Abstract
Expressed on epidermal Langerhans cells, CD1a presents a range of self-lipid antigens found within the skin. However, the extent to which CD1a presents microbial ligands from bacteria colonizing the skin is unclear. Here, we identified CD1a-dependent T cell responses to phosphatidylglycerol (PG), a ubiquitous bacterial membrane phospholipid, as well as to lysyl phosphatidylglycerol (lysylPG), a modified PG, present in several gram-positive bacteria, and highly abundant in Staphylococcus aureus. The crystal structure of the CD1a-PG complex showed that the acyl chains were buried within the A′- and F′-pockets of CD1a, while the phosphoglycerol headgroup remained solvent-exposed in the F′-portal and was available for T cell receptor contact. Using lysylPG and PG-loaded CD1a tetramers, we identified T cells in peripheral blood and in skin that respond to these lipids in a dose dependent manner. Tetramer+ CD4+ T cell lines secreted Th2 cytokines in response to phosphatidylglycerols as well as to co-cultures of CD1a+ dendritic cells and Staphylococcus bacteria. The expansion in atopic dermatitis (AD) patients of CD4+ CD1a-(lysyl)PG tetramer+ T cells suggest a response to lipids made by bacteria associated with atopic dermatitis, and provide a link supporting involvement of PG-based lipid-activated T cells in AD pathogenesis.
INTRODUCTION
The lipid antigen presenting molecule CD1a is constitutively expressed at extremely high density on Langerhans cells1, the resident dendritic cell type in stratified epithelia of the skin, mouth, and genital mucosa2. The prominent presence of CD1a specifically localized within human epithelia suggests a possible role for lipid antigen recognition by T cells in barrier immunity and homeostasis. In addition, CD1a-expressing Langerhans cells are in close proximity to the epithelial microbiota, suggesting that CD1a may have routine access to microbial lipids from skin surface resident bacteria and fungi and present these to CD1a-restricted T cells3. Despite these anatomical links between CD1a and microbiota, as well as rapidly growing insights into skin microbiome4, these connections have not been addressed experimentally. To date, only one CD1a-presented bacterial lipid antigen has been identified: a lipopeptide known as dideoxymycobactin (DDM) from the lung pathogen, Mycobacterium tuberculosis 5, 6. Since CD1a is most abundantly expressed on Langerhans cells, which survey the skin microbiome7, we wanted to use CD1a tetramers to see whether the human T cell repertoire also contains T cells that recognize lipids present in common skin commensals and pathogens.
We focused on phosphatidylglycerols (PG), which are highly abundant phospholipids in the membranes of essentially all bacteria 8, 9. Further, PG can be modified to form lysyl-phosphatidylglycerol (lysylPG), which is the product of the lysine addition to PG by the bacterial multi peptide resistance factor (MprF) enzyme, expressed in several gram-positive bacteria10, 11. The addition of a lysine moiety to PG creates a positive charge on the molecule, rendering the bacterial membrane more resistant to cationic anti-microbial peptides10, 12, and lysylPG is therefore considered a virulence factor. The membrane of the important human pathogen and common skin colonizer Staphylococcus aureus (S. aureus), is comprised mainly of PG, lysyl PG, cardiolipin and diglucosyldiacylglycerol (DGDG), where lysylPG and PG are the major structural lipids13, 14. Depending on the strain, lysylPG can constitute between 20–40% of all phospholipids in the S. aureus membrane 11. Although abundant in S. aureus, lysylPG is not unique to this pathogen and can also be produced by other gram-positive bacteria, including other MprF-expressing Staphylococcus species, such as S. epidermidis. Given that Staphylococcus species make up approximately 25% of bacteria on the skin15, both PG and lysylPG are major components of the host skin microbiome. Since MprF is not expressed in mammalian cells, lysylPG can be considered a true foreign lipid. Furthermore, in mammalian cells, PG is present only at trace levels and localized to mitochondrial membranes16, supporting the notion that PG is also primarily a foreign lipid. Therefore, we sought to determine whether lysylPG and unmodified PG can be presented in the context of CD1a and elicit a T cell response.
Using lysylPG and PG-loaded CD1a tetramers, we identified a previously undescribed population of human T cells that responds to these phospholipids in a CD1a-dependent manner. The crystal structures of CD1a-PG confirmed that PG-based lipids are bona fide CD1a ligands bound in the cleft. Using sorted CD1a-lysylPG and PG tetramer+ T cell populations ex vivo and in vitro, we gain insight in the gene expression profiles of these T cells, their response to Staphylococcus bacteria, and expansion in atopic dermatitis patients. Overall, this study identifies a novel bacterial phospholipid-reactive T cell population that likely represents part of the human T cell response to common gram-positive bacteria and is implicated in inflammatory skin disease.
RESULTS
PG and lysylPG are CD1a binding ligands
To investigate if CD1a-restricted T cells respond to bacteria, we sought proof of principle that CD1a can present known lipids from the skin microbiome. We chose the lipids PG and lysylPG because of their abundance in the membranes of Staphylococcal species. We developed and validated fluorescently-labeled CD1a tetramers by first testing with a CD1a-autoreactive T cell line originally isolated from human skin (DermT)17, 18. This T cell line responds to CD1a-transfected K562 cells in a CD1a-dependent manner that does not require exogenous lipid (Figure 1a). A recent study demonstrated that CD1a-endo tetramers stained CD1a-autoreactive T cells from human skin 18. We confirmed this observation by staining the DermT cell line with CD1a-mock tetramers, revealing a population of CD4+ tetramer+ T cells. (Figure 1b). Using fluorescence-activated cell sorting (FACS), this tetramer+ population was purified from the line and expanded in vitro, resulting in a cell population highly enriched for tetramer positive cells. Prior data showed that small hydrophobic lipids, including free fatty acids, nest inside CD1a and allow a CD1a-autoreactive TCR to interact with CD1a 19, 20. Consistent with these data, CD1a tetramers treated with a short chain C16:1 fatty acid bound the CD1a-reactive DermT cell population, at a similar mean fluorescence intensity (MFI) as the CD1a-mock (Figure 1c).
Figure 1. CD1a tetramers identify populations of human T cells.
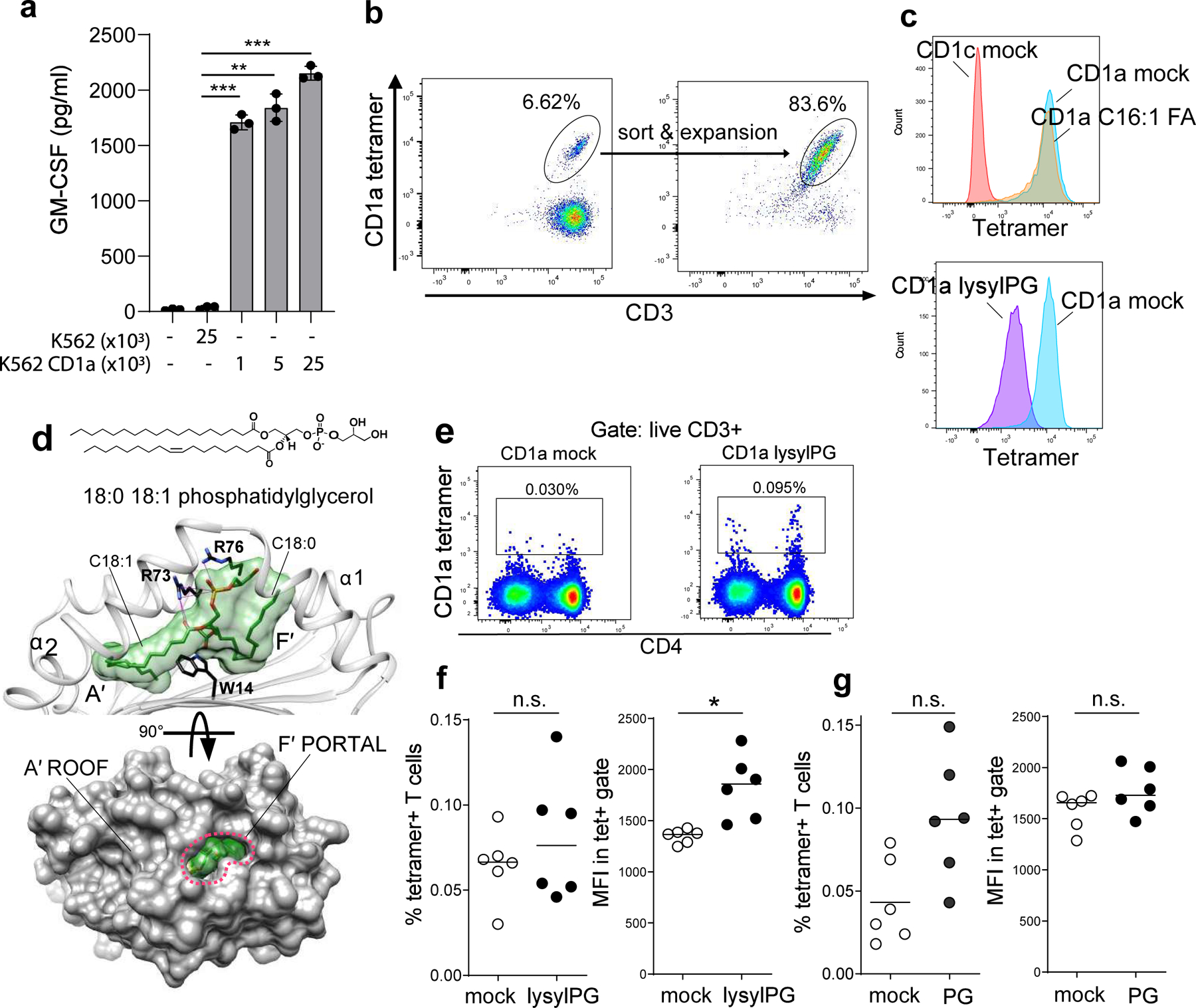
(a) GM-CSF release measured by ELISA in the supernatant of co-culture of a CD1a-autoreactive T cell line (DermT) with CD1a transfected K562 cells (K562 CD1a) and K562 transfected with empty vector (K562, control). Depicted is the mean ± SD of triplicate cultures. Results are representative of >3 independent experiments. P-values were based on the paired two-tailed t-test. ** p<0.01, *** p<0.001 (p-values: p=0.0008, p=0.002, p=0.003). (b) Flow cytometric analysis of DermT cell line stained with mock-loaded CD1a tetramer before and after FACS sorting and expansion. Percentage of tetramer+ T cells indicated. (c, upper panel) Histograms of DermT tetramer staining with mock loaded CD1c tetramers (CD1c mock), CD1a mock and C16:1 fatty acid loaded CD1a tetramers. (c, lower panel) Histograms of DermT tetramer staining with CD1a mock and lysylPG-loaded CD1a tetramers. (d, upper panel) Crystal structure of CD1a-PG at 2.0 Å. The position of phosphatidylglycerol (green) in the binding cleft of CD1a (grey) formed by the α1 and α2 helices. A’ and F’ pockets are filled with the oleyl and stearyl acyl tails, respectively. The side chain of Trp14 at the bottom of the cleft is shown in black. The side chains of Arg73 and Arg76 are shown in black and their electrostatic contacts with PG are depicted as magenta lines. (d, lower panel) Surface representation of the membrane-distal segment of CD1a showing the F’ portal (marked with red dashed lines) and the solvent-exposed part of the lipid ligand surface (green). (e) Representative dot plot example of staining of PBMC ex vivo with mock-treated CD1a and lysylPG-treated CD1a tetramers, gated on live CD14-/CD19-/CD3+ T cells according to gating strategy in Supplementary Figure 1a. (f, left panel) Percentage of CD1a tetramer+ T cell among total live T cells in 6 healthy donors, and (f, right panel) mean fluorescence intensity (MFI) of the tetramer+ T cells in the tetramer gate. Horizontal bars indicate median values. P-values were based on the Wilcoxon matched-pairs signed rank test, two-sided *p<0.05 (p-value: p=0.03. n.s. = not significant. (g, left panel) Percentage of tetramer+ T cells staining with mock-treated CD1a and PG-treated CD1a tetramers, and (g, right panel) MFI of the T cell in the tetramer gate, in an additional set of 6 donors. Horizontal bars indicate median values. P-values were based on the Wilcoxon matched-pairs signed rank test, two-sided n.s. = not significant.
We next treated the CD1a monomer with lysylPG, and subsequently tetramerized the lipid-treated biotinylated monomers. CD1a tetramer staining of the DermT cell line was significantly decreased (MFI from 10,003 to 1,848) when CD1a was treated with lysylPG. The decreased tetramer binding suggests that lysylPG binds to CD1a and reduces the DermT cell TCR interaction with CD1a. PG loading of CD1a tetramers similarly reduced the binding of the tetramer to DermT (Extended Data Figure 1a). In contrast to this result, but in line with prior observations 18, phosphatidylcholine (PC) did not affect the CD1a tetramer binding of the CD1a-autoreactive TCR. Together these observations with CD1a-autoreactive T cells suggest that PG and lysylPG bind CD1a and displace permissive self lipids from the antigen binding cleft.
Crystal structure confirms binding of PG in the CD1a antigen binding cleft
We solved the crystal structure of CD1a-PG with intermediate chain fatty acyl groups (18:0, 18:1) (Figure 1d, Table 1, Extended Data Figure 1b). The oleoyl (C18:1) acyl tail at the position sn-2 was anchored deep inside the cleft of CD1a fully occupying the available volume of the hydrophobic A’ pocket. The stearyl (C 18:0) chain at the position sn-1 sharply turned 90° around Trp14 at the bottom of the cleft and partially filled the F’ pocket (Figure 1d). In the CD1a-PG complex, the phosphate remained buried within the neck of the F′ pocket, near the edge of the A′ roof, establishing electrostatic contacts with Arg73 and Arg76. The remaining part of the polar headgroup occupied the F′ portal, the only solvent accessible segment of the binding cleft of CD1a. Due to the modest size of the glycerol moiety only ~3% of the lipid surface area was solvent exposed (. Overall, the crystal structure of the CD1a-PG complex confirmed that PG-based lipids are bona fide CD1a ligands.
Table 1.
Data collection and refinement statistics on CD1a-phosphatidylgycerol binary complex structure.
| CD1a-PG 18:0 18:1 | |
|---|---|
| Data collection statistics | |
| Temperature (K) | 100 |
| Wavelength (Å) | 0.954 |
| Resolution Range (Å) | 44.9 – 2.0 (2.1 – 2.0) |
| Space group | P 21 21 21 |
| Unit cell | |
| a, b, c (Å) | 42.2 89.8 105.2 |
| α,β,γ(°) | 90.0, 90.0, 90.0 |
| Total reflections | 373755 (27820) |
| Unique reflections | 27852 (1996) |
| Multiplicity | 13.4 (13.9) |
| Completeness (%) | 100 (100) |
| Mean I/σ1 | 11.9 (1.8) |
| Rp.i.m.a (%) | 5.1 (46.3) |
| Wilson B-factor (Å2) | 28 |
| Refinement statistics | |
| Rworkb (%) | 19.3 |
| Rfreec (%) | 23.0 |
| Non-hydrogen atoms | 3469 |
| macromolecules | 3061 |
| ligands | 101 |
| solvent | 307 |
| Protein residues | 378 |
| rmsd bonds (Å) | 0.005 |
| rmsd angles (°) | 0.82 |
| Ramachandran | |
| favored (%) | 98.1 |
| allowed (%) | 1.6 |
| outliers (%) | 0.3 |
| Average B-factor (Å2) | 40 |
| protein | 39 |
| ligand | 54 |
| water | 39 |
Statistics for the highest-resolution shell are shown in parentheses.
Rp.i.m = Σhkl [1/(N-1)]1/2 Σi | Ihkl, i - <Ihkl> | / Σhkl <Ihkl>
Rwork = ( Σ | |Fo| - |Fc| | ) / ( Σ |Fo| ) - for all data except as indicated in footnote 3.
4% of data was used for the Rfree calculation
CD1a tetramers identify a population of antigen-specific human T cells.
We next used the CD1a-lysylPG and CD1-PG tetramers to determine if the peripheral blood T cell repertoire contains T cells that specifically bind these tetramers. CD1a-mock tetramer stained a small percentage of T cells (range 0.03 – 0.093% of all T cells) (Figure 1e.), which is consistent with the presence of CD1a-autoreactive T cells in peripheral blood 17, 21. Staining with the lysylPG-treated CD1a tetramer also stained a fraction of the peripheral blood T cells (range 0.046 – 0.14%), which was not significantly larger than the percentage of T cells binding the CD1a-mock tetramers, but showed higher mean fluorescence intensity (MFI), where majority of tetramer+ T cells expressed the CD4 co-receptor (Figure 1f, Extended Data Figure 1c). PG-treated CD1a tetramers also stained a population of tetramer+ T cells, as detected in PBMC from a second set of 6 donors (range 0.043 – 0.149%, Figure 1g). Overall, the detection of human T cells binding lysylPG and PG-treated CD1a tetramers, suggests that, human blood contains CD1a-restricted T cells that recognize bacterial phospholipids as antigens.
We sought to confirm if the observed tetramer+ population indeed represented T cells that are activated by CD1a-lysylPG complexes. As a first step, we screened additional blood bank donors (n = 21) for the presence of a detectable (> 0.02% of T cells) tetramer+ population ex vivo using lysylPG-treated CD1a tetramers. From the donors with the highest percentage of CD1a tetramer staining T cells (n=14) we sorted the tetramer+ T cells by FACS, which resulted in the successful enrichment and expansion of CD1a-lysylPG staining T cell lines from half of these donors (n = 7, donors 213, 325, 350, 834, 921, 966, 003, Figure 2a). The tetramer+ T cell population was enriched by a second round of sorting and expansion, yielding T cell cultures with up to >90% of the T cells binding the lysylPG-treated CD1a tetramers (Figure 2b). Similar to the ex vivo analysis (Extended Data Figure 1c), the majority of the enriched and purified tetramer+ T cell populations expressed the CD4 co-receptor. Consequently, we focused on the CD4+ tetramer+ T cell subset for follow up experiments.
Figure 2. CD1a-lysylPG tetramer+ T cells functionally respond to lysylPG antigen.
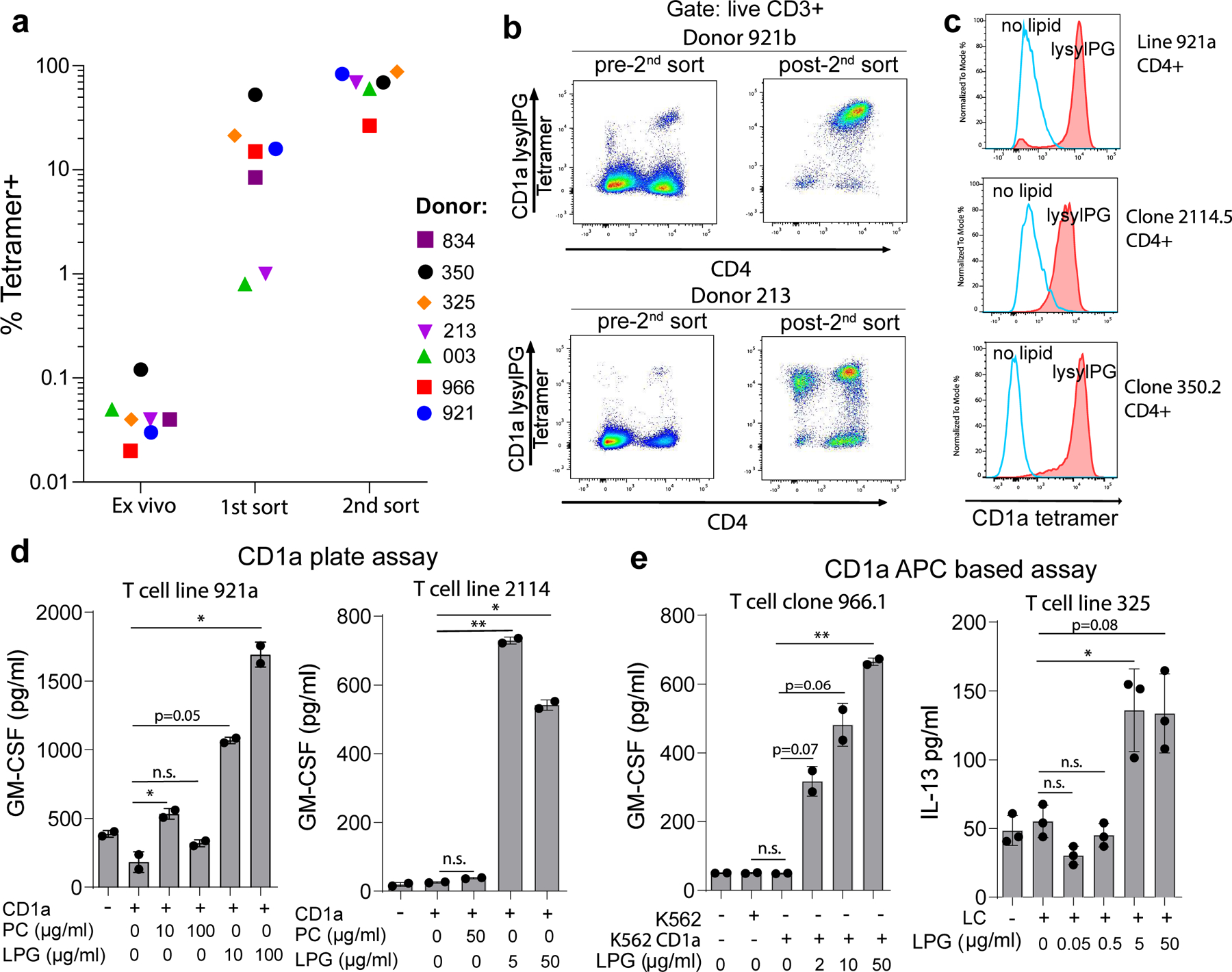
(a) The frequency of T cells staining with lysylPG-treated CD1a tetramers among live T cells from healthy donors (n=7) is indicated directly ex vivo, after 1st sort and expansion, and after a 2nd sort and expansion. Gating strategy for ex vivo tetramer staining according to Supplementary Figure 1a. (b) Two examples of tetramer-based enrichment show tetramer staining prior to the 2nd sort and after sort and expansion, gated on live CD3+ T cells. (c) Histograms of CD1a tetramer staining of sorted CD4+ T cell line (921a) and clones (clones 2114.5 and 350.2): staining with lysylPG treated CD1a tetramers (closed histograms) and mock-treated CD1a tetramers (open histograms). (d) T cell lines 921a and 2114 were incubated with plate bound CD1a loaded with indicated lipids: phosphatidylcholine (PC) and lysylPG. GM-CSF was measured in the supernatant after 24 hrs by ELISA. Assays were set up in duplicate, and depicted is the mean ± SD per condition. Results are representative of 2 independent experiments. P-values were based on the paired two-tailed t-test, * p<0.05, ** p<0.01, n.s. not significant (p-values assay 921a: p=0.048, p=0.17, p=0.050, p=0.049. P-values assay 2114: p=0.16, p=0.008, p=0.011) (e) CD1a transfected K562 cells incubated with increasing concentrations of lysylPG were co-cultured with T cell clone 966.1.4+ and T cell line 325. GM-CSF and IL-13 were measured in supernatant after 24 hrs by ELISA. Assays were set up in duplicate (clone 966.1.4+) or triplicate (T cell line 325), and depicted is the mean ± SD per condition. Results are representative of 2 independent experiments. P-values were based on the paired two-tailed t-test, * p<0.05, ** p< 0.01, n.s. not significant. (P-values assay 966.1: p=0.69, p=0.069, p=0.064, p=0.009. P-values 325: p=0.12, p=0.42, p=0.045, p=0.078).
CD1a tetramer staining of the sorted CD4+ T cell lines and clones revealed a staining pattern opposite to that of the CD1a-autoreactive T cells (Figure 2c). The specificity of the CD1a restriction was confirmed by the absence of staining of T cells with lysylPG treated CD1b, CD1c or CD1d tetramers, while the loading of lysylPG on these isoforms was confirmed by isoelectric focusing (Extended Data Figure 2a, 2b).
To determine if direct functional recognition of lysylPG underlies the binding of lysylPG-treated CD1a tetramers to the T cells, we performed an assay in which recombinant biotinylated CD1a was bound to neutravidin plates and loaded with increasing concentrations of lysylPG. Tetramer+ T cells specifically released cytokine in response to lysylPG treated CD1a, and not to untreated CD1a, nor CD1a treated with a control phospholipid (PC) (Figure 2d). Similarly, in antigen presenting cell based assays, tetramer+ T cells responded to lysylPG when loaded on K562 CD1a cells or on Langerhans cells (Figure 2e), suggesting that lysylPG is efficiently loaded onto and presented by CD1a on an antigen presenting cell line. Overall, CD1a-lysylPG reactive CD4+ T cells can be reproducibly isolated from the peripheral blood of unrelated donors, and therefore represent a bona fide CD1a-restricted T cell subset.
Phospholipid specificity of CD1a-lysylPG tetramer+ T cells
To gain more insight in the specificity and potential phospholipid cross-reactivity of the CD1a lysylPG tetramer+ T cells, we developed a set of CD1a tetramers loaded with common phospholipids present in mammalian cells, including phosphatidic acid (PA), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI) and phosphatidylserine (PS). We further included CD1a-PG tetramers (Figure 3a), which have the core phosphoglycerol epitope but not the lysyl moiety in lysyl-PG. Using these tetramers, we screened the 921a T cell line (pre 2nd sort), which contained a population of CD1a-lysylPG staining CD4+ T cells. As expected, the line did not stain with CD1a-mock tetramers, nor did it stain with PC, PE or PS loaded tetramers (Figure 3b). CD1a tetramers loaded with PA appeared to show weak binding of all the cells, and was therefore not considered antigen-specific. PI loaded tetramers did appear to stain the subset of CD4+ T cells, however, at a MFI lower than lysylPG. PG loaded tetramers stained the cells at similar MFI as lysylPG loaded tetramers .
Figure 3. Phospholipid reactivity of T cells binding lysylPG-treated CD1a tetramers.
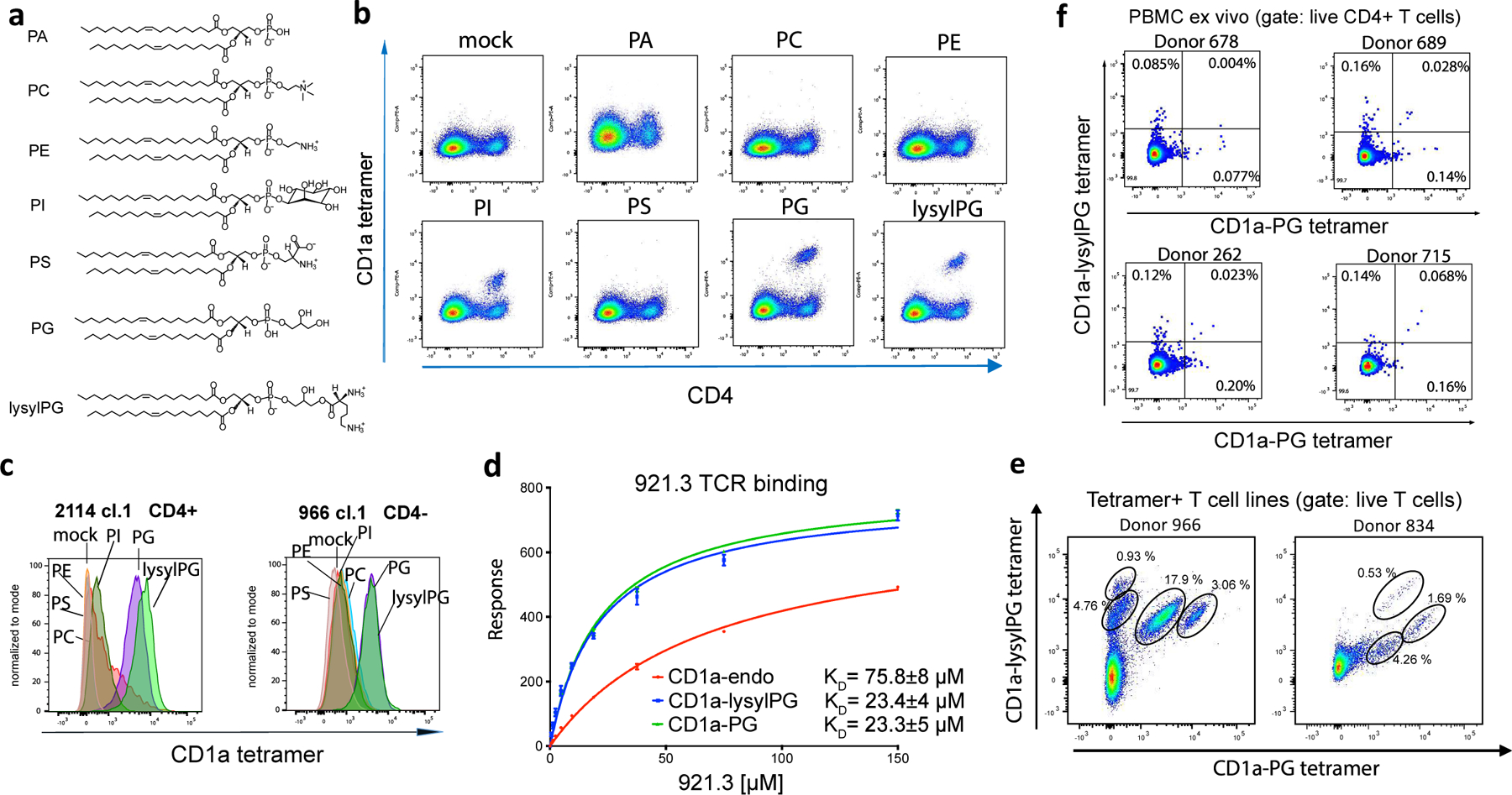
(a) Phospholipid structures: phosphatidic acid (PA), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidylserine (PS), phosphatidylglycerol (PG), and lysylphosphatidylglycerol (lysylPG). (b) Flowcytometric analysis of CD1a-lysylPG reactive T cell line (921a) stained with CD1a tetramers loaded with indicated phospholipids. Gated for live cells. Results were representative of >3 independent experiments. (c) Overlapping histograms of flowcytometric analysis of CD1a-lysylPG reactive T cell clones (2114.1, 966.1.4-) stained with CD1a tetramers loaded with indicated phospholipids. (d) Surface plasmon resonance was used to measure the affinity of the interaction between soluble 921.3 TCR and immobilized untreated CD1a (CD1a-endo, red) and PG (green) or lysylPG (blue)- treated CD1a. Shown binding curves correspond to one experiment with two injections for each concentration of the TCR (from 0 to 150 μM). Equilibrium KD was calculated from two independent measurements. (e) Flowcytometric analysis of T cell lines from donors 834 and 966 co-stained with CD1a-lysylPG PE-labeled tetramers and CD1a-PG APC-labeled tetramers. Cells were gated for live CD4+ T cells, and distinct populations of tetramer+ T cells are indicated. (f) Flowcytometric analysis of PBMC stained ex vivo with CD1a-lysylPG PE-labeled tetramers and CD1a-PG APC-labeled tetramers. Cells were gated for live CD14-/CD19-/CD3+/CD4+ T cells according to the gating strategy in Supplementary Figure 1a. Percentages of single and double tetramer staining cells are indicated in the quadrants.
This pattern of tetramer binding was not unique to this T cell line, since an independently isolated CD1a-lysylPG reactive CD4+ T cell clone (2114.1) from a different donor, showed a similar phospholipid recognition hierarchy (Figure 3c). Also, a CD4-negative T cell clone (from donor 966) bound both the PG and lysylPG loaded tetramers . The tetramer staining with CD1a-PG also translated into functional reactivity, namely in an assay with recombinant CD1a bound to a plate, the, lysylPG reactive T cell line (921a) responded to both lysylPG as well as PG (Extended Data Figure 2c). We obtained the T cell receptor (TCR) sequence of a T cell clone from the 921 line and generated a recombinant soluble 921.3 αβ TCR. Using surface plasmon resonance we confirmed that 921.3 TCR was directly recognizing CD1a loaded with PG-based lipid antigens in vitro manifested by nearly identical binding affinity towards CD1a-lysylPG (KD = 23.4±4 μM) and CD1a-PG (KD= 23.3±5 μM) (Figure 3d).
Altogether our data suggested that the phosphoglycerol headgroup was required for recognition, and the lysine head group present on the added antigen was tolerated but not required. We also considered an alternate explanation, namely that the lysine group is hydrolyzed during loading or storage, yielding PG. To distinguish between the two possibilities, we investigated the stability of lysylPG during and after loading onto CD1a.
pH dependent stability of lysylPG
Prior studies have suggested that the lysine modification of PG is relatively stable at acidic pH, whereas at higher pH (pH > 7) the lysine headgroup is sensitive to hydrolysis22. We confirmed this pH dependent stability by high performance liquid chromatography coupled with Quadrupole-Time-of Flight mass spectrometry (HPLC-QToF-MS) (Extended Data Figure 3a). The pH dependent stability of free lysylPG suggested that lipid loading of CD1a must be performed at low pH to minimize hydrolysis and ensure the formation of intact lysylPG-CD1a complexes. Indeed, isoelectric focusing (IEF) data showed that loading at pH 8 resulted in a band corresponding to the CD1a-lipid complex shifting towards the -1 charge, similar to CD1a-PG complexes (Extended Data Figure 3b). This shift was not observed when the lysylPG was loaded at pH 5.5, suggesting that loading at low pH resulted in intact CD1a-lysylPG complexes. This cleavage reaction was more quantitatively measured by HPLC-MS analysis of lipids eluted from CD1a, showing that the low pH loading has no significant effect on lysylPG/PG ratio, similar to the input lipids, in which lysylPG was >90% intact (Extended Data Figure 3c).
To determine if lysylPG is stable once bound to CD1a, we incubated CD1a protein that was loaded with lysylPG at pH 5.5 and was confirmed to contain high lysylPG/PG ratio, overnight at pH 5.0, pH 7.0, and pH 9.0 and analyzed by IEF (Extended Data Figure 3d). There was no shift observed in CD1a-lipid complex, and IEF did not reveal a band at -1 charge, which would correspond to CD1a-PG. This suggested that once bound to CD1a, lysylPG is not readily hydrolyzed to PG, even at neutral or high pH.
Dual tetramer staining of T cell lines with CD1a-lysylPG and CD1a-PG tetramers loaded at low pH and each labeled with a different fluorophore, revealed multiple tetramer+ populations, each with distinct tetramer binding intensities. Specifically, the T cell line from donor 966 showed that, although the majority of the tetramer+ T cells bound both CD1a PG as well as CD1a lysylPG, there were also populations that specifically bound CD1a-lysylPG with no binding to CD1a-PG (Figure 3e). In the line from donor 834, three different populations bound both tetramers, albeit at different staining intensities.
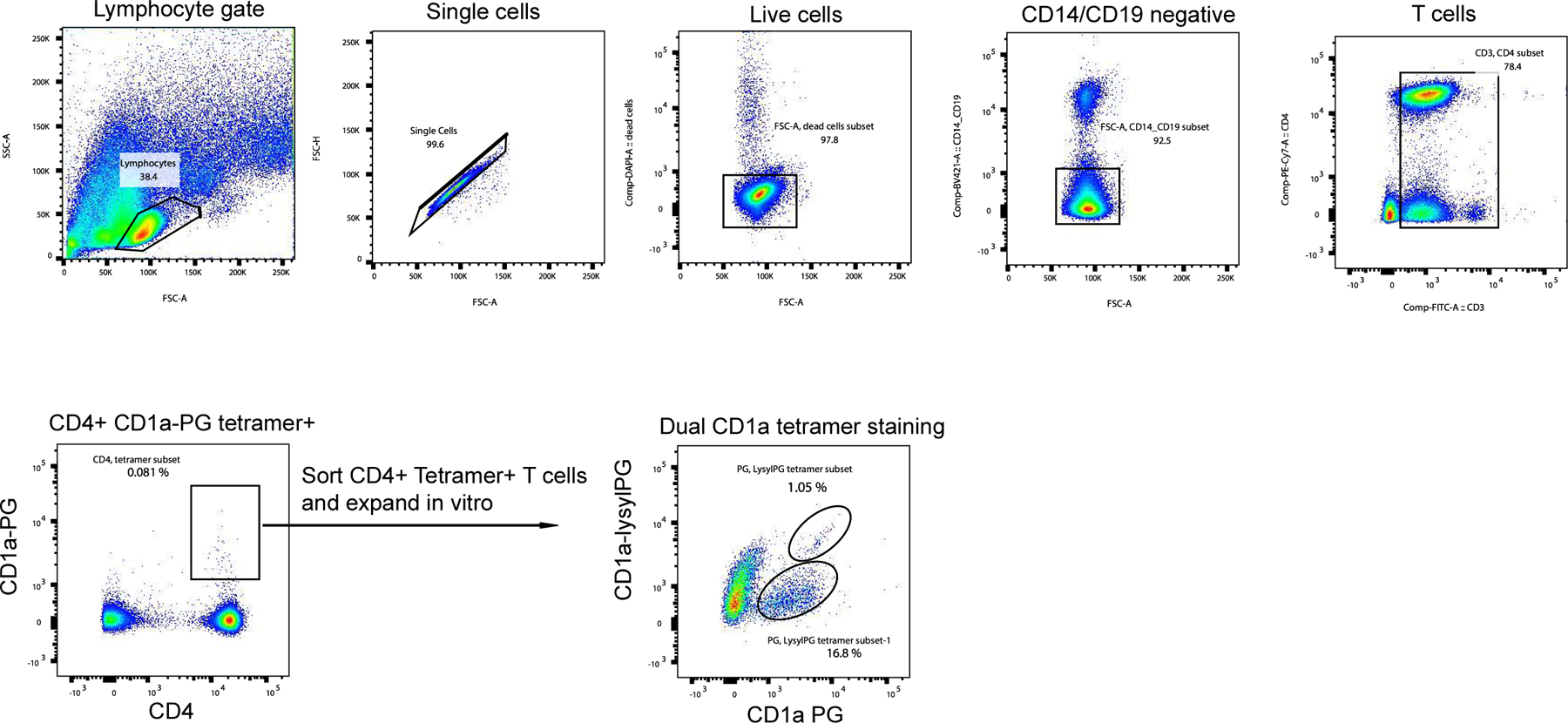
To determine if the repertoire contains T cells that specifically bind CD1a-PG, but not CD1a-lysylPG tetramers, we sorted and expanded CD1a-PG tetramer+ CD4+ T cells ex vivo, and performed dual tetramer staining. This approach indeed revealed a large population of T cells that stained with CD1a-PG tetramer only, as well as a small population of T cells binding both tetramers (Extended Data Figure 4). When staining four PBMC samples directly ex vivo with the dual tetramers, we detected three tetramer+ populations in 3 out of 4 donors (Figure 3f). Overall this analysis demonstrates that the T cell repertoire contains cross-reactive T cells for whom the lysine group is redundant for recognition, but also T cells for which the lysine modification either impairs or promotes TCR binding. The pH dependent sensitivity of lysylPG to lysine hydrolysis further suggests that CD1a tetramers loaded at neutral pH with lysylPG likely capture a polyclonal mixture of T cells that recognize lysylPG, PG or both. We further refer to these tetramers as CD1a-(lysyl)PG tetramers.
Gene expression profiles of CD1a-restricted CD4+ T cells
To gain insight into the cytokine and chemokine gene expression profiles of CD1a-restricted T cells specific for phosphatidylglycerols, we performed gene expression analysis of three independently derived CD4+ polyclonal CD1a-(lysyl)PG tetramer+ T cell lines from donors 921 and 966. We analyzed gene expression profiles in response to anti-CD3/CD28 stimulation by bulk RNA-sequencing (RNA-seq). All three independently derived T cell lines showed a very similar cytokine/chemokine expression profile, with a dominant upregulation of Th2 cytokines IL-13, IL-5, and IL-4 (Figure 4a). Cytokine genes associated with other T-helper subsets, such as IL17 / IL22 (Th17/Th22), IL21 (Tfh) and TGFB/ IL10 (Treg) were not expressed or weakly upregulated, although there was some IFNG (Th1) upregulation in response to stimulation.
Figure 4. Cytokine gene expression of CD4+ CD1a-(lysyl)PG tetramer+ T cells.
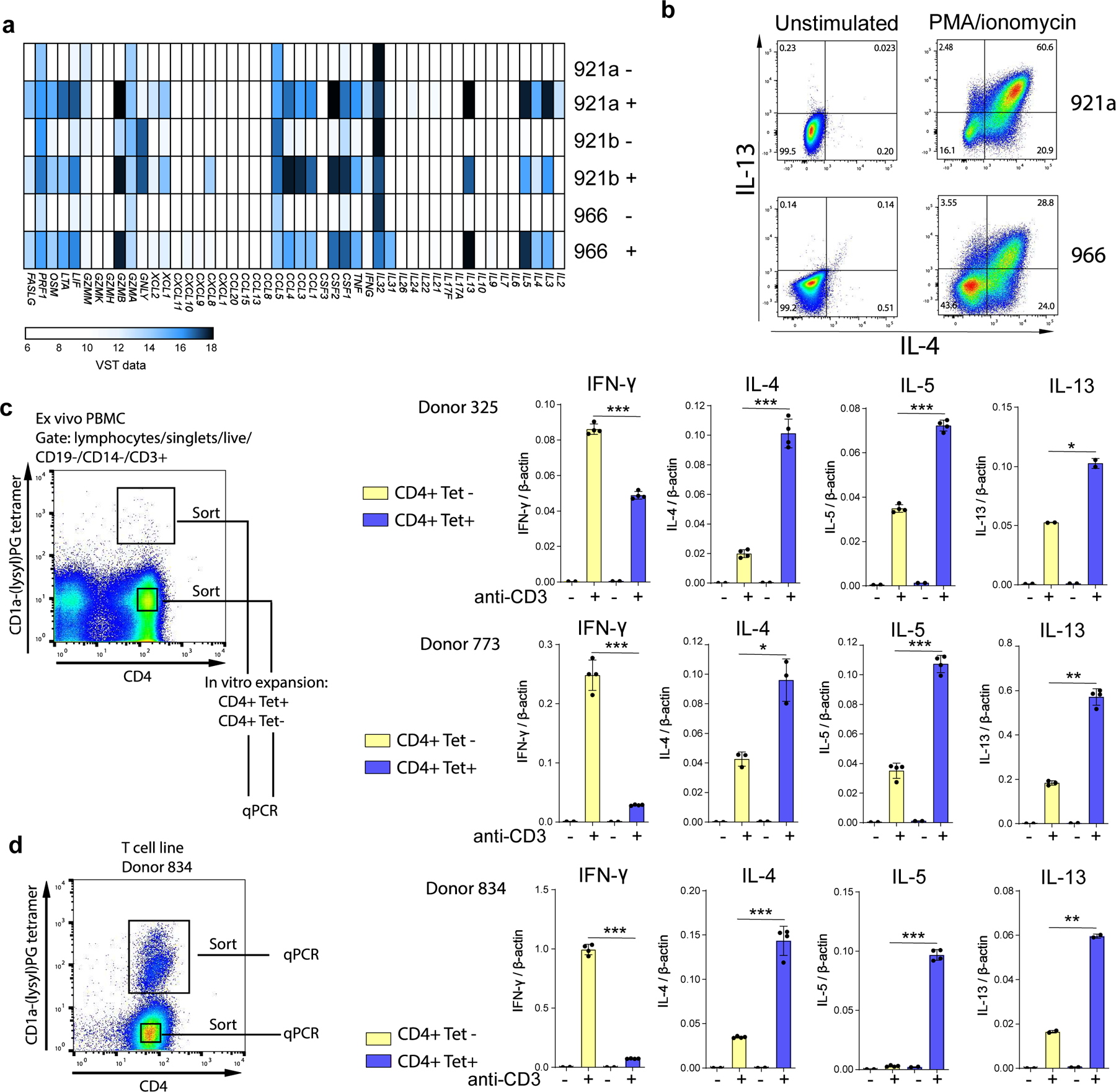
(a) Bulk RNA sequencing analysis of three CD1a-(lysyl)PG reactive T cell lines, unstimulated (-) and stimulated (+) with anti-CD3 / CD28 beads. Heatmap displays cytokine and chemokine expression after Variance Stabilizing Transformation of the count data. (b) Flowcytometric analysis of intracellular IL-4 and IL-13 staining of CD1a-(lysyl)PG reactive T cell lines (921a and 966) after 6 hr stimulation with PMA-ionomycin. (c) PBMC from donors 325 and 773 were FACS sorted ex vivo into CD4+ CD1a-(lysyl)PG tetramer+ and CD4+ tetramer- cells (gating strategy according to Supplementary Figure 1a). After one round of in vitro expansion, cytokine mRNA upregulation in response to plate-bound anti-CD3 (OKT3) was measured using qPCR. Expression values were normalized to β-actin. Depicted is the mean ± SD of 2–4 replicate qPCR reactions. P-values were based on the paired two-tailed t-test. *p<0.05, **p<0.01, ***p<0.001. (P-values donor 325: p=0.0005, p=0.0002, p=0.0002, p=0.035. P-values donor 773: p=0.0005, p=0.011, p=0.0006, p=0.0027) (d) CD1a-(lysyl)PG tetramer+ and tetramer- CD4+ T cells from T cell line 834 were FACS sorted, and cytokine mRNA upregulation in response to plate-bound anti-CD3 (OKT3) was measured using qPCR. Expression values normalized to β-actin. Depicted is the mean ± SD of 2–4 replicate qPCR reactions. P-values were based on the paired two-tailed t-test. ** p<0.01, ***p<0.001. (P-values donor 834: p<0.0001, p=0.0009, p<0.0001, p=0.0027).
We sought to confirm the transcriptional profile at the protein level by intracellular cytokine staining for IL-13 and IL-4 (Figure 4b). Beyond Th2 cytokines, the CD1a-lysylPG reactive T cell lines upregulated CSF2 (GM-CSF), CSF1 (M-CSF), IL3, LTA (lymphotoxin alpha), TNF as well as chemokine genes CCL1, CCL3, and CCL4. (Figure 4a). The RNA-seq data additionally showed a strong upregulation of GZMB (Granzyme B), FASLG (Fas Ligand) and expression of PRF1 (perforin), suggesting, beyond Th2, a potential cytotoxic function for lysylPG-reactive T cells.
The expression of genes associated with cytotoxicity together with the observed chemokine profile corresponds to previously described cytotoxic CD4+ T cells 23, 24.. Although cytotoxicity is generally thought to coincide with a Th1 cytokine profile, the combination with Th2 cytokines has also been described 25. Also, since these T cell lines contained more than one clone, the Th2 and cytotoxic profile could be associated with different individual clones.Next we sought to confirm whether the observed Th2 cytokine profile is more prominent in CD1a-(lysyl)PG tetramer+ than in tetramer- CD4+ T cells by sorting tetramer+ and tetramer- CD4+ T cells from the same donor ex vivo. After one round of in vitro expansion, cytokine gene upregulation was measured by qPCR. In both donors, tetramer+ CD4+ showed relatively lower expression of the Th1 cytokine IFN-γ and higher expression of Th2 cytokines IL-4, IL-5 and IL-13 (Figure 4c). Similar results were obtained in a more stringent experiment, where we analyzed tetramer- and tetramer+ CD4+ T cells sorted from a single T cell line, in which all cells been subjected to identical culture conditions (Figure 4d). Again these data suggest that CD1a-(lysyl)PG tetramer+ T cells are more likely to be skewed towards Th2 compared to tetramer- CD4+ T cells.
Single cell transcriptional analysis
To better understand the gene expression profiles of individual cells ex vivo, wperformed single cell RNA-seq using Plate-seq (ref) of CD1a-(lysyl)PG tetramer+ CD4+ T cells isolated directly ex vivo from two donors from whom we had previously confirmed the presence of CD1a-(lysyl)PG reactive T cells (donors 325 and 921). Unsupervised Uniform Manifold Approximation and Projection (UMAP) analysis revealed six clusters among the sorted CD4+ T cells (Figure 5a). Embedding of the tetramer- and tetramer+ on the UMAP showed that these populations largely overlapped, with the majority of all T cells falling in clusters 0 and 1. However, differences in the distribution of tetramer- and tetramer+ CD4+ T cells were clear among other clusters, in particular clusters 4 and 5, where CD1a-(lysyl)PG tetramer+ T cells were overrepresented (Figure 5b, 5c).
Figure 5. Single cell RNA-sequencing of CD1a-(lysyl)PG tetramer+ CD4+ T cells.
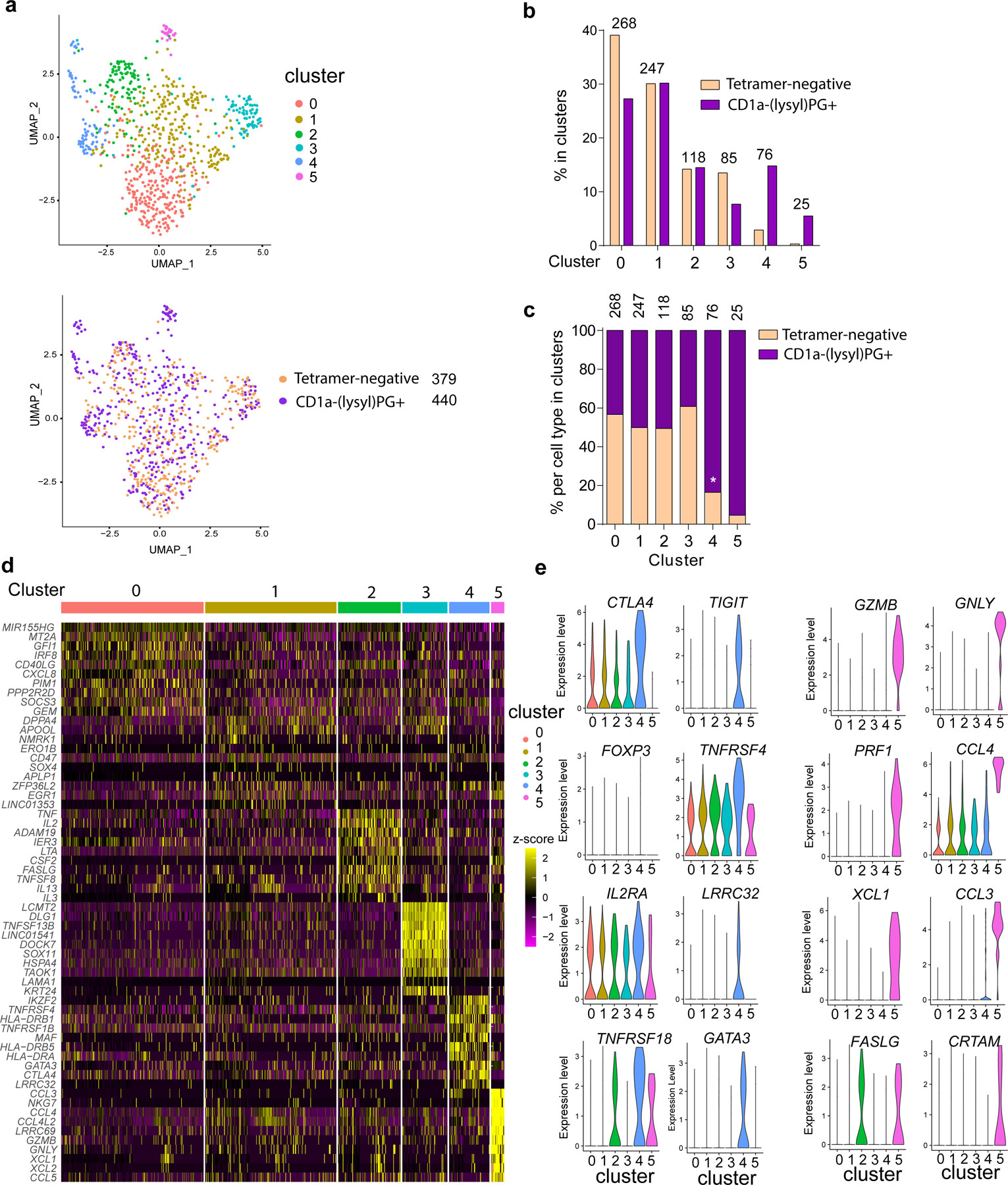
(a, top graph) UMAP of single-cell RNA-seq data from CD1a-tetramer+ and tetramer- CD4+ T cells. The colors represent cells in the six clusters defined using top variable genes and unsupervised clustering. (a, bottom graph) The tetramer+ and tetramer- CD4+ T cells subsets embedded in the UMAP. The total number of tetramer- and tetramer+ T cells are indicated. (b) Bar graph representing the proportions of tetramer+ and tetramer- T cell subsets in each of the six different clusters, (c) within each of the six clusters. The number of cells in each cluster is indicated above the bars in the graphs. P-values were based on the one-sample Z-test for proportion. *indicates observed proportion differs significantly from 50%. (d) Heatmap of unsupervised clustering analysis featuring top 10 discriminative genes per cluster. Log normalized counts were scaled based on z-score distribution. (e) Violin plots depict expression levels of markers per cluster. Selected for discriminative genes in clusters 4 and 5.
The 10 most variable genes in each cluster are depicted as scaled log-normalized expression values (Figure 5d), providing clues regarding the phenotypes and functions of the cell subsets in the clusters. Cluster 5, which contains primarily tetramer+ T cells, has features of cytotoxic CD4+ T cells with expression of granzyme B (GZMB), Perforin (PRF1), granulysin (GNLY), CD94 (KLRD1) as well as CRTAM and NKG7. In addition, chemokines CCL3, CCL4, CCL5, as well as XCL1 and XCL2 were increased in cluster 5 (Figure 5d and 5e and Supplementary Figure 3). This cluster is similar in gene expression to a population of CD4+ T cells recently identified in single cell RNA-seq experiments as cytotoxic CD4+ T cells or CD4+ TEMRA 23, 26, 27 28, as well as more innate T cell populations 29. Furthermore, the cytotoxicity and chemokine gene expression profile of cluster 5 largely overlapped with that of the CD1a-(lysyl)PG tetramer+ T cell lines (Figure 4a), suggesting that the gene expression profiles of the in vitro T cell lines correspond with those of tetramer+ T cells ex vivo.
Since all our tetramer+ T cell lines tested produced Th2 cytokines (Supplementary Table 1), we investigated the expression of Th2 associated genes in the single cell data set. IL4 was minimally detected in the entire RNA-seq dataset, whereas IL13 gene expression was primarily detected in cluster 2 (Supplementary Figure 3). Interestingly, GATA3, which is considered a master transcription factor of Th2 cells30, 31, was differentially expressed in cluster 4, in which CD1a-(lysyl)PG tetramer+ were also overrepresented compared to tetramer-negative T cells (Figures 5b and 5c). This increased expression of GATA3 in CD1a-(lysyl)PG tetramer+ T cells may explain the preferential expression of Th2 cytokines in this cell population.
Other genes that were differentially expressed in Cluster 4 included CTLA4, TIGIT, IL2RA, LRRC32 (GARP), TNFRSF18 (GITR) and HLA class II genes (Figure 5d, 5e and Supplementary Figure 3), suggesting that this cluster contained CD4+ T cells with features of regulatory T cells. There was some detection of FOXP3 expression in this cluster, though overall there was limited detection of FOXP3 in the entire dataset. Interestingly, IKZF2 (Helios), which is a member of the Ikaros transcription factor family expressed in Tregs32, was also differentially expressed in cluster 4. Consistent with known percentages of Tregs in peripheral blood CD4+ T cells, previously measured at 1–5%33, approximately 3% of tetramer-negative T cells fell within cluster 4, yet almost 15% of the tetramer+ CD4+ T cells fell in this Treg like cluster (Figure 5b). This outcome suggested an overrepresentation of T cells with Treg markers among these CD1a-restricted T cells. Accordingly, gene expression profiles reveal that CD1a (lysyl)PG tetramer+ CD4+ T cells are not a single homogenous subset but instead are found among distinct CD4+ subsets, including those expressing cytotoxic effector molecules, Treg markers, as well as the Th2 transcription factor GATA3.
CD1a-dependent activation of tetramer+ T cells by Staphylococcus bacteria
To test natural PG/lysylPG compounds, we set up an assay in which dendritic cells (DC) were incubated with live S. aureus (SA113), the membranes of which are primarily composed of PG, lysylPG, cardiolipin, and diglucosyl-diacylglycerol 13. Next we confirmed the presence of PG and lysylPG in S. aureus, and analyzed the major species of PGs and lysylPGs by negative-mode, reversed-phase HPLC-QToF-MS. (Figure 6a, b, Extended Data Figure 5, 6, and Supplementary Figure 4). Overall, PG and lysylPG species present in in vitro cultured S. aureus consisted of a broad range of chain lengths, with C15:0 primarily found in the sn-2 position, whereas acyl chains in the sn-1 position ranged from C14:0 to C20:0 (Supplementary Figure 4). We also included S. epidermidis in our co-cultures and lipid analyses, which was confirmed to contain both PG and lysylPG, most commonly harboring C20:0 in combination with C15:0 in the acyl chains (Extended Data Figure 6, Supplementary Figure 4), thereby containing on average somewhat longer acyl chains than S. aureus.
Figure 6. Response to Staphylococcal bacteria and CD1a-(lysyl)PG tetramer+ T cells in Atopic Dermatitis.
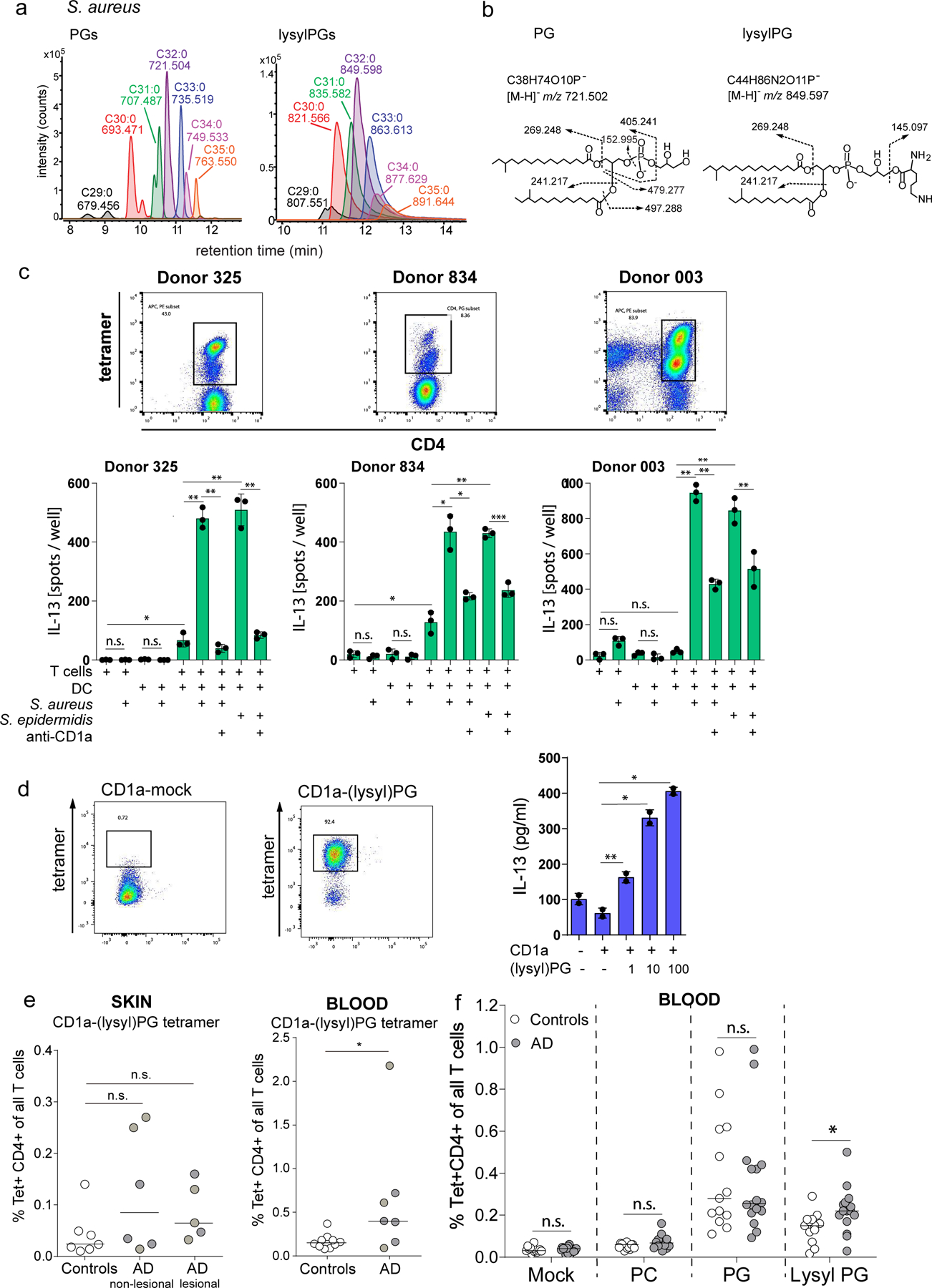
(a) Mass chromatograms of major species of PGs and lysylPGs from S. aureus SA113 were analyzed by negative-mode, reversed-phase HPLC-QToF-MS. The length and saturation of the combined fatty acyl chains were deduced by the detected m/z matching the structural formula. (b) CID-MS of the most abundant species in PG (left) and lysylPG (right) showed the diagnostic fragments of phosphoglycerol (m/z 152.995) and lysine (m/z 145.097) for PG and lysylPG, respectively. (c) IL-13 ELISpot of purified CD1a-PG tetramer-sorted T cells from lines 325, 834 and 003, co-cultured with monocyte-derived dendritic cells pre-incubated with media or live S. aureus or S. epidermidis bacteria. CD1a-restriction was determined by pre-incubating DC and bacteria with anti-CD1a (OKT6). Depicted is the mean ± SD of triplicate co-cultures. P-values were based on the paired two-tailed t-test. n.s. = not significant, * p<0.05, ** p<0.01, *** p<0.001. (P-values donor 325: p=0.036, p=0.0059, p=0.0033, p=0.0014, 0.0038. P-values donor 834: p=0.017, p=0.023, p=0.0032, p=0.018, p=0.0002. P-values donor 003: p=0.0010, p=0.0028, p=0.0058, p=0.0017) (d) CD1a-(lysyl)PG tetramer sorted skin T cell line (skinT4) stained with mock-treated and lysylPG-treated CD1a tetramers and analyzed by flow cytometry, gate: live cells. IL-13 release measured by ELISA in supernatant of co-cultures of skinT4 with plate bound CD1a loaded with indicated concentrations of lysylPG (µg/ml). Depicted is the mean ± SD of duplicate cultures. P-values were based on the paired two-tailed t-test, *p<0.05, **p<0.01 (P-values: p=0.006, p=0.014, p=0.033). Results are representative of 3 independent experiments. (e, left graph) CD1a-(lysyl)PG tetramer staining of expanded skin T cells (controls n=7, AD non-lesional n=6, AD lesional n=5). e, right graph) CD1a-(lysyl)PG tetramer staining of PBMC (controls n=10, AD n=7). Gating strategy according to Supplementary Figure 1a). Plotted are the percentages of CD1a-(lysyl)PG tetramer+ CD4+ T cells among all T cells. Medians are indicated with horizontal bars. P-values were based on the exact Wilcoxon rank sum test, two-sided. *P<0.05, n.s. = not significant. (P-values: p=0.11, p=0.22, p=0.027) (f) In a separate cohort of AD patients and healthy controls % of CD1a tetramer+ CD4+ T cells was measured using indicated tetramers (Gating strategy according to Supplementary Figure 1b). Plotted are the percentages of CD1a-tetramer+ CD4+ T cells among all T cells for controls (n=13) and AD patients (n=16). Medians are indicated with horizontal bars. For each tetramer, comparison between AD and controls was performed using the exact Wilcoxon rank sum test, two-sided. *p<0.05, n.s. = not significant (p=0.72, p=0.094, p=0.87, p=0.012).
After incubation with S. aureus or S. epidermidis, the DC were co-cultured with tetramer-sorted CD1a-(lysyl)PG reactive CD4+ T cell lines in media in the presence of antibiotics. Three sorted T cell lines were tested (Figure 6c, and Supplementary Figure 5) and all three showed IL-13 release in response to S. aureus and S. epidermidis treated DCs. S. aureus did not appear to have a direct effect on the T cells in the absence of DCs, and anti-CD1a blocked the response to DCs, indicating that CD1a was required for T cell activation. Thus, CD1a-(lysyl)PG tetramer+ T cell lines that respond to (lysyl)PG lipid antigen in the context of CD1a, also respond to antigen-containing bacteria. Since the Elispot data specifically measured IL-13, we broadened the cytokine analysis of the T cell lines in response to S. aureus and S. epidermidis bacteria to measuring multiple T cell cytokines by multiplex cytokine assay (LEGENDplex™). This experiment revealed that beyond IL-13, other Th2 cytokines IL-5 and IL-4 were produced in response to bacteria (Extended Data Figure 7).
Next, we aimed to more directly determine the recognition of natural S. aureus PGs. We purified the S. aureus PG using thin layer chromatography (TLC) followed by reversed-phase HPLC, and quantified the resultant PG lipids (Extended Data Figure 6). S. aureus PG was incubated with CD1a monomers, and tetramers were made in parallel to CD1a tetramers with C18:1 PG. Although the S. aureus PG treated CD1a tetramers did bind the T cell lines, the MFI was lower than with pure C18:1 PG CD1a tetramers (Extended Data Figure 8).
Overall, the CD1a-dependent activation of phosphatidylglycerol-specific CD1a-restricted T cells in response to Staphylococcus bacteria, supports the notion that these CD1a-(lysyl)PG tetramer+ T cells functionally respond to common skin pathogens and commensals.
CD1a-(lysyl)PG tetramer+ CD4+ T cells in atopic dermatitis patients.
Next, we aimed to determine if CD1a-(lysyl)PG-reactive T cells could be detected in human skin. T cells isolated from discarded skin tissue taken at surgery were screened for the presence of tetramer+ T cells by flow cytometry. This revealed the presence of CD1a-(lysyl)PG tetramer staining T cells among polyclonal skin T cells. Similar to the tetramer+ T cells isolated from peripheral blood, a tetramer-enriched T cell line from the skin released IL-13 in a dose-dependent manner in response to (lysyl)PG loaded onto plate-bound CD1a (Figure 6d). Thus, normal skin contains CD1a (lysyl)PG-reactive T cells.
Atopic dermatitis (AD) is associated with colonization of the skin with S. aureus, and in patients, flares coincide with an increase in S. aureus bacterial burden 34, 35. In addition, the skin barrier is often impaired in AD, possibly increasing exposure of the epidermal Langerhans cells to bacteria and bacterial products36. Last, Th2 cytokines are thought to underlie much of AD pathogenesis, prompting the question whether CD1a-lysylPG reactive Th2 type CD4 T cells are expanded in individuals with AD. To investigate these questions, we initially performed a small pilot study for which we enrolled AD patients (n=7) and healthy controls (n=10). We obtained peripheral blood samples, as well as 4 mm skin biopsies from lesional and non-lesional skin sites from AD patients, and matched skin sites from healthy controls.
We investigated the frequencies of tetramer binding T cells in the lesional and non-lesional skin of AD patients with that of healthy controls. In control skin, the numbers of T cells extracted from a 4 mm punch biopsy were too low to allow for an accurate enumeration of low frequency tetramer+ populations directly ex vivo. Therefore, for both healthy and AD skin T cells, we included an in vitro expansion step prior to analysis. We generated sufficient T cell numbers to analyse the frequency of tetramer+ T cells from 7 control, 6 AD non-lesional, and 5 AD lesional skin biopsies. There was a trend for increased frequency of CD1a (lysyl)PG tetramer+ CD4+ T cells in both lesional and non-lesional AD skin compared to control skin, although this difference did not reach statistical significance (Figure 6e). It is important to note though, that absolute numbers of CD4+ T cells in AD dermis have been quantified to be >5x that of normal dermis37, 38, which suggests that in absolute numbers CD1a-(lysyl)PG reactive CD4+ T cells are increased in AD skin.
Next we measured frequencies of CD4+ CD1a-(lysyl)PG tetramer+ T cells in PBMC samples from AD patients and healthy controls. Compared to healthy controls, there was a higher percentage of CD4+ CD1a-(lysyl)PG tetramer+ T cells in peripheral blood of AD patients (Figure 6e), suggesting that CD4+ T cells specific for phosphatidylglycerols are expanded in AD. Because this was a small pilot study with a large range in specific T cell frequency (<0.1% - >2%), we initiated a larger study including 16 AD patients and 13 healthy controls. Also, for the follow up study, we made CD1a tetramers that were incubated with lipid at low pH to maintain intact lysylPG. Screening of the PBMC samples from patients and controls showed that, whereas there was not a significant difference in frequency of CD1a-PG tetramer+ CD4+ T cells, CD1a-lysylPG tetramer+ CD4+ T cells were slightly higher in AD patients than in controls (Figure 6f).
This result suggests that CD4+ CD1a-lysylPG specific T cells are expanded in the peripheral blood of AD patients. Together with the presence of CD1a-lysylPG tetramer+ T cells in AD skin, this finding prompts the question whether these T cells reflect a response to increased bacterial exposure in AD, and whether they contribute to Th2 mediated pathology in this inflammatory skin disease.
DISCUSSION
The investigation of lipid antigen specificity, diversity and functions of human CD1a-restricted T cells is important to understand their role in within the human immune system. Here, we have identified bacterially-derived phosphatidylglycerols, lysylPG and PG as bona fide CD1a-presented lipid antigens, recognized by populations of human T cells. Our data show that lipid antigen loaded CD1a tetramers are a powerful tool to identify and quantify these T cells in peripheral blood and skin, and isolate them for transcriptomic analysis directly ex vivo.
Previously, studies in human and mouse CD1-restricted T cells provided precedent that phosphatidylglycerol-based lipids can function as antigens. Unmodified phosphatidylglycerols were identified as antigens for human CD1b-restricted T cells and for murine CD1d-restricted non-invariant NKT cells 39, 40, 41, 42, 43. Also, a recent study using a human CD1b and CD1c-transgenic mouse strain and systemic infection with methicillin resistant S. aureus (MRSA) provided evidence that CD1-restricted T cells reduced bacterial burden and responded to S. aureus lipid fractions containing cardiolipin, PG and lysylPG44. Although our study did not address direct anti-bacterial effects of CD1a-(lysyl)PG specific human T cells, follow up work will address whether CD1-(lysyl)PG-reactive T cells may play a protective role against bacterial infection in humans.
In the cell membranes of Staphylococcus species, which make up approximately 25% of bacteria on human skin15, PG and lysylPG are the two most dominant phospholipids 13, 14. Therefore, these lipids are ubiquitous in the human skin microbiome, suggesting that all individuals have likely been exposed to PG and lysylPG during their lifetime. The ubiquitous nature of these lipids in the skin microbiome, together with the constitutive expression of CD1a is in line with the fact that CD1a-(lysyl)PG tetramer+ T cells were detected in most donors tested. Possibly, activation of these T cells occurs only when there is a damaged skin barrier, an increased skin bacterial burden, or both.
LysylPG is uniquely foreign, since the aminoacylation of phospholipids does not occur in mammalian cells. The requirement of low pH to prevent lysine hydrolysis, suggests that lysylPG is a relatively labile antigen. However, in the slightly acidic early endosomal system (pH 6.0) of Langerhans cells as well as in the stratum corneum, where the pH is on average 5.5, lysylPG is likely to remain intact. Dual tetramer staining with PG and lysylPG loaded CD1a tetramers indicated that lysylPG and PG epitopes can be differentially recognized. Thus, the bacteria-specific lysine modification generates an epitope that can affect the T cell response, and therefore, the relative amounts of PG versus lysylPG encountered may shape the CD1a-dependent T cell repertoire against bacteria. This is further corroborated by the observation that CD1a-lysylPG but not CD1a-PG tetramer+ CD4+ T cells were increased in AD patients compared to controls.
The crystal structure of the CD1a-PG complex showed that PG occupies the cleft of CD1a in a manner distinct from the previously solved structures of CD1a carrying blocking antigens20, 45, 46. PG is not, strictly speaking, foreign in its biochemical structure, but it is expressed at much higher levels in bacteria compared to mammalian cells. The abundance of PG in most bacterial membranes predicts that T cells that respond to CD1a-PG complexes will likely broadly respond to bacteria besides Staphylococcus species. Beyond bacterial membranes, PG is also present in trace amounts in mammalian cells, in mitochondria 16, functioning as a high turnover intermediate for cardiolipin. In addition, PG is present in human lung surfactant where it represents between 5 and 10% of phospholipids 47. The fact that PG is not uniquely a foreign lipid prompts the question of what the precise functions are of CD1a-PG specific T cells in the immune system, and in which situations they respond to endogenous PG.
The combined results from the RNA-sequencing of the T cell lines and the ex vivo single cell RNA-seq show that among CD1a tetramer+ T cells several subsets exist that differ significantly in their gene expression profiles and their presumed effector functions. This outcome implies that to understand the role of these CD1a-restricted T cells in inflammatory skin disease, it will become important to analyze not only the frequency of these T cells by tetramer staining, but also their cytokine profiles and surface marker expression. Given prior studies of IL-22 predominance among skin homing17 and skin resident T cells18 that are autoreactive to CD1a, the Th2 phenotypes seen in multiple lines of experiments here is particularly striking and provides a possible link of CD1a to Th2-mediated inflammatory skin disease. Future investigation into proinflammatory versus regulatory populations among the tetramer+ T cells will shed light on their functions in disease.
In summary, this study has provided insights in a previously unknown subset of CD1a-restricted T cells that responds to phosphatidylglycerols and S. aureus bacteria, and can be detected in the circulation of most individuals tested. Ongoing investigations aim to understand the physiological role of this T cell subset and their contribution to epithelial and anti-bacterial immunity.
MATERIALS & METHODS
Subject enrolment and sample collection
Skin swabs, 4 mm skin biopsies, and blood samples were collected from AD patients and healthy controls enrolled in our study. This study was approved by the Institutional Review Board at Columbia University under protocol # AAAQ9797, and informed consent was obtained from all participants. For the pilot study depicted in Figure 6e, 10 controls and 7 AD patients were enrolled and provided a blood draw and skin biopsies. The age range of the AD patients was 23–63 years (Average: 38 years. Males 71% Females 29%). The age range of the controls was 22 – 34 (Average 29 years. Males 63% Females 37%). AD patients had a confirmed diagnosis of atopic dermatitis, were not on systemic immunosuppressive of immunomodulatory therapy, and did not use topical steroids for 7 days prior to the biopsy. Participants were reimbursed $200. A separate cohort of atopic dermatitis patient and healthy volunteer samples were collected under good clinical practice guidance with ethics approval at Oxford University Hospitals NHS dermatology clinic (NRES 14/SC/0106). For this study, depicted in Figure 6f, 13 controls and 16 AD patients were enrolled and provided a blood draw. The age range of the AD patients was 23–86 years (Average: 45 years. Males 25% Females 75%). The age range of the controls was 26–77 (Average: 51 years. Males: 31% Females 69%). AD patients had a confirmed diagnosis of atopic dermatitis and were not on systemic immunosuppressive or immunomodulatory therapy. Informed consent was obtained from all participants. No participant compensation was provided. In addition, we obtained buffy coats from healthy blood donors through the NY blood center, as well as skin tissue as discarded tissue from plastic surgery procedures.
Synthetic Lipids
Certain lipids used in for loading on the CD1 monomers, as well as for functional assays, were purchased from Avanti Polar lipids, Inc. C18:1 lysylPG (#840521), C18:1 PC (#850375), C18:1 PE (#850725), C18:1 PA (#840875), C18:1 PS (#840035), C18:1 PI (#850149), C18:1 PG (#840475).
Bacterial lipid extraction
Bacterial pellets were washed in sodium acetate buffer (20 mM, pH 4.5) and flash frozen and stored at -80C until extracted. To extract total lipids, 1 ml of frozen S. aureus (SA113) or S. epidermidis (ATCC 12228) pellet was resuspended in 6 ml of methanol and equally distributed to three 15-ml conical glass tubes. Chloroform was added to each tube followed by more methanol to give the chloroform/methanol ratio of 1:2 (v/v). After 1 hr of orbital shaking, extracts were collected after centrifugation. The organic solvent was evaporated under nitrogen onto glass and the dried extracts were redissolved in C/M (1:1) at 1 mg/ml and stored at -20 °C.
1D TLC purification for S. aureus PG and lysylPG.
Described in Supplementary Notes
PBMC isolation and lymphocyte isolation from skin samples.
Peripheral blood mononuclear cells (PBMCs) were isolated using Ficoll-paque (GE lifesciences - 17–5442-02) and following manufacturer’s instruction. PBMCs were cryopreserved in FBS supplemented with 10% dimethyl sulfoxide (DMSO) and stored in liquid nitrogen.
For the estimation of CD1a-(lysyl)PG tetramer+ T cell frequencies in AD patient and control biopsies, lymphocytes were isolated from 4 mm skin punch biopsies using Whole skin dissociation kit (human, Miltenyi Biotec, 130–101-540) according to the manufacturer’s protocol. After skin dissociation, the cells were washed in RPMI 1640 (Thermo Fisher Scientific – 61870127) with 10% FBS, L-glutamine, 2-mercaptoethanol, and Antibiotic-Antimycotic (100X, Gibco), then cryopreserved in FBS 10% DMSO, and stored in liquid nitrogen until T cell expansion and analysis.
For the recovery of T cells from discarded skin tissue samples for the isolation of CD1a-(lysyl)PG tetramer+ T cells, we used a culture-based method modified from48 . Briefly, after removal of subcutaneous fat tissue full thickness skin was cut into pieces of ~1mm3. Tissue pieces were let to adhere to tissue culture plate (6 well plate), after which complete T cell media was added (RPMI-1640 with L-glutamine, 8% human AB serum, non-essential and essential aminoacids, penicillin-streptomycin, 2-mercaptoethanol, Antibiotic-Antimycotic) supplemented with recombinant IL-2 (100 IU/ml) and IL-15 10 ng/ml. After 7–14 days, lymphocytes start spilling out of the tissue, and were harvested, washed and cryopreserved until analysis.
Recombinant CD1a expression and purification
Human CD1a/β2m dimers were expressed in human embryonic kidney (HEK) 293S GnTI− cells and purified as previously described18. The lipid loading was performed at 20C overnight with C18:1 lysylPG (Avanti 840521) or 18:0–18:1 PG (Avanti 840503) solubilized in 0.5%CHAPS/20mM Tris pH 8, 150mM NaCl. Unbound lipid and detergent were separated from the protein by ion exchange chromatography using a MonoQ 10/100 GL column (GE Healthcare). For crystallization the expression tags were removed using thrombin (Sigma-Aldrich) and the protein was de-glycosylated with endoglycosidase H (New England BioLabs). For SPR, the protein was biotinylated using BirA ligase. The stability of lysylPG during lipid loading was assessed by incubating CD1a-endo with 10x molar excess of lysylPG or PG with different buffers (50mM citrate pH 5.5 or 50mM MES pH 5.5 or 20mM Tris pH 8) at 20C overnight. To confirm lysylPG stability once bound to CD1a CD1a-lysylPG was incubated at 20C overnight in 0.1M MMT buffer (MES, DL-Malic acid, Tris) at different pH (5 or 7 or 9). Lipid loading and degradation was assessed by isoelectric focusing where 1 μg of protein was run on a 5–8 PhastGel (Cytiva).
Crystallization, structure determination, and refinement
The protein sample was up-concentrated to 2mg/ml and crystallized at 20°C in 20 to 25% polyethylene glycol 1500, 10% DL-Malic acid, MES monohydrate, Tris (MMT) buffer (pH 5 to 6) using previously obtained CD1a-lipid seeds46. The crystals were flash-frozen in liquid nitrogen and diffracted at the MX1 beamline (Australian Synchrotron). Data processing and scaling was performed in XDS and Aimless, respectively49, 50. Previously solved CD1a binary structure was used as a search model for molecular replacement phasing in PHASER51. Upon rigid body refinement in phenix.refine the unbiased electron in the cleft of CD1a was used to build the ligand. The model fit was improved using phenix.refine and performing manual refinement in the program COOT52, 53. The final refinement round led to the R/R-free factors of 19/23% for CD1a-PG complex structure. The quality of the data was confirmed by the Protein Data Bank (PDB) validation server and the final model and structure factor files were deposited in the PDB under PDB ID: 7SH4.
Expression, refolding, and purification of recombinant TCRs
The 921.3 and BK6 TCRs were produced using a previously described method20. Briefly, individual α and β chains of the TCR, with an engineered disulfide bond between the TCRα and TCRβ constant domains were expressed in BL21 Escherichia coli cells as inclusion bodies and solubilized in buffer containing 8 M urea, 10 mM tris-HCl (pH 8), 0.5 mM Na-EDTA, and 1 mM dithiothreitol. The TCR was then refolded in 5M urea, 50 mM tris-HCl (pH 8), 2 mM Na-EDTA, 400 mM L-Arg-HCl, 0.5 mM oxidized glutathione, and 5 mM reduced glutathione. The refolded solution was dialyzed three times against 10 mM tris-HCl (pH 8.0) for a total of 24h. The dialyzed samples were then purified through DEAE cellulose, hydrophobic interaction chromatography (HIC) and gel filtration chromatography. The quality and purity of the samples were analyzed via SDS–polyacrylamide gel electrophoresis.
Surface plasmon resonance
Soluble monomers of biotinylated CD1a carrying endogenous lipids (CD1a-endo) or loaded with specific lipids (CD1a-lysylPG or CD1a-PG) were coupled onto a streptavidin chip (Cytiva) to a mass concentration of ~2500 resonance units per flow cell. Increasing concentrations of the 921.3 or BK6 TCRs (0 to 150 μM) were injected over all flow cells for 60 s at a rate of 5 μl/min on Biacore T200 in 20 mM tris-HCl (pH 8), 150 mM NaCl, 0.5% bovine serum albumin (BSA) buffer. The total binding response was calculated in Scrubber2 after subtracting the non-specific binding to an unrelated protein. The KD was calculated using one-site specific binding model in GraphPad.
CD1a Tetramers
Biotinylated CD1a monomers were provided by the NIH tetramer facility. Lipids were solubilized in pre-warmed phosphate-buffered saline (PBS, pH 7.4) containing 0.5% 3-cholamidopropyl dimethylammonio 1-propanesulfonate (CHAPS) in a glass tube, incubated at 56°C for 30min and then sonicated at 37°C for 30min to solubilize the lipid. CD1a monomer (molar ratio lipid : CD1a = 50 : 1) was then added to the solution with as well as sodium azide (0.02%). The sample was incubated overnight (16 hrs) at 37°C. The next day, the monomers were tetramerized by adding streptavidin-PE (Thermo Fisher Scientific - s866A SAPE) in 6 x 1.8 µl in 10 minute intervals or adding streptavidin-APC (Thermo Fischer Scientific –s868) in 6 x 1.2 µl in 10 minute intervals to 48μl of CD1a-lipid solution. Final concentration was ~ 0.15 µg/µl of CD1a monomer. To minimize the lysine hydrolysis of lysylPG during loading, we established an alternative protocol, which we used to make CD1a-PG and CD1a-lysylPG tetramers for stainings in Figures 3e, 3f. 6f. Lipids were solubilized in MES buffer (20mM, pH 5.6) containing 0.5% CHAPS and sonicated for 30 minutes at 37°C, then incubated with CD1a monomer (ratio lipid ; CD1a 50 : 1) for 1 hr at 37°C. Excess lipid and detergent were removed using Amicon Ultra-0.5 centrifugal filter unit, 30 kDa by washing of the loaded CD1a monomer with PBS. The monomers were then tetramerized as described above.
Tetramer staining and isolation of CD1a tetramer+ T cells by FACS
PBMCs were thawed and plated at a density of ~ 1.5 x 106 cells/ml in complete T cell media (Fisher Scientific - 61870127) supplemented with 8% human AB serum (Sigma-Aldrich H4522) and 1 ng/ml IL-15 (Peprotech - 200–15). The following day, cells were washed with staining buffer (PBS 2%FBS), incubated at a concentration of max. 0.5 x 106 cells in 25 µl of buffer, and 1 µl of CD1a tetramer for 30 minutes at 20–25°C. Without washing, 0.1 µg of anti-CD3 monoclonal antibody (OKT3, BioLegend - 317325) was added in a 10 µl volume and the sample was incubated at 37°C for 10 minutes. Finally, fluorescently-labelled surface antibodies were added to the staining mix: CD14 BV421 (Biolegend - 301830), CD19 BV421 (Biolegend 302234), CD3 BB515 (BD 564466), CD4 Pe-cy7 (Biolegend 300515) and incubated on ice for 20 minutes. 4’,6-Diamidino-2-Phenylindole, Dilactate (DAPI) (Biolegend, 422801) was used to exclude dead cells. A BD Influx analyzer was used for the FACS-sorting of the tetramer positive cells. Flowjo v10 was used to analyze flowcytometry data.
Culture and expansion of CD1a-tetramer+ T cells
Tetramer+ T cells were sorted directly in U-bottom 96-well plates containing 105 PBMC irradiated at 2500 rad, 100 IU/ml IL-2 (PeproTech - 200–02), 10ng/ml IL-15 (Peprotech - 200–15), and 30ng/ml OKT-3 (BioLegend - 317325), and incubated in cell culture incubator (37°C, 5% CO2). Fresh medium (RPMI 8% Human AB serum) supplemented with 20 ng/ml IL-15 was added every 2–3 days. After 10–14 days sorted and expanded T cells were tested for enrichment by CD1a tetramer staining.
Supplementary Table 1 provides an overview of all donors, polyclonal T cell lines, and T cell clones as well as the experiments in which they were used.
CD1a plate Assay
Plate-binding assays were performed as previously described19. Briefly, neutravidin plates (Thermo Fisher Scientific, 15129) were coated for 6 hours at 20–25°C with 2.5μg/ml anti-CD11a (Biorad, MCA1848) and 10μg/ml biotinylated CD1a monomers (NIH). Lipid of interest was sonicated in 37°C PBS 0.05% CHAPS (37°C) in a glass tube and sonicated for 30 minutes at 37°C. After coating, plates were washed 3x with warm PBS. 50μl of lipid solution per well was added for overnight incubation at 37°C. The next day, plates were washed 3x with warm PBS, and 1x105 T cells per well were added. Supernatant was collected after 24h for ELISA.
CD1a antigen presenting assay using CD1a transfected antigen presenting cells
Lipids of interest were suspended in complete T cell medium (RPMI 1640) supplemented with 10% FBS and sonicated at 37°C for 20 minutes. As antigen presenting cells, K562 cells were used, which were stably transfected K562 cells with CD1a17. Sonicated lipid solution and 5,000 antigen presenting cells (APCs) were combined in a 96-well plate and incubated for 1h at 37°C. T cell lines were added at a 5:1 or 10:1 T cell:APC ratio. Supernatant was collected at 24hr and cytokine was measured in the supernatant by ELISA.
Dendritic cell (DC) and S. aureus / S. epidermidis co-culture assay
CD1a-(lysyl)PG tetramer+ T cell lines were thawed and rested overnight (16hrs) in culture media supplemented with recombinant human IL-15 (1 ng/ml). The following day, the CD4+ tetramer+ T cells were sorted by flow cytometry from the T cell lines and were again incubated overnight with IL-15 (1 ng/ml). Staphylococcus aureus (SA113) and Staphylococcus epidermidis (ATCC 12228) were grown for 16 hrs in LB broth at 37°C on a shaker. A 96-well filtration plate with 0.45μm hydrophobic high protein binding immobilon-P-membrane from Millipore was prepared for IL-13 ELISpot assay (Mabtech) as per the manufacturer’s protocol. Approximately, 6000–8000 DCs in a volume of 50 µl/well were co-cultured in a 96-well U-bottom plate with 50 µl of 105/ml concentration of S. aureus or S.epidermidis in RPMI 1640 (Thermo Fisher Scientific - 61870127) supplemented with 10% FBS without antibiotics. The cells and bacteria were co-cultured in the cell incubator at 37°C for 30 minutes. Final concentration of 100 µg/ml gentamicin in complete RMPI 1640 media 10% FBS was added to all the wells. Anti-CD1a blocking antibody (clone OKT6) was added at a final concentration of 20 µg/ml to the lanes that required the CD1a blocking antibody. The plate was then incubated at 37°C for 60 minutes. After incubation, the cells were transferred from the 96-well U-bottom plate to the coated and blocked IL-13 ELISpot plate and approximately 15,000 T cells in a volume of 50 µl/well were then added to the wells. The Elispot plate was incubated for 24 h at 37°C after which the supernatant was harvested for multiplex cytokine analysis and the IL-13 ELISpot assay (Mabtech) completed according manufacturer’s protocol. Cytokine analysis in the supernatant was performed using LEGENDplex™ human T helper cytokine panel (Biolegend 741027) according to manufacturer’s protocol.
Intracellular cytokine staining (ICS)
Tetramer+ T cell lines were stimulated with PMA-ionomycin (Cell Stimulation Cocktail, eBioscience 00–4970-03), in cell culture incubator. After 30 minute Golgistop (monensin) was added and cells were incubated for an additional 4 hrs and 30 minutes. Cell were stained with fixable live/dead marker (Thermo Fisher Scientific, L23105), and then fixed and permeabilized using Fix/Perm buffer and further processed and stained according to the protocol for intracellular cytokine staining using Fixation and Permeabilization Solution Kit with BD GolgiStop™ (Cat. No. 554715). Antibodies used for ICS and surface stains: CD4-PE (Biolegend, 300508), CD3 APC (Biolegend, 300412), IL-4 Alexa fluor 488 (Biolegend, 500817), IL-13 V450 (BD Bioscience, 561158).
LysylPG stability test and HPLC-MS analysis
For lysylPG stability test at different pH values, 10 µg of lysylPG was incubated in the tetramer loading buffer at pH 5.6, 7.4 or 8.0 either at 20–25°C (RT) overnight (o/n) or 37 °C for 1 hour. For lysyPG recovery from lipid-loaded CD1a proteins purified CD1a was incubated at 20°C overnight with 15X molar excess of lysylPG or PG dissolved in 0.5% CHAPS, 150mM NaCl and either 50mM MES pH 5.5 or 20mM Tris pH 8. The protein was separated from the excess lipid and detergent by size exclusion chromatography run in 150mM NaCl and either 50mM MES pH 5.5 or 20mM Tris pH8. 50 μg samples of lipid loaded CD1a were used for mass spectrometry analyses. These samples were extracted by chloroform, methanol, and water 54. The organic phase from each sample was collected and dried under nitrogen gas. Before the injection for HPLC-MS analysis, the lipid extract from the tetramer buffer was normalize to 2.5 µM in the starting mobile phase, whereas the eluent residue from the CD1a protein was normalized to 25 µM based on the protein concentration. These samples were run on an Agilent Poroshell 120 A, EC-C18, 3 x 50 mm, 1.9 µm reverse phase column coupled with a 3 x 5 mm, 2.7 µm reverse phase guard column and analyzed by an Agilent 6546 Accurate-Mass Q-ToF/1260 series HPLC instrument. The binary solvent systems were used as described 55. For PG and lysylPG quantification, a series of concentrations of these two synthetic standards were prepared. The chromatogram areas from the HPLC-MS analysis were plotted against the corresponding concentrations to generate the standard curves. The unknown lipid concentrations of lysylPG or PG were estimated by external standard curve fitting (Extended Data Figure 3).
Fatty acid methyl ester (FAME) preparation and HPLC-MS analysis.
Described in Supplementary Notes.
Nanoelectrospray CID-MS analysis of PGs.
50 µg of the total lipids from S. aureus or S. epidermidis were dissolved in 100 µl of methanol. 10 ul of the methanol solution was loaded onto a nanospray tip for negative-mode ESI-MS and CID-MS using LXQ Linear Ion Trap Mass Spectrometer (Thermo Scientific). The spray voltage was set to 0.7 kV and the capillary temperature was set at 200 °C. For the CID-MS, collision energy was 20–30% of maximum and product ions were trapped with a q value of 0.25.
Single cell RNA-sequencing using PLATE-seq (pooled library amplification for transcriptome expression sequencing)56
PBMCs were thawed and incubated in cell culture incubator overnight in complete RPMI (Thermo Fisher Scientific - 61870127) supplemented with 8% human AB serum (Sigma-Aldrich H4522) and 1 ng/ml IL-15 (Peprotech - 200–15). The next day, PMA_ionomycin based cell stimulation cocktail (eBioscience 00–4970-03) was added to the cells for 2.5 hours prior to CD1a tetramer staining.. The gating strategy included: lymphocyte gate, gating out of doublets, dead cells, CD14+, CD19+ cells, and positive selection for CD3+ CD4+ (Supplementary Figure 1a). To avoid confounding plate effects, we designed the plates so that tetramer+ and tetramer- T cell subsets were represented on each plate, rather than separate plates per subset 57. Individual tetramer+ and tetramer- CD4+ T cells were sorted with a BD Influx analyzer into the wells of a 96-well plate, each containing 7.5 µl of lysis buffer [0.2% Triton X-100 (Sigma), SUPERaseIN (1 U/μl) (Thermo Fisher Scientific), 2 mM deoxyribonucleotides (dNTPs) (Thermo Fisher Scientific), and 2 μM reverse transcriptase (RT) primer (Integrated DNA Technologies)]. Primer annealing was performed at 72°C for 3 min. Reverse transcription was performed by adding 7.5 μl of RT mix to each well [containing: 40U Maxima H- (Thermo Fisher Scientific), 2X Maxima H- Buffer (Thermo Fisher Scientific), 4U SUPERaseIN (Thermo Fisher Scientific), 15% polyethylene glycol, 2µM Template Switching Oligo (Integrated DNA Technologies)]. Further sample processing and library preparation was performed by the Single Cell Analysis Core of Columbia University according to published protocol 58, and libraries were sequences on Illumina NextSeq 500 using the 75-cycle High Outpit Kit [read lengths 26(R1) × 8(i) × 58(R2)]. Custom sequencing primers were used for Read 1 (SMART_R1seq and ILMN_R1seq; 56). With each plate, ~70 million reads were targeted. Library pools were loaded at 1.8 pM with 30% PhiX (Illumina).
Analysis of single cell RNA-seq data
Reads were aligned to the human genome reference GRCh38 using STAR (version 2.5) 59. During alignment, reads with more than one mapping were recorded as unmapped using the arguments ‘—out FilterMultimap N max 1’ and ‘—out SAMunmapped Within’ . Alignment counts were generated using featureCounts function of the Subread package (version 1.6)60 . Reads were assigned to cell and unique molecular indicator (UMI) barcodes using UMI-tools 61. The resulting gene-abundance files were analyzed using the Seurat V4 workflow. Briefly, the gene-count matrices were first screened for initial Quality control (QC) checkpoints. In short, genes that were expressed in less than 3 cells were removed from the analysis, as well as cells that had less than 200 genes expressed. Moreover, cells that had a total UMI count of less than 200 or more than 5000 or a percentage of mitochondrial reads higher than 11% were removed from the analysis62. After the initially QC and pre-processing to remove the unwanted, dying cells or doublets, RNA counts were log-normalized and then merged using identified anchors (Supplementary Figure 2). Further, contaminating APCs forming a separate cluster and overexpressing DCs markers were also removed from the analysis. The dataset from donor 325 included a separately sorted population of CD1a mock staining CD4+ T cells, which was also removed from the analysis. The datasets from two donors were merged using ‘merge’ followed by clusters computation, data scaling and data normalization using the ‘FindClusters’, ‘Normalizedata’ and ‘ScaleData’ functions respectively. Cluster markers were identified using ‘FindMarkers’ function in Seurat with default parameters for differential expression test. Dimensionality reduction was performed using PCA and UMAP using RunPCA and RunUMAP functions. The heatmaps were generated using DoHeatmap.
Bulk RNA-sequencing of CD1a-lysylPG tetramer+ CD4+ T cells
Described in Supplementary Notes
Cell lines used
K562 CD1 stable transfectants were used in antigen presentation assays. The basis for this cell line is the K562, which is not on the list of commonly misidentified cell lines. Although we have not performed genetic testing of this cell line, we routinely check surface expression of HLA class I (absent/low), HLA DR (absent) and CD1 (positive on the transfected cells) by flow cytometry.
Statistics
Paired data were compared using the paired t-test with equal or unequal variance, depending on the result of the variance test. Data from independent samples were compared using the exact Wilcoxon rank sum test.
Data availability
The CD1a-PG final model and structure factor files were deposited in the Protein Data Base (PDB) under PDB ID: 7SH4.
Single-cell and bulk RNA-sequencing files are available in Gene Expression Omnibus (GEO) under accession number GSE186459
Code availability
The Seurat object (RDs file format), intermediate output files and differential expression files (csv format) along with the code used in this manuscript for single-cell and bulk RNA-seq analysis is available in GitHub project repository (https://github.com/IshaMonga/CD1a-single-cell)
Extended Data
Extended data Figure 1.
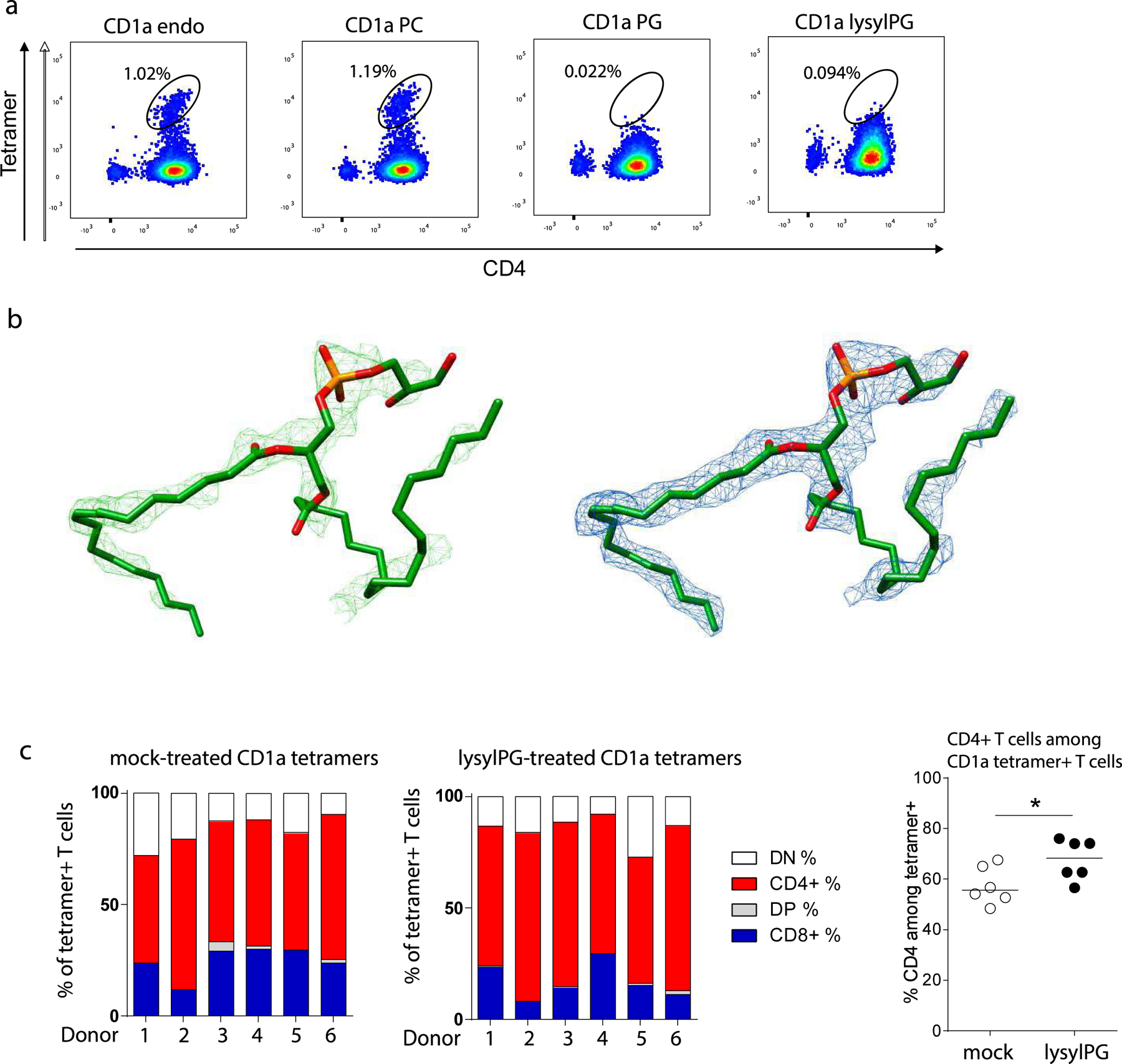
(a) Flowcytometric analysis of CD1a-autoreactive DermT cell line stained with the indicated CD1a tetramers: unloaded (endo), PC, PG, and lysylPG, co-stained for CD4 and gated on live cells. Indicated in the dotplots is the percentage of tetramer+ T cells in the tetramer gate. (b) Electron density maps of phosphatidylglycerol C18:1, C18:0 in the cleft of CD1a. Left: Unbiased (green, Fo-Fc at 2.5 σ. Right: Refined (blue, 2Fo-Fc at 0.9 σ) (c) Co-receptor expression determined on CD1a tetramer+ T cells measured in Figure 1f. Indicated are the percentages of tetramer+ T cells expressing CD4 (red), CD8 (blue), neither (white) of both (grey) of the co-receptors. Right graph: Percentage of CD4+ T cells among tetramer+ cells. *p<0.05 as determined by Wilcoxon matched-pairs signed rank test (2-sided).
Extended data Figure 2.
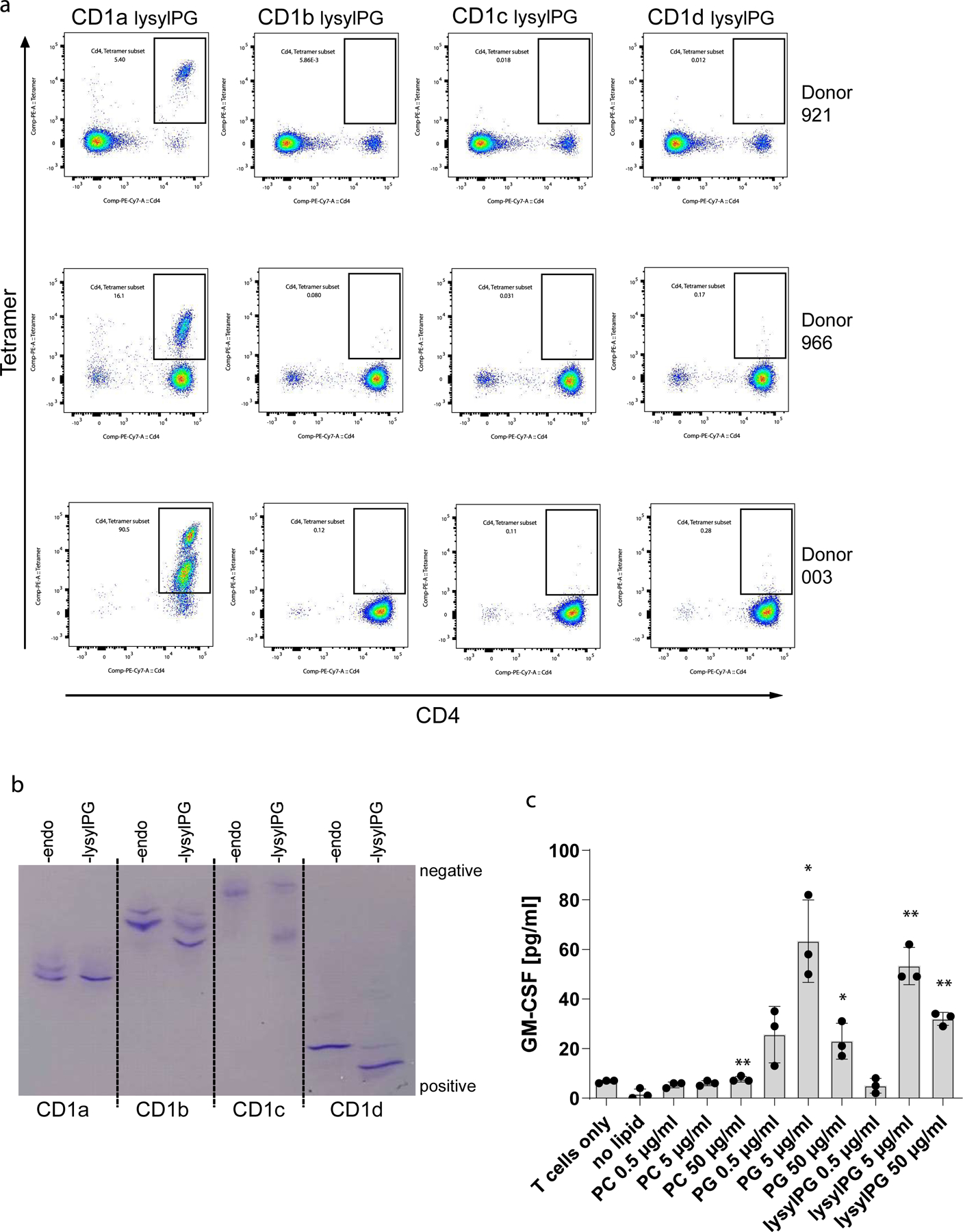
(a) Three T cells lines containing CD1a-lysylPG tetramer+ T cells were staining with lysylPG-treated CD1b, CD1c, and CD1d tetramers co-stained with anti-CD4 and analyzed by flow cytometry. Plots are gated on live T cells. (b) Human CD1/β2m isoforms were expressed in Expi293F GnTI- cells and purified by nickel-affinity and size exclusion chromatography. CD1 proteins carrying endogenous lipids (CD1-endo) were incubated with lysylPG 16 hrs at 25°C in 0.1 M MES pH5.5, 150mM NaCl, 0.05% CHAPS. 1 µg of CD1-endo and CD1-lysylPG samples were loaded on a 3–9 gel (Cytiva), and displacement of endogenous lipid and/or shifts towards more electropositive values indicate efficient loading of lysylPG. (c) GM-CSF concentration in supernatant of CD1a-lysylPG tetramer+ T cell line (921a) after 24hrs of incubation with plate-bound CD1a and indicated concentrations of PC, PG and lysylPG. Indicated are mean ± SD of triplicate values. P values were based on paired two-tailed t-test. * p<0.05 ** p<0.01
Extended data Figure 3.
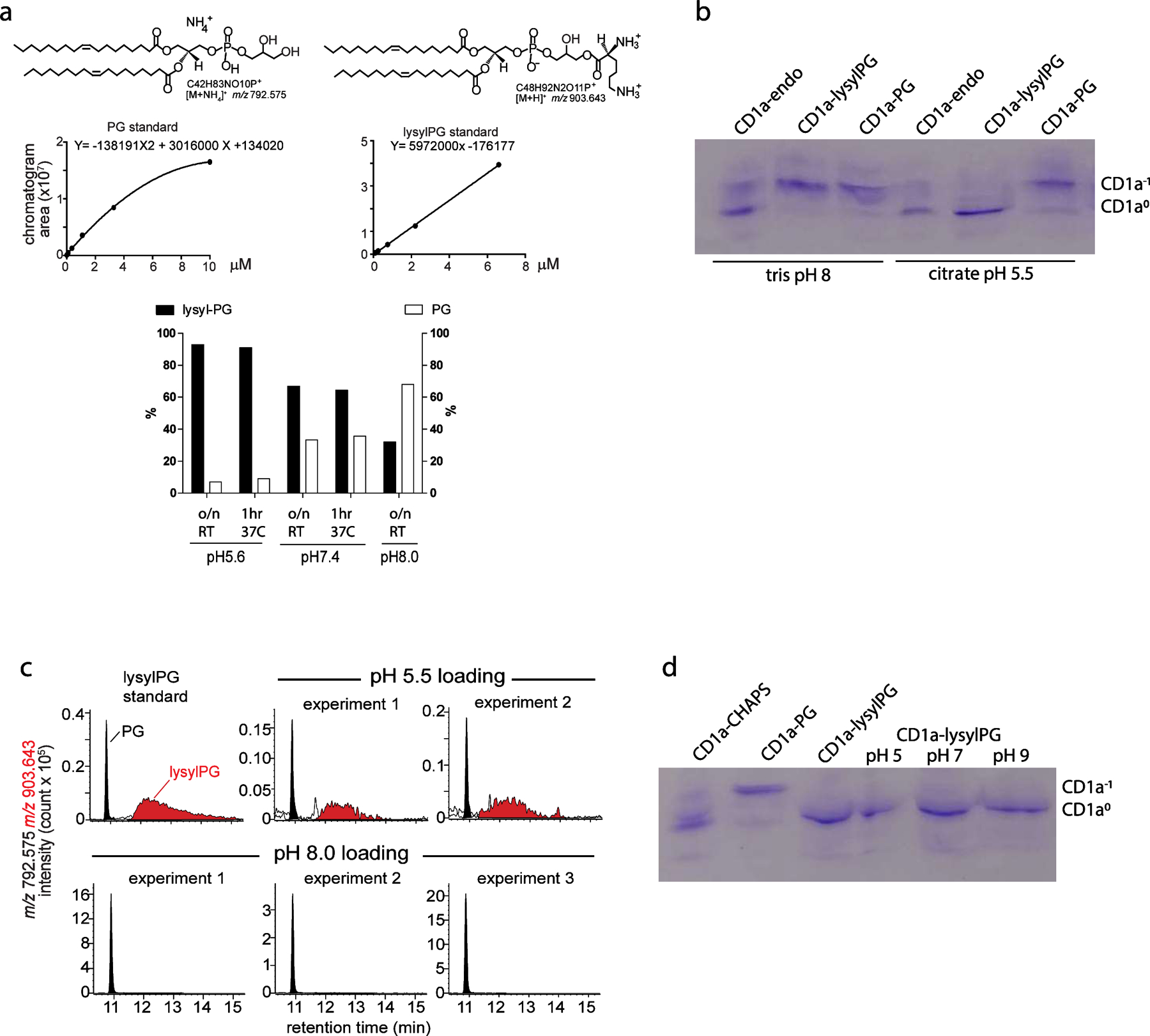
(a) LysylPG was treated at the indicated pH at 25°C overnight (16 hr) or 37°C for 1 hr, followed by lipid extraction and HPLC-MS analysis. The PG and lysylPG were quantified as percentage of the sum of both lipids, based on the curve fitting of external standards. (b) Isoelectric focusing gel showing the migration pattern of human CD1a carrying heterogenous mixture of lipids derived from the mammalian expression system (CD1a-endo) upon incubation with lysylPG or PG at low (citrate pH 5.5) or high (tris pH 8) pH. Presence of negatively charged lipids (e.g. PG) in the cleft of CD1a shifts the PI of the protein towards electronegative values (CD1a-1). Degradation of lysylPG to PG at high pH results in migration pattern identical to CD1a-PG. (c) CD1a proteins treated with lysylPG (pH 5.5 or pH 8) described in (b) were extracted and the eluted lipids, lysylPG and PG were analyzed by HPLC-MS. (d) Isoelectric focusing gel showing distinct migration pattern of CD1a incubated with CHAPS, PG or lysylPG based on the pI of the protein. LysylPG-loaded CD1a was further incubated overnight in MMT buffer at different pH to assess the stability of the lysylated headgroup. The lack of band corresponding to more negatively charged species of CD1a (CD1a-1) suggests that lysylPG is stable once bound to CD1a.
Extended data Figure 4.
FACS sorting of CD4+ CD1a-PG tetramer+ T cells from donor 211 PBMC. Sorted T cells were expanded in vitro for 14 days after which the cells were tested for specificity using dual tetramer staining with CD1a-PG (APC-labeled) and CD1a-lysylPG (PE-labeled).
Extended data Figure 5.
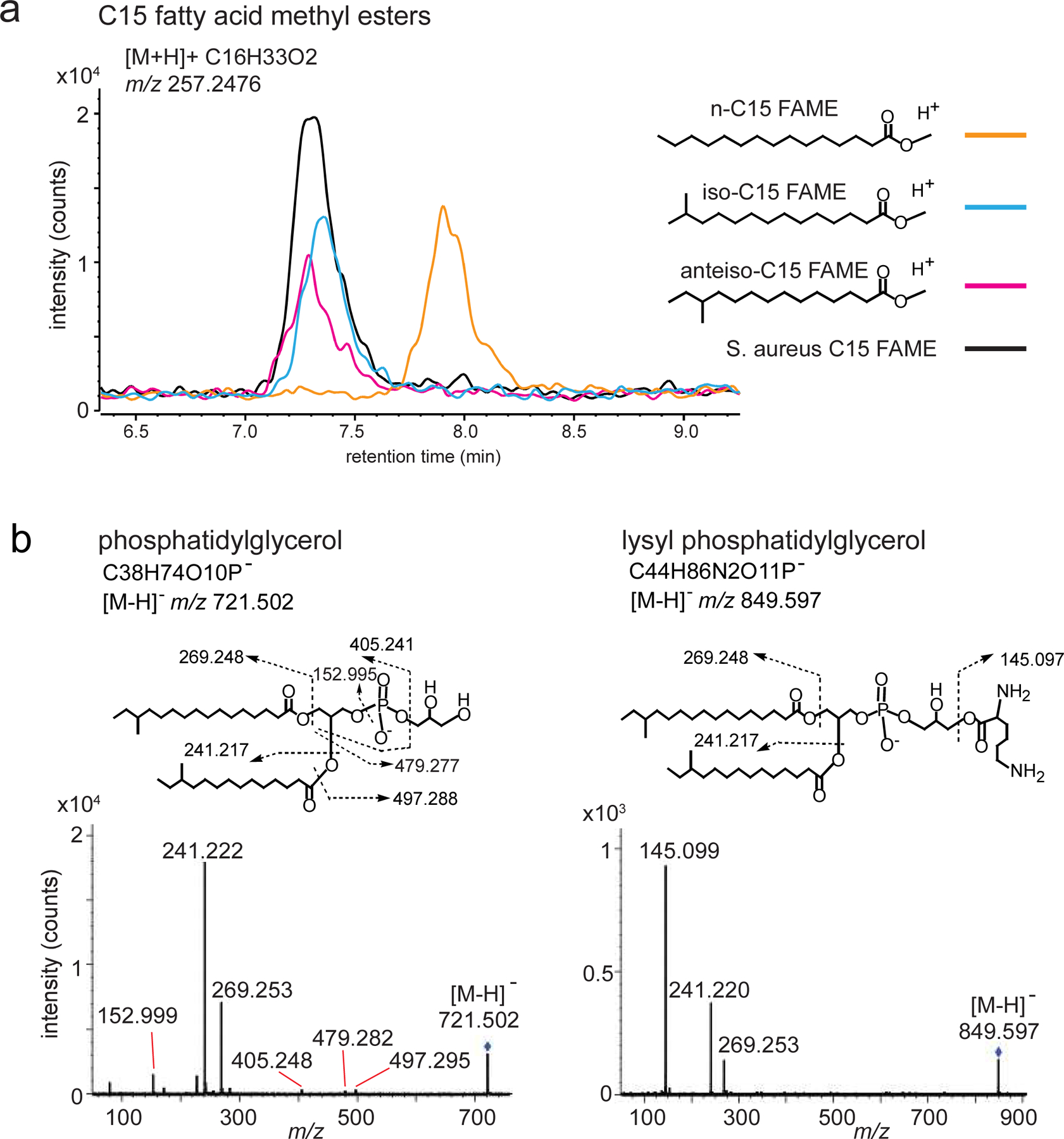
(a) S. aureus PGs were saponified and methylated to form FAMEs. The bacterial C15 fatty acid was determined as branched by co-elution of its FAME with iso-C15 FAME and anteiso-C15 FAME external standards, but not with straight chain n-C15 FAME. Data are presented as an overlay of mass chromatograms of m/z. 257.2476 [C15 FAME+H]+ by the positive mode reversed phase HPLC-QTOF-MS analysis. (b) CID_MS spectra for PG and lysylPG supports assignments in Figure 6b.
Extended data Figure 6.
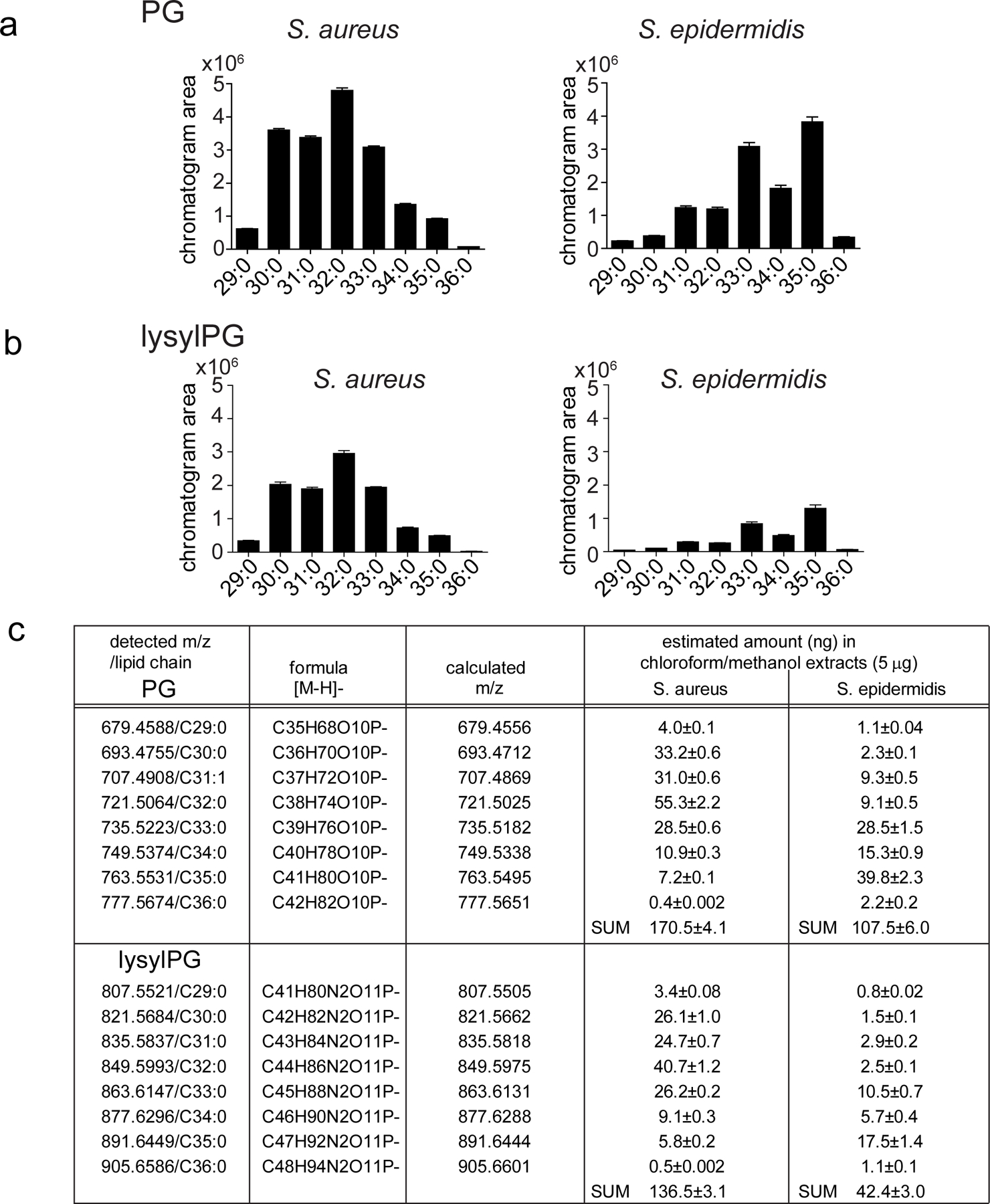
(a, b) Mass chromatogram of eight most abundant PG and lysylPG species from S. aureus and S. epidermidis are depicted. (c) The detected PGs and lysylPGs by the QToF mass spectrometer are summarized. The quantity (ng) of each lipid species in injected total lipid extracts (5µg) was determined by curve fitting of the chromatogram area to the external standard curve. The measurements were performed in triplicate (mean ± SD).
Extended data Figure 7.
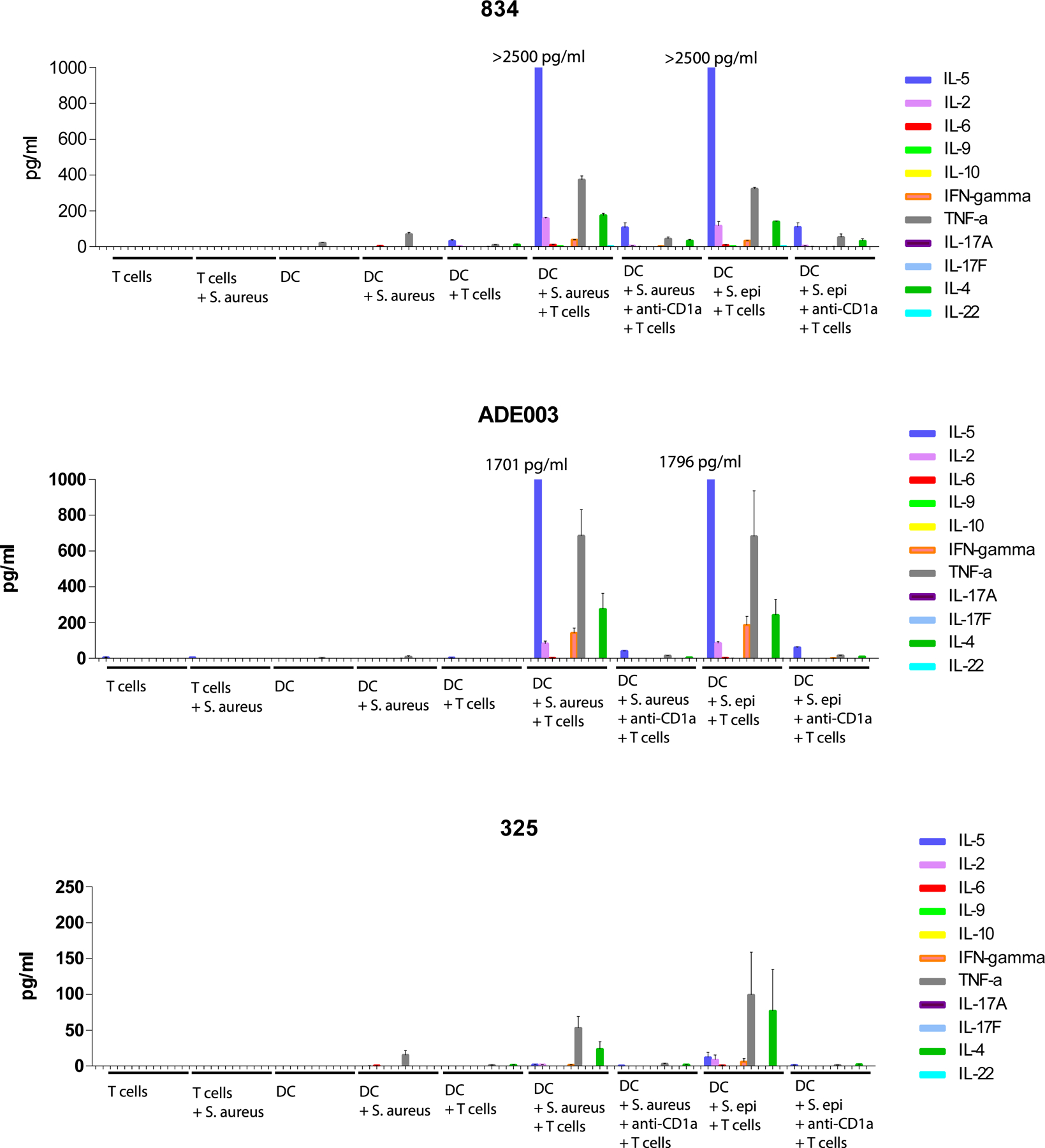
Supernatants from the IL-13 Elispot assays of DC – S. aureus/ S. epidermidis co-cultures with CD1a-(lysyl)PG tetramer+ CD4+ T cells (Figure 6c) were analyzed by LEGENDplex in duplicate. This panel measured 12 different human T cell cytokines, 11 of which are depicted (IL-13 was measured using Elispot and could therefore not be reliably measured in the supernatant). Concentrations in the supernatant for indicated cytokines are depicted for each culture condition (mean ± SD).
Extended data Figure 8.
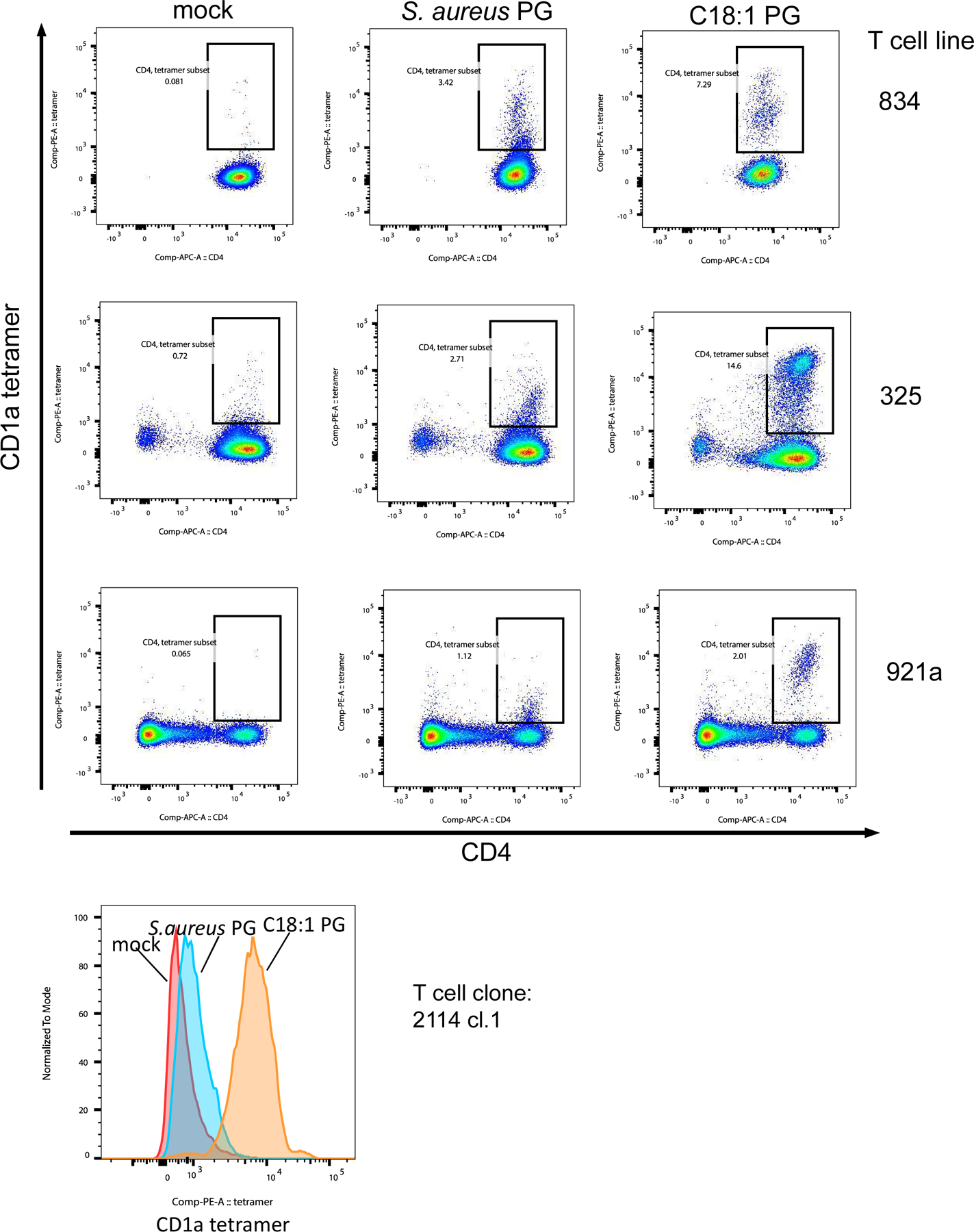
Three T cell lines (834, 325, 921a) and one T cell clone (2114.1) were stained with mock-treated, S.aureus PG-treated and C18:1 PG-treated CD1a tetramers. The lines were co-stained with CD4 antibody and DAPI, and live cells were analyzed for tetramer staining by flow cytometry.
Supplementary Material
ACKNOWLEDGEMENTS
We thank the NIH Tetramer Core Facility for CD1 proteins, Peter Sims and Michael Finlayson for their advice regarding the analysis of single cell RNA-sequencing data, Andreas Peschel for providing S. aureus strain, Quinten Cremers for assistance with ChemDraw, and Ildiko van Rhijn for critical reading of the manuscript. The work is supported by the NIAMS (R01 AR074037, K01 AR068475, P30 AR069632 to A.d.J. and, R01 AR048632 to D.B.M.) and the Wellcome Trust Collaborative Award (to D.B.M., G.O. and J.R.) as well as an Irving Scholarship (to A.d.J.). J.R. is supported by an NHMRC Investigator award. G.O. receives funding from the Medical Research Council UK and NIHR Oxford Biomedical Research Centre. This work was supported by the NCI Cancer Center Support Grant (P30 CA013696 and used the Genomics and High Throughput Screening Shared Resource, and by the NCATS (UL1 TR001873), as well as funding to CUIMC flow core facilities through S10RR027050 and S10OD020056. We thank the staff of the MX1 beamline of the Australian Synchrotron, part of Australian Nuclear Science and Technology (ANSTO).
Footnotes
COMPETING INTERESTS STATEMENT
A.d.J. and D.B.M. provide consulting to Pfizer. G.O. and Y.L.C. have relevant research collaborations with UCB and Janssen and a patent related to CD1a. The remaining authors declare no competing interests.
REFERENCES
- 1.Murphy GF, Bhan AK, Sato S, Mihm MC Jr. & Harrist TJ A new immunologic marker for human Langerhans cells. The New England journal of medicine 304, 791–792 (1981). [DOI] [PubMed] [Google Scholar]
- 2.de Fraissinette A, Schmitt D & Thivolet J Langerhans cells of human mucosa. The Journal of dermatology 16, 255–262 (1989). [DOI] [PubMed] [Google Scholar]
- 3.Kubo A, Nagao K, Yokouchi M, Sasaki H & Amagai M External antigen uptake by Langerhans cells with reorganization of epidermal tight junction barriers. The Journal of experimental medicine 206, 2937–2946 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Byrd AL, Belkaid Y & Segre JA The human skin microbiome. Nature reviews. Microbiology 16, 143–155 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Moody DB et al. T cell activation by lipopeptide antigens. Science 303, 527–531 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Kasmar AG et al. Cutting Edge: CD1a tetramers and dextramers identify human lipopeptide-specific T cells ex vivo. Journal of immunology 191, 4499–4503 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouchi T et al. Langerhans cell antigen capture through tight junctions confers preemptive immunity in experimental staphylococcal scalded skin syndrome. The Journal of experimental medicine 208, 2607–2613 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sohlenkamp C & Geiger O Bacterial membrane lipids: diversity in structures and pathways. FEMS microbiology reviews 40, 133–159 (2016). [DOI] [PubMed] [Google Scholar]
- 9.Dugail I, Kayser BD & Lhomme M Specific roles of phosphatidylglycerols in hosts and microbes. Biochimie 141, 47–53 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Peschel A et al. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. The Journal of experimental medicine 193, 1067–1076 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slavetinsky C, Kuhn S & Peschel A Bacterial aminoacyl phospholipids - Biosynthesis and role in basic cellular processes and pathogenicity. Biochim Biophys Acta Mol Cell Biol Lipids 1862, 1310–1318 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Ernst CM & Peschel A MprF-mediated daptomycin resistance. International journal of medical microbiology : IJMM 309, 359–363 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Kuhn S, Slavetinsky CJ & Peschel A Synthesis and function of phospholipids in Staphylococcus aureus. International journal of medical microbiology : IJMM 305, 196–202 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Hines KM et al. Lipidomic and Ultrastructural Characterization of the Cell Envelope of Staphylococcus aureus Grown in the Presence of Human Serum. mSphere 5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grice EA et al. Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morita SY & Terada T Enzymatic measurement of phosphatidylglycerol and cardiolipin in cultured cells and mitochondria. Scientific reports 5, 11737 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Jong A et al. CD1a-autoreactive T cells are a normal component of the human alphabeta T cell repertoire. Nature immunology 11, 1102–1109 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cotton RN et al. Human skin is colonized by T cells that recognize CD1a independently of lipid. The Journal of clinical investigation 131, e140706 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Jong A et al. CD1a-autoreactive T cells recognize natural skin oils that function as headless antigens. Nature immunology 15, 177–185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birkinshaw RW et al. alphabeta T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nature immunology 16, 258–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Lalla C et al. High-frequency and adaptive-like dynamics of human CD1 self-reactive T cells. European journal of immunology 41, 602–610 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Danner S, Pabst G, Lohner K & Hickel A Structure and thermotropic behavior of the Staphylococcus aureus lipid lysyl-dipalmitoylphosphatidylglycerol. Biophysical journal 94, 2150–2159 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hashimoto K et al. Single-cell transcriptomics reveals expansion of cytotoxic CD4 T cells in supercentenarians. Proceedings of the National Academy of Sciences of the United States of America 116, 24242–24251 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeuchi A et al. CRTAM determines the CD4+ cytotoxic T lymphocyte lineage. The Journal of experimental medicine 213, 123–138 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeuchi A & Saito T CD4 CTL, a Cytotoxic Subset of CD4(+) T Cells, Their Differentiation and Function. Frontiers in immunology 8, 194 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patil VS et al. Precursors of human CD4(+) cytotoxic T lymphocytes identified by single-cell transcriptome analysis. Science immunology 3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szabo PA et al. Single-cell transcriptomics of human T cells reveals tissue and activation signatures in health and disease. Nature communications 10, 4706 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cano-Gamez E et al. Single-cell transcriptomics identifies an effectorness gradient shaping the response of CD4(+) T cells to cytokines. Nature communications 11, 1801 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gutierrez-Arcelus M et al. Lymphocyte innateness defined by transcriptional states reflects a balance between proliferation and effector functions. Nature communications 10, 687 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zheng W & Flavell RA The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89, 587–596 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Flavell RA et al. Molecular basis of T-cell differentiation. Cold Spring Harbor symposia on quantitative biology 64, 563–571 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Thornton AM et al. Expression of Helios, an Ikaros transcription factor family member, differentiates thymic-derived from peripherally induced Foxp3+ T regulatory cells. Journal of immunology 184, 3433–3441 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gregg R et al. The number of human peripheral blood CD4+ CD25high regulatory T cells increases with age. Clinical and experimental immunology 140, 540–546 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leyden JJ, Marples RR & Kligman AM Staphylococcus aureus in the lesions of atopic dermatitis. The British journal of dermatology 90, 525–530 (1974). [DOI] [PubMed] [Google Scholar]
- 35.Kong HH et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome research 22, 850–859 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.De Benedetto A et al. Tight junction defects in patients with atopic dermatitis. The Journal of allergy and clinical immunology 127, 773–786 e771–777 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bos JD et al. Predominance of “memory” T cells (CD4+, CDw29+) over “naive” T cells (CD4+, CD45R+) in both normal and diseased human skin. Archives of dermatological research 281, 24–30 (1989). [DOI] [PubMed] [Google Scholar]
- 38.Guttman-Yassky E et al. Major differences in inflammatory dendritic cells and their products distinguish atopic dermatitis from psoriasis. The Journal of allergy and clinical immunology 119, 1210–1217 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Van Rhijn I et al. Human autoreactive T cells recognize CD1b and phospholipids. Proceedings of the National Academy of Sciences of the United States of America 113, 380–385 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shahine A et al. A molecular basis of human T cell receptor autoreactivity toward self-phospholipids. Science immunology 2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shahine A et al. A T-cell receptor escape channel allows broad T-cell response to CD1b and membrane phospholipids. Nature communications 10, 56 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wolf BJ et al. Identification of a Potent Microbial Lipid Antigen for Diverse NKT Cells. Journal of immunology 195, 2540–2551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tatituri RV et al. Recognition of microbial and mammalian phospholipid antigens by NKT cells with diverse TCRs. Proceedings of the National Academy of Sciences of the United States of America 110, 1827–1832 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Visvabharathy L et al. Group 1 CD1-restricted T cells contribute to control of systemic Staphylococcus aureus infection. PLoS pathogens 16, e1008443 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zajonc DM et al. Molecular mechanism of lipopeptide presentation by CD1a. Immunity 22, 209–219 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Cotton RN et al. CD1a selectively captures endogenous cellular lipids that broadly block T cell response. The Journal of experimental medicine 218 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Veldhuizen R, Nag K, Orgeig S & Possmayer F The role of lipids in pulmonary surfactant. Biochimica et biophysica acta 1408, 90–108 (1998). [DOI] [PubMed] [Google Scholar]
References Methods
- 48.Clark RA et al. A novel method for the isolation of skin resident T cells from normal and diseased human skin. The Journal of investigative dermatology 126, 1059–1070 (2006). [DOI] [PubMed] [Google Scholar]
- 49.Kabsch W. Xds. Acta crystallographica. Section D, Biological crystallography 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winn MD et al. Overview of the CCP4 suite and current developments. Acta crystallographica. Section D, Biological crystallography 67, 235–242 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCoy AJ et al. Phaser crystallographic software. Journal of applied crystallography 40, 658–674 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG & Cowtan K Features and development of Coot. Acta crystallographica. Section D, Biological crystallography 66, 486–501 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Afonine PV et al. Towards automated crystallographic structure refinement with phenix.refine. Acta crystallographica. Section D, Biological crystallography 68, 352–367 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bligh EG & Dyer WJ A rapid method of total lipid extraction and purification. Canadian journal of biochemistry and physiology 37, 911–917 (1959). [DOI] [PubMed] [Google Scholar]
- 55.van ‘t Klooster JS et al. Periprotein lipidomes of Saccharomyces cerevisiae provide a flexible environment for conformational changes of membrane proteins. eLife 9 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bush EC et al. PLATE-Seq for genome-wide regulatory network analysis of high-throughput screens. Nature communications 8, 105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lun ATL & Marioni JC Overcoming confounding plate effects in differential expression analyses of single-cell RNA-seq data. Biostatistics 18, 451–464 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Snyder ME et al. Generation and persistence of human tissue-resident memory T cells in lung transplantation. Science immunology 4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Granot T et al. Dendritic Cells Display Subset and Tissue-Specific Maturation Dynamics over Human Life. Immunity 46, 504–515 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liao Y, Smyth GK & Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Osorio D & Cai JJ Systematic determination of the mitochondrial proportion in human and mice tissues for single-cell RNA-sequencing data quality control. Bioinformatics 37, 963–967 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The CD1a-PG final model and structure factor files were deposited in the Protein Data Base (PDB) under PDB ID: 7SH4.
Single-cell and bulk RNA-sequencing files are available in Gene Expression Omnibus (GEO) under accession number GSE186459


