Abstract
Actinomorphic flowers usually orient vertically (relative to the horizon) and possess symmetric nectar guides, while zygomorphic flowers often face horizontally and have asymmetric nectar guides, indicating that floral symmetry, floral orientation, and nectar guide patterning are correlated. The origin of floral zygomorphy is dependent on the dorsoventrally asymmetric expression of CYCLOIDEA (CYC)-like genes. However, how horizontal orientation and asymmetric nectar guides are achieved remains poorly understood. Here, we selected Chirita pumila (Gesneriaceae) as a model plant to explore the molecular bases for these traits. By analyzing gene expression patterns, protein–DNA and protein–protein interactions, and encoded protein functions, we identified multiple roles and functional divergence of 2 CYC-like genes, i.e. CpCYC1 and CpCYC2, in controlling floral symmetry, floral orientation, and nectar guide patterning. CpCYC1 positively regulates its own expression, whereas CpCYC2 does not regulate itself. In addition, CpCYC2 upregulates CpCYC1, while CpCYC1 downregulates CpCYC2. This asymmetric auto-regulation and cross-regulation mechanism might explain the high expression levels of only 1 of these genes. We show that CpCYC1 and CpCYC2 determine asymmetric nectar guide formation, likely by directly repressing the flavonoid synthesis-related gene CpF3′5′H. We further suggest that CYC-like genes play multiple conserved roles in Gesneriaceae. These findings shed light on the repeated origins of zygomorphic flowers in angiosperms.
CYCLOIDEA-like genes play multiple roles in determining floral symmetry, floral orientation, and nectar guide patterning in the model plant Chirita pumila and likely other Gesneriaceae species.
In a Nutshell.
Background: Three types of floral symmetry can be distinguished based on the number of symmetry planes: polysymmetry (with several symmetry planes), monosymmetry (with only 1 symmetry plane), and asymmetry. Early angiosperms have polysymmetric floral organs, while monosymmetry originated many times from polysymmetry, and several large clades produce predominantly or entirely monosymmetric flowers. In addition to having differential morphologies and sizes in the second and third whorls of floral organs, monosymmetric flowers usually possess horizontal orientation and asymmetric nectar guides. CYCLOIDEA (CYC)-like TCP transcription factors control floral monosymmetry in many species, but little is known about how horizontal orientation and asymmetric nectar guides are achieved.
Question: Are floral symmetry, floral orientation, and nectar guide patterning correlated traits, and are they determined by the same set of master regulators, such as CYC-like genes?
Findings: We selected Chirita pumila (Gesneriaceae) as a model system to address this issue. Plants overexpressing CpCYC1 and CpCYC2 generated dorsalized flowers, with a change in floral orientation from horizontal to upward and the loss of yellow nectar guides in the ventral corolla tube. By contrast, the cyc1 cyc2 double mutant produced ventralized flowers with upward orientation and uniform yellow nectar guides. Therefore, CpCYC1 and CpCYC2 not only determine floral symmetry, but also regulate floral orientation and nectar guide patterning. CpCYC1 positively regulates itself and downregulates CpCYC2, while CpCYC2 upregulates CpCYC1. We also identified the flavonoid biosynthesis-related gene CpF3′5′H, which regulates yellow nectar guide formation. CpCYC1 and CpCYC2 repress yellow pigment formation in nectar guides out the ventral region of the flower, likely by directly repressing CpF3′5′H.
Next steps: Further studies are needed to elucidate how CpCYC1 and CpCYC2 control these distinct floral traits by regulating different target genes or interacting with different cofactors.
Introduction
Three key innovations in angiosperms have been identified based on the fossil records of early flowering plants, including the showy radially symmetrical (polysymmetric or actinomorphic) flower with closed carpels, the bilaterally symmetrical (monosymmetric or zygomorphic) flower, and fleshy fruits and nutritious nuts and seeds (Dilcher 2000). While actinomorphic flowers have 2 or more symmetry planes, zygomorphic flowers only have 1 (Endress 2001) due to the different morphologies and sizes of floral organs, especially those in the second and third whorls. In addition, zygomorphic flowers usually have a long and narrow corolla tube that restricts certain types of pollinators and a specific entrance indicated by a nectar or pollen guide that helps pollinators access and forage nectar or pollen deep in the flower (Neal et al. 1998). The complexity of zygomorphic flowers enhances the efficiency of precision pollination by directing specialized pollinators to approach and visit them. In the model plant snapdragon (Antirrhinum majus), CYCLOIDEA (CYC) and DICHOTOMA (DICH), 2 paralogous genes encoding the plant-specific TCP family of transcription factors, have been identified as key regulators of floral symmetry due to their dorsal-specific expression patterns in flowers (Luo et al. 1996, 1999). A growing number of studies in many zygomorphic species have indicated that CYC-like genes are widely involved in controlling floral zygomorphy in angiosperms, especially in core eudicots (Hileman et al. 2003; Feng et al. 2006; Busch and Zachgo 2007; Gao et al. 2008; Wang et al. 2008; Song et al. 2009; Yang et al. 2012, 2015; Su et al. 2017; Chen et al. 2018; Dong et al. 2018; Hsin et al. 2019; Tong et al. 2022).
The origin of floral zygomorphy is usually accompanied by the occurrence of other floral traits, such as floral orientation, because natural selection acts on whole organisms rather than isolated parts in order to generate organisms with integrated suites of traits or integrated phenotypes (Santos and Cannatella 2011; Murren 2012; Armbruster et al. 2014; Kern et al. 2016; Bawa et al. 2019; Sinnott-Armstrong et al. 2020). Floral orientation, defined as the angle between the main axis of the corolla and the horizon, can be divided into 3 basic types: upward, horizontal, and downward (Hodges et al. 2002). While actinomorphic flowers often face vertically, zygomorphic ones usually orient horizontally (Neal et al. 1998; Ushimaru and Hyodo 2005; Ushimaru et al. 2009), which is indicative of the correlated evolution of floral symmetry and floral orientation. Horizontal orientation might restrict the movement of visiting insects to facilitate their recognition of complex floral patterns (the recognition–facilitation hypothesis) or might direct the landing of insect pollinators to flowers (the landing–controlling hypothesis) (Neal et al. 1998; Ushimaru and Hyodo 2005; Ushimaru et al. 2007, 2009). Fenster et al. (2009) proposed that the horizontal orientation of flowers represents the first step toward the evolution of floral zygomorphy. However, how horizontal orientation is established at the molecular level is still poorly understood (Dong et al. 2018).
In addition to floral orientation, the change of floral symmetry is also associated with the elaboration of floral color patterns. Floral color patterns, which are common in angiosperms, are believed to enhance the pollinator's ability to detect flowers, move into the flower center, and orient toward floral rewards to increase pollination efficiency (Dafni and Kevan 1996; Hansen et al. 2012; Koski and Ashman 2014). Flower color that appears uniform to humans may appear patterned to insects due to spatial variation in ultraviolet (UV) reflectance on petals. A circular “bullseye” pattern (either in the human-visible or UV spectrum), with petal bases and apices displaying different spectral features, is frequently adopted by actinomorphic species or Asteraceae plants, which produce seemly “actinomorphic” capitula (Koski and Ashman 2014; Moyers et al. 2017; Koski et al. 2020). This “bullseye” pattern, which usually acts as a nectar guide, can attract pollinators and direct their access to the flower center where nectar is secreted. By contrast, zygomorphic flowers usually have an asymmetric nectar guide pattern due to the exclusively distributed nectar guides in specific floral regions, usually in ventral petals (Yuan et al. 2013; Jachuła et al. 2018; Hsu and Kuo 2021), pointing to a general correlation between floral symmetry and nectar guide patterns. Given the importance of nectar guides in mediating plant–pollinator interactions, we still know little about the molecular mechanism underlying their formation (Yuan et al. 2013; Demarche et al. 2015), let alone how the asymmetric nectar guide pattern is achieved at the molecular level.
The Gesneriaceae family, 1 of basal-most groups in Lamiales, is characterized by having numerous members with zygomorphic flowers due to the dorsoventrally asymmetric expression of CYC-like genes (Gao et al. 2008; Song et al. 2009; Yang et al. 2012, 2015; Dong et al. 2018; Hsin et al. 2019). We recently reported that CYC-like genes function in horizontal orientation and floral pigmentation in Gesneriaceae (Dong et al. 2018; Liu, Wang, et al. 2021), suggesting that they would also be ideal candidates for deciphering the molecular mechanisms of floral orientation and nectar guide patterning. We reasoned that an integrated study of floral symmetry, floral orientation, and nectar guide patterns could provide comprehensive functional annotation of CYC-like genes in these distinct floral traits.
In this study, we selected Chirita pumila, a model for the evolutionary biology of floral development (Liu et al. 2014; Liu, Wang, et al. 2021; Liu, Wu, et al. 2021), to assess the molecular mechanism underlying the association of floral symmetry, floral orientation, and nectar guide patterning. We demonstrate that 2 CYC-like genes have undergone expression divergence in that CpCYC1 is specifically transcribed in dorsal petals and dorsal/lateral stamens, the dorsal corolla tube, and the dorsal part of the receptacle, while CpCYC2 has a narrower expression domain and a much lower expression level. The expression divergence of these genes is in line with their functional differences, as confirmed by overexpression, dominant repression using the SRDX motif, and CRISPR/Cas9-mediated gene editing. We further show that CpCYC1 and CpCYC2 undergo asymmetric auto-regulation and cross-regulation, as CpCYC1 positively regulates itself and is activated by CpCYC2, while CpCYC2 is negatively regulated by CpCYC1. CpCYC1 and CpCYC2 inhibit the formation of yellow nectar guides out of the ventral region of the flower, likely by directly repressing a flavonoid synthesis-related gene. Comparative expression and functional analyses in a relative of C. pumila indicated that CYC-like genes share a conserved function in controlling the association of floral symmetry, floral orientation, and nectar guide patterning in Gesneriaceae. These findings provide important insights into how the repeated origins of zygomorphic flowers are achieved by recruiting 1 type of pleiotropic master regulators.
Results
C. pumila produces zygomorphic flowers with horizontal orientation and an asymmetric nectar guide pattern
C. pumila generates typical zygomorphic flowers with monosymmetric floral organs in the second and third whorls (Figs. 1 and S1). In the second whorl, dorsal and ventral petals are shorter than lateral ones, while dorsal petals are wider than lateral and ventral ones (Figs. 1A and S1, A and B; Supplemental Data Set 1). As a result, both dorsal and lateral petals are larger than ventral ones (Supplemental Fig. S1B). Scanning electron microscopy (SEM) showed that dorsal petal cells are larger than lateral and ventral petal cells in the adaxial epidermis (Supplemental Fig. S1, C and D). In the third whorl, the development of both dorsal and lateral stamens is delayed to different degrees (Fig. 1, E and F). SEM showed that floral zygomorphy is established gradually, as petal and stamen primordia in dorsal, lateral, and ventral regions are similar in size during early stages of flower development (Fig. 1, I to L). Subsequently, the growth rates of dorsal and lateral stamens are greatly reduced once the second whorl of floral organs starts to form a corolla tube (Fig. 1M). The difference among dorsal, lateral, and ventral stamens becomes increasingly evident during flower development (Fig. 1, N to R). As a result, only 2 ventral stamens can develop normally and generate fertile anthers (Fig. 1R).
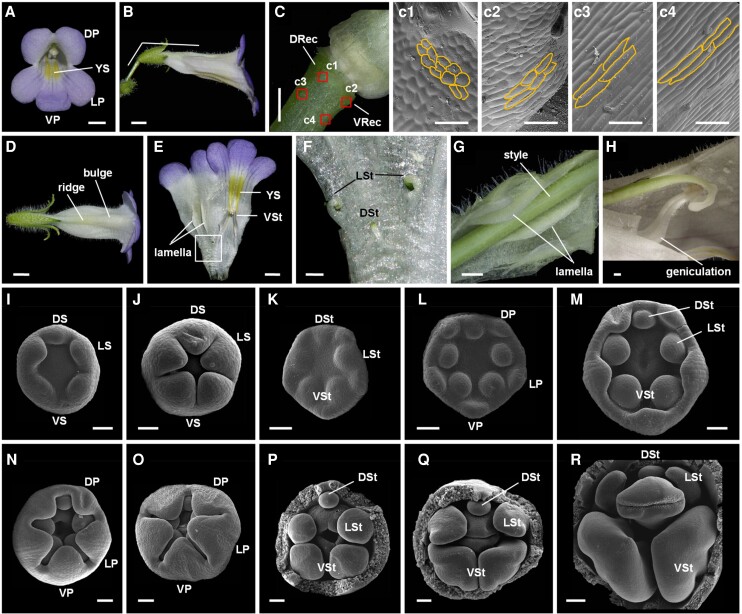
Figure 1.
Flowers of wild-type C. pumila. A) Front view of a flower showing dorsal/lateral/ventral petals (DP/LP/VP) and yellow spots (YS). Scale bar: 0.5 cm. B) The lateral view of the flower showing the horizontal orientation. Scale bar: 0.5 cm. C) The asymmetric development of dorsal and ventral parts of the receptacle (DRec and VRec) with c1 to c4 showing cells in different regions. Scale bars: C) 0.5 cm; c1 to c4, 50 µm. D) Dorsal view of the flower showing the ridge and bulge structures in the dorsal corolla tube. Scale bar: 0.5 cm. E and F) The inner structure of the flower showing 2 lamellae, YS, ventral stamens (VSt), and dorsal/lateral staminodes (DSt/LSt). Scale bars: E) 0.5 cm; F) 0.1 cm. G) The style fits perfectly between 2 lamellae. Scale bar: 0.1 cm. H) The geniculate filament of the ventral stamens. Scale bar: 0.1 cm. I to R) Floral development revealed by SEM. DS/LS/VS, dorsa/lateral/ventral sepals. Scale bars: 50 µm.
We observed that C. pumila zygomorphic flowers always orientate horizontally due to the asymmetrical development of dorsal and ventral parts of the receptacle (an enlarged structure just below the flower, distinguished from the pedicel; Fig. 1, B and C). The ventral part of the receptacle contains short rod-like cells that are regularly arranged in the direction of the pedicle (Fig. 1, c2 to c4). On the contrary, the dorsal part of the receptacle contains rounder and shorter cells that arrange almost perpendicularly to the pedicle (Fig. 1, c1). Therefore, the horizontal orientation of C. pumila zygomorphic flowers is likely due to the different directions of cell expansion in the dorsal and ventral parts of the receptacle.
Zygomorphic flowers of C. pumila are also characterized by yellow pigments exclusively distributed near the throat of ventral corolla tube (Fig. 1, A and E), leading to an asymmetric nectar guide pattern. Interestingly, the yellow spots absorbed UV light, sharply contrasting with their surrounding regions (Supplemental Fig. S2, A and B). Given that most insects can perceive UV light (Briscoe and Chittka 2001), the asymmetric yellow spots should be perceptible to insects under both visible and UV light to direct them to the nectar secreted from the circular nectary located at the base of the ovary (Supplemental Fig. S2C).
In addition, C. pumila flowers contain a ridge structure in the middle region of the dorsal corolla tube and a bulge structure near the dorsal petal lobe (Fig. 1D). Inside the dorsal corolla tube, 2 lamellae form, corresponding to the outside ridge structure. Both the inside lamellae and the outside ridge generate a groove structure in which the style perfectly fits and is therefore fixed (Fig. 1, E and G), and the bulge structure provides a relatively wide space for the stigma (Fig. 1A). The filaments of 2 ventral stamens are geniculate at the middle region and thus push the 2 cohesive anthers to the abaxial side of the stigma lobe (Fig. 1H).
Expression patterns of CpCYC1 and CpCYC2
Given the series of traits that accompany zygomorphy in C. pumila flowers, it is important to examine whether these traits are determined by the same set of genes controlling floral symmetry, e.g. CYC-like TCP genes. We previously isolated only 1 CYC-like gene from C. pumila by similarity-based cloning (Liu, Wang, et al. 2021; Liu, Wu, et al. 2021). Here, using the local BLAST tool, we retrieved a total of 2 CYC-like genes from the whole genome sequence of C. pumila. Phylogenetic analysis revealed that these 2 CYC-like genes are orthologs of snapdragon AmCYC/DICH genes. These genes respectively clustered with the Gesneriaceae CYC1 (GCYC1) and GCYC2 lineages (Supplemental Fig. S3 and Data Set 2). Therefore, we named these genes CpCYC1 and CpCYC2. In addition, it seems that C. pumila has lost orthologs of GCYC1C and GCYC2B. A similar phenomenon was reported in Bournea leiophylla (Gesneriaceae) in which GCYC1D and GCYC2A were lost (Zhou et al. 2008; Supplemental Fig. S3). CpCYC1 and CpCYC2 encode proteins with 357 and 330 amino acid residues, respectively, both of which exclusively localize to the nucleus (Supplemental Fig. S4). Both proteins contain a DNA-binding TCP domain and an arginine-rich R domain, which are typical of CYC-like TCP transcription factors (Cubas et al. 1999; Supplemental Fig. S5). Notably, both the TCP and R domains of CpCYC1 and CpCYC2 are different, with 2 and 1 linage-specific amino acid residue difference, respectively, suggesting these proteins might have divergent functions (Supplemental Fig. S5).
CpCYC1 transcripts were first detected across the apex of young floral meristems via in situ hybridization (Fig. 2A). When sepal primordia emerged, CpCYC1 transcripts were detected in both dorsal and ventral sepal primordia (Fig. 2B). Subsequently, CpCYC1 signals were distributed extensively in all petal and stamen primordia (Fig. 2C). With the differentiation of dorsal/lateral/ventral stamens, CpCYC1 transcription signals in petals were greatly reduced and were mainly restricted to the adaxial epidermis of dorsal petals (Fig. 2, D and E). In the third whorl, CpCYC1 transcript levels were lower in ventral stamens than in lateral and dorsal stamens and were ultimately undetectable (Fig. 2, D and E). The expression patterns of CpCYC2 were basically similar to those of CpCYC1, except for a much weaker difference in signal among dorsal/lateral/ventral stamens (Fig. 2, G to K). As negative controls, sense probes for both CpCYC1 and CpCYC2 generated no signal (Fig. 2, F and L).
Figure 2.
Expression patterns of CpCYC1 and CpCYC2. A to E) The results of in situ hybridization using CpCYC1 antisense probe. CpCYC1 was expressed across floral meristems (FM; A and B) and the dorsal/lateral/ventral petal and stamen primordia (DP/LP/VP, DSt/LSt/VSt; C). Its signal then decreased in VSt D) and finally was specifically distributed in DP and DSt/LSt E). Scale bars: 50 µm. F) No signal was detected using CpCYC1 sense probe. Scale bar: 50 µm. G to K) The expression pattern of CpCYC2 was similar to that of CpCYC1 at early stages G to J), but its signal did not disappear in VSt at late stages of floral development K). Scale bars: 50 µm. L) No signal was detected using the CpCYC2 sense probe. Scale bar: 50 µm. M to O)CpCYC1 and CpCYC2 expression patterns in dissected floral organs, as revealed by RT-qPCR. The expression levels were normalized to those of CpACTIN. The error bars indicate the SD from 3 independent samples (except for sterile stamens). DS/VS, dorsal/ventral sepals; DRec/VRec, dorsal/ventral parts of the receptacle; YS, yellow spots. Asterisks indicate significant differences between samples (2-tailed Student's t test, **P < 0.01).
Reverse transcription quantitative PCR (RT-qPCR) showed that both CpCYC1 and CpCYC2 were specifically expressed in reproductive organs, including inflorescences and young floral buds (Supplemental Fig. S6, A and B). CpCYC2 was strongly downregulated with the development of floral buds (Supplemental Fig. S6B). In dissected floral organs, CpCYC1 was specifically expressed in dorsal petals, dorsal ridge, and bulge structures, as well as dorsal and lateral staminodes (Fig. 2, M and N). In addition, CpCYC1 expression levels were significantly higher in the dorsal region of the receptacle than in the ventral region (Fig. 2O). Unlike CpCYC1, CpCYC2 transcripts were mainly detected in dorsal and lateral staminodes, with much lower expression levels in dorsal ridge and bulge structures (Fig. 2, M and N). No difference in CpCYC2 expression was detected between dorsal and ventral parts of the receptacle (Fig. 2O). Taken together, the expression patterns of CpCYC1 and CpCYC2 are similar during early stages of flower development but later become divergent in that CpCYC1 has much higher expression levels and wider expression domains than CpCYC2.
Overexpression of CpCYC1 and CpCYC2 generates dorsalized flowers with upward orientation and the loss of yellow nectar guides
We investigated the functions of CpCYC1 and CpCYC2 by overexpressing these genes in C. pumila under the control of the duplicated CaMV 35S constitutive promoter (d35Spro). We obtained 31 independent d35Spro:CpCYC1 lines, 18 of which produced dorsalized flowers to varying degrees (Figs. 3 and S7). For example, some lines produced flowers with the loss of yellow pigmentation and the shortening of ventral stamens, some generated flowers with increasing lamella formation and the repressed development of 1 ventral stamen, while some generated sterile stamens in all regions (Supplemental Fig. S7, A to C). In line 26, all petals adopted the dorsal identity, and the dorsal lamella structure expanded into all regions (Fig. 3, A, C, D, F, H, and I). In concert with the dorsalization of corolla, yellow spots disappeared and all stamens became sterile (Fig. 3, C, E, H, and J). Furthermore, floral orientation changed from horizontal to somewhat upward (Fig. 3, B and G).
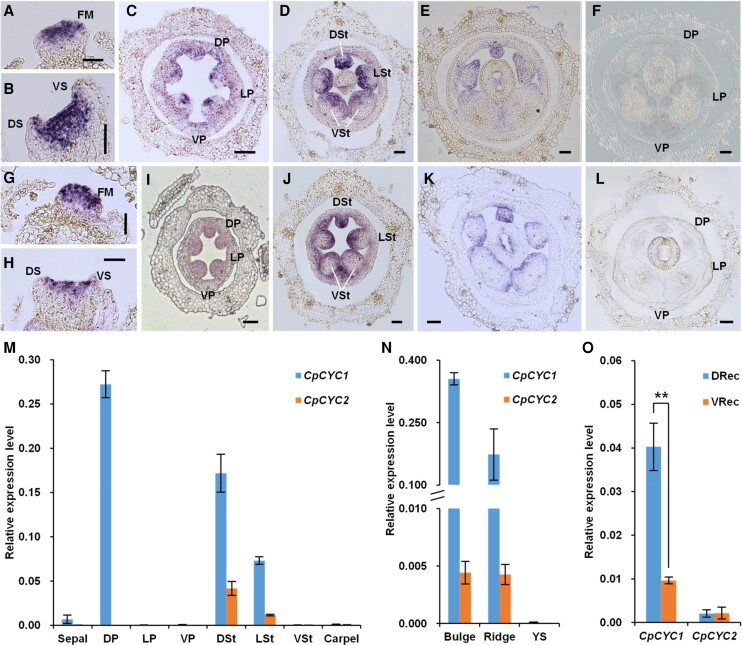
Figure 3.
Ectopic expression of CpCYC1 and CpCYC2 generates dorsalized actinomorphic flowers. A to J) Representative flowers of d35Spro:CpCYC1 with 5 A to E) or 6 F to J) petals. Scale bars: A to D) and F to I), 0.5 cm; E) and J), 0.1 cm. K to T) Representative flowers of d35Spro:CpCYC2 with 5 K to O) or 6 P to T) petals. Scale bars: K to N) and P to S), 0.5 cm; O and T), 0.1 cm. U) Ectopic expression of CpCYC1-All and CpCYC2-All (including both exogenous and endogenous) in d35Spro:CpCYC1 and d35Spro:CpCYC2 transgenic lines. V) Expression of endogenous CpCYC1 (CpCYC1-En) and CpCYC2 (CpCYC2-En) in d35Spro:CpCYC1 and d35Spro:CpCYC2 transgenic lines. The expression levels were normalized to those of CpACTIN. The error bars indicate the SD from 3 independent samples. DP/LP/VP, dorsal/lateral/ventral petals; DTube/VTube, dorsal/ventral corolla tubes.
Of the 21 independent d35Spro:CpCYC2 transgenic lines produced, 14 generated dorsalized flowers of varying degrees, including the weakening of yellow pigmentation and the shortening of ventral stamens (line 3), the impaired development of 2 ventral stamens (line 16), and the developmental delay of ventral stamens and increased number of lamella (line 7) (Supplemental Fig. S7, E to G). In line 21, all petals adopted the dorsal identity in terms of shape, all stamens became sterile, and the dorsal lamella expanded into all regions (Fig. 3, K, M to P, and R to T). In addition, line 21 generated upward-oriented flowers (Fig. 3, L and Q). Taken together, the d35Spro:CpCYC1 and d35Spro:CpCYC2 transgenic lines produced largely similar dorsalized flowers, including the dorsalization of all petals, the developmental delay of all stamens, the expansion of dorsal lamella, and the upward orientation of flowers. However, while d35Spro:CpCYC1 transgenic lines produced purple petals, d35Spro:CpCYC2 generated pale petals. Furthermore, d35Spro:CpCYC2 transgenic lines with strong phenotypes generated “open” flowers from early stages on, likely due to the developmental delay of both the corolla tube and petal lobes (Supplemental Fig. S7, D and H).
SEM showed that both transgenic lines, especially d35Spro:CpCYC1, contained much larger petal cells in the adaxial epidermis than wild-type plants (Supplemental Fig. S1, C and D and Data Set 1), indicating that both CpCYC1 and CpCYC2 promote cell expansion. The d35Spro:CpCYC2 transgenic lines with strong dorsalized phenotypes produced much smaller flowers with much smaller petal lobes than wild-type plants (compare Fig. 3 with Fig. 1), suggesting that CpCYC2 overexpression also inhibited cell proliferation in addition to promoting cell expansion.
RT-qPCR showed that CpCYC1 and CpCYC2 were highly expressed in all petal lobes and corolla tubes in the respective transgenic plants with the strongest phenotypes (Fig. 3U). We designed a pair of primers aimed to amplify a region including the 3′ UTR of CpCYC1, finding that the endogenous CpCYC1 gene was upregulated in lateral and ventral petals as well as the ventral corolla tube in both d35Spro:CpCYC1 and d35Spro:CpCYC2 transgenic plants. This result indicates that the overproduction of both CpCYC1 and CpCYC2 proteins ectopically activated CpCYC1. On the contrary, the overexpression of CpCYC1 and CpCYC2 did not upregulate the endogenous CpCYC2 (Fig. 3V). Therefore, the formation of fully dorsalized actinomorphic flowers in d35Spro:CpCYC2 transgenic plants could be also attributed to the ectopic production of the endogenous CpCYC1 protein, but the decoloration of petals and the delayed corolla expansion could only be due to the overexpression of the exogenous CpCYC2 gene.
Downregulation of CpCYC1 and CpCYC2 generates ventralized flowers with upward orientation and expanded yellow nectar guides
To further investigate the functions of CpCYC1 and CpCYC2, we generated 29 and 23 independent 35Spro:CpCYC1-SRDX and 35Spro:CpCYC2-SRDX transgenic lines, respectively, by expressing these genes in-frame with the SRDX repression domain (Hiratsu et al. 2003; Fig. 4). Of these, 9 and 6 lines generated ventralized actinomorphic flowers, respectively, with all petals acquiring the ventral identity in both the T1 and T2 generations (Fig. 4, A to D). Accordingly, yellow spots extended throughout the corolla tube, and all stamens became fertile (Fig. 4, A to D). However, in most cases, floral orientation did not change as expected, likely due to the failure of the CaMV 35S promoter to trigger the expression of the transgenes in the corresponding regions. RT-qPCR showed that the exogenous CpCYC1-SRDX and CpCYC2-SRDX transgenes were overexpressed in the respective transgenic plants (Fig. 4, E and F). Interestingly, the endogenous CpCYC1 and CpCYC2 genes were downregulated in both transgenic plants (Fig. 4, G and H), suggesting that the overproduction of the strong repressors CpCYC1-SRDX and CpCYC2-SRDX in turn repressed the expression of these endogenous genes.
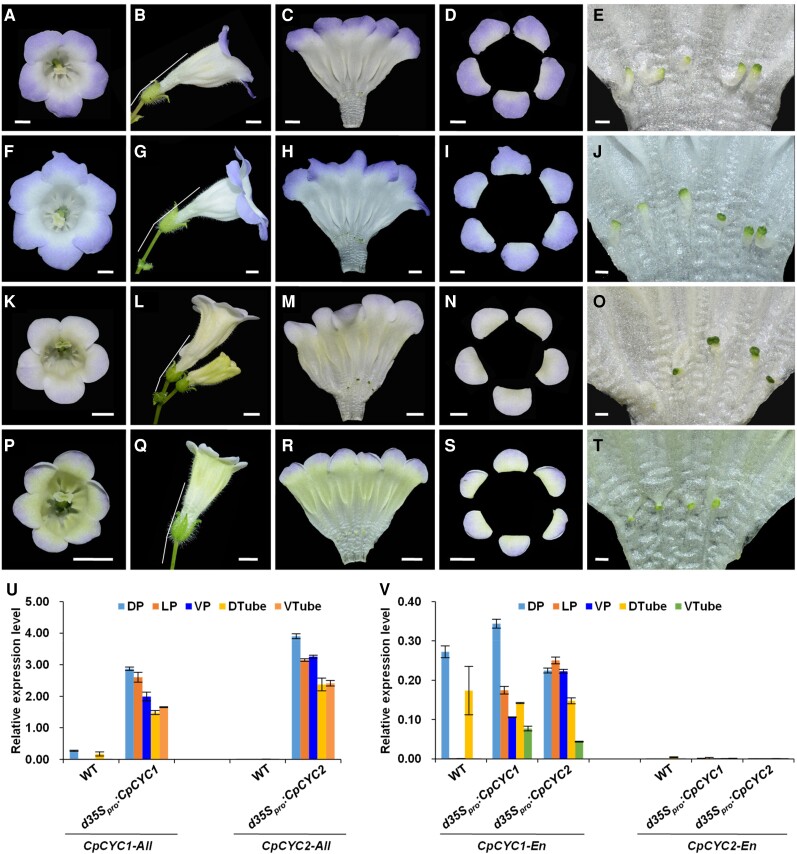
Figure 4.
Knockdown of CpCYC1 and CpCYC2 generates ventralized actinomorphic flowers. A and B) Flowers of 35Spro:CpCYC1-SRDX with 5 A) or 6 B) petals. Scale bars: 0.5 cm. C and D) Flowers of 35Spro:CpCYC2-SRDX with 5 C) or 6 D) petals. Scale bars: 0.5 cm. E and F) Ectopic expression of exogenous CpCYC1-SRDX and CpCYC2-SRDX in 35Spro:CpCYC1-SRDXE) and 35Spro:CpCYC2-SRDXF) plants. G and H) Expression levels of endogenous CpCYC1 (CpCYC1-En, G) and CpCYC2 (CpCYC2-En, H) were greatly reduced in both transgenic plants. The expression levels were normalized to those of CpACTIN. The error bars indicate the SD from 3 independent samples (except for sterile stamens). DP/LP/VP, dorsal/lateral/ventral petals; DSt/LSt/VSt, dorsal/lateral/ventral stamens.
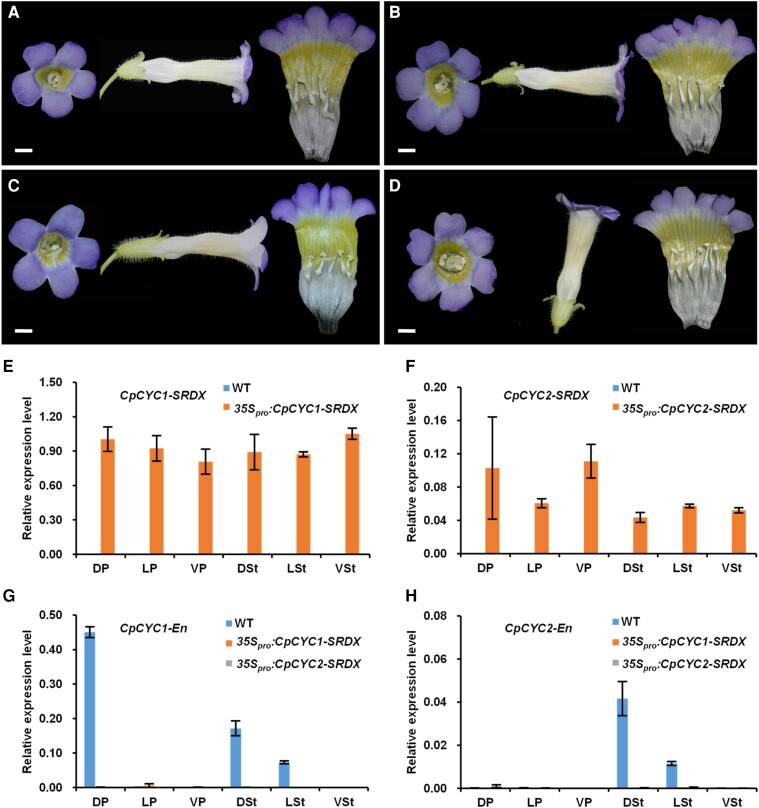
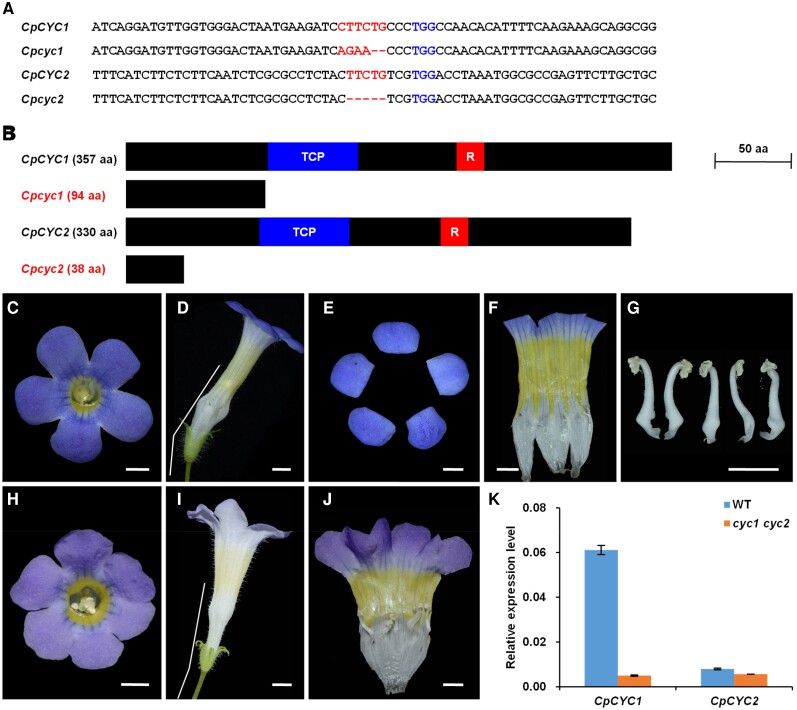
We further investigated the functions of CpCYC1 and CpCYC2 genes by constructing a CRISPR/Cas9 plasmid containing 2 guide RNAs (gRNAs), i.e. gRNA1 and gRNA2, targeting CpCYC1 and CpCYC2, respectively (Supplemental Fig. S8). Among the 14 positive T1 transgenic lines, 8 lines showed the loss of floral zygomorphy to varying degrees (Supplemental Fig. S9). For example, line 10 produced flowers with all petals adopting the morphology of ventral petals, 1 lateral stamen that become fertile, and weak yellow spots in the dorsal region. However, the corolla was still zygomorphic (Supplemental Fig. S9D). Line 3 produced an actinomorphic corolla, but the dorsal stamen was still sterile, and the yellow pigmentation was still weaker in the dorsal region than the ventral region (Supplemental Fig. S9A). Lines 7 and 8 produced fully actinomorphic corollas with extensive yellow pigmentation and 5 fertile stamens (Supplemental Fig. S9, B and C). Lines 3, 8, and 10 were chimeric for both CpCYC1 and CpCYC2, while in line 7, the CpCYC1 locus appeared to be homozygous (Supplemental Fig. S10A).
Therefore, line 7 was allowed to self to generate a T2 population. By genotyping 192 of the T2 plants, we obtained several homozygous mutants for CpCYC1, CpCYC2, and CpCYC1/2. However, only 1 mutant (lines 7 to 70) was shown to be Cas9-free (Supplemental Fig. S10B) in which a premature stop codon was introduced into both the cyc1 and cyc2 alleles (Fig. 5A). Therefore, if possible, both the cyc1 and cyc2 alleles would translate truncated proteins lacking the entire TCP domain, which is involved in both DNA-binding and protein–protein interactions (Cubas et al. 1999; Fig. 5B). As a result, lines 7 to 70 (the cyc1 cyc2 double mutant) should be a null mutant for both CpCYC1 and CpCYC2. In support of this notion, cyc1 cyc2 produced fully ventralized flowers with upward orientation and uniform yellow pigmentation (Fig. 5, C to J). RT-qPCR showed that both genes (especially CpCYC1) were strongly downregulated in the double mutant (Fig. 5K), likely due to the impaired ability for auto-regulation and cross-regulation.
Figure 5.
The cyc1 cyc2 double mutant generated by CRISPR/Cas9 produces ventralized actinomorphic flowers. A) The deletion and/or insertion mutations in CpCYC1 and CpCYC2 in the double mutant. B) The mutated CpCYC1 and CpCYC2 might generate truncated proteins. C to J) The double mutant produces fully ventralized flowers with upward orientation and uniform yellow spots. Scale bars: 0.5 cm. K) Expression levels of CpCYC1 and CpCYC2 were strongly reduced in the double mutant. Floral buds <0.5 cm long were used. The expression levels were normalized to those of CpACTIN. The error bars indicate the SD from 3 independent samples.
To identify possible functional divergence between CpCYC1 and CpCYC2, we crossed cyc1 cyc2 with wild-type plants. F1 plants generated zygomorphic flowers with horizontal orientation. However, dorsal petals in the flowers of F1 plants were narrower than those in wild-type plants, dorsal lamella formation was weakened, yellow spot formation in the ventral corolla tube was strengthened, and weak yellow pigmentation appeared in the dorsal region (Supplemental Fig. S11). Therefore, CpCYC1 and CpCYC2 have a dosage effect on floral symmetry and asymmetric nectar guide formation. By genotyping the F2 population, we obtained the cyc1 and cyc2 single mutants. The cyc1 single mutant generated partially ventralized flowers with only 1 staminode in the dorsal region and the disappearance of the dorsal lamella structure. The yellow spots were distributed evenly and the flower oriented upward (Supplemental Fig. S11). The cyc1 single mutant also produced fully ventralized flowers, probably due to the downregulation of CpCYC2 by nonfunctional cyc1 protein. The cyc2 single mutant also produced fully ventralized flowers but at a lower frequency. In most cases, cyc2 produced partially ventralized flowers with 3 or 4 fertile stamens and the lack of the dorsal lamella structure (Supplemental Fig. S11). Therefore, CpCYC1 and CpCYC2 have undergone functional divergence after gene duplication. We suggest that CpCYC1 might retain almost all functions of the progenitor gene in determining floral symmetry, floral orientation, and nectar guide patterning, while CpCYC2 has minor effects on these processes.
Protein–DNA and protein–protein interaction assays of CpCYC1 and CpCYC2
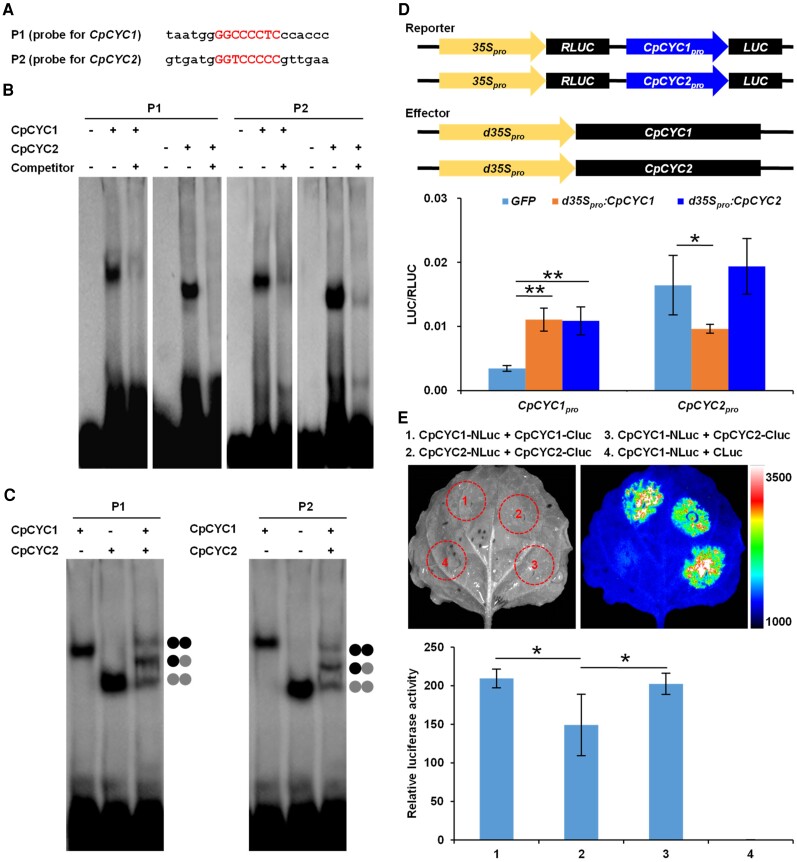
The above functional evidence suggests that CpCYC1 and CpCYC2 undergo auto-regulation and cross-regulation. To further clarify the regulatory relationship between these genes, we isolated and analyzed their promoter sequences. In the proximate promoter region (<1.0-kb upstream of the start codon), each gene possesses a DNA motif that perfectly matches the consensus DNA-binding site for CYC-like TCP genes, i.e. GGNCCCNC (Costa et al. 2005; Yang et al. 2012; Figs. 6A and S12). To determine whether these DNA motifs can be bound by CpCYC1 and CpCYC2, we performed electrophoresis mobility shift assays (EMSAs) using recombinant CpCYC1 and CpCYC2 proteins heterologously expressed and purified from Escherichia coli cells. Biotin-labeled probes containing the CYC-binding site from either CpCYC1 or CpCYC2 were bound by both proteins and therefore migrated much more slowly than free DNA probes (Fig. 6B). When excess amounts of unlabeled probes were added, the bands corresponding to the protein–DNA complexes disappeared. When both proteins were added to the reaction, 3 slowly migrating bands formed, which corresponded to the complex formed between the DNA probe and CpCYC1 homodimers, CpCYC2 homodimers, and CpCYC1/2 heterodimers (Fig. 6C). Therefore, both CpCYC1 and CpCYC2 can be bound by their own and each other's proteins.
Figure 6.
CpCYC1 and CpCYC2 undergo asymmetric auto-regulation and cross-regulation. A to C) EMSA showing that CpCYC1 and CpCYC2 recombinant proteins can bind to their own gene promoters by forming homodimers and heterodimers. The shift band was abolished when excessive amounts of unlabeled probes were added. D) Transient gene expression assays showing that overexpression of CpCYC1 and CpCYC2 enhances CpCYC1 promoter activity. Asterisks indicate significant differences (2-tailed Student's t test, *P < 0.05, **P < 0.01). E) LCI assays showing the quantification of homodimerization and heterodimerization between CpCYC1 and CpCYC2. Asterisks indicate significant differences (2-tailed Student's t test, *P < 0.05).
We then examined whether the binding of CpCYC1 and CpCYC2 to their promoters is functional by performing transient gene expression assays using the dual-luciferase reporter gene system. In this system, the expression of the firefly luciferase reporter gene (LUC) was specifically controlled by the CpCYC1 or CpCYC2 promoter, while the internal reference RLUC (Renilla reniformis luciferase reporter gene) was driven by the constitutive CaMV 35S promoter. As an effector, Agrobacterium tumefaciens containing either the d35Spro:CpCYC1 or d35Spro:CpCYC2 construct was coinfiltrated into Nicotiana benthamiana leaves with the reporter strain. In the presence of the effector, the LUC/RLUC ratio was significantly higher for the CpCYC1 reporter plasmid than the control plasmid, i.e. the empty pCAMBIA1302 vector carrying the GFP gene (Fig. 6D; Supplemental Data Set 1), indicating that the overproduction of both CpCYC1 and CpCYC2 can enhance the expression of CpCYC1. However, the activity of the CpCYC2 promoter decreased rather than increased in the presence of CpCYC1 and did not change in the presence of CpCYC2 (Fig. 6D; Supplemental Data Set 1). Therefore, CpCYC1, but not CpCYC2, activates itself. In addition, while CpCYC1 is positively regulated by CpCYC2, CpCYC2 is negatively regulated by CpCYC1. The regulatory relationship between CpCYC1 and CpCYC2 is totally in line with their changes in expression in d35Spro:CpCYC1 and d35Spro:CpCYC2 transgenic lines, as the ectopic production of both gene products activated endogenous CpCYC1 but not CpCYC2 (Fig. 3U).
We performed yeast 2-hybrid experiments to confirm the protein–protein interaction detected by EMSA. Both CpCYC1 and CpCYC2 were expressed as fusion proteins with the transcription activation domain (AD) and DNA-binding domain (BD) of yeast (Saccharomyces cerevisiae) GAL4. CpCYC1-AD interacted with CpCYC1-BD in yeast cells. CpCYC2-AD interacted with CpCYC2-BD but much less strongly. In addition, CpCYC1-AD interacted with CpCYC2-BD (Supplemental Fig. S13). As negative controls, CpCYC1-AD and BD, CpCYC2-AD and BD, CpCYC1-BD and AD, and CpCYC2-BD and AD failed to interact with each other. Taken together, these results indicate that CpCYC1 and CpCYC2 can form homodimers and heterodimers in yeast.
We performed a firefly luciferase complementation imaging (LCI) assay (Chen et al. 2008) to confirm and quantify the protein–protein interaction between CpCYC1 and CpCYC2 in planta. We inserted both the CpCYC1 and CpCYC2 coding sequences (CDSs) into pCAMBIA1300-NLuc and pCAMBIA1300-CLuc to construct 4 different plasmids: CpCYC1-NLuc, CpCYC1-CLuc, CpCYC2-NLuc, and CpCYC2-CLuc, and coinfiltrated N. benthamiana leaf epidermal cells with Agrobacterium cultures harboring CpCYC1-NLuc and CpCYC1-CLuc, CpCYC2-NLuc and CpCYC2-CLuc, and CpCYC1-NLuc and CpCYC2-CLuc. Coinfiltration of CpCYC1 and CpCYC2 generated strong LUC signals, indicating that these proteins can form heterodimers. In addition, both CpCYC1 and CpCYC2 formed homodimers (Fig. 6E). As a negative control, coinfiltration of the empty vector pCAMBIA1300-CLuc with CpCYC1-NLuc led to no LUC signal. The interactions of CpCYC1-NLuc and CpCYC1-CLuc, as well as CpCYC1-NLuc and CpCYC2-CLuc, were significantly stronger than that of CpCYC2-NLuc and CpCYC2-CLuc (Fig. 6E; Supplemental Data Set 1).
CpCYC1 and CpCYC2 control asymmetric nectar guide patterning by inhibiting the expression of flavonoid synthesis-related genes
A conspicuous characteristic of C. pumila zygomorphic flowers is the yellow pigmentation in the ventral corolla tube, but not in the dorsal region, where CpCYC1 and CpCYC2 normally carry out their functions. Most importantly, the expansion or loss of yellow spots is tightly associated with loss- or gain-of-function mutation of CpCYC1 and CpCYC2. We reasoned that CpCYC1 and CpCYC2 might repress yellow pigmentation out of the ventral region by inhibiting the accumulation of carotenoids or flavonoids, 2 important compounds in yellow nectar guides in plants (Jorgensen and Geissmann 1955; Sasakia and Takahashib 2002; Yuan et al. 2013; Jachula et al. 2018), by downregulating related genes. Carotenoids, which absorb visible light but not UV light (Yuan et al. 2015), were precluded from further analysis because a UV-absorbance feature was detected in nectar guides (Supplemental Fig. S2B), a typical characteristic of various flavonoid compounds (Thompson et al. 1972). Biochemical assays indicated that flavonoids, probably flavanones or flavonols, are responsible for the yellow pigmentation in the ventral corolla tube. Specifically, ethanol extract from yellow spots turned luminous yellow in 1% AlCl3 solution and generated green fluorescence under UV light, whereas it turned orange–red in 1% Na3CO3 solution and generated yellow fluorescence under UV light (Supplemental Fig. S2, D and E).
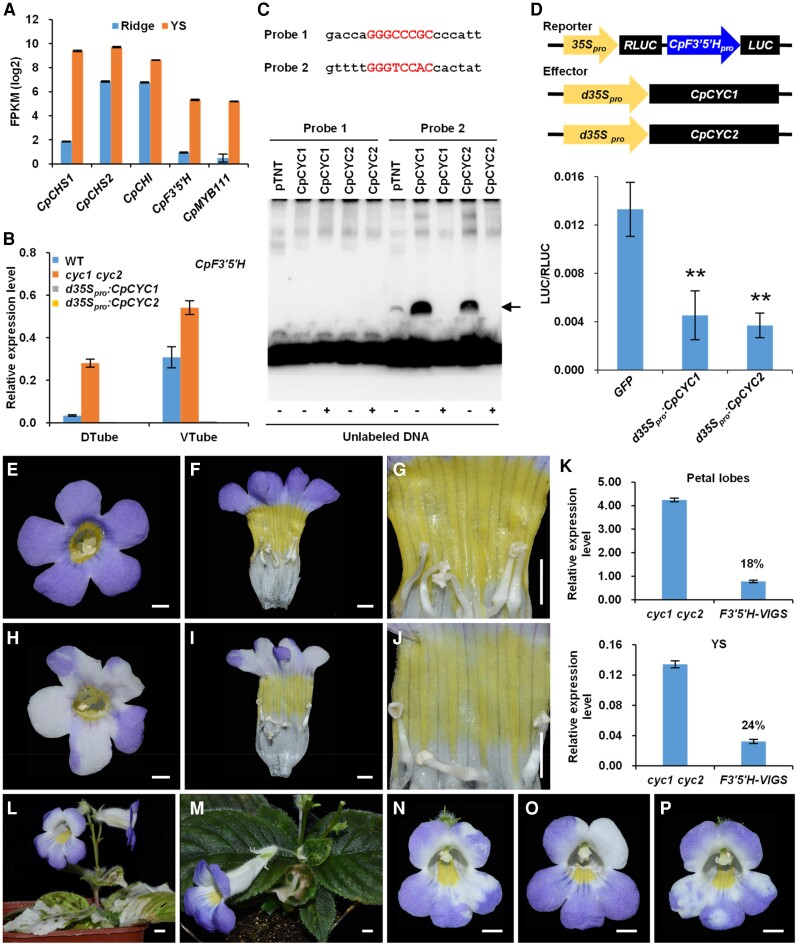
RNA-sequencing (RNA-seq) showed that 5 key flavonoid synthesis-related genes, including 2 putative paralogs encoding chalcone synthase (CpCHS1 and CpCHS2), 1 chalcone isomerase encoding gene (CpCHI), 1 flavonoid 3′,5′-hydroxylase encoding gene (CpF3′5′H), and 1 MYB transcription factor (CpMYB111), were expressed at much higher levels in yellow spots than in dorsal ridges (Figs. 7A and S14; Supplemental Data Set 2). RT-qPCR suggested that only CpF3′5′H is strongly regulated by CpCYC1 and CpCYC2, as it exhibited much higher expression levels in cyc1 cyc2 than the wild type but an undetectable expression signal in d35Spro:CpCYC1 and d35Spro:CpCYC2 transgenic plants (Figs. 7B and S15, A to D). By isolating and analyzing its promoter sequence, we found that CpF3′5′H contains 2 cis-regulatory elements that perfectly match the CYC-binding site in its proximate regulatory region (Supplemental Fig. S12). In an EMSA, 1 of the 2 elements was bound by CpCYC1 and CpCYC2 proteins translated using the wheat germ protein expression system (Fig. 7C). Transient gene expression assays in N. benthamiana showed that overexpression of both CpCYC1 and CpCYC2 led to the significantly downregulated activity of the LUC gene when placed under the control of the CpF3′5′H promoter (Fig. 7D; Supplemental Data Set 1). Therefore, CYC-like genes in C. pumila inhibit the formation of yellow pigmentation out of the ventral region of the flower, likely by directly repressing CpF3′5′H.
Figure 7.
CpCYC1 and CpCYC2 repress yellow spot formation by negatively regulating CpF3′5′H. A) Expression difference of 5 genes involved in flavonoid synthesis between the dorsal ridge and ventral yellow spots in C. pumila revealed by RNA-seq. B) Expression changes of CpF3′5′H in CpCYC1 and CpCYC2 gain- and loss-of-function mutants. The expression levels were normalized to those of CpACTIN. The error bars indicate the SD from 3 independent samples. C) One of the 2 CYC-binding site in the CpF3′5′H promoter can be directly bound by CpCYC1 and CpCYC2 proteins translated using the wheat germ protein expression system (the arrow indicates the shifted band). D)CpF3′5′H is directly repressed by CpCYC1 and CpCYC2. Asterisks indicate significant differences between samples (2-tailed Student's t test, **P < 0.01). E to J) The downregulation of CpF3′5′H in the cyc1 cyc2 double mutant using VIGS leads to light yellow spots and bleached petal lobes H to J) compared to flowers without any mutant phenotype E to G) from the same plant. Scale bars: 0.5 cm. K) The expression level of CpF3′5′H is greatly reduced in both bleached petals and light yellow spots (18% and 24% relative to the control, respectively) collected from open flowers. The expression levels were normalized to those of CpACTIN. The error bars indicate the SD from 3 independent experiments. L and M) Knockdown of CpPDS only bleaches leaves and sepals. Scale bars: 0.5 cm. N to P) Knockdown of CpANS only decolors petal lobes. Scale bars: 0.5 cm.
To further investigate whether CpF3′5′H contributes to the formation of yellow spots in the ventral corolla tube, we downregulated its expression using virus-induced gene silencing (VIGS). Downregulating CpF3′5′H decreased yellow pigment production in both wild-type and cyc1 cyc2 plants (Figs. 7, E to K, and S15, E to K). As negative controls, the knockdown of CpPDS, encoding phytoene desaturase in the carotenoid biosynthetic pathway, only led to bleached leaves and sepals (Fig. 7, L and M), while the reduced expression of CpANS, encoding anthocyanidin synthase, only led to bleaching of purple petals (Fig. 7, N to P). The downregulation of CpF3′5′H also led to the bleaching of purple petals (Fig. 7, E to K). This result is totally expected because the flavonoid 3′,5′-hydroxylase gene is located upstream of the anthocyanidin synthase gene in the anthocyanidin biosynthetic pathway (Nishihara and Nakatsuka 2011).
PhCYC1C and PhCYC1D share similar functions with CpCYC1
As described above, CYC-like genes in C. pumila, especially CpCYC1, have multiple functions in floral symmetry, floral orientation, and nectar guide patterning. We therefore explored whether CYC-like genes in other Gesneriaceae plants have similar functions by turning our attention to the relatives of C. pumila with zygomorphic flowers (Figs. 8 and S16). We found that Clostera anachoreta, the sister group of C. pumila in the genus Chirita, produces zygomorphic flowers with horizontal orientation and an asymmetric nectar guide pattern attributed to the orange pigmentation in the ventral corolla tube, in sharp contrast to the pure white corolla (Supplemental Fig. S16, A to C). C. anachoreta flowers also share other features with C. pumila, such as dorsal ridge and bulge structures, the geniculate structure of ventral stamens, and so on (Supplemental Fig. S16, C to E). Primulina heterotricha zygomorphic flowers also share most features with C. pumila, including dorsal ridge and bulge structures, ventral yellow spots, and horizontal orientation (Fig. 8, A to F). Taken together, the concerted evolution of floral zygomorphy, horizontal orientation, and asymmetric nectar guide patterns might be commonplace in Gesneriaceae.
Figure 8.
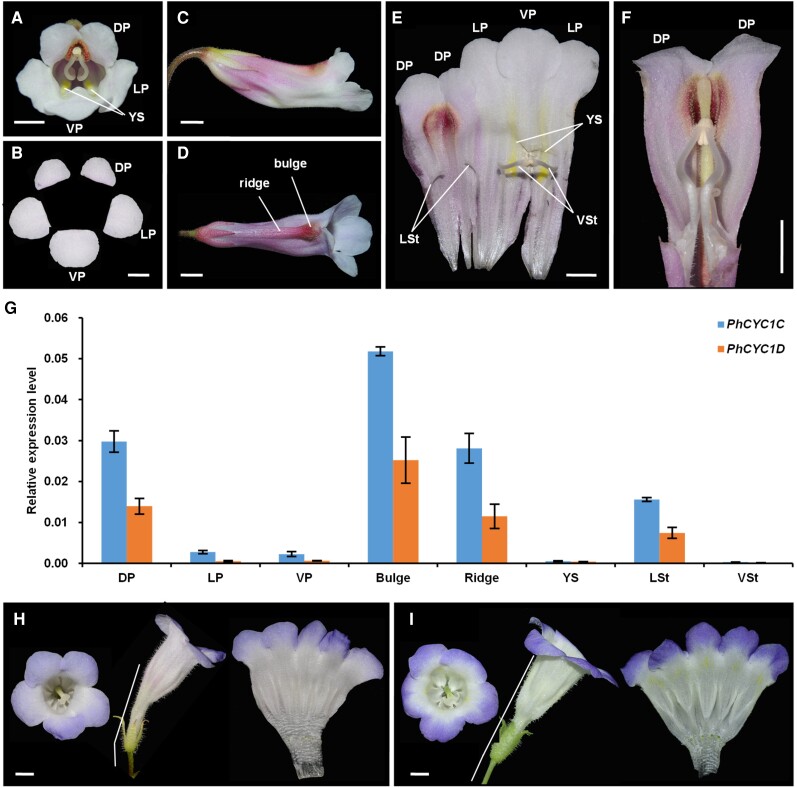
P. heterotricha CYC1C and CYC1D genes have similar functions to CpCYC1. A and B)P. heterotricha produces zygomorphic flowers with 5 petals that differ in both size and morphology. Scale bars: 0.5 cm. C) Side view of a flower in the horizontal orientation. Scale bar: 0.5 cm. D) Dorsal view of a flower showing the bulge and ridge structures in the dorsal corolla tube. Scale bar: 0.5 cm. E) The inner structure of the flower showing 2 ventral fertile stamens and 2 lateral staminodes. Scale bar: 0.5 cm. F) The dorsal part of a corolla tube showing the inside of the bulge and ridge structures and the geniculate filaments of the ventral stamens. Scale bar: 0.5 cm. G) Expression patterns of PhCYC1C and PhCYC1D normalized to those of PhACTIN. The error bars indicate the SD from 3 independent samples. H and I) Overexpression of PhCYC1CH) and PhCYC1DI) generates dorsalized actinomorphic flowers. White bars: 0.5 cm. DP/LP/VP, dorsal/lateral/ventral petals; LSt/VSt, lateral/ventral stamens; YS, yellow spots.
We selected P. heterotricha to investigate whether CYC-like genes in other Gesneriaceae species carry out similar functions to CpCYC1 and CpCYC2 genes because all 4 CYC-like genes from this species have been isolated and analyzed (Gao et al. 2008; Yang et al. 2012). Here, we only focused on PhCYC1C and PhCYC1D because the 2 other paralogs (PhCYC2A and PhCYC2B) are only transiently expressed in young inflorescences, with undetectable transcripts in floral buds of any developmental stage (Gao et al. 2008). RT-qPCR showed that both PhCYC1C and PhCYC1D were specifically expressed in dorsal petals and lateral staminodes, as reported previously (Gao et al. 2008; Yang et al. 2012; Fig. 8G). Importantly, these genes were highly expressed in dorsal ridge and bulge structures (Fig. 8G). Therefore, PhCYC1C and PhCYC1D share similar expression patterns with CpCYC1 and might be also responsible for the establishment of floral zygomorphy, horizontal orientation, and asymmetric nectar guide patterning in P. heterotricha. To support this notion, we ectopically expressed PhCYC1C and PhCYC1D in C. pumila because P. heterotricha is currently not amenable to genetic transformation. The overexpression of both PhCYC1C and PhCYC1D in C. pumila generated dorsalized actinomorphic flowers with upward orientation and the loss of yellow spots, mimicking CpCYC1 overexpressor (Fig. 8, H and I). Therefore, CYC-like genes play conserved roles in determining floral symmetry, floral orientation, and nectar guide patterning in Gesneriaceae.
Discussion
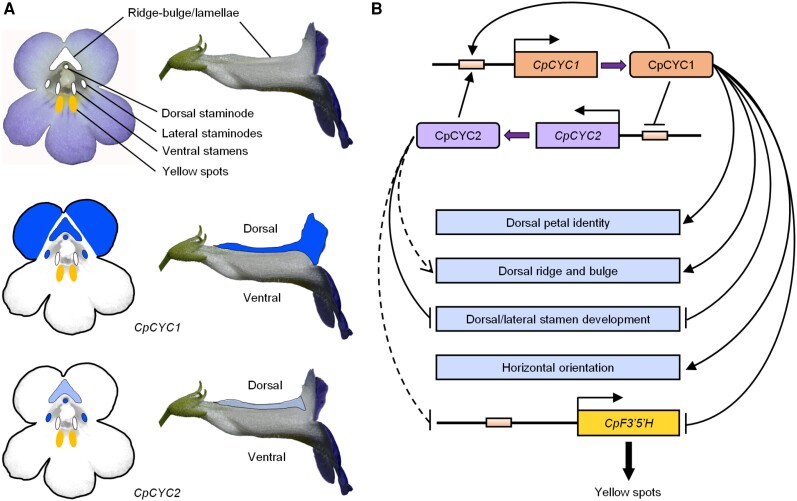
In this study, we showed that C. pumila zygomorphic flowers have a series of integrated traits adapted to specific pollinators, including bilaterally symmetric corolla and stamens, dorsal ridge and bulge structures, an asymmetric nectar guide pattern, and horizontal orientation. We confirmed that CYC-like genes, which are widely involved in controlling floral symmetry in angiosperms (Luo et al. 1996, 1999; Hileman et al. 2003; Feng et al. 2006; Busch and Zachgo 2007; Song et al. 2009; Yang et al. 2012, 2015; Chen et al. 2018; Dong et al. 2018), also help determine these floral traits in C. pumila (Fig. 9, A and B) and its close relative P. heterotricha. Therefore, CYC-like genes in Gesneriaceae have conserved functions in controlling multiple floral symmetry-related traits.
Figure 9.
Model showing the expression and functional divergence as well as the pleiotropic functions of CpCYC1 and CpCYC2. A)CpCYC1 and CpCYC2 have divergent expression patterns in the second and third whorls of floral organs. Dark blue represents higher expression levels, while light blue represents lower expression levels. B) Model showing the multiple functions, functional divergence, and the asymmetric auto-regulation and cross-regulation of CpCYC1 and CpCYC2. CpCYC1 independently controls dorsal petal identity and floral horizontal orientation and determines the development of the dorsal ridge and bulge together with CpCYC2. Both CpCYC1 and CpCYC2 are required to inhibit the development of dorsal and lateral stamens. They determine asymmetric nectar guide formation likely by directly repressing the flavonoid synthesis-related gene CpF3′5′H. Solid lines display strong promoting or repressive activity, while dashed lines represent weak activity.
Functional and regulatory divergence of paralogous CYC-like genes in Gesneriaceae
We demonstrated that CYC1 and CYC2 in C. pumila have undergone expression and functional divergence (Fig. 9, A and B). CpCYC1 plays a major role, while CpCYC2 only plays a minor role in controlling floral zygomorphy. Nevertheless, both genes are indispensable for the stable formation of fully zygomorphic flowers, as the mutation of either gene led to the impairment of floral zygomorphy. The expression and functional divergence of paralogous CYC-like genes has also been reported in many other zygomorphic species (Luo et al. 1996, 1999; Feng et al. 2006; Wang et al. 2008; Song et al. 2009; Yang et al. 2012; Chen et al. 2018; Hsin et al. 2019). For example, in A. majus, CYC determines the identity of dorsal petals and represses the development of dorsal stamens, while DICH mainly controls the internal asymmetry of dorsal petals (Luo et al. 1996, 1999). In Opithandra dinghushanensis (Gesneriaceae), OpdCYC2A mainly controls the morphology of dorsal petals and inhibits the development of dorsal stamens, while OpdCYC1C represses ventral stamen development (Song et al. 2009) by evolving a new expression domain (Hileman and Cubas 2009), likely due to cis-regulatory changes. Such changes might also have occurred in CpCYC1 and CpCYC2 considering their expression divergence. Furthermore, the different phylogenetic positions (belonging to the GCYC1 and GCYC2 lineages, respectively) and amino acid changes (occurred in both highly conserved TCP and R domains and variable regions) suggest that they also have divergent protein functions.
In parallel with their functional divergence, CpCYC1 and CpCYC2 showed distinctively different protein–DNA and protein–protein interactions. While CpCYC1 positively regulates itself and is activated by CpCYC2, CpCYC2 has no auto-regulation ability even though has retained a perfect auto-regulatory element in its promoter region. The lack of auto-regulation ability of CpCYC2 might be due to cis-regulatory changes in surrounding regions of the CYC-binding site rather than a lesion in CpCYC2 protein because the overproduction of CpCYC2 led to the activation of CpCYC1 and repression of CpF3′5′H, similar to CpCYC1. Most importantly, we showed that CpCYC2 is negatively regulated by CpCYC1. Therefore, the auto-regulation and cross-regulation of 2 CYC-like genes in C. pumila is asymmetric (Fig. 9B) in order to maintain the strong expression of only 1 gene. By contrast, PhCYC1C and PhCYC1D form a double positive auto-regulatory loop to maintain the strong expression of both genes throughout floral development (Yang et al. 2012). We reasoned that the extremely low expression levels of PhCYC2A and PhCYC2B in P. heterotricha (Gao et al. 2008) might also attribute to asymmetric cross-regulation between PhCYC1C/1D and PhCYC2A/2B. While symmetric auto-regulation and cross-regulation has been widely documented among duplicated genes with redundant functions in plants (Schwarz-Sommer et al. 1992; Hill et al. 1998; Tilly et al. 1998; Honma and Goto 2001; Crews and Pearson 2009), asymmetric cross-regulation has rarely been reported in animals or plants. One example is the asymmetric cross-regulation of UPSTREAM STIMULATORY FACTOR 1 (USF1) and USF2 reported in mouse (Mus musculus) in which USF1 normally represses USF2, whereas USF2 increases the expression of USF1 (Sirito et al. 1998). In Arabidopsis thaliana, the regulatory relationship between APETALA1 (AP1) and CAULIFLOWER (CAL) is somewhat similar to that of CpCYC1 and CpCYC2 in that AP1 can undergo auto-regulation as well as cross-regulation by CAL. However, AP1 cannot regulate CAL due to the lack of the CArG-box in its promoter region (Ye et al. 2016). Therefore, the asymmetric auto-regulation and cross-regulation between CpCYC1 and CpCYC2 reported in this study provides valuable insights into the complex regulatory relationship between paralogous genes in plants.
Roles of CYC-like genes in the correlated evolution of floral symmetry, floral orientation, and nectar guide patterns
Angiosperms are complex systems composed of developmentally and functionally related traits. The genetic correlation of multiple traits is common in plants, such as the evolution of color, nutritional content, and morphology in bird-dispersed fleshy Viburnum fruits (Sinnott-Armstrong et al. 2020), flower size and seed number in monocots (Bawa et al. 2019), stem and leaf functional traits in sunflower (Helianthus) (Pilote and Donovan 2016), and fruit size, color, and spines in Neotropical palms (Nascimento et al. 2020). The origin of floral zygomorphy, a key innovation during angiosperm radiation, is also frequently accompanied by the evolution of other floral traits, such as horizontal orientation (Neal et al. 1998; Ushimaru and Hyodo 2005; Ushimaru et al. 2009). In Gesneriaceae, most zygomorphic species produce flowers with horizontal orientation. The change in floral symmetry from zygomorphy to actinomorphy, either dorsalized or ventralized, is usually accompanied by a change in floral orientation from horizontal to upward (Yang et al. 2015; Dong et al. 2018; Liu, Wang, et al. 2021; this study), pointing to the genetic correlation of floral symmetry and floral orientation. Interestingly, zygomorphic flowers usually exhibit asymmetric nectar guide patterns that are significantly associated with changes in pollination type in Gesneriaceae (Hsu and Kuo 2021). Here, we observed that several zygomorphic species in Gesneriaceae are characterized by yellow spots exclusively distributed in the ventral corolla tube (Figs. 1, 8, and S16). Most importantly, in naturally occurred peloric (uncharacteristically radially symmetric) mutant flowers of both P. heterotricha and C. pumila, yellow spots expand into all tube regions (Yang et al. 2012; Liu, Wu, et al. 2021). Taken together, our findings shed light on the concerted evolution of floral symmetry, floral orientation, and nectar guide patterns.
Genetic correlations usually result from gene pleiotropy (Kern et al. 2016; Wessinger and Hileman 2016). However, the mutation of pleiotropic genes has the potential to generate deleterious effects that scale with the number of traits affected by these mutations. Functional modularity (in which multiple related traits share a common function) was proposed as a mechanism to reduce such potential deleterious effects (Wagner et al. 2007; Wagner and Zhang 2011). It is clear that floral symmetry, floral orientation, and nectar guide patterns share a common function in mediating plant–pollinator interactions. Therefore, it is likely that the same set of genes (or even a single one) with pleiotropy helps determine all these floral traits, such as the single SsCYC gene, which controls both floral symmetry and orientation in Sinningia speciosa (Gesneriaceae) (Dong et al. 2018). We demonstrated that 2 CYC-like genes, especially CpCYC1, function in floral zygomorphy, floral orientation, and nectar guide patterning in C. pumila.
Given that the association between floral zygomorphy and horizontal orientation is widespread in Gesneriaceae (Song et al. 2009; Yang et al. 2012, 2015; Dong et al. 2018), we suggest that CYC-like genes have a conserved function in controlling floral orientation in zygomorphic flowers in this family. However, while C. pumila produces horizontal flowers due to the asymmetric development of dorsal and ventral parts of the receptacle, S. speciosa generates horizontal flowers due to the gibbous structure at the base of the dorsal corolla tube, indicating that the horizontal orientation of zygomorphic flowers in various Gesneriaceae species might have different structural bases (Dong et al. 2018; this study). CYC-like genes might control horizontal orientation in diversified lineages of Gesneriaceae by interacting with different cofactors or recruiting different downstream genes; this concept deserves further detailed investigation.
In this study, the ectopic expression of CpCYC1 and CpCYC2 led to the disappearance of yellow spots, while their loss of function resulted in the extension of yellow spots, indicating that these genes directly or indirectly control this type of pigmentation. Expression analysis showed that the flavonoid 3′,5′-hydroxylase encoding gene (CpF3′5′H) was downregulated in CpCYC1 and CpCYC2 overexpressors but upregulated in cyc1 cyc2. EMSA and transient gene expression assays suggested that CpF3′5′H might be directly repressed by CpCYC1 and CpCYC2. The flavonoid 3′,5′-hydroxylase, belonging to the Cytochrome P450 superfamily, was required to produce flavonols (such as quercetin and myricetin) in engineered E. coli strains (Leonard et al. 2006). In tea plant (Camellia sinensis), the flavonoid 3′,5′-hydroxylase catalyzes the conversion of flavones, flavanones, dihydroflavonols, and flavonols into 3′,4′,5′-hydroxylated derivatives (Wang et al. 2014). Here, the downregulation of CpF3′5′H by VIGS significantly reduced the contents of flavonoid pigments in nectar guides in both wild-type and cyc1 cyc2 plants (Figs. 7 and S15). Therefore, CYC-like genes might negatively regulate the production of yellow pigments by repressing flavonoid synthesis-related genes, thus inhibiting the synthesis of certain flavonoids in spots outside the ventral corolla tube in C. pumila.
Since PhCYC1C and PhCYC1D overproduction also repressed the accumulation of yellow pigments (this study) and their loss-of-function mutations led to the expansion of yellow spots in P. heterotricha peloric flowers (Yang et al. 2012), we suggest that CYC-like genes are widely involved in controlling asymmetric nectar guide patterning in Gesneriaceae. However, whether CpCYC1 and CpCYC2 directly or indirectly repress CpF3′5′H expression, thus controlling asymmetric nectar guide patterning, still needs to be verified by further functional investigations, such as CRISPR/Cas9-mediated targeting of the CpF3′5′H promoter to disrupt its CYC-binding site. In addition, it would be interesting to further explore how yellow flavonoid pigments are specifically restricted to nectar guides in the ventral corolla tube, while other colored pigments are broadly distributed in petals, more or less asymmetrically along the dorsoventral axis; this process may involve other factors that directly or indirectly interact with CYC-like genes in the complex floral symmetry regulatory network.
Different plants may generate yellow nectar guides by accumulating distinct pigments, primarily carotenoids and flavonoids. For example, in A. majus, the color of yellow nectar guides, as well as yellow flowers, is due to the accumulation of the flavonoids aurones (Jorgensen and Geissmann 1955; Ono et al. 2006), while in purple monkey flower (Mimulus lewisii; Scrophulariaceae), this coloration is due to the accumulation of carotenoid pigments (Yuan et al. 2013). In this study, we determined that certain flavonoids are responsible for the yellow nectar guides in C. pumila, which are related to CYC-like gene activities. Further investigation of flavonoid compounds in the yellow spots and functional investigations of related genes would provide more insights into how the asymmetric nectar guide pattern in C. pumila is established by recruiting flavonoid synthesis-related genes into the regulatory pathway of CYC-like genes. Taken together, this study revealed that CYC-like genes are highly pleiotropic in controlling the concerted evolution of floral zygomorphy, horizontal orientation, and asymmetric nectar guide patterning in Gesneriaceae. These findings greatly broaden our understanding of the functions of CYC-like genes beyond controlling the identity of dorsal floral organs, as initially reported in A. majus (Luo et al. 1996, 1999).
The repeated origins of zygomorphy from actinomorphy have led to the formation of several large, successful clades of plants, predominantly with zygomorphic flowers (Endress 2001). How zygomorphic flowers have originated repeatedly is a central question in the evolutionary developmental biology of floral symmetry. We suggest that a single CYC-like gene pair regulates the correlated evolution of multiple traits in zygomorphic flowers in Gesneriaceae. CYC-like genes might act as “hotspot” genes (Richardson and Brakefield 2003; Papa et al. 2008), which can be repeatedly recruited to regulate the evolution of complex traits, thereby facilitating the repeated origination of zygomorphic flowers and the explosive radiation of major clades of angiosperms.
Materials and methods
Plant material and culture conditions
C. pumila D. Don (Wang, HK01), collected from Hekou County, Yunnan, China, was used in this study. The plants were grown in 7-cm pots containing a mixture of vermiculite and commercially available humus soil (1:2) in the growth chamber. The seeds were surface-sterilized as described before (Liu et al. 2014) and germinated on half-strength Murashige and Skoog medium containing 10 g/L sucrose and 0.02 mg/L α-naphthylacetic acid (Liu, Wang, et al. 2021). Growth conditions were as described (Liu et al. 2014; Liu, Wang, et al. 2021).
SEM and Cryo-SEM
For SEM, floral buds of different stages were immediately fixed in FAA (3.7% formalin: 50% ethanol: 5% acetic acid) fixative via vacuum infiltration for 20 min. After dissecting the materials with a needle in 70% ethanol under a light microscope, the samples were treated with a graded ethanol series and subjected to critical point drying with CO2. The dissected floral buds were imaged under a Hitachi S-4800 scanning electron microscope (Tokyo).
To observe the surface morphology of petals and receptacles, fresh materials were collected and immediately observed under a Quorum PP3010T-Hitachi Regulus 8100 cryo-electron microscope (Tokyo). To quantify petal cell size, 30 cells per sample were measured using Fiji.
Gene isolation and phylogenetic analysis
Genomic DNA (gDNA) was extracted from fresh C. pumila leaves using a Plant DNA Extraction Kit (Tiangen). Total RNA was extracted from young flower buds using an SV Total RNA Isolation System (Promega) following the manufacturer's instructions, and complementary DNA (cDNA) was synthesized using a RevertAid H Minus First-Strand cDNA Synthesis Kit (Fermentas). Gene-specific primers (Supplemental Data Set 3) were used to amplify full-length CYC-like genes from gDNA or cDNA using high-fidelity PrimeSTAR Max DNA Polymerase (TaKaRa). The PCR products were cloned using a pEASY-Blunt Simple Cloning Kit (TransGen Biotech) and sequenced (Beijing Genomics Institute Tech).
DNAMAN software (Lynnon Biosoft) was used to translate CpCYC1, CpCYC2, and flavonoid synthesis-related genes into amino acid sequences. The amino acid sequences of related genes were retrieved from the National Center for Biotechnology Information (NCBI) database (www.ncbi.nlm.nih.gov/). The sequences were aligned using Clustal X (Thompson et al. 1997) and adjusted manually with BioEdit (Hall 1999). MEGA6 (Tamura et al. 2013) was utilized to construct phylogenetic trees, and the evolutionary history was inferred by using the maximum likelihood method based on the Jones-Taylor-Thornton matrix-based (Jones et al. 1992) or Le and Gascuel (2008) model. Bootstrap values were calculated for 1,000 replicates.
RNA in situ hybridization
Young inflorescences and floral buds were immediately fixed in ice-cold FAA as described above. After embedding in wax, 8-µm sections were prepared using a rotary microtome (Leica RM2165). Hybridization and detection were conducted as previously described (Bradley et al. 1993) with some modifications. Briefly, after incubation in xylene and drying though an ethanol series, the sections were treated with Proteinase K (Roche) for 30 min at 37 °C and postfixed with FAA. Hybridization was carried out at 50 °C overnight. Posthybridization washes were performed using 0.1 × SSC. For immunological detection, anti-DIG antibody (Roche) was used at a 1:3,000 dilution. After removing the nonspecific antibody, the slides were incubated in NBT/NCIP solution (Roche) at room temperature overnight.
For probe synthesis, cDNA fragments of CpCYC1 and CpCYC2 were amplified using gene-specific primers (Supplemental Data Set 3). Probes were synthesized with a DIG RNA Labeling Kit (Roche) using purified PCR products as templates.
RNA-seq and RT-qPCR
To identify genes that might control yellow spot formation, pooled samples of dorsal ridges and ventral yellow spots were dissected from at least 5 wild-type flowers ∼1.5 cm in length. Each sample included 3 biological replicates from at least 3 plants. Total RNA was extracted from the samples and sequenced (Beijing Genomics Institute Tech). The expression level of each gene was normalized by fragments per kilobase (FPKM of exon per million fragments mapped). Genes with a <0.01 false discovery rate and a >2-fold change were regarded as differentially expressed. We mainly focused on differentially expressed genes related to flavonoid synthesis.
For RT-qPCR, roots were collected from tissue-cultured seedlings, while stems and young leaves were collected from plants before flowering. Flowers of different stages were sampled as shown in Supplemental Fig. S6. Briefly, whole young flower buds 0.5, 1.0, and 1.5 cm in length were collected, while 2-cm flowers were dissected into sepals, dorsal/lateral/ventral petal lobes, dorsal/lateral/ventral stamens, bulges, ridges, yellow spots, and carpels. For tiny dorsal and lateral staminodes, each comprised only 1 pooled sample but was collected from more than 20 flowers from several plants. Other pooled materials included 3 biological replicates from at least 3 plants, and each replicate was collected from 3 to 5 flowers. For P. heterotricha, flower buds ∼2 cm long were dissected into dorsal/lateral/ventral petal lobes, lateral/ventral stamens, bulges, ridges, and yellow spots, each with 3 biological replicates. Gene-specific primers were used to amplify all genes, and CpACTIN was amplified as a reference (Supplemental Data Set 3). Primers for PhCYC1C, PhCYC1D, and PhACTIN were described before (Yang et al. 2012). The specificity of all primers was confirmed by sequencing the PCR products. RT-qPCR was performed using TB Green Premix Ex Taq (TaKaRa) in a StepOne Plus Real-Time PCR System (AB Applied Biosystems). The PCR conditions were as follows: initial denaturation at 95 °C for 30 s, 40 cycles of 95 °C for 5 s, and 60 °C for 30 s. Dissociation curves were recorded using 1 cycle of 95 °C for 15 s, 60 °C for 60 s, and 95 °C for 15 s. Relative expression levels were determined by normalizing the PCR threshold cycle number of each gene with that of ACTIN using the 2−ΔCt method.
Genetic transformation of C. pumila
For overexpression and dominant repression experiments, GFP was removed from pCAMBIA1302 to exclude its possible interference of target gene function. To overexpress CpCYC1 and CpCYC2, the CaMV 35S promoter (35Spro) was replaced by the duplicated 35S promoter (d35Spro). The full-length CDSs of CpCYC1, CpCYC2, PhCYC1C, and PhCYC1D were amplified and inserted into the respective vectors to obtain d35Spro:CpCYC1, d35Spro:CpCYC2, 35Spro:PhCYC1C, and 35Spro:PhCYC1D using an In-Fusion HD Cloning Kit (TaKaRa). For dominant repression, CpCYC1 and CpCYC2 were fused in-frame with the SRDX motif (Hiratsu et al. 2003) to generate 35Spro:CpCYC1-SRDX and 35Spro:CpCYC2-SRDX, respectively. For CRISPR/Cas9, gRNA1 and gRNA2 were designed, which respectively target CpCYC1 and CpCYC2. Reverse complementary oligonucleotides, CpCYC1-gRNA-F/R for gRNA1 and CpCYC2-gRNA-F/R for gRNA2 (Supplemental Fig. S8A), were synthesized, annealed, and inserted into the VK005-14 vector (Viewsolid Biotech) to obtain dpCas9-gRNA1 and dpCas9-gRNA2, respectively. dpCas9-gRNA1 was digested with Asc I and Spe I and inserted into the Asc I and Avr II sites of dpCas9-gRNA2 to obtain dpCas9-gRNA1-gRNA2 (Cas9-gRNA12; Supplemental Fig. S8B). All primers and oligonucleotides are listed in Supplemental Data Set 3. All plasmids were introduced into Agrobacterium strain LBA4404 or EHA105 and transformed into C. pumila according to Liu, Wang, et al. (2021) with only 1 modification: 5 mg/L rather than 10 mg/L of hygromycin was used in the selection medium.
VIGS assays
Gene-specific fragments of CpPDS (409 bp), CpANS (441 bp), and CpF3′5′H (400 bp) were amplified and inserted into pTRV2. The resulting plasmids, as well as pTRV1, were transformed into Agrobacterium strain GV3101. A 5-mL culture was grown at overnight 28 °C in YEB medium containing 50 mg/L kanamycin and 50 mg/L rifampicin. The next day, the culture was inoculated into 50-mL YEB medium containing antibiotics, 10 mm MES, and 20 mm acetosyringone. The cells were grown to an OD600 of ∼2.0. The Agrobacterium cells were harvested and resuspended in infiltration medium (10 mm MgCl2, 10 mm MES, 100 mm acetosyringone, and pH 5.8), adjusted to the OD600 value of 2.0, and incubated at room temperature in the dark for 3 h. Agrobacterium cells harboring pTRV1 and the newly constructed pTRV2 plasmids were mixed at a ratio of 1:1 just before infiltration. After adding 0.01% Silwet, the cell suspensions were infiltrated into fresh cuts in C. pumila inflorescence stems using 1-mL pipette tips. The plants were covered, incubated in the dark for 2 days, and grown under normal conditions.
Subcellular protein localization assays
The full-length CpCYC1 and CpCYC2 CDSs were inserted into the HBT-GFP vector to generate CpCYC1-GFP and CpCYC2-GFP plasmids, respectively. The plasmids were transformed into C. pumila mesophyll protoplast as described before (Liu, Wang, et al. 2021).
Production of recombinant proteins and EMSA
Partial CpCYC1 and CpCYC2 CDSs were respectively amplified using gene-specific primers (Supplemental Data Set 3), digested with BamH I and EcoR I, and inserted into the pET30a vector (Novagen). Recombinant proteins were expressed in BL21 E. coli cells and purified from the soluble fractions using a His SpinTrap Kit (GE Healthcare). To analyze the interaction of CpCYC1 and CpCYC2 proteins with 2 cis-regulatory elements in the CpF3′5′H promoter, the full-length CpCYC1 and CpCYC2 CDSs were inserted into the pTNT vector. CpCYC1 and CpCYC2 proteins were translated using the TNT SP6 High-Yield Wheat Germ Protein Expression System (Promega).
To prepare probes, 20 bp of biotin-labeled oligonucleotides were synthesized (Sangon) and annealed to form double-stranded DNA probes. EMSA was performed using a LightShift Chemiluminescent EMSA Kit (Pierce). To determine whether CpCYC1 and CpCYC2 could form homodimers or heterodimers, CpCYC2 protein was added to the reaction mixture after CpCYC1 had been incubated with the probe at room temperature for 15 min. The mixture was incubated for another 30 min as described before (Yang et al. 2012).
Yeast 2-hybrid assay
The full-length CDSs of CpCYC1 and CpCYC2 were amplified and inserted into pGADT7 and pGBKT7 to obtain the CpCYC1-AD, CpCYC1-BD, CpCYC2-AD, and CpCYC2-BD plasmids. Different combinations of plasmids were cotransformed into yeast (S. cerevisiae) strain Y2HGold (Clontech) using the lithium acetate/polyethylene glycol transformation method. The transformed yeast cells were cultured on SD/-Trp/-Leu medium to select positive transformants. Protein–protein interactions were identified by inoculating positive transformants onto SD/-Trp/-Leu/-His/-Ade and SD/-Trp/-Leu/-His/-Ade/X-α-gal plates.
Transient gene expression assays
The promoter sequences of CpCYC1 (1,435 bp), CpCYC2 (1,196 bp), and CpF3′5′H (2,194 bp) were amplified and inserted into the pGreenII0800 vector using an In-Fusion HD Cloning Kit to obtain CpCYC1pro:LUC, CpCYC2pro:LUC, and CpF3′5′Hpro:LUC. A. tumefaciens GV3101 pSoup containing each reporter plasmid and EHA105 harboring d35Spro:CpCYC1 or d35Spro:CpCYC2 were respectively resuspended in MMA induction medium (10 mm MgCl2, 10 mm MES, 200 µm acetosyringone, and pH 5.6) to a final OD600 of 0.6, mixed, and infiltrated into N. benthamiana leaves. The LUC activity was measured 2 days later using the Dual-Luciferase Reporter Assay System (Promega).
LCI assays
The full-length CDSs of CpCYC1 and CpCYC2 were fused in-frame to the N-terminus of NLuc in pCAMBIA1300-NLuc and the C-terminus of CLuc in the pCAMBIA1300-CLuc (Chen et al. 2008). The resulting constructs were respectively transformed into A. tumefaciens strains EHA105 and GV3101. For coinfiltration, A. tumefaciens containing each plasmid was resuspended in MMA induction medium to a final OD600 value of 1.0. N. benthamiana coinfiltration experiments were performed as described above. LUC signals were detected under a low-light cooled CCD camera (Tanon) (Chen et al. 2008). For quantification, 6 leaves were coinfiltrated for all combinations, and the signal intensity was measured using Fiji.
Statistical analysis
For simple pair-wise data analysis, 2-tailed Student's t test was performed using Microsoft Excel. For multiple data comparison, ANOVA followed by a post hoc Tukey's test was performed using SPSS 16.0 software.
Accession numbers
Sequence data from this article can be found in the NCBI database under the following accession numbers: CpCYC1 (OQ515463), CpCYC2 (OQ515464), CpCHS1 (OQ515465), CpCHS2 (OQ515466), CpCHI (OQ515467), CpF3′5′H (OQ515468), and CpMYB111 (OQ515469).
Supplementary Material
Acknowledgments
We greatly appreciate Prof. Jian-Min Zhou (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) for sharing the pCAMBIA1300-NLuc and pCAMBIA1300-CLuc vectors. We thank Dr. Peng-Wei Li (Guangxi Institute of Botany, Chinese Academy of Sciences) for providing photographs of C. anachoreta, Dr. Pi-Chang Gong (Institute of Botany, Chinese Academy of Sciences; IBCAS) for help in LCI assays, and Meng-Qi Han as well as Quan Yuan (IBCAS) for taking the UV photos. We also appreciate Dr. Xiu-Ping Xu from Plant Science Facility (IBCAS) for her excellent technical assistance with SEM and Cryo-SEM.
Contributor Information
Xia Yang, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China.
Yang Wang, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Tian-Xia Liu, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Qi Liu, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Jing Liu, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China.
Tian-Feng Lü, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China.
Rui-Xue Yang, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Feng-Xian Guo, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Yin-Zheng Wang, State Key Laboratory of Systematic and Evolutionary Botany, Institute of Botany, Chinese Academy of Sciences, Beijing 100093, China; China National Botanical Garden, Beijing 100093, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Author contributions
Y.-Z.W. initiated and supervised the project, and revised the manuscript. X.Y. designed and performed most of the experiments except for in situ hybridization, analyzed the data, prepared the figures, and wrote the manuscript. Y.W. established the VIGS system using the reference genes PDS and ANS. T.-X.L. performed in situ hybridization. Q.L. and J.L. were involved in genotyping. F.-T.L. constructed Y2H vectors and helped with RNA-seq analysis. R.-X.Y. and F.-X.G. helped with the transgenic experiments. All authors read and approved the manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Measurement of C. pumila petal size.
Supplemental Figure S2. Flavonoids are responsible for yellow spot formation in C. pumila.
Supplemental Figure S3. Phylogenetic analysis of CYC-like proteins by the maximum likelihood method.
Supplemental Figure S4. Subcellular localization of CpCYC1 and CpCYC2 proteins via transient gene expression in C. pumila mesophyll protoplasts.
Supplemental Figure S5. Amino acid sequence alignment of TCP and R domains of GCYC1-like proteins.
Supplemental Figure S6. CpCYC1 and CpCYC2 are specifically expressed in inflorescences and flowers.
Supplemental Figure S7. Different d35Spro:CpCYC1 and d35Spro:CpCYC2 transgenic lines show varying degrees of dorsalized flowers.
Supplemental Figure S8. Diagram showing how the Cas9-gRNA12 plasmid was constructed.
Supplemental Figure S9. Different T1 lines of Cas9-gRNA1/2 show varying degrees of ventralized flowers.
Supplemental Figure S10. Genotyping results of Cas9-gRNA1/2 T1 plants.
Supplemental Figure S11. Phenotypes of cyc1 and cyc2 single mutants from a cross between wild-type plants and the cyc1 cyc2 double mutant.
Supplemental Figure S12. Gene structures of CpCYC1, CpCYC2, and CpF3′5′H.
Supplemental Figure S13. CpCYC1 and CpCYC2 can form homodimers and heterodimers in yeast.
Supplemental Figure S14. Phylogenetic analysis of flavonoid synthesis-related proteins by the maximum likelihood method.
Supplemental Figure S15. CpCYC1 and CpCYC2 repress yellow spot formation by negatively regulating CpF3′5′H.
Supplemental Figure S16. C. anachoreta flowers possess similar morphology to C. pumila.
Supplemental Data Set 1. Raw data for statistical analysis.
Supplemental Data Set 2. Sequence alignments of proteins for phylogenetic analysis.
Supplemental Data Set 3. Primers used in this study.
Funding
This work was supported by the National Natural Science Foundation of China (grant nos. 31970239 to Y.-Z.W. and 31670217 to X.Y.), the Youth Innovation Promotion Association of the Chinese Academy of Sciences (2016079 to X.Y.), and the K.C. Wong Education Foundation (GJTD-2020-05).
Data availability
Source data are provided with this paper. All additional data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Armbruster WS, Pélabon C, Bolstad GH, Hansen TF. Integrated phenotypes: understanding trait covariation in plants and animals. Phil Trans R Soc Lond B Biol Sci. 2014:369(1649):20130245. 10.1098/rstb.2013.0245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bawa KS, Ingty T, Revell LJ, Shivaprakash KN. Correlated evolution of flower size and seed number in flowering plants (monocotyledons). Annals Bot. 2019:123(1):181–190. 10.1093/aob/mcy154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley D, Carpenter R, Sommer H, Hartley N, Coen E. Complementary floral homeotic phenotypes result from opposite orientations of a transposon at the plena-locus of Antirrhinum. Cell 1993:72(1):85–95. 10.1016/0092-8674(93)90052-R [DOI] [PubMed] [Google Scholar]
- Briscoe AD, Chittka L. The evolution of color vision in insects. Ann Rev Entomol. 2001:46(1):471–510. 10.1146/annurev.ento.46.1.471 [DOI] [PubMed] [Google Scholar]
- Busch A, Zachgo S. Control of corolla monosymmetry in the Brassicaceae Iberis amara. Proc Natl Acad Sci U S A. 2007:104(42):16714–16719. 10.1073/pnas.0705338104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Shen C-Z, Guo Y-P, Rao G-Y. Patterning the Asteraceae capitulum: duplications and differential expression of the flower symmetry CYC2-like genes. Front Plant Sci. 2018:9:551–514. 10.3389/fpls.2018.00551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou JM. Firefly luciferase complementation imaging assay for protein–protein interactions in plants. Plant Physiol. 2008:146(2):368–376. 10.1104/pp.107.111740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa MMR, Fox S, Hanna AI, Baxter C, Coen E. Evolution of regulatory interactions controlling floral asymmetry. Development 2005:132(22):5093–5101. 10.1242/dev.02085 [DOI] [PubMed] [Google Scholar]
- Crews ST, Pearson JC. Transcriptional autoregulation in development. Curr Biol. 2009:19(6):R241–R246. 10.1016/j.cub.2009.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubas P, Lauter N, Doebley J, Coen E. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 1999:18(2):215–222. 10.1046/j.1365-313X.1999.00444.x [DOI] [PubMed] [Google Scholar]
- Dafni A, Kevan PG. Floral symmetry and nectar guides: ontogenetic constraints from floral development, colour pattern rules and functional significance. Bot J Linn Soc. 1996:120(4):371–377. 10.1111/j.1095-8339.1996.tb00487.x [DOI] [Google Scholar]
- Demarche ML, Miller TJ, Kay KM. An ultraviolet floral polymorphism associated with life history drives pollinator discrimination in Mimulus guttatus. Amer J Bot. 2015:102(3):396–406. 10.3732/ajb.1400415 [DOI] [PubMed] [Google Scholar]
- Dilcher D. Toward a new synthesis: major evolutionary trends in the angiosperm fossil record. Proc Natl Acad Sci U S A. 2000:97(13):7030–7036. 10.1073/pnas.97.13.7030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Liu J, Li P-W, Li C-Q, Lü T-F, Yang X, Wang Y-Z. Evolution of Darwin's Peloric gloxinia (Sinningia speciosa) is caused by a null mutation in a pleiotropic TCP gene. Mol Biol Evol. 2018:35(8):1901–1915. 10.1093/molbev/msy090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress PK. Evolution of floral symmetry. Curr Opin Plant Biol. 2001:4(1):86–91. 10.1016/S1369-5266(00)00140-0 [DOI] [PubMed] [Google Scholar]
- Feng X, Zhao Z, Tian Z, Xu S, Luo Y, Cai Z, Wang Y, Yang J, Wang Z, Weng L, et al. Control of petal shape and floral zygomorphy in Lotus japonicus. Proc Natl Acad Sci U S A. 2006:103(13):4970–4975. 10.1073/pnas.0600681103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenster CB, Armbruster WS, Dudash MR. Specialization of flowers: is floral orientation an overlooked first step? New Phytol. 2009:183(3):502–506. 10.1111/j.1469-8137.2009.02852.x [DOI] [PubMed] [Google Scholar]
- Gao Q, Tao J-H, Yan D, Wang Y-Z, Li Z-Y. Expression differentiation of CYC-like floral symmetry genes correlated with their protein sequence divergence in Chirita heterotricha (Gesneriaceae). Dev Genes Evol. 2008:218(7):341–351. 10.1007/s00427-008-0227-y [DOI] [PubMed] [Google Scholar]
- Hall TA. Bioedit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999:41:95–98. https://www.researchgate.net/publication/200028981 [Google Scholar]
- Hansen DM, Van der Niet T, Johnson SD. Floral signposts: testing the significance of visual ‘nectar guides’ for pollinator behaviour and plant fitness. Proc Biol Sci. 2012:279(1729):634–639. 10.1098/rspb.2011.1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman LC, Cubas P. An expanded evolutionary role for flower symmetry genes. J Biol. 2009:8(10):90. 10.1186/jbiol193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hileman LC, Kramer EM, Baum DA. Differential regulation of symmetry genes and the evolution of floral morphologies. Proc Natl Acad Sci U S A. 2003:100(22):12814–12819. 10.1073/pnas.1835725100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TA, Day CD, Zondlo SC, Thackeray AG, Irish VF. Discrete spatial and temporal cis-acting elements regulate transcription of the Arabidopsis floral homeotic gene APETALA3. Development 1998:125(9):1711–1721. 10.1242/dev.125.9.1711 [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. Plant J. 2003:34(5):733–739. 10.1046/j.1365-313X.2003.01759.x [DOI] [PubMed] [Google Scholar]
- Hodges SA, Whittall JB, Fulton M, Yang JY. Genetics of floral traits influencing reproductive isolation between Aquilegia formosa and Aquilegia pubescens. Am Nat. 2002:159(S3):S51S60. 10.1086/338372 [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K. Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 2001:409(6819):525–529. 10.1038/35054083 [DOI] [PubMed] [Google Scholar]
- Hsin K-T, Lu J-Y, Möller M, Wang C-N. Gene duplication and relaxation from selective constraints of GCYC genes correlated with various floral symmetry patterns in Asiatic Gesneriaceae tribe Trichosporeae. PLoS One 2019:14(1):e0210054. 10.1371/journal.pone.0210054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu H-C, Kuo Y-F. Nectar guide patterns on developmentally homologous regions of the subtribe Ligeriinae (Gesneriaceae). Front Plant Sci. 2021:12:1–11. 10.3389/fpls.2021.650836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jachuła J, Konarska A, Denisow B. Micromorphological and histochemical attributes of flowers and floral reward in Linaria vulgaris (Plantaginaceae). Protoplasma 2018:255(6):1763–1776. 10.1007/s00709-018-1269-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput App Biosci. 1992:8(3):275–282. 10.1093/bioinformatics/8.3.275 [DOI] [PubMed] [Google Scholar]
- Jorgensen EC, Geissmann TA. The chemistry of flower pigmentation in Antirrhinum majus color genotypes. III. Relative anthocyanin and aurone concentration. Arch Biochem Biophys. 1955:55(2):389–402. 10.1016/0003-9861(55)90420-3 [DOI] [Google Scholar]
- Kern EMA, Robinson D, Gass E, Godwin J, Langerhans RB. Correlated evolution of personality, morphology and performance. Anim Behav. 2016:117:79–86. 10.1016/j.anbehav.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski MH, Ashman T-L. Dissecting pollinator responses to a ubiquitous ultraviolet floral pattern in the wild. Func Ecol. 2014:28(4):868–877. 10.1111/1365-2435.12242 [DOI] [Google Scholar]
- Koski MH, MacQueen D, Ashman T-L. Floral pigmentation has responded rapidly to global change in ozone and temperature. Curr Biol. 2020:30(22):4425–4431. 10.1016/j.cub.2020.08.077 [DOI] [PubMed] [Google Scholar]
- Le SQ, Gascuel O. An improved general amino acid replacement matrix. Mol Biol Evol. 2008:25(7):1307–1320. 10.1093/molbev/msn067 [DOI] [PubMed] [Google Scholar]
- Leonard E, Yan Y, Koffas MAG. Functional expression of a P450 flavonoid hydroxylase for the biosynthesis of plant-specific hydroxylated flavonols in Escherichia coli. Metab Eng. 2006:8(2):172–181. 10.1016/j.ymben.2005.11.001 [DOI] [PubMed] [Google Scholar]
- Liu J, Wang J-J, Wu J, Wang Y, Liu Q, Liu F-P, Yang X, Wang Y-Z. An optimized transformation system and functional test of CYC-like TCP gene CpCYC in Chirita pumila (Gesneriaceae). Int J Mol Sci. 2021:22(9):4544. 10.3390/ijms22094544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Wu J, Yang X, Wang Y-Z. Regulatory pathways of CYC-like genes in patterning floral zygomorphy exemplified in Chirita pumila. J Syst Evol. 2021:59(3):567–580. 10.1111/jse.12574 [DOI] [Google Scholar]
- Liu B-L, Yang X, Liu J, Dong Y, Wang Y-Z. Characterization, efficient transformation and regeneration of Chirita pumila (Gesneriaceae), a potential evo-devo model plant. Plant Cell Tiss Organ Cult. 2014:118(2):357–371. 10.1007/s11240-014-0488-2 [DOI] [Google Scholar]
- Luo D, Carpenter R, Copsey L, Vincent C, Clark J, Coen E. Control of organ asymmetry in flowers of Antirrhinum. Cell 1999:99(4):367–376. 10.1016/S0092-8674(00)81523-8 [DOI] [PubMed] [Google Scholar]
- Luo D, Carpenter R, Vincent C, Copsey L, Coen E. Origin of floral asymmetry in Antirrhinum. Nature 1996:383(6603):794–799. 10.1038/383794a0 [DOI] [PubMed] [Google Scholar]
- Moyers BT, Owens GL, Baute GJ, Rieseberg LH. The genetic architecture of UV floral patterning in sunflower. Ann Bot. 2017:120(1):39–50. 10.1093/aob/mcx038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murren CJ. The integrated phenotype. Integr Compar Biol. 2012:52(1):64–76. 10.1093/icb/ics043 [DOI] [PubMed] [Google Scholar]
- Nascimento LFD, Guimarães PR, Onstein RE, Kissling WD, Pires MM. Associated evolution of fruit size, fruit colour and spines in Neotropical palms. J Evol Biol. 2020:33(6):858–868. 10.1111/jeb.13619 [DOI] [PubMed] [Google Scholar]
- Neal PR, Dafni A, Giurfa M. Floral symmetry and its role in plant–pollinator systems: terminology, distribution, and hypotheses. Annu Rev Ecol Syst. 1998:29(1):345–373. 10.1146/annurev.ecolsys.29.1.345 [DOI] [Google Scholar]
- Nishihara M, Nakatsuka T. Genetic engineering of flavonoid pigments to modify flower color in floricultural plants. Biotechnol Lett. 2011:33(3):433–441. 10.1007/s10529-010-0461-z [DOI] [PubMed] [Google Scholar]
- Ono E, Fukuchi-Mizutani M, Nakamura N, Fukui Y, Yonekura-Sakakibara K, Yamaguchi M, Nakayama T, Tanaka T, Kusumi T, Tanaka Y. Yellow flowers generated by expression of the aurone biosynthetic pathway. Proc Natl Acad Sci U S A. 2006:103(29):11075–11080. 10.1073/pnas.0604246103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa R, Martin A, Reed RD. Genomic hotspots of adaptation in butterfly wing pattern evolution. Curr Opin Genet Dev. 2008:18(6):559–564. 10.1016/j.gde.2008.11.007 [DOI] [PubMed] [Google Scholar]
- Pilote AJ, Donovan LA. Evidence of correlated evolution and adaptive differentiation of stem and leaf functional traits in the herbaceous genus, Helianthus. Amer J Bot. 2016:103(12):2096–2104. 10.3732/ajb.1600314 [DOI] [PubMed] [Google Scholar]
- Richardson MK, Brakefield PM. Hotspots for evolution. Nature 2003:424(6951):894–895. 10.1038/424894a [DOI] [PubMed] [Google Scholar]
- Santos JC, Cannatella DC. Phenotypic integration emerges from aposematism and scale in poison frogs. Proc Natl Acad Sci U S A. 2011:108(15):6175–6180. 10.1073/pnas.1010952108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasakia K, Takahashib T. A flavonoid from Brassica rapa flower as the UV-absorbing nectar guide. Phytochemistry 2002:61(3):339–343. 10.1016/S0031-9422(02)00237-6 [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer Z, Hue I, Huijser P, Flor PJ, Hansen R, Tetens F, Lonnig W-E, Saedler H, Sommer H. Characterization of the Antirrhinum floral homeotic MADS-box gene deficiens: evidence for DNA binding and autoregulation of its persistent expression throughout flower development. EMBO J. 1992:1(1):251–263. 10.1002/j.1460-2075.1992.tb05048.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinnott-Armstrong MA, Lee C, Clement WL, Donoghue MJ. Fruit syndromes in Viburnum: correlated evolution of color, nutritional content, and morphology in bird-dispersed fleshy fruits. BMC Evol Biol. 2020:20(1):7. 10.1186/s12862-019-1546-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirito M, Lin Q, Deng JM, Behringer RR, Sawadogo M. Overlapping roles and asymmetrical cross-regulation of the USF proteins in mice. Proc Natl Acad Sci U S A. 1998:95(7):3758–3763. 10.1073/pnas.95.7.3758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C-F, Lin Q-B, Liang R-H, Wang Y-Z. Expressions of ECE-CYC2 clade genes relating to abortion of both dorsal and ventral stamens in Opithandra (Gesneriaceae). BMC Evol Biol. 2009:9(1):244. 10.1186/1471-2148-9-244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S, Xiao W, Guo W, Yao X, Xiao J, Ye Z, Wang N, Jiao K, Lei M, Peng Q, et al. The CYCLOIDEA–RADIALIS module regulates petal shape and pigmentation, leading to bilateral corolla symmetry in Torenia fournieri (Linderniaceae). New Phytol. 2017:215(4):1582–1593. 10.1111/nph.14673 [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013:30(12):2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997:25(24):4876–4882. 10.1093/nar/25.24.4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WR, Meinwald J, Aneshansley D, Eisner T. Flavonols: pigments responsible for ultraviolet absorption in nectar guide of flower. Science 1972:177(4048):528–530. 10.1126/science.177.4048.528 [DOI] [PubMed] [Google Scholar]
- Tilly JJ, Allen DW, Jack T. The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 1998:125(9):1647–1657. 10.1242/dev.125.9.1647 [DOI] [PubMed] [Google Scholar]
- Tong J, Knox EB, Morden CW, Cellinese N, Mossolem F, Zubair AS, Howarth DG. Duplication and expression patterns of CYCLOIDEA-like genes in Campanulaceae. EvoDevo. 2022:13(1):5. 10.1186/s13227-021-00189-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushimaru A, Dohzono I, Takami Y, Hyodo F. Flower orientation enhances pollen transfer in bilaterally symmetrical flowers. Oecologia 2009:160(4):667–674. 10.1007/s00442-009-1334-9 [DOI] [PubMed] [Google Scholar]
- Ushimaru A, Hyodo F. Why do bilaterally symmetrical flowers orient vertically? Flower orientation influences pollinator landing behaviour. Evol Ecol Res. 2005:7:252–260. https://www.researchgate.net/publication/43695340 [Google Scholar]
- Ushimaru A, Watanabe T, Nakata K. Colored floral organs influence pollinator behavior and pollen transfer in Commelina communis. Amer J Bot. 2007:94(2):249–258. 10.3732/ajb.94.2.249 [DOI] [PubMed] [Google Scholar]
- Wagner GP, Pavlicev M, Cheverud JM. The road to modularity. Nat Rev Genet. 2007:8(12):921–931. 10.1038/nrg2267 [DOI] [PubMed] [Google Scholar]
- Wagner GP, Zhang J. The pleiotropic structure of the genotype–phenotype map: the evolvability of complex organisms. Nat Rev Genet. 2011:12(3):204–213. 10.1038/nrg2949 [DOI] [PubMed] [Google Scholar]
- Wang Z, Luo Y, Li X, Wang L, Xu S, Yang J, Weng L, Sato S, Tabata S, Ambrose M, et al. Genetic control of floral zygomorphy in pea (Pisum sativum L). Proc Natl Acad Sci U S A. 2008:105(30):10414–10419. 10.1073/pnas.0803291105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y-S, Xu Y-J, Gao L-P, Yu O, Wang X-Z, He X-J, Jiang X-L, Liu Y-J, Xia T. Functional analysis of flavonoid 3′,5′-hydroxylase from tea plant (Camellia sinensis): critical role in the accumulation of catechins. BMC Plant Biol. 2014:14(1):347. 10.1186/s12870-014-0347-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessinger CA, Hileman LC. Accessibility, constraint, and repetition in adaptive floral evolution. Dev Biol. 2016:419(1):175–183. 10.1016/j.ydbio.2016.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Pang H-B, Liu B-L, Qiu Z-J, Gao Q, Wei L, Dong Y, Wang Y-Z. Evolution of double positive autoregulatory feedback loops in CYCLOIDEA2 clade genes is associated with the origin of floral zygomorphy. Plant Cell 2012:24(5):1834–1847. 10.1105/tpc.112.099457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zhao X-G, Li C-Q, Liu J, Qiu Z-J, Dong Y, Wang Y-Z. Distinct regulatory changes underlying differential expression of TEOSINTE BRANCHED1-CYCLOIDEA- PROLIFERATING CELL FACTOR genes associated with petal variations in zygomorphic flowers of Petrocosmea spp. of the family Gesneriaceae. Plant Physiol. 2015:169(3):2138–2151. 10.1104/pp.15.01181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Wang B, Zhang W, Shan H, Kong H. Gains and losses of cis-regulatory elements led to divergence of the Arabidopsis APETALA1 and CAULIFLOWER duplicate genes in the time, space, and level of expression and regulation of one paralog by the other. Plant Physiol. 2016:171(2):1055–1069. 10.1104/pp.16.00320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y-W, Sagawa JM, Di Stilio VS, Bradshaw HD Jr. Bulk segregant analysis of an induced floral mutant identifies a MIXTA-like R2R3 MYB controlling nectar guide formation in Mimulus lewisii. Genetics 2013:194(2):523–528. 10.1534/genetics.113.151225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Zhang J, Nageswaran D, Li L. Carotenoid metabolism and regulation in horticultural crops. Horticul Res. 2015:2:1–11. 10.1038/hortres.2015.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X-R, Wang Y-Z, Smith JF, Chen R. Altered expression patterns of TCP and MYB genes relating to the floral developmental transition from initial zygomorphy to actinomorphy in Bournea (Gesneriaceae). New Phytol. 2008:178(3):532–543. 10.1111/j.1469-8137.2008.02384.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data are provided with this paper. All additional data that support the findings of this study are available from the corresponding author upon reasonable request.