Abstract
Background & Aims
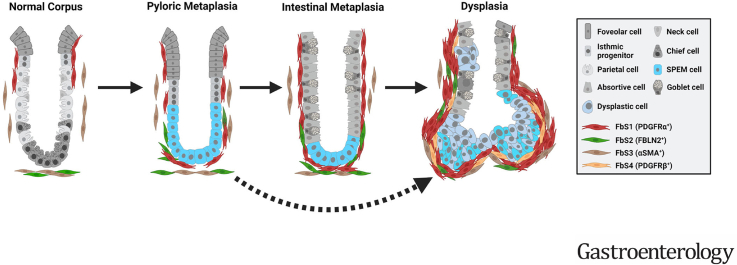
Elements of field cancerization, including atrophic gastritis, metaplasia, and dysplasia, promote gastric cancer development in association with chronic inflammation. However, it remains unclear how stroma changes during carcinogenesis and how the stroma contributes to progression of gastric preneoplasia. Here we investigated heterogeneity of fibroblasts, one of the most important elements in the stroma, and their roles in neoplastic transformation of metaplasia.
Methods
We used single-cell transcriptomics to evaluate the cellular heterogeneity of mucosal cells from patients with gastric cancer. Tissue sections from the same cohort and tissue microarrays were used to identify the geographical distribution of distinct fibroblast subsets. We further evaluated the role of fibroblasts from pathologic mucosa in dysplastic progression of metaplastic cells using patient-derived metaplastic gastroids and fibroblasts.
Results
We identified 4 subsets of fibroblasts within stromal cells defined by the differential expression of PDGFRA, FBLN2, ACTA2, or PDGFRB. Each subset was distributed distinctively throughout stomach tissues with different proportions at each pathologic stage. The PDGFRα+ subset expanded in metaplasia and cancer compared with normal, maintaining a close proximity with the epithelial compartment. Co-culture of metaplasia- or cancer-derived fibroblasts with gastroids showing the characteristics of spasmolytic polypeptide-expressing metaplasia–induced disordered growth, loss of metaplastic markers, and increases in markers of dysplasia. Culture of metaplastic gastroids with conditioned media from metaplasia- or cancer-derived fibroblasts also promoted dysplastic transition.
Conclusions
These findings indicate that fibroblast associations with metaplastic epithelial cells can facilitate direct transition of metaplastic spasmolytic polypeptide-expressing metaplasia cell lineages into dysplastic lineages.
Keywords: Gastric Carcinogenesis, Fibroblasts, Metaplasia, SPEM, PDGFRA
Graphical abstract
Altered stromal fibroblasts in the precancerous gastric lining promote the transition of precancerous metaplastic cells into dysplasia, supporting the development of gastric cancer.
What You Need to Know.
Background and Context
Gastric cancer develops in a field of precancerous metaplasia and alterations in stromal elements. Little is known about the influence of fibroblast populations on the progression of metaplasia to dysplasia.
New Findings
Normal and pathologic gastric tissues showed 4 populations of fibroblasts, and co-culture of metaplasia-derived or cancer-derived fibroblasts with metaplastic gastroids induced progression to dysplasia.
Limitations
The mode of action for each fibroblast subset involved in precancerous progression needs to be clarified further.
Clinical Research Relevance
These investigations demonstrate that fibroblasts in the precancerous gastric mucosa can promote the development of dysplasia from metaplasia.
Basic Research Relevance
The results show that PDGFR + fibroblasts intimately associated with gastric metaplasia cells. Co-culture of metaplastic gastroids with the characteristics of spasmolytic polypeptide-expressing metaplasia with either metaplasia-derived or cancer-derived fibroblasts caused loss of metaplastic markers CD44v9 and AQP5 expression and gain of dysplasia markers TROP2 and CEACAM5 expression.
Intestinal-type gastric cancer usually develops through well-defined intermediate stages leading to carcinogenesis.1,2 Mucosal injury, characterized by loss of glandular elements, especially parietal cells, triggers chronic atrophic gastritis with alterations in immune cells and fibroblasts at the injured site, followed by sequential precancerous transitions, including metaplasia and dysplasia. The following 2 types of metaplasia have been identified in the corpus of the stomach: intestinal metaplasia (characterized by the presence of intestinal goblet cells)3 and pyloric metaplasia (characterized by the presence of spasmolytic polypeptide-expressing metaplasia [SPEM] lineage cells with features of deep antral gland cells).4 Intestinal metaplasia can be further subclassified as complete or incomplete, with incomplete intestinal metaplasia representing the highest risk for progression to cancer.5 Several studies have elucidated how normal stomach transitions to premalignancy in mouse models in response to genetic modification, pharmacologically induced parietal cell loss, or chronic Helicobacter spp infection.6, 7, 8
In most of the experimental models and patients with gastric cancer, interactions between elements of the microenvironment and epithelial cells seem to be crucial in disease initiation as well as progression. The microenvironment is composed of a field of heterogenous cell types, including immune cells, fibroblasts, and endothelial cells, which coordinate homeostasis in the normal gastric mucosa.9, 10, 11 Fibroblasts play an important role in establishing and organizing extracellular matrix, and are also a main source of soluble factors, such as WNT ligands, several cytokines and chemokines, and growth factors.12,13 Cancer-associated fibroblasts (CAFs) are thought to promote the expansion and migration of cancer cells in pancreatic ductal adenocarcinoma12 and breast cancer.14 Recent transcriptomic analyses at the single-cell level have revealed the cellular and molecular heterogeneity of epithelial and stromal lineages and the dramatic alterations in cell composition during gastric carcinogenesis in both humans and mice.15, 16, 17, 18 However, no previous studies have described the geographical distribution of diverse fibroblast subsets (FbSs) and their precise roles in development and progression of preneoplastic lesions in human stomach.
In this study, we analyzed mucosal cells from patients with gastric cancer. Based on single-cell RNA-sequencing (scRNA-seq) data analysis of fibroblasts from patients and immunostaining, 4 distinct FbSs were identified across normal, metaplasia, and dysplasia, or cancer with differential geographic distributions within tissues. We have also directly evaluated the role of fibroblasts in the progression of gastric preneoplasia using 2 different methodologies for 3-dimensional and Transwell air–liquid interface (ALI) co-cultures of metaplastic gastroids with fibroblasts isolated from tissues of patients with gastric cancer. These functional studies revealed that metaplasia- or cancer-derived fibroblasts promote metaplasia progression to dysplasia.
Methods
Ethics Declaration
The research protocol governing management and stewardship of patient tissue detailed in this article was approved by the McGill University Health Care Research Ethics Board (2007-856). All patient information was anonymized and deidentified before experiments and the clinicopathologic information, such as tumor grade and clinical stage, is provided in Supplementary Table 1.
Bioinformatic Methods
The entire workflow was implemented using Singularity containers for reproducibility (all codes available, please contact authors for information).
Three-Dimensional Co-Culture and Transwell Air–Liquid Interface Co-Culture
For 3-dimensional co-culture in Matrigel, separately maintained fibroblasts and gastroids were collected as described above and suspended in 30 μL of Matrigel/type I collagen mixture. Approximately 60,000 fibroblasts in 15 μL of Matrigel/collagen mixture were mixed with small chunks of gastroids in 15 μL of Matrigel/collagen mixture. A total 30 μL of final mixture per well was seeded on a 48-well plate for live imaging or an 8-well chamber slide for whole-mount staining, and then 300 μL of IntestiCult medium was added to each well after polymerization of Matrigel/collagen mixture for 30 minutes at 37°C. Phase-contrast live imaging was performed on the JuLI stage, a Real-Time Cell History Recorder (NanoEntek), and an EVOS M7000 inverted microscope (ThermoFisher).
ALI cultures were performed in 24-well plate Transwells (Corning). Transwell filters were previously coated by adding 300 μL of type I collagen (2.7 mg/cm2) and incubating for 30 minutes at 37°C. After the incubation, the remaining collagen liquid was removed to air dry the filters. Fibroblasts derived from normal, metaplasia, or cancer were harvested and seeded (approximately 60,000 cells in 200 μL of fibroblast medium) on the collagen-coated filters. Three hundred microliters of fibroblast medium were added to the well under the Transwell filter. Fibroblasts were incubated at 37°C, 5% CO2 in a humidified incubator for 6 hours. During that time, gastroids were incubated with Organoid Harvesting Solution (R&D Systems) for 30 minutes at 4°C to remove Matrigel (ECM; Corning). After incubation, gastroids were centrifuged and the supernatant was removed. The pellet was resuspended in TrypLE (Gibco) to obtain single-cell suspensions. A total of 90,000 cells of gastroids in approximately 200 μL of human IntestiCult medium (StemCell Technology) were seeded onto the top of a collagen-coated Transwell filter with or without fibroblasts (gastroid only). Fibroblast medium under the Transwell filter was replaced with 300 μL of human IntestiCult medium. Top and bottom IntestiCult medium were changed after 48 hours. Seeded gastroid cells were left for 7 days to grow on top of collagen-coated Transwells layered with or without fibroblasts. On day 7, the media overlying the cells were removed to expose the surface of the cells to air and the ALI process lasted 14 days. The media at the bottom wells were replaced every 3 days and fluid secreted from the cells was removed every day from the top Transwells.
Further detailed methods are included in the Supplementary Methods.
Results
Single-Cell RNA Sequencing Reveals Cellular Heterogeneity of Human Gastric Mucosal Cell Lineages
To profile cellular and molecular heterogeneity during gastric carcinogenesis, we performed scRNA-seq on the adjacent normal (gastritis), metaplastic, dysplastic, or cancerous stomach tissues from 5 patients with gastric cancer. We analyzed a total of 72,126 cells after quality control from freshly prepared tissues, based on the following lineage-specific genes: CLDN18 and EPCAM for gastric epithelial cells; PTPRC (encoding CD45; total hematopoietic cells), CD3D (T cells), CD79A (B cells), CSF1R (macrophages), CSF3R (granulocytes), and TPSAB1 (mast cells) for immune cells; COL1A1 and ACTA2 for fibroblasts and myofibroblasts; vWF and PECAM1 for vascular endothelial cells; and NRXN1 and GPM6B for neural cells (Supplementary Figure 1A–G).
We next examined subsets within gastric epithelial cells. As shown in Supplementary Figure 2A and B and Supplementary Figure 3A, CLDN18- and/or EPCAM-expressing epithelial clusters in the Uniform Manifold Approximation and Projection of total cells were further divided into 9 different groups with several undefined clusters, including gastric, intestinal, and dysplastic or cancer cell lineages. We also identified a dual AQP5+/MUC6+ SPEM-lineage population in cluster 2, a major component of pyloric metaplasia, as reported previously.7 Notably, compared with the chief cell cluster (cluster 3), transdifferentiating (cluster 16) and SPEM cells in cluster 2 displayed graded decreases in expression of PGA3, PGA4, and PGA5 with sequential increases of AQP5, MUC6, and CLU (Supplementary Figure 2C). Intestinal metaplasia lineage clusters showed the characteristics of normal intestinal cell types, including VIL1+ absorptive cells (cluster 6), DMBT1+ intestinal metaplastic intermediate cells (clusters 5 and 7), TFF3+ and/or MUC2+ goblet cells (cluster 18), and 2 different types of enteroendocrine cells (clusters 12 and 13), but DEFA5+ Paneth cells were rarely observed (Supplementary Figure 3A). In addition, we identified a distinctive intestinal cell lineage purely composed of CFTR+ and BEST4+ cells (cluster 21), known to reside in the villus of human small intestine, but whose function is not well-understood (Supplementary Figure 3A).19,20 Compared with other lineages, the intestinal metaplasia clusters 5, 6, 7, and 18 highly expressed DMBT1, CLDN7, MUC17, ANPEP, REG4, MUC3A, KRT20, PHGR1, CDHR2, and CDHR5 (Supplementary Figure 2C).
Dysplastic Cell Lineages Specifically Express CEACAM5
To understand the molecular heterogeneity of the cells at the final step in gastric carcinogenesis and identify potential markers for dysplastic cell lineages, we focused on clusters 11 and 14, which were designated as dysplastic or cancer cell clusters on the basis of the expression of TACSTD2, the transcript for TROP2 protein (Supplementary Figure 2C and Supplementary Figure 3B).21 These 2 clusters were also enriched with OLFM4, CLDN3, CLDN4, EPCAM, CD9, MUC13, AQP3, CEACAM5, and CEACAM6 (Supplementary Figure 2C and Supplementary Figure 3B). A previous report described increased expression of CLDN4 during gastric carcinogenesis.22 We observed that CLDN3, CLDN4, and CLDN7 transcripts were up-regulated in most of the dysplastic or cancer cells, although these genes were also expressed in some of the intestinal cell lineages (Supplementary Figure 2C and Supplementary Figure 3B and C). In addition, we found several markers that can more specifically define dysplastic or cancer clusters. Matrix metallopeptidase 7 has been implicated in invasion and metastasis of tumor cells and is mainly expressed in epithelial cells.23 Accordingly, MMP7 transcripts in the present data set very specifically marked a portion of cluster 11 (Supplementary Figure 3C). CEACAM5 and CEACAM6 identically defined the remaining portion of cluster 11 and the whole cluster 14 (Supplementary Figure 3C).
We next evaluated these markers for dysplastic lineages at the protein level in tissue microarrays containing 24 cores, including the following 36 regions of interest in human early gastric cancer resections: 5 regions for adjacent normal, 7 regions for incomplete intestinal metaplasia, 11 regions for dysplasia, and 13 regions for intestinal-type gastric cancer. Although TROP2, CLDN7, and CEACAM5 were completely negative in adjacent normal regions, more than one-half of the regions of incomplete intestinal metaplasia, dysplasia, and gastric cancer expressed TROP2 (71.4%, 54.5%, and 53.8%, respectively) and/or CLDN7 (85.7%, 63.6%, and 63.6%, respectively) (Supplementary Figure 2D and E). CEACAM5, known as a marker indicating poor prognosis of gastric cancer,24 was weakly expressed in only 28.6% of incomplete intestinal metaplasia regions. However, in accordance with our transcriptomic data, dysplastic and cancerous cells more strongly and frequently showed apical CEACAM5 expression (90.9% and 76.9%, respectively) (Supplementary Figure 2D and E).
Single-Cell RNA Sequencing Identifies Distinct Fibroblast Subsets at Each Stage of Gastric Carcinogenesis
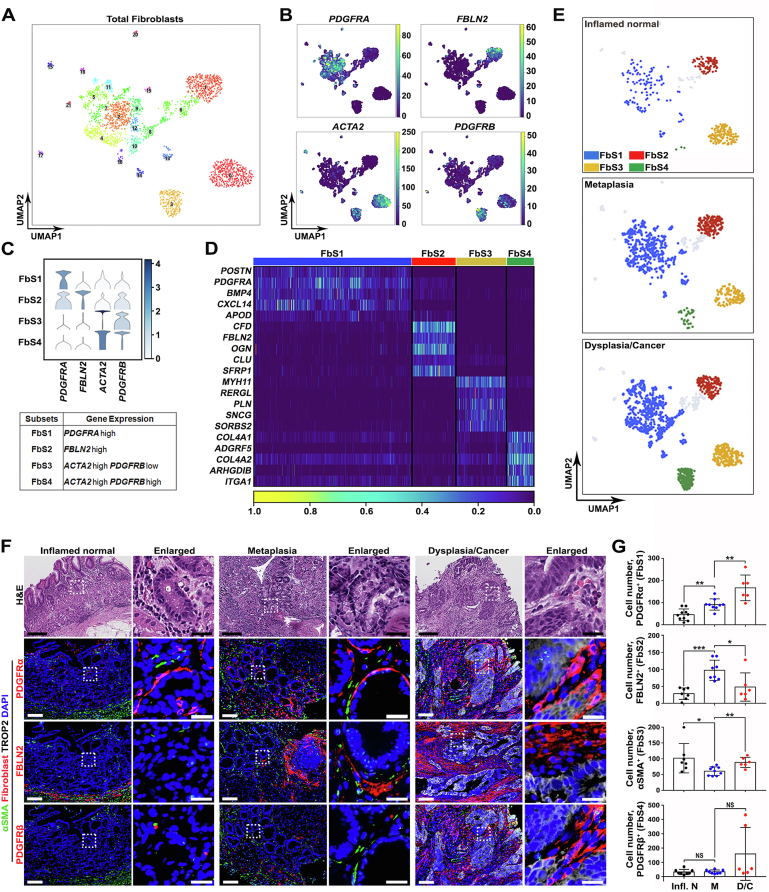
We analyzed the transcriptomic profiles of fibroblasts to identify whether distinct subsets exist in the stomach, as described in pancreas or breast tissues.25,26 Unsupervised clustering resolved a total of 21 fibroblast clusters that could be assigned to 4 large subsets—FbS1, PDGFRAhi; FbS2, FBLN2hi; FbS3, ACTA2hi PDGFRBlo; and FbS4, ACTA2hi PDGFRBhi (Figure 1A–C)—which were consistently observed in different pathologic conditions (Supplementary Figure 4A and B). Although FbS1 and FbS2 were linked by Uniform Manifold Approximation and Projection analysis of the scRNA-seq results, the FbS3 and FbS4 cells defined clearly separate clusters (Figure 1A and B). Notably, each subset had a distinctive gene signature (Figure 1D and Supplementary Figure 4C). POSTN and TGFBI, encoding important extracellular matrix components during inflammation, were abundant in FbS1 (Supplementary Figure 4C and D). IL1R1, which encodes the interleukin (IL) receptor type 1 responsible for proinflammatory IL1β signaling, was mostly enriched in FbS1, but less abundant in FbS2 and FbS4 (Supplementary Figure 4D). In our dataset, gene expression of various cytokines and chemokines typically up-regulated in inflammation was not significantly detected. However, some cells of FbS1 and FbS2 were enriched in CXCL14, CCL2, CCL11, CXCL12, or IL33 transcripts (Supplementary Figure 4D). Other inflammatory response-related genes, such as SERPINF1, FCGRT, and DCN, were also mainly expressed in FbS1 and FbS2 (Supplementary Figure 4D).
Figure 1.
scRNA-seq defines 4 different fibroblasts subsets (FbSs) with a distinct geographic distribution. (A) Uniform Manifold Approximation and Projection (UMAP) of 2709 fibroblasts in 21 color-coded clusters. Each dot in the UMAP indicates an individual cell. (B) UMAPs representing expression of selected markers for fibroblasts and myofibroblasts. Based on the marker genes, the total fibroblast population can be divided into 4 subsets. (C) Violin plot of normalized expression of PDGFRA, FBLN2, ACTA2, and PDGFRB in the different subsets; PDGFRAhi FbS1, FBLN2hi FbS2, ACTA2hiPDGFRBlo FbS3, and ACTA2hiPDGFRBhi FbS4. (D) Heatmap of selected genes enriched in each of the 4 FbSs according to scRNA-seq data. Columns indicate single cells. (E) UMAPs color-coded according to pathologic condition, based on the expression of PDGFRA, FBLN2, ACTA2, and PDGFRB. Representative images (F) and quantification (G) of immunofluorescence staining for PDGFRα for FbS1, FBLN2 for FbS2, αSMA for FbS3, PDGFRβ for FbS4, and dysplastic marker TROP2 with nuclear 4′,6-diamidino-2-phenylindole (DAPI) in gastric cancer patient–derived tissue sections of inflamed normal, metaplasia, or dysplasia/cancer regions. Data are presented as mean ± SD (n = 6–10 tissue sections of 7 patients). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. Scale bar: 100 μm and 20 μm for enlarged.
Gene Ontology analysis of differentially expressed genes also revealed that each FbS is involved in different biological processes during gastric carcinogenesis (Supplementary Figure 5A). The differentially expressed genes in the PDGFRAhi subset (FbS1) were enriched in pathways related to extracellular matrix organization and cell motility (Supplementary Figure 5A). The differentially expressed genes in the FBLN2hi subset (FbS2) were uniquely enriched in complement- or neutrophil-associated inflammatory pathways (Supplementary Figure 5A). The ACTA2hi PDGFRBlo subset (FbS3) demonstrated the characteristics of myofibroblasts, while the ACTA2hi PDGFRBhi FbS4 subset had a gene signature consistent with a pericyte population, including increased expression of ACTA2, RGS5, NDUFA4L2, CSPG4, NOTCH3, and ENPEP (Supplementary Figures 4C and D and 5A).
Because fibroblast populations are key sources of WNT ligands,27 we also sought to evaluate the expression of WNT signaling-related genes in the 4 subsets. FbS1 was the main source of BMP4, WNT4, and WNT5A, whereas FbS2 highly expressed WNT2B and RSPO3 compared with other subsets (Supplementary Figure 5B and C). WNT pathway-associated gene expression was rare in FbS3, and FbS4 expressed WNT6 and BMP8A (Supplementary Figure 5B and C).
Pathologic diagnosis-based analysis revealed that the proportion of FbS2 and FbS3 was most abundant in the non-metaplastic inflamed mucosa with normal lineages (subsequently referred to as inflamed normal) and decreased in metaplasia and dysplasia or cancer (Figure 1E). PDGFRα, encoded by PDGFRA, is a well-known marker for CAFs in pancreas and has been used for identification of telocytes in the intestine.26,28 Interestingly, the proportion of PDGFRAhi FbS1 cells increased in more advanced lesions compared with the inflamed normal (Figure 1E), implying that expansion of FbS1 may be associated with progression of gastric carcinogenesis. In contrast, the FbS4 pericyte population was only prominently observed in dysplastic or cancer samples (Figure 1E).
Each Fibroblast Subset Is Differentially Distributed Throughout Stomach Tissues
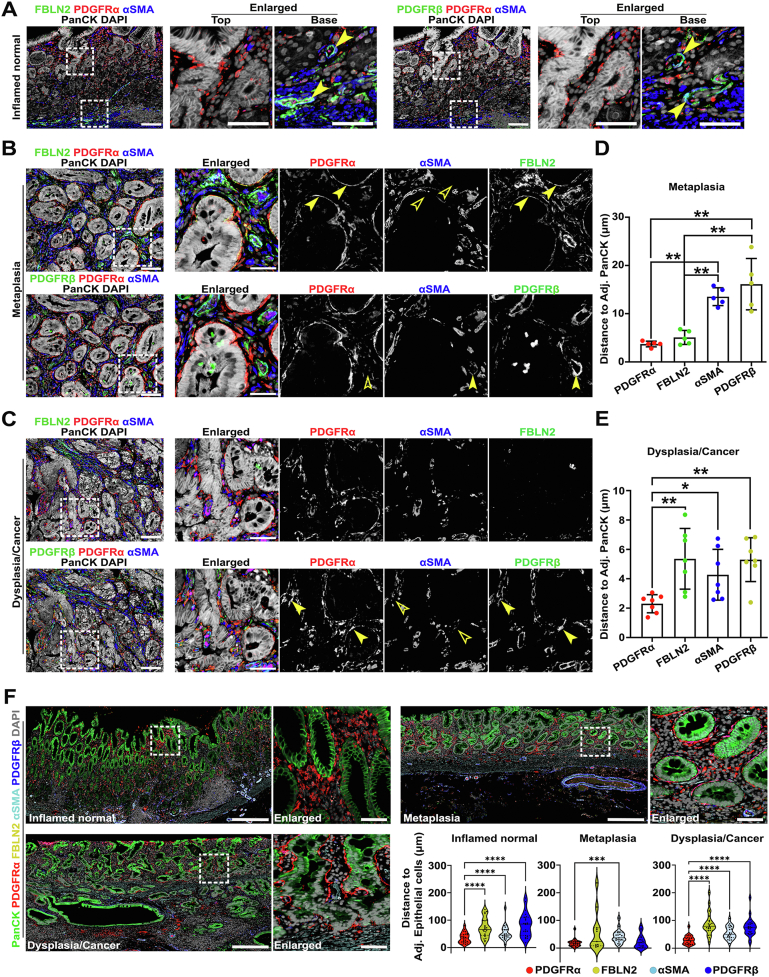
Based on the transcriptomic data, we investigated geographical distribution and population changes of FbSs during carcinogenesis through immunostaining for subset-specific markers. In the adjacent normal gastric corpus tissues with or without inflammation, PDGFRα+ FbS1 mostly resided in the isthmus region surrounding corpus glands (Figures 1F and 2A). Notably, this subset was morphologically similar to telocytes in the intestine, with elongated extension of cytoplasmic processes.27 In metaplastic tissues, FbS1 fibroblasts were prominently expanded throughout entire metaplastic glands in close proximity to the metaplastic lineages, often observed cupping SPEM cells at the bases of metaplastic glands (Figures 1F and G and 2B). FBLN2 expression was occasionally observed in the PDGFRα-expressing cells (Figure 2B). The PDGFRα+ FbS1 in dysplastic or cancerous tissues also wrapped around the epithelial compartment, but with a more disorganized pattern (Figures 1F and G and 2C). In contrast to FbS1 in metaplastic tissues, in dysplasia or cancer regions, these cells frequently co-expressed PDGFRβ (Figure 2C). FBLN2+ FbS2 fibroblasts in the adjacent normal and metaplastic tissues were mainly observed in the submucosa associated with inflammatory cell infiltration or vascular structures (Figures 1F and 2A). In more severely inflamed metaplastic lesions, this subset expanded around adjacent metaplastic lineages, as shown for FbS1, but was slightly set back from the gland compared with FbS1 (Figures 1F and 2B). Although the FbS2 population was considerably enhanced in the metaplastic tissues compared with inflamed normal tissues (Figure 1G), this subset exhibited variation in the cell number in the dysplastic or cancer tissues, depending on the extent of inflammation (Figures 1G and 2C).
Figure 2.
Multiplexed immunofluorescence staining revealed geographic distribution of FbSs during gastric carcinogenesis. (A–C) Representative images of immunofluorescence staining for PDGFRα for FbS1, FBLN2 for FbS2, αSMA for FbS3, PDGFRβ for FbS4 fibroblasts, and epithelial marker pan-cytokeratin (PanCK) with nuclear 4′,6-diamidino-2-phenylindole (DAPI) in human gastric cancer patient–derived tissue sections. (A) Inflamed normal tissue. PDGFRα+ cells are present in the isthmus region of the corpus mucosa. Arrowheads indicate αSMA+ vascular structure with FBLN2 (left panel) or PDGFRβ (right panel) expression. (B) Metaplastic tissue. PDGFRα+ cells are observed in close apposition to metaplastic gland cells. Arrowheads in the upper panel indicate fibroblasts with strong PDGFRα+ staining and weaker FBLN2+ without αSMA expression (empty arrowheads) surrounding metaplastic glands. Arrowheads in the lower panel indicate αSMA+PDGFRβ+ vascular structures without PDGFRα expression (empty arrowheads). (C) Cancer tissue. PDGFRα+ cells are expanded between cancerous glands. Arrowheads in the lower panel indicate PDGFRα+PDGFRβ+ fibroblasts surrounding cancerous epithelial compartment without αSMA expression (empty arrowheads). Scale bar: 100 μm and 50 μm for enlarged. (D, E) Quantification of the distance between epithelial cells and neighboring FbSs in metaplastic (D) and cancer-bearing stomach tissues (E). Data are presented as mean ± SD (n = 5 patients). ∗P < .05; ∗∗P < .01. (F) Representative images and quantification of multiplexed immunofluorescence staining for PDGFRα, FBLN2, αSMA, and PDGFRβ cells and their relationship with epithelial cells (positive for PanCK, CD44v9, or TROP2) with nuclear 4′,6-diamidino-2-phenylindole (DAPI) staining from 3 human tissue microarray slides (n = 24 cores per each pathologic condition). ∗∗∗P < .001; ∗∗∗∗P < .0001. Scale bar: 500 μm and 50 μm for enlarged.
α–Smooth muscle actin ([αSMA]; encoded by ACTA2)-positive FbS3 myofibroblasts were mostly present in the submucosa of the inflamed normal, metaplastic, and dysplastic/cancer tissues (Figure 1F and Figure 2A–C). Although the distribution pattern of this subset was not significantly different among most of the samples, cell numbers were slightly decreased in the metaplastic stomachs (Figure 1G). Furthermore, there were no significant differences in PDGFRβ+ FbS4 pericytes throughout most of the samples, although a few cancerous tissues showed a prominent expansion of the FbS4 population with or without PDGFRα co-expression surrounding the epithelial compartment (Figure 1F and G and Figure 2A–C).
We also examined the geographic association of FbSs with epithelial cells. Although FbS1 and FbS2 surrounded metaplastic glands, FbS3 and FbS4 were located significantly farther from the metaplastic epithelial cells (Figure 2D). In the dysplastic or cancer tissues, most of the fibroblasts were closer to the epithelial compartment compared with the subsets in the metaplastic tissues (Figure 2E). In particular, PDGFRα+ fibroblasts with or without expression of other markers were positioned adjacent to epithelial cells compared with other FbSs (Figure 2E). To confirm these relationships, we also examined 3 human tissue microarrays with samples of gastritis, metaplasia, or dysplasia or early gastric cancer using multiplexed immunostaining and unbiased digital quantitation and analysis. Figure 2F demonstrates that, as seen for the original patients from McGill, PDGFRα+ fibroblasts showed significantly closer apposition to epithelial cells compared with αSMA+ myofibroblasts in all samples.
Fibroblasts Derived From Different Lesions in a Human Gastric Cancer Patient Stomach Have Distinctive Characteristics
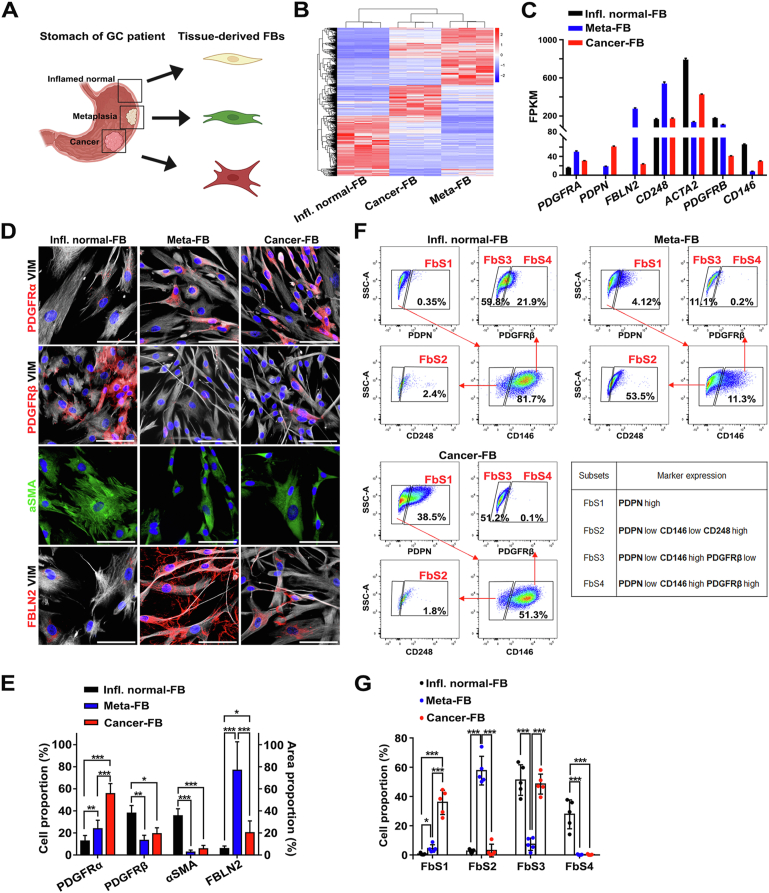
To investigate the role of fibroblasts in progression of gastric preneoplastic lesions, we freshly isolated and established gastroid and fibroblast lines from inflamed normal, metaplasia, and cancerous tissues of human patients (Figure 3A) and profiled their phenotypic and genetic features. First, bulk RNA-seq of 3 fibroblast lines derived from inflamed normal, metaplastic, or cancer-bearing tissues demonstrated that each line had a quite different gene signature (Figure 3B and Supplementary Figure 6A–C). Although the inflamed normal-derived fibroblasts prominently expressed ACTA2, PDGFRB, and CD146, which were hallmarks of FbS3 myofibroblasts and FbS4 pericytes in the scRNA-seq data analysis, the metaplasia-derived fibroblasts were enriched instead for genes up-regulated in FbS2, such as FBLN2 and CD248. Of note, the cancer-derived fibroblasts had significantly increased expression of PDGFRA and PDPN compared with inflamed normal-derived cells, indicating these fibroblasts were transcriptionally similar to FbS1 (Figure 3C). Compared with inflamed normal-derived fibroblasts, genes up-regulated in the metaplasia- or cancer-derived fibroblasts were enriched for “channel activity,” “receptor-ligand activity,” and “extracellular matrix structural constituent” pathways (Supplementary Figure 6D and E). These pathway-related gene signatures were further up-regulated in cancer-derived fibroblasts compared with metaplasia-derived fibroblasts (Supplementary Figure 6F).
Figure 3.
Specific pathologic condition-derived fibroblasts (FBs) have distinctive genetic characteristics and cell composition. (A) Schematic illustration of FB isolation from human gastric cancer (GC) patient samples. (B) Heatmap of differentially expressed genes (DEGs) in inflamed normal- (Infl. Normal-FB), metaplasia- (Meta-FB), and cancer-derived FBs (Cancer-FB). Rows and columns represent individual genes and replicates per each group, respectively. (C) Bar graph representing the normalized counts of selected marker genes in different lesion-derived FBs, confirmed by scRNA-seq data from human patient samples. Representative images (D) and quantification (E) of immunofluorescence staining for the markers for each FB subset in different lesion-derived FBs. Data are presented as mean ± SD (n = 4 independent experiments). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. Scale bar: 50 μm. Fluorescence-activated cell sorting plots showing the proportion of 4 FB subsets, described in the table, in different lesion-derived FB populations (F) and quantification (G). Data are presented as mean ± SD (n = 5 independent experiments). ∗P < .05; ∗∗∗P < .001.
We also examined protein expression levels for markers of each FbS in patient-derived fibroblasts. In inflamed normal-derived fibroblasts, almost 40% of the cells showed positivity for PDGFRβ or αSMA, whereas PDGFRα- or FBLN2-expressing cells were rare (Figure 3D and E). The metaplasia-derived fibroblasts were enriched in intracellular and extracellular FBLN2, and 20% of the cells were positive for PDGFRα (Figure 3D and E). As shown in the transcriptome analysis, cancer-derived fibroblasts showed an increased proportion of PDGFRα-expressing cells compared with other fibroblasts. While seen more prominently in metaplasia-derived fibroblasts, some of the fibroblasts from cancer-bearing mucosa also showed intracellular and extracellular FBLN2 positivity (Figure 3D).
To identify the composition of patient-derived fibroblast populations more precisely, we next examined expression of several surface markers (podoplanin [PDPN], CD248, CD146 [MCAM] and PDGFRβ) through fluorescence-activated cell sorting, using a gating strategy based on the transcriptomic data analyses (Supplementary Figure 7A and B). PDPN is one of the major surface markers for CAFs in pancreatic cancer.12 We confirmed that PDPN was uniquely enriched in the FbS1 cluster. The proportion of PDPNhi FbS1 fibroblasts was increased 10- or 100-fold in metaplasia- or cancer-derived fibroblasts, respectively, compared with inflamed normal-derived fibroblasts (Figure 3F and G). Although the inflamed normal-derived fibroblasts were enriched for PDPNlo CD146hi FbS3 and FbS4 cells, more than one-half of the metaplasia-derived fibroblasts were PDPNlo CD146lo CD248hi FbS2 cells (Figure 3F and G), as observed with the level of CD248 transcript expression and FBLN2 protein staining.
Isolation of Patient-Derived Gastroids With the Characteristics of Spasmolytic Polypeptide-Expressing Metaplasia
For characterization of gastroids derived from metaplastic tissues of 2 different patients, immunostaining for several lineage-specific markers was performed. The tissue sections-derived from cancerous or adjacent noncancerous regions of patients 1–3 demonstrated morphologic features of inflamed normal, metaplastic, or dysplastic or cancer in histopathologic examination (Supplementary Figure 8A). The gastroids demonstrated phenotypic features similar to the tissue of origin with positivity for SPEM (CD44v9, AQP5) and/or gastric antral and intestinal lineage markers (PDX1 and CDX1). However, expression of TROP2 or CEACAM5, markers for more advanced preneoplastic lesions, such as incomplete intestinal metaplasia or dysplasia (Supplementary Figure 8B), was weak or rare in these gastroids (Supplementary Figure 8C). These staining data indicated that the gastroids established from patient tissues were SPEM-lineage predominant metaplastic gastroids.7
Co-Culture of Metaplastic Gastroids With Human Metaplasia- or Cancer-Derived Fibroblasts Promotes Progression Into Dysplasia
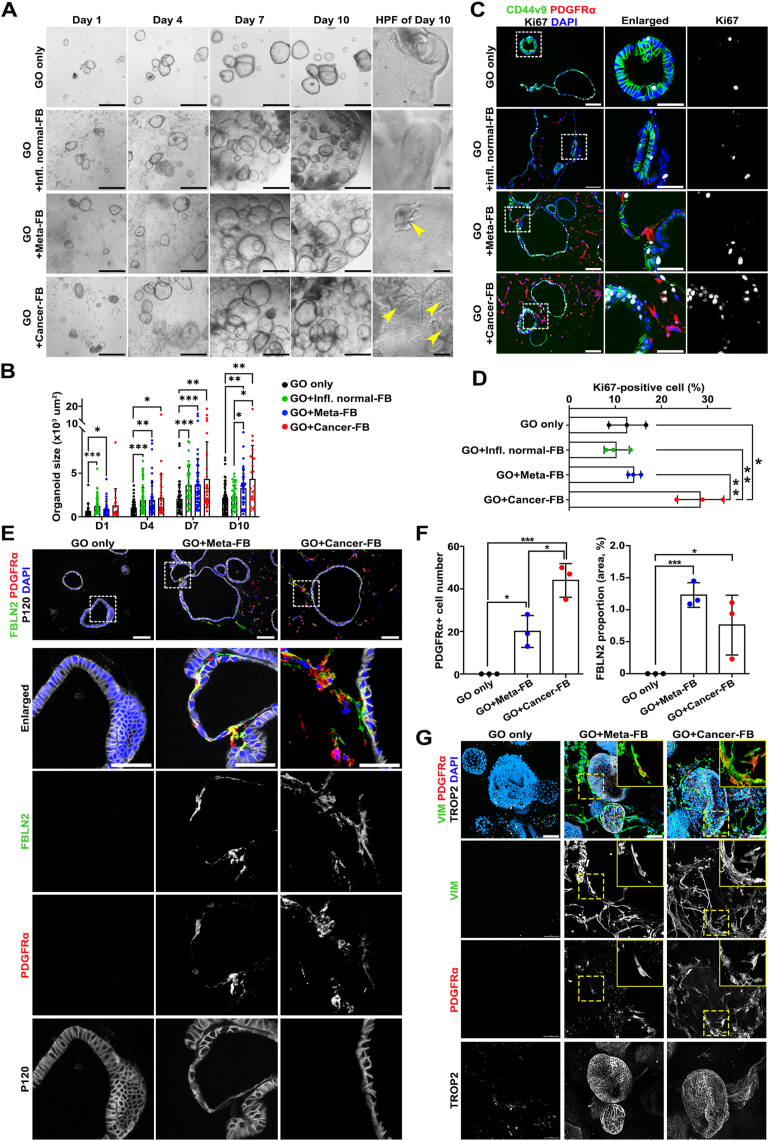
Based on these characteristics of fibroblasts and gastroids, we first performed a 3-dimensional co-culture of the metaplastic gastroids with metaplasia- or cancer-derived fibroblasts to investigate whether gastroid phenotypes can be altered by different types of fibroblasts. Initially, fibroblasts were irregularly distributed in Matrigel domes with gastroids, but began to migrate toward the gastroids at day 3, and some finally were situated close to gastroids, similar to the pattern seen in human tissues (Figure 4A, C, and E). After 7 days, gastroids co-cultured with either metaplasia- or cancer-derived fibroblasts grew faster than gastroids cultured alone and reached maximal size (Figure 4A and B). However, proliferative activity was significantly enhanced by co-culture with cancer-derived fibroblasts, but not with inflamed normal- or metaplasia-derived fibroblasts (Figure 4C and D). We next sought to examine the cell composition of fibroblasts and gastroids after co-culture. In accordance with profiles of patient-derived fibroblasts cultured alone, metaplasia- or cancer-derived fibroblasts in co-culture with gastroids remained mostly positive for PDGFRα and FBLN2 and abundantly expressed extracellular transforming growth factor–β-induced (TGFBI), whereas inflamed normal-derived fibroblasts in co-culture mainly consisted of αSMA+ cells (Figure 4E and F and Supplementary Figure 9A and B). Furthermore, cancer-derived fibroblasts showed a 2-fold higher number of PDGFRα+ fibroblasts compared with metaplasia-derived fibroblasts, and intra- or extracellular FBLN2 positivity was slightly up-regulated in metaplasia-derived fibroblasts compared with cancer-derived fibroblasts (Figure 4F).
Figure 4.
Three-dimensional co-culture with metaplasia-derived and cancer-derived fibroblast (FB) enhances growth of metaplastic gastroids. (A) Representative brightfield images of gastroids (GOs) cultured for 10 days with or without FBs isolated from inflamed normal-, metaplastic-, or cancer-bearing mucosae. Arrowheads indicate budding formation. Scale bar: 500 μm and 100 μm for high-power field. (B) Quantification of GO size at each time point. Data are presented as mean ± SD (n = 20–90 organoids from 2 independent experiments). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. (C) Representative images of immunofluorescence staining for metaplasia marker CD44v9, PDGFRα for FbS1, and Ki67 for proliferative activity with nuclear 4′,6-diamidino-2-phenylindole (DAPI). (D) Quantification of proliferative GO cells. Data are presented as mean ± SD (n = 3 independent experiments). ∗P < .05; ∗∗P < .01. Representative images (E) and quantification (F) of immunofluorescence staining for FBLN2 for FbS2, PDGFRα for FbS1 and epithelial membrane marker P120 with nuclear DAPI. Data are presented as mean ± SD (n = 3 independent experiments). ∗P < .05; ∗∗∗P < .001. (G) Representative images of whole-mount staining for vimentin (VIM) for pan-FBs, PDGFRα for FbS1, and dysplasia marker TROP2 with nuclear DAPI. Boxes are enlarged insets showing VIM+PDGFRα+ FbS1 close by GOs.
In terms of geographical correlation between fibroblasts and gastroids, while αSMA+ cells from inflamed normal-derived fibroblasts were irregularly scattered between gastroids, several PDGFRα+, FBLN2+, and/or TGFBI+ cells from metaplasia- or cancer-derived fibroblasts directly contacted with gastroids (Figure 4E and Supplementary Figure 9A and B), similar to FbS1 and FbS2 that closely surrounded epithelial compartments in various preneoplastic lesions of human stomach (Figure 1F and Figure 2A–C). Some of the PDGFRα+ cancer-derived fibroblasts surrounding gastroids also expressed PDGFRβ (Supplementary Figure 9B), as observed in fibroblasts adjacent to epithelial cells in cancer tissues. Whole-mount staining also revealed that a portion of VIM+ fibroblasts with extended cytoplasmic processes came directly in contact with gastroids (Figure 4G and Supplementary Figure 9C). Given that FbS1 fibroblasts were enriched for genes involved in “cell migration,” these data suggest that the different FbSs might display distinct migratory activity toward epithelial cells. As shown in paraffin section staining, PDGFRα+ fibroblast populations were more prominently observed in cancer-derived fibroblasts compared with metaplasia-derived fibroblasts in 3-dimensional whole-mount immunostaining (Supplementary Figure 9C).
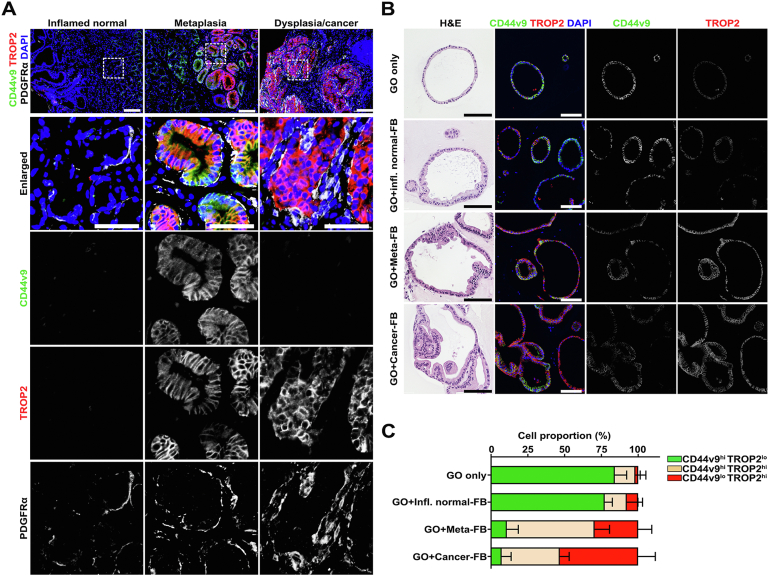
We previously reported that TROP2 is a marker for incomplete intestinal metaplasia and dysplasia, and that CD44v9 is a metaplasia marker in stomach mucosa.21,29 In the present study, we found differential expression of CD44v9 and TROP2 in metaplastic or dysplastic or cancer tissues. As shown in Figure 5A, adjacent normal tissues with or without inflammation did not express either CD44v9 or TROP2, whereas metaplastic glands were positive for CD44v9 and/or TROP2. Of note, dysplastic/cancer tissues showed only TROP2 expression. Taken together, altered expression of CD44v9 and TROP2 in a gland can therefore be considered as a hallmark of dysplastic transition in metaplastic glands.21 Accordingly, almost 80% of cells in cultured metaplastic tissue–derived gastroids strongly expressed CD44v9 with low TROP2 levels. Although inflamed normal-derived fibroblasts did not induce alterations in marker expression, the proportion of TROP2-expressing cells with similar or lower CD44v9 expression increased by almost 90% in gastroids co-cultured with metaplasia- or cancer-derived fibroblasts (Figure 5B and C).
Figure 5.
Recruited metaplasia-derived and cancer-derived fibroblasts facilitate dysplastic progression in metaplastic gastroids (GOs). (A) Immunofluorescence staining in human stomach sections for metaplasia marker CD44v9, dysplasia marker TROP2, and FbS1 marker PDGFRα with nuclear 4′,6-diamidino-2-phenylindole (DAPI). Scale bar: 100 μm and 50 μm for enlarged. Representative images of immunofluorescence staining in GO sections for metaplasia marker CD44v9 and dysplasia marker TROP2 (B) and quantification of positive GO cells for each marker (C). Scale bar: 100 μm.
Direct Contact With Metaplasia- or Cancer-Derived Fibroblasts Induces Dysplastic Transition in Metaplasia
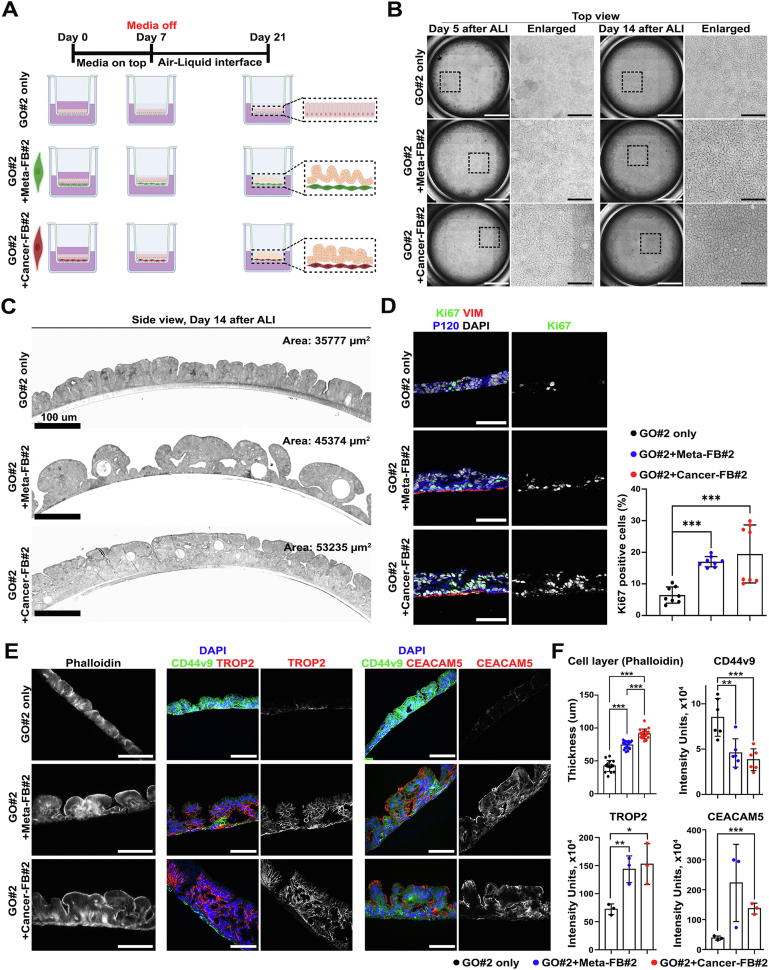
To facilitate the direct interaction of fibroblasts in contact with metaplastic gastroid cells, we employed an ALI co-culture system on Transwell filters composed of metaplastic gastroids from patient 2 with metaplasia- or cancer-derived fibroblasts from the same patient (Figure 6A). At day 5 after media removal for initiation of the ALI condition, gastroid cells without fibroblasts predominantly displayed a single layer in the center of the well (Figure 6B). Interestingly, gastroid cells co-cultured with either metaplasia- or cancer-derived fibroblasts for 5 days after ALI initiation formed large numbers of protrusions from the monolayer throughout the well and maintained polypoid structures at day 14 (Figure 6B). At this time point, projecting polyps of co-cultured gastroid cells covered the entire area of the well and coalesced with each other, resulting in a “gyri and sulci-like structure” (Figure 6B and C). Accordingly, proliferative activity in gastroid cells increased in co-culture conditions with either metaplasia- or cancer-derived fibroblasts compared with gastroids cultured without fibroblasts (Figure 6D).
Figure 6.
ALI co-culture demonstrates that metaplasia (Meta)- or cancer-derived fibroblasts (FBs) can induce dysplastic transition in precancerous metaplastic cells. (A) Schematic illustration of ALI co-culture of metaplastic gastroids (GOs) with Meta- or cancer-derived FBs for a total of 21 days (created with BioRender.com). Representative brightfield images (top view; B) and confocal images of plastic sections stained with toluidine blue (side view; C) of ALI co-culture of metaplastic GO cells with Meta- or cancer-derived FBs, both from patient 2. Scale bar: 2000 μm and 500 μm for enlarged. Note the multiple-layered disorganized cells after co-culture with Meta- or cancer-derived FBs in (C). (D) Representative images of immunofluorescence staining for proliferation marker Ki67, fibroblast marker vimentin (VIM), and epithelial membrane marker P120 with nuclear 4′,6-diamidino-2-phenylindole (DAPI) and quantification of Ki67-positive cells. Data are presented as mean ± SD (n = 8 images from 2 different sections). ∗∗∗P <.001. (E) Representative images of immunofluorescence staining for phalloidin to identify cell borders, metaplasia marker CD44v9, and dysplasia markers TROP2 or CEACAM5 with nuclear DAPI. Scale bar: 100 μm. Note the loss of CD44v9 and the gain of TROP2 and CEACAM5 expression when GO cells were cultured in the presence of Meta- or cancer-derived FBs. (F) Quantification of the thickness of GO cell layer determined by phalloidin staining and extent of protein marker expression measured by intensity units. Data are presented as mean ± SD (n = 5 or 3 of different sections for phalloidin or other markers, respectively). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001.
We next investigated CD44v9, TROP2, and CEACAM5 expression in gastroid cells in each condition to evaluate whether the morphologic changes induced by co-culture with fibroblasts correlated with metaplasia progression. Metaplastic gastroid cells cultured alone mostly displayed strong CD44v9 expression (Figure 6E and F). In contrast, co-culture with metaplasia- or cancer-derived fibroblasts enhanced TROP2 expression in cells within polypoid projections, whereas CD44v9 expression was rarely observed or showed residual expression at the basal cell layer (Figure 6E and F). We then evaluated CEACAM5 expression in these gastroid cells based on the findings that CEACAM5 was prominently and specifically expressed in dysplastic tissues compared with metaplastic tissues (Supplementary Figure 2D and E and Supplementary Figure 8A). Gastroid cells strongly expressed CEACAM5 only after co-culture with either metaplasia- or cancer-derived fibroblasts, especially in the polypoid regions (Figure 6E and F and Supplementary Figure 10A–C). Finally, metaplastic gastroid cells derived from patient 1 or 2 were co-cultured with metaplasia- or cancer-derived fibroblasts from different patients (patient 1, 2, or 3) to assess whether these effects of fibroblasts on epithelial cells were restricted to co-culture with the same patient-derived cells. All patient-derived fibroblasts from either metaplasia- or cancer-bearing tissues also induced polypoid protrusions of metaplastic gastroid cells from the Transwell membranes (Supplementary Figure 11A and C).
Expression of the metaplasia marker CD44v9 was more intensely and consistently observed in the gastroid cells without fibroblasts than in co-cultured gastroid cells (Supplementary Figure 11B and D). The SPEM cell lineage-specific marker AQP5 was continuously and strongly expressed at the apical side of single-layered cells in metaplastic gastroids cultured alone, but was only intermittently and faintly expressed in cells within polypoid projections in gastroids co-cultured with metaplasia- or cancer-derived fibroblasts (Supplementary Figure 11B and D). Co-cultured gastroid cells exclusively exhibited TROP2 and CEACAM5 expression, especially in protruding polyps, with preparations of fibroblasts from 3 different patients (Supplementary Figure 11B and D). In contrast with the effects of metaplasia- or cancer-derived fibroblasts on metaplastic cells, true normal-derived fibroblasts, isolated from histologically normal corpus mucosa without metaplasia or neoplasia obtained from the distal margins of esophagogastrectomies, did not promote polyp formation or expression of TROP2 or CEACAM5 compared with gastroid cells alone (Supplementary Figure 12B–D). These normal fibroblast populations were mainly composed of FbS3 and FbS4 (Supplementary Figure 12A).
Conditioned Media From Metaplasia- or Cancer-Derived Fibroblasts Induce Dysplastic Transition in Metaplasia
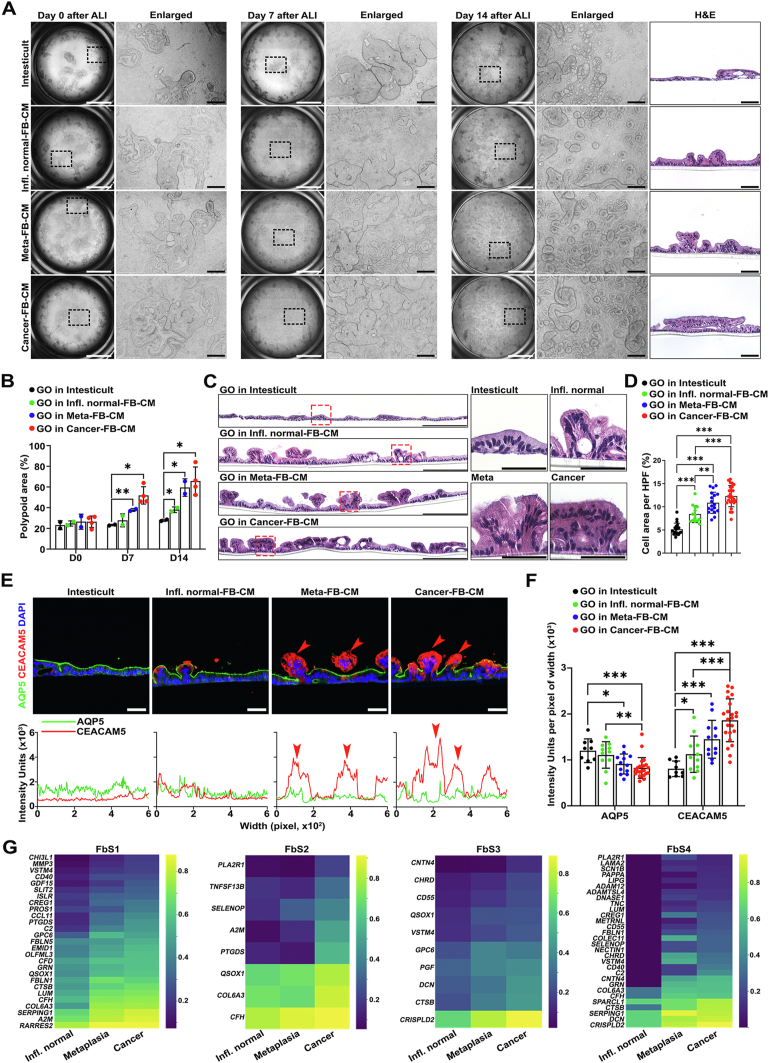
To investigate whether secreted factors from fibroblasts could promote dysplastic progression of metaplasia without direct contact, we cultured metaplastic gastroids in ALI conditions with regular media or with addition of conditioned media from inflamed normal-, metaplasia-, or cancer-derived fibroblasts. Although gastroid cells in regular media did not show any significant changes in morphology at day 7 after ALI, the polypoid area of gastroid cells grown in conditioned media expanded (Figure 7A and B). In addition, conditioned media from metaplasia- or cancer-derived fibroblasts not only promoted polyp formation in gastroid cells, but also increased cellularity in the polyps at day 14 after ALI, compared with regular or conditioned media from inflamed normal-derived fibroblasts (Figure 7A–C). Based on these structural changes, we next evaluated lineage marker expression for AQP5 or CEACAM5, which define metaplasia or dysplasia, respectively. Monolayered gastroid cells in regular media were consistently positive for AQP5, and negative for CEACAM5. However, the epithelial cells in polypoid structures observed in cultures with conditioned media from metaplasia- or cancer-derived fibroblasts strongly expressed CEACAM5 at the apical side with weak or negative AQP5 expression (Figure 7D and E). Although AQP5+ monolayered gastroid cells also expressed MUC6, a mucin observed in SPEM, CEACAM5+ cells in polyps were essentially negative for MUC6 or MUC2, a type of intestinal mucin, suggesting direct conversion from SPEM cells to dysplastic cells (Supplementary Figure 13A). We also examined the effect of fibroblast-derived conditioned media on epithelial proliferation. In contrast to the results from direct co-culture with fibroblasts, conditioned media even from cancer-derived fibroblasts did not promote gastroid cell proliferation (Supplementary Figure 13A).
Figure 7.
ALI culture with fibroblast (FB)-derived conditioned media (CM) demonstrates that secreted factors from FBs can induce dysplastic transition in metaplastic (Meta) cells. (A) Representative brightfield images of ALI co-culture of Meta gastroid cells in CM from inflamed normal- (Infl. normal-), Meta-, or cancer-derived FBs at each time point, all from patient 2. Paired H&E images show morphologic differences in polyp formation. Scale bars: 2000 μm and 200 μm for enlarged or 100 μm for H&E. (B) Quantification of the proportion of projected area from the base of filters shown in (A). Data are presented as mean ± SD (n = 2 independent experiments composed of CM from inflamed normal-, Met-, or cancer-derived FBs from patient 2 and cancer-derived FB from patient 1). ∗P < 0.05; ∗∗P < 0.01. (C) Representative H&E images of an entire filter of each ALI culture condition. Red boxes are enlarged on the right side, demonstrating morphologic alterations and different cell density in the polyps for each condition. Scale bars: 200 μm and 50 μm for enlarged. (D) Quantification of the area occupied with gastroid cells under high-power fields. Data are presented as mean ± SD (n = 2 independent experiments). ∗∗P < 0.01; ∗∗∗P < .001. (E) Representative images of immunofluorescence staining for metaplasia marker AQP5 and dysplasia marker CEACAM5 with nuclear 4′,6-diamidino-2-phenylindole (DAPI) and matched profiling of the expression of 2 markers. Arrowheads indicate polyps expressing CEACAM5. Scale bars: 50 μm. (F) Quantification of the staining shown in (E), presenting total expression levels for the 2 markers in each condition. Data are presented as mean ± SD (n = 2 independent experiments). ∗P < .05; ∗∗P < .01; ∗∗∗P < .001. (G) Heatmaps of up-regulated genes in each FB subset from metaplasia- or cancer-bearing tissues compared with the same subset from inflamed normal tissues, derived from the candidate genes described in Supplementary Figure 13B–E.
We next sought to identify candidate secreted factors from fibroblasts that may be responsible for dysplastic progression. Among up-regulated genes in metaplasia- or cancer-derived fibroblasts compared with inflamed normal-derived fibroblasts (fold-change cutoff = 2, false discovery rate < 0.05), 79 or 96 candidate genes, respectively, were sorted from a database (http://genomics.cicbiogune.es/VerSeDa) as candidates encoding secreted molecules (Supplementary Table 2). Among them, 58 genes were observed in both groups.30 Construction of a protein–protein interaction network of the candidates revealed that most of the genes were functionally connected to each other. Furthermore, 2 networks from metaplasia- or cancer-derived fibroblasts shared several hubs (Supplementary Figure 13B). In patient-derived scRNA-seq, these candidate genes were more frequently expressed in FbS1 and FbS2, which were the main components of metaplasia- or cancer-derived fibroblasts (Supplementary Figure 13C and D). Notably, metaplasia or cancer tissue–derived FbSs showed higher expression levels of several candidate genes compared with the same subset from inflamed normal tissues (Figure 7F). Gene Ontology analysis demonstrated that the candidate genes were functionally enriched for “extracellular matrix,” “cell proliferation,” “cell adhesion,” or “secretion”-related pathways (Supplementary Figure 13E). These results imply that specific subsets of fibroblast secreted factors may account for distinct phenotypes at different pathologic stages.
Discussion
Previous investigations in a number of systems have noted the importance of fibroblasts in the maintenance of both physiological and pathophysiological mucosae. In the present study, we identified 4 FbSs through scRNA-seq data analysis that were present in normal stomach and displayed distinctive geographical distributions with differential expansion in the context of metaplastic or dysplastic mucosae. Based on the prominent expression of PDGFRα, the close proximity to the epithelium and the morphologic features of extended cytoplasmic processes, FbS1 likely represents a telocyte-like population that can be distinguished from other fibroblasts, such as αSMA+ myofibroblasts.31,32 The PDGFRα+ telocytes in human and mouse intestine are located in the subepithelial regions, playing important roles in maintaining epithelial integrity, transduction of sensory signals, and serving as a component of the stem cell niche.27,31 PDGFRα+ telocytes in the pericryptal region of the colonic mucosa are a major source of several WNT ligands and BMP, which promote a balance between proliferation and differentiation of colonic epithelial cells.33 In accordance with these features, FbS1 cells expressing PDGFRα in adjacent normal stomach were mainly located in the isthmus region of the stomach corpus, where progenitor cells reside, which was distinct from the location of αSMA+ FbS3 myofibroblasts. In addition to the topography, FbS1 in single-cell transcriptomics uniquely expressed WNT4, WNT5A, BMP2, and BMP4, implicating FbS1 fibroblasts as the major functional telocyte population in stomach. Nevertheless, the levels of plasticity among fibroblast populations remain unclear.12
Previous studies have reported the association of fibroblasts, also called CAFs, with gastric cancer cell physiology, including promotion of proliferation, migration, and metastasis.34 A previous study using a bone marrow transplantation mouse model also found that Helicobacter infection–induced recruitment of bone marrow–derived mesenchymal stem cells can give rise to niche for gastric carcinogenesis.35 Recruitment of the cells to gastric mucosa is known to be dependent on CXCL12, CXCL16, or TGFβ secreted by epithelial or immune cells, and the recruited cells usually express high levels of soluble molecules, such as IL-6, WNT5A, BMP4, or CCL5.36 Nevertheless, the subsets of fibroblasts or CAFs and their functions during the gastric carcinogenic process had not been elucidated in detail. Kumar et al17 recently suggested that the presence of a FAP+ and INHBA+ CAF subset is associated with poor prognosis of gastric cancer through single-cell transcriptomic analysis. Our present studies indicate that the patterns of fibroblasts promoting early carcinogenesis in the stomach are quite different from previous reports in other organ systems. In contrast with CAFs associated with pancreatic ductal adenocarcinoma, myofibroblasts (FbS3) do not seem to be strongly associated with promotion of gastric carcinogenesis. Contrarily, telocyte-like fibroblasts (FbS1) appear to play a major role in establishing a metaplastic and dysplastic niche in gastric carcinogenesis, with expansion of the FbS1 population in close contact with metaplastic and dysplastic cell lineages at the bases of preneoplastic glands. These FbS1 cells are the most significant source of WNT ligands in the metaplastic and dysplastic stomach, and recent studies have demonstrated that dysplastic stem cell expansion and progression is driven by WNT activation.37 In addition, we also found that several factors facilitating cell growth and remodeling of extracellular matrix are enriched in FbS1 and FbS2 cells derived from metaplastic or cancer tissues. Thus, the gastric precancerous milieu may be driven by the combined action of telocyte-like FbS1 cells and inflammatory CAF-like FbS2 cells rather than myofibroblasts.
We have sought to identify markers that define the transitions between metaplasia and dysplasia, particularly for incomplete intestinal metaplasia, which represents the pathologic finding most associated with progression to gastric cancer.5 Previous studies have shown that AQP5 and CD44v9 are absent in the normal stomach corpus, but are up-regulated in SPEM lineage cells.7,21,38 AQP5 is positive in SPEM lineage cells at the bases of glands of both pyloric metaplasia and incomplete intestinal metaplasia,7 but AQP5 is down-regulated in dysplasia. TROP2 is up-regulated in intestinal lineage cells in incomplete intestinal metaplasia and dysplasia, but is absent from SPEM cells or complete intestinal metaplasia glands.21 Our present studies indicate that CEACAM5 is expressed in the intestinal lineage cells within incomplete intestinal metaplasia and more strongly in dysplasia. Previous studies have noted the up-regulation of CEACAM5 in gastric cancers.24 We have used these marker transitions to define the effects of fibroblasts on human metaplastic gastroids that predominantly display the characteristics of SPEM lineages, with prominent expression of CD44v9 and AQP5, but without significant expression of TROP2 or CEACAM5. In ALI Transwell cultures, the SPEM gastroids grew mostly as monolayers with prominent expression of CD44v9 and AQP5. When they were co-cultured with fibroblasts from true normal gastric mucosa, they maintained their monolayer growth phenotype with expression of CD44v9 and AQP5. However, co-culture of SPEM gastroids with fibroblasts derived from either metaplastic or cancer-bearing mucosae induced a profound change in growth pattern with development of multiple polypoid lesions with multiple lumens. Most importantly, the polypoid lesions showed a marked decrease in CD44v9 and AQP5 expression, while also demonstrating prominent increases in the expression of TROP2 and CEACAM5. Because we did not see evidence for intestinalization during the entire period of co-culture, these findings indicate that SPEM lineage cells under the influence of altered fibroblast populations may evolve to dysplasia without intermediate conversion to fully intestinalized cell lineages. Furthermore, SPEM gastroids under the influence of conditioned media from metaplastic- or cancer-derived fibroblasts recapitulated dysplastic transition with an increase in the dysplasia marker CEACAM5 and a concomitant loss of the metaplasia marker AQP5. However, an increase in proliferation was not induced with conditioned media in the gastroids. Thus, while some stable secreted factors and matrix components are likely present in the conditioned medium, fibroblasts likely secrete labile factors that can promote metaplasia to dysplasia transition and proliferation.12 In addition, gradients of factor concentration may be important, and thus apposition of fibroblasts adjacent to epithelial cells is likely important for the full manifestation of dysplastic progression.
In summary, we have performed a directed analysis of the precancerous milieu in patients with gastric cancer. Studies using scRNA-seq have established discrete subsets of fibroblasts in the normal and pathologic gastric mucosae that demonstrate distinct geographical patterns for association with normal, metaplastic, and dysplastic epithelial cell lineages. Furthermore, isolated fibroblasts from metaplastic and cancer-containing mucosae can directly promote progression of metaplastic SPEM gastroid cells to dysplasia. These findings demonstrate the critical role of distinct FbSs and their interaction with epithelial cells in promoting dysplastic transitions in the stomach.
Acknowledgments
The authors would like to thank Yu Chang Wang, Spyridon Oikonomopoulos, Janick St-Cyr, and Haig Djambazian for performing singe-cell captures, sequencing data production, and data quality control.
CRediT Authorship Contributions
Su-Hyung Lee, DVM, PhD (Conceptualization: Equal; Data curation: Equal; Formal analysis: Lead; Investigation: Lead; Methodology: Equal; Validation: Lead; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Lead).
Ela W. Contreras Panta, BS (Conceptualization: Supporting; Data curation: Equal; Formal analysis: Lead; Investigation: Lead; Methodology: Equal; Validation: Lead; Visualization: Lead; Writing – original draft: Lead; Writing – review & editing: Lead).
David Gibbs, PhD (Data curation: Lead; Formal analysis: Equal; Investigation: Lead; Methodology: Equal; Resources: Equal; Software: Supporting; Writing – original draft: Lead; Writing – review & editing: Lead).
Yoonkyung Won, PhD (Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Validation: Supporting; Visualization: Equal; Writing – review & editing: Equal).
Jimin Min, PhD (Data curation: Equal; Formal analysis: Equal; Investigation: Equal; Methodology: Equal; Visualization: Supporting; Writing – review & editing: Supporting).
Changqing Zhang, BS (Formal analysis: Equal; Investigation: Equal; Methodology: Lead; Validation: Supporting; Writing – review & editing: Supporting).
Joseph T. Roland, PhD (Data curation: Equal; Formal analysis: Equal; Methodology: Equal; Software: Lead; Validation: Supporting; Visualization: Supporting; Writing – review & editing: Supporting).
Se-Hoon Hong, PhD (Formal analysis: Supporting; Investigation: Supporting; Writing – review & editing: Supporting).
Yoojin Sohn, BS (Formal analysis: Supporting; Investigation: Supporting; Writing – review & editing: Supporting).
Evan Krystofiak, PhD (Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Methodology: Equal; Visualization: Equal; Writing – review & editing: Supporting).
Bogun Jang, DVM, PhD (Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting).
Lorenzo Ferri, MD (Conceptualization: Supporting; Funding acquisition: Equal; Investigation: Equal; Project administration: Equal; Supervision: Equal; Writing – review & editing: Supporting).
Veena Sangwan, PhD (Formal analysis: Supporting; Investigation: Supporting; Writing – review & editing: Supporting).
Jiannis Ragoussis, PhD (Data curation: Supporting; Investigation: Supporting; Methodology: Equal; Writing – review & editing: Supporting).
Sophie Camilleri-Broët, MD (Formal analysis: Equal; Investigation: Supporting; Writing – review & editing: Supporting).
Joseph Caruso, PhD (Investigation: Supporting; Methodology: Supporting; Writing – review & editing: Supporting).
Chira Chen-Tanyolac, BS (Investigation: Supporting; Writing – review & editing: Supporting).
Michael Strasser, PhD (Conceptualization: Supporting; Data curation: Supporting; Formal analysis: Supporting; Investigation: Supporting; Methodology: Supporting; Supervision: Supporting; Writing – review & editing: Supporting).
Philippe Gascard, PhD (Investigation: Supporting; Writing – review & editing: Supporting).
Thea D. Tlsty, PhD (Conceptualization: Equal; Funding acquisition: Lead; Supervision: Supporting; Writing – review & editing: Supporting).
Sui Huang, PhD (Conceptualization: Equal; Formal analysis: Supporting; Funding acquisition: Equal; Methodology: Supporting; Supervision: Supporting; Writing – review & editing: Supporting).
Eunyoung Choi, PhD (Formal analysis: Equal; Investigation: Equal; Supervision: Equal; Validation: Supporting; Writing – review & editing: Supporting).
James R. Goldenring, MD, PhD (Conceptualization: Lead; Data curation: Supporting; Formal analysis: Supporting; Funding acquisition: Equal; Investigation: Supporting; Methodology: Equal; Project administration: Lead; Supervision: Equal; Validation: Supporting; Visualization: Supporting; Writing – original draft: Supporting).
Footnotes
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by a Cancer Research UK Grand Challenge Award funding to James R. Goldenring (29075), Lorenzo Ferri (29071), Sui Huang (29068) and Thea D. Tlsty (lead principal investigator; 27145). This work was also supported by grants to James R. Goldenring from a Department of Veterans Affairs (VA) Merit Review Award IBX000930, Department of Defense CA190172, and National Institutes of Health (NIH) R01 DK101332 and to Eunyoung Choi from NIH R37 CA244970, National Cancer Institute (NCI) R01 CA272687, and an American Gastroenterological Association Research Foundation Funderburg Award (AGA2022-32-01). The Vanderbilt Flow Cytometry Shared Resource, the Vanderbilt Cell Imaging Shared Resource, and the Vanderbilt Digital Histology Shared Resource are supported by the Vanderbilt Ingram Cancer Center (P30 CA068485) and the Vanderbilt Digestive Disease Research Center (DK058404). Single-cell sequencing at the McGill Genome Center, Victor Phillip Dahdaleh Institute of Genomic Medicine is supported by the Canadian Foundation for Innovation (CFI) as part of Canada’s Genome Enterprise (Jiannis Ragoussis), CFI 33408, 40104 and CFI-MSI35444. The Translational Pathology Shared Resource is supported by NCI/NIHCancer Center Support Grant (P30 CA68485). The Digital Histology Shared Resource is supported by VA Shared Equipment grant 1IS1BX003097.
Data Availability Data, code, analytical methods, and study materials will be made available to other researchers upon request.
Author names in bold designate shared co-first authorship.
Note: To access the supplementary material accompanying this article, visit the online version of Gastroenterology at www.gastrojournal.org, and at https://doi.org/10.1053/j.gastro.2023.04.038.
Supplementary Material
References
- 1.Correa P. Human gastric carcinogenesis: a multistep and multifactorial process-first American Cancer Society Award Lecture on Cancer Epidemiology and Prevention. Cancer Res. 1992;52:6735–6740. [PubMed] [Google Scholar]
- 2.Tan P., Yeoh K.G. Genetics and molecular pathogenesis of gastric adenocarcinoma. Gastroenterology. 2015;149:1153–1162.e3. doi: 10.1053/j.gastro.2015.05.059. [DOI] [PubMed] [Google Scholar]
- 3.Correa P. A human model of gastric carcinogenesis. Cancer Res. 1988;48:3554–3560. [PubMed] [Google Scholar]
- 4.Schmidt P.H., Lee J.R., Joshi V., et al. Identification of a metaplastic cell lineage associated with human gastric adenocarcinoma. Lab Invest. 1999;79:639–646. [PubMed] [Google Scholar]
- 5.Shao L., Li P., Ye J., et al. Risk of gastric cancer among patients with gastric intestinal metaplasia. Int J Cancer. 2018;143:1671–1677. doi: 10.1002/ijc.31571. [DOI] [PubMed] [Google Scholar]
- 6.Choi E., Hendley A.M., Bailey J.M., et al. Expression of activated ras in gastric chief cells of mice leads to the full spectrum of metaplastic lineage transitions. Gastroenterology. 2016;150:918–930.e13. doi: 10.1053/j.gastro.2015.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee S.H., Jang B., Min J., et al. Up-regulation of Aquaporin 5 defines spasmolytic polypeptide-expressing metaplasia and progression to incomplete intestinal metaplasia. Cell Mol Gastroenterol Hepatol. 2022;13:199–217. doi: 10.1016/j.jcmgh.2021.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meyer A.R., Engevik A.C., Madorsky T., et al. Group 2 innate lymphoid cells coordinate damage response in the stomach. Gastroenterology. 2020;159:2077–2091.e8. doi: 10.1053/j.gastro.2020.08.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Busada J.T., Ramamoorthy S., Cain D.W., et al. Endogenous glucocorticoids prevent gastric metaplasia by suppressing spontaneous inflammation. J Clin Invest. 2019;129:1345–1358. doi: 10.1172/JCI123233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sigal M., Logan C.Y., Kapalczynska M., et al. Stromal R-spondin orchestrates gastric epithelial stem cells and gland homeostasis. Nature. 2017;548:451–455. doi: 10.1038/nature23642. [DOI] [PubMed] [Google Scholar]
- 11.Senol K., Ozkan M.B., Vural S., et al. The role of inflammation in gastric cancer. Adv Exp Med Biol. 2014;816:235–257. doi: 10.1007/978-3-0348-0837-8_10. [DOI] [PubMed] [Google Scholar]
- 12.Biffi G., Tuveson D.A. Diversity and biology of cancer-associated fibroblasts. Physiol Rev. 2021;101:147–176. doi: 10.1152/physrev.00048.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.LeBleu V.S., Neilson E.G. Origin and functional heterogeneity of fibroblasts. FASEB J. 2020;34:3519–3536. doi: 10.1096/fj.201903188R. [DOI] [PubMed] [Google Scholar]
- 14.Dumont N., Liu B., Defilippis R.A., et al. Breast fibroblasts modulate early dissemination, tumorigenesis, and metastasis through alteration of extracellular matrix characteristics. Neoplasia. 2013;15:249–262. doi: 10.1593/neo.121950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang P., Yang M., Zhang Y., et al. Dissecting the single-cell transcriptome network underlying gastric premalignant lesions and early gastric cancer. Cell Rep. 2019;27:1934–1947.e5. doi: 10.1016/j.celrep.2019.04.052. [DOI] [PubMed] [Google Scholar]
- 16.Min J., Vega P.N., Engevik A.C., et al. Heterogeneity and dynamics of active Kras-induced dysplastic lineages from mouse corpus stomach. Nat Commun. 2019;10:5549. doi: 10.1038/s41467-019-13479-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar V., Ramnarayanan K., Sundar R., et al. Single-cell atlas of lineage states, tumor microenvironment, and subtype-specific expression programs in gastric cancer. Cancer Discov. 2022;12:670–691. doi: 10.1158/2159-8290.CD-21-0683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sathe A., Grimes S.M., Lau B.T., et al. Single-cell genomic characterization reveals the cellular reprogramming of the gastric tumor microenvironment. Clin Cancer Res. 2020;26:2640–2653. doi: 10.1158/1078-0432.CCR-19-3231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito G., Okamoto R., Murano T., et al. Lineage-specific expression of bestrophin-2 and bestrophin-4 in human intestinal epithelial cells. PLoS One. 2013;8 doi: 10.1371/journal.pone.0079693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakab R.L., Collaco A.M., Ameen N.A. Characterization of CFTR high expresser cells in the intestine. Am J Physiol Gastrointest Liver Physiol. 2013;305:G453–G465. doi: 10.1152/ajpgi.00094.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riera K.M., Jang B., Min J., et al. Trop2 is upregulated in the transition to dysplasia in the metaplastic gastric mucosa. J Pathol. 2020;251:336–347. doi: 10.1002/path.5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cunningham S.C., Kamangar F., Kim M.P., et al. Claudin-4, mitogen-activated protein kinase kinase 4, and stratifin are markers of gastric adenocarcinoma precursor lesions. Cancer Epidemiol Biomarkers Prev. 2006;15:281–287. doi: 10.1158/1055-9965.EPI-05-0539. [DOI] [PubMed] [Google Scholar]
- 23.McDonnell S., Navre M., Coffey R.J., et al. Expression and localization of the matrix metalloproteinase pump-1 (MMP-7) in human gastric and colon carcinomas. Mol Carcinog. 1991;4:527–533. doi: 10.1002/mc.2940040617. [DOI] [PubMed] [Google Scholar]
- 24.Zhou J., Fan X., Chen N., et al. Identification of CEACAM5 as a biomarker for prewarning and prognosis in gastric cancer. J Histochem Cytochem. 2015;63:922–930. doi: 10.1369/0022155415609098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costa A., Kieffer Y., Scholer-Dahirel A., et al. Fibroblast heterogeneity and immunosuppressive environment in human breast cancer. Cancer Cell. 2018;33:463–479.e10. doi: 10.1016/j.ccell.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 26.Helms E., Onate M.K., Sherman M.H. Fibroblast heterogeneity in the pancreatic tumor microenvironment. Cancer Discov. 2020;10:648–656. doi: 10.1158/2159-8290.CD-19-1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shoshkes-Carmel M., Wang Y.J., Wangensteen K.J., et al. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature. 2018;557:242–246. doi: 10.1038/s41586-018-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bahar Halpern K., Massalha H., Zwick R.K., et al. Lgr5+ telocytes are a signaling source at the intestinal villus tip. Nat Commun. 2020;11:1936. doi: 10.1038/s41467-020-15714-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wada T., Ishimoto T., Seishima R., et al. Functional role of CD44v-xCT system in the development of spasmolytic polypeptide-expressing metaplasia. Cancer Sci. 2013;104:1323–1329. doi: 10.1111/cas.12236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cortazar AR, Oguiza JA, Aransay AM, et al. VerSeDa: vertebrate secretome database. Database (Oxford) 2017;2017:baw171. [DOI] [PMC free article] [PubMed]
- 31.Vannucchi M.G. The telocytes: ten years after their introduction in the scientific literature. an update on their morphology, distribution, and potential roles in the gut. Int J Mol Sci. 2020;21:4478. doi: 10.3390/ijms21124478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kurahashi M., Nakano Y., Peri L.E., et al. A novel population of subepithelial platelet-derived growth factor receptor alpha-positive cells in the mouse and human colon. Am J Physiol Gastrointest Liver Physiol. 2013;304:G823–G834. doi: 10.1152/ajpgi.00001.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCarthy N., Manieri E., Storm E.E., et al. Distinct mesenchymal cell populations generate the essential intestinal BMP signaling gradient. Cell Stem Cell. 2020;26:391–402.e5. doi: 10.1016/j.stem.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ham I.H., Lee D., Hur H. Role of cancer-associated fibroblast in gastric cancer progression and resistance to treatments. J Oncol. 2019;2019 doi: 10.1155/2019/6270784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Quante M., Tu S.P., Tomita H., et al. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer Cell. 2011;19:257–272. doi: 10.1016/j.ccr.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kobayashi H., Enomoto A., Woods S.L., et al. Cancer-associated fibroblasts in gastrointestinal cancer. Nat Rev Gastroenterol Hepatol. 2019;16:282–295. doi: 10.1038/s41575-019-0115-0. [DOI] [PubMed] [Google Scholar]
- 37.Min J., Zhang C., Bliton R.J., et al. Dysplastic stem cell plasticity functions as a driving force for neoplastic transformation of precancerous gastric mucosa. Gastroenterology. 2022;163:875–890. doi: 10.1053/j.gastro.2022.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan S.H., Swathi Y., Tan S., et al. AQP5 enriches for stem cells and cancer origins in the distal stomach. Nature. 2020;578:437–443. doi: 10.1038/s41586-020-1973-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.