Abstract
Genome-wide significant loci for metformin response in type 2 diabetes reported elsewhere have not been replicated in the Diabetes Prevention Program (DPP). To assess pharmacogenetic interactions in prediabetes, we conducted a genome-wide association study (GWAS) in the DPP. Cox proportional hazards models tested associations with diabetes incidence in the metformin (MET; n = 876) and placebo (PBO; n = 887) arms. Multiple linear regression assessed association with 1-year change in metformin-related quantitative traits, adjusted for baseline trait, age, sex, and 10 ancestry principal components. We tested for gene-by-treatment interaction. No significant associations emerged for diabetes incidence. We identified four genome-wide significant variants after correcting for correlated traits (P < 9 × 10−9). In the MET arm, rs144322333 near ENOSF1 (minor allele frequency [MAF]AFR = 0.07; MAFEUR = 0.002) was associated with an increase in percentage of glycated hemoglobin (per minor allele, β = 0.39 [95% CI 0.28, 0.50]; P = 2.8 × 10−12). rs145591055 near OMSR (MAF = 0.10 in American Indians) was associated with weight loss (kilograms) (per G allele, β = −7.55 [95% CI −9.88, −5.22]; P = 3.2 × 10−10) in the MET arm. Neither variant was significant in PBO; gene-by-treatment interaction was significant for both variants [P(G×T) < 1.0 × 10−4]. Replication in individuals with diabetes did not yield significant findings. A GWAS for metformin response in prediabetes revealed novel ethnic-specific associations that require further investigation but may have implications for tailored therapy.
Introduction
Metformin is the most commonly used agent for initial treatment of type 2 diabetes (T2D), with more than 100 million users worldwide. Over the years, it has remained the foundation of clinical practice guidelines for the management of diabetes globally because it is cheap, safe, effective, well-tolerated, orally administered, and does not promote weight gain (1–4). However, despite its widespread use, metformin does not work well for everyone; in the ADOPT study, which examined the glycemic durability of three oral glucose-lowering agents, each as monotherapy for T2D, 21% of individuals on metformin failed to meet glycemic goals within the first 5 years of treatment (5). Moreover, treatment failure rates are as high as 50% in children and adolescents, as shown by the TODAY study (6).
The reasons for metformin treatment failure remain unclear and could be related to genetic, pharmacologic, pathophysiologic, or environmental factors. Using the genome-wide complex trait analysis method, the heritability of metformin response has been estimated to be between 20 and 34%, a value comparable to other complex phenotypes (7). Indeed, genome-wide association studies (GWAS) have identified common genetic variants associated with metformin response (8–10). In the first GWAS for metformin response, a variant around the gene encoding the ataxia-telangiectasia mutated kinase (ATM) was found to be associated with metformin response in GoDARTS and UKPDS participants (8). Subsequently, in a meta-analysis of >10,000 participants with a harmonized glycemic measure of metformin response, a genome-wide significant association was observed in an intron of the SLC2A2 gene encoding the GLUT2 glucose transporter (9). However, neither of these findings was reproduced in the Diabetes Prevention Program (DPP) (9,11). While the difference in findings could be related to statistical power, one key distinction between the DPP and other studies is that DPP participants have prediabetes and not established T2D. Therefore, it is possible that the influence of genetics on drug response could vary in the prediabetes state compared with the more advanced T2D state.
Thus, we performed a GWAS in the DPP to uncover novel variation associated with metformin response in this distinctive multiethnic cohort that was selected to be at high risk of T2D by virtue of elevated 2-h glucose, fasting glucose, and BMI. Our objective was to identify genetic variants associated with metformin response, as measured by both diabetes incidence and change in quantitative glycemic and metabolic traits. Through this approach, we were able to examine the genetics of metformin response as it relates to both prediabetes and T2D development as well as glycemic response.
Research Design and Methods
Description of Participants and the DPP Study Design
The DPP study design and baseline characteristics of the participants have been described previously (12,13). Briefly, the DPP was a multicenter, randomized controlled trial designed to test the effects of intensive lifestyle modification and pharmacologic intervention on preventing progression to T2D in high-risk individuals. Enrolled participants had a fasting plasma glucose ranging from 95 to 125 mg/dL (5.3–6.9 mmol/L) and a 2-h plasma glucose level between 140 and 199 mg/dL (7.8–11.0 mmol/L) on a 75-g oral glucose tolerance test (OGTT). A total of 3,819 participants were randomly assigned to intensive lifestyle modification (goal weight loss ≥7% and ≥150 min/week of physical activity), standard lifestyle recommendations plus metformin (850 mg twice daily), standard lifestyle recommendations plus troglitazone (400 mg daily), or standard lifestyle recommendations plus placebo. The primary end point was diabetes incidence, diagnosed by a fasting glucose of ≥126 mg/dL (7.0 mmol/L) or a 2-h glucose of ≥200 mg/dL (11.1 mmol/L) after OGTT and confirmed subsequently on a second test within 6 weeks. The primary study demonstrated that over a mean follow-up of 2.8 years, there was a 58% (95% CI 48, 66) reduction of diabetes incidence in the intensive lifestyle intervention group and a 31% (95% CI 17, 43) reduction in the metformin group compared with placebo (14).
Institutional review board approval was obtained by each participating clinical center. All participants included in this analysis provided written informed consent for the main investigation and for subsequent genetic studies.
Genome-Wide Genotyping and Quality Control
DNA was extracted from peripheral blood leukocytes. Genotyping was performed on 3,227 samples using the Human CoreExome genome-wide array (Illumina, San Diego, CA). We excluded single nucleotide polymorphisms (SNPs) with a call rate <95% or if they failed Hardy-Weinberg equilibrium (P < 1.0 × 10−8) within each ethnic group. Samples with discrepant sex, call rate <95%, inbreeding coefficient <−1, and identity-by-state as measured by pi-hat close to 1 were discarded. As 9,730 SNPs and 3,222 samples were additionally genotyped on the MetaboChip (Illumina), we performed a concordance check, excluding SNPs and samples with a concordance rate <95%. After all quality checks were performed, 3,168 samples remained (Supplementary Fig. 1).
Imputation
We performed a two-stage imputation procedure, which consisted of prephasing the genotypes into whole chromosome haplotypes followed by imputation itself. The prephasing was performed using SHAPEIT2 (15), and IMPUTE2 was used for genotype imputation (16). GWIMp-COMPSs can incorporate the contribution of several reference panels (17), and in this work we used 1000 Genomes Phase 3 haplotypes (October 2014) (18). Supplementary Table 1 summarizes the distribution of the 3,168 samples by self-reported race/ethnicity and the DPP treatment arm that underwent imputation.
Statistical Analyses
Because the study was conceived as a pharmacogenetic study on the influence of genetics on metformin response, we focused our primary analyses on the standard lifestyle recommendations plus metformin (MET) group and the placebo (PBO) group. Continuous variables are presented as mean ± SD and categorical variables as frequency (percentage). Baseline characteristics were compared with ANOVA tests for quantitative variables and χ2 tests for qualitative variables.
Diabetes Incidence
A Cox proportional hazards model tested the association between genetic variants and diabetes incidence under an additive genetic model in the MET and PBO arms. For any genome-wide significant findings, we planned to check and ensure that the proportionality assumptions were met. We evaluated the impact of genetic variation in the MET arm only, and, in a second model, we included a formal interaction test between the MET and PBO arms. Models were adjusted for age at randomization, sex, and 10 ancestry principal components (PCs) for population structure; secondary models were also adjusted for waist circumference or BMI. We addressed common variants (minor allele frequency [MAF] >1%), both those directly genotyped and imputed. For imputed markers, we excluded variants that had an imputation quality score <0.7, as well as variants that did not pass a filter test (2 × MAF × [1 − MAF] × n.events > 75, where n.events is the number of events), as described in the GWASTools package (19). We evaluated for genome-wide significance, defined as P < 5 × 10−8.
Change in Quantitative Traits
We used a multiple linear regression model to test allelic associations, assuming additive effects, with the change in quantitative traits relevant to metformin action at 1 year; traits included fasting glucose, 2-h glucose after 75-g OGTT, fasting insulin, insulin sensitivity index (ISI), hemoglobin A1c (HbA1c), and weight. One-year change in each quantitative outcome was defined as 1-year minus baseline value. Non-normally distributed traits were natural log transformed. To minimize the influence of outliers, winsorization was performed (at percentiles of 0.5 and 99.5 for normally distributed traits and percentiles of 1 and 99 for natural log-transformed traits) (20). Analyses were adjusted for age, sex, first 10 ancestry PCs, and the baseline value of the trait. Similar to the diabetes incidence analyses, we evaluated the impact of genetic variation in the MET arm only for each of the six outcomes, and, in a second model, we tested for a gene-by-treatment (G×T) interaction for the MET and PBO arms. We filtered our results to a study-wide MAF >1% and imputation quality ≥0.7. To account for multiple testing and the possible correlation between quantitative traits, we calculated that there were 5.47 effective outcomes, using a previously described method (21), and we set an experiment-wide significance threshold of P < 9 × 10−9 (5 × 10−8/5.47). For top findings that emerged, we performed an exploratory analysis that evaluated these findings in the intensive lifestyle treatment arm to understand whether the variants act through a shared pathway between metformin and lifestyle intervention.
Replication
For each of the quantitative trait outcomes, we compiled a list of the top 500 genotyped and imputed variants with an MAF >1% and imputation score ≥0.7 for imputed markers, for a total of ∼6,000 variants (500 markers × 6 traits × 2 models). We explored their relevance in the Metformin Genetics (MetGen) Consortium, which has data from >10,000 participants from >12 observational studies and clinical trials, in which a pharmacogenetic meta-analysis of metformin response was performed (9). We did additional filtering of variants, excluding those with a minor allele count <10 and with imputation score <0.7 from the Pharmacogenomics of Metformin (PMET) study, which represents the largest cohort comprising MetGen participants. We then calculated the number of effective variants after accounting for correlation between genetic variants (21) and determined the replication significance threshold. In addition, we attempted to replicate our top variant in the Million Veteran Program (MVP) and the Diabetes Multiomic Investigation of Drug Response (DIAMOND). Full details of replication cohorts and the statistical models used in the replication analyses are described in the Supplementary Material and Supplementary Table 2.
Data and Resource Availability
Genetic data generated and analyzed in the current study are available in dbGap (dbGaP Study Accession: phs000681.v2.p1, https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000681.v2.p1). Phenotype data for the DPP are available through the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Data Repository. No applicable resources were generated or analyzed during the current study.
Results
Baseline Demographics
Table 1 describes the characteristics of the subset of DPP participants in the MET and PBO arms in whom genome-wide genotyping was available. The mean baseline age was 50.7 ± 10.4 years, 67% were women, and ∼44% self-reported as non-White. Baseline measurements were consistent with a population at risk for developing T2D. There were no significant differences in baseline measurements by treatment arm. The event rate for diabetes incidence is reported for each self-reported race/ethnicity group for both the MET and PBO arms in Supplementary Table 3.
Table 1.
Baseline characteristics of 1,768 participants in the DPP in the MET and PBO arms with genome-wide genotyping
| Baseline characteristic | Total | PBO | MET | P value** |
|---|---|---|---|---|
| n | 1,768 | 888 | 880 | |
| Age, years | 50.7 ± 10.4 | 50.6 ± 10.4 | 50.9 ± 10.3 | 0.538 |
| Men | 578 (32.7) | 281 (31.6) | 297 (33.8) | 0.345 |
| Women | 1,190 (67.3) | 607 (68.4) | 583 (66.3) | |
| Self-reported race/ethnicity | ||||
| White | 997 (56.4) | 490 (55.2) | 507 (57.6) | 0.746 |
| Black | 364 (20.6) | 186 (20.9) | 178 (20.2) | |
| Hispanic/Latino | 290 (16.4) | 147 (16.6) | 143 (16.3) | |
| Asian/Pacific Islander | 69 (3.9) | 38 (4.3) | 31 (3.5) | |
| American Indian | 48 (2.7) | 27 (3.0) | 21 (2.4) | |
| Weight (kg) | 94.7 ± 19.8 | 94.9 ± 20.0 | 94.5 ± 19.6 | 0.692 |
| HbA1c (%)† | 5.917 ± 0.501 | 5.920 ± 0.495 | 5.915 ± 0.507 | 0.832 |
| Fasting insulin (pmol/L)†# | 167 (111, 236) | 167 (111, 229) | 167 (111, 236) | 0.595*** |
| Fasting glucose (mmol/L) | 5.940 ± 0.462 | 5.956 ± 0.466 | 5.923 ± 0.458 | 0.138 |
| ISI†# | 0.161 (0.112, 0.235) | 0.161 (0.113, 0.236) | 0.161 (0.110, 0.234) | 0.762*** |
Data are means ± SD for continuous variables and n (%) for categorical variables unless otherwise indicated.
P values from F test for continuous variables and χ2 for categorical variables.
P value from Kruskal-Wallis test.
n = 1,766 for total; n = 887 for PBO; n = 879 for MET.
Median (25th, 75th percentile).
Association of Genetic Variation With Diabetes Incidence
In the MET arm only (n = 876) (Supplementary Table 4), we performed association analysis of genetic variants with diabetes incidence during the main DPP study, which had a mean follow-up of 3.2 years. While we did not observe any genome-wide significant findings (Supplementary Fig. 2A), we identified a genetic locus near HIF1AN that reached suggestive significance. The C effect allele of rs2489017 had a frequency of 0.59 in the entire DPP cohort and was notably more common in non-White populations (White: 0.49, African American: 0.83, Asian/Pacific Islander: 0.62, Hispanic/Latino: 0.62, and American Indian: 0.78). Per copy of the variant, participants had a decreased risk of progressing to diabetes (hazard ratio 0.54 [95% CI 0.43, 0.67]; P = 9.2 × 10−8). In the placebo group, the variant did not influence diabetes incidence (P = 0.72), and we noted the presence of an interaction with treatment arm [P(G×T) = 5.9 × 10−5].
We subsequently combined the MET and PBO arms (n = 1,763) (Supplementary Table 4) and used a Cox proportional hazards model to assess for a gene-by-treatment arm interaction for the outcome of diabetes incidence. We did not identify any genome-wide significant findings or variants that met a suggestive significance threshold of P < 1 × 10−6 (Supplementary Fig. 2B).
Association of Genetic Variation With 1-Year Change in Quantitative Traits
With respect to the quantitative trait outcomes, we performed association analysis of genetic variants with 1-year change in any of six quantitative traits (fasting glucose, 2-h glucose after OGTT, fasting insulin, ISI, HbA1c, and weight) across two separate models, one examining the MET arm only and another testing for a gene-by-treatment arm interaction. Supplementary Table 3 presents the sample size for each model and quantitative trait outcome tested. Table 2 summarizes the 14 independent signals that met genome-wide significance (P < 5 × 10−8) for at least 1 quantitative trait, 4 of which met experiment-wide significance (P < 9 × 10−9): 2 for change in HbA1c (Supplementary Fig. 3) and 2 for change in weight (Supplementary Fig. 4). These results remained the same after additionally adjusting the analyses for BMI (data not shown). The effect sizes and P values for the top findings, stratified by self-reported race/ethnicity, are also presented in Supplementary Table 5. Because we report the effect allele frequencies stratified by self-reported ethnicity/race, we compared these groupings with the ancestry groupings determined by PC analysis and illustrated that the self-identified groups tended to cluster tightly on the PC analysis plot (Supplementary Fig. 5).
Table 2.
Fourteen independent genome-wide significant findings (P < 5 × 10−8) for 1-year change in quantitative traits in the DPP
| rsID | Chr | Position* | Nearest gene | EA | NEA | Trait | N | β (95% CI) | P | Effect allele frequency | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All† | AfrAm‡ | AsnPI‡ | Hisp‡ | AI‡ | White‡ | ||||||||||
| Metformin only model | |||||||||||||||
| rs144322333 | 18 | 705550 | ENOSF1 | C | CTGTT | HbA1c, % | 818 | 0.39 (0.28, 0.50) | 2.9 × 10−12 | 0.013 | 0.065 | 0.001 | 0.009 | 0.0001 | 0.002 |
| rs145591055 | 5 | 38849463 | OSMR | G | A | Weight, kg | 829 | −7.55 (−9.87, −5.22) | 3.2 × 10−10 | 0.014 | 0.001 | 0.018 | 0.064 | 0.103 | 0.007 |
| rs13401282§ | 2 | 207810690 | CPO | A | T | ISI, ln | 808 | 0.44 (0.29, 0.59) | 1.7 × 10−8 | 0.029 | 0.119 | 0.003 | 0.015 | 0.006 | 0.002 |
| rs186681623 | 13 | 81546608 | LINC00377 | C | T | Weight, kg | 829 | −2.66 (−3.58, −1.74) | 2.0 × 10−8 | 0.068 | 0.121 | 0.074 | 0.122 | 0.161 | 0.036 |
| rs17083791 | 5 | 93863813 | KIAA0825 | G | A | Weight, kg | 829 | −1.85 (−2.50, −1.20) | 3.3 × 10−8 | 0.149 | 0.052 | 0.194 | 0.210 | 0.367 | 0.137 |
| rs9931871 | 16 | 19928315 | GPRC5B | G | A | Fasting glucose, mmol/L | 821 | 0.68 (0.44, 0.93) | 3.5 × 10−8 | 0.011 | 0.049 | <0.0001 | 0.003 | <0.0001 | <0.0001 |
| rs549305231 | 6 | 19497504 | LOC101928519 | A | G | HbA1c, % | 818 | 0.35 (0.23, 0.48) | 3.9 × 10−8 | 0.012 | 0.046 | <0.0001 | 0.005 | <0.0001 | 0.0003 |
| rs73944532 | 2 | 104691634 | LOC100287010 | A | G | Fasting insulin, ln | 802 | −0.47 (−0.63, −0.30) | 4.1 × 10−8 | 0.020 | 0.075 | 0.005 | 0.007 | 0.001 | 0.002 |
| Gene × treatment model‖ | |||||||||||||||
| rs6838493 | 4 | 185879789 | LINC01093 | A | T | HbA1c, % | 1,621 | −0.66 (−0.87, −0.44) | 1.6 × 10−9 | 0.011 | 0.041 | 0.004 | 0.005 | 0.004 | 0.002 |
| rs148219263 | 18 | 58808522 | CDH20 | C | T | Weight, kg | 1,673 | 10.73 (7.15, 14.30) | 4.8 × 10−9 | 0.012 | 0.007 | 0.011 | 0.011 | 0.0009 | 0.017 |
| rs75147163 | 14 | 57626769 | EXOC5 | A | G | Fasting glucose, mmol/L | 1,633 | −0.73 (−0.98, −0.48) | 1.4 × 10−8 | 0.022 | 0.053 | 0.018 | 0.022 | 0.0002 | 0.015 |
| rs78075715 | 4 | 6613716 | MAN2B2 | C | T | HbA1c, % | 1,621 | −0.51 (−0.68, −0.33) | 1.4 × 10−8 | 0.013 | 0.063 | <0.0001 | 0.002 | <0.0001 | 0.0004 |
| rs12314996 | 12 | 24567467 | SOX5 | A | G | HbA1c, % | 1,621 | −0.47 (−0.63, −0.30) | 3.9 × 10−8 | 0.016 | 0.062 | <0.0001 | 0.007 | 0.001 | 0.002 |
| rs143203347 | 7 | 107384199 | CBLL1 | C | G | Fasting glucose, mmol/L | 1,633 | −0.92 (−1.24, −0.59) | 4.1 × 10−8 | 0.015 | 0.062 | 0.0002 | 0.011 | <0.0001 | <0.0001 |
AfrAm, African American; AI, American Indian; AsnPI, Asian/Pacific Islander; Chr, chromosome; EA, effect allele; Hisp, Hispanic/Latino; ISI, insulin sensitivity index; ln, natural log transformed; NEA, noneffect allele; rsID, reference SNP ID.
GRCh37 assembly.
This is the effect allele frequency for the number of participants in the model (N) that was calculated based on imputation.
Effect allele frequency breakdown by self-reported race/ethnicity is for all 3,168 participants with genome-wide genotyping, calculated based on imputation.
rs13401282 is also associated with fasting insulin (β = −0.39 [95% CI −0.53, −0.25]; P = 4.65 × 10−8). G×T models contain an exposure term for the treatment arm and a G×T interaction term. Results are filtered to P(G×T) < 5 × 10−8 and P(SNP) < 1 × 10−4.
β Estimates and CIs are reported for the interaction term rather than the main effect of metformin.
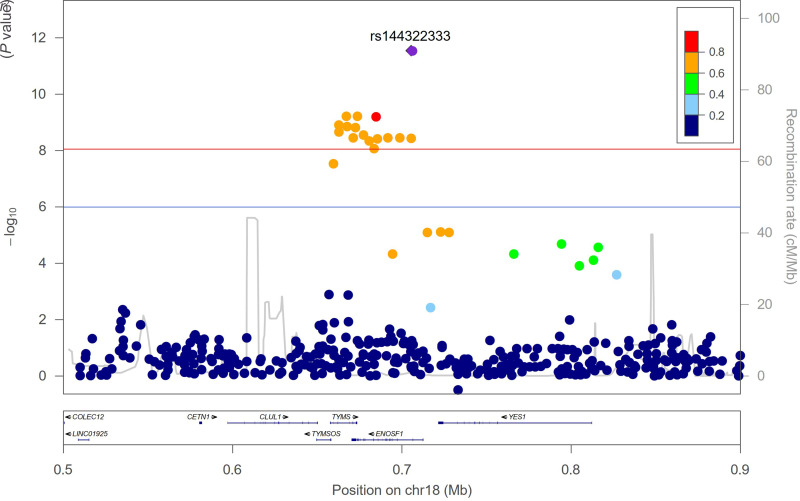
The top hit, rs144322333, which was associated with change in HbA1c in the metformin treatment group, is a deletion polymorphism near ENOSF1 (Fig. 1) and was mainly found in African American participants with an MAF of 6%, whereas it was present in under 0.2% in White individuals (Table 2). Carriers had a 0.39% increase in 1-year change in HbA1c per copy of the deletion, consistent with a worse response to metformin (P = 2.9 × 10−12) (Fig. 2A). The proportion of variance in 1-year change in HbA1c explained by the variant was 5.2%, compared with 10.8% for baseline HbA1c, the strongest predictor of this quantitative outcome in the model.
Figure 1.
Regional association plot of rs144322333 for 1-year change in HbA1c. The red line indicates the experiment-wide significance threshold of P < 9 × 10−9, and the blue line indicates a suggestive significance threshold of P < 1 × 10−6.
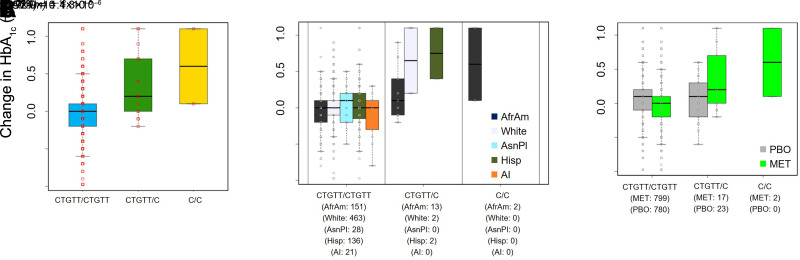
Figure 2.
A: Box plot illustrating the mean change in HbA1c (1-year minus baseline) by rs144322333 genotype in the MET only arm. n = 818. B: Stratified analyses by self-reported race/ethnicity. P value was calculated for the subgroup of African American individuals. n = 166. C: Comparison of the influence of rs144322333 genotype on the mean change in HbA1c in all subjects in the MET (n = 818) and PBO (n = 803) arms. The interaction P value is reported. For the purposes of generating the box plots, fractional alleles were converted to “hard calls” by rounding to the nearest integer. AfrAm, African American; AI, American Indian; AsnPI, Asian/Pacific Islander; Hisp, Hispanic/Latino.
Since rs144322333 is more common in African American individuals, we performed a subgroup analysis and found that the effect was detectable in this group (P = 1.3 × 10−6) (Fig. 2B). In fact, African American individuals without any copies of the deletion experienced a decrease in HbA1c of 0.03% after a year of metformin, whereas those who carried one or two copies of the deletion experienced increases of 0.2% and 0.6%. respectively. Finally, since the association was discovered in the MET arm only, we also evaluated its impact in the PBO arm, in which there was no significant effect (P = 0.32). The interaction with treatment arm was notably significant [P(G×T) = 1.4 × 10−6] (Fig. 2C), reflecting the differential impact of this genetic variant.
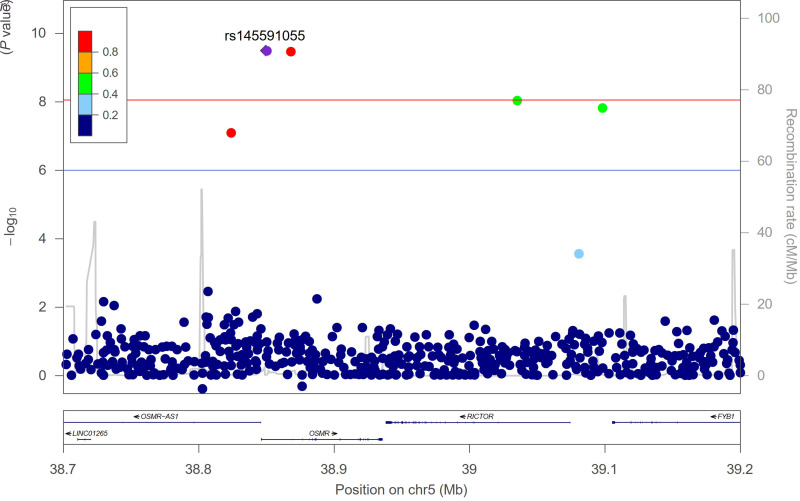
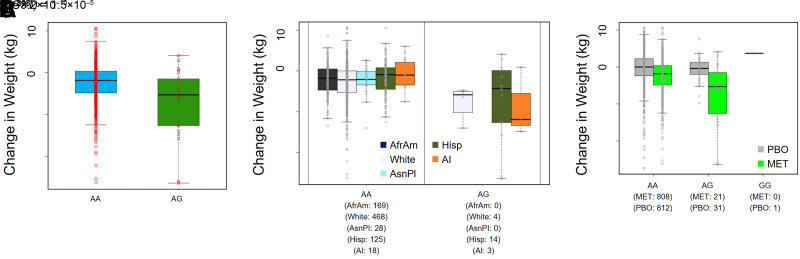
Another novel experiment-wide significant variant was rs145591055 in chromosome 5 near OMSR (Fig. 3), a gene that encodes the oncostatin M receptor (Table 2). Carriers of the G effect allele had a 7.6-kg greater decline in weight at 1 year following initiation of metformin treatment in adjusted analyses (P = 3.2 × 10−10) (Fig. 4A). The variant has an effect allele frequency of 10% in American Indians, calculated from the entire DPP study (Table 2). While a subgroup analysis in American Indians was limited by sample size (n = 21) (Fig. 4B), we noted that noncarriers experienced a decrease in weight of 0.5 kg at 1 year, compared with a decrease of 8.6 kg in heterozygous carriers. We also evaluated the influence of this variant in the placebo arm and found that there was no significant effect on weight (per G allele, β = −0.69; P = 0.50). Again, there was a significant gene-by-treatment interaction [P(G×T) = 1.5 × 10−5] (Fig. 4C).
Figure 3.
Regional association plot of rs145591055 for 1-year change in weight. The red line indicates the experiment-wide significance threshold of P < 9 × 10−9, and the blue line indicates a suggestive significance threshold of P < 1 × 10−6.
Figure 4.
A: Box plot illustrating the mean change in weight (1-year minus baseline) by rs145591055 genotype in the MET only arm. n = 829. B: Box plot stratified by self-reported race/ethnicity. C: Comparison of the influence of rs144322333 genotype on the mean change in weight in all subjects in the MET (n = 829) and PBO (n = 844) arms. The interaction P value is reported. For the purposes of generating the box plots, fractional alleles were converted to “hard calls” by rounding to the nearest integer. AfrAm, African American; AI, American Indian; AsnPI, Asian/Pacific Islander; Hisp, Hispanic/Latino.
In the gene × treatment models, we found that rs6838493, in chromosome 4 near LINC01093 (Supplementary Fig. 6), had a differential influence on change in HbA1c [P(G×T) = 1.6 × 10−9] (Supplementary Fig. 7), in which heterozygous carriers experienced a rise in HbA1c at 1 year in the PBO arm (β = 0.5; P = 1.3 × 10−8) in comparison with a decline in HbA1c in the MET arm (β = −0.1; P = 0.03). The impact of rs148219263, located in chromosome 18 near CDH20 (Supplementary Fig. 8), on 1-year change in weight was also different by treatment arm [P(G×T) = 4.8 × 10−9] (Supplementary Fig. 9). In this study, in comparison with their TT counterparts, TC carriers experienced weight loss in the placebo arm (β = −7.0; P = 2.4 × 10−8) but did not achieve weight loss after receiving metformin (β = 3.9; P = 4.0 × 10−3).
In an exploratory analysis, we evaluated our top findings from Table 2 in the intensive lifestyle arm. Of the nine variants that emerged from the analysis completed in the MET arm only, one variant (rs13401282) appeared to have a similar effect on ISI in the lifestyle arm as in the MET arm (P = 0.004) (Supplementary Table 6); the other eight variants were nonsignificant in the lifestyle arm. For the five variants identified through the gene × treatment model that combined the MET and PBO arms, we tested their relevance in a similar interaction model that combined the PBO and lifestyle arms. We found that four of the five variants demonstrated similar findings between the two gene × treatment models, as illustrated by the magnitude and direction of the β estimates, as well as the P values (Supplementary Table 6).
We searched our lead findings in the Genotype-Tissue Expression (GTex) project (https://gtexportal.org) (22) and eQTLGen Consortium (https://eqtlgen.org) (23) to evaluate for expression quantitative trait loci (eQTL) associations. We observed that rs17083791, which was associated with a greater decline in weight at 1 year following metformin, is a cis-eQTL associated with higher expression of KIAA0825, the nearest gene, in fibroblasts (normalized effect size [NES] 0.411; P = 2.7 × 10−12), subcutaneous adipose tissue (NES 0.285; P = 7.9 × 10−7), skeletal muscle (NES 0.241; P = 5.0 × 10−5), and visceral adipose tissue (NES 0.229; P = 1.6 × 10−4). In blood samples in eQTLGen Consortium, we noted that the variant was also a cis-eQTL for MCTP1 (P = 7.3 × 10−189). We also evaluated the top variants in the pancreatic islet genotype tissue-expression resource (Translational Human Pancreatic Islet Genotype Tissue-Expression Resource [TIGER]; http://tiger.bsc.es), which aggregates >500 human islet genomic data sets (24), but there were no notable findings. Finally, we searched top variants against phenotypes associated with T2D using the Type 2 Diabetes Knowledge Portal (https://t2d.hugeamp.org). We found that the G allele of rs145591055, which was associated with greater 1-year weight change on metformin, was marginally associated with a higher BMI-adjusted waist-to-hip ratio (β = 0.079; P = 0.001).
Independent Replication
We sought to replicate our pharmacogenetic findings in additional cohorts in which metformin response has been defined. As there are few cohorts of patients with prediabetes, our first approach was to explore the relevance of the variants in MetGen, in which the outcome was glycemic response, as measured by baseline minus minimum on-treatment HbA1c within 18 months after metformin initiation (9). Out of ∼6,000 variants, only 3,050 had available genotype information in MetGen, resulting in 2,610 effective variants; we thus set a significance threshold of P < 1.9 × 10−5 (0.05/2,610). After filtering, results were available in MetGen for only 6 of the 14 genome-wide significant variants from our DPP GWAS, none of which replicated (Table 3).
Table 3.
Attempt at replication of genome-wide significant findings from the DPP in the MetGen Consortium
| DPP model | Trait | rsID | DPP β* | MetGen (n) | MetGen β* | MetGen P value |
|---|---|---|---|---|---|---|
| MET | Weight, kg | rs186681623 | −2.66 (greater weight loss) | 10,519 | −0.05 (decreased HbA1c reduction) | 0.10 |
| MET | Weight, kg | rs17083791 | −1.85 (greater weight loss) | 12,578 | 0.004 (greater HbA1c reduction) | 0.79 |
| MET | Fasting insulin, ln | rs73944532 | −0.47 (greater decrease in fasting insulin) | 7,048 | 0.03 (greater HbA1c reduction) | 0.84 |
| G×T | Weight, kg | rs148219263 | 10.73 (reduced weight loss) | 10,519 | −0.04 (decreased HbA1c reduction) | 0.39 |
| G×T | Fasting glucose, mmol/L | rs75147163 | −0.73 (greater decrease in fasting glucose) | 10,288 | −0.02 (decreased HbA1c reduction) | 0.70 |
| G×T | HbA1c, % | rs12314996 | −0.47 (greater decrease in HbA1c) | 7,048 | 0.19 (greater HbA1c reduction) | 0.24 |
ln, natural log transformed; rsID, reference SNP ID.
In the DPP, the β estimates represent 1-year change in the quantitative trait as calculated by follow-up value minus baseline value. In MetGen, the outcome was change in HbA1c (%), defined as baseline value minus follow-up value within 18 months (will have opposite signs of β estimates for change in HbA1c compared with that in the DPP). For each analysis and cohort, the impact of the variant on the metformin response outcome is indicated in parentheses.
Because the study cohorts in MetGen were of predominately European ancestry, we sought to replicate our top HbA1c finding in African American cohorts. We first examined 2,733 participants with established T2D from MVP who self-reported as non-Hispanic African Americans and had received metformin monotherapy for up to 15 months. The top variant, rs144322333, which had an MAF of 7% and an imputation score of 0.89 in MVP, was not significantly associated with change in HbA1c (β = 0.05; P = 0.15).
We also sought replication in DIAMOND, an observational population-based cohort with electronic medical record–linked clinical data on individuals with T2D on metformin monotherapy. We identified a subset of 471 individuals who self-reported as African American and were calculated to have a reliable exposure of at least 500 mg metformin daily as monotherapy for treatment of T2D in the 120 days preceding the follow-up HbA1c measurement. In DIAMOND, rs144322333 had an MAF of 8% and an imputation score of 0.98. Per copy of the deletion, carriers had a 0.13% reduction in HbA1c lowering over 1 year in DIAMOND, indicating a worse metformin response, which was similar in direction to that seen in the DPP but smaller in magnitude and not reaching statistical significance (P = 0.19).
Discussion
To date, GWAS have uncovered several genetic loci influencing the glycemic response to metformin, but these were largely performed in European populations with established T2D (8–10). When these variants were assessed in the DPP, the reported associations were not confirmed for the outcomes of diabetes incidence and other relevant physiologic parameters, such as insulin sensitivity, fasting glucose, HbA1c, or oral disposition index (9,11). One possible explanation for this inconsistency is that metformin response is defined differently in a disease cohort, in which the measure of response is the achievement of an HbA1c ≤7%, compared with a cohort with prediabetes, in which diabetes incidence or quantitative traits are the studied outcomes. Furthermore, genetic variation may influence drug response differently in individuals with prediabetes, who likely have better pancreatic β-cell function.
Thus, we undertook a GWAS in the DPP examining the outcomes of diabetes prevention as well as 1-year change in six quantitative traits known to be affected by metformin therapy. The design of the DPP is unique for studying metformin pharmacogenetics, as it includes both a PBO and MET arm, permitting the use of an interaction test to assess genetic variants that may have a differential response based on drug exposure. In assessing diabetes incidence, we did not identify any genome-wide significant variants. We acknowledge that the relatively modest sample size of the DPP with a limited range of impaired glucose tolerance may have restricted our ability to detect an interaction effect. However, we did observe an association that met a suggestive threshold that could be investigated in the future or meta-analyzed with other cohorts.
After correcting for multiple testing, our quantitative trait analyses yielded several genome-wide significant associations. ENOSF1, the closest gene to the top variant rs144322333, encodes the enolase superfamily member 1, a mitochondrial enzyme involved in the catabolism of l-fucose(a sugar found on cellular glycoproteins). Interestingly, ENOSF1 is next to a kinase, YES1, which has been shown to regulate the activity of organic cation transporters OCT1 (SLC22A1) and OCT2 (SLC22A2) that play a critical role in metformin disposition and elimination in the kidney and liver, respectively (25,26). Moreover, LINC01093 near rs6838493 has been shown to be related to several liver phenotypes, including fibrosis (27) and hepatocellular carcinoma (28), and may play a role in modulating metformin’s action on the liver.
Many of our genome-wide significant findings were ancestry-specific (rs144322333 near ENOSF1, rs145591055 near OSMR, and rs6838493 near LINC01093). We further illustrate this through subgroup analyses and demonstrate the persistent effect of top variants of interest after stratifying by self-reported ethnicity/race. Based on these stratified analyses, it appeared that the ancestry specificity of our findings was mainly driven by allele frequency, since similar trends were observed across subgroups. We acknowledge that this may have been a consequence of conducting our analyses in a multi-ancestry cohort, which could have biased our results toward variants with concordant effects across subgroups. One limitation of this approach is that variants with opposite effects in different subgroups could be potentially missed. We also estimated that our top genetic finding (rs144322333) explained 5.2% of the variance in 1-year change in HbA1c, which was half of that of baseline HbA1c, supporting the notion that the contribution of genetics is modest compared with a traditional predictor of drug response.
In the MET arm of the DPP, the average weight loss was 2.1 kg and was relatively stable throughout the study follow-up period (14). In comparison, the association of rs145591055 (near OSMR) variation with 1-year change in weight was 7.6 kg, more than three times greater in magnitude. Individuals who carry this variant, which has an effect allele frequency of 10% in American Indians, appear to derive a significant weight-loss benefit from metformin, which is especially relevant given the high incidence and prevalence of T2D in this population (29,30). Moreover, the G allele was reported in the T2D Knowledge Portal to be marginally associated with a higher BMI-adjusted waist-to-hip ratio, also suggesting that this is a group of individuals who would benefit from metformin. However, the underlying mechanism responsible for this weight effect is not clear and requires further investigation. We also evaluated our top findings in the GTex project, the eQTLGen Consortium, and the TIGER portal. Interestingly, we observed that the variant rs17083791 was a cis-eQTL for the nearest gene KIAA0825 in several relevant tissues, suggesting that changes in KIAA0825 gene expression may modulate metformin-related weight loss. The same variant was also a strong cis-eQTL for MCTP1, which codes for a protein involved in calcium ion binding, though the significance of this finding is unknown. While these genomic resources can be useful for functional follow-up of variants, the majority of participants are of European ancestry, and thus, their utility is limited for variants that are common only in specific populations.
In an exploratory analysis, we evaluated the top findings from the quantitative trait analysis using the intensive lifestyle arm of the DPP. There were five variants for which the association with the quantitative trait was similar in the lifestyle arm as in the MET arm. Prior studies have suggested that the intensive lifestyle intervention of the DPP worked similarly to metformin in improving insulin sensitivity (31). Therefore, it is possible that the mechanism of action of these variants may be shared between metformin and lifestyle. In contrast, the remaining nine variants that met genome-wide significance were nonsignificant when evaluated in the lifestyle arm, illustrating they may be specific to metformin response.
Finally, we took a comprehensive approach to replicate findings that emerged from our GWAS, though this was a major challenge due to the lack of a suitable replication cohort of individuals with prediabetes who received longitudinal metformin exposure. We evaluated approximately the top 500 variants for each quantitative trait analyses in MetGen, the largest meta-analysis examining glycemic response to metformin in T2D (9). Our efforts did not yield replication, but this could be explained by the composition of MetGen as previously discussed. We subsequently identified two African American cohorts (MVP and DIAMOND) for replication of our top variant rs144322333. Though the study populations again largely reflected individuals with T2D, we constructed our models to reflect a similar length of metformin exposure as in the DPP and included similar covariates as in our quantitative trait analyses. Despite imputation on different reference panels in these cohorts, we reassuringly observed similar MAFs for rs14432233; however, neither replication produced a nominally significant result. We acknowledge the possibility that given the low study-wide MAF of our top findings and the small sample size, our findings could represent false-positive results.
Our work underscores the importance of conducting multi-ancestry pharmacogenetic studies to improve upon treatment algorithms across diverse populations in the future. A prior study based on pharmacy fill data reported that African American adults appear to have a better glycemic response to metformin compared with European Americans (32), and the TODAY study showed that African American youth with T2D on metformin had poorer glycemic response to metformin (6). Though these studies were based on self-reported race/ethnicity information, we demonstrated in the DPP that there is clustering of the self-reported ethnicity/race groups along the genetic ancestry PCs. Thus, the identification of variants in our study that are more prevalent in African American populations suggests that there may be a heritable component to these differences in response to metformin.
Strengths of our study include the diverse composition of the DPP, which includes nearly 45% non-White individuals by self-report and represents a cohort of individuals at a higher risk of developing T2D in the U.S. Participants were well-characterized under standardized clinical trial protocols, and the genetic data were obtained using the highest standards for genotyping and imputation at the time. As described, limitations of our study are the small sample size, which is a frequent problem in pharmacogenomic GWAS (33), and the lack of replication of our findings in patients with diabetes, though this may reflect differences in genetic modulation of metformin response between the prediabetes and diabetes state. However, we were exhaustive in our attempts to identify independent cohorts, including the largest African American cohorts with diabetes known to us. Future directions should include additional follow-up and meta-analyses in additional independent ancestry-specific cohorts as they arise in order to advance work in this area.
In conclusion, we have identified several genome-wide significant associations with metformin response, as measured by change in quantitative traits, in a well-defined multi-ancestry clinical trial cohort. We believe that we have generated a valuable resource that can be used for future genetic investigation and to gain insight into the genetic underpinnings of interindividual differences in metformin response in a population at risk for developing T2D. Finally, we illustrate the peremptory need to generate suitable pharmacogenomic and transcriptomic resources in diverse populations.
Article Information
Acknowledgments. The DPP Research Group gratefully acknowledges the commitment and dedication of the participants of the DPP and DPP Outcomes Study (DPPOS).
Funding. Research reported in this publication was supported by the NIDDK of the National Institutes of Health under award numbers U01 DK048489, U01 DK048339, U01 DK048377, U01 DK048349, U01 DK048381, U01 DK048468, U01 DK048434, U01 DK048485, U01 DK048375, U01 DK048514, U01 DK048437, U01 DK048413, U01 DK048411, U01 DK048406, U01 DK048380, U01 DK048397, U01 DK048412, U01 DK048404, U01 DK048387, U01 DK048407, U01 DK048443, and U01 DK048400, by providing funding during DPP and DPPOS to the clinical centers and the Coordinating Center for the design and conduct of the study, and collection, management, analysis, and interpretation of the data. Funding was also provided by the National Institute of Child Health and Human Development, the National Institute on Aging, the National Eye Institute, the National Heart, Lung, and Blood Institute, the National Cancer Institute, the Office of Research on Women’s Health, the National Institute on Minority Health and Health Disparities, the Centers for Disease Control and Prevention, and the American Diabetes Association. The Southwestern American Indian Centers were supported directly by the NIDDK, including its Intramural Research Program, and the Indian Health Service. The General Clinical Research Center Program, the National Center for Research Resources, and the Department of Veterans Affairs supported data collection at many of the clinical centers. Merck KGaA provided medication for DPPOS. DPP/DPPOS also received donated materials, equipment, or medicines for concomitant conditions from Bristol-Myers Squibb, Parke-Davis, and LifeScan Inc., Health-O-Meter, Hoechst Marion Roussel, Inc., Merck-Medco Managed Care, LLC, Merck and Co., Nike Sports Marketing, Slimast Foods Co., and Quaker Oats Co. McKesson BioServices Corp., Matthews Media Group, Inc., and the Henry M. Jackson Foundation provided support services under subcontract with the Coordinating Center. This work was also supported by the NIDDK grant R01DK072041 to J.C.F. and K.A.J. J.H.L. received individual support from National Institutes of Health grant T32DK007028. S.S. was funded by National Institutes of Health grant K23DK120932. A.G. was supported by NIDDK grant 1K01DK120631 when this work was performed. S.E.K. was supported in part by the Department of Veterans Affairs.
The sponsor of this study, the NIH/NIDDK, was represented on the steering committee and played a part in study design, how the study was done, and publication. All authors in the writing group had access to all data. The opinions expressed are those of the study group and do not necessarily reflect the views of the funding agencies or the National Institutes of Health.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. J.H.L., J.A.P., K.A.J., S.S., T.I.P., and J.C.F. contributed to the conception and design of the study. Genotyping, quality control, and imputation of the genetic data were performed by L.C., M.H., and J.M.M. GWAS analyses were performed by J.A.P., K.A.J., and T.I.P. J.H.L., J.A.P., K.A.J., S.S., J.N.T., J.M.M., Q.P., P.W.F., R.L.H., S.E.K., W.C.K., T.I.P., and J.C.F. contributed to data interpretation. A.Y.D., S.W.Y., E.R.P., K.M.G., A.G., A.M.H., S.X., and L.K.W. contributed to the replication analyses described in the manuscript. J.H.L., J.A.P., K.A.J., S.S., and J.C.F. contributed to manuscript writing, with editing by all authors and approval of submission of the final version. J.C.F. is the guarantor of this work and, as such, had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Portions of this study were presented in oral form at the 81st Scientific Sessions of the American Diabetes Association (Virtual), 25–29 June 2021.
Footnotes
Clinical trial reg. no. NCT00004992, clinicaltrials.gov
See accompanying article, p. 1057.
This article contains supplementary material online at https://doi.org/10.2337/figshare.21719837.
J.N.T. is currently affiliated with the Division of Endocrinology, Department of Pediatrics, University of Vermont Children’s Hospital, Burlington, VT.
A complete list of the members of the Diabetes Prevention Program Research Group and Diabetes Prevention Program Outcomes Study 1, 2, and 3A can be found in the supplementary material online.
Contributor Information
Diabetes Prevention Program Research Group::
George A. Bray, Kishore M. Gadde, Iris W. Culbert, Annie Chatellier, Jennifer Arceneaux, Amber Dragg, Catherine M. Champagne, Crystal Duncan, Barbara Eberhardt, Frank Greenway, Fonda G. Guillory, April A. Herbert, Michael L. Jeffirs, Betty M. Kennedy, Erma Levy, Monica Lockett, Jennifer C. Lovejoy, Laura H. Morris, Lee E. Melancon, Donna H. Ryan, Deborah A. Sanford, Kenneth G. Smith, Lisa L. Smith, Julia A. St. Amant, Richard T. Tulley, Paula C. Vicknair, Donald Williamson, Jeffery J. Zachwieja, Kenneth S. Polonsky, Janet Tobian, David A. Ehrmann, Margaret J. Matulik, Karla A. Temple, Bart Clark, Kirsten Czech, Catherine DeSandre, Brittnie Dotson, Ruthanne Hilbrich, Wylie McNabb, Michael T. Quinn, Ann R. Semenske, Jose F. Caro, Kevin Furlong, Barry J. Goldstein, Pamela G. Watson, Kellie A. Smith, Jewel Mendoza, Wendi Wildman, Marsha Simmons, Genine Jensen, Renee Liberoni, John Spandorfer, Constance Pepe, Richard P. Donahue, Ronald B. Goldberg, Ronald Prineas, Patricia Rowe, Anna Giannella, Jeanette Calles, Juliet Sanguily, Paul Cassanova-Romero, Sumaya Castillo-Florez, Hermes J. Florez, Rajesh Garg, Lascelles Kirby, Olga Lara, Carmen Larreal, Valerie McLymont, Jadell Mendez, Arlette Perry, Patrice Saab, Bertha Veciana, Steven M. Haffner, Helen P. Hazuda, Maria G. Montez, Juan Isaac, Kathy Hattaway, Carlos Lorenzo, Arlene Martinez, Monica Salazar, Tatiana Walker, Richard F. Hamman, Dana Dabelea, Patricia V. Nash, Sheila C. Steinke, Lisa Testaverde, Jennifer Truong, Denise R. Anderson, Larry B. Ballonoff, Alexis Bouffard, Rebecca S. Boxer, Brian Bucca, B. Ned Calonge, Lynne Delve, Martha Farago, James O. Hill, Shelley R. Hoyer, Tonya Jenkins, Bonnie T. Jortberg, Dione Lenz, Marsha Miller, Thomas Nilan, Leigh Perreault, David W. Price, Judith G. Regensteiner, Emily B. Schroeder, Helen Seagle, Carissa M. Smith, Brent VanDorsten, Edward S. Horton, Medha Munshi, Kathleen E. Lawton, Catherine S. Poirier, Kati Swift, Sharon D. Jackson, Ronald A. Arky, Marybeth Bryant, Jacqueline P. Burke, Enrique Caballero, Karen M. Callaphan, Barbara Fargnoli, Therese Franklin, Om P. Ganda, Ashley Guidi, Mathew Guido, Alan M. Jacobsen, Lyn M. Kula, Margaret Kocal, Lori Lambert, Kathleen E. Lawton, Sarah Ledbury, Maureen A. Malloy, Roeland J.W. Middelbeek, Maryanne Nicosia, Cathryn F. Oldmixon, Jocelyn Pan, Marizel Quitingon, Riley Rainville, Stacy Rubtchinsky, Ellen W. Seely, Jessica Sansoucy, Dana Schweizer, Donald Simonson, Fannie Smith, Caren G. Solomon, Jeanne Spellman, James Warram, Steven E. Kahn, Brenda K. Montgomery, Basma Fattaleh, Celeste Colegrove, Wilfred Fujimoto, Robert H. Knopp, Edward W. Lipkin, Michelle Marr, Ivy Morgan-Taggart, Anne Murillo, Kayla O’Neal, Dace Trence, Lonnese Taylor, April Thomas, Elaine C. Tsai, Abbas E. Kitabchi, Samuel Dagogo-Jack, Mary E. Murphy, Laura Taylor, Jennifer Dolgoff, Ethel Faye Hampton, William B. Applegate, Michael Bryer-Ash, Debra Clark, Sandra L. Frieson, Uzoma Ibebuogu, Raed Imseis, Helen Lambeth, Lynne C. Lichtermann, Hooman Oktaei, Harriet Ricks, Lily M.K. Rutledge, Amy R. Sherman, Clara M. Smith, Judith E. Soberman, Beverly Williams-Cleaves, Avnisha Patel, Ebenezer A. Nyenwe, Ethel Faye Hampton, Boyd E. Metzger, Mark E. Molitch, Amisha Wallia, Mariana K. Johnson, Sarah VanderMolen, Daphne T. Adelman, Catherine Behrends, Michelle Cook, Marian Fitzgibbon, Mimi M. Giles, Monica Hartmuller, Cheryl K.H. Johnson, Diane Larsen, Anne Lowe, Megan Lyman, David McPherson, Samsam C. Penn, Thomas Pitts, Renee Reinhart, Susan Roston, Pamela A. Schinleber, David M. Nathan, Charles McKitrick, Heather Turgeon, Mary Larkin, Marielle Mugford, Nopporn Thangthaeng, Fernelle Leander, Kathy Abbott, Ellen Anderson, Laurie Bissett, Kristy Bondi, Enrico Cagliero, Jose C. Florez, Linda Delahanty, Valerie Goldman, Elaine Grassa, Lindsey Gurry, Kali D’Anna, Fernelle Leandre, Peter Lou, Alexandra Poulos, Elyse Raymond, Valerie Ripley, Christine Stevens, Beverly Tseng, Jerrold M. Olefsky, Elizabeth Barrett-Connor, Sunder Mudaliar, Maria Rosario Araneta, Mary Lou Carrion-Petersen, Karen Vejvoda, Sarah Bassiouni, Madeline Beltran, Lauren N. Claravall, Jonalle M. Dowden, Steven V. Edelman, Pranav Garimella, Robert R. Henry, Javiva Horne, Marycie Lamkin, Simona Szerdi Janesch, Diana Leos, William Polonsky, Rosa Ruiz, Jean Smith, Jennifer Torio-Hurley, F. Xavier Pi-Sunyer, Blandine Laferrere, Jane E. Lee, Susan Hagamen, Kim Kelly-Dinham, David B. Allison, Nnenna Agharanya, Nancy J. Aronoff, Maria Baldo, Jill P. Crandall, Sandra T. Foo, Jose A. Luchsinger, Carmen Pal, Kathy Parkes, Mary Beth Pena, Julie Roman, Ellen S. Rooney, Gretchen E.H. Van Wye, Kristine A. Viscovich, Melvin J. Prince, David G. Marrero, Kieren J. Mather, Mary de Groot, Susie M. Kelly, Marcia A. Jackson, Gina McAtee, Paula Putenney, Ronald T. Ackermann, Carolyn M. Cantrell, Yolanda F. Dotson, Edwin S. Fineberg, Megan Fultz, John C. Guare, Angela Hadden, James M. Ignaut, Marion S. Kirkman, Erin O’Kelly Phillips, Kisha L. Pinner, Beverly D. Porter, Paris J. Roach, Nancy D. Rowland, Madelyn L. Wheeler, Robert E. Ratner, Vanita Aroda, Michelle Magee, Gretchen Youssef, Sue Shapiro, Natalie Andon, Catherine Bavido-Arrage, Geraldine Boggs, Marjorie Bronsord, Ernestine Brown, Holly Love Burkott, Wayman W. Cheatham, Susan Cola, Cindy Evans, Peggy Gibbs, Tracy Kellum, Lilia Leon, Milvia Lagarda, Claresa Levatan, Milajurine Lindsay, Asha K. Nair, Jean Park, Maureen Passaro, Angela Silverman, Gabriel Uwaifo, Debra Wells-Thayer, Renee Wiggins, Mohammed F. Saad, Karol Watson, Maria Budget, Sujata Jinagouda, Medhat Botrous, Anthony Sosa, Sameh Tadros, Khan Akbar, Claudia Conzues, Perpetua Magpuri, Kathy Ngo, Amer Rassam, Debra Waters, Kathy Xapthalamous, Julio V. Santiago, Samuel Dagogo-Jack, Neil H. White, Angela L. Brown, Ana Santiago, Samia Das, Prajakta Khare-Ranade, Tamara Stich, Edwin Fisher, Emma Hurt, Jackie Jones, Tracy Jones, Michelle Kerr, Sherri McCowan, Lucy Ryder, Cormarie Wernimont, Christopher D. Saudek, Sherita Hill Golden, Vanessa Bradley, Emily Sullivan, Tracy Whittington, Caroline Abbas, Adrienne Allen, Frederick L. Brancati, Sharon Cappelli, Jeanne M. Clark, Jeanne B. Charleston, Janice Freel, Katherine Horak, Alicia Greene, Dawn Jiggetts, Delois Johnson, Hope Joseph, Rita Kalyani, Kimberly Loman, Nestoras Mathioudakis, Nisa Maruthur, Henry Mosley, John Reusing, Richard R. Rubin, Alafia Samuels, Thomas Shields, Shawne Stephens, Kerry J. Stewart, LeeLana Thomas, Evonne Utsey, Paula Williamson, David S. Schade, Karwyn S. Adams, Carolyn Johannes, Claire Hemphill, Penny Hyde, Janene L. Canady, Leslie F. Atler, Patrick J. Boyle, Mark R. Burge, Lisa Chai, Kathleen Colleran, Ateka Fondino, Ysela Gonzales, Doris A. Hernandez-McGinnis, Patricia Katz, Carolyn King, Julia Middendorf, Amer Rassam, Sofya Rubinchik, Willette Senter, Debra Waters, Harry Shamoon, Jill Crandall, Janet O. Brown, Gilda Trandafirescu, Danielle Powell, Elsie Adorno, Liane Cox, Helena Duffy, Samuel Engel, Allison Friedler, Angela Goldstein, Crystal J. Howard-Century, Jennifer Lukin, Stacey Kloiber, Nadege Longchamp, Helen Martinez, Dorothy Pompi, Jonathan Scheindlin, Norica Tomuta, Elissa Violino, Elizabeth A. Walker, Judith Wylie-Rosett, Elise Zimmerman, Joel Zonszein, Rena R. Wing, Trevor Orchard, Elizabeth Venditti, Gaye Koenning, M. Kaye Kramer, Marie Smith, Susan Jeffries, Valarie Weinzierl, Susan Barr, Catherine Benchoff, Miriam Boraz, Lisa Clifford, Rebecca Culyba, Marlene Frazier, Ryan Gilligan, Stephanie Guimond, Susan Harrier, Louann Harris, Andrea Kriska, Qurashia Manjoo, Monica Mullen, Alicia Noel, Amy Otto, Jessica Pettigrew, Bonny Rockette-Wagner, Debra Rubinstein, Linda Semler, Cheryl F. Smith, Katherine V. Williams, Tara Wilson, Richard F. Arakaki, Marjorie K. Mau, Renee W. Latimer, Mae K. Isonaga, Narleen K. Baker-Ladao, Ralph Beddow, Nina E. Bermudez, Lorna Dias, Jillian Inouye, John S. Melish, Kathy Mikami, Pharis Mohideen, Sharon K. Odom, Raynette U. Perry, Robin E. Yamamoto, William C. Knowler, Robert L. Hanson, Vallabh Shah, Charlotte Dodge, Mary A. Hoskin, Carol A. Percy, Norman Cooeyate, Camille Natewa, Charlotte Dodge, Alvera Enote, Harelda Anderson, Kelly J. Acton, Vickie L. Andre, Rosalyn Barber, Shandiin Begay, Peter H. Bennett, Mary Beth Benson, Evelyn C. Bird, Brenda A. Broussard, Brian C. Bucca, Marcella Chavez, Sherron Cook, Jeff Curtis, Tara Dacawyma, Matthew S. Doughty, Roberta Duncan, Cyndy Edgerton, Jacqueline M. Ghahate, Justin Glass, Martia Glass, Dorothy Gohdes, Wendy Grant, Ellie Horse, Louise E. Ingraham, Merry Jackson, Priscilla Jay, Roylen S. Kaskalla, Karen Kavena, David Kessler, Kathleen M. Kobus, Jonathan Krakoff, Jason Kurland, Catherine Manus, Cherie McCabe, Sara Michaels, Tina Morgan, Yolanda Nashboo, Julie A. Nelson, Steven Poirier, Evette Polczynski, Christopher Piromalli, Mike Reidy, Jeanine Roumain, Debra Rowse, Robert J. Roy, Sandra Sangster, Janet Sewenemewa, Miranda Smart, Chelsea Spencer, Darryl Tonemah, Rachel Williams, Charlton Wilson, Michelle Yazzie, Raymond Bain, Sarah Fowler, Michael D. Larsen, Kathleen Jablonski, Marinella Temprosa, Tina Brenneman, Sharon L. Edelstein, Solome Abebe, Julie Bamdad, Melanie Barkalow, Joel Bethepu, Tsedenia Bezabeh, Anna Bowers, Nicole Butler, Jackie Callaghan, Caitlin E. Carter, Costas Christophi, Gregory M. Dwyer, Mary Foulkes, Yuping Gao, Robert Gooding, Adrienne Gottlieb, Kristina L. Grimes, Nisha Grover-Fairchild, Lori Haffner, Heather Hoffman, Steve Jones, Tara L. Jones, Richard Katz, Preethy Kolinjivadi, John M. Lachin, Yong Ma, Pamela Mucik, Robert Orlosky, Qing Pan, Susan Reamer, James Rochon, Alla Sapozhnikova, Hanna Sherif, Charlotte Stimpson, Ashley Hogan Tjaden, Fredricka Walker-Murray, Elizabeth M. Venditti, Andrea M. Kriska, Linda Semler, Valerie Weinzierl, Santica Marcovina, F. Alan Aldrich, Jessica Harting, John Albers, Greg Strylewicz, Anthony Killeen, Deanna Gabrielson, R. Eastman, Judith Fradkin, Sanford Garfield, Christine Lee, Edward Gregg, Ping Zhang, Dan O’Leary, Gregory Evans, Matthew Budoff, Chris Dailing, Elizabeth Stamm, Ann Schwartz, Caroline Navy, Lisa Palermo, Pentti Rautaharju, Ronald J. Prineas, Elsayed Z. Soliman, Teresa Alexander, Charles Campbell, Sharon Hall, Yabing Li, Margaret Mills, Nancy Pemberton, Farida Rautaharju, Zhuming Zhang, Julie Hu, Susan Hensley, Lisa Keasler, Tonya Taylor, Barbara Blodi, Ronald Danis, Matthew Davis, Larry Hubbard, Barbara Blodi, Ryan Endres, Deborah Elsas, Samantha Johnson, Dawn Myers, Nancy Barrett, Heather Baumhauer, Wendy Benz, Holly Cohn, Ellie Corkery, Kristi Dohm, Amitha Domalpally, Vonnie Gama, Anne Goulding, Andy Ewen, Cynthia Hurtenbach, Daniel Lawrence, Kyle McDaniel, Jeong Pak, James Reimers, Ruth Shaw, Maria Swift, Pamela Vargo, Sheila Watson, Jose A. Luchsinger, Jennifer Manly, Elizabeth Mayer-Davis, Robert R. Moran, Ted Ganiats, Kristin David, Andrew J. Sarkin, Erik Groessl, Naomi Katzir, Helen Chong, William H. Herman, Michael Brändle, Morton B. Brown, Jose C. Florez, David Altshuler, Liana K. Billings, Ling Chen, Maegan Harden, Robert L. Hanson, William C. Knowler, Toni I. Pollin, Alan R. Shuldiner, Kathleen Jablonski, Paul W. Franks, Marie-France Hivert, Josephine H. Li, James A. Perry, Shylaja Srinivasan, Josep M. Mercader, and Jennifer N. Todd
References
- 1. American Diabetes Association . 9. Pharmacologic approaches to glycemic treatment: Standards of Medical Care in Diabetes—2021. Diabetes Care 2021;44(Suppl. 1):S111–S124 [DOI] [PubMed] [Google Scholar]
- 2. Davies MJ, D’Alessio DA, Fradkin J, et al. Management of hyperglycaemia in type 2 diabetes, 2018. A consensus report by the American Diabetes Association (ADA) and the European Association for the Study of Diabetes (EASD). Diabetologia 2018;61:2461–2498 [DOI] [PubMed] [Google Scholar]
- 3. Chawla R, Madhu SV, Makkar BM, Ghosh S, Saboo B; RSSDI-ESI Consensus Group . RSSDI-ESI clinical practice recommendations for the management of type 2 diabetes mellitus 2020. Indian J Endocrinol Metab 2020;24:1–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jia W, Weng J, Zhu D, et al.; Chinese Diabetes Society . Standards of medical care for type 2 diabetes in China 2019. Diabetes Metab Res Rev 2019;35:e3158. [DOI] [PubMed] [Google Scholar]
- 5. Kahn SE, Haffner SM, Heise MA, et al.; ADOPT Study Group . Glycemic durability of rosiglitazone, metformin, or glyburide monotherapy. N Engl J Med 2006;355:2427–2443 [DOI] [PubMed] [Google Scholar]
- 6. Zeitler P, Hirst K, Pyle L, et al.; TODAY Study Group . A clinical trial to maintain glycemic control in youth with type 2 diabetes. N Engl J Med 2012;366:2247–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhou K, Donnelly L, Yang J, et al.; Wellcome Trust Case Control Consortium 2 . Heritability of variation in glycaemic response to metformin: a genome-wide complex trait analysis. Lancet Diabetes Endocrinol 2014;2:481–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhou K, Bellenguez C, Spencer CC, et al.; GoDARTS and UKPDS Diabetes Pharmacogenetics Study Group; Wellcome Trust Case Control Consortium 2; MAGIC investigators . Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat Genet 2011;43:117–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhou K, Yee SW, Seiser EL, et al.; MetGen Investigators; DPP Investigators; ACCORD Investigators . Variation in the glucose transporter gene SLC2A2 is associated with glycemic response to metformin. Nat Genet 2016;48:1055–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rotroff DM, Yee SW, Zhou K, et al.; MetGen Investigators; ACCORD/ACCORDion Investigators . Genetic variants in CPA6 and PRPF31 are associated with variation in response to metformin in individuals with type 2 diabetes. Diabetes 2018;67:1428–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Florez JC, Jablonski KA, Taylor A, et al.; Diabetes Prevention Program Research Group . The C allele of ATM rs11212617 does not associate with metformin response in the Diabetes Prevention Program. Diabetes Care 2012;35:1864–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. The Diabetes Prevention Program Research Group . The Diabetes Prevention Program. Design and methods for a clinical trial in the prevention of type 2 diabetes. Diabetes Care 1999;22:623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. The Diabetes Prevention Program Research Group . The Diabetes Prevention Program: baseline characteristics of the randomized cohort. Diabetes Care 2000;23:1619–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Knowler WC, Barrett-Connor E, Fowler SE, et al.; Diabetes Prevention Program Research Group . Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 2002;346:393–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Delaneau O, Marchini J, Zagury JF. A linear complexity phasing method for thousands of genomes. Nat Methods 2011;9:179–181 [DOI] [PubMed] [Google Scholar]
- 16. Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009;5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sánchez F, Bonàs S, Guindo M, et al. GWImp-COMPSs: an integrated framework for large-scale genome-wide imputation and association testing on parallel computing platforms. Accessed 30 July 2021. Available at cg.bsc.es/guidance/
- 18. 1000 Genomes Project Consortium; Auton A, Brooks LD, Durbin RM, et al. A global reference for human genetic variation. Nature 2015;526:68–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gogarten SM, Bhangale T, Conomos MP, et al. GWASTools: an R/Bioconductor package for quality control and analysis of genome-wide association studies. Bioinformatics 2012;28:3329–3331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dixon WJ. Simplified estimation from censored normal samples. Ann Math Stat 1960;31:385–391 [Google Scholar]
- 21. Li J, Ji L. Adjusting multiple testing in multilocus analyses using the eigenvalues of a correlation matrix. Heredity 2005;95:221–227 [DOI] [PubMed] [Google Scholar]
- 22. GTEx Consortium . The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 2020;369:1318–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Võsa U, Claringbould A, Westra HJ, et al.; BIOS Consortium; i2QTL Consortium . Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat Genet 2021;53:1300–1310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alonso L, Piron A, Morán I, et al.; MAGIC . TIGER: the gene expression regulatory variation landscape of human pancreatic islets. Cell Rep 2021;37:109807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sprowl JA, Ong SS, Gibson AA, et al. A phosphotyrosine switch regulates organic cation transporters. Nat Commun 2016;7:10880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Uddin ME, Garrison DA, Kim K, et al. Influence of YES1 kinase and tyrosine phosphorylation on the activity of OCT1. Front Pharmacol 2021;12:644342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tang Y, Ma N, Luo H, Chen S, Yu F. Downregulated long non-coding RNA LINC01093 in liver fibrosis promotes hepatocyte apoptosis via increasing ubiquitination of SIRT1. J Biochem 2020;167:525–534 [DOI] [PubMed] [Google Scholar]
- 28. Li L, Huang K, Lu Z, et al. Bioinformatics analysis of LINC01554 and its co-expressed genes in hepatocellular carcinoma. Oncol Rep 2020;44:2185–2197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Acton KJ, Burrows NR, Moore K, Querec L, Geiss LS, Engelgau MM. Trends in diabetes prevalence among American Indian and Alaska native children, adolescents, and young adults. Am J Public Health 2002;92:1485–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pavkov ME, Hanson RL, Knowler WC, Bennett PH, Krakoff J, Nelson RG. Changing patterns of type 2 diabetes incidence among Pima Indians. Diabetes Care 2007;30:1758–1763 [DOI] [PubMed] [Google Scholar]
- 31. Kitabchi AE, Temprosa M, Knowler WC, et al.; Diabetes Prevention Program Research Group . Role of insulin secretion and sensitivity in the evolution of type 2 diabetes in the diabetes prevention program: effects of lifestyle intervention and metformin. Diabetes 2005;54:2404–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Williams LK, Padhukasahasram B, Ahmedani BK, et al. Differing effects of metformin on glycemic control by race-ethnicity. J Clin Endocrinol Metab 2014;99:3160–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McInnes G, Yee SW, Pershad Y, Altman RB. Genomewide association studies in pharmacogenomics. Clin Pharmacol Ther 2021;110:637–648 [DOI] [PMC free article] [PubMed] [Google Scholar]