SUMMARY
Poorly immunogenic small molecules pose challenges for the production of clinically efficacious vaccines and antibodies. To address this, we generate an immunization platform derived from the immunogenic surface coat of the African trypanosome. Through sortase-based conjugation of the target molecules to the variant surface glycoprotein (VSG) of the trypanosome surface coat, we develop VSG-immunogen array by sortase tagging (VAST). VAST elicits antigen-specific memory B cells and antibodies in a murine model after deploying the poorly immunogenic molecule fentanyl as a proof of concept. We also develop a single-cell RNA sequencing (RNA-seq)-based computational method that synergizes with VAST to specifically identify memory B cell-encoded antibodies. All computationally selected antibodies bind to fentanyl with picomolar affinity. Moreover, these antibodies protect mice from fentanyl effects after passive immunization, demonstrating the ability of these two coupled technologies to elicit therapeutic antibodies to challenging immunogens.
In brief
Triller et al. exploit the unique immunogenicity of Trypanosoma brucei surface coats to generate an antibody elicitation platform tailored to difficult targets including small molecules. They use this platform to elicit high-affinity monoclonal antibodies to the synthetic opioid fentanyl, which could be used as overdose-preventing prophylactics.
Graphical abstract
INTRODUCTION
The mammalian immune system struggles to generate antibody responses against small molecules unless they are fused to immunogenic carrier proteins to facilitate CD4+ T cell-dependent B cell activation.1 Classical hapten-carrier systems have been extensively tested in animals.2–4 More recently, the desire to elicit antibodies to small-molecule drugs of abuse has propelled the implementation of classical hapten-carrier systems into human trials with limited success and some prominent failures, mostly due to the high individual variability in polyclonal antibody responses.5,6 While additional opioid vaccines are currently being tested in humans (e.g., ClinicalTrials.gov: NCT04458545) or readied for clinical trials (e.g., 3UG3DA047711–02S1), recent clinical studies have demonstrated safety and efficacy of monoclonal antibodies (mAbs) against methamphetamine (ClinicalTrials.gov: NCT03336866 and NCT04715230), paving the way for this alternative strategy, provided that efficacious mAbs can be generated.
To accelerate mAb development, we developed an antibody elicitation platform that avoids the need for exogenous adjuvants. The system exploits the inherent immunogenicity of the blood-resident parasitic protozoan Trypanosoma brucei, which largely depends on the densely patterned array of variant surface glycoprotein (VSG) molecules that coat the surface of the organism (~10 million copies of a specific VSG per trypanosome, forming the overwhelming majority of the cell’s total surface protein7). This repetitive array facilitates epitope presentation to the immune system, driving both T-dependent and T-independent protective antibody responses.8,9 Furthermore, antibody responses to VSG arrays can be remarkably restricted in variable gene usage through an “epitope-focusing” effect that is potentially unique to this organism.10
VSG arrays elicit long-lasting responses: antibodies raised to a clonal VSG array protect the animal from infection with the same VSG-coated parasite for life.11 Counterintuitively, long-standing evolutionary pressures have likely selected for VSG coats that elicit strong, but highly VSG-specific, antibody responses. This is due to the remarkable system of antigenic variation utilized by this pathogen. Trypanosomes possess a large genomic cache of antigenically distinct VSGs. During infection, the pathogen produces successive populations of “switched” cells that express different VSGs on the surface. VSG proteins with unique and immunodominant epitopes generate antibody repertoires that are highly focused on a given VSG10 but that are unlikely to recognize switched cells. The host immune system thus invests considerable resources into a given VSG only to find itself naive to switched cells. Therefore, the VSG array has been selected precisely for the ability to generate robust serological responses.
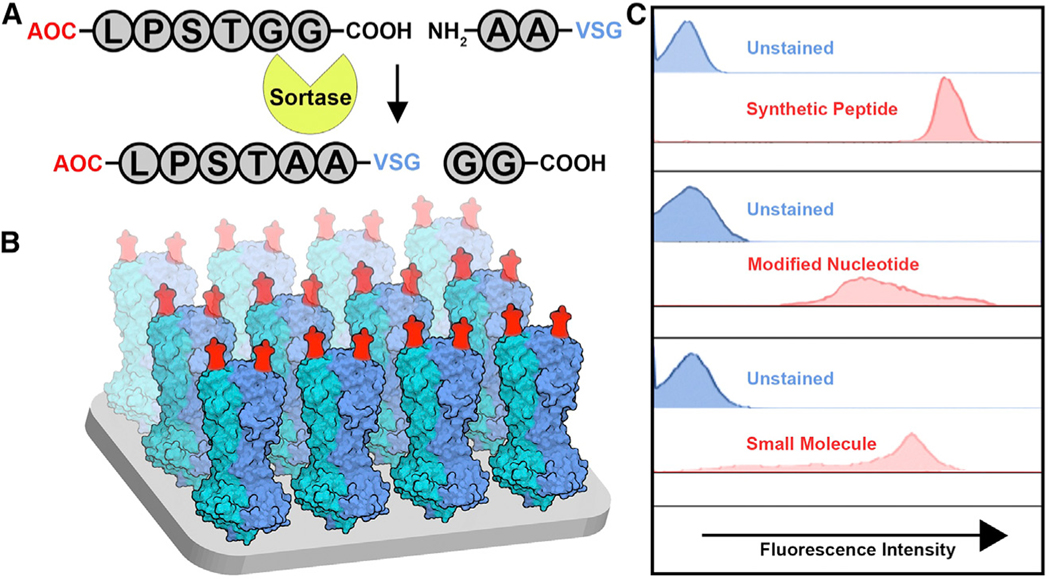
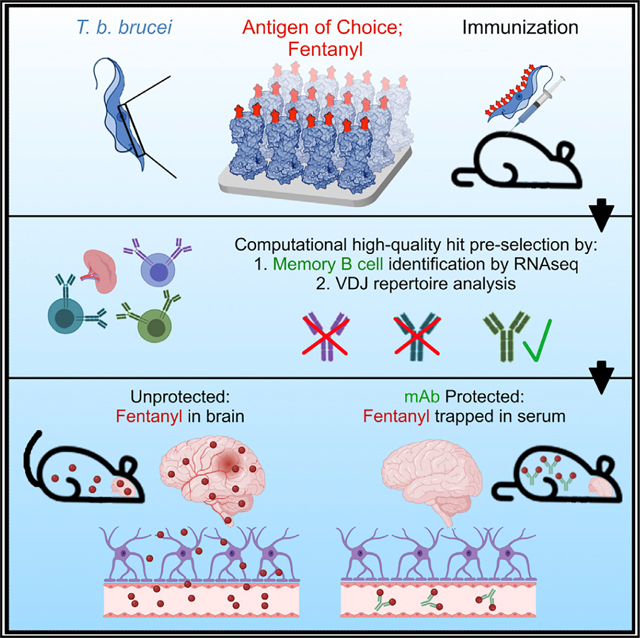
We initially exploited the exceptionally antigenic nature of trypanosomes by genetically engineering VSGs to incorporate short peptides (e.g., FLAG) into their solvent-exposed loops.12 To then broaden the applicability of the system to other types of antigens, we instead engineered VSGs to become substrates of the transpeptidation reaction known as “sortagging.” The term refers to the activity of the bacterial enzymes called sortases.13 By genetically engineering sortaggable VSGs14 and chemically synthesizing sortaggable antigens of interest (Figure 1A), the surface envelope of the trypanosome was effectively converted into a molecular display platform (Figure 1B). We can thus exhibit a wide variety of antigens against which antibodies are to be elicited (Figure 1C). We have named this platform VSG-immunogen array by sortase tagging (VAST).
Figure 1. The VSG-immunogen array by sortase tagging (VAST) platform: A trypanosome-based antigen display platform.
(A) An antigen of choice (AOC) is shown as a conjugate to the sortase recognition motif LPXTGG (X = S in this case). The sortase enzyme conjugates the AOC to modified VSG proteins containing an N-terminally extended AA motif.
(B) Model of the VAST platform. Red moieties represent the sortagged antigens. The blue and teal structures are the VSG homodimers (each monomer represented by one color).
(C) Flow cytometry plots revealing the results of sortagging experiments using live trypanosomes sortagged with AOCs representing the indicated molecule classes. Synthetic peptide: peptide derived from the SARS-CoV-2 spike. Modified nucleotide: inosine. Small molecule: 6-FAM.
VAST addresses the difficulties in generating antibodies against difficult targets; however, subsequent candidate antibody identification remains a notable hurdle. Indeed, the process of identifying high-quality mAbs from the sea of mixed-affinity splenocytes after immunization often invokes many months of large-scale screening. In some cases, thousands of candidate antibodies are functionally interrogated before the top hits are selected.15 We sought to address this financial and technical challenge by developing a computational strategy that could identify the best-quality B cells within a splenocyte pool. These high-quality B cells define the memory compartment, i.e., cells that have possibly undergone multiple rounds of affinity maturation during the immunization process. Unfortunately, these cells cannot yet be reliably identified based on flow cytometry-compatible surface markers due to the lack of suitable markers and staining panels in mice. However, memory B cell transcriptomes have been described,16–18 revealing the most reproducibly differentially regulated genes in the memory population. We reasoned that a transcriptomics approach to candidate anti-body identification would be suitable given that it would also simultaneously provide the raw data required for antibody sequence assembly and BCR repertoire analysis. The VAST system is therefore a combined suite of two separate components: a trypanosome-based antibody generation platform, and a computational antibody identification platform.
As a proof of concept, we sought to apply the VAST system toward the elicitation of antibodies to the small-molecule drug fentanyl, a clinically relevant model antigen. Fentanyl is a synthetic opioid 50- to 100-fold more potent than morphine. Although used clinically as a pain reliever and anesthetic, fentanyl is repeatedly mixed with recreationally used heroin or cocaine to mask impurities. This means that individuals are often unaware that they are self-administering the more potent fentanyl.19 Fentanyl is currently a leading cause of overdose and is the strongest driver of the increasing opioid-associated death rate. Death by opioid overdose was listed as the sixth largest cause of mortality in the United States in 2017, with figures having risen substantially during the COVID-19 pandemic and recently eclipsing the mark of 100,000 fatal overdoses per year (CDC, November 2021; https://www.cdc.gov/nchs/pressroom/nchs_press_releases/2021/20211117.htm).
The current development status of immunotherapeutics in the context of substance use disorder is reviewed here.20 Their development reflects both the acute need for relapse protection measures (the rate of relapse is >80% after exiting rehabilitation clinics21,22) and the need for an adjunct to treatment with naloxone, the opioid receptor antagonist that is the current standard of care for overdose reversal. To date, several vaccine formulations have shown efficacy against the pharmacological effects of fentanyl and its analogs.23–28 However, all these efforts have employed the classical adjuvanted hapten-carrier protein combinations that have yet to produce a clinically approved product. These strategies have employed monovalent prime-boost combinations (i.e., using the same hapten-carrier protein conjugate for each injection), bivalent combinations employing the co-administration of admixed individual conjugates,29 and bivalent homologous and heterologous prime-boost combinations28 to target individual or multiple opioid compounds simultaneously. These strategies have also been used to generate mAbs against a variety of target drugs of abuse including fentanyl.30 Notably, fentanyl-specific mAbs are effective in both preventing and reversing fentanyl’s pharmacological effects in rodent models, further highlighting the promise of immunotherapeutics in the opioid space.31–34 While promising, there remains a lack of clinical output in this space and a desperate need for additional intervention options.
RESULTS
The VAST platform induces antibody titers against fentanyl
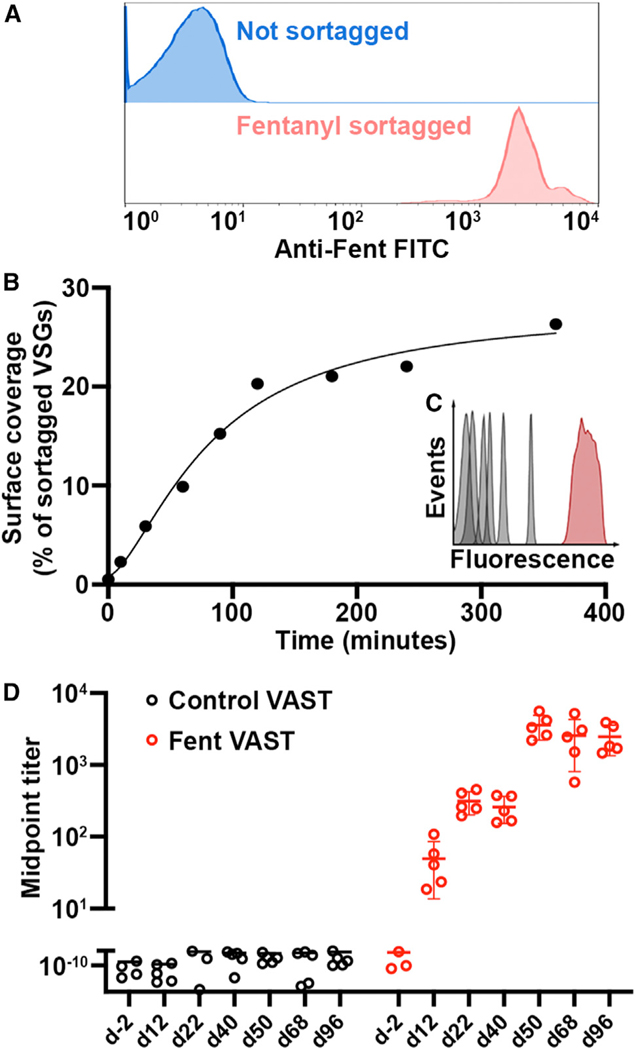
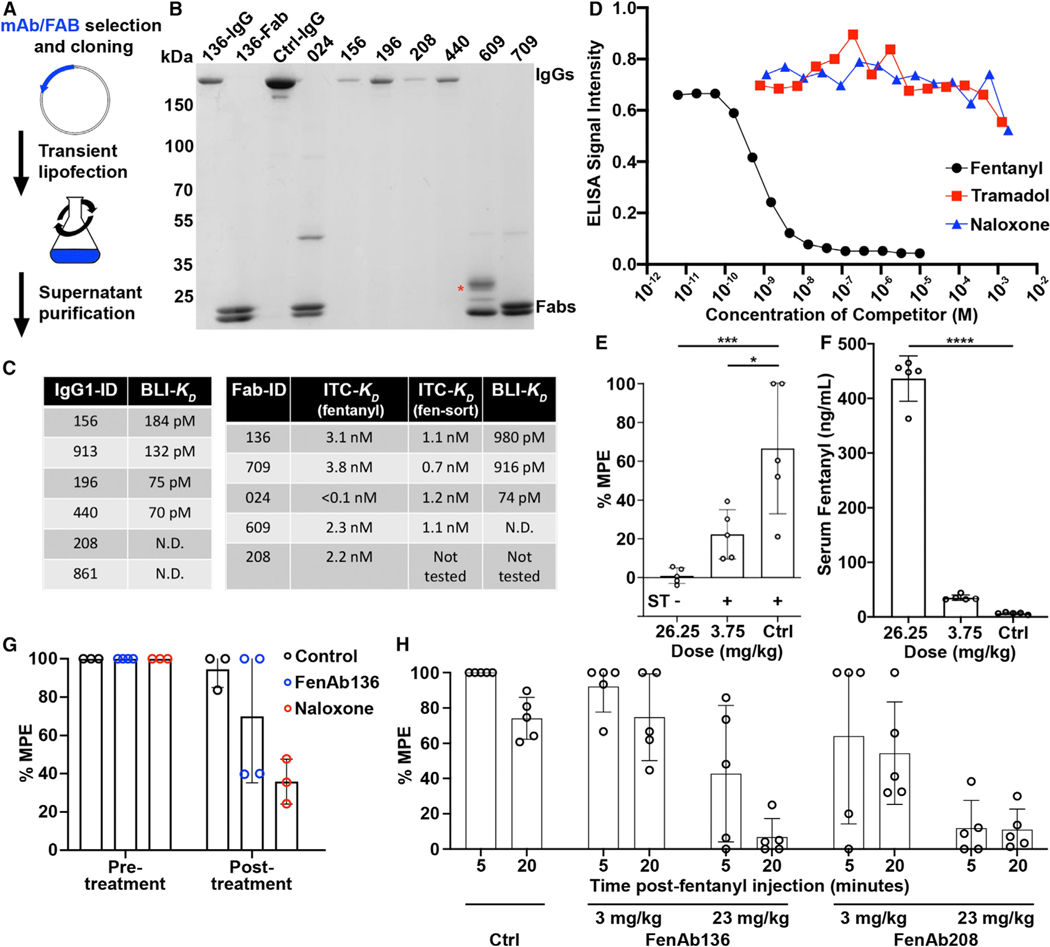
A sortaggable version of fentanyl (fen-sort) was produced using a modified synthesis of a fentanyl derivative previously described in Raleigh et al.26 and Robinson et al.27 (Figures S1A and S1B).26–28 After sortagging fen-sort to trypanosomes, 100% of the cells were decorated with fen-sort (Figure 2A) with VSG coverage per cell estimated at 20% (Figures 2B and 2C).
Figure 2. The VAST platform induces antibody titers against fentanyl.
(A) Representative (assay repeated >10 times) flow cytometry plot depicting a Fent-VAST sortagging reaction after anti-fentanyl fluorescent antibody staining.
(B) The graph depicts a sortagging reaction time course (non-linear curve fit R2 = 0.968). Cells were sortagged with FITC, and fluorescence was quantified at the indicated time points. The y axis quantifies the approximate percentage of the total VSGs sortagged per cell at each time point, as determined using the assay in (C).
(C) The flow cytometry histograms show the fluorescence of 6 bead populations (gray) coated with known copy numbers of FITC and the fluorescence of FITC-sortagged trypanosomes at the final time point in (B) (red). The most fluorescent bead is coated with 1e6 FITC molecules, which would be equivalent to approximately 10% of the total number of sortaggable sites on a trypanosome.
(D) ELISA measurements of serum IgG against fentanyl are shown. The mice immunized with control (not sortagged) VAST are shown in black dots, with Fent-VAST in red dots. Each dot represents antiserum from 1 mouse (n = 5 mice per immunogen; means ± SD are shown). In this particular experiment, the first boost was delivered at day 42. All titers below the scale break are equivalent to baseline. Some mice with baseline-level ELISA signal are excluded due to erroneous outliers that arise when calculating the midpoint titer.
See also Figures S1 and S2.
Immunizing mice with UV-crosslinked (inactivated) trypanosomes recapitulates the immune response launched by the live organism.14 Thus, mice were first primed with either fentanyl (Fent)-VAST (comprising the material in Figure 1B in UV-inactivated form) or control-VAST (UV-inactivated but not sortagged). However, compelling evidence suggests that while B cells normally encounter antigens in a membrane-bound format during initial activation,35 memory recall after initial activation can be quickly stimulated by soluble antigens since smaller entities are more easily able to penetrate B cell follicles.36,37 Therefore, we designed a two-step injection paradigm where priming was followed by boosting with a 10- to 20-fold higher dose of antigen-conjugated VSG in a soluble format (i.e., after cleavage from the membrane and biochemical purification of the VSG protein; Figures S2A–S2C). This led to the generation of high antibody titers against fentanyl in the mice immunized with Fent-VAST that matured with each injection (Figure 2D). Injection of membrane-bound Fent-VAST alone led to the generation of only modest anti-fentanyl titers (Figure 2D, days 12–40), while injecting soluble-Fent-VAST in the absence of a priming step did not elicit high fentanyl titers (Figure S2D). However, the combination of both steps resulted in the recalling of antigen-specific memory B cells that were produced after priming, where recall is represented by the marked increase in titers after the first boost administered on day 42 (Figure 2D). This ability to recall with soluble-Fent-VAST was maintained until at least 4 months post-priming (Figure S2E), confirming that long-term immunological memory is established and then recalled by the VAST platform. Recalled memory cells will have most likely undergone affinity maturation by the time that they differentiate into antibody-producing plasma cells. It is therefore reasonable to hypothesize that the great majority (>90% given the logarithmic titer jump from day 40 to 50, Figure 2D) of circulating antibodies post-boost are affinity-matured memory-derived antibodies. We compared the ability of VAST to elicit such a high proportion of high-quality antibodies with that of a conventional globular carrier, keyhole limpet hemocyanin (KLH), adjuvanted with aluminum salts. Many adjuvanted globular carriers elicit tremendous priming responses upon initial injection, which we also observed after immunizing mice with Fent-KLH (Figure S2F). However, subsequent boost injections triggered relatively weak increases in titer, while VAST triggers a noticeable boost-dependent titer jump, indicative of memory recall (Figure S2F).
Anti-fentanyl titers protect from fentanyl effects
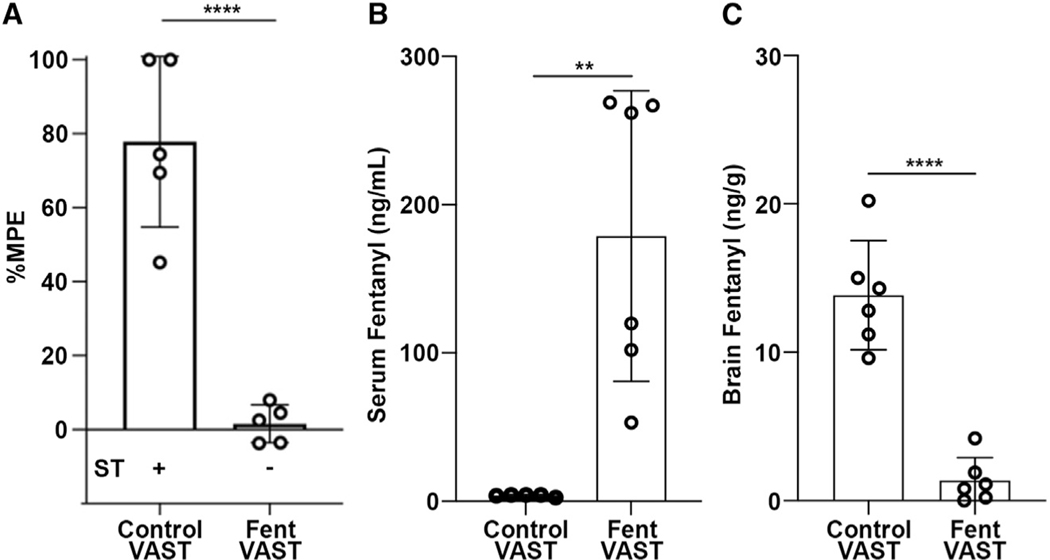
Next, we tested whether the strong antibody response induced by Fent-VAST immunization could protect mice from fentanyl. The pharmacologic activity of 100 mg/kg fentanyl was assessed using the hot plate assay of nociception and the Straub tail reaction test (assays described in Figures S3A–S3C) 10 days after the final boost (vaccination schedule in Figure S2C). Fent-VAST immunization reproducibly ablated any detectable fentanyl-induced antinociceptive effects via the hot plate assay and prevented the typical Straub tail reaction (Figures 3A and S3C; Video S1). We then injected naloxone immediately after the behavioral assessments (Figures S3A and S3B). Fentanyl-induced antinociception was reversed in all mice (Figure S3B), indicating that the antibodies do not interfere with rescue of overdose via naloxone treatment.
Figure 3. Anti-fentanyl titers protect from fentanyl pharmacological effects.
(A) Analgesic activity at day 100 was tested by using the hot plate antinociception assay as described by Cox and Weinstock.38 Mice were dosed with a cumulative dose of 100 μg/kg fentanyl. The percentage maximum possible effect (%MPE) is shown. A positive (+) or negative (−) reaction in the Straub tail (ST) test is indicated below and representative of the whole group. Means ± standard deviation of 5 mice per group are shown. ****p < 0.0001 by unpaired t test.
(B-C) LC-MS analysis of the distribution of fentanyl in the sera (B) and brains (C) of the mice in (A) is shown. Means ± standard deviation of 6 mice per group are shown. ****p < 0.001, **p < 0.01 by unpaired t test.
See also Figure S3.
Antibodies are hypothesized to protect against fentanyl effects by binding the drug in the bloodstream, decreasing circulating free drug, and thereby preventing fentanyl from entering a multitude of tissues,28 including the central nervous system. We thus measured the fentanyl concentration in the brain and serum of mice after fentanyl challenge by liquid chromatography-mass spectrometry (LC-MS). While control mice exhibited levels of 10– 20 ng/mL fentanyl in the brain and less than 5 ng/mL in the serum, the Fent-VAST-vaccinated mice exhibited less than 5 ng/mL in the brain and 80–280 ng/mL fentanyl in the serum (Figures 3B and 3C). This demonstrates that the Fent-VAST-induced antibodies could trap the fentanyl in the serum of immunized mice, preventing the drug’s blood-brain-barrier penetration.
The VAST platform elicits six different B cell subsets
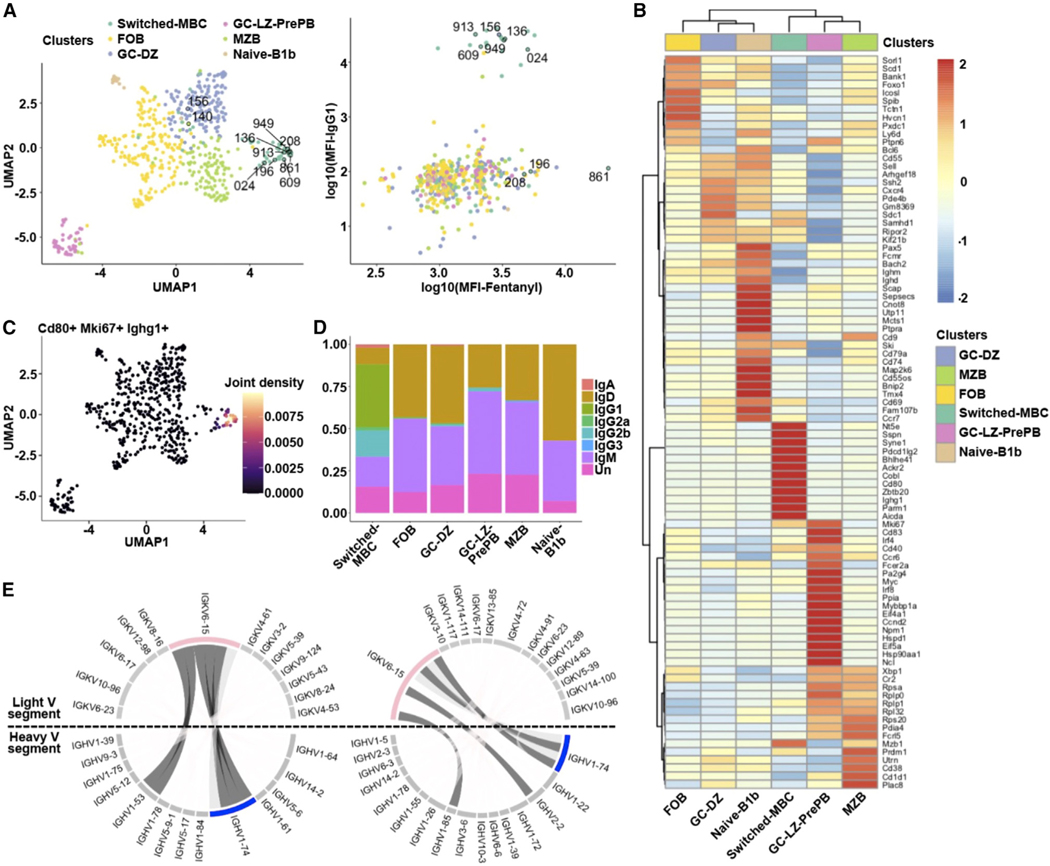
We sought to assess whether the platform establishes bona fide memory B cells and to sequence their encoded antibodies given that antigen-primed memory B cell receptors (BCRs) are likely to be of high affinity. To this end, we used single-cell RNA sequencing (RNA-seq) to precisely identify the different B cell subsets induced by the VAST platform. We fluorescence-activated cell sorted (FACS) fentanyl-binding B cells from two mice immunized with Fent-VAST (the gating strategy and baiting results are shown in Figures S4A and S4B) and performed singlecell RNA-seq. We identified six distinct B cell subpopulations among the sorted fentanyl-specific B cells (Figure 4A). The subpopulations were annotated based on the expression levels of the top 10 most differentially expressed genetic markers for each subcluster (Figure 4B), among them Myc, Cd55, Cd21 (or Cr2), Cd80, and Fcrl5.39–44 The subpopulations included a germinal center light-zone pre-plasma (GC-LZ-pre-plasma) population; a GC dark-zone (GC-DZ) population; a follicular B cell (FOB) population, a marginal zone-like atypical memory B cell (MZB) population, and a naive B1b cell population.45–48 Importantly, there was also a recognizable CD40-expressing isotype-switched memory B cell (switched-MBC) population identified with this analysis. These data further support earlier preclinical evidence that opioid-carrier conjugate vaccines induce CD4+ T cell-dependent B cell processes to elicit opioid-specific immunoglobulin G (IgG) antibodies, including GC formation as assessed by antigen-specific GC B cells, T follicular helper (Tfh) cells, GC-Tfh cells, and subsequent switched-MBCs.49–52
Figure 4. The VAST platform elicits six different B cell subsets.
(A) Uniform manifold approximation and projection (UMAP) visualization of the B cell subclusters and scatterplot of the log10(mean fluorescence intensity) (MFI) for the fentanyl binding vs. the IgG1 surface expression is shown. The color code indicates the different subclusters. The numerically labeled cells correspond to the B cells from which the BCRs were cloned to produce Fabs and full-length antibodies.
(B) Heatmap visualizing the top 10 statistically significantly upregulated genes based on the MAST statistical framework for each subcluster along with several known B cell markers. Genes with an adjusted p value <0.05 have been identified as markers. The columns correspond to the different subclusters (the same color code used in A) and the rows to the average gene expression for the selected genes. Red indicates relatively high gene expression, while blue indicates low expression.
(C) UMAP visualization of the joint expression of the Cd80, Mki67, and Ighg1 genes. Beige indicates a relatively high simultaneous expression pattern, while purple indicates low simultaneous expression.
(D) Barplot depicts the heavy-chain isotype distribution for each B cell subpopulation. There are cells (labeled as Un [unknown]) for which we were unable to identify the isotype via BLAST.
(E) Circos plots of the switched memory B cell subpopulation for each of two mice are shown. The expanded heavy (in blue, IGHV1–74) and light (in pink, IGKV6–15) variable chain genes are highlighted. Dark gray: BCRs that were selected for cloning.
See also Figure S4.
Pairing each cell’s transcriptomically identified subtype with data from FACS index sorting, we observed that the cells with the highest affinity to fentanyl were from the memory subpopulation (Figure 4A, right panel), which was also enriched for Cd80 and Cd73, two T-dependent MBC markers that are absent from the other subpopulations.53 The Mki67 proliferation marker was also upregulated in the switched-MBC and pre-plasma populations, suggesting the reactivation of MBCs immediately after the boost administered 10 days prior to cell collection, which is consistent with the known kinetics of the recall response (Figures 4C and S4C).54 Additionally, the subpopulation expresses longevity markers such as Zbdb20, traditionally expressed in long-lived plasma cells.55,56 The unswitched populations produced BCRs mainly of the IgM isotype, while the memory population predominantly expressed IgG1 and IgG2b (Figure 4D).
To further characterize these BCRs from the switched-MBCs, we performed BCR repertoire analysis based on VDJ usage of the heavy and light chains of the BCRs. We found that many of the B cells from the switched-MBC subpopulation showed a similar VDJ profile for paired heavy and light chains (Figure 4E). Heavy-and light-chain pairing was dominated by the IGHV1–74/IGHJ4 or IGHV1–53/IGHJ2 genes in combination with IGKV6–15/IGKJ2 (Figure 4E), pairs that are not at all represented within the cumulative repertoire of the five non-memory subsets (Figure S4D). Notably, these pairs are also not overrepresented in naive mouse BCR repertoires or repertoires from mice infected with live T. brucei.10 In addition, some BCRs had similar or even identical heavy- and light-chain CDR3 regions and shared several somatic hypermutations, suggesting that the cells were clonally related (Table 1).
Table 1.
Monoclonal anti-fentanyl antibodies cloned from BCRs of B cells (MBC ID) belonging to the switched memory B cell population
| Mouse | MBC ID | Heavy V chain | Heavy D chain | Heavy J chain | Heavy CDR3 aa | Heavy constant | Heavy SHM | Light V chain | Light J chain | Light CDR3 aa | Light constant | Light SHM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 196 | IGHV1–53 | IGHD1–1 | IGHJ2 | AIEVGYYDY | Ighg2b | 6 | IGKV6–15 | IGKJ2 | EQYNS YPYT | Igkc | 3 |
| 1 | 208 | IGHV1–53 | IGHD1–1 | IGHJ2 | AIEVGYYDY | Ighg2b | 6 | IGKV6–15 | IGKJ2 | EQYNS YPYT | Igkc | 3 |
| 1 | 24 | IGHV1–74 | IGHD2–3 | IGHJ4 | AIEIYDGYY AMDY | Ighg1 | 2 | IGKV6–15 | IGKJ5 | QQYNS YPLT | Igkc | 1 |
| 1 | 136 | IGHV1–74 | IGHD2–3 | IGHJ4 | AIEIYDGYN TMDY | Ighg1 | 2 | IGKV6–15 | IGKJ5 | QQYNN YPLT | Igkc | 1 |
| 1 | 156 | IGHV1–75 | IGHD1–1 | IGHJ2 YFDY | ARRDYGSS | Ighg1 | 0 | IGKV1–99 | IGKJ4 | FQSN YLPLT | Igkc | 0 |
| 6 | 609 | IGHV1–74 | IGHD1–1 | IGHJ2 | AMEDYYGS SYEDY | Ighg1 | 3 | IGKV6–15 | IGKJ2 | QQYNT YPYT | Igkc | 1 |
| 6 | 949 | IGHV1–74 | IGHD1–1 | IGHJ2 | AIERDYYGS REDY | Ighg1 | 0 | IGKV6–15 | IGKJ2 | QQYNS YPYT | Igkc | 2 |
| 6 | 913 | IGHV1–85 | IGHD1–1 | IGHJ1 | AIEGFTTVV ARNFDV | Ighg1 | 4 | IGKV6–15 | IGKJ2 | QQYNT YPYT | Igkc | 3 |
| 6 | 861 | IGHV2–2 | IGHD1–3 | IGHJ2 | ATEVGYFDY | Ighg2b | 2 | IGKV6–15 | IGKJ2 | QQYNS YPYT | Igkc | 1 |
The gene usage of the V(D)J combination is indicated, along with the CDR3 amino acid sequence, and the constant region for each heavy and light chain combination. In the “SHM” column, the number of silent and missense mutations detected along the full-length of the heavy or light chain is indicated.
Fentanyl-specific antibodies are of high affinity and specificity and protect mice from fentanyl effects
We cloned and expressed the selected BCRs as recombinant full-length mAbs or Fabs and purified them for functional characterization (Figures 5A and 5B). Affinity and thermodynamic characteristics of purified Fabs binding to fentanyl and fen-sort were first analyzed using isothermal titration calorimetry (ITC). All fentanyl-binding reactions were exothermic with nanomolar and subnanomolar ranges of affinities (Figure 5C). Large enthalpy changes (ΔH ~ –68–100 kJ/mol) attenuated by unfavorable entropy inputs (–TΔS = 13–47 kJ/mol) contributed to binding energies DG of approximately –50–60 kJ/mol. The binding reactions of fen-sort were similarly enthalpy driven (ΔH ~ –132– 156 kJ/mol, –TDS~ 80–105 kJ/mol) and resulted in 0.7–1 nM KDs. The subnanomolar affinities fell below the optimal measurement range for ITC, and indeed the raw data suggested that the actual affinities may be higher than what had been calculated. We therefore used biolayer interferometry (BLI), a technique with a wider dynamic range. These affinities were in the picomolar range, spanning from 980 pm to less than the measurable limit of the experimental apparatus (Figure 5C). These limitations were often driven by the exceptionally long Koff rates of the antibodies, especially for the bivalent IgGs.
Figure 5. Fentanyl-specific antibodies are of high affinity and specificity and protect mice from fentanyl pharmacological effects.
(A) Schematic of the expression system used to produce recombinant antibodies.
(B) Coomassie-stained non-reducing SDS-PAGE after affinity purification of a panel of the expressed IgGs (running near 150 kDa) and Fabs (running as separate heavy and light chains near 25 kDa). The red asterisk marks a band produced by a glycosylated Fab, while the remaining Fabs are not glycosylated. The first lane marked “136” is the full-length IgG version of this antibody, while the second lane is the Fab. 156, 196, 208, and 440 are IgG protein samples, while 024, 609, and 709 are Fab protein samples. Ctrl-IgG is a control IgG sample as an additional molecular weight reference.
(C) Binding affinities determined for each antibody using both ITC and BLI. Antibodies whose affinity were too high to measure are here reported as “not determinable” (N.D.).
(D) Competition ELISA data generated with haptenated fentanyl-coated plates. The indicated soluble competitors are serially diluted into the assay to determine the relative efficiency of cross-binding to other opioid molecules, with soluble fentanyl serving as a positive control.
(E) Behavioral protection from fentanyl after prophylactic passive immunization with FenAb136. A positive (+) or negative (−) reaction in the ST test is indicated below and representative of the whole group. Means ± SD of 5 mice per group are shown. *p < 0.05, ***p < 0.005 by Dunnett’s multiple comparisons test.
(F) Sera from the mice in (E) were collected and analyzed for fentanyl content by mass spectrometry. Means ± SD of 5 mice per group are shown. ****p < 0.001 by Dunnett’s multiple comparisons test.
(G) Behavioral protection from fentanyl after therapeutic (rescue) passive immunization with FenAb136. Hot plate assay recordings were made at T = 15 and 30 min post-fentanyl injection. Means ± SD of >3 mice per group are shown.
(H) Behavioral protection from fentanyl after prophylactic passive immunization with FenAb136 or FenAb208. Means ± SD of 5 mice per group are shown.
See also Figure S5.
Further characterization by competition ELISA confirmed that these antibodies were fentanyl specific and, importantly, did not interact with naloxone (Figure 5D), consistent with in vivo data (Figure S3B). Every individual mAb cloned and expressed after selection by this MBC-focused computational selection method has displayed the above properties (picomolar affinity and fentanyl class binding specificity).
To investigate if the MBCs contributed to protection, we first selected FenAb136 to determine if the antibody was able to directly protect mice from fentanyl challenge. Mice were passively immunized by intraperitoneal injection with two different concentrations of the full-length antibody 24 h prior to fentanyl challenge. Although only 37% (higher dose) and 25% (lower dose) of the originally injected amount of FenAb136 could be detected in the blood at the time of challenge (Figure S5A), mice were protected from fentanyl effects in a dose-dependent manner (Figure 5E). LC-MS revealed that fentanyl was trapped in the serum of the antibody-administered mice as expected (Figure 5F). In fact, we detected equimolar serum-fentanyl and serum-FenAb136 levels (approximately 1.2 μM for the high-dose group and 0.1 mM for the low-dose group; Figure S5B).
We also investigated if FenAB136 could be used as a therapeutic post-overdose, where fentanyl was administered prior to a mAb (experimental design in Figure S5C). All groups responded rapidly to the injection of fentanyl, while mice subsequently treated with naloxone then returned to baseline (Figure 5G). The antibody-injected group also trended toward baseline at T = 15 min, while the control group continued to experience fentanyl effects. As an additional assessment of fentanyl effect inhibition, we employed the laboratory animal behavior observation registration and analysis system (LABORAS), an automated approach that longitudinally records mouse movement and a number of additional movement-associated parameters. LABORAS data revealed that mice experiencing fentanyl effects moved in a noticeably circular fashion around the periphery of the cage (Figure S5D). Naloxone-administered mice appeared to exhibit normal behavior, while the antibody-injected mice trended toward normal behavior more rapidly than the control group. Quantifiable measurements of total recorded travel distance supported antibody efficacy (Figure S5E).
We next interrogated the role that antibody affinity plays in fentanyl protection. We compared the prophylactic protective capacity of FenAb136 (our lowest-affinity antibody) with that of FenAb208 (one of the highest-affinity antibodies). In the hot plate assay, the lower-affinity FenAb136 was indistinguishable from control at the low dose of 3 mg/kg, while some protection was afforded by the higher-affinity FenAb208 (Figure 5H). A similar trend was observed with the high dose, where FenAb208 kept all mice at baseline even immediately after fentanyl injection (the 5 min time point), while FenAb136 was only partially protective (Figure 5H). LABORAS data revealed that control animals continuously run in a circular fashion around the periphery of their cage post-fentanyl, while only the FenAb208 groups and the high dose of FenAb136 group displayed more normal mouse behavior (Figure S5F).
VAST-elicited antibodies bind fentanyl in a deep, enveloping pocket
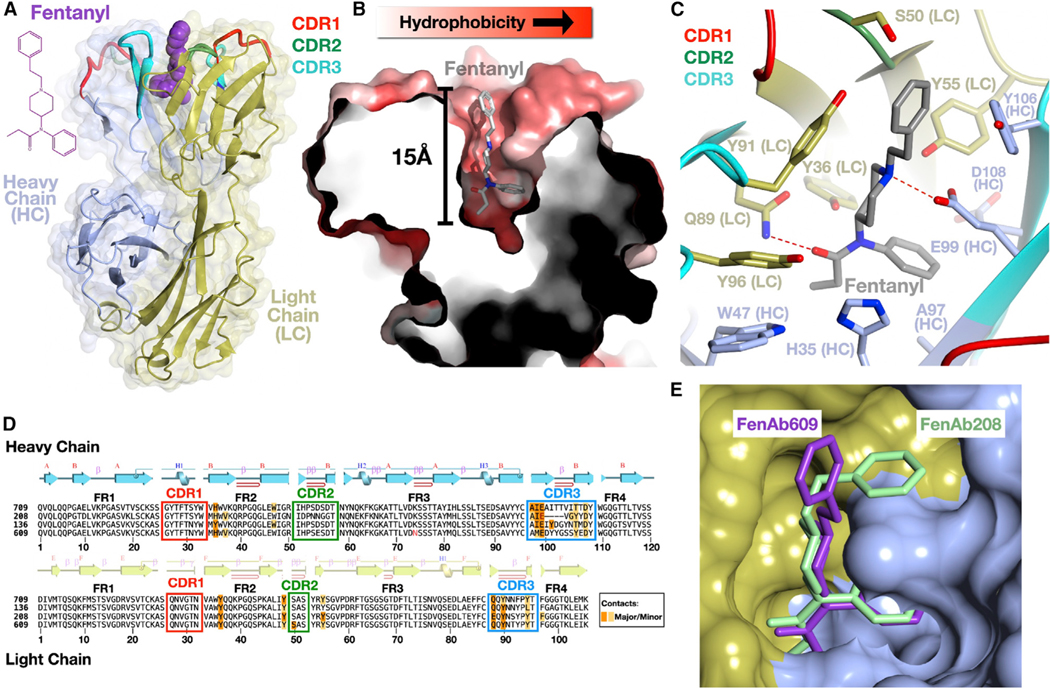
We co-purified drug:Fab complexes (two with fentanyl and two with fen-G4) and determined four high-resolution structures by X-ray crystallography (Figures 6 and S6; Table S1). All the antibodies share a similar binding mode consisting of a deep, invaginated pocket formed by residues from both the heavy and the light chains of the immunoglobulin. While the upper portions of the pocket consist of CDR regions of the chains, the pocket is so deep (approximately 15 Å from the upper portions to the lower regions) that the bottom of the pocket is lined with residues from the beta-sheet framework regions, with several amino acids from these segments making contact with the drug.
Figure 6. VAST-elicited antibodies bind fentanyl in a deep, enveloping pocket.
(A) Overall structure of a complex of a Fab (FenAb609, heavy chain colored light blue, light chain colored gold) with fentanyl (purple, space filling depiction) shown as a ribbon diagram with the two-dimensional chemical structure of fentanyl on the left.
(B) Illustration of the fentanyl binding pocket as a thin slice through the molecular surface of the protein (colored in a gradient from white to red to reflect increasing hydrophobicity of the surface) colored using the method of Eisenberg.57 Fentanyl is shown as a stick model with atoms of carbon, nitrogen, and oxygens in gray, blue, and red, respectively.
(C) Contacts of the protein to fentanyl shown with side chains as stick models and the mainchain as a ribbon diagram. “HC” denotes heavy chain, and “LC” denotes the light chain. Hydrogen bonds are shown as dashed red lines between bonded atoms.
(D) Sequences of four fentanyl-binding Fab molecules where the alignment was generated by superimposing the crystal structures. The secondary structure of a representative Fab (FenAb609) is shown above the sequence, colored as per (A). Disulfide bonds are shown as lines connecting cysteine residues. Major and minor contacts are indicated in orange and yellow, respectively. The framework regions are denoted as FR1, −2, −3, and −4.
(E) Superposition of FenAb609 and FenAb208 crystal structures with the molecular surface of FenAb208 shown (heavy and light chain regions colored as in A) along with the stick figures of fentanyl in both molecules.
See also Figure S6.
Fentanyl (N-(1-(2-phenylethyl)-4-piperidinyl)-N-phenyl-propanamide) inserts deeply into the cavity in a mostly elongated conformation (Figure 6). Projecting only slightly out from the molecular surface of the Fab is the phenylethyl ring (the group absent in the fen-sort construct used to immunize mice; Figures S1A vs. S1B). This ring is mostly buried (Figure 6B), and the remaining chemical structure plunges straight into the pocket so that the piperidine ring is fitted tightly about halfway into the cavity, locked in place with numerous hydrophobic van der Waals contacts and a hydrogen bond (Figure 6C). The drug descends further into the pocket bottom, where the two groups of the N-phenyl-propanamide group each plug into a snug series of contacts. A cavity is carved into the surface at the pocket bottom, into which the benzene ring inserts, while the propionyl group projects in the opposite direction and is held tightly by numerous contacts including a deeply buried hydrogen bond. Several contacts from framework residues occur in this region at the bottom of the pocket (Figures 6C and 6D). The network of interactions observed in the structures support the ITC data, which suggest that the net formation of hydrogen bonds between fentanyl moiety and Fab is a major driving force of binding and antigen specificity. Interestingly, the binding of a ligand (e.g., testosterone, steroids, musk odorant) to a deep hydrophobic pocket in a Fab is often hypothesized to be regulated by shape complementarity and the concomitant desolvation of the complementary surfaces with a subsequent rise in system entropy.58 In the case of the fentanyl-specific antibodies described here, the entropy change was unfavorable, indicating that conformational rearrangements of Fab loops were likely required to form the pocket around fentanyl. Unbound Fab should exhibit higher degrees of freedom, which are lost upon ligand binding, leading to an entropy drop. This hypothesis correlates well with the observation from crystallization trials that the unliganded Fab could not be crystalized.
As noted, the pocket structure is common to all the antibodies identified, and while many contacts are conserved between the different Fabs, there are differences that could contribute to the range of affinities measured. In addition, the “top” aromatic group of fentanyl is seen in FenAb208 to adopt a different conformation than in some of the other fentanyl-bound antibodies (Figure 6E). In particular, the CDR3 loop of the heavy chain in FenAb208 is significantly shorter and adopts a very different conformation, which creates a small socket not present in the other structures into which the phenylethyl ring inserts. Because this ring is absent in fen-sort, it demonstrates that the antibody repertoire is able to produce a collection of different immune+globulins that can engage this chemical group effectively, even without any pressure to select such variants.
DISCUSSION
Many obstacles impede the production of specific and high-affinity mAbs against several different categories of antigens, one being small molecules. VAST can serve as a mAb-producing tool, particularly in contexts where more conventional methods have failed. VAST does not require the addition of adjuvants, in contrast to most standard immunization methods. Thus, VAST serves as a “self-adjuvanting” array that harnesses the natural immunogenicity of the trypanosome coat to elicit an immune reaction against even poorly immunogenic antigens, which is hypothetically powered primarily by the high density of the displayed antigen. In this study, only 20% surface coverage was sufficient to elicit picomolar-affinity antibodies. More recently, we have observed that VAST can be sortagged with at least up to 75% coverage, although it remains unclear if coverage values above 20% actually provide any marked improvement in anti-body elicitation.
Small molecules that are rapidly purged from the body may evade B cell detection for temporal reasons, while the molecules themselves are also unlikely to be presented on the major histocompatibility complex (MHC) or act as T cell epitopes. However, the use of chronic opioids has been reported to elicit low levels of anti-opioid antibodies,59 with little known about the underlying mechanisms permitting antibody elicitation. Thus, a core concept in the conventional design of small-molecule immunizations is to conjugate the small molecule hapten to a larger immunogenic carrier that drives a CD4+ T cell response in order to sustain B cell activation. In fact, depletion of CD4+ T cells, use of TCR knockout (KO) mice, and T cell-independent carriers such as ficoll and dextran49,50 show that T cell help is required for the generation of antibody responses in the context of opioid immunizations. However, decades’ worth of literature show that trypanosomes drive a strong T-independent antibody response,8,9 and so we cannot rule out the roles that T-independent pathways may have in the immune response to VAST-carried haptens. Generally, we hypothesize that immune stimulation by VAST is distinct from that which is driven by conventional protein carriers. For example, the VAST platform priming steps use an enormous antigenic carrier. The inherent antigenicity of the trypanosome notwithstanding, the size of the organism is substantially larger compared with that of smaller carriers like KLH (an entire eukaryote vs. a large protein). Classically, material of that size is likely to be cleared through phagocytosis, while we hypothesize that smaller carriers are unlikely to stimulate that type of clearance mechanism. The different mechanism by which antigens are recognized, cleared, and processed is likely to lead to noticeably different immune stimulating outcomes (e.g., different types of cytokine and chemokine responses). These differences may facilitate the strong responses driven by VAST that we have observed in the absence of adjuvants. Further, indeed, the VSG proteins (and the rest of the organism) must contain a plethora of immunostimulatory T cell epitopes. Some VSGs may be slightly more or less immunostimulatory than others (which has not yet been carefully explored to our knowledge), although we hypothesize that any VSG with a surface-accessible N terminus for sortagging will be able to stimulate antigen-specific antibody responses.
Importantly, we show that the VAST platform can induce MBCs, which is a crucial quality checkpoint for successful immunizations. Previous studies have shown that conjugate vaccines elicit opioid-specific B cell population subsets, including switched-MBCs,49 and that B cell formation is dependent upon GC formation and involvement of cognate CD4+ T cells.2,49 Furthermore, vaccination could boost anti-bodies well beyond the disappearance of the first antibody response.60 Here, VAST-elicited MBCs are recalled in mice through at least 16 weeks after the second prime injection, which renders this platform flexible for different immunization regimens and experimental setups.
The single-cell RNA-seq technique used here is not only able to identify the MBC population of interest but also five additional B cell subsets based on subset-specific markers, implying both T cell-dependent and T cell-independent formation. More importantly, it samples deeply enough to pick up several highly similar MBCs based on the usage of the same IGHV and IGKV combinations that appear reproducibly even between mice. In combination with the high expression levels of the Mki67 proliferation marker, this suggests that clonal expansion of the switched-MBC population had occurred. Additionally, we were able to detect two MBCs with nearly identical BCR sequences except for a couple of point mutations (FenAb196 and FenAb208), which also share some somatic hypermutations compared with the germline sequence, strongly suggesting that these cells originated from the same progenitor cell, which is another line of evidence suggesting that the VAST platform is indeed able to induce true immunological memory. Memory cell establishment and memory cell identification are thus both critical components to successful hit identification. We therefore hypothesize that the VAST immunogen and the computational selection method both contribute to the hit identification rate reported here. The VAST immunogen elicits notably boost-responsive titer jumps (Figure 2D), revealing that the overwhelming majority of the titers present at the time of splenocyte harvest are produced by recalled/expanded memory cells. A key advantage associated with the VAST system, therefore, is the ease with which memory cells can be selected through the combined effects of the overall abundance of expanded memory cells relative to naive and through their specific identification by RNA-seq.
Although anti-fentanyl mAbs have been described to effectively protect from fentanyl challenge before,31–33 the modality of fentanyl binding of these antibodies has only recently been subjected to investigation. Ban et al.32 reported molecular modeling results indicating the presence of a deep pocket in one of two mAbs elicited by immunization with a similar hapten, although these results were generated in silico, while here we report crystal structures. Notably, additional crystal structures of fentanyl-bound mAbs were published during this article’s revision process.61 The crystals reveal important differences related to the residues involved in ligand binding, the overall topology of the antibody-ligand interaction (we observe an approximately 180° inversion in docking mode relative to the published models), and the conformational changes involved in ligand binding (identified through the combination of the crystallography and ITC-based thermodynamics). We hypothesize that this conformational change that “traps” the fentanyl is key to high affinity and subsequent protection. The trapping effect correlates nicely with the slow Koff that we observed by BLI. Without a deep pocket trapping the drug, it is possible that the drug may be bound in a less stable fashion and thus more readily leech back out into circulation.
The deep pocket is also most likely responsible for the high specificity of the antibodies, as only very similar small and elongated structures would be able to fit. Antibodies elicited by this immunization scheme show no evidence of being “pan-opioid” and thereby remain promising preclinical candidates. For instance, antibodies previously raised against the fentanyl hapten used in our studies showed selectivity for fentanyl and the closely related sufentanil and acetylfentanyl but not methadone, buprenorphine, naloxone, naltrexone, and other critical care medications such as anesthetics.27 This specificity also has positive implications for eventual human use from a safety perspective, as we hypothesize that mAbs whose binding is dependent on this structural motif would be less likely to non-specifically interact with human proteins and lead to toxicity at a variety of mAb dosages. It remains difficult to predict mAb doses required for protection in the field, however, since the fentanyl doses will vary from case to case. A therapeutic of this nature, such as FenAb208 delivered routinely as a prophylactic, would thereby need to be combined with additional existing substance use disorder mitigation strategies. We maintain that immunotherapeutics have an important role to play in the future of this field as an adduct to all options currently in development. Nevertheless, there may remain concerns that the bound drug could have a biologically negative impact in vivo over time, i.e., through prolonged neural adaptation driven by a slow leeching effect from the antibody if the drug-bound mAb remains in circulation. Longitudinal studies will need to be performed in the future in order to identify the mechanism by which antibody-trapped fentanyl is ultimately cleared from the system and whether any long-term neural toxicity can be observed.
In conclusion, the VAST is a platform that elicits and identifies high-quality mAbs for direct therapeutic or research applications. This epitope-focused platform may serve as a key to help unlock certain fields of therapeutic antibody development that have yet to be successfully explored. Future studies will address the hypothesis that the VAST platform can elicit high-affinity antibodies for a wide variety of different antigen classes, many of which are already underway (Figure 1C).
Limitations of the study
The antibodies elicited in this study have murine variable domains and will thus need to eventually be humanized/de-immunized by any one of various technology platforms prior to human use. There is a risk associated with this process; humanization may result in a decrease in antibody efficacy, likely through a drop in binding affinity. Ideally, we would avoid this limitation in the future by using genetically modified mice with humanized antibody systems during the immunization process. However, we have yet to validate the effectiveness of the VAST platform in these humanized mice, which remains a key future direction.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Dr. Joseph P. Verdi (joey.verdi@hepionetx.com or j.verdi@dkfz-heidelberg.de).
Materials availability
All newly generated materials produced by these studies are available for distribution under certain conditions. The VAST platform, and all key reagents associated with it or derived from its use in animals, are licensed for commercial purposes to Panosome GmbH and/or Hepione Therapeutics. These companies must authorize any material distributions to third parties and be privy to the associated material transfer agreements established therein.
Data and code availability
Crystal structure files have been deposited at the Protein DataBank and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Single-cell RNA-seq data have been deposited at GEO. To request access, contact lead contact. Original western blot images reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE Antibodies | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| mouse anti-fentanyl monoclonal antibody | M. Pravetoni, University of Washington | N/A |
| mouse anti-fentanyl-FITC monoclonal antibody | this paper | N/A |
| mouse anti-VSG3 monoclonal antibody | Pingeretal., 201714 | N/A |
| goat anti-mouse IgG-HRP | Jackson Immuno Research | Cat#115-035-062; RRID: AB_2338504 |
| streptavidin-BV785 | BioLegend | Cat#405249 |
| rat anti-mouse CD19-BV412 | BioLegend | Cat#115537; RRID: AB_10895761 |
| rat anti-mouse IgG1-BV650 | BioLegend | Cat#406629; RRID: AB_2716013 |
| rat anti-mouse CD138-BV510 | BioLegend | Cat#142521; RRID: AB_2562727 |
| rat anti-mouse GL-7-FITC | BD Pharmingen | Cat#553666; RRID: AB_394981 |
| rat anti-mouse CD38-PE-Cy7 | BioLegend | Cat#102718; RRID AB_2275531 |
| goat anti-mouse IgM-biotin | Jackson ImmunoResearch | Cat#115-065-075, RRID AB_2338566 |
| rat anti-mouse IgD-APC-Cy7 | BioLegend | Cat#405716; RRID AB_10662544 |
| streptavidin-HRP | ThermoFisher Scientific | Cat#N100 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| E. coli DH5a | Thermofisher | Cat#18265017 |
| E. coli BL21 (DE3) | Thermofisher | Cat#C600003 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| fentanyl-LPSTGG | this paper | N/A |
| FITC | Abcam | Cat#ab102884 |
| FITC-LPSTGG | this paper | N/A |
| fen-G4 peptide | this paper | N/A |
| fentanyl-citrate | Sigma-Aldrich | Cat#990-73-8 |
| Fentanyl-citrate | West-Ward | N/A |
| fentanyl solution | Sigma-Aldrich | Cat#F-013 |
| fentanyl-hapten-biotin | this paper | N/A |
| fentanyl | LGC | LGCAMP0528.00-02 |
| fentanyl-d5 | LGC | LGCAMP0528.80-02 |
| fentanyl-PE | Marco Pravetoni, University of Washington | N/A |
| decoy-PE-AF647 | Marco Pravetoni, University of Washington | N/A |
| EDC crosslinker | ThermoFisher Scientific | Cat#22980 |
| ABTS tablets | Roche | Cat#11112422001 |
| Srt-VSG3 protein | this paper | N/A |
| Recombinant RNase Inhibitor | Takara Bio Europe S.A.S | Cat#2313A |
| Penta-His-biotin conjugate | Qiagen | Cat#34440 |
| Sortase A from S. pyogenes | this paper | N/A |
| Trizma® Hydrochlorid -hydrochlorid | Sigma-Aldrich | Cat#T3253 |
| Sodium chloride | Carl-Roth | Cat# 3957.1 |
| Imidazole | Carl-Roth | Cat#X998.4 |
| 2-Mercaptoethanol | Sigma-Aldrich | Cat#M6250 |
| 1,4-Dithiothreitol (DTT) | Carl-Roth | Cat#6908.1 |
| LB-broth (Miller) | Sigma-Aldrich | Cat#L3522-250G |
| HEPES | Carl-Roth | Cat#9105 |
| Dulbecco’s PBS | Sigma-Aldrich | Cat#D8537 |
| Ammonium sulfate | Carl-Roth | Cat#3746 |
| N-(2-Acetamido)-iminodiacetic acid | Sigma-Aldrich | Cat#00307 |
| Polyethelene glycol 400 | Merck | Cat#8.07485.1000 |
| Polyethelene glycol 6000 | VWR | Cat#26603-293 |
| Glycerol | Carl-Roth | Cat#3783 |
| Ethanol | Sigma-Aldrich | Cat#32205-M |
| Sodium acetate | Carl-Roth | Cat#6773 |
| Triethanolamine | Sigma-Aldrich | Cat#90279 |
| Magnesium sulfate heptahydrate | Merck | Cat#1.05886.0500 |
| Sodium citrate tribasic dihydrate | Sigma-Aldrich | Cat#71402 |
| Ethelene glycol | MP | Cat#151089 |
| Hexylene glycol (MPD) | Sigma-Aldrich | Cat#112100 |
| HMI-9 Medium | PAN Biotech | SO-15701 |
| FreeStyle 293 Expression Medium | Gibco | Cat#12338-018 |
| Carbonate-bicarbonate buffer capsules | Sigma-Aldrich | Cat#C3041 |
| 20x PBS Tween 20 | Thermo Scientific | Cat#PI28352 |
| Gelatin from porcine skin | Sigma-Aldrich | Cat#G2500 |
| Octet Streptavidin biosensors | Sartorius | Cat#18-5019 |
| Tramadol hydrochloride | Fagron | 20L09-U10-009586 |
| Naloxone hydrochloride dihydrate | Sigma-Aldrich | N7758 |
| Acetonitrile (LC-MS Grade) | Carl Roth | Cat#AE70.2 |
| Phosphoric acid | Carl Roth | Cat#6366.1 |
| Methanol (ROTISOLV®) | Carl Roth | Cat#AE71.1 |
| Formic acid | Carl Roth | Cat#1EHK.1 |
| Ammonium hydroxide | Carl Roth | Cat#6774.1 |
| Ammonium formate | Carl Roth | Cat#5093.1 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Quantum™ FITC-5 MESF kit | Bangs Laboratories | CAT#555 |
| LIVE/DEAD™ Fixable Blue Dead Cell Stain Kit | Invitrogen | Cat#L23105 |
| Maxima H Minus Reverse Transcriptase, 10000U | ThermoScientific | Cat#EP0752 |
| Kapa High Fidelity Hot Start | Roche | Cat#7958935001 |
| Nextera XT DNA Library Preparation Kit (96SMP) | illumina GmbH | Cat#FC-131-1096 |
| QuikChange Lightning site-directed mutagenesis kit | Agilent technologies | Cat#210513 |
| Pierce™ Protein G Agarose | ThermoFisher Scientific | Cat#20397 |
| Gibco™ 293fectin™ Transfection Reagent | ThermoFisher Scientific | Cat#12347750 |
| Ni-NTA agarose | Qiagen | Cat#30230 |
| SigmaFAST OPD | Sigma-Aldrich | Cat#P9187-5SET |
| Superdex 200 Increase 10/300 GL column | Cytiva | Cat#28-9909-44 |
| Q-Sepharose Fast-Flow | Cytiva | Cat#17-0510-01 |
| NEBuilder Master Mix | New England Biolabs | Cat#M5520AA |
| Phusion HiFi DNA Polymerase | New England Biolabs | Cat#M0530L |
| FreeStyle™ MAX Reagent | Gibco | Cat#16447100 |
| GeneJET Plasmid Miniprep Kit | Thermo Scientific | Cat#K0502 |
| Monarch® DNA Gel Extraction Kit | New England Biolabs | Cat#T1020S |
| Q5® High-Fidelity 2x Master Mix | New England Biolabs | Cat#M0492S |
| AgeI-HF® | New England Biolabs | Cat#R3552S |
| SalI-HF® | New England Biolabs | Cat#R3138S |
| BsiWI-HF® | New England Biolabs | Cat#R3553S |
| EcoRI-HF® | New England Biolabs | Cat#R3101S |
|
| ||
| Deposited data | ||
|
| ||
| Crystal structures | Protein DataBank | PDB codes: 7QT0, 7QT2, 7QT3, 7QT4 |
| RNA-seq data | GEO | Upon request |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| FreeStyle 293F | ThermoFisher Scientific | Cat#R79007 |
|
| ||
| Experimental models: Organisms/Strains | ||
|
| ||
| C57BL/6JRj | Javier-labs | Cat#C57BL/6JRj |
| Trypanosoma brucei brucei Lister 427 2T1 Srt-VSG3-S317A GPI-PLC WT | Pingeretal., 201714 | N/A |
| Trypanosoma brucei brucei Lister 427 Srt-VSG3-S317A GPI-PLC −/− | this paper | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| dNTP Mix (10 mM each)-1 mL | ThermoFisher Scientific | Cat#R0192 |
| Oligonucleotides | See Table S2 | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| IGγ1-, IGκ- or IGλ-expression vectors | Tiller et al., 200862 | N/A |
| Fab heavy and light chain expression vectors | Mirjana Lilic, The Rockefeller University | N/A |
| Sortase A from Streptococcus pyogenes with N-terminal His6-Tag in pET28a expression vector | this paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| FlowJo v10.0.8 | Tree Star | https://www.flowjo.com/solutions/flowjo/downloads |
| GraphPad Prism v6.07 | GraphPad | http://www.graphpad.com |
| R v4.0.3 and v4.1 | The R project for statistical computing | http://www.R-project.org/ |
| FACSDiVa v8.0.1 | Becton Dickinson | Cat#659528 |
| Adobe Illustrator CS6 v16.0.3 | Adobe | http://www.adobe.com/de/products/illustrator.html |
| Adobe Photoshop 2021 v22.0.0 | Adobe | https://www.adobe.com/de/products/photoshop |
| Microsoft PowerPoint v16 | Microsoft | https://www.microsoft.com/de-de/microsoft-365/powerpoint |
| Microsoft Excel v16 | Microsoft | https://www.microsoft.com/de-de/microsoft-365/excel |
| CCP4MG v2.10.11 | McNicholas et al., 201163; de Beeretal., 201 464; https://pymol.org/2/ | https://www.ccp4.ac.uk/MG/ |
| Biorender | Biorender | https://biorender.com/ |
| XDS | Kabschetal., 201065 | http://xds.mpimf-heidelberg.mpg.de |
| Phaser | McCoy et al., 200766 | https://www.phenix-online.org |
| Phenix | Adams et al., 201067 | https://www.phenix-online.org |
| Coot | Emsleyetal., 201068 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PDB-redo | Joosten et al., 201469 | https://pdb-redo.eu/ |
| PyMol v2.4.2 | N/A | https://pymol.sourceforge.net |
| TrimGalore v0.6.4_dev | Krueger et al., 201570 | http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| MultiQC v0.4 | Ewelsetal., 201671 | https://multiqc.info/ |
| FastQC v0.11.9 | N/A | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Samtools v1.9 | Danecek et al., 202172 | http://www.htslib.org/ |
| STAR v2.6.0a | Dobinetal., 201373 | https://github.com/alexdobin/STAR |
| Bioconductor v3.12 | Bioconductor Software Packages | https://bioconductor.org/packages/3.12/bioc/ |
| Bioconductor v3.14 | Bioconductor Software Packages | https://bioconductor.org/packages/3.14/bioc/ |
| GenomicFeatures v1.42.3 | Lawrence et al., 201374 | https://bioconductor.org/packages/3.12/bioc/html/GenomicFeatures.html |
| Ensembldb v2.14.1 | Raineretal., 201975 | https://bioconductor.org/packages/3.12/bioc/html/ensembldb.html |
| GenomicAlignments v1.26.0 | Lawrence et al., 201374 | https://bioconductor.org/packages/3.12/bioc/html/GenomicAlignments.html |
| AnnotationDbi v1.52.0 | Pagès et al., 202076 | https://bioconductor.org/packages/3.12/bioc/html/AnnotationDbi.html |
| SingleCellExperiment v1.16.0 | Amezquita et al., 202077 | https://bioconductor.org/packages/3.14/bioc/html/SingleCellExperiment.html |
| scater v1.22 | McCarthy et al., 201778 | https://bioconductor.org/packages/3.14/bioc/html/scater.html |
| scran v1.22.1 | Lunetal., 201679 | https://bioconductor.org/packages/3.14/bioc/html/scran.html |
| org.Mm.eg.db v3.12.0 | Carlson etal.,201980 | https://bioconductor.statistik.tu-dortmund.de/packages/3.12/data/annotation/html/org.Mm.eg.db.html |
| rLiger v1.0.0 | Welch etal., 201941 | https://www.rdocumentation.org/packages/rliger/versions/1.0.0 |
| MAST v1.16.0 | Finaketal., 201581 | https://bioconductor.org/packages/3.12/bioc/html/MAST.html |
| Seurat v4.0 | Haoetal., 202182 | https://satijalab.org/seurat/ |
| BASIC v1.5.0 | Canzar et al., 201783 | https://people.cs.uchicago.edu/~aakhan/BASIC/ |
| Blast v2.11.0 | Alquicira-Hernandez et al., 202184 | https://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastNews |
| Igblast v1.16.0 | Ye etal., 201385 | https://ncbi.github.io/igblast/ |
| Change-O v1.0.0 | Gupta etal.,201586 | https://changeo.readthedocs.io/en/stable/ |
| Shazam v1.1.0 | Gupta etal.,201586 | https://shazam.readthedocs.io/en/stable/#shazam |
| ggplot2 v3.3.5 | Wickham, 200987 | https://github.com/tidyverse/ggplot2/releases/tag/v3.3.5 |
| gridExtra v2.3 | B. Auguie | https://cran.r-project.org/web/packages/gridExtra/index.html |
| ggrepel v0.9.1 | K. Slowikowski | https://cran.r-project.org/web/packages/ggrepel/index.html |
| Circlize vO.4.13 | Gu etal., 201488 | https://mran.revolutionanalytics.com/snapshot/2021-11-01/web/packages/circlize/index.html |
| Octet DataAnalysis v12.0.2.3 | Sartorius | N/A |
| LABORAS v2 | Metris | https://www.metris.nl/en/products/laboras/laboras2/ |
| Analyst v1.6 | Sciex | https://de-store.sciex.com/de/EUR/software |
| MassHunter | Agilent | https://www.agilent.com/en/product/software-informatics/mass-spectrometry-software |
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Female healthy wildtype C57BL/6J mice (6–8-weeks old) were purchased from Janvier Labs and handled in accordance with the German Animal Protection Law (§8 Tierschutzgesetz) and their use was approved by the Regierungspräsidium Karlsruhe, Germany (project numbers Aktenzeichen 35–9185.81/G-285/18). Mice were randomly assigned to experimental groups which were kept in separate cages.
T. brucei brucei parasites
All trypanosome cell lines used in this study were bloodstream-form trypanosomes ultimately derived from the Lister 427 cell line.89 Trypanosomes were cultivated in HMI-9 medium with 10% fetal bovine serum at 37°C and 5% CO2. All trypanosomes used in this study expressed a modified version of VSG3 (S317A). The S317A variant was genetically engineered to lack an O-linked glycan present at the 317th residue of the wild-type counterpart. The newly engineered variant (Srt-VSG3) additionally contains the sortagging motif AA on the N-terminus of the VSG that has been extended above the top of the surface coat through the addition of a linker region consisting of 3 Gly-Gly-Gly-Gly-Ser motifs. They were either expressing (GPI-PLC WT) or lacking GPI-PLC (GPI-PLC −/−). The GPI-PLC WT cells used were the 2T1 cell line,90 while the GPI-PLC −/− counterparts were derived from the 2T1s in-house using knockout vectors kindly provided by Dr. M. Carrington. GPI-PLC −/− cells lack the ability to shed their VSG and thus stay intact during UV irradiation.
Cell lines used for antibody production
Human Embryonic Kidney (HEK) “FreeStyle” 293F cells (ThermoFisher Scientific, Cat# R79007) were grown in tissue culture flasks in FreeStyle™ 293 Expression Medium (ThermoFisher Scientific, Cat# 12338018). Cells were incubated at 37°C with 5–8% CO2 on a shaking platform operating at 120–130 rpm.
Bacteria
E. coli strain DH5a was grown in LB-broth (Miller) medium (Sigma, Cat# L3522–250G) for cultivation and plasmid purification. E. coli strain BL21 was also grown in LB-broth (Miller) medium for cultivation and sortase expression. Bacteria were cultivated at 37°C in shaking incubators rotating at 180 rpm.
METHOD DETAILS
Experimental design
Female mice were used in all experiments and were randomly assigned to the various experimental groups. All experiments were performed using age-matched mice from 6 to 8 weeks old at the start of the experiment. Similar vaccination experiments to those shown in Figure 2 were performed extensively with similar results. The single-cell sequencing approach for B cell repertoire analysis was performed after consulting the literature for known markers and ensuring that population identification was robust. Statistical analyses are detailed throughout the manuscript.
Synthesis of fentanyl derivatives
Lithium 5-oxo-5-((2-(4-(N-phenylpropionamido)piperidin-1-yl)ethyl)amino)pentanoate (3): To a solution of the bis trifluoroacetate salt of amine 1 (compound 5 in Raleigh et al.26; 16.513 g, 32.8 mol) in anhydrous CH2Cl2 (250 mL) was added pyridine (15.9 mL, 197 mmol, 6.0 equiv.) followed by glutaric acid monomethyl ester chloride (4.54 mL, 32.8 mmol, 1.0 equiv.) at 0°C under argon. After addition, the cooling bath was removed and after 16 h, the reaction was diluted with CH2Cl2 (150 mL), washed with saturated aqueous NaHCO3 (3 × 200 mL) and brine (100 mL). The organic layer was dried (MgSO4), filtered, and concentrated in vacuo. The product was purified by column chromatography (5%–7.5% gradient of MeOH in CH2Cl2) to give ester 2 (9.79 g, 74%) as a colorless oil. 1H NMR (400 MHz, CDCl3, CHCl3 referenced to 7.26 ppm) δ 7.40–7.32 (m, 3H), 7.06–7.02 (m, 2H), 6.00 (br s, 1H), 4.58 (tt, J = 12.2, 3.9 Hz, 1H), 3.58 (s, 3H), 3.22 (q, J = 5.8 Hz, 2H), 2.83 (m, 2H), 2.36 (t, J = 6.0 Hz, 2H), 2.28 (t, J = 7.2 Hz, 2H), 2.12 (t, 7.2 Hz, 2H), 2.07 (m, 2H), 1.89–1.81 (m, 4H), 1.76–1.70 (m, 2H), 1.32 (m, 2H), 0.95 (t, J = 7.5 Hz, 3H) ppm; 13C NMR (101 MHz, CDCl3, CHCl3 referenced to 77.16 ppm) δ 173.7, 173.6, 172.1, 139.0, 130.4, 129.4, 128.4, 56.7, 53.0, 52.3, 51.6, 36.2, 35.4, 33.1, 30.5, 28.6, 20.9, 9.7 ppm; LC/MS (ESI+) m/z = 404.2. To a solution of methyl ester 2 (9.65 g, 23.9 mmol) in MeOH (120 mL) was added a solution of LiOH (1.72 g, 71.7 mmol, 3.0 equiv.) in H2O (30 mL) at room temperature. After 22 h, the reaction was partially concentrated in vacuo and completely dried via lyophilization to give unpurified lithium carboxylate 3 as a white powder (9.06 g, ~85%). 1H NMR (400 MHz, D2O) d 7.53–7.47 (m, 3H), 7.25–7.21 (m, 2H), 4.45 (tt, J = 12.2, 3.9 Hz, 1H), 3.25 (t, J = 7Hz, 2H), 2.91 (m, 2H), 2.45 (t, J = 7.0 Hz, 2H), 2.25–2.11 (m, 6H), 1.97 (q, J = 7.6 Hz, 2H), 1.82–1.73 (m, 4H), 1.32 (dq, J = 12.5, 3.5, 2H), 0.92 (t, J = 7.5 Hz, 3H) ppm; 13C NMR (101 MHz, D2O, referenced to external TSP = 0.0 ppm) d 185.3, 179.8, 179.1, 140.7, 132.6, 132.4, 131.8, 58.4, 55.7, 54.9, 39.6, 39.1, 38.3, 32.0, 31.2, 25.2, 12.1 ppm; LC/MS (ESI–) m/z = 388.2. This material was used without further purification for solid phase synthesis.
Solid-phase peptide synthesis procedure definitions:
WASH:
The resin is suspended in DMF (3 mL) with agitation for 2 min, followed by disposal of the solvent. This process is repeated two more times for a total of three washes.
Fmoc-OFF:
The resin is suspended in a 20% solution of piperidine in DMF (3 mL) with agitation for 5 min, followed by disposal of the solution. This process is repeated one more time, but with an incubation time of 7 min.
COUPLING (name of coupling partner):
A DMF solution (3 mL) containing “name of coupling partner” (4.0 M equivalents relative to the initial resin loading), PyBOP (4.0 equiv.), and i-Pr2NEt (8.0 equiv) is prepared and pre-activated for 10 min. The resin is then suspended and agitated in this cocktail for 1 h (unless otherwise noted), followed by disposal of the solution.
Fen-G4:
Briefly, fen-G4 was prepared by solid-phase peptide synthesis starting with a glycine-labeled resin, followed by coupling to Fmoc-GGG-OH (BACHEM) and then compound 3. In detail, a syringe (5 mL) for peptide synthesis (frit made of PE) was charged with pre-loaded Fmoc-G-SASRIN resin (417 mg, loading: 0.72 mmol/g, BACHEM) and swelled for 1 h in DMF (3 mL). The synthesis procedure then proceeded according to the above definitions: WASH, Fmoc-OFF, COUPLING (Fmoc-GGG-OH, BACHEM, for 2 h), WASH, Fmoc-OFF, WASH, COUPLING (compound 3), WASH. The resin was then washed (2 min) with CH2Cl2 (3 mL) three times. For cleavage, the resin was treated (5 min) with 80% trifluoroacetic (TFA) in water (3 mL) twice, wherein the resin beads became dark purple. The two solutions were combined and concentrated in vacuo. This residue was dissolved in water (ca. 25 mL) and lyophilized. The product was purified via reverse-phase chromatography (C18 silica, 1 to 40% gradient of MeCN in H2O) and then lyophilized with a few drops of added TFA to obtain fen-G4 as a trifluoroacetate salt (49 mg, 0.067 mmol, 22%). 1H NMR (400 MHz, D2O, HOD referenced to 3.79 ppm) d 7.57–7.48 (m, 3H), 7.29–7.23 (m, 2H), ~4.75 (1H, obscured by solvent, confirmed by HSQC), 3.99 (s, 6H), 3.93 (s, 2H), 3.66 (d, J = 12.2 Hz, 2H), 3.54 (m, 2H), 3.23 (m, 2H), 3.16 (t, J = 12.4 Hz, 2H), 2.33–2.22 (m, 4H), 2.13 (d, J = 13.8 Hz, 2H), 2.01 (q, J = 7.4 Hz, 2H), 1.84 (app pent, J = 7.2 Hz, 2H), 1.64 (q, J = 12.5 Hz, 2H), 0.94 (t, 7.2 Hz, 3H) ppm; 13C NMR (101 MHz, D2O, referenced to internal MeOD = 49.5 ppm) d 178.9, 178.2, 177.9, 174.7, 173.8, 173.5, 173.2, 164.4 (q, 2JCF = 36.4 Hz), 138.5, 131.2 (2C), 130.7, 117.7 (q, 1JCF = 291.3 Hz), 57.4, 53.8, 51.2, 43.9 (2C), 43.7, 42.5, 35.8 (2C), 35.6, 29.7, 28.8, 22.4, 10.5 ppm; HRMS (ESI–) m/z calcd for [C29H42N7O8–]: 616.3100; found: 616.3104.
Sortaggable Fentanyl (fen-sort):
Briefly, fen-sort was prepared by solid-phase peptide synthesis starting with a glycine-labeled resin, followed by sequential coupling to Fmoc-protected G, T, S, P, L, S, G, G, G (all from BACHEM), and then compound 3. In detail, a syringe (5 mL) for peptide synthesis (frit made of PE) was charged with pre-loaded Fmoc-G-SASRIN resin (417 mg, loading: 0.72 mmol/g, BACHEM) and swelled for 1 h in DMF (3 mL). The synthesis procedure then proceeded according to the above definitions: WASH, Fmoc-OFF, COUPLING (Fmoc-G-OH), WASH, Fmoc-OFF, WASH, COUPLING (Fmoc-T(t-Bu)-OH), WASH, Fmoc-OFF, WASH, COUPLING (Fmoc-S(t-Bu)-OH; Note: collidine was used instead of i-Pr2NEt in this step), WASH, Fmoc-OFF, WASH, COUPLING (Fmoc-P-OH), WASH, Fmoc-OFF, WASH, COUPLING (Fmoc-L-OH), WASH, Fmoc-OFF, WASH, COUPLING (Fmoc-S(t-Bu)-OH; Note: collidine was used instead of i-Pr2NEt in this step), WASH, Fmoc-OFF, WASH, COUPLING (Fmoc-G-OH), WASH, Fmoc-OFF, WASH, COUPLING (Fmoc-G-OH), WASH, Fmoc-OFF, WASH, COUPLING (Fmoc-G-OH), WASH, Fmoc-OFF, WASH, COUPLING (compound 3), repeat COUPLING (compound 3), WASH. The resin was then washed (2 min) with CH2Cl2 (3 mL) three times. For cleavage the resin was treated (5 min) with a cocktail of 90% TFA, 5% water and 5% triisopropylsilane (3 mL) three times, during which the resin beads became dark purple. The three washings were combined and stirred for 1 h, and then added dropwise to cold Et2O (50 mL). The obtained precipitate was filtered and washed thoroughly with cold Et2O. The solid residue was taken up in water and lyophilized. The product was purified via reverse-phase MPLC (C18 silica, 1 to 40% gradient of MeCN in H2O) and then lyophilized with a few drops of added TFA to obtain fen-sort as a trifluoroacetate salt (99 mg, 0.078 mmol, 26%). 1H NMR (400 MHz, D2O, HOD referenced to 3.79 ppm) d 7.56–7.49 (m, 3H), 7.29–7.24 (m, 2H), ~4.75 (1H, obscured by solvent, confirmed by HSQC), 4.65 (dd, J = 9.9, 4.5 Hz, 1H), 4.52–4.44 (m, 3H), 4.41 (d, J = 3.8 Hz, 1H), 4.31 (dq, J = 6.4, 3.8 Hz, 1H), 4.03–3.97 (m, 7H), 3.96–3.91 (m, 3H), 3.90–3.81 (m, 4H), 3.70–3.63 (m, 3H), 3.55 (d, J = 6.2 Hz, 2H), 3.24 (d, J = 6.2 Hz, 2H), 3.20–3.12 (m, 2H), 2.35–2.23 (m, 5H), 2.14 (d, J = 13.8 Hz, 2H), 2.08–1.90 (m, 5H), 1.85 (sept, J = 7.3 Hz, 2H), 1.72–1.55 (m, 5H), 1.22 (d, J = 6.4 Hz, 3H), 0.97–0.90 (m, 9H) ppm; 13C NMR (101 MHz, D2O, referenced to internal MeOD = 49.5 ppm) d 178.9, 178.2, 177.9, 175.8, 174.5, 174.1, 173.8 (3C), 173.5, 173.1, 172.8 (2C), 164.3 (q, 2JCF = 35.6 Hz), 138.5, 131.2 (2C), 130.7, 117.8 (q, 1JCF = 291.6 Hz), 68.4, 62.6, 62.3, 61.9, 60.5, 57.4, 57.0, 56.8, 53.8, 51.8, 51.2, 49.3, 43.9 (2C), 43.8 (2C), 42.4, 40.5, 35.8 (2C), 35.6, 30.8, 29.7, 28.8, 26.1, 25.8, 23.9, 22.4, 22.1, 20.2, 10.5 ppm; HRMS (ESI–) m/z calcd. for [C52H80N13O17–]: 1158.5801; found: 1158.5810.
Purification of Srt-VSG3 protein
Srt-VSG3 protein was purified from T. brucei PLC WT expressing Srt-VSG3 as described elsewhere.14 Briefly, cells were cultured in vitro in HMI-9 media to a density of 4 × 106 cells/mL. Cells were pelleted, lysed in 0.2 mM ZnCl2 + HALT protease inhibitor and then the lysis mixture was centrifuged at 10,000xg for 10 min. The pellet which contained the membrane material was resuspended in pre-warmed (40°C) 20 mM HEPES pH 7.5 with 150 mM sodium chloride (NaCl), enabling the activation of endogenous lipases and resulting in the efficient release of surface VSG protein from the membrane. The membranous material was then pelleted two more times, while supernatants (containing soluble VSG) were collected. Supernatants were loaded onto an anion-exchange column (Q-Sepharose Fast-Flow, GE Healthcare), which had been equilibrated with 20 mM HEPES buffer with 150 mM NaCl (the VSG does not bind to the column, while contaminating proteins are trapped). Srt-VSG3 was then separated from remaining contaminants and aggregation products via a gel filtration column (Superdex 200, GE Healthcare) equilibrated in 20 mM HEPES buffer with 150 mM NaCl. Aliquots from the gel filtration runs were analyzed on SDS–PAGE for visual inspection (Figure S2B).
Sortagging of soluble Srt-VSG3
Sortase A was expressed and purified as described.14 Sortagging solutions containing 100 μM purified sortase A and 300 μM of fentanyl-LPSTGG in PBS or 20 mM HEPES, 150 mM NaCl, pH 8.0 were incubated on ice for 30 min. Purified Srt-VSG3 protein was added to the sortagging solution at a concentration of not higher than 2 mg/mL and incubated for 2 h at 37°C while gently shaking and then at 4°C rotating overnight. Fentanyl-sortagged Srt-VSG3 protein was re-purified via gel filtration (Superdex 200, GE Healthcare), concentrated to not higher than 2 mg/mL, and flash frozen for storage at −80°C until further use.
Sortagging of intact T. brucei brucei
In order to avoid the naturally occurring shedding of the VSG coat, a protection mechanism of trypanosomes from antibody attachment in the host, Srt-VSG3 PLC−/− T. brucei was used throughout the whole paper. Srt-VSG3 PLC−/− T. brucei expresses VSG3 on the surface and lacks the GPI-PLC enzyme that is otherwise responsible for the natural VSG shedding. For sortagging whole trypanosomes, the cells were incubated in a sortagging solution containing 100 mM purified sortase A and 300 mM of fen-sort in HMI-9 media on ice for 30 min. PLC −/− trypanosomes expressing Srt-VSG3 were pelleted, resuspended in the sortagging solution at a concentration of 108 cells/mL, and incubated for 2h at 4°C on an inversion rotator. Cells were then pelleted, washed once with HMI-9 media, and pelleted again before resuspension in HMI-9 or staining solution containing an anti-fentanyl monoclonal antibody (provided by M. Pravetoni, University of Minnesota) conjugated to FITC (Abcam, ab102884) for analyzing sortagging success. Trypanosomes were immediately analyzed using a BD FACS Calibur and FlowJo v10 software.
Estimation of the sortagging surface coverage
Trypanosomes were sortagged with FITC-LPSTGG as a proxy for fentanyl and other haptens. Cells were sortagged in a time course at 4°C, and the fluorescence readings were recorded by flow cytometry. For the standard curve, the Quantum™ MESF kit was obtained from Bangs Laboratories, containing one blank bead population and a series of five fluorescent bead populations labeled with varying amounts of FITC. The FACS signal from each bead population was recorded with the same PMT settings as the cells in order to establish a calibration curve that relates instrument channel values and standardized fluorescence intensity units (MESF). The background value of the reaction without sortase was subtracted from the corresponding signal of the FITC sortagged trypanosome samples for each time point. The subtracted values were then read against the MESF beads curve for determination of conjugation efficiency (i.e., quantitation of the fluorescent signal from each time point). The sortagging percentage was then calculated assuming 107 as the maximum number of VSG molecules on the surface of each individual cell corresponding to 100% labeling.
UV irradiation of sortagged T. brucei brucei
After sortagging with fentanyl, the trypanosomes were pelleted, washed with irradiation buffer (PBS-glucose (10 g/L)) and resuspended in irradiation buffer to a density of 108 cells/mL. A volume of 1 mL of this resuspension was aliquoted into each well of a non-treated six-well tissue culture plate. Plates were UV-irradiated for four times in 1 min intervals using an FB-UVXL-1000 UV cross-linker. Plates were swirled between 1 min intervals to ensure equal irradiation of trypanosomes. Complete irradiation was confirmed using a microscope and cells were counted and diluted for immunization.
Mouse immunizations
Five 6–8 weeks old female C57BL/6J mice per group were primed at day 0 and 14 or 30 with 5 × 106 intact UV-irradiated trypanosomes, either sortagged or not sortagged with fentanyl, and without adjuvant via subcutaneous injection. The mice then received a booster of 100 μg of soluble Srt-VSG3, either sortagged or not sortagged with fentanyl (without adjuvant) at days 42 and 70 or days 60 and 90. Serum samples were taken 2 days before and one week after injection.
Serum ELISA
The fen-G4 peptide was conjugated to BSA via EDC crosslinking as a heterologous carrier protein and used to coat 96-well ELISA plates at 10 μg/mL overnight at 4°C. Similarly, plates were coated with FPLC-purified Srt-VSG3 protein at 5 μg/well. Plates were blocked for 1.5 h at RT with 4% BSA in PBS. Coated plates were incubated with the serum for 1.5 h at RT. As a control, an anti-VSG3 monoclonal antibody or an anti-fentanyl monoclonal antibody were used at a starting concentration of 1 μg/mL or 5 μg/mL, respectively, and diluted 4-fold. Bound antibodies were detected by goat anti-mouse IgG secondary antibody coupled to horse-radish peroxidase (HRP) (Jackson Immuno Research) diluted 1:3,000 in PBS with 1% BSA, which was then resolved using an ABTS substrate solution complimented with H2O2 (Roche). The optical density (OD) at 405 nm was determined on an M1000Pro plate reader (Tecan) after 40 min. Graphs depicting the calculated midpoint titers were generated with GraphPad Prism v9.
Antinociception
Analgesia was tested by using the hot plate assay as described by Cox and Weinstock.38 Baseline nociception of the mice was measured by placing them on a hot plate at 54°C and measuring the latency to a response (in seconds) like flipping or licking a paw or jumping. In order to avoid tissue damage, the test was aborted (in the absence of an observable reaction) after 60 s. Baseline measurements were always recorded prior to any manipulation. Mice were then injected subcutaneously with a cumulative dose of 100 mg/kg fentanyl (split between two individual injections of 50 μg/kg). The effect of fentanyl was measured as latency to response (seconds) as well as the percentage maximum possible effect (%MPE). %MPE was calculated as the post-test latency minus the baseline latency divided by the maximum effect time (10 s) minus the baseline latency times 100.26 All experimental studies were conducted multiple times using a blinded observational style of data collection.
Straub-Tail reaction
The Straub tail reaction is a dorsiflexion of the tail that is often almost vertical to the orientation of the body or curling back over the animal, often coupled with stereotyped walking behavior.91 This phenomenon was first described as a response to opiates in mice (Straub (1911) cited by (Bilbey, Salem and Grossmann, 196091)), and is thought to be mediated by activation of the opioid receptor system because opioid receptor antagonists such as naloxone block the phenomenon.92–94 The Straub-Tail reaction was recorded as present (+) or absent (−).
Analysis of fentanyl concentrations in tissue
Preparation of brain samples
Brain tissue was homogenized using Agilent ceramic beads for 4 min with a Beadblaster 24 homogenizer (Benchmark Scientific, Sayreville, NJ) at a speed of 7 m/s, then centrifuged briefly to reduce bubbles. The homogenate was transferred to a cryogenic tube and placed at −20°C until time of extraction.
Sample extraction
Extraction of serum, brain homogenate, and standards was performed at 4°C. For standards, 20 μL of stock calibrator solution was added to 180 μL of fetal bovine serum. 200 μL of sample was used for extraction with 20 μL of internal standard solution added to all samples. 600 μL of cold LC-MS grade acetonitrile was added to 1.6 mL microcentrifuge tubes to precipitate proteins and then centrifuged at 8,609xg for 10 min. Supernatant was transferred to a 2 mL 96-well collection plate, evaporated to 200 μL on a Mini Vap (Porvair Sciences, Wrexham, UK) and then diluted 1:4 with 2% phosphoric acid. Extraction was performed using Bond Elut 96, Plexa PCX, 1 mL, 30 mg (Agilent, Santa Clara, CA). Cartridges were first washed with 500 mL methanol followed by 500 mL water and then samples were loaded onto the plate. The plate was washed in series, first using 600 μL of 2% formic acid followed by 600 μL 1:1 methanol/acetonitrile. The plate was dried on a Positive Pressure Manifold (Agilent, Santa Clara, CA), placed above a fresh round bottom 1 mL 96-well collection plate to elute samples using 750 μL of 5% ammonium hydroxide in 1:1 methanol/acetonitrile, and dried on the Mini Vap. Samples were reconstituted in 200 μL LC-MS grade water, 0.1% ammonium formate, 0.01% LC-MS grade formic acid (mobile phase A).
LC-MS/MS conditions
2 μL of sample were injected onto a reversed phase Agilent (Santa Clara, CA) Poroshell 120 SB-C18 column (2.1 mm × 50 mm 2.7 μm). The LC-MS/MS system consisted of an Agilent G6470A triple quadrupole with an Infinity II 1290 G7116B Multicolumn Thermostat, G7120A High Speed Quad Pumps, G7267B Multisampler. The samples were kept at 4°C during injection. Gradient elution was performed with a mixture of mobile phase A and methanol, 0.01% formic acid (mobile phase B) as follows: 0–0.5 min 5% mobile phase B, 0.5–2.25 min 15 → 50% mobile phase B, 2.25–4.0 min 50 → 95% mobile phase B, 4.0–6.0 95% mobile phase B. The flow rate was kept at a constant 0.40 mL/min and the total run time was 6 min. Electrospray ionization was achieved by Agilent Jet Stream high sensitivity ion source in the positive ion mode. Instrument settings were: gas temperature 325°C, gas flow 9 L/min, nebulizer pressure 40 psi, sheath gas temperature 380°C, sheath gas flow 10 L/min, capillary 2500 V, and pos nozzle 0V. Data acquisition and peak integration were interfaced to a computer workstation using Mass Hunter (Tokyo, Japan). High-performance liquid chromatography-tandem mass spectrometry (LC-MS/MS) and their specific mode was used for the mass spectrometric analysis to identify the appropriate ions to monitor m/z: fentanyl 337.2–188.1, secondary 337.2–105.1, fentanyl-d5 342.3–188.1, secondary 342.3–105.1. The analysis of extracted serum samples from the mice after dosing (26.25 and 3.75 mg/kg) as well as the controls were also measured by applying LC-MS/MS and fentanyl concentrations were determined after appropriate sample preparation following the above-described method.
Flow cytometry and single cell sorting
Fentanyl-reactive B cells were analyzed on an LSR II instrument and isolated using an ARIA II cell sorter (BD Bioscience). Cells were stained using fentanyl-PE (1:1000; 1 μM stock), a decoy-PE-AF647 (1:50; 1 mM stock)49,50,95 and the following antibodies: rat anti-mouse CD19-BV412 (1:100, Biolegend), rat anti-mouse IgG1-BV650 (1:100, Biolegend), rat anti-mouse CD138-BV510 (1:300, Bio-legend), rat anti-mouse GL-7-FITC (1:1000, BD Pharmingen), rat anti-mouse CD38-PE-Cy7 (1:400, Biolegend), goat anti-mouse IgM-biotin (1:400, Jackson), rat anti-mouse IgD-APC-Cy7 (1:1000, Biolegend), streptavidin-BV785 (1:400, Biolegend). The LIVE/DEAD™ Fixable Blue Dead Cell Stain Kit (1:1000, Invitrogen) was included in all stainings to exclude dead cells. The data were analyzed using FlowJo v10 software. For single-cell sorting, fentanyl-reactive B cells were defined as live fentanyl + decoy-CD19+ and sorted into 384-well plates containing lysis buffer using an ARIA II cell sorter (BD Bioscience and using the index-sort function).
SMART-seq 2.5 library preparations
Single-cell RNA sequencing was performed following the SMART-seq 2.5 library preparation protocol as described in Picelli et al.,96 and modified by the DKFZ scOpenLab in Bioquant, Heidelberg. Plate preparation was assisted by the BRAVO Agilent and Mosquito LV liquid handling platforms. Quality control was performed on the TapeStation provided by Agilent Technology, using a D5000 Tape. Single cell library preparation was performed using the Nextera XT DNA SMP Prep Kit (96 SMP). DNA was cleaned-up using a magnetic size selection. The samples were finally quantified and assessed on a D1000 TapeStation.
Single cell RNA sequencing
Single cell RNA sequencing was performed on the Illumina sequencing platform of the DKFZ Genomics and Proteomic sequencing facility. The samples were pooled and delivered as multiplexed. Next-generation sequencing (NGS) of 75 bp single reads was performed using the NextSeq 550 Sequencing System. Quality control was performed before and after the sequencing process. The quality of the raw reads was assessed using fastqc and MultiQC.71 Adapter (Nextera transposase sequence) and quality trimming (Phred score cutoff of 20, overlap of 3 bp) was performed with TrimGalore version: v0.6.4_dev (Cutadapt v1.18),70 keeping the sequencing reads with a length of at least 36 bps. Genome reference-based alignment was performed using the slice-aware STAR (v2.6.0a) alignment tool73 with the default parameters with Release M25 (GENCODE-GRCm38.p6) of the mouse genome. The reference genome was indexed with a bp overhang of 100–1. The generated bam files were sorted using Samtools (v1.9)72 based on the chromosome coordinates. Unmapped reads, PCR duplicates, and reads with an alignment p.score <20 were removed by filtering the sorted bam files.
Transcriptome analysis
Downstream preprocessing analysis was performed separately for the sorted plates in the R programming language using the Bioconductor packages SingleCellExperiment (v1.16.0),77 scater (v1.22),78 and scran (v1.22.1).79 The count matrices with only the exonic sequences were generated using the summarizeOverlaps function of the GenomicAlignments package (v1.3074). Gene annotation was performed with the makeTxDbFromGFF function of the GenomicFeatures package (v1.46.1),74 the AnnotationDbi (v1.52.0),76 the Ensembldb (v2.14.1),75 and the org.Mm.eg.db (v3.12.0)80 package. The mitochondrial genes were identified by extracting the genomic coordinates according to the TxDb.Mmusculus.UCSC.mm10.ensGene dataset.
Quality control, normalization, and feature
Outlier cells with high mitochondrial percentage, low library numbers and low unique features were identified and removed based on a median absolute deviation (MAD) = 3. Additionally, gene filtering was applied by removing genes with <3 reads in <5 cells. Further, we also eliminated transcripts that map to the VDJ immunoglobulin genes, while keeping the reads aligning to the constant regions of the heavy and light chains. The gene expression was normalized by deconvolution by applying the quickCluster and ComputeSum-Factors functions of scran. Next, we computed the log-transformed normalized expression values. Feature selection was performed with modelGeneVar, which models the variance of the log-expression profiles for each gene on a fitted mean-variance trend. Genes with a variance of >0 and FDR <0.1 are maintained (~13,000 for each plate).
Data integration
The sorted plates (262 and 288 cells for each plate respectively) were integrated based on non-negative matrix factorization using the rLiger package (v1.0.0), by asserting all the genes that are either shared or unique for each plate and have a variance >0.1 and an alpha threshold of 0.0540. The lambda and kappa values were fixed to 5 and 20, respectively. Genes were normalized and scaled, and the cells were clustered using a resolution of 0.9. Finally, Uniform Manifold Approximation and Projection (UMAP)97 was applied by computing the ‘cosine’ distance of 30 neighboring cells with a minimum distance of 0.1.
Marker gene identification
A first characterization of the B cell subclusters was performed by using a set of known B cell markers. UMAP visualizations of the joint expression of 3 known memory B cell markers were created using the Nebulosa package (v1.484). Differential gene expression between clusters was performed with the MAST algorithm81 integrated in the Seurat:4 package82 after transforming the liger object into a Seurat object of 550 cells, using the default parameters. For the subcluster characterized as “switched memory B cells”, more stringent parameters were applied (at least <0.5 LFC and the presence of the markers in at least 30% of the cells) to reassure the identification of robust marker genes. For visualization purposes (Figure 4B), the Z score of the top 10 genes from each subcluster with a positive log fold change along with several known B cell markers were plotted together. However, to strengthen our confidence in the subcluster identification, we also examined the expression levels of 35 of the most widely recognized B cell markers such as Fcer2a (or Cd23), Mzb1, Cd9, Cd38, Ccr4, Fos, Cd40, and several transcription factors such as Irf4, Irf8, Blimp1, Bach2, and Bcl6 (Figure 4B).39–44 Using the pheatmap package, hierarchical clustering between both the selected genes (rows) and the identified subclusters (columns) was applied. Columns were clustered based on the “correlation” distance and the “ward.D” method, while rows were based on the “euclidean” distance.
BCR sequencing analysis
The trimmed reads of the raw fastq files were analyzed using the BASIC vdj assemble package (v1.5.0).83 Annotation of the VDJ segments was performed with the IgBLAST (v1.16.0)85 using a database of V(D)J genes from IMGT as a reference while BLAST (v2.11.0) was used for the identification of the constant chains. Downstream analysis was performed in R, using the Change-O - Repertoire clonal assignment toolkit from the “Immcantation” portal (v1.0.086) and Shazam (v1.1.0) for the mutational load analysis.86
Monoclonal antibody (mAB) cloning
For IgGs, VDJ regions were synthesized via Twist Biosciences with flanking restriction sites matching the expression vectors taken from Tiller et al. . Heavy chain VDJ regions were flanked with a 5′ AgeI restriction site and 3′ SalI restriction site. Light chain VDJ regions were flanked with a 5′ AgeI restriction site and 3′ BsiWI restriction site. Heavy and light chain vectors and VDJ region-containing plasmids were digested as per Tiller et al.62 using enzymes obtained from NEB. Linear inserts were ligated into the appropriate linear vector using T4 DNA Ligase (ThermoFisher Scientific EL0014). The resulting expression vectors encode antibodies under the control of a CMV promoter. For Fabs, VDJ regions of the Fab heavy chain FenAB136, 609, 709 and FenAB136 light chain were synthesized commercially (Thermo Fisher Scientific, GeneArt), while the VDJ regions of FenAB208 were cloned out of the IgG vector templates. Mammalian expression vectors containing the various Fab heavy chain genes, as well as those containing light chain FenAB136 and FenAB208, were generated via DNA Assembly (NEBuilder) using heavy and light chain vectors kindly provided by Mirjana Lilic. Heavy chains have a C-terminal hexahistidine tag. Vectors encoding the Fab light chains FenAB609 and FenAB709 were generated by QuikChange site-directed mutagenesis (Agilent QuikChange Lightning site-directed mutagenesis kit) using the FenAB136 light chain as a template. The resulting expression vectors encode Fabs under the control of a CMV promoter. All oligonucleotides used for PCR reactions during the above cloning processes are listed in the key resources table.
mAB expression and purification
For IgGs, Human Embryonic Kidney (HEK) 293F cells (ThermoFisher Scientific R79007) grown in 125 mL flasks in FreeStyle™ 293 Expression Medium (ThermoFisher Scientific 12338018) were transfected at cell densities between 8.0 × 105 to 1.0 × 106 cells/mL with equal masses of plasmids containing the heavy chain and light chain genes using FreeStyle™ MAX Reagent (ThermoFisher Scientific 16447100); both plasmids were diluted in OptiPRO™ SFM (ThermoFisher Scientific 12309050). Transfected cells were incubated at 37°C with 5% CO2 on a Digital Orbital Shaker (Southwest Science SBT300) operating at 120 rpm for 6 days. Supernatants were collected after 6 days and passed through a gravity chromatography column containing 1 mL of Pierce™ Protein G Agarose resin (ThermoFisher Scientific 20397). The column was washed with 20 mL of wash buffer (150 mM NaCl, 50 mM Tris, pH 7.4) and eluted in 1 mL fractions with elution buffer (150 mM NaCl, 50 mM Glycine, pH 2.8) prior to neutralization with neutralization buffer (1 M Tris, pH 8.0). Samples were analyzed by SDS-PAGE.
Fabs were produced in 293F cells by co-transfections of these cells with equal amounts of purified heavy and light chain plasmid, using 293fectin (Thermo Fisher 12347–019). Cells were incubated at 37°C, 8% CO2 on an orbital shaker at 130 rpm for 5 days. Supernatants were harvested by centrifugation at 1500xg for 15 min, filtered, concentrated in an Amicon stirred cell and diluted (2x) in binding buffer (50 mM Sodium Phosphate, 20 mM Imidazole, 300 mM NaCl, pH8). Supernatant was then incubated with Ni-NTA agarose (Qiagen) for 1–2 h at room temperature or overnight at 4°C. Next, gravity flow columns were used to collect the Ni-NTA agarose and columns were washed with binding buffer. Fabs were eluted with elution buffer (50 mM Sodium Phosphate, 300 mM NaCl, 250 mM Imidazole, pH8), concentrated in centrifugal units and re-suspended in Dulbecco’s PBS. Samples were analyzed by SDS-PAGE.
BLI and ITC
Biolayer interferometry (BLI) was performed on an Octet Red 96e system (Sartorius). Streptavidin-coated biosensors were pre-hydrated with PBS-T (phosphate buffered saline pH 7.5, 0.05% Tween 20), and baseline responses were measured in PBS-T. Biosensors were loaded with biotinylated fentanyl-hapten (where the biotin replaces the poly-gly sequence from fen-G4; biotinylated fentanyl derivative syntheses are described in33), 0.2 μg/mL for 60 s. Association of antibody with biotinylated hapten was measured in PBS-T with 5 sample concentrations ranging from 5 nM–200 nM for 180 s, followed by dissociation in PBS-T for 300 s. Isothermal titration calorimetry (ITC) experiments were performed using Microcal PEAQ-ITC (Malvern, UK). For all experiments, PBS was used to dissolve fentanyl-citrate (Sigma, 2 mg/mL), fentanyl-hapten, and for the final purification step of Fabs. Titrations were performed at 25°C by injecting consecutive (1–3 μL) aliquots of fentanyl or fentanyl-hapten (both 50 μM) into Fab fragment (4–7 μM) with 120 s intervals. The binding data was corrected for the heat of dilution and fit to a one-site binding model to calculate the Kd, and the binding parameters, N and ΔH. Binding sites were assumed to be identical.
Relative affinity by competition ELISA
Relative affinity was determined by competition ELISA. 96-well plates (Costar polystyrene high-binding plates, Corning) were coated overnight with fen-G4 conjugated to BSA, 50 ng/mL in a 50 mM carbonate buffer, pH 9.6 (Sigma). Plates were blocked with 1% gelatin for 1 h, washed with PBS-T, and free drug was loaded onto plates as competitor. Plates were incubated with 20 ng/mL HIS-tagged Fab for 2 h, washed, and incubated with Penta-His-biotin conjugate 1:5000 (Qiagen) overnight. Streptavidin-HRP 1:5000 (Thermo Fisher) was added, and plates were incubated for 1 h and washed. Fab bound to plates was measured using SigmaFAST OPD substrate (Sigma), and quantitated by absorbance at 492 nm on a microplate reader (Tecan). The fentanyl-hapten structure used for this particular assay differs from the structures presented in Figure S1. Specifically, we here used the “F3” hapten structure from Robinson et al., 2020,27 with the exact same BSA-conjugation methodology described in that publication.
Passive immunization
To collect the data shown in Figures 5E, 5F, 5H and S5A + F: Five 6–8 weeks old female C57BL/6J mice per group were passively immunized by intraperitoneal injection of monoclonal antibody in 200 μL of PBS. One day later, mice were challenged in antinociception and movement tracking assays as described above.
To collect the data shown in Figures 5G, S5D and S5E, mice were first administered fentanyl and then subsequently injected intravenously (after approximately 9 min) with monoclonal antibody in 120 μL of PBS. Mice were challenged in antinociception and movement tracking assays as described above.
LABORAS
The automated laboratory animal behavior observation registration and analysis system (LABORAS) was used to monitor the behavior of the experimental mice. In general, the system utilizes a home cage environment, it is non-invasive and able to track a wide range of genuine behaviors, such as resting, locomotion, traveled distance, climbing, grooming, eating and drinking, without the need for human observers. It measures the vibrations created by the movement of an animal and converts them into behaviors and tracking data using pattern recognition and signal analysis technology. The software can detect each behavior by its distinct signature of signal characteristics, as well as extract the position and speed of the animal.
In the experiments descripted in this manuscript, mice were individually placed in a LABORAS cage and provided with food and water. A baseline measurement of 15–30 min was performed for each mouse before the start of the experiment, followed by 15-min-sessions after fentanyl injections. For each session, the software tracks the movement of the animals depicted in heat maps.
Crystallography of Fab-ligand complexes
Fabs (2–4 mg/mL) were mixed with a 2 to 3-fold molar excess of fen-G4 or fentanyl (Sigma F-013) in Dulbecco’s PBS and incubated for 1–2 h at 4°C. Complexes were purified by gel filtration (GE Healthcare, Superdex 200 Increase 10/300 GL column) in 10 mM HEPES, 150 mM NaCl, pH 8 and concentrated in centrifugal units to about 5 mg/mL for crystallization. All datasets were collected at the Paul Scherrer Institut (Beamline X06DA) at a wavelength of 1.0 Å, using a nitrogen stream at 100 K. For FenAb709 iMosflm98 was used to remove reflections overlapping with ice rings. All the other datasets were processed using the XDS package.65 The structure of FenAb136 was solved by molecular replacement with PHENIX67,99 (PHASER module66) using PDB ID 5H2B as a search model.100 All the other structures were solved using FenAb136 as a starting model. The AUTOBUILD module of PHENIX was used for constructing the initial models. PHENIX, Coot,68 and PDB-redo69 were used for subsequent model building and refinement. The fentanyl or fen-sort was built after refinement of the protein structure. In all cases the density for the ligand was clearly visible in the electron density maps.
Plots and figures
The plots shown in Figure 4 as a part of the sequencing analyses were created in R (v3.6.3 and v4.0.3) using ggplot2 (v3.3.5),87 grid-Extra (v2.3) (https://cran.r-project.org/web/packages/gridExtra/index.html) and ggrepel (v0.9.1) (https://cran.r-project.org/web/packages/ggrepel/index.html), joint density plots with Nebulosa (v1.4), heatmap with pheatmap (v1.0.1.) and circos plots were formed using the circlize package (v0.4.13).88 The images represented throughout the rest of the manuscript were generated using Adobe Photoshop 20201 (v22.0.0) and Adobe Illustrator CS6 (v16.0.3) after rendering plots and tables in GraphPad Prism (v9), Microsoft Powerpoint (v16), Microsoft Excel (v16), and/or FlowJo (v10.7.2). Structures and molecular surfaces are illustrated with CCP4MG (v2.10.11) (https://pymol.org/2/).63,64 The images shown in Figures 1B and S2C were generated with the assistance of BioRender.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical details of experiments can be found in the figures and figure legends. For data related to animal experiments, statistics were analyzed using GraphPad Prism v9. Parametric or non-parametric tests were applied accordingly and are stated in the figure legends. Tests were chosen after consulting similarly designed experiments present in the literature. p values < 0.05 were considered significant (****: p < 0.0001, ***: p < 0.005, **: p < 0.01, *: p < 0.05) and indicated in the figures. Statistical details of the RNA-seq analysis are described elsewhere throughout the STAR Methods section.
Supplementary Material
Highlights.
Antibody elicitation platform based on the surface coat of Trypanosoma brucei
Adjuvant-free immunizations elicit antibodies against fentanyl
Memory B cell identification by RNA-seq facilitates high-affinity mAb discovery
Picomolar-affinity anti-fentanyl mAbs protect from fentanyl pharmacological effects
ACKNOWLEDGMENTS
We acknowledge synchrotron time at the Paul Scherrer Institut, Villingen, Switzerland (SLS, beamline PXIII, Vincent Olieric and colleagues). We acknowledge the DKFZ Flow Cytometry core facility and the DKFZ Single-Cell Open Lab (scOpenLab). We acknowledge Dr. Claudia Pitzer and Barbara Kurpiers of the Interdisciplinary Neurobehavioral Core (INBC) facility, Heidelberg, for the use of the LABORAS system and the hot plate and their assistance. We thank Dustin Hicks, Jenny Vigliaturo (University of Minnesota), Elias Amro (DKFZ), and Vanessa Hofmann for key experimental assistance. This project was funded by grants from the NIH (R01AI097127 to F.N.P. and 1R43DA052960-01 to J.P.V.) and funds from the Helmholtz Foundation (to F.N.P. and C.E.S.). We also acknowledge early contributions by P. Stavropoulos and J. Pinger, who helped initiate the work presented here.
Footnotes
DECLARATION OF INTERESTS
G.T., C.E.S., F.N.P., and J.P.V. report being shareholders of Panosome GmbH and Hepione Therapeutics and having patents planned, pending, or issued broadly relevant to the work. C.E.S. and F.N.P. report being the managing directors of Panosome GmbH. S.K. reports being a shareholder of Panosome GmbH and Hepione Therapeutics. S.R. and K.U. report being employees of Panosome GmbH. P.V., K.U., M.v.S., J.P.Z., Y.K., A.S., A.K.M, A.B., and H.H. report having patents planned, pending, or issued broadly relevant to the work. M.P. and C.B. have patents pending related to fentanyl haptens, conjugates, and mAbs against fentanyl-like compounds.
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2023.112049.
REFERENCES
- 1.Charles A Janeway J, Travers P, Walport M, and Shlomchik MJ (2001). Appendix I. Immunologists’ Toolbox, 5th Ed (Immunobiol. Immune Syst. Health Dis.). [Google Scholar]
- 2.Baruffaldi F, Raleigh MD, King SJ, Roslawski MJ, Birnbaum AK, Hassler C, Carroll FI, Runyon SP, Winston S, Pentel PR, and Pravetoni M. (2019). Formulation and characterization of conjugate vaccines to reduce opioid use disorders suitable for pharmaceutical manufacturing and clinical evaluation. Mol. Pharm. 16, 2364–2375. 10.1021/acs.molpharmaceut.8b01296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng K, Xu Z, Chen M, and Liu X. (2021). Keyhole limpet hemocyanin-conjugated peptides from hepatitis C virus glycoproteins elicit neutralizing antibodies in BALB/c mice. J. Immunol. Res. 2021, 3108157. 10.1155/2021/3108157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sen-Kilic E, Blackwood CB, Boehm DT, Witt WT, Malkowski AC, Bevere JR, Wong TY, Hall JM, Bradford SD, Varney ME, et al. (2019). Intranasal peptide-based FpvA-KLH conjugate vaccine protects mice from Pseudomonas aeruginosa acute murine pneumonia. Front. Immunol. 10, 2497. 10.3389/fimmu.2019.02497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoogsteder PHJ, Kotz D, van Spiegel PI, Viechtbauer W, Brauer R, Kessler PD, Kalnik MW, Fahim REF, and van Schayck OCP (2012). The efficacy and safety of a nicotine conjugate vaccine (NicVAX®) or placebo co-administered with varenicline (Champix®) for smoking cessation: study protocol of a phase IIb, double blind, randomized, placebo controlled trial. BMC Publ. Health 12, 1052. 10.1186/1471-2458-12-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kosten TR, Domingo CB, Shorter D, Orson F, Green C, Somoza E, Sekerka R, Levin FR, Mariani JJ, Stitzer M, et al. (2014). Vaccine for cocaine dependence: a randomized double-blind placebo-controlled efficacy trial. Drug Alcohol Depend. 140, 42–47. 10.1016/j.drugalcdep.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shimogawa MM, Saada EA, Vashisht AA, Barshop WD, Wohls-chlegel JA, and Hill KL (2015). Cell surface proteomics provides insight into stage-specific remodeling of the host-parasite interface in trypanosoma brucei. Mol. Cell. Proteomics 14, 1977–1988. 10.1074/mcp.M114.045146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reinitz DM, and Mansfield JM (1990). T-cell-independent and T-cell-dependent B-cell responses to exposed variant surface glycoprotein epitopes in trypanosome-infected mice. Infect. Immun. 58, 2337–2342. 10.1128/iai.58.7.2337-2342.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verdi J, Zipkin R, Hillman E, Gertsch RA, Pangburn SJ, Thomson R, Papavasiliou N, Sternberg J, and Raper J. (2020). Inducible germline IgMs bridge trypanosome lytic factor assembly and parasite recognition. Cell Host Microbe 28, 79–88.e4. 10.1016/j.chom.2020.04.012. [DOI] [PubMed] [Google Scholar]
- 10.Gkeka A, Aresta-Branco F, Triller G, Vlachou EP, Lilic M, Olinares PDB, Perez K, Chait BT, Blatnik R, Ruppert T, et al. (2021). Immunodominant surface epitopes power immune evasion in the African trypanosome. Prepritnt at bioRxiv. 10.1101/2021.07.20.453071. [DOI] [PubMed] [Google Scholar]
- 11.Mugnier MR, Cross GAM, and Papavasiliou FN (2015). The in vivo dynamics of antigenic variation in Trypanosoma brucei. Science 347, 1470–1473. 10.1126/science.aaa4502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stavropoulos P, and Papavasiliou FN (2010). Using T. brucei as a biological epitope-display platform to elicit specific antibody responses. J. Immunol. Methods 362, 190–194. 10.1016/j.jim.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Susmitha A, Bajaj H, and Madhavan Nampoothiri K. (2021). The divergent roles of sortase in the biology of Gram-positive bacteria. Cell Surf. 7, 100055. 10.1016/j.tcsw.2021.100055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinger J, Chowdhury S, and Papavasiliou FN (2017). Variant surface glycoprotein density defines an immune evasion threshold for African trypanosomes undergoing antigenic variation. Nat. Commun. 8, 828. 10.1038/s41467-017-00959-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hansen J, Baum A, Pascal KE, Russo V, Giordano S, Wloga E, Fulton BO, Yan Y, Koon K, Patel K, et al. (2020). Studies in humanized mice and convalescent humans yield a SARS-CoV-2 anti-body cocktail. Science 369, 1010–1014. 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brioschi S, Wang W-L, Peng V, Wang M, Shchukina I, Greenberg ZJ, Bando JK, Jaeger N, Czepielewski RS, Swain A, et al. (2021). Heterogeneity of meningeal B cells reveals a lymphopoietic niche at the CNS borders. Science 373, eabf9277. 10.1126/science.abf9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nguyen HTT, Guevarra RB, Magez S, and Radwanska M. (2021). Single-cell transcriptome profiling and the use of AID deficient mice reveal that B cell activation combined with antibody class switch recombination and somatic hypermutation do not benefit the control of experimental trypanosomosis. PLoS Pathog. 17, e1010026. 10.1371/journal.ppat.1010026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgan D, and Tergaonkar V. (2022). Unraveling B cell trajectories at single cell resolution. Trends Immunol. 43, 210–229. 10.1016/j.it.2022.01.003. [DOI] [PubMed] [Google Scholar]
- 19.Seth P, Scholl L, Rudd RA, and Bacon S. (2018). Overdose deaths involving opioids, cocaine, and psychostimulants — United States, 2015–2016. MMWR Morb. Mortal. Wkly. Rep. 67, 349–358. 10.15585/mmwr.mm6712a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pravetoni M, and Comer SD (2019). Development of vaccines to treat opioid use disorders and reduce incidence of overdose. Neuropharmacology 158, 107662. 10.1016/j.neuropharm.2019.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smyth BP, Barry J, Keenan E, and Ducray K. (2010). Lapse and relapse following inpatient treatment of opiate dependence. Ir. Med. J. 103, 176–179. [PubMed] [Google Scholar]
- 22.Zanda MT, Floris G, and Sillivan SE (2021). Drug-associated cues and drug dosage contribute to increased opioid seeking after abstinence. Sci. Rep. 11, 14825. 10.1038/s41598-021-94214-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Torten M, Miller CH, Eisele JH, Henderson GL, and Benjamini E. (1975). Prevention of the effects of fentanyl by immunological means. Nature 253, 565–566. 10.1038/253565a0. [DOI] [PubMed] [Google Scholar]
- 24.Bremer PT, Kimishima A, Schlosburg JE, Zhou B, Collins KC, and Janda KD (2016). Combatting synthetic designer opioids: a conjugate vaccine ablates lethal doses of fentanyl class drugs. Angew. Chem. Int. Ed. Engl. 55, 3772–3775. 10.1002/anie.201511654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang CS, Smith LC, Natori Y, Ellis B, Zhou B, and Janda KD (2018). Efficacious vaccine against heroin contaminated with fentanyl. ACS Chem. Neurosci. 9, 1269–1275. 10.1021/acschem-neuro.8b00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raleigh MD, Baruffaldi F, Peterson SJ, Le Naour M, Harmon TM, Vigliaturo JR, Pentel PR, and Pravetoni M. (2019). A fentanyl vaccine alters fentanyl distribution and protects against fentanyl-induced effects in mice and rats. J. Pharmacol. Exp. Ther. 368, 282–291. 10.1124/jpet.118.253674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robinson C, Gradinati V, Hamid F, Baehr C, Crouse B, Averick S, Kovaliov M, Harris D, Runyon S, Baruffaldi F, et al. (2020). Therapeutic and prophylactic vaccines to counteract fentanyl use disorders and toxicity. J. Med. Chem. 63, 14647–14667. 10.1021/acs.jmedchem.0c01042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Crouse B, Wu MM, Gradinati V, Kassick AJ, Song D, Jahan R, Averick S, Runyon S, Comer SD, and Pravetoni M. (2022). Efficacy and selectivity of monovalent and bivalent vaccination strategies to protect against exposure to carfentanil, fentanyl, and their mixtures in rats. ACS Pharmacol. Transl. Sci. 5, 331–343. 10.1021/acsptsci.1c00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barrientos RC, Whalen C, Torres OB, Sulima A, Bow EW, Komla E, Beck Z, Jacobson AE, Rice KC, and Matyas GR (2021). Bivalent conjugate vaccine induces dual immunogenic response that attenuates heroin and fentanyl effects in mice. Bioconjug. Chem. 32, 2295–2306. 10.1021/acs.bioconjchem.1c00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hicks D, Baehr C, Silva-Ortiz P, Khaimraj A, Luengas D, Hamid FA, and Pravetoni M. (2022). Advancing humanized monoclonal antibody for counteracting fentanyl toxicity towards clinical development. Hum. Vaccin. Immunother. 18, 2122507. 10.1080/21645515.2022.2122507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith LC, Bremer PT, Hwang CS, Zhou B, Ellis B, Hixon MS, and Janda KD (2019). Monoclonal antibodies for combating synthetic opioid intoxication. J. Am. Chem. Soc. 141, 10489–10503. 10.1021/jacs.9b04872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ban B, Barrientos RC, Oertel T, Komla E, Whalen C, Sopko M, You Y, Banerjee P, Sulima A, Jacobson AE, et al. (2021). Novel chimeric monoclonal antibodies that block fentanyl effects and alter fentanyl biodistribution in mice. mAbs 13, 1991552. 10.1080/19420862.2021.1991552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baehr C, Kelcher AH, Khaimraj A, Reed DE, Pandit SG, AuCoin D, Averick S, and Pravetoni M. (2020). Monoclonal antibodies counteract opioid-induced behavioral and toxic effects in mice and rats. J. Pharmacol. Exp. Ther. 375, 469–477. 10.1124/jpet.120.000124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baehr CA, Wu MM, Pandit SG, Arias-Umana J, AuCoin D, and Pravetoni M. (2022). Pharmacological profiling of anti-fentanyl monoclonal antibodies in combination with naloxone in pre- and post-exposure models of fentanyl toxicity. J. Pharmacol. Exp. Ther. 381, 129–136. 10.1124/jpet.121.001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carrasco YR, and Batista FD (2006). B cell recognition of membranebound antigen: an exquisite way of sensing ligands. Curr. Opin. Immunol. 18, 286–291. 10.1016/j.coi.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 36.Roozendaal R, Mempel TR, Pitcher LA, Gonzalez SF, Verschoor A, Mebius RE, von Andrian UH, and Carroll MC (2009). Conduits mediate transport of low-molecular-weight antigen to lymph node follicles. Immunity 30, 264–276. 10.1016/j.immuni.2008.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akkaya M, Kwak K, and Pierce SK (2020). B cell memory: building two walls of protection against pathogens. Nat. Rev. Immunol. 20, 229–238. 10.1038/s41577-019-0244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cox BM, and Weinstock M. (1964). Quantitative studies of the antagonism by nalorphine of some of the actions of morphine-like analgesic drugs. Br. J. Pharmacol. Chemother. 22, 289–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Riedel R, Addo R, Ferreira-Gomes M, Heinz GA, Heinrich F, Kummer J, Greiff V, Schulz D, Klaeden C, Cornelis R, et al. (2020). Discrete populations of isotype-switched memory B lymphocytes are maintained in murine spleen and bone marrow. Nat. Commun. 11, 2570. 10.1038/s41467-020-16464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi W, Liao Y, Willis SN, Taubenheim N, Inouye M, Tarlinton DM, Smyth GK, Hodgkin PD, Nutt SL, and Corcoran LM (2015). Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat. Immunol. 16, 663–673. 10.1038/ni.3154. [DOI] [PubMed] [Google Scholar]
- 41.Welch JD, Kozareva V, Ferreira A, Vanderburg C, Martin C, and Macosko EZ (2019). Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell 177, 1873–1887.e17. 10.1016/j.cell.2019.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Meng Q-H, and White HN (2017). CD21int CD23+ follicular B cells express antigen-specific secretory IgM mRNA as primary and memory responses. Immunology 151, 211–218. 10.1111/imm.12724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.King HW, Orban N, Riches JC, Clear AJ, Warnes G, Teichmann SA, and James LK (2021). Single-cell analysis of human B cell maturation predicts how antibody class switching shapes selection dynamics. Sci. Immunol. 6, eabe6291. 10.1126/sciimmunol.abe6291. [DOI] [PubMed] [Google Scholar]
- 44.Laidlaw BJ, and Cyster JG (2021). Transcriptional regulation of memory B cell differentiation. Nat. Rev. Immunol. 21, 209–220. 10.1038/s41577-020-00446-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cerutti A, Cols M, and Puga I. (2013). Marginal zone B cells: virtues of innate-like antibody-producing lymphocytes. Nat. Rev. Immunol. 13, 118–132. 10.1038/nri3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gonzales SJ, Bol S, Braddom AE, Sullivan R, Reyes RA, Ssewanyana I, Eggers E, Greenhouse B, and Bunnik EM (2021). Longitudinal analysis of FcRL5 expression and clonal relationships among classical and atypical memory B cells following malaria. Malar. J. 20, 435. 10.1186/s12936-021-03970-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mathew NR, Jayanthan JK, Smirnov IV, Robinson JL, Axelsson H, Nakka SS, Emmanouilidi A, Czarnewski P, Yewdell WT, Schö n K, et al. (2021). Single-cell BCR and transcriptome analysis after influenza infection reveals spatiotemporal dynamics of antigen-specific B cells. Cell Rep. 35, 109286. 10.1016/j.celrep.2021.109286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pé rez-Mazliah D, Gardner PJ, Schweighoffer E, McLaughlin S, Hosking C, Tumwine I, Davis RS, Potocnik AJ, Tybulewicz VL, and Langhorne J. (2018). Plasmodium-specific atypical memory B cells are short-lived activated B cells. Elife 7, e39800. 10.7554/eLife.39800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laudenbach M, Baruffaldi F, Vervacke JS, Distefano MD, Titcombe PJ, Mueller DL, Tubo NJ, Griffith TS, and Pravetoni M. (2015). The frequency of naive and early-activated hapten-specific B cell subsets dictates the efficacy of a therapeutic vaccine against prescription opioid abuse. J. Immunol. 194, 5926–5936. 10.4049/jimmunol.1500385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baruffaldi F, Kelcher AH, Laudenbach M, Gradinati V, Limkar A, Roslawski M, Birnbaum A, Lees A, Hassler C, Runyon S, and Pravetoni M. (2018). Preclinical efficacy and characterization of candidate vaccines for treatment of opioid use disorders using clinically viable carrier proteins. Mol. Pharm. 15, 4947–4962. 10.1021/acs.molpharmaceut.8b00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robinson C, Baehr C, Schmiel SE, Accetturo C, Mueller DL, and Pravetoni M. (2019). Alum adjuvant is more effective than MF59 at prompting early germinal center formation in response to peptide-protein conjugates and enhancing efficacy of a vaccine against opioid use disorders. Hum. Vaccin. Immunother. 15, 909–917. 10.1080/21645515.2018.1558697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crouse B, Robinson C, Huseby Kelcher A, Laudenbach M, Abrahante JE, and Pravetoni M. (2020). Mechanisms of interleukin 4 mediated increase in efficacy of vaccines against opioid use disorders. NPJ Vaccines 5, 99. 10.1038/s41541-020-00247-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tomayko MM, Steinel NC, Anderson SM, and Shlomchik MJ (2010). Cutting edge: hierarchy of maturity of murine memory B cell subsets. J. Immunol. 185, 7146–7150. 10.4049/jimmunol.1002163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Palm A-KE, and Henry C. (2019). Remembrance of things past: long-term B cell memory after infection and vaccination. Front. Immunol. 10, 1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chevrier S, Emslie D, Shi W, Kratina T, Wellard C, Karnowski A, Erikci E, Smyth GK, Chowdhury K, Tarlinton D, and Corcoran LM (2014). The BTB-ZF transcription factor Zbtb20 is driven by Irf4 to promote plasma cell differentiation and longevity. J. Exp. Med. 211, 827–840. 10.1084/jem.20131831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Y, and Bhattacharya D. (2014). Adjuvant-specific regulation of long-term antibody responses by ZBTB20. J. Exp. Med. 211, 841–856. 10.1084/jem.20131821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eisenberg D, Schwarz E, Komaromy M, and Wall R. (1984). Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J. Mol. Biol. 179, 125–142. 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 58.Langedijk AC, Spinelli S, Anguille C, Hermans P, Nederlof J, Butenandt J, Honegger A, Cambillau C, and Plu€ckthun A. (1999). Insight into odorant perception: the crystal structure and binding characteristics of antibody fragments directed against the musk odorant traseolide. J. Mol. Biol. 292, 855–869. 10.1006/jmbi.1999.3101. [DOI] [PubMed] [Google Scholar]
- 59.Kyzer JL, McGuire M, Park H, Belz TF, Bonakdar R, Janda KD, and Wenthur CJ (2020). Anti-opioid antibodies in individuals using chronic opioid therapy for lower back pain. ACS Pharmacol. Transl. Sci. 3, 896–906. 10.1021/acsptsci.0c00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Raleigh MD, King SJ, Baruffaldi F, Saykao A, Hamid FA, Winston S, LeSage MG, Pentel PR, and Pravetoni M. (2021). Pharmacological mechanisms underlying the efficacy of antibodies generated by a vaccine to treat oxycodone use disorder. Neuropharmacology 195, 108653. 10.1016/j.neuropharm.2021.108653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodarte JV, Baehr C, Hicks D, Liban TL, Weidle C, Rupert PB, Jahan R, Wall A, McGuire AT, Strong RK, et al. (2023). Structures of drug-specific monoclonal antibodies bound to opioids and nicotine reveal a common mode of binding. Structure 31, 20–32.e5. 10.1016/j.str.2022.11.008. [DOI] [PubMed] [Google Scholar]
- 62.Tiller T, Meffre E, Yurasov S, Tsuiji M, Nussenzweig MC, and Wardemann H. (2008). Efficient generation of monoclonal antibodies fromsingle human B cells by single cell RT-PCR and expression vector cloning. J. Immunol. Methods 329, 112–124. 10.1016/j.jim.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McNicholas S, Potterton E, Wilson KS, and Noble MEM (2011). Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. D Biol. Crystallogr. 67, 386–394. 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.de Beer TAP, Berka K, Thornton JM, and Laskowski RA (2014). PDBsum additions. Nucleic Acids Res. 42, D292–D296. 10.1093/nar/gkt940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kabsch W. (2010). XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adams PD, Afonine PV, Bunkó czi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221. 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Emsley P, Lohkamp B, Scott WG, and Cowtan K. (2010). Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Joosten RP, Long F, Murshudov GN, and Perrakis A. (2014). The PDB_REDO server for macromolecular structure model optimization. IUCrJ 1, 213–220. 10.1107/S2052252514009324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Krueger F. (2015). Trim Galore!. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/. [Google Scholar]
- 71.Ewels P, Magnusson M, Lundin S, and Kä ller M. (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, and Li H. (2021). Twelve years of SAMtools and BCFtools. GigaScience 10, giab008. 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lawrence M, Huber W, Pagè s H, Aboyoun P, Carlson M, Gentleman R, Morgan MT, and Carey VJ (2013). Software for computing and annotating genomic ranges. PLoS Comput. Biol. 9, e1003118. 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rainer J, Gatto L, and Weichenberger CX (2019). ensembldb: an R package to create and use Ensembl-based annotation resources. Bioinformatics 35, 3151–3153. 10.1093/bioinformatics/btz031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pagè s H, Carlson M, Falcon S, and Li N. (2020). AnnotationDbi: Manipulation of SQLite-Based Annotations in Bioconductor. 10.18129/B9.Bioc.AnnotationDbi. [DOI] [Google Scholar]
- 77.Amezquita RA, Lun ATL, Becht E, Carey VJ, Carpp LN, Geistlinger L, Marini F, Rue-Albrecht K, Risso D, Soneson C, et al. (2020). Orchestrating single-cell analysis with Bioconductor. Nat. Methods 17, 137–145. 10.1038/s41592-019-0654-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McCarthy DJ, Campbell KR, Lun ATL, and Wills QF (2017). Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186. 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lun ATL, McCarthy DJ, and Marioni JC (2016). A step-by-step workflow for low-level analysis of single-cell RNA-seq data with Bioconductor. F1000Res. 5, 2122. 10.12688/f1000research.9501.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Carlson M. (2019). org.Mm.eg.db: Genome Wide Annotation for Mouse. R Package Version 3.8.2. Bioconductor. http://bioconductor.org/packages/org.Mm.eg.db/. [Google Scholar]
- 81.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, et al. (2015). MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 16, 278. 10.1186/s13059-015-0844-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zager M, et al. (2021). Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29. 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Canzar S, Neu KE, Tang Q, Wilson PC, and Khan AA (2017). BASIC: BCR assembly from single cells. Bioinformatics 33, 425–427. 10.1093/bioinformatics/btw631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alquicira-Hernandez J, and Powell JE (2021). Nebulosa recovers single cell gene expression signals by kernel density estimation. Bioinformatics 37, 2485–2487. 10.1093/bioinformatics/btab003. [DOI] [PubMed] [Google Scholar]
- 85.Ye J, Ma N, Madden TL, and Ostell JM (2013). IgBLAST: an immunoglobulin variable domain sequence analysis tool. Nucleic Acids Res. 41, W34–W40. 10.1093/nar/gkt382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gupta NT, Vander Heiden JA, Uduman M, Gadala-Maria D, Yaari G, and Kleinstein SH (2015). Change-O: a toolkit for analyzing largescale B cell immunoglobulin repertoire sequencing data. Bioinformatics 31, 3356–3358. 10.1093/bioinformatics/btv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wickham H. (2009). ggplot2 (Springer). 10.1007/978-0-387-98141-3. [DOI] [Google Scholar]
- 88.Gu Z, Gu L, Eils R, Schlesner M, and Brors B. (2014). Circlize implements and enhances circular visualization in R. Bioinformatics 30, 2811–2812. 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 89.Wirtz E, Leal S, Ochatt C, and Cross GA (1999). A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89–101. 10.1016/s0166-6851(99)00002-x. [DOI] [PubMed] [Google Scholar]
- 90.Alsford S, and Horn D. (2008). Single-locus targeting constructs for reliable regulated RNAi and transgene expression in Trypanosoma brucei. Mol. Biochem. Parasitol. 161, 76–79. 10.1016/j.molbio-para.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bilbey DL, Salem H, and Grossman MH (1960). The anatomical basis of the straub phenomenon. Br. J. Pharmacol. Chemother. 15, 540–543. 10.1111/j.1476-5381.1960.tb00277.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Aceto MD, McKEAN DB, and Pearl J. (1969). Effects of opiates and opiate antagonists on the Straub tail reaction in mice. Br. J. Pharmacol. 36, 225–239. 10.1111/j.1476-5381.1969.tb09500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nath C, Gupta MB, Patnaik GK, and Dhawan KN (1994). Morphine-induced straub tail response: mediated by central mu2-opioid receptor. Eur. J. Pharmacol. 263, 203–205. 10.1016/0014-2999(94)90543-6. [DOI] [PubMed] [Google Scholar]
- 94.Zarrindast MR, Alaei-Nia K, and Shafizadeh M. (2001). On the mechanism of tolerance to morphine-induced Straub tail reaction in mice. Pharmacol. Biochem. Behav. 69, 419–424. 10.1016/s0091-3057(01)00519-6. [DOI] [PubMed] [Google Scholar]
- 95.Pravetoni M, Pentel PR, Potter DN, Chartoff EH, Tally L, and LeS-age MG (2014). Effects of an oxycodone conjugate vaccine on oxycodone self-administration and oxycodone-induced brain gene expression in rats. PLoS One 9, e101807. 10.1371/journal.pone.0101807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Picelli S, Faridani OR, Bjö rklund AK, Winberg G, Sagasser S, and Sandberg R. (2014). Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181. 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 97.McInnes L, Healy J, Saul N, and Großberger L. (2018). UMAP: Uniform Manifold approximation and projection. J. Open Source Softw. 3, 861. 10.21105/joss.00861. [DOI] [Google Scholar]
- 98.Battye TGG, Kontogiannis L, Johnson O, Powell HR, and Leslie AGW (2011). iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 67, 271–281. 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liebschner D, Afonine PV, Baker ML, Bunkó czi G, Chen VB, Croll TI, Hintze B, Hung LW, Jain S, McCoy AJ, et al. (2019). Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in Phenix. Acta Crystallogr. D Struct. Biol. 75, 861–877. 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tatsumi K, Sakashita G, Nariai Y, Okazaki K, Kato H, Obayashi E, Yoshida H, Sugiyama K, Park S-Y, Sekine J, and Urano T. (2017). G196 epitope tag system: a novel monoclonal antibody, G196, recognizes the small, soluble peptide DLVPR with high affinity. Sci. Rep. 7, 43480. 10.1038/srep43480. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Crystal structure files have been deposited at the Protein DataBank and are publicly available as of the date of publication. Accession numbers are listed in the key resources table. Single-cell RNA-seq data have been deposited at GEO. To request access, contact lead contact. Original western blot images reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE Antibodies | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
|
| ||
| mouse anti-fentanyl monoclonal antibody | M. Pravetoni, University of Washington | N/A |
| mouse anti-fentanyl-FITC monoclonal antibody | this paper | N/A |
| mouse anti-VSG3 monoclonal antibody | Pingeretal., 201714 | N/A |
| goat anti-mouse IgG-HRP | Jackson Immuno Research | Cat#115-035-062; RRID: AB_2338504 |
| streptavidin-BV785 | BioLegend | Cat#405249 |
| rat anti-mouse CD19-BV412 | BioLegend | Cat#115537; RRID: AB_10895761 |
| rat anti-mouse IgG1-BV650 | BioLegend | Cat#406629; RRID: AB_2716013 |
| rat anti-mouse CD138-BV510 | BioLegend | Cat#142521; RRID: AB_2562727 |
| rat anti-mouse GL-7-FITC | BD Pharmingen | Cat#553666; RRID: AB_394981 |
| rat anti-mouse CD38-PE-Cy7 | BioLegend | Cat#102718; RRID AB_2275531 |
| goat anti-mouse IgM-biotin | Jackson ImmunoResearch | Cat#115-065-075, RRID AB_2338566 |
| rat anti-mouse IgD-APC-Cy7 | BioLegend | Cat#405716; RRID AB_10662544 |
| streptavidin-HRP | ThermoFisher Scientific | Cat#N100 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| E. coli DH5a | Thermofisher | Cat#18265017 |
| E. coli BL21 (DE3) | Thermofisher | Cat#C600003 |
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| fentanyl-LPSTGG | this paper | N/A |
| FITC | Abcam | Cat#ab102884 |
| FITC-LPSTGG | this paper | N/A |
| fen-G4 peptide | this paper | N/A |
| fentanyl-citrate | Sigma-Aldrich | Cat#990-73-8 |
| Fentanyl-citrate | West-Ward | N/A |
| fentanyl solution | Sigma-Aldrich | Cat#F-013 |
| fentanyl-hapten-biotin | this paper | N/A |
| fentanyl | LGC | LGCAMP0528.00-02 |
| fentanyl-d5 | LGC | LGCAMP0528.80-02 |
| fentanyl-PE | Marco Pravetoni, University of Washington | N/A |
| decoy-PE-AF647 | Marco Pravetoni, University of Washington | N/A |
| EDC crosslinker | ThermoFisher Scientific | Cat#22980 |
| ABTS tablets | Roche | Cat#11112422001 |
| Srt-VSG3 protein | this paper | N/A |
| Recombinant RNase Inhibitor | Takara Bio Europe S.A.S | Cat#2313A |
| Penta-His-biotin conjugate | Qiagen | Cat#34440 |
| Sortase A from S. pyogenes | this paper | N/A |
| Trizma® Hydrochlorid -hydrochlorid | Sigma-Aldrich | Cat#T3253 |
| Sodium chloride | Carl-Roth | Cat# 3957.1 |
| Imidazole | Carl-Roth | Cat#X998.4 |
| 2-Mercaptoethanol | Sigma-Aldrich | Cat#M6250 |
| 1,4-Dithiothreitol (DTT) | Carl-Roth | Cat#6908.1 |
| LB-broth (Miller) | Sigma-Aldrich | Cat#L3522-250G |
| HEPES | Carl-Roth | Cat#9105 |
| Dulbecco’s PBS | Sigma-Aldrich | Cat#D8537 |
| Ammonium sulfate | Carl-Roth | Cat#3746 |
| N-(2-Acetamido)-iminodiacetic acid | Sigma-Aldrich | Cat#00307 |
| Polyethelene glycol 400 | Merck | Cat#8.07485.1000 |
| Polyethelene glycol 6000 | VWR | Cat#26603-293 |
| Glycerol | Carl-Roth | Cat#3783 |
| Ethanol | Sigma-Aldrich | Cat#32205-M |
| Sodium acetate | Carl-Roth | Cat#6773 |
| Triethanolamine | Sigma-Aldrich | Cat#90279 |
| Magnesium sulfate heptahydrate | Merck | Cat#1.05886.0500 |
| Sodium citrate tribasic dihydrate | Sigma-Aldrich | Cat#71402 |
| Ethelene glycol | MP | Cat#151089 |
| Hexylene glycol (MPD) | Sigma-Aldrich | Cat#112100 |
| HMI-9 Medium | PAN Biotech | SO-15701 |
| FreeStyle 293 Expression Medium | Gibco | Cat#12338-018 |
| Carbonate-bicarbonate buffer capsules | Sigma-Aldrich | Cat#C3041 |
| 20x PBS Tween 20 | Thermo Scientific | Cat#PI28352 |
| Gelatin from porcine skin | Sigma-Aldrich | Cat#G2500 |
| Octet Streptavidin biosensors | Sartorius | Cat#18-5019 |
| Tramadol hydrochloride | Fagron | 20L09-U10-009586 |
| Naloxone hydrochloride dihydrate | Sigma-Aldrich | N7758 |
| Acetonitrile (LC-MS Grade) | Carl Roth | Cat#AE70.2 |
| Phosphoric acid | Carl Roth | Cat#6366.1 |
| Methanol (ROTISOLV®) | Carl Roth | Cat#AE71.1 |
| Formic acid | Carl Roth | Cat#1EHK.1 |
| Ammonium hydroxide | Carl Roth | Cat#6774.1 |
| Ammonium formate | Carl Roth | Cat#5093.1 |
|
| ||
| Critical commercial assays | ||
|
| ||
| Quantum™ FITC-5 MESF kit | Bangs Laboratories | CAT#555 |
| LIVE/DEAD™ Fixable Blue Dead Cell Stain Kit | Invitrogen | Cat#L23105 |
| Maxima H Minus Reverse Transcriptase, 10000U | ThermoScientific | Cat#EP0752 |
| Kapa High Fidelity Hot Start | Roche | Cat#7958935001 |
| Nextera XT DNA Library Preparation Kit (96SMP) | illumina GmbH | Cat#FC-131-1096 |
| QuikChange Lightning site-directed mutagenesis kit | Agilent technologies | Cat#210513 |
| Pierce™ Protein G Agarose | ThermoFisher Scientific | Cat#20397 |
| Gibco™ 293fectin™ Transfection Reagent | ThermoFisher Scientific | Cat#12347750 |
| Ni-NTA agarose | Qiagen | Cat#30230 |
| SigmaFAST OPD | Sigma-Aldrich | Cat#P9187-5SET |
| Superdex 200 Increase 10/300 GL column | Cytiva | Cat#28-9909-44 |
| Q-Sepharose Fast-Flow | Cytiva | Cat#17-0510-01 |
| NEBuilder Master Mix | New England Biolabs | Cat#M5520AA |
| Phusion HiFi DNA Polymerase | New England Biolabs | Cat#M0530L |
| FreeStyle™ MAX Reagent | Gibco | Cat#16447100 |
| GeneJET Plasmid Miniprep Kit | Thermo Scientific | Cat#K0502 |
| Monarch® DNA Gel Extraction Kit | New England Biolabs | Cat#T1020S |
| Q5® High-Fidelity 2x Master Mix | New England Biolabs | Cat#M0492S |
| AgeI-HF® | New England Biolabs | Cat#R3552S |
| SalI-HF® | New England Biolabs | Cat#R3138S |
| BsiWI-HF® | New England Biolabs | Cat#R3553S |
| EcoRI-HF® | New England Biolabs | Cat#R3101S |
|
| ||
| Deposited data | ||
|
| ||
| Crystal structures | Protein DataBank | PDB codes: 7QT0, 7QT2, 7QT3, 7QT4 |
| RNA-seq data | GEO | Upon request |
|
| ||
| Experimental models: Cell lines | ||
|
| ||
| FreeStyle 293F | ThermoFisher Scientific | Cat#R79007 |
|
| ||
| Experimental models: Organisms/Strains | ||
|
| ||
| C57BL/6JRj | Javier-labs | Cat#C57BL/6JRj |
| Trypanosoma brucei brucei Lister 427 2T1 Srt-VSG3-S317A GPI-PLC WT | Pingeretal., 201714 | N/A |
| Trypanosoma brucei brucei Lister 427 Srt-VSG3-S317A GPI-PLC −/− | this paper | N/A |
|
| ||
| Oligonucleotides | ||
|
| ||
| dNTP Mix (10 mM each)-1 mL | ThermoFisher Scientific | Cat#R0192 |
| Oligonucleotides | See Table S2 | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| IGγ1-, IGκ- or IGλ-expression vectors | Tiller et al., 200862 | N/A |
| Fab heavy and light chain expression vectors | Mirjana Lilic, The Rockefeller University | N/A |
| Sortase A from Streptococcus pyogenes with N-terminal His6-Tag in pET28a expression vector | this paper | N/A |
|
| ||
| Software and algorithms | ||
|
| ||
| FlowJo v10.0.8 | Tree Star | https://www.flowjo.com/solutions/flowjo/downloads |
| GraphPad Prism v6.07 | GraphPad | http://www.graphpad.com |
| R v4.0.3 and v4.1 | The R project for statistical computing | http://www.R-project.org/ |
| FACSDiVa v8.0.1 | Becton Dickinson | Cat#659528 |
| Adobe Illustrator CS6 v16.0.3 | Adobe | http://www.adobe.com/de/products/illustrator.html |
| Adobe Photoshop 2021 v22.0.0 | Adobe | https://www.adobe.com/de/products/photoshop |
| Microsoft PowerPoint v16 | Microsoft | https://www.microsoft.com/de-de/microsoft-365/powerpoint |
| Microsoft Excel v16 | Microsoft | https://www.microsoft.com/de-de/microsoft-365/excel |
| CCP4MG v2.10.11 | McNicholas et al., 201163; de Beeretal., 201 464; https://pymol.org/2/ | https://www.ccp4.ac.uk/MG/ |
| Biorender | Biorender | https://biorender.com/ |
| XDS | Kabschetal., 201065 | http://xds.mpimf-heidelberg.mpg.de |
| Phaser | McCoy et al., 200766 | https://www.phenix-online.org |
| Phenix | Adams et al., 201067 | https://www.phenix-online.org |
| Coot | Emsleyetal., 201068 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| PDB-redo | Joosten et al., 201469 | https://pdb-redo.eu/ |
| PyMol v2.4.2 | N/A | https://pymol.sourceforge.net |
| TrimGalore v0.6.4_dev | Krueger et al., 201570 | http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ |
| MultiQC v0.4 | Ewelsetal., 201671 | https://multiqc.info/ |
| FastQC v0.11.9 | N/A | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ |
| Samtools v1.9 | Danecek et al., 202172 | http://www.htslib.org/ |
| STAR v2.6.0a | Dobinetal., 201373 | https://github.com/alexdobin/STAR |
| Bioconductor v3.12 | Bioconductor Software Packages | https://bioconductor.org/packages/3.12/bioc/ |
| Bioconductor v3.14 | Bioconductor Software Packages | https://bioconductor.org/packages/3.14/bioc/ |
| GenomicFeatures v1.42.3 | Lawrence et al., 201374 | https://bioconductor.org/packages/3.12/bioc/html/GenomicFeatures.html |
| Ensembldb v2.14.1 | Raineretal., 201975 | https://bioconductor.org/packages/3.12/bioc/html/ensembldb.html |
| GenomicAlignments v1.26.0 | Lawrence et al., 201374 | https://bioconductor.org/packages/3.12/bioc/html/GenomicAlignments.html |
| AnnotationDbi v1.52.0 | Pagès et al., 202076 | https://bioconductor.org/packages/3.12/bioc/html/AnnotationDbi.html |
| SingleCellExperiment v1.16.0 | Amezquita et al., 202077 | https://bioconductor.org/packages/3.14/bioc/html/SingleCellExperiment.html |
| scater v1.22 | McCarthy et al., 201778 | https://bioconductor.org/packages/3.14/bioc/html/scater.html |
| scran v1.22.1 | Lunetal., 201679 | https://bioconductor.org/packages/3.14/bioc/html/scran.html |
| org.Mm.eg.db v3.12.0 | Carlson etal.,201980 | https://bioconductor.statistik.tu-dortmund.de/packages/3.12/data/annotation/html/org.Mm.eg.db.html |
| rLiger v1.0.0 | Welch etal., 201941 | https://www.rdocumentation.org/packages/rliger/versions/1.0.0 |
| MAST v1.16.0 | Finaketal., 201581 | https://bioconductor.org/packages/3.12/bioc/html/MAST.html |
| Seurat v4.0 | Haoetal., 202182 | https://satijalab.org/seurat/ |
| BASIC v1.5.0 | Canzar et al., 201783 | https://people.cs.uchicago.edu/~aakhan/BASIC/ |
| Blast v2.11.0 | Alquicira-Hernandez et al., 202184 | https://blast.ncbi.nlm.nih.gov/Blast.cgi?CMD=Web&PAGE_TYPE=BlastNews |
| Igblast v1.16.0 | Ye etal., 201385 | https://ncbi.github.io/igblast/ |
| Change-O v1.0.0 | Gupta etal.,201586 | https://changeo.readthedocs.io/en/stable/ |
| Shazam v1.1.0 | Gupta etal.,201586 | https://shazam.readthedocs.io/en/stable/#shazam |
| ggplot2 v3.3.5 | Wickham, 200987 | https://github.com/tidyverse/ggplot2/releases/tag/v3.3.5 |
| gridExtra v2.3 | B. Auguie | https://cran.r-project.org/web/packages/gridExtra/index.html |
| ggrepel v0.9.1 | K. Slowikowski | https://cran.r-project.org/web/packages/ggrepel/index.html |
| Circlize vO.4.13 | Gu etal., 201488 | https://mran.revolutionanalytics.com/snapshot/2021-11-01/web/packages/circlize/index.html |
| Octet DataAnalysis v12.0.2.3 | Sartorius | N/A |
| LABORAS v2 | Metris | https://www.metris.nl/en/products/laboras/laboras2/ |
| Analyst v1.6 | Sciex | https://de-store.sciex.com/de/EUR/software |
| MassHunter | Agilent | https://www.agilent.com/en/product/software-informatics/mass-spectrometry-software |