Summary
Direct diagnosis and accurate assessment of metabolic syndrome (MetS) allow for prompt clinical interventions. However, traditional diagnostic strategies overlook the complex heterogeneity of MetS. Here, we perform metabolomic analysis in 13,554 participants from the natural cohort and identify 26 hub plasma metabolic fingerprints (PMFs) associated with MetS and its early identification (pre-MetS). By leveraging machine-learning algorithms, we develop robust diagnostic models for pre-MetS and MetS with convincing performance through independent validation. We utilize these PMFs to assess the relative contributions of the four major MetS risk factors in the general population, ranked as follows: hyperglycemia, hypertension, dyslipidemia, and obesity. Furthermore, we devise a personalized three-dimensional plasma metabolic risk (PMR) stratification, revealing three distinct risk patterns. In summary, our study offers effective screening tools for identifying pre-MetS and MetS patients in the general community, while defining the heterogeneous risk stratification of metabolic phenotypes in real-world settings.
Keywords: metabolic syndrome, plasma metabolic fingerprint, LDI-MS, machine learning, risk stratification
Graphical abstract
Highlights
-
•
Advanced ferric particle-assisted LDI-MS enables plasma metabolomic analysis of MetS
-
•
Plasma metabolic fingerprints (PMFs) highlight the diagnosis of MetS and pre-MetS
-
•
PMFs prompt the assessment of the four significant risk factors contributing to MetS
-
•
Personalized three-dimensional stratification reveals three metabolic risk patterns
Chen et al. develop diagnostic models that are robust and can be used for the personalized three-dimensional plasma metabolic risk (PMR) stratification of metabolic syndrome (MetS) as well as its early identification (pre-MetS) via plasma metabolic fingerprints. The results show that there are heterogeneous risk patterns in metabolic phenotypes.
Introduction
The Centers for Disease Control and Prevention estimates that the prevalence of metabolic syndrome (MetS) is 34.7% in the United States1 and 33.9% in China,2 with increasing severity in recent decades and an annual growth rate of >8% among subjects >60 years old due to increasing rates of diabetes and obesity.1,3 MetS is recognized as a progressive chronic pathophysiological state that significantly increases the risks of cardiovascular (odds ratio [OR], 1.80) and cerebrovascular events (OR, 2.05),4 as well as liver (relative risk [RR], 1.43)5 and colorectal cancers (RR, 1.25).6 MetS is diagnosed primarily by the coincident identification of three or more of the following known risk factors, including obesity, hypertension, hyperglycemia, and dyslipidemia.7,8,9 As an early sign of MetS, one or two risk factors for MetS are required for it to qualify as a pre-MetS (pre-MS). Although there are at least five different international MetS guidelines,10,11,12,13,14 none of these definitions have a decisive advantage in predicting cardiovascular events,15 and the different guidelines showed a discordance of 20%–41% in the same population.16,17 Thus, there is a need to develop an effective and useful screening platform to promote individualized strategies for the management of MetS.18,19
A blood test serves as a common diagnostic method for detecting molecules inside the body. Notably, the comprehensive profiling of small-molecule metabolites, termed the plasma metabolome,20 provides predictive biomarkers for MetS and a valuable source for understanding the underlying pathophysiologic mechanisms.21,22 Given the increasing concern about the burden of MetS and the healthcare costs in an aging population, the development of rapid and portable community-based diagnostic screening will provide a powerful window for therapeutic intervention.
Recent studies have promoted the role of metabolomics research in the revolutionary progress of discovering biomarkers for MetS prediction.23 For example, a study of 58 donors confirmed that 36 metabolites were closely associated with MetS using traditional 1H nuclear magnetic resonance (NMR).24 Another study of 311 subjects found that plasma 5-hydroxyindole-3-acetic acid concentrations were higher in MetS donors than in those without MetS using liquid chromatography-mass spectrometry (LC-MS).25 In addition, a study of 163 participants found that 2-hydroxybutyric acid, inositol, and D-glucose served as potential biomarkers for MetS using gas chromatography-mass spectrometry (GC-MS).26 However, these traditional spectrometry methods require strict pretreatment procedures to separate and enrich the metabolites, limiting their use for large-scale screening (over 10,000 samples) in general population-based cohorts.27 Despite the importance of these findings, these methods may not precisely reflect the real-world state of MetS, with several potentially important signaling molecules being missed. Recent technological advances have introduced the use of nanoparticle-assisted laser desorption/ionization mass spectrometry (LDI-MS) as a high-throughput screening (≈300 samples/h) tool for accurate metabolic analysis (mass error <50 ppm) with easier sample preparation, thus allowing the identification of specific biomarkers of MetS to reflect the status of the patients. This approach detects MetS metabolites in ultralow volumes without enrichment or purification and produces results after a short processing time.
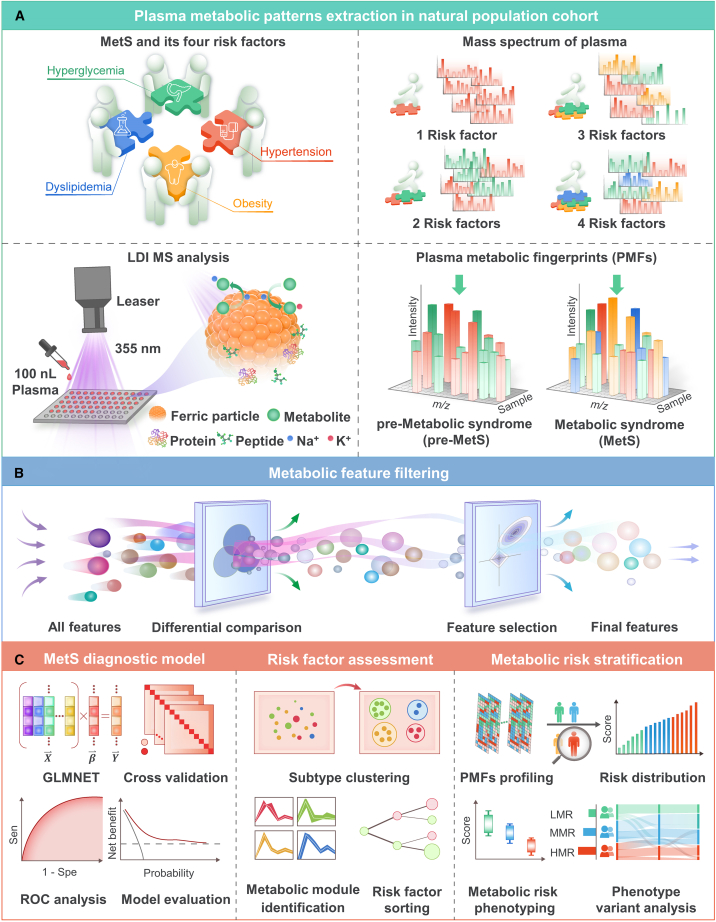
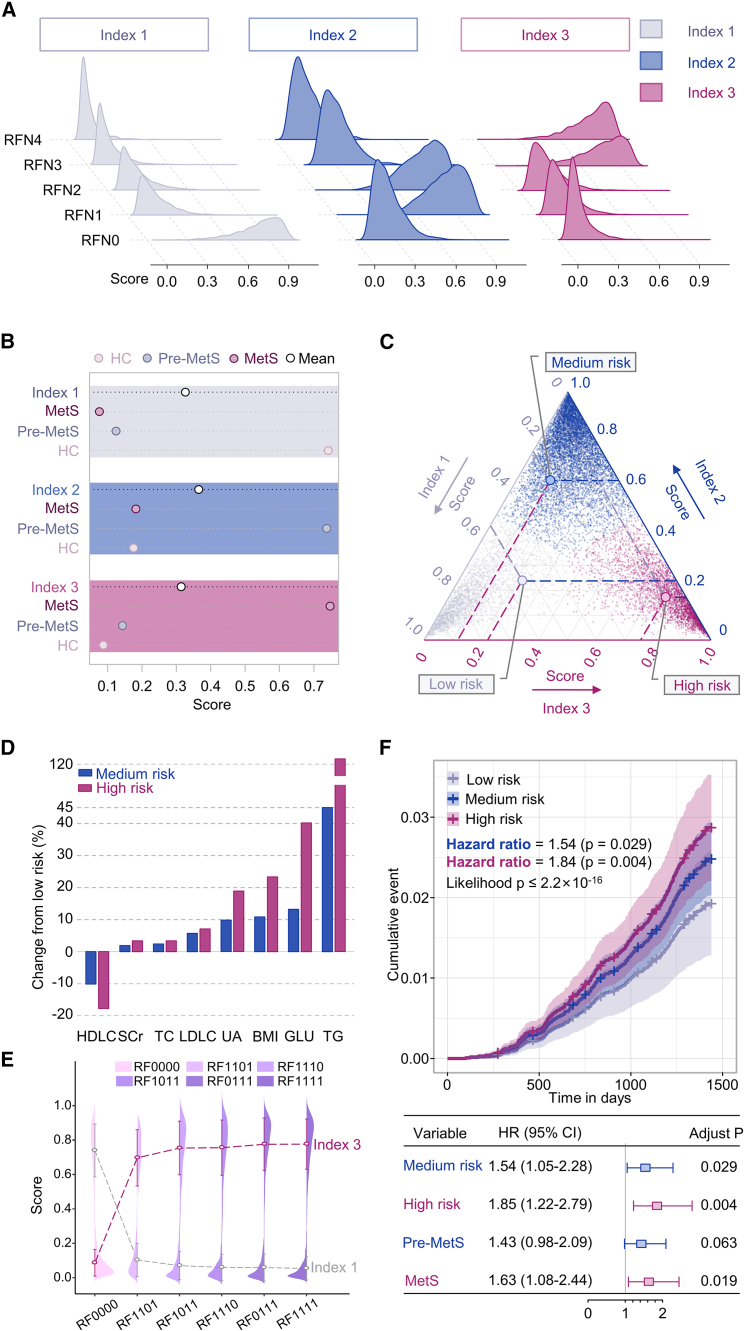
Figure 1 illustrates how the overall experiment was conducted. We obtained plasma metabolic fingerprints (PMFs), including low-molecular-weight metabolites (100–300 Da), from 13,554 participants, including healthy volunteers, pre-MetS patients (having one or two MetS risk factors), and MetS patients (having at least three MetS risk factors). Through combined LDI-MS detection, we filtered out 26 hub PMFs to develop precise and efficient machine-learning (ML)-based diagnostic platforms for large-scale screening of pre-MetS and MetS, with areas under curves (AUCs) of 0.867 (95% confidence interval [CI], 0.854–0.881) and 0.864 (95% CI, 0.843–0.885) for healthy control (HC) vs. pre-MetS, 0.849 (95% CI, 0.838–0.860) and 0.835 (95% CI, 0.817–0.853) for pre-MetS vs. MetS, and 0.891 (95% CI, 0.880–0.902) and 0.886 (95% CI, 0.868–0.903) for HC vs. MetS in the discovery and validation sets, respectively. We reclassified the 16 subclusters (RF0000–RF1111) into four metabolic phenotypes (MPs) with 26 hub PMFs. By evaluating the contribution of each risk factor to MetS, we highlighted the importance of glycemic control for prevention and intervention of MetS. Finally, we constructed a three-dimensional plasma metabolic risk (PMR) stratification adjusted for age and gender to classify metabolic heterogeneity into three risk patterns. Consistent with the change in conventional clinical parameters and the cumulative all-cause death events by 4-year follow-up adjusted by gender and age (for medium risk, hazard ratio [HR] 1.54, 95% CI 1.05–2.28, p for log-rank test = 0.029. For high risk, HR 1.85, 95% CI 1.22–2.79, p for log-rank test = 0.004), this PMR assessment provided an alternative approach to reflect individual actual metabolic status.
Figure 1.
Overall schematics for evaluating MetS among the general aging population based on the combination of PMFs and ML
(A) Schematic workflow for the extraction of PMFs using ferric particle-assisted laser desorption/ionization mass spectrometry (LDI-MS). One hundred nanoliters of native plasma was used for direct analysis without pretreatment procedures. Only Na+- and K+-adducted metabolites can be selectively detected with the coexistence of high concentrations of peptides and proteins.
(B) Feature filtering for hub PMFs was carried out according to difference comparisons among the healthy control (HC), pre-MetS, and MetS groups.
(C) PMF-based diagnostic models were constructed for HC vs. pre-MetS, pre-MetS vs. MetS, and HC vs. MetS. Traditional risk factors were assessed by unsupervised clustering of various subtypes of MetS. Finally, PMF-based metabolic risk stratification was computed to define three metabolic risk patterns: low metabolic risk (LMR), moderate metabolic risk (MMR), and high metabolic risk (HMR).
Results
Comprehensive clinical and metabolic characterization of MetS
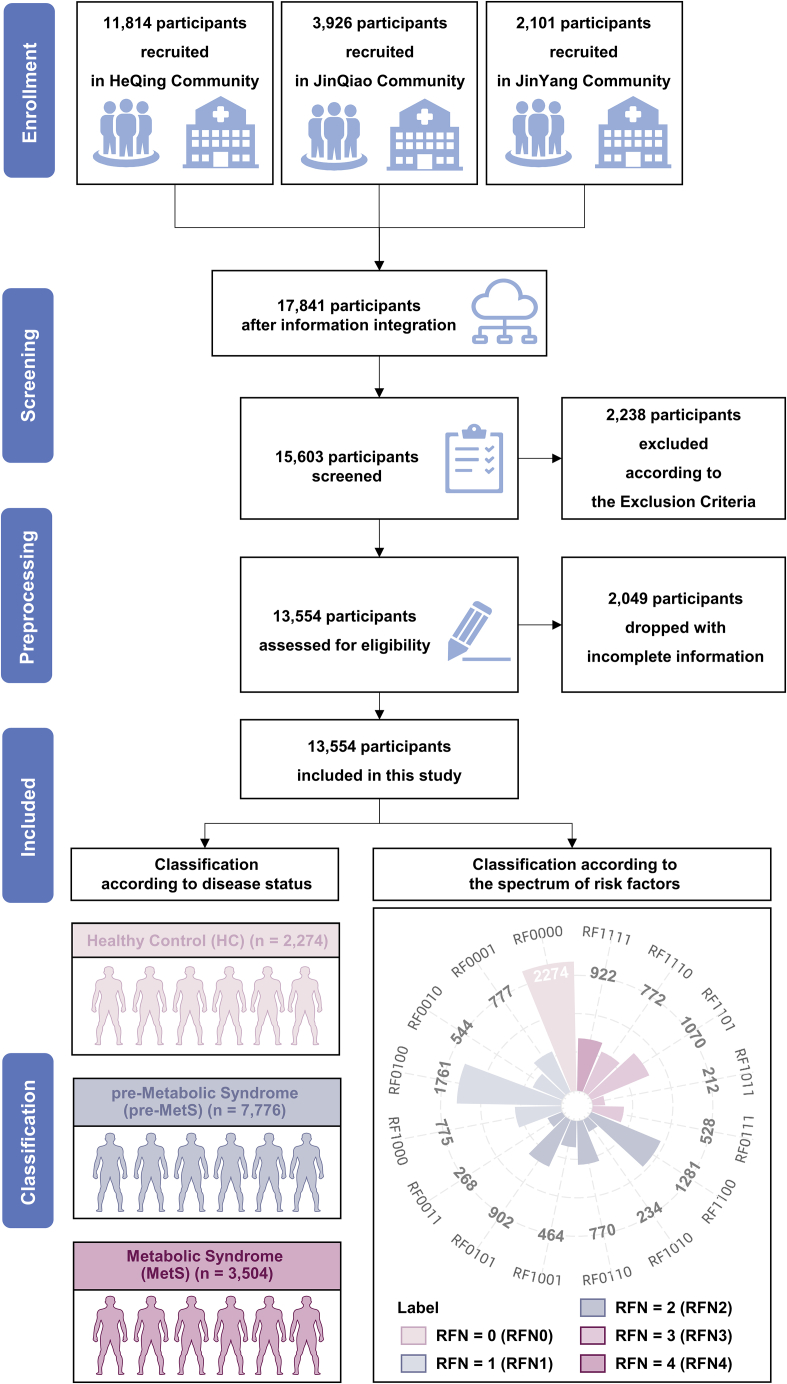
There were 17,841 participants enrolled in the prospective Shanghai Community Cohort Establishment and Follow-up (NCT04517513) from January to March 2019. After filtering out participants (n = 4,287) according to the flow chart (Figure 2 and STAR Methods), a total of 13,554 (76%) participants (mean age ± standard deviation 67.84 ± 5.90 years; 53.0% females) were finally eligible for inclusion in the cohort study. In the study, we also adopted the statement of the Chinese Diabetes Society 2004 (Table S1)28,29 to better reflect the Chinese population. To identify potential biomarkers for MetS and enable more granular analysis of the relationship between cardiovascular risk factors and the severity of the syndrome, the current study divided the participants into three groups: a MetS group (with at least three MetS risk factors), a pre-MetS group (with one or two MetS risk factors), and an HC group, including 3,504 (25.9%; age 68.29 ± 5.66 years; 52.9% females), 7,776 (57.4%; age 67.88 ± 5.99 years; 53.0% females), and 2,274 participants (16.8%; age 67.01 ± 5.85 years; 53.2% females), respectively (Figure 2). By this classification, we could better understand the nuanced differences among HC, pre-MetS, and MetS. Table 1 summarizes the demographic and clinical characteristics of the participants. The gender distribution was not significantly different among the three groups. Demographically, there was a stepwise increase across the groups in the proportions of obesity, hypertension, hyperglycemia, and dyslipidemia (p < 0.001) (Table 1). Based on the number of MetS risk factors, we further investigated the heterogeneity of MetS development by dividing the participants into five subgroups (RFN0–RFN4), including RFN0 (no risk factors), RFN1(one risk factor), RFN2 (two risk factors), RFN3 (three risk factors), and RFN4 (four risk factors) (Figure 2). Moreover, we classified the cohort into 16 subclusters (RF0000–RF1111) according to the permutations and combinations of the four MetS risk factors, each binary coded by "l" or "0" to indicate their presence or absence in the conditions (Figure 2 and Table 2).
Figure 2.
Overview flow chart of the study design and enrollment
The complete study design consisted of five steps, including enrollment, screening, preprocessing, inclusion, and classification. For enrollment, 11,814, 3,926, and 2,101 individuals were initially recruited in the HeQing, JinQiao, and JinYang communities based on the prospective Shanghai Community Cohort Establishment and Follow-up (NCT04517513). For screening, 2,238 participants were excluded according to the exclusion criteria (see STAR Methods). For preprocessing, 2,049 participants with incomplete information were excluded. Finally, 13,554 participants were included in this study. Participants were classified into the HC (n = 2,274; no use of lipid-lowering, antidiabetic, or antihypertensive drugs), pre-MetS (n = 7,776; clustering of <3 traditional risk factors, including obesity, hypertension, dyslipidemia, and dyslipidemia), and MetS (n = 3,504; clustering of ≥3 traditional risk factors) groups according to the statement of the Chinese Diabetes Society 2004. Participants were also classified into five subgroups (RFN1–5) according to the present number of four MetS risk factors or into 16 subclusters (RF0000–RF111) considering a combination of risk factors (0/1 = absence/presence of each risk factor). For more details, see Table 2. RFN indicates the number of traditional MetS risk factors.
Table 1.
Baseline characteristics of the analyzed participants in the general population
| HC | pre-MetS | MetS | Overall | p valuea | |
|---|---|---|---|---|---|
| Number (%) | 2,274 (16.8) | 7,776 (57.4) | 3,504 (25.9) | 13,554 (100.0) | <0.001 |
| Obesity (%) | 0 (0.0) | 2,754 (35.4) | 2,976 (84.9) | 5,730 (42.3) | <0.001 |
| Hypertension (%) | 0 (0.0) | 4,714 (60.6) | 3,292 (93.9) | 8,006 (59.1) | <0.001 |
| Hyperglycemia (%) | 0 (0.0) | 1,816 (23.4) | 2,434 (69.5) | 4,250 (31.4) | <0.001 |
| Dyslipidemia (%) | 0 (0.0) | 2,411 (31.0) | 2,732 (78.0) | 5,143 (37.9) | <0.001 |
| Male (%) | 1,064 (46.8) | 3,651 (47.0) | 1,649 (47.1) | 6,364 (47.0) | 0.98 |
| Age, mean (SD) | 67.01 (5.85) | 67.88 (5.99) | 68.29 (5.66) | 67.84 (5.90) | <0.001 |
| BMI, mean (SD) | 21.69 (2.11) | 24.26 (2.98) | 27.15 (2.86) | 24.58 (3.34) | <0.001 |
| TC, mean (SD) | 4.77 (0.72) | 4.92 (0.95) | 4.96 (1.05) | 4.91 (0.94) | <0.001 |
| GLU, mean (SD) | 5.10 (0.52) | 5.78 (1.59) | 7.16 (2.28) | 6.02 (1.83) | <0.001 |
| HDLC, mean (SD) | 1.56 (0.38) | 1.40 (0.36) | 1.27 (0.38) | 1.39 (0.38) | <0.001 |
| LDLC, mean (SD) | 2.75 (0.63) | 2.93 (0.82) | 2.97 (0.87) | 2.91 (0.81) | <0.001 |
| TGs, mean (SD) | 1.02 (0.31) | 1.58 (1.04) | 2.44 (1.62) | 1.71 (1.24) | <0.001 |
| UA, mean (SD) | 302.44 (70.21) | 333.46 (83.11) | 362.62 (86.86) | 335.80 (84.35) | <0.001 |
HC, healthy control; pre-MetS, pre-metabolic syndrome; MetS, metabolic syndrome; SD, standard deviation; BMI, body mass index; TC, serum total cholesterol; GLU, glucose; HDLC, high-density lipoprotein cholesterol; LDLC, low-density lipoprotein cholesterol; TGs, triglycerides; UA, uric acid.
p values calculated by χ2 tests for categorical data and one-way analysis of variance for parametric data.
Table 2.
Distribution and prevalence of 16 subcluster conditions in this study
| Obesity | Hypertension | Hyperglycemia | Dyslipidemia | Number (%)a | |
|---|---|---|---|---|---|
| RF0000 | 0 | 0 | 0 | 0 | 2,274 (16.78) |
| RF0001 | 0 | 0 | 0 | 1 | 777 (5.73) |
| RF0010 | 0 | 0 | 1 | 0 | 544 (4.01) |
| RF0100 | 0 | 1 | 0 | 0 | 1,761 (12.99) |
| RF1000 | 1 | 0 | 0 | 0 | 775 (5.72) |
| RF0011 | 0 | 0 | 1 | 1 | 268 (1.97) |
| RF0101 | 0 | 1 | 0 | 1 | 902 (6.65) |
| RF1001 | 1 | 0 | 0 | 1 | 464 (3.42) |
| RF0110 | 0 | 1 | 1 | 0 | 770 (5.68) |
| RF1010 | 1 | 0 | 1 | 0 | 234 (1.73) |
| RF1100 | 1 | 1 | 0 | 0 | 1,281 (9.45) |
| RF0111 | 0 | 1 | 1 | 1 | 528 (3.90) |
| RF1011 | 1 | 0 | 1 | 1 | 212 (1.56) |
| RF1101 | 1 | 1 | 0 | 1 | 1,070 (7.89) |
| RF1110 | 1 | 1 | 1 | 0 | 772 (5.69) |
| RF1111 | 1 | 1 | 1 | 1 | 922 (6.80) |
0/1 = absence/presence of each risk factor; RF0000–RF1111, different combinations of four risk factors.
Data are presented as the number/total number (percentage) of subjects unless otherwise indicated.
We applied a recently developed ferric particle-assisted LDI-MS technique, followed by an in-house computational pipeline (STAR Methods).30 This procedure provided an effective detection platform with superior-quality and large-scale biospecimens for subsequent analysis. First, we prepared tailored ferric particles and observed rough-surfaced nanoparticles of uniform size 300 ± 20 nm in diameter (polydispersity index [PDI] of 0.358) by transmission electron microscopy (TEM) (Figure S1A) and dynamic light scattering (DLS) (Figure S1Bi). The ferric particles with elements (including Fe and O) were uniformly distributed in the elemental mapping and scanning electron microscopy (SEM) images shown in Figure S1A. The zeta potential of −15.13 ± 0.15 mV indicated that these ferric particles had a negative charge (Figure S1Bii). Accordingly, ferric particles had a unique extinction spectrum at 350–450 nm (Figure S1Biii) compatible with the LDI-MS laser. Subsequently, we tested different contents of standards, including lysine, D-glucose, and sucrose, and showed reliable results, affording R2 values of 0.98, 0.92, and 0.92, respectively (Figure S1C). To further confirm the detection performance, we also compared the results of six standards using ferric particles and organic matrices by LDI-MS, including 2,5-dihydroxybenzoic acid (DHB) and α-cyano-4-hydroxycinnamic acid (CHCA). The results showed that the detection limit of standard metabolites for standards obtained by ferric particles was significantly lower than those obtained by DHB and CHCA (Figures S2 and S3 and Table 2). Furthermore, we compared the LDI-MS results of the plasma sample before and after preprocessing among the ferric particles, DHB, and CHCA. The results showed that ferric particle-assisted LDI-MS, with or without pretreatment, could detect more peaks of plasma samples than DHB and CHCA (Figure S4). Regarding detection speed, we achieved rapid LDI-MS detection of 13,554 samples at a speed of ∼2 s per sample (2,000 shots at a laser frequency of 1,000 Hz) using an automatic scanning model with a 1 s interval between samples. Overall, the LDI-MS of detection achieved a leap in performance with ferric nanoparticles' help in analytical reproducibility, speed, and throughput.
Top discriminating PMFs associated with pre-MetS and MetS
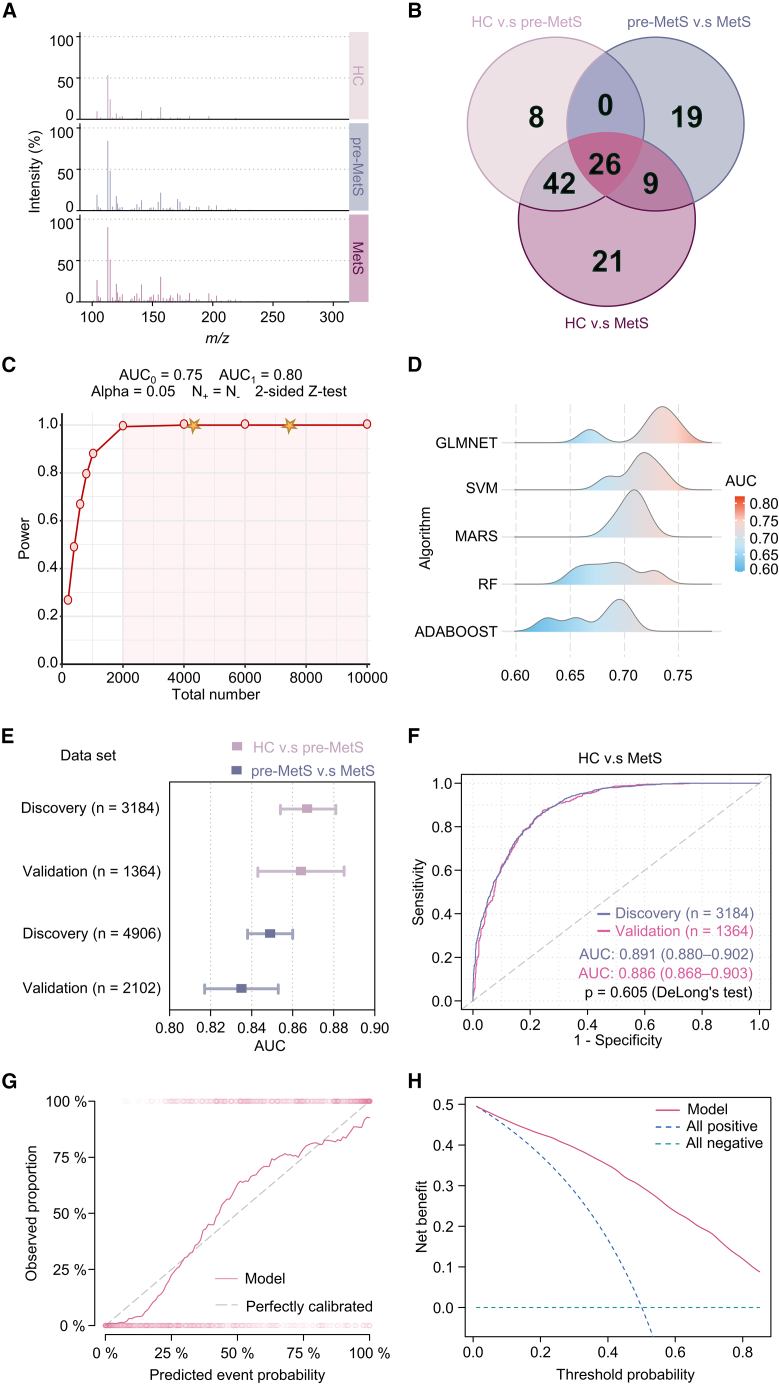
Exploiting the high-throughput metabolic profiling provided by LDI-MS, we developed a metabolic screening platform for the one-step diagnosis of MetS using small plasma samples. This study recorded the typical mass spectra results for 100 nL of plasma from participants in the HC, pre-MetS, and MetS groups (Figure 3A). For further analysis, we extracted 303 m/z signals as PMFs with stable uniformity from the raw mass spectrum (Figure S5).
Figure 3.
Diagnostic models for pre-MetS and MetS by 26 hub PMFs adjusted for age and gender using ML
(A) Typical mass spectrometry spectra within an m/z range of 100–300 obtained by ferric particle-assisted LDI-MS of plasma samples from the HC, pre-MetS, and MetS groups.
(B) The final 26 hub PMFs were filtered out by comparison of differential PMFs among HC vs. pre-MetS, pre-MetS vs. MetS, and HC vs. MetS groups using Kruskal-Wallis rank-sum test and Bonferroni/Dunnett correction in the discovery cohorts.
(C) Power analysis of the diagnosis of pre-MetS and MetS using a two-sided Z test. AUC0 and AUC1 are the areas under the receiver operating characteristic (ROC) curves (AUCs) for the null and alternative hypotheses, respectively. N+ and N− are the numbers of items sampled from cases and controls, respectively. The stars indicate the numbers in our datasets (n = 4,548, 4,548, and 7,008) for classification among HC vs. pre-MetS, HC vs. MetS, and pre-MetS vs. MetS, respectively.
(D) Distribution of the AUC using generalized linear models via least absolute shrinkage and selection operator and elastic-net regularization (GLMNET), support vector machine (SVM), multivariate adaptive regression splines (MARS), random forest (RF), and adaptive boosting (ADABOOST) to distinguish between HC and MetS groups in the validation cohort (n = 1,364).
(E) Distribution of the AUCs of HC vs. pre-MetS and pre-MetS vs. MetS in both the discovery and validation sets (HC vs. pre-MetS in pink and pre-MetS vs. MetS in purple).
(F) ROC curves for the PMF-based MetS diagnostic model using the GLMNET algorithm to distinguish between MetS and HC in the discovery (n = 3,184) and validation sets (n = 1,364).
(G) Calibration curves for our model showed good correlation between predicted and observed outcomes. The calibration curve was close to the 45° perfectly calibrated line.
(H) DCA plot depicting the standardized net benefit of our model.
To balance the sample categories, we randomly selected 4,548, 4,548, and 7,008 participants with 1:1 case-control matching from the general population to build diagnostic models for HC vs. MetS, HC vs. pre-MetS, and pre-MetS vs. MetS, respectively. We randomly split this cohort at a ratio of 0.7 to 0.3 into the discovery (for HC vs. pre-MetS n = 3,184; for pre-MetS vs. MetS n = 4,906; for HC vs. MetS n = 3,184) and validation (for HC vs. pre-MetS n = 1,364; for pre-MetS vs. MetS n = 2,102; for HC vs. MetS n = 1,364) sets, respectively. There were no statistical differences between the gender of controls and cases (Table S3).
Ninety-eight PMFs significantly differed between the HC and MetS groups in the discovery set (two-sided Kruskal-Wallis rank-sum test, p < 0.05; Figure 3B). Similarly, 76 differential PMFs were screened between the HC and pre-MetS groups in the discovery set, and 54 differential PMFs were screened between the MetS and pre-MetS groups in the discovery set (two-sided Kruskal-Wallis rank-sum test, p < 0.05; Figure 3B). Following Bonferroni correction and Dunnett’s test for multiple comparisons among these three groups, 26 differential PMFs were further identified as the top discriminating metabolic patterns (adjusted p < 0.05; Figure 3B) and might be potential plasma metabolic biomarkers for diagnosing pre-MetS and MetS.
Hub PMFs optimizing the metabolic classification of MetS through ML
The current large sample sizes (for HC vs. MetS n = 4,548; for HC vs. pre-MetS n = 4,548; for pre-MetS vs. MetS n = 7,008) achieved 99.99% diagnostic power to detect statistical significance and thus produce reliable, valid, and generalizable results (Figure 3C).
We compared the model performance for HC vs. MetS through 5-fold cross-validation among five ML algorithms, including generalized linear models via least absolute shrinkage and selection operator and elastic-net regularization (GLMNET) (mean AUC 0.72), support vector machine (SVM) (mean AUC 0.71), multivariate adaptive regression splines (MARS) (mean AUC 0.71), random forest (RF) (mean AUC 0.69), and adaptive boosting (ADABOOST) (mean AUC 0.67) (Figure 3D). Although the SVM and MARS models showed a high sensitivity (0.70 and 0.64), the SVM model had the lowest specificity (0.60) compared to the MARS and glmnet models (both 0.69) (Figure S6 and Table S4). Due to its combined AUC, sensitivity, and specificity performance, the glmnet algorithm was identified as an appropriate ML algorithm to build diagnostic models for pre-MetS and MetS. Then, we sequentially optimized parameters for HC vs. pre-MetS, pre-MetS vs. MetS, and HC vs. MetS using all 26 hub PMFs adjusted for age and gender under 5-fold cross-validation. The AUCs for HC vs. pre-MetS were 0.867 (95% CI, 0.854–0.881) and 0.864 (95% CI, 0.843–0.885) (one-sided DeLong test; p = 0.813) in the discovery (n = 3,184) and validation sets (n = 1,364), respectively (Figures 3E and S7A). The AUCs for pre-MetS vs. MetS were 0.849 (95% CI, 0.838–0.860) and 0.835 (95% CI, 0.817–0.853) (one-sided DeLong test; p = 0.195) in the discovery (n = 4,906) and validation sets (n = 2,012), respectively (Figures 3E and S7C). Other evaluation metrics, including accuracy, F1 score, negative predictive rate, positive predictive rate, specificity, and sensitivity, are shown in Figures S7B and S7D. The specificity of the HC vs. pre-MetS model was 0.99 in both the discovery and validation sets, and the F1 score was 0.88 in the discovery set and 0.87 in the validation set. The specificity of the pre-MetS vs. MetS model was 0.76 and 0.75 in the discovery and validation sets, while its F1 score was 0.78 and 0.77, respectively.
The model for HC vs. MetS (Figures S8A and S8B) showed good diagnostic performance in both the discovery (n = 3,184) and validation (n = 1,364) sets, with AUCs of 0.891 (95% CI, 0.880–0.902) and 0.886 (95% CI, 0.868–0.903) (one-sided DeLong test; p = 0.280), respectively (Figure 3F). In the HC vs. MetS model, the specificity was 0.79 in both the discovery and validation sets, while the F1 scores were 0.79 and 0.80, respectively (Figure S8C). The confusion matrix for the validation set is shown in Figure S8D. Furthermore, we compared the role of 26 hub PMFs with 303 features for HC vs. MetS to verify the representativeness of these hub features. The results showed that the AUCs were 0.885 (95% CI, 0.867–0.903) and 0.886 (95% CI, 0.868–0.903) (one-sided DeLong test; p = 0.967) in the validation sets of the model with 303 hub PMFs and 26 features, respectively (Figure S8E). Comparisons of other model evaluation metrics are shown in Figure S8F. The p values did not differ significantly between the two groups, demonstrating that hub PMFs were representative.
To effectively utilize the entire dataset to evaluate the performance of the PMFs-based diagnostic model, we conducted 100 independent and randomized 1:1 case-control matching sample selections for model evaluation. In the 100 repeated trials, each sample from the MetS group was selected at least 47 times (Figure S9A); 95% of the discovery set had AUC values in the range of [0.842, 0.869], and 95% of the validation set had AUC values in the range of [0.828, 0.867] (Figure S9B). Moreover, we randomly and independently split the discovery and validation sets 100 times using the same dataset as in Figure 3F. The results showed that 95% of the discovery set had AUC values in the range of [0.884, 0.896], and 95% of the validation set had AUC values in the range of [0.871, 0.899] (Figure S9C). These results indicate that the PMFs-based MetS diagnostic model is robust through the evaluation using the entire dataset.
Moreover, we compared the performance of GLMNET and categorical boosting (CatBoost) models in distinguishing between HC and MetS. The CatBoost model showed a significant difference between discovery and validation AUC (p < 0.001) (Figure S9D), suggesting overfitting. However, the advantage of CatBoost models is their ability to analyze multi-class labels. We also compared the AUC of the GLMNET model and the CatBoost model on the discovery set and found no statistically significant difference between the two models (p = 0.756) in diagnosing MetS (Figure S9E). Furthermore, we separately analyzed the importance of each feature using the coefficients of the GLMNET model and the Shapley Additive Explanation (SHAP) values of the CatBoost model (Figures S10A and S10B). We also compared the coefficients and SHAP values, finding that five variables showed positive effects in both evaluation methods, while four showed negative effects (Figure S10C).
Given the confounding effects of drugs, we conducted a sensitivity analysis to eliminate the potential impact of medication on our results. We collected medication information on participants in both the discovery and validation sets. In the discovery set, medication information was available for 58.4%, and it was available for 56.4% in the validation set (Figure S11A). Conventional principal-component analysis (PCA) was unable to differentiate between MetS patients who were using medication and those who were not (Figure S11B). We also evaluated the performance of our diagnostic model separately for the medication and non-medication groups. The AUC for distinguishing MetS from HC in the medication group was 0.920 (95% CI, 0.903–0.936) in the discovery set and 0.922 (95% CI, 0.898–0.946) in the validation set, with a p value of 0.890 (Figure S11C). The AUC for the non-medication group was 0.861 (95% CI, 0.843–0.879) in the discovery set and 0.863 (95% CI, 0.836–0.890) in the validation set, with a p value of 0.923 (Figure S11C). Other model evaluation indicators also show good performance (Figure S11D). Previous literature has suggested that patients adhering to medication regimens exhibit alterations in metabolic profiles.31,32,33 In this study, we observed a significant difference in the AUC within the validation set between the non-medication and medication groups, yielding a p value of 0.002, which implies a potential influence of medication on metabolism. Nevertheless, this effect does not facilitate a distinct PCA-based separation between MetS patients in the medication and non-medication groups (Figure S11B). These results underscore the robustness of the PMF-based diagnostic model for MetS, accounting for diverse real-world scenarios and including both medication and non-medication groups, thereby facilitating clinical diagnosis in practical healthcare settings.
Next, we examined whether the diagnostic model based on PMFs could serve as a clinical tool to define MetS. To that end, we introduced calibration curves to summarize model calibration over the observed range of predicted probabilities and assess the accuracy of the predictions.34,35 MetS cases predicted by this PMF-based model were in high agreement with the actual observations in the calibration plot (Figure 3G). Additionally, this model increased the net benefit rate through decision-curve analysis (DCA) (Figure 3H). Overall, by comparing the plasma profiles of donors with distinct risk factors, we constructed reliable ML-based classifiers for pre-MetS and MetS using 26 hub PMFs. These PMF-based diagnostic models exhibited excellent discrimination, calibration, and clinical utility independent of traditional biochemical examinations.
Re-clustering MetS subtypes optimized the assessment of different MetS risk factors
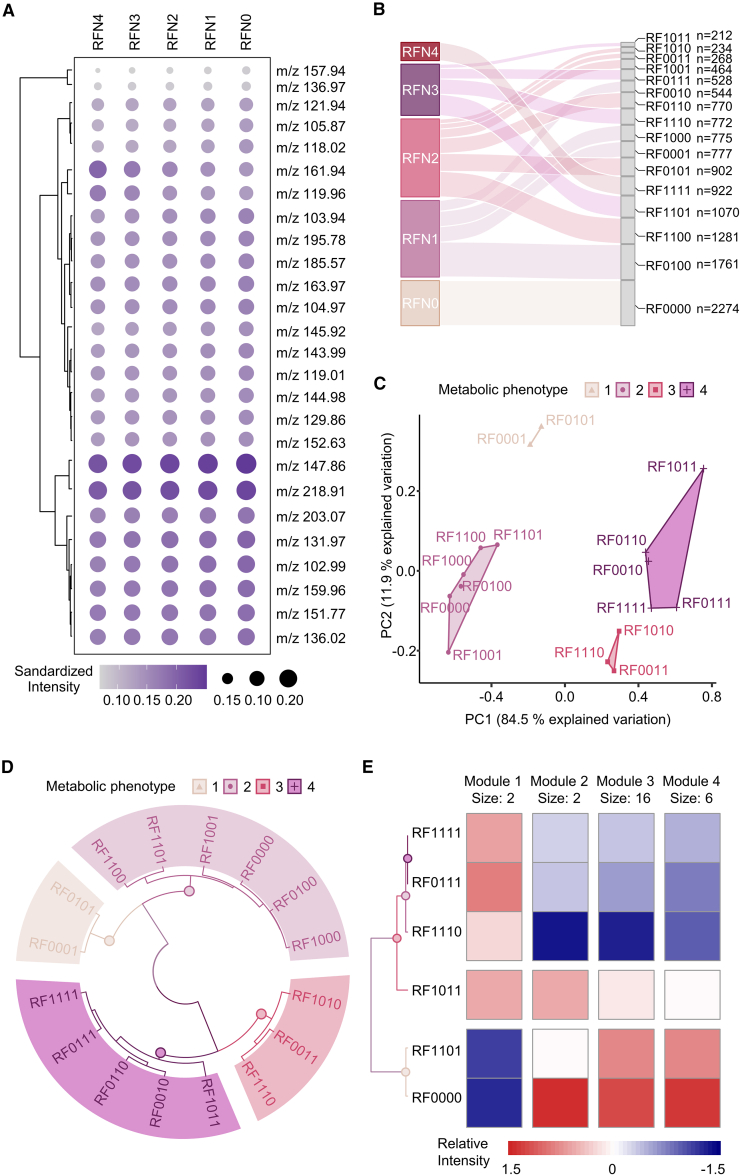
To investigate the heterogeneity of four risk factors and their combinations in MetS, we calculated the average intensity of 26 hub PMFs in five subgroups (RFN0–RFN4) sorted by the number of risk factors (Figure 4A). However, using only the mean intensity did not distinguish the heterogeneity of the subgroups well. Therefore, we divided five subgroups into 16 subclusters (RF0000–RF1111) according to the composition of MetS risk factors. The population numbers for each subcluster are summarized in Figure 4B. We used two unsupervised ML strategies, K-means and hierarchical clustering, to reclassify the metabolic heterogeneity of 16 subclusters after dimensionality reduction by PCA. The first two principal components (PC1 and 2) explained over 96.4% of the total variation in the entire cohort (Figure 4C). The results showed that both analytical strategies classified these 16 subclusters into the same four MPs (Figures 4C and 4D). The optimal number of clusters was calculated according to the gap statistic (Figure S12 and Table S5). The subclusters of unitary dyslipidemia (RF0001) and dyslipidemia combined with hypertension (RF0101) clustered together (MP1 in Figures 4C and 4D). All subclusters containing obesity, except those with the complication of hyperglycemia (RF1000, RF1001, RF1101, RF1100), were highly consistent with the HC subcluster (RF0000) and the subcluster of unitary hypertension (RF0100) (MP2 in Figures 4C and 4D). MP3 contained the subclusters of obesity combined with hyperglycemia (RF1010), obesity combined with hypertension and hyperglycemia (RF1110), and hyperglycemia combined with dyslipidemia (RF0011) (MP3 in Figures 4C and 4D). The remaining five subclusters, including unitary hyperglycemia (RF0010), hyperglycemia combined with hypertension (RF0110), hyperglycemia combined with obesity and dyslipidemia (RF1011), and a combination of all four risk factors (RF111), were classified as MP4 (MP4 in Figures 4C and 4D). Based on these results validated with two different algorithms, our PMF-based metabolic reclassification provided the first real-world evidence of the metabolic phenotypic diversity of the traditional composition of four MetS risk factors based on a large community cohort study of over 10,000 people.
Figure 4.
PMF-based unsupervised ML revealed the heterogeneity of MetS
(A) The standardized intensity of all hub PMFs among five subgroups (RFN0–RFN4) according to the number of risk factors.
(B) The distribution of five subgroups (RFN0–RFN4) and 16 subclusters (RF0000–RF1111) classified according to traditional risk factors in the general population.
(C) K-means clustering analysis scatter diagram regrouping 16 subclusters into four metabolic phenotypes (MPs) based on the results of PCA. Each point represents a subcluster condition containing (or not) the risk factor according to Table 2. MPs are indicated by colored shading.
(D) Circular hierarchical cluster analysis dendrogram grouping 16 subclusters into the same four MPs based on phenotypic similarity. Colors of subcluster names based on the MPs.
(E) Relative risk assessment among the five MetS subgroups (RF0111, RF1011, RF1101, RF1110, RF1111) and HC group through K-means clustering analysis based on the relative intensity of all 26 hub PMFs. These PMFs were divided into four metabolic feature modules (modules 1–4).
Moreover, to precisely interpret the proportion of each risk factor involved in MetS, we re-clustered the five MetS subtypes (RF1110, RF1011, RF1101, RF0111, and RF1111) as well as the HC group (RF0000) using the K-means algorithm to clarify the contributions of the different risk factors to the pathogenesis of MetS. The MetS subtype with risk factors for obesity, hypertension, and dyslipidemia (RF1101) showed the highest similarity to the HC group (RF0000), suggesting that the missing risk factor, hyperglycemia, had the greatest impact on chronic metabolic disorders in the general population (Figure 4E). Similarly, hypertension had a greater effect on MetS than dyslipidemia (RF1011 vs. RF1110) or obesity (RF1011 vs. RF0111), as demonstrated by comparing groups in turn according to the degree of clustering similarity (Figure 4E). The contribution of dyslipidemia to MetS was similar to that of obesity (RF1110 vs. RF0111) (Figure 4E). The clustering results also divided the 26 hub PMFs into four intensity-specific metabolic modules (modules 1–4), which contained 2, 2, 16, and 6 hub PMFs, respectively (Figure 4E). The relative intensity of these modules among the five MetS subtypes and the HC group is depicted in Figure S13. Based on these results, the contributions of all the MetS risk factors could be sorted in descending order: hyperglycemia, hypertension, dyslipidemia, and obesity.
Individualized PMR stratification mirroring actual metabolic dysfunction
Compared to clinical parameters, plasma metabolites could better reflect an individual’s disease progression and metabolic dysfunction.36,37 This study constructed a personalized three-dimensional PMR stratification using 26 hub PMFs and CatBoost (an algorithm for gradient boosting on decision trees) in the general population (Figure 5). The composite PMR scores comprised three-dimensional indexes (indexes 1–3). To calculate indexes 1–3, we first determined each participant’s mean predicted probability scores for HC, pre-MetS, and MetS. Then, we used these mean probability scores as cutoff values to categorize each participant into one of the three metabolic patterns.
Figure 5.
Construction of the three-dimensional PMR stratification for evaluating individual metabolic heterogeneity
(A) Distribution of the three PMR indexes (index 1–3) among the five subgroups (RFN0–RFN4) classified according to the present number of traditional risk factors.
(B) Mean scores of these three indexes (index 1–3) in the HC, pre-MetS, and MetS groups.
(C) Specific PMR patterns for all participants in the general population cohort. Gray, blue, and purple dots indicate individual status ranked by LMR, MMR, and HMR patterns, respectively.
(D) The relative changes in high-density lipoprotein cholesterol (HDLC), serum creatinine (SCr), serum total cholesterol (TC), low-density lipoprotein cholesterol (LDLC), uric acid (UA), body mass index (BMI), glucose (GLU), and triglycerides (TGs) in the MMR and HMR groups compared with those in the LMR group.
(E) Mean scores for index 1 (in gray) and index 3 (in purple) in the five MetS subtypes (RF1110, RF1011, RF1101, RF0111, and RF1111) and the HC group (RF0000).
(F) Cumulative curves and forest plots of the 4-year mortality events for 13,554 patients with three PMR statuses stratified by gender and age. The p value for multivariate Cox regression analysis models was calculated by the likelihood test. The p value for variables was obtained by a log-rank test and hazard ratio (HR).
The score of index 1 was closely related to the subgroup with no risk factors (RFN0) (Figure 5A) and the HC group (Figure 5B). The score of index 2 was extremely high in the subgroup with one or two risk factors (RFN1 and 2) (Figure 5A) and the pre-MetS group (Figure 5B), while, in other groups, it was close to zero. The score of index 3 was only sensitive to the subgroup with three or four risk factors (RFN3 and 4) (Figure 5A) and the MetS group (Figure 5B). These results suggested that the three indexes characterized different aspects of MetS. By calculating the priority of the three index scores, we reclassified the cohort (n = 13,554) into three metabolic risk patterns: low (LMR), medium (MMR), and high metabolic risk (HMR) patterns (Figure 5C). The LMR (n = 2,371), MMR (n = 7,674), and HMR (n = 3,509) patterns had the highest index 1, index 2, and index 3 scores, respectively.
Compared to individuals with the MMR pattern, those with the HMR pattern showed greater relative increases in serum creatinine (SCr), serum total cholesterol (TC), low-density lipoprotein cholesterol (LDLC), uric acid (UA), body mass index (BMI), glucose (GLU), and triglycerides (TGs) when the expression in the LMR pattern was taken as the baseline (Figure 5D and Table S6), which is consistent with clinical practice. Additionally, the HMR group exhibited a more pronounced relative decrease in high-density lipoprotein cholesterol (HDLC) (Figure 5D and Table S6), suggesting that HDLC is a protective factor for metabolic disorders. We also calculated scores for index 1, which reflects health status, and index 3, which reflects disease status, in the five MetS subtypes (RF1110, RF1011, RF1101, RF0111, and RF1111) as well as in the HC group (RF0000) to reassess the metabolic heterogeneity of MetS. The results showed that index 1 scores increased sequentially in the RF0000, RF1101, RF1011, RF1110, RF0111, and RF1111 subtypes (Figures 4C and 5E), while index 3 scores decreased sequentially in the same subtypes (Figure 5E). This ordering of MetS subtypes was consistent with the results of the K-means cluster analysis described above (Figure 5E), which further supports reliability and robustness of our PMR-based stratification model.
To investigate the clinical relevance of PMR-based stratification, we collected all-cause death events in the entire cohort within 4 years, including 291 (2.1%) all-cause death events as major points and 28 (0.2%) accidental death events as censors. Based on the traditional disease classification, the status was found to be significantly associated with a higher incidence of adverse events without adjusting for gender and age (for pre-MetS, HR 1.58, 95% CI 1.08–2.30, p for log-rank test = 0.018; for MetS, HR 1.78, 95% CI 1.18–2.66, p for log-rank test = 0.006) (Figure S14B). However, statistical significance disappeared for pre-MetS (adjust p = 0.063) (Figure 5F) after excluding age and gender as confounders. This finding implies that our conventional disease staging, which heavily relies on age and gender, may not adequately reflect the metabolic status of the patient. Notably, PMR-based stratification was also significantly related to worse event accumulation without gender and age correction (for medium risk, HR 1.76, 95% CI 1.19–2.59, p for log-rank test = 0.004; for high risk, HR 2.06, 95% CI 1.36–3.11, p for log-rank test = 0.001) (Figure S14A). Using the above PMR-based stratification model, we also revealed that patients with medium risk had a significantly worse event accumulation than those with low risk adjusted for gender and age (HR 1.54, 95% CI 1.05–2.28, p for log-rank test = 0.029). The patients with high risk had a significantly worse event accumulation than those with low risk adjusted for gender and age (HR 1.85, 95% CI 1.22–2.79, p for log-rank test = 0.004) (Figure 5F), suggesting the independent relationship between this PMR-based stratification and all-cause death events.
We further investigated the association between the traditional risk grouping and PMR-based metabolic patterns using variant analysis to illustrate the changes (Table S7). The RFN0 subgroup (n = 2,274) could be reclassified into LMR (n = 2,188, 96.2%), MMR (n = 74 3.3%), and HMR (n = 12 0.5%) patterns (Table S7). Similarly, the variant analysis of novel risk patterns and existing traditional classifications based on disease states is summarized in Table S7. The PMF-based metabolic risk patterns were at least 95.58% consistent with the traditional classification following the number of risk factors. These results indicated that the conventional determination of MetS status was too ambiguous to precisely reflect an individual’s metabolic dysfunction, especially in the general population. Even a homogeneous subgroup of MetS patients (concerning traditional risk factors) had heterogeneous risk patterns of metabolic dysfunction. Therefore, individualized risk stratification and preventive measures are necessary for MetS patients. Our PMR stratification with personalized metabolic patterns and three-dimensional PMR scores provides a robust and feasible solution for accurate metabolic risk stratification.
To identify the specific metabolic pathways driving these risk factors in MetS, we matched 26 hub PMFs to the Human Metabolome Database (HMDB) and classified them into organic acids (n = 12), organoheterocyclic compounds (n = 4), benzenoids (n = 3), fatty acyls (n = 2), carbohydrates (n = 2), and nucleic acids (n = 1) (Figure S15A and Table S8). Recent studies have highlighted the role of lipid metabolites with large molecular weights in the pathological process of MetS.38,39 However, our advanced LDI-MS revealed the importance of small-molecule metabolites in the diagnosis and risk stratification of MetS, thereby promoting the potential clinical application in the early identification of pre-MetS.
Using pathway topology analysis, we further computed the metabolic pathway enrichment of the 26 hub PMFs to regulate the process of MetS. Four differential metabolic pathways (enrichment ratio >5 and p < 0.05), including taurine and hypotaurine metabolism, phenylacetate metabolism, homocysteine degradation, and phosphatidylethanolamine biosynthesis, were identified to be involved in the development of MetS (Figure S15B and Table S9). These findings suggest that small-molecule metabolites play potential roles in MetS development and could serve as new targets for attenuating MetS risk.
Discussion
Metabolic risk factors present significant global challenges, necessitating effective early intervention strategies.40 In this study, we employed ferric particle-assisted LDI-MS to identify potential biomarkers for pre-MetS and MetS in a community-based setting. While prior research primarily focused on discovering new metabolites associated with MetS, our approach emphasized the heterogeneity of metabolic dysfunctions through ML-based integrated PMRs scoring rather than relying solely on traditional risk factors. Using unsupervised clustering, we successfully characterized MetS subtypes based on four distinct metabolic modules, revealing the varying contributions of hyperglycemia, hypertension, dyslipidemia, and obesity (Figure 4). This finding was further supported by the correlation between clinical parameters and the reclassified metabolic risk patterns (Figure 5). These results provide a novel and scalable screening approach for identifying MetS patients, mainly targeting elderly individuals at risk of metabolic dysfunction and facilitating interventions that effectively address modifiable risk factors.
Metabolomics is a powerful tool for precision medicine in biomarkers. However, metabolomics-based biomarkers for MetS are still in their infancy. Previous studies have primarily focused on analyzing MetS patients in bulk using small sample sizes23,41 and employing univariate analysis methods.42 These studies aimed to identify distinct population-level risk factors rather than investigating the phenotypic consequences from the perspective of integrated PMFs. Our study represents the largest investigation of PMFs in MetS patients, utilizing a general community cohort of 13,554 individuals and achieving a remarkable diagnostic power of 99.99% (Table 1 and Figure 3C). A previous cross-sectional serum metabolomic study on MetS in 2021 included only 1,800 participants.43 To ensure generalizability, we employed independent discovery and validation sets (Table S3).
Most MetS analyses have utilized NMR,24 LC‒MS,25 or GC-MS18 methods. However, conventional spectrometric techniques have been reported27 to be challenging to achieve the required diagnostic performance using high-throughput quantitative metabolomic analysis of ultralow-volume samples when using these individual platforms separately to detect large numbers of samples (Table S10). NMR requires a pretreatment step and large sample volume (∼microliters) to enhance sensitivity (∼picomoles) by expanding nuclear-level rotation. On the other hand, GC-MS/LC-MS methods necessitate longer sophisticated sample handling (∼hours) to reduce sample complexity.44,45,46 In contrast, LDI-MS, as an alternative metabolite recognition method, offers a high-performance technique that enables large-scale MetS screening.
The traditional multi-process diagnosis of MetS is challenging to promote in community screening due to its need for cumbersome physical examination and clinical indicators. In addition, complex examinations of four risk factors using different instruments can result in significant measurement errors and low efficiency, particularly in large-scale screening programs.16 Plasma metabolites provide a fundamental view into the dynamics of MetS.47 Recent studies41,48,49 focused on comparing MetS and non-MetS and therefore did not reveal the heterogeneity of metabolic disorders. Unlike the traditional classification, the current study divided the participants into three groups, a MetS group (n = 3,504), a pre-MetS group (n = 7,776), and an HC group (n = 2,274), to achieve widespread screening of MetS in the community. We systematically validated PMF-based classifiers for pre-MetS and MetS with high performance. The AUC for pre-MetS vs. HC was 0.867 (95% CI, 0.854–0.881) and 0.864 (95% CI, 0.843–0.885) in the discovery (n = 3,184) and validation (n = 1,364) set (Figures 3E and S7A). The AUC for MetS vs. HC was 0.891 (95% CI, 0.880–0.902) and 0.886 (95% CI, 0.868–0.903) in the discovery (n = 3,184) and validation (n = 1,364) set, respectively (Figure 3F). The AUC for pre-MetS vs. MetS was 0.849 (95% CI, 0.838–0.860) and 0.835 (95% CI, 0.817–0.853) in the discovery (n = 4,906) and validation (n = 2,012) sets (Figures 3E and S7C). These results were superior to prior research of 1,800 participants in 2021 (AUC 0.750, 95% CI 0.698–0.802).43 Moreover, we further confirmed the favorable consistency through the calibration plot (Figure 3G), and the DCA suggested that our model had a reasonable clinical net benefit (Figure 3H). Therefore, these advanced classifiers based on PMFs demonstrated excellent discrimination, calibration, and clinical utility, which can pave the way for a future community screening of pre-MetS and MetS.
Obesity, hypertension, hyperglycemia, and dyslipidemia represent dominant risk factors for MetS development.50 However, the specific contributions of each risk factor to metabolic impairment progression remain largely unknown.51 Here, we proposed a metabolomics-based approach to evaluate the contribution of various risk factors using different unsupervised clustering methods. Through unsupervised analysis, we found that hyperglycemia emerged as the dominant risk factor in MetS. Metabolic disturbance phenotypes observed in MetS patients with hyperglycemia were similar to those with four risk factors (Figure 4E). We selected the 26 PMFs to explain more than 96.4% of total variation (Figure 4C). These selected PMFs effectively captured the underlying patterns and aligned with the theory of MetS development, where hyperglycemia plays a crucial role as a direct indicator of insulin resistance.52 Additionally, the result shows obesity with smaller contributions to MetS (Figure 4E), suggesting that obesity is not synonymous, and metabolically benign obesity in MetS may be induced by insulin resistance.53,54 These results provide systematic evidence from a metabolomics perspective and offer a solid theoretical foundation for optimizing cost-effective interventions tailored to patients with different risk factors.
The lack of consistent cutoff points among different diagnostic criteria55 and the significant heterogeneity in coronary heart disease (CHD) risk among individuals with MetS56,57 indicate that current diagnostic criteria do not correlate with an individual’s metabolic status. Here, we aimed to focus on individual-level metabolic diversity and identify personalized metabolic heterogeneity. We established an integrated three-dimensional PMR stratification that considered age and gender (Figure 5). Thus, significant increases in SCr, TC, LDLC, UA, BMI, GLU, and TGs were observed in the HMR pattern compared to the MMR pattern (Figure 5D and Table S6), which aligned with clinical practices. Additionally, HDLC alteration declined in the HMR pattern (Figure 5D and Table S6), indicating that HDLC play a protective role to some extent against metabolic disorders.
To verify PMR stratification, we conducted a 4-year death event follow-up in large prospective cohort. Compared with the traditional disease classification, PMR stratification was independently related to all-cause death events in this large retrospective cohort of 13,554 participants (for medium risk, HR 1.54, 95% CI 1.05–2.28, p for log-rank test = 0.029; for high risk, HR 1.85, 95% CI 1.22–2.79, p for log-rank test = 0.004). Thus, our study highlights that risk stratification and preventive measures are person specific in the development of MetS.
We conducted a comparative analysis between PMF-based molecular typing and traditional typing of MetS using flow analysis (Table S7). The results indicated that the conventional determination of MetS status based on different risk factors was too ambiguous to reflect an individual’s metabolic dysfunction. Even a homogeneous subgroup of MetS patients (concerning traditional risk factors) had heterogeneous risk patterns of metabolic dysfunction. The integrated PMR stratification with personalized metabolic patterns and three-dimensional PMR scores provides a robust and feasible solution for accurate metabolic risk stratification, which could not only mirror the actual individual MPs but also replace the traditional classification method based on the number of risks.
Limitations of the study
Several limitations should also be considered in our study. First, potential bias might limit the results of our study from being replicated in other settings. Therefore, we involved a relatively large number of participants among four communities controlling for sampling error to minimize drawbacks. Moreover, the data were randomly partitioned into discovery and validation sets in a 7:3 ratio. Another limitation is that the study lacks potential experimental validation, which is important in establishing the reliability and robustness of the findings. Future efforts will focus on validating the models in different centers and conducting experimental verification through animal experiments.
Conclusions
Using PMF of MetS with LDI-MS, we established portable and efficient diagnostic models for pre-MetS and MetS with improved performance for clinical promotion in the general population. Furthermore, we assessed the contributions of different MetS risk factors based on specific metabolic modules using unsupervised clustering. We demonstrated the importance of glycemic control for the prevention and treatment of MetS. Finally, we devised a personalized three-dimensional PMR stratification to decode the individual metabolic heterogeneity into three metabolic patterns related to all-cause death in our large prospective cohort. Overall, our results introduce novel paradigms and interventional directions to accelerate the application of precision medicine for pre-MetS and MetS.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Biological samples | ||
| Human plasma samples | Shanghai Renji Hospital, China | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Bovine serum albumin (BSA, purity>98%) | Sigma, USA | Cat #V900933 |
| 2,5-dihydroxybenzoic acid (DHB, purity>98%) | Sigma, USA | Cat # 149357 |
| α-cyano-4-hydroxycinnamic acid (CHCA, purity>99%) | Sigma, USA | Cat # 476870 |
| acetonitrile (ACN, purity>99%) | Sigma, USA | Cat # 110086 |
| cetyltrimethylammonium chloride (CTAC, purity>99%) | Sigma, USA | Cat # 52366 |
| dithiothreitol (DTT, purity>99%) | Sigma, USA | Cat #D9163 |
| L-lysine (purity>98%) | Sigma, USA | Cat #L5501 |
| D-glucose (purity>99.5%) | Sigma, USA | Cat #G8270 |
| glycine (purity>99%) | Sigma, USA | Cat #W328707 |
| Sucrose | Sigma, USA | Cat # 1.07687 |
| L-tryptophan (purity>99%) | Sigma, USA | Cat # 574597 |
| L-glutamine (purity>99%) | Sigma, USA | Cat #G3126 |
| Ethanol absolute (99.7%) | Sinopharm Chemical Reagent Beijing Co. Ltd. (Beijing, China) | Cat # 10009218 |
| sodium acetate (purity>99%) | Sinopharm Chemical Reagent Beijing Co. Ltd. (Beijing, China) | Cat # 10018718 |
| trisodium citrate (purity>99%) | Sinopharm Chemical Reagent Beijing Co. Ltd. (Beijing, China) | Cat # 10019408 |
| ethylene glycol | Sinopharm Chemical Reagent Beijing Co. Ltd. (Beijing, China) | Cat # 10009818 |
| ferric chloride (purity>97%) | Sinopharm Chemical Reagent Beijing Co. Ltd. (Beijing, China) | Cat # 81015228 |
| trifluoroacetic acid (TFA, purity>99%) | Sinopharm Chemical Reagent Beijing Co. Ltd. (Beijing, China) | Cat # 80134716 |
| Resorcinol (purity>99%) | Sinopharm Chemical Reagent Beijing Co. Ltd. (Beijing, China) | Cat # 81012016 |
| Thiol-aptamers | Sangon Biotech Co., Ltd. (Shanghai, China) | Cat # A100958 |
| Formaldehyde solution (CH2O, purity >36.0%) | Shanghai Aladdin Reagent Co. Ltd. (Shanghai, China) | Cat #F140859 |
| Deposited data | ||
| LDI-MS based metabolomics | This paper | NMDR: ST002733, Project DOI: https://doi.org/10.21228/M8BT6J |
| Software and algorithms | ||
| R version 4.0.5 | R project | https://www.r-project.org/ |
| PASS 15.0.5 | NCSS, LLC. Kaysville, Utah, USA | https://www.ncss.com/download/pass/updates/pass15/ |
| MATLAB (R2016a) | The Mathworks, Natick, MA, USA | https://ww2.mathworks.cn/products/matlab.html |
| MetaboAnalyst 5.0 | MetaboAnalyst | http://www.metaboanalyst.ca |
| mlbench 2.1 | N/A | https://cran.r-project.org/web/packages/mlbench/index.html |
| pROC 1.18.2 | N/A | https://cran.r-project.org/web/packages/pROC/index.html |
| ModelGood 1.0.9 | N/A | https://cran.r-project.org/web/packages/ModelGood/index.html |
| glmnet 4.1 | N/A | https://cran.r-project.org/web/packages/glmnet/index.html |
| caret 6.0 | N/A | https://cran.r-project.org/web/packages/caret/index.html |
| cluster 2.1.4 | N/A | https://cran.r-project.org/web/packages/cluster/index.html |
| catboost 1.2 | N/A | https://pypi.org/project/catboost/ |
| ggalluvial 0.12.5 | N/A | https://cran.r-project.org/web/packages/ggalluvial/index.html |
| ggpubr 0.6.0 | N/A | https://cran.r-project.org/web/packages/ggpubr/index.html |
| survival 3.5 | N/A | https://cran.r-project.org/web/packages/survival/index.html |
| ggplot2 3.4.2 | N/A | https://cran.r-project.org/web/packages/ggplot2/index.html |
| pheatmap 1.0.12 | N/A | https://cran.r-project.org/web/packages/pheatmap/index.html |
| circlize 0.4.15 | N/A | https://cran.r-project.org/web/packages/circlize/index.html |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Jun Pu (pujun310@hotmail.com).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Participants and ethics
The Shanghai Community Cohort Establishment and Follow-up (NCT04517513) is registered with ClinicalTrials.gov as a large general population cohort in Pudong New District, Shanghai, China. As a part of the series of studies on this cohort, all subjects (n = 17,841) with different degrees of metabolic syndrome (MetS) were enrolled from three different communities including HeQing, JinQiao, and JinYang communities between January 2019 and March 2019, with approval by the Ethics Committee of Renji Hospital (KY2019-136), School of Medicine, Shanghai Jiao Tong University. To avoid potential interference from other confounding factors, 2,238 participants were excluded using the following exclusion criteria. After excluding subjects with missing clinical information, 13,554 volunteers were included in the current study. To better reflect the Chinese population, pre-MetS and MetS were diagnosed by the following standard criteria according to the statement of the Chinese Diabetes Society 200428 (Table S1). According to the guidelines of the Declaration of Helsinki (2013),59 written informed consent was obtained from all participants and their information was anonymized to protect their privacy.
Exclusion criteria
Patients with any acute and infectious clinical symptoms, including but not limited to fever, headache, cough, malaise, sore throat, loss of smell, runny nose, abdominal pain, and diarrhea, within 3 weeks before sampling. Patients with chronic inflammatory diseases, such as rheumatoid arthritis, systemic lupus erythematosus, and inflammatory bowel disease.
Definition of metabolic syndrome (MetS)
MetS is defined as the presence of three or more of the four risk factors established by the Chinese Diabetes Society 200428: 1) obesity (BMI≥25 kg/m2); 2) hypertension (blood pressure≥140/90mmHg and/or have been confirmed and treated as hypertension); 3) hyperglycemia (fasting plasma glucose≥6.1 mmol/L [110 mg/dl] and/or 2-h postprandial blood glucose≥7.8 mmol/L [140 mg/dl], and/or have been diagnosed and treated as diabetes); 4) dyslipidemia (high triglyceride≥1.7 mmol/L [150 mg/dl], and/or low high-density lipoprotein cholesterol< 0.9 mmol/L [35 mg/dl] in men or <1.0 mmol/L [39 mg/dl] in women) (Table S1).
Definition of pre-metabolic syndrome (pre-MS)
pre-MS is defined as having one or two risk factors of MetS but does not meet the criteria for the diagnosis of MetS.60,61,62
Baseline examination
All community subjects underwent physical examinations and biochemical tests after overnight fasting without medication. Current regular medication, previous medical history, sleep quality, mental condition, and smoking habits were recorded based on questionnaires. Waist/hip circumference, height, and weight were measured by tape, calibrated stadiometer, and scale, respectively. Body mass index (BMI) was calculated as weight (kg)/height (m2). The average of three systolic and diastolic blood pressure recordings was noted by qualified staff to the nearest 1 mmHg using cuff-based blood pressure measurement after at least 15 min of rest in the sitting position. Triglycerides, high-density lipoprotein cholesterol, and fasting blood glucose were detected using a standard spectrophotometer.
Follow-up
The follow-up period is 4 years and its deadline was December 2022. Our predefined primary outcome measure is the full cause of death within 4 years. For each participant, the death event would be verified through the official death-certificates by Shanghai Pudong New Area Public Security Bureau on 9th December 2022. To ensure complete and accurate registration of death information, highly trained clinicians coded the causes of death and each record was cross-checked by a local community physician and further validated by the Center for Disease Control and Prevention (CDC) at the municipal and district levels. If there were any discrepancies, we would re-check medical records, family reports or official death-certificates. All the causes of death were coded and classified according to the International Statistical Classification of Diseases and Related Health Problems 10th Revision (ICD-10) in 2010.
Sample collection, preparation, and storage
Samples were provided by Renji Hospital Biobank, Shanghai Jiaotong University School of Medicine. All human peripheral venous blood samples were obtained following the protocols approved by the Shanghai Jiao Tong University Institutional Review Board. In order to prevent any interference from dietary factors, peripheral vein blood samples were obtained from individuals following an overnight fast of at least 8 h. The collected blood was gently mixed in EDTA anticoagulant tubes. The plasma was separated after centrifugation for 10min (4000 rpm; 4°C) within 2 h of collection. All plasma samples were stored at −80°C for future assays.
Method details
Matrix synthesis
A modified solvo-thermal method was adopted to prepare the ferric particles (FPs), which provided a cost-effective approach for large-scale production. Initially, ethylene glycol solution was used to dissolve ferric chloride. Then, we added trisodium citrate (weights from 0 to 0.8g) to tune the surface charge of the mixture. After adding 1.8g of sodium acetate, we sonicated the products at room temperature for 30min. To form the FPs, we held the products at 200°C for 10h after transferring them into a Teflon-lined stainless-steel autoclave (capacity 50mL). After being washed with ethanol and deionized water, the final particles were dried at 60°C before use.
Material characterization methods
A JEM-2100F instrument (JEOL, Japan) was used to collect transmission electron microscopy (TEM) images by depositing 10μL of material suspension onto a copper grid. An S-4800 instrument (Hitachi, Japan) was used to record scanning electron microscopy (SEM) images by placing a drop of material suspension on aluminum foil. A UV1900 spectrophotometer (AuCy, China) was used to obtain the optical absorption spectrum of the matrixes at room temperature. An ASAP 2020M (Micromeritics, USA) was used to test the nitrogen adsorption isotherm of the samples degassed in a vacuum. A Nano ZS90 instrument (Malvern, Worcestershire, UK) was used to perform zeta potential and dynamic light scattering (DLS) size measurements in water at 25°C.
Nano-assisted laser desorption/ionization mass spectrometry (LDI MS) profiling
LDI MS experiments were conducted as reported previously.30 All MS measurements were performed on an Autoflex speed TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). In a typical LDI MS experiment, ferric particles were dispersed in deionized water at a concentration of 1 mg mL−1 for use as a matrix. The CHCA and DHB organic matrices were dissolved in TA30 solution (acetonitrile/0.1% TFA in water, 7/3, v/v) at a concentration of 10 mg/mL. For the detection of the standards, 100nL of analyte solution (each standard listed in the part of chemicals and reagents) with different densities (100 ng mL−1, 1 μg mL−1, 10 μg mL−1, 100 μg mL−1, 1 mg mL−1) was mixed with 100nL of matrix slurry on the plate and dried for LDI MS analysis. For plasma sample detection, the samples were first prepared through protein precipitation, centrifugation, and supernatant filtration according to a commonly applied procedure. Then, a volume of 100nL of plasma solution was spotted on the plate and dried in air at room temperature, followed by adding 100nL of matrix slurry and drying for LDI MS analysis. Then, mass spectra were performed on a 5800 Proteomics Analyzer (Applied Biosystems, Framingham, MA, USA) equipped with a Nd:YAG laser (2 kHz, 355 nm). The acquisitions were extracted in positive reflector ion mode employing delayed extraction with a repetition rate of 1,000Hz and an acceleration voltage of 20 kV. The delay time for this experiment was optimized to 250 ns. The 2,000 laser shots per analysis were for all LDI MS experiments. All the original mass spectra data were visualized in DataExplorer (Version 4.5). Only the m/z signals within 100–300Da and with a signal-to-noise ratio over 3 were then acquired without smoothing processes. For pre-processing, we applied a “home-developed” program using MATLAB (R2016a, The Mathworks, Natick, MA, USA) to normalize and standardize the mass spectra data after peak extraction and alignment.63 And standard molecules for the accurate mass measurement (±0.05Da) of both Na+-adducted and K+-adducted signals were used to perform the mass calibration. The detection limit of standard metabolites (listed in chemicals and reagents’ part) obtained by ferric particle, DHB, and CHCA-assisted LDI MS were calculated as previously reported.30
Hub differential plasma metabolic fingerprints (PMFs) screening
After testing for a normal distribution, significant differential PMFs among the three states of HC, pre-MetS, and MetS in the discovery cohorts were assessed using the Kruskal-Wallis rank-sum test through the primary function in R. The significance level was set as p < 0.05. For multiple comparisons, the final MetS-specific hub PMFs were filtered according to the Bonferroni correction and Dunnett’s test in R version 4.0.5.
Power analysis
Power analysis for MetS diagnosis was calculated using PASS 15.0.5 (NCSS, LLC. Kaysville, Utah, USA) to determine the appropriate sample size required to detect an effect of a given size with a given degree of confidence.64 For HC vs. pre-MetS and HC vs. MetS, samples of 2,274 from the positive group and 2,274 from the negative group achieved 99.99% power to detect a difference of 0.05 between the area under the receiver operating characteristic (ROC) curve (AUC) under the null hypothesis of 0.75 and an AUC under the alternative hypothesis of 0.80 using a two-sided Z-test at a significance level of 0.05. For pre-MetS vs. MetS, samples of 3,504 from the positive group and 3,504 from the negative group achieved 99.99% power to detect a difference of 0.05 between AUC under the null hypothesis of 0.75 and an AUC under the alternative hypothesis of 0.80 using a two-sided Z-test at a significance level of 0.05.
Data sets splitting
To improve the generalization of the diagnostic models, we randomly selected 4,548, 7,008, and 4,548 participants (1:1 case-control matched) for HC vs. pre-MetS, pre-MetS vs. MetS and HC vs. MetS, respectively. And all these data sets were split at a ratio of 0.7 to 0.3 into the discovery (n = 3,184, 4,906, 3,184 70%) and validation (n = 1,364, 2,102, 1,364 30%) sets for HC vs. pre-MetS, pre-MetS vs. MetS and HC vs. MetS, respectively.
Model selection
All 26 PMFs were included in the model construction as biomarker panels with age and gender adjustment. To choose the suitable machine learning (ML) algorithm for model construction in this study, five machine-learning algorithms including generalized linear models via least absolute shrinkage and selection operator and elastic-net regularization (GLMNET), support vector machine (SVM), multivariate adaptive regression splines (MARS), random forest (RF), and adaptive boosting (ADABOOST) were adopted in to build diagnostic model for HC vs. MetS by 5-fold cross-validation through the “caret” package and randomized searching through the “mlbench” package in R version 4.0.5. After comprehensively comparing the sensitivity, specificity, and AUCs in the validation cohort (n = 1,364), the GLMNET algorithm was used to construct the final classifiers for pre-MetS and MetS with powerful performance.
PMFs-based diagnostic models
The final PMF-based diagnostic models were constructed using all 60 hub PMFs, with age and gender correction, by 5-fold cross validation through the “glmnet” package. And the parameter optimization was performed by randomized searching and nested cross-validation through the “mlbench” package in R version 4.0.5. To further evaluate the classification performance of our models, ROC curves and AUCs were generated using the “pROC” package in R version 4.0.5. We also compared the model performance of 26 hub features with all the features of 303 PMFs for HC vs. MetS. Similarly, calibration curves35 were drafted according to the GLMNET regression results to assess the model reliability using the “ModelGood” package in R version 4.0.5. The individual benefits of our model were evaluated by decision curve analyses65 in R version 4.0.5.
Metabolomic clustering and assessment of MetS risk factors
Four were identified according to the gap-statistic calculated using the “cluster” packages in R version 4.0.5 to optimize the number of clusters. Unsupervised learning of 16 sub-clusters (RF0000–RF1111) containing different MetS risk factors with standardized 26 hub PMFs was then conducted using the K-means algorithm and hierarchical clustering though the “cluster” and “circlize” packages in R version 4.0.5 after dimensionality deduction with principal component (PC) analysis. The five MetS subgroups (RF0111, RF1011, RF1101, RF1110, and RF1111) and the HC group (RF0000) were clustered and visualized with the K-means algorithm with the “pheatmap” packages in R version 4.0.5. And the 26 hub PMFs were clustered into four metabolic modules (MP).
Construction and validation of plasma metabolic risk (PMR) stratification
The average intensity of 26 hub PMFs among all the five subgroups (RFN0-RFN4) according to the numbers of MetS risk factors were shown using the “ggplot2” package in R version 4.0.5. The variant analysis was performed using the “ggalluvial” package in R version 4.0.5 to track differences between the five subgroups (RFN0-RFN4) and sixteen sub-clusters (RF0000-RF1111) on behalf of the Sankey diagram. In order to ascertain the true metabolic status of individuals within a general population cohort, a PMR model was constructed using candidate factors such as age, gender, and 26 hub PMFs. The model was developed using the categorical boosting (CatBoost) algorithm, a widely used method for gradient boosting on decision trees.66 Three indexes (Index1-3) were calculated for each participant. By comparing the three index scores of everyone, all participants in the general population cohort were re-classified into three metabolic patterns: low-(LMR), medium-(MMR), and high (HMR) metabolic risk patterns, respectively. The relative change in different clinical parameters from the LMR pattern was calculated according to the following formula:
| (Equation 1) |
where represented the mean expression of each clinical parameter in the MMR or HMR pattern. Variant analysis was performed using the “ggalluvial” package in R version 4.0.5 to track differences between individual metabolic risk subgroups and traditional MetS classifications on behalf of the Sankey diagram. To validate the predictive significance of this PMR stratification, the scores of these three indexes were compared among the five subgroups (RFN0-RFN4) and three groups (HC, pre-MetS, and MetS) in the general population cohort using the Kruskal-Wallis test in the “ggpubr” package in R version 4.0.5. For the survival analysis of the 4-year mortality events, we conducted univariate Cox regression, multivariate Cox regression and Kaplan-Meier analysis using the “survival” package in R version 4.0.5. The univariate and multivariate Cox proportional hazards models were used to determine the Hazard Ratio (HR) of the variables. Models were tested statistically using the Likelihood test. Cumulative curves were calculated using the Kaplan-Meier method and log-rank test was used to examine the intergroup differences. p < 0.05 was considered to indicate a statistically significant difference.
Pathway analysis
The 26 hub PMFs were validated as a metabolite feature panel by accurate mass measurement through the human metabolome database Version 5.0 (http://www.hmdb.ca).67 Pathway topology analysis was conducted to explore the respective biochemical pathways among these differential metabolomic profiles using MetaboAnalyst 5.0 (http://www.metaboanalyst.ca).68
Quantification and statistical analysis
Statistical analysis was performed using R version 4.0.5. Descriptive data were presented as mean and standard deviation. The Shapiro-Wilk test was applied to test for a normal distribution and Levene’s test was used to test for homogeneity of variance. Differences among groups were tested by one-way analysis of variance (ANOVA) for parametric data such as age, and the Kruskal-Wallis rank-sum test was applied for non-parametric data, such as m/z signals. The Bonferroni test and Dunnett’s test were applied for multiple comparisons. Categorical data, such as gender, were presented as numbers and percentages and analyzed by χ2 tests. AUC was compared using a one-sided DeLong test. All significance levels were set at 5%.
Additional resources
This study is based the prospective Shanghai Community Cohort Establishment and Follow-up (NCT04517513): https://classic.clinicaltrials.gov/ct2/show/NCT04517513.
Acknowledgments
The authors gratefully thank the National Natural Science Foundation of China (82230014, 81930007, 81930007, 81800307, 81971771), Medical-Engineering Joint Funds of Shanghai Jiao Tong University (YG2021ZD09, YG2022QN107, YG2023ZD08), National Science Fund for Distinguished Young Scholars (81625002), Joint Project of Health Commission of Shanghai Pudong District (PW2019D-11), Science and Technology Commission of Shanghai Municipality (22JC1402100, 22QA1405400, 22DZ2292400, 201409005200), Shanghai Municipal Health Commission (2022JC013), Shanghai Cancer Institute (ZZ-20-22SYL), Shanghai Hospital Development Center (SHDC12022102), Shanghai Institutions of Higher Learning project (2021-01-07-00-02-E00083), National Research Center for Translational Medicine Shanghai (TMSK-2021-124, NRCTM(SH)-2021-06), and National High Level Hospital Clinical Research Funding (2022-PUMCH-C-023) for financial support. This work was also financially supported by grants 2022YFE0103500, 2021YFF0703500, 2021YFA0910100, and 2022YFC2502801 from the National Ministry of Science and Technology (MOST). We thank all the participants, the physicians, nurses, and research staff for their contributions to the Shanghai Community Cohort Establishment and Follow-up (NCT04517513). The authors extend their heartfelt appreciation to the staff members, including Xiaonan Kang, Zhuqing Wang, Kai Luo, Yumeng Liu, and others, from the Biobank of Renji Hospital.
Author contributions
Conceptualization, J.P., K.Q., and A.C.; cohort construction and investigation, W.Z., R.T., A.Y., Z.L., H.J., and L.H.; data analysis, Y.C., W.X., L.H., Y.X., Z.Z., and M.S.; resources, Y.C. and W.X.; data curation, B.M. and X.Z.; writing – original draft, Y.C., X.W., and W.Z.; writing – review & editing, J.P., K.Q., and A.C.; visualization, Y.C. and W.X.; supervision, J.P., K.Q., and A.C.; project administration, J.P.; funding acquisition, J.P.
Declaration of interests
The authors have filed patents for both the technology and the use of the technology to analyze biofluid samples.
Inclusion and diversity
We worked to ensure gender balance in the recruitment of human subjects. We worked to ensure ethnic or other types of diversity in the recruitment of human subjects. We worked to ensure that the study questionnaires were prepared in an inclusive way. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list. We avoided “helicopter science” practices by including the participating local contributors from the region where we conducted the research as authors on the paper.
Published: July 18, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2023.101109.
Contributor Information
Alex F. Chen, Email: chenfengyuan@xinhuamed.com.cn.
Kun Qian, Email: k.qian@sjtu.edu.cn.
Jun Pu, Email: pujun310@hotmail.com.
Supplemental information
Data and code availability
The standardized metabolomics data have been deposited to the NIH Common Fund’s National Metabolomics Data Repository (NMDR) Website, the Metabolomics Workbench, https://www.metabolomicsworkbench.org58 where it has been assigned Study ID: ST002733 (as listed in the key resources table). This paper does not report original code. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.
References
- 1.Hirode G., Wong R.J. Trends in the Prevalence of Metabolic Syndrome in the United States, 2011-2016. JAMA. 2020;323:2526–2528. doi: 10.1001/jama.2020.4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu J., Wang L., Li M., Xu Y., Jiang Y., Wang W., Li J., Mi S., Zhang M., Li Y., et al. Metabolic Syndrome Among Adults in China: The 2010 China Noncommunicable Disease Surveillance. J. Clin. Endocrinol. Metab. 2017;102:507–515. doi: 10.1210/jc.2016-2477. [DOI] [PubMed] [Google Scholar]
- 3.Gurka M.J., Golden S.H., Musani S.K., Sims M., Vishnu A., Guo Y., Cardel M., Pearson T.A., DeBoer M.D. Independent associations between a metabolic syndrome severity score and future diabetes by sex and race: the Atherosclerosis Risk In Communities Study and Jackson Heart Study. Diabetologia. 2017;60:1261–1270. doi: 10.1007/s00125-017-4267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang Y., Li J., Zheng X., Jiang Z., Hu S., Wadhera R.K., Bai X., Lu J., Wang Q., Li Y., et al. Risk Factors Associated With Major Cardiovascular Events 1 Year After Acute Myocardial Infarction. JAMA Netw. Open. 2018;1 doi: 10.1001/jamanetworkopen.2018.1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Esposito K., Chiodini P., Colao A., Lenzi A., Giugliano D. Metabolic syndrome and risk of cancer: a systematic review and meta-analysis. Diabetes Care. 2012;35:2402–2411. doi: 10.2337/dc12-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H., Zheng X., Zong X., Li Z., Li N., Hur J., Fritz C.D., Chapman W., Jr., Nickel K.B., Tipping A., et al. Metabolic syndrome, metabolic comorbid conditions and risk of early-onset colorectal cancer. Gut. 2021;70:1147–1154. doi: 10.1136/gutjnl-2020-321661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grundy S.M. Metabolic syndrome update. Trends Cardiovasc. Med. 2016;26:364–373. doi: 10.1016/j.tcm.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 8.Huang P.L. A comprehensive definition for metabolic syndrome. Dis. Model. Mech. 2009;2:231–237. doi: 10.1242/dmm.001180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saklayen M.G. The Global Epidemic of the Metabolic Syndrome. Curr. Hypertens. Rep. 2018;20:12. doi: 10.1007/s11906-018-0812-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grundy S.M., Cleeman J.I., Daniels S.R., Donato K.A., Eckel R.H., Franklin B.A., Gordon D.J., Krauss R.M., Savage P.J., Smith S.C., Jr., et al. Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation. 2005;112:2735–2752. doi: 10.1161/CIRCULATIONAHA.105.169404. [DOI] [PubMed] [Google Scholar]
- 11.Alberti K.G.M.M., Eckel R.H., Grundy S.M., Zimmet P.Z., Cleeman J.I., Donato K.A., Fruchart J.C., James W.P.T., Loria C.M., Smith S.C., Jr., et al. Harmonizing the metabolic syndrome: a joint interim statement of the International Diabetes Federation Task Force on Epidemiology and Prevention; National Heart, Lung, and Blood Institute; American Heart Association; World Heart Federation; International Atherosclerosis Society; and International Association for the Study of Obesity. Circulation. 2009;120:1640–1645. doi: 10.1161/CIRCULATIONAHA.109.192644. [DOI] [PubMed] [Google Scholar]
- 12.Alberti K.G.M.M., Zimmet P., Shaw J., IDF Epidemiology Task Force Consensus Group The metabolic syndrome--a new worldwide definition. Lancet. 2005;366:1059–1062. doi: 10.1016/S0140-6736(05)67402-8. [DOI] [PubMed] [Google Scholar]
- 13.Expert Panel on Detection, E., and Treatment of High Blood Cholesterol in, Expert Panel on Detection Evaluation and Treatment of High Blood Cholesterol in Adults (2001). Executive Summary of The Third Report of The National Cholesterol Education Program (NCEP) Expert Panel on Detection, Evaluation, And Treatment of High Blood Cholesterol In Adults (Adult Treatment Panel III). JAMA 285, 2486-2497. 10.1001/jama.285.19.2486. [DOI] [PubMed]
- 14.Alberti K.G., Zimmet P.Z. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet. Med. 1998;15:539–553. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 15.Hosseinpanah F., Asghari G., Barzin M., Golkashani H.A., Azizi F. Prognostic impact of different definitions of metabolic syndrome in predicting cardiovascular events in a cohort of non-diabetic Tehranian adults. Int. J. Cardiol. 2013;168:369–374. doi: 10.1016/j.ijcard.2012.09.037. [DOI] [PubMed] [Google Scholar]
- 16.Athyros V.G., Ganotakis E.S., Tziomalos K., Papageorgiou A.A., Anagnostis P., Griva T., Kargiotis K., Mitsiou E.K., Karagiannis A., Mikhailidis D.P. Comparison of four definitions of the metabolic syndrome in a Greek (Mediterranean) population. Curr. Med. Res. Opin. 2010;26:713–719. doi: 10.1185/03007991003590597. [DOI] [PubMed] [Google Scholar]
- 17.Herath H.M.M., Weerasinghe N.P., Weerarathna T.P., Amarathunga A. A Comparison of the Prevalence of the Metabolic Syndrome among Sri Lankan Patients with Type 2 Diabetes Mellitus Using WHO, NCEP-ATP III, and IDF Definitions. Int. J. Chronic Dis. 2018;2018 doi: 10.1155/2018/7813537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kassi E., Pervanidou P., Kaltsas G., Chrousos G. Metabolic syndrome: definitions and controversies. BMC Med. 2011;9:48. doi: 10.1186/1741-7015-9-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sperling L.S., Mechanick J.I., Neeland I.J., Herrick C.J., Després J.P., Ndumele C.E., Vijayaraghavan K., Handelsman Y., Puckrein G.A., Araneta M.R.G., et al. The CardioMetabolic Health Alliance: Working Toward a New Care Model for the Metabolic Syndrome. J. Am. Coll. Cardiol. 2015;66:1050–1067. doi: 10.1016/j.jacc.2015.06.1328. [DOI] [PubMed] [Google Scholar]
- 20.Holmes E., Loo R.L., Stamler J., Bictash M., Yap I.K.S., Chan Q., Ebbels T., De Iorio M., Brown I.J., Veselkov K.A., et al. Human metabolic phenotype diversity and its association with diet and blood pressure. Nature. 2008;453:396–400. doi: 10.1038/nature06882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alshehry Z.H., Mundra P.A., Barlow C.K., Mellett N.A., Wong G., McConville M.J., Simes J., Tonkin A.M., Sullivan D.R., Barnes E.H., et al. Plasma Lipidomic Profiles Improve on Traditional Risk Factors for the Prediction of Cardiovascular Events in Type 2 Diabetes Mellitus. Circulation. 2016;134:1637–1650. doi: 10.1161/CIRCULATIONAHA.116.023233. [DOI] [PubMed] [Google Scholar]
- 22.Ussher J.R., Elmariah S., Gerszten R.E., Dyck J.R.B. The Emerging Role of Metabolomics in the Diagnosis and Prognosis of Cardiovascular Disease. J. Am. Coll. Cardiol. 2016;68:2850–2870. doi: 10.1016/j.jacc.2016.09.972. [DOI] [PubMed] [Google Scholar]
- 23.Monnerie S., Comte B., Ziegler D., Morais J.A., Pujos-Guillot E., Gaudreau P. Metabolomic and Lipidomic Signatures of Metabolic Syndrome and its Physiological Components in Adults: A Systematic Review. Sci. Rep. 2020;10:669. doi: 10.1038/s41598-019-56909-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Y., Wang Y., Zhuang Y., Zhang P., Chen S., Asakawa T., Gao B. Serum Metabolomic Profiles Associated With Untreated Metabolic Syndrome Patients in the Chinese Population. Clin. Transl. Sci. 2020;13:1271–1278. doi: 10.1111/cts.12817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fukui M., Tanaka M., Toda H., Asano M., Yamazaki M., Hasegawa G., Imai S., Nakamura N. High plasma 5-hydroxyindole-3-acetic acid concentrations in subjects with metabolic syndrome. Diabetes Care. 2012;35:163–167. doi: 10.2337/dc11-1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin Z., Vicente Gonçalves C.M., Dai L., Lu H.M., Huang J.H., Ji H., Wang D.S., Yi L.Z., Liang Y.Z. Exploring metabolic syndrome serum profiling based on gas chromatography mass spectrometry and random forest models. Anal. Chim. Acta. 2014;827:22–27. doi: 10.1016/j.aca.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 27.Wishart D.S. Emerging applications of metabolomics in drug discovery and precision medicine. Nat. Rev. Drug Discov. 2016;15:473–484. doi: 10.1038/nrd.2016.32. [DOI] [PubMed] [Google Scholar]
- 28.Xiang K., Xiang H., Yang W., Jia W., Qian R., Weng J. Diagnosis and management of the metabolic syndrome: a Chinese Diabetes Society Scientific Statement. Chin J Diabetes. 2004:156–161. (in Chinese) [Google Scholar]
- 29.Xu H., Song Y., You N.C., Zhang Z.F., Greenland S., Ford E.S., He L., Liu S. Prevalence and clustering of metabolic risk factors for type 2 diabetes among Chinese adults in Shanghai, China. BMC Publ. Health. 2010;10:683. doi: 10.1186/1471-2458-10-683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang L., Wang L., Hu X., Chen S., Tao Y., Su H., Yang J., Xu W., Vedarethinam V., Wu S., et al. Machine learning of serum metabolic patterns encodes early-stage lung adenocarcinoma. Nat. Commun. 2020;11:3556. doi: 10.1038/s41467-020-17347-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forslund S.K., Chakaroun R., Zimmermann-Kogadeeva M., Markó L., Aron-Wisnewsky J., Nielsen T., Moitinho-Silva L., Schmidt T.S.B., Falony G., Vieira-Silva S., et al. Combinatorial, additive and dose-dependent drug-microbiome associations. Nature. 2021;600:500–505. doi: 10.1038/s41586-021-04177-9. [DOI] [PubMed] [Google Scholar]
- 32.Guo L., Milburn M.V., Ryals J.A., Lonergan S.C., Mitchell M.W., Wulff J.E., Alexander D.C., Evans A.M., Bridgewater B., Miller L., et al. Plasma metabolomic profiles enhance precision medicine for volunteers of normal health. Proc. Natl. Acad. Sci. USA. 2015;112:E4901–E4910. doi: 10.1073/pnas.1508425112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matsumoto M., Harada S., Iida M., Kato S., Sata M., Hirata A., Kuwabara K., Takeuchi A., Sugiyama D., Okamura T., Takebayashi T. Validity Assessment of Self-reported Medication Use for Hypertension, Diabetes, and Dyslipidemia in a Pharmacoepidemiologic Study by Comparison With Health Insurance Claims. J. Epidemiol. 2021;31:495–502. doi: 10.2188/jea.JE20200089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Amini B., Bassett R.L., Jr., Haygood T.M., McEnery K.W., Richardson M.L. Confidence Calibration: An Introduction With Application to Quality Improvement. J. Am. Coll. Radiol. 2020;17:620–628. doi: 10.1016/j.jacr.2019.12.009. [DOI] [PubMed] [Google Scholar]
- 35.Alba A.C., Agoritsas T., Walsh M., Hanna S., Iorio A., Devereaux P.J., McGinn T., Guyatt G. Discrimination and Calibration of Clinical Prediction Models: Users' Guides to the Medical Literature. JAMA. 2017;318:1377–1384. doi: 10.1001/jama.2017.12126. [DOI] [PubMed] [Google Scholar]
- 36.Fan Y., Li Y., Chen Y., Zhao Y.J., Liu L.W., Li J., Wang S.L., Alolga R.N., Yin Y., Wang X.M., et al. Comprehensive Metabolomic Characterization of Coronary Artery Diseases. J. Am. Coll. Cardiol. 2016;68:1281–1293. doi: 10.1016/j.jacc.2016.06.044. [DOI] [PubMed] [Google Scholar]
- 37.Chen X.W., Ding G., Xu L., Li P. A glimpse at the metabolic research in China. Cell Metabol. 2021;33:2122–2125. doi: 10.1016/j.cmet.2021.09.014. [DOI] [PubMed] [Google Scholar]
- 38.Wang-Sattler R., Yu Z., Herder C., Messias A.C., Floegel A., He Y., Heim K., Campillos M., Holzapfel C., Thorand B., et al. Novel biomarkers for pre-diabetes identified by metabolomics. Mol. Syst. Biol. 2012;8:615. doi: 10.1038/msb.2012.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suvitaival T., Bondia-Pons I., Yetukuri L., Pöhö P., Nolan J.J., Hyötyläinen T., Kuusisto J., Orešič M. Lipidome as a predictive tool in progression to type 2 diabetes in Finnish men. Metabolism. 2018;78:1–12. doi: 10.1016/j.metabol.2017.08.014. [DOI] [PubMed] [Google Scholar]
- 40.Yifan C., Jun P. Understanding the Clinical Features of Coronavirus Disease 2019 From the Perspective of Aging: A Systematic Review and Meta-Analysis. Front. Endocrinol. 2020;11 doi: 10.3389/fendo.2020.557333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shim K., Gulhar R., Jialal I. Exploratory metabolomics of nascent metabolic syndrome. J. Diabet. Complicat. 2019;33:212–216. doi: 10.1016/j.jdiacomp.2018.12.002. [DOI] [PubMed] [Google Scholar]
- 42.Antonio L., Wu F.C.W., O'Neill T.W., Pye S.R., Carter E.L., Finn J.D., Rutter M.K., Laurent M.R., Huhtaniemi I.T., Han T.S., et al. Associations between sex steroids and the development of metabolic syndrome: a longitudinal study in European men. J. Clin. Endocrinol. Metab. 2015;100:1396–1404. doi: 10.1210/jc.2014-4184. [DOI] [PubMed] [Google Scholar]
- 43.Wu Q., Li J., Sun X., He D., Cheng Z., Li J., Zhang X., Xie Y., Zhu Y., Lai M. Multi-stage metabolomics and genetic analyses identified metabolite biomarkers of metabolic syndrome and their genetic determinants. EBioMedicine. 2021;74 doi: 10.1016/j.ebiom.2021.103707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garcia-Perez I., Posma J.M., Serrano-Contreras J.I., Boulangé C.L., Chan Q., Frost G., Stamler J., Elliott P., Lindon J.C., Holmes E., Nicholson J.K. Identifying unknown metabolites using NMR-based metabolic profiling techniques. Nat. Protoc. 2020;15:2538–2567. doi: 10.1038/s41596-020-0343-3. [DOI] [PubMed] [Google Scholar]
- 45.Bian Y., Zheng R., Bayer F.P., Wong C., Chang Y.C., Meng C., Zolg D.P., Reinecke M., Zecha J., Wiechmann S., et al. Robust, reproducible and quantitative analysis of thousands of proteomes by micro-flow LC-MS/MS. Nat. Commun. 2020;11:157. doi: 10.1038/s41467-019-13973-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Alseekh S., Aharoni A., Brotman Y., Contrepois K., D'Auria J., Ewald J., C Ewald J., Fraser P.D., Giavalisco P., Hall R.D., et al. Mass spectrometry-based metabolomics: a guide for annotation, quantification and best reporting practices. Nat. Methods. 2021;18:747–756. doi: 10.1038/s41592-021-01197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu B., Chen G., Zhao R., Huang D., Tao L. Temporal trends in the prevalence of metabolic syndrome among middle-aged and elderly adults from 2011 to 2015 in China: the China health and retirement longitudinal study (CHARLS) BMC Publ. Health. 2021;21:1045. doi: 10.1186/s12889-021-11042-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Surowiec I., Noordam R., Bennett K., Beekman M., Slagboom P.E., Lundstedt T., van Heemst D. Metabolomic and lipidomic assessment of the metabolic syndrome in Dutch middle-aged individuals reveals novel biological signatures separating health and disease. Metabolomics. 2019;15:23. doi: 10.1007/s11306-019-1484-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olszanecka A., Kawecka-Jaszcz K., Czarnecka D. Association of free testosterone and sex hormone binding globulin with metabolic syndrome and subclinical atherosclerosis but not blood pressure in hypertensive perimenopausal women. Arch. Med. Sci. 2016;12:521–528. doi: 10.5114/aoms.2016.59925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Asgharnezhad M., Joukar F., Naghipour M., Nikbakht H.A., Hassanipour S., Arab-Zozani M., Mansour-Ghanaei F. Exploratory factor analysis of gender-based metabolic syndrome components: Results from the PERSIAN Guilan cohort study (PGCS) Clin. Nutr. ESPEN. 2020;40:252–256. doi: 10.1016/j.clnesp.2020.09.011. [DOI] [PubMed] [Google Scholar]
- 51.Nichols G.A., Moler E.J. Metabolic syndrome components are associated with future medical costs independent of cardiovascular hospitalization and incident diabetes. Metab. Syndr. Relat. Disord. 2011;9:127–133. doi: 10.1089/met.2010.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Diabetes Canada Clinical Practice Guidelines Expert Committee. Punthakee Z., Goldenberg R., Katz P. Definition, Classification and Diagnosis of Diabetes, Prediabetes and Metabolic Syndrome. Can. J. Diabetes. 2018;42(Suppl 1):S10–S15. doi: 10.1016/j.jcjd.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 53.Engin A. The Definition and Prevalence of Obesity and Metabolic Syndrome. Adv. Exp. Med. Biol. 2017;960:1–17. doi: 10.1007/978-3-319-48382-5_1. [DOI] [PubMed] [Google Scholar]
- 54.Ortega F.B., Lee D.C., Katzmarzyk P.T., Ruiz J.R., Sui X., Church T.S., Blair S.N. The intriguing metabolically healthy but obese phenotype: cardiovascular prognosis and role of fitness. Eur. Heart J. 2013;34:389–397. doi: 10.1093/eurheartj/ehs174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Suzuki T., Zeng Z., Zhao B., Wei Z., Tanabe M., Shimbo T., Kajio H., Kato N., Naruse M. Comparison of coronary heart disease risk among four diagnostic definitions of metabolic syndrome. J. Endocrinol. Invest. 2016;39:1337–1346. doi: 10.1007/s40618-016-0538-1. [DOI] [PubMed] [Google Scholar]
- 56.Hoang K.C., Ghandehari H., Lopez V.A., Barboza M.G., Wong N.D. Global coronary heart disease risk assessment of individuals with the metabolic syndrome in the. Diabetes Care. 2008;31:1405–1409. doi: 10.2337/dc07-2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou J., Gao Q., Wang J., Zhang M., Ma J., Wang C., Chen H., Peng X., Hao L. Comparison of coronary heart disease risk assessments among individuals with metabolic syndrome using three diagnostic definitions: a cross-sectional study from China. BMJ Open. 2018;8 doi: 10.1136/bmjopen-2018-022974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sud M., Fahy E., Cotter D., Azam K., Vadivelu I., Burant C., Edison A., Fiehn O., Higashi R., Nair K.S., et al. Metabolomics Workbench: An international repository for metabolomics data and metadata, metabolite standards, protocols, tutorials and training, and analysis tools. Nucleic Acids Res. 2016;44:D463–D470. doi: 10.1093/nar/gkv1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.World Medical Association World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA. 2013;310:2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 60.de las Fuentes L., Brown A.L., Mathews S.J., Waggoner A.D., Soto P.F., Gropler R.J., Dávila-Román V.G. Metabolic syndrome is associated with abnormal left ventricular diastolic function independent of left ventricular mass. Eur. Heart J. 2007;28:553–559. doi: 10.1093/eurheartj/ehl526. [DOI] [PubMed] [Google Scholar]
- 61.Vidigal F.d.C., Ribeiro A.Q., Babio N., Salas-Salvadó J., Bressan J. Prevalence of metabolic syndrome and pre-metabolic syndrome in health professionals: LATINMETS Brazil study. Diabetol. Metab. Syndrome. 2015;7:6. doi: 10.1186/s13098-015-0003-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gesteiro E., Megía A., Guadalupe-Grau A., Fernandez-Veledo S., Vendrell J., González-Gross M. Early identification of metabolic syndrome risk: A review of reviews and proposal for defining pre-metabolic syndrome status. Nutr. Metabol. Cardiovasc. Dis. 2021;31:2557–2574. doi: 10.1016/j.numecd.2021.05.022. [DOI] [PubMed] [Google Scholar]
- 63.Zhu Y., Qiao L., Prudent M., Bondarenko A., Gasilova N., Möller S.B., Lion N., Pick H., Gong T., Chen Z., et al. Sensitive and fast identification of bacteria in blood samples by immunoaffinity mass spectrometry for quick BSI diagnosis. Chem. Sci. 2016;7:2987–2995. doi: 10.1039/c5sc04919a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Obuchowski N.A., McClish D.K. Sample size determination for diagnostic accuracy studies involving binormal ROC curve indices. Stat. Med. 1997;16:1529–1542. doi: 10.1002/(sici)1097-0258(19970715)16:13<1529::aid-sim565>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 65.Vickers A.J., van Calster B., Steyerberg E.W. A simple, step-by-step guide to interpreting decision curve analysis. Diagn. Progn. Res. 2019;3:18. doi: 10.1186/s41512-019-0064-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hancock J.T., Khoshgoftaar T.M. CatBoost for big data: an interdisciplinary review. J. Big Data. 2020;7:94. doi: 10.1186/s40537-020-00369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wishart D.S., Guo A., Oler E., Wang F., Anjum A., Peters H., Dizon R., Sayeeda Z., Tian S., Lee B.L., et al. HMDB 5.0: the Human Metabolome Database for 2022. Nucleic Acids Res. 2022;50:D622–D631. doi: 10.1093/nar/gkab1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pang Z., Chong J., Zhou G., de Lima Morais D.A., Chang L., Barrette M., Gauthier C., Jacques P.É., Li S., Xia J. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49:W388–W396. doi: 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The standardized metabolomics data have been deposited to the NIH Common Fund’s National Metabolomics Data Repository (NMDR) Website, the Metabolomics Workbench, https://www.metabolomicsworkbench.org58 where it has been assigned Study ID: ST002733 (as listed in the key resources table). This paper does not report original code. Any additional information required to reanalyze the data reported in this work paper is available from the lead contact upon request.