Summary
Population-based genome-wide association studies (GWAS) normally require a large sample size, which can be labor intensive and costly. Recently, we reported a human induced pluripotent stem cell (hiPSC) array-based GWAS method, identifying NDUFA4 as a host factor for Zika virus (ZIKV) infection. In this study, we extended our analysis to trophectoderm cells, which constitute one of the major routes of mother-to-fetus transmission of ZIKV during pregnancy. We differentiated hiPSCs from various donors into trophectoderm cells. We then infected cells carrying loss of function mutations in NDUFA4, harboring risk versus non-risk alleles of SNPs (rs917172 and rs12386620) or having deletions in the NDUFA4 cis-regulatory region with ZIKV. We found that loss/reduction of NDUFA4 suppressed ZIKV infection in trophectoderm cells. This study validated our published hiPSC array-based system as a useful platform for GWAS and confirmed the role of NDUFA4 as a susceptibility locus for ZIKV in disease-relevant trophectoderm cells.
Subject areas: Stem cells research, Virology
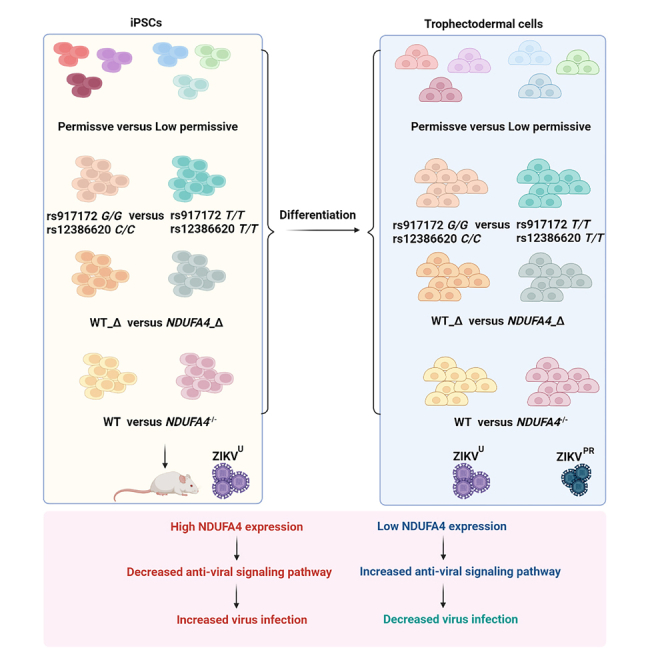
Graphical abstract
Highlights
-
•
Trophectoderm from various hiPSCs show variable susceptibility to ZIKV
-
•
A deficiency of NDUFA4 in trophectoderm causes reduced ZIKV infection
-
•
Trophectoderm carrying risk alleles show increased ZIKV infection
-
•
Deletion of NDUFA4 regulatory elements leads to decreased ZIKV infection
Stem cells research; Virology
Introduction
Genome-wide association studies (GWAS) have been successful at identifying specific genetic variations that associate with particular diseases.1 GWAS are typically based on human cohorts, members of whom have diverse lifestyles, variable co-morbidities, and different environmental exposures. Human population based GWAS, therefore, usually requires recruitment of a large cohort, which is expensive and labor-intensive.2 Recently, we performed GWAS using a human induced pluripotent stem cell (hiPSC) array, containing 77 hiPSC lines with distinct genetic backgrounds. We identified NDUFA4 as an important host dependency factor for Zika virus (ZIKV) replication in hiPSCs and hiPSC-derived cerebral organoids.3 In this study, we explore the relevance of NDUFA4 to ZIKV infection in trophectoderm cells, which constitute a route of ZIKV transmission from mother to fetus and therefore represent an important disease-relevant cell type.
ZIKV is a member of the Flaviviridae family. Vertical transmission of ZIKV from mother to fetus is linked to increased incidence of congenital ZIKV syndrome in babies, including microcephaly, congenital malformation, and fetal demise.4,5,6,7 Among mothers with confirmed or suspected infection of ZIKV, about 5% give birth to babies suffering from ZIKV-associated birth defects. Unfortunately, there are no vaccines or drugs to prevent or treat ZIKV infection. Therefore, it is urgent to explore the mechanism and find a potential treatment for ZIKV infection during pregnancy.8 The outer layer of the blastocyst-stage embryo is trophectoderm, which is the precursor of all trophoblast cells of the placenta. Recent studies using pre-implantation human and mouse embryos demonstrated that trophectoderm cells can be infected with and propagate ZIKV, causing neural progenitor cell death.9 In addition, ZIKV can infect the trophectoderm of in vitro fertilized rhesus macaque embryos, which negatively impacts the attachment, growth, and survival of the blastocysts.10 Studies also showed that ZIKV can infect both cytotrophoblast and syncytiotrophoblast derived from placental villi at term and colonies of trophoblast differentiated from embryonic stem cells (ESC).11 ZIKV infection of the trophoblast cells affects cell migration, metabolism and immune homeostasis control,12 endoplasmic reticulum stress, apoptosis13 and pyroptosis.14 Type III interferons produced by human placental trophoblasts protect the non-placental cells against ZIKV infection.15 In addition, ZIKV infected trophoblasts become a target of decidual natural killer cells, which release their granules and kill ZIKV-infected trophoblasts.16 ZIKV infected decidual cells exaggerate the infection of trophoblasts.17 Here, we systematically examine the impact of NDUFA4 and associated SNPs on the permissiveness of trophectoderm cells to ZIKV infection. In addition, we created a xenograft model to examine the impact of the loss of NDUFA4 on ZIKV infection in vivo. Together, our studies show that NDUFA4 and associated SNPs affect the ZIKV infection of trophectoderm cells in vitro and in vivo.
Results
Trophectoderm cells originating from different hiPSC lines show variable permissiveness to ZIKV infection
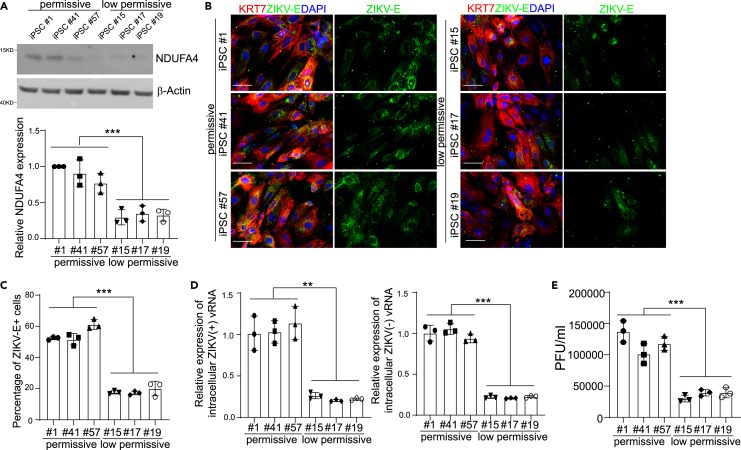
In our previous study, we screened 77 hiPSC lines for their ability to support ZIKV infection, identifying 39 “low-permissive” lines and 38 “permissive” lines.3,18To determine whether trophectoderm cells derived from these hiPSC lines also show differential permissiveness to ZIKV infection, three permissive lines (hiPSC lines #1, #41, and #57) and three low-permissive lines (hiPSC lines #15, #17 and #19) were differentiated into trophectoderm cells3,9 (Figure S1A). All hiPSC lines showed a similar capacity to generate keratin-7-positive (KRT7+) trophectoderm cells (∼90%, Figures S1B and S1C). We performed several assays to test the permissiveness of trophectoderm cells to ZIKV infection. Consistent with the expression of NDUFA4 in hiPSCs, trophectoderm cells derived from permissive hiPSC lines expressed higher levels of NDUFA4 compared to those derived from low-permissive lines (Figure 1A). As shown in Figures 1B and 1C, the percentage of ZIKV-E+ cells was significantly higher for trophectoderm cells derived from permissive hiPSC lines compared to those derived from low-permissive lines (ZIKVPR, Puerto Rico strain, PRVABC59). In addition, significantly higher levels of both (+) and (−) strands of ZIKV vRNA were detected in trophectoderm cells derived from permissive hiPSC lines (Figure 1D). A similar pattern was seen when the infection efficiency was monitored by measuring the release of infectious particles in the supernatant of infected cells. (Figure 1E). In all, these findings indicate that ZIKV infection in trophectoderm cells follows the same pattern seen in their progenitor hiPSCs.
Figure 1.
Trophectoderm cells derived from different hiPSC lines display distinct ZIKV permissiveness
(A) Western blotting analysis and the quantification of NDUFA4 protein expression in trophectoderm cells derived from permissive cell lines: iPSC #1, iPSC #41 and iPSC #57 and low permissive cell lines: iPSC #15, iPSC #17 and iPSC #19. β-Actin was used as a loading control.
(B and C) Representative confocal images (B) and the quantification (C) of ZIKV-E staining in KRT7+ trophectoderm cells derived from permissive cell lines: iPSC #1, iPSC #41 and iPSC #57 and low permissive cell lines: iPSC #15, iPSC #17 and iPSC #19 at 72 hpi (ZIKVPR, MOI = 1). Scale bar = 50 μm.
(D) qRT-PCR analysis of (+) or (−) ZIKV vRNA strands in trophectoderm cells derived from permissive cell lines: iPSC #1, iPSC #41 and iPSC #57 and low permissive cell lines: iPSC #15, iPSC #17 and iPSC #19 at 72 hpi (ZIKVPR, MOI = 1).
(E) Viral titers of ZIKV in the supernatant of trophectoderm cells derived from permissive cell lines: iPSC #1, iPSC #41 and iPSC #57 and low permissive cell lines: iPSC #15, iPSC #17 and iPSC #19 (ZIKVPR, MOI = 1) quantified by plaque assay. Data are representative of at least three independent experiments. For p values, we averaged the 3 technical replicates within each cell line, then used the averages for an unpaired two-tailed Student’s t test. ∗∗p < 0.01, and ∗∗∗p < 0.001.
See also Figure S1.
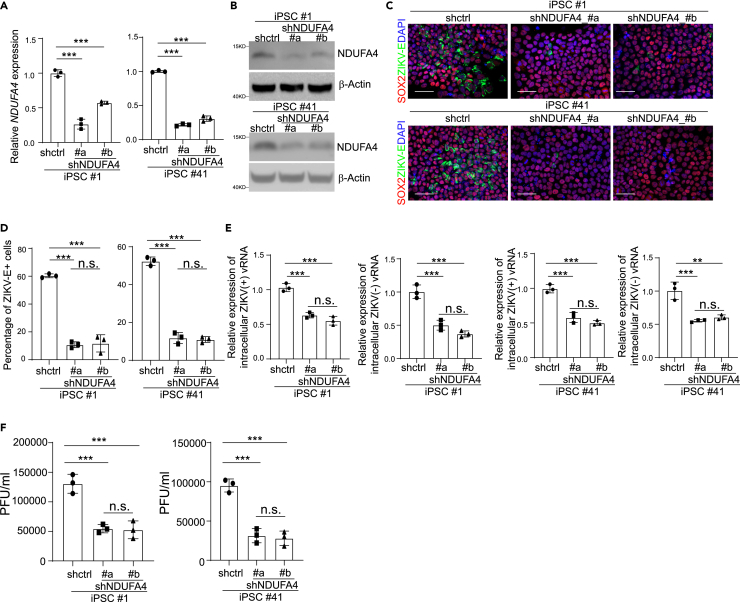
Our previous study established a link between NDUFA4 expression and permissiveness to ZIKV infection using one iPSC line.3 To further validate these findings with multiple iPSC lines from different gender and race backgrounds, we knocked down NDUFA4 in two permissive iPSC lines (#1 and #41) using shRNAs. qRT-PCR (Figure 2A) and western blotting assays (Figure 2B) confirmed the reduction of NDUFA4 levels in iPSCs expressing shNDUFA4 #a or shNDUFA4 #b. Following ZIKV infection, the iPSCs carrying shNDUFA4 #a or shNDUFA4 #b showed a lower percentage of ZIKV-E+ cells at 72 hpi (ZIKVPR: Figures 2C and 2D; ZIKVU: West Africa strain, MR766: Figures S2A and S2B) and decreased levels of ZIKV (+) and (−) vRNA strands (ZIKVPR: Figure 2E; ZIKVU: Figure S2C) compared to control iPSCs carrying scrambled shRNA. Consistently, the decreased yields of infectious ZIKV were detected in the supernatant of the iPSCs carrying shNDUFA4 #a or shNDUFA4 #b (ZIKVPR: Figure 2F; ZIKVU: Figure S2D).
Figure 2.
Knockdown of NUDFA4 decreases ZIKV infection permissiveness
(A) qRT-PCR analysis of NDUFA4 mRNA expression levels in the NDUFA4 knockdown cell lines of iPSC #1 (shctrl, shNDUFA4 #a, shNDUFA4 #b) and iPSC 41 (shctrl, shNDUFA4 #a, shNDUFA4 #b). The shctrl is a scrambled shRNA.
(B) Western blotting analysis of NDUFA4 protein expression levels in the NDUFA4 knockdown cell lines of iPSC #1 (shctrl, shNDUFA4 #a, shNDUFA4 #b) and iPSC #41 (shctrl, shNDUFA4 #a, shNDUFA4 #b). β-Actin was used as a loading control.
(C and D) Representative confocal images (C) and the quantification (D) of SOX2 and ZIKV-E staining in NDUFA4 knockdown cell lines of iPSC #1 (shctrl, shNDUFA4 #a, shNDUFA4 #b) or iPSC #41 (shctrl, shNDUFA4 #a, shNDUFA4 #b) at 72 hpi (ZIKVPR, MOI = 1). Scale bar = 50 μm.
(E) qRT-PCR analysis of (+) and (−) ZIKV vRNA strands in NDUFA4 knockdown cell lines of iPSC #1 (shctrl, shNDUFA4 #a, shNDUFA4 #b) or iPSC #41 (shctrl, shNDUFA4 #a, shNDUFA4 #b) at 72 hpi (ZIKVPR, MOI = 1).
(F) Viral titers of ZIKV in the supernatant of NDUFA4 knockdown cell lines of iPSC #1 (shctrl, shNDUFA4 #a, shNDUFA4 #b) or iPSC #41 (shctrl, shNDUFA4 #a, shNDUFA4 #b) at 72 hpi (ZIKVPR, MOI = 1) quantified by plaque assay. Data are representative of at least three independent experiments. p values were calculated by one-way ANOVA followed by a Dunnett’s post hoc test with a common control for multiple testing correction. n.s. no significance and ∗∗∗p < 0.001.
See also Figure S2.
Deficiency of NDUFA4 in trophectoderm cells leads to decreased ZIKV infection
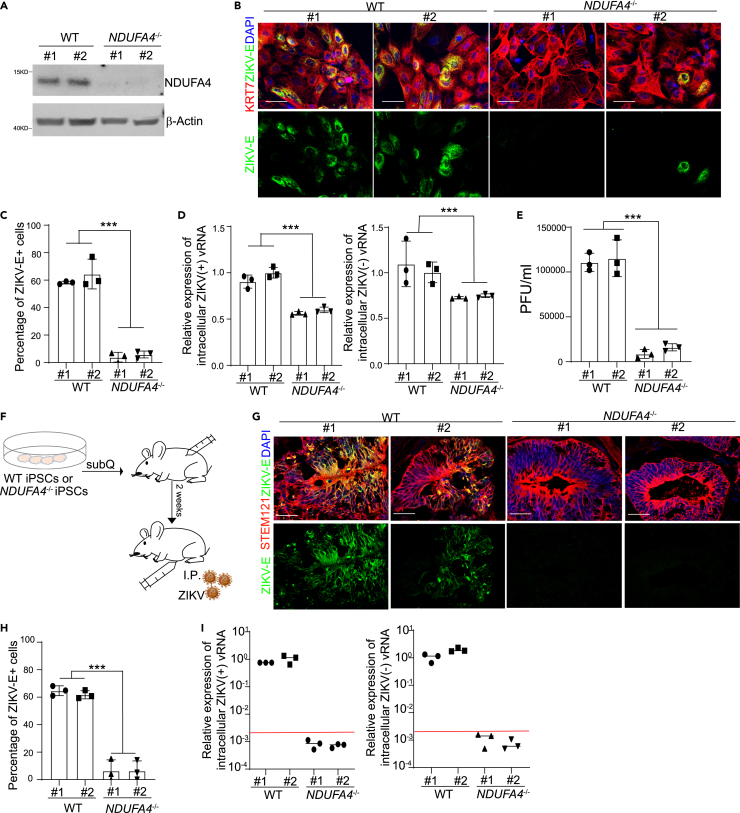
In our previous study, we generated NDUFA4−/− iPSC lines using a CRISPR-based gene knockout strategy.3 To determine the role of NDUFA4 in ZIKV infection in trophectoderm cells, WT and NDUFA4−/− iPSCs were first differentiated into trophectoderm cells. Western blot analysis confirmed the loss of NDUFA4 expression in NDUFA4−/− trophectoderm cells (Figure 3A). WT and NDUFA4−/− iPSCs demonstrated similar capacities to differentiate into KRT7+ trophectoderm cells (Figures S3A and S3B), suggesting that the loss of NDUFA4 does not affect iPSC differentiation to trophectoderm cells. Following infection of WT or NDUFA4−/− iPSC-derived trophectoderm cells with ZIKV, we found a lower percentage of ZIKV-E+ cells in NDUFA4−/− trophectoderm cells (ZIKVPR: Figures 3B and 3C; ZIKVU: Figures S3C and S3D). Consistent with this observation, we found lower levels of (+) and (−) strand ZIKV vRNA in NDUFA4−/− trophectoderm cells (ZIKVPR: Figure 3D; ZIKVU: Figure S3E), as well as lower viral titers in the supernatant of NDUFA4−/− trophectoderm cells (ZIKVPR: Figure 3E; ZIKVU: Figure S3F).
Figure 3.
Deficiency of NDUFA4 leads to decreased ZIKV infection in vitro and in vivo
(A) Western blotting analysis of NDUFA4 protein expression in trophectoderm cells derived from WT or NDUFA4−/− hiPSCs. β-Actin was used as a loading control.
(B and C) Representative confocal images (B) and the quantification (C) of ZIKV-E staining in KRT7+ trophectoderm cells derived from WT or NDUFA4−/− hiPSCs at 72 hpi (ZIKVPR, MOI = 1). Scale bar = 50 μm.
(D) qRT-PCR analysis of (+) or (−) ZIKV vRNA strands in trophectoderm cells derived from WT or NDUFA4−/− hiPSCs at 72 hpi (ZIKVPR, MOI = 1).
(E) Viral titers of ZIKV in the supernatant of trophectoderm cells derived from WT or NDUFA4−/− hiPSCs at 72 hpi (ZIKVPR, MOI = 1) quantified by plaque assay.
(F) Scheme of the in vivo xeno-transplantation and ZIKV infection.
(G and H) Representative images (G) and the quantification (H) of ZIKV-E and STEM121 staining in WT or NDUFA4−/− hiPSCs derived xenografts at 5 dpi (ZIKVU, 5 × 105 PFU). Scale bar = 50 μm.
(I) qRT-PCR analysis of (+) or (−) ZIKV vRNA strands in WT or NDUFA4−/− hiPSCs derived xenografts at 5 dpi (ZIKVU, 5 × 105 PFU). The viral RNA level in NDUFA4−/− hiPSCs derived xenografts is below detection. The red line indicates the detection limit. Data are representative of at least three independent experiments. p values were calculated by two-way ANOVA analysis; ∗∗∗p < 0.001.
See also Figure S3.
To examine the permissiveness of NDUFA4−/− and WT hiPSCs to ZIKV infection in vivo, hiPSCs were transplanted subcutaneously into immune-deficient SCID-beige mice, and the mice were inoculated with ZIKVU by intraperitoneal injection two weeks post-transplantation (Figure 3F). Five days post infection, the xenografts were collected and stained with antibodies against human antigen (STEM121) and ZIKV-E. The percentage of ZIKV-E+ cells in NDUFA4−/− iPSC-derived xenografts was significantly lower compared with WT hiPSC-derived xenografts (Figures 3G and 3H). We also found decreased viral RNA in NDUFA4−/− iPSC-derived xenografts examined by qRT-PCR (Figure 3I).
Trophectoderm cells derived from hiPSC lines carrying the risk alleles of rs917172 and rs12386620 show increased ZIKV infection
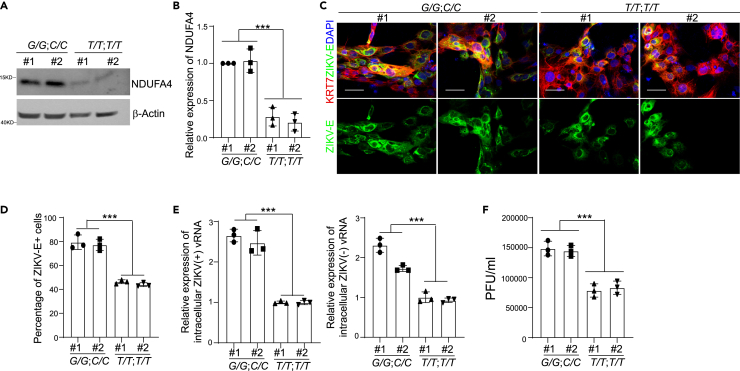
Our hiPSC-based GWAS analysis identified several SNPs that are associated with permissiveness to ZIKV infection, including rs917172 and rs12386620 located in an approximately 1 kb region downstream of NDUFA4. Using CRISPR based gene editing, we created isogenic hiPSC lines with risk G allele for rs917172 and risk C allele for rs12386620. Two hiPSC clones with non-risk T alleles for both rs917172 and rs12386620 loci were used as controls. To determine the role of genetic variants in disease-relevant trophectoderm cells, the isogenic hiPSC lines carrying risk alleles (G/G; C/C) or non-risk alleles (T/T; T/T) were differentiated into trophectoderm cells. Trophectoderm cells derived from hiPSC lines carrying risk alleles (G/G; C/C) showed higher NDUFA4 expression than non-risk alleles (T/T; T/T) (Figures 4A and 4B). The iPSC lines carrying risk alleles (G/G; C/C) or non-risk alleles (T/T; T/T) equally efficiently differentiated into KRT7+ trophectoderm cells (Figures S4A and S4B). 72h after ZIKV infection, a higher percentage of ZIKV-E+ cells were found in trophectoderm cells carrying risk alleles (G/G; C/C) compared to those carrying non-risk alleles (T/T; T/T) (ZIKVPR: Figures 4C and 4D; ZIKVU: Figures S4C and S4D). Similarly, increased levels of ZIKV (+) and (−) vRNA strands (ZIKVPR: Figure 4E; ZIKVU: Figure S4E) and infectious virus particles (ZIKVPR: Figure 4F; ZIKVU: Figure S4F) were detected in trophectoderm cells harboring risk alleles (G/G; C/C) than non-risk alleles (T/T; T/T).
Figure 4.
Trophectoderm cells derived from iPSCs with risk alleles of rs917172 and rs12386620 show increased sensitivity to ZIKV infection
(A and B) Western blotting analysis (A) and quantification (B) of NDUFA4 protein expression in trophectoderm cells derived from hiPSCs carrying risk (G/G; C/C) or non-risk (T/T; T/T) alleles. β-Actin was used as a loading control.
(C and D) Representative confocal images (C) and the quantification (D) of ZIKV-E staining in KRT7+ trophectoderm cells derived from hiPSCs carrying risk (G/G; C/C) or non-risk (T/T; T/T) alleles at 72 hpi (ZIKVPR, MOI = 1). Scale bar = 50 μm.
(E) qRT-PCR analysis of (+) or (−) ZIKV vRNA strands in trophectoderm cells derived from hiPSCs carrying risk (G/G; C/C) or non-risk (T/T; T/T) alleles at 72 hpi (ZIKVPR, MOI = 1).
(F) Viral titers of ZIKV in the supernatant of trophectoderm cells derived from hiPSCs carrying risk (G/G; C/C) or non-risk (T/T; T/T) alleles at 72 hpi (ZIKVPR, MOI = 1) quantified by plaque assay. Data are representative of at least three independent experiments. p values were calculated by two-way ANOVA analysis; ∗∗∗p < 0.001.
See also Figure S4.
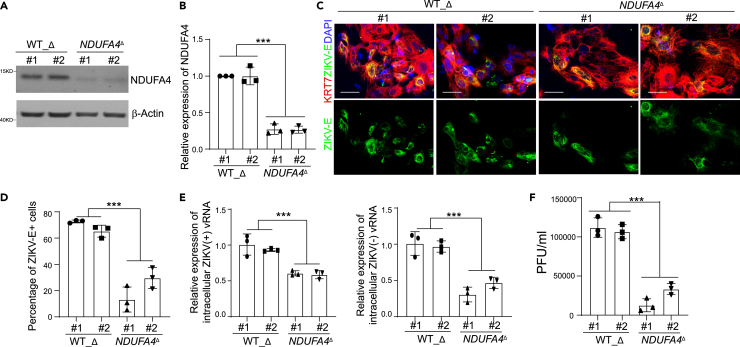
Trophectoderm cells carrying deleted cis-regulatory elements of NDUFA4 show decreased ZIKV infection
Rs917172 and rs12386620 are located in an approximately 1 kb region downstream of NDUFA4. We previously knocked out this putative cis-regulatory region in hiPSCs to obtain NDUFA4Δ cells. To determine whether this regulatory region is associated with permissiveness to ZIKV infection in trophectoderm cells, we differentiated NDUFA4Δ and WT_Δ hiPSC lines into trophectoderm cells. Trophectoderm cells derived from WT_Δ hiPSC lines showed higher NDUFA4 expression than NDUFA4Δ hiPSC lines (Figures 5A and 5B). No difference in differentiation potential was seen between NDUFA4Δ and WT_Δ iPSCs (Figures S5A and S5B). After ZIKV infection, a lower percentage of ZIKV-E+ cells were detected in NDUFA4Δ hiPSC-derived trophectoderm cells compared with WT_Δ hiPSC-derived trophectoderm cells (ZIKVPR: Figures 5C and 5D; ZIKVU: Figures S5C and S5D). In addition, the levels of ZIKV (+) and (−) vRNA strands were significantly lower in NDUFA4Δ hiPSC-derived trophectoderm cells compared with WT_Δ hiPSC-derived trophectoderm cells (ZIKVPR: Figure 5E; ZIKVU: Figure S5E). Consistently, a lower yield of infectious ZIKV was detected in the supernatant of trophectoderm cells derived from NDUFA4Δ hiPSCs (ZIKVPR: Figure 5F; ZIKVU: Figure S5F). In summary, these data suggest that the genomic region containing the cluster of SNPs functions as a cis-regulatory region for NDUFA4 expression relevant to ZIKV infection of trophectoderm cells.
Figure 5.
Trophectoderm cells derived from iPSCs with deleted cis-regulatory region of NDUFA4 show decreased ZIKV infection
(A and B) Western blotting analysis (A) and quantification (B) of NDUFA4 protein expression in trophectoderm cells derived from WT_Δ or NDUFA4Δ hiPSCs. β-Actin was used as a loading control.
(C and D) Representative confocal images (C) and the quantification (D) of ZIKV-E staining in KRT7+ trophectoderm cells derived from WT_Δ or NDUFA4Δ hiPSCs at 72 hpi (ZIKVPR, MOI = 1). Scale bar = 50 μm.
(E) qRT-PCR analysis of (+) or (−) ZIKV vRNA strands of trophectoderm cells derived from WT_Δ or NDUFA4Δ hiPSCs at 72 hpi (ZIKVPR, MOI = 1).
(F) Viral titers of ZIKV in the supernatant of trophectoderm cells derived from WT_Δ or NDUFA4Δ hiPSCs at 72 hpi (ZIKVPR, MOI = 1) quantified by plaque assay. Data are representative of at least three independent experiments. p values were calculated by two-way ANOVA analysis; ∗∗∗p < 0.001.
See also Figure S5.
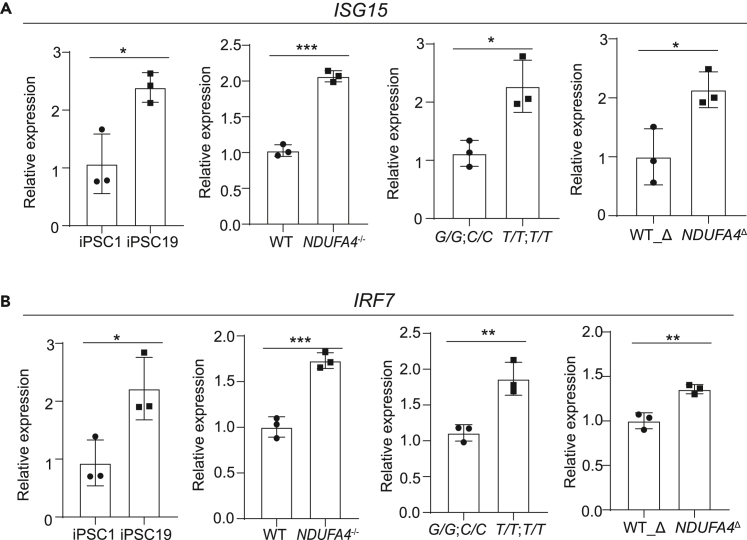
Our previous study showed that NDUFA4 deficiency leads to mitochondrial DNA leakage, which can trigger type I interferon signaling in hiPSCs, leading to upregulation of interferon-stimulated genes (ISGs).3 To test if NDUFA4 exerts a similar effect in trophectoderm cells, we examined the expression of two ISGs, ISG15 and interferon regulatory factor 7 (IRF7), in hiPSC-derived trophectoderm cells. Increased expression of these ISGs was seen in cells derived from NDUFA4−/− iPSCs versus WT iPSCs, iPSC lines carrying non-risk (T/T; T/T) versus risk (G/G; C/C) alleles, NDUFA4Δ iPSCs versus WT_Δ iPSCs and iPSC #19 versus iPSC #1 lines under ZIKV infected conditions (Figures 6A and 6B). Altogether, we showed that NDUFA4 deficiency leads to increased type I interferon signaling to limit ZIKV infection in trophectoderm cells.
Figure 6.
Loss or reduction of NDUFA4 leads to interferon-stimulated gene expression
(A and B) qRT-PCR analysis of ISG15 (A) and IRF7 (B) mRNA expression levels in trophectoderm cells derived from iPSC #1 v.s. iPSC #19; WT v.s. NDUFA4−/− hiPSCs; risk (G/G; C/C) v.s. non-risk (T/T; T/T) hiPSCs and WT_Δ v.s. NDUFA4Δ hiPSCs at 72 hpi (ZIKVPR, MOI = 1). Data are representative of at least three independent experiments. p values were calculated by unpaired two-tailed Student’s t test; ∗p < 0.05, ∗∗p < 0.01 and ∗∗∗p < 0.001.
Discussion
In our previous study, we conducted a hiPSC-based GWAS and identified NDUFA4 as a locus associated with ZIKV susceptibility.3 Through CRISPR-Cas9 technology, we also showed that the SNPs (rs917172 and rs12386620) and a cis-regulatory region associated with NDUFA4 expression impact ZIKV infection. Yet, our previous study only focused on the role of NDUFA4 in hiPSCs and cerebral organoids. An advantage of hiPSC-based approach is that these cells can be differentiated into multiple disease-relevant cell types. In this study, we extended our analysis to trophectoderm cells, which act as one of the major routes of vertical transmission of ZIKV during pregnancy. We differentiated WT hiPSC and CRISPR-based gene-edited hiPSC into disease-relevant trophectoderm cells and validated the role of NDUFA4 and its associated variants in ZIKV infection. It is interesting that we detected ZIKV envelope protein in some cells not expressing KRT7 following the knockdown of NDUFA4 or with deletion of the cis-regulatory region. We hypothesize that the few non-KRT7-positive cells could be undifferentiated hiPSCs, which were previously reported to be susceptible to ZIKV infection.3
NDUFA4 is a mitochondrial complex associated protein. Recent studies reported that NDUFA4 might be a component of the cytochrome c oxidase (Balsa et al., 2012; Kadenbach, 2017). Here we found that the loss of NDUFA4 leads to decreased susceptibility to ZIKV infection in trophectoderm cells. Mechanistically, lower expression of NDUFA4 induces mitochondrial stress and leakage of mitochondrial DNA, which can induce upregulation of type-1 interferon signaling, blocking viral infection.3,19 Our data suggest that the trophectoderm cells with relatively high expression levels of NDUFA4 may have lower type I interferon signaling, resulting in the increased capacity for viral replication.
In summary, our study establishes the role of NDUFA4 in ZIKV infection of human trophectoderm cells. We showed that the loss of the NDUFA4 cis-regulatory region and the correction of risk-associated SNPs in the NDUFA4 gene led to decreased ZIKV infection in trophectoderm cells. These findings are consistent with our previous report in which we investigated the role of NDUFA4 in hiPSC-derived neuronal cells, suggesting that the function of NDUFA4 is conserved across cell types. Overall, this study highlights the value of our hiPSC-GWAS platform for identifying human genes essential for virus replication in different cellular backgrounds.
Limitations of the study
In this study, we showed that NDUFA4 and its associated genetic variants have crucial roles in ZIKV infection in trophectoderm cells. Further investigation is required to ascertain if these genetic variants correlate with human susceptibility to infection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Flavivirus group antigen [D1-4G2-4-15 (4G2)] | GeneTex | Cat #GTX57154; RRID: AB_2887950 |
| Anti-Flavivirus group antigen [D1-4G2-4-15 (4G2)] | Millipore | Cat #MAB10216-I; RRID: AB_827205 |
| Sox2 Antibody | Cell Signaling | Cat #3579; RRID: AB_2195767 |
| Anti-NDUFA4 antibody | Abcam | Cat #ab129752; RRID: AB_11155881 |
| Anti-Keratin 7 (D1E4) XP® | Cell Signaling | Cat #4465; RRID: AB_11178382 |
| Anti-STEM121 human antigen | Takara | Cat #Y40410; RRID: AB_2801314 |
| Donkey anti-Mouse IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat #A-21202; RRID: AB_141607 |
| Donkey anti-Mouse IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 594 | Thermo Fisher Scientific | Cat #A-21203; RRID: AB_141633 |
| Donkey anti-Rabbit IgG (H + L) Secondary Antibody, Alexa Fluor 594 conjugate | Thermo Fisher Scientific | Cat #A-21207; RRID: AB_141637 |
| Donkey anti-Rabbit IgG (H + L) Secondary Antibody, Alexa Fluor 647 conjugate | Thermo Fisher Scientific | Cat #A-31573; RRID: AB_2536183 |
| Donkey anti-Mouse IgG (H + L) Secondary Antibody, Alexa Fluor 647 | Thermo Fisher Scientific | Cat #A-31571; RRID: AB_162542 |
| DAPI | Santa Cruz | Cat# D1306; RRID: AB_2629482 |
| Bacterial and virus strains | ||
| ZIKVU (West Africa strain, MR766) | ZeptoMetrix | # 0810521CF |
| ZIKVPR (Puerto Rico strain, PRVABC59) | ZeptoMetrix | # 0810521CF |
| Chemicals, peptides, and recombinant proteins | ||
| Y-27632 | MedchemExpress | #HY-10583 |
| Recombinant Human bFGF Protein | Peprotech | #100-18B-500UG |
| Recombinant Human BMP-4 Protein | R & D Systems | #314-BP |
| StemFlex | Gibco Thermo Fisher | #A3349401 |
| Knockout Serum Replacement | Gibco Thermo Fisher | #10828028 |
| DMEM (high glucose) | Gibco Thermo Fisher | #11-965-118 |
| Penicillin-Streptomycin (5,000 U/mL) | Gibco Thermo Fisher | #15070063 |
| GlutaMAX Supplement | Thermo Fisher Scientific | #35050079 |
| MEM Non-Essential Amino Acids Solution (100X) | Gibco Thermo Fisher | #11140050 |
| Accutase | Stem Cell Technologies | # 07920 |
| ReleSR | Stem Cell Technologies | # 05872 |
| Matrigel | Corning | #354234 |
| Deposited data | ||
| Original western blot images | This paper (Elsevier’s Mendeley Data repository) | https://doi.org/10.17632/9y6cbwtwbg.1 |
| Experimental models: Cell lines | ||
| 293T | ATCC | #CRL-11268 |
| Vero E6 | ATCC | #CRL-1586 |
| irradiated CF1 Mouse Embryonic Fibroblasts | Thermo Fisher Scientific | #A34180 |
| Experimental models: Organisms/strains | ||
| SCID-beige mice | The Jackson Laboratory | NA |
| Recombinant DNA | ||
| pLV[Exp]-EGFP:T2A:Puro-EF1A > hNDUFA4 | VectorBuilder | VB900004-0599xvh |
| pLV[shRNA]-EGFP:T2A:Puro-U6>hNDUFA4[shRNA#1] | VectorBuilder | VB900052-8762rzm |
| pLV[shRNA]-EGFP:T2A:Puro-U6>hNDUFA4[shRNA#2] | VectorBuilder | VB900052-8767wwk |
| Software and algorithms | ||
| Adobe illustrator CC2017 | Adobe | https://www.adobe.com/product/photoshop.html |
| Graphpad Prism 8 | Graphpad software | https://www.graphpad.com |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Shuibing Chen (shc2034@med.cornell.edu).
Materials availability
NDUFA4−/−, WT iPSCs, iPSCs carrying risk (G/G; C/C) or non-risk (T/T; T/T) alleles, and NDUFA4Δ, WT_Δ iPSCs are available upon request under an appropriate Material Transfer Agreement.
Experimental model and subject details
The age and species/strains of mice
Six to eight weeks of age matched mice were used for mouse studies. Males were used for animal studies.
In vivo animal studies
All animal work was conducted in agreement with NIH guidelines and approved by the Weill Cornell Medicine Institutional Animal Care and Use Committee and the Institutional Biosafety Committee.
iPSC cell lines
All hiPSC experiments have been approved by Weill Cornell Medicine Human Embryonic Stem Cell Research Oversight Committee. The gender of iPSCs are: iPSC#1: Female; iPSC#41: Male; iPSC#44: Female; iPSC#15: Male; iPSC#17: Female; iPSC#19: Female. Cells were tested negative for mycoplasma contamination.
Method details
hiPSC culture and infection
hiPSCs were grown on matrigel-coated plates using StemFlex medium (Thermo Fisher Scientific, #A3349401). Cells were maintained at 37°C with 5% CO2. ZIKVPR (PRVABC59) strain and ZIKVU (MR766) strain were obtained from ZeptoMetrix and titered by plaque assay using Vero cells. iPSCs were infected with ZIKV (ZIKVU, MOI =0.15; ZIKVPR strain MOI = 1) for 2 h and changed to virus-free medium. At 72 hpi, cells were fixed, stained with the antibody against the ZIKV envelope protein (ZIKV-E), and analyzed by qRT-PCR.
Construction of NDUFA4 knockdown cell lines
The control shRNA and NDUFA4 specific shRNAs were obtained from VectorBuilder (VB900052-8767wwk and VB900052-8775ugm). The shRNA plasmids were transfected into 293T cells together with pMD2G and psPAX2 plasmids using calcium phosphate transfection. Lentivirus was collected and concentrated using Lenti-X Concentrator (Clontech) at 48 and 72 hpi. Then, iPSC line #1 and iPSC line #41 were infected with lentivirus for 48 h and sorted based on GFP expression.
Animal transplantation and infection
3.0 × 106 WT or NDUFA4−/− iPSCs were subcutaneously injected into 6-8-week-old male SCID-beige mice. Two weeks later, the mice were inoculated with ZIKV virus (5 × 105 PFU) by intraperitoneal injection. Five days after virus infection, the mice were euthanized. Xenografts were isolated, fixed in 4% PFA for two days and then transferred to 30% sucrose, followed by snap-freezing in O.C.T (Fisher Scientific).
Generation and infection of trophectoderm cells
Human iPSCs were differentiated into trophectoderm cells using our previously reported protocol.9 Briefly, hiPSCs were cultured to reach 80% confluent and dissociated to single cells with Accutase (Innovative Cell Technologies, #AT-104). Then, cells were seeded at 2.0 × 105 cells/well on 6-well plates coated with matrigel (100X, Corning, #354277) and cultured in StemFlex medium (Thermo Fisher Scientific, #A3349401) supplemented with 10 μM ROCK inhibitor Y-27632 (R&D Systems, #1254). The day after plating, trophectoderm cell differentiation was initiated by changing the culture medium to MEF conditional medium supplemented with 4 ng/mL bFGF (Peprotech, 100-18B). Then, cells were cultured in MEF conditional medium supplemented with 100 ng/mL BMP4 for another 4 days. On day 4, trophectoderm cells were harvested enzymatically with Accutase (Innovative Cell Technologies, #AT-104) and replated onto matrigel-coated 96-well plates or 24-well plates. iPSC-derived trophectoderm cells were infected with ZIKV (ZIKVU, MOI =0.15; ZIKVPR strain MOI = 1) for 2 h and changed to virus-free medium. At 72 hpi, cells were collected for analysis.
MEF conditional medium was prepared using irradiated CF1 Mouse Embryonic Fibroblasts (Thermo Fisher Scientific, #A34180). MEF conditional medium contains DMEM high glucose (Gibco Thermo Fisher, #11-965-118) supplemented with 10% Knockout Serum Replacement (Thermo Fisher, #10828028), 1×NEAA (Thermo Fisher, #11140050), 1×Glutamax (Thermo Fisher, #35050079) and 1×PenStrep (Penicillin, #15140163).
qRT-PCR
Total RNA samples were prepared from cells using TRIzol reagent (Qiagen, #74136) and reverse transcribed (RT) with high-capacity cDNA reverse transcription Kit supplemented with RNase inhibitor (Thermo Fisher, # 4374966). For the quantification of (+) and (−) strands of ZIKV vRNAs, strand specific primers were used in RT. Human β-Actin was employed as an internal reference. PrimeTime Gene Expression 2X Master Mix (IDT, #1055772) and probes for ZIKV vRNA and human β-Actin were used to perform qPCR reactions. For quantification of genes, random primers were used in RT. Human β-Actin was employed as an internal reference. The sequences of primers/probes are provided in Table S1.
Virus titer assay
Supernatants were collected from ZIKV infected cell cultures and then diluted serially from 10- to 108-fold. Vero cells were maintained in DMEM medium plus 10% fetal bovine serum. Cells were plated into 96-well plates at a density of 25,000 cells/well and then were infected with diluted supernatants in 96-well plates for 2 h. After infection, medium was replaced with semi-solid medium of alpha-MEM containing 10% fetal bovine serum and 1% methylcellulose. Two days after infection, cells were fixed with 4% PFA and stained with anti-ZIKV-E antibody.
Immunohistochemistry
Cells were fixed in 4% PFA for 20 min at room temperature, blocked in Mg2+/Ca2+ free PBS plus 5% horse serum and 0.2% Triton-X for 1 hour at room temperature, and then incubated with primary antibody at 4°C overnight. The information for primary antibodies is provided in Table S2. Secondary antibodies included donkey anti-mouse, goat, rabbit or chicken antibodies conjugated with Alexa-Fluor-488, Alexa-Fluor-594 or Alexa-Fluor-647 fluorophores (1:500, Life Technologies). Nuclei were counterstained by DAPI. Images were acquired using an LSM 880 Laser Scanning Confocal Microscope and processed with Zen software. Quantification was performed using ImageJ (NIH) software.
Western blotting
Cells were collected in Pierce RIPA buffer (Thermo Fisher Scientific) plus HALT protease inhibitor cocktail (1:100) (Thermo Fisher Scientific) and lysates loaded on 12% NuPage Bis-Tris pre-cast gels (Thermo Fisher Scientific). After separation by electrophoresis, proteins were transferred to 0.2 μm nitrocellulose membranes (Thermo Fisher Scientific). Membranes were blocked with 5% milk in TBS +0.1% Tween and incubated with primary antibody overnight. Information for primary antibodies is provided in Table S2. Membranes were washed and incubated with secondary antibody for 1 h at room temperature in 5% milk-TBS-0.1% Tween and developed using SuperSignal West Pico PLUS chemiluminescent substrate (Thermo Fisher Scientific). Human β-Actin was employed as an internal reference.
Flow cytometry analysis
hiPSC-derived trophectoderm cells were dissociated with Acctuase into single cells and washed with PBS twice, then cells were fixed and permeabilized using Fixation/Permeabilization Solution Kit (BD Biosciences). Cells were incubated with primary antibody at 4°C overnight and secondary antibodies at RT for 1 h, followed by washing twice and flow cytometry analysis. The information for primary antibodies and secondary antibodies were provided in Table S2.
Quantification and statistical analysis
Data are shown as mean ± SD. For comparisons with a common control, we used the one-way ANOVA followed by a Dunnett’s post hoc test for multiple testing correction. For low permissive and permissive cell line comparisons, we averaged the 3 technical replicates within each cell line, then used the averages for an unpaired two-tailed Student’s t-test. For the comparisons of two phenotypes and two colonies, we used two-way ANOVA analysis followed by Tukey’s post hoc test for multiple testing correction. N=3 independent biological replicates were used for all experiments unless otherwise indicated. n.s. indicates a non-significant difference. ∗p<0.05, ∗∗p<0.01 and ∗∗∗p<0.001. For statistical analyses, we used GraphPad Prism 8.4.1 software.
Acknowledgments
This work was supported by NIDKK (R01 DK119667-02S1, R01 DK124463, R01DK130454-01 and U01 DK127777, S.C.), Department of Surgery, Weill Cornell Medicine (T.E., S.C.), Bill and Melinda Gates Foundation (S.C., T.E.), NIAID (1R01AI124690, 1R01AI091707, C.R.), and Eunice Kennedy Shriver NICHD (1F32HD096810-01A1, L.A.L.). Y.H. is an NYSTEM Stem Cell Biology Scholar. L.A.L is funded by the Mastercard Diversity-Mentorship Collaborative’s Research Assistance for Primary Parent’s Award.
Author contributions
L.Y., Y.H., T.Z, L.L., C.M.R., S.A.N., Q.Q., T.E, and S.C. conceived and designed the experiments. L.Y., Y.H., T.Z., L.L., M.S., C.T., R.D., C.C., J.Z., Z.Z., A.M.G., Y.L., X.D., D.M., D.P., and A.Z., carried out the iPSCs preparation, ZIKV infection and related assays. Z.C. performed statistics analysis. L.Y., Y.H., T.Z., T.E. and S.C. wrote the article.
Declaration of interests
S.C. and T.E. are the co-founders of OncoBeat, LLC. S.C. is a consultant of Vesalius Therapeutics.
Published: May 29, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107001.
Supplemental information
Data and code availability
-
•
Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
There is no newly generated code to report.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- 1.Graham S.E., Clarke S.L., Wu K.H.H., Kanoni S., Zajac G.J.M., Ramdas S., Surakka I., Ntalla I., Vedantam S., Winkler T.W., et al. The power of genetic diversity in genome-wide association studies of lipids. Nature. 2021;600:675–679. doi: 10.1038/s41586-021-04064-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mozzi A., Pontremoli C., Sironi M. Genetic susceptibility to infectious diseases: Current status and future perspectives from genome-wide approaches. Infect. Genet. Evol. 2018;66:286–307. doi: 10.1016/j.meegid.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Han Y., Tan L., Zhou T., Yang L., Carrau L., Lacko L.A., Saeed M., Zhu J., Zhao Z., Nilsson-Payant B.E., et al. A human iPSC-array-based GWAS identifies a virus susceptibility locus in the NDUFA4 gene and functional variants. Cell Stem Cell. 2022;29:1475–1490.e6. doi: 10.1016/j.stem.2022.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.van der Eijk A.A., van Genderen P.J., Verdijk R.M., Reusken C.B., Mögling R., van Kampen J.J.A., Widagdo W., Aron G.I., GeurtsvanKessel C.H., Pas S.D., et al. Miscarriage associated with Zika virus infection. N. Engl. J. Med. 2016;375:1002–1004. doi: 10.1056/NEJMc1605898. [DOI] [PubMed] [Google Scholar]
- 5.Soares de Souza A., Moraes Dias C., Braga F.D.C.B., Terzian A.C.B., Estofolete C.F., Oliani A.H., Oliveira G.H., Brandão de Mattos C.C., de Mattos L.C., Nogueira M.L., Vaz-Oliani D.C.M. Fetal infection by Zika virus in the third trimester: report of 2 Cases. Clin. Infect. Dis. 2016;63:1622–1625. doi: 10.1093/cid/ciw613. [DOI] [PubMed] [Google Scholar]
- 6.Pacheco O., Beltrán M., Nelson C.A., Valencia D., Tolosa N., Farr S.L., Padilla A.V., Tong V.T., Cuevas E.L., Espinosa-Bode A., et al. Zika virus disease in Colombia - preliminary report. N. Engl. J. Med. 2020;383:e44. doi: 10.1056/NEJMoa1604037. [DOI] [PubMed] [Google Scholar]
- 7.Honein M.A., Dawson A.L., Petersen E.E., Jones A.M., Lee E.H., Yazdy M.M., Ahmad N., Macdonald J., Evert N., Bingham A., et al. Birth defects among fetuses and infants of US women with evidence of possible Zika virus infection during pregnancy. JAMA. 2017;317:59–68. doi: 10.1001/jama.2016.19006. [DOI] [PubMed] [Google Scholar]
- 8.Roth N.M., Reynolds M.R., Lewis E.L., Woodworth K.R., Godfred-Cato S., Delaney A., Akosa A., Valencia-Prado M., Lash M., Elmore A., et al. Zika-associated birth defects reported in pregnancies with laboratory evidence of confirmed or possible Zika virus infection - U.S. Zika pregnancy and infant registry, December 1, 2015-March 31, 2018. MMWR Morb. Mortal. Wkly. Rep. 2022;71:73–79. doi: 10.15585/mmwr.mm7103a1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tan L., Lacko L.A., Zhou T., Tomoiaga D., Hurtado R., Zhang T., Sevilla A., Zhong A., Mason C.E., Noggle S., et al. Pre- and peri-implantation Zika virus infection impairs fetal development by targeting trophectoderm cells. Nat. Commun. 2019;10:4155. doi: 10.1038/s41467-019-12063-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Block L.N., Aliota M.T., Friedrich T.C., Schotzko M.L., Mean K.D., Wiepz G.J., Golos T.G., Schmidt J.K. Embryotoxic impact of Zika virus in a rhesus macaque in vitro implantation model†. Biol. Reprod. 2020;102:806–816. doi: 10.1093/biolre/ioz236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheridan M.A., Yunusov D., Balaraman V., Alexenko A.P., Yabe S., Verjovski-Almeida S., Schust D.J., Franz A.W., Sadovsky Y., Ezashi T., Roberts R.M. Vulnerability of primitive human placental trophoblast to Zika virus. Proc. Natl. Acad. Sci. USA. 2017;114:E1587–E1596. doi: 10.1073/pnas.1616097114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vota D., Torti M., Paparini D., Giovannoni F., Merech F., Hauk V., Calo G., Ramhorst R., Garcia C., Pérez Leirós C. Zika virus infection of first trimester trophoblast cells affects cell migration, metabolism and immune homeostasis control. J. Cell. Physiol. 2021;236:4913–4925. doi: 10.1002/jcp.30203. [DOI] [PubMed] [Google Scholar]
- 13.Muthuraj P.G., Sahoo P.K., Kraus M., Bruett T., Annamalai A.S., Pattnaik A., Pattnaik A.K., Byrareddy S.N., Natarajan S.K. Zika virus infection induces endoplasmic reticulum stress and apoptosis in placental trophoblasts. Cell Death Discov. 2021;7:24. doi: 10.1038/s41420-020-00379-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Z., Li Q., Ashraf U., Yang M., Zhu W., Gu J., Chen Z., Gu C., Si Y., Cao S., Ye J. Zika virus causes placental pyroptosis and associated adverse fetal outcomes by activating GSDME. Elife. 2022;11 doi: 10.7554/eLife.73792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bayer A., Lennemann N.J., Ouyang Y., Bramley J.C., Morosky S., Marques E., Jr., Cherry S., Sadovsky Y., Coyne C.B. Type III interferons produced by human placental trophoblasts confer protection against Zika virus infection. Cell Host Microbe. 2016;19:705–712. doi: 10.1016/j.chom.2016.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sen Santara S., Crespo Â.C., Mulik S., Ovies C., Boulenouar S., Strominger J.L., Lieberman J. Decidual NK cells kill Zika virus-infected trophoblasts. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2115410118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guzeloglu-Kayisli O., Guo X., Tang Z., Semerci N., Ozmen A., Larsen K., Mutluay D., Guller S., Schatz F., Kayisli U.A., Lockwood C.J. Zika virus–infected decidual cells elicit a gestational age–Dependent innate immune response and exaggerate trophoblast Zika permissiveness: implication for vertical transmission. J. Immunol. 2020;205:3083–3094. doi: 10.4049/jimmunol.2000713. [DOI] [PubMed] [Google Scholar]
- 18.Paull D., Sevilla A., Zhou H., Hahn A.K., Kim H., Napolitano C., Tsankov A., Shang L., Krumholz K., Jagadeesan P., et al. Automated, high-throughput derivation, characterization and differentiation of induced pluripotent stem cells. Nat. Methods. 2015;12:885–892. doi: 10.1038/nmeth.3507. [DOI] [PubMed] [Google Scholar]
- 19.Moretton A., Morel F., Macao B., Lachaume P., Ishak L., Lefebvre M., Garreau-Balandier I., Vernet P., Falkenberg M., Farge G. Selective mitochondrial DNA degradation following double-strand breaks. PLoS One. 2017;12 doi: 10.1371/journal.pone.0176795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Original western blot images have been deposited at Mendeley and are publicly available as of the date of publication. The DOI is listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request.
-
•
There is no newly generated code to report.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.