Abstract
RNA synthesis during viral replication requires specific recognition of RNA promoters by the viral RNA-dependent RNA polymerase (RdRp). Four nucleotides (−17, −14, −13, and −11) within the brome mosaic virus (BMV) subgenomic core promoter are required for RNA synthesis by the BMV RdRp (R. W. Siegel et al., Proc. Natl. Acad. Sci. USA 94:11238–11243, 1997). The spatial requirements for these four nucleotides and the initiation (+1) cytidylate were examined in RNAs containing nucleotide insertions and deletions within the BMV subgenomic core promoter. Spatial perturbations between nucleotides −17 and −11 resulted in decreased RNA synthesis in vitro. However, synthesis was still dependent on the key nucleotides identified in the wild-type core promoter and the initiation cytidylate. In contrast, changes between nucleotides −11 and +1 had a less severe effect on RNA synthesis but resulted in RNA products initiated at alternative locations in addition to the +1 cytidylate. The results suggest a degree of flexibility in the recognition of the subgenomic promoter by the BMV RdRp and are compared with functional regions in other DNA and RNA promoters.
Accurate and efficient RNA synthesis during replication is a key feature of viral pathogenesis. Viral RNA replication is mediated by RNA-dependent RNA polymerases (RdRps) which direct synthesis from RNA templates without DNA intermediates. Unlike the well-studied DNA-dependent polymerases (DdRps), the mechanism by which RdRps direct RNA synthesis is only now being elucidated, and little is known about how RdRps recognize RNA promoters. Discerning the mechanism by which RdRps interact with their cognate RNAs will allow a comparison of the mechanisms of RNA synthesis by different polymerases and contribute to understanding of the features of protein-RNA interaction.
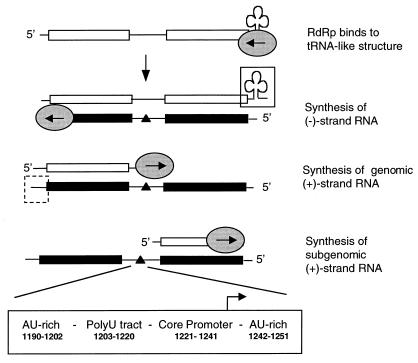
Brome mosaic virus (BMV) is a model well suited to studies of the mechanisms of RNA synthesis. BMV is a single-stranded, positive-sense RNA virus that belongs to the bromovirus group of plant viruses in the alphavirus-like virus superfamily (10). The tripartite BMV genome is composed of RNAs designated RNA1 (3.2 kb), RNA2 (2.8 kb), and RNA3 (2.1 kb) (3). A subgenomic RNA4 (0.9 kb) is synthesized during infection to direct translation of the viral capsid (3). There are three classes of promoters found within the BMV genome (Fig. 1). A tRNA-like structure found on the 3′ end of all positive-sense BMV RNAs directs the synthesis of the complementary minus-strand RNA (5, 18, 21). A second promoter located at the 3′ end of the newly synthesized minus-strand RNA directs genomic plus-strand RNA synthesis (22). The third, subgenomic promoter is located within the minus-strand RNA3 (1, 17, 28). The BMV RdRp utilizes all three distinct promoters in vitro, initiating RNA synthesis at the nucleotides used in vivo (2, 13, 17, 31).
FIG. 1.
BMV RdRp utilizes three RNA promoters during RNA replication. The viral RNA replication process is illustrated with BMV RNA3. The replicase complex and the direction of RNA synthesis are denoted by an oval and the enclosed arrow. Replication begins with recognition of the minus [(−)]-strand promoter (solid box) located at the 3′ tRNA-like (cloverleaf) end of the plus [(+)]-strand genomic RNA. The newly synthesized, complementary minus-strand RNA is depicted in black. Minus-strand RNA is then used as template for both full-length, genomic plus-strand synthesis and subgenomic plus-strand synthesis. The approximate location of the genomic plus-strand promoter is indicated with a dashed box. The subgenomic promoter is located internal to RNA3 and is responsible for RNA4 synthesis. The four domains of the BMV subgenomic promoter characterized by Marsh et al. (16) are illustrated as an expansion within the minus-strand RNA. The nucleotides spanning each domain are listed as well.
The subgenomic promoter has been previously characterized as containing four components: an upstream A-U rich sequence, a polyuridylate tract, the core promoter, and the downstream A-U rich sequence (16) (Fig. 1). Work from our lab has determined that only the core promoter is required and sufficient for accurate and efficient synthesis of runoff RNA products in vitro (1, 28). These RNAs are named proscripts since they contain both promoter and template sequences. Mutational analysis of the subgenomic core promoter revealed that four essential nucleotides at positions −17, −14, −13, and −11 relative to the initiation cytidylate (+1) are recognized by the RdRp in a sequence-specific manner (28). The base moieties of these key nucleotides and riboses responsible for RdRp recognition were predicted by mutational analysis (28) and subsequently identified by using RNAs containing chemically synthesized nucleotide analogs (29).
The present investigation examines the spatial requirements in the BMV core promoter. Insertions and deletions that perturb the spacing within the nucleotides required for recognition by RdRp, and between the required nucleotides and the initiation site, were engineered and examined in a quantitative in vitro RNA synthesis assay. The results of these assays give insights to the mechanisms by which the BMV RdRp recognizes the subgenomic promoter.
MATERIALS AND METHODS
Synthesis of proscripts.
Oligonucleotides (Operon Technologies Inc.) containing the sequence of the BMV subgenomic promoter which contain various insertion or deletions were annealed to a second partially complementary oligonucleotide containing the T7 promoter (5′ TAATACGACTCACTATAGGATTATTAATACGCTG 3′). The single-stranded portions of the annealed oligonucleotides were extended by Taq polymerase and deoxynucleoside triphosphates, creating double-stranded DNA products. RNA proscripts used in RdRp assays were generated from the PCR products by using T7 polymerase (Ampliscribe kit; Epicenter Technologies). Proscript RNAs were purified from nucleoside triphosphates and proteins from the T7 reaction by 20% denaturing polyacrylamide gel electrophoresis (PAGE). RNAs were excised from the crushed gels, eluted with 0.3 M ammonium acetate overnight, extracted with phenol-chloroform, ethanol precipitated, and resuspended in water. RNAs were quantified by spectrophotometry and visualized by toluidine blue staining following denaturing PAGE.
RdRp activity assay and product analysis.
BMV RdRp was prepared from infected barley as previously described (33). Standard RdRp activity assays consisted of a 43-μl reaction mixture containing 1 pmol of proscript template, 10 μl of BMV RdRp, 20 mM sodium glutamate (pH 8.2), 4 mM MgCl2, 12 mM dithiothreitol, 0.5% Triton X-100, 2 mM MnCl2, 200 μM ATP, 200 μM UTP, 500 μM GTP, and 242 nM [α-32P]CTP (400 Ci/mmol, 10 mC/ml; Amersham). The addition of MnCl2 to the reaction has been found to stimulate product synthesis but does not affect the specificity of the BMV RdRp for its promoter (12a). After incubation for 90 min at 30°C, reactions were terminated by phenol-chloroform extraction followed by ethanol precipitation using 10 μg of glycogen and 0.4 M ammonium acetate. Products were suspended in 1× denaturing loading buffer (45% [vol/vol] deionized formamide, 1.5% [vol/vol] glycerol, 0.04% [vol/vol] bromophenol blue, 0.04% [wt/vol] xylene cyanol), heated for 3 min at 60°C, and separated by 20% denaturing (8 M urea) PAGE (19:1 acrylamide-bisacrylamide gel). Gels were wrapped in plastic and exposed to film at −80°C. RdRp products were quantified with a PhosphorImager (Molecular Dynamics), and amounts of synthesis were quantified relative to the wild-type (WT) proscript level. Each value represents a mean of at least three independent experiments with at least two replicates for each proscript. All trends observed per experiment were consistent with those of independent assays.
RESULTS
Effects of nucleotide insertions and deletions within the core promoter.
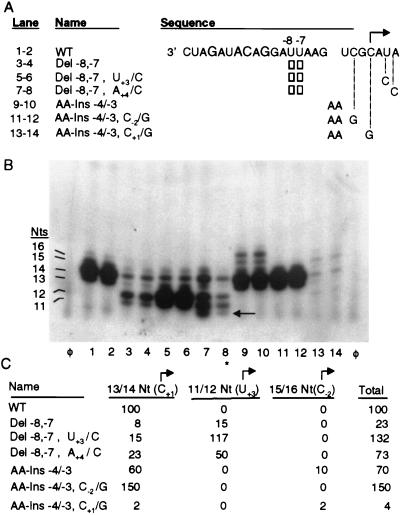
The correct positioning of the RdRp on the promoter is important to establish efficient initiation of subgenomic RNA synthesis. To facilitate analysis of the RdRp-promoter recognition, we decided to treat the core promoter as four regions delineated by the nucleotides recognized by RdRp: −17 to −14, −14 to −13, −13 to −11, and −11 to +1. Proscripts containing insertions and deletions within each of the four regions were assayed for the synthesis of a 13-nucleotide (nt) RNA product. The first 11 nt in the product correspond to the complement of the 5′-most sequence of the BMV subgenomic RNA. The last two guanylates of the product allows for incorporation of radiolabeled CTPs by the BMV RdRp.
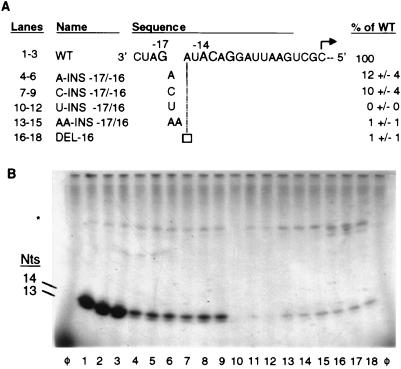
The first region to be examined was between nt −17 and −14. Insertions of an adenylate, cytidylate, or uridylate between nt −17 and −16 as well as insertions of two adenylates and the removal of nt −16, were tested (Fig. 2A). The products of the RdRp assay were resolved on a gel as two bands of 13 and 14 nt (Fig. 2B). The 14-nt RNA is the result of a nontemplated nucleotide addition to the 13-nt product (28). Terminal nucleotide addition is an activity of the RdRp preparation and is observed with other polymerases (4, 20). A terminal nucleotide is also inefficiently placed on the input proscripts, as seen by the faint bands running above the major products. The amount of synthesis from the WT proscript was normalized to 100% (Fig. 2B, lanes 1 to 3). Proscripts containing either insertions of one or two nucleotides or the deletion of one nucleotide suffered severe reductions in the efficiency of synthesis in comparison to a WT subgenomic promoter (Fig. 2B, lanes 4 to 18). However, all of the mutations between nt −17 and −14 gave rise to products of 13 and 14 nt, indicating that the accuracy of the initiation was unaffected.
FIG. 2.
Nucleotide insertions and deletions within the −17/−14 region. (A) Sequence of the WT subgenomic core promoter. The four nucleotides required to interact with RdRp are in a larger font; the initiation cytidylate is marked with an arrow pointed in the direction of RNA synthesis. Proscripts are named for the changes from the WT sequence. Nucleotides which are changed from WT are denoted in the gap in the WT sequence between nt −17 and −16. The empty box denotes the removal of nt −16. Nucleotides which are not changed from WT are not shown. The RdRp products generated by each proscript and the values for 1 standard deviation from the mean are listed at the right. All values represent at least three independent assays. The column on the left provides the key for the lanes in the autoradiograph below. (B) Autoradiograph of RdRp products synthesized from proscripts containing insertions and deletions within the −17/−14 region of the subgenomic promoter analyzed by 20% denaturing PAGE. Terminal nucleotide addition to the input proscript is denoted with an asterisk. Three independent reactions are shown for each proscript. The positions of the 14- and 13-nt products are indicated on the left. Here and in Fig. 3 and 4, lanes φ represent products of control reactions without added templates.
The spatial requirements between nt −14 and −13 and nt −13 and −11 were examined next. A summary of the data along with the sequences and activities of the proscripts relative to WT is shown in Table 1. Insertions of an adenylate or a uridylate within the −14/−13 region reduced RNA synthesis to less than 25% of the WT level. Insertions of two adenylates further decreased synthesis in comparison with the insertion of one adenylate. Proscripts containing single insertions and deletions within the −13/−11 region retained 10 to 40% of WT activity. Again, the insertions of two nucleotides within this region had a more detrimental effect (11 and 15% of WT activity) than the insertion of one nucleotide (36 and 41% of WT activity). These results are similar to those from changes within the −17/−14 region in regard to the qualitative effect on RNA synthesis. Furthermore, even though insertions and deletions in either the −14/−13 or −13/−11 region reduced RNA synthesis, all products synthesized were 13 and 14 nt in length, reflecting that RNA synthesis initiated at the authentic +1 cytidylate.
TABLE 1.
Insertions and deletions of nucleotides between positions −17 and −11
| Proscript | % of WT | Sequence (3′-5′)a |
|---|---|---|
| WT | 100 | cuaGauACaGgauuaagucgCau |
| Mutant | ||
| −17/−14 | ||
| A-INS −17/−16 | 12 ± 4 | cuaGAauACaGgauuaagucgCau |
| C-INS −17/−16 | 10 ± 4 | cuaGCauACaGgauuaagucgCau |
| U-INS −17/−16 | 0 ± 0 | cuaGUauACaGgauuaagucgCau |
| AA-INS −17/−16 | 1 ± 1 | cuaGAAauACaGgauuaagucgCau |
| DEL −16 | 1 ± 1 | cuaG*uACaGgauuaagucgCau |
| A-INS −17/−16, −17 G to U | 1 ± 1 | cuaUAauACaGgauuaagucgCau |
| −14 to −13 | ||
| A-INS −14/−13 | 25 ± 8 | cuaGauAaCaGgauuaagucgCau |
| U-INS −14/−13 | 4 ± 3 | cuaGauAuCaGgauuaagucgCau |
| AA-INS −14/−13 | 10 ± 4 | cuaGauAaaCaggauuaaguccCau |
| −13 to −11 | ||
| A-INS −13/−12 | 36 ± 10 | cuaGauACaaGgauuaagucgcau |
| U-INS −13/−12 | 41 ± 21 | cuaGauACuaGgauuaagucgcau |
| AA-INS −13/−12 | 11 ± 5 | cuaGauACaaaGgauuaagucgcau |
| UU-INS −13/−12 | 15 ± 4 | cuaGauACuuaGgauuaagucgcau |
| DEL −12 | 27 ± 7 | cuaGauAC*Ggauuaagucgcau |
Nucleotides inserted are indicated in boldface, letters in larger font size represent those previously found to be required for recognition by RdRp, and asterisks denote deleted nucleotides.
The −13/−11 region.
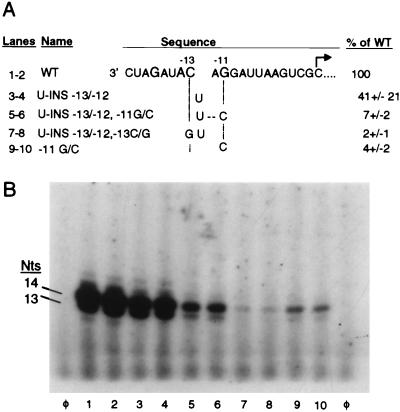
The observation that single nucleotide insertions within the −13/−11 region retained a moderate level of activity suggests two possibilities by which synthesis is maintained: (i) despite the insertions, RdRp maintains some recognition with the original −13 cytidylate and −11 guanylate residues. (ii) RdRp recognizes the presence of nucleotides at the correct spatial positions despite the changes in identities of these nucleotides. The latter possibility is less likely since substitutions at the key contact sites resulted in little or no RNA synthesis (28). To address these two possibilities, proscripts containing an insertion of a uridylate between positions −13 and −12 (U-INS −13/−12) were engineered to contain a second change at either nt −13 or nt −11 (Fig. 3A). The proscripts were subsequently assayed for the relative ability to direct RNA synthesis. Proscript U-INS −13/−12 showed 41% of WT activity (Fig. 3B, lanes 3 and 4). An additional change of the authentic −11 guanylate to a cytidylate resulted in 7% relative RNA synthesis (lanes 5 and 6). This level of synthesis is similar to a proscript which has only a −11 guanylate-to-adenylate substitution (lanes 9 and 10). Proscript U-INS −13/12, with a transversion of the authentic −13 cytidylate to a cytidylate, reduced RNA synthesis to 2% (lanes 7 and 8). Taken together, the data suggest that RdRp maintains recognition of the −13 cytidylate as well as of the −11 guanylate even in proscripts with single nucleotide insertions between positions −13 and −12. The moderate levels of activity displayed by these proscripts in comparison to WT may be a result of difficulty in maintaining recognition in the presence of a nucleotide insertion.
FIG. 3.
RdRp recognition of key nucleotides is required even in the presence of a nucleotide insertion. (A) The sequence of the WT proscript is presented as described in the legend to Fig. 2A. The identities of the insertions between nt −13 and −12 are shown in the area below the gap in the WT sequence. Additional changes at specific positions are indicated by the letters in the dashed lines. (B) Autoradiograph of RdRp products synthesized from proscripts containing insertions and substitutions within the −13/−11 region of the subgenomic promoter analyzed by 20% denaturing PAGE. Duplicate independent reactions are shown. Positions of the 14- and 13-nt products are indicated on the left.
A similar mechanism would explain the activity of proscripts with single nucleotide insertions within the region between nt −17 and −14 (Table 1). In this case, the 10% activity observed with proscripts A-INS −17/−16 and C-INS −17/−16 may be due to RdRp retaining a limited recognition of the −17 guanylate. To test this hypothesis, proscript A-INS −17/−16, which also changed the −17 guanylate to an uridylate, was assayed. It resulted in a decrease in RNA synthesis to 1% activity (Table 1), further suggesting that RdRp maintains some recognition with the −17 guanylate within the single insertion proscripts.
Insertions and deletions within nt −11 to +1.
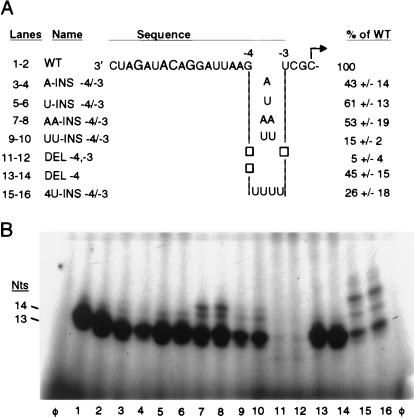
Over a dozen mutant proscripts were engineered to examine the spatial requirements for the large region between the nt −11 and +1. Single or multiple insertions or deletions positioned along the length of the sequence between nt −11 and +1 were assayed (Table 2). In contrast to results from the −17/−14, −14/−13, and −13/12 regions, changes between nt −11 and +1 had generally less severe effects on the efficiency of RNA synthesis (Fig. 4A; Table 2). Constructs with insertions and deletions of one nucleotide between −11 and +1 all retained more than 40% of WT activity. In contrast to results from region between −17 and −11, insertion of two nucleotides between −11 and +1 all retained more than 15% of WT activity. In fact, a proscript that contained an insertion of four uridylates between nt −4 and −3 (Fig. 4, lanes 15 and 16) retained 26% of WT activity. The most severe perturbation within this region was a deletion of nt −4 and −3, which resulted in only 5% of WT activity (Table 2; Fig. 4B, lanes 11 and 12). This mutation may be more severe in comparison to other multiple deletions within the −11/+1 region due to the removal of the −4 residue, a change that had a moderate affect on RNA synthesis (28). While changes between −11 and +1 had only moderate effects on the efficiency of RNA synthesis, products of anomalous size were observed with several of the mutant proscripts (Fig. 4B, lanes 7, 8, 15, and 16). These products may be the result of initiation of RNA synthesis from sites other than the authentic cytidylate.
TABLE 2.
Insertions and deletions of nucleotides between positions −11 and +1
| Proscript | % of WT | Sequence (3′-5′)a |
|---|---|---|
| WT | 100 | cuaGauACaGgauuaagucgcau |
| Mutant | ||
| U-INS −11/−10 | 69 ± 15 | cuaGauACaGugauuaagucgcau |
| UU-INS −11/10 | 52 ± 18 | cuaGauACaGuugauuaagucgcau |
| DEL −10 | 65 ± 7 | cuaGauACaG*auuaagucgcau |
| DEL −10,−9 | 24 ± 19 | cuaGauACaG**uuaagucgcau |
| DEL −8 | 71 ± 15 | cuaGauACaGga*uaagucgcau |
| Del −8,−7 | 23 ± 9 | cuaGauACaGga**aagucgcau |
| U-INS −7/−6 | 114 ± 15 | cuaGauACaGgauuuaagucgcau |
| UU-Ins −7/−6 | 59 ± 17 | cuaGauACaGgauuuuaagucgcau |
| A-INS −4/−3 | 43 ± 14 | cuaGauACaGgauuaagaucgcau |
| U-INS −4/−3 | 61 ± 13 | cuaGauACaGgauuaaguucgcau |
| AA-INS −4/−3 | 70 ± 19 | cuaGauACaGgauuaagaaucgcau |
| UU-INS −4/−3 | 15 ± 2 | cuaGauACaGgauuaaguuucgcau |
| 4U-INS −4/−3 | 26 ± 18 | cuaGauACaGgauuaaguuuuucgcau |
| DEL −4 | 45 ± 15 | cuaGauACaGgauuaa*ucgcau |
| DEL −4,−3 | 5 ± 4 | cuaGauACaGgauuaa**cgcau |
Nucleotides inserted are indicated in boldface, letters in larger font size denote those previously found to be required for recognition by RdRp, and asterisks denote deleted nucleotides.
FIG. 4.
Nucleotide insertions and deletions within the −11/+1 region. (A) The sequence of the WT proscript is presented as described in the legend to Fig. 2A. Changes from the WT sequence between nt −4 and −3 are illustrated in the area below the gap in the sequence. An empty box indicate that a nucleotide has been deleted. The RdRp products generated by each proscript and the values for 1 standard deviation from the mean are listed on the right. All values represent at least six independent assays. Quantified values in reactions with multiple products represent the sum of all products made. (B) Autoradiograph of RdRp products synthesized from proscripts containing insertions and deletions within the −11/+1 region of the subgenomic promoter. Duplicate independent reactions are shown, and positions of the 14- and 13-nt products are indicated on the left. Anomalous-size products are clearly visible as bands of greater than 14 nt in lanes 7, 8, 15, and 16.
Insertions and deletions between −11 and +1 alter initiation specificity.
Initiation of subgenomic BMV RNA synthesis takes place at the complement of the cytidylate at nt 1242 in plus-strand RNA 3 both in vivo and in vitro. The potential misinitiation of RNA synthesis is of interest since it could reveal the mechanisms underlying promoter recognition and initiation during RNA synthesis. Therefore, a more careful examination of the products from proscripts DEL −8,−7 and AA-INS −4/−3 was undertaken, since both of these proscripts produced RNAs that deviated from the expected sizes of 14 and 13 nt. The most abundant products of proscript DEL−8,−7 are 11 and 12 nt, in length as determined by comparison to RdRp products of corresponding sizes (data not shown), and were present at a combined 15% of WT activity; DEL−8,−7 also synthesized the 14/13-nt products at 8% of the WT level, presumably by initiating synthesis from the authentic cytidylate (Fig. 5, lanes 3 and 4). The 12/11-nt products most likely initiated at the +3 uridylate. Previous work has shown that RdRp is able to initiate RNA synthesis from a cytidylate moved one position away from the initiation cytidylate and inefficiently from a uridylate residue (28). To examine the initiation nucleotide used to generate the 12/11-nt products, a DEL−8,−7 proscript which also contains a change of the +3 uridylate to a cytidylate was constructed with the rationale that a change to a preferred initiation cytidylate would increase the synthesis of the 12/11-nt product. Indeed, proscript DEL−8,−7, U+3/C synthesized the 12/11-nt product at 117% of the level from the WT proscript, an approximately 10-fold increase from DEL−8,−7 proscript (Fig. 5B, lanes 5 and 6; Fig. 5C). In addition, DEL−8,−7 with a substitution of the +4 adenylate to a potential initiation cytidylate may direct synthesis of a 11/10-nt product (Fig. 5B, lanes 7 and 8). A 10-nt product unique to this proscript was clearly observed (Fig. 5B, arrow), but the 11-nt product was obscured by comigration with the product that likely initiated at the +3 position.
FIG. 5.
Nucleotide insertions and deletions within the −11/+1 region can alter the site for initiation. (A) The sequence of the WT proscript is presented as described in the legend to Fig. 2A. Nucleotides inserted in mutant proscripts are denoted in the gap in the WT sequence. Empty boxes denote deletions of specific nucleotides. Changes of nucleotides in the template sequence are indicated by letters at the ends of the dashed lines. (B) Autoradiograph of RdRp products synthesized from proscripts containing insertions and deletions within the −11/+1 region of the subgenomic promoter. Duplicate independent reactions are shown, and the sizes of the products are indicated on the left. The arrow points to a 10-nt product visible in lanes 7 and 8. Lane 8 is marked with an asterisk because the level of products in this lane is lower due to loss of sample during the assay. (C) Quantitation of the 13/14-nt, 11/12-nt, and 15/16-nt products initiated from nt +1, +3, and −2. Products are quantitated relative to synthesis from WT. The column on the right represents the value of the total amount of all products synthesized. Values for 1 standard deviation from the mean are listed and represent at least three independent assays.
Proscript AA-INS −4/−3, containing insertions of two nucleotides within the −11/+1 region, predominantly produced 14/13-nt RNAs which likely initiated from the authentic cytidylate. In addition, products of 16 or 15 nt were also observed at 10% of the WT level (Fig. 5B, lanes 9 and 10). The 16/15-nt products were likely generated by initiation at the cytidylate at the authentic nt −2. To test this hypothesis, proscript AA-INS −4/−3 was modified to contain a guanylate at either the −2 or +1 cytidylate position. A guanylate present at position +1 had been previously demonstrated to decrease RNA synthesis to 4% (28). Proscript AA-INS −4/−3, with a guanylate at the −2 position, abolished synthesis of the 16/15-nt products, demonstrating that the 16/15-nt products were the result of inefficient initiation taking place at the original −2 position (Fig. 5B, lanes 11 and 12). Furthermore, preventing initiation from the −2 position increased synthesis of the 14/13-nt products from 60 to 150% of the WT level. Proscript AA-INS −4/−3, which also has the authentic +1 cytidylate changed to a guanylate, reduced synthesis of the correctly initiated 14/13-nt product from 60 to 2% while reducing synthesis of the 16/15-nt product from 10 to 2% (Fig. 5C). The reduction of the amount of the 16/15-nt product may be due to a guanylate being present at both the +2 and +3 positions relative to the initiation of the 16/15-nt product (2).
DISCUSSION
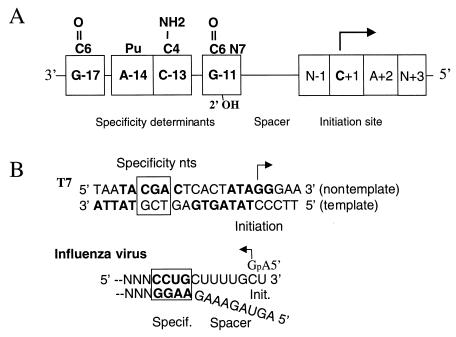
Using the BMV RdRp as a model system, we are attempting to elucidate the mechanism of RNA synthesis by RdRp (2, 13, 33–35), including how RdRp recognizes the viral RNA promoters. The short length (<33 nt) of the subgenomic promoter required for specific and accurate initiation of RNA synthesis in vitro allows easy manipulation of the BMV subgenomic promoter for biochemical analyses (1, 28). Previous work demonstrated that the BMV RdRp recognizes the subgenomic core promoter in a sequence-specific manner (28). Moieties in the nucleotide bases and in the riboses required for efficient RNA synthesis have also been recently identified (29). In this work, we used proscripts which are affected in the normal spacing of the key nucleotides recognized by RdRp to elucidate the requirements of RdRp-RNA promoter interaction. We have determined that spacing in the regions spanning nt −17 to −13 are important for efficient RNA synthesis by RdRp, while nt −11 to +1 are less important for directing efficient RNA synthesis but can affect the selection of the initiation site. This and previous work suggest that the BMV subgenomic core promoter can be functionally divided into three domains: the specificity domain which is presumed to be contacted by the RdRp, a spacer sequence which is more tolerant of changes in nucleotide identity and length, and the initiation site (Fig. 6A). These three functional domains will be discussed in turn along with comparisons to the T7 DNA promoter and the influenza virus RNA promoter.
FIG. 6.
Schematics summarizing features in DNA and RNA promoters. (A) The BMV subgenomic core promoter and initiation site. Nucleotides required for recognition by the BMV RdRp are boxed; the base moieties important for the recognition by RdRp (28) are shown above and below the boxes. The arrow denotes the initiation nucleotide. (B) Consensus promoter recognized by the T7 RNA polymerase (24) and the influenza virus promoter for minus-strand RNA synthesis (6). The nucleotides important for polymerase specificity are boxed, and the nucleotides determined to be contacted by the respective polymerases are in boldface. An arrow is placed over the initiation nucleotide to denote the direction of RNA synthesis.
Specificity determinants.
Nucleotides −17 and −11 of the BMV subgenomic promoter contains the domain which confers specific binding by RdRp. Nucleotide substitutions and changes in the relative spatial distances within this region will severely decrease RNA synthesis without affecting the site of RNA initiation (Table 1). In template competition assays for RdRp-RNA binding, substitutions at positions −17, −14, −13, and −11 abolished binding to RdRp (29). For recognition by the influenza virus RdRp, key specificity nucleotides include −9, −10, and −11 relative to the initiation site at the 3′ end of the viral minus strand (6, 26) (Fig. 6B). However, the 5′ end of the influenza virus RNA is needed to interact with this region to form a double-stranded region recognized by the influenza virus RdRp (7, 36). Changes in the nucleotides important for the influenza virus promoter all resulted in severe reductions in RNA synthesis (7, 36). The best-characterized specificity region are bp −10 to −12 of the consensus T7 and T3 DNA promoters (11, 14, 24, 25). Changes of one or more of these base pairs may abolish its use by the homologous polymerase and also confer recognition by the heterologous polymerase (14, 23, 26).
Spacer.
The sequence between nt −10 and −2 may represent a molecular spacer needed to maintain an optimal distance between the specificity and initiation regions. Sequences within the spacer affect interaction with RdRp, as evidenced by the observation that some nucleotide substitutions in this region decreased RNA synthesis, although the effects were less severe than those in the specificity region (Table 2 and reference 28). Perhaps the most intriguing result with the putative spacer region is that insertions and deletions in this region, but not in the specificity region, will affect the choice of the initiation site by RdRp.
The spacer region may need to adapt conformations acceptable to the polymerase. The 17-nt spacer sequence in the Escherichia coli sigma 70 promoter which lies between the −35 and −10 specificity domains has been demonstrated to not specifically bind the E. coli holoenzyme in DNA footprinting (27). Also an acceptable B-form DNA structure in this spacer must be maintained or else polymerase-promoter interaction would be adversely affected (31, 37). In the corresponding domain of the T7 promoter, insertion of flexible polyanionic spacers in place of nucleotide positions −2 or −4 had little detrimental effect on the fidelity or overall amount of RNA synthesis. However, replacing the −1 nt with a polyanionic spacer or an abasic residue increased the frequency of products initiated from the +2 nt (38).
The role of local RNA structure in the RNA promoters remains to be determined experimentally. However, in both the BMV and influenza virus RNA promoters, insertions and deletions in the putative spacer region had similar effects. One or two nucleotide insertions only weakly inhibited influenza virus RNA synthesis in vitro, while a deletion of one nucleotide or the insertion of three or more nucleotides in the influenza virus promoter caused significant decrease in RNA synthesis (26). Nucleotides −8 to −3 of the influenza virus promoter and nt −10 to −1 of the BMV subgenomic core promoter may be present to keep the proper distance between the specificity and initiation.
Initiation site.
In addition to the specificity domain in the BMV core promoter, the initiation site spanning nucleotides −1 to +2 is another domain specifically recognized by RdRp (Fig. 6) (29). We speculated that the recognition of the initiation site by the BMV RdRp requires a trimolecular interaction between RdRp, the template RNA, and the primer nucleotide, GTP. The base pairing between the initiation template nucleotide and the GTP may be responsible for enhancing the stability of RdRp-RNA interaction. The +2 nucleotide, usually an adenylate in the promoters directing plus-strand RNA synthesis, is required for efficient initiation of RNA synthesis (2). The requirement for an appropriate initiation and +2 nucleotide may explain the preference for the authentic initiation site of the subgenomic promoter over other cytidylates which may act as the initiation site (Fig. 5).
Flexibility in RdRp recognition sites.
An unexpected result from this work is that RdRp interaction with the preferred recognition nucleotides in the subgenomic core promoter can take place even when the spacing between the nucleotides is altered. In contrast, observations from analyses of the T7 and T3 DdRps indicate that changes in the specificity domain will abolish recognition by the homologous polymerase (14, 24). Our data suggest that RdRp has the ability to adjust its RNA binding in a manner analogous to having independent suspension in the wheels of an all-terrain automobile. This flexibility is demonstrated in two sets of changes in the specificity domain. (i) In the presence of an insertion between −17 and −16, the contact between RdRp and the −17 guanylate occurs, resulting in approximately 10% synthesis (Fig. 2). Mutation of the authentic −17 guanylate in the presence of the insertion will abolish RNA synthesis (Table 1). (ii) The recognition between RdRp and key nt −13 and −11 is required in the presence of an insertion, resulting in RNA synthesis at approximately 40% of the WT promoter level. Changes of the −11 cytidylate and the −13 guanylate to their Watson-Crick transversions in addition to the insertion will drastically decrease RNA synthesis, demonstrating that sequences required in the context of the WT spacing are also required in the presence of an insertion.
A third demonstration of flexibility in RdRp may be the selection of the initiation sites when the length of the spacer has been altered. Deletions of nt −8 and −7 and double insertions between nt −4 and −3 preferentially retained the use of the authentic initiation site (Fig. 5). This result is consistent with our previous observation that RdRp could use a cytidylate as the initiation nucleotide when it was one nucleotide to either side of the authentic position (28). In this case, it is difficult to rule out the possibility that the length of the spacer allows the RNA to alter its conformation rather than induce an adjustment by the RdRp. It is also possible that both RdRp and the RNA must adjust to each other during their initial recognition. Examples of similar changes in protein-RNA interaction (generally called induced fit) have recently been reviewed by Frankel and Smith (8). Similar interpretations for the flexibility of the influenza virus RdRp can be made from the mutations in the spacer of the influenza virus promoter (26).
The ability of RdRp to adjust to the required recognition sites provides a testable hypothesis for how one enzyme complex can direct RNA synthesis from all three classes of RNA promoters found in the BMV genome BMV RNAs (Fig. 1). The three BMV promoters are very different in sequence and predicted RNA secondary structures, with no obvious features in common except that the initiation nucleotide is always a cytidylate. We hypothesize that RdRp recognition of the core promoter takes place through only a few nucleotides placed at acceptable spatial positions. This flexible RdRp is then able to adjust its promoter contact sites or alter the local structure of the promoter as dictated by key nucleotides in the promoter. Alternatively, this mode of RdRp-promoter interaction would permit species-specific RNA synthesis while allowing the individual sequences to gain, through evolutionary selection, other features desirable for viral infection such as RNA stability and efficiency of translation. We note that this model does not preclude the contributions of the local RNA structure which may exist in this region. However, previous results from our lab indicates that the primary mode of recognition of the core promoter in vitro is through a sequence-specific mechanism (28, 29).
In addition to similarities in the promoters recognized by DdRps and RdRps, evidence is also rapidly accumulating on the similarities in other processes of RNA synthesis, including the recent realizations that RNA syntheses by DdRp and by RdRps both go through a highly ordered and parallel series of steps (reference 2 and references therein) and that host factors associated with viral RNA replication are functionally analogous to the factors which direct basal and activated eukaryotic transcription (15). Recent work by Siegel et al. (29a) suggests that RdRp has at least two RNA binding domains. This finding is consistent with the current models for DdRps where both upstream and downstream template binding sites have been demonstrated (9, 19). Other similarities of the different polymerases, including the overall similar in tertiary structure and mechanism of nucleotidyl transfer, have been recently emphasized by Steitz (32) and Joyce (12). These developments suggest that future studies in viral RNA replication would directly benefit from the lessons learned from the better-characterized polymerases.
ACKNOWLEDGMENTS
We thank members of the IU Cereal Killer group for helpful discussions during the course of this work, especially Scott Adkins and Matt Chapman for editing the manuscript.
Funding was provided by U.S. Department of Agriculture grant 9702126. Scott Stevenson Stawicki also acknowledges support from a plant biology Floyd summer fellowship.
REFERENCES
- 1.Adkins S, Siegel R W, Sun J H, Kao C C. Minimal templates directing accurate initiation of subgenomic RNA synthesis in vitro by the brome mosaic virus RNA-dependent RNA polymerase. RNA. 1997;3:634–647. [PMC free article] [PubMed] [Google Scholar]
- 2.Adkins S, Stawicki S S, Faurote G, Siegel R, Kao C C. Mechanistic analysis of RNA synthesis by RNA-dependent RNA polymerase from two promoters reveals similarities to DNA-dependent RNA polymerases. RNA. 1998;4:455–470. [PMC free article] [PubMed] [Google Scholar]
- 3.Ahlquist P. Bromovirus RNA replication and transcription. Curr Opin Genet Dev. 1992;2:271–276. doi: 10.1016/s0959-437x(05)80325-9. [DOI] [PubMed] [Google Scholar]
- 4.Bausch J, Kramer F, Miale E, Dobkin C, Mills D. Terminal adenylation in the synthesis of RNA by Qβ replicase. J Biol Chem. 1983;258:1978–1984. [PubMed] [Google Scholar]
- 5.Dreher T W, Hall T C. Mutational analysis of the sequence and structural requirements in brome mosaic virus RNA for minus strand promoter activity. J Mol Biol. 1988;201:31–40. doi: 10.1016/0022-2836(88)90436-6. [DOI] [PubMed] [Google Scholar]
- 6.Fodor E, Seong B L, Brownlee G G. Photochemical crosslinking of influenza A polymerase to its virion RNA promoter defines a polymerase binding site at residues 9 to 12 of the promoter. J Gen Virol. 1993;74:1327–1333. doi: 10.1099/0022-1317-74-7-1327. [DOI] [PubMed] [Google Scholar]
- 7.Fodor E, Pritlove D C, Brownlee G G. Characterization of the RNA-fork model of virion RNA in the initiation of transcription in influenza A virus. J Virol. 1995;69:4012–4019. doi: 10.1128/jvi.69.7.4012-4019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frankel A, Smith A. Induced folding in RNA-protein recognition: more than a simple molecular handshake. Cell. 1998;92:149–151. doi: 10.1016/s0092-8674(00)80908-3. [DOI] [PubMed] [Google Scholar]
- 9.Gelles J, Landick R. RNA polymerase as a molecular motor. Cell. 1998;93:13–16. doi: 10.1016/s0092-8674(00)81140-x. [DOI] [PubMed] [Google Scholar]
- 10.Goldbach R, LeGall O, Wellink J. Alpha-like viruses in plants. Semin Virol. 1991;2:19–25. [Google Scholar]
- 11.Ikeda R, Chang L L, Warshamana G S. Selection and characterization of a mutant T7 RNA polymerase that recognizes an expanded range of T7 promoter-like sequences. Biochemistry. 1993;32:9115–9124. doi: 10.1021/bi00086a016. [DOI] [PubMed] [Google Scholar]
- 12.Joyce C M. Choosing the right sugar: how polymerases select a nucleotide substrate. Proc Natl Acad Sci USA. 1997;94:1619–1622. doi: 10.1073/pnas.94.5.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12a.Kao, C. C. Unpublished data.
- 13.Kao C C, Sun J H. Initiation of minus-strand RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: use of oligoribonucleotide primers. J Virol. 1996;70:6826–6830. doi: 10.1128/jvi.70.10.6826-6830.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klement J F, Moorefield M B, Jorgensen E, Brown J E, Risman S, McAllister W T. Discrimination between bacteriophage T3 and T7 promoters by the T3 and T7 RNA polymerases depend primarily upon a three base-pair region located 10–12 base-pairs upstream from the start site. J Mol Biol. 1990;215:21–29. doi: 10.1016/s0022-2836(05)80091-9. [DOI] [PubMed] [Google Scholar]
- 15.Lai M M. Cellular factors in the transcription and replication of viral RNA genomes. A parallel to DNA-dependent RNA transcription. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 16.Marsh L E, Dreher T W, Hall T C. Mutational analysis of the core and modulator sequences of the BMV RNA3 subgenomic promoter. Nucleic Acids Res. 1988;16:981–995. doi: 10.1093/nar/16.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller W A, Dreher T W, Hall T C. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (−)-sense genomic RNA. Nature. 1985;313:68–70. doi: 10.1038/313068a0. [DOI] [PubMed] [Google Scholar]
- 18.Miller W A, Bujarski J J, Dreher T W, Hall T C. Minus-strand initiation by brome mosaic virus replicase within the 3′ tRNA-like structure of native and modified RNA templates. J Mol Biol. 1986;187:537–546. doi: 10.1016/0022-2836(86)90332-3. [DOI] [PubMed] [Google Scholar]
- 19.Nudler E, Avetissova E, Markovtsov V, Goldfarb A. Transcription processivity: protein-DNA interaction holding together the elongation complex. Science. 1996;273:211–214. doi: 10.1126/science.273.5272.211. [DOI] [PubMed] [Google Scholar]
- 20.Nuefeld K, Galarza J, Richards O, Summers D, Ehrenfeld E. Identification of terminal adenyl transferase activity of the poliovirus polymerase 3Dpol. J Virol. 1994;68:5811–5818. doi: 10.1128/jvi.68.9.5811-5818.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perret V, Florentz C, Dreher T, Giege R. Structural analogies between the 3′ tRNA-like structure of the brome mosaic virus RNA and yeast tRNAtyr revealed by protection studies with yeast tyrosyl tRNA synthase. Eur J Biochem. 1989;185:331–339. doi: 10.1111/j.1432-1033.1989.tb15120.x. [DOI] [PubMed] [Google Scholar]
- 22.Pogue G, Hall T. The requirement for a 5′ stem-loop structure in the brome mosaic virus replication supports a new model for viral positive-strand RNA synthesis. J Virol. 1992;66:674–684. doi: 10.1128/jvi.66.2.674-684.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raskin C A, Diaz G, Joho K, McAllister W T. Substitution of a single bacteriophage T3 residue in bacteriophage T7 RNA polymerase at position 748 results in a switch in promoter specificity. J Mol Biol. 1992;228:506–515. doi: 10.1016/0022-2836(92)90838-b. [DOI] [PubMed] [Google Scholar]
- 24.Schick C, Martin C T. Identification of specific contacts in T3 RNA polymerase-promoter interactions: kinetic analysis using small synthetic promoters. Biochemistry. 1993;32:4275–4280. doi: 10.1021/bi00067a016. [DOI] [PubMed] [Google Scholar]
- 25.Schick C, Martin C T. Tests of a model of specific contacts in T7 RNA polymerase-promoter interactions. Biochemistry. 1995;34:666–672. doi: 10.1021/bi00002a034. [DOI] [PubMed] [Google Scholar]
- 26.Seong B L, Brownlee G G. Nucleotides 9 to 11 of the influenza A virion RNA promoter are crucial for activity in vitro. J Gen Virol. 1992;73:3115–3124. doi: 10.1099/0022-1317-73-12-3115. [DOI] [PubMed] [Google Scholar]
- 27.Siebenlist U, Simpson R B, Gilbert W. E. coli RNA polymerase interacts homologously with two different promoters. Cell. 1980;20:269–281. doi: 10.1016/0092-8674(80)90613-3. [DOI] [PubMed] [Google Scholar]
- 28.Siegel R W, Adkins S, Kao C C. Sequence-specific recognition of a subgenomic promoter by a viral RNA polymerase. Proc Natl Acad Sci USA. 1997;94:11238–11243. doi: 10.1073/pnas.94.21.11238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siegel R W, Bellon L, Beigelman L, Kao C C. Moieties in an RNA promoter specifically recognized by a viral RNA-dependent RNA polymerase. Proc Natl Acad Sci USA. 1998;95:11613–11618. doi: 10.1073/pnas.95.20.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29a.Siegel, R. W., et al. Unpublished data.
- 30.Sivakumaran, K., and C. C. Kao. Unpublished data.
- 31.Stefano J E, Gralla J D. Spacer mutations in the lacPs promoter. Proc Natl Acad Sci USA. 1982;79:1069–1072. doi: 10.1073/pnas.79.4.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steitz T. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 33.Sun J H, Adkins S, Faurote G, Kao C C. Initiation of (−)-strand RNA synthesis catalyzed by the BMV RNA-dependent RNA polymerase: synthesis of oligoribonucleotides. Virology. 1996;225:1–12. doi: 10.1006/viro.1996.0622. [DOI] [PubMed] [Google Scholar]
- 34.Sun J H, Kao C C. RNA synthesis by the brome mosaic virus RNA-dependent RNA polymerase: transition from initiation to elongation. Virology. 1997a;233:63–73. doi: 10.1006/viro.1997.8583. [DOI] [PubMed] [Google Scholar]
- 35.Sun J H, Kao C C. Characterization of RNA products associated with or aborted by a viral RNA-dependent RNA polymerase. Virology. 1997b;236:348–353. doi: 10.1006/viro.1997.8742. [DOI] [PubMed] [Google Scholar]
- 36.Tiley L S, Hagan M, Mathews J T, Krystal M. Sequence specific binding of the influenza RNA polymerase to sequences located at the 5′ end of the viral RNAs. J Virol. 1994;68:5108–5116. doi: 10.1128/jvi.68.8.5108-5116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warner S, DeHaseth P L. Promoter recognition by Escherichia coli RNA polymerase. Effects of single basepair deletions and insertions in the spacer DNA separating the −10 and −35 region are dependent on spacer DNA sequence. Biochemistry. 1993;32:6134–6140. doi: 10.1021/bi00075a003. [DOI] [PubMed] [Google Scholar]
- 38.Weston B F, Kuzmine I, Martin C T. Position of the start site of initiation of transcription by bacteriophage T7 RNA polymerase. J Mol Biol. 1997;272:21–30. doi: 10.1006/jmbi.1997.1199. [DOI] [PubMed] [Google Scholar]