Abstract
The visna virus Tat protein is required for efficient viral transcription from the visna virus long terminal repeat (LTR). AP-1 sites within the visna virus LTR, which can be bound by the cellular transcription factors Fos and Jun, are also necessary for Tat-mediated transcriptional activation. A potential mechanism by which the visna virus Tat protein could target the viral promoter is by protein-protein interactions with Fos and/or Jun bound to AP-1 sites in the visna virus LTR. Once targeted to the visna virus promoter, the Tat protein could then interact with basal transcription factors to activate transcription. To examine protein-protein interactions with cellular proteins at the visna virus promoter, we used an in vitro protein affinity chromatography assay and electrophoretic mobility shift assay, in addition to an in vivo two-hybrid assay, to show that the visna virus Tat protein specifically interacts with the cellular transcription factors Fos and Jun and the basal transcription factor TBP (TATA binding protein). The Tat domain responsible for interactions with Fos and Jun was localized to an alpha-helical domain within amino acids 34 to 69 of the protein. The TBP binding domain was localized to amino acids 1 to 38 of Tat, a region previously described by our laboratory as the visna virus Tat activation domain. The bZIP domains of Fos and Jun were found to be important for the interactions with Tat. Mutations within the basic domains of Fos and Jun abrogated binding to Tat in the in vitro assays. The visna virus Tat protein was also able to interact with covalently cross-linked Fos and Jun dimers. Thus, the visna virus Tat protein appears to target AP-1 sites in the viral promoter in a mechanism similar to the interaction of human T-cell leukemia virus type 1 Tax with the cellular transcription factor CREB, by binding the basic domains of an intact bZIP dimer. The association between Tat, Fos, and Jun would position Tat proximal to the viral TATA box, where the visna virus Tat activation domain could contact TBP to activate viral transcription.
Visna virus is a retrovirus of the lentivirus family, whose members include the primate viruses human and simian immunodeficiency viruses (HIV and SIV), equine infectious anemia virus (EIAV), and caprine arthritis encephalitis virus (CAEV). Visna virus causes chronic-progressive pneumonitis, arthritis, and encephalitis in sheep and is characterized by an extended period of clinical latency (9, 18, 31, 37). The targets of visna virus infection in vivo are cells of the monocyte/macrophage lineage. The pathogenesis of the disease is due to an activation of a cytokine cascade in macrophages, leading to an acute inflammatory response in target organs (30, 32, 48). Although visna virus can infect monocyte precursors in bone marrow and peripheral blood monocytes, little or no viral gene expression is detected in these cells (15, 16, 47). Differentiation of the monocyte into a macrophage is necessary to stimulate visna virus expression; activation of viral transcription, upon differentiation, requires both viral and cellular factors (8, 10, 12, 14–16, 19, 31).
The visna virus tat gene product encodes a 94-amino-acid (aa), 10-kDa protein that is necessary for activation of viral transcription (10). Tat-mediated transcriptional activation also requires the presence of AP-1 and AP-4 sites within the U3 region of the visna virus long terminal repeat (LTR) (14). Mutational analysis identified a consensus AP-1 site, proximal to the visna virus promoter TATA box, that is the most important element for induction of transcription in response to cellular differentiation, and studies have shown that visna virus Tat can act through heterologous promoters containing AP-1 sites (12, 14, 36, 40). Although AP-1 sites appear to be a necessary element for Tat-mediated transcriptional activation, visna virus Tat does not bind AP-1 sequences directly, nor does it bind any DNA sequences within the viral LTR (14). Additionally, visna virus does not contain any sequences 3′ of the transcription start site that are important for transcriptional activation. Therefore, a cis-acting transactivation response (TAR) element, such as that found in the HIV LTR, does not appear to play a role in visna virus Tat-mediated transcriptional activation (19).
In previous studies, we have shown that the visna virus Tat protein contains a potent acidic activation domain between aa 1 to 38 of the protein (Tat 1-38) (5). The visna virus Tat activation domain was found to have a pattern of critical hydrophobic residues similar to those of other acidic activation domains, such as the herpesvirus VP16 activation domain. Acidic activators, such as VP16, have been found to interact with a number of general transcription factors, including the TATA box binding protein (TBP) (21, 42). The current model of transcriptional activation contends that once a transactivating protein is targeted to a specific promoter, the activation domain stimulates transcription through these specific interactions with RNA polymerase II-associated general transcription factors (3, 46).
In addition to the activation domain, a specific domain important in mediating the AP-1 responsiveness of the Tat protein was identified (6). This AP-1-responsive domain, contained within aa 34 to 62 of the Tat protein, is characterized by four leucine residues that are highly conserved among visna virus strains and the related lentivirus CAEV. The leucine residues within this domain do not form a heptad repeat, typical of leucine zipper domains. There have, however, been characterized a number of proteins that contain domains important in protein-protein interactions that contain leucine residues but do not conform to a leucine zipper motif (11). It is possible, therefore, that this region of the Tat protein is important in making contacts with other proteins and that these protein-protein interactions are critical in mediating the AP-1 responsiveness of the Tat protein. In competition experiments, the leucine domain alone was able to competitively inhibit Tat transactivation of a visna virus LTR-chloramphenicol acetyltransferase (CAT) reporter containing AP-1 binding sites (6). This suggests that the leucine domain is able to bind cellular factors important for Tat-mediated activation of the visna virus promoter.
To determine how the visna virus Tat protein is targeted to the promoter and how it activates transcription, we have examined interactions between Tat and cellular proteins that recognize visna virus promoter elements. By both in vitro and in vivo analysis, we demonstrate that the Tat protein interacts with both cellular AP-1 transcription factors, Fos and Jun. This interaction would be expected to target the visna virus Tat protein to the viral promoter by way of the AP-1 sites located upstream of the transcriptional initiation site. In addition, the visna virus Tat protein was able to bind to TBP. This characteristic is shared by a number of other transcriptional activators and potentially provides a mechanism for activating transcription. Additionally, we identify the region of the Tat protein responsible for the interactions with Fos and Jun. This region is contained within aa 34 to 69 of the visna virus Tat protein, which is the same region previously shown to be important in mediating the Tat protein’s responsiveness to AP-1 DNA sequences. Furthermore, we show that the bZIP domains within Fos and Jun are involved in the interaction between the visna virus Tat protein and the AP-1 binding factors and that amino acids within the basic region of the bZIP domain are critical for this interaction.
MATERIALS AND METHODS
Plasmids. (i) Construction of GST-Tat fusion proteins.
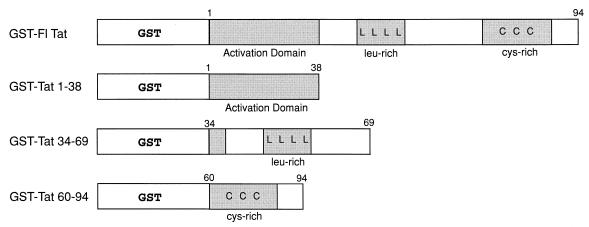
Glutathione S-transferase (GST)-Tat, GST-Tat 1-38, GST-Tat 34-69, and GST-Tat 60-94 were constructed by cloning fragments generated by PCR amplification of regions in the visna virus tat gene into the GST expression vector pGEX-3X (Pharmacia). GST-Tat, GST-Tat 1-38, and GST-Tat 60-94 were cloned into a BglII site of the modified pGEX-3X vector GH418, obtained from Gary Haywood (Johns Hopkins University Medical School). The remaining GST-Tat constructs were cloned into the BamHI and EcoRI sites of pGEX-3X. All constructs were confirmed by nucleotide sequence analysis.
(ii) Construction of fos and jun in vitro expression vectors.
The wild-type fos and fos and jun mutant constructs (described in reference 17) were obtained from Tom Kerppola (University of Michigan Medical Center). The fos mutants were excised from the plasmid pDS56 by digestion with BamHI and HindIII and cloned into pGEM-7Z (Promega) for in vitro expression. jun mutant constructs were excised from pGEM4 by digestion with EcoRI and cloned into pGEM-7Z. The wild-type jun construct was obtained from Daniel Nathans (Johns Hopkins University Medical School). Constructs were confirmed by restriction digest.
(iii) Construction of eukaryotic expression vectors for two-hybrid experiments.
The VP/FosZip and VP/JunZip vectors were constructed as follows. The Fos bZIP region (amino acids [aa] 131 to 208) was PCR amplified from the wild-type fos construct (described above), and the Jun bZIP region (aa 222 to 331) was digested from the GST-cJun (bZIP) construct obtained from Michael Green (University of Massachusetts Medical Center). Sequences encoding the VP16 activation domain (aa 1 to 47) were amplified from plasmid GH322, obtained from Gary Hayward and described by Hardwick et al. (18a). The VP16 activation domain was then ligated to the Fos and Jun bZIP domains, and these constructs were ligated into the vector pCDNA 3.1 (Promega). Gal4-Tat constructs were prepared as previously described (5). All constructs were confirmed by nucleotide sequence analysis.
Expression of GST fusion proteins.
DNA for each GST expression vector was used to transform Escherichia coli JM109 cells. Two milliliters of an overnight culture of transformed bacteria was used to seed 20 ml of LB medium containing ampicillin (100 μg/ml) and grown at 30°C until an optical density of 0.7 to 0.8 was reached. The cells were then induced with 0.05 mM isopropyl-β-d-thiogalactoside for 2 h at room temperature. The cells were then centrifuged and resuspended in 1 ml of phosphate-buffered saline (PBS; pH 7.3) containing 1% Triton X-100 (Sigma) and lysozyme (100 μg/ml, final concentration) for 30 min at room temperature. The cells were then sonicated twice with a 5-mm tip for 10 s. The lysate was spun in a microcentrifuge at full speed for 10 min, and the supernatant was collected. A 50% suspension (50 μl) of glutathione-Sepharose 4B beads (Pharmacia) in PBS was then added and incubated at room temperature with rocking for 30 min. The beads were washed three times with 250 ml of PBS and then suspended in 25 μl of PBS for use in the GST binding assays.
Purification of bacterially expressed full-length Tat protein.
GST-Tat protein was expressed in bacterial cells as described above. The full-length Tat protein was cleaved from GST by digestion with factor Xa as specified by the manufacturer (Pharmacia) except that 0.1% Triton X-100 and 5 mM dithiothreitol were included in both the PBS wash buffers and factor Xa cleavage buffer. Factor Xa protein was then removed by incubation of the eluted Tat protein solution with 65 μl of 50% benzamidine-Sepharose beads (Pharmacia) per 2.5 liters of starting culture for 15 min. with rocking. The beads were pelleted by ultracentrifugation for 5 min at 500 × g for 5 min, and supernatant containing the purified Tat protein was removed, aliquoted, and stored at −80°C.
GST binding assays.
Five micrograms of fusion protein bound to Sepharose beads was incubated with 2 × 105 cpm of [35S]methionine-labeled in vitro-translated proteins synthesized in a coupled transcription-translation reaction (Promega) in 200 μl of binding buffer (40 mM HEPES [pH 7.55], 100 mM KCl, 10 μM ZnCl2, 0.1% Nonidet P-40, 20 mM 2-mercaptoethanol, 2 μg of bovine serum albumin per ml). The proteins were incubated for 2 h at 4°C and then washed four times with 500 μl of binding buffer; 30 μl of 2× sodium dodecyl sulfate (SDS) loading buffer was then added, and 15 μl of sample was assayed by SDS-polyacrylamide electrophoresis (SDS-PAGE). Alternatively, 5 μg of purified fusion protein was incubated with in vitro-translated proteins in 200 μl of binding buffer for 2 h and then incubated with 50 μl of GST beads for 30 min before being washed as described above. Either method produced the same results.
Transfections.
Sheep choroid plexus (SCP) cells (32) were maintained in modified Eagle medium supplemented with 10% fetal bovine serum and incubated at 5% CO2. Cells were transfected with the Lipofectamine reagent (Promega) according to the manufacturer’s protocol; 0.5 μg of each DNA and 5 μl of Lipofectamine were used per reaction. The cells were incubated with the DNA and the Lipofectamine reagent for 7 h and were harvested for CAT assay analysis 48 h after the start of transfection.
Cross-linking.
In vitro-translated proteins (4 × 105 cpm) were incubated with 0.25 mM BS3 (Pierce) in 20 μl of 1× PBS for 5 min at room temperature. The reaction was then quenched with 50 mM Tris (pH 7.5) for 15 min at room temperature.
CAT assays.
CAT assays were performed by using the Promega CAT enzyme assay system.
Electrophoretic mobility shift assay (EMSA).
Binding reaction mixtures contained 1 μl of 32P-labeled DNA probe, 1.6 μl of 5× binding buffer (125 mM HEPES [pH 7.9], 25 mM KCl, 5 mM EDTA, 5 mg of bovine serum albumin per ml, 50% glycerol, 1.25 mM dithiothreitol), 1 μl of dI-dC (300 ng), 100 ng of each protein, and H2O to 8 μl. Reaction mixtures were incubated at 37°C for 20 min, chilled on ice, then loaded on a 4% Tris-glycine-EDTA gel, and run at 200 V for 3 h at 4°C. The LTR probe was a SalI/NcoI-digested fragment from the visna virus LTR, including sequences from −65 to +56. The AP-1 probe (Santa Cruz Biotechnology Inc.) contains the sequence 5′-CGCTTGATGACTCAGCCGGAA-3′.
The Fos bZIP (aa 139 to 200) and Jun bZIP (aa 199 to 334) domain proteins were kindly provided by Tom Kerppola. These proteins were purified from E. coli overexpression strains to greater than 90% homogeneity by nickel chelate affinity chromotography.
RESULTS
The visna virus Tat protein interacts with Fos, Jun, and TBP.
The visna virus Tat protein has been shown to transactivate its viral promoter and heterologous promoters through cis-acting AP-1 sites in the DNA. However, Tat does not activate transcription from AP-1 sites through direct interactions with the DNA (14). One mechanism by which Tat could target the promoters through an AP-1 site would be via interactions with the cellular AP-1 binding proteins, Fos and Jun. Additionally, once targeted to the visna virus promoter, Tat functions to activate transcription through its activation domain. Other viral transcriptional activating proteins, such as herpesvirus VP16 and HIV Tat, have been found to interact with TBP, which is thought to be a mechanism for activating transcription (21, 22). To examine potential protein-protein interactions of the visna virus Tat protein with the AP-1 binding proteins Fos and Jun and with TBP, full-length visna virus Tat was expressed as a GST-Tat fusion protein in bacteria and then immobilized on glutathione-Sepharose beads (Fig. 1). As controls, GST alone and a fusion protein of GST and the VP16 activation domain (GST-VP16) were also immobilized on glutathione-Sepharose beads. GST alone, GST-Tat, and GST-VP16 were then incubated with 35S-labeled Fos, Jun, and TBP synthesized by in vitro transcription-translation in a rabbit reticulocyte lysate and washed as described in Materials and Methods. The 35S-labeled proteins that were retained on the immobilized GST proteins were then analyzed by SDS-PAGE.
FIG. 1.
GST-visna virus Tat fusion protein constructs used in in vitro affinity chromatography experiments. Fusion proteins were expressed in E. coli JM109, purified, and used for in vitro affinity chromatography as described in Materials and Methods. Shaded areas refer to regions of the visna virus Tat protein.
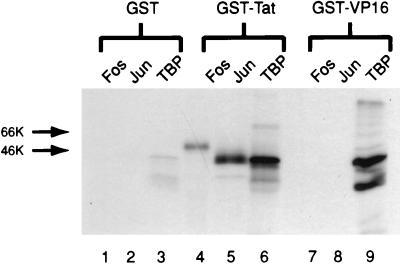
The control GST resin did not bind a significant amount of Fos, Jun, or TBP in this assay (Fig. 2, lanes 1 to 3). GST-Tat, however, specifically retained the AP-1 binding factors Fos and Jun in this in vitro chromatography assay, as well as the basal transcription factor TBP (Fig. 2, lanes 3 to 5). GST-VP16 also bound TBP in the assay (Fig. 2, lane 9), which has previously been reported as a functional interaction (21). There was, however, no retention of Fos or Jun by GST-VP16 (Fig. 2, lanes 7 and 8), demonstrating that the binding of Fos and Jun in this assay is specific for the Tat protein.
FIG. 2.
Interactions of visna virus Tat protein with Fos, Jun, and TBP. GST alone (lanes 1 to 3), GST-Tat (lanes 4 to 6), and GST-VP16 (lanes 7 to 9) were immobilized on glutathione-Sepharose beads and incubated with in vitro-translated, [35S]methionine-labeled Fos, Jun, and TBP for 2 h at 4°C. Following incubation, bound proteins were washed extensively, eluted, and analyzed by SDS-PAGE on a 10% gel followed by autoradiography.
Specific domains within the Tat protein are responsible for binding the AP-1 factors and to TBP.
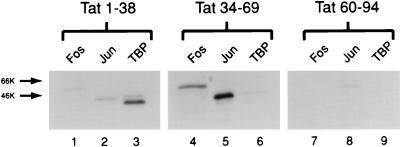
To define domains within the visna virus Tat protein that interact with Fos, Jun, and TBP, we made a number of constructs that expressed portions of Tat as GST fusion proteins (Fig. 1). When the visna virus Tat activation domain was expressed as a GST fusion protein (GST-Tat 1-38) and incubated with in vitro-translated Fos, Jun, and TBP, there was significant retention only of TBP (Fig. 3, lanes 1 to 3). This result is consistent with studies with other acidic transcriptional activators, in that the activation domains of these proteins are responsible for contacting general transcription factors, such as TBP (20–22).
FIG. 3.
Interactions of different domains of the visna virus Tat protein with Fos, Jun, and TBP. GST-Tat 1-38 (lanes 1 to 3), GST-Tat 34-69 (lanes 4 to 6), and GST-Tat 60-94 (lanes 7 to 9) were immobilized on glutathione-Sepharose beads and incubated with in vitro-translated, [35S]methionine-labeled Fos, Jun, and TBP, and bound proteins were analyzed as described for Fig. 2.
A leucine-containing domain within aa 34 to 62 of the visna virus Tat protein has been shown to be important in mediating the AP-1 responsiveness of Tat. In studies using computational analysis and circular dichroism (data not shown), it was found that visna virus Tat contains an alpha-helical region within aa 34 to 69. Based on these observations, this entire helical region was included within a fusion protein containing the leucine domain (33, 34). The GST-Tat 34-69 construct, containing this helical domain, was found to specifically interact with Fos and Jun but not TBP (Fig. 3, lanes 4 to 6).
An additional domain within the C-terminal region of the Tat protein contains three cysteines that are also conserved among visna virus strains and CAEV. Although this region is necessary for Tat function in a number of in vivo assays, GST-Tat 60-94, containing this domain, did not bind Fos, Jun, or TBP in this assay (Fig. 3, lanes 7 to 9) (14).
Specific regions within the Fos and Jun bZIP domains are responsible for interacting with visna virus Tat.
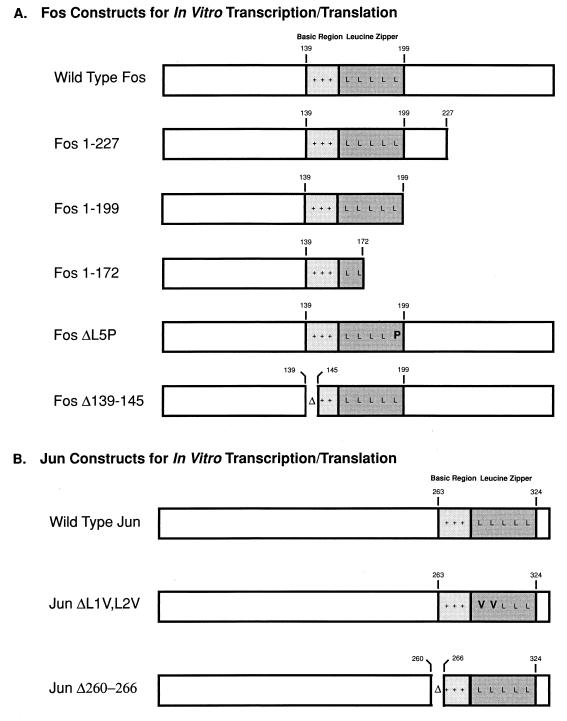
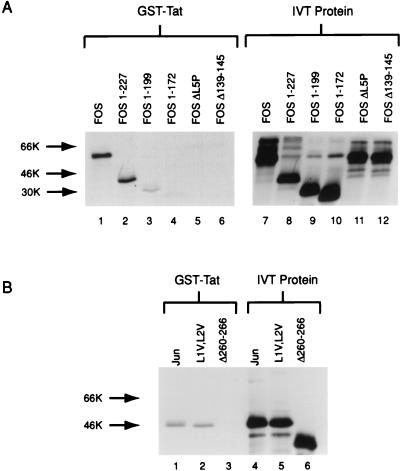
To identify which regions of Fos and Jun were important for interactions with the visna virus Tat protein, we used a panel of Fos and Jun 35S-labeled proteins (Fig. 4) in the in vitro protein affinity chromatography experiments (Fig. 5). GST-Tat was incubated with a number of in vitro-translated Fos proteins (Fig. 5A). The full-length Fos protein (Fig. 5A, lane 1) and a truncated Fos protein (aa 1 to 227) (lane 2) bound Tat efficiently. The Fos 1-227 protein contains an intact bZIP region of Fos, which is located between aa 139 and 200. When the Fos protein was truncated at the C-terminal bZIP boundary (Fos 1-199), binding to GST-Tat was greatly diminished (lane 3). Further truncation into the bZIP domain (Fos 1-172) almost completely eliminates any binding to GST-Tat (lane 4). Additionally, a proline substitution of the fifth leucine in the leucine zipper (Fos ΔL5P), a mutation that destabilizes the bZIP alpha helix, eliminates binding to GST-Tat (lane 5). Interestingly, a deletion of aa 139 to 145, within the basic region of the bZIP domain (Fos Δ139-145), also eliminated the ability of the in vitro-translated Fos protein to bind GST-Tat (lane 6). This deletion does not affect the ability of Fos to interact with Jun through their leucine zippers (17). The relative amount of each in vitro-translated Fos protein incubated with GST-Tat is shown in Fig. 5A, lanes 7 to 12.
FIG. 4.
Fos and Jun proteins used in in vitro affinity chromatography experiments. Plasmids containing Fos and Jun wild-type and mutated sequences were transcribed, translated, and labeled in vitro with [35S]methionine as described in Materials and Methods. Leucine zipper regions of the Fos and Jun bZIP domains of the proteins are marked by darkly shaded areas; basic domains of the bZIP domains are represented by lightly shaded regions. The point mutations in Fos ΔL5P and JunΔL1V,ΔL2V are in boldface type.
FIG. 5.
Interactions of GST-Tat with Fos and Jun. Fos and Jun proteins were translated in vitro with [35S]methionine, and GST-Tat was incubated with equivalent counts per minute of each Fos and Jun protein. (A) In vitro-translated Fos proteins were incubated with GST-Tat as described for Fig. 2 (lanes 1 to 6). In vitro-translated Fos proteins (IVT Protein) were loaded directly onto the gel, without interaction with GST-Tat, in lanes 7 to 12 to show that equal counts were loaded. Lanes 7 to 12 were exposed to radiography roughly 1/10 as long as lanes 1 to 6. (B) In vitro-translated Jun proteins were incubated with GST-Tat as described for Fig. 2 (lanes 1 to 3). In lanes 4 to 6, a 1/10 dilution of the amount of each Jun protein incubated GST-Tat was loaded directly on the gel to show that equal counts were loaded.
Two constructs that express Jun proteins with mutations in the bZIP domain were also tested for the ability to bind GST-Tat (Fig. 5B). JunΔL1V,L2V has the first and second leucines in the leucine zipper substituted with valines. These mutations do not disturb the overall structure of the bZIP domain, in contrast to the proline substitution in the Fos leucine domain, although they do inhibit Fos dimerization to Jun (17). This substitution was not, however, able to disrupt the interaction with Tat in this assay; it bound GST-Tat as well as wild-type Jun (Fig. 5B, lanes 1 and 2). A deletion in the basic region of Jun does, however, abrogate binding to GST-Tat (lane 3), much like the same mutation in the bZIP domain of the Fos protein. As is the case for the Fos basic domain deletion, this Jun deletion mutant does not affect its ability to form a leucine zipper dimer (17). The relative amount of Jun in vitro-translated protein incubated with GST-Tat is shown (lanes 4 to 6).
Visna virus Tat protein interacts with the bZIP domains of Fos and Jun in mammalian cells.
The bZIP domains of Fos and Jun were found to be important in mediating protein-protein interactions with Tat in the in vitro binding assay. Many viral transcriptional activators, such as human T-cell leukemia virus type 1 (HTLV-1) Tax and the hepatitis B virus X protein, bind the bZIP domains of cellular transcription factors to activate viral and cellular promoters (1, 43–45).
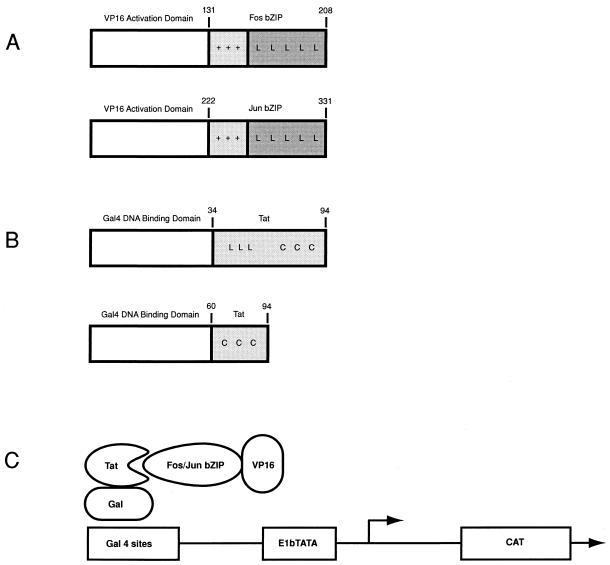
To determine if Tat interacts with the bZIP domains of Fos and Jun in the context of the cellular environment, a mammalian two-hybrid system was used (Fig. 6). The Tat protein lacking the activation domain was fused to the DNA binding domain of Gal4 (Gal4-Tat 34-94). A Tat construct lacking the leucine domain found to be important in binding Fos and Jun (Gal4-Tat 60-94) was also fused to Gal4. The Fos and Jun bZIP domains were then fused to the transcriptional activating region of VP16 (VP/FosZip and VP/JunZip). The Tat-Gal4 constructs were transfected into SCP cells, a primary cell line that visna virus replicates in productively, along with the bZIP-VP16 constructs and a CAT reporter containing five Gal4 DNA binding sites. The interaction of the Tat domain of the Tat-Gal4 fusions with either the Fos bZIP or Jun bZIP domains of the bZIP-VP16 fusions would target the VP16 activation domain to the Gal4 sites in the reporter and activate transcription of the reporter gene.
FIG. 6.
Mammalian two-hybrid analysis. (A) The Fos and Jun bZIP domains were fused to the activation domain of VP16. (B) Portions of visna virus Tat were fused to the DNA binding domain of Gal4. (C) Schematic showing that the interaction of visna virus Tat with the bZIP domains of Fos or Jun will target the VP16 activation domain to a reporter construct containing Gal4 binding sites and drive transcription of the CAT gene when cotransfected into SCP cells.
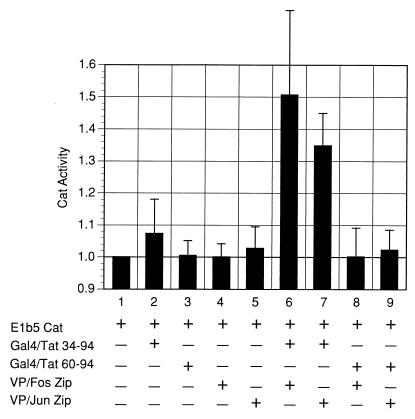
With this assay, the Gal4-Tat and VP16-bZIP fusions were not able to significantly activate transcription from the CAT reporter containing the Gal4 DNA binding sites above the background level when transfected into cells alone (Fig. 7, bars 1 to 5). When cotransfected with either the Fos bZIP or Jun bZIP VP-16 fusion, Gal4-Tat 34-94 could effectively interact in the SCP cells, recruiting the VP16 activation domain to the CAT reporter and driving transcription of the reporter gene (bars 6 and 7). Gal4-Tat 60-94, which lacks the region found to be important in interacting with Fos and Jun in the in vitro binding assays, does not interact with the bZIP domains in this assay to activate the CAT reporter gene (bars 8 and 9).
FIG. 7.
Visna virus Tat interaction with the Fos and Jun bZIP domains in vivo by mammalian two-hybrid analysis. SCP cells were transfected with the E1b5CAT plasmid (lanes 1 to 9) and cotransfected with Gal4 DNA binding domain-Tat fusion constructs and with VP16 activation domain-bZIP constructs (shown in Fig. 6) as described in Materials and Methods. The activity of the E1b5CAT vector alone was assigned an activity of 1, and the CAT activities of the other samples are plotted relative to this background activity. This result is representative of at least three independent experiments, and the standard error is shown.
Dimerized Fos and Jun can interact with the visna virus Tat protein.
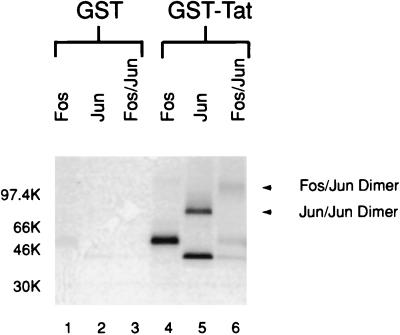
Fos and Jun are found in the cell as dimer pairs. If the visna virus Tat protein contacts the basic region of the proteins, then it is possible that it interacts with Jun-Jun or Fos-Jun dimers. To examine this possibility, Fos and Jun were cross-linked with an irreversible cross-linker to form covalently linked dimers. These dimers were then tested for the ability to bind GST-Tat in an in vitro affinity chromatography assay (Fig. 8). To control for nonspecific interactions with the dimerized Fos and Jun proteins, cross-linked protein was also incubated with GST alone (Fig. 8, lanes 1 to 3). In this assay, the covalently cross-linked Jun-Jun and Fos-Jun dimers are able to interact with GST-Tat (lanes 4 to 6).
FIG. 8.
Visna virus Tat interaction with Fos and Jun dimers. In vitro-translated Fos and Jun were cross-linked and then tested for the ability to bind GST by in vitro affinity chromatography as described in Materials and Methods. Chemically cross-linked in vitro-translated Fos and Jun proteins were tested for the ability to bind GST alone (lanes 1 to 3) and GST-Tat (lanes 4 to 6) as described for Fig. 2. The samples were analyzed by electrophoresis on an SDS–4 to 20% gradient polyacrylamide gel and autoradiography.
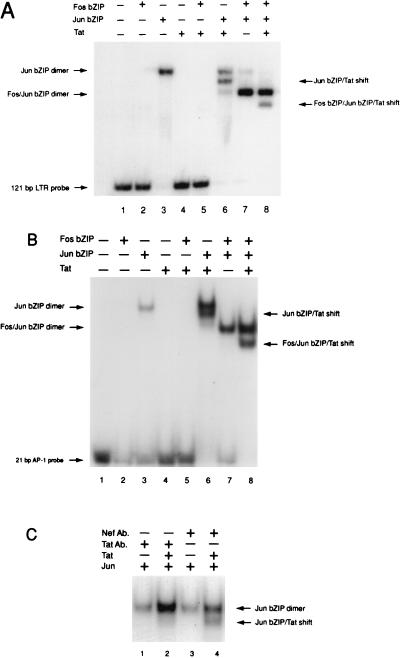
Visna virus Tat interacts with Fos and Jun to form a faster-migrating band in an EMSA.
Having established that visna virus Tat can interact with the bZIP domains of Fos and Jun in vitro and in vivo, we used an EMSA to determine if these interactions take place in the context of an AP-1 binding site on DNA. In the EMSA, bacterially expressed, purified visna virus Tat protein, the Fos bZIP domain (aa 139 to 200), and Jun bZIP domain (aa 199 to 334) were incubated alone and in combination with a 32P-labeled restriction fragment of the visna virus LTR containing the proximal AP-1 site and an AP-4 site. In addition, a synthetic oligonucleotide containing a consensus AP-1 site was used in the EMSA. Neither the Fos bZIP or the Tat protein alone shifted the LTR probe (Fig. 9A, lanes 2, 4, and 5). The Jun bZIP domain alone forms a distinct shifted band in the EMSA (lane 3), as does the Fos-Jun bZIP heterodimer (lane 7). Interestingly, when the visna virus Tat protein is incubated with either the Jun bZIP dimer or the Fos-Jun bZIP heterodimer, there appears in each case a distinct novel band that migrates more rapidly than either complex (lanes 6 and 8). These results were also observed with a synthetic oligonucleotide containing the consensus AP-1 site (Fig. 9B). In an additional EMSA analyzing full-length Fos and Jun protein heterodimers, it was observed that the addition of the visna virus Tat protein once again resulted in the formation of a novel, more rapidly migrating band (data not shown).
FIG. 9.
(A) EMSA using a fragment of the visna virus LTR containing the TATA proximal AP-1 site (−65 to +56) and bacterially expressed and purified Jun bZIP (aa 199 to 334), Fos bZIP (aa 199 to 334), and full-length Tat proteins. The indicated proteins (100 ng of each) were incubated with the 32P-labeled visna virus LTR fragment as indicated in Materials and Methods. (B) EMSA using a 32P-labeled oligonucleotide probe containing a consensus AP-1 site. Bacterially expressed and purified Jun bZIP, Fos bZIP, and Tat proteins were incubated with the 32P-labeled oligonucleotide by the procedure used for panel A. (C) Antibody (Ab) specific to the visna virus Tat N-terminal region (lane 2) or a nonspecific antibody (antibody to SIV Nef; lane 4) was preincubated with the Tat protein on ice for 1 h and then run in an EMSA with the Jun protein and labeled AP-1 probe as for panel B. The antibody to visna virus Tat and the control antibody were also included in the reactions with Jun alone (lanes 1 and 3).
Aberrant migration of bZIP proteins bound to DNA binding sites, resulting in more rapid migration in the EMSA, has been observed with Fos and Jun and other bZIP proteins. The mechanism behind this shift in mobility has been described as either an effect of the bZIP proteins on the bending of the DNA probe or a change in the conformation of the bZIP dimer (23–25, 28, 38, 39). It is possible, therefore, that the Tat protein exerts an effect on the bZIP dimers either by altering their interaction with the DNA or by altering the conformation of the complex, causing it to migrate more rapidly in the gel. The small size of the AP-1 oligonucleotide, however, argues against any effect due to altered DNA bending, as differences in bending of an oligonucleotide of this size would not influence complex mobility.
To further determine whether the effect on the Fos and Jun complexes was due to the Tat protein itself, and eliminate any possibility that the shift may have been due to a nonspecific effect, an antibody to the N terminus of visna virus Tat was used in the EMSA. When preincubated with the anti-Tat antibody, the purified visna virus Tat protein did not affect the formation of the Jun-Jun dimer (Fig. 9C, lane 1), but it blocked the downshifted Jun-Tat complex from forming (lane 2). In contrast, a nonspecific antibody (SIV Nef) had no effect on formation of either the Jun-Jun dimer or the Tat-Jun complex.
DISCUSSION
Transcriptional activators utilize a variety of methods to target the appropriate promoter. They may contain DNA binding domains that directly contact sequences within the targeted promoter, as do many cellular transcriptional activators, such as SP1 or NFκB. The lentivirus transcriptional activator HIV Tat, which contains an RNA binding motif, uses another mechanism that can tether the transactivator to the promoter by way of a nascent RNA transcript (2, 35, 41). Protein-protein interactions also target activators to a specific promoter. This last mechanism is utilized by a number of viral transcriptional activators, including adenovirus E1A and HTLV-1 Tax (1, 7, 27, 44, 45). The visna virus Tat protein does not bind DNA directly, nor does it bind to nascent RNA via a TAR-like element found in HIV or EIAV Tat (13, 14, 19). It is therefore most likely that Tat is targeted to the promoter through direct protein-protein interactions with cellular factors binding to the AP-1 sites in the visna virus LTR.
In this study, protein-protein interactions with cellular factors important for activation of the visna virus promoter were examined by using both in vitro (protein affinity chromatography assays and EMSAs) and in vivo (two-hybrid analysis) approaches. Using these techniques, we have demonstrated that the visna virus Tat protein can specifically interact with both Fos and Jun, which would target Tat to AP-1 sites located in the visna virus LTR. Additionally, we have shown that Tat binds TBP. This interaction is shared among a number of other transcriptional activators, including herpesvirus VP16 and p53, and has been correlated with increasing the level TBP at the TATA box (4, 26).
The domains within Tat responsible for these protein-protein interactions were also identified. The TBP binding domain was localized to Tat aa 1 to 38, a region previously identified as being the activation domain of the protein (5). The activation domains of other transcriptional activators have also been found to bind general transcription factors such as TBP (4, 5, 26). Mutation of two phenyalanine residues that had been found to be important for the Tat activation domain in in vivo transcription assays did not affect the visna virus Tat protein’s ability to interact with TBP (data not shown). Therefore, although the ability of the visna virus activation domain to bind TBP may be important in activating transcription, other mechanisms involving other transcriptional machinery may also be involved. In fact, recent data from our laboratory indicate that the C-terminal domain of RNA polymerase II is necessary for visna virus Tat transcriptional activation, and transcriptional elongation mechanisms may be involved in Tat activity (29).
Utilizing in vitro binding assays, we also determined that a helical, leucine-rich domain (aa 34 to 69), previously associated with targeting Tat activity to AP-1 sites (6), was responsible for Tat interactions with Fos and Jun. Mutagenesis of the leucine residues in the visna virus Tat AP-1 binding domain (aa 34 to 69) did not affect the interaction with Fos or Jun in in vitro chromatography experiments (data not shown). The leucine residues within this domain, therefore, although necessary for Tat activity in vivo, are not critical for individual interactions with Fos or Jun in vitro.
The bZIP domains alone of Fos and Jun are able to interact with Tat in SCP cells, as demonstrated in the two-hybrid experiments (Fig. 7). Furthermore, mutations within the bZIP domains of Fos and Jun severely affect binding to Tat in vitro. The targeting of bZIP domains of cellular proteins is a strategy utilized by a number of other viral proteins to target promoters. HTLV-1 Tax, hepatitis B X protein, and the adenovirus E1a protein all interact with the bZIP domains of cellular proteins to target specific promoters (1, 7, 27, 44, 45).
Although the leucine residues in the Tat domain that interacts with Fos and Jun do not form a heptad repeat, we speculated that visna virus Tat may interact with Fos and Jun through a coiled-coil interaction characteristic of leucine zipper domains. Our data would argue against this possibility, since visna virus Tat can interact with Jun when the first and second leucines of the leucine zipper are mutated to valine. This double mutation inhibits Jun dimerization through the leucine zipper (17). Additionally, our finding that residues within the basic domain of both Fos and Jun are important for binding visna virus Tat protein indicates that the Tat protein may not bind Fos and Jun through a leucine zipper interaction. This finding, coupled with the fact that the leucines within the AP-1 targeting domain of Tat are not critical for interacting with Fos and Jun in vitro, suggests that the interaction of visna virus Tat with Fos and Jun is not through a Fos-Tat or Jun-Tat leucine zipper interaction. In HTLV-1, the Tax protein interacts with a CREB dimer, contacting the dimer within the basic region of the bZIP domains (1, 45). It is possible that the visna virus Tat protein binds the AP-1 factors in a similar fashion. The additional finding that Tat is able to interact with crosslinked Jun-Jun and Fos-Jun dimers would indicate that Tat may bind similarly to HTLV-1 Tax.
Our experiments indicate that the basic region of Fos and Jun are important in interactions with the visna virus Tat protein. Mutations within the leucine zipper region of the bZIP domain, however, can also decrease interaction with the visna virus Tat protein. This was demonstrated when Fos was truncated into the leucine zipper or when a proline residue was substituted for a leucine in the zipper region. These mutations would, however, tend to profoundly affect the local secondary structure of the region. Therefore, even though our data suggest that it may be the basic regions of the bZIP domains of Fos and Jun that mediate the interaction with visna virus Tat, our data also suggest that the structural integrity of the bZIP domain as a whole is important in this interaction.
From the results of the above-described in vitro and in vivo binding experiments, a model for Tat transactivation emerges. Visna virus Tat would be targeted to the visna virus promoter through interactions with the DNA binding basic region of a Fos-Jun heterodimer or possibly a Jun-Jun homodimer at the AP-1 sites in the visna virus promoter. The ability of visna virus Tat to interact with bZIP dimers complexed to DNA in the EMSA strengthens this argument. Once targeted, Tat would then contact TBP through its activation domain to increase transcriptional initiation from the visna virus promoter. The recent data that visna virus Tat has an effect on transcriptional elongation mechanisms, in addition to initiation, indicates that the exact mechanism of transcriptional activation may be quite complex.
This model of Tat transactivation corresponds well with what is known about visna virus replication in the host. The restricted replication of visna virus in monocytes, versus permissive replication when monocytes are activated to macrophages, could be accounted for by the increase in Fos and Jun levels in macrophages, which would increase the amount of Tat targeted to the visna virus promoter. It is possible that this mechanism of transactivation also has an effect on the host cell itself. Interaction with Fos and Jun may target visna virus Tat not only to its own promoter but also to cellular promoters containing AP-1 sites. This could lead to activation of a number of cellular genes, which could profoundly affect cell and viral growth and thus play a role in the pathogenesis of visna virus.
ACKNOWLEDGMENTS
We thank Maryann Brooks for assistance with the manuscript. Special thanks go to Thomas Kerppola for his generous contribution of plasmids and purified Fos and Jun proteins.
These studies were supported by grants NS23039 and NS07392 from the National Institutes of Health to Janice E. Clements.
REFERENCES
- 1.Adya N, Zhao L J, Huang W, Boros I, Giam C Z. Expansion of CREB’s DNA recognition specificity by Tax results from interaction with Ala-Ala-Arg at positions 282-284 near the conserved DNA-binding domain of CREB. Proc Natl Acad Sci USA. 1994;91:5642–5646. doi: 10.1073/pnas.91.12.5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berkhout B, Silverman R H, Jeang K T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 3.Buratowski S. The basics of basal transcription by RNA polymerase II. Cell. 1994;77:1–3. doi: 10.1016/0092-8674(94)90226-7. [DOI] [PubMed] [Google Scholar]
- 4.Caron C, Rousset R, Beraud C, Moncollin V, Egly J M, Jalinot P. Functional and biochemical interaction of the HTLV-1 Tax1 transactivator with TBP. EMBO J. 1993;12:4269–4278. doi: 10.1002/j.1460-2075.1993.tb06111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carruth L M, Hardwick J M, Morse B A, Clements J E. Visna virus Tat protein: a potent transcription factor with both activator and repressor domains. J Virol. 1994;68:6137–6146. doi: 10.1128/jvi.68.10.6137-6146.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carruth L M, Morse B A, Clements J E. The leucine domain of the visna virus Tat protein mediates targeting to an AP-1 site in the viral long terminal repeat. J Virol. 1996;70:4338–4344. doi: 10.1128/jvi.70.7.4338-4344.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatton B, Bocco J L, Gaire M, Hauss C, Reimund B, Goetz J, Kedinger C. Transcriptional activation by the adenovirus larger E1a product is mediated by members of the cellular transcription factor ATF family which can directly associate with E1a. Mol Cell Biol. 1993;13:561–570. doi: 10.1128/mcb.13.1.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clements J E, Zink M C, Narayan O, Gabuzda D H. Lentivirus infection of macrophages. Immunol Ser. 1994;60:589–600. [PubMed] [Google Scholar]
- 9.Cork L C, Hadlow W J, Gorham J R, Pyper R C, Crawford T B. Infectious leukoencephalomyelitis of goats. J Infect Dis. 1974;129:134–141. doi: 10.1093/infdis/129.2.134. [DOI] [PubMed] [Google Scholar]
- 10.Davis J L, Clements J E. Characterization of a cDNA clone encoding the visna virus transactivating protein. Proc Natl Acad Sci USA. 1989;86:414–418. doi: 10.1073/pnas.86.2.414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Flemington E, Speck S H. Evidence for coiled-coil dimer formation by an Epstein-Barr virus transactivator that lacks a heptad repeat of leucine residues. Proc Natl Acad Sci USA. 1990;87:9459–9463. doi: 10.1073/pnas.87.23.9459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabuzda D H, Hess J L, Small J A, Clements J E. Regulation of the visna virus long terminal repeat in macrophages involves cellular factors that bind sequences containing AP-1 sites. Mol Cell Biol. 1989;9:2728–2733. doi: 10.1128/mcb.9.6.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gdovin, S. L., L. M. Carruth, and J. E. Clements. Personal communication.
- 14.Gdovin S L, Clements J E. Molecular mechanisms of visna virus tat: identification of the targets for transcriptional activation and evidence for a post-transcriptional effect. Virology. 1992;188:438–450. doi: 10.1016/0042-6822(92)90497-d. [DOI] [PubMed] [Google Scholar]
- 15.Gendelman H E, Narayan O, Kennedy-Stoskopf S, Kennedy P G, Ghotbi Z, Clements J E, Stanley J, Pezeshkpour G. Tropism of sheep lentiviruses for monocytes: susceptibility to infection and virus gene expression increase during maturation of monocytes to macrophages. J Virol. 1986;58:67–74. doi: 10.1128/jvi.58.1.67-74.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gendelman H E, Narayan O, Molineaux S, Clements J E, Ghotbi Z. Slow, persistent replication of lentiviruses: role of tissue macrophages and macrophage precursors in bone marrow. Proc Natl Acad Sci USA. 1985;82:7086–7090. doi: 10.1073/pnas.82.20.7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gentz R, den Rauscher F J, Abate C, Curran T. Parallel association of Fos and Jun leucine zippers juxtaposes DNA binding domains. Science. 1989;243:1695–1699. doi: 10.1126/science.2494702. [DOI] [PubMed] [Google Scholar]
- 18.Gudnadottir M. Visna-maedi in sheep. Prog Med Virol. 1974;18:336–349. [PubMed] [Google Scholar]
- 18a.Hardwick J M, Tse L, Applegren N, Nicholas J, Veliuona M A. The Epstein-Barr virus R transactivator (Rta) contains a complex, potent activation domain with properties different from those of VP16. J Virol. 1992;66:5500–5508. doi: 10.1128/jvi.66.9.5500-5508.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hess J L, Small J A, Clements J E. Sequences in the visna virus long terminal repeat that control transcriptional activity and respond to viral trans-activation: involvement of AP-1 sites in basal activity and trans-activation. J Virol. 1989;63:3001–3015. doi: 10.1128/jvi.63.7.3001-3015.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horikoshi N, Maguire K, Kralli A, Maldonado E, Reinberg D, Weinmann R. Direct interaction between adenovirus E1A protein and the TATA box binding transcription factor IID. Proc Natl Acad Sci USA. 1991;88:5124–5128. doi: 10.1073/pnas.88.12.5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ingles C J, Shales M, Cress W D, Triezenberg S J, Greenblatt J. Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature. 1991;88:588–590. doi: 10.1038/351588a0. [DOI] [PubMed] [Google Scholar]
- 22.Kashanchi F, Piras G, Radonovich M F, Duvall J F, Fattaey A, Chiang C M, Roeder R G, Brady J N. Direct interaction of human TFIID with the HIV-1 transactivator Tat. Nature. 1994;367:295–299. doi: 10.1038/367295a0. [DOI] [PubMed] [Google Scholar]
- 23.Kerppola T K, Curran T. DNA bending by Fos and Jun: the flexible hinge model. Science. 1991;254:1210–1214. doi: 10.1126/science.1957173. [DOI] [PubMed] [Google Scholar]
- 24.Kerppola T K, Curran T. Fos-Jun heterodimers and Jun homodimers bend DNA in opposite orientations: implications for transcription factor cooperativity. Cell. 1991;66:317–326. doi: 10.1016/0092-8674(91)90621-5. [DOI] [PubMed] [Google Scholar]
- 25.Kerppola T K, Curran T. Selective DNA bending by a variety of bZIP proteins. Mol Cell Biol. 1993;13:5479–5489. doi: 10.1128/mcb.13.9.5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lieberman P M, Berk A J. The Zta trans-activator protein stablizies TFIID association with promoter DNA by direct protein-protein interaction. Genes Dev. 1991;5:2441–2454. doi: 10.1101/gad.5.12b.2441. [DOI] [PubMed] [Google Scholar]
- 27.Liu F, Green M R. Promoter targeting by adenovirus E1a through interaction with different cellular DNA-binding domains. Nature. 1994;368:520–525. doi: 10.1038/368520a0. [DOI] [PubMed] [Google Scholar]
- 28.McCormick R J, Badalian T, Fisher D E. The leucine zipper may induce electrophoretic mobility anomalies without DNA bending. Proc Natl Acad Sci USA. 1996;93:14434–14439. doi: 10.1073/pnas.93.25.14434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morse, B. A., J. L. Corden, J. Pyper, and J. E. Clements. 1998. Unpublished data.
- 30.Narayan O, Clements J E. Biology and pathogenesis of lentiviruses of ruminant animals. New York, N.Y: Marcel Dekker, Inc.; 1988. [Google Scholar]
- 31.Narayan O, Cork L C. Lentiviral diseases of sheep and goats Chronic pneumonia, leukoencephalomyelitis and arthritis. Rev Infect Dis. 1985;7:89–98. doi: 10.1093/clinids/7.1.89. [DOI] [PubMed] [Google Scholar]
- 32.Narayan O, Griffin D E, Silverstein A M. Slow virus infection: replication and mechanisms of persistence of visna virus in sheep. J Infect Dis. 1977;135:800–806. doi: 10.1093/infdis/135.5.800. [DOI] [PubMed] [Google Scholar]
- 33.Rost B, Sander C. Conservation and prediction of solvent accessibility in protein families. Proteins. 1994;20:216–226. doi: 10.1002/prot.340200303. [DOI] [PubMed] [Google Scholar]
- 34.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 35.Selby M J, Peterlin B M. Transactivation by HIV-1 Tat via a heterologous RNA binding protein. Cell. 1990;62:769–776. doi: 10.1016/0092-8674(90)90121-t. [DOI] [PubMed] [Google Scholar]
- 36.Shih D S, Carruth L M, Anderson M, Clements J E. Involvement of Fos and Jun in the activation of visna virus gene expression in macrophages through an AP-1 site in the viral LTR. Virology. 1992;190:84–91. doi: 10.1016/0042-6822(92)91194-y. [DOI] [PubMed] [Google Scholar]
- 37.Sigurdsson B, Palsson P A. Visna of sheep A slow demyelination infection. Br J Exp Pathol. 1958;39:519–528. [PMC free article] [PubMed] [Google Scholar]
- 38.Sitlani A, Crothers D M. DNA-binding domains of Fos and Jun do not induce DNA curvature: an investigation with solution and gel methods. Proc Natl Acad Sci USA. 1998;95:1404–1409. doi: 10.1073/pnas.95.4.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sitlani A, Crothers D M. Fos and Jun do not bend the AP-1 recognition site. Proc Natl Acad Sci USA. 1996;93:3248–3252. doi: 10.1073/pnas.93.8.3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Small J A, Bieberich C, Ghotbi Z, Hess J, Scangos G A, Clements J E. The visna virus long terminal repeat directs expression of a reporter gene in activated macrophages, lymphocytes, and the central nervous systems of transgenic mice. J Virol. 1989;63:1891–1896. doi: 10.1128/jvi.63.5.1891-1896.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Southgate C, Zapp M L, Green M R. Activation of transcription by HIV-1 Tat protein tethered to nascent RNA through another protein. Nature. 1990;345:640–642. doi: 10.1038/345640a0. [DOI] [PubMed] [Google Scholar]
- 42.Stringer K F, Ingles C J, Greenblatt J. Direct and selective binding of an acidic transcriptional activation domain to the TATA box factor TFIID. Nature. 1990;345:783–786. doi: 10.1038/345783a0. [DOI] [PubMed] [Google Scholar]
- 43.Wagner S, Green M R. HTLV-I Tax protein stimulation of DNA binding of bZIP proteins by enhancing dimerization. Science. 1993;262:395–399. doi: 10.1126/science.8211160. [DOI] [PubMed] [Google Scholar]
- 44.Williams J S, Andrisani O M. The hepatitis B virus X protein targets the basic region-leucine zipper domain of CREB. Proc Natl Acad Sci USA. 1995;92:3819–3823. doi: 10.1073/pnas.92.9.3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin M J, Paulssen E, Seeler J, Gaynor R B. Chimeric proteins composed of Jun and CREB define domains required for interaction with the human T-cell leukemia virus type 1 Tax protein. J Virol. 1995;69:6209–6218. doi: 10.1128/jvi.69.10.6209-6218.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- 47.Zink M C, Narayan O. Lentivirus-induced interferon inhibits maturation and proliferation of monocytes and restricts the replication of caprine arthritis-encephalitis virus. J Virol. 1989;63:2578–2584. doi: 10.1128/jvi.63.6.2578-2584.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zink M C, Yager J A, Myers J D. Pathogenesis of lentiviruses: cellular localization of viral transcripts in tissues of goats infected with caprine arthritis-encephalitis virus. Am J Pathol. 1990;136:843–854. [PMC free article] [PubMed] [Google Scholar]