Abstract
Gliding motility is observed in a large variety of phylogenetically unrelated bacteria. Gliding provides a means for microbes to travel in environments with a low water content, such as might be found in biofilms, microbial mats, and soil. Gliding is defined as the movement of a cell on a surface in the direction of the long axis of the cell. Because this definition is operational and not mechanistic, the underlying molecular motor(s) may be quite different in diverse microbes. In fact, studies on the gliding bacterium Myxococcus xanthus suggest that two independent gliding machineries, encoded by two multigene systems, operate in this microorganism. One machinery, which allows individual cells to glide on a surface, independent of whether the cells are moving alone or in groups, requires the function of the genes of the A-motility system. More than 37 A-motility genes are known to be required for this form of movement. Depending on an additional phenotype, these genes are divided into two subclasses, the agl and cgl genes. Videomicroscopic studies on gliding movement, as well as ultrastructural observations of two myxobacteria, suggest that the A-system motor may consist of multiple single motor elements that are arrayed along the entire cell body. Each motor element is proposed to be localized to the periplasmic space and to be anchored to the peptidoglycan layer. The force to glide which may be generated here is coupled to adhesion sites that move freely in the outer membrane. These adhesion sites provide a specific contact with the substratum. Based on single-cell observations, similar models have been proposed to operate in the unrelated gliding bacteria Flavobacterium johnsoniae (formerly Cytophaga johnsonae), Cytophaga strain U67, and Flexibacter polymorphus (a filamentous glider). Although this model has not been verified experimentally, M. xanthus seems to be the ideal organism with which to test it, given the genetic tools available. The second gliding motor in M. xanthus controls cell movement in groups (S-motility system). It is dependent on functional type IV pili and is operative only when cells are in close proximity to each other. Type IV pili are known to be involved in another mode of bacterial surface translocation, called twitching motility. S-motility may well represent a variation of twitching motility in M. xanthus. However, twitching differs from gliding since it involves cell movements that are jerky and abrupt and that lack the organization and smoothness observed in gliding. Components of this motor are encoded by genes of the S-system, which appear to be homologs of genes involved in the biosynthesis, assembly, and function of type IV pili in Pseudomonas aeruginosa and Neisseria gonorrhoeae. How type IV pili generate force in S-motility is currently unknown, but it is to be expected that ongoing physiological, genetic, and biochemical studies in M. xanthus, in conjunction with studies on twitching in P. aeruginosa and N. gonorrhoeae, will provide important insights into this microbial motor. The two motility systems of M. xanthus are affected to different degrees by the MglA protein, which shows similarity to a small GTPase. Bacterial chemotaxis-like sensory transduction systems control gliding motility in M. xanthus. The frz genes appear to regulate gliding movement of individual cells and movement by the S-motility system, suggesting that the two motors found in this bacterium can be regulated to result in coordinated multicellular movements. In contrast, the dif genes affect only S-system-dependent swarming.
Motility is arguably the most impressive feature of a microbe’s physiology. Active movement ultimately uncovered microbes as living organisms to the first microscopic inspection by Antonie van Leeuwenhoek more than 300 years ago (31) and since this time has attracted the curiosity of innumerable scientists. In general, motile prokaryotic microorganisms move in aqueous environments by swimming or by control of buoyancy or along surfaces by using distinct modes of surface translocation. Most research has focused on understanding swimming motility in prokaryotes. Fundamental insights have been gained from thorough studies of the molecular architecture of the flagellum, from the energy transduction mechanism and mode of force generation, and from the control of motility, all of which serve now as paradigms for motility in biology (for a review, see reference 14). Notably, as shown by Waterbury et al. for a cyanobacterium (128), not all swimming bacteria are flagellated. Swimming motility is advantageous only for microorganisms living in aqueous habitats. Many microbes, however, live in environments with a low water content or changing humidity. These environments include biofilms, microbial mats, and soil, where the exploration of a new food source by means of swimming motility is unfeasible. These organisms are faced with the challenge of how to move on surfaces that are covered with only a few layers of water molecules. One solution of some swimming bacteria is to produce excessive lateral flagella that enable them to swarm in a thin fluid layer on a solid surface (for a review, see reference 46). However, many other prokaryotes employ one of the two modes of active cell translocation on a solid surface: gliding or twitching.
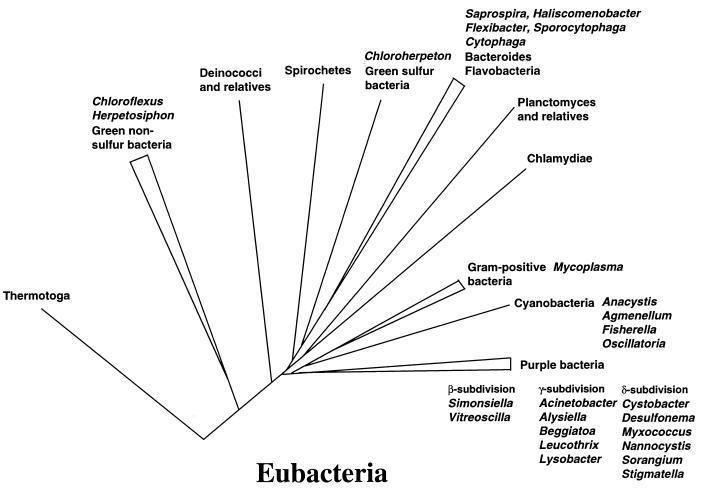
Historically, gliding is defined as the movement of a nonflagellated cell in the direction of its long axis on a surface (51). This rather broad definition, which does not specify a molecular apparatus or a mode of force generation, has therefore been used to describe movements by many phylogenetically unrelated bacteria (Fig. 1). As will be shown in this review, gliding movement can be generated by fundamentally different molecular mechanisms that can operate simultaneously even in a single microorganism. Consequently, use of the term “gliding” should not be considered to imply the operation of a specific motility mechanism. Several models for gliding have been proposed for different organisms to explain the seemingly smooth advancement of cells on solid surfaces. These models include operation of contractile elements (22), directional propagation of waves along the cell surface (60), directional extrusion of slime (59, 99), rotary motors (91), controlled release of surfactants from poles of cells (34, 67), and movement of adhesion sites in the outer membrane along tracks fixed to the peptidoglycan of the cell wall (72). In most cases, the models were postulated as a result of observations on motility behavior of single cells. However, to date, no model has been verified by biochemical, molecular, and ultrastructural studies, and it seems unlikely that all gliding bacteria harbor the same motility mechanism. The focus of this review is on recent research which seeks to provide a mechanistic and molecular understanding for gliding. The single-cell gliding bacteria Myxococcus xanthus, Flavobacterium, and Cytophaga strain U67, as well as the filamentous organism Flexibacter polymorphus, serve as model organisms in these studies. Interestingly, gliding motility has so far not been reported for archaea. Diversity and ecological aspects of gliding motility have been reviewed recently (95, 96).
FIG. 1.
Occurrence of gliding bacteria among the eubacteria. The figure depicts examples of bacteria (indicated in italics) that have been reported to move by “gliding.” Note that “gliding” is an operational definition, and not all bacteria may glide by the same mechanism. No gliding archaea have been reported. Modified from reference 58.
GLIDING MOTILITY IN MYXOCOCCUS XANTHUS
Most of the research on gliding motility has been conducted with M. xanthus (50, 127, 143, 145). This microorganism is a Gram-negative soil bacterium which belongs to the delta subdivision of proteobacteria and is specialized to mineralize insoluble organic matter, specifically proteins (33, 95, 104). In common with other myxobacteria, M. xanthus exhibits an unusually complex life cycle during which gliding motility plays a crucial role (110). Under vegetative conditions, cells move as coordinated swarms. These swarms may contain thousands of cells, which secrete hydrolytic enzymes into their environment that lyse other cells and convert insoluble proteins into soluble, transportable amino acids. Metabolism of these amino acids and other compounds is strictly aerobic. The feeding strategy of moving in swarms on the metabolizable substrate has been termed the “wolf pack” effect to reflect the fact that large numbers of organized cells undoubtedly utilize insoluble nutrients more efficiently than a single cell (104). Furthermore, coordination of vegetative cells in swarms provides the basis for survival when nutrients become limiting. When cells are starved of nutrients, tens of thousands aggregate to form a fruiting body, within which cells differentiate into spores. The organized movement of vegetative cells ensures that sufficient cells are present to form this fruiting body. Subsequent dispersal of mature, spore-containing fruiting bodies guarantees that after spore germination, cells are present at a sufficiently high density to allow the formation of new vegetative swarms. It is therefore apparent that the motility of M. xanthus is a crucial prerequisite for this lifestyle and thus has attracted much research effort.
Over the few past decades, research has focused on genetic and molecular approaches, as well as on high-resolution motion analyses, to develop a cellular and mechanistic understanding of gliding motility in M. xanthus. Hodgkin and Kaiser initiated a genetic analysis by screening chemical- and UV-induced mutants for visible defects in colony swarming (56, 57). Subsequent analysis of these mutants revealed that motility in M. xanthus is controlled by two multigene systems: the A (adventurous) system, which controls gliding motility of individual, isolated cells, and the S (social) motility system, which is essential for cell movement in swarms and during aggregation and fruiting-body formation (56, 57). Both motility systems are required for wild-type swarming, because swarming is completely abolished in any A−S− double mutant. It should be noted that although gliding of isolated cells is observed only in A+ cells, cells in swarms can move by either A motility or S motility or by both systems at a time. The use of single-cell observations to infer involvement of a specific type of motility system can be misleading, and in a strict sense, A motility and S motility are defined only by the macroscopically visible nonswarming phenotype of a double-mutant colony (56).
As a result of significant progress in recent years, a refined picture of how M. xanthus moves is becoming apparent. Experimental evidence that is reviewed below suggests that A and S motility do not represent different modes of a single gliding mechanism but, instead, comprise two distinct motility mechanisms: pilus-independent single-cell gliding (A motility) and pilus-dependent movements (S motility), which may be related to another surface translocation mechanism, called twitching. Twitching is described as intermittent, jerky cell movement that seems to lack the degree of organization seen in gliding. Twitching, unlike S motility, can occur in directions other than the long axis of the cell (53). However, functional type IV pili are essential for both twitching motility and S motility. Genes of the S system have recently been shown to include genes necessary for the synthesis, processing, export, assembly, and function of type IV pili (102, 125, 139–141).
Gliding of M. xanthus Cells as Single Cells or in Swarms
In this section, observations on the movement of wild-type swarms and on isolated single cells are summarized. Colonies of wild-type M. xanthus (DK1622, DZ2) expand as flat swarms on a 1.5% agar surface (Fig. 2A). Microscopy at the edges of these swarms shows that the advancing front of the swarm is composed predominantly of individual cells, with groups of cells forming behind (56) (Fig. 2A). The rate of swarm expansion depends on cell density in a type of first-order kinetics with a maximal rate of approximately 1.6 μm/min and a half-saturation cell density of approximately 2 × 108 cells per ml (62). The swarming rate also depends on the concentration of the agar support (57). When the rate is measured as the expansion of a colony area, swarming appears to be faster on 0.4% agar and drops to about 1/10 the efficiency when the agar concentration increases to 2% (107). M. xanthus colonies also respond to stress forces in the (agar) surface, a phenomenon called elasticotaxis (37, 119). Small compressions in the agar surface lead M. xanthus colonies to swarm preferentially in the direction perpendicular to the stress force, which results in an elongated elliptical colony (37).
FIG. 2.
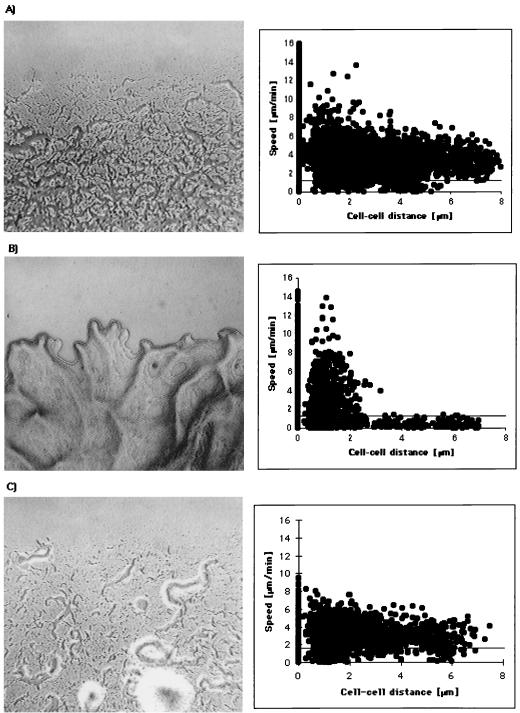
Two gliding motility systems are operative in M. xanthus. (Left) Colony morphology. (Right) Gliding speed plotted against cell-cell distance of individual cells (the detection limit for active movement was 1 μm/min [117]). (A) Wild-type DK1622 (A+S+). Gliding-speed data are from reference 117. (B) JZ315 (cglB, A−S+). Gliding-speed data are from reference 101. Note that when the cells were separated by more than 2 μm, no active single-cell movement was observed. When the cells were in contact, the velocities observed were similar to those found when wild-type cells moved in close proximity (A). (C) DK3473 (pilR, A+S−); cell movement of 50 individual DK3473 cells was examined as described previously (117). A total of 4,531 speed and cell-cell distance values were obtained and plotted. The speed of isolated cells was similar to that of wild-type cells. However, cells did not exhibit high-speed movements, as observed for wild-type and cglB cells (A and B) when in close proximity.
Videomicroscopy motion analyses have been used to determine the rates at which individual isolated cells glide on a 1.5% agar surface. The studies showed that the translocation velocity is highly variable for a given cell but also varies between different cells (117). Most cells were found to move at velocities between 1.5 and 6 μm/min, although occasionally speeds of up to 20 μm/min were observed (117). Cells change the direction of movement by reversing every 5 to 7 min, so that the leading end becomes the lagging end (12, 118). Bending of the highly flexible cell body during movement frequently results in deviations from the original track of less than 90°, thus also contributing to directional changes. Individual M. xanthus cells do not have a preferred direction of movement; i.e., a cell does not have a “head or tail” (106, 118). This is indicated by measurements of cellular gliding velocity taken in both the forward and backward directions, as well as by studies that timed the duration of movement in one or the other direction. Gliding speed is also independent of cell length. When M. xanthus cells were treated with cephalexin, which increases the cell length to up to 20 μm (compared to 6 μm untreated), no difference in gliding speed compared to untreated cells was noticeable (118a), suggesting that the gliding motor(s) operates at constant speed. Interestingly, the translocation velocity of individual cells varied with cell-cell distance (117) (Fig. 2A). When individual cells were separated by more than one cell diameter (0.5 μm) from the nearest neighbor, they moved with an average velocity of 3.8 μm/min. However, in close proximity, cells moved with an average velocity of 5.0 μm/min (117). These kinetically distinguishable modes of single-cell gliding can be separated genetically into A- and S-motility-related movements, respectively (Fig. 2B and C).
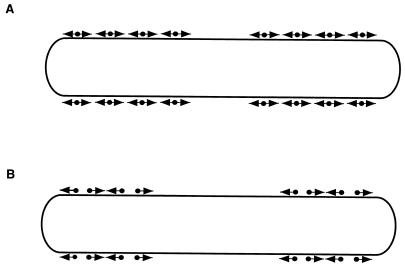
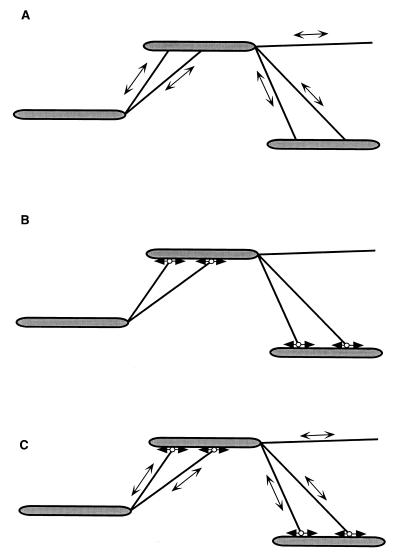
On rare occasions, individual wild-type cells have been observed to glide while one cell pole is fortuitously fixed on an agar surface. This event results in flexing of the cell as the end which is not fixed glides backward and forward (118). Long cells have also been observed to bend into a “U” and to move in one direction in this configuration. Because the cell poles would move away from each other if the cell was not a bent “U,” this observation suggests that each cell pole can move independently in both directions. In addition, the region between the poles can exhibit movement, indicating that the subcellular elements that promote motility are localized at both cell poles and along the length of the cell body (117) (Fig. 3). Therefore, M. xanthus cells are proposed to contain not one single gliding motor which is localized to a specific site but, instead, multiple motor elements positioned along the entire cell surface (117) (Fig. 3). The activities of these multiple motor elements would normally be coordinated to generate overall movement in either the forward or the reverse direction. Currently, no data are available to differentiate between the possibility that cells (i) contain a single set of motor elements where each element is capable of operating in both directions or (ii) contain two sets of unidirectional motors that are arranged opposite to each other and differentially regulated (Fig. 3).
FIG. 3.
Models for localization of motility elements on the surface of M. xanthus cells. Multiple motor units are located along the cell body. (A) In this model, a single motor unit can generate displacement in two directions that are opposite to each other (◂ • ▸). (B) In this model, two types of unidirectional motors exist (• ▸ and ◂ •), each capable of generating force in only one direction. The motor elements are arranged opposite each other, so that selected activation of one or the other results in movement in one or the other direction.
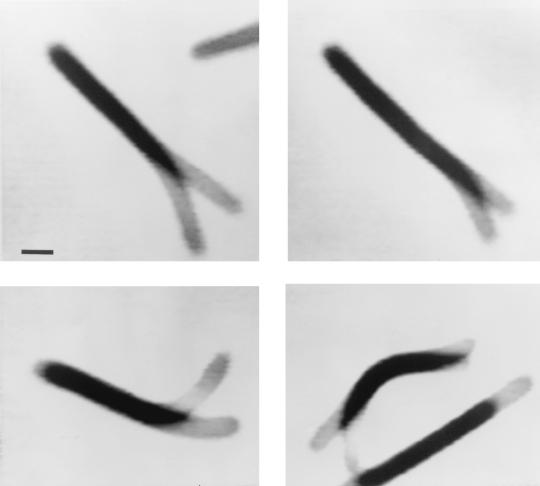
Under certain conditions, M. xanthus cells have also been found to bend in the absence of any translocation of the cell body. Such bending was observed when dsp cells (see below) attached to nitrocellulose-coated coverslips (Fig. 4). While the majority of the cell body did not show any displacement, the ends of individual cells were found to flex until a certain degree of bending was achieved, when the end would “snap back” to form a straight cell. The flexing proceeded at a rate similar to that of gliding. These observations suggest that force can be generated relative to distal body sections in the absence of translocation of the entire cell.
FIG. 4.
Flexing of an M. xanthus cell. Cells of DK1680 (dsp) were grown in CTT liquid medium (117) to a density of about Klett 100 (∼5 × 108 cells/ml), and 10-μl drops were placed on nitrocellulose-coated coverslips. Cell movement was monitored under an inverted microscope, and images were recorded during the observation period. In the absence of net movement, cells were observed to flex. This can be easily shown when superimposing two images that were recorded at different times. Dark areas indicate overlap of the cell during both recordings, and lighter areas show displacement of cell sections. The time difference between the two superimposed images was 1 min 48 s for the picture at top left (size bar included), 1 min 10 s for the picture at bottom left, 7 s for the picture at top right, and 27 s for the picture at bottom right. In the last picture, the lower cell was gliding in direction of the long axis of the cell while the upper cell flexed. Bar, 0.5 μm.
In contrast to other gliding bacteria, such as Cytophaga, M. xanthus cells have not been reported to rotate during translocation. However, cells have been observed to pivot around one cell pole which is tethered to a surface in a wet mount. Similar to studies with other gliding bacteria (72, 91), movement of beads along the surface of a cell has been examined in M. xanthus (118a). It appears that the direction of a moving bead does not always correlate with the direction of cell gliding. While a cell is moving forward, beads may be moving forward, backward, or not at all. Bead movement was also observed on cells which were not moving. Two beads on the surface of a single cell could be found to move in opposite directions. These observations are very similar to those made previously by Lapidus and Berg with Cytophaga strain U67 (72). Beads frequently seem to become “trapped” at cell poles. The velocity of bead movement is on the order of 2 to 4 μm/min (118a), which is comparable to the velocity of individual gliding cells, suggesting that bead movement along the cell surface may be related to gliding movement (72).
No study that identifies a source of energy for gliding movement in M. xanthus has been reported. The electrochemical membrane potential was proposed to power gliding motility in Flexibacter (see below). Because two gliding mechanisms operate in M. xanthus, each mechanism may utilize a different energy source (e.g., ATP versus electrochemical ion potential), thus complicating bioenergetic studies.
Subcellular Structures with Proposed Roles in Motility
Several subcellular structures in M. xanthus are believed to be involved in promoting cell movement: (i) periodic chain-like structures associated with the outer membrane (38, 75) and (ii) polar type IV pili (61, 79).
Chain-like structures.
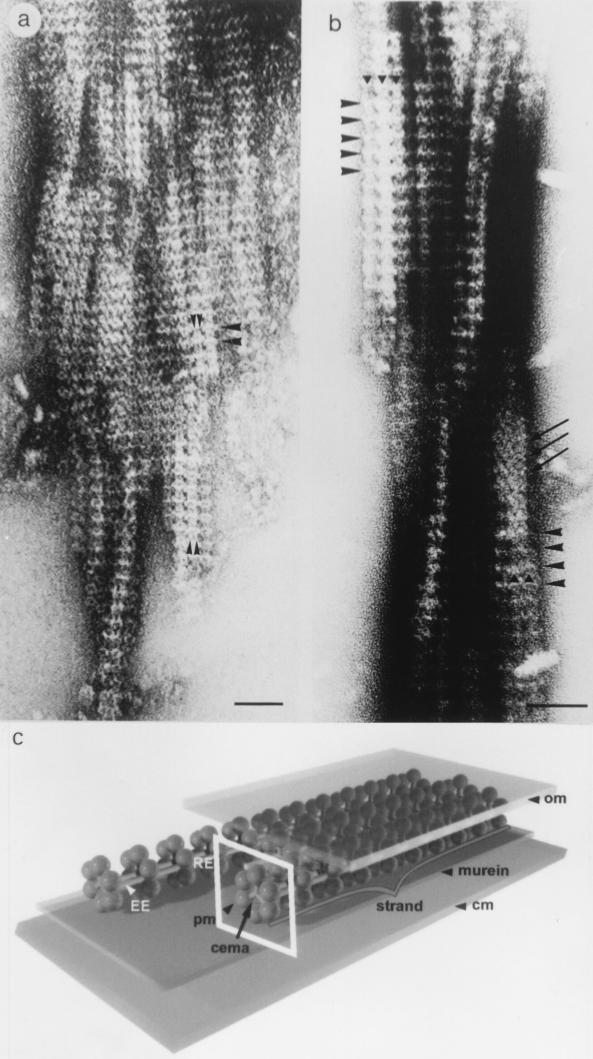
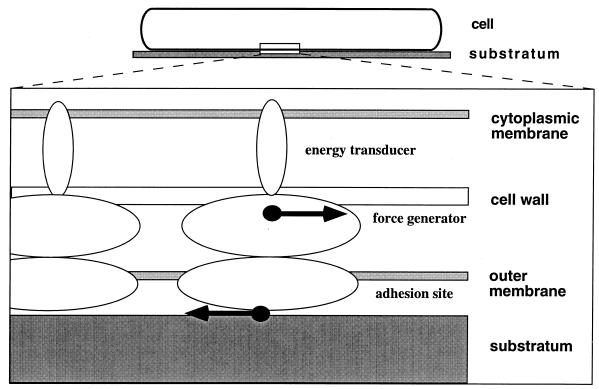
Reichenbach and coworkers conducted detailed electron microscopy studies with whole cells and subcellular fractions of the gliding myxobacteria Myxococcus fulvus and M. xanthus (strains Mx f65-9 and DK1622). In both organisms, similar structures that are believed to be components of the gliding apparatus were identified (Fig. 5) (38, 75). The basic elements of this proposed gliding apparatus in M. xanthus DK1622 are linear chain-like strands. These strands are composed of multiple ring-like structures, which are threaded evenly along elongated elements (Fig. 5). Each ring element in a strand has an average outer diameter of about 16.4 nm and is composed of six or more peripheral protein masses and possibly three small central masses. The rings are connected to each other by two parallel strings of filamentous proteins, the elongated elements, which attach at the inner side of a ring. They separate two neighboring rings evenly at a distance of about 15.5 nm. Often, three chain-like strands appear to assemble into parallel, ribbon-like structures where rings of neighboring strands are in lateral contact. Several ribbons form a belt which wraps helically around the cell. The cells appear to be completely covered by these belts. The strand structures are located in the periplasmic space and are associated with the outer membrane. A significant amount of strand-like structures was released only after lysozyme treatment, which suggests that the strands are also associated with the thin peptidoglycan layer. It is hypothesized that the connection between a ring element and the elongated element may be flexible and may be the site of force generation during gliding (Fig. 5) (38, 75).
FIG. 5.
Chain-like structures proposed to be involved in gliding motility. The images are from Myxococcus fulvus, and similar structures were observed in M. xanthus. (A) Isolated aggregates of chain-like strands showing different orientations of periodic structural elements. Arrowheads indicate rings. Bar, 50 nm. (B) Regular spacing and positioning of rings normal to the longitudinal axis of the strands (large arrowheads). Three strands form a morphological unit, the ribbon (solid triangles). Long arrows indicate proposed contracted units resulting in a herringbone pattern. Bar, 50 nm. (C) Three-dimensional model of the architecture of strands localized in the periplasmic space and anchored to the peptidoglycan layer and outer membrane. Picture courtesy of Freese et al. Three strands are presented. Conformation of ring elements which are located normal to the outer membrane is shown. The model is not drawn to scale. RE, ring element, framed; EE elongated element; CM, cytoplasmic membrane; OM, outer membrane; cema, central mass; pm peripheral mass. For a description, see the text. Panels A and B reprinted from reference 75 with permission. Panel C reprinted from reference 38 with permission.
Type IV pili.
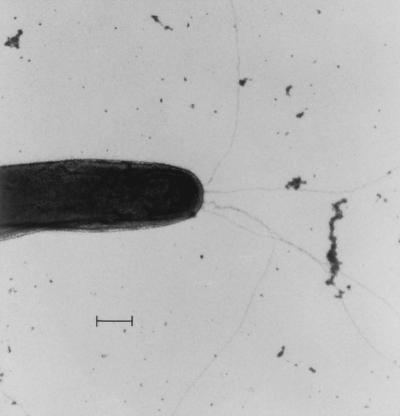
In early motility studies on M. xanthus, pili were recognized to be required for S motility (61). Pili, which are comprised of proteinacious filaments 10 nm in diameter and 3 to 10 μm long, are localized predominantly at one cell pole (61, 78) (Fig. 6). Recent studies revealed that these pili belong to the class of type IV pili which are also required for twitching motility. Twitching has been observed in many gram-negative microorganisms, including Eiknella corrodens, Moxarella osloensis, Neisseria gonorrhoeae (52, 73), enteropathogenic Escherichia coli (114), Pseudomonas aeruginosa (28, 132), and many other pseudomonads (16, 17, 52, 53, 54). Type IV pili are also referred to as type 4 fimbriae. In addition to their role as potential motility organelles, the type IV pili are essential components of several important cellular functions including adhesion to surfaces and phage binding. Molecular details of assembly and function of type IV pili in M. xanthus are discussed below in the context of S motility.
FIG. 6.
Polar pili on an M. xanthus cell. Picture kindly provided by Dale Kaiser. Bar, 0.5 μm. Reprinted from reference 61 with permission.
Motility Mutants of M. xanthus
In the following section, genes and the corresponding mutant phenotypes that are involved in M. xanthus motility are discussed. Emphasis is placed on the function of the A- and S-system genes and on how the function of the frz and mgl genes relate to single-cell gliding and movement of cell swarms.
The A-motility system controls single-cell gliding.
Single cells are visible at the edge of A-motile colonies (Fig. 2C). Videomicroscopy analysis of A-motile cells shows that individual cells glide with wild-type speed when well isolated from other cells (Fig. 2A and C), suggesting that the A-motility system includes all components needed for the machinery and the control of gliding of single, isolated cells. However, compared to the wild type, fewer cells appear to organize into small groups at the perimeter (57) (Fig. 2C). The rate of swarm expansion of A+S− cells depends on the cell density to a similar extent to that of A+S+ cells (half-saturation cell density about 108 cells/ml). The maximal swarming rate is only 0.65 μm/min on 1.5% agar (62). A+S− cells are capable of performing elasticotaxis (37). In fact, the elasticotaxis response in these strains is more pronounced that in the wild-type strain DK1622. The swarming of growing A+S− colonies is strongly reduced on a low-percentage (0.3 to 0.5%) agar surface, and the swarming rate increases with increasing agar concentration (107).
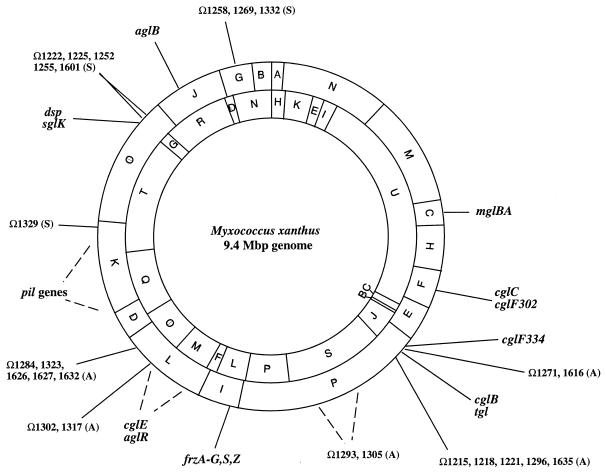
By definition, genes belong to the A-motility system only if they result in a nonswarming double-mutant colony when they are mutated and introduced into an S-motility mutant (57). Colonies of cells with mutations in the A-motility system (A−S+) have flares with a smooth edge where no isolated, individual cells are visible (Fig. 2B). Table 1 summarizes the genes of the A motility system as well as the genes that affect the cellular movement pattern of individual isolated cells. More than 37 different loci of the A-motility system have been identified and appear to map to at least seven clusters on the 9.4-Mbp M. xanthus chromosome (26, 56, 63, 76) (Fig. 7). Considering the loci identified by the independent mutant hunts, it appears that the A-motility system has not been saturated by mutagenesis. While mutations in the A-motility system result in obvious defects in vegetative swarming, with some exceptions they do not affect developmental aggregation (56, 57, 77).
TABLE 1.
Genes affecting A motility and/or gliding of individual, isolated cellsa
| Gene, locus or gene cluster | Molecular properties | Function (putative or proposed) in motility | Assignmentb | Reference(s) |
|---|---|---|---|---|
| agl genes | ||||
| aglA-R | Unknown | Unknown | g | 56 |
| A1 cluster (Ω1215, Ω1218, Ω1221, Ω1296, Ω1635) | Unknown | Unknown | g | 76 |
| A2 cluster (Ω1272, Ω1616) | Unknown | Unknown | g | 76 |
| A3 cluster (Ω1293, Ω1305) | Unknown | Unknown | g | 76 |
| A4 cluster (Ω1284, Ω1323, Ω1632, Ω1626) | Unknown | Unknown | g | 76 |
| A5 cluster (Ω1302, Ω1317) | Unknown | Unknown | g | 76 |
| mglA | pras-like GTPase | Control of single-cell reversals and gliding speed | g | 48, 77, 118 |
| mglB | Protein with Ca2+ binding motif | Control of single-cell reversals and gliding speed | c | 49, 118 |
| cgl genes | ||||
| cglB | Lipoprotein with high cysteine content | Unknown | g | 63, 76, 101 |
| cglC | Unknown | Unknown | g | 76 |
| cglD | Unknown | Unknown | g | 76 |
| cglE | Unknown | Unknown | g | 76 |
| cglF | Unknown | Unknown | g | 76 |
| Ω302 | Unknown | Unknown | g | 63 |
| Ω334 | Unknown | Unknown | g | 63 |
| Other genes | ||||
| frzA | CheW homolog | Control of single-cell reversals, coupling of FrzCD and FrzE | c | 12 |
| frzB | No homology to Che protein | Control of single-cell reversals | c | 12 |
| frzCD | MCP homolog | Control of single-cell reversals | c | 12 |
| frzE | Response regulator containing a CheA-like histidine autokinase and a CheY-like receiver domain | Control of single-cell reversals | c | 12 |
| frzF | CheR homolog | Control of single-cell reversals | c | 12 |
| frzZ | Protein with two CheY-like domains | Control of single-cell reversals | c | 123 |
Movements of isolated single cells are movements when cells are >0.5 μm apart from each other.
Indicates the criterion used to assign a gene to the A-motility system or to gliding as individual, isolated cells. g, in a genetic cross with an S− mutant the resulting colony was nonswarming; c, the single-cell motility behavior was different from wild type.
FIG. 7.
Localization of motility genes on the M. xanthus genome. AseI (outer circle) and SpeI (inner circle) restriction maps of the DK1622 chromosome are shown. Modified from references 26 and 76 with permission.
All A-motility mutants can be divided into two subclasses, the agl and the cgl mutants (55, 56) (Table 1, Fig. 8). Currently, 32 agl genes and five cgl loci (cglB, cglC, cglD, cglE, and cglF) have been identified (56, 57, 76). If the swarming defect of an A-motility mutant can be complemented by extracellular rescue, i.e., upon mixing mutant cells with wild-type cells or cells of another motility class, the gene is called a cgl gene (for contact or conditional gliding). Rescue of A motility is detected in these mutants when single cells and flares of cells emerge from an area where donor cells and recipient cgl mutants are mixed (55) (Fig. 8). A-motility mutants that are not rescued by this extracellular complementation are termed agl (for adventurous gliding) mutants. Stimulation of A motility in cgl mutants requires cell-cell contact between donor and recipient cells on agar. Addition of cell extract or culture supernatant does not rescue single-cell gliding of cgl mutants. However, cells that were killed by brief treatment with formaldehyde do stimulate A-motility movements in cgl mutants (with the exception of cglB and cglF) (55). Interestingly, stimulation is most effective if S motility is retained in the recipient strain.
FIG. 8.
Stimulation of motility of the agl and cgl A-motility mutants. Drops (on the left of each image) of a liquid culture of A-motility mutants (A−S+) (recipient cells) were spotted on CTT agar plates and allowed to dry, and another drop of DK6204 (donor cells), a nonswarming ΔmglBA mutant, was added to intersect with the former spot. (A) cgl recipient strain (A−S+). After a few hours, single cells of DK1219 are visibly gliding as individual cells. (B) agl recipient strain (A−S+). No stimulation of single-cell movement is observed. Bar, 20 μm.
Studies of Tn5 insertion mutants suggested that some A-motility mutants are defective in biosynthesis of the O antigen of lipopolysaccharides (36). Recent findings, however, provide evidence that the motility defects of those lps mutants are due to defects in S motility (15). TnphoA mutagenesis resulted in the identification of three mutants that exhibit defects in A motility and that express alkaline phosphatase activity (63). Linkage analysis mapped two of these mutations to the cglB and cglC loci, respectively, suggesting that these proteins are components of the cell envelope and that they function outside of the cytoplasm.
To date, a detailed molecular analysis of an A-motility gene has been conducted only on cglB (101). Videomicroscopy studies of cells with mutations in cglB have also been performed (101, 118). Gliding movement of individual cglB cells is abolished when the cells are more than 0.5 μm apart from each other (101) (Fig. 2C). When they are in close proximity, extensive cell movement is apparent which is presumably due to S motility (see below), suggesting that A motility controls the gliding of isolated individual cells. The cglB gene encodes a 412-amino-acid putative outer membrane protein which contains a typical N-terminal signal peptidase II leader sequence and cleavage site. These findings suggest that the mature protein is localized to the outer membrane, an observation consistent with the TnphoA studies (63). The mature CglB protein is predicted to contain 16 cysteine (excluding the N-terminal) residues, which is an unusually large number for an outer membrane protein. Comparison of the predicted amino acid sequence with sequences in public protein databases suggests that CglB is a unique protein (101). Studies with a cglB allele which encodes a CglB protein with 6 histidine residues tagged at the C terminus (CglB-His) showed that this protein is active and promotes single-cell gliding at wild-type velocity (101). However, cells that carry only the CglB-His protein do not stimulate ΔcglB mutants to glide, presumably due to a reduced level of CglB-His protein. Because of the cglB phenotype, at least two distinct functions of CglB are possible: (i) CglB may be involved in the gliding motor of the A-system as part of the force-generating complex or in the process of force transmission to the surface, or (ii) CglB may function as a signaling molecule and may stimulate single cells to move, e.g., by suppressing reversals. The above-discussed observation on the cglB-His allele may suggest that a regulatory role of CglB is less likely than a mechanistic function and that the observed stimulation may reflect localization of CglB to the outer membrane (101).
The S-motility system controls gliding of cells in swarms.
S-motile colonies show a clearly defined, undulating edge which is indicative of movement of cells within the colony (Fig. 2B). The cell density dependence of the swarming rate is decreased in these mutants compared to the wild type. The maximal swarming rate of three A−S+ strains tested is approximately 0.45 μm/min, with a half-saturation cell density of 7 × 108 cells per ml (62). Cells that are motile by S motility only are unable to respond to physical stress forces in the substratum (37). Accordingly, the absence of S motility causes a dramatic reduction in swarming of growing A+S− colonies on a 0.3% agar surface, and the swarming rate increases with increasing agar concentration (107). Most of the S-motility mutants are also defective in fruiting-body formation (57, 77).
In contrast to A-motile colonies, no single cells are visible at the perimeter of S-motile colonies. To develop an understanding of cellular motility that is due to S motility exclusively, translocation of single cells of a cglB::TnphoA strain (A−S+) was studied (118). Individual cells were found to move only when they were no more than 2 μm apart (56, 118) (Fig. 2B). The movements, most of which occur in the direction of the long axis of the cell, are jerky but occur at similar high speeds to those observed in wild-type cells (118) (Fig. 2B). Additionally, these A−S+ cells reverse the direction of movement at a frequency of approximately 2.7 reversals per min, which is more than 10-fold higher than in wild-type cells (0.17 per min). In general, the high-reversal mode of cglB mutants is observed mostly at the edge of a group of cells (118). However, these A−S+ cells are not locked in a high-reversal mode, because cells are able to switch quickly into a low-reversal, high-velocity mode when they are located inside a group of cells. This motility behavior may represent the type IV pilus-dependent gliding in M. xanthus and may be related to twitching. As indicated above, pili were recognized to be required for S motility (61).
(i) pil genes.
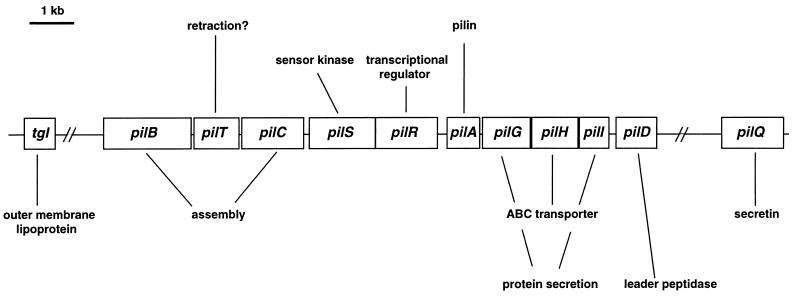
Analogous to defining A motility, genes belong to the S system if they result in a nonswarming double-mutant colony when they are mutated in an A−S+ mutant (57). Numerous S-motility genes have been identified in mutant screens (Table 2). In the original genetic analysis by Hogkin and Kaiser, two major regions of S-motility genes were identified, the sglI and sglII regions (57). In recent research, it became clear that most of the sgl genes of the sglI region showed significant similarity to genes involved in pilin expression and processing and in export, assembly, and function of type IV pili in other bacteria. Therefore, these genes were concluded to be pil genes (126, 137–140, 141) (Fig. 9). Twitching motility is a type IV pilus-based mode of surface translocation and has been observed in many gram-negative microorganisms (16, 17, 28, 52, 53, 54, 132). Our current understanding of type IV pilus structure and biogenesis rests mostly on studies conducted with P. aeruginosa and N. gonorrhoeae (for recent reviews, see references 3, 30, and 39). Genes involved in type IV pilus export and assembly have multiple homologs to components of DNA uptake and protein secretion systems. These systems have in common that they catalyze a vectorial transport of larger polymers (polypeptides, DNA) across the outer and inner membranes of gram-negative bacteria. These transport systems can function either for import (DNA) or for export (pili, proteins). The strong similarity between the gene products in these systems suggests that these different systems may operate by a common mechanism. In the following discussion, only the model for type IV pilus formation and function is discussed, without elaborating the experimental evidence in support of that model (see volume 192 of Gene [1997] for numerous specific reviews).
TABLE 2.
Genes affecting S motility and/or gliding of cells in groupsa
| Gene, locus, or gene cluster | Molecular properties | Function (putative or proposed) in motility | Assignmentb | Reference(s) |
|---|---|---|---|---|
| sgl genes | ||||
| pilA | PilA protein | Major pilin subunit | g | 57, 140 |
| pilG | No homolog | Involved in protein secretion | g | 57 |
| pilH | ABC transporter | Involved in protein secretion | g | 57, 141 |
| pilI | No homolog | Involved in protein secretion | g | 57 |
| pilD | Leader peptidase/N-methylase | Processing of pre-PilA protein | g | 57 |
| pilQ | Secretin | Synthesis, export, assembly, and function of type IV pili | g | 125 |
| pilB | Membrane-associated protein with nucleotide binding site | Involved in pilus assembly | g | 57 |
| pilC | Inner membrane protein | Involved in pilus assembly | g | 57 |
| pilT | Membrane-associated protein with nucleotide binding site | Involved in pilus retraction? | g | 57 |
| pilS | Sensor kinase | Regulation of pilA transcription | g | 57, 140 |
| pilR | Transcriptional regulator | Regulation of pilA transcription | g | 57, 140 |
| S1 cluster (Ω1222, Ω1225, Ω1601, Ω1252, Ω1255) | Unknown | Unknown | g | 76 |
| sglK | DnaK chaperone homolog | Protein and polysaccharide secretion | g | 76, 129 |
| S2 cluster (Ω1258, Ω1269, Ω1332) | Unknown | Unknown | g | 76 |
| S3 cluster (Ω1329) | Unknown | Unknown | g | 76 |
| dsp | Unknown | Required for fibril production | g | 6, 7, 88a |
| mglA | pras-like GTPase | Unknown | g | 48, 77 |
| mglB | Protein with Ca2+ binding motif | Unknowng | 49 | |
| tgl gene | ||||
| tgl | Outer membrane lipoprotein with six tandem tetratricopeptide repeats | Involved in pilus assembly? | g | 57, 102 |
| Other genes | ||||
| frzA | CheW homolog | Control of S motility, coupling FrzCD and FrzE | s | 12 |
| frzB | No homology to Che protein | Control of S motility | s | 12 |
| frzCD | MCP homolog | Control of S motility | s | 12 |
| frzE | Response regulator containing a CheA-like histidine autokinase and a CheY-like receiver domain | Control of S motility | s | 12 |
| frzF | CheR homolog | Control of S motility | s | 12 |
| frzZ | Protein with two CheY-like domains | Control of S motility | s | 123 |
| frzS | Protein contains a receiver domain | Control of S motility | g | 127a |
| difA | MCP homolog | Control of S-motility | g | 142 |
| difC | CheW homolog | Control of S motility | 142 | |
| difD | CheY homolog | Control of S motility | 142 | |
| difE | CheA homolog | Control of S motility | g | 142 |
| wzm wzt wbgA | ABC transporter | Export of O antigen | g | 15 |
| lps-1, lps-2, lps-4, lps-5 | Unknown | O-antigen biosynthesis | g | 15 |
Movement of cells in groups are movements when cells are <2 μm apart from each other. Note that if a strain contains functional A motility, some aspect of the movement observed in close proximity may also be due to some aspect of A motility.
Indicates the criterion used to assign a gene to the S-motility system or to gliding in groups. g, in a genetic cross with an A− mutant, the resulting colony was nonswarming; s, swarming or other behavior was similar to that of S− mutants.
FIG. 9.
Organization of M. xanthus pil genes. Compiled from references 102, 125, and 139 to 141. For details, see the text.
In P. aeruginosa, the pilA gene encodes the prepilin protein, which, upon cleavage of the 6-amino-acid leader sequence and subsequent N-methylation by the membrane-bound PilD protein, forms the major structural subunit of the pilus filament. For pilin export and assembly, a large multimeric protein complex is thought to reach from the inner to the outer membrane. PilC is a polytopic inner membrane protein (68, 90), and PilQ is an outer membrane protein which forms an oligomeric complex. This complex acts as a gated channel to facilitate the exit of the PilA subunits. A pilus grows by assembling pilin subunits at its base at the export apparatus. Export of pilin requires the activity of PilB, a protein containing a putative nucleotide binding site, which is associated with the inner face of the cytoplasmic membrane. Mutants with mutations in pilD are unable to assemble pili even though pilA is expressed. PilT and PilU are two PilB homologs. Of particular interest for type IV pilus function in twitching motility is pilT. The PilT protein is predicted to be a member of a family of nucleotide-binding proteins (132, 139). PilB is required for pilus extension, whereas PilT may catalyze pilus retraction. In P. aeruginosa, a set of pil genes (pilGHIJK genes) that show strong homology to the enteric chemotaxis genes and to the frz genes of M. xanthus were identified (28–30, 144). Mutations in these P. aeruginosa genes abolish pilus formation or impair pilus function in twitching (29).
Table 2 and Figure 9 summarize the known M. xanthus pil genes and the proposed functions of the gene products. The pilin protein, the major constituent of the pilus filament, is encoded by the pilA gene. Expression of pilA is under control of a ς54 promoter, which is regulated by the two-component regulatory system pilS and pilR (140). Expression of pilA requires the response regulator pilR as a transcriptional activator but is negatively regulated by the putative sensor kinase pilS. Additionally, pilA expression is autoregulated (140). Under high-nutrient conditions as well as under developmental conditions, expression of pilA is induced but is independent of pilSR (140). The homologs of the M. xanthus PilB and PilC proteins in P. aeruginosa are believed to be involved in pilus assembly (90). PilH is highly similar to an ATP binding cassette (ABC) transporter protein, but PilG and PilI do not reveal homology to known proteins. These proteins are hypothesized to function with PilH (141). In P. aeruginosa, pilD is a bifunctional leader peptidase and N-methylase. M. xanthus pilT mutants are piliated (139). P. aeruginosa (133) and E. coli (114) pilT mutants are hyperpiliated, and twitching motility in these organisms is abolished. Also, M. xanthus dsp mutants (see below) carry pili but are defective in S motility. Similar to P. aeruginosa and N. gonorrhoeae, M. xanthus contains a pilQ gene that is essential for pilus biogenesis (125). The PilQ protein belongs to the secretin superfamily of proteins, which multimerize in the outer membrane to form a channel for uptake of macromolecules (11, 69, 125). Before the molecular similarity to pilQ was known, the gene was referred to as sglA in M. xanthus (125). pilQ mutants are frequently isolated as dispersed growing mutants which still form fruiting bodies (e.g., DK101 and DZ1 [125]).
The molecular basis of how type IV pili generate displacement is unknown. The cellular movement patterns observed in the A−S+ mutant (cglB [see above and reference 118] [Fig. 2]) may reveal the type IV pilus-dependent motility in M. xanthus. For nongliding microorganisms, a hypothesis of controlled pilus retraction and elongation was proposed earlier as a mechanism of pilus function which may result in twitching movements (16–18) (Fig. 10). According to that model, a pilus which is attached by its tip to another cell or to a surface could depolymerize and polymerize at the membrane export apparatus, which would result in shortening and extending of the filament and thereby in displacement of the cell (Fig. 10). A depolymerization could be caused by nucleoside triphosphate hydrolysis, e.g., catalyzed by PilT, which could release the energy for pilus retraction. Depolymerization of polar pili would generate movement in the direction of the long axis of the cell. A pilus receptor that could serve as an attachment point specifically for pili from neighboring cells has not been identified, although the fibrils (see below) are certainly good candidates for such function. In gliding microorganisms such as M. xanthus, pili of neighboring cells may also attach to surface attachment sites of the A-system gliding apparatus. When these attachment sites undergo gliding-dependent displacements parallel to the cell axis, they can “pull” the pilus and, thus, the neighboring cell. It also seems plausible that in M. xanthus, a combination of pilus retraction-extension and pilus displacement along gliding tracks may result in S motility (Fig. 10C). An argument in favor of the latter model is that the A- and S-motility systems do not operate independently. This is indicated by the cell density dependence of the swarm expansion rates in wild-type and in A- and S-system mutants. The rate of expansion of wild-type swarms is higher than the sum of the rates of A−S+ and A+S− swarms (62). These observations suggest a synergistic effect of the two motility systems.
FIG. 10.
Models for generation of cell movement by type IV pili in M. xanthus. To illustrate the model of pilus-dependent S motility, individual cells are not drawn to be in contact with other cells. It should be noted that cell-cell contact is required for S motility. (A) A depolymerization-polymerization (↔) shortens a pilus, thus generating displacement mostly in the direction of the long axis of the cell. (B) Pili attach to neighboring cells at “adhesion sites” (◂ ○ ▸), e.g., which are part of the gliding motor of A motility and are involved in A-motility-dependent gliding. Linked to the moving adhesion sites by a pilus, a neighboring cell is “dragged” along, thus performing a translocation in the direction of the long axis of the cell. (C) A combination of pilus retraction-extension and adhesion site-dependent displacement (A plus B).
(ii) tgl gene.
In one S-motility mutant, tgl (for transient gliding), S motility and type IV pilus assembly can be stimulated by aligning tgl cells with cells of a tgl+ strain (57, 61, 102, 103, 124, 126). Because of the stimulation phenotype, tgl is, in a sense, the counterpart in S motility to the cgl genes in A motility. Similar to CglB, Tgl appears to be a lipoprotein which localizes to the outer membrane (102, 103). Tgl contains six tandem tetratricopeptide repeats, a motif which is known to be involved in protein-protein interactions. The observed stimulation by Tgl is believed to be due to Tgl acting as a pilus assembly factor rather than as a cell-cell signal (124).
(iii) Other S-motility genes.
Next to the pil genes and tgl, there is a third group of S-motility genes that affects cell surface structures in M. xanthus: the genes of the dsp locus and the linked sglK locus. Both genes are also required for formation of extracellular fibrils (7, 108, 109, 111, 129). Mutants defective in dsp were isolated as S− mutants and grow as dispersed cultures in liquid medium (88a). When crossed with an A− mutant, A−dsp double-mutant colonies show some reduced swarming after prolonged incubation. dsp mutants form pili but carry aberrant fibrils and are defective in cell cohesion and development (6, 7). Fibrils are extracellular, irregular branched structures of variable width and length (5, 7) and are required for cell-cell cohesion (5, 6). They are composed of polysaccharides and contain a class of integral fibril protein, IFP-1, which is defined as being released from fibril material only after treatment with sodium dodecyl sulfate and β-mercaptoethanol (8, 9, 129). It has been suggested that the different sizes of IFP-1 proteins result from multimer formation of a single small-molecular-size subunit whose amino acid composition and N-terminal sequence were determined (9). Fibrils are found mostly on cells in groups where they establish cell-cell and cell-substratum contacts (7). A link between exopolysaccharides (fibrils) and type IV pilus-dependent S motility may exist, similar to the link between alginate production and twitching motility in P. aeruginosa (131). AlgR and FimS, which are a regulator and a sensor, respectively, are required for alginate production and for twitching motility. This sensor-regulator couple does not appear to regulate PilA production in P. aeruginosa (131), raising the possibility that the produced exopolysaccharides are mechanistically required for twitching movements. Accordingly, fibrils may have an analog function in S-motility gliding of M. xanthus.
The developmental defect of dsp mutants can be complemented by extracellular addition of isolated fibrils (23). Furthermore, dsp mutants are unable to bind Congo red, a dye that binds to extracellular polysaccharides in wild-type M. xanthus cells (6). Wild-type cells behave phenotypically as dsp mutants upon incubation with Congo red, suggesting that fibrils are required for cell cohesion (5, 6). However, fibrils per se do not seem to be required for S motility and development, because dsp second-site suppressor mutants that carry no fibrils exhibit motility and development (24). Also, some recently isolated cgs mutants that lack fibrils and are defective in agglutination and developmental aggregation were shown to be proficient in swarming on low-percentage agar (94). Therefore, dsp may have several functions including one in fibril production and one in S-motility control. Disruption of the stk gene, which presumably is a negative regulator of fibril synthesis, causes overexpression of fibrils as well as increased cell cohesion and dye binding in wild-type cells but not in dsp mutants (27). Recently, it was shown that mutants with mutations in the S-motility gene sglK are unable to produce fibrils (129). The sglK gene product is predicted to resemble a DnaK chaperone homolog (129). It is cotranscribed with another gene that is predicted to encode another chaperone homolog, GrpE. Both chaperones are believed to be involved in protein and polysaccharide secretion (129).
The dif genes were recently isolated in a screen for mutants defective in fruiting-body formation (142). Molecular analyses of the dif locus revealed four open reading frames, called difA, difC, difD, and difE, whose products show significant identity to the bacterial chemotaxis proteins methyl-accepting chemotaxis protein (MCP), CheW, CheY, and CheA, respectively. Interestingly, DifA, a tentative homolog of MCP, shows the strongest identity to the Bacillus subtilis TlpB. Mutations in tlpB cause B. subtilis cells to stick together. Based on the genetic double-mutant test, the difA and difE genes belong to the S-motility system (142). Very recently, another gene that is believed to be involved in a signal transduction pathway, frzS, was shown by the genetic double-mutant test to belong to the S-motility system (127a) (see below). It is tempting to speculate that the dif genes, and some frz genes, represent a signal transduction pathway involved in the control of S motility that affects the activity of type IV pili (e.g., by controlling retraction, extension, or attachment of the pili) or their expression.
Some O antigens of lipopolysaccharide were recently shown to be required for S motility (15). A null mutant with mutations of the wzm, wzt, and wbgA genes, which map to the sasA locus, behaved as an S− mutant (15). These mutants were previously isolated as mutants that suppress a developmental defect of asgA mutants (64).
As indicated above, no direct observation of pilus-mediated cell movement has been reported. However, with many S-motility genes available (Table 2) and in conjunction with high-resolution videomicroscopy, specific models of pilus function can now be tested, and it should be possible in the near future to uncover the molecular mechanism(s) of type IV pilus-mediated movements in twitching and in S-motility gliding.
A−S− double mutants.
The nonswarming phenotype of A−S− double-mutant colonies has served as the conceptual basis for defining the two gliding systems in M. xanthus (56, 57). Motility mutants identified by the double-mutant test will most likely include those defective in structure, assembly, and function of the gliding motor elements (e.g., pil genes). Mutations in genes whose products play a modulating role on the activity of the gliding motors may only partially affect the swarming pattern of the A- or S-motility system when observed in a null mutant background of the other motility system. These mutants may exhibit reduced but not completely abolished movements. For example, mutations in frzCD, frzF, or frzE, when crossed into an A− mutant, show a reduced S-motility swarming (37) (Tables 1 and 2; also see below). Also, genes of the dsp locus may play a regulatory role in S motility, because S-system swarming is severely reduced in A−dsp double mutants.
Motion analysis of single cglB pilR (A−S−) cells revealed that the cells are not completely nonmotile, as might have been suggested from the nonswarming colony morphology, but are able to conduct a few single-stroke movements (118). A stroke movement consists of a small (∼1- to 1.5-μm) displacement, followed by a pause for at least 20 min. Most cells did not move during the observation period. One possible rationalization of this residual movement is that because of the pilR mutation, no functional pili are present, which eliminates any pilus-dependent S motility as a cause of the residual movement (118, 140). The mutation in the A system abolished the gliding of isolated single cells. In the absence of a clear understanding of the A-system gliding apparatus, however, it is not obvious that a null mutation in A motility, such as ΔcglB, results in a knockout mutant in the A-system gliding motor. It is conceivable, for example, that some A-system mutants, like the cgl mutants, are defective in transmission of force between the cell body and the substratum, e.g., because they lack gliding-specific adhesion sites. Such mutants would behave as A-motility mutants in swarming and single-cell assays but would still have an A-system gliding motor that is proficient in force generation. In the absence of such a specific adhesion site for A motility, other components of the cell envelope may be poor substitutes in force transmission, thus resulting in ineffective displacement. This could be one scenario to explain the observed residual movement in that particular A−S− mutant. Many agl genes are awaiting molecular characterization. One imminent question is also whether the large collection of agl and cgl mutants contains a mutant that represents a “true” motor mutant. If such a mutant cannot be isolated, this may suggest either that M. xanthus carries multiple copies of these genes or that a complete loss of the complex motility apparatus, caused by a knockout mutation, may destabilize the cell wall and result in a lethal phenotype.
Control of M. xanthus motility by the frz genes.
The frz locus was identified by a set of mutants which display a unique developmental defect in aggregation and fruiting-body formation (144). Under starvation conditions, these frz mutants form entangled, “frizzy” aggregates. To date, seven frz genes have been identified within this locus: frzA, frzB, frzCD, frzE, frzF, frzG, and frzZ (13, 83, 123). In addition to the aggregation defect, the frz mutants carry a defect in colony swarming on low-percentage agar that somewhat resembles that seen in S-motility mutants (37, 107). Mutations in the frz genes also reduce the level of sporulation (105, 123) although this effect is strain dependent. Interestingly, single cells of most frz mutants have an altered frequency of reversing their direction of movement; wild-type cells reverse their direction once every 5 to 7 min (0.17 reversal min−1), while most frz mutant cells reverse on average once per hour (<0.02 reversal min−1). One notable exception was caused by a Tn5 insertion at the 3′ end of the frzCD gene (a frzD mutant). Single cells bearing this mutation exhibit an increased (1.5 reversals min−1) rather than a decreased reversal frequency (12, 118). Gliding velocities, however, are unaffected in frzE mutants, as shown by high-resolution videomicroscopy studies (12, 118).
The amino acid sequences of the predicted Frz proteins show striking similarities to those of the Che proteins involved in chemotaxis of enteric bacteria (13, 83, 87) (Tables 1 and 2). The enteric che signal transduction system is a two-component regulatory system, where the input stimulus is the concentration of an attractant or repellent in the environment and the output is a change in swimming behavior (for a recent review, see reference 14). The basic signal relay consists of two interacting proteins, CheA, a histidine kinase that autophosphorylates a histidine residue at position 48, and CheY, a response regulator protein that receives the activated phosphate from CheA at a conserved aspartate residue (Asp 57) in a CheA-dependent transphosphorylation reaction. The phosphorylated form of CheY then interacts with the FliM protein of the flagellar switch to cause a bias of flagellar rotation toward clockwise, resulting in an increased tumbling frequency. Input of environmental signals (attractant or repellent) into the Che signal transduction cascade is mediated by MCPs. These receptors are transmembrane proteins that interact either directly with attractants or repellents or indirectly with periplasmic binding proteins on their periplasmic side and with CheA at their cytoplasmic interface. The interaction between a chemoattractant and its cognate MCP induces a conformational change in the MCP which results in inhibition of the CheA autokinase activity. CheW is involved in mediating the interaction between the MCPs and CheA. Another protein, CheZ, catalyzes the dephosphorylation of the response regulator, CheY, ensuring that only the most recently sensed change in stimulus concentration is integrated into a motility response. Adaptation, or the control of sensor sensitivity, is regulated by the extent of methylation of specific glutamate residues on the cytoplasmic domain of the MCP proteins. The activities of two enzymes control the state of methylation; the CheR methyltransferase uses S-adenosylmethionine as a methyl group donor to methylate the cytoplasmic domain of a MCP, while the phosphorylated form of CheB, which has methylesterase activity, demethylates the protein. In some nonenteric bacteria (e.g., Rhizobium meliloti and Rhodobacter sphaeroides), Che homologs are involved in chemokinetic rather than chemotactic behavior, and cells respond to an absolute concentration of chemoattractant rather than to a gradient, by swimming at different speeds (4, 115, 116).
In M. xanthus, the frz genes include homologs of cheA, cheY, cheW, cheR, and cheB, as well as genes containing combinations of different domains of che genes and genes which show no match with che genes (Table 2). For example, the M. xanthus FrzE protein contains a CheA-like histidine kinase domain and a CheY-like regulator domain, while FrzZ is predicted to contain two CheY domains. The proposed receptor, FrzCD, which is considered to be a MCP homolog, does not contain either a membrane-spanning region or the periplasmic domain of the enteric MCPs and is a cytoplasmic protein (82, 83). Since frz genes are defined by the “frizzy” aggregation phenotype under developmental conditions which involves A- and S-motility movements, the control of M. xanthus motility by frz genes appears to be more complex than the Che cascade, which regulates only one motor in swimming bacteria.
Biochemical studies have demonstrated that the Frz proteins can act as a signal transduction system operating by a phosphorelay mechanism. In vitro experiments with recombinant FrzE and [γ-32P]ATP demonstrated that FrzE is capable of autophosphorylation, presumably at a histidine residue (87). This finding, in conjunction with the predicted histidine kinase response regulator fusion of the protein, suggests that FrzE contains both autokinase and transphosphorylation activity (87) and thus is capable of functioning as a signal relay module (87). Since adaptation in the enteric bacteria requires the reversible methylation of the MCPs, the methylation state of FrzCD has been examined under both vegetative and developmental conditions. Such an analysis was possible since the methylated receptors migrate faster than the unmethylated form during electrophoretic separation on sodium dodecyl sulfate-polyacrylamide gels (82, 86, 89). Similar to the situation in enteric bacteria, methylation of FrzCD is dependent on the methyltransferase FrzF (86).
In recent years, research on the function of frz genes has been directed to address two fundamental questions: (i) the identity of the signal input into the Frz cascade, and (ii) the cellular apparatus on which the output signal acts. Due to the similarity between the Frz proteins and the enteric Che system, it was initially hypothesized that motility of vegetative and developmental M. xanthus cells could be controlled by regulation of the reversal frequency of the cell. Movement with a low reversal frequency was considered a response to an attractant, and movement with a high reversal frequency was considered a response to a repellent (81). Extensive studies have been conducted to identify physiological attractant or repellent molecules which could cause altered, Frz-dependent motility behavior and would affect the methylation state of FrzCD. It was hypothesized that by analogy to enteric MCPs, an increase in the amount of the methylated form of FrzCD might represent an adaptive response to an attractant stimulus and, similarly, an increased level of demethylated FrzCD may indicate adaptation to a repellent (81). Since chemotaxis requires an adaptive response, the ratio of methylated and unmethylated FrzCD was used to test a large number of molecules as potential chemoattractants or repellents (84, 105). Casitone and yeast extract was found to increase the level of methylated FrzCD, whereas some short-chain alcohols, including isoamyl alcohol, were found to decrease the level of methylated FrzCD (84). Isoamyl alcohol, which has not been reported to be an intermediate in M. xanthus metabolism, increased the reversal frequency of single wild-type cells when added at a high concentration (ca. 30 mM) (105). This response of vegetative cells required frzA, frzCD, and frzE (105). However, single cells exposed to gradients of potential attractants did not show changes in motility behavior (32, 122). While the observed responses to repellents are consistent with the enteric paradigm, responses to attractant stimuli are more complex and are now believed to include chemokinetic behavior in response to some self-generated stimulus (127). Recently, phosphatidylethanolamines, including those found in M. xanthus, were reported to behave as chemoattractants in swarm and single-cell motility assays (66). The motility response is specific to a particular composition of fatty acid and correlates with a decrease in reversal frequency of individual cells. Over a period of 1 h, the suppressed reversal frequency returned to the “prestimulus” level. This observation was interpreted as adaptation, suggesting that cells may indeed respond chemotactically to these compounds. Interestingly, these behaviors were only partially dependent on intact frz genes, which suggests the existence of an additional signal transduction cascade required for responses to phosphatidylethanolamines (66).
The frz genes are developmentally regulated (130), and the Frz proteins are activated during development, as indicated by increased FrzCD methylation (84). One developmentally regulated molecule has been identified as a positive input signal to the frz system: the extracellular C-factor signaling protein required for fruiting-body formation and sporulation. During an analysis of the C-factor signaling pathway, Tn5lac mutants which arrested at the aggregation stage were identified (113). One subclass of these transposon insertions mapped to the frz locus, while a second subclass mapped to the fruA locus (previously the class II gene). These mutants were blocked at a similar stage in aggregation to csgA mutants but sporulated at a level higher than that of csgA mutants, which do not produce C factor (113). It was suggested that C factor is a component of two separate pathways, one that regulates developmental aggregation and one that regulates sporulation. The ratio of methylated to unmethylated FrzCD protein was used to examine whether C factor can function as an input signal to the frz cascade. Addition of purified C factor to developing cells of a csgA strain resulted in an increase in the methylated form of FrzCD as detected by Western blot analysis (112). This methylation was dependent on the C-factor concentration and did require FrzF, the putative methyltransferase in M. xanthus. Other developmental mutants that are defective in production of other development-essential extracellular signals were found to be defective in FrzCD methylation as well (40). These observations demonstrate that the frz signaling cascade is activated under developmental conditions of coordinated cell movements during aggregation and fruiting-body maturation in response to cell-cell signals.
While several recent studies have identified compounds that could provide input to the Frz cascade, some advances relating to the output of the system have occurred. The recently discovered dif genes, which also show strong similarity to bacterial chemotaxis proteins, seem to affect only S motility, because the reversal frequency of difA and difE mutant cells is unaltered from that of the wild type (142). This is in contrast to frz mutant cells. Because of the complexity of the Frz components and the motility responses, it seems possible that the Frz output interacts with both the A- and S-motility systems. An output of Frz into the A-motility system is indicated by an altered reversal frequency of isolated frz cells. No mutations that suppress the low-reversal phenotype of frzE mutant cells have been reported. Many observations also hint at S motility as an output of Frz. (i) The swarming motility of vegetative M. xanthus depends on the agar support; low-percentage agar (0.3%) is almost exclusively conducive to S motility of swarms (37, 57, 62, 107). Vegetative swarming in response to Casitone and yeast extract, and also to isoamyl alcohol, was most dramatic on 0.3% agar (105). This response depends on the frz genes (frzA, frzB, frzCD, frzE, and frzF). (ii) Suppression of the swarming defect of some frz mutants (frzF, frzCD) is specific to the sglA1 (pilQ) allele (65). Transposon insertions in frzF and frzCD render starving M. xanthus cells unable to form aggregates at low cell density, and sporulation is reduced to only 1% of the wild-type level in a sglA+ background. However, in a sglA1 (pilQ) mutant background, the low-cell-density aggregation defect is suppressed and double-mutant colonies form frizzy aggregates. Notably, frz mutants were identified in a sglA1 mutant background (144). At high cell density, the partial sporulation defect of the frz mutants is suppressed by sglA1 (pilQ). This suppression may not be due to S motility per se as indicated by the specificity; only a sglA1 (pilQ) allele but not pilC (sglG) promotes suppression. (iii) Similar to S− mutants, frz mutants are not defective in elasticotaxis, a response that appears to be specific to the A-motility system (37). (iv) A frzD mutation can partially suppress the swarming defect of an mglBA mutant (118) (see below). (v) In P. aeruginosa, a set of genes required for pilus biogenesis and twitching motility that shows strong homology to che genes, specifically the frz genes of M. xanthus, was found (29). Since mutations in some of these genes block pilus production and a pilH mutant exhibits a “frizzy”-like swarming pattern, it was suggested that these Che-like proteins control twitching motility in P. aeruginosa (29). Because functional type IV pili are required for S motility in M. xanthus, it is tempting to speculate that one mode of action of the frz gene products is to control type IV pilus-dependent movement. The postulated mode of pilus action, i.e., retraction and/or extension, could result in phenotypic reversals (see also Fig. 10). Also recently, it was postulated that in addition to the frz genes, other signaling cascades that are necessary for developmental motility behavior may exist (66). (vi) frzS was recently identified as a new frz gene which, when mutated, results in a “frizzy” phenotype (127a). In contrast to the other frz mutants, no effect on the cellular reversal frequency was found. Moreover, frzS was shown to belong to the S-motility system as indicated by the genetic double-mutant test (127a). (vii) The dif genes, which also show homology to the enteric che genes, were recently identified in M. xanthus and appear to also affect S motility (105). Considering these observations, it seems likely that Frz affects S motility.
Motility and the mgl genes.
In addition to isolating A- and S-motility mutants, Hodgkin and Kaiser identified one locus that, with a single mutation, rendered a colony completely nonswarming (57). Ten independent mutations were identified in this locus. Because of the colony-swarming defect, this locus appeared to be essential to both A and S motility and was called mgl (for mutual function for gliding). Therefore, it was reasoned that the mgl gene(s) might encode components of the gliding motor. In addition to abolishing swarming, mgl mutants are defective in fruiting-body formation and sporulation, presumably due to their inability to conduct C-factor signaling during development (70, 120). A molecular analysis of the mgl locus revealed that the mgl operon contains two genes, mglA and mglB (120, 121). A mutation in mglA causes the severe swarming and developmental defect, whereas mglB mutant colonies exhibit only partially reduced swarming, aggregation, and sporulation (49). This reduced swarming correlates with the reduced cellular level of the cytoplasmic MglA protein (49). A stabilizing interaction between MglA and MglB is suggested by the finding that in an mglB mutant, the cellular level of MglA is only 15 to 20% of that in wild-type cells but the level of mglBA mRNA is unaffected (49). MglA is not essential for growth.
High-resolution motion analysis of single ΔmglBA cells revealed that despite the nonswarming colony phenotype, individual cells are motile (118). The movement pattern of ΔmglBA cells, however, is distinctly different from that of the wild type and all other M. xanthus motility mutants that were examined. Individual ΔmglBA cells translocate by abrupt, jerky displacements and can reverse the direction of movement about 2.9 times per min, which is more than 10-fold higher than in wild-type cells. The average translocation speed is reduced to 1.9 μm/min. As a result, cells perform a net movement of less than 1 μm in 9 min. These movement patterns are different from those of wild-type cells; of A−S+ cells, which reverse similarly often but also translocate by extended, unidirectional movement at high speed (4.7 μm/min) (118; see above) (Fig. 2B); of A+S− cells, which glide with wild-type speed when separated from other cells (Fig. 2C) and exhibit normal reversal frequencies; and of A−S− double mutants, which conduct only short, single-stroke displacements and which do not move most of the time (118). Interestingly, the high-reversal phenotype of ΔmglBA mutant cells as well as the movement activity of the overall population is dependent on the presence of pili (118); in ΔmglBA pilR double-mutant cells, the high-reversal pattern and the jerky movement are reduced (118). Single cells of an mglB mutant exhibit an intermediate phenotype; the reversal frequency is 1.8 times min−1 and the average translocation speed is 2.6 μm/min (118). Such an intermediate phenotype correlates with a reduced level of cellular MglA protein in mglB mutants. The above observations suggest that a correct level of MglA is required for M. xanthus cells to move with wild-type speed and wild-type reversal frequency. Because the movement pattern of ΔmglBA cells is dependent on S motility and because ΔmglBA cells have pili (although at reduced levels), it seems that ΔmglBA mutant cells behave more like a strong A-system mutant (abolished single-cell movement, high-reversal mode when cells are in close proximity) with an only partially defective S-motility system (no extended runs in one direction at high speed, requirement for pili, and movement distinguishable from that of a A−S− double mutants). Therefore, the cellular motility phenotype suggests that mgl function affects the A- and S-motility system differently (118).
The predicted amino acid sequence of the 195-amino-acid MglA protein reveals homology to the nucleotide binding site of Sar1 and p21ras, both of which belong to the class of small eukaryotic GTPases (48, 49). These GTPases are regulatory proteins which play crucial roles in signal transduction, cytoskeleton organization, protein trafficking, and organelle functions (80). Small GTPases, such as p21ras, have two protein conformations depending on whether they are in the GTP-bound or the GDP-bound state. Interaction with GTPase-activating proteins and guanine nucleotide release factor proteins are believed to regulate the transition between the conformational states. MglB is predicted to be a 159-amino-acid protein that includes a region resembling a calcium binding site of yeast calmodulin (49). Calcium is required for gliding in M. xanthus and in Stigmatella aurantiaca based on Ca2+ ionophore and inhibitor studies (136). To examine whether MglA may function in M. xanthus in a mode similar to that of Sar1, genetic complementation studies of a ΔmglBA mutant with Ha-ras and SAR1 of Saccharomyces cerevisiae were conducted (47). When inserted in the M. xanthus genome, SAR1 complemented the sporulation defect of the parent ΔmglBA mutant (47). A SAR1 allele with defective GTPase activity did not complement the sporulation defect, demonstrating that sporulation requires a functional GTPase activity. Neither Ha-ras nor SAR1 complemented the motility defect. Interestingly, a second-site mutation in a gene called rpm (for restore partial motility), is necessary to partially suppress the swarming defect of a ΔmglBA SAR1 strain. This suppression seems to be allele specific. These findings support the notion that SAR1 affects sporulation and motility differently and that SAR1 may interact with another protein to control motility. The ΔmglBA SAR1 rpm strain swarms on 0.3 and 1.5% agar, suggesting that A and S motility, as indicated by enhanced swarming, was restored. Interestingly, the S-motility system swarming defect of mglBA colonies can be partially suppressed by a frzD mutation (118).
In summary, MglA appears to be a small GTPase that is crucial for several independent cellular functions, i.e., development, sporulation, and motility, and may affect these cellular functions in different ways. With respect to motility, MglA appears to represent, next to the A- and S-motility systems, a novel element that controls motility. Work at the molecular level is required to uncover the mode of action of this remarkable protein.
Model for Single-Cell Gliding (A Motility) in M. xanthus
Considering the above-discussed genetic, molecular, ultrastructural, and behavioral studies of M. xanthus, it is tempting to summarize these observations in the following speculative model for A-motility gliding of isolated M. xanthus cells (Fig. 11). This model also rests on observations and models, formulated for Cytophaga strain U67 and Flexibacter, that were conducted by Lapidus and Berg (72), Ridgway and Lewis (100), and other authors (see below) on other gliding bacteria. It should be kept in mind that because of the operational definition of gliding, the molecular mechanism(s) to achieve gliding movement of well-isolated single cells can differ among microorganisms.
FIG. 11.
Speculative model for single-cell gliding (A motility) in M. xanthus. The picture is a close-up of the interface of the surface of a gliding cell with the substratum and shows one of the numerous motor units in detail. The cell is moving from left to right. In this model, the force generator (molecular motor) is anchored to the cell wall (rigid peptidoglycan layer), which functions as a skeleton of a bacterial cell. A force of action is generated mainly in the direction of the long axis of the cell and is coupled to an adhesion site in the outer membrane, which interacts with the substratum. Because the coupling of the adhesion site to the surface is tight, the force generator moves to the right relative to the adhesion site, and the relative displacement is indicated by the arrows. The chemical energy that the force generator transforms into mechanical work is delivered by an energy transducer and is derived from the electrochemical ion potential across the cytoplasmic membrane.
Based on observations on the motility behavior of individual cells as well as on bead movement along the cell surface, it is generally believed that multiple motor elements exist along the cell body (see above) (45, 72, 100, 117). Because a gliding cell moves only when the force of action is generated at a rigid cellular structure such as the cytoskeleton or the cell wall, it is expected that the force-generating elements for gliding are connected with the cell wall as well as with the substratum (Fig. 11). This predicts that the motility apparatus catalyzing the conversion of chemical energy into mechanical energy may not be localized inside the cytosol. In general, ATP is not believed to be present in the periplasmic space, and in Flexibacter, gliding is most likely to be powered by the proton motive force (98). It is tempting to speculate that in M. xanthus, the energy for A-motility gliding is also directly derived from the proton motive force. A model for energy transduction between the cytoplasmic membrane and the periplasmic motor elements can be developed by analogy to the function of the energy transducer TonB. TonB, in a complex with ExbB and ExbD, mediates the active transport of iron siderophores across the outer membrane in E. coli (for a recent review, see reference 88). TonB is a cytoplasmic membrane protein that extends through the periplasmic space to contact the iron chelate receptor in the outer membrane. Energy is transduced to the receptor via proton motive force-induced conformational changes of the proteins involved. Accordingly, in A-motility system gliding, the energy of the proton motive force could be transduced by a TonB-like protein or protein complex to induce a conformational change in the gliding force generator.
It can be speculated that the products of A-motility genes include those that specify the structure of the gliding motor and gliding-specific adhesion sites, as well as those required for assembly of the structure, energy transduction, and regulation of the motor activity. A motility operates best on high-percentage agar surfaces, and A-motile cells respond to stress forces in the substrate matrix. These observations are consistent with the notion that cellular motor elements interact with rigid polymers in the substratum surface, e.g., in the process of force transmission. On a low-percentage agar surface, only few contact points are available, which could explain the reduced swarming by A motility in A+S− cells on that surface. CglB could be required for force transmission to the substratum. The residual movement observed in the A−S− (cglB pilR) mutant could be due to ineffective force transmission by other, less specific molecules. Such function of the CglB protein is consistent with its property of being transferred between cells under certain conditions.
Although presently no experimental evidence exists that the chain-like structures are the motor elements for gliding, they nevertheless fit certain requirements expected for the gliding motor (Fig. 5 and 11). For example, the superstructure is anchored to the peptidoglycan layer and is in contact with the outer membrane. The chain-like strands that assemble in larger periodic ribbon-like and belt-like structures cover the complete cell in a longitudinal fashion. A ring could, in a sense, represent a single motor element. The fundamental step in force generation of this motor element could be a conformational change of the ring structure with respect to the elongated filaments in form of a tilt. Such a tilt would result in a small displacement of the outer perimeter of a ring with respect to the elongated filament. Because the outer perimeter of a ring is postulated to be in contact with the adhesion sites in the outer membrane, the vectorial force along the long axis of the cell is transduced to the substratum. Alternatively, a tilting of the rings in a way like that described by Freese et al. (38) could lead to a local contraction of the superstructure and thus transduce the force as well. A “relaxation” of that conformational state would be required for the molecular motor to return in its prestroke state. Because each strand contains a large number of ring-like structures, a M. xanthus cell carries multiple motor elements along the complete cell body. The presence of multiple motor elements were predicted from studies of U-shaped cells and of cells that are fortuitously attached with only one cell pole. The bead movement observed along the entire cell body may also support this hypothesis. Bead movement in the opposite direction of the cell translocation may indicate that the beads tagged the relaxation to the prestroke state.
Although the above-described thoughts are very hypothetical, some specific hypotheses can be derived and tested in experiments. These questions include the following. Is the superstructure involved in gliding? Is the observed cellular structure the molecular machinery that converts chemical energy into mechanical work? How is force transmitted to the substratum? How is the activity of a motor element regulated? How is gliding speed regulated? Is it due to adjustable activities of individual motor elements or to activation of different quantities of motors that operate in an on-only/off-only mode? Furthermore, how is the gliding movement of isolated single cells coordinated to result in macroscopic A motility, and how is type IV pilus-dependent movement coordinated to result in S motility?
GLIDING MOTILITY IN FLAVOBACTERIUM JOHNSONIAE AND CYTOPHAGA STRAIN U67
Early studies on gliding motility focused on Cytophaga spp., since these cells are among the fastest of gliding bacteria, moving at speeds of up to 2 μm/s on a glass surface. Therefore, these gliders can be easily observed under the microscope. Although many similar gliding properties have been observed between Cytophaga johnsonae (which was recently reclassified as Flavobacterium johnsoniae [10]) and Cytophaga strain U67, the gliding mechanisms used by these species may be distinct. Based on studies of sensitivity to phage infection, Cytophaga strain U67 may be a close relative of F. johnsoniae (97).
Using video microscopy, Lapidus and Berg (72) conducted thorough behavioral studies on gliding movements of isolated Cytophaga strain U67 cells on the surface of glass slides. Slime trails, which were often postulated to be involved in active gliding in other organisms, were not detected in association with Cytophaga strain U67 movement. During gliding in either the forward or reverse direction, cells were observed to suddenly enter into abrupt clockwise or counterclockwise rotations (pivoting) around either cell pole at a rate of approximately 0.5 Hz; occasionally, faster pivoting was observed, sometimes for extended periods. Incomplete pivots, or flippings, where one cell pole is lifted and deposited somewhere else in the absence of complete rotations, have also been described. Cytophaga strain U67 cells were not observed to flex or to rotate during gliding. The gliding speed in Cytophaga strain U67 was demonstrated to be independent of the cell length, an observation which has also subsequently been made for M. xanthus cells (118a). Polystyrene latex beads that were found to adhere to these cells (72, 91) were used as tools to help identify potential subcellular motility elements. These spheres were observed to move from one cell pole to the other at speeds similar to the normal gliding speed of a cell. These events were independent of whether cells were in suspension or were attached to a surface (72). Bead movements could also stop and then resume in the opposite direction before they had reached the cell end. One particularly interesting observation was that multiple beads on the cell surface could move independently of each other. While one bead could be seen moving along the cell body, a second bead on the same cell could stop while a third could move in the opposite direction to the first bead. The speed of bead movement along the cell was independent of bead diameter, which ranged from 0.1 to 1.3 μm. Depletion of oxygen resulted in cessation of both gliding movements and movements of beads on cells, although, interestingly, attached spheres did not exhibit any noticeable Brownian motion. From these observations, a model of gliding for Cytophaga strain U67, in which adhesion sites in the outer membrane, which attach to the surface (or to polystyrene beads), are moved in the outer membrane on tracks that are fixed to the rigid cell wall, was proposed (72).
More recently, interference reflection microscopy was used to examine cell-substratum contact during gliding movement in Cytophaga strain U67. The entire length of the cell body was rarely seen in contact with the substratum (42). The observed cell-substratum sites were variable and moved relative to the glass substratum. Surface-exposed proteins that form physical contact with a solid substratum were reported after their identification by using substratum-immobilized 125iodide (21). This set of currently uncharacterized proteins may contain those that mediate gliding-associated cell-substratum adhesion. Cytophaga strain U67 shows some degree of curvature in cell shape and was observed in this study to rotate during movement (42). The rotation of Cytophaga strain U67 cells around the long axis was sinistral when gliding on a glass surface, and a pitch of the helix about 79° ± 3° for every 8.3 μm of translational gliding movement was observed (42). Surfactants were shown to inhibit swarming and gliding (20).
In F. johnsoniae, sulfonolipids were detected as an unusual component of the cell membrane (1). The role of these sulfonolipids was speculated to be in presenting specific polysaccharides to the outer cell surface (41, 44). An uncharacterized mutant defective in sulfonolipid formation was unable to glide or to move polystyrene beads along the cell body. In addition, the mutant was insensitive to phage infection (45). When the sulfonolipid precursor cysteate was provided to the mutant, gliding was rapidly restored. However, bead movement and phage sensitivity took longer to be restored, suggesting that the cell surface features required for bead movement and phage sensitivity are different from those required for gliding (45). A causal relationship between phage sensitivity and bead movement, on the one hand, and gliding motility, on the other, was considered unlikely (43, 45). Recently, negative chemotaxis in F. johnsoniae in response to the repellents H2O2 and OCl−, and N-chlorotaurine was reported (74).
Many motility mutants of F. johnsoniae that are defective in colony swarming have been isolated (25). One subclass of these mutants were nonmotile (“truly nonmotile”) in wet-mount preparations and exhibited other pleiotropic phenotypes such as defects in chitin digestion, phage absorption, and cell adherence (25, 135). However, research on these and other F. johnsoniae mutants was hampered by the absence of a genetic system to facilitate further molecular analyses. A breakthrough came in 1996, when McBride and Kempf reported successful mutagenesis of F. johnsoniae by using Bacteroides transposon Tn4351 (85). A plasmid, based on the cryptic plasmid pCP1 (isolated from the closely related F. psychrophilus), that stably replicates in F. johnsoniae was developed. This plasmid served as the basis for constructing a genomic library of this microorganism. Using this library, a single open reading frame, named gldA, was identified and was found to be sufficient to complement the motility defect of 4 of 61 independently isolated nonmotile mutants (2). The predicted amino acid sequence of GldA shows strong homology to ABC transport proteins. GldB and GldD are predicted to be membrane proteins. Other mutations rendering cells unable to glide map to genes encoding proteins involved in cell surface lipopolysaccharide synthesis (85a). Further use of these tools for genetic investigations of gliding in F. johnsoniae will undoubtedly uncover new insights in the gliding mechanism of this bacterium. Comparing the molecular components identified in F. johnsoniae with those in M. xanthus will significantly enhance our understanding of this translocation process and its diversity.
GLIDING MOTILITY IN FLEXIBACTER POLYMORPHUS
In contrast to the above gliding bacteria, Flexibacter polymorphus is a filamentous bacterium that forms multicellular filaments which are enclosed by a common outer membrane. Extensive studies of gliding movements of F. polymorphus filaments were conducted by Ridgway and Lewin (100). Similar to M. xanthus and Cytophaga strain U67, F. polymorphus filaments start to glide immediately once in contact with a surface (100). When suspended in liquid medium and when forming contact only with each other, F. polymorphus filaments were observed to glide relative to each other (100). Gliding of filaments on an agar surface was accompanied by a sinistral rotation of the cell body. The gliding speed of a filament is approximately 12 μm/s and is independent of the filament length. F. polymorphus filaments were observed to spin or pivot around the long axis of the cell when only one cell end was attached to the substratum. By using dextran to manipulate the viscosity of the liquid medium, it was found that the gliding speed was inversely proportional to the viscosity, suggesting that the gliding motor operates at constant torque (100). Filaments exhibited a tactile response and reversed the direction of movement by gliding backward upon contact with an object. The frequency of reversal was observed to be inversely correlated with the filament length. Polystyrene beads were observed to move across the entire cell length at approximately the same speed as the filaments glide. Bead movement occurred in a more irregular fashion than did analogous movements in Cytophaga strain U67. Small beads tended to move in a more helical fashion, whereas particles of >0.3 μm were translocated less uniformly and did not circumvent the filament. Movement of multiple beads on a single filament seemed to be independent of each other. Bead movement was also observed on nongliding filaments and filaments suspended in liquid. Because bead movement was observed in nongliding filaments, it was concluded that bead movement may not necessarily be coupled to gliding movements. Extracellular fibrils that originate laterally from the cell body are involved in cell-substratum adhesion. The chemical composition of the F. polymorphus fibrils and associated exopolysaccharide is unknown, and these fibrils may very well be different from those observed in M. xanthus. The energy to power the gliding machinery is believed to be derived from the electrochemical proton potential (98). No genetic studies have been reported for F. polymorphus.
MOTILITY IN CYANOBACTERIA
As indicated above, “gliding” is an operational definition, and considering the diversity of microorganisms capable of this type of surface translocation, different mechanisms may operate in different microbes. Two interesting observations on motility in cyanobacteria are important to mention when considering non-flagellum-based motility: swimming of nonflagellated Synechococcus, and slime extrusion in gliding of Phormidium unicatum and Anabena variabilis. In 1985, Waterbury et al. reported the isolation of several strains of the unicellular cyanobacterium Synechococcus that are capable of rapid swimming motility in the absence of flagella (128). Subsequent studies showed that their motility is dependent on a sodium gradient as the energy source (134). Furthermore, Ca2+ was found to be required for motility, and a highly abundant Ca2+ binding protein that is essential for motility was identified in the cell surface (19, 93). Self-electrophoresis was ruled out as a mechanism for this very interesting mode of movement (92). Based on theoretical considerations, a mechanism of traveling (longitudinal) surface waves was considered instead (35). A similar mechanism of wave propagation along the cell surface also presents an attractive model for single-cell gliding motility. Most behavioral observations made on gliding microorganisms (single-cell studies, bead movement, etc.), as well as the ultrastructural findings in M. xanthus, are consistent with such mechanism. However, gliding microorganisms have to be able to adhere reversibly to the substratum. Notably, M. xanthus, F. johnsoniae, Cytophaga strain U67, and F. polymorphus have not been reported to move by swimming. Research on this unusual mode of swimming in unicellular Synechococcus may provide unexpected insights into the diversity of bacterial translocation mechanisms that may affect our thinking on gliding motility as well.
Recently, the slime extrusion hypothesis of Ridgway (99) was revived by reports by Hoiczyk and Baumeister while studying the filamentous gliding cyanobacteria P. unicatum and A. variabilis (59). Using electron microscopy studies, these authors reported finding “junctional pore complexes” at the cross wall or septa of the filaments. These structures show some similarity to the type III secretion system apparatus in S. typhimurium (71). The junctional pore complexes seem to be involved in slime secretion in these cyanobacteria. As in many other gliding microorganisms, slime production is often associated with gliding. It was speculated that directional slime extrusion through the junctional pore complexes may be the motor for gliding in these cyanobacteria (59). However, no physiological evidence supports this hypothesis, and is it not clear whether slime formation is the result or cause of gliding motility.
CONCLUSIONS
The mechanism of gliding motility in prokaryotes is one of the few remaining enigmas in microbiology. Research in the past years has provided a wealth of information and revealed that several mechanistically unrelated gliding motors (pilus- dependent versus pilus-independent gliding) are involved in gliding, even in a single microorganism. Although the pilus-independent mechanisms may have different molecular architecture in the microorganisms described here, certain strikingly similar features have been observed in all the microbes. Considering the genetic, biochemical, and high-resolution optical tools available, it seems certain that in the near future we will begin to understand the molecular mechanics of single-cell gliding (pilus-independent gliding) and type IV pilus-dependent gliding.
ACKNOWLEDGMENTS
I thank Dale Kaiser, Dan Wall, and Mandy Ward for many valuable and stimulating discussions. In addition to these individuals, I am indebted to Mark McBride, Mitch Singer, Bettina Rosner, Rolf Thauer, and unknown reviewers for critical comments on the manuscript.
This work was supported by an NSF CAREER Award and Terman Fellowship.
REFERENCES
- 1.Abbant D R, Leadbetter E R, Godchaux W I, Escher A. Sulfonolipids are molecular determinants of gliding motility. Nature. 1986;324:367–369. [Google Scholar]
- 2.Agarwal S, Hunnicutt D W, McBride M J. Cloning and characterization of the Flavobacterium johnsoniae (Cytophaga johnsonae) gliding motility gene, gldA. Proc Natl Acad Sci USA. 1997;94:12139–12144. doi: 10.1073/pnas.94.22.12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alm R A, Mattick J S. Genes involved in the biogenesis and function of type-4 fimbriae in Pseudomonas aeruginosa. Gene. 1997;192:89–98. doi: 10.1016/s0378-1119(96)00805-0. [DOI] [PubMed] [Google Scholar]
- 4.Armitage J P, Schmitt R. Bacterial chemotaxis: Rhodobacter sphaeroides and Sinorhizobium meliloti: variations on a theme. Microbiology. 1997;143:3671–3682. doi: 10.1099/00221287-143-12-3671. [DOI] [PubMed] [Google Scholar]
- 5.Arnold J W, Shimkets L J. Cell surface properties correlated with cohesion in Myxococcus xanthus. J Bacteriol. 1988;170:5771–5777. doi: 10.1128/jb.170.12.5771-5777.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arnold J W, Shimkets L J. Inhibition of cell-cell interactions in Myxococcus xanthusby congo red. J Bacteriol. 1988;170:5765–5770. doi: 10.1128/jb.170.12.5765-5770.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Behmlander R M, Dworkin M. Extracellular fibrils and contact-mediated cell interactions in Myxococcus xanthus. J Bacteriol. 1991;173:7810–7821. doi: 10.1128/jb.173.24.7810-7820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Behmlander R M, Dworkin M. Biochemical and structural analyses of the extracellular matrix fibrils of Myxococcus xanthus. J Bacteriol. 1994;176:6295–6303. doi: 10.1128/jb.176.20.6295-6303.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Behmlander R M, Dworkin M. Integral proteins of the extracellular matrix fibrils of Myxococcus xanthus. J Bacteriol. 1994;176:6304–6311. doi: 10.1128/jb.176.20.6304-6311.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernardet J F, Segers P, Vancanneyt M, Kersters B F, K, Vandamme P. Cutting the Gordian knot: emended classification and description of the genus Flavobacterium, emended description of the family Flavobacteriacae, and proposal of Flavobacterium hydatis nom nov (Basonym, Cytophaga aquatilisStrohl and Tait 1978) Int J Syst Bacteriol. 1996;46:128–148. [Google Scholar]
- 11.Bitter W, Koster M, Latijnhouwers M, de Cock H, Tommassen J. Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol Microbiol. 1998;27:209–219. doi: 10.1046/j.1365-2958.1998.00677.x. [DOI] [PubMed] [Google Scholar]
- 12.Blackhart B D, Zusman D R. “Frizzy” genes of Myxococcus xanthusare involved in control of frequency of reversal of gliding motility. Proc Natl Acad Sci USA. 1985;82:8767–8770. doi: 10.1073/pnas.82.24.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blackhart B D, Zusman D R. Analysis of the products of the Myxococcus xanthus frzgenes. J Bacteriol. 1986;166:673–678. doi: 10.1128/jb.166.2.673-678.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blair D F. How bacteria sense and swim. Annu Rev Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 15.Bowden M G, Kaplan H B. The Myxococcus xanthuslipopolysaccharide O-antigen is required for social motility and multicellular development. Mol Microbiol. 1998;30:275–284. doi: 10.1046/j.1365-2958.1998.01060.x. [DOI] [PubMed] [Google Scholar]
- 16.Bradley D E. Stimulation of pilus formation in Pseudomonas aeruginosaby RNA bacteriophage adsorption. Biochem Biophys Res Commun. 1972;47:1080–1087. doi: 10.1016/0006-291x(72)90944-8. [DOI] [PubMed] [Google Scholar]
- 17.Bradley D E. A function of Pseudomonas aeruginosaPAO polar pili: twiching motility. Can J Microbiol. 1980;26:146–154. doi: 10.1139/m80-022. [DOI] [PubMed] [Google Scholar]
- 18.Bradley D E, Pitt T L. Pilus dependence of 4 Pseudomonas aeruginosabacteriophages with noncontractile tails. J Gen Virol. 1974;24:1–15. doi: 10.1099/0022-1317-24-1-1. [DOI] [PubMed] [Google Scholar]
- 19.Brahamsha B. An abundant cell-surface polypeptide is required for swimming by the nonflagellated marine cyanobacterium Synechococcus. Proc Natl Acad Sci USA. 1996;93:6504–6509. doi: 10.1073/pnas.93.13.6504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burchard R. The effect of surfactants on the motility and adhesion of gliding bacteria. Arch Microbiol. 1986;146:147–150. [Google Scholar]
- 21.Burchard R, Bloodgood R. Surface proteins of the gliding bacterium Cytophagasp. strain U67 and its mutants defective in adhesion and motility. J Bacteriol. 1990;172:3379–3387. doi: 10.1128/jb.172.6.3379-3387.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burchard R P. Gliding motility of prokaryotes: ultrastructure, physiology, and genetics. Annu Rev Microbiol. 1981;35:497–529. doi: 10.1146/annurev.mi.35.100181.002433. [DOI] [PubMed] [Google Scholar]
- 23.Chang B Y, Dworkin M. Isolated fibrils rescue cohesion and development in the dsp mutant of Myxococcus xanthus. J Bacteriol. 1994;176:7190–7196. doi: 10.1128/jb.176.23.7190-7196.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang B Y, Dworkin M. Mutants of Myxococcus xanthus dspdefective in fibril binding. J Bacteriol. 1996;178:697–700. doi: 10.1128/jb.178.3.697-700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang L Y E, Pate J L, Betzig R J. Isolation and characterization of nonspreading mutants of the gliding bacterium Cytophaga johnsonae. J Bacteriol. 1984;159:26–35. doi: 10.1128/jb.159.1.26-35.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H W, Kuspa A, Keseler I M, Shimkets L J. Physical map of the Myxococcus xanthuschromosome. J Bacteriol. 1991;173:2109–2115. doi: 10.1128/jb.173.6.2109-2115.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dana J R, Shimkets L J. Regulation of cohesion-dependent cell interactions in Myxococcus xanthus. J Bacteriol. 1993;175:3636–3647. doi: 10.1128/jb.175.11.3636-3647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darzins A. The pilG genes product required for Pseudomonas aeruginosa pilus production and twitching motility is homologous to the enteric single domain response regulator cheY. J Bacteriol. 1993;175:5934–5944. doi: 10.1128/jb.175.18.5934-5944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Darzins A. Characterization of a Pseudomonas aeruginosa gene cluster involved in pilus biosynthesis and twitching motility: sequence similarity to the chemotaxis proteins of enterics and the gliding bacterium Myxococcus xanthus. Mol Microbiol. 1994;11:137–153. doi: 10.1111/j.1365-2958.1994.tb00296.x. [DOI] [PubMed] [Google Scholar]
- 30.Darzins A, Russell M A. Molecular genetic analysis of type-4 pilus biogenesis and twitching motility using Pseudomonas aeruginosaas a model system: a review. Gene. 1997;192:109–115. doi: 10.1016/s0378-1119(97)00037-1. [DOI] [PubMed] [Google Scholar]
- 31.Dobell C E. Antoni van Leeuwenhoek and his “little animals.”. N.Y.: Dover Publications, New York; 1960. [Google Scholar]
- 32.Dworkin M, Eide D. Myxococcus xanthusdoes not respond chemotactically to moderate concentration gradients. J Bacteriol. 1983;154:437–442. doi: 10.1128/jb.154.1.437-442.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dworkin M, Kaiser D. Myxobacteria II. Washington, D.C: ASM Press; 1993. [Google Scholar]
- 34.Dworkin M, Keller K H, Weisberg D. Experimental observations consistent with a surface tension model of gliding motility of Myxococcus xanthus. J Bacteriol. 1983;155:1367–1371. doi: 10.1128/jb.155.3.1367-1371.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ehlers K M, Samuel A D T, Berg H C, Montgomery R. Do cyanobacteria swim using traveling surface waves? Proc Natl Acad Sci USA. 1996;93:8340–8343. doi: 10.1073/pnas.93.16.8340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fink J M, Zissler J F. Defects in motility and development of Myxococcus xanthuslipopolysaccharide mutants. J Bacteriol. 1989;171:2042–2048. doi: 10.1128/jb.171.4.2042-2048.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fontes M, Kaiser D. Myxococcuscells respond to elastic forces in their substrate. Proc Natl Acad Sci USA. 1999;96:8052–8057. doi: 10.1073/pnas.96.14.8052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Freese A, Reichenbach H, Lünsdorf H. Further characterization and in situ localization of chain-like aggregates of the gliding bacteria Myxococcus fulvus and Myxococcus xanthus. J Bacteriol. 1997;179:1246–1252. doi: 10.1128/jb.179.4.1246-1252.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fussenegger M, Rudel T, Barten R, Ryll R, Meyer T F. Transformation competence and type-1 pilus biogenesis in Neisseria gonorrhoeae: a review. Gene. 1997;192:125–134. doi: 10.1016/s0378-1119(97)00038-3. [DOI] [PubMed] [Google Scholar]
- 40.Geng Y, Yang Z, Downard J, Zusman D, Shi W. Methylation of FrzCD defines a discrete step in the developmental program of Myxococcus xanthus. J Bacteriol. 1998;180:5765–5768. doi: 10.1128/jb.180.21.5765-5768.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Godchaux W, Gorski L, Leadbetter E R. Outer membrane polysaccharide deficiency in two nongliding mutants of Cytophaga johnsonae. J Bacteriol. 1990;172:1250–1255. doi: 10.1128/jb.172.3.1250-1255.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Godwin S L, Fletcher M, Burchard R P. Interference reflection microscopic study of sites of association between gliding bacteria and glass substrata. J Bacteriol. 1989;171:4589–4594. doi: 10.1128/jb.171.9.4589-4594.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorski L, Godchaux W, Leadbetter E R, Wagner R. Diversity in surface features of Cytophaga johnsonaemotility mutants. J Gen Microbiol. 1992;138:1767–1772. [Google Scholar]
- 44.Gorski L, Godchaux W, Leadbetter E R. Structural specificity of sugars that inhibit gliding motility of Cytophaga johnsonae. Arch Microbiol. 1993;160:121–125. [Google Scholar]
- 45.Gorski L, Leadbetter E R, Godchaux W. Temporal sequence of the recovery of traits during phenotypic curing of a Cytophaga johnsonaemotility mutant. J Bacteriol. 1991;173:7534–7539. doi: 10.1128/jb.173.23.7534-7539.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harshey R M. Bees aren’t the only ones: swarming in Gram-negative bacteria. Mol Microbiol. 1994;13:389–394. doi: 10.1111/j.1365-2958.1994.tb00433.x. [DOI] [PubMed] [Google Scholar]
- 47.Hartzell P. Complementation of sporulation and motility defects in a prokaryote by a eukaryotic GTPase. Proc Natl Acad Sci USA. 1997;94:9881–9886. doi: 10.1073/pnas.94.18.9881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hartzell P, Kaiser D. Function of MglA, a 22-kilodalton protein essential for gliding in Myxococcus xanthus. J Bacteriol. 1991;173:7615–7624. doi: 10.1128/jb.173.23.7615-7624.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hartzell P, Kaiser D. Upstream gene of the mgl operon controls the level of MglA protein in Myxococcus xanthus. J Bacteriol. 1991;173:7625–7635. doi: 10.1128/jb.173.23.7625-7635.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hartzell P L, Youderian P. Genetics of gliding motility and development in Myxococcus xanthus. Arch Microbiol. 1995;164:309–323. doi: 10.1007/BF02529977. [DOI] [PubMed] [Google Scholar]
- 51.Henrichsen J. Bacterial surface translocation: survey and a classification. Bacteriol Rev. 1972;36:478–503. doi: 10.1128/br.36.4.478-503.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henrichsen J. The occurrance of twitching motility among Gram negative bacteria. Acta Pathol Microbiol Scand Sect B. 1975;83:171–178. doi: 10.1111/j.1699-0463.1975.tb00089.x. [DOI] [PubMed] [Google Scholar]
- 53.Henrichsen J. Twitching motility. Annu Rev Microbiol. 1983;37:81–93. doi: 10.1146/annurev.mi.37.100183.000501. [DOI] [PubMed] [Google Scholar]
- 54.Henrichsen J, Froholm L O, Bovre K. Studies on bacterial surface translocation part 2: Correlation of twitching motility and fimbriation in colony variants of Moraxella nonliquifaciens, Moraxella bovis, and Moraxella kingii. Acta Pathol Microbiol Scand Sect B. 1972;80:445–452. [PubMed] [Google Scholar]
- 55.Hodgkin J, Kaiser D. Cell-to-cell stimulation of movement in nonmotile mutants of Myxococcus. Proc Natl Acad Sci USA. 1977;74:2938–2942. doi: 10.1073/pnas.74.7.2938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus(Myxobacterales): genes controlling movement of single cells. Mol Gen Genet. 1979;171:167–176. [Google Scholar]
- 57.Hodgkin J, Kaiser D. Genetics of gliding motility in Myxococcus xanthus(Myxobacterales): two gene systems control movement. Mol Gen Genet. 1979;171:177–191. [Google Scholar]
- 58.Hodgson D A. Bacterial diversity: the range of interesting things that bacteria do. In: Hopwood D A, Chater K F, editors. Genetics of bacterial diversity. London, United Kingdom: Academic Press Ltd.; 1989. pp. 1–22. [Google Scholar]
- 59.Hoiczyk E, Baumeister W. The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria. Curr Biol. 1998;8:1161–1168. doi: 10.1016/s0960-9822(07)00487-3. [DOI] [PubMed] [Google Scholar]
- 60.Humphrey B A, Dickson M R, Marshall K C. Physiochemical and in situ observations on the adhesion of gliding bacteria to surfaces. Arch Microbiol. 1979;120:231–238. [Google Scholar]
- 61.Kaiser D. Social gliding is correlated with the presence of pili in Myxococcus xanthus. Proc Natl Acad Sci USA. 1979;76:5952–5956. doi: 10.1073/pnas.76.11.5952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kaiser D, Crosby C. Cell movement and its coordination in swarms of Myxococcus xanthus. Cell Motil. 1983;3:227–245. [Google Scholar]
- 63.Kalos M, Zissler J. Transposon tagging of genes for cell-cell interactions in Myxococcus xanthus. Proc Natl Acad Sci USA. 1990;87:8316–8320. doi: 10.1073/pnas.87.21.8316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaplan H B, Kuspa A, Kaiser D. Suppressors that permit A-signal-independent developmental gene expression in Myxococcus xanthus. J Bacteriol. 1991;173:1460–1470. doi: 10.1128/jb.173.4.1460-1470.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kashefi K, Hartzell P. Genetic suppression and phenotypic masking of a Myxococcus xanthus frzF−defect. Mol Microbiol. 1995;15:483–494. doi: 10.1111/j.1365-2958.1995.tb02262.x. [DOI] [PubMed] [Google Scholar]
- 66.Kearns D B, Shimkets L J. Chemotaxis in a gliding bacterium. Proc Natl Acad Sci USA. 1998;95:11957–11962. doi: 10.1073/pnas.95.20.11957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Keller K H, Grady M, Dworkin M. Surface tension gradient: feasible model for gliding motility of Myxococcus xanthus. J Bacteriol. 1983;155:1358–1366. doi: 10.1128/jb.155.3.1358-1366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Koga T, Ishimoto K, Lory S. Genetic and functional characterization of the gene cluster specifying expression of Pseudomonas aeruginosapili. Infect Immun. 1993;61:1371–1377. doi: 10.1128/iai.61.4.1371-1377.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Koster M, Bitter W, De Cock H, Allaoui A, Cornelis G R, Tommassen J. The outer membrane component, YscC, of the Yop secretion machinery of Yersinia enterocoliticaforms a ring-shaped multimeric complex. Mol Microbiol. 1997;26:789–797. doi: 10.1046/j.1365-2958.1997.6141981.x. [DOI] [PubMed] [Google Scholar]
- 70.Kroos L, Hartzell P, Stephens K, Kaiser D. A link between cell movement and gene expression argues that motility is required for cell-cell signaling during fruiting body development. Genes Dev. 1988;2:1677–1685. doi: 10.1101/gad.2.12a.1677. [DOI] [PubMed] [Google Scholar]
- 71.Kubori T, Matsushima Y, Nakamura D, Uralil J, Lara-Tejero M, Sukhan A, et al. Supramolecular structure of the Salmonella typhimuriumtype III protein secretion system. Science. 1998;280:602–605. doi: 10.1126/science.280.5363.602. [DOI] [PubMed] [Google Scholar]
- 72.Lapidus I R, Berg H C. Gliding motility of Cytophagasp. strain U67. J Bacteriol. 1982;151:384–398. doi: 10.1128/jb.151.1.384-398.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lauer P, Albertson N H, Koomey M. Conservation of genes encoding components of a type IV pilus assembly-two-step protein export pathway in Neisseria gonorrhoeae. Mol Microbiol. 1993;8:357–368. doi: 10.1111/j.1365-2958.1993.tb01579.x. [DOI] [PubMed] [Google Scholar]
- 74.Liu Z, Fridovich I. Negative chemotaxis in Cytophaga johnsonae. Can J Microbiol. 1996;42:515–518. doi: 10.1139/m96-069. [DOI] [PubMed] [Google Scholar]
- 75.Lünsdorf H, Reichenbach H. Ultrastructural details of the apparatus of gliding motility of Myxococcus fulvus(Myxobacterales) J Gen Microbiol. 1989;135:1633–1641. [Google Scholar]
- 76.MacNeil S D, Calara F, Hartzell P L. New clusters of genes required for gliding motility in Myxococcus xanthus. Mol Microbiol. 1994;14:61–71. doi: 10.1111/j.1365-2958.1994.tb01267.x. [DOI] [PubMed] [Google Scholar]
- 77.MacNeil S D, Mouzeyan A, Hartzell P L. Genes required for both gliding motility and development in Myxococcus xanthus. Mol Microbiol. 1994;14:785–795. doi: 10.1111/j.1365-2958.1994.tb01315.x. [DOI] [PubMed] [Google Scholar]
- 78.MacRae T H, McCurdy D. Evidence for motility-related fimbriae in the gliding microorganism Myxococcus xanthus. Can J Microbiol. 1976;22:1589–1593. doi: 10.1139/m76-234. [DOI] [PubMed] [Google Scholar]
- 79.MacRae T H, Dobson W J, McCurdy H D. Fimbriation in gliding bacteria. Can J Microbiol. 1977;23:1096–1108. doi: 10.1139/m77-165. [DOI] [PubMed] [Google Scholar]
- 80.Maraca I G, Lounsbury K M, Richards S A, McKiernan C, Bar-Sagi D. The Ras superfamily of GTPases. FASEB J. 1996;10:625–630. doi: 10.1096/fasebj.10.5.8621061. [DOI] [PubMed] [Google Scholar]
- 81.McBride M J, Hartzell P L, Zusman D A. Motility and tactic behavior of Myxococcus xanthus. In: Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, D.C: ASM Press; 1993. pp. 285–305. [Google Scholar]
- 82.McBride M J, Kohler T, Zusman D R. Methylation of FrzCD, a methyl-accepting taxis protein of Myxococcus xanthus, is correlated with factors affecting cell behavior. J Bacteriol. 1992;174:4246–4257. doi: 10.1128/jb.174.13.4246-4257.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.McBride M J, Weinberg R A, Zusman D R. “Frizzy” aggregation genes of the gliding bacterium Myxococcus xanthusshow sequence similarities to the chemotaxis genes of enteric bacteria. Proc Natl Acad Sci USA. 1989;86:424–428. doi: 10.1073/pnas.86.2.424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McBride M J, Zusman D R. FrzCD, a methyl-accepting taxis protein from Myxococcus xanthus, shows modulated methylation during fruiting body formation. J Bacteriol. 1993;175:4936–4940. doi: 10.1128/jb.175.15.4936-4940.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.McBride M L, Kempf M J. Development of techniques for the genetic manipulation of the gliding bacterium Cytophaga johnsonae. J Bacteriol. 1996;178:583–590. doi: 10.1128/jb.178.3.583-590.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85a.McBride, M. L. Personal communication.
- 86.McCleary W R, McBride M J, Zusman D R. Developmental sensory transduction in Myxococcus xanthusinvolves methylation and demethylation of FrzCD. J Bacteriol. 1990;172:4877–4887. doi: 10.1128/jb.172.9.4877-4887.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McCleary W R, Zusman D R. Purification and characterization of the Myxococcus xanthusFrzE protein shows that it has autophosphorylation activity. J Bacteriol. 1990;172:6661–6668. doi: 10.1128/jb.172.12.6661-6668.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moeck G S, Coulton J W. TonB-dependent iron acquisition: mechanisms of siderophore-mediated active transport. Mol Microbiol. 1998;28:675–681. doi: 10.1046/j.1365-2958.1998.00817.x. [DOI] [PubMed] [Google Scholar]
- 88a.Morandi, D. Personal communication.
- 89.Nowlin D M, Bollinger J, Hazelbauer G L. Site-directed mutations altering methyl-accepting residues of a sensory transducer protein. Proteins Struct Funct Genet. 1988;3:102–112. doi: 10.1002/prot.340030205. [DOI] [PubMed] [Google Scholar]
- 90.Nunn D, Bergman S, Lory S. Products of three accessory genes, pilB, pilC, and pilD, are required for biogenesis of Pseudomonas aeruginosapili. J Bacteriol. 1990;172:2911–2919. doi: 10.1128/jb.172.6.2911-2919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Pate J L, Chang L-Y E. Evidence that gliding motility in prokaryotic cells is driven by rotary assemblies in the cell envelope. Curr Microbiol. 1979;2:59–64. [Google Scholar]
- 92.Pitta T P, Berg H C. Self-electrophoresis is not the mechanism for motility in swimming cyanobacteria. J Bacteriol. 1995;177:5701–5703. doi: 10.1128/jb.177.19.5701-5703.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pitta T P, Sherwood E E, Kobel A M, Berg H C. Calcium is required for swimming by the nonflagellated cyanobacterium Synechococcusstrain WH8113. J Bacteriol. 1997;179:2524–2528. doi: 10.1128/jb.179.8.2524-2528.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ramaswamy S, Dworkin M, Downard J. Identification and characterization of Myxococcus xanthusmutants deficient in calcofluor white binding. J Bacteriol. 1997;179:2863–2871. doi: 10.1128/jb.179.9.2863-2871.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Reichenbach H. Biology of myxobacteria: ecology and taxonomy. In: Dworkin M, Kaiser D, editors. Myxobacteria II. Washington, D.C: ASM Press; 1993. pp. 13–62. [Google Scholar]
- 96.Reichenbach H. The ecology of the myxobacteria. Environ Microbiol. 1999;1:15–21. doi: 10.1046/j.1462-2920.1999.00016.x. [DOI] [PubMed] [Google Scholar]
- 97.Richter C A, Pate J L. Temperate phages and bacteriocins of the gliding bacterium Cytophaga johnsonae. J Gen Microbiol. 1988;134:253–262. doi: 10.1099/00221287-134-2-253. [DOI] [PubMed] [Google Scholar]
- 98.Ridgway H F. Source of energy for gliding motility in Flexibacter polymorphus: effects of metabolic and respiratory inhibitors on gliding movement. J Bacteriol. 1977;131:544–556. doi: 10.1128/jb.131.2.544-556.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ridgway H F. Ultrastructural characterization of goblet shaped particles from the cell wall of Flexibacter polymorphus. Rev Can Microbiol. 1977;23:1201–1213. doi: 10.1139/m77-181. [DOI] [PubMed] [Google Scholar]
- 100.Ridgway H F, Lewin R A. Characterization of gliding motility in Flexibacter polymorphus. Cell Motil Cytoskeleton. 1988;11:46–63. doi: 10.1002/cm.970110106. [DOI] [PubMed] [Google Scholar]
- 101.Rodriguez A, Spormann A M. Genetic and molecular characterization of cglB, a gene essential for single-cell gliding in Myxococcus xanthus. J Bacteriol. 1999;181:4381–4390. doi: 10.1128/jb.181.14.4381-4390.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rodriguez-Soto J P, Kaiser D. Identification and localization of the Tgl protein, which is required for Myxococcus xanthussocial motility. J Bacteriol. 1997;179:4372–4381. doi: 10.1128/jb.179.13.4372-4381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rodriguez-Soto J P, Kaiser D. The tgl gene: social motility and stimulation in Myxococcus xanthus. J Bacteriol. 1997;179:4361–4371. doi: 10.1128/jb.179.13.4361-4371.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rosenberg E, Keller K H, Dworkin M. Cell density-dependent growth of Myxococcus xanthuson casein. J Bacteriol. 1977;129:770–777. doi: 10.1128/jb.129.2.770-777.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shi W, Kohler T, Zusman D R. Chemotaxis plays a role in the social behaviour of Myxococcus xanthus. Mol Microbiol. 1993;9:601–611. doi: 10.1111/j.1365-2958.1993.tb01720.x. [DOI] [PubMed] [Google Scholar]
- 106.Shi W, Ngok F, Zusman D R. Cell density regulates cellular reversal frequency in Myxococcus xanthus. Proc Natl Acad Sci USA. 1996;93:4142–4146. doi: 10.1073/pnas.93.9.4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shi W, Zusman D R. The two motility systems of Myxococcus xanthusshow different selective advantages on various surfaces. Proc Natl Acad Sci USA. 1993;90:3378–3382. doi: 10.1073/pnas.90.8.3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Shimkets L J. Correlation of energy-dependent cell cohesion with social motility in Myxococcus xanthus. J Bacteriol. 1986;166:837–841. doi: 10.1128/jb.166.3.837-841.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shimkets L J. Role of cell cohesion in Myxococcus xanthusfruiting body formation. J Bacteriol. 1986;166:842–848. doi: 10.1128/jb.166.3.842-848.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shimkets L J. Social and developmental biology of the myxobacteria. Microbiol Rev. 1990;54:473–501. doi: 10.1128/mr.54.4.473-501.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Shimkets L J, Kaiser D. Induction of coordinated movement of Myxococcus xanthuscells. J Bacteriol. 1982;152:451–461. doi: 10.1128/jb.152.1.451-461.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sogaard-Andersen L, Kaiser D. C factor, a cell-surface-associated intercellular signaling protein, stimulates the cytoplasmic Frz signal transduction system in Myxococcus xanthus. Proc Natl Acad Sci USA. 1996;93:2675–2679. doi: 10.1073/pnas.93.7.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sogaard-Andersen L, Slack F J, Kimsey H, Kaiser D. Intercellular C-signaling in Myxococcus xanthusinvolves a branched signal transduction pathway. Genes Dev. 1996;10:740–754. doi: 10.1101/gad.10.6.740. [DOI] [PubMed] [Google Scholar]
- 114.Sohel I, Puente J L, Ramer S W, Bieber D, Wu C Y, Schoolnik G K. Enteropathogenic Escherichia coli: identification of a gene cluster coding for bundle-forming pilus morphogenesis. J Bacteriol. 1996;178:2613–2628. doi: 10.1128/jb.178.9.2613-2628.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sourjik V, Schmitt R. Different roles of CheY1 and CheY2 in the chemotaxis of Rhizobium meliloti. Mol Microbiol. 1996;22:427–436. doi: 10.1046/j.1365-2958.1996.1291489.x. [DOI] [PubMed] [Google Scholar]
- 116.Sourjik V, Schmitt R. Phosphotransfer between CheA, CheY1, and CheY2 in the chemotaxis signal transduction chain of Rhizobium meliloti. Biochemistry. 1998;37:2327–2335. doi: 10.1021/bi972330a. [DOI] [PubMed] [Google Scholar]
- 117.Spormann A M, Kaiser D. Gliding movements in Myxococcus xanthus. J Bacteriol. 1995;177:5846–5852. doi: 10.1128/jb.177.20.5846-5852.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Spormann A M, Kaiser D. Gliding mutants of Myxococcus xanthuswith high reversal frequency and small displacements. J Bacteriol. 1999;181:2593–2601. doi: 10.1128/jb.181.8.2593-2601.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118a.Spormann, A. M. Unpublished data.
- 119.Stanier R. A note on elasticotaxis in myxobacteria. J Bacteriol. 1942;44:405–412. doi: 10.1128/jb.44.4.405-412.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Stephens K, Hartzell P, Kaiser D. Gliding motility in Myxococcus xanthus: mgllocus, RNA, and predicted protein products. J Bacteriol. 1989;171:819–830. doi: 10.1128/jb.171.2.819-830.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stephens K, Kaiser D. Genetics of gliding motility in Myxococcus xanthus: Molecular cloning of the mgllocus. Mol Gen Genet. 1987;207:256–266. [Google Scholar]
- 122.Tieman S, Koch A, White D. Gliding motility in slide cultures of Myxococcus xanthusin stable and steep chemical gradients. J Bacteriol. 1996;178:3480–3485. doi: 10.1128/jb.178.12.3480-3485.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Trudeau K G, Ward M J, Zusman D R. Identification and characterization of frzZ, a novel response regulator necessary for swarming and fruiting-body formation in Myxococcus xanthus. Mol Microbiol. 1996;20:645–655. doi: 10.1046/j.1365-2958.1996.5521075.x. [DOI] [PubMed] [Google Scholar]
- 124.Wall D, Kaiser D. Alignment enhances the cell-to-cell transfer of pilus phenotype. Proc Natl Acad Sci USA. 1998;95:3054–3058. doi: 10.1073/pnas.95.6.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wall D, Kohlenbrander P E, Kaiser D. The Myxococcus xanthus pilQ (sglA) gene encodes a sectretin homolog required for type IV pilus biogenesis, social motility and development. J Bacteriol. 1999;181:24–33. doi: 10.1128/jb.181.1.24-33.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wall D, Wu S S, Kaiser D. Contact stimulation of Tgl and type IV pili in Myxococcus xanthus. J Bacteriol. 1998;180:759–761. doi: 10.1128/jb.180.3.759-761.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ward M J, Zusman D R. Regulation of directed motility in Myxococcus xanthus. Mol Microbiol. 1997;24:885–893. doi: 10.1046/j.1365-2958.1997.4261783.x. [DOI] [PubMed] [Google Scholar]
- 127a.Ward, M. J. Personal communication.
- 128.Waterbury J B, Willey J M, Frank D G, Valois F W, Watson S W. A cyanobacterium capable of swimming motility. Science. 1985;230:74–76. doi: 10.1126/science.230.4721.74. [DOI] [PubMed] [Google Scholar]
- 129.Weimer R M, Creighton C, Stassinopoulos A, Youderian P, Hartzell P. A chaperone in the HSP70 family controls production of extracellular fibrils in Myxococcus xanthus. J Bacteriol. 1998;180:5357–5368. doi: 10.1128/jb.180.20.5357-5368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Weinberg R A, Zusman D R. Evidence that the Myxococcus xanthus frzgenes are developmentally regulated. J Bacteriol. 1989;171:6174–6186. doi: 10.1128/jb.171.11.6174-6186.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Whitchurch C B, Alm R A, Mattick J S. The alginate regulator AlgR and an associated sensor FimS are required for twitching motility in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1996;93:18. doi: 10.1073/pnas.93.18.9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Whitchurch C B, Hobbs M, Livingston S P, Krishnapillai V, Mattick J S. Characterization of a Pseudomonas aeruginosatwitching motility gene and evidence for a specialised protein export system widespread in eubacteria. Gene. 1990;101:33–44. doi: 10.1016/0378-1119(91)90221-v. [DOI] [PubMed] [Google Scholar]
- 133.Whitchurch C B, Mattick J S. Characterization of a gene, pilU, required for twitching motility but not phage sensitivity in Pseudomonas aeruginosa. Mol Microbiol. 1994;13:1079–1091. doi: 10.1111/j.1365-2958.1994.tb00499.x. [DOI] [PubMed] [Google Scholar]
- 134.Willey J M, Waterbury J B, Greenberg E P. Sodium-dependent motility in a swimming cyanobacterium. J Bacteriol. 1987;169:3429–3434. doi: 10.1128/jb.169.8.3429-3434.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wolkin R, Pate J. Phage adsorption and cell adherence are motility-dependent characteristics of the gliding bacterium Cytophaga johnsonae. J Gen Microbiol. 1986;132:355–367. [Google Scholar]
- 136.Womack B J, Gilmore D F, White D. Calcium requirement for gliding motility in myxobacteria. J Bacteriol. 1989;171:6093–6096. doi: 10.1128/jb.171.11.6093-6096.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wu S S, Kaiser D. Genetic and functional evidence that type IV pili are required for social motility in Myxococcus xanthus. Mol Microbiol. 1995;18:547–558. doi: 10.1111/j.1365-2958.1995.mmi_18030547.x. [DOI] [PubMed] [Google Scholar]
- 138.Wu S S, Kaiser D. Markerless deletion of pil genes in Myxococcus xanthus generated by counterselection with the Bacillus subtilis sacBgene. J Bacteriol. 1996;178:5817–5821. doi: 10.1128/jb.178.19.5817-5821.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Wu S S, Kaiser D. The Myxococcus xanthus pilTlocus is required for social gliding although pili are still produced. Mol Microbiol. 1997;23:109–121. doi: 10.1046/j.1365-2958.1997.1791550.x. [DOI] [PubMed] [Google Scholar]
- 140.Wu S S, Kaiser D. Regulation of expression of the pilA gene of Myxococcus xanthus. J Bacteriol. 1997;179:7748–7758. doi: 10.1128/jb.179.24.7748-7758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wu S S, Wu J, Cheng Y L, Kaiser D. The pilH gene encodes an ABC transporter homologue required for type IV pilus biogenesis and social gliding motility in Myxococcus xanthus. Mol Microbiol. 1998;29:1249–1261. doi: 10.1046/j.1365-2958.1998.01013.x. [DOI] [PubMed] [Google Scholar]
- 142.Yang Z, Geng Y, Xu D, Kaplan H B, Shi W. A new set of chemotaxis homologues is essential for Myxococcus xanthussocial motility. Mol Microbiol. 1998;22:528–536. doi: 10.1046/j.1365-2958.1998.01160.x. [DOI] [PubMed] [Google Scholar]
- 143.Youderian P. Bacterial motility: secretory secrets of gliding bacteria. Curr Biol. 1998;8:R408–R411. doi: 10.1016/s0960-9822(98)70264-7. [DOI] [PubMed] [Google Scholar]
- 144.Zusman D R. “Frizzy” mutants: a new class of aggregation-defective developmental mutants of Myxococcus xanthus. J Bacteriol. 1982;150:1430–1437. doi: 10.1128/jb.150.3.1430-1437.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zusman D R, McBride M J. Sensory transduction in the gliding bacterium Myxococcus xanthus. Mol Microbiol. 1991;5:2323–2329. doi: 10.1111/j.1365-2958.1991.tb02077.x. [DOI] [PubMed] [Google Scholar]