Abstract
Species of Pestalotiopsis were mainly introduced as endophytes, plant pathogens or saprobes from various hosts. In this study, ten strains were isolated from Ficus macrocarpa, Phoebe zhennan and Spatholobus suberectus in China. Based on multilocus phylogenies from the internal transcribed spacer (ITS), the partial translation elongation factor 1-alpha gene (tef1α) and the partial beta-tubulin gene (tub2), in conjunction with morphological characteristics, we describe three new species, viz., Pestalotiopsis ficicola sp. nov., P. phoebes sp. nov. and P. spatholobi sp. nov.
Keywords: Pestalotiopsis, morphology, new species, phylogeny
1. Introduction
Pestalotiopsis Steyaert, belonging to Sporocadaceae (Amphisphaeriales, Ascomycota), was introduced by Steyaert in 1949 (type species: Pestalotiopsis guepinii De Not.) [1]. Species of Pestalotiopsis are endophytic, plant pathogenic or saprobic and are associated with a wide range of host plants [2,3,4,5,6]. Currently, a total of 402 names are documented for Pestalotiopsis in the Index Fungorum (http://www.indexfungorum.org/, accessed on 16 May 2023). Initially, Pestalotiopsis resembling those taxa having affinities with Pestalotia were also known as pestalotioid fungi. Pestalotioid fungi are easily characterized by multiseptate and more or less fusiform conidia with appendages at one or both ends, frequently with some melanized cells.
Pestalotia, isolated from Vitis sp., was introduced by De Not. in 1841 [7]. Subsequently, Steyaert split Pestalotia into three genera, viz., Pestalotia (with six-celled conidia), Pestalotiopsis (with five-celled conidia) and Truncatella (with four-celled conidia), based on the number of cells [1]. Although Guba [8,9] initiated some controversy, Steyaert [10,11,12] provided further evidence in support of splitting Pestalotia. Sutton [13] accepted most of the genera discussed here (Pestalotia, Pestalotiopsis and Truncatella) which fit into fairly well-defined groups and cited the electron microscope investigation of Griffiths and Swart [14] to support Steyaert’s division of Pestalotiopsis. Molecular phylogenetic analysis largely promoted the development of taxonomy. Jeewon et al. [15] established a phylogenetic tree by analyzing some ITS sequences of Pestalotiopsis species and found that Pestalotiopsis species could be divided into three large branches, with the color type of colored cells and the morphology of the end of the apical accessory filaments as the main distinguishing characteristics. Hu et al. analyzed the ITS and tubulin gene of Pestalotiopsis, combined with morphological characteristics and molecular data, and analyzed the relationship between species of Pestalotiopsis [16]. Maharachch. et al. split Pestalotiopsis sensu lato into three genera—Pestalotiopsis sensu stricto, Neopestalotiopsis and Pseudopestalotiopsis—based on phylogeny of multiple genes and conidial morphology; Pseudopestalotiopsis and Pestalotiopsis can be easily distinguished from Neopestalotiopsis by its versicolorous median cells [6]. Liu et al. included Pestalotiopsis into Sporocadaceae using morphological characteristics and on the basis of a multilocus phylogenetic analysis [5]. Some genera of the Sporocadaceae can be divided into three categories based on the number of appendages, viz., genera with a single apical and basal appendage (Monochaetia, Seiridium), other genera which do not form appendages (Nonappendiculata) or genera which have 2–4 appendages (Pestalotiopsis, Ciliochorella, Neopestalotiopsis and Pseudopestalotiopsis) [17]. Recently, Jiang et al. obtained 43 isolates of Pestalotiopsis from diseased leaf tissues of Fagaceae based on combined morphology and phylogeny between 2016 and 2021 [18].
In this study, we produced a collection of the genera Pestalotiopsis species from leaves of Ficus macrocarpa, Phoebe zhennan and Spatholobus suberectus in East Harbour National Nature Reserve, Hainan Province, China. Three new species were described based on unique morphological characters and distinct phylogenetic placement, viz., Pestalotiopsis ficicola sp. nov., P. phoebes sp. nov. and P. spatholobi sp. nov.
2. Materials and Methods
2.1. Isolation and Morphology
Samples of Ficus macrocarpa L. f., Phoebe zhennan S. Lee et F. N. Wei and Spatholobus suberectus Dunn showing obvious disease spots were collected from Hainan Province during 2021 in East Harbour National Nature Reserve (110°32′~110°37′ E, 19°51′~20°1′ N), China. The cultures of Pestalotiopsis were isolated from diseased tissues of the sample leaves using tissue isolation methods [19]. Fragments (5 × 5 mm) were taken from the edges of the leaf lesions, surface sterilized for 30 s in 75% ethanol, rinsed in sterile deionized water for 30 s, rinsed in 5% sodium hypochlorite solution for 1 min and then rinsed four times in sterile deionized water for 30 s [17]. The pieces were blotted on sterile filter paper to dry, transferred onto PDA flats (PDA medium: potato 200 g, agar 15–20 g, dextrose 15–20 g, deionized water 1 L, pH~7.0, available after sterilization) and incubated at 25 °C for 2–3 days. The edges of hyphal growth were then transferred to new PDA flats to obtain pure cultures; simultaneously, they were inoculated on PDA and incubated at 23 °C under continuous near-ultraviolet light to promote sporulation [20].
All Pestalotiopsis plates were incubated at 25 °C for 14 days and morphological characters including graphs of the colonies were documented on the 14th day using a digital camera (Canon G7X, Canon, Tokyo, Japan). Morphological characters of conidiomata were studied using a stereomicroscope (Olympus SZX10, Olympus Corporation, Tokyo, Japan) while the micromorphological structures were observed using a microscope (Olympus BX53, Olympus Corporation, Tokyo, Japan). All cultures were deposited in 10% sterilized glycerin and sterile water at 4 °C for future studies. Micromorphological structural measurements were taken using the Digimizer software v. 5.6.0 (https://www.digimizer.com/, accessed on 16 May 2023), with 20 measurements taken for each structure [17]. Voucher specimens were deposited in the Herbarium Mycologicum Academiae Sinicae, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China (HMAS) and the Herbarium of the Department of Plant Pathology, Shandong Agricultural University, Taian, China (HSAUP). Ex-holotype living cultures were deposited in the Shandong Agricultural University Culture Collection (SAUCC). Taxonomic information of the new taxa was submitted to MycoBank (http://www.mycobank.org, accessed on 16 May 2023).
2.2. DNA Extraction and Amplification
Genomic DNA was extracted from the colonies grown on PDA using a kit method (OGPLF-400, GeneOnBio Corporation, Changchun, China) [21]. Gene sequences were obtained from five loci including the internal transcribed spacer regions with the intervening 5.8S nrRNA gene (ITS), the partial translation elongation factor 1-alpha gene (tef1α) and the partial beta-tubulin gene (tub2). These were amplified by the primer pairs and polymerase chain reaction (PCR) programs listed in Table 1. Amplification reactions were performed in a 25 μL reaction volume which contained 10 μL 2 × Hieff Canace® Plus PCR Master Mix (With Dye) (Yeasen Biotechnology, Cat No. 10154ES03, Shanghai, China), 0.5 μL of each forward and reverse primer (10 μM) (TsingKe, Qingdao, China) and 1 μL template genomic DNA, adjusted with distilled deionized water to a total volume of 25 μL. PCR amplification products were visualized on 2% agarose electrophoresis gel. DNA sequencing was performed using an Eppendorf Master Thermocycler (Hamburg, Germany) at the TsingKe Company Limited (Qingdao, China) bidirectionally. Consensus sequences were obtained using MEGA 7.0 [22]. All sequences generated in this study were deposited in GenBank (Table S1: See Supplementary File S1).
Table 1.
Molecular markers and their PCR primers and programs used in this study.
| Loci | PCR Primers | Sequence (5′—3′) | PCR Cycles | References |
|---|---|---|---|---|
| ITS |
ITS5 ITS4 |
GGA AGT AAA AGT CGT AAC AAG G TCC TCC GCT TAT TGA TAT GC |
(95 °C: 30 s, 55 °C: 30 s, 72 °C: 1 min) × 35 cycles | [23] |
| tef1α |
EF1-728F EF-2 |
CAT CGA GAA GTT CGA GAA GG GGA RGT ACC AGT SAT CAT GTT |
(95 °C: 30 s, 48 °C: 30 s, 72 °C: 1 min) × 35 cycles | [24,25] |
| tub2 |
Bt-2a Bt-2b |
GGT AAC CAA ATC GGT GCT GCT TTC ACC CTC AGT GTA GTG ACC CTT GGC |
(95 °C: 30 s, 53 °C: 30 s, 72 °C: 1 min) × 35 cycles | [26] |
2.3. DNA Extraction and Amplification
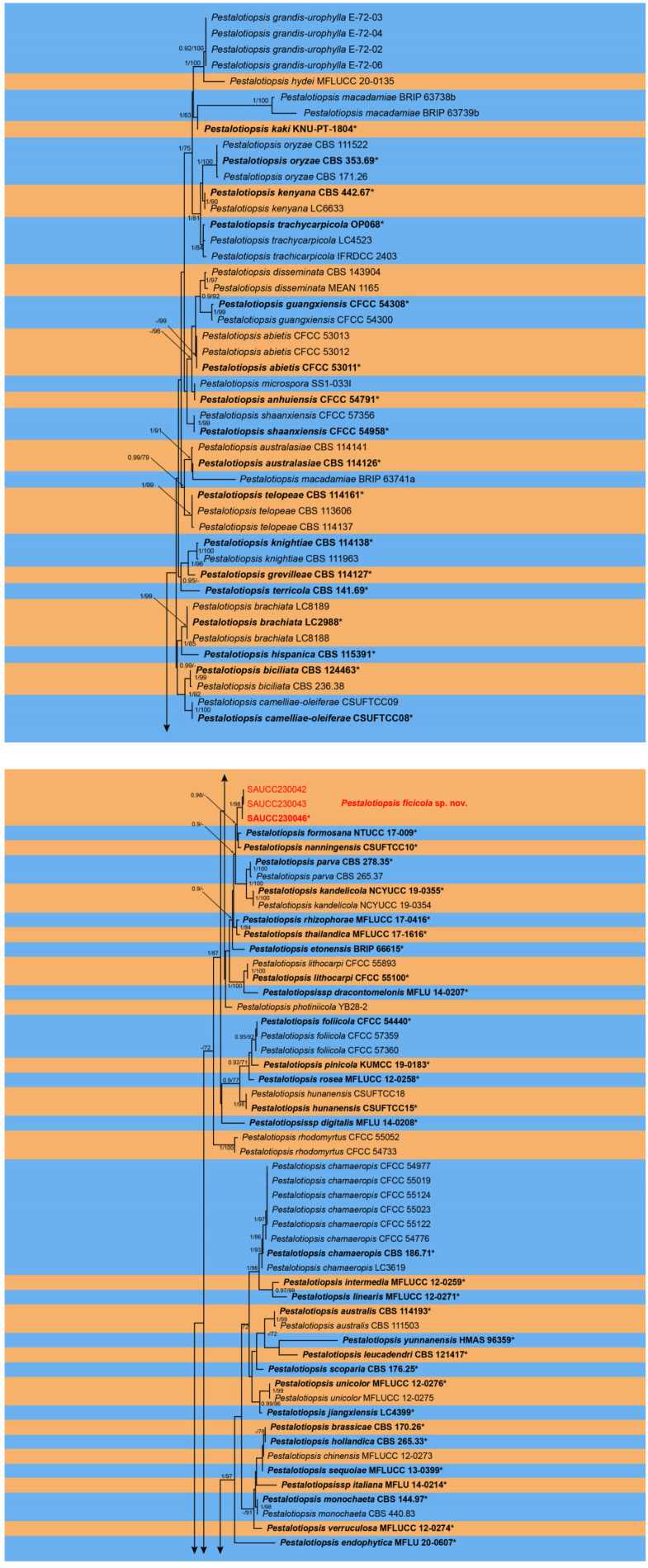
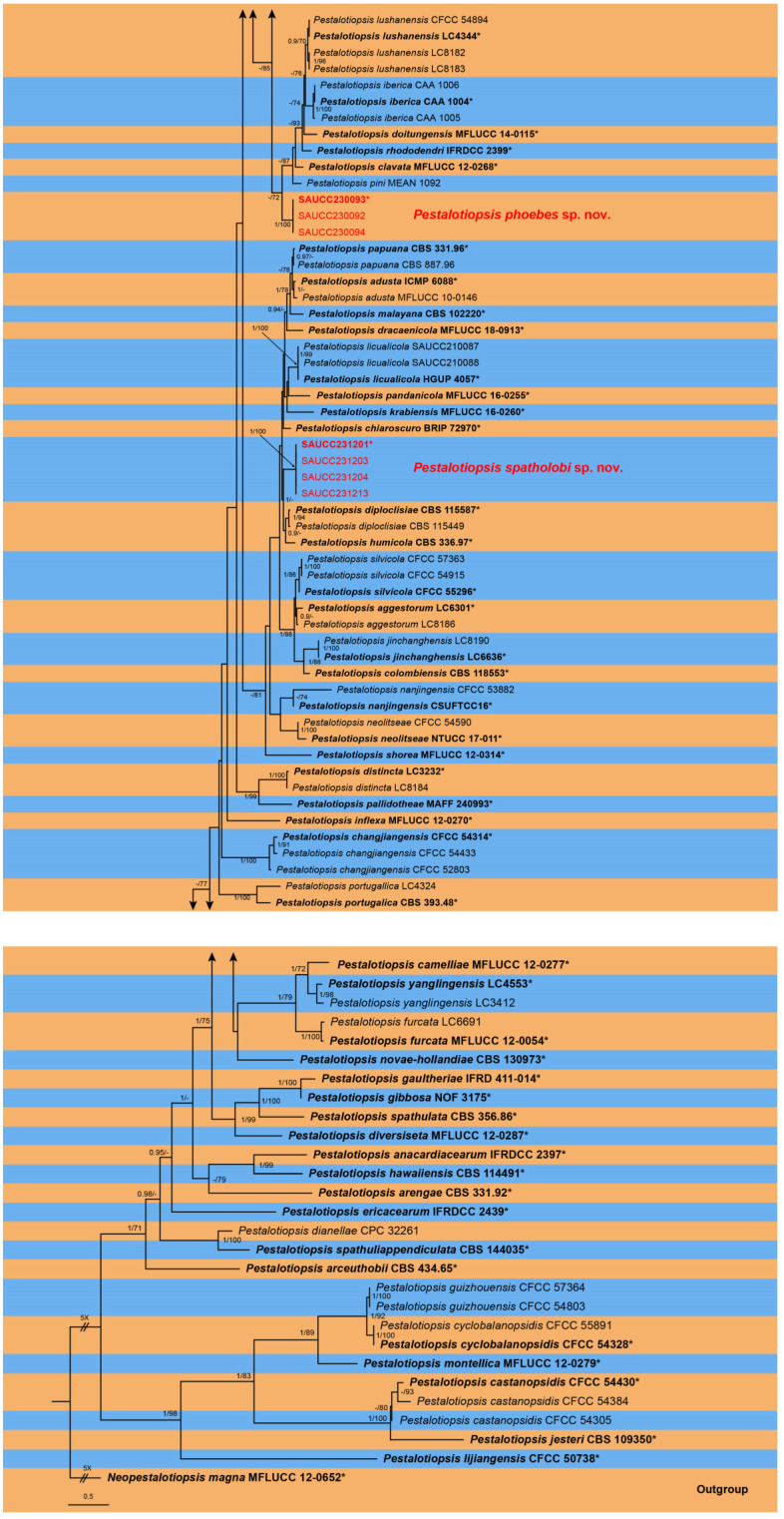
Novel sequences obtained in this study and related sets of sequences from Jiang et al. [18] were aligned with MAFFT v. 7 and corrected manually using MEGA 7 [27]. Multilocus phylogenetic analyses were based on the algorithms maximum likelihood (ML) and Bayesian inference (BI) methods. The ML was run on the CIPRES Science Gateway portal (https://www.phylo.org, accessed on 16 May 2023) [28] using RAxML–HPC2 on XSEDE v. 8.2.12 [29] and employed a GTRGAMMA substitution model with 1000 bootstrap replicates. Other parameters were default. For Bayesian inference analyses, the best model of evolution for each partition was determined using ModelTest v. 2.3 [30] and included the analyses. The BI was performed using MrBayes on XSEDE v. 3.2.7a [31,32,33] and two Markov chain Monte Carlo (MCMC) chains were run, starting from random trees, for 2,000,000 generations (average standard deviation of split frequencies < 0.01). Additionally, a sampling frequency of 100 generations was used. The first 25% of trees were discarded as burn-in and BI posterior probabilities (PPs) were conducted from the remaining trees. The consensus trees were optimized using FigTree v. 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree, accessed on 16 May 2023) and embellished with Adobe Illustrator CC 2019 (Figure 1).
Figure 1.
A phylogram of the Pestalotiopsis with Neopestalotiopsis magna (MFLUCC 12-0652) as outgroup based on a concatenated ITS, tef1α and tub2 sequence alignment (only shows the topology of the ML tree). BI posterior probabilities and maximum likelihood bootstrap support values above 0.70 and 90% are shown at the first and second position, respectively. Ex-type cultures are marked in bold face and *. Strains obtained in the present study are in red. Some branches are shortened for layout purposes—these are indicated by two diagonal lines with the number of times. The orange and blue areas are used to distinguish different species. The scale bar at the left–bottom represents 0.5 substitutions per site.
3. Results
3.1. Phylogenetic Analyses
The alignment contained 182 strains representing Pestalotiopsis and the strain MFLUCC 12-0652 of Neopestalotiopsis magna was used as outgroup [6]. The dataset had an aligned length of 2011 characters including gaps, viz., ITS: 1–565, tef1α: 566–1149 and tub2: 1150–2011 (Supplementary File S1). Of these, 1172 were constant, 281 were parsimony uninformative and 558 were parsimony informative. The ModelTest suggested that the BI used the Dirichlet base frequencies, the GTR + I + G evolutionary mode for ITS and tub2, and HKY + I + G for tef1α.
The topology of the ML tree was consistent with that of the Bayesian tree; therefore, it only showed the topology of the ML tree as a representative for summarizing the evolutionary relationship within the genus Pestalotiopsis. The final ML optimization likelihood was −17,300.001718. The 181 strains were assigned to 109 species clades on the phylogram (Figure 1). Based on the phylogenetic resolution and morphological analyses, the present study introduced three novel species of the Pestalotiopsis, viz., Pestalotiopsis ficicola sp. nov., P. phoebes sp. nov. and P. spatholobi sp. nov.
3.2. Taxonomy
3.2.1. Pestalotiopsis ficicola Z.X. Zhang, J.W. Xia and X.G. Zhang, sp. nov.
MycoBank: No. MB848628
Etymology: The epithet “ficicola” pertains to the generic name of the host plant Ficus microcarpa.
Type: China, Hainan Province, East Harbour National Nature Reserve, on diseased leaves of Ficus microcarpa, 23 May 2021, Z.X. Zhang, holotype HMAS 352477, ex-holotype living culture SAUCC230046.
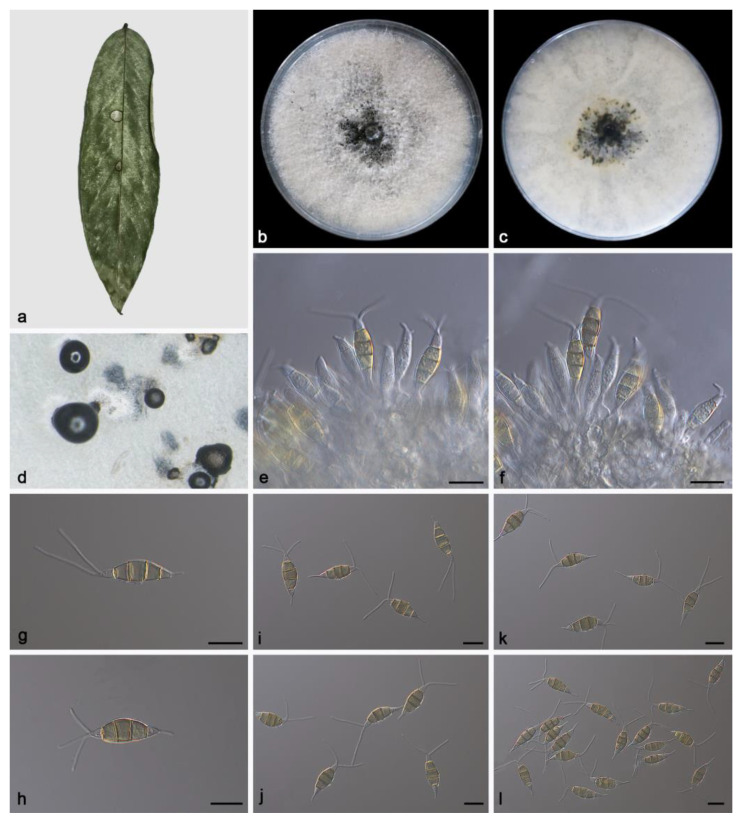
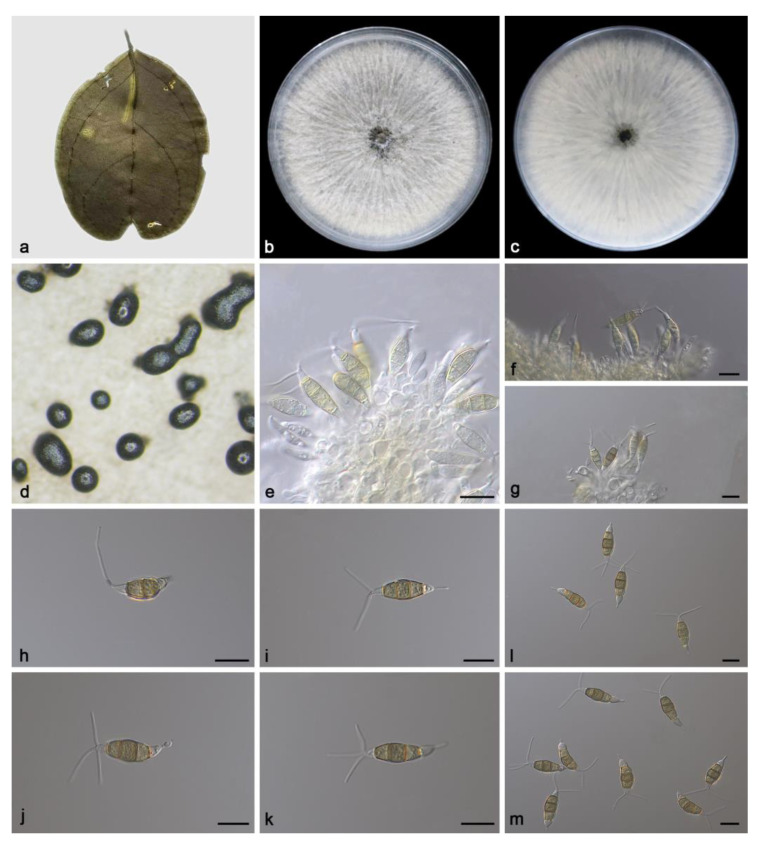
Description: Conidiomata in culture sporodochial, aggregated or solitary, erumpent, pulvinate, straw yellow, exuding black conidial masses. Conidiophores simple, hyaline, subcylindrical, usually reduced to conidiogenous cells. Conidiogenous cells aggregative, hyaline, smooth, cylindrical, 16.5–22.4 × 4.1–6.0 μm. Conidia fusoid, straight or slightly curved, 4-septate, smooth, slightly constricted at the septa, 18.1–22.7 × 5.6–7.9 μm; basal cell obconic with a truncate base, thin-walled, hyaline, 3.6–5.2 μm; median cells 3, trapezoidal or subcylindrical, concolorous, pale brown to brown, thick-walled, the first median cell from base 3.6–4.3 μm long, the second cell 3.4–5.7 μm long, the third cell 3.5–5.6 μm long, together 11.8–14.2 μm long; apical cell conic with an acute apex, thin-walled, hyaline, 2.0–4.0 μm long; basal appendage single, unbranched, tubular, centric, straight or slightly bent, 3.0–6.7 μm long; apical appendages 2–3, unbranched, tubular, straight or slightly bent, 10.8–25.5 μm long. Sexual morph unknown, see Figure 2.
Figure 2.
Pestalotiopsis ficicola (holotype HMAS 352477). (a) Leaf of Ficus microcarpa; (b,c) inverse and reverse sides of colony after 15 days on PDA; (d) colony overview; (e,f) conidiogenous cells with conidia; (g–l) conidia. Scale bars: (e–l) 10 μm.
Culture characteristics: The colonies diameter reached 90 mm after 14 days of dark culture at 25 °C on PDA, flat, aerial mycelium flocculent fluffy, white; reverse center pale yellow, edge white.
Additional specimen examined: China, Hainan Province, East Harbour National Nature Reserve, on diseased leaves of Ficus microcarpa, 23 May 2021, Z.X. Zhang, HSAUP230043, living culture SAUCC230043, on diseased leaves of Ficus microcarpa, 23 May 2021, Z.X. Zhang, HSAUP230042, living culture SAUCC230042.
Notes: Phylogenetic analyses of three combined genes (ITS, tef1α and tub2) showed Pestalotiopsis ficicola sp. nov. was closely related to P. formosana and P. nanningensis. In detail, P. ficicola was distinguished from P. formosana by 7/540 bp in ITS, 3/478 bp in tef1α and 3/388 bp in tub2 and from P. nanningensis by 5/540 in ITS, 4/478 in tef1α and 8/440 in tub2. In morphology, the conidia of P. ficicola was longer than P. formosana (18.1–22.7 × 5.6–7.9 vs. 8–22 × 6–7 µm) and shorter than P. nanningensis (18.1–22.7 × 5.6–7.9 vs. 24–26.5 × 7–8 µm). What is more, the apical appendages of P. ficicola were longer than P. formosana (18–22.5 vs. 8–20 µm) and shorter than P. nanningensis (10.8–25.5 vs. 13.5–26.5 µm) [34,35].
3.2.2. Pestalotiopsis phoebes Z.X. Zhang, J.W. Xia and X.G. Zhang, sp. nov.
MycoBank: No. MB848629
Etymology: The epithet “phoebes” pertains to the generic name of the host plant Phoebe zhennan.
Type: China. Hainan Province, East Harbour National Nature Reserve, on diseased leaves of Phoebe zhennan, 23 May 2021, Z.X. Zhang, holotype HMAS 352478, ex-holotype living culture SAUCC230093.
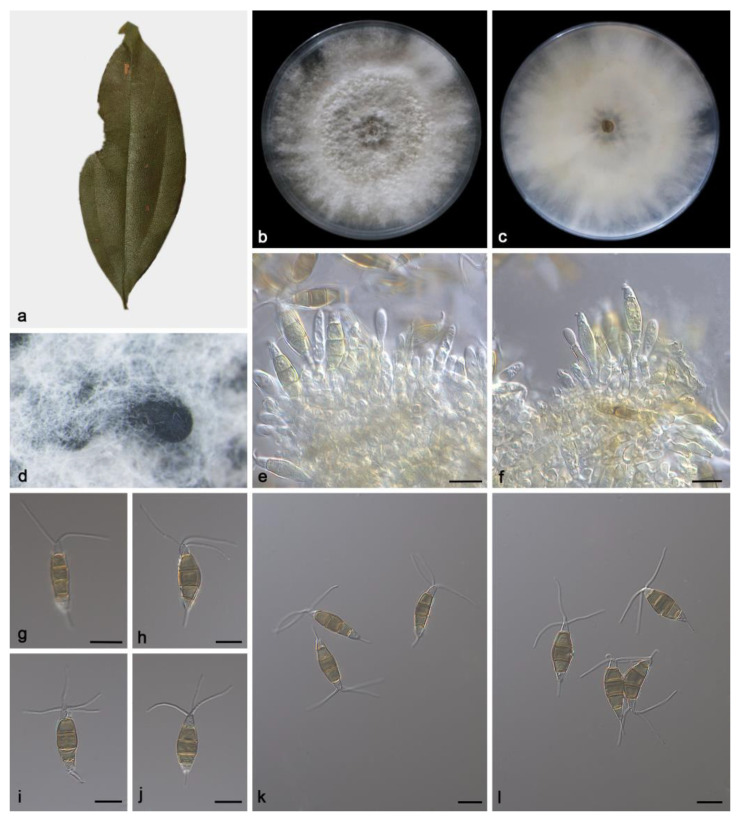
Description: Conidiomata in culture sporodochial, aggregated or solitary, erumpent, pulvinate, black, exuding black conidial masses. Conidiophores simple, hyaline, usually reduced to conidiogenous cells. Conidiogenous cells aggregative, hyaline, smooth, cylindrical, 16.4–36.6 × 3.1–5.3 μm. Conidia fusoid, straight or slightly curved, 4-septate, smooth, slightly constricted at the septa, 20.2–23.5 × 6.4–8.6 μm; basal cell obconic with a truncate base, thin-walled, hyaline, 2.9–4.3 μm; median cells 3, trapezoidal or subcylindrical, concolorous, pale brown to brown, thick-walled, the first median cell from base 4.5–6.1 μm long, the second cell 5–5.9 μm long, the third cell 3.5–4.9 μm long, together 13.7–15.7 μm long; apical cell conic with an acute apex, thin-walled, hyaline, 2.9–4.1 μm long; basal appendage single, unbranched, tubular, centric, straight or slightly bent, 4.7–6.2 μm long; apical appendages 2–4, unbranched, tubular, straight or slightly bent, 15.2–19.6 μm long. Sexual morph unknown, see Figure 3.
Figure 3.
Pestalotiopsis phoebes (holotype HMAS 352478). (a) Leaf of Phoebe zhennan; (b,c) inverse and reverse sides of colony after 15 days on PDA; (d) colony overview; (e,f) conidiogenous cells with conidia; (g–l) conidia. Scale bars: (e–l) 10 μm.
Culture characteristics: The colonies diameter reached 90 mm after 14 days of dark culture at 25 °C on PDA, flat, aerial mycelium flocculent fluffy, grows irregularly, white; reverse center had a light brown ring, edge white.
Additional specimen examined: China, Hainan Province, East Harbour National Nature Reserve, on diseased leaves of Phoebe zhennan, 23 May 2021, Z.X. Zhang, HSAUP230092, living culture SAUCC230092, on diseased leaves of Phoebe zhennan, 23 May 2021, Z.X. Zhang, HSAUP230094, living culture SAUCC230094.
Notes: Phylogenetic analyses of three combined genes (ITS, tef1α and tub2) showed Pestalotiopsis phoebes sp. nov. was closely related to P. clavata and P. pini. In detail, P. phoebes was distinguished from P. clavata by 2/540 bp in ITS, 7/259 bp in tef1α and 11/441 bp in tub2 and from P. pini by 3/540 in ITS, 9/254 in tef1α and 10/762 in tub2. In morphology, the conidia of P. phoebes was shorter than P. clavate (20.2–23.5 × 6.4–8.6 vs. 20–27 × 6.5–8 µm) and P. pini (20.2–23.5 × 6.4–8.6 vs. 20.0–27.6 × 4.7–8.2 µm). What is more, the apical appendages of P. phoebes were shorter than P. clavate (15.2–19.6 vs. 20–25 µm) and P. pini (15.2–19.6 vs. 9.7–27.8 µm) [36,37].
3.2.3. Pestalotiopsis spatholobi Z.X. Zhang, J.W. Xia and X.G. Zhang, sp. nov.
MycoBank: No. MB848630
Etymology: The epithet “spatholobi” pertains to the generic name of the host plant Spatholobus suberectus.
Type: China. Hainan Province, East Harbour National Nature Reserve, on diseased leaves of Spatholobus suberectus, 23 May 2021, Z.X. Zhang, holotype HMAS 352479, ex-holotype living culture SAUCC231201.
Description: Conidiomata in culture sporodochial, aggregated or solitary, erumpent, exuding black conidial masses, covered with mycelium. Conidiophores simple, hyaline, usually reduced to conidiogenous cells. Conidiogenous cells aggregative, hyaline, smooth, cylindrical, 11.4–26.8 × 2.3–6.2 μm. Conidia fusoid, straight or slightly curved, 4-septate, smooth, slightly constricted at the septa, 16.7–23.0 × 5.9–7.4 μm; basal cell obconic with a truncate base, thin-walled, hyaline, 3.0–5.4 μm; median cells 3, trapezoidal or subcylindrical, concolorous, pale brown to brown, thick-walled, the first median cell from base 3.5–5.0 μm long, the second cell 3.7–5.5 μm long, the third cell 3.6–4.9 μm long, together 11.2–14.9 μm long; apical cell conic with an acute apex, thin-walled, hyaline, 3.1–5.0 μm long; basal appendage single, unbranched, tubular, centric, straight or slightly bent, 0.9–3.1 μm long; apical appendages 1–3, unbranched, tubular, straight or slightly bent, 8.4–15.3 μm long. Sexual morph unknown, see Figure 4.
Figure 4.
Pestalotiopsis spatholobi (holotype HMAS 352479). (a) Leaf of Spatholobus suberectus; (b,c) inverse and reverse sides of colony after 15 days on PDA; (d) colony overview; (e–g) conidiogenous cells with conidia; (h–m) conidia. Scale bars: (e–m) 10 μm.
Culture characteristics: The colonies diameter reached 90 mm after 14 days of dark culture at 25 °C on PDA, flat, aerial mycelium flocculent fluffy, radial stripes from the middle to the periphery, white; reverse center was black, edge white.
Additional specimen examined: China, Hainan Province, East Harbour National Nature Reserve, on diseased leaves of Spatholobus suberectus, 23 May 2021, Z.X. Zhang, HSAUP231203, living culture SAUCC231203, on diseased leaves of Spatholobus suberectus, 23 May 2021, Z.X. Zhang, HSAUP231204, living culture SAUCC231204, on diseased leaves of Spatholobus suberectus, 23 May 2021, Z.X. Zhang, HSAUP231213, living culture SAUCC231213.
Notes: Phylogenetic analyses of three combined genes (ITS, tef1α and tub2) showed Pestalotiopsis spatholobi sp. nov. formed an independent clade and was closely related to P. diploclisia and P. humicola. In detail, P. spatholobi was distinguished from P. diploclisia by 2/539 bp in ITS, 11/258 bp in tef1α and 12/762 bp in tub2 and from P. humicola by 3/539 in ITS, 10/260 in tef1α and 15/762 in tub2. In morphology, the conidia of P. spatholobi was shorter than P. diploclisia (16.7–23.0 × 5.9–7.4 vs. 20–28 × 5–7 µm) and P. humicola (16.7–23.0 × 5.9–7.4 vs. 17–23 × 5–7.5 µm). What is more, the apical appendages of P. spatholobi were shorter than P. diploclisia (8.4–15.3 vs. 10–22 µm) and P. humicola (8.4–15.3 vs. 6–13 µm) [6].
4. Discussion
In the present study, ten strains from three host genera (Ficus macrocarpa, Phoebe zhennan and Spatholobus suberectus) were split into three new species (Pestalotiopsis ficicola sp. nov., P. phoebes sp. nov. and P. spatholobi sp. nov.) based on phylogeny and morphology. The Global Biodiversity Information Facility (https://www.gbif.org/, accessed on 16 May 2023) contains 4174 georeferenced records of Pestalotiopsis species reported around the world. Most species are distributed in countries such as the United States of American, Europe, Australia and China where suitable climates and the environment favors growth of unusual microbial species [38].
Historically, traditional identification of Pestalotiopsis species has long been a complicated endeavor [8,9,10,11,12]. Fortunately, with the development of molecular technology and the efforts of many taxonomists, the taxonomic status of Pestalotiopsis (Sporocadaceae, Amphisphaeriales) has become increasingly apparent [5,6,13,18,19]. Recent research showed that the same Pestalotiopsis species can be found on hosts belonging to multiple plant families, for example, P. chamaeropis was isolated from Chamaerops humilis (Arecaceae), Quercus sp., Castanopsis sp. (Fagaceae) and Camellia sp. (Theaceae) [6,18,39]. On the other hand, a variety of Pestalotiopsis species were isolated from the same plant host, for example, Pestalotiopsis chamaeropis, P. kenyana, P. nanjingensis and P. rhodomyrtus were isolated from the Quercus aliena (Fagaceae) [18]. These findings indicate that Pestalotiopsis species have the ability to infect a wide range of host plants rather than showing restricted host preferences.
In the pestalotioid species, apical or basal appendages differ in number, origin, position, number of branches and branching pattern, and these characters can be divided into some genera of the Sporocadaceae [5,17]. Monochaetia and Seiridium possess single apical and basal appendages, Nonappendiculata do not possess any appendages, and Pestalotiopsis, Ciliochorella, Neopestalotiopsis, Pseudopestalotiopsis possess 2–4 appendages. In previous studies, some members of Sporocadaceae were shown to contain secondary metabolites with significant biological activity, e.g., the sterol constituents of the marine fungus Pestalotiopsis sp. XWS03F09 exhibit selective inhibitory activities against antitumor cells [40,41,42,43]. Therefore, it is necessary to carry out genome sequencing of Pestalotiopsis species and fully explore, through structural and functional annotation, the diversity of secondary metabolites which might be useful for various biotechnological applications. This shows the importance of taxonomy and deposition of specimens.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11071627/s1, Supplementary File S1: Table S1 and the combined ITS, tef1 and tub2 sequences.
Author Contributions
Conceptualization, methodology, software, Z.Z.; validation, formal analysis, J.Z.; investigation, resources, D.L.; data curation, writing—original draft preparation, Z.Z.; writing—review and editing, visualization, J.X.; supervision, J.X.; project administration, X.Z.; funding acquisition, X.Z. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
The sequences from the present study were submitted to the NCBI database (https://www.ncbi.nlm.nih.gov/, accessed on 16 May 2023) and the accession numbers were listed in Table S1.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was supported by the Science and Technology Fundamental Resources Investigation Program (Grant No. 2019FY100700) and the National Natural Science Foundation of China (No. U2002203 and No. 32270024).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Steyaert R.L. Contribution a l’etude monographique de Pestalotia de Not. et Monochaetia Sacc. (Truncatella gen. nov. et Pestalotiopsis gen. nov.) Bull. Jard. Bot. Etat Brux. 1949;19:285–354. doi: 10.2307/3666710. [DOI] [Google Scholar]
- 2.Strobel G., Li J.Y., Ford E., Worapong J., Hess W.M. Pestalotiopsis jesteri sp. nov. an endophyte from Fragraea bodenii Wernh., a Commaon plant in the southern highlands of Papua New Guinea. Mycotaxon. 2000;76:257–266. [Google Scholar]
- 3.Das R., Chutia M., Das K., Jha D.K. Factors affecting sporuiation of Pestalotiopsis disseminata causing grey bligiht disease of Persea bombycina Kost., the primary food plant of muga silkwom. Crap. Prot. 2010;29:963–968. doi: 10.1016/j.cropro.2010.05.012. [DOI] [Google Scholar]
- 4.Jayawardena R.S., Zhang W., Liu M., Maharachchikumbura S.S.N., Zhou Y., Huang J.B., Nilthong S., Wang Z.Y., Li X.H., Yan J.Y., et al. Identification and characterization of Pestalotiopsis-like fungi related to grapevine diseases in China. Fungal Biol. 2015;119:348–361. doi: 10.1016/j.funbio.2014.11.001. [DOI] [PubMed] [Google Scholar]
- 5.Liu F., Bonthond G., Groenewald J.Z., Cai L., Crous P.W. Sporocadaceae, a family of coelomycetous fungi with appendage-bearing conidia. Stud. Mycol. 2019;92:287–415. doi: 10.1016/j.simyco.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maharachchikumbura S.S.N., Hyde K.D., Groenewald J.Z., Xu J., Crous P.W. Pestalotiopsis revisited. Stud. Mycol. 2014;79:121–186. doi: 10.1016/j.simyco.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Notaris G. Micromycetes italiei Dec II. Mere R. Acad. Sci. Torino II. 1839;3:80–81. [Google Scholar]
- 8.Guba E.F. Monochaetia and Pestalotia vs. Truncatella, Pestalotiopsis and Pestalotia. Ann. Microbiol. 1956;7:74–76. [Google Scholar]
- 9.Guba E.F. Monograph of Pestalotia and Monochaetia. Harvard University Press; Cambridge, MA, USA: 1961. [Google Scholar]
- 10.Steyaert R.L. New and old species of Pestalotiopsis. Trans. Br. Mycol. Soc. 1953;36:81–89. doi: 10.1016/S0007-1536(53)80052-5. [DOI] [Google Scholar]
- 11.Steyaert R.L. Type specimens of Spegazzini’s collections in the Pestalotiopsis and related genera (Fungi Imperfecti: Melanconiales) Darwinia. 1961;12:157–190. [Google Scholar]
- 12.Steyaert R.L. Complementary informations concerning Pestalotiopsis guepini (Desmazieres) Steyaert and designation of its lectotype. Bull. Jard. Bot. l’Etat Brux. 1963;33:369–373. doi: 10.2307/3667200. [DOI] [Google Scholar]
- 13.Sutton B.C. The Coelomycetes. Fungi Imperfecti with Pycnidia, Acervuli and Stromata. Commonwealth Mycological Institute; Surrey, UK: 1980. [Google Scholar]
- 14.Griffiths D.A., Swart H.J. Conidial structure in two species of Pestalotiopsis. Trans. Br. Mycol. Soc. 1974;62:295–304. doi: 10.1016/S0007-1536(74)80038-0. [DOI] [Google Scholar]
- 15.Jeewon R., Liew E.C.Y., Simpson J.A., Hodgkiss I.J., Hyde K.D. Phylogenetic significance of morphological characters in the taxonomy of Pestalotiopsis species. Mol. Phylogenetics Evol. 2003;27:372–383. doi: 10.1016/S1055-7903(03)00010-1. [DOI] [PubMed] [Google Scholar]
- 16.Hu H.L., Jeewon R., Zhou D.Q., Zhou T.X., Hyde K.D. Phylogenetic diversity of endophytic Pestalotiopsis species in Pious armandii and Ribes spp.: Evidence from rDNA and β-tubulin gene phylogenies. Fungal Divers. 2007;24:1–22. [Google Scholar]
- 17.Zhang Z.X., Liu R.Y., Liu S.B., Mu T.C., Zhang X.G., Xia J.W. Morphological and phylogenetic analyses reveal two new species of Sporocadaceae from Hainan, China. MycoKeys. 2022;88:171–192. doi: 10.3897/mycokeys.88.82229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang N., Voglmayr H., Xue H., Piao C.G., Li Y. Morphology and Phylogeny of Pestalotiopsis (Sporocadaceae, Amphisphaeriales) from Fagaceae Leaves in China. Microbiol. Spectr. 2022;10:e03272-22. doi: 10.1128/spectrum.03272-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang N., Voglmayr H., Ma C.Y., Xue H., Piao C.G., Li Y. A new Arthrinium-like genus of Amphisphaeriales in China. MycoKeys. 2022;92:27–43. doi: 10.3897/mycokeys.92.86521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braun U., Nakashima C., Crous P.W., Groenewald J.Z., Moreno-Rico O., Rooney-Latham S., Blomquist C.L., Haas J., Marmolejo J. Phylogeny and taxonomy of the genus Tubakia s. lat. Fungal Syst. Evol. 2018;1:41–99. doi: 10.3114/fuse.2018.01.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao H., Zhou M., Liu X.Y., Wu F., Dai Y.C. Phylogeny, Divergence Time Estimation and Biogeography of the Genus Onnia (Basidiomycota, Hymenochaetaceae) Front. Microbiol. 2022;13:907961. doi: 10.3389/fmicb.2022.907961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.White T.J., Bruns T., Lee S., Taylor J.W. Amplification and direct sequencing of fungal ribosomal rna genes for phylogenetics. In: Innis M.A., Gelfand D.H., Sninsky J.J., White T.J., editors. PCR Protocols: A Guide to Methods and Applications. Academic Press Inc.; New York, NY, USA: 1990. pp. 315–322. [DOI] [Google Scholar]
- 24.O’Donnell K., Kistler H.C., Cigelnik E., Ploetz R.C. Multiple Evolutionary Origins of the Fungus Causing Panama Disease of Banana: Concordant Evidence from Nuclear and Mitochondrial Gene Genealogies. Proc. Natl. Acad. Sci. USA. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carbone I., Kohn L.M. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia. 1999;91:553–556. doi: 10.1080/00275514.1999.12061051. [DOI] [Google Scholar]
- 26.Glass N.L., Donaldson G.C. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl. Environ. Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019;20:1160–1166. doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller M.A., Pfeiffer W., Schwartz T. The CIPRES science gateway: Enabling high-impact science for phylogenetics researchers with limited resources; Proceedings of the 1st Conference of the Extreme Science and Engineering Discovery Environment: Bridging from the Extreme to the Campus and Beyond; Chicago, IL, USA. 16–20 July 2012; p. 8. [DOI] [Google Scholar]
- 29.Stamatakis A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nylander J.A.A. Evolutionary Biology Centre. Uppsala University; Uppsala, Sweden: 2004. MrModelTest v. 2. Program distributed by the author. [Google Scholar]
- 31.Huelsenbeck J.P., Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 32.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian Phylogenetic Inference under Mixed Models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 33.Ronquist F., Teslenko M., van der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ariyawansa H.A., Hyde K.D. Additions to Pestalotiopsis in Taiwan. Mycosphere. 2018;9:999–1013. doi: 10.5943/mycosphere/9/5/4. [DOI] [Google Scholar]
- 35.Li L.L., Yang Q., Li H. Morphology, Phylogeny, and Pathogenicity of Pestalotioid Species on Camellia oleifera in China. J. Fungi. 2021;7:1080. doi: 10.3390/jof7121080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maharachchikumbura S.S.N., Guo L.D., Cai L., Chukeatirote E., Wu W.P., Sun X., Crous P.W., Bhat D.J., McKenzie E.H.C., Bahkali A.H., et al. A multi-locus backbone tree for Pestalotiopsis, with a polyphasic characterization of 14 new species. Fungal Divers. 2012;56:95–129. doi: 10.1007/s13225-012-0198-1. [DOI] [Google Scholar]
- 37.Silva A.C., Diogo E., Henriques J., Ramos A.P., Sandoval-Denis M., Crous P.W., Bragança H. Pestalotiopsis pini sp. nov., an Emerging Pathogen on Stone Pine (Pinus pinea L.) Forests. 2020;11:805. doi: 10.3390/f11080805. [DOI] [Google Scholar]
- 38.Zhang Z.X., Liu X.Y., Zhang X.G., Meng Z. Morphological and phylogenetic analyses reveal two new species and a new record of Phyllosticta (Botryosphaeriales, Phyllostictaceae) from Hainan, China. MycoKeys. 2022;91:1–23. doi: 10.3897/mycokeys.91.84803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu F., Hou L.W., Raza M., Cai L. Pestalotiopsis and allied genera from Camellia, with description of 11 new species from China. Sci. Rep. 2017;7:866. doi: 10.1038/s41598-017-00972-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collado J., Platas G., Bills G.F., Basilio Á., Vicente F., Tormo J.R., Hernández P., Teresa Díez M., Peláez F. Studies on Morinia: Recognition of Morinia longiappendiculata sp. nov. as a new endophytic fungus, and a new circumscription of Morinia pestalozzioides. Mycologia. 2006;98:616–627. doi: 10.1080/15572536.2006.11832665. [DOI] [PubMed] [Google Scholar]
- 41.Gangadevi V., Muthumary J. Taxol, an anticancer drug produced by an endophytic fungus Bartalinia robillardoides Tassi, isolated from a medicinal plant, Aegle marmelos Correa ex Roxb. World J. Microbiol. Biotechnol. 2008;24:717–724. doi: 10.1007/s11274-007-9530-4. [DOI] [Google Scholar]
- 42.Liu L., Li Y., Liu S.C., Zheng Z.H., Chen X.L., Zhang H., Guo L.D., Che Y.S. Chloropestolide A, an antitumor metabolite with an unprecedented spiroketal skeleton from Pestalotiopsis fici. Org. Lett. 2009;11:2836–2839. doi: 10.1021/ol901039m. [DOI] [PubMed] [Google Scholar]
- 43.Lei H., Xia T., Wang J., Xiong X., Liu L. Separation and purification of sterols from marine fungus Pestalotiopsis sp. and study on its anti-tumor activity. China Meas. Test. 2023;49:60–64. doi: 10.11857/j.issn.1674-5124.2021120105. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequences from the present study were submitted to the NCBI database (https://www.ncbi.nlm.nih.gov/, accessed on 16 May 2023) and the accession numbers were listed in Table S1.