105–107. doi: 10.1126/science.7939627. [DOI] [PubMed] [Google Scholar]
- 540.Stetter K O. Hyperthermophilic procaryotes. FEMS Microbiol Rev. 1996;18:149–158. [Google Scholar]
- 541.Stock J B, Surette M G, Levit M, Park P. Two-component signal transduction systems: structure-function relationships and mechanisms of catalysis. In: Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: American Society for Microbiology; 1995. pp. 25–51. [Google Scholar]
- 542.Stoeckenius W, Lozier R H. Light energy conversion in Halobacterium halobium. J Supramol Struct. 1974;2:769–774. doi: 10.1002/jss.400020519. [DOI] [PubMed] [Google Scholar]
- 543.Stoeckenius W, Wolff E K, Hess B. A rapid population method for action spectra applied to Halobacterium halobium. J Bacteriol. 1988;170:2790–2795. doi: 10.1128/jb.170.6.2790-2795.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Storch K F, Rudolph J, Oesterhelt D. Car: a cytoplasmic sensor responsible for arginine chemotaxis in the archaeon Halobacterium salinarum. EMBO J. 1999;18:1146–1158. doi: 10.1093/emboj/18.5.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 545.Subramaniam S, Faruqi A R, Oesterhelt D, Henderson R. Electron diffraction studies of light-induced conformational changes in the Leu-93 → Ala bacteriorhodopsin mutant. Proc Natl Acad Sci USA. 1997;94:1767–1772. doi: 10.1073/pnas.94.5.1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.Subramaniam S, Gerstein M, Oesterhelt D, Henderson R. Electron diffraction analysis of structural changes in the photocycle of bacteriorhodopsin. EMBO J. 1993;12:1–8. doi: 10.1002/j.1460-2075.1993.tb05625.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 547.Subramaniam S, Marti T, Khorana G. Protonation state of Asp(Glu)-85 regulates the purple-to-blue transition in bacteriorhodopsin mutants Arg-82 → Ala and Asp-85 → Glu: the blue form is inactive in proton translocation. Proc Natl Acad Sci USA. 1990;87:1013–1017. doi: 10.1073/pnas.87.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 548.Sulzner M, Stan-Lotter H, Hochstein L I. Nucleotide-protectable labeling of sulfhydryl groups in subunit I of the ATPase from Halobacterium saccharovorum. Arch Biochem Biophys. 1992;296:347–349. doi: 10.1016/0003-9861(92)90582-h. [DOI] [PubMed] [Google Scholar]
- 549.Sumi M, Yohda M, Koga Y, Yoshida M. F0F1-ATPase genes from an archaebacterium, Methanosarcina barkeri. Biochem Biophys Res Commun. 1997;241:427–433. doi: 10.1006/bbrc.1997.7809. [DOI] [PubMed] [Google Scholar]
- 550.Swaffield J C, Purugganan M D. The evolution of the conserved ATPase domain (CAD): reconstructing the history of an ancient protein module. J Mol Evol. 1997;45:549–563. doi: 10.1007/pl00006259. [DOI] [PubMed] [Google Scholar]
- 551.Sykes A. Plastocyanin and the blue copper proteins. Struct Bonding. 1990;75:177–224. [Google Scholar]
- 552.Száraz S, Oesterhelt D, Ormos P. pH-induced structural changes in bacteriorhodopsin studied by Fourier transform infrared spectroscopy. Biophys J. 1994;67:1706–1712. doi: 10.1016/S0006-3495(94)80644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Takahashi T, Tomioka H, Kamo N, Kobatake Y. A photosystem other than PS370 also mediates the negative phototaxis of Halobacterium halobium. FEMS Microbiol Lett. 1985;28:161–164. [Google Scholar]
- 554.Takahashi T, Yan B, Mazur P, Derguini F, Nakanishi K, Spudich J L. Color regulation in the archaebacterial phototaxis receptor phoborhodopsin (sensory rhodopsin II) Biochemistry. 1990;29:8467–8474. doi: 10.1021/bi00488a038. [DOI] [PubMed] [Google Scholar]
- 555.Takase K, Kakinuma S, Yamato I, Konishi K, Igarashi K, Kakinuma Y. Sequencing and characterization of the ntp gene cluster for vacuolar-type Na+ translocating ATPase of Enterococcus hirae. J Biol Chem. 1994;269:11037–11044. [PubMed] [Google Scholar]
- 556.Takeda K, Sato H, Hino T, Kono M, Fukuda K, Sakurai I, Okada T, Kouyama T. A novel three-dimensional crystal of bacteriorhodopsin obtained by successive fusion of the vesicular assemblies. J Mol Biol. 1998;283:463–474. doi: 10.1006/jmbi.1998.2103. [DOI] [PubMed] [Google Scholar]
- 557.Tarasov V Y, Kostyukova A S, Tiktopulo E I, Pyatibratov M G, Fedorov O V. Unfolding of tertiary structure of Halobacterium halobium flagellins does not result in flagella destruction. J Protein Chem. 1995;14:27–31. doi: 10.1007/BF01902841. [DOI] [PubMed] [Google Scholar]
- 558.Teixeira, M. 1998. Personal communication.
- 559.Teixeira M, Batista R, Campos A P, Gomes C, Mendes J, Pacheco I, Anemüller S, Hagen W R. A seven-iron ferredoxin from the thermoacidophilic archaeon Desulfurolobus ambivalens. Eur J Biochem. 1995;227:322–327. doi: 10.1111/j.1432-1033.1995.tb20392.x. [DOI] [PubMed] [Google Scholar]
- 560.Terlesky K C, Ferry J G. Ferredoxin requirement for electron transport from the carbon monoxide dehydrogenase complex to a membrane-bound hydrogenase in acetate-grown Methanosarcina thermophila. J Biol Chem. 1988;263:4075–4079. [PubMed] [Google Scholar]
- 561.Thauer R K. Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiology. 1998;144:2377–2406. doi: 10.1099/00221287-144-9-2377. [DOI] [PubMed] [Google Scholar]
- 562.Thauer R K, Jungermann K, Decker K. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Thomas J W, Lemieux L J, Alben J O, Gennis R B. Site-directed mutagenesis of highly conserved residues in helix VIII of subunit I of the cytochrome bo ubiquinol oxidase from Escherichia coli: an amphipathic transmembrane helix that may be important in conveying protons to the binuclear center. Biochemistry. 1993;32:11173–11180. doi: 10.1021/bi00092a029. [DOI] [PubMed] [Google Scholar]
- 564.Thomas J W, Puustinen A, Alben J O, Gennis R B, Wikström M. Substitution of asparagine for aspartate-135 in subunit I of the cytochrome bo ubiquinol oxidase of Escherichia coli eliminates proton-pumping activity. Biochemistry. 1993;32:10923–10928. doi: 10.1021/bi00091a048. [DOI] [PubMed] [Google Scholar]
- 565.Tiedge H, Luensdorf H, Schäfer G, Schairer H U. Subunit stoichiometry and juxtaposition of the photosynthetic coupling factor 1: immunoelectron microscopy using monoclonal antibodies. Proc Natl Acad Sci USA. 1985;82:7874–7878. doi: 10.1073/pnas.82.23.7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 566.Tindall B J. Procaryotic life in the alkaline, saline, athalassic environment. In: Rodriguez-Valera F, editor. Halophilic bacteria. Boca Raton, Fla: CRC Press, Inc.; 1988. pp. 31–67. [Google Scholar]
- 567.Tindall B J. The family halobacteriaceae. In: Balows A, Tüber H G, Dworkin M, Harder W, Schleifer K H, editors. The procaryotes. New York, N.Y: Springer; 1991. pp. 768–808. [Google Scholar]
- 568.Tittor J, Oesterhelt D. The quantum yield of bacteriorhodopsin. FEBS Lett. 1990;263:269–273. [Google Scholar]
- 569.Tittor J, Schweiger U, Oesterhelt D, Bamberg E. Inversion of proton translocation in bacteriorhodopsin mutants D85N, D85T, and D85,96N. Biophys J. 1994;67:1682–1690. doi: 10.1016/S0006-3495(94)80642-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 570.Tittor J, Wahl M, Schweiger U, Oesterhelt D. Specific acceleration of de- and reprotonation steps by azide in mutated bacteriorhodopsins. Biochim Biophys Acta. 1994;1187:191–197. [Google Scholar]
- 571.Tolner B, Poolman B, Konings W N. Adaptation of microorganisms and their transport systems to high temperatures. Comp Biochem Physiol. 1997;118A:423–428. doi: 10.1016/s0300-9629(97)00003-0. [DOI] [PubMed] [Google Scholar]
- 572.Trewhella J, Anderson S, Fox R, Gogol E, Khan S, Engelman D M, Zaccai G. Assignment of segments of the bacteriorhodopsin sequence to positions in the structural map. Biophys J. 1983;42:233–241. doi: 10.1016/S0006-3495(83)84391-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Trincone A, Nicolaus B, Palmieri G, De Rosa M, Huber R, Stetter K O, Gambacorta A. Distribution of complex core lipids within new hyperthermophilic members of the archaea domain. Syst Appl Microbiol. 1992;15:11–17. [Google Scholar]
- 574.Trumpower B L. Cytochrome bc1 complexes of microorganisms. Microbiol Rev. 1990;54:101–129. doi: 10.1128/mr.54.2.101-129.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 575.Trumpower B L. The protonmotive Q-cycle. J Biol Chem. 1990;265:11409–11412. [PubMed] [Google Scholar]
- 576.Trumpower B L, Gennis R B. Energy transduction by cytochrome complexes in mitochondrial and bacterial respiration: the enzymology of coupling electron transfer reactions to transmembrane proton translocation. Annu Rev Biochem. 1994;63:675–716. doi: 10.1146/annurev.bi.63.070194.003331. [DOI] [PubMed] [Google Scholar]
- 577.Tsuda M, Nelson B, Chang C H, Govindjee R, Ebrey T G. Characterization of the chromophore of the third rhodopsin-like pigment of Halobacterium halobium and its photoproduct. Biophys J. 1985;47:721–724. doi: 10.1016/S0006-3495(85)83969-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 578.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Ito K, Ryosuke N, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Angström. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- 579.Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8 Å. Science. 1995;269:1069–1074. doi: 10.1126/science.7652554. [DOI] [PubMed] [Google Scholar]
- 580.Van Alebeek G-J W M, Keltjens J T, Van der Drift C. Purification and characterization of inorganic pyrophosphatase from Methanobacterium thermoautotrophicum (strain DeltaH) Biochim Biophys Acta. 1994;1206:231–239. doi: 10.1016/0167-4838(94)90213-5. [DOI] [PubMed] [Google Scholar]
- 581.Van Raaij M J, Abrahams J P, Leslie A G W, Walker J E. The structure of bovine F1-ATPase complexed with the antibiotic inhibitor aurovertin B. Proc Natl Acad Sci USA. 1996;93:6913–6917. doi: 10.1073/pnas.93.14.6913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 582.Váró G, Brown L S, Needleman R, Lanyi J K. Proton transport by halorhodopsin. Biochemistry. 1996;35:6604–6611. doi: 10.1021/bi9601159. [DOI] [PubMed] [Google Scholar]
- 583.Váró G, Brown L S, Sasaki J, Kandori H, Maeda A, Needleman R, Lanyi J K. Light-driven chloride ion transport by halorhodopsin from Natronobacterium pharaonis. 1. The photochemical cycle. Biochemistry. 1995;34:14490–14499. doi: 10.1021/bi00044a027. [DOI] [PubMed] [Google Scholar]
- 584.Váró G, Zimányi L, Fan X, Sun L, Needleman R, Lanyi J K. Photocycle of halorhodopsin from Halobacterium salinarum. Biophys J. 1995;68:2062–2072. doi: 10.1016/S0006-3495(95)80385-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.Vetriani C, Maeder D L, Tolliday N, Yip K S P, Stillman T J, Britton K L, Rice D W, Klump H H, Robb F T. Protein thermostability above 100°C: a key role for ionic interactions. Proc Natl Acad Sci USA. 1998;95:12300–12305. doi: 10.1073/pnas.95.21.12300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 586.Völkl P, Huber R, Drobner E, Rachel R, Burggraf S, Trincone A, Stetter K O. Pyrobaculum aerophilum sp. nov., a novel nitrate-reducing hyperthermophilic archaeum. Appl Environ Microbiol. 1993;59:2918–2926. doi: 10.1128/aem.59.9.2918-2926.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 587.Vonck J. A three-dimensional difference map of the N intermediate in the bacteriorhodopsin photocycle: part of the F helix tilts in the M to N transition. Biochemistry. 1996;35:5870–5878. doi: 10.1021/bi952663c. [DOI] [PubMed] [Google Scholar]
- 588.Vonrhein C, Bönisch H, Schäfer G, Schulz G E. The structure of a trimeric archaeal adenylate kinase. J Mol Biol. 1998;282:167–179. doi: 10.1006/jmbi.1998.2003. [DOI] [PubMed] [Google Scholar]
- 589.Wächtershäuser G. Before enzymes and templates: theory of surface metabolism. Microbiol Rev. 1988;52:452–484. doi: 10.1128/mr.52.4.452-484.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 590.Wakagi T, Lee C-H, Oshima T. An extremely stable inorganic pyrophosphatase purified from the cytosol of a thermoacidophilic archaebacterium, Sulfolobus acidocaldarius strain 7. Biochim Biophys Acta. 1992;1120:289–296. doi: 10.1016/0167-4838(92)90250-h. [DOI] [PubMed] [Google Scholar]
- 591.Wakagi T, Oshima T. Membrane-bound ATPase of a thermoacidophilic archaebacterium, Sulfolobus acidocaldarius. Biochim Biophys Acta. 1985;817:33–41. doi: 10.1016/0005-2736(85)90065-3. [DOI] [PubMed] [Google Scholar]
- 592.Wakagi T, Oshima T. Membrane-bound ATPase and electron transport system of Sulfolobus acidocaldarius. Syst Appl Microbiol. 1986;7:342–345. [Google Scholar]
- 593.Wakagi T, Oshima T. Energy metabolism of a thermoacidophilic archaebacterium Sulfolobus acidocaldarius. Origins Life. 1987;17:391–399. [Google Scholar]
- 594.Wakagi T, Yamauchi T, Oshima T, Mueller M, Azzi A. A novel a-type terminal oxidase from Sulfolobus acidocaldarius with cytochrome c oxidase activity. Biochem Biophys Res Commun. 1989;165:1110–1114. doi: 10.1016/0006-291x(89)92717-4. [DOI] [PubMed] [Google Scholar]
- 595.Wakao H, Wakagi T, Oshima T. Purification and properties of NADH dehydrogenase from a thermoacidophilic archaebacterium, Sulfolobus acidocaldarius. J Biochem (Japan) 1987;102:255–262. doi: 10.1093/oxfordjournals.jbchem.a122049. [DOI] [PubMed] [Google Scholar]
- 596.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related gene sequences in the a- and b-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 597.Walter T J, Braiman M S. Anion-protein interactions during halorhodopsin pumping: halide binding at the protonated schiff base. Biochemistry. 1994;33:1724–1733. doi: 10.1021/bi00173a015. [DOI] [PubMed] [Google Scholar]
- 598.Weber J, Senior A E. Catalytic mechanism of F1-ATPase. Biochim Biophys Acta. 1997;1319:19–58. doi: 10.1016/s0005-2728(96)00121-1. [DOI] [PubMed] [Google Scholar]
- 599.Weiss D S, Gärtner P, Thauer R K. The energetics and sodium-ion dependence of N5-methyltetrahydromethanopterin:coenzyme M methyltransferase studied with cob(I)alamin as methyl acceptor and methylcob(III)alamin as methyl donor. Eur J Biochem. 1994;226:799–809. doi: 10.1111/j.1432-1033.1994.00799.x. [DOI] [PubMed] [Google Scholar]
- 600.Westenberg D, Braune A, Ruppert C, Müller V, Herzberg C, Gottschalk G, Blaut M. The F420 dehydrogenase from Methanolobus tindarius: cloning of the ffd operon and expression of the genes in E. coli. FEMS Microbiol Lett. 1999;170:389–398. doi: 10.1111/j.1574-6968.1999.tb13399.x. [DOI] [PubMed] [Google Scholar]
- 601.Wilkens S, Capaldi R A. Electron microscopic evidence of two stalks linking the F1 and F0 parts of the E. coli ATP synthase. Biochim Biophys Acta. 1998;1365:93–97. doi: 10.1016/s0005-2728(98)00048-6. [DOI] [PubMed] [Google Scholar]
- 602.Wilms R, Freiberg C, Wegerle E, Meier I, Mayer F, Müller V. Subunit structure and organization of the genes of the A1A0 ATPase from the archaeon Methanosarcina mazei Go1. J Biol Chem. 1996;271:18843–18852. doi: 10.1074/jbc.271.31.18843. [DOI] [PubMed] [Google Scholar]
- 603.Winner C, Gottschalk G. H2 and CO2 production from methanol and formaldehyde by the methanogenic bacterium strain GÖ1 treated with 2-bromoethanesulfonic acid. FEMS Microbiol Lett. 1989;65:259–264. [Google Scholar]
- 604.Wittenberg R. Charakterisierung der Elektronentransportkette und Untersuchungen zur Bioenergetik in Natronobacterium pharaonis. Ph.D. thesis. Bochum, Germany: Ruhr-Universität Bochum; 1995. [Google Scholar]
- 605.Woese C. The universal ancestor. Proc Natl Acad Sci USA. 1998;95:6854–6859. doi: 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606.Woese C R. p. vvii–xxix. In M. Kates, D. J. Kushner, and A. T. Matheson (ed.), The biochemistry of Archaea. Elsevier, Amsterdam, The Netherlands. 1993. The Archaea: their history and significance. [Google Scholar]
- 607.Woese C R, Fox G E. Phylogenetic structure of the procaryotic domain: the primary kingdoms. Proc Natl Acad Sci USA. 1977;74:5088–5090. doi: 10.1073/pnas.74.11.5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 608.Woese C R, Kandler O, Wheelis M L. Towards a natural system of organisms: proposal for the domains archaea, bacteria, and eucarya. Proc Natl Acad Sci USA. 1990;87:4576–4579. doi: 10.1073/pnas.87.12.4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 609.Wood H G, Goss N H. Phosphorylation enzymes of the propionic acid bacteria and the roles of ATP, inorganic pyrophosphate, and polyphosphates. Proc Natl Acad Sci USA. 1985;82:312–315. doi: 10.1073/pnas.82.2.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.Xia D, Yu C A, Kim H, Xian J Z, Kachurin A M, Zhang L, Yu L, Deisenhofer J. Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science. 1997;277:60–66. doi: 10.1126/science.277.5322.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 611.Xu D, Martin C, Schulten K. Molecular dynamics study of early picosecond events in the bacteriorhodopsin photocycle: dielectric response, vibrational cooling and the J, K intermediates. Biophys J. 1996;70:453–460. doi: 10.1016/S0006-3495(96)79588-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 612.Xu D, Sheves M, Schulten K. Molecular dynamics study of the M412 intermediate of bacteriorhodopsin. Biophys J. 1995;69:2745–2760. doi: 10.1016/S0006-3495(95)80146-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 613.Yamazaki Y, Hatanaka M, Kandori H, Sasaki J, Karstens W F J, Raap J, Lugtenburg J, Bizounok M, Herzfeld J, Needleman R, Lanyi J K, Maeda A. Water structural changes at the proton uptake site (the Thr46-Asp96 domain) in the L intermediate of bacteriorhodopsin. Biochemistry. 1995;34:7088–7093. doi: 10.1021/bi00021a021. [DOI] [PubMed] [Google Scholar]
- 614.Yamazaki Y, Tuzi S, Saitô H, Kandori H, Needleman R, Lanyi J K, Maeda A. Hydrogen bonds of water and C ⩵ O groups coordinate long-range structural interactions in the L photointermediate of bacteriorhodopsin. Biochemistry. 1996;35:4063–4068. doi: 10.1021/bi9524530. [DOI] [PubMed] [Google Scholar]
- 615.Yan B, Spudich J L. Evidence that the repellent receptor form of sensory rhodopsin I is an attractant signaling state. Photochem Photobiol. 1991;54:1023–1026. doi: 10.1111/j.1751-1097.1991.tb02125.x. [DOI] [PubMed] [Google Scholar]
- 616.Yan B, Takahashi T, Johnson R, Derguini F, Nakanishi K, Spudich J L. All-trans/13-cis isomerization of retinal is required for phototaxis signaling by sensory rhodopsins in Halobacterium halobium. Biophys J. 1990;57:807–814. doi: 10.1016/S0006-3495(90)82600-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 617.Yan B, Takahashi T, Johnson R, Spudich J L. Identification of signaling states of a sensory receptor by modulation of lifetimes of stimulus-induced conformations: the case of sensory rhodopsin II. Biochemistry. 1991;30:10686–10692. doi: 10.1021/bi00108a012. [DOI] [PubMed] [Google Scholar]
- 618.Yao V J, Spudich E N, Spudich J L. Identification of distinct domains for signaling and receptor interaction of the sensory rhodopsin I transducer, HtrI. J Bacteriol. 1994;176:6931–6935. doi: 10.1128/jb.176.22.6931-6935.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 619.Yao V J, Spudich J L. Primary structure of an archaebacterial transducer, a methyl-accepting protein associated with sensory rhodopsin I. Proc Natl Acad Sci USA. 1992;89:11915–11919. doi: 10.1073/pnas.89.24.11915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 620.Yokoyama K, Oshima T, Yoshida M. Thermus thermophilus membrane-associated ATPase: indication of a eubacterial v-type ATPase. J Biol Chem. 1990;265:21946–21950. [PubMed] [Google Scholar]
- 621.Zhang Q, Iwasaki T, Wakagi T, Oshima T. 2-Oxoacid:ferredoxin oxidoreductase from the thermoacidophilic archaeon, Sulfolobus sp. strain 7. J Biochem (Tokyo) 1996;120:587–599. doi: 10.1093/oxfordjournals.jbchem.a021454. [DOI] [PubMed] [Google Scholar]
- 622.Zhang W S, Brooun A, McCandless J, Banda P, Alam M. Signal transduction in the archaeon Halobacterium salinarum is processed through three subfamilies of 13 soluble and membrane-bound transducer proteins. Proc Natl Acad Sci USA. 1996;93:4649–4654. doi: 10.1073/pnas.93.10.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 623.Zhang W S, Brooun A, Mueller M M, Alam M. The primary structures of the archaeon Halobacterium salinarum blue light receptor sensory rhodopsin II and its transducer, a methyl-accepting protein. Proc Natl Acad Sci USA. 1996;93:8230–8235. doi: 10.1073/pnas.93.16.8230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 624.Zhang X N, Spudich J L. HtrI is a dimer whose interface is sensitive to receptor photoactivation and His-166 replacements in sensory rhodopsin I. J Biol Chem. 1998;273:19722–19728. doi: 10.1074/jbc.273.31.19722. [DOI] [PubMed] [Google Scholar]
- 625.Zhang Z, Huang L, Shulmeister V M, Chi Y, Kim K K, Hung L, Crofts A R, Berry E A, Kim S. Electron transfer by domain movement in cytochrome bc1. Nature. 1998;392:677–684. doi: 10.1038/33612. [DOI] [PubMed] [Google Scholar]
- 626.Zillig W, Palm P, Klenk H-P, Langer D, Hüdepohl U, Hain J, Lanzendörfer M, Holz I. Transcription in Archaea. In: Kates M, Kushner D J, Matheson A T, editors. The biochemistry of Archaea. Amsterdam, The Netherlands: Elsevier; 1993. pp. 367–392. [Google Scholar]
- 627.Zimanyi L, Keszthelyi L, Lanyi J K. Transient spectroscopy of bacterial rhodopsins with an optical multichannel analyzer. 1. Comparison of the photocycles of bacteriorhodopsin and halorhodopsin. Biochemistry. 1989;28:5165–5172. doi: 10.1021/bi00438a038. [DOI] [PubMed] [Google Scholar]
- 628.Zimanyi L, Lanyi J K. Fourier transform Raman study of retinal isomeric composition and equilibration in halorhodopsin. J Phys Chem B. 1997;101:1930–1933. [Google Scholar]