Abstract
The simian immunodeficiency virus (SIV) Mne envelope undergoes genetic changes that alter tropism, syncytium-inducing capacity, and antigenic properties of the emerging variant virus population during the course of an infection. Here we investigated whether the mutations in envelope of SIVMne also influence coreceptor usage. The data demonstrate that the infecting macrophage-tropic SIVMne clone as well as the envelope variants that are selected during the course of disease progression all recognize both CCR5 and Bob (GPR15) but not Bonzo (STRL33), CXCR4, or CCR3. Although it remains to be determined if there are other coreceptors specific for dualtropic or T-cell-tropic variants of SIVMne that emerge during late stages of infection, these data suggest that such SIV variants that evolve in pathogenic infections do not lose the ability to recognize CCR5 or Bob/GPR15.
Primary isolates of human immunodeficiency virus type 1 (HIV-1) obtained from peripheral blood at different stages of infection often display distinct in vitro biological characteristics. Viruses commonly found early in infection are typically slowly replicating, macrophage-tropic (M-tropic), and non-syncytium inducing (NSI), while variant viruses that emerge late in association with CD4+ T cell decline and progression to AIDS may be rapidly replicating and syncytium inducing (SI) and may have the ability to infect certain T-cell lines (10, 15, 16, 23, 29–31, 51, 62, 67, 68). Regulation of HIV-1 tropism and replication potential has primarily been ascribed to mutations that lead to changes in variable regions V1 through V4 of the envelope extracellular protein (Env SU) (3–5, 11, 20, 33, 42, 44). Indeed, the discovery that several members of the chemokine and orphan receptor family, in conjunction with CD4, directly interact with the Env SU protein of HIV-1 to mediate entry into target cells provided an explanation for the differences in tropism between M-tropic NSI variants and T-cell line-adapted SI variants. The M-tropic NSI viruses principally use CCR5 for infection, whereas the T-cell line-adapted SI viruses use CXCR4 (1, 13, 22–24, 28). Primary HIV-1 SI variants that emerge late during progressive infections typically have the capacity to use a greater number of coreceptors than the NSI viruses from which they evolved, presumably allowing them to escape the inhibitory effects of C-C chemokines and infect new target cells (18, 35, 61). Also, despite having an expanded coreceptor repertoire, many primary HIV-1 SI variants have been shown to maintain the ability to infect macrophages (15, 18, 64, 70). However, the ability of primary SI viruses to infect macrophages is controversial, and other reports find a lack of M-tropism for the majority of primary SI strains (17, 36, 62, 63). Since the identification of CCR5 and CXCR4 as HIV-1 coreceptors, the number of coreceptors for HIV-1, as well as HIV-2, has increased and now includes Apj, CCR2b, CCR3, CCR8, CCR9, Bonzo (STRL33), and Bob (GPR15) (2, 12–14, 21, 23, 26, 27, 45, 50). Together, these data suggest that the selection of viruses with specific Env SU mutations in HIV-1 in vivo may, in part, result from adaptation to additional coreceptors or new target cell populations or from selective pressure of chemokines that block entry.
The simian immunodeficiency viruses (SIVs), as a group, have been shown to utilize a set of coreceptors that overlaps those used by HIV-1. To date, coreceptors for SIV infection include Apj, CCR5, CCR8, Bonzo, Bob, and GPR1 but not CCR2b, CCR3, or CXCR4 (2, 7, 9, 12, 14, 21, 27, 48, 57, 59). Envelope proteins from related clones or distinct strains of SIV may differ in the ability to utilize specific coreceptors, such as Apj or CCR8, for fusion and entry (12, 57). However, whether a switch or expansion of coreceptor usage by primary SIV variants occurs during infections in a manner analogous to the changes reported for HIV-1 variants is unknown. In previous studies, we generated a panel of chimeric and full-length proviral clones encoding envelope gene sequences representative of viruses at various stages of infection and disease in macaques inoculated with an M-tropic NSI SIVMne clone (39, 40, 54, 55, 58–60). These sequential variants differ in their tropism, antigenic properties, and SI capacities compared to the infecting virus. Here, we investigated whether the SIVMne envelope variants from various stages of infection show differences in their recognition of previously identified SIV coreceptors.
To determine whether or not SIV evolves to utilize an expanded repertoire of coreceptors during the course of a pathogenic infection, we examined the ability of several previously characterized temporal variants to infect a panel of CD4+ indicator cell lines that express different known HIV and SIV coreceptors. The viruses we studied included (i) chimeric proviruses generated by the amplification and cloning of variant envelope sequences, present in peripheral blood mononuclear cells at different stages of infection, into the parental virus SIVMneCL8 (59); (ii) uncloned isolates from symptomatic infection (58); and (iii) two full-length molecular clones from late-stage disease, one from blood (SIVMne170) and one from lymph node tissue (SIVMne027) (39, 40). Each of these viruses displayed distinct genetic and phenotypic properties compared to the parent M-tropic NSI virus, SIVMneCL8, from which they evolved in vivo (summarized in Table 1 and Fig. 1). The Env SU sequences of the chimeras are representative of the virus population found at sequential stages of infection of pig-tailed macaques inoculated with SIVMneCL8 (54, 55, 58, 60), and each of the chimeric viruses is capable of using CCR5 for entry into CD4+ cells (59). Coreceptors that support infection by the cytopathic molecular clones SIVMne170 and SIVMne027 are unknown. Infectious virus stocks of each clone were produced by previously described methods (39, 40, 59). The titers of the virus stocks were determined by using the sMAGI assay developed by Chackerian et al.; they generally reflect titers obtained by limiting dilution analysis (e.g., 50% tissue culture infectious doses [TCID50]) with CEMx174 or macaque peripheral blood mononuclear cells used as target cells for infection (6).
TABLE 1.
Summary of viral phenotypesa
| Virus | Tropismb | Cytopathic effectc | SI capacityd |
|---|---|---|---|
| SIVMneCL8 | M, P, T | Minimal | − |
| 35wkSU | T, P | Minimal | − |
| 81wkSU | T, P | Minimal | +/− |
| 121wkSU | NDe | Minimal | +/− |
| 170wkPBSU | ND | Minimal | ND |
| 170wkCESU | T, P | Minimal | + |
| 170wkJKSU | M, P, T | Minimal | + |
| MixVar83 | M, P, T | Moderate | ND |
| MixVar170 | P, T | High | + |
| SIVMne170 | M, P, T | High | + |
| SIVMne027 | M, P, T | High | − |
M, macaque monocyte-derived macrophages; P, macaque peripheral blood mononuclear cells; T, human cell lines such as the T-cell lines MT4, C8166, and Molt4 clone 8 and the T-B hybrid cell line CEMx174.
Cytopathic effects were determined with macaque peripheral blood mononuclear cells or CEMx174 cells.
SI capacities were determined with CEMx174 cells. −, NSI; +/−, minimally SI; +, highly SI.
ND, not determined.
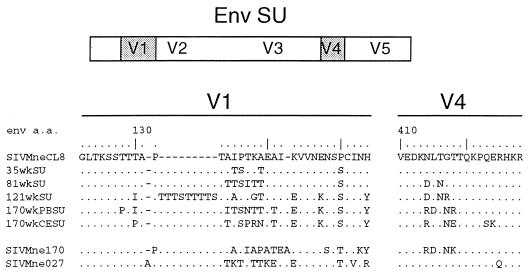
FIG. 1.
Predicted amino acid (a.a.) sequences of Env SU variable regions V1 and V4 from early and late molecular variants of SIVMne. Each of the variant Env SU sequences is shown with respect to the sequence of SIVMneCL8, the parent virus. Amino acids are shown in single-letter code. Amino acids found in the variant sequences that are identical to the reference sequence are represented by dots, specific differences are shown, and a dash indicates the absence of an amino acid. The envelope genes for 35wkWU, 81wkSU, 121wkSU, 170wkPBSU, and 170wkCESU were previously inserted into the background of the parent proviral clone, SIVMneCL8, to construct replication-competent chimeric viruses (59). The Env SU sequences for SIVMne170 and SIVMne027 are derived from infectious full-length variant molecular clones (39, 40).
The indicator cells (GHOST4 coreceptor HIV indicator cells) used as targets to analyze coreceptor usage were derived from HOS cells, a CD4− cell line that is not susceptible to SIV or HIV infection (37, 47, 69). The modified HOS cells were stably cotransfected with a vector expressing the human CD4 gene and a reporter plasmid for HIV or SIV infection, which contains the humanized green fluorescent protein gene (hGFP) (19) under the control of the HIV-2 long terminal repeat (LTR). Introduction of each coreceptor was achieved by retroviral transduction with the pBabe-puro vector containing one of a set of human chemokine or orphan receptor genes, including CCR3, CCR5, CXCR4, Bob, and Bonzo. The indicator cells were maintained in Dulbecco modified Eagle medium (DMEM) containing 10% fetal bovine serum, glutamine, penicillin, streptomycin, 500 μg of G418 per ml, 100 μg of hygromycin per ml, and 1 μg of puromycin per ml. For infections, approximately 1 × 104 infectious virions, as determined by the sMAGI assay (6), were applied to 5 × 104 cells of each GHOST4 coreceptor cell line in the presence of 10 μg of polybrene per ml. Two days postinfection, the cells were removed from the plates by using 1 mM EDTA in phosphate-buffered saline (PBS) and fixed with 2% paraformaldehyde in PBS solution. Following 1 h of fixation, the cells were recovered by centrifugation, rinsed free of paraformaldehyde with PBS, and resuspended in PBS containing 2% fetal bovine serum. Approximately 10,000 cells were analyzed for GFP expression with a FACScalibur analyzer (Becton Dickinson). Infected GHOST4 indicator cells were identified by a 20-fold shift in mean GFP fluorescence relative to the uninfected cell population. The indicator cells were scored positive for infection if greater than 1% of the cells demonstrated a 20-fold increase in mean GFP fluorescence.
The parental virus, SIVMneCL8, and each chimeric virus, viral isolate, and molecular variant were capable of infecting the GHOST4 indicator cell lines that expressed either Bob or CCR5 but not the parental GHOST4 cell line or GHOST4 cells expressing either Bonzo or CXCR4 (Table 2). GHOST4 cells expressing CCR3 also did not support infection by any of the viruses in this panel (data not shown). Importantly, there was no difference in coreceptor usage between clones and isolates capable of infecting macrophages (SIVMneCL8, 170wkJKSU, SIVMne170, SIVMne027, and MixVar83) and the T-cell-tropic viruses (35wkSU, 81wkSU, 170wkCESU, and MixVar170) or between viruses with an NSI or SI phenotype. To demonstrate functional coreceptor expression on each of the GHOST4 indicator cell lines, we also infected the panel of reporter cells with clones of HIV-1, HIV-2, and SIVmac. As previously described (21), HIV-2ROD was able to use Bob, Bonzo, CCR5, and CXCR4 for entry; HIV-1JRFL infected only cells expressing CCR5; SIVmac1A11 infected Bob- and CCR5-expressing cells; and SIVagmTYO infected Bob-, Bonzo-, and CCR5-expressing cells. None of these viruses infected the parental cell line, demonstrating that the expression of an appropriate coreceptor was essential for entry. Furthermore, HIV-1 particles pseudotyped with the amphotropic murine leukemia envelope protein were able to induce GFP expression from each GHOST4 coreceptor cell line, including the parental GHOST4 cells, demonstrating that each cell line retained the HIV-2 LTR-hGFP indicator construct. CD4 expression on each of the cell lines was verified by FACS analysis (data not shown). Together these data demonstrate that coreceptor recognition by SIVMne variants may be limited to Bob and CCR5 and that mutations in envelope that alter macrophage tropism, SI capacity, and antigenicity do not affect the range of coreceptors utilized, at least among those examined.
TABLE 2.
Infection of GHOST4 indicator cells with variants of SIVMnea
| Virus | Parent | Coreceptor expressed
|
|||
|---|---|---|---|---|---|
| Bob | Bonzo | CCR5 | CXCR4 | ||
| Mock | − | − | − | − | − |
| SIVMneCL8 | − | + | − | + | − |
| 35wkSU | − | + | − | + | − |
| 81wkSU | − | + | − | + | − |
| 121wkSU | − | + | − | + | − |
| 170wkSU | − | + | − | + | − |
| MixVar83 | − | + | − | + | − |
| MixVar170 | − | + | − | + | − |
| SIVMne170 | − | + | − | + | − |
| SIVMne027 | − | + | − | + | − |
| HIV-2ROD | − | + | + | + | + |
| HIV-1JRFL | − | − | − | + | − |
| SIVmac1A11 | − | + | − | + | − |
| SIVagmTYO | − | + | + | + | − |
| HIV/A-MLV | + | + | + | + | + |
GHOST4 coreceptor cells expressing the indicated receptor were scored positive (+) if greater than 1% of the cells expressed GFP at a high level relative to the uninfected cells. A negative sign (−) indicates that there was no induction of GFP expression.
To verify that the SIVMne variant viruses could use either CCR5 or Bob as a coreceptor for entry into CD4+ cells, we determined whether MAGI indicator cells could be infected if they expressed either human CCR5 or Bob. Previous studies demonstrated that MAGI indicator cells, which express endogenous CXCR4, were able to support SIV or M-tropic HIV-1 infection if a CCR5 expression vector was introduced into the cells (7, 71). To produce a MAGI indicator cell line that expressed Bob, we transduced the MAGI cell line with a pBabe-puro-Bob expression vector. Twenty-four hours postinfection, cells carrying the pBabe-puro-Bob vector were selected in DMEM supplemented with 10% calf serum, glutamine, penicillin/streptomycin, 200 μg of G418 per ml, and 50 U of hygromycin per ml (MAGI medium) plus 1 μg of puromycin per ml. The parental MAGI (41), MAGI-CCR5 (7), or MAGI-Bob cell lines were infected with each SIV variant and monitored for infection, as described by Kimpton and Emerman (41). Titers of each stock were compared with those determined by the sMAGI assay (Table 3). Because each virus stock contained different amounts of infectious virus when titered with the sMAGI assay (TCIDsMAGI), we determined the total amount of virions in each stock by measuring viral RNA (vRNA) with an established quantitative competitive reverse transcription (RT)-PCR technique (72). The vRNA/TCIDsMAGI ratios were similar for each virus stock, suggesting that the differences in titers generally corresponded to the total amount of virions but not to large differences in infectivity for the sMAGI cells. Exceptions were the 121wkSU virus, which appeared to be less infectious and has been shown to replicate poorly (59), and variant SIVMne170, which may be slightly more infectious and which has been shown to replicate efficiently (40).
TABLE 3.
Titers of virus stocks on different indicator cell lines
| Virus | Infectious virions/mla
|
vRNA and infectivityb
|
||||
|---|---|---|---|---|---|---|
| sMAGI | MAGI | MAGI-CCR5 | MAGI-Bob | RNA/ml | RNA/TCIDsMAGI | |
| SIVMneCL8 | 2.0 × 105 | 0 | 2.9 × 105 | 3.3 × 104 | 3.8 × 109 | 1.9 × 104 |
| 35wkSU | 2.1 × 105 | 0 | 4.9 × 104 | 2.3 × 104 | 1.0 × 1010 | 4.9 × 104 |
| 82wkSU | 2.0 × 105 | 0 | 2.0 × 105 | 7.3 × 104 | 3.4 × 109 | 1.7 × 104 |
| 121wkSU | 6.4 × 104 | 0 | 3.7 × 104 | 2.1 × 104 | 1.0 × 1010 | 1.6 × 105 |
| 170wkPBSU | 1.4 × 103 | 0 | 1.7 × 103 | 1.3 × 103 | 1.0 × 107 | 7.1 × 103 |
| 170wkCESU | 2.5 × 105 | 0 | 2.4 × 105 | 1.1 × 105 | 2.6 × 109 | 1.0 × 104 |
| 170wkJKSU | 2.2 × 105 | 0 | 2.3 × 105 | 1.6 × 105 | 1.0 × 1010 | 4.5 × 104 |
| MixVar170 | 3.2 × 105 | 0 | 5.0 × 105 | 1.4 × 105 | 5.7 × 109 | 1.8 × 104 |
| SIVMne170 | 1.3 × 105 | 0 | 3.3 × 105 | 1.9 × 104 | 6.4 × 108 | 5.0 × 103 |
| SIVMne027 | 3.2 × 105 | 0 | 1.1 × 106 | 4.6 × 105 | 5.3 × 109 | 1.7 × 104 |
Data are averages of infections performed in duplicate. MAGI cells were infected with 102 to 104 infectious virions, as determined by sMAGI assay. A value of 0 indicates that no cells were infected when cultured with a minimum of 104 infectious virions.
RNA, virions associated with vRNA; TCIDsMAGI, titers of virus stocks with sMAGI indicator cells (infectious virions/ml).
As we found with the GHOST4-CCR5 and GHOST4-Bob cell lines, each of the SIVMne molecular variants and the late variant isolate (MixVar170) infected MAGI-CCR5 and MAGI-Bob cells but not the parental MAGI cell line, which expresses endogenous CXCR4 (Table 3). Furthermore, the titers of each virus stock with the MAGI-CCR5 cells were similar to those determined with the sMAGI indicator cells, suggesting that they were able to infect indicator cells expressing CCR5 as efficiently as they infected sMAGI cells. These data with the chimeric viruses and MAGI-CCR5 cells are consistent with our previous observations (7, 59). Additionally, the results extend to the late variant full-length proviral clones, which are both dualtropic and cytopathic variants of SIVMne. Interestingly, with different passages of the MAGI-CCR5 cell line, the titer of certain chimeric virus stocks was variably decreased by as much as 10- to 20-fold (data not shown). By contrast, both late variants SIVMne170 and SIVMne027 remained highly infectious for all cell passages. One explanation for these preliminary findings is that the later viruses are better adapted to use of low levels of coreceptor than the earlier variants, but this model will require more extensive analysis. All of the viruses infected MAGI-Bob cells. However, some of the viruses tended to be less infectious for the MAGI-Bob cell line than for the sMAGI cells. For example, the titers of the parent virus SIVMneCL8, early variant 35wkSU chimera, and late variant clone SIVMne170 were 6- to 10-fold lower on MAGI-Bob cells than on sMAGI cells. In contrast, the late variant viruses 170wkPBSU, 170wkCESU, 170wkJKSU, MixVar170, and SIVMne027 had similar titers (within twofold) with both MAGI-Bob and sMAGI cells. Thus, while there may be subtle differences in the ability of the viruses to infect MAGI cells expressing CCR5 or Bob, these data confirm that CCR5 and Bob, but not CXCR4, are coreceptors for entry by SIVMne.
Because CCR5 is believed to be a major coreceptor for SIV and because all of the SIVMne variants were cloned or isolated from pig-tailed macaques (M. nemestrina), we further examined the ability of SIVMne variants to utilize pig-tailed macaque CCR5 (ptCCR5) for entry into MAGI cells. The gene for ptCCR5 was cloned by PCR with primers that amplified the coding region in the second exon. ptCCR5 is 97.4% identical to human CCR5 and 99.7% identical to the rhesus CCR5 protein (32). To express ptCCR5 in MAGI cells, the gene was inserted into the pBabe-puro vector and introduced into MAGI cells by retroviral transduction. As before, cells containing pBabe-puro-ptCCR5 were selected in MAGI medium plus 1 μg of puromycin per ml. All SIVMne variants examined were infectious for MAGI cells expressing ptCCR5 at a level similar to that found with MAGI cells expressing human CCR5, demonstrating that ptCCR5 functions as a coreceptor for SIVMne variant viruses (Table 4).
TABLE 4.
Infection of MAGI cells expressing ptCCR5
| Virus | Infectious virions/ml
|
|
|---|---|---|
| MAGI-CCR5 | MAGI-ptCCR5 | |
| SIVMneCL8 | 3.8 × 104 | 5.1 × 104 |
| 35wkSU | 6.8 × 104 | 1.2 × 105 |
| 81wkSU | NDa | 3.4 × 105 |
| 121wkSU | 6.8 × 103 | 8.1 × 103 |
| 170wkCESU | ND | 1.3 × 105 |
| MixVar170 | 3.0 × 105 | 5.4 × 105 |
| SIVMne170 | ND | 2.8 × 104 |
| SIVMne027 | 3.3 × 105 | 4.3 × 105 |
Not determined.
Although several studies have identified SIV coreceptors, none have addressed the coreceptor specificity of SIV variants that evolve over the course of disease progression. We have examined for the first time the ability of temporal variants of SIV that predominate during different stages of infection to utilize different coreceptors for entry into CD4+ cells. The data demonstrate that at least two coreceptors, CCR5 and Bob, can be used for entry by all variants of SIVMne. Furthermore, at least one additional coreceptor appears to be recognized by both early- and late-stage SIVMne variants because sMAGI cells, which are highly susceptible to infection by all of the SIVMne variants examined, do not express either Bob or CCR5, at least at levels detectable by Northern blot analysis (32). Importantly, our data suggest that a shift in the repertoire of coreceptors recognized by variants of SIVMne may not occur with progression to simian AIDS and that there is a strong selective pressure for recognizing CCR5 and Bob. Whether this repertoire of coreceptors is maintained because each receptor is essential for SIV replication in vivo or because of functional constraints in the envelope protein structure is unknown but will be important to determine.
The Env SU chimeric and full-length variants of SIVMne were genetically and phenotypically distinguishable from the parent virus, SIVMneCL8, from which they evolved. The mutations in V1 of the chimeric viruses have previously been shown to be sufficient for reducing replication in macaque monocyte-derived macrophages and for altering antigenicity, while mutations located in V3 and V4 likely contribute to the SI phenotype (8, 59). Interestingly, none of these mutations appears to alter utilization of CCR5 or Bob for entry into CD4+ cells. Both CCR5 and Bob are expressed in monocytes/macrophages, and CCR5 has been directly shown to be important for monocyte/macrophage infection by HIV-1 (14, 21, 27, 56). Thus, the inability of some variants of SIVMne to replicate in macaque monocyte-derived macrophages may be determined by postentry blocks in reverse transcription or poor viral protein processing similar to that reported for SIVmac239 (52, 66), suggesting the possibility that coreceptor recognition may not be the sole determinant of macrophage tropism. Alternatively, another coreceptor may be functionally important for productive infection of monocytes/macrophages by M-tropic SIV variants. Of note, Edinger et al. demonstrated that an M-tropic SIVmac Env SU protein interacts with different domains of CCR5 than does the T-tropic SIVmac239 Env SU (25). How differences in envelope-CCR5 interaction may influence postentry events or viral protein processing in macrophages is unclear, and the specific domains within Env SU that are involved in these events remain to be defined.
The significance of the expansion in HIV-1 coreceptor usage during infections is unclear but may serve as a mechanism for broadening the target cell population during persistent infections. Because the emergence of HIV-1 variants with increased in vitro virulence are associated with a change in coreceptor usage, resistance to inhibition by C-C chemokines, and disease progression, it has been hypothesized that changes in coreceptor usage may influence pathogenicity (18, 35, 44, 61). However, even though there is an expansion of the coreceptor repertoire, to date only CCR5 has been shown to be consistently used by most primary HIV-1 isolates from early and late stages of infection regardless of their NSI or SI phenotype (18, 61, 64). Furthermore, epidemiological data provide strong evidence for the importance of CCR5, and perhaps CCR2, in disease progression (34, 43, 46, 49, 53, 65). Thus, it may be that few of the identified coreceptors for HIV-1 infection are significant for viral replication and disease progression. SIV infection of macaques provides a relevant model system to investigate the influence of coreceptor usage on viral pathogenicity. Our preliminary data suggests that changes in coreceptor specificity may not be important for differences in pathogenicity of variant SIVs. Two of the late variant molecular clones of SIVMne tested here, SIVMne170 and SIVMne027, replicate more efficiently in vivo and appear to be more pathogenic than the early parent virus, SIVMneCL8, despite the conservation of their coreceptor specificities (38). One model that could account for the differences is that envelope variants that evolve over time may have a higher affinity for specific coreceptors. This hypothesis would allow for a mechanism of expanding the target cell population and increasing the efficiency of viral replication while maintaining a constant repertoire of coreceptors that the early and late viruses can recognize.
Acknowledgments
We thank Nancy Haigwood’s laboratory for performing the quantitative competitive RT-PCR assays.
This work was supported by NIH grant AI34251 to J.O. J.T.K. was supported by NRSA individual postdoctoral fellowship F32 AI09337. J.J.G. was supported by NIH training grant T32 CA09229. V.N.K. is the recipient of a fellowship from the Damon Runyon-Walter Winchell Foundation. D.R.L. is an investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, Mip-1α, Mip-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Alkhatib G, Liao F, Berger E A, Farber J M, Peden K W C. A new SIV co-receptor STRL33. Nature. 1997;388:238. doi: 10.1038/40789. [DOI] [PubMed] [Google Scholar]
- 3.Boyd M T, Simpson G R, Cann A J, Johnson M A, Weiss R A. A single amino acid substitution in the V1 loop of human immunodeficiency virus type 1 gp120 alters cellular tropism. J Virol. 1993;67:3649–3652. doi: 10.1128/jvi.67.6.3649-3652.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carrillo A, Ratner L. Cooperative effects of the human immunodeficiency virus type 1 envelope variable loops V1 and V3 in mediating infectivity for T cells. J Virol. 1996;70:1310–1316. doi: 10.1128/jvi.70.2.1310-1316.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carrillo A, Ratner L. Human immunodeficiency virus type 1 tropism for T-lymphoid cell lines: role of the V3 loop and C4 envelope determinants. J Virol. 1996;70:1301–1309. doi: 10.1128/jvi.70.2.1301-1309.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chackerian B, Haigwood N L, Overbaugh J. Characterization of a CD4-expressing macaque cell line that can detect virus after a single replication cycle and can be infected by diverse simian immunodeficiency virus isolates. Virology. 1995;213:386–394. doi: 10.1006/viro.1995.0011. [DOI] [PubMed] [Google Scholar]
- 7.Chackerian B, Long E M, Luciw P A, Overbaugh J. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J Virol. 1997;71:3932–3939. doi: 10.1128/jvi.71.5.3932-3939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chackerian B, Rudensey L M, Overbaugh J. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J Virol. 1997;71:7719–7727. doi: 10.1128/jvi.71.10.7719-7727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Z, Ho D D, Landau N R, Marx P A. Genetically divergent strains of simian immunodeficiency virus use CCR5 as a coreceptor for entry. J Virol. 1997;71:2705–2714. doi: 10.1128/jvi.71.4.2705-2714.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng-Mayer C, Seto D, Tateno M, Levy J A. Biologic features of HIV-1 that correlate with virulence in the host. Science. 1988;240:80–82. doi: 10.1126/science.2832945. [DOI] [PubMed] [Google Scholar]
- 11.Cheng-Mayer C, Shioda T, Levy J A. Host range, replicative, and cytopathic properties of human immunodeficiency virus type 1 are determined by very few amino acid changes in tat and gp120. J Virol. 1991;65:6931–6941. doi: 10.1128/jvi.65.12.6931-6941.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choe H, Farzan M, Konkel M, Martin K, Sun Y, Marcon L, Cayabyab M, Berman M, Dorf M E, Gerard N, Gerard C, Sodroski J. The orphan seven-transmembrane receptor Apj supports the entry of primary T-cell-line-tropic and dualtropic human immunodeficiency virus type 1. J Virol. 1998;72:6113–6118. doi: 10.1128/jvi.72.7.6113-6118.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The b-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 14.Clapham P R, Weiss R A. Spoilt for choice of co-receptors. Nature. 1997;388:230–231. doi: 10.1038/40758. [DOI] [PubMed] [Google Scholar]
- 15.Connor R I, Ho D D. Human immunodeficiency virus type 1 variants with increased replicative capacity develop during the asymptomatic stage before disease progression. J Virol. 1994;68:4400–4408. doi: 10.1128/jvi.68.7.4400-4408.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Connor R I, Mohri H, Cao Y, Ho D D. Increased viral burden and cytopathicity correlate temporally with CD4+ T-lymphocyte decline and clinical progression in human immunodeficiency virus type 1-infected individuals. J Virol. 1993;67:1772–1777. doi: 10.1128/jvi.67.4.1772-1777.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Connor R I, Paxton W A, Sheridan K E, Koup R A. Macrophages and CD4+ T lymphocytes from two multiply exposed, uninfected individuals resist infection with primary non-syncytium-inducing isolates of human immunodeficiency virus type 1. J Virol. 1996;70:8758–8764. doi: 10.1128/jvi.70.12.8758-8764.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 20.de Jong J-J, Goudsmit J, Keulen W, Klaver B, Krone W, Tersmette M, de Ronde A. Human immunodeficiency virus type 1 clones chimeric for the envelope V3 domain differ in syncytium formation and replication oapacity. J Virol. 1992;66:757–765. doi: 10.1128/jvi.66.2.757-765.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng H-K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 22.Deng H K, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 23.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the b-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 24.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 25.Edinger A L, Amedee A, Miller K, Doranz B J, Endres M, Sharron M, Samson M, Lu Z-H, Clements J E, Murphey-Corb M, Peiper S C, Parmentier M, Broder C C, Doms R W. Differential utilization of CCR5 by macrophage and T cell tropic simian immunodeficiency virus strains. Proc Natl Acad Sci USA. 1997;94:4005–4010. doi: 10.1073/pnas.94.8.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Endres M J, Clapham P R, Marsh M, Ahuja M, Davis Turner J, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 27.Farzan M, Choe H, Martin K, Marcon L, Hofmann W, Karlsson G, Sun Y, Barrett P, Marchand N, Sullivan N, Gerard N, Gerard C, Sodroski J. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J Exp Med. 1997;186:405–411. doi: 10.1084/jem.186.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane domain, G-protein coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 29.Fenyö, E. M., J. Albert, and B. Åsjö. 1989. Replicative capacity, cytopathic effects and cell tropism of HIV. AIDS 3(Suppl.):S5–S12. [DOI] [PubMed]
- 30.Fenyö E M, Morfeldt-Manson L, Chiodi F, Lind B, von Gegerfelt A, Albert J, Olausson E, Åsjö B. Distinct replicative and cytopathic characteristics of human immunodeficiency virus isolates. J Virol. 1988;62:4414–4419. doi: 10.1128/jvi.62.11.4414-4419.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiore J R, Bjorndal A, Peipke K A, Di Stefano M, Angarano G, Pastore G, Gaines H, Fenyö E M, Albert J. The biological phenotype of HIV-1 is usually retained during and after sexual transmission. Virology. 1994;204:297–303. doi: 10.1006/viro.1994.1534. [DOI] [PubMed] [Google Scholar]
- 32.Gosink, J., and J. Overbaugh. Unpublished observations.
- 33.Groenink M, Fouchier R A, Broersen S, Baker C H, Koot M, van’t Wout A B, Huisman H G, Miedema F, Tersmette M, Schuitemaker H. Relation of phenotype evolution of HIV-1 to envelope V2 configuration. Science. 1993;260:1513–1516. doi: 10.1126/science.8502996. [DOI] [PubMed] [Google Scholar]
- 34.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Cerandini D, Jin Z, Yazdanbakhsh K, Kunstman K, Erickson D, Dragon E, Landau N, Phair J, Ho D D, Koup R A. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 35.Jansson M, Popovic M, Karlsson A, Cocchi F, Rossi P, Albert J, Wigzell H. Sensitivity to inhibition by b-chemokines correlates with biological phenotypes of primary HIV-1 isolates. Proc Natl Acad Sci USA. 1996;93:15382–15387. doi: 10.1073/pnas.93.26.15382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karita E, Nkengasong J N, Willems B, Vanham G, Fransen K, Heyndrickx L, Janssens W, Piot P, van der Groen G. Macrophage-tropism of HIV-1 isolates of different genetic subtypes. AIDS. 1997;11:1303–1304. doi: 10.1097/00002030-199710001-00010. [DOI] [PubMed] [Google Scholar]
- 37.KewalRamani, V. N., B. Volsky, D. S. Kwon, W.-K. Xiang, J. Gao, D. Unutmaz, C. M. Hill, R. E. Sutton, and D. R. Littman. 1998. Unpublished data.
- 38.Kimata, J. T., L. Kuller, D. Anderson, P. Dailey, and J. Overbaugh. Unpublished data. [DOI] [PubMed]
- 39.Kimata J T, Mozaffarian A, Overbaugh J. A lymph node-derived cytopathic simian immunodeficiency virus Mne variant replicates in nonstimulated peripheral blood mononuclear cells. J Virol. 1998;72:245–256. doi: 10.1128/jvi.72.1.245-256.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kimata J T, Overbaugh J. The cytopathicity of a simian immunodeficiency virus Mne variant is determined by mutations in Gag and Env. J Virol. 1997;71:7629–7639. doi: 10.1128/jvi.71.10.7629-7639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated β-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koito A, Harrowe G, Levy J A, Cheng-Mayer C. Functional role of the V1/V2 region of human immunodeficiency virus type 1 envelope glycoprotein gp120 in infection of primary macrophages and soluble CD4 neutralization. J Virol. 1994;68:2253–2259. doi: 10.1128/jvi.68.4.2253-2259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kostrikis L G, Huang Y, Moore J P, Wolinsky S M, Zhang L, Guo Y, Deutsch L, Phair J, Neumann A U, Ho D D. A chemokine receptor CCR2 allele delays HIV-1 disease progression and is associated with a CCR5 promoter mutation. Nat Med. 1998;4:350–353. doi: 10.1038/nm0398-350. [DOI] [PubMed] [Google Scholar]
- 44.Levy J A. Pathogenesis of human immunodeficiency virus infection. Microbiol Rev. 1993;57:183–289. doi: 10.1128/mr.57.1.183-289.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liao F, Alkhatib G, Peden W, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-topic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 47.Mack M, Luckow B, Nelson P J, Cihak J, Simmons G, Clapham P R, Signoret N, Marsh M, Stangassinger M, Borlat F, Wells T N C, Schlöndorff D, Proudfoot A E I. Aminooxypentane-RANTES induces CCR5 internalization but inhibits recycling: a novel inhibitory mechanism of HIV infectivity. J Exp Med. 1998;187:1215–1224. doi: 10.1084/jem.187.8.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marcon L, Choe H, Martin K A, Farzan M, Ponath P D, Newman W L W, Gerard N, Gerard C, Sodroski J. Utilization of C-C chemokine receptor 5 by the envelope glycoproteins of a pathogenic simian immunodeficiency virus, SIVmac239. J Virol. 1997;71:2522–2527. doi: 10.1128/jvi.71.3.2522-2527.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McDermott D H, Zimmerman P A, Guignard F, Kleeberger C A, Leitman S F, Murphy P M. CCR5 promoter polymorphism and HIV-1 disease progression. Multicenter AIDS cohort study (MACS) Lancet. 1998;352:866–870. doi: 10.1016/s0140-6736(98)04158-0. [DOI] [PubMed] [Google Scholar]
- 50.McKnight A, Dittmar M T, Moniz-Periera J, Ariyoshi K, Reeves J D, Hibbits S, Whitby D, Aarons E, Proudfoot A E I, Whittle H, Clapham P R. A broad range of chemokine receptors are used by primary isolates of human immunodeficiency virus type 2 as coreceptors with CD4. J Virol. 1998;72:4065–4071. doi: 10.1128/jvi.72.5.4065-4071.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miedema F, Meyaard L, Koot M, Klein M R, Roos M T L, Groenink M, Fouchier R A M, Van’t Wout A B, Tersmette M, Schellekens P T A, Schuitemaker H. Changing virus-host interactions in the course of HIV-1 infection. Immunol Rev. 1994;140:35–72. doi: 10.1111/j.1600-065x.1994.tb00864.x. [DOI] [PubMed] [Google Scholar]
- 52.Mori K, Ringler D J, Desrosiers R C. Restricted replication of simian immunodeficiency virus strain 239 in macrophages is determined by env but is not due to restricted entry. J Virol. 1993;67:2807–2814. doi: 10.1128/jvi.67.5.2807-2814.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mummidi S, Ahuja S S, Gonzalez E, Anderson S A, Santiago E N, Stephan K T, Craig F E, O’Connell P, Tryon V, Clark R A, Dolan M J, Ahuja S K. Genealogy of the CCR5 locus and chemokine system gene variants associated with altered rates of HIV-1 disease progression. Nat Med. 1998;4:786–793. doi: 10.1038/nm0798-786. [DOI] [PubMed] [Google Scholar]
- 54.Overbaugh J, Rudensey L M. Alterations in potential sites for glycosylation predominate during evolution of the simian immunodeficiency virus envelope gene in macaques. J Virol. 1992;66:5937–5948. doi: 10.1128/jvi.66.10.5937-5948.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Overbaugh J, Rudensey L M, Papenhausen M D, Benveniste R E, Morton W R. Variation in simian immunodeficiency virus env is confined to V1 and V4 during progression to simian AIDS. J Virol. 1991;65:7025–7031. doi: 10.1128/jvi.65.12.7025-7031.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rana S, Besson G, Cook D G, Rucker J, Smyth R J, Yi Y, Turner J D, Guo H-H, Du J-G, Peiper S C, Lavi E, Samson M, Libert F, Liesnard C, Vassart G, Doms R W, Parmentier M, Collman R G. Role of CCR5 in infection of primary macrophages and lymphocytes by macrophage-tropic strains of human immunodeficiency virus: resistance to patient-derived and prototype isolates resulting from Δccr5 mutation. J Virol. 1997;71:3219–3227. doi: 10.1128/jvi.71.4.3219-3227.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rucker J, Edinger A L, Sharron M, Samson M, Lee B, Berson J F, Yi Y, Margulies B, Collman R G, Doranz B J, Parmentier M, Doms R W. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J Virol. 1997;71:8999–9007. doi: 10.1128/jvi.71.12.8999-9007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rudensey L M, Kimata J T, Benveniste R E, Overbaugh J. Progression to AIDS in macaques is associated with changes in the replication, tropism, and cytopathic properties of the simian immunodeficiency virus variant population. Virology. 1995;207:528–542. doi: 10.1006/viro.1995.1113. [DOI] [PubMed] [Google Scholar]
- 59.Rudensey L M, Kimata J T, Long E M, Chackerian B, Overbaugh J. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of simian immunodeficiency virus SIVMne infection affect neutralizing antibody recognition, syncytium formation, and macrophage tropism but not replication, cytopathicity, or CCR-5 coreceptor recognition. J Virol. 1998;72:209–217. doi: 10.1128/jvi.72.1.209-217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rudensey L M, Papenhausen M D, Overbaugh J. Replication and persistence of simian immunodeficiency virus variants after passage in macaque lymphocytes and established human cell lines. J Virol. 1993;67:1727–1733. doi: 10.1128/jvi.67.3.1727-1733.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Scarlatti G, Tresoldi E, Björndal Å, Fredricksson R, Colognesi C, Deng H K, Malnati M S, Plebani A, Siccardi A G, Littman D R, Fenyo E M, Lusso P. In vivo evolution of HIV-1 co-receptor usage and sensitivity to chemokine-mediated suppression. Nat Med. 1997;3:1259–1265. doi: 10.1038/nm1197-1259. [DOI] [PubMed] [Google Scholar]
- 62.Schuitemaker H, Koot M, Kootstra N A, Dercksen M W, de Goede R E Y, van Steenwijk R P, Lange J M A, Schattenkerk J K M E, Miedema F, Tersmette M. Biological phenotype of human immunodeficiency virus type 1 clones at different stages of infection: progression of disease is associated with a shift from monocytotropic to T-cell-tropic virus populations. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schuitemaker H, Kootstra N A, de Goede R E, de Wolf F, Miedema F, Tersmette M. Monocytotropic human immunodeficiency virus type 1 (HIV-1) variants detectable in all stages of HIV-1 infection lack T-cell line tropism and syncytium-inducing ability in primary T-cell culture. J Virol. 1991;65:356–363. doi: 10.1128/jvi.65.1.356-363.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simmons G, Wilkinson D, Reeves J D, Dittmar M T, Beddows S, Weber J, Carnegie G, Desselberger U, Gray P W, Weiss R A, Clapham P R. Primary, syncytium-inducing human immunodeficiency virus type 1 isolates are dual-tropic and most can use either Lestr or CCR5 as coreceptors for virus entry. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smith M W, Dean M, Carrington M, Winkler C, Huttley G A, Lomb D A, Goedert J J, O’Brien T R, Jacobson L P, Kaslow R, Buchbinder S, Vittinghoff E, Vlahov D, Hoots K, Hilgartner M W, O’Brien S J. Contrasting genetic influence of CCR2 and CCR5 variants on HIV-1 infection and disease progression. Hemophilia Growth and Development Study (HGDS), Multicenter AIDS Cohort Study (MACS), Multicenter Hemophilia Cohort Study (MHCS), San Francisco City Cohort Study (SFCC), ALIVE Study. Science. 1997;277:959–965. doi: 10.1126/science.277.5328.959. [DOI] [PubMed] [Google Scholar]
- 66.Stephens E B, McClure H M, Narayan O. The proteins of lymphocyte- and macrophage-tropic strains of simian immunodeficiency virus are processed differently in macrophages. Virology. 1995;206:535–544. doi: 10.1016/s0042-6822(95)80070-0. [DOI] [PubMed] [Google Scholar]
- 67.Tersmette M, Gruters R A, De Wolf F, De Goede R E Y, Lange J M A, Schellekens P T A, Goudsmit J, Huisman H G, Miedema F. Evidence for a role of virulent human immunodeficiency virus (HIV) variants in the pathogenesis of acquired immunodeficiency syndrome: studies on sequential HIV isolates. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tersmette M, Lange J M A, DeGoede R E Y, DeWolf F, Eeftink-Schattenkerk J K M, Schellekens P T A, Coutinho R A, Huisman J G, Goudsmit J, Miedema F. Association between biological properties of human immunodeficiency virus variants and risk for AIDS and AIDS mortality. Lancet. 1989;i:983–985. doi: 10.1016/s0140-6736(89)92628-7. [DOI] [PubMed] [Google Scholar]
- 69.Trkola A, Ketas T, Kewalramani V N, Endorf F, Binley J M, Katinger H, Robinson J, Littman D R, Moore J P. Neutralization sensitivity of human immunodeficiency virus type 1 primary isolates to antibodies and CD4-based reagents is independent of coreceptor usage. J Virol. 1998;72:1876–1885. doi: 10.1128/jvi.72.3.1876-1885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Valentin A, Albert J, Fenyo E M, Asjo B. Dual tropism for macrophages and lymphocytes is a common feature of primary human immunodeficiency virus type 1 and 2 isolates. J Virol. 1994;68:6684–6689. doi: 10.1128/jvi.68.10.6684-6689.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vodicka M A, Goh W C, Wu L I, Rogel M E, Bartz S R, Schweickart V L, Raport C J, Emerman M. Indicator cell lines for detection of primary strains of human and simian immunodeficiency viruses. Virology. 1997;233:193–198. doi: 10.1006/viro.1997.8606. [DOI] [PubMed] [Google Scholar]
- 72.Watson A, Ranchalis J, Travis B, McClure J, Sutton W, Johnson P R, Hu S L, Haigwood N L. Plasma viremia in macaques infected with simian immunodeficiency virus: plasma viral load early in infection predicts survival. J Virol. 1997;71:284–290. doi: 10.1128/jvi.71.1.284-290.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]