Abstract
Triple-negative breast cancer (TNBC) is highly aggressive with a poor prognosis because of a lack of cell markers as drug targets. α9-Nicotinic acetylcholine receptor (nAChR) is expressed abundantly in TNBC; thus, it is a valuable biomarker for TNBC detection and treatment. In this study, we utilized thermodynamically stable three-way junction (3WJ) packaging RNA (pRNA) as the core to construct RNA nanoparticles with an α9-nAChR RNA aptamer as a targeting ligand and an anti-microRNA-21 (miR-21) as a therapeutic module. We compared the configuration of the two RNA nanoparticles and found that 3WJ-B-α9-nAChR-aptamer fluorescent RNA nanoparticles (3WJ-B-α9-apt-Alexa) exhibited better specificity for α9-nAChR in TNBC cells compared with 3WJ-C-α9-nAChR. Furthermore, 3WJ-B-α9-apt-Alexa bound more efficiently to TNBC patient-derived xenograft (PDX) tumors than 3WJ fluorescent RNA nanoparticles (3WJ-Alexa) with little or no accumulation in healthy organs after systemic injection in mice. Moreover, 3WJ-B-α9-nAChR-aptamer RNA nanoparticles carrying anti-miR-21 (3WJ-B-α9-apt-anti-miR-21) significantly suppressed TNBC-PDX tumor growth and induced cell apoptosis because of reduced miR-21 gene expression and upregulated the phosphatase and tensin homolog (PTEN) and programmed cell death 4 (PDCD4) proteins. In addition, no pathological changes were detected upon toxicity examination of treated mice. In conclusion, the 3WJ-B-α9-nAChR-aptamer RNA nanoparticles established in this study efficiently deliver therapeutic anti-miR-21, indicating their potential as a novel TNBC therapy.
Keywords: MT: Oligonucleotides: Therapies and Applications; α9-nicotinic acetylcholine receptor; RNA aptamer; TNBC-PDX, triple-negative breast cancer, patient-derived xenograft; microRNA-21; fluorescence resonance energy transfer, FRET, microscopy
Graphical abstract
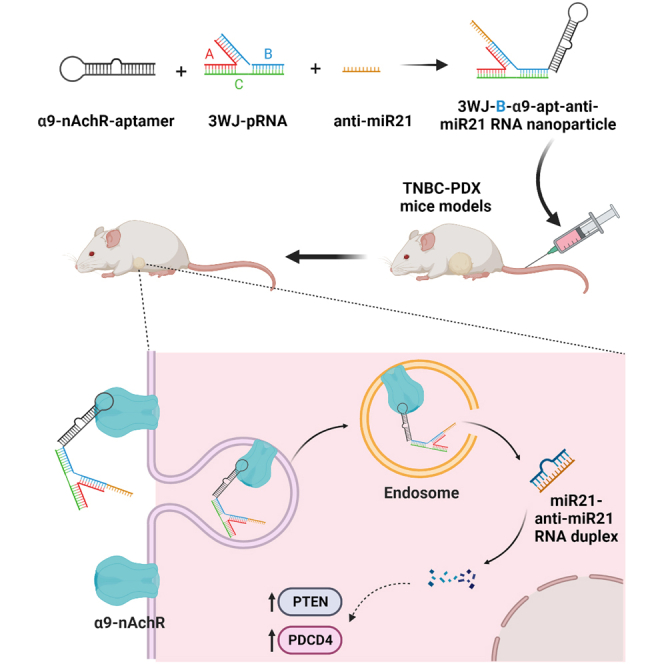
Dr. Ho and his colleagues developed novel 3WJ-B-α9-nAChR aptamer RNA nanoparticles with specificity for the α9-nicotinic acetylcholine receptor (α9-nAChR), a membrane protein abundantly expressed in TNBC. This nanoparticle effectively binds TNBC patient-derived xenograft (PDX) tumors and delivers therapeutic anti-microRNA-21 (miR-21) to suppress tumor growth without toxicity, indicating its potential for clinical application.
Introduction
Breast cancer (BC) is the most common cancer diagnosis in women and the leading cause of cancer death worldwide.1 Among the four BC subtypes determined by the presence of hormone receptor (HR) and epidermal growth factor receptor 2 (HER2), triple-negative BC (TNBC) accounts for 10%–15% of the total number of BC patients and has the worst prognosis.2,3 Therefore, there is an urgent clinical need for a valuable biomarker combined with more effective targeted therapy to improve the survival rate of TNBC patients. Accumulating evidence links high BC risk to the smoking status of active and passive smoking in females, even if there is a long literature controversy.4,5,6,7 However, nicotine, the primary addictive substance in tobacco, has been shown to promote tumorigenesis, metastasis, and drug resistance by interacting with nicotinic acetylcholine receptors (nAChRs) to transduce various signaling cascades.8,9 Our previous study showed that the mRNA of α9-nAChR was overexpressed in BC tissue, especially in patients with smoking-related and advanced BC, suggesting that α9-nAChR plays a crucial role in breast tumorigenesis.10,11 An in vivo study also showed that the high expression of α9-nAChR in TNBC tumors is closely related to the propensity for distant metastasis, implying that α9-nAChR is critical for mediating TNBC metastasis.12 Furthermore, specific α9-nAChR antagonists show therapeutic potential to inhibit BC growth and metastasis by inhibiting α9-nAChR activity.11 These experimental results endow α9-nAChR with an essential role as a biomarker and potential therapeutic target for BC therapy.
The biological function and regulation of ribonucleic acid (RNA), including coding RNA (mRNA) and non-coding RNA (ncRNA), play critical roles in living organisms.13,14 RNAs are characterized by sequence simplicity, structural diversity, and functional diversity, which are critical for constituting multiple RNA moieties such as receptor-binding aptamers, small interfering RNAs (siRNAs), microRNAs (miRNAs), ribozymes, and riboswitches.15,16 With advances in nanotechnology, many synthetic RNAs have been generated as drug carriers, making them attractive candidates for therapeutic purposes.17,18,19 RNA nanotechnology was first proposed in 1998, demonstrating that redesigned phage phi29 packaging RNA (pRNA) molecules can self-assemble from the bottom up to form RNA dimers, trimers, tetramers, and hexameric complexes to construct RNA nanoparticles.20 Nowadays, the dynamic, motile, and deformative properties of RNA are well known and used in extensive applications; many RNA nanoparticles with different functions in drug delivery, gene regulation, and molecule sensing can be structurally computed and designed in advance, easily constructed at nanoscale, and rapidly modified by several innovative methods.21,22,23 The elastic and rubber properties of RNA nanoparticles enable them to easily squeeze out the leaky blood vessels within tumor angiogenesis and rapidly across the glomerular in the kidney to excrete in urine.24 The incorporated ligands in RNA nanoparticles further improve biodistribution, which enables more efficient targeting for solid tumors.25 These advantages of RNA nanoparticles contribute to the mature field of therapeutic RNA nanotechnology and position RNA therapeutics as the third milestone in pharmaceutical drug development. However, there are still some concerns, with barrier penetration, material toxicity, immunogenicity, scalability, and unified guidance documents of synthesis and characterization remaining challenges in this field.26,27,28,29,30
RNA aptamers are single-stranded RNA oligonucleotide sequences capable of binding to specific targets with high affinity and specificity because of their three-dimensional structure.31,32 nAChR RNA aptamers isolated from human muscle cells using the systematic evolution of ligands by exponential enrichment (SELEX) technique were initially divided into two classes based on their function: class I aptamers directly inhibit nAChRs, and class II aptamers alleviate noncompetitive inhibition of the receptor.33,34,35 Direct blockade of α9-nAChR activity by specific antagonists has an inhibitory effect on BC progression, suggesting that class I aptamers may be more valuable than class II aptamers for treating BC. However, the major disadvantage of RNA aptamers is that they are susceptible to rapid degradation, leading to poor Pharmacokinetic/Pharmacodynamic(PK/PD) properties in vivo.36 To overcome this challenge, we developed a chemically and thermodynamically stable three-way junction (3WJ) motif derived from 2′-deoxy fluoro (2′-F)-modified pRNA as a core scaffold to connect different functional modules and constructed various polymerizations of RNA nanoparticles, rendering them biologically stable and able to retain their native folding properties in animals.24,37,38 Furthermore, through specific tumor-targeting RNA aptamers, these ultra-stable 3WJ-pRNA nanoparticles can effectively deliver different cargoes, such as fluorophores, chemical drugs, ribozyme, siRNA, and anti-miRNA, into the desired cells without affecting the function of incorporated modules.39,40,41,42,43,44 Therefore, this critical property allows 3WJ-pRNA RNA nanoparticles to be developed quickly and be incorporated extensively into various cancer therapies, especially novel RNAi therapeutics.45,46,47,48
miRNAs are small endogenous ncRNAs that regulate multiple target genes by binding to imperfectly complementary sites in the 3′ untranslated region (3′ UTR).49 miR-21 is an oncogenic miRNA that plays a vital role in tumor growth, apoptosis, and metastasis by downregulating tumor suppressors such as phosphatase and tensin homolog (PTEN) and programmed cell death 4 (PDCD4).50,51,52 Furthermore, miR-21 inactivation has also been demonstrated to induce tumor regression and apoptosis, which offers excellent potential for cancer therapy strategies.53,54,55 However, a major barrier to developing therapeutic anti-miRNAs as anticancer drugs is precise delivery into tumors without adverse effects on healthy tissues.56 To address these issues, an 8-nt (5′-GATAAGCT-3′) locked nucleic acid (LNA) complementary to the miR-21 seed region has been used to improve the stability and affinity of anti-miRNA.57 Moreover, the additional folate acid and specific aptamers, such as EGFR, linked to 3WJ-pRNA nanoparticles have been demonstrated to facilitate delivery of anti-miR-21 LNA and silence miR-21 in various solid tumors, including TNBC,41,44,58 GBM,59,60 and prostate cancer.43
In this study, we used the stable pRNA-3WJ motif as a core scaffold combined with the α9-nAChR RNA aptamer to construct RNA nanoparticles to accurately deliver fluorescent moieties and therapeutic anti-miRNAs to TNBC cells and patient-derived xenografts (PDXs). We observed that the α9-nAChR RNA aptamer combined with 3WJ RNA nanoparticles could effectively direct therapeutic anti-miR-21 LNA into TNBC tumors and therefore holds promise as a novel BC therapy for clinical application.
Results
The α9-nAChR RNA aptamer specifically targets α9-nAChR in TNBC cells
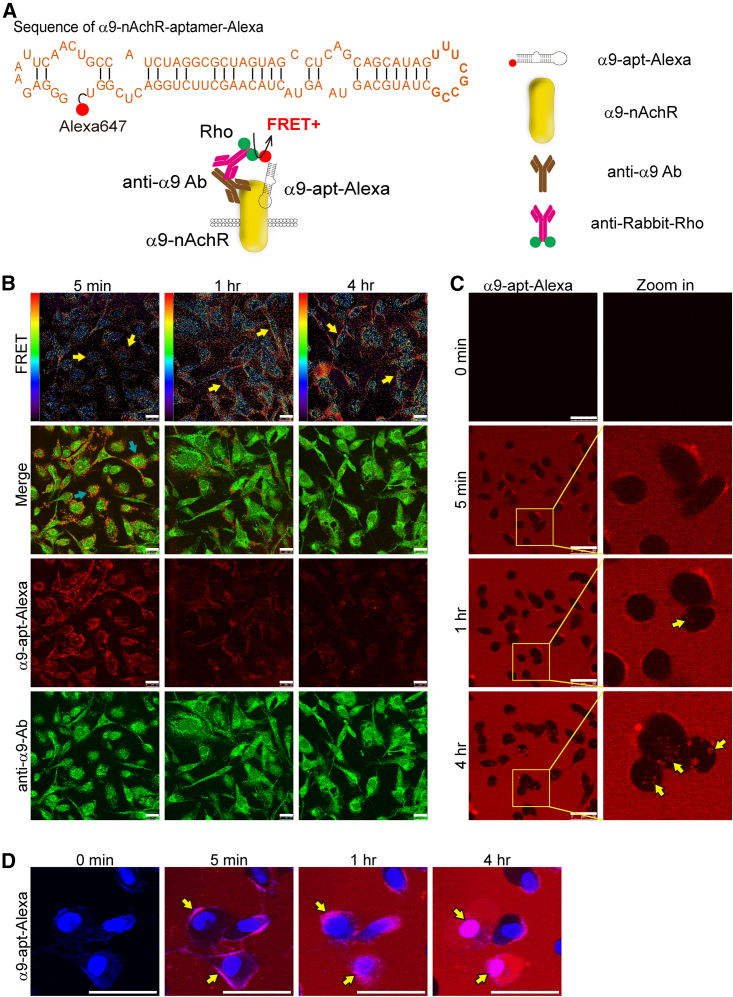
Our previous study demonstrated that overexpression of α9-nAChR protein in BC cells can induce BC tumorigenesis. Therefore, specific targeting of α9-nAChR protein by an RNA aptamer has potential for developing novel TNBC drugs. The designed α9-nAChR-RNA aptamer33 was labeled with a red-emitting fluorophore (Alexa 647) to form a fluorescent α9-nAChR RNA aptamer (named α9-apt-Alexa), and we evaluated its ability to target TNBC cells (Figure 1A). Human TNBC cells (MDA-MB-231) were treated with α9-apt-Alexa for various times. Results from confocal microscopy showed that α9-apt-Alexa bound to α9-nAChR on the membrane of TNBC cells (MDA-MB-231) as early as 5 min after treatment (Figure 1B, green arrow). To confirm the specific binding of α9-apt-Alexa to α9-nAChR, cells treated with α9-apt-Alexa were harvested for immunofluorescence using an α9-nAChR-specific antibody and detected by a goat anti-rabbit rhodamine-labeled secondary antibody. The strength of the interaction between α9-apt-Alexa and the α9-nAChR-specific antibody demonstrated specific binding of α9-apt-Alexa to α9-nAChR, as detected by fluorescence resonance energy transfer (FRET) (Figure 1B). α9-apt-Alexa could be seen interacting with the α9-nAChR-specific antibody, and a significant FRET signal was detected when α9-apt-Alexa was added to MDA-MB-231 cells for 1–4 h (Figure 1B, yellow arrow). The process of α9-apt-Alexa targeting living cells was observed by live-cell real-time laser confocal microscopy. The results showed that α9-apt-Alexa could bind to the TNBC cell membrane (MDA-MB-231) within 5 min, enter the cell for about 1 h, and remain there for at least 4 h (Figure 1C, yellow arrows; video at https://youtu.be/PnPB7tDGYrs). Under the same experimental conditions, we stained the nuclei of living cells with Hochest 33342, and the results showed that α9-apt-Alexa bound to the cell membrane in 5 min; noticeable observation results could be obtained in 1 h, and it entered the cell after 4 h (Figure 1D, yellow arrows; video at https://youtube.com/v=NLOttMXyAsA). These results indicated that the designed α9-apt-Alexa could specifically bind to α9-nAChR on the TNBC cell membrane.
Figure 1.
Targeting and internalization of the α9-nAChR RNA aptamer in TNBC cells
(A) The schematic shows the sequence of the α9-nAChR RNA aptamer labeled with Alexa 647 (top) and the fluorescent staining strategy for detecting the FRET signal between α9-nAChR and α9-apt-Alexa (bottom). The α9-nAChR proteins are detected using antibodies specific to α9nAchR and labeled by a secondary rabbit antibody with rhodamine (Rho; green dots). (B) Time-dependent fluorescence and FRET images in MDA-MB-231 cells after α9-apt-Alexa treatment. Scale bar, 25 μm. The red/blue spectrum represents the intensity of FRET efficiency. Yellow arrows indicate a positive FRET signal. (C and D) Time-lapse fluorescence live-cell images of MDA-MB-231 cells after α9-apt-Alexa treatment. Yellow arrows indicate internalization of α9-apt-Alexa. Hochest 33342 is a nuclear indicator (blue) (D). Scale bars, 50 μm (C and D).
Synthesis of multivalent 3WJ-α9-nAChR aptamer RNA nanoparticles targeting α9-nAChR in TNBC cells
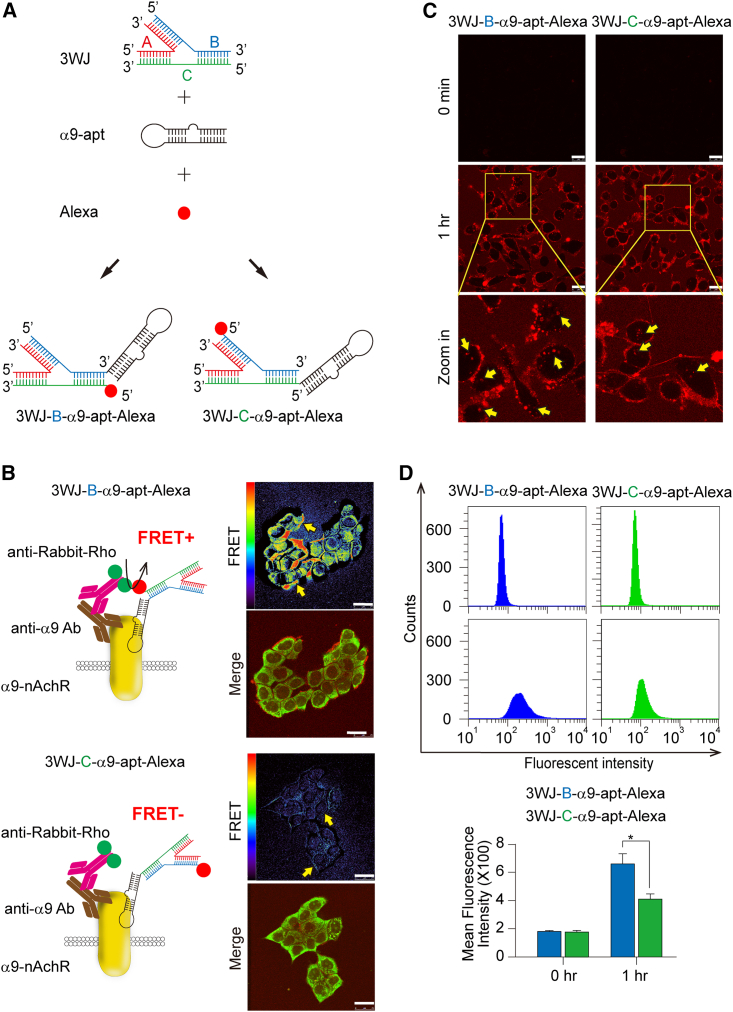
As mentioned above, α9-apt-Alexa has a specific molecular targeting effect, but the stability of unmodified single-stranded RNA is usually poor. According to our previous findings, the stability, penetration, and biosafety of 3WJ RNA nanoparticles to conjugate various functional RNA molecules with chemical drugs are superior to single-stranded RNA.37 We then synthesized a multivalent 3WJ-α9-nAChR aptamer RNA nanoparticle using a red-emitting fluorophore (Alexa 647) as a biodistribution indicator. We verified the binding affinity of the 3WJ-α9-nAChR aptamer RNA nanoparticles based on binding of the α9-nAChR RNA aptamer to the 3WJ branch position. We named them 3WJ-B-α9-apt-Alexa (Figure 2A, left) and 3WJ-C-α9-apt-Alexa (Figure 2A, right). MDA-MB-231 cells were treated with 3WJ-B-α9-apt-Alexa or 3WJ-C-α9-apt-Alexa for 1 h. Cells were harvested and subjected to immunofluorescence (IF) staining and FRET analysis using an α9-nAChR-specific antibody according to the technical principle in Figure 1. The results showed that, compared with 3WJ-C-α9-apt-Alexa, 3WJ-B-α9-apt-Alexa bound to α9-nAChR presented a significantly stronger FRET signal on the TNBC cell membrane (Figure 2B, right, yellow arrow). Further observations by live-cell system and real-time laser confocal microscopy showed that both RNA nanoparticles could bind to the cell membrane and stay for more than 1 h (Figure 2C, left, yellow arrow; videos at https://youtu.be/Wld9UzT4TFM, https://youtu.be/3xANv9h9JVs). However, 3WJ-B-α9-apt-Alexa entered more efficiently and stayed in the cell for 4 h (Figure S1). Flow cytometry analysis was performed to confirm whether the efficiency of these two RNA nanoparticles binding to α9-nAChR and entering cells is different within 1 h, and the results showed that 3WJ-B-α9-apt-Alexa is more efficient at entering living cells (3WJ-B-α9-apt-Alexa vs. 3WJ-C-α9-apt-Alexa, p = 0.0363) (Figure 2D). Our results showed that 3WJ-B-α9-apt-Alexa RNA nanoparticles are the optimal structure because they have more efficient entry into TNBC cells and better α9-nAChR-specific targeting on cell membranes compared with 3WJ-C-α9-apt-Alexa.
Figure 2.
The specificity and binding affinity of multivalent 3WJ-α9-nAChR aptamer RNA nanoparticles in TNBC cells
(A) The schematic demonstrates the synthesis of multivalent 3WJ-α9-nAChR RNA aptamer-Alexa RNA nanoparticles. (B) Left: Schematic showing the fluorescent staining strategy for detecting FRET signals between α9-nAChR and 3WJ-B-α9-apt-Alexa (top) and 3WJ-C-α9-apt-Alexa (bottom), respectively. Right: FRET and fluorescence images in MDA-MB-231 cells after treatment with 3WJ-B-α9-apt-Alexa (top) or 3WJ-C-α9-apt-Alexa (bottom). Yellow arrows indicate a positive FRET signal. The red/blue color spectrum represents the intensity of FRET efficiency. (C) Time-lapse fluorescence live-cell images of MDA-MB-231 cells after treatment with 3WJ-B-α9-apt-Alexa (left) or 3WJ-C-α9-apt-Alexa (right). Scale bars, 25 μm (B and C). Yellow arrows indicate internalization of RNA nanoparticles. (D) The binding affinity of 3WJ-B-α9-apt-Alexa (blue) and 3WJ-C-α9-apt-Alexa (green) to α9-nAChR proteins in treated MDA-MB-231 cells was detected by flow cytometry. n = 3 independent samples. Data are presented as mean ± SEM. Statistical analysis was performed using a two-tailed unpaired t test. ∗p < 0.05.
The 3WJ-B-α9-apt-Alexa RNA nanoparticles specifically target the N-terminal domain of α9-nAChR in TNBC cells
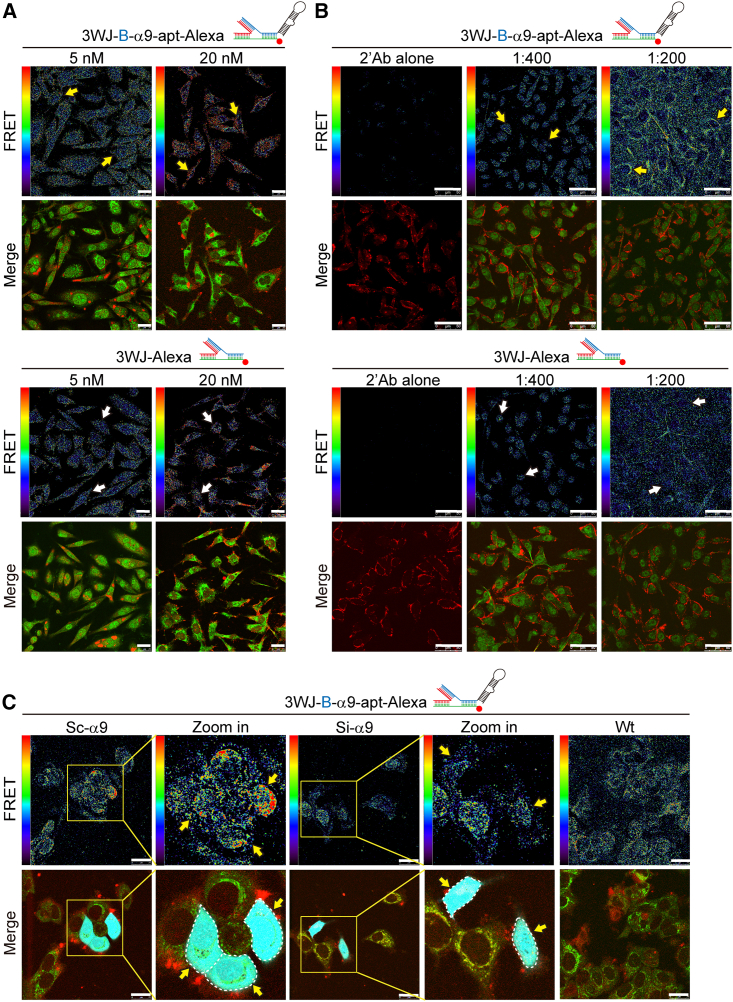
To verify the specificity of 3WJ-B-α9-apt-Alexa RNA nanoparticles targeting α9-nAChR in TNBC cells, we synthesized 3WJ-Alexa RNA nanoparticles (3WJ-Alexa) as a negative control group for FRET analysis. To confirm the specific binding site between 3WJ-B-α9-apt-Alexa and α9-nAChR protein in TNBC cells, we selected a commercially available α9-nAChR antibody that can recognize amino acids 1–50 at the N terminus. After the TNBC cells were exposed to 3WJ-B-α9-apt-Alexa (5–20 nM) for 1 h and stained by an α9-nAChR-specific antibody, the interaction between 3WJ-B-α9-apt-Alexa and the α9-nAChR-specific antibody was detected by FRET analysis using laser confocal microscopy (Figure 3A). The results showed that the increase in 3WJ-B-α9-apt-Alexa concentration significantly intensified TNBC cells’ FRET signal (Figure 3A, top, yellow arrow) compared with 3WJ-Alexa, which had no significant change (Figure 3A, bottom, white yellow). We then adjusted the ratio of the α9-nAChR-specific antibody (Figure 3B) during IF staining. The interaction strength between 3WJ-B-α9-apt-Alexa and α9-nAChR-specific antibodies on TNBC cells was then evaluated by the FRET technique (Figure 3B). The results showed that, when the concentration ratio of the α9-nAChR-specific antibody increased, it enhanced the FRET activity signal (Figure 3B, top, yellow arrow), while 3WJ-Alexa remained unchanged (Figure 3B, bottom, white arrow). After inhibiting expression of α9-nAChR in TNBC cells by siRNA, the FRET activity signal between 3WJ-B-α9-apt-Alexa and the α9-nAChR-specific antibody in the TNBC cell membrane was significantly inhibited (Figure 3C, yellow arrow), which indirectly indicates that there is an interaction between 3WJ-B-α9-apt-Alexa and α9-nAChR in TNBC cells. In addition, 3WJ-B-α9-apt-Alexa could also specifically target α9-nAChR on HER2+ (SKBR-3) BC cell membranes compared with 3WJ-Alexa (Figure S2). Therefore, these results confirmed the specificity of 3WJ-B-α9-apt-Alexa RNA nanoparticles targeting α9-nAChR in TNBC cells.
Figure 3.
The 3WJ-B-α9-apt-Alexa RNA nanoparticles specifically target α9-nAChR in TNBC cells
(A and B) FRET and fluorescence images in MDA-MB-231 cells after treatment with 3WJ-B-α9-apt-Alexa (top) or 3WJ-Alexa (bottom) according to the specified dose of nanoparticle (A) or anti-α9 antibody (B). Scale bar, 25 μm. Yellow and white arrows indicate the FRET signals between α9-nAChR and 3WJ-B-α9-apt-Alexa or 3WJ-Alexa, respectively. The red/blue color spectrum represents the intensity of FRET efficiency. (C) FRET images in MDA-MB-231 wild-type, α9-nAChR-siRNA and α9-nAChR-scRNA transfected cells after treatment with 3WJ-B-α9-apt-Alexa. Green, α9-nAChR; red, 3WJ-B-α9-apt-Alexa; cyan, GFP+ cells. Scale bars, 50 μm (B and C).
Direct observation of 3WJ-B-α9-apt-Alexa RNA nanoparticles targeting TNBC-PDX tumors in vivo
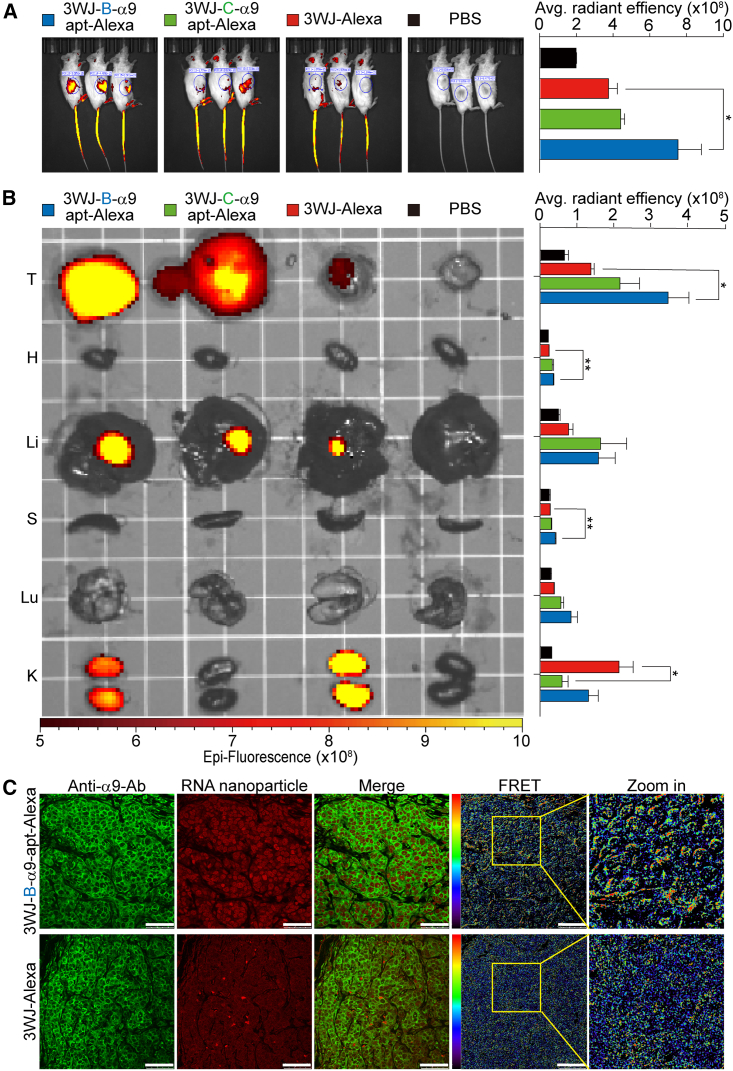
Next, the in vivo tumor targeting and quantitative biodistribution of the 3WJ-B-α9-apt-Alexa RNA nanoparticles were evaluated. After 3WJ-B-α9-apt-Alexa, 3WJ-C-α9-apt-Alexa, and 3WJ-Alexa were systemically injected into non-obese diabetic, severe combined immunodeficiency, interleukin-2R (IL-2R), gamma-null (NSG) mice (n = 3) with TNBC-PDX tumors by tail vein, optical imaging was performed using the In Vivo Imaging System (IVIS) 200 to continuously observe the distribution in the whole body. The results showed that 3WJ-B-α9-apt-Alexa had a stronger fluorescent signal than 3WJ-Alexa in the tumor area of the mice 2 h post injection (3WJ-B-α9-apt-Alexa vs. 3WJ-Alexa, p = 0.0495) (Figure 4A, the blue circle indicates the tumor). Ex vivo images of the tumor and healthy organs in each group harvested from mice 8 h post injection were used to analyze the accumulation of fluorescent RNA nanoparticles by IVIS using the following excitation and emission filters: 640 and 680 nm, respectively. Quantitative analysis of each organ in fluorescence images was performed by measuring the average radiant efficiency in the defined area. The images and quantitative analysis showed that 3WJ-B-α9-apt-Alexa accumulated significantly in the tumors and were higher than in other organs, while 3WJ-Alexa remained heavily in the kidneys (tumor: 3WJ-B-α9-apt-Alexa vs. 3WJ-Alexa, p = 0.0218; kidneys: 3WJ-Alexa vs. 3WJ-C-α9-apt-Alexa, p = 0.0216) (Figure 4B). Additionally, we normalized the radiance efficiency of each RNA nanoparticle-treated organ against its organ weight to ensure an equivalent comparison between each group to more accurately verify our findings (tumor: 3WJ-B-α9-apt-Alexa vs. 3WJ-Alexa, p = 0.0293; kidneys: 3WJ-Alexa vs. 3WJ-B-α9-apt-Alexa, p = 0.0307; 3WJ-Alexa vs. 3WJ-B-α9-apt-Alexa, p = 0.0169) (Figure S3). These tumors were stained with α9-nAChR-specific antibodies and then analyzed by FRET to measure the interaction strength of 3WJ-B-α9-apt-Alexa and α9-nAChR. The results showed that 3WJ-B-α9-apt-Alexa could specifically bind to α9-nAChR in TNBC tumors (Figure 4C). The overall results confirmed that the 3WJ-B-α9-apt-Alexa RNA nanoparticles could effectively and specifically target TNBC tumors in vivo.
Figure 4.
In vivo biodistribution of 3WJ-B-α9-apt-Alexa RNA nanoparticles in mice with TNBC-PDX tumors
(A) Whole-body images (left) and quantitative analysis of tumor area (right) in TNBC-PDX tumor-bearing mice 2 h after tail vein injection of 3WJ-B-α9-apt-Alexa, 3WJ-C-α9-apt-Alexa, and 3WJ-Alexa (2 μmol/L solution at 100 μL) with PBS as a negative control. (B) Representative ex vivo images (left) and quantitative analysis (right) in tumors and major organs of 3WJ-B-α9-apt-Alexa, 3WJ-C-α9-apt-Alexa, 3WJ-Alexa, and PBS 8 h post-injection into mice. T, tumor; H, heart; Li, liver; S, spleen; Lu, lung; K, kidney. Red-yellow color scale: radiant efficiency (p s−1 cm−2 sr−1) (μW cm−2)−1. n = 3 biologically independent animals; statistics were calculated by two-tailed unpaired t test and presented as mean ± SEM. ∗p < 0.05, ∗∗p < 0.001 (A and B). (C) Fluorescence and FRET images of ex vivo RNA nanoparticle-treated TNBC-PDX tumors stained with α9-nAChR antibodies (secondary antibodies labeled with Rho, green). The red/blue color spectrum represents the intensity of FRET efficiency. Scale bars, 50 μm.
3WJ-B-α9-nAChR aptamer RNA nanoparticles delivered anti-tumor drugs to inhibit TNBC cell growth
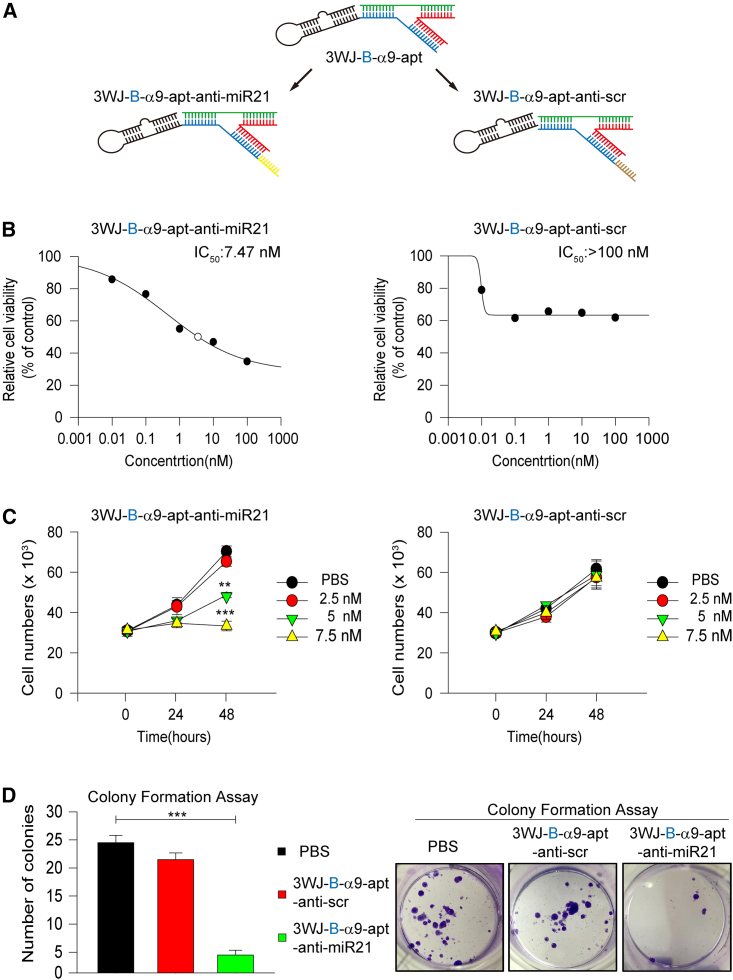
We further tested whether 3WJ-B-α9-nAChR aptamer RNA nanoparticles can be used as a specific targeting molecule in vivo to carry anticancer drugs to attack TNBC tumors and produce therapeutic effects. Many studies have shown that miR-21 plays a crucial role in human tumorigenesis.50,51,52 Many studies have also shown that antisense oligonucleotides targeting miR-21 inhibit tumor growth.53,54,55 Therefore, in this study, an anti-miR-21 RNA sequence was incorporated into the B chain of 3WJ-B-α9-apt to construct 3WJ-B-α9-apt-anti-miR-21 RNA nanoparticles (3WJ-B-α9-apt-anti-miR-21; Figure 5A, left). A scrambled sequence of anti-miR-21 RNA was designed as a control (3WJ-B-α9-apt-anti-scr; Figure 5A, right). Then, we assessed the cytotoxic effect by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenylterazolium (MTT) assay to determine the effective inhibitory concentration of these RNA nanoparticles. The results showed that the half-maximal inhibitory concentration (IC50) of 3WJ-B-α9-apt-anti-miR-21 was about 7.5 nM compared with more than 100 nM for 3WJ-B-α9-apt-anti-scr as a control (Figure 5B). We then assessed the effect of 3WJ-B-α9-apt-anti-miR-21 on cell viability inhibition using a trypan blue exclusion assay. The results showed that 3WJ-B-α9-apt-anti-miR-21(>5 nM) significantly inhibited the survival of MDA-MB-231 cells after 48 h of treatment compared with the control group (3WJ-B-α9-apt-anti-scr) (control vs. 5 nM, p = 0.002; control vs. 7.5 nM, p < 0.0001) (Figure 5C). To confirm no adverse effect in normal breast epithelial cells by 3WJ-B-α9-apt-anti-m-R-21 treatment, we evaluated the cytotoxic effect of 3WJ-B-α9-apt-anti-miR-21 in the MCF-10A, MCF-12A, and 184A1 normal breast epithelial cell lines using an MTT assay. The results showed no cytotoxic effect in normal breast epithelial cells, even with a concentration of 3WJ-B-α9-apt-anti-miR-21 of 100 nM. However, only 1 nM 3WJ-B-α9-apt-anti-miR-21 is enough to significantly inhibit proliferation of MDA-MB-231 cells (control vs. 1 nM, p = 0.0135) (Figure S4). Furthermore, a colony formation assay was performed to evaluate the long-term effect of 3WJ-B-α9-apt-anti-miR-21 treatment, and the results showed that the number of colonies of αMDA-MB-231 cells treated by 3WJ-B-α9-apt-anti-miR-21 was reduced significantly (3WJ-B-α9-apt-anti-miR-21 vs. PBS, p < 0.0001) (Figure 5D). To further verify the results of successful silencing of miR-21 by 3WJ-B-α9-apt-anti-miR-21 treatment, downstream target protein changes of miR-21 were detected by western blot analysis. The result showed that PTEN, PDCD4, p27, and p21 increased significantly and phospho-Akt decreased in 3WJ-B-α9-apt-anti-miR-21-treated MDA-MB-231 cells compared with the control group, but there was no noticeable protein change in the same three treated normal breast epithelial cell lines (Figure S5). We confirmed that 3WJ-B-α9-apt-anti-miR-21 RNA nanoparticles have a practical growth-inhibitory effect on TNBC cells in vitro.
Figure 5.
In vitro growth and survival inhibition effect of 3WJ-B-α9-apt-anti-miR-21 RNA nanoparticles in TNBC cells
(A) Schematic of 3WJ-B-α9-nAChR aptamer RNA nanoparticles carrying anti-miR-21 LNA or anti-scramble RNA. (B) Evaluation of the half-maximal inhibitory concentration (IC50) of 3WJ-B-α9-apt-anti-miR-21 (left) and 3WJ-B-α9-apt-anti-scr (right) in MDA-MB-231 cells via MTT assay. (C) In vitro cell growth inhibition effects of 3WJ-B-α9-apt-anti-miR-21 (left) and 3WJ-B-α9-apt-anti-scr (right) in MDA-MB-231 cells via cell counting. (D) In vitro cell survival inhibition of 3WJ-B-α9-apt-anti-miR-21 (green) and 3WJ-B-α9-apt-anti-scr (red) in MDA-MB-231 via colony formation assay. The calculated number of colonies (left) and images (right) are shown. n = 3 independent samples. Statistics were calculated by two-tailed unpaired t test and presented as mean ± SEM (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
RNA nanoparticles specifically targeting the α9-nAChR aptamer deliver anti-miR-21 for tumor growth inhibition in the TNBC-PDX model
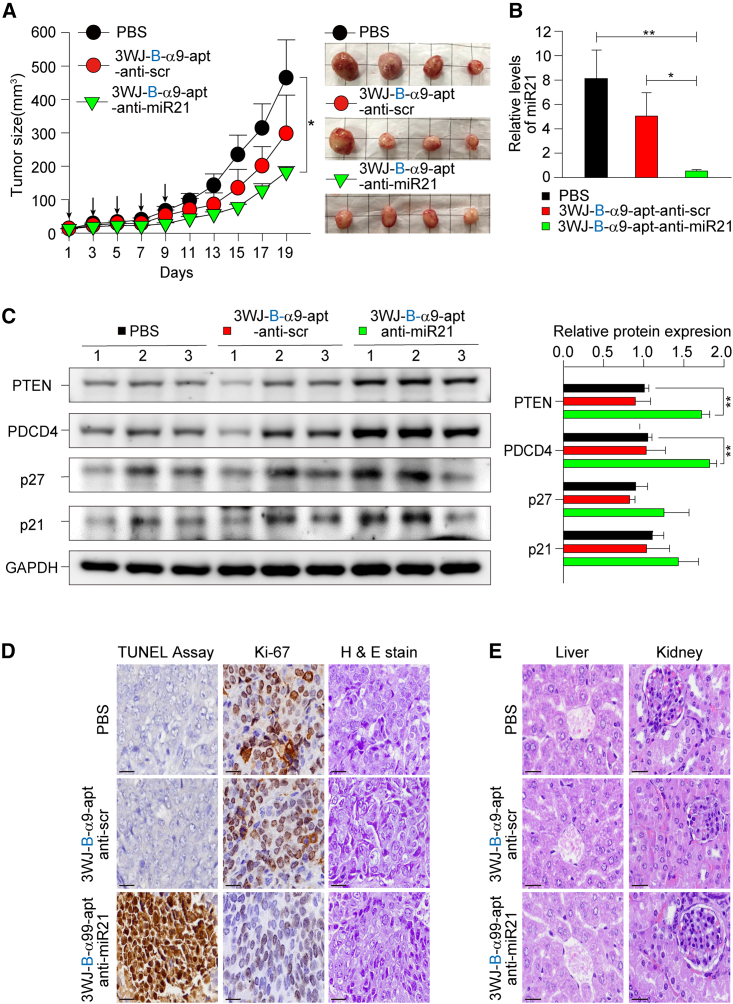
Bringing together the data above, we aimed to demonstrate the ability of RNA nanoparticles to deliver and release miRNA to a solid tumor for growth inhibition. The in vivo therapeutic effects of 3WJ-B-α9-apt-anti-miR-21 were evaluated using the TNBC-PDX mouse model. NSG mice with TNBC-PDX tumors were administered 3WRNAJ-B-α9-apt-anti-miR21 and 3WJ-B-α9-apt-anti-scr at a dose of 8 mg kg−1 every 2 days for a total of five doses by tail vein injection. The result showed that 3WJ-B-α9-apt-anti-miR-21 had an excellent inhibitory effect on tumor growth, while control groups formed faster-growing tumors (467.8 mm3 vs. 183.8 mm3, p = 0.049) (Figure 6A). After RNA nanoparticle treatment, mice without weight loss were sacrificed, and tumors were removed for analysis (Figure S6). The results of qRT-PCR analysis show that the 3WJ-B-α9-apt-anti-miR-21 group had significantly lower miR-21 gene expression than the control group (Figure 6B). Western blot analysis of the downstream related protein changes of miR-21 showed that PTEN and PDCD4 increased significantly and p27 and p21 increased only a little (Figure 6C), suggesting that 3WJ-B-α9-apt-anti-miR-21 could effectively deliver anti-miR-21 into TNBC tumors. In addition, Ki67 IHC staining and a terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) assay showed that the expression of Ki-67 and positive cells in the 3WJ-B-α9-apt-anti-miR-21 group decreased compared with the PBS group, while the TUNEL-positive area increased (Figure 6D), demonstrating RNA nanoparticle drug-induced cell death effects. To evaluate in vivo safety, the cardiac blood was collected from the mice for biochemical analysis at the TMU hospital according to standard protocols. The mice weight in each group was not changed, whereas the mice in the 3WJ-B-α9-apt-anti-miR-21 group had high aspartate aminotransferase (AST) (Table 1). Therefore, the liver and kidneys were dissected, and the pathological morphology was observed by hematoxylin and eosin (H&E) staining in each group; the results indicated no toxic changes (Figure 6E).
Figure 6.
In vivo therapeutic effect of 3WJ-B-α9-apt-anti-miR-21 RNA nanoparticles in TNBC-PDX mice
(A) NSG mice bearing TNBC-PDX xenografts were treated i.v. with 3WJ-B-α9-apt-anti-miR-21 (green), 3WJ-B-α9-apt-anti-scr (red), and PBS (black) every other day five times in total (20 μmol/L solution, 100 μL, indicated by arrows). Tumor volume (mm3) and body weight (g) were monitored during treatment. Representative images of BC tumors harvested from mice after treatment are shown on the right. (B) In vivo delivery of anti-miR-21 was analyzed by quantitative real-time PCR on mRNA isolated from 3WJ-B-α9-apt-anti-miR-21-treated tumors. The endogenous control U6 snRNA normalized the mRNA levels of miR-21. n = 4 independent samples. Statistics were calculated by two-tailed unpaired t test and presented as mean ± SD (∗p < 0.05, ∗∗p < 0.01, comparing 3WJ-B-α9-apt-anti-miR-21 with 3WJ-B-α9-apt-anti-scr and PBS, respectively). (C) Western blot examined the downstream protein expression of PTEN, PDCD4, p21, and p27 resulting from miR-21 silencing in nanoparticle-treated tumor extracts. Quantitative analyses are shown on the right, using GAPDH as an internal control. n = 3 independent samples. Statistics were determined by two-tailed unpaired t test and presented as mean ± SEM (∗∗p < 0.01, ∗∗p = 3.78 × 10−3 of PTEN and 1.67 × 10−3 of PDCD4 comparing 3WJ-Bα9-apt-anti-miR-21 with PBS). (D) Tumor growth inhibition and apoptosis induction of 3WJ-B-α9-apt-anti-miR-21 in TNBC-PDX tumor tissues. Cells stained positively by Ki-67 IHC and TUNEL assay are brown. (E) Histological examination of the organ toxicity of 3WJ-B-α9-apt-anti-miR-21 in the liver and kidneys by H&E staining. Scale bars, 10 μm (D and E).
Table 1.
Serum biochemical analysis of mice treated with RNA nanoparticles
| Markers(units) | 3WJ-B-α9-apt-anti-scr | 3WJ-B-α9-apt-anti-miR-21 | PBS |
|---|---|---|---|
| Body weight (g) | 21.03 ± 0.00 | 21.10 ± 0.25 | 21.44 ± 0.68 |
| Albumin (g/dL) | 4.43 ± 0.05 | 4.28 ± 0.08 | 4.35 ± 0.05 |
| Globulin (g/dL) | 1.28 ± 0.08 | 1.5 ± 0.13 | 1.33 ± 0.08 |
| Total protein (g/dL) | 5.70 ± 0.08 | 5.78 ± 0.16 | 5.68 ± 0.08 |
| BUN (mg/dL) | 19.25 ± 0.63 | 20.00 ± 1.41 | 18.25 ± 0.48 |
| Creatinine (mg/dL) | 0.30 ± 0.00 | 0.35 ± 0.03 | 0.30 ± 0.00 |
| Uric acid (mg/dL) | 2.20 ± 0.24 | 2.55 ± 0.22 | 2.18 ± 0.10 |
| ALP (U/L) | 151.50 ± 6.74 | 142.25 ± 3.97 | 135.50 ± 3.33 |
| AST (U/L) | 263.25 ± 42.80 | 732.00 ± 188.20∗ | 221.75 ± 48.84 |
| ALT (U/L) | 37.00 ± 5.26 | 143.50 ± 60.63 | 37.75 ± 2.96 |
Values are mean ± SEM (n = 4/each group); ∗p\0.05, ∗∗p\0.01, ∗∗∗p\0.001 versus the PBS group, non-paired t test. BUN, blood urea nitrogen; ALP, alkaline phosphatase; AST, aspartate aminotransferase; ALT alanine aminotransferase.
AST: 3WJ-B-α9-apt-anti-miR-21 vs PBS ∗p = 0.003936
In summary, this study confirmed that 3WJ-B-α9-nAChR aptamer RNA nanoparticles exhibit a favorable ability to specifically target α9-nAChR in TNBC in vitro and in vivo. Furthermore, α9-nAChR targeting with an anti-miRNA or siRNA strategy provided a significant therapeutic effect to inhibit growth of TNBC and had no adverse effects on essential organs in vivo.
Discussion
TNBC patients have no specific target biomarkers compared with patients with other BC subtypes, resulting in non-response to most small-molecule drug treatments, rapid deterioration, and shortened survival.3,61 α9-nAChR is considered an oncogenic transmembrane protein in BC.10 In our past studies, we found that α9-nAChR protein was highly expressed in TNBC tumor tissues,10 lung adenocarcinoma,62 and melanoma.63 Therefore, in this study, for the first time, we used an α9-nAChR RNA aptamer as a specific targeting ligand combined with 3WJ pRNA nanoparticles to develop a novel therapeutic strategy against TNBC tumors. Our previous research shows that α9-nAChR interacts with many oncogenic proteins on the membrane of BC cells64 and plays a role in nicotine’s proliferative and anti-apoptotic effects by activating multiple oncogenic pathways, such as Akt and ERK signaling.65 In addition, recent research has provided evidence that chronic exposure to nicotine can stimulate lung pre-metastatic niche formation to promote lung metastasis of TNBC.66 According to our previous studies, α9-nAChR is the most validated nAChR subtype in BC and plays an essential role in TNBC metastasis.10,12 Therefore, selective antagonists targeting α9-nAChR may provide a promising therapeutic strategy for preventing nicotine-induced lung metastasis. Based on this rationale, our experiments found that the United States Food and Drug Administration (FDA)-approved drug bupropion may be applied in combination with existing therapeutic drugs and to design novel treatment strategies for smoking-induced lung metastasis of TNBC patients.64 In this study, we used screening methods to search potent inhibitors of α9-nAChR (e.g., fencamfamine, mitotane, and bupropion), and bupropion was chosen because of its docking into an allosteric binding site with the lowest binding energy in a binding model. We also found that it effectively suppressed nicotine-induced cell invasion and migration in vitro and reduced TNBC lung metastasis in vivo, making it an attractive anti-metastasis agent for combination with chemotherapy agents or targeted therapies. More experimental studies on its optimal dosage, safety, and potential drug interactions are still under investigation. A similar study also found that the αO-conotoxin GeXIVA inhibits TNBC cell proliferation by interacting with α9-nAChR to cause cell-cycle arrest.67 These studies highlight the importance of α9-nAChR as a biomarker and therapeutic target in BC, especially for aggressive TNBC subtypes.68 In view of the results confirming the carcinogenesis of α9-nAChR, this study used RNA aptamers to specifically target α9-nAChR in BC cells and designed 3WJ-pRNA nanoparticles containing therapeutic components. Our results suggest that this drug is effective against TNBC and has therapeutic potential in patients with α9-nAChR-overexpressing tumors.
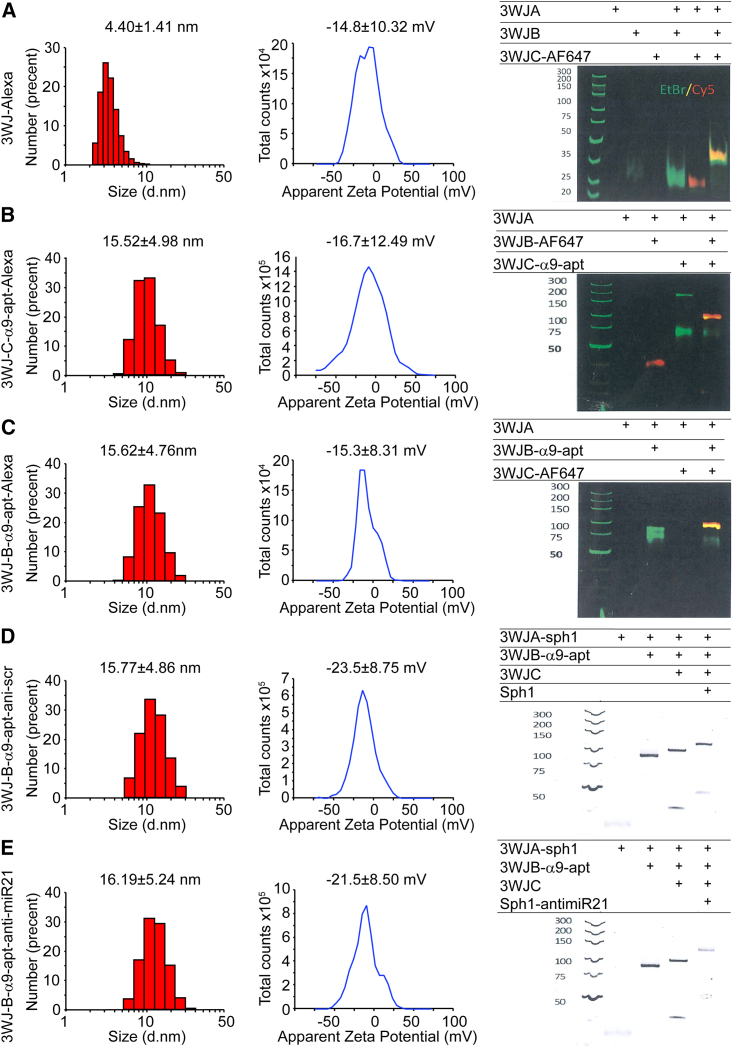
For drug-resistant tumors, in addition to the specific targeting effect, the biosafety of the drug must also be a factor of consideration. The α9-nAChR RNA aptamer was discovered initially from the electroplax of Torpedo californica to compete with the cocaine-binding site of muscle nAChR to attenuate the toxicity of drugs of abuse.33,34 Further studies found that the truncated α9-nAChR RNA aptamer inhibited α9-nAChR activity or alleviated the inhibitory effect of MK-801 and cocaine on nAChR. However, the reduction in sequence length led to a significant reduction in the biological activity of the RNA aptamer.35 A recent study reported that short nAChR DNA aptamers also have activity similar to RNA aptamers to displace cocaine from the muscle-type nAChR.69 Unlike the measurement method of the dissociation constant in previous studies, we directly verified the specific interaction between TNBC cells’ α9-nAChR and the full-length α9-nAChR RNA aptamer by FRET assay under laser confocal microscopy. The FRET assay can be used to measure molecular dynamic interactions to detect complex formation between protein-protein and protein-nucleic acids, whose principle is similar to an electrophoretic mobility shift assay (EMSA), and this method has been applicated in recent studies by other research groups.70,71 We further confirmed that RNA aptamer internalization into TNBC cells increases with time (Figures 1B–1D). However, according to past studies, although synthetic RNA aptamers can precisely target receptors, their biological stability still needs to be improved.36 Therefore, to improve this defect, our team confirms that stable 3WJ-pRNA nanoparticles can be an excellent platform to improve RNA molecules’ thermodynamic and chemical stability.37,38,72 In addition, the 2′-F chemical modification of 3WJ-pRNA-aptamer nanoparticles further enhanced the biostability against RNase degradation in vivo.73,74 Apart from considering the biological safety and stability of the drug in vivo, it is necessary to precisely design the configuration of the RNA nanodrug so that it has a specific targeting effect on α9-nAChR and establish a theoretical basis for future drug design. Based on these principles, two different structures of 3WJ-pRNA nanoparticles were designed in this study, which contain α9-nAChR RNA aptamers and fluorophores linking different branches to the 3WJ core (Figure 2A). By mixing each strand at an equal molar concentration in a one-stop self-assembly method, these RNA nanoparticles were constructed with high efficiency, as shown in gel electrophoresis with stepwise assembly of these complexes (Figure 7, right). Our dynamic light scattering (DLS) analysis showed that the average hydrodynamic diameter of 3WJ-C-α9-apt-Alexa and 3WJ-B-α9-apt-Alexa were 15.52 ± 4.98 nm and 15.62 ± 4.76 nm, respectively, compared with 4.40 ± 1.41 nm for 3WJ-Alexa (Figures 7A–7C, left). The larger nanoparticle sizes align with predictions because of the added α9-nAChR aptamer but are still within a reasonable size range for in vivo biodistribution. Furthermore, the zeta potential of 3WJ-Alexa, 3WJ-C-α9-apt-Alexa, and 3WJ-B-α9-apt-Alexa was determined to be −14.8 ± 10.32, −16.7 ± 12.49, and −15.3 ± 8.31, respectively (Figures 7A–7C, center). The negative charge of these RNA nanoparticles can prevent non-specific binding to normal cells because of the anionic nature of the cell membrane.
Figure 7.
Characterization and construction of 3WJ RNA nanoparticles carrying the α9-nAChR aptamer linked to Alexa 647 or anti-miR-21
The size distribution (left) and zeta potential (center) of 3WJ-Alexa (A), 3WJ-C-α9-apt-Alexa (B), 3WJ-B-α9-apt-Alexa (C), 3WJ-B-α9-anti-scr (D), and 3WJ-B-α9-anti-miR-21 (E) RNA nanoparticles by DLS measurements are shown. The stepwise self-assembly (right) of all of these RNA nanoparticles was evaluated by 8% TBE native PAGE.
A previous study proposed that the orientation of molecularly targeted RNA aptamers on 3WJ-pRNA nanoparticles affects their binding to specific cells and alters their intracellular cargo transport.75 Our study also found that the α9-nAChR RNA aptamer linked to the core B chain of 3WJ exhibited higher specificity and faster internalization of α9-nAChR on TNBC cells than the aptamer linked to the core C chain (Figures 2B–2D). This study found that the 3WJ-B-α9-apt configuration is more specific to TNBC. As the 3WJ-EGFRapt/anti-miR-21 and 3WJ/CD133apt/anti-miR-21 RNA nanoparticles in previous studies, the emerging drug showed high specificity and affinity for TNBC cell binding.41,44 Although these RNA aptamers as targeting ligands confer high cellular binding and specificity to the 3WJ-pRNA nanoparticles, there were still some concerns about the undesired non-specific targeting to receptor-negative or -low cells.76 To address these concerns, reducing the expression of α9-nAChR in TNBC cells by siRNA technology in TNBC cells inhibited the specific binding between 3WJ-B-α9-apt-Alexa and α9-nAChR (Figure 3C). Interestingly, 3WJ-B-α9-apt-Alexa showed excellent specificity for α9-nAChR in TNBC and HER2+ BC cells (Figure S2). The most crucial properties of RNAi-based therapeutic agents are their in vivo tumor-targeting ability and safety.77,78 3WJ-pRNA nanoparticles carrying tumor-targeting RNA aptamers and fluorophores have been shown to successfully bind solid tumors without accumulating in healthy organs of treated mice in past studies.41,42,43,44,60. Our results in this study also indicated that 3WJ-B-α9-apt-Alexa RNA nanoparticles had binding specificity to TNBC-PDX tumors with little or no accumulation in healthy organs compared with 3WJ-Alexa RNA nanoparticles (Figures 4B and S3). Given that the configuration of 3WJ-B-α9-apt RNA nanoparticles can specifically bind to α9-nAChR, 3WJ-B-α9-apt RNA nanoparticles will be suitable for delivering therapeutic elements to tumors that highly express α9-nAChR.
Elevated miR-21 expression in TNBC has been reported to be associated with increased tumor growth, metastasis, and poor clinical outcome, attributed to downregulation of tumor suppressors and upregulation of anti-apoptotic genes.50,79,80 Previous studies have proven that antisense inhibition of endogenous miR-21 by anti-miR-21 effectively impairs tumor growth and induces cell apoptosis in several cancers, including glioblastoma,52 prostate cancer,81 and TNBC.82 Although the seed-targeting 8-nt LNA has a favorable oncogenic miRNA binding affinity,57 a safe delivery platform such as RNA nanoparticles carrying a tumor-targeting aptamer is still required to enhance its specificity for silencing miRNA and reducing off-target effects. In this study, the 3WJ-B-α9-apt-anti-miR-21 along with control 3WJ-B-α9-apt-anti-scr displayed highly efficient stepwise assembly in gel electrophoresis (Figures 7D and 7E, right). In addition, they were measured for their average hydrodynamic diameter using DLS and were shown to be 16.19 ± 5.24 nm and 15.77 ± 4.86 nm in size with a zeta potential of −21.5 ± 8.50 and −23.5 ± 8.75 mV (Figures 7D and 7E, left and center), respectively, which is similar to previous RNA nanoparticles. For therapeutic assessment, 3WJ-B-α9-nAChR aptamer RNA nanoparticles carrying anti-miR-21 LNA significantly suppressed TNBC-PDX tumor growth and induced cell apoptosis (Figure 6A and 6D). Furthermore, our results here demonstrated that 3WJ-B-α9-apt-anti-miR-21 RNA nanoparticle treatment significantly reduced the gene level of miR-21 with upregulated PTEN and PDCD4 expression in TNBC tumor cells but did not affect normal breast epithelial cells (Figures 6B, 6C, S4, and S5). As in our study on α9-nAChR,10 the expression level of miR-21 is lower in normal breast tissue compared with invasive BC.83,84,85 Thus, it is reasonable that 3WJ-B-α9-apt RNA-anti-miR-21 RNA nanoparticles have no adverse effect on normal breast epithelial cells. Importantly, no pathological changes were observed in normal organs of 3WJ-B-α9-apt-anti-miR-21 RNA nanoparticle-treated mice by biochemistry analysis of cardiac blood and histological examination of major organs. (Figure 6E; Table 1). These findings revealed that 3WJ-B-α9-apt-anti-miR-21 RNA nanoparticles accurately silenced miR-21 in TNBC tumors without affecting health.
In conclusion, we demonstrated that α9-nAChR RNA aptamers accurately bind and internalize into TNBC cells. The α9-nAChR RNA aptamers conjugated to 3WJ-pRNA motif core to form 3WJ-B-α9-apt RNA nanoparticle provide 3WJ-pRNA nanoparticles' functionality of high specific binding and rapid internalization into TNBC cells. Compared with 3WJ-Alexa RNA nanoparticles without α9-nAChR RNA aptamers, 3WJ-B-α9-apt-Alexa RNA nanoparticles effectively targeted TNBC-PDX tumors and caused more substantial tumor accumulation without spreading to other healthy organs in mice. Moreover, in vitro and in vivo therapeutic evaluation further found successful delivery of anti-miR-21 LNA for miR-21 inhibition to suppress TNBC tumor growth by 3WJ-B-α9-apt-anti-miR-21 RNA nanoparticle treatment. In summary, 3WJ-B-α9-apt RNA nanoparticles have specificity for α9-nAChR to deliver therapeutic miRNA into tumors, indicating their clinical potential for TNBC treatment.
Materials and methods
Cell line
MDAMB-231 (American Type Culture Collection [ATCC] HTB-26), SKBR-3(ATCC HTB-30), MCF-10A (ATCC CRL-10317), MCF-12A (ATCC CRL-10782), and 184A1 (ATCC CRL-8798) cells were purchased from the ATCC (VA, USA) and cultured in Dulbecco’s modified Eagle’s medium/nutrient mixture F-12 medium (DMEM/F12; Gibco, CA, USA) supplemented with 10% heat-inactivated fetal bovine serum (FBS; Gibco, Amarillo, TX, USA) and 50 U/mL penicillin/streptomycin/neomycin (PSN; Invitrogen, Waltham, MA, USA) in a 37°C humidified incubator under 5% CO2.
Design and source of RNA nanoparticles
All RNA nanoparticles were synthesized and bought from ExonanoRNA (Columbus, OH, USA) and its subsidiary ExonanoRNA (Fo-shan) Biomedicine. These 2′F-modified RNA nanoparticles included α9-apt-Alexa (190618-APT14-Alexa647, FR-353), 3WJ-B-α9-apt-Alexa (3WJ-Balpha9, RNP-F40), 3WJ-C-α9-apt-Alexa (3WJ-alpha9C, RNP-F41), 3WJ-Alexa(3WJsf5-FL, RNP-F44), 3WJ-B-α9-apt-anti-scr (3WJsf5-Balpha9-antiscr, RNP-Msc-14), and 3WJ-B-α9-apt-anti-miR-21 (3WJsf5-Balpha9-anti21, RNP-M21-20). Detailed information on each RNA sequence in these RNA nanoparticles is provided in Table S1. The final synthesized RNA nanoparticles were assembled in alpha9 aptamer binding buffer (145 mM NaCl, 5.3 mM KCl, 1.8 mM CaCl2, 1.7 mM MgCl2, 25 mM HEPES [pH 7.4]) by mixing each strand at the same molar concentration, heated to 95°C for 5 min, cooled slowly to 4°C over 40 min, and then vacuum dried. They can then be resuspended in DEPC-treated water for use.
Construction and characterization of assembled RNA nanoparticles
Stepwise assembly of the RNA nanoparticles was verified by loading for native 8% native polyacrylamide gel electrophoresis (PAGE) running in 1× TBE (89 mM Tris-borate, 2 mM EDTA) buffer, stained by ethidium bromide (EtBr), and visualized by Typhoon FLA 7000 (GE Healthcare) under a Cy5 channel.
The apparent hydrodynamic sizes and zeta potential of the assembled RNA nanoparticles (1 μM) in alpha9 aptamer binding buffer were measured by a Zetasizer Nano ZSP (Malvern Instruments, DKSH, Taipei, Taiwan) with a laser wavelength of 633 nm at 25°C.
Binding and internalization assay
For the cell binding assay, 3WJ-B-α9-apt-Alexa and 3WJ-C-α9-apt-Alexa RNA nanoparticles were incubated with MDA-MB-231 cells at 37°C for 1 h before analysis by flow cytometry. The contents of nanoparticles in live cells were detected with excitation at 640 nm and emission at 680 nm. MDA-MB-231 cells were seeded in a 3-cm glass Petri dish overnight to study internalization. Then, 3WJ-B-α9-apt-Alexa and 3WJ-B-α9-apt-Alexa RNA nanoparticles were added directly to cells in the live-cell system with a real-time laser confocal microscope (DMI 6000B CS, Leica, Wetzlar, Germany). Hochest 33342 was added for cell nucleus staining. The temperature was set to 37°C throughout the experiment in a 5% CO2 atmosphere, following the manufacturer’s instructions. Live-cell images were acquired synchronously using a 63× oil objective to observe internalization and cell entry with excitation at 640 nm and emission at 680 nm. Consecutive images were concatenated into video clips using PowerDirector software and uploaded to YouTube.
IF staining and FRET assay
Cells were first seeded on glass coverslips and cultured at 37°C overnight. Then, 3WJ-Alexa, 3WJ-B-α9-apt-Alexa, and 3WJ-C-α9-apt-Alexa RNA nanoparticles (10 nM) were added and incubated at 37°C for 1 h. After washing twice with PBS buffer, cells were fixed with 4% formaldehyde for 20 min and blocked for 30 min with 2% bovine serum albumin (BSA) at room temperature. Primary antibodies against CHRNA9 (PA5-46826, Thermo Fisher Scientific, IL, USA) were diluted 1:100 in PBS with 1% BSA and then incubated with the cells for 2 h at room temperature. Subsequent AffiniPure goat anti-rabbit rhodamine (111-025-144, Jackson ImmunoResearch, PA, USA) secondary antibodies were diluted at 1:50 and incubated for 1 h at room temperature. Samples were mounted with Vectashield Antifade Mounting Medium (H-1000, Vector Laboratories, CA, USA) and imaged via confocal microscopy (DMI 6000B CS, Leica). For FRET analysis, the photobleaching FRET method was established according to the manufacturer’s instructions (FRET Wizards in the Leica Application Suite) as described in our previous paper.64 After acquiring the pre-bleached fluorescence images of RNA nanoparticles and α9-nAChR-specific antibodies by 633- and 561-nm light lasers to excite fluorescent dyes, we then photobleached the field at 633 nm by high-intensity light laser (100%) for 1 min and obtained a set of post-bleached fluorescence images. The FRET efficiency can be estimated as follows: FRETeff= (Dpost − Dpre)/Dpost, where Dpre and Dpost represent the donor fluorescence intensity before and after photobleaching, respectively. The presence of FRET signals was eventually adjusted to FRET images within the FRET efficiency spectrum model based on the same background signals through the Leica TCS SP5 confocal spectral microscope imaging system.
RNA interference
An α9-nAChR shRNA plasmid were used to knockdown the α9-nAchR protein expression in MDA-MB-231 cells by electroporation transfection with a scrambled sequences of shRNA as a negative control. Primer sequences were inserted into Bgl II and Hin dIII-cut pSUPER vectors to generate the pSUPER-Si α9-nAChR and pSUPER-scramble plasmids. After trypsinization of cells, 1 × 106 cells were mixed with 5 μg pSUPER-Sh-CHRNA9 and pSUPER-Sc-CHRNA9 plasmids for electroporation. Two pulses were applied for 30 ms under a fixed voltage of 1.2 kV on an MP-100 pipette-type microporator (Digital Bio, Seoul, Korea). The transfected cells were seeded on glass coverslips for IF staining. The primer sequences were as follows: pSUPER-Sh-CHRNA9 sense sequence, 5′-GATCCCCCACCAATGTGGTCCTGCGGTTCAAGAGACCGCAGGACCACATTGGTGTTTTTA-3′; pSUPER-Sh-CHRNA9 antisense sequence, 5′-AGCTTAAAAACACCAATGTGGTCCTGCGGTCTCTTGAACCGCAGGACCACATTGGTGGGG-3′; pSUPER-Sc-CHRNA9 sense sequence, 5′-GATCCCCGAGAACACGAATACACCCCTTCAAGAGAGGGGTGTCGTGTTCTCTTTTTA-3′; pSUPER-Sc-CHRNA9 sense sequence antisense sequence, 5′-AGCTTAAAAAGAGAACACGAATACACCCCTCTCTTGAAGGGGTGTATTCGTGTTCTCGGG-3ʹ.
MTT assay
An MTT assay was carried out to assess the cytotoxic effect of nanoparticles. MDA-MB-231 cells were seeded at a density of 8,000 cells per well in 96-well plates and treated with 3WJ-B-α9-apt-anti-miR-21 and 3WJ-B-α9-apt-anti-scr RNA nanoparticles (0.01, 0.1, 1, 10, and 100 μM) for 48 h. Then, the MTT reagent (5 mg/mL) was added to the wells and incubated for 4 h at 37°C in the dark. The medium was removed, and 200 μL DMSO and 25 μL Sorensens’s glycine buffer were added to the wells for 15 min. The absorbance of the samples was measured using an ELISA microplate reader at a wavelength of 570 nm. Cell proliferation was calculated as the ratio of the absorbance of the samples to the absorbance of an untreated control. The results of the dose-response curve at different concentrations per drug were plotted in SigmaPlot software 12.0. (System Software, San Jose, CA, USA). The percentage of growth inhibition was calculated, and statistical analysis of IC50 values (the concentration of each drug that achieves 50% growth inhibition) was obtained graphically from the survival curve. All experiments were performed in triplicate wells for each condition.
Cell proliferation assay
Briefly, the cells were seeded at a density of 1 × 104 cells/mL in 6-well Petri dishes. Cells were treated with different concentrations of 3WJ-B-α9-apt-anti-miR-21 and 3WJ-B-α9-apt-anti-scr RNA nanoparticles (2.5, 5, and 7.5 nM) for 48 h. At 24 and 48 h, the treated cells were trypsinized, washed, and resuspended in a medium containing 0.4% trypan blue. The viable cells were counted under the microscope (DMI 4000B, Leica). The calculation was repeated in three independent experiments. Data were statistically analyzed by two-tailed unpaired t test and presented as mean ± SD (∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001).
Colony formation assay
MDA-MB-231 cells were seeded at 200 cells per 6-well Petri dish and maintained at 37°C. Cells were treated with 7.5 nM 3WJ-B-α9-apt-anti-miR-21 and 3WJ-B-α9-apt-anti-scr RNA nanoparticles for an additional 14 days, and we renewed these nanoparticles every 2 days. After treatment for 2 weeks, the visible colonies were fixed with 4% formaldehyde for 10 min and stained with crystal violet solution (0.05% crystal violet, 1% formaldehyde, 1% methanol, and 1× PBS) for 30 min. Images of the colonies were collected by microscope (DMI 4000B, Leica) and analyzed using ImageJ software. The calculation was repeated in three independent experiments. Data were statistically analyzed by two-tailed unpaired t test and presented as mean ± SD (∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001).
Establishment of the TNBC-PDX mouse model
NSG mice and PDX tumor-bearing mice (J000100674) were obtained from breeding pairs (JAX 4659679) originally purchased from Jackson Laboratories (005557, Bar Harbor, ME, USA). NSG mice were bred in a pathogen-free unit and maintained in sterile cages. Mice were handled and cared for with strict adherence to the guidelines established by the Animal Resource Center and following study protocols approved by the Laboratory Animal Center and Use Committee at National Defense Medical Center (NDMC-LAC) (IACUC protocol LAC- 20–264). PDX tumor specimens from PDX tumor-bearing mice (J000100674) were transplanted into female NSG mice. The animals were monitored for engraftment via routine palpation, and the tumors were harvested when they reached a volume of 0.5 cm3.
In vivo tumor therapeutic assessment
TNBC-PDX tumors were implanted in the fat pad area near the mammary gland tissue. When PDX tumors reached an average volume of 20 mm3, PDX tumor-bearing mice were randomly divided into three groups (n = 4, biologically independent animals). Each group was administered 3WJ-B-α9-apt-anti-miR-21 and 3WJ-B-α9-apt-anti-scr RNA nanoparticles (100 μL of 20 μmol/L) throughout 5 intravenous (i.v.) injections every other day. PBS-treated mice served as controls. During the experiment, the tumor size was measured using calipers every 2 days post injection for up to 19 days. The tumor volume was calculated as (length × width2)/2. On day 19, the mice were sacrificed, followed by tumor extraction. Data were statistically analyzed by two-tailed unpaired t test and presented as mean ± SD (∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001).
In vivo biodistribution assay
Female NSG mice with TNBC-PDX tumors (n = 3) were systemically administered 3WJ-Alexa, 3WJ-B-α9-apt-Alexa, and 3WJ-C-α9-apt-Alexa RNA nanoparticles (100 μL of 20 μmol/L) via tail vein injection. Mice were administered PBS as a negative fluorescence control. Whole-body fluorescence imaging of mice was conducted at time points of 2 h using an IVIS Spectrum (PerkinElmer) with excitation at 640 nm and emission at 680 nm to assess biodistribution profiles. The mice were sacrificed at 8 h post injection by CO2 inhalation, followed by cervical dislocation. Ex vivo fluorescence images for excised tumors and organs, including the heart, kidneys, liver, spleen, and lungs, were also obtained by the IVIS Spectrum (PerkinElmer) using the following excitation and emission filters: 640 and 680 nm, respectively. To maximize signal sensitivity, images were captured under the same field of view using automatic exposure corresponding to pixel binning of 8 and f/stop of 2 in the IVIS system program. Regions of interest (ROIs) were then drawn in the defined area and quantified in the physical, calibrated unit “average radiant efficiency [p/s/sr]/[μW/cm2]” using Living Image 4.3.1 software. The Living Image software can automatically normalize sensitivity differences resulting from different acquisition parameters without any user input when ROI values are expressed in a calibrated, physical unit. Data were statistically analyzed by two-tailed unpaired t test and presented as mean ± SD (∗p < 0.05, ∗∗p < 0.01, ∗∗∗∗p < 0.0001).
Quantitative real-time PCR
Total RNA, including miRNA, was isolated from PDX tumor tissues using TRIzol (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Reverse transcription was performed using the TaqMan MicroRNA Reverse Transcription Kit (4366596, Applied Biosystems, Life Technologies, Grand Island, NY, USA), and single-stranded cDNA was synthesized from total RNA using specific miRNA primers (4427975, Applied Biosystems, Life Technologies). Specific miRNA primers for miR-21 and U6 small nuclear RNA (snRNA) in the TaqMan miRNA assays (4427975-000397 and 4427975-001973) were also used in quantitative real-time PCR step. Next, quantitative PCR (qPCR) was carried out using the above primers with TaqMan Universal PCR Master Mix (4366596, Applied Biosystems), and the fluorescence intensity of the PCR product was detected using a LightCycler thermocycler (Roche Molecular Biochemicals, Mannheim, Germany). The expression of miR-21 in each sample was normalized to U6 snRNA as an endogenous control by the built-in software (Roche LightCycler v.4).
Western blot
For protein extraction, cells were washed twice with ice-cold PBS and lysed on ice in Golden lysis buffer (20 mM Tris-HCl [pH 8.0], 137 mM NaCl, 5.95 mM EDTA, 5 mM EGTA, 10 mM NaF, 1% Triton X-100, and 10% glycerol) supplemented with protease inhibitors (Roche, Indianapolis, IN, USA) and phosphatase inhibitors (Sigma-Aldrich, St. Louis, MO, USA), and the tumors tissues from the PDX model (n = 3) were homogenized three times at setting 3 (18,000 rpm) in Golden lysis buffer using a PRO 200 homogenizer (PRO Scientific, Monroe, CT, USA). Protein lysates (50 μg) were separated via 12% sodium dodecyl sulfate (SDS)-PAGE and transferred to polyvinylidene fluoride membranes. Specific antibodies against PTEN (1:800 dilution, ab31392, Abcam, CA, USA), PDCD-4 (1:1,000 dilution, GTX104901, GeneTex, Irvine, CA, USA), p21 (1:1,000 dilution, GTX629627, GeneTex), p27 (1:1,000 dilution, GTX100446, GeneTex), T-Akt (1:1,000 dilution, GTX629627, GeneTex), p-Akt (1:1,000 dilution, 9271, Cell Signaling Technology, Beverly, MA, USA), and GAPDH (sc-47724, Santa Cruz Biotechnology, Beverly, CA, USA) as a control were diluted 1:2,000 in Tris-buffered saline/Tween 20, and the membranes were incubated overnight at 4°C. Horseradish peroxidase-conjugated anti-mouse immunoglobulin G (IgG) (sc-2344, Santa Cruz Biotechnology) and anti-rabbit IgG (sc-2004, Santa Cruz Biotechnology) secondary antibodies were diluted 1:4,000 and incubated with the membranes for 1 h at room temperature.
Immunohistochemistry staining
Protein expression in PDX tumor tissues was detected using IHC. Paraffin-embedded PDX tumor tissues were cut into 8-μm sections and preincubated in 3% H2O2 and 0.3% Triton X-100 before microwaving for antigen retrieval. For detection of Ki-67 protein immunostaining, sections were microwaved in Tris buffer (pH 6) for 30 min. The antigenicity of the tumor cells in sections was blocked in 5% horse serum (Chemicon, Temecula, CA, USA) for 30 min and incubated with a diluted (1:200) Ki-67 (GTX16667, GeneTex)-specific antibody for 2 h at room temperature. Staining was developed using the streptavidin-biotin-peroxidase method and an LSAB 2 kit purchased from Dako (Carpinteria, CA, USA). Briefly, sections were washed in phosphate-buffered saline (PBS) and incubated with a biotinylated anti-rabbit secondary antibody. The samples were rewashed in the same buffer and incubated with a streptavidin-biotin-peroxidase complex. Staining was completed after incubation with substrate-chromogen solution. The incubation duration in solution with 3,3′-diaminobenzidine was determined using low-power microscopic inspection. Slides were washed, dehydrated, and coverslipped using a mixture of di-styrene, plasticizer, and xylene (DPX mounting medium) (44581, Sigma-Aldrich). Adjacent sections on the same slides were counterstained with hematoxylin for general histological orientation.
TUNEL assay
Apoptotic cells in PDX tumor tissues were detected using a TUNEL assay. The TUNEL assay was performed using the DeadEnd Colorimetric TUNEL System following the manufacturer’s instructions (Promega, Madison, WI, USA). 8-μm paraffin sections of PDX tumor tissues were de-paraffinized by xylene, rehydrated using serial ethanol concentrations, and then washed in PBS. After being washed in PBS, tissue sections were fixed in 4% paraformaldehyde and permeabilized using 20 μg/mL Proteinase K solution. For fragmented DNA labeling, tissue sections were incubated with equilibration buffer and TdT enzyme in a humidified chamber at 37°C for 60 min. They were subsequently put into pre-warmed working-strength stop-wash buffer for 10 min. Endogenous peroxidase of tissue sections was blocked by incubation in 3% hydrogen peroxide solution for 5 min, followed by washing in PBS. Briefly, staining was developed using streptavidin-conjugated HRP at room temperature for 30 min. Visualization of apoptotic cells was performed by application of substrate-chromogen (3,3′-diaminobenzidine) solution. Adjacent sections on the same slides were counterstained with hematoxylin for general histological orientation.
Biological toxicity assessment
After treated mice (n = 4) were sacrificed at the end of the experiment, the major organs, including the liver, spleen, kidneys, heart, lungs, and brain, were harvested, and sections were stained with H&E. Blood samples were harvested from each mouse by cardiac puncture and centrifugated at 1,200 × g for 10 min. Biochemical indicators in serum supernatant were further analyzed at the TMU hospital according to standard protocols using a Roche Cobas c702 analyzer, a quantitative, multiple-parameter, automated biochemistry analyzer supplied by Roche Diagnostics (Taiwan). Data were statistically analyzed by two-tailed unpaired t test and presented as mean ± SD (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001).
Statistics
Each experiment for each tested sample was repeated at least three times independently, and the results were presented as mean ± standard deviation (SD). Statistical differences were evaluated using an unpaired t test with SigmaPlot software, and p < 0.05 was considered statistically significant. No blinding or randomization was used. No samples or animals were excluded from the analysis. All criteria were pre-established.
Acknowledgments
This study was supported by the Health and Welfare Surcharge of Tobacco Products grant (MOHW110-TDU-B-212-144014) and by the Ministry of Science and Technology, Taiwan (MOST 110-2320-B-039-079) awarded to L.-C.C. and by the National Science and Technology Council, Taiwan (NSTC 111-2320-B-039-068, NSTC 112-2320-B-039-057 and MOST 111-2320-B-039-067) awarded to Y.-S.H.
Author contributions
L.-C.C. and Y.-S.H. conceived and supervised the project and acquired funding. Y.-C.L. and T.-C.C. conducted experimental work and data analyses. L.-C.C., J.C., P.G., and Y.-S.H. edited the manuscript. Y.-C.L., L.-C.C., and Y.-S.H. wrote the manuscript and compiled it with contributions from the other authors. All authors discussed the results and implications and reviewed the manuscript at all stages. All authors have read and agreed to the published version of the manuscript.
Declaration of interests
P.G. is the cofounder and chairman of the Board of Directors of ExonanoRNA, LLC and its subsidiary ExonanoRNA (Fo-shan) Biomedicine Co., Ltd.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2023.07.013.
Contributor Information
Peixuan Guo, Email: guo.1091@osu.edu.
Li-Ching Chen, Email: lcchen@mail.cmu.edu.tw.
Yuan-Soon Ho, Email: hoyuansn@mail.cmu.edu.tw.
Supplemental information
Data availability
The data supporting the findings of this study are available within the article and its supplemental information.
References
- 1.Siegel R.L., Miller K.D., Wagle N.S., Jemal A. Cancer statistics, 2023. CA A Cancer J. Clin. 2023;73:17–48. doi: 10.3322/caac.21763. [DOI] [PubMed] [Google Scholar]
- 2.Foulkes W.D., Smith I.E., Reis-Filho J.S. Triple-Negative Breast Cancer. N. Engl. J. Med. 2010;363:1938–1948. doi: 10.1056/NEJMra1001389. [DOI] [PubMed] [Google Scholar]
- 3.Derakhshan F., Reis-Filho J.S. Pathogenesis of Triple-Negative Breast Cancer. Annu. Rev. Pathol. 2022;17:181–204. doi: 10.1146/annurev-pathol-042420-093238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Reiner A.S., Watt G.P., John E.M., Lynch C.F., Brooks J.D., Mellemkjær L., Boice J.D., Knight J.A., Concannon P., Smith S.A., et al. Smoking, Radiation Therapy, and Contralateral Breast Cancer Risk in Young Women. J. Natl. Cancer Inst. 2022;114:631–634. doi: 10.1093/jnci/djab047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dai X., Gil G.F., Reitsma M.B., Ahmad N.S., Anderson J.A., Bisignano C., Carr S., Feldman R., Hay S.I., He J., et al. Health effects associated with smoking: a Burden of Proof study. Nat. Med. 2022;28:2045–2055. doi: 10.1038/s41591-022-01978-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He Y., Si Y., Li X., Hong J., Yu C., He N. The relationship between tobacco and breast cancer incidence: A systematic review and meta-analysis of observational studies. Front. Oncol. 2022;12:961970. doi: 10.3389/fonc.2022.961970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones M.E., Schoemaker M.J., Wright L.B., Ashworth A., Swerdlow A.J. Smoking and risk of breast cancer in the Generations Study cohort. Breast Cancer Res. 2017;19:118. doi: 10.1186/s13058-017-0908-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grando S.A. Connections of nicotine to cancer. Nat. Rev. Cancer. 2014;14:419–429. doi: 10.1038/nrc3725. [DOI] [PubMed] [Google Scholar]
- 9.Schuller H.M. Is cancer triggered by altered signalling of nicotinic acetylcholine receptors? Nat. Rev. Cancer. 2009;9:195–205. doi: 10.1038/nrc2590. [DOI] [PubMed] [Google Scholar]
- 10.Lee C.-H., Huang C.-S., Chen C.-S., Tu S.-H., Wang Y.-J., Chang Y.-J., Tam K.-W., Wei P.-L., Cheng T.-C., Chu J.-S., et al. Overexpression and Activation of the α9-Nicotinic Receptor During Tumorigenesis in Human Breast Epithelial Cells. J. Natl. Cancer Inst. 2010;102:1322–1335. doi: 10.1093/jnci/djq300. [DOI] [PubMed] [Google Scholar]
- 11.Wu C.-H., Lee C.-H., Ho Y.-S. Nicotinic Acetylcholine Receptor-Based Blockade: Applications of Molecular Targets for Cancer Therapy. Clin. Cancer Res. 2011;17:3533–3541. doi: 10.1158/1078-0432.CCR-10-2434. [DOI] [PubMed] [Google Scholar]
- 12.Huang L.-C., Lin C.-L., Qiu J.-Z., Lin C.-Y., Hsu K.-W., Tam K.-W., Lee J.-Y., Yang J.-M., Lee C.-H. Nicotinic Acetylcholine Receptor Subtype Alpha-9 Mediates Triple-Negative Breast Cancers Based on a Spontaneous Pulmonary Metastasis Mouse Model. Front. Cell. Neurosci. 2017;11:336. doi: 10.3389/fncel.2017.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sharp P.A. The Centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 14.Licatalosi D.D., Darnell R.B. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breaker R.R. Natural and engineered nucleic acids as tools to explore biology. Nature. 2004;432:838–845. doi: 10.1038/nature03195. [DOI] [PubMed] [Google Scholar]
- 16.Dykstra P.B., Kaplan M., Smolke C.D. Engineering synthetic RNA devices for cell control. Nat. Rev. Genet. 2022;23:215–228. doi: 10.1038/s41576-021-00436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Isaacs F.J., Dwyer D.J., Collins J.J. RNA synthetic biology. Nat. Biotechnol. 2006;24:545–554. doi: 10.1038/nbt1208. [DOI] [PubMed] [Google Scholar]
- 18.Yu A.-M., Choi Y.H., Tu M.-J. RNA Drugs and RNA Targets for Small Molecules: Principles, Progress, and Challenges. Pharmacol. Rev. 2020;72:862–898. doi: 10.1124/pr.120.019554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Childs-Disney J.L., Yang X., Gibaut Q.M.R., Tong Y., Batey R.T., Disney M.D. Targeting RNA structures with small molecules. Nat. Rev. Drug Discov. 2022;21:736–762. doi: 10.1038/s41573-022-00521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo P., Zhang C., Chen C., Garver K., Trottier M. Inter-RNA Interaction of Phage φ29 pRNA to Form a Hexameric Complex for Viral DNA Transportation. Mol. Cell. 1998;2:149–155. doi: 10.1016/s1097-2765(00)80124-0. [DOI] [PubMed] [Google Scholar]
- 21.Guo P. The emerging field of RNA nanotechnology. Nat. Nanotechnol. 2010;5:833–842. doi: 10.1038/nnano.2010.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shu Y., Pi F., Sharma A., Rajabi M., Haque F., Shu D., Leggas M., Evers B.M., Guo P. Stable RNA nanoparticles as potential new generation drugs for cancer therapy. Adv. Drug Deliv. Rev. 2014;66:74–89. doi: 10.1016/j.addr.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jasinski D., Haque F., Binzel D.W., Guo P. Advancement of the Emerging Field of RNA Nanotechnology. ACS Nano. 2017;11:1142–1164. doi: 10.1021/acsnano.6b05737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X., Bhullar A.S., Binzel D.W., Guo P. The dynamic, motile and deformative properties of RNA nanoparticles facilitate the third milestone of drug development. Adv. Drug Deliv. Rev. 2022;186:114316. doi: 10.1016/j.addr.2022.114316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Panigaj M., Johnson M.B., Ke W., McMillan J., Goncharova E.A., Chandler M., Afonin K.A. Aptamers as Modular Components of Therapeutic Nucleic Acid Nanotechnology. ACS Nano. 2019;13:12301–12321. doi: 10.1021/acsnano.9b06522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Afonin K.A., Dobrovolskaia M.A., Ke W., Grodzinski P., Bathe M. Critical review of nucleic acid nanotechnology to identify gaps and inform a strategy for accelerated clinical translation. Adv. Drug Deliv. Rev. 2022;181:114081. doi: 10.1016/j.addr.2021.114081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rolband L., Beasock D., Wang Y., Shu Y.-G., Dinman J.D., Schlick T., Zhou Y., Kieft J.S., Chen S.-J., Bussi G., et al. Biomotors, viral assembly, and RNA nanobiotechnology: Current achievements and future directions. Comput. Struct. Biotechnol. J. 2022;20:6120–6137. doi: 10.1016/j.csbj.2022.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Afonin K.A., Dobrovolskaia M.A., Church G., Bathe M. Opportunities, Barriers, and a Strategy for Overcoming Translational Challenges to Therapeutic Nucleic Acid Nanotechnology. ACS Nano. 2020;14:9221–9227. doi: 10.1021/acsnano.0c04753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panigaj M., Skelly E., Beasock D., Marriott I., Johnson M.B., Salotti J., Afonin K.A. Therapeutic immunomodulation by rationally designed nucleic acids and nucleic acid nanoparticles. Front. Immunol. 2023;14:1053550. doi: 10.3389/fimmu.2023.1053550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weng Y., Huang Q., Li C., Yang Y., Wang X., Yu J., Huang Y., Liang X.-J. Improved Nucleic Acid Therapy with Advanced Nanoscale Biotechnology. Mol. Ther. Nucleic Acids. 2020;19:581–601. doi: 10.1016/j.omtn.2019.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 32.Tuerk C., Gold L. Systematic Evolution of Ligands by Exponential Enrichment: RNA Ligands to Bacteriophage T4 DNA Polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 33.Ulrich H., Ippolito J.E., Pagán O.R., Eterović V.A., Hann R.M., Shi H., Lis J.T., Eldefrawi M.E., Hess G.P. In vitro selection of RNA molecules that displace cocaine from the membrane-bound nicotinic acetylcholine receptor. Proc. Natl. Acad. Sci. USA. 1998;95:14051–14056. doi: 10.1073/pnas.95.24.14051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hess G.P., Ulrich H., Breitinger H.-G., Niu L., Gameiro A.M., Grewer C., Srivastava S., Ippolito J.E., Lee S.M., Jayaraman V., Coombs S.E. Mechanism-based discovery of ligands that counteract inhibition of the nicotinic acetylcholine receptor by cocaine and MK-801. Proc. Natl. Acad. Sci. USA. 2000;97:13895–13900. doi: 10.1073/pnas.240459497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sivaprakasam K., Pagán O.R., Hess G.P. Minimal RNA Aptamer Sequences That Can Inhibit or Alleviate Noncompetitive Inhibition of the Muscle-Type Nicotinic Acetylcholine Receptor. J. Membr. Biol. 2010;233:1–12. doi: 10.1007/s00232-009-9215-z. [DOI] [PubMed] [Google Scholar]
- 36.Zhou J., Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shu D., Shu Y., Haque F., Abdelmawla S., Guo P. Thermodynamically stable RNA three-way junction for constructing multifunctional nanoparticles for delivery of therapeutics. Nat. Nanotechnol. 2011;6:658–667. doi: 10.1038/nnano.2011.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Binzel D.W., Li X., Burns N., Khan E., Lee W.-J., Chen L.-C., Ellipilli S., Miles W., Ho Y.S., Guo P. Thermostability, Tunability, and Tenacity of RNA as Rubbery Anionic Polymeric Materials in Nanotechnology and Nanomedicine—Specific Cancer Targeting with Undetectable Toxicity. Chem. Rev. 2021;121:7398–7467. doi: 10.1021/acs.chemrev.1c00009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shu D., Khisamutdinov E.F., Zhang L., Guo P. Programmable folding of fusion RNA in vivo and in vitro driven by pRNA 3WJ motif of phi29 DNA packaging motor. Nucleic Acids Res. 2014;42:e10. doi: 10.1093/nar/gkt885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang Y., Leonard M., Shu Y., Yang Y., Shu D., Guo P., Zhang X. Overcoming Tamoxifen Resistance of Human Breast Cancer by Targeted Gene Silencing Using Multifunctional pRNA Nanoparticles. ACS Nano. 2017;11:335–346. doi: 10.1021/acsnano.6b05910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shu D., Li H., Shu Y., Xiong G., Carson W.E., III, Haque F., Xu R., Guo P. Systemic Delivery of Anti-miRNA for Suppression of Triple Negative Breast Cancer Utilizing RNA Nanotechnology. ACS Nano. 2015;9:9731–9740. doi: 10.1021/acsnano.5b02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pi F., Zhang H., Li H., Thiviyanathan V., Gorenstein D.G., Sood A.K., Guo P. RNA nanoparticles harboring annexin A2 aptamer can target ovarian cancer for tumor-specific doxorubicin delivery. Nanomedicine. 2017;13:1183–1193. doi: 10.1016/j.nano.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Binzel D.W., Shu Y., Li H., Sun M., Zhang Q., Shu D., Guo B., Guo P. Specific Delivery of MiRNA for High Efficient Inhibition of Prostate Cancer by RNA Nanotechnology. Mol. Ther. 2016;24:1267–1277. doi: 10.1038/mt.2016.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yin H., Xiong G., Guo S., Xu C., Xu R., Guo P., Shu D. Delivery of Anti-miRNA for Triple-Negative Breast Cancer Therapy Using RNA Nanoparticles Targeting Stem Cell Marker CD133. Mol. Ther. 2019;27:1252–1261. doi: 10.1016/j.ymthe.2019.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pang H.-H., Huang C.-Y., Chen P.-Y., Li N.-S., Hsu Y.-P., Wu J.-K., Fan H.-F., Wei K.-C., Yang H.-W. Bioengineered Bacteriophage-Like Nanoparticles as RNAi Therapeutics to Enhance Radiotherapy against Glioblastomas. ACS Nano. 2023;17:10407–10422. doi: 10.1021/acsnano.3c01102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo L., Shi D., Shang M., Sun X., Meng D., Liu X., Zhou X., Li J. Utilizing RNA nanotechnology to construct negatively charged and ultrasound-responsive nanodroplets for targeted delivery of siRNA. Drug Deliv. 2022;29:316–327. doi: 10.1080/10717544.2022.2026532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang L., Mu C., Zhang T., Yang D., Wang C., Chen Q., Tang L., Fan L., Liu C., Shen J., Li H. Development of targeted therapy therapeutics to sensitize triple-negative breast cancer chemosensitivity utilizing bacteriophage phi29 derived packaging RNA. J. Nanobiotechnol. 2021;19:13. doi: 10.1186/s12951-020-00758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pang L., Shah H., Wang H., Shu D., Qian S.Y., Sathish V. EpCAM-Targeted 3WJ RNA Nanoparticle Harboring Delta-5-Desaturase siRNA Inhibited Lung Tumor Formation via DGLA Peroxidation. Mol. Ther. Nucleic Acids. 2020;22:222–235. doi: 10.1016/j.omtn.2020.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 50.Zhu S., Wu H., Wu F., Nie D., Sheng S., Mo Y.-Y. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 51.Meng F., Henson R., Wehbe-Janek H., Ghoshal K., Jacob S.T., Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gaur A.B., Holbeck S.L., Colburn N.H., Israel M.A. Downregulation of Pdcd4 by mir-21 facilitates glioblastoma proliferation in vivo. Neuro Oncol. 2011;13:580–590. doi: 10.1093/neuonc/nor033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kwak G., Kim H., Park J., Kim E.H., Jang H., Han G., Wang S.Y., Yang Y., Chan Kwon I., Kim S.H. A Trojan-Horse Strategy by In Situ Piggybacking onto Endogenous Albumin for Tumor-Specific Neutralization of Oncogenic MicroRNA. ACS Nano. 2021;15:11369–11384. doi: 10.1021/acsnano.1c00799. [DOI] [PubMed] [Google Scholar]
- 54.Pulito C., Mori F., Sacconi A., Goeman F., Ferraiuolo M., Pasanisi P., Campagnoli C., Berrino F., Fanciulli M., Ford R.J., et al. Metformin-induced ablation of microRNA 21-5p releases Sestrin-1 and CAB39L antitumoral activities. Cell Discov. 2017;3:17022. doi: 10.1038/celldisc.2017.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Medina P.P., Nolde M., Slack F.J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature. 2010;467:86–90. doi: 10.1038/nature09284. [DOI] [PubMed] [Google Scholar]
- 56.Winkle M., El-Daly S.M., Fabbri M., Calin G.A. Noncoding RNA therapeutics — challenges and potential solutions. Nat. Rev. Drug Discov. 2021;20:629–651. doi: 10.1038/s41573-021-00219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Obad S., dos Santos C.O., Petri A., Heidenblad M., Broom O., Ruse C., Fu C., Lindow M., Stenvang J., Straarup E.M., et al. Silencing of microRNA families by seed-targeting tiny LNAs. Nat. Genet. 2011;43:371–378. doi: 10.1038/ng.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang T., Wu Y., Yang D., Wu C., Li H. Preparation, characterization, and in vitro tumor-suppressive effect of anti-miR-21-equipped RNA nanoparticles. Biochem. Biophys. Res. Commun. 2021;558:107–113. doi: 10.1016/j.bbrc.2021.04.040. [DOI] [PubMed] [Google Scholar]
- 59.Yoo J.Y., Yeh M., Kaur B., Lee T.J. Targeted delivery of small noncoding RNA for glioblastoma. Cancer Lett. 2021;500:274–280. doi: 10.1016/j.canlet.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee T.J., Yoo J.Y., Shu D., Li H., Zhang J., Yu J.-G., Jaime-Ramirez A.C., Acunzo M., Romano G., Cui R., et al. RNA Nanoparticle-Based Targeted Therapy for Glioblastoma through Inhibition of Oncogenic miR-21. Mol. Ther. 2017;25:1544–1555. doi: 10.1016/j.ymthe.2016.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bianchini G., De Angelis C., Licata L., Gianni L. Treatment landscape of triple-negative breast cancer — expanded options, evolving needs. Nat. Rev. Clin. Oncol. 2022;19:91–113. doi: 10.1038/s41571-021-00565-2. [DOI] [PubMed] [Google Scholar]
- 62.Mucchietto V., Fasoli F., Pucci S., Moretti M., Benfante R., Maroli A., Di Lascio S., Bolchi C., Pallavicini M., Dowell C., et al. α9- and α7-containing receptors mediate the pro-proliferative effects of nicotine in the A549 adenocarcinoma cell line. Br. J. Pharmacol. 2018;175:1957–1972. doi: 10.1111/bph.13954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nguyen H.D., Liao Y.-C., Ho Y.-S., Chen L.-C., Chang H.-W., Cheng T.-C., Liu D., Lee W.-R., Shen S.-C., Wu C.-H., Tu S.H. The α9 Nicotinic Acetylcholine Receptor Mediates Nicotine-Induced PD-L1 Expression and Regulates Melanoma Cell Proliferation and Migration. Cancers. 2019;11:1991. doi: 10.3390/cancers11121991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin C.-Y., Lee C.-H., Chuang Y.-H., Lee J.-Y., Chiu Y.-Y., Wu Lee Y.H., Jong Y.J., Hwang J.K., Huang S.H., Chen L.C., et al. Membrane protein-regulated networks across human cancers. Nat. Commun. 2019;10:3131. doi: 10.1038/s41467-019-10920-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pucci S., Zoli M., Clementi F., Gotti C. α9-Containing Nicotinic Receptors in Cancer. Front. Cell. Neurosci. 2022;15:805123. doi: 10.3389/fncel.2021.805123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tyagi A., Sharma S., Wu K., Wu S.-Y., Xing F., Liu Y., Zhao D., Deshpande R.P., D’Agostino R.B., Watabe K. Nicotine promotes breast cancer metastasis by stimulating N2 neutrophils and generating pre-metastatic niche in lung. Nat. Commun. 2021;12:474. doi: 10.1038/s41467-020-20733-9. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 67.Sun Z., Bao J., Zhangsun M., Dong S., Zhangsun D., Luo S. αO-Conotoxin GeXIVA Inhibits the Growth of Breast Cancer Cells via Interaction with α9 Nicotine Acetylcholine Receptors. Mar. Drugs. 2020;18:195. doi: 10.3390/md18040195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee K.-L., Kuo Y.-C., Ho Y.-S., Huang Y.-H. Triple-Negative Breast Cancer: Current Understanding and Future Therapeutic Breakthrough Targeting Cancer Stemness. Cancers. 2019;11:1334. doi: 10.3390/cancers11091334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sivaprakasam K., Hess G.P. In Vitro Selection of Short DNA Aptamers that Can Inhibit or Alleviate Cocaine and MK-801 Inhibition of Muscle-Type Nicotinic Acetylcholine Receptors. J. Membr. Biol. 2022;255:41–53. doi: 10.1007/s00232-021-00202-0. [DOI] [PubMed] [Google Scholar]
- 70.Wijesinghe K.M., Sabbih G., Algama C.H., Syed R., Danquah M.K., Dhakal S. FRET-Based Single-Molecule Detection of Pathogen Protein IsdA Using Computationally Selected Aptamers. Anal. Chem. 2023;95:9839–9846. doi: 10.1021/acs.analchem.3c00717. [DOI] [PubMed] [Google Scholar]
- 71.Sampedro Vallina N., McRae E.K.S., Hansen B.K., Boussebayle A., Andersen E.S. RNA origami scaffolds facilitate cryo-EM characterization of a Broccoli–Pepper aptamer FRET pair. Nucleic Acids Res. 2023;51:4613–4624. doi: 10.1093/nar/gkad224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Piao X., Wang H., Binzel D.W., Guo P. Assessment and comparison of thermal stability of phosphorothioate-DNA, DNA, RNA, 2'-F RNA, and LNA in the context of Phi29 pRNA 3WJ. RNA. 2018;24:67–76. doi: 10.1261/rna.063057.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Li H., Lee T., Dziubla T., Pi F., Guo S., Xu J., Li C., Haque F., Liang X.-J., Guo P. RNA as a stable polymer to build controllable and defined nanostructures for material and biomedical applications. Nano Today. 2015;10:631–655. doi: 10.1016/j.nantod.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shu Y., Shu D., Haque F., Guo P. Fabrication of pRNA nanoparticles to deliver therapeutic RNAs and bioactive compounds into tumor cells. Nat. Protoc. 2013;8:1635–1659. doi: 10.1038/nprot.2013.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pi F., Binzel D.W., Lee T.J., Li Z., Sun M., Rychahou P., Li H., Haque F., Wang S., Croce C.M., et al. Nanoparticle orientation to control RNA loading and ligand display on extracellular vesicles for cancer regression. Nat. Nanotechnol. 2018;13:82–89. doi: 10.1038/s41565-017-0012-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kelly L., Maier K.E., Yan A., Levy M. A comparative analysis of cell surface targeting aptamers. Nat. Commun. 2021;12:6275. doi: 10.1038/s41467-021-26463-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kara G., Calin G.A., Ozpolat B. RNAi-based therapeutics and tumor targeted delivery in cancer. Adv. Drug Deliv. Rev. 2022;182:114113. doi: 10.1016/j.addr.2022.114113. [DOI] [PubMed] [Google Scholar]
- 78.Guo S., Li H., Ma M., Fu J., Dong Y., Guo P. Size, Shape, and Sequence-Dependent Immunogenicity of RNA Nanoparticles. Mol. Ther. Nucleic Acids. 2017;9:399–408. doi: 10.1016/j.omtn.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chi L.H., Cross R.S.N., Redvers R.P., Davis M., Hediyeh-zadeh S., Mathivanan S., Samuel M., Lucas E.C., Mouchemore K., Gregory P.A., et al. MicroRNA-21 is immunosuppressive and pro-metastatic via separate mechanisms. Oncogenesis. 2022;11:38. doi: 10.1038/s41389-022-00413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.MacKenzie T.A., Schwartz G.N., Calderone H.M., Graveel C.R., Winn M.E., Hostetter G., Wells W.A., Sempere L.F. Stromal expression of miR-21 identifies high-risk group in triple-negative breast cancer. Am. J. Pathol. 2014;184:3217–3225. doi: 10.1016/j.ajpath.2014.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kanagasabai T., Li G., Shen T.H., Gladoun N., Castillo-Martin M., Celada S.I., Xie Y., Brown L.K., Mark Z.A., Ochieng J., et al. MicroRNA-21 deficiency suppresses prostate cancer progression through downregulation of the IRS1-SREBP-1 signaling pathway. Cancer Lett. 2022;525:46–54. doi: 10.1016/j.canlet.2021.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Devulapally R., Sekar N.M., Sekar T.V., Foygel K., Massoud T.F., Willmann J.K., Paulmurugan R. Polymer nanoparticles mediated codelivery of antimiR-10b and antimiR-21 for achieving triple negative breast cancer therapy. ACS Nano. 2015;9:2290–2302. doi: 10.1021/nn507465d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yan L.-X., Huang X.-F., Shao Q., Huang M.-Y., Deng L., Wu Q.-L., Zeng Y.-X., Shao J.-Y. MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA. 2008;14:2348–2360. doi: 10.1261/rna.1034808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Si M.L., Zhu S., Wu H., Lu Z., Wu F., Mo Y.Y. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- 85.Iorio M.V., Ferracin M., Liu C.-G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M., et al. MicroRNA Gene Expression Deregulation in Human Breast Cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available within the article and its supplemental information.