Preclinical models of germline BRCA–associated pancreatic ductal adenocarcinoma (glBRCA PDAC) that reflect different clinical time points along the response spectrum of platinum/PARP inhibitor treatment were developed and used to evaluate mechanisms of resistance to these therapies.
Abstract
Germline BRCA–associated pancreatic ductal adenocarcinoma (glBRCA PDAC) tumors are susceptible to platinum and PARP inhibition. The clinical outcomes of 125 patients with glBRCA PDAC were stratified based on the spectrum of response to platinum/PARP inhibition: (i) refractory [overall survival (OS) <6 months], (ii) durable response followed by acquired resistance (OS <36 months), and (iii) long-term responders (OS >36 months). Patient-derived xenografts (PDX) were generated from 25 patients with glBRCA PDAC at different clinical time points. Response to platinum/PARP inhibition in vivo and ex vivo culture (EVOC) correlated with clinical response. We deciphered the mechanisms of resistance in glBRCA PDAC and identified homologous recombination (HR) proficiency and secondary mutations restoring partial functionality as the most dominant resistant mechanism. Yet, a subset of HR-deficient (HRD) patients demonstrated clinical resistance. Their tumors displayed basal-like molecular subtype and were more aneuploid. Tumor mutational burden was high in HRD PDAC and significantly higher in tumors with secondary mutations. Anti–PD-1 attenuated tumor growth in a novel humanized glBRCA PDAC PDX model. This work demonstrates the utility of preclinical models, including EVOC, to predict the response of glBRCA PDAC to treatment, which has the potential to inform time-sensitive medical decisions.
Significance:
glBRCA PDAC has a favorable response to platinum/PARP inhibition. However, most patients develop resistance. Additional treatment options for this unique subpopulation are needed. We generated model systems in PDXs and an ex vivo system (EVOC) that faithfully recapitulate these specific clinical scenarios as a platform to investigate the mechanisms of resistance for further drug development.
This article is highlighted in the In This Issue feature, p. 1749
INTRODUCTION
Pancreatic ductal adenocarcinoma (PDAC) is a dismal disease, with the majority of patients diagnosed at an advanced, metastatic stage. Combination regimens such as FOLFIRINOX or gemcitabine and nab-paclitaxel have improved patient outcomes, yet the median overall survival (mOS) for patients with advanced disease remains less than 1 year (1, 2).
Comprehensive genomic analysis has identified an unstable PDAC genome subtype associated with genomic alterations in DNA damage repair (DDR) genes. This group is predominantly enriched in patients harboring germline BRCA1/2 (glBRCA) mutations. BRCA1/2 proteins are key mediators of the DDR pathway, including DNA double-strand break repair via homologous recombination (HR; ref. 3). The frequency of glBRCA mutations is 4.6% to 6.2% in most PDAC cases, and 10% to 15% in high-risk populations (e.g., Ashkenazi Jewish; refs. 4, 5).
PDAC tumors with glBRCA mutations display enhanced response to platinum-based therapy and PARP inhibitors (PARPi; refs. 4, 6). Importantly, glBRCA is one of the first biomarker-driven PDAC subtypes to be targeted in a phase III trial (7). Patients with glBRCA PDAC treated in the PARPi maintenance setting following systemic therapy with platinum analogues demonstrated improved progression-free survival (PFS; ref. 7). Indeed, the PARPi olaparib has recently received FDA and European Medicines Agency approval for maintenance therapy for platinum-sensitive stage IV BRCA-associated pancreatic cancer. Despite these promising results, patients with glBRCA PDAC display a spectrum of responses to platinum-based and/or PARPi treatment; a subset of patients are refractory to first-line platinum-based therapy, most patients are initially responsive but eventually develop resistance, and a third group demonstrates complete response or stable oligometastatic disease for over 36 months (8). However, the mechanisms that underlie these differing responses are unknown.
Preclinical models that recapitulate specific clinical scenarios and the pathologic, cellular, and molecular properties of human PDAC have been well described (8). For example, patient-derived xenograft (PDX) models from glBRCA PDAC patient tumors sustain a high mutational load and may serve as surrogate model systems (9, 10). Moreover, PDX models have been shown to be predictive of drug sensitivity and to correlate with clinical scenarios. In order to model the response to treatments in an alternative ex vivo system that retains the architecture, tumor–stromal interactions, and genetic heterogeneity of PDAC (11), we chose to utilize an ex vivo organ culture (EVOC) system that has been optimized for culturing tumors ex vivo for 5 or more days (12). The EVOC method enables testing of drug efficacy on tumor slices from both human biopsies and PDXs.
In this study, we aimed to recapitulate the glBRCA PDAC population spectrum of responses to platinum and PARPi therapy, to explore the genomic and clinical features that may lead to different responses, and to investigate the mechanisms of resistance.
RESULTS
Clinical Characteristics
The Sheba Medical Center (SMC) PDAC patient cohort harboring glBRCA mutations included a total of 125 patients. Germline mutational distribution was as follows: BRCA1 n = 43 (34.4%), BRCA2 n = 81 (64.8%), with one patient harboring both BRCA1 and BRCA2 germline mutations. Thirty-six patients (28.8%) had previous malignancy, with 72.2% BRCA-associated tumors. Moreover, 94.4% of patients had a family history of malignancy—the majority of those BRCA-associated tumors (79.6%)—and 17.8% had a family history of PDAC (Table 1).
Table 1.
glBRCA PDAC clinical characterization
| Characteristics | n = 125 (%) |
|---|---|
| Age at diagnosis | |
| Mean ± SD | 66 ± 12 |
| Range | 26–88 |
| Sex | |
| Male | 75 (59.5) |
| Female | 50 (40.5) |
| AJCC stage at diagnosis | |
| I/II | 47 (37.6) |
| III | 11 (8.8) |
| IV | 66 (52.8) |
| UNK | 1 (0.8) |
| Tumor location | |
| Head | 63 (50.4) |
| Body | 18 (14.4) |
| Tail | 28 (22.4) |
| Head + body/body + tail | 4 (3.2)/7 (5.6) |
| Multiple | 2 (1.6) |
| UNK | 3 (2.4) |
| Mutation distribution | |
| BRCA1 | 43 (34.4) |
| 185delAG | 29 (67.4) |
| 5285insC | 7 (16.3) |
| Other | 7 (16.3) |
| BRCA2 | 81 (64.8) |
| 6174delT | 60 (74.1) |
| Other | 21 (25.9) |
| BRCA1 + BRCA2 | 1 (0.008) |
| Personal history of malignancy | 36 (28.8) |
| BRCA-associated malignancya | 26 (72.2) |
| Family history of malignancy | 118 (94.4) |
| BRCA-associated malignancy | 94 (79.6) |
| PDAC | 21 (17.8) |
| Treatments (all stages) | |
| Plt/PARPi | 119 (95.2) |
| Plt only | 56 (44.8) |
| PARPi only | 3 (2.4) |
| Other/no treatments | 6 (4.8) |
| Immunotherapy | 6 (4.8) |
Abbreviations: AJCC, American Joint Committee on Cancer; PARPi, PARP inhibitor; Plt, platinum therapy; UNK, unknown.
aBRCA-associated malignancies include breast, ovarian, prostate, and pancreatic cancers.
Spectrum of Response of glBRCA PDAC to Platinum and PARPi Treatment
We explored the clinical response and OS of 125 patients with glBRCA-mutated PDAC treated at the SMC over a 12-year period (2010–2022). Ninety-five percent of patients were exposed to standard-of-care platinum-based treatment during their disease (neoadjuvant/adjuvant/1–5 lines), with 82.5% of patients with metastatic disease treated with platinum as first-line treatment. Fifty-seven metastatic patients (58.8%) were treated with PARPi. Six metastatic patients (6.2%) were treated with immune checkpoint inhibitors (Table 1).
Among all patients who were treated for metastatic disease (at diagnosis or from recurrence; n = 97), the mOS was 13 months [range, 0–138 months (95% confidence interval [CI], 15.84–24.76)]. At the time of analysis (censor date), 11 patients (11.3%) were responding to platinum/PARPi treatment and excluded from categorization. We classified clinical response into three groups using the following definitions:
Refractory: patients who demonstrated short-term or lack of response to platinum agents, with OS ≤6 months [mOS = 3 months; range, 0–6 months, n = 20 (20.6%)].
Acquired resistance: patients with durable response to platinum agents and PARPi maintenance treatment, followed by the emergence of clinical resistance (n = 54; 55.7%). mOS for this group was 14 months (range, 7–36 months).
Long-term responders: patients responding to platinum and maintenance PARPi for >36 months (n = 12; 12.3%). mOS for this unique population was 68 months (range, 43–138 months). Nine patients had no evidence of disease for over 4 years.
The mOS for all metastatic patients, excluding the refractory group, was 18 months [range, 7–138 (95% CI, 20.06–30.05)]. The clinical response of the metastatic patients (stage IV at the time of diagnosis/recurrence) is presented in a swimmer plot (Fig. 1).
Figure 1.
Spectrum of response for patients with stage IV glBRCA-mutated PDAC. Swimmer plot of glBRCA patients with stage IV PDAC: time from diagnosis until last follow-up or death, with time from diagnosis until last follow-up or death. Blue: time on platinum/PARPi treatment. Red: time off platinum/PARPi treatment. Green: time from last platinum/PARPi treatment to censor date. Red infinity symbol: patients with complete response. Black arrow: patients alive at censor date. Inset, Kaplan–Meier curves of patients by clinical subgroup (red, refractory; gray, acquired resistance; green, long-term responders). Plt/PARPi tx, platinum/PARPi treatment.
The long-term responding cohort was enriched with patients who had a significant deep response to platinum-based chemotherapy (mostly FOLFIRINOX). Additionally, most of these patients had oligometastatic disease, predominantly lung or liver metastasis at the initiation of PARPi maintenance therapy. These data exemplify the spectrum of response to platinum and PARPi therapy of glBRCA PDAC patients.
To explore the mechanisms underlying the varying responses, we established PDX models that recapitulate the heterogeneous nature of glBRCA PDAC tumors and performed whole-genome sequencing (WGS) and RNA sequencing (RNA-seq; refs. 8, 13).
glBRCA-Mutated PDAC PDX Models
Twenty-five PDX models from 24 different patients with glBRCA PDAC at different clinical time points were established as previously described (9, 13). From the glBRCA cohort, we transplanted 48 tumor samples, of which 25 (54%) engrafted to establish PDX models. Tissue origin of established tumors was as follows: primary tumors, liver metastases, peritoneum, and ascites malignant fluid (Supplementary Fig. S1A). Liver biopsies had the highest engraftment rate (Supplementary Fig. S1B). To capture different clinical time points, some tumor samples were acquired before treatment initiation (naive; n = 14) and others at tumor progression following exposure to platinum and/or PARPi treatment (exposed; n = 11). In a retrospective analysis, we categorized the PDXs based on the patient clinical responses at the time point of PDX generation into three groups: sensitive/responding (n = 7), refractory (n = 12), and acquired resistance (n = 5). The clinical response for one patient (SPC_285) was unknown. A smaller proportion of successful engraftment was noted in the tissues obtained from the responding population (40%) compared with 70% engraftment in the refractory/resistant population (Supplementary Fig. S1C).
In Vivo Efficacy of Cisplatin and Olaparib (PARPi)
The therapeutic response to olaparib and cisplatin was tested in vivo in 11 different glBRCA PDAC PDX models and correlated to clinical response at the time of tissue acquisition. In vivo average tumor volume was calculated after 28 to 52 days of cisplatin (n = 11 models) or olaparib (n = 8 models) treatment. Six PDX models were resistant to cisplatin (>0.7-fold change in treatment vs. control; range, 0.7–1.15), and four of these were also resistant to olaparib (0.74–2.15-fold change). Five PDX models demonstrated sensitivity to cisplatin (<0.7-fold change; 0.05–0.41) and olaparib (0.1–0.68; Fig. 2A). Representative tumor growth kinetics for sensitive (SPC_467) and resistant (SPC_187) models are presented in Fig. 2B and Supplementary Fig. S1D.
Figure 2.
Efficacy of cisplatin and olaparib in preclinical models and correlation to clinical response. A,In vivo antitumor activity of PARPi (olaparib) and cisplatin in glBRCA PDAC PDXs. Tumor growth (y-axis) for each PDX model after 28 to 52 days (x-axis) of cisplatin (2 mg/kg; i.p.; once weekly) or olaparib (50 mg/kg; i.p.; 5 days on/2 days off) treatment. The dashed line represented a 70% threshold for sensitivity. Columns color code models by the clinical subgroup of response to platinum/PARPi at the time of tissue acquisition (green, sensitive; yellow, refractory; red, acquired resistance; and gray, unknown). B, Tumor growth curves of sensitive (SPC_467; top) and acquired resistance (SPC_187) models. Average relative tumor volume (y-axis) of control, cisplatin-treated (2 mg/kg; i.p; once weekly), and olaparib-treated (50 mg/kg; i.p.; 5 days on/2 days off) mice (n = 6–8 mice/group). C, EVOC H&E representative images of sensitive (SPC_467) and resistant (SPC_187) PDX models. Scale bar indicated in the figure. Insert, magnification ×2.5. D, A 2 × 2 table showing the distribution of responders and nonresponders based on EVOC prediction and clinical actual response to PARPi and platinum agents. E, Correlation between the in vivo and EVOC efficacy for each model and treatment; y-axis, in vivo tumor growth (%); x-axis, EVOC score (0–60 resistant; 61–100 sensitive); black circles [cisplatin (Cis)]; and gray circles [olaparib (Olap)]. r2 = 0.59; P < 0.001.
EVOC as a Predictive Diagnostic Tool for Clinical Response
To test whether the ex vivo (EVOC) system could accurately predict the clinical responses and recapitulate the in vivo efficacy data, 18 glBRCA PDX models were treated with cisplatin or olaparib (total n = 36 tumor samples) in the EVOC system. Sensitivity to treatment was evaluated based on histopathologic [hematoxylin and eosin (H&E)] characterization and Ki-67 index staining and scored from zero (resistant) to 100 (sensitive). Twenty-three tumors (n = 23) were graded as resistant to olaparib or cisplatin, and 13 were sensitive to both. Representative images of sensitive and resistant models (SPC_467 and SPC_187, respectively) are shown in Fig. 2C.
We next compared the EVOC scores with the patient clinical responses to platinum agents (FOLFIRNOX/cisplatin) and PARPi for 17 and 10 patients, respectively (Fig. 2D). EVOC predicted patient responses with a specificity of 89% (17/19) and sensitivity of 100% (8/8). Specifically, in eight out of 10 PDXs generated from diagnostic biopsies at a “naive” clinical time point, EVOC accurately predicted clinical outcomes. Next, EVOC scores were compared with the in vivo response for each model and treatment. A significant correlation between EVOC sensitivity to cisplatin and olaparib and in vivo tumor growth was noted (R2 = 0.59, n = 16, P < 0.001). Concordance was observed between EVOC and PDX response in 13/16 models, with a sensitivity of 100% and specificity of 89% (Fig. 2E). Notably, in the three patients with discordance, each showed limited clinical response to platinum and the EVOC score was borderline for classification as responsive (Supplementary Table S1). Overall, these results demonstrate a significant correlation between the clinical response to platinum agents and PARPi and the preclinical PDX and in EVOC responses.
Preclinical Olaparib Maintenance Therapy Model and Establishment of “In Vivo Acquired Resistance” Models
To compare sensitive versus resistant tumors in the same patient, and to recapitulate the clinical setting of olaparib maintenance therapy after platinum, as recommended for glBRCA patients (7), we developed a complementary “olaparib maintenance therapy preclinical” and an acquired resistant model. We utilized clinical treatment-naive PDX models—SPC_467, SPC_123, and SPC_285—and treated them with cisplatin followed by PARPi until resistance emergence. For PDX SPC_285, mice were initially randomized to control (n = 6) and cisplatin (n = 7) treatment groups, and tumor volume was monitored twice a week. Control mice reached a maximum tumor volume (∼1,206 mm3) 28 days after treatment initiation, at which point they were sacrificed (Fig. 3A). On day 28, the average tumor volume for the cisplatin group was significantly lower than control (311 mm3; P < 0.001). At this time point, cisplatin-treated mice were divided into groups to continue cisplatin (n = 2) or initiate olaparib (n = 5) maintenance treatment. A durable response was demonstrated in the cisplatin:olaparib group, with maximum tumor growth inhibition on day 63. From this time point, tumors started to regrow in the olaparib-treated mice (range, 63–171 days after treatment initiation). Mice continued to receive daily olaparib treatment to generate an “in vivo olaparib acquired resistance” model. Tumors were excised when they reached maximum volume (1,500–2,000 mm3) or upon study termination. Two mice treated with continuous cisplatin demonstrated complete response until study termination (day 185; Fig. 3A, part i, and B). No viable cells were observed in the remaining tissue excised from cisplatin-treated mice. In both the acquired resistance tumors with large volume tumors and in mice with small tumors, viable glandular cells were detected (Fig. 3C). The two clones (original naive PDX and “in vivo acquired resistance” clone) were tested by EVOC for sensitivity to cisplatin and olaparib. A profound response with a score of 26 (sensitive) was seen in the original PDX model (Supplementary Fig. S1B, left). In contrast, viable cancer cells were observed in the “acquired resistance” clone, which scored 45 (resistant; Supplementary Fig. S1B, right).
Figure 3.
Preclinical olaparib maintenance therapy and establishment of an in vivo acquired resistant model. A, Tumor growth kinetics of two BRCA2-mutated PDAC PDXs with biallelic inactivation. i, SPC_285: Mice were initially randomized to control (vehicle; i.p.; once weekly) or cisplatin (2 mg/kg; i.p.; once weekly); n = 6–7/group. Cisplatin was administrated for 28 days, and mice were then split to continue cisplatin (n = 2) or to switch to maintenance olaparib (50 mg/kg; i.p.; 5 days on/2 days off). ii, SPC_467: Mice were randomized to control (n = 7), olaparib (n = 5), or cisplatin (n = 13). On day 50, cisplatin-treated mice were switched to olaparib (50 mg/kg/i.p.; 5 days on/2 days off). B, Tumor images of SPC_285 for each treatment group (top, control; middle, cisplatin → olaparib; and bottom, cisplatin) on sacrifice day as indicated. C, H&E representative images of tumors treated in control, cisplatin, and cisplatin → olaparib groups. AR, acquired resistance. D, Top: Venn diagrams showing SNV overlap between platinum-sensitive baseline and AR samples. Scatter plots comparing single-nucleotide variant (SNV) variant allele frequencies (VAF) of AR xenograft samples to paired baseline. Bottom, dotted black line represents y = x, where SNV VAF of AR xenograft is equal to SNV VAF of the baseline. Baseline unique variants are shown in gray, whereas AR unique variants are in red and shared variants in orange. E, Double nucleotide alterations characterized by doublet base substitutions of CT>AA/AC in the acquired resistance exposed to the platinum/PARPi model.
Growth kinetics for SPC_467 can be observed in Fig. 3A, part ii. Mice were initially randomized to cisplatin (n = 13), olaparib (n = 7), and control (n = 7). Both cisplatin and olaparib treatments significantly decreased tumor growth. On day 49, nine cisplatin-treated mice started olaparib maintenance therapy (n = 9). Six of the nine mice demonstrated complete response. Two mice, one with complete response and one with partial response, developed acquired resistance on continuous olaparib treatment (Fig. 3A, part ii).
To investigate the mechanisms of resistance in the in vivo acquired resistance models, we performed WGS on three paired samples: baseline PDX (“platinum sensitive”) and the acquired resistance PDX tumor. In all models, WGS demonstrated that the majority of the mutations were shared rather than private (66%–82%; Fig. 3D and Supplementary Fig. S2A), with similar variant allele frequency (VAF) at baseline and in acquired resistance tumors. Interestingly, we found the presence of the platinum-specific mutational signature DBS5 (ref. 14; characterized by doublet base substitutions of CT>AA/AC) in all acquired resistance models and complete absence of this score in baseline tumors (Fig. 3E).
Mechanism of Response and Resistance to Platinum and PARPi in glBRCA PDAC
Multiple mechanisms of resistance in BRCA-mutated tumors have been described, mainly for ovarian and breast tumors (15–17). These can be directed to the DDR pathway, non–DDR-related mechanisms, and clonal selection during exposure to platinum/PARPi. To identify relevant resistance mechanisms in our cohort, we analyzed the genomic and clinical data in the sensitive and resistant groups based on the time point of tissue acquisition (Supplementary Table S2).
BRCA Allelic Status
Inactivation of the second BRCA allele [biallelic; HR-deficient (HRD)] correlates with the BRCA mutation signature (18) and is associated with response to platinum/PARPi therapy (19). Correspondingly, lack of BRCA inactivation [monoallelic; HR-proficient (HRP)] is associated with a lack of response (20, 21). Somatic BRCA status was available for 37 patients [WGS n = 26, BRCA restriction analysis (refs. 22, 23) n = 11]. In our cohort, the BRCA monoallelic and biallelic fractions were 24% and 73%, respectively. The biallelic fraction was significantly higher in BRCA2- (n = 24; 87.5%) versus BRCA1-mutated tumors (n = 12; 50%; P < 0.02). Monoallelic tumors were significantly associated with resistance (45% in resistant vs. 6% in sensitive tumors; P < 0.001; Supplementary Fig. S2B). The second hits in BRCA genes leading to biallelic inactivation were predominantly somatic loss of heterozygosity of the wild-type allele (n = 23); the next most common alterations were due to frameshift mutations (n = 3), structural variants (n = 2), and a splicing mutation (n = 1). The WGS summary is presented in Fig. 4A. In two patients, somatic structural variant inactivations involved large-scale chromothripsis events that shattered and rearranged chr13/BRCA2 and chr17/BRCA1, respectively (Supplementary Fig. S2C).
Figure 4.
WGS and mechanisms of resistance. A, Genomic mutational profiles of 26 whole-genome sequenced tumors. Tumor mutational burden (TMB), HR status, BRCA gene allelic state, mutated BRCA gene, patient clinical response, patient platinum exposure, presence of platinum mutational signature, HRDetect score, HRD mutational signature proportion, tumor ploidy, and mutations observed in PDAC driver genes are shown. HRD−SM, HRDsecond_mut; SV, structural variant. B, Table of resistant samples with specific information. WT, wild-type. C, Illustration of secondary mutation reversing reading frame back to WT in patient SPC_187. D, Integrative Genomics Viewer (IGV) snapshot of SPC_291 WGS reads mapping to the BRCA2 locus. Reads from normal are shown in the middle, where the blue box highlights a 1-bp germline frameshift with a zoomed-in view on the right. Reads from tumor are shown on top, where the orange box highlights a somatic SV that deletes the region around the germline frameshift. Zoomed-in view of somatic SV breakpoints at the bottom shows BRCA2 codons reverted to inframe as a result of SV. E, IGV snapshot capturing the BRCA2 germline (left) and the somatic (right) insertion or deletion (indel) frameshift in SPC285. Reads mapping to this region are shown for the germline reference (bottom), platinum-sensitive baseline xenograft (middle), and acquired resistance (AR) xenograft (top). The AR clone was analyzed by WGS and compared with baseline PDX. F, Distance from diploid [absolute(ploidy-2)] in resistant and sensitive samples (P < 0.05). G, Molecular subtype by response—basal-like score–classical score in resistant and sensitive samples (P < 0.05). H, SHAP analysis. A detailed perspective on the contribution of the various factors to the resistance classification score. Each line corresponds to one factor and each dot in it to a single sample (patient). The color of the dot represents the value of the factor for that sample (high BRCA allelic state value corresponds to monoallelic, high reversion-mutation value corresponds to the presence of the reverse mutation, high subtype value corresponds to “basal-like,” high sex value corresponds to female, and high BRCA value corresponds to BRCA2). The location of the dot on the x-axis is the relevant SHAP value of that factor for that sample. As an example, the red dots on the far right at the “Reversion mutation” line indicate that the presence of this mutation (in five patients) is a very strong indicator of resistance. The features on the y-axis are ordered by their average importance.
Secondary Mutations
In the resistant group (Fig. 4B), secondary mutations were identified by WGS in five tumors (31.2%; termed HRDsecond_mut). These secondary mutations may partially restore BRCA function, thus leading to HR proficiency and lack of response to platinum/PARPi therapy. This is in line with our previous findings demonstrating secondary mutations in 35% of acquired resistance samples (20). Depiction of the secondary mutation (deletion) restoring the reading frame back to wild-type is presented in Fig. 4C. In one patient (SPC_291), the reversion occurred through a large chromothripsis-associated deletion that restored the germline BRCA2 reading frame and potentially HR function (Fig. 4D).
Additional genomic analysis demonstrated that HRD and HRDsecond_mut tumors were both dominated by the COSMIC (24) signature 3, whereas HRP tumors largely showed age-related signatures 1, 8, and 5, consistent with previous literature (14). Importantly, we were able to detect the presence of platinum signatures DBS5, SBS31, and SBS35 in 10 out of 12 tumors treated with platinum prior to WGS, including all five HRDsecond_mut tumors (Fig. 4A).
There was no difference in the frequency of driver gene mutations (KRAS, TP53, CDKN2A, and SMAD4) between the sensitive, refractory, and acquired resistance groups. We in-depth analyzed the alterations, translocations, and copy-number gain/loss specifically of 22 DDR genes known to be associated with platinum/PARPi resistance, including TP53BP1, RIF1, and CHD4 (25). No pathogenic alterations or copy-number loss/gain was identified in this gene set (Supplementary Table S3).
Clonality
Tumor cell clonality has been shown to contribute to resistance (26). In the context of BRCA-deficient tumors, sparse evidence in ovarian cancer exists for BRCA1 wild-type copies positively selected under treatment pressure and dominant at resistance (27). To determine subclonality in our cohort, baseline BRCA VAF was evaluated in six tumors harboring somatic biallelic inactivation. Somatic second hits were clonal in these cases. However, bulk tumor WGS of one tumor sample per patient cannot accurately estimate subclonal heterogeneity; such analysis could be best addressed by single-cell sequencing.
Subclonality was identified in one in vivo acquired resistance model (SPC_285; biallelic inactivated by a somatic frameshift mutation). The VAF of the somatic BRCA2 frameshift dropped from 26.5% at baseline to 4.5% in the acquired resistance model (Fig. 4E). The VAF of this somatic frameshift suggests the presence of two subclones at baseline—an HRD subclone carrying the somatic BRCA2 frameshift and an HRP subclone retaining the wild-type allele. Furthermore, the drop in VAF from baseline to acquired resistance implies that treatment selected for the HRP subclone retains the wild-type allele, potentially leading to acquired resistance.
Aneuploidy
Tumor aneuploidy is known to be associated with tumor aggressiveness and chemoresistance (10, 28). In 25 samples with available data on tumor ploidy, we determined the degree of aneuploidy based on the distance from a diploid genome [absolute(ploidy-2)]. The resistant tumors were significantly more aneuploid than the sensitive ones (0.58 vs. 0.27; P < 0.01). Specifically, this was most significant in the acquired resistance tumors (n = 5; average distance from diploidy = 0.75; Fig. 4F).
Expression Subtype
We profiled 121 PDX models by bulk RNA-seq, including 23 glBRCA tumors. The basal-like phenotype is known to be associated with chemoresistance and worse survival (29, 30). Refractory samples displayed significantly more “basal-like” subtype (Fig. 4G; P < 0.05), which was independently predictive of worse survival (Supplementary Fig. S2D). Importantly, all five clinical resistant patients with HRD (biallelic) and no secondary mutations displayed a basal-like phenotype.
We performed DESeq2 (31) analysis to identify differentially expressed genes between resistant and sensitive models, and then applied gene set enrichment analysis (32) to find pathways with a significant enrichment score (FDR corrected P < 0.05). DNA repair, focal adhesion, reactive oxygen species, and the p53 pathways were upregulated in the resistant samples (Supplementary Fig. S2E). These pathways are correlated with the basal-like phenotype (33, 34). We utilized the entire cohort of non–BRCA-mutated tumors (n = 121) as a validation set; these pathways were upregulated in basal-sensitive tumors compared with classical resistance tumors, further supporting the strong impact of the basal-like phenotype in platinum/PARPi resistance in the BRCA cohort.
To evaluate the prediction value of each variable mentioned above on the probability of resistance to PARPi/platinum, we performed a multivariable logistic regression analysis (SHAP analysis: A Unified Approach to Interpreting Model Predictions; arXiv:1705.07874v2), including sex, age, BRCA1/2, BRCA allelic status, transcriptomic subtype, and tumor ploidy. Patient outcome was determined as sensitive or resistant based on the clinical response at tumor acquisition. Monoallelic status and the existence of secondary mutations were the strongest predictors for resistance (mean absolute SHAP values 0.15 and 0.12, respectively; Fig. 4H; Supplementary Fig. S2F). Expression of the “basal-like” subtype also showed a strong predictive value for resistance (0.11), and tumor cell aneuploidy had a predictive value of 0.08. The type of germline mutation (BRCA1/BRCA2) had a very low contribution (0.01). AUC for the dataset was 0.89 (0.73–0.99; Supplementary Fig. S2G).
Correlation of Clinical Data to Resistance
We reviewed the clinical data from the patients from whom resistant samples were acquired (Fig. 4B). These patients were enriched with poor Eastern Cooperative Oncology Group performance status, multiple comorbidities, and widespread extensive metastatic disease—clinical findings that have been shown to compromise the dose intensity of doublet/triplet chemotherapy treatment and contribute to lower OS (35, 36). These poor prognostic parameters seen in resistant patients with biallelic, HRD tumors may contribute to clinical resistance, in addition to the tumor polyploidy and basal-like subtype.
Full clinical information, including demographics, tissue analyzed, genomics, and clinical parameters of this cohort, is presented in Supplementary Table S2.
ENLIGHT: Treatment Predictor Based on Artificial Intelligence and Omics Analysis
We applied a computational tool called ENLIGHT (37), which identifies synthetic lethal/rescue interactions between the gene targets of a specific drug and other genes, using big pan-cancer data, and uses them as a biomarker to predict patient response to that drug based on the tumor transcriptome. We used the ENLIGHT PARPi network to retrospectively analyze response to platinum/PARPi. ENLIGHT accurately predicted which models would be resistant (objective response: 1.375, average precision: 0.474; Supplementary Fig. S2H). We explored the genes that constitute the ENLIGHT PARPi network, which highlighted the upregulation of genes involved in the G2–M cell-cycle checkpoint and focal adhesion pathways (both pathways are statistically enriched in the biomarker; FDR corrected P value of hypergeometric test: 0.004 and 0.02, respectively). Activation of these pathways is known to be associated with resistance and is highly correlated to the basal-like subtype, but the ENLIGHT predictive biomarker was derived from a pan-cancer dataset, providing independent support for the hypothesis that these pathways play a role in resistance to platinum and PARPi therapy.
Immune Checkpoint Inhibition Efficacy in a Humanized BRCA2 PDX Model
A significantly higher tumor mutational burden (TMB) was noted in biallelic tumors with HRDsecond_mut [9.7 mutations/megabase (Mb), P = 0.003] vs. HRD (4.1 mutations/Mb) and HRP (2.5 mutations/Mb) tumors (Fig. 4A). The high TMB and neoantigen load in the BRCA-mutated tumors and specifically tumors with HRD may indicate that patients in this subgroup may benefit from immune checkpoint inhibition. In our cohort, six patients were treated with immune checkpoint blockade agents alone or in combination in clinical trial settings. Five of these patients did not respond, but one patient displayed complete response for over 2 years.
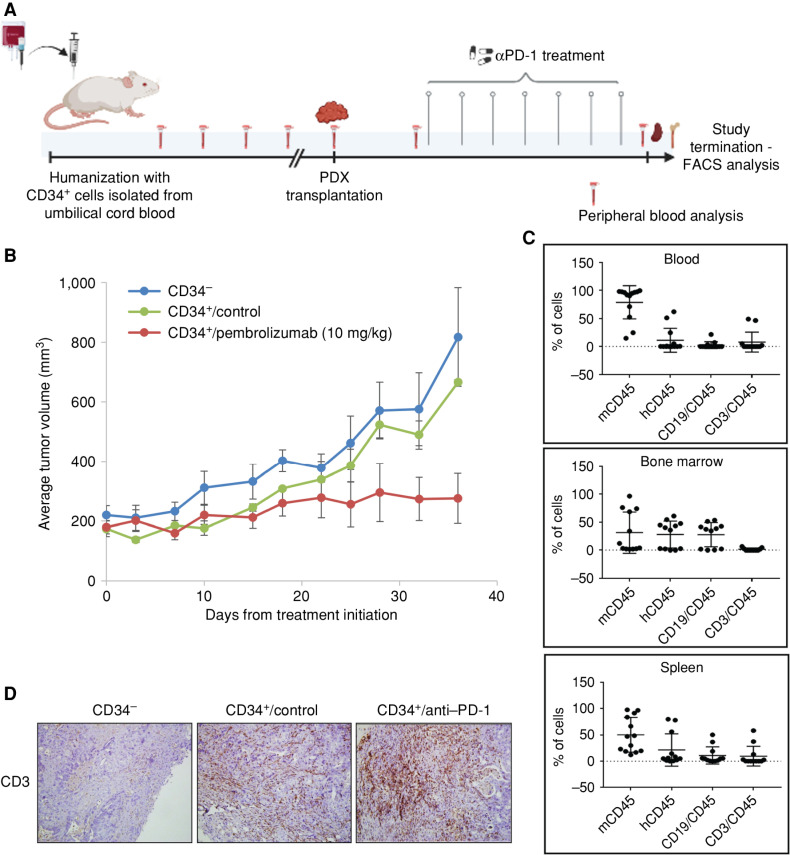
To test the potential of anti–PD-1 treatment in the preclinical setting, we established a novel humanized glBRCA2 PDAC PDX utilizing an acquired resistant HRDsecond_mut tumor with the highest TMB in our cohort (SPC_126; 12.2 mutations/Mb). We isolated CD34+ stem cells from umbilical cord blood and injected them into 3- to 4-week-old NOD/SCID/ILgRKO (NSG) mice (38). Human cell engraftment (hCD45) was detected in the peripheral blood from week 12 onward. At week 18, cryopreserved tumor chunks from the aforementioned PDX model were subcutaneously transplanted to hCD45-engrafted and nonengrafted mice, and treatment with anti–PD-1 was initiated when tumors reached an average volume of 100 mm3 (Fig. 5A). A significant attenuation of the tumor growth rate was observed in the hCD45-engrafted anti–PD-1–treated mice compared with hCD45-engrafted control untreated mice, with one mouse demonstrating complete response (Fig. 5B). hCD45+ cells, B (CD19) cells, and T (CD4 and CD8) cells were detected in the spleen, bone marrow, and blood of engrafted mice (Fig. 5C). IHC analysis demonstrated tumor T-cell infiltration (Fig. 5D). These preliminary data demonstrate the potential of immune checkpoint blockade in glBRCA2-mutated PDAC with high mutational load.
Figure 5.
Anti–PD-1 efficacy in a humanized glBRCA2 PDAC PDX model. A, Preclinical illustration of humanized mouse model generation. CD34+ cells were isolated from umbilical cord blood by magnetic beads. Purity was validated by FACS analysis. CD34+ cells or PBS control was injected into 3- to 4-week-old sublethal irradiated NSG mice. Peripheral blood was obtained every 2 to 3 weeks, and human CD45 was assessed. On week 18, a PDX tumor chunk was subcutaneously transplanted to mice and treated with pembrolizumab (10 mg/kg; i.p.) or vehicle control. B, Average tumor volume in nonhumanized mice (blue); humanized control-treated (green) mice; and humanized/pembrolizumab-treated mice (red). C, Immune cell characterization: mouse CD45 (mCD45), human CD45 (hCD45), B cells (CD19), and T cells (CD3) in the blood, spleen, and bone marrow of all mice at study termination. D, IHC staining of tumors demonstrating T-cell infiltrate in the CD34+ engrafted mice. Created with BioRender.com.
DISCUSSION
PDAC patients with glBRCA mutations are a clinical outlier cohort with extended OS, and BRCA mutation status is an important biomarker in PDAC (4, 5). Several studies have shown the effectiveness of PARPi treatment in glBRCA PDAC, and it has become a standard of care for this population. However, there is a spectrum of responses of glBRCA PDAC to platinum and PARPi treatment. Here, we sought to clinically characterize a large cohort of patients with glBRCA PDAC, develop preclinical models reflecting the spectrum of response, and decipher the mechanisms of sensitivity and resistance. Based on clinical observations, we stratified patients into three groups of response: refractory, durable response, followed by acquired resistance, and long-term responders (8, 9).
The clinical spectrum of response is reflected in clinical trials (7, 39, 40). In the phase III POLO maintenance PARPi trial, approximately 30% of patients did not meet inclusion criteria due to disease progression on platinum-based chemotherapy (41). These patients are likely refractory to treatment and signify the underlying aggressive nature of pancreatic cancer even in the setting of glBRCA. Specific markers predictive for resistance to platinum/PARPi in glBRCA PDAC include monoallelic tumors (42), secondary mutations (10, 20), basal-like subtype (29, 30), and tumor aneuploidy (10, 28).
PDXs derived from metastatic refractory patients are a unique opportunity to study the resistance mechanisms in glBRCA PDAC. These tumor samples are underrepresented in large genomic databases, which are fueled mainly by primary tumors (19) and lately by metastatic samples (43). In a cohort of 19 resistant samples, we found that the BRCA monoallelic status and the presence of secondary mutations were the strongest predictors of platinum/PARPi resistance. Somatic secondary mutations are the most prevalent described mechanism of resistance in glBRCA-associated tumors (20, 25, 44). Recently, reversion mutations were identified by cell-free DNA (cfDNA) analysis in ovarian cancer (45). In future studies, we will utilize our PDAC blood biobank for cfDNA analysis, which may detect secondary mutations at an earlier stage before clinical progression by imaging.
The basal-like subtype has been associated with poor response to chemotherapy and shorter survival in PDAC (43, 46, 47). Accordingly, in our HRD PDAC series, we found that a basal-like transcriptomic subtype was independently predictive of resistance and worse survival. Furthermore, tumor aneuploidy predicted resistance. This is consistent with previous reports showing that polyploid cells are resistant to cytotoxic and biological drugs (48, 49). In our cohort, we specifically identified an association between aneuploidy and DDR resistance. It was recently shown that aneuploid clones activate the DDR and are consequently more resistant to further DNA damage induction (https://www.biorxiv.org/content/10.1101/2023.01.27.525822v1). Resistance to DNA-damaging agents is associated with the degree of aneuploidy. Interestingly, the highest polyploidy in our cohort was noted in the acquired resistance samples. In the future, it will be important to dissect the evolutionary process of aneuploidy and examine tumor ploidy status at baseline.
Resistance can develop due to subclonal selection. A study in ovarian cancer demonstrated the existence of BRCA1 wild-type copies present at baseline and positively selected under treatment pressure, resulting in the dominance of clones with wild-type copies at resistance (50). In our resistant preclinical models, we demonstrate evidence of subclonality during treatment leading to platinum/PARPi resistance. This mechanism of resistance is sparsely described in the literature, but subclone analysis at initial diagnosis and at progression by single-cell and deep analysis may reveal low prevalence subpopulations (15–17) that could have prognostic and predictive value.
In an analysis of the resistant patient population, we found common and direct disruption of BRCA1/2 function, such as monoallelic tumors or secondary mutations. However, an interesting small subset of patients harbored biallelic tumors and HR deficiency. These tumors display the basal-like phenotype and are more aneuploid. Combined with patient poor prognostic clinical parameters, this may influence their clinical resistance. Further investigation of the basal-like phenotype and discovery of more effective treatment options for these patients may improve outcome. We hypothesize that the tumor biology is primarily driven by the basal-like phenotype, and the anticipated synthetic lethality in HR deficiency to PARPi has limited clinical impact.
To recapitulate the clinical spectrum of the disease we generated PDX models at different clinical time points. A strong correlation was observed between the in vivo PDX response to cisplatin and olaparib and the clinical response at the time of PDX generation (tumor naive to treatment and patient responsive/refractory vs. tumor sample obtained at time of clinical resistance). We demonstrate that EVOC predicts the in vivo PDX response and reflects the clinical response. EVOC offers several advantages, with the main ones being the short turnaround time and the potential of performing several treatment combinations in parallel. The main limitation of the EVOC system is it reflects of one time point, which in the clinical setting would be at diagnosis or change of the line of treatment. Furthermore, the EVOC score cannot predict the durability of response or how rapidly resistance will emerge.
Over the past decade, our research team has invested substantial efforts in obtaining adequate paired tumor samples for model establishment. However, in PDAC, low cellularity and engraftment rate have limited the establishment of preclinical models from all patients and extensive genomic analysis (9, 13). Therefore, to compare sensitive versus resistant tumors in the same patient we generated “in vivo acquired resistance” models. Comparative WGS analysis demonstrated high concordance between the sensitive and acquired resistance models and no evidence for secondary mutations in BRCA genes. This preclinical acquired resistance model system can be used to investigate combinatorial treatment strategies at the time of acquired resistance.
There are several limitations to PDX models, such as differential response to drugs observed in the same model. This exemplifies the heterogeneity within a tumor and can indicate a bias to a specific clone that selectively grows at PDX generation. Indeed, intratumoral PDAC heterogeneity, in which both classical and basal-like cells coexist, has been described in several publications (51, 52). Furthermore, in clinical practice and in our cohort, mixed clinical response is observed, perhaps reflecting the apparent heterogeneity. Several studies have shown the resemblance of PDXs to primary tumors in different cancer types (53, 54) and the use of PDX as a tool for predicting treatment (55).
Interestingly, in our PDX cohort, we do not have a representative model of a superresponder patient. Tumor samples from such patients were transplanted, but the engraftment rate for the “sensitive” tumor samples is low (40%) compared with 70% in the “resistant” tumor samples. This can be attributed to genomic features, tumor cellularity, or tumor cell turnover. The sensitive samples were enriched with biallelic tumors, classic phenotype, and diploid tumors. Clinically, these patients tend to demonstrate a deep response to first-line platinum therapy and typically display oligometastatic disease to liver or lungs. Additional genomic and immune analysis is currently being performed from those patient tumor samples and is beyond the scope of this paper.
Approximately 50% of glBRCA patients demonstrate an initial good response to platinum followed by PARPi, and in some cases, the tumor mass load reaches a minimal state with oligometastatic disease. It is important to identify patients who are likely to progress prior to clinical manifestation of resistance and seek novel treatment options in this unique disease state. Whole-genome analysis has demonstrated that the majority of the glBRCA PDXs harbor a high TMB load, specifically the HRDsecond_mut tumors (9.7 mutations/Mb). Increased TMB in these tumors may be partially attributed to neoantigens as a result of new secondary mutations, usually caused by deletions/insertions in the vicinity of the original pathogenic germline mutations. In the case of certain founder pathogenic mutations (e.g., BRCA2:c.5946delT, all HRDsecond_mut cases presented herein), it was shown that the reversions that arise are generally localized to the 3′ flanking sequence of the original frameshift mutation (56). Compensatory frameshift secondary mutations introduce novel amino acid sequences, which differ from the original wild-type protein, and can thus constitute neoantigens. This may open a window of opportunity to treat with alternative treatments such as immunotherapy. A small clinical cohort demonstrating the potential efficacy of PD-1 inhibition and CTLA4 inhibition has recently been published (57).
Limited clinical data support this hypothesis (58–61). Studies have shown that in addition to the effect of PARPi on cancer cell death, PARPi can enhance the immune response. PARPi can induce accumulation of cytosolic DNA damage and trigger the interferon pathways (62, 63), and thus activation of immune cells. PARPi can also induce PD-L1 expression (64). The high mutational load of BRCA-mutated tumors and the PARPi effect on the tumor microenvironment and priming of the immune system provide a strong rationale for the integration of immune checkpoint inhibition in glBRCA PDAC. Clinical assessment of the combination of PARPi with immune checkpoint inhibition in pancreatic cancer is ongoing (NCT: 04548752, 04753879, 03851614, and 03637491).
To summarize, herein we characterize the spectrum of response of patients with glBRCA PDAC to platinum/PARPi into three subgroups. Such clinical distinction of patients may facilitate more informative treatment decisions. We precisely recapitulated various clinical scenarios of glBRCA PDAC to platinum/PARPi in the preclinical setting. Further analyses to decipher additional mechanisms of resistance and develop alternative treatments for BRCA-associated PDAC are ongoing.
METHODS
Clinical Data and Survival Analysis
Patients with PDAC harboring glBRCA mutations, treated at the SMC during a 10-year period (2010–2022), were included in this cohort. All clinical data on participants were extracted from patient records or an existing institutional review board (IRB)–approved (4474 and 5073-18) institutional database. Written informed consent was obtained from all patients prior to study enrollment. The protocols were approved by the IRB at the SMC, and the studies were conducted in accordance with the Good Clinical Practice guidelines and the Declaration of Helsinki.
Preclinical Efficacy Experiments in PDX Models
Animal studies were performed in specific pathogen–free (SPF) conditions and an IRB-approved study. PDXs were generated and monitored as previously described (13). Olaparib was purchased from Selleckchem (Tivan Biotech) and provided by AstraZeneca. Cisplatin was provided by the SMC pharmacy.
PDX tumor chunks were transplanted to the lower flank of 6-week-old female nude mice. When tumors reached palpable size (∼100 mm3), mice were randomized into treatment groups (n = 6–8): (a) PARPi (olaparib, Selleckhem) dissolved in 10% Hydroxypropyl-b-Cyclodextrin (Sigma), 50 mg/kg, i.p., 5 days on/2 days off; (b) cisplatin (2 mg/kg, i.p., once a week); and (c) vehicle control (solvent only, i.p.). Tumor volumes were measured twice a week using a caliper according to the formula (length 3 width2)/2. Mice were weighed twice a week. Mice were euthanized if the tumor burden reached 1.5 cm3 or if they presented over 20% of the initial body weight loss. At study termination, animals were sacrificed by CO2 inhalation. Tumor growth curves are presented as mean relative tumor volume (RTV) ± SE at each measurement point, for each treatment group. Tumor growth inhibition was calculated for each treatment group compared with the control (average RTV treatment group/average RTV control group). Mice were purchased from Envigo (Israel) and maintained in SPF rooms in a temperature-controlled room at 208°C with a 12-hour–12-hour dark–light cycle. Animals had free access to water and commercial chow. Differences between treatment groups were analyzed by the Mann–Whitney test using GraphPad Prism (GraphPad Software). Error bars represent SE.
Establishment of a Humanized CD34+ Engrafted NSG PDX Model System
NSG mice were purchased from The Jackson Laboratory. Mice were bred and housed in SPF conditions. All animal experiments were approved by the Animal Care Committee at SMC (IRB 1007/15). Fresh cord blood units were obtained from the SMC cord blood bank under IRB-approved protocols. CD34+ cells were isolated using magnetic beads (Miltenyi) following the conventional method. Cells were cultured in SFEM media (STEMCELL) supplemented with hSCF (100 ng/μL), IL6 (20 ng/μL), hFLT3 ligand (100 ng/μL), and TPO (20 ng/μL) for 5 days; 100,000 cells/50 μL PBS or vehicle control (50 μL PBS) were injected (i.v.) into 3- to 4-week-old sublethal irradiated (125 rad) NSG mice. Mice were bled once every 2 to 3 weeks starting 8 weeks after transplantation to monitor engraftment. At ∼15 to 18 weeks after CD34+ transplantation, mice harboring >25% hCD45 cell engraftment underwent subcutaneous transplantation of PDX tumor chunks as described previously (8, 9, 13, 38). Mice were randomized to treatment groups based on tumor volume and hCD45 engraftment: (i) CD34+ engrafted/pembrolizumab (10 mg/kg, twice a week, i.p.); (ii) CD34+ engrafted/vehicle control; and (iii) CD34−/vehicle control. When control mice reached the maximum allowed tumor volume, the study was terminated. Upon sacrifice, blood, tumor, bone, and spleen were excised. Spleen and peripheral blood mononuclear cells were isolated and cleaned with red blood cell lysis buffer. Bone marrow was flushed from the femur. Cells were characterized by FACS. Tumors were snap-frozen and fixed in 4% formaldehyde. Paraffin-embedded tumor sections were stained with H&E and immunostained for CD45, CD3, CD4, and CD8.
EVOC System
Technique.
PDX tumors were sliced to a size of 250-μm thick using a vibratome (VF300, Precisionary Instruments) and placed in 6-well plates on titanium grids (Alabama R&D) with 4 mL of DMEM/F12 medium (supplemented with 5% FCS, penicillin 100 IU/mL with streptomycin 100 μg/mL, amphotericin B 2.5 μg/mL, gentamicin sulfate 50 mg/mL, and L-glutamine 100 μL/mL). Tissues were cultured at 37°C, 5% CO2, and 80% O2 on an orbital shaker (TOU-120N, MRC) at 70 rpm. The following day, tissues were treated with cisplatin (100 μmol/L) or olaparib (80 μmol/L) for 96 hours, with drug change after 48 hours. Upon completion, tissues were fixed overnight (4% PFA), followed by formalin-fixed paraffin embedding (FFPE).
Outcome Evaluation.
Tissue IHC was performed on 4-μm sections from FFPE tissue samples from the EVOC. H&E staining was performed using an automated stainer. Ki-67 staining [Thermo Fisher Antibody (RM-9106); 1:500 dilution] was performed using an automated stainer (BOND RX, Leica Biosystems). Tissue viability and scoring were performed using pathologic criteria grading tumor cell death (70%), the viability of live tumor cells (20%), and Ki-67 proliferation (10%). A scale of 0 to 100 was created, with a score of zero representing completely viable cancer cells (“resistance”) and a score of 100 representing no viable cancer cells (“sensitive”).
WGS
Whole-genome analysis was performed on snap-frozen PDX tumor chunks or fresh frozen tumor samples as previously described (9, 14). Briefly, raw genome sequencing reads were aligned to human reference genome build hg38 using Burrow-Wheeler-Aligner (BWA; v0.7.17; ref. 15). Xenograft genome sequencing reads were aligned to mouse reference, where contaminating mouse reads were removed using XenofilteR (16) with default settings. Germline variant calling was performed using the Genome Analysis Tool Kit (GATK4; v4.1.2; refs. 17, 18). Somatic single-nucleotide variations (SNV) were identified as the intersection of calls by two separate tools—“Tier 1 SNVs” from Strelka2 (v2.9.10; ref. 19) and “PASS” variants from MuTect2 (v4.1.2)—with both tools run using default settings. Insertions or deletions (indel) were identified as the overlap between two out of four indel variant callers—Strelka2, MuTect2, SVaBA (v134; ref. 20), and DELLY2 (v0.8.1; ref. 21). Copy-number segments, tumor cellularity, and ploidy were obtained by using an in-house algorithm, Celluloid (22). Somatic structural rearrangements were called as the consensus from two out of three variant callers—SVaBA, DELLY2, and Manta (v1.6.0; ref. 23).
BRCA Somatic Allelic Status
BRCA allelic state was retrieved from WGS (n = 26) and target sequencing (n = 1), and determined by PCR-based restriction analysis (24, 25).
RNA-seq Analysis
PDX tumors were profiled by bulk RNA-seq (65, 66). Each model (i.e., all PDX generations of a single patient specimen progeny) was profiled with up to three biological replicates (i.e., samples from different PDX generation of the same progeny). The cohort encompassed 301 samples from 121 models, which passed all quality control, including 23 glBRCA. We separated the malignant and stromal compartments by aligning each read to both mouse (stromal) and human (malignant) references, keeping only the best match in cases where a read aligned to both genomes. The biological replicates highly resemble each other compared with other models (R = 0.93 on average), suggesting that the patient's unique expression patterns are preserved across PDX generations.
Statistical Analysis
In vivo tumor responses and differences between treatment groups were calculated by nonparametric Mann–Whitney U test comparing the treatment group to the control group (n = 5–10 tumors/treatment group). OS was calculated using the Kaplan–Meier analysis and log-rank test in python. Pearson correlation between EVOC score and in vivo efficacy data was performed using Prism software. SHAP value (A Unified Approach to Interpreting Model Predictions; arXiv:1705.07874v2) analysis was performed to estimate the contribution of each factor to the classification score. Analysis was performed on the full dataset (n = 38) and included sex, age, germline BRCA mutation, BRCA allelic state, subtype, and tumor ploidy. Patient outcome was determined as sensitive or resistant based on the clinical response at the time of tumor acquisition.
The AUC of the logistic regression model was estimated using the leave-one-out (LOO) approach. As there were 11 patients with partial genomic information, we also examined the LOO performance of the model on the remaining 27 samples.
ENLIGHT (37) was applied to the data to look for additional evidence of mechanisms of resistance. ENLIGHT identifies synthetic lethal and synthetic rescue interactions between the gene targets of a specific drug and other genes, using big pan-cancer data, and uses them as a biomarker to predict patient response to that drug based on the tumor transcriptome.
Data Availability Statement
PDX models generated at SMC will be available upon completion of material transfer agreements.
Raw sequencing data are housed at the Gene Expression Omnibus portal under accession number GSE235843 (RNA-seq data) and the European Genome-phenome Archive under accession number EGAD00001011129 (WGS data), and will be available to researchers upon completion of a data access agreement.
Supplementary Material
In vivo, EVOC and clinical response
Clinical data of analyzed BRCA PDAC patients
20 genes in DDR resistance
In vivo growth kinetics and EVOC efficacy in acquired resistance model
Mechanisms of resistance in glBRCA PDAC
Acknowledgments
The study was conducted with support from the Sheba Medical Center ARC innovation center and a charitable donation from the Canadian Friends of the Hebrew University (Alex U. Soyka). This work was supported in part by the Cooperation Program in Cancer Research of the Deutsches Krebsforschungszentrum (DKFZ) and Israel's Ministry of Science, Technology and Space (MOST). Olaparib was provided by AstraZeneca as part of a collaborative research project. This work partially fulfills the PhD degree requirements for C. Stossel, Faculty of Medicine, Tel Aviv University, Israel. Figure 5 was created with BioRender.com.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Discovery Online (http://cancerdiscovery.aacrjournals.org/).
Authors’ Disclosures
J.M. Wilson reports other support from the Government of Ontario during the conduct of the study. G.M. O'Kane reports personal fees from AstraZeneca, Incyte, Servier, and MSD, and grants and personal fees from Roche outside the submitted work. S.J. Salpeter reports other support from Immunyx Pharma during the conduct of the study, as well as a patent for the EVOC platform pending. V. Bar reports personal fees from Curesponse Ltd. during the conduct of the study; personal fees from Curesponse Ltd. outside the submitted work; and a patent for EVOC issued. G. Dinstag reports personal fees from Pangea Biomed Ltd. during the conduct of the study, as well as personal fees from Pangea Biomed Ltd. outside the submitted work. U. Ben-David reports grants from Novocure outside the submitted work. N.S. Gavert reports other support from Curesponse Ltd. outside the submitted work. R. Straussman reports personal fees from Curesponse Ltd. during the conduct of the study, as well as personal fees from Curesponse Ltd. outside the submitted work. R. Berger reports personal fees from Pangea Biomed Ltd., and personal fees and other support from Curesponse Ltd. during the conduct of the study, as well as personal fees from Aummune and Immunai outside the submitted work. T. Golan reports grants and personal fees from AstraZeneca, and personal fees and other support from Curesponse Ltd. during the conduct of the study, as well as grants and personal fees from Merck, and personal fees from AbbVie, BiolineRx, Bayer, and Teva outside the submitted work. No disclosures were reported by the other authors.
Authors’ Contributions
C. Stossel: Conceptualization, resources, data curation, software, formal analysis, supervision, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. M. Raitses-Gurevich: Conceptualization, resources, data curation, formal analysis, validation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. D. Atias: Conceptualization, resources, data curation, validation, investigation, visualization, methodology, writing–original draft, writing–review and editing. T. Beller: Conceptualization, data curation, validation, investigation, visualization, methodology, writing–review and editing. Y. Glick Gorman: Conceptualization, data curation, validation, investigation, methodology, writing–review and editing. S. Halperin: Data curation, methodology. E. Peer: Data curation, software, validation, methodology, writing–review and editing. R.E. Denroche: Data curation, software, formal analysis, validation, methodology, writing–review and editing. A. Zhang: Data curation, software, formal analysis, investigation, methodology, writing–review and editing. F. Notta: Data curation, software, formal analysis, validation, methodology, writing–review and editing. J.M. Wilson: Resources, data curation, visualization. G.M. O'Kane: Visualization. E. Haimov Talmoud: Data curation, writing–review and editing. N. Amison: Data curation, methodology. M. Schvimer: Data curation, formal analysis, investigation, methodology, writing–review and editing. S.J. Salpeter: Data curation, formal analysis, investigation, writing–review and editing. V. Bar: Data curation, formal analysis, methodology, writing–review and editing. A. Zundelevich: Data curation, methodology. I. Tirosh: Resources, data curation, investigation. R. Tal: Data curation, software, formal analysis, investigation. G. Dinstag: Software, formal analysis. Y. Kinar: Software, formal analysis. Y. Eliezer: Formal analysis. U. Ben-David: Formal analysis. N.S. Gavert: Data curation, investigation, visualization, methodology. R. Straussman: Data curation, investigation, visualization, methodology. S.J. Gallinger: Resources, data curation, formal analysis, validation, methodology, writing–review and editing. R. Berger: Resources, supervision, validation, writing–review and editing. T. Golan: Conceptualization, resources, supervision, funding acquisition, validation, investigation, visualization, methodology, writing-original draft, project administration, writing–review and editing.
References
- 1. Conroy T, Desseigne F, Ychou M, Bouche O, Guimbaud R, Becouarn Y, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med 2011;364:1817–25. [DOI] [PubMed] [Google Scholar]
- 2. Rahib L, Smith BD, Aizenberg R, Rosenzweig AB, Fleshman JM, Matrisian LM. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014;74:2913–21. [DOI] [PubMed] [Google Scholar]
- 3. Venkitaraman AR. Cancer susceptibility and the functions of BRCA1 and BRCA2. Cell 2002;108:171–82. [DOI] [PubMed] [Google Scholar]
- 4. Golan T, Kanji ZS, Epelbaum R, Devaud N, Dagan E, Holter S, et al. Overall survival and clinical characteristics of pancreatic cancer in BRCA mutation carriers. Br J Cancer 2014;111:1132–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Golan T, Kindler HL, Park JO, Reni M, Macarulla T, Hammel P, et al. Geographic and ethnic heterogeneity of germline BRCA1 or BRCA2 mutation prevalence among patients with metastatic pancreatic cancer screened for entry into the POLO trial. J Clin Oncol 2020;38:1442–54. [DOI] [PubMed] [Google Scholar]
- 6. Kaufman B, Shapira-Frommer R, Schmutzler RK, Audeh MW, Friedlander M, Balmana J, et al. Olaparib monotherapy in patients with advanced cancer and a germline BRCA1/2 mutation. J Clin Oncol 2015;33:244–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Golan T, Hammel P, Reni M, Van Cutsem E, Macarulla T, Hall MJ, et al. Maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. N Engl J Med 2019;381:317–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Golan T, Atias D, Stossel C, Raitses-Gurevich M. Patient-derived xenograft models of BRCA-associated pancreatic cancers. Adv Drug Deliv Rev 2021;171:257–65. [DOI] [PubMed] [Google Scholar]
- 9. Golan T, Stossel C, Atias D, Buzhor E, Halperin S, Cohen K, et al. Recapitulating the clinical scenario of BRCA-associated pancreatic cancer in pre-clinical models. Int J Cancer 2018;143:179–83. [DOI] [PubMed] [Google Scholar]
- 10. Wang Y, Park JYP, Pacis A, Denroche RE, Jang GH, Zhang A, et al. A preclinical trial and molecularly annotated patient cohort identify predictive biomarkers in homologous recombination-deficient pancreatic cancer. Clin Cancer Res 2020;26:5462–76. [DOI] [PubMed] [Google Scholar]
- 11. Hickman JA, Graeser R, de Hoogt R, Vidic S, Brito C, Gutekunst M, et al. Three-dimensional models of cancer for pharmacology and cancer cell biology: capturing tumor complexity in vitro/ex vivo. Biotechnol J 2014;9:1115–28. [DOI] [PubMed] [Google Scholar]
- 12. Gavert N, Zwang Y, Weiser R, Greenberg O, Halperin S, Jacobi O, et al. Ex vivo organotypic cultures for synergistic therapy prioritization identify patient-specific responses to combined MEK and Src inhibition in colorectal cancer. Nat Cancer 2022;3:219–31. [DOI] [PubMed] [Google Scholar]
- 13. Golan T, Stossel C, Schvimer M, Atias D, Halperin S, Buzhor E, et al. Pancreatic cancer ascites xenograft—an expeditious model mirroring advanced therapeutic resistant disease. Oncotarget 2017;8:40778–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Alexandrov LB, Kim J, Haradhvala NJ, Huang MN, Tian Ng AW, Wu Y, et al. The repertoire of mutational signatures in human cancer. Nature 2020;578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li H, Liu ZY, Wu N, Chen YC, Cheng Q, Wang J. PARP inhibitor resistance: the underlying mechanisms and clinical implications. Mol Cancer 2020;19:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rose M, Burgess JT, O'Byrne K, Richard DJ, Bolderson E. PARP inhibitors: clinical relevance, mechanisms of action and tumor resistance. Front Cell Dev Biol 2020;8:564601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burdett NL, Willis MO, Alsop K, Hunt AL, Pandey A, Hamilton PT, et al. Multiomic analysis of homologous recombination-deficient end-stage high-grade serous ovarian cancer. Nat Genet 2023;55:437–50. [DOI] [PubMed] [Google Scholar]
- 18. Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, et al. Signatures of mutational processes in human cancer. Nature 2013;500:415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Waddell N, Pajic M, Patch AM, Chang DK, Kassahn KS, Bailey P, et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015;518:495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Golan T, O'Kane GM, Denroche RE, Raitses-Gurevich M, Grant RC, Holter S, et al. Genomic features and classification of homologous recombination deficient pancreatic ductal adenocarcinoma. Gastroenterology 2021;160:2119–32. [DOI] [PubMed] [Google Scholar]
- 21. Maxwell KN, Wubbenhorst B, Wenz BM, De Sloover D, Pluta J, Emery L, et al. BRCA locus-specific loss of heterozygosity in germline BRCA1 and BRCA2 carriers. Nat Commun 2017;8:319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abeliovich D, Kaduri L, Lerer I, Weinberg N, Amir G, Sagi M, et al. The founder mutations 185delAG and 5382insC in BRCA1 and 6174delT in BRCA2 appear in 60% of ovarian cancer and 30% of early-onset breast cancer patients among Ashkenazi women. Am J Hum Genet 1997;60:505–14. [PMC free article] [PubMed] [Google Scholar]
- 23. Schayek H, De Marco L, Starinsky-Elbaz S, Rossette M, Laitman Y, Bastos-Rodrigues L, et al. The rate of recurrent BRCA1, BRCA2, and TP53 mutations in the general population, and unselected ovarian cancer cases, in Belo Horizonte, Brazil. Cancer Genet 2016;209:283–4. [DOI] [PubMed] [Google Scholar]
- 24. ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020;578:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Waks AG, Cohen O, Kochupurakkal B, Kim D, Dunn CE, Buendia Buendia J, et al. Reversion and non-reversion mechanisms of resistance to PARP inhibitor or platinum chemotherapy in BRCA1/2-mutant metastatic breast cancer. Ann Oncol 2020;31:590–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Farkkila A, Rodriguez A, Oikkonen J, Gulhan DC, Nguyen H, Dominguez J, et al. Heterogeneity and clonal evolution of acquired PARP inhibitor resistance in TP53- and BRCA1-deficient cells. Cancer Res 2021;81:2774–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hill SJ, Decker B, Roberts EA, Horowitz NS, Muto MG, Worley MJ Jr, et al. Prediction of DNA repair inhibitor response in short-term patient-derived ovarian cancer organoids. Cancer Discov 2018;8:1404–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Replogle JM, Zhou W, Amaro AE, McFarland JM, Villalobos-Ortiz M, Ryan J, et al. Aneuploidy increases resistance to chemotherapeutics by antagonizing cell division. Proc Natl Acad Sci U S A 2020;117:30566–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O'Kane GM, Grunwald BT, Jang GH, Masoomian M, Picardo S, Grant RC, et al. GATA6 expression distinguishes classical and basal-like subtypes in advanced pancreatic cancer. Clin Cancer Res 2020;26:4901–10. [DOI] [PubMed] [Google Scholar]
- 30. Dreyer SB, Upstill-Goddard R, Paulus-Hock V, Paris C, Lampraki EM, Dray E, et al. Targeting DNA damage response and replication stress in pancreatic cancer. Gastroenterology 2021;160:362–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guo W, Zhang Y, Guo S, Mei Z, Liao H, Dong H, et al. Tumor microbiome contributes to an aggressive phenotype in the basal-like subtype of pancreatic cancer. Commun Biol 2021;4:1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Puleo F, Nicolle R, Blum Y, Cros J, Marisa L, Demetter P, et al. Stratification of pancreatic ductal adenocarcinomas based on tumor and microenvironment features. Gastroenterology 2018;155:1999–2013. [DOI] [PubMed] [Google Scholar]
- 35. Bicakli DH, Uslu R, Guney SC, Coker A. The relationship between nutritional status, performance status, and survival among pancreatic cancer patients. Nutr Cancer 2020;72:202–8. [DOI] [PubMed] [Google Scholar]
- 36. Ettrich TJ, Seufferlein T. Systemic therapy for metastatic pancreatic cancer. Curr Treat Options Oncol 2021;22:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dinstag G, Shulman ED, Elis E, Ben-Zvi DS, Tirosh O, Maimon E, et al. Clinically oriented prediction of patient response to targeted and immunotherapies from the tumor transcriptome. Med 2023;4:15–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Steinkamp MP, Lagutina I, Brayer KJ, Schultz F, Burke D, Pankratz VS, et al. Humanized patient-derived xenograft models of disseminated ovarian cancer recapitulate key aspects of the tumor immune environment within the peritoneal cavity. Cancer Res Commun 2023;3:309–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. O'Reilly EM, Lee JW, Zalupski M, Capanu M, Park J, Golan T, et al. Randomized, multicenter, phase II trial of gemcitabine and cisplatin with or without veliparib in patients with pancreas adenocarcinoma and a germline BRCA/PALB2 mutation. J Clin Oncol 2020;38:1378–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Reiss KA, Mick R, O'Hara MH, Teitelbaum U, Karasic TB, Schneider C, et al. Phase II study of maintenance rucaparib in patients with platinum-sensitive advanced pancreatic cancer and a pathogenic germline or somatic variant in BRCA1, BRCA2, or PALB2. J Clin Oncol 2021;39:2497–505. [DOI] [PubMed] [Google Scholar]
- 41. Golan THP, Reni M, Cutsem EV, Macarulla T, Hall MJ, Park JO, et al. Overall survival from the phase 3 POLO trial: maintenance olaparib for germline BRCA-mutated metastatic pancreatic cancer. J Clin Oncol 39, 2021(suppl 3; abstr 378). [Google Scholar]
- 42. Momtaz P, O'Connor CA, Chou JF, Capanu M, Park W, Bandlamudi C, et al. Pancreas cancer and BRCA: a critical subset of patients with improving therapeutic outcomes. Cancer 2021;127:4393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Aung KL, Fischer SE, Denroche RE, Jang GH, Dodd A, Creighton S, et al. Genomics-driven precision medicine for advanced pancreatic cancer: early results from the COMPASS trial. Clin Cancer Res 2018;24:1344–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tobalina L, Armenia J, Irving E, O'Connor MJ, Forment JV. A meta-analysis of reversion mutations in BRCA genes identifies signatures of DNA end-joining repair mechanisms driving therapy resistance. Ann Oncol 2021;32:103–12. [DOI] [PubMed] [Google Scholar]
- 45. Lin KK, Harrell MI, Oza AM, Oaknin A, Ray-Coquard I, Tinker AV, et al. BRCA reversion mutations in circulating tumor DNA predict primary and acquired resistance to the PARP inhibitor rucaparib in high-grade ovarian carcinoma. Cancer Discov 2019;9:210–9. [DOI] [PubMed] [Google Scholar]
- 46. Martinelli P, Carrillo-de Santa Pau E, Cox T, Sainz B Jr, Dusetti N, Greenhalf W, et al. GATA6 regulates EMT and tumour dissemination, and is a marker of response to adjuvant chemotherapy in pancreatic cancer. Gut 2017;66:1665–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Moffitt RA, Marayati R, Flate EL, Volmar KE, Loeza SG, Hoadley KA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet 2015;47:1168–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee AJ, Endesfelder D, Rowan AJ, Walther A, Birkbak NJ, Futreal PA, et al. Chromosomal instability confers intrinsic multidrug resistance. Cancer Res 2011;71:1858–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Passerini V, Ozeri-Galai E, de Pagter MS, Donnelly N, Schmalbrock S, Kloosterman WP, et al. The presence of extra chromosomes leads to genomic instability. Nat Commun 2016;7:10754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sokolenko AP, Savonevich EL, Ivantsov AO, Raskin GA, Kuligina ES, Gorodnova TV, et al. Rapid selection of BRCA1-proficient tumor cells during neoadjuvant therapy for ovarian cancer in BRCA1 mutation carriers. Cancer Lett 2017;397:127–32. [DOI] [PubMed] [Google Scholar]
- 51. Chan-Seng-Yue M, Kim JC, Wilson GW, Ng K, Figueroa EF, O'Kane GM, et al. Transcription phenotypes of pancreatic cancer are driven by genomic events during tumor evolution. Nat Genet 2020;52:231–40. [DOI] [PubMed] [Google Scholar]
- 52. Nicolle R, Blum Y, Duconseil P, Vanbrugghe C, Brandone N, Poizat F, et al. Establishment of a pancreatic adenocarcinoma molecular gradient (PAMG) that predicts the clinical outcome of pancreatic cancer. EBioMedicine 2020;57:102858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pan CX, Zhang H, Tepper CG, Lin TY, Davis RR, Keck J, et al. Development and characterization of bladder cancer patient-derived xenografts for molecularly guided targeted therapy. PLoS One 2015;10:e0134346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Song HN, Lee C, Kim ST, Kim SY, Kim NK, Jang J, et al. Molecular characterization of colorectal cancer patients and concomitant patient-derived tumor cell establishment. Oncotarget 2016;7:19610–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gao H, Korn JM, Ferretti S, Monahan JE, Wang Y, Singh M, et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med 2015;21:1318–25. [DOI] [PubMed] [Google Scholar]
- 56. Pettitt SJ, Frankum JR, Punta M, Lise S, Alexander J, Chen Y, et al. Clinical BRCA1/2 reversion analysis identifies hotspot mutations and predicted neoantigens associated with therapy resistance. Cancer Discov 2020;10:1475–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Terrero G, Datta J, Dennison J, Sussman DA, Lohse I, Merchant NB, et al. Ipilimumab/nivolumab therapy in patients with metastatic pancreatic or biliary cancer with homologous recombination deficiency pathogenic germline variants. JAMA Oncol 2022;8:1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Seeber A, Puccini A, Xiu J, Goldberg RM, Grothey A, Shields AF, et al. Association of BRCA-mutant pancreatic cancer with high tumor mutational burden (TMB) and higher PD-L1 expression. J Clin Oncol 2019;37:4133. [Google Scholar]
- 59. Terrero G, Pollack T, Sussman DA, Lockhart AC, Hosein PJ. Exceptional responses to ipilimumab/nivolumab (ipi/nivo) in patients (pts) with refractory pancreatic ductal adenocarcinoma (PDAC) and germline BRCA or RAD51 mutations. J Clin Oncol 2022;38:754. [Google Scholar]
- 60. Hosein AN, Dougan SK, Aguirre AJ, Maitra A. Translational advances in pancreatic ductal adenocarcinoma therapy. Nat Cancer 2022;3:272–86. [DOI] [PubMed] [Google Scholar]
- 61. Samstein RM, Krishna C, Ma X, Pei X, Lee KW, Makarov V, et al. Mutations in BRCA1 and BRCA2 differentially affect the tumor microenvironment and response to checkpoint blockade immunotherapy. Nat Cancer 2021;1:1188–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bakhoum SF, Ngo B, Laughney AM, Cavallo JA, Murphy CJ, Ly P, et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018;553:467–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Shen J, Zhao W, Ju Z, Wang L, Peng Y, Labrie M, et al. PARPi triggers the STING-dependent immune response and enhances the therapeutic efficacy of immune checkpoint blockade independent of BRCAness. Cancer Res 2019;79:311–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Jiao S, Xia W, Yamaguchi H, Wei Y, Chen MK, Hsu JM, et al. PARP inhibitor upregulates PD-L1 expression and enhances cancer-associated immunosuppression. Clin Cancer Res 2017;23:3711–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Neftel C, Laffy J, Filbin MG, Hara T, Shore ME, Rahme GJ, et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 2019;178:835–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Raghavan S, Winter PS, Navia AW, Williams HL, DenAdel A, Lowder KE, et al. Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer. Cell 2021;184:6119–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
In vivo, EVOC and clinical response
Clinical data of analyzed BRCA PDAC patients
20 genes in DDR resistance
In vivo growth kinetics and EVOC efficacy in acquired resistance model
Mechanisms of resistance in glBRCA PDAC
Data Availability Statement
PDX models generated at SMC will be available upon completion of material transfer agreements.
Raw sequencing data are housed at the Gene Expression Omnibus portal under accession number GSE235843 (RNA-seq data) and the European Genome-phenome Archive under accession number EGAD00001011129 (WGS data), and will be available to researchers upon completion of a data access agreement.



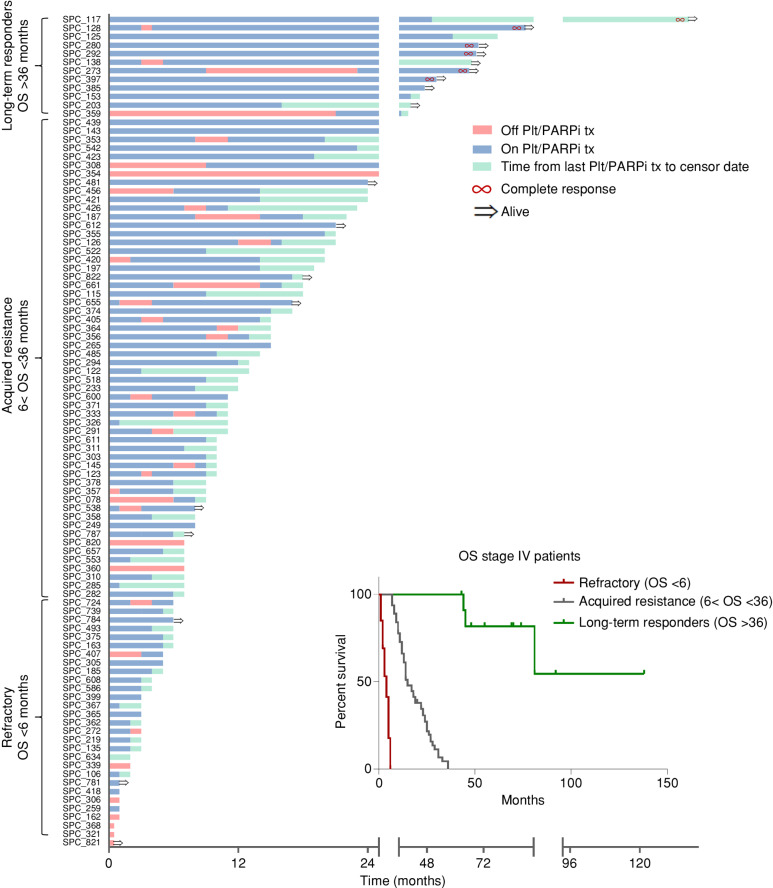
![Figure 2. Efficacy of cisplatin and olaparib in preclinical models and correlation to clinical response. A, In vivo antitumor activity of PARPi (olaparib) and cisplatin in glBRCA PDAC PDXs. Tumor growth (y-axis) for each PDX model after 28 to 52 days (x-axis) of cisplatin (2 mg/kg; i.p.; once weekly) or olaparib (50 mg/kg; i.p.; 5 days on/2 days off) treatment. The dashed line represented a 70% threshold for sensitivity. Columns color code models by the clinical subgroup of response to platinum/PARPi at the time of tissue acquisition (green, sensitive; yellow, refractory; red, acquired resistance; and gray, unknown). B, Tumor growth curves of sensitive (SPC_467; top) and acquired resistance (SPC_187) models. Average relative tumor volume (y-axis) of control, cisplatin-treated (2 mg/kg; i.p; once weekly), and olaparib-treated (50 mg/kg; i.p.; 5 days on/2 days off) mice (n = 6–8 mice/group). C, EVOC H&E representative images of sensitive (SPC_467) and resistant (SPC_187) PDX models. Scale bar indicated in the figure. Insert, magnification ×2.5. D, A 2 × 2 table showing the distribution of responders and nonresponders based on EVOC prediction and clinical actual response to PARPi and platinum agents. E, Correlation between the in vivo and EVOC efficacy for each model and treatment; y-axis, in vivo tumor growth (%); x-axis, EVOC score (0–60 resistant; 61–100 sensitive); black circles [cisplatin (Cis)]; and gray circles [olaparib (Olap)]. r2 = 0.59; P < 0.001.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/ed78/10401074/12f11c1b4308/1826fig2.jpg)
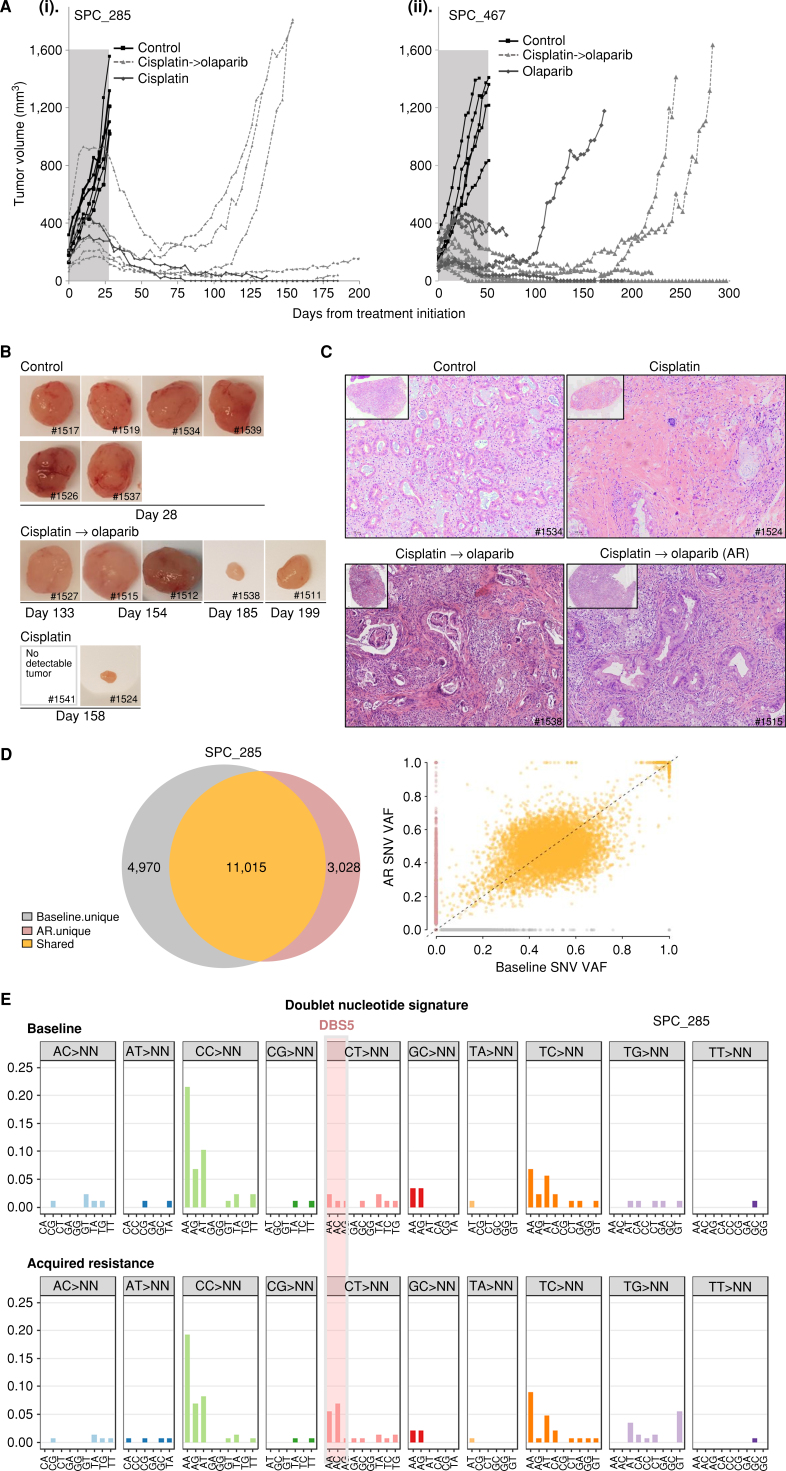
![Figure 4. WGS and mechanisms of resistance. A, Genomic mutational profiles of 26 whole-genome sequenced tumors. Tumor mutational burden (TMB), HR status, BRCA gene allelic state, mutated BRCA gene, patient clinical response, patient platinum exposure, presence of platinum mutational signature, HRDetect score, HRD mutational signature proportion, tumor ploidy, and mutations observed in PDAC driver genes are shown. HRD−SM, HRDsecond_mut; SV, structural variant. B, Table of resistant samples with specific information. WT, wild-type. C, Illustration of secondary mutation reversing reading frame back to WT in patient SPC_187. D, Integrative Genomics Viewer (IGV) snapshot of SPC_291 WGS reads mapping to the BRCA2 locus. Reads from normal are shown in the middle, where the blue box highlights a 1-bp germline frameshift with a zoomed-in view on the right. Reads from tumor are shown on top, where the orange box highlights a somatic SV that deletes the region around the germline frameshift. Zoomed-in view of somatic SV breakpoints at the bottom shows BRCA2 codons reverted to inframe as a result of SV. E, IGV snapshot capturing the BRCA2 germline (left) and the somatic (right) insertion or deletion (indel) frameshift in SPC285. Reads mapping to this region are shown for the germline reference (bottom), platinum-sensitive baseline xenograft (middle), and acquired resistance (AR) xenograft (top). The AR clone was analyzed by WGS and compared with baseline PDX. F, Distance from diploid [absolute(ploidy-2)] in resistant and sensitive samples (P < 0.05). G, Molecular subtype by response—basal-like score–classical score in resistant and sensitive samples (P < 0.05). H, SHAP analysis. A detailed perspective on the contribution of the various factors to the resistance classification score. Each line corresponds to one factor and each dot in it to a single sample (patient). The color of the dot represents the value of the factor for that sample (high BRCA allelic state value corresponds to monoallelic, high reversion-mutation value corresponds to the presence of the reverse mutation, high subtype value corresponds to “basal-like,” high sex value corresponds to female, and high BRCA value corresponds to BRCA2). The location of the dot on the x-axis is the relevant SHAP value of that factor for that sample. As an example, the red dots on the far right at the “Reversion mutation” line indicate that the presence of this mutation (in five patients) is a very strong indicator of resistance. The features on the y-axis are ordered by their average importance.](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/ed78/10401074/fff883e132b1/1826fig4.jpg)