Abstract
Lignocellulosic biomass is a significant source of sustainable fuel and high-value chemical production. However, due to the complex cross-linked three-dimensional network structure, lignin is highly rigid to degradation. In natural environments, the degradation is performed by wood-rotting fungi. The process is slow, and thus, the use of lignin degradation by fungi has not been regarded as a feasible technology in the industrial lignocellulose treatment. Fungi produce a wide variety of ligninolytic enzymes that can be directly introduced in industrial processing of lignocellulose. Within this study, screening of ligninolytic enzyme production using decolorization of ABTS and Azure B dyes was performed for 10 fungal strains with potentially high enzyme production abilities. In addition to standard screening methods, media containing lignin and hay biomass as carbon sources were used to determine the change in enzyme production depending on the substrate. All selected fungi demonstrated the ability to adapt to a carbon source limitation; however, four strains indicated the ability to secrete ligninolytic enzymes in all experimental conditions—Irpex lacteus, Pleurotus dryinus, Bjerkandera adusta, and Trametes versicolor—respectively displayed a 100%, 82.7%, 82.7%, and 55% oxidation of ABTS on lignin-containing media and 100%, 87.9%, 78%, and 70% oxidation of ABTS on hay-containing media after 168 h of incubation. As a result, the most potent strains of fungi were selected to produce lignocellulose-degrading enzymes and to demonstrate their potential application in biological lignocellulose pretreatment.
Keywords: lignocellulosic biomass, biomass pretreatment, ligninolytic enzymes, white rot fungi
1. Introduction
The rapid growth of the global population over the past half-century has led to an increasing demand not only for fresh water and food but also for petroleum products [1]. At present, energy consumption and production contribute to two-thirds of global emissions, and 81% of the global energy system is still based on fossil fuels, the same percentage as 30 years ago [2,3]. The current aim of the European Union (EU) is to become carbon neutral with net-zero greenhouse gas emissions by 2050 [4]. Renewable biomass will continue to have an important role in EU energy, covering approximately 5% of the primary energy supply of the EU-27 [5]. Generally, lignocellulosic biomass consists of three main components—cellulose (40–50%), hemicellulose (25–30%), and lignin (15–20%), as well as extractives (0–15%), and the proportions of these vary depending on the biomass source [6,7]. From these, lignin is an aromatic biopolymer found in the vascular tissues of plants, and together with cellulose and hemicellulose, forms a natural structural bio-composite, which provides rigidity and mechanical strength to the plant’s cell and structures [8]. The composition of lignocellulosic biomass varies based on the source of the biomass and type of the plant (Table 1); it is also released as a by-product in the pulp, paper, and bioethanol industries. The pulp industry produces around 30 million tons of lignin per year [9], which makes the industry a large producer of lignocellulosic and lignin waste. Lignin, both from plant biomass and industrial waste sources, can serve as an important renewable raw material source in a variety of other commercial applications. Biofuel production, as well as the biological production of chemicals, are potential applications for the biodegradation of lignin.
Most of the applications of lignocellulosic biomass require pretreatment to partly or completely degrade lignin [10]. However, in addition to providing plant stems the rigidity and waterproofing vascular tissues for sap circulation, the role of lignin is the protection of the cellulose polymer towards hydrolytic attack by saprotrophic organisms, which makes the lignin treatment and degradation process difficult. Nevertheless, some microorganisms have developed a strategy to be able to degrade lignin based on unspecific one-electron oxidation of the benzenic rings in the different lignin substructures by extracellular ligninolytic enzymes [11]. Given the role of ligninolytic enzymes in the lignocellulosic biomass degradation and detoxification in the environment [6,12], the biological pretreatment could be a possible environmentally friendly and chemical-free method for the degradation of lignin and lignin biowaste in many industries [6].
Table 1.
| Lignocellulosic Materials | Cellulose (%) | Hemicellulose (%) | Lignin (%) |
|---|---|---|---|
| Natural biomass sources | |||
| Hardwood | 40–55 | 24–40 | 18–25 |
| Oak | 43.2 | 21.9 | 35.4 |
| Pine | 45.6 | 24 | 26.8 |
| Natural hay | 44.9 | 31.4 | 12 |
| Leaves | 15–20 | 80–85 | 0 |
| Reed | 34–36 | 26–27 | 21 |
| Switchgrass | 31–45 | 20–31 | 12–18 |
| Agricultural biomass by-products or residues | |||
| Barley straw | 31–45 | 27–38 | 14–19 |
| Wheat straw | 33–38 | 26–32 | 17–19 |
| Rye straw | 33–35 | 27–30 | 16–19 |
| Oat straw | 31–37 | 27–38 | 16–19 |
| Silage | 39.27 | 25.96 | 9.02 |
| Hemp | 53.86 | 10.6 | 8.76 |
| Rapeseed | 20–35 | 15–22 | 15–23 |
| Industrial residues and waste | |||
| Willow sawdust | 35.6 | 21.5 | 28.7 |
| Paper | 85–99 | 0 | 0–15 |
| Newspaper | 40–55 | 25–40 | 18–30 |
| Waste papers from chemical pulps | 60–70 | 10–20 | 5–10 |
| Primary wastewater solids | 8–15 | NA | 24–29 |
| Sugarcane bagasse | 44 | 28 | 21 |
Numerous fungi, as well as bacterial species, can cause lignin degradation (Table 2); however, fungi are more efficient in the breakdown of lignin than bacteria, which are more limited in their enzyme secretion abilities and lignin degradation rates [6]. It has been reported that around 1600–1700 wood-rot fungal species identified in North America are capable of biodegrading lignin [19]. Wood-degrading species are mostly saprotrophs or weak parasites in forest ecosystems. Saprotrophic fungi associated with lignin degradation have been divided into the following three major groups depending on their morphology and enzymes associated with the lignin degradation mechanism: white rot, brown rot, and soft rot fungi. All three groups of fungi are able to degrade lignin, but only Basidiomycota (aerobic white rot fungi) are able to decompose lignin completely to CO2 and H2O [20,21]. In nature, white rot fungi mostly occur on hardwoods and are primary lignin degraders, whereas brown rot fungi more often are found on softwoods in coniferous ecosystems [22].
Table 2.
Bacteria and fungi degradation of lignin on different feedstocks.
| Species | Feedstock Used | Lignin Degradation (%) | Time (Days) | Reference |
|---|---|---|---|---|
| Bacteria | ||||
| Pseudomonas spp. | Kraft lignin | 39 | 52 | [23] |
| Poplar wood | 40–52 | 30 | [24] | |
| Kraft lignin | 20 | 40–60 | [23] | |
| Acinetobacter spp. | Poplar wood | 47–57 | 30 | [24,25] |
| Xanthomonas spp. | Poplar wood | 39–48 | 30 | [24,25] |
| Streptomyces badius | Indulin lignin | 3–4 | 35 | [26] |
| Streptomyces viridosporous | Indulin lignin | 3–4 | 35 | [26] |
| Streptomyces cyaneus | Barley straw | 29–52 | 21 | [27] |
| Thermomonospora mesophila | Barley straw | 36–48 | 21 | [24,27] |
| Fungi | ||||
| Pleurotus ostreatus | Cotton stalks | 40 | 30 | [28] |
| Beech wood | 56.5 | 120 | [28] | |
| Phanerochaete chrysosporium | Cotton stalks | 60 | 30 | [28] |
| Cotton stalks | 28 | 14 | [23] | |
| Trametes versicolor spp. | Beech wood | 57.4 | 120 | [29] |
| Bamboo culms | 9–24 | 28 | [30,31] | |
| Irpex lacteus | Cornstalks | 15 | 11.84 | [32] |
| Echinodontium taxodii 2538 | Bamboo culms | 24 | 28 | [30] |
| Phlebia sp. MG-60 | Oak wood | 40.6 | 56 | [33] |
| Ceriporia lacerata | Red pine | 13 | 56 | [34] |
| Stereum hirsutum | Red pine | 15 | 56 | [31] |
| Ceriporiopsis subvermispora | Corn stover | 39.2 | 42 | [35,36] |
The enzymes involved in lignin degradation have been divided into two groups—lignin-modifying enzymes (LME) and lignin-degrading auxiliary (LDA) enzymes [37]. LMEs are also called ligninolytic enzymes and have gained attention as biological agents for the degradation of lignocellulosic waste-containing compounds and other organic pollutants [12]. These enzymes also have a role in industrial waste treatment and other xenobiotic compounds through the biodegradation and decolorization process [37]. The LDA enzymes are unable to degrade lignin on their own and need additional enzyme involvement for complete lignin degradation; however, these enzymes enable the process of lignin degradation through the sequential action of several proteins that may include oxidative H2O2. This group includes cellobiose dehydrogenase, aryl alcohol oxidases, glyoxal oxidase, glucose oxidase, and pyranose 2-oxydase [12].
LME produced by microorganisms are classified as phenol oxidases (laccases) and heme-containing peroxidases—lignin, manganese, and versatile (multifunctional) peroxidase [38,39]. Recently, a new superfamily of heme peroxidases called dye-peroxidases (DyP, originally named dye-decolorizing peroxidases) was identified in fungi and later in bacteria. DyP may also have a role in the lignin degradation process; however, the clear mechanisms behind their abilities are yet to be discovered [40].
The lignin peroxidases (LiP) are capable of attacking lignin polymers and are relatively non-specific to their substrate. These enzymes are characterized by their ability to oxidase different phenolic aromatic compounds as well as a variety of non-phenolic lignin model compounds and other organic molecules [38]. LiPs were first discovered in the 1980s in Phanerochaete chrysosporium and later in Trametes versicolor, Bjerkandera sp., and Phlebia tremellosa, which are well-known white rot fungi species [37]. Microorganism-secreted enzymes are usually a family of isozymes whose relative composition and isoelectric points vary depending on growth conditions, culture media, and nutrients provided in the cultivation process [41].
Similarly to LiP, manganese peroxidases (MnP), as a family of isozymes, were first discovered over 30 years ago and were first detected in P. chrysosporium. MnPs are another important LME and have also been found in other Basidiomycota species, including Panus tigrinus, Lenzites betulinus, Agaricus bisporus, Bjerkandera sp., and Nematoloma frowardii. Details about MnPs presence in bacteria, yeast, and mold are emerging in the scientific literature, and the presence and activity of these enzymes have been studied in several species under the absence and presence of enzyme inducers [37].
Versatile peroxidases (VPs) combine the molecular architecture of LiP and MnP and oxidize typical LiP substrates as well as Mn2+, yet they also oxidize azo-dyes and other non-phenolic compounds with high redox potentials in the absence of mediators [42,43]. VPs were first found in members of the genera Pleurotus (P. eryngii, P. ostreatus) and Bjerkandera (B. adusta, B. fumosa) [37].
Dye-decolorizing peroxidases (DyP) are a new family of heme peroxidases, phylogenetically unrelated to other LME peroxidases [44]. These enzymes were first discovered in a culture of the fungus B. adusta and, as the name suggests, are able to decolorize a wide range of dyes [40]. The ligninolytic activity of DyP has been reported in other fungi (Termitomyces albuminosus, Auricularia auricula-judae, and Irpex lacteus) and several bacterial species [37]. The presence of DyP-expressing genes is more common in bacteria, and a smaller number of genes are reported in fungi and higher eukaryotes and archaea, suggesting that these enzymes are the bacterial equivalent of fungal LME [45]. DyPs are relatively non-specific to their substrate and oxidase all typical peroxidase substrates as well as have an additional hydrolase or oxygenase activity [46]. DyP are active at lower pH values (pH range 3–4) and are able to degrade different dyes; however, the physiological role of these enzymes is still unclear [47].
Laccases are considered the most important components of the lignin degradation process and are widely distributed in plants, fungi, bacteria, and insects; however, the role of laccases in these processes is not known in detail. All laccases oxidize a range of aromatic compounds, phenolic components also found in lignin, aromatic amines, benzenothiols, and hydroxyindols using molecular oxygen as an electron acceptor, bypassing a stage of hydrogen peroxide production [37,48]. These enzymes are extracellular, periplasmic, and intracellular proteins, and a majority of fungi produce mostly extracellular as well as some intracellular laccases [48]. In plants, intracellular laccases participate in the synthesis of lignin, intracellular fungal laccases, and periplasmic bacterial laccases most likely participate in the transformation of phenolic compounds in the cell, while extracellular laccases participate in lignin degradation [49,50]. Fungal laccases also participate in the pathogenesis, detoxification, and development of higher fungi [51]. Laccases belong to the group of polyphenol oxidases and are also called blue multicopper oxidases due to containing copper atoms in the catalytic site of the enzyme [51], and mainly react with free phenolic fragments of lignin due to the random polymer nature of lignin and laccases lower redox potential; however, mediators can cause a reaction to non-phenolic compounds with higher redox potential [52]. Similarly, as peroxidases, laccases are also secreted as several isoforms in most fungi, originating from the same or different genes. The number and properties of isozymes secreted vary depending on the growth conditions, fungal species, as well as nutrients and inducers found in the growth media [37,48].
In recent years, ligninolytic enzymes have gained applications in the fields of the food industry, textile industry, synthetic chemistry, cosmetics, soil bioremediation and biodegradation of environmental phenolic pollutants, and removal of endocrine disruptors [53,54]. These enzymes are also used for paper and pulp delignification, where they can be used in the enzymatic adhesion of fibers in the manufacturing of lignocellulose-based composite materials, such as fiberboards [54]. Using fungal strains, which can produce the enzymes needed for biomass conversion and to produce ethanol, improved biorefinery efficiency can also be achieved [55,56].
To evaluate the ability of fungal growth and lignocellulosic biomass degradation, mostly culture media or pure lignin in culture media has been used [40,57,58,59]. Here we report a screening study of 10 biotechnologically relevant fungal isolates on their ability to produce lignin-degrading enzymes in the presence of untreated lignocellulosic biomass.
2. Materials and Methods
2.1. Microorganisms
In this study, commercially available cultures of Irpex lacteus DSM 9595, Pleurotus dryinus (Pers.) P. Kumm, Pleurotus ostreatus DSM 1020, Bjerkandera adusta DSM 23426, Trametes versicolor DSM 6401, Pycnoporus cinnabarinus (Fr.) P. Karst, Aspergillus brasiliensis ATCC®16404™, and cultures isolated from pine forests of Latvia—Fusarium graminearum, Fomitopsis pinicola, Trichoderma paraviridescens, which were maintained on potato dextrose agar (PDA) (Oxoid Ltd., Basingstoke, Hants, UK) medium at 2–8 °C, were used in ligninolytic enzyme screening tests.
2.2. Media Conditions and Screening of Ligninolytic Enzymes
To detect the ability of selected fungal species for ligninolytic enzyme production, 0.1% (w/v) ABTS (2,2′-Azino-bis(3-ethylbonzotiazoline-6-sulfonic acid)) diammonium salt) [60], and 0.01% (w/v) Azure B [61] were used as reaction substrates. ABTS is a non-phenolic dye that is oxidized by laccase to the more stable and preferred state of ABTS cation radical. The radical is responsible for the distinct blue-green color and can be correlated to laccase activity [62]. A distinct purple color formation has been described when the laccase content was equal to or higher than needed for the reduction of ABTS present [63]. Azure B is a triarylmethane dye and has a similar structure to lignin, thus is usually used as the substrate to measure the ligninolytic enzyme activity. Decolorization of this dye illustrates the presence of LiP, MnP, or laccase produced by the fungi [64].
To prepare agar medium containing lignin or hay, 2 g of the respective substrate, 0.8 g KH2PO4, 0.4 g K2HPO4, 0.5 g MgSO4·7H2O, 2 g NH4NO3, 2 g yeast extract, and 15 g agar (pH 5.5 ± 0.2) were added per L of distilled water. The prepared agar media were sterilized by autoclaving at 121 °C for 15 min. The chemical composition of the hay biomass from grasslands, which includes approximately 22–26% cellulose, 14–25% hemicellulose, and 1–13% lignin, has been adopted for this study [65].
After media preparation, 1 cm2 mycelial disk of each fungal species was placed on 9 types of agars (Table 3) containing PDA, lignin (Sigma-Aldrich, Darmstadt, Germany) or hay (dry weight (DW): 92.8 ± 1.3%, collected from semi-natural grassland in Latvia) as biomass substrate. The specific initial color of the media was recorded (Table 3).
Table 3.
Color of uninoculated agar used in ligninolytic enzyme screening tests.
| Substrate | - | 0.1% w/v ABTS | 0.01% w/v Azure B |
|---|---|---|---|
| Potato dextrose agar |

|
|

|
| Lignin |
|
|
|
| Hay |
|

|

|
During the screening tests, agar plates were incubated for 168 h at 25 °C and 80% rH in constant climate chamber (KBF 115, BINDER GmbH, Tuttlingen, Germany). Oxidation zone and dark color formation around the mycelium indicated on the presence of ligninolytic enzymes. The diameter and intensity of the color change were used as an indicator of the lignocellulosic enzyme production. Color zone formation and color change intensity were monitored and captured daily using NIKON D3300 (NIKON, Tokyo, Japan). Visual analysis was performed to determine the qualitative changes in the agar plates. Quantitative analysis was conducted to determine the percentage of the agar plate area that was covered by fungal mycelium and underwent oxidation induced in the presence of fungal enzymes by measuring the diameter of the fungal mycelium and the oxidation zone. The experiments were performed in 3 independent repetitions.
3. Results and Discussion
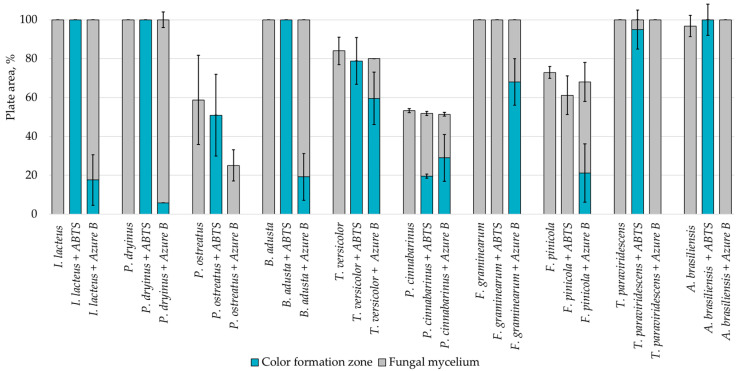
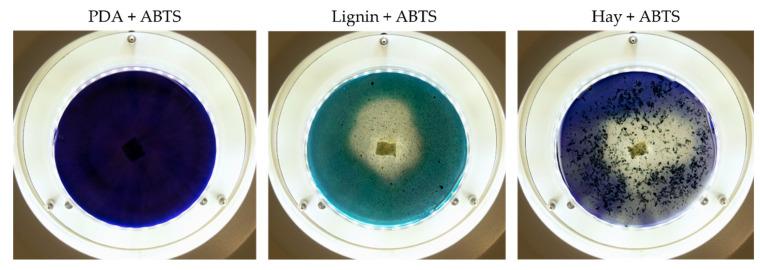
Fungal growth and substrate oxidation results show that on PDA, within 168 h, four fungal strains—I. lacteus, P. dryinus, B. adusta, and T. paraviridescens—fully covered the agar plate and simultaneously showed the most intense ABTS oxidation (Figure 1). However, only one white rot fungus—T. versicolor—caused significant color change on both ABTS (78.8% of plate area) and Azure B (59.6% of plate area) agar.
Figure 1.
Percentage of grown fungal mycelium and formed oxidation zone of the total PDA agar area after 168 h of incubation.
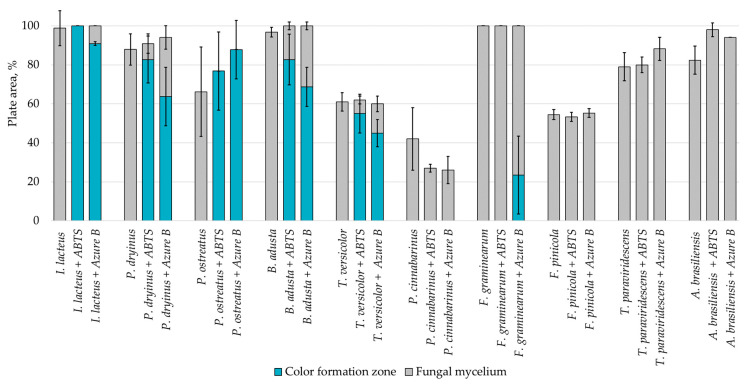
Agar containing only lignin as a carbon source limited the fungal growth rate, and only F. graminearum was able to fill the agar plates completely (Figure 2) in both PDA and lignin-containing medium. The mycelium growth rate of I. lacteus and B. adusta was less influenced by a change in carbon source. The growth activity of I. lacteus was decreased only by 1.2% and B. adusta—only by 3.2%. A more significant decrease in mycelium growth was observed in T. paraviridescens (21.0%), P. cinnabarinus (21.2%), F. pinicola (25.2%), T. versicolor (27.4%), and A. brasiliensis (48%) cultures. This suggests that different fungal species have varying adaptability and metabolic strategies in response to carbon source limitations.
Figure 2.
Percentage of grown fungal mycelium and formed oxidation zone of the total lignin agar area after 168 h of incubation.
Despite the limitations imposed by the carbon source, significant color change and ABTS oxidation were still observed in five white rot fungal cultures—I. lacteus, P. drynus, P. osteatus, B. adusta, and T. versicolor. In contrast to the PDA plates, decolorization of Azure B was found in different intensities in the previously mentioned cultures depending on the media composition, which suggests that carbon source limitation to agar only intensifies enzyme secretion in fungal cultures leading to more efficient decolorization and lignin degradation.
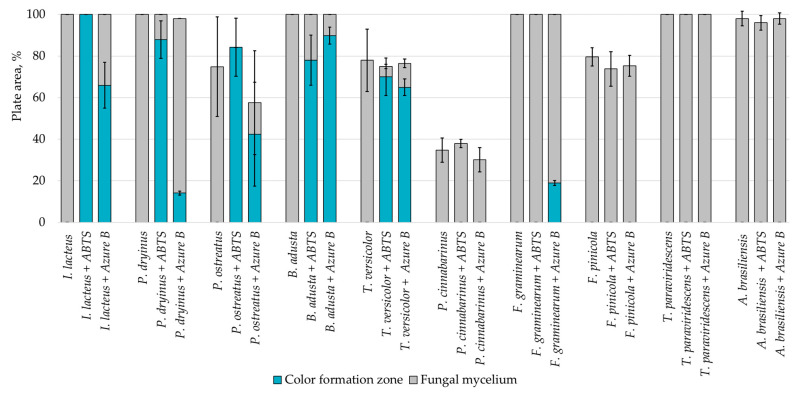
Hay biomass provides a more easily available carbon source for fungal growth when compared to lignin, and numerous cultures fully covered the plate surface within 168 h of incubation (Figure 3). The mycelium growth rate of all white rot fungi was increased—by 1.2% in the I. lacteus culture, 3.2% in B. adusta, 11.6% in P. ostreatus, 12.1% in P. dryinus, and up to 21.8% in the T. versicolor culture. The most significant increase in the growth activity was observed in F. pinicola (31.5%). I. lacteus, P. dryinus, B. adusta, F. graminearum, and T. paraviridescens were able to fully grow in the given time frame, but F. graminearum and T. paraviridescens failed to demonstrate significant color zone formation with ABTS or Azure B. Significant color formation zones and decolorization of Azure B were observed with I. lacteus, P. dryinus, B. adusta, and T. versicolor. P. ostreatus was also able to ensure intense oxidation of both ABTS and Azure B, although, the growth rate of P. ostreatus was limited and very variable between experiment repetitions. Previous studies describe P. ostreatus as a well-known fungal laccase producer with a high catalytic potential and one of the first cultures to produce a significant amount of DyP enzymes. However, it has been tested only in lignin systems or fungal gene expressions in other organisms to produce these enzymes [40,48]. Therefore, the results gathered in this study suggest that for lignocellulose biomass degradation, other fungal cultures seem more promising, given the screening tests with the hay biomass substrate offer a model closer to possible industrial applications for lignocellulosic biomass treatment.
Figure 3.
Percentage of grown fungal mycelium and formed oxidation zone of the total hay agar area after 168 h of incubation.
The screening results on lignin and hay biomass agar plates suggest that cultures of I. lacteus, P. dryinus, B. adusta, and T. versicolor have the potential to develop technologies for lignin or lignocellulose biomass degradation.
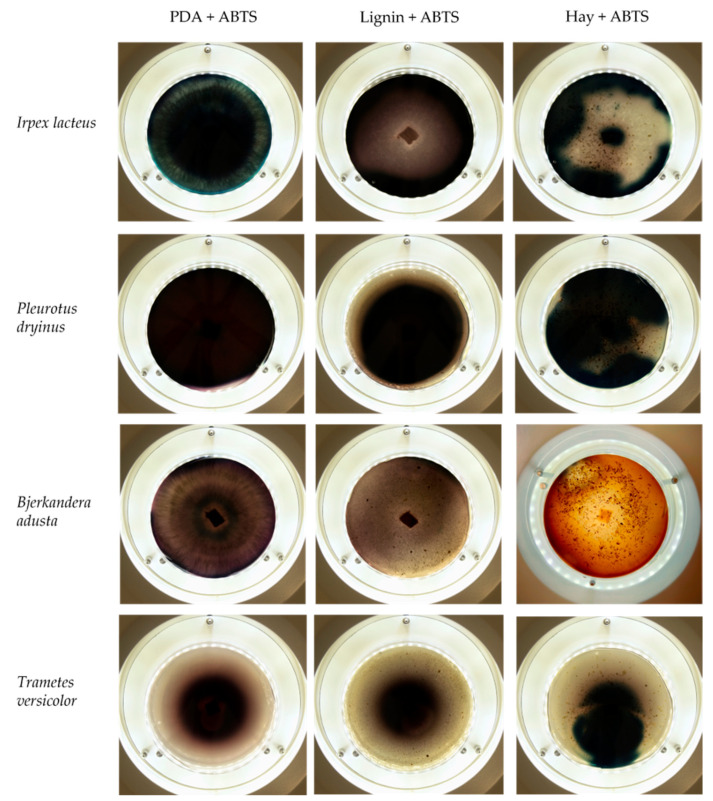
Based on the growth rate, color formation zone, and the oxidation intensity of the selected fungi during screening tests, I. lacteus, P. dryinus, B. adusta, and T. versicolor were selected as potent fungal species for lignin and lignocellulose biomass treatment. All the selected strains also showed intense color formation during the screening tests using ABTS (Figure 4). However, the most significant color formation on all types of agars was performed by P. dryinus and T. versicolor. Although, compared to I. lacteus and B. adusta, the mentioned fungi were characterized by a lower growth rate of the fungal mycelium. The most significant Azure B dye decolorization results were observed using B. adusta, the only fungal strain that degraded the added dye completely in the oxidation zone (Figure 5).
Figure 4.
Color formation on ABTS-containing agar by selected fungal strains after 168 h of incubation.
Figure 5.
The decolorization of Azure B by B. adusta after 168 h of incubation.
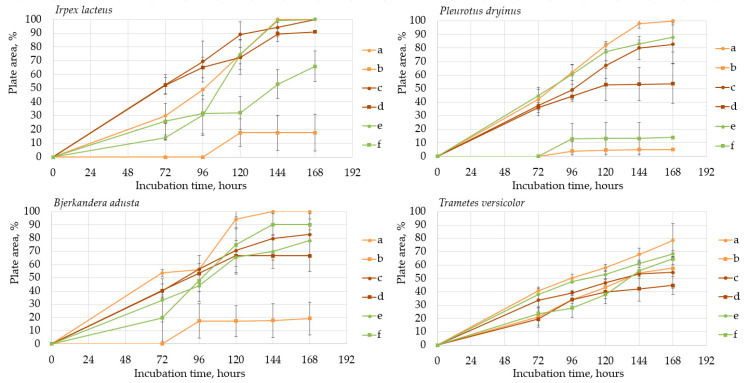
When comparing the degradation process of ABTS and Azure B (Figure 6), all four selected fungi caused a faster color formation on the ABTS-containing agar, regardless of the added carbon source. The potential explanation is that the ABTS used in this study for the screening tests has also been studied and used as a mediator that facilitates and promotes the release of lignin-degrading enzymes [59]. The highest rate of color formation occurred on PDA and hay-containing agar. Hay biomass and PDA contain more easily obtainable carbon compounds necessary for fungal growth, which facilitate more rapid fungal growth and enzyme release.
Figure 6.
Oxidation zone formation on (a) PDA + 0.1% (w/v) ABTS; (b) PDA + 0.01% (w/v) Azure B; (c) lignin agar + 0.1% (w/v) ABTS; (d) lignin + 0.01% (w/v) Azure B; (e) hay agar + 0.1% (w/v) ABTS; (f) lignin + 0.01% (w/v) Azure B.
Comparing the ABTS oxidation rate and color formation zone diameter, the fastest color formation was performed by I. lacteus, which filled 100% of the plate area within 144 h. The color formation and oxidation of ABTS by B. adusta occurred more slowly; however, this culture degraded the color most intensively and efficiently on agar-containing Azure B. This is explained by the fact that B. adusta was discovered as a dye-decolorizing peroxidase-secreting fungus and is able to decolorize a wide range of dyes [40]. T. versicolor was characterized by the slowest growth compared to I. lacteus, B. adusta, and P. dryinus; however, it showed a high efficiency of the oxidation of both ABTS and Azure B and linear growth and oxidation zone formation rate on all types of agars. P. dryinus was characterized by a relatively rapid growth and oxidation zone formation rate on all types of agars, except glucose- and hay-containing medium with Azure B, where the oxidation zone did not exceed 15% of the plate area during 168 h of screening.
4. Conclusions
I. lacteus, P. dryinus, B. adusta, and T. versicolor demonstrated the most promising results in terms of ABTS and Azure oxidation. These ligninolytic enzyme-producing white rot fungi were identified as potent fungal to offer an environmentally friendly, sustainable, and cost-efficient technology for lignin pretreatment, which could improve lignocellulosic biomass degradation and attain in production of high-value products. No change in the mycelium growth rate of I. lacteus, P. dryinus, or B. adusta on the lignocellulose biomass media was observed, and a decrease of 2.4–8.2% was observed on lignin-containing media when compared to PDA. T. versicolor demonstrated a more significant decrease in growth rate; however, all four selected fungal strains formed intense ABTS oxidation zones. Moreover, B. adusta displayed efficient decolorization of Azure B dye.
Given the screening results with lignin and lignocellulose substrates, the selected fungal strains can be characterized by the intense release of ligninolytic enzymes and dye oxidation abilities. These findings suggest that these fungal cultures have the potential for developing technologies aimed at lignin or lignocellulose biomass degradation. Further investigation is needed to explore their enzymatic capabilities and optimize their performance for industrial applications in lignocellulosic biomass treatment.
Acknowledgments
The authors thank Santa Vaivare for the initiation of fungal laccase screening studies.
Author Contributions
Conceptualization, A.C., A.A.S.-J. and L.M.; methodology, A.C. and A.A.S.-J.; formal analysis, A.C.; investigation, A.C. and A.A.S.-J.; writing—original draft preparation, A.C., A.A.S.-J. and L.M.; writing—review and editing, L.M. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was supported by EEA Grants project No. EEA-RESEARCH-173 “NoviCo” (Agreement No. EEZ/BPP/VIAA/2021/7).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Moshood T.D., Nawanir G., Mahmud F. Microalgae biofuels production: A systematic review on socioeconomic prospects of microalgae biofuels and policy implications. Environ. Chall. 2021;5:100207. doi: 10.1016/j.envc.2021.100207. [DOI] [Google Scholar]
- 2.Tsita K.G., Pilavachi P.A. Evaluation of next generation biomass derived fuels for the transport sector. Energy Policy. 2013;62:443–455. doi: 10.1016/j.enpol.2013.07.114. [DOI] [Google Scholar]
- 3.Statistical Review of World Energy 2022. BP p.l.c. [(accessed on 3 February 2023)]. Available online: https://www.bp.com/en/global/corporate/energy-economics/statistical-review-of-world-energy.html.
- 4.European Commission 2050 Long-Term Strategy. 2020. [(accessed on 3 February 2023)]. Available online: https://climate.ec.europa.eu/eu-action/climate-strategies-targets/2050-long-term-strategy_en.
- 5.Internation Energy Agency . Statistics Report Key World Energy Statistics 2021. Internation Energy Agency; Paris, France: 2021. [Google Scholar]
- 6.Atiwesh G., Parrish C.C., Banoub J., Le T.T. Lignin degradation by microorganisms: A review. Biotechnol. Prog. 2022;38:e3226. doi: 10.1002/btpr.3226. [DOI] [PubMed] [Google Scholar]
- 7.Yang C., Lü X. Chapter 5—Composition of plant biomass and its impact on pretreatment. In: Lü X., editor. Advances in 2nd Generation of Bioethanol Production. Woodhead Publishing; Cambridge, UK: 2021. pp. 71–85. Woodhead Publishing Series in Energy. [DOI] [Google Scholar]
- 8.Kirk T. Lignin Biodegradation: Microbiology, Chemistry, and Potential Applications. CRC Press; Boca Raton, FL, USA: 2018. [DOI] [Google Scholar]
- 9.Hatakeyama H., Hatakeyama T. Lignin Structure, Properties, and Applications. Biopolymers. 2009;232:1–63. [Google Scholar]
- 10.Sjulander N., Kikas T. Two-Step Pretreatment of Lignocellulosic Biomass for High-Sugar Recovery from the Structural Plant Polymers Cellulose and Hemicellulose. Energies. 2022;15:8898. doi: 10.3390/en15238898. [DOI] [Google Scholar]
- 11.Ruiz-Dueñas F.J., Martínez Á.T. Microbial degradation of lignin: How a bulky recalcitrant polymer is efficiently recycled in nature and how we can take advantage of this. Microb. Biotechnol. 2009;2:164–177. doi: 10.1111/j.1751-7915.2008.00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kumar A., Chandra R. Ligninolytic enzymes and its mechanisms for degradation of lignocellulosic waste in environment. Heliyon. 2020;6:e03170. doi: 10.1016/j.heliyon.2020.e03170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bajpai P., editor. Lignocellulosic Biomass in Biotechnology. Elsevier; Amsterdam, The Netherlands: 2022. Chapter 3—Sources of lignocellulosic biomass; pp. 25–36. [DOI] [Google Scholar]
- 14.Ferdeș M., Dincă M.N., Moiceanu G., Zăbavă B.Ș., Paraschiv G. Microorganisms and Enzymes Used in the Biological Pretreatment of the Substrate to Enhance Biogas Production: A Review. Sustainability. 2020;12:7205. doi: 10.3390/su12177205. [DOI] [Google Scholar]
- 15.Rezania S., Din M.F., Mohamad S.E., Sohaili J., Taib S.M., Yusof M.B.M., Kamyab H., Darajeh N., Ahsan A. Review on pretreatment methods and ethanol production from cellulosic water hyacinth. Bioresources. 2017;12:2108–2124. doi: 10.15376/biores.12.1.Rezania. [DOI] [Google Scholar]
- 16.Tayyab M. Bioethanol production from lignocellulosic biomass by enviornment-friendly pretreatment methods: A review. Appl. Ecol. Environ. Res. 2018;16:225–249. doi: 10.15666/aeer/1601_225249. [DOI] [Google Scholar]
- 17.Madadi M., Penga C., Abbas A. Advances in Genetic Manipulation of Lignocellulose to Reduce Biomass Recalcitrance and Enhance Biofuel Production in Bioenergy Crops. J. Plant Biochem. Physiol. 2017;5:182. doi: 10.4172/2329-9029.1000182. [DOI] [Google Scholar]
- 18.Ajala E.O., Ighalo J.O., Ajala M.A., Adeniyi A.G., Ayanshola A.M. Sugarcane bagasse: A biomass sufficiently applied for improving global energy, environment and economic sustainability. Bioresour. Bioprocess. 2021;8:87. doi: 10.1186/s40643-021-00440-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbertson R.L. Wood-Rotting Fungi of North America. Mycologia. 1980;72:1–49. doi: 10.1080/00275514.1980.12021153. [DOI] [Google Scholar]
- 20.Blanchette R.A. Degradation of the lignocellulose complex in wood. Can. J. Bot. 1995;73:999–1010. doi: 10.1139/b95-350. [DOI] [Google Scholar]
- 21.Otjen L., Blanchette R., Effland M., Leatham G. Assessment of 30 White Rot Basidiomycetes for Selective Lignin Degradation. Holzforschung. 1987;41:343–349. doi: 10.1515/hfsg.1987.41.6.343. [DOI] [Google Scholar]
- 22.Sigoillot J.-C., Berrin J.-G., Bey M., Lesage-Meessen L., Levasseur A., Lomascolo A., Record E., Uzan-Boukhris E. Fungal Strategies for Lignin Degradation. Adv. Bot. Res. 2012;61:263–308. doi: 10.1016/b978-0-12-416023-1.00008-2. [DOI] [Google Scholar]
- 23.Shi J., Chinn M.S., Sharmashivappa R. Microbial pretreatment of cotton stalks by solid state cultivation of Phanerochaete chrysosporium. Bioresour. Technol. 2008;99:6556–6564. doi: 10.1016/j.biortech.2007.11.069. [DOI] [PubMed] [Google Scholar]
- 24.Dionisi D., Anderson J.A., Aulenta F., McCue A., Paton G. The potential of microbial processes for lignocellulosic biomass conversion to ethanol: A review. J. Chem. Technol. Biotechnol. 2015;90:366–383. doi: 10.1002/jctb.4544. [DOI] [Google Scholar]
- 25.Odier E., Odier E., Janin G., Janin G., Monties B., Monties B., Odier E., Odier E., Janin G., Janin G., et al. Poplar Lignin Decomposition by Gram-Negative Aerobic Bacteria. Appl. Environ. Microbiol. 1981;41:337–341. doi: 10.1128/aem.41.2.337-341.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giroux H., Vidal P., Bouchard J., Lamy F. Degradation of Kraft Indulin Lignin by Streptomyces viridosporus and Streptomyces badius. Appl. Environ. Microbiol. 1988;54:3064–3070. doi: 10.1128/aem.54.12.3064-3070.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zimmermann W., Broda P. Utilization of lignocellulose from barley straw by actinomycetes. Appl. Microbiol. Biotechnol. 1989;30:103–109. doi: 10.1007/BF00256005. [DOI] [Google Scholar]
- 28.Kerem Z., Friesem D., Hadar Y. Lignocellulose Degradation during Solid-State Fermentation: Pleurotus ostreatus versus Phanerochaete chrysosporium. Appl. Environ. Microbiol. 1992;58:1121–1127. doi: 10.1128/aem.58.4.1121-1127.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bari E., Taghiyari H.R., Naji H.R., Schmidt O., Ohno K.M., Clausen C.A., Bakar E.S. Assessing the destructive behaviors of two white-rot fungi on beech wood. Int. Biodeterior. Biodegrad. 2016;114:129–140. doi: 10.1016/j.ibiod.2016.06.010. [DOI] [Google Scholar]
- 30.Suhara H., Kodama S., Kamei I., Maekawa N., Meguro S. Screening of selective lignin-degrading basidiomycetes and biological pretreatment for enzymatic hydrolysis of bamboo culms. Int. Biodeterior. Biodegrad. 2012;75:176–180. doi: 10.1016/j.ibiod.2012.05.042. [DOI] [Google Scholar]
- 31.Shirkavand E., Baroutian S., Gapes D.J., Young B.R. Pretreatment of radiata pine using two white rot fungal strains Stereum hirsutum and Trametes versicolor. Energy Convers. Manag. 2017;142:13–19. doi: 10.1016/j.enconman.2017.03.021. [DOI] [Google Scholar]
- 32.Du W., Yu H., Song L., Zhang J., Weng C., Ma F., Zhang X. The promoting effect of byproducts from Irpex lacteus on subsequent enzymatic hydrolysis of bio-pretreated cornstalks. Biotechnol. Biofuels. 2011;4:37. doi: 10.1186/1754-6834-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamei I., Hirota Y., Meguro S. Integrated delignification and simultaneous saccharification and fermentation of hard wood by a white-rot fungus, Phlebia sp. MG-60. Bioresour. Technol. 2012;126:137–141. doi: 10.1016/j.biortech.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 34.Lee J.-W., Gwak K.-S., Park J.-Y., Park M.-J., Choi D.-H., Kwon M., Choi I.-G. Biological pretreatment of softwood Pinus densiflora by three white rot fungi. J. Microbiol. 2007;45:485–491. [PubMed] [Google Scholar]
- 35.Öhgren K., Bura R., Lesnicki G., Saddler J., Zacchi G. A comparison between simultaneous saccharification and fermentation and separate hydrolysis and fermentation using steam-pretreated corn stover. Process. Biochem. 2007;42:834–839. doi: 10.1016/j.procbio.2007.02.003. [DOI] [Google Scholar]
- 36.Liu X., Hiligsmann S., Gourdon R., Bayard R. Anaerobic digestion of lignocellulosic biomasses pretreated with Ceriporiopsis subvermispora. J. Environ. Manag. 2017;193:154–162. doi: 10.1016/j.jenvman.2017.01.075. [DOI] [PubMed] [Google Scholar]
- 37.Janusz G., Pawlik A., Sulej J., Świderska-Burek U., Jarosz-Wilkołazka A., Paszczyński A. Lignin degradation: Microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 2017;41:941–962. doi: 10.1093/femsre/fux049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wong D.W.S. Structure and Action Mechanism of Ligninolytic Enzymes. Appl. Biochem. Biotechnol. 2009;157:174–209. doi: 10.1007/s12010-008-8279-z. [DOI] [PubMed] [Google Scholar]
- 39.Weng C., Peng X., Han Y. Depolymerization and conversion of lignin to value-added bioproducts by microbial and enzymatic catalysis. Biotechnol. Biofuels. 2021;14:84. doi: 10.1186/s13068-021-01934-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fernández-Fueyo E., Linde D., Almendral D., López-Lucendo M.F., Ruiz-Dueñas F.J., Martínez A.T. Description of the first fungal dye-decolorizing peroxidase oxidizing manganese(II) Appl. Microbiol. Biotechnol. 2015;99:8927–8942. doi: 10.1007/s00253-015-6665-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vivanco J.M., Baluška F., editors. Secretions and Exudates in Biological Systems. Volume 12. Springer; Berlin/Heidelberg, Germany: 2012. [DOI] [Google Scholar]
- 42.Garcia-Ruiz E., Mate D.M., Gonzalez-Perez D., Molina-Espeja P., Camarero S., Martínez A.T., Ballesteros A.O., Alcalde M. Cascade Biocatalysis. Wiley-VCH Verlag GmbH & Co. KGaA; Weinheim, Germany: 2014. Directed Evolution of Ligninolytic Oxidoreductases: From Functional Expression to Stabilization and Beyond; pp. 1–22. [DOI] [Google Scholar]
- 43.Hofrichter M., Ullrich R., Pecyna M.J., Liers C., Lundell T. New and classic families of secreted fungal heme peroxidases. Appl. Microbiol. Biotechnol. 2010;87:871–897. doi: 10.1007/s00253-010-2633-0. [DOI] [PubMed] [Google Scholar]
- 44.Zámocký M., Hofbauer S., Schaffner I., Gasselhuber B., Nicolussi A., Soudi M., Pirker K.F., Furtmüller P.G., Obinger C. Independent evolution of four heme peroxidase superfamilies. Arch. Biochem. Biophys. 2015;574:108–119. doi: 10.1016/j.abb.2014.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.I Colpa D., Fraaije M.W., van Bloois E. DyP-type peroxidases: A promising and versatile class of enzymes. J. Ind. Microbiol. Biotechnol. 2014;41:1–7. doi: 10.1007/s10295-013-1371-6. [DOI] [PubMed] [Google Scholar]
- 46.Sugano Y., Matsushima Y., Tsuchiya K., Aoki H., Hirai M., Shoda M. Degradation pathway of an anthraquinone dye catalyzed by a unique peroxidase DyP from Thanatephorus cucumeris Dec 1. Biodegradation. 2009;20:433–440. doi: 10.1007/s10532-008-9234-y. [DOI] [PubMed] [Google Scholar]
- 47.Liers C., Pecyna M.J., Kellner H., Worrich A., Zorn H., Steffen K.T., Hofrichter M., Ullrich R. Substrate oxidation by dye-decolorizing peroxidases (DyPs) from wood- and litter-degrading agaricomycetes compared to other fungal and plant heme-peroxidases. Appl. Microbiol. Biotechnol. 2013;97:5839–5849. doi: 10.1007/s00253-012-4521-2. [DOI] [PubMed] [Google Scholar]
- 48.Morozova O.V., Shumakovich G.P., Gorbacheva M.A., Shleev S.V., Yaropolov A.I. “Blue” laccases. Biochemistry. 2007;72:1136–1150. doi: 10.1134/S0006297907100112. [DOI] [PubMed] [Google Scholar]
- 49.Baldrian P. Fungal laccases—Occurrence and properties. FEMS Microbiol. Rev. 2006;30:215–242. doi: 10.1111/j.1574-4976.2005.00010.x. [DOI] [PubMed] [Google Scholar]
- 50.Youn H.-D., Hah Y.C., Kang S.-O. Role of laccase in lignin degradation by white-rot fungi. FEMS Microbiol. Lett. 1995;132:183–188. doi: 10.1111/j.1574-6968.1995.tb07831.x. [DOI] [Google Scholar]
- 51.Leonowicz A., Cho N., Luterek J., Jarosz-Wilkolazka A., Wojtas-Wasilewska M., Matuszewska A., Hofrichter M., Wesenberg D., Rogalski J. Fungal laccase: Properties and activity on lignin. J. Basic Microbiol. 2001;41:185–227. doi: 10.1002/1521-4028(200107)41:3/4<185::AID-JOBM185>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 52.Kunamneni A., Ballesteros A., Plou F.J., Alcalde M. In: Fungal laccase—A versatile enzyme for biotechnological applications. Communicating Current Research and Educational Topics and Trends in Applied Microbiology. Mendez-Vilas A., editor. Volume 1. Formex; Badajoz, Spain: 2007. pp. 233–245. [Google Scholar]
- 53.Shraddha, Shekher R., Sehgal S., Kamthania M., Kumar A. Laccase: Microbial Sources, Production, Purification, and Potential Biotechnological Applications. Enzym. Res. 2011;2011:217861. doi: 10.4061/2011/217861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rodriguez Couto S., Toca-Herrera J.L. Industrial and biotechnological applications of laccases: A review. Biotechnol. Adv. 2006;24:500–513. doi: 10.1016/j.biotechadv.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 55.Ragauskas A.J., Beckham G.T., Biddy M.J., Chandra R., Chen F., Davis M.F., Davison B.H., Dixon R.A., Gilna P., Keller M., et al. Lignin Valorization: Improving Lignin Processing in the Biorefinery. Science. 2014;344:1246843. doi: 10.1126/science.1246843. [DOI] [PubMed] [Google Scholar]
- 56.Lange L. Fungal Enzymes and Yeasts for Conversion of Plant Biomass to Bioenergy and High-Value Products. Microbiol. Spectr. 2017;5:1029–1048. doi: 10.1128/microbiolspec.FUNK-0007-2016. [DOI] [PubMed] [Google Scholar]
- 57.Erden E., Ucar M.C., Gezer T., Pazarlioglu N.K. Screening for ligninolytic enzymes from autochthonous fungi and applications for decolorization of remazole marine blue. Braz. J. Microbiol. 2009;40:346–353. doi: 10.1590/S1517-83822009000200026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dhouib A., Hamza M., Zouari H., Mechichi T., Hmidi R., Labat M., Martinez M.J., Sayadi S. Screening for Ligninolytic Enzyme Production by Diverse Fungi from Tunisia. World J. Microbiol. Biotechnol. 2005;21:1415–1423. doi: 10.1007/s11274-005-5774-z. [DOI] [Google Scholar]
- 59.Longe L.F., Couvreur J., Grandchamp M.L., Garnier G., Allais F., Saito K. Importance of Mediators for Lignin Degradation by Fungal Laccase. ACS Sustain. Chem. Eng. 2018;6:10097–10107. doi: 10.1021/acssuschemeng.8b01426. [DOI] [Google Scholar]
- 60.Matsumura E., Shin T., Murao S., Yamamoto E., Kawano T. New Enzymatic Colorimetric Reactions of Benzoic Acid Derivatives with ABTS [2,2′-Azino-di-(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt] in the Presence of Laccase. Biosci. Biotechnol. Biochem. 1987;51:2743–2750. doi: 10.1080/00021369.1987.10868460. [DOI] [Google Scholar]
- 61.Sharma A., Aggarwal N.K., Yadav A. First Report of Lignin Peroxidase Production from Alternaria alternata ANF238 Isolated from Rotten Wood Sample. Bioeng. Biosci. 2016;4:76–87. doi: 10.13189/bb.2016.040502. [DOI] [Google Scholar]
- 62.More S.S., Renuka P.S., Pruthvi K., Swetha M., Malini S., Veena S.M. Isolation, Purification, and Characterization of Fungal Laccase from Pleurotus sp. Enzym. Res. 2011;2011:248735. doi: 10.4061/2011/248735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kut K., Stefaniuk I., Bartosz G., Sadowska-Bartosz I. Formation of a Purple Product upon the Reaction of ABTS Radicals with Proteins. Int. J. Mol. Sci. 2023;24:8912. doi: 10.3390/ijms24108912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang J., Zhao J., Jiang J., Xu H., Zhang N., Xie J., Wei M. Isolation and Characterization of Bacillus Sp. Capable of Degradating Alkali Lignin. Front. Energy Res. 2021;9:807286. doi: 10.3389/fenrg.2021.807286. [DOI] [Google Scholar]
- 65.French K.E. Assessing the bioenergy potential of grassland biomass from conservation areas in England. Land Use Policy. 2019;82:700–708. doi: 10.1016/j.landusepol.2018.12.001. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.