Abstract
Polypyrimidine tract binding protein 1 (PTBP1) is thought to be expressed only at embryonic stages in central neurons. Its down-regulation triggers neuronal differentiation in precursor and non-neuronal cells, an approach recently tested for generation of neurons de novo for amelioration of neurodegenerative disorders. Moreover, PTBP1 is replaced by its paralog PTBP2 in mature central neurons. Unexpectedly, we found that both proteins are coexpressed in adult sensory and motor neurons, with PTBP2 restricted mainly to the nucleus, while PTBP1 also shows axonal localization. Levels of axonal PTBP1 increased markedly after peripheral nerve injury, and it associates in axons with mRNAs involved in injury responses and nerve regeneration, including importin β1 (KPNB1) and RHOA. Perturbation of PTBP1 affects local translation in axons, nociceptor neuron regeneration and both thermal and mechanical sensation. Thus, PTBP1 has functional roles in adult axons. Hence, caution is required before considering targeting of PTBP1 for therapeutic purposes.
PTBP1 functions in translation and sensation in adult sensory axons, expanding its roles beyond embryonic neuron development.
INTRODUCTION
Axonal regeneration requires elongating growth from the proximal nerve segment after injury. Although functional regeneration occurs in the peripheral nervous system (PNS), central nervous system (CNS) neurons must contend with a lower intrinsic growth capacity as well as a growth inhibitory extracellular environment (1). What are the intrinsic differences that enable more efficient regeneration in the PNS?
PNS regeneration requires activation of intrinsic growth programs by retrograde signaling from injured axons (2). An extensive series of studies have shown that intra-axonal protein synthesis is required for both retrograde injury signaling and for subsequent axonal regeneration (3). The RNA binding proteins (RBPs) for regeneration associated mRNAs in axons are a critical regulatory node in the system (3–5). Sensory axons contain a plethora of RBPs (6); however, it is still unclear how dozens of RBPs associated with thousands of transcripts are localized and regulated to support axonal growth and maintenance. Misregulation of RBPs can cause serious damage in the nervous system and is a contributing factor in diverse neurological diseases (7, 8).
The current study originated in a search for the RBPs required for axonal localization of the mRNA encoding importin β1 (KPNB1), a critical regulatory factor for retrograde injury signaling (9–11). Polypyrimidine tract binding protein 1 (PTBP1) was one of the most prominent candidates we identified, and this was particularly intriguing since previous studies have established that PTBP1 is absent from mature CNS neurons, where it is replaced by its paralog PTBP2 (12). Both paralogs have partially redundant roles in pre-mRNA splicing (13). PTBP1 is highly expressed in proliferating CNS progenitors during development but is down-regulated upon differentiation and is absent in mature CNS neurons. This effect can be attributed to the action of the neuron-specific microRNA miR-124, which promotes neuronal differentiation by reducing PTBP1 levels, with concomitant increase in PTBP2 (14). Notably, we found both proteins coexpressed in adult sensory neurons. Levels of axonal PTBP1 increased markedly after peripheral nerve injury, and perturbation of PTBP1 affects axonal local translation, neuronal injury responses, axon outgrowth, and sensation. These findings suggest that PTBP1 has functional roles in axons of adult neurons.
RESULTS
PTBP1 is associated with KPNB1 mRNA in adult peripheral neurons
We identified PTBP1 by RNA affinity chromatography for RBPs associated with the MAIL RNA localization motif (11) of KPNB1 in axons (Fig. 1A). Pull-downs were conducted from rat sciatic nerve axoplasm extracted as previously described (15), since this enabled examination of an axon-enriched source material without membrane lysis or detergent-induced perturbations (fig. S1A). Intriguingly, PTBP1 was one of the most prominent specific candidates identified by mass spectrometry (MS) from pull-downs with MAIL versus control motifs (Fig. 1B and data S1). Another recent study had also found mass spectrometric evidence for PTBP1 in RNA pull-downs from rat sciatic nerve axoplasm (6). We validated specificity of a series of anti-PTBP1 Abs (Abs; fig. S1, B to D, and table S1) and then verified the interaction in mouse sciatic nerve axoplasm by pull-down followed by Western blots (WBs), using mutated motifs as controls (Fig. 1C). The mutated motifs include GMAIL (G mutated MAIL), with four U to G mutations in the predicted loop, and IMAIL (inactive MAIL), with additional mutations described (11). Similar results were obtained from bovine axoplasm (fig. S1E). A direct interaction of PTBP1 with the MAIL motif was confirmed by in vitro pull-downs using recombinant protein (Fig. 1D), with comparison to recombinant nucleolin, which was previously identified as a MAIL-binding RBP (11).
Fig. 1. Identification of KPNB1 RBPs in the sciatic nerve.
(A) Secondary structure predictions of RNA motifs used in RNA affinity chromatography as shown. Secondary structures for MAIL (Motif for Axonal Importin Localization), and mutated GMAIL and IMAIL control motifs, were generated using Mfold (61) with modified bases shown in red. (B) Proteins identified by MS on MAIL versus GMAIL pull-downs from rat axoplasm. Enrichment score (cutoff in red = 0.667) versus peptide count (PC)/molecular weight (MW) shown in log scale (cutoff in red = 0.0004), for details see data S1. (C) WBs for PTBP1 from mouse sciatic nerve (SN) axoplasm after pull-down with MAIL or control RNA motifs. Note that PTBP1 is enriched in the MAIL pull-down while depleted from the supernatant. (D) In vitro binding assay using recombinant PTBP1 or nucleolin (NCL) protein with MAIL, IMAIL or the zipcode motif (ZIP) of β-actin, with detection by WB, shows direct association of PTBP1 protein with the MAIL RNA localization motif of KPNB1.
PTBP1 is a multifunctional RBP with high expression levels in many cell types, except for mature neurons where it is typically replaced by its neuronal paralog PTBP2 (12, 14, 16, 17). PTBP1 levels were found to be low in extracts of adult mouse brain, but levels in dorsal root ganglia (DRG) were notably high (Fig. 2A). Immunostaining on lumbar DRG sections detected PTBP1 expression in both glial cells and neurons (Fig. 2, B to D), and immunostaining on sciatic nerve sections detected PTBP1 in axons (Fig. 2, E and F). PTBP1 protein was present in all DRG neurons, independent of their cell body or axonal sizes (fig. S2, A and B). Comparison with other PTB family members across different tissues revealed robust coexpression of PTBP2, but not PTBP3, in brain and DRG (fig. S2C). A battery of immunostaining approaches, including cleared tissue staining using immunolabeling-enabled three-dimensional imaging of solvent-cleared organs (iDISCO) (fig. S2D), sciatic nerve cross-section staining (fig. S2E), and cultured neurons immunostaining (fig. S2F), revealed strict neuronal localization of PTBP2, while PTBP1 was found in both neurons and glia. PTBP2 was restricted to neuronal nuclei, while PTBP1 was found throughout in nuclei, cell body cytoplasm, and axons.
Fig. 2. PTBP1 is expressed in adult sensory neurons.
(A) WB analysis for PTBP1 protein levels in liver, brain, and DRG from adult mice. PTBP1 levels were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and shown as fold change (FC) from liver. Individual data points with mean ± SEM, unpaired t test for brain versus DRG, ***P < 0.001, n = 6 mice. (B) Tissue sections of adult lumbar DRG ganglia, stained for DAPI, PTBP1, and TUJ1 (tubulin beta 3 class III). Scale bars, 20 μm. Merged images show outlined neuronal nuclei (white line) and cytoplasm (white dashed line), and glia nuclei are marked by a black cross. (C) Quantification of PTBP1 signal in nuclei shows PTBP1 expression in both cell types with somewhat higher expression in glia. Individual data points with mean ± SEM, ratio paired t test, **P < 0.01, n = 4 mice (>100 nuclei for each mouse). (D) Quantification of PTBP1 in nucleus and cytoplasm of TUJ1-positive neurons. Individual data points with mean ± SEM, ratio paired t test, **P < 0.01, n = 4 mice (>60 cells for each mouse). (E) Immunohistochemistry for PTBP1 on cross sections of sciatic nerve with or without blocking peptide for the PTBP1 Ab. Axonal PTBP1 is shown in magenta hot scale (0 to 255), and borders of TUJ1-positive axonal profiles are indicated with white lines. TL, transmission light. Scale bar, 5 μm. (F) Quantification of PTBP1 in TUJ1-positive axons. Individual data points with mean ± SEM, paired t test, **P < 0.01, n = 3 mice (>1000 axons for each mouse). Gray dashed lines indicate paired data points. AU, arbitrary units.
PTBP1 associates with regeneration-associated mRNAs in injured axons
As KPNB1 is a key injury response mRNA (9, 10), we monitored PTBP1 levels in sciatic nerve axons over time after nerve crush. We observed an increase in PTBP1 levels in axons 3 days after injury, reaching maximal elevation after 1 week (Fig. 3, A to C). PTBP1 was maintained at elevated levels for up to 21 days and then declined back to baseline levels at 56 days (fig. S3A). Significant up-regulation was observed within sensory axons of all diameters at the 1 week time point (Fig. 3, B and C, and fig. S3, B and C). Motor axons and glial cells also expressed PTBP1, with more modest or variable changes upon injury (fig. S3, D and E). There was also no significant increase in PTBP1 in DRG neuron cell bodies after sciatic nerve injury (Fig. 3, D to F, and fig. S3, F to H). Together, these data showing an increase in PTBP1 in injured axons without significant increase in cell bodies might suggest that PTBP1 is preferentially up-regulated in or transported to axons of injured sensory neurons.
Fig. 3. PTBP1 levels increase in sensory axons following nerve crush.
(A) PTBP1 protein expression in sciatic nerve (SN) axoplasm under naive (na) and injury (inj) conditions. Detection by WB. PTBP1 levels were normalized to ERK1/2 (ERK), and the data are shown as FC to naive (red dashed line). Individual data points with mean ± SEM, one-way analysis of variance (ANOVA) with Dunnett’s multiple comparisons test against naive control condition, **P < 0.01, n = 3 mice for each time point. (B) Representative images of SN cross sections from naive nerve and 7 days after crush injury. Axonal PTBP1 is shown in magenta hot scale (0 to 255), axonal profiles positive for TUJ1 indicated with white lines. Scale bars, 5 μm. (C) Quantification of axonal PTBP1 signal in TUJ1-positive axons of different sizes (axon area bins: 0 to 1, 1 to 2, 2 to 5, and 5 to 10μm2). PTBP1 intensity is shown as FC to naive (red dashed line), individual data points with mean ± SEM, two-way repeated measure (RM) ANOVA, ****P < 0.0001, Sidak’s multiple comparisons test, **P< 0.01, *P < 0.05, n = 3 (>700 axons per mouse for each size category). (D) PTBP1 protein expression in cell bodies of DRG. WB of PTBP1 with normalization to GAPDH, shown as FC to naive (red dashed line). Individual data points with mean ± SEM, one-way ANOVA with Tukey’s multiple comparisons test, not significant, n = 3 to 4 mice for each time point. (E) Representative images of DRG tissue sections from naive, 3 and 7 days after SN crush. Scale bars, 20 μm. (F) Quantification of nuclear PTBP1 signal in DRG neurons (summary of all cell sizes, NHF+ and NHF− cells). Individual data points with mean ± SEM, one-way ANOVA with Tukey’s multiple comparisons test, not significant, n = 3 to 7 mice (>95 cells per mouse).
PTBP1 up-regulation in injured axons raises the possibility that it functions to regulate axonal RNAs required for neuronal regeneration. Hence, we carried out RNA sequencing on PTBP1 immuno-precipitates from sciatic nerve axoplasm before and 7 days after injury (Fig. 4A). Axoplasm was harvested in physiological buffers without detergents as previously described (15), ensuring minimal disruption of native complexes. Specificity of the pull-downs was verified by use of the Ab-blocking peptide as control (fig. S4A). We identified 1069 mRNAs specifically associated with PTBP1 in axoplasm, of which 348 were enriched in injured nerve axoplasm and 825 in naive nerve (Fig. 4, B and C; fig. S4, B to D; and data S2). 3′ Untranslated region (3′UTR) motif analyses on both these gene sets revealed an enrichment of CU motifs (Fig. 4D), which are highly similar to the canonical pyrimidine-rich PTBP1 binding repeats (18). The KPNB1 MAIL motif also contains a CU-rich sequence stretch (Fig. 1A), which might account for its PTBP1 binding. Ingenuity pathway analysis highlighted specific signaling networks enriched after injury, suggesting that PTBP1-associated RNAs encode proteins involved in cytoskeleton remodeling and axon growth after nerve injury (Fig. 4E).
Fig. 4. Identification of PTBP1 associated mRNAs in axoplasm.
(A) Schematic of RNA coimmunoprecipitation (RNA-IP) with PTBP1 from naive (na) sciatic nerve axoplasm or 7 days after crush injury (inj). A blocking peptide specific for the PTBP1 Ab was used as a negative control (Ctrl). RNA-seq, RNA sequencing. (B) Bar plot showing the total number of transcripts identified as differentially and nondifferentially expressed [false discovery rate (FDR) < 0.1]. Gray = not significant (ns), blue = enriched in na, red = enriched in inj. (C) Heatmap of the differentially expressed genes (DEG). Only the 1069 significantly enriched transcripts are shown (IP versus Ctrl: logFC > 0, FDR < 0.1) with three replicates for each group. (D) Motif enrichment analysis on the 3′UTR of significant mRNAs in naive and injury conditions using HOMER (63). Top three ranked motifs in each condition are shown with all P values < e−20. HOMER de novo motif results show enrichment of CU- motifs in the PTBP1 IP samples. (E) Comparison of PTBP1 associated mRNAs between naive and injury. Venn diagram shows the number of unique or overlapping mRNAs in naive/injury (1069 total). Naive, blue; injury, red; overlap, purple. Side panels show top 15 canonical pathways enriched in each section [Ingenuity Pathway Analysis (IPA), Qiagen]. Heatmap shows −log10 (P value) from minimum to maximum value (spectrum from white to saturated color for each section). A complete list of mRNAs can be found in data S2.
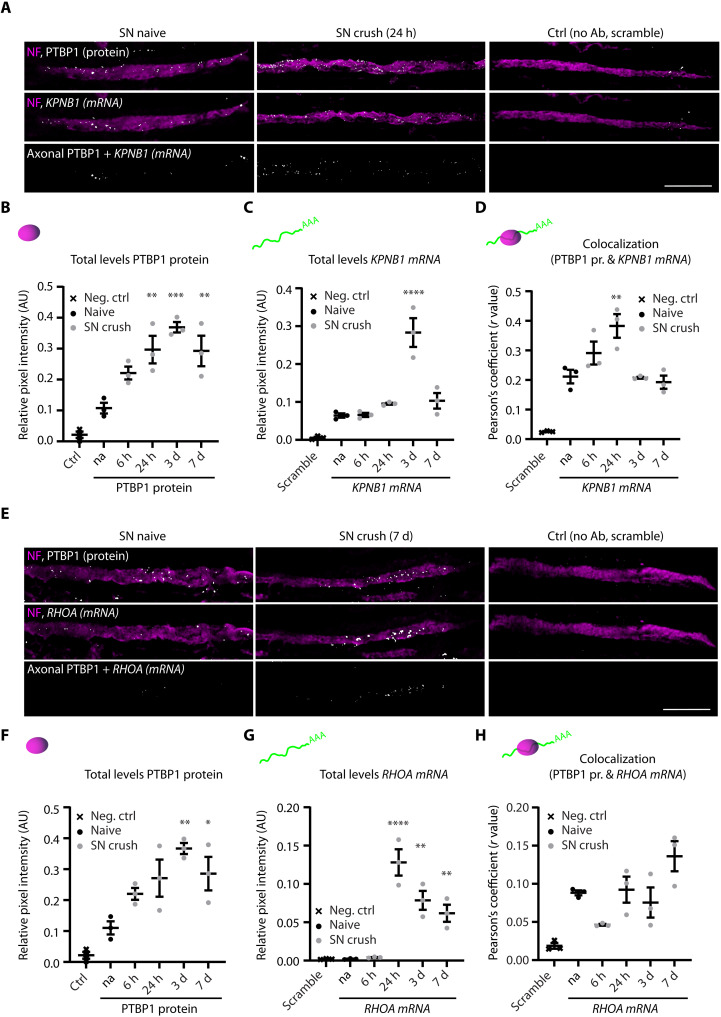
We proceeded to validate axonal colocalization of PTBP1 protein with key mRNAs from the dataset, focusing on KPNB1, which, as noted above, is a central injury response mRNA, and RHOA, a well-characterized nerve regeneration regulator (19). We combined RNA fluorescence in situ hybridization (FISH) for the target mRNAs with immunostaining for PTBP1 in sciatic nerve sections (Fig. 5 and fig. S5). There was significant axonal colocalization between KPNB1 mRNA and PTBP1 protein with a marked increase 24-hour postlesion in injured nerve (Fig. 5, A to D, and fig. S5, A and B), while for RHOA axonal colocalization was similar under both naive and injury conditions (Fig. 5, E to H, and fig. S5, C and D). However, overall levels of RHOA mRNA were extremely low in naive sciatic nerve and during the first few hours after crush, with markedly increased levels from 24 hours onward, as also noted for PTBP1 (Fig. 5, F and G).
Fig. 5. FISH validation of PTBP1 associated mRNAs.
(A and E) Images of longitudinal sections from sciatic nerve (SN) collected at the designated time points after crush injury and stained for NF (neurofilament) and PTBP1 protein, combined with FISH for KPNB1 (A) or RHOA (E) mRNA. No primary Ab was used as control for PTBP1 staining (Ctrl) and scramble mRNA probe for FISH. Top: Overlay of NF (magenta) with PTBP1 protein (gray), single plain (XY). Middle: NF (magenta) with mRNA (gray), single plane (XY). Bottom: Colocalization of axonal PTBP1 with mRNA, maximum projection of z-stack (XYZ). Images are overexposed for illustration purposes. Scale bars, 10 μm. (B to D and F to H) Quantification of different FISH parameters showing individual data points with mean ± SEM, one-way ANOVA with Sidak’s multiple comparisons test comparing the different time points after SN crush against naïve, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, n = 3 mice for each condition. Corresponding M1 and M2 values can be found in fig. S5. (B) Quantification of the total levels of PTBP1 protein in the SN. (C) Quantification of the total levels of KPNB1 mRNA in the SN. (D) Quantification of axonal colocalization by Pearson’s correlation with r values as individual data points. (F) Quantification of the total levels of PTBP1 protein in the SN. (G) Quantification of the total levels of RHOA mRNA in the SN. (H) Quantification of axonal colocalization by Pearson’s correlation with r values as individual data points. Please see fig. S5 for supplementary colocalization analyses.
PTBP1 regulates injury responses of adult nociceptive neurons
The findings above implicate PTBP1 in injury responses after nerve lesion. We tested this by using adeno-associated virus (AAV)–encoded short hairpin RNA (shRNA) for knockdown of PTBP1 in DRG in vivo (Fig. 6, A and B). We validated two different PTBP1 shRNAs, targeted to either the open reading frame or the 3′UTR region of PTBP1 mRNA (fig. S6, A to C). shRNAs were transduced via intrathecal injections of appropriate AAV9 or PhP.S constructs 3 weeks before a conditioning sciatic nerve lesion. Neurons were then cultured from L4/L5 lumbar DRG 7 days after the lesion. PTBP1 knockdown with either PTBP1 shRNA attenuated neuronal outgrowth after conditioning lesion (Fig. 6, C to E, and fig. S6, D and E).
Fig. 6. PTBP1 knockdown affects sensation and injury responses in adult sensory neurons.
(A) Viral constructs used for knockdown experiments. (B) Schematic of in vivo knockdown experiments. At day 7 after crush, both hindlimbs (naive left and injury right) were tested in CatWalk gait analysis, or DRG ganglia were collected for neuron culture. IT, intrathecal. (C) Representative images for DRG neurons 20 hours in culture for naive and injury conditioned cells from shCtrl and shPTBP1 UTR treated mice. TUJ1 (tubulin beta 3 class III). Scale bar, 100 μm. (D and E) Quantification of neuron growth. Mean ± SEM, two-way ANOVA with Sidak’s multiple comparisons test (comparing naive versus injury in each condition), *P < 0.05, **P < 0.01, n = 5 to 9 mice per condition. (D) Percent of growing neurons. (E) Total outgrowth in μm. (F) CatWalk gait analysis after knockdown of PTBP1, naive or 7 days after SN crush. Mean ± SEM, two-way RM ANOVA with Sidak’s multiple comparisons test, not significant, n = 11 to 13 mice. (G and H) Naive animals were tested in the hot plate test (G) and von Frey mechanosensation (H) assay before (baseline) and 3 weeks after AAV injection. Mean ± SEM, two-way RM ANOVA with Sidak’s multiple comparisons test. ****P < 0.0001, n = 8 to 14 mice, PWT (paw withdrawal threshold). (I) SN longitudinal sections 3 days after sciatic nerve crush for shCtrl and shPTBP1 UTR-treated mice. Merged image of CGRP staining in magenta and EYFP from shRNA in green, injury site is indicated by the yellow dashed line, left side proximal, right side distal. Scale bar, 500 μm. (J) Quantification of the number of double positive regenerating axons (CGRP and GFP) 1 mm distal to the injury site. Mean ± SEM, two-tailed unpaired t test, **P < 0.01, n = 10 to 17 images from three to four mice.
We next used behavioral assays to investigate the functional effects of PTBP1 depletion in baseline conditions and after injury in vivo. Knockdown of PTBP1 had no effect on baseline or injury-associated gait parameters assessed on a CatWalk apparatus (Fig. 6F and fig. S7A). On the other hand, treatment with either PTBP1 shRNA increased mechanical and thermal sensitivity under both baseline conditions (Fig. 6, G and H) and at different time points after sciatic nerve crush (fig. S7, B and C). We subsequently tested the impact of PTBP1 knockdown on axonal regeneration after sciatic nerve lesion in vivo, by examining expression of two different regeneration associated marker genes, calcitonin gene-related peptide (CGRP) for nociceptors (20, 21) and superior cervical ganglion 10 (SCG10) for general regenerating fibers (22). We observed a significant reduction in CGRP-positive but not SCG10-positive, regenerating enhanced yellow fluorescent protein [(EYFP) labeled] fibers distal to the lesion site in PTBP1 knockdown nerves (Fig. 6, I and J, and fig. S7, D and E). Thus, PTBP1 effects on both basal sensation and injury responses are primarily seen in nociceptor neurons.
PTBP1 regulates axonal translation of RHOA mRNA
To better characterize the relationship between PTBP1 and its associated mRNAs, we next used puromycin–proximity ligation assay (PLA) labeling (23) to evaluate the impact of PTBP1 knockdown on translation of RHOA mRNA. Knockdown of PTBP1 significantly reduced RHOA mRNA translation in DRG neuron cell bodies in culture, in parallel with a marked and significant increase in local translation in axons (Fig. 7, A to C). These changes suggest a critical role for PTBP1 in regulating axonal translation of RHOA mRNA. We further analyzed axonal RHOA mRNA by live imaging of a molecular beacon (MB) probe (24) in parallel with AAV-mediated expression of PTBP1 fused with EYFP. After validating the RHOA MB (Fig. 7, D and E), we examined association of PTBP1-EYFP protein and RHOA mRNA in live neurons over time. As shown in Fig. 7, D to F, there is clear and significant association of RHOA mRNA with PTBP1 in live neurons, but the colabeled puncta are essentially stationary or oscillatory (note velocity ranges shown in Fig. 7F). We did not observe effective long distance processive movement of PTBP1 together with RHOA mRNA in axons. Together, these data suggest that PTBP1 is a local repressor of RHOA mRNA translation in axons.
Fig. 7. PTBP1 knockdown affects RhoA translation.
(A) Representative images of cultured DRG neurons, transduced as indicated with shControl or shPTBP1-UTR, and imaged after puromycin and/or anisomycin pulse. EYFP signal in red, RhoA-puromycin PLA puncta in white. Zoom-in on indicated axon segment below. Scale bars, 20 μm. (B and C) Number of RhoA-puromycin PLA puncta in cell bodies (B) or axons (C) of puromycin (Puro)– or puromycin/anisomycin (Aniso)–treated cells. Individual data points with median, one-way ANOVA with Tukey’s multiple comparisons test (B), or Kruskal-Wallis with Dunn’s multiple comparisons test (C). *P < 0.05, **P < 0.01, ****P < 0.0001, n = 30 to 40 neurons per condition. (D) Representative images from live tracking, TYE665-scramble or TYE665-RhoA MB in magenta, PTBP1-EYFP in green. Colocalizing particles in white. Scale bar, 5 μm. (E and F) Quantification of live imaging data. (E) Pearson’s coefficient for PTBP1-EYFP protein and TYE665-scramble /TYE665-RhoA MB in axons (gray and magenta, respectively, n ≥ 12), individual data points with mean ± S.E.M, unpaired t test. **P < 0.01. (F) Velocities of TYE665-scramble or TYE665-RhoA puncta cotransported with PTBP1 (gray and magenta, respectively, n ≥ 48 puncta acquired), individual data points with median and 95% confidence interval, Mann-Whitney test, ****P < 0.0001.
Axonal perturbation of PTBP1 affects functional responses
The findings above support roles for PTBP1 in the regulation of sensation and injury responses in adult sensory neurons. Are these due to canonical nuclear roles of PTBP1, or do they arise from axonal mechanisms? We sought to test whether local perturbation of PTBP1 in axons might affect thermal/mechanical sensitivity and/or injury responses, using a decoy RNA oligonucleotide for acute and local block of PTBP1 (Fig. 8A) (25). PTBP1-blocking (PTBP1i) or control (SCRMi) oligonucleotides were injected locally to the sciatic nerve just before lesion (Fig. 8B). This experimental paradigm enables local perturbation of PTBP1 in the sciatic nerve, as the decoy oligo accumulates in axons but does not reach neuronal cell bodies in the DRG (fig. S8, A to C). The decoy oligo clearly disrupted colocalization of PTBP1 with RHOA and KPNB1 mRNA. The effect was most prominent at 7 days postinjury for RHOA and at 1 day for KPNB1 (Fig. 8, C to E). The effect was clearly specific for colocalization of PTBP1 with RHOA, without affecting expression levels of either component, while in the case of KPNB1, total mRNA levels were also somewhat affected (fig. S8, D to F). The postinjury effects of the decoy oligonucleotide were similar to those observed previously by shRNA knockdown of PTBP1, namely, no effects on gait parameters (Fig. 8F and fig. S8G) and increases in mechanical and thermal sensitivity after injury (Fig. 8, G and H, and fig. S8, H and I). Together, these data indicate that PTBP1 effects on mechanical and thermal sensitivity of injured adult DRG neurons are mediated in the axonal compartment.
Fig. 8. Functional effects of local inhibition of PTBP1 in crushed nerve.
(A) The RNA decoy oligo for PTBP1 (PTBP1i) prevents PTBP1 protein from binding to endogenous mRNA targets. (B) Timeline for functional assays after inhibition of PTBP1 in the sciatic nerve (SN). (C) Representative images of longitudinal sections from SN 7 days after crush injury and RNA oligos injections, stained for NF (neurofilament) and PTBP1 protein, and FISH for RHOA mRNA. Top: Single plane showing overlay of NF (magenta) with PTBP1 protein (gray). Middle: Single plane showing NF (magenta) with FISH for RHOA mRNA (gray). Bottom: Maximum z-stack projection showing colocalization of axonal PTBP1 with RHOA mRNA. Images overexposed for illustration purposes. Scale bar, 5 μm. (D) Quantification of axonal colocalized PTBP1 protein and RHOA mRNA. Pearson’s correlation analysis, r value of individual data points with mean ± SEM, two-way ANOVA with Sidak’s multiple comparisons test to compare between different groups (SCRMi versus PTBP1i), ***P < 0.001, n = 3 mice for each condition. (E) Quantification of axonal colocalized PTBP1 protein and KPNB1 mRNA. Pearson’s correlation analysis, r value of individual data points with mean ± SEM, two-way ANOVA with Sidak’s multiple comparisons test to compare between different groups (SCRMi versus PTBP1i), ***P < 0.001, ****P< 0.0001, n = 3 mice for each condition. (F to H) Behavior after inhibition of PTBP1, 7 days after SN crush. Individual data points with mean ± SEM, two-way RM ANOVA with Sidak’s multiple comparisons test, **P < 0.01, ****P < 0.0001, n = 9 mice. (F) CatWalk gait analysis, (G) heat probe, and (H) von Frey mechanosensation.
Axon-specific rescue of PTBP1 perturbation
We sought to generate a PTBP1 mutant deficient in axonal localization, to allow further examination of its site of action. To this end, we generated a series of chimeras, substituting different domains of PTBP1 with the corresponding region in PTBP2 (fig. S9A), and sought chimeras that would be restricted to the nucleus in a similar manner as wild-type PTBP2. The different chimeras were screened for axonal localization by plasmid transfection in cultured hippocampal neurons (fig. S9B). Substitution of the RNA recognition motifs (RRMs) of PTBP1 with those of PTBP2 affected axonal localization, and the most effective substitution proved to be replacement of the first two RRMs together with their connecting linker sequence (R1-L1-R2) (fig. S9C). A previous study has shown that PTBP1 nucleocytoplasmic shuttling is encoded in a very similar region (26). The PTBP1 (PTBP2 R1 + L1 + R2) chimera, henceforth termed PTBP1R1R2, was restricted to the nucleus to the same degree as wild-type PTBP2 (fig. S9C). We further confirmed equivalent overall expression with lower axonal localization of the chimera in comparison to wild-type PTBP1, after viral transduction in DRG neurons (Fig. 9, A to C).
Fig. 9. Axonal PTBP1 is required for normal mechanosensitivity.
(A) AAV constructs rescue experiments. PTBP1 regions in green, PTBP2 in magenta. HSyn1 (human synapsin promoter), WPRE (Woodchuck hepatitis virus posttranscriptional regulatory element). (B) Representative images for DRG neurons transduced as indicated. TUJ1 in green, FLAG in magenta, merge. Scale bar, 20 μm. Growth cone inserts scale bar, 1 μm. (C) FLAG intensity in growth cones of transduced neurons (n ≥ 80), mean ± SEM, unpaired t test, ****P < 0.0001. (D and E) Mechanosensitivity effects of transduction with wild-type PTBP1WT or PTBP1R1R2 in PTBP1 knockdown animals. Animals underwent von Frey testing 3 weeks after AAV transduction in uninjured animals (D) and at the indicated time points after sciatic nerve crush (E). Four treatment groups were compared as shown. shCtrl in solid gray, shPTBP1 UTR in solid magenta, shPTBP1 UTR + PTBP1wt in blue, shPTBP1 UTR + PTBP1R1R2 in red. Individual data points with mean ± SEM shown in (D), mean ± SEM in (E), one-way ANOVA with Tukey’s multiple post hoc test. *P < 0.05, ***P < 0.001, ****P < 0.0001. n = 3 to 8 mice per group.
Last, we compared the efficacy of PTBP1wt and PTBP1R1R2 in ameliorating the increase in sensitivity induced by PTBP1 knockdown. Animals were transduced with AAV expressing either shCtrl, shPTBP1 UTR, or a mixture of shPTBP1 UTR with PTBP1wt or PTBP1R1R2. Notably, only PTBP1wt was able to rescue the increase in mechanical and thermal sensitivity induced by the PTBP1 knockdown (Fig. 9D and fig. S10A). Similar results were obtained after sciatic nerve crush, with monitoring of mechanical and thermal sensitivity at different time points after the lesion. As shown in Fig. 9E and fig. S10B, PTBP1wt effectively ameliorated the increases in mechanical and thermal sensitivity over time after injury, while PTBP1R1R2 had little or no effect. Thus, the activities of PTBP1 are mediated at axonal sites in adult sensory neurons.
DISCUSSION
The findings that PTBP1 is expressed in adult sensory and motor neurons and that it has functional roles in sensation, injury response, and regeneration in adult neurons are unexpected in light of the well-established model that PTBP1 down-regulation triggers neuronal differentiation, and it is not expressed in adult CNS neurons (13, 17). A previous bioinformatics study reported that mature sensory neurons have splicing profiles consistent with continued expression of PTBP1 in adulthood (27) but did not provide experimental evidence to confirm that prediction. Our study provides experimental data on multiple levels for PTBP1 expression in adult sensory neurons and moreover highlights roles for PTBP1 in axonal regulation of mRNAs required for both injury response and regenerative growth.
We identified PTBP1 in adult axons due to its binding to the KPNB1 MAIL axonal localization motif. KPNB1 is a key injury response mRNA in sensory axons, and its local translation is required in retrograde injury signaling (9, 10) and in regulation of axon length sensing (5, 11, 28). These functions are mediated by an RNP comprising the RBP nucleolin and other canonical growth regulating mRNAs, including mTOR (11, 29). Apart from KPNB1 itself, there is limited overlap between the mRNA ensembles bound by nucleolin and PTBP1 in sensory axons; hence, nucleolin and PTBP1 are not likely to participate in the same transport RNP. Rather, the dynamics of the interactions suggest that PTBP1 may be involved in regulating KPNB1 in the axon itself, perhaps as part of the temporal regulation of injury response versus regeneration promoting mRNAs in axons (30). A previous study implicated PTBP1 in regulation of mRNAs in focal adhesions of mouse embryonic fibroblasts (31); hence, PTBP1 may have conserved cytoplasmic roles in diverse cellular extensions and extremities.
Ras homolog family member A (RhoA), on the other hand, is a well-characterized cytoskeleton regulating factor with extensive roles in neuronal survival and in axon growth and regeneration (19, 32, 33). Local translation of RHOA has been reported in embryonic axon growth cone collapse and in inhibitory axon growth pathways in embryonic and postnatal neurons (34, 35), with a tightly regulated dynamics of local synthesis coupled with localized degradation to control RhoA activation in the growth cone (36). RhoA signaling has been identified as a key regulatory junction in inhibition of axon growth by multiple external factors (37). In this context, our data showing increased translation of RHOA mRNA in axons after PTBP1 knockdown, with concomitant impaired axonal regeneration of nociceptive neurons, suggest that PTBP1 repression of local translation of its associated mRNAs in axons is required for effective nociceptor regeneration. Previous studies have implicated PTBP1 in either enhancing (38) or repressing (39) translation of different mRNAs in other systems. If PTBP1 is a key factor for determining the composition of the nascent axonal proteome, then it is intriguing to speculate that a lack of PTBP1 in CNS axons may contribute to CNS regeneration failure.
A number of groups recently took advantage of the canonical role of PTBP1 in neuronal differentiation (40–42) to assess the feasibility of PTBP1 knockdown for in vivo differentiation of new neurons to ameliorate neurodegeneration. Despite initial optimistic reports of disease-ameliorating conversion of endogenous glia to different neuronal subtypes (43–45), subsequent studies suggested that the purported glia-converted neurons were actually mislabeled endogenous neurons (46–50). Moreover, detailed lineage tracing did not support in vivo conversion of endogenous glia to neurons, despite efficient PTBP1 knockdown (51). Regardless, a basic assumption in all those studies is the notion that PTBP1 is not expressed and does not have functions in adult neurons. Our current work clearly shows that this is not the case for peripheral sensory neurons. PTBP1 has roles in regulating basal sensation, injury-induced local translation, and regenerative growth of nociceptors and might have similar functions in other neuronal subtypes. Additional studies on the roles of PTBP1 in adult neurons will be required before revisiting PTBP1 targeting approaches in the clinic.
MATERIALS AND METHODS
Animals
All animal procedures were performed in accordance with the guidelines of the Weizmann Institute of Science and University of South Carolina Institutional Animal Care and Use Committees. Adult C57BL/6 and Hsd:ICR mice were purchased from Envigo Ltd. (Israel) and maintained at the Veterinary Resources Department of the Weizmann Institute with free access to food and water. Wistar rats (Envigo Ltd., Israel) were used for MS analyses. Hsd:ICR mice were used only for WB analyses (fig. S3A), all other experiments were performed on C57BL/6. All experiments were performed on male animals between 2 and 5 months of age.
SN crush model
Animals were anesthetized with ketamine/xylazine intraperitoneal (10 mg/kg body weight). Sciatic nerve (SN) crush was performed at mid-thigh level using a fine jewelers’ forceps in two adjacent positions for 30 s each. Only the right side was subjected to the crush injury procedure (inj), the contralateral side served as uninjured, naive (na) control.
Behavior
All mice that were used for behavioral testing were kept on a reverse dark/light cycle so that the assays could be performed during the “dark” active phase of the animal. One hour of habituation was carried out in the test room before each session. All behavioral tests were done in a blinded manner.
CatWalk
Gait analysis was performed using CatWalk XT 10.6 (Noldus Information Technology, The Netherlands) as previously described (10). This setup allows for quantification of footprints and gait in unforced moving animals. Motivation was achieved by placing the home cage at the end of the runway. At least three compliant runs per animal were recorded in each session.
Hot plate
Naïve mice are tested for heat sensitivity. Animals are placed on a 52°C metal flat surface covered with an additional thermal insulating plastic surface. A transparent plastic box (25 cm by 15 cm by 25 cm) is placed on top of the surface to restrict the mouse in the test area. Slowly, the plastic surface is removed to expose the mouse to the hot surface. The latency time to hind paw licking and/or shaking, or jumping, is recorded, and at this point, the animal is immediately removed from the hot plate (52).
Heat probe
This test was used to evaluate the response to noxious heat, as previously described (53). While holding the animal, a metal probe, heated to 58°C, was applied to both hind paws consecutively (middle part of the plantar side, left paw naive, and right paw after crush injury). The paw withdrawal latency was recorded in seconds (s) for three trials, with 20-min intervals between repeats.
von Frey
Mechanical sensitivity was tested in the von Frey paradigm as previously described (53). Mice were placed in an elevated setup of transparent chambers with a wire mesh grid on the bottom. Following a 1-hour habituation period, nylon monofilaments of different diameter (von Frey Hairs #37450-275, Ugo Basile) were pressed against the plantar surface of the hind paw until bending of the filament and held for a maximum of 3 s. A positive response was noted if the paw was sharply withdrawn upon application of the filament. The test was started with a stimulus of 1.4 g [filament target force of 13.7 millinewtons (mN)] and continued in an up-down testing paradigm for at least five representations of the stimulus. The paw withdrawal threshold (PWT) was calculated for each animal using UDMAP (V3.0) (54).
Abs and validation
Primary Abs used in this study for immunoprecipitation (IP), immuno-cytochemistry (ICC), immunohistochemistry (IHC), immunofluorescence (IF: ICC and IHC), and WB were as follows: anti-PTBP1 (Santa Cruz Biotechnology, sc-16547) or anti-PTBP1 (Abcam, ab5642): both recognize the same epitope and were used for IP at 2.5 μg/25 μl beads, WB 1:500, and IF 1:200. The blocking peptide for PTBP1 Abs (sc-16547 and ab5642) was synthesized by Sigma-Aldrich ([H]-DGIVPDIAVGTKRGSDELFS-[OH]). In addition, we used anti-PTBP1 (Abcam, ab133734, WB 1:1000) and anti-PTBP1 (Thermo Fischer Scientific, 32-4800, WB 1:1000). For PTBP2, we primarily used anti-PTBP2 (Abnova, H00058155-A01, IF 1:1000, WB 1:2000). Other anti-PTBP2 Abs tested were from Abcam (ab154787, WB 1:1000) or Santa Cruz Biotechnology (sc-103847, WB 1:500). Details of the precise Ab used for each figure are provided in table S1.
To test cross reactivity of PTBP1 and PTBP2 Abs, we generated plasmids containing FLAG or MYC-tagged versions of PTBP1 and PTBP2 for overexpression. Constructs were based on Ensembl IDs ENSRNOT00000044865.7 (PTBP1-001, rat) and ENSRNOT00000015036.7 (PTBP2, rat), inserted into a pcDNA3.1 backbone. Plasmids were transfected into HeLa cells (229, American Type Culture Collection) using jetPEI (Polyplus). After 48 hours, cells were fixed for staining or used for protein extraction. Abs for PTBP1 and PTBP2 were tested to specifically detect either recombinant PTBP1 or PTBP2, respectively. MYC/FLAG served as a positive control for recombinant proteins in both samples.
We also validated our main PTBP1 and PTBP2 Abs (PTBP1-g: ab5642 and PTBP2-m: H00058155-A01) in knockdown conditions using commercially available small interfering RNAs (siRNAs). siRNA transfections were performed using specific siRNA probes (siGENOME, Mouse, siRNA SMARTpool, Dharmacon) according to the standard protocols with DharmaFECT 4 Transfection Reagent (T-2004-02). DRG neurons were transfected 2 hours after plating with 100 nM siRNA for Ctrl (D-001210-05-20), PTBP1 (M-042865-01-0005), PTBP2 (M-049626-00-0005) or a combination of both. Medium was changed after 24 hours, and proteins were extracted 48 hours later (total time in culture, 72 hours) and analyzed by WB.
Additional Abs used in this study were as follows: anti-ROD1 (regulator of differentiation 1) (Santa Cruz, sc-100845, WB 1:200), anti–green fluorescent protein (GFP) (Abcam, ab6556, IHC 1:500, ICC and WB 1:5000), anti-NFH (neurofilament heavy chain) (Abcam, ab72996, IHC 1:1000, ICC 1:2000), anti-NF (neurofilament) (Dev Hybrid Studies Bank, RT97, mouse, IHC 1:100), anti–glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Santa Cruz Biotechnology, sc-32233, WB 1:5000), anti-ERK1/2 (extracellular signal-regulated kinase 1/2) (Sigma-Aldrich, M5670, WB 1:30000), anti–β-III tubulin TUJ1 (Abcam, ab18207, IF 1:1000, WB: 1:6000), anti-CGRP (AbD SEROTEC 1720-9007, IF 1:1000), anti-FLAG M2 (Sigma-Aldrich, F1804, IF 1:500), anti Stathamin-2/SCG-10 (Novus Biologicals, NBP1-49461 IF: 1:1000), anti-Nucleolin (Abcam, ab50279, IF & WB 1:1000), anti-RhoA (Abcam, Ab187027, IF 1:150), and anti-puromycin (Millipore, 12D10 clone, MABE343, IF 1:3500). 4′,6-Diamidino-2-phenylindole (DAPI; Thermo Fisher Scientific, D1306) was used at 1 μg/ml.
Secondary Abs used for IF are donkey anti-mouse/rabbit/goat/chicken Alexa Fluor 488/594/647 (Jackson ImmunoResearch, IHC 1:500, ICC 1:1000). Secondary horseradish peroxidase (HRP)–conjugated mouse and rabbit Abs for WB were purchased from Bio-Rad Laboratories, secondary bovine anti-goat immunoglobulin G–HRP (Santa Cruz Biotechnology, sc-2350). All HRP-conjugated Abs were used 1:10,000.
Protein extraction and SDS-PAGE
For total protein extraction, cells or tissues were collected in radioimmunoprecipitation assay (RIPA) buffer [150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, and 50 mM tris (pH 8.0.)], supplemented with protease/phosphatase/ribonuclease (RNase) inhibitors. Tissue samples were homogenized in Dounce Tissue Grinders (WHEATON 33, 1 ml #357538, 7 ml #357542) or using plastic pestles in an Eppendorf tube.
To obtain neuron-enriched protein extracts from DRG tissue, ganglia were dissociated according to the DRG culture protocol described below. After the Percoll gradient (depletion of non-neuronal cells), cell pellets were washed in phosphate-buffered saline (PBS) and then lysed in RIPA buffer.
Axoplasm for biochemical analysis was extracted from mouse or rat SN as previously described (11, 15, 55). To minimize glia contamination, transport buffer [20 mM Hepes, 110 mM KAc, and 5 mM MgAc (pH 7.4) supplemented with protease/phosphatase/RNase inhibitors] was used in this extraction protocol. All protein extracts were incubated on ice for 20 min, followed by a spin down at 10,000g for 10 min at 4°C. The supernatant was used for protein electrophoresis (Mini-PROTEAN Tetra Cell Systems, Bio-Rad) and blotting (Trans-Blot Turbo Transfer System, Bio-Rad). Chemiluminescence was detected with ImageQuant LAS 4000 (GE Healthcare), and band intensities were quantified using built-in software (ImageQuant TL).
Pull-down of RBPs
Biotinylated RNA probes for pull-down assays were synthesized by Integrated DNA Technologies (Syntezza, Israel): MAIL: 5′-Biotin-TEG-UCACAAACAAGCUCUCUCCUGACUUGUAUUGUGG-3′; GMAIL: 5′-Biotin-TEG-UCACAAACAAGCGCGCGCCGGACUUGUAUUGUGG-3′; IMAIL: 5′-Biotin-TEG-UGAGAAAGAAGGGCGCGCCGGACUUCUAUUCUCC-3′; ZIP (ACTB): 5′-Biotin-TEG ACCGGACUGUUACCAACACCCACACCCCUGUGAUGAAACAAAACCCAUAAAUGC-3′.
RNA affinity chromatography was performed as previously described (55). Briefly, streptavidin magnetic beads were washed several times and incubated with the biotinylated probe for 1 hour at 4°C, followed by three additional washes. Meanwhile, freshly prepared axoplasm was applied to beads without a probe for 1 hour to deplete unspecific proteins. Afterward, the unbound fraction was added to the specific probes for 1 hour at 4°C.
For the in vitro binding assay, purified recombinant human PTBP1 protein was purchased from Abnova (H00005725-P01). One microgram of recombinant PTBP1 was used for each probe (MAIL, IMAIL, and ZIP with 20-μl streptavidin beads). Bound material was washed and eluted from the beads using SDS sample buffer (WB) or RNase A (for MS). The samples were loaded onto 10% SDS–polyacrylamide gel electrophoresis gels, followed by WB or MS analysis.
RNA binding protein identification using MS
Sample preparation for MS
Proteins binding to MAIL versus control RNA motifs were fractionated on SDS-PAGE (Bio-Rad tris-glycine precast gels 4 to 15%) and stained using the Colloidal Blue Staining Kit (Invitrogen). Each lane was cut into 10 pieces, and these pieces were subjected to in-gel digestion (http://ms-facility.ucsf.edu/protocols.html). Briefly, the SDS and CBB were removed using 25 mM ammonium bicarbonate (ABC) in 50% acetonitrile/water, then the disulfide bridges were reduced with dithiothreitol in 25 mM ABC buffer, and the free −SH groups were alkylated with iodoacetamide. Gels were subsequently dehydrated and reconstituted with the trypsin solution in 25 mM ABC buffer. After 4-hour digestion at 37°C, the resulting peptides were extracted with 5% formic acid in 50% acetonitrile/water. The peptide solutions were concentrated and submitted to liquid chromatography–tandem MS (LC/MS/MS).
Mass spectrometry analysis
LC/MS/MS was performed using a NanoACQUITY- LTQ-Orbitrap XL system (Waters and Thermo Fisher Scientific, respectively). The peptide fractionation was performed on a C18 column (75 μm by 150 mm) at a flowrate of ~400 μl; solvent A was 0.1% formic acid in water, and solvent B was 0.1% formic acid in acetonitrile. A linear gradient was applied from 2 to 35% organic in 40 min. Data were acquired for 60 min following sample injection, in a data-dependent manner. The precursor ions were measured in the Orbitrap, and the six most abundant multiply charged ions were selected for collision-induced dissociation experiments performed and measured in the linear trap. The trigger threshold was 1000, and dynamic exclusion was enabled.
Data processing
PAVA in-house script (56) was used for peak picking, and database searches were performed with the 10 peak lists representing the same sample combined, using Protein Prospector. Search parameters: Only tryptic peptides were permitted, with two missed cleavages; mass accuracy within 20 parts per million and 0.6 Da for precursors and fragments, respectively; fixed modification: Cys carbamidomethylation; variable modifications: Met(O); protein N-acetylation; N-terminal Gln to pyroGlu. The database was UniProtKB.2013.6.17, concatenated with random sequences for each entry, Rattus and Mus musculus proteins were selected, and human keratins were added to the list (108,456 entries were searched). Acceptance criteria: Minimum score: 20 and 15; max E value: 0.05 and 0.1 for proteins and peptides, respectively. False discovery rate (FDR) < 1%, based on the number of decoy hits. Peptide counts (PC) were normalized by the molecular weight (MW) of the identified protein. Enrichment of MAIL versus GMAIL (control) was calculated as
Proteins (peptides) only identified in the MAIL pull-down have an enrichment score of 1, whereas peptides only identified in GMAIL control have an enrichment score of 0. To identify potential candidates, a twofold enrichment of MAIL versus GMAIL was used (enrichment score > 0.6667), as well as a cutoff for high coverage with PC/MW > 0.0004. A complete list of identified proteins can be found in data S1.
DRG culture
The procedure for DRG neuron culture was performed as previously described (28). Briefly, adult mouse DRGs were dissociated with 100 U of papain (P4762, Sigma-Aldrich) followed by collagenase II (1 mg/ml; 11179179001, Roche) and dispase II (1.2 mg/ml; 04942078001, Roche). The ganglia were then triturated in Hanks’ balanced salt solution, 10 mM glucose, and 5 mM Hepes (pH 7.35) using a fire-coated Pasteur pipette. Neurons were layered on 20% Percoll in L15 media and recovered through centrifugation at 1000g for 8 min. Cells were washed briefly in growth media [F12; 10% fetal bovine serum (FBS) and Primocin (100 μg/ml; InvivoGen #ant-pm-1)] and plated on poly-l-lysine (P4832, Sigma-Aldrich)– and laminin (23017-015, Invitrogen)–coated glass cover slips. Culture media and serum were from Thermo Fisher Scientific.
Histology, IF, and image analysis
Immunocytochemistry
Cultured cells were washed in PBS and fixed in 4% paraformaldehyde (PFA) for 20 min. Blocking was performed in 10% donkey serum, bovine serum albumin (1 mg/ml; BSA), and 0.2% Triton in PBS for 1 hour at room temperature (RT, ~25°C). Primary Abs were incubated in AB solution [5% donkey serum, BSA (1 mg/ml), in PBS] for 1 hour at RT or overnight at 4°C. Secondary Abs (raised in donkey, 1:1000, Jackson ImmunoResearch) were applied in AB solution for 2 hours at RT. For imaging, coverslips were mounted with Fluoromount Aqueous Mounting Medium (F4680, Sigma-Aldrich). Sample images were acquired using an Olympus FV1000 confocal laser-scanning microscope at ×60 magnification with oil-immersion objective [Olympus UPLSAPO, (numerical aperture) NA 1.35]
Immunohistochemistry for cryo-sections
Tissues (DRG or sciatic nerve) were fixed in 4% PFA for 24 hours at 4°C and then washed in PBS, followed by incubation in 30% sucrose for 48 hours at 4°C. Tissues were embedded in O.C.T. Compound (Scigen, 4583), and 12- or 15-μm sections were obtained (cross sections for sciatic nerve). Blocking was performed in 10% horse serum and 0.2% Triton X-100 in PBS for 2 hours at RT. Primary Abs were incubated in AB solution (5% horse serum and 0.2% Triton X-100 in PBS) overnight at 4°C. Secondary Abs were applied in AB solution for 2 hours at RT. Slides were mounted in Fluoromount (F4680, Sigma-Aldrich), and confocal images were taken in the following days. Samples were imaged with a DMi8 Leica (Leica Microsystems, Mannheim, Germany) confocal laser-scanning microscope, using a HC PL APO 40×/1.3 oil-immersion objective and HyD SP GaAsP detectors. Imagestacks of 4 μm were collected for each sample, with 0.5-μm-thick optical sections. Images were acquired by maintaining a pixel size of 0.142 μm and an image dimension of 2048 by 2048 pixels.
Quantification of PTBP1 levels in DRG sections
For each mouse, L4 and L5 ganglia were quantified from both sides (left naive and right after SN crush injury), and 8 to 10 non-overlapping sections were imaged and used for analysis. Neuronal or glial nuclei were segmented with a machine learning–based pixel classification workflow implemented in Ilastik (57). PTBP1 levels were also evaluated using a customized protocol. For this analysis, neuronal cell bodies were segmented on the basis of TUJ1 or NFH staining. Only neurons showing a clear nuclear signal (DAPI) were selected for quantification. Measurements of average PTBP1 intensity were obtained using ImageJ/Fiji (58). DAPI was used to identify the nuclear compartment and was subtracted from the total cell body area to obtain the cytoplasmic compartment. For analysis based on cell size, the cell body area (square micrometers) was quantified, and neurons were divided in size bins: 0 to 200, 200 to 300, 300 to 500, and >500 μm2.
Quantification of PTBP1 levels in sciatic nerve sections
Per each mouse, eight images (each one containing hundreds of axons) were acquired from nonconsecutive sections, from naive or crushed nerve (1 mm proximal to the injury site). Axons were identified using different markers: TUJ1, NFH, or ChAT. Measurements of average PTBP1 intensity were obtained using ImageJ/Fiji, only the intensity of PTBP1 inside the axons was considered for the analysis. For analysis based on axon size, the axon area (square micrometers) was measured, and neurons were divided in size bins: 0 to 1, 1 to 2, 2 to 5, and 5 to 10 μm2. Clusters of small diameter axons were identified on the basis of TUJ1 signal and segmented with a machine learning–based pixel classification workflow implemented in Ilastik (57). The software was trained to identify clusters of small diameter axons based on morphology, pattern, and intensity of TUJ1 signal and ignore single axons or structures that differ from the above. PTBP1 levels were also examined in nuclei of non-neuronal cells based on DAPI positivity. A separate set of sections served as negative control, where we used a blocking peptide ab23105, specific for the PTBP1 Ab (ab5642), at a concentration of 0.011 μg/μl, corresponding to 200× molar excess compared to the primary Ab (0.002 μg/μl). The primary Ab, alone or together with the blocking peptide, was incubated in the staining solution for 2 hours at RT, before proceeding with the protocol.
Immunohistochemistry for paraffin sections
SNs were fixed in 4% PFA overnight at 4°C. The next day, nerve segments were dehydrated by increasing concentrations of ethanol, embedded in paraffin, and 5-μm cross sections were taken. The slides then underwent deparaffinization with xylene and ethanol, followed by antigen retrieval in tris-EDTA buffer [10 mM tris base, 1 mM EDTA, and 0.05% Tween 20 (pH 9.0)] using a pressure cooker at 125°C for 1 min with subsequent cooling to RT for 1 hour. After two washes in PBS, blocking was performed in 20% horse serum and 0.2% Triton in PBS for 1 hour at RT. Primary Abs (TUJ1, PTBP1, and PTBP2) were incubated in AB solution (2% horse serum and 0.2% Triton in PBS) overnight at RT. Secondary Abs (donkey, 1:500, Jackson ImmunoResearch) were applied in AB solution for 2 hours at RT. Slides were mounted in Fluoromount (F4680, Sigma-Aldrich), and confocal images were taken using an Olympus FV1000 Confocal laser-scanning microscope at ×60 magnification with oil-immersion objective (Olympus UPLSAPO, NA 1.35).
Whole tissue staining (iDISCO)
Whole tissue staining was performed as described (59) with minor adjustments. Tissue was fixed in 4% PFA o/n at 4°C and washed in PBS. Subsequently, samples were dehydrated in methanol/H2O series (20, 40, 60, 80. and 100 for 1 hour each), bleached in fresh 5% H2O2 in methanol o/n at 4°C and rehydrated the next day. After two washes in PTx.2 buffer (0.2% Triton X-100 in PBS), samples were permeabilized in PTx.2 containing glycine (23 mg/ml) and 20% (v/v) dimethyl sulfoxide (DMSO) for 2 days at RT. Blocking was performed in PTx.2 containing 6% donkey serum and 10% DMSO for 3 days at RT. Abs for PTBP1, PTBP2, and NFH were applied in in PTwH buffer [0.2% Tween 20 and heparin (0.01 mg/ml) in PBS] supplemented with 5% DMSO and 3% donkey serum for 3 days at RT. After five washes (each 30 min) in PTwH buffer, tissue samples were incubated with secondary Abs in PTwH with 3% donkey serum for 2 days at RT, followed 1 hour DAPI in PTwH and five washes in PTwH (30 min). Last, samples were transferred to 75% glycerol for 1 week before imaging. For imaging, an Olympus FV1000 Confocal laser-scanning microscope at ×60 magnification with oil-immersion objective (Olympus UPLSAPO, NA 1.35) was used.
Fluorescence in situ hybridization
Single-molecule FISH combined with IF was used to detect and quantitate RHOA and KPNB1 mRNA and PTBP1 protein in mouse sciatic nerve sections as described (60). 5′ Labeled Stellaris probes were designed against mouse RHOA and KPNB1 mRNA. SN segments were fixed overnight in 2% PFA at 4°C and then cryoprotected overnight in 30% sucrose at 4°C. Cryosections (25 μm thick) were prepared and stored at −20°C until use. Slides were dried at 37°C for 1 hour and then brought to RT; all subsequent steps were performed at RT unless indicated otherwise. Sections were washed 10 min in PBS once and then 10 min in 20 mM glycine three times followed by 5 min in fresh 0.25 M NaBH4 three times. After a 0.1 M triethanolamine (TEA) rinse, sections were incubated in 0.25% acetic anhydride in 0.1 M TEA for 10 min, followed by two washes in 2× saline-sodium citrate (SSC) buffer and dehydration in in graded ethanol solutions (70, 95, and 100% 3 min each). Sections were then delipidated in chloroform for 5 min, rehydrated in graded ethanol (100 and 95% 3 min each), and equilibrated in 2× SSC. Samples were then incubated at 37°C for 5 min in a humidified chamber in hybridization buffer [10% dextran sulfate, Escherichia coli transfer RNA (1 mg/ml), 2 mM vanadyl ribonucleosides, BSA (200 μg/ml), 2× SSC, 10% formamide, Roche Blocking buffer] followed by overnight in hybridization buffer with probes (7 μM each), goat anti-PTBP1 (1:100; Abcam, ab5642), and RT97 mouse anti-NF (1:100; Dev Hybrid Studies Bank). Sections were washed twice in 2× SSC + 10% formamide at 37°C for 30 min and once in 2× SSC for 5 min. Tissues were permeabilized in PBS + 1% Triton X-100 for 5 min and then incubated for 1 hour in either Cy3- or Cy5-conjugated donkey anti-goat and fluorescein isothiocyanate–conjugated donkey anti-mouse Abs (1:200 for each; Jackson ImmunoResearch) in 1× blocking buffer (Roche) plus 0.3% Triton X-100. After washing with PBS for 5 min, sections were postfixed in buffered 2% PFA for 15 min, washed in PBS three times for 5 min, rinsed in diethyl pyrocarbonate–treated water, and mounted using Prolong Gold Antifade.
For analyses of FISH signals, images were acquired using a Leica SP8X confocal microscope with HyD detectors and Lightening deconvolution. Scramble (SCRM) probe signals were used to set the image acquisition parameters such that all acquisitions were set at the scramble probe parameters that generated least signals. Xyz image stacks were obtained using 63× oil-immersion objective (1.4 NA) at two random locations along each nerve section. NIH ImageJ colocalization plug-in (https://imagej.nih.gov/ij/plugins/colocalization.html) was used to extract RNA signals in each optical plane that overlapped with neurofilament. Quantification of this “axon only” mRNA signal was done by analysis of pixel intensity across each xy plane of the extracted axon only channels using ImageJ. FISH signal intensity was normalized as pixels/μm2 of NF signal within each xy plane. The average of FISH signal intensity to NF immunoreactivity in each xy plane was averaged across the image z stack of tile image. The relative mRNA signal intensity was averaged for all tiles in each biological replicate. Using NIH ImageJ plugin JACoP (https://imagej.nih.gov/ij/plugins/track/jacop.html), Pearson’s coefficient for colocalization of axon-only PTBP1 protein with either RHOA or KPNB1 mRNA was generated.
Live imaging of axonal PTBP1 and RhoA mRNA
Molecular beacon design
The MB sequence was manually designed to bind to a predicted single-stranded region on mouse RhoA mRNA with favorable MB: mRNA stability, using two secondary structure-prediction algorithms: m-Fold (61) and OLIGOWALK (62). GC was added to each end to ensure loop formation. The sequence was confirmed to be unique by NCBI BLAST analysis (https://blast.ncbi.nlm.nih.gov/Blast.cgi), confirming unannotated gene sequences by direct comparison with RhoA mRNA. The MB was designed as a locked nucleic acid (+N)/2′-O-methyl RNA (mN) chimera (24) with a GC-rich stem, with 5′TYE665 fluorophore, and a 3′ Iowa Black RQ-Sp quencher and was synthesized by Integrated DNA Technologies. Sequences (5′ to 3′) are as follows: RhoA MB: /5TYE665/mGmC mUmGmG mUmUmU + CmUmA mC + CmU mCmA + C mUmCmU mCmUmC mCmAmG mC/3IAbRQSp/; Scramble TYE665: /5TYE665/mCmC mGmCmG mGmCmG mGmA + A mAmCmU mU + AmU mA + CmA + CmUmU mAmAmC mGmCmC mGmCmG mG/3IAbRQSp/.
DRG culture and MB transfection
For primary DRG culture, DRGs were isolated from 2 to 3 months old C57BL/6J mice and then dissociated as described previously (30). After centrifugation and washing in Dulbecco’s modified Eagle’s medium (DMEM)/F12 (Life Technologies), cells were resuspended in 50 μl of growth media containing DMEM/F12, 1× N1 supplement (Sigma-Aldrich), 10% FBS (Hyclone), and 10 μM cytosine arabinoside (Sigma-Aldrich). For each four DRGs, 1 μl of AAV-PhP.s-GFP-PTBP1 (titer 7 × 1010/μl) virus was added, resuspended, and incubated in ice for 30 min. Virus-transduced dissociated DRGs were then plated immediately on laminin/poly-l-lysine–coated live cell imaging dishes. DRGs were cultured for 7 days in vitro (DIV). MB transfection was performed on the seventh day of DRG culture; 5 nM of RhoA or Scrambled MB was transfected using DharmaFECT 3 reagent (Dharmacon), and cells were then incubated for 24 hours before imaging. Growth medium was changed to no-phenol red containing DMEM/F12 (Life Technologies) 1 hour before starting live cell imaging.
Live cell imaging
Time lapses of RhoA-TYE665/ PTBP1 or scramble-TYE665/ PTBP1 for the cotrafficking analysis consisted of 200 consecutive frames, for a total time of 20 min with a 6.01-s delay between two consecutive frames.
Live cell imaging quantification
Kymographs were generated from the acquired movies using ImageJ. The “KymoReslicedWide” FIJI/ImageJ plugin was used to generate kymographs from time-lapse movies. The Multi Kymograph plugin for FIJI/ImageJ (https://biii.eu/multi-kymograph) was used to extract particle velocity and spatial directionality from the tracked traces. Particle velocities were calculated by considering the average speed of their segmental components.
Colocalization analyses
Colocalization analysis was performed using ImageJ. An axon length of 50 to 100 μm was analyzed for colocalization of RhoA-TYE665/ PTBP1 or scramble-TYE665/ PTBP1. The number of colocalized puncta was analyzed using JACoP plugin (https://imagej.net/plugins/jacop).
RNA IP and RNA sequencing
Mice were subjected to sciatic nerve crush and allowed to recover for 7 days before axoplasm collection from injured (inj) and naive (na) nerves, using 20 animals per biological replicate. Before splitting the samples to the experimental conditions (na PTBP1-IP, na Ctrl, inj PTBP1-IP, and inj Ctrl), 10% of naive and injury extract was taken as input sample. IP was performed as described previously (55). RNA from input and IP samples was extracted using the RNeasy Micro Kit (Qiagen, catalog no. 74004) with on-column deoxyribonuclease treatment according to the manufacturer’s instructions. We performed WB on supernatants to validate successful PTBP1-IP (depletion of PTBP1 in supernatants of IP samples, no depletion in Ctrl; extended data; Fig. 3A).
RNA sequencing was performed for three biological replicates, with three experimental conditions (input, PTBP1-IP, Ctrl) and two time points: naive and 7 days after SN crush injury. RNA sequencing libraries were prepared using the Nugen Ovation RNA Ultra Low Input (500 ng) with TrueSeq Nano kit (Illumina). Libraries were indexed and sequenced by HiSeq4000 with 50-bp paired-end reads, and at least 59 M reads (average, 74.5 M) were obtained for each sample.
Quality control was performed on base qualities and nucleotide composition of sequences, mismatch rate, mapping rate to the whole genome, repeats, chromosomes, key transcriptomic regions (exons, introns, UTRs, genes), insert sizes, AT/GC dropout, transcript coverage, and GC bias to identify problems in library preparation or sequencing. Reads were aligned to the mouse mm10 reference genome (GRCm38.75) using the STAR spliced read aligner (ver. 2.4.0). Average percentage of uniquely aligned reads was 66.3%. Total counts of read fragments aligned to known gene regions within the mouse (mm10) ensembl (GRCm38.p6) transcript reference annotation were used as the basis for quantification of gene expression. Fragment counts were derived using HTSeq program (ver. 0.6.0). Genes with minimum of 5 counts for at least one condition (all replicates) were selected, and differentially expressed transcripts were determined by Bioconductor package EdgeR (ver. 3.14.0). Scripts used in the RNA sequencing analyses are available at https://doi.org/10.5281/zenodo.8006804. RNA sequencing data are available under Gene Expression Omnibus (GEO) no. GSE142576.
Motif enrichment analysis (RNA sequence motifs) was performed using HOMER (63) (v4.11, 10-24-2019); http://homer.ucsd.edu/homer/motif/rnaMotifs.html. The 3′UTR sequences of the significantly enriched mRNA transcripts [PTBP1 IP versus Ctrl: log fold change (FC) > 0, FDR < 0.1] were used as target sequences and compared to a custom background set (not significantly enriched genes for each condition: PTBP1 IP versus Ctrl: logFC < 0, FDR > 0.1). Canonical pathway analysis was performed using QIAGEN Ingenuity Pathway Analysis (https://digitalinsights.qiagen.com/IPA) (64). A complete list of identified transcripts and IPA analysis can be found in data S2.
shRNA-mediated knockdown of PTBP1
The design of the shRNA constructs was based on AAV-shRNA-ctrl (Addgene, plasmid #85741) (65). shRNA expression is driven from a U6 promoter and the plasmid also contains an EYFP reporter. AAV serotype 9/PhP.S (66) were chosen due to their preference for peripheral neurons in adult mice. shCtrl: (5′-GTTCAGATGTGCGGCGAGTGAAGCTTGACTCGCCGCACATCTGAAC-3′); shPTBP1 ORF: (5′-GGGAAGTCAGTGCGCATTAGAAGCTTGTAATGCGCACTGACTTCCC-3′); and shPTBP1 UTR: (5′-AGCCGCTTTCTGTGCCTTAGAAGCTTGTAAGGCACAGAAAGCGGCT-3′) shRNA sequences were subcloned using Bam HI and Xba I restriction sites.
Knockdown was validated by transduction of DRG neurons in vivo followed by immunostaining analyses (fig. S6, A to C). Cells were fixed in 4% PFA, stained for PTBP1 or PTBP2, and quantified using ImageJ/Fiji.
AAV-mediated expression of PTBP1wt or PTBP1chimera
pcDNA3.1 Flag-PTBP1 and PTBP2 constructs are based on Ensembl IDs ENSRNOT00000044865.7 (PTBP1-001, rat) and ENSRNOT00000015036.7 (PTBP2, rat). All the different PTBP1 chimeras were generated in house with restriction free cloning (67). Each domain of PTBP1 was replaced with the corresponding fragment of PTBP2 and tested for axonal localization. Both single and multiple domains were replaced. The main chimeras of interest are shown in fig. S9.
Chimera screen for localization
P0 rat hippocampal neurons were cultured as previously described (68). After 4 days in culture, cells were transfected with 2 μg of FLAG-PTBP1, FLAG-PTBP2 or FLAG-chimera plasmids using calcium phosphate. Cells were fixed after 48 hours, followed by staining for FLAG, TUJ1, and DAPI. Quantification of FLAG staining in neurites is based on overlap with TUJ1 (excluding DAPI-positive regions) using CellProfiler. The most interesting chimera (PTBP1 R1-L1-R2) was tested for axonal localization in vitro in DRG neurons and for the behavioral rescue experiments.
AAV production
Low-passage human embryonic kidney (HEK) 293T cells were maintained at 37°C with 5% CO2 in DMEM supplemented with 10% FBS. To produce rAAV-Php.S, a triple cotransfection procedure was used to introduce rAAV vector plasmids together with pPhp.S and pXX6-80, at a 1:1:1 molar ratio (66, 69). HEK293T cells were transfected using poly-ethylenimine (linear; MW, 25,000) (Polysciences Inc., Warrington, PA), and medium was replaced at 18 hours posttransfection. Cells were harvested at 72 hours posttransfection, subjected to three rounds of freeze-thawing, and then digested with benzonase (100 U/ml; EMD Millipore, Billerica, MA) at 37°C for 1 hour. Viral vectors were purified by iodixanol (Sigma-Aldrich, Israel) gradient ultracentrifugation (70), followed by further concentration using Amicon ultra-15 100K (100,000-MW cutoff; Merck Millipore, Ireland) and washed with PBS.
Purified AAV9 was produced in HEK 293T cells (Takara Bio, #632273), with the AAVpro Purification Kit (All Serotypes) from TaKaRa (Takara Bio, #6666). For each construct, 10 plates (15 cm) were transfected with 20 μg of DNA (AAV-plasmid containing the construct of interest and two AAV9 helper plasmids) using jetPEI (Polyplus) in DMEM medium without serum or antibiotics. pAAV2/9n and pAdDeltaF6 helper vectors were obtained from the University of Pennsylvania Vector Core. Medium [DMEM, 20% FBS, 1 mM sodium pyruvate, penicillin (100 U/ml), and streptomycin (100 mg/ml)] was added on the following day to a final concentration of 10% FBS, and extraction was done 3 days posttransfection. Purification was performed according to the manufacturer’s instructions (Takara Bio, #6666). For both constructs, we obtained titers of ~5 × 1014 viral genomes/ml, which were used undiluted for intrathecal injections (5 μl per animal) or transduction in culture (1 μl per well).
Knockdown and rescue in vivo
To knockdown PTBP1 in DRG neurons in vivo, we delivered shCtrl, shPTBP1 ORF, or shPTBP1 UTR via intrathecal injection of AAV9/PhP.S vectors. To rescue PTBP1 in DRG neurons in vivo, we delivered an equimolar mixture of shPTBP1 UTR and PTBP1wt or shPTBP1 UTR and PTBP1chimera via intrathecal injection of AAV9/PhP.S vectors. A total of 5 μl (~5 × 1014 viral genomes/ml) was injected using a sterile 10-μl Hamilton micro syringe fitted with a 30-gauge needle. Baseline behavior was assessed 3 weeks after injection. Mice were subjected to behavioral testing (CatWalk, hot plate, von Frey). Subsequently, we performed sciatic nerve crush injury on the right hindleg, the left side was used as an uninjured, naive control. At distinct time points after nerve crush, mice were subjected to behavioral testing (CatWalk, heat probe, von Frey).
Conditioning lesion culture
For growth assays after conditioning lesion (71), L4 and L5 DRG were extracted separately from both sides (left side: naive; right side: 7 days after sciatic nerve crush) from both experimental groups. Cells were allowed to grow for 20 hours after plating. Imaging and analysis were performed using ImageXpress Micro (Molecular Devices) automated microscopy system with MetaXpress analysis software (Molecular Devices, version 5.1). Cells, with the longest neurite > 2× the diameter of the cell body, were considered as “growing”; and only EYFP-positive neurons were included in the outgrowth analysis.
SN longitudinal sections
Tissues were fixed in 4% PFA for 24 hours at 4°C and then washed in PBS, followed by incubation in 30% sucrose for 48 hours at 4°C. Tissues were embedded in O.C.T. Compound (Scigen, 4583), and 15-μm sections were obtained. Blocking was performed in 10% horse serum and 0.2% Triton X-100 in PBS for 2 hours at RT. Primary Abs were incubated in AB solution (5% horse serum and 0.2% Triton X-100 in PBS) overnight at 4°C. Secondary Abs were applied in AB solution for 2 hours at RT. Slides were mounted in Fluoromount (F4680, Sigma-Aldrich). Samples were acquired using ImageXpress Micro (Molecular Devices) automated microscopy system equipped with a 20× objective, with MetaXpress analysis software (Molecular Devices, version 5.1). For the regeneration analysis, the number of double positive axons (EYFP and CGRP or SCG-10) reaching 1 mm distal from the injury site was counted.
RhoA translation
To monitor RHOA spatial translation, DRG neurons were cultured for 7 DIV and then replated on a glass coverslip for imaging as previously described (11). After 24 hours, the neurons were incubated for 2 hours with anisomycin (200 μg/ml; Sigma-Aldrich, A9789) + 20′ puromycin (100 μg/ml; Sigma-Aldrich, P8833) for background signal control or for 20′ with puromycin. Cells were then fixed with 4% PFA and stained with the standard staining protocol (as described previously in the “Immunocytochemistry” section) with RhoA and puromycin Ab. PLA (72) between RhoA and Puromycin was performed according to the manufacturer’s instructions using Duolink (Sigma-Aldrich; PLA probe anti-mouse minus DUO92004, anti-rabbit plus DUO92002, and detection kit red DUO92008). Cells were acquired using an Olympus FV1000 Confocal laser-scanning microscope at ×60 magnification with oil-immersion objective (Olympus UPLSAPO, NA 1.35). The PLA signal was quantified with an in-house script with ImageJ software, using a mask based on intensity of EYFP staining and normalizing the number of PLA-positive puncta by the EYFP-positive area in the cell body or in the axons.
PTBP1 inhibition via decoy RNA oligo
Inhibitory decoy RNA oligonucleotides for PTBP1 (PTBP1i) and scramble control (SCRMi) were as previously described (25). These oligonucleotides are single-stranded RNA molecules of three/four tandem motif repeats with a 2′-O-methyl modification on the ribose of each nucleotide. To increase stability for in vivo experiments, we modified the oligos with a phosphorothioate backbone and also added a fluorophore (TYE665) to the 5′ end to allow for imaging. Oligos were synthesized by Integrated DNA Technologies (Syntezza, Israel). SCRMi: 5′TYE665-GCAAUCCGCAAUCCGCAAUCC-3′; PTBP1i: 5′TYE665-CUCUCUCUCUCUCUCUCUCUCUCU-3′.
Decoy RNA oligos were used to locally inhibit PTBP1 in the SN and to verify its role after SN crush injury. One set of mice was injected with 865 ng of PTBP1i or SCRMi oligo (in a volume of 2 μl) directly into the SN using a sterile 10-μl Hamilton micro syringe fitted with a 34-gauge needle. Immediately after, we crushed the sciatic nerve in the same position. The other side served as a naive control. Mice were subjected to behavioral testing (CatWalk, heat probe, von Frey) in a similar manner as in the knockdown experiment.
A separate set of mice was injected for FISH analysis of PTBP1 protein colocalization with RHOA or KPNB1 mRNA. In this case, each mouse was injected bilaterally with either PTBP1i or SCRMi oligo (865 ng for each nerve as above). Only one side of the animal was subjected to the crush injury. One day or 7 days after the injection, sciatic nerves from both sides were isolated and processed for longitudinal cryosections and RNA-FISH as described above. Since treatment of the samples according to the FISH protocol leads to quenching of the fluorescent signal (TYE665) of the decoy RNA oligos, a separate set of sections were imaged to show successful injection into the SN (fig. S8).
Statistical analyses
All analyses were performed using GraphPad Prism version 8.43 for Windows (GraphPad Software, La Jolla, CA, USA; www.graphpad.com). Statistically significant P values are shown as *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001. Outliers were discarded using the ROUT method with a Q (maximum desired false discovery rate) of 1%, as detailed in the figure legends. All data underwent normality testing (Anderson-Darling, D’Agostino-Pearson omnibus, Shapiro-Wilk, and Kolmogorov-Smirnov). Normally distributed data were subjected to parametric analysis. Two-tailed, Student’s t test was used for analysis with two groups, ordinary one-way analysis of variance (ANOVA) was used to compare multiple groups. Two-way ANOVA or mixed-effects analysis was used to compare two variables (PTBP1 perturbation and SN injury). Paired t test or repeated measure (RM) analysis was done on data coming from the same animal and is indicated in the corresponding figure legends. Unpaired analysis was performed in all other cases (independent samples). Tukey’s, Dunnett’s, or Sidak’s multiple comparisons tests were used in the follow-up analyses as specified in the figure legends. Datasets that did not pass the normality test were subjected to nonparametric analysis using Mann-Whitney test for two groups or Kruskal-Wallis test for multiple group evaluation, followed by Dunn’s multiple comparisons test. The results are shown displaying individual values whenever possible, together with mean ± SEM. Individual values (n) correspond to the number of mice or cells or sections used in the experiment. All statistical parameters for each analysis are stated in the corresponding figure legends.
Acknowledgments
We are thankful to V. Kiss and M. Mahn for help and advice on imaging and analysis, M. Tsoory for guidance with behavioral assays, D. Gordon and R. Perry for helpful discussions, and G. Lambiase for the help in the tissue collection and processing. We thank R. Levy for help with preparing the hippocampal neuron cultures and O. Singer and the viral core facility of the Weizmann Institute for the AAV production.
Funding: We gratefully acknowledge funding from the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (A.L.B., G.C., J.L.T., and M.F.), the International Foundation for Research in Paraplegia (IRP grant no. P177 to M.F.), the Minerva Foundation (M.F.), the Weizmann Center for Research on Neurodegeneration (M.F.), the Swiss Society Center for Research on Perception and Action at the Weizmann Institute of Science (M.F.), the National Institutes of Health (R01-NS117821 to J.L.T., P41GM103481, and 1S10OD016229 to A.L.B.), and the South Carolina Spinal Cord Injury Research Fund (2019-PD-02 to P.K.S.). M.F. is the incumbent of the Chaya Professorial Chair in Molecular Neuroscience, and J.L.T. is the incumbent of the Smartstate Chair in Childhood Neurotherapeutics.
Author contributions: S.A., P.D.M., and M.F. designed the study; S.A., P.D.M., A.D.P., N.P., P.F., L.M., E.D.-M., N.O., S.J.L., and P.K.S. conducted experiments; M.D.Z., I.D.C., and P.K.S. carried out FISH and MB analyses; R.K. performed RNA sequencing; K.F.M. performed MS; R.N. conducted imaging analyses; I.R., A.L.B., G.C., J.L.T., and M.F. supervised research; S.A., P.D.M., and M.F. wrote the manuscript draft with input from all the co-authors.
Competing interests: S.A. is currently an employee of F. Hoffmann-La Roche Ltd. (Basel, Switzerland). G.C. is currently an employee of Regeneron Pharmaceuticals (Tarrytown, NY). He receives his salary from and owns options and stock in Regeneron Pharmaceuticals. S.J.L. is currently an employee of Biogen (Cambridge, MA). The other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Details for all proteins identified by MS are provided in data S1 and are further available under accession number MSV000092006 in MassIVE (https://massive.ucsd.edu). RNA sequencing data are available under accession number GSE142576 in NCBI’s GEO (https://ncbi.nlm.nih.gov/geo/) and are also provided in data S2. Scripts used in the RNA sequencing analyses are available at https://doi.org/10.5281/zenodo.8006804.
Supplementary Materials
This PDF file includes:
Figs. S1 to S10
Table S1
Legends for data S1 and S2
Other Supplementary Material for this manuscript includes the following:
Data S1 and S2
REFERENCES AND NOTES
- 1.M. Mahar, V. Cavalli, Intrinsic mechanisms of neuronal axon regeneration. Nat. Rev. Neurosci. 19, 323–337 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.I. Rishal, M. Fainzilber, Axon-soma communication in neuronal injury. Nat. Rev. Neurosci. 15, 32–42 (2014). [DOI] [PubMed] [Google Scholar]
- 3.I. Dalla Costa, C. N. Buchanan, M. D. Zdradzinski, P. K. Sahoo, T. P. Smith, E. Thames, A. N. Kar, J. L. Twiss, The functional organization of axonal mRNA transport and translation. Nat. Rev. Neurosci. 22, 77–91 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.B. Turner-Bridger, C. Caterino, J. M. Cioni, Molecular mechanisms behind mRNA localization in axons. Open Biol. 10, 200177 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.E. Doron-Mandel, I. Koppel, O. Abraham, I. Rishal, T. P. Smith, C. N. Buchanan, P. K. Sahoo, J. Kadlec, J. A. Oses-Prieto, R. Kawaguchi, S. Alber, E. E. Zahavi, P. Di Matteo, A. Di Pizio, D. A. Song, N. Okladnikov, D. Gordon, S. Ben-Dor, R. Haffner-Krausz, G. Coppola, A. L. Burlingame, P. Jungwirth, J. L. Twiss, M. Fainzilber, The glycine arginine-rich domain of the RNA-binding protein nucleolin regulates its subcellular localization. EMBO J. 40, e107158 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.S. J. Lee, J. A. Oses-Prieto, R. Kawaguchi, P. K. Sahoo, A. N. Kar, M. Rozenbaum, D. Oliver, S. Chand, H. Ji, M. Shtutman, S. Miller-Randolph, R. J. Taylor, M. Fainzilber, G. Coppola, A. L. Burlingame, J. L. Twiss, hnRNPs interacting with mRNA localization motifs define axonal RNA regulons. Mol. Cell. Proteomics 17, 2091–2106 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.J. K. Nussbacher, R. Tabet, G. W. Yeo, C. Lagier-Tourenne, Disruption of RNA metabolism in neurological diseases and emerging therapeutic interventions. Neuron 102, 294–320 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.K. E. Cosker, S. J. Fenstermacher, M. F. Pazyra-Murphy, H. L. Elliott, R. A. Segal, The RNA-binding protein SFPQ orchestrates an RNA regulon to promote axon viability. Nat. Neurosci. 19, 690–696 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.S. Hanz, E. Perlson, D. Willis, J.-Q. Zheng, R. Massarwa, J. J. Huerta, M. Koltzenburg, M. Kohler, J. van-Minnen, J. L. Twiss, M. Fainzilber, Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron 40, 1095–1104 (2003). [DOI] [PubMed] [Google Scholar]
- 10.R. B. Perry, E. Doron-Mandel, E. Iavnilovitch, I. Rishal, S. Y. Dagan, M. Tsoory, G. Coppola, M. K. McDonald, C. Gomes, D. H. Geschwind, J. L. Twiss, A. Yaron, M. Fainzilber, Subcellular knockout of importin β1 perturbs axonal retrograde signaling. Neuron 75, 294–305 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.R. B. Perry, I. Rishal, E. Doron-Mandel, A. L. Kalinski, K. F. Medzihradszky, M. Terenzio, S. Alber, S. Koley, A. Lin, M. Rozenbaum, D. Yudin, P. K. Sahoo, C. Gomes, V. Shinder, W. Geraisy, E. A. Huebner, C. J. Woolf, A. Yaron, A. L. Burlingame, J. L. Twiss, M. Fainzilber, Nucleolin-mediated RNA localization regulates neuron growth and cycling cell size. Cell Rep. 16, 1664–1676 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.N. Keppetipola, S. Sharma, Q. Li, D. L. Black, Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit. Rev. Biochem. Mol. Biol. 47, 360–378 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.J. K. Vuong, C. H. Lin, M. Zhang, L. Chen, D. L. Black, S. Zheng, PTBP1 and PTBP2 serve both specific and redundant functions in neuronal pre-mRNA splicing. Cell Rep. 17, 2766–2775 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.E. V. Makeyev, J. Zhang, M. A. Carrasco, T. Maniatis, The MicroRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435–448 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.I. Rishal, I. Michaelevski, M. Rozenbaum, V. Shinder, K. F. Medzihradszky, A. L. Burlingame, M. Fainzilber, Axoplasm isolation from peripheral nerve. Dev. Neurobiol. 70, 126–133 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.K. Lillevali, A. Kulla, T. Ord, Comparative expression analysis of the genes encoding polypyrimidine tract binding protein (PTB) and its neural homologue (brPTB) in prenatal and postnatal mouse brain. Mech. Dev. 101, 217–220 (2001). [DOI] [PubMed] [Google Scholar]
- 17.J. Hu, H. Qian, Y. Xue, X.-D. Fu, PTB/nPTB: Master regulators of neuronal fate in mammals. Biophys. Rep. 4, 204–214 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.F. M. Hamid, E. V. Makeyev, Regulation of mRNA abundance by polypyrimidine tract-binding protein-controlled alternate 5′ splice site choice. PLOS Genet. 10, e1004771 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.T. Kalpachidou, L. Spiecker, M. Kress, S. Quarta, Rho GTPases in the physiology and pathophysiology of peripheral sensory neurons. Cell 8, 591 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.S. Meltzer, C. Santiago, N. Sharma, D. D. Ginty, The cellular and molecular basis of somatosensory neuron development. Neuron 109, 3736–3757 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.C. C. Toth, D. Willis, J. L. Twiss, S. Walsh, J. A. Martinez, W. Q. Liu, R. Midha, D. W. Zochodne, Locally synthesized calcitonin gene-related peptide has a critical role in peripheral nerve regeneration. J. Neuropathol. Exp. Neurol. 68, 326–337 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.J. E. Shin, S. Geisler, A. DiAntonio, Dynamic regulation of SCG10 in regenerating axons after injury. Exp. Neurol. 252, 1–11 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.S. tom Dieck, L. Kochen, C. Hanus, M. Heumuller, I. Bartnik, B. Nassim-Assir, K. Merk, T. Mosler, S. Garg, S. Bunse, D. A. Tirrell, E. M. Schuman, Direct visualization of newly synthesized target proteins in situ. Nat. Methods 12, 411–414 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.I. E. Catrina, S. A. Marras, D. P. Bratu, Tiny molecular beacons: LNA/2'-O-methyl RNA chimeric probes for imaging dynamic mRNA processes in living cells. ACS Chem. Biol. 7, 1586–1595 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.P. Denichenko, M. Mogilevsky, A. Clery, T. Welte, J. Biran, O. Shimshon, G. D. Barnabas, M. Danan-Gotthold, S. Kumar, E. Yavin, E. Y. Levanon, F. H. Allain, T. Geiger, G. Levkowitz, R. Karni, Specific inhibition of splicing factor activity by decoy RNA oligonucleotides. Nat. Commun. 10, 1590 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.R. V. Kamath, D. J. Leary, S. Huang, Nucleocytoplasmic shuttling of polypyrimidine tract-binding protein is uncoupled from RNA export. Mol. Biol. Cell 12, 3808–3820 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.S. M. Weyn-Vanhentenryck, H. Feng, D. Ustianenko, R. Duffie, Q. Yan, M. Jacko, J. C. Martinez, M. Goodwin, X. Zhang, U. Hengst, S. Lomvardas, M. S. Swanson, C. Zhang, Precise temporal regulation of alternative splicing during neural development. Nat. Commun. 9, 2189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.I. Rishal, N. Kam, R. B. Perry, V. Shinder, E. M. Fisher, G. Schiavo, M. Fainzilber, A motor-driven mechanism for cell-length sensing. Cell Rep. 1, 608–616 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.M. Terenzio, S. Koley, N. Samra, I. Rishal, Q. Zhao, P. K. Sahoo, A. Urisman, L. Marvaldi, J. A. Oses-Prieto, C. Forester, C. Gomes, A. L. Kalinski, A. Di Pizio, E. Doron-Mandel, R. B. Perry, I. Koppel, J. L. Twiss, A. L. Burlingame, M. Fainzilber, Locally translated mTOR controls axonal local translation in nerve injury. Science 359, 1416–1421 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.P. K. Sahoo, A. N. Kar, N. Samra, M. Terenzio, P. Patel, S. J. Lee, S. Miller, E. Thames, B. Jones, R. Kawaguchi, G. Coppola, M. Fainzilber, J. L. Twiss, A Ca2+-dependent switch activates axonal casein kinase 2α translation and drives G3BP1 granule disassembly for axon regeneration. Curr. Biol. 30, 4882–4895.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.I. Babic, S. Sharma, D. L. Black, A role for polypyrimidine tract binding protein in the establishment of focal adhesions. Mol. Cell. Biol. 29, 5564–5577 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.S. Stern, B. J. Hilton, E. R. Burnside, S. Dupraz, E. E. Handley, J. M. Gonyer, C. Brakebusch, F. Bradke, RhoA drives actin compaction to restrict axon regeneration and astrocyte reactivity after CNS injury. Neuron 109, 3436–3455.e9 (2021). [DOI] [PubMed] [Google Scholar]
- 33.M. A. Hintermayer, S. S. Drake, A. E. Fournier, To grow and to stay, both controlled by RhoA: Opposing cellular effects on axon regeneration. Neuron 109, 3395–3397 (2021). [DOI] [PubMed] [Google Scholar]
- 34.K. Y. Wu, U. Hengst, L. J. Cox, E. Z. Macosko, A. Jeromin, E. R. Urquhart, S. R. Jaffrey, Local translation of RhoA regulates growth cone collapse. Nature 436, 1020–1024 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.B. A. Walker, S. J. Ji, S. R. Jaffrey, Intra-axonal translation of RhoA promotes axon growth inhibition by CSPG. J. Neurosci. 32, 14442–14447 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.A. Deglincerti, Y. Liu, D. Colak, U. Hengst, G. Xu, S. R. Jaffrey, Coupled local translation and degradation regulate growth cone collapse. Nat. Commun. 6, 6888 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.J. W. Fawcett, The struggle to make CNS axons regenerate: Why has it been so difficult? Neurochem. Res. 45, 144–158 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.S. Galban, Y. Kuwano, R. Pullmann Jr., J. L. Martindale, H. H. Kim, A. Lal, K. Abdelmohsen, X. Yang, Y. Dang, J. O. Liu, S. M. Lewis, M. Holcik, M. Gorospe, RNA-binding proteins HuR and PTB promote the translation of hypoxia-inducible factor 1alpha. Mol. Cell. Biol. 28, 93–107 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.F. Besse, S. Lopez de Quinto, V. Marchand, A. Trucco, A. Ephrussi, Drosophila PTB promotes formation of high-order RNP particles and represses oskar translation. Genes Dev. 23, 195–207 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Y. Xue, K. Ouyang, J. Huang, Y. Zhou, H. Ouyang, H. Li, G. Wang, Q. Wu, C. Wei, Y. Bi, L. Jiang, Z. Cai, H. Sun, K. Zhang, Y. Zhang, J. Chen, X. D. Fu, Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 152, 82–96 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Y. Xue, H. Qian, J. Hu, B. Zhou, Y. Zhou, X. Hu, A. Karakhanyan, Z. Pang, X. D. Fu, Sequential regulatory loops as key gatekeepers for neuronal reprogramming in human cells. Nat. Neurosci. 19, 807–815 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.K. H. Yeom, S. Mitchell, A. J. Linares, S. Zheng, C. H. Lin, X. J. Wang, A. Hoffmann, D. L. Black, Polypyrimidine tract-binding protein blocks miRNA-124 biogenesis to enforce its neuronal-specific expression in the mouse. Proc. Natl. Acad. Sci. U.S.A. 115, E11061–E11070 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.H. Zhou, J. Su, X. Hu, C. Zhou, H. Li, Z. Chen, Q. Xiao, B. Wang, W. Wu, Y. Sun, Y. Zhou, C. Tang, F. Liu, L. Wang, C. Feng, M. Liu, S. Li, Y. Zhang, H. Xu, H. Yao, L. Shi, H. Yang, Glia-to-neuron conversion by CRISPR-CasRx alleviates symptoms of neurological disease in mice. Cell 181, 590–603.e16 (2020). [DOI] [PubMed] [Google Scholar]
- 44.H. Qian, X. Kang, J. Hu, D. Zhang, Z. Liang, F. Meng, X. Zhang, Y. Xue, R. Maimon, S. F. Dowdy, N. K. Devaraj, Z. Zhou, W. C. Mobley, D. W. Cleveland, X. D. Fu, Reversing a model of Parkinson's disease with in situ converted nigral neurons. Nature 582, 550–556 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.R. Maimon, C. Chillon-Marinas, C. E. Snethlage, S. M. Singhal, M. McAlonis-Downes, K. Ling, F. Rigo, C. F. Bennett, S. Da Cruz, T. S. Hnasko, A. R. Muotri, D. W. Cleveland, Therapeutically viable generation of neurons with antisense oligonucleotide suppression of PTB. Nat. Neurosci. 24, 1089–1099 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.L. L. Wang, C. Serrano, X. Zhong, S. Ma, Y. Zou, C. L. Zhang, Revisiting astrocyte to neuron conversion with lineage tracing in vivo. Cell 184, 5465–5481.e16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.W. Chen, Q. Zheng, Q. Huang, S. Ma, M. Li, Repressing PTBP1 fails to convert reactive astrocytes to dopaminergic neurons in a 6-hydroxydopamine mouse model of Parkinson's disease. eLife 11, e75636 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.T. Hoang, D. W. Kim, H. Appel, N. A. Pannullo, P. Leavey, M. Ozawa, S. Zheng, M. Yu, N. S. Peachey, S. Blackshaw, Genetic loss of function of Ptbp1 does not induce glia-to-neuron conversion in retina. Cell Rep. 39, 110849 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Y. Xie, J. Zhou, B. Chen, Critical examination of Ptbp1-mediated glia-to-neuron conversion in the mouse retina. Cell Rep. 39, 110960 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.T. Guo, X. Pan, G. Jiang, D. Zhang, J. Qi, L. Shao, Z. Wang, H. Xu, Y. Zhao, Downregulating PTBP1 fails to convert astrocytes into hippocampal neurons and to alleviate symptoms in Alzheimer's mouse models. J. Neurosci. 42, 7309–7317 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.L. L. Wang, C. L. Zhang, In vivo glia-to-neuron conversion: Pitfalls and solutions. Dev. Neurobiol. 82, 367–374 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.D. J. Langford, J. S. Mogil, Pain testing in the laboratory mouse. Am. Coll. Lab., 549–560 (2008). [Google Scholar]
- 53.L. Marvaldi, N. Panayotis, S. Alber, S. Y. Dagan, N. Okladnikov, I. Koppel, A. Di Pizio, D. A. Song, Y. Tzur, M. Terenzio, I. Rishal, D. Gordon, F. Rother, E. Hartmann, M. Bader, M. Fainzilber, Importin α3 regulates chronic pain pathways in peripheral sensory neurons. Science 369, 842–846 (2020). [DOI] [PubMed] [Google Scholar]
- 54.S. R. Chaplan, F. W. Bach, J. W. Pogrel, J. M. Chung, T. L. Yaksh, Quantitative assessment of tactile allodynia in the rat paw. J. Neurosci. Methods 53, 55–63 (1994). [DOI] [PubMed] [Google Scholar]
- 55.E. Doron-Mandel, S. Alber, J. A. Oses, K. F. Medzihradszky, A. L. Burlingame, M. Fainzilber, J. L. Twiss, S. J. Lee, Isolation and analyses of axonal ribonucleoprotein complexes. Methods Cell Biol. 131, 467–486 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.S. Guan, J. C. Price, S. B. Prusiner, S. Ghaemmaghami, A. L. Burlingame, A data processing pipeline for mammalian proteome dynamics studies using stable isotope metabolic labeling. Mol. Cell. Proteomics 10, M111.010728 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.S. Berg, D. Kutra, T. Kroeger, C. N. Straehle, B. X. Kausler, C. Haubold, M. Schiegg, J. Ales, T. Beier, M. Rudy, K. Eren, J. I. Cervantes, B. Xu, F. Beuttenmueller, A. Wolny, C. Zhang, U. Koethe, F. A. Hamprecht, A. Kreshuk, ilastik: Interactive machine learning for (bio)image analysis. Nat. Methods 16, 1226–1232 (2019). [DOI] [PubMed] [Google Scholar]
- 58.J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J. Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak, A. Cardona, Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.N. Renier, Z. Wu, D. J. Simon, J. Yang, P. Ariel, M. Tessier-Lavigne, iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 159, 896–910 (2014). [DOI] [PubMed] [Google Scholar]
- 60.M. Spillane, A. Ketschek, T. T. Merianda, J. L. Twiss, G. Gallo, Mitochondria coordinate sites of axon branching through localized intra-axonal protein synthesis. Cell Rep. 5, 1564–1575 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.M. Zuker, Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Z. J. Lu, D. H. Mathews, OligoWalk: An online siRNA design tool utilizing hybridization thermodynamics. Nucleic Acids Res. 36, W104–W108 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.S. Heinz, C. Benner, N. Spann, E. Bertolino, Y. C. Lin, P. Laslo, J. X. Cheng, C. Murre, H. Singh, C. K. Glass, Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.A. Kramer, J. Green, J. Pollard Jr., S. Tugendreich, Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 30, 523–530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.H. Yu, Y. Su, J. Shin, C. Zhong, J. U. Guo, Y. L. Weng, F. Gao, D. H. Geschwind, G. Coppola, G. L. Ming, H. Song, Tet3 regulates synaptic transmission and homeostatic plasticity via DNA oxidation and repair. Nat. Neurosci. 18, 836–843 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.K. Y. Chan, M. J. Jang, B. B. Yoo, A. Greenbaum, N. Ravi, W. L. Wu, L. Sanchez-Guardado, C. Lois, S. K. Mazmanian, B. E. Deverman, V. Gradinaru, Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat. Neurosci. 20, 1172–1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.S. R. Bond, C. C. Naus, RF-Cloning.org: An online tool for the design of restriction-free cloning projects. Nucleic Acids Res. 40, W209–W213 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.M. Mahn, M. Prigge, S. Ron, R. Levy, O. Yizhar, Biophysical constraints of optogenetic inhibition at presynaptic terminals. Nat. Neurosci. 19, 554–556 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.C. Zincarelli, S. Soltys, G. Rengo, J. E. Rabinowitz, Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 16, 1073–1080 (2008). [DOI] [PubMed] [Google Scholar]
- 70.S. Zolotukhin, B. J. Byrne, E. Mason, I. Zolotukhin, M. Potter, K. Chesnut, C. Summerford, R. J. Samulski, N. Muzyczka, Recombinant adeno-associated virus purification using novel methods improves infectious titer and yield. Gene Ther. 6, 973–985 (1999). [DOI] [PubMed] [Google Scholar]
- 71.D. S. Smith, J. H. Skene, A transcription-dependent switch controls competence of adult neurons for distinct modes of axon growth. J. Neurosci. 17, 646–658 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.M. Gullberg, S. M. Gustafsdottir, E. Schallmeiner, J. Jarvius, M. Bjarnegard, C. Betsholtz, U. Landegren, S. Fredriksson, Cytokine detection by antibody-based proximity ligation. Proc. Natl. Acad. Sci. U.S.A. 101, 8420–8424 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S10
Table S1
Legends for data S1 and S2
Data S1 and S2