Abstract
BACKGROUND
Coronavirus disease 2019 (COVID-19) has demonstrated several clinical manifestations which include not only respiratory system issues but also liver, kidney, and other organ injuries. One of these abnormalities is coagulopathies, including thrombosis and disseminated intravascular coagulation. Because of this, the administration of low molecular weight heparin is required for patients that need to be hospitalized. In addition, Remdesivir is an antiviral that was used against Middle East Acute Respiratory Syndrome, Ebola, Acute Respiratory Syndrome, and other diseases, showing satisfactory results on recovery. Besides, there is evidence suggesting that this medication can provide a better prognosis for patients with COVID-19.
AIM
To investigate in silico the interaction between Remdesivir and clotting factors, pursuing a possibility of using it as medicine.
METHODS
In this in silico study, the 3D structures of angiotensin-converting enzyme 2 (ACE2), Factor I (fibrinogen), Factor II (prothrombin), Factor III (thromboplastin), Factor V (proaccelerin), Factor VII (proconvertin), Factor VIII (antihemophilic factor A), Factor IX (antihemophilic factor B), Factor X (Stuart-Prower factor), and Factor XI (precursor of thromboplastin (these structures are technically called receptors) were selected from the Protein Data Bank. The structures of the antivirals Remdesivir and Osetalmivir (these structures are called ligands) were selected from the PubChem database, while the structure of Atazanavir was selected from the ZINC database. The software AutoDock Tools (ADT) was used to prepare the receptors for molecular docking. Ions, peptides, water molecules, and other ones were removed from each ligand, and then, hydrogen atoms were added to the structures. The grid box was delimited and calculated using the same software ADT. A physiological environment with pH 7.4 is needed to make the ligands interact with the receptors, and still the software Marvin sketch® (ChemAxon®) was used to forecast the protonation state. To perform molecular docking, ADT and Vina software was connected. Using PyMol® software and Discovery studio® software from BIOVIA, it was possible to analyze the amino acid residues from receptors that were involved in the interactions with the ligands. Ligand tortions, atoms that participated in the interactions, and the type, strength, and duration of the interactions were also analyzed using those software.
RESULTS
Molecular docking analysis showed that Remdesivir and ACE2 had an affinity energy of -8.8 kcal/moL, forming a complex with eight hydrogen bonds involving seven atoms of Remdesivir and five amino acid residues of ACE2. Remdesivir and prothrombin had an interaction with six hydrogen bonds involving atoms of the drug and five amino acid residues of the clotting factor. Similar to that, Remdesivir and thromboplastin presented interactions via seven hydrogen bonds involving five atoms of the drug and four residues of the clotting factor. While Remdesivir and Factor V established a complex with seven hydrogen bonds between six antiviral atoms and six amino acid residues from the factor, and Factor VII connected with the drug by four hydrogen bonds, which involved three atoms of the drug and three residues of amino acids of the factor. The complex between Remdesivir and Factor IX formed an interaction via 11 hydrophilic bonds with seven atoms of the drug and seven residues of the clotting factor, plus one electrostatic bond and three hydrophobic interactions. Factor X and Remdesivir had an affinity energy of -9.6 kcal/moL, and the complex presented 10 hydrogen bonds and 14 different hydrophobic interactions which involved nine atoms of the drug and 16 amino acid residues of the clotting factor. The interaction between Remdesivir and Factor XI formed five hydrogen bonds involving five amino acid residues of the clotting factor and five of the antiviral atoms.
CONCLUSION
Because of the in silico significant affinity, Remdesivir possibly could act in the severe acute respiratory syndrome coronavirus 2 infection blockade by interacting with ACE2 and concomitantly act in the modulation of the coagulation cascade preventing the hypercoagulable state.
Keywords: Clotting factors, Coagulating blood cascade, COVID-19 treatment, Remdesivir, SARS-CoV-2
Core Tip: In the initial period of the emergence of coronavirus disease 2019, bioinformatics tools filled the need for rapid knowledge about the severe acute respiratory syndrome coronavirus 2 and research in search of drug treatment, since experimental research takes time that was not available. Bioinformatics continues to be used for the same purpose in this study, remembering that this methodology does not rule out either in vivo or in vitro testing.
INTRODUCTION
At the end of 2019, the health system of Wuhan, China reported an alarming growth in the number of patients with severe acute respiratory syndrome (SARS) triggered by a new coronavirus, the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2)[1-3]. A highly contagious disease, coronavirus disease 2019 (COVID-19), spread across several countries[4] in the following months, culminating in the decree of a pandemic by the World Health Organization on March 11, 2020[1]. Due to the high transmissibility of the disease and the increase in the number of deaths, several governments imposed measures of using face masks, isolation of new cases and social distancing, and restriction of circulation, with the closure of schools and universities, prohibition of agglomerations, as well as decreeing the paralyzing of non-essential activities in an attempt to slow down the spread of the disease[2].
The most widespread form of contagion of SARS-CoV-2 is through the contact with droplets of oropharynx secretions from an infected person. In addition, the transmission of the virus is aggravated by its incubation time, which varies from 3 to 14 d, and by the probability of asymptomatic people or those with mild symptoms transmitting the disease[5]. COVID-19 may not manifest symptoms (asymptomatic patients), manifest mild, moderate, or severe symptoms, and may progress to severe pneumonia and death[6,7]. In addition to the respiratory system, other organs and systems may also present injury from viral infection, such as the liver, kidneys, heart, and neural system[8]. Thus, although respiratory failure is the main factor leading to death, causes linked to organ failure, thrombosis, and disseminated intravascular coagulation (DIC), accompanied by intense inflammatory response, are also responsible for a great decline in the health status of patients[9]. Of the COVID-19 positive patients who had DIC, only 0.6% survived, which demonstrates the high lethality at this stage of the disease[10,11].
The immune response to SARS-CoV-2 infection is associated with the prevalence of coagulation pathway activation and the large increase in the production of pro-inflammatory cytokines, tumor necrosis factor (TNF)-alpha and IFN-gamma, a condition called "cytokine storm". In addition to acting in the conversion reaction of fibrinogen into fibrin to form the clot, thrombin can also act in increasing inflammation, since these inflammatory events are related to high concentrations of this enzyme and low concentrations of proteinase activated receptors, antithrombin III, and protein C[12]. Although the coagulopathies observed in these patients are reminiscent of DIC and other thrombotic events, they have characteristics inherent to COVID-19[9]. D-dimer, a product of fibrin degradation, when elevated, has been associated with a higher mortality rate[13]. Some studies have reported that the increase in D-dimer and fibrinogen in the bloodstream is associated with a poor prognosis in patients with COVID-19[14,15]. At other times, these parameters are not consistent with the severity of pathological conditions[9,10,16]. In any case, the severity of COVID-19 promotes the imbalance of coagulation and anticoagulation factors, in addition to precipitating the formation of thrombosis, DIC, respiratory deterioration, and multiple organ failure[11].
The International Society of Thrombosis and Hemostasis recommends that low molecular weight heparin (LMWH) be administered in patients in need of hospitalization, that prophylactic doses of LMWH should be administered in all hospitalized patients, and that D-dimer, prothrombin, activated partial thromboplastin, and fibrinogen levels be monitored daily[17]. The adoption of LMWH was recommended since the tests in patients treated with prophylactic doses had better prognoses and a reduction of mortality when compared to the untreated ones[18].
Near the end of 2020, studies were underway for the development of prophylactic vaccines against COVID-19. Currently, several vaccines are available to the world population, with different vaccination strategies in relation to the number of doses. As for the present time, the epidemic is controlled and vaccine prophylaxis contributed to this scenario. However, the situation of the COVID-19 pandemic is still unpredictable. There is no effective vaccine in fact, given reinfections, or specific antiviral drugs to treat patients with severe COVID-19[19].
The treatment of severe cases, accompanied by DIC, represents a challenge in the therapeutic process of safeguarding the lives of patients. Although there are studies on experimental drugs considered promising, the lack of proven and effective treatment for severe cases of COVID-19 raises the possibility of repurposing drugs, i.e., the reuse of existing drugs, in order to optimize time in controlling disease progression and reduce the cost spent by health systems[20,21].
Remdesivir (GS-5734) is a broad-spectrum antiviral that has been shown to be effective in containing the replication of pathogens that cause Middle East Acute Respiratory Syndrome, Acute Respiratory Syndrome (SARS-CoV), Ebola, and other infectious agents[22-24]. This drug is a prodrug of adenosine and its metabolic product, the active triphosphate GS-443902, is analogous to ATP and inhibits by competition the enzyme RNA-dependent RNA polymerase[25-28]. Studies conducted with COVID-19-positive patients, who were treated with Remdesivir, showed satisfactory results of faster recovery when compared to patients not treated or those who received placebo[29-32]. In addition, a report validated results of hospitalized patients with severe COVID-19 who were treated with Remdesivir, suggesting that the drug can prevent the progression of the severity of infection, since in the treated group there was less evolution of cases[30].
The analysis of data and hypotheses about the action and effectiveness of drugs, such as Remdesivir, can be aided by bioinformatics tools that can act as a method of understanding and predicting the molecular interactions of the drug with possible molecules that may be involved in the process of viral infection or in the development of symptoms and pathological conditions. In this context, some in silico studies have proposed candidate drugs against some target structures of SARS-CoV-2 that have been deposited in the databases until then[33,34]. Based on the hypothesis that Remdesivir acts in symptomatic events of COVID-19 and according to the premises that patients' coagulopathies are of great severity and that Remdesivir is an approved and useful drug in the treatment of patients affected by COVID-19, this study aimed to verify by molecular docking the interaction of Remdesivir with coagulation cascade factors, seeking to contribute to the best understanding of its action and application of COVID-19 pharmacological therapy.
MATERIALS AND METHODS
Preparation of receptors and ligands
The three-dimensional structures of the human proteins were obtained from the Protein Data Bank (PDB) (rcsb.org/pdb): ACE2 (PDB:6VW1), Factor I (fibrinogen; PDB:3GHG), Factor II (prothrombin; PDB:3U69), Factor III (thromboplastin; PDB:5W06), Factor V (proaccelerin; PDB:1CZT), Factor VII (proconvertin; PDB:1KLJ), Factor VIII (antihemophilic factor A; PDB:2R7E), Factor IX (antihemophilic factor B; PDB:6MV4), Factor X clotting factor (Stuart-Prower factor; PDB:4Y71), and Factor XI (thromboplastin precursor; PDB:5QTU). These structures, technically called receptors, were prepared for molecular docking using the AutoDock Tools 4 (ADT) software (autodock.scripps.edu). Any ions, peptides, and molecules previously attached to the receptor, as well as water molecules derived from the crystallization technique for resolution of the structure by X-ray diffraction, were removed at this stage. Subsequently, missing hydrogen atoms were added to the receptors, and the location and dimensions of the grid box were determined. The grid box features x, y, and z coordinates were built with grid spacing of 1.0 Å using the AutoGrid component of the ADT software. Grid box data and coordinates were used in molecular fitting.
The three-dimensional structures of the ligands (drugs) were obtained from two different sources. In the PubChem database (pubchem.ncbi.nlm.nih.gov), the three-dimensional structures of the antivirals Remdesivir (PubChem 121304016) and Oseltamivir (PubChem 65028) were obtained. Oseltamivir was administered with good results in patients with COVID-19 without hypoxia[35].
In the ZINC database (zinc12.docking.org), the three-dimensional structure of the antiviral Atazanavir (ZINC 3941496) was obtained. The ligands interact with their receptors in a physiological environment with pH 7.4. Thus, to reliably simulate their interaction, a calculation was performed to predict the protonation state using Marvin sketch® software (ChemAxon®). Protonated ligands were prepared for molecular docking using ADT software which detects the torsion points of the inhibitors and calculates their torsion angle.
Molecular docking
Molecular docking simulations were performed using ADT software connected to AutoDock Vina v.1.2.0. software[36]. This software associates two components - a search algorithm and a scoring function. The algorithm is responsible for searching for possible combinations in the bindings, and exploring the degrees of rotational, translational, and conformational freedom of the ligand as well as proteins. Then the scoring function is used to choose the best binding modes. These functions are obtained according to the strength fields of molecular mechanics and empirical parameters from free energy calculations, so an affinity energy is calculated. An affinity energy < -6.0 kcal/moL is considered the cutoff point for stable interactions[37]. The results are based on the first conformer of the ligands, with the reference root-mean-square deviation of atomic position equaling zero[38]. The analyses of amino acid residues of the receptors involved in the interactions with the ligands, as well as the ligand tortions, atoms involved in the interactions, and the type, strength, and duration of the interactions, were performed using the PyMol® (Schrödinger, Inc.) and BIOVIA Discovery studio® (Dassault Systemes) software.
RESULTS
Among the complexes formed between protein and antiviral drugs, the most stable ones were selected, according to the affinity energy value. Simulations between Atazanavir, Remdesivir, and Osetalmivir with ACE2, Factors I (fibrinogen) and II (protrombin) indicated significantly greater stability between the three proteins and Remdesivir (Table 1). Due to the most promising data of Remdesivir, the analyses followed with this drug. Molecular docking was performed between Remdesivir and other clotting factors and the drug interacted with all tested factors in silico (Table 2). Remdesivir and ACE2 formed a complex through eight hydrogen bonds involving seven atoms of Remdesivir and five amino acid residues of ACE2 (Table 3); affinity energy was -8.8 kcal/moL. In molecular docking simulations, Remdesivir was used in its protonation state at pH 7.4, in which the amount of the drug is 99.85% protonated.
Table 1.
Energy of affinity of ligand/receptor complexes obtained in molecular docking and characteristics of interactions between Remdesivir and angiotensin-converting enzyme 2
|
Receptor/ligand complex
|
Energy of affinity (kcal/moL)
|
| ACE2/Remdesivir | -8.8 |
| ACE2/Osetalmivir | -5.7 |
| ACE2/Atazanavir | -3.9 |
| Fibrinogen/Remdesivir | -8.5 |
| Fibrinogen/Osetalmivir | -6.3 |
| Fibrinogen/Atazanavir | -4 |
| Prothrombin/Remdesivir | -8.4 |
| Prothrombin/Osetalmivir | -6 |
| Prothrombin/Atazanavir | -4.1 |
ACE2: Angiotensin-converting enzyme 2.
Table 2.
Characteristics of interactions between Remdesivir and angiotensin-converting enzyme 2
|
|
Interaction1
|
Distance (Å)
|
Bond type
|
| ACE2/Remdesivir | N-18:GLU-398 | 3.3 | Hydrogen bond |
| O-29:HIS-378 | 2.5 | Hydrogen bond | |
| O-30:ALA-348 | 2.4 | Hydrogen bond | |
| O-30:ALA-348 | 3.3 | Hydrogen bond | |
| O-34:ALA-348 | 3.2 | Hydrogen bond | |
| O-36:ASP-350 | 2.3 | Hydrogen bond | |
| O-37:ASP-350 | 3.4 | Hydrogen bond | |
| O-40:ARG-393 | 3.5 | Hydrogen bond |
Remdesivir: Residue. ACE2: Angiotensin-converting enzyme 2; O: Oxygen; N: Nitrogen; GLU: Glutamate; ALA: Alanine; ASP: Aspartate; ARG: Arginine.
Table 3.
Energy of affinity of ligand/receptor complexes obtained in silico
|
Ligand/receptor complex
|
Energy of affinity (kcal/moL)
|
| Remdesivir/tissue factor (thromboplastin) | -8.2 |
| Remdesivir/Factor V (proacelerin) | -6.8 |
| Remdesivir/Factor VII (proconvertin) | -7.0 |
| Remdesivir/Factor IX (anti-hemophilic B) | -6.4 |
| Remdesivir/Factor X | -9.6 |
| Remdesivir/Factor XI | -8.3 |
The association between Remdesivir and prothrombin occurred through six hydrogen bonds involving atoms of the drug and five amino acid residues of the clotting factor (Table 4); similarly, Remdesivir/thromboplastin complex was stabilized by seven hydrogen bonds involving five atoms of the drug and four residues of the clotting factor.
Table 4.
Binding characteristics of Remdesivir/Factor II and Remdesivir/Factor VII complexes
| Complex | Interaction 1 | Distance (Å) | Bond type |
| Remdesivir/Factor II II (protrombin) | O-23: GLY-548 | 2 | Hydrogen bond |
| O-3: GLY-548 | 3.2 | Hydrogen bond | |
| N-15: ASN-463 | 2.6 | Hydrogen bond | |
| O-35: SER-525 | 2.4 | Hydrogen bond | |
| O-35: HIS-363 | 2.6 | Hydrogen bond | |
| O-3: GLY-550 | 2 | Hydrogen bond | |
| Remdesivir/Factor VII (Proconvertin) | O-3: ASP-60 | 3.5 | Hydrogen bond |
| O-3: THR-98 | 3.1 | Hydrogen bond | |
| O-2: THR-98 | 3.6 | Hydrogen bond | |
| O-2: GLY-97 | 3.2 | Hydrogen bond | |
| N-17: GLY-216 | 3.1 | Hydrogen bond |
Remdesivir: Residue. O: Oxygen; N: Nitrogen; ASP: Aspartate; ASN: Asparagine; THR: Threonine; SER: Serine; HIS: Histidine; GLY: Glycine.
Factor V and Remdesivir formed a complex through seven hydrogen bonds between six antiviral atoms and six amino acid residues from the factor. Four hydrogen bonds were observed between Remdesivir and Factor VII. These bonds involved three atoms of the drug and three residues of amino acids of the protein (Table 4). The Remdesivir/Factor IX complex was stabilized by 11 hydrophilic bonds (hydrogen bonds) involving seven atoms of the drug and seven residues of the clotting factor, one electrostatic bond, and three hydrophobic interactions.
Factor X and Remdesivir were associated with an energy of affinity equal to -9.6 kcal/moL. The complex was established and maintained by hydrogen bonds (10) and hydrophobic interactions (14) of varied types involving nine atoms of the drug and 16 amino acid residues of the clotting factor (Table 5). Remdesivir and Factor XI formed a complex by the interaction between five amino acid residues of the clotting factor and five antiviral atoms by means of five hydrogen bonds. Complexes with significant affinity energy, whose physiological behavior is relevant to explain the supposed action of Remdesivir as a treatment of symptoms resulting from COVID-19, are shown in Figure 1.
Table 5.
Binding characteristics of Remdesivir/Factor X complex
| Interaction 1 | Distance (Å) | Bond type |
| N-18: ALA-190 | 3.33 | Hydrogen bond |
| N-18: GLY-219 | 3.44 | Hydrogen bond |
| N-23: GLY-193 | 2.3 | Hydrogen bond |
| N-23: GLN-192 | 3.35 | Hydrogen bond |
| O-29: SER-195 | 2.92 | Hydrogen bond |
| O-29: SER-195 | 2.29 | Hydrogen bond |
| O-29: SER-214 | 2.78 | Hydrogen bond |
| O-29: HIS-57 | 2.92 | Hydrogen bond |
| O-29: HIS-57 | 2.53 | Hydrogen bond |
| O-30: HIS-57 | 2.93 | Hydrogen bond |
| Aromatic ring C-3: TRP-215 | 4.21 | Pi-stacking |
| Aromatic ring C-3: TRP-215 | 4.03 | Pi-stacking |
| Aromatic ring C-3: TYR-99 | 5.06 | Pi-stacking |
| Aromatic ring C-3: PHE-174 | 5.13 | Pi-stacking |
| Aromatic ring C-8: CYS-191 and GLN-192 | 4.57 | Amide pi-stacking |
| Aromatic ring C-31: CYS-191 and GLN-192 | 4.31 | Amide pi-stacking |
| Aromatic ring C-8: TRP-215 and GLY-216 | 4.53 | Amide pi-stacking |
| Aromatic ring C-31: TRP-215 and GLY-216 | 4.89 | Amide pi-stacking |
| C-2: LYS-96 | 3.96 | Alkyl |
| C-2: TYR-60 | 5.05 | Pi-alkyl |
| C-2: PHE-94 | 5.39 | Pi-alkyl |
| C-2: PHE-94 | 4.87 | Pi-alkyl |
| Aromatic ring C-8: CYS-220 | 5.47 | Pi-alkyl |
| Aromatic ring C-31: ALA-190 | 5.29 | Pi-alkyl |
Remdesivir: Residue. O: Oxygen; N: Nitrogen; C: Carbon; ALA: Alanine; GLY: Glycine; SER: Serine; HIS: Histidine; TRP: Tryptophan; TYR: Tyrosine; PHE: Phenylalanine; CYS: Cysteine; LYS: Lysine.
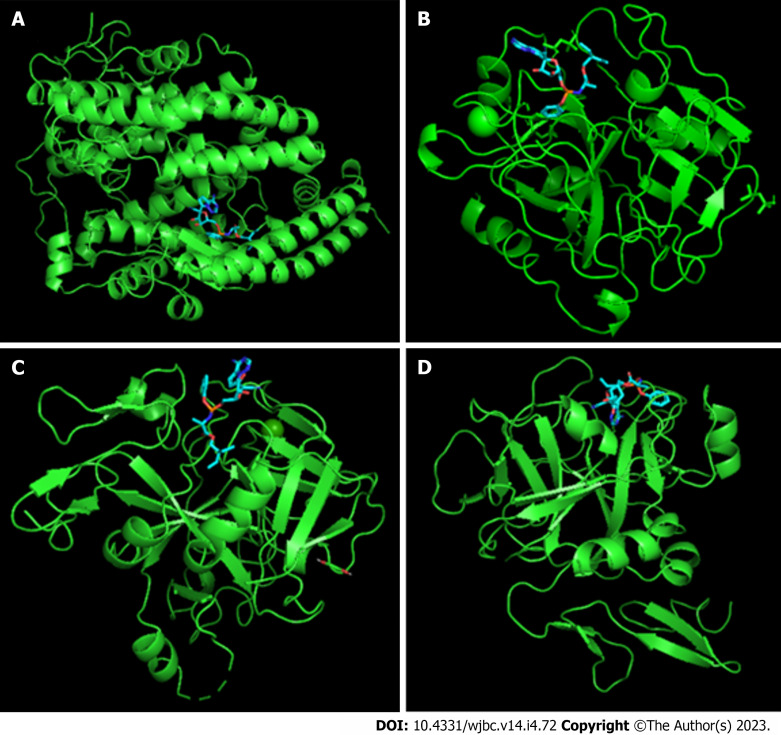
Figure 1.
Remdesivir/receptor complexes. A: Remdesivir/angiotensin-converting enzyme 2; B: Remdesivir/Factor II; C: Remdesivir/Factor VII; D: Remdesivir/Factor X. Receptors are shown in green and Remdesivir is shown in blue/red.
DISCUSSION
Since the affinity energy value between two structures must be less than −6.0 kcal/moL for the interaction to be considered stable[37], it is possible to point out that Remdesivir was the only antiviral that presented itself within this premise. Remdesivir is a pro-drug, analogous to adenosine, which acts by inhibiting the viral replication of several viral families, including the Coronaviridae family. With regard to its mechanism of action, it is observed that it competes with the endogenous ATP for incorporation at viral RNA through RNA-dependent RNA polymerase, triggering the termination of the chain[39].
The in silico analyses indicated that Remdesivir interacts with clotting factors, so in the investigation of the consequences of its use in patients with COVID-19, it is important to analyze the possible routes of action of the drug, as well as the related effects, since during infection by SARS-CoV-2, a broad spectrum of changes from bleeding to thrombus formation can occur[40]. Therefore, for the understanding of the interference of Remdesivir in the coagulation, it is essential to understand its physiological functioning.
It is important to highlight that the pathways generated by the cadence action of the different agents that are part of the hemostatic process are not fixed to a single series, occurring simultaneously and in ways not completely understood in vivo[41]. As described in an article by a researcher of University Hospital of Wales (Cardiff, United Kingdom), hemostasis is triggered by endothelial damage, with collagen exposure and tissue factor, and this process is accompanied by endothelial secretion that results in localized vasoconstriction reflex, reducing local blood flow[42].
Based on the results of molecular docking, it was possible to observe that in addition to the stable interaction with ACE2, Remdesivir also interacted, with significant affinity energy, with clotting factors of the intrinsic, extrinsic, and common pathways. It is emphasized that Factors V (common pathway) and IX (intrinsic pathway) had less significant interactions (> -7 kcal/moL) with the drug in relation to the other clotting factors, which may result in a lower interference in the cascade from the inhibition of these two factors and, consequently, in anticoagulation.
However, there was significant interaction (-7.0 kcal/moL) between Remdesivir and Factor VII, an initial factor of the cascade. Activated Factor VII (Factor VIIa) joins the tissue factor, forming a complex that activates Factor IX, leading to thrombin production; this same complex activates more Factor VII molecules, so even with a low initial level of this factor, its effectiveness is satisfactory due to the action of the factor/tissue factor complex[43]. In this respect, exclusively for the possible inhibition of Factor VII by Remdesivir, a significant alteration in the cascade does not necessarily occur, because the release of tissue factor is an essential condition for the formation of the complex and occurs in case of endothelial damage, increased during the pro-inflammatory state induced by SARS-CoV-2 infection[44]. In addition, low levels of Factor VIIa are sufficient to feed back the track.
Between Remdesivir and Factor X (common pathway), an interaction with an affinity energy value of -9.6 kcal/moL was recorded, which is the most significant interaction observed in this study, so this would be the most stable and most favorable interaction to occur in vivo and may be an important means of inhibiting the coagulation cascade. Secondary hemostasis is formed by the extrinsic and intrinsic pathways, activated by different factors, which converge in the activation of Factor X, resulting in a common pathway, and inhibition of this factor has been shown to prevent the progression of the cascade in baboon models[45]. In the common pathway, Factor Xa associated with Factor Va (activated by thrombin) cleaves prothrombin into thrombin. Thrombin (Factor IIa), which potentiates secondary hemostasis, along with Ca2+, activates Factor XIII and also cleaves fibrinogen into fibrin, forming polymers. Associated with Factor XIIIa, fibrin polymers form cross-links. In the convergence of the two pathways, the activation of Factor X allows the proteolytic cleavage of prothrombin into thrombin, which converts fibrinogen (soluble) into fibrin (insoluble); activates Factor XIII, which establishes cross-links in the fibrin network; potentiates the coagulation cascade by catalyzing Factors VIII and V; potentiates the coagulation cascade by catalyzing Factors VIII and V; and promotes platelet activation[46]. Thus, additional anticoagulatory effects could occur due to the relevance of Factor X in hemostasis and its interaction with Remdesivir.
Meanwhile, Factors I, II, III, and XI also connected to Remdesivir with significant affinity energies (between -9.0 and -8.0 kcal/moL), so these factors would be possible sites of action of the drug in the coagulation cascade. Factor XIa in the presence of Ca2+ and, separately, Factor VIIa activate factor IX, which, together with Factor VIIIa, activates Factor X[47]. Thus, combined with inhibition of Factor X, all these possible interactions with the drug would result in anticoagulation effects, which would be additional to the main mechanism of antiviral action of Remdesivir.
In view of the possibility of interaction of Remdesivir with components of hemostasis, it is necessary to discuss the possible pharmacodynamic interactions between this drug and anticoagulants that have been widely used for the treatment of patients with COVID-19, including heparins, warfarin, and new oral anticoagulants (NOAC).
About the class of heparins, represented by unfractionated heparin (UFH), LMWH, and Fondaparinux, it is contextualized that these compounds do not present intrinsic anticoagulant activity, but they bind to antithrombin to trigger a higher rate of inhibition of coagulation proteases[48]. Considering the three representatives, among the pharmacodynamic differences, the influence of variation on the number of chains of saccharides that make up the molecules stands out, in which the length of these molecules influences the binding and inactivation capacity of specific components of the coagulation cascade, conferring particularities to each molecule. For example, in the case of UFH, to potentiate thrombin inhibition (Factor IIa), chains composed of at least 18 units of saccharides (approximate molecular weight of 5400 Da) are required. By presenting in their composition molecules with an average weight of 15000 Da, virtually all UFH molecules are long enough for this function. On the other hand, LMWH, advantageously, has a greater capacity to potentiate the inhibition of Factor Xa than thrombin, and this results from its average molecular weight being between 4500 to 5000 Da, resulting in the preference of inhibition of Factor Xa in relation to thrombin. In relation to Fondaparinux, it accelerates only the inhibition of Factor Xa by antithrombin, and its length is insufficient to bind antithrombin to thrombin, which justifies the selectivity of action[49].
The NOAC, represented by the drugs Dabigatran, Rivaroxabana, Apixabana, and Edoxabana, target the same clotting factors, with the first being an active compound that binds competitively and reversibly to the active thrombin site (Factor IIa), preventing procoagulant activity, while the others acting as direct inhibitors of Factor Xa.
Considering these particularities, it can be inferred that Remdesivir has the potential for synergistic interaction with the aforementioned drugs. In this sense, due to the stable affinity energy (-9.6 kcal/moL) of Remdesivir/Factor Xa complex, UFH, LMWH, Fondaparinux, Rivaroxaban, Apixaban, and Edoxabana may have their effects pronounced when associated with the antiviral[50]. Similarly, this effect can occur with Dabigatran and heparins, since they are connected to Factor IIa, but with less emphasis on LMWH. This results from the occurrence of a stable binding complex from Remdesivir to thrombin (-8.4 kcal/moL), resulting in a possible therapeutic contribution.
Regarding warfarin, its mechanism of action involves the inhibition of coagulation cascade components, Factors II, VII, IX, and X, which are dependent on vitamin K for its activation. Inhibition is the result of blocking the enzyme vitamin K epoxide reductase, and warfarin prevents the conversion of oxidized vitamin K epoxide to its reduced form, vitamin K hydroquinone. As a consequence, there is inhibition of vitamin K-dependent carboxylation, since the hydroquinone form would act as a cofactor for gamma-glutamyl carboxylase, an enzyme responsible for the carboxylation process, dependent on vitamin K, of clotting factors[51,52]. After carboxylation, Factors II, VII, IX, and X are able to bind to Ca2+ and interact with the anionic surface of phospholipids. With conversion blockage, there is no link between the factors and calcium, resulting in the absence of interaction with platelets, preventing the continuity of the cascade.
Considering the mechanism of action of warfarin, possible synergistic drug interactions between warfarin and Remdesivir can be inferred. The possibility lies in the observation, in silico, of stable binding between Remdesivir and Factors II, VII, IX, and X, with energy of affinity values, respectively, of -8.4 kcal/moL, -7.0 kcal/moL, -6.5 kcal/moL, and -9.6 kcal/moL.
Similar to the in silico data, case reports were observed in the medical literature, despite the low level of evidence, and these studies strengthen the hypothesis of a drug interaction between Remdesivir and warfarin, due to the performance of both in the coagulation cascade. In the first observation of changes in the international normalized ratio (INR), a marker used for analysis of the extrinsic and common pathways, resulting from a possible interaction between the two drugs, a case report describes an INR that increased 2 d after the start of Remdesivir administration. The laboratory parameter remained elevated during remdesivir therapy and began to decrease after completion of treatment, reaching the therapeutic target of 2-3 after 5 d of the last dose of Remdesivir. In addition, the authors mentioned that the probability score calculated on the Naranjo scale was 6, suggesting that the increase in INR observed was secondary to a probable drug interaction between warfarin and Remdesivir[53].
In addition, Landayan and colleagues reported a potential unwell-understood interaction between warfarin and Remdesivir with dexamethasone. The application of the Drug Interaction Probability Scale to this case resulted in a score of 5, highlighting a potential drug interaction. According to the authors, this probable interaction is demonstrated by a marked increase of INR within 24 to 48 h after the beginning of the combination (Remdesivir plus dexamethasone) in two patients with a history of stable INR[54].
In another case report, Yao et al[55] described an increase in INR after the onset of Remdesivir administration to treat pneumonia due to COVID-19 in a patient on stable chronic warfarin therapy. The INR remained supratherapeutic after discontinuation of Remdesivir and returned to therapeutic levels approximately 8 d after discontinuation. The Naranjo Drug Adverse Reaction Probability Scale was 4, indicating a possible adverse reaction to the drug.
CONCLUSION
In silico analyses indicate that Remdesivir behaves as an antiviral by interacting with ACE2 with considerable affinity, and thus competing with the virus and hindering cellular infection. At the same time, this drug is able to bind with affinity and stability to several clotting factors, which would prevent the activation of the blood coagulation cascade. In cases of severe COVID-19 with the possibility of DIC, Remdesivir could act synergistically with anticoagulants to modulate hypercoagulation states and prevent further harm to patients.
ARTICLE HIGHLIGHTS
Research background
The need for a treatment for coronavirus disease 2019 (COVID-19) has prompted some commercial drugs to be tested off-label. The evaluation of off-label drugs has provided incomplete information about their efficacy and gaps about long-term and chronic adverse effects. Remdesivir is an example of an off-label drug that showed promising results in the treatment of COVID-19, but may have other benefits that have not been explored and could be studied in silico.
Research motivation
The motivation for this study arose from the need to provide data that could help inform studies on Remdesivir, since this drug has not been evaluated through standard clinical trials for COVID-19. Remdesivir has met clinical trials for the treatment of another viral disease, and when used for the treatment of a different disease, both benefits and side effects can arise. Since our team observed the affinity of this antiviral for clotting factors in silico, we hypothesized that it might have benefits not only for preventing severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, but also for treating symptoms of severe COVID-19, such as disseminated intravascular coagulation.
Research objectives
The aim of the research was to evaluate whether Remdesivir has interaction affinity with clotting factors and to compare these interactions with the interaction with key receptors in SARS-CoV-2 infection to infer possible pharmacological activities.
Research methods
The study was performed in a computational environment using molecular docking and bioinformatics tools that allow to mimic the rational behavior of Remdesivir based on its chemical characteristics. The affinity energy and chemical bonds of Remdesivir with target molecules were analyzed and used to extrapolate in silico information to the human physiological environment.
Research results
The in silico results on the activity of Remdesivir in the coagulation cascade are promising when considering severe cases of COVID-19, but they need to be evaluated in vivo. In silico data show that Remdesivir can bind to various clotting factors with different affinities; this suggests that this drug can prevent coagulation by acting at several distinct points of the cascade, which increases its effectiveness when disseminated intravascular coagulation is triggered by viral infection.
Research conclusions
Our research team suggests that Remdesivir may be an off-label drug used as an antiviral for the treatment of COVID-19, but it can also be used to treat severe symptoms of the disease; however, this needs to be confirmed by in vivo testing.
Research perspectives
The research perspective is to evaluate, in silico, other off-label drugs suggested for the treatment of COVID-19.
Footnotes
Institutional review board statement: This study used data from three-dimensional structures deposited in public online databases.
Conflict-of-interest statement: The authors state that there is no conflict of interest of any kind with regard to this study.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Peer-review started: December 30, 2022
First decision: April 13, 2023
Article in press: July 3, 2023
Specialty type: Chemistry, medicinal
Country/Territory of origin: Brazil
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Jian X, China; Meng FZ, China S-Editor: Liu JH L-Editor: Wang TQ P-Editor: Cai YX
Contributor Information
Luis Gustavo Pagliarin, Campus Toledo, Universidade Federal do Paraná, Toledo 85.919-899, Paraná, Brazil.
Lucca Miketen de Oliveira, Campus Toledo, Universidade Federal do Paraná, Toledo 85.919-899, Paraná, Brazil.
Valentina Nunes Fontoura dos Anjos, Campus Toledo, Universidade Federal do Paraná, Toledo 85.919-899, Paraná, Brazil.
Cristiano de Bem Torquato de Souza, Campus Toledo, Universidade Federal do Paraná, Toledo 85.919-899, Paraná, Brazil.
Gabrielle Caroline Peiter, Programa Multicêntrico de Pós-graduação em Bioquímica e Biologia Molecular - Setor Palotina, Universidade Federal do Paraná, Palotina 85.950-000, Paraná, Brazil.
Cinthia Façanha Wendel, Campus Toledo, Universidade Federal do Paraná, Toledo 85.919-899, Paraná, Brazil.
Anderson Dillmann Groto, Campus Toledo, Universidade Federal do Paraná, Toledo 85.919-899, Paraná, Brazil.
Fabrício Freire de Melo, Instituto Multidisciplinar em Saúde - Campus Anísio Teixeira, Universidade Federal da Bahia, Vitória da Conquista 45.029-094, Bahia, Brazil.
Kádima Nayara Teixeira, Campus Toledo, Universidade Federal do Paraná, Toledo 85.919-899, Paraná, Brazil; Programa Multicêntrico de Pós-graduação em Bioquímica e Biologia Molecular - Setor Palotina, Universidade Federal do Paraná, Palotina 85.950-000, Paraná, Brazil. kadimateixeira@ufpr.br.
Data sharing statement
This study was developed in silico and no patient data or data from medical records were used in this study.
References
- 1. WHO Director-General’s opening remarks at the media briefing on COVID-19 - 11 March 2020. Available from: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---11-march-2020 .
- 2.Kupferschmidt K, Cohen J. Can China's COVID-19 strategy work elsewhere? Science. 2020;367:1061–1062. doi: 10.1126/science.367.6482.1061. [DOI] [PubMed] [Google Scholar]
- 3.Lai CC, Liu YH, Wang CY, Wang YH, Hsueh SC, Yen MY, Ko WC, Hsueh PR. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): Facts and myths. J Microbiol Immunol Infect. 2020;53:404–412. doi: 10.1016/j.jmii.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Munster VJ, Koopmans M, van Doremalen N, van Riel D, de Wit E. A Novel Coronavirus Emerging in China - Key Questions for Impact Assessment. N Engl J Med. 2020;382:692–694. doi: 10.1056/NEJMp2000929. [DOI] [PubMed] [Google Scholar]
- 5.Bai Y, Yao L, Wei T, Tian F, Jin DY, Chen L, Wang M. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA. 2020;323:1406–1407. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, Xing F, Liu J, Yip CC, Poon RW, Tsoi HW, Lo SK, Chan KH, Poon VK, Chan WM, Ip JD, Cai JP, Cheng VC, Chen H, Hui CK, Yuen KY. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/S0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Read JM, Bridgen JRE, Cummings DAT, Ho A, Jewell CP. Novel coronavirus 2019-nCoV: early estimation of epidemiological parameters and epidemic predictions. medRxiv. 2020 doi: 10.1098/rstb.2020.0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Almeida JO, de Oliveira VRT, Avelar JLS, Moita BS, Lima LM. COVID-19: Fisiopatologia e Alvos para Intervenção Terapêutica . Rev Virtual Quim. 2020;2020:12. [Google Scholar]
- 9.Iba T, Levy JH, Levi M, Connors JM, Thachil J. Coagulopathy of Coronavirus Disease 2019. Crit Care Med. 2020;48:1358–1364. doi: 10.1097/CCM.0000000000004458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asakura H, Ogawa H. COVID-19-associated coagulopathy and disseminated intravascular coagulation. Int J Hematol. 2021;113:45–57. doi: 10.1007/s12185-020-03029-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jose RJ, Manuel A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir Med. 2020;8:e46–e47. doi: 10.1016/S2213-2600(20)30216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.José RJ, Williams AE, Chambers RC. Proteinase-activated receptors in fibroproliferative lung disease. Thorax. 2014;69:190–192. doi: 10.1136/thoraxjnl-2013-204367. [DOI] [PubMed] [Google Scholar]
- 13.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, Qiu Y, Wang J, Liu Y, Wei Y, Xia J, Yu T, Zhang X, Zhang L. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang N, Li D, Wang X, Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J Thromb Haemost. 2020;18:844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, Guan L, Wei Y, Li H, Wu X, Xu J, Tu S, Zhang Y, Chen H, Cao B. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Asakura H. Classifying types of disseminated intravascular coagulation: Clinical and animal models. J Intensive Care. 2014;2:1–7. doi: 10.1186/2052-0492-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thachil J, Tang N, Gando S, Falanga A, Cattaneo M, Levi M, Clark C, Iba T. ISTH interim guidance on recognition and management of coagulopathy in COVID-19. J Thromb Haemost. 2020;18:1023–1026. doi: 10.1111/jth.14810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang N, Bai H, Chen X, Gong J, Li D, Sun Z. Anticoagulant treatment is associated with decreased mortality in severe coronavirus disease 2019 patients with coagulopathy. J Thromb Haemost. 2020;18:1094–1099. doi: 10.1111/jth.14817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tsang HF, Chan LWC, Cho WCS, Yu ACS, Yim AKY, Chan AKC, Ng LPW, Wong YKE, Pei XM, Li MJW, Wong SC. An update on COVID-19 pandemic: the epidemiology, pathogenesis, prevention and treatment strategies. Expert Rev Anti Infect Ther. 2021;19:877–888. doi: 10.1080/14787210.2021.1863146. [DOI] [PubMed] [Google Scholar]
- 20.Cheng F, Murray JL, Rubin DH. Drug Repurposing: New Treatments for Zika Virus Infection? Trends Mol Med. 2016;22:919–921. doi: 10.1016/j.molmed.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 21.Zhou Y, Hou Y, Shen J, Huang Y, Martin W, Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020;6:14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agostini ML, Andres EL, Sims AC, Graham RL, Sheahan TP, Lu X, Smith EC, Case JB, Feng JY, Jordan R, Ray AS, Cihlar T, Siegel D, Mackman RL, Clarke MO, Baric RS, Denison MR. Coronavirus Susceptibility to the Antiviral Remdesivir (GS-5734) Is Mediated by the Viral Polymerase and the Proofreading Exoribonuclease. mBio. 2018;9 doi: 10.1128/mBio.00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lo MK, Jordan R, Arvey A, Sudhamsu J, Shrivastava-Ranjan P, Hotard AL, Flint M, McMullan LK, Siegel D, Clarke MO, Mackman RL, Hui HC, Perron M, Ray AS, Cihlar T, Nichol ST, Spiropoulou CF. GS-5734 and its parent nucleoside analog inhibit Filo-, Pneumo-, and Paramyxoviruses. Sci Rep. 2017;7:43395. doi: 10.1038/srep43395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheahan TP, Sims AC, Graham RL, Menachery VD, Gralinski LE, Case JB, Leist SR, Pyrc K, Feng JY, Trantcheva I, Bannister R, Park Y, Babusis D, Clarke MO, Mackman RL, Spahn JE, Palmiotti CA, Siegel D, Ray AS, Cihlar T, Jordan R, Denison MR, Baric RS. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci Transl Med. 2017;9 doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Committee for Medicinal Products for Human Use – CHMP opinion. Remdesivir Gilead, INN-remdesivir 2020. Available from: https://www.ema.europa.eu/en/documents/other/conditions-use-conditions-distribution-patients-targeted-conditions-safety-monitoring-addressed_en-3.pdf .
- 26.Pizzorno A, Padey B, Dubois J, Julien T, Traversier A, Dulière V, Brun P, Lina B, Rosa-Calatrava M, Terrier O. In vitro evaluation of antiviral activity of single and combined repurposable drugs against SARS-CoV-2. Antiviral Res. 2020;181:104878. doi: 10.1016/j.antiviral.2020.104878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh AK, Singh A, Singh R, Misra A. Remdesivir in COVID-19: A critical review of pharmacology, pre-clinical and clinical studies. Diabetes Metab Syndr. 2020;14:641–648. doi: 10.1016/j.dsx.2020.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spinner CD, Gottlieb RL, Criner GJ, Arribas López JR, Cattelan AM, Soriano Viladomiu A, Ogbuagu O, Malhotra P, Mullane KM, Castagna A, Chai LYA, Roestenberg M, Tsang OTY, Bernasconi E, Le Turnier P, Chang SC, SenGupta D, Hyland RH, Osinusi AO, Cao H, Blair C, Wang H, Gaggar A, Brainard DM, McPhail MJ, Bhagani S, Ahn MY, Sanyal AJ, Huhn G, Marty FM GS-US-540-5774 Investigators. Effect of Remdesivir vs Standard Care on Clinical Status at 11 Days in Patients With Moderate COVID-19: A Randomized Clinical Trial. JAMA. 2020;324:1048–1057. doi: 10.1001/jama.2020.16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beigel JH, Tomashek KM, Dodd LE, Mehta AK, Zingman BS, Kalil AC, Hohmann E, Chu HY, Luetkemeyer A, Kline S, Lopez de Castilla D, Finberg RW, Dierberg K, Tapson V, Hsieh L, Patterson TF, Paredes R, Sweeney DA, Short WR, Touloumi G, Lye DC, Ohmagari N, Oh MD, Ruiz-Palacios GM, Benfield T, Fätkenheuer G, Kortepeter MG, Atmar RL, Creech CB, Lundgren J, Babiker AG, Pett S, Neaton JD, Burgess TH, Bonnett T, Green M, Makowski M, Osinusi A, Nayak S, Lane HC ACTT-1 Study Group Members. Remdesivir for the Treatment of Covid-19 - Final Report. N Engl J Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grein J, Ohmagari N, Shin D, Diaz G, Asperges E, Castagna A, Feldt T, Green G, Green ML, Lescure FX, Nicastri E, Oda R, Yo K, Quiros-Roldan E, Studemeister A, Redinski J, Ahmed S, Bernett J, Chelliah D, Chen D, Chihara S, Cohen SH, Cunningham J, D'Arminio Monforte A, Ismail S, Kato H, Lapadula G, L'Her E, Maeno T, Majumder S, Massari M, Mora-Rillo M, Mutoh Y, Nguyen D, Verweij E, Zoufaly A, Osinusi AO, DeZure A, Zhao Y, Zhong L, Chokkalingam A, Elboudwarej E, Telep L, Timbs L, Henne I, Sellers S, Cao H, Tan SK, Winterbourne L, Desai P, Mera R, Gaggar A, Myers RP, Brainard DM, Childs R, Flanigan T. Compassionate Use of Remdesivir for Patients with Severe Covid-19. N Engl J Med. 2020;382:2327–2336. doi: 10.1056/NEJMoa2007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holshue ML, DeBolt C, Lindquist S, Lofy KH, Wiesman J, Bruce H, Spitters C, Ericson K, Wilkerson S, Tural A, Diaz G, Cohn A, Fox L, Patel A, Gerber SI, Kim L, Tong S, Lu X, Lindstrom S, Pallansch MA, Weldon WC, Biggs HM, Uyeki TM, Pillai SK Washington State 2019-nCoV Case Investigation Team. First Case of 2019 Novel Coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanville B, Corbett R, Pidcock W, Hardin K, Sebat C, Nguyen MV, Thompson GR, Haczku A, Schivo M, Cohen S. A Community-transmitted Case of Severe Acute Respiratory Distress Syndrome (SARS) Due to SARS-CoV-2 in the United States. Clin Infect Dis. 2020;71:2222–2226. doi: 10.1093/cid/ciaa347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chauhan N. Possible Drug Candidates for COVID-19. ChemRxiv. Cambridge: Cambridge Open Engage; 2020. [Google Scholar]
- 34.Deshpande RR, Tiwari AP, Nyayanit N, Modak M. In silico molecular docking analysis for repurposing therapeutics against multiple proteins from SARS-CoV-2. Eur J Pharmacol. 2020;886:173430. doi: 10.1016/j.ejphar.2020.173430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiba S. Effect of early oseltamivir on outpatients without hypoxia with suspected COVID-19. Wien Klin Wochenschr. 2021;133:292–297. doi: 10.1007/s00508-020-01780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wade M, Li YC, Wahl GM. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coutsias EA, Seok C, Dill KA. Using quaternions to calculate RMSD. J Comput Chem. 2004;25:1849–1857. doi: 10.1002/jcc.20110. [DOI] [PubMed] [Google Scholar]
- 39.Aleissa MM, Silverman EA, Paredes Acosta LM, Nutt CT, Richterman A, Marty FM. New Perspectives on Antimicrobial Agents: Remdesivir Treatment for COVID-19. Antimicrob Agents Chemother. 2020;65 doi: 10.1128/AAC.01814-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohamed A. Coagulation Inhibitors in COVID-19 Leading to Compressive Airway Hematoma. Cureus. 2021;13:e12580. doi: 10.7759/cureus.12580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riddel JP Jr, Aouizerat BE, Miaskowski C, Lillicrap DP. Theories of blood coagulation. J Pediatr Oncol Nurs. 2007;24:123–131. doi: 10.1177/1043454206298693. [DOI] [PubMed] [Google Scholar]
- 42.Bloom AL. Physiology of blood coagulation. Haemostasis. 1990;20 Suppl 1:14–29. doi: 10.1159/000216159. [DOI] [PubMed] [Google Scholar]
- 43.Franco RF. Overview of coagulation, anticoagulation and fibrinolysis. Medicina (B Aires) 2001;34:229–237. [Google Scholar]
- 44.Birnhuber A, Fließer E, Gorkiewicz G, Zacharias M, Seeliger B, David S, Welte T, Schmidt J, Olschewski H, Wygrecka M, Kwapiszewska G. Between inflammation and thrombosis: endothelial cells in COVID-19. Eur Respir J. 2021;58 doi: 10.1183/13993003.00377-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schöchl H, van Griensven M, Heitmeier S, Laux V, Kipman U, Roodt J, Bahrami S, Redl H. Dual inhibition of thrombin and activated factor X attenuates disseminated intravascular coagulation and protects organ function in a baboon model of severe Gram-negative sepsis. Crit Care. 2017;21:51. doi: 10.1186/s13054-017-1636-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Camire RM, Bos MH. The molecular basis of factor V and VIII procofactor activation. J Thromb Haemost. 2009;7:1951–1961. doi: 10.1111/j.1538-7836.2009.03622.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fredenburgh JC, Weitz JI. Factor XI as a Target for New Anticoagulants. Hamostaseologie. 2021;41:104–110. doi: 10.1055/a-1384-3715. [DOI] [PubMed] [Google Scholar]
- 48.Gallagher JT. Heparin - A Century of Progress. Handb Exp Pharmacol. 2012;207:347–360. doi: 10.1007/978-3-642-23056-1_15. Accessed 28 December 2022. Available from: http://Link.springer.com/10.1007/978-3-642-23056-1 . [DOI] [PubMed] [Google Scholar]
- 49.Rezaie AR. Heparin chain-length dependence of factor Xa inhibition by antithrombin in plasma. Thromb Res. 2007;119:481–488. doi: 10.1016/j.thromres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 50.Hu Y, Meng X, Zhang F, Xiang Y, Wang J. The in vitro antiviral activity of lactoferrin against common human coronaviruses and SARS-CoV-2 is mediated by targeting the heparan sulfate co-receptor. Emerg Microbes Infect. 2021;10:317–330. doi: 10.1080/22221751.2021.1888660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weiss P, Soff GA, Halkin H, Seligsohn U. Decline of proteins C and S and factors II, VII, IX and X during the initiation of warfarin therapy. Thromb Res. 1987;45:783–790. doi: 10.1016/0049-3848(87)90088-0. [DOI] [PubMed] [Google Scholar]
- 52.Paul B, Oxley A, Brigham K, Cox T, Hamilton PJ. Factor II, VII, IX and X concentrations in patients receiving long term warfarin. J Clin Pathol. 1987;40:94–98. doi: 10.1136/jcp.40.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manigaba K, Hawks J, Kima M. Remdesivir-Warfarin Interaction: A Case Report. HCA Healthcare Journal of Medicine. 2020;1:15. doi: 10.36518/2689-0216.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Landayan RP, Saint-Felix S, Williams A. Probable Interaction Between Warfarin and the Combination of Remdesivir With Dexamethasone for Coronavirus Disease 2019 (COVID-19) Treatment: A 2 Case Report. J Pharm Pract. 2022;35:1039–1043. doi: 10.1177/08971900211008623. [DOI] [PubMed] [Google Scholar]
- 55.Yao L, Byas W, Li M, Saliaj M. A case of INR elevation with remdesivir and warfarin in a hospitalized patient with COVID-19. Case Rep Intern Med. 2021;8:9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
This study was developed in silico and no patient data or data from medical records were used in this study.