Summary
Type 1 diabetes (T1D) results from autoimmune destruction of β cells. Insufficient availability of biomarkers represents a significant gap in understanding the disease cause and progression. We conduct blinded, two-phase case-control plasma proteomics on the TEDDY study to identify biomarkers predictive of T1D development. Untargeted proteomics of 2,252 samples from 184 individuals identify 376 regulated proteins, showing alteration of complement, inflammatory signaling, and metabolic proteins even prior to autoimmunity onset. Extracellular matrix and antigen presentation proteins are differentially regulated in individuals who progress to T1D vs. those that remain in autoimmunity. Targeted proteomics measurements of 167 proteins in 6,426 samples from 990 individuals validate 83 biomarkers. A machine learning analysis predicts if individuals would remain in autoimmunity or develop T1D 6 months before autoantibody appearance, with areas under receiver operating characteristic curves of 0.871 and 0.918, respectively. Our study identifies and validates biomarkers, highlighting pathways affected during T1D development.
Keywords: type 1 diabetes, biomarkers, plasma proteomics, autoimmune response
Graphical abstract
Highlights
-
•
Untargeted proteomics across 184 individuals identifies 376 regulated proteins
-
•
Extracellular matrix and antigen presentation are regulated pre-type 1 diabetes
-
•
Targeted proteomics validates 83 biomarkers in 990 individuals
-
•
Machine learning predicts islet autoimmunity and type 1 diabetes development
Nakayasu et al. perform a biomarker discovery and validation analysis to identify plasma proteins that can predict the onset of autoimmunity and type 1 diabetes. They find biomarkers that can predict both aspects of the disease with high accuracy. The study also reveals pathways that are regulated during disease development.
Introduction
Type 1 diabetes (T1D) is a chronic metabolic condition that affects approximately 20 million people worldwide. Its associated morbidities (e.g., cardiovascular disease, blindness, and kidney failure) reduce life expectancy of individuals by 11 years,1 and there is no cure yet for this disease. T1D results from a gradual destruction of insulin-producing β cells by an autoimmune response, which is associated with the appearance of autoantibodies against pancreatic islet proteins (hereafter referred to as “seroconversion”).2,3 However, the cause(s) that triggers and the mechanisms that govern this autoimmune response are still poorly understood. The Environmental Determinants of Diabetes in the Young (TEDDY) study has an ambitious goal of identifying factors that contribute to islet autoimmunity (IA) or T1D, toward enabling the development of therapeutic interventions.4 A key bottleneck in this process is the lack biomarkers that can accurately predict each step of T1D development.
Plasma proteomics analysis is a promising approach for discovering protein biomarkers,5,6,7 and it has been applied to identify biomarkers of T1D onset.8,9,10,11 Proteomics analysis can also provide important insights on the mechanism(s) of disease. Despite previous efforts,10,11 there is still an urgent need for biomarkers that can predict the different stages of T1D development. Islet autoantibodies are excellent diagnostic biomarkers for IA, and multipositivity to islet autoantibodies predicts an almost inevitable development of T1D. However, there is a desperate need for biomarkers that predict and can be used to monitor the onset of IA. Moreover, it is also important to be able to distinguish between individuals that develop T1D vs. individuals that develop IA but not hyperglycemia to appropriately focus potential treatments to the relevant stage of disease development.
Biomarker development is a long process, and many studies fall short due to the lack of systematic validation of candidates.12 Here, we conducted a robust T1D plasma protein biomarker discovery and validation study13 in the TEDDY cohort. We performed machine learning analysis to identify biomarker panels that can predict either the development of T1D or if individuals would remain in IA until the age of 6 years both with high accuracy and as early as 6 months before the appearance of the autoimmune response. By comparing them with previously published proteomics models of insulitis using human islets and cultured β cells treated with cytokines, our results also provide insights on the mechanism of T1D development.
Results
Experimental design and discovery phase analysis
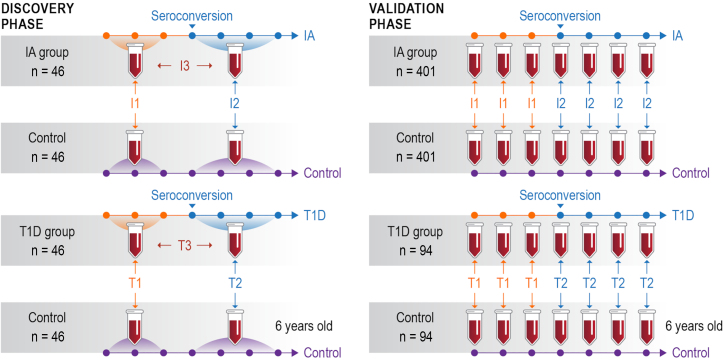
The study was based on a nested case-control design4 and aimed to identify biomarkers predictive of IA and T1D development, with samples divided into 8 groups: pre- and post-seroconversion for individuals that developed T1D (T1D group) or remained in IA (IA group) by the age of 6 years, each paired with respective control groups. The following comparisons were considered: I1, IA group vs. control pre-seroconversion; T1, T1D group vs. control pre-seroconversion; I2, IA group vs. control post-seroconversion; T2, T1D group vs. control post-seroconversion; I3, pre-vs. post-seroconversion of IA group; and T3, pre-vs. post-seroconversion of T1D group (Figure 1).
Figure 1.
Study design: A two-phase study design to discover and validate biomarkers in human blood plasma
Individual plasma samples from a smaller number (n = 46) of individuals were pooled from pre- and post-seroconversion visits and analyzed by in-depth untargeted proteomics in the discovery phase (left panel) (n = 401 and 94). Individual plasma samples from several collection time points (represented by the dots in the timeline) were analyzed in a larger cohort by targeted proteomics in the validation phase (right panel). Comparison I1: time point(s) before seroconversion of the group that remained in autoimmunity by the age of 6 years (IA group) paired against matched controls. Comparison T1: time point(s) before seroconversion of the group that developed type 1 diabetes (T1D) by the age of 6 years (T1D group) paired against matched controls. Comparisons I2 and T2 have the same group of individuals as I1 and T1, respectively, but after seroconversion. Comparisons I3 and T3 compare IA and T1D groups before vs. after seroconversion, respectively. See Figure S1 for details on sample collection time points.
The study was comprised of two phases: a discovery phase focused on a deep proteomics analysis of pooled samples from a limited number of individuals5 (n = 184) and a subsequent validation phase with selected biomarker candidates analyzed by targeted proteomics in many samples from a much larger cohort14 (n = 990) across multiple time points (Figures 1, S1, and S2). The characteristics and demographic information for both discovery and validation phase cohorts are presented in Table 1. A total of 1,488 mass spectrometry analyses from 62 multiplexed proteomics sets were performed in the discovery phase. To ensure quality across 18 months of data collection, we developed and implemented an automated quality control system named QC-ART (quality control analysis in real time).15 This tight quality control analysis assured that consistent data were collected across the study. The data profile had very similar distributions of peptide abundances across different multiplexed sets (Figure S3A) and numbers of identified peptides in each group (Figure S3B). A total of 36,252 peptides derived from 1,720 proteins were identified, and after normalizing to a reference sample that was included in each multiplexed proteomics set, peptides were sequentially removed from the dataset based on the following criteria: (1) detected in 2 or fewer samples across any group, (2) coefficient of variance greater than 150%, (3) detected in fewer than 2 matched case-control pairs, and (4) p value >0.05 across different comparisons. These criteria resulted in a final discovery phase proteomics dataset that included 376 significant proteins (373 with ≥2 peptides and 3 with 1 peptide) at a p value threshold of ≤0.05 (Figure 2A; Table S1).
Table 1.
Characteristics of the study cohort
| Discovery |
Validation |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| T1D |
IA |
T1D |
IA |
||||||
| Cases | Matched controls | Cases | Matched controls | Cases | Matched controls | Cases | Matched controls | ||
| Number | 46 | 46 | 46 | 46 | 94 | 94 | 401 | 401 | |
| Case seroconversion age (months) | median | 12 | – | 23 | – | 12 | – | 22 | – |
| Q1 | 9 | – | 14 | – | 10 | – | 12 | – | |
| Q3 | 18 | – | 33 | – | 19 | – | 33 | – | |
| Gender | female | 25 | 25 | 17 | 17 | 43 | 43 | 179 | 179 |
| male | 21 | 21 | 29 | 29 | 51 | 51 | 222 | 222 | |
| Clinical center | Colorado | 8 | 8 | 4 | 4 | 13 | 13 | 57 | 57 |
| Georgia/Florida | – | – | 1 | 1 | 6 | 6 | 28 | 28 | |
| Washington | 4 | 4 | 5 | 5 | 6 | 6 | 37 | 37 | |
| Finland | 23 | 23 | 23 | 23 | 31 | 31 | 113 | 113 | |
| Germany | 6 | 6 | 1 | 1 | 13 | 13 | 33 | 33 | |
| Sweden | 5 | 5 | 12 | 12 | 25 | 25 | 133 | 133 | |
| HLA-DR-DQ genotypes | HLA ineligible | 1 | 1 | – | – | 1 | 3 | 1 | 2 |
| DR3/4 | 26 | 20 | 25 | 17 | 55 | 39 | 211 | 152 | |
| DR4/4 | 7 | 6 | 10 | 9 | 13 | 18 | 65 | 71 | |
| DR4/8 | 7 | 5 | 8 | 6 | 14 | 10 | 61 | 67 | |
| DR3/3 | 1 | 6 | 3 | 11 | 5 | 9 | 46 | 81 | |
| FDR specific | 4 | 8 | – | 3 | 6 | 13 | 17 | 28 | |
| Family history of T1D | GP | 28 | 28 | 42 | 42 | 61 | 61 | 309 | 309 |
| FDR: mother | 3 | 11 | 1 | 3 | 4 | 15 | 16 | 37 | |
| FDR: father | 11 | 5 | 2 | 1 | 20 | 14 | 54 | 40 | |
| FDR: both parents | – | – | – | – | 1 | – | 1 | – | |
| FDR: sibling | 4 | 2 | 1 | – | 8 | 4 | 21 | 15 | |
| Type of first autoantibody | not IA+ | – | 44 | – | 45 | – | 89 | – | 392 |
| IAA only | 27 | 2 | 27 | – | 53 | 4 | 194 | 0 | |
| GADA only | 8 | – | 11 | 1 | 14 | 1 | 132 | 3 | |
| IA-2A only | – | – | 1 | – | – | – | 6 | – | |
| two or more autoantibodies | 11 | – | 7 | – | 27 | – | 69 | – | |
FDR, first-degree relative; GADA, glutamic acid decarboxylase autoantibody; GP, general population; HLA, human leukocyte antigen; IA, islet autoimmunity group (remained in autoimmunity by the age of 6); IA-2A, islet antigen-2 autoantibody; IAA, insulin autoantibody; T1D, type 1 diabetes group (progressed to autoimmune diabetes by the age of 6).
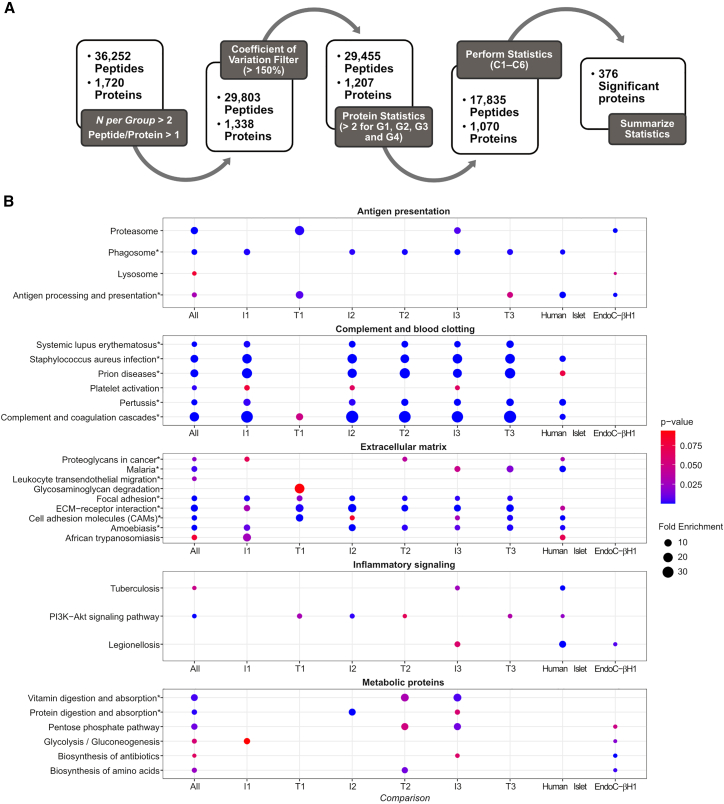
Figure 2.
Discovery phase data analysis
(A) Discovery phase data quality and statistical analysis workflow (N = 46). Sequential pre-filtering steps focused on identification and removal of high-variability (steps 1–2) and low-coverage (step 3) peptides and proteins. Resulting proteins and peptides were submitted to multiple statistical comparisons in the context of autoimmunity and T1D development (steps 4–5).
(B) Functional enrichment analysis. The 376 differentially abundant proteins identified from the discovery phase were submitted to function enrichment analysis with DAVID using the KEGG annotation. Pathways were plotted as circles, with sizes based on their fold enrichment and colors based on p values. Individual pathways were grouped into larger biological processes based on the overlapping proteins between each pathway. Pathways that were also enriched among the 167 targets of the validation phase are marked with asterisks. Comparison I1: time point(s) before seroconversion of the group that remained in autoimmunity by the age of 6 years (IA group) paired against matched controls. Comparison T1: time point(s) before seroconversion of the group that developed T1D by the age of 6 years (T1D group) paired against matched controls. Comparisons I2 and T2 have the same group of individuals as I1 and T1, respectively, but after seroconversion. Comparisons I3 and T3 compare IA and T1D groups before vs. after seroconversion, respectively.
Biological pathways regulated in IA and T1D development
A functional-enrichment analysis of the discovery phase data showed that 22 pathways were overrepresented among the 376 differentially abundant proteins and their proteoforms (Figure 2B). To facilitate the interpretation, we further grouped these pathways into fewer biological processes based on the components of each pathway that were regulated in the different comparisons. We plot the pathways as circles, with their sizes being proportional to the fold enrichment and colored based on the enrichment significance (Figure 2B). Complement and blood clotting, antigen presentation, extracellular matrix, nutrient digestion and absorption, cellular metabolism, and inflammatory signaling processes were significantly enriched with differentially abundant proteins (Figure 2B). To investigate if these processes might also occur in islets during T1D development, we compared the results of the functional enrichment analysis of the TEDDY proteomics data to published proteomics analyses of human islets16 and the β cell line EndoC-βH117 from the Human Islet Research Network (HIRN). Each sample type was treated with pro-inflammatory cytokines interleukin-1β (IL-1β)+interferon γ (IFNγ) as a model of insulitis during IA. Proteins related to complement and blood clotting, antigen presentation, extracellular matrix, and inflammatory signaling were also enriched among the IL-1β+IFNγ regulated proteins in the human islet study (Figure 2B). In EndoC-βH1 cells, pathways related to antigen presentation, inflammatory signaling, and cell metabolism were regulated similarly to the plasma signatures (Figure 2B). This shows that similar inflammatory signatures that occur in plasma of individuals during T1D development also occur in islets during insulitis.
Extracellular matrix
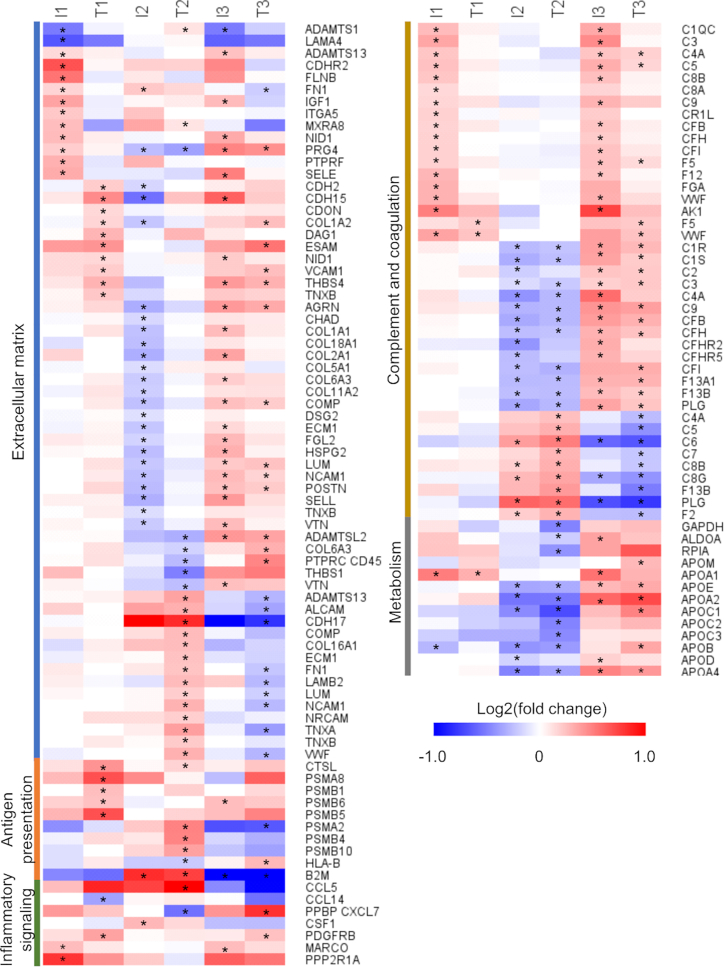
Pathways related to the extracellular matrix were commonly enriched among the different comparisons. However, there was only a small overlap of significant proteins between the different comparisons, as shown in the heatmap. At pre-seroconversion, the IA group had 14 regulated proteins (12 upregulated) (comparison I1, Figure 3), while the T1D group had 10 regulated proteins (all upregulated) (comparison T1, Figure 3). Post-seroconversion, the scenario became more distinct, with the IA group having 24 out of 25 regulated proteins downregulated, while the T1D group had 17 out of the 17 regulated proteins upregulated (comparisons I2 and T2, respectively, Figure 3).
Figure 3.
Regulated pathways
The 376 differentially abundant proteins identified from the discovery phase were submitted to function enrichment analysis with DAVID using the KEGG annotation. The heatmap shows the proteins enriched for each pathway. Asterisks mark those proteins in specific comparisons that were statistically significant. Comparison I1: time point(s) before seroconversion of the group that remained in autoimmunity by the age of 6 years (IA group) paired against matched controls. Comparison T1: time point(s) before seroconversion of the group that developed T1D by the age of 6 years (T1D group) paired against matched controls. Comparisons I2 and T2 have the same group of individuals as I1 and T1, respectively, but after seroconversion. Comparisons I3 and T3 compare IA and T1D groups before vs. after seroconversion, respectively.
Antigen presentation
Antigen processing and presentation was the most distinctive pathway at the pre-seroconversion time point when comparing the T1D group vs. the IA group. In the T1D group, a higher level of antigen-processing proteins was observed, including cathepsin L1 (CTSL) (protein names are abbreviated using their UniProt gene names) and proteasome subunits PSMA8, PSMB1, PSBM5, and PSBM6 (comparison T1, Figure 3). Cathepsin L1 and proteasome subunits PSMA2, PSBM4, and PSBM10 were also higher after seroconversion but were accompanied also by the antigen-presenting complex human leukocyte antigen class I (HLA-B) and β-2-macroblobulin (B2M) (comparison T2, Figure 3).
Inflammatory signaling
Four cytokines and chemokines were regulated across different comparisons. C-C motif chemokine 14 (CCL14) was downregulated pre-seroconversion in the T1D group compared with the control (comparison T1, Figure 3). CCL5 and proplatelet basic protein (PPBP; or CXCL7) were up- and downregulated, respectively, in the T1D group compared with the control at the post-seroconversion time point (comparison T2, Figure 3). Receptors, such as platelet-derived growth factor receptor β (PDGFRB) and macrophage receptor MARCO, and signaling transduction proteins, such as serine/threonine-protein phosphatase 2A 65 kDa regulatory subunit A alpha isoform (PPP2R1A), were also regulated (Figure 3).
Complement and coagulation
At pre-seroconversion, complement factors C1QC, C3 C4A, C5, C8A, C8B, C9, CR1L, CFB, CFH, and CFI and coagulation factors F5, F12, fibrinogen α and γ, von Willebrand, and adenylate kinase were higher in the IA group vs. respective controls (comparison I1, Figure 3), while proteoforms of F5 and von Willebrand factors were upregulated in the T1D group (comparison T1, Figure 3). Post-seroconversion, both groups had lower levels of most coagulation and complement factors compared with their respective controls. However, specific proteoforms were regulated in the opposite way (comparisons I2 and T2, Figure 3), probably reflecting processing or post-translational modifications of these proteins.
Metabolic proteins
Among the central carbon metabolism enzymes, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), fructose-bisphosphate aldolase A, and ribose-5-phosphate isomerase were reduced post-seroconversion in the T1D group but not in the IA group (comparisons I2 and T2, Figure 3), suggesting an abnormal sugar metabolism. Lipoproteins represent another class of metabolic proteins regulated in plasma. Apolipoprotein (Apo) A1 was increased in both groups pre-seroconversion but had similar levels to the control after seroconversion (Figure 3). Conversely, Apo A2, A4, B, C1, C2, C3, D, E, H, and J had similar levels compared with the controls in both groups pre-seroconversion but declined after seroconversion (Figure 3). Overall, these data indicate changes in metabolic proteins that precede hyperglycemia.
Validation of protein biomarker candidates
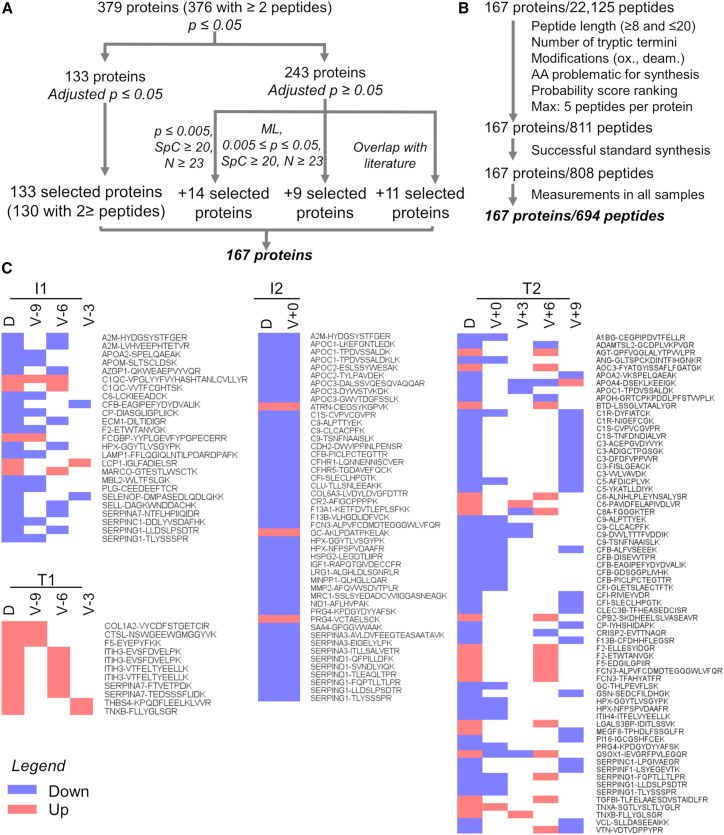
We performed a systematic prioritization of the candidate biomarkers from the discovery phase based on the following criteria: (1) statistical significance at Benjamini-Hochberg adjusted p value ≤0.05; (2) ≥2 peptides identified per protein, a spectral count (SpC) ≥20, and an unadjusted p value <0.005; (3) ≥2 peptides identified per protein, an SpC ≥20, detected in more than 23 samples, and machine learning (ML) to determine the group of proteins that are the most predictive of each of the 6 comparisons; or (4) proteins that were previously described as potential T1D onset biomarkers in the literature8,9,10,11 and had an unadjusted p value ≤ 0.05 (Figure 4A). This analysis led to the selection of 167 proteins for the validation phase, of which 811 peptides were selected for targeted proteomics assay development (as described in STAR Methods). Similar to the discovery phase, we developed an informatics tool named Q4SRM (quality control analysis for selected reaction monitoring)18 to systematically track data quality across 29 months of analyses. A total of 694 peptides from all 167 proteins were successfully monitored until the end of the study (Figure 4B; Table S2). An additional post hoc quality control analysis showed a strikingly high correlation (>95%) for almost all the 6,426 targeted proteomics analyses performed (Figure S4). From the measured peptides, 127 peptides from 83 (50%) proteins were significant and showed similar abundance patterns to the discovery phase across comparisons I1, T1, I2, and T2, validating them as biomarkers (Figure 4; Tables S3, S4, and S5). The 83 validated proteins belong to all major biological processes observed as regulated in T1D development in the discovery phase: antigen presentation, complement and blood clotting, extracellular matrix, inflammatory signaling, and metabolic proteins.
Figure 4.
Validation phase data analysis
(A) Biomarker candidates were selected first based on statistical test with p value correction. Additional candidates were selected for validation based on the p value, number of samples in which the peptide was detected, spectral count, machine learning, and previous reports in the literature.
(B) Up to 5 peptides for each candidate protein were selected based on their physicochemical properties and probability ranking for a likely successful measurement by targeted proteomics.
(C) Cross-validated proteins across discovery (D) and validation (V) phases. Only significant, validated proteins are represented in the heatmap and are colored based on their regulation. Time points are represented by months prior (−) or post (+) seroconversion. Comparison I1: time point(s) before seroconversion of the group that remained in autoimmunity by the age of 6 years (IA group) paired against matched controls (n = 401). Comparison T1: time point(s) before seroconversion of the group that developed T1D by the age of 6 years (T1D group) paired against matched controls (n = 94). Comparisons I2 and T2 have the same group of individuals as I1 and T1, respectively, but after seroconversion. Comparisons I3 and T3 compare IA and T1D groups before vs. after seroconversion, respectively.
ML models for predicting T1D onset
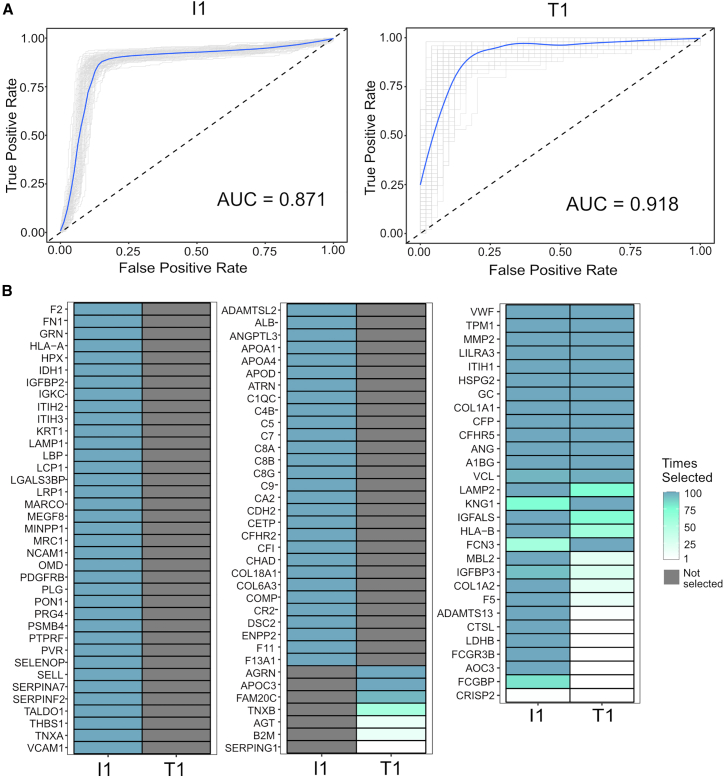
ML is a powerful approach to identify individual or combinations of biomarkers that can predict a phenotype. Therefore, we performed ML analysis to identify biomarkers that can predict if the individual will remain in IA or develop T1D by the age of 6 years prior to seroconversion. We used logistic regression with a LASSO penalization to build ML models that can predict the different outcomes. This analysis can identify models based on panels of peptides that best predict the different outcomes, and they were tested by cross-validation repeated for 100 bootstrap iterations. The receiver operating characteristic curves from this analysis show that both IA and T1D states at the age of 6 years can be predicted with high accuracy at 6 months prior to the seroconversion time with average areas under the curves of 0.871 and 0.918 and bootstrapped 95% confidence intervals of (0.826, 0.912) and (0.830, 0.942), respectively (Figure 5A). Figure 5B shows the proteins that correlate with the panel of peptides that were selected by the ML analysis to build the models. Among the most important proteins, i.e., the ones that appeared with more frequency across the training models, there were proteins from the complement and coagulation cascades (e.g., C4B, C5, C6, C8B, C9, F2 and F5), extracellular matrix (e.g. MMP2, COL1A1, COL1A2, WVF, and ADAMTS13), and antigen processing and presentation (HLA-A, HLA-B, and B2M) (Figure 5B; Table S6), suggesting that they are important processes in the disease development. A total of 28 out of the 116 selected peptides were commonly selected across both I1 and T1 comparisons, while 81 were selected only in the I1 comparison and 7 only in T1 (Figure 5B; Table S6), showing that both IA and T1D groups have some overlapping but also distinct signatures. Overall, the ML analysis showed that IA and T1D states at the age of 6 years can be predicted even 6 months prior to the onset of IA.
Figure 5.
Prediction of autoimmunity with normoglycemia or T1D onset prior to seroconversion by machine learning analysis
(A) The panels show receiver operating characteristic (ROC) curves of peptide panels that predict normoglycemia (comparison I1) (n = 247) and T1D onset (comparison T1) (n = 49) at 6 months prior to the seroconversion. The numbers (n) of case-control pairs used at each time point are shown at the top of each ROC curve. Individual bootstrap curves are shown in gray with the mean curve given in blue.
(B) Heatmaps showing the selected proteins and their frequencies of being kept in the model over the 100 bootstrap iterations for the most important peptide features used to predict the model. The left two panels contain proteins that were selected in only one comparison, whereas the right panel shows proteins that were commonly selected. Proteins are named based on UniProt gene names.
Discussion
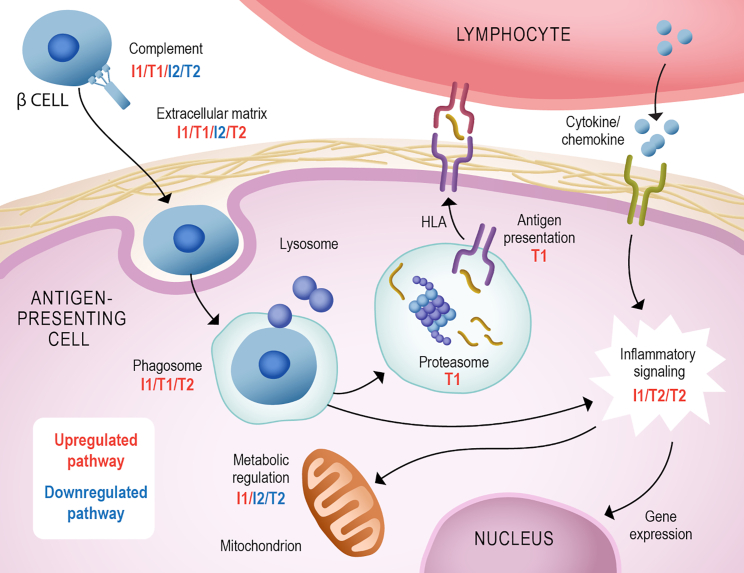
We initially identified 376 differentially abundant proteins among the varying points of IA with normoglycemia and T1D development in a cohort of the TEDDY study. These proteins were overrepresented in processes related to T1D development such as complement and blood clotting, antigen presentation, extracellular matrix, nutrient digestion and absorption, cellular metabolism, and inflammatory signaling (Figure 6). Importantly, these processes were also regulated in human islets and cultured β cells stimulated with pro-inflammatory cytokines to mimic the insulitis process. This suggests that some of these processes also occur in the pancreas during T1D development. Overall, our data showed a regulation in the complement and coagulation cascades. Polymorphism in complement has been associated with a higher risk of T1D development.19,20 Increased complement activation and deposition have been shown in pancreata from individuals with T1D.21 Patients with T1D also have increased clotting condition, including upregulation in platelet aggregation and coagulation activity and reduction in fibrinolysis.22 Complement can also participate in opsonization of pathogens or dead cells, probably β cells, toward phagocytosis (Figure 6).
Figure 6.
Summary of pathways regulated in autoimmunity and T1D development
Many components of the complement cascade were found to be increased pre-seroconversion (comparisons T1/I1) and decreased post-seroconversion (T2/I2). An increase in phago/lysosome components was observed in comparisons T1/I1/T2. However, an increase in proteasome and antigen presentation components was only observed in T1. This process can trigger cellular signaling along with the stimulation of cytokine/chemokine receptors, regulating gene expression and cell metabolism (I1/T2/I2). We also observed a regulation of the extracellular matrix proteins (up in T1/I1/T2 and down in I2), which can regulate the interaction with immune cells. Comparison I1: time point(s) before seroconversion of the group that remained in autoimmunity by the age of 6 years (IA group) paired against matched controls. Comparison T1: time point(s) before seroconversion of the group that developed T1D by the age of 6 years (T1D group) paired against matched controls. Comparisons I2 and T2 have the same group of individuals as I1 and T1, respectively, but after seroconversion. Comparisons I3 and T3 compare IA and T1D groups before vs. after seroconversion, respectively.
The phagocytosis and lysosome components were also shown to be regulated in our data (Figure 6). This process is involved in pathogen and dead cell destruction and antigen presentation. The subsequent processes of proteasome antigen processing and presentation with HLA were only upregulated at the pre-seroconversion stage of individuals that developed T1D (comparison T1, Figure 6), reinforcing the importance of this process in the disease development. It is possible that higher proteasome levels result in abnormal antigen presentation and autoimmunity development. Polymorphism on the antigen presentation gene HLA is indeed the major risk factor for developing T1D.23 The HLA variants can differentially present islet self-antigens and are believed to be involved in autoimmunity development.24 During IA, pro-inflammatory cytokines and chemokines are produced, triggering β cell apoptosis and helping to recruit leukocytes and leading to insulitis.25 This signaling also leads to regulation in gene expression and cell metabolism, which is observed in our data (Figure 6).
Our data show shifts in metabolic proteins even pre-seroconversion (Figure 6). Changes in metabolite profiles have been shown to predict development of autoantibodies 6 months prior to seroconversion.26 In addition, metabolite profiles detected in 3- to 9-month-old children from the TEDDY study are predictive of their developing T1D by the age of 6 years.27 In addition, abnormal pro-insulin-to-C-peptide ratio can be detected 12 months prior to the onset of T1D,28 suggesting a dysfunction in insulin processing that may affect the body metabolism even before causing hyperglycemia. Similarly, bile acid metabolism is dysregulated prior to seroconversion in T1D development.29 Furthermore, several components of plasma lipoproteins were downregulated after seroconversion. Triacylglycerols, which are major components of plasma lipoproteins, have been shown to be lower in children that developed T1D compared with children that had IA but who remained normoglycemic.30 Lipoprotein subunits, such as Apo CIII, have been linked to T1D development. Apo CIII has been shown to trigger β cell apoptosis.31 Overall, changes in metabolism precede the disease onset and may also be involved in T1D development.
Another process highly regulated in our data was the extracellular matrix (Figure 6). Circulating extracellular matrix proteins are good indicators of tissue damage32 and may indicate damage on the pancreatic islets. In addition, during recruitment of leukocytes, the islet extracellular matrix undergoes major remodeling to allow cell infiltration.33 Our data show a different profile on plasma extracellular proteins between the individuals with IA that developed T1D or had normoglycemia, possibly enabling or impeding β cell destruction.34
In clinical diagnosis, T1D is diagnosed by blood glucose levels or glycated hemoglobin.3 For predictive biomarkers, HLA genotype and autoantibodies against islet proteins have been used, but they lack enough discriminative power due to the heterogeneity of the disease.35 Biomarkers based on T cells are currently being developed but require further validation.36 Proteomics has been applied to identify T1D biomarkers, but some of these were focused on disease diagnosis after onset.8,9 In biomarker studies prior to T1D onset, von Toerne et al.10 performed a proteomics discovery and validation study on samples from individuals after seroconversion to identify biomarkers that can diagnose the onset of IA and T1D development. They identified several circulating biomarkers of IA and found that a protein panel composed of hepatocyte growth factor activator, complement factor H, ceruloplasmin, and age can predict progression time to T1D.10 Moulder et al. performed untargeted proteomics analysis in a longitudinal study from 3 months to 12 years of age for 13 individuals that developed T1D vs. age-matched controls and found that the profile of proteins such as complement proteins and Apos can predict the onset of T1D.11 Here, we performed a study to identify and validate biomarkers of different stages of the disease and the likelihood of developing T1D. Unlike the study by Moulder et al. that matched case-control pairs based on age, we make our comparisons in relation to seroconversion. We identified and validated 83 biomarkers of IA and T1D development prior to the onset of the disease. Furthermore, we performed ML analysis and identified panels of proteins that can predict both the development of persistent autoantibodies with normoglycemia and T1D even 6 months prior to the appearance of the autoimmune response. We believe evaluation of these promising predictive protein panels in other ongoing prospective studies of development of autoimmunity and T1D in human cohorts could aide in the development of prognostics and therapeutics.
Limitations of the study
One limitation of our study is that the validation was not performed in an independent cohort of samples. Validation in independent cohorts of samples can eliminate some confounding factors based on geographical and populational biases. However, our cohort includes individuals from 7 different centers in the US and Europe, which can reduce some of the regional confounding factors. Another limitation of our study is that the ML models were also not validated in an independent cohort of samples. However, they have gone through 100 bootstrap iterations of repeated cross-validation for the robustness of the analysis. The ML analysis also requires the development of a baseline value for control individuals before being put in practice. Therefore, these two limitations are among the points that need to be further evaluated in additional studies of independent cohorts before implementing our findings in clinical practice. Finally, baseline model performance based on traditional risk factors, such as gender or metabolic markers, was either used in pairing case and control subjects or was not available and thus was not explicitly evaluated. However, model performance area under the curve (AUC) values in this study were found to outperform previously published baseline metrics.37,38 Despite these limitations, our results provided biological insights on the molecular pathways regulated in T1D development and identified biomarker candidates for the disease.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Human plasma | The Environmental Determinants of Diabetes in the Young (TEDDY) study | https://teddy.epi.usf.edu/ |
| Chemicals, peptides, and recombinant proteins | ||
| Custom Synthesized Heavy Isotope-Labeled Peptides | New England Peptides, now Vivitide | N/A |
| 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) | Sigma - Aldrich | H3375 |
| Acetonitrile, HPLC grade | J.T. Baker | 9829–03 |
| Acetonitrile anhydrous | Sigma - Aldrich | 271004 |
| Ammonium hydroxide solution | Sigma - Aldrich | 338818 |
| Aprotinin | Sigma - Aldrich | A6103 |
| Buffer A for Multiple Affinity Removal LC Column | Agilent | 5185–5987 |
| Buffer B for Multiple Affinity Removal LC Column | Agilent | 5185–5988 |
| Chloroform | Sigma - Aldrich | C2432 |
| Dithiothreitol | Thermo Scientific | 20291 |
| Ethylenediaminetetraacetic acid | Sigma - Aldrich | E7889 |
| Formic acid | Sigma - Aldrich | 33015 |
| Iodoacetamide | Thermo Scientific | 90034 |
| HPLC Grade Water | J.T. Baker | 4218–03 |
| Hydroxylamine Solution 50% | Sigma - Aldrich | 467804 |
| Methanol, HPLC grade | Fluka | 34966 |
| Sequencing grade modified trypsin | Promega | V5117 |
| Tris (hydroxymethyl)aminomethane hydrochloride pH 8.0 | Sigma - Aldrich | T2694 |
| Trifluoroacetic acid | Sigma - Aldrich | 91707 |
| Urea | Sigma - Aldrich | U0631 |
| Critical commercial assays | ||
| 8-plex iTRAQ kit | Applied Biosystems | 4390811 |
| Deposited data | ||
| Discovery phase mass spectrometry data | MassIVE | MSV000091560 |
| Validation phase mass spectrometry data | MassIVE | MSV000091562 |
| Software and algorithms | ||
| R package (v3.2.3) | The R Project for Statistical Computing | https://www.r-project.org/ |
| Decon2LS_V2 | Pacific Northwest National Laboratory | https://github.com/PNNL-Comp-Mass-Spec |
| DTA Refinery | Pacific Northwest National Laboratory | https://github.com/PNNL-Comp-Mass-Spec |
| MSGF+ | University of California – San Diego | https://msgfplus.github.io/ |
| MASIC | Pacific Northwest National Laboratory | https://github.com/PNNL-Comp-Mass-Spec |
| QC-ART | Pacific Northwest National Laboratory | https://github.com/PNNL-Comp-Mass-Spec |
| Q4SRM | Pacific Northwest National Laboratory | https://github.com/PNNL-Comp-Mass-Spec |
| Skyline | University of Washington | https://skyline.ms/ |
| DAVID | National Institutes of Health | https://david.ncifcrf.gov/ |
| Other | ||
| Hu-14 4.6 × 100 mm MARS column | Agilent | 5188–6558 |
| 3-kDa MWCO Amicon centrifugal filters | Millipore | UFC5003BK |
| Reversed phase tC18 SepPak SPE columns | Waters | WAT054925 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Thomas O. Metz (thomas.metz@pnnl.gov).
Materials availability
For materials availability, please reference the TEDDY access page: https://teddy.epi.usf.edu/research/. There are restrictions on the availability of samples, and they are subject to approval by the TEDDY Ancillary Studies Committee.
Experimental model and subject details
Study design, sample cohort, batching and randomization
The study was conducted after approval from the Institutional Review Boards of the University of South Florida (USF) and the Pacific Northwest National Laboratory (PNNL) in accordance with federal regulations. The study analyzed samples at pre- and post-seroconversion from individuals that developed T1D (T1D group) or remained in IA (IA group) by the age of 6 years, each paired with respective control groups. The following comparisons were made: I1: IA group vs. controls pre-seroconversion; T1: T1D group vs. controls pre-seroconversion; I2: IA group vs. controls post-seroconversion; T2: T1D group vs. controls post-seroconversion; I3: pre-vs. post-seroconversion within the IA group; and T3: pre-vs. post-seroconversion within the T1D group (Figure 1).
TEDDY study participants have higher genetic risk of developing T1D, and to reduce the study to a manageable size, samples were previously matched based on clinical center, gender, and family history of T1D.4 Samples were collected from September 2004 to May 2012. Individuals were treated for 30–40 min with topical EMLA anesthetic, and blood was drawn by venipuncture of the antecubital vein in EDTA tubes. Plasma was separated from cells by centrifugation and stored at −70°C at the NIDDK repository until the start of the proteomics project in September 2014. Seroconversion was determined by screening for autoantibodies against islet cells (IA-2A), insulin (IAA), glutamic acid decarboxylase (GADA) and zinc transporter 8A (ZnT8A) as previously described.39 T1D was diagnosed based on blood glucose levels and oral glucose tolerance test following the recommendations of the World Health Organization and American Diabetes Association.40
In this nested case-control study, TEDDY monitored over 8,000 individuals from 7 centers (Germany, Sweden, and Finland in Europe; and Denver, Georgia, Florida, and Washington in the USA) from the ages of 0–6 years old. From these, 418 developed IA and 114 progressed to T1D.4 For the proteomics analysis, we selected 401 individuals for the IA group and 94 individuals for the T1D group, each paired to a matched control. The characteristics of the subset of case-control samples are listed in Table 1.
The proteomics study was designed with a discovery and a validation phase to ensure an in-depth and robust analysis and was also conducted in a blinded fashion until the conclusion of the validation phase. Sample selection, batching, and randomization were performed at USF, whereas proteomics measurements were conducted at PNNL. Randomization was performed to assure that the study endpoints and patient time points were appropriately dispersed across the study and that the nested case-control pairs were analyzed within the same batch during processing to match the statistical design.
Method details
Discovery phase - Untargeted proteomics analysis
A statistical power analysis was performed to determine the number of case-control pairs needed in each study group, using a similar plasma proteomics dataset collected in the authors’ laboratory and consisting of 16,928 peptides measured in 12 individuals across multiple time points. Using the power.t.test function in R package (v3.2.3), it was determined that 23 case-control pairs were required to reach 80% power to detect a 2-fold difference utilizing a variance estimate associated with the 75-th percentile of all measured peptides from the proteomics data. This number was doubled to 46 case-control pairs to account for missing data frequently encountered during untargeted proteomics analysis. This resulted in 2252 plasma samples (considering multiple time points) that were then combined per donor within pre- or post-seroconversion into 368 pooled samples due to costs and logistics. For a detailed distribution of the samples used, see Figure S1. In the analysis, fourteen of the most abundant proteins in each sample were depleted using a Hu-14 4.6 × 100 mm MARS column (Agilent Technologies, Palo Alto, CA) coupled to a 1200 series HPLC (Agilent) and concentrated in Amicon centrifugal filters (3-kDa MWCO, Millipore, Burlington, MA). Proteins were digested in 96-well plates,41 and peptides were labeled with 8-plex iTRAQ reagent (Applied Biosystems, Foster City, CA) following manufacturer recommendations. A pooled reference sample was created by mixing aliquots of each sample and was used for normalization across different datasets. The multiplexed iTRAQ-labeled samples were fractionated by high pH reversed phase chromatography and analyzed on a nanoAquity UPLC® system (Waters) connected to an LTQ Orbitrap Velos mass spectrometer (Thermo Scientific).15,42 Mass spectra were processed using Decon2LS_V2 and DTA Refinery,43,44 with peptides identified using MSGF+45 by searching against the human SwissProt sequences of the Uniprot Knowledgebase. The parameters included: (1) 6 ppm parent ion mass tolerance, (2) partial tryptic digestion, (3) cysteine carbamidomethylation (+57.0215) and N-terminal/lysine 8-plex iTRAQ (+304.2053) addition as static modifications, and (4) oxidation (+15.9949 Da) on methionine, cysteine, tyrosine, and tryptophan, dioxidation (+31.9898 Da) on cysteine, and deamidation/deamination (+0.9840 Da) on asparagine, glutamine, and arginine residues as variable modifications. Identifications were filtered with MSGF probability scores of ≤1.0 × 10−9, ≤7 × 10−11 and ≤2 × 10−12 at spectral, peptide and protein levels, respectively, resulting in <1% false-discovery rate. iTRAQ reporter ion intensities were extracted with MASIC,46 and the intensities of multiple MS/MS spectra from the same peptide were summed together to remove redundancy.
Validation phase - Targeted proteomics analysis
Up to 5 peptides were selected as surrogates for candidate biomarker proteins identified in the discovery phase based on their physical-chemical properties (between 8 and 20 amino acid residues, derived from trypsin digestion at both termini, and lack of post-translationally modified amino acid residues or residues that are problematic for chemical synthesis), and a Bayesian network-generated probabilistic score was used to select the peptides more likely to be successfully developed into targeted proteomics assays. To account for possible proteoforms, peptides from the same proteins that were not statistically significant were also included. Samples were prepared and analyzed in batches of approximately 80 samples in 96-well plates. The case and control pairs were restricted to the same batch and randomized across the plate. Time points from each individual is shown in Figure S2. Quality control samples were comprised of 6 pooled plasma samples from TEDDY and 1 commercial pooled plasma sample from BioIVT (Westbury, NY); these were also randomized within each batch. Whole plasma of 6,426 individual samples were digested in 80 batches in 96-well plates41 and spiked with custom synthesized peptides (New England Peptides, now Vivitide) containing heavy isotopes in the C-terminal residues. Targeted proteomics analyses were performed using a Nano M-class UPLC (Waters) interfaced to a TSQ Altis triple quadrupole mass spectrometer (Thermo Fisher Scientific). The linearity of the assays was checked by diluting human plasma into chicken plasma. Data quality was assessed using the Q4SRM tool.18 Data were analyzed with the Skyline software and were manually inspected for proper alignment and background threshold.
Quantification and statistical analysis
Statistical analysis
Statistical quality control of untargeted proteomics data involved removing peptides that were observed in only one sample per group and outlier identification using a Mahalanobis distance method.47,48 Protein quantification from the peptide-level data was based on standard and scaled median quantification48,49 and statistics were performed on proteins and proteoforms (different forms of the same proteins resulting from gene isoforms, processing or post-translational modifications) based on their abundance profiles using an analysis of variance model, while accounting for sample pairing and batch in the model. The p values were subsequently corrected with a Benjamini-Hochberg multiple comparison adjustment50 within each comparison to account for the multiple tests being performed. Participant gender and age were incorporated as potential covariates to account for any effects not removed by pairing. After p value correction for multiple testing, no significant evidence of either factor was found. Machine learning was also performed to identify possible validation candidate proteins that did not meet the p value threshold but were predictive of outcome in a multi-variate model. This was done using R and consisted of data imputation with Random Forest,51 risk association via Probabilistic Conditional Logistic Regression integrated with least absolute shrinkage and selection operator (LASSO) for feature selection (clogitLasso).52
Machine learning analysis to identify early biomarker panels predictive of disease onset
Validation phase data was filtered to remove 3 peptides observed in less than 50% of samples for at least one of the three time points prior to seroconversion. Remaining missing values were imputed with Random Forest51 imputation. A pairing correction53 was applied to the data to account for the case-control study design. Logistic regression with a LASSO penalization function was fit to the data with case/control status as the explanatory variable. The machine learning model was fit separately to each time point’s data using 4-fold cross-validation repeated for 100 bootstrap iterations.
Function-enrichment analysis
Differentially abundant proteins were filtered for function-enrichment analysis using DAVID,54 and only pathways containing KEGG annotation were used. The biological interpretations were only performed after the targeted proteomics data analysis were completed to avoid unconscious bias in sample and data analysis.
Acknowledgments
The authors thank Mr. Nathan Johnson and Mrs. Shannon Colson of PNNL for their help with illustrations and Drs. Jennifer Van Eyk (Cedars-Sinai Medical Center) and Michael McCoss (University of Washington) for insightful discussion. Part of the work was performed at the Environmental Molecular Sciences Laboratory, a US DOE national scientific user facility at the Pacific Northwest National Laboratory (PNNL) in Richland, WA (USA). Battelle operates PNNL for the DOE under contract DE-AC05-76RLO01830. The TEDDY Study is funded by U01 DK63829, U01 DK63861, U01 DK63821, U01 DK63865, U01 DK63863, U01 DK63836, U01 DK63790, UC4 DK63829, UC4 DK63861, UC4 DK63821, UC4 DK63865, UC4 DK63863, UC4 DK63836, UC4 DK95300, UC4 DK100238, UC4 DK106955, UC4 DK112243, UC4 DK117483, U01 DK124166, U01 DK128847, and contract no. HHSN267200700014C from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK); the National Institute of Allergy and Infectious Diseases (NIAID); the Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD); the National Institute of Environmental Health Sciences (NIEHS); the Centers for Disease Control and Prevention (CDC); and JDRF. This work is supported in part by the NIH/NCATS Clinical and Translational Science Awards to the University of Florida (UL1 TR000064) and the University of Colorado (UL1 TR002535). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. T.O.M., B.-J.M.W.-R., E.S.N., and W.-J.Q. were also supported by NIDDK projects U01 DK127505, U01 DK127786, and U01 DK124020. Proteomics measurements were obtained using capabilities developed partially under National Institutes of Health grant P41GM103493.
Author contributions
E.S.N., C.A., M.A.G., A.A.S., L.M.B., P.D.P., W.-J.Q., B.I.F., S.S., J.T., A.-G.Z., A.L., W.H., B.A., R.D.S., B.-J.M.W.-R., M.J.R., T.O.M., and the TEDDY Study Group (TSG) conceived the study and participated in the study design. The TSG collected the samples. M.A.G., A.A.S., T.R.C., P.D.P., T.L.F., D.J.O., and R.J.M. prepared the samples and performed the liquid chromatography-mass spectrometry (LC-MS) analyses. E.S.N., C.A., L.M.B., P.D.P., Y.G., B.A.S., D.J.O., W.J.Q., B.I.F., R.D.S., B.-J.M.W.-R., M.J.R., T.O.M., and T.S.G. performed the data analysis. E.S.N., C.A., M.A.G., A.A.S., L.M.B., P.D.P., D.W.E., T.L.F., W.-J.Q., R.D.S., B.-J.M.W.-R., M.J.R., T.O.M., and the TSG developed the targeted MS assays for validating the biomarker candidates. E.S.N., L.M.B., A.A.S., P.D.P., T.R.C., B.-J.M.W.-R., and T.O.M. wrote the manuscript. All authors read, revised, and approved the final manuscript for publication.
Declaration of interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: June 29, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101093.
Supplemental information
Data availability
For data availability, please reference the TEDDY data summary: https://teddy.epi.usf.edu/research/. The mass spectrometry raw data files were deposited into MassIVE (https://massive.ucsd.edu/) under accession numbers MSV000091560 (untargeted proteomics) and MSV000091562 (targeted proteomics).
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
References
- 1.Livingstone S.J., Levin D., Looker H.C., Lindsay R.S., Wild S.H., Joss N., Leese G., Leslie P., McCrimmon R.J., Metcalfe W., et al. Estimated life expectancy in a Scottish cohort with type 1 diabetes, 2008-2010. JAMA. 2015;313:37–44. doi: 10.1001/jama.2014.16425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkinson M.A., Eisenbarth G.S., Michels A.W. Type 1 diabetes. Lancet. 2014;383:69–82. doi: 10.1016/S0140-6736(13)60591-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.DiMeglio L.A., Evans-Molina C., Oram R.A. Type 1 diabetes. Lancet. 2018;391:2449–2462. doi: 10.1016/S0140-6736(18)31320-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee H.S., Burkhardt B.R., McLeod W., Smith S., Eberhard C., Lynch K., Hadley D., Rewers M., Simell O., She J.X., et al. Biomarker discovery study design for type 1 diabetes in the Environmental Determinants of Diabetes in the Young (TEDDY) study. Diabetes. Metab. Res. Rev. 2014;30:424–434. doi: 10.1002/dmrr.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keshishian H., Burgess M.W., Specht H., Wallace L., Clauser K.R., Gillette M.A., Carr S.A. Quantitative, multiplexed workflow for deep analysis of human blood plasma and biomarker discovery by mass spectrometry. Nat. Protoc. 2017;12:1683–1701. doi: 10.1038/nprot.2017.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geyer P.E., Kulak N.A., Pichler G., Holdt L.M., Teupser D., Mann M. Plasma proteome profiling to assess human Health and disease. Cell Syst. 2016;2:185–195. doi: 10.1016/j.cels.2016.02.015. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y., Buil A., Collins B.C., Gillet L.C.J., Blum L.C., Cheng L.Y., Vitek O., Mouritsen J., Lachance G., Spector T.D., et al. Quantitative variability of 342 plasma proteins in a human twin population. Mol. Syst. Biol. 2015;11:786. doi: 10.15252/msb.20145728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Q., Fillmore T.L., Schepmoes A.A., Clauss T.R.W., Gritsenko M.A., Mueller P.W., Rewers M., Atkinson M.A., Smith R.D., Metz T.O. Serum proteomics reveals systemic dysregulation of innate immunity in type 1 diabetes. J. Exp. Med. 2013;210:191–203. doi: 10.1084/jem.20111843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhi W., Sharma A., Purohit S., Miller E., Bode B., Anderson S.W., Reed J.C., Steed R.D., Steed L., Hopkins D., She J.X. Discovery and validation of serum protein changes in type 1 diabetes patients using high throughput two dimensional liquid chromatography-mass spectrometry and immunoassays. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M111.012203. M111.012203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.von Toerne C., Laimighofer M., Achenbach P., Beyerlein A., de Las Heras Gala T., Krumsiek J., Theis F.J., Ziegler A.G., Hauck S.M. Peptide serum markers in islet autoantibody-positive children. Diabetologia. 2017;60:287–295. doi: 10.1007/s00125-016-4150-x. [DOI] [PubMed] [Google Scholar]
- 11.Moulder R., Bhosale S.D., Erkkilä T., Laajala E., Salmi J., Nguyen E.V., Kallionpää H., Mykkänen J., Vähä-Mäkilä M., Hyöty H., et al. Serum proteomes distinguish children developing type 1 diabetes in a cohort with HLA-conferred susceptibility. Diabetes. 2015;64:2265–2278. doi: 10.2337/db14-0983. [DOI] [PubMed] [Google Scholar]
- 12.MacLean E., Broger T., Yerlikaya S., Fernandez-Carballo B.L., Pai M., Denkinger C.M. A systematic review of biomarkers to detect active tuberculosis. Nat. Microbiol. 2019;4:748–758. doi: 10.1038/s41564-019-0380-2. [DOI] [PubMed] [Google Scholar]
- 13.Nakayasu E.S., Gritsenko M., Piehowski P.D., Gao Y., Orton D.J., Schepmoes A.A., Fillmore T.L., Frohnert B.I., Rewers M., Krischer J.P., et al. Tutorial: best practices and considerations for mass-spectrometry-based protein biomarker discovery and validation. Nat. Protoc. 2021;16:3737–3760. doi: 10.1038/s41596-021-00566-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shi T., Fillmore T.L., Sun X., Zhao R., Schepmoes A.A., Hossain M., Xie F., Wu S., Kim J.S., Jones N., et al. Antibody-free, targeted mass-spectrometric approach for quantification of proteins at low picogram per milliliter levels in human plasma/serum. Proc. Natl. Acad. Sci. USA. 2012;109:15395–15400. doi: 10.1073/pnas.1204366109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stanfill B.A., Nakayasu E.S., Bramer L.M., Thompson A.M., Ansong C.K., Clauss T.R., Gritsenko M.A., Monroe M.E., Moore R.J., Orton D.J., et al. QC-ART: a tool for real-time quality control assessment of mass spectrometry-based proteomics data. Mol. Cell. Proteomics. 2018;17:1824–1836. doi: 10.1074/mcp.RA118.000648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nakayasu E.S., Syed F., Tersey S.A., Gritsenko M.A., Mitchell H.D., Chan C.Y., Dirice E., Turatsinze J.V., Cui Y., Kulkarni R.N., et al. Comprehensive proteomics analysis of stressed human islets identifies GDF15 as a target for type 1 diabetes intervention. Cell Metab. 2020;31:363–374.e6. doi: 10.1016/j.cmet.2019.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramos-Rodríguez M., Raurell-Vila H., Colli M.L., Alvelos M.I., Subirana-Granés M., Juan-Mateu J., Norris R., Turatsinze J.V., Nakayasu E.S., Webb-Robertson B.J.M., et al. The impact of proinflammatory cytokines on the beta-cell regulatory landscape provides insights into the genetics of type 1 diabetes. Nat. Genet. 2019;51:1588–1595. doi: 10.1038/s41588-019-0524-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gibbons B.C., Fillmore T.L., Gao Y., Moore R.J., Liu T., Nakayasu E.S., Metz T.O., Payne S.H. Rapidly assessing the quality of targeted proteomics experiments through monitoring stable-isotope labeled standards. J. Proteome Res. 2019;18:694–699. doi: 10.1021/acs.jproteome.8b00688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcelli-Barge A., Poirier J.C., Chantome R., Deschamps I., Hors J., Colombani J. Marked shortage of C4B DNA polymorphism among insulin-dependent diabetic patients. Res. Immunol. 1990;141:117–128. doi: 10.1016/0923-2494(90)90131-h. [DOI] [PubMed] [Google Scholar]
- 20.Törn C., Liu X., Hagopian W., Lernmark Å., Simell O., Rewers M., Ziegler A.G., Schatz D., Akolkar B., Onengut-Gumuscu S., et al. Complement gene variants in relation to autoantibodies to beta cell specific antigens and type 1 diabetes in the TEDDY Study. Sci. Rep. 2016;6 doi: 10.1038/srep27887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rowe P., Wasserfall C., Croker B., Campbell-Thompson M., Pugliese A., Atkinson M., Schatz D. Increased complement activation in human type 1 diabetes pancreata. Diabetes Care. 2013;36:3815–3817. doi: 10.2337/dc13-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Targher G., Chonchol M., Zoppini G., Franchini M. Hemostatic disorders in type 1 diabetes mellitus. Semin. Thromb. Hemost. 2011;37:58–65. doi: 10.1055/s-0030-1270072. [DOI] [PubMed] [Google Scholar]
- 23.Nejentsev S., Howson J.M.M., Walker N.M., Szeszko J., Field S.F., Stevens H.E., Reynolds P., Hardy M., King E., Masters J., et al. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature. 2007;450:887–892. doi: 10.1038/nature06406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Lummel M., van Veelen P.A., de Ru A.H., Pool J., Nikolic T., Laban S., Joosten A., Drijfhout J.W., Gómez-Touriño I., Arif S., et al. Discovery of a selective islet peptidome presented by the highest-risk HLA-DQ8trans molecule. Diabetes. 2016;65:732–741. doi: 10.2337/db15-1031. [DOI] [PubMed] [Google Scholar]
- 25.Eizirik D.L., Colli M.L., Ortis F. The role of inflammation in insulitis and beta-cell loss in type 1 diabetes. Nat. Rev. Endocrinol. 2009;5:219–226. doi: 10.1038/nrendo.2009.21. [DOI] [PubMed] [Google Scholar]
- 26.Webb-Robertson B.J.M., Bramer L.M., Stanfill B.A., Reehl S.M., Nakayasu E.S., Metz T.O., Frohnert B.I., Norris J.M., Johnson R.K., Rich S.S., Rewers M.J. Prediction of the development of islet autoantibodies through integration of environmental, genetic, and metabolic markers. J. Diabetes. 2021;13:143–153. doi: 10.1111/1753-0407.13093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Webb-Robertson B.J.M., Nakayasu E.S., Frohnert B.I., Bramer L.M., Akers S.M., Norris J.M., Vehik K., Ziegler A.G., Metz T.O., Rich S.S., Rewers M.J. Integration of infant metabolite, genetic and islet autoimmunity signatures to predict type 1 diabetes by 6 Years of age. J. Clin. Endocrinol. Metab. 2022;107:2329–2338. doi: 10.1210/clinem/dgac225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sims E.K., Chaudhry Z., Watkins R., Syed F., Blum J., Ouyang F., Perkins S.M., Mirmira R.G., Sosenko J., DiMeglio L.A., Evans-Molina C. Elevations in the fasting serum proinsulin-to-C-peptide ratio precede the onset of type 1 diabetes. Diabetes Care. 2016;39:1519–1526. doi: 10.2337/dc15-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamichhane S., Sen P., Dickens A.M., Alves M.A., Härkönen T., Honkanen J., Vatanen T., Xavier R.J., Hyötyläinen T., Knip M., Orešič M. Dysregulation of secondary bile acid metabolism precedes islet autoimmunity and type 1 diabetes. Cell Rep. Med. 2022;3 doi: 10.1016/j.xcrm.2022.100762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamichhane S., Ahonen L., Dyrlund T.S., Kemppainen E., Siljander H., Hyöty H., Ilonen J., Toppari J., Veijola R., Hyötyläinen T., et al. Dynamics of plasma lipidome in progression to islet autoimmunity and type 1 diabetes - type 1 diabetes prediction and prevention study (DIPP) Sci. Rep. 2018;8 doi: 10.1038/s41598-018-28907-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Valladolid-Acebes I., Berggren P.O., Juntti-Berggren L. Apolipoprotein CIII is an important piece in the type-1 diabetes jigsaw puzzle. Int. J. Mol. Sci. 2021;22 doi: 10.3390/ijms22020932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morillas P., Quiles J., de Andrade H., Castillo J., Tarazón E., Roselló E., Portolés M., Rivera M., Bertomeu-Martínez V. Circulating biomarkers of collagen metabolism in arterial hypertension: relevance of target organ damage. J. Hypertens. 2013;31:1611–1617. doi: 10.1097/HJH.0b013e3283614c1c. [DOI] [PubMed] [Google Scholar]
- 33.Medina C.O., Nagy N., Bollyky P.L. Extracellular matrix and the maintenance and loss of peripheral immune tolerance in autoimmune insulitis. Curr. Opin. Immunol. 2018;55:22–30. doi: 10.1016/j.coi.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu G., Rausell-Palamos F., Zhang J., Zheng Z., Zhang T., Valle S., Rosselot C., Berrouet C., Conde P., Spindler M.P., et al. Dextran sulfate protects pancreatic beta-cells, reduces autoimmunity, and ameliorates type 1 diabetes. Diabetes. 2020;69:1692–1707. doi: 10.2337/db19-0725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mathieu C., Lahesmaa R., Bonifacio E., Achenbach P., Tree T. Immunological biomarkers for the development and progression of type 1 diabetes. Diabetologia. 2018;61:2252–2258. doi: 10.1007/s00125-018-4726-8. [DOI] [PubMed] [Google Scholar]
- 36.Ahmed S., Cerosaletti K., James E., Long S.A., Mannering S., Speake C., Nakayama M., Tree T., Roep B.O., Herold K.C., Brusko T.M. Standardizing T-cell biomarkers in type 1 diabetes: challenges and recent advances. Diabetes. 2019;68:1366–1379. doi: 10.2337/db19-0119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Frohnert B.I., Webb-Robertson B.J., Bramer L.M., Reehl S.M., Waugh K., Steck A.K., Norris J.M., Rewers M. Predictive modeling of type 1 diabetes stages using disparate data sources. Diabetes. 2020;69:238–248. doi: 10.2337/db18-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Webb-Robertson B.J.M., Nakayasu E.S., Frohnert B.I., Bramer L.M., Akers S.M., Norris J.M., Vehik K., Ziegler A.G., Metz T.O., Rich S.S., Rewers M.J. Integration of infant metabolite, genetic, and islet autoimmunity signatures to predict type 1 diabetes by age 6 years. J. Clin. Endocrinol. Metab. 2022;107:2329–2338. doi: 10.1210/clinem/dgac225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vehik K., Bonifacio E., Lernmark Å., Yu L., Williams A., Schatz D., Rewers M., She J.X., Toppari J., Hagopian W., et al. Hierarchical order of distinct autoantibody spreading and progression to type 1 diabetes in the TEDDY study. Diabetes Care. 2020;43:2066–2073. doi: 10.2337/dc19-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vehik K., Cuthbertson D., Boulware D., Beam C.A., Rodriguez H., Legault L., Hyytinen M., Rewers M.J., Schatz D.A., Krischer J.P., et al. Performance of HbA1c as an early diagnostic indicator of type 1 diabetes in children and youth. Diabetes Care. 2012;35:1821–1825. doi: 10.2337/dc12-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Piehowski P.D., Petyuk V.A., Orton D.J., Xie F., Moore R.J., Ramirez-Restrepo M., Engel A., Lieberman A.P., Albin R.L., Camp D.G., et al. Sources of technical variability in quantitative LC-MS proteomics: human brain tissue sample analysis. J. Proteome Res. 2013;12:2128–2137. doi: 10.1021/pr301146m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gritsenko M.A., Xu Z., Liu T., Smith R.D. Large-Scale and deep quantitative proteome profiling using isobaric labeling coupled with two-dimensional LC-MS/MS. Methods Mol. Biol. 2016;1410:237–247. doi: 10.1007/978-1-4939-3524-6_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mayampurath A.M., Jaitly N., Purvine S.O., Monroe M.E., Auberry K.J., Adkins J.N., Smith R.D. DeconMSn: a software tool for accurate parent ion monoisotopic mass determination for tandem mass spectra. Bioinformatics. 2008;24:1021–1023. doi: 10.1093/bioinformatics/btn063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Petyuk V.A., Mayampurath A.M., Monroe M.E., Polpitiya A.D., Purvine S.O., Anderson G.A., Camp D.G., 2nd, Smith R.D. DtaRefinery, a software tool for elimination of systematic errors from parent ion mass measurements in tandem mass spectra data sets. Mol. Cell. Proteomics. 2010;9:486–496. doi: 10.1074/mcp.M900217-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim S., Pevzner P.A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Monroe M.E., Shaw J.L., Daly D.S., Adkins J.N., Smith R.D. MASIC: a software program for fast quantitation and flexible visualization of chromatographic profiles from detected LC-MS(/MS) features. Comput. Biol. Chem. 2008;32:215–217. doi: 10.1016/j.compbiolchem.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Matzke M.M., Waters K.M., Metz T.O., Jacobs J.M., Sims A.C., Baric R.S., Pounds J.G., Webb-Robertson B.J.M. Improved quality control processing of peptide-centric LC-MS proteomics data. Bioinformatics. 2011;27:2866–2872. doi: 10.1093/bioinformatics/btr479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Webb-Robertson B.J.M., Matzke M.M., Datta S., Payne S.H., Kang J., Bramer L.M., Nicora C.D., Shukla A.K., Metz T.O., Rodland K.D., et al. Bayesian proteoform modeling improves protein quantification of global proteomic measurements. Mol. Cell. Proteomics. 2014;13:3639–3646. doi: 10.1074/mcp.M113.030932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Matzke M.M., Brown J.N., Gritsenko M.A., Metz T.O., Pounds J.G., Rodland K.D., Shukla A.K., Smith R.D., Waters K.M., McDermott J.E., Webb-Robertson B.J. A comparative analysis of computational approaches to relative protein quantification using peptide peak intensities in label-free LC-MS proteomics experiments. Proteomics. 2013;13:493–503. doi: 10.1002/pmic.201200269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 51.Liaw A., Wiener M. Classification and regression by random forest. R. News. 2002;2:18–22. [Google Scholar]
- 52.Avalos M., Pouyes H., Grandvalet Y., Orriols L., Lagarde E. Sparse conditional logistic regression for analyzing large-scale matched data from epidemiological studies: a simple algorithm. BMC Bioinf. 2015;16(Suppl 6):S1. doi: 10.1186/1471-2105-16-S6-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stanfill B., Reehl S., Bramer L., Nakayasu E.S., Rich S.S., Metz T.O., Rewers M., Webb-Robertson B.J., TEDDY Study Group Extending classification algorithms to case-control studies. Biomed. Eng. Comput. Biol. 2019;10 doi: 10.1177/1179597219858954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang D.W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
For data availability, please reference the TEDDY data summary: https://teddy.epi.usf.edu/research/. The mass spectrometry raw data files were deposited into MassIVE (https://massive.ucsd.edu/) under accession numbers MSV000091560 (untargeted proteomics) and MSV000091562 (targeted proteomics).
-
•
Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.