Summary
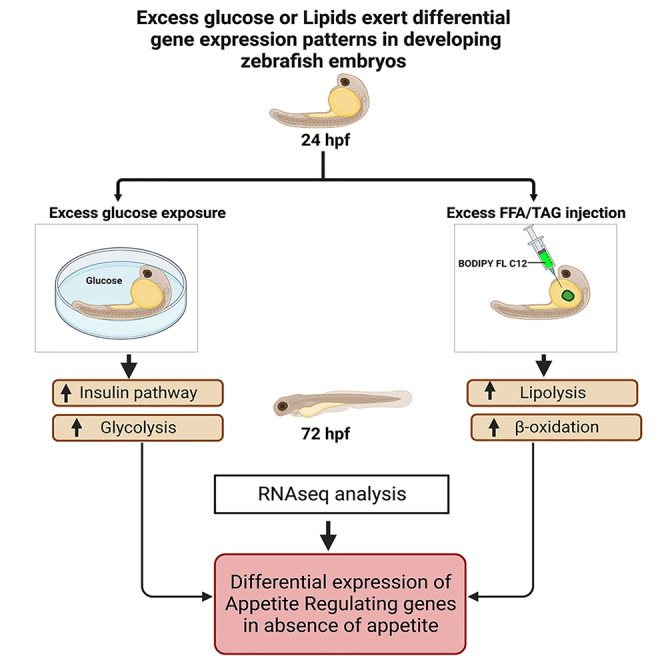
Zebrafish embryos use their yolk sac reserve as the sole nutrient source during embryogenesis. The two main forms of energy fuel can be found in the form of glucose or fat. Zebrafish embryos were exposed to glucose or injected with free fatty acid/Triacylglycerol (FFA/TAG) into the yolk sac at 24 hpf. At 72 hpf, glucose exposed or FFA/TAG injected had differential effects on gene expression in embryos, with fat activating lipolysis and β-oxidation and glucose activating the insulin pathway. Bulk RNA-seq revealed that more gene expression was affected by glucose exposure compared to FFA/TAGs injection. Appetite-controlling genes were also differently affected by glucose exposure or FFA/TAG injections. Because the embryo did not yet feed itself at the time of our analysis, gene expression changes occurred in absence of actual hunger and revealed how the embryo manages its nutrient intake before active feeding.
Subject areas: Embryology, Transcriptomics, Model organism
Graphical abstract
Highlights
-
•
Embryo response to excess glucose varies with developmental stages
-
•
FFA/TAG excess promotes fat use, impacting embryo metabolism
-
•
Excess glucose or FFA/TAG elicits distinct gene expression responses
-
•
Nutrient sources influence appetite gene expression during development
Embryology; Transcriptomics; Model organism
Introduction
The availability of maternal nutrients has a direct impact on fetal growth.1 Fuel sources have an important role in energy balance, as well as cellular and physiological processes. This may play a key role in regulating active food intake after conception in growing embryos.1,2 Although all physiological processes necessitate energy during development, the thrifty hypothesis of developmental programming postulates that intrauterine exposure to unfavorable environments, such as excess nutrients, contributes to the pathophysiology of metabolic diseases later in life.3 Energy fuel can be found in the form of sugar (glucose) and fat (triacylglycerides (TAGs)). Each of these fuel sources uses a different pathway; insulin-dependent pathway for glucose and lipolysis and β oxidation for TAGs. The main source of energy for the developing fetus is glucose oxidation, however, mitochondrial fatty acid oxidation also plays a role.4 Excess in-utero glucose exposure stimulates glucose transfer from the mother to the fetus through the maternal-fetal gradient, resulting in increased energy storage and birth weight in neonates.5 Sustained maternal-fetal glucose flux leads to lipid substrate accumulation, which together with free fatty acid flux from the placenta in the fetus, promotes a neonatal risk factor.6 In response to excess fuel energy sources during development, transcriptional processes are modulated to provide metabolic homeostasis through the regulation of gene expression.7,8,9
Increased birth weight has been linked to uterine glucose exposure, which has previously been associated with obesity and cardiovascular risk.6,7,9 Furthermore, weight gain during gestation has been suggested to exacerbate fetal outcomes and is linked with childhood obesity.10 More importantly, intrauterine exposure to a high-fat diet causes hyperglycemia in mice, as well as post-translational modification of genes such as insulin receptor substrate 2 (Irs2) which is involved in insulin signaling and genes involved in appetite regulation, such as hypothalamic proopiomelanocortin (POMC) and the melanocortin 4 receptor (Mc4r).11,12,13,14
Insulin increases glycolytic and lipogenic enzyme activity.15,16 Glucose diffusion into and out of cells is dependent on insulin-mediated activation of GLUT4 glucose transporters.17 Insulin primarily promotes glucose utilization through insulin-induced glucose absorption in muscle and adipose tissue.18 Glucose transporter 2 (GLUT2) is present in the liver and pancreas. It plays an insulin-independent role in glucose sensing and homeostasis,19 whereas insulin primarily promotes GLUT4 transfer and improves glucose absorption across the plasma membrane of muscle and adipose tissue membrane.20 When insulin is withdrawn, the amount of GLUT4 on the plasma membrane decreases and the rate of translocation to the cell membrane returns to its baseline.21 Insulin signaling in target tissues is impaired by persistent high glucose levels.22 In insulin resistance, insulin signaling is disrupted, resulting in an even lower insulin response.23
Lipolysis has also been found to be increased in developing fetuses as a result of impaired fetal insulin response.24 The stimulatory effects of insulin on lipogenesis suggest that fetal hyperinsulinemia promotes fat storage.22,25 The key regulators of lipogenesis have also been shown to be active early in fetal development.26 In-utero glucose exposure increases lipolysis in fetal adipose tissue because of reduced insulin sensitivity, which is mediated by an increase in lipoprotein lipase (LPL) activity.24 Fetal glycerol utilization increases significantly during development, allowing for the re-esterification of glycerol released by lipolysis in fetal adipose tissue.27 As a result, the restriction on acetyl-CoA-carboxylase (ACC) activity is reduced, resulting in a higher rate of lipogenesis and greatly contributing to fetal body fat accretion.28 Changes in lipogenic and lipolytic factors during development help to increase the synthesis of extra adipocytes. In terms of appetite and energy balance, adipocyte fat generates both known and unknown signals.29 To address the impact of an excess fuel energy source in regulating gene expression and appetite during development, we used the zebrafish model supplied with an excess of glucose or free fatty acid (FFA)/TAGs as an energy source during embryogenesis to investigate the transcriptional regulation of genes. The zebrafish embryo relies on the maternal deposited yolk until the end of embryogenesis (5 days post fertilization).30 The yolk sac is composed of different lipids and the composition changes throughout embryogenesis.31,32 In zebrafish, homologous genes that regulate lipid and glucose use are conserved in humans.33,34 At the yolk-embryo contact, referred to as the yolk syncytial layer (YSL), lipids enter the developing embryo.35 Lipoproteins (such as vitellogenin) and a complex of lipid-regulating factors carry lipids from the yolk sac to the embryo in the YSL. The yolk is depleted by 5–6 days post fertilization (dpf), and larvae must feed by itself to absorb its own nutrients. Because the embryonic YSL and the larval digestive organs have similar gene expression patterns, yolk utilization during early zebrafish embryogenesis has been found to be a tightly controlled mechanism similar to lipid transport mediated by intestinal and hepatic lipoproteins.35
The hypothalamic mechanisms that regulate appetite may be irreversibly altered during early development.36 Excess in-utero energy exposure has been shown to have an effect on the central appetite-regulating system’s response to peripheral cues because of increased lipids.37 Peripheral insulin signals are also important in the appetite gene network.38 Insulin receptor ablation in POMC neurons has been linked to a loss of glucose homeostasis, resulting in systemic insulin resistance.39 Zebrafish have recently been used to study the regulation of appetite genes during embryonic development and larval stages.40 To date, very limited data exist regarding how a change in fuel usage, i.e. excess lipid or excess glucose affects the expression of metabolic and appetite-regulating genes in a developing embryo, which at this stage, does not yet feed on its own and therefore does not experience appetite per se. To address this gap of knowledge in this paper, we used a zebrafish model of excess glucose or excess FFA/TAGs to identify transcriptome differences to provide molecular insight into nutrient impact on appetite. Sequence analysis revealed a variety of genes/pathways whose expression is modified under excess glucose or fat during embryonic development and identified a differential role for glucose or fat in controlling appetite-regulating genes when the organism does not feed on its own.
Results
Glucose exposure during embryonic development activates the insulin pathway
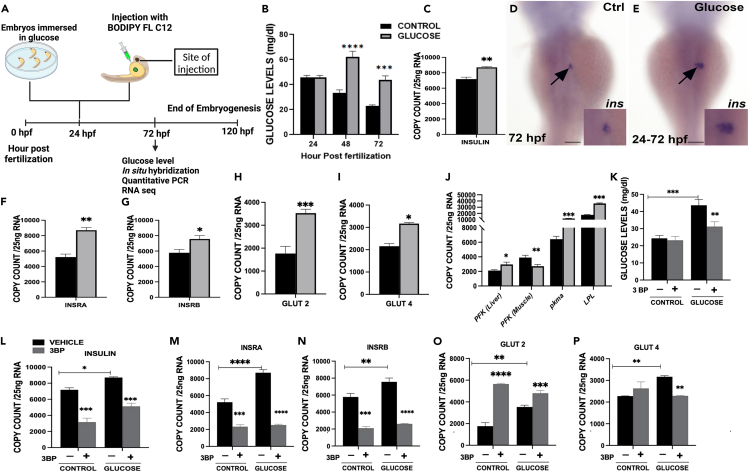
To determine whether glucose exposure mediates changes in embryonic glucose levels, embryos were exposed to glucose (3% w/v) during development at 24 hpf (Figure 1A) and glucose levels were compared to control untreated embryos (Figure 1B) from 24 to 72 hpf. Glucose exposure induced a significant increase of free glucose levels after 24 and 48 h exposure compared to controls (p < 0.0001, p = 0.0003 respectively) (Figure 1B). We determined that the increased glucose levels observed in Figure 1B resulted in an increased insulin secretion. We also quantified the expression of the preproinsulin gene (Figure 1C) and detected that the increase in the domain of insulin expression after 48 h glucose exposure (Figures 1D and 1E) correlated with an over-expression after 48 h glucose exposure (Figure 1C). Insulin receptor A and B expression was also increased at 72 hpf (Figures 1F and 1G). Of interest, a 24 h 3% w/v glucose exposure does not increase the expression levels of preproinsulin or the insulin receptors, nor does it increase the domain of expression of insulin mRNA in the islet (Figure S1).
Figure 1.
Effect of glucose on zebrafish embryos
(A) Timeline of treatment for embryos exposed to glucose or injected with free fatty acid/TAG at 24 hpf.
(B) Embryonic glucose concentrations at different time points during development following glucose treatment at 24 hpf.
(C) Insulin mRNA expression at 72 hpf. WISH of insulin mRNA expression 48 h after glucose exposure (arrows), insets are close up of insulin expression in the developing islet.
(D) Control.
(E) Glucose exposed.
(F–I) mRNA expression of insulin receptors (INSRA and INSRB), Glucose transporters (Glut 2 and Glut 4) at 72 hpf.
(J–P) mRNA expression of tissue specific genes phosphofructokinase (liver and muscle isoform), pyruvate kinase M1/2a and lipoprotein lipase K. Quantification glucose levels for embryos co-exposed with glucose and 3-Bromopyruvate (3-BP), (−) vehicle (control, glucose) without 3-BP, (+) (control, glucose) with 3-BP. mRNA expression of: L. insulin; M-N. insulin receptors (A, B); O-P. Glucose transporters (Glut 2 and Glut 4) of embryos treated with 3-BP. ∗ Represent statistically significant threshold levels (∗p < 0.05, ∗∗p < 0.01,∗∗∗p < 0.001,∗∗∗∗p < 0.0001). Scale bar 100 μm.
The presence of glucose transporters, which make glucose available to cells, facilitates the translocation of glucose across the cell membrane.18 Therefore, at 72 hpf, mRNA expression levels of Glut2 and Glut4 glucose transporters were measured, and we found that the expression of both genes was significantly higher (p = 0.0008, p = 0.02) compared to controls (Figures 1H and 1I). However, Glut4 expression was down-regulated at 48 hpf after 24 h glucose exposure (Figure S1). With these findings in hand, we decided to focus our subsequent investigations on the implications of 3% w/v glucose exposure on 72 hpf embryos during development.
To determine whether glucose will promote tissue specific physiological response, we measured the expression levels of liver and muscle isoforms of phosphofructokinase (PFK) and liver specific lipoprotein lipase (Figure 1, J) at 72 hpf. Our findings show that glucose has a differential effect on phosphofructokinase because the liver isoform is significantly upregulated, whereas the muscle isoform is significantly downregulated (Figure 1, J). This may be related to metabolic oscillatory expression in the muscle isoform of phosphofructokinase (PFK-M).41 We also observed an insulin mediated increase of lipoprotein lipase at 72 hpf and an increase in the pyruvate kinase M1/2a (pkma), which is mainly expressed in the brain at 72 hpf42 (Figure 1, J).
Inhibition of glycolysis during glucose exposure induces a reduction in preproinsulin and insulin receptors but modulates glucose transporters
Our data show that excess glucose leads to an increase in glycolysis. To investigate how glycolysis inhibition affects the fate of insulin, insulin receptors, and glucose transporters in the presence of high glucose levels, we used 3-Bromopyruvate (3-BP), a known potent inhibitor of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and hexokinase 2 (HK2).43 Embryos were co-exposed to 3% w/v glucose and 75 μM 3-BP for 12 h from 60 to 72 hpf and compared to controls that received only 3-BP. In comparison to controls, 3-BP caused a significant reduction in glucose levels in embryos co-treated with glucose (Figure 1K). Our findings also revealed that 3-BP induced a downregulation in preproinsulin and insulin receptors expression at 72 hpf (Fig. L-N). Of interest, we observed that 3-BP promoted a downregulation of Glut 4, but not Glut2 expression when co-treated with glucose (Figures 1O and 1P).
Injection of exogenous FFA/TAG into the yolk sac during development induces lipogenesis and β-oxidation
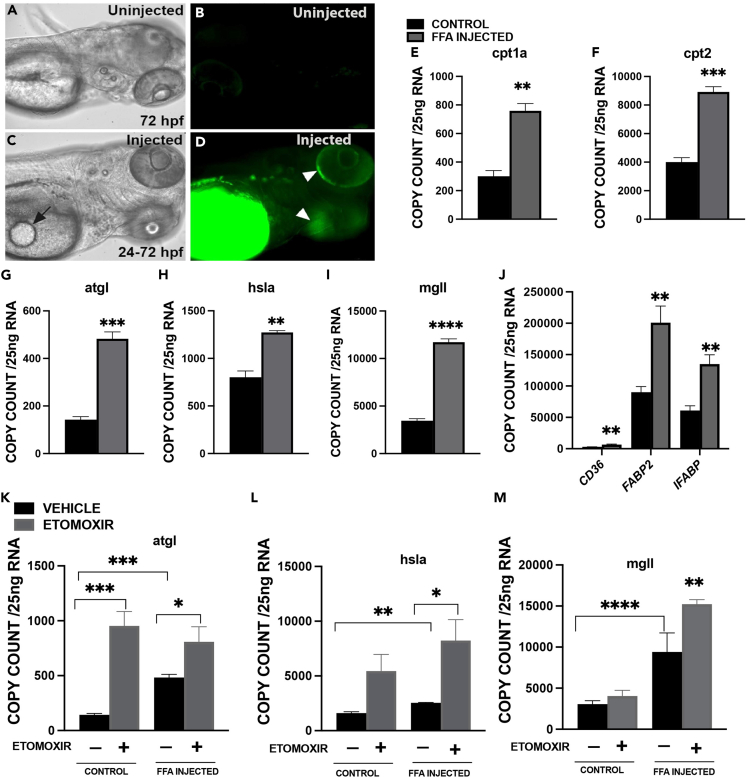
To investigate the effects of excess FFA and TAG in embryos during development, 24 hpf embryos were injected with approximately 5 nL of a BODIPY-labeled fluorescent lipid analog diluted in canola oil (1 mg/mL). Injected embryos were monitored after 24 and 48 h post-injection, and the most fluorescence was detected in the embryo’s body after 48 h post injection (Figure S2). Therefore, the time point of 72 hpf was selected for studying the effects of FFA/TAG injections in the yolk sac at 24 hpf. In injected embryos at 72 hpf, the FFA labeled BODIPY spread throughout the yolk sac, as indicated by the black arrow from the site of injection (Figure 2C). Fluorescence was also detected in the embryo’s body and around the eyes (shown with arrows in Figure 2D), indicating that excess FFA/TAG reached the developing embryo’s entire body. However, no fluorescence was detected in non-injected embryos (Figures 2A and 2B). As seen in Figure 2C, the overall development of the embryos was not affected by FFA/TAG injection, similar to previous data by our group and others.31,33 Injection of FFA/TAG in the yolk sac led to an increased expression of genes implicated in β-oxidation such as cpt1a and cpt2. β-oxidation is the process of using fatty acid as an energy source in the cell.44 Moreover, the expression of genes implicated in the breakdown of TAGs to produce FFA are: adipose triglyceride lipase (agtl) producing DAG from TAG, hormone-sensitive lipase a (hsla) producing MAG and FFA from DAG, and monoglyceride lipase (mgll) producing glycerol. FFA is increased in FFA/TAG injected embryos (Figures 2G–2I).
Figure 2.
Effects of FFA/TAG injection on zebrafish embryos. BODIPY-labeled fluorescent FLC12 FFA/TAG injected into the yolk sac of 72 hpf embryos
(A) Control in bright field (BF).
(B) Control- Fluorescent image.
(C) FFA/TAG injected embryo displaying the site of injection (arrow).
(D–F) Fluorescent FFA injected embryos with diverging fluorescence (white arrowheads). Bar plots showing mRNA expression of Carnitine Palmitoyl transferase (cpt1a (E), cpt2 (F)).
(G) Adipocyte triglyceride lipase (atgl).
(H) Hormone sensitive lipase a (hsla).
(I–L) monoglyceride lipase mgll and J. lipid transporter or binding protein, (cluster of differentiation 36 (CD36), fatty acid-binding protein 2 (FABP2) and intestinal fatty acid-binding protein 2 (IFABP). Exposure of FFA/TAG injected embryos to etomoxir or vehicle (control, FFA/TAG injected) for the quantification of mRNA expression of K. Adipocyte triglyceride lipase (atgl); L. Hormone sensitive lipase a (hsla).
(M) monoglyceride lipase (mgll). In panels G-I, a significant difference between group means was determined using the Student’s t test, whereas in panels K-M, a two-way ANOVA test was performed. (∗p < 0.05, ∗∗p < 0.01∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). Scale bar 100 μm.
To explore the effect of injected FFA/TAG on lipid transport and binding, we measured the expression of CD36, a long chain free fatty acid transporter,45 and fatty acid binding proteins (FABP2, IFABP) that facilitate lipid transport to particular compartments in the cell.46 All lipid-mediated transport genes (CD36, FABP2, IFABP) showed significant increase in expression (Figure 2J) in FFA/TAG injected embryos compared to controls.
Inhibition of β-oxidation in FFA/TAG injected embryos modulates lipolytic genes
To better understand how β-oxidation affects the expression of lipolytic genes during development when FFA/TAG is in excess, we utilized etomoxir, a cpt1 inhibitor47 to inhibit β-oxidation in FFA/TAG injected embryos. Embryos were injected with FFA/TAG and exposed to etomoxir in experimental media for 12 h from 60 hpf to 72 hpf and compared to etomoxir-exposed controls. Blocking β-oxidation revealed a significant increase in expression of lipolytic genes atgl, hsla, and mgll in FFA/TAG injected embryos exposed to etomoxir compared to control embryos (Figures 2K–2M).
Transcriptome profiling of 72 hpf embryos with excess glucose or FFA/TAG reveals distinct gene expression profiles
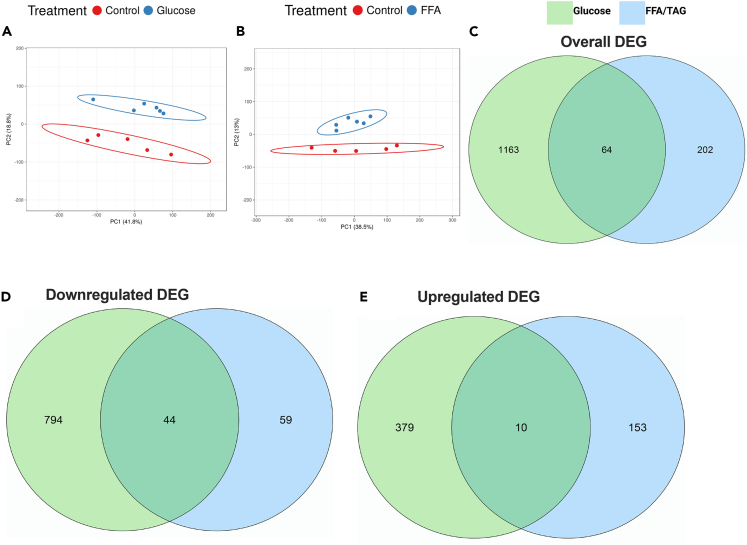
The raw reads of our RNA-seq dataset were aligned to the zebrafish reference genome assembly GRCz11 release 105 using STAR (an ultrafast universal RNA-seq aligner) which achieved an average mapping rate of approximately 95%. Between 86.6% and 92.3% of these reads were uniquely mapped. All samples showed adequate quality after normalization. Normalized data were used as input for statistical hypothesis testing, which identified genes that differed significantly between sample groups. Principal component analysis (PCA) showed that the treatment groups accounted for ∼50% of the variance observed in the normalized dataset and where data points distinctly clustered from the control (Figures 3A and 3B). At a statistical threshold of false discovery rate (FDR) adjusted p value <0.01, between 266 and 2,535 genes were found to be significantly differentially expressed. In general, FFA/TAG injected embryos exhibited fewer significant differentially expressed genes (DEG, 266) than glucose exposed embryos (DEG,1227) (Figure 3C). Of interest, glucose exposed embryos and FFA/TAG injected embryos shared only a small number of genes (64) whose expression significantly changed in both conditions compared to controls (Figure 3C). Overall, 838 genes were downregulated in glucose-exposed embryos compared to controls, whereas 103 genes were downregulated in FFA-induced embryos (Figure 3D). Furthermore, 389 genes were significantly upregulated in glucose-exposed embryos when compared to controls, whereas 163 genes were significantly upregulated in FFA/TAG injected embryos when compared to controls (Figure 3E).
Figure 3.
Sample population clusters and the number of differentially expressed genes in glucose-exposed and FFA/TAG-injected embryos
PCA plot displaying discrete and distinct population clusters between treatment groups.
(A) Glucose exposed versus Control.
(B) FFA/TAG injected versus Control. Venn Diagrams summarizing the intersection of genes between treatment groups.
(C) Overall genes that were significantly differentially expressed between glucose-exposed versus control and FFA/TAG-injected versus control.
(D) Downregulated genes differentially expressed in glucose-exposed versus control and FFA/TAG-injected versus control.
(E) Upregulated genes differentially expressed in glucose-exposed versus control and FFA/TAG-injected versus control.
Gene association studies reveal significant distinct gene expression patterns between all sample groups
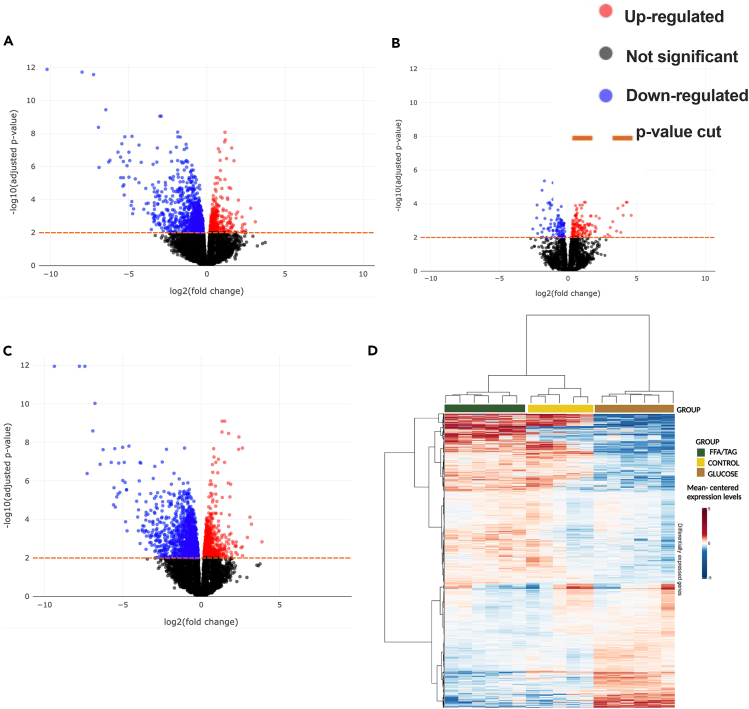
The number of overlapping DEGs between glucose and FFA/TAG embryos was defined using an adjusted p value of 0.01. The volcano plots (Figures 4A–4C) demonstrate that there are about five times more DEGs in glucose versus control compared to FFA/TAG versus control in all comparisons across a range of statistical significance thresholds. This is consistent with our Venn diagram findings (Figures 3C–3E). Of interest, the volcano plot analysis shows that the most DEGs are found in the glucose exposed versus FFA/TAG injected groups of all the significant genes identified among the three groups. This means that the difference in terms of gene expression is higher in the glucose exposed (Figure 3A) versus FFA/TAG injected (Figure 3B) than in each condition individually compared to controls. This points to the fact that activation of the glucose/glycolysis pathway and activation of the lipolysis/β-oxidation pathway share very limited common genes. The strength of association between overlapping DEGs (defined using adjusted p value <0.01) between all pairwise combinations of comparisons, as revealed by the Heatmap depicts distinct upregulation and downregulation of gene expression in both glucose and FFA/TAG samples (Figure 4D). However, in the FFA/TAG injected samples, a distinct pattern of expression is observed, which is opposed to the gene expression pattern observed in the glucose exposed samples (Figure 4D).
Figure 4.
Gene association test
(A–D) Volcano plots for all comparisons showing significance (as -log10 transformed p value) against magnitude (log2 (fold change). Genes identified as significantly differentially expressed are represented as red (upregulated) or blue (downregulated) or black (non-significant) dots. The horizontal orange line represents the applied p value threshold A. Glucose versus Control; B. FFA/TAG versus Control; C. Glucose versus FFA/TAG injected D. Heatmap showing gene intensity per sample relative to the average level across all samples. Individual genes are shown on the Y axis while samples are shown along the X axis. Red and blue cells correspond to higher and lower RNA-seq levels, respectively.
Differential gene analysis reveals modulation of metabolic genes in response to glucose exposure or FFA/TAG injection
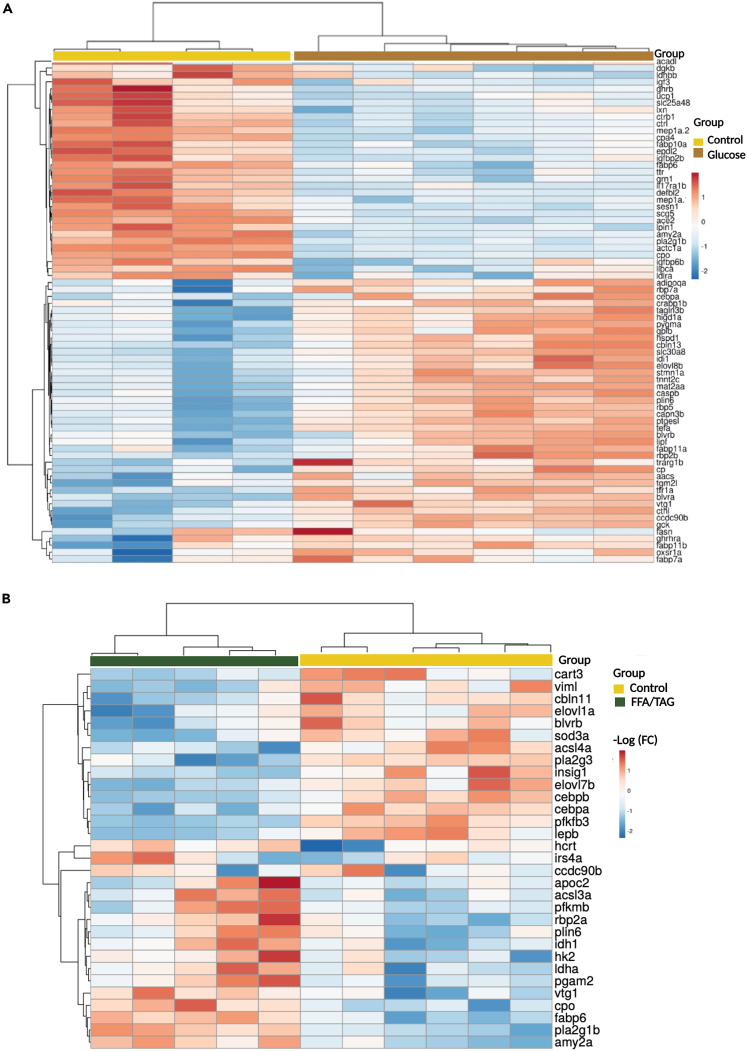
Using a hypothesized based approach, genes expected to change with glucose or FFA/TAG exposure were investigated. Glucose exposure and activation of the insulin pathway results in the downregulation of relevant lipolytic genes such as phospholipase A (pla2g1b) (phospholipid hydrolysis) and lipid binding proteins (fabp6, fabp10a) (Figure 5A). Glucose exposure increases the expression of genes involved in the glycolytic pathway, such as glucokinase (gck) and glucose-6-phosphate isomerase (gpib), and genes involved in adipogenesis such as perilipin 6 (plin6), adiponectin (adipoqa), and lipogenesis [(fatty acid synthase (fasn), transgelin (tagln3b) (involved in LDL endocytosis), CCAAT/enhancer-binding-protein-α (cepba) (induction of lipogenesis and regulation of glucose homeostasis) and ELOVL fatty acid elongase (evol8b)] (Figure 5A). FFA/TAG injection induces a downregulation of glycolysis-related genes such as hexokinase (hk2), phosphofructokinase (pfkmb), phosphoglycerate mutase 2 (pgam2), insulin receptor substrate 4a (irs4a), lipolytic genes such as phospholipase A2 (pla2g1b) (phospholipid hydrolysis), apolipoprotein C-II (apoc2) (Figure 5B). Besides that, FFA/TAG injection increases the expression of gluconeogenic genes such as 6-phosphofructo-2-kinase/fructose-2 (pfkfb3), and genes involved in lipid synthesis such as insulin-induced gene 1(insig1) (a new regulator of lipid synthesis 47), ELOVL fatty acid elongase 1a (elovl1a), phospholipase A2 group III (pla2g3). Genes involved in adipogenesis and appetite regulation such as leptin b (lep b), cocaine- and amphetamine-regulated transcript 3 (cart3) are also upregulated (Figure 5B, see also Table S3). When we compare DEGs between glucose exposed and FFA/TAG injected embryos, we find a downregulation of lipid metabolism genes such as phospholipase A2 group IB (pancreas) (pla2g1b), carnitine palmitoyltransferase 1b (cpt1b), insulin induced gene 1(insig1), chymotrypsin-like (ctrl), fatty acid binding protein 6 ileal (gastrotropin), (fabp6), ELOVL fatty acid elongase 7b (evol7b), carboxypeptidase O (cpo) (lipid droplet-associated enzyme), CCAAT enhancer binding protein (cebpb) and appetite regulating genes such as leptin b, (lepb), leptin receptor (lepr), cocaine- and amphetamine-regulated transcript 3 (cart3) (Figure S4). In addition, we also observe an upregulation of genes that are associated with glucose metabolism [(Hexokinase (hk2), glucokinase (gck), phosphofructokinase (pfkma), glycogen synthase (gys1), phosphoglycerate mutase (pgam2), insulin receptor 4a (irs4a)], mitochondrion biogenesis [(peroxisome proliferator-activated receptor gamma coactivator 1 alpha (ppargc1a)], lipid metabolism (fatty acid synthase (fasn), acetyl-CoA carboxylase alpha (acaca), carnitine palmitoyl transferase 1Cb (cpt1b), perilipin 6 (plin6)) and appetite regulating genes (hypocretin (orexin) neuropeptide precursor (hcrt), proopiomelanocortin a (pomca)) (See also Figure S4).
Figure 5.
Heatmap analysis of differentially expressed genes
Heatmap showing gene intensity per sample relative to the average level across all samples. Individual genes are shown on the Y-axis while samples are shown along the X axis. The first 50 upregulated or downregulated genes represented within a range of (−2 to 2) of log 2 of the fold change were chosen.
(A) Shows downregulated (blue) and upregulated (red) genes of treatment glucose compared to controls.
(B) Shows downregulated (blue) and upregulated (red) genes of FFA/TAG injected samples compared to controls.
Enrichment of Gene Ontology terms reveals modulation of lipogenic and appetite regulating genes in response to glucose exposure or FFA/TAG injection
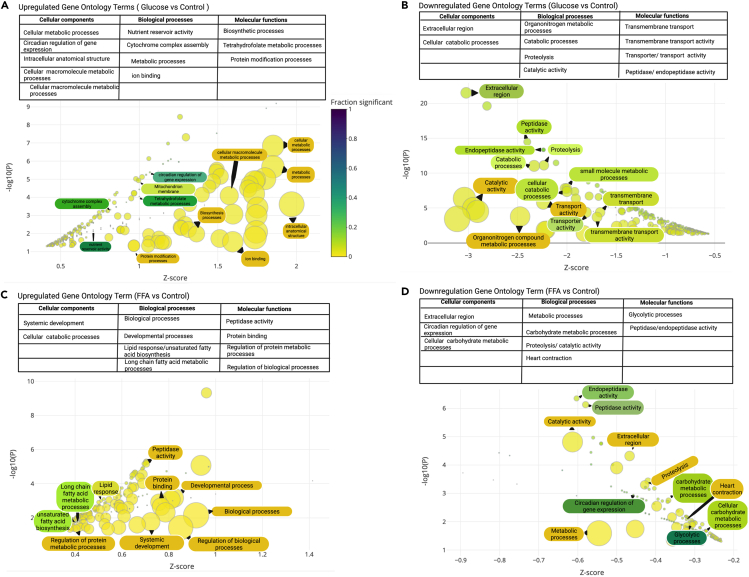
To improve biological interpretation and investigate the DEGs using unbiased approach studies, pathway enrichment analysis was performed to determine the DEGs within zebrafish Gene Ontology (GO) terms, where a statistical threshold of FDR-adjusted p < 0.05 was applied as a cut-off and only top 50 terms are displayed (Figures 6A–6D). Only enrichment terms with two or more genes were considered. In addition, only GO terms with an enrichment p value <0.05 were included for display. As shown in Figure 6, the various genes associated with a given GO term can be both up-and down-regulated within a single comparison. Several metabolism and digestion-related pathways (e.g., metabolism) were found to be enriched in upregulated DEGs, as well as catalytic activity and peptidase-related GO terms in the glucose versus control group (Figure 6B), In addition, a handful of epigenetic modification terms, such as DNA methylation and DNA demethylation (GO:0044728) were found enriched in up-regulated DEGs in the glucose-exposed versus control contrast (Figure S5). Notably, in the FFA/TAG injected versus control contrast, lipid biosynthetic and peptidase activity enriched in the upregulated DEGs (Figure 6C) as well as downregulation of carbohydrate metabolic and glycolytic processes enriched DEGs (Figure 6D). Also, both biotic and abiotic stress response terms were found enriched in the up-regulated DEGs, such as response to stress (GO:0006950) and response to heat (GO:0009408) (Figure S5). Analysis of the glucose-exposed versus FFA/TAG-injected contrast highlighted the pathways and terms enriched in DEGs between the two treatment groups. This included the enrichment of DNA repairing pathways in the up-regulated DEGs and stress-response related terms in the down-regulated DEGs (Figure S5). Because our two zebrafish models of excess nutrients: glucose or lipid, selectively activates different pathways: insulin pathway or liploysis/β-oxidation, we were interested in measuring the effects of excess nutrients on appetite regulation before the organism experiences an actual appetite (i.e., by active feeding). We and others have shown that during embryogenesis, before the zebrafish embryos are able to ingest food, appetite-regulating genes expression is tightly regulated.40,48 The dogmatic view of glucose signaling is that insulin secretion will suppress appetite by regulating the expression of genes associated with orexigenic signals.49
Figure 6.
Bubble plots depicting significantly enriched Gene Ontology terms
Enrichment analyses with enrichment Z score on the X axis and -log10(p value) on the y axis. Point size represents term size and point color represents the fraction of genes that were significant. Gene ontology terms range from highly significant (blue) to less significant (yellow). Only GO terms with an enrichment p value less than 0.05, were included. Furthermore, only the top 50 terms are displayed. In addition, terms were restricted to include only those with 2 or more genes.
(A) Upregulated Gene ontology terms between glucose exposed embryos and their controls with GO terms classified in the table above.
(B) Downregulated Gene Ontology term between glucose exposed and controls with GO terms classified in the table above.
(C) Upregulated Gene ontology terms between FFA injected and controls with GO terms classified in the table above.
(D) Downregulated Gene Ontology term between FFA injected and controls with GO terms classified in the table above.
Modulation of expression of appetite-regulating gene in response to glucose exposure or FFA/TAG injection
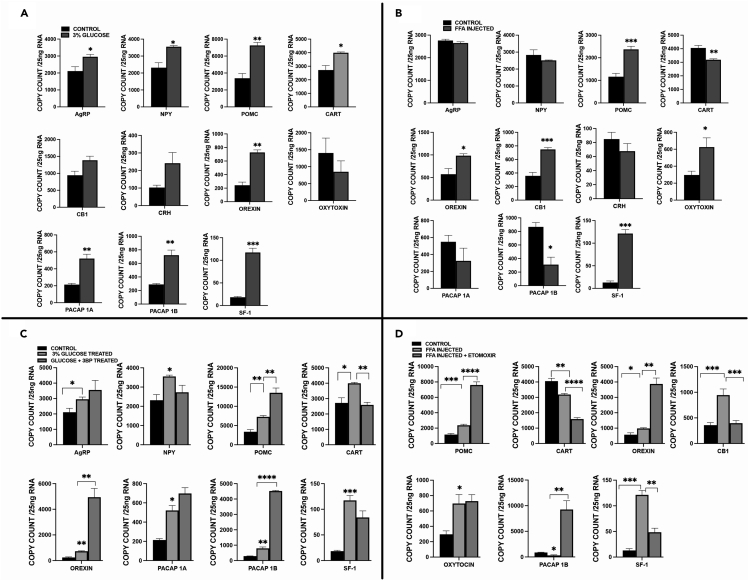
qPCR analysis of glucose exposed embryos revealed that the expression of several genes or peptides that are known to be involved in satiety is significantly increased as expected: POMC (Proopiomelanocortin), CART3 (cocaine and amphetamine regulated transcript), PACAP1 (Pituitary adenylate cyclase-activating peptide 1(A and B) and SF-1 (steroidogenic factor 1) (Figure 7A). Surprisingly, the expression of genes or peptides that are known to stimulate appetite is also found to be significantly increased under excess glucose exposure: AgRP (Agouti-Related Protein), NPY (Neuropeptide Y), Orexin (Hypocretin) (Figure 7A). The area of CB1 transcript in the hypothalamus in glucose-exposed embryos is lower than that in controls as detected by WISH (Figure S6). When excess calories are induced by excess FFA/TAG injection, the situation is substantially different. In this insulin-independent scenario, POMC, CB1, Orexin, Oxytocin, and SF-1 are significantly up-regulated, as one would expect in an excess food situation (Figure 7B), but the area of CB1 transcript in the hypothalamus is unchanged (not shown). To note, a zebrafish POMC mutant does not become obese as is the case in mammals48 and the role of SF-1 in appetite regulation acts via the regulation of leptin.50 In FFA/TAG injected embryos, CART, PACAP1A, and B, NPY, and CRH expression is decreased (Figure 7B).
Figure 7.
Differential expression of appetite regulation genes at 72 hpf in glucose exposed or FFA/TAG injected embryos
(A) mRNA expression of genes in glucose exposed embryos compared to controls (Agouti-related Peptide (AgRP), Neuropeptide Y(NPY), Proopiomelanocortin (POMC), Cocaine- and Amphetamine-regulated Transcript (CART), Orexin, cannabinoid receptor type 1 (CB1), Corticotropin-releasing hormone (CRH), Oxytocin, Pituitary adenylate-cyclase activating peptide (PACAP 1A/1B), Steroidal factor-1 (SF-1).
(B) Expression of genes in FFA/TAG injected embryos compared to controls (Agouti-related Peptide (AgRP), Neuropeptide Y(NPY), Proopiomelanocortin (POMC), Cocaine- and Amphetamine-regulated Transcript (CART), Orexin, cannabinoid receptor type 1 (CB1), Corticotropin-releasing hormone (CRH), Oxytocin, Pituitary adenylate-cyclase activating peptide (PACAP 1A/1B), Steroidal factor 1 (SF-1).
(C) Compares mRNA expression in glucose exposed embryos and controls to the expression in embryos co-exposed to glucose and 3-Bromopyruvate in significant expressed appetite-regulating genes in Figure 7A.
(D) Compares mRNA expression in FFA/TAG injected embryos and controls to the expression in FFA/TAG embryos co-exposed to etomoxir in significant expressed appetite-regulating genes in Figure 7B. ∗ Represent statistically significant threshold levels (∗p < 0.05, ∗∗p < 0.01∗∗∗, p < 0.001,∗∗∗∗p < 0.0001).
Blocking glycolysis or β-oxidation in embryos exposed to glucose or FFA/TAG similarly modulates expression of appetite regulating gene
In Figure 1L, inhibiting glycolysis resulted in a significant decrease in preproinsulin expression. Hence, we reasoned to investigate the effects of glycolysis inhibition on the expression of appetite regulating genes in glucose exposed embryos that showed statistical significance in Figure 7A. Embryos were co-exposed with 3-Bromopyruvate and glucose for 12 h. Our findings revealed exacerbation of POMC, Orexin, and PACAP 1B expression when insulin and insulin receptors were downregulated (Figure 7C). However, interestingly, we observed an attenuation of CART expression to control expression levels in the absence of insulin and its receptors (Figure 7C).
Moreover, FFA/TAG injected embryos triggered a significant increase in β-oxidation as shown in Figures 2E and 2F. Therefore, we wanted to explore the effects of blocking β-oxidation on the expression of appetite regulating genes that revealed statistical significance in Figure 7B. Our findings revealed in FFA/TAG injected embryos exposed to etomoxir that etomoxir exacerbated the expression of POMC known to be controlled by β-oxidation,51 Orexin, and PACAP 1B (Figure 7D). In addition, we observed an attenuation of CART, CB1 and SF-1 expression in these embryos (Figure 7D).
Discussion
A zebrafish model of excess glucose or excess fat during development
To provide insight into the fundamental actions of excess glucose or excess fat during embryonic development in a vertebrate organism, we either exposed zebrafish embryos to high glucose levels (3% w/v) or injected FFA/TAG into the yolk sac. To validate our model in terms of excess glucose or fat, we analyzed the consequences of our intervention.
As expected, high glucose exposure leads to free glucose accumulation of 60 mg/dL (or 3.36 mmol/L) in the embryo’s body compared to 37 mg/dL (or 2.07 mmol/L) in controls after 24 h. A concentration of free glucose level of 3.36 mmol/L is classified as a low range on the diabetic scale.52 Between the first and third day of zebrafish development, free glucose levels of the embryos dropped from 2.40 mmol/L at 24 hpf to 1.23 mmol/L at 72 hpf (Figure 1B). This might indicate that the embryo has used up most of the glucose deposited by the mother in the yolk sac and that gluconeogenesis is not yet fully active during that point in embryogenesis. As a comparison, human babies have a glycemia average of just under 2 mmol/L at birth.53 Of interest, although the embryos are still raised in 3% w/v glucose in medium, the concentration of free glucose levels is less after 48 h exposure compared to 24 h exposure (40 mg/dL at 72 hpf compared to 60 mg/dL at 48 hpf). We hypothesized that somehow the embryos’ physiology is adapting to high glucose levels and the embryos are able to utilize this excess glucose better either through increased glycolysis, mitochondrial respiration or de novo lipogenesis. The levels of free glucose observed in 3% w/v glucose exposed embryos are not at a normal physiological range for the zebrafish.54 However, they are within or below the pathological range observed in the blood of pregnant mothers suffering from diabetes with glycemia values of 95 mg/dL or more.52 As a consequence of the high free glucose levels in the embryo’s body, we observed an increase in insulin synthesis, activating the insulin pathway; the expression of both insulin receptors is higher in glucose-exposed embryos (Figure 1). Excitingly, no significant glucose mediated insulin response after 24 h post exposure was observed, which could be because of the pancreas’s developmental stage at the time.55
Excess FFA/TAG activates lipolysis and β-oxidation during embryogenesis
Embryos injected with the FFA/TAG BODIPY-labeled fluorescent lipid analog at 72 hpf showed increased expression of genes involved in β-oxidation and lipolysis. Although it has been reported that FFA transport is extremely efficient after a few hours of BODIPY injection, and that lipid metabolism occurs independently of the TAG from the injected canola oil,33 we show that after 2 days post injection, TAG hydrolysis may play a significant role in lipid homeostasis. Contrary to what was observed in the glucose exposed embryos, excess FFA/TAG in the body will not activate the insulin pathway.56 However, injection of FFA/TAG in the yolk sac affects the physiology of the embryos by activating β-oxidation detected by an increase in cpt1a and cpt1b expression and by increasing lipolysis because of excess TAG in the body (Figure 2). Therefore, our nutrient excess models work by activating two independent pathways: the insulin pathway for glucose exposed embryos and lipolysis and β-oxidation for FFA/TAG injected embryos.
Excess FFA bind to the lipid transporter CD36 to facilitate the cellular uptake of long-chain fatty acids57 and plasma membrane-associated FABP mediate diverse lipid signals and acts as central lipid chaperone for lipid homeostasis.46,58,59 As expected, in the current study, excess FFA/TAG mediated an increase in CD36 and FABP/IFABP expression. β-oxidation serves as an intracellular fatty acid sensor and regulates lipolysis.60 Our data showed that, inhibiting β-oxidation induced a feedback response in which the expression of adipocyte triglyceride lipase (atgl), hormone sensitive lipase (hsla) and monoacylglycerol lipase (mgll) was significantly increased to produce more FFA that should be utilized by β-oxidation if it was not pharmacologically blocked by etomoxir.
Differential transcriptomic response to excess glucose or FFA/TAG during development
Bulk RNA-seq analysis revealed that we only have a limited number of genes with significant changes in expression that are common to glucose exposure and FFA/TAG injection (Figures 3C–3E) (Figures 4A–4C). This re-enforces the point that each intervention is acting on different pathways. Indeed, heatmap analysis of glucose exposed and FFA/TAG injected embryos show an almost mirror image of each other (Figure 4D). Of interest, the glucose exposed embryos clade is closer in terms of gene expression to the control than they are to the FFA/TAG injected clade. One of the common genes that is highly down-regulated in both glucose-exposed and FFA/TAG injected embryos is phospholipase A2 (pla2g1b). In adult rats, excess glucose activates phospholipase A2 which increases arachidonic acid levels which seem to contribute to insulin secretion,61 whereas the excess of unsaturated fatty acid can inhibit PLA2 in vitro and may act as endogenous suppressor of lipolysis.62 As expected, several genes implicated directly in glycolysis or the insulin pathway are up-regulated in glucose-exposed embryos such as glucokinase (gck), biliverdin reductase A and B (blvra/b), glucose-6-phosphate isomerase B (gpib), glucose transporter (slc30a8) (Figure 5). Comparison of glucose exposed embryos and FFA/TAG injected embryos shows that the gene expression changes observed when each condition was compared to control embryos remain true and are even more exacerbated. Meaning that in terms of gene expression changes, untreated control embryos are closer than glucose exposed embryos and TAG/FFA injected embryos than the two conditions are with each other. This can be detected either by heatmap analysis, gene expression changes or GO and pathway analysis, confirming that glucose exposure and FFA/TAG injection have the dramatic opposite effect on gene expression.
The age of zebrafish feeding varies from one lab to another but it is generally accepted that free-feeding larvae will be able to ingest food and develop an active feeding behavior between 5 and 6 dpf and delay of initial feeding at 8 dpf does not affect the survival and growth of the juvenile.63 This means that the zebrafish can effectively survive on its yolk sac reserve until 8 dpf. In our experimental settings, intervention from 1 to 3 dpf, the embryos solely rely on their yolk sac reserve for energy. How much of an impact does a drastic modification of glucose levels or lipid availability in the yolk sac have on active nutrient uptake from the yolk sac to the embryo’s body? Several appetite-regulating genes have been detected in our bulk RNA-seq analysis, therefore to answer this question we perform a systematic quantification of appetite-regulating genes in our two conditions in the whole embryo extract. This analysis (Figures 7A and 7B) revealed some expected and unexpected results. For example, excess glucose and activation of the insulin pathway led to a significant increase in POMC, CART and pituitary adenylate cyclase-activating peptide (PACAP) 1A and 1B expression, which are all considered as appetite suppressor genes.64 Surprisingly, AgRP/NPY and Orexin expression, which stimulates food intake, is also upregulated during insulin secretion. Although activation of insulin has been reported to inhibit AgRP/NPY neurons via the PI3K pathway,65 chronical elevated insulin levels such as in insulin resistance states can increase hypothalamic NPY levels66,67 suggesting a development of insulin resistance to the feedback effects of insulin on hypothalamic NPY. Also, fasting in zebrafish increases the expression of AgRP/NPY.68 Although these findings are unexpected, it is possible that high insulin secretion has resulted in a state of insulin resistance at 72 hpf, where the embryos begin to signal a fasting state, resulting in the insulin-mediated increase expression of AgRP/AgRP. In addition, whereas it is expected that activation of the AgRP/NPY neuronal axis will counteract the interaction of the α-MSH/MC4R to inhibit POMC, it has been demonstrated that insulin activation can also activate POMC genes because of the synaptic plasticity of POMC neurons, particularly in response to feeding and fasting.69,70 Moreover, overfed rats show increase POMC expression relative to their controls.71 Furthermore, glucose mediated insulin release induces orexin expression72 and Pacap 1a expression, which are neurons associated with the pancreatic release of insulin.73 Similarly, in glucose exposed embryos, the expression of orexin (appetite stimulator) was significantly increased.74 Our data also shows that CB1, an appetite stimulator have hypothalamic transcript levels significantly reduced as detected by in situ hybridization (Figure S5). Of interest, insulin activation significantly increased SF-1 expression (Figure 7A). This is expected because it is reported that hyperactivating of the insulin signaling in mice results in the increased expression of SF-1 expression.75
Inhibiting glycolysis in embryos for 12 h before mRNA extraction in our model had no effect on AgRP but exacerbated the increase in POMC, Orexin and PACAP-IB expression despite lower glucose availability and insulin level (Figure 7C). Furthermore, our results show an attenuation of NPY, CART and SF-1 expression levels. These differential gene expressions may agree with reports that indicate that glycolysis in the hypothalamic neurons control glucose sensing that modulates appetite gene expression.76 In addition, CART for instance, displays a differential expression in control of energy homeostasis.77
The situation is more complex in FFA/TAG injected embryos when β-oxidation is increased. Although the appetite suppressor genes POMC and Oxytocin expressions are increased, the expression of other genes implicated in appetite stimulation, orexin and CB1, is also significantly increased. Surprisingly, we observed CART and PACAP 1B both appetite suppressor genes are downregulated in FFA/TAG excess.78 Of interest, PACAP 1B is increased in the presence of insulin in Figure 7A, but is downregulated in an insulin independent mechanism in Figure 7B, showing that PACAP expression is predominantly dependent on insulin availability.
Inhibition of β-oxidation with the cpt1 specific inhibitor etomoxir reverses energy expenditure and enhances glucose oxidation.79,80 Similar to what happens in glucose excess, blocking β-oxidation significantly upregulates POMC and PACAP1B (appetite suppressors) as well as Orexin, a sensor of changes in energy balance.81 CART, CB1, and SF-1 expression levels were significantly attenuated. Inhibiting fatty acid synthase increases CART expression,82 which promotes β-oxidation as seen in FFA-injected embryos. In agreement with this mechanism, we find that inhibiting β-oxidation reduces CART expression. CB1 and SF-1 downregulation could be because of additional mechanistic interactions.
These expression patterns suggest that although the embryo’s body is full of energy in the form of TAGs and FFA, at the molecular level the brain continuing to send a signal to stimulate nutrient uptake from the yolk may be to increase the levels of glucose or other carbohydrates in the body. Alternatively, one cannot exclude that the function of some of the genes referenced as appetite stimulators or appetite-suppressors, when an actual appetite is present, may have a slightly different function when no active feeding occurs. This is the case for two well-known appetite-regulating genes: leptin and ghrelin. It is widely accepted that the function of leptin produced by the adipocytes is to decrease appetite.83 However, during zebrafish embryogenesis, adipocytes are not yet formed and will be first detected at around 8 dpf.84 Moreover, it seems that leptin signaling in zebrafish has a limited effect on food intake and adipostasis.85 In post-fetal life, ghrelin is expressed by the gut to stimulate food intake and store fat.86 However, during embryonic developments, before the gut acts as a nutrient-processing and absorbing organ, ghrelin is expressed in the epsilon cells of the endocrine pancreas.87 Brain transcriptomic of ghrelin zebrafish mutants did not reveal any gene expression changes in appetite-regulating genes,88 confirming that during embryonic development, well-characterized appetite-regulating genes may have a different function than when the organism is feeding on its own. Further studies will be required to link the effects of appetite related gene expression changes during zebrafish embryogenesis on actual changes in appetite and/or nutrient absorption by the organism.
Conclusion
This project aimed at understanding how modifying fuel supply (i.e. excess glucose or fat) during embryonic development when an actual appetite based on food ingestion is not yet occurring will affect the transcriptome of the developing embryos, especially genes known to be implicated in appetite regulation. Our zebrafish models revealed that changing the fuel source during embryonic development induces dramatic different changes in terms of gene expression if excess glucose or excess fat is implemented with more transcriptomic changes detected when excess glucose is implemented compare to excess fat. Moreover, our data also confirm an idea that was previously presented: nutrients uptake from the yolk sac (embryogenesis) or through active feeding (larvae and older) are actively regulated by several genes, but the regulation of the expression of these genes depends on the status of the organism, actively feeding or relying on the yolk sac. Further studies will be needed to understand how nutrient uptakes from the yolk sac is actively regulated by the zebrafish embryo during the first 5 days of life.
Limitation of the study
This study’s drawback could be the fact that embryos were directly exposed to glucose in the medium while FFA/TAG injections were being given to embryos. Despite the possibility of bias, embryos were injected at 24 hpf and allowed time to reach 72 hpf before being selected for mRNA extraction. Although we were able to measure the amount of glucose incorporated by the embryo at various times, the velocity of diffusion of the fluorescent BODIPY tag was so strong that fluorescent quantification could not be performed because of excess fluorescence in the yolk or the embryo’s body.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-Dig antibody1 | Roche | RRID: AB_514497 |
| Dig RNA labelling kit for in. situ hybridization1 | Roche | CAT# 186033-10-3 |
| Chemicals, peptides, and recombinant proteins | ||
| D-glucose | Fisher scientific | Cas# 50.99-7 |
| BODIPY FL C12 | Thermo Fisher Scientific | REF D3822 |
| Etomoxir | Cayman Chemicals | Cas# 828934-41-4 |
| 3-Bromopyruvate | Cayman Chemicals | 1113-59-3 |
| Critical commercial assays | ||
| Insulin kit | Crystal Chem Ultra-sensitive mouse insulin ELISA kit | CAT#90080 |
| Glucometer | Contour Next blood Glucose kit | SN97059827358 |
| TRI reagent | Zymo Research | CAT# R2050-1-200 |
| Iscript reverse transcription supermix for qPCR | Biorad | CAT# 1708841 |
| Qx 200 ddPCR Evagreen supermix | Biorad | CAT#1864034 |
| Deposited data | ||
| RNA Sequencing data | GEO | GSE213859 |
| Experimental models: Organisms/strains | ||
| Zebrafish Danio rerio | UMMC zebrafish facility | AB WILD TYPE STRAIN |
| Oligonucleotides | ||
| See Table S1 | ||
| Software and algorithms | ||
| Photoshop 2023 | NA | NA |
| GraphPad Prism Vs 9.0.0 | NA | NA |
Resource availability
Lead contact
Further information and request for resources and reagent should be directed to and will be fulfilled by the Lead Contact, Yann Gibert (ygibert@umc.edu).
Material availability
-
•
This study did not generate new unique reagents.
-
•
All probes generated for in-situ hybridization are accessible for request by the zebrafish community.
Experimental model and study participant details
-
•
Zebrafish (Dani rerio) AB wild type strain were used.
-
•
Embryos were 48 and 72 hours postfertilization of developmental age.
-
•
The influence of sex is not reported in embryos at this point in development because embryos are not sexually differentiated.
All animal husbandry and experimental techniques were reviewed and approved by the UMMC institutional animal care and use committee (IACUC). The zebrafish were handled in accordance with the specified guidelines of the IACUC. Embryos were generated using an AB wildtype strain received from the Zebrafish International Resource Center (ZIRC) and raised in embryonic medium (E3) at 28.5°C under standard conditions with a 14 h light/10 h dark cycle.
Method details
Glucose exposure
Following our previously reported methodology, zebrafish Wild-Type AB embryos were exposed to 3% w/v glucose (176 mmol/L) dissolved in water from 24 to 72 hours postfertilization (hpf).54 Hours depicted in the figures are all in hpf, not in hours post exposure.
Free glucose assay
Zebrafish wild types (AB) embryos at 24 hpf were collected and raised in embryonic E3 media supplemented with 3% w/v D-glucose media at a density of 50 embryos per dish at 28.5°C± 2°C. Embryos were exposed to 3% w/v D-glucose media from 24 to 72 hpf to induce hyperglycemia. Control embryos were kept under the same timelines and conditions in embryonic E3 media. Upon removing all media, embryos were rinsed three times with embryonic E3 media to eliminate all residual glucose. At 72 hpf, 10 embryos were homogenized in 5 μl of saline using a pestle homogenizer and centrifuged at 14000 rev/min for 2 mins. A volume of 2 μl of supernatant was placed on the strips of the glucometer. Free glucose was measured three times for each replicate among the four biological replications. The mean and standard error were determined using the student t-test and used to measure statistical significance.
FFA/TAG microinjection
24 hpf zebrafish embryos were injected with BODIPY™ FL C12, a fluorescently labeled lipid analog dissolved in commercial canola oil solution (1 mg/ml) into the centre of the yolk cell with injection volume kept between 3 and 5 nl31. As we previously reported, embryonic medium injected did not alter embryogenesis.31 Canola oil is a TAG, composed of 62% oleic acid (C18:1, ω-9), 19% linoleic acid (C18:2, ω-6), 9% α-linolenic acid (C18:3, ω-3) and 7% saturated fatty acid.33 Capillary tips were pulled for the injection using the flaming/brown type micropipette puller (Sutter Instrument Co. USA). A pressure injector (Tritech Research Inc.) was used to deliver the solution into the embryo's yolk cell. The delivered volume was determined by varying the injection pressure, time, and diameter of the microcapillary tip. The embryos were arranged in rows on agar furrows, and injections were made directly into the yolk from the vegetal pole. Embryos were fixed at 72 hpf for analysis. Hours depicted in the figures are all in hpf, not in hours post injection.
Insulin quantification
At 72 hpf, 3%w/v glucose treated embryos as well as standards were carefully washed with PBS. Embryos were separated into four sets of ten embryos each. Each set was homogenized in 60 μl of manufacturer-supplied sample diluent (Crystal Chem). Insulin standards were diluted in accordance to the manufacturer's instructions. A volume of 100 μl of prepared standards was aliquoted into antibody coated microplate wells as well as 50 μl of supernatant from embryo homogenate. Wells were covered and incubated at 4°C for 2 hrs. After aspirating and discarding the well contents, the wells were washed five times with wash buffer. In each standard and sample well, 100 μl of anti-insulin enzyme conjugate was dispensed and incubated at room temperature for 30 minutes. Well content was taken and discarded once again, and the wells were washed seven times. After 40 minutes of incubation at room temperature with enzyme substrate solution, the reaction stopped with enzyme reaction stop solution. Background absorbance at 630nm was measured and subtracted from the absorbance at 450nm. A linear plot of absorbance of standard against concentration of standards was used to calculate insulin concentration. To determine the difference between the means of the two groups, the student t test was utilized.
RNA extraction and cDNA synthesis
At 72 hpf, embryonic media were carefully removed until samples were dried entirely, and embryos were then flash frozen in liquid nitrogen. Total RNA was extracted from zebrafish embryos using TRI® reagent (Zymo Research, USA) according to the manufacturer’s protocol. Total RNA was used as a template to generate cDNA. One microgram of total RNA from each sample was reverse transcribed using the iScript reverse transcription super mix (Biorad) and iScript supermix with no reverse transcriptase was used to make controls according to the manufacturer’s protocol. The cDNA was generated thermal cycler (Biorad-T100) with an annealing temperature of 46°C.
Digital droplet PCR (ddPCR)
cDNA was diluted 1:2 and used as the PCR template for the subsequent qPCR. To make a 20 μl reaction mix, 5 μl of nuclease-free water, 10 μl of QX200TM ddPCR EvaGreen Supermix (Bio-Rad), 4 μl of cDNA, and 1 μl of each gene-specified primer (forward and reverse primers, Table S1) were added to the ddPCR mixture. To eliminate genomic DNA contamination, a negative control with no reverse transcriptase enzyme was included for cDNA synthesis and was used as a no-template control during DNA amplification. Droplets were created using a Biorad Qx 200 Droplet reader, and DNA amplification was performed at an annealing temperature of 60°C. The QuantaSoft software was used to capture the fluorescent intensity of the droplets after DNA amplification (Bio-Rad Laboratories Ltd). Data were analyzed using a two-tail unpaired student t-test to detect differences between the expression means of treated and controls.
Whole-mount in-situ hybridization (WISH)
Embryos that were exposed to glucose or injected with FFA/TAG and their respective controls were fixed overnight in 4% paraformaldehyde (PFA) at 4°C. Digoxigenin-labeled riboprobes were transcribed in vitro using the Roche Diagnostics kit. Whole-mount in situ hybridization (WISH) was performed as described by Fraher et al.31
RNA isolation and bulk RNA sequencing
Embryos were treated with 3%w/v glucose or injected into a yolk sac with BODIPY-labeled fluorescent lipid dissolved in a canola oil solution (1 mg/ml) at 24 hpf. At 72 hpf, samples were flash-frozen. 10 embryos were used per condition with a total of six replicates (60 embryos total) for our experimental groups: glucose-exposed or FFA/TAG injected embryos. We used a total of five replicates for the control group. After flash freezing, zebrafish embryos were sent to the UMMC Molecular and Genomics Core Facility (www.umc.edu/genomicscore) for RNA isolation, QC, and bulk RNAseq. RNA was extracted using TRIzol Reagent along with Pure Link RNA Mini Kit (Invitrogen) according to manufacturer instructions and assessed for quality control parameters of minimum concentration and fidelity (i.e. 18S and 28S bands, RIS >8). Libraries were developed using the Illumina TruSeq mRNA Stranded Library Prep Kit (Set-A-indexes), quantified with a Qubit fluorimeter (Invitrogen), and assessed for quality and size using the QIAxcel Advanced System as done previously.89,90 Samples were pooled into a single library (n=10 pooled samples per library) and sequenced using the NextSeq 500 High Output Kit (150 cycles, paired-end 75bp) on the Illumina NextSeq 500 platform. The run generated 522 million reads (%QC30=91.7) or ∼30 million reads passing the filter per sample. Sequenced reads were assessed for quality using the Illumina BaseSpace Cloud Computing Platform and FASTQ sequence files were used to align reads to the Danio Rerio genome [DANRER7 (REFSEQ)] using RNA-Seq Alignment Application (using STAR aligner). On average, >95% of reads per sample are mapped to the Danio Rerio genome. Differential expression was determined using the RNA-Differential Expression Application (v1.0.1) and DESeq2. Gene expression differences are denoted as log2 (ratio) and q > 0.05. RNAseq data have been deposited to the GEO repository: [NCBI tracking system #23327218].
Drug treatments
Zebrafish embryos were grouped into two set, one set was exposed to 75 μM of 3- bromopyruvate dissolved in experimental media as the control and other set were exposed to 75 μm of 3- bromopyruvate (Cayman chemicals) dissolved in 3%w/v glucose for 12 hours from 60 hpf up to 72 hpf to block glycolysis. Alternatively, Zebrafish embryos were again divided into two sets where the first group were controls exposed to 75 μM etomoxir, a cpt1 specific inhibitor (Cayman chemicals) dissolved in experimental media whiles the other set of FFA/TAG injected embryos were exposed to 75 μM etomoxir dissolved in experimental media.
Quantification and statistical analysis
Each experiment was carried out at least three times on different occasions. Mean with SEM is used to represent the results of the analysis. To examine differences between two groups, the Student's t test was utilized. A two-tailed t test was used to test against a control as ∗p <0.05, ∗∗p< 0.01, ∗∗∗p<0.001, ∗∗∗∗p<0.0001. Two-way ANOVA was used to compare more than two groups. GraphPad Prism 9.0 software was used for the analyses.
Acknowledgments
The author would like to thank Goldie Faircloth at the UMMC zebrafish facility for providing excellent husbandry care. YG is supported by a National Institute of Health grant (P20 GM104357), a COBRE/MS CEPR National Institute of Health grant (P20GM121334) and by a National Institutes of Health grant (R01DE029803) and, is supported by the Cancer Center and Research Institute. MRG is supported in part by the NIGMS (P20GM103476) and the Obesity, Cardiorenal, and Metabolic Diseases- COBRE (P20 GM104357). The work performed through the UMMC Molecular and Genomics Facility is supported, in part, by funds from the NIGMS, including Mississippi INBRE (P20GM103476) and Obesity, Cardiorenal, and Metabolic Diseases-COBRE (P20GM104357). The authors also would like to thank Fios Genomics (Edinburgh, Scotland) for the help they provided with RNA seq analysis.
YG is supported by a National Institutes of Health grant (P20 GM104357), by a COBRE/MS CEPR National Institute of Health grant (P20GM121334), and by a National Institutes of Health grant (R01DE029803). MRG is supported in part by the NIGMS (P20GM103476) and the Obesity, Cardiorenal, and Metabolic Diseases - COBRE (P20 GM104357).
Author contributions
Y.G. supervised the project; B.K., M.R.G., and Y.G. designed the experiments; M.R.G. and Y.G. provided the project resources; B.K., C.K.C., M.R.G., and Y.G. performed the experiments; B.K., M.R.G., and Y.G. analyzed the datasets; B.K. and Y.G. wrote the original manuscript draft; B.K., C.K.C., M.R.G., and Y.G. critically revised the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
Published: June 7, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107063.
Supplemental information
Data and code availability
-
•
RNA Sequencing data generated in this study has been deposited at GEO. Accession number are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact
-
•
This paper does not report original code.
References
- 1.Wu Y., Zhang Q., Xiao X. The effect and potential mechanism of maternal micronutrient intake on offspring glucose metabolism: an emerging field. Front. Nutr. 2021;8:763809. doi: 10.3389/fnut.2021.763809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hales C.N., Barker D.J. Type 2 (non-insulin-dependent) diabetes mellitus: the thrifty phenotype hypothesis. Diabetologia. 1992;35:595–601. doi: 10.1007/BF00400248. [DOI] [PubMed] [Google Scholar]
- 3.Brett K.E., Ferraro Z.M., Yockell-Lelievre J., Gruslin A., Adamo K.B. Maternal-fetal nutrient transport in pregnancy pathologies: the role of the placenta. Int. J. Mol. Sci. 2014;15:16153–16185. doi: 10.3390/ijms150916153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gluckman P.D., Hanson M.A., Cooper C., Thornburg K.L. Effect of in utero and early-life conditions on adult health and disease. N. Engl. J. Med. 2008;359:61–73. doi: 10.1056/NEJMra0708473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Desoye G., Nolan C.J. The fetal glucose steal: an underappreciated phenomenon in diabetic pregnancy. Diabetologia. 2016;59:1089–1094. doi: 10.1007/s00125-016-3931-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kong L., Nilsson I.A.K., Gissler M., Lavebratt C. Associations of maternal diabetes and body mass index with offspring birth weight and prematurity. JAMA Pediatr. 2019;173:371–378. doi: 10.1001/jamapediatrics.2018.5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Basu M., Garg V. Maternal hyperglycemia and fetal cardiac development: clinical impact and underlying mechanisms. Birth Defects Res. 2018;110:1504–1516. doi: 10.1002/bdr2.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haro D., Marrero P.F., Relat J. Nutritional regulation of gene expression: carbohydrate-fat- and amino acid-dependent modulation of transcriptional activity. Int. J. Mol. Sci. 2019;20:1386. doi: 10.3390/ijms20061386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landon M.B., Mele L., Varner M.W., Casey B.M., Reddy U.M., Wapner R.J., Rouse D.J., Tita A.T.N., Thorp J.M., Chien E.K., et al. The relationship of maternal glycemia to childhood obesity and metabolic dysfunction. J. Matern. Fetal Neonatal Med. 2020;33:33–41. doi: 10.1080/14767058.2018.1484094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Josey M.J., McCullough L.E., Hoyo C., Williams-DeVane C. Overall gestational weight gain mediates the relationship between maternal and child obesity. BMC Publ. Health. 2019;19:1062. doi: 10.1186/s12889-019-7349-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dodd G.T., Michael N.J., Lee-Young R.S., Mangiafico S.P., Pryor J.T., Munder A.C., Simonds S.E., Brüning J.C., Zhang Z.-Y., Cowley M.A., et al. Insulin regulates POMC neuronal plasticity to control glucose metabolism. Elife. 2018;7 doi: 10.7554/eLife.38704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernandez-Twinn D.S., Hjort L., Novakovic B., Ozanne S.E., Saffery R. Intrauterine programming of obesity and type 2 diabetes. Diabetologia. 2019;62:1789–1801. doi: 10.1007/s00125-019-4951-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin A.C., Filatova N., Lindtner C., Chi T., Degann S., Oberlin D., Buettner C. Insulin receptor signaling in POMC, but not AgRP, neurons controls adipose tissue insulin action. Diabetes. 2017;66:1560–1571. doi: 10.2337/db16-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q., Xiao X., Zheng J., Li M., Yu M., Ping F., Wang T., Wang X. A maternal high-fat diet induces DNA methylation changes that contribute to glucose intolerance in offspring. Front. Endocrinol. 2019;10:871. doi: 10.3389/fendo.2019.00871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Girard J., Ferré P., Foufelle F. Mechanisms by which carbohydrates regulate expression of genes for glycolytic and lipogenic enzymes. Annu. Rev. Nutr. 1997;17:325–352. doi: 10.1146/annurev.nutr.17.1.325. [DOI] [PubMed] [Google Scholar]
- 16.Sauerheber R.D. Perspectives on insulin stimulation of glucose transport. Am. J. Biomed. Sci. Res. 2019;4:356–361. doi: 10.34297/AJBSR.2019.04.000834. [DOI] [Google Scholar]
- 17.Wang T., Wang J., Hu X., Huang X.-J., Chen G.-X. Current understanding of glucose transporter 4 expression and functional mechanisms. World J. Biol. Chem. 2020;11:76–98. doi: 10.4331/wjbc.v11.i3.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chadt A., Al-Hasani H. Glucose transporters in adipose tissue, liver, and skeletal muscle in metabolic health and disease. Pflügers Archiv. 2020;472:1273–1298. doi: 10.1007/s00424-020-02417-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Panahi M., Rodriguez P.R., Fereshtehnejad S.-M., Arafa D., Bogdanovic N., Winblad B., Cedazo-Minguez A., Rinne J., Darreh-Shori T., Hase Y., et al. Insulin-independent and dependent glucose transporters in brain mural cells in CADASIL. Front. Genet. 2020;11:1022. doi: 10.3389/fgene.2020.01022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galicia-Garcia U., Benito-Vicente A., Jebari S., Larrea-Sebal A., Siddiqi H., Uribe K.B., Ostolaza H., Martín C. Pathophysiology of type 2 diabetes mellitus. Int. J. Mol. Sci. 2020;21:E6275. doi: 10.3390/ijms21176275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stöckli J., Fazakerley D.J., James D.E. GLUT4 exocytosis. J. Cell Sci. 2011;124:4147–4159. doi: 10.1242/jcs.097063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Katsoulieris E.N., Drossopoulou G.I., Kotsopoulou E.S., Vlahakos D.V., Lianos E.A., Tsilibary E.C. High glucose impairs insulin signaling in the glomerulus: an in vitro and ex vivo approach. PLoS One. 2016;11 doi: 10.1371/journal.pone.0158873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boucher J., Kleinridders A., Kahn C.R. Insulin receptor signaling in normal and insulin-resistant states. Cold Spring Harbor Perspect. Biol. 2014;6:a009191. doi: 10.1101/cshperspect.a009191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schaefer-Graf U.M., Meitzner K., Ortega-Senovilla H., Graf K., Vetter K., Abou-Dakn M., Herrera E. Differences in the implications of maternal lipids on fetal metabolism and growth between gestational diabetes mellitus and control pregnancies. Diabet. Med. 2011;28:1053–1059. doi: 10.1111/j.1464-5491.2011.03346.x. [DOI] [PubMed] [Google Scholar]
- 25.Desoye G., Herrera E. Adipose tissue development and lipid metabolism in the human fetus: the 2020 perspective focusing on maternal diabetes and obesity. Prog. Lipid Res. 2021;81 doi: 10.1016/j.plipres.2020.101082. [DOI] [PubMed] [Google Scholar]
- 26.Herrera E., Desoye G. Maternal and fetal lipid metabolism under normal and gestational diabetic conditions. Horm. Mol. Biol. Clin. Invest. 2016;26:109–127. doi: 10.1515/hmbci-2015-0025. [DOI] [PubMed] [Google Scholar]
- 27.Dunlop M., Court J.M. Lipogenesis in developing human adipose tissue. Early Hum. Dev. 1978;2:123–130. doi: 10.1016/0378-3782(78)90004-x. [DOI] [PubMed] [Google Scholar]
- 28.Enzi G., Zanardo V., Caretta F., Inelmen E.M., Rubaltelli F. Intrauterine growth and adipose tissue development. Am. J. Clin. Nutr. 1981;34:1785–1790. doi: 10.1093/ajcn/34.9.1785. [DOI] [PubMed] [Google Scholar]
- 29.Trayhurn P., Bing C. Appetite and energy balance signals from adipocytes. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006;361:1237–1249. doi: 10.1098/rstb.2006.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pelegri∗ F. DEVELOPMENTAL DYNAMICS; 2003. Maternal Factors in Zebrafish Development. [DOI] [PubMed] [Google Scholar]
- 31.Fraher D., Sanigorski A., Mellett N.A., Meikle P.J., Sinclair A.J., Gibert Y. Zebrafish embryonic lipidomic analysis reveals that the yolk cell is metabolically active in processing lipid. Cell Rep. 2016;14:1317–1329. doi: 10.1016/j.celrep.2016.01.016. [DOI] [PubMed] [Google Scholar]
- 32.Yoganantharjah P., Byreddy A.R., Fraher D., Puri M., Gibert Y. Rapid quantification of neutral lipids and triglycerides during zebrafish embryogenesis. Int. J. Dev. Biol. 2017;61:105–111. doi: 10.1387/ijdb.160209yg. [DOI] [PubMed] [Google Scholar]
- 33.Miyares R.L., de Rezende V.B., Farber S.A. Zebrafish yolk lipid processing: a tractable tool for the study of vertebrate lipid transport and metabolism. Dis. Model. Mech. 2014;7:915–927. doi: 10.1242/dmm.015800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlegel A., Gut P. Metabolic insights from zebrafish genetics, physiology, and chemical biology. Cell. Mol. Life Sci. 2015;72:2249–2260. doi: 10.1007/s00018-014-1816-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anderson J.L., Carten J.D., Farber S.A. Zebrafish lipid metabolism: from mediating early patterning to the metabolism of dietary fat and cholesterol. Methods Cell Biol. 2011;101:111–141. doi: 10.1016/B978-0-12-387036-0.00005-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barrand S., Crowley T.M., Wood-Bradley R.J., De Jong K.A., Armitage J.A. Impact of maternal high fat diet on hypothalamic transcriptome in neonatal Sprague Dawley rats. PLoS One. 2017;12 doi: 10.1371/journal.pone.0189492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kirk S.L., Samuelsson A.-M., Argenton M., Dhonye H., Kalamatianos T., Poston L., Taylor P.D., Coen C.W. Maternal obesity induced by diet in rats permanently influences central processes regulating food intake in offspring. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Timper K., Brüning J.C. Hypothalamic circuits regulating appetite and energy homeostasis: pathways to obesity. Dis. Model. Mech. 2017;10:679–689. doi: 10.1242/dmm.026609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varela L., Horvath T.L. Leptin and insulin pathways in POMC and AgRP neurons that modulate energy balance and glucose homeostasis. EMBO Rep. 2012;13:1079–1086. doi: 10.1038/embor.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishio S.-I., Gibert Y., Berekelya L., Bernard L., Brunet F., Guillot E., Le Bail J.-C., Sánchez J.A., Galzin A.M., Triqueneaux G., Laudet V. Fasting induces CART down-regulation in the zebrafish nervous system in a cannabinoid receptor 1-dependent manner. Mol. Endocrinol. 2012;26:1316–1326. doi: 10.1210/me.2011-1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boscá L., Corredor C. Is phosphofructokinase the rate-limiting step of glycolysis? Trends Biochem. Sci. 1984;9:372–373. [Google Scholar]
- 42.Kuwabara S., Yamaki M., Yu H., Itoh M. Notch signaling regulates the expression of glycolysis-related genes in a context-dependent manner during embryonic development. Biochem. Biophys. Res. Commun. 2018;503:803–808. doi: 10.1016/j.bbrc.2018.06.079. [DOI] [PubMed] [Google Scholar]
- 43.Bragato C., Carra S., Blasevich F., Salerno F., Brix A., Bassi A., Beltrame M., Cotelli F., Maggi L., Mantegazza R., Mora M. Glycogen storage in a zebrafish Pompe disease model is reduced by 3-BrPA treatment. Biochim. Biophys. Acta, Mol. Basis Dis. 2020;1866 doi: 10.1016/j.bbadis.2020.165662. [DOI] [PubMed] [Google Scholar]
- 44.Houten S.M., Violante S., Ventura F.V., Wanders R.J.A. The biochemistry and physiology of mitochondrial fatty acid β-oxidation and its genetic disorders. Annu. Rev. Physiol. 2016;78:23–44. doi: 10.1146/annurev-physiol-021115-105045. [DOI] [PubMed] [Google Scholar]
- 45.Chen Y., Zhang J., Cui W., Silverstein R.L. CD36, a signaling receptor and fatty acid transporter that regulates immune cell metabolism and fate. J. Exp. Med. 2022;219 doi: 10.1084/jem.20211314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Furuhashi M., Hotamisligil G.S. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat. Rev. Drug Discov. 2008;7:489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Renquist B.J., Zhang C., Williams S.Y., Cone R.D. Development of an assay for high-throughput energy expenditure monitoring in the zebrafish. Zebrafish. 2013;10:343–352. doi: 10.1089/zeb.2012.0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shi C., Lu Y., Zhai G., Huang J., Shang G., Lou Q., Li D., Jin X., He J., Du Z., et al. Hyperandrogenism in POMCa-deficient zebrafish enhances somatic growth without increasing adiposity. J. Mol. Cell Biol. 2020;12:291–304. doi: 10.1093/jmcb/mjz053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woods S.C., Lutz T.A., Geary N., Langhans W. Pancreatic signals controlling food intake; insulin, glucagon and amylin. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2006;361:1219–1235. doi: 10.1098/rstb.2006.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim C.-W., Addy C., Kusunoki J., Anderson N.N., Deja S., Fu X., Burgess S.C., Li C., Ruddy M., Chakravarthy M., et al. Acetyl CoA carboxylase inhibition reduces hepatic steatosis but elevates plasma triglycerides in mice and humans: a bedside to bench investigation. Cell Metabol. 2017;26:394–406.e6. doi: 10.1016/j.cmet.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jo Y.-H., Su Y., Gutierrez-Juarez R., Chua S. Oleic acid directly regulates POMC neuron excitability in the hypothalamus. J. Neurophysiol. 2009;101:2305–2316. doi: 10.1152/jn.91294.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.American Diabetes Association Professional Practice Committee 2. Classification and diagnosis of diabetes: standards of medical care in diabetes—2022. Diabetes Care. 2022;45:S17–S38. doi: 10.2337/dc22-S002. [DOI] [PubMed] [Google Scholar]
- 53.Thompson-Branch A., Havranek T. Neonatal hypoglycemia. Pediatr. Rev. 2017;38:147–157. doi: 10.1542/pir.2016-0063. [DOI] [PubMed] [Google Scholar]
- 54.Singh A., Castillo H.A., Brown J., Kaslin J., Dwyer K.M., Gibert Y. High glucose levels affect retinal patterning during zebrafish embryogenesis. Sci. Rep. 2019;9:4121. doi: 10.1038/s41598-019-41009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tiso N., Moro E., Argenton F. Zebrafish pancreas development. Mol. Cell. Endocrinol. 2009;312:24–30. doi: 10.1016/j.mce.2009.04.018. [DOI] [PubMed] [Google Scholar]
- 56.Gulseth H.L., Gjelstad I.M.F., Tiereny A.C., McCarthy D., Lovegrove J.A., Defoort C., Blaak E.E., Lopez-Miranda J., Dembinska-Kiec A., Risérus U., et al. Effects of dietary fat on insulin secretion in subjects with the metabolic syndrome. Eur. J. Endocrinol. 2019;180:321–328. doi: 10.1530/EJE-19-0022. [DOI] [PubMed] [Google Scholar]
- 57.Glatz J.F.C., Nabben M., Luiken J.J.F.P. CD36 (SR-B2) as master regulator of cellular fatty acid homeostasis. Curr. Opin. Lipidol. 2022;33:103–111. doi: 10.1097/MOL.0000000000000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu B., Li M.-Y., Lin Q., Liang Z., Xin Q., Wang M., He Z., Wang X., Wu X., Chen G.G., et al. Lipid oversupply induces CD36 sarcolemmal translocation via dual modulation of PKCζ and TBC1D1: an early event prior to insulin resistance. Theranostics. 2020;10:1332–1354. doi: 10.7150/thno.40021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Howie D., Ten Bokum A., Necula A.S., Cobbold S.P., Waldmann H. The role of lipid metabolism in T lymphocyte differentiation and survival. Front. Immunol. 2017;8:1949. doi: 10.3389/fimmu.2017.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ding L., Sun W., Balaz M., He A., Klug M., Wieland S., Caiazzo R., Raverdy V., Pattou F., Lefebvre P., et al. Peroxisomal β-oxidation acts as a sensor for intracellular fatty acids and regulates lipolysis. Nat. Metab. 2021;3:1648–1661. doi: 10.1038/s42255-021-00489-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Konrad R.J., Jolly Y.C., Major C., Wolf B.A. Inhibition of phospholipase A2 and insulin secretion in pancreatic islets. Biochim. Biophys. Acta. 1992;1135:215–220. doi: 10.1016/0167-4889(92)90139-3. [DOI] [PubMed] [Google Scholar]
- 62.Franson R.C., Harris L.K., Raghupathi R. Fatty acid oxidation and myocardial phospholipase A2 activity. Mol. Cell. Biochem. 1989;88:155–159. doi: 10.1007/BF00223437. [DOI] [PubMed] [Google Scholar]
- 63.Hernandez R.E., Galitan L., Cameron J., Goodwin N., Ramakrishnan L. Delay of initial feeding of zebrafish larvae until 8 Days postfertilization has No impact on survival or growth through the juvenile stage. Zebrafish. 2018;15:515–518. doi: 10.1089/zeb.2018.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ueno H., Nakazato M. Mechanistic relationship between the vagal afferent pathway, central nervous system and peripheral organs in appetite regulation. J. Diabetes Investig. 2016;7:812–818. doi: 10.1111/jdi.12492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roh E., Song D.K., Kim M.-S. Emerging role of the brain in the homeostatic regulation of energy and glucose metabolism. Exp. Mol. Med. 2016;48:e216. doi: 10.1038/emm.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Loh K., Herzog H., Shi Y.-C. Regulation of energy homeostasis by the NPY system. Trends Endocrinol. Metabol. 2015;26:125–135. doi: 10.1016/j.tem.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 67.Könner A.C., Brüning J.C. Selective insulin and leptin resistance in metabolic disorders. Cell Metabol. 2012;16:144–152. doi: 10.1016/j.cmet.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 68.Opazo R., Plaza-Parrochia F., Cardoso dos Santos G.R., Carneiro G.R.A., Sardela V.F., Romero J., Valladares L. Fasting upregulates npy, agrp, and ghsr without increasing ghrelin levels in zebrafish (Danio rerio) larvae. Front. Physiol. 2018;9:1901. doi: 10.3389/fphys.2018.01901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mizuno T.M., Kleopoulos S.P., Bergen H.T., Roberts J.L., Priest C.A., Mobbs C.V. Hypothalamic pro-opiomelanocortin mRNA is reduced by fasting and [corrected] in ob/ob and db/db mice, but is stimulated by leptin. Diabetes. 1998;47:294–297. doi: 10.2337/diab.47.2.294. [DOI] [PubMed] [Google Scholar]
- 70.Horvath T.L., Sarman B., García-Cáceres C., Enriori P.J., Sotonyi P., Shanabrough M., Borok E., Argente J., Chowen J.A., Perez-Tilve D., et al. Synaptic input organization of the melanocortin system predicts diet-induced hypothalamic reactive gliosis and obesity. Proc. Natl. Acad. Sci. USA. 2010;107:14875–14880. doi: 10.1073/pnas.1004282107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hagan M.M., Rushing P.A., Schwartz M.W., Yagaloff K.A., Burn P., Woods S.C., Seeley R.J. Role of the CNS melanocortin system in the response to overfeeding. J. Neurosci. 1999;19:2362–2367. doi: 10.1523/JNEUROSCI.19-06-02362.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsuneki H., Wada T., Sasaoka T. Role of orexin in the regulation of glucose homeostasis. Acta Physiol. 2010;198:335–348. doi: 10.1111/j.1748-1716.2009.02008.x. [DOI] [PubMed] [Google Scholar]
- 73.Ahrén B. Role of pituitary adenylate cyclase-activating polypeptide in the pancreatic endocrine system. Ann. N. Y. Acad. Sci. 2008;1144:28–35. doi: 10.1196/annals.1418.003. [DOI] [PubMed] [Google Scholar]
- 74.Liu L., Wang Q., Liu A., Lan X., Huang Y., Zhao Z., Jie H., Chen J., Zhao Y. Physiological implications of orexins/hypocretins on energy metabolism and adipose tissue development. ACS Omega. 2020;5:547–555. doi: 10.1021/acsomega.9b03106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kinyua A.W., Doan K.V., Yang D.J., Huynh M.K.Q., Choi Y.-H., Shin D.M., Kim K.W. Insulin regulates adrenal steroidogenesis by stabilizing SF-1 activity. Sci. Rep. 2018;8:5025. doi: 10.1038/s41598-018-23298-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guo X., Li H., Xu H., Woo S., Dong H., Lu F., Lange A.J., Wu C. Glycolysis in the control of blood glucose homeostasis. Acta Pharm. Sin. B. 2012;2:358–367. doi: 10.1016/j.apsb.2012.06.002. [DOI] [Google Scholar]
- 77.Lau J., Farzi A., Qi Y., Heilbronn R., Mietzsch M., Shi Y.-C., Herzog H. CART neurons in the arcuate nucleus and lateral hypothalamic area exert differential controls on energy homeostasis. Mol. Metabol. 2018;7:102–118. doi: 10.1016/j.molmet.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Merriam L.A., Roman C.W., Baran C.N., Girard B.M., May V., Parsons R.L. Pretreatment with nonselective cationic channel inhibitors blunts the PACAP-induced increase in Guinea pig cardiac neuron excitability. J. Mol. Neurosci. 2012;48:721–729. doi: 10.1007/s12031-012-9763-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thupari J.N., Landree L.E., Ronnett G.V., Kuhajda F.P. C75 increases peripheral energy utilization and fatty acid oxidation in diet-induced obesity. Proc. Natl. Acad. Sci. USA. 2002;99:9498–9502. doi: 10.1073/pnas.132128899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schlaepfer I.R., Glodé L.M., Hitz C.A., Pac C.T., Boyle K.E., Maroni P., Deep G., Agarwal R., Lucia S.M., Cramer S.D., et al. Inhibition of lipid oxidation increases glucose metabolism and enhances 2-deoxy-2-[18F]Fluoro-d-Glucose uptake in prostate cancer mouse xenografts. Mol. Imag. Biol. 2015;17:529–538. doi: 10.1007/s11307-014-0814-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Milbank E., López M. Orexins/hypocretins: key regulators of energy homeostasis. Front. Endocrinol. 2019;10:830. doi: 10.3389/fendo.2019.00830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hu Z., Cha S.H., Chohnan S., Lane M.D. Hypothalamic malonyl-CoA as a mediator of feeding behavior. Proc. Natl. Acad. Sci. USA. 2003;100:12624–12629. doi: 10.1073/pnas.1834402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Klok M.D., Jakobsdottir S., Drent M.L. The role of leptin and ghrelin in the regulation of food intake and body weight in humans: a review. Obes. Rev. 2007;8:21–34. doi: 10.1111/j.1467-789X.2006.00270.x. [DOI] [PubMed] [Google Scholar]
- 84.Flynn E.J., Trent C.M., Rawls J.F. Ontogeny and nutritional control of adipogenesis in zebrafish (Danio rerio) J. Lipid Res. 2009;50:1641–1652. doi: 10.1194/jlr.M800590-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Michel M., Page-McCaw P.S., Chen W., Cone R.D. Leptin signaling regulates glucose homeostasis, but not adipostasis, in the zebrafish. Proc. Natl. Acad. Sci. USA. 2016;113:3084–3089. doi: 10.1073/pnas.1513212113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pradhan G., Samson S.L., Sun Y. Ghrelin: much more than a hunger hormone. Curr. Opin. Clin. Nutr. Metab. Care. 2013;16:619–624. doi: 10.1097/MCO.0b013e328365b9be. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lavergne A., Tarifeño-Saldivia E., Pirson J., Reuter A.-S., Flasse L., Manfroid I., Voz M.L., Peers B. Pancreatic and intestinal endocrine cells in zebrafish share common transcriptomic signatures and regulatory programmes. BMC Biol. 2020;18:109. doi: 10.1186/s12915-020-00840-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Blanco A.M., Cortés R., Bertucci J.I., Soletto L., Sánchez E., Valenciano A.I., Cerdá-Reverter J.M., Delgado M.J. Brain transcriptome profile after CRISPR-induced ghrelin mutations in zebrafish. Fish Physiol. Biochem. 2020;46:1–21. doi: 10.1007/s10695-019-00687-6. [DOI] [PubMed] [Google Scholar]
- 89.Dungan W.C., Garrett M.R., Welch B.A., Lawson W.J., Himel A.R., Dungey A., Vick K.D., Grayson B.E. Whole genome transcriptome analysis of the stomach resected in human vertical sleeve gastrectomy: cutting more than calories. Physiol. Genom. 2021;53:193–205. doi: 10.1152/physiolgenomics.00082.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Johnson A.C., Wu W., Attipoe E.M., Sasser J.M., Taylor E.B., Showmaker K.C., Kyle P.B., Lindsey M.L., Garrett M.R. Loss of Arhgef11 in the dahl salt-sensitive rat protects against hypertension-induced renal injury. Hypertension. 2020;75:1012–1024. doi: 10.1161/HYPERTENSIONAHA.119.14338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
RNA Sequencing data generated in this study has been deposited at GEO. Accession number are listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact
-
•
This paper does not report original code.