MdMAPK3 promotes apple fruit softening during storage by regulating the phosphorylation status and degradation of an apple transcription factor, MdNAC72.
Abstract
The phytohormone ethylene plays an important role in promoting the softening of climacteric fruits, such as apples (Malus domestica); however, important aspects of the underlying regulatory mechanisms are not well understood. In this study, we identified apple MITOGEN-ACTIVATED PROTEIN KINASE 3 (MdMAPK3) as an important positive regulator of ethylene-induced apple fruit softening during storage. Specifically, we show that MdMAPK3 interacts with and phosphorylates the transcription factor NAM-ATAF1/2-CUC2 72 (MdNAC72), which functions as a transcriptional repressor of the cell wall degradation-related gene POLYGALACTURONASE1 (MdPG1). The increase in MdMAPK3 kinase activity was induced by ethylene, which promoted the phosphorylation of MdNAC72 by MdMAPK3. Additionally, MdPUB24 functions as an E3 ubiquitin ligase to ubiquitinate MdNAC72, resulting in its degradation via the 26S proteasome pathway, which was enhanced by ethylene-induced phosphorylation of MdNAC72 by MdMAPK3. The degradation of MdNAC72 increased the expression of MdPG1, which in turn promoted apple fruit softening. Notably, using variants of MdNAC72 that were mutated at specific phosphorylation sites, we observed that the phosphorylation state of MdNAC72 affected apple fruit softening during storage. This study thus reveals that the ethylene–MdMAPK3–MdNAC72–MdPUB24 module is involved in ethylene-induced apple fruit softening, providing insights into climacteric fruit softening.
Introduction
Softening is a common physiological stage of most fleshy fruits after harvest and can lead to a decline in fruit quality (Paniagua et al. 2014). Therefore, inhibiting softening can effectively prolong the shelf life of fruit. The degradation and remodeling of cell wall polymers are considered important factors in fruit softening (Paniagua et al. 2014). Approximately 90% of the dry weight from the cell wall consists of polysaccharides (cellulose, hemicellulose, and pectin), while the remaining 10% includes proteins, fatty acids, phenylpropanoids, and inorganic compounds (Wolf-Dieter 1998). Cellulose is composed of unbranched β-1,4-glucan chains and forms the basic skeleton of the cell wall. In addition, dozens of glucan chains are entangled and interwoven into microfibrils, which constitute the higher order structure of cellulose (Römling and Galperin 2015). The hemicellulose backbone is composed of β-1,4-glucan chains, which are modified by various sugar branches, mainly xylan, xyloglucan, mannan, and mixed-linkage glucan. Hemicellulose binds to the surface of the microfibrils to maintain the higher order structure of cellulose (Khodayari et al. 2021). Pectin is located in the primary wall and middle lamella and is a complex polysaccharide that plays an important role in cell-to-cell adhesion. Three major classes of pectin have been identified: homogalacturonan, rhamnogalacturonan I, and rhamnogalacturonan II. Furthermore, pectin is intertwined with cellulose and hemicellulose to maintain cell strength (Wang et al. 2018).
Fruit softening is mainly caused by the breakdown of the primary cell wall and middle lamella. A series of hydrolytic enzymes catalyzes these steps, including pectin methylesterase (PME), which catalyzes the removal of methyl ester groups from pectin; polygalacturonase (PG), which hydrolyzes α-1,4 glycosidic bonds in galactosyluronic; pectate lyase (PL), which causes eliminative cleavage of polymers of α-1,4 galactosyluronic acid molecules to yield oligosaccharides with 4-deoxy-α-d-galact-4-enuronosyl groups at their nonreducing ends; β-galactosidase (β-Gal), which removes galactosyl residues from pectin; arabinofuranosidase (α-AFase), which releases terminal arabinofuranosyl residues from a variety of pectic and hemicellulosic polymers; xyloglucan endotransglycosylase/hydrolases (XETs), which catalyze the cleavage and polymerization of xyloglucan molecules; and expansin (EXP), which catalyzes the reversible breakage of hemicellulose–cellulose hydrogen bonds (Tucker et al. 2017; Wang et al. 2018).
Apples (Malus domestica) are an economically important crop consumed worldwide. During storage, apples undergo physiological and biochemical changes. Of these complex changes, softening is one of the most obvious changes affecting the visual quality of the fruit. Excessive softening not only makes the fruit less attractive to consumers but also increases transportation and storage costs due to higher pathogen susceptibility. Softening in apples is associated with an increase in the expression of a number of cell wall degradation-related genes, such as MdPG1, Mdβ-Gal, MdEXP3, and MdXET1 (Wakasa et al. 2006; Tacken et al. 2010). Of these, the best-characterized gene thus far is MdPG1. The correlation between PG expression and softening rate, together with quantitative trait locus analysis, has suggested that PG is involved in the regulation of softening and textural changes during postharvest storage (Costa et al. 2010; Atkinson et al. 2012; Longhi et al. 2012). Consistently, MdPG1 is specifically expressed during postharvest storage of fruit, and overexpressing MdPG1 in apples was shown to promote intercellular separation in leaves (Atkinson et al. 2002). Conversely, silencing MdPG1 in apples inhibited pectin degradation and softening during postharvest storage (Atkinson et al. 2012), making MdPG1 an extensively studied gene as a marker of apple fruit softening (Tacken et al. 2010).
Apple fruits typically exhibit climacteric ripening (Velasco et al. 2010). During postharvest storage, there is a peak in ethylene release and a simultaneous decrease in firmness (Yang et al. 2013). Regulation of apple fruit softening by ethylene has been demonstrated, with ethylene treatment promoting MdPG1 expression, coinciding with accelerated fruit softening, whereas treatment with the competitive ethylene signaling inhibitor 1-methylcyclopropene (1-MCP) has the opposite effect (Wakasa et al. 2006). In other studies, transcriptional regulation has been shown to be integral to ethylene-induced softening of apples. For example, the transcription factors MdEIL2 (ETHYLENE-INSENSITIVE3 [EIN3]-LIKE2) and MdCBF2 (C-REPEAT/DRE BINDING FACTOR 2) directly regulate MdPG1 transcript levels and together participate in regulating ethylene- and low-temperature-promoted apple fruit softening during postharvest storage (Tacken et al. 2010). In addition, the transcription factors MADS6, MADS8, and MADS9 directly regulate the expression of MdPG1 (Ireland et al. 2013). Furthermore, in tomato (Solanum lycopersicum), the presence of several NAC (NAM-ATAF1/2-CUC2)-binding sites in the promoters of cell wall degradation-related genes has been reported, and Non-ripening (NOR, encoding a NAC transcription factor) mutant fruit exhibited decreased expression of these genes, suggesting that NAC is an important transcription factor that regulates fruit softening (Kosma et al. 2010; Osorio et al. 2011). In addition, a recent study showed that ETHYLENE TRANSCRIPTION FACTOR.F12 (SlERF.F12) inhibits tomato fruit softening by regulating multiple fruit ripening-related genes, including SlPG2a and SlPL, via epigenetic modifications affecting the acetylation levels of their promoter regions (Deng et al. 2022). However, little is known about the role of posttranslational mechanisms in modulating the activity of softening-associated genes and their transcriptional regulation.
Phosphorylation of 1 key posttranslational modification plays an important role in many aspects of gene/protein action, including the regulation of protein activity, subcellular location, protein–protein interactions, and stability (Trewavas 1976; Kwon et al. 2006). Protein phosphorylation mediated by mitogen-activated protein kinases (MAPK) is a common protein phosphorylation pathway in plants and is highly conserved. A typical MAPK signaling cascade consists of 3 protein kinases, MAPKKK (MAPK kinase kinase), MAPKK (MAPK kinase), and MAPK, which are sequentially activated by an upstream kinase. As the terminal molecule of the signaling cascade, MAPK specifically recognizes substrate proteins, including enzymes, structural proteins, and transcription factors, and phosphorylates them, thus playing a critical role in various signaling networks (Tena et al. 2001). Recently, phosphorylation modifications of transcription factors have been extensively studied because of the important roles of transcription factors as key regulatory proteins in many signal transduction pathways. For example, in rice (Oryza sativa), phosphorylation of OsbHLH002 (also named OsICE1 [INDUCER OF CBF EXPRESSION 1]) by OsMAPK3 inhibits its degradation and increases its transcriptional activation ability during acclimation to chilling stress (Zhang et al. 2017). In banana (Musa acuminata), phosphorylation of MabZIP21 (leucine zipper 21) by MaMPK6-3 promotes the transcriptional activation of fruit ripening- and softening-related genes, thus accelerating the physiological responses they control (Wu et al. 2022a). A recent study showed that light-induced MdMAPK4-mediated phosphorylation of MdMYB1 promotes its transcriptional activation and inhibits ubiquitination-mediated degradation of MdMYB1, which, in turn, promotes the accumulation of anthocyanins in fruits (Yang et al. 2021). However, the role of MAPKs in ethylene-induced fruit softening is poorly understood.
In this study, we aimed to elucidate the role of MAPK signaling in ethylene-induced softening of apple fruits. We showed that postharvest ethylene treatment of apple fruit promotes phosphorylation of MdNAC72 by MdMAPK3, which promotes MdNAC72 ubiquitination by the E3 ligase MdPUB24 (PLANT U-BOX 24) and subsequent degradation. MdNAC72 is a negative regulator of MdPG1 transcription; therefore, reduced accumulation of MdNAC72 resulted in increased MdPG1 transcript abundance and accelerated softening of apple fruit during postharvest storage. These results provide insights into the molecular mechanism by which ethylene induces fruit softening.
Results
Ethylene promotes apple fruit softening and MdMAPK3 activity during storage
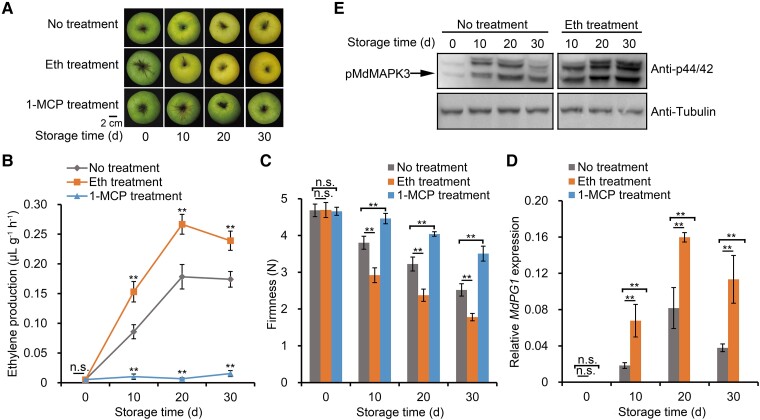
To investigate the effects of ethylene on apple fruit softening during storage, we treated ripe Golden Delicious (M. domestica cv. GD) fruit with ethylene or 1-MCP (an ethylene signal inhibitor). Ethylene treatment accelerated ethylene biosynthesis and fruit softening, whereas 1-MCP treatment had the opposite effect (Fig. 1, A–C). Fruit firmness decreased by approximately 50% when fruits were stored for 30 d, whereas softening was significantly accelerated by treatment with the ethylene precursor ethephon and inhibited after 1-MCP treatment (Fig. 1C). Furthermore, the expression of the cell wall degradation-related gene, MdPG1, increased following ethephon treatment, but was repressed upon 1-MCP treatment (Fig. 1D). These results are consistent with ethylene promoting MdPG1 expression, causing increased degradation of pectin, leading to fruit softening.
Figure 1.
Ethylene promotes apple fruit softening and MdMAPK3 activity during storage. A) Apple fruits were collected 140 d after full bloom (DAFB, the commercial harvest day). No treatment, apple fruit not receiving treatment; Eth treatment, apple fruit treated with 0.1% [v/v] ethephon; 1-MCP treatment, apple fruit treated with 1 μL L–1 of the ethylene inhibitor 1-MCP. Scale bar, 2 cm. B) Ethylene production and (C) fruit firmness in fruits receiving the indicated treatments. Fruits sampled at each sampling point were divided into 3 subgroups (10 fruit per subgroup) and each subgroup was considered 1 biological replicate. Three biological replicates were analyzed. Values are means ± standard error (Se). Statistical significance was determined using Student's t-test (**P < 0.01). n.s., no significant difference. D) RT-qPCR analysis of relative MdPG1 expression during apple fruit storage. Storage after harvest at RT is shown in days. For RT-qPCR, fruits sampled at each sampling point were divided into 3 subgroups (5 fruits per subgroup). Fruit flesh from each subgroup was evenly mixed for RNA isolation. RNA isolation from each group was considered 1 biological replicate. Three biological replicates were analyzed. Values are means ± Se. Statistical significance was determined using Student's t-test (**P < 0.01). n.s., no significant difference. E) Immunoblot analysis of MdMAPK activity levels during fruit storage using an anti-phospho-p44/42 antibody. Tubulin was used as a loading control.
The MAPK cascade is an important component of the ethylene signal transduction pathway and is known to affect postharvest fruit physiology (Ouaked et al. 2003; Wu et al. 2022a). A previous study showed that the phospho-p44/42 MAPK antibody specifically recognizes the phosphorylated/active forms of MAPKs that contain a TEY motif (p-tEpY) (Guo et al. 2020; Mao et al. 2022), which is the case for MdMAPK3, MdMAPK4, MdMAPK6, MdMAPK7, and MdMAPK13 in apple fruit (Supplemental Fig. S1, A and B). We confirmed that these MdMAPK-TEY proteins are recognized by the phospho-p44/42 antibody as 3 distinct bands (Supplemental Fig. S1C). Based on their molecular weights, we assigned the lowest band in the immunoblot to phosphorylated MdMAPK3 (Supplemental Fig. S1C, left panel). We also used an anti-FLAG antibody to determine the position of MdMAPK3 in immunoblots of protein extracts from 35S:FLAG-MdMAPK3 transgenic apple calli; MdMAPK3 again corresponded to the lowest band (Supplemental Fig. S1C, right panel). In addition, the phosphorylation of the lowest band was lower in MdMAPK3-silenced fruit than in wild-type fruit, indicative of lower kinase activity in MdMAPK3-silenced fruit (Supplemental Fig. S1D). These results indicate that the lowest band identified in the immunoblot analysis is phosphorylated MdMAPK3.
To determine whether MAPKs are involved in ethylene-induced fruit softening in apples, we used the phospho-p44/42 antibody to examine MAPK phosphorylation status and to infer activity in response to ethylene treatment. Immunoblot analysis showed that MdMAPK3 activity in ethylene-treated apples is indeed significantly higher than that in untreated fruit during storage (Fig. 1E). In addition, to determine whether MAPK signaling is involved in the rapid response of apple fruit to ethylene (de Zelicourt et al. 2016), we treated apple fruit that had not been stored with ethylene for 2, 4, 6, or 8 h and measured their MAPK activity (Supplemental Fig. S2). These results demonstrate that ethylene promotes MdMAPK3 activity within 2 to 8 h, indicating that MAPK signaling responds rapidly to ethylene.
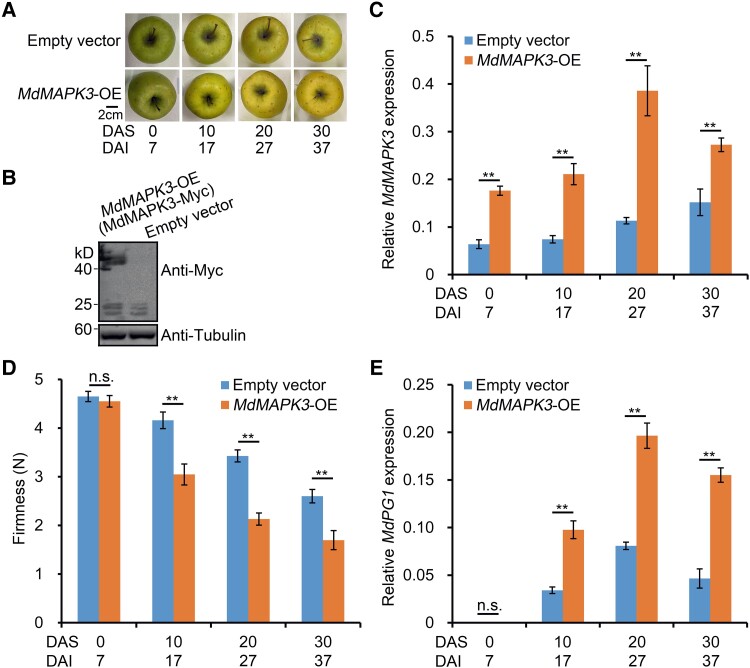
To characterize the function of MdMAPK3 (Sequence ID: MDP0000237742) in apple fruit softening, we transiently overexpressed the gene in GD apples. We harvested apple fruit transiently injected with a 35S:Myc-MdMAPK3 construct 7 d after infiltration and stored them at room temperature (RT) for up to 30 d (Fig. 2A), along with control fruit injected with an empty Myc vector. We confirmed the overexpression of MdMAPK3 by immunoblot analysis with an anti-Myc antibody (Fig. 2B). In addition, we conducted reverse transcription-quantitative PCR (RT-qPCR) analysis, which indicated that MdMAPK3 expression is significantly higher in fruit overexpressing MdMAPK3 (MdMAPK3-OE) than in control fruit (Fig. 2C). After injection, MdMAPK3-OE fruit exhibited a rapid decrease in firmness compared with fruit injected with the empty vector (Fig. 2D). In addition to fruit softening, MdPG1 expression was significantly higher in MdMAPK3-OE fruit than in control fruit (Fig. 2E). Conversely, we transiently silenced MdMAPK3 in fruit and observed that fruit firmness and MdPG1 expression in MdMAPK3-silenced (MdMAPK3-AS) fruit exhibit the phenotype opposite to MdMAPK3-OE fruit during storage (Supplemental Fig. S3). These results indicate that MdMAPK3 promotes the softening of apples during storage.
Figure 2.
MdMAPK3 promotes apple fruit softening during storage. A) Apple fruit transiently overexpressing MdMAPK3 (MdMAPK3-OE) or empty vector (control) during storage. MdMAPK3 was overexpressed in apple fruit using Agrobacterium-mediated injection. MdMAPK3-OE fruits were harvested 7 d after injection and stored at RT for 30 d. DAI, days after infiltration; DAS, days after storage. Scale bar, 2 cm. B) Immunoblot analysis of proteins from apple fruit injected with empty vector or MdMAPK3-Myc and detected using an anti-Myc antibody. Fruit tissue at the injection region was used for protein extraction. Tubulin was used as a loading control. C) Relative MdMAPK3 expression in the fruits shown in (A) by RT-qPCR analysis. D) Apple fruit firmness during storage. Fruits sampled at each sampling point were divided into 3 subgroups (5 fruits per subgroup) and each subgroup was considered 1 biological replicate. Three biological replicates were analyzed. Values are means ± Se. Statistical significance was determined using Student's t-test (**P < 0.01). n.s., no significant difference. E) Relative MdPG1 expression by RT-qPCR analysis. For RT-qPCR, 3 biological replicates were used as described in Fig. 1. Values represent means ± Se. Statistical significance was determined using Student's t-test (**P < 0.01). n.s., no significant difference.
MdMAPK3 interacts with MdNAC72
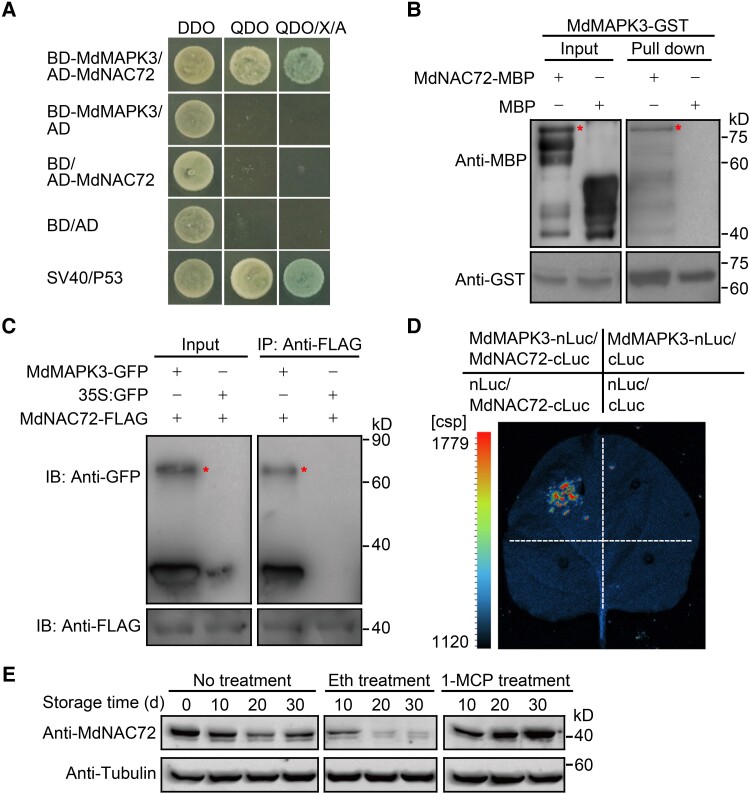
To elucidate the role of MdMAPK3 in fruit softening, we performed a yeast two-hybrid (Y2H) screen of a cDNA library derived from genes expressed during apple fruit storage to identify putative MdMAPK3 interacting partners. We identified a total of 42 positive clones (Supplemental Data Set 1), 5 of which encoded the MdNAC72 transcription factor (Sequence ID: LOC103407991). Since the NAC transcription factor family has been reported to be involved in fruit softening in both apples and tomatoes (Gao et al. 2018; Migicovsky et al. 2021), we hypothesized that the interaction of MdMAPK3 and MdNAC72 might play a role in apple fruit softening. To test this idea, we first confirmed that MdMAPK3 and MdNAC72 interact with each other using a directed Y2H assay (Fig. 3A) and an in vitro glutathione S-transferase (GST) pull-down assay (Fig. 3B). Furthermore, a co-immunoprecipitation (co-IP) assay showed that MdMAPK3 tagged with a green fluorescent protein (GFP) tag interacts with MdNAC72 fused to a FLAG tag in apple fruit calli. We detected no interaction between GFP and MdNAC72 (Fig. 3C). Finally, an in vivo firefly luciferase complementation imaging (LCI) assay showed a strong luminescence signal during the co-expression of MdMAPK3-nLuc and MdNAC72-cLuc, indicating an interaction (Fig. 3D). Together, these results support the MdMAPK3–MdNAC72 interaction in vitro and in vivo. Notably, MdNAC72 was expressed in roots, stems, leaves, and flowers of unripe and ripe apple fruits, and its expression levels remained unchanged after ethylene and 1-MCP treatment during apple fruit storage (Supplemental Fig. S4). Immunoblot analysis revealed that MdNAC72 protein abundance substantially decreases after ethylene treatment, however, while it remained unchanged after 1-MCP treatment (Fig. 3E), and this expression pattern was opposite to that of MdPG1 (Fig. 1D). This result indicates that MdNAC72 may be involved in regulating MdPG1 in a posttranslational manner.
Figure 3.
MdMAPK3/MdNAC72 interaction and MdNAC72 protein abundance. A) MdMAPK3 and MdNAC72 interact in a Y2H assay. DDO, synthetic defined (SD) medium lacking Trp and Leu; QDO, SD medium lacking Trp, Leu, His, and Ade; QDO/X/A, QDO medium containing X-α-gal and aureobasidin A. BD and AD vectors were used as negative controls. SV40/P53 was used as the positive control. B) Pull-down assay showing that MdMAPK3 interacts with MdNAC72 in vitro. Recombinant MdNAC72-MBP was incubated with immobilized MdMAPK3-GST and detected for immunoblot analysis using an anti-MBP antibody (upper panel) or an anti-GST antibody (bottom panel). The asterisk represents the target protein. C) Confirmation of the interaction between MdMAPK3 and MdNAC72 by a co-IP assay. MdMAPK3-GFP and MdNAC72-FLAG were overexpressed in apple fruit calli; an anti-FLAG antibody was used for immunoprecipitation. Anti-GFP and anti-FLAG antibodies were used for immunoblotting. D) A firefly LCI assay confirms the interaction between MdMAPK3 and MdNAC72 in N. benthamiana leaves. E) Immunoblot analysis of MdNAC72 abundance during fruit storage using a specific anti-MdNAC72 antibody. Tubulin was used as a loading control.
MdNAC72 directly suppresses MdPG1 transcription
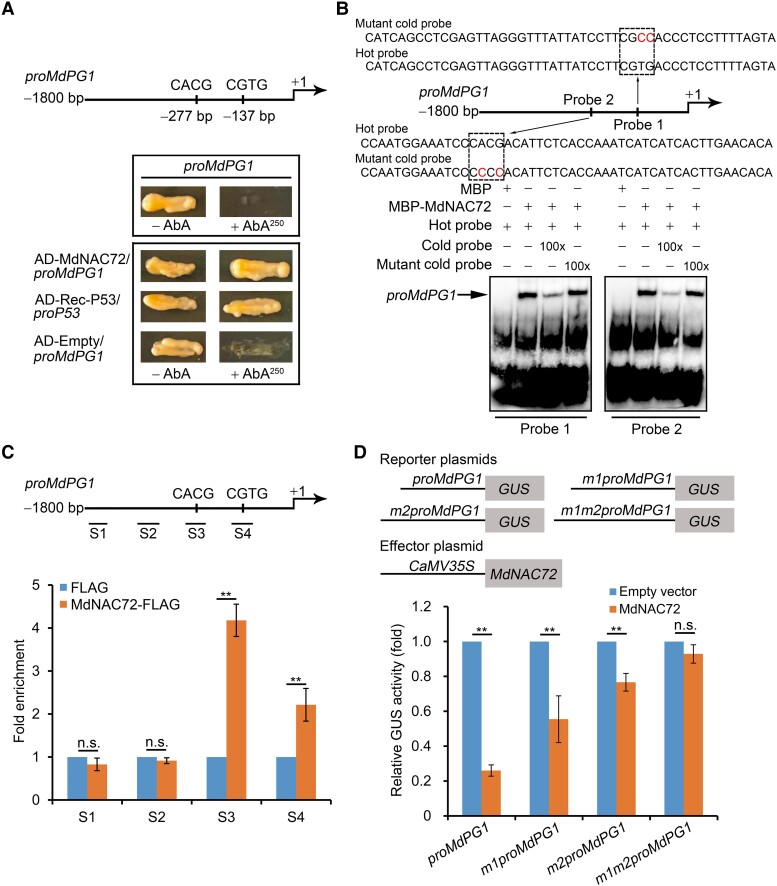
Because MdNAC72 interacts with MdMAPK3, we investigated its role in fruit softening. First, we silenced MdNAC72 in apple calli using Agrobacterium (Agrobacterium tumefaciens)-mediated transformation. We generated 8 transgenic lines, of which Lines #1, #4, and #5 exhibited substantially lower MdNAC72 transcript levels (Supplemental Fig. S5A). MdPG1 expression was markedly higher in these MdNAC72-silenced calli compared with the control, suggesting that MdNAC72 may act as a negative regulator of MdPG1 (Supplemental Fig. S5B). We noticed that the MdPG1 promoter contains NAC-binding sites (CACG and CGTG); in agreement, yeast one-hybrid (Y1H) assays revealed that MdNAC72 binds directly to the MdPG1 promoter (Fig. 4A). Electrophoresis mobility shift assays (EMSAs) showed that MdNAC72 binds directly to both NAC-binding sites (CACG and CGTG) (Fig. 4B). An in vivo chromatin immunoprecipitation (ChIP)-qPCR assay in transgenic apple calli showed that the S3 and S4 regions of the MdPG1 promoter, containing the NAC-binding sites, are highly enriched in immunoprecipitated chromatin compared with chromatin purified from transgenic calli harboring the empty FLAG vector (Fig. 4C). These findings demonstrate that MdNAC72 directly binds to the MdPG1 promoter.
Figure 4.
MdNAC72 represses MdPG1 transcription by binding to its promoter. A) Y1H assay showing that MdNAC72 directly binds to the MdPG1 promoter. The basal concentration of AbA (aureobasidin A) used was 250 ng mL–1. The empty vector and the MdPG1 promoter were used as negative controls. The Rec-P53+P53-promoter was used as a positive control. B) Electrophoretic mobility shift assay (EMSA) to assess the binding of MdNAC72 to the CACG or CGTG binding sites in the MdPG1 promoter. The hot probes (probe 1 and probe 2, Supplemental Data Set 4) were biotin-labeled fragments of the MdPG1 promoter containing the NAC binding site. The cold probes were unlabeled and used as competitive probes (100×). The mutant cold probe consisted of an unlabeled hot probe with 2 nucleotides mutated. Recombinant MdNAC72-MBP was purified from E. coli and used for DNA-binding assays. C) ChIP-qPCR assay showing that MdNAC72 binds to the MdPG1 promoter in vivo. The various crosslinked chromatin samples were extracted from MdNAC72-FLAG apple fruit calli and immunoprecipitated with an anti-FLAG antibody. The eluted DNA was used to amplify the sequences neighboring the NAC binding site using qPCR. Four different regions (S1–S4) were investigated. Fruit calli injected with the empty vector (35S:FLAG) were used as a negative control. The ChIP-qPCR assay was performed 3 times, and the enriched DNA fragments in each ChIP were considered 1 biological replicate for qPCR. Values are means ± Se. Statistical significance was determined using Student's t-test (**P < 0.01). n.s., no significant difference. D) β-glucuronidase activity assay showing that MdNAC72 represses MdPG1 promoter activity. The 35S:MdNAC72 effector vector together with the proMdPG1:GUS promoter, or a mutated promoter (m1proMdPG1, m2proMdPG1, and m1m2proMdPG1), were infiltrated into N. benthamiana leaves to analyze GUS activity. The 35S:LUC was included as an internal control for normalization of transformation efficiency. Three independent infiltration experiments were performed; values are means ± Se. Statistical significance was determined using Student's t-test (**P < 0.01). n.s., no significant difference.
We next investigated the effect of MdNAC72 on MdPG1 expression in Nicotiana benthamiana leaves using a β-glucuronidase (GUS) transactivation assay. When we co-transformed 35S:MdNAC72 with the MdPG1 promoter fused to the GUS reporter gene (proMdPG1:GUS), we detected a significantly lower GUS signal than when proMdPG1:GUS alone was transformed, indicating that MdNAC72 suppresses the transcriptional activity of the MdPG1 promoter. By contrast, mutating the 2 NAC-binding sites from CACG and CGTG to CCCC and CGCC restored high GUS activity when co-transformed with 35S:MdNAC72 (Fig. 4D). These findings indicate that MdNAC72 directly suppresses MdPG1 transcription.
To characterize MdNAC72 function during fruit storage, we transiently silenced its expression in fruits. We harvested transgenic fruit 7 d after transformation using the Agrobacterium-mediated transient transformation method. We determined that MdNAC72-silenced (MdNAC72-AS) fruits soften more rapidly than control fruits (empty vector, pRI101). MdNAC72 transcript levels were significantly lower in MdNAC72-AS fruits than in control fruits. As the expression of MdPG1 was significantly higher in MdNAC72-AS fruits during storage (Supplemental Fig. S6), we conclude that MdNAC72 suppresses apple fruit softening by regulating MdPG1 transcription during storage.
However, based on the expression patterns of cell wall degradation-related genes, other cell wall degradation-related genes have been speculated to be involved in the regulation of apple fruit softening besides MdPG1 (Goulao et al. 2008). To identify cell wall degradation-related genes regulated by ethylene during apple fruit storage, we reanalyzed previous transcriptome sequencing of 0-fruit (fruits that were not stored), 15-fruit (fruits stored for 15 d), and 15-MCP-fruit (fruits stored for 15 d after 1-MCP treatment) (Li et al. 2022). We detected an upregulation in transcript levels for the 6 cell wall degradation-related genes MdPG1, MdPL5, Mdβ-Gal9, Mdα-AFase2, MdXET1, and MdEXP8, in 0-fruit compared with 15-fruit and downregulated in 15-fruit relative to 15-MCP-fruit (Supplemental Data Set 2). We confirmed this result by RT-qPCR, showing that the expression of these genes is promoted by ethylene treatment during fruit storage (Supplemental Fig. S7). Interestingly, the promoters of these genes have binding sites for NAC transcription factors. Furthermore, as shown above, the expression of these genes was regulated by ethylene treatment. Therefore, MdNAC72 may regulate their expression, which, in turn, affects fruit softening. Finally, an in vivo ChIP-qPCR assay in transgenic 35S:FLAG-MdNAC72 apple calli showed that MdNAC72 directly binds to the promoters of MdPL5, Mdβ-Gal9, Mdα-AFase2, MdXET1, and MdEXP8 (Supplemental Fig. S8).
MdMAPK3 phosphorylates MdNAC72, resulting in suppression of its transcriptional repression activity
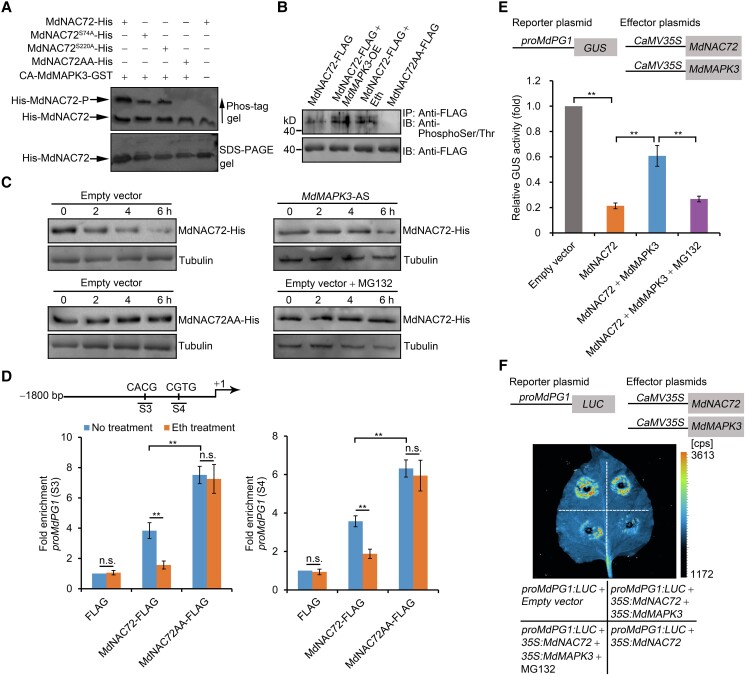
The interaction between MdMAPK3 and MdNAC72 raised the question of whether MdNAC72 might be a direct substrate of MdMAPK3. Based on bioinformatics predictions (http://gps.biocuckoo.org/), MdNAC72 has 2 putative MAPK3 phosphorylation sites: S74 and S220. We employed an in vitro phosphorylation assay to test this possibility using a variant of MdMAPK3 (CA-MdMAPK3), which had been shown in a previous study to be constitutively active (Berriri et al. 2012; Wang et al. 2022). In contrast to the negative control, we observed a phosphorylation signal in the presence of recombinant GST-fused CA-MdMAPK3 and His- MdNAC72 (Fig. 5A). To verify whether S74 and S220 are potential phosphorylation sites, we generated MdNAC72 variants harboring alanine mutations at S74 and/or S220 to form partial nonphosphorylated (MdNAC72S74A, MdNAC72S220A) or fully nonphosphorylated (MdNAC72AA: MdNAC72S74A,S220A) MdNAC72, respectively. We then assessed the phosphorylation status of MdNAC72S74A, MdNAC72S220A, and MdNAC72AA in an in vitro phosphorylation assay. Immunoblot analysis showed that the phosphorylation signals of MdNAC72S74A and MdNAC72S220A are weaker than that of MdNAC72, while we detected no phosphorylation signal when we used MdNAC72AA as the substrate (Fig. 5A). We also performed in vivo phosphorylation assays using transgenic apple calli harboring MdNAC72-FLAG, MdNAC72-FLAG MdMAPK3-OE, MdNAC72-FLAG MdMAPK3-AS, and MdNAC72AA-FLAG. An immunoblot analysis of proteins immunoprecipitated with an anti-FLAG antibody showed that MdMAPK3 enhances phosphorylation of intact MdNAC72-FLAG, while we detected no phosphorylation for MdNAC72AA. Furthermore, ethylene induced the phosphorylation of MdNAC72 in planta (Fig. 5B). In addition, the intensity of the slow-migrating band corresponding to phosphorylated MdNAC72 decreased in phos-tag gels when MdMAPK3 was silenced (Supplemental Fig. S9). These results show that MdMAPK3 phosphorylates MdNAC72 at the S74 and S220 sites.
Figure 5.
MdMAPK3 phosphorylates MdNAC72 to suppress its transcriptional repression activity. A) Phos-tag mobility shift assay showing that MdMAPK3 phosphorylates MdNAC72, but not MdNAC72AA, in vitro. CA-MdMAPK3, constitutively active form. MdNAC72 variants harboring alanine substitutions at S74 and/or S220 (MdNAC72S74A; MdNAC72S220A; MdNAC72AA, MdNAC72S74A,S220A). Proteins were separated on a phos-tag gel and an immunoblot analysis was performed using an anti-His antibody to detect MdNAC72. The slowly migrating band on the phos-tag gel represents phosphorylated MdNAC72-His (upper panel). Proteins were also separated on a normal SDS-PAGE gel to verify equal loading (bottom panel). B) MdMAPK3 phosphorylates MdNAC72 in vivo. Proteins were immunoprecipitated from transgenic apple calli (35S:FLAG-MdNAC72, 35S:FLAG-MdNAC72+35S:MdMAPK3, or 35S:FLAG-MdNAC72AA) using an anti-FLAG antibody and separated using SDS-PAGE for immunoblot analysis. An anti-phosphoSer/Thr antibody was used to detect MdNAC72 phosphorylation. Eth, apple calli treated with ethylene. C) In vitro cell-free degradation assay showing that MdNAC72 degradation rate is slower in protein extracts from MdMAPK3-silenced (MdMAPK3-AS) transgenic apple calli compared with a control calli (transformed with the empty vector). Recombinant MdNAC72-His was added to total protein extracts, incubated for the indicated times, and subjected to immunoblot analysis using an anti-His antibody. Transgenic apple calli transformed with the empty vector were treated with 50 μm MG132. Tubulin was used as a loading control. D) ChIP-qPCR assay showing that ethylene treatment decreases the binding of MdNAC72 to the MdPG1 promoter, but not of MdNAC72AA. Crosslinked chromatin samples were extracted from 35S:FLAG, 35S:FLAG-MdNAC72, and 35S:FLAG-MdNAC72AA fruit calli (no treatment) or treated with ethylene and precipitated using an anti-FLAG antibody. ProPG1-S3 and ProPG1-S4 refer to the MdPG1 promoter regions from Fig. 4C. For ChIP-qPCR, 3 biological replicates were analyzed as described in Fig. 4. Values are means ± Se. Statistical significance was determined using Student's t-test (**P < 0.01). n.s., no significant difference. E) GUS activity assay showing that MdNAC72 is phosphorylated by MdMAPK3 to upregulate MdPG1 transcript levels. The proMdPG1:GUS reporter, together with the effector constructs 35S:MdNAC72 or 35S:MdNAC72 and 35S:MdMAPK3, was separately infiltrated into N. benthamiana leaves to assess GUS activity. Three independent infiltrations were performed as described in Fig. 4. MG132, N. benthamiana leaves treated with 50 μm MG132. F) Dual-luciferase (LUC) reporter assay showing that MdNAC72 phosphorylation by MdMAPK3 upregulates MdPG1 transcription. The reporter proMdPG1:LUC, together with the effectors 35S:MdNAC72 or 35S:MdNAC72 and 35S:MdMAPK3, was separately infiltrated into N. benthamiana leaves to assess LUC activity. MG132, N. benthamiana leaves treated with 50 μm MG132.
We performed an in vitro cell-free degradation assay, which revealed that MdNAC72 degradation is strongly attenuated when recombinant MdNAC72 is incubated with total proteins extracted from the MdMAPK3-AS transgenic apple calli compared with that from the empty vector (Fig. 5C). However, degradation of intact MdNAC72 was inhibited by treatment with the proteasome inhibitor MG132 and this degradation also disappeared on MdNAC72AA, suggesting that the phosphorylation of MdNAC72, which is dependent on MdMAPK3, affects MdNAC72 degradation. These results also indicated that MdMAPK3 phosphorylates MdNAC72, causing it to undergo 26S proteasome-mediated protein degradation, thereby changing the abundance of MdNAC72 bound to the MdPG1 promoter and effectively regulating MdPG1 transcript levels. To verify this hypothesis, we performed a ChIP-qPCR assay using an anti-FLAG antibody in transgenic 35S:FLAG-MdNAC72, 35S:FLAG-MdNAC72AA, and 35S:FLAG apple calli with and without ethylene treatment. We observed that MdNAC72AA-FLAG shows a greater enrichment at the S3 and S4 fragments from the MdPG1 promoter than did MdNAC72-FLAG; moreover, the enrichment of these S3 and S4 fragments mediated by MdNAC72-FLAG was inhibited by ethylene treatment, whereas MdNAC72AA enrichment was not affected and remained high regardless of ethylene treatment (Fig. 5D). This observation suggests that ethylene regulates MdPG1 transcript levels through MdNAC72, which is affected by MdNAC72 phosphorylation by MdMAPK3. In addition, a GUS transactivation assay showed that co-expression of MdNAC72 with the MdPG1 promoter results in a significant decrease in GUS activity, which was reversed when MdMAPK3 was co-expressed, consistent with our hypothesis. Notably, treatment with MG132 resulted in sustained MdNAC72-dependent repression of MdPG1 transcription (Fig. 5E). A dual-luciferase reporter assay with the MdPG1 promoter driving firefly LUC transcription confirmed these results (Fig. 5F), indicating that ethylene promotes the phosphorylation of MdNAC72 by MdMAPK3, impairing its binding to the MdPG1 promoter and resulting in the suppression of its transcriptional repression activity.
MdPUB24 ubiquitinates MdNAC72 for proteasomal degradation
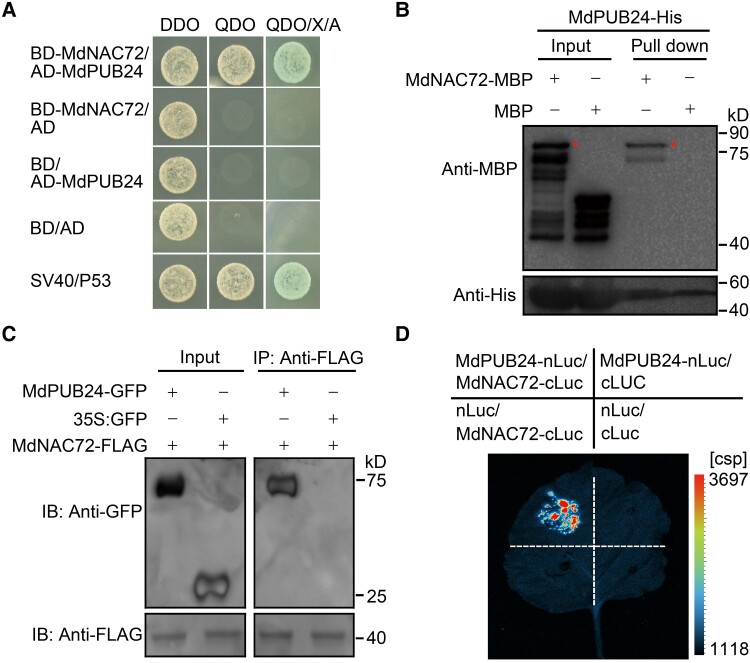
To determine the basis of MdNAC72 degradation following phosphorylation by MdMAPK3, we screened an apple fruit Y2H library using MdNAC72 as bait. Among the 54 positive clones (Supplemental Data Set 3), we identified a U-box-type E3 ubiquitin ligase, named MdPUB24 (Sequence ID: LOC103421753), as an interactor of MdNAC72. We confirmed this interaction in a directed Y2H assay (Fig. 6A) and an in vitro maltose-binding protein (MBP) pull-down assay (Fig. 6B). We also detected an interaction in planta using a co-IP assay in transgenic calli (35S:GFP-MdPUB24 35S:FLAG-MdNAC72 and 35S:GFP 35S:FLAG-MdNAC72) (Fig. 6C). Finally, an LCI assay further demonstrated that MdNAC72 interacts with MdPUB24 (Fig. 6D).
Figure 6.
MdPUB24 interacts with MdNAC72. A) Y2H assay showing that MdPUB24 interacts with MdNAC72 in yeast; the assay was performed as described in Fig. 3. B) A pull-down assay showing that MdPUB24 interacts with MdNAC72 in vitro. Recombinant MdNAC72-MBP was incubated with immobilized MdPUB24-His and detected for immunoblot analysis using an anti-MBP antibody (upper panel) or an anti-His antibody (bottom panel). The asterisk represents the target protein. C) Co-IP assay showing that MdPUB24 interacts with MdNAC72 in vivo. MdPUB24-GFP and MdNAC72-FLAG were overexpressed in apple calli. An anti-FLAG antibody was used for immunoprecipitation analysis. Anti-GFP and anti-FLAG antibodies were used for immunoblotting. D) Firefly LCI assay verifying that MdPUB24 and MdNAC72 interact in N. benthamiana leaves.
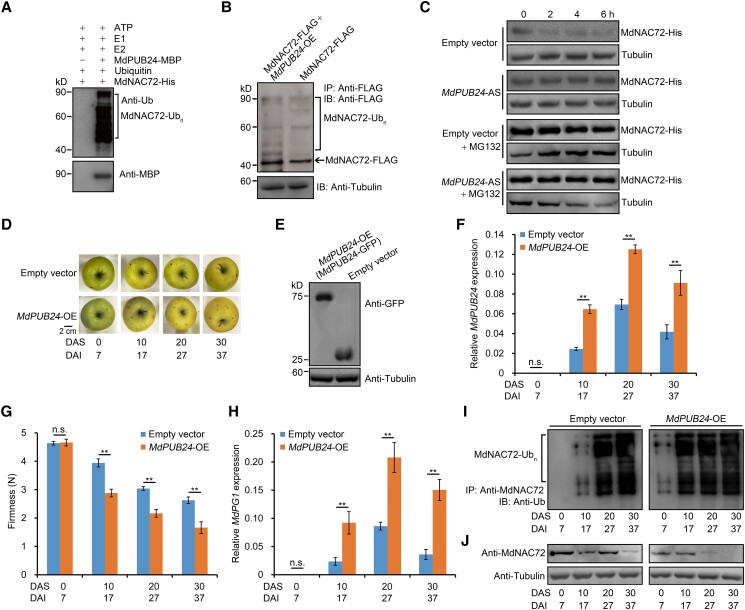
We asked whether MdPUB24 affected the stability of MdNAC72 by altering its ubiquitination status using an in vitro ubiquitination assay. Following the incubation of recombinant MdNAC72-His and MdPUB24-MBP in a reaction mixture containing ATP, ubiquitin (Ub), a ubiquitin-activating enzyme (E1), and a ubiquitin-conjugating enzyme (E2), we analyzed the reaction mixture by immunoblotting using an anti-Ub antibody. Co-incubation of MdNAC72-His and MdPUB24-MBP in the presence of Ub, E1, and E2 led to the appearance of high molecular-mass bands corresponding to ubiquitinated MdNAC72. In the absence of MdPUB24-MBP, these bands were absent (Fig. 7A). This result demonstrated that MdNAC72 can be directly ubiquitinated by MdPUB24. Moreover, an in vivo ubiquitination detection assay using MdNAC72-FLAG and MdNAC72-FLAG MdPUB24-OE transgenic apple calli showed that MdPUB24 enhances MdNAC72-FLAG ubiquitination (Fig. 7B), suggesting that MdPUB24 ubiquitinates MdNAC72 in vivo. Next, we performed an in vitro protein degradation assay to investigate whether MdPUB24 also affected MdNAC72 stability. To this end, we incubated protein extracts from transgenic apple calli transformed with empty vector or MdPUB24-AS (MdPUB24 silenced) with recombinant MdNAC72-His. We observed that silencing of MdPUB24 prevents MdNAC72 degradation and that this degradation is substantially inhibited by MG132 treatment (Fig. 7C). Taken together, these results indicate that MdPUB24 accelerates MdNAC72 degradation by regulating its ubiquitination.
Figure 7.
MdPUB24 ubiquitinates MdNAC72 and promotes apple fruit softening. A) MdPUB24 ubiquitinates MdNAC72 in vitro. MdNAC72-His was used to detect potential E3 ubiquitin ligase activity, in the presence of ATP, ubiquitin, E1, E2, and recombinant MdPUB24-MBP. An immunoblot analysis was used to detect the ubiquitination of MdNAC72 using an anti-ubiquitin (Ub) antibody. B) Ubiquitination of MdNAC72 in MdNAC72-FLAG and MdPUB24-OE+MdNAC72-FLAG pretreated (50 μm MG132) calli. MdNAC72 was immunoprecipitated using an anti-FLAG antibody and the anti-FLAG antibody was used to detect ubiquitinated MdNAC72-FLAG. C) Cell-free degradation assay for recombinant MdNAC72-His in protein extracts from MdPUB24-silenced (MdPUB24-AS) transgenic apple calli. MdNAC72-His abundance was determined by immunoblot analysis using an anti-His antibody. Empty vector or MdPUB24-AS transgenic apple calli were separately treated with 50 μm MG132, and tubulin was used as the loading control. D) Apple fruit transiently overexpressing MdPUB24 (MdPUB24-OE) or empty vector (control) during storage. MdPUB24 was overexpressed in apple fruit using Agrobacterium-mediated transient injection. MdPUB24-OE fruits were harvested 7 d after injection and stored at RT for 30 d. DAI, days after infiltration; DAS, days after storage. Scale bar, 2 cm. E) Immunoblot analysis using an anti-GFP antibody of proteins from apple fruit injected with an empty vector or MdPUB24-GFP. Fruit tissues at the injection region were used for protein detection. Tubulin was used as the loading control. F) Relative MdPUB24 expression by RT-qPCR. G) Firmness of injected apple fruit during storage. Three biological replicates were performed as described in Fig. 2D. H) Relative MdPG1 expression in MdPUB24-OE apple fruit during storage as detected by RT-qPCR. Fruits injected with empty vectors were used as control. For RT-qPCR, 3 biological replicates were performed as described in Fig. 1. I and J) The ubiquitination level of MdNAC72 increases and MdNAC72 abundance decreases in MdPUB24-OE apple fruit during storage. Tubulin was used as the loading control.
To elucidate the function of MdPUB24 in apple fruit softening during storage, we transiently overexpressed MdPUB24 in apple fruits (Fig. 7D). Accordingly, we harvested fruits 7 d after injection with Agrobacterium harboring 35S:GFP-MdPUB24; an immunoblot analysis revealed that MdPUB24 overexpression in apple fruits was successful (Fig. 7E). We also detected higher MdPUB24 transcript levels in MdPUB24-OE fruits relative to control fruit (Fig. 7F). In addition, MdPUB24-OE fruits softened more rapidly than control fruits (Fig. 7G). In addition to fruit softening, MdPG1 transcript levels were significantly higher in MdPUB24-OE apples during storage (Fig. 7H). Moreover, MdNAC72 ubiquitination was enhanced in MdPUB24-OE fruit, and MdNAC72 protein abundance was lower than in controls (Fig. 7, I and J). In addition, we transiently silenced MdPUB24 in fruits and observed that fruit firmness and MdPG1 expression in MdPUB24-AS fruits exhibit a change opposite to that seen in MdPUB24-OE fruits during storage (Supplemental Fig. S10). Taken together, our results are consistent with MdPUB24-mediated ubiquitination and degradation of MdNAC72, which contributes to fruit softening.
MdMAPK3 phosphorylates MdNAC72, which enhances ubiquitination of MdNAC72 by MdPUB24, promoting MdPG1 expression
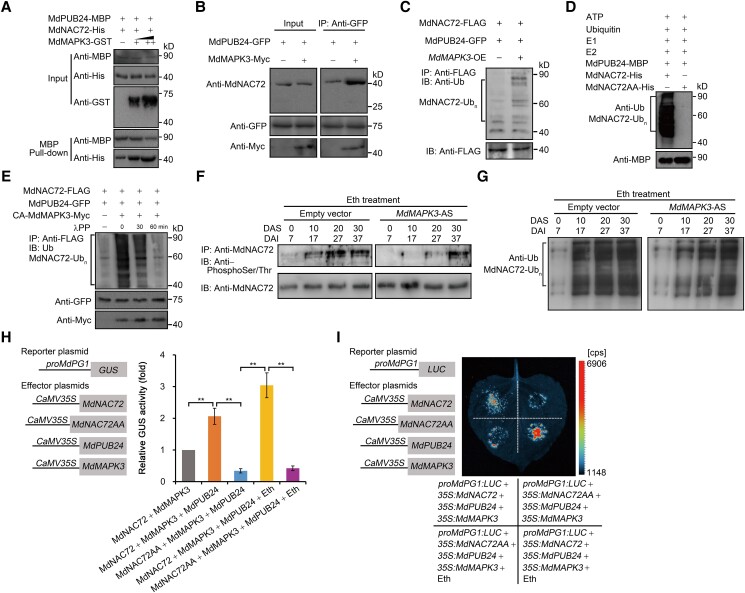
Our data showed that MdMAPK3 mediates the phosphorylation of MdNAC72 (Fig. 5), and MdPUB24 mediates the ubiquitination of MdNAC72 (Fig. 7). Moreover, MdNAC72 directly binds to the MdPG1 promoter and negatively regulates its transcription (Fig. 4). Therefore, we hypothesized that MdMAPK3 mediates the phosphorylation of MdNAC72, which is in turn ubiquitinated by MdPUB24, thereby altering the binding of MdNAC72 to the MdPG1 promoter. In a pull-down assay with the recombinant proteins MdPUB24-MBP, MdNAC72-His, and MdMAPK3-GST, we determined that MdMAPK3-GST enhances the abundance of MdNAC72-His immunoprecipitated by MdPUB24-MBP (Fig. 8A). This result indicated that MdMAPK3 promotes the interaction between MdNAC72 and MdPUB24 in vitro. Furthermore, an in vivo co-IP assay using transgenic apple calli (35S:GFP-MdPUB24 and 35S:GFP-MdPUB24 35S:Myc-MdMAPK3) showed that MdPUB24-GFP immunoprecipitates a greater amount of MdNAC72 when MdMAPK3 is overexpressed (Fig. 8B), indicating that this enhancement also takes place in vivo. Because MdMAPK3 enhances the interaction between MdNAC72 and MdPUB24, we hypothesized that it may promote the ubiquitination of MdNAC72 by MdPUB24. To test this hypothesis, we performed an in vivo ubiquitination assay using N. benthamiana leaves transiently expressing MdNAC72-FLAG and MdPUB24-GFP or MdNAC72-FLAG and MdPUB24-GFP and MdMAPK3. Immunoblot analysis showed that overexpression of MdMAPK3 enhances the ubiquitination of MdNAC72 by MdPUB24 (Fig. 8C). In addition, an in vitro ubiquitination assay with recombinant MdPUB24-MBP and MdNAC72-His revealed the presence of high molecular-mass bands corresponding to ubiquitinated MdNAC72-His, whereas co-incubation of MdPUB24-MBP with MdNAC72AA-His resulted in no bands (Fig. 8D). This result indicates that nonphosphorylated MdNAC72 cannot be ubiquitinated by MdPUB24 and that phosphorylation of MdNAC72 is a prerequisite for its ubiquitination by MdPUB24. Overall, these results indicate that MdMAPK3 promotes the ubiquitination of MdNAC72 by MdPUB24.
Figure 8.
MdMAPK3-mediated phosphorylation of MdNAC72 enhances its ubiquitination by MdPUB24, which upregulates MdPG1 transcription. A) In vitro pull-down assay showing that the interaction between MdPUB24 and MdNAC72 is enhanced by MdMAPK3. Recombinant MdNAC72-His and MdMAPK3-GST were incubated with immobilized MdPUB24-MBP. The pulled-down fractions were visualized using an anti-His antibody. B) Co-IP assay showing that MdMAPK3 promotes the interaction between MdPUB24 and MdNAC72 in vivo. An anti-GFP antibody was used for immunoprecipitation from protein extracts of transgenic apple calli (MdPUB24-GFP MdMAPK3-Myc and MdPUB24-GFP). Anti-MdNAC72 and anti-Myc antibodies were used for immunoblotting. C) In vivo ubiquitination assay showing that MdMAPK3 promotes the ubiquitination of MdNAC72 by MdPUB24. Proteins were extracted from N. benthamiana leaves transiently expressing MdNAC72-FLAG and MdPUB24-GFP or MdNAC72-FLAG and MdPUB24-GFP and MdMAPK3 pretreated with 50 μm MG132. MdNAC72 was immunoprecipitated using an anti-FLAG antibody. Immunoblot analysis was used to detect the ubiquitination of MdNAC72 by MdPUB24 using an anti-Ub antibody. D) In vitro ubiquitination assay showing that phosphorylation of MdNAC72 is a prerequisite for its ubiquitination by MdPUB24. Recombinant MdNAC72-His and MdNAC72AA-His were analyzed for ubiquitination in the presence of ATP, ubiquitin, E1, E2, and recombinant MdPUB24-MBP. An immunoblot analysis was used to detect the ubiquitination of MdNAC72 or MdNAC72AA using an anti-Ub antibody. E) In vivo ubiquitination assay showing that MdMAPK3 phosphorylates MdNAC72, which enhances the ubiquitination of MdNAC72 by MdPUB24. Proteins were extracted from N. benthamiana leaves transiently expressing MdNAC72-FLAG and MdPUB24-GFP or MdNAC72-FLAG and MdPUB24-GFP and CA-MdMAPK3 pretreated with 50 μm MG132. MdNAC72 protein immunoprecipitated using an anti-FLAG antibody was incubated with λPP at 30°C for 30 or 60 min. Immunoblot analysis was used to detect the ubiquitination of MdNAC72 by MdPUB24 using an anti-Ub antibody. Immunoblot analysis for MdPUB24-GFP or CA-MdMAPK3-Myc proteins using anti-GFP or anti-Myc antibodies, respectively, indicated equal loading. F and G) Ethylene promotes the phosphorylation and ubiquitination of MdNAC72, which is suppressed in MdMAPK3-silenced (MdMAPK3-AS) apples. MdMAPK3-AS fruits were harvested 7 d after injection and stored at RT for 30 d. The fruits were treated with ethylene. Empty vector-infected fruits were used as controls. DAI, days after infiltration; DAS, days after storage. Total protein was extracted from each sample to analyze MdNAC72 phosphorylation and ubiquitination. MdNAC72 was immunoprecipitated using an anti-MdNAC72 antibody. Anti-phosphoSer/Thr and anti-Ub antibodies were used to detect the phosphorylation (F) and ubiquitination (G) of MdNAC72. H) GUS activity assay showing that MdMAPK3 phosphorylates MdNAC72 to promote its ubiquitination by MdPUB24, which suppresses the transcriptional repression of the MdPG1 promoter by MdNAC72. The proMdPG1:GUS reporter, together with the effectors 35S:MdNAC72, 35S:MdPUB24, and 35S:MdMAPK3 or 35S:MdNAC72AA, 35S:MdPUB24, and 35S:MdMAPK3, was separately infiltrated into N. benthamiana leaves to assess GUS activity. The reporter vector proMdPG1:GUS, together with the effectors 35S:MdNAC72 and 35S:MdMAPK3, was infiltrated into N. benthamiana leaves as a control. Eth, N. benthamiana leaves treated with ethylene. Three independent infiltration experiments were performed, as described in Fig. 4. I) Dual-LUC reporter assay showing that MdMAPK3 phosphorylates MdNAC72 to promote its ubiquitination by MdPUB24, thereby suppressing the transcriptional repression of the MdPG1 promoter by MdNAC72. The reporter proMdPG1:LUC, together with the effectors 35S:MdNAC72, 35S:MdPUB24, and 35S:MdMAPK3 or 35S:MdNAC72AA, 35S:MdPUB24, and 35S:MdMAPK3, was separately infiltrated into N. benthamiana leaves to assess LUC activity. Eth, N. benthamiana leaves treated with ethephon.
To investigate how MdMAPK3 affects the ubiquitination of MdNAC72 by MdPUB24, we performed an in vivo ubiquitination assay using N. benthamiana leaves transiently expressing MdNAC72-FLAG and MdPUB24-GFP or MdNAC72-FLAG and MdPUB24-GFP and CA-MdMAPK3. Immunoblot analysis indicated that overexpression of CA-MdMAPK3 enhances the ubiquitination of MdNAC72 by MdPUB24; however, incubation of the protein extracts with lambda protein phosphatase (λPP) eliminated the signal corresponding to ubiquitinated MdNAC72 with increasing reaction time (Fig. 8E). This finding indicates that MdMAPK3 phosphorylates MdNAC72, which enhances the ubiquitination of MdNAC72 by MdPUB24.
We established that the ethylene-promoted phosphorylation and ubiquitination of MdNAC72 is suppressed when silencing MdMAPK3 (Fig. 8, F and G), indicating that MdMAPK3 is a key component of ethylene-induced ubiquitination and degradation of MdNAC72. Moreover, a GUS transactivation assay in N. benthamiana leaves showed that when 35S:MdMAPK3 and 35S:MdNAC72 or 35S:MdMAPK3 and 35S:MdNAC72 and 35S:MdPUB24 constructs were separately co-infiltrated with proMdPG1:GUS, the transcriptional repression of the MdPG1 promoter by MdNAC72 is significantly suppressed in the presence of MdPUB24, which mediates MdNAC72 degradation. Furthermore, when we co-infiltrated 35S:MdMAPK3 and 35S:MdNAC72 and 35S:MdPUB24 or 35S:MdMAPK3 and 35S:MdNAC72AA and 35S:MdPUB24 together with proMdPG1:GUS, the transcriptional repression of the MdPG1 promoter by MdNAC72AA was significantly stronger than by MdNAC72. Ethylene treatment suppressed the ability of MdNAC72 to transcriptionally repress the MdPG1 promoter; however, the effect of MdNAC72AA on the MdPG1 promoter was not affected by ethylene treatment (Fig. 8H). A dual-luciferase reporter assay confirmed these results (Fig. 8I). In conclusion, ethylene-induced increases in MdPG1 transcript levels involve phosphorylation of MdNAC72 mediated by MdMAPK3 and subsequent ubiquitination and degradation by MdPUB24.
Phosphorylation of MdNAC72 is involved in the regulation of apple fruit softening
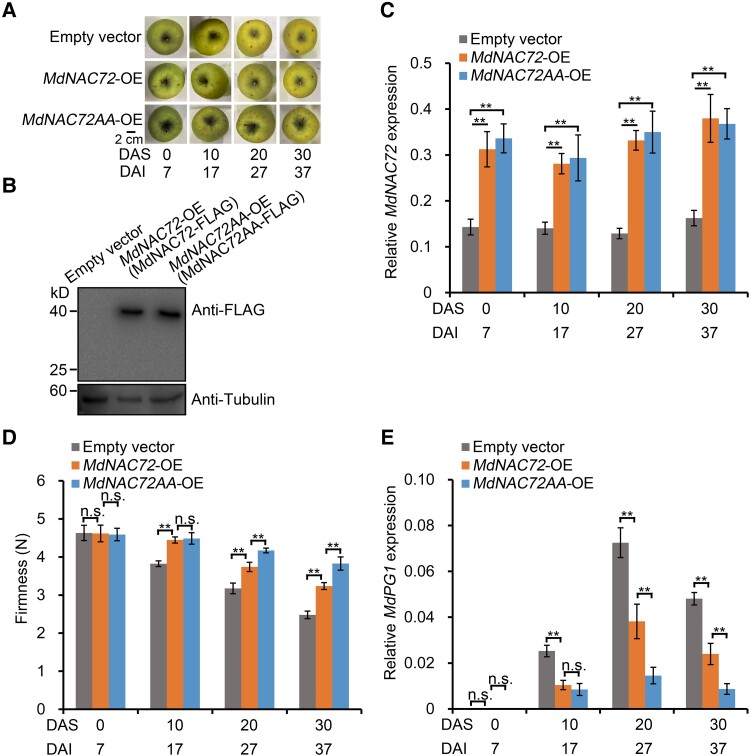
To further characterize the significance of MdMAPK3-mediated phosphorylation of MdNAC72 in fruit softening, we transiently injected the constructs 35S:FLAG-MdNAC72 and 35S:FLAG-MdNAC72AA individually in GD fruit. We harvested injected apples 7 d after injection with Agrobacterium (Fig. 9A). We confirmed the successful infection and overexpression of MdNAC72 or MdNAC72AA by immunoblot analysis (Fig. 9B); RT-qPCR analysis indicated that the expression of MdNAC72 in MdNAC72-OE transgenic fruit is significantly higher than that in control fruits injected with the empty vector (Fig. 9C). Notably, MdNAC72-OE fruits exhibited delayed softening (Fig. 9D), indicating that MdNAC72 negatively regulates apple fruit softening. Moreover, we noticed that MdNAC72AA-OE transgenic fruit display a stronger effect on suppressing softening than MdNAC72-OE transgenic fruit (Fig. 9D), likely because nonphosphorylated MdNAC72AA cannot be ubiquitinated and degraded by MdPUB24, which maintains its inhibitory effect on fruit softening. In addition, MdPG1 transcript levels were lower in MdNAC72AA-OE fruit compared with MdNAC72-OE and control fruit (Fig. 9E). Furthermore, we transiently overexpressed FLAG-MdNAC72DD (encoding a phosphomimic version of MdNAC72, MdNAC72DD: MdNAC72S74D,S220D) in GD fruit and verified successful injection and overexpression of MdNAC72DD by immunoblot and RT-qPCR assays (Supplemental Fig. S11, A–C). We observed that MdPG1 expression and fruit firmness do not change significantly compared with control fruits (Supplemental Fig. S11, D and E). We speculate that the phosphomimic variant of MdNAC72 may be ubiquitinated and degraded after MdPUB24 is initiated, and is thus unstable during apple fruit storage. Overall, these results indicate that the phosphorylation status of MdNAC72, mediated by MdMAPK3, affects apple fruit softening during storage.
Figure 9.
Effect of the MdNAC72 phosphorylation state on apple fruit softening. A) Apple fruit transiently overexpressing various MdNAC72 variants (MdNAC72 or MdNAC72AA) or an empty vector (control) during storage. Transient overexpression in apple fruit was accomplished using Agrobacterium-mediated injection. Overexpressing fruits (MdNAC72-OE or MdNAC72AA-OE) were harvested 7 d after injection and stored at RT for 30 d. DAI, days after infiltration; DAS, days after storage. B) Immunoblot analysis of proteins from apple fruit transformed with empty vector, MdNAC72-FLAG, or MdNAC72AA-FLAG using an anti-FLAG antibody. The fruit tissues at the injection region were used for protein detection. Tubulin was used as the loading control. C) Relative MdNAC72 expression as detected by RT-qPCR in the fruits shown in (A). D) Firmness of injected apple fruit during storage. Three biological replicates were performed as described in Fig. 2D. E) Relative MdPG1 expression as determined by RT-qPCR. For RT-qPCR, 3 biological replicates were used, as described in Fig. 1. Values are means ± Se. Statistical significance was determined using Student's t-test (**P < 0.01). n.s., no significant difference.
Discussion
The main factor that determines postharvest deterioration of fruit crops is their softening rate, which directly affects nutritional quality and shelf life. An apple is a typical climacteric fruit in that it exhibits a peak of ethylene production that is accompanied by softening during storage (Fig. 1, B and C). Ethylene is considered a key regulator in the promotion of apple softening during storage (Tucker et al. 2017), but despite recent progress, our current understanding of the molecular mechanisms controlling ethylene-induced fruit softening is limited. In this study, we dissected a regulatory network involving the MdMAPK3–MdNAC72–MdPUB24 module and its association with the cell wall degradation-related gene, MdPG1, in ethylene-induced apple fruit softening. We demonstrated that a protein kinase, MdMAPK3, activated by ethylene, phosphorylated MdNAC72, which acted as a negative regulator of MdPG1 transcription, resulting in the ubiquitination and degradation of MdNAC72 by the E3 ligase MdPUB24, which promoted MdPG1 transcription and fruit softening during storage.
Previous studies have shown that cell wall degradation-related genes likely play an important role in ethylene-induced apple fruit softening (Wakasa et al. 2006; Costa et al. 2008). Although multiple cell wall degradation-related genes are involved in fruit softening, the only example supported by strong genetic evidence is MdPG1. Silencing MdPG1 in apples attenuates pectin degradation and extends fruit firmness during postharvest storage (Atkinson et al. 2012). Therefore, we chose MdPG1 as a marker gene for fruit softening in this study to explore the connection between the MdMAPK3–MdNAC72–MdPUB24 module and fruit softening in postharvest apples. Interestingly, MdNAC72 directly bound to the promoters of MdPL5, Mdβ-Gal9, Mdα-AFase2, MdXET1, and MdEXP8 (Supplemental Fig. S8). This finding indicates that the ethylene–MdMAPK3–MdNAC72–MdPUB24 module not only regulates MdPG1 transcription but also regulates that of other cell wall degradation-related genes through MdNAC72, which plays a critical role in controlling fruit softening.
Transcription factors, including EIL2, MADS-box, ETHYLENE RESPONSE FACTORs (ERFs), and NACs, have been shown to directly bind to the promoters of cell wall degradation-related genes, such as PME, PG, PL, XET, EXP, and EGase, and regulate their transcription, consistent with their involvement in the regulation of fruit softening (Tacken et al. 2010; Ireland et al. 2013; Zhu et al. 2014; Gao et al. 2018, 2020). Interestingly, SlERF.F12 was shown to interact with the co-repressor TOPLESS-LIKE2 (TPL2) via its C-terminal EAR motif to recruit HISTONE DEACETYLASE1 (SlHDA1) and SlHDA2 to form a tripartite complex that represses the transcription of ripening genes, including SlPG2a and SlPL, by decreasing the level of acetylation at their promoter regions, thereby inhibiting fruit softening in tomato (Deng et al. 2022). Furthermore, in persimmon (Diospyros kaki), DkNAC9, a direct transcriptional activator of DkEGase1, interacts with DkERF8 and DkERF16 to promote fruit post-deastringency softening (Wu et al. 2020). This observation underscores the complex regulatory network involving transcription factors in fruit softening, which relies on interactions between transcription factors and epigenetic modifications. Here, we showed that ethylene promoted the phosphorylation of MdNAC72 by MdMAPK3 to enhance its ubiquitination, thus implicating phosphorylation and ubiquitination of a transcription factor in the posttranslational regulation of apple fruit softening. Our results expand the ways as to how transcription factors regulate fruit softening.
In this study, because we were unable to obtain an antibody specific to phosphorylated MAPK in apples, we used the phospho-p44/42 antibody, which specifically recognizes phosphorylated MAPK proteins that contain a TEY motif, to investigate the relationship between MAPK activity and ethylene during apple fruit storage. MdMAPK-TEY proteins were recognized using the phospho-p44/42 antibody as 3 bands in apple, one of which was phosphorylated MdMAPK3. The same method was previously applied to identify FvMAPK3 activity in strawberries (Fragaria × ananassa) (Mao et al. 2022). Compared with MdMAPK3, the phosphorylation of other MdMAPK-TEY proteins was also weakly promoted by ethylene treatment during apple fruit storage. We speculate that ethylene may promote the activity of MdMAPKs through their upstream regulators MAPKKK and MAPKK. Here, we demonstrated that MdMAPK3 was involved in ethylene signal transduction during ethylene-induced apple fruit softening during storage and identified the transcription factor MdNAC72 as an interaction partner of MdMAPK3 that is phosphorylated by MdMAPK3. NAC family transcription factors have been shown to bind to the promoters of some cell wall degradation-related genes, including PG, thereby regulating their transcription (Gao et al. 2018). Indeed, we showed that MdNAC72 directly suppressed MdPG1 transcription. Most transcription factors acting as suppressors contain a conserved EAR motif in their C termini (Nakano et al. 2006; Deng et al. 2022); however, MdNAC72 lacks this motif. This situation is analogous to that in wheat (Triticum aestivum), in which TaNAC019 lacks this motif but still acts as a repressor (Liu et al. 2020). Furthermore, we found that MdMAPK3 activity, induced by ethylene, mediates the degradation of MdNAC72, but not of the nonphosphorylatable variant MdNAC72AA, indicating that the degradation of MdNAC72 is dependent on MdMAPK3-mediated phosphorylation of MdNAC72. Interestingly, MdMAPK3, whose activity was promoted by ethylene, phosphorylated MdNAC72 to enhance its ubiquitination by MdPUB24 (Figs. 1, 5, and 8), thus implicating the MdMAPK3–MdNAC72–MdPUB24 module in the regulation of apple fruit softening in response to ethylene.
The classic ethylene signaling pathway (ethylene–CTR1–EIN2–EIN3) has been well documented in the model plant Arabidopsis (Arabidopsis thaliana) (Guo and Ecker 2004). In this pathway, ethylene is sensed by the ethylene receptor CONSTITUTIVE TRIPLE RESPONSE 1 (CTR1), which transmits ethylene signals to ETHYLENE INSENSITIVE 2 (EIN2). EIN2 further transmits signals to EIN3, which ultimately regulates the expression of ethylene-responsive genes. A previous study reported that ethylene can enhance the transcriptional activation activity of MdEIL2, thereby promoting MdPG1 transcription and apple fruit softening (Tacken et al. 2010). This finding suggests that apples can sense ethylene signaling through the ethylene–CTR1–EIN2–EIN3 module during ethylene-induced fruit softening. Notably, Ouaked et al. (2003) identified a MAPK cascade as another pathway for ethylene signal transduction in alfalfa (Medicago sativa) and Arabidopsis, with the ethylene-induced MAPK pathway activating ethylene-induced target gene expression independently of EIN2 and EIN3 (Chang 2003; Ouaked et al. 2003). Here, we also discovered that a protein kinase, MdMAPK3, can be induced by ethylene to enhance MdNAC72 phosphorylation, thereby regulating MdPG1 transcript levels to promote fruit softening. Our results indicate that a MAPK cascade is involved in ethylene signal transmission and that 2 ethylene signal transduction pathways exist in apple fruits to regulate ethylene-induced ripening: the ethylene–CTR1–EIN2–EIN3 and ethylene–MAPK cascade pathways. However, whether these 2 pathways are connected in climacteric fruits requires further investigation.
Interestingly, the same mechanism also exists in the response of rice to the plant hormone abscisic acid (ABA), as ABA-activated SAPK8 (STRESS/ABA-ACTIVATED PROTEIN KINASE 8) and GSK2 (GLYCOGEN SYNTHASE KINASE 2) phosphorylate OsNAC016, leading to its ubiquitination and degradation by OsPUB43 (Wu et al. 2022b). This observation led to the suggestion that phosphorylation/dephosphorylation and stabilization/destabilization of NACs posttranslationally fine-tune their functions, and that the ubiquitination of NACs mediated by E3 ligases from the PUB family is a conserved mechanism in plant hormone signaling pathways. In addition, our previous study showed that MdPUB24 is specifically expressed during apple fruit storage and can be promoted by ethylene treatment and that MdPUB24-mediated ubiquitination of the transcription factor MdBEL7 (BEL1-LIKE HOMEODOMAIN transcription factor 7) causes degreening during apple fruit storage (Wei et al. 2021). This study builds on our previous work to show that MdPUB24-mediated ubiquitination of MdNAC72 causes apple fruit softening during storage. This result suggests that MdPUB24-mediated ubiquitination is specifically initiated after fruit harvest (in artificial orchard settings or fruit dropping from the tree in a natural orchard) and can simultaneously regulate fruit coloration and softening. This, in turn, may make fruits more attractive for frugivores, thus increasing seed dispersal.
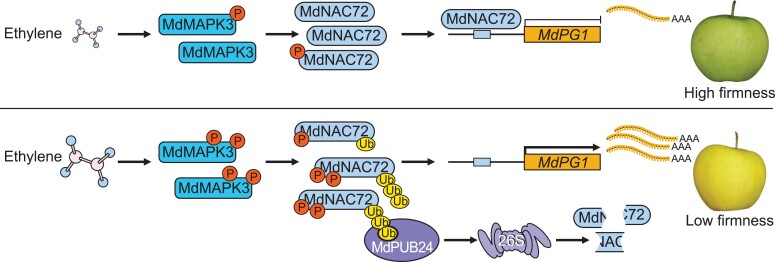
In summary, we discovered that ethylene promotes MdMAPK3 phosphorylation of MdNAC72, which is a negative regulator of the cell wall degradation-related gene MdPG1. Phosphorylated MdNAC72 can be ubiquitinated by the E3 ligase MdPUB24, leading to its degradation. This, in turn, upregulates MdPG1 transcription and results in apple fruit softening during storage (Fig. 10). Our data provide insights into the molecular basis by which ethylene promotes fruit softening, which may lead to the development of strategies for extending the shelf life of climacteric fruits.
Figure 10.
A model showing the molecular mechanism of MdMAPK3-mediated phosphorylation of MdNAC72 in ethylene-induced fruit softening. When a ripe apple fruit with low ethylene levels enters storage conditions, MdNAC72 abundance is maintained at high levels, which inhibits MdPG1 transcription, and the apple fruit is firm. Following storage, high levels of ethylene production promote MdMAPK3 phosphorylation of MdNAC72, leading to its ubiquitination and degradation by ethylene-induced MdPUB24. Consequently, MdNAC72 protein abundance declines and MdPG1 transcript levels increase, leading to pectin degradation and fruit softening.
Materials and methods
Plant materials and treatments
Apple fruits (M. domestica cv. GD) were collected from the Liaoning Pomology Institute orchard (Xiongyue, China). The commercial GD harvest date in this region is 140 d after full bloom (Li et al. 2015) and the apple fruits harvested at that time were stored. Fruits were divided into 3 groups and kept at RT (24°C) for 30 d, with sampling every 10 d. The first group was the untreated control; the second group was treated with ethylene; and the third group was treated with 1-MCP (an ethylene antagonist), as previously described (Tan et al. 2013). Briefly, for ethylene treatment, fruits were dipped into 0.1% [v/v] ethephon (A600453, Sangon Biotech, China) solution for 15 s and then kept in a closed container for 24 h at RT. For 1-MCP treatment, fruits were exposed to 1 μL L–1 of 1-MCP (Fresh Doctor, China) for 12 h at RT in a closed container. To investigate whether MAPK signaling is involved in the rapid response of apple fruit to ethylene, fruits that had just been harvested were treated with 1 mL L–1 ethylene gas for 2, 4, 6, or 8 h in a closed container, or not treated for the no ethylene control. Apple (cv. Orin) calli and N. benthamiana plants were grown in our laboratory, as previously described (Li et al. 2016). For MG132 treatment, calli were immersed in liquid MS medium containing 50 μm MG132 and gently shaken at 24°C for 12 h. N. benthamiana leaves were infiltrated with 50 μm MG132 and then placed in the dark for 12 h. Apple fruits were sectioned (1 mm thick and 10 mm diameter) and immersed in liquid MS medium containing 50 μm MG132 for 12 h. For ethylene treatment, calli and N. benthamiana leaves were placed in a closed container for 12 h at RT.
RNA isolation, cDNA synthesis, RT-qPCR, and ethylene production rate
Total RNA was extracted from apple fruit and calli using a modified cetyltrimethylammonium bromide method (Gasic et al. 2004). First-strand cDNAs were reverse transcribed from 1 μg of total RNA using PrimeScript RT reagent Kit (Cat. no. RR047, TaKaRa, Japan) with gDNA Eraser. qPCR was performed using 2 × UltraSYBR Mixture (Cat. no. CW2601, CWBIO, China) on a qTOWER3 G PCR System (Analytik Jena, Germany) as previously described (Li et al. 2015). The primers used for qPCR are listed in Supplemental Data Set 4. MdACTIN (EB136338) was used as the reference gene for normalization (Ji et al. 2021). qPCR was performed with 3 biological replicates. Ethylene production in apple fruits was measured as previously described (Tan et al. 2013). Briefly, apple fruits were enclosed in an airtight container (860 mL) equipped with septa at 24°C for 1h and 1 mL gas was sampled using a syringe. The ethylene concentration was measured with a gas chromatograph (Agilent 7890A, USA) equipped with a flame ionization detector.
Measurement of apple fruit firmness
Apple fruit firmness was measured using a TA.XT Plus Texture Analyzer (Stable Micro Systems, UK) according to previous methods (Zhang et al. 2018). The measurement points were located near the equator of the fruit and each fruit was measured at 4 points. The parameters were set as follows: probe diameter, 2 mm; penetration depth, 10 mm; descent speed, 1 mm/s; penetration speed, 1 mm/s; and post-measurement speed, 10 mm/s.
Protein isolation and immunoblot analysis
Protein isolation from apple fruit and calli and immunoblot analysis were performed as previously described (Li et al. 2015). The protein concentration of each extract was measured using a BCA protein assay kit (Cat. no. P0012S, CWBIO). Purified recombinant MdNAC72 was used to generate a specific antibody in rabbit (Abmart, China, http://ab-mart.com.cn/). Anti-His (Cat. no. HT501; TransGen Biotech, China), anti-GST (Cat. no. HT601; TransGen Biotech), anti-MBP (Cat. no. HT701; TransGen Biotech, Beijing, China), anti-GFP (Cat. no. HT801; TransGen Biotech, Beijing, China), anti-FLAG (Cat. no. 14793S; Cell Signaling Technology, USA), anti-Myc (Cat. no. HT101; TransGen Biotech), anti-Ub (Cat. no. ST1200, Sigma, USA), anti-phospho-p44/42 (Cat. no. 4730; Cell Signaling Technology), anti-phosphoSer/Thr (Cat. no. ab117253, Abcam, UK), and anti-tubulin (Cat. no. M20045, Abmart) antibodies were diluted 1:1,000 with Tris-buffered saline with Tween 20 (TBST buffer, 20 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.1% [v/v] Tween 20) and incubated with nitrocellulose membranes (Cat. no. S80209, Pall Corporation, USA). Secondary antibodies (goat anti-mouse or anti-rabbit horseradish peroxidase-conjugated; Cat. no. HS201 or HS101, TransGen Biotech) were diluted to 1:3,000 in TBST.
Protein production and purification
The coding sequences of MdNAC72, MdNAC72AA, and MdPUB24 (CDS) were separately cloned into the pEASY-E1 vector (Cat. no. CE111; Transgen Biotech) to produce the corresponding 6×histidine (His) fusion proteins in Escherichia coli. The MdNAC72 and MdPUB24 CDSs were individually cloned into the pMAL-c2x vector (Cat. no. E8201S; New England Biolabs, USA) to produce the corresponding MBP fusion proteins. The MdMAPK3 and CA-MdMAPK3 CDSs were cloned into the pGEX-4T-1 vector (Cat. no. CW2198, CWBIO) to produce the corresponding GST fusion proteins. These recombinant plasmids were separately transferred into E. coli BL21 (DE3) cells to produce the target proteins, as previously described (Li et al. 2015). The temperature used for MdNAC72-His, MdNAC72AA-His, and MdPUB24-His production was 16°C; 23°C for MdMAPK3-GST and CA-MdMAPK3-GST; and 16°C for MdPUB24-MBP and MdNAC72-MBP. The final isopropylthio-β-galactoside concentration used to induce protein production was 0.5 mm.
Y2H and pull-down assay
A cDNA library was constructed using mRNA extracted from ripe apples (GD) and stored for 0, 5, 10, 15, or 20 d using the Make Your Own Mate & Plate Library System (Cat. no. 630489, Clontech, USA) (Wei et al. 2021). The MdMAPK3 and MdNAC72 CDSs were individually cloned into the pGBKT7 vector. The resulting plasmids were used as bait to screen a cDNA library according to the Matchmaker Y2H Library Screening System kit manual (Cat. no. 630489; Clontech). For protein interaction assays, the MdMAPK3 and MdNAC72 CDSs were cloned into the pGBKT7 vector harboring the sequence encoding the GAL4 DNA-binding domain (BD). The MdNAC72 and MdPUB24 coding sequences were cloned into the pGADT7 vector carrying the sequence encoding the GAL4 activation domain (AD). The resulting AD and BD plasmids were co-transformed into the Y2H-gold yeast strain. Interaction analysis between proteins was performed using a Yeastmaker Yeast Transformation Kit (Cat. no. 630489; Clontech, Mountain View, CA). Pull-down assays to test for interaction between MdMAPK3 and MdNAC72 were performed as previously described (Li et al. 2017). Thirty micrograms of recombinant MdMAPK3-GST were captured on GST-binding resin (Cat. no. DP201, Transgen Biotech) and then incubated with 10 μg prey protein (MdNAC72-MBP or MBP) at 4°C for 2 h in buffer (137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4). Subsequently, nonspecifically bound proteins were removed by washing 5 times with buffer. The precipitated prey proteins eluted with elution buffer (EB) (50 mm Tris-HCl, pH 8.0, 10 mm reduced glutathione) were detected by immunoblot analysis using an anti-MBP antibody. In addition, a pull-down assay for the MdNAC72–MdPUB24 interaction was performed as previously described (Li et al. 2016). Thirty micrograms of bait MdPUB24-His was captured using Ni-NTA His-binding resin (Cat. no. DP101, Transgen Biotech) and then incubated with 10 μg prey protein (MdNAC72-MBP or MBP) at 4°C for 2 h in buffer (20 mm Tris-HCl, 0.5 M NaCl, 10 mm imidazole, pH 7.9). Subsequently, nonspecifically bound proteins were removed by washing 5 times with buffer. The precipitated prey proteins eluted with EB (20 mm Tris-HCl, 0.5 m NaCl, 500 mm imidazole, pH 7.9) were detected by immunoblot analysis using an anti-MBP antibody. Furthermore, in the competitive pull-down assays, 10 or 20 μg MdMAPK3-GST was added to the buffer (20 mm Tris-HCl, 0.2 M NaCl, 10 mm β-mercaptoethanol, 1 mm EDTA, pH 8.0) incubation of MdNAC72-His protein and MdPUB24-MBP protein, respectively. Subsequently, the interaction between MdNAC72-his and MdPUB24 was detected as previously described (Yuan et al. 2014).
Co-IP assay
The MdMAPK3 and MdPUB24 CDSs were separately cloned into a pRI101-GFP vector (pRI101 [Cat. no. 3262, TaKaRa] vector as the backbone) in-frame and downstream of the GFP sequence and driven by the cauliflower mosaic virus (CaMV) 35S promoter to generate 35S:GFP-MdMAPK3 and 35S:GFP-MdPUB24 plasmids. The MdNAC72 CDS was cloned into a pRI101-FLAG vector downstream of the sequence encoding the FLAG tag and the 35S promoter to obtain the 35S:FLAG-MdNAC72 plasmid. The resulting plasmids 35S:GFP-MdMAPK3 and 35S:FLAG-MdNAC72 or 35S:GFP-MdPUB24 and 35S:FLAG-MdNAC72 were co-transformed into apple calli, as previously described with minor modifications (Xie et al. 2012; Li et al. 2020). Briefly, 2-wk-old apple calli were collected and suspended in a liquid medium containing Agrobacterium strain EHA105 harboring the corresponding plasmid for 15 min with shaking (140 rpm) at 25°C. The calli were co-cultured on solid MS medium (4.34 g L–1 MS salts, 0.225 mg L–1 of 6-Benzylaminopurine [6-BA], 1.0 mg L–1 of 2, 4-Dichlorophenoxyacetic acid [2, 4-D], 30 g L–1 sucrose, and 7 g L–1 of agar at pH 5.8) at 25°C in the dark. After 3 d, the transgenic calli were washed 3 times with sterile water and then cultured on solid MS medium containing 250 mg L–1 carbenicillin and 30 mg L–1 kanamycin for selection. Transgenic calli were treated with 50 μm MG132 for 12 h to maintain MdNAC72 protein stability. A callus co-transformed with 35S:FLAG-MdNAC72 and 35S:GFP was used as a negative control. Transgenic calli were used for co-IP analysis as previously described (Li et al. 2016). Briefly, a Pierce Co-immunoprecipitation (IP) Kit (Cat. no. 26149, Thermo Scientific, USA) was used to immunoprecipitate MdNAC72-FLAG using an anti-FLAG antibody. The precipitated proteins were analyzed by immunoblot using an anti-GFP antibody. For competitive co-IP assays, changes in the enrichment of MdNAC72 protein by co-immunoprecipitated MdPUB24-GFP detected using an anti-GFP antibody were analyzed when MdMAPK3 was overexpressed in apple fruit calli. Overexpression and co-IP methods were performed as described above.
Firefly LCI assay
The MdMAPK3 or MdPUB24 CDSs were separately cloned into the pCAMBIA1300-nLuc vector (Chen et al. 2008) to construct the 35S:MdMAPK3-nLuc and 35S:MdPUB24-nLuc plasmids. The MdNAC72 CDS was cloned into the pCAMBIA1300-cLuc vector to generate the 35S:MdNAC72-cLuc plasmid. 35S:MdMAPK3-nLuc, 35S:MdNAC72-cLuc, 35S:MdPUB24-nLuc, and 35S:MdNAC72-cLuc plasmids were transformed into Agrobacterium strain EHA105 using electroporation, and cultures harboring each plasmid were infiltrated into N. benthamiana leaves in the appropriate combinations as previously described (Li et al. 2017). Twelve hours before LUC imaging, N. benthamiana leaves were treated with 50 μm of MG132. Luciferase activity was measured as described previously (Li et al. 2017; Ji et al. 2021).
Y1H assay and EMSA
For the Y1H assay, the MdNAC72 CDS was cloned into the pGADT7 vector (Cat. no. 630491; Clontech), and the MdPG1 promoter fragment was ligated into the pAbAi vector (Cat. no. 630491; Clontech, Mountain View, CA). The Y1H assay was performed as previously described (Li et al. 2016). EMSA was performed using a Chemiluminescent EMSA Kit (cat. no. 20148; Thermo Fisher Scientific, Waltham, MA). Recombinant MdNAC72-MBP was prepared as described above. Biotin-labeled probes were synthesized by annealing complementary oligonucleotides, and the proteins and probes were incubated at 24°C for 30 min. The sequences of the 2 MdPG1 promoter probes are listed in Supplemental Data Set 4.
ChIP-qPCR assay
The MdNAC72 and MdNAC72AA CDSs were cloned into the pRI101 vector downstream of the sequence encoding a FLAG tag and the 35S promoter to generate the 35S:FLAG-MdNAC72 and 35S:FLAG-MdNAC72AA plasmids. The resulting constructs were transformed into apple calli as described above. Transgenic calli were treated with or without ethylene. For ethylene treatment, calli were placed in a clean sealed container containing 1 mL L–1 of ethylene for 12 h at RT. ChIP-qPCR assays were performed as previously described (Qin et al. 2012) using an anti-FLAG antibody (Cat. no. 14793S; Cell Signaling Technology, Danvers, MA). The amount of immunoprecipitated chromatin was determined using qPCR as previously described (Yue et al. 2020).
GUS assay
The MdPG1 promoter (1,849 bp upstream of the ATG start codon) was inserted into the pCAMBIA1300-GUS vector to construct the proMdPG1:GUS reporter plasmid. Mutations were introduced into the NAC binding sites (from CACG or CGTG to CCCC or CGCC) of the MdPG1 promoter using a QuickMutation Plus Kit (Cat. no. D0208S, Beyotime Biotechnology, China). The MdNAC72, MdNAC72AA, MdMAPK3, and MdPUB24 CDSs were separately cloned into the pRI101 vector containing the 35S promoter to generate effector constructs. The reporter and effector plasmids were transformed into N. benthamiana leaves via Agrobacterium-mediated infiltration as above and 35S:LUC was included as an internal control for normalization of transformation efficiency. The relative GUS activity was calculated as the ratio of GUS to LUC activity as previously described (Li et al. 2016; Yan et al. 2018). MG132 (50 μm) or ethylene (1 mL L–1) treatment was performed 12 h before detection.
Dual-luciferase reporter assay
The MdPG1 promoter (1,849 bp upstream of the ATG start codon) was introduced into the pGreen II 0800-Luc vector to construct the proMdPG1:LUC reporter plasmid. The MdNAC72, MdNAC72AA, MdMAPK3, and MdPUB24 CDSs were separately cloned into the pRI101 vector containing the 35S promoter to generate effector constructs. Transformation of reporter and effector plasmids into N. benthamiana leaves and measurement of luciferase activity were carried out as previously described (Wang et al. 2020). For MG132 or ethephon treatment, MG132 (50 μm) or ethephon (0.1% [v/v]) was infiltrated into N. benthamiana leaves 12 h before detection.
In vitro and in vivo phosphorylation assay
For the in vitro phosphorylation assays, phosphorylation reactions containing 5 μg of recombinant substrate protein (MdNAC72-His, MdNAC72AA-His, MdNAC72S74A, or MdNAC72S220A) and 10 μg of kinase (CA-MdMAPK3-GST) were performed in 100 μL kinase buffer (25 mm Tris-HCl pH 7.5, 10 mm MgCl2, 1 mm CaCl2, 10 mm ATP, and 1 mm DTT) at 30°C for 2 h. The phosphorylated proteins were separated on a 10% SDS-PAGE gel containing 50 μm Phos-tag and μM 100 MnCl2. After the proteins were electroblotted onto a polyvinylidene fluoride membrane, MdNAC72 and MdNAC72AA were detected using the anti-His antibody generated in this study. As a loading control, phosphorylated proteins were fractionated on a 10% SDS-PAGE gel without phos-tag and MnCl2 and detected using the anti-His antibody. The in vivo phosphorylation assay was performed as previously described (Wang et al. 2022; Li et al. 2023). Apple fruit proteins (empty vector or 35S:MdMAPK3-AS transgenic fruit) and apple calli proteins (35S:FLAG-MdNAC72 alone, 35S:FLAG-MdNAC72 and MdMAPK3-OE, 35S:FLAG-MdNAC72AA, or 35S:FLAG-MdNAC72 and MdMAPK3-AS transgenic calli) were isolated in buffer (50 mm Tris-HCl pH 7.4, 150 mm NaCl, 1 mm EDTA, 0.1% [v/v] Triton X-100, 0.1% [v/v] Nonidet P-40, 1 mm PMSF, 1 × phosphatase inhibitor cocktail [MedChem Express, USA], and 1 × protease inhibitor cocktail [MedChemExpress]). Twenty microliters of anti-FLAG magnetic beads (cat. no. 14793S, Cell Signaling Technology) or anti-MdNAC72 magnetic beads (Abmart) were added to 1 mg total protein extract and incubated at 4°C for 4 h to immunoprecipitate the corresponding proteins, as described in the Pierce classic IP Kit manual (Cat. no. 26146; Thermo Scientific), using anti-FLAG and anti-MdNAC72 antibodies. After 5 washes with buffer, the concentration of the precipitated proteins was determined using a BCA Protein Assay Kit (Cat. no. P0012S, CWBIO), and the proteins were eluted by boiling in SDS sample buffer for 10 min. Immunoprecipitated proteins were separated on a 10% SDS-PAGE gel for immunoblotting using an anti-phosphoSer/Thr antibody (Cat. no. ab117253, Abcam). As a control, immunoprecipitated proteins were detected using anti-FLAG or anti-MdNAC72 antibody. For phos-tag mobility shift analysis, the immunoprecipitated proteins were investigated as described above.
In vitro protein degradation assay
MdNAC72-His or MdNAC72AA-His proteins were produced and purified as described above. Total proteins from 35S:MdMAPK3-AS or 35S:MdPUB24-AS transgenic apple calli were extracted in degradation buffer (25 mm Tris-HCl, pH 7.5, 10 mm NaCl, 10 mm MgCl2, 4 mm PMSF, 5 mm DTT, and 10 mm ATP), as previously described (Wei et al. 2021). A transgenic empty vector calli was used as a control. Recombinant MdNAC72-His or MdNAC72AA-His protein (10 μg) was added to total protein extracts (500 μg) containing MG132 (50 μm) or dimethyl sulfoxide as control and incubated for 2, 4, or 6 h at 25°C. MdNAC72-His or MdNAC72AA-His was separated on an SDS-PAGE gel and detected using an anti-His antibody.
In vitro and in vivo ubiquitination assay
The in vitro ubiquitination assay was performed as previously described (Li et al. 2012). Briefly, 50 ng of human E1 (Boston Biochem), 50 ng of human E2 (Boston Biochem), 200 ng of E3 (MdPUB24), 10 μg of His-6-ubiquitin (Boston Biochem), and 100 ng of MdNAC72 (or MdNAC72AA) were incubated in 30 μL of ubiquitination reaction buffer (50 mm Tris-HCl pH 7.5, 10 mm MgCl2, 10 mm ATP, 1 mm DTT) at 30°C for 4 h. The proteins were separated by SDS-PAGE, and ubiquitinated MdNAC72 or MdNAC72AA was detected using an anti-Ub (Cat. no. ST1200, Sigma) antibody. In vivo ubiquitination assays were performed as previously described (An et al. 2017). Briefly, 35S:FLAG-MdNAC72 transgenic calli and 35S:FLAG-MdNAC72 35S:MdPUB24-OE co-transgenic calli were pretreated with 50 μm MG132 for 12 h. A Pierce classic imm unoprecipitation (IP) Kit (Cat. no. 26146, Thermo Scientific) was used to immunoprecipitate MdNAC72 using an anti-FLAG antibody, and the eluted proteins were analyzed by immunoblotting using an anti-FLAG antibody (Cat. no. 14793S; Cell Signaling Technology, Danvers, MA). In addition, the anti-MdNAC72 antibody was used to immunoprecipitate ubiquitinated MdNAC72 from transgenic apple fruit (35S:MdPUB24-OE or 35S:MdMAPK3-AS), which were treated with MG132 (50 μm) for 12 h before immunoprecipitation. Transgenic empty vector fruits were used as controls. To explore whether MdMAPK3 affects the ubiquitination of MdNAC72 by MdPUB24, an in vivo ubiquitination assay was performed in N. benthamiana leaves as previously described (Shan et al. 2020). Briefly, N. benthamiana leaves were transiently infiltrated with the plasmids 35S:FLAG-MdNAC72 and 35S:GFP-MdPUB24 or 35S:FLAG-MdNAC72 and 35S:GFP-MdPUB24 and 35S:MdMAPK3 via Agrobacterium-mediated transient infiltration. Leaves were pretreated with 50 μm MG132 proteasome inhibitor for 12 h before immunoprecipitation. A Pierce classic IP Kit (Thermo Scientific, USA) was used to immunoprecipitate MdNAC72-FLAG using an anti-FLAG antibody. The precipitated proteins were analyzed by immunoblotting using an anti-Ub antibody. To explore how MdMAPK3 affects the ubiquitination of MdNAC72 by MdPUB24, an in vivo ubiquitination assay was performed on N. benthamiana leaves infiltrated with 35S:FLAG-MdNAC72 and 35S:GFP-MdPUB24 or 35S:FLAG-MdNAC72 and 35S:GFP-MdPUB24 and 35S:CA-MdMAPK3, as described above. MdNAC72 immunoprecipitated using an anti-FLAG antibody was incubated with λPP (New England Biolabs) at 30°C for 30 min or 60 min, as previously described (Wang et al. 2022).
Agrobacterium-mediated transformation of apple calli or fruits
To transiently overexpress MdMAPK3, MdPUB24, MdNAC72, MdNAC72AA, or MdNAC72DD, Agrobacterium-mediated transformation was performed as previously described (Ji et al. 2022). The MdMAPK3, MdPUB24, MdNAC72, MdNAC72AA, and MdNAC72DD CDSs were separately cloned into the pRI101 vector to construct overexpression constructs. To silence MdMAPK3, MdPUB24, or MdNAC72 expression in apple fruits, a partial CDS fragment of MdMAPK3 (587 to 809 bp, with +1 being the start of the ATG), MdPUB24 (162 to 406 bp), or MdNAC72 (285 to 589 bp) was inserted into the pRI101 vector in the reverse direction to generate antisense silencing constructs (35S:MdMAPK3-AS, 35S:MdMPUB24-AS, and 35S:MdMNAC72-AS, respectively). Each plasmid was introduced into Agrobacterium strain EHA105 using the heat-shock method (Hofgen and Willmitzer 1988), with pRI101-GFP and pRI101-Myc empty vectors used as controls. The suspension and injection solutions for silencing or overexpression in apple fruit were prepared as previously described (Li et al. 2016; Wei et al. 2021). One hundred microliters of Agrobacterium cell suspension harboring the above plasmids was collected with a sterile 1 mL syringe and injected approximately 0.5 cm deep into fruits at 7 d before commercial harvest time.
Statistical analysis
Two-tailed Student's t-tests were used on 2 groups of data points in GraphPad Prism version 9.5. Asterisks indicate significant differences between the 2 data groups (*P < 0.05, **P < 0.01). n.s. (P > 0.05) represents no significant differences between the 2 groups of data. The summary of these analyses is shown in Supplemental Data Set 5.
Accession numbers
Sequence data from this study can be found at NCBI GenBank (https://www.ncbi.nlm.nih.gov/), GDR (https://www.rosaceae.org/), and TAIR (https://www.arabidopsis.org/index.jsp) under the accession numbers MdACTIN (EB136338), MdNAC72 (LOC103407991), MdPUB24 (LOC103421753), MdPG1 (MDP0000326734), MdPL5 (MDP0000277149), Mdβ-Gal9 (MDP0000416548), Mdα-AFase2 (MDP0000140483), MdXET1 (MDP0000398765), MdEXP8 (MDP0000431696), MdMAPK3 (MDP0000237742), MdMAPK4 (MDP0000251955), MdMAPK6 (MDP0000340624), MdMAPK7 (MDP0000807889), MdMAPK13 (MDP0000422421), MdMAPK9 (MDP0000294142), MdMAPK16 (MDP0000170804), MdMAPK17 (MDP0000277562), MdMAPK19 (MDP0000121116), MdMAPK20 (MDP0000188369), AtMAPK3 (At3G45640), AtMAPK4 (At4G01370) and AtMAPK6 (At2G43790).
Supplementary Material
Contributor Information
Yun Wei, Key Laboratory of Fruit Postharvest Biology (Liaoning Province), Shenyang Agricultural University, Shenyang 110866, China; Key Laboratory of Protected Horticulture (Ministry of Education), Shenyang Agricultural University, Shenyang 110866, China; National & Local Joint Engineering Research Center of Northern Horticultural Facilities Design & Application Technology (Liaoning), Shenyang Agricultural University, Shenyang 110866, China; College of Horticulture, Shenyang Agricultural University, Shenyang 110866, China.
Zhi Liu, Liaoning Institute of Pomology, Xiongyue 115009, China.
Tianxing Lv, Liaoning Institute of Pomology, Xiongyue 115009, China.
Yaxiu Xu, Key Laboratory of Fruit Postharvest Biology (Liaoning Province), Shenyang Agricultural University, Shenyang 110866, China; Key Laboratory of Protected Horticulture (Ministry of Education), Shenyang Agricultural University, Shenyang 110866, China; National & Local Joint Engineering Research Center of Northern Horticultural Facilities Design & Application Technology (Liaoning), Shenyang Agricultural University, Shenyang 110866, China; College of Horticulture, Shenyang Agricultural University, Shenyang 110866, China.
Yajing Wei, Key Laboratory of Fruit Postharvest Biology (Liaoning Province), Shenyang Agricultural University, Shenyang 110866, China; Key Laboratory of Protected Horticulture (Ministry of Education), Shenyang Agricultural University, Shenyang 110866, China; National & Local Joint Engineering Research Center of Northern Horticultural Facilities Design & Application Technology (Liaoning), Shenyang Agricultural University, Shenyang 110866, China; College of Horticulture, Shenyang Agricultural University, Shenyang 110866, China.
Weiting Liu, Key Laboratory of Fruit Postharvest Biology (Liaoning Province), Shenyang Agricultural University, Shenyang 110866, China; Key Laboratory of Protected Horticulture (Ministry of Education), Shenyang Agricultural University, Shenyang 110866, China; National & Local Joint Engineering Research Center of Northern Horticultural Facilities Design & Application Technology (Liaoning), Shenyang Agricultural University, Shenyang 110866, China; College of Horticulture, Shenyang Agricultural University, Shenyang 110866, China.
Li Liu, Key Laboratory of Fruit Postharvest Biology (Liaoning Province), Shenyang Agricultural University, Shenyang 110866, China; Key Laboratory of Protected Horticulture (Ministry of Education), Shenyang Agricultural University, Shenyang 110866, China; National & Local Joint Engineering Research Center of Northern Horticultural Facilities Design & Application Technology (Liaoning), Shenyang Agricultural University, Shenyang 110866, China; College of Horticulture, Shenyang Agricultural University, Shenyang 110866, China.
Aide Wang, Key Laboratory of Fruit Postharvest Biology (Liaoning Province), Shenyang Agricultural University, Shenyang 110866, China; Key Laboratory of Protected Horticulture (Ministry of Education), Shenyang Agricultural University, Shenyang 110866, China; National & Local Joint Engineering Research Center of Northern Horticultural Facilities Design & Application Technology (Liaoning), Shenyang Agricultural University, Shenyang 110866, China; College of Horticulture, Shenyang Agricultural University, Shenyang 110866, China.
Tong Li, Key Laboratory of Fruit Postharvest Biology (Liaoning Province), Shenyang Agricultural University, Shenyang 110866, China; Key Laboratory of Protected Horticulture (Ministry of Education), Shenyang Agricultural University, Shenyang 110866, China; National & Local Joint Engineering Research Center of Northern Horticultural Facilities Design & Application Technology (Liaoning), Shenyang Agricultural University, Shenyang 110866, China; College of Horticulture, Shenyang Agricultural University, Shenyang 110866, China.
Author contributions
T.L. and A.W. conceived and designed the study. Y.W. performed the majority of the experiments. Y.X. and Y.W. performed the protein prokaryotic expression and purification. W.L. and Y.J.W. performed the protein isolation from fruits and calli. L.L. performed the gene expression analysis. Z.L. and T.X.L. provided apples. T.L. and Y.W. wrote the manuscript. All authors analyzed the data and discussed the article.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Identification of an MdMAPK3 activity band using SDS-PAGE.
Supplemental Figure S2. Analysis of MdMAPK activity levels during fruit storage.
Supplemental Figure S3. Silencing MdMAPK3 suppresses apple fruit softening during storage.
Supplemental Figure S4. Relative expression of MdNAC72 during apple fruit storage.
Supplemental Figure S5. Functional analysis of MdNAC72 in apple fruit calli.
Supplemental Figure S6. Silencing MdNAC72 promotes apple fruit softening during storage.
Supplemental Figure S7. Relative expression of cell wall degradation-related genes during apple fruit storage.
Supplemental Figure S8. Analysis of MdNAC72 binding to the promoters of cell wall degradation-related genes in vivo.
Supplemental Figure S9. Phos-tag mobility shift analysis showing that MdMAPK3 phosphorylates MdNAC72 in vivo.
Supplemental Figure S10. Silencing MdPUB24 suppresses apple fruit softening during storage.
Supplemental Figure S11. Effect of MdNAC72DD on apple fruit softening.
Supplemental Data Set 1. MdMAPK3 potential interactors obtained by screening a yeast cDNA library.
Supplemental Data Set 2. List of genes upregulated in 0-fruit (fruit stored for 0 d) compared with 15-fruit (fruit stored for 15 d) or downregulated in 15-fruit relative to 15-MCP-fruit (fruit stored for 15 d after 1-MCP treatment).
Supplemental Data Set 3. Potential MdNAC72 interactors obtained by screening a yeast cDNA library.
Supplemental Data Set 4. List of primers used in this study.
Supplemental Data Set 5. Summary of statistical tests.
Funding
This work was supported by the National Natural Science Foundation of China (32125034, 32002006), the National Key Research and Development Program of China (SQ2022YFD2100005), and the Natural Science Foundation of Liaoning Province (2022-YQ-19).
References
- An J-P, Liu X, Li H-H, You C-X, Wang X-F, Hao Y-J. Apple RING E3 ligase MdMIEL1 inhibits anthocyanin accumulation by ubiquitinating and degrading MdMYB1 protein. Plant and Cell Physiol. 2017:58(11):1953–1962. 10.1093/pcp/pcx129 [DOI] [PubMed] [Google Scholar]
- Atkinson RG, Schröder R, Hallett IC, Cohen D, MacRae EA. Overexpression of polygalacturonase in transgenic apple trees leads to a range of novel phenotypes involving changes in cell adhesion. Plant Physiol. 2002:129(1):122–133. 10.1104/pp.010986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson RG, Sutherland PW, Johnston SL, Gunaseelan K, Hallett IC, Mitra D, Brummell DA, Schröder R, Johnston JW, Schaffer RJ. Down-regulation of POLYGALACTURONASE1 alters firmness, tensile strength and water loss in apple (Malus x domestica) fruit. BMC Plant Biol. 2012:12(1):129. 10.1186/1471-2229-12-129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriri S, Garcia AV, dit Frey NF, Rozhon W, Pateyron S, Leonhardt N, Montillet J-L, Leung J, Hirt H, Colcombet J. Constitutively active mitogen-activated protein kinase versions reveal functions of Arabidopsis MPK4 in pathogen defense signaling. Plant Cell. 2012:24(10):4281–4293. 10.1105/tpc.112.101253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C. Ethylene signaling: the MAPK module has finally landed. Trends Plant Sci. 2003:8(8):365–368. 10.1016/S1360-1385(03)00156-0 [DOI] [PubMed] [Google Scholar]
- Chen H, Zou Y, Shang Y, Lin H, Wang Y, Cai R, Tang X, Zhou J-M. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008:146(2):323–324. 10.1104/pp.107.111740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa F, Peace CP, Stella S, Serra S, Musacchi S, Bazzani M, Sansavini S, Van de Weg WE. QTL dynamics for fruit firmness and softening around an ethylene-dependent polygalacturonase gene in apple (Malus×domestica Borkh). J Exp Bot. 2010:61(11):3029–3039. 10.1093/jxb/erq130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa F, Van de Weg WE, Stella S, Dondini L, Pratesi D, Musacchi S, Sansavini S. Map position and functional allelic diversity of Md-Exp7, a new putative expansin gene associated with fruit softening in apple (Malus × domestica Borkh.) and pear (Pyrus communis). Tree Genet Genomes. 2008:4(3):575–586. 10.1007/s11295-008-0133-5 [DOI] [Google Scholar]
- Deng H, Chen Y, Liu Z, Liu Z, Shu P, Wang R, Hao Y, Su D, Pirrello J, Liu Y, et al. SlERF.F12 modulates the transition to ripening in tomato fruit by recruiting the co-repressor TOPLESS and histone deacetylases to repress key ripening genes. Plant Cell. 2022:34(4):1250–1272. 10.1093/plcell/koac025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Zelicourt A, Colcombet J, Hirt H. The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 2016:21(8):677–685. 10.1016/j.tplants.2016.04.004 [DOI] [PubMed] [Google Scholar]
- Gao Y, Wei W, Zhao X, Tan X, Fan Z, Zhang Y, Jing Y, Meng L, Zhu B, Zhu H, et al. A NAC transcription factor, NOR-like1, is a new positive regulator of tomato fruit ripening. Hortic Res. 2018:5:75. 10.1038/s41438-018-0111-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Zhang Y, Li Z, Liu M. Role of ethylene response factors (ERFs) in fruit ripening. Food Qual Saf. 2020:4(1):15–20. 10.1093/fqsafe/fyz042 [DOI] [Google Scholar]
- Gasic K, Hernandez A, Korban SS. RNA Extraction from different apple tissues rich in polyphenols and polysaccharides for cDNA library construction. Plant Mol Biol Rep. 2004:22(4):437–438. 10.1007/BF02772687 [DOI] [Google Scholar]
- Goulao LF, Cosgrove DJ, Oliveira CM. Cloning, characterisation and expression analyses of cDNA clones encoding cell wall-modifying enzymes isolated from ripe apples. Postharvest Biol Technol. 2008:48(1):37–51. 10.1016/j.postharvbio.2007.09.022 [DOI] [Google Scholar]
- Guo H, Ecker JR. The ethylene signaling pathway: new insights. Curr Opin Plant Biol. 2004:7(1):40–49. 10.1016/j.pbi.2003.11.011 [DOI] [PubMed] [Google Scholar]
- Guo T, Lu Z-Q, Shan J-X, Ye W-W, Dong N-Q, Lin H-X. ERECTA1 acts upstream of the OsMKKK10-OsMKK4-OsMPK6 cascade to control spikelet number by regulating cytokinin metabolism in rice. Plant Cell. 2020:32(9):2763–2779. 10.1105/tpc.20.00351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofgen R, Willmitzer L. Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res. 1988:16(20):9877–9877. 10.1093/nar/16.20.9877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireland HS, Yao J-L, Tomes S, Sutherland PW, Nieuwenhuizen N, Gunaseelan K, Winz RA, David KM, Schaffer RJ. Apple SEPALLATA1/2-like genes control fruit flesh development and ripening. Plant J. 2013:73(6):1044–1056. 10.1111/tpj.12094 [DOI] [PubMed] [Google Scholar]
- Ji Y, Qu Y, Jiang Z, Yan J, Chu J, Xu M, Su X, Yuan H, Wang A. The mechanism for brassinosteroids suppressing climacteric fruit ripening. Plant Physiol. 2021:185(4):1875–1893. 10.1093/plphys/kiab013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Xu M, Liu Z, Yuan H, Lv T, Li H, Xu Y, Si Y, Wang A. NUCLEOCYTOPLASMIC shuttling of ETHYLENE RESPONSE FACTOR 5 mediated by nitric oxide suppresses ethylene biosynthesis in apple fruit. New Phytol. 2022:234(5):1714–1734. 10.1111/nph.18071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodayari A, Thielemans W, Hirn U, Van Vuure AW, Seveno D. Cellulose-hemicellulose interactions—a nanoscale view. Carbohydr Polym. 2021:270:118364. 10.1016/j.carbpol.2021.118364 [DOI] [PubMed] [Google Scholar]
- Kosma DK, Parsons EP, Isaacson T, Lü S, Rose JKC, Jenks MA. Fruit cuticle lipid composition during development in tomato ripening mutants. Physiol Plant. 2010:139(1):107–117. 10.1111/j.1399-3054.2009.01342.x [DOI] [PubMed] [Google Scholar]
- Kwon SJ, Choi EY, Choi YJ, Ahn JH, Park OK. Proteomics studies of post-translational modifications in plants. J Exp Bot. 2006:57(7):1547–1551. 10.1093/jxb/erj137 [DOI] [PubMed] [Google Scholar]
- Li T, Jiang Z, Zhang L, Tan D, Wei Y, Yuan H, Li T, Wang A. Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J. 2016:88(5):735–748. 10.1111/tpj.13289 [DOI] [PubMed] [Google Scholar]
- Li T, Liu Zhi, Lv T, Xu Y, Wei Y, Liu W, Wei Y, Liu L, Wang A. Phosphorylation of MdCYTOKININ RESPONSE FACTOR4 suppresses ethylene biosynthesis during apple fruit ripening. Plant Physiol. 2023:191(1):694–714. 10.1093/plphys/kiac498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y-Y, Mao K, Zhao C, Zhao X-Y, Zhang H-L, Shu H-R, Hao Y. MdCOP1 ubiquitin E3 ligases interact with MdMYB1 to regulate light-induced anthocyanin biosynthesis and red fruit coloration in apple. Plant Physiol. 2012:160(2):1011–1022. 10.1104/pp.112.199703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Meng D, Piñeros MA, Mao Y, Dandekar AM, Cheng L. A sugar transporter takes up both hexose and sucrose for sorbitol-modulated in vitro pollen tube growth in apple. Plant Cell. 2020:32(2):449–469. 10.1105/tpc.19.00638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Tan D, Liu Z, Jiang Z, Wei Y, Zhang L, Li X, Yuan H, Wang A. Apple MdACS6 regulates ethylene biosynthesis during fruit development involving ethylene-responsive factor. Plant Cell Physiol. 2015:56(10):1909–1917. 10.1093/pcp/pcv111 [DOI] [PubMed] [Google Scholar]
- Li T, Xu Y, Zhang L, Ji Y, Tan D, Yuan H, Wang A. The jasmonate-activated transcription factor MdMYC2 regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell. 2017:29(6):1316–1334. 10.1105/tpc.17.00349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Zhang X, Wei Y, Xu Y, Liu W, Li H, Yang G, Wang A, Wang X. Comparative transcriptome analysis of the climacteric of apple fruit uncovers the involvement of transcription factors affecting ethylene biosynthesis. Hortic Plant J. 2022. 10.1016/j.hpj.2022.12.002 [DOI] [Google Scholar]
- Liu Y, Hou J, Wang X, Li T, Majeed U, Hao C, Zhang X. The NAC transcription factor NAC019-A1 is a negative regulator of starch synthesis in wheat developing endosperm. J Exp Bot. 2020:71(19):5794–5807. 10.1093/jxb/eraa333 [DOI] [PubMed] [Google Scholar]
- Longhi S, Moretto M, Viola R, Velasco R, Costa F. Comprehensive QTL mapping survey dissects the complex fruit texture physiology in apple (Malus x domestica Borkh). J Exp Bot. 2012:63(3):1107–1121. 10.1093/jxb/err326 [DOI] [PubMed] [Google Scholar]
- Mao W, Han Y, Chen Y, Sun M, Feng Q, Li L, Liu L, Zhang K, Wei L, Han Z, et al. Low temperature inhibits anthocyanin accumulation in strawberry fruit by activating FvMAPK3-induced phosphorylation of FvMYB10 and degradation of Chalcone Synthase 1. Plant Cell. 2022:34(4):1226–1249. 10.1093/plcell/koac006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migicovsky Z, Yeats TH, Watts S, Song J, Forney CF, Burgher-MacLellan K, Somers DJ, Gong Y, Zhang Z, Vrebalov J, et al. Apple ripening is controlled by a NAC transcription factor. Front Genet. 2021:12:671300. 10.3389/fgene.2021.671300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006:140(2):411–432. 10.1104/pp.105.073783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio S, Alba R, Damasceno CMB, Lopez-Casado G, Lohse M, Zanor MI, Tohge T, Usadel B, Rose JKC, Fei Z, et al. Systems biology of tomato fruit development: combined transcript, protein, and metabolite analysis of tomato transcription factor (nor, rin) and ethylene receptor (Nr) mutants reveals novel regulatory interactions. Plant Physiol. 2011:157(1):405–425. 10.1104/pp.111.175463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouaked F, Rozhon W, Lecourieux D, Hirt H. A MAPK pathway mediates ethylene signaling in plants. EMBO J. 2003:22(6):1282–1288. 10.1093/emboj/cdg131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniagua C, Posé S, Morris VJ, Kirby AR, Quesada MA, Mercado JA. Fruit softening and pectin disassembly: an overview of nanostructural pectin modifications assessed by atomic force microscopy. Ann Bot. 2014:114(6):1375–1383. 10.1093/aob/mcu149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin G, Wang Y, Cao B, Wang W, Tian S. Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J. 2012:70(2):243–255. 10.1111/j.1365-313X.2011.04861.x [DOI] [PubMed] [Google Scholar]
- Römling U, Galperin MY. Bacterial cellulose biosynthesis: diversity of operons, subunits, products, and functions. Trends Microbiol. 2015:23(9):545–557. 10.1016/j.tim.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan W, Kuang JF, Wei W, Fan ZQ, Deng W, Li ZG, Bouzayen M, Pirrello J, Lu WJ, Chen JY. MaXB3 modulates MaNAC2, MaACS1, and MaACO1 stability to repress ethylene biosynthesis during banana fruit ripening. Plant Physiol. 2020:184(2):1153–1171. 10.1104/pp.20.00313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tacken E, Ireland H, Gunaseelan K, Karunairetnam S, Wang D, Schultz K, Bowen J, Atkinson RG, Johnston JW, Putterill J, et al. The role of ethylene and cold temperature in the regulation of the apple POLYGALACTURONASE1 gene and fruit softening. Plant Physiol. 2010:153(1):294–305. 10.1104/pp.109.151092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan D, Li T, Wang A. Apple 1-aminocyclopropane-1-carboxylic acid synthase genes, MdACS1 and MdACS3a, are expressed in different systems of ethylene biosynthesis. Plant Mol Biol Rep. 2013:31(1):204–209. 10.1007/s11105-012-0490-y [DOI] [Google Scholar]
- Tena G, Asai T, Chiu W-L, Sheen J. Plant mitogen-activated protein kinase signaling cascades. Curr Opin Plant Biol. 2001:4(5):392–400. 10.1016/S1369-5266(00)00191-6 [DOI] [PubMed] [Google Scholar]
- Trewavas A. Post-translational modification of proteins by phosphorylation. Annu Rev Plant Physiol. 1976:27(1):349–374. 10.1146/annurev.pp.27.060176.002025 [DOI] [Google Scholar]
- Tucker G, Yin X, Zhang A, Wang M, Zhu Q, Liu X, Xie X, Chen K, Grierson D. Ethylene and fruit softening. Food Qual Saf. 2017:1(4):253–267. 10.1093/fqsafe/fyx024 [DOI] [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, Dhingra A, Cestaro A, Kalyanaraman A, Fontana P, Bhatnagar SK, Troggio M, Pruss D, et al. The genome of the domesticated apple (Malus × domestica Borkh). Nat Genet. 2010:42(10):833–839. 10.1038/ng.654 [DOI] [PubMed] [Google Scholar]
- Wakasa Y, Kudo H, Ishikawa R, Akada S, Senda M, Niizeki M, Harada T. Low expression of an endopolygalacturonase gene in apple fruit with long-term storage potential. Postharvest Biol Technol. 2006:39(2):193–198. 10.1016/j.postharvbio.2005.10.005 [DOI] [Google Scholar]
- Wang N, Liu W, Yu L, Guo Z, Chen Z, Jiang S, Xu H, Fang H, Wang Y, Zhang Z, et al. HEAT SHOCK FACTOR A8a modulates flavonoid synthesis and drought tolerance. Plant Physiol. 2020:184(3):1273–1290. 10.1104/pp.20.01106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wang T, Li Q, Xu C, Tian J, Wang Y, Zhang X, Xu X, Han Z, Wu T. Phosphorylation of MdERF17 by MdMPK4 promotes apple fruit peel degreening during light/dark transitions. Plant Cell. 2022:34(5):1980–2000. 10.1093/plcell/koac049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Yeats TH, Uluisik S, Rose JKC, Seymour GB. Fruit softening: revisiting the role of pectin. Trends Plant Sci. 2018:23(4):302–310. 10.1016/j.tplants.2018.01.006 [DOI] [PubMed] [Google Scholar]
- Wei Y, Jin J, Xu Y, Liu W, Yang G, Bu H, Li T, Wang A. Ethylene-activated MdPUB24 mediates ubiquitination of MdBEL7 to promote chlorophyll degradation in apple fruit. Plant J. 2021:108(1):169–182. 10.1111/tpj.15432 [DOI] [PubMed] [Google Scholar]
- Wolf-Dieter R. The molecular analysis of cell wall components. Trends Plant Sci. 1998:3(1):27–32. 10.1016/S1360-1385(97)01169-2 [DOI] [Google Scholar]
- Wu Q, Liu Y, Xie Z, Yu B, Sun Y, Huang J. OsNAC016 regulates plant architecture and drought tolerance by interacting with the kinases GSK2 and SAPK8. Plant Physiol. 2022b:189(3):1296–1313. 10.1093/plphys/kiac146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-J, Shan W, Liu X-C, Zhu L-S, Wei W, Yang Y-Y, Guo Y-F, Bouzayen M, Chen J-Y, Lu W-J, et al. Phosphorylation of transcription factor bZIP21 by MAP kinase MPK6-3 enhances banana fruit ripening. Plant Physiol. 2022a:188(3):1665–1685. 10.1093/plphys/kiab539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu W, Wang M-M, Gong H, Liu X-F, Guo D-L, Sun N-J, Huang J-W, Zhu Q-G, Chen K-S, Yin X-R. High CO2/hypoxia-induced softening of persimmon fruit is modulated by DkERF8/16 and DkNAC9 complexes. J Exp Bot. 2020:71(9):2690–2700. 10.1093/jxb/eraa009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X-B, Li S, Zhang R-F, Zhao J, Chen Y-C, Zhao Q, Yao Y-X, You C-X, Zhang X-S, Hao Y-J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012:35(11):1884–1897. 10.1111/j.1365-3040.2012.02523.x [DOI] [PubMed] [Google Scholar]
- Yan C, Fan M, Yang M, Zhao J, Zhang W, Su Y, Xiao L, Deng H, Xie D. Injury activates Ca2+/calmodulin-dependent phosphorylation of JAV1-JAZ8-WRKY51 complex for jasmonate biosynthesis. Mol Cell. 2018:70(1):136–149.e7. 10.1016/j.molcel.2018.03.013 [DOI] [PubMed] [Google Scholar]
- Yang T, Ma H, Li Y, Zhang Y, Zhang J, Wu T, Song T, Yao Y, Tian J. Apple MPK4 mediates phosphorylation of MYB1 to enhance light-induced anthocyanin accumulation. Plant J. 2021:106(6):1728–1745. 10.1111/tpj.15267 [DOI] [PubMed] [Google Scholar]
- Yang X, Song J, Campbell-Palmer L, Fillmore S, Zhang Z. Effect of ethylene and 1-MCP on expression of genes involved in ethylene biosynthesis and perception during ripening of apple fruit. Postharvest Biol Technol. 2013:78:55–66. 10.1016/j.postharvbio.2012.11.012 [DOI] [Google Scholar]
- Yuan H, Meng D, Gu Z, Li W, Wang A, Yang Q, Zhu Y, Li T. A novel gene, MdSSK1, as a component of the SCF complex rather than MdSBP1 can mediate the ubiquitination of S-RNase in apple. J Exp Bot. 2014:65(12):3121–3131. 10.1093/jxb/eru164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P, Lu Q, Liu Z, Lv T, Li X, Bu H, Liu W, Xu Y, Yuan H, Wang A. Auxin-activated MdARF5 induces the expression of ethylene biosynthetic genes to initiate apple fruit ripening. New Phytol. 2020:226(6):1781–1795. 10.1111/nph.16500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Li J, Li F, Liu H, Yang W, Chong K, Xu Y. OsMAPK3 phosphorylates OsbHLH002/OsICE1 and inhibits its ubiquitination to activate OsTPP1 and enhances rice chilling tolerance. Dev Cell. 2017:43(6):731–743.e5. 10.1016/j.devcel.2017.11.016 [DOI] [PubMed] [Google Scholar]
- Zhang AD, Wang WQ, Tong Y, Li MJ, Grierson D, Ferguson I, Chen KS, Yin XR. Transcriptome analysis identifies a zinc finger protein regulating starch degradation in kiwifruit. Plant Physiol. 2018:178(2):850–863. 10.1104/pp.18.00427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Chen G, Zhou S, Tu Y, Wang Y, Dong T, Hu Z. A new tomato NAC (NAM/ATAF1/2/CUC2) transcription factor, SlNAC4, functions as a positive regulator of fruit ripening and carotenoid accumulation. Plant Cell Physiol. 2014:55(1):119–135. 10.1093/pcp/pct162 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.