Abstract
Salinity is one of the most significant environmental factors limiting legumes development and productivity. Salt stress disturbs all developmental stages of legumes and affects their hormonal regulation, photosynthesis and biological nitrogen fixation, causing nutritional imbalance, plant growth inhibition and yield losses. At the molecular level, salt stress exposure involves large number of factors that are implicated in stress perception, transduction, and regulation of salt responsive genes’ expression through the intervention of transcription factors. Along with the complex gene network, epigenetic regulation mediated by non-coding RNAs, and DNA methylation events are also involved in legumes’ response to salinity. Different alleviation strategies can increase salt tolerance in legume plants. The most promising ones are Plant Growth Promoting Rhizobia, Arbuscular Mycorrhizal Fungi, seed and plant’s priming. Genetic manipulation offers an effective approach for improving salt tolerance. In this review, we present a detailed overview of the adverse effect of salt stress on legumes and their molecular responses. We also provide an overview of various ameliorative strategies that have been implemented to mitigate/overcome the harmful effects of salt stress on legumes.
Keywords: Legumes, Salinity, Salt injury, Alleviation strategies, Molecular responses, Tolerance
Introduction
Grain legumes offer the main source of calories and proteins for a large proportion of the world’s population due to their relatively cheap sources of dietary protein, vitamins and minerals for humans and animals mostly in developing countries around the Mediterranean region (Jha et al. 2019). Legumes occupy 12–15% of arable land worldwide to produce 27% of major crop production (Mishra et al. 2014a, b).
Environmental abiotic stresses severely affect plant growth and productivity worldwide. In Mediterranean countries, salinity is considered one of the most important environmental stresses hampering legume growth and yield, particularly in semi-arid and arid regions (Hellal et al. 2012). Most legume plants are sensitive to high concentrations of salts in the soil. Salinity affects almost all aspects of plant development (Shrivastava and Kumar 2015). Salinity adversely affects plant growth by decreasing plant's ability to absorb water from soil along with the accumulation of toxic ions (Na+ and Cl−) in cell tissues (Dell’Aversana et al. 2021). The imposed osmotic and ionic stress occurring reduces cell expansion, hinders tissue growth leading to the reduction of grain yield and quality (Farooq et al. 2017, Dell'Aversana et al. 2021). Roots exposure to high chloride (Cl− ions) triggers a long-distance signal resulting in leaf apoplastic pH alkalinization, leaf Abscisic acid (ABA) redistribution, stomata closure and photosynthesis impairement (Geilfus 2018). The soil salinity also decreases nodulation, nodule weight, nitrogen fixation, nitrogenase activity, N content and nitrogen fixation (Cordovilla et al. 1994).
It is well established that salt stress alters plant growth and development by reducing nutrient uptake, which eventually affects cell function and alters cellular, biochemical and metabolic activities (Abid et al. 2017; Assaha et al. 2017). To cope with this, legumes have developed complex mechanisms to overcome the harmful effect of Na+ on cells (Tester and Davenport 2003; Hassan et al. 2016).
Several strategies have been developed in order to improve legume growth under salinity stress and mitigate the toxic effect of salt at different stages of plant development (Ashraf and Akram 2009). Breeding for salt-tolerant genotypes and the identification of novel salinity tolerance loci can be an approach to alleviate the harmful effect of salt stress (Atieno et al. 2021). Genetic transformation technology offers the possibility to increase plant tolerance to salinity, through the genetic manipulation of genes known to be involved in plant response and/or tolerance to saltn stress (Mishra et al. 2014a, b). Additionally, the combination of genetic engineering and traditional breeding tools appears to be more efficient for developing crops adapted to salt stress (Anwar and Kim 2020). The Plant Growth Promoting Bacteria (PGPB)-legume can be used as a cost-effective way to increase salinity tolerance and boost plant growth. Particularly, the Plant Growth Promoting Rhizobia (PGPR)-legumes symbiosis is naturally occurring and it contribute to salt stress tolerance (Wang et al. 2016a, b). AMF is another alternative that acts as growth regulator and mitigates the detrimental effects of salt stress on plants, as well as, enhancing plant growth and yield (Hashem et al. 2014). Whereas, the synergistic interactions of PGPR and AMF with legumes can multiply the ameliorative capacity under salinity stress (Nadeem et al. 2014). Salt stress can also be alleviated by the application of seed or plant priming (Azooz 2009; Dawood and El-Awadi 2015; Sagervanshi et al. 2021). In this review, we describe (1) the effect of salinity on legume growth and development, (2) the molecular responses to salt stress in legume plants and (3) the different strategies to alleviate salinity stress in this family. The tolerance mechanisms conferred by legumes' association with Rhizobium (Rhizobium-legume symbiosis) are also highlighted.
Adverse effects of salt stress on the growth and development of legumes
Effects of salt on legume seed germination
Seed germination is one of the most critical stages in seedling establishment, representing the first contact with the encounting environment, particularly water and soil (Tlahig et al. 2021). Numerous studies have reported reductions in seed germination along with a delay in this process in legumes with increasing salinity levels (Sidari et al. 2008; Farissi et al. 2011; Bouallègue et al. 2017; Anaya et al. 2018; Tlahig et al. 2021). The impact of salt stress on seed’s germination was highly variable depending on plant species (Table 1). High sodium chloride (NaCl) concentrations limit cell division and expansion, which obstruct seed germination and induce cell death (Keshavarzi 2011). Salt inhibition of seed germination is mainly attributed to the restriction of water uptake and ion toxicity on the embryo. This results in the obstruction of seed’ reserves mobilization due to the inhibition of hydrolytic enzyme activities mainly α-amylase, β-amylase and α-glucosidase (Sidari et al. 2008; Farissi et al. 2011; Farooq et al. 2015). Other authors explained this inhibition as the suppression of ethylene production during imbibition. While others have linked it with the decrease in the gibberellin (GA) content due to the negative regulation of GA biogenesis pathway by salt stress along with the activation of ABA biosynthesis pathway. Consequently, this variation leads to a reduction in the GA/ ABA ratios showing that these two phytohormones are key determinants of seed germination (Meng et al. 2016; Shu et al. 2017; Chang et al. 2010).
Table 1.
Effect of salt stress on seed's germination of some legume’s species
| Plant species | Salt concentration | Reduction (%) | References |
|---|---|---|---|
| Arachis hypogaea L. | 200 mM | 45 | Desheva et al. (2020) |
| Acacia longifolia subsp. longifolia | 200 mM | 80–85 | Welgama et al. (2019) |
| Centrosema pubescens | 16 dS/m | 55 | Sevanayak et al. (2020) |
| Cicer arietinum | 7.47 g/L | ≈ 87 | Lavrenko et al. (2019) |
| Clitoria ternatea | 16 dS/m | 70 | Sevanayak et al. (2020) |
| Glycine max L. | 300 mM | 70–80 | Kumar (2017) |
| Lathyrus odoratus L. | 21.6 dS/m | ≈ 15 | El-Serafy et al. (2021) |
| Lotus ornithopodioides L. | 250 mM | 20–44 | Hajri et al. (2018) |
| Macroptilium atroperpureum | 16 dS/m | 35 | Sevanayak et al. (2020) |
| Macrotyloma uniflorum | 20 dS/m | ≈ 25 | Pantola et al. (2017) |
| Medicago ciliaris L. | 200 mM | ≈ 64–78 | Mbarki et al. (2020) |
| Medicago intertexta L. | 150–200 mM | 97–98 | Mbarki et al. (2020) |
| Medicago sativa | 200–250 mM | 50–79 | Bicakci et al. (2018) |
| Medicago scutellata L. | 150–200 mM | 97 | Mbarki et al. (2020) |
| Phaseolus vulgaris L. | 85.5 M | 42 | Alsaeedi et al. (2017) |
| Phaseolus vulgaris L. | 300 mM | 40 | Mansouri et al. (2019) |
| Phaseolus vulgaris L. | 150 mM | 12–34 | Yu et al. (2019) |
| Sulla coronaria (L.) | 150–200 mM | 60–76 | De Rossi et al. (2021) |
| Trifolium rubens | 40 mM | 80 | Kołodziejek (2018) |
| Vachellia karroo | 600 mM | 100 | Kheloufi et al. (2017) |
| Vicia faba L. | 200 mM | 55 | Anaya et al. (2018) |
| Vigna unguiculata (L.) Walp | 12 dS/m | 15–40 | Islam et al. (2019) |
| Vigna unguiculata (L.) Walp | 250–300 mM | 100 | Osman et al. (2019) |
| Vigna radiata L. | 15.6 dS/m | ≈ 50 | Dutta and Bera (2014) |
| Vigna radiata L. | 160 mM | 48–49 | Podder et al. (2020) |
| Vigna radiata L. | 150 mM | 70 | Ghanbari et al. (2020) |
Effects of salt on legume growth
It is well established that salt stress inhibits plant growth in many plant species, even in legumes’ species (Luo et al. 2006). Salt induced growth arrest was reported in many Fabaceae plants. Pitann et al. (2011) have found that salt stress application induced a significant decrease in legume growth attributes. Similar findings were also underlined by Abdul Qados (2011). They reported that increasing NaCl negatively affected plant height, number of leaves and leaf area of faba bean cultivars. Deleterious effects of increased salinity have also been reported in pea and chickpea plants’ growth and development (Yousef et al. 2020). In soybean, exposure to 150 mM of NaCl for 7 days resulted in the inhibition of plant growth (Ning et al. 2018). Mung bean (Vigna radiata) seedlings’ growth inhibition in response to increased NaCl concentrations as been reported by Sehrawat et al. (2019). Bojović et al. (2010) explained the growth arrest by the disturbance of the ionic and osmotic balance caused by excessive salinity, which ultimately leads to plant destruction. Indeed, salt stress induces a decrease in soil water potential, disruption of nutrient uptake, disturbance of ionic balance and alteration of photosynthetic enzymes (Sheidaei et al. 2011; Saghari et al. 2020). Na+ and Cl− accumulation induced a decrease in photosynthesis and quantum yield due to chlorophyll degradation. The growth arrest due to salt stress can also be attributed to the lack of cell wall acidification (Pitann et al. 2011). The aforementioned structure is mediated by the activity of plasmalemma H+-ATPase and cell-wall-loosening enzymes that stimulate plant growth by prompting cell growth and enlargement (Pitann et al. 2011). Therefore, the disturbance in this process decreases cell growth rate, which ultimately results in plant growth reduction (Pitann et al. 2011; Farooq et al. 2017).
Effects of salt on legume physiology
Plant exposure to salt stress triggers a wide range of physiological changes. Several studies have underlined the inhibitory effect of salt stress on biochemical processes, of which photosynthesis is the most important (Qados 2011). Salt stress alters photosynthetic pigments, which influence the photosynthesis activity (Garg and Singla 2004; Sheidaei et al. 2011; Hniličková et al. 2019; Nadeem et al. 2019; Najar et al. 2019; Saghari et al. 2020). Ahmad et al. (2018) explained the alteration in plant photosynthesis activity by Mg2+ uptake restriction and the alteration in photosynthetic pigments mostly chlorophyll by the activation of chlorophyllase enzymes. NaCl stress triggers the synthesis of reactive oxygen species (ROS) (superoxide, hydrogen peroxide, etc.), which are components of oxidative stress. However, the ROS over-accumulation harms the chloroplast layers, inhibits Rubisco and leads to stomatal closure which ultimately result in a significant reduction in plant growth and productivity (Ahmad et al. 2017; Alzahrani et al. 2019b).
Photosynthesis impairment induced by osmotic stress that occurred as a result of salinity causes irreversible metabolic imbalances and prompts the synthesis of ROS (Geilfus et al. 2015). Normal concentration of ROS is required for cell signaling. However, their overproduction induces distinct changes in cell biochemistry in terms of membrane permeability. Indeed, ROS accumulation constrains cell metabolism through the stimulation of DNA, proteins and lipids oxidation which can cause serious damage to cellular processes such as lipid peroxidation, protein degradation, inactivation of enzymes, damage to nucleic acids, disruption in normal cell metabolism, and damage to cell membrane, which leads to cell death (Rohman et al. 2020; Sarker and Oba 2020). ROS accumulation has been reported in many legume species in response to salinity (Geilfus et al. 2015; El-Esawi et al. 2019). Hydrogen peroxide (H2O2) content subsequently increased in faba bean plants upon salinization (Geilfus et al. 2015). Alqarawi et al. (2014) explained this hazardous effect to the deleterious impacts of salt stress on the composition of polyunsaturated fatty acids, which ultimately cause membrane dysfunction.
Effects of salt on legume yield
Salt stress inhibits legumes yield by 12–100% due to the alteration of plant morphological and physiological attributes (Farooq et al. 2017). A significant decrease in yield attributes namely the number of branches, the number of pods, the number of seeds per plant and the seed’s yield was recorded in lentil (Lens culinaris Medik.) when cultivated in the presence of 100 mM of NaCl (Yasir et al. 2021). In faba bean, yield components including number of pods per plant, number of seeds per pod and seed weight was adversely affected by increasing concentrations of NaCl (Qados and Moftah 2015). Indeed, the number of pods, the number of seeds and seed weight decreased by 54%, 45% and 46% respectively when plants were grown in the presence of 200 mM of NaCl (Qados and Moftah 2015). This reduction is attributed to the negative effect of salt on pollen viability, stigma receptivity and photo-assimilates supply during reproductive stages (Farooq et al. 2017; Khan et al. 2017). The exposure to salinity causes ovarian disruption and injuries which lead to premature fruit drop and yield decrease as reported in Mungbean (V. radiata) (HanumanthaRao et al. 2016).
Salt stress adversely affects grain legume composition and quality. Salt stress significantly reduces protein content, carbohydrate and polysaccharide. The decrease in carbohydrate and polysaccharide contents is mainly attributed to ion toxicity, reduced photosynthesis and nutritional imbalance while the reduction in protein content is the result of the decline in nitrogen supply for soil (Swaraj and Bishnoi 1999; Qados 2011).
Effects of salt on biological nitrogen fixation
Symbiotic nitrogen fixation by rhizobia bacteria provides a source of biologically available nitrogen to legume plants (Vance et al. 2015). However, salt stress obstructs nodule formation and, therefore, the nitrogen fixation process (Bruning and Rozema 2013). It was previously reported that faba bean irrigation with 35% sea water inhibits nodule formation (Fahmi et al. 2011). This decrease was estimated to be 50% in Lotus japonicus cultivated in the presence of 50 mM NaCl (López et al. 2008). Similar inhibition rates have been recorded by Fahmi et al. (2011) in salt stressed faba bean plants. Katejri et al. (2003) have estimated the decline of nitrogen fixation in the same species under saline conditions by 16–24% In Medicago truncaluta, the reduction in nitrogen fixation rate was less pronounced (14% at 50 mM NaCl) (López et al. 2008). Manchanda and Garg (2008) explained this phenomenon by root hair formation obstruction due to salt stress injuries which ultimately reduce the nodule’s number per plant and the amount of fixed nitrogen per unit of nodules. Furthermore, root-hair curling, a main step in rhizobia colonization was negatively impacted by NaCl-induced stress (Zahran and Sprent 1986).
Salt stress disturbs nitrogenase activity, the key enzyme involved in atmospheric N2 fixation by Rhizobia strains. Furthermore, the total as well as specific nitrogenase activity of root nodules decreases with salinity stress (Yousef and Sprent 1983; Fernández-Pascual et al. 1996). Nevertheless, glutamine synthetase and glutamate synthase, which are required for ammonium assimilation display a different tolerance pattern in salt stress conditions (Bernard and Habash 2009; Betti et al. 2012). It was reported that glutamine synthetase is more resistant to salinity than glutamate synthase, which ultimately reduces the assimilation of ammonium under saline conditions (Cordovilla et al. 1999).
The set of symbiotic interaction relies on the activation of a sequence of events coordinated by the host plant and the symbiotic rhizobia (Zhang et al. 2019). The signaling process starts with the exudation of flavonoids from the leguminous roots, which stimulates the secretion of lipo-chito oligosaccharides nodulation factors (NOD). The NOD is defined as the fundamental molecular signal that trigger the nodulation program (Bruning and Rozema 2013). However, salt stress adversely affects the signal exchange. Furthermore, salt stress increases the biosynthesis of nod factors (NFs) and induces changes in their structures as previously reported by Hasanuzzaman et al. (2020). These newly generated NFs induce morphological and biochemical changes namely, root hair deformation, intra-and extra-cellular alkalinization, ROS accumulation, phosphatidic acid and diacylglycerol formation (Hasanuzzaman et al. 2020). The root hair deformation has an inhibitive action on rhizobia colonization, which ultimately reduces the infection rate, weight and number of nodules (Egamberdieva et al. 2014).
The harsh effects of salt stress on plant growth, physiology, productivity, and nitrogen fixation are the direct result of the modification of molecular machinery. Thus, the thorough understanding of plant stress response mechanisms will provide valuable information for improving crop engineering for salt stress tolerance.
Molecular responses to salt stress in legume plants
Legumes' genetic response to salt
Salt stress affects plants in multiple ways: (1) osmotic injury due to high concentrations of solutes which ultimately leads to a water deficit; (2) ionic stress as a result of the disturbance of K+/Na+ contents and (3) nutritional disorders, which arise from the availability, absorption and transport of nutrients within the plant (Assaha et al. 2017). In response to these harmful conditions, legumes evolve complexed mechanisms to mitigate the deleterious effects of stress and ensure plant survival under these harmful conditions. These mechanisms aim to lower the toxic effect of Na+ and preserve a low cytosolic Na+ concentration along with a high K+/Na+ ratio by reducing Na+ influx into root cells, Na+ compartmentation into vacuoles and Na+ exclusion from root cells (Tester and Davenport 2003; Hassan et al. 2016).
Salt stress sensing and signal transduction in legumes
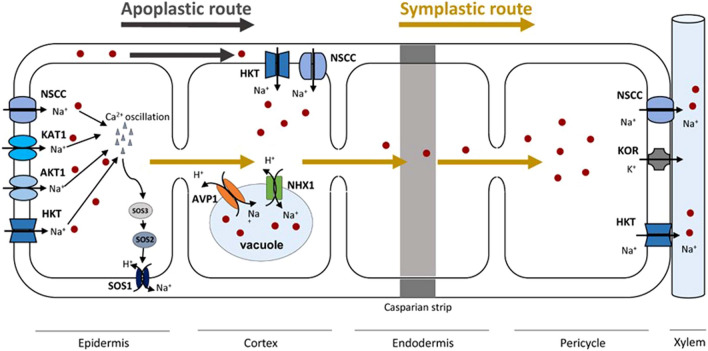
The high amounts of Na+ detected in soil under salt stress conditions modify the electrochemical gradient which prompts Na+ influx from the soil into root cells (Blumwald et al. 2000). Na+ entry occurs via the symplastic and apoplastic pathways (Isayenkov and Maathuis 2019). The apoplastic pathway (through cell walls) which represents only 1% of the transpirational volume flow, is described in the literature as a direct and continuous flow from the outside of the root cell to the xylem (Kronzucker and Britto 2011; Hossain et al. 2016). In faba bean, it has been demonstrated that short term exposure to NaCl concentrations enhanced Na+ uptake through the apoplastic pathway, which strongly underlines the importance of this transport route in Na+ uptake and signaling in salt stressed beans (Shahzad et al. 2013). In turn, the root endodermis interrupts the apoplastic transport of Na+ ions due to the deposition of hydrophobic compounds (lignin and suberin). It thus allows the formation of the framework of the Casparian strip, which constitutes a physical barrier for water and ion movement. At this level, ions and water are forced to undergo membrane control and to take the symplastic path (Ketehouli et al. 2019) (Fig. 1).
Fig. 1.
General mechanism of Na+ entry into root cells of legume plants. Na+ toxic ions enter to root cell through apoplastic or sympastic paths. The apoplastic route (grey narrow) occurs as a direct and continuous flow from the outside of root cell to the xylem. Within this route, Na+ transport is interrupted in the endodermis by the casparian strip. The symplastic pathway is mainly operated through transporters. Within the latter path, Na+ absorption by epidermis cells is promoted by Nonselective Cation Channels (NSCCs), High Affinity Potassium Transporters (HKT) proteins and the Shaker Type K+ channels: AKT1 and KAT1. Na+ influx cause membrane depolarization, Ca2+ activation and the generation of Ca2+ oscillations that serve as a second messenger that activate SOS pathway that trigger Na+ exclusion to the apoplast. Apoplastic Na+ ions reach the cortex through the following transporters: NSCC and HKT. At this level, Na+ ions can be stocked in vacuoles promoted by NHX1 and AVP1 transporters. Non sequestrated ions reach the xylem trough NSCC and HKT transporters
The Na+ root uptake through the symplastic pathway requires the intervention of specific transporters (Kronzucker and Britto 2011). Initial entrance of Na+ ions into the cytoplasm of root cortical cells mainly occurs either via non-selective voltage-dependent cation channels (NSCCs) or through the intervention of sodium transporters (Demidchik and Maathuis 2007). Nonselective cation channels (NSCCs) are encoded by two gene families: glutamate receptor-like channels (GLRs) and cyclic nucleotide-gated channels (CNGCs) (Zhang et al. 2002). Several studies have shown that DA-NSCCs (Depolarization-activated nonselective cation channel) transporters are involved in K+ loading into xylem and its redistribution in Phaseolus vulgare plants (Zhang et al. 2004; Yang et al. 2005). Na+ influx into cells induces membrane depolarization, which instigate the K+ leak through the intervention of depolarization-activated K+-selective outwardly rectifying KOR channels. Na+ entry and accumulation in the cytosol prompted ROS production which can lead to the activation of NSCC channels providing an additional avenue for further K+ leakage (Percey et al. 2014). Other transporters like the High Affinity Potassium Transporters (HKT) proteins and the Shaker Type K+ channels (AKT1 and KAT1), have been described to be involved in Na+ uptake in many plant species, including Arabidopsis thaliana, Oryza sativa and the halophyte Suaeda maritima (Kader and Lindberg 2005; Wang et al. 2007; Horie et al. 2009). However, no previous work had demonstrated their presence in legume plants and their implication in sodium transport or salt stress signaling.
The pronounced increase of cytoplasmic Na+ occurring because of Na+ influx disturbs the enzymatic functions of plant cells. Na+ internal concentration can be reduced mainly through three distinctive processes that enable: (1) reduction if Na+ entry into cells; (2) trigger of Na+ compartmentation into cell vacuoles and (3) stimulation of Na+ efflux out of cells (Ji et al. 2013). The NSCCs-orchestrated influx of Na+ ions causes membrane depolarization, Ca2+ activation and the generation of Ca2+ oscillations and a stress signal (Tester and Davenport 2003). The Ca2+-generated signal serves as a second messenger and activates specific transporters and channels involved in sodium extrusion. This signal is perceived by SOS3 (Salt Overly Sensitive 3), a sensor protein that interacts with the serine/threonine protein kinase SOS2 (Manchanda and Garg 2008). This interaction causes the phosphorylation of the plasma membrane Na+/H+ exchanger SOS1, known for its involvement in Na+ retrieval from cells. In roots, about 70–95% of the Na+ ions entering in the roots via the symplastic route are extruded to the apoplast (Tester and Davenport 2003). The SOS1 mechanism of Na+ efflux from roots and its loading into xylem has been intensively studied in Arabidopsis (Shi et al. 2002). SOS homologs had been identified in many plant species including legumes (Liu et al. 2015a; Quan et al. 2016). SOS1 and SOS2 gene transcripts increased substantially and subsequently in salt stressed alfalfa (Medicago sativa) genotypes (Quan et al. 2016). SOS1 expression was significantly induced after 2 days of salt stress exposure which contribute to Na+ extrusion in soybean (Glycine max) S111-9 genotype (He et al. 2015). In salt-tolerant Medicago falcata, SOS1 expression was upregulated in response to salt stress. This salt-enhanced tolerance was correlated with the higher expression of the SOS1 gene, required for more efficient Na+ retrieval from root cells (Liu et al. 2015a).
Sodium vacuolar compartmentation is considered one of the key strategies used by plants against salinity (Yang et al. 2005). It allows the removal of potentially toxic Na+ ions from the cytoplasm and cellular osmolarity increase, which are required for a better tolerance to osmotic stress (Ahmad and Rasool 2014). Sodium/hydrogen exchanger (NHX) proteins, which belong to the Na+/H+ antiporter family of transporters, play a crucial role in salt tolerance in many plant species through prompting Na+ accumulation in vacuoles and pH regulation (Zahran et al. 2007). A number of genes encoding vacuolar Na+/H+ antiporters have been isolated from diverse legume plants including VrNHX1 from mung bean (Mishra et al. 2014b), TrNHX1 from Trifolium repens and MtNHX1 and its ortholog’ from Medicago truncaluta (Al-Farsi et al. 2020). In legume plants, several reports have stated the involvement of the NHX gene family in plant response to salt (Tang et al. 2010; Mishra et al. 2014b; Quan et al. 2016). For instance, it has been found that MsNHX1, an Na+/H+ antiporter of M. sativa, was highly expressed in leaf tissue under salt stress, underlying its putative role in Na+ compartmentation in leaf vacuoles (Yang et al. 2005). The constitutive expression of AtNHX1 in soybean (G. max) confers a better tolerance to salt stress by prompting the accumulation of high amounts of toxic Na+ ions (Li et al. 2010). Similar findings were also reported in the model legume Lotus corniculatus, in which the soybean NHX1 gene was overexpressed (Sun et al. 2006). In mung bean, the increased salt tolerance was correlated with a higher K+/Na+ ratio in the aerial parts through the accumulation of high amounts of Na+ ions in roots, which is considered an efficient strategy of salt tolerance through the restriction of Na+ movement in roots (Mishra et al. 2014b).
The tonoplast H+- pyrophosphatases are H+ pumps that allowed the acidification of vacuoles through pyrophosphate hydrolysis and orchestrate the active proton transport (Segami et al. 2014). They ensure the generation of H+ motive force required for ion entry into vacuoles and thus the regulation of cell turgor (Bao et al. 2016). Genetic manipulation of H+-pyrophosphatases in many plant species including legumes conferred a better tolerance to salt as a result of increased Na+ accumulation in leaves and roots (Bao et al. 2009; Jha et al. 2013; Bassil and Blumwald 2014). The co-expression of Zygophyllum xanthoxylum ZxVP1-1 and ZxNHX genes in alfalfa (M. sativa) resulted in a significant accumulation of Na+ in leaves and roots. Transgenic alfalfa plants also showed less injury from NaCl-induced stress, which ultimately conferred a better tolerance to salt (Bao et al. 2016). The overexpression of the AtAVP1 gene in transgenic alfalfa plants exhibited high Na+ amounts, which was likely related to the enhanced transport efficiency of the vacuolar Na+/H+ antiporter resulting from the overexpression of the AtAVP1 gene (Bao et al. 2009). In soybean, co-expression of both A. thaliana AtNHX1 and AtAVP1 genes confers better salt tolerance to transgenic plants. This increased tolerance to salt was mainly assessed due to the ability of transgenic plants to sequester toxic Na+ ions in their vacuoles as a result of the activity of both NHX1 and AVP1 transporters (Nguyen et al. 2019a) (Fig. 1).
Transcriptional regulation of salt responsive genes in legumes
Transcription Factors (TFs) play key roles in the transcriptional regulation of plant responses to salinity and other abiotic stresses. Many transcription factors involved in the salt stress response had been identified (Fig. 2). The most studied ones are Zinc Finger Proteins (ZFPs), AP2/EREBP (APETALA2/Ethylene-Responsive Element Binding Proteins), NAC TFs, bZIP (basic leucine zipper domain) and WRKYs family of TFs, which are involved in plant response to salt stress. The overexpression of genes encoding for these TFs confers a better tolerance to salt stress. The expression of ZFP protein was highly up-regulated in alfalfa plants when subjected to salt stress (Chao et al. 2009).
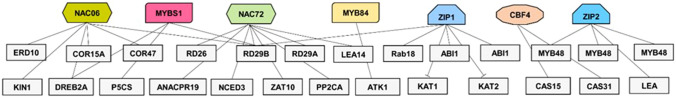
Fig. 2.
A schematic representation of the transcriptional regulatory network involved in the transcriptional regulation of salt responsive genes in legume plants. Salt-responsive genes (symbolized by grey boxes) can be regulated by one or several transcription factors (represented in colored box) which underline the complexity of the molecular response of legume species to salt stress
The AP2/ERF are TFs belonging to the superfamily of AP2/EREBP regulate the transcription of target genes through direct interaction with dehydration responsive elements found at the promoters of target genes and instigate salt stress response (Sakuma et al. 2006). The expression of AP2/EREBP subfamily members were significantly induced by salt in legume plants (Li et al. 2005; Pennycooke et al. 2008). The CCAAT motif-binding factor (CBF)gene expression was significantly regulated by salt stress in soybean (Li et al. 2005). Ethylene- responsive transcription factor (ERF-WIN1), which also belongs to the AP2/EREBP subfamily has been reported to be highly induced in response to salt and osmotic stress in chickpea (Kaashyap et al. 2018). In M. sativa, the expression of CBF4 gene was highly induced in salt tolerant genotype in response to high NaCl concentrations, which might explain the increased tolerance observed in this genotype. The overexpression of Medicago truncatula CBF4 enhanced plant tolerance to salt stress in transgenic plants by regulating the expression of downstream genes (Li et al. 2011a). Li et al. (2011a) identified two of the MtCBF4 regulated genes; MtCAS15 (cold acclimation-specific 15) and MtCAS31 (cold acclimation-specific 31), encoding for dehydrin proteins. MtCAS31 was later proposed as a key actor in the autophagic degradation pathway. Li et al. (2020) have demonstrated that MtCAS31 interacts with the plasma-membrane intrinsic protein MtPIP2;7, a plant aquaporin protein, and induces the degradation of this latter. In Cicer arietinum, the dehydration responsive element (DRE/CRT) showed differential expression in the salt-tolerant genotype in comparison with the sensitive genotype (Kaashyap et al. 2018).
Another important transcription factors family that has been widely associated with salt tolerance in legumes such as soybean (G. max) and peanut (Arachis hypogaea) is the NAC family. In peanut, several NAC genes were differentially expressed in response to salt which strongly suggests that these TFs are involved in the response to salt (Yuan et al. 2020). In chickpea, RNAseq analysis revealed that the expression of many NAC genes was significantly up-regulated in the salt-tolerant genotype (Kaashyap et al. 2018). In soybean, the overexpression of GmNAC06 enhanced plant tolerance to NaCl excess, by controlling the K+/Na+ ratio prompting proline and glycine betaine accumulation in transgenic plants (Li et al. 2021). The introduction of OoNAC72, a NAC-Type Oxytropis ochrocephala transcription factor in A. thaliana, increased transgenic plants tolerance to salt and drought. This enhanced resistance was correlated with the up-regulation of the expression of many stress-responsive genes (RD29A, RD29B, RD26, LEA14, ANACPR19, ZAT10, PP2CA and NCED3), suggesting that this transcription factor regulates the expression of these genes (Guan et al. 2019). Constitutive expression of chickpea CarNAC4 in A. thaliana enhanced salt tolerance in transgenic plants (Yu et al. 2016). CarNAC4 overexpression enhanced the expression of stress-responsive genes, including RD29A, ERD10, COR15A, COR47, KIN1 and DREB2A which clearly indicated the involvement of CarNAC4 as a transcription factor regulating salt-related genes.
The basic leucine zipper (bZIP) family of TFs is involved in many aspects of plant development and interaction with environment. They regulate gene expression by interacting with specific cis-elements that include the ABRE (ABA-responsive element) (Ayra et al. 2018). Previous reports had demonstrated the responsiveness of bZIP genes to salt stress in legume species such as common bean (Phaseolus vulgaris), soybean (G. max) and luserne (Medicago truncatula) (Liao et al. 2008; Hiz et al. 2014; Wang et al. 2015a). In P. vulgaris, many bZIP genes displayed a differential expression in response to salt excess. The authors also suggested that PvZIP genes are key regulators that interact with other TFs to control the expression of stress-responsive genes in common bean (Ayra et al. 2018). The soybean GmbZIP2 expression was significantly up-regulated in salt stressed plants and triggered the expression of several stress response genes required for ion homeostasis, ROS scavenging or scaffolding molecule that ensure protein proper functioning (Yang et al. 2020). Among the regulated genes, the authors identified the following genes: GmMYB48, GmWD40, GmDHN15, GmGST1 and GmLEA. Another member of the bZIP transcriptional regulator family; GmbZIP1 was found to be involved in the set of stress responses in soybean. For instance, Gao et al. (2011) have demonstrated that the overexpression of GmbZIP1 conferred multiple stress tolerance mainly to salt, drought and cold. This increased tolerance was linked to the regulation of ABA-stress regulated target genes, namely abscisic acid insensitive 1 and 2 (ABI1 and ABI2), Desiccation-Responsive RD29B, and rab-related (responsive to ABA) Rab18 genes. The GmbZIP1 factor was also reported as a negative regulator of stomatal closure by controlling the expression of KAT1 and KAT2 which encode for inward-rectifying K+ channel subunits, involved in stomatal aperture (Kim et al. 2004; Gao et al. 2011).
The WRKY transcription factors play a main role in the transcriptional regulation during legume response to salt stress. WRKY72 and WRKY73 were differentially expressed in salt tolerant genotype of chickpea. Thus, these two TFs were proposed as key actors in chickpea tolerance to salt stress (Kaashyap et al. 2018). The expression of MsWRKY11 was highly up regulated in alfalfa plants suffering from salt stress injuries. The overexpression of this TF in soybean enhanced transgenic plants tolerance and improved plant physiological attributes under salt stress conditions. This increased tolerance was linked to the accumulation of proline and the activation of ROS scavenging enzymes (Wang et al. 2018a). Similar findings were also recorded in A. thaliana overexpressing the G. max GmWRKY54 gene (Zhou et al. 2008).
Another family of transcriptional regulators that seems to be involved in legumes response to salinity is the MYB (v-myb avian myeloblastosis viral oncogene homolog) family. The expression of MtMYBS1 gene is inducible by NaCl (Dong et al. 2017). Its introduction in A. thaliana enabled the transgenic plants to overcome salt stress through the regulation of the expression of P5CS, a key gene involved in the proline biosynthesis pathway (Dong et al. 2017). The constitutive expression of M. sativa MsMYB4 in A. thaliana improved the plants’ salinity tolerance in an ABA-dependent manner (Dong et al. 2018). In soybean, GmMYB84 gene was highly induced in salt stressed plants. The heterologous expression of the GmMYB84-encoding gene in A. thaliana confers a better tolerance to salt stress through a direct interaction with the promoter of GmATK1, the homolog of Arabidopsis K+ Transporter 1 (AKT1) involved in K+ acquisition and homeostasis under saline conditions (Zhang et al. 2020). Constitutive expression of M. truncaluta MYBS1 in A. thaliana unraveled a positive regulation of the expression of salt stress responsive genes mainly, RD22, RD29A, RD29B, P5CS, and DREB2A under saline conditions (Dong et al. 2017). The up-regulated genes are associated with proline biosynthesis (P5CS), Dehydration-Responsive proteins (DREB2A, MYB2) and Desiccation-Responsive proteins (RD22, RD29A and RD29B).
Expression of salt stress responsive genes
To prevent stress damages and repair stress-induced injuries, plants have evolved several pathways to ensure cell survival even at metabolically inhibitory levels of ionic and osmotic stresses. These pathways mainly include ion homeostasis (Na+ exclusion and Na+ sequestration), osmolytes accumulation, induction of proteins involved in stress responses, and restoration of osmotic balance (Manchanda and Garg 2008). The induction of stress proteins, more likely Late Embryogenesis Abundant (LEA), and Heat Shock Protein (HSP) families, and their involvement in legumes’ response to salt excess will be discussed in this sub-section.
LEA (Late Embryogenesis Abundant) proteins are members of a large group of hydrophilic, glycine-rich proteins found in a wide range of plant species (Magwanga et al. 2018). LEA proteins had been studied in soybean plants exposed to abiotic stresses (Phang et al. 2008). It was previously reported that the constitutive expression of either Soybean PM11 or PM30 in E. coli enabled the transgenic bacteria to grow in saline growth conditions which strongly suggests that these genes play an important role in soybean tolerance to salinity (Lan et al. 2005). In M. truncatula, salt stress induced the accumulation of transcripts from LEA genes belonging to groups 2, 3, 4, 6 and 7 (Battaglia and Covarrubias 2013). The overexpression of G. max GmLEA2-A in transgenic Arabidopsis conferred tolerance to salt stress suggesting that this gene is a key actor in plant response to salt (Wang et al. 2018a, b).
Another family of stress proteins that plays a crucial role in legumes’ response to salt excess is the HSP family. HSPs are involved in refolding misfolded proteins and degrading damaged proteins under stress conditions (Zhou et al. 2013). Büyük et al. (2016) showed that PvHSP70 was inducible by salt stress in common bean (P. vulgaris). Other HSPs proteins have been reported as involved in P. vulgaris response to salt stress. This is the case of PvHSP90 and PvDnaJ3, whose expression was significantly regulated in salt stressed plants (Hernández-Lucero et al. 2014). The expression of GmHsp90A2, GmHsp90A4, GmHsp90B1, GmHsp90C1.1 and GmHsp90C2.1 was highly induced by salt stress in G. max. Their overexpression in A. thaliana conferred increased tolerance to salinity by minimizing the deleterious effects of salt (Xu et al. 2013).
Epigenetic regulation of legumes' response to salt
Epigenetic regulation of gene expression plays a crucial role in plant response to salinity. This regulatory process, operates through different mechanisms involving DNA methylation, histone modifications and non-coding RNA that induce gene activation or knock out (Salgotra and Gupta 2019).
DNA methylation
DNA methylation is the foremost epigenetic mode of regulation observed in eukaryotes. This process can be simply defined as the addition of a methyl group on C5 of the cytosine base to form 5-methylcytosine (Salgotra and Gupta 2019). Plant DNA methylation is found in three different contexts; CG, CHH and CHG, wherein H can be any base except for guanine (Windels et al. 2021). In plants, DNA methylation is basically catalyzed by a group of methyltransferase enzymes (Al-Lawati et al. 2016). DNA methylation is usually associated with gene silencing, whereas DNA demethylation allows gene activation (Salgotra and Gupta 2019). In pigeon pea, salinity induced a global decrease of DNA methylation, while 26% increase in global DNA methylation has been recorded in alfalfa (M. truncatula) plants irrigated with 20 dS/m (Al-Lawati et al. 2016; Awana et al. 2019). Under salt stress conditions, a positive correlation was demonstrated between global DNA methylation and methyltransferase genes transcripts. The authors also underlined the preponderant role of DNA methylation in salt tolerance acquisition in alfafa by applying a DNA methylation inhibitor that increased plant susceptibility to salt (Al-Lawati et al. 2016). Soybean exposure to salt enhanced global DNA demethylation mainly in salt tolerant genotype. Profound demethylation analysis showed that CG and CHG contexts were more critical than CHH in gene regulation of soybean adaptability to salinity (Liang et al. 2019). This increase was positively correlated with an increase in DNA demethylases transcripts (Al-Lawati et al. 2016). Van Dam et al. (2009) suggested that the high incidence of the DNA demethylation process under stressful conditions could be linked to the role of chromatin demethylation as a transcriptional switch for several stress-regulated genes. In soybean salt-tolerant genotype, some differentially methylated genes are involved in gene transcription, DNA repair, RNA splicing, protein processing in the endoplasmic reticulum processes (Liang et al. 2019). Overall, the epigenetic changes occurring at the DNA methylation level appears to be a key regulatory process in plant response to salt stress. This epigenetic process of regulation can occur either through demethylation or methylation depending on plant species and stress period.
Non-coding RNAs
Non-coding RNAs (ncRNAs) are functional RNAs with low-protein coding potential. They can be classified according to their length into small ncRNAs (sRNAs) (18–30 nucleotides), medium-sized ncRNAs (31–200 nucleotides) and long non-coding RNAs (LncRNAs) (more than 200 nucleotides) (Wang et al. 2017). microRNAs (miRNAs) are usually 21–23 nucleotides sRNAs deriving from intergenic regions and produced from single-stranded primary miRNAs (Bartel 2004; Alzahrani et al. 2019a). With a unique hairpin structure, they are known for their interaction with messenger RNAs (mRNAs) 3ʹ untranslated regions (3ʹUTRs), which results in the down-regulation of target genes (Windels et al. 2021). miRNAs have recently emerged as a key regulator of gene expression at the transcriptional and post-transcriptional levels (Long et al. 2015). To date, with the development of high-throughput sequencing technology, a great number of small RNAs have been discovered in many plant species including legumes, particularly, Medicago truncatula and Medicago sativa (Lelandais-Brière et al. 2009; Long et al. 2015). Lelandais et al. (2009) have identified several salt responsive miRNAs in M. truncatula roots using a high-throughput sequencing strategy. Among the regulated miRNAs, miR393 expression was significantly repressed in M. truncatula salt stressed roots (Long et al. 2015). miR393 is involved in the regulation of auxin signaling pathway actors: Transport Inhibitore Receptor 1 (TIR1) and Auxin-related F-Box 2 (AFB2) and F-box protein genes encoding for auxin receptors, which underlines the role of miR393 in response to salt stress through the regulation of auxin action (Sunkar et al. 2007). In Cicer aestivum, three miRNAs were found to be upregulated in response to salt stress. This is the case of miR156, miR396 and miR319 (Kohli et al. 2014). Those miRNAs were reported to be involved in Arabidopsis response to high salinity (Liu et al. 2008). Three novel legume-specific miRNAs, miR008, miR015 and miR015 were also identified by the same authors through high-throughput sequencing. Evaluating their expression by Real-Time-PCR revealed their high responsiveness to salt stress which strongly supports their involvement in salt stress response in chickpea (Kohli et al. 2014). Target prediction of miRNAs targets revealed that miR156 is involved in the regulation of squamosa promoter-binding protein; a transcriptional activator (Williams et al. 2005; Preston and Hileman 2013). miR159 target encodes for a key enzyme involved in ester biosynthesis named acyltransferase, while miR319 and miR396 can be involved in the regulation of serine/threonine protein kinases and Mitogen Activated Protein Kinase (MAPK) protein, known for their involvement in salt stress signaling pathways (Kohli et al. 2014). Jatan et al. (2019) found that seven distinctive miRNAs were differentially expressed in chickpea in response to salinity. Interestedly, they displayed different expression patterns. For instance, miR160, miR166, miR169, miR396, miR167 and miR171 expression was notably downregulated by opposition to miR159, whose expression was significantly induced. The regulated miRNAs mostly targeted transcriptional factors. Other miRNAs displaying a differential expression in response to high salinity have been identified in Chickpea by Khandal et al. (2017). This was the case for miR397, miR398 and miR164 whose expression was highly induced in chickpea salt stressed roots, while the expression level of miR399 was downregulated (Khandal et al. 2017). Throughout studying soybean roots response to salt stress, Sun et al. (2016) identified a total of 71 miRNAs candidates, of which 46 were responsive to salt stress. Among the regulated miRNA, miR399 was suggested to be involved in soybean root development and plasticity. Comparative expression analysis of miRNA in salt-tolerant genotype of faba bean (Vicia faba) revealed the responsiveness of 665 known miRNAs belonging to 31 miRNA families and 28 novel miRNA families. The expression pattern of most regulated miRNAs was downregulated. Target prediction showed that the regulated miRNAs modulate the expression of salt stress-related genes, namely those involved in plant hormone signal transduction, flavonoid biosynthesis, ATP Binding Cassette (ABC) transporter activity, ubiquitin-mediated proteolysis, flavonoid biosynthesis and DNA repair (Alzahrani et al. 2019a).
Besides miRNAs, other non-coding RNAs, known as long non-coding RNAs or LncRNAs are also involved in salt stress response. Lacking of protein-coding capacity, LncRNAs are typically about 200 nucleotides long and mainly located in the cytoplasm, with crapped 5'-ends and merged introns as well as poly(A) tails (Chen et al. 2019). Recent studies have underlined the importance of these non-coding RNAs in salt stress response in legumes including soybean (Glycine max.), groundnut (A. hypogaea) and Medicago truncatula (Wang et al. 2015a, b, c; Chen et al. 2019; Tian et al. 2020). For instance, soybean strand-specific transcriptome sequencing analysis allowed the identification of over 3030 LncRNAs in salt-stressed roots (Chen et al. 2019). In M. truncatula, Wang et al. (2015a, b, c) discovered that LncRNAs regulate Medicago's response to salt stress through the alleviation of ROS-induced oxidative stress. Within the same work, the authors discovered the function of several LncRNAs in salt stress response. Among the regulated lncRNAs, a functional analysis was conducted for TCONS_00116877, a LncRNA targeting the glutathione peroxidase-encoding gene (Medtr7g094600) (Wang et al. 2015a, b, c). Besides targeting genes involved in ROS scavenging, differentially expressed LncRNAs from chickpea (C. arietinum) act as regulators of several salt responses related genes, namely potassium transporter family genes, Tonoplast Intrinsic Protein (TIP) and PIP aquaporin-encoding genes, serine/threonine-protein kinase and several transcriptional regulators (AP2, bZIP, MYB, WRKY, and NAC) (Kumar et al. 2021).
Histone modifications
Post translational regulation of histones alters the expression of genes by inducing chromatin restructuration or regulatory protein recruitment. Histone-occurring modifications can be the result of acetylation, methylation, ubiquitination, phosphorylation and syccinylation reactions, etc. (Hashiguchi and Komatsu 2016; Yung et al. 2021). This process has been previously described for many plant species including A. thaliana, O. sativa, Brassica napus and Solanum lycopersicum. However, to our knowledge, histone methylation has not yet been studied in legumes.
Tolerance mechanisms in the Rhizobium-legume symbiosis
Rhizobium has a positive effect on legume subjected to salt stress by improving the activity of several molecules responsible for salt tolerance. Rhizobia synthesize 1-aminocyclopropane-1-carboxylase (ACC) deaminase, produce They synthesize ACC deaminase and produce various types of phytohormones and secondary compounds such as exopolysaccharides and regulate plant defense systems by activating plant’s antioxidative enzymes (Fig. 3). ACC synthase and ACC oxidase transcripts increase under salt stress conditions leading to an increase in ethylene production in plants. The rhizobia have mechanisms that regulate plant ACC and, consequently, ethylene levels ACC deaminase (Okazaki et al. 2004), which is the key to bacterial plant growth-promotion. The ACC deaminase cleaves ACC, the immediate precursor of ethylene in plants, to form ammonia and α-ketobutyrate (Glick et al. 2007). This multimeric enzyme belongs to the tryptophan synthase (beta superfamily) of pyridoxal phosphate-binding proteins (Nascimento et al. 2016). The gene AcdS encodes the ACC deaminase under the transcriptional control of the regulatory gene acdR which encodes a Leucine-responsive Regulatory Protein (LRP)-like protein. The acdR is a common regulator of acdS gene transcription and is present in most strain possessing the acdS gene (including Azorhizobium, Bradyrhizobium, Methylobacterium, Rhizobium, Sinorhizobium, Burkholderia, and Cupriavidus) (Nascimento et al. 2014). For example, the R. leguminosarum acdR gene deletion resulted in a loss of ACC deaminase activity (Ma et al 2003), indicating that acdR is the main gene controlling acdS transcription in R. leguminosarum. The ACC deaminase has a crucial role in symbiotic conditions. The expression of exogenous ACC deaminase from Rhizobia species increased the ability to nodulate in several plant legumes such as Medicago sativa (Ma et al. 2004), Cicer arietinum (Brígido et al. 2013), Medicago lupulina (Kong et al. 2015), Pisum sativum (Ma et al 2003) and Lotus spp. (Conforte et al. 2010) under stress conditions. Brígido et al. (2013) studied the symbiotic performance of two Mesorhizobium ciceri strains (salt sensitive and salt tolerant), transformed with an exogenous 1-aminocyclopropane-1-carboxylate deaminase gene (acdS), in chickpea plants under salinity stress. They demonstrated that by expressing an exogenous acdS gene, a salt sensitive Mesorhizobium strain was able to induce nodules in chickpea plants to the same extent as a salt-tolerant strain. Furthermore, the use of acdS-expressing rhizobia protected chickpea plants from salinity stress-induced symptoms. Kumari and Khanna (2015) showed that Mesorhizobium ciceris isolates producing ACC-deaminase enhanced chickpea growth especially under salt stress. Overall, as described by Singh et al. (2015), the ACC deaminase is a natural weapon produced by diverse bacteria against “stress ethylene”. The role of ACC deaminase-producing bacteria in protecting plants from the harmful effects of salt as well as improving plant growth is widely reported suggesting the importance of their application in the future as bio-fertilizers.
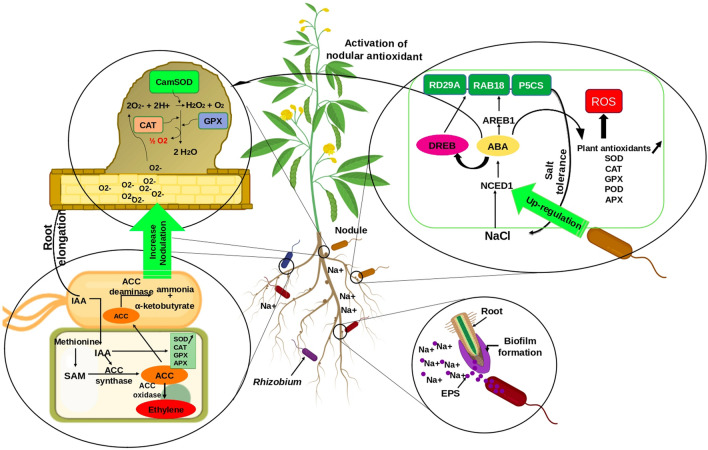
Fig. 3.
Illustration of Rhizobium sp. molecular actions leading to legume tolerance and growth promotion under salinity stress. Rhizobium strains have the ability to synthetize ACC deaminase, phytohormones (e.g. IAA) and secondary metabolites (e.g. exopolysaccharides) which can reduce the deleterious effects of salt stress and activate plant defense mechanisms through the activation of plant's antioxidative enzymes
The increased level of ethylene produced by the stressed legumes inhibits the IAA (indole-3-acetic acid, auxin) signal transduction thereby limiting IAA synthesis and transport (Sanyal and Bangerth 1998), which consequently inhibit root elongation. It has been demonstrated that plant mutants with defects in auxin transport were more sensitive to salt stress (Korver et al. 2018). Furthermore, salt stress affects auxin transport by altering the expression of PIN genes involved in auxin polar transport leading to a reduction in root meristem size (Liu et al. 2015a, b). In soybean, the GmPIN (a legume-specific PIN gene) is down-regulated by salt (Wang et al. 2015b), suggesting that IAA transport between plant cells affects plant response to saline conditions. Many rhizobacteria can synthetize IAA. Pseudomonas, Bacillus, Rhizobium, and Microbacterium are among the most active IAA producers (Tsavkelova 2011). The synthesized and secreted IAA is taken up by plant cells, which can stimulate plant cell proliferation (Glick et al. 2007). It can also promote the growth of primary and lateral roots (Ivanchenko et al. 2010) and alleviate some of the adverse effects of salt stress. The exogenous IAA enhanced the yield of faba beans under high salinity conditions (Abdel Latef et al. 2021). The IAA interferes with lipid peroxidation and/or dissociates malondialdehyde accumulated by salt-induced oxidative damage by improving antioxidant enzymes, including Superoxide dismutase (SOD), Catalase (CAT), Glutathion Peroxidase (GPX), and Ascorbate Peroxidase (APX) (Abdel Latef et al. 2021). It has been reported that increased IAA can stimulate ACC synthase (VR-ACS1) transcription and reduce the gene expression level of ACC oxidase (VR-ACO1) transcripts in mung bean (Kim et al. 2001), suggesting that IAA participate in the regulation of ethylene production. The bacteria that produce both IAA and ACC deaminase possess a significant advantage over those producing only IAA, since they can decrease ACC due to increased IAA action. IAA enhances the transcription of ACC synthase. A large amount of ACC is liberated by root, taken up by bacterial cells and finally cleaved by ACC deaminase (Gamalero and Glick 2015). As a result, IAA improves plant growth and ACC deaminase decreases ethylene production.
ABA is a key phytohormone involved in signaling pathways against abiotic stress. When plants face salinity stress, ABA regulates stress-response genes, such as Response-to-Dehydration 29A (RD29A), which originates from enhancement of DREB2 activity, the ABA-responsive gene (RAB18) and delta 1-pyrroline-5-carboxylate synthetase (P5CS) (Kaushal and Wani 2016). The abscisic acid-response element-binding proteins (AREBs) and bZIP transcriptional factors, are known to mediate gene activation pathway related to abiotic stress tolerance by recognizing ABA in plants (Uno et al. 2000). Some rhizobacteria such as Azospirillum sp. and Pseudomonas sp. were reported to increase further ABA accumulation in leaves, thus conferring better tolerance to plants (Naz and Bano 2015). Those bacteria upregulate 9-cisepoxycarotenoid dioxygenase 1 (NCED1) and abscisic acid-response element-binding proteins 1 (AREB1) genes (Yoo et al. 2019). The enzyme 9-cisepoxycarotenoid dioxygenase (NCED) is key in the biosynthesis of ABA in plants (Liu et al. 2016); highly induced by abiotic stresses leading to ABA accumulation. The overexpression of the NCED gene caused over-production of ABA, and enhanced abiotic stress tolerance (Thompson et al. 2000). Furthermore, the ABA induce antioxidant enzymes production in root nodules by stimulating the expression of antioxidant genes encoding Cu/Zn-SOD, Mn/Fe-SOD and CAT, and increasing SOD, CAT, GPX and APX activities in plant tissues and root nodules (Palma et al. 2014). This induction is a result of ABA accumulation in plants tissues subjected to salinity stress. Asensio et al. (2012) demonstrated that ABA stimulates Fe-SOD synthesis in all plant tissues of soybean under stress, underscoring the important role of ABA as a signal molecule in the activation of the nodular antioxidant metabolism.
Antioxidants in plants and nodules include a host of enzymes and metabolites that function to eliminate ROS (synthetized by stressed host legumes). Higher tolerance is associated with reduced lipid peroxidation, higher activities of SOD, CAT, peroxidase (POD), and APX as well as higher concentrations of reduced glutathione (GSH) and soluble sugar in nodulated roots under salt stress (Wang et al. 2016a, b; Irshad et al. 2021). Superoxide dismutase (SOD) acts as a first line of defense against superoxide radical (O2−). SOD catalyzes the conversion or dismutation of toxic O2 radicals to H2O2 and molecular oxygen (O2). The H2O2 is subsequently detoxified to water (H2O) by CAT or GPX (Wang et al. 2016a, b). Four groups of SOD exist including copper-zinc superoxide dismutase (Cu/Zn-SOD), manganese superoxide dismutase (Mn-SOD) and iron superoxide dismutase (FeSOD) (Miao and St. Clair 2009). Cu/Zn-SOD are located within the cytosol and plastids, whereas Fe/Mn-SODs are usually located within organelles, such as mitochondria. So far, cambialistic SOD (Cam-SOD) has been discovered (Asensio et al. 2012). Cam-SOD may have either Fe or Mn as a ligand and is of bacterial origin. They are the most active SOD in response to oxidative stress and have principally Rhizobium endosymbiont origin (Asensio et al. 2012). Catalase (CAT) is a common antioxidant enzyme present in almost all living tissues that utilize oxygen. The enzyme uses either iron or manganese as a cofactor and catalyzes the reduction of hydrogen peroxide (H2O2) to water and molecular oxygen, consequently completing the detoxification process initiated by SOD. CAT is highly efficient; it can break down millions of hydrogen peroxide molecules in one second (Ighodaro and Akinloye 2018). Glutathione Peroxidase (GPX) is an important intracellular enzyme that breakdown hydrogen peroxide (H2O2) to water; and lipid peroxide to their corresponding alcohols mainly in the mitochondria and sometimes in the cytosol (Ighodaro and Akinloye 2018). Several studies demonstrated that the presence of Rhizobium sp. under salinity stress decreased APX and GPX activity in the plant, while activity of CAT increased (Matamoros et al. 2003; Rabiei et al. 2020). In nodules, CAT was found to be the main enzyme involved in H2O2 scavenging in faba bean (Fatnassi et al. 2015). The antioxidant machinery in legume plants helps them to overcome the adverse effect of salinity by protecting them from oxidative stress. In addition, the rhizobia play an important role in increasing the activity of those antioxidants, which confer tolerance to salt and promote plant growth under stressful conditions.
Bacterial exopolysaccharides (EPS) are necessary for a functional Rhizobium-legume symbiosis in both favorable and salt stress conditions. It helps plants to mitigate salinity stress by fixing sodium ions in the soil, reducing their absorption by the plant and preventing these ions from reaching the stem, thereby increasing nutrient uptake by roots (Bhagat et al. 2021). EPS are high-molecular weight polymers attached to the outer surface of bacteria. Those acidic polysaccharides are responsible for bacterial cells attachement to surfaces including plant roots and soil particles and enhance the soil fertility and nutrient transport to roots (Forni et al. 2017). Another mechanism used by EPS to impart salt-tolerance to plants is their ability to establish a biofilm (Benidire et al. 2020). Increased concentration of salt induces an increase in EPS production, which triggers biofilm formation and sodium chelation, thus reducing the adverse effect of salt stress on plant growth (Bhagat et al. 2021). EPS production is co-regulated with Nod factors, but the type of co-regulation varies depending on the rhizobial strain (Acosta-Jurado et al. 2021). RosR, a gene encoding a positive transcriptional regulator of EPS synthesis in R. leguminosarum (Janczarek and Skorupska 2007). The RosR mutants produced three times less exopolysaccharide than wild type, decreased attachment and colonization of root hairs and were defective in biofilm formation (Janczarek et al. 2010). In addition, prsD and prsE genes are responsible for secretion of the exopolysaccharide (EPS)-glycanases PlyA and PlyB and are involved in biofilm formation by Rhizobium leguminosarum (Russo et al. 2006). The mutant disrupted in prsD and prsE genes engendered an immature biofilm formation with an atypical structure. A mutation or deletion in the pssA genes, which encode the first IP-glucosyl transferase abolished the ability of R. leguminosarum to develop a biofilm (Russo et al. 2006). A number of studies have shown that the expression of succinoglycan (EPS I) and galactoglucan (EPS II) of Rhizobium meliloti, is regulated at the transcriptional level by ion concentrations. ExpR regulates genes that play a role in salt tolerance (Lloret et al. 1998; Miller-Williams et al. 2006). The role executed by EPS argue the potential of EPS-producing bacteria and recommends their use for salinity-stress management strategies.
Salt stress alleviation strategies in legume plants
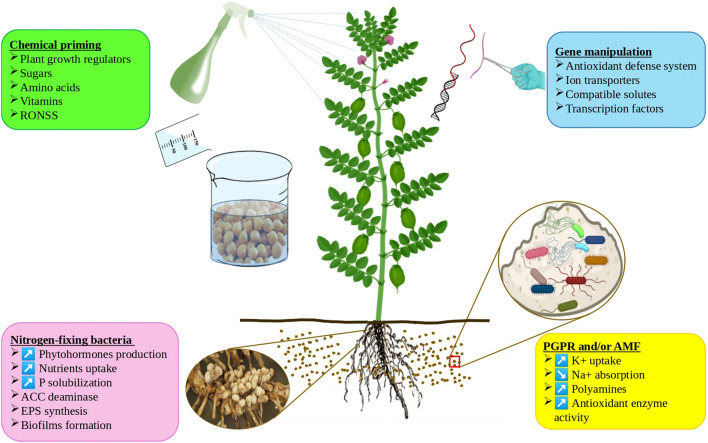
Numerous strategies were applied to improve legume growth, particularly under salinity stress and to mitigate the adverse effect of salt at different stages of plant development (Fig. 4). Biological, chemical, and physical treatments were successfully applied to seeds, seedlings, or plants to enhance tolerance to salt stress as well as the identification of salt-adapted cultivars in saline areas, breeding for plant salt tolerance and introducing new genes for salt tolerance into legume plants (Table 2).
Fig. 4.
Schematic representation of the main strategies for salt stress alleviation in legumes. These approaches include chemical priming of seeds and plants using natural and/or synthetic substances, the use of nitrogen-fixing bacteria, PGPR alone or in association with AMF or gene manipulation (Knock-out, knock-down and overexpression strategies). At the end, these alleviation approaches increase plant survival and productivity under salt stress conditions by: (i) improving nutrient uptake, (ii) stimulating the antioxidant defense machinery, (iii) enhancing the production of compatible solutes and (iv) activating plant defense mechanisms
Table 2.
Overview of the main salt alleviation strategies applied to legume plants that have been reported in previous studies
| Alleviation strategy | Legume plants | Applied treatment/approach | Notable effects | References |
|---|---|---|---|---|
| Plant growth promoting bacteria and rhizobia | Arachis hypogaea L. | Inoculation with Brachybacterium saurashtrense, haererohalobacter, and Brevibacterium casei | Increased plant growth and biomass under saline conditions | Shukla et al. (2012) |
| Cajanus cajan | Inoculation with Bradyrhizobium, and Burkholderia cepacia |
Pigeon pea co-inoculation with Bradyrhizobium and Burkholderia cepacia strains reduced the accumulation of Na+ ions This increased growth when exposed to salt stress |
Bano et al. (2015) | |
| Cicer arietinum | Triple inoculation with Rhizobium, mycorrhizal fungi, and Stenotrophomonas maltophilia |
Improved nutritional status of chickpea plants occurring as a result of the enhancement of K, P, carbohydrate, and protein contents Increased leghamoglobin content and nitrogenase activity under salt stress conditions |
Abd-Alla et al. (2019) | |
| Glycine max | Inoculation with Sinorhizobium meliloti |
Chickpea plants' nutritional status has improved as a result of higher K, P, carbohydrate, and protein contents Leghemoglobin concentration and nitrogenase activity were elevated when exposed to salt stress |
Qu et al. (2016) | |
| Inoculation with Arthrobacter woluwensis, Microbacterium oxydans, Arthrobacter aurescens, Bacillus megaterium, and Bacillus aryabhattai | Increased growth attributes and chlorophyll content in salt-stressed soybean plants | Khan et al. (2019) | ||
| Co-inoculation with Pseudomonas putida and Bradyrhizobium japonicum | increased ability to withstand salt stress by altering the architecture of the root system, which is still advantageous for nutrient uptake and nodule production | Egamberdieva et al. (2017) | ||
| Co-inoculation of Rhizobium sp. and Hydrogenophaga sp. with Bradyrhizobium | Improved growth under salt stress conditions. The shoot and root growth, shoot biomass, seed weight, and grain yield all increased | Ilangumaran et al. (2021) | ||
| Inoculation with Pseudomonas putida | Improved salt tolerance of soybean plants through the stimulation of plant growth attributes and nodulation | Kang et al. (2014a, b) | ||
| Inoculation with Penicullium funiculosum | Improved characteristics of seed germination and plant growth, and promoted isoflavone biosynthesis | Khan et al. (2011) | ||
| Lens culinaris | Inoculation with Pseudomonas putida, Pseudomonas fluorescens, and Serratia ficaria |
Growth attributes, namely shoot and root fresh weights and lengths improved with the bacterial inoculation Even under salt stress situations, higher chlorophyll concentration |
Muscolo et al. (2019) | |
| Medicago sativa | Inoculation with Klebsiella sp., Kosakonia cowanii, and Sinorhizobium meliloti |
Increased growth indicators for alfalfa Reduced Na+ accumulation and increased K+ absorption, resulting in a decrease in K+/Na+ Higher proline content and a strong CAT and SOD antioxidant activities |
Noori et al. (2018) | |
| Co-inoculation of Rhizobium meliloti and Pseudomonas fluorescens | Increased N and P amounts and decreased Na+ contents, resulting in a better salt tolerance | Younesi et al. (2013) | ||
| Phaseolus mungo | 15 rhizobia strains tested for the inoculation | Improved chlorophyll content, plant growth and biomass | Yasin et al. (2018) | |
| Pisum sativum | Inoculation with Arthrobacter protophormiae | Increased plant tolerance to salinity by improving colonization of a diverse bacterial population and increasing ACC deaminase activity | Barnawal et al. (2014) | |
| Vicia faba | Inoculation with Rhizobium leguminosarum |
Reduced Na+, Ca2+, and K+ absorption by faba bean plants Increased plant biomass, nitrogen content and nodule number |
Benidire et al. (2017) | |
| Pseudomonas putida co-inoculation with Pseudomonas fluorescens and Bacillus subtilis | Enhanced plant growth attributes, mainly plant height, fresh shoot weight, and leaf area | Metwali et al. (2015) | ||
| Inoculation with Azotobacter chrococcum | Enhanced soil fertility and improved plant growth under salt stress conditions due to biofilm formation and EPS production by bacterial strain | Mohammed (2018) | ||
| Combined action of Azotobacter chroococcum and melatonin |
Increased faba bean growth attributes and yield components under salt stress Enhanced N, P, and K amounts, proline content, and K+/Na+ ratio |
Abd El-Ghany and Attia (2020) | ||
| Vigna radiata L. | Inoculation with Pseudomonas syringae, Pseudomonas fluorescens, and Rhizobium phaseoli | Reduced ethylene synthesis and promoted nodulation under salt stress conditions | Ahmad et al. (2011) | |
| Inoculation with Pantoa sp. and Enterococcus | Increased tolerance to salt stress as a result of the bacteria's powerful ACC deaminase and plant growth-promoting abilities | Panwar et al. (2016) | ||
| Vigna unguiculata | Inoculation with Rhizobia strains | Increased cowpea growth attributes and yield | Nyaga and Njeru (2020) | |
| Arbuscular mycorrhizal fungi | Cajanus cajan L. | Inoculation with Rhizophagus irregularis and Funneliformis mossseae |
Increased plant biomass, yield, and nutrient absorption under salt stress conditions Enhanced membrane stability achieved by maintaining proper K+/Na+ and Ca2+/Na+ ratios |
Garg and Pandey (2015) |
| Inoculation with Glomus mosseae |
Improved nodulation and leghemoglobin content along with the SOD, APOX, GR, POX, and CAT activities Reduced membrane permeability and lipid peroxidation Enhanced plant tolerance to salt stress |
Garg and Manchanda (2008) | ||
| Inoculation with AMF strains |
Improvements in nodule dry mass, nitrogenase activity, and phosphorus amount under salt stress Enhanced antioxidant enzyme activities, thus alleviating the toxic effects of salt stress |
Manchanda and Garg (2011) | ||
| Cicer arietinum L. | Inoculation with Glomus mosseae and Acaulospora laevis | Enhanced host plant mineral concentration and increased plant growth and yield under salt stress conditions | Kadian et al. (2013) | |
| Lens culinaris | Inoculation with AMF isolates | Enhanced seed germination percentage, seed and stover yield under saline conditions | Rahman et al. (2017) | |
| Medicago sativa | Inoculation with Glomus mosseae | Increased nodule number, dry weights, and nitrogenase activity, resulting in better plant growth and nitrogen fixation under salt stress | Moradi (2016) | |
| Inoculation with Glomus mosseae |
Increased proline and soluble protein concentrations, plant height, biomass, and plant survival rate Increased the amount of osmotic control compounds, which helped plants grow better in saline-alkaline soils |
Zhao and Bao (2015) | ||
| Vicia faba |
Inoculation with Funneliformis mosseae, Rhizophagus intraradices, and Claroideoglomus etunicatum |
Increased nodule activities and pigment contents, and improved K and Ca accumulation under salt stress Improved growth and plant yield under salt stress conditions |
Abeer et al. (2014) | |
| Vigna unguiculata | Inoculation with AMF strains |
Salt stress led to a rise in the activity of several enzymes, including catalase, superoxide dismutase, ascorbate peroxidase, peroxidase, and glutathione reductase A higher level of salt stress resistance in plants as a result of enhanced nitrogen uptake |
Abeer et al. (2015) | |
| Seed and plant priming | Glycine max L. | Seed priming with Potassium nitrate |
Enhanced emergence and germination percentage Improved plant dry weight, radical and plumule length |
Ahmadvand et al. (2012) |
| Seed priming with Polyethylene glycol | Increased germination percentage | Khalil et al. (2001) | ||
| Pisum sativum | Seed priming with Salicylic acid | Improved photosynthetic efficiency and stronger plant antioxidant defense system result in a better tolerance to salt stress | Ahmad et al. (2017) | |
| Vicia faba L. | Seed priming with Salicylic acid |
Reduced the inhibitory effects of salt stress on seed germination Increased growth attributes, total chlorophyll content, soluble carbohydrate content, and antioxidant activities |
Azooz (2009) | |
| Seed priming with Salicylic acid |
Prevented the negative effects of salt stress Enhanced IAA and IBA amounts and decreased ABA levels |
Ahmad et al. (2018) | ||
| Seed priming with Salicylic acid |
Decreased the salt stress-induced growth interruption Improved seeds' fresh, dry weights and germination |
Anaya et al. (2018) | ||
| Seed priming with Salicylic acid and hydrogen peroxide | An increase in sugar accumulation, alpha amylase, and antioxidant activities, which promotes primary root elongation and seed germination | Bouallègue et al. (2017) | ||
| Vigna radiata L. | Seed priming with sodium chloride |
IIncreased chlorophyll content and osmolytes buildup A strengthened antioxidant defense mechanism, which reduces the damaging effects of salinity on mung bean plants |
Saha et al. (2010) | |
| Breeding | Cicer arietinum L. | Image based phenotyping | The primary factor of chickpea salt tolerance has been identified: seed number | Atieno et al. (2017) |
| Comparative transcriptome analysis (HiSeq-2500) | Up-regulation of transcripts encoding potassium transporter family HAK/KUP proteins, MIP/aquaporin protein family, NADH dehydrogenase, pectinesterase, and PP2C family proteins under salt stress | Kumar et al. (2021) | ||
| Lens culinaris | GWAS | Potassium transporters have been identified as the most likely indicator of salt stress tolerance | Dissanayake et al. (2021) | |
| Medicago sativa | Comparative transcriptome analysis | Differentially expressed genes by salt stress have been identified. The DEG are associated with calcium, redox, and hormone signaling | Kaundal et al. (2021) | |
| Medicago trunculata | Comparative transcriptome analysis | Identification of specific gene clusters associated with the auxin pathway, modified histone variant isoforms, and selectively controlled by salt in root apices | Zahaf et al. (2012) | |
| Phaseolus vulgaris | Comparative transcriptome analysis | Identification 441 transcription factors and differentially expressed genes in response to salt stress | Hiz et al. (2014) | |
| Pisum sativum L. | QTL mapping | Identification salt tolerance QTLs on linkage groups Ps III and VII with flanking SNP markers? | Leonforte et al. (2013) | |
| Vicia faba L. | Conventional breeding | Given that salt stress had no impact on plant growth or nitrogen fixation, the VF112 line was identified as salt tolerant | del Pilar et al. (1995) | |
| Conventional breeding | Identification of the salt tolerant genotypes: Fiesta VF, Acc 1512/2 and Acc 1487/7/ based on yield-related characteristics | Tavakkoli et al. (2012) | ||
| Identification of "water-stress-day index" as the best predictor for yield | Katerji et al. (2003) | |||
| Conventional breeding | Identification of salt-tolerant cultivar "Giza 843" with greater osmotic adjustment ability and higher proline and nutrient contents | Orabi and Abdelhamid (2016) | ||
| Identification of leaf osmotic potential and photosynthesis rate as the main determinants of salt tolerance | Tavakkoli et al. (2010) | |||
| Vigna radiata L. | SNP-based genome wide association | Identification of salt tolerance-associated SNPs on chromosomes 7 and 9 | Breria et al. (2020) | |
| Gene manipulation | Arachis hypogaea | Heterologous expression of Arabidopsis DREB1A gene | Enhanced plant growth attributes include proline, chlorophyll contents, electrolyte leakage, and osmotic potential | Sarkar et al. (2014) |
| Glycine max L. | Introduction of Arabidopsis vacuolar Na+/H+ antiporter gene (AtNHX1) |
improved plant growth under salt stress circumstances by reducing the harmful effects of salt stress Improved Na+ homeostasis |
Li et al. (2010) | |
| Overexpression of GB1 gene | Increased glycine betaine levels in salt stressed soybean plants | Castiglioni et al. (2018) | ||
| Overexpression of GmNAC085 gene | Increased antioxidant enzyme activities, proline contents, and dehydrin accumulation in salt stress conditions | Hoang et al. (2021) | ||
| Overexpression of soybean GmMYB84 gene | Increased seed germination, primary root elongation, proline accumulation, membrane integrity, antioxidant enzyme activity, and K+ levels | Zhang et al. (2020) | ||
| Overexpression of soybean GmDREB6 gene | Improved proline accumulation and salt tolerance in genetically modified soybean plants | Nguyen et al. (2019b) | ||
| Knock-out of the ABA-induced transcriptional repressors; AITRs genes | Improved plant growth under salt stress in AITR mutants generated using CRISPR-Cas9 technology | Wang et al. (2021) | ||
| CRISPR-Cas9 silencing of GmNAC06 gene | Enhanced proline and glycine betaine accumulation along with better regulation of Na+/K+ ratio | Li et al. (2021) | ||
| Overexpression of GmNHX5 gene using CRISPR/Cas9 technology |
Enhanced K+/Na+ ratio under salt stress conditions Up-regulation of GmSOS1 and GmSKOR genes, encoding the plasma membrane Na+/H+ antiporter and sodium/hydrogen exchanger, respectively |
Sun et al. (2021) | ||
| Medicago sativa | Overexpression of a plastid glycogen synthase kinase 3 encoding gene (MsK3) | Enhanced starch and glucose levels result in better tolerance to salt stress | Andersen et al. (2007) | |
| Overexpression of Alfin1 gene | Enhanced endogenous MsPRP2 levels result in better tolerance to salt stress | Winicov and Bastola (1999) | ||
| Medicago trunculata | Introduction of cyanobacterial flavodoxin | Improved the symbiotic performance under salt stress by inducing significant changes in enzymatic activities that control nodule redox balance | Coba de la Pena et al. (2010) | |
| Vicia faba | Heterologous expression of potato PR10a |
Increased plant survival under salt stress conditions Improved tolerance to salt resulting from a strong accumulation of proline under stressful conditions |
Hanafy et al. (2013) | |
| Constitutive expression of Arabidopsis vacuolar Na+/H+ antiporter gene (AtNHX1) |
Improved plant growth under salt stress conditions This alleviated the harmful effects of salinity |
Hassanein et al. (2019) | ||
| Vigna radiata L. | Constitutive expression of Arabidopsis vacuolar Na+/H+ antiporter gene (AtNHX1) |
Improved plant growth attributes under salt stress, namely plant height, foliage, dry mass and seed yield Enhanced Na+ sequestration in roots and prevented Na+ influx to shoots, resulting in a better intracellular ion homeostasis and osmoregulation |
Kumar et al. (2017) | |
| Overexpression of Arabidopsis vacuolar Na+/H+ antiporter gene (AtNHX1) |
Reduced damages imposed by salt stress exposure through maintaining higher K+/Na+ in the aerial parts and higher Na+ levels in roots Reduced lipid peroxidation and ROS accumulation due to the increased antioxidant activity |
Sahoo et al. (2016) | ||
| Vigna unguiculata L. | Overexpression of Vu NHX1 gene in Arabidopsis |
Improved plant tolerance to salinity, enhanced proline and chlorophyll contents This maintained the K+/Na+ ratio at a high level, resulting in better ion homeostasis |
Mishra et al. (2015) |
Plant growth promoting bacteria
Soil is an abundant source for microorganisms, particularly in the rhizosphere, the layer of soil influenced by plant root. Among these different microorganisms, bacteria are the most common (Glick 2012). In unstressed soils, up to 108 or 109 microbial cells per gram of soil can exist. However, the number of bacteria may be as low as 104 cells per gram of soil in stressed soils (Timmusk et al. 2011). The term plant growth-promoting bacteria (PGPB) is employed to encompass all these bacteria that enhance plant growth (Kang et al. 2014a). The PGPB occupy mainly roots sur-rounding soil (Orozco-Mosqueda et al. 2020). The most widely exploited and studied group of PGPB are plant growth-promoting rhizobacteria (PGPR) that have the capacity to colonize the root surfaces and closely adhere to the rhizosphere (Kloepper et al. 1999).
The PGPB is an alternative strategy to alleviate salt stress and boost the growth of salt-stressed crops. PGPB can be used as a cost effective way to increase salinity tolerance and promote plant growth (Numan et al. 2018). The PGPB can stimulate plant growth by secreting plant growth substances in the absence of biotic or abiotic stress, facilitating resource acquisition or modulating plant hormone levels. Some PGPB also have specialized mechanisms that play a key role in salt stress tolerance and plant growth promotion. These bacteria trigger plants to produce different plant growth hormones like auxin, cytokinin and gibberellin as well as volatile organic compounds. The PGPB fix nitrogen, solubilize organic and inorganic phosphate and also produce siderophore, which improve plant Fe nutrition (Numan et al. 2018). Besides, PGPB can operate as biocontrol, a widely recognized mechanisms based on a competition for an ecological niche or a substrate, the production of inhibitory allelochemicals (iron-chelating, siderophores, antibiotics, biocidal volatiles, lytic enzymes, and detoxification enzymes), and the induction of systemic resistance (ISR) in hostplants to a broad spectrum of pathogens and/or abiotic stresses (Compant et al. 2005).
The ameliorative effects of PGPR on legume growth under saline conditions have been proven in multiple legume species including faba bean (Benidire et al. 2017), soybean (Qu et al. 2016), alfalfa (Noori et al. 2018), pigeon pea (Bano et al. 2020), cowpea (Nyaga and Njeru 2020), lentil (Muscolo et al. 2019) and chickpea (Abd-Alla et al. 2019) (Table 2). PGPR display their beneficial features to overcome the toxic effects of high NaCl on morphological, physiological, and biochemical processes in plants, resulting in a significant rescue of yield loss (Ha-Tran et al. 2021). The inoculation of Arthrobacter woluwensis, Microbacterium oxydans, Arthrobacter aurescens, Bacillus megaterium, and Bacillus aryabhattai increased plant growth attributes and chlorophyll content in soybean plants under 200 mM NaCl stress (Khan et al. 2019). The use of salt-tolerant species of Pseudomonas has been reported to improve legume health. For instance, Pseudomonas putida co-inoculated with Pseudomonas fluorescens and Bacillus subtilis on the growth of six faba bean cultivars under salinity stress enhanced plant height (10.66%), shoot fresh weight (9.52%), and plant leaf area (61.86%) (Metwali et al. 2015). In addition, Egamberdieva et al. (2017) indicated that the inoculation of salt-tolerant P. putida with Bradyrhizobium japonicum synergistically improved soybean salt tolerance through the modification of the root system architecture, which can facilitate nitrogen and phosphorus acquisition, and nodule formation.
PGPB can stimulate plant growth during salt stress through several mechanisms including N2 fixation, phytohormone production, amelioration of nutrient uptake, aminocyclopropane-1-carboxylic acid deaminase production, phosphorus solubilization, exopolysaccharide (EPS) synthesis, iron acquisition, and biofilm formation (Mokrani et al. 2020). Mohamed et al. (2018) studied the production of biofilm and EPS by plant growth-promoting rhizobacteria (PGPR) under different salt concentrations. They demonstrated that biofilm formation and EPS-production by Azotobacter chroococcum significantly contribute to soil fertility and improve faba bean growth. Furthermore, the combination of EPS-producing bacteria (Az. chroococcum) and melatonin significantly increased the growth parameters and yield components in faba bean grown in the presence of salt stress (Abd El-Ghany and Attia 2020). Both bacteria inoculation and melatonin application enhanced N, P, and K concentrations; proline content; relative water content (RWC); and the K+/Na+ ratio. In addition, Na+ and Cl− concentrations decreased significantly in salt-stressed faba beans (Abd El-Ghany and Attia 2020). Furthermore, PGPR can stop the expression of plant genes that increase plant sensitivity under salinity stress. For example, Pi et al. (2019) demonstrated that PGPR inhibited the phosphorylation of the gene encoding a cytochrome P450 monooxygenase, which contributes to the accumulation of monohydroxy B-ring flavonoids that negatively regulate soybean tolerance to salinity. The application of PGPR inoculants is among the best way to overcome the adverse effect of salinity and increase tolerance to this stress. Almost success of PGPR application was proven in the laboratories and green house. Nevertheless, the challenge now is its large-scale use in the field, its adoption as a future biofertilizer as well as its sustainability in agriculture. Therefore, the research should focus more on the widespread application of PGPR in the field.
Rhizobia–Legume symbiosis
Symbiosis is one of the alternatives to mitigate the adverse effect of salinity and to discover new mechanisms involved in stress tolerance (Dodd and Pérez-Alfocea 2012). Several studies have demonstrated that plant–microbe interactions prompt abiotic stress tolerance along with growth and biomass (Kumar and Verma 2018). The Legume-Rhizobium symbiosis is a highly integrated system that leads to biological nitrogen fixation (Cordovilla et al. 1995). About 40 rhizobia species within seven genera known to nodulate and fix nitrogen with legumes have been identified and includes Rhizobium, Allorhizobium, Bradyrhizobium, Sinorhizobium, Azorhizobium, Mesorhizobium, and Methylobacterium (Lemaire et al. 2015). The legume–rhizobia association is highly specific. For instance, Rhizobium strains are largely associated with pea (Pisum sativum), lentil (L. culinaris), faba bean (V. faba) and common bean (P. vulgaris), while Bradyrhizobium strains often nodulate soybean (G. max), cowpea (Vigna unguiculata), lupine (Lupinus albus) and peanut (A. hypogaea) plants. Finally, chickpea (C. arietinum) is mostly nodulated by different species belonging to the Mesorhizobium genus (Silva et al. 2017; Koskey et al. 2018). In faba bean, the identification of rhizobia from nodules indicates that the most common species are Rhizobium leguminosarum bv. Viciae (Mutch and Young 2004), Rhizobium fabae (Tian et al. 2008), Rhizobium laguerreae (Saïdi et al. 2014) and Rhizobium anhuiense (Zhang et al. 2015).
It is known that rhizobia strains vary in their salt tolerance. For example, B. japonicum, Rhizobium etli, and R. leguminosarum are salt sensitive with their growth inhibited completely at 100 mM NaCl (Boncompagni et al. 1999). Nevertheless, some rhizobacteria can be halotolerant to salt stress. For instance, Rhizobium spp. isolated from nodules of Leucaena, Acacia, Prosopis and Hedysarum plants can tolerate up to 500 mM NaCl (Zhang et al. 1991). Rhizobacteria isolated from saline habitats can be more efficient at enhancing plant tolerance to salt than those isolated from non-saline habitats (Etesami and Beattie 2018). In saline soils in Morocco, Benidire et al. (2017) isolated two indigenous strains of R. leguminosarum (RhOF34 and RhOF125). The isolated strains improved nodulation, increased plant biomass and N content in faba bean and induced plant protection against salinity. The co-inoculation of rhizobia with other rhizobacteria improves also the growth and development of the host-legume. Ilangumaran et al. (2021) co-inoculated Rhizobium sp. SL42 and Hydrogenophaga sp. SL48 with Bradyrhizobium which resulted in higher growth soybean plants than the Bradyrhizobium control. Rhizobium and Pseudomonas co-inoculation moderated the negative effects of salinity in alfalfa (M. sativa) and significantly increased P and N contents while Na+ accumulation decreased (Younesi et al. 2013). Several studies reported that the inoculation of different genera and species of rhizobia in several grain legumes affected host plant metabolism and increased antioxidant and other compatible solutes accumulation, which can ultimately lead to better tolerance under salinity stress. Qu et al. (2016) found that Sinorhizobium meliloti-1021 played significant roles in regulating the transcription of several key enzymes related to flavonoids metabolism, which can lead to a significant increase in soybean’s tolerance to salt. In addition, Irshad et al. (2021) explained the improved salt tolerance in inoculated M. sativa plants with R. meliloti with higher activities of enzymatic and nonenzymatic antioxidants and higher compatible solutes synthesis (proline, free amino acids, glycine betaine, soluble sugars, and proteins) when compared with non-inoculated plants.
Arbuscular mycorrhizal fungi
Many reports solicited other microorganisms such as Mycorrhiza to be used as inoculants under unfavorable and stressful environmental conditions. The symbiotic relationship between mycorrhiza and roots is abundant in nature. The AMF is the most common type of this symbiosis, which has a great ecological and economic importance. The AMF symbiosis is a key component in helping plants to cope with salt stress and in increasing salinity tolerance, as demonstrated in a number of host plant and fungal species (Yang et al. 2014; Evelin et al. 2019; Wang et al. 2020). In faba bean, AMF acts as a stimulator of plant growth under salt stress. It helps plant to overcome the negative effects of NaCl by increasing nodules activities and pigments contents, which improves plant growth and yield. Furthermore, AMF also restores K and Ca contents and maintains their ratios under salt stress (Hashem et al. 2014). When plants form a symbiotic relationship with fungi, changes occur in their morphology, nutrition, and physiology, increasing their resistance to abiotic stress. AMF supply mineral nutrients to plants, especially phosphorus, which is precipitated by ions such as Ca2+, Mg2+, and Zn2+ (Al-Karaki et al. 2001). AMF can also improve photosynthesis in plants exposed to salt stress (Chen et al. 2017). In addition, mycorrhizal symbiosis facilitates K+ uptake and intercept Na+ absorption and translocation to the shoots (Chang et al. 2018; Liu et al. 2019). Application of AMF also increases polyamines: nitrogenous compounds that have been proven effective in alleviating NaCl stress (Hashem et al. 2014). Antioxidant enzyme activities like SOD, CAT, POD and APX are the main defense for legumes against NaCl stress and mycorrhiza treated legumes showed greater increase in antioxidant activities (Hashem et al. 2014). In addition, plants' inoculation with AMF showed enhancement in nodule activity and pigment content under salt stress conditions (Hashem et al. 2014).
The positive impact on plant growth and stimulation of stress tolerance by synergistic interactions of PGPR and AMF under hostile environments was also mentioned. An improvement in faba bean-rhizobia symbiotic performance by AMF has been reported (Yinsuo et al. 2004), as well as other legume species such as lentil (Xavier and Germida 2002), common bean (Tajini et al. 2012), alfalfa (Ashrafi et al. 2014) and chickpea (Abd-Alla et al. 2019). The synergistic interactions among the components of the tripartite symbiotic association (Rhizobium–AMF–faba bean) increased the photosynthetic rate and plant productivity. Accordingly, Dubova et al. (2015) have underlined that rhizobia-mycorrhizae inoculation showed better improvement of faba bean growth in saline soil compared to simple inoculation with mycorrhizae (Dubova et al. 2015). Furthermore, Rabie and Almadini (2005) examined tripartite interactions among a bacterium (Azospirillum brasilens), an arbuscular mycorrhizal fungi (Glomus clarum), and faba bean under increased NaCl levels. Significant positive effects of inoculation were observed in plants with respect to salinity tolerance, mycorrhizal dependence, phosphorus level, phosphatase enzymes, nodule number, nitrogen uptake, protein content and nitrogenase activity. AMF can be an effective bio-fertilizer to improve legume cultivation on saline agricultural lands. Moreover, the consideration of rhizobacteria and AMF interactions could be an exciting alternative to guarantee and increase tolerance to this kind of stress.
Seed and plant priming
Seed and plant priming is a promising, efficient, and low-cost approach to enhance plant tolerance under various abiotic stresses including salinity. Seed priming is defined as a pre-sowing treatment, applied before germination, that allows a partial hydration of seeds and leads to an improvement in seed germination potential, growth and crop’s productive capability (Paparella et al. 2015). However, plant priming is a mechanism leading to a physiological state that enables plants to respond more rapidly and/or more robustly after exposure to biotic or abiotic stresses (Aranega-Bou et al. 2014; Costa et al. 2018).
The adverse effect of salinity stress can be alleviated by the use of many chemical priming agents that have a significant impact on plant growth (Ashraf et al. 2018). In faba bean, various priming agents were reported to be efficient in salinity stress alleviation. Some of them are naturally occurring metabolites but also help in stress tolerance when applied exogenously such as sugars, amino acids and their derivatives, plant growth regulators and vitamins (Costa et al. 2018). Under salt stress conditions, they could also play the role of osmoprotectants or antioxidants. Another group of chemical agents called Reactive Oxygen, Nitrogen, and Sulfur Species (RONSS) is also effective in inducing plant tolerance to abiotic stresses. This kind of composite includes NO (nitric oxide), H2S (hydrogen sulfide) and H2O2 (hydrogen peroxide) and plays an important part in plant adaptation to abiotic stress due to their direct impact on gene regulation and signal transduction (Fotopoulos et al. 2015; Ashraf et al. 2018).
It has been reported that seedlings of faba bean primed seeds emerge faster, grow more vigorously and perform better under suboptimal conditions such as salinity stress (Azooz 2009; Anaya et al. 2018). Hormone priming or plant growth regulators have been widely used to increase synchronized seed germination and seedling growth under salinity stress (Ma et al. 2018). It has also been demonstrated that Salicylic Acid (SA) increases salinity tolerance and seed germination and plant productivity in faba bean (Azooz 2009). Indeed, faba bean seeds primed with salicylic acid stimulated CAT, APX and glutathione reductase (GR) activities under salinity stress (Azooz 2009). Furthermore, the application of SA enhanced the IAA and Indole-3-butyric acid (IBA) and decreased the ABA concentration (Ahmad et al. 2018). The SA supplementation mitigates the negative effects of NaCl toxicity in faba bean seedlings through the modulation of different osmoprotectants, antioxidants and nutrients uptake (Ahmad et al. 2018), and the regulation of the expression of genes involved in germination (Li et al. 2017), resulting in an increase in seedling quality. It is also possible that SA stimulated amylase activity and starch mobilization, thereby, stimulating germination and cell integrity (Bouallègue et al. 2017). Besides, this could be due to the improvement in metabolic activities (including α-amylase activity) caused by SA application resulting in better production of radicle and plumule for seedling development (Anaya et al. 2018).
Melatonin can alleviate the toxicity of oxygen and nitrogen species of faba bean seeds (Moustafa-Farag et al. 2020). Seeds priming with melatonin increased photosynthetic pigments, total carbohydrate, total phenolic content, indole acetic acid, K+, Ca2+ as well as K+/Na+ and Ca2+/Na+ ratios in the leaves under salt stress (Dawood and El-Awadi 2015).
Vitamins priming was also applied to faba bean. Semida et al. (2014) suggested that α-tocopherol could activate antioxidants in plants and decrease oxidative damage, resulting in physiological modifications in plants grown in saline soil. Exogenous application of 200 or 400 mg/L of nicotinamide (vitamin B3) increased nutrient concentrations, sucrose, soluble sugars, free amino acids, oxidative enzymes, photosynthetic pigments, plant growth, seed yield, and seed quality of faba bean salt-stressed plants, while decreasing lipid peroxidation products and the oxidative enzymes (Abdelhamid et al. 2013). Ascorbic acid (AA) priming was highly effective to increase vegetative growth, soluble carbohydrates, proline, free amino acids, K+/Na+ ratio, and photosynthesis-related pigments in faba bean. AA application improved bean seed germination and seedling growth under salinity stress (Azooz et al. 2013).
Amino acids are fundamental metabolites and plant growth stimulator that significantly mitigate abiotic stress injuries. Amino acid mixture foliar spraying on faba bean plants increased nucleic acid DNA and RNA, total carbo-hydrates, polysaccharides, photosynthetic pigments, fresh and dry shoot weight, leaf number per plant and shoot length (Sadak et al. 2015). In addition, amino acid application at a rate of 500 or 1000 or 1500 mg/L increased osmotic solutes, phenolic content, IAA, and endogenous polyamine content, while it reduced antioxidant enzymes and lipid peroxidation, thereby enhancing faba bean tolerance to salt. Exogenous amino acids enhanced free amino acids, proline, and soluble sugars in salt-stressed faba bean (Sadak et al. 2015). Proline provides osmoprotection and facilitates the growth of salt-stressed faba bean plants (Dawood and El-Awadi 2015).
Besides natural priming agent, other agents were also tested. Hellal et al. (2012) showed that Silicon (SiO2) foliar application alleviates the toxic effects caused by abiotic stresses. The authors observed an increase of the growth parameters of faba bean under salinity. Particularly, the application of 1000 ppm of Silicon significantly improved chlorophyll and carotene content, pod yield and seed number per plant. According to these authors, increased K content and reduced Na in shoots and seeds may be one of the possible mechanisms of the increased salinity tolerance. The improvement of salt tolerance after NSi and Si treatments was due to the improvement of membrane stability, chloroplast formation and sugar accumulation as well as a significant increase in APX, CAT and POD activities in leaves. In addition, the oxidative damage in faba bean, produced by salinity stress, seemed to decrease in accordance with the increase in antioxidant enzymes activity under NSi and Si treatments (Qados 2015).
KNO3 pre-soaking of faba bean seeds increased the germination percentage and had a significant increase in proline content especially in shoots regardless of the salinity levels used due to a notable decrease in Na+ and increased K+ contents (Abdel-Baki et al. 2018). Bouallègue et al. (2017) reported that exogenous application of H2O2 to faba bean seeds improved the germination rate and increased primary root elongation under salt stress. This increase was associated with enhanced total amylase activity and total sugar levels and reduced starch content in germinating seeds. For the large-scale use of these approaches in the field, seeds and plant priming can be easily applied and could be a sustainable method to alleviate salinity stress, especially seed priming that only needs seeds' presoaking. A large quantity of seeds can be pre-treated with a little volume of the desired chemical, which will be easy and less expensive in agriculture and can be a promising strategy compared to plant priming that require huge space for foliar application.
Breeding for salt tolerance improvement
Breeding for salt-tolerant genotypes that can grow under high salinity is among the indispensable strategies to alleviate the harmful effects of salt stress. In order to develop legumes with salt stress tolerance, a consistent approach involving the estimation of existing genetic variation, exploiting diverse and novel sources to create new variations, and the use of breeding strategies with several traits instead of a single trait is needed (Duc et al. 2015; Smýkal et al. 2015). For example, Atieno et al. (2017) reported broad genetic variation for growth rate, days to flower, plant height, leaf senescence, shoot Na+ and K+ content, shoot biomass, pod number and seed number under salinity in 245 accessions of chickpea using image-based phenotyping. Similarly, a significant variation was noted among the varieties (Atieno et al. 2017). Furthermore, high coefficients of variation (CV) were registered among four cultivars of faba bean for the different indices of germination under salt stress (El-Bastawisy et al. 2018).
In the last century, plant breeders performed various breeding programs wherein they well implemented the genetic variation of crops at intra-specific, inter-specific, and inter-generic levels to produce salt-tolerant lines and cultivars. As a result, they somewhat succeeded in developing a few salt-tolerant lines or cultivars of several potential crops via traditional breeding (Ashraf and Akram 2009). One example in faba bean is the line “VF112,” which has been reported as salt-tolerant because salt stress had no effect on its growth or nitrogen fixation (del Pilar et al. 1995). Other examples of salinity tolerant genotypes such as Fiesta VF, Acc 1487/7 and Acc 1512/2 were also reported (Tavakkoli et al. 2012).
Since the ultimate aim of salt stress tolerance is yield under stress, the traits used for evaluating salt stress tolerance must be correlated with yield and its components (Flowers et al. 2010). Tavakkoli et al. (2012) noted significant variations in salt tolerance degree in eleven faba bean genotypes for yield-related characteristics. Atieno et al. (2017) added that seed number is the major determinant for salinity tolerance measured as yield in chickpea and proposed this trait as a selection trait in breeding salt tolerant chickpea cultivars. Katerji et al. (2003) evaluated nine legume species, including faba bean for salinity tolerance. They found that the best predictor for yield was the ‘water-stress-day index’, which was the average of the difference in leaf-water potential between the saline and non-saline treatments through the growing season. However, this screening model was clearly not suitable for screening large numbers of genotypes (Stoddard et al. 2006).
Besides, various traits have been used to screen for salt stress tolerance such as seedling emergence, leaf soluble proline, leaf Ca2+ /Na+, and K+ /Na+ ratio, stomatal conductance, photosynthetic activity, nodulation and osmotic adjustment. In faba bean, physiological tolerance characterized by the accumulation of high quantities of inorganic osmotic N, P, K+, Ca2+, and Mg2+ and lower quantities of Na+ and Cl–, as well as higher K+/Na+ and Ca2+/Na+ ratios (Orabi and Abdelhamid 2016). Moreover, they found that the salt-tolerance of the faba bean cultivar “Giza 843” was correlated with a superior capacity of osmotic adjustment by building up proline and P, K+, Ca2+, and Mg2+ ions, compared to the salt-sensitive cultivar “Giza 3”. Many studies have considered tissue Na+ concentration in NaCl-stressed plants as a measurement of the mechanism of tolerance (Tavakkoli et al. 2010; Jaarsma et al. 2013; Assaha et al. 2017; Muchate et al. 2019). In parallel, other studies were interested in the high Cl– concentrations associated with to salt tolerance and the Cl– tolerance mechanism (Teakle and Tyerman 2010; Wu and Li 2019). An experiment was conducted to compare the responses to Na+ and to Cl– separately with the response to NaCl in a soil-based system using two varieties of faba bean, that differ in salinity tolerance (Tavakkoli et al. 2010). Consistently with the report of Tavakkoli et al. (2010), they compare the behavior of two faba bean varieties tested under salinity, the variety Nura as salt sensitive and 1487/7 as salt tolerant. The variety 1487/7 exhibited a higher leaf K+/Na+ ratio, a significant Na+ exclusion and better maintenance of leaf K+ concentrations under Na+ and NaCl stress compared with Nura. In addition, the variety 1487/7 had a higher leaf osmotic potential as well as a higher photosynthesis rate and higher K+ and Ca2+ concentrations and lower Na+ concentrations in the shoots. In addition, the variety 1487/7 maintained a higher capacity of the PSII system compared with Nura. Those parameters were proven to be associated with salt tolerance and can be exploited in breeding program for selecting tolerant varieties. There is always a need to develop cultivars, that can tolerate salinity stress. Furthermore, to accelerate breeding cycles, there is a need to use marker-assisted selection, which had not been widely used in legumes.
Genomic tools and plant molecular breeding techniques could accelerate the legume breeding process by understanding plant genetic and genomic (Gnanasambandam et al. 2012; Sallam and Ul-Allah 2019). To improve the breeding for salt tolerance in legumes, further identification of stress resistance Quantitative Trait Loci (QTLs) by using comparative linkage mapping is required (Lavania et al. 2014). Several genetic linkage maps have been developed in faba bean in the last decade using bi-parental populations, derived from crosses between two inbred lines (Khazaei et al. 2020). Consequently, two genes VfWRKY1 and VfWRKY2 were identified in faba bean and conferred salinity and drought tolerance (Abid et al. 2017). The comparative linkage mapping was also used in field pea (P. sativum), where QTLs for salinity tolerance were identified on linkage groups Ps III and VII, with flanking SNP markers suitable for the selection of resistant cultivars (Leonforte et al. 2013). Furthermore, comparative genomic analysis with other legume species showed higher levels of conserved synteny with the genomes of M. truncatula Gaertn. and chickpea (C. arietinum L.) than with soybean (G. max [L.] Merr.), L. japonicus L. and pigeon pea (Cajanus cajan [L.] Millsp.) QTLs for salinity tolerance were identified on linkage groups Ps III and VII, with flanking SNP markers suitable for selection of resistant cultivars (Leonforte et al. 2013). A genome-wide association study (GWAS) was conducted using a mixed linear model on 276 accessions of lentil. A range of candidate genes was identified with the most plausible being potassium transporters, which are known to be involved in salt tolerance in related species (Dissanayake et al. 2021). The most marker-trait associations were observed on chromosome 2 as well as chromosome 4. This study also revealed a salt tolerance mechanism in lentils. Tolerant accessions do not transport Na+ ions around the plant; instead, they are localized within root tissues (Dissanayake et al. 2021). Besides, a SNP-based genome-wide association study to mine genetic loci associated with salinity tolerance was applied in mungbean (V. radiata L.) (Breria et al. 2020). SNPs associated with salt-stress tolerance were mostly identified on chromosomes 7 and 9. The associated region at chromosome 7 contains the gene Vradi07g01630, which was annotated as ammonium transport protein (AMT). While, the associated region in chromosome 9 contained the genes Vradi09g0951 and Vradi09g09600, annotated as OsGrx_S16-glutaredoxin subgroup II and dnaJ domain proteins respectively and having functions related to salt-stress tolerance (Breria et al. 2020). However, due to the complexity of the genome of some legume species like faba bean, neither QTL mapping nor GWAS were reported earlier for salt tolerance.
Next-generation sequencing (NGS), especially high-throughput RNA sequencing (RNA–seq) technology, one of the most powerful tools currently available for transcriptome profiling can enhance the efficiency and speed of gene discovery in legumes. For example, the RNA-seq method was used to investigate genome-wide transcription profiles of two faba bean varieties with contrasted salt-tolerance during seed germination under salinity (Zhang et al. 2020). A total of 4,486 differentially expressed genes (DEGs) were identified by the comparison of the salt-tolerant variety Y134 and the salt-sensitive variety Y078 treated with salinity or not. Out of these, 1,410 candidate DEGs were identified as salt-stress response genes. Furthermore, 623 DEGs were identified as variety-specific response genes during seed germination at 16 h or 24 h with salt treatment (Zhang et al. 2020). The obtained results are helpful for the understanding of salt tolerance mechanism of crops during seed germination, and provide more genetic resources for future exploitation in faba bean breeding. Another recent example in chickpea, Kumar et al. (2021) utilized a comparative transcriptome analysis of tolerant and sensitive chickpea genotypes in control and salt-stressed conditions. Using Illumina HiSeq-2500, 21,698 differentially expressed genes (DEGs) were identified, of which 11,456 and 10,242 were up- and down-regulated, respectively. They found a significant up-regulation of transcripts encoding potassium transporter family HAK/KUP proteins, MIP/aquaporin protein family, NADH dehydrogenase, pectinesterase, and PP2C family proteins occurred under salt stress. The identification of differentially expressed genes (DEGs) and related pathways by comprehensive analysis of transcriptomes was also applied in Phaseolus vulgaris (Hiz et al. 2014), Medicago sativa (Kaundal et al. 2021), Sophora alopecuroides (Yan et al. 2020) and Medicago trunculata (Zahaf et al. 2012). This will help to understand the salt tolerance mechanism during seeds germination and plant development. The data generated from all the previous studies will accelerate functional and applied genomics research in legumes for their genetic enhancement. Furthermore, it will provide valuable genetic resource for the breeding of salt-tolerant legumes in the future.
Gene manipulation for salt stress tolerance in legumes
While salinity tolerance is a polygenic trait, the breeding approaches in such a case are still very limited, which can explain the scarcity of commercial salt-tolerant crops. Therefore, engineering crops with improved salt stress tolerance traits is one of the most important challenges for modern agriculture (Hanin et al. 2016). Mutagenesis allows the diversification and genetic diversity of plant species (Arriagada et al. 2022). According to the joint FAO/IAEA Division of Nuclear Techniques in Food and Agriculture, a total of 21 mutants of soybean with increased tolerance to abiotic stresses, have been developed to date (https://mvd.iaea.org/, accessed on 12 April, 2023). The success of mutagenesis was also reported in other legume species, such as chickpea (Toker 2014), groundnut (Azad et al. 2013), Clitoria ternatea (Talukdar 2011), faba bean (El-Awadi et al. 2017) and Lathyrus sativus (Talukdar 2011). For example, Toker, (2014) reported an increased tolerance to salt, drought and heat stresses in gamma irradiated Cicer species. According to the same author, all the mutant lines displayed a better tolerance to a broad number of abiotic stresses, mainly salinity.
Genetic transformation can also be applied to improve salt tolerance in legume plants (Table 2). For instance, constitutive expression of the pathogenesis-related gene PR10a from potato in transgenic faba bean enhanced plant tolerance to salt stress and osmotic stress (Hanafy et al. 2013). The overexpression of Arabidopsis vacuolar Na+/H+ antiporter AtNHX1 in transgenic plants exhibited a higher ability for vacuolar sequestration of Na+, maintaining osmotic balance in vacuoles using Na+ as an ionic osmolyte and reducing detrimental effects of excess Na+ in the cytosol (Hanin et al. 2016; Kumar et al. 2017). Afterwards, it was also applied to legumes such as alfalfa (Li et al. 2011b), soybean, cowpea (Mishra et al. 2015) and mungbean (Sahoo et al. 2016; Kumar et al. 2017). Recently, AtNHX1 was transferred into the shoot apices of three Egyptian faba bean cultivars via Agrobacterium-mediated transformation. The gene was highly expressed and enhances salt tolerance in transgenic plants (Hassanein et al. 2019).
Transgenic plants overexpressing antioxidants or detoxification molecules such as ascorbate, glutathione, carotenoids, anthocyanins osmolytes, peroxiredoxin and tocopherol can be used to enhance defense in plants (Wang et al. 2003). The transfer of one or several genes that encode for those protecting molecules could be a valuable strategy for increasing salt tolerance. Other molecules could also play a protective role under salt stress. For example, the betaines, proline, mannitol, and trehalose are compatible molecules that may protect plants against salt stress and can act as osmoprotective compounds by directly stabilizing membranes and/or proteins (Slama et al. 2015). A recent study in soybean reported overexpression of a novel GB1 gene that effectively increased the content of glycine betaine in transgenic plants, which could confer a better tolerance to abiotic stress including salinity (Castiglioni et al. 2018). Always in the soybean, Hoang et al. (2021) reported that transgenic plants overexpressing GmNAC085 showed a better defense system against salinity-induced oxidative stress, with higher activities of antioxidant enzymes responsible for scavenging hydrogen peroxide or superoxide radicals. Adding that the key stress-responsive of the gene GmNAC085 is involved in the proline biosynthetic pathway, sodium ion transporter and accumulation of dehydrins. In M. sativa, the over-expression of MsK4 in transgenic plants resulted in the overproduction of sugar metabolism and the accumulation of higher levels of starch and glucose which led to a better tolerance to salt stress (Andersen et al. 2007). Furthermore, the expression of cyanobacterial flavodoxin in transgenic M. trunculata induced significant changes in enzymatic activities that involve nodule redox balance, which improved the symbiotic performance under salt stress (Peña et al. 2010). The regulated expression of AtDREB1A gene, a class of DREB from A. thaliana was studied in transgenic peanut (A. hypogaea) lines. The transgenic plants carrying AtDREB1A showed improved growth parameters that were correlated with physio-biochemical parameters such as proline content, total chlorophyll content, osmotic potential, electrolytic leakage and relative water content under drought or salinity stress (Sarkar et al. 2014). In addition to the antioxidant defense system, ion transporters and compatible solutes, salinity tolerance mechanism also depend on transcription factors. The transcriptional regulator, Alfin1, over-expressed in alfalfa (M. sativa) regulates endogenous NaCl-inducible gene expression, resulting in salinity tolerance (Winicov and Bastola 1999). In soybean, the transcription factor-encoding gene GmMYB84 was modulated by DNA methylation, which confers salinity stress tolerance (Zhang et al. 2020). Similarly, the overexpression of the GmDREB6 gene improve proline accumulation and salt tolerance in transformed soybean plants (Nguyen et al. 2019b). Although more research is required to identify more genes involved in legume salt tolerance and their expression in transgenic plants.
Genome-engineering using CRISPR-Cas9 technology (clustered regularly interspaced short palindromic repeats-CRISPR associated protein 9) has demonstrated broad potential in developing salt-resilient plants for several plant species (Razzaq et al. 2022). CRISPR-Cas9 offers a precise genetic modification of crops, resulting in a notable increase of varieties obtained in a short period of time. However, few studies have been conducted on legume plants with regard to the improvement of salt tolerance. This advanced technology has mostly been applied in soybean for salt tolerance studies. For instance, the knock-out of GmAITR (ABA-induced transcriptional repressors) genes using the CRISPR/CAS9 genome editing tool to soybean (G. max) resulted in better plant growth under saline conditions (Wang et al. 2021). Besides soybean, its silencing in Arabidopsis also enhanced salinity and drought tolerance (Chen et al. 2021). CRISPR-Cas9 mediated silencing of GmNAC06 confers a better salt tolerance to soybean plants (Li et al. 2021). The increased tolerance was associated with a notable accumulation of proline and glycine betaine along with a better regulation of Na+/K+ ratio (Li et al. 2021). Salt tolerance has been achieved by overexpressing sodium/hydrogen exchanger GmHNX5 in soybean, using CRISPR-Cas9 technology. The GmNHX5 overexpression reduced organelle injuries occurring as a result of salt stress exposure by enhancing the K+/Na+ ratio (Sun et al. 2021). At the molecular level, the overexpression of GmNHX5 was accompanied with an up-regulation of GmSOS1 and GmSKOR genes, encoding for plasma membrane Na+/H+ antiporter and sodium/hydrogen exchanger respectively (Sun et al. 2021). With the ability to readily enhance salt tolerance in legume species, genome-editing techniques have the potential to become a powerful tool in crop improvement programs. This is supported by the promising results described above. The CRISPR toolkit is a recent addition to legume study, with a particular emphasis on investigating salinity stress.
Conclusion
Salt stress is one of the major environmental constraints, hampering plant growth, development, and productivity. It negatively affects legume germination and physiology resulting in a significant decrease in plant yield and eventually, plant death. To cope with salt excess occurring in soils and irrigation water, legumes have developed complex mechanisms to overcome the harmful effects of salt stress and to guarantee plant survival and productivity under these growth-limiting conditions. These elaborated mechanisms rely on the involvement of several molecular actors, ensuring stress signal perception, integration, and signaling, leading to the activation of salt response genes by specific transcriptional regulators. Besides the genetic regulation of salt stress response, epigenetic regulation represents a key component in salt stress response in legume species. An in-depth understanding of the different molecular mechanisms involved in plant response to salt stress undoubtebly facilitates the development of salt-tolerat crops, thereby positively impacting global food security. The comparison between salt-tolerant and salt-sensitive species or cultivars using omics technologies for global molecular profiling (genomics, transcriptomics, proteomics, and metabolomics) offers the possibility to extend our well-understanding of the molecular mechanisms associated with salt tolerance acquisition and thus, developing new strategies for plant breeding for salt-tolerant species. The generated data will hasten/accelerate functional and applied genomics research in legume species for their genetic improvement. Moreover, this will also provide relevant genetic resources for the breeding of salt-tolerance legumes in the future.
Legumes have the great ability to develop synergistic associations with rhizospheric microorganisms, most likely bacteria and arbuscular mycorhizal fungi, that remain beneficial for plant growth and productivity under optimal and sub-optimal growth conditions. Exploring these worthwhile associations has proven to be one of the most promising alleviation strategies that can be applied to enhance legumes' yield and productivity under salt-stress conditions. Several examples of the great potential of PGPBs and AMF have been addressed in the present review, thereby underlining the possibility of the adoption of bacterial and AMF-derived biofertilizers for sustainable agriculture. However, the scale-up use of these "biofertilizers" in the field represents the only challenge ahead. Seed and plant priming can also be applied to neutralize the negative effects of salt stress on legumes' growth and development. The use of chemical substances such as sodium nitrate, salicylic acid, polyethylene glycol, vitamins, or even low to moderate concentrations of NaCl increases plant survival under salt stress conditions, it has been well-illustrated within this review of the literature. This strategy displays many advantages, mainly the easy implementation and low material requirements compared to the other alleviation strategies. Overall, the different alleviation approaches reviewed above applied alone or in combination with sustainable agriculture practices can be implemented to reduce the deleterious effects of salt stress on legume species and ensure food security worldwide, especially for local populations under the threat of salt excess.
Author contributions
Writing-Original draft preparation, SB and FH; Writing-Review and editing, MF and AS; Supervision, MF and AS. All authors have read and agreed the proof version to be published.
Funding
This article is being produced in the framework of the PRIMA DIVICIA project.
Data availability
Data sharing not applicable to this article as no data sets were generated or analyzed during the present study.
Declarations
Conflict of interest
The authors have declared that no competing interests exist.
Footnotes
Sarah Bouzroud and Fatima Henkrar contributed equally to this review.
Contributor Information
Sarah Bouzroud, Email: a.smouni@um5r.ac.ma.
Abdelaziz Smouni, Email: s.bouzroud@um5r.ac.ma.
References
- Abd El-Ghany M, Attia M. Effect of exopolysaccharide-producing bacteria and melatonin on faba bean production in saline and non-saline soil. Agronomy. 2020;10:316. doi: 10.3390/agronomy10030316. [DOI] [Google Scholar]
- Abd-Alla M, Nafady N, Bashandy S, Hassan A. Mitigation of effect of salt stress on the nodulation, nitrogen fixation and growth of chickpea (Cicer arietinum L.) by triple microbial inoculation. Rhizosphere. 2019;10:100148. doi: 10.1016/j.rhisph.2019.100148. [DOI] [Google Scholar]
- Abdel Latef AAH, Tahjib-Ul-Arif M, Rhaman MS. Exogenous auxin-mediated salt stress alleviation in Faba Bean (Vicia faba L.) Agronomy. 2021;11:547. doi: 10.3390/agronomy11030547. [DOI] [Google Scholar]
- Abdel-Baki G, Shaddad MAK, Mostafa D, Rafat A-S. The effect of seed presoaking with KNO3 on seed germination, proline, protein pattern, ß-amylase and mineral composition of two faba bean cultivars treated with NaCl. Egypt J Bot. 2018;58:445–461. doi: 10.21608/ejbo.2018.3423.1166. [DOI] [Google Scholar]
- Abdelhamid M, Mervat ShS, Urs S, Abdel-Kareem ME. Interactive effects of salinity stress and nicotinamide on physiological and biochemical parameters of faba bean plant. Acta Biol Colomb. 2013;18:499–510. [Google Scholar]
- Abeer H, Abd Allah E, Alqarawi A, et al. Alleviation of adverse impact of salinity on Faba bean (Vicia faba L.) by arbuscular mycorrhizal fungi. Pak J Bot. 2014;46:2003–2013. [Google Scholar]
- Abeer H, Abd Allah E, Alqarawi A, Egamberdieva D. Induction of salt stress tolerance in cowpea [Vigna unguiculata (L.) Walp.] by arbuscular mycorrhizal fungi. Legume Res Int J. 2015;38:579–588. [Google Scholar]
- Abid G, Muhovski Y, Mingeot D, et al. Identification and characterization of two faba bean (Vicia faba L.) WRKY transcription factors and their expression analysis during salt and drought stress. J Agric Sci. 2017;155:791–803. doi: 10.1017/S0021859616000885. [DOI] [Google Scholar]
- Acosta-Jurado S, Fuentes-Romero F, Ruiz-Sainz J-E, et al. Rhizobial exopolysaccharides: genetic regulation of their synthesis and relevance in symbiosis with legumes. Int J Mol Sci. 2021;22:6233. doi: 10.3390/ijms22126233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad P, Rasool S. Emerging technologies and management of crop stress tolerance: volume 1-biological techniques. Cambridge: Academic Press; 2014. [Google Scholar]
- Ahmad F, Singh A, Kamal A. Ameliorative effect of salicylic acid in salinity stressed Pisum sativum by improving growth parameters, activating photosynthesis and enhancing antioxidant defense system. Biosci Biotech Res Comm. 2017;10:481–489. doi: 10.21786/bbrc/10.3/22. [DOI] [Google Scholar]
- Ahmad M, Zahir ZA, Asghar HN, Asghar M. Inducing salt tolerance in mung bean through coinoculation with rhizobia and plant-growth-promoting rhizobacteria containing 1-aminocyclopropane-1-carboxylate deaminase. Can J Microbiol. 2011;57:578–589. doi: 10.1139/w11-044. [DOI] [PubMed] [Google Scholar]
- Ahmad P, Alyemeni MN, Ahanger MA, et al. Salicylic acid (SA) induced alterations in growth, biochemical attributes and antioxidant enzyme activity in Faba Bean (Vicia faba L.) seedlings under NaCl toxicity. Russ J Plant Physiol. 2018 doi: 10.1134/S1021443718010132. [DOI] [Google Scholar]
- Ahmadvand G, Soleimani F, Saadatian B, Pouya M. Effects of seed priming on germination and emergence traits of two soybean cultivars under salinity stress. J Basic Appl Sci Res. 2012;3:234–241. [Google Scholar]
- Al-Farsi SM, Nawaz A, Nadaf SK, et al. Effects, tolerance mechanisms and management of salt stress in lucerne (Medicago sativa) Crop Pasture Sci. 2020;71:411–428. doi: 10.1071/CP20033. [DOI] [Google Scholar]
- Al-Karaki GN, Hammad R, Rusan M. Response of two tomato cultivars differing in salt tolerance to inoculation with mycorrhizal fungi under salt stress. Mycorrhiza. 2001;11:43–47. doi: 10.1007/s005720100098. [DOI] [Google Scholar]
- Al-Lawati A, Al-Bahry S, Victor R, et al. Salt stress alters DNA methylation levels in alfalfa (Medicago spp.) Genet Mol Res. 2016;15:15018299. doi: 10.4238/gmr.15018299. [DOI] [PubMed] [Google Scholar]
- Alsaeedi AH, El-Ramady H, Alshaal T, et al. Engineered silica nanoparticles alleviate the detrimental effects of Na+ stress on germination and growth of common bean (Phaseolus vulgaris) Environ Sci Pollut Res. 2017;24:21917–21928. doi: 10.1007/s11356-017-9847-y. [DOI] [PubMed] [Google Scholar]
- Alqarawi A, Hashem A, Abd Allah E, et al. Effect of salinity on moisture content, pigment system, and lipid composition in Ephedra alata Decne. Acta Biol Hung. 2014;65:61–71. doi: 10.1556/ABiol.65.2014.1.6. [DOI] [PubMed] [Google Scholar]
- Alzahrani SM, Alaraidh IA, Khan MA, et al. Identification and characterization of salt-responsive microRNAs in Vicia faba by high-throughput sequencing. Genes. 2019;10:303. doi: 10.3390/genes10040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzahrani SM, Alaraidh IA, Migdadi H, et al. Physiological, biochemical, and antioxidant properties of two genotypes of Vicia faba grown under salinity stress. Pak J Bot. 2019 doi: 10.30848/PJB2019-3(3). [DOI] [Google Scholar]
- Anaya F, Fghire R, Wahbi S, Loutfi K. Influence of salicylic acid on seed germination of Vicia faba L. under salt stress. J Saudi Soc Agric Sci. 2018;17:1–8. doi: 10.1016/j.jssas.2015.10.002. [DOI] [Google Scholar]
- Andersen S, Rozhon W, Šamaj J, et al. A plastid-localized glycogen synthase kinase 3 modulates stress tolerance and carbohydrate metabolism. Plant J. 2007;49:1076–1090. doi: 10.1111/j.1365-313X.2006.03025.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar A, Kim J-K. Transgenic breeding approaches for improving abiotic stress tolerance: recent progress and future perspectives. Int J Mol Sci. 2020;21:2695. doi: 10.3390/ijms21082695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranega-Bou P, de la Leyva MO, Finiti I, et al. Priming of plant resistance by natural compounds. Hexanoic acid as a model. Front Plant Sci. 2014 doi: 10.3389/fpls.2014.00488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arriagada O, Cacciuttolo F, Cabeza RA, et al. A comprehensive review on chickpea (Cicer arietinum L.) breeding for abiotic stress tolerance and climate change resilience. Int J Mol Sci. 2022;23:6794. doi: 10.3390/ijms23126794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asensio AC, Gil-Monreal M, Pires L, et al. Two Fe-superoxide dismutase families respond differently to stress and senescence in legumes. J Plant Physiol. 2012;169:1253–1260. doi: 10.1016/j.jplph.2012.04.019. [DOI] [PubMed] [Google Scholar]
- Ashraf M, Akram N. Improving salinity tolerance of plants through conventional breeding and genetic engineering: an analytical comparison. Biotechnol Adv. 2009;27:744–752. doi: 10.1016/j.biotechadv.2009.05.026. [DOI] [PubMed] [Google Scholar]
- Ashrafi E, Zahedi M, Razmjoo J. Co-inoculations of arbuscular mycorrhizal fungi and rhizobia under salinity in alfalfa. Soil Sci Plant Nutr. 2014;60:619–629. doi: 10.1080/00380768.2014.936037. [DOI] [Google Scholar]
- Ashraf M, Akbar A, Askari S, et al. Recent advances in abiotic stress tolerance of plants through chemical priming: an overview. In: Rakshit A, Singh H, et al., editors. Advances in seed priming. Singapore: Springer; 2018. pp. 51–79. [Google Scholar]
- Assaha D, Ueda A, Saneoka H, et al. The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front Physiol. 2017;8:509. doi: 10.3389/fphys.2017.00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atieno J, Li Y, Langridge P, et al. Exploring genetic variation for salinity tolerance in chickpea using image-based phenotyping. Sci Rep. 2017;7:1300. doi: 10.1038/s41598-017-01211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atieno J, Colmer TD, Taylor J, et al. Novel salinity tolerance loci in chickpea identified in glasshouse and field environments. Front Plant Sci. 2021 doi: 10.3389/fpls.2021.667910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awana M, Yadav K, Rani K, et al. Insights into salt stress-induced biochemical, molecular and epigenetic regulation of spatial responses in Pigeonpea (Cajanus cajan L.) J Plant Growth Regul. 2019;38:1545–1561. doi: 10.1007/s00344-019-09955-4. [DOI] [Google Scholar]
- Ayra L, Ramírez M, Íñiguez LP, et al. The common bean (Phaseolus vulgaris) Basic Leucine Zipper (bZIP) transcription factor family: response to salinity stress in fertilized and symbiotic N2-fixing plants. Agriculture. 2018;8:160. doi: 10.3390/agriculture8100160. [DOI] [Google Scholar]
- Azad M, Alam M, Hamid M. Modification of salt tolerance level in groundnut (Arachis hypogaea L.) through induced mutation. Legume Res Int J. 2013;36:224–233. [Google Scholar]
- Azooz M. Salt stress mitigation by seed priming with salicylic acid in two faba bean genotypes differing in salt tolerance. Int J Agric Biol. 2009;11:343–350. [Google Scholar]
- Azooz M, Alzahrani A, Youssef M. The potential role of seed priming with ascorbic acid and nicotinamide and their interactions to enhance salt tolerance in broad bean (Vicia faba L.) Aust J Crop Sci. 2013;7:2091–2100. [Google Scholar]
- Bano DA, Singh R, Waza SA, Singh N. Effect of cowpea Bradyrhizobium (RA-5) and Burkholderia cepacia (RRE-5) on growth parameters of pigeonpea under salt stress conditions. J Pure Appl Microbiol. 2015;9:2539–2546. [Google Scholar]
- Bano DA, Singh RK, Waza SA, Singh NP (2020) Effect of cowpea bradyrhizobium (RA-5) and Burkholderiacepacia (RRE-5) on growth parameters of pigeonpea under salt stress conditions. In: J. Pure Appl. Microbiol. https://microbiologyjournal.org/effect-of-cowpea-bradyrhizobium-ra-5-and-burkholderia-cepacia-rre-5-on-growth-parameters-of-pigeonpea-under-salt-stress-conditions/. Accessed 29 Aug 2021
- Bao A-K, Wang S-M, Wu G-Q, et al. Overexpression of the Arabidopsis H+-PPase enhanced resistance to salt and drought stress in transgenic alfalfa (Medicago sativa L.) Plant Sci. 2009;176:232–240. doi: 10.1016/j.plantsci.2008.10.009. [DOI] [Google Scholar]
- Bao A, Du B, Touil L, et al. Co-expression of tonoplast Cation/H+ antiporter and H+-pyrophosphatase from xerophyte Zygophyllum xanthoxylum improves alfalfa plant growth under salinity, drought and field conditions. Plant Biotechnol J. 2016;14:964–975. doi: 10.1111/pbi.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnawal D, Bharti N, Maji D, et al. ACC deaminase-containing Arthrobacter protophormiae induces NaCl stress tolerance through reduced ACC oxidase activity and ethylene production resulting in improved nodulation and mycorrhization in Pisum sativum. J Plant Physiol. 2014;171:884–894. doi: 10.1016/j.jplph.2014.03.007. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bassil E, Blumwald E. The ins and outs of intracellular ion homeostasis: NHX-type cation/H+ transporters. Curr Opin Plant Biol. 2014;22:1–6. doi: 10.1016/j.pbi.2014.08.002. [DOI] [PubMed] [Google Scholar]
- Battaglia M, Covarrubias AA. Late embryogenesis abundant (LEA) proteins in legumes. Front Plant Sci. 2013;4:190. doi: 10.3389/fpls.2013.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benidire L, Lahrouni M, El Khalloufi F, et al. Effects of Rhizobium leguminosarum inoculation on growth, nitrogen uptake and mineral assimilation in Vicia faba plants under salinity stress. J Agric Sci Technol. 2017;19:889–901. [Google Scholar]
- Benidire L, El Khalloufi F, Oufdou K, et al. Phytobeneficial bacteria improve saline stress tolerance in Vicia faba and modulate microbial interaction network. Sci Total Environ. 2020;729:139020. doi: 10.1016/j.scitotenv.2020.139020. [DOI] [PubMed] [Google Scholar]
- Bernard SM, Habash DZ. The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 2009;182:608–620. doi: 10.1111/j.1469-8137.2009.02823.x. [DOI] [PubMed] [Google Scholar]
- Betti M, García-Calderón M, Pérez-Delgado CM, et al. Glutamine synthetase in legumes: recent advances in enzyme structure and functional genomics. Int J Mol Sci. 2012;13:7994–8024. doi: 10.3390/ijms13077994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhagat N, Raghav M, Dubey S, Bedi N. Bacterial exopolysaccharides: insight into their role in plant abiotic stress tolerance. J Microbiol Biotechnol. 2021;31:1045–1059. doi: 10.4014/jmb.2105.05009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicakci T, Aksu E, Arslan M. Effect of seed coating on germination, emergence and early seedling growth in Alfalfa (Medicago sativa L.) under salinity conditions. Fresenius Env Bull. 2018;27:6978–6984. [Google Scholar]
- Blumwald E, Aharon GS, Apse MP. Sodium transport in plant cells. Biochim Biophys Acta BBA-Biomembr. 2000;1465:140–151. doi: 10.1016/S0005-2736(00)00135-8. [DOI] [PubMed] [Google Scholar]
- Bojović B, Đelić G, Topuzović M, Stanković M. Effects of NaCl on seed germination in some species from families Brassicaceae and Solanaceae. Kragujev J Sci. 2010;32:83–87. [Google Scholar]
- Boncompagni E, Østerås M, Poggi M, le Rudulier D. Occurrence of choline and glycine betaine uptake and metabolism in the family rhizobiaceae and their roles in osmoprotection. Appl Environ Microbiol. 1999;65:2072–2077. doi: 10.1128/AEM.65.5.2072-2077.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouallègue A, Souissi F, Nouairi I, et al. Salicylic acid and hydrogen peroxide pretreatments alleviate salt stress in faba bean (Vicia faba) seeds during germination. Seed Sci Technol. 2017;45:675–690. [Google Scholar]
- Breria CM, Hsieh C-H, Yen T-B, et al. A SNP-based genome-wide association study to mine genetic loci associated to salinity tolerance in mungbean (Vigna radiata L.) Genes. 2020;11:759. doi: 10.3390/genes11070759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brígido C, Nascimento FX, Duan J, et al. Expression of an exogenous 1-aminocyclopropane-1-carboxylate deaminase gene in Mesorhizobium spp. reduces the negative effects of salt stress in chickpea. FEMS Microbiol Lett. 2013;349:46–53. doi: 10.1111/1574-6968.12294. [DOI] [PubMed] [Google Scholar]
- Bruning B, Rozema J. Symbiotic nitrogen fixation in legumes: perspectives for saline agriculture. Environ Exp Bot. 2013;92:134–143. doi: 10.1016/j.envexpbot.2012.09.001. [DOI] [Google Scholar]
- Büyük İ, Inal B, Ilhan E, et al. Genome-wide identification of salinity responsive HSP70 s in common bean. Mol Biol Rep. 2016;43:1251–1266. doi: 10.1007/s11033-016-4057-0. [DOI] [PubMed] [Google Scholar]
- Castiglioni P, Bell E, Lund A, et al. Identification of GB1, a gene whose constitutive overexpression increases glycinebetaine content in maize and soybean. Plant Direct. 2018;2:e00040. doi: 10.1002/pld3.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C, Wang B, Shi L, et al. Alleviation of salt stress-induced inhibition of seed germination in cucumber (Cucumis sativus L.) by ethylene and glutamate. J Plant Physiol. 2010;167:1152–1156. doi: 10.1016/j.jplph.2010.03.018. [DOI] [PubMed] [Google Scholar]
- Chang W, Sui X, Fan X, et al. Arbuscular mycorrhizal symbiosis modulates antioxidant response and ion distribution in salt-stressed Elaeagnus angustifolia seedlings. Front Microbiol. 2018 doi: 10.3389/fmicb.2018.00652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y, Kang J, Sun Y, et al. Molecular cloning and characterization of a novel gene encoding zinc finger protein from Medicago sativa L. Mol Biol Rep. 2009;36:2315. doi: 10.1007/s11033-009-9450-5. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhang H, Zhang X, Tang M. Arbuscular mycorrhizal symbiosis alleviates salt stress in black locust through improved photosynthesis, water status, and K+/Na+ homeostasis. Front Plant Sci. 2017 doi: 10.3389/fpls.2017.01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Li M, Zhang H, et al. Continuous salt stress-induced long non-coding RNAs and DNA methylation patterns in soybean roots. BMC Genom. 2019;20:1–12. doi: 10.1186/s12864-019-6101-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhang N, Zhou G, et al. Knockout of the entire family of AITR genes in Arabidopsis leads to enhanced drought and salinity tolerance without fitness costs. BMC Plant Biol. 2021;21:1–15. doi: 10.1186/s12870-021-02907-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coba de la Pena T, Redondo FJ, Manrique E, et al. Nitrogen fixation persists under conditions of salt stress in transgenic Medicago truncatula plants expressing a cyanobacterial flavodoxin. Plant Biotechnol J. 2010;8:954–965. doi: 10.1111/j.1467-7652.2010.00519.x. [DOI] [PubMed] [Google Scholar]
- Compant S, Reiter B, Sessitsch A, et al. Endophytic colonization of Vitis vinifera L. by plant growth-promoting bacterium Burkholderia sp. strain PsJN. Appl Environ Microbiol. 2005;71:1685–1693. doi: 10.1128/AEM.71.4.1685-1693.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforte VP, Echeverria M, Sánchez C, et al. Engineered ACC deaminase-expressing free-living cells of Mesorhizobium loti show increased nodulation efficiency and competitiveness on Lotus spp. J Gen Appl Microbiol. 2010;56:331–338. doi: 10.2323/jgam.56.331. [DOI] [PubMed] [Google Scholar]
- Cordovilla MP, Ligero F, Lluch C. The effect of salinity on N fixation and assimilation in Vicia faba. J Exp Bot. 1994;45:1483–1488. doi: 10.1093/jxb/45.10.1483. [DOI] [Google Scholar]
- Cordovilla M, Ocaña A, Ligero F, Lluch C. Growth stage response to salinity in simbiosis Vicia faba-Rhizobium leguminosarum bv. viciae. Life Sci Adv. 1995;14:105–111. [Google Scholar]
- Cordovilla MDP, Ligero F, Lluch C. Effect of salinity on growth, nodulation and nitrogen assimilation in nodules of faba bean (Vicia faba L.) Appl Soil Ecol. 1999;11:1–7. doi: 10.1016/S0929-1393(98)00132-2. [DOI] [Google Scholar]
- Costa A, Navazio L, Szabo I. The contribution of organelles to plant intracellular Calcium signalling. J Exp Bot. 2018 doi: 10.1093/jxb/ery185. [DOI] [PubMed] [Google Scholar]
- Dawood M, El-Awadi M. Alleviation of salinity stress on Vicia faba L. plants via seed priming with metatonin. Acta Biológica Colomb. 2015;20:223–235. doi: 10.15446/abc.v20n2.43291. [DOI] [Google Scholar]
- De Rossi S, Di Marco G, Bruno L, et al. Investigating the drought and salinity effect on the redox components of Sulla Coronaria (L.) Medik. Antioxidants. 2021;10:1048. doi: 10.3390/antiox10071048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pilar CM, Ligero F, Lluch C. Influence of host genotypes on growth, symbiotic performance and nitrogen assimilation in faba bean (Vicia faba L.) under salt stress. Plant Soil. 1995;172:289–297. doi: 10.1007/BF00011331. [DOI] [Google Scholar]
- Dell’Aversana E, Cirillo V, Van Oosten MJ, et al. Ascophyllum nodosum based extracts counteract salinity stress in tomato by remodeling leaf nitrogen metabolism. Plants. 2021;10:1044. doi: 10.3390/plants10061044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V, Maathuis FJM. Physiological roles of nonselective cation channels in plants: from salt stress to signalling and development. New Phytol. 2007;175:387–404. doi: 10.1111/j.1469-8137.2007.02128.x. [DOI] [PubMed] [Google Scholar]
- Desheva G, Desheva GN, Stamatov SK. Germination and early seedling growth characteristics of Arachis hypogaea L. under salinity (NaCl) stress. Agric Conspec Sci. 2020;85:113–121. [Google Scholar]
- Dissanayake R, Cogan NOI, Smith KF, Kaur S. Application of genomics to understand salt tolerance in lentil. Genes. 2021;12:332. doi: 10.3390/genes12030332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodd I, Pérez-Alfocea F. Microbial amelioration of crop salinity stress. J Exp Bot. 2012;63:3415–3428. doi: 10.1093/jxb/ers033. [DOI] [PubMed] [Google Scholar]
- Dong W, Song Y, Zhao Z, et al. The Medicago truncatula R2R3-MYB transcription factor gene MtMYBS1 enhances salinity tolerance when constitutively expressed in Arabidopsis thaliana. Biochem Biophys Res Commun. 2017;490:225–230. doi: 10.1016/j.bbrc.2017.06.025. [DOI] [PubMed] [Google Scholar]
- Dong W, Liu X, Li D, et al. Transcriptional profiling reveals that a MYB transcription factor MsMYB4 contributes to the salinity stress response of alfalfa. PLoS ONE. 2018;13:e0204033. doi: 10.1371/journal.pone.0204033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubova L, Šenberga A, Alsiņa I (2015) The effect of double inoculation on the broad bean (Viciafaba L.) yield quality. https://www.semanticscholar.org/paper/THE-EFFECT-OF-DOUBLE-INOCULATION-ON-THE-BROAD-BEANS-Dubova-%C5%A0enberga/f44c75d49a5b09d4c3b86b2e7c90a7eeb88336f7. Accessed 4 Jul 2021
- Duc G, Agrama H, Bao S, et al. Breeding annual grain legumes for sustainable agriculture: new methods to approach complex traits and target new cultivar ideotypes. Crit Rev Plant Sci. 2015;34:381–411. doi: 10.1080/07352689.2014.898469. [DOI] [Google Scholar]
- Dutta P, Bera A. Effect of NaCl salinity on seed germination and seedling growth of mungbean cultivars. Legume Res Int J. 2014;37:161–164. doi: 10.5958/j.0976-0571.37.2.024. [DOI] [Google Scholar]
- Egamberdieva D, Shurigin V, Gopalakrishnan S, Sharma R. Growth and symbiotic performance of chickpea (Cicer arietinum) cultivars under saline soil conditions. J Biol Chem Res. 2014;2014:1–10. [Google Scholar]
- Egamberdieva D, Wirth S, Jabborova D, et al. Coordination between Bradyrhizobium and Pseudomonas alleviates salt stress in soybean through altering root system architecture. J Plant Interact. 2017;12:100–107. doi: 10.1080/17429145.2017.1294212. [DOI] [Google Scholar]
- El-Awadi M, Sadak M, Dawood MG, et al. Amelioration the adverse effects of salinity stress by using γ-radiation in faba bean plants. Bull NRC. 2017;41:293–310. [Google Scholar]
- El-Bastawisy ZM, El-Katony TM, Abd El-Fatah SN. Genotypic variability in salt tolerance of Vicia faba during germination and early seedling growth. J King Saud Univ Sci. 2018;30:270–277. doi: 10.1016/j.jksus.2017.04.004. [DOI] [Google Scholar]
- El-Esawi MA, Al-Ghamdi AA, Ali HM, Alayafi AA. Azospirillum lipoferum FK1 confers improved salt tolerance in chickpea (Cicer arietinum L.) by modulating osmolytes, antioxidant machinery and stress-related genes expression. Environ Exp Bot. 2019 doi: 10.1016/j.envexpbot.2018.12.001. [DOI] [Google Scholar]
- El-Serafy RS, El-Sheshtawy A-NA, Atteya AK, et al. Seed priming with silicon as a potential to increase salt stress tolerance in Lathyrus odoratus. Plants. 2021;10:2140. doi: 10.3390/plants10102140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etesami H, Beattie G. Mining halophytes for plant growth-promoting halotolerant bacteria to enhance the salinity tolerance of non-halophytic crops. Front Microbiol. 2018 doi: 10.3389/fmicb.2018.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evelin H, Devi T, Gupta S, Kapoor R. Mitigation of salinity stress in plants by arbuscular mycorrhizal symbiosis: current understanding and new challenges. Front Plant Sci. 2019 doi: 10.3389/fpls.2019.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahmi AI, Nagaty HH, Eissa RA, Hassan MM. Effects of salt stress on some nitrogen fixation parameters in faba bean. Pak J Biol Sci PJBS. 2011;14:385–391. doi: 10.3923/pjbs.2011.385.391. [DOI] [PubMed] [Google Scholar]
- Farissi M, Bouizgaren A, Faghire M, et al. Agro-physiological responses of Moroccan alfalfa (Medicago sativa L.) populations to salt stress during germination and early seedling stages. Seed Sci Technol. 2011;39:389–401. doi: 10.15258/sst.2011.39.2.11. [DOI] [Google Scholar]
- Farooq M, Hussain M, Wakeel A, Siddique KHM. Salt stress in maize: effects, resistance mechanisms, and management A Review. Agron Sustain Dev. 2015;35:461–481. doi: 10.1007/s13593-015-0287-0. [DOI] [Google Scholar]
- Farooq M, Gogoi N, Hussain M, et al. Effects, tolerance mechanisms and management of salt stress in grain legumes. Plant Physiol Biochem. 2017;118:199–217. doi: 10.1016/j.plaphy.2017.06.020. [DOI] [PubMed] [Google Scholar]
- Fatnassi IC, Chiboub M, Saadani O, et al. Impact of dual inoculation with Rhizobium and PGPR on growth and antioxidant status of Vicia faba L. under copper stress. C R Biol. 2015;338:241–254. doi: 10.1016/j.crvi.2015.02.001. [DOI] [PubMed] [Google Scholar]
- Fernández-Pascual M, De Lorenzo C, De Felipe M, et al. Possible reasons for relative salt stress tolerance in nodules of white lupin cv. Multolupa. J Exp Bot. 1996;47:1709–1716. doi: 10.1093/jxb/47.11.1709. [DOI] [Google Scholar]
- Flowers T, Gaur P, Gowda C, et al. Salt sensitivity in chickpea. Plant Cell Environ. 2010;33:490–509. doi: 10.1111/j.1365-3040.2009.02051.x. [DOI] [PubMed] [Google Scholar]
- Forni C, Duca D, Glick BR. Mechanisms of plant response to salt and drought stress and their alteration by rhizobacteria. Plant Soil. 2017;410:335–356. doi: 10.1007/s11104-016-3007-x. [DOI] [Google Scholar]
- Fotopoulos V, Christou A, Antoniou C, Manganaris GA. REVIEW ARTICLE Hydrogen sulphide: a versatile tool for the regulation of growth and defence responses in horticultural crops. J Hortic Sci Biotechnol. 2015;90:227–234. doi: 10.1080/14620316.2015.11513176. [DOI] [Google Scholar]
- Gamalero E, Glick BR. Bacterial modulation of plant ethylene levels. Plant Physiol. 2015;169:13–22. doi: 10.1104/pp.15.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S-Q, Chen M, Xu Z-S, et al. The soybean GmbZIP1 transcription factor enhances multiple abiotic stress tolerances in transgenic plants. Plant Mol Biol. 2011;75:537–553. doi: 10.1007/s11103-011-9738-4. [DOI] [PubMed] [Google Scholar]
- Garg N, Manchanda G. Effect of arbuscular mycorrhizal inoculation on salt-induced nodule senescence in Cajanus cajan (pigeonpea) J Plant Growth Regul. 2008;27:115–124. doi: 10.1007/s00344-007-9038-z. [DOI] [Google Scholar]
- Garg N, Pandey R. Effectiveness of native and exotic arbuscular mycorrhizal fungi on nutrient uptake and ion homeostasis in salt-stressed Cajanus cajan L. (Millsp.) genotypes. Mycorrhiza. 2015;25:165–180. doi: 10.1007/s00572-014-0600-9. [DOI] [PubMed] [Google Scholar]
- Garg N, Singla R. Growth, photosynthesis, nodule nitrogen and carbon fixation in the chickpea cultivars under salt stress. Braz J Plant Physiol. 2004;16:137–146. doi: 10.1590/S1677-04202004000300003. [DOI] [Google Scholar]
- Geilfus C-M. Chloride: from nutrient to toxicant. Plant Cell Physiol. 2018;59:877–886. doi: 10.1093/pcp/pcy071. [DOI] [PubMed] [Google Scholar]
- Geilfus CM, Niehaus K, Gödde V, et al. Fast responses of metabolites in Vicia faba L. to moderate NaCl stress. Plant Physiol Biochem. 2015 doi: 10.1016/j.plaphy.2015.04.008. [DOI] [PubMed] [Google Scholar]
- Ghanbari M, Mokhtassi-Bidgoli A, Mansour Ghanaei-Pashaki K, Karamniya S. Germination characteristics and enzyme activity of mung bean (Vigna radiata) in response to methyl jasmonate and salinity treatments. Iran J Seed Res. 2020;7:83–97. doi: 10.29252/yujs.7.1.83. [DOI] [Google Scholar]
- Glick B. Plant growth-promoting bacteria: mechanisms and applications. Scientifica. 2012;2012:e963401. doi: 10.6064/2012/963401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BR, Todorovic B, Czarny J, et al. Promotion of plant growth by bacterial ACC deaminase. Crit Rev Plant Sci. 2007;26:227–242. doi: 10.1080/07352680701572966. [DOI] [Google Scholar]
- Gnanasambandam A, Paull J, Torres A, et al. Impact of molecular technologies on faba bean (Vicia faba L.) breeding strategies. Agronomy. 2012;2:132–166. doi: 10.3390/agronomy2030132. [DOI] [Google Scholar]
- Guan H, Liu X, Niu F, et al. OoNAC72, a NAC-type Oxytropis ochrocephala transcription factor, conferring enhanced drought and salt stress tolerance in Arabidopsis. Front Plant Sci. 2019;10:890. doi: 10.3389/fpls.2019.00890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajri R, Ouhibi C, Mechri M, et al. Salinity and water deficit effects on seed germination and recovery of lotus populations from northern Tunisia. Pak J Bot. 2018;50:2085–2090. [Google Scholar]
- Hanafy M, El-Banna A, Schumacher H, et al. Enhanced tolerance to drought and salt stresses in transgenic faba bean (Vicia faba L.) plants by heterologous expression of the PR10a gene from potato. Plant Cell Rep. 2013;32:663–674. doi: 10.1007/s00299-013-1401-x. [DOI] [PubMed] [Google Scholar]
- Hanin M, Ebel C, Ngom M, et al. New insights on plant salt tolerance mechanisms and their potential use for breeding. Front Plant Sci. 2016;7:1787. doi: 10.3389/fpls.2016.01787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HanumanthaRao B, Nair RM, Nayyar H. Salinity and high temperature tolerance in mungbean [Vigna radiata (L.) Wilczek] from a physiological perspective. Front Plant Sci. 2016;7:957. doi: 10.3389/fpls.2016.00957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasanuzzaman M, Araújo S, Gill SS. The plant family fabaceae. Berlin: Springer; 2020. [Google Scholar]
- Hashem A, Abd Allah E, Alqarawi A, et al. Alleviation of adverse impact of salinity on faba bean (Vicia faba L.) by arbuscular mycorrhizal fungi. Pak J Bot. 2014;46:2003–2013. [Google Scholar]
- Hashiguchi A, Komatsu S. Impact of post-translational modifications of crop proteins under abiotic stress. Proteomes. 2016;4:42. doi: 10.3390/proteomes4040042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan MA, Pacurar A, López-Gresa MP, et al. Effects of salt stress on three ecologically distinct plantago species. PLoS ONE. 2016 doi: 10.1371/journal.pone.0160236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanein R, El-Kazzaz A, Hashem H, et al. Transformation with the sodium/proton antiporter’AtNHX1’enhances salt tolerance in faba bean ('Vicia faba’ L.) Plant Omics. 2019;12:48. [Google Scholar]
- Ha-Tran DM, Nguyen TTM, Hung S-H, et al. Roles of plant growth-promoting rhizobacteria (PGPR) in stimulating salinity stress defense in plants: a review. Int J Mol Sci. 2021;22:3154. doi: 10.3390/ijms22063154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Fu J, Yu C, et al. Increasing cyclic electron flow is related to Na+ sequestration into vacuoles for salt tolerance in soybean. J Exp Bot. 2015;66:6877–6889. doi: 10.1093/jxb/erv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellal FA, Abdelhameid M, Abo-Basha DM, Zewainy RM. Alleviation of the adverse effects of soil salinity stress by foliar application of silicon on Faba bean (Vica faba L.) J Appl Sci Res. 2012;2012:4428–4433. [Google Scholar]
- Hernández-Lucero E, Rodríguez-Hernández AA, Ortega-Amaro MA, Jiménez-Bremont JF. Differential expression of genes for tolerance to salt stress in common bean (Phaseolus vulgaris L.) Plant Mol Biol Report. 2014;32:318–327. doi: 10.1007/s11105-013-0642-8. [DOI] [Google Scholar]
- Hiz MC, Canher B, Niron H, Turet M. Transcriptome analysis of salt tolerant common bean (Phaseolus vulgaris L.) under saline conditions. PLoS ONE. 2014;9:e92598. doi: 10.1371/journal.pone.0092598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hniličková H, Hnilička F, Orsák M, Hejnák V. Effect of salt stress on growth, electrolyte leakage, Na+ and K+ content in selected plant species. Plant Soil Environ. 2019;65:90–96. doi: 10.17221/620/2018-PSE. [DOI] [Google Scholar]
- Hoang XLT, Chuong NN, Hoa TTK, et al. The drought-mediated soybean GmNAC085 functions as a positive regulator of plant response to salinity. Int J Mol Sci. 2021;22:8986. doi: 10.3390/ijms22168986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horie T, Hauser F, Schroeder JI. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 2009;14:660–668. doi: 10.1016/j.tplants.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain MR, Pritchard J, Ford-Lloyd BV. Qualitative and quantitative variation in the mechanisms of salinity tolerance determined by multivariate assessment of diverse rice (Oryza sativa L.) genotypes. Plant Genet Resour. 2016;14:91. doi: 10.1017/S1479262115000118. [DOI] [Google Scholar]
- Ighodaro OM, Akinloye OA. First line defence antioxidants-superoxide dismutase (SOD), catalase (CAT) and glutathione peroxidase (GPX): Their fundamental role in the entire antioxidant defence grid. Alex J Med. 2018;54:287–293. doi: 10.1016/j.ajme.2017.09.001. [DOI] [Google Scholar]
- Ilangumaran G, Schwinghamer TD, Smith DL. Rhizobacteria from root nodules of an indigenous legume enhance salinity stress tolerance in soybean. Front Sustain Food Syst. 2021;4:308. doi: 10.3389/fsufs.2020.617978. [DOI] [Google Scholar]
- Irshad A, Rehman RNU, Abrar MM, et al. Contribution of rhizobium-legume symbiosis in salt stress tolerance in medicago truncatula evaluated through photosynthesis, antioxidant enzymes, and compatible solutes accumulation. Sustainability. 2021;13:3369. doi: 10.3390/su13063369. [DOI] [Google Scholar]
- Isayenkov SV, Maathuis FJM. Plant salinity stress: many unanswered questions remain. Front Plant Sci. 2019;10:80. doi: 10.3389/fpls.2019.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MM, Haque MS, Sarwar AG. Salt tolerance of cowpea genotypes during seed germination and seedling growth. J Bangl Agric Univ. 2019;17:39–44. doi: 10.3329/jbau.v17i1.40661. [DOI] [Google Scholar]
- Ivanchenko MG, Napsucialy-Mendivil S, Dubrovsky JG. Auxin-induced inhibition of lateral root initiation contributes to root system shaping in Arabidopsis thaliana. Plant J Cell Mol Biol. 2010;64:740–752. doi: 10.1111/j.1365-313X.2010.04365.x. [DOI] [PubMed] [Google Scholar]
- Jaarsma R, Vries R, Boer A. Effect of salt stress on growth, Na+ accumulation and proline metabolism in potato (Solanum tuberosum) Cultivars. PLoS ONE. 2013;8:e60183. doi: 10.1371/journal.pone.0060183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janczarek M, Skorupska A. The Rhizobium leguminosarum bv. trifolii RosR: transcriptional regulator involved in exopolysaccharide production. Mol Plant-Microbe Interact MPMI. 2007;20:867–881. doi: 10.1094/MPMI-20-7-0867. [DOI] [PubMed] [Google Scholar]
- Janczarek M, Kutkowska J, Piersiak T, Skorupska A. Rhizobium leguminosarum bv. trifolii rosR is required for interaction with clover, biofilm formation and adaptation to the environment. BMC Microbiol. 2010;10:284. doi: 10.1186/1471-2180-10-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jatan R, Chauhan PS, Lata C. Pseudomonas putida modulates the expression of miRNAs and their target genes in response to drought and salt stresses in chickpea (Cicer arietinum L.) Genomics. 2019;111:509–519. doi: 10.1016/j.ygeno.2018.01.007. [DOI] [PubMed] [Google Scholar]
- Jha B, Mishra A, Jha A, Joshi M. Developing transgenic Jatropha using the SbNHX1 gene from an extreme halophyte for cultivation in saline wasteland. PLoS ONE. 2013;8:e71136. doi: 10.1371/journal.pone.0071136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jha UC, Bohra A, Jha R, Parida SK. Salinity stress response and ‘omics’ approaches for improving salinity stress tolerance in major grain legumes. Plant Cell Rep. 2019;38:255–277. doi: 10.1007/s00299-019-02374-5. [DOI] [PubMed] [Google Scholar]
- Ji H, Pardo JM, Batelli G, et al. The Salt Overly Sensitive (SOS) pathway: established and emerging roles. Mol Plant. 2013;6:275–286. doi: 10.1093/mp/sst017. [DOI] [PubMed] [Google Scholar]
- Kaashyap M, Ford R, Kudapa H, et al. Differential regulation of genes involved in root morphogenesis and cell wall modification is associated with salinity tolerance in chickpea. Sci Rep. 2018;8:1–19. doi: 10.1038/s41598-018-23116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader MA, Lindberg S. Uptake of sodium in protoplasts of salt-sensitive and salt-tolerant cultivars of rice, Oryza sativa L. determined by the fluorescent dye SBFI. J Exp Bot. 2005;56:3149–3158. doi: 10.1093/jxb/eri312. [DOI] [PubMed] [Google Scholar]
- Kadian N, Yadav K, Badda N, Aggarwal A. AM fungi ameliorates growth, yield and nutrient uptake in Cicer arietinum L. under salt stress. Russ Agric Sci. 2013;39:321–329. doi: 10.3103/S1068367413040058. [DOI] [Google Scholar]
- Kang S, Waqas M, Khan A, Lee I. Use of microbes for the alleviation of soil stresses. New York: Springer; 2014. Plant-growth-promoting rhizobacteria: potential candidates for gibberellins production and crop growth promotion; pp. 1–19. [Google Scholar]
- Kang S-M, Radhakrishnan R, Khan AL, et al. Gibberellin secreting rhizobacterium, Pseudomonas putida H-2-3 modulates the hormonal and stress physiology of soybean to improve the plant growth under saline and drought conditions. Plant Physiol Biochem. 2014;84:115–124. doi: 10.1016/j.plaphy.2014.09.001. [DOI] [PubMed] [Google Scholar]
- Katerji N, van Hoorn JW, Hamdy A, Mastrorilli M. Salinity effect on crop development and yield, analysis of salt tolerance according to several classification methods. Agric Water Manag. 2003;62:37–66. doi: 10.1016/S0378-3774(03)00005-2. [DOI] [Google Scholar]
- Kaushal M, Wani SP. Rhizobacterial-plant interactions: strategies ensuring plant growth promotion under drought and salinity stress. Agric Ecosyst Environ. 2016;231:68–78. doi: 10.1016/j.agee.2016.06.031. [DOI] [Google Scholar]
- Kaundal R, Duhan N, Acharya BR, et al. Transcriptional profiling of two contrasting genotypes uncovers molecular mechanisms underlying salt tolerance in alfalfa. Sci Rep. 2021;11:5210. doi: 10.1038/s41598-021-84461-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavarzi MHB. Effect of salt stress on germination and early seedling growth of savory (Satureja hortensis) Aust J Basic Appl Sci. 2011;5:3274–3279. [Google Scholar]
- Ketehouli T, Idrice Carther KF, Noman M, et al. Adaptation of plants to salt stress: characterization of Na+ and K+ transporters and role of CBL gene family in regulating salt stress response. Agronomy. 2019;9:687. doi: 10.3390/agronomy9110687. [DOI] [Google Scholar]
- Khalil SK, Mexal JG, Murray LW. Germination of soybean seed primed in aerated solution of polyethylene glycol 8000. OnLine J Biol Sci. 2001;1:105–107. doi: 10.3923/jbs.2001.105.107. [DOI] [Google Scholar]
- Khan HA, Siddique KHM, Colmer TD. Vegetative and reproductive growth of salt-stressed chickpea are carbon-limited: sucrose infusion at the reproductive stage improves salt tolerance. J Exp Bot. 2017;68:2001–2011. doi: 10.1093/jxb/erw177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AL, Hamayun M, Kim Y-H, et al. Ameliorative symbiosis of endophyte (Penicillium funiculosum LHL06) under salt stress elevated plant growth of Glycine max L. Plant Physiol Biochem. 2011;49:852–861. doi: 10.1016/j.plaphy.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Khan MA, Asaf S, Khan AL, et al. Halotolerant rhizobacterial strains mitigate the adverse effects of NaCl stress in soybean seedlings. BioMed Res Int. 2019;2019:e9530963. doi: 10.1155/2019/9530963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandal H, Parween S, Roy R, et al. MicroRNA profiling provides insights into post-transcriptional regulation of gene expression in chickpea root apex under salinity and water deficiency. Sci Rep. 2017;7:1–14. doi: 10.1038/s41598-017-04906-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazaei H, O’Sullivan D, Stoddard F, et al. Recent advances in Faba Bean genetic and genomic tools for crop improvement. Legume Sci. 2020 doi: 10.2094/preprints202012.0372.v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kheloufi A, Chorfi A, Mansouri L-M. Germination kinetics in two Acacia karroo hayne ecotypes under salinity conditions. Open Access Libr J. 2017;4:1–11. [Google Scholar]
- Kim JH, Kim WT, Kang BG. IAA and N6-benzyladenine inhibit ethylene-regulated expression of ACC oxidase and ACC synthase genes in mungbean hypocotyls. Plant Cell Physiol. 2001;42:1056–1061. doi: 10.1093/pcp/pce133. [DOI] [PubMed] [Google Scholar]
- Kim S, Kang J, Cho D, et al. ABF2, an ABRE-binding bZIP factor, is an essential component of glucose signaling and its overexpression affects multiple stress tolerance. Plant J. 2004;40:75–87. doi: 10.1111/j.1365-313X.2004.02192.x. [DOI] [PubMed] [Google Scholar]
- Kloepper JW, Rodríguez-Kábana R, Zehnder GW, et al. Plant root-bacterial interactions in biological control of soilborne diseases and potential extension to systemic and foliar diseases. Australas Plant Pathol. 1999;28:21. doi: 10.1071/AP99003. [DOI] [Google Scholar]
- Kohli D, Joshi G, Deokar AA, et al. Identification and characterization of wilt and salt stress-responsive microRNAs in chickpea through high-throughput sequencing. PLoS ONE. 2014;9:e108851. doi: 10.1371/journal.pone.0108851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kołodziejek J. Seed germination responses to some environmental factors in the red feather (Trifolium rubens) Pak J Bot. 2018;50:59–65. [Google Scholar]
- Kong Z, Glick BR, Duan J, et al. Effects of 1-aminocyclopropane-1-carboxylate (ACC) deaminase-overproducing Sinorhizobium meliloti on plant growth and copper tolerance of Medicago lupulina. Plant Soil. 2015;391:383–398. doi: 10.1007/s11104-015-2434-4. [DOI] [Google Scholar]
- Korver RA, Koevoets IT, Testerink C. Out of shape during stress: a key role for auxin. Trends Plant Sci. 2018;23:783–793. doi: 10.1016/j.tplants.2018.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koskey G, Mburu SW, Kimiti JM, et al. Genetic characterization and diversity of rhizobium isolated from root nodules of mid-altitude climbing bean (Phaseolus vulgaris L.) varieties. Front Microbiol. 2018;9:968. doi: 10.3389/fmicb.2018.00968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronzucker HJ, Britto DT. Sodium transport in plants: a critical review. New Phytol. 2011;189:54–81. doi: 10.1111/j.1469-8137.2010.03540.x. [DOI] [PubMed] [Google Scholar]
- Kumar A. Germination behaviour of soybean varieties under different salinity stress. Int J Appl Agric Res. 2017;12:69–76. [Google Scholar]
- Kumar A, Verma J. Does plant–microbe interaction confer stress tolerance in plants: a review? Microbiol Res. 2018;207:41–52. doi: 10.1016/j.micres.2017.11.004. [DOI] [PubMed] [Google Scholar]
- Kumar S, Kalita A, Srivastava R, Sahoo L. Co-expression of Arabidopsis NHX1 and bar improves the tolerance to salinity, oxidative stress, and herbicide in transgenic mungbean. Front Plant Sci. 2017;8:1896. doi: 10.3389/fpls.2017.01896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar N, Bharadwaj C, Sahu S, et al. Genome-wide identification and functional prediction of salt-stress related long non-coding RNAs (lncRNAs) in chickpea (Cicer arietinum L.) Physiol Mol Biol Plants. 2021;27:2605–2619. doi: 10.1007/s12298-021-01093-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari P, Khanna V. (PDF) ACC-deaminase and EPS production by salt tolerant rhizobacteria augment growth in Chickpea under salinity stress. Int J Bio-Resour Stress Manag. 2015 doi: 10.5958/0976-4038.2015.00084.6. [DOI] [Google Scholar]
- Lan Y, Cai D, Zheng Y. Expression in Escherichia coli of three different soybean late embryogenesis abundant (LEA) genes to investigate enhanced stress tolerance. J Integr Plant Biol. 2005;47:613–621. doi: 10.1111/j.1744-7909.2005.00025.x. [DOI] [Google Scholar]
- Lavania D, Siddiqui M, Al-Whaibi M, et al. Genetic approaches for breeding heat stress tolerance in faba bean (Vicia faba L.) Acta Physiol Plant. 2014;37:1737. doi: 10.1007/s11738-014-1737-z. [DOI] [Google Scholar]
- Lavrenko S, Lavrenko N, Lykhovyd P. Effect of degree of salinity on seed germination and initial growth of chickpea (Cicer arietinum) Biosyst Divers. 2019;27:101–105. doi: 10.15421/011914. [DOI] [Google Scholar]
- Lelandais-Brière C, Naya L, Sallet E, et al. Genome-wide Medicago truncatula small RNA analysis revealed novel microRNAs and isoforms differentially regulated in roots and nodules. Plant Cell. 2009;21:2780–2796. doi: 10.1105/tpc.109.068130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire B, Dlodlo O, Chimphango S, et al. Symbiotic diversity, specificity and distribution of rhizobia in native legumes of the Core Cape Subregion (South Africa) FEMS Microbiol Ecol. 2015;91:1–17. doi: 10.1093/femsec/fiu024. [DOI] [PubMed] [Google Scholar]
- Leonforte A, Sudheesh S, Cogan NO, et al. SNP marker discovery, linkage map construction and identification of QTLs for enhanced salinity tolerance in field pea (Pisum sativum L.) BMC Plant Biol. 2013;13:161. doi: 10.1186/1471-2229-13-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-P, Tian A-G, Luo G-Z, et al. Soybean DRE-binding transcription factors that are responsive to abiotic stresses. Theor Appl Genet. 2005;110:1355–1362. doi: 10.1007/s00122-004-1867-6. [DOI] [PubMed] [Google Scholar]
- Li TY, Zhang Y, Liu H, et al. Stable expression of Arabidopsis vacuolar Na+/H+ antiporter gene AtNHX1, and salt tolerance in transgenic soybean for over six generations. Chin Sci Bull. 2010;55:1127–1134. doi: 10.1007/s11434-010-0092-8. [DOI] [Google Scholar]
- Li D, Zhang Y, Hu X, et al. Transcriptional profiling of Medicago truncatula under salt stress identified a novel CBF transcription factor MtCBF4 that plays an important role in abiotic stress responses. BMC Plant Biol. 2011;11:1–19. doi: 10.1186/1471-2229-11-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Wang D, Jin T, et al. The vacuolar Na+/H+ antiporter gene SsNHX1 from the halophyte salsola soda confers salt tolerance in transgenic alfalfa (Medicago sativa L.) Plant Mol Biol Report. 2011;29:278–290. doi: 10.1007/s11105-010-0224-y. [DOI] [Google Scholar]
- Li Z, Xu J, Gao Y, et al. The synergistic priming effect of exogenous salicylic acid and H2O2 on chilling tolerance enhancement during maize (Zea mays L.) seed germination. Front Plant Sci. 2017;8:1153. doi: 10.3389/fpls.2017.01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu Q, Feng H, et al. Dehydrin MtCAS31 promotes autophagic degradation under drought stress. Autophagy. 2020;16:862–877. doi: 10.1080/15548627.2019.1643656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Chen R, Jiang Q, et al. GmNAC06, a NAC domain transcription factor enhances salt stress tolerance in soybean. Plant Mol Biol. 2021;105:333–345. doi: 10.1007/s11103-020-01091-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Hou X, Li J, et al. High-resolution DNA methylome reveals that demethylation enhances adaptability to continuous cropping comprehensive stress in soybean. BMC Plant Biol. 2019;19:1–17. doi: 10.1186/s12870-019-1670-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Zou H-F, Wei W, et al. Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta. 2008;228:225–240. doi: 10.1007/s00425-008-0731-3. [DOI] [PubMed] [Google Scholar]
- Liu H-H, Tian X, Li Y-J, et al. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. RNA. 2008;14:836–843. doi: 10.1261/rna.895308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Wang T-Z, Zhang W-H. Sodium extrusion associated with enhanced expression of SOS1 underlies different salt tolerance between Medicago falcata and Medicago truncatula seedlings. Environ Exp Bot. 2015;110:46–55. doi: 10.1016/j.envexpbot.2014.09.005. [DOI] [Google Scholar]
- Liu W, Li R-J, Han T-T, et al. Salt stress reduces root meristem size by nitric oxide-mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiol. 2015;168:343–356. doi: 10.1104/pp.15.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Li M, Su L, et al. Negative feedback regulation of ABA biosynthesis in peanut (Arachis hypogaea): a transcription factor complex inhibits AhNCED1 expression during water stress. Sci Rep. 2016;6:37943. doi: 10.1038/srep37943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Liu J, Liu J, et al. The potassium transporter SlHAK10 is involved in mycorrhizal potassium uptake1[OPEN] Plant Physiol. 2019;180:465–479. doi: 10.1104/pp.18.01533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloret J, Wulff BBH, Rubio JM, et al. Exopolysaccharide II production is regulated by salt in the halotolerant strain Rhizobium meliloti EFB1. Appl Environ Microbiol. 1998;64:1024. doi: 10.1128/AEM.64.3.1024-1028.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long R, Li M, Kang J, et al. Small RNA deep sequencing identifies novel and salt-stress-regulated microRNAs from roots of Medicago sativa and Medicago truncatula. Physiol Plant. 2015;154:13–27. doi: 10.1111/ppl.12266. [DOI] [PubMed] [Google Scholar]
- López M, Herrera-Cervera JA, Iribarne C, et al. Growth and nitrogen fixation in Lotus japonicus and Medicago truncatula under NaCl stress: nodule carbon metabolism. J Plant Physiol. 2008;165:641–650. doi: 10.1016/j.jplph.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Luo G, Wang Y, Xie Z, et al. The putative Ser/Thr protein kinase gene GmAAPK from soybean is regulated by abiotic stress. J Integr Plant Biol. 2006;48:327–333. doi: 10.1111/j.1744-7909.2006.00169.x. [DOI] [Google Scholar]
- Ma W, Guinel FC, Glick BR. Rhizobium leguminosarum biovar viciae 1-aminocyclopropane-1-carboxylate deaminase promotes nodulation of pea plants. Appl Environ Microbiol. 2003;69:4396–4402. doi: 10.1128/AEM.69.8.4396-4402.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W, Charles TC, Glick BR. Expression of an exogenous 1-aminocyclopropane-1-carboxylate deaminase gene in sinorhizobium meliloti increases its ability to nodulate alfalfa. Appl Environ Microbiol. 2004;70:5891–5897. doi: 10.1128/AEM.70.10.5891-5897.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Zhao D, Ning Q, et al. A multi-year beneficial effect of seed priming with gibberellic acid-3 (GA 3) on plant growth and production in a perennial grass, Leymus chinensis. Sci Rep. 2018;8:13214. doi: 10.1038/s41598-018-31471-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwanga RO, Lu P, Kirungu JN, et al. Characterization of the late embryogenesis abundant (LEA) proteins family and their role in drought stress tolerance in upland cotton. BMC Genet. 2018;19:1–31. doi: 10.1186/s12863-017-0596-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchanda G, Garg N. Salinity and its effects on the functional biology of legumes. Acta Physiol Plant. 2008;30:595–618. doi: 10.1007/s11738-008-0173-3. [DOI] [Google Scholar]
- Manchanda G, Garg N. Alleviation of salt-induced ionic, osmotic and oxidative stresses in Cajanus cajan nodules by AM inoculation. Plant Biosyst. 2011;145:88–97. doi: 10.1080/11263504.2010.539851. [DOI] [Google Scholar]
- Mansouri L, Heleili N, Boukhatem Z, Kheloufi A. Seed germination and radicle establishment related to type and level of salt in common bean (Phaseolus vulgaris L. var. djedida) Cercet Agron În Mold Agron Res Mold. 2019;52:262–277. [Google Scholar]
- Matamoros MA, Dalton DA, Ramos J, et al. Biochemistry and molecular biology of antioxidants in the rhizobia-legume symbiosis. Plant Physiol. 2003;133:499–509. doi: 10.1104/pp.103.025619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbarki S, Skalicky M, Vachova P, et al. Comparing salt tolerance at seedling and germination stages in local populations of Medicago ciliaris L. to Medicago intertexta L. and Medicago scutellata L. Plants. 2020;9:526. doi: 10.3390/plants9040526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng Y, Chen F, Shuai H, et al. Karrikins delay soybean seed germination by mediating abscisic acid and gibberellin biogenesis under shaded conditions. Sci Rep. 2016;6:1–12. doi: 10.1038/srep22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metwali EMR, Abdelmoneim TS, Bakheit MA, Kadasa NMS. Alleviation of salinity stress in faba bean (Vicia faba L.) plants by inoculation with plant growth promoting rhizobacteria (PGPR) Plant Omics. 2015;8:449–460. [Google Scholar]
- Miao L, St. Clair DK. Regulation of superoxide dismutase genes: implications in diseases. Free Radic Biol Med. 2009;47:344–356. doi: 10.1016/j.freeradbiomed.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Williams M, Loewen PC, Oresnik IJ. Isolation of salt-sensitive mutants of Sinorhizobium meliloti strain Rm1021. Microbiol Read Engl. 2006;152:2049–2059. doi: 10.1099/mic.0.28937-0. [DOI] [PubMed] [Google Scholar]
- Mishra S, Behura R, Awasthi JP, et al. Ectopic overexpression of a mungbean vacuolar Na+/H+ antiporter gene (VrNHX1) leads to increased salinity stress tolerance in transgenic Vigna unguiculata L. Walp Mol Breed. 2014;34:1345–1359. doi: 10.1007/s11032-014-0120-5. [DOI] [Google Scholar]
- Mishra S, Panda SK, Sahoo L. Transgenic Asiatic grain legumes for salt tolerance and functional genomics. Rev Agric Sci. 2014;2:21–36. doi: 10.7831/ras.2.21. [DOI] [Google Scholar]
- Mishra S, Alavilli H, Lee B, et al. Cloning and characterization of a novel vacuolar Na+/H+ antiporter gene (VuNHX1) from drought hardy legume, cowpea for salt tolerance. Plant Cell Tissue Organ Cult PCTOC. 2015;120:19–33. doi: 10.1007/s11240-014-0572-7. [DOI] [Google Scholar]
- Mohammed A. Effectiveness of exopolysaccharides and biofilm forming plant growth promoting rhizobacteria on salinity tolerance of faba bean (Vicia faba L.) Afr J Microbiol Res. 2018;12:399–404. doi: 10.5897/AJMR2018.8822. [DOI] [Google Scholar]
- Mokrani S, Nabti E, Cruz C. Current advances in plant growth promoting bacteria alleviating salt stress for sustainable agriculture. Appl Sci. 2020;10:7025. doi: 10.3390/app10207025. [DOI] [Google Scholar]
- Moradi A. Effect of mycorrhizal inoculation on growth, nitrogen fixation and nutrient uptake in alfalfa (Medicago sativa) under salt stress. Cercetări Agronomice în Moldova. 2016;1(165):67–80. doi: 10.1515/cerce-2016-0006. [DOI] [Google Scholar]
- Moustafa-Farag M, Elkelish A, Dafea M, et al. Role of melatonin in plant tolerance to soil stressors: salinity, pH and heavy metals. Mol Basel Switz. 2020;25:E5359. doi: 10.3390/molecules25225359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchate N, Rajurkar N, Suprasanna P, Nikam T. NaCl induced salt adaptive changes and enhanced accumulation of 20-hydroxyecdysone in the in vitro shoot cultures of Spinacia oleracea (L.) Sci Rep. 2019;9:12522. doi: 10.1038/s41598-019-48737-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muscolo A, Panuccio MR, Zhair Z, et al. Use of plant growth-promoting rhizobacteria to ameliorate the performance of lentil under salinity: rhizobium and lentil under salinity. J Appl Bot Food Qual. 2019;92:179–186. doi: 10.5073/JABFQ.2019.092.024. [DOI] [Google Scholar]
- Mutch L, Young J. Diversity and specificity of Rhizobium leguminosarum biovar viciae on wild and cultivated legumes. Mol Ecol. 2004;13:2435–2444. doi: 10.1111/j.1365-294X.2004.02259.x. [DOI] [PubMed] [Google Scholar]
- Nadeem S, Ahmad M, Zahir Z, et al. The role of mycorrhizae and plant growth promoting rhizobacteria (PGPR) in improving crop productivity under stressful environments. Biotechnol Adv. 2014;32:429–448. doi: 10.1016/j.biotechadv.2013.12.005. [DOI] [PubMed] [Google Scholar]
- Nadeem M, Li J, Yahya M, et al. Grain legumes and fear of salt stress: focus on mechanisms and management strategies. Int J Mol Sci. 2019;20:799. doi: 10.3390/ijms20040799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najar R, Aydi S, Sassi-Aydi S, et al. Effect of salt stress on photosynthesis and chlorophyll fluorescence in Medicago truncatula. Plant Biosyst Int J Deal Asp Plant Biol. 2019;153:88–97. [Google Scholar]
- Nascimento FX, Rossi MJ, Soares CRFS, et al. New insights into 1-aminocyclopropane-1-carboxylate (ACC) deaminase phylogeny, evolution and ecological significance. PLoS ONE. 2014;9:e99168. doi: 10.1371/journal.pone.0099168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nascimento FX, Rossi MJ, Glick BR. Role of ACC deaminase in stress control of leguminous plants. In: Subramaniam G, Arumugam S, Rajendran V, editors. Plant growth promoting actinobacteria: a new avenue for enhancing the productivity and soil fertility of grain legumes. Singapore: Springer; 2016. pp. 179–192. [Google Scholar]
- Naz R, Bano A. Molecular and physiological responses of sunflower (Helianthus Annuus L.) to pgpr and sa under salt stress. Pak J Bot. 2015;47:35–42. [Google Scholar]
- Nguyen NT, Vu HT, Nguyen TT, et al. Co-expression of Arabidopsis AtAVP1 and AtNHX1 to improve salt tolerance in soybean. Crop Sci. 2019;59:1133–1143. doi: 10.2135/cropsci2018.10.0640. [DOI] [Google Scholar]
- Nguyen Q, Vu L, Nguyen L, et al. Overexpression of the GmDREB6 gene enhances proline accumulation and salt tolerance in genetically modified soybean plants. Sci Rep. 2019;9:19663. doi: 10.1038/s41598-019-55895-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning L, Kan G, Shao H, Yu D. Physiological and transcriptional responses to salt stress in salt-tolerant and salt-sensitive soybean (Glycine max [L.] Merr.) seedlings. Land Degrad Dev. 2018;29:2707–2719. doi: 10.1002/ldr.3005. [DOI] [Google Scholar]
- Noori F, Etesami H, Najafi Zarini H, et al. Mining alfalfa (Medicago sativa L.) nodules for salinity tolerant non-rhizobial bacteria to improve growth of alfalfa under salinity stress. Ecotoxicol Environ Saf. 2018;162:129–138. doi: 10.1016/j.ecoenv.2018.06.092. [DOI] [PubMed] [Google Scholar]
- Numan M, Bashir S, Khan Y, et al. Plant growth promoting bacteria as an alternative strategy for salt tolerance in plants: a review. Microbiol Res. 2018;209:21–32. doi: 10.1016/j.micres.2018.02.003. [DOI] [PubMed] [Google Scholar]
- Nyaga JW, Njeru EM. Potential of native rhizobia to improve cowpea growth and production in semiarid regions of Kenya. Front Agron. 2020;2:28. doi: 10.3389/fagro.2020.606293. [DOI] [Google Scholar]
- Okazaki S, Nukui N, Sugawara M, Minamisawa K. Rhizobial strategies to enhance symbiotic interactions: rhizobitoxine and 1-aminocyclopropane-1-carboxylate deaminase. Microb Environ. 2004;19:99–111. doi: 10.1264/jsme2.19.99. [DOI] [Google Scholar]
- Orabi S, Abdelhamid M. Protective role of α-tocopherol on two Vicia faba cultivars against seawater-induced lipid peroxidation by enhancing capacity of anti-oxidative system. J Saudi Soc Agric Sci. 2016;15:145–154. doi: 10.1016/j.jssas.2014.09.001. [DOI] [Google Scholar]
- Orozco-Mosqueda M, Glick B, Santoyo G. ACC deaminase in plant growth-promoting bacteria (PGPB): an efficient mechanism to counter salt stress in crops. Microbiol Res. 2020;235:126439. doi: 10.1016/j.micres.2020.126439. [DOI] [PubMed] [Google Scholar]
- Osman ME, Mohsen AA, Nessim AA, et al. Evaluation of biochar as a soil amendment for alleviating the harmful effect of salinity on Vigna unguiculata (L.) Walp. Egypt J Bot. 2019;59:617–631. [Google Scholar]
- Palma F, López-Gómez M, Tejera NA, Lluch C. Involvement of abscisic acid in the response of Medicago sativa plants in symbiosis with Sinorhizobium meliloti to salinity. Plant Sci Int J Exp Plant Biol. 2014;223:16–24. doi: 10.1016/j.plantsci.2014.02.005. [DOI] [PubMed] [Google Scholar]
- Pantola S, Bargali K, Bargali S. Effects of NaCl on germination and seedling growth in macrotyloma uniflorum and Vigna mungo. Curr Agric Res J. 2017;5:169. doi: 10.12944/CARJ.5.2.03. [DOI] [Google Scholar]
- Panwar M, Tewari R, Nayyar H. Native halo-tolerant plant growth promoting rhizobacteria Enterococcus and Pantoea sp. improve seed yield of Mungbean (Vigna radiata L.) under soil salinity by reducing sodium uptake and stress injury. Physiol Mol Biol Plants. 2016;22:445–459. doi: 10.1007/s12298-016-0376-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paparella S, Araújo SS, Rossi G, et al. Seed priming: state of the art and new perspectives. Plant Cell Rep. 2015;34:1281–1293. doi: 10.1007/s00299-015-1784-y. [DOI] [PubMed] [Google Scholar]
- de la Peña TC, Redondo FJ, Manrique E, et al. Nitrogen fixation persists under conditions of salt stress in transgenic Medicago truncatula plants expressing a cyanobacterial flavodoxin. Plant Biotechnol J. 2010;8:954–965. doi: 10.1111/j.1467-7652.2010.00519.x. [DOI] [PubMed] [Google Scholar]
- Pennycooke JC, Cheng H, Stockinger EJ. Comparative genomic sequence and expression analyses of Medicago truncatula and alfalfa subspecies falcata COLD-ACCLIMATION-SPECIFIC genes. Plant Physiol. 2008;146:1242–1254. doi: 10.1104/pp.107.108779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Percey WJ, Shabala L, Breadmore MC, et al. Ion transport in broad bean leaf mesophyll under saline conditions. Planta. 2014;240:729–743. doi: 10.1007/s00425-014-2117-z. [DOI] [PubMed] [Google Scholar]
- Phang T, Shao G, Lam H. Salt tolerance in soybean. J Integr Plant Biol. 2008;50:1196–1212. doi: 10.1111/j.1744-7909.2008.00760.x. [DOI] [PubMed] [Google Scholar]
- Pi E, Xu J, Li H, et al. Enhanced salt tolerance of rhizobia-inoculated soybean correlates with decreased phosphorylation of the transcription factor GmMYB183 and altered flavonoid biosynthesis*. Mol Cell Proteom. 2019;18:2225–2243. doi: 10.1074/mcp.RA119.001704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitann B, Kranz T, Zörb C, et al. Apoplastic pH and growth in expanding leaves of Vicia faba under salinity. Environ Exp Bot. 2011;74:31–36. doi: 10.1016/j.envexpbot.2011.04.015. [DOI] [Google Scholar]
- Podder S, Ray J, Das D, Sarker BC. Effect of salinity (NaCl) on germination and seedling growth of mungbean (Vigna radiata L.) J Biosci Agric Res. 2020;24:2012–2019. doi: 10.18801/jbar.240220.246. [DOI] [Google Scholar]
- Preston JC, Hileman L. Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Front Plant Sci. 2013;4:80. doi: 10.3389/fpls.2013.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qados AMSA. Effect of salt stress on plant growth and metabolism of bean plant Vicia faba (L.) J Saudi Soc Agric Sci. 2011;10:7–15. [Google Scholar]
- Qados AMSA. Mechanism of nanosilicon-mediated alleviation of salinity stress in faba bean (Vicia faba L.) plants. Am J Exp Agric. 2015;7:78–95. [Google Scholar]
- Qados A, Moftah A. Influence of silicon and nano-silicon on germination, growth and yield of Faba Bean (Vicia faba L.) under salt stress conditions. Am J Exp Agric. 2015 doi: 10.9734/ajea/2015/14109. [DOI] [Google Scholar]
- Qu L, Huang Y, Zhu C, et al. Rhizobia-inoculation enhances the soybean’s tolerance to salt stress. Plant Soil. 2016;400:209–222. doi: 10.1007/s11104-015-2728-6. [DOI] [Google Scholar]
- Quan W, Liu X, Wang H, Chan Z. Physiological and transcriptional responses of contrasting alfalfa (Medicago sativa L.) varieties to salt stress. Plant Cell Tissue Organ Cult. 2016 doi: 10.1007/s11240-016-0981-x. [DOI] [Google Scholar]
- Rabie GH, Almadini AM. Role of bioinoculants in development of salt-tolerance of Vicia faba plants under salinity stress. Afr J Biotechnol. 2005;4:210–222. doi: 10.5897/AJB2005.000-3041. [DOI] [Google Scholar]
- Rabiei Z, Hosseini SJ, Pirdashti H, Hazrati S. Physiological and biochemical traits in coriander affected by plant growth-promoting rhizobacteria under salt stress. Heliyon. 2020;6:e05321. doi: 10.1016/j.heliyon.2020.e05321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman M, Bhuiyan M, Ali M, et al. Effect of arbuscular mycorrhizal fungi on the tolerance to sodium chloride levels, and on growth and yield of lentil (Lens culinaris) Agricult. 2017;15:156–169. doi: 10.3329/agric.v15i1.33439. [DOI] [Google Scholar]
- Razzaq MK, Akhter M, Ahmad RM, et al. CRISPR-Cas9 based stress tolerance: new hope for abiotic stress tolerance in chickpea (Cicer arietinum) Mol Biol Rep. 2022;49:8977–8985. doi: 10.1007/s11033-022-07391-4. [DOI] [PubMed] [Google Scholar]
- Rohman MM, Molla MR, Akhi AH, et al. The plant family Fabaceae. Berlin: Springer; 2020. Use of osmolytes for improving abiotic stress tolerance in Fabaceae plants; pp. 181–222. [Google Scholar]
- Russo DM, Williams A, Edwards A, et al. Proteins exported via the PrsD-PrsE type I secretion system and the acidic exopolysaccharide are involved in biofilm formation by Rhizobium leguminosarum. J Bacteriol. 2006;188:4474–4486. doi: 10.1128/JB.00246-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagervanshi A, Naeem A, Geilfus C-M, et al. One-time abscisic acid priming induces long-term salinity resistance in Vicia faba: Changes in key transcripts, metabolites, and ionic relations. Physiol Plant. 2021;172:146–161. doi: 10.1111/ppl.13315. [DOI] [PubMed] [Google Scholar]
- Saghari M, Khoshrou V, Alahmadi MJ, Foroughifar H. The effect of salinity stress on germination and growth characteristics of haloxylon aphyllum and halothamnus subaphyllus. Plant Arch. 2020;20:3664–3668. [Google Scholar]
- Saha P, Chatterjee P, Biswas AK. NaCl pretreatment alleviates salt stress by enhancement of antioxidant defense system and osmolyte accumulation in mungbean (Vigna radiata L. Wilczek) Indian J Exp Bio. 2010;48:593–600. [PubMed] [Google Scholar]
- Sahoo D, Kumar S, Mishra S, et al. Enhanced salinity tolerance in transgenic mungbean overexpressing Arabidopsis antiporter (NHX1) gene. Mol Breed. 2016;36:144. doi: 10.1007/s11032-016-0564-x. [DOI] [Google Scholar]
- Saïdi S, Ramírez-Bahena M, Santillana N, et al. Rhizobium laguerreae sp. nov. nodulates Vicia faba on several continents. Int J Syst Evol Microbiol. 2014;64:242–247. doi: 10.1099/ijs.0.052191-0. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Maruyama K, Osakabe Y, et al. Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell. 2006;18:1292–1309. doi: 10.1105/tpc.105.035881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salgotra RK, Gupta M. Epigenetics in plants of agronomic importance: fundamentals and applications. Cham: Springer; 2019. Exploring the role of epigenetics in cereal and leguminous crops exposed to abiotic stress; pp. 149–170. [Google Scholar]
- Sallam A, Ul-Allah S. Genomics-aided breeding for climate-smart traits in Faba Bean. In: Kole C, editor. Genomic designing of climate-smart pulse crops. Cham: Springer; 2019. pp. 359–395. [Google Scholar]
- Sanyal D, Bangerth F. Stress induced ethylene evolution and its possible relationship to auxin-transport, cytokinin levels, and flower bud induction in shoots of apple seedlings and bearing apple trees. Plant Growth Regul. 1998;24:127–134. doi: 10.1023/A:1005948918382. [DOI] [Google Scholar]
- Sarkar T, Thankappan R, Kumar A, et al. Heterologous expression of the AtDREB1A gene in transgenic peanut-conferred tolerance to drought and salinity stresses. PLoS ONE. 2014;9:e110507. doi: 10.1371/journal.pone.0110507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarker U, Oba S. The response of salinity stress-induced A. tricolor to growth, anatomy, physiology, non-enzymatic and enzymatic antioxidants. Front Plant Sci. 2020 doi: 10.3389/fpls.2020.559876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segami S, Makino S, Miyake A, et al. Dynamics of vacuoles and H+-pyrophosphatase visualized by monomeric green fluorescent protein in Arabidopsis: artifactual bulbs and native intravacuolar spherical structures. Plant Cell. 2014;26:3416–3434. doi: 10.1105/tpc.114.127571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semida WM, Taha RS, Abdelhamid MT, Rady MM. Foliar-applied α-tocopherol enhances salt-tolerance in Vicia faba L. plants grown under saline conditions. South Afr J Bot. 2014;95:24–31. doi: 10.1016/j.sajb.2014.08.005. [DOI] [Google Scholar]
- Sevanayak D, Edna A, Koti R. Salinity tolerance of forage range legumes during germination and early seedling growth. Progressive Res. 2020;12:1357–1360. [Google Scholar]
- Sadak M, Abdelhamid M, Schmidhalter U. Effect of foliar application of aminoacids on plant yield and some physiological parameters in bean plants irrigated with seawater. Acta Biológica Colomb. 2015;20:141–152. doi: 10.15446/abc.v20n1.42865. [DOI] [Google Scholar]
- Sehrawat N, Yadav M, Sharma AK, et al. Salt stress and mungbean [Vigna radiata (L.) Wilczek]: effects, physiological perspective and management practices for alleviating salinity. Arch Agron Soil Sci. 2019;65:1287–1301. doi: 10.1080/03650340.2018.1562548. [DOI] [Google Scholar]
- Shahzad M, Zörb C, Geilfus C, Mühling KH. Apoplastic Na+ in Vicia faba leaves rises after short-term salt stress and is remedied by silicon. J Agron Crop Sci. 2013;199:161–170. doi: 10.1111/jac.12003. [DOI] [Google Scholar]
- Sheidaei S, Zahedi M, Meibodo S. Effect of salinity stress on dry matter accumulation and ion distribution pattern in five safflower (Carthamus tinctorius L.) genotypes. Iran J Field Crop Sci. 2011;41:811–819. [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu J-K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell. 2002;14:465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrivastava P, Kumar R. Soil salinity: a serious environmental issue and plant growth promoting bacteria as one of the tools for its alleviation. Saudi J Biol Sci. 2015;22:123–131. doi: 10.1016/j.sjbs.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu K, Qi Y, Chen F, et al. Salt stress represses soybean seed germination by negatively regulating GA biosynthesis while positively mediating ABA biosynthesis. Front Plant Sci. 2017;8:1372. doi: 10.3389/fpls.2017.01372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla PS, Agarwal PK, Jha B. Improved salinity tolerance of Arachis hypogaea (L.) by the interaction of halotolerant plant-growth-promoting rhizobacteria. J Plant Growth Regul. 2012;31:195–206. doi: 10.1007/s00344-011-9231-y. [DOI] [Google Scholar]
- Sidari M, Santonoceto C, Anastasi U, et al. Variations in four genotypes of lentil under NaCl-salinity stress. Am J Agric Biol Sci. 2008;3:410–416. doi: 10.3844/ajabssp.2008.410.416. [DOI] [Google Scholar]
- Silva LR, Bento C, Gonçalves AC, et al. Legume bioactive compounds: influence of rhizobial inoculation. AIMS Microbiol. 2017;3:267–278. doi: 10.3934/microbiol.2017.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Shelke G, Kumar A, Jha P. Biochemistry and genetics of ACC deaminase: a weapon to “stress ethylene” produced in plants. Front Microbiol. 2015;6:937. doi: 10.3389/fmicb.2015.00937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slama I, Abdelly C, Bouchereau A, et al. Diversity, distribution and roles of osmoprotective compounds accumulated in halophytes under abiotic stress. Ann Bot. 2015;115:433–447. doi: 10.1093/aob/mcu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smýkal P, Coyne C, Ambrose M, et al. Legume crops phylogeny and genetic diversity for science and breeding. Crit Rev Plant Sci. 2015;34:43–104. doi: 10.1080/07352689.2014.897904. [DOI] [Google Scholar]
- Souana K, Taïbi K, Abderrahim LA, et al. Salt-tolerance in Vicia faba L. is mitigated by the capacity of salicylic acid to improve photosynthesis and antioxidant response. Sci Hortic. 2020;273:109641. doi: 10.1016/j.scienta.2020.109641. [DOI] [Google Scholar]
- Stoddard FL, Balko C, Erskine W, et al. Screening techniques and sources of resistance to abiotic stresses in cool-season food legumes. Euphytica. 2006;147:167–186. doi: 10.1007/s10681-006-4723-8. [DOI] [Google Scholar]
- Sun Y, Wang D, Bai Y, et al. Studies on the overexpression of the soybean GmNHX1 in Lotus corniculatus: the reduced Na+ level is the basis of the increased salt tolerance. Chin Sci Bull. 2006;51:1306–1315. doi: 10.1007/s11434-006-1306-y. [DOI] [Google Scholar]
- Sun Z, Wang Y, Mou F, et al. Genome-wide small RNA analysis of soybean reveals auxin-responsive microRNAs that are differentially expressed in response to salt stress in root apex. Front Plant Sci. 2016;6:1273. doi: 10.3389/fpls.2015.01273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun T, Ma N, Wang C, et al. A golgi-localized sodium/hydrogen exchanger positively regulates salt tolerance by maintaining higher K+/Na+ ratio in soybean. Front Plant Sci. 2021;12:638340. doi: 10.3389/fpls.2021.638340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R, Chinnusamy V, Zhu J, Zhu J-K. Small RNAs as big players in plant abiotic stress responses and nutrient deprivation. Trends Plant Sci. 2007;12:301–309. doi: 10.1016/j.tplants.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Swaraj K, Bishnoi NR (1999) Effect of salt stress on nodulation and nitrogen fixation in legumes. IJEB 37(09): 843-848 [PubMed]
- Tajini F, Trabelsi M, Drevon J. Combined inoculation with Glomus intraradices and Rhizobium tropici CIAT899 increases phosphorus use efficiency for symbiotic nitrogen fixation in common bean (Phaseolus vulgaris L.) Saudi J Biol Sci. 2012;19:157–163. doi: 10.1016/j.sjbs.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talukdar D. Isolation and characterization of NaCl-tolerant mutations in two important legumes, Clitoria ternatea L. and Lathyrus sativus L.: induced mutagenesis and selection by salt stress. J Med Plants Res. 2011;5:3619–3628. [Google Scholar]
- Tang R, Li C, Xu K, et al. Isolation, functional characterization, and expression pattern of a Vacuolar Na+/H+ antiporter Gene TrNHX1 from Trifolium repens L. Plant Mol Biol Report. 2010;28:102–111. doi: 10.1007/s11105-009-0135-y. [DOI] [Google Scholar]
- Tavakkoli E, Rengasamy P, McDonald G. High concentrations of Na+ and Cl– ions in soil solution have simultaneous detrimental effects on growth of faba bean under salinity stress. J Exp Bot. 2010;61:4449–4459. doi: 10.1093/jxb/erq251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavakkoli E, Paull J, Rengasamy P, McDonald G. Comparing genotypic variation in faba bean (Vicia faba L.) in response to salinity in hydroponic and field experiments. Field Crops Res. 2012;127:99–108. doi: 10.1016/j.fcr.2011.10.016. [DOI] [Google Scholar]
- Teakle N, Tyerman S. Mechanisms of Cl- transport contributing to salt tolerance. Plant Cell Environ. 2010;33:566–589. doi: 10.1111/j.1365-3040.2009.02060.x. [DOI] [PubMed] [Google Scholar]
- Tester M, Davenport R. Na+ tolerance and Na+ transport in higher plants. Ann Bot. 2003;91:503–527. doi: 10.1093/aob/mcg058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson AJ, Jackson AC, Symonds RC, et al. Ectopic expression of a tomato 9-cis-epoxycarotenoid dioxygenase gene causes over-production of abscisic acid. Plant J Cell Mol Biol. 2000;23:363–374. doi: 10.1046/j.1365-313x.2000.00789.x. [DOI] [PubMed] [Google Scholar]
- Tian C, Wang E, Wu L, et al. Rhizobium fabae sp. nov., a bacterium that nodulates Vicia faba. Int J Syst Evol Microbiol. 2008;58:2871–2875. doi: 10.1099/ijs.0.2008/000703-0. [DOI] [PubMed] [Google Scholar]
- Tian H, Guo F, Zhang Z, et al. Discovery, identification, and functional characterization of long noncoding RNAs in Arachis hypogaea L. BMC Plant Biol. 2020;20:1–16. doi: 10.1186/s12870-020-02510-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmusk S, Paalme V, Pavlicek T, et al. Bacterial distribution in the rhizosphere of wild barley under contrasting microclimates. PLoS ONE. 2011;6:e17968. doi: 10.1371/journal.pone.0017968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker C. Mutagenesis: exploring novel genes and pathways. Wageningen: Wageningen Academic Publishers; 2014. Mutagenesis for resistance to abiotic stresses: chickpea as model crop; pp. 78–81. [Google Scholar]
- Tlahig S, Bellani L, Karmous I, et al. Response to salinity in legume species: an insight on the effects of salt stress during seed germination and seedling growth. Chem Biodivers. 2021;18:e2000917. doi: 10.1002/cbdv.202000917. [DOI] [PubMed] [Google Scholar]
- Tsavkelova E. Bacteria Associated with Orchid Roots. In: Maheshwari DK, editor. Bacteria in agrobiology: plant growth responses. Berlin: Springer; 2011. pp. 221–258. [Google Scholar]
- Uno Y, Furihata T, Abe H, et al. Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci. 2000;97:11632–11637. doi: 10.1073/pnas.190309197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dam J, Faaij AP, Hilbert J, et al. Large-scale bioenergy production from soybeans and switchgrass in Argentina: Part B. Environmental and socio-economic impacts on a regional level. Renew Sustain Energy Rev. 2009;13:1679–1709. doi: 10.1016/j.rser.2009.03.012. [DOI] [Google Scholar]
- Vance ME, Kuiken T, Vejerano EP, et al. Nanotechnology in the real world: redeveloping the nanomaterial consumer products inventory. Beilstein J Nanotechnol. 2015 doi: 10.3762/bjnano.6.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahab AMA, Zahran HH. Effects of salt stress on nitrogenase activity and growth of four legumes. Biol Plant. 1981;23:16. doi: 10.1007/BF02909205. [DOI] [Google Scholar]
- Wang W, Vinocur B, Altman A. Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta. 2003;218:1–14. doi: 10.1007/s00425-003-1105-5. [DOI] [PubMed] [Google Scholar]
- Wang S-M, Zhang J-L, Flowers TJ. Low-affinity Na+ uptake in the halophyte Suaeda maritima. Plant Physiol. 2007;145:559–571. doi: 10.1104/pp.107.104315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T-Z, Liu M, Zhao M-G, et al. Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing. BMC Plant Biol. 2015;15:1–13. doi: 10.1186/s12870-015-0530-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Chai C, Valliyodan B, et al. Genome-wide analysis and expression profiling of the PIN auxin transporter gene family in soybean (Glycine max) BMC Genom. 2015;16:951. doi: 10.1186/s12864-015-2149-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Cheng K, Wan L, et al. Genome-wide analysis of the basic leucine zipper (bZIP) transcription factor gene family in six legume genomes. BMC Genom. 2015;16:1–15. doi: 10.1186/s12864-015-2258-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Xia MX, Chen J, et al. Gene expression characteristics and regulation mechanisms of superoxide dismutase and its physiological roles in plants under stress. Biochem Mosc. 2016;81:465–480. doi: 10.1134/S0006297916050047. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhang Z, Zhang P, et al. Rhizobium symbiosis contribution to short-term salt stress tolerance in alfalfa (Medicago sativa L.) Plant Soil. 2016;402:247–261. doi: 10.1007/s11104-016-2792-6. [DOI] [Google Scholar]
- Wang J, Meng X, Dobrovolskaya OB, et al. Non-coding RNAs and their roles in stress response in plants. GenomicsProteom Bioinform. 2017;15:301–312. doi: 10.1016/j.gpb.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Jiang L, Chen J, et al. Overexpression of the alfalfa WRKY11 gene enhances salt tolerance in soybean. PLoS ONE. 2018;13:e0192382. doi: 10.1371/journal.pone.0192382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Yang Q, Shao Y, et al. GmLEA2-1, a late embryogenesis abundant protein gene isolated from soybean (Glycine max (L.) Merr.), confers tolerance to abiotic stress. Acta Biol Hung. 2018;69:270–282. doi: 10.1556/018.68.2018.3.4. [DOI] [PubMed] [Google Scholar]
- Wang H, Liang L, Liu B, et al. Arbuscular mycorrhizas regulate photosynthetic capacity and antioxidant defense systems to mediate salt tolerance in maize. Plants. 2020;9:1430. doi: 10.3390/plants9111430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Xun H, Wang W et al (2021) Mutation of GmAITR genes by CRISPR/Cas9 genome editing results in enhanced salinity stress tolerance in soybean. Front Plant Sci 2752 [DOI] [PMC free article] [PubMed]
- Welgama A, Florentine S, Marchante H, et al. The germination success of Acacia longifolia subsp. longifolia (Fabaceae): a comparison between its native and exotic ranges. Aust J Bot. 2019;67:414–424. doi: 10.1071/BT19018. [DOI] [Google Scholar]
- Williams L, Grigg SP, Xie M et al (2005) Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. 132(16):3657–3668 [DOI] [PubMed]
- Windels D, Dang TT, Chen Z, Verdier J. Snapshot of epigenetic regulation in legumes. Legume Sci. 2021;3:e60. doi: 10.1002/leg3.60. [DOI] [Google Scholar]
- Winicov I, Bastola D. Transgenic overexpression of the transcription factor Alfin1 enhances expression of the endogenous MsPRP2 gene in alfalfa and improves salinity tolerance of the plants. Plant Physiol. 1999;120:473–480. doi: 10.1104/pp.120.2.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Li Z. The importance of Cl− exclusion and vacuolar Cl− sequestration: revisiting the role of Cl− transport in plant salt tolerance. Front Plant Sci. 2019 doi: 10.3389/fpls.2019.01418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier LJC, Germida JJ. Response of lentil under controlled conditions to co-inoculation with arbuscular mycorrhizal fungi and rhizobia varying in efficacy. Soil Biol Biochem. 2002;34:181–188. doi: 10.1016/S0038-0717(01)00165-1. [DOI] [Google Scholar]
- Xu J, Xue C, Xue D, et al. Overexpression of GmHsp90s, a heat shock protein 90 (Hsp90) gene family cloning from soybean, decrease damage of abiotic stresses in Arabidopsis thaliana. PLoS ONE. 2013;8:e69810. doi: 10.1371/journal.pone.0069810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F, Zhu Y, Zhao Y, et al. De novo transcriptome sequencing and analysis of salt-, alkali-, and drought-responsive genes in Sophora alopecuroides. BMC Genom. 2020;21:423. doi: 10.1186/s12864-020-06823-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Wu M, Wang P, et al. Cloning and expression analysis of a vacuolar Na+/H+ antiporter gene from Alfalfa. DNA Seq. 2005;16:352–357. doi: 10.1080/10425170500272742. [DOI] [PubMed] [Google Scholar]
- Yang S, Zhang Z, Xue Y, et al. Arbuscular mycorrhizal fungi increase salt tolerance of apple seedlings. Bot Stud. 2014;55:70. doi: 10.1186/s40529-014-0070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yu T-F, Ma J, et al. The soybean bZIP transcription factor gene GmbZIP2 confers drought and salt resistances in transgenic plants. Int J Mol Sci. 2020;21:670. doi: 10.3390/ijms21020670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasin NA, Khan WU, Ahmad SR, et al. Imperative roles of halotolerant plant growth-promoting rhizobacteria and kinetin in improving salt tolerance and growth of black gram (Phaseolus mungo) Environ Sci Pollut Res. 2018;25:4491–4505. doi: 10.1007/s11356-017-0761-0. [DOI] [PubMed] [Google Scholar]
- Yasir TA, Khan A, Skalicky M, et al. Exogenous sodium nitroprusside mitigates salt stress in lentil (Lens culinaris Medik.) by affecting the growth, yield, and biochemical properties. Molecules. 2021;26:2576. doi: 10.3390/molecules26092576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yinsuo J, Gray V, Straker C. The influence of Rhizobium and arbuscular mycorrhizal fungi on nitrogen and phosphorus accumulation by Vicia faba. Ann Bot. 2004;94:251–258. doi: 10.1093/aob/mch135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S-J, Weon H-Y, Sang J, Sang MK. Induced tolerance to salinity stress by halotolerant bacteria Bacillus aryabhattai H19-1 and B mesonae H20-5 in tomato plants. J Microbiol Biotechnol. 2019;29:1124–1136. doi: 10.4014/jmb.1904.04026. [DOI] [PubMed] [Google Scholar]
- Younesi O, Baghbani A, Namdari A. The effects of Pseudomonas fluorescence and Rhizobium meliloti co-inoculation on nodulation and mineral nutrient contents in alfalfa (Medicago sativa) under salinity stress. Int J Agric Crop Sci IJACS. 2013;5:1500–1507. [Google Scholar]
- Yousef A, Sprent J. Effects of NaCl on growth, nitrogen incorporation and chemical composition of inoculated and NH4NO3 fertilized Vicia faba (L.) plants. J Exp Bot. 1983;34:941–950. doi: 10.1093/jxb/34.8.941. [DOI] [Google Scholar]
- Yousef F, Shafique F, Ali Q, Malik A. Effects of salt stress on the growth traits of chickpea (Cicer arietinum L.) and pea (Pisum sativum L.) seedlings. Biol Clin Sci Res J. 2020;2020:1. doi: 10.54112/bcsrj.v2020i1.29. [DOI] [Google Scholar]
- Yu X, Liu Y, Wang S, et al. CarNAC4, a NAC-type chickpea transcription factor conferring enhanced drought and salt stress tolerances in Arabidopsis. Plant Cell Rep. 2016;35:613–627. doi: 10.1007/s00299-015-1907-5. [DOI] [PubMed] [Google Scholar]
- Yu S, Yu L, Hou Y, et al. Contrasting effects of NaCl and NaHCO3 stresses on seed germination, seedling growth, photosynthesis, and osmoregulators of the common bean (Phaseolus vulgaris L.) Agronomy. 2019;9:409. doi: 10.3390/agronomy9080409. [DOI] [Google Scholar]
- Yuan C, Li C, Lu X, et al. Comprehensive genomic characterization of NAC transcription factor family and their response to salt and drought stress in peanut. BMC Plant Biol. 2020;20:1–21. doi: 10.1186/s12870-020-02678-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yung W, Li M, Sze C, et al. Histone modifications and chromatin remodelling in plants in response to salt stress. Physiol Plant. 2021;173:1495–1513. doi: 10.1111/ppl.13467. [DOI] [PubMed] [Google Scholar]
- Zahaf O, Blanchet S, de Zélicourt A, et al. Comparative transcriptomic analysis of salt adaptation in roots of contrasting medicago truncatula genotypes. Mol Plant. 2012;5:1068–1081. doi: 10.1093/mp/sss009. [DOI] [PubMed] [Google Scholar]
- Zahran HH, Marín-Manzano MC, Sánchez-Raya AJ, et al. Effect of salt stress on the expression of NHX-type ion transporters in Medicago intertexta and Melilotus indicus plants. Physiol Plant. 2007;131:122–130. doi: 10.1111/j.1399-3054.2007.00940.x. [DOI] [PubMed] [Google Scholar]
- Zahran HH, Sprent JI. Effects of sodium chloride and polyethylene glycol on root-hair infection and nodulation of Vicia faba L. plants by Rhizobium leguminosarum. Planta. 1986;167:303–309. doi: 10.1007/BF00391332. [DOI] [PubMed] [Google Scholar]
- Zhang X, Harper R, Karsisto M, Lindström K. Diversity of rhizobium bacteria isolated from the root nodules of leguminous trees. Int J Syst Evol Microbiol. 1991;41:104–113. doi: 10.1099/00207713-41-1-104. [DOI] [Google Scholar]
- Zhang W-H, Skerrett M, Walker NA, et al. Nonselective currents and channels in plasma membranes of protoplasts from coats of developing seeds of bean. Plant Physiol. 2002;128:388–399. doi: 10.1104/pp.010566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Walker NA, Patrick JW, Tyerman SD. Calcium-dependent K current in plasma membranes of dermal cells of developing bean cotyledons. Plant Cell Environ. 2004;27:251–262. doi: 10.1046/j.1365-3040.2004.01152.x. [DOI] [Google Scholar]
- Zhang Y, Zheng W, Everall I, et al. Rhizobium anhuiense sp. nov., isolated from effective nodules of Vicia faba and Pisum sativum. Int J Syst Evol Microbiol. 2015;65:2960–2967. doi: 10.1099/ijs.0.000365. [DOI] [PubMed] [Google Scholar]
- Zhang H, Yasmin F, Song BH. Neglected treasures in the wild—legume wild relatives in food security and human health. Curr Opin Plant Biol. 2019;49:17–20. doi: 10.1016/j.pbi.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Wang N, Yang J, et al. The salt-induced transcription factor GmMYB84 confers salinity tolerance in soybean. Plant Sci. 2020;291:110326. doi: 10.1016/j.plantsci.2019.110326. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Bao Y. Effect of arbuscular mycorrhizal fungi on growth and two phenolic acids of Medicago sativa under various mixed salt-alkaline stresses. Acta Bot Boreali-Occident Sin. 2015;35:1829–1836. [Google Scholar]
- Zhou Q, Tian A, Zou H, et al. Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol J. 2008;6:486–503. doi: 10.1111/j.1467-7652.2008.00336.x. [DOI] [PubMed] [Google Scholar]
- Zhou SJ, Jing Z, Shi JL. Genome-wide identification, characterization, and expression analysis of the MLO gene family in Cucumis sativus. Genet Mol Res. 2013;12:6565–6578. doi: 10.4238/2013.December.11.8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing not applicable to this article as no data sets were generated or analyzed during the present study.